Key Points
Autophagy, an essential degradation pathway, is constitutively active in resting platelets and is induced upon platelet activation.
Platelet autophagy is indispensable for hemostasis and thrombus formation.
Abstract
Autophagy is important for maintaining cellular homeostasis, and thus its deficiency is implicated in a broad spectrum of human diseases. Its role in platelet function has only recently been examined. Our biochemical and imaging studies demonstrate that the core autophagy machinery exists in platelets, and that autophagy is constitutively active in resting platelets. Moreover, autophagy is induced upon platelet activation, as indicated by agonist-induced loss of the autophagy marker LC3II. Additional experiments, using inhibitors of platelet activation, proteases, and lysosomal acidification, as well as platelets from knockout mouse strains, show that agonist-induced LC3II loss is a consequence of platelet signaling cascades and requires proteases, acidic compartments, and membrane fusion. To assess the physiological role of platelet autophagy, we generated a mouse strain with a megakaryocyte- and platelet-specific deletion of Atg7, an enzyme required for LC3II production. Ex vivo analysis of platelets from these mice shows modest defects in aggregation and granule cargo packaging. Although these mice have normal platelet numbers and size distributions, they exhibit a robust bleeding diathesis in the tail-bleeding assay and a prolonged occlusion time in the FeCl3-induced carotid injury model. Our results demonstrate that autophagy occurs in platelets and is important for hemostasis and thrombosis.
Introduction
Autophagy, one of the major degradation pathways in eukaryotes, is important for cellular homeostasis and is implicated in a broad spectrum of human diseases including cancer, neurodegeneration, and cardiovascular diseases.1 There are 3 main types: chaperone-mediated autophagy,2 microautophagy,3,4 and the primary form, macroautophagy (referred to hereafter as autophagy).5 Autophagy involves de novo synthesis of double-membraned organelles called autophagosomes that contain cytosolic constituents including damaged organelles (eg, mitochondria) and protein aggregates. Autophagosomes fuse with multivesicular bodies, late endosomes, and lysosomes to form autolysosomes,6 in which waste elimination, energy production, and recycling of cellular components take place.
Autophagosome biogenesis and maturation use several protein complexes.7-10 In mammals, cellular signals (eg, starvation) activate the Ulk1 complex (Ulk1, FIP200, Atg13, and Atg101),11-14 which together with syntaxin 17,15 localizes Atg14/Atg14L16-19 to the endoplasmic reticulum.15,20 This recruits the Beclin 1-Vps34 core complex (Beclin 1, Vps34, and Vps15) to autophagosome initiation sites and promotes phosphatidylinositol 3-phosphate production for recruitment of additional autophagy pathway components.20,21 Two ubiquitin-like conjugation systems are also important for autophagosome biogenesis. One uses Atg7 and Atg10 (E1 and E2 enzymes, respectively) to form an Atg12–Atg5 conjugation.22-24 A second system uses Atg7, Atg3, and the Atg12–Atg5–Atg16L1 complex (E1, E2, and E3 enzymes, respectively) to catalyze the addition of phosphatidylethanolamine to the microtubule-associated protein 1A/B light chain 3 (LC3), producing LC3II.25,26 LC3II is attached to both inner and outer autophagosome membranes,27 and thus is a marker for isolation membranes (ie, phagophores), autophagosomes, and autolysosomes. During maturation, autophagosomes fuse with multivesicular bodies, late endosomes, and lysosomes, forming acidic compartments in which LC3II on the inner autophagosome membrane is degraded, along with the luminal contents. This fusion process is mediated by syntaxin 17, SNAP29, VAMP3, VAMP8, and Atg14/Atg14L.6,28-30
Although autophagy is important in nucleated cells, its role in anucleate cells is less recognized. In the eye, deletion of Atg5 or Vps34, although causing cataracts, does not impair organelle clearance in the lens organelle-free zone.31 The role of autophagy in differentiated anucleate red blood cells is unclear, despite its importance for mitochondrial clearance during erythroid maturation.32-38 Autophagy in platelets is understudied. Only recently, a role for autophagy in megakaryopoiesis and thrombopoiesis was indicated in a study on hematopoietic lineage-specific Atg7 deletion mice carrying Vav promoter-driven Cre.39 Feng et al reported starvation/rapamycin-induced autophagy in platelets.40 They also found that platelets isolated from whole-body heterozygous Becn1 knockout mice are defective in ex vivo collagen-induced aggregation, as well as flow-based platelet adhesion and thrombus formation.40 However, they did not detect autophagy induction in response to platelet agonists, nor were they able to distinguish the unique contribution of platelet autophagy deficiency to hemostasis impairment, given Becn1-deficiency in other cell types (eg, endothelial cells). Our studies extend these initial observations and show that autophagy not only occurred constitutively in resting platelets but also was induced during platelet activation. We further show that megakaryocyte- and platelet-specific deletion of Atg7 (using platelet factor 4 [PF4] promoter-driven Cre) caused modest defects in platelet aggregation and granule cargo packaging, but had a profound effect on hemostasis and thrombosis, despite normal platelet counts and mean platelet volumes. Our results demonstrate a critical role for autophagy in platelet activation during hemostasis and thrombosis.
Method
Mouse strains
Animal procedures were approved by the Institutional Animal Care and Use Committees at the relevant institutions. GFP-LC3/+ mice, in which GFP-LC3 expression is controlled by an actin promoter, were obtained from Noboru Mizushima (University of Tokyo, Japan).41,42 Becn1-EGFP/+43 and EGFP-Atg5/+ mice were generated using bacterial artificial chromosome (BAC) transgenics.44 To make EGFP-Atg5/+ mice, BAC clone (#RP24-117F24) containing the entire mouse Atg5 gene was purchased from Children's Hospital Oakland Research Institute (Oakland, CA). The sequence for EGFP was inserted after the Atg5 start codon, and the modified BAC was injected into B6C3 F1 hybrid eggs at the Mount Sinai School of Medicine core facility (New York, NY). GFP-LC3/+, Becn1-EGFP/+, and EGFP-Atg5/+ mice were genotyped using 5′- CCT ACG GCG TGC AGT GCT TCA GC-3′ (forward) and 5′-CGG CGA GCT GCA CGC TGC GTC CTC-3′ (reverse).
VAMP8−/− mice were described previously.45 Platelet-specific VAMP2/3−/− mice were generated by PF4-Cre-mediated46 expression of tetanus neurotoxin endopeptidase, which cleaves both VAMP2 and VAMP3.47 VAMP2/3/8−/− mice were generated from VAMP2/3−/− and VAMP8−/− mice and will be described elsewhere. Atg7f/f;PF4-Cre/+ mice were generated by crossing Atg7f/f mice48 with PF4-Cre mice.46 Floxed Atg7 alleles were genotyped using 5′- TGG CTG CTA CTT CTG CAA TGA TGT-3′ (forward) and 5′-CAG GAC AGA GAC CAT CAG CTC CAC-3′ (reverse). PF4-Cre was genotyped using 5′-CCC ATA CAG CAC ACC TTT TG-3′ (forward) and 5′-TGC ACA GTC AGC AGG TT-3′ (reverse).
Ex vivo assays
Human platelets were prepared as described49 from platelet-rich plasma (obtained from the Kentucky Blood Center). Mouse platelets were prepared as described.49 Blood was drawn via heart puncture into 0.38% (final) sodium citrate, diluted (1:1) with Tyrode’s buffer (pH 6.5; 20 mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid/KOH, 128 mM NaCl, 2.8 mM KCl, 1 mM MgCl2, 5 mM d-glucose, 12 mM NaHCO3, 0.4 mM NaH2PO4) containing 0.2 U/mL apyrase and 10 ng/mL PGI2 and separated at 215g for 10 minutes at room temperature. Platelet-rich plasma was separated at 675g for 10 minutes. Platelet pellets were resuspended in Tyrode’s buffer (pH 6.5) containing apyrase, PGI2, and 1 mM EGTA. After washing, platelets were resuspended in Tyrode’s buffer (pH 7.4) with 0.02 U/mL apyrase and 1 mM CaCl2. Platelet concentrations were measured using a Z2 Counter (Beckman Coulter, Miami, FL).
For western blot analysis, washed platelets (1.2 × 109/mL) were preincubated with the indicated inhibitors and stimulated with the indicated agonists. Proteins were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis on 4% to 12% (or 12% for LC3) Bis-Tris gels (Invitrogen, Grand Island, NY) before western blotting, using appropriate antibodies (see the supplemental Methods, available on the Blood Web site). Films were exposed for different periods to optimize signals. Images of films were scanned in positive mode and analyzed using ImageJ (National Institutes of Health).
Platelet aggregation and adenosine triphosphate release were measured with washed platelets (2.5 × 108/mL), as described,45 in a Lumi-Dual Aggregometer (Model 460VS) with Aggro/Link interface (Model 810) and software (Chrono-Log, Havertown, PA).
Dense granule, α-granule, and lysosome secretion were assayed as described.45,50-53 Specifically, PF4 was assessed by anti-PF4/CXCL4 ELISA, [3H]-serotonin levels determined by scintillation counting, and β-hexosaminidase activity evaluated using a colorimetric assay.54 Total PF4 levels, total uptake of [3H]-serotonin, and total β-hexosaminidase activity were recorded, and the percentage secretion was calculated.53
In vivo tail-bleeding assay
This assay was performed as described.55 To measure bleeding, tails of 5- to 6-week-old sedated (ketamine, 75 mg/kg, intraperitoneally) mice were transected 3 mm from the tip and immersed in saline at 37°C. The time required for bleeding cessation was recorded, and the mice were observed for an additional minute. Experiments were terminated at 10 minutes. The transected tail tips were used for genotyping.
In vivo FeCl3-induced carotid injury model
This assay was performed as described.56 Mice at 21 to 32 weeks of age were anesthetized with avertin (tribromoethanol; 200 mg/kg; intraperitoneally). The left carotid artery was exposed and visualized through a dissecting microscope. A Doppler, microvascular probe was placed on the exposed artery to measure vascular blood flow, and the artery was injured by placing a small piece of filter paper saturated with 6% FeCl3 directly on the artery for 3 minutes. Times to form stable, occlusive thrombi were recorded. Experiments were terminated at 3 minutes postocclusion or 30 minutes postinjury, whichever occurred first.
Platelet count and volume measurements
Mice were anesthetized with isoflurane, and blood (50∼200 µL) was collected from the retroorbital plexus into heparinized capillary tubes containing EDTA (10 mM). Platelet counts and mean platelet volumes were analyzed with a HEMAVET HV950FS analyzer (Drew Scientific, Waterbury, CT).
Results
Autophagy machinery is present in murine and human platelets
Autophagy gene transcripts57,58 and proteins59-62 have been detected in platelets. To confirm, we probed murine and human platelet extracts with antibodies to autophagy-related proteins. We detected components of the Ulk1 complex (eg, Ulk1 and FIP200), the Atg12-Atg5 and LC3II conjugation systems (eg, Atg7, Atg3, Atg12, Atg5, and LC3II), and the Beclin 1-Vps34 complex (eg, Beclin 1, Vps34, Vps15, Atg14L, Nrbf2, and UVRAG) (Figure 1A; supplemental Figure 1, summarized in supplemental Table 1). We next examined platelets from transgenic mice expressing GFP-fused autophagy markers (LC3,41 Atg5, and Beclin 143) to visualize isolation membranes, autophagosomes, and autolysosomes in resting platelets. GFP-LC3, expressed at higher levels than endogenous LC3II (supplemental Figure 2, lane 2), displayed both diffuse and punctate localization patterns (Figure 1B). EGFP-Atg5 and Beclin 1-EGFP, both expressed at lower than endogenous protein levels (supplemental Figure 2, lanes 3 and 4), were also localized to puncta (Figure 1B). These data confirm the presence of the autophagy machinery in murine and human platelets and suggest that autophagosome-related structures are present in resting platelets.
Figure 1.
Autophagy machinery is present in murine and human platelets. (A) Western blot analyses of the autophagy machinery in freshly prepared mouse and human platelets and in banked human platelets. Extracts from mouse and human platelets were loaded in each lane and probed with antibodies to the indicated proteins. Atg5+/+ and Atg5−/− mouse embryonic fibroblasts (MEFs) were used as additional molecular weight markers for LC3I (unconjugated form) and LC3II (phosphatidylethanolamine-conjugated form), as Atg5−/− MEFs produced LC3I but not the faster-migrating LC3II. Atg5−/− MEFs also had higher p62 levels than Atg5+/+ MEFs because of impaired autophagy. Note that the loading amounts for the MEFs were much lower than those for platelets to have comparable LC3II levels. As a result, and as platelets have high actin content than the MEFs, both actin and p62 levels in the MEF samples are much lower than in platelets. The images shown are representative of at least 3 independent experiments. Full images of all blots are shown in supplemental Figure 1. (B) Platelets from the autophagy reporter mice (ie, GFP-LC3/+, Becn1-EGFP/+, EGFP-Atg5/+) but not wild-type mice show GFP-LC3-positive, EGFP-Atg5-positive, and Beclin 1-EGFP-positive puncta, respectively (arrows), which are autophagosome-related structures including isolation membranes, autophagosomes, and autolysosomes. Samples were visualized for GFP or EGFP fluorescence (fluorescein isothiocyanate [FITC] channel) and differential interference (DIC). The images shown are representative of at least 2 independent experiments.
Basal autophagy occurs in resting platelets
To determine whether basal autophagy occurred in resting platelets, we monitored LC3II levels in unstimulated platelets in the presence or absence of lysosomal inhibitors (eg, NH4Cl and chloroquine). Treatment with NH4Cl and, less effectively, with chloroquine, increased LC3II levels over time in unstimulated human platelets (Figure 2A). Consistently, unstimulated GFP-LC3/+ platelets containing GFP-LC3-positive puncta increased from 21 ± 8% in the absence of NH4Cl to 64 ± 10% in the presence of NH4Cl (Figure 2B; P < 10−7). Together, these findings indicate the presence of a basal autophagic flux (ie, turnover of autophagosomes) in resting platelets, which can be inhibited by lysosomal inhibitors.
Figure 2.
Basal autophagy occurs in resting platelets. (A) Western blot analysis of LC3II in unstimulated human platelets treated with either NH4Cl (20 mM) or chloroquine (50 µM) to inhibit lysosomal activity. Two exposure times, short and long, are shown. Note that LC3I is visible only with longer exposures in this experiment. (B) GFP-LC3 puncta in unstimulated platelets without and with NH4Cl treatment (20 mM, 2 h). Samples were visualized for DIC and GFP fluorescence (GFP-LC3). Ten images (17∼74 platelets/field) were obtained at random and quantified for each condition. Platelets containing GFP-LC3-positive puncta are 21 ± 8% and 64 ± 10% without and with NH4Cl treatment, respectively. Statistical significance was evaluated with the Student t test. (C) Western blot analysis of LC3II in human platelets pretreated for 1 h with cycloheximide (100 µg/mL) or puromycin (10 µg/mL), in combination with pretreatment of NH4Cl (20 mM, 1 h) or protease inhibitor cocktail (1 h). The images shown are representative of at least 3 independent experiments.
Because anucleate platelets have few means for transcription, we asked whether translation contributed to basal autophagy, and thus accounted for the changes in LC3II seen in lysosomal inhibitor-treated resting platelets. Neither cycloheximide nor puromycin affected LC3II levels, regardless of whether lysosomal proteolysis was inhibited or not (Figure 2C). These results imply that in resting platelets, LC3II and autophagosomes are continuously produced, independent of LC3 translation.
Platelet activation leads to LC3II reduction and altered autophagic flux
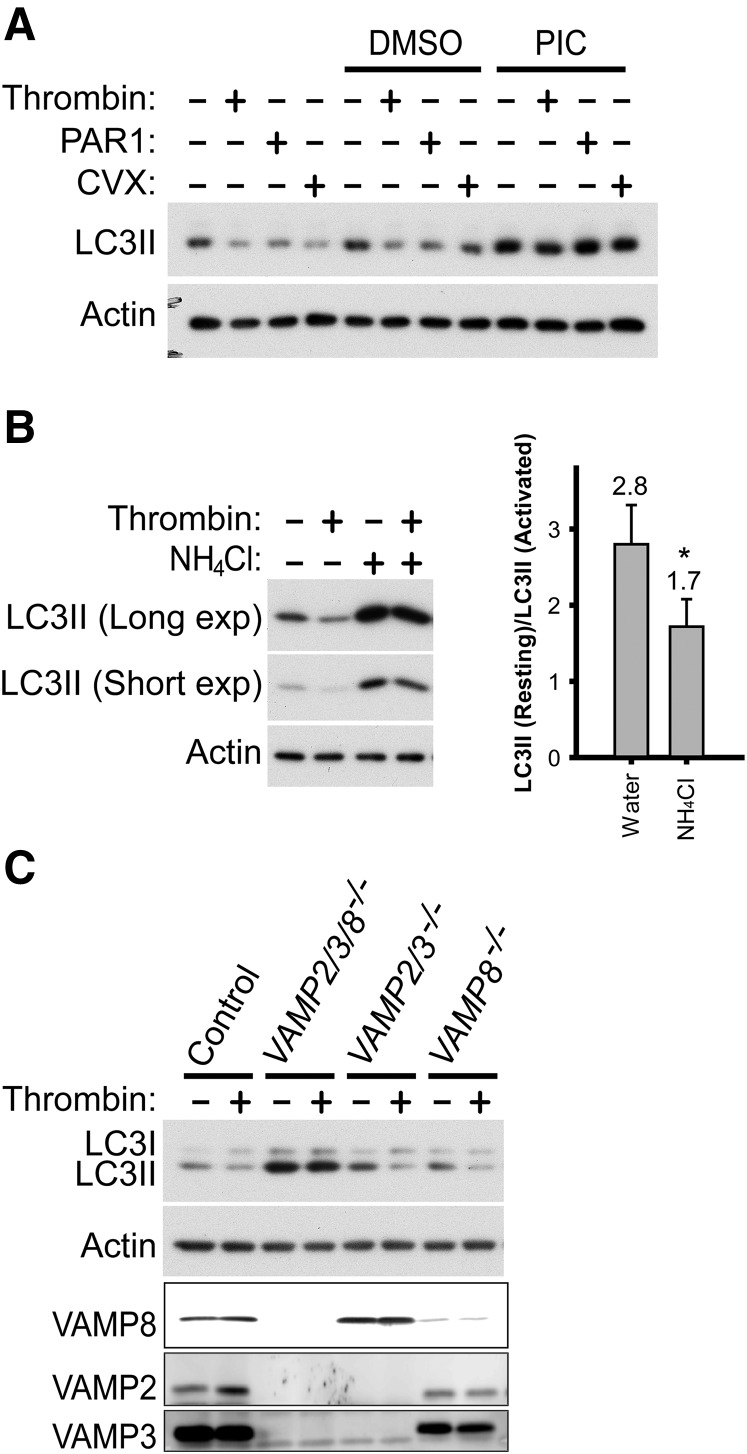
Given that autophagic flux appeared to occur in resting platelets, we sought to determine whether it changed upon platelet activation. We found that LC3II levels decreased in response to thrombin in both murine and human platelets; this observation was confirmed using 4 different anti-LC3 antibodies (Figure 3A). We tested other platelet agonists and found that LC3II levels in human platelets were reduced by ∼2.5-fold in response to strong activators (thrombin, convulxin [CVX], and PAR1 peptide), whereas weaker activators (ADP, collagen, and U46619) led to modest LC3II reduction (∼1.5-fold) (Figure 3B). Similar agonist-induced LC3II reductions were observed in murine platelets (supplemental Figure 3). These results indicate that platelet activation results in a loss of LC3II, indicating an alteration in autophagic flux.
Figure 3.
Platelet activation leads to LC3II reduction. (A) Western blot analysis of LC3II in the murine and human platelets before and after stimulation (0.1 U/mL thrombin, 30 minutes), with various anti-LC3 antibodies. Cell extracts from Atg5+/+ and Atg5−/− MEFs were used as additional molecular weight markers to indicate the migration positions of LC3I and LC3II. Note that the loading volumes for the MEFs were optimized to have comparable LC3II levels, and platelets have high actin content. Thus, both actin and p62 levels in the MEF samples were below detection limits. The images shown are representative of at least 2 independent experiments. (B) Western blot analyses of LC3II in human platelets before and after stimulation with various agonists. Platelets were stimulated for 30 minutes with thrombin (0.1 U/mL), CVX (0.1 µg/mL), PAR1 or PAR4 peptide (100 µM), ADPβS (10 µM), collagen (1 µg/mL), A23187 (10 µM in DMSO), and U46619 (20 µM in DMSO). N = 2∼8 for various inhibitors. (C) Western blot analysis of LC3II, before and after stimulation, in human platelets pretreated with inhibitors of platelet activation. Platelets were pretreated for 60 minutes with U-73122 (20 µM), U-73343 (inactive analog of U-73122, 20 µM), BAPTA-AM (100 µM), Ro-31-8220 (10 µM), or PP2 (100 µM) and then stimulated for 30 minutes with either thrombin (0.1 U/ml) or CVX (0.1 µg/mL). Of note, U-73343, Ro-31-8220, and PP2 increased LC3II levels in resting platelets for an unknown reason. All inhibitor stock solutions were prepared in DMSO. N = 3 for all inhibitors. For B and C, *P < .05 by the Student t test.
To delineate which platelet activation step or steps are required for agonist-induced LC3II reduction and to determine whether autophagy is downstream of the known platelet signaling cascades, we tested the effects of platelet activation inhibitors on agonist-induced LC3II reduction. The phospholipase C inhibitor, U-73122, but not its inactive analog, U-73343, blocked thrombin-induced LC3II reduction (Figure 3C). The permeant calcium chelator BAPTA-AM and the protein kinase C inhibitor Ro-31-8220 also blocked thrombin-induced LC3II reduction (Figure 3C). In addition, PP2, the inhibitor of Src-family tyrosine kinases, blocked CVX-induced LC3II reduction (Figure 3C). Together, our results demonstrate that platelet agonists induce LC3II reduction, which requires known elements of platelet activation signaling cascades; for example, phospholipase C, Ca2+, protein kinase C, and Src-family kinases.
Agonist-induced LC3II reduction involves autolysosomal proteolysis
To determine whether agonist-induced LC3II reduction involved proteolysis, we preincubated human platelets with a protease inhibitor cocktail (PIC) before stimulation. The PIC pretreatment blocked thrombin-, PAR1-, and CVX-induced LC3II reduction in platelets (Figure 4A). Although inhibition of thrombin-induced reduction in LC3II could be attributed to a direct effect of serine protease inhibitors in the PIC on thrombin, inhibition of PAR1- and CVX-induced LC3II reduction was likely a result of inhibition of proteolysis unrelated to thrombin. The ubiquitin–proteasome system and autophagy are 2 major degradation pathways in eukaryotes. The ubiquitin–proteasome system has been reported to be important for thrombopoiesis and platelet function.63,64 We asked whether agonist-induced LC3II reduction involved proteasomes. Inhibition of proteasomes with MG132 had no effect on CVX-induced LC3II reduction in mouse platelets (supplemental Figure 4A), suggesting agonist-induced LC3II degradation is not a result of proteasomal activity. In contrast, treatment of mouse platelets with NH4Cl to neutralize lysosomal pH suppressed thrombin-induced LC3II reduction (Figure 4B), suggesting active proteolysis in acidic compartments contributes to agonist-induced LC3II reduction.
Figure 4.
Agonist-induced LC3II reduction involves autolysosomal proteolysis. (A) Western blot analysis of LC3II changes in response to agonists in banked human platelets pretreated with a PIC. Platelets were incubated with the PIC for 60 minutes before 30-minute agonist stimulation with thrombin (0.1 U/mL), PAR1 peptide (100 µM), and CVX (0.1 µg/mL), respectively. The images shown are representative of at least 3 independent experiments. (B) Western blot analysis of LC3II in thrombin-stimulated (0.1 U/mL, 30 minutes) mouse platelets after preincubation with NH4Cl (20 mM, 2 hours). The images shown are representative of 4 independent experiments. *P < .05. (C) Western blot analysis of LC3II in thrombin-stimulated (0.1 U/mL, 30 minutes) platelets isolated from mice lacking VAMP2, VAMP3, and/or VAMP8, and from wild-type control mice. The images shown are representative of 2 independent experiments.
In nucleated cells, proteolysis of LC3II requires autophagosome–lysosome fusion. To determine whether this is also required for agonist-induced LC3II reduction in platelets, we examined platelets from mice with defective membrane fusion machinery. Platelets lacking 3 of the 4 major VAMPs (ie, VAMP2/3/8−/−) showed greatly impaired thrombin-induced LC3II reduction, whereas those deficient in either VAMP2/VAMP3 or VAMP8 showed no overt impairment (Figure 4C). These results suggest that VAMP2, VAMP3, and VAMP8 have redundant function in agonist-induced autophagosome–lysosome fusion. Notably, the resting platelets purified from VAMP8−/−, VAMP2/3−/−, and VAMP2/3/8−/− mice had increased LC3II levels relative to control platelets, which expressed normal levels of VAMP2, VAMP3, and VAMP8, with VAMP2/3/8−/− platelets having the highest LC3II levels (Figure 4C). This is consistent with VAMP-mediated autophagosome–lysosome fusion being required for basal autophagy in resting platelets. Together, our results suggest that agonist-induced LC3II reduction likely results from platelet autolysosomal proteolysis; that is, canonical degradative autophagy is induced upon platelet activation.
Of note, a new role of autophagy in unconventional secretion is emerging recently.65 We asked whether exocytosis of LC3II-decorated exosomes occurred upon agonist stimulation. We activated mouse platelets with CVX, PAR4 peptide, or thrombin before separating platelet-containing pellets from releasate-containing supernatants. PF4, a known α-granule cargo, was the releasate control. Despite the appearance of PF4, LC3II was not detected in the supernatants of activated platelets (supplemental Figure 4B), suggesting the absence of agonist-induced exocytosis of LC3II-decorated exosomes.
Atg7-deficient platelets are defective in ex vivo platelet aggregation and granule cargo packaging
Given the observed effects activation exerted on autophagy, we asked whether platelet autophagy had a functional role. We generated a mouse strain with a megakaryocyte- and platelet-specific deletion of Atg7 (ie, Atg7f/f;PF4-Cre/+). Although Atg7 levels were only partially reduced in platelets from Atg7f/f;PF4-Cre/+ mice, LC3II was largely depleted (Figure 5A), suggesting Atg7 is likely the rate-limiting enzyme for LC3II production. We then systematically characterized platelets from these Atg7-deficient mice, using standard ex vivo assays. First, we monitored platelet aggregation by lumi-aggregometry and found that Atg7 deficiency caused defects in aggregation in response to modest-to-low concentrations of thrombin and collagen (Figure 5B). Second, we assayed thrombin-dependent platelet exocytosis by monitoring secretion of PF4 from α-granules, [3H]-serotonin from dense granules, and β-hexosaminidase from lysosomes. Our data show that the total levels of α-granule cargo PF4, the total uptake of [3H]-serotonin into dense granules, and the total lysosomal β-hexosaminidase activity were slightly, but significantly, reduced in platelets from Atg7f/f;PF4-Cre/+ mice (by 15%, 26%, and 18%, respectively; P < .05), suggesting modestly impaired granule cargo packing (Figure 5C). However, Atg7 deficiency did not impair percentage release of PF4, [3H]-serotonin, or β-hexosaminidase (supplemental Figure 5A). Consistently, we also found no overt alterations in the levels of the fusion machinery (eg, STX11, SNAP23, VAMP3, and VAMP8) and the α-granule cargo von Willebrand factor (supplemental Figure 5B). Third, to determine the cause of the observed granule cargo packaging defects, we assessed overall morphology of Atg7f/f and Atg7f/f;PF4-Cre/+ platelets by electron microscopy. Our micrographs show that Atg7-proficient platelets (Figure 5Di-iii) and Atg7-deficient platelets (Figure 5Div-vi) had no overt difference in morphology. Of note, we observed isolation membranes in platelets (Figure 5Dii-iii, arrowheads and inset). Fourth, we examined integrin inside-out and outside-in signaling and found that Atg7 deficiency had no effect on activation-dependent Jon/A (supplemental Figure 5C) or fibrinogen (data not shown) binding, platelet spreading (data not shown), or clot retraction (either in the absence or presence of glucose; supplemental Figure 5D). In summary, we detected modest aggregation and granule cargo packaging defects, but normal cargo release and integrin signaling in the Atg7f/f;PF4-Cre/+ platelets.
Figure 5.
Atg7-deficient platelets showed modest defects in ex vivo platelet function assays. (A) Representative western blot analysis showing reduced Atg7 and depleted LC3II levels in platelets from Atg7f/f;PF4-Cre/+ mice compared with platelets from Atg7f/f mice. For each genotype, results from 2 different mice are shown. (B) Lumi-aggregometry measurements of platelets from Atg7f/f (red traces) and Atg7f/f;PF4-Cre/+ (blue traces) mice when stimulated with thrombin (upper) or collagen (lower). Different agonist concentrations were titrated. The images shown are representative of at least 3 independent experiments. (C) Total levels of PF4 (left), total [3H]-serotonin uptake (middle), and total β-hexosaminidase activity (right) in platelets purified from Atg7f/f and Atg7f/f;PF4-Cre/+ mice. Data represent an average of 21 measurements. *P < .05, **P < .001, Student t test. (D) EM images of platelets from Atg7f/f (i-iii) and Atg7f/f;PF4-Cre/+ (iv-vi) mice. Samples were prepared by the freeze substitution method. α, α granules; arrows, dense granules; arrowheads and inset, isolation membranes. Scale bars, 500 nm. The images shown are representative micrographs from 2 independent experiments.
Deletion of Atg7 in platelets leads to impaired hemostasis and thrombus formation
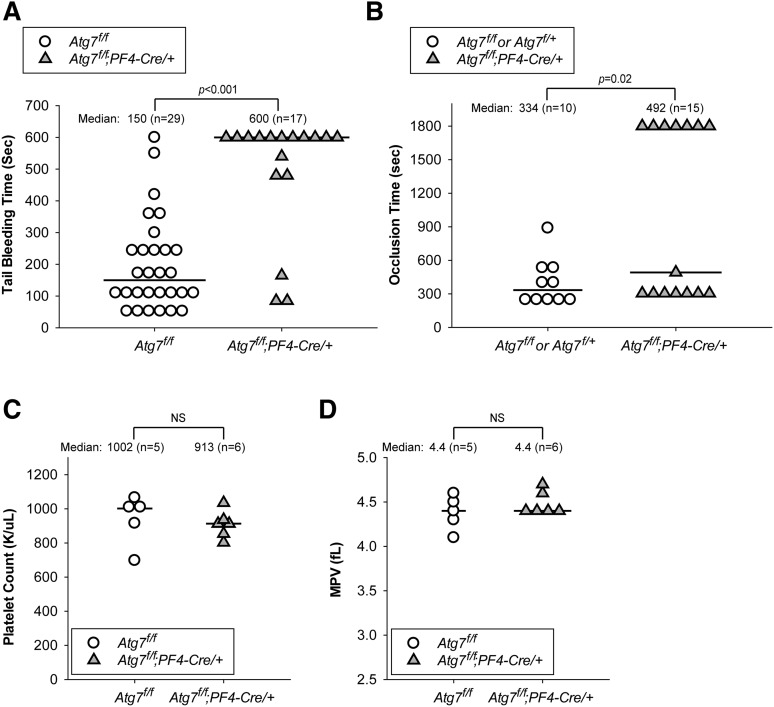
As we observed defects in the Atg7-deficient platelets ex vivo, we next determined the consequence of deleting Atg7 in platelets in vivo. Using the tail-bleeding assay for hemostasis, we showed that 11 (∼65%) of 17 of Atg7f/f;PF4-Cre/+ mice were unable to stop bleeding within 10 minutes after tail transection, whereas Atg7f/f littermates demonstrated normal bleeding time (median, ∼2.5 minutes) (Figure 6A). Moreover, using the FeCl3-induced carotid injury model, we further showed that 7 (∼47%) of 15 of Atg7f/f;PF4-Cre/+ mice were unable to form occlusive thrombi within 30 minutes after injury, whereas all Atg7f/f littermates could do so (median occlusion time, ∼5.6 minutes) (Figure 6B). Of note, the occlusion times of Atg7f/f;PF4-Cre/+ mice appeared to show a bimodal distribution for an unknown reason (Figure 6B). Because PF4-Cre expression deletes the floxed Atg7 gene at a later stage of megakaryocyte differentiation,46 it is possible that bleeding in Atg7f/f;PF4-Cre/+ mice is a result of defective platelet production, especially given the thrombocytopenia phenotype of the Atg7f/f;Vav-Cre/+ mice, where Atg7 is deleted at an earlier stage of the hematopoietic cell lineage.39 To investigate this possibility, we measured platelet counts and mean platelet volumes as metrics of thrombocytopenia and faulty platelet production. Compared with Atg7f/f mice, Atg7f/f;PF4-Cre/+ mice had indistinguishable platelet counts (Figure 6C) and mean platelet volumes (Figure 6D), suggesting that defects in Atg7-mediated platelet function or functions, but not thrombocytopenia, accounted for the severe bleeding in Atg7f/f;PF4-Cre/+ mice. Together, our results show that even partial reduction of Atg7 protein levels in Atg7f/f;PF4-Cre/+ platelets profoundly impairs hemostasis and thrombus formation. As both LC3II depletion and impaired hemostasis and thrombus formation were observed in Atg7f/f;PF4-Cre/+ platelets, our results suggest that Atg7 may be a rate-limiting factor in the role that platelet autophagy plays during hemostasis and thrombosis.
Figure 6.
Megakaryocyte- and platelet-specific Atg7 deletion causes impaired hemostasis and thrombus formation without thrombocytopenia. (A) Tail-bleeding time measurements taken from Atg7f/f (○) and Atg7f/f;PF4-Cre/+ (filled gray triangles) mice. (B) Occlusion time measurements taken from Atg7f/f (○) and Atg7f/f;PF4-Cre/+ (filled gray triangles) mice in the FeCl3 carotid injury model of thrombosis. For A and B, each data point represents an individual mouse, with the horizontal line marking the median. Statistical significance was evaluated with the log-rank test using SigmaPlot12 software. (C) Platelet counts and (D) mean platelet volumes from Atg7f/f (○) and Atg7f/f;PF4-Cre/+ (filled gray triangles) mice. For C and D, statistical significance was evaluated with the Student t test. NS, not significant.
Discussion
Although autophagy-related mRNAs and proteins have been reported in transcriptomic57,58 and proteomic59-62 studies, the functional ramifications of autophagy in platelets have only recently begun to be addressed.39,40 Here we confirm the presence of many autophagy-related proteins in platelets. Using transgenic mice expressing GFP-fused LC3, Beclin 1, and Atg5, we further show that these autophagy markers are localized to punctate structures in platelets. We demonstrate that autophagic flux occurs in resting platelets and is enhanced in response to various platelet agonists. Using mice with PF4-Cre-driven, megakarcyte/platelet-specific Atg7 deletion, we further show that platelet autophagy contributes to hemostasis and thrombus formation. Collectively, our data demonstrate physiological relevance of autophagy in platelets and add autophagy to a growing list of cellular processes (eg, translation, secretion, cytoskeletal rearrangements) that are unexpectedly important for platelet functions.
Feng et al reported that platelet autophagy was independent of activation.40 We tested several anti-LC3 antibodies, including the one used by Feng et al (ie, Sigma L7543). All but the L7543 antibody detected thrombin-induced LC3II reduction in both human and murine platelets; even the L7543 antibody detected thrombin-induced LC3II reduction in human platelets. We postulate that the L7543 antibody may recognize a subpopulation of mouse LC3II that is less affected by platelet activation. As LC3II reduction could result from decreased biogenesis, increased degradation, or both, a standard assay for evaluating autophagic flux is to monitor changes in LC3II levels in the absence vs presence of lysosomal inhibitors.66,67 Using NH4Cl, we confirmed agonist-induced LC3II reduction as a readout for agonist-induced enhancement of autophagic flux. Other potential metrics of autophagic flux, however, appear to be inappropriate for monitoring platelet activation-induced autophagy. A classical autophagic flux assay measures the long-lived protein degradation rates through pulse-chase labeling. This assay is not applicable to platelets, as chase time may exceed the time during which cultured platelets are fully functional. The levels of p62, an adaptor protein that delivers substrates to autophagosomes and is itself degraded in autolysosomes,68 have also been used as indicators of autophagic flux in certain cell types.18,69,70 However, p62 did not accumulate in Atg7-deficient platelets (supplemental Figure 5B). As p62 is one of many adaptors that bring selected cargoes to autophagosomes, we cannot rule out the possibility that agonist-induced platelet autophagy uses adaptors other than p62. Together, our work establishes agonist-induced LC3II reduction as a useful measure for platelet activation-induced autophagy. To our knowledge, this is the first report of autophagy induction by platelet activation.
Deleting Atg7 early in platelet production by Vav-Cre39 or prolonged platelet proteasome inhibition by genetic means63 have been shown to cause abnormal megakaryopoiesis or thrombopoiesis, suggesting key degradation processes may be needed for proper thrombocytogenesis. In contrast, although PF4-Cre-driven Atg7 deletion caused severe hemostasis and thrombosis defects, Atg7f/f;PF4-Cre/+ mice showed normal platelet counts, mean platelet volumes, and ultrastructure (Figures 6C-D and 5D, respectively), indicating proper platelet production. Thus, the use of the PF4-Cre system allows us to examine the effects of autophagy deficiency on platelets and hemostasis without the complication of deficient platelet production.
The use of PF4-Cre also allows us to examine the effects of autophagy deficiency on platelets and hemostasis without the complication caused by lack of autophagy in other cell types. The bleeding phenotype reported by Feng et al, in a global heterozygous Becn1 mouse,40 is likely the culmination of Beclin 1 depletion in several cell types; for example, endothelial cells and platelets. Consistently, other studies have shown that loss of autophagy in endothelial cells, although it causes no abnormality in platelets, decreases secretion of von Willebrand factor, prolongs thrombus formation times, and yields smaller thrombi.71,72 Our work is the first direct report of platelet autophagy being essential for hemostasis and thrombosis. Our work clearly demonstrates that platelet Atg7 is indispensable and likely a rate-limiting factor for platelet autophagy’s role in hemostasis and thrombosis. This is confirmed by our unpublished data showing similar bleeding diatheses in mice with megakaryocyte/platelet-specific deletion of other key autophagy genes; for example, Becn1f/f;PF4-Cre/+ and Pik3c3f/f;PF4-Cre/+ (data not shown).
Despite robust in vivo phenotypes, megakaryocyte/platelet-specific Atg7 deficiency only modestly affected aggregation and granule cargo packaging and had limited effects on cargo release or integrin signaling. We performed additional ex vivo platelet assays to examine effects of platelet Atg7 loss on phosphatidylserine (PS) exposure and energetics. Both Atg5 and Beclin 1 are essential for apoptotic cell clearance during development by mediating expression of extracellular PS.73 As PS exposure is also important for hemostasis,74,75 we thought that autophagy-mediated PS exposure in platelets might be required for hemostasis. Unfortunately, this hypothesis is unlikely, as our studies with annexin V (data not shown) and lactadherin (supplemental Figure 5E) binding showed no impairment in activation-dependent PS exposure on Atg7f/f;PF4-Cre/+ platelets. As the conventional role of autophagy is for adenosine triphosphate production and nutrient recycling through catabolism, and autophagy is essential for the clearance of damaged organelles such as mitochondria, we speculated that platelet autophagy may be critical for energetics. However, there was no clot retraction defect observed in the Atg7-deficient platelets, even when glucose was withheld (supplemental Figure 5D), suggesting Atg7-deficiency does not impair glucose use in platelets.
At this stage, we have insufficient data to define the mechanism by which Atg7-deficient mice demonstrate severe hemostasis and thrombosis defects. There are ample published examples in which deletion of a signaling cascade protein causes only modest defects in platelet activation in ex vivo assays, yet the protein contributes significantly to in vivo hemostasis and thrombosis.56,76-79 In a recent example, STXBP5 deletion yielded modest defects on dense granule release but was absolutely required for hemostasis in the tail transection model and for thrombosis in the FeCl3-induced carotid injury model.80 Therefore, it is possible that the sensitivity of the ex vivo assays we employed are insufficient to fully delineate the effects of deleting the autophagy machinery. There may be an additive or even synergistic effect resulting from several singularly mild impairments.
Alternatively, it is possible that autophagy deficiency may cause some unidentified platelet dysfunction, leading to the observed severe hemostasis and thrombosis defects. Our current hypothesis is that platelet autophagy-mediated degradation of a specific substrate or substrates is required for effective hemostasis and thrombosis. Consistent with this concept, there are functional similarities between the 2 major degradation systems: the ubiquitin–proteasome system and autophagy in platelets. Both occur in resting platelets and are activated upon stimulation64,81-83 (Figures 2-4), and both are needed for occlusive thrombus formation after FeCl3 injury64 (Figure 6B). In addition, the severity of the hemostasis and thrombosis defects caused by inhibiting either proteasome or autophagy exceed the expectation for modest defects based on ex vivo assay phenotypes. This implies that some degradative processes must occur in situ, in the growing thrombi for effective hemostasis and thrombosis. For the proteasome, some of the important targets have been identified; for example, filamin, talin, and Syk.64,84 For autophagy, the substrates are unknown. Proteomic analysis of extracts and releasates from autophagy-deficient platelets will be required to test this hypothesis. In short, our data and those of others clearly demonstrate that some regulated degradation steps are important for platelet function. What remains is to determine what targets must be degraded and why their removal is important.
Note in added proof
While this manuscript was under review, a case report was published illustrating autophagosome-like structures in platelets from a dog with severe nonregenerative anemia.85 Moreover, another work using hematopoietic system conditional Atg7 knockout (ie, Atg7f/f;Vav-Cre) mice was published that demonstrates that in addition to severe defects in megakaryogenesis, megakaryocyte differentiation and thrombopoiesis observed in these mice, platelets isolated from these mice are defective in activation and aggregation.39 Our work using Atg7f/f;PF4-Cre mice, which deletes the Atg7 genes at a later stage in platelet production, distinguishes the roles of autophagy in platelets and in hemostasis from roles in megakaryogenesis and in megakaryocyte differentiation.
Acknowledgments
We thank the Kentucky Blood Center for provision of platelet-rich plasma. Dr Noboru Mizushima (University of Tokyo, Japan) generously provided the GFP-LC3/+ transgenic mice. We also thank the University of Kentucky Imaging Facility and the Imaging Core of the National Institutes of Health Center of Biomedical Research Excellence (P20GM103486) for providing the access to microscopes and the University of Kentucky and Veterans Affairs Fluorescence-Activated Cell Sorting facilities for providing the access to flow cytometers.
This work was supported by National Institutes of Health Grants P20GM103486 (National Institute of General Medical Sciences; to Q.J.W.), HL56652 and HL091893 (National Heart, Lung, and Blood Institute; to S.W.W.), R01NS060123 (National Institute of Neurological Disorders; to Z.Y.), and R01HL119393 (National Heart, Lung, and Blood Institute; to B.S.); the Ellison Medical Foundation (Q.J.W.); University of Kentucky Research Support Grant and start-up fund (Q.J.W.); and American Heart Association Predoctoral Fellowship (Y.H.).
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: Q.J.W. conceived the project and coordinated the study. M.M.O., Y.H., Z.L., B.S., S.W.W., and Q.J.W. designed the experiments. M.M.O., Y.H., M.B., S.J., L.M., Y.Z., H.L., X.L., B.X., G.Z., M.K., Z.Y., Z.L., S.W.W., and Q.J.W. carried out the experiments. M.M.O., Y.H., M.B., B.S., S.W.W., and Q.J.W. analyzed the data. Q.J.W. wrote the manuscript, and S.W.W. helped with the editing. For experiments, both M.M.O. and Y.H. contributed to all figures; M.B. contributed to the aggregometry and cargo release assays; S.J. and L.M. contributed to the electron microscopic analysis; S.J. also contributed to the generation of the VAMP-deficient platelets; H.L. and S.W.W. contributed to the in vivo tail bleeding assay; G.Z. and H.L. contributed to the FeCl3 carotid injury model. Y.Z. contributed to Figures 1-3; H.L., Z.L., B.X., and G.Z. contributed to Figures 5-6; X.L., Q.J.W., and Z.Y. generated the EGFP-Atg5/+ BAC transgenic mice; Z.Y. also provided the Becn1-EGFP/+ BAC transgenic mice; and M.K. and M.M.O. generated the Atg7f/f;PF4-Cre/+ mice.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Qing Jun Wang, Department of Molecular and Cellular Biochemistry, University of Kentucky College of Medicine, B163 BBSRB, 741 South Limestone, Lexington, KY 40536; e-mail: qingjun.wang@uky.edu.
References
- 1.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451(7182):1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Massey AC, Kaushik S, Cuervo AM. Lysosomal chat maintains the balance. Autophagy. 2006;2(4):325–327. doi: 10.4161/auto.3090. [DOI] [PubMed] [Google Scholar]
- 3.Kunz JB, Schwarz H, Mayer A. Determination of four sequential stages during microautophagy in vitro. J Biol Chem. 2004;279(11):9987–9996. doi: 10.1074/jbc.M307905200. [DOI] [PubMed] [Google Scholar]
- 4.Uttenweiler A, Schwarz H, Neumann H, Mayer A. The vacuolar transporter chaperone (VTC) complex is required for microautophagy. Mol Biol Cell. 2007;18(1):166–175. doi: 10.1091/mbc.E06-08-0664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Suzuki K, Ohsumi Y. Molecular machinery of autophagosome formation in yeast, Saccharomyces cerevisiae. FEBS Lett. 2007;581(11):2156–2161. doi: 10.1016/j.febslet.2007.01.096. [DOI] [PubMed] [Google Scholar]
- 6.Itakura E, Kishi-Itakura C, Mizushima N. The hairpin-type tail-anchored SNARE syntaxin 17 targets to autophagosomes for fusion with endosomes/lysosomes. Cell. 2012;151(6):1256–1269. doi: 10.1016/j.cell.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 7.Nakatogawa H, Suzuki K, Kamada Y, Ohsumi Y. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat Rev Mol Cell Biol. 2009;10(7):458–467. doi: 10.1038/nrm2708. [DOI] [PubMed] [Google Scholar]
- 8.Lamb CA, Yoshimori T, Tooze SA. The autophagosome: origins unknown, biogenesis complex. Nat Rev Mol Cell Biol. 2013;14(12):759–774. doi: 10.1038/nrm3696. [DOI] [PubMed] [Google Scholar]
- 9.Mizushima N, Yoshimori T, Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol. 2011;27:107–132. doi: 10.1146/annurev-cellbio-092910-154005. [DOI] [PubMed] [Google Scholar]
- 10.Burman C, Ktistakis NT. Autophagosome formation in mammalian cells. Semin Immunopathol. 2010;32(4):397–413. doi: 10.1007/s00281-010-0222-z. [DOI] [PubMed] [Google Scholar]
- 11.Chang YY, Neufeld TP. An Atg1/Atg13 complex with multiple roles in TOR-mediated autophagy regulation. Mol Biol Cell. 2009;20(7):2004–2014. doi: 10.1091/mbc.E08-12-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jung CH, Jun CB, Ro SH, et al. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol Biol Cell. 2009;20(7):1992–2003. doi: 10.1091/mbc.E08-12-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ganley IG, Lam H, Wang J, Ding X, Chen S, Jiang X. ULK1.ATG13.FIP200 complex mediates mTOR signaling and is essential for autophagy. J Biol Chem. 2009;284(18):12297–12305. doi: 10.1074/jbc.M900573200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hosokawa N, Hara T, Kaizuka T, et al. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol Biol Cell. 2009;20(7):1981–1991. doi: 10.1091/mbc.E08-12-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hamasaki M, Furuta N, Matsuda A, et al. Autophagosomes form at ER-mitochondria contact sites. Nature. 2013;495(7441):389–393. doi: 10.1038/nature11910. [DOI] [PubMed] [Google Scholar]
- 16.Itakura E, Kishi C, Inoue K, Mizushima N. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol Biol Cell. 2008;19(12):5360–5372. doi: 10.1091/mbc.E08-01-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun Q, Fan W, Chen K, Ding X, Chen S, Zhong Q. Identification of Barkor as a mammalian autophagy-specific factor for Beclin 1 and class III phosphatidylinositol 3-kinase. Proc Natl Acad Sci USA. 2008;105(49):19211–19216. doi: 10.1073/pnas.0810452105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhong Y, Wang QJ, Li X, et al. Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1-phosphatidylinositol-3-kinase complex. Nat Cell Biol. 2009;11(4):468–476. doi: 10.1038/ncb1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matsunaga K, Saitoh T, Tabata K, et al. Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat Cell Biol. 2009;11(4):385–396. doi: 10.1038/ncb1846. [DOI] [PubMed] [Google Scholar]
- 20.Matsunaga K, Morita E, Saitoh T, et al. Autophagy requires endoplasmic reticulum targeting of the PI3-kinase complex via Atg14L. J Cell Biol. 2010;190(4):511–521. doi: 10.1083/jcb.200911141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Itakura E, Mizushima N. Characterization of autophagosome formation site by a hierarchical analysis of mammalian Atg proteins. Autophagy. 2010;6(6):764–776. doi: 10.4161/auto.6.6.12709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mizushima N, Noda T, Yoshimori T, et al. A protein conjugation system essential for autophagy. Nature. 1998;395(6700):395–398. doi: 10.1038/26506. [DOI] [PubMed] [Google Scholar]
- 23.Mizushima N, Sugita H, Yoshimori T, Ohsumi Y. A new protein conjugation system in human. The counterpart of the yeast Apg12p conjugation system essential for autophagy. J Biol Chem. 1998;273(51):33889–33892. doi: 10.1074/jbc.273.51.33889. [DOI] [PubMed] [Google Scholar]
- 24.Shintani T, Mizushima N, Ogawa Y, Matsuura A, Noda T, Ohsumi Y. Apg10p, a novel protein-conjugating enzyme essential for autophagy in yeast. EMBO J. 1999;18(19):5234–5241. doi: 10.1093/emboj/18.19.5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ichimura Y, Kirisako T, Takao T, et al. A ubiquitin-like system mediates protein lipidation. Nature. 2000;408(6811):488–492. doi: 10.1038/35044114. [DOI] [PubMed] [Google Scholar]
- 26.Hanada T, Noda NN, Satomi Y, et al. The Atg12-Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy. J Biol Chem. 2007;282(52):37298–37302. doi: 10.1074/jbc.C700195200. [DOI] [PubMed] [Google Scholar]
- 27.Kabeya Y, Mizushima N, Ueno T, et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19(21):5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takáts S, Nagy P, Varga Á, et al. Autophagosomal Syntaxin17-dependent lysosomal degradation maintains neuronal function in Drosophila. J Cell Biol. 2013;201(4):531–539. doi: 10.1083/jcb.201211160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fader CM, Sánchez DG, Mestre MB, Colombo MI. TI-VAMP/VAMP7 and VAMP3/cellubrevin: two v-SNARE proteins involved in specific steps of the autophagy/multivesicular body pathways. Biochim Biophys Acta. 2009;1793(12):1901–1916. doi: 10.1016/j.bbamcr.2009.09.011. [DOI] [PubMed] [Google Scholar]
- 30.Diao J, Liu R, Rong Y, et al. ATG14 promotes membrane tethering and fusion of autophagosomes to endolysosomes. Nature. 2015;520(7548):563–566. doi: 10.1038/nature14147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morishita H, Eguchi S, Kimura H, et al. Deletion of autophagy-related 5 (Atg5) and Pik3c3 genes in the lens causes cataract independent of programmed organelle degradation. J Biol Chem. 2013;288(16):11436–11447. doi: 10.1074/jbc.M112.437103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kundu M, Lindsten T, Yang CY, et al. Ulk1 plays a critical role in the autophagic clearance of mitochondria and ribosomes during reticulocyte maturation. Blood. 2008;112(4):1493–1502. doi: 10.1182/blood-2008-02-137398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schweers RL, Zhang J, Randall MS, et al. NIX is required for programmed mitochondrial clearance during reticulocyte maturation. Proc Natl Acad Sci USA. 2007;104(49):19500–19505. doi: 10.1073/pnas.0708818104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sandoval H, Thiagarajan P, Dasgupta SK, et al. Essential role for Nix in autophagic maturation of erythroid cells. Nature. 2008;454(7201):232–235. doi: 10.1038/nature07006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang J, Randall MS, Loyd MR, et al. Mitochondrial clearance is regulated by Atg7-dependent and -independent mechanisms during reticulocyte maturation. Blood. 2009;114(1):157–164. doi: 10.1182/blood-2008-04-151639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nishida Y, Arakawa S, Fujitani K, et al. Discovery of Atg5/Atg7-independent alternative macroautophagy. Nature. 2009;461(7264):654–658. doi: 10.1038/nature08455. [DOI] [PubMed] [Google Scholar]
- 37.Matsui M, Yamamoto A, Kuma A, Ohsumi Y, Mizushima N. Organelle degradation during the lens and erythroid differentiation is independent of autophagy. Biochem Biophys Res Commun. 2006;339(2):485–489. doi: 10.1016/j.bbrc.2005.11.044. [DOI] [PubMed] [Google Scholar]
- 38.Mortensen M, Ferguson DJ, Edelmann M, et al. Loss of autophagy in erythroid cells leads to defective removal of mitochondria and severe anemia in vivo. Proc Natl Acad Sci USA. 2010;107(2):832–837. doi: 10.1073/pnas.0913170107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cao Y, Zhang A, Cai J, et al. Autophagy regulates the cell cycle of murine HSPCs in a nutrient-dependent manner. Exp Hematol. 2015;43(3):229–242. doi: 10.1016/j.exphem.2014.11.002. [DOI] [PubMed] [Google Scholar]
- 40.Feng W, Chang C, Luo D, et al. Dissection of autophagy in human platelets. Autophagy. 2014;10(4):642–651. doi: 10.4161/auto.27832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell. 2004;15(3):1101–1111. doi: 10.1091/mbc.E03-09-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kuma A, Mizushima N. Chromosomal mapping of the GFP-LC3 transgene in GFP-LC3 mice. Autophagy. 2008;4(1):61–62. doi: 10.4161/auto.4846. [DOI] [PubMed] [Google Scholar]
- 43.Arsov I, Li X, Matthews G, et al. BAC-mediated transgenic expression of fluorescent autophagic protein Beclin 1 reveals a role for Beclin 1 in lymphocyte development. Cell Death Differ. 2008;15(9):1385–1395. doi: 10.1038/cdd.2008.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Heintz N. BAC to the future: the use of bac transgenic mice for neuroscience research. Nat Rev Neurosci. 2001;2(12):861–870. doi: 10.1038/35104049. [DOI] [PubMed] [Google Scholar]
- 45.Ren Q, Barber HK, Crawford GL, et al. Endobrevin/VAMP-8 is the primary v-SNARE for the platelet release reaction. Mol Biol Cell. 2007;18(1):24–33. doi: 10.1091/mbc.E06-09-0785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tiedt R, Schomber T, Hao-Shen H, Skoda RC. Pf4-Cre transgenic mice allow the generation of lineage-restricted gene knockouts for studying megakaryocyte and platelet function in vivo. Blood. 2007;109(4):1503–1506. doi: 10.1182/blood-2006-04-020362. [DOI] [PubMed] [Google Scholar]
- 47.Kim JC, Cook MN, Carey MR, Shen C, Regehr WG, Dymecki SM. Linking genetically defined neurons to behavior through a broadly applicable silencing allele. Neuron. 2009;63(3):305–315. doi: 10.1016/j.neuron.2009.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Komatsu M, Waguri S, Ueno T, et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol. 2005;169(3):425–434. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Choi W, Karim ZA, Whiteheart SW. Arf6 plays an early role in platelet activation by collagen and convulxin. Blood. 2006;107(8):3145–3152. doi: 10.1182/blood-2005-09-3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ren Q, Wimmer C, Chicka MC, et al. Munc13-4 is a limiting factor in the pathway required for platelet granule release and hemostasis. Blood. 2010;116(6):869–877. doi: 10.1182/blood-2010-02-270934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen D, Lemons PP, Schraw T, Whiteheart SW. Molecular mechanisms of platelet exocytosis: role of SNAP-23 and syntaxin 2 and 4 in lysosome release. Blood. 2000;96(5):1782–1788. [PubMed] [Google Scholar]
- 52.Lemons PP, Chen D, Whiteheart SW. Molecular mechanisms of platelet exocytosis: requirements for alpha-granule release. Biochem Biophys Res Commun. 2000;267(3):875–880. doi: 10.1006/bbrc.1999.2039. [DOI] [PubMed] [Google Scholar]
- 53.Schraw T, Whiteheart S. The development of a quantitative enzyme-linked immunosorbent assay to detect human platelet factor 4. Transfusion. 2005;45(5):717–724. doi: 10.1111/j.1537-2995.2005.04243.x. [DOI] [PubMed] [Google Scholar]
- 54.Chen D, Bernstein AM, Lemons PP, Whiteheart SW. Molecular mechanisms of platelet exocytosis: role of SNAP-23 and syntaxin 2 in dense core granule release. Blood. 2000;95(3):921–929. [PubMed] [Google Scholar]
- 55.Schraw TD, Rutledge TW, Crawford GL, et al. Granule stores from cellubrevin/VAMP-3 null mouse platelets exhibit normal stimulus-induced release. Blood. 2003;102(5):1716–1722. doi: 10.1182/blood-2003-01-0331. [DOI] [PubMed] [Google Scholar]
- 56.Zhang G, Xiang B, Dong A, et al. Biphasic roles for soluble guanylyl cyclase (sGC) in platelet activation. Blood. 2011;118(13):3670–3679. doi: 10.1182/blood-2011-03-341107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rowley JW, Oler AJ, Tolley ND, et al. Genome-wide RNA-seq analysis of human and mouse platelet transcriptomes. Blood. 2011;118(14):e101–e111. doi: 10.1182/blood-2011-03-339705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dittrich M, Birschmann I, Pfrang J, et al. Analysis of SAGE data in human platelets: features of the transcriptome in an anucleate cell. Thromb Haemost. 2006;95(4):643–651. [PubMed] [Google Scholar]
- 59.Craig R, Cortens JP, Beavis RC. Open source system for analyzing, validating, and storing protein identification data. J Proteome Res. 2004;3(6):1234–1242. doi: 10.1021/pr049882h. [DOI] [PubMed] [Google Scholar]
- 60.Lewandrowski U, Wortelkamp S, Lohrig K, et al. Platelet membrane proteomics: a novel repository for functional research. Blood. 2009;114(1):e10–e19. doi: 10.1182/blood-2009-02-203828. [DOI] [PubMed] [Google Scholar]
- 61.Burkhart JM, Vaudel M, Gambaryan S, et al. The first comprehensive and quantitative analysis of human platelet protein composition allows the comparative analysis of structural and functional pathways. Blood. 2012;120(15):e73–e82. doi: 10.1182/blood-2012-04-416594. [DOI] [PubMed] [Google Scholar]
- 62.Zeiler M, Moser M, Mann M. Copy number analysis of the murine platelet proteome spanning the complete abundance range. Mol Cell Proteomics. 2014;13(12):3435–3445. doi: 10.1074/mcp.M114.038513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shi DS, Smith MC, Campbell RA, et al. Proteasome function is required for platelet production. J Clin Invest. 2014;124(9):3757–3766. doi: 10.1172/JCI75247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gupta N, Li W, Willard B, Silverstein RL, McIntyre TM. Proteasome proteolysis supports stimulated platelet function and thrombosis. Arterioscler Thromb Vasc Biol. 2014;34(1):160–168. doi: 10.1161/ATVBAHA.113.302116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ponpuak M, Mandell MA, Kimura T, Chauhan S, Cleyrat C, Deretic V. Secretory autophagy. Curr Opin Cell Biol. 2015;35:106–116. doi: 10.1016/j.ceb.2015.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Menzies FM, Moreau K, Puri C, Renna M, Rubinsztein DC. Measurement of autophagic activity in mammalian cells. Curr Protoc Cell Biol. 2012;Chapter 15:16. doi: 10.1002/0471143030.cb1516s54. [DOI] [PubMed] [Google Scholar]
- 67.Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140(3):313–326. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bjørkøy G, Lamark T, Brech A, et al. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol. 2005;171(4):603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang QJ, Ding Y, Kohtz DS, et al. Induction of autophagy in axonal dystrophy and degeneration. J Neurosci. 2006;26(31):8057–8068. doi: 10.1523/JNEUROSCI.2261-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhong Y, Morris DH, Jin L, et al. Nrbf2 protein suppresses autophagy by modulating Atg14L protein-containing Beclin 1-Vps34 complex architecture and reducing intracellular phosphatidylinositol-3 phosphate levels. J Biol Chem. 2014;29(38):26021–26037. doi: 10.1074/jbc.M114.561134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Torisu T, Torisu K, Lee IH, et al. Autophagy regulates endothelial cell processing, maturation and secretion of von Willebrand factor. Nat Med. 2013;19(10):1281–1287. doi: 10.1038/nm.3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yau JW-H, Hou Y, Lei X, Singh K, Verma S. Autophagy regulates thrombus formation in mice. Presented at International Society on Thrombosis and Haemostasis 2015 Congress. June 22, 2015. Toronto, Ontario, Canada. [Google Scholar]
- 73.Qu X, Zou Z, Sun Q, et al. Autophagy gene-dependent clearance of apoptotic cells during embryonic development. Cell. 2007;128(5):931–946. doi: 10.1016/j.cell.2006.12.044. [DOI] [PubMed] [Google Scholar]
- 74.Zwaal RF, Comfurius P, Bevers EM. Platelet procoagulant activity and microvesicle formation. Its putative role in hemostasis and thrombosis. Biochim Biophys Acta. 1992;1180(1):1–8. doi: 10.1016/0925-4439(92)90019-j. [DOI] [PubMed] [Google Scholar]
- 75.de Witt SM, Verdoold R, Cosemans JMEM, Heemskerk JWM. Insights into platelet-based control of coagulation. Thromb Res. 2014;133(Suppl 2):S139–S148. doi: 10.1016/S0049-3848(14)50024-2. [DOI] [PubMed] [Google Scholar]
- 76.Woulfe D, Jiang H, Morgans A, Monks R, Birnbaum M, Brass LF. Defects in secretion, aggregation, and thrombus formation in platelets from mice lacking Akt2. J Clin Invest. 2004;113(3):441–450. doi: 10.1172/JCI20267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Konopatskaya O, Gilio K, Harper MT, et al. PKCalpha regulates platelet granule secretion and thrombus formation in mice. J Clin Invest. 2009;119(2):399–407. doi: 10.1172/JCI34665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Adam F, Kauskot A, Nurden P, et al. Platelet JNK1 is involved in secretion and thrombus formation. Blood. 2010;115(20):4083–4092. doi: 10.1182/blood-2009-07-233932. [DOI] [PubMed] [Google Scholar]
- 79.Canobbio I, Cipolla L, Consonni A, et al. Impaired thrombin-induced platelet activation and thrombus formation in mice lacking the Ca(2+)-dependent tyrosine kinase Pyk2. Blood. 2013;121(4):648–657. doi: 10.1182/blood-2012-06-438762. [DOI] [PubMed] [Google Scholar]
- 80.Ye S, Huang Y, Joshi S, et al. Platelet secretion and hemostasis require syntaxin-binding protein STXBP5. J Clin Invest. 2014;124(10):4517–4528. doi: 10.1172/JCI75572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kraemer BF, Weyrich AS, Lindemann S. Protein degradation systems in platelets. Thromb Haemost. 2013;110(5):920–924. doi: 10.1160/TH13-03-0183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yukawa M, Sakon M, Kambayashi J, et al. Proteasome and its novel endogeneous activator in human platelets. Biochem Biophys Res Commun. 1991;178(1):256–262. doi: 10.1016/0006-291x(91)91807-o. [DOI] [PubMed] [Google Scholar]
- 83.Yukawa M, Sakon M, Kambayashi J, et al. Purification and characterization of endogenous protein activator of human platelet proteasome. J Biochem. 1993;114(3):317–323. doi: 10.1093/oxfordjournals.jbchem.a124174. [DOI] [PubMed] [Google Scholar]
- 84.Dangelmaier CA, Quinter PG, Jin J, Tsygankov AY, Kunapuli SP, Daniel JL. Rapid ubiquitination of Syk following GPVI activation in platelets. Blood. 2005;105(10):3918–3924. doi: 10.1182/blood-2004-09-3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pieczarka EM, Yamaguchi M, Wellman ML, Judith Radin M. Platelet vacuoles in a dog with severe nonregenerative anemia: evidence of platelet autophagy. Vet Clin Pathol. 2014;43(3):326–329. doi: 10.1111/vcp.12170. [DOI] [PubMed] [Google Scholar]