Abstract
Very little is known about the effect of gut microbiota on the ontogeny of drug-processing genes (DPGs) in liver. In this study, livers were harvested from conventional (CV) and germ-free (GF) male and female mice from 1 to 90 days of age. RNA-Seq in livers of 90-day-old male mice showed that xenobiotic metabolism was the most downregulated pathway within the mRNA transcriptome in absence of intestinal bacteria. In male livers, the mRNAs of 67 critical DPGs partitioned into 4 developmental patterns (real-time-quantitative polymerase chain reaction): Pattern-1 gradually increased to adult levels in livers of CV mice and were downregulated in livers of GF mice, as exemplified by the major drug-metabolizing enzymes cytochrome 3a (Cyp3a) family, which are prototypical pregnane X receptor (PXR)-target genes. Genes in Pattern-2 include Cyp1a2 (aryl hydrocarbon receptor-target gene), Cyp2c family, and Cyp2e1, which were all upregulated mainly at 90 days of age; as well as the peroxisome proliferator-activated receptor α (PPARα)-target genes Cyp4a family and Aldh3a2, which were upregulated not only in 90-days adult age, but also between neonatal and adolescent ages (from 1 to 30 days of age). Genes in Pattern-3 were enriched predominantly in livers of 15-day-old mice, among which the sterol-efflux transporter dimers Abcg5/Abcg8 were downregulated in GF mice. Genes in Pattern-4 were neonatal-enriched, among which the transporter Octn1 mRNA tended to be lower in GF mice at younger ages but higher in adult GF mice as compared with age-matched CV mice. Protein assays confirmed the downregulation of the PXR-target gene Cyp3a protein (Western-blot and liquid chromatography tandem mass spectroscopy), and decreased Cyp3a enzyme activities in male GF livers. Increased microsomal-Cyp4a proteins and nuclear-PPARα were also observed in male GF livers. Interestingly, in contrast to male livers, the mRNAs of Cyp2c or Cyp4a were not readily upregulated in female GF livers approaching adult age, suggesting the maturation of female-specific hormones interferes with the interactions between intestinal microbiota and DPG ontogeny. In conclusion, intestinal microbiota markedly impacts the ontogeny of many hepatic DPGs in a gender-specific manner.
Keywords: germ-free, drug-processing genes, liver development
Infants are not miniature adults, and profound changes occur in the expression patterns and functional capacities of various drug-metabolizing enzymes and transporters (drug-processing genes [DPGs]) after birth (Hines, 2013; Mooij et al., 2014; Strolin Benedetti et al., 2005), leading to marked pharmacokinetic differences between newborns and adults (Anderson, 2002; Fernandez-Fernandez et al., 2011). Drug-metabolizing enzymes include phase-1 enzymes that perform hydrolysis, reduction, and oxidation, and phase-2 enzymes that perform conjugation. DPGs are regulated by xenobiotic-sensing transcription factors such as the aryl hydrocarbon receptor (AhR), constitutive androstane receptor (CAR, NR1I3), pregnane X receptor (PXR, NR1I2), and peroxisome proliferator-activated receptor α (PPARα, NR1C1), as well as the oxidative stress sensor nuclear factor erythroid 2-related factor 2 (Nrf2, Nfe2L2).
Liver is a major organ for drug metabolism and transport, and the hepatic DPGs display unique developmental patterns (Mooij et al., 2014), with newborns usually having reduced hepatic clearance (Crom et al., 1991; Klaassen, 1973). Because very little is known regarding the regulation of the ontogeny of DPGs, newborns and children are at a much higher risk of adverse drug reactions. Therefore, there is a need to characterize the ontogeny patterns of DPGs and the differences in the pharmacokinetics in children as compared with adults (Kearns et al., 2003). Recently, we and others have unveiled distinct developmental patterns of various hepatic phase-1 and phase-2 enzymes as well as transporters (Cui et al., 2012a,b; Fattah et al., 2014; Hart et al., 2009 ; Lee et al., 2011; Lu et al., 2013; Peng et al., 2012, 2013).
At the same time that these ontogeny changes of DPGs are occurring in the liver, there are distinct changes in the composition and number of intestinal microbiota as part of the development process (Koenig et al., 2011). Intestinal microbiota of infants undergoes rapid changes and are adult-like in composition and number at around 3 years of age (Park et al., 2005; Palmer et al., 2007; Yatsunenko et al., 2012). However, it is not known whether the intestinal microbiota alters the ontogeny of DPGs in the host liver. This is because most previous reports have focused on determining the regulation of DPGs by transcription factors and epigenetic modifiers (Aleksunes and Klaassen, 2012; Cui et al., 2012a; Jover et al., 2009; Kamiya et al., 2003; Liddle and Goodwin, 2002). However, recent studies have suggested that intestinal microbiota may serve as an additional mechanism for regulating DPGs in liver, at least in adult mice (Bjorkholm et al., 2009; Toda et al., 2009b). For example, adult germ-free (GF) mice have decreased expression of hepatic cytochrome P450 (Cyp) 3a, which encodes the major phase-I drug-metabolizing enzyme (Toda et al., 2009b).
Intestinal microbiota are altered by various factors such as diet, environmental exposure, host genetics, and microbial exposure (Human Microbiome Project Consortium, 2012). Changes in diet take as little as 3 days to change the composition of intestinal microbiota (David et al., 2014). Antibiotic-treatment in early ages of life alters the intestinal microbiota, and is associated with increased adiposity and other pediatric diseases such as necrotizing enterocolitis, allergy, and autism-spectrum disorder (Arrieta et al., 2014; Cho et al., 2012). Probiotic-supplementation during weaning leads to altered metabolic profile, improved immune response, and reduces the incidence of eczema (Chorell et al., 2013; West et al., 2009, 2012). This illustrates that the intestinal microbiota is an important regulator of host physiology in children as much as in adults. However, there are essentially no studies in the literature that systematically characterize the effect of intestinal microbiota on the ontogeny of drug processing genes in liver. Therefore, this study analyzed the ontogeny of critical hepatic DPGs in GF mice and CV mice at various developmental ages to determine how intestinal microbiota affects the ontogeny of DPGs and the xeno-sensor signaling pathways. To quantitatively determine the proteins of important DPGs in liver, a highly sensitive and quantitative liquid chromatography tandem mass spectroscopy (LC-MS/MS) method was utilized as described previously (Prasad and Unadkat, 2014a,b; Prasad et al., 2014).
MATERIALS AND METHODS
Animal Procedures
All mice were housed in an AAALAC (Association for Assessment and Accreditation of Laboratory Animal Care International)-accredited facility at the University of Kansas Medical Center, with a 14-h light/10-h dark-cycle, in a temperature and humidity-controlled environment, with ad libitum access to water. The initial breeding colony of GF C57BL/6J/UNC mice was established with mice purchased from the National Gnotobiotic Rodent Resource Center (University of North Carolina, Chapel Hill). All conventional (CV) mice were purchased from the Jackson Laboratories. GF mice were housed in vinyl flexible film isolators purchased from Class Biologically Clean Ltd. (Madison, Wisconsin). The condition of GF status at the end of the study was confirmed by 16S rRNA assay targeting the universal bacteria in the DNA extract from the intestinal content (data not shown). All animal experiments were approved by the Institutional Animal Care and Use Committee at the University of Kansas Medical Center.
RNA Isolation
Total RNA was isolated from tissues using RNA Bee reagent (Tel-Test Inc., Friendswood, Texas) following the manufacturer's protocol. The concentration of total RNA in each sample was quantified spectrophotometrically at 260 nm using a NanoDrop 1000 Spectrophotometer (Thermo Scientific, Waltham, Massachusetts). RNA integrity was confirmed using a dual Agilent 2100 Bioanalyzer (Agilent Technologies Inc. Santa Clara, California).
RNA-Sequencing
The cDNA library preparation and sequencing of male CV- and GF-mouse livers at 90 days of age were performed using an Illumina TruSeq RNA sample prep kit (Illumina, San Diego, California). The cDNA libraries were clustered onto a TruSeq paired-end flow cell and sequenced (2 × 50 bp) using a TruSeq SBS kit (Illumina) on the Illumina HiSeq2000 sequencer. Data were analyzed using a similar method as we described previously (Cui et al., 2012a). The differentially regulated genes were determined using CuffDiff (false discovery rate (Benjamini Hockberg) < 0.05), and these genes were subjected to Ingenuity Pathway Analysis (IPA) for the significantly altered networks between the CV- and GF-liver transcriptome.
Reverse Transcription and Real-time Polymerase Chain Reaction Analysis
Total RNA was transcribed to single-stranded cDNA using a High Capacity cDNA Reverse Transcription Kit 1001073 (Applied Biosystems, Foster City, California). The cDNAs were amplified by polymerase chain reaction (PCR), using SsoAdvanced Universal SYBR Green Supermix in a Bio-Rad CFX384 Real-Time PCR Detection System (Bio-Rad, Hercules, California). The PCR primers were synthesized by Integrated DNA Technologies (Coralville, Iowa) and the sequences are shown in Supplementary Table 1. The ddCq values were calculated for each target gene and were normalized to the expression of the geometric means of the housekeeping genes β-actin, Gapdh, and 18 S rRNA.
Western Blotting
Primary antibody against mouse Cyp3a11 (anti-rat CYP3A1/2 mAb, clone 2-13-1, 1:500) was a generous gift from Dr Frank Gonzalez at the National Cancer Institute. Primary antibody against Cyp4a14 (goat polyclonal IgG, 1:500) was purchased from Santa Cruz Biotechologies (SC-46087). Primary antibodies against mouse histone H3 and β-actin were purchased from Abcam (ab12079 and ab8227, respectively, 1:500). Protein concentrations were determined using a Qubit Protein Assay Kit (Thermo Fisher Scientific, Life Technologies, Grand Island, New York). The samples were subjected to polyacrylamide gel electrophoresis and transferred onto a polyvinylidene difluoride membrane. (horseradish peroxidase) HRP-linked secondary antibodies were purchased from Sigma Aldrich, and applied at 1:2000 to the proteins to be detected (namely rabbit anti-mouse A9044 for Cyp3a11 and PPARα, rabbit anti-goat A5420 for Cyp4a14, and goat anti-rabbit A6154 for H3). Proteins were detected using chemiluminescence (Thermo Fisher Scientific, Life Technologies). Intensities of the protein bands were quantified using the Image J Software (National Institutes of Health, Bethesda, Maryland).
Cyp Enzyme Activity Assay
Cyp3a enzyme activity in liver microsomes from 90-day-old CV- and GF mice was determined using the Promega P450-Glo CYP3A4 Luciferin-IPA Assay (which also recognizes the mouse Cyp3a isoform) per the manufacturer’s instructions with minor modifications. Luciferin-IPA (7907MB) is a highly sensitive and selective substrate for CYP3A (Doshi and Li, 2011). Briefly, hepatic microsomes and Luciferin-isopropyl acetal were combined in potassium phosphate (KPO4) buffer with the addition of an (Nicotinamide adenine dinucleotide phosphate) NADPH-regeneration system, and were incubated at 37°C for 10 min. A standard curve with various concentrations of pooled hepatic microsomes was generated before the individual assays, to ensure the loaded amount was within the linear range of detection. A negative control (with the addition of the CYP3A inhibitor Ketoconazole) and a minus-P450 negative control were performed in parallel to ensure specificity of the assay. An equal volume of Luciferin detection reagent was added to simultaneously stop the Cyp reaction and initiate luminescent signaling that is proportional to the amount of Cyp activity. Signals were then allowed to stabilize for 20 min at room temperature before determining the luminescence.
Serum ALT
Serum samples were analyzed by standard enzymatic-colorimetric assays using ALT kits according to the manufacturer’s protocol (Pointe Scientific, Inc.) as described previously (Cui et al., 2009).
LC/MS-MS Protein Quantification
Light peptide standards (Supplementary Table 3) for various proteins were obtained from New England Peptides (Boston, Massachusetts) or Thermo Fisher Scientific (Rockford, Illinois). The corresponding stable isotope-labeled heavy peptides (internal standards) were obtained from Thermo Fisher Scientific. High-performance liquid chromatography–grade acetonitrile was purchased from Fischer Scientific (Fair Lawn, New Jersey), and formic acid was purchased from Sigma-Aldrich (St. Louis, Missouri). All reagents were analytical grade. Other reagents used for trypsin digestion of membrane proteins and sample preparations include iodoacetamide and dithiothreitol (Pierce Biotechnology, Rockford, Illinois), and as well as ammonium bicarbonate (Acros Organics, New Jersey).
Briefly, the procedure is similar as published in a recent paper with few modifications (Prasad et al., 2014). For LC-MS/MS transporter/enzyme quantification unique signature surrogate peptides were selected using the published approach (Prasad, AAPS Journal, 2014) (Supplementary Table 3). The membrane proteins of the mouse livers were isolated using the ProteoExtract native membrane protein extraction kit (Calbiochem, Temecula, California). The proteins were quantified using a bicinchoninic acid kit, followed by digestion using an in-solution trypsin digestion kit (Pierce Biotechnology). Briefly, 80 µl of the tissue extract (2 mg/ml total protein concentration) was mixed with 20 µl dithiothreitol (100 mM) and 50 µl ammonium bicarbonate buffer (100 mM, pH 7.8), and incubated at 95°C for 5 min (denaturation and reduction). Subsequently, 20 µl iodoacetamide (200 mM) was added and the sample was incubated for 20 min at ambient temperature in the dark (alkylation). Ice-cold methanol (0.5 ml), chloroform (0.2 ml), and water (0.4 ml) were added to each sample. After centrifugation at 16,000 × g for 5 min at 4°C, the upper layer was removed and the pellet was washed with ice-cold methanol (0.5 ml) and centrifuged at 16,000 × g for 5 min at 4°C. The pellet was resuspended with 40 µl of reconstitution buffer (mixture of equal volume of 3% sodium deoxycholate (wt/vol) and 50 mM ammonium bicarbonate buffer). Trypsin (20 µl) was added at 1:25 trypsin: protein ratio (wt/wt) and samples were incubated for 18 h at 37°C. Trypsin digestion was stopped by adding 30 µl of chilled quenching solvent (50% acetonitrile, with 0.1% formic acid) containing heavy peptide internal standard, and samples were centrifuged for 5 min at 4000 × g and 4°C. Specific transporter/enzyme proteins were determined by comparing to gene-specific light-calibrator peptide standards (Supplementary Table 3, New England Peptides and Thermo Fisher Scientific. The corresponding stable isotope-labeled heavy peptides were used as internal standards (Thermo Fisher Scientific). LC-MS/MS protein quantification was carried out on a Xevo TQ-S, Triple Quadrupole Mass Spectrometer (Waters Corporation, Milford, Massachusetts) coupled to an ACQUITY Ultra Performance LCT (UPLC) System. The supernatant was analyzed by LC-MS/MS using the ACQUITY UPLC HSS T3 1.8 µm, 2.1 × 100 mm column (Waters, Hertfordshire, UK) at a flow rate of 0.3 ml/min at 23°C ± 2°C column temperature and 5 µl injection volume. The Mobile phase and gradient conditions are listed in Supplementary Table 4.
Clustering Analysis
A hierarchical clustering dendrogram (Ward’s minimum variance method, distance scale) was performed based on the mean mRNA expression of 62 DPGs with known importance in hepatic drug metabolism and transport functions.
Statistical Analysis
Data are presented as mean ± SEM. Asterisks (*) represent significant differences between CV and GF mice at the same age, as determined by the Student’s t test (P < .05).
RESULTS
There were no apparent differences in the body weight, liver weight, or serum ALT levels between CV and GF mice (data not shown).
Xenobiotic Metabolism Is the Most Downregulated Pathway Evidenced by mRNA Transcriptome Sequencing of Livers From 90-Day-Old GF Mice
To determine the contribution of the intestinal microbiota on regulating the host xenobiotic metabolism pathway in liver, as compared with its contribution to many other pathways on a genomic scale, using 90-day-old mice as a starting point, we performed RNA-Seq in male livers of CV and GF mice (90 days of age, n = 3/group). Among approximately 900 genes that are differentially regulated in livers of GF relative to CV mice, IPA reported that xenobiotic metabolism ranks among the most differentially regulated network in livers of GF mice. The mRNAs of many DPGs were decreased in livers of GF mice (Fig. 1A), including several Cyps in phase-I and certain glutathione S-transferases (Gsts) in phase-II metabolism. Therefore, the absence of gut microbiota appears to markedly impact xenobiotic metabolism, at least in livers of 90-day-old mice. To further investigate whether the intestinal microbiota also downregulates the ontogenic expression of Cyp3a and Gsts, as well as other DPGs, we quantified the mRNAs of 62 critical DPGs at various ages during development, as well as 5 xenobiotic-sensing receptors in CV and GF mice by RT-quantitative PCR (qPCR).
FIG. 1.
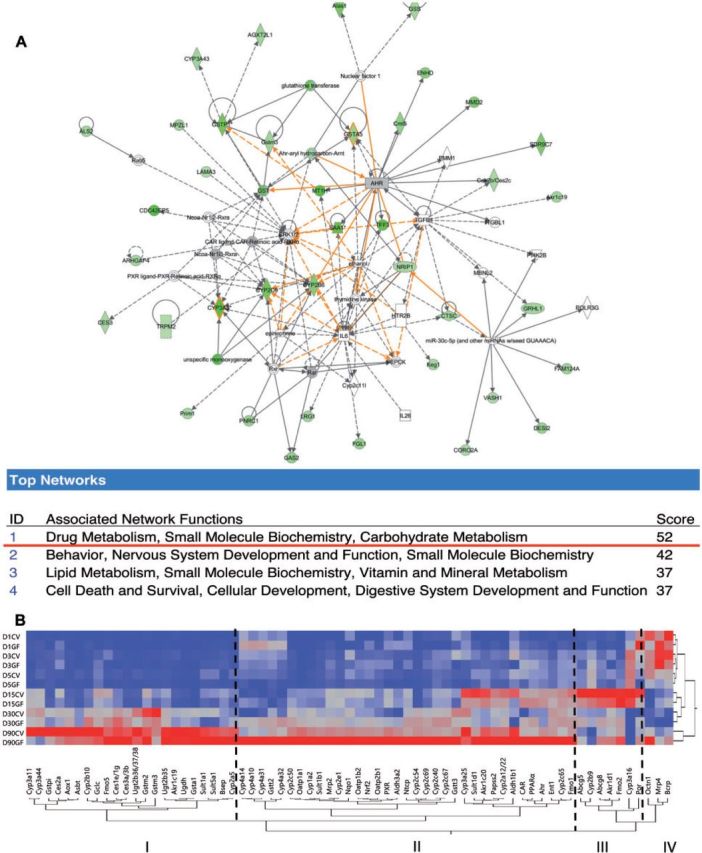
A, IPA analysis of genes expression changes in male GF mice at 90 days of age compared with male CV mice. B, Two-way hierarchical clustering dendrogram of critical DPGs gene expression at 1, 3, 5, 15, 30, and 90 days of age in the livers of male CV and GF mice. (n = 4/group). IPA, Ingenuity Pathway Analysis. Data are expressed at mean % of the geometric means of the housekeeping genes β-actin, Gapdh, and 18S rRNA.
As shown in Figure 1B, a two-way hierarchical clustering dendrogram of the average gene expression (from n = 4 in both CV and GF mice) revealed 4 distinct developmental patterns of the 67 DPGs in livers of CV and GF mice at 1, 3, 5, 15, 30, and 90 days of age. From left to right, Pattern 1 includes 22 DPGs, the mRNAs of these DPGs are lowly expressed at younger ages, and gradually increase to adult levels in both CV and GF mice. However, in livers of GF mice, the developmental increase of most of these genes was attenuated between 15 and 90 days of age, exemplified by Cyp3a11, which encodes the major drug-metabolizing enzyme for the biotransformation of over 75% of prescribed drugs. Pattern 2 is also an adult-enriched pattern, which includes 35 DPGs. Interestingly, contrary to our hypothesis that other DPGs are downregulated in absence of gut microbiota in liver, these DPGs were upregulated in livers of GF mice between 15 and 90 day of age. In general, many of the DPGs in this pattern are not only important for processing drugs, but also play critical roles in nutrient homeostasis, exemplified by the Cyp4 family members that are targets of the lipid-sensing nuclear receptor PPARα, as well as Ntcp and Oatp1b2 that are major uptake transporters for conjugated and unconjugated bile acids, respectively. In addition, many Cyp2c family members were also increased markedly in livers of GF mice.
Pattern 3 includes 7 DPGs that were enriched predominantly at 15 days of age in both CV and GF mice and were regulated by intestinal microbiota in an age-specific pattern. For example, the sterol efflux transporters Abcg5 and Abcg8 were both downregulated at 15 days of age, but were upregulated thereafter compared with CV mice. Pattern 4 includes 3 DPGs that were neonatal-enriched in both CV and GF livers; however, the carnitine transporter Octn1 was markedly upregulated in livers of 90-day-old GF mice.
Hepatic Ontogeny of Cyps in Male Mouse Livers
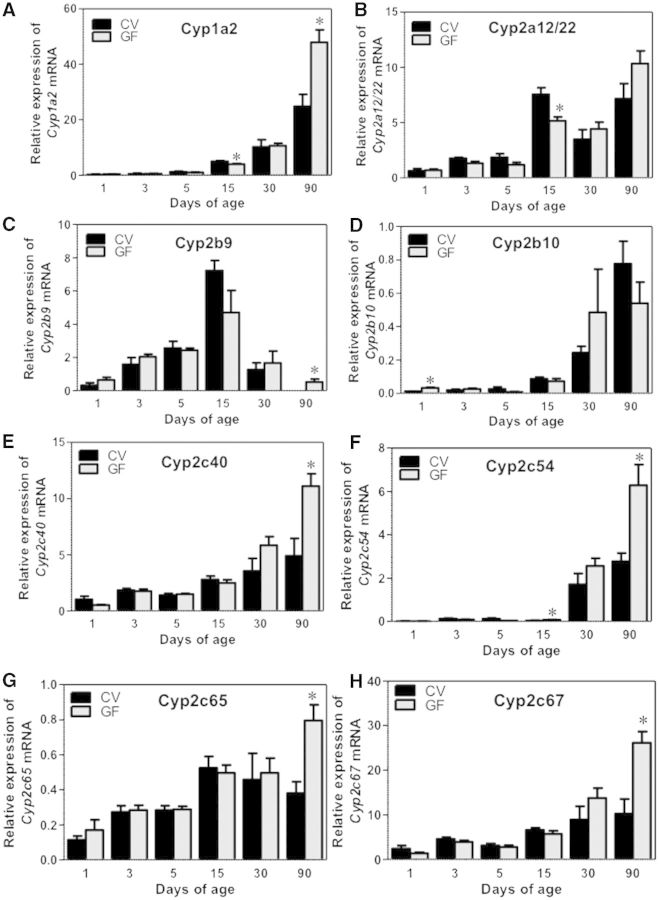
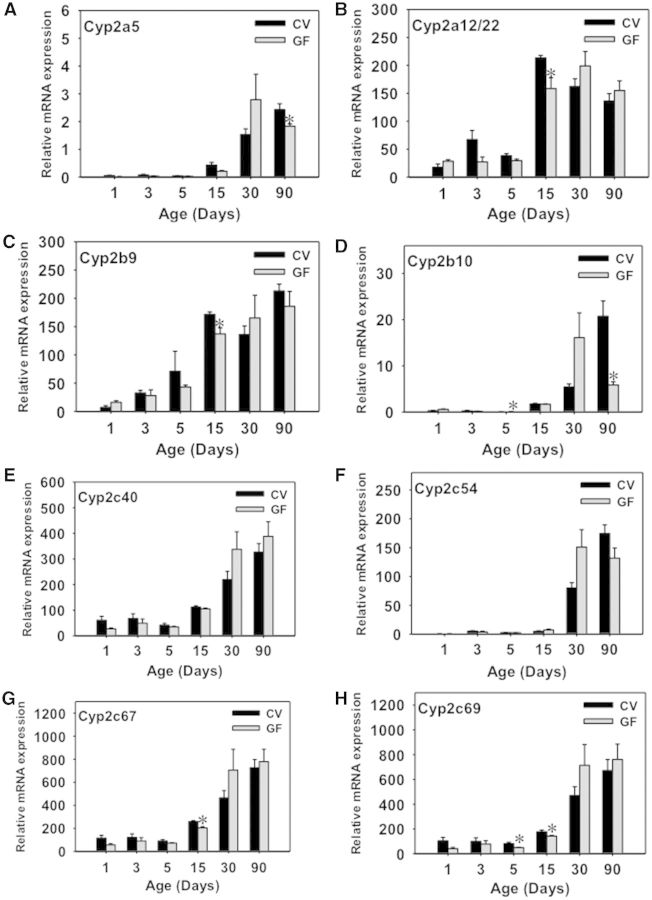
The ontogeny of 19 critical Cyp genes was analyzed in livers of CV and GF mice. The mRNA of Cyp1a2 increases gradually from birth to adulthood in livers of both CV and GF mice. Although the mRNA of Cyp1a2 was slightly lower in GF mice at 15 days of age, it increased 2-fold in GF mice at 90 days of age, compared with age-matched CV mice (Fig. 2A). There were no differences in the mRNA of Cyp1a2 between CV and GF mouse livers at other ages. Regarding the Cyp2a gene family, Cyp2a12/22 mRNA was moderately lower in GF than in CV livers at 15 days of age, but there were no differences between CV and GF levels at other ages. In CV-mouse livers, the peak expression of Cyp2a12/22 was observed at 15 days of age, however, in GF-mouse livers, the peak expression of Cyp2a12/22 was observed at 90 days of age (Fig. 2B). Cyp2a5 mRNA gradually increased to adult levels in both CV- and GF-mouse livers, and there was no difference in the expression between the two mouse models (Supplementary Fig. 1A). Regarding the Cyp2b family, Cyp2b9 mRNA displayed an adolescent-enriched pattern with peak expression observed at 15 days of age in both CV and GF livers, whereas Cyp2b10 displayed an adult-enriched pattern with peak expression observed at 90 days of age (Fig. 2C and D). CV-mouse livers had minimal Cyp2b9 mRNA at 90 days of age, whereas Cyp2b9 mRNA increased markedly in age-matched GF-mouse livers (Fig. 2C). At 1 day of age, moderately higher Cyp2b10 mRNA expression was observed in GF mouse livers as compared with age-matched CV-mouse livers (Fig. 2D). There was no difference in the mRNA expression of these Cyp2b genes at other ages.
FIG. 2.
Ontogeny of Phase-I enzymes cytochrome P450s Cyp1a2 to Cyp2c67 gene expression in male livers of CV and GF mice. Data are presented as means ± SEM, n = 4/group. Asterisks (*) represent statistically significant differences between CV and GF mice (P < .05) by Student’s t test in that age group. Data are expressed at mean % of the geometric means of the housekeeping genes β-actin, Gapdh, and 18S rRNA.
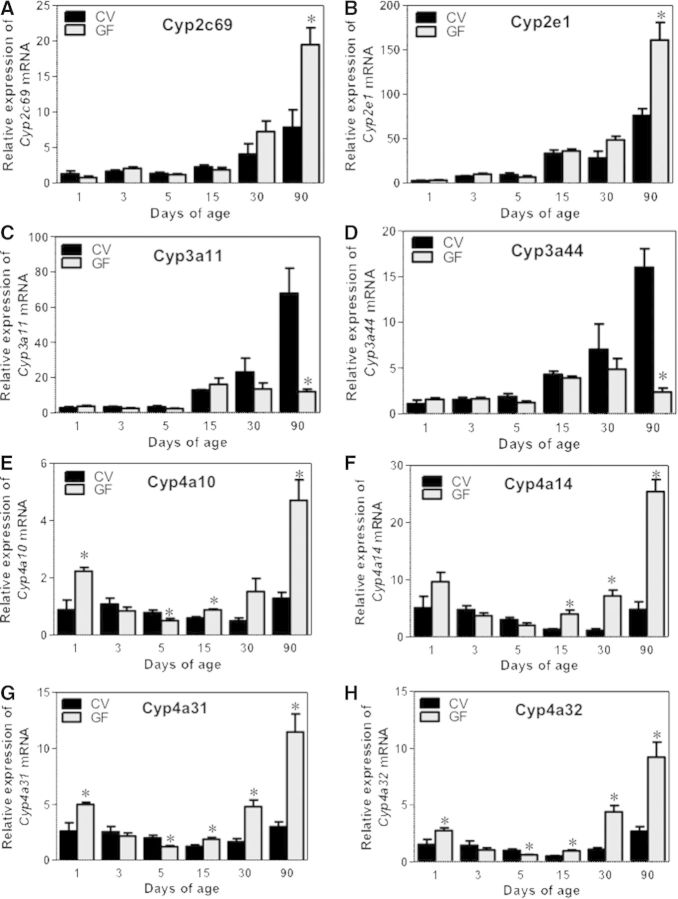
The Cyp2c gene family members all appeared to gradually increase to adult levels in CV- and GF-mouse livers. Interestingly, all of the Cyp2c genes that were determined, namely Cyp2c40, 2c54, 2c65, 2c50, and 2c69, were markedly upregulated at least 50% in livers of GF mice at 90 days of age (Figs. 2E–H and 3A). The Cyp2c50 mRNA also appeared to be higher in livers of 90-day-old GF mice, although a statistical significance was not achieved (Supplementary Fig. 1B). There was no difference in the Cyp2c mRNAs between age-matched livers of CV and GF mice at other ages. Cyp2e1 mRNA, which gradually increased to adult levels in both mouse models, was also higher in GF-mouse livers at 90 days of age (Fig. 3B).
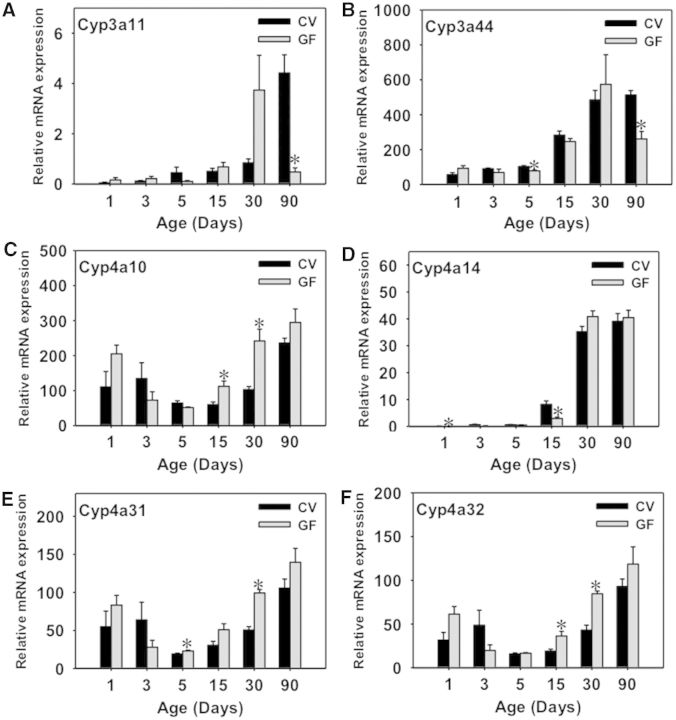
FIG. 3.
Ontogeny of Phase-I enzymes cytochrome P450s Cyp2c69 to Cyp4a32 gene expression in male livers of CV and GF mice. Data are presented as means ± SEM, n = 4/group. Asterisks (*) represent statistically significant differences between CV and GF mice (P < .05) by Student’s t test in that age group. Data are expressed as mean % of the geometric means of the housekeeping genes β-actin, Gapdh, and 18S rRNA.
Regarding the Cyp3a genes, Cyp3a11 mRNA increases gradually after birth in both CV and GF mice, but GF mice had lower Cyp3a11 mRNA most notably at 90 days of age (an 80% decrease as compared with CV mice; Fig. 3C). A similar pattern was also observed for Cyp3a44 mRNA (Fig. 3D). Cyp3a25 mRNA was lower in GF livers at 15 days of age, but remained at similar levels between livers of CV- and GF-mice at other ages (Supplementary Fig. 1C).
Regarding the Cyp4a genes, interestingly, among all of the Cyp4a family members examined, namely Cyp4a10, 4a14, 4a31, and 4a32, a marked upregulation in their mRNAs were observed in of 90-day-old GF mice as compared with age-matched livers of CV mice. (Fig. 3E–H). In addition, Cyp4a10 mRNA was higher at 1- and 15-day-old GF-mouse livers; Cyp4a14 mRNA was higher in 15- and 30-day-old GF-mouse livers; Cyp4a31 and 4a32 mRNAs were higher at 1-, 15-, and 30-day-old GF-mouse livers, compared with age-matched CV mouse livers. At 5 days of age, a moderate decrease was observed in Cyp4a10, 4a31, and 4a32 mRNAs in GF-mouse livers compared with CV livers (Fig. 3E–H). For Cyp P450 reductase (Por), which is required for the function of all microsomal Cyp enzymes, its mRNA was moderately downregulated in GF mice livers at 15 days of age, but was upregulated at 30 days of age, as compared with age-matched CV-mouse livers (Supplementary Fig. 1C).
Hepatic Ontogeny of Other Phase-1 Enzymes in Male Mouse Livers
The ontogeny of 13 other critical Phase-1 enzymes were also analyzed in the livers of CV and GF mice.
Aldo-keto Reductases
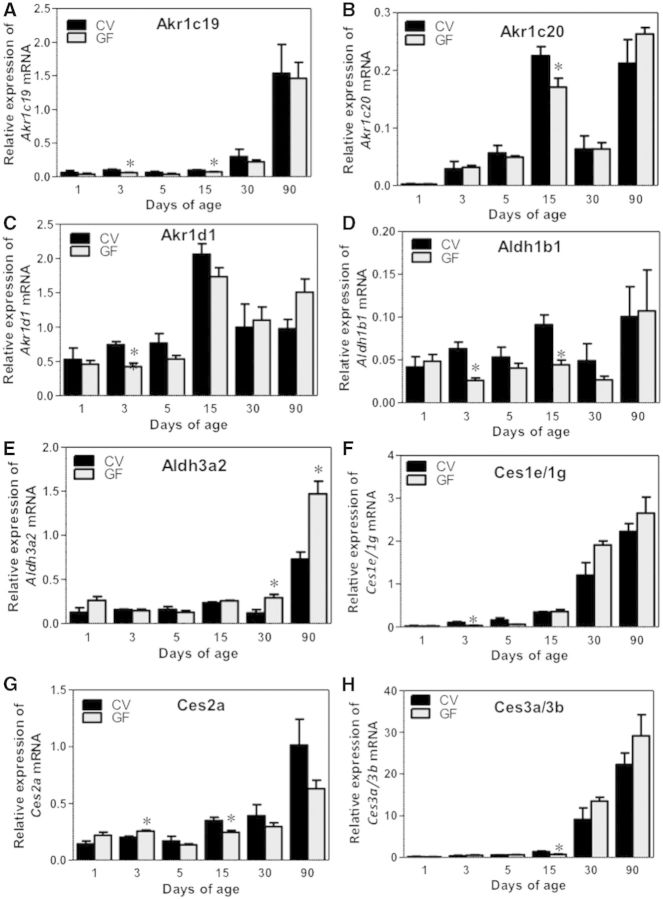
The mRNA of Akr1c19 was lowly expressed before 30 days of age, and reached a peak at 90 days of age, whereas Akr1c20 and 1d1 were enriched around 15 days of age in livers of CV mice. The absence of intestinal microbiota generally had minimal effect on the expression of these Aldo-keto reductases, except for a moderate decrease in the Akr1c19 mRNA at 3 and 15 days of age, Akr1d1 mRNA at 3 days of age, and Akr1c20 mRNA at 15 days of age (Fig. 4A–D).
FIG. 4.
Ontogeny of other Phase-I enzymes, including Ahrs (Akr1c19, 1c20, and), Aldhs (Aldh1b1 and 3a2), as well as Cess (Ces1e/1g, 2a, and 3a/3b) gene expression in male livers of CV and GF mice. Data are presented as means ± SEM, n = 4/group. Asterisks (*) represent statistically significant differences between CV and GF mice (P < .05) by Student’s t test in that age group. Data are expressed as mean % of the geometric means of the housekeeping genes β-actin, Gapdh, and 18S rRNA.
Aldehyde Dehydrogenases (Aldhs)
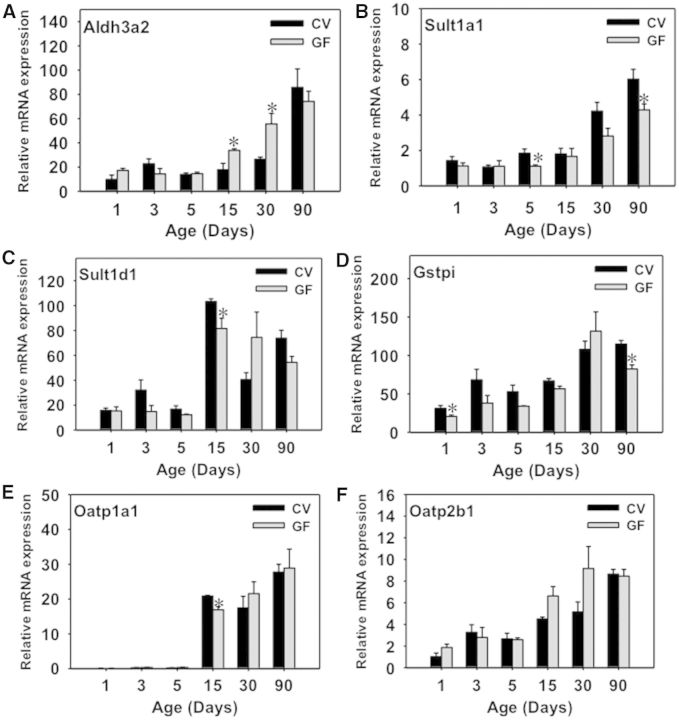
The mRNA of Aldh1b1, which is involved in the oxidative metabolism of aldehydes, was lower in livers of GF mice at 3 and 15 days of age compared with CV mice (Fig. 4D), however, the mRNA of Aldh3a2 was higher in livers of GF mice at 30 and 90 days of age (Fig. 4E). The mRNAs of both Aldh1b1 and Aldh3a2 gradually increased to adult levels during liver development. Interestingly, the absence of intestinal microbiota tended to attenuate the ontogenic increase of Aldh1d1 mRNA, but had the opposite effect on the Aldh3a2 mRNA.
Carboxyesterases and Aldehyde Oxidase 1
Carboxyesterase 1e (Ces1e)/1g and 3a/3b mRNAs were minimally expressed before 30 days of age, and the GF condition appeared to further decrease Ces1e1/1g mRNA at 3 days of age, and Ces3a/3b mRNA at 15 days of age (Fig. 4F and H). The mRNA of the hydrolytic enzyme Ces2a was higher in livers of GF mice at 3 days of age, but lower in livers of GF mice at 15 days of age (Fig. 4G). Aldehyde oxidase 1 mRNA was expressed minimally and increased with age in both CV and GF mice (Supplementary Fig. 1D).
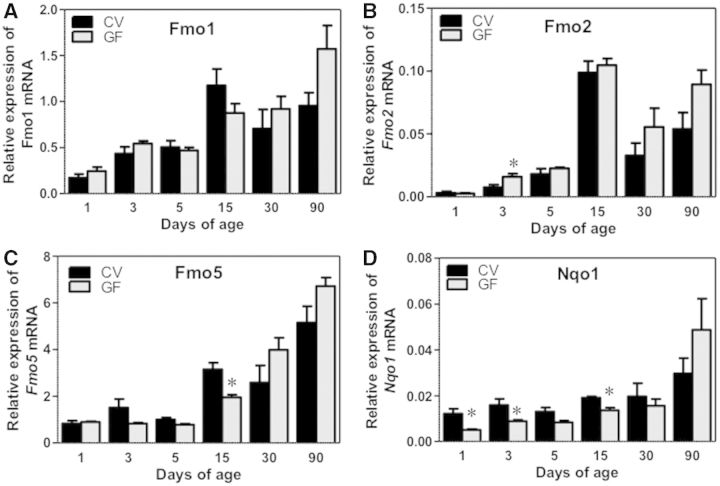
Flavin-Containing Monooxygenases and Nqo1
The mRNAs of flavin-containing monooxygenase 1 (Fmo1), 2, and 5 all gradually increased to adult levels between 15 and 90 days of age in both CV and GF mice; whereas the GF condition had no effect on the ontogeny of Fmo1 mRNA, a moderate increase of Fmo2 mRNA at 3 days of age, and a decrease in the mRNA of Fmo5 at 3 and 15 days of age was observed (Fig. 5A–C). Interestingly, the mRNA of the prototypical Nrf2-target gene NAD(P)H dehydrogenase, quinone 1 (Nqo1), which is a marker of oxidative stress, was not altered at 90 days of age, but was consistently downregulated in livers of GF mice at young ages (1, 3, 5, and 15 days of age) compared with age-matched CV mice, (Fig. 5D), suggesting the age-specific contribution of gut microbiota on Nrf2-signaling and antioxidative stress pathways in liver.
FIG. 5.
Ontogeny of other Phase-I enzymes, including Fmos (Fmo1, 2, and 5) as well as Nqo1 gene expression in male livers of CV and GF mice. Data are presented as means ± SEM, n = 4/group. Asterisks (*) represent statistically significant differences between CV and GF mice (P < .05) by Student’s t test in that age group. Data are expressed as mean % of the geometric means of the housekeeping genes β-actin, Gapdh, and 18S rRNA.
The Ontogeny of Phase-II Enzymes in Male Mouse Livers
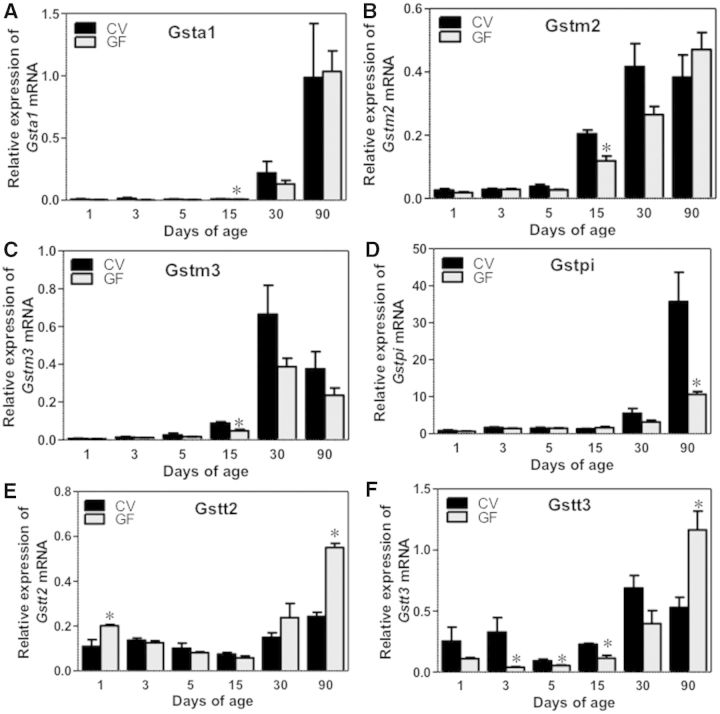
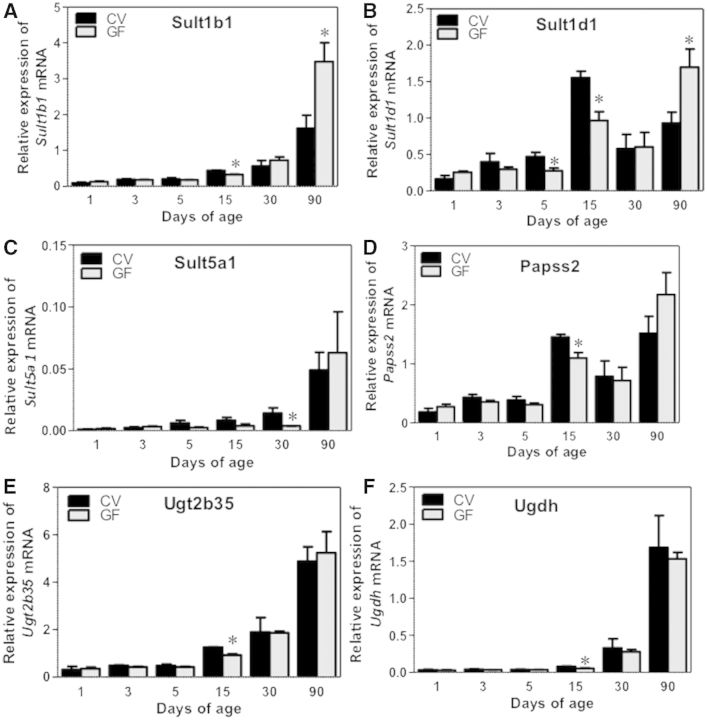
Gsts, sulfotransferases (Sults) and UDP-glucuronyltransferases (Ugts) are 3 major classes of phase-II enzymes. The gene expression of most of the phase-II enzymes were low at birth and increased with age in both CV and GF mice. Regarding Gsts, Gsta1 mRNA was in general not altered between livers of CV and GF mice at any age, except for a slight decrease in 15-day-old GF-mouse livers (Fig. 6A). Both Gstm2 and m3 mRNAs were lower in 15-day-old GF-mouse livers compared with age-matched GF-mouse livers, but there was no difference in their expression at other ages (Fig. 6B and C). For Gstpi, there was a marked downregulation in Gstpi mRNA in livers of GF mice at 90 days of age (Fig. 6D), but there was no difference in its expression between CV- and GF-mouse livers at other ages. Both Gstt2 and Gstt3 were markedly upregulated in livers of 90-day-old GF-mice compared with age-matched GF mice; Gstt2 mRNA was also higher in livers of GF- than in CV-mice at 1 day of age, however Gstt3 mRNA was generally downregulated in neonatal GF mice (before 15 days old) (Fig. 6E and F). GF conditions had no effect on the ontogeny of Gclc mRNA, which gradually increased to adult levels in both mouse models (Supplementary Fig. 1F). Regarding Sults, Sult1a1 mRNA gradually increased to adult levels in both mouse models, and it was similar in GF-mouse livers (Supplementary Fig. 1G). Sult1b1 mRNA was moderately downregulated in 15-day-old GF-mouse livers, but was markedly upregulated in 90-day-old GF-mouse livers, as compared with age-matched CV mice (Fig. 7A). Sult1d1 mRNA was lower in 5- and 15-day-old GF-mouse livers, but was higher in 90-day-old GF-mouse livers, compared with age-matched CV-mouse livers (Fig. 7B). Sult5a1 mRNA was lower in 30-day-old GF-mouse livers than CV-mouse livers, but was not altered by the GF condition at other ages (Fig. 7C). Regarding the enzyme that synthesizes the co-substrate of sulfation reactions, namely 3′-phosphoadenosine 5′-phosphosulfate synthase 2 (Papss2), the Papss2 mRNA was moderately downregulated in 15-day-old GF-mouse livers, but was not altered by the GF condition at other developmental ages (Fig. 7D). Regarding Ugts, Ugt2b35 as well as the enzyme that synthesizes the cosubstrate UDP-GA, namely Udgh, were both downregulated in 15-day-old GF-mouse livers, but remained unchanged at other ages as compared with age-matched CV-mouse livers (Fig. 7E and F).
FIG. 6.
Ontogeny of Phase-II glutathione-S-transferases (Gsta1, m2, m3, pi, t2, and t3) gene expression in male livers of CV and GF mice. Data are presented as means ± SEM, n = 4/group. Asterisks (*) represent statistically significant differences between CV and GF mice (P < .05) by Student’s t test in that age group. Data are expressed as mean % of the geometric means of the housekeeping genes β-actin, Gapdh, and 18S rRNA.
FIG. 7.
Ontogeny of Phase-II Sults and cosubstrates gene expression in male livers of CV and GF mice. Data are presented as means ± SEM, n = 4/group. Asterisks (*) represent statistically significant differences between CV and GF mice (P < .05) by Student’s t test in that age group. Data are expressed as mean % of the geometric means of the housekeeping genes β-actin, Gapdh, and 18S rRNA.
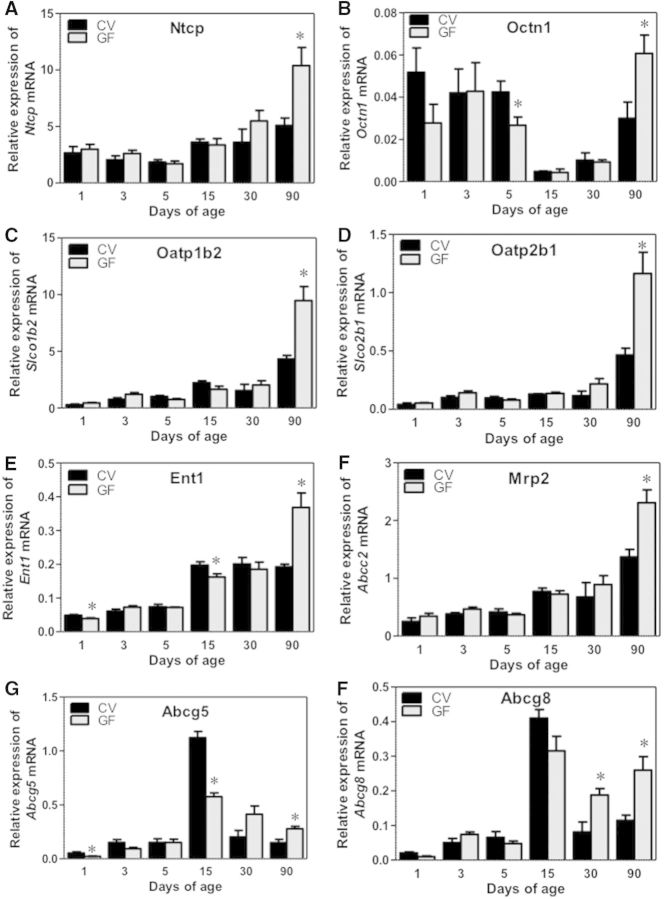
The Ontogeny of Uptake Transporters in Male-Mouse Livers
The gene expression of most of the hepatic uptake transporters (Fig. 8A–E) were low at birth and increased with age in both CV and GF mice, except for Ntcp and Octn1. The gene expression of Ntcp is high at 1 day of age, and its expression remains relatively stable at all ages analyzed in livers of CV mice. In contrast, for Octn1, the gene expression is high at birth but decreases by 15 days of age and increases by 90 days of age in CV mice. The mRNAs of uptake transporters, including Ntcp, Octn1, Oatp1b2, Oatp2b1, and equilibrative transporter Ent1, were all higher in livers of GF mice at 90 day of age compared with CV mice. Oatp1a1 mRNA also tended to be higher in 90-day-old GF livers, although a statistical significance was not achieved (Supplementary Fig. 2A). The expression of the hepatic carnitine transporter Octn1, was lower in the livers of GF mice at 5 days of age compared with CV mice. The mRNA of Ent1 was slightly lower in livers of GF mice at 1 and 15 days of age. The ontogeny pattern of most of these transporters remained unaffected by the absence of intestinal microbiota throughout the developmental ages in this study. The mRNA of the bile-acid uptake transporter Asbt, which is present at low levels in liver and is specifically localized on cholangiocytes, also increases with age in livers of both CV and GF mice, and at 1 day of age the mRNA is higher in GF mice compared with CV mice (Supplementary Fig. 2B).
FIG. 8.
Ontogeny of hepatic transporters gene expression in male livers of CV and GF mice. Data are presented as means ± SEM, n = 4/group. Asterisks (*) represent statistically significant differences between CV and GF mice (P < .05) by Student’s t test in that age group. Data are expressed as mean % of the geometric means of the housekeeping genes β-actin, Gapdh, and 18S rRNA.
The Ontogeny of Efflux Transporters in Male-Mouse Livers
The mRNA of the apical efflux transporter Mrp2, which is responsible for the biliary efflux of glutathione-conjugates, was lowly expressed after birth and increased with age in both CV and GF mice (Fig. 8F), although at 90 days of age, the mRNA of Mrp2 was higher in livers of GF mice compared with CV mice. The mRNAs of the apical sterol-efflux transporters Abcg5 and Abcg8 were low at birth, peaked at 15 days of age, and then decreased to adult levels in livers of both CV and GF mice. The mRNA of Abcg5 in livers of GF mice was lower at 1 and 15 days of age but higher at 90 days of age than the CV mice. The mRNA of Abcg8 was higher in livers of GF mice at 30 and 90 days of age than the CV mice (Fig. 8G and H). The mRNA of Bsep was low at birth and increased with age in livers of both CV and GF mice, whereas the mRNA of Bcrp and Mrp4 were relatively high at birth and remained relatively stable across the ages in livers of both CV and GF mice (Supplementary Fig. 2C–E).
Western-Blotting Analysis of Prototypical PXR- and PPARα-Target Genes in CV- and GF-Male Mice
Because the mRNA of the prototypical target genes of PXR (Cyp3a11) and PPARα (Cyp4a family members) were differentially regulated in the absence of gut microbiota in an age-specific manner, Western-blotting analysis was performed to determine whether similar changes occur at the protein level. Due to limited amount of protein, livers from 1 to 5 days of age were pooled at each age. As shown in Figure 9A, there was an apparent decrease in Cyp3a11 protein in age-matched GF livers most notably after 15 days of age; conversely, there was an apparent increase in Cyp4a14 protein in livers of GF mice throughout liver development (Fig. 9A and B).
FIG. 9.
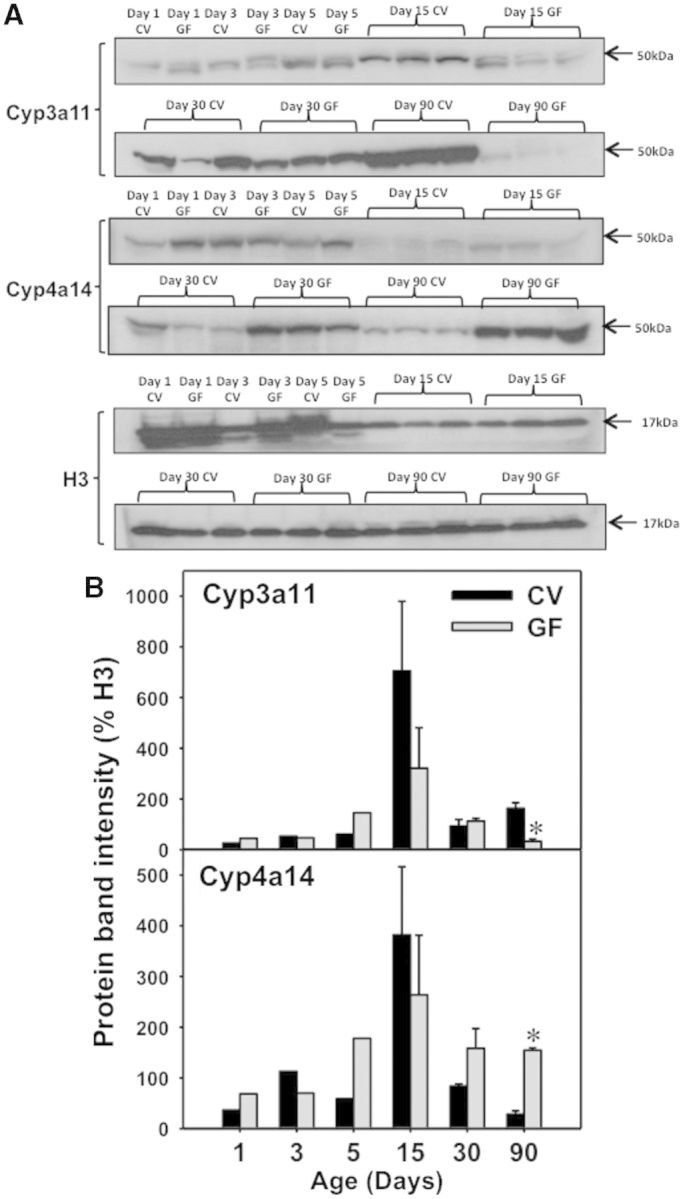
Protein expression by Western blots in male livers of CV and GF mice. A, Western blotting of Cyp3a11, Cyp4a14, and histone H3 proteins in CV and GF male livers from 1 to 90 days of age. B, Quantification of protein band intensities after normalization to H3 using Image J software. Asterisk (*) represents statistically significant differences between CV and GF mice (P < .05) by Student’s t test in that age group.
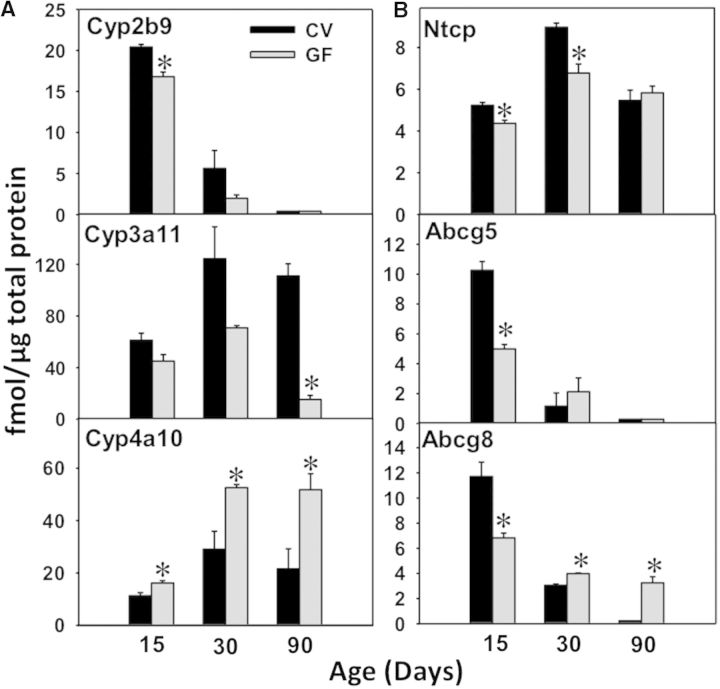
LC-MS/MS Proteomic Quantification of DPGs in Male Mouse Livers
To further confirm the mRNA and protein expression of important DPGs in livers of CV and GF mice during development, a LC-MS/MS proteomic approach was used as described in Materials and Methods. The following proteins were quantified based on either their importance in drug-processing or lack of availability of antibodies for Western-blotting analysis: Cyp2b9, Cyp3a11, 4a10, Ntcp, Abcg5, and Abcg8 (n = 3 per group). Due to limited protein amount, the proteins at earlier developmental ages were not determined using this approach. As shown in Figure 8, Cyp2b9 protein was highest at 15 days of age, but markedly decreased thereafter in both CV and GF livers. In addition, there was a moderate decrease in the Cyp2b9 protein expression in livers of GF mice at 15 days of age. The Cyp3a11 protein was also downregulated most prominently at 90 days of age, but it also tended to decrease at the other ages (although statistical significance was not achieved). Conversely, there was a marked increase in Cyp4a10 protein in livers of GF mice at all 3 ages examined. For transporters, Ntcp protein was moderately lower in livers of GF mice at 15 and 30 days of age, but there was no difference in its protein expression between CV- and GF-mouse livers at 90 days of age. Abcg5 and Abcg8 proteins were both highest at 15 days of age and were downregulated in livers of GF mice at this age. After 15 days of age, the proteins of both Abcg5 and Abcg8 were lower, however in age-matched CV livers, the Abcg8 protein was upregulated in GF livers, whereas Abcg5 protein remained low in both CV and GF conditions (Fig. 10A and B).
FIG. 10.
UPLC-MS quantification of protein levels in male livers of CV and GF mice (n = 3/group). Data are presented as means ± SEM, n = 3/group. Asterisk (*) represents statistically significant differences between CV and GF mice (P < .05) by Student’s t test in that age group.
Cyp3a-Enzyme Activities in Hepatic Microsomes of CV- and GF-Male Livers of 90-Day-Old Mice
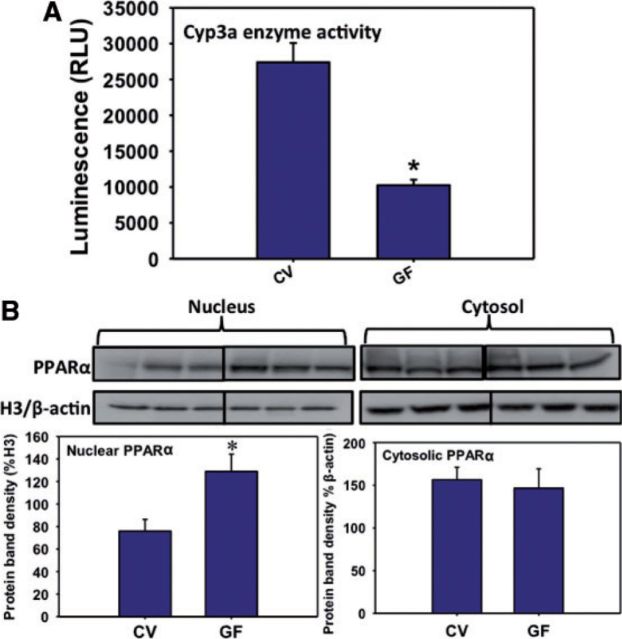
Because the most prominent decrease in the Cyp3a11 mRNA was observed at 90 days of age, the hepatic Cyp3a microsomal activity was quantified in CV- and GF-mouse livers at this age (n = 3). Consisent with the mRNA and Western-blot data, there was also a marked decrease in the enzyme activity of Cyp3a in GF livers (Fig. 11A).
FIG. 11.
A, Cyp3a enzyme activity in 90-day-old CV and GF male liver microsomes as determined using the Promega P450-Glo CYP3A4 Luciferin-IPA Assay per the manufacturer’s instructions. Asterisk (*) represents statistically significant differences between CV and GF mice (P < .05) by Student’s t test. B, Western blotting of PPARα, and loading controls (histone H3 for nucleus, and β-actin for cytosol) in cytosolic and nuclear fractions of the male livers. Data are expressed as protein band density normalized to the corresponding loading controls (H3 for nucleus and β-actin for cytosol). Asterisk (*) represents statistically significant differences between CV and GF mice (P < .05) by Student’s t test.
PPARα Nuclear and Cytosolic Protein Expression in CV- and GF Livers of 90-Day-Old Mice
Because the most prominent upregulation in Cyp4a mRNAs was observed at 90 days of age, the protein of PPARα, which is the prototypical transcriptional activator of Cyp4a genes, was quantified in nucleus and cytosol of CV- and GF-mouse livers at this age. Interestingly, there was a marked increase in the nuclear PPARα protein in GF-mouse livers, suggesting increased PPARα signaling in GF mice; however, there was no difference in cytosolic PPARα-protein expression (Fig. 11B). This indicates that the upregulated PPARα-signaling may be due to an overall increase in PPARα gene expression as well as ligand activation. Indeed, increased PPARα mRNA expression was confirmed in Figure 12D.
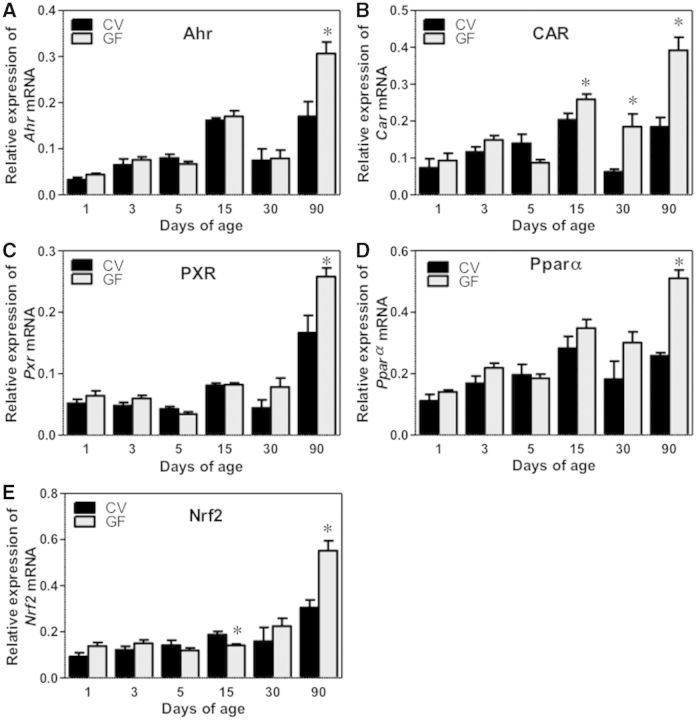
FIG. 12.
Ontogeny of xenobiotic-sensing transcription factors gene expression in male livers of CV and GF mice. Data are presented as means ± SEM, n = 4/group. Asterisks (*) represent statistically significant differences between CV and GF mice (P < .05) by Student’s t test in that age group.
The Ontogeny of Xeno-Sensing Transcription Factors
In general, the mRNAs of the transcription factors AhR, CAR, PXR, PPARα, and Nrf2 increased with age in livers of both CV and GF mice (Fig. 12A–E). Interestingly, the mRNAs of all transcription factors were higher in livers of GF mice compared with CV mice at 90 days of age. The mRNA of CAR was also higher in livers of GF mice compared with CV mice at 15 and 30 days of age. The mRNA of Nrf2 was lower in the livers of GF mice compared with CV mice at 15 days of age (Fig. 12E).
Hepatic Expression of DPGs in Female CV- and GF-Mouse Livers
Because certain DPGs have been reported to have gender-divergent expression patterns at least in adult livers (Alnouti and Klaassen, 2011; Alnouti et al., 2006; Buckley and Klaassen, 2007, 2009; Cheng et al., 2005, 2006; Lickteig et al., 2008; Renaud et al., 2011), the expression of these genes (selected based on the literature mentioned above, as well as differential expression between CV and GF mice in male livers as unveiled in this study) was quantified in CV and GF livers at various developmental ages. For Cyps, except for Cyp3a11, which showed no gender differences in adult livers (Renaud et al., 2011), all other Cyps included in Figures 13 and 14 are known to be female-predominant (Renaud et al., 2011). Compared with male livers, the ontogenic expression of Cyp2a5 and Cyp2a12/22 were similar, in that both of their mRNAs gradually increased to adult levels in CV- and GF-mouse livers. Similar to Cyp2a5 mRNA ontogeny data from male mice, Cyp2a5 mRNA in female livers also appeared to be only minimally influenced by the GF condition (except for a slight decrease in its expression at 90 days of age). Cyp2a12/22 mRNA in both male and female livers was downregulated in 15-day-old GF mice compared with age-matched CV mice (Fig. 13A and B). In contrast, regarding the Cyp2 genes, there was a marked gender difference regarding how the GF condition modifies their mRNA ontogenic expression patterns: for Cyp2b genes, Cyp2b9 was minimally expressed in male livers but was expressed highest in livers of adult females, whereas Cyp2b9 mRNA was upregulated only in male-GF livers at 90 days of age but was not altered in female-GF livers at this age, compared with age-matched GF mice of the same gender (Figs. 2C and 13C). At 15 days of age, there appears to be a decrease in Cyp2b9 mRNA in GF livers of both genders (although males did not reach statistical significance). For Cyp2b10, there was no difference in its mRNA expression at 90 days of age between male CV and male GF mice, however, there was a marked decrease in its mRNA between female CV- and female GF-mice (Fig. 13D). Regarding the Cyp2c mRNAs, which were all upregulated in 90-day-old male-GF livers, interestingly, none of them were altered between female-CV and GF mice at 90 days of age (Fig. 13E–H). In addition, some Cyp2c genes appeared to be downregulated around adolescent ages in female-GF livers (such as Cyp2c67 at 15 days of age, and Cyp2c69 at 5 and 15 days of age), however, there was no change in the mRNAs at these ages in male-GF livers as compared with age-matched CV livers.
FIG. 13.
Ontogeny of Phase-I cytochrome P450s Cyp2a5 to Cyp2c69 gene expression in female livers of CV and GF mice. Data are presented as means ± SEM, n = 4/group. Asterisks (*) represent statistically significant differences between CV and GF mice (P < .05) by Student’s t test in that age group. Data are expressed at % of β-actin.
FIG. 14.
Ontogeny of Phase-I cytochrome P450s Cyp3a11 to Cyp4a32 gene expression in female livers of CV and GF mice. Data are presented as means ± SEM, n = 4/group. Asterisks (*) represent statistically significant differences between CV and GF mice (P < .05) by Student’s t test in that age group. Data are expressed at % of β-actin.
Regarding the Cyp3a genes, both Cyp3a11 and 3a44 were downregulated in 90-day-old female-GF livers as compared with age-matched female-CV livers (Fig. 14A and B), and this finding was similar to that in male livers. Regarding the Cyp4a genes, which were upregulated in 90-day-old male-GF mice (Fig. 3E–H) as well as in many earlier ages, interestingly, none of these Cyp4a genes were altered between GF and CV livers at 90 days of age. However, an upregulation in female-GF livers as compared with age-matched female-CV mice was observed at earlier developmental ages, such as Cyp4a10 at 15 and 30 days of age, Cyp4a31 at 5 and 30 days of age, and 4a32 at 15 and 30 days of age (Fig. 14C–F).
In summary, the ontogeny of Cyps in female-CV and -GF livers indicates that the maturation of female-specific hormones, or the absence of male-specific hormones, or the female-specific secretion patterns in growth hormones, may have an inhibitory effect on GF-mediated upregulation of Cyp2b, Cyp2c, and Cyp4a genes, but the GF condition has minimal effect on the Cyp2a/3a ontogeny.
Regarding the female expression of other DPGs that have known gender-divergent patterns in adult livers (Alnouti and Klaassen, 2008, 2011; Cheng et al., 2005; Knight et al., 2007), Aldh3a2, Sult1a1, Sult1d1, and Oatp2b1 were previously shown to be female-predominant in adult livers (Alnouti and Klaassen, 2008, 2011; Cheng et al., 2005), whereas Gstpi is male-predominant in adult livers (Cheng et al., 2005; Knight et al., 2007). For Aldh3a2, which was upregulated in livers of male GF-livers at 30 and 90 days of age compared with age-matched male CV livers (Fig. 4E), it is also upregulated in female-GF livers at 15 and 30 days of age, but not 90 days of age (Fig. 15A). For Sult1a1, which was not altered by GF at any developmental age in male livers (Supplementary Fig. 1F), its mRNA was downregulated in livers of 5- and 90-day-old female GF mice compared with age-matched female CV mice (Fig. 15B). For Sult1d1, which was downregulated by the GF condition at 5 and 15 days of age but upregulated at 90 days of age in male livers, its mRNA tended to be downregulated by the GF condition at younger ages of female livers, but remained unchanged at 90 days of age between CV- and GF-female livers (Fig. 15C). Regarding Gstpi, it was downregulated markedly only by the GF condition in livers of 90-day-old male mice (Fig. 6D), however, in female livers, there was a slight decrease in the Gstpi in GF-mouse livers at 90 days of age compared with age-matched female-CV mice (Fig. 15D). Regarding Oatp1a1, GF-mediated regulation of its ontogeny appeared to be similar between male and females, except that a slight decrease by the GF condition was observed at 15 days of age in females (Fig. 15E), but there was no change in the Oatp1a1 mRNA in males by the GF condition at this age (Supplementary Fig. 2A). Oatp2b1, was markedly upregulated by the GF condition at 90 days of age in male livers (Fig. 8C), but there was no difference in Oatp2b1 mRNA in between livers of female GF and CV mice at this age (Fig. 15F).
FIG. 15.
Ontogeny of other DPGs gene expression in female livers of CV and GF mice. Data are presented as means ± SEM, n = 4/group. Asterisks (*) represent statistically significant differences between CV and GF mice (P < .05) by Student’s t test in that age group. Data are expressed at % of β-actin.
In summary, the ontogeny of other DPGs in female CV and GF livers indicate that, for female-predominant DPGs such as Aldh3a2, Sult1a1, Sult1d1, and Oatp2b1, the maturation of female-specific hormones (or the absence of male-specific hormones as well as the secretion patterns of growth hormones) likely has an inhibitory effect on GF-mediated upregulation of these genes. Although for the male-predominant Gstpi, the maturation of female-specific hormones (or the absence of male-specific hormones as well as growth hormone secretion patterns) likely disinhibits the GF-mediated downregulation of Gstpi mRNA.
DISCUSSION
This study has illustrated that the absence of gut microbiota regulates the mRNA expression of many important DPGs, among which the most notable findings are (1) a marked downregulation of the prototypical PXR-target Cyp3a gene family (such as Cyp3a11 and 3a44) in postweaning ages and (2) a marked upregulation of the prototypical PPARα-target genes of the Cyp4a gene family (such as Cyp4a10 and 4a14) at both neonatal and adult ages in male livers. In addition, we have observed a neonatal to adolescent-specific downregulation of the prototypical Nrf2-target gene Nqo1 (from Days 1 to 15), as well as an adolescent-specific downregulation of LXR target genes Abcg5 and Abcg8 (at Day 15), suggesting that intestinal microbiota modulates multiple transcription factor signaling pathways of the host liver in an age-specific manner.
Results from this study have added knowledge to the existing paradigm regarding the roles of intestinal microbiota in drug metabolism. Many studies in the literature have demonstrated that intestinal microbiota can metabolize drugs using their own enzymes, and this may interfere with the metabolism of drugs by the host. For example, intestinal microbiota can metabolize digoxin, and the intestinal bacterial metabolite p-cresol competes with acetaminophen for sulfation in the liver (Clayton et al., 2009). However, relatively less is known regarding how intestinal microbiota impacts the host xenobiotic-sensing transcription factors to indirectly modulate the chemical metabolism and transport during development. In adults, intestinal microbiota has been shown to regulate the levels of certain host drug-metabolizing enzymes (Bjorkholm et al., 2009; Toda et al., 2009b). A recent study demonstrated that Cyp3a11 mRNA levels are lower in livers of adult-GF mice and they increase to levels seen in CV mice within 20 days of colonization (Claus et al., 2011). Our results on Cyp3a11 gene expression at 90 days of age at both the mRNA and protein levels are consistent with the previous reports in the literature, and this study has also shown that Cyp3a11 tends to be downregulated at the postweaning adolescent age Day-30 in GF livers.
The downregulation of Cyp3a11 and Cyp3a44 in the absence of intestinal microbiota is likely due to attenuated PXR-signaling, because PXR is a well-known transcription activator of Cyp3a genes. The decrease in PXR-signaling in livers of GF mice is likely due to decreased endogenous ligands for PXR. First, secondary BAs, such as lithoholic acid, which are produced by certain gut bacteria, are known PXR activators (Staudinger et al., 2001). Indeed, replacement of lithocholic acid to GF-adult mice has been shown to restore the hepatic Cyp3a11 gene expression (Toda et al., 2009b). Our observation of decreased Cyp3a11 expression in GF mice is also similar to the previous observations in CV mice treated with the antibiotic ciprofloxacin, and the antibiotic-mediated decrease in hepatic Cyp3a11 has been attributed to the decrease in the amount of lithocholic acid producing bacteria such as Bacteroides fragilis (Toda et al., 2009a) and because intraperitoneal injection of lithocholic acid increases hepatic Cyp3a11 mRNA in mice (Ishii et al., 2014). Second, it is also possible that lack of intestinal microbial metabolites, namely indole 3-propionic acid, contributes to decreased PXR-signaling and its target gene expression in liver, because it has recently been shown that indole 3-propionic acid is an endogenous PXR-ligand (Venkatesh et al., 2014). It has been reported that the exogenous PXR ligand PCN can also upregulate other PXR-target genes in adult CV mouse livers, such as Gsta1, m1-m3, Oatp1a4, and Mrp3 (Aleksunes and Klaassen, 2012). However, these genes were not readily downregulated in adult GF mice livers, which is likely due to the possibility that the constitutive expression of these PXR-target genes under basal conditions are not dependent on PXR, whereas the basal Cyp3a expression relies on endogenous PXR signaling.
Interestingly, probiotics also appear to regulate Cyp3a gene expression, as rats treated with probiotics have decreased expression of Cyp3a9 in the intestine (Matuskova et al., 2011). Therefore, intestinal microbiota modifiers, including those that decrease intestinal microbiota (GF or antibiotics) and altering the composition of gut microbiota (probiotics), appear to be important regulators of the Cyp3a gene expression in the host. Further studies need to be performed using intestinal microbiome sequencing of CV mice during development to determine which bacteria are most important in modulating the ontogeny of host DPGs in liver.
Intestinal microbiota has been well established as a player contributing to intermediary metabolism. The composition of the intestinal microbiota is altered in obesity (Ley et al., 2005) and is associated with the capacity to harvest energy (Turnbaugh et al., 2006). GF mice are resistant to high-fat diet induced obesity, even though they consume more food, they are more active, excrete more lipids in feces and is more insulin sensitive (Backhed et al., 2007; Rabot et al., 2010). Colonization of GF mice increases body weight (Claus et al., 2011). Intestinal bacteria can metabolize dietary phosphatidylcholine to trimethylamine-N-oxide (TMAO), and increased TMAO is associated with cardiovascular diseases in humans (Tang et al., 2013; Wang et al., 2011). Antibiotic-treated mice and GF mice do not metabolize phosphatidylcholine to TMAO (Wang et al., 2011). Intestinal microbiota metabolize L-carnitine found in red meat to TMAO and this results in increased atherosclerosis (Koeth et al., 2013). However, relatively less is known regarding how intestinal microbiota modulates the lipid-sensing nuclear receptors of the host during development.
Our results have provided the first evidence that the prototypical PPARα-target genes (ie the Cyp4a genes) are markedly upregulated in livers of GF mice at both mRNA and protein levels from neonatal to adulthood, suggesting that hepatic PPARα-signaling is enhanced in absence of intestinal microbiota. Aldh3a2, which is another known PPARα-target gene in liver (Aleksunes and Klaassen, 2012), was also upregulated in livers of male-GF mice at 90 days of age. Because dietary lipids and fatty acids are major ligands of PPARα, our observations have unveiled that intestinal microbiota play significant roles in repressing endogenous PPARα activities in liver during development, likely by metabolizing various PPARα activators from the diet or other intermediary metabolites. Certain intestinal microbiota modifiers may be used therapeutically to treat hyperlipidemia and hypertriglyceridemia. Future studies need to be performed to quantify the lipid metabolome profiles in CV and GF mice during development, to confirm this hypothesis.
Another interesting finding is that the interactions between intestinal microbiota and the ontogeny of DPGs appear to be gender-specific. For example, many female-predominantly expressed DPGs in adults are upregulated in livers of 90-day-old GF mice, such as Cyp2c and Cyp4a family. However, such GF-mediated upregulation is not observed in female-GF livers. It has been reported that sex hormones and STAT5b mediated differences in the growth hormone secretion patterns between males and females are important in determining the gender-divergent expression of many DPGs in adult liver (Buckley and Klaassen, 2009; Cheng et al., 2007; Waxman, 2000). It is possible that gender-specific hormones interfere with the interactions between the transcription factors (such as PPARα) and the intestinal microbial metabolites, which serve as the ligands of the receptors. Additional mechanistic studies are needed to confirm this hypothesis.
Conventional protein quantification approaches, such as Western-blotting analysis, rely on the availability of species- and gene-specific antibodies and are not absolute quantitative method to determine the actual abundance of targeted proteins. In comparison, LC-MS/MS proteomics has a much broader applicability because it does not require selective antibodies for various proteins, and allows high-throughput screening of multiple protein targets in a quantitative mode (Prasad and Unadkat, 2014b). In this study, the LC-MS/MS data on Cyp3a11 is consistent with the Western blot results (Figs. 7 and 8). Cyp4a10 (LC-MS/MS) and Cyp4a14 (Western blot) both show an age-specific increase. In addition, LC-MS/MS allows the quantification of other DPGs where highly selective anti-mouse antibodies are not readily available (such as Cyp2b9, Cyp4a10, Abcg5, and Abcg8). Therefore, LC-MS/MS is superior for protein quantification of DPGs. Most of the protein expression data are consistent with our observations on mRNAs, except for the hepatic uptake transporters Ntcp and Oatp1b2, for which we observed an increase in the mRNA expression at 90 days of age, however there was no change in protein expression. Surprisingly, Ntcp protein was even downregulated at 15 and 30 days of age in the livers of GF mice. This discrepancy is likely due to posttranscriptional modifications, such as microRNAs and protein ubiquitination pathways, and future studies need to be performed to characterize the expression of these mediators.
Intestinal microbiota are altered by factors immediately after birth, such as mode of delivery and early environmental exposure (Palmer et al., 2007), breast-milk or formula-fed (Ardeshir et al., 2014), and antibiotic administration (Antonopoulos et al., 2009; Dethlefsen and Relman, 2011; Dethlefsen et al., 2008). In addition, intestinal microbiota modifiers such as antibiotics and probiotics are used in pediatric populations for prevention or treatment of diseases. For example, obese children receiving the probiotic Lactobacillus rhamnosus strain GG for 8 weeks have a significant decrease in alanine aminotransferase (Vajro et al., 2011), suggesting that this probiotic is beneficial for treating obesity-related liver diseases. Exposure to antibiotics in the first year of life is associated with childhood obesity (Azad et al., 2014; Bailey et al., 2014), with potentially additional long-term health consequences. Early antibiotic exposure may also alter the expression of DPGs, which can result in adverse drug reactions in the pediatric population (Strolin Benedetti et al., 2005). Using GF mice as a research model, our study has characterized for the first time the effect of intestinal microbiota on the ontogeny of DPGs during development, and has laid the foundation for further understanding the “bug-drug interactions” and improving the safety and efficacy of using drugs in pediatric patients. Future studies will utilize GF mice and humanized ex-GF mice to investigate the effect of intestinal bacteria on the host metabolism of various drugs and environmental chemicals in liver.
SUPPLEMENTARY DATA
Supplementary data are available online at http://toxsci.oxfordjournals.org/.
FUNDING
This work was supported by National Institutes of Health (NIH) GM111381 and ES019487, as well as start-up funds from University of Washington Center of Ecogenetics and Environmental Health (CEEH) (P30 ES0007033) provided to Dr J.Y.C.
Supplementary Material
ACKNOWLEDGMENTS
The authors would like to acknowledge all members of the UW DEOHS Biomarker Laboratory for their technical assistance in the qPCR analysis at the beginning of this project, and Ms. Shirley Shi-Yu Chang in Dr David Eaton’s laboratory for her help in Cyp enzyme activity assays.
REFERENCES
- Aleksunes L. M., Klaassen C. D. (2012). Coordinated regulation of hepatic phase I and II drug-metabolizing genes and transporters using AhR-, CAR-, PXR-, PPARalpha-, and Nrf2-null mice. Drug Metab. Dispos. 40, 1366-1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alnouti Y., Klaassen C. D. (2008). Tissue distribution, ontogeny, and regulation of aldehyde dehydrogenase (Aldh) enzymes mRNA by prototypical microsomal enzyme inducers in mice. Toxicol. Sci. 101, 51-64. [DOI] [PubMed] [Google Scholar]
- Alnouti Y., Klaassen C. D. (2011). Mechanisms of gender-specific regulation of mouse sulfotransferases (Sults). Xenobiotica 41, 187-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alnouti Y., Petrick J. S., Klaassen C. D. (2006). Tissue distribution and ontogeny of organic cation transporters in mice. Drug Metab. Dispos. 34, 477-482. [DOI] [PubMed] [Google Scholar]
- Anderson G. D. (2002). Children versus adults: pharmacokinetic and adverse-effect differences. Epilepsia 43(Suppl 3), 53-59. [DOI] [PubMed] [Google Scholar]
- Antonopoulos D. A., Huse S. M., Morrison H. G., Schmidt T. M., Sogin M. L., Young V. B. (2009). Reproducible community dynamics of the gastrointestinal microbiota following antibiotic perturbation. Infect. Immun. 77, 2367-2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardeshir A., Narayan N. R., Mendez-Lagares G., Lu D., Rauch M., Huang Y., Van Rompay K. K., Lynch S. V., Hartigan-O'Connor D. J. (2014). Breast-fed and bottle-fed infant rhesus macaques develop distinct gut microbiotas and immune systems. Sci. Transl. Med. 6, 252ra120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrieta M. C., Stiemsma L. T., Amenyogbe N., Brown E. M., Finlay B. (2014). The intestinal microbiome in early life: health and disease. Front. Immunol. 5, 427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azad M. B., Bridgman S. L., Becker A. B., Kozyrskyj A. L. (2014). Infant antibiotic exposure and the development of childhood overweight and central adiposity. Int. J. Obes. 38, 1290-1298. [DOI] [PubMed] [Google Scholar]
- Backhed F., Manchester J. K., Semenkovich C. F., Gordon J. I. (2007). Mechanisms underlying the resistance to diet-induced obesity in germ-free mice. Proc. Natl. Acad. Sci. U.S.A. 104, 979-984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey L. C., Forrest C. B., Zhang P., Richards T. M., Livshits A., DeRusso P. A. (2014). Association of antibiotics in infancy with early childhood obesity. JAMA Pediatr. 168, 1063-1069. [DOI] [PubMed] [Google Scholar]
- Bjorkholm B., Bok C. M., Lundin A., Rafter J., Hibberd M. L., Pettersson S. (2009). Intestinal microbiota regulate xenobiotic metabolism in the liver. PLoS One 4, e6958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley D. B., Klaassen C. D. (2007). Tissue- and gender-specific mRNA expression of UDP-glucuronosyltransferases (UGTs) in mice. Drug Metab. Dispos. 35, 121-127. [DOI] [PubMed] [Google Scholar]
- Buckley D. B., Klaassen C. D. (2009). Mechanism of gender-divergent UDP-glucuronosyltransferase mRNA expression in mouse liver and kidney. Drug Metab. Dispos. 37, 834-840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X., Buckley D., Klaassen C. D. (2007). Regulation of hepatic bile acid transporters Ntcp and Bsep expression. Biochem. Pharmacol. 74, 1665-1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X., Maher J., Chen C., Klaassen C. D. (2005). Tissue distribution and ontogeny of mouse organic anion transporting polypeptides (Oatps). Drug Metab. Dispos. 33, 1062-1073. [DOI] [PubMed] [Google Scholar]
- Cheng X., Maher J., Lu H., Klaassen C. D. (2006). Endocrine regulation of gender-divergent mouse organic anion-transporting polypeptide (Oatp) expression. Mol. Pharmacol. 70, 1291-1297. [DOI] [PubMed] [Google Scholar]
- Cho I., Yamanishi S., Cox L., Methe B. A., Zavadil J., Li K., Gao Z., Mahana D., Raju K., Teitler I., et al. (2012). Antibiotics in early life alter the murine colonic microbiome and adiposity. Nature 488, 621-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chorell E., Karlsson Videhult F., Hernell O., Antti H., West C. E. (2013). Impact of probiotic feeding during weaning on the serum lipid profile and plasma metabolome in infants. Br. J. Nutr. 110, 116-126. [DOI] [PubMed] [Google Scholar]
- Claus S. P., Ellero S. L., Berger B., Krause L., Bruttin A., Molina J., Paris A., Want E. J., de Waziers I., Cloarec O., et al. (2011). Colonization-induced host-gut microbial metabolic interaction. MBio 2, e00271-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton T. A., Baker D., Lindon J. C., Everett J. R., Nicholson J. K. (2009). Pharmacometabonomic identification of a significant host-microbiome metabolic interaction affecting human drug metabolism. Proc. Natl. Acad. Sci. U.S.A. 106, 14728-14733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crom W. R., Relling M. V., Christensen M. L., Rivera G. K., Evans W. E. (1991). Age-related differences in hepatic drug clearance in children: studies with lorazepam and antipyrine. Clin. Pharmacol. Ther. 50, 132-140. [DOI] [PubMed] [Google Scholar]
- Cui J. Y., Gunewardena S. S., Yoo B., Liu J., Renaud H. J., Lu H., Zhong X. B., Klaassen C. D. (2012a). RNA-Seq reveals different mRNA abundance of transporters and their alternative transcript isoforms during liver development. Toxicol. Sci. 127, 592-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J. Y., Renaud H. J., Klaassen C. D. (2012b). Ontogeny of novel cytochrome P450 gene isoforms during postnatal liver maturation in mice. Drug Metab. Dispos. 40, 1226-1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y. J., Aleksunes L. M., Tanaka Y., Goedken M. J., Klaassen C. D. (2009). Compensatory induction of liver efflux transporters in response to ANIT-induced liver injury is impaired in FXR-null mice. Toxicol. Sci. 110, 47-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David L. A., Maurice C. F., Carmody R. N., Gootenberg D. B., Button J. E., Wolfe B. E., Ling A. V., Devlin A. S., Varma Y., Fischbach M. A., et al. (2014). Diet rapidly and reproducibly alters the human gut microbiome. Nature 505, 559-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dethlefsen L., Relman D. A. (2011). Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc. Natl. Acad. Sci. U.S.A. 108(Suppl 1), 4554-4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dethlefsen L., Huse S., Sogin M. L., Relman D. A. (2008). The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 6, e280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doshi U., Li A. P. (2011). Luciferin IPA-based higher throughput human hepatocyte screening assays for CYP3A4 inhibition and induction. J. Biomol. Screen 16, 903-909. [DOI] [PubMed] [Google Scholar]
- Fattah S., Augustijns P. P., Annaert P. P. (2014). Age-dependent Activity of the Uptake Transporters Ntcp and Oatp1b2 in Male Rat hepatocytes: from birth till adulthood. Drug Metab. Dispos. 43, 1-8. [DOI] [PubMed] [Google Scholar]
- Fernandez-Fernandez B., Montoya-Ferrer A., Sanz A. B., Sanchez-Nino M. D., Izquierdo M. C., Poveda J., Sainz-Prestel V., Ortiz-Martin N., Parra-Rodriguez A., Selgas R., et al. (2011). Tenofovir nephrotoxicity: 2011 update. AIDS Res. Treat. 2011, 354908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart SN, Cui Y, Klaassen CD, Zhong XB. (2009). Three patterns of cytochrome P450 gene expression during liver maturation in mice. Drug Metab. Dispos. 37, 116-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hines R. N. (2013). Developmental expression of drug metabolizing enzymes: impact on disposition in neonates and young children. Int. J. Pharm. 452, 3-7. [DOI] [PubMed] [Google Scholar]
- Human Microbiome Project Consortium. (2012). Structure, function and diversity of the healthy human microbiome. Nature 486, 207-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii M., Toda T., Ikarashi N., Kusunoki Y., Kon R., Ochiai W., Machida Y., Sugiyama K. (2014). Gastrectomy increases the expression of hepatic cytochrome P450 3A by increasing lithocholic acid-producing enteric bacteria in mice. Biol. Pharm. Bull. 37, 298-305. [DOI] [PubMed] [Google Scholar]
- Jover R., Moya M., Gomez-Lechon M. J. (2009). Transcriptional regulation of cytochrome p450 genes by the nuclear receptor hepatocyte nuclear factor 4-alpha. Curr. Drug Metab. 10, 508-519. [DOI] [PubMed] [Google Scholar]
- Kamiya A., Inoue Y., Gonzalez F. J. (2003). Role of the hepatocyte nuclear factor 4alpha in control of the pregnane X receptor during fetal liver development. Hepatology 37, 1375-1384. [DOI] [PubMed] [Google Scholar]
- Kearns G. L., Abdel-Rahman S. M., Alander S. W., Blowey D. L., Leeder J. S., Kauffman R. E. (2003). Developmental pharmacology—drug disposition, action, and therapy in infants and children. N. Engl. J. Med. 349, 1157-1167. [DOI] [PubMed] [Google Scholar]
- Klaassen C. D. (1973). Comparison of the toxicity of chemicals in newborn rats to bile duct-ligated and sham-operated rats and mice. Toxicol. Appl. Pharmacol. 24, 37-44. [DOI] [PubMed] [Google Scholar]
- Knight T. R., Choudhuri S., Klaassen C. D. (2007). Constitutive mRNA expression of various glutathione S-transferase isoforms in different tissues of mice. Toxicol. Sci. 100, 513-524. [DOI] [PubMed] [Google Scholar]
- Koenig J. E., Spor A., Scalfone N., Fricker A. D., Stombaugh J., Knight R., Angenent L. T., Ley R.E. (2011). Succession of microbial consortia in the developing infant gut microbiome. Proc. Natl. Acad. Sci. U.S.A.108(Suppl 1), 4578-4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koeth R. A., Wang Z., Levison B. S., Buffa J. A., Org E., Sheehy B. T., Britt E. B., Fu X., Wu Y., Li L., et al. (2013). Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat. Med. 19, 576-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. S., Ward W. O., Liu J., Ren H., Vallanat B., Dekler D., Corton J. C. (2011). Hepatic xenobiotic metabolizing enzymes and transporters gene expression through the life stages of the mouse. PloS One 6, e24381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley R. E., Backhed F., Turnbaugh P., Lozupone C. A., Knight R. D., Gordon J. I. (2005). Obesity alters gut microbial ecology. Proc. Natl. Acad. Sci. U.S.A. 102, 11070-11075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lickteig A. J., Cheng X., Augustine L. M., Klaassen C. D., Cherrington N. J. (2008). Tissue distribution, ontogeny and induction of the transporters multidrug and toxin extrusion (MATE) 1 and MATE2 mRNA expression levels in mice. Life Sci. 83, 59-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liddle C., Goodwin B. (2002). Regulation of hepatic drug metabolism: role of the nuclear receptors PXR and CAR. Semin. Liver Dis. 22, 115-122. [DOI] [PubMed] [Google Scholar]
- Lu H., Gunewardena S., Cui J. Y., Yoo B., Zhong X. B., Klaassen C. D. (2013). RNA-sequencing quantification of hepatic ontogeny and tissue distribution of mRNAs of phase II enzymes in mice. Drug Metab. Dispos. 41, 844-857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matuskova Z., Siller M., Tunkova A., Anzenbacherova E., Zacharova A., Tlaskalova-Hogenova H., Zidek Z., Anzenbacher P. (2011). Effects of Lactobacillus casei on the expression and the activity of cytochromes P450 and on the CYP mRNA level in the intestine and the liver of male rats. Neuro. Endocrinol. Lett. 32(Suppl 1), 8-14. [PubMed] [Google Scholar]
- Mooij M. G., Schwarz U. I., de Koning B. A., Leeder J. S., Gaedigk R., Samsom J. N., Spaans E., van Goudoever J. B., Tibboel D., Kim R. B., et al. (2014). Ontogeny of human hepatic and intestinal transporter gene expression during childhood: age matters. Drug Metab. Dispos. 42, 1268-1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer C., Bik E. M., DiGiulio D. B., Relman D. A., Brown P. O. (2007). Development of the human infant intestinal microbiota. PLoS Biol. 5, e177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H. K., Shim S. S., Kim S. Y., Park J. H., Park S. E., Kim H. J., Kang B. C., Kim C. M. (2005). Molecular analysis of colonized bacteria in a human newborn infant gut. J. Microbiol. 43, 345-353. [PubMed] [Google Scholar]
- Peng L., Cui J. Y., Yoo B., Gunewardena S. S., Lu H., Klaassen C. D., Zhong X. B. (2013). RNA-sequencing quantification of hepatic ontogeny of phase-I enzymes in Mice. Drug Metab. Dispos. 12, 2175-2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng L., Yoo B., Gunewardena S. S., Lu H., Klaassen C. D., Zhong X. B. (2012). RNA sequencing reveals dynamic changes of mRNA abundance of cytochromes P450 and their alternative transcripts during mouse liver development. Drug Metab. Dispos. 40, 1198-1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad B., Unadkat J. D. (2014a). Comparison of Heavy Labeled (SIL) Peptide versus SILAC Protein Internal Standards for LC-MS/MS Quantification of Hepatic Drug Transporters. Int. J. Proteomics 2014, 451510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad B., Unadkat J. D. (2014b). Optimized approaches for quantification of drug transporters in tissues and cells by MRM proteomics. AAPS J. 16, 634-648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad B., Evers R., Gupta A., Hop C. E., Salphati L., Shukla S., Ambudkar S. V., Unadkat J. D. (2014). Interindividual variability in hepatic organic anion-transporting polypeptides and P-glycoprotein (ABCB1) protein expression: quantification by liquid chromatography tandem mass spectroscopy and influence of genotype, age, and sex. Drug Metab. Dispos. 42, 78-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabot S., Membrez M., Bruneau A., Gerard P., Harach T., Moser M., Raymond F., Mansourian R., Chou C. J. (2010). Germ-free C57BL/6J mice are resistant to high-fat-diet-induced insulin resistance and have altered cholesterol metabolism. FASEB J. 24, 4948-4959. [DOI] [PubMed] [Google Scholar]
- Renaud H. J., Cui J. Y., Khan M., Klaassen C. D. (2011). Tissue distribution and gender-divergent expression of 78 cytochrome P450 mRNAs in mice. Toxicol. Sci. 124, 261-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staudinger J. L., Goodwin B., Jones S. A., Hawkins-Brown D., MacKenzie K. I., LaTour A., Liu Y., Klaassen C. D., Brown K. K., Reinhard J., et al. (2001). The nuclear receptor PXR is a lithocholic acid sensor that protects against liver toxicity. Proc. Natl. Acad. Sci. U.S.A. 98, 3369-3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strolin Benedetti M., Whomsley R., Baltes E. L. (2005). Differences in absorption, distribution, metabolism and excretion of xenobiotics between the paediatric and adult populations. Expert Opin. Drug Metab. Toxicol. 1, 447-471. [DOI] [PubMed] [Google Scholar]
- Tang W. H., Wang Z., Levison B. S., Koeth R. A., Britt E. B., Fu X., Wu Y., Hazen S. L. (2013). Intestinal microbial metabolism of phosphatidylcholine and cardiovascular risk. N. Engl. J. Med. 368, 1575-1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda T., Ohi K., Kudo T., Yoshida T., Ikarashi N., Ito K., Sugiyama K. (2009a). Ciprofloxacin suppresses Cyp3a in mouse liver by reducing lithocholic acid-producing intestinal flora. Drug Metab. Pharmacokinet. 24, 201-208. [DOI] [PubMed] [Google Scholar]
- Toda T., Saito N., Ikarashi N., Ito K., Yamamoto M., Ishige A., Watanabe K., Sugiyama K. (2009b). Intestinal flora induces the expression of Cyp3a in the mouse liver. Xenobiotica 39, 323-334. [DOI] [PubMed] [Google Scholar]
- Turnbaugh P. J., Ley R. E., Mahowald M. A., Magrini V., Mardis E. R., Gordon J. I. (2006). An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444, 1027-1031. [DOI] [PubMed] [Google Scholar]
- Vajro P., Mandato C., Licenziati M. R., Franzese A., Vitale D. F., Lenta S., Caropreso M., Vallone G., Meli R. (2011). Effects of Lactobacillus rhamnosus strain GG in pediatric obesity-related liver disease. J. Pediatr. Gastroenterol. Nutr. 52, 740-743. [DOI] [PubMed] [Google Scholar]
- Venkatesh M., Mukherjee S., Wang H., Li H., Sun K., Benechet A. P., Qiu Z., Maher L., Redinbo M. R., Phillips R. S., et al. (2014). Symbiotic bacterial metablites regulate gastrointestinal barrier function via the xenobiotic sensor PXR and Toll-like receptor 4. Immunity 41, 296-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Klipfell E., Bennett B. J., Koeth R., Levison B. S., Dugar B., Feldstein A. E., Britt E. B., Fu X., Chung Y. M., et al. (2011). Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature 472, 57-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waxman D. J. (2000). Growth hormone pulse-activated STAT5 signalling: a unique regulatory mechanism governing sexual dimorphism of liver gene expression. Novartis Found. Symp. 227, 61-74; discussion 75-81. [DOI] [PubMed] [Google Scholar]
- West C. E., Hammarstrom M. L., Hernell O. (2009). Probiotics during weaning reduce the incidence of eczema. Pediatr. Allergy Immunol. 20, 430-437. [DOI] [PubMed] [Google Scholar]
- West C. E., Hernell O., Andersson Y., Sjostedt M., Hammarstrom M. L. (2012). Probiotic effects on T-cell maturation in infants during weaning. Clin. Exp. Allergy 42, 540-549. [DOI] [PubMed] [Google Scholar]
- Yatsunenko T., Rey F. E., Manary M. J., Trehan I., Dominguez-Bello M. G., Contreras M., Magris M., Hidalgo G., Baldassano R. N., Anokhin A. P., et al. (2012). Human gut microbiome viewed across age and geography. Nature 486, 222-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.