Abstract
Diisocyanates, reactive chemicals used to produce polyurethane products, are the most common causes of occupational asthma. The aim of this study is to identify susceptibility gene variants that could contribute to the pathogenesis of diisocyanate asthma (DA) using a Genome-Wide Association Study (GWAS) approach. Genome-wide single nucleotide polymorphism (SNP) genotyping was performed in 74 diisocyanate-exposed workers with DA and 824 healthy controls using Omni-2.5 and Omni-5 SNP microarrays. We identified 11 SNPs that exceeded genome-wide significance; the strongest association was for the rs12913832 SNP located on chromosome 15, which has been mapped to the HERC2 gene (p = 6.94 × 10−14). Strong associations were also found for SNPs near the ODZ3 and CDH17 genes on chromosomes 4 and 8 (rs908084, p = 8.59 × 10−9 and rs2514805, p = 1.22 × 10−8, respectively). We also prioritized 38 SNPs with suggestive genome-wide significance (p < 1 × 10−6). Among them, 17 SNPs map to the PITPNC1, ACMSD, ZBTB16, ODZ3, and CDH17 gene loci. Functional genomics data indicate that 2 of the suggestive SNPs (rs2446823 and rs2446824) are located within putative binding sites for the CCAAT/Enhancer Binding Protein (CEBP) and Hepatocyte Nuclear Factor 4, Alpha transcription factors (TFs), respectively. This study identified SNPs mapping to the HERC2, CDH17, and ODZ3 genes as potential susceptibility loci for DA. Pathway analysis indicated that these genes are associated with antigen processing and presentation, and other immune pathways. Overlap of 2 suggestive SNPs with likely TF binding sites suggests possible roles in disruption of gene regulation. These results provide new insights into the genetic architecture of DA and serve as a basis for future functional and mechanistic studies.
Keywords: asthma, diisocyanate, genetic polymorphism, genome-wide association study
Diisocyanates are the leading causes of occupational asthma (OA), estimated to cause asthma in 5%–15% of chronically exposed workers (Bakerly et al., 2008; Bernstein, 2011; Redlich and Karol, 2002). The most common isomers used in industry are: the aliphatic agent 1,6 hexamethylene diisocyanate (HDI), 4, 4-diphenylmethane diisocyanate (MDI), and toluene diisocyanate (TDI). Despite improved industrial hygiene efforts, new cases of diisocyanate asthma (DA) continue to occur (Booth et al., 2009; Campo et al., 2010; Kenyon et al., 2012). The National Institute for Occupational Safety and Health (NIOSH) National Occupational Exposure Survey database showed that at least 280 000 workers were potentially exposed to some form of diisocyanates in the United States alone (NIOSH, 1983). TDI was reported to account for between 2.9% and 13% of all cases of occupational asthma in Korea (Park et al., 2002). Considering the extent of workplace exposure to diisocyanates, identification of genetic variants increasing susceptibility to DA could be important in developing new risk assessment and preventive strategies for chronically exposed workers. In addition, incorporation of genetic data into human health risk assessment will be important for refining occupational exposure limits for diisocyanates to improve protection of worker health.
We have previously identified a number of DA associated variants of: HLA genes (HLA-B, HLA-E, HLA-DOA, HLA-DPB1, and HLA-DQA2) (Yucesoy et al., 2014), Th2 immune response-associated genes (IL-4Rα, IL-13, and CD14) (Bernstein et al., 2006, 2011) and antioxidant defense genes (SOD2, GST, and EPHX1) (Yucesoy et al., 2012). Although genome-wide association studies (GWAS) have been performed in various asthma phenotypes, only one GWAS has investigated workers with OA. This study was conducted in 84 diisocyanate-exposed Korean workers with TDI-asthma confirmed by specific inhalation challenge (SIC) and 263 unexposed healthy controls and identified 4 CTNNA3 (alpha-T catenin) single nucleotide polymorphisms (SNPs) associated with DA (Kim et al., 2009).
In this study, we aimed to increase our understanding of genetic susceptibility to DA by conducting a GWAS in a different background population of Caucasian subjects. Although population admixture, specific diisocyanate exposure, and genotyping platforms differed between the 2 GWAS studies, we searched for common overlapping genetic loci between the Korean population and ours. Our findings revealed several genes/SNPs that may offer promising avenues for future genetic and functional studies of DA susceptibility genes.
MATERIALS AND METHODS
Study participants
The study population included 88 diisocyanate-exposed workers with DA and 832 self-reported healthy Caucasian individuals (Prahalad et al., 2000). Diisocyanate-exposed workers, referred for clinical evaluation of possible DA by occupational pulmonary disease clinics located in Canada (Sacré-Coeur Hospital, Montreal; Laval Hospital, Sainte-Foy; University Health Network, Toronto) and Spain (Fundación Jiménez Díaz, Madrid and Hospitals Vall D’Hebron, Barcelona), were recruited for participation in this study. To confirm or exclude DA, controlled SIC testing was performed according to published protocols with the relevant diisocyanate chemical to which the worker was exposed (Malo et al., 1999; Sastre et al., 2003). A positive SIC response was defined as a fall in forced expiratory volume in 1 second (FEV1) of at least 20% from prechallenge baseline following a controlled exposure to a diisocyanate chemical generated in a challenge chamber. For subjects recruited and evaluated in Toronto, lung function data collected during diisocyanate exposure at work was used to establish a DA diagnosis. Data pertinent to demographics, smoking status, duration, and nature of chemical exposure were collected in all subjects. Atopic status was evaluated by skin prick testing to commercial aeroallergen extracts. Atopy was defined by a positive response with one or more common aeroallergens defined by a wheal of 3 mm ≥ saline control. Whole blood was collected for genetic testing.
Ethics
All subjects gave written informed consent. The study protocol conformed to the ethical guidelines of the 1975 Declaration of Helsinki and was approved and renewed annually by the Ethical Research Committee of Hôpital du Sacré Cœur de Montréal (Montréal, Québec, Canada), the Research Ethics Committee of the Institut Universitaire de Cardiologie et de Pneumologie de Québec (Sainte Foy, Québec, Canada), University Health Network Research Ethics Board (Toronto, Ontario, Canada), the Ethics Committee of Clinical Research (IRB) of Fundación Jiménez Díaz-UTE (Madrid, Spain), and Val d’Hebron University Ethics Committee (Barcelona, Spain). The comparator group was recruited from the Cincinnati Genomic Control Cohort (Kovacic et al., 2011). This study was approved by the Institutional Review Board at Cincinnati Children’s Hospital Medical Center. Written informed consent for the purpose of DNA collection and genotyping was obtained from all subjects. Parents gave written informed consent for the children’s participation, and children gave assent.
Genotyping
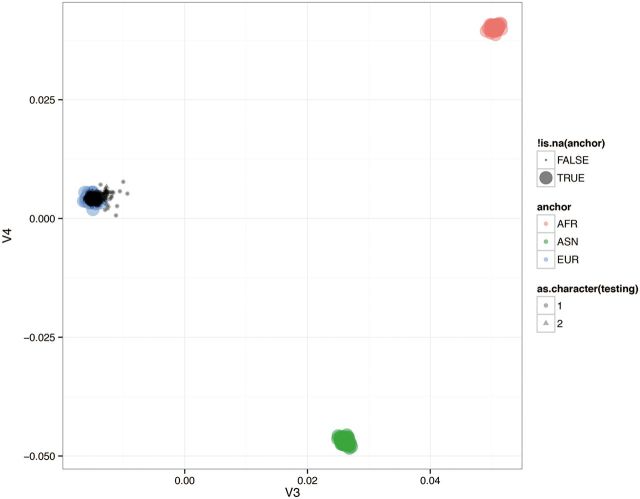
Whole blood samples were collected for genetic analysis and genomic DNA was extracted using the QIAamp blood kit (QIAGEN, Chatsworth, California). Genome-wide SNP genotyping was performed on 88 cases using Omni-2.5 SNP microarray (Illumina, San Diego, California). The chip contains approximately 2.5 million SNP markers with an average call frequency of >99% and is unbiased with respect to coding and noncoding regions of the genome. 832 controls were run on the Omni-5 microarray which contains approximately 4.3 million SNPs. A total of approximately 2.4 million markers were common to both arrays and used for analysis. As starting material, 250 ng of genomic DNA was used for the assay. Internal quality control measures were used for the data obtained from each chip. Genotypes were auto called using GenomeStudio software (Illumina). A total of 7 individuals with genotyping call rate <95% were removed and the genotyping rate in the remaining individuals was 97%. Genome-wide data were used to infer the top 6 principal components of genetic variation and correct for population stratification using Goldenhelix SVS (Bozeman, Montana). Samples were removed from analysis if they segregated >4 SDs outside of the mean of the first 4 principal components. Figure 1 shows the principal component analysis (PCA) plot of the first 2 components based on 100 000 randomly selected SNPs. Minor allele frequencies of SNPs in the control group were also compared with those reported in the phases 1 and 3 of the 1000 Genomes Project. These analyses indicated that both cases and controls were of European origin and there was no population stratification. We also examined LD patterns around the top-ranked SNPs and identified multiple linked SNPs. LocusZoom association plots showing local LD patterns for the top-ranked SNPs are presented in Supplementary Figure 1.
FIG. 1.
PCA plot showing the first two components based on 100 K randomly selected SNPs.
Of the SNPs assayed on the chip, approximately 835 000 SNPs were excluded because of a low call rate, low minor allele frequency (MAF), or deviation from Hardy–Weinberg equilibrium (HWE) in the controls (P < .0001). About 74 cases and 824 controls satisfying all QC measurements were used for analysis. Markers were excluded if they had a MAF < 0.01 in controls and <0.03 in cases. As a result of this basic QC, 1 556 551 SNPs were included in the final analysis with an average sample call rate of 99.98% and a SNP call rate of 99.84%.
Statistical analyses
HWE was carried out using a Pearson goodness-of-fit test. The association between each SNP and DA was assessed using a chi-square test using Goldenhelix SVS (Bozeman, Montana). In order to investigate previously identified regions within the Korean population, we examined SNPs within 25 kb of reported associations.
TF Binding Site Analysis
For identifying SNPs that might impact TF binding events in cell types relevant to DA, we first compiled functional genomics data from several sources, including the ENCODE Project Consortium (2012), Roadmap Epigenomics (Bernstein et al., 2010), and the UCSC Genome Browser (Dreszer et al., 2012). Custom software was used to restrict to cell types relevant to asthma (including cell lines and cells derived from lung tissues) and identify SNPs that intersect these datasets. We then identified those among the remaining SNPs that are located within ChIP-seq binding peaks for TFs. The resulting SNPs were further filtered to those likely to impact TF binding by scanning each ChIP-seq peak for the most likely binding site for the associated TF using position weight matrix (PWM) models taken from Weirauch et al. (2014), and the standard log likelihood scoring system (Stormo, 1990), and restricting to those SNPs that are located within these binding sites. The resulting SNPs are therefore (1) located in likely regulatory regions in relevant cell types; (2) located within a ChIP-seq binding peak for a TF; and (3) located within the most likely binding site for the corresponding TF within that binding peak.
RESULTS
Clinical Characteristics of the Study Subjects
The demographic characteristics of the study groups included in the statistical analyses are described in Table 1. The DA group consisted of 88 subjects of European Caucasian descent and 1 Asian with a mean age of 40.7 years (between 22 and 64 years). The control group included 832 self-reported non-Hispanic Caucasian healthy individuals ranging in age from 2 to 62 years with a mean age of 15.8 years, and consisted of 406 males and 426 females.
Table 1.
Demographic Characteristics of the Study Groups
| DA+ | Controls | |
|---|---|---|
| N | 88 | 832 |
| Sex, M/F | 77/11 | 406/426 |
| Mean age ± SD range | 40.7 + 11.9 (22–64) | 15.8 ± 12.5 (2–62) |
| Diisocyanate exposure (HDI/MDI/TDI) | 44/19/25 | N/A |
| Duration of exposure mean months ± SD | 147.9 ± 137.8 | N/A |
| Skin prick test Positive/negative/unknown | 53/31/4 | N/A |
| Smoker (Current/ex/never) | 16/34/38 | N/A |
| Mean pack-years ± SD (All subjects) | 10.7 ± 14.0 | N/A |
| Mean pack-years ± SD (Current & Ex-smokers) | 19.0 ± 13.3 | N/A |
| Race White/non-White | 87/1 | 832/0 |
| Ethnicity (French Canadian/Othera) | 60/28 | N/A |
Abbreviations: DA, diisocyanate asthma; N/A, not applicable.
aOther includes English Canadian (3), Spanish (18), Italian (3), Polish (1), Moroccan (1), Ecuadorian (1), Indian (1).
Genetic Associations
Top-ranked genes are presented in Table 2. Eleven SNPs exceeded the threshold for genome-wide significance (p < 10−7) and 3 of them were mapped to the HERC2, CDH17, and ODZ3 genes. As seen in Table 3, the most significant association was observed for the rs12913832 SNP mapping to the HERC2 gene (p = 6.94 × 10−14), located at position 28365618 on chromosome 15q13. The second most significant signal was for the rs12568266 SNP (p = 6.20 × 10−10). This is an intergenic variant and located on chromosome 1 between the hCG_2036596 (left) and LOC644357 (right) genes. The next 2 SNPs (rs74380195 and rs76966929, p = 9.58 × 10−10) were located on chromosome 11 and were not mapped to any gene. The next significant variant (rs7588010, p = 4.08 × 10−9) was also not mapped to any gene but is located between the TACR1 (left) and FAM176A (right) genes on chromosome 2. Following 2 markers were intron variants and mapped to the ODZ3 (rs908084, p = 8.59 × 10−9) and CDH17 (rs2514805, p = 1.22 × 10−8) genes. The rest of the markers reaching a genome-wide significance were rs16867528 (p = 1.65 × 10−8), rs12143327 (p = 4.57 × 10−8), rs79252495 (p = 5.85 × 10−8), and rs76684306 (p = 9.90 × 10−8) SNPs. They were intergenic variants and located on chromosomes 5, 1, 15, and 5, respectively. Except HERC2 rs12913832 SNP, none of these top SNPs have been previously implicated in any disease.
Table 2.
Top-Ranked Genes
| Entrez Gene ID | Gene Symbol | Gene Name | Number of SNPs | p Values (range) |
|---|---|---|---|---|
| 8924 | HERC2 | HECT and RLD domain containing E3 ubiquitin protein ligase 2 | 1 | 6.94 × 10−14 |
| 55714 | ODZ3 (TENM3) | Teneurin transmembrane protein 3 | 3 | 8.59 × 10−9–5.64 × 10−7 |
| 1015 | CDH17 | Cadherin 17, liver-intestine cadherin | 4 | 1.22 × 10−8–4.74 × 10−7 |
| 130013 | ACMSD | Aminocarboxymuconate semialdehyde decarboxylase | 1 | 6.35 × 10−7 |
| 26207 | PITPNC1 | Phosphatidylinositol transfer protein, cytoplasmic 1 | 6 | 6.33 × 10−7–7.82 × 10−7 |
| 7704 | ZBTB16 | Zinc finger and BTB domain containing 16 | 5 | 1.68 × 10−7–7.03 × 10−7 |
| Suggestive genes | ||||
| 64067 | NPAS3 | Neuronal PAS domain protein 3 | 3 | 2.42 × 10−6–1.69 × 10−5 |
| 5578 | PRKCA | Protein kinase C, alpha | 1 | 2.55 × 10−6 |
| 6539 | SLC6A12 | Solute carrier family 6 (neurotransmitter transporter), member 12 | 3 | 1.47 × 10−6–1.67 × 10−5 |
| 6869 | TACR1 | Tachykinin receptor 1 | 13 | 4.08 × 10−9–7.95 × 10−5 |
Table 3.
Genome-Wide Association Results for Significant (p < 1 × 10−7) and Suggestive (p < 1 × 10−6) SNPs
| SNP | Chr | Position | Gene | Feature | Risk Allele | MAF Cases | MAF Controls | p value | OR | 95% L | 95% U |
|---|---|---|---|---|---|---|---|---|---|---|---|
| rs12913832 | 15 | 28365618 | HERC2 | Intron | A | 0.513 | 0.233 | 6.94 × 10−14 | 3.475 | 2.468 | 4.891 |
| rs12568266 | 1 | 4193551 | NA | NA | A | 0.127 | 0.029 | 6.20 × 10−10 | 5.017 | 2.859 | 8.803 |
| rs74380195 | 11 | 102887229 | NA | NA | G | 0.173 | 0.050 | 9.58 × 10−10 | 4.018 | 2.493 | 6.478 |
| rs76966929 | 11 | 102891066 | NA | NA | A | 0.173 | 0.050 | 9.58 × 10−10 | 4.018 | 2.493 | 6.478 |
| rs7588010 | 2 | 75473916 | NA | NA | A | 0.593 | 0.357 | 4.08 × 10−9 | 2.719 | 1.928 | 3.836 |
| rs908084 | 4 | 183543648 | ODZ3 | Intron | A | 0.153 | 0.044 | 8.59 × 10−9 | 3.970 | 2.401 | 6.564 |
| rs2514805 | 8 | 95167247 | CDH17 | Intron | G | 0.28 | 0.118 | 1.22 × 10−8 | 2.952 | 2.004 | 4.349 |
| rs16867528 | 5 | 15607448 | NA | NA | A | 0.068 | 0.010 | 1.65 × 10−8 | 7.391 | 3.291 | 16.598 |
| rs12143327 | 1 | 112610895 | NA | NA | A | 0.213 | 0.081 | 4.57 × 10−8 | 3.142 | 2.045 | 4.829 |
| rs79252495 | 15 | 26722016 | NA | NA | A | 0.12 | 0.032 | 5.85 × 10−8 | 4.250 | 2.415 | 7.477 |
| rs76684306 | 5 | 15616676 | NA | NA | G | 0.073 | 0.011 | 9.90 × 10−8 | 6.562 | 2.971 | 14.492 |
| Suggestive associations | |||||||||||
| rs118079879 | 12 | 41713306 | NA | NA | A | 0.073 | 0.013 | 1.25 × 10−7 | 5.927 | 2.815 | 12.478 |
| rs10769691 | 11 | 6375809 | NA | NA | A | 0.193 | 0.072 | 1.57 × 10−7 | 3.131 | 2.003 | 4.895 |
| rs1672691 | 11 | 113942151 | ZBTB16 | intron | A | 0.38 | 0.200 | 1.68 × 10−7 | 2.498 | 1.756 | 3.553 |
| rs7115199 | 11 | 102871623 | NA | NA | A | 0.1 | 0.024 | 1.85 × 10−7 | 4.534 | 2.441 | 8.421 |
| rs74609360 | 15 | 31540395 | NA | NA | A | 0.113 | 0.030 | 2.08 × 10−7 | 4.142 | 2.323 | 7.386 |
| rs72974161 | 2 | 136802456 | NA | NA | A | 0.24 | 0.102 | 2.11 × 10−7 | 2.832 | 1.883 | 4.258 |
| rs2340023 | 8 | 95157476 | CDH17 | Intron | A | 0.267 | 0.120 | 2.47 × 10−7 | 2.712 | 1.833 | 4.014 |
| rs76314368 | 5 | 116492833 | NA | NA | A | 0.093 | 0.022 | 2.63 × 10−7 | 4.672 | 2.459 | 8.879 |
| rs3018331 | 11 | 113940469 | ZBTB16 | Intron | A | 0.347 | 0.178 | 2.83 × 10−7 | 2.505 | 1.747 | 3.592 |
| rs11722354 | 4 | 183565618 | ODZ3 | Intron | A | 0.122 | 0.035 | 2.86 × 10−7 | 3.925 | 2.243 | 6.870 |
| rs2513797 | 8 | 95143138 | CDH17 | CS | G | 0.267 | 0.121 | 3.39 × 10−7 | 2.681 | 1.812 | 3.967 |
| rs13017967 | 2 | 75461177 | NA | NA | A | 0.54 | 0.338 | 3.46 × 10−7 | 2.368 | 1.687 | 3.325 |
| rs6716987 | 2 | 136963494 | NA | NA | A | 0.48 | 0.287 | 4.28 × 10−7 | 2.353 | 1.676 | 3.305 |
| rs6442708 | 3 | 1994644 | NA | NA | A | 0.155 | 0.053 | 4.41 × 10−7 | 3.351 | 2.043 | 5.496 |
| rs2446824 | 8 | 95127574 | NA | NA | A | 0.267 | 0.123 | 4.64 × 10−7 | 2.651 | 1.792 | 3.922 |
| rs2513791 | 8 | 95128529 | NA | NA | A | 0.267 | 0.123 | 4.64 × 10−7 | 2.651 | 1.792 | 3.922 |
| rs2446823 | 8 | 95127612 | NA | NA | C | 0.267 | 0.123 | 4.64 × 10−7 | 2.651 | 1.792 | 3.922 |
| rs2262592 | 8 | 95155550 | CDH17 | Intron | G | 0.267 | 0.122 | 4.74 × 10−7 | 2.678 | 1.802 | 3.980 |
| rs11731869 | 4 | 183559826 | ODZ3 | Intron | A | 0.12 | 0.035 | 5.64 × 10−7 | 3.796 | 2.172 | 6.633 |
| rs2846629 | 11 | 113941756 | ZBTB16 | Intron | G | 0.38 | 0.207 | 5.72 × 10−7 | 2.401 | 1.689 | 3.413 |
| rs4852364 | 2 | 75457783 | NA | NA | A | 0.3 | 0.512 | 5.92 × 10−7 | 0.404 | 0.280 | 0.582 |
| rs12328696 | 2 | 75460555 | NA | NA | A | 0.3 | 0.512 | 5.93 × 10−7 | 0.404 | 0.280 | 0.582 |
| rs1784684 | 11 | 113939539 | ZBTB16 | Intron | G | 0.347 | 0.182 | 6.20 × 10−7 | 2.44 | 1.702 | 3.498 |
| rs7224314 | 17 | 65386214 | PITPNC1 | Intron | C | 0.413 | 0.223 | 6.33 × 10−7 | 2.372 | 1.675 | 3.359 |
| rs4954192 | 2 | 135632981 | ACMSD | Intron | A | 0.627 | 0.410 | 6.35 × 10−7 | 2.362 | 1.671 | 3.340 |
| rs2309284 | 4 | 181103729 | NA | NA | A | 0.093 | 0.023 | 6.63 × 10−7 | 4.427 | 2.340 | 8.375 |
| rs62084082 | 17 | 65388046 | PITPNC1 | Intron | G | 0.413 | 0.224 | 6.98 × 10−7 | 2.364 | 1.669 | 3.348 |
| rs2735199 | 11 | 113942191 | ZBTB16 | Intron | G | 0.38 | 0.208 | 7.03 × 10−7 | 2.385 | 1.677 | 3.390 |
| rs60244812 | 9 | 7267614 | NA | NA | A | 0.107 | 0.029 | 7.05 × 10−7 | 4.035 | 2.230 | 7.302 |
| rs1491921 | 5 | 21259138 | NA | NA | C | 0.068 | 0.013 | 7.41 × 10−7 | 5.690 | 2.626 | 12.327 |
| rs62085810 | 17 | 65372855 | PITPNC1 | NG-5 | A | 0.40 | 0.219 | 7.63 × 10−7 | 2.363 | 1.667 | 3.352 |
| rs62084078 | 17 | 65386886 | PITPNC1 | Intron | G | 0.413 | 0.225 | 7.82 × 10−7 | 2.355 | 1.663 | 3.335 |
| rs62084077 | 17 | 65386837 | PITPNC1 | Intron | A | 0.413 | 0.225 | 7.82 × 10−7 | 2.355 | 1.663 | 3.335 |
| rs62084080 | 17 | 65387014 | PITPNC1 | Intron | A | 0.413 | 0.225 | 7.82 × 10−7 | 2.355 | 1.663 | 3.335 |
| rs4954572 | 2 | 136926719 | NA | NA | A | 0.473 | 0.286 | 8.19 × 10−7 | 2.307 | 1.643 | 3.241 |
| rs75646498 | 15 | 29876649 | NA | NA | A | 0.115 | 0.030 | 8.29 × 10−7 | 3.996 | 2.211 | 7.224 |
| rs6858365 | 4 | 12172669 | NA | NA | A | 0.073 | 0.015 | 8.72 × 10−7 | 5.213 | 2.511 | 10.820 |
| rs116146467 | 6 | 169558013 | NA | NA | A | 0.067 | 0.013 | 9.32 × 10−7 | 5.614 | 2.592 | 12.160 |
Abbreviations: ACMSD, aminocarboxymuconate semialdehyde decarboxylase; CS, coding synonymous; CDH17, cadherin 17; HERC2, HECT and RLD domain containing E3 ubiquitin protein ligase 2; MAF, minor allele frequency; NG-5, near gene-5; ODZ3 (TENM3), teneurin transmembrane protein 3; PITPNC1, phosphatidylinositol transfer protein, cytoplasmic 1; ZBTB16, zinc finger and BTB domain containing 16; 95% U, upper bound; 95% L, lower bound.
OR, odds ratio in risk genotype relative to common genotype according to each analysis model.
We also prioritized 38 suggestive genome-wide significant SNPs (p < 1 × 10−6) based on their potential involvement in DA (Table 3). Among them, 17 SNPs map to the PITPNC1, ACMSD, ZBTB16, ODZ3, and CDH17 genes. None of the prioritized candidates have been previously implicated in asthma. The full set of results is available in Supplementary Table 1.
In addition, 4 suggestive genes NPAS3, PRKCA, SLC6A12, and TACR1 were identified based on previous reports related to airway diseases and proximities to our significant SNPs (Table 4). Supplementary Table 2 presents complete genetic association results for these genes.
Table 4.
Suggestive Associations Based on Previous Reports and Proximity to Significant SNPs
| SNP ID | Chr | Position | Gene | Chi-squared p value |
|---|---|---|---|---|
| rs28564912 | 14 | 33798766 | NPAS3 | 2.42 × 10−6 |
| rs56962758 | 14 | 33769003 | NPAS3 | 1.67 × 10−5 |
| rs9635191 | 14 | 33746767 | NPAS3 | 1.69 × 10−5 |
| rs4636942 | 17 | 64793346 | PRKCA | 2.55 × 10−6 |
| rs188610 | 12 | 313839 | SLC6A12 | 1.47 × 10−6 |
| rs7138605 | 12 | 320510 | SLC6A12 | 1.67 × 10−5 |
| rs16928441 | 12 | 321353 | SLC6A12 | 1.67 × 10−5 |
| Gene left to the significant SNPs | ||||
| rs7588010 | 2 | 75473916 | TACR1 | 4.08 × 10−9 |
| rs13017967 | 2 | 75461177 | TACR1 | 3.46 × 10−7 |
| rs4852364 | 2 | 75457783 | TACR1 | 5.92 × 10−7 |
| rs12328696 | 2 | 75460555 | TACR1 | 5.93 × 10−7 |
| rs4853124 | 2 | 75449986 | TACR1 | 2.14 × 10−6 |
| rs7583283 | 2 | 75478007 | TACR1 | 4.70 × 10−6 |
| rs4853128 | 2 | 75491046 | TACR1 | 2.14 × 10−5 |
| rs1861430 | 2 | 75470713 | TACR1 | 4.31 × 10−5 |
| rs7576919 | 2 | 75471057 | TACR1 | 4.31 × 10−5 |
| rs10490307 | 2 | 75482223 | TACR1 | 4.35 × 10−5 |
| rs4508624 | 2 | 75479258 | TACR1 | 4.69 × 10−5 |
| rs7605785 | 2 | 75453787 | TACR1 | 6.43 × 10−5 |
| rs11684394 | 2 | 75444073 | TACR1 | 7.95 × 10−5 |
Abbreviation: SNP, single nucleotide polymorphism.
Cut-off, p < 1 × 10−4.
We also investigated chromosome X and identified 8 SNPs that exceed nominal significance (p < 1 × 10−4). Top-ranked SNPs did not map to any gene. These results are presented in Supplementary Table 3.
Functional Analysis of Genetic Variants
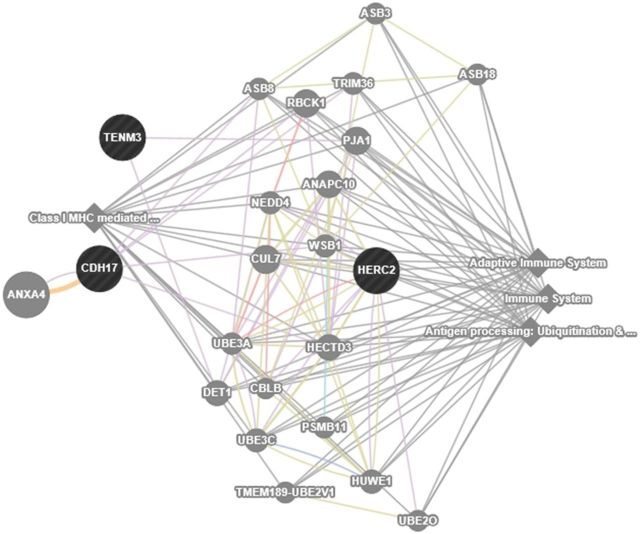
In order to gain insight into molecular mechanisms, we examined whether genes cluster into particular biological pathways, and searched for their expression profiles in the lung using publicly available databases. We also predicted functional relationships between the top 3 genes using the GeneMANIA webserver (Warde-Farley et al., 2010). This analysis suggested that these genes are associated with adaptive immunity, antigen processing, ubiquitination and proteasome degradation, and class I MHC mediated antigen processing and presentation. Figure 2 shows interactive functional association network related to genome-wide significant loci. Considering significant and suggestive genes, we found a significant clustering of genes involved in protein binding, metabolic processes, response to stimulus, and biological regulation using WEB-based GEne SeT AnaLysis Toolkit (Wang et al., 2013). We also used the CGAP EST cDNA library (http://cgap.nci.nih.gov) and the BioGPS gene expression portal to explore the expression profiles of our top-ranked genes in normal human lung tissues (Wu et al., 2013) All candidate genes were expressed in the lung to a varying extent [the number of expressed sequence tag (EST) per 200 000 tags in the CGAP EST cDNA library were 4–7 tags for HERC2, <2 tags for ODZ3, CDH17, AMCSD, ZBTB16, and 2–3 tags for PITPNC1].
FIG. 2.
Interactive functional association network among the top-ranked genes (HERC2, CDH17, and ODZ3). The relationship between the genes in the network includes coexpression, physical and genetic interactions, pathways, colocalization, protein domain similarity, and predicted interactions. The network was filtered by removing all the interactions where weights <0.1.
Effects of SNPs on TF Binding Sites
We used a large collection of TF DNA binding models to predict specific TFs whose binding might be affected by the alleles of each SNP (Weirauch et al., 2014). Based on multiple types of data (including histone marks, ChIP-seq for TFs and chromatin binding proteins, and DNase-seq peaks), this procedure identified a locus 3′ of the CDH17 gene encompassing the rs2446823 and rs2446824 SNPs. This region is likely to play an important regulatory role in several lung cell lines (ie, A549, AG04450, IMR90, and NHLF), as well as lung and fetal lung tissues. These SNPs, which are separated by 37 bases, were located within ChIP-seq peaks for the CEBPA/B (CCAAT/Enhancer Binding Protein) (in U937, A549, and IMR90 cells) and HNF4A (Hepatocyte Nuclear Factor 4, Alpha) (in Caco-2 cells) TFs, respectively.
Overlaps Between Korean and Caucasian GWAS
We detected association signals within 25 kb of significant SNPs in the Korean DA GWAS. These SNPs were mapped to CTNNA1, CTNNA3, TRPM8, KCNIP4, DOCK2, TUSC3, SAMD12, ASTN2, CRTAC1, LHPP, PDGFD, and PCNX genes. Figure 3 shows Manhattan plot of the p values obtained from our and Korean GWAS. 146 SNPs were mapped to chromosome 10 where top hit CTNNA3 gene was located. This gene has been identified as a susceptibility locus in the Korean study and replicated by our group (Bernstein et al., 2013). The overlapping regions have been presented in Supplementary Table 4.
FIG. 3.
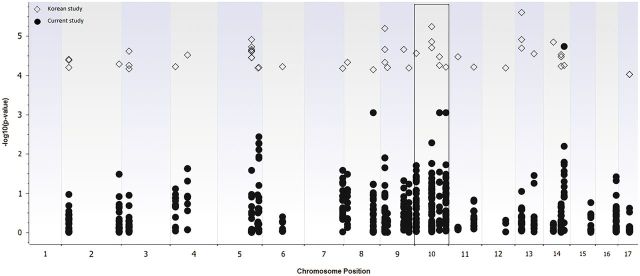
Manhattan plot of the p values obtained from Korean (open diamonds) and Caucasian (filled circles) GWAS studies. A total of 146 SNPs were mapped to chromosome 10 (marked in a square) where the top hit gene (CTNNA3) in the Korean study was located.
DISCUSSION
In this study, we conducted a GWAS for DA in a Caucasian population of European background and identified SNPs reaching genome wide significant associations with confirmed occupational asthma caused by isocyanates. The top ranked SNP mapped to the HERC2 (HECT and RLD domain containing E3 ubiquitin protein ligase 2) gene. Although there were 3 SNPs mapping to this gene, only 1 (rs12913832) exceeded genome-wide significance (p = 6.9 × 10−14). As one of the E3 ubiquitin ligases, HERC2 is involved in the addition of ubiquitin to proteins undergoing degradation. It has also been implicated in DNA replication and DNA damage response (Bekker-Jensen et al., 2010; Izawa et al., 2011). Protein ubiquitination is an important regulatory posttranslational process that controls intracellular signaling and antigen processing. Due to its fundamental role in cell signaling, the ubiquitin system plays an important role in cancer immunity (Sun, 2006). The HERC2 rs12913832 SNP has been implicated in immune (eg, ulcerative colitis) and developmental conditions (eg, pigmentation, iris color) (Franke et al., 2008; Visser et al., 2012). In a recent study, N-succinimidyl N-methylcarbamate (NSNM), a surrogate chemical containing a functional isocyanate group, was used to understand the mechanism behind isocyanate toxicity in the model organism Saccharomyces cerevisiae. The biological effect of NSNM on the growth of yeast deletion mutants of various pathways was investigated. The deletion mutants of ubiquitylation factors and mitochondrial chaperones were found to be hypersensitive to NSNM in a growth assay suggesting that isocyanates specifically target these pathways (Azad et al., 2014). Our results support this observation and suggest an interaction between genetic variability within this gene involved in the ubiquitylation pathway and DA. Elucidation of the functional role of this SNP may provide new insights into the incompletely defined pathogenesis of DA.
The next marker (rs908084) reaching genome-wide significance (p = 8.59 × 10−9) mapped to the ODZ3 (teneurin transmembrane protein 3, also called TENM3) gene. The protein encoded by ODZ3 (4q34.3) belongs to the teneurin subfamily and is involved in homophilic cell adhesion and regulation of neuronal development. The role of this gene in the asthmatic process is unknown. Due to its role in cellular adhesion, it is possible that ODZ3 variants could play a functional role in the airway epithelium response to diisocyanate exposure. An external search of the catalog of published genome-wide association studies did not reveal associations of this SNP with other asthma phenotypes (http://www.genome.gov/gwastudies).
The other genome-wide significant marker (rs2514805, p = 1.22 × 10−8) mapped to the CDH17 (cadherin 17) gene. CDH17 gene is located on chromosome 8q22.1 and encodes liver–intestine–cadherin. Cadherins are cell–cell adhesion molecules that play an important role in maintaining tissue structure and morphology. CDH17 functions as a peptide transporter and cell adhesion molecule to maintain tissue integrity in epithelia. In an Australian GWAS, another CDH17 SNP (rs11776675) was found to be 1 of 5 independent SNPs that were associated with heightened asthma risk at a pre-defined cut-off of p ≤ 5 × 10−6 (OR = 1.21, p = 2.7 × 10−6). However, this result was not replicated in a follow up meta-analysis of 4 independent asthma studies (Ferreira et al., 2011). We found 9 SNPs in this region with only 1, rs2514805, reaching genome-wide significance (p = 1.22 × 10−8). Although expression of CDH17 is low in the lungs, significant markers mapping to the CDH17 gene in Australian and current GWAS studies suggests potential involvement of this gene in the asthmatic process by possibly impacting epithelial barrier integrity. Existence of additional linked SNPs around the top-ranked variants further supports the association of these genomic regions with DA (see Supplementary Fig. 1).
Based on previously reported associations with other asthma phenotypes and proximities to our significant and suggestive genes in Table 3, we identified 4 additional suggestive genes, NPAS3, PRKCA, SLC6A12, and TACR1. NPAS3 SNPs have been identified in asthmatics of different backgrounds as one of the shared genetic factors (Ding et al., 2013). NPAS3 is known to activate or suppress multiple distinct signaling pathways in lung development and repair, and its deficiency was reported to be associated with emphysema and asthma (Zhou et al., 2009). In our analysis, we identified 7 SNPs mapping to this gene and only 3 of them achieved suggestive p values. PRKCA that plays a role in cellular transformation has been identified as a positional candidate gene for asthma (Murphy et al., 2009). We obtained one significant signal mapping to the PRKCA gene with a suggestive p value. SLC6A12 SNPs have been associated with aspirin-intolerant asthma in a Korean population (Pasaje et al., 2010). We identified 3 suggestive SNPs mapping to this gene which is known to play a critical role in mucus production in asthma. TACR1 (tachykinin receptor 1) gene was located left of 13 SNPs that were not mapped to any gene (Table 3). Tachykinins have been found to activate NF-kB and stimulate proinflammatory cytokine expression in epithelial cells following lung injury (Williams et al., 2007). In animal models for TDI-induced asthma, tachykinins were found to play a role in the development of airway hyperresponsiveness (Mapp et al., 1998; Scheerens et al., 1996). Our results provide novel insights about potential involvement of these genes in the DA process.
Results from recent studies suggest that as many as 93% of disease- and trait-associated SNPs are located in regulatory regions (Hindorff et al., 2009; Maurano et al., 2012), indicating that many SNPs might act by affecting the binding of TFs. We therefore used functional genomics data to identify SNPs located in likely regulatory regions in cell types relevant to DA. We found that 2 suggestive intergenic SNPs (rs2446823 and rs2446824) were located within ChIP-seq peaks for the CEBPA/B and HNF4A TFs. Both SNPs are likely located within the specific region bound by each TF, suggesting that each might function by impacting the binding of their respective TF(s). Intriguingly, CEBPB and HNF4A are well known interaction partners in a variety of cellular contexts (Schmidt et al., 2010) and previous studies have linked asthma to impaired translation of CEBPs (Borger et al., 2007; Miglino et al., 2012). It has been suggested that reduced CEBP levels stimulate cell proliferation and the release of proinflammatory cytokines, resulting in airway inflammation.
Based on systems-level functional analysis (Warde-Farley et al., 2010), our top 3 genes (CDH17, HERC2, and ODZ3) are all involved in pathways related to antigen presentation/binding and the immune response (Fig. 2). Although the pathology of DA has been considered similar to common environmental asthma, recent evidence suggests that immune responses elicited by low molecular weight agents such as diisocyanates are distinct from those induced by common environmental allergens (high molecular weight) (Wisnewski et al., 2008). Associations of DA with variants of genes involved in antigen processing and adaptive immunity is biologically plausible considering the different nature of mechanisms involved. We also searched our genes to see whether they are related to a particular molecular/biological function. The findings showed that significant genes (genome-wide and suggestive) were mainly related to protein binding and biological regulation. Although unproven, genetic variations influencing their ability to either bind or transport small (usually charged) molecules could be mechanistically relevant to poorly understood immune mechanisms contributing to DA (Redlich and Karol, 2002).
In addition, we searched GWAS results from a Korean population for any overlapping signals. This study reported a novel locus on chromosome 10q21 encompassing the CTNNA3 gene (encoding catenin alpha 3), and an additional 54 suggestive SNPs. Although there were no exactly overlapping SNPs with our study, we detected significant signals in the proximity of their top ranked SNPs (Supplementary Table 4). The sheer number of signals in this same region is highly unlikely to be a chance observation, and suggests that this genomic region might contain common susceptibility variants. Mapping these regions in detail to identify causal variants, followed by exploration of causal mechanisms may yield new insights into the pathogenesis of DA.
It should be noted that this study has several limitations. Although this was the largest GWAS study of DA to date, our sample size is small due to the relative rarity of DA compared to other types of asthma. However, rigorous phenotypic characterization of this population helps to maximize the discriminatory potential between study groups and with a larger control group, this study was better powered to identify statistically significant signals. Although the controls were genetically homozygous to the cases based on PCA, they were from a younger, healthy population not exposed to diisocyanates. DA occurs as a consequence of exposure to diisocyanates and our control population had negligible potential for future exposure. Considering this and the difficulty in recruiting age-matched workplace controls, we took advantage of an existing large cohort recruited from the general population. Another limitation of this study is the lack of a replication stage for those loci that were most highly associated with DA. As such, the highly associated loci remain suggestive until confirmed in future studies.
Although we detected signals around significant SNPs, we were unable to replicate results of the previous GWAS in Korean workers with DA. Most GWAS findings do not replicate consistently, which may be related to ancestry variations in the study populations, differences in asthma phenotype definitions, type of exposure, unaccounted environmental factors, and the coverage of genotyping platform used. The previous GWAS platform (Affymetrix 500) had a lower coverage as compared to Illumina Omni-2.5 and Omni-5 SNP microarrays which may result in missing several genes that might interact with diisocyanate exposure. In addition, the Korean population was exposed only to TDI whereas our population was exposed to HDI, TDI, and MDI.
In summary, this study identified 3 novel genome-wide significant loci and prioritized an additional 5 genome-wide suggestive candidate genes for DA. In addition to several novel candidate genes, we found that rs2446823 and rs2446824 SNPs could affect binding of CEBP and HNF4A TFs. Although this study is exploratory and novel signals need to be followed up in a large independent sample, these results offer new avenues for future studies of genes contributing to DA susceptibility.
FUNDING
NIOSH/CDC (R01 OH 008795), University of Cincinnati Research Council, Interdisciplinary Faculty Research Support Grant (2012).
SUPPLEMENTARY DATA
Supplementary data are available online at http://toxsci.oxfordjournals.org/.
REFERENCES
- Azad G. K., Singh V., Tomar R. S. (2014). Assessment of the biological pathways targeted by isocyanate using N-succinimidyl N-methylcarbamate in budding yeast Saccharomyces cerevisiae. PLoS One 9, e92993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakerly N. D., Moore V. C., Vellore A. D., Jaakkola M. S., Robertson A. S., Burge P. S. (2008). Fifteen-year trends in occupational asthma: data from the Shield surveillance scheme. Occup. Med. 58, 169–174. [DOI] [PubMed] [Google Scholar]
- Bekker-Jensen S., Rendtlew Danielsen J., Fugger K., Gromova I., Nerstedt A., Lukas C., Bartek J., Lukas J., Mailand N. (2010). HERC2 coordinates ubiquitin-dependent assembly of DNA repair factors on damaged chromosomes. Nat. Cell. Biol. 12, 80–86. [DOI] [PubMed] [Google Scholar]
- Bernstein B. E., Stamatoyannopoulos J. A., Costello J. F., Ren B., Milosavljevic A., Meissner A., Kellis M., Marra M. A., Beaudet A. L., Ecker J. R., et al. (2010). The NIH Roadmap Epigenomics Mapping Consortium. Nat. Biotechnol. 28, 1045–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein D. I. (2011). Genetics of occupational asthma. Curr. Opin. Allergy. Clin. Immunol. 11, 86–89. [DOI] [PubMed] [Google Scholar]
- Bernstein D. I., Kashon M., Lummus Z. L., Johnson V. J., Fluharty K., Gautrin D., Malo J. L., Cartier A., Boulet L. P., Sastre J., et al. (2013). CTNNA3 (alpha-catenin) gene variants are associated with diisocyanate asthma: a replication study in a Caucasian worker population. Toxicol. Sci. 131, 242–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein D. I., Kissling G. E., Khurana Hershey G., Yucesoy B., Johnson V. J., Cartier A., Gautrin D., Sastre J., Boulet L. P., Malo J. L., et al. (2011). Hexamethylene diisocyanate asthma is associated with genetic polymorphisms of CD14, IL-13, and IL-4 receptor alpha. J. Allergy Clin. Immunol. 128, 418–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein D. I., Wang N., Campo P., Chakraborty R., Smith A., Cartier A., Boulet L. P., Malo J. L., Yucesoy B., Luster M., et al. (2006). Diisocyanate asthma and gene-environment interactions with IL4RA, CD-14, and IL-13 genes. Ann. Allergy Asthma Immunol. 97, 800–806. [DOI] [PubMed] [Google Scholar]
- Booth K., Cummings B., Karoly W. J., Mullins S., Robert W. P., Spence M., Lichtenberg F. W., Banta J. (2009). Measurements of airborne methylene diphenyl diisocyanate (MDI) concentration in the U.S. workplace. J. Occup. Environ. Hyg. 6, 228–238. [DOI] [PubMed] [Google Scholar]
- Borger P., Matsumoto H., Boustany S., Gencay M. M., Burgess J. K., King G. G., Black J. L., Tamm M., Roth M. (2007). Disease-specific expression and regulation of CCAAT/enhancer-binding proteins in asthma and chronic obstructive pulmonary disease. J. Allergy Clin. Immunol. 119, 98–105. [DOI] [PubMed] [Google Scholar]
- Campo P., Aranda A., Rondon C., Donia I., Diaz-Perales A., Canto G., Lisbona F. J., Pineda F., Blanca M. (2010). Work-related sensitization and respiratory symptoms in carpentry apprentices exposed to wood dust and diisocyanates. Ann. Allergy Asthma Immunol. 105, 24–30. [DOI] [PubMed] [Google Scholar]
- ENCODE Project Consortium. (2012). An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L., Abebe T., Beyene J., Wilke R. A., Goldberg A., Woo J. G., Martin L. J., Rothenberg M. E., Rao M., Hershey G. K., et al. (2013). Rank-based genome-wide analysis reveals the association of ryanodine receptor-2 gene variants with childhood asthma among human populations. Hum. Genomics 7, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreszer T. R., Karolchik D., Zweig A. S., Hinrichs A. S., Raney B. J., Kuhn R. M., Meyer L. R., Wong M., Sloan C. A., Rosenbloom K. R., et al. (2012). The UCSC Genome Browser database: extensions and updates 2011. Nucleic Acids Res. 40, D918–D923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira M. A., Matheson M. C., Duffy D. L., Marks G. B., Hui J., Le Souef P., Danoy P., Baltic S., Nyholt D. R., Jenkins M., et al. (2011). Identification of IL6R and chromosome 11q13.5 as risk loci for asthma. Lancet 378, 1006–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke A., Balschun T., Karlsen T. H., Hedderich J., May S., Lu T., Schuldt D., Nikolaus S., Rosenstiel P., Krawczak M., et al. (2008). Replication of signals from recent studies of Crohn's disease identifies previously unknown disease loci for ulcerative colitis. Nat. Genet. 40, 713–715. [DOI] [PubMed] [Google Scholar]
- Hindorff L. A., Sethupathy P., Junkins H. A., Ramos E. M., Mehta J. P., Collins F. S., Manolio T. A. (2009). Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc. Natl. Acad. Sci .USA 106, 9362–9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izawa N., Wu W., Sato K., Nishikawa H., Kato A., Boku N., Itoh F., Ohta T. (2011). HERC2 Interacts with Claspin and regulates DNA origin firing and replication fork progression. Cancer. Res. 71, 5621–5625. [DOI] [PubMed] [Google Scholar]
- Kenyon N. J., Morrissey B. M., Schivo M., Albertson T. E. (2012). Occupational asthma. Clin. Rev. Allergy Immunol. 43, 3–13. [DOI] [PubMed] [Google Scholar]
- Kim S. H., Cho B. Y., Park C. S., Shin E. S., Cho E. Y., Yang E. M., Kim C. W., Hong C. S., Lee J. E., Park H. S. (2009). Alpha-T-catenin (CTNNA3) gene was identified as a risk variant for toluene diisocyanate-induced asthma by genome-wide association analysis. Clin. Exp. Allergy 39, 203–212. [DOI] [PubMed] [Google Scholar]
- Kovacic M. B., Myers J. M., Wang N., Martin L. J., Lindsey M., Ericksen M. B., He H., Patterson T. L., Baye T. M., Torgerson D., Roth L. A., et al. (2011). Identification of KIF3A as a novel candidate gene for childhood asthma using RNA expression and population allelic frequencies differences. PLoS One 6, e23714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malo J. L., Ghezzo H., Elie R. (1999). Occupational asthma caused by isocyanates: patterns of asthmatic reactions to increasing day-to-day doses. Am. J. Respir. Crit. Care. Med. 159, 1879–1883. [DOI] [PubMed] [Google Scholar]
- Mapp C. E., Lucchini R. E., Miotto D., Chitano P., Jovine L., Saetta M., Maestrelli P., Springall D. R., Polak J., Fabbri L. M. (1998). Immunization and challenge with toluene diisocyanate decrease tachykinin and calcitonin gene-related peptide immunoreactivity in guinea pig central airways. Am. J. Respir. Crit. Care. Med. 158, 263–269. [DOI] [PubMed] [Google Scholar]
- Maurano M. T., Humbert R., Rynes E., Thurman R. E., Haugen E., Wang H., Reynolds A. P., Sandstrom R., Qu H., Brody J., et al. (2012). Systematic localization of common disease-associated variation in regulatory DNA. Science 337, 1190–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miglino N., Roth M., Tamm M., Borger P. (2012). Asthma and COPD - The C/EBP Connection. Open Respir. Med. J 6, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy A., Tantisira K. G., Soto-Quiros M. E., Avila L., Klanderman B. J., Lake S., Weiss S. T., Celedon J. C. (2009). PRKCA: a positional candidate gene for body mass index and asthma. Am. J. Hum. Genet. 85, 87–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NIOSH (1983). National Occupational Exposure Survey (NOES), 1981–1983. [Google Scholar]
- Park H. S., Cho S. H., Hong C. S., Kim Y. Y. (2002). Isocyanate-induced occupational asthma in far-east Asia: pathogenesis to prognosis. Clin. Exp. Allergy 32, 198–204. [DOI] [PubMed] [Google Scholar]
- Pasaje C. F., Kim J. H., Park B. L., Cheong H. S., Chun J. Y., Park T. J., Lee J. S., Kim Y., Bae J. S., Park J. S., et al. (2010). Association of SLC6A12 variants with aspirin-intolerant asthma in a Korean population. Ann. Hum. Genet. 74, 326–334. [DOI] [PubMed] [Google Scholar]
- Prahalad S., Ryan M. H., Shear E. S., Thompson S. D., Giannini E. H., Glass D. N. (2000). Juvenile rheumatoid arthritis: linkage to HLA demonstrated by allele sharing in affected sibpairs. Arthritis Rheum. 43, 2335–2338. [DOI] [PubMed] [Google Scholar]
- Redlich C. A., Karol M. H. (2002). Diisocyanate asthma: clinical aspects and immunopathogenesis. Int. Immunopharmacol. 2, 213–224. [DOI] [PubMed] [Google Scholar]
- Sastre J., Vandenplas O., Park H. S. (2003). Pathogenesis of occupational asthma. Eur. Respir. J 22, 364–373. [DOI] [PubMed] [Google Scholar]
- Scheerens H., Buckley T. L., Muis T., Van Loveren H., Nijkamp F. P. (1996). The involvement of sensory neuropeptides in toluene diisocyanate-induced tracheal hyperreactivity in the mouse airways. Br. J. Pharmacol. 119, 1665–1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt D., Wilson M. D., Ballester B., Schwalie P. C., Brown G. D., Marshall A., Kutter C., Watt S., Martinez-Jimenez C. P., Mackay S., et al. (2010). Five-vertebrate ChIP-seq reveals the evolutionary dynamics of transcription factor binding. Science 328, 1036–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stormo G. D. (1990). Consensus patterns in DNA. Methods. Enzymol. 183, 211–221. [DOI] [PubMed] [Google Scholar]
- Sun Y. (2006). E3 ubiquitin ligases as cancer targets and biomarkers. Neoplasia 8, 645–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visser M., Kayser M., Palstra R. J. (2012). HERC2 rs12913832 modulates human pigmentation by attenuating chromatin-loop formation between a long-range enhancer and the OCA2 promoter. Genome. Res. 22, 446–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Duncan D., Shi Z., Zhang B. (2013). WEB-based GEne SeT AnaLysis Toolkit (WebGestalt): update 2013. Nucleic Acids Res. 41, W77–W83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warde-Farley D., Donaldson S. L., Comes O., Zuberi K., Badrawi R., Chao P., Franz M., Grouios C., Kazi F., Lopes C. T., et al. (2010). The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 38, W214–W220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weirauch M. T., Yang A., Albu M., Cote A. G., Montenegro-Montero A., Drewe P., Najafabadi H. S., Lambert S. A., Mann I., Cook K., et al. (2014). Determination and inference of eukaryotic transcription factor sequence specificity. Cell 158, 1431–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams R., Zou X., Hoyle G. W. (2007). Tachykinin-1 receptor stimulates proinflammatory gene expression in lung epithelial cells through activation of NF-kappaB via a G(q)-dependent pathway. Am. J. Physiol. Lung Cell. Mol. Physiol. 292, L430–437. [DOI] [PubMed] [Google Scholar]
- Wisnewski A.V., Liu Q., Liu J., Redlich C.A. (2008). Human innate immune responses to hexamethylene diisocyanate (HDI) and HDI-albumin conjugates. Clin. Exp. Allergy, 38, 957–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C., Macleod I., Su A. I. (2013). BioGPS and MyGene.info: organizing online, gene-centric information. Nucleic Acids Res. 41, D561–D565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yucesoy B., Johnson V. J., Lummus Z. L., Kashon M. L., Rao M., Bannerman-Thompson H., Frye B., Wang W., Gautrin D., Cartier A., et al. (2014). Genetic variants in the major histocompatibility complex class i and class ii genes are associated with diisocyanate-induced asthma. J. Occup. Environ. Med. 56, 382–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yucesoy B., Johnson V. J., Lummus Z. L., Kissling G. E., Fluharty K., Gautrin D., Malo J. L., Cartier A., Boulet L. P., Sastre J., et al. (2012). Genetic variants in antioxidant genes are associated with diisocyanate-induced asthma. Toxicol. Sci. 129, 166–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S., Degan S., Potts E. N., Foster W. M., Sunday M. E. (2009). NPAS3 is a trachealess homolog critical for lung development and homeostasis. Proc. Natl. Acad. Sci. USA 106, 11691–11696. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.