Abstract
Alcoholism is a complex disease exhibiting a multifactorial mode of transmission. To simplify the genetic and phenotypic complexity of the alcoholic phenotype, alcohol-preferring (P) and -non-preferring (NP) rats were developed on the basis of alcohol preference and consumption as an animal model of alcoholism. Total gene expression analysis (TOGA) and quantitative trait loci (QTL) analysis were applied to selectively bred, inbred P and NP rats as complementary studies to identify genetic factors that contribute to alcohol preference and consumption. TOGA analysis was utilized to screen for differential expression in several brain regions involved in the mesocorticolimbic dopamine (DA) system. Genes exhibiting differences in expression were then screened for an association to the alcohol preference phenotype, the quantitative trait of a previously identified QTL. By evaluating differences in gene expression for linkage to a quantitative trait, this combined approach was implemented to identify α-synuclein, a candidate gene for alcohol preference.
Introduction
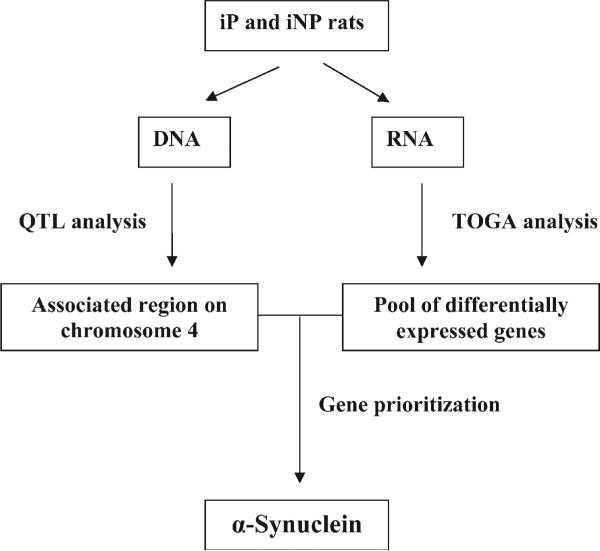
The identification of genetic components contributing to alcoholism is an important aspect of alcohol research. Knowing the genes and pathways that influence alcohol-seeking behaviour will advance the understanding of this complex disease by providing genetic markers that can be used to screen individuals who are predisposed to alcoholism and to develop new treatments to prevent excessive alcohol consumption. However, the genes contributing to alcoholism are difficult to pinpoint due to both the genetic complexity of alcoholism and environmental variability among alcoholics. While various techniques can be utilized to identify possible molecular and physiological patterns associated with alcoholism, localizing the primary genetic determinants of alcoholism still proves to be difficult. Combining selective breeding, quantitative trait locus (QTL) analysis and total gene expression analysis (TOGA) provides a systematic strategy that integrates differences in a specific gene's expression with chromosomal regions that are associated with alcoholism (Fig. 1). This combined approach dramatically increases the probability that a differentially expressed gene is contributing to a specific phenotype such as alcohol preference.
Figure 1.
Complementary experimental approaches. The flow chart depicts the incorporation of rats selectively bred for alcohol preference [inbred alcohol-preferring (iP) and -non-preferring (iNP)], quantitative trait loci analysis (QTL), and total gene expression analysis (TOGA) to identify a candidate gene(s) for alcohol preference (Sutcliff et al., 2000).
Alcoholism is a complex disease with a multifactorial mode of transmission. Multiple genes have been implicated in the predisposition to alcoholism, and the environmental component also plays a substantial role in the development of alcoholism (Devor and Cloniger, 1989). Evidence from twin, family and adoption studies suggests that the heritability of alcoholism ranges from 50 to 60% (Heath et al., 1997). Previous studies have utilized a multifaceted approach to study the genetic components that influence the predisposition to alcoholism, integrating various molecular and behavioural techniques (Murphy et al., 2002; Tabakoff et al., 2003).
Complementary approach to candidate gene identification
Selective breeding for alcohol preference
The identification of specific genes contributing to alcohol dependence is under way in various human studies; however, only a limited number of genes have been identified that predispose a person to alcoholism (Edenberg, 2002; Dick et al., 2004; Edenberg et al., 2004). Thus far, only the protective effects of the alcohol metabolizing enzymes have been replicated consistently (Thomasson et al., 1993; Foroud and Li, 1999). Both the heterogeneity of the human population and the complexity of the alcoholic phenotype complicate the identification of specific genetic markers that influence alcoholism. To limit the environmental component and simplify the genetic complexity of alcoholism, animal models have been developed to study various aspects of alcohol dependence, including alcohol-seeking behaviour (Tabakoff and Hoffman, 2000). These models are advantageous to the study of complex diseases because animal models display greater phenotypic homogeneity than the extremely complex phenotypes of human populations, and these specific pheno-types can be assessed readily using specific quantitative measures.
The alcohol-preferring (P) and nonpreferring (NP) rat lines were developed through bidirectional selective breeding from a randomly bred closed colony of Wistar rats on the basis of alcohol consumption and preference (Li et al., 1991). In these lines, P rats display the phenotypic characteristics considered necessary for an animal model of alcoholism (Cicero, 1979). Subsequently, inbred P (iP) and NP (iNP) strains have been established that maintain highly divergent alcohol consumption scores. Due to the physiological and genetic similarity between humans and this model, iP and iNP rats can be studied to identify important genetic factors that influence alcoholic predisposition in humans.
QTL analysis using iP/iNP animals
QTL analysis was implemented in the iP/iNP animals to identify chromosomal regions that are associated with alcohol preference and consumption, specific quantitative measures of alcoholism (Carr et al., 1998; Lander and Botstein, 1989). To conduct QTL analysis, reciprocal crosses of the iP and iNP animals were performed to produce F2 progeny that displayed quantitative trait measures at opposite extremes of the phenotypic distribution. The F2 population was then tested to identify QTLs or candidate regions that segregate with the phenotype. These regions are likely to harbour relevant genes affecting the alcohol preference phenotype (Lander et al., 1987; Lincoln et al., 1992).
QTL analysis of the iP/iNP F2 progeny resulted in the identification of a highly significant QTL on chromosome 4 with a maximum lod score of 9.2 and suggestive QTLs on chromosomes 3 and 8 (Bice et al., 1998; Carr et al., 1998).
The chromosome 4 QTL acts in an additive fashion and accounts for approximately 11% of the phenotypic variability. Due to the strong association between the chromosome 4 QTL region and alcohol preference, this region is likely to harbour a gene(s) that contribute(s) directly to alcohol preference. However, because this region is broad, the task of identifying a specific gene(s) contributing to this QTL remains difficult.
TOGA analysis using several brain regions implicated in alcohol preference
Total gene expression analysis (TOGA) was integrated with the QTL analysis to identify genes that are differentially expressed between the iP and iNP rats and map to the chromosome 4 QTL region. The TOGA approach is based on a variation of the 3′ Expressed Sequence Tag (EST) method in which the polymerase chain reaction (PCR) is used to amplify specific mRNA sequences. TOGA is a gene-profiling method that identifies and isolates both known and unknown mRNAs that are expressed differentially between two experimental samples. Using the TOGA method, both the presence and relative concentration of nearly every mRNA in an individual sample is determined.
The TOGA method provides several advantages for the identification of target genes. Because TOGA is based on PCR, only a small amount of RNA is necessary for the analysis (Lo et al., 2001). This increased efficiency permits an effective comparison using only small amounts of tissue. Because less than 1 μg of total RNA is generally required to obtain consistent results, TOGA increases the resolution of mRNA expression patterns by requiring less tissue and, thereby, permitting more specificity in dissection. In addition, the peaks generated using the TOGA method are predictable and can be easily isolated for cloning and sequence analysis. Therefore, combined with informatics, the TOGA method has been established as a powerful tool for research. However, like microarray analysis, the ability to localize a specific gene(s) influencing a phenotype from TOGA is difficult due to the large number of differentially expressed genes that are identified resulting from inherent physiological differences and gene – gene interactions. Even in inbred rats selected for alcohol preference, various phenotypic and physiological differences exist that are not directly related to alcohol preference (Smith et al., 1996).
A gene's expression profile is influenced largely by tissue selection and sample dissection. These experimental components can affect both the quality and relevance of expression data. The hypothalamus, the hippocampus, the caudateputamen, the nucleus accumbens and olfactory tubercles and the cortex were selected because these regions have been implicated in the mesocorticolimbic dopamine (DA) system. Previous studies suggest that these brain subregions interact reciprocally with the VTA to regulate alcohol-drinking behaviour (McBride and Li, 1998).
To carry out the TOGA analysis (Sutcliffe et al., 2000) in the iP and iNP animals, cytoplasmic RNA was isolated from four subregions microdissected from 10 iP and 10 iNP rats: (1) the hypothalamus, (2) the hippocampus, (3) the caudateputamen, the nucleus accumbens and olfactory tubercles and (4) the prefrontal, frontal and parietal cortices (Liang et al., 2003). Each set of samples was pooled to generate a representative sample containing the total RNA isolated from each respective subregion for both the iP and the iNP rats. cDNA was generated from these samples and digested with the restriction endonuclease MspI, and an oligonucleo-tide adapter was ligated to the 5′-end MspI overhang, integrating a start site for in vitro transcription within the 3′ fragments. Next, the 3′ fragments were amplified using T3 polymerase, and 256 primers were paired with a fluorescent 3′ primer to produce 256 nonoverlapping pools of products. These primers correspond to all of the possible permutations for the four nucleotides immediately adjacent to the MspI recognition site (N1N2N3N4). The PCR products were then separated using electrophoresis, and each product was assigned an eight-nucleotide sequence (derived from the four MspI recognition nucleotides and the adjacent four parsing nucleotides) and a length, attributes of the individual mRNAs.
The amplitudes of the fluorescent PCR products correspond to the initial concentrations of their parent mRNAs, and these amplitudes were collated automatically into a database and indexed. An electronic query was used to identify experimental samples that displayed different concentrations of mRNA. A differentially regulated Digital Sequence Tag (DST) was then verified by the ‘extended’ TOGA®™ method using a primer generated from the cloned PCR product. This PCR product, corresponding to a specific fragment length, was cloned, and a new 5′ PCR primer was built from the cloned DST. This primer and the universal 3′ PCR primer were utilized to produce a PCR product. The length of this product was compared to the original PCR product that was produced in the TOGA®™ reaction using mRNA extracted from the iP and iNP samples.
Coupling QTL and TOGA analyses to identify a candidate gene: α-synuclein
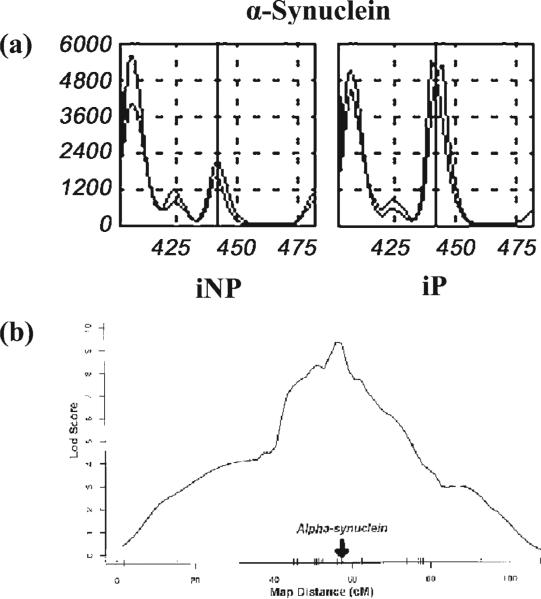
From the four discrete brain regions of iP and iNP rats, TOGA® detected more than 19 900 mRNAs. Twenty-eight of these genes displayed a difference greater than twofold and were evaluated further using quantitative real-time PCR to confirm the TOGA results. Confirmed genes were then screened for any relationship to the previously identified QTL regions (Bice et al., 1998) and for their relevance to alcohol metabolism and physiology. α-Synuclein was among the mRNAs identified by TOGA® as being expressed at higher levels in iP vs. iNP rat hippocampus (Liang et al., 2003). The differentially amplified peaks are illustrated in Fig. 2a.
Figure 2.
TOGA and QTL analysis yield a candidate gene: α-synuclein. (a) TOGA profile for the product corresponding to rat α-synuclein. α-synuclein was among the mRNAs identified by TOGA® as being expressed at higher levels in iP vs. iNP rat hippocampus (Liang et al., 2003). A vertical solid line is drawn through the PCR product corresponding to rat α-synuclein. (b) Multipoint lod score computed for alcohol preference with the program Mapmaker/QTL (Lander et al., 1987). α-synuclein mapped to the peak of this chromosome 4 QTL region (arrow) (Liang et al., 2003).
In addition to displaying differential expression between iP and iNP rats, α-synuclein mapped to the chromosome 4 QTL region (Fig. 2b). By comparing differentially expressed genes with the chromosomal 4 QTL region, these complementary experimental approaches provided a screen for detecting differentially expressed genes that are likely to be associated with the alcohol preference phenotype. The association of α-synuclein to alcohol preference dramatically increases the likelihood that its differential expression may be contributing to alcohol preference in iP and iNP rats.
The relationship between α-synuclein expression and dopaminergic neurotransmission has been established in previous literature, further supporting α-synuclein as a candidate gene for alcohol preference. α-Synuclein is expressed throughout the central nervous system and is particularly abundant in presynaptic nerve terminals (Maroteaux et al., 1988; Iwai et al., 1995; Mori et al., 2002). α-Synuclein may interact with tyrosine hydroxylase by inhibiting its activity and ultimately reducing dopamine synthesis (Perez et al., 2002). Furthermore, α-synuclein has been shown to decrease the activity of the dopamine transporter in cultured cells (Wersinger and Sidhu, 2003). α-Synuclein overexpression is lethal to dopaminergic neurones in human primary culture (Zhou et al., 2002). In addition, α-synuclein reactive inclusions were observed in the hippocampus, neocortex and substantia nigra of transgenic mice that exhibited degeneration of dopaminergic terminals (Masliah et al., 2000). Therefore, α-synuclein may be modulating dopaminergic neurotransmission in the mesolimbic pathway, a mechanism involved in both alcoholism and alcohol preference.
Dopaminergic projections from the VTA to the nucleus accumbens are implicated in the rewarding properties associated with drugs of abuse, including alcohol (Koob et al., 1998). When ethanol is administered an increase in dopamine release is observed in the nucleus accumbens of rats, while dopamine antagonists cause a reduction in the self-administration of ethanol (Pfeffer and Samson, 1988; Weiss et al., 1993). P rats display a deficiency in this dopaminergic pathway; P rats and high drinking iP X iNP F2 rats exhibit a 25 – 30% reduction in dopamine levels in important limbic structures (Murphy et al., 1987, 2002). Furthermore, a selective reduction in the number of dopaminergic projections has been documented from the VTA to the nucleus accumbens in P rats relative to NP rats (Zhou et al., 1995). Therefore, due to its involvement in the modulation of dopamine neurotransmission (Lee et al., 2002; Miranda et al., 2003), these previous studies support a possible mechanism whereby α-synuclein may influence alcohol-seeking behaviour.
These combined data support a role for α-synuclein in the modulation of alcohol preference between P and NP rats and define α-synuclein as a candidate gene of interest for alcohol-seeking behaviour. However, additional studies were necessary to characterize the molecular differences between αsynuclein in P and NP rats (Liang et al., 2003). Quantitative real-time PCR was implemented to confirm the TOGA analysis. Western blot analysis was also utilized to confirm that increased levels of α-synuclein mRNA translated into increased protein expression in the iP hippocampus and the caudate putamen. Comparative sequence analysis was conducted on α-synuclein cDNA between the iP and iNP lines. Two polymorphisms at + 439 and + 679 were identified in the 3′UTR and subsequently assessed for functionality using in vitro expression analysis. In vitro expression analysis indicated that the + 679 polymorphism reduced expression in iNP animals consistent with lower α-synuclein mRNA and protein levels detected in the iNP hippocampus. In addition, in situ hybridization was performed to localize α-synuclein expression in several brain regions. These additional data further support the initial prioritization of α-synuclein as a candidate gene for alcohol preference.
Further studies are necessary to determine α-synuclein's mechanism of action and solidify its role as a candidate gene for alcohol preference in iP and iNP rats. The development of transgenic and knock-out animals allows direct evaluation of a candidate gene's action on the phenotype of interest. In knockout mice, a deletion of α-synuclein has already been shown to decrease ethanol consumption (Miranda et al., 2003). The creation of a congenic line exhibiting the QTL region should be informative for both genetic and phenotypic analyses. These future studies should further characterize α-synuclein's role in alcohol preference in iP and iNP rats.
Summary
To expedite the search for novel candidate genes, QTL analysis can be applied to selectively bred animals to localize chromosomal regions harboring genes that contribute to a quantitative trait. QTLs create the foundation for further study by defining a finite chromosomal interval influencing a quantitative trait, thereby limiting the number of possible candidate genes. By employing detection methods such as TOGA, chromosomal regions can then be targeted systematically by screening these regions for differentially expressed genes. Thus, this complementary approach refines the search for candidate genes by integrating physiological differences (TOGA) with genomic associations (QTLs). In short, combining these analyses limits false positives that can result from random inheritance and gene – gene interactions.
The advancement of molecular techniques and bioinformatics has increased data generation and analysis exponentially. Applying these resources separately to a complex disease such as alcoholism often results in interesting, but specifically uninformative data. The combination of QTL and TOGA analyses provides a systematic approach to data analysis, greatly reducing the potential candidate genes of interest. Combining these analyses generated a specific gene of interest in α-synuclein from a broad QTL region and a multitude of differentially expressed genes.
Acknowledgments
This study was supported by US Public Service Grants AA10707, AA07611, AA00285 and Digital Gene Technologies.
References
- Bice P, Foroud T, Bo R, Castelluccio P, Lumeng L, Li TK, Carr LG. Genomic screen for QTLs underlying alcohol consumption in the P and NP rat lines. Mamm Genome. 1998;9:949–955. doi: 10.1007/s003359900905. [DOI] [PubMed] [Google Scholar]
- Carr LG, Foroud T, Bice P, Gobbett T, Ivashina J, Edenberg H, Lumeng L, Li TK. A quantitative trait locus for alcohol consumption in selectively bred rat lines. Alcohol Clin Exp Res. 1998;22:884–887. [PubMed] [Google Scholar]
- Cicero TJ. A critique of animal analogues of alcoholism. In: Majchrowicz E, Noble EP, editors. Biochemistry and pharmacology of ethanol. Vol. 2. Plenum Press; New York: 1979. pp. 533–560. [Google Scholar]
- Devor EJ, Cloninger CR. Genetics of alcoholism. Ann Rev Genet. 1989;23:19–36. doi: 10.1146/annurev.ge.23.120189.000315. [DOI] [PubMed] [Google Scholar]
- Dick DM, Edenberg HJ, Xuei X, Goate A, Kuperman S, Schuckit M, Crowe R, Smith TL, Porjesz B, Begleiter H, Foroud T. Association of GABRG3 with alcohol dependence. Alcohol Clin Exp Res. 2004;28:4–9. doi: 10.1097/01.ALC.0000108645.54345.98. [DOI] [PubMed] [Google Scholar]
- Edenberg HJ. The collaborative study on the genetics of alcoholism: an update. Alcohol Res Health. 2002;26:214–218. [PMC free article] [PubMed] [Google Scholar]
- Edenberg HJ, Dick DM, Xuei XL, Tian H, Almasy L, Bauer LO, Crowe RR, Goate A, Hesselbrock V, Jones K, Kwon J, Li TK, Nurnberger JI, O'Connor SJ, Reich T, Rice J, Schuckit MA, Porjesz B, Foroud T, Begleiter H. Variations in GABRA2, encoding the β2 subunit of the GABAA receptor, are associated with alcohol dependence and with brain oscillations. Am J Hum Genet. 2004;74:705–714. doi: 10.1086/383283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foroud T, Li TK. Genetics of alcoholism: a review of recent studies in human and animal models. Am J Addict. 1999;8:261–278. doi: 10.1080/105504999305677. [DOI] [PubMed] [Google Scholar]
- Heath AC, Bucholz KK, Madden PAF, Dinwiddie SH, Slutske WS, Statham DJ, Dunne MP, Whitfield J, Martin NG. Genetic and environmental contributions to alcohol dependence risk in a national twin sample: consistency of findings in women and men. Psychol Med. 1997;27:1381–1396. doi: 10.1017/s0033291797005643. [DOI] [PubMed] [Google Scholar]
- Iwai A, Masliah E, Yoshimoto M, Ge N, Flanagan L, de Silva HA, Kittel A, Saitoh T. The precursor protein of non-A beta component of Alzheimer's disease amyloid is a presynaptic protein of the central nervous system. Neuron. 1995;14:467–475. doi: 10.1016/0896-6273(95)90302-x. [DOI] [PubMed] [Google Scholar]
- Koob GF, Sanna PP, Bloom FE. Neuroscience of addiction. Neuron. 1998;21:467–476. doi: 10.1016/s0896-6273(00)80557-7. [DOI] [PubMed] [Google Scholar]
- Lander ES, Green P, Abrahamson J, Barlow A, Daly MJ, Lincoln SE, Newburg L. MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics. 1987;1:174–181. doi: 10.1016/0888-7543(87)90010-3. [DOI] [PubMed] [Google Scholar]
- Lander ES, Botstein D. Mapping mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics. 1989;121:185–199. doi: 10.1093/genetics/121.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MK, Stirling W, Xu Y, Xu X, Qui D, Mandir AS, Dawson TM, Copeland NG, Jenkins NA, Price DL. Human alpha-synuclein-harboring familial Parkinson's disease-linked Ala-53 > Thr mutation causes neurodegenerative disease with alpha-synuclein aggregation in transgenic mice. Proc Natl Acad Sci USA. 2002;99:8968–8973. doi: 10.1073/pnas.132197599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li TK, Lumeng L, Doolittle DP, Carr LG. Molecular associations of alcohol-seeking behaviour in rat lines selectively bred for high and low voluntary ethanol drinking. Alcohol Alcohol Suppl. 1991;1:121–124. [PubMed] [Google Scholar]
- Liang T, Spence J, Liu L, Strother WN, Chang HW, Ellison JA, Lumeng L, Li TK, Foroud T, Carr LG. alpha-Synuclein maps to a quantitative trait locus for alcohol preference and is differentially expressed in alcohol-preferring and -nonpreferring rats. Proc Natl Acad Sci USA. 2003;100:4690–4695. doi: 10.1073/pnas.0737182100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo D, Hilbush B, Sutcliffe JG. TOGA analysis of gene expression to accelerate target development. Eur J Pharm Sci. 2001;14:191–196. doi: 10.1016/s0928-0987(01)00174-9. [DOI] [PubMed] [Google Scholar]
- Maroteaux L, Campanelli JT, Scheller RH. Synuclein: a neuron-specific protein localized to the nucleus and presynaptic nerve terminal. J Neurosci. 1988;8:2804–2815. doi: 10.1523/JNEUROSCI.08-08-02804.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masliah E, Rockenstein E, Veinbergs I, Mallory M, Hashimoto M, Takeda A, Sagara Y, Sisk A, Mucke L. Dopaminergic loss and inclusion body formation in alpha-synuclein mice: implications for neurodegenerative disorders. Science. 2000;287:1265–1269. doi: 10.1126/science.287.5456.1265. [DOI] [PubMed] [Google Scholar]
- McBride WJ, Li TK. Animal models of alcoholism: neurobiology of high alcohol-drinking behaviour in rodents. Crit Rev Neurobiol. 1998;12:339–369. doi: 10.1615/critrevneurobiol.v12.i4.40. [DOI] [PubMed] [Google Scholar]
- Miranda C, Walker D, Alva H, Blednov YA, Harris RA. Deletion of alpha-synuclein decreases ethanol consumption in mice. Alcohol Clin Exp Res. 2003;27:84A. [Google Scholar]
- Mori F, Tanji K, Yoshimoto M, Takahashi H, Wakabayashi K. Immunohistochemical comparison of alpha- and beta-synuclein in adult rat central nervous system. Brain Res. 2002;941:118–126. doi: 10.1016/s0006-8993(02)02643-4. [DOI] [PubMed] [Google Scholar]
- Murphy JM, McBride WJ, Lumeng L, Li TK. Contents of monoamines in forebrain regions of alcohol-preferring (P) and -nonpreferring (NP) lines of rats. Pharmacol Biochem Behav. 1987;26:389–392. doi: 10.1016/0091-3057(87)90134-1. [DOI] [PubMed] [Google Scholar]
- Murphy JM, Stewart RB, Bell RL, Badia-Elder NE, Carr LG, McBride WJ, Lumeng L, Li TK. Phenotypic and genotypic characterization of the Indiana University rat lines selectively bred for high and low alcohol preference. Behav Genet. 2002;32:363–388. doi: 10.1023/a:1020266306135. [DOI] [PubMed] [Google Scholar]
- Perez RG, Waymire JC, Lin E, Liu JJ, Guo F, Zigmond MJ. A role for alpha-synuclein in the regulation of dopamine biosynthesis. J Neurosci. 2002;22:3090–3099. doi: 10.1523/JNEUROSCI.22-08-03090.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer AO, Samson HH. Haloperidol and apomorphine effects on ethanol reinforcement in free feeding rats. Pharmacol Biochem Behav. 1988;29:343–350. doi: 10.1016/0091-3057(88)90167-0. [DOI] [PubMed] [Google Scholar]
- Smith SV, Lumeng L, Read MS, Parise LV, Reddick RL, Sigman JL, Boudignon-Proudhon C, Smith JS, Li TK, Brinkhous KM. Characterization of a new hereditary thrombopathy in a closed colony of Wistar rats. J Lab Clin Med. 1996;128:601–611. doi: 10.1016/s0022-2143(96)90133-x. [DOI] [PubMed] [Google Scholar]
- Sutcliffe JG, Foye PE, Erlander MG, Hilbush BS, Bodzin LJ, Durham JT, Hasel KW. TOGA: an automated parsing technology for analyzing expression of nearly all genes. Proc Natl Acad Sci USA. 2000;97:1976–1981. doi: 10.1073/pnas.040537997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabakoff B, Hoffman PL. Animal models in alcohol research. Alcohol Res Health. 2000;24:77–84. [PMC free article] [PubMed] [Google Scholar]
- Tabakoff B, Bhave SV, Hoffman PL. Selective breeding, quantitative trait locus analysis, and gene arrays identify candidate genes for complex drug-related behaviours. J Neurosci. 2003;23:4491–4498. doi: 10.1523/JNEUROSCI.23-11-04491.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomasson HR, Crabb DW, Edenberg HJ, Li TK. Alcohol and aldehyde dehydrogenase polymorphisms and alcoholism. Behav Genet. 1993;23:131–136. doi: 10.1007/BF01067417. [DOI] [PubMed] [Google Scholar]
- Weiss F, Lorang MT, Bloom FE, Koob GF. Oral alcohol self-administration stimulates dopamine release in the rat nucleus accumbens: genetic and motivational determinants. J Pharmacol Exp Ther. 1993;267:250–258. [PubMed] [Google Scholar]
- Wersinger C, Sidhu A. Attenuation of dopamine transporter activity by alpha-synuclein. Neurosci Lett. 2003;340:189–192. doi: 10.1016/s0304-3940(03)00097-1. [DOI] [PubMed] [Google Scholar]
- Zhou FC, Zhang JK, Lumeng L, Li TK. Mesolimbic dopamine system in alcohol-preferring rats. Alcohol. 1995;12:403–412. doi: 10.1016/0741-8329(95)00010-o. [DOI] [PubMed] [Google Scholar]
- Zhou W, Schaack J, Zawada WM, Freed CR. Overexpression of human alpha-synuclein causes dopamine neuron death in primary human mesencephalic culture. Brain Res. 2002;926:42–50. doi: 10.1016/s0006-8993(01)03292-9. [DOI] [PubMed] [Google Scholar]