Abstract
In nuclear transgenic plants, expression of multiple genes requires introduction of individual genes and time-consuming subsequent backcrosses to reconstitute multi-subunit proteins or pathways, a problem that is compounded by variable expression levels. In order to accomplish expression of multiple genes in a single transformation event, we have introduced several genes into the chromoplast genome. We confirmed stable integration of the cry2Aa2 operon by PCR and Southern blot analyses in T0 and T1 transgenic plants. Foreign protein accumulated at 45.3% of the total soluble protein in mature leaves and remained stable even in old bleached leaves (46.1%), thereby increasing the efficacy and safety of transgenic plants throughout the growing season. This represents the highest level of foreign gene expression reported in transgenic plants to date. Insects that are normally difficult to control (10-day old cotton bollworm, beet armyworm) were killed 100% after consuming transgenic leaves. Electron micrographs showed the presence of the insecticidal protein folded into cuboidal crystals. Formation of crystals of foreign proteins (due to hyperexpression and folding by the putative chaperonin, ORF 2) provides a simple method of purification by centrifugation and enhances stability by protection from cellular proteases. Demonstration of expression of an operon in transgenic plants paves the way to engineering new pathways in plants in a single transformation event.
Keywords: polycistrons, plastid transformation, GM crops, Bt resistance
In plant and animal cells, the monocistronic translation of nuclear messenger RNAs (mRNAs) poses problems in engineering multiple genes in plants1. To express the polyhydroxybutyrate polymer or Guy’s 13 antibody, for example, single genes were first introduced into individual transgenic plants, then these plants were back-crossed to reconstitute the entire pathway or the complete protein2,3. Similarly, in a seven year long effort, Ye et al.4 recently introduced a set of three genes for a short biosynthetic pathway that resulted in β-carotene expression in rice. In contrast, most chloroplast genes of higher plants are co-transcribed1. Multiple steps of chloroplast mRNA processing are involved in the formation of mature mRNAs. Expression of polycistrons via the chloroplast genome provides a unique opportunity to express entire pathways in a single transformation event. Additionally, chloroplast genetic engineering is an environmentally friendly approach resulting in containment of foreign genes and hyperexpression5,6.
In this study, the Bacillus thuringiensis (Bt) cry2Aa2 operon is used as a model system to demonstrate operon expression and crystal formation via the chloroplast genome. The cry2Aa2 is the distal gene of a three-gene operon. The open reading frame (ORF) immediately upstream of cry2Aa2 codes for a putative chaperonin that facilitates the folding of Cry2Aa2 (and other proteins) to form proteolytically stable cuboidal crystals7–9. Because Cry protein levels decrease in plant tissues late in the growing season or under physiological stress10, a more stable protein expressed at high levels in the chloroplast throughout the growing season should increase toxicity of Bt transgenic plants to target insects and help eliminate the development of Bt resistance. Therefore, the cry2Aa2 bacterial operon is expressed in tobacco chloroplasts to test the resultant transgenic plants for increased expression and improved persistence of the accumulated insecticidal protein(s).
Results
Chloroplast vector
The 4.0 kb cry2Aa2 operon was inserted into the universal chloroplast expression vector pLD CtV2 (5.8 kb) to form the final Escherichia coli and tobacco shuttle vector pLD-BD Cry2Aa2 operon (9.8 kb) (Fig. 1A). This vector should be able to transform chloroplast genomes of several plant species because the flanking sequences are highly conserved among higher plants11,12. This vector contains the 16S ribosomal RNA (rRNA) promoter (Prrn) driving the aadA gene (aminoglycoside 3′-adenylyltransferase) for spectinomycin selection and the three genes of the cry2Aa2 operon. The terminator is the psbA 3′ region from the tobacco chloroplast genome from a gene coding for the photosystem II reaction center component. The 16S rRNA promoter is one of the strong chloroplast promoters recognized by both nuclear and plastid-encoded RNA polymerases in tobacco, and the psbA 3′ region stabilizes the transcript of foreign genes. This construct integrates both genes into the spacer region between the chloroplast transfer RNA genes coding for isoleucine and alanine within the inverted repeat (IR) region of the chloroplast genome by homologous recombination. The integration into these transcribed spacer regions allows the gene to be inserted without interfering with gene coding regions. Also, each genome will contain two gene copies as a result of integration into the two IR regions, resulting in a higher copy number (7,000–8,000 copies/cell) and higher levels of expression.
Figure 1.
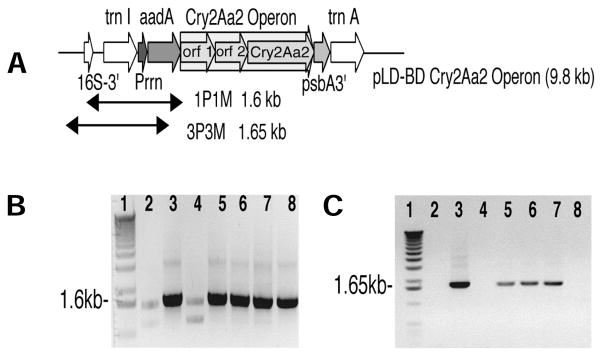
Chloroplast expression vector and PCR analysis. (A) pLD-BD Cry2Aa2 operon (9.8 kb) with PCR primer binding sites and expected fragment sizes. PCR analysis of untransformed and putative chloroplast transformants using two primer sets: (B) 1P1M and (C) 3P3M. Lane 1, 1 kb ladder; lane 2, untransformed; lanes 3–7, pLD-BD Cry2Aa2 operon putative transformants; lane 8, pLD-BD Cry2Aa2 operon plasmid DNA.
Chloroplast integration of foreign genes
Chloroplast transgenic plants were obtained as described13,14. Foreign gene integration into the chloroplast genome was determined by PCR screening of chloroplast transformants (Fig. 1 A–C). Primers were designed to eliminate spectinomycin mutants and nuclear integration. The first primer set, 1P1M, targets one primer (1P) to the 3′ end of the 16S rRNA flanking sequence and another primer (1M) to aadA (Fig. 1A). This is to distinguish between spectinomycin mutants and true spectinomycin transformants. A 1.6 kb fragment is seen in true transformants (Fig. 1B, lanes 3, 5–7). Lane 4 shows a spectinomycin mutant with no PCR product. Untransformed tobacco DNA (lane 2), as expected, shows no product, whereas pLD-BD Cry2Aa2 operon plasmid DNA in lane 8 produced the 1.6 kb fragment. The second primer set, 3P3M, targets one primer (3P) to the native chloroplast genome adjacent to the point of integration, and another primer (3M) on the aadA gene (Fig. 1A). This primer set generated a 1.65 kb PCR product in chloroplast transformants (Fig. 1C, lanes 3, 5–7). Untransformed tobacco DNA (lane 2) showed no PCR product, and pLD-BD Cry2Aa2 operon plasmid DNA in lane 8 also showed no PCR product because 3P binds to native chloroplast DNA. Lane 4 was negative for chloroplast integration, again proving this transformant to be a spectinomycin mutant.
Southern blot analysis was done to further demonstrate site-specific chloroplast integration of the 4.0 kb cry2Aa2 operon and to determine heteroplasmy or homoplasmy (Fig. 2). BglII digested DNA from transformed plants produce 8.42 and 1.4 kb fragments when probed with the 0.81 kb probe that hybridizes to the trnI and trnA flanking sequences. Transgenic plant DNA (T0 and T1) produced the 8.42 and 1.4 kb fragments (lanes 3–9). A 4.47 kb fragment is seen in untransformed plant DNA (lane 2). T0 plant DNA also shows this native untransformed 4.47 kb fragment (lanes 3–7), thereby showing heteroplasmy in the T0 generation. This 4.47 kb native band is absent from the T1 generation (lanes 8, 9), thus indicating homoplasmy. If only a fraction of the genomes were transformed, the gene copy number should be <8,000 per cell. Confirmation of homoplasmy in T1 transgenic lines indicates that the cry2Aa2 operon gene copy number could be as many as 7,000–8,000 per cell.
Figure 2.

Southern blot analysis of T0 and T1 generations. Lane 1, 1 kb ladder; lane 2, untransformed; lanes 3–7, T0 transgenic lines; lanes 8 and 9, T1 transgenic lines.
CRY2Aa2 protein expression and quantification
Expression profile of the operon-derived (OD) Cry2Aa2 and single gene-derived (SG) Cry2Aa2 (ref. 15) is shown on a Coomassie-stained sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) gel (Fig. 3). The primary goal of this experiment is to investigate the location of the operon-derived Cry2Aa2 protein (the pellet or supernatant) and correlate with cuboidal crystals observed in electron micrographs (see Fig. 6). Lane 2 contains partially purified 65 kDa Cry2Aa2 from E. coli. Because crystalline Cry2Aa2 inclusion bodies are solubilized at high alkaline pH (ref. 16), the 50 mM sodium hydroxide-solubilized pellet was analyzed from each plant sample after centrifugation for 20 min at 13,000 g (lanes 3, 5, 7). Results show that OD Cry2Aa2 expression forms crystalline inclusion bodies because the protein is found mostly in the pellet after centrifugation (lanes 5, 6). In contrast, expression of SG Cry2Aa2 is observed in both the pellet and the supernatant (lanes 3, 4). No Cry2Aa2 expression was seen in untransformed tobacco in either the supernatant or the pellet (lanes 7, 8).
Figure 3.

10% SDS–PAGE gel stained with R-250 Coomassie blue. Loaded protein concentrations are provided in parentheses. Lane 1, prestained protein standard; lane 2, partially purified Cry2Aa2 protein from E. coli (5 μg); lane 3, single gene-derived Cry2Aa2 pellet extract solubilized in 50 mM NaOH (22.4 μg); lane 4, single gene-derived Cry2Aa2 supernatant (66.5 μg); lane 5, operon-derived Cry2Aa2 pellet extract solubilized in 50 mM NaOH (22.9 μg); lane 6, operon-derived Cry2Aa2 supernatant (58.6 μg); lane 7, untransformed tobacco pellet extract solubilized in 50 mM NaOH (29.8 μg); lane 8, untransformed tobacco supernatant (30.4 μg). Colored compounds observed in the supernatant of transgenic plants interfered with the DC Bio-Rad protein assays.
Figure 6.
Transmission electron micrographs. Operon-derived Cry2Aa2 leaf sections in young (A), mature (B, D), and old, bleached leaf (C). (E) Single gene-derived Cry2Aa2 mature leaf; (F) mature untransformed leaf.
Cry 2Aa2 polypeptides (lanes 3, 5) were scanned using Storm 840 Gel Scanner and Image Quant Software (Molecular Dynamics, Sunnyvale, CA). The OD expression results only in a 2.5-fold greater accumulation of Cry2Aa2 than that of SG-derived Cry2Aa2 in the pellet fraction; this does not correlate with more than 100-fold difference observed in enzyme-linked immunosorbent assay (ELISA; Fig. 4). The reason for this discrepancy is the extreme difference in solubilization between SG Cry2Aa2-derived amorphous inclusion bodies and the OD Cry2Aa2-derived cuboidal crystals, as reported previously17,18. Despite the large difference in protein accumulation (as shown by ELISA and electron micrographs, Figs 4, 6), the concentration of solubilized protein loaded in the pellet fraction was similar in SG Cry2Aa2 and OD Cry2Aa2 (Fig. 3, lanes 3, 5). Attempts to completely solubilize crystalline inclusion bodies for SDS–PAGE analysis were not successful because higher pH interfered with gel electrophoresis and repeated dilution decreased protein concentration below detectable levels in Coomassie-stained gels.
Figure 4.
Protein quantification by ELISA in young, mature, and old transgenic leaves. (A) Single gene-derived Cry2Aa2 expression shown as a percentage of total soluble protein. (B) Operon-derived Cry2Aa2 expression shown as a percentage of total soluble protein.
However, for quantification using ELISA it was possible to completely solubilize crystalline inclusion bodies under optimal conditions and dilute the protein to fit within the linear range of the Cry2aA2 standard. Therefore, protein expression levels of SG Cry2Aa2 and OD Cry2Aa2 were quantified using ELISA (Fig. 4). Additionally, Cry protein accumulation in young, mature, and old transgenic leaves derived from a single gene or operon was compared to investigate their stability over time. Young, mature, and old leaves expressed SG Cry2Aa2 at 0.014%, 0.36%, and 0.03% respectively (Fig. 4A). Cry2Aa2 levels peaked in the mature leaf (0.36%) and drastically declined to 0.03% as the plant senesced. However, young, mature, and old leaves containing OD Cry2Aa2 accumulated at 34.9%, 45.3%, and 46.1%, respectively (Fig. 4B). As these transgenic plants aged, OD Cry2Aa2 concentrations remained stable and did not decline like the SG Cry2Aa2. The presence of the operon-expressed putative chaperonin should enable the toxin to be folded into stable crystalline structures that are protected from degradation. Based on quantitative expression, the cry2Aa2 operon-derived expression levels are comparable to those of the RuBisCo, the most abundant protein on earth that composes up to 65% of leaf soluble protein19.
Insect bioassays
Five-day-old tobacco budworm (Heliothis virescens), 10-day-old cotton bollworm (Helicoverpa zea), and beet armyworm (Spodoptera exigua) insects consumed the entire leaf after 24 h on the untransformed control (Fig. 5A, D, G). When feeding on SG Cry2Aa2 leaves, H. virescens insects died after five days (Fig. 5B) whereas insects died after three days on OD Cry2Aa2 leaves (Fig. 5C). For SG Cry2Aa2, H. zea insects had consumed considerable leaf material after 24 h, stopped feeding after three days, and died after five days (Fig. 5E). For OD Cry2Aa2, H. zea insects had consumed very little material after 24 h, stopped feeding, and died after five days (Fig. 5F). When feeding on SG Cry2Aa2 (Fig. 5H) or OD Cry2Aa2 (Fig. 5I), S. exigua were lethargic after 24 h and died after 48 h.
Figure 5.
Insect bioassays. (A, D, G) Untransformed tobacco leaves; (B, E, H) single gene-derived Cry2Aa2 transformed leaves; (C, F, I) operon-derived Cry2Aa2 transformed leaves. (A–C) Bioassays with Heliothis virescens; (D–F) bioassays with Helicoverpa zea; (G–I) bioassays with Spodoptera exigua. All leaf samples for each replicate were from the same leaf. Two samples were evaluated per treatment, and observed daily for mortality and leaf damage for five days. Treatments were replicated three times. Insects were tested at 5 or 10 days old (see text for details).
Milkweed leaves dusted with OD Cry Aa2 transgenic pollen were not toxic to monarch butterfly insects (data not shown), confirming earlier observations that foreign proteins are not present in tobacco pollen5.
Electron microscopic (EM) analysis
Untransformed and transgenic leaf sections were immunogold-labeled with a Cry2A polyclonal antibody (Fig. 6). Figure 6A–C shows developmental OD Cry2Aa2 in chloroplasts in young, mature, and old leaves, respectively. In a young green OD Cry2Aa2 transgenic leaf (Fig. 6A), labeled Cry2Aa2 occupies a significant amount of the chloroplast, but no crystalline structures are observed. In a mature green OD Cry2Aa2 transgenic leaf (Fig. 6B), labeled Cry2Aa2 occupies a larger amount of the chloroplast than the younger leaf, resulting in crystals. These cuboidal crystals are essentially identical to those expressed in wild-type Cry2Aa2 crystals, or recombinantly in Bt or E. coli18. In an old bleached OD Cry2Aa2 transgenic leaf (Fig. 6C), labeled Cry2Aa2 maintains the crystalline structure and occupies the highest volume of the chloroplast observed, despite being bleached and senescent. These findings correlate with OD Cry2Aa2 ELISA results. In young developing leaves, OD Cry2Aa2 begins accumulating (34.9%), folds Cry2Aa2 into a cuboidal configuration in mature leaves occupying more cell volume (45.3%), and maintains this cuboidal structure and volume in old leaves (46.1%). Essentially, as the transgenic OD Cry2Aa2 plant ages, OD Cry2Aa2 is accumulated, folded, and maintained.
Figure 6D is a mature green OD Cry2Aa2 transgenic leaf showing crystal formation with no immunogold label. This probably occurs because as the Cry2Aa2 is folded by the putative chaperonin, epitopes are concealed thereby decreasing labeling. Crystal formation in Figure 6D would cause the OD Cry2Aa2 to pellet after centrifugation as seen in SDS–PAGE (Fig. 3, lane 5). In EM analysis of mature leaves expressing SG Cry2Aa2 (Fig. 6E), protein aggregation is observed, although no crystalline folding is seen. Cry2Aa2 immunogold labeling occurs in an area of much lower density than is seen in OD Cry2Aa2 transgenic plants suggesting lower expression. These results also correlate with ELISA (0.36% in SG Cry2Aa2 in mature leaves). There is no localized antibody observed in untransformed tobacco (Fig. 6F).
Transgenic phenotypes
Phenotypes of OD Cry2Aa2 transgenic plants are not morphologically different from SG Cry2Aa2 transgenic plants (data not included). Therefore, higher levels of expression and accumulation of Cry proteins did not visibly influence their phenotype. Both transgenic plants flowered and set seeds. Characterization of OD Cry2Aa2 T1 transgenic plants for stable integration and transmission of foreign genes has been shown earlier (Fig. 2).
Discussion
Introducing blocks of foreign genes in a single operon would avoid complications inherent in putting one gene at a time into random locations in the nuclear genome1. Cloning several genes into a single T-DNA does not avoid the compounded variable expression problem encountered in nuclear transgenic plants1. This study shows that a bacterial operon can be expressed in a single integration event. Expression of multiple genes through a single transformation event opens the possibility of expressing foreign pathways or pharmaceutical proteins involving multiple genes. Also, formation of crystals of foreign proteins provides a simple method of purification by centrifugation. Plants transformed with the cry2Aa2 operon show a large accumulation and improved persistence of the expressed insecticidal protein(s) throughout the life of the plant. This is most likely because of the folding of the insecticidal protein into cuboidal crystals, thereby protecting it from proteases. The folded crystals may improve the safety of the Bt transgenic plants. In contrast to currently marketed transgenic plants that contain soluble Cry proteins, folded protoxin crystals will be processed only by those target insects that have a highly alkaline gut environment. For example, Bradley et al.17 have shown that there is a more than 30-fold concentration difference in the activity of soluble and crystalline Cry protein and that this difference was due to the host midgut alkaline environment. In addition, absence of insecticidal protein in transgenic pollen eliminates toxicity to nontarget insects through pollen. Expression of the cry2Aa2 operon in chloroplasts provides a model system for hyperexpression of foreign proteins in a folded configuration, which should enhance their stability and facilitate single-step purification.
Experimental protocol
Bombardment and selection of transgenic plants
Sterile leaves were bombarded using the Bio-Rad PDS-1000/He biolistic device as described13,14. Bombarded leaves were subjected to two rounds of selection on RMOP medium containing 500 μg/ml of spectinomycin to regenerate transformants.
PCR Analysis
DNA was extracted from leaves using the Qiagen DNeasy Plant Mini Kit (Qiagen, Valencia, CA). PCR was done using the Perkin Elmer Gene Amp PCR System 2400 (Perkin Elmer, Chicago, IL). All PCR reactions were performed using the Qiagen Taq DNA Polymerase Kit and with primers reported earlier11,12,15. Samples were run for 30 cycles with the following sequence: 94°C for 1 min, 65°C for 1.5 min, and 72°C for 3 min. PCR products were separated on 0.8% agarose gels.
Southern blot analysis
Plant DNA was digested with BglII and transferred to a nylon membrane by capillary action. The 0.81 kb probe was generated by digesting pLD-CtV2 vector DNA with BamH1/BglII and was labeled with 32P using the ProbeQuant G-50 Micro Columns (Amersham, Arlington Heights, IL). The probe was hybridized with the nylon membrane using the Stratagene QUICK-HYB hybridization solution and protocol (Stratagene, La Jolla, CA).
SDS–PAGE Analysis
Leaf material (600 mg) was ground to a powder in liquid nitrogen. Protein extraction buffer from the Cry2Aa2 plate kit from Envirologix (Portland, ME) used for quantification was added to the powder, and further grinding was done. The mixture was centrifuged at 4°C at 13,000 g for 20 min. The supernatant was removed, boiled in sample buffer, and loaded on a 10% SDS–PAGE gel. The pellet was resuspended in 50 mM NaOH and centrifuged at 4°C at 5,000 g for 5 min to pellet cell debris. The supernatant was removed, boiled in sample buffer, and loaded on a 10% SDS–PAGE gel at 200 V for 4 h. The DC protein assay by Bio-Rad (Hercules, CA) was used to determine total soluble and pellet protein concentration following the manufacturer’s protocol.
ELISA
A Cry2Aa2 plate kit from Envirologix was used for this experiment. Approximately 20 mg of leaf were ground in 100 μl of 50 mM NaOH to solubilize Cry proteins. Transgenic leaf extracts were diluted to fit in the linear range of the provided Cry2aA2 standard. The μQuant microtiter plate reader from Bio-Tek (Highland Park, VT) read the plate at 450 nm. A 1 p.p.m. Cry2Aa2 standard was supplied by the kit and was used in the linear range between 200 and 1000 ng for quantification. The DC protein assay by Bio-Rad was used to determine total soluble protein concentration following the manufacturer’s protocol.
Insect bioassays and transmission electron microscopy
Leaf disk bioassays were conducted as reported15. All insects were reared on typical lepidopteran artificial diet before use20, 21. Immunogold-labeled EM was carried out as described22. Sections were examined in a Zeiss EM 10 transmission electron microscope at 60 kV.
Acknowledgments
We thank F. Gould (North Carolina State University) for providing Heliothis virescens. This study was supported in part by the USDA-NRICGP grants 95-37500-2664, 97-35504-5297, and 98-35300-6973 to H.D.
References
- 1.Bogorad L. Engineering chloroplasts: an alternative site for foreign genes, proteins, reactions, and products. Trends Biotechnol. 2000;18:257–263. doi: 10.1016/s0167-7799(00)01444-x. [DOI] [PubMed] [Google Scholar]
- 2.Navrath C, Poirier Y, Somerville C. Targeting of the polyhydroxybutyrate biosynthetic pathway to the plastids of Arabidopsis thaliana results in high levels of polymer accumulation. Proc Natl Acad Sci USA. 1994;91:12760–12764. doi: 10.1073/pnas.91.26.12760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ma J, et al. Generation and assembly of secretory antibodies in plants. Science. 1995;268:716–719. doi: 10.1126/science.7732380. [DOI] [PubMed] [Google Scholar]
- 4.Ye X, et al. Engineering the provitamin A (β-carotene) biosynthetic pathway into (carotenoid-free) rice endosperm. Science. 2000;287:303–305. doi: 10.1126/science.287.5451.303. [DOI] [PubMed] [Google Scholar]
- 5.Daniell H. New tools for chloroplast genetic engineering. Nat Biotechnol. 1999;17:855–856. doi: 10.1038/12841. [DOI] [PubMed] [Google Scholar]
- 6.Daniell H. GM crops: public perception and scientific solutions. Trends Plant Sci. 1999;4:467–469. doi: 10.1016/s1360-1385(99)01503-4. [DOI] [PubMed] [Google Scholar]
- 7.Crickmore N, Ellar D. Involvement of a possible chaperonin in the efficient expression of a cloned cryIIA δ-endotoxin gene in Bacillus thuringiensis. Mol Microbiol. 1992;6:1533–1537. doi: 10.1111/j.1365-2958.1992.tb00874.x. [DOI] [PubMed] [Google Scholar]
- 8.Ge B, et al. Differential effects of helper proteins encoded by the cry2A and cry11A operons on the formation of Cry2A inclusions in Bacillus thuringiensis. FEMS Microbiol Lett. 1998;165:35–41. doi: 10.1111/j.1574-6968.1998.tb13124.x. [DOI] [PubMed] [Google Scholar]
- 9.Crickmore N, Wheeler V, Ellar D. Use of an operon fusion to induce expression and crystallisation of a Bacillus thuringiensis δ-endotoxin encoded by a cryptic gene. Mol Gen Genet. 1994;242:365–368. doi: 10.1007/BF00280428. [DOI] [PubMed] [Google Scholar]
- 10.Greenplate J. Quantification of Bacillus thuringiensis insect control protein Cry1Ac over time in bollgard cotton fruit and terminals. J Econ Entomol. 1999;92:1377–1383. [Google Scholar]
- 11.Daniell H, Datta R, Gray S, Varma S, Lee S. Containment of a herbicide resistance through genetic engineering of the chloroplast genome. Nat Biotechnol. 1998;16:345–348. doi: 10.1038/nbt0498-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guda C, Lee SB, Daniell H. Stable expression of a biodegradable protein-based polymer in tobacco chloroplasts. Plant Cell Rep. 2000;19:257–262. doi: 10.1007/s002990050008. [DOI] [PubMed] [Google Scholar]
- 13.Daniell H. Foreign gene expression in chloroplasts of higher plants mediated by tungsten particle bombardment. Methods Enzymol. 1993;217:536–556. doi: 10.1016/0076-6879(93)17088-m. [DOI] [PubMed] [Google Scholar]
- 14.Daniell H. Transformation and foreign gene expression in plants mediated by microprojectile bombardment. Methods Mol Biol. 1997;62:463–489. doi: 10.1385/0-89603-480-1:463. [DOI] [PubMed] [Google Scholar]
- 15.Kota M, et al. Overexpression of the Bacillus thuringiensis (Bt) Cry2Aa2 protein in chloroplasts confers resistance to plants against susceptible and Bt-resistant insects. Proc Natl Acad Sci USA. 1999;96:1840–1845. doi: 10.1073/pnas.96.5.1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yamamoto T, Iizuka T. Two types of entomocidal toxins in the parasporal crystals of Bacillus thuringiensis var. kurstaki. Arch Biochem Biophys. 1983;227:233–241. doi: 10.1016/0003-9861(83)90366-1. [DOI] [PubMed] [Google Scholar]
- 17.Bradley D, Harkey M, Kim M, Biever K, Bauer L. The insecticidal CryIB crystal protein of Bacillus thuringiensis ssp thuringiensis has dual specificity to coleopteran and lepidopteran larvae. J Invert Pathol. 1995;65:162–173. doi: 10.1006/jipa.1995.1024. [DOI] [PubMed] [Google Scholar]
- 18.Daniell H, Dessai P, Prakash C, Moar W. Engineering plants for stress tolerance via organelle genomes. NATO ASI Series. 1994;86:589–592. [Google Scholar]
- 19.Roy H, Nierzwicki-Bauer S. RuBisCo: genes, structure, assembly and evolution. In: Bogorad L, Vasil I, editors. The photosynthetic apparatus. Academic Press; New York: 1994. pp. 347–364. [Google Scholar]
- 20.Moar W, Trumble J, Hice R, Backman P. Insecticidal activity of the CryIIA protein from the NRD-12 isolate of Bacillus thuringiensis subsp kurstaki expressed in Escherichia coli and Bacillus thuringiensis and in a leaf-colonizing strain of Bacillus cereus. Appl Environ Microbiol. 1994;60:896–902. doi: 10.1128/aem.60.3.896-902.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moar W, et al. Development of Bacillus thuringiensis Cry1C resistance by Spodoptera exigua. Appl Environ Microbiol. 1995;61:2086–2092. doi: 10.1128/aem.61.6.2086-2092.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vrekleij AJ, Leunissen JM, editors. Immuno-gold labeling in cell biology. CRC Press; Boca Raton, FL: 1989. [Google Scholar]





