Abstract
Target-mediated toxicity is a major limitation in the development of chimeric antigen T cell receptors (CAR) for adoptive cell therapy of solid tumors. In this study, we developed a strategy to adjust the affinities of the scFv component of CAR to discriminate tumors overexpressing the target from normal tissues which express it at physiologic levels. A CAR-expressing T cell panel was generated with target antigen affinities varying over three orders of magnitude. High-affinity cells recognized target expressed at any level, including at levels in normal cells that were undetectable by flow cytometry. Affinity-tuned cells exhibited robust antitumor efficacy similar to high-affinity cells, but spared normal cells expressing physiologic target levels. The use of affinity-tuned scFvs offers a strategy to empower wider use of CAR T cells against validated targets widely overexpressed on solid tumors, including those considered undruggable by this approach.
Introduction
Adoptive immunotherapy with CAR engineered T (CART) cells can target and kill malignant cells, thereby inducing durable clinical responses in hematopoietic malignancies (1–3). However, many commonly targeted tumor antigens are also expressed by healthy tissues and on-target, off-tumor toxicity from T cell–mediated destruction of normal tissue has limited the development of this otherwise promising type of cancer therapy. Recent reports on severe adverse events associated with treatment of cancer patients with CAR- or TCR-engineered T lymphocytes further illustrate the critical importance of target selection for safe and efficient therapy (4–7). In specific, the targeting of ErbB2 (Her2/neu or CD340) with high affinity CARTs led to serious toxicity due to target recognition on normal cardiopulmonary tissue (8), and similarly, the presence of relatively high levels of EGFR in healthy skin leads to dose-limiting skin toxicity (9).
Selecting highly tissue-restricted antigens, cancer testis antigens, mutated gene products or viral proteins as targets could significantly improve the safety profile of using CART cells. However, none of these antigens is present with high frequency in common cancers. Most of the top-ranked target antigens that could be targeted by CART are expressed in potentially important normal tissues, such as ErbB2, EGFR, MUC1, PSMA, and GD2 (10). Current strategies for generating CARs consist of selecting scFvs with high affinity, as previous studies have shown that the activation threshold is inversely correlated with the affinity of the scFv (11, 12). However, it was found that after TCR stimulation there is a narrow window of affinity for optimal T cell activation, and increasing the affinity of the TCR does not necessarily improve treatment efficacy (13, 14).
Here we have tested the hypothesis that equipping T cells with high affinity scFv may limit the utility of CARs, due to poor discrimination of the CART for tumors and normal tissues that express the same antigen at lower levels. We sought to determine if fine-tuning the affinity of the scFv could increase the ability of CART cells to discriminate tumors from normal tissues expressing the same antigen at lower levels. In this study, CARs with affinities against two validated targets, ErbB2 and EGFR, which are amplified or overexpressed in variety of cancers but are also expressed, at lower levels by normal tissues were tested against multiple tumor lines, as well as primary cell lines from normal tissues and organs. We found that decreasing the affinity of the scFv could significantly increase the therapeutic index of CARs while maintaining robust antitumor efficacy both in vitro and in xenogeneic mouse tumor models.
Materials and Methods
Cell lines and primary human lymphocytes
SK-BR3, SK-OV3, BT-474, MCF7, MDA231, MDA468, HCC2281, MDA-361, MDA-453, HCC-1419, HCC-1569, UACC-812, LnCap, MDA-175, MCF-10A, HCC38 and HG261 cell lines were purchased from American Type Culture Collection and cultured as instructed. Primary cell lines (keratinocytes, osteoblast, renal epithelial, pulmonary artery endothelial cells, pulmonary artery smooth muscle, neural progenitor, CD34+ enriched PBMC) were obtained from Promocell and cultured according to their protocols. Primary lymphocytes were isolated from normal donors provided by the University of Pennsylvania Human Immunology Core and cultured in R10 medium (RPMI 1640 supplemented with 10% fetal calf serum; Invitrogen). Primary lymphocytes were stimulated with microbeads coated with CD3 and CD28 stimulatory antibodies (Life Technologies, Grand Island, NY, Catalog) as described (15). T cells were cryopreserved at day 10 in a solution of 90% fetal calf serum and 10% dimethylsulfoxide (DMSO) at 1 × 108 cells/vial.
Generation of CAR constructs for mRNA electroporation and lentiviral transduction
CAR scFv domains against ErbB2 or EGFR were synthesized and/or amplified by PCR, based on sequencing information provided by the relevant publications (16, 17), linked to CD8 transmembrane domain and 4-1BB and CD3 zeta intracellular signaling domains, and subcloned into pGEM.64A RNA based vector (18) or pTRPE lentiviral vectors (19).
mRNA in vitro transcription and T cell electroporation
T7 mscript systems kit (CellScript) was used to generate in vitro transcription (IVT) RNA. CD3/CD28 bead stimulated T cells were electroporated with IVT RNA using BTX EM830 (Harvard Apparatus BTX) as previously described (18).
Flow cytometry analysis
Antibodies were obtained from the following suppliers: anti-human CD3 (BD Biosciences, 555335), anti-human CD8 (BD Biosciences 555366), anti-human CD107a (BD Biosciences 555801), anti-human CD137 (BD Biosciences 555956). Cell surface expression of ErbB2 was detected by biotylated anti-ErbB2 Affibody (Abcam, ab31890), and EGFR by FITC conjugated anti-EGFR affibody (Abcam, ab81872). EGFR, ErbB2 and CD19 specific CAR expression were detected by biotin-labeled polyclonal anti-human F(ab)2 antibody for (EGFR CAR) or anti-mouse F(ab)2 antibody (for ErbB2 and CD19 CARs)(Jackson Immunoresearch). Samples were then stained with PE-conjugated anti-human IgG Fc Ab (eBioscience, 12-4998-82) or phycoerythrin-labeled streptavidin (eBioscience, 17-4317-82). Flow cytometry acquisition was performed on either a BD FacsCalibur or Accuri C6 Cytometer (BD Biosciences). Analysis was performed using FlowJo software (Treestar).
ELISA assays
Target cells were washed and suspended at 1× 106 cells/ml in R10 medium. 100ul each target cell type were added in triplicate to a 96 well round bottom plate (Corning). Effector T cells were washed, and re-suspended at 1× 106 cells/ml in R10 medium and then 100μl of T cells were combined with target cells in the indicated wells. The plates were incubated at 37°C for 18 to 24 hours. After the incubation, supernatant was harvested and subjected to an ELISA assay (eBioscience).
CD107a staining
Cells were plated at an E:T of 1:2 (1 × 105 effectors: 2 × 105 targets) in 160 μl of R10 medium in a 96 well plate. 20μl of phycoerythrin-labeled anti-CD107a Ab was added and the plate was incubated at 37°C for 1 hour before adding Golgi Stop (2ul Golgi Stop in 3ml R10 medium, 20ul/well; BD Biosciences, 51-2092KZ ) and incubating for another 2.5 hours. Then 5 μl FITC-anti-CD8 and 5ul APC-anti-CD3 was added and incubated at 37°C for 30 min. After incubation, the samples were washed with FACS buffer and analyzed by flow cytometry.
CFSE based T cells proliferation assay
Resting CD4 T cells were washed and suspend at concentration of 1 × 107 cells/ml in PBS. Then 120 ul CFSE working solution (25 μM CFSE) was added to 1 × 107 cells for 3.5 min at 25°C. The labeling was stopped with 5% FBS (in PBS), washed twice with 5% FBS and cultured in R10 with 10 IU/ml IL2. After overnight culture, the CFSE labeled T cells were electroporated with indicated CAR RNA. Two to four hours after electroporation, T cells were suspended at concentration of 1 × 106/ml in R10 medium (with 10 IU/ml IL2). Tumor or K562 cell lines were irradiated and suspended at 1 × 106/mL in R10 medium. Cells were plated at an E:T of 1:1 (5 × 105 effectors: 5 × 105 targets) in 1ml of complete RPMI medium in a 48 well plate. T cells were then counted and fed every 2 days from day 3. CFSE dilution was monitored by flow cytometry at day 3, day 5 and day 7.
Luciferase based CTL assay
Nalm6-CBG tumor cells were generated and employed in a modified version of a luciferase based CTL assay(20). Briefly, Click beetle green luciferase (CBG)-T2A-eGFP was lentivirally transduced into Nalm6 tumor cells and sorted for GFP expression. Resulting Nalm6-CBG cells were resuspended at 1 × 105 cells/ml in R10 medium and incubated with different ratios of T cells (e.g. 30:1, 15:1, etc.) overnight at 37°C. 100μl of the mixture was transferred to a 96 well white luminometer plate, 100ul of substrate was added and the luminescence was immediately determined. Results are reported as percent killing based on luciferase activity in wells with tumor, but no T cells. (% killing = 100-((RLU from well with effector and target cell coculture)/(RLU from well with target cells)×100)).
Mouse xenograft studies
Studies were performed as previously described with certain modifications (15, 19). Briefly, 6–10 week old NOD-SCID-γ −/− (NSG) mice were injected subcutaneously with 1 × 106 PC3-CBG tumors cells on the right flank at day 0 and the same mice were given SK-OV3-CBG tumor cells (5 × 106 cells/mouse, s.c.) on the left flank at day 5. The mice were treated with T cells via the tail vein at day 23 post PC3-CBG tumor inoculation such that both tumors were approximately 200 mm3 in volume. Lentivirally transduced T cells were given as indicated.
Results
Lowering the affinity of the anti-ErbB2 scFv improves the therapeutic index of ErbB2 CART cells in vitro
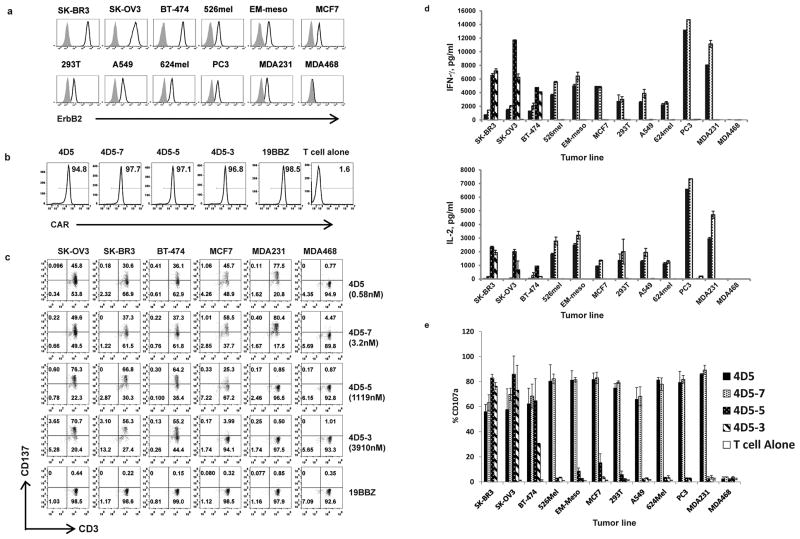
We compiled a panel of tumor lines with a wide range of ErbB2 expression as measured by flow cytometry (Fig. 1a). SK-OV3 (ovarian cancer), SK-BR3 (breast cancer), BT-474 (breast cancer) over-express ErbB2, while EM-Meso (mesothelioma), MCF7 (breast cancer), 293T (embryonic kidney 293 cell), A549 (lung cancer), 624mel (melanoma), PC3 (prostate cancer), MDA231 (breast cancer) express ErbB2 at lower levels and ErbB2 was not detected in MDA468 (breast cancer). ErbB2 mRNA levels were also measured by real time PCR and there was a strong correlation between the two techniques (Fig. S1).
Figure 1. Characterization of ErbB2 affinity-tuned CART cells and tumor cell lines.
(a) Detection of ErbB2 surface expression on tumors and cell lines. Cells were stained with anti-ErbB2 Affibody-biotin and detected with streptavidin-allophycocyanin (APC) (open histograms); cells incubated with APC alone indicate background (grey filled histograms). (b) FACS analysis of affinity-tuned CAR expression in mRNA electroporated T cells. T cells were electroporated with indicated CAR mRNA and one day after the electroporation, the CAR expression was detected using an anti-mouse IgG Fab antibody (for both CD19.BBZ and ErbB2.BBZ CARs). T cells without electroporation were used as a negative control. (c) The induction of CD137 (4-1BB) expression on CART cells after stimulation by tumor cells was measured. One day after electroporation the various CART cells (KD, nM) were co-cultured with the indicated tumor cell lines. CD137 and CD3 expression was measured after 24 hr (CD3+ gated). (d) Cytokine secretion was measured (ELISA) in culture supernatants. T cells were electroporated with affinity-tuned ErbB2 CAR mRNA as indicated. One day after the electroporation, the CART cells were co-cultured with indicated tumor cell lines for 24h. Bar charts show results from a representative experiment (values represent the average ± SD of triplicates) for IFN-gamma (upper panel) and IL-2 (lower panel). (e) CD107a up-regulation on CART cells stimulated by tumors. T cells were electroporated with ErbB2 CAR mRNAs encoding the indicated scFv and one day later the CART cells were co-cultured with the indicated cell line for 4 hr. CD107a expression CD3+ T cells was measured (values represent the average ± SD of 4 similar independent experiments).
A panel of ErbB2 CARs was constructed making use of scFvs derived from the published mutations of 4D5-8 antibody (4D5) (16). The monovalent affinities of the ErbB2 scFvs varied by approximately 3 orders of magnitude (Table S1), in contrast to the corresponding mutant antibodies that retained binding affinities within 10-fold of each other (16). CARs were constructed by linking the various scFvs to the CD8 alpha hinge and transmembrane domain followed by the 4-1BB and CD3 zeta intracellular signaling domains. The CARs were expressed by lentiviral vector technology or by cloning into an RNA-based vector (18). After production of mRNA by in vitro transcription and electroporation into T cells, the surface expression of the panel of affinity-modified ErbB2 RNA CARs was similar (Fig. 1b). To compare recognition thresholds, the panel of ErbB2 CART cells was stimulated with ErbB2 high expressing (SK-BR3, SK-OV3 and BT-474) or low expressing tumor cell lines (MCF7, 293T, A549, 624Mel, PC3, MDA231 and MDA468) and T cell activation was assessed by upregulation of CD137 (4-1BB; Fig. 1c), secretion of IFN-γ and IL-2 (Fig. 1d) and induction of surface CD107a expression (Fig. 1e); the data in Fig. S2 shows a representative experiment. T cells expressing a CD19-specific CAR or untransfected T cells served as control for allogeneic reactivity. Lower affinity CART cells (4D5-5 and 4D5-3) were strongly reactive to tumors with amplified ErbB2 expression and exhibited undetectable or low reactivity to the tumor lines that expressed ErbB2 at lower levels. In contrast, higher affinity CART cells (4D5 and 4D5-7) showed strong reactivity to tumor lines expressing high and low levels of ErbB2. These results were extended by assaying additional ErbB2-expressing cell lines (Fig. S3). Interestingly, higher affinity CART cells secreted greater levels of IFN-γ and IL-2 when exposed to targets expressing low levels of ErbB2, while lower affinity CART cells secreted more cytokines when exposed to cells expressing high levels of target (Fig. 1d). As expected, the CD19.BBZ CAR was not reactive against ErbB2-expressing cell lines. In summary, higher affinity 4D5.BBZ or 4D5-7.BBZT cells recognized all the ErbB2 expressing lines tested, whereas CARs with lower affinity scFvs, 4D5-5-BBζ or 4D5-3-BBζ, were highly reactive to all tumor lines with overexpressed ErbB2, but displayed negligible reactivity to cell lines expressing low or undetectable levels of ErbB2.
ErbB2 CARs with lower affinity scFvs discriminate between tumor cells expressing low and high levels of ErbB2
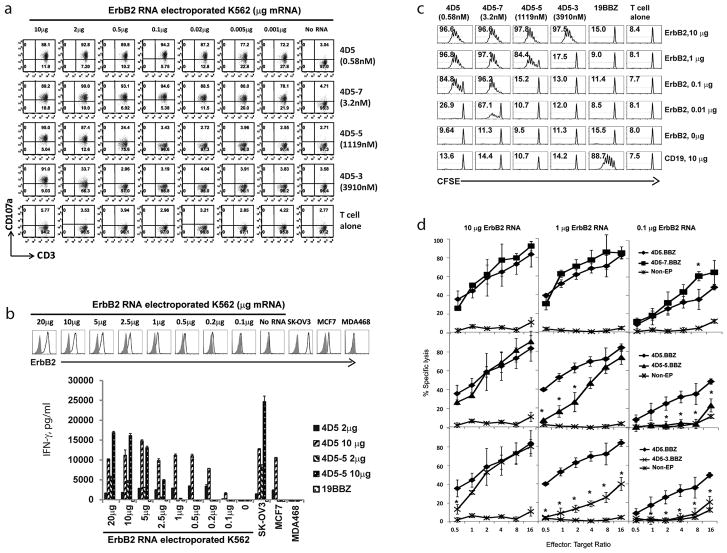
To exclude any tumor-specific effects that might contribute to the above results, we assayed the activity of the panel of ErbB2.BBZ CART cells against a single tumor line expressing varying levels of ErbB2. We observed that T cells expressing higher affinity scFvs (4D5 and 4D5-7) recognized K562 cells electroporated with ErbB2 RNA at doses as low as 0.001μg, which is 100-fold lower than the flow cytometrically detectable level of 0.1μg mRNA (Fig. 2a, b). In contrast the CARs with lower affinity scFvs (4D5-5 and 4D5-3) only recognized K562 electroporated ErbB2 RNA at doses of 0.5 μg (4D5-5; Fig. 2a) or higher, indicating that CART cell sensitivity was decreased by 500- (4D5-5) to 2000- fold (4D5-3) compared to the high affinity 4D5 CART cells. Moreover, the antigen dose associated reactivity observed with lower affinity ErbB2 CARs (4D5-5 and 4D5-3; Fig. 2a,b), was confirmed by performing a CFSE-based proliferation assay (Fig. 2c). Interestingly, decreasing the CAR RNA dose 5 fold (from 10μg RNA/100μl T cells to 2μg RNA/100 μl T cells), further increased the antigen recognition threshold of the T cells with lower affinity CARs as assessed by cytokine secretion (Fig. 2b), suggesting that fine tuning of CAR density on the surface of the T cells is also an important variable.
Figure 2. Control of ErbB2 target recognition density by scFv affinity tuning.
(a) K562 cells were electroporated with indicated amounts of ErbB2 mRNA and CART cells expressing the indicated scFv (KD, nM) were co-cultured with target for 4h and the percentage CD107a expression was quantified on CD3+ cells. (b) IFN-gamma secretion by the panel of ErbB2 CART cells stimulated by ErbB2 mRNA electroporated K562 cells. K562 cells were electroporated with the indicated amount of ErbB2 mRNA (upper panel). T cells were electroporated with 2ug or 10ug ErbB2 CAR mRNA as indicated. CART cells were co-cultured with indicted K562 targets and IFN-gamma secretion was measured by ELISA after 24hrs (lower panel). (c) Proliferation of the panel of affinity-tuned CART cells after stimulation by ErbB2 mRNA electroporated K562 cells. Resting CD4 T cells were labeled with CFSE and electroporated with 10ug CAR mRNA. K562 cells were electroporated with the indicated amount of ErbB2 mRNA or control CD19 mRNA. The T cells and irradiated targets were cultured (1:1 ratio) for 7 days and CFSE dilution was measured by flow cytometry (CD3 gated); the % divided T cells is shown. (d) Cytolytic activity of affinity-tuned CART cells, using non-electroporated T cells (Non-EP) as control, against Nalm6-CBG ErbB2-expressing target cells was measured. T cells were electroporated with ErbB2 CAR mRNA as indicated. Nalm6-CBG (click beetle green) target cells were electroporated with ErbB2 mRNA at the indicated dose. One day after the electroporation, the CART cells were co-cultured with Nalm6-CBG cells at indicated E:T ratio and % specific lysis calculated after 8hr. All groups were compared with 4D5.BBZ. The results shown here are the averages of three independent experiments (*, P<0.05, Student t test).
We used a luciferase based cytolytic T cell (CTL) assay to determine whether T cells with affinity decreased CARs could maintain potent killing activity against ErbB2 over expressing targets while sparing cells expressing lower ErbB2 levels. When Nalm6 target cells were transfected with 10μg ErbB2 RNA, T cells with either higher or lower affinity ErbB2 CARs effectively lysed target cells. CARs with higher affinity scFvs (4D5 and 4D5-7) exhibit potent lytic activity against target cells transfected with 1μg ErbB2 RNA, but lower affinity scFvs (4D5-5 and 4D5-3) showed decreased killing activity. Finally, only CARs with higher affinity scFvs were able to kill target cells expressing very low amounts of target after electroporation with 0.1 μg ErbB2 RNA (Fig. 2d). These data support that fine-tuning the affinity of ErbB2 CART cells enhances discrimination of ErbB2 over-expressing tumor from cells that have low or undetectable levels of ErbB2 expression.
Affinity decreased ErbB2 CART cells fail to recognize physiologic levels of ErbB2
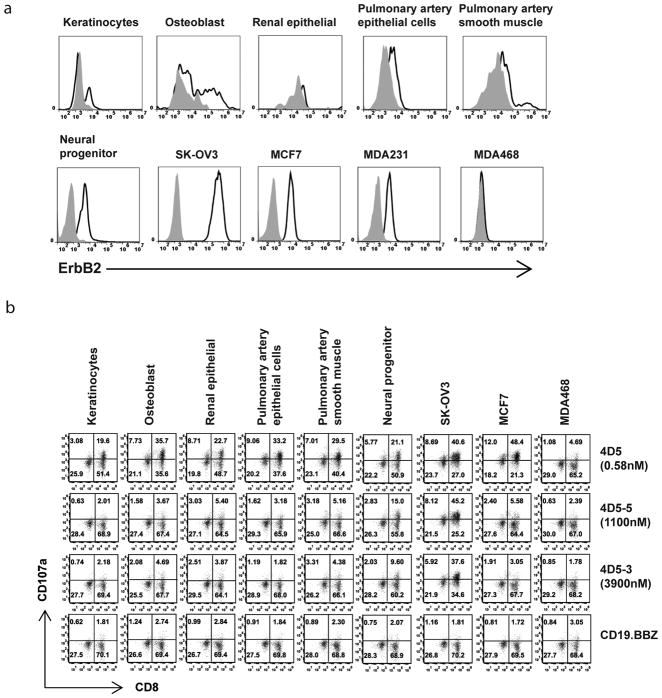
Given the previous serious adverse event which occurred upon administration of the high affinity ErbB2 CAR that incorporated the scFv from the parental 4D5 trastuzumab antibody (8), it is of paramount importance to evaluate potential reactivity of the reduced affinity ErbB2 CART cells to physiologic levels of ErbB2 expression. To address this, six primary cell lines isolated from different organs were tested for ErbB2 expression. Most of the primary lines had detectable levels of surface ErbB2, with the neural progenitor line expressing the highest levels of target (Fig. 3a). T cells expressing the high affinity 4D5 CAR were strongly reactive to all primary lines tested, as evidenced by levels of CD107a up-regulation (Fig. 3b). However, T cells expressing the affinity decreased ErbB2 CARs 4D5-5 and 4D5-3 exhibited no reactivity to the primary lines with the exception of weak reactivity to the neural progenitor line.
Figure 3. Selective targeting of ErbB2 on primary cell lines.
(a) Cell lines were stained using anti-ErBb2 Affibody-biotin and detected using streptavidin-allophycocyanin (APC) (open histograms); cells stained with APC only were used as control (grey filled histograms). (b) The panel of CART cells was stimulated with the indicated cell line for 4hr and the % CART cells expressing CD107a was measured by gating on CD3+ cells.
Comparable effects with affinity-tuned ErbB2 CARs expressed using lentiviral transduction or RNA electroporation
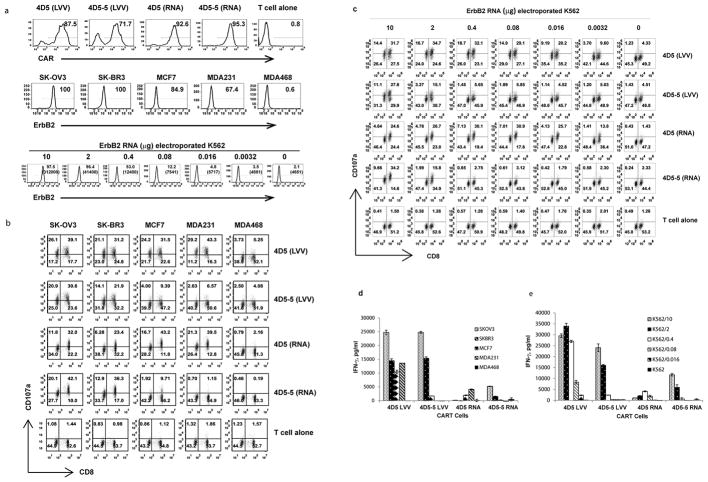
To establish comparability between T cells permanently expressing CARs by lentiviral transduction with mRNA electroporated CART cells, the panel of affinity-tuned CARs was expressed in T cells from the same normal donor using either lentiviral transduction or mRNA electroporation (Fig 4a, upper panel). T cells were stimulated with tumor cell lines (Fig. 4a, middle panel), or K562 cells, expressing varying amounts of ErbB2 (Fig.4a, lower panel). CART cell recognition and activation were monitored by CD107a upregulation (Fig. 4b and c), CD137 upregulation (Fig. S4) and IFN-γ secretion (Fig. 4d and e). In agreement with the previous ErbB2 mRNA CART cell results, T cells that constitutively expressed high affinity CARs showed strong reactivity to all cell lines expressing ErbB2; no correlation was observed between antigen expression levels and T cell activity. In contrast, T cells with low affinity CARs expressed by lentiviral technology demonstrated a robust correlation between target antigen expression and activation (Fig. 4b–e), similar to RNA CART cells. These results confirm that the sensitivity of ErbB2 antigen recognition is dependent on scFv affinity using both mRNA electroporated and lentiviral transduced CART cells.
Figure 4. Comparison of the activity of ErbB2 CART cells engineered with lentiviral vector or mRNA electroporation.
(a) T cells were modified with high (4D5) or low (4D5-5) affinity ErbB2 CAR using lentiviral transduction (LVV) or mRNA electroporation (RNA) as indicated. The % CAR expression and brightness was measured using anti-mouse IgG Fab antibody (upper panel). ErbB2 expression on a panel of tumor lines and K562 cells electroporated with ErbB2 mRNA was detected by flow cytometry (middle and lower panels); % of cells with surface expression of ErB2 and (MFI) shown for K562 cells. (b) CD107a up-regulation was measured on lentiviral transduced or mRNA electroporated CART cells after 4hr stimulation with indicated tumor lines (gated on CD3+ cells). (c) Induction of CD107a expression was measured on lentiviral transduced or mRNA electroporated CART cells after 4hr stimulation with ErbB2 electroporated K562 cells by gating on CD3+ cells (d) IFN-gamma secretion by lentiviral transduced or RNA electroporated CART cells was measured by Elisa after 24hr. (e) IFN-gamma production by CART cells measured 24hr after stimulation with K562 cells electroporated with indicated amount of ErbB2 mRNA.
Affinity decreased ErbB2 CART cells eliminate tumor in vivo and significantly reduced the toxicity against tissues expressing physiological levels of ErbB2
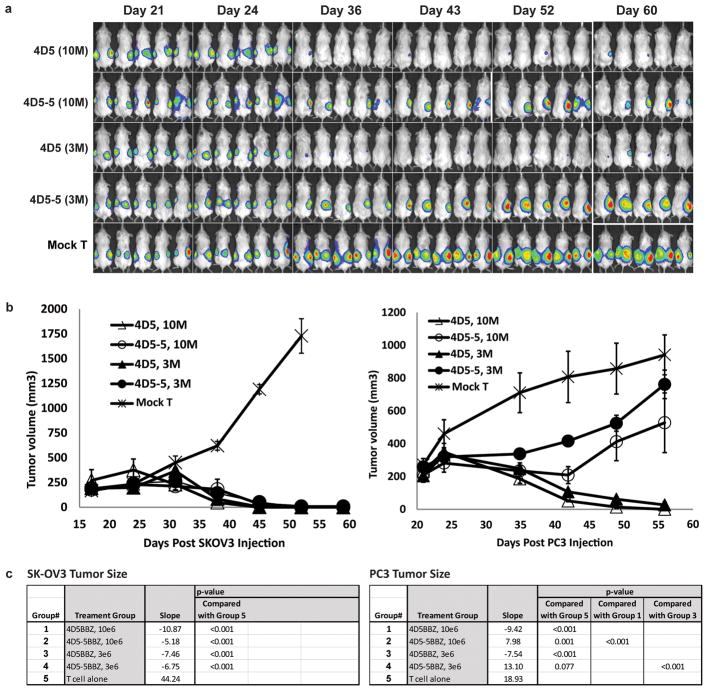
To extend the above in vitro results, a series of experiments were conducted in NSG mice with advanced vascularized tumor xenografts. Based on data above in figure 1a, the human ovarian cancer cell line SK-OV3 was selected as a representative ErbB2 over-expressing tumor and PC3, a human prostate cancer line, was chosen to model normal tissue ErbB2 levels. We first compared the antitumor efficacy of ErbB2 CART cells expressing either the high affinity 4D5 scFv or the low affinity 45D-5 scFv in NSG mice with day 18 established flank SK-OV3 tumors (Fig. S5). Serial bioluminescence imaging revealed that both the high and low affinity CART cells resulted in the rapid elimination of the tumors.
To further evaluate the therapeutic index of the low affinity ErbB2 CART cells in vivo, a mouse model was designed to simultaneously compare the efficacy and normal tissue toxicity of the high affinity (4D5.BBZ) and low affinity (4D5-5.BBZ) ErbB2 CARs. SK-OV3 and PC3 tumor cell lines were injected subcutaneously into opposite flanks of the same NSG mouse and T cells were administered when tumor volumes reached approximately 200 mm3. Mice were injected (i.v.) with either 3×106 or 1×107 CART cells on day 22 and serial bioluminescence imaging and tumor size assessments were conducted. Mice treated with either dose of the CART cells exhibited nearly complete regression of the ErbB2 overexpressing SK-OV3 tumor (Fig. 5a,b). In addition, almost complete regression of the PC3 tumor expressing ErbB2 at low levels on the opposite flank was also seen for the mice treated with high affinity 4D5-based CART cells. In contrast, the progressive tumor growth of PC3 was observed in the mice treated with low affinity 4D5-5-based CART cells, indicating that whereas the lower affinity CART cells were efficacious against ErbB2 overexpressing tumor, they show limited or no significant reactivity against cells expressing ErbB2 at physiological levels. Moreover, the selective tumor elimination was observed in mice treated at both high and low doses of CART cells. The above effects were not due to allorecognation because progressive tumor growth of both tumors was observed in mice treated with mock transduced T cells.
Figure 5. Affinity-tuned ErbB2 CART cells increase the therapeutic index and induce regression of advanced vascularized tumors in mice.
(a) In vivo discrimination of high ErbB2 (SK-OV3) and low ErbB2 (PC3) expressing tumors by affinity tuned CARs. T cells modified with different affinity ErbB2 CARs by lentiviral transduction were tested in dual-tumor engrafted NSG mice. Mice (n=5) were implanted with PC3-CBG tumor cells (1×106 cells/mouse, s.c.) on the right flank on day 0. On day 5 the same mice were given SK-OV3-CBG tumor cells (5×106 cells/mouse, s.c.) on the left flank. The mice were treated with T cells (i.v.) at day 23 after PC3 tumor inoculation. CART cells were given as a single injection of 1×107/mouse (10M), or 3×106/mouse (3M) as indicted. Mice treated with non-transduced T cells served as control. Animals were imaged at the indicated time post PC3 tumor inoculation. (b) SK-OV3 tumor size (left panel) or PC3 tumor sizes (right panel) in dual-tumor grafted NSG mice treated with the indicated ErbB2 CAR. Tumor sizes were measured, and the tumor volume was calculated and plotted. (c) Biostatistics analysis results for the tumor size. Data (not transformed) was analyzed for day < 56 (PC3) or day < 52 (SKOV3). Method was mixed models, with day, group, and day x group interaction as fixed effects, and mouse as random effect. For random effect, each mouse had a separate intercept and slope. (Method also known as random coefficients.) p-values presented are interaction p-values.
Affinity tuning of scFv increases the therapeutic index of EGFR CART cells
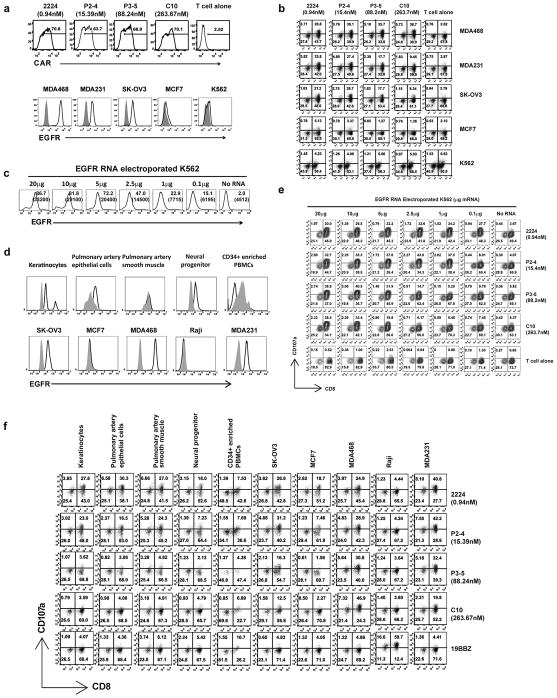
To test the broader applicability the strategy to fine tune the affinity of the scFv, we evaluated a panel of EGFR CARs. EGFR.BBZ CARs were constructed from scFvs derived from the parental human anti-EGFR antibody C10 (21). The monovalent affinities of the panel of EGFR-specific scFvs varied over a range of approximately 300-fold (17). The 2224, P2-4, P3-5 and C10 scFvs were cloned into an RNA-based vector and in vitro transcribed for T cell mRNA electroporation. Levels of CAR surface expression were assayed and found to be similar among the EGFR CAR constructs (Fig. 6a, upper panel). To compare reactivities of the panel of EGFR CARs, CART cells were stimulated with EGFR-expressing tumor cell lines that have a broad range of EGFR expression at the cell surface (Fig. 6a, lower panel). CART cell activation was evaluated by levels of CD107a up-regulation; the data is summarized in Fig. 6b. Higher affinity EGFR CARs (2224.BBZ and P2-4.BBZ) responded to all EGFR positive tumor lines (MDA468, MDA231 and SK-OV3) regardless of EGFR expression levels (Fig. 6b). However, the reactivity exhibited by lower affinity EGFR CARs (P3-5.BBZ and C10.BBZ) against EGFR-expressing tumor lines correlated with the levels of EGFR expression. Furthermore, lower affinity EGFR CARs displayed more potent reactivity to the EGFR overexpressing tumor, MDA468, than the higher affinity EGFR CARs, while provoking a much weaker response to EGFR low expressing cells (Fig. 6b). None of the EGFR CART cells reacted to the EGFR negative tumor line K562.
Figure 6. Characterization of EGFR affinity tuned CART cells and tumor cell lines.
(a) CAR expression on T cells electroporated with EGFR CAR mRNA were stained by an anti-human IgG Fab and detected by flow cytometry staining (upper panel); the affinity of the scFv is indicated (nM). Tumor lines (lower panel) were stained with anti-EGFR Affibody-FITC (open histograms), the same cells were stained with mouse IgG1-FITC as isotype control (grey filled histograms). (b) EGFR CAR recognition sensitivity is correlated with affinity. A panel of EGFR CART cells with the indicated affinity of the scFv (KD, nM) was stimulated with the panel of tumors expressing EGFR at the density shown in (a). After 4h stimulation, CD107a up-regulation on the CART cells was detected by gating on CD3+ cells. (c) K562 cells were electroporated with the indicated amount of EGFR mRNA and EGFR expression was detected using anti-EGFR Affibody-FITC staining 14h post electroporation. (d) EGFR expression on a panel of primary cell lines and tumor lines was measured by FACs using anti-EGFR Affibody-FITC (open histograms), with the same cells stained with mouse IgG1-FITC as isotype control (grey filled histograms). (e) EGFR CAR recognition sensitivity is correlated with affinity. T cells were electroporated with the panel of EGFR CARs with different affinities as indicated and stimulated with K562 electroporated with EGFR mRNA at different levels as shown in (d). After 4hr stimulation, CD107a expression on CART cells was measured by gating on CD3+ cells. (f) Affinity dependent recognition of primary cell lines and tumor cells using affinity-tuned EGFR CARs. T cells were electroporated with the indicated EGFR CAR mRNA. One day after electroporation, the CART cells were stimulated with the panel of cells for 4 hr and the induction CD107a expression on the CART cells was quantified (CD3+ gated).
To confirm that the level of response was related to scFv affinity and the level of EGFR expression, and to exclude tumor-specific effects, the panel of EGFR CART cells was co-cultured with K562 cells expressing varying levels of EGFR after electroporation with mRNA (Fig. 6c). The higher affinity EGFR CARs did not discriminate between target cells with different levels of EGFR expression (Fig. 6e). For example, T cells expressing EGFR CAR 2224.BBZ responded equally well to K562 cells electroporated with a 200-fold difference in EGFR mRNA (0.1 μg to 20μg). However in agreement with the above ErbB2 CAR results, the lower affinity EGFR CARs (P3-5 and C10) exhibited a high correlation between T cell responses and EGFR expression levels (6e).
To confirm the increased safety profile of the lower affinity EGFR CARs, we tested the reactivities of EGFR CARs against primary cells derived from different organs. Five primary cell lines and five tumor cell lines were tested for both surface levels of EGFR (Fig. 6d) and ability to trigger CART cell reactivity (Fig. 6f). Three of the primary cell lines examined express detectable levels of EGFR and two did not (pulmonary artery smooth muscle and PBMC). Two of the tumor cell lines (MCF7 and Raji) did not express detectable EGFR on the cell surface. Comparing EGFR CART cells to CD19 CART cells, T cells with higher EGFR affinity CARs (2224 and P2-4) reacted to all the primary lines tested and all of the tumors except Raji (Fig. 6f). However, T cells with the affinity decreased EGFR CART cells P3-5 and C10 were not reactive to any of the five primary cells tested (Fig. 6f). CD19 specific CART cells reacted to the CD19+ line Raji, and to PBMCs, presumably to the B cells in PBMC, but did not respond to any of the tumor lines or other primary cell lines. These data demonstrate that our strategy to affinity tune scFv can increase the therapeutic index for CART cells that target either ErbB2 or EGFR.
Discussion
The efficacy of CART cells is dictated in part by the differential expression of the target antigen in tumor versus normal tissue. Our results demonstrate that CARs with known severe on-target toxicities can be reengineered by affinity tuning, retaining potent in vivo efficacy while eliminating or reducing toxicity. In particular, the 4D5 CAR based on trastuzumab had lethal toxicity (8), due to recognition of physiologic levels of ErbB2 expressed in cardiopulmonary tissues (22). Here we demonstrate that by reducing the KD of scFv employed in CART cells by 2- to 3-log, a substantial improvement in the therapeutic index for ErbB2 and EGFR CART cells. CART cells with lower affinity scFv showed equally robust anti-tumor activity against ErbB2 overexpressing tumors as compared to the high affinity CARs, but displayed significantly reduced reactivity against physiologic levels of ErbB2.
CARs specific for the B cell lineage antigens CD19 and CD20 have been tested by a variety of groups and have displayed potent efficacy in B cell malignancies (23). However in solid tumors, with the exception of tumor-specific isoforms such as EGFRviii (24), on target toxicity is anticipated to be a severe limitation for CART cells. This limitation is expected to be a more serious with CARs than with antibody therapies using intact antibodies or antibody drug conjugates, due to the lower limit of target sensitivity for CART cells compared to antibody based therapies that differs by several orders of magnitude. Our present studies using target cells electroporated with ErbB2 or EGFR mRNA are consistent with previous studies indicating that CART cells can recognize tumor cells with ~100 targets per cell (25). In contrast, amplification of ErbB2 occurs in approximately 20% to 25% of primary human breast cancers and typically results in overexpression of ErbB2 protein at > 1 million copies per cell (26, 27). At present, available data indicate that cancer cells do not lose ErbB2 expression when they become refractory to ErbB2 directed therapies (28).
Our findings support previous work from Chmielewski (11), suggesting that the high affinity CARs exhibit less discrimination between target cells with high or low target expression levels. However, the present results differ from Chmielewski and coworkers in that none of the higher affinity CARs (with KD ranging from 15pM to 16nM) in their report were reactive to cells with low level expression of ErbB2 and their lower affinity CAR that only recognized tumors with amplified ErbB2 showed a substantial reduction in T cell efficacy compared to the higher affinity CARs. In contrast, we found that the ErbB2 CAR using the 4D5 scFv with KD at 0.3nM was strongly reactive to keratinocytes and even to cell lines transfected with extremely low amounts ErbB2 mRNA that were 100 times below detectable levels, while affinity-tuned CART cells retained reactivity to ErbB2 amplified tumors that was at least as potent as the high affinity CAR, both in vitro and in aggressive mouse tumor models. Some variables that may explain these differences include the use of different scFvs (C5.6 versus 4D5) that may recognize different epitopes, distinct CAR signaling domain configuration (zeta alone versus 4-1BB-zeta), and different gene transfer approaches (retroviral transduction versus RNA electroporation or lentiviral transduction) that may affect CAR surface expression levels on the T cells. Together, this suggests that each of these factors should be considered when selecting the affinity of a CAR in relevant clinical situations.
The advent of more potent adoptive transfer strategies has prompted a reassessment of targets previously considered as safe using weaker immunotherapeutic strategies (29). Strategies to maximize the therapeutic index of CART cells include target selection, CAR design, cell manufacturing and gene transfer techniques. In addition to affinity tuning, other strategies being developed to manage on target toxicity include the use of dual CART cell approaches (30, 31), conditional deletion and suicide systems (32, 33), and repeated infusions of T cells having mRNA CARs that have transient expression and self-limiting toxicity (34).
Efficient T cell activation by pMHC on the surface of APCs requires an optimal dwell time of TCR-pMHC interaction that is sufficiently long to complete the signal cascade, but also short enough to permit serial engagement of multiple TCR molecules by pMHC, especially when pMHC density is low (35, 36). Increasing TCR affinity above “natural” affinity range (1μM-100μM) could reduce the ability to recognize low density pMHC (37–39). The reactivity of high affinity TCRs against low density cognate pMHC could be rescued by decreasing MHC expression (39), or decreasing TCR-pMHC interaction by using mutated MHC molecules (35)or stimulating with plate-bound pMHC molecules (38). These findings suggest that the activation of T cells with very high affinity TCR is not only determined by the density of pMHC on the APCs, but the interaction of TCR with MHC and the presence of endogenous pMHC profoundly impact T cell activation as well. CAR imitates TCR by using TCR/CD3 zeta signaling moiety to activate T cells. However, CART cells are largely different from conventional alpha/beta T cells by the facts that 1), CART recognizes antigen independent of target MHC and CD8 expression ; 2), CART activates through only zeta chain without involvement of gamma, delta and epsilon chains; and 3), co-stimulator signal is incorporated in CART. Despite the differences between CART and conventional T cells, our finding that decreasing the affinity of a CAR to “natural” high affinity range (1μM-10μM) increased CART activities against antigen high expression target is consistent with the finding for conventional T cells whose optimal activation requires TCR affinity in the “natural” affinity range. As mentioned above, decreasing overall avidity of TCR/pMHC led to improved serial engagement of TCR against low density pMHC for high affinity TCR. Similarly, we found in our current study that high affinity CART cells showed improved activities against low levels of target antigen, which presumably is caused by decreased binding half-life of CAR/antigen interaction and improved serial engagement due to overall decreased avidity. Therefore, the signals that T cells receive from high affinity CAR against low enough surface antigen might be the same as that from lower affinity CAR against high surface antigen.
There are several limitations of our study that will require evaluation in a phase I pilot trial. This includes the cell to cell variation in expression of ErbB2 and EFGR (40), and the unknown variation that may occur in the setting of inflammation. In addition, while we have demonstrated that affinity-tuning can increase the therapeutic index for ErbB2 and EGFR it remains unknown if this is a general strategy. In addition to scFv affinity, other variables that require examination on a case by case basis include the location of the target epitope, the length of the hinge and the nature of the signaling domain (12, 41).
In summary, ErbB2 and EGFR have previously been considered as undruggable targets for CART cells. Given that dysregulation of the expression of ErbB2 and EGFR occurs frequently in multiple human carcinomas including breast, glioblastoma, lung, pancreatic, ovarian, head and neck squamous cell cancer and colon cancer, our findings have considerable clinical importance. Our strategy has the potential not only to improve the safety profile and clinical outcome of CARs directed against validated targets but also to expand the landscape to targets not previously druggable with CART cells because of on-target toxicities. More generally, our findings suggest that safer and more potent CARs can be designed by employing affinity-tuned scFvs for a variety of common carcinomas.
Supplementary Material
Acknowledgments
We thank our colleagues at Novartis J Brogdon, P. Gotwals, S. Ettenberg and W. Sellers for insightful discussions and L. Johnson for primary cell lines.
Funding: This work was supported in part by grants from the National Institutes of Health (P01CA066726; 2R01CA120409) and by Novartis.
Footnotes
Conflicts of interest: X.L., C.H.J. and Y.Z. have financial interests due to intellectual property and patents in the field of cell and gene therapy. Conflicts of interest are managed in accordance with University of Pennsylvania policy and oversight. The other authors declare that they have no competing interests.
Author contributions: The manuscript was written by X.L. and Y.Z. and edited by C.H.J, R. Y. All authors discussed and interpreted results.
Data and materials availability: Data and materials are available from the authors under a material transfer agreement.
References
- 1.Kochenderfer JN, Wilson WH, Janik JE, Dudley ME, Stetler-Stevenson M, Feldman SA, et al. Eradication of B-lineage cells and regression of lymphoma in a patient treated with autologous T cells genetically engineered to recognize CD19. Blood. 2010;116:4099–4102. doi: 10.1182/blood-2010-04-281931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Porter DL, Levine BL, Kalos M, Bagg A, June CH. Chimeric antigen receptor-modified T cells in chronic lymphoid leukemia. The New England journal of medicine. 2011;365:725–733. doi: 10.1056/NEJMoa1103849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brentjens RJ, Davila ML, Riviere I, Park J, Wang X, Cowell LG, et al. CD19-Targeted T Cells Rapidly Induce Molecular Remissions in Adults with Chemotherapy-Refractory Acute Lymphoblastic Leukemia. Science translational medicine. 2013;5:177ra138. doi: 10.1126/scitranslmed.3005930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lamers CH, Sleijfer S, Vulto AG, Kruit WH, Kliffen M, Debets R, et al. Treatment of metastatic renal cell carcinoma with autologous T-lymphocytes genetically retargeted against carbonic anhydrase IX: first clinical experience. J Clin Oncol. 2006;24:e20–e22. doi: 10.1200/JCO.2006.05.9964. [DOI] [PubMed] [Google Scholar]
- 5.Parkhurst MR, Yang JC, Langan RC, Dudley ME, Nathan DA, Feldman SA, et al. T cells targeting carcinoembryonic antigen can mediate regression of metastatic colorectal cancer but induce severe transient colitis. Molecular therapy: the journal of the American Society of Gene Therapy. 2011;19:620–626. doi: 10.1038/mt.2010.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morgan RA, Chinnasamy N, Abate-Daga D, Gros A, Robbins PF, Zheng Z, et al. Cancer Regression and Neurological Toxicity Following Anti-MAGE-A3 TCR Gene Therapy. Journal of immunotherapy (Hagerstown, Md: 1997) 2013;36:133–151. doi: 10.1097/CJI.0b013e3182829903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Linette GP, Stadtmauer EA, Maus MV, Rapoport AP, Levine BL, Emery L, et al. Cardiovascular toxicity and titin cross-reactivity of affinity enhanced T cells in myeloma and melanoma. Blood. 2013;122:863–871. doi: 10.1182/blood-2013-03-490565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morgan R, Yang J, Kitano M, Dudley M, Laurencot C, Rosenberg S. Case Report of a Serious Adverse Event Following the Administration of T Cells Transduced With a Chimeric Antigen Receptor Recognizing ERBB2. Molecular Therapy. 2010;18:843–851. doi: 10.1038/mt.2010.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Perez-Soler R, Saltz L. Cutaneous adverse effects with Her1/EGFR-targeted agents: Is there a silver lining? J Clin Oncol. 2005;23:5235–5246. doi: 10.1200/JCO.2005.00.6916. [DOI] [PubMed] [Google Scholar]
- 10.Cheever MA, Allison JP, Ferris AS, Finn OJ, Hastings BM, Hecht TT, et al. The prioritization of cancer antigens: a national cancer institute pilot project for the acceleration of translational research. Clinical cancer research: an official journal of the American Association for Cancer Research. 2009;15:5323–5337. doi: 10.1158/1078-0432.CCR-09-0737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chmielewski M, Hombach A, Heuser C, Adams GP, Abken H. T cell activation by antibody-like immunoreceptors: increase in affinity of the single-chain fragment domain above threshold does not increase T cell activation against antigen-positive target cells but decreases selectivity. J Immunol. 2004;173:7647–7653. doi: 10.4049/jimmunol.173.12.7647. [DOI] [PubMed] [Google Scholar]
- 12.Hudecek M, Lupo-Stanghellini MT, Kosasih PL, Sommermeyer D, Jensen MC, Rader C, et al. Receptor affinity and extracellular domain modifications affect tumor recognition by ROR1-specific chimeric antigen receptor T cells. Clinical cancer research: an official journal of the American Association for Cancer Research. 2013;19:3153–3164. doi: 10.1158/1078-0432.CCR-13-0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhong S, Malecek K, Johnson LA, Yu Z, Vega-Saenz de Miera E, Darvishian F, et al. T-cell receptor affinity and avidity defines antitumor response and autoimmunity in T-cell immunotherapy. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:6973–6978. doi: 10.1073/pnas.1221609110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schmid DA, Irving MB, Posevitz V, Hebeisen M, Posevitz-Fejfar A, Sarria JC, et al. Evidence for a TCR affinity threshold delimiting maximal CD8 T cell function. J Immunol. 2010;184:4936–4946. doi: 10.4049/jimmunol.1000173. [DOI] [PubMed] [Google Scholar]
- 15.Barrett DM, Zhao Y, Liu X, Jiang S, Carpenito C, Kalos M, et al. Treatment of advanced leukemia in mice with mRNA engineered T cells. Human gene therapy. 2011;22:1575–1586. doi: 10.1089/hum.2011.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carter P, Presta L, Gorman CM, Ridgway JB, Henner D, Wong WL, et al. Humanization of an anti-p185HER2 antibody for human cancer therapy. Proceedings of the National Academy of Sciences of the United States of America. 1992;89:4285–4289. doi: 10.1073/pnas.89.10.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou Y, Drummond DC, Zou H, Hayes ME, Adams GP, Kirpotin DB, et al. Impact of single-chain Fv antibody fragment affinity on nanoparticle targeting of epidermal growth factor receptor-expressing tumor cells. Journal of molecular biology. 2007;371:934–947. doi: 10.1016/j.jmb.2007.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao Y, Moon E, Carpenito C, Paulos CM, Liu X, Brennan AL, et al. Multiple injections of electroporated autologous T cells expressing a chimeric antigen receptor mediate regression of human disseminated tumor. Cancer research. 2010;70:9053–9061. doi: 10.1158/0008-5472.CAN-10-2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carpenito C, Milone MC, Hassan R, Simonet JC, Lakhal M, Suhoski MM, et al. Control of large, established tumor xenografts with genetically retargeted human T cells containing CD28 and CD137 domains. Proc Natl Acad Sci U S A. 2009;106:3360–3365. doi: 10.1073/pnas.0813101106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moon EK, Wang LC, Dolfi DV, Wilson CB, Ranganathan R, Sun J, et al. Multifactorial T-cell hypofunction that is reversible can limit the efficacy of chimeric antigen receptor-transduced human T cells in solid tumors. Clinical cancer research: an official journal of the American Association for Cancer Research. 2014;20:4262–4273. doi: 10.1158/1078-0432.CCR-13-2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heitner T, Moor A, Garrison JL, Marks C, Hasan T, Marks JD. Selection of cell binding and internalizing epidermal growth factor receptor antibodies from a phage display library. J Immunol Methods. 2001;248:17–30. doi: 10.1016/s0022-1759(00)00340-9. [DOI] [PubMed] [Google Scholar]
- 22.Press MF, Cordon-Cardo C, Slamon DJ. Expression of the HER-2/neu proto-oncogene in normal human adult and fetal tissues. Oncogene. 1990;5:953–962. [PubMed] [Google Scholar]
- 23.Maus MV, Grupp SA, Porter DL, June CH. Antibody modified T cells: CARs take the front seat for hematologic malignancies. Blood. 2014;123:2625–2635. doi: 10.1182/blood-2013-11-492231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morgan RA, Johnson LA, Davis J, Zheng Z, Woolard K, Feldman SA, et al. Recognition of glioma stem cells by genetically modified T cells targeting EGFRvIII and development of adoptive cell therapy for glioma. Human gene therapy. 2012 doi: 10.1089/hum.2012.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stone JD, Aggen DH, Schietinger A, Schreiber H, Kranz DM. A sensitivity scale for targeting T cells with chimeric antigen receptors (CARs) and bispecific T-cell Engagers (BiTEs) Oncoimmunology. 2012;1:863–873. doi: 10.4161/onci.20592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robertson KW, Reeves JR, Smith G, Keith WN, Ozanne BW, Cooke TG, et al. Quantitative Estimation of Epidermal Growth Factor Receptor and c-erbB-2 in Human Breast Cancer. Cancer research. 1996;56:3823–3830. [PubMed] [Google Scholar]
- 27.Vogel CL, Cobleigh MA, Tripathy D, Gutheil JC, Harris LN, Fehrenbacher L, et al. Efficacy and safety of trastuzumab as a single agent in first-line treatment of HER2-overexpressing metastatic breast cancer. Journal of Clinical Oncology. 2002;20:719–726. doi: 10.1200/JCO.2002.20.3.719. [DOI] [PubMed] [Google Scholar]
- 28.Ritter CA, Perez-Torres M, Rinehart C, Guix M, Dugger T, Engelman JA, et al. Human breast cancer cells selected for resistance to trastuzumab in vivo overexpress epidermal growth factor receptor and ErbB ligands and remain dependent on the ErbB receptor network. Clinical cancer research: an official journal of the American Association for Cancer Research. 2007;13:4909–4919. doi: 10.1158/1078-0432.CCR-07-0701. [DOI] [PubMed] [Google Scholar]
- 29.Hinrichs CS, Restifo NP. Reassessing target antigens for adoptive T-cell therapy. Nature biotechnology. 2013;31:999–1008. doi: 10.1038/nbt.2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kloss CC, Condomines M, Cartellieri M, Bachmann M, Sadelain M. Combinatorial antigen recognition with balanced signaling promotes selective tumor eradication by engineered T cells. Nature biotechnology. 2013;31:71–75. doi: 10.1038/nbt.2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lanitis E, Poussin M, Klattenhoff AW, Song D, Sandaltzopoulos R, June CH, et al. Chimeric antigen receptor T cells with dissociated signaling domains exhibit focused anti-tumor activity with reduced potential for toxicity. Cancer immunology research. 2013;1:43–53. doi: 10.1158/2326-6066.CIR-13-0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Di Stasi A, Tey SK, Dotti G, Fujita Y, Kennedy-Nasser A, Martinez C, et al. Inducible apoptosis as a safety switch for adoptive cell therapy. The New England journal of medicine. 2011;365:1673–1683. doi: 10.1056/NEJMoa1106152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang X, Chang WC, Wong CW, Colcher D, Sherman M, Ostberg JR, et al. A transgene-encoded cell surface polypeptide for selection, in vivo tracking, and ablation of engineered cells. Blood. 2011;118:1255–1263. doi: 10.1182/blood-2011-02-337360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Beatty GL, Haas AR, Maus MV, Torigian DA, Soulen MC, Plesa G, et al. Mesothelin-specific chimeric antigen receptor mRNA-engineered T cells induce anti-tumor activity in solid malignancies. Cancer immunology research. 2014;2:112–120. doi: 10.1158/2326-6066.CIR-13-0170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kalergis AM, Boucheron N, Doucey MA, Palmieri E, Goyarts EC, Vegh Z, et al. Efficient T cell activation requires an optimal dwell-time of interaction between the TCR and the pMHC complex. Nature immunology. 2001;2:229–234. doi: 10.1038/85286. [DOI] [PubMed] [Google Scholar]
- 36.Valitutti S. The Serial Engagement Model 17 Years After: From TCR Triggering to Immunotherapy. Frontiers in immunology. 2012;3:272. doi: 10.3389/fimmu.2012.00272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thomas S, Xue SA, Bangham CR, Jakobsen BK, Morris EC, Stauss HJ. Human T cells expressing affinity-matured TCR display accelerated responses but fail to recognize low density of MHC-peptide antigen. Blood. 2011;118:319–329. doi: 10.1182/blood-2010-12-326736. [DOI] [PubMed] [Google Scholar]
- 38.Gonzalez PA, Carreno LJ, Coombs D, Mora JE, Palmieri E, Goldstein B, et al. T cell receptor binding kinetics required for T cell activation depend on the density of cognate ligand on the antigen-presenting cell. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:4824–4829. doi: 10.1073/pnas.0500922102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao Y, Bennett AD, Zheng Z, Wang QJ, Robbins PF, Yu LY, et al. High-affinity TCRs generated by phage display provide CD4+ T cells with the ability to recognize and kill tumor cell lines. J Immunol. 2007;179:5845–5854. doi: 10.4049/jimmunol.179.9.5845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Szöllösi J, Balázs M, Feuerstein BG, Benz CC, Waldman FM. ERBB-2 (HER2/neu) Gene Copy Number, p185HER-2 Overexpression, and Intratumor Heterogeneity in Human Breast Cancer. Cancer research. 1995;55:5400–5407. [PubMed] [Google Scholar]
- 41.Guedan S, Chen X, Madar A, Carpenito C, McGettigan SE, Frigault MJ, et al. ICOS-based chimeric antigen receptors program bipolar TH17/TH1 cells. Blood. 2014;124:1070–1080. doi: 10.1182/blood-2013-10-535245. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.