Abstract
Background
Transplantation of hematopoietic cells from unrelated donors can cure blood disorders but carries a significant risk of acute graft-versus-host disease (GVHD). The risk is higher when the recipient and donor are HLA-DPB1–mismatched, but the mechanisms leading to GVHD are unknown. The HLA-DPB1 regulatory region variant rs9277534 is associated with HLA-DPB1 expression. We tested the hypothesis that the GVHD risk correlates with the rs9277534 allele linked to the mismatched HLA-DPB1 in the recipient.
Methods
We genotyped rs9277534 in 3505 persons to define rs9277534-DPB1 haplotypes. Among 1441 recipients of transplants from HLA-A,B,C,DRB1,DQB1–matched unrelated donors with only one HLA-DPB1 mismatch, linkage of the rs9277534 A and G alleles to the mismatched HLA-DPB1 was determined. HLA-DPB1 expression was assessed by means of a quantitative polymerase-chain-reaction assay. The risk of acute GVHD among recipients whose mismatched HLA-DPB1 allele was linked to rs9277534G (high expression) was compared with the risk among recipients whose mismatched HLA-DPB1 allele was linked to rs9277534A (low expression).
Results
The mean HLA-DPB1 expression was lower with rs9277534A than with rs9277534G. Among recipients of transplants from donors with rs9277534A-linked HLA-DPB1, the risk of acute GVHD was higher for recipients with rs9277534G-linked HLA-DPB1 mismatches than for recipients with rs9277534A-linked HLA-DPB1 mismatches (hazard ratio, 1.54; 95% confidence interval [CI], 1.25 to 1.89; P<0.001), as was the risk of death due to causes other than disease recurrence (hazard ratio, 1.25; 95% CI, 1.00 to 1.57; P = 0.05).
Conclusions
The risk of GVHD associated with HLA-DPB1 mismatching was influenced by the HLA-DPB1 rs9277534 expression marker. Among recipients of HLA-DPB1–mismatched transplants from donors with the low-expression allele, recipients with the high-expression allele had a high risk of GVHD. (Funded by the National Institutes of Health and others.)
Hematopoietic-cell Transplantation from unrelated donors can cure blood disorders; however, graft-versus-host disease (GVHD) remains a major impediment to successful outcomes.1 GVHD can occur after HLA-matched transplantation when the donor cells recognize polymorphic peptides (“minor histocompatibility antigens”) presented by the recipient’s HLA.2 In HLA-mismatched transplantation, direct recognition of the recipient’s mismatched HLA by the donor’s cells provides a potent stimulus for graft-versus-host allorecognition3–7; recognition of the donor’s mismatched HLA by the recipient’s immune system leads to graft rejection.8,9
The importance of HLA class II alloantigens was established early in the history of clinical transplantation.10 Advances in understanding the biologic relevance of HLA-DP were hampered by the need for an in vitro cellular assay to assess HLA-DP incompatibility and by the overall lower cell-surface expression of HLA-DP relative to other classical HLAs.11,12 The introduction of polymerase-chain-reaction (PCR) assays in the late 1980s ushered in a new era of HLA typing.6 DNA-based typing of HLA-A,B,C,DRB1,DQB1–matched recipients and donors uncovered HLA-DPB1 mismatching in 85% of transplantations, and undetected HLA-DPB1 mismatching was shown to be associated with life-threatening GVHD.13–15 The immunogenicity of HLA-DPB1 mismatches may be defined by the strength of in vitro T-cell–mediated cytotoxicity against polymorphic amino acid residues of HLA-DPβ.16
HLA expression plays an important role in human disease. In the class I region, HLA-C expression influences the clinical course of human immunodeficiency virus infection and the acquired immunodeficiency syndrome (HIV–AIDS), susceptibility to Crohn’s disease, and the risk of GVHD after hematopoietic-cell transplantation.17,18 Variation in the 3′ untranslated region of HLA-DPB1 is associated with spontaneous clearance of hepatitis B virus in both Japanese and U.S. populations.19,20 The mechanism facilitating viral clearance may be related to the A→G single-nucleotide polymorphism rs9277534, which marks HLA-DP cell-surface expression.20 The rs9277534G allele is associated with high expression of HLA-DP, and the rs9277534A allele is associated with low expression. These data suggest that information about the regulation of HLA expression levels may increase our understanding of the role of HLA in disease.
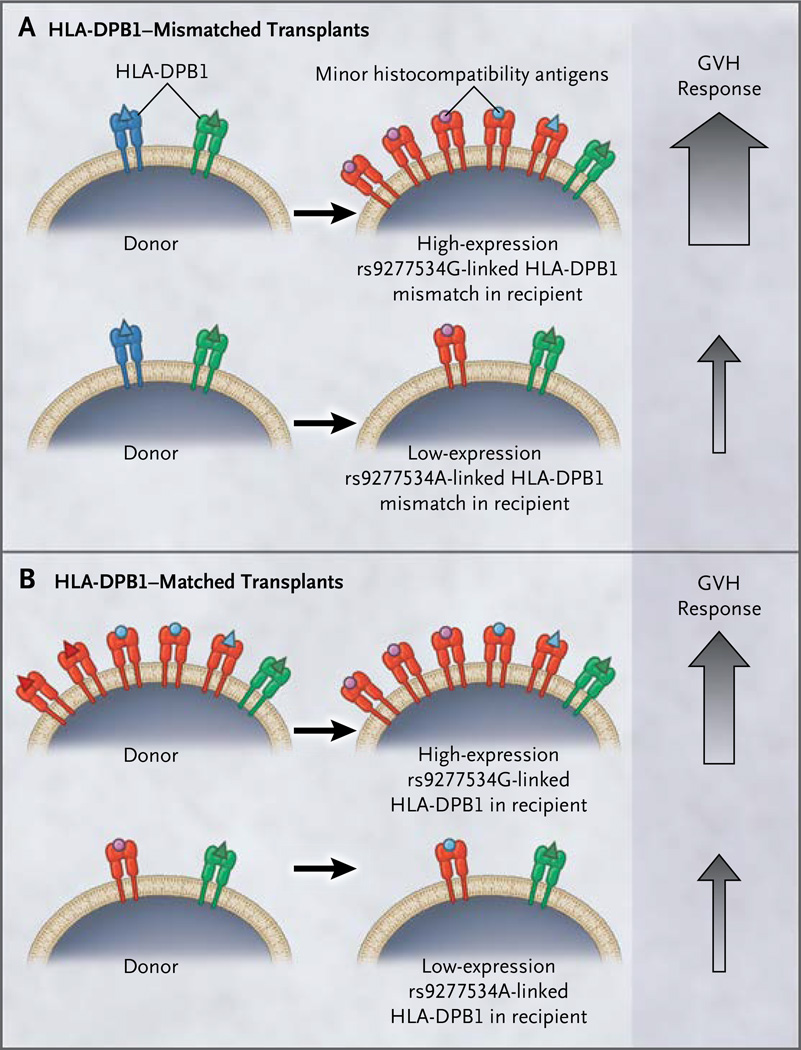
We previously identified an association of rs2281389 with acute GVHD and replicated this finding in an independent validation cohort.21 The rs2281389 variant resides in a noncoding region of DNA and consequently is unlikely to be a direct mediator of GVHD. Located only 4989 bp from HLA-DPB1, rs2281389 is strongly associated with rs9277534 in the HLA-DPB1 regulatory region. The rs9277534 variant, in turn, is linked to the HLA-DPB1 exon 2 allele that defines the HLA-DPβ protein and tissue type. The close relationship between rs2281389 and rs9277534 across haplotypes makes the rs9277534 expression variant a plausible marker for the risk of GVHD. We hypothesized that rs9277534G-linked HLA-DPB1 in a transplant recipient that is not shared by the donor cells (constituting a high-expression graft-versus-host mismatch) is more visible to the donor cells than is rs9277534A-linked HLA-DPB1 (constituting a low-expression graft-versus-host mismatch) and leads to a higher risk of acute GVHD (Fig. 1A). If the level of HLA-DPB1 expression provides information about HLA-DPB1 mismatches that are well tolerated (i.e., associated with favorable outcomes), then rs9277534 can be used prospectively to screen potential unrelated donors before transplantation in order to lower the risk of acute GVHD.
Figure 1. Proposed Schema for the Role of HLA-DPB1 Expression in Graft-versus-Host Recognition.
In the context of an HLA-DPB1 mismatch between a recipient and a donor (Panel A), an rs9277534G-linked (high-expression) mismatch is hypothesized to be a visible target for donor-mediated graft-versus-host (GVH) recognition. Red HLA molecules represent the mismatched HLA-DPB1 in the recipient, blue molecules represent the donor’s mismatched HLA-DPB1, and green molecules are shared (matched) between the recipient and the donor. Black arrows pointing from the donor toward the recipient represent the GVH vector of allorecognition. The size of the upward-pointing gray arrows indicates the magnitude of the GVH response. In the context of an HLA-DPB1 match between a recipient and a donor (Panel B), peptides (minor histocompatibility antigens) presented by an rs9277534G-linked (high-expression) HLA-DPB1 molecule in a recipient may be a more visible target for donor-mediated GVH recognition. Diverse peptides presented by HLA are shown as purple and blue circles and red, blue, and green triangles.
The nonrandom association between rs2281389 and rs9277534 on haplotypes (in a test of linkage disequilibrium, D′ = 1 and r2=0.4) (see Table S1 in the Supplementary Appendix, available with the full text of this article at NEJM.org) led us to hypothesize that rs9277534-linked HLA-DPB1 mismatches have a role in GVHD. We defined the linkage of rs9277534 alleles to HLA-DPB1 mismatches, confirmed the level of expression of rs9277534G-linked and rs9277534A-linked HLA-DPB1 among reference cells, and examined the GVHD risk associated with rs9277534G-linked HLA-DPB1 mismatches as compared with rs9277534A-linked mismatches. Finally, we determined the likelihood of identifying donors with rs9277534A-linked HLA-DPB1 mismatches.
Methods
Study Population
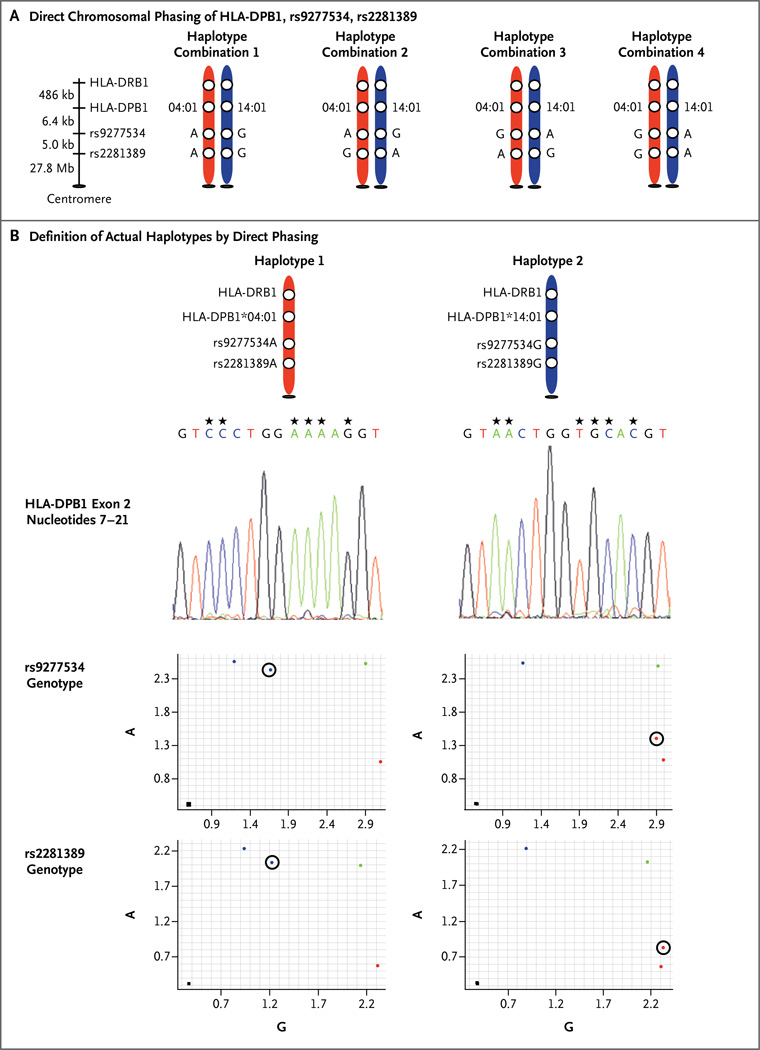
We genotyped rs9277534 in 3505 persons (Table S2 in the Supplementary Appendix). Cell lines from 18 persons were a source of high-quality genomic DNA used to physically separate the two HLA haplotypes from each other and to determine the linkage of HLA-DPB1 with rs9277534 and rs2281389 alleles carried on each haplotype (Fig. 2, and the Materials and Methods section in the Supplementary Appendix). HLA-DPB1 expression was determined in 49 persons with rs9277534AA and 32 with rs9277534GG.
Figure 2. Direct Chromosomal Phasing of HLA-DPB1, rs9277534, rs2281389.
A person with the genotype DPB1*04:01,14:01; rs9277534AG; rs2281389AG is heterozygous at all three genetic markers. The diploid genotype (both chromosomes together) can be produced by four theoretical haplotypes (Panel A). The physical linkage of HLA-DPB1, rs9277534, and rs2281389 alleles on each haplotype in a diploid sample requires separation of the chromosomes (a process known as phasing) (Panel B). The DNA was phased with the rs9277534A probe (red chromosome) and the rs9277534G probe (blue chromosome). The hap-loid DNA (one chromosome by itself) captured by the rs9277534A probe carries the HLA-DPB1*04:01 sequence (shown in reverse orientation for nucleotides 7 through 21, corresponding to residues 8 through 11 of HLA-DPβ; asterisks above the nucleotides indicate positions that distinguish DPB1*04:01 from 14:01) and is physically linked to rs9277534A (circled) and rs2281389A (circled), as ascertained by means of TaqMan genotyping. This is the HLA-DPB1*04:01-A-A haplotype. The haploid DNA captured by the rs9277534G probe carries the HLA-DPB1*14:01 allele and is physically linked to rs9277534G (circled) and rs2281389G (circled). This is the HLA-DPBl* 14:01-G-G haplotype. Controls shown for TaqMan genotyping include rs9277534AA (blue dot), rs9277534AG (green dot), rs9277534GG (red dot), and rs2281389AA (blue dot), rs2281389AG (green dot), rs2281389GG (red dot) cells from the International HLA Workshop panel (see the Materials and Methods section in the Supplementary Appendix). Black squares indicate the absence of template controls.
Clinical outcomes were assessed in 2029 recipients of transplants from unrelated donors who underwent transplantation for the treatment of acute leukemia, chronic myeloid leukemia, or the myelodysplastic syndrome between 1988 and 2008 and whose clinical data were reported to the Center for International Blood and Marrow Transplant Research (Table S3 in the Supplementary Appendix). Of the 2029 recipients, 1441 received an HLA-A,B,C,DRB1,DQB1–matched transplant from an unrelated donor with only one HLA-DPB1 mismatch in the graft-versus-host vector of incompatibility (recipient HLA-DPB1 not present in the donor). The remaining 588 recipients received a transplant from an HLA-A,B,C,DRB1,DQB1,DPB1–matched unrelated donor. Most recipients were prepared for transplantation with a myeloablative regimen (83.2% of HLA-DPB1–mismatched recipients) and received cyclophosphamide with total-body irradiation (64.2%) or busulfan with cyclophosphamide (22.9%) (Table S3 in the Supplementary Appendix). We determined the likelihood of avoiding HLA-DPB1 mismatching for 146 transplant recipients who were referred to the Fred Hutchinson Cancer Research Center for an unrelated-donor search and for whom two or more otherwise equivalent donors were identified. All participants provided written informed consent for participation in this study.
Study Oversight
Protocols were approved by the institutional review boards of the National Institutes of Health Office for Human Research Protections, the Fred Hutchinson Cancer Research Center, and the National Marrow Donor Program. The funding agencies had no role in study design, data collection and analysis, the decision to submit the manuscript for publication, or the preparation of the manuscript.
Genotyping and Linkage Analysis
We genotyped rs9277534 using a TaqMan PCR assay (Table S4 in the Supplementary Appendix).21,22 The linkage of rs9277534 to HLA-DPB1 was determined in 3505 persons, of whom 1893 were homozygous (i.e., had two copies of the same allele) at rs9277534, HLA-DPB1, or both, providing unambiguous haplotypes. A total of three unique haplotypes were found among the 1893 homozygous persons. The three haplotypes were validated in 18 reference cell lines by first separating chromosomes at rs9277534 and rs2281389 and then genotyping HLA-DPB1, rs9277534, and rs2281389 on each haplotype,23 a process known as phasing (Fig. 2, and the Materials and Methods section in the Supplementary Appendix). These phased haplotypes showed that the combination of two haplotypes in persons who are heterozygous (i.e., have two different alleles) at both rs9277534 and HLA-DPB1 can be accurately inferred.
All 1441 HLA-DPB1–mismatched transplant recipients included in the analysis of clinical outcomes were heterozygous at HLA-DPB1. Of these recipients, 796 were also heterozygous at rs9277534, and the linkage of rs9277534 to HLA-DPB1 was inferred as described above. We genotyped rs9277534 in 833 donors with available DNA samples, and we inferred the genotype from the HLA-DPB1 type in 604 donors without available DNA samples. The linkage of rs9277534 to HLA-DPB1 was inferred in the 588 HLA-A,B,C,DRB1,DQB1,DPB1–matched pairs.
HLA-DPB1 Expression
HLA-DPB1 expression was determined in 49 persons with rs9277534AA and 32 with rs9277534GG by means of a quantitative PCR assay (see the Materials and Methods section in the Supplementary Appendix).24
Statistical Analysis
The primary end point of this study was acute GVHD (assessed as grade II, III, or IV and as grade III or IV); secondary end points were chronic GVHD, relapse, death not preceded by relapse, and death from any cause. For each of these end points, Cox regression models were fit to compare the risk of GVHD among recipients whose mismatched HLA-DPB1 allele was linked to rs9277534G with the risk among recipients whose mismatched HLA-DPB1 allele was linked to rs9277534A. All regression models were adjusted for age, source of stem cells, disease severity, T-cell depletion (yes or no), transplantation regimen, sex of recipient and donor (male–male, male–female, female–male, or female–female), cytomegalovirus serostatus, and year of transplantation. P values from Cox regression were obtained with the Wald test. All reported P values are two-sided.
Results
Association of rs9277534 and HLA-DPB1 Alleles
Among 537 samples that were homozygous for HLA-DPB1 and rs9277534, HLA-DPB1*02, 04, and 17 were associated with rs9277534AA, whereas HLA-DPB1*01, 03, 05, 06, 10, 11, 13, 15, 16, and 19 were linked to rs9277534GG. Likewise, when the number of copies (0, 1, or 2) of each HLA-DPB1 allele was examined in all samples, HLA-DPB1*02, 04, and 17 alleles were associated with rs9277534A, and HLA-DPB1*01, 03, 05, 06, 09, 10, 11, 13, 14, 15, 16, 19, and 20 were associated with rs9277534G (P<0.001 for both).
To confirm that the HLA-DPB1–rs9277534 linkage can be predicted with high accuracy in persons who are heterozygous at HLA-DPB1, rs9277534, rs2281389, or a combination of these markers, we physically separated chromosomes in 18 cell lines (see the Materials and Methods section in the Supplementary Appendix) and linked rs9277534 with its HLA-DPB1 allele and rs2281389 (Fig. 2). Phasing produced haplotypes identical to the predicted linkage in the 3505 persons in whom we genotyped rs9277534. On the basis of unphased and phased samples, 3254 rs9277534A-positive HLA-DPB1 haplotypes and 254 rs9277534G-positive haplotypes were identified (Table S5 in the Supplementary Appendix).
Effect of rs9277534 Allele on HLA-DPB1 Expression
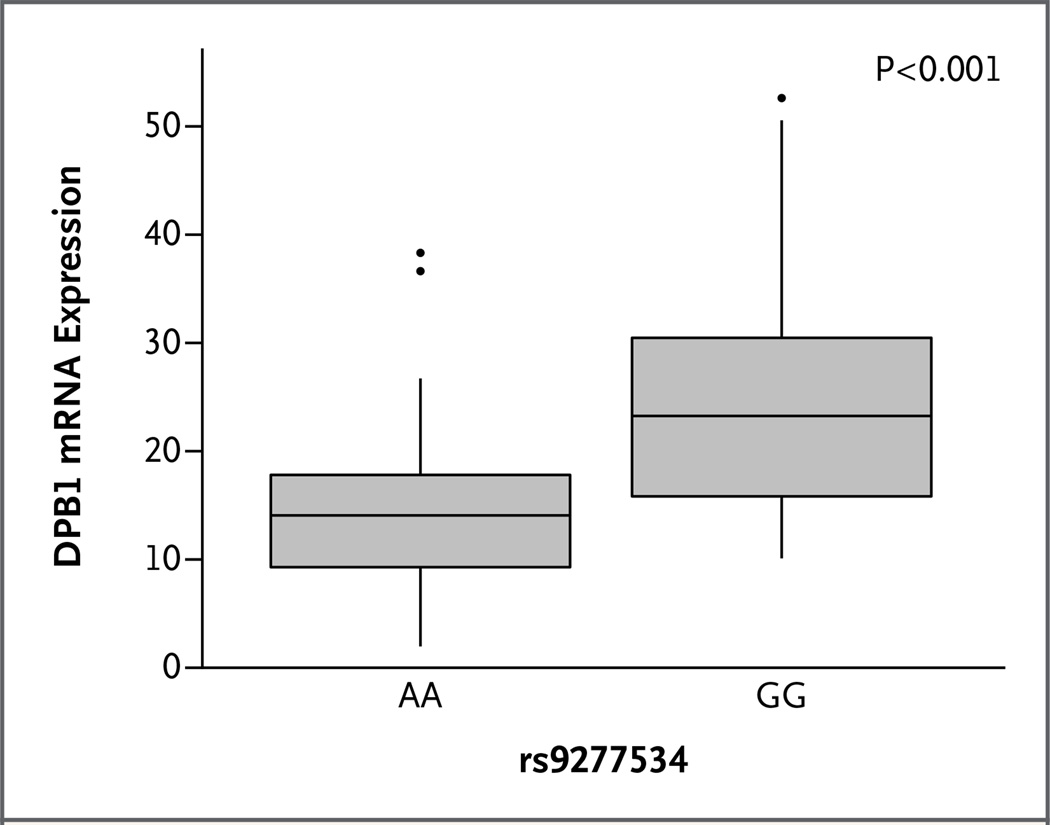
The mean normalized HLA-DPB1 expression was 14.62 normalized individual data points for rs9277534AA samples, as compared with 24.95 for rs9277534GG samples (mean difference in expression, −10.33; 95% confidence interval [CI], −14.95 to −5.70; P<0.001) (Fig. 3). These results validate previous observations that rs9277534A-linked HLA-DPB1 alleles are expressed, on average, at lower levels than are rs9277534G-linked alleles.20
Figure 3. Correlation of HLA-DPB1 Expression with the rs9277534 Allele in the 3′ Untranslated Region of HLA-DPB1.
HLA-DPB1 messenger RNA (mRNA) expression was determined by means of a quantitative polymerase-chain-reaction assay (TaqMan Gene Expression assay) in 81 persons: 49 with the rs9277534AA genotype and 32 with the rs9277534GG genotype. The data are presented as normalized individual data points (2−deltaCt [delta Ct = Ct gene of interest − Ct endogenous control]) in box-and-whisker plots. The horizontal line within each box indicates the median expression value; vertical lines indicate the smallest and the largest non-outliers in these data.
Association between Clinical Outcome and rs9277534 Allele
Transplant recipients with rs9277534G-linked HLA-DPB1 mismatches had higher risks of grade II, III, or IV acute GVHD and of grade III or IV disease (Table S6 in the Supplementary Appendix), as compared with recipients who had rs9277534A-linked mismatches, with hazard ratios of 1.32 (95% CI, 1.13 to 1.55; P<0.001) and 1.34 (95% CI, 1.08 to 1.68; P =0.009), respectively. Recipients with rs9277534G-linked mismatches also had a lower risk of relapse, as compared with recipients who had rs9277534A-linked mismatches (hazard ratio, 0.80; 95% CI, 0.64 to 0.99; P = 0.04), which is consistent with a graft-versus-leukemia effect.25 The higher risk of GVHD was balanced by the lower risk of relapse, leading to similar overall mortality (hazard ratio for death from any cause with rs9277534G-linked mismatches, 1.03; 95% CI, 0.90 to 1.18; P = 0.70) (Table S6 in the Supplementary Appendix). Among the 753 recipients who died without having had recurrent disease, 31.6% had infection, 29.3% had organ toxicity or failure, 19.8% had GVHD, and 19.3% died from other causes.
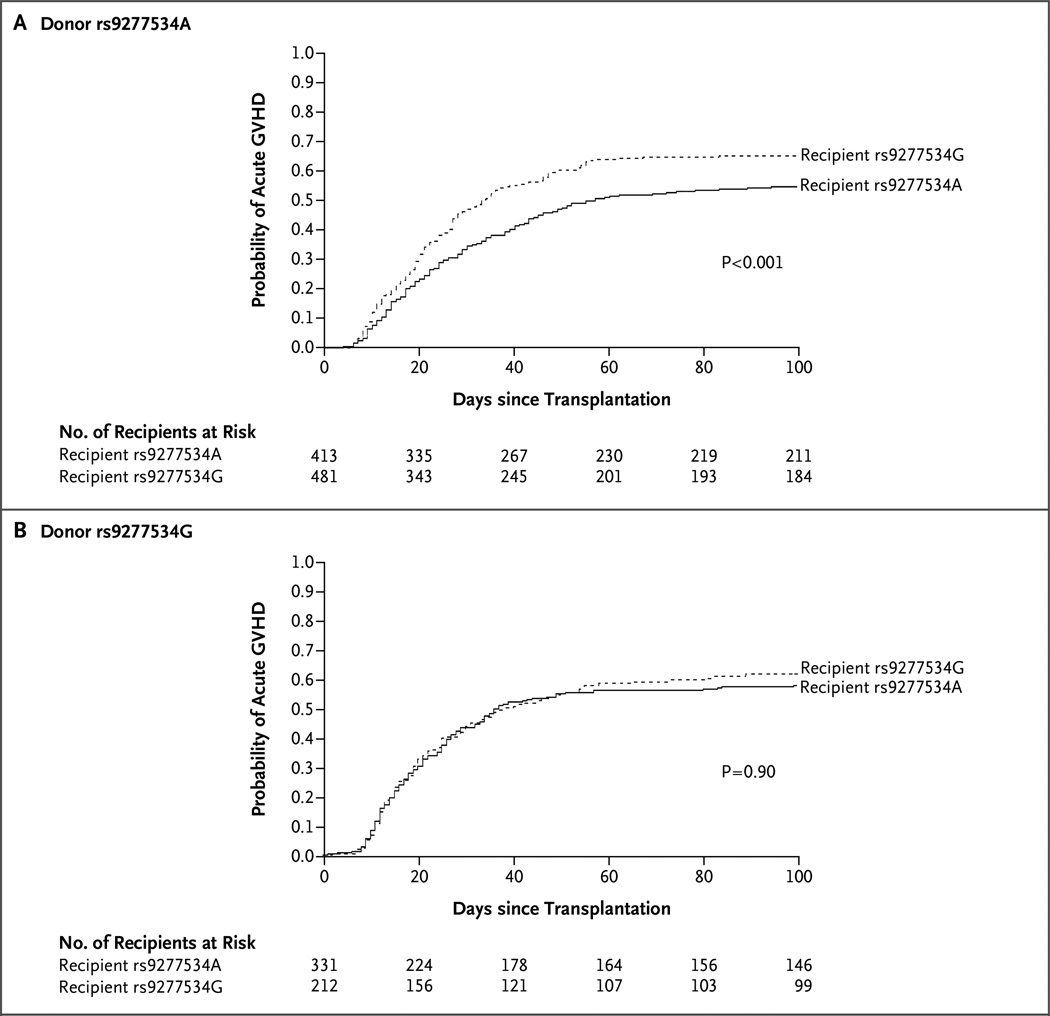
However, tests of interaction between recipient and donor rs9277534 suggested that the mismatches linked to rs9277534G in recipients have a deleterious effect on the risk of GVHD that differed according to the rs9277534 linkage of the donor’s HLA-DPB1 (Table 1). We therefore compared rs9277534G-linked mismatches in recipients with rs9277534A-linked mismatches in recipients separately for recipients whose donors had rs9277534G-linked HLA-DPB1 and recipients whose donors had rs9277534A-linked HLA-DPB1 (Table 1). We found that rs9277534G-linked mismatches (high HLA-DPB1 expression) had a similar detrimental effect with respect to the risk of grade II, III, or IV acute GVHD and the risk of grade III or IV disease, but this effect was observed only when the mismatch was linked to rs9277534A in the donor (low HLA-DPB1 expression) (Fig. 4 and Table 1). Among recipients of transplants from donors with rs9277534A-linked HLA-DPB1, recipients with rs9277534G had higher risks of grade II, III, or IV acute GVHD and of grade III or IV disease than did recipients with rs9277534A (hazard ratio for grade II, III, or IV GVHD, 1.54; 95% CI, 1.25 to 1.89; P<0.001 for interaction; and hazard ratio for grade III or IV disease, 1.50; 95% CI, 1.12 to 2.01; P =0.007 for interaction). Secondary outcomes are shown in Table 1.
Table 1.
Hazard Ratios for Outcomes of HLA-DPB1 Mismatches in Transplant Recipients, According to the rs9277534 Allele Linked to the Mismatch*
| Clinical End Point | HLA-DPB1-linked rs9277534 Allele |
G Allele vs. A Allele in Recipient | P Value for Interaction† |
||
|---|---|---|---|---|---|
| Donor | Recipient | Hazard Ratio (95% CI) | P Value | ||
| Grade II, III, or IV acute GVHD |
A | G A |
1.54 (1.25–1.89) | <0.001 | 0.01 |
| G | G A |
1.01 (0.78–1.32) | 0.90 | ||
| Grade III or IV acute GVHD |
A | G A |
1.50(1.12–2.01) | 0.007 | 0.28 |
| G | G A |
1.15 (0.80–1.66) | 0.43 | ||
| Chronic GVHD | A | G A |
1.09 (0.89–1.34) | 0.39 | 0.66 |
| G | G A |
1.01 (0.78–1.32) | 0.92 | ||
| Relapse | A | G A |
0.89(0.68–1.17) | 0.40 | 0.15 |
| G | G A |
0.63 (0.43–0.94) | 0.02 | ||
| Death not preceded by relapse |
A | G A |
1.25 (1.00–1.57) | 0.05 | 0.30 |
| G | G A |
1.04 (0.79–1.37) | 0.80 | ||
| Death from any cause | A | G A |
1.13 (0.95–1.35) | 0.16 | 0.13 |
| G | G A |
0.91 (0.72–1.14) | 0.39 | ||
The rs9277534 allele linked to the donor’s and recipient’s HLA-DPB1 mismatch was used to define four recipient-donor groups: A-A (413 recipients and donors), A-G (481), G-A (331), and G-G (212). All hazard ratios are for the comparison of recipients who had rs9277534G-linked HLA-DPB1 mismatches with recipients who had rs9277534A-linked mismatches. For each end point, data are based on the number of recipients (and donors) for whom complete clinical information was available for the end point of interest. GVHD denotes graft-versus-host disease.
Tests for statistical interaction were performed to determine whether the effect of rs9277534G or rs9277534A in a transplant recipient was the same whether the donor had rs9277534G or rs9277534A. Thus, the P values provide information about whether the hazard ratio for a deleterious effect of HLA-DPB1 mismatching in a recipient with rs9277534G or rs9277534A when the donor had rs9277534G was the same as the hazard ratio when the donor had rs9277534A.
Figure 4. Probability of Grade II, III, or IV Acute Graft-versus-Host Disease.
The probability of grade II, III, or IV acute graft-versus-host disease (GVHD) is shown for transplant recipients with an rs9277534A-linked HLA-DPB1 mismatch (solid line) or an rs9277534G-linked HLA-DPB1 mismatch (dashed line) when the donor’s HLA-DPB1 was linked to rs9277534A (Panel A) or rs9277534G (Panel B). The effects of the mismatch were deleterious in recipients with rs9277534G-linked HLA-DPB1 (high-expression) only when the donor had rs9277534A-linked (low expression) HLA-DPB1. P values were estimated from multivariable regression models adjusted for age, source of stem cells, disease severity, T-cell depletion (yes or no), transplantation regimen, sex of recipient and donor (male–male, male-female, female–male, or female–female), cytomegalovirus serostatus, and year of transplantation.
In HLA-A,B,C,DRB1,DQB1,DPB1–matched transplantation, the presence of rs9277534G was also associated with a higher risk of acute GVHD than was the absence of rs9277534G (hazard ratio for 146 HLA-DPB1–matched rs9277534AG transplants vs. 420 rs9277534AA transplants, 1.86; 95% CI, 1.21 to 2.84; P = 0.004; and hazard ratio for 22 rs9277534GG transplants vs. 420 rs9277534AA transplants, 1.53; 95% CI, 0.55 to 4.25; P =0.41). These data suggest a schema in which minor histocompatibility antigens presented by highly expressed HLA molecules provoke robust graft-versus-host alloreactivity from the donor cells, as compared with HLA molecules expressed at low levels on the cell surface2 (Fig. 1B).
The rs2281389G allele is always observed on a haplotype with rs9277534G, yet rs2281389A can be linked to either rs9277534G or rs9277534A. Therefore, the presence of rs2281389A provides insufficient information for determining whether the haplotype encodes a high- or low-expression HLA-DPB1 allele. Among rs2281389A recipients of transplants from rs9277534A donors, recipients with rs9277534G-linked mismatches had a higher risk of GVHD than did recipients with rs9277534A-linked mismatches (hazard ratio for grade II, III, or IV acute GVHD, 1.60; 95% CI, 1.25 to 2.06; P<0.001; and hazard ratio for grade III or IV disease, 1.66; 95% CI, 1.16 to 2.35; P = 0.005), showing that rs9277534 is a better predictor of the risk of GVHD than is rs2281389, possibly because rs9277534 is a better marker of expression than is rs2281389.
Donor-derived T-cell clones isolated after graft rejection react against immunogenic T-cell epitopes of the recipient’s HLA-DPB19 and can be used to predict the immunogenic potential of mismatched HLA-DPB1 alleles between recipients and donors that are associated with a high risk of GVHD (T-cell epitope “nonpermissive”) or a low risk of GVHD (T-cell epitope “permissive”).16 We examined the rs9277534 alleles linked to the mismatched recipient–donor HLA-DPB1 alleles to assess whether outcomes after epitope-permissive and epitope-nonpermissive mismatched transplantation are associated with rs9277534. Among the transplant pairs for which the HLA-DPB1 mismatch was linked to rs9277534A in both recipient and donor (the A-A group), 90.7% of the mismatches were also epitope-permissive. In the G-G group, 31% of the mismatches were epitope-permissive, showing some correlation between permissiveness indicated by T-cell epitope and permissiveness indicated by rs9277534. However, among epitope-permissive mismatches, the risk of grade III or IV acute GVHD was higher for recipients in the A-G, G-A, and G-G (recipient–donor) groups than for those in the A-A group (hazard ratio, 1.50, 1.38, and 1.53, respectively). Likewise, among mismatches that were epitope-nonpermissive, the hazard ratios for recipients in the A-G, G-A, and G-G rs9277534 groups were 1.03, 1.72, and 1.70, respectively. These data show that after isolation of the T-cell epitope effect, rs9277534 contributes additional information about the risk of GVHD. However, within the A-A, A-G, G-A, and G-G recipient–donor mismatch groups, epitope-nonpermissive mismatches were not associated with a higher risk of grade III or IV acute GVHD, as compared with epitope-permissive mismatches (hazard ratio, 0.91, 0.61, 1.04, and 0.98, respectively), suggesting that the T-cell epitope does not contribute additional information about rs9277534 status.
Probability of Finding Donors with a Permissible HLA-DPB1 Mismatch
We found that a mismatch for the transplant recipient’s rs9277534G-linked HLA-DPB1 was associated with a high risk of acute GVHD. Among recipients with one or two rs9277534G-linked HLA-DPB1 alleles, we estimated the likelihood of avoiding a mismatch for the G-linked allotype in recipients with rs9277534AG (55 recipients) or rs9277534GG (15 recipients) and their donors for whom DNA samples were available. Among the 55 recipients with rs9277534AG, 30 (55%) had acceptable donors — that is, either an HLA-DPB1–matched donor (17 recipients) or a donor mismatched with the recipient’s rs9277534A-linked HLA-DPB1 (13 recipients); the other 25 recipients (45%) had only donors with a mismatch for the recipient’s rs9277534G-linked HLA-DPB1. None of the 15 recipients with rs9277534GG had HLA-DPB1–matched donors. Hence, prospective genotyping of donors for recipients with rs9277534AA or rs9277534GG provides a means to identify matches and avoid mismatching for rs9277534G-linked HLA-DPB1. Recipients with rs9277534AA have good donor choices. Of the 76 recipients with rs9277534AA, 38 (50%) had donors who were either completely matched (HLA 12/12 match, 25 recipients) or matched for graft-versus-host allorecognition (13 recipients), 32 (42%) had donors who were mismatched for one HLA-DPB1 allele, and 6 (8%) had only donors with two HLA-DPB1 mismatches.
Discussion
HLA-DPB1 mismatching occurs for more than 80% of otherwise HLA-matched transplant recipients and unrelated donors and contributes to substantial morbidity and mortality associated with GVHD.13,14 The potential mechanisms that give rise to GVHD in the context of HLA-DPB1 mismatching are unknown. The current study shows that an HLA-DPB1 mismatch is not sufficient by itself to lead to GVHD but that it is immunogenic when the recipient has a high-expression allele and the donor has a low-expression allele. In our transplantation model, the classic notion that genotype influences phenotype is reflected by the effects of a genetic regulatory variant on the physiological graft-versus-host response. The model suggests that the HLA-DPB1–rs9277534 haplotype is a marker for GVHD, with important implications for lowering the risk of GVHD and for leveraging antileukemic effects in transplant recipients.26,27 The collective experience from the HIV–AIDS, Crohn’s disease, hepatitis B, and transplantation models firmly establishes the importance of HLA expression in immunologically mediated diseases.17–20
A key to the successful elucidation of rs9277534-associated GVHD was the development of a direct phasing method for determining the physical linkage between rs2281389, rs9277534, and HLA-DPB1 exon 2 that provided new insight into haplotype structure in this region of the human major histocompatibility complex (MHC). GVHD is a physiological consequence of rs9277534-DPB1 haplotypes and, together with the difference in mean HLA-DPB1 expression between the rs9277534G and rs9277534A alleles, aligns with functional data that implicate recognition of HLA-DPB1 by CD4+ T cells in GVHD and relapse, notably in the context of induced expression of MHC class II by viral infections after transplantation.28,29 Furthermore, the observation of an increased risk of GVHD among recipients of HLA-DPB1–matched transplants with rs9277534G-linked HLA-DPB1 alleles provides supporting evidence for enhanced donor recognition of minor histocompatibility antigens presented by the recipient’s HLA-DP as a clinically relevant source of variation.
Analysis of DNA samples from transplant recipients and their donors provided an understanding of the potential role of pretransplantation rs9277534 screening in minimizing the risk of GVHD. Our study shows that prospective selection of donors with low-expression mismatches is entirely feasible and can be used to substantially reduce the number of transplantations involving high-risk mismatches. Our data provide new information on the role of HLA-DPB1 expression in transplantation-associated risks that can be used to guide the selection of donors for future transplant recipients in order to minimize the risk of acute GVHD. We discovered that if rs9277534 genotypes had been known at the time of the search for an unrelated donor, almost 55% of recipients with rs9277534AG would have had suitable donors, and mismatching for their G-linked HLA-DPB1 allele could have been avoided. Fully HLA-compatible donors remain the donors of choice for recipients with rs9277534GG. Taken together, these data support rs9277534 and HLA-DPB1 genotyping for donor selection.
The importance of levels of HLA-DP and HLA-C expression in influencing the strength of alloimmune responses in transplant recipients has direct implications for transplants mismatched at multiple HLA loci. Synergistic effects of multilocus HLA mismatching on transplantation outcomes have been well described.3,4,8,13,15 Whether mismatching for multiple low-expression HLA alleles carries a lower risk of GVHD than mismatching for many high-expression alleles remains an important question for future studies.
Supplementary Material
Acknowledgments
Supported by grants from the National Institutes of Health (AI069197, to Drs. Petersdorf, Malkki, Gooley, Haagenson, Horowitz, Spellman, and Wang; CA100019, to Drs. Petersdorf, Malkki, and Gooley; CA18029, to Drs. Petersdorf, Malkki, Gooley, and Stevenson; CA76518, to Dr. Horowitz; HHSH234200637015C, to Drs. Haagenson, Horowitz, and Spellman; U24-CA76518, to Drs. Haagenson, Horowitz, Spellman, and Wang; and 5U01HL069294, to Drs. Haagenson, Horowitz, Spellman, and Wang) and from the Office of Naval Research (N00014-12-1-0142, to Drs. Haagenson, Horowitz, and Spellman; N00014-13-1-0039, to Drs. Haagenson, Horowitz, and Spellman; and N00014-14-1-0028, to Dr. Horowitz) and by a contract from the Frederick National Laboratory for Cancer Research, National Institutes of Health (HHSN261200800001E).
We thank Mark Gatterman and Dawn Miller for technical assistance and Gary Schoch for database support.
Footnotes
The views expressed in this article do not necessarily reflect the official policy or position of the National Institutes of Health, the Department of the Navy, the Department of Defense, or any other agency of the U.S. government.
Disclosure forms provided by the authors are available with the full text of this article at NEJM.org.
References
- 1.Cutler C, Antin JH. Manifestations and treatment of acute graft-versus-host disease. In: Appelbaum FR, Forman SJ, Negrin RS, Blume KG, editors. Thomas’ hematopoietic cell transplantation. 4th ed. Chichester, United Kingdom: Wiley-Black-well; 2009. pp. 1287–1303. [Google Scholar]
- 2.Martin PJ. Increased disparity for minor histocompatibility antigens as a potential cause of increased GVHD risk in marrow transplantation from unrelated donors compared with related donors. Bone Marrow Transplant. 1991;8:217–223. [PubMed] [Google Scholar]
- 3.Sasazuki T, Juji T, Morishima Y, et al. Effect of matching of class I HLA alleles on clinical outcome after transplantation of hematopoietic stem cells from an unrelated donor. N Engl J Med. 1998;339:1177–1185. doi: 10.1056/NEJM199810223391701. [DOI] [PubMed] [Google Scholar]
- 4.Lee SJ, Klein J, Haagenson M, et al. High-resolution donor-recipient HLA matching contributes to the success of unrelated donor marrow transplantation. Blood. 2007;110:4576–4583. doi: 10.1182/blood-2007-06-097386. [DOI] [PubMed] [Google Scholar]
- 5.Kawase T, Morishima Y, Matsuo K, et al. High-risk HLA allele mismatch combinations responsible for severe acute graft-versus-host disease and implication for its molecular mechanism. Blood. 2007;110:2235–141. doi: 10.1182/blood-2007-02-072405. [DOI] [PubMed] [Google Scholar]
- 6.Mickelson E, Petersdorf EW. Histocompatibility. In: Appelbaum FR, Forman SJ, Negrin RS, Blume KG, editors. Thomas’ hematopoietic cell transplantation. 4th ed. Chichester, United Kingdom: Wiley-Black-well; 2009. pp. 145–162. [Google Scholar]
- 7.Pidala J, Lee SJ, Ahn KW, et al. Non-permissive HLA-DPB1 mismatch increases mortality after myeloablative unrelated allogeneic hematopoietic cell transplantation. Blood. 2014;124:2596–2606. doi: 10.1182/blood-2014-05-576041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Petersdorf EW, Hansen JA, Martin PJ, et al. Major-histocompatibility-complex class I alleles and antigens in hemato-poietic-cell transplantation. N Engl J Med. 2001;345:1794–1800. doi: 10.1056/NEJMoa011826. [DOI] [PubMed] [Google Scholar]
- 9.Zino E, Frumento G, Marktel S, et al. A T-cell epitope encoded by a subset of HLA-DPB1 alleles determines nonpermis-sive mismatches for hematologic stem cell transplantation. Blood. 2004;103:1417–1424. doi: 10.1182/blood-2003-04-1279. [DOI] [PubMed] [Google Scholar]
- 10.Van Rood JJ, van Leeuwen A. Major and minor histocompatibility systems in man and their importance in bone marrow transplantation. In: Dupont B, Good RA, editors. Immunobiology of bone marrow transplantation. New York: Gruen & Strat-ton; 1976. pp. 103–110. [PubMed] [Google Scholar]
- 11.Edwards JA, Durant BM, Jones DB, Evans PR, Smith JL. Differential expression of HLA class II antigens in fetal human spleen: relationship of HLA-DP, DQ, and DR to immunoglobulin expression. J Immunol. 1986;137:490–497. [PubMed] [Google Scholar]
- 12.Guardiola J, Maffei A. Control of MHC class II gene expression in autoimmune, infectious, and neoplastic diseases. Crit Rev Immunol. 1993;13:247–268. [PubMed] [Google Scholar]
- 13.Petersdorf EW, Gooley T, Malkki M, et al. The biological significance of HLA-DP gene variation in haematopoietic cell transplantation. Br J Haematol. 2001;112:988–994. doi: 10.1046/j.1365-2141.2001.02655.x. [DOI] [PubMed] [Google Scholar]
- 14.Shaw BE, Mayor NP, Russell NH, et al. Diverging effects of HLA-DPB1 matching status on outcome following unrelated donor transplantation depending on disease stage and the degree of matching for other HLA alleles. Leukemia. 2010;24:58–65. doi: 10.1038/leu.2009.239. [DOI] [PubMed] [Google Scholar]
- 15.Fernández-Viña MA, Klein JP, Haagenson M, et al. Multiple mismatches at the low expression HLA loci DP, DQ, and DRB3/4/5 associate with adverse outcomes in hematopoietic stem cell transplantation. Blood. 2013;121:4603–4610. doi: 10.1182/blood-2013-02-481945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fleischhauer K, Shaw BE, Gooley T, et al. Effect of T-cell-epitope matching at HLA-DPB1 in recipients of unrelated-donor haemopoietic-cell transplantation: a retrospective study. Lancet Oncol. 2012;13:366–374. doi: 10.1016/S1470-2045(12)70004-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Apps R, Qi Y, Carlson JM, et al. Influence of HLA-C expression level on HIV control. Science. 2013;340:87–91. doi: 10.1126/science.1232685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Petersdorf EW, Gooley TA, Malkki M, et al. HLA-C expression levels define permissible mismatches in hematopoietic cell transplantation. Blood. 2014;124:3996–4003. doi: 10.1182/blood-2014-09-599969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kamatani Y, Wattanapokayakit S, Ochi H, et al. A genome-wide association study identifies variants in the HLA-DP locus associated with chronic hepatitis B in Asians. Nat Genet. 2009;41:591–595. doi: 10.1038/ng.348. [DOI] [PubMed] [Google Scholar]
- 20.Thomas R, Thio CL, Apps R, et al. A novel variant marking HLA-DP expression levels predicts recovery from hepatitis B virus infection. J Virol. 2012;86:6979–6985. doi: 10.1128/JVI.00406-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Petersdorf EW, Malkki M, Gooley TA, et al. MHC-resident variation affects risks after unrelated donor hematopoietic cell transplantation. Sci Transl Med. 2012;4 doi: 10.1126/scitranslmed.3003974. ra101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Malkki M, Petersdorf EW. Genotyping of single nucleotide polymorphisms by 5’ nuclease allelic discrimination. Methods Mol Biol. 2012;882:173–182. doi: 10.1007/978-1-61779-842-9_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moran D, Morishima S, Malkki M, Petersdorf EW. Identification of the MICA*070 allele by sequencing and phasing. Hum Immunol. 2013;74:557–561. doi: 10.1016/j.humimm.2012.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 25.Horowitz MM, Gale RP, Sondel PM, et al. Graft-versus-leukemia reactions after bone marrow transplantation. Blood. 1990;75:555–562. [PubMed] [Google Scholar]
- 26.Thus KA, Ruizendaal MT, de Hoop TA, et al. Refinement of the definition of permissible HLA-DPB1 mismatches with predicted indirectly recognizable HLA-DPB1 epitopes. Biol Blood Marrow Transplant. 2014;20:1705–1710. doi: 10.1016/j.bbmt.2014.06.026. [DOI] [PubMed] [Google Scholar]
- 27.Rutten CE, van Luxemburg-Heijs SAP, Halkes CJM, et al. Patient HLA-DP-specific CD4+ T cells from HLA-DPB1-mismatched donor lymphocyte infusion can induce graft-versus-leukemia reactivity in the presence or absence of graft-versus-host disease. Biol Blood Marrow Transplant. 2013;19:40–48. doi: 10.1016/j.bbmt.2012.07.020. [DOI] [PubMed] [Google Scholar]
- 28.Rutten CE, van Luxemburg-Heijs SA, van der Meijden ED, et al. Both permissive and nonpermissive HLA-DPB1 mismatches can induce polyclonal HLA-DPB1 specific immune responses in vivo and in vitro. Blood. 2010;115:151–153. doi: 10.1182/blood-2009-10-249821. [DOI] [PubMed] [Google Scholar]
- 29.Stevanovic S, van Bergen CA, van Luxemburg-Heijs SA, et al. HLA class II upregulation during viral infection leads to HLA-DP-directed graft-versus-host disease after CD4+ donor lymphocyte infusion. Blood. 2013;122:1963–1973. doi: 10.1182/blood-2012-12-470872. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.