Abstract
Somatic mutations in the epidermal growth factor receptor (EGFR) kinase domain drive lung adenocarcinoma. We have previously identified MIG6, an inhibitor of ERBB signaling and a potential tumor suppressor, as a target for phosphorylation by mutant EGFRs. Here we demonstrate that Mig6 is a tumor suppressor for the initiation and progression of mutant EGFR-driven lung adenocarcinoma in mouse models. Mutant EGFR-induced lung tumor formation was accelerated in Mig6-deficient mice, even with Mig6 haploinsufficiency. We demonstrate that constitutive phosphorylation of MIG6 at Y394/395 in EGFR-mutant human lung adenocarcinoma cell lines is associated with an increased interaction of MIG6 with mutant EGFR, which may stabilize EGFR protein. MIG6 also fails to promote mutant EGFR degradation. We propose a model whereby increased tyrosine phosphorylation of MIG6 decreases its capacity to inhibit mutant EGFR. Nonetheless, the residual inhibition is sufficient for Mig6 to delay mutant EGFR-driven tumor initiation and progression in mouse models.
Keywords: Mig6, EGFR, Mice, Lung Adenocarcinoma, Recycling
Introduction
Lung cancer is the leading cause of cancer mortality in the United States, accounting for about 27% of cancer-related deaths. EGFR and KRAS are among the most commonly mutated genes associated with the initiation and maintenance of lung adenocarcinomas. The most prevalent EGFR mutations associated with lung cancer are two hotspot mutations, a Leucine to Arginine substitution at position 858 (L858R, 40–45%) and an in-frame deletion mutation eliminating the conserved sequence LREA in exon 19 (e.g. Del E746-A750, 45%) (1–4). These mutations render the EGFR protein-tyrosine kinase constitutively active. Lung adenocarcinomas harboring these mutations are sensitive to EGFR-directed tyrosine kinase inhibitors (TKIs), such as erlotinib and gefitinib. Unfortunately, patients undergoing TKI treatment eventually develop acquired resistance. A mutation in the gatekeeper residue, T790M, accounts for 50–60% of acquired drug resistance (5, 6). Other mechanisms of resistance to TKIs include MET amplification, with or without concomitant T790M mutation (7, 8), HER2 amplification (9), CRKL amplification (10), NF1 loss (11), small cell lung cancer (SCLC) transformation (12, 13), epithelial mesenchymal transformation (EMT) (14–16) and low frequency mutations in BRAF (17) and HER2 (18). It is therefore important to understand the signaling pathways activated downstream of mutant EGFRs in TKI-sensitive and resistant lung adenocarcinoma cells.
Aberrant EGFR signaling that leads to activation of downstream signaling components such as AKT and ERK is associated with increased cellular proliferation and development of cancer (19–21). Recently, several groups, including ours, have performed global phosphoproteomic profiling of lung adenocarcinoma tumor tissue from patients and in cell lines, particularly TKI-sensitive lung adenocarcinoma cell lines, and have identified a large number of sites that are tyrosine phosphorylated (22, 23). We previously employed stable isotope labeling with amino acids in cell culture (SILAC) and quantitative phosphoproteomics to elucidate the differences in use of phorphorylation targets of wild type and mutant EGFRs in isogenic human bronchial epithelial cells (24). One of the candidates that was hyper-phosphorylated on tyrosines in cells expressing mutant EGFRs was MIG6 (gene symbol ERRFI1, also known as RALT, Gene 33), an immediate early response gene that is induced by growth factors, including EGF and stress stimuli (25, 26). MIG6 functions as a negative feedback regulator of ERBB family members, including EGFR and ERBB2 (27). Ablation of Mig6 in mice leads to tumors of various tissues, including lung, implicating Mig6 as a potential tumor suppressor gene (28–30). Several studies have reported that Mig6 inhibits EGFR by blocking its kinase activity, as well as by promoting its degradation (29, 31, 32). It has also been demonstrated that MIG6 RNA is increased in EGFR mutant lung adenocarcinoma cell lines (33). These observations raise the questions as to whether MIG6 is a tumor suppressor for mutant EGFR-driven lung adenocarcinoma and, if so, how mutant EGFR induces lung adenocarcinomas in the presence of MIG6.
In this study we sought to establish whether Mig6 deficiency would accelerate tumorigenesis induced by the common mutant alleles of EGFR, thus demonstrating its tumor suppressive role. We generated doxycycline-inducible mutant EGFR transgenic mice on different Mig6 genetic backgrounds and demonstrate that Mig6 deficiency accelerates the initiation and progression of mutant EGFR-driven tumorigenesis in vivo. Mig6 also functions as a haploinsufficient tumor suppressor in this model. To further examine the mechanisms of tumor suppression by MIG6 and to elucidate how mutant EGFR can circumvent MIG6 function in human lung tumors, we studied the consequences of tyrosine phosphorylation of MIG6 in human cancer cell lines. Using global quantitative mass spectrometry-based phosphoproteomics we identified Y394/395 as constitutively phosphorylated sites on MIG6 in lung adenocarcinoma cells expressing mutant EGFRs; these sites are inhibited by erlotinib in TKI-sensitive lung adenocarcinoma cells but not in drug-resistant cells. Increased phosphorylation of MIG6 increases the interaction of mutant EGFRs and MIG6. However, contrary to its effects on wild-type EGFR, MIG6 does not promote degradation of mutant EGFR. We propose a model in which mutant EGFR may circumvent the tumor suppressor function of MIG6 by constitutively phosphorylating Y394/395. However, the attenuated inhibitory function of MIG6 in the context of mutant EGFRs is still sufficient to delay tumorigenesis in a mouse model of mutant EGFR-driven lung adenocarcinoma.
Results
Ablation of Mig6 accelerates formation of mutant EGFR-induced adenocarcinomas and decreases survival of transgenic mice expressing mutant EGFR
Tissue-specific knockout of Mig6 increases EGFR signaling and the proliferation of epithelial cells in mouse lungs, suggesting that Mig6 is essential for lung homeostasis (34). Deletion of Mig6 in mice also promotes adenomas and adenocarcinomas in the lung, gallbladder, and bile duct, albeit at low penetrance (30). However, the role of Mig6 in mutant EGFR-driven lung tumorigenesis has not been studied. To test this, we crossed Mig6 heterozygous mice (Mig6+/−) (30) with doxycycline-inducible mutant EGFR transgenic mice (tetO-EGFRmut) (35) and CCSPrtTA mice (36). The resulting tetO-EGFRmut/Mig6+/− and CCSPrtTA/Mig6+/− mice were further bred to generate transgenic mice with conditional, doxycycline-inducible expression of EGFRL858R or EGFRDel in type II lung epithelial cells in Mig6+/+, Mig6+/−, and Mig6−/− backgrounds. After induction of transgenic mutant EGFRs, we monitored mice for the appearance of lung tumors by serial magnetic resonance imaging (MRI). CC10rtTA/EGFRL858R/Mig6−/− mice developed tumors earlier than CC10rtTA/EGFRL858R/Mig6+/+ mice (Fig. 1A, Supplementary Fig. S1A). The same was true for CC10rtTA/EGFRDel/Mig6−/− mice (Supplementary Fig. S1B). The Mig6−/− mice carrying mutant EGFR transgenes were euthanized earlier than mice without EGFR transgenes because of progressive disease. The Mig6−/− mice without the EGFR transgene had to be euthanized between 3–6 months of age, not due to lung tumor formation, but because of osteoarthritis affecting food intake (data not shown). Although there were transgenic line-specific differences, histopathology of the tumors at the survival endpoint indicated a higher incidence of adenocarcinoma in Mig6−/− mice compared to Mig6+/+ mice (Fig. 1B and Supplementary Fig. S1C- Table). Lungs of CC10rtTA/EGFRL858R/Mig6+/+ mice showed only pulmonary adenomas or adenomas with infrequent adenocarcinomas. There were no signs of invasion. The surrounding alveolar compartment showed type II cellular hyperplasia and variable amounts of macrophages (Supplementary Fig. S1D; A–C). The neoplastic lesions induced by both EGFR mutants in Mig6+/− or Mig6−/− mice were more advanced with features of adenocarcinoma (Supplementary Fig. S1E; A–C). Lungs were often completely effaced with hyper- and dysplastic-alveolar type II epithelial cells and had intense infiltration of macrophages and other inflammatory cells. These mice also demonstrated marked abnormalities of the airway lining epithelium with Clara cell hyperplasia or dysplasias and proliferation at bronchioalveolar duct junctions of the terminal bronchioles (Supplementary Fig. S1E; D–E).
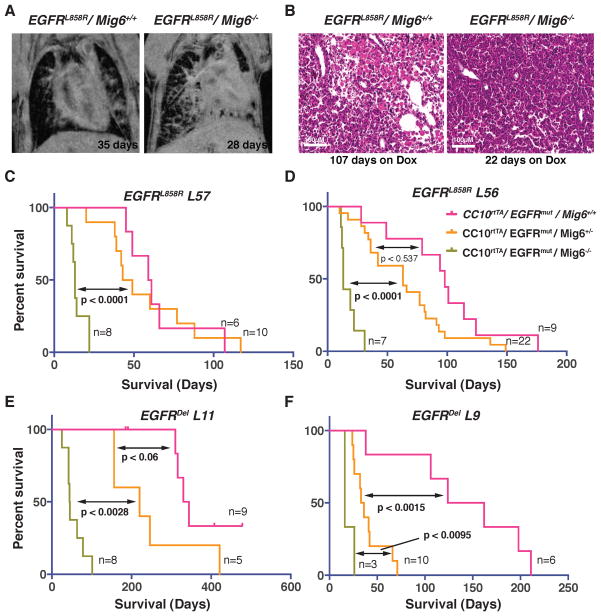
Figure 1. Accelerated initiation and progression of tumorigenesis and decreased overall survival in Mig6−/− mice.
(A) CC10rtTA/EGFRL858R/Mig6+/+ and CC10rtTA/EGFRL858R/Mig6−/− mice were treated with doxycycline to induce expression of the EGFRL858R transgene and serial MRI imaging was performed to monitor tumor initiation and progression. Representative images at 35 and 28 days of doxycycline induction show that there is more tumor burden at earlier time periods in CC10rtTA/EGFRL858R/Mig6−/− mice. (B) H&E staining of lung tissue sections from CC10rtTA/EGFRL858R/Mig6+/+ and CC10rtTA/EGFRL858R/Mig6−/− mice treated with doxycycline for 107 and 22 days at survival end point. (C–F) Kaplan-Meier survival curves of doxycycline-treated CC10rtTA/EGFRL858R/Mig6+/+, CC10rtTA/EGFRL858R/Mig6+/−, and CC10rtTA/EGFRL858R/Mig6−/− mice from line 57 (L57) (C) and line 56 (L56) (D), and CC10rtTA/EGFRDEL/Mig6+/+, CC10rtTA/EGFRDEL/Mig6+/−, and CC10rtTA/EGFRDEL/Mig6−/− mice from line 11 (L11) (E) and line 9 (L9) (F) show reduced survival of Mig6-deficient mice. The number (n) of mice used in each group for survival analysis is indicated in each graph. p value was calculated between two groups (double arrowhead) separately and shown in each panel, p<0.05 indicates a significant difference in survival.
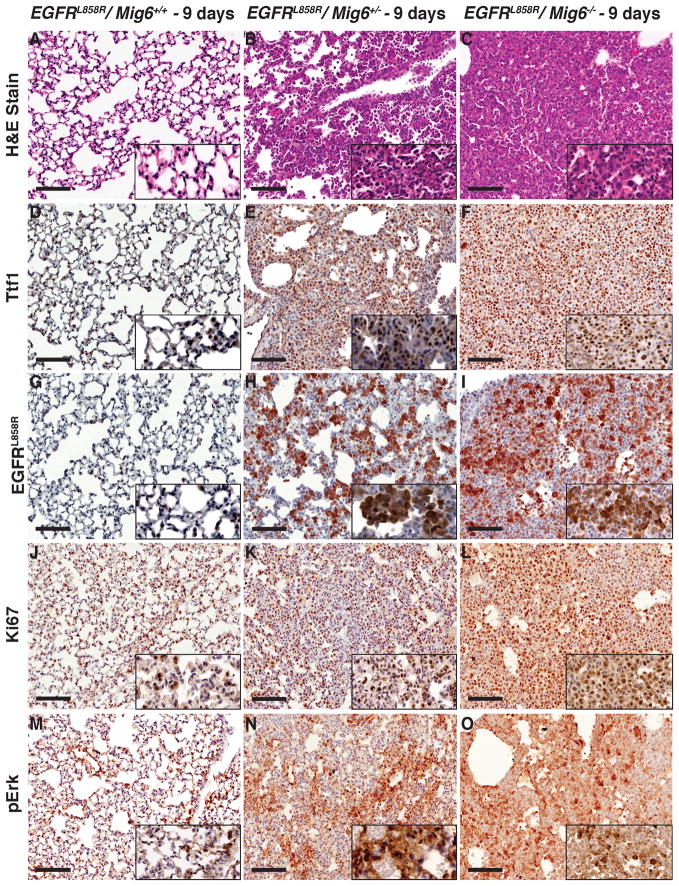
To examine the effect of Mig6 deletion on the survival of mice harboring mutant EGFRs (EGFRL858R and EGFRDel), we generated tumors by doxycycline induction of mutant EGFR transgenes and euthanized the mice when they displayed specific criteria related to lung tumor burden, such as labored breathing, weight loss, and failure to thrive. We performed Kaplan–Meier survival analyses in two separate transgenic lines of each of the mutant EGFRs in Mig6+/+, Mig6+/−, and Mig6−/− backgrounds (Fig. 1C–F). In all the tested lines of mutant EGFR mice, the survival time of Mig6−/− mice was significantly shorter than that of Mig6+/+ mice. The median survival of CCSP-rtTA/TetO-EGFRL858R/Mig6−/− mice after doxycycline induction was 13 days (both lines) compared to 60 (Line 57) to 100 days (Line 56) for CCSP-rtTA/TetO-EGFRL858R/Mig6+/+ mice (Fig. 1C–D). The median survival of CCSP-rtTA/TetO-EGFRDel/Mig6−/− mice after doxycycline induction was between 16 (Line 9) to 45 days (Line 11) compared to 143 (Line 9) to 337 days (Line 11) for CCSP-rtTA/TetO-EGFRDel/Mig6+/+ mice (Fig. 1E–F). Interestingly, the median survival of Mig6+/− mice expressing Del EGFR in EGFRDel L9 (Line 9) was only 34.5 days after doxycycline induction and was significantly shorter than the Mig6+/+ mice. A similar trend was observed in EGFRDel L11 (Line 11) mice, although the EGFRDel-driven tumors appeared later than EGFRL858R-driven tumors. Thus the survival of mice with EGFRDel-induced tumors in a Mig6+/− background appears more curtailed than in a Mig6+/+ background due to the longer latency of tumor induction by transgenic EGFRDel. We euthanized littermates with various genotypes 9 days after doxycycline induction to demonstrate the possible early appearance of tumors in Mig6 deficient mice. Although there was mild hyperplasia of type II cells in Mig6+/+ mice (Fig. 2A), Mig6+/− mice showed intermediate histopathology with increased type II hyperplasia, adenomas and adenocarcinomas (Fig. 2B, S2A; A–D), and Mig6−/− littermates exhibited dramatic effacement of lung alveoli with diffuse adenocarcinoma (Fig. 2C). We performed immunohistochemistry on lung tissue sections from Mig6+/+ Mig6+/− and Mig6−/− mice 9 days following doxycycline induction, using antibodies against Ttf1 (a Type-II epithelial cell marker and hence also a marker for lung cancer cells), EGFRL858R, the proliferation marker Ki67, and pErk. There was an increase in Ttf1, EGFRL858R, Ki67, and pErk immunoreactive cells in both Mig6+/− and Mig6−/− mice, compared to lungs of Mig6+/+ mice (Fig. 2D–O), confirming significantly increased lung tumor burden 9 days following doxycycline induction of mutant EGFRs in Mig6 deficient background.
Figure 2. Rapid progression of tumorigenesis at 9 days after doxycycline induction of mutant EGFR in Mig6+/− and Mig6−/− mice.
Immunohistochemistry of lung tissue sections from CC10rtTA/EGFRL858R/Mig6+/+, CC10rtTA/EGFRL858R/Mig6+/−, and CC10rtTA/EGFRL858R/Mig6−/− littermates after 9 days of doxycycline induction. Staining with H & E (A–C), Ttf1 (D–F), EGFRL858R (G–I), Ki67 (J–L), and pErk (M–O) shows significantly increased tumor burden in Mig6+/− and Mig6−/− mice. (Scale bar: 100 μm).
Loss of Mig6 cooperates with loss of Pten for endometrial cancer initiation and progression in a mouse model (37). This acceleration of tumorigenesis was shown to be due to the prevention of apoptosis. Normal mammary gland development in Mig6 null mice demonstrated that Mig6 promotes apoptosis in terminal end buds (38). Mig6 has also been shown to be an inducer of replicative or oncogene-induced senescence in fibroblasts (39–41). To further investigate whether mutant EGFR-driven tumor cells in Mig6−/− mice escape apoptosis and/or senescence, we performed TUNEL staining and senescence specific p19ARF staining (42, 43) of lung tissue sections from Mig6+/+, Mig6+/− and Mig6−/− mice 9 days after doxycycline induction. However, we did not observe any significant apoptosis or senescence in mutant EGFR-induced mouse lung tumorigenesis (Supplementary Fig. S2B; A–H), further confirming that the rapid progression of mutant EGFR-driven tumorigenesis is likely a result of increased proliferation as a result of increased Erk/Mapk pathway activation.
Since we observed that median survival of EGFRL858R/Mig6−/− mice is about thirteen days, we performed H&E staining and immunohistochemistry on lung tissue sections from Mig6+/+, Mig6+/− and Mig6−/− mice 7 days following doxycycline induction, using antibodies against Ttf1 and EGFRL858R. We also observed an increase in Ttf1 and EGFRL858R immunoreactive cells in Mig6−/− mice, relative to lungs of Mig6+/− and Mig6+/+ mice both in EGFRL858R Line 56 (Supplementary Fig. S3A; A–I) and EGFRL858R Line 57 (Supplementary Fig. S3B; A–I).
We then investigated whether there was a loss of heterozygosity (LOH) of the Mig6 gene in mutant EGFR-driven lung tumors developing in Mig6+/− mice. We performed quantitative genomic PCR for Mig6 copy number in lung tumor DNA and compared it to that of germline copy number in tail DNA. Our results showed that the Mig6 copy number did not change in lung tumors from a group of Mig6+/− mice that had to be euthanized early, arguing against LOH (Supplementary Fig. S4). Hence, Mig6 is a haploinsufficient tumor suppressor for initiation and progression of tumorigenesis at least in the context of expression of transgenic EGFRDel in mice.
Increased EGFR-MAPK signaling in Mig6-deficient tumors
To examine whether accelerated tumor growth in Mig6−/− mice is correlated with increased phosphorylation and activity of transgenic EGFR mutants, we measured the levels and phosphorylation status of mutant EGFR proteins in mouse lung tissue lysates and lung tissue sections by immunoblotting and immunohistochemistry. Early after doxycycline induction, cells reactive with Ttf1 and EGFRL858R antisera were significantly more abundant in lungs from Mig6−/− and Mig6+/− as opposed to Mig6+/+ mice (Fig. 2D–I), indicating early tumor initiation due to Mig6 deficiency. This was also associated with increased immunoreactivity to Ki67 and pErk, suggesting increased proliferation due to activation of the MAPK pathway (Fig. 2J–O). However, we found both by immunohistochemistry and Western blotting that EGFRL858R levels were paradoxically reduced in tumors collected at survival endpoint from Mig6-deficient animals (Fig. 3A–B). When adjacent tumor sections were stained with anti-Ttf1 and anti-EGFR antibodies, levels of EGFRL858R were significantly lower in Ttf1+ cells from EGFRL858R/Mig6−/− mice than in those from EGFRL858R/Mig6+/+ mice (Fig. 3B).
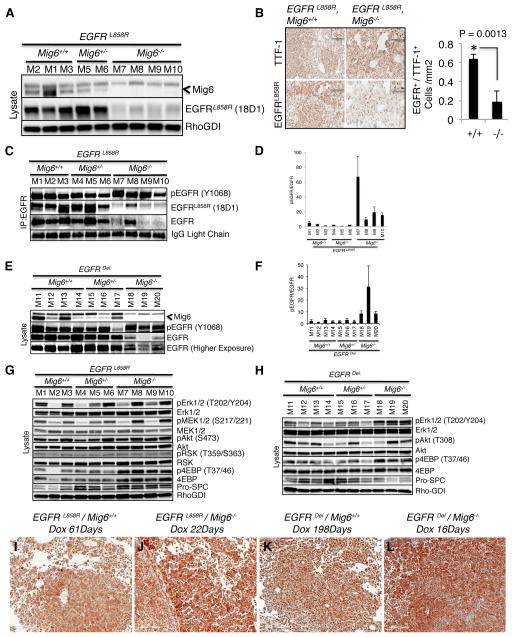
Figure 3. Expression of mutant EGFR and downstream signaling components in tumors of mice at survival endpoint.
(A) Immunoblot analysis of protein lysates from lungs of Mig6+/+, Mig6+/−, and Mig6−/− mice expressing transgenic doxycycline-induced EGFRL858R. Lysates from mice designated as M1–M10 (Supplemental Table 1) were probed with Mig6, EGFRL858R, and Rho-GDI (control) specific antibodies. (B) Immunohistochemical staining of tumor-bearing sections from the lungs of CC10rtTA/EGFRL858R/Mig6+/+ and CC10rtTA/EGFRL858R/Mig6−/− mice with EGFRL858R and Ttf-1 specific antibodies shows reduced expression of mutant EGFR in EGFRL858R/Mig6−/− mice. The intensities of L858R and Ttf-1 were quantified and shown in the graph as EGFR/Ttf-1 ratio. EGFR/Ttf-1 ratio was significantly lower in Mig6−/− mice compared to Mig6+/+ mice. (C) Immunoprecipitation of tumor-bearing mouse lung lysates using EGFR antibody followed by immunoblotting with pY1068-EGFR, EGFRL858R (clone 18D1), and EGFR to detect the expression of phospho-EGFR and mutant EGFR. (D) The band intensities for pEGFR (Y1068) and EGFR from the above experiment were quantified and plotted as pEGFR/EGFR ratios. The graph represents average value ± SE from four experiments. (E) Immunoblot analysis of tumor-bearing mouse lung lysates from Mig6+/+, Mig6+/−, and Mig6−/− mice expressing EGFRDel using specific antibodies against Mig6, pY1068-EGFR, and EGFR. (F) The band intensities for pEGFR and EGFR in the above experiment were quantified and plotted as pEGFR/EGFR ratio. The graph represents average value ± SE from four experiments. (G–H) Immunoblot analyses of lung extracts from Mig6+/+, Mig6+/−, and Mig6−/− mice expressing EGFRL858R (G) or EGFRDel (H) using both phospho-specific and total antibodies against signaling components downstream of EGFR. Expression analyses of lung epithelial cell specific pro-surfactant C (Pro-SPC) and Rho-GDI (loading control) were also performed. (I–L) Immunohistochemical analysis of pERK expression performed on tumor tissue sections from CC10rtTA/EGFRL858R/Mig6+/+ and CC10rtTA/EGFRDel/Mig6+/+ mice (I, K) and from CC10rtTA/EGFRL858R/Mig6−/− and CC10rtTA/EGFRDel/Mig6−/− mice (J, L) shows increased pErk immunoreactivity in Mig6−/− tumors.
To confirm this unexpected result, we immunoprecipitated EGFR from mouse tumor lysates and examined the immunoprecipitated EGFR with anti-pEGFR (Y1068), anti EGFRL858R, and anti-EGFR antibodies (Fig. 3C–D). Although the levels of EGFRL858R were reduced in established tumors, the levels of pEGFR remained high; the ratio of pEGFR to total EGFR was higher in tumors from Mig6−/− mice than in tumors from Mig6+/+ or Mig6+/− mice (Fig. 3D). We also observed decreased EGFRDel levels in tumors from EGFRDel/Mig6−/− mice compared with protein levels in tumors from EGFRDel/Mig6+/+ or EGFRDel/Mig6+/− mice at the survival endpoint (Fig. 3E). Likewise, as with Mig6−/− mice expressing the EGFRL858R transgene, the pEGFR/EGFR ratio was higher in the EGFRDel/Mig6−/− mice (Fig. 3F). Thus, the higher proportion of pEGFR appears to be sufficient for tumor maintenance in Mig6−/− mice.
We further measured components of the MAPK and PI3K signaling pathways in established tumors. Immunoblots of whole lung extracts from Mig6+/+, Mig6+/−, and Mig6−/− mice expressing either EGFRL858R or EGFRDel showed highly variable levels of pERK and pMEK. The level of pERK was higher in whole lung lysates from Mig6−/− mice than from Mig6+/+ or Mig6+/− mice, particularly those with the transgene encoding EGFRDel (Fig. 3G–H). Furthermore, immunohistochemistry demonstrated increased pERK reactivity in distinct, comparable areas of lung tumors in Mig6−/− mice compared with those from Mig6+/+ mice (Fig. 3I–L). Interestingly, we observed increased levels of both p-4EBP and 4EBP protein synthesis factor in Mig6−/− mice (Fig. 3G–H).
The reduced levels of mutant EGFR in established lung tumors at survival endpoint was surprising. We next determined whether this reduction in protein levels could be due to reduced levels of EGFR RNA. We examined whole lung lysates for levels of Mig6, transgenic human EGFR, mouse Egfr, rtTA, and mouse Sftpc RNAs in tumors obtained at the survival endpoint (Supplementary Fig. S5). We confirmed complete loss of Mig6 transcripts in Mig6−/− mice (Fig. S5A). Although the transcript level of endogenous mouse Egfr was the same regardless of Mig6 status, at this late time-point, the levels of the human EGFR transgene (EGFRL858R or EGFRDel) RNAs were slightly lower in tumors from Mig6−/− mice (Fig. S5B–C). This was accompanied by a slight decrease in transgenic rtTA RNA levels (Fig. S5D) but no significant change in levels of Sftpc RNA (Fig. S5E). We also examined transcript levels in lung lysates obtained early (9 or 14 days) after doxycycline induction of mutant EGFRs and found no significant differences in EGFRL858R or rtTA expression at these early time-points among mice with different Mig6 genotypes (Supplementary Fig. S6A–E).
Identification of TKI-regulated major sites for tyrosine phosphorylation of MIG6 by phosphoproteomic analysis
As part of a larger screen to identify proteins that may be regulated by tyrosine phosphorylation downstream of mutant EGFRs in lung adenocarcinoma cells, we employed a global phosphoproteomic approach in PC9 and H1975 cells that express TKI-sensitive EGFRDel(E746−A750) and TKI-resistant EGFRL858R+T790M, respectively. Although EGFR mutants are constitutively active in these cells, they can be further stimulated by EGF treatment. The cells were metabolically labeled with “light, medium, or heavy” isotope forms of arginine and lysine to measure the relative abundance of tyrosine phosphorylation in cells expressing the EGFR mutants in three states: serum-starved (“light”), EGF-stimulated (“medium”), or EGF-stimulated with prior erlotinib treatment (“heavy”) (Fig. 4A). We performed this triple-SILAC experiment to identify the changes in tyrosine phosphorylation by maximal activation of mutant EGFR signaling and inhibition by TKIs. We expected increased tyrosine phosphorylation upon EGF stimulation in both PC9 and H1975 cells. However, while the identified phospho-sites were expected to show decreased phosphorylation after erlotinib pre-treatment in the sensitive PC9 cells, they were predicted to remain unchanged in the erlotinib-resistant H1975 cells. Phospho-sites showing such changes in phosphorylation are the likely targets for phosphorylation by mutant EGFR. We identified a total of 309 and 488 unique tyrosine phospho-sites with quantitative phosphorylation data in PC9 and H1975 cells, respectively (data not shown). Based on this screen, a large number of phosphopeptides were identified as potential targets of mutant EGFRs.
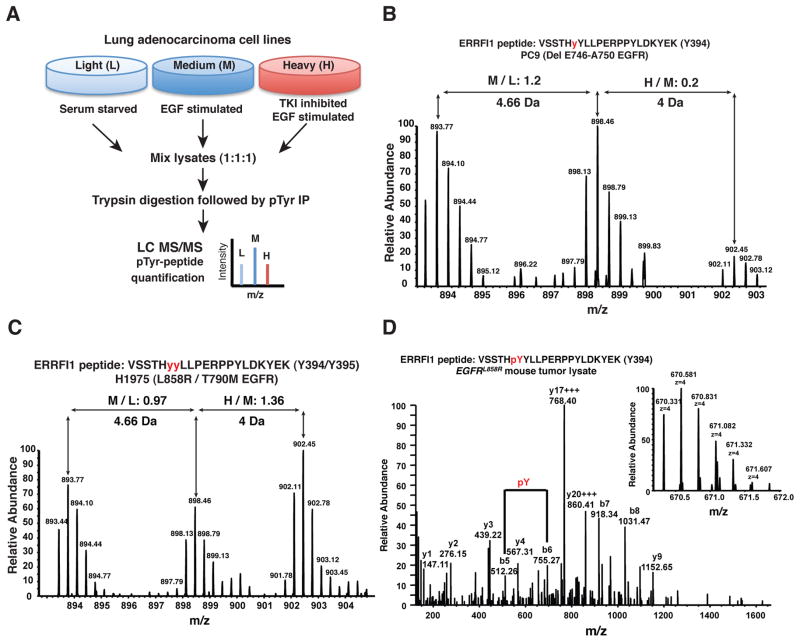
Figure 4. Identification of Y394/395 phosphorylation in human lung adenocarcinoma cells and mutant EGFR-driven mouse lung tumors in vivo by mass spectrometry.
(A–D) SILAC-based quantitative phosphoproteomics reveals constitutive phosphorylation of MIG6 at Y394/395 and significant reduction of phosphorylation upon erlotinib treatment in TKI-sensitive lung adenocarcinoma cells, but not in TKI resistant cells (A) Schematic of experimental design for SILAC-based quantitative phosphoproteome analysis of lung adenocarcinoma cells. (B) A representative MS spectrum of a MIG6/ERRFI1 peptide from PC9 cells containing tyrosine 394/395 residues indicates that phosphorylation of Mig6 (Y394) is not altered in the presence of EGF, but is significantly inhibited by erlotinib (TKI) treatment. The relative abundances of individual labeled peptides under different treatment conditions (EGF/serum starved:M/L, and EGF+Erlotinib/EGF:H/M) is quantified by SILAC ratios and is shown at the top of the spectrum. (C) A representative MS spectrum of the same peptide from H1975 cells demonstrates that tyrosine phosphorylation at Y394/395 is unchanged upon erlotinib. (D) Representative MS and MS/MS spectra of the Mig6 Y394/395-containing peptide identified from EGFRL858R mouse tumor lysate indicating in vivo phosphorylation of Mig6 at Y394.
MIG6 was identified as a target of mutant EGFR kinase in both lung adenocarcinoma cell lines examined. Y394 or both Y394/395 sites were found to be constitutively phosphorylated in these cells. A representative MS spectrum demonstrates that the relative abundance of the phosphorylated Y394-containing MIG6 peptide from PC9 cells decreases significantly upon erlotinib treatment (Fig. 4B). There was no significant change in phosphorylation at Y394/Y395 sites in H1975 cells upon erlotinib treatment (Fig. 4C). MS/MS analysis of this peptide revealed that the Y394 residue was phosphorylated in PC9 cells while both Y394 and Y395 were phosphorylated in H1975 cells (Supplementary Fig. S7). To validate whether Y394/395 is the major site of Mig6 phosphorylation in vivo, we carried out phosphoproteomic analysis of p-Tyr peptides from tumor lysates of EGFRL858R transgenic mice and identified the Mig6 phosphopeptide with phosphorylation at the Y394 residue (Fig. 4D). Using purified proteins for in vitro kinase assays, it has been shown that EGFR can phosphorylate MIG6 directly (44). Our results in this study and in HBECs expressing mutant EGFRs (24) provide in vivo evidence consistent with three conclusions: that MIG6 is a direct target of mutant EGFR; that Y394 and Y395 are sites constitutively phosphorylated by mutant EGFRs; and that erlotinib inhibits such phosphorylation in TKI-sensitive, but not resistant cells.
Phosphorylation of MIG6 at Y394 and Y395 residues promotes the interaction of MIG6 and EGFR
We have demonstrated that MIG6 residues Y394 and Y395 are constitutively phosphorylated in mutant EGFR-expressing lung adenocarcinoma cells. Using in vitro kinase assays with purified proteins, others have shown that EGFR can directly phosphorylate MIG6 on tyrosines (44). A 77 amino acid region of MIG6 segment 1 (aa336-412) has been shown to be necessary for EGFR inhibition (31). The structural determinants of MIG6 required for binding to EGFR have been previously mapped to an EGFR binding region (EBR), spanning residues 323 to 411 at the C-terminus of MIG6 protein (31, 45). The binding domain contains six tyrosine residues at positions 341, 358, 394, 395, 403, and 407, all of which have been shown to be phosphorylated in mass spectrometry-based experiments (data from Phosphosite Plus) (24, 46). The crystal structure of the EGFR kinase domain bound to part of segment 1 of Mig6 indicates that Y358 resides in the binding interface; and mutation of this residue to alanine (Y358A) disrupts binding (31). Frosi et al also showed that, unlike wild-type (WT) MIG6, the Y358A mutant failed to promote endocytosis of EGFR, indicating that the Y358 residue is important for MIG6 function (32). But Y358 is not a major site of phosphorylation in vivo. Moreover, the effect of phosphorylation of the major sites, Y394/395, on MIG6 function has not been studied in detail. Recently, using purified proteins and in vitro kinase assays, it has been shown that phosphorylation of MIG6-Y394 reduces the inhibitory function of MIG6 on the EGFR kinase (47). We postulate that phosphorylation of Y394/395 and other tyrosine sites within the EBR domain of MIG6 affects the binding of EGFR and MIG6 and regulation of EGFR kinase activity by MIG6.
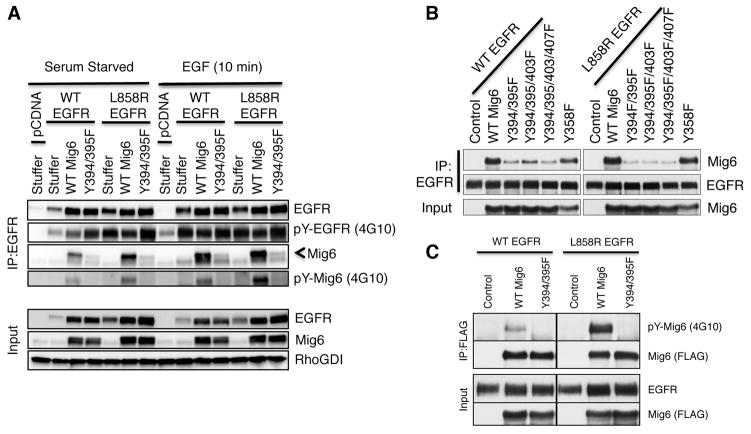
To determine whether phosphorylation of MIG6 at Y394 and Y395 is important for the interaction of MIG6 with EGFR, we replaced these tyrosines with phenylalanine to mimic unphosphorylated tyrosine. Expression vectors containing WT or mutant MIG6 cDNAs were cotransfected into HEK 293 cells with vectors containing WT EGFR, EGFRL858R, or an empty vector as control. After serum starvation for 18 h, some cultures were stimulated with EGF for 10 min. Cell extracts were examined for interacting proteins by immunoprecipitation with EGFR-specific antibodies, followed by Western blotting with second antibodies against MIG6 and other proteins (Fig. 5A). These studies indicate that MIG6 interacts with both WT and mutant EGFR. EGF stimulation increases the interaction of MIG6 with WT EGFR. Furthermore, MIG6 interacts more efficiently with L858R EGFR than WT EGFR in unstimulated cells, and the increase is associated with increased Tyr phosphorylation of MIG6. More importantly, mutation of residues Y394/Y395 to phenylalanine, abolishing phosphorylation of these sites, impairs the ability of MIG6 protein to bind both WT and mutant EGFR proteins. However, it is possible that Y-F mutants may affect hydrogen bonding interactions due to the loss of a hydroxyl in the phenylalanine and hence influence interactions beyond just the loss of phosphorylation.
Figure 5. Tyrosine phosphorylation of MIG6 Y394/395 is critical for its interaction with EGFR.
(A) HEK293 cells were transiently co-transfected with wild-type or L858R EGFR together with either wild-type or Y394/395F Mig6-expressing plasmids. Cells were serum starved for 18 h then treated with 100 ng/ml EGFR for 10 mins. Cell lysates were immunoprecipitated with anti-EGFR antibody and analyzed on Western immunoblots for total and phosphorylated (4G10) EGFR and MIG6 levels. (B–C) H322M cells, stably expressing wild-type or mutant MIG6 were transiently transfected with either wild-type or L858R EGFR. (B) Cell lysates were immunoprecipitated with EGFR and analyzed on Western immunoblots for EGFR and MIG6. (C) Cell lysates were immunoprecipitated with Flag antibody (MIG6-Flag) and analyzed on Western immunoblots for total (FLAG) and phosphorylated (4G10) MIG6. Input lysates were immunoblotted with EGFR, MIG6, or RhoGDI.
Since HEK293 cells express endogenous MIG6, we also performed co-immunoprecipitation studies in H322M cells, a lung adenocarcinoma cell line that does not express detectable levels of endogenous MIG6 because of a homozygous MIG6 nonsense mutation, E83Stop (30). H322M cells stably producing wild-type or Y-F MIG6 mutants were transiently transfected with either WT EGFR or EGFRL858R expression vectors and analyzed by EGFR co-immunoprecipitation assays (Fig. 5B). These experiments demonstrated a reduced interaction of MIG6 Y-F mutants with both WT EGFR and L858R EGFR except for the MIG6-Y358F mutant. To examine the overall tyrosine phosphorylation of MIG6, we immunoprecipitated MIG6 from lysates of H322M cells transfected with WT EGFR or L858R EGFR and probed the blots with pTyr and FLAG (Mig6) antibodies. There was increased tyrosine phosphorylation of MIG6 in L858R EGFR-expressing cells. Y394/395 were again demonstrated to be the predominant sites of tyrosine phosphorylation in MIG6 (Fig. 5C).
MIG6 does not promote degradation of mutant EGFR, and may also stabilize activated WT EGFR
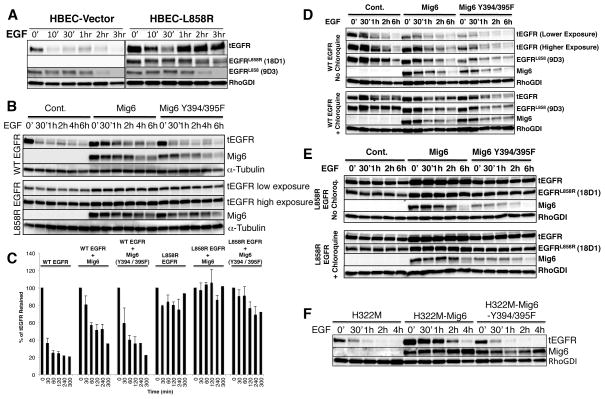
Previous studies have shown that EGF promotes activation-dependent endocytosis and degradation of EGFR, potentially regulating the duration of downstream signaling (48, 49). However, mutant EGFRs are ineffectively internalized (50, 51) and exhibit diminished down-regulation following ligand activation (52, 53). To assess differences in the degradation of WT EGFR and mutant EGFR, we first used isogenic human bronchial epithelial cells (HBECs). Lysates from HBECs treated with EGF and cycloheximide for 0 min, 10 min, 30 min, 1 h, 2 h, and 3 h were immunoblotted with antibodies that specifically recognize pan-EGFR (both wild-type and L858R EGFR), EGFRL858R, and EGFRL858 (epitope surrounding the L858 residue in WT EGFR) (Supplementary Fig. S8). Substantial degradation of WT EGFR occurred within two hours of EGF-induced activation of EGFR. In contrast, degradation of L858R EGFR was significantly reduced in HBECs expressing the mutant EGFR (Fig. 6A), indicating that L858R EGFR is more resistant to degradation than WT EGFR upon EGF stimulation.
Figure 6. Delayed degradation of EGFRL858R upon EGF stimulation in presence or absence of wild-type or Y394/395F mutant MIG6.
(A) HBECs with endogenous WT EGFR or stably transduced L858R EGFR expression were grown in serum-free medium for 18 h followed by cycloheximide (100 μM) treatment for 1 h to inhibit new protein synthesis, then treated with 100 ng/ml of EGF for indicated time points. RIPA cell lysates were immunoblotted with tEGFR (Clone 13), EGFRL858R and EGFRL858-specific antibodies to detect total, mutant, and corresponding wild-type EGFR. RhoGDI antibody was used for cell lysate loading controls. (B) HEK293 cells stably expressing wild-type or L858R EGFR alone or together with wild-type or Y394/395F MIG6 were serum starved for 18h and treated with 100 μM of cycloheximide for 1h followed by 100 ng/mL of EGF for indicated time points to induce receptor degradation. Lysates from treated cells were immunoblotted and probed with specific antibodies against tEGFR and Mig6. Alpha-tubulin specific antibody was used to probe cell lysates for loading control. (C) The band intensities for tEGFR in the above experiment were quantified and plotted as % of tEGFR retained following EGF treatment. The graph represents average value + SE from three experiments. (D–E) Stably transfected HEK293 cells, as described above, were serum starved for 18 h and treated with cycloheximide (100 μM). After 45 mins, chloroquine (100 μM) was added, then 15 mins later EGF (100 ng/ml) was added for the indicated time points. Lysates from stably transduced WT EGFR expressing cells were immunoblotted with EGFRL858-specific (D) and those expressing L858R EGFR were immunoblotted with EGFRL858R-specific antibodies (E). All lysates were also probed with tEGFR, Mig6 and RhoGDI-specific antibodies. (F) H322M cells stably expressing wild-type or Y394/395F MIG6 and endogenous wild-type EGFR were serum starved for 18 h and treated with 100 μM of cycloheximide for 1 h followed by 100 ng/mL of EGF for indicated time points to induce receptor degradation. Lysates from treated cells were immunoblotted with tEGFR and Mig6 antibodies and RhoGDI-specific antibody as a loading control.
MIG6 inhibits EGFR kinase activity and promotes WT EGFR trafficking to the degradation pathway (29, 32). Kinetic modeling based on EGFR endocytosis experiments performed on lung adenocarcinoma cells expressing WT and mutant (Del 746-750) EGFRs have suggested that MIG6 promotes wild-type EGFR, but not mutant EGFR internalization (54, 55). To investigate the effect of MIG6 on EGF-induced degradation of EGFR, we performed an EGFR degradation assay with cycloheximide on HEK293 cells stably transfected with wild-type or Y394/395F MIG6, together with WT EGFR or L858R EGFR (Fig. 6B), and quantified retained EGFRs (Fig. 6C). As predicted, WT EGFR was degraded upon EGF stimulation. However, there was little effect of EGF on mutant EGFR degradation. Furthermore, expression of MIG6 had no effect on the degradation of mutant EGFR, and it slowed the degradation of activated WT EGFR. To ascertain whether WT EGFR degradation utilizes the lysosomes, we performed these degradation experiments in presence of chloroquine, a lysosomotropic inhibitor. We observed that chloroquine significantly delayed degradation of WT EGFR (Fig. 6D). However, there was no additional effect of chloroquine on the retention of L858R EGFR suggesting that the lysosomal pathway is utilized for WT EGFR, but probably not for mutant EGFR degradation (Fig. 6E). We further performed these degradation assays in wild-type or Y394/395F MIG6-transduced H322M lung adenocarcinoma cells that contain WT EGFR and no endogenous MIG6. WT EGFR is efficiently degraded upon EGF stimulation in absence or the presence of Y394/395F MIG6; however, WT EGFR appears more stable in the presence of MIG6, findings similar to those observed in HEK293 cells (Fig. 6F).
Discussion
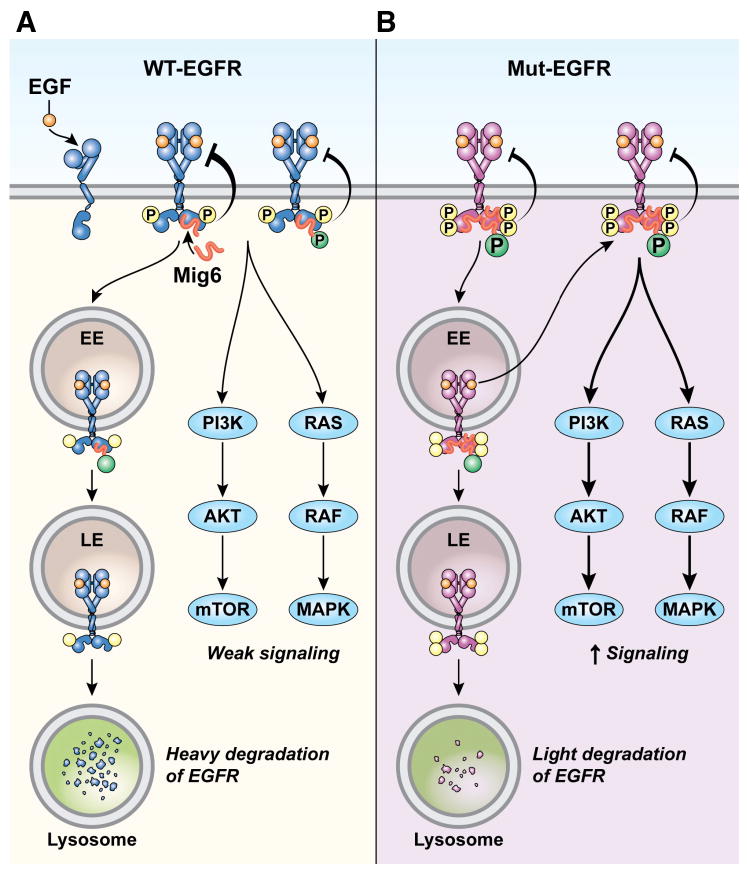
MIG6 is known to inhibit EGFR kinase activity and promote the degradation of WT EGFR, so it is considered a suppressor of tumors with active WT EGFR signaling. However, the role of MIG6 in regulating lung cancer-specific mutant EGFRs has not been studied in detail. Here we show for the first time that Mig6 deficiency, even haploinsufficiency, accelerates the initiation and progression of tumorigenesis and lethality driven by mutant EGFR in mouse models. We have previously shown in isogenic HBECs that mutant EGFR enhances tyrosine phosphorylation of MIG6 more efficiently than WT EGFR (24). Here we demonstrate constitutive phosphorylation of MIG6 at Y394/395 in lung adenocarcinoma cells harboring EGFR mutations. Phosphorylation at these sites is inhibited by erlotinib in TKI-sensitive, but not resistant cells, suggesting mutant EGFRs directly phosphorylate and possibly regulate MIG6. This conclusion is consistent with published evidence that purified EGFR protein can directly phosphorylate MIG6 in vitro (44). We have further examined the functional consequences of tyrosine phosphorylation of Y394/395 on MIG6 and how this impacts its tumor suppressor function. We show that phosphorylation increases binding of MIG6 to mutant EGFRs; but, in contrast to wild-type EGFR, the increased interaction does not direct mutant EGFR to the degradation pathway. Our observation is consistent with another study in which MIG6 was shown to be a poor inhibitor of the kinase activity nearly full length mutant EGFR in vitro (44). A phosphorylated Y394-containing fragment of MIG6 was also shown to be a poor kinase inhibitor of WT EGFR compared to its unphosphorylated counterpart using kinase assays with purified proteins in vitro (47). Taken together, our findings of constitutive phosphorylation of MIG6 Y394/395 in lung adenocarcinoma cell lines and the increased binding of MIG6 with mutant EGFRs (possibly leading to stability of mutant EGFR) along with the published in vitro studies of inadequate EGFR kinase inhibition by phosphorylated MIG6 provide strong evidence that mutant EGFRs can partially circumvent inhibition by MIG6 in lung adenocarcinoma cells through tyrosine phosphorylation of MIG6 on key residues. However, most importantly, we show that the residual inhibitory activity of MIG6 is still tumor suppressive in mutant EGFR-driven lung tumor models, since MIG6-deficiency reduces survival of mice due to accelerated tumorigenesis. This was not expected based on results of enzymologic studies published to date (44, 47). It is also possible that Mig6 is capable of inhibiting the formation of heterodimers of mouse wild-type Egfr and human mutant EGFR, a likely scenario in early stages of doxycycline induction in our mouse model, especially when levels of transgenic mutant EGFR are low. A schematic of our model of inhibitory activity of MIG6 against WT and mutant EGFRs is depicted in Fig. 7A–B.
Figure 7. Model depicting the regulation of WT EGFR and mutant EGFRs by MIG6.
(A) MIG6 binds to and inhibits kinase activity of activated WT EGFR. MIG6 also promotes WT EGFR trafficking to the degradation pathway. However, once activated, EGFR phosphorylates MIG6 Y394/395 residues to increase MIG6 binding and decrease inhibition of WT EGFR kinase. This may be a feedback mechanism for reversing the inhibitory role of MIG6 on WT EGFR. (B) Mutant EGFRs are constitutively active. This results in constitutive and increased phosphorylation at MIG6-Y394/395. This increases the interaction of MIG6 with mutant EGFRs. Increased tyrosine phosphorylation of MIG6 results in decreased kinase inhibition of mutant EGFRs. Furthermore, MIG6 cannot promote mutant EGFR degradation. Hence mutant EGFRs undergo relatively attenuated inhibition by MIG6. However, this incomplete inhibition of mutant EGFRs is still sufficient for Mig6 to function as a potent tumor suppressor for the initiation and progression of tumorigenesis in mouse models of mutant EGFR-driven lung tumorigenesis.
The accelerated initiation and progression of mutant EGFR-driven tumorigenesis in Mig6 deficient background was quite striking. The manifestation of early tumorigenesis was dramatic in the early time periods of doxycycline induction of mutant EGFRs. There was almost complete effacement of normal alveoli by type II cells 7–9 days after doxycycline induction in Mig6−/− mice, the earliest times at which tissue was analyzed in this study, at a time when only focal type II cellular hyperplasia was observed in Mig6+/+ littermates. Increased proliferation was associated with increased pERK immunoreactivity in the lungs of Mig6−/− mice. Interestingly, our experiments also demonstrate a statistically significant difference in the survival of Mig6+/− compared to Mig6+/+ mice in the presence of an EGFRDel transgene, at least in line 9, with a strong trend towards decreased survival in EGFRDel line 11 mice. In two EGFRL858R mouse lines, we saw a statistically insignificant trend towards decreased survival for Mig6+/− mice. The EGFRDel mice develop tumors much later than EGFRL858R mice, which may allow for the difference in survival between the Mig6+/+ and Mig6+/− mice to manifest. We did not observe Mig6 LOH in a select group of Mig6+/− mice we studied. However, it is still possible that there could be LOH in individual mice that we did not analyze. The fact that there is strong evidence of Mig6 being a haploinsufficient tumor suppressor in our studies has implications in mutant EGFR-driven human lung adenocarcinoma biology. A recent study evaluated the relative expression level of MIG6 and EGFR in a small cohort of lung cancer patients treated prospectively with gefitinib. This study concluded that a lower MIG6/EGFR ratio is associated with sensitivity to TKIs, whereas a higher MIG6/EGFR ratio is a predictor of TKI resistance (56). In another study, ratio of MIG6 and miR200c RNA levels correlated with EMT and resistance to erlotinib (57). However, these studies were performed primarily with patients harboring WT EGFR. Further prospective clinical studies are warranted to ascertain whether absolute MIG6 levels can influence the initiation, progression, and EGFR-TKI response in mutant EGFR-driven human lung adenocarcinoma.
We saw comparable induction of steady-state levels of transgenic mutant EGFR transcripts in the early time periods of doxycycline induction regardless of Mig6 status. However, there were more Ttf-1 and EGFRL858R immunostained cells at these time periods, consistent with enhanced stimulation of growth by mutant EGFR and early tumorigenesis in Mig6-null transgenic mice. Surprisingly, mutant EGFR protein levels in tumor lysates collected at the survival endpoint were significantly lower in Mig6−/− mice as compared to Mig6+/+ mice. We demonstrated that the low level of transgenic EGFR protein was still hyperphosphorylated in these late-stage tumors and the pEGFR/EGFR ratio was higher in tumors from Mig6−/− mice than in tumors from Mig6+/+ mice, suggesting that the residual mutant EGFR activity was sufficient to maintain these aggressive tumors. We did not find any difference in the mutant EGFR and rtTA transcript levels at earlier time periods. However, there was a slight decrease of rtTA and mutant EGFR transcript expression in the late-stage tumors. This could be a result of decreased CCSP promoter activity in the lungs of older Mig6−/− mice, reducing mutant EGFR transcripts. This is corroborated by the fact that Mig6 is essential for normal lung development (34). However, the modest decrease in rtTA or mutant EGFR mRNA does not explain the reduced levels of mutant EGFR protein. We speculate that during the progression of tumorigenesis there is selection for lower transgenic EGFR-expressing cells. However, the residual mutant EGFR signal-strength is still enough to maintain these aggressive tumors. We also speculate that MIG6 inhibits heterodimers of mutant and WT EGFR and promotes their degradation in the early stages of doxycycline induction of mutant EGFRs in these models, thus explaining the dramatic tumor suppressive role. At later stages, because of increased transgenic mutant EGFR levels, the mutant EGFRs exist predominantly as homodimers. MIG6 is unable to traffic these homodimers to degradation pathways because of increased feedback tyrosine phosphorylation, instead MIG6 binds more strongly and stabilizes mutant EGFR homodimers at this stage. Hence Mig6 deficiency results in lower levels of transgenic mutant EGFRs in end-stage tumors.
We postulate a two-pronged mechanism by which mutant EGFRs dampen inhibition by MIG6; one acts to regulate the levels and the other modulates the function of MIG6. The first is by a down-regulation of MIG6 protein levels similar to the regulation of a classic tumor suppressor. In a recent study, lung cancer-specific EGFR mutations correlated with loss of MIG6 protein; 12 out of 16 EGFR mutant tumors lacked MIG6 protein (58). At least one lung adenocarcinoma cell line, H322M harbors a homozygous nonsense mutation in MIG6, with undetectable MIG6 protein in the context of WT EGFR expression (30). Around 50% of primary glioblastoma multiforme (GBM) tumor samples and cell lines have reduced MIG6 RNA and protein expression (29). MIG6 levels may also be regulated by epigenetic mechanisms. MIG6 promoter methylation was observed in 79% of papillary thyroid carcinomas (59) and histone deacetylation (HDAC) inhibition upregulates MIG6 in lung cancer cell lines (60). We confirmed that MIG6 functions as a potent tumor suppressor in the initiation and progression of tumorigenesis induced by mutant EGFRs in mouse models. Furthermore, our studies indicate that MIG6 can act as a haploinsufficient tumor suppressor in the context of mutant EGFRs.
The second mechanism of reduced MIG6 inhibitory function is the increased tyrosine phosphorylation of Y394/395 by mutant EGFRs, leading to decreased kinase inhibition (44, 47) and increased constitutive binding of MIG6 to mutant EGFRs, possibly stabilizing mutant EGFRs presented in this study. Recently, Ying et. al., also observed that the EGFRvIII mutant does not undergo MIG6 mediated endocytosis and degradation in lysosomes of GBM cell lines unlike WT EGFR (29). A recent study showed a modest decrease in EGFR internalization upon MIG6 knockdown in PC9 cells, a lung adenocarcinoma cell line harboring the EGFRDel mutant (54). The Walsh et al. study does not distinguish between mutant EGFR and WT EGFR in PC9 cells, which are heterozygous for the EGFRDel mutant. Furthermore, we also noticed that mutant EGFRs could be effectively internalized into early endosomes in HBECs (Supplementary Fig. S9 A–B). Interestingly, we found strong co-localization of mutant or WT EGFR and MIG6 in discrete vesicles upon EGF stimulation of HBECs. However, there was less colocalization of mutant EGFR compared to WT EGFR and LAMP1, a lysosomal marker even after 2 hours of EGF stimulation suggesting mutant EGFR may not traffic through the lysosomal degradation pathway (Supplementary Fig. S9 C–D). Our data suggest that mutant EGFR degradation is inhibited in-spite of the increased interaction of MIG6 and mutant EGFRs (Fig. 6 and Fig. S9).
Prospective biomarker-validation studies are warranted to establish the role of MIG6 expression or phosphorylation in the overall prognosis of patients harboring WT-EGFR or mutant EGFRs. Such clinical studies are needed to ascertain whether absolute MIG6 levels can influence the initiation, progression, and EGFR-TKI response in mutant EGFR-driven lung adenocarcinoma.
Materials and Methods
Additional materials and methods are described in the Supplementary Materials and Methods section.
Reagents and antibodies
RPMI and DMEM tissue culture media and fetal bovine serum (FBS) were obtained from Invitrogen. Defined FBS for H3255 adenocarcinoma cell culture was obtained from Hyclone. All chemicals were obtained from Sigma-Aldrich, except if mentioned otherwise. Fugene X-tremeGENE 9 DNA transfection reagent, complete minitab protease inhibitor, and PhosStop phosphatase inhibitor were obtained from Roche Applied Science. Nitrocellulose western transfer sandwich was obtained from Invitrogen and nitrocellulose membrane was obtained from GE Healthcare Life Sciences. EGF was obtained from Millipore or Peprotech. Tyrosine kinase inhibitor erlotinib was obtained from Beta Pharma, Inc. Mouse anti-Mig6 mAb was a kind gift from Dr. Oreste Segatto, Regina Elena Cancer Institute, Italy and was also obtained from Abnova. Mouse anti-EGFR mAb was obtained from BD Bioscience. Rabbit polyclonal antibodies to EGFRDel E746-A750, pEGFR (Y1068), EGFR, AKT, ERK, RSK, 4EBP, pAkt, pERK, pRSK, and p4EBP as indicated in the figures were obtained from Cell Signaling Technology. Rabbit polyclonal antibodies to Mig6 (H125) was obtained from Santacruz Biotech. Mouse mAbs to EGFRL858R (18D1), EGFRL858 (9D3), EGFRE746 (13D6) were made in collaboration with nanoTools. Anti-EEA1 and anti-LAMP1 antibodies were obtained from Abcam and Cell Signaling Technology, respectively. Anti-TTF1 antibody was obtained from Dako, Inc., and Rabbit anti-Rho-GDI polyclonal antibodies, protein A and G sepharose were obtained from Sigma. Ki67 and p19ARF specific antibodies were obtained from Abcam. TUNEL staining was performed using ApopTag Peroxidase In Situ Apoptosis Detection Kit (Millipore).
Cell lines
H1975 and HEK293 cell lines were purchased from American Type Culture Collection (ATCC), PC9 was obtained from the Varmus Laboratory, and H322M cells were obtained from the DCTD Tumor Cell Line Repository, National Cancer Institute at Frederick, Maryland. All human lung adenocarcinoma cells were maintained in RPMI supplemented with 10% FBS, 100 units/mL penicillin, and 100μg/mL streptomycin. Human embryonic kidney cell line HEK293 was cultured in DMEM supplemented with 10% FBS, 100 units/mL penicillin, and 100 μg/mL streptomycin. The HBECs were a kind gift from Dr. John D. Minna, UTSW, and were maintained in KSFM media (Invitrogen) supplemented with bovine pituitary extract (BPE) and EGF. Cells were authenticated by short tandem repeat (STR) profiling using the AmpFℓSTR® Identifiler kit at the Protein Expression Laboratory, NCI, Frederick in February 2015.
Plasmids
Site directed mutagenesis of human EGFR, MIG6, and subcloning of wild type and mutant constructs for lentivirus production were performed at the Protein Expression Laboratory, an NCI/SAIC core facility.
Cell extract and mouse tissue extract preparation, immunoprecipitation, and immunoblot analysis
Tissue culture or mouse tissue lysates used for immunoblot was prepared in RIPA lysis buffer (150 mM NaCl, 1.0% IGEPAL CA-630, 0.5% sodium deoxycholate, 0.1% SDS, and 50 mM Tris, pH 8.0). For immunoprecipitation, cell extracts were prepared in NP-40 lysis buffer. Mouse tissue extracts were prepared in RIPA buffer using a tissue lyser (Qiagen) following the manufacturer’s protocol. For phosphoproteomic analysis, mouse tissue extracts were prepared in Urea lysis buffer (20 mM Hepes pH 8.0, 9 M urea, 1 mM sodium orthovanadate, 2.5 mM sodium pyrophosphate, and 1 mM β-glycerophosphate). All lysis buffers contained protease and phosphatase inhibitor cocktails from Roche, and 100 μM sodium orthovanadate to inhibit protease and phosphatase activities. Protein concentrations were quantified using a modified Lowry method (BioRad). For immunoprecipitation 800 to 1000 μg of lysate was incubated overnight at 4°C with 2 to 5 μg of mouse anti-EGFR (MAB108) or mouse anti-Flag (MIG6) monoclonal antibody. The antigen-antibody complex was then captured by incubating the mixture with protein G beads for an additional 1 h. The immunocomplexes were washed with NP-40 lysis buffer twice and once with PBS buffer containing 100 μM sodium orthovanadate. The bound proteins were then extracted with 2x SDS-loading buffer (65.8 mM Tris-HCl, pH 6.8, 2.1% SDS, 26.3% (w/v) glycerol, 0.01% bromophenol blue, 5% 2-mercaptoethanol) by incubating at 95°C for 5 min and fractionated by SDS-polyacrylamide (4–15%) gel electrophoresis. The proteins were transferred to nitrocellulose membrane using either the semi-dry or wet transfer method, and probed with the specified antibody.
EGFR degradation assays
Cells were serum starved for 18 h and treated with 100 μM cycloheximide (Clontech) for 1 h to inhibit new protein synthesis. Cells were either mock-stimulated or stimulated with EGF (100 ng/mL) for different time points at 37°C. Following EGF treatment, the cells were quickly chilled and washed with cold PBS, and lysed in RIPA buffer supplemented with protease and phosphatase inhibitors as previously described. For the degradation assay in the presence of chloroquine, cells were serum starved and treated with cycloheximide as described above. Fifteen minutes prior to EGF addition, cells were treated with or without 100 μM of chloroquine. Extracts for Western blot analysis were prepared at various time points after EGF stimulation.
Mouse strains
The doxycycline-inducible EGFRL858R and EGFRDel transgenic models have been described previously (35). All mice were maintained in a pathogen-free facility approved by NCI and Memorial Sloan-Kettering Cancer Center (MSKCC) Animal Care and Use Committees (ACUC). Animal studies were carried out with the approval of research protocols by the ACUC. We bred Mig6+/− mice with CCSP-rtTA mice, and Mig6+/− mice with TetO-EGFRmut mice. Resulting Mig6+/−/CCSP-rtTA and Mig6+/−/TetO-EGFRmut offsprings were then crossed to generate mutant EGFR expressing mice in a Mig6 wild-type (Mig6+/+), Mig6 heterozygous (Mig6+/−) or Mig6 null (Mig6−/−) background.
Tumor monitoring
Transgenic mice were fed with doxycycline-impregnated food pellets (625 ppm; Harlan-Teklad) to induce mutant forms of human EGFR from doxycycline-regulated promoter. Mice were monitored for EGFR-driven tumor development by Magnetic Resonance Imaging (MRI) of the lungs. MRI was carried out with respiratory gating at the MRI Core Facility of NCI or MSKCC. Serial MRI analyses were performed and the tumor burden was quantified by ImageJ software. Regions of interest tool (ROI) was used to outline the lung and tumor within the lung. ROI measurements provided area and mean intensity of lung and tumor. For each MRI time point % tumor burden was calculated from the total lung and tumor measurements obtained. Mice were selected to be euthanized for survival analysis primarily using clinical criteria such as hunched posture, trachypnea, weight loss, and decreased movement. Lungs were perfused with PBS, and a representative portion of the tumor tissue was frozen in liquid nitrogen for further analysis. Another representative lung tumor tissue was processed by perfusion with phosphate buffered 4% paraformaldehyde for histopathology evaluation.
Statistical Analysis
For replicate experiments, standard deviation (SD) or standard error (SE) was calculated to indicate the variation between experiments, and values given represent the mean ± SD. Statistical analyses of the results to assess the significance of differences were performed using an unpaired Student’s t-test. A threshold of P≤0.05 was used for significance. Kaplan-Meier survival analyses were performed on tumor bearing mice using Graphpad Prism.
Supplementary Material
Statement of Significance.
This study demonstrates that Mig6 is a potent tumor suppressor for mutant EGFR-driven lung tumor initiation and progression in mice and provides a possible mechanism by which mutant EGFR can partially circumvent this tumor suppressor in human lung adenocarcinoma.
Acknowledgments
We thank George Vande Woude for Mig6 knockout mice, Oreste Segatto for Mig6 antibody, and helpful discussions, and Philip Cole for helpful discussions. This work was supported by US National Cancer Institute, Center for Cancer Research (NCI-CCR) Intramural Research Program ZIA BC 011259 (U.G), K99 Career Transition Award (K99CA140792) (U.G), and PO1 CA129243 (HV).
Footnotes
Conflict of interest statement: Authors declare no conflict of interest.
References
- 1.Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–39. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 2.Paez JG, Janne PA, Lee JC, Tracy S, Greulich H, Gabriel S, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 3.Pao W, Miller V, Zakowski M, Doherty J, Politi K, Sarkaria I, et al. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci U S A. 2004;101:13306–11. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shigematsu H, Lin L, Takahashi T, Nomura M, Suzuki M, Wistuba II, et al. Clinical and biological features associated with epidermal growth factor receptor gene mutations in lung cancers. J Natl Cancer Inst. 2005;97:339–46. doi: 10.1093/jnci/dji055. [DOI] [PubMed] [Google Scholar]
- 5.Kobayashi S, Boggon TJ, Dayaram T, Janne PA, Kocher O, Meyerson M, et al. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med. 2005;352:786–92. doi: 10.1056/NEJMoa044238. [DOI] [PubMed] [Google Scholar]
- 6.Pao W, Miller VA, Politi KA, Riely GJ, Somwar R, Zakowski MF, et al. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2005;2:e73. doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Engelman JA, Zejnullahu K, Mitsudomi T, Song Y, Hyland C, Park JO, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316:1039–43. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- 8.Bean J, Brennan C, Shih JY, Riely G, Viale A, Wang L, et al. MET amplification occurs with or without T790M mutations in EGFR mutant lung tumors with acquired resistance to gefitinib or erlotinib. Proc Natl Acad Sci U S A. 2007;104:20932–7. doi: 10.1073/pnas.0710370104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takezawa K, Pirazzoli V, Arcila ME, Nebhan CA, Song X, de Stanchina E, et al. HER2 amplification: a potential mechanism of acquired resistance to EGFR inhibition in EGFR-mutant lung cancers that lack the second-site EGFRT790M mutation. Cancer Discov. 2012;2:922–33. doi: 10.1158/2159-8290.CD-12-0108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheung HW, Du J, Boehm JS, He F, Weir BA, Wang X, et al. Amplification of CRKL induces transformation and epidermal growth factor receptor inhibitor resistance in human non-small cell lung cancers. Cancer Discov. 2011;1:608–25. doi: 10.1158/2159-8290.CD-11-0046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Bruin EC, Cowell C, Warne PH, Jiang M, Saunders RE, Melnick MA, et al. Reduced NF1 expression confers resistance to EGFR inhibition in lung cancer. Cancer Discov. 2014;4:606–19. doi: 10.1158/2159-8290.CD-13-0741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zakowski MF, Ladanyi M, Kris MG. EGFR mutations in small-cell lung cancers in patients who have never smoked. N Engl J Med. 2006;355:213–5. doi: 10.1056/NEJMc053610. [DOI] [PubMed] [Google Scholar]
- 13.Sequist LV, Waltman BA, Dias-Santagata D, Digumarthy S, Turke AB, Fidias P, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med. 2011;3:75ra26. doi: 10.1126/scitranslmed.3002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thomson S, Buck E, Petti F, Griffin G, Brown E, Ramnarine N, et al. Epithelial to mesenchymal transition is a determinant of sensitivity of non-small-cell lung carcinoma cell lines and xenografts to epidermal growth factor receptor inhibition. Cancer Res. 2005;65:9455–62. doi: 10.1158/0008-5472.CAN-05-1058. [DOI] [PubMed] [Google Scholar]
- 15.Uramoto H, Iwata T, Onitsuka T, Shimokawa H, Hanagiri T, Oyama T. Epithelial-mesenchymal transition in EGFR-TKI acquired resistant lung adenocarcinoma. Anticancer Res. 2010;30:2513–7. [PubMed] [Google Scholar]
- 16.Zhang Z, Lee JC, Lin L, Olivas V, Au V, LaFramboise T, et al. Activation of the AXL kinase causes resistance to EGFR-targeted therapy in lung cancer. Nat Genet. 2012;44:852–60. doi: 10.1038/ng.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ohashi K, Sequist LV, Arcila ME, Moran T, Chmielecki J, Lin YL, et al. Lung cancers with acquired resistance to EGFR inhibitors occasionally harbor BRAF gene mutations but lack mutations in KRAS, NRAS, or MEK1. Proc Natl Acad Sci U S A. 2012;109:E2127–33. doi: 10.1073/pnas.1203530109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang SE, Narasanna A, Perez-Torres M, Xiang B, Wu FY, Yang S, et al. HER2 kinase domain mutation results in constitutive phosphorylation and activation of HER2 and EGFR and resistance to EGFR tyrosine kinase inhibitors. Cancer Cell. 2006;10:25–38. doi: 10.1016/j.ccr.2006.05.023. [DOI] [PubMed] [Google Scholar]
- 19.Greulich H, Chen T-H, Feng W, Janne PA, Alvarez JV, Zappaterra M, et al. Oncogenic transformation by inhibitor-sensitive and -resistant EGFR mutants. Plos Med. 2005;2:e313. doi: 10.1371/journal.pmed.0020313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang J, Greulich H, Janne PA, Sellers WR, Meyerson M, Griffin JD. Epidermal growth factor-independent transformation of Ba/F3 cells with cancer-derived epidermal growth factor receptor mutants induces gefitinib-sensitive cell cycle progression. Cancer Res. 2005;65:8968–74. doi: 10.1158/0008-5472.CAN-05-1829. [DOI] [PubMed] [Google Scholar]
- 21.Roberts PJ, Der CJ. Targeting the Raf-MEK-ERK mitogen-activated protein kinase cascade for the treatment of cancer. Oncogene. 2007;26:3291–310. doi: 10.1038/sj.onc.1210422. [DOI] [PubMed] [Google Scholar]
- 22.Rikova K, Guo A, Zeng Q, Possemato A, Yu J, Haack H, et al. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell. 2007;131:1190–203. doi: 10.1016/j.cell.2007.11.025. [DOI] [PubMed] [Google Scholar]
- 23.Guo A, Villen J, Kornhauser J, Lee KA, Stokes MP, Rikova K, et al. Signaling networks assembled by oncogenic EGFR and c-Met. Proc Natl Acad Sci U S A. 2008;105:692–7. doi: 10.1073/pnas.0707270105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guha U, Chaerkady R, Marimuthu A, Patterson AS, Kashyap MK, Harsha HC, et al. Comparisons of tyrosine phosphorylated proteins in cells expressing lung cancer-specific alleles of EGFR and KRAS. Proc Natl Acad Sci U S A. 2008;105:14112–7. doi: 10.1073/pnas.0806158105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fiorentino L, Pertica C, Fiorini M, Talora C, Crescenzi M, Castellani L, et al. Inhibition of ErbB-2 mitogenic and transforming activity by RALT, a mitogen-induced signal transducer which binds to the ErbB-2 kinase domain. Mol Cell Biol. 2000;20:7735–50. doi: 10.1128/mcb.20.20.7735-7750.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Makkinje A, Quinn DA, Chen A, Cadilla CL, Force T, Bonventre JV, et al. Gene 33/Mig-6, a transcriptionally inducible adapter protein that binds GTP-Cdc42 and activates SAPK/JNK. A potential marker transcript for chronic pathologic conditions, such as diabetic nephropathy. Possible role in the response to persistent stress. J Biol Chem. 2000;275:17838–47. doi: 10.1074/jbc.M909735199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang YW, Vande Woude GF. Mig-6, signal transduction, stress response and cancer. Cell Cycle. 2007;6:507–13. doi: 10.4161/cc.6.5.3928. [DOI] [PubMed] [Google Scholar]
- 28.Ferby I, Reschke M, Kudlacek O, Knyazev P, Pante G, Amann K, et al. Mig6 is a negative regulator of EGF receptor-mediated skin morphogenesis and tumor formation. Nat Med. 2006;12:568–73. doi: 10.1038/nm1401. [DOI] [PubMed] [Google Scholar]
- 29.Ying H, Zheng H, Scott K, Wiedemeyer R, Yan H, Lim C, et al. Mig-6 controls EGFR trafficking and suppresses gliomagenesis. Proc Natl Acad Sci U S A. 2010;107:6912–7. doi: 10.1073/pnas.0914930107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang YW, Staal B, Su Y, Swiatek P, Zhao P, Cao B, et al. Evidence that MIG-6 is a tumor-suppressor gene. Oncogene. 2007;26:269–76. doi: 10.1038/sj.onc.1209790. [DOI] [PubMed] [Google Scholar]
- 31.Zhang X, Pickin KA, Bose R, Jura N, Cole PA, Kuriyan J. Inhibition of the EGF receptor by binding of MIG6 to an activating kinase domain interface. Nature. 2007;450:741–4. doi: 10.1038/nature05998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Frosi Y, Anastasi S, Ballaro C, Varsano G, Castellani L, Maspero E, et al. A two-tiered mechanism of EGFR inhibition by RALT/MIG6 via kinase suppression and receptor degradation. J Cell Biol. 2010;189:557–71. doi: 10.1083/jcb.201002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nagashima T, Ushikoshi-Nakayama R, Suenaga A, Ide K, Yumoto N, Naruo Y, et al. Mutation of epidermal growth factor receptor is associated with MIG6 expression. FEBS J. 2009;276:5239–51. doi: 10.1111/j.1742-4658.2009.07220.x. [DOI] [PubMed] [Google Scholar]
- 34.Jin N, Cho SN, Raso MG, Wistuba I, Smith Y, Yang Y, et al. Mig-6 is required for appropriate lung development and to ensure normal adult lung homeostasis. Development. 2009;136:3347–56. doi: 10.1242/dev.032979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Politi K, Zakowski MF, Fan PD, Schonfeld EA, Pao W, Varmus HE. Lung adenocarcinomas induced in mice by mutant EGF receptors found in human lung cancers respond to a tyrosine kinase inhibitor or to down-regulation of the receptors. Genes Dev. 2006;20:1496–510. doi: 10.1101/gad.1417406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perl AK, Tichelaar JW, Whitsett JA. Conditional gene expression in the respiratory epithelium of the mouse. Transgenic Res. 2002;11:21–9. doi: 10.1023/a:1013986627504. [DOI] [PubMed] [Google Scholar]
- 37.Kim TH, Franco HL, Jung SY, Qin J, Broaddus RR, Lydon JP, et al. The synergistic effect of Mig-6 and Pten ablation on endometrial cancer development and progression. Oncogene. 2010;29:3770–80. doi: 10.1038/onc.2010.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hopkins S, Linderoth E, Hantschel O, Suarez-Henriques P, Pilia G, Kendrick H, et al. Mig6 is a sensor of EGF receptor inactivation that directly activates c-Abl to induce apoptosis during epithelial homeostasis. Dev Cell. 2012;23:547–59. doi: 10.1016/j.devcel.2012.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xie B, Zhao L, Chen H, Jin B, Mao Z, Yao Z. The mitogen-inducible gene-6 is involved in regulation of cellular senescence in normal diploid fibroblasts. Biol Cell. 2013;105:488–99. doi: 10.1111/boc.201200052. [DOI] [PubMed] [Google Scholar]
- 40.Milewska M, Kolch W. Mig-6 participates in the regulation of cell senescence and retinoblastoma protein phosphorylation. Cell Signal. 2014;26:1870–7. doi: 10.1016/j.cellsig.2014.05.003. [DOI] [PubMed] [Google Scholar]
- 41.Anastasi S, Castellani L, Alema S, Segatto O. A pervasive role for MIG6 in restraining cell proliferation. Cell Death Differ. 2013;21:345–347. doi: 10.1038/cdd.2013.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen Z, Trotman LC, Shaffer D, Lin HK, Dotan ZA, Niki M, et al. Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigenesis. Nature. 2005;436:725–30. doi: 10.1038/nature03918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Collado M, Serrano M. The power and the promise of oncogene-induced senescence markers. Nat Rev Cancer. 2006;6:472–6. doi: 10.1038/nrc1884. [DOI] [PubMed] [Google Scholar]
- 44.Wang Z, Longo PA, Tarrant MK, Kim K, Head S, Leahy DJ, et al. Mechanistic insights into the activation of oncogenic forms of EGF receptor. Nat Struct Mol Biol. 2011;18:1388–93. doi: 10.1038/nsmb.2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Anastasi S, Baietti MF, Frosi Y, Alema S, Segatto O. The evolutionarily conserved EBR module of RALT/MIG6 mediates suppression of the EGFR catalytic activity. Oncogene. 2007;26:7833–46. doi: 10.1038/sj.onc.1210590. [DOI] [PubMed] [Google Scholar]
- 46.Hornbeck PV, Kornhauser JM, Tkachev S, Zhang B, Skrzypek E, Murray B, et al. PhosphoSitePlus: a comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucleic Acids Res. 2012;40:D261–70. doi: 10.1093/nar/gkr1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang Z, Raines LL, Hooy RM, Roberson H, Leahy DJ, Cole PA. Tyrosine phosphorylation of mig6 reduces its inhibition of the epidermal growth factor receptor. ACS Chem Biol. 2013;8:2372–6. doi: 10.1021/cb4005707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yarden Y, Sliwkowski MX. Untangling the ErbB signalling network. Nat Rev Mol Cell Biol. 2001;2:127–37. doi: 10.1038/35052073. [DOI] [PubMed] [Google Scholar]
- 49.Sorkin A, Goh LK. Endocytosis and intracellular trafficking of ErbBs. Exp Cell Res. 2008;314:3093–106. doi: 10.1016/j.yexcr.2008.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hendriks BS, Griffiths GJ, Benson R, Kenyon D, Lazzara M, Swinton J, et al. Decreased internalisation of erbB1 mutants in lung cancer is linked with a mechanism conferring sensitivity to gefitinib. IEE Proc Syst Biol. 2006;153:457–66. doi: 10.1049/ip-syb:20050108. [DOI] [PubMed] [Google Scholar]
- 51.Lazzara MJ, Lane K, Chan R, Jasper PJ, Yaffe MB, Sorger PK, et al. Impaired SHP2-mediated extracellular signal-regulated kinase activation contributes to gefitinib sensitivity of lung cancer cells with epidermal growth factor receptor-activating mutations. Cancer Res. 2010;70:3843–50. doi: 10.1158/0008-5472.CAN-09-3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang S, Qu S, Perez-Tores M, Sawai A, Rosen N, Solit DB, et al. Association with HSP90 inhibits Cbl-mediated down-regulation of mutant epidermal growth factor receptors. Cancer Res. 2006;66:6990–7. doi: 10.1158/0008-5472.CAN-06-1042. [DOI] [PubMed] [Google Scholar]
- 53.Padron D, Sato M, Shay JW, Gazdar AF, Minna JD, Roth MG. Epidermal growth factor receptors with tyrosine kinase domain mutations exhibit reduced Cbl association, poor ubiquitylation, and down-regulation but are efficiently internalized. Cancer Res. 2007;67:7695–702. doi: 10.1158/0008-5472.CAN-07-0484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Walsh AM, Lazzara MJ. Regulation of EGFR trafficking and cell signaling by Sprouty2 and MIG6 in lung cancer cells. J Cell Sci. 2013;126:4339–48. doi: 10.1242/jcs.123208. [DOI] [PubMed] [Google Scholar]
- 55.Walsh AM, Lazzara MJ. Differential parsing of EGFR endocytic flux among parallel internalization pathways in lung cancer cells with EGFR-activating mutations. Integrative biology: quantitative biosciences from nano to macro. 2014;6:312–23. doi: 10.1039/c3ib40176f. [DOI] [PubMed] [Google Scholar]
- 56.Chang X, Izumchenko E, Solis LM, Kim MS, Chatterjee A, Ling S, et al. The relative expression of Mig6 and EGFR is associated with resistance to EGFR kinase inhibitors. PLoS One. 2013;8:e68966. doi: 10.1371/journal.pone.0068966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Izumchenko E, Chang X, Michailidi C, Kagohara L, Ravi R, Paz K, et al. The TGFbeta-miR200-MIG6 pathway orchestrates the EMT-associated kinase switch that induces resistance to EGFR inhibitors. Cancer Res. 2014;74:3995–4005. doi: 10.1158/0008-5472.CAN-14-0110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li Z, Dong Q, Wang Y, Qu L, Qiu X, Wang E. Downregulation of Mig-6 in nonsmall-cell lung cancer is associated with EGFR signaling. Mol Carcinog. 2012;51:522–34. doi: 10.1002/mc.20815. [DOI] [PubMed] [Google Scholar]
- 59.Lin CI, Barletta JA, Nehs MA, Morris ZS, Donner DB, Whang EE, et al. Thyroid-specific knockout of the tumor suppressor mitogen-inducible gene 6 activates epidermal growth factor receptor signaling pathways and suppresses nuclear factor-kappaB activity. Surgery. 2011;150:1295–302. doi: 10.1016/j.surg.2011.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang Y, Sun Y, Pan Y, Li C, Shen L, Li Y, et al. Frequency of driver mutations in lung adenocarcinoma from female never-smokers varies with histologic subtypes and age at diagnosis. Clin Cancer Res. 2012;18:1947–53. doi: 10.1158/1078-0432.CCR-11-2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.