Abstract
Background
Inuit are considered to be vulnerable to cardiovascular disease (CVD) as their lifestyles become more Westernized. During sequence analysis of Inuit individuals at extremes of lipid traits, we identified two nonsynonymous variants in LDLR encoding the low-density lipoprotein (LDL) receptor, namely p.G116S and p.R730W.
Methods and Results
Genotyping these variants in 3,324 Inuit from Alaska, Canada and Greenland showed they were common, with allele frequencies 10–15%. Only p.G116S was associated with dyslipidemia: the increase in LDL cholesterol was 0.54 mmol/L (20.9 mg/dL) per allele (P = 5.6 × 10−49), which was >3-times larger than the largest effect sizes seen with common variants in other populations. Carriers of p.G116S had a 3.02-fold increased risk of hypercholesterolemia (CI 2.34 – 3.90, P = 1.7 × 10−17), but did not have classical familial hypercholesterolemia. In vitro, p.G116S showed 60% reduced ligand binding activity compared to wild-type receptor. In contrast, p.R730W was associated with neither LDL cholesterol nor altered in vitro activity.
Conclusions
LDLR p.G116S is thus unique: a common dysfunctional variant in Inuit whose large effect on LDL cholesterol may have public health implications.
Keywords: lipoprotein, indigenous population, cardiovascular risk, DNA polymorphism, low-density lipoprotein receptor, familial hypercholesterolemia
Inuit were long-believed to have lower CVD risk than non-indigenous populations.1–3 However, re-evaluation of population studies indicates that ischemic heart disease rates are similar between Inuit and non-Indigenous people.4 Furthermore, ongoing Westernization in many Inuit communities has intensified their exposure to CVD risk factors such as smoking, calorie-dense processed foods, and a more comfortable but also sedentary lifestyle, all of which affect CVD risk and prevalence.4–10 Among classical CVD risk factors, Inuit adults tend to have higher plasma concentrations of LDL cholesterol than non-indigenous populations.11–15
The predominant monogenic cause of elevated LDL cholesterol concentration in most global populations is familial hypercholesterolemia (FH, Online Mendelian Inheritance in Man [OMIM] 143890).16 Heterozygous FH (HeFH) prevalence may be as high as 1:200 in certain European populations, and it is a potent predisposition state for early CVD.11–13 To date, DNA sequencing and biochemical studies have identified >1,600 rare loss-of-function mutations in the gene encoding the LDL receptor (LDLR), which can increase LDL cholesterol levels by 100% or more, and underlie >95% of cases of molecularly diagnosed FH.16 But despite the relatively high levels of LDL cholesterol observed in some Inuit, the role of LDLR gene variation has not been systematically studied.13–15 We thus investigated the LDLR locus in Inuit and tested for association of variants therein with plasma lipids. Through Sanger sequencing and targeted genotyping, we found two new LDLR variants common to five Inuit subgroups from across North America and Greenland: 1) p.G116S was both dysfunctional in vitro and associated with a relatively large increase in plasma LDL cholesterol levels; while 2) p.R730W had minimal dysfunction and impact on the lipid profile.
Methods
For the purposes of this study, we referred to all participants as “Inuit”; however, acknowledge that the circumpolar north is inhabited by a spectrum of diverse indigenous people.
Participants included Inuit >18 years of age (N=3,324) residing in arctic communities across North America and Greenland. North American population-based samples were collected as part of regional health surveys that included: 1) The Center for Alaska Native Health Research study of 2007, which covered 11 Southwest (SW) Alaska Yup’ik communities (N=1,222);17 2) the “Qanuippitaa” Health Survey of 2004, which covered 14 coastal communities in Nunavik, Quebec (N=429);6 3) the Keewatin Health Assessment Study of 1990–1991 which surveyed the Keewatin (Kivalliq) region of Nunavut (N=210);18 and 4) the Adult Inuit Health Survey of 2008 which surveyed the Inuvialuit region of the Northwest Territories (N=281).19 Inuit living in West Greenland and Denmark (N=1,182) were also included as part of our study cohort from a regional survey conducted in 1993–1994.20
The study was approved by the appropriate institutional research ethics boards including the Laval University Ethical Board and Comité Provincial de Santé Publique for use of Nunavik, Quebec samples; the University of Manitoba for use of Kivalliq samples; and McGill University for use of Inuvialuit samples. Yup’ik participants provided written informed consent using protocols approved by the University of Alaska Review Board, the National and Alaska Area Indian Health Service Institutional Review Boards, and the Yukon Kuskokwim Human Studies Committee. The Greenland population study was ethically approved by the Commission for Scientific Research in Greenland. Participants gave their written consent after being informed about the study both orally and in writing.
The LDLR promoter region and exons were Sanger sequenced within a discovery subset of 10 healthy Greenland Inuit with extreme plasma LDL cholesterol concentrations >6.0 mmol/L (>95th percentile for non-Inuit adults). Two novel variants, p.G116S and p.R730W, were identified in LDLR exons 4 and 15 respectively (Supplemental Figure 1). Both variants were then genotyped in independent Inuit samples from five different regions (Table 1) with custom TaqMan SNP genotyping assays (Applied Biosystems; Foster City, CA). As a comparator, we genotyped a common polymorphism with a relatively large effect on LDL cholesterol levels, namely the apolipoprotein (apo) E gene (APOE) protein isoforms using TaqMan SNP genotyping assays for SNPs rs429358 and rs7412 (Applied Biosystems; Foster City, CA).21 Genotypes were tested for association with blood lipid traits, including total cholesterol (TC), LDL cholesterol, HDL cholesterol, non-HDL cholesterol and triglyceride as well as apo B concentration, where available.
Table 1.
Demographics and LDLR variant frequencies for select circumpolar populations.
| n | Age | Female (%) |
BMI (kg/m2) |
TC | LDL-C | HDL-C | Non- HDL-C |
apo B | TG | MAF (%) | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| p.G116S | p.R730W | |||||||||||
| Greenland | 1182 | 44±14 | 56 | 26±5 | 5.91±1.13 | 3.82±1.04 | 1.57±0.44 | 4.33±1.14 | 0.92±0.23 | 1.16±0.67 | 0.13 | 0.11 |
| Kivalliq | 210 | 37±16 | 54 | 27±4 | 5.00±1.03 | 3.09±0.92 | 1.45±0.41 | 3.55±1.01 | 0.98±0.26 | 1.03±0.57 | 0.02 | 0.17 |
| Inuvialuit | 281 | 45±16 | 67 | 30±7 | 5.05±0.99 | 2.91±0.89 | 1.37±0.42 | 3.68±1.03 | 0.91±0.25 | 1.74±1.27 | 0.05 | 0.13 |
| Nunavik | 429 | 37±14 | 56 | 27±6 | 4.99±0.99 | 2.79±0.86 | 1.63±0.43 | 3.33±1.02 | 0.96±0.24 | 1.23±0.72 | 0.09 | 0.13 |
| SW Alaska | 1222 | 38±16 | 53 | 28±6 | 5.20±1.15 | 3.20±0.98 | 1.64±0.44 | 3.61±1.08 | n.d. | 0.94±0.56 | 0.10 | 0.16 |
| Combined | 3324 | 40±16 | 56 | 27±6 | 5.40±1.16 | 3.30±1.05 | 1.58±0.44 | 3.83±1.15 | 0.93±0.24 | 1.13±0.74 | 0.10 | 0.14 |
All demographics are reported ± standard deviation. Lipid-related traits are all reported in mmol/L except apo B which is in g/L.
BMI, body mass index; LDL-C, low-density lipoprotein cholesterol; HDL-C, high-density lipoprotein cholesterol; MAF, minor allele frequency; n.d., no data; TC, total cholesterol concentration; TG, triglyceride concentration.
Detailed descriptions of methods are provided in the Supplemental Materials section.
Results
LDLR p.G116S and p.R730W are common and exclusive to Inuit
To evaluate the genetic basis for elevated LDL cholesterol in Inuit, we used candidate sequencing of the LDL receptor gene (LDLR) to screen 10 Inuit with plasma LDL cholesterol concentrations >6.0 mmol/L (>95th percentile for non-Inuit adults). We found two heterozygous LDLR gene variants, namely p.G116S and p.R730W, in three and four Inuit with high LDL cholesterol, respectively (Supplemental Figure 1). We then genotyped these variants in 3,324 Inuit samples from Southwest (SW) Alaska, Northern Canada (Inuvialuit, Kivalliq and Nunavik), and Greenland (Table 1). The p.G116S variant frequency ranged from 2% in Kivalliq to 13% in Greenland, with an overall frequency of 10% across all regions. The p.R730W variant frequency ranged from 11% in Greenland to 17% in Kivalliq, with an overall frequency of 14% across all regions. The variants were not in linkage disequilibrium (r2 = 0.017; P= NS). Both variants were absent from other indigenous population samples and neither was observed in 4281 European samples and 2193 African American samples from the NHLBI Exome Sequencing Project database. However, p.G116S was previously reported in a single hypercholesterolemic subject of unspecified ethnic background ascertained in a lipid clinic in Denmark.22
LDLR p.G116S is robustly associated with higher plasma LDL cholesterol concentration
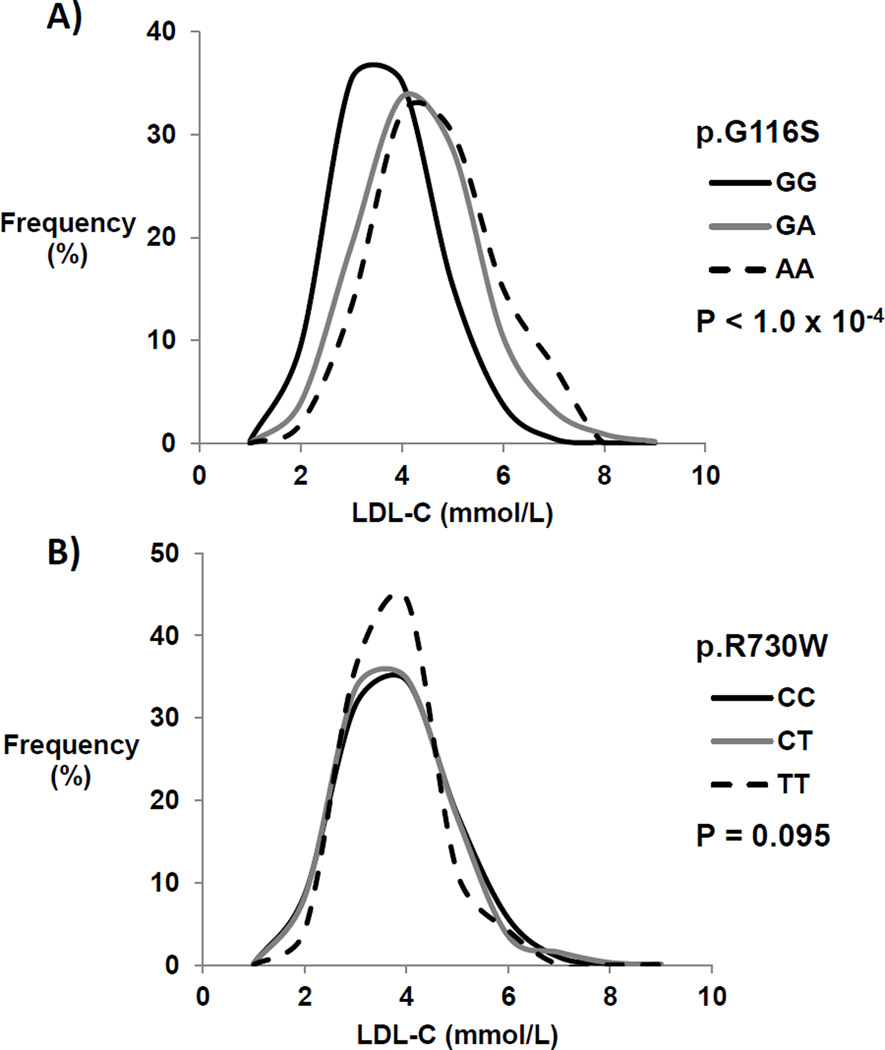
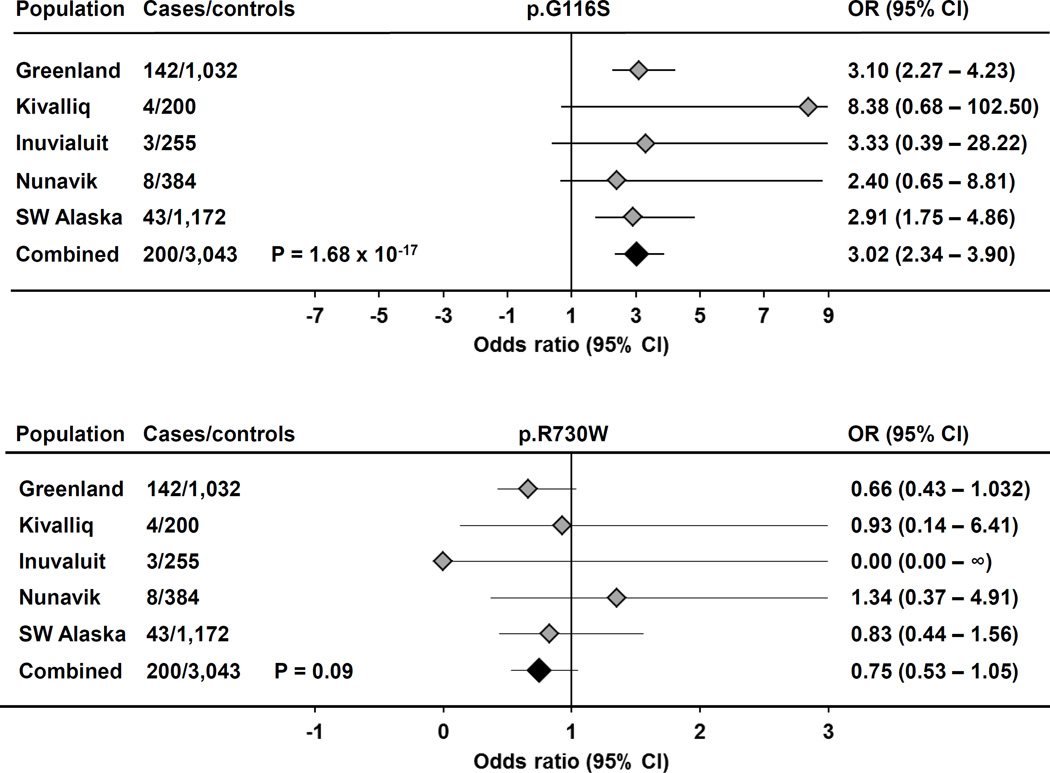
We stratified plasma lipoprotein profiles according to LDLR p.G116S or p.R730W genotype (Table 2). In each sample, p.G116S carriers had significantly higher total, non-HDL, and LDL cholesterol concentrations compared to non-carriers (Supplemental Tables 1A and 1B). In the overall sample p.G116S was associated with a ~0.54 mmol/L (20.9 mg/dL) increase in LDL cholesterol per copy (Table 3; P = 5.6 × 10−49); mean plasma apolipoprotein (apo) B and non-HDL cholesterol concentrations were also proportionately higher per copy of p.G116S. In contrast, p.R730W was not significantly associated with LDL cholesterol overall (P = 0.13). In the combined Inuit samples, LDLR p.G116S genotype had an additive (co-dominant) effect on LDL cholesterol concentration (Figure 1): mean LDL cholesterol concentration was significantly higher in p.G116S heterozygotes than in p.G116 homozygotes (P=2.0×10−34), and tended to be higher still in p.G116S homozygotes compared to heterozygotes (P=0.058). In contrast, the relationship between p.R730W and plasma LDL cholesterol concentrations was not significant overall. Each copy of p.G116S was associated with increased risk of hypercholesterolemia, defined as a plasma LDL cholesterol >5.0 mmol/L, which Canadian dyslipidemia guidelines23 suggest as the cutpoint for prescription of lipid-lowering treatment (Figure 2; OR 3.02, 95% CI 2.34 – 3.90, P = 1.7 × 10−17). In contrast, p.R730W was not associated with increased risk of clinically actionable hypercholesterolemia.
Table 2.
Mean lipid traits based on p.G116S and p.R730W genotype in a combined Inuit cohort.
| p.G116S genotype (A) | p.R730W genotype (T) | |||||
|---|---|---|---|---|---|---|
| Lipid traits | GG | GA | AA | CC | CT | TT |
| TC | 5.29±1.11 | 5.94±1.24 | 6.20±1.25** | 5.43±1.17 | 5.37±0.17 | 5.47±1.07 |
| LDL-C | 3.22±0.98 | 3.88±1.14 | 4.21±1.14** | 3.37±1.06 | 3.28±1.04 | 3.30±0.82 |
| apo B | 0.90±0.23 | 1.03±0.24 | 1.11±0.20** | 0.93±0.23 | 0.91±0.24 | 0.97±0.27 |
| HDL-C | 1.57±0.44 | 1.59±0.43 | 1.58±0.49 | 1.56±0.44 | 1.61±0.46 | 1.68±0.45* |
| TG | 1.13±0.75 | 1.09±0.62 | 0.97±0.38 | 1.12±0.76 | 1.11±0.67 | 1.11±0.68 |
| Non-HDL-C | 3.71±1.09 | 4.35±1.24 | 4.62±1.20** | 3.86±1.15 | 3.76±0.16 | 3.80±1.10* |
indicates P<0.05 and
indicates P<0.0001 using ANOVA adjusted for age, sex and BMI.
Abbreviations as in Table 1.
Table 3.
Associations between LDLR variants and plasma LDL cholesterol.
| Variant | Population | β (mmol/L) | SE | P-value |
|---|---|---|---|---|
| p.G116S | Greenland | 0.64 | 0.05 | 1.8×10−30 |
| Kivalliq | 1.02 | 0.27 | 1.7×10−4 | |
| Inuvialuit | 0.52 | 0.16 | 0.0011 | |
| Nunavik | 0.40 | 0.09 | 3.7×10−5 | |
| SW Alaska | 0.41 | 0.06 | 9.4×10−12 | |
| Combined | 0.54 | 0.04 | 5.6×10−49 | |
| p.R730W | Greenland | −0.11 | 0.06 | 0.077 |
| Kivalliq | 0.003 | 0.09 | 0.97 | |
| Inuvialuit | −0.13 | 0.11 | 0.24 | |
| Nunavik | −0.05 | 0.08 | 0.52 | |
| SW Alaska | −0.01 | 0.05 | 0.85 | |
| Combined | −0.05 | 0.03 | 0.13 | |
Effect sizes and P-values are based on the minor alleles p.G116S or p.R730W. SE, standard error. Greenland (N=1162), Kivalliq (N=204), Inuvialuit region (N=253), Nunavik (N=389), SW Alaska (N=1113), Combined (N=3121).
Figure 1. LDLR variant associations with LDL cholesterol (C) in a combined Inuit cohort.
Inuit participants were separated based on LDLR p.G116S or p.R730W genotype and LDL cholesterol concentration. A) Frequency distribution of Inuit participants based on p.G116S carrier status and LDL cholesterol concentration. Mean LDL cholesterol concentrations between non-carriers (GG, n=2585), heterozygotes (GA, n=559) and homozygotes (AA, n=53) were compared using one-way ANOVA with the P-value as indicated. B) Frequency distributions were similarly constructed for p.R730W non-carriers (CC, n=2408), heterozygotes (CT, n=717) and homozygotes (TT, n=72) and were also compared using one-way ANOVA.
Figure 2. Association between Inuit LDLR variants and very elevated LDL cholesterol (C) concentration.
Forest plots indicate odds ratios (OR) and 95% confidence intervals (95% CI) from association testing between both LDLR p.G116S or p.R730W variant genotypes and severely elevated LDL cholesterol status (LDL cholesterol >5.0 mmol/L).
LDLR p.G116S has a larger effect size on LDL cholesterol than the APOE E4 isoform
We compared the effect size of p.G116S to that of the APOE E4 isoform, a well-established common variant associated with increased LDL cholesterol.24, 25 In Inuit, APOE E4 allele frequencies ranged from 21% to 27% (Supplemental Table 2) and each copy of E4 increased LDL cholesterol by 0.18 mmol/L (7.0 mg/dL; P = 9.0 × 10−11). Furthermore, the top LDL cholesterol-associated variants from genome-wide association (GWA) studies had effect sizes per allele ranging from 0.05 to 0.18 mmol/L.26 Thus, LDLR p.G116S in Inuit is unique, with >3-fold larger effect on LDL cholesterol than any other common variant.
LDLR p.G116S impairs LDLR ligand binding activity in vitro
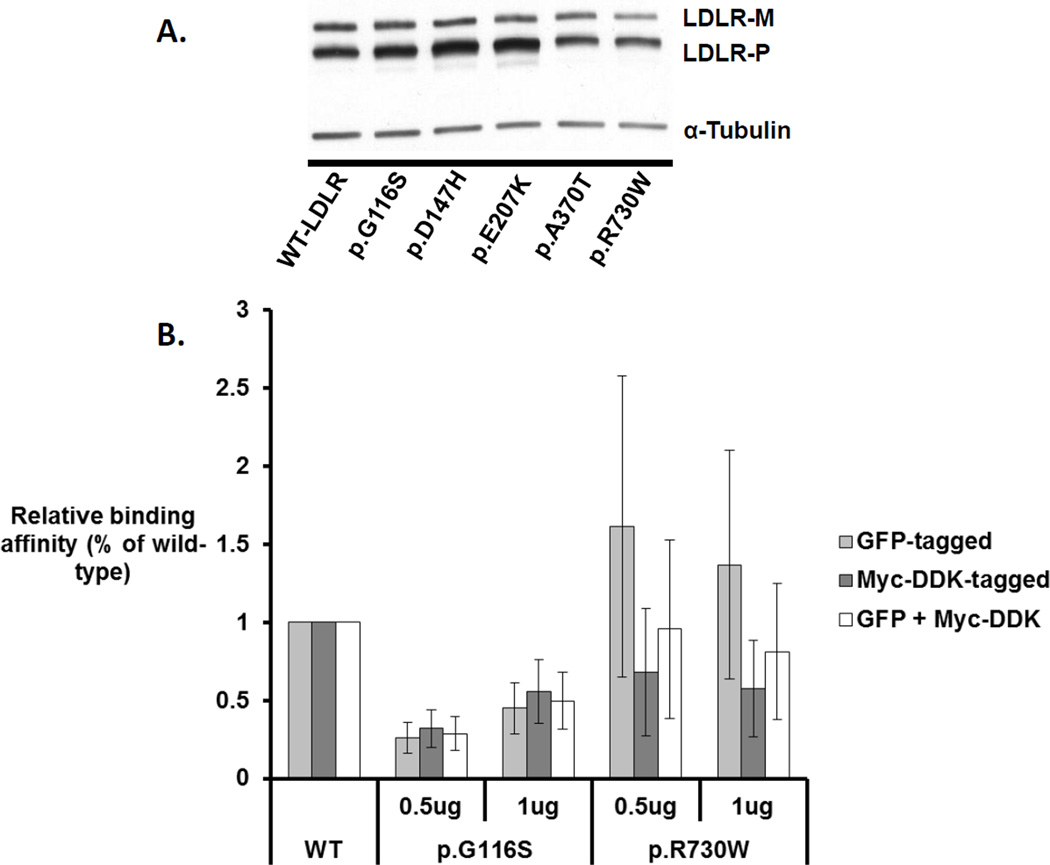
Finally, we investigated the function of both variants in vitro, using cell-based models transfected with plasmid constructs encoding wild-type, p.G116S, or p.R730W LDLR variants. Overall, p.G116S tended to show increased mean mature LDLR expression by 31%, while p.R730W had reduced mean mature LDLR expression by 63% relative to the wild-type LDLR constructs. In vitro LDL-binding assays adjusted for total LDLR expression linked p.G116S with a significant 61% reduction in LDL binding ability, while p.R730W had a non-significant 12% reduction in binding ability (Figure 3).
Figure 3. Comparison of LDLR expression and ligand binding ability between wild-type and Inuit-identified variants.
A) COS7 cells were transfected with expression vectors encoding Myc-DDK-tagged LDLR and LDLR variants mutated to recreate the Inuit-identified polymorphisms or established LDLR variants previously reported. Total cell lysates were analysed and transfections for this model were performed twice. B) LDLR binding activity was measured in HepG2 cells and normalized based on expression data from the corresponding mature LDLR isoforms. Binding activity experiments were performed in triplicate at two doses of LDL and were averaged. Average binding activity estimates were then normalized based on the expression data determined from either the Myc-DDK-tagged, GFP-tagged, or combined GFP- and Myc-DDK-tagged expression models. Error bars represent normalized standard error. All binding read-outs for p.G116S transfectants are significantly different from wild-type (all P<0.05) while none of the read-outs for p.R730W transfectants are significantly different from wild-type (all P >0.05).
The LDLR p.G116S variant in exon 4 resides within the ligand binding domain.27 Of missense or nonsense mutations in LDLR that cause monogenic FH, ~ 20% reside within exon 4, which is considered to be a mutational hot-spot.28 The pathogenic relevance of p.G116 in receptor function was supported by identification of the p.G116C variant in a Polish patient with hypercholesterolemia.29 In contrast, p.R730W is within in exon 15, which encodes an attachment site for O-linked carbohydrate chains; this domain has no clear functional role.27 Fewer than 1% of disease-causing LDLR mutations reside within exon 15.28 Sequence conservation analysis suggested stronger evolutionary conservation at p.G116 compared to p.R730 (Supplemental Figure 2) while multiple algorithms predicted a more deleterious effect for p.G116S than p.R730 on LDL receptor function (Supplemental Table 3). The role of p.R730 in LDLR function remained unclear as a different mutation at p.R730, namely p.R730Q, was found in a sample from a Dutch FH cohort but was predicted to be benign and was reported as likely not disease-causing.30
Discussion
LDLR p.G116S thus appears to be an example of the hypothesized but so far elusive entity in lipoprotein genetics, namely a common genetic variant whose LDL cholesterol raising effect is intermediate between the modest effects attributable to GWAS alleles and the large effects of rare LDLR mutations causing monogenic disease (FH). While bioinformatic predictions further supported a functional consequence of p.G116S, the functional studies ultimately corroborate the observed phenotypic effect of p.G116S. The ~ 60% reduced ligand-binding ability of cells expressing the p.G116S is intermediate between that of wild-type LDLR and of rare FH-causing mutations, which show up to 100% reductions of ligand-binding ability.27
Our discovery of the association between p.G116S and LDL cholesterol concentration is of particular interest from a public health perspective, as Inuit communities may currently be at the tipping point of environment-related increased risk of CVD and metabolic disorders. In other populations, every 1 mmol/L increase in LDL cholesterol corresponds to a ~20% increase in CVD and ~15% increase in all-cause mortality.31 Thus, the ~0.5 mmol/L increase in LDL cholesterol per p.G116S allele could potentially lead to ~10% and ~7.5% increased risk of CVD and all-cause mortality, respectively. Our analyses indicated that p.G116S carriers were at a ~3-fold increased risk of high LDL cholesterol (>5 mmol/L) which suggested that p.G116S carriers were also more likely to be candidates for pharmacologic intervention than non-carriers (OR 3.02, 95% CI 2.34 – 3.90, p = 1.7 × 10−17). Unfortunately, data on CVD end points were not systematically collected in the surveys that comprised this study, so the possible impact of p.G116S on metabolic and CVD risk among the Inuit cannot be directly inferred at this time. A link between this genetic variant and CVD risk would need to be formally evaluated, for instance using Mendelian randomization or another appropriate prospective study design. Furthermore, it would be of interest to detect possible interactions between lifestyle factors, other risk factors and the phenotypic impact of LDLR p.G116S. While baseline between-population differences in lipid profiles might be consistent with environmental effects (Table 1), we have not systematically collected comprehensive diet and lifestyle data; while we would like to do this in the future, such an analysis is beyond the scope of the present report.
As with all association studies, a potential risk of population stratification artefacts exists. However, there are several reasons why we believe that this is not a major issue here. First, we adjusted for geographic location in the association and correlation analyses for the combined Inuit cohort; the association of G116S with LDL-C was highly significant with this adjustment variable included. Second, while the minor allele of G116S varies by geographic region, the directionality of the association by genotype is the same, and is individually significant, in each of the 5 subpopulations for LDL-C, and the related traits of TC, non-HDL-C and apo B (see Supplemental Tables 1A and 1B). Third, we have functionally evaluated the variants in vitro in 2 different cell lines and show a significant loss of binding function for the variant that is significantly associated with LDL cholesterol levels, but no functional impact of the variant that is not associated with LDL cholesterol levels. The findings for dysfunction of G116S are similar in quality, although smaller in magnitude, than those that we have seen for our patients with clinical FH with mutations in the LDLR gene. Finally, principal component analysis performed using genome-wide markers from the exome array on three of the five Inuit subpopulations show a distinctive clustering, with no overlap at all with Caucasian or African American clusters (data not shown). However, some small stratification artefacts are still possible.
While we studied LDLR variation, we did not screen the additional FH genes APOB and PCSK9, so we cannot rule out similar additional effects on this quantitative trait. Furthermore, GWA studies in other global populations have recently implicated >30 genes that modulate LDL cholesterol concentration; cumulatively these might have a larger impact than the 0.54 mmol/L per allele effect of p.G116S.26 A comprehensive genetic screen for LDL cholesterol-related variants using multi-locus high-density genotyping strategies or microarrays, while of potential interest in these samples, is far beyond the scope of the studies reported here. Also, the reason that these distinct variants arose in circumpolar peoples in the first place cannot be determined or even reasonably speculated upon at this time. The effect size on LDL cholesterol is not consistent with any known or obvious survival advantage, nor does there seem to be any potential for negative selection, since CVD onset typically follows decades after the onset of the reproductive years. Finally, while the p.R730W variant appeared to have minimal impact on LDL cholesterol at the population level, a possible impact on other pathways or networks cannot be ruled out from the studies performed here.
Thus, our screen for FH-related variation in the Inuit uncovered a unique genetic variant among global populations: LDLR p.G116S is a common, dysfunctional variant that is strongly associated with a large LDL cholesterol-raising effect, although not causing classical FH. It seems to embody the type of variant that has been long sought-after in the post-GWAS era and warrants consideration in evaluating clinical and public health implications as part of the fabric of CVD risk in the circumpolar north.
Supplementary Material
Acknowledgments
We thank Cynthia G. Sawyez for technical support and advice.
Sources of Funding
Dr. Hegele holds the Edith Schulich Vinet Canada Research Chair (Tier I) in Human Genetics, the Martha G. Blackburn Chair in Cardiovascular Research, and the Jacob J. Wolfe Distinguished Medical Research Chair at the University of Western Ontario. This work was supported by CIHR (MOP-13430 and MOP-79533), the Heart and Stroke Foundation of Ontario (T-6066 and 000353) and Genome Canada through Genome Quebec.
Footnotes
Disclosures
None.
References
- 1.Bjerregaard P, Dyerberg J. Mortality from ischaemic heart disease and cerebrovascular disease. Greenland Int J Epidemiol. 1988;17:514–519. doi: 10.1093/ije/17.3.514. [DOI] [PubMed] [Google Scholar]
- 2.Middaugh JP. Cardiovascular deaths among Alaskan Natives. 1980–86. Am J Public Health. 1990;80:282–285. doi: 10.2105/ajph.80.3.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Young TK, Moffatt ME, O'Neil JD. Cardiovascular diseases in a Canadian Arctic population. Am J Public Health. 1993;83:881–887. doi: 10.2105/ajph.83.6.881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bjerregaard P, Young TK, Hegele RA. Low incidence of cardiovascular disease among the Inuit--what is the evidence? Atherosclerosis. 2003;166:351–357. doi: 10.1016/s0021-9150(02)00364-7. [DOI] [PubMed] [Google Scholar]
- 5.Bjerregaard P, Mulvad G, Pedersen HS. Cardiovascular risk factors in Inuit of Greenland. Int J Epidemiol. 1997;26:1182–1190. doi: 10.1093/ije/26.6.1182. [DOI] [PubMed] [Google Scholar]
- 6.Chateau-Degat ML, Dewailly E, Louchini R, Counil E, Noel M, Ferland A, et al. Cardiovascular burden and related risk factors among Nunavik (Quebec) Inuit: insights from baseline findings in the circumpolar Inuit health in transition cohort study. Can J Cardiol. 2010;26:190–196. doi: 10.1016/s0828-282x(10)70398-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ebbesson SO, Adler AI, Risica PM, Ebbesson LO, Yeh JL, Go OT, et al. Cardiovascular disease and risk factors in three Alaskan Eskimo populations: the Alaska-Siberia project. Int J Circumpolar Health. 2005;64:365–386. doi: 10.3402/ijch.v64i4.18014. [DOI] [PubMed] [Google Scholar]
- 8.Howard BV, Comuzzie A, Devereux RB, Ebbesson SO, Fabsitz RR, Howard WJ, et al. Cardiovascular disease prevalence and its relation to risk factors in Alaska Eskimos. Nutr Metab Cardiovasc Dis. 2010;20:350–358. doi: 10.1016/j.numecd.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jernigan VB, Duran B, Ahn D, Winkleby M. Changing patterns in health behaviors and risk factors related to cardiovascular disease among American Indians and Alaska Natives. Am J Public Health. 2010;100:677–683. doi: 10.2105/AJPH.2009.164285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kellett S, Poirier P, Dewailly E, Sampasa H, Chateau-Degat ML. Is severe obesity a cardiovascular health concern in the Inuit population? Am J Hum Biol. 2012;24:441–445. doi: 10.1002/ajhb.22241. [DOI] [PubMed] [Google Scholar]
- 11.Haase A, Goldberg AC. Identification of people with heterozygous familial hypercholesterolemia. Curr Opin Lipidol. 2012;23:282–289. doi: 10.1097/MOL.0b013e3283556c33. [DOI] [PubMed] [Google Scholar]
- 12.Jorgensen ME, Bjerregaard P, Kjaergaard JJ, Borch-Johnsen K. High prevalence of markers of coronary heart disease among Greenland Inuit. Atherosclerosis. 2008;196:772–778. doi: 10.1016/j.atherosclerosis.2007.01.008. [DOI] [PubMed] [Google Scholar]
- 13.Redwood DG, Lanier AP, Johnston JM, Asay ED, Slattery ML. Chronic disease risk factors among Alaska Native and American Indian people, Alaska, 2004–2006. Prev Chronic Dis. 2010;7:A85. [PMC free article] [PubMed] [Google Scholar]
- 14.Bjerregaard P, Jorgensen ME, Borch-Johnsen K. Serum lipids of Greenland Inuit in relation to Inuit genetic heritage, westernisation and migration. Atherosclerosis. 2004;174:391–398. doi: 10.1016/j.atherosclerosis.2004.02.010. [DOI] [PubMed] [Google Scholar]
- 15.Ebbesson SO, Schraer C, Nobmann ED, Ebbesson LO. Lipoprotein profiles in Alaskan Siberian Yupik Eskimos. Arctic Med. Res. 1996;55:165–173. [PubMed] [Google Scholar]
- 16.Goldstein JL, Brown MS. The LDL receptor. Arterioscler Thromb Vasc Biol. 2009;29:431–438. doi: 10.1161/ATVBAHA.108.179564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mohatt GV, Plaetke R, Klejka J, Luick B, Lardon C, Bersamin A, et al. The Center for Alaska Native Health Research Study: a community-based participatory research study of obesity and chronic disease-related protective and risk factors. Int J Circumpolar Health. 2007;66:8–18. doi: 10.3402/ijch.v66i1.18219. [DOI] [PubMed] [Google Scholar]
- 18.Moffatt ME, Young TK, O'Neil JD, Eidelheit S, Fish I, Mollins J. The Keewatin Health Assessment Study, NWT, Canada. Arctic Med Res. 1993;52:18–21. [PubMed] [Google Scholar]
- 19.Saudny H, Leggee D, Egeland G. Design and methods of the Adult Inuit Health Survey 2007–2008. Int J Circumpolar Health. 2012;71 doi: 10.3402/ijch.v71i0.19752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bjerregaard P, Curtis T, Borch-Johnsen K, Mulvad G, Becker U, Andersen S, et al. Inuit health in Greenland: a population survey of life style and disease in Greenland and among Inuit living in Denmark. Int J Circumpolar Health. 2003;62(Suppl 1):3–79. doi: 10.3402/ijch.v62i0.18212. [DOI] [PubMed] [Google Scholar]
- 21.Fullerton SM, Clark AG, Weiss KM, Nickerson DA, Taylor SL, Stengard JH, et al. Apolipoprotein E variation at the sequence haplotype level: implications for the origin and maintenance of a major human polymorphism. Am J Hum Genet. 2000;67:881–900. doi: 10.1086/303070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Damgaard D, Larsen ML, Nissen PH, Jensen JM, Jensen HK, Soerensen VR, et al. The relationship of molecular genetic to clinical diagnosis of familial hypercholesterolemia in a Danish population. Atherosclerosis. 2005;180:155–160. doi: 10.1016/j.atherosclerosis.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 23.Anderson TJ, Gregoire J, Hegele RA, Couture P, Mancini GB, McPherson R, et al. 2012 update of the Canadian Cardiovascular Society guidelines for the diagnosis and treatment of dyslipidemia for the prevention of cardiovascular disease in the adult. Can J Cardiol. 2013;29:151–167. doi: 10.1016/j.cjca.2012.11.032. [DOI] [PubMed] [Google Scholar]
- 24.Khan TA, Shah T, Prieto D, Zhang W, Price J, Fowkes GR, et al. Apolipoprotein E genotype, cardiovascular biomarkers and risk of stroke: systematic review and meta-analysis of 14,015 stroke cases and pooled analysis of primary biomarker data from up to 60,883 individuals. Int J Epidemiol. 2013;42:475–492. doi: 10.1093/ije/dyt034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ward H, Mitrou PN, Bowman R, Luben R, Wareham NJ, Khaw KT, et al. APOE genotype, lipids, and coronary heart disease risk: a prospective population study. Arch Intern Med. 2009;169:1424–1429. doi: 10.1001/archinternmed.2009.234. [DOI] [PubMed] [Google Scholar]
- 26.Teslovich TM, Musunuru K, Smith AV, Edmondson AC, Stylianou IM, Koseki M, et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hobbs HH, Russell DW, Brown MS, Goldstein JL. The LDL receptor locus in familial hypercholesterolemia: mutational analysis of a membrane protein. Annu Rev Genet. 1990;24:133–170. doi: 10.1146/annurev.ge.24.120190.001025. [DOI] [PubMed] [Google Scholar]
- 28.Stenson PD, Ball EV, Howells K, Phillips AD, Mort M, Cooper DN. The Human Gene Mutation Database: providing a comprehensive central mutation database for molecular diagnostics and personalized genomics. Hum Genomics. 2009;4:69–72. doi: 10.1186/1479-7364-4-2-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chmara M, Wasag B, Zuk M, Kubalska J, Wegrzyn A, Bednarska-Makaruk M, et al. Molecular characterization of Polish patients with familial hypercholesterolemia: novel and recurrent LDLR mutations. J Appl Genet. 2010;51:95–106. doi: 10.1007/BF03195716. [DOI] [PubMed] [Google Scholar]
- 30.Fouchier SW, Kastelein JJ, Defesche JC. Update of the molecular basis of familial hypercholesterolemia in The Netherlands. Hum Mutat. 2005;26:550–556. doi: 10.1002/humu.20256. [DOI] [PubMed] [Google Scholar]
- 31.Cholesterol Treatment Trialists' (CTT) Collaborators. Mihaylova B, Emberson J, Blackwell L, Keech A, Simes J, et al. The effects of lowering LDL cholesterol with statin therapy in people at low risk of vascular disease: meta-analysis of individual data from 27 randomised trials. Lancet. 2012;380;9841:581–590. doi: 10.1016/S0140-6736(12)60367-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.