ABSTRACT
A rising theme among intracellular microbes is the delivery of ankyrin repeat-containing effectors (Anks) that interact with target proteins to co-opt host cell functions. Orientia tsutsugamushi, an obligate intracellular bacterium and the etiologic agent of scrub typhus, encodes one of the largest Ank repertoires of any sequenced microorganism. They have been previously identified as type 1 secretion system substrates. Here, in silico and manual sequence analyses revealed that a large proportion of O. tsutsugamushi strain Ikeda Anks bear a eukaryotic/poxvirus-like F-box motif, which is known to recruit host cell SCF1 ubiquitin ligase machinery. We assessed the Anks for the ability to serve as F-box proteins. Coimmunoprecipitation assays demonstrated that F-box-containing Anks interact with overexpressed and/or endogenous SCF1 components. When coexpressed with FLAG-Ank4_01 or FLAG-Ank9, a glutathione S-transferase (GST)-tagged version of the SCF1 component SKP1 localized to subcellular sites of FLAG-Ank accumulation. The abilities of recombinant Anks to interact and colocalize with SKP1 were F-box dependent. GST-SKP1 precipitated O. tsutsugamushi-derived Ank9 from infected host cells, verifying both that the pathogen expresses Ank9 during infection and the protein's capability to bind SKP1. Aligning O. tsutsugamushi, poxviral, and eukaryotic F-box sequences delineated three F-box residues that are highly conserved and likely to be functionally important. Substitution of these residues ablated the ability of GFP-Ank9 to interact with GST-SKP1. These results demonstrate that O. tsutsugamushi strain Ikeda Anks can co-opt host cell polyubiquitination machinery, provide the first evidence that an O. tsutsugamushi Ank does so during infection, and advance overall understanding of microbial F-box proteins.
IMPORTANCE Ankyrin repeat-containing proteins (Anks) are important virulence factors of intracellular bacteria that mediate protein-protein interactions with host cell targets. Orientia tsutsugamushi, which causes a debilitating infection called scrub typhus in one of the most densely populated regions of the world, encodes one of the largest Ank armamentariums of any sequenced bacterium. This study demonstrates that O. tsutsugamushi strain Ikeda Anks also bear F-box motifs that interact with host cell polyubiquitination machinery. By proving that an Orientia-derived Ank interacts with SKP1 in infected cells, this evidences the first bona fide Orientia effector and the first example of an endogenous F-box-containing Ank–mammalian-host ligand interaction for any intracellular bacterium. Also, importantly, this work identifies key residues that are essential for microbial F-box function.
INTRODUCTION
The obligate intracellular bacterium Orientia tsutsugamushi is the causative agent of scrub typhus, a potentially fatal disease transmitted to humans through the bite of an infected trombiculid mite. O. tsutsugamushi invades phagocytic cells of the immune system, as well as endothelial cells of the skin and major organs (1–4). Diagnosis of scrub typhus is complicated by its nonspecific symptoms, which include fever, rash, pneumonitis, and meningitis. If left untreated, the disease can progress to disseminated intravascular coagulation, circulatory collapse, organ failure, and death. Depending on the bacterial strain, patient immune competence, and antibiotic intervention, the mortality rate can be as high as 50% (2, 5–7). Scrub typhus is endemic to a 13 million-km2 area of the Asia-Pacific region and threatens over 1 billion people, with an estimated 1 million cases annually (2, 5, 8). Military personnel serving in areas of endemicity are at risk for contracting scrub typhus, as evidenced by over 7 decades of military disease incidence. Hundreds of American and Allied soldiers died from and several thousand became ill with scrub typhus during World War II (5, 8–10), and scrub typhus was the second leading cause of febrile illness among troops during the Vietnam conflict (5, 8, 11–13).
How O. tsutsugamushi modulates host cell functions is poorly understood. Ankyrin repeat-containing proteins (Anks) are important virulence factors of intracellular bacterial pathogens (14–26). These effectors contain one or more ankyrin repeats, each of which consists of a 33-amino-acid motif that mediates protein-protein interactions (15). Recently, we characterized Anks from O. tsutsugamushi strain Ikeda (NCBI accession number NC_010793.1) (23). The strain's genome includes 9 ank pseudogenes and 38 ank open reading frames (ORFs), 8 of which occur as multiple identical or nearly identical copies and 12 of which exist as single-copy genes (27). We previously selected all of the single-copy Anks and one representative member of each multicopy Ank group to arrive at a subset of 20 full-length distinguishable Anks for characterization (23). O. tsutsugamushi expresses all 20 of the representative ank genes during infection of mammalian host cells. The encoded proteins are type 1 secretion system (T1SS) substrates that, when ectopically expressed, localize to distinct subcellular locales, including the endoplasmic reticulum, Golgi apparatus, and nucleus, suggesting their potential for modulating diverse host cellular processes (23).
In this study, further examination revealed that 16 of the 20 Anks carry sequences that are homologous to eukaryotic/poxviral F-box motifs known to interact with SKP1 (S-phase kinase-associated protein 1) of the SCF1 (SKP1-Cullin 1-F-box) E3 ubiquitin ligase complex (28). F-box-containing proteins (FBPs) also possess a repetitive sequence, such as an ankyrin repeat, that promotes protein-protein interactions. With this pair of interaction motifs, FBPs bridge SCF1 components with protein substrates to be polyubiquitinated and subsequently destroyed in the 26S proteasome. Eukaryotic polyubiquitination of target proteins occurs via three sequential steps: (i) E1 ubiquitin ligase activates ubiquitin, (ii) ubiquitin is transferred to the E2 ubiquitin conjugase, and (iii) ubiquitin is linked to the substrate by the E3 ligase complex. The SCF1 complex, one of the most studied E3 ligases, consists of SKP1, Cullin 1 (CUL1), and RING box 1 (RBX1) subunits, in addition to the FBP. CUL1 serves as the scaffold for the catalytic core, binding both RBX1 and SKP1, with the FBP binding SKP1 via its F-box. As the final SCF1 component, the FBP interacts with the target protein via its protein-protein interaction domain, acting as a substrate specificity module to direct SCF1-mediated polyubiquitination of the target (29, 30). Numerous intracellular bacterial pathogens encode FBPs (20–22, 31–37), evidencing their potential to exploit the SCF1 complex.
Considering that the majority of Ikeda Anks carry putative F-boxes, their abilities to interact with SCF1 components were investigated. The results presented here demonstrate that Ikeda Anks can co-opt host cell polyubiquitination machinery. They also demonstrated that Orientia-derived Ank9 interacts with SKP1 in infected cells, thereby providing the first evidence that an O. tsutsugamushi Ank, or any effector, does so during infection. Analysis of the Ank F-box motifs showed homology with canonical F-box sequences, though the Ikeda Anks resemble poxviral F-boxes in terms of arrangement and length. Aligning the O. tsutsugamushi F-box sequences revealed conservation of three key residues that are essential for F-box function, as demonstrated by the inability of a recombinant Ank with these residues replaced to interact with SKP1. Overall, this study advances the understanding of microbial F-box proteins, delineates the first bona fide O. tsutsugamushi Ank effector that is expressed and interacts with SKP1 during infection, and underscores the importance of studying virulence factor function in more than one scrub typhus bacterial strain.
MATERIALS AND METHODS
Cell culture and infection.
HeLa human cervical epithelial cells (CCL-2; ATCC, Manassas, VA) were maintained in RPMI 1640 (Gibco, Grand Island, NY) medium supplemented with 10% fetal bovine serum (FBS) at 37°C in a humidified incubator with 5% CO2. O. tsutsugamushi strain Ikeda was maintained in HeLa cells and passaged via addition of O. tsutsugamushi-laden supernatant to uninfected HeLa cells treated with 0.4 μg/ml daunorubicin hydrochloride (Sigma-Aldrich, St. Louis, MO) (38).
Plasmids.
Constructs for expressing mammalian codon-optimized Anks N-terminally fused to FLAG or green fluorescent protein (GFP) tags, respectively, have been previously described (23). Ank4_01ΔF-box constructs were made by PCR amplification of Ank4_01 nucleotides 4 to 945 (corresponding to amino acids 2 to 315) using pBMH-Ank4_01 (23) as the template and primers containing KpnI and XbaI restriction sites (5′-TGCTGGTACCAAACAATGGGAACCTGCTGCAC-3′ and 5′-GATCTCTAGATCATTCTTTCATCCACAGCAGTCTC-3′ [boldface indicates extra nucleotides upstream of restriction sites; restriction sites are underlined]). Ank9ΔF-box constructs were made by PCR amplification of Ank9 nucleotides 4 to 1149 (corresponding to amino acids 2 to 383) using pBMH-Ank9 (23) as the template and primers containing the EcoRI and SalI restriction sites (5′-ATCGGAATTCGGGGAGATTCACCAG-3′ and 5′-ATCGATTGGTCGACTCAATTGTTCCAGCTAGGGGTGG-3′). Ank PCR products were subsequently cloned into the pEGFP-C1 and p3XFLAG-CMV-14 vectors. PCR amplification, restriction digestion, and ligation were performed as previously described (23). All generated clones were sequenced to ensure nucleotide fidelity. pApex-3-FLAG-SKP2, pApex-3-GST-SKP1, pApex-3-GST-CUL1, and pApex-3-GST-RBX1 were kind gifts from Andrew Mercer (University of Otago, Dunedin, New Zealand) (39). pFLAG-BAP, which encodes the negative control, FLAG-tagged bacterial alkaline phosphatase (BAP), was purchased from Sigma-Aldrich. Genes corresponding to Ank9 with the key conserved F-box residues L384, I392, and E400 converted to alanines (LIE-AAA) or to alanines and asparagine (LIE-AAN) were synthesized with flanking EcoRI and SalI restriction cloning sites by Genewiz (South Plainfield, NJ). These Ank9 mutant genes were cloned into pEGFP-C1 as previously described (23) to generate pGFP-Ank9-LIE-AAA and pGFP-Ank9-LIE-AAN.
In silico analyses of O. tsutsugamushi Anks.
Identification of putative F-box domains was accomplished as previously noted with the SMART (Simple Modular Architecture Research Tool) algorithm (http://smart.embl-heidelberg.de/) (23) and by manual sequence scanning using Geneious R6 v.6.1.8 software.
Antiserum generation.
Affinity-purified rabbit polyclonal antiserum targeting Ank9 amino acids 12 to 28 was generated by New England Peptide (Gardner, MA). Recombinant O. tsutsugamushi outer membrane protein A (OmpA; OTT_1320) was generated using primers 5′-GACGACGACAAGATATGTTTATGGCAAAGATCTAAACATAGTAAC-3′ and 5′-GAGGAGAAGCCCGGTTATTTATGTTTCCCATGTATAGCTTGTAAAAACTG-3′ (sequences that are compatible with ligation-independent cloning are italicized) to amplify the region corresponding to amino acids 22 to 93, which is unique to OmpA, as assessed by NCBI BLAST (Basic Local Alignment Search Tool) (http://blast.ncbi.nlm.nih.gov/) searches. The primers harbored ligase-independent cloning (LIC) tails, and the amplicon was annealed with pET46 Ek/LIC (Novagen, EMD Millipore, Billerica, MA). Plasmids were propagated in Escherichia coli DE3 NovaBlue cells (EMD Millipore). Protein was expressed by inoculation of 100 ml Luria-Burtani (LB) broth supplemented with 100 mg liter−1 of ampicillin with a colony from freshly plated E. coli, and the culture was allowed to autoinduce protein expression overnight. The final recombinant protein, which carried an N-terminal 6×His tag of 1.7 kDa, was purified (>95% homogeneity) by immobilized metal affinity chromatography (IMAC) (HisTrap; GE Healthcare Biosciences, Pittsburgh, PA) according to the manufacturer's protocol and concentrated using Amicon Ultra filters (EMD Millipore). The protein concentration was determined using the bicinchoninic acid assay. Two 8-week-old male Sprague-Dawley rats were immunized with 40 μg His-OmpA protein emulsified in a 1:1 ratio with complete Freund's adjuvant (CFA). The recombinant protein was intraperitoneally administered in a total volume of 400 μl. At weeks 2 and 4, the rats were boosted with 40 μg protein in Freund's incomplete adjuvant. At week 6, the rats were humanely sacrificed, blood was recovered by cardiac puncture, and the sera were isolated. Antiserum from one of the rats was used at 1:1,000 for Western blot analysis. These studies were performed in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals. The Virginia Commonwealth University Institutional Animal Care and Use Committee approved the protocol.
Pulldown and Western blot analyses.
HeLa cells were seeded in wells of a six-well plate for 90 to 95% confluence the next day. A total of 8 μg of plasmid DNA was transfected per well using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) according to the manufacturer's directions and incubated for 18 to 24 h. In some instances, culture media from heavily infected (≥90%) HeLa cell medium containing naturally liberated O. tsutsugamushi organisms was applied, and infection was allowed to proceed for 48 h. The cells were released from well surfaces by the addition of 0.05% trypsin-EDTA. The cells were pelleted at 300 × g for 5 min and subsequently lysed in 300 μl of lysis buffer (20 μM Tris, pH 7.4, 0.5 M NaCl, 0.7% Tween 20, with EDTA-free protease inhibitor [Roche Diagnostics GmBH, Mannheim, Germany]) for 40 min on ice. The insoluble material was pelleted at 10,000 × g for 10 min at 4°C and discarded. Glutathione-Sepharose (GE Healthcare) or anti-FLAG affinity agarose resin (Sigma-Aldrich) was incubated with the cell lysate supernatants overnight at 4°C with rotation to precipitate glutathione S-transferase (GST)- or FLAG-tagged proteins, respectively. The resin was washed three or four times in lysis buffer prior to elution of proteins with 30 μl 2× SDS sample buffer. Samples were resolved by SDS-PAGE and transferred to nitrocellulose as previously described (23). The resulting blots were screened with antibodies specific for GST (1:10,000; Santa Cruz Biotechnology, Dallas, TX), FLAG (1:1,000; Sigma-Aldrich), GFP (1:1,000; Invitrogen), SKP1 (1:500; Cell Signaling, Beverly, MA), RBX1 (ROC1; 1:250; AbCam, Cambridge, MA), CUL1 (1:5,000; AbCam), O. tsutsugamushi strain Ikeda Ank9 (1:1,000), or O. tsutsugamushi OmpA (1:1,000). Horseradish peroxidase-conjugated secondary antibodies to mouse, rat, or rabbit IgG were used (1:10,000; Cell Signaling), followed by addition of Dura or Femto chemiluminescent substrate (Thermo Scientific, Rockford, IL) and imaging using standard blue film or the ChemiDoc Touch Imaging System with Image Lab 5.2 software (Bio-Rad, Hercules, CA).
Ectopic expression and confocal microscopic analysis of O. tsutsugamushi Ank9 with SKP1 in mammalian host cells.
HeLa cells were seeded onto 12-mm glass coverslips in a 24-well plate with 400 μl of RPMI medium containing 10% (vol/vol) FBS per well. After 24 h, the medium was removed and the cells were washed once with PBS prior to addition of 500 μl RPMI medium to serum starve the cells for 24 h. The resulting synchronized cells were serum released by the addition of 400 μl RPMI containing 10% FBS. At 1 h after serum release, the cells were cotransfected with 0.4 μg of plasmid encoding GFP-Ank9, GFP-Ank9ΔFBox, GFP-Ank4_01, GFP-Ank4_01ΔF-box, or GFP and 0.13 μg of plasmid GST-SKP1 using Lipofectamine 2000. The cells were fixed in 4% (vol/vol) paraformaldehyde (PFA) (Electron Microscopy Science, Hatfield, PA) in PBS at 10 h posttransfection. Coverslips were blocked for 1 h at room temperature in 5% (vol/vol) bovine serum albumin (BSA) in PBS. The coverslips were then stained with chicken GFP-IgY antibody (Invitrogen) and rabbit GST antibody (Invitrogen) at a 1:500 dilution in 1% BSA-PBS for 1 h. The wells were washed three times in PBS prior to addition of Alexa Fluor 488-conjugated goat anti-chicken IgG antibody (1:1,000; Invitrogen) and Alexa Fluor 594-conjugated goat anti-rabbit IgG antibody (1:1,000; Invitrogen) in 1% BSA-PBS for 1 h. The coverslips were washed three times with PBS prior to mounting on slides using Prolong Gold Antifade plus 4′,6-diamidino-2-phenylindole (DAPI) (Molecular Probes, Life Technologies, Grand Island, NY). Samples were imaged using a Zeiss LSM 700 laser scanning confocal microscope. ImageJ (40) was used for image profiling analysis with the RGB_Profiler plug-in (http://rsb.info.nih.gov/ij/plugins/rgb-profiler.html).
F-box sequence alignments.
The 42- to 44-amino-acid segment comprising the putative F-box region of each Ank was globally aligned with canonical poxviral and eukaryotic F-box sequences using Geneious R6 v.6.1.8 software. Residues were considered similar if they fell within the following groups: K and R; D and E; A, V, L, I, and M; P and G; F, Y, and W (39). The sequence logo was generated with WebLogo version 2.8.2 (41; http://weblogo.berkeley.edu/logo.cgi). NCBI BLAST was used to compare O. tsutsugamushi strain Ikeda and Boryong Ank amino acid sequences.
RESULTS
O. tsutsugamushi Anks with F-box motifs interact with SKP1.
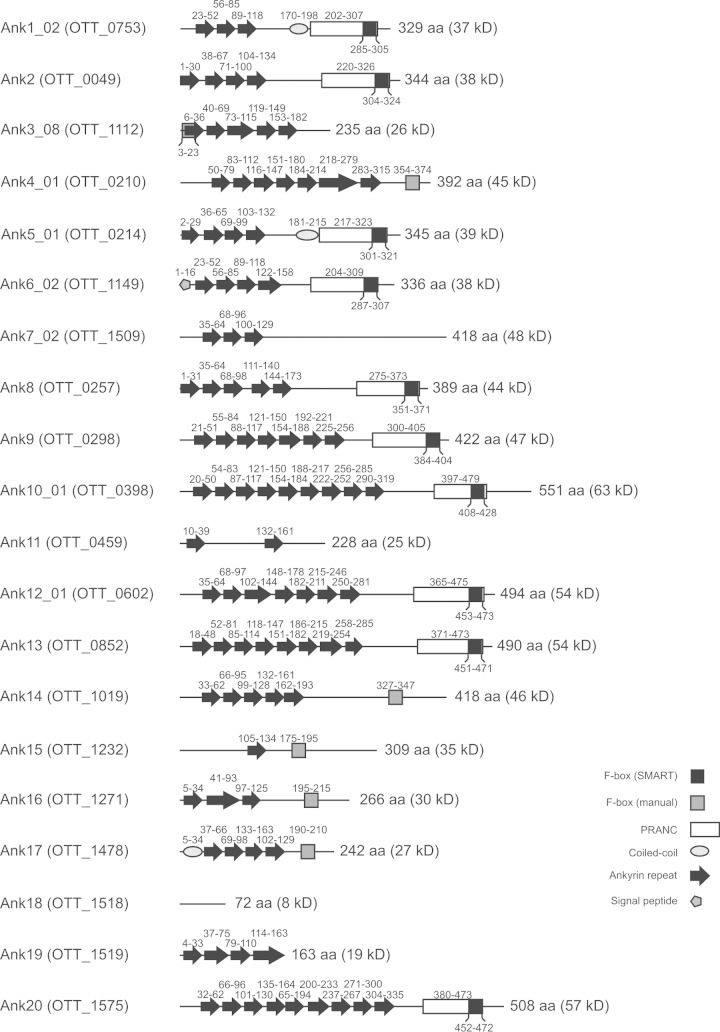
FBPs recruit the SCF1 ubiquitin ligase complex via interaction between the F-box motif and SKP1 (28). SMART analysis determined that 10 of the 20 selected O. tsutsugamushi strain Ikeda Anks (Ank1_02, Ank2, Ank5_01, Ank6_02, Ank8, Ank9, Ank10_01, Ank12_01, Ank13, and Ank20) possess sequences that share similarity with pox protein repeats of ankyrin C-terminal (PRANC) domains (Fig. 1), which are domains that include an F-box and are commonly found within the C termini of poxviral Anks (39, 42). Additionally, manual sequence analysis in which Ikeda Ank residues were compared with those of canonical eukaryotic (43) and poxviral (39, 42) F-boxes revealed that Ank3_08, Ank4_01, Ank14, Ank15, Ank16, and Ank17 carried F-boxes that were not detected by the SMART algorithm. Ank18 carries no ankyrin repeats but is annotated as an Ank because it is homologous to the non-ankyrin repeat portion of the O. tsutsugamushi strain Boryong Ank1u7, which has an ankyrin repeat domain (44). Because it lacks both ankyrin repeats and an F-box (Fig. 1), we excluded Ank18 from further study.
FIG 1.
Schematic of domains found within O. tsutsugamushi strain Ikeda Anks. Each Ank is listed by name, followed by its gene annotation in parentheses. The amino acid locations of ankyrin repeats, putative F-boxes, coiled-coil domains, signal peptides, and PRANC domains are noted above or below the corresponding shape. F-boxes predicted using the SMART algorithm are found within PRANC domains, while those found via manual sequence searches are shown as separate boxes. The length of each Ank is noted to the right of its respective schematic, with the estimated protein size. Though Ank18 does not contain any ankyrin repeats itself, it is annotated as an Ank because it shares homology with nonrepeat regions of O. tsutsugamushi strain Boryong Ank1u7 (OTBS_1195), which does contain ankyrin repeats. aa, amino acids.
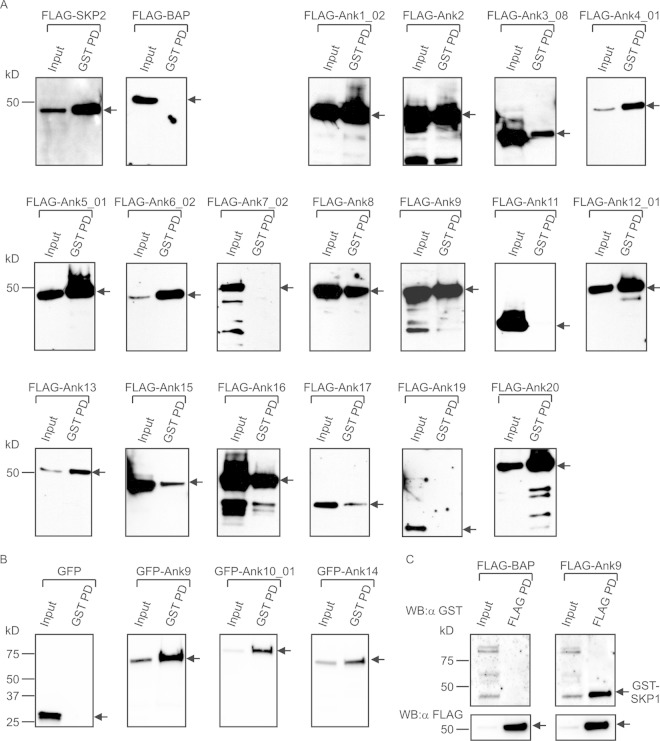
With 16 O. tsutsugamushi Anks bearing putative F-boxes, we tested the abilities of recombinant forms of these proteins to interact with human SKP1. HeLa cells were transfected to coexpress GST-SKP1, together with Ank proteins N-terminally fused to FLAG or GFP (23). GST-SKP1 and protein binding partners were coprecipitated with glutathione-Sepharose, eluted, and Western blotted. The blots were probed with FLAG or GFP antibody to detect Anks that coprecipitated with GST-SKP1. SKP2, a mammalian FBP and a known binding partner of SKP1 (43, 45), was a positive control, while FLAG-BAP and GFP were negative controls. As shown in Fig. 2, all of the tagged Anks were expressed in the transfected HeLa cells (input lanes). GST-SKP1 coprecipitated all 16 F-box-containing Anks but failed to pull down F-box-deficient Ank7_02, Ank11, and Ank19 (Fig. 1 and 2A). In most instances, the F-box-containing Anks robustly precipitated with GST-SKP1. However, FLAG-tagged Ank3_08, Ank15, and Ank17 precipitated poorly with GST-SKP1 relative to the amount of FLAG-Ank protein expressed (Fig. 2A), suggesting that these Anks facilitate tenuous F-box–SKP1 interactions. GFP-tagged Ank10 and Ank14 were evaluated for SKP1 interactions because FLAG-tagged versions of these proteins expressed poorly. GFP-Ank9 was included as a positive control. GFP-tagged Ank9, Ank10, and Ank14 were each coprecipitated by GST-SKP1 (Fig. 2B). Thus, recombinant Ank proteins are capable of binding SKP1 regardless of their fusion tags. As a complementary approach, a reciprocal pulldown was performed in which FLAG-binding resin was used to precipitate FLAG-Ank9 and thereby coprecipitate GST-SKP1 (Fig. 2C). FLAG-BAP failed to coprecipitate GST-SKP1. Overall, these results demonstrate that all 16 O. tsutsugamushi Ikeda F-box-containing Anks interact with SKP1 and the 3 that lack F-boxes do not.
FIG 2.
O. tsutsugamushi Anks interact with GST-SKP1. (A) GST-SKP1 coprecipitates FLAG-tagged Anks. Lysates of transfected HeLa cells expressing GST-SKP1 together with FLAG-tagged Anks, positive-control FLAG-SKP2, or negative control FLAG-BAP were incubated with glutathione-Sepharose to precipitate GST-SKP1 and thereby coprecipitate interacting proteins. The resulting Western blots were screened with FLAG antibody. (B) GST-SKP1 coprecipitates GFP-tagged Anks. Lysates of transfected HeLa cells expressing GST-SKP1 together with GFP-tagged Ank9, Ank10, Ank14, or negative-control GFP alone were Western blotted and screened with GFP antibody. (C) FLAG-Ank9 coimmunoprecipitates GST-SKP1. Lysates of transfected HeLa cells expressing GST-SKP1 together with FLAG-Ank9 or FLAG-BAP were incubated with FLAG antibody-conjugated agarose beads to immunoprecipitate FLAG-tagged proteins and coprecipitate interacting proteins. The resulting Western blots (WB) were probed with GST antibody (α-GST). Precipitation of FLAG-tagged proteins was confirmed by probing the stripped blots with FLAG antibody (α-FLAG). (A to C) For each experiment, expression of the protein of interest was confirmed in the “Input” lane containing 3% of the sample. The pulldown (PD) lanes represent proteins that coprecipitated with GST-SKP1 (A and B) or coimmunoprecipitated with FLAG-tagged protein (C). Precipitation of GST-SKP1 was confirmed, but is not shown, for each sample in panels A and B. The arrows indicate the expected sizes of the prey proteins. All the blots are aligned at the 50-kDa marker, as indicated on the leftmost blot in each row. The data presented are representative of two to five experiments with similar results.
Ank-SKP1 interactions are F-box dependent.
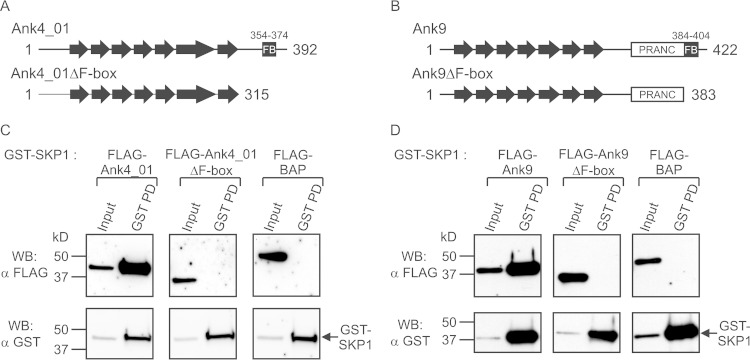
To confirm that the Ank-SKP1 interactions are F-box dependent, additional GST-SKP1 pulldowns were performed with FLAG-tagged Anks that had their F-box motifs deleted. Ank4_01 and Ank9, containing a manually predicted and a SMART algorithm-predicted F-box, respectively, were chosen as representative Anks to have their F-boxes deleted via C-terminal truncations (Fig. 3A and B). Removal of the last 77 residues from FLAG-Ank4_01 (FLAG-Ank4_01ΔF-box) was sufficient to ablate interaction with GST-SKP1 (Fig. 3C). Similarly, removing the C-terminal 39 amino acids of FLAG-Ank9 containing the F-box, but leaving the remainder of the PRANC domain intact (FLAG-Ank9ΔF-box), was sufficient to prevent interaction with GST-SKP1 (Fig. 3D). These results confirm that interactions of O. tsutsugamushi Anks with SKP1 are indeed dependent upon residues within their F-box motifs.
FIG 3.
Ank-SKP1 interactions are F-box dependent. (A and B) Schematics of wild-type Ank4_01 and Ank9 proteins and corresponding Anks with their F-box-containing C termini deleted (Ank4_01ΔF-box and Ank9ΔF-box). Ankyrin repeats are depicted as arrows, with the PRANCs and F-boxes (FB) indicated. The length of each protein is marked with amino acid numbers at its N and C termini, and residues comprising the F-boxes are denoted above. (C and D) GST-SKP1 and interacting proteins precipitated from lysates of HeLa cells coexpressing GST-SKP1 and FLAG-tagged Ank4_01 or Ank9, Ank4_01ΔF-box or Ank9ΔF-box, or the BAP negative control were Western blotted and screened with FLAG antibody to determine which FLAG-tagged proteins coprecipitated with GST-SKP1. GST-SKP1 pulldown was confirmed by probing the stripped blots with GST antibody. The input lanes represent 3% of the total protein input. The data are representative of two or three experiments with similar results.
F-box-containing Anks interact with both ectopically expressed and endogenous SCF1 components.
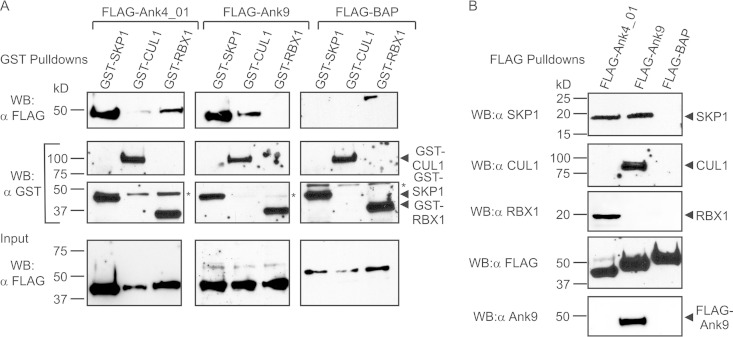
Because F-box-containing Anks interact with SKP1, we investigated if they are also capable of recruiting CUL1 or RBX1. GST pulldown assays were performed on lysates of HeLa cells coexpressing FLAG-tagged Ank4_01 or Ank9 together with GST-tagged SKP1 (positive control), CUL1, or RBX1. As expected, GST-SKP1 coprecipitated FLAG-tagged Ank4_01 and Ank9 (Fig. 4A). GST-RBX1 and, to a lesser degree, GST-CUL1 each coprecipitated FLAG-Ank4_01. Conversely, GST-CUL1, but not GST-RBX1, coprecipitated FLAG-Ank9. GST-tagged SKP1, CUL1, and RBX1 each failed to coprecipitate the negative control, FLAG-BAP. Additional pulldowns were performed to determine if FLAG-tagged Ank4_01 and Ank9 interact with native SKP1, CUL1, and RBX1. FLAG-Ank4_01 precipitated endogenous SKP1 and RBX1, while FLAG-Ank9 precipitated endogenous SKP1 and CUL1 (Fig. 4B). FLAG-BAP failed to pull down any SCF1 component. These data imply that O. tsutsugamushi F-box-containing Anks are capable of recruiting multiple components of the host cell SCF1 ubiquitin ligase complex.
FIG 4.
FLAG-tagged Ank4_01 and Ank9 interact with ectopically expressed and endogenous SCF1 ubiquitin ligase complex components. (A) GST-tagged SCF1 ubiquitin ligase components coprecipitate FLAG-Ank4_01 and FLAG-Ank9. FLAG-tagged Ank4_01, Ank9, and BAP (negative control) were each coexpressed in HeLa cells with GST-tagged SKP1, CUL1, or RBX1. (Top row) GST pulldowns were performed, and the resulting Western blots were probed with FLAG antibody to detect coprecipitated FLAG proteins. (Middle rows) Precipitation of GST-SKP1, GST-CUL1, and GST-RBX1 was confirmed by stripping and reprobing the blots with GST antibody. The asterisk denotes a nonspecific background band that was detected by GST antibody. (Bottom row) FLAG-tagged-protein expression was confirmed using FLAG antibody to screen input samples. (B) FLAG-tagged Ank4_01 and Ank9 coprecipitate endogenous SCF1 components. FLAG-tagged Ank4_01, Ank9, and BAP were precipitated from lysates of transfected HeLa cells using FLAG affinity resin. Native SCF1 ubiquitin ligase components that coprecipitated with FLAG-tagged proteins were detected with SKP1, CUL1, and RBX1 antibodies. Expression and precipitation of FLAG-tagged proteins was confirmed using FLAG antibody. The specificity of Ank9 antiserum was verified by probing the FLAG pulldown blots to detect FLAG-Ank9 but neither FLAG-Ank4_01 nor FLAG-BAP. The data presented are representative of at least two experiments with similar results.
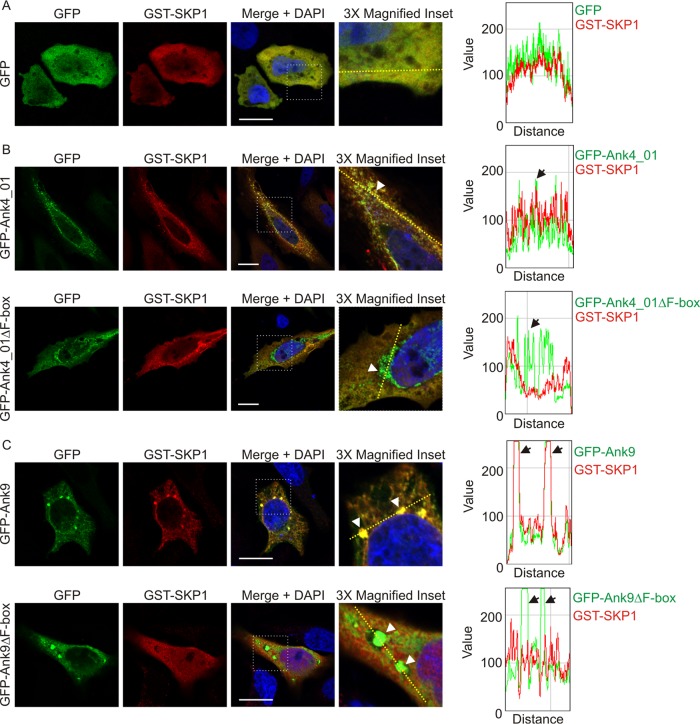
Colocalization of GFP-tagged Anks with GST-SKP1 in host cells is F-box dependent.
The ability of F-box-containing Anks to precipitate with SKP1 in an F-box-dependent manner suggests that they recruit SKP1 in an F-box-dependent fashion to their sites of accumulation in host cells. To investigate this phenomenon, HeLa cells expressing GFP-tagged Ank4_01, Ank4_01ΔF-box, Ank9, Ank9ΔF-box, or GFP together with GST-SKP1 were examined for GFP and GST signal colocalization using laser scanning confocal microscopy (LSCM) and post-data-acquisition image profiling. For the latter method, a line was drawn through a confocal micrograph, and the pixel intensities for each signal were graphed for all points along the line. Ank4_01 and Ank9, and their respective F-box deletions, were chosen for this experiment because when ectopically expressed as GFP fusions, they localize to easily discernible vesicular aggregates (23). In cells expressing GST-SKP1 and GFP, both proteins exhibited diffuse, uniform cytosolic distribution (Fig. 5A). However, upon coexpression with either GFP-Ank4_01 (Fig. 5B) or GFP-Ank9 (Fig. 5C), GST-SKP1 localized to sites of GFP-Ank accumulation. Though GFP-tagged Ank4_01ΔF-box and Ank9ΔF-box retained the aggregative patterns exhibited by their full-length counterparts, GST-SKP1 did not localize with either (Fig. 5B and C). Thus, the ability of Ank4_01 and Ank9 to recruit SKP1 in host cells is F-box dependent.
FIG 5.
Colocalization of GFP-tagged Anks with GST-SKP1 is F-box dependent. HeLa cells expressing GST-SKP1 together with GFP (A), GFP-Ank4_01 or GFP-Ank4_01ΔF-box (B), or GFP-Ank9 or GFP-Ank9ΔF-box (C) were screened with GFP and GST antibodies, stained with DAPI, and examined using LSCM. Scale bars, 20 μm. The areas demarcated by dotted boxes in the “Merge + DAPI” column are magnified 3-fold in the right column. The graphs represent the levels of green and red pixels at each point along the dashed line (moving left to right) in each image in the right column. The arrowheads in the inset images point to a region of interest that correlates with the color peaks denoted by arrows on each respective graph. The images are representative of multiple cells imaged over two to four experiments with similar results.
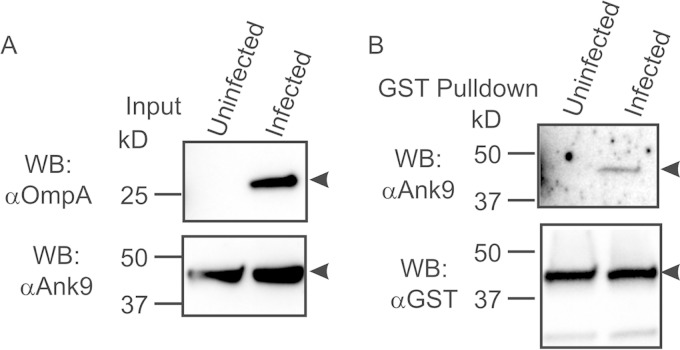
GST-SKP1 precipitates Ank9 expressed by O. tsutsugamushi during infection of mammalian host cells.
Because the studies reported here had thus far examined interactions of ectopically expressed Anks with SKP1, we next examined if an O. tsutsugamushi-derived Ank interacts with SKP1 during infection. First, antiserum against a peptide corresponding to Ank9 amino acids 12 to 28 was generated. Screening Western-blotted lysates of HeLa cells expressing FLAG-Ank4_01, FLAG-Ank9, and FLAG-BAP with the antiserum verified its specificity for Ank9 (Fig. 4B). Next, the Ank9 antiserum was evaluated for the ability to detect native Ank9 in lysates of O. tsutsugamushi-infected HeLa cells. Infection was confirmed using serum against O. tsutsugamushi OmpA (Fig. 6A). Unfortunately, Ank9 antiserum detected a host cell protein with an apparent molecular weight that was similar to that of Ank9 and consequently obscured its detection. Accordingly, GST-SKP1 was ectopically expressed in and subsequently precipitated from O. tsutsugamushi-infected HeLa cells to enrich for Ank9 bound to GST-SKP1 in the absence of the host cell background protein. Ank9 antiserum detected a single protein of the expected size for Ank9 in eluates of GST-SKP1-coprecipitated proteins from infected but not uninfected cells (Fig. 6B). Thus, Ank9 expressed by O. tsutsugamushi during infection of host cells interacts with SKP1.
FIG 6.
GST-SKP1 precipitates O. tsutsugamushi-derived Ank9. HeLa cells expressing GST-SKP1 were infected with O. tsutsugamushi for 48 h prior to lysis and GST pulldown analysis. Uninfected HeLa cells served as a negative control. (A) Ank9 antiserum recognizes a host cell background protein with an apparent molecular mass similar to that of Ank9. To confirm infection, lysates (Input) were subjected to Western blotting with O. tsutsugamushi OmpA antibody. Probing the lysates with Ank9 antiserum detected a background band in both uninfected and infected cell lysates that was the same size as that expected for Ank9 (∼47 kDa). (B) Ank9 antiserum detects O. tsutsugamushi-derived Ank9 coprecipitated by GST-SKP1. To verify that O. tsutsugamushi expresses Ank9 during infection and that it is capable of interacting with SKP1, Western blots of proteins coprecipitated by GST affinity resin were probed with Ank9 antiserum. Screening with GST antibody confirmed expression and precipitation of GST-SKP1. Protein sizes are indicated on the left. The arrowheads point to bands of interest in each blot. The data are representative of at least two experiments with similar results.
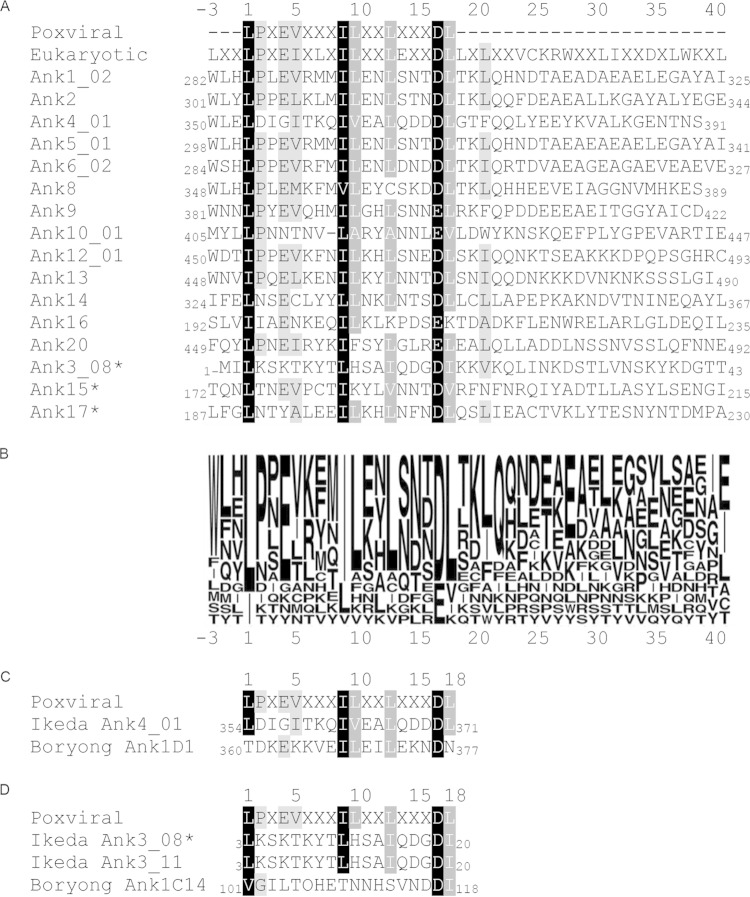
Residues that are highly conserved among poxviral and eukaryotic F-boxes are conserved among O. tsutsugamushi Ikeda Ank F-boxes.
Canonical poxviral and eukaryotic F-boxes share several residues, which implies their importance in mediating interactions with SKP1 (39, 42, 43). To determine if such residues are conserved among O. tsutsugamushi Anks, the F-box sequences of the 16 representative Ikeda F-box-containing Anks were aligned with canonical poxviral (39, 42) and eukaryotic (43) F-box sequences (Fig. 7A). Residues were considered similar if they fell within the following groups: A, V, L, I, and M; D and E; K and R; P and G; or F, Y, and W (39). Branched side chain amino acids (L, I, and V) at positions 1 and 9 in the alignment and acidic residues (D/E) at position 17 were conserved among all 16 O. tsutsugamushi F-box motifs and the canonical sequences (Fig. 7A and B). Branched side chain residues at positions 10, 13, and 18 were conserved among 83.3 to 94.4% of the aligned F-boxes, while residues at positions 2, 4, 5, and 21 were conserved among 72.2 to 77.8% of the sequences. Position 21 was shared between the canonical eukaryotic F-box and 12 of the O. tsutsugamushi Ank F-boxes but not the canonical poxviral F-box sequence. The F/W at position −3 and the Q at position 22 were present in 10 of the Orientia Anks but were absent in both canonical F-box motifs. Ank3, Ank15, and Ank17, each of which inefficiently coprecipitated with GST-SKP1 relative to other F-box-containing Anks (Fig. 2A), maintained conservation of positions 1, 9, and 17 but uniformly lacked conservation at position 2 and, additionally, lacked conservation at positions 4, 5, 10, and/or 21 (Fig. 7A and B).
FIG 7.
Identification of highly conserved O. tsutsugamushi strain Ikeda Ank F-box residues. (A) Alignment of F-boxes from the 16 O. tsutsugamushi strain Ikeda Anks that bind SKP1 with canonical poxviral and eukaryotic F-box sequences. Regions containing each Ank protein's F-box domain were aligned to canonical poxviral and eukaryotic F-box sequences using Geneious software. The shaded residues indicate amino acid similarity, with black highlighting residues that are similar among 100% of the aligned sequences, dark gray highlighting residues that are similar among 83.3 to 94.4% of the aligned sequences, and light gray highlighting residues that are similar among 72.2 to 77.8% of the aligned sequences. Residues were considered similar if they fell within the following groups: A, V, L, I, and M; D and E; K and R; P and G; or F, Y, and W (39). The three Anks marked with asterisks contain a predicted F-box sequence but interacted weakly with SKP1 in pulldown assays compared to the other 13 Anks, as shown in Fig. 2. The numbers at the N and C termini of each sequence indicate the amino acid positions of the F-box motif within the full-length Ank. (B) Sequence logo representation corresponding to the alignment in panel A, showing Ank amino acid conservation at each position. The letter size indicates the relative frequency of a given residue at its respective position in the alignment. (C) Alignment of the Ikeda Ank4_01 F-box sequence with that of its Boryong homolog, Ank1D1. (D) Alignment of the sequence of the Ikeda Ank3_08 F-box, which was shown to bind weakly to SKP1 in this study (Fig. 2), with that of its Ikeda paralog, Ank3_11, and its homolog, Boryong Ank1C14. The numbers above (A, C, and D) and below (B) the alignment serve as references for amino acid positions. The number 1 denotes the first position of the canonical poxviral F-box residue.
To further infer the functional importance of O. tsutsugamushi Ank F-box residues, alignments were performed with two Anks from O. tsutsugamushi strain Boryong that are known not to interact with SKP1 (46). These Anks have homologs in the Ikeda genome that were demonstrated in this study to have functional F-boxes. The F-box of Boryong Ank1D1, which is homologous to Ikeda Ank4_01 (27, 44), lacks the L/I at position 1, the L/I/V/A at position 5, and the L/I/V at position 18 (Fig. 7C). An alignment of the F-boxes of Boryong Ank1C14, its Ikeda homolog, Ank3_11 (which was not studied here but has an F-box identical to that of Ank3_08) (27, 44), and Ank3_08 and the poxviral canonical F-box showed that Ank1C14 lacks the highly conserved I/L/V at position 9, as well as conservation at residues 4, 5, 10, and 13. Taken together, these data indirectly suggest that varying combinations of the 9 residues that exhibit high conservation among O. tsutsugamushi Ank and canonical eukaryotic and canonical poxviral F-boxes, especially those at positions 1, 9, and 17, have additive effects that likely contribute to SKP1 binding. Based on F-box sequence conservation among the O. tsutsugamushi strain Ikeda Ank paralogs (Table 1), we can extend our findings of the 16 functionally validated F-box-containing Anks to include 13 identical or nearly identical paralogs to arrive at a total of 29 F-box-containing Anks.
TABLE 1.
Validated O. tsutsugamushi F-box Anks and their paralogs
| F-box Ank | Paralog(s) (% similarity to F-box of validated F-box-containing Ank)a |
|---|---|
| Ank1_02 | Ank1_01 (95) |
| Ank2 | Single copy |
| Ank3_08b | Ank3_01 (100); Ank3_04 (100); Ank3_05 (no F-box)c; Ank3_06 (no F-box); Ank3_09 (no F-box); Ank3_11 (100); Ank3_13 (100); Ank3_15 (86) |
| Ank4_01 | Ank4_02 (100) |
| Ank5_01 | Ank5_02 (100), Ank5_03 (100) |
| Ank6_02 | Ank6_01 (100), Ank6_03 (100), Ank6_04 (100) |
| Ank8 | Single copy |
| Ank9 | Single copy |
| Ank10_01 | Ank10_02 (100) |
| Ank12_01 | Ank12_02 (100) |
| Ank13 | Single copy |
| Ank14 | Single copy |
| Ank15b | Single copy |
| Ank16 | Single copy |
| Ank17b | Single copy |
| Ank20 | Single copy |
Pseudogenes were excluded. Similarity is based on sequence comparison between the paralog's analogous F-box residues and the F-box of confirmed F-box-containing Anks, as designated in Fig. 7 for reference residues 1 to 21.
F-box-containing Ank that exhibited weak interaction with GST-SKP1 in coprecipitation assays.
Ank3 paralogs with the “no F-box” designation do not possess F-boxes.
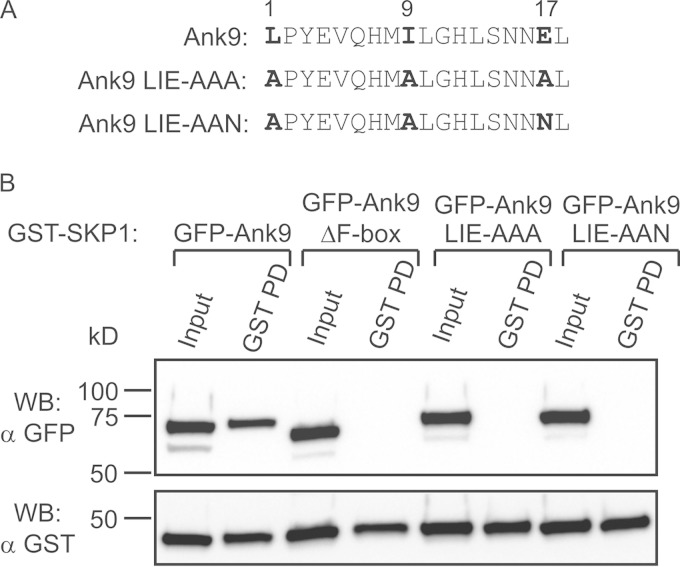
Identification of residues essential for the O. tsutsugamushi F-box motif to interact with SKP1.
It was next verified whether the conserved residues at positions 1, 9, and 17 of the O. tsutsugamushi Ank proteins' consensus F-box motif are indeed necessary for interacting with SKP1. Ank9 amino acids L384, I392, and E400, which correspond to the three F-box consensus residues of interest, were each replaced with alanine (LIE-AAA) (Fig. 8A). To determine if the side chain charge of consensus residue 17 contributes to SKP1 binding, a second Ank9 mutant protein was generated in which E400 was replaced with asparagine and L384 and I392 were each replaced with alanine (LIE-AAN). GST-SKP1 was coexpressed in HeLa cells with GFP-tagged Ank9, Ank9ΔF-box (negative control), Ank9 LIE-AAA, or Ank9 LIE-AAN. GST-SKP1 coprecipitated GFP-Ank9 but failed to precipitate GFP-Ank9ΔF-box, GFP-Ank9 LIE-AAA, or GFP-Ank9 LIE-AAN (Fig. 8B). These data demonstrate that L384, I392, and E400 of the Ank9 F-box, and presumably the corresponding residues of all O. tsutsugamushi Ank F-boxes, are essential for binding SKP1.
FIG 8.
Mutation of conserved F-box residues abolishes the ability of GFP-Ank9 to interact with GST-SKP1. (A) Alignment of the F-box motifs of Ank9, Ank9 LIE-AAA, and Ank9 LIE-AAN. The numbers above the alignment indicate the relative positions of the O. tsutsugamushi Ank consensus F-box motif. The boldface residues of Ank9 correspond to those that are mutated to alanine or asparagine in Ank9 LIE-AAA and Ank9 LIE-AAN. (B) Lysates of transfected HeLa cells expressing GST-SKP1 together with GFP-tagged Ank9, Ank9 LIE-AAA, Ank9 LIE-AAN, or the negative-control Ank9ΔF-box were Western blotted and screened with GFP antibody. Stripping and probing the blot with GST antibody confirmed precipitation of GST-SKP1 in all samples. The results are representative of three experiments with similar results.
DISCUSSION
Although numerous eukaryotic F-box proteins have been identified, FBPs have only recently been reported in microbes, including numerous viruses (particularly poxviruses) (39, 42, 47–59); various plant bacterial pathogens (22, 60, 61); and several intracellular bacteria that infect mammals, such as Legionella pneumophila (20–22, 31, 34, 35), Coxiella burnetii (22, 32, 37), Salmonella enterica serovar Typhimurium (36), and Chlamydia spp. (22, 32, 33). C. burnetii encodes three F-box-containing proteins, one of which, CpeC, is secreted into host cells and colocalizes with ubiquitin-rich structures (37), though its specific function is not yet known. L. pneumophila encodes and secretes at least five F-box proteins, the best-characterized of which is AnkB, which exploits polyubiquitination to target proteins for proteasomal degradation, yielding amino acids that are essential for bacterial growth (20, 21, 34). The S. Typhimurium F-box effector, GogB, interacts with SKP1 to disrupt ubiquitin regulation of NF-κB, thereby blocking activation of the proinflammatory response (36). Poxviral PRANC proteins, which possess both ankyrin repeats and PRANC/F-box domains, are proposed to subvert the host SCF ubiquitin ligase complex for degradation of antiviral host factors (39, 42, 53, 62). Thus, the use of F-box proteins is a conserved strategy among diverse microbes for hijacking host ubiquitination pathways to benefit pathogen survival.
Of the bacterial F-box proteins described thus far, most have their F-boxes N-terminally located with additional protein-protein interaction domains downstream, similar to the arrangement of eukaryotic F-box proteins (22, 32, 33). One exception is GogB, which has a C-terminal F-box positioned downstream from a non-ankyrin repeat domain (36). Regardless of whether they are N- or C-terminally located, most bacterial and viral F-box motifs are located in close proximity to another protein-protein interaction motif, such as an ankyrin, WD40, or leucine-rich repeat, which specifically binds the substrate that will become ubiquitinated by the F-box-recruited SCF1 complex (28, 43). Unlike the other reported intracellular bacterial F-box effectors, most O. tsutsugamushi Anks have the domain orientation found in poxviruses, where the F-box lies downstream of the ankyrin repeats. The only exceptions to this are Ikeda Ank3_08 and some of its paralogs, which have the eukaryotic-like arrangement of an N-terminal F-box.
Also like the poxviral F-boxes, the O. tsutsugamushi Ank F-box motifs are short, comprising ∼21 residues as opposed to longer eukaryotic motifs (∼45 residues). Thus, rather than forming three alpha helices, as is found in eukaryotic F-box domains, the Orientia Ank F-boxes likely resemble the truncated two-helix structure modeled for many poxviral FBPs (63). Interestingly, the conserved amino acids at positions 1 and 9 among O. tsutsugamushi Ank F-boxes are also conserved in poxviral and eukaryotic sequences. These positions flank the edges of the first alpha-helical structure (corresponding to residues 2 to 11) (39, 63). The conserved acidic residue at Orientia Ank F-box position 17, which lies within the interior of helix 2 (corresponding to residues 14 to 21), however, varies among eukaryotic and poxviral sequences (39). Lastly, though obviously part of the F-box protein family, O. tsutsugamushi Anks share some F-box sequence patterns that set them apart from other characterized FBPs, such as the F/W and Q residues at positions −3 and 22, respectively.
Until recently, the number of F-box proteins that O. tsutsugamushi encodes has been underappreciated. A previous scan of 22 bacterial species for putative F-box proteins predicted that only one O. tsutsugamushi strain Boryong candidate, Ank1U9 (OTBS_2178), possessed a C-terminal F-box downstream of its ankyrin repeats (22). Further examination of Boryong Anks revealed that Ank1U9 and at least six other Anks possessed F-boxes and, when ectopically expressed in recombinant form, were capable of binding SCF1 components in pulldown assays (46), underscoring the necessity for manual sequence evaluation in searching for putative F-box proteins. Here, 16 O. tsutsugamushi strain Ikeda Anks were identified and confirmed to serve as FBPs. Inclusion of the Ikeda Ank paralogs, which have F-box sequences identical or nearly identical to their assayed counterparts, yields a total of 29 Ikeda F-box-containing Anks. Though Neochlamydia and Parachlamydia spp. boast upwards of 129 to 158 and 29 putative F-box proteins (33), respectively, O. tsutsugamushi is among the bacterial species encoding the largest FBP armamentariums. Consequently, this study is the first of its kind to validate F-box activity of such a sizable protein family in one organism and to show that a bacterially encoded FBP-SKP1 interaction is detectable during the course of infection in mammalian cells.
It is important to study bacterial virulence factor genotypic and phenotypic diversity. The data presented here complement a recent study by Min et al. that examined nine Anks from O. tsutsugamushi strain Boryong and identified F-boxes within Ank1A2, Ank1F1, Ank1U5, Ank1U9, Ank1U4, and Ank1B1 (46). These Boryong proteins share sequence identity (shown as percent identity) to Ikeda Ank10_01 (83%), Ank12_01 (91%), Ank2 (84%), Ank20 (90%), Ank6_02 (66%), and Ank8 (93%), respectively, which are reported to have F-boxes in this study. Boryong Ank1E2 was also noted as having an F-box (46), though its ORF is annotated as a pseudogene and it does not have an Ikeda protein homolog (44). Of note, Boryong Ank1D1 and Ank1C14 do not interact with SKP1 (46), though their Ikeda homologs, Ank4_01 (63%) and Ank3_11 (74%), respectively, interacted with SKP1 and other SCF1 components in this study. Comparison of the F-box motifs from these Anks revealed the loss of conserved residues at position 1 of the Ank1D1 F-box and position 9 of the Ank1C14 F-box, with additional variations at less conserved positions. Ikeda Ank3_08, Ank15, and Ank17 also differ from the other Ank F-boxes at less conserved positions but maintained SKP1 binding, suggesting the importance of conservation at positions 1, 9, and 17 in SKP1 interactions. Together, the combined Boryong and Ikeda F-box data support the idea that at least three key residues are important in determining whether an Ank participates in SKP1 interactions. Indeed, replacing these residues with alanine and/or asparagine ablated the ability of GFP-Ank9 to interact with GST-SKP1.
Aside from SCF1 components, the host cell proteins targeted by O. tsutsugamushi strain Ikeda Anks, which may become ubiquitinated and proteasomally degraded, are unknown. Several ectopically expressed Boryong F-box-containing Anks examined by Min and colleagues were suggested to interact with and promote ubiquitination and proteasomal destruction of eukaryotic elongation factor 1α (46). Despite numerous attempts, however, we were unable to recapitulate these phenomena with any ectopically expressed Ikeda Ank, even when the proteasome was pharmacologically inhibited. O. tsutsugamushi possesses a huge capacity to exploit host ubiquitination and various cell functions with the number of F-box Ank effectors that it encodes. Therefore, it is imperative to elucidate F-box Ank binding partners and to determine if they become ubiquitinated/proteasomally degraded and the functional benefit of these phenomena to Orientia in order to understand the mechanisms by which this poorly understood pathogen modulates host cell functions.
ACKNOWLEDGMENTS
We thank Andrew Mercer at the University of Otago, Dunedin, New Zealand, for reagents and helpful discussions.
This work was supported by National Institutes of Health grants R03 AI101666 and R21 AI103606 (J.A.C.), American Heart Association (AHA) Grant-in-Aid 13GRNT16810009 (J.A.C.), AHA Predoctoral Fellowship 13PRE16840032 (L.V.), the VCU Presidential Research Quest Fund (J.A.C.), and George and Lavinia Blick Scholar funds (J.A.C.). A.R.B. is a fellowship recipient supported by VCU Institutional Research and Academic Career Development Award grant K12 GM093857. LSCM was performed at the VCU Department of Anatomy and Neurobiology Microscopy Facility, which is supported in part by funding from NIH-NINDS Center core grant 5P30NS047463.
REFERENCES
- 1.Paris DH, Phetsouvanh R, Tanganuchitcharnchai A, Jones M, Jenjaroen K, Vongsouvath M, Ferguson DP, Blacksell SD, Newton PN, Day NP, Turner GD. 2012. Orientia tsutsugamushi in human scrub typhus eschars shows tropism for dendritic cells and monocytes rather than endothelium. PLoS Negl Trop Dis 6:e1466. doi: 10.1371/journal.pntd.0001466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chattopadhyay S, Richards AL. 2007. Scrub typhus vaccines: past history and recent developments. Hum Vaccin 3:73–80. doi: 10.4161/hv.3.3.4009. [DOI] [PubMed] [Google Scholar]
- 3.Ge Y, Rikihisa Y. 2011. Subversion of host cell signaling by Orientia tsutsugamushi. Microbes Infect 13:638–648. doi: 10.1016/j.micinf.2011.03.003. [DOI] [PubMed] [Google Scholar]
- 4.Moron CG, Popov VL, Feng HM, Wear D, Walker DH. 2001. Identification of the target cells of Orientia tsutsugamushi in human cases of scrub typhus. Mod Pathol 14:752–759. doi: 10.1038/modpathol.3880385. [DOI] [PubMed] [Google Scholar]
- 5.Kelly DJ, Richards AL, Temenak J, Strickman D, Dasch GA. 2002. The past and present threat of rickettsial diseases to military medicine and international public health. Clin Infect Dis 34:S145–S169. doi: 10.1086/339908. [DOI] [PubMed] [Google Scholar]
- 6.Richards AL, Soeatmadji DW, Widodo MA, Sardjono TW, Yanuwiadi B, Hernowati TE, Baskoro AD, Roebiyoso Hakim L, Soendoro M, Rahardjo E, Putri MP, Saragih JM, Strickman D, Kelly DJ, Dasch GA, Olson JG, Church CJ, Corwin AL. 1997. Seroepidemiologic evidence for murine and scrub typhus in Malang, Indonesia. Am J Trop Med Hyg 57:91–95. [DOI] [PubMed] [Google Scholar]
- 7.Rapmund G. 1984. Rickettsial diseases of the Far East: new perspectives. J Infect Dis 149:330–338. doi: 10.1093/infdis/149.3.330. [DOI] [PubMed] [Google Scholar]
- 8.Kelly DJ, Fuerst PA, Ching WM, Richards AL. 2009. Scrub typhus: the geographic distribution of phenotypic and genotypic variants of Orientia tsutsugamushi. Clin Infect Dis 48(Suppl 3):S203–S230. doi: 10.1086/596576. [DOI] [PubMed] [Google Scholar]
- 9.Browning JS, Raphael M, Kline EF, Coblenz A. 1945. Scrub-typhus. Am J Trop Med Hyg 25:481–492. [DOI] [PubMed] [Google Scholar]
- 10.Philip CB. 1948. Tsutsugamushi disease in World War II. J Parasitol 34:169–191. doi: 10.2307/3273264. [DOI] [PubMed] [Google Scholar]
- 11.Berman SJ, Kundin WD. 1973. Scrub typhus in South Vietnam. A study of 87 cases. Ann Intern Med 79:26–30. [DOI] [PubMed] [Google Scholar]
- 12.Gormley TS. 1996. A diagnosis of scrub typhus. Navy Med 87:20–22. [Google Scholar]
- 13.Deller JJ Jr, Russell PK. 1967. An analysis of fevers of unknown origin in American soldiers in Vietnam. Ann Intern Med 66:1129–1143. doi: 10.7326/0003-4819-66-6-1129. [DOI] [PubMed] [Google Scholar]
- 14.Al-Khodor S, Price CT, Habyarimana F, Kalia A, Abu Kwaik Y. 2008. A Dot/Icm-translocated ankyrin protein of Legionella pneumophila is required for intracellular proliferation within human macrophages and protozoa. Mol Microbiol 70:908–923. doi: 10.1111/j.1365-2958.2008.06453.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Al-Khodor S, Price CT, Kalia A, Abu Kwaik Y. 2010. Functional diversity of ankyrin repeats in microbial proteins. Trends Microbiol 18:132–139. doi: 10.1016/j.tim.2009.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Habyarimana F, Al-Khodor S, Kalia A, Graham JE, Price CT, Garcia MT, Kwaik YA. 2008. Role for the Ankyrin eukaryotic-like genes of Legionella pneumophila in parasitism of protozoan hosts and human macrophages. Environ Microbiol 10:1460–1474. doi: 10.1111/j.1462-2920.2007.01560.x. [DOI] [PubMed] [Google Scholar]
- 17.IJdo JW, Carlson AC, Kennedy EL. 2007. Anaplasma phagocytophilum AnkA is tyrosine-phosphorylated at EPIYA motifs and recruits SHP-1 during early infection. Cell Microbiol 9:1284–1296. doi: 10.1111/j.1462-5822.2006.00871.x. [DOI] [PubMed] [Google Scholar]
- 18.Lin M, den Dulk-Ras A, Hooykaas PJ, Rikihisa Y. 2007. Anaplasma phagocytophilum AnkA secreted by type IV secretion system is tyrosine phosphorylated by Abl-1 to facilitate infection. Cell Microbiol 9:2644–2657. doi: 10.1111/j.1462-5822.2007.00985.x. [DOI] [PubMed] [Google Scholar]
- 19.Price CT, Al-Khodor S, Al-Quadan T, Abu Kwaik Y. 2010. Indispensable role for the eukaryotic-like ankyrin domains of the ankyrin B effector of Legionella pneumophila within macrophages and amoebae. Infect Immun 78:2079–2088. doi: 10.1128/IAI.01450-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Price CT, Al-Khodor S, Al-Quadan T, Santic M, Habyarimana F, Kalia A, Kwaik YA. 2009. Molecular mimicry by an F-box effector of Legionella pneumophila hijacks a conserved polyubiquitination machinery within macrophages and protozoa. PLoS Pathog 5:e1000704. doi: 10.1371/journal.ppat.1000704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Price CT, Al-Quadan T, Santic M, Rosenshine I, Abu Kwaik Y. 2011. Host proteasomal degradation generates amino acids essential for intracellular bacterial growth. Science 334:1553–1557. doi: 10.1126/science.1212868. [DOI] [PubMed] [Google Scholar]
- 22.Price CT, Kwaik YA. 2010. Exploitation of host polyubiquitination machinery through molecular mimicry by eukaryotic-like bacterial F-box effectors. Front Microbiol 1:122. doi: 10.3389/fmicb.2010.00122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.VieBrock L, Evans SM, Beyer AR, Larson CL, Beare PA, Ge H, Singh S, Rodino K, Heinzen RA, Richards AL, Carlyon JA. 2014. Orientia tsutsugamushi ankyrin repeat-containing protein family members are type 1 secretion system substrates that traffic to the host cell endoplasmic reticulum. Front Cell Infect Microbiol 4:186. doi: 10.3389/fcimb.2014.00186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Voth DE. 2011. ThANKs for the repeat: intracellular pathogens exploit a common eukaryotic domain. Cell Logist 1:128–132. doi: 10.4161/cl.1.4.18738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Voth DE, Howe D, Beare PA, Vogel JP, Unsworth N, Samuel JE, Heinzen RA. 2009. The Coxiella burnetii ankyrin repeat domain-containing protein family is heterogeneous, with C-terminal truncations that influence Dot/Icm-mediated secretion. J Bacteriol 191:4232–4242. doi: 10.1128/JB.01656-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wakeel A, den Dulk-Ras A, Hooykaas PJ, McBride JW. 2011. Ehrlichia chaffeensis tandem repeat proteins and Ank200 are type 1 secretion system substrates related to the repeats-in-toxin exoprotein family. Front Cell Infect Microbiol 1:22. doi: 10.3389/fcimb.2011.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakayama K, Yamashita A, Kurokawa K, Morimoto T, Ogawa M, Fukuhara M, Urakami H, Ohnishi M, Uchiyama I, Ogura Y, Ooka T, Oshima K, Tamura A, Hattori M, Hayashi T. 2008. The whole-genome sequencing of the obligate intracellular bacterium Orientia tsutsugamushi revealed massive gene amplification during reductive genome evolution. DNA Res 15:185–199. doi: 10.1093/dnares/dsn011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bai C, Sen P, Hofmann K, Ma L, Goebl M, Harper JW, Elledge SJ. 1996. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell 86:263–274. doi: 10.1016/S0092-8674(00)80098-7. [DOI] [PubMed] [Google Scholar]
- 29.Ardley HC, Robinson PA. 2005. E3 ubiquitin ligases. Essays Biochem 41:15–30. doi: 10.1042/EB0410015. [DOI] [PubMed] [Google Scholar]
- 30.Petroski MD, Deshaies RJ. 2005. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol 6:9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- 31.Al-Quadan T, Kwaik YA. 2011. Molecular characterization of exploitation of the polyubiquitination and farnesylation machineries of Dictyostelium discoideum by the AnkB F-box effector of Legionella pneumophila. Front Microbiol 2:23. doi: 10.3389/fmicb.2011.00023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Angot A, Vergunst A, Genin S, Peeters N. 2007. Exploitation of eukaryotic ubiquitin signaling pathways by effectors translocated by bacterial type III and type IV secretion systems. PLoS Pathog 3:e3. doi: 10.1371/journal.ppat.0030003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Domman D, Collingro A, Lagkouvardos I, Gehre L, Weinmaier T, Rattei T, Subtil A, Horn M. 2014. Massive expansion of ubiquitination-related gene families within the Chlamydiae. Mol Biol Evol 31:2890–2904. doi: 10.1093/molbev/msu227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ensminger AW, Isberg RR. 2010. E3 ubiquitin ligase activity and targeting of BAT3 by multiple Legionella pneumophila translocated substrates. Infect Immun 78:3905–3919. doi: 10.1128/IAI.00344-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lomma M, Dervins-Ravault D, Rolando M, Nora T, Newton HJ, Sansom FM, Sahr T, Gomez-Valero L, Jules M, Hartland EL, Buchrieser C. 2010. The Legionella pneumophila F-box protein Lpp2082 (AnkB) modulates ubiquitination of the host protein parvin B and promotes intracellular replication. Cell Microbiol 12:1272–1291. doi: 10.1111/j.1462-5822.2010.01467.x. [DOI] [PubMed] [Google Scholar]
- 36.Pilar AV, Reid-Yu SA, Cooper CA, Mulder DT, Coombes BK. 2012. GogB is an anti-inflammatory effector that limits tissue damage during Salmonella infection through interaction with human FBXO22 and Skp1. PLoS Pathog 8:e1002773. doi: 10.1371/journal.ppat.1002773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Voth DE, Beare PA, Howe D, Sharma UM, Samoilis G, Cockrell DC, Omsland A, Heinzen RA. 2011. The Coxiella burnetii cryptic plasmid is enriched in genes encoding type IV secretion system substrates. J Bacteriol 193:1493–1503. doi: 10.1128/JB.01359-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hanson B. 1987. Factors influencing Rickettsia tsutsugamushi infection of cultured cells. Am J Trop Med Hyg 36:621–630. [DOI] [PubMed] [Google Scholar]
- 39.Sonnberg S, Seet BT, Pawson T, Fleming SB, Mercer AA. 2008. Poxvirus ankyrin repeat proteins are a unique class of F-box proteins that associate with cellular SCF1 ubiquitin ligase complexes. Proc Natl Acad Sci U S A 105:10955–10960. doi: 10.1073/pnas.0802042105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Crooks GE, Hon G, Chandonia JM, Brenner SE. 2004. WebLogo: a sequence logo generator. Genome Res 14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mercer AA, Fleming SB, Ueda N. 2005. F-box-like domains are present in most poxvirus ankyrin repeat proteins. Virus Genes 31:127–133. doi: 10.1007/s11262-005-1784-z. [DOI] [PubMed] [Google Scholar]
- 43.Schulman BA, Carrano AC, Jeffrey PD, Bowen Z, Kinnucan ER, Finnin MS, Elledge SJ, Harper JW, Pagano M, Pavletich NP. 2000. Insights into SCF ubiquitin ligases from the structure of the Skp1-Skp2 complex. Nature 408:381–386. doi: 10.1038/35042620. [DOI] [PubMed] [Google Scholar]
- 44.Cho NH, Kim HR, Lee JH, Kim SY, Kim J, Cha S, Darby AC, Fuxelius HH, Yin J, Kim JH, Lee SJ, Koh YS, Jang WJ, Park KH, Andersson SG, Choi MS, Kim IS. 2007. The Orientia tsutsugamushi genome reveals massive proliferation of conjugative type IV secretion system and host-cell interaction genes. Proc Natl Acad Sci U S A 104:7981–7986. doi: 10.1073/pnas.0611553104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zheng N, Schulman BA, Song L, Miller JJ, Jeffrey PD, Wang P, Chu C, Koepp DM, Elledge SJ, Pagano M, Conaway RC, Conaway JW, Harper JW, Pavletich NP. 2002. Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature 416:703–709. doi: 10.1038/416703a. [DOI] [PubMed] [Google Scholar]
- 46.Min CK, Kwon YJ, Ha NY, Cho BA, Kim JM, Kwon EK, Kim YS, Choi MS, Kim IS, Cho NH. 2014. Multiple Orientia tsutsugamushi ankyrin repeat proteins interact with SCF1 ubiquitin ligase complex and eukaryotic elongation factor 1 alpha. PLoS One 9:e105652. doi: 10.1371/journal.pone.0105652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang Q, Tao T, Han Y, Chen X, Fan Z, Li D, Yu J, Han C. 2013. Nonstructural protein P7-2 encoded by Rice black-streaked dwarf virus interacts with SKP1, a core subunit of SCF ubiquitin ligase. Virol J 10:325. doi: 10.1186/1743-422X-10-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mitchell JK, Byers NM, Friesen PD. 2013. Baculovirus F-box protein LEF-7 modifies the host DNA damage response to enhance virus multiplication. J Virol 87:12592–12599. doi: 10.1128/JVI.02501-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Baumberger N, Tsai CH, Lie M, Havecker E, Baulcombe DC. 2007. The Polerovirus silencing suppressor P0 targets ARGONAUTE proteins for degradation. Curr Biol 17:1609–1614. doi: 10.1016/j.cub.2007.08.039. [DOI] [PubMed] [Google Scholar]
- 50.Pazhouhandeh M, Dieterle M, Marrocco K, Lechner E, Berry B, Brault V, Hemmer O, Kretsch T, Richards KE, Genschik P, Ziegler-Graff V. 2006. F-box-like domain in the polerovirus protein P0 is required for silencing suppressor function. Proc Natl Acad Sci U S A 103:1994–1999. doi: 10.1073/pnas.0510784103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bortolamiol D, Pazhouhandeh M, Marrocco K, Genschik P, Ziegler-Graff V. 2007. The Polerovirus F box protein P0 targets ARGONAUTE1 to suppress RNA silencing. Curr Biol 17:1615–1621. doi: 10.1016/j.cub.2007.07.061. [DOI] [PubMed] [Google Scholar]
- 52.Sonnberg S, Fleming SB, Mercer AA. 2011. Phylogenetic analysis of the large family of poxvirus ankyrin-repeat proteins reveals orthologue groups within and across chordopoxvirus genera. J Gen Virol 92:2596–2607. doi: 10.1099/vir.0.033654-0. [DOI] [PubMed] [Google Scholar]
- 53.Sperling KM, Schwantes A, Schnierle BS, Sutter G. 2008. The highly conserved orthopoxvirus 68k ankyrin-like protein is part of a cellular SCF ubiquitin ligase complex. Virology 374:234–239. doi: 10.1016/j.virol.2008.02.018. [DOI] [PubMed] [Google Scholar]
- 54.van Buuren N, Couturier B, Xiong Y, Barry M. 2008. Ectromelia virus encodes a novel family of F-box proteins that interact with the SCF complex. J Virol 82:9917–9927. doi: 10.1128/JVI.00953-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Burles K, van Buuren N, Barry M. 2014. Ectromelia virus encodes a family of Ankyrin/F-box proteins that regulate NFkappaB. Virology 468-470:351–362. doi: 10.1016/j.virol.2014.08.030. [DOI] [PubMed] [Google Scholar]
- 56.Werden SJ, Lanchbury J, Shattuck D, Neff C, Dufford M, McFadden G. 2009. The myxoma virus m-t5 ankyrin repeat host range protein is a novel adaptor that coordinately links the cellular signaling pathways mediated by Akt and Skp1 in virus-infected cells. J Virol 83:12068–12083. doi: 10.1128/JVI.00963-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Blanie S, Gelfi J, Bertagnoli S, Camus-Bouclainville C. 2010. MNF, an ankyrin repeat protein of myxoma virus, is part of a native cellular SCF complex during viral infection. Virol J 7:56. doi: 10.1186/1743-422X-7-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chang SJ, Hsiao JC, Sonnberg S, Chiang CT, Yang MH, Tzou DL, Mercer AA, Chang W. 2009. Poxvirus host range protein CP77 contains an F-box-like domain that is necessary to suppress NF-kappaB activation by tumor necrosis factor alpha but is independent of its host range function. J Virol 83:4140–4152. doi: 10.1128/JVI.01835-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Noel EA, Kang M, Adamec J, Van Etten JL, Oyler GA. 2014. Chlorovirus Skp1-binding ankyrin repeat protein interplay and mimicry of cellular ubiquitin ligase machinery. J Virol 88:13798–13810. doi: 10.1128/JVI.02109-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schrammeijer B, Risseeuw E, Pansegrau W, Regensburg-Tuink TJ, Crosby WL, Hooykaas PJ. 2001. Interaction of the virulence protein VirF of Agrobacterium tumefaciens with plant homologs of the yeast Skp1 protein. Curr Biol 11:258–262. doi: 10.1016/S0960-9822(01)00069-0. [DOI] [PubMed] [Google Scholar]
- 61.Magori S, Citovsky V. 2011. Hijacking of the host SCF ubiquitin ligase machinery by plant pathogens. Front Plant Sci 2:87. doi: 10.3389/fpls.2011.00087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Barry M, van Buuren N, Burles K, Mottet K, Wang Q, Teale A. 2010. Poxvirus exploitation of the ubiquitin-proteasome system. Viruses 2:2356–2380. doi: 10.3390/v2102356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sonnberg S, Fleming SB, Mercer AA. 2009. A truncated two-alpha-helix F-box present in poxvirus ankyrin-repeat proteins is sufficient for binding the SCF1 ubiquitin ligase complex. J Gen Virol 90:1224–1228. doi: 10.1099/vir.0.009324-0. [DOI] [PubMed] [Google Scholar]