ABSTRACT
The ubiquitous aquatic bacterium Caulobacter crescentus is highly resistant to uranium (U) and facilitates U biomineralization and thus holds promise as an agent of U bioremediation. To gain an understanding of how C. crescentus tolerates U, we employed transposon (Tn) mutagenesis paired with deep sequencing (Tn-seq) in a global screen for genomic elements required for U resistance. Of the 3,879 annotated genes in the C. crescentus genome, 37 were found to be specifically associated with fitness under U stress, 15 of which were subsequently tested through mutational analysis. Systematic deletion analysis revealed that mutants lacking outer membrane transporters (rsaFa and rsaFb), a stress-responsive transcription factor (cztR), or a ppGpp synthetase/hydrolase (spoT) exhibited a significantly lower survival rate under U stress. RsaFa and RsaFb, which are homologues of TolC in Escherichia coli, have previously been shown to mediate S-layer export. Transcriptional analysis revealed upregulation of rsaFa and rsaFb by 4- and 10-fold, respectively, in the presence of U. We additionally show that rsaFa mutants accumulated higher levels of U than the wild type, with no significant increase in oxidative stress levels. Our results suggest a function for RsaFa and RsaFb in U efflux and/or maintenance of membrane integrity during U stress. In addition, we present data implicating CztR and SpoT in resistance to U stress. Together, our findings reveal novel gene targets that are key to understanding the molecular mechanisms of U resistance in C. crescentus.
IMPORTANCE Caulobacter crescentus is an aerobic bacterium that is highly resistant to uranium (U) and has great potential to be used in U bioremediation, but its mechanisms of U resistance are poorly understood. We conducted a Tn-seq screen to identify genes specifically required for U resistance in C. crescentus. The genes that we identified have previously remained elusive using other omics approaches and thus provide significant insight into the mechanisms of U resistance by C. crescentus. In particular, we show that outer membrane transporters RsaFa and RsaFb, previously known as part of the S-layer export machinery, may confer U resistance by U efflux and/or by maintaining membrane integrity during U stress.
INTRODUCTION
Uranium (U) contamination is widespread and poses significant concerns for environmental ecology and human health (1). Chemical and physical techniques for waste treatment or removal of U are challenging and expensive. A promising, microbially mediated method for U remediation, in situ U immobilization, is more cost-effective and environmentally friendly than conventional approaches (2). If we seek to task microbes with the cleanup of contaminated sites, an understanding of how microbes defend against U toxicity is crucial. Understanding these mechanisms is necessary for the optimization of strains intended for use as U biosensors (3) or for the purpose of bioremediation (2, 4, 5).
A great deal is known about various strategies used by microbes to transform and defend against extracellular U. These include reductive precipitation by outer membrane cytochromes, conductive pili, or spores (6–10), surface adsorption by exopolysaccharide (EPS) or S-layers (9, 11), or precipitation with phosphate (4, 5, 12, 13). However, mechanisms used by cells for combating internal U toxicity are poorly understood (14). Previous studies revealed upregulation of metal efflux pumps, NADH quinone oxidoreductases, or other reactive oxygen species (ROS) scavenging enzymes upon exposure to U in the sulfate-reducing bacterium Desulfotomaculum reducens (15), Escherichia coli grown at low pH (16), and the plant Arabidopsis thaliana (17). General and membrane stress responses were particularly pronounced when Shewanella oneidensis was exposed to U (18), and nucleic acid and protein damage have been shown to represent the primary modes of U toxicity in Desulfovibrio alaskensis G20 (19). Phosphate transporters and cell wall proteins are known to be important for survival following U exposure for the budding yeast Saccharomyces cerevisiae (20), whereas thermoacidophilic archaea employ temporary degradation of cellular RNA as a dynamic mechanism for resisting U toxicity (21). However, there is still a significant knowledge gap regarding the persistence of microbes in U-contaminated environments (9, 22) as the mechanisms for responding to acute U toxicity are not necessarily the same as those that enable survival and growth over a longer term.
We have chosen to focus on the aquatic organism Caulobacter crescentus, a widely distributed nonpathogenic bacterium that can survive in low-nutrient environments with great potential to be exploited for the purpose of bioremediation (3, 23). C. crescentus is known to tolerate high levels of U(VI) and is able to facilitate U biomineralization through the formation of uranium phosphate precipitates (4). Transcriptomic and proteomic studies during U exposure in C. crescentus revealed dozens of genes with significant changes in expression in response to U; these genes do not appear to overlap substantially with those that are variably expressed in response to other heavy metal stresses, such as cadmium (Cd) and chromium, suggesting a divergent cellular response to U(VI) (23, 24). This response manifests, in part, as a temporary arrest in cell cycle progression and DNA replication, along with some cell filamentation (a possible consequence of U-induced DNA damage) observed during growth recovery following U detoxification (25). Proteins upregulated in response to U include the periplasmic protein UrcA, a phytase enzyme, two-component signaling factors, and an ABC transporter. However, individual deletion of these upregulated genes did not increase U susceptibility (23, 24), indicating that these genes are not required for U tolerance. These results further suggest that the cellular response to U is complex and includes more than a direct response to acute toxicity.
Here, we use transposon mutagenesis paired with deep sequencing (Tn-seq) to identify genes and gene products involved in U tolerance in C. crescentus. Compared to conventional transcriptomic and proteomic techniques, Tn-seq represents a novel tool for functional genomics that utilizes high-throughput, massively parallel sequencing to uncover genes that contribute to cell fitness in a condition of interest (Fig. 1) (26, 27). The results presented here elucidate potential toxicity mechanisms of U and reveal novel genes and pathways important for growth and survival of C. crescentus in the presence of U.
FIG 1.
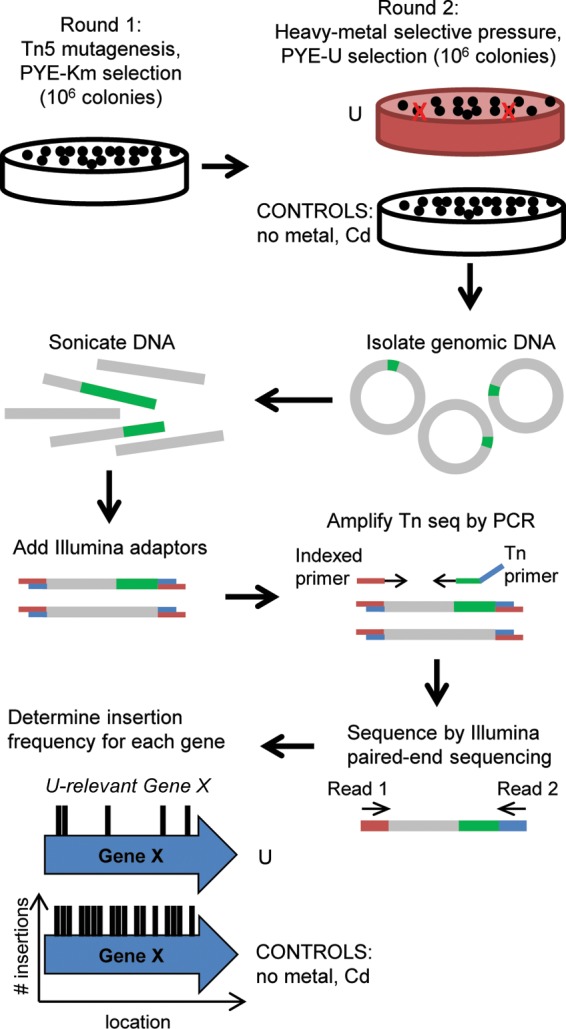
Scheme for Tn-seq method. A hypersaturated Tn5 mutant library was first generated by kanamycin selection (round 1), after which the library was exposed to U, Cd, or no-metal selective pressure (round 2). Genomic DNA was then extracted and sheared by sonication, and Illumina adaptors were added to the sheared fragments. Genomic regions adjacent to the Tn5 insertions were PCR amplified using an indexed primer and a Tn5-specific primer. PCR fragments were subsequently sequenced by Illumina paired-end sequencing. The Tn insertion site was determined for each mutant, and the insertion frequency for each gene was calculated. Genes important for U fitness are expected to have a lower insertion frequency under the U condition than under the no-metal or Cd control condition.
MATERIALS AND METHODS
Materials, bacterial strains, and growth conditions.
All chemicals were purchased from Sigma-Aldrich (St. Louis, MO) unless otherwise noted. Peptone, yeast extract, and agar were purchased from Amresco (Solon, OH). Uranyl nitrate hexahydrate [(UO2)(NO3)2 · 6H2O] was obtained from SPI Supplies (West Chester, PA). A stock solution of uranyl nitrate (100 mM) was prepared in 0.1 N nitric acid. All PCRs were conducted using iProof polymerase from Bio-Rad (Hercules, CA) supplemented with 5% dimethyl sulfoxide (DMSO) according to the manufacturer's instructions. E. coli WM3064 (see Table S1 in the supplemental material) was maintained on solid LB medium supplemented with 300 μM diaminopimelic acid (DAP) and 1.5% agar. Liquid cultures were prepared in LB medium supplemented with 300 μM DAP. Where appropriate, LB medium was supplemented with kanamycin at 50 μg/ml for both liquid and solid media. Caulobacter crescentus NA1000 was maintained on solid PYE medium (0.2% peptone, 0.1% yeast extract, 0.5 mM MgSO4, and 1 mM CaCl2) supplemented with 1.5% agar (28). Liquid cultures were prepared in PYE medium. Buffered PYE medium with 50 mM morpholineethanesulfonic acid (MES; pH 6.1) added was used for U spike-in experiments. Where applicable, PYE medium was supplemented with antibiotics at the following concentrations (for liquid and solid media, respectively, in μg/ml): kanamycin (5 and 20), spectinomycin (25 and 100), streptomycin (5 and 5), chloramphenicol (1 and 1), and tetracycline (Tet) (1 and 2).
Construction of Tn-seq library and growth under U and Cd conditions.
To construct the master library of Tn5 mutants, the transposon delivery plasmid pXMCS2::Tn5Pxyl (29) was conjugated from an E. coli WM3064 donor strain into a C. crescentus ΔrecA strain. Transposition events were selected on PYE-agar plates containing kanamycin (which selects specifically for mutants that carry the Tn5Pxyl transposon) and 0.1% xylose (which circumvents polar effects by allowing expression of genes downstream of the transposon from the C. crescentus xylX promoter). We note that genes downstream from the transposon are no longer under the control of their native promoter and are constitutively expressed from the xylX promoter. Plates were incubated at 30°C for 7 days. Two master libraries, each containing ∼1.5 × 106 Tn5Pxyl insertion mutants, were prepared. Sets of ∼10,000 colonies were pooled, resuspended in PYE medium containing 10% glycerol, and stored at −80°C for subsequent selection on U and Cd plates.
For U or Cd selection, the frozen glycerol stocks were diluted in PYE medium and plated on PYE-agar plates containing 0.1% xylose and supplemented with either 250 μM uranyl nitrate (U library 1), 275 μM uranyl nitrate (U libraries 2 and 3), 12 μM cadmium sulfate, or no metal. Each stock was plated on enough plates to generate ∼15,000 colonies with ∼300 to 500 colonies on each plate. The PYE-agar plates were incubated at 30°C for 7 days. Each library contained ∼1.5 × 106 Tn5 mutants, except U libraries 2 and 3, which had ∼2.5 × 106 and ∼5 × 105 Tn5 mutants, respectively. Colonies were pooled and stored as described above.
DNA processing for sequencing.
From each library, 10 μl of each frozen glycerol aliquot was pooled, and cells were harvested by centrifugation at 10,000 × g for 10 min. Genomic DNA was extracted from the cells using phenol-chloroform extraction according to standard procedures (30). One to three replicate DNA extractions were performed for each library (master, U, Cd, and no metal) and were processed and sequenced as described below.
One microgram of genomic DNA (gDNA) was sheared in a 120-μl volume using a Covaris E210 machine (Woburn, MA) and a Covaris AFA microTube. Each sample was sheared for 150 s under the following conditions: 10% duty cycle, intensity 5, and 200 cycles per burst. Sheared samples were then cleaned and size selected with a double Agencourt Ampure XP bead cleanup (Beckman-Coulter, Brea, CA). The first round of the cleanup used a 0.85:1 volume ratio of beads to sample; the second round used a 1.4:1 volume ratio. One microliter of the size-selected gDNA was run on an Agilent Bioanalyzer DNA 1000 chip (Santa Clara, CA) to confirm a mean fragment size distribution of 200 to 250 bp. Each sample was then treated with end-repair and A-tailing enzymes as described in the NEBNext DNA Library Prep protocol for Illumina sequencing (New England BioLabs, Ipswich, MA). Following cleanup after A-tailing reactions, modified Illumina paired-end adapters were ligated to the DNA fragments. After adapter ligation, excess adapter and adapter dimer were removed with a 1:1 volume ratio of the Ampure XP beads. Samples were then amplified with Tn-specific and indexed primers (see Table S2 in the supplemental material) using JumpStart Taq DNA polymerase (Sigma-Aldrich) in a 100-μl volume. This step selected for and amplified DNA fragments containing transposon insertion sites. Thermal cycling conditions were as follows: 94°C for 2 min, followed by 30 cycles of 94°C for 30 s, 65°C for 20 s, and 72°C for 30 s, with a final step at 72°C for 10 min and a 4°C hold. PCRs were cleaned a final time with a 0.9:1 volume ratio of the Ampure XP beads and eluted in 25 μl of TE buffer (10 mM Tris, 1 mM EDTA, pH 8.0). One microliter of each sample was run on an Agilent Bioanalyzer DNA 1000 chip to confirm the presence of a library and the absence of adapter/primer dimer. When excess dimer was observed, an additional 0.85:1 volume ratio of Ampure XP bead cleanup was performed with Agilent chip quality control.
DNA sequencing and mapping to the genome.
DNA was sequenced on an Illumina HiSeq 2000 (150-bp read length, paired end; San Diego, CA). Reads from desired transposon insertions into the C. crescentus genome have an expected sequence composition consisting of (i) 5 random nucleotides, (ii) 35 bp corresponding to the terminal inverted repeat of the Tn5 transposon, and (iii) 110 bp derived from the C. crescentus genome and/or read-through into the Illumina sequencing adapter. Reads beginning with the transposon Tn5 sequence were identified using Cutadapt (31). Tn5 sequence and any detectable sequences derived from Illumina sequencing adapters were removed using Cutadapt, leaving DNA corresponding to insertion sites in the C. crescentus genome. Trimmed sequences were subject to quality checking using Trimmomatic (32) to remove trailing sequences with a sliding-window average quality score of 15 or less. Trimmed sequences were then aligned to the Caulobacter crescentus NA1000 reference genome sequence (GenBank accession number NC_011916.1) using the Burrows-Wheeler Aligner (BWA) (33). BAM alignment files were converted to BedGraph (reads versus genome position) for viewing using custom Perl scripts. Transposon insertion sites were identified as the mapped location of the first base position of sequencing strings after the Tn5 end sequences were trimmed. Statistics from the sequencing analysis are summarized in Table 1.
TABLE 1.
Statistics for Tn-seq analysis
| Library | Replicate no.a | Total no. of reads | No. of reads (%) after filtering | No. of reads (%) mapped to the genome | No. of unique insertions | Insertion resolution (bp/insertion) |
|---|---|---|---|---|---|---|
| Master Tn library 1 | 1 | 4,622,651 | 3,879,040 (84) | 1,128,991 (29) | 254,837 | 14 |
| Master Tn library 2 | 1 | 6,821,312 | 5,648,392 (83) | 1,655,997 (29) | 299,848 | 12 |
| Uranium library 1b | 1 | 5,258,684 | 4,475,230 (85) | 1,407,024 (31) | 172,227 | 21 |
| 2 | 4,533,632 | 4,105,593 (91) | 471,566 (11) | 102,123 | 36 | |
| Uranium library 2c | 1 | 4,345,587 | 3,535,408 (81) | 1,570,567 (44) | 229,517 | 16 |
| 2 | 3,769,970 | 3,073,357 (82) | 1,421,032 (46) | 214,127 | 17 | |
| 3 | 5,484,725 | 4,452,924 (81) | 2,078,630 (47) | 233,053 | 16 | |
| Uranium library 3c | 1 | 5,281,822 | 4,168,258 (79) | 2,382,532 (57) | 172,368 | 21 |
| No-metal library | 1 | 10,718,904 | 9,297,151 (87) | 1,160,988 (12) | 150,231 | 25 |
| 2 | 8,977,647 | 7,058,714 (79) | 2,848,953 (40) | 215,835 | 17 | |
| Cadmium library | 1 | 4,771,097 | 3,685,040 (77) | 2,342,281 (64) | 213,944 | 17 |
Replicates are different DNA isolations from the same Tn mutant library.
Uranium library 1 was conducted at 250 μM uranyl nitrate.
Uranium libraries 2 and 3 were conducted at 275 μM uranyl nitrate.
Data analysis for U-specific essentiality.
The open-source, web-based software ESSENTIALS (34) was used to analyze and compare data from the test libraries (U, Cd, and no-metal) to the control master libraries. For each pairwise analysis, BedGraph files for replicates originating from a single test library of mutants were compared to BedGraph files from the control master libraries. The BedGraph files were analyzed using the genome of Caulobacter crescentus NA1000 (GenBank accession number NC_011916.1) with default parameters, including use of “truncated.ppt” for truncation of the 3′ end of genes, use of LOESS genomic position bias removal, TMM read count normalization, unpaired (qCML) analysis, tag-wise dispersion modeling of variance with a value of 5 for prior.n smoothing, and Benjamini-Hochberg (BH) P value adjustment. Library sizes were inputted as 1,500,000. Normalized reads/gene, log2 fold change in normalized total insertions per gene compared to the levels in the master libraries (logFC), and adjusted P values were outputted from the software (see Table S3 in the supplemental material).
For heat map analysis, the raw counts of total insertions per gene were normalized using the ESSENTIALS program with the master libraries as the reference data sets and all the test libraries as the test sets. The heat map was generated using the default setting in the R heatmap function for unsupervised hierarchical clustering.
In order to identify genes that have lower fitness under each test condition than in the master libraries, one needs to assign significant logFC and P value cutoffs. Based on known genes that are required for Cd survival (35) (see Table S4 in the supplemental material), we used the Cd data set to determine the logFC (<−0.9) and P value (<0.05) cutoffs. Genes previously identified as high fitness or essential in the genome of C. crescentus (29) were removed from further analysis due to potential complications in analyzing whether U susceptibility for mutants in these genes is due to poor fitness in general or to a U-specific defect.
Genes with significantly lower fitness in U library 2 but not in the no-metal or Cd library were identified as U-specific gene candidates (Table 2; see also Table S5 in the supplemental material). U library 2 was used as the primary library because it had the most number of replicates and the most reproducible data among replicates (data not shown). The U-specific gene candidates identified based on U library 2 were further divided into three regimes based on their degrees of prevalence in the other two U libraries: the top regime comprised genes that had significantly lower fitness in all three U mutant libraries, while the middle and bottom regimes had genes that had significantly lower fitness in two of the U libraries and in only one library (U library 2), respectively.
TABLE 2.
Select gene candidates with low U fitness
| Gene | Annotation | LogFCa |
||||
|---|---|---|---|---|---|---|
| Uranium library 1 | Uranium library 2 | Uranium library 3 | No-metal library | Cadmium library | ||
| CCNA_00080 | LexA-like transcriptional repressor | −1.17* | −2.43** | −1.59** | −0.70 | −0.31 |
| CCNA_03858 | secB, protein translocase subunit | −1.67** | −1.97** | −3.32** | −0.14 | 1.46** |
| CCNA_03624 | cztA, sodium bicarbonate cotransporter | −1.79** | −1.54** | −2.58** | −0.14 | −0.79* |
| CCNA_00285 | Acetylglutamate kinase | −0.97* | −1.50** | −2.00** | −0.87 | 0.55 |
| CCNA_01379 | rsaFb, type I secretion outer membrane protein | −0.93* | −1.41** | −1.91** | −0.13 | 0.20 |
| CCNA_03625 | cztR, LysR family transcriptional regulator | −1.29** | −1.41** | −2.26** | −0.06 | −0.70 |
| CCNA_03498 | MarR/EmrR family transcriptional regulator | −0.98* | −1.36** | −2.23** | −0.42 | 0.42 |
| CCNA_01521 | Hypothetical protein | −0.98** | −1.20** | −3.29** | −0.69 | −0.12 |
| CCNA_00290 | Autotransporter protein | −0.98** | −1.04** | −1.74** | −0.54 | −0.77** |
| CCNA_02552 | clpS, Clp protease adaptor protein | −0.14 | −1.39** | −1.99** | −0.54 | 1.19** |
| CCNA_02553 | clpA, Clp protease ATP-binding subunit | 0.21 | −1.31** | −1.33** | −0.36 | 1.85** |
| CCNA_02140 | fliM, flagellar motor switch protein | −1.14** | −1.03** | −0.58* | −0.90 | −0.77** |
| CCNA_01067 | rsaFa, type I secretion outer membrane protein | −0.24 | −1.03** | −0.91** | −0.37 | 1.54** |
| CCNA_01061 | rsaE, type I secretion adaptor protein RsaE | −0.06 | −1.32** | 0.23 | 0.03 | 2.46** |
| CCNA_01622 | ppGpp hydrolase-synthetase RelA/SpoT | −0.37 | −0.98** | −0.12 | −0.25 | −0.02 |
Log2 fold change in normalized total insertions per gene compared to values in the master libraries. *, P < 0.05; **, P < 0.01.
Construction of deletion mutants.
For construction of clean deletion mutants, ∼500-bp regions upstream and downstream of a given gene were PCR amplified using the primers listed in Table S2 in the supplemental material. A three-way Gibson assembly (In-Fusion HD Cloning Plus kit; Clontech, Mountain View, CA) of the upstream and downstream fragments into HindIII and EcoRI sites of pNPTS138 generated the plasmids listed in Table S1. The sequences of the plasmids were confirmed by DNA sequencing. In-frame deletion of a gene was generated using the respective plasmid through a standard homologous recombination method as previously described (36, 37).
For construction of mutants lacking rsaA, a Tetr cassette was amplified from pKO3 (38) using rsaAtet_for and rsaAtet_rev primers (see Table S2 in the supplemental material) and inserted into the NdeI site of pMCY04 (between the upstream and downstream flanking regions of rsaA) using Gibson assembly to generate pMCY04a. In-frame replacement of rsaA with the Tetr cassette was generated using pMCY04a as previously described (38). This method was used because of difficulties in obtaining clean deletions of rsaA in the mutant backgrounds of rsaFa and rsaFb due to unknown reasons.
U survival assay.
Strains of C. crescentus were precultured in 2 ml of PYE medium supplemented with the appropriate antibiotic(s) from single colonies at 30°C overnight. Cells were then diluted to an initial optical density at 600 nm (OD600) of 0.2 in PYE medium and cultured until the OD600 reached ∼0.4 to 0.6 (late exponential phase). Cells were harvested, washed, and resuspended in 50 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)] buffer (pH 7.0) to a final OD600 of 0.5. Uranyl nitrate was added to the suspensions to a final concentration of 50 μM and incubated at room temperature (RT). A negative control without U was also included. After 1 h of U exposure, aliquots were removed, serial dilutions of 101 to 106 of the aliquots were prepared, and 5 μl of each dilution was spotted onto PYE-agar medium. After 2 days of incubation at 30°C, CFU counts were determined. The fold change in cell death was calculated by dividing the number of CFU/ml from assays without U by that with U. At least four biological replicates were included for each assay.
Growth in buffered PYE medium spiked with U.
C. crescentus cells were precultured from single colonies in 500 μl of PYE medium supplemented with the appropriate antibiotic(s) at 30°C for 8 h. Cells were then diluted to an initial OD600 of 0.001 in PYE medium with the appropriate antibiotic(s) and cultured for 16 h until the OD600 was about 0.6 (late exponential phase). Cells were harvested, spent medium was removed, and cells were inoculated into PYE medium supplemented with a final concentration of 50 mM MES, pH 6.1, to an initial OD600 of 0.02. MES buffer was added to maintain pH; without buffering, the medium pH would decrease upon addition of uranyl nitrate due to U hydrolysis and the nitric acid present in the U stock solution. Cells were cultured at 30°C and 220 rpm until the OD600 reached 0.06 (early exponential phase), at which point uranyl nitrate was added to a final concentration of 350 μM. Controls without U addition were included. Cell density was monitored by the OD600, and biological triplicates were included. Given that no significant cell elongation was observed during growth of C. crescentus in buffered PYE medium containing U (data not shown), doubling times were estimated based on OD600 values.
Gene expression by NanoString.
C. crescentus NA1000 and rsaFa mutant cells were grown in MES-buffered PYE medium as described above with U spiked in at early exponential phase. After 30 min of U exposure, 10 ml of cells was removed, and transcription was immediately stopped by addition of 1.25 ml of a solution of 5% phenol–95% ethanol on ice. Cells were harvested by centrifugation at 4°C, and total RNA was extracted from cells using TRIzol (Life Technologies, Carlsbad, CA) according to the manufacturer's instructions. Samples were prepared in biological triplicates. NanoString analysis to determine log2 expression levels was conducted as described elsewhere (39). NanoString probes used for mRNA detection are listed in Table S6 in the supplemental material.
Cellular accumulation of U.
Cells were grown, washed, and resuspended in PIPES buffer as described for the U survival assay above. Uranyl nitrate was added to the suspensions to a final concentration of 100 μM and incubated at RT for 30 min. The cell suspension was centrifuged at 20,000 × g for 5 min, and the supernatant was collected and analyzed for U content using an Arsenazo III colorimetric assay as described previously (4, 40). An abiotic control was also performed without addition of cells to control for abiotic U precipitation. All assays were performed in biological triplicates. The amount of U accumulated by the cells was calculated as follows: U accumulation = total U − abiotic U precipitate − U in the supernatant.
Determination of antimicrobial sensitivity by agar disk diffusion assays.
C. crescentus NA1000, rsaFa mutant, rsaFb mutant, and rsaFa rsaFb (rsaFaFb) double mutant cells were grown in PYE medium to exponential phase. Cultures were normalized and adjusted to an OD600 of 0.3, and 100 μl was plated on PYE-agar medium. Filter paper disks with a diameter of 7 mm were placed on the agar with the following compounds pipetted onto a disk (volume and concentration, respectively): tetracycline (5 μl, 50 μg/ml), cadmium sulfate (10 μl, 40 mM), uranyl nitrate (10 μl, 200 mM), SDS (5 μl, 5% [wt/vol]), or hydrogen peroxide (5 μl, 100 mM). Plates were incubated at 30°C for 2 days. Pictures of the agar plates were taken using a ChemiDoc imager (Bio-Rad), and diameters of zones of inhibition were measured using ImageJ (41).
U biomineralization assay.
The U biomineralization assay was conducted as previously described (4). Briefly, cells grown in PYE medium to late exponential phase were harvested, washed once with 10 mM NaCl, and resuspended in PIPES buffer to a final OD600 of 0.5. Biomineralization assays were started by the addition of glycerol-2-phosphate and uranyl nitrate to final concentrations of 5 mM and 500 μM, respectively, and incubated at 30°C. Aliquots were removed at specific time points to quantitate soluble U using the Arsenazo III colorimetric assay and to determine CFU counts.
RESULTS
Tn-seq screen for U-sensitive mutants.
In our Tn-seq screening scheme (Fig. 1), master libraries of Tn5 mutants were first generated by selection on PYE-kanamycin plates according to Christen et al. (29). These master libraries were subsequently exposed to each of three conditions on PYE plates to generate the experimental libraries: (i) uranyl nitrate (U) at 250 or 275 μM, (ii) cadmium sulfate (Cd) at 12 μM, or (iii) no metal. The concentrations of U and Cd were chosen as the maximum concentrations in which the wild-type C. crescentus strain did not exhibit a significant growth defect on PYE-agar medium supplemented with the given metal (data not shown). Tn5 mutant generation and selection on U were performed in two steps due to observed incompatibility between U and kanamycin selection, presumably due to chemical interactions that block antibiotic activity when U and kanamycin are used concurrently (data not shown). The no-metal condition identifies mutants that have lower fitness due to conditions endured during the second plating step, such as freeze-thaw stress. The Cd condition controls for genes that are involved in tolerance to heavy metal stress in general rather than U-specific stress. Each of the libraries obtained in this study contained ∼106 colonies in order to obtain enough mutants to saturate the genome (29).
An Illumina HiSeq system was used to determine the genomic distributions of Tn5 insertions for each library (29). One to three replicates of genomic DNA from a single library were isolated and sequenced to test the reproducibility of our method. Of the 4 to 10 million reads generated for each replicate, 77 to 91% passed initial filtering, demonstrating that the reads from the HiSeq were of high quality (Table 1). However, only 11 to 64% of the total reads were mapped to the C. crescentus genome; for reasons unclear to us, the majority of the remaining reads mapped to the pXMCS2::Tn5Pxyl plasmid used for transposon mutagenesis. Less than 1% of reads were unmapped, representing nonspecific PCR products. Despite the low proportion of mapped reads, ∼100,000 to 300,000 unique insertion sites (i.e., one insertion for every 12 to 36 bp on average) are represented in each data set, which is comparable to the insertion frequencies reported in other Tn-seq studies (42–44). By plotting the distribution of unique insertions as a function of gene length for every C. crescentus open reading frame (ORF), we were able to determine whether transposon insertions that are underrepresented in each data set correspond to the set of essential or fitness-relevant genes previously identified for C. crescentus (29) and thereby evaluate the reliability of the transposon mutagenesis scheme. We found that 96% ± 4% of C. crescentus ORFs identified as essential in the Christen et al. Tn-seq screen (29) are found within the quartile of C. crescentus genes with the lowest proportion of unique insertions per unit gene length in each data set (see Fig. S1 in the supplemental material). These results demonstrate the integrity of each data set and validate the set of genes previously found to contribute substantially to fitness or viability.
Using ESSENTIALS, an open-source, web-based algorithm (34), we compared and normalized data from each experimental library (U libraries 1, 2, and 3; Cd; no-metal) to data obtained from the master libraries in a pairwise fashion (test library versus master libraries). This analysis enabled determination of insertion frequency (number of transposon insertions/gene) for each gene and the fold change in insertion frequency for every gene under each experimental condition relative to the frequencies of the master libraries. Heat map representation of all normalized data sets revealed that replicates from the same experimental library clustered together, showing high reproducibility among biological replicates (see Fig. S2 in the supplemental material). The three U libraries were only loosely clustered, which is likely attributed to the differences in U concentrations (250 μM used in U library 1 compared to 275 μM in U libraries 2 and 3) and mutant library sizes (0.5 × 106 mutants in U library 3 compared to 1.5 × 106 to 2.5 × 106 mutants in U libraries 1 and 2, respectively).
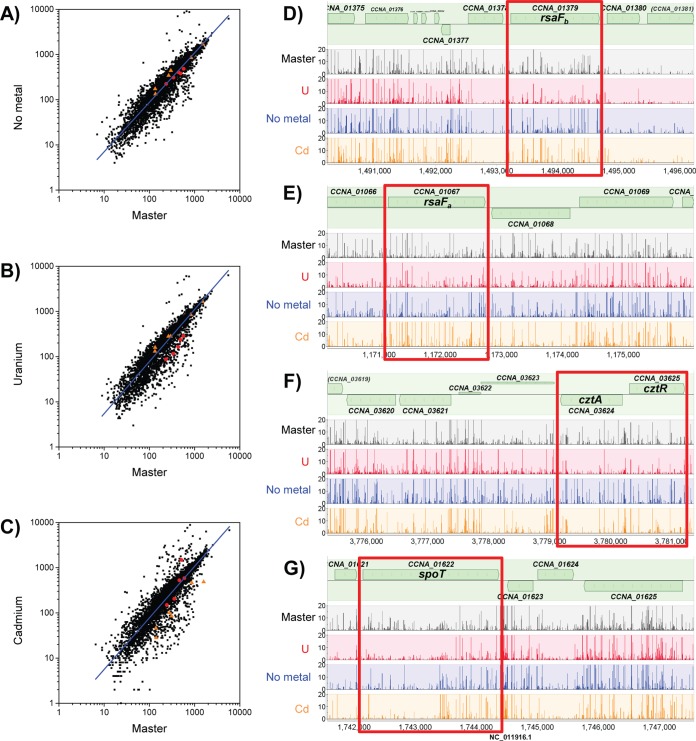
We define U-specific fitness genes as those ORFs that exclusively exhibit lower insertion frequencies in the U libraries than in the master libraries (Fig. 2). The Cd data set was used to assign cutoffs for logFC (<−0.9) and P values (<0.05) representing lower insertion frequencies based on genes that are known to be required for fitness in the presence of Cd (35) (see Table S4 in the supplemental material). Using these criteria, we identified 37 U-specific candidates (see Table S5). We note that these candidates are not those with the largest differences in insertion frequencies between the master and U libraries; the more extreme outliers were eliminated because they were also extreme outliers in the Cd and no-metal data sets and likely represent a category of genes that affect survival during the second plating step. From the list of 37 U-specific candidates, we selected 15 candidate genes for further study with priority toward those genes identified in all three U libraries (Table 2). Mutants of each of the 15 genes were obtained (see Table S1 in the supplemental material) and first tested for their ability to grow in the presence of PYE medium supplemented with 300 μM uranyl nitrate without pH buffering (see Fig. S3), conditions similar to those used in the Tn-seq screen. Results revealed that 10 of the 15 mutants exhibited a growth defect in the presence of U relative to growth of the wild type. We later discovered that the pH of PYE medium decreases from 6.5 to 5.7 with addition of 350 μM uranyl nitrate, affecting growth phenotypes (25). To uncouple the U response from changes in pH and growth conditions, we tested the ability of each mutant to survive U exposure under buffered conditions (Fig. 3) and found that 4 of the 15 mutants showed significantly higher U susceptibility than the wild type. These four mutants will be discussed in the following sections.
FIG 2.
Comparison of insertion frequency among mutants under different selective pressures. (A to C) Plots of log(total insertions/gene length) for the no-metal, U library 2, or Cd condition versus the master library prior to selection. Genes with lower fitness under the U condition (rsaFa, rsaFb, cztR, cztA, and spoT) are highlighted with red circles. Genes with lower fitness under the Cd condition (ccna_02805-ccna_02811; Ni/Co/Cd resistance cassette) are highlighted with orange triangles. Linear regression analyses of the data are shown as blue lines. (D to G) Total insertion counts at representative genome loci with low U fitness are compared in the master libraries, U library 2, no-metal library, and Cd library, as indicated. Regions containing genes of interest are boxed in red. Plots were generated using MochiView (67), with insertion counts averaged among replicates from the same library.
FIG 3.
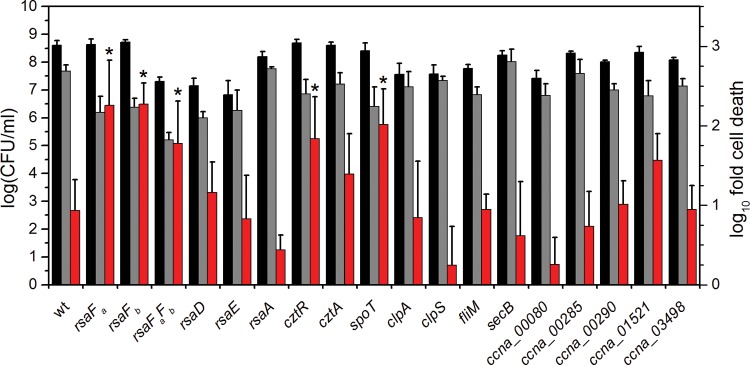
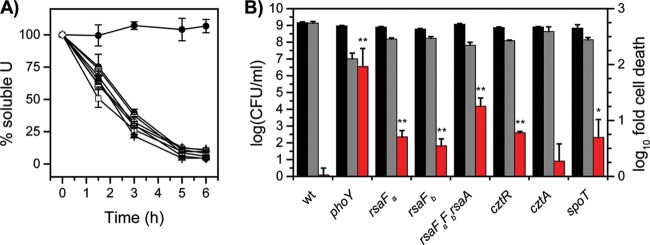
Comparison of U survival by mutants identified by Tn-seq. Strains were exposed to a no-U control (black bars) or 50 μM uranyl nitrate (gray bars) in 50 mM PIPES buffer at pH 7.0 for 1 h, after which CFU/ml were counted (left axis). Red bars represent the fold change in CFU/ml counts between the U condition and the control (right axis). Error bars represent standard deviations from at least four biological replicates. Mutants showing a lower U survival rate (i.e., higher susceptibility to U) than the wild type are highlighted with asterisks (*, P < 0.01).
rsaFa and rsaFb are important for U fitness.
Of the candidates tested, rsaFa and rsaFb mutants were observed to have the most significant reduction in CFU/ml in the U survival assay, with an ∼10-fold-lower survival rate than that of the wild type when exposed to U. While the rsaFaFb double mutant showed a reduction in survival rate similar to reductions of the single mutants, it exhibited a survival defect even in the absence of U, unlike the single mutants, suggesting that the double mutant had poor fitness in general (Fig. 3). rsaFa and rsaFb are two homologous genes encoding the outer membrane proteins RsaFa and RsaFb that are part of the type I protein translocation pathway for exporting the highly abundant S-layer protein RsaA to the cell surface of C. crescentus (45). The fact that rsaFa and rsaFb are located 322 kb apart in the genome and that we were able to identify both genes in our Tn-seq screen suggest that they are individually critical for U tolerance. Furthermore, expression analysis revealed that rsaFa and rsaFb transcripts are upregulated ∼4-fold and ∼10-fold, respectively, during growth in buffered PYE medium by the addition of 350 μM U (Fig. 4), implying a concerted regulatory response for RsaFa and RsaFb in U resistance.
FIG 4.
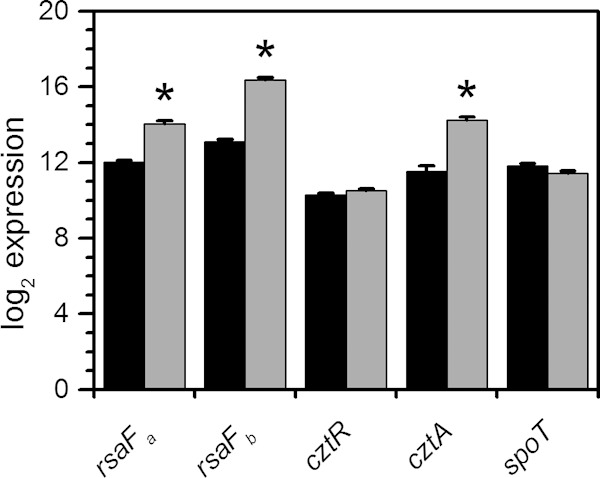
Transcript levels of select gene candidates in response to U. C. crescentus cells were grown in buffered PYE medium to early exponential phase and were either untreated (black bars) or spiked with 350 μM uranyl nitrate (gray bars). Samples were collected 30 min after spike-in, and transcript levels were determined by NanoString analysis. Log2 expression values are presented, and error bars represent standard deviations from three biological replicates. Transcripts significantly upregulated are highlighted with asterisks (*, P < 0.01).
To confirm that these survival defects are not the result of a compromised S-layer, we tested U susceptibility of rsaA, rsaD, and rsaE null mutants, which are deficient in various factors required for S-layer biogenesis (Fig. 3). RsaA is the structural protein that comprises the S-layer, and RsaD and RsaE are the ATP-binding ABC transporter and membrane fusion proteins, respectively, that, together with RsaF, form the channel to export RsaA across the cell envelope (46); all mutants were previously shown to be deficient in S-layer production (45, 46). In contrast to the RsaF mutants, the rsaA, rsaD, and rsaE null mutants showed similar CFU/ml counts as the wild type in the presence of U, suggesting (i) that the S-layer per se does not confer U resistance and (ii) that RsaD and RsaE are not involved in U tolerance, indicating that the mechanism of U tolerance does not rely on components of the S-layer transport system other than RsaF.
In addition to cell survival, rsaF mutants were tested for U tolerance during growth in buffered PYE medium in a spike-in experiment, where U was added during early exponential phase (Fig. 5; see also Fig. S4 in the supplemental material). We observed that the rsaFb mutant did not exhibit any significant change in doubling time with or without U, whereas the rsaFa mutant exhibited an increase in doubling time compared to that of the wild type whether or not U was added, indicating a general (U-independent) growth defect. Unexpectedly, the rsaFaFb double mutant reversed this defect; in the absence of U, the double mutant grew at a rate only slightly lower than that of the wild type. The double mutant still exhibited a significant increase in doubling time in U-containing medium compared to that of the wild type, suggesting that RsaFa, but not RsaFb, is needed for U tolerance during growth. The unexpected lack of direct correlation between growth and survival phenotypes in the rsaF mutants (compare Fig. 3 and 5) indicates that mechanisms of U toxicity during active growth and the survival assay are different. The cause of the general growth defect observed for the rsaFa mutant is unclear.
FIG 5.
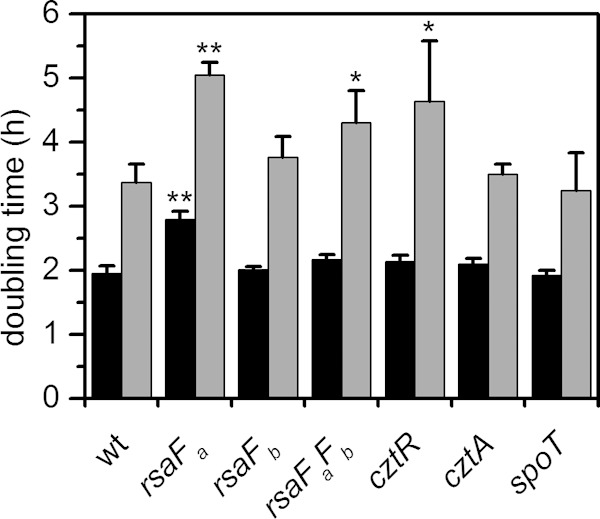
Comparison of doubling times of mutant strains in the presence or absence of U. Mutants were grown in buffered PYE medium to early exponential phase and were either untreated (black bars) or spiked with 350 μM uranyl nitrate (gray bars). Doubling times were estimated based on OD600 values during growth. Error bars represent standard deviations from three biological replicates. Mutants showing a significant increase in doubling time compared to that of the wild type (wt) under the same conditions (without or with U) are highlighted with asterisks (*, P < 0.02; **, P < 0.0001).
To determine whether the increased U sensitivity observed in mutants lacking one or both RsaF subunits is an indirect consequence of intracellular accumulation of RsaA, we prepared rsaA gene deletions in the rsaFa, rsaFb, and rsaFaFb mutant backgrounds and measured the change, if any, in U tolerance in survival and growth assays. In the survival assay, all three of the deletion strains constructed (rsaFa rsaA, rsaFb rsaA, and rsaFaFb rsaA strains) showed survival rates in the presence of U similar to those of their respective rsaA+ parents (∼102- to 103-fold more cell death relative to the level of the no-U control), suggesting that U sensitivity is not caused or exacerbated by RsaA accumulation (see Fig. S5A in the supplemental material). However, we did observe that the U-independent survival defect of the rsaFaFb double mutant (Fig. 3; see also Fig. S5A) was completely suppressed by the deletion of rsaA. This result demonstrates that intracellular RsaA accumulation contributes to the general survival defect of the rsaFaFb mutant. In the growth assay, the rsaFa rsaA mutant displayed a prolonged doubling time relative to that of the rsaFa single mutant regardless of U addition, suggesting that the slow growth of this strain is not caused by cytoplasmic RsaA (see Fig. S5B). Interestingly, the rsaFaFb rsaA triple mutant grew slightly better than the rsaFaFb mutant in both the presence and the absence of U (see Fig. S5B), indicating that cytoplasmic RsaA accumulation may, indeed, slow growth in cells lacking RsaF activity. We therefore used the rsaFaFb rsaA triple mutant in place of the rsaFaFb double mutant in subsequent U-uptake experiments in order to avoid complications in interpretation due to the effects of intracellular RsaA accumulation.
Finally, to confirm the results of our Tn-seq experiment, we examined the ability of the rsaFaFb rsaA triple mutant to survive on solid PYE-agar medium containing either 300 μM uranyl nitrate, 12 μM cadmium sulfate, or no metal (see Fig. S5C in the supplemental material). While wild-type C. crescentus exhibited full survival under all conditions tested, the rsaFaFb rsaA mutant exhibited a survival defect in the presence of U (∼102-fold increase in cell death) but not in the presence of Cd or the no-metal control. In contrast, a deletion mutant of the Cd efflux transporter ccna_02807 exhibited a survival defect in the presence of Cd but not in the presence of U or the no-metal control. These results validated the results of the Tn-seq screen from which mutants in the rsaF system were identified.
RsaF resembles TolC from E. coli.
Smit and coworkers have previously reported that the two RsaF proteins are both homologues of TolC, a multifunctional E. coli outer membrane protein known to interact with several efflux pump partners in transporting a variety of compounds (including antibiotics and metals) from the cytoplasm to outside the cell (45). A BLAST search of the C. crescentus genome indicates that RsaFa and RsaFb have the most significant identity to TolC, with sequence identities of 23% and 26% (87% coverage), respectively (47). Given the evolutionary relationship with the TolC efflux pump, we hypothesized that RsaFa and RsaFb may confer U tolerance by negotiating the export of U through an RsaF-dependent pathway that can be genetically separated from S-layer biogenesis.
To test this hypothesis, we quantified U accumulation in the wild type and the rsaF mutants (Fig. 6). We reasoned that higher levels of U accumulation should be observed in the rsaF mutants than in the wild type if the TolC-like RsaF proteins play a role in U efflux. Consistent with this prediction, the rsaFa and rsaFaFb rsaA mutants exhibited ∼30% and ∼20% higher U accumulation, respectively, than the wild type after 30 min of U exposure. The increase in U accumulation in these strains cannot be attributed to S-layer perturbation since the rsaA single mutant exhibited the same amount of U accumulation as the wild type. Since the rsaFb mutant did not exhibit increased U accumulation, the observations together suggest that RsaFa may play a specific role in the prevention of U accumulation in C. crescentus. However, we cannot rule out the possibility that the higher U accumulation observed in the rsaFa and rsaFaFb rsaA mutants was due to defects in outer membrane integrity. Attempts to measure U efflux were not successful (data not shown), which may be due to potential U precipitation or cell wall adsorption.
FIG 6.
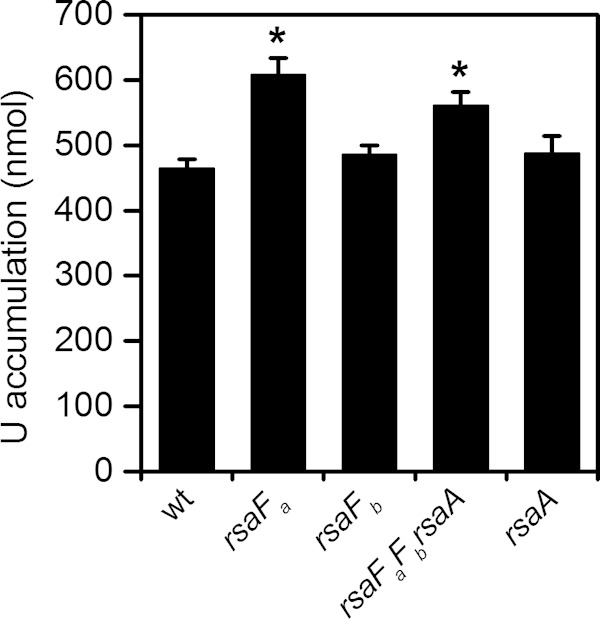
Comparison of U accumulation by wild type (wt) and rsaF mutants. Amounts represent U accumulated by 8 × 107 cells after 1 h of U exposure in 50 mM PIPES buffer at pH 7.0. Error bars represent standard deviations from three biological replicates. Mutants showing higher U accumulation than that of the wild type are highlighted with asterisks (*, P < 0.002).
To determine whether the C. crescentus rsaF system could behave similarly to a TolC-like efflux system, the susceptibility of the rsaF mutant strains against several antimicrobial substrates of TolC-utilizing type I secretion systems (48) was determined using a disk diffusion antimicrobial sensitivity assay (Fig. 7). All three rsaF mutants showed larger zones of growth inhibition in the presence of tetracycline or U than the wild-type strain. The rsaFa and rsaFaFb mutants but not the rsaFb mutant showed larger inhibition zones in the presence of Cd or SDS. Thus, RsaFa appears to be the primary factor involved in resistance to these two compounds. We note that Cd sensitivity was observed only in the disk diffusion assay but not under the conditions of the Tn-seq screen (see Fig. S5C in the supplemental material), which is likely due to higher Cd concentrations in the disk diffusion assay, resulting in increased sensitivity. Together, these results demonstrate that rsaF mutants show broad antimicrobial susceptibility that is not limited to U, similar to TolC in E. coli. The rsaF mutants were also tested for susceptibility to chloramphenicol, ethidium bromide, zinc, and chromium and exhibited the same degree of susceptibility as the wild type (data not shown). The mechanisms of antimicrobial selectivity by the rsaF system, however, are currently unclear.
FIG 7.
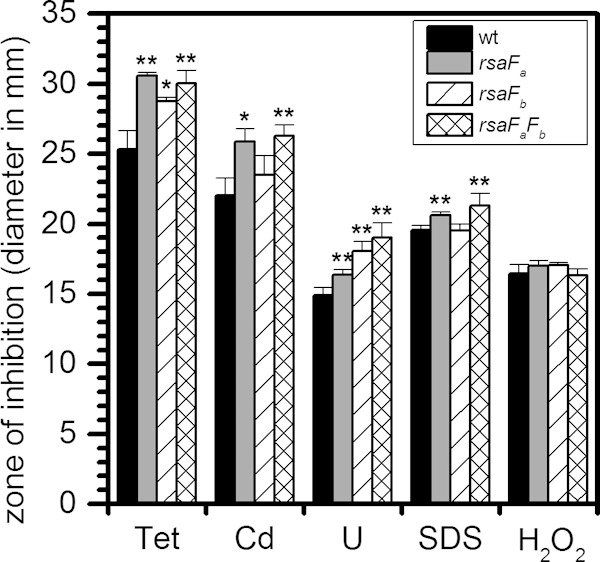
Comparison of sensitivity of wild type and rsaF mutants to different antimicrobials using an agar disk diffusion assay. Diameters of zones of inhibition are shown in millimeters. Tet, tetracycline; Cd, cadmium sulfate; U, uranyl nitrate; SDS, sodium dodecyl sulfate; H2O2, hydrogen peroxide; wt, wild type. Error bars represent standard deviations from three biological replicates. Mutants showing a significant increase in diameter compared to that of the wild type are highlighted with asterisks (*, P < 0.02; **, P < 0.01).
Previous reports indicate that the lack of TolC can induce oxidative stress in E. coli (49). Reasoning that this confounding effect could at least partially account for the apparent U sensitivity of rsaF mutants, we tested whether the deletion of rsaFa leads to an increase in oxidative stress in C. crescentus. To do this, we measured the transcription of oxidative stress genes in the wild type and the rsaFa mutant grown in buffered PYE medium with and without U (see Fig. S6A in the supplemental material). Results indicated that the expression levels of key oxidative stress genes (sodB, katG, oxyR, and dps) are not significantly changed in the rsaFa mutant, suggesting that deletion of rsaFa is not sufficient to induce an oxidative stress response and that U toxicity is not an indirect effect of increased oxidative stress in the rsaFa mutant. Consistently, all rsaF mutants tested showed susceptibility to hydrogen peroxide stress similar to that of the wild type in the disk diffusion assay (Fig. 7). In addition, we note that genes involved in heat shock, DNA damage, and outer membrane stress (e.g., LpxC) (see Fig. S6B, C, and D, respectively, in the supplemental material) also exhibited similar expression levels between the wild type and the rsaFa mutant, indicating that the loss of RsaFa does not cause activation of common cytoplasmic or envelope stress responses. Significantly, the expression of LpxC, which is involved in lipopolysaccharide (LPS) biosynthesis (50), is also unchanged, arguing against significant remodeling of the LPS. However, we cannot conclusively rule out the possibility that the lack of rsaFa itself in some way affects the composition and/or properties of the outer membrane.
cztR-cztA.
The cztR and cztA genes were also significant hits in our Tn-seq screen. The cztR gene encodes a LysR family transcriptional regulator that, along with the putative CztA transporter, has been implicated in response to Zn limitation and Cd exposure (51). To characterize the contribution of CztRA to U tolerance, we performed growth and survival assays in strains lacking cztR or cztA. A mutant lacking cztR exhibited lower fitness upon U exposure, with ∼10-fold increase in cell death compared to that in the wild type (Fig. 3). Consistently, the cztR mutant exhibited a U-dependent growth defect, with a significant increase in doubling time in the presence of U compared to that in the wild type (Fig. 5; see also Fig. S4 in the supplemental material). Conversely, no discernible changes in cell survival or U sensitivity in growth were apparent following deletion of cztA (Fig. 3 and 5; see also Fig. S4). There was no significant change in cztR expression upon U exposure (Fig. 4). An ∼8-fold increase in cztA expression occurs in the presence of uranyl nitrate; however, this increase may be due to nitrate presence since an ∼8-fold increase in cztA expression also occurred in the presence of potassium nitrate (data not shown). Although the CztRA system may not be specifically induced by U, it clearly plays a role in U fitness under both survival and growth conditions. Given its previously reported function in Cd resistance (35, 51), the CztRA system may be involved in a general heavy metal stress response.
spoT.
A mutant lacking spoT was observed to have a lower fitness with U, with an additional ∼10-fold reduction in the number of CFU/ml upon U exposure compared to that of the wild type (Fig. 3). The spoT gene encodes a ppGpp hydrolase/synthetase that was previously shown to be involved in the stringent response in many bacteria including C. crescentus and E. coli (52, 53). Deletion of spoT in C. crescentus resulted in cell death upon carbon starvation due to unconstrained chromosome replication (54). In order to test whether the cell death observed with the spoT mutant upon U exposure is related to carbon starvation, we repeated the U survival assay in the presence of glucose (Fig. 8). The spoT mutant showed a survival rate similar to that of the wild type upon U exposure in the presence of glucose, suggesting that the observed U defect in the absence of glucose is a consequence of carbon starvation rather than of U-specific toxicity. We note that U toxicity is somewhat mitigated by the presence of glucose, presumably due to complexation with U. Consistently, the growth assay indicated that the doubling time of the spoT mutant is similar to that of the wild type, regardless of the presence of U (Fig. 5), and in the wild-type strain, the expression level of spoT remained unaltered by U (Fig. 4). Thus, spoT appears to be important for U tolerance only under carbon starvation conditions, likely resulting from enhanced U toxicity to cells with unchecked DNA replication (54).
FIG 8.
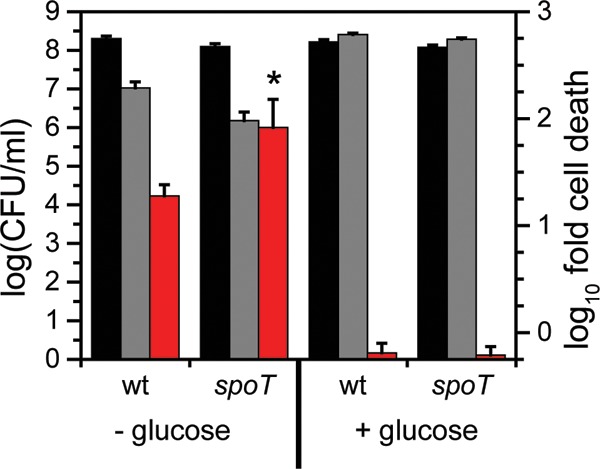
Comparison of cell survival of wild type and spoT mutant in response to U in the presence or absence of glucose. Strains were untreated (no U; black bars) or exposed to 50 μM uranyl nitrate (gray bars) and either not supplemented or supplemented with 1% glucose in 50 mM PIPES buffer at pH 7.0 for 1 h, after which CFU/ml were counted (left axis). Red bars represent the fold change in CFU/ml counts between the U condition and the control (right axis). Error bars represent standard deviations from four biological replicates. Higher susceptibility of the mutant to U than of the wild type is highlighted with an asterisk (*, P < 0.01).
Biomineralization activity of mutants.
We have previously found that C. crescentus actively biomineralizes U via its native phosphatase activity and that perturbed biomineralization activity causes survival defects during U exposure (4). To test whether the U-sensitive mutants identified above retain their U biomineralization activity, we performed a U biomineralization assay using glycerol-2-phosphate as the phosphate source for the rsaF, cztR, cztA, and spoT mutants (Fig. 9A). The ΔphoY strain, which lacks biomineralization activity and exhibits a severe survival defect in the presence of U (4), was included as a negative control. All mutant strains tested had activity similar to that of the wild type, with more than 85% of the U mineralized after 6 h, demonstrating that none of the strains had perturbed U biomineralization activity. Cell survival analysis under the U biomineralization condition (Fig. 9B) showed that all strains, with the exception of the cztA mutant, had an ∼10-fold reduction in the number of CFU/ml compared to the wild-type level, indicating that U sensitivity in these strains is independent of U biomineralization. These cell survival trends are similar to those observed under nonmineralizing conditions (Fig. 3). We note that the effective U toxicity under mineralizing conditions is much lower than that under nonmineralizing conditions, presumably due to U complexation with glycerol-2-phosphate (4). Cell death exhibited by the ΔphoY control strain was highest (∼100-fold reduction in CFU/ml), showing that lack of biomineralization activity caused the most severe susceptibility to U under mineralizing conditions.
FIG 9.
Comparison of U biomineralization activity and cell survival of mutant strains under U-mineralizing conditions. (A) U biomineralization activity is shown for the wild type (■), phoY mutant (●), rsaFa mutant (▲), rsaFb mutant (◆), rsaFaFb rsaA triple mutant (□), cztR mutant (○), cztA mutant (△), and spoT mutant (♢). Error bars represent standard deviations from three biological replicates. (B) Cell survival under U-mineralizing conditions after 6 h of incubation. CFU/ml counts (left axis) at 0 h (black bars) and 6 h (gray bars) are shown. Fold change in CFU/ml counts between 0 and 6 h (red bars; right axis) is also shown. Mutants showing significantly higher susceptibility to U than the wild type are highlighted with asterisks (*, P < 0.03; **; P < 0.01).
DISCUSSION
Tn-seq is a powerful whole-genome genotypic screening method that has recently been widely employed to identify genes that are critical for survival upon exposure to antibiotics and various other chemical stresses (27, 42, 55). We have performed Tn-seq and identified transposon insertions in 37 distinct genes that result in reduced fitness under U stress, 15 of which were chosen for further study. Individual single mutants lacking each of these 15 genes were tested for U susceptibility, and 10 of these mutants were found to exhibit a growth defect in unbuffered PYE medium supplemented with U, with only 4 mutants confirmed to have a U survival defect in a buffered solution. Besides the potential difference in U susceptibility under different testing conditions, the low correlation between survival phenotype and Tn-seq fitness could also be due to the low stringency used in selecting candidate genes from our screen. Almost all of the genes that we identified had logFC values between 1 and 2 compared to the insertion frequencies of the master libraries, corresponding to 2- to 4-fold reductions in fitness. These values are significantly lower than values found in other Tn-seq studies examining antibiotic resistance and other environmental stresses (27, 42, 55). Gallagher et al. reported that genes with weak, 2.5- to 5-fold, negative selection have a low correlation between phenotype and Tn-seq fitness (42). Given that similar numbers of unique insertions are present in our data sets compared to those of other Tn-seq studies, we believe that the low fitness values are indicative of the fact that C. crescentus has a broad and relatively graded response to U toxicity. Consistent with this notion, previous proteomic and transcriptomic studies (23, 24) also indicate that a small group of specialized genes cannot account for U resistance. Instead, the response occurs on a global level, involving many genes from different pathways serving functions that may act redundantly to mitigate U toxicity. These results highlight the need to conduct detailed phenotypic analysis of mutants following a Tn-seq screen.
Despite the weak negative selection by U, the Tn-seq screen was successful in identifying outer membrane transporters RsaFa and RsaFb, homologues of TolC in E. coli, as factors important for U resistance. Deletion of rsaFa resulted in higher U susceptibility as well as higher levels of U accumulation. In contrast, the rsaFb mutant did not show a significant U growth defect or higher U accumulation. We also observed that the single rsaFa mutant had higher U susceptibility and accumulation of U than the rsaFaFb double mutant. The reason for these results is unclear but may reflect differential gene expression when rsaFa alone is disrupted versus disruption of both genes, leading to different pleiotropic effects. Although RsaFa and RsaFb appear to have redundant functions in S-layer export in C. crescentus (45), our results suggest that RsaFa plays a dominant role in U resistance. RsaFa and RsaFb likely exist as trimers in the outer membrane, similar to TolC, although it is currently unclear whether they preferentially form homotrimers or heterotrimers. Our ongoing efforts focus on building functional RsaF structures computationally and experimentally and testing the preference of different RsaF models/compositions toward drugs/cations.
Given the results from our study, we speculate that RsaF behaves like TolC and acts as an outer membrane efflux pump to enable excretion of a variety of antimicrobials and metals, including U. The increased susceptibility of rsaF mutants to tetracycline, cadmium, and SDS, compounds known to be exported via type I secretion systems (48, 56, 57), suggests that RsaF may be involved in active export of these antimicrobial compounds via a mechanism similar to that of TolC (48). TolC proteins are known to interact with various inner membrane translocases/pumps, consisting of an adaptor protein and an energy-providing protein, typically an ATP-binding cassette protein or proton antiporter of the RND (resistance-nodulation-cell division) system or major facilitator superfamily (MFS) (58), which provide specificity to export particular substrates. In E. coli, TolC is multifunctional and known to interact with several translocases/pumps, including HlyBD for hemolysin toxin secretion and AcrAB for multidrug efflux (58). In C. crescentus, Smit and coworkers previously identified RsaFa and RsaFb as the TolC-like proteins responsible for export of the S-layer protein RsaA and, recently, the S-layer-associated metalloprotease SapA via the translocase comprised of RsaDE (45, 46, 59). U is unlikely to use the same export system as the S-layer protein, however, given that deletion of RsaD and RsaE does not produce increased U susceptibility. Instead, we hypothesize that RsaF may interact with other translocases/pumps to export compounds other than proteins, such as antibiotics and metals. There are several annotated type I pumps for multidrug (ccna_01261, ccna_01865, and ccna_03218) and metal (ncc and czc systems for Ni/Cd/Co/Zn) export in the C. crescentus genome for which the outer membrane export partner is unknown (57, 60). In addition, our Tn-seq screen identified putative multidrug resistance protein B (CCNA_03831) as one of the targets with low U fitness (see Table S5 in the supplemental material). These proteins serve as potential inner membrane module candidates that interface with RsaF to actively export antibiotics and metals via TolC-like systems although further work is needed to determine if they interact with RsaF and have export activities.
Alternatively, RsaF may play a role in maintaining the integrity and function of the outer membrane that acts as a protective layer against antimicrobial compounds. Thus, the absence of RsaF could result in outer membrane perturbation and leakage, leading to increased U permeability, accumulation, and susceptibility. A defective outer membrane could also cause increased sensitivity to antibiotics if a defective outer membrane no longer acts as a barrier to the antibiotics. It is well known that outer membrane integrity affects susceptibility of microbes to antimicrobial compounds (61), especially those with hydrophobic properties. The fact that the rsaFa mutant is sensitive to the detergent SDS is a strong indication that outer membrane barrier function is perturbed in these mutants, directly or indirectly caused by RsaF disruption. Studies in E. coli have also shown that TolC mutants have a variety of pleiotropic effects, including oxidative stress (49, 62, 63), perturbed peptidoglycan biosynthesis (64), misregulation of outer membrane composition (62, 63, 65), and aberrant cell morphology under Fe limitation (66). While it is feasible that these pleiotropic effects occur in RsaF mutants, we note that genes involved in oxidative stress or membrane stress were not significantly differentially regulated in the rsaFa mutant compared to levels in the wild type although our analysis was limited to a small subset of genes.
Finally, another promising set of genes that we identified in our Tn-seq screen are cztR and cztA, which are annotated as a LysR family regulator and sodium bicarbonate transporter, respectively. Braz et al. found that CztR autoregulates its own expression and positively regulates expression of CztA, demonstrating a link between these two proteins (51). They propose that the CztRA system is involved in zinc transport under zinc-limited conditions and that CztR may be involved in regulation of other oxidative stress genes (51). We found that a cztR mutant, but not a cztA mutant, is more susceptible to U in terms of both growth and survival, suggesting that CztR does not confer U resistance by activating expression of cztA. We suspect that the identification of the cztA gene in the Tn-seq screen is an artifactual consequence of the organization of cztA and cztR into a single operon. As a transcriptional regulator, CztR may facilitate U resistance by activating genes that are required for U detoxification, including oxidative stress genes which have been demonstrated to be upregulated in response to U (23) (D. M. Park, unpublished data). Interestingly, these two genes were also previously identified in a transposon mutagenesis screen of C. crescentus under cadmium stress (35). Supporting these prior results, we found that cztR and cztA also exhibited significantly lower fitness under Cd selection in our Tn-seq screen (Table 2), suggesting that the system may be involved in a general heavy metal stress response. The magnitude of the logFC values for these genes in the U libraries, however, were even higher than in the Cd libraries, indicating that CztR may be more important for growth and survival in U than in Cd.
In summary, our Tn-seq screen for genes involved in detoxification and stress regulation during U exposure by C. crescentus has yielded several novel targets whose identity and function had previously remained elusive through other types of omics analyses (23, 24). The results highlight that genes necessary for longer-term growth and survival under U stress do not significantly overlap those that are upregulated in response to acute U stress. Our results thus demonstrate the power of using Tn-seq to conduct genotypic profiling to identify novel genes required for fitness under stress conditions. These novel genes are crucial leads to a detailed mechanistic understanding as to how bacteria are able to resist U in the environment and thus have potential utility in informing future bioremediation strategies.
Supplementary Material
ACKNOWLEDGMENTS
We thank Lucy Shapiro and Harley McAdams for providing ΔrecA, ΔspoT, and fliM::kan strains and for helpful discussions. We thank Marilis Marques for providing the ΔcztA and ΔcztR strains and for helpful suggestions. We thank Urs Jenal for providing ΔclpA and ΔclpS strains. We thank Rong Jiang for assistance with ESSENTIALS and heat map analysis.
This work was performed under the auspices of the U.S. Department of Energy by Lawrence Livermore National Laboratory under contract DE-AC52-07NA27344 (LLNL-JRNL-670205). This study was supported by a Department of Energy Early Career Research Program award from the Office of Biological and Environmental Sciences (to Y.J.) and the JGI Community Science Program.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00382-15.
REFERENCES
- 1.Markich SJ. 2002. Uranium speciation and bioavailability in aquatic systems: an overview. ScientificWorldJournal 2:707–729. doi: 10.1100/tsw.2002.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gadd GM. 2010. Metals, minerals and microbes: geomicrobiology and bioremediation. Microbiology 156:609–643. doi: 10.1099/mic.0.037143-0. [DOI] [PubMed] [Google Scholar]
- 3.Hillson NJ, Hu P, Andersen GL, Shapiro L. 2007. Caulobacter crescentus as a whole-cell uranium biosensor. Appl Environ Microbiol 73:7615–7621. doi: 10.1128/AEM.01566-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yung MC, Jiao Y. 2014. Biomineralization of uranium by PhoY phosphatase activity aids cell survival in Caulobacter crescentus. Appl Environ Microbiol 80:4795–4804. doi: 10.1128/AEM.01050-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liang X, Hillier S, Pendlowski H, Gray N, Ceci A, Gadd GM. 2015. Uranium phosphate biomineralization by fungi. Environ Microbiol 17:2064–2075. doi: 10.1111/1462-2920.12771. [DOI] [PubMed] [Google Scholar]
- 6.Marshall MJ, Beliaev AS, Dohnalkova AC, Kennedy DW, Shi L, Wang Z, Boyanov MI, Lai B, Kemner KM, McLean JS, Reed SB, Culley DE, Bailey VL, Simonson CJ, Saffarini DA, Romine MF, Zachara JM, Fredrickson JK. 2006. c-Type cytochrome-dependent formation of U(IV) nanoparticles by Shewanella oneidensis. PLoS Biol 4:e268. doi: 10.1371/journal.pbio.0040268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cologgi DL, Lampa-Pastirk S, Speers AM, Kelly SD, Reguera G. 2011. Extracellular reduction of uranium via Geobacter conductive pili as a protective cellular mechanism. Proc Natl Acad Sci U S A 108:15248–15252. doi: 10.1073/pnas.1108616108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vecchia ED, Veeramani H, Suvorova EI, Wigginton NS, Bargar JR, Bernier-Latmani R. 2010. U(VI) reduction by spores of Clostridium acetobutylicum. Res Microbiol 161:765–771. doi: 10.1016/j.resmic.2010.08.001. [DOI] [PubMed] [Google Scholar]
- 9.Llorens I, Untereiner G, Jaillard D, Gouget B, Chapon V, Carriere M. 2012. Uranium interaction with two multi-resistant environmental bacteria: Cupriavidus metallidurans CH34 and Rhodopseudomonas palustris. PLoS One 7:e51783. doi: 10.1371/journal.pone.0051783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shelobolina ES, Coppi MV, Korenevsky AA, DiDonato LN, Sullivan SA, Konishi H, Xu H, Leang C, Butler JE, Kim BC, Lovley DR. 2007. Importance of c-type cytochromes for U(VI) reduction by Geobacter sulfurreducens. BMC Microbiol 7:16. doi: 10.1186/1471-2180-7-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Merroun ML, Raff J, Rossberg A, Hennig C, Reich T, Selenska-Pobell S. 2005. Complexation of uranium by cells and S-layer sheets of Bacillus sphaericus JG-A12. Appl Environ Microbiol 71:5532–5543. doi: 10.1128/AEM.71.9.5532-5543.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Merroun ML, Nedelkova M, Ojeda JJ, Reitz T, Fernandez ML, Arias JM, Romero-Gonzalez M, Selenska-Pobell S. 2011. Bio-precipitation of uranium by two bacterial isolates recovered from extreme environments as estimated by potentiometric titration, TEM and X-ray absorption spectroscopic analyses. J Hazard Mater 197:1–10. doi: 10.1016/j.jhazmat.2011.09.049. [DOI] [PubMed] [Google Scholar]
- 13.Reitz T, Rossberg A, Barkleit A, Selenska-Pobell S, Merroun ML. 2014. Decrease of U(VI) immobilization capability of the facultative anaerobic strain Paenibacillus sp. JG-TB8 under anoxic conditions due to strongly reduced phosphatase activity. PLoS One 9:e102447. doi: 10.1371/journal.pone.0102447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nies DH. 1999. Microbial heavy-metal resistance. Appl Microbiol Biotechnol 51:730–750. doi: 10.1007/s002530051457. [DOI] [PubMed] [Google Scholar]
- 15.Junier P, Dalla Vecchia E, Bernier-Latmani R. 2011. The response of Desulfotomaculum reducens MI-1 to U(VI) exposure: a transcriptomic study. Geomicrobiol J 28:483–496. doi: 10.1080/01490451.2010.512031. [DOI] [Google Scholar]
- 16.Khemiri A, Carriere M, Bremond N, Ben Mlouka MA, Coquet L, Llorens I, Chapon V, Jouenne T, Cosette P, Berthomieu C. 2014. Escherichia coli response to uranyl exposure at low pH and associated protein regulations. PLoS One 9:e89863. doi: 10.1371/journal.pone.0089863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vanhoudt N, Vandenhove H, Smeets K, Remans T, Van Hees M, Wannijn J, Vangronsveld J, Cuypers A. 2008. Effects of uranium and phosphate concentrations on oxidative stress related responses induced in Arabidopsis thaliana. Plant Physiol Biochem 46:987–996. doi: 10.1016/j.plaphy.2008.06.003. [DOI] [PubMed] [Google Scholar]
- 18.Bencheikh-Latmani R, Williams SM, Haucke L, Criddle CS, Wu L, Zhou J, Tebo BM. 2005. Global transcriptional profiling of Shewanella oneidensis MR-1 during Cr(VI) and U(VI) reduction. Appl Environ Microbiol 71:7453–7460. doi: 10.1128/AEM.71.11.7453-7460.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li X, Zhang H, Ma Y, Liu P, Krumholz LR. 2014. Genes required for alleviation of uranium toxicity in sulfate reducing bacterium Desulfovibrio alaskensis G20. Ecotoxicology 23:726–733. doi: 10.1007/s10646-014-1201-2. [DOI] [PubMed] [Google Scholar]
- 20.Sakamoto F, Nankawa T, Ohnuki T, Fujii T, Iefuji H. 2012. Yeast genes involved in uranium tolerance and uranium accumulation: a functional screening using the nonessential gene deletion collection. Geomicrobiol J 29:470–476. doi: 10.1080/01490451.2011.581330. [DOI] [Google Scholar]
- 21.Mukherjee A, Wheaton GH, Blum PH, Kelly RM. 2012. Uranium extremophily is an adaptive, rather than intrinsic, feature for extremely thermoacidophilic Metallosphaera species. Proc Natl Acad Sci U S A 109:16702–16707. doi: 10.1073/pnas.1210904109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Choudhary S, Islam E, Kazy SK, Sar P. 2012. Uranium and other heavy metal resistance and accumulation in bacteria isolated from uranium mine wastes. J Environ Sci Health A Tox Hazard Subst Environ Eng 47:622–637. doi: 10.1080/10934529.2012.650584. [DOI] [PubMed] [Google Scholar]
- 23.Hu P, Brodie EL, Suzuki Y, McAdams HH, Andersen GL. 2005. Whole-genome transcriptional analysis of heavy metal stresses in Caulobacter crescentus. J Bacteriol 187:8437–8449. doi: 10.1128/JB.187.24.8437-8449.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yung MC, Ma J, Salemi MR, Phinney BS, Bowman GR, Jiao Y. 2014. Shotgun proteomic analysis unveils survival and detoxification strategies by Caulobacter crescentus during exposure to uranium, chromium, and cadmium. J Proteome Res 13:1833–1847. doi: 10.1021/pr400880s. [DOI] [PubMed] [Google Scholar]
- 25.Park DM, Jiao Y. 2014. Modulation of medium pH by Caulobacter crescentus facilitates recovery from uranium-induced growth arrest. Appl Environ Microbiol 80:5680–5688. doi: 10.1128/AEM.01294-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Opijnen T, Bodi KL, Camilli A. 2009. Tn-seq: high-throughput parallel sequencing for fitness and genetic interaction studies in microorganisms. Nat Methods 6:767–772. doi: 10.1038/nmeth.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Deutschbauer A, Price MN, Wetmore KM, Shao W, Baumohl JK, Xu Z, Nguyen M, Tamse R, Davis RW, Arkin AP. 2011. Evidence-based annotation of gene function in Shewanella oneidensis MR-1 using genome-wide fitness profiling across 121 conditions. PLoS Genet 7:e1002385. doi: 10.1371/journal.pgen.1002385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ely B. 1991. Genetics of Caulobacter crescentus. Methods Enzymol 204:372–384. doi: 10.1016/0076-6879(91)04019-K. [DOI] [PubMed] [Google Scholar]
- 29.Christen B, Abeliuk E, Collier JM, Kalogeraki VS, Passarelli B, Coller JA, Fero MJ, McAdams HH, Shapiro L. 2011. The essential genome of a bacterium. Mol Syst Biol 7:528. doi: 10.1038/msb.2011.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Green MR, Sambrook J. 2012. Molecular cloning: a laboratory manual, 4th ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 31.Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17:10–12. [Google Scholar]
- 32.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zomer A, Burghout P, Bootsma HJ, Hermans PW, van Hijum SA. 2012. ESSENTIALS: software for rapid analysis of high-throughput transposon insertion sequencing data. PLoS One 7:e43012. doi: 10.1371/journal.pone.0043012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Braz VS, Marques MV. 2005. Genes involved in cadmium resistance in Caulobacter crescentus. FEMS Microbiol Lett 251:289–295. doi: 10.1016/j.femsle.2005.08.013. [DOI] [PubMed] [Google Scholar]
- 36.Ried JL, Collmer A. 1987. An nptI-sacB-sacR cartridge for constructing directed, unmarked mutations in gram-negative bacteria by marker exchange-eviction mutagenesis. Gene 57:239–246. doi: 10.1016/0378-1119(87)90127-2. [DOI] [PubMed] [Google Scholar]
- 37.Stephens C, Reisenauer A, Wright R, Shapiro L. 1996. A cell cycle-regulated bacterial DNA methyltransferase is essential for viability. Proc Natl Acad Sci U S A 93:1210–1214. doi: 10.1073/pnas.93.3.1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Skerker JM, Prasol MS, Perchuk BS, Biondi EG, Laub MT. 2005. Two-component signal transduction pathways regulating growth and cell cycle progression in a bacterium: a system-level analysis. PLoS Biol 3:e334. doi: 10.1371/journal.pbio.0030334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pena J, Plante JA, Carillo AC, Roberts KK, Smith JK, Juelich TL, Beasley DW, Freiberg AN, Labute MX, Naraghi-Arani P. 2014. Multiplexed digital mRNA profiling of the inflammatory response in the West Nile Swiss Webster mouse model. PLoS Negl Trop Dis 8:e3216. doi: 10.1371/journal.pntd.0003216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fritz JS, Bradford EC. 1958. Detection of thorium and uranium. Anal Chem 30:1021–1022. doi: 10.1021/ac60138a002. [DOI] [Google Scholar]
- 41.Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH image to ImageJ: 25 years of image analysis. Nat Methods 9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gallagher LA, Shendure J, Manoil C. 2011. Genome-scale identification of resistance functions in Pseudomonas aeruginosa using Tn-seq. mBio 2(1):e00315-10. doi: 10.1128/mBio.00315-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Palace SG, Proulx MK, Lu S, Baker RE, Goguen JD. 2014. Genome-wide mutant fitness profiling identifies nutritional requirements for optimal growth of Yersinia pestis in deep tissue. mBio 5(4):e01385-14. doi: 10.1128/mBio.01385-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Johnson CM, Grossman AD. 2014. Identification of host genes that affect acquisition of an integrative and conjugative element in Bacillus subtilis. Mol Microbiol 93:1284–1301. doi: 10.1111/mmi.12736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Toporowski MC, Nomellini JF, Awram P, Smit J. 2004. Two outer membrane proteins are required for maximal type I secretion of the Caulobacter crescentus S-layer protein. J Bacteriol 186:8000–8009. doi: 10.1128/JB.186.23.8000-8009.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Awram P, Smit J. 1998. The Caulobacter crescentus paracrystalline S-layer protein is secreted by an ABC transporter (type I) secretion apparatus. J Bacteriol 180:3062–3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Boratyn GM, Camacho C, Cooper PS, Coulouris G, Fong A, Ma N, Madden TL, Matten WT, McGinnis SD, Merezhuk Y, Raytselis Y, Sayers EW, Tao T, Ye J, Zaretskaya I. 2013. BLAST: a more efficient report with usability improvements. Nucleic Acids Res 41:W29–W33. doi: 10.1093/nar/gkt282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li XZ, Plesiat P, Nikaido H. 2015. The challenge of efflux-mediated antibiotic resistance in gram-negative bacteria. Clin Microbiol Rev 28:337–418. doi: 10.1128/CMR.00117-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dhamdhere G, Zgurskaya HI. 2010. Metabolic shutdown in Escherichia coli cells lacking the outer membrane channel TolC. Mol Microbiol 77:743–754. doi: 10.1111/j.1365-2958.2010.07245.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Barb AW, Zhou P. 2008. Mechanism and inhibition of LpxC: an essential zinc-dependent deacetylase of bacterial lipid A synthesis. Curr Pharm Biotechnol 9:9–15. doi: 10.2174/138920108783497668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Braz VS, da Silva Neto JF, Italiani VC, Marques MV. 2010. CztR, a LysR-type transcriptional regulator involved in zinc homeostasis and oxidative stress defense in Caulobacter crescentus. J Bacteriol 192:5480–5488. doi: 10.1128/JB.00496-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Braeken K, Moris M, Daniels R, Vanderleyden J, Michiels J. 2006. New horizons for (p)ppGpp in bacterial and plant physiology. Trends Microbiol 14:45–54. doi: 10.1016/j.tim.2005.11.006. [DOI] [PubMed] [Google Scholar]
- 53.Boutte CC, Crosson S. 2011. The complex logic of stringent response regulation in Caulobacter crescentus: starvation signalling in an oligotrophic environment. Mol Microbiol 80:695–714. doi: 10.1111/j.1365-2958.2011.07602.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lesley JA, Shapiro L. 2008. SpoT regulates DnaA stability and initiation of DNA replication in carbon-starved Caulobacter crescentus. J Bacteriol 190:6867–6880. doi: 10.1128/JB.00700-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.van Opijnen T, Camilli A. 2012. A fine scale phenotype-genotype virulence map of a bacterial pathogen. Genome Res 22:2541–2551. doi: 10.1101/gr.137430.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nies DH. 2003. Efflux-mediated heavy metal resistance in prokaryotes. FEMS Microbiol Rev 27:313–339. doi: 10.1016/S0168-6445(03)00048-2. [DOI] [PubMed] [Google Scholar]
- 57.Valencia EY, Braz VS, Guzzo C, Marques MV. 2013. Two RND proteins involved in heavy metal efflux in Caulobacter crescentus belong to separate clusters within proteobacteria. BMC Microbiol 13:79. doi: 10.1186/1471-2180-13-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Koronakis V, Eswaran J, Hughes C. 2004. Structure and function of TolC: the bacterial exit duct for proteins and drugs. Annu Rev Biochem 73:467–489. doi: 10.1146/annurev.biochem.73.011303.074104. [DOI] [PubMed] [Google Scholar]
- 59.Gandham L, Nomellini JF, Smit J. 2012. Evaluating secretion and surface attachment of SapA, an S-layer-associated metalloprotease of Caulobacter crescentus. Arch Microbiol 194:865–877. doi: 10.1007/s00203-012-0819-9. [DOI] [PubMed] [Google Scholar]
- 60.Nierman WC, Feldblyum TV, Laub MT, Paulsen IT, Nelson KE, Eisen JA, Heidelberg JF, Alley MR, Ohta N, Maddock JR, Potocka I, Nelson WC, Newton A, Stephens C, Phadke ND, Ely B, DeBoy RT, Dodson RJ, Durkin AS, Gwinn ML, Haft DH, Kolonay JF, Smit J, Craven MB, Khouri H, Shetty J, Berry K, Utterback T, Tran K, Wolf A, Vamathevan J, Ermolaeva M, White O, Salzberg SL, Venter JC, Shapiro L, Fraser CM. 2001. Complete genome sequence of Caulobacter crescentus. Proc Natl Acad Sci U S A 98:4136–4141. doi: 10.1073/pnas.061029298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pages JM, James CE, Winterhalter M. 2008. The porin and the permeating antibiotic: a selective diffusion barrier in Gram-negative bacteria. Nat Rev Microbiol 6:893–903. doi: 10.1038/nrmicro1994. [DOI] [PubMed] [Google Scholar]
- 62.Corbalán NS, Adler C, de Cristóbal RE, Pomares MF, Delgado MA, Vincent PA. 2010. The tolC locus affects the expression of sbmA through sigma E activity increase. FEMS Microbiol Lett 311:185–192. doi: 10.1111/j.1574-6968.2010.02090.x. [DOI] [PubMed] [Google Scholar]
- 63.Rosner JL, Martin RG. 2013. Reduction of cellular stress by TolC-dependent efflux pumps in Escherichia coli indicated by BaeSR and CpxARP activation of spy in efflux mutants. J Bacteriol 195:1042–1050. doi: 10.1128/JB.01996-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Humnabadkar V, Prabhakar KR, Narayan A, Sharma S, Guptha S, Manjrekar P, Chinnapattu M, Ramachandran V, Hameed SP, Ravishankar S, Chatterji M. 2014. UDP-N-acetylmuramic acid l-alanine ligase (MurC) inhibition in a tolC mutant Escherichia coli strain leads to cell death. Antimicrob Agents Chemother 58:6165–6171. doi: 10.1128/AAC.02890-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fralick JA, Burns-Keliher LL. 1994. Additive effect of tolC and rfa mutations on the hydrophobic barrier of the outer membrane of Escherichia coli K-12. J Bacteriol 176:6404–6406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vega DE, Young KD. 2014. Accumulation of periplasmic enterobactin impairs the growth and morphology of Escherichia coli tolC mutants. Mol Microbiol 91:508–521. doi: 10.1111/mmi.12473. [DOI] [PubMed] [Google Scholar]
- 67.Homann OR, Johnson AD. 2010. MochiView: versatile software for genome browsing and DNA motif analysis. BMC Biol 8:49. doi: 10.1186/1741-7007-8-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.