ABSTRACT
Escherichia coli microcin C (McC) consists of a ribosomally synthesized heptapeptide attached to a modified adenosine. McC is actively taken up by sensitive Escherichia coli strains through the YejABEF transporter. Inside the cell, McC is processed by aminopeptidases, which release nonhydrolyzable aminoacyl adenylate, an inhibitor of aspartyl-tRNA synthetase. McC is synthesized by the MccB enzyme, which terminally adenylates the MccA heptapeptide precursor MRTGNAN. Earlier, McC analogs with shortened peptide lengths were prepared by total chemical synthesis and were shown to have strongly reduced biological activity due to decreased uptake. Variants with longer peptides were difficult to synthesize, however. Here, we used recombinant MccB to prepare and characterize McC-like molecules with altered peptide moieties, including extended peptide lengths. We find that N-terminal extensions of E. coli MccA heptapeptide do not affect MccB-catalyzed adenylation and that some extended-peptide-length McC analogs show improved biological activity. When the peptide length reaches 20 amino acids, both YejABEF and SbmA can perform facilitated transport of toxic peptide adenylates inside the cell. A C-terminal fusion of the carrier maltose-binding protein (MBP) with the MccA peptide is also recognized by MccB in vivo and in vitro, allowing highly specific adenylation and/or radioactive labeling of cellular proteins.
IMPORTANCE Enzymatic adenylation of chemically synthesized peptides allowed us to generate biologically active derivatives of the peptide-nucleotide antibiotic microcin C with improved bioactivity and altered entry routes into target cells, opening the way for development of various McC-based antibacterial compounds not found in nature.
INTRODUCTION
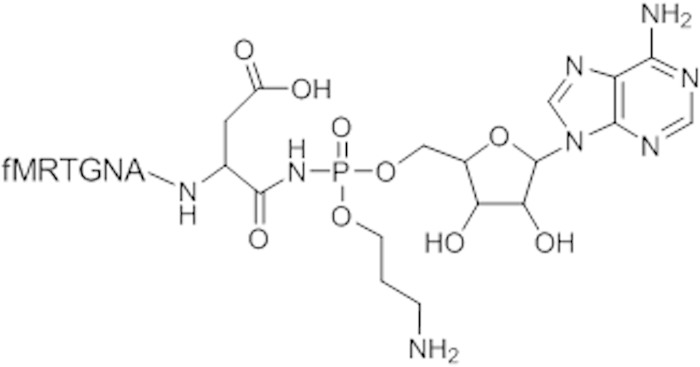
The antibiotic microcin C (McC) (Fig. 1) is produced by Escherichia coli strains harboring a plasmid-borne mcc operon. McC consists of a ribosomally synthesized heptapeptide that is covalently linked through a phosphoramidate bond to adenosine; the phosphoramidate linker is esterified with an aminopropyl moiety (1). Inside a sensitive cell, the N-terminal formyl group of the McC peptide is removed by peptide deformylase, after which the peptide part is processed by aminopeptidases (2). As a result, processed McC, a modified nonhydrolyzable aspartyl-adenylate, is released. Processed McC is a potent inhibitor of aspartyl-tRNA synthetase (AspRS), an essential enzyme (3).
FIG 1.
Structure of microcin C. The chemical structure of the part of the molecule corresponding to toxic processed McC (modified aspartyl-adenylate) released inside the cell is shown. The transport part, i.e., the first 6 residues of the MccA peptide, are indicated in a single-letter amino acid code. The N-terminal methionine is formylated (f).
McC penetrates the inner membrane of E. coli cells through the YejABEF transporter (4). While intact McC inhibits the growth of sensitive E. coli cells at low micromolar concentrations (5, 6), processed McC does not affect cell growth, even at millimolar concentrations (3, 7). Thus, the peptide chain enables processed McC function through a Trojan horse mechanism by promoting its active uptake via YejABEF. YejABEF is uniquely responsible for McC transport, since yej mutants are fully resistant to McC (4). The biological function of YejABEF (other than McC transport) is presently unknown. In Salmonella, it confers resistance to some antimicrobial peptides and allows proliferation inside activated macrophages, thus contributing to virulence (8). The latter property may be related to the fact that YejABEF interferes with peptide presentation on major histocompatibility complex (MHC) class I molecules (9).
The peptide part of McC is encoded by the mccA gene. At seven codons, this gene is considered to be the shortest natural gene known (10). The MccA peptide sequence is MRTGNAN. During McC maturation, MccA is C-terminally adenylated by the MccB synthetase (11). Bioinformatics searches identified MccB homologs in diverse bacteria, and in some cases, short nearby genes that could code for substrate peptides were also predicted (12, 13). Apart from the C-terminal asparagine residue, most predicted MccA-like peptides have no sequence similarity to each other or to E. coli MccA. However, many predicted MccA-like peptides (from Helicobacter pylori, Bartonella, Lactobacillus, and Streptococcus) have the same length as the E. coli peptide. On the other hand, a 56-amino-acid-long cyanobacterial MccA peptide and a 42-amino-acid-long peptide from Yersinia pseudotuberculosis are subject to terminal adenylation by their cognate MccB enzymes (13), suggesting that the length of MccB target peptides can exceed the seven amino acids characteristic of most mcc-like operons.
In vitro adenylation of MRTGNAN peptide by recombinant E. coli MccB is very efficient (11). The reaction proceeds in two steps. First, one ATP molecule is consumed to convert the terminal asparagine into succinimide. This activated intermediate is coupled with the second ATP molecule, resulting in a biologically active peptide adenylate with terminal aspartate (11). In the presence of the MccD and MccE enzyme pair, an aminopropyl group is attached to the product of MccB-catalyzed adenylation using S-adenosylmethionine (SAM) as a donor (14). The presence of aminopropyl increases the biological activity severalfold, probably by increasing the avidity of modified nonhydrolyzable aspartate adenylate to the target enzyme (6).
Structure-activity analysis of the McC peptide was carried out by introduction of point substitutions in the mccA gene (15). Using this approach, codons 2 to 7 of mccA were each systematically substituted for codons coding for 19 remaining standard amino acids, and the effects of these substitutions on the ability of cells harboring mutated mcc operons to produce mutant microcins were determined. The role of the first methionine of the MccA peptide could not be studied using this approach, as this residue is required for translation initiation. The analysis of the seventh codon of mccA was also not informative, since only peptides with a terminal asparagine can be adenylated by the MccB enzyme (11). Despite these limitations, a series of McC derivatives, including some with increased bioactivity, was obtained using this approach, indicating that E. coli MccB is not strictly specific for its peptide substrates.
Total chemical synthesis of McC analogs yielded several active McC variants with nonnatural residues at the seventh position of the peptide, targeting aminoacyl-tRNA synthetases other than the AspRS targeted by natural McC (16). The chemical approach was also used to investigate peptide length requirements for facilitated E. coli McC transport. The results indicated that shortening of McC peptide by even one amino acid strongly decreased the biological activity by affecting YejABEF-facilitated transport (7). Further decrease of the peptide length abolished facilitated transport, resulting in bioactivity levels comparable to that of processed McC. McC variants with longer wild-type MccA-based peptides proved to be impossible to obtain due to synthesis complications. Yet, a small amount of adenylated MTRGNAAG peptide, with an additional alanine (highlighted in bold) inserted after position 6, was prepared. This compound targeted GlyRS and was slightly more active than control chemically synthesized heptapeptide MRTGNAG adenylate (7). Overall, these results provided a lower bound for the peptide moiety of YejABEF substrates, which appears to coincide with the wild-type McC peptide length, and suggested that increasing the length of the transport peptide may lead to more potent McC-based antibacterials.
In this study, we used enzymatic synthesis of peptide adenylates by E. coli MccB to study McC structure-function. This approach is free from the limitations of both the molecular genetics and chemical synthesis approaches and allowed us to prepare and characterize McC derivatives with substitutions of the N-terminal methionine and with extended peptide lengths. We show that the N-terminal amino acid of MccA plays a critical role in the binding to the MccB enzyme. We confirm that extension of McC peptide length up to a certain point increases bioactivity. The biological activity of longer peptide adenylates can be further increased by aminopropylation. We finally show that MccA fusions to full-size proteins such as 43-kDa maltose-binding protein (MBP) are also subject to adenylation by MccB in vivo and in vitro, allowing efficient labeling of fusion proteins.
MATERIALS AND METHODS
DNA and molecular cloning.
The E. coli mccB gene was cloned between the NcoI and BamHI sites of the pET32b (pETMccB) vector or between the NcoI and SalI sites of MCS1 of vector pCOLADuet-1. The E. coli MBP gene fused with C-terminal sequence encoding GGGGMRTGNAN (E. coli MccA with N-terminal tetraglycine linker) was next cloned between the NdeI and XhoI sites of the MCS2 pCOLADuet-1 vector containing mccB (yielding plasmid pColMccB-MBP-MccA). A sequence encoding GGGGMRTGNAN was also cloned between the EcoRI and HindIII sites of the pMAL-c2X vector to generate pMAL-MBP-MccA.
Strains and strain construction.
E. coli DH5α was used for molecular cloning; for susceptibility tests, we used E. coli BL21(DE3) and its ΔyejB, ΔsbmA, and ΔyejB ΔsbmA derivatives. The ΔyejB and ΔsbmA derivatives are described in reference 13. The ΔyejB ΔsbmA double mutant was constructed from ΔsbmA and ΔyejB single mutants (the latter marked by chloramphenicol resistance) using P1 transduction (17). E. coli 0256 and 0193 are clinical isolates obtained from St. Petersburg Research Institute of Children's Infections of the Federal Medical and Biological Agency of Russia.
Protein expression and purification.
E. coli BL21(DE3) cells were used as the expression host. The BL21(DE3) cells transformed with appropriate expression plasmids were grown at 37°C in 200 ml LB medium supplemented with 1% glucose and necessary antibiotics until the optical density at 600 nm (OD600) reached 0.6. Cells were harvested by centrifugation, washed thrice with fresh LB medium, and resuspended in 200 ml of fresh LB with antibiotics and 0.1 to 0.3 mM isopropyl β-d-1-thiogalactopyranoside (IPTG). Cells harboring MccB protein plasmid were grown for 20 h at 18°C with vigorous agitation; cells harboring MBP plasmid were grown for 4 h at 30°C. Cells were harvested and resuspended in 8 ml of appropriate loading buffer (MccB loading buffer, consisting of 20 mM Tris-HCl [pH 8.0], 500 mM NaCl, and 10 mM MgCl2, or MBP loading buffer, consisting of 20 mM Tris-HCl [pH 7.4], 200 mM NaCl, 1 mM EDTA, 1 mM sodium azide, and 10 mM β-mercaptoethanol) and disrupted by sonication. The lysates were centrifuged at 30,000 × g for 30 min at 4°C. Supernatants were mixed with 200 to 300 μl of appropriate resin (for MccB, His Bind resin [Novagen]; for MBP, amylose resin [NEB]) equilibrated in the same buffer, and proteins were allowed to bind for 2 to 4 h at 4°C with gentle agitation. The resin was allowed to settle by gravity and washed with 15 ml of MBP loading buffer with (MccB) or without (MBP) 50 mM imidazole, and bound proteins were eluted with 0.5 ml of elution buffer (MccB elution buffer, consisting of 20 mM Tris-HCl [pH 8.0], 500 mM NaCl, 10 mM MgCl2, and 200 mM imidazole, or MBP elution buffer, consisting of 20 mM Tris-HCl [pH 7.4], 200 mM NaCl, 1 mM EDTA, 1 mM sodium azide, 10 mM β-mercaptoethanol, and 10 mM maltose). Five consecutive elutions were performed with each resin sample. Fractions were supplemented with glycerol up to 50% and stored at −20°C until further use. Proteins were at least 90% pure as judged by visual inspection of overloaded Coomassie blue-stained SDS gels.
His-tagged MccD, MccE, and Mtn proteins were prepared as described in reference 14.
Synthesis of peptide substrates.
All peptides were synthesized by solid-phase synthesis by Syneuro LLC, Russia (at least 98% purity by high-pressure liquid chromatography [HPLC] and mass spectrometry [MS]), or by GenScript USA Inc. (at least 85% purity).
In vitro enzymatic assays.
Standard peptide adenylation reactions were performed in a total volume of 100 μl. MccB buffer (1×; 75 mM Tris-HCl [pH 8.0], 5 mM MgCl2) was supplemented with 5 μM MccB, 2.5 mM tris(2-carboxyethyl)phosphine (TCEP), and 4 mM ATP. As the substrate, 4 μM MBP-MccA or 225 μM synthetic peptide was used. The control reaction did not contain ATP. Reaction mixtures were incubated at 23°C for 20 h and were terminated by freezing at −20°C.
To set up competition assays, standard adenylation reaction mixtures were preincubated with 7.5 mM competitor peptide for 40 min and then were supplemented with 150 μM MRTGNAN substrate peptide. Reactions were processed as described above, and reaction products were separated by HPLC.
Aminopropylation of 40 μM HPLC-purified adenylated peptides was carried out with 2.7 μM MccD, 0.5 μM MccE, and 0.8 μM Mtn in 20 mM Tris-HCl (pH 8.0) buffer supplemented with 50 mM NaCl, 500 μM SAM, and 20 μM pyridoxal phosphate (PLP).
In vitro radiolabeling.
Reactions were performed in the total volume of 50 μl containing 75 mM Tris-HCl (pH 8.0), 10 mM MgCl2 4 μM MccB, 4 mM dithiothreitol (DTT), 2.5 mM ATP, and 0.75 μM [α-32P]ATP (3,000 Ci mmol−1). The reactions were supplemented with either 4 μM MBP-MccA or 200 μM MRTGNAN peptide. Reaction mixtures were incubated at 23°C for 24 h. Ten-microliter reaction aliquots were deposited on an E. coli BL21(DE3) sensitive lawn to confirm peptide conversion into a toxic adenylated compound. The products were also resolved by 10% SDS-PAGE. The gel was stained with Coomassie brilliant blue R-250 and radioactive proteins revealed by autoradiography.
In vivo adenylation assays.
E. coli BL21(DE3) cells were transformed with pColMBP-MccA or pColMccB-MBP-MccA. Cells were grown at 37°C in 200 ml LB supplemented with 1% glucose and 50 μg ml−1 kanamycin until the OD600 reached 0.6. Cells were harvested by centrifugation, washed thrice with fresh LB medium, and resuspended in 200 ml of fresh LB with kanamycin and 0.3 mM IPTG. Cells were grown for 5 h at 37°C with vigorous agitation and harvested, and MBP-MccA was purified using amylose resin. MBP-MccA protein fusion was incubated with protease factor Xa in appropriate buffer for 3 h. Reaction mixtures were purified from cleaved MBP using amylose resin and analyzed by matrix-assisted laser desorption ionization (MALDI)-MS.
Identification of adenylation reaction products.
The products of the MccB reaction were identified by reverse-phase HPLC. Completed reaction mixtures were mixed with an equal volume of acetonitrile (MeCN) supplemented with 0.1% trifluoroacetic acid (TFA) and incubated for 20 min on ice. Precipitated protein was removed by centrifugation at 30,000 × g for 10 min at +4°C. Supernatants were collected, dried, resuspended in 100 to 200 μl of deionized water, and applied to a 4.6- by 150-mm XSelect HSS C18 3.5-μm column (Waters) equilibrated with buffer A (0.1% TFA). The column was developed at 1 ml min−1 using the following gradient: 0 to 10% buffer B (100% acetonitrile) for 5 min followed by 10 to 40% or 10 to 20% B in 40 min. Individual chromatographic peaks were analyzed by MALDI-MS.
MALDI-MS and MS/MS analysis.
Mass spectra were recorded on an Ultraflextreme MALDI–time of flight (MALDI-TOF)/TOF mass spectrometer (Bruker Daltonik) equipped with an Nd laser (355 nm). The MH+ molecular ions were measured in reflector mode; the accuracy of monoisotopic mass peak measurement was 50 ppm. One-microliter aliquots of desalted adenylation reaction mixtures were mixed on a steel target with 0.5 μl of 2,5-dihydroxybenzoic acid (Aldrich) solution (20 mg ml−1 in 30% MeCN–0.5% TFA).
In vivo sensitivity tests.
Wild-type or mutant (ΔyejB, ΔsbmA, or ΔyejB ΔsbmA) cell cultures were grown in 10 ml of M63 broth without Fe2+ salts and supplemented with yeast extract at 37°C to an OD600 of ∼0.8. One milliliter of cell culture was added to 20 ml of melted top agar (0.65 g liter−1 of agar in M63 broth) cooled to ∼50°C. The mixture was poured on the surfaces of LB agar plates. After the agar solidified, 10-μl drops of completed adenylation reaction mixtures (see above), aminopropylation reaction mixtures, or HPLC-purified adenylated peptides were placed on the plate surface and allowed to dry. Plates were incubated for 4 to 6 h at 37°C, and growth inhibition zones around the sites where samples were applied were visually detected.
RESULTS
Use of in vitro adenylation by MccB to prepare mutant McC variants.
Several derivatives of MccA heptapeptide MRTGNAN were obtained, and their ability to be adenylated by the recombinant E. coli MccB was studied in vitro using wild-type peptide as a control. Several peptides matched mccA mutants previously tested in vivo (15). These included derivatives with substitutions of the terminal residue (MRTGNAD and MRTGNAQ) and substitutions in the third (MRAGNAN, MRLGNAN, MRKGNAN, MRSGNAN, and MRCGNAN) and fourth (MRTWNAN) positions of the MccA peptide. For each reaction, the products were analyzed by MALDI-MS. As expected, the wild-type peptide was fully converted into the adenylated form (Fig. 2). Both peptides with substituted C-terminal residues remained intact; neither the attachment of AMP nor the succinimide intermediate of the adenylation reaction was detected (data not shown). The results agree with earlier data, since cells harboring genetic constructs expressing the corresponding mccA mutant genes did not produce adenylated peptides (15). The MRKGNAN peptide was not modified, in agreement with earlier in vivo data. The MRLGNAN and MRCGNAN peptides were adenylated, also in agreement with in vivo data (Fig. 2). MRAGNAN, which according to in vivo data was not modified, was poorly modified by the MccB enzyme in vitro, though small amounts of succinimide intermediate and trace amounts of adenylated product were detected (Fig. 2). MRSGNAN presents an interesting case, as the corresponding adenylate was not detected in vivo but the in vitro adenylation reaction proceeded to completion (Fig. 2). The MRTWNAN peptide with substitution at position 4 was modified in agreement with the published data.
FIG 2.
In vitro adenylation of E. coli MccA peptide mutants by MccB. Chemically synthesized peptides corresponding to 7-amino-acid-long wild-type MccA MRTGNAN, mutants bearing the indicated single-amino-acid substitutions (underlined), or an MRTGNAN peptide containing a d-stereoisomer of threonine at position 3 were combined with recombinant MccB in the absence (top) and the presence (bottom) of ATP. Reaction products were analyzed by MALDI-MS. The m/z values of mass peaks corresponding to starting peptides and adenylation products are indicated. Adenylation by MccB adds 329 Da to the substrate peptide. Asterisks indicate peaks corresponding to adenylation reaction intermediates containing a terminal succinimide.
We also tested a peptide containing the nonnatural d-enantiomer of threonine at position 3. The peptide was not modified by MccB (Fig. 2).
To determine if the products of MccB-catalyzed reactions are biologically active, aliquots of completed reaction mixtures were deposited on lawns of McC-sensitive E. coli cells, and formation of growth inhibition zones around the deposited drops was monitored after overnight growth (Table 1). As controls, biological activities of reaction aliquots were also tested on lawns of McC-resistant yejB mutant cells. For reaction mixtures containing wild-type MccA peptide, robust growth inhibition zones on wild-type but not mutant cell lawns were observed. No inhibition zones on either cell lawn were produced around deposited aliquots of reaction mixtures containing peptides with substituted terminal residues or with MRAGNAN or MRKGNAN, as expected, since these peptides were either not subject to adenylation or were very poorly modified (MRAGNAN). Reaction mixtures containing adenylated MRLGNAN, MRCGNAN, and MRTWNAN inhibited bacterial growth, in agreement with earlier in vivo data (15). None of the active variants inhibited the growth of yejB mutant cells. The adenylate of MRSGNAN that was not produced in vivo was also biologically active. Overall, the results of this analysis show that there is a good correspondence between results obtained by the in vivo and the enzymatic in vitro approaches. Yet, as suggested by the case of serine substitution at position 3, certain MccA peptide variants that are recognized and modified by MccB were missed during the in vivo screen, possibly because of modified stability of mutant peptides in the cell. The in vitro approach is free of such limitations and thus allows more detailed structure-function analysis of McC variants.
TABLE 1.
In vitro adenylation and in vivo activity of MccA heptapeptide point mutantsa
| Peptide sequence | Adenylation by MccB in vitro |
In vivo activity |
|
|---|---|---|---|
| Wild-type cells | ΔyejB cells | ||
| MRTGNAN | + | + | − |
| MRTGNAD | − | − | − |
| MRTGNAQ | − | − | − |
| MRAGNAN | −/+ | − | − |
| MRLGNAN | +/− | + | − |
| MRSGNAN | + | + | − |
| MRCGNAN | +/− | + | − |
| MRKGNAN | − | − | − |
| MR(d-Thr)GNAN | − | − | − |
| MRTWNAN | +/− | + | − |
| GRTGNAN | − | − | − |
| ARTGNAN | − | − | − |
| IRTGNAN | +/− | + | − |
| VRTGNAN | − | − | − |
| LRTGNAN | +/− | + | − |
| FRTGNAN | + | + | − |
| NRTGNAN | − | − | − |
| QRTGNAN | − | − | − |
| DRTGNAN | − | − | − |
| ERTGNAN | − | − | − |
| KRTGNAN | − | − | − |
The table presents the results of in vitro adenylation by MccB of each peptide listed as detected by MALDI-MS analysis of reactions conducted under standard conditions (see, for example, Fig. 2 and Materials and Methods) and the results of bioactivity testing of 10-μl aliquots of products of completed adenylation reactions on lawns of wild-type or yejB mutant E. coli cell lawns. +, a clear growth inhibition zone was detected; −, no growth inhibition zone was observed; +/−, incomplete conversion.
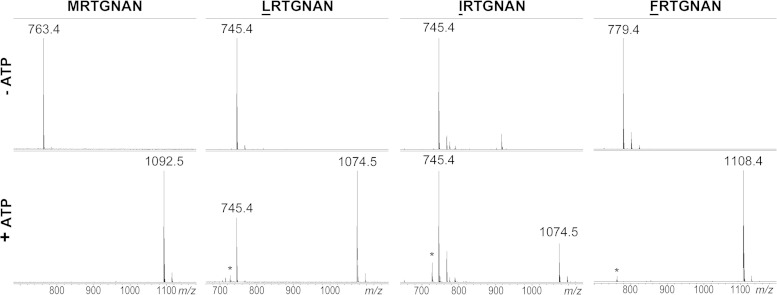
The N-terminal MccA residue is critical for the binding to MccB.
We next prepared several MccA peptide variants with substitutions of the N-terminal methionine. This residue is encoded by the starting codon of mccA, and so its significance could not have been studied before by using the molecular genetic approach. Structural analysis of the MccB-MccA complex suggests that the side chain of MccA Met1 contributes to peptide binding to the enzyme (18). A total of 11 variants were tested (GRTGNAN, ARTGNAN, IRTGNAN, VRTGNAN, LRTGNAN, FRTGNAN, NRTGNAN, QRTGNAN, DRTGNAN, ERTGNAN, and KRTGNAN), introducing side chains with different properties instead of Met1. Among the peptides tested, LRTGNAN and IRTGNAN were partially adenylated, while FRTGNAN was adenylated to completion under our conditions (Fig. 3). The rest of the mutant peptides were not modified by the enzyme. This included VRTGNAN, an unexpected result considering that LRTGNAN and IRTGNAN were good substrates for adenylation. Structural modeling using the existing MccB-MccA complex structure as a template (18) shows that the valine side chain is too small to make van der Waals contacts with the binding pocket in the protein (in contrast to the case for leucine and isoleucine side chains [data not shown]). All adenylated MccA peptides with a substituted first amino acid were active against the wild-type E. coli but not the yejB mutant cells (Table 1).
FIG 3.
In vitro adenylation of E. coli MccA peptide mutants with substitution of the N-terminal methionine residue. Reactions were set up and analyzed as described in the Fig. 2 legend. Results are shown only for peptides that were modified by MccB. Other tested peptides bearing N-terminal substitutions are listed in Table 1.
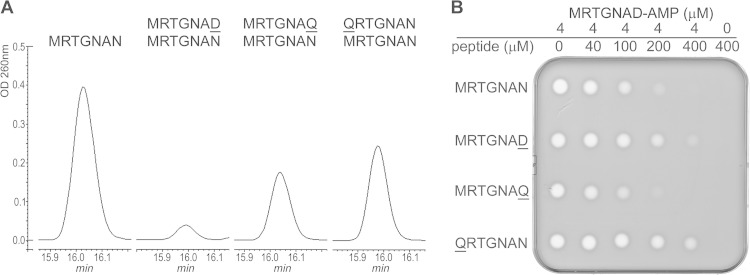
The results obtained with N-terminally substituted MccA variants point to the importance of the first residue of MccA for the binding to MccB and/or proper presentation of the C-terminal residue of the bound peptide in the enzyme catalytic center. The following experiment was performed to determine if a bulky N-terminal hydrophobic residue of MccA is required for MccB binding. Wild-type MccB adenylation reactions were conducted in the presence of a 50-fold excess of the QRTGNAN or MRTGNAQ peptide (Fig. 4), neither of which is subject to adenylation. As control, we used the MRGTNAD peptide, which is also not adenylated and was previously shown to inhibit adenylation of wild-type MccB (11). Reaction products were resolved by HPLC, and the size of an HPLC peak containing adenylated MccA was determined (Fig. 4A). As can be seen, MRGTNAD inhibited MccA adenylation, as expected. Both QRTGNAN and MRTGNAQ were much less effective inhibitors of the MccA adenylation reaction (Fig. 4A), suggesting that they are equally compromised for binding to MccB.
FIG 4.
Effect of MccA variants that are not subject to adenylation on wild-type MccA adenylation and on bioactivity of adenylated MccA. (A) The adenylation reaction of wild-type MccA was conducted in the presence of a 50-fold excess of QRTGNAN, MRTGNAQ, or MRTGNAD. Reaction products were separated by reverse-phase HPLC, and adenylation of wild-type MccA was determined by monitoring absorbance at 260 nm. The presence of adenylated MccA in the peak was confirmed by MALDI-MS. (B) The product of wild-type MccA adenylation was purified and combined with the indicated concentrations of MccA peptide variants, and 10-μl reaction aliquots were deposited onto cell lawns formed by E. coli cells. The results of overnight growth at 37°C are shown. The plate shown is representative of one of three independent experiments.
The experiment was next modified such that competing peptides were added to purified adenylated MccA just prior to biological activity tests. Previously, it was shown that MccA and longer peptides containing the MccA sequence at their C termini interfere with McC antibacterial activity by competing for entry through the YejABEF transporter (7). The MRTGNAQ peptide attenuated bioactivity of adenylated MccA as well as control wild-type MccA MRTGNAN (Fig. 4B), indicating that both peptides were recognized by YejABEF. In contrast, the addition of an excess of QRTGNAN had little or no effect on the biological activity of adenylated MccA. This observation confirms the important role of Met1 not just in the binding to the MccB enzyme but also in recognition by the YejABEF transporter. However, the MRTGNAD peptide, whose sequence is identical to that of the peptide part of McC, was also a poor competitor, indicating that the YejABEF specificity determinants are not limited by the nature of the N-terminal amino acid.
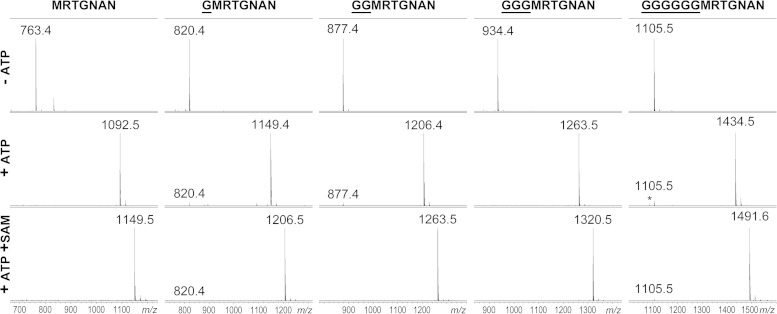
Enzymatic synthesis and bioactivity of McC variants with longer peptide parts.
We next wondered if enzymatic synthesis can be used to obtain McC-like compounds with extended peptide lengths. We tested a series of wild-type MccA-based peptides extended by one, two, three, or six glycine residues at the N terminus. Each peptide was readily adenylated by MccB (Fig. 5). The oligo-glycine tail by itself did not appear to increase the binding properties of MccA substrates, since a G6RTGNAN peptide lacking a residue corresponding to MccA Met1 was not adenylated (data not shown), further underscoring the significance of the methionine residue for binding to MccB.
FIG 5.
In vitro adenylation of extended-length MccA peptide variants. Chemically synthesized peptides corresponding to wild-type MccA MRTGNAN and the indicated N-terminally extended variants were combined with recombinant MccB in the absence (top) and the presence (middle) of ATP. Reaction mixtures shown at the bottom were incubated in the presence of ATP, recombinant MccD, MccE, and Mtn enzymes, SAM substrate, and PLP cofactor under conditions promoting aminopropylation of peptidyl adenylates. The addition of aminopropyl adds 57 Da to adenylated peptides.
Reaction mixtures containing elongated MccA adenylates inhibited the growth of wild-type but not of yejB mutant E. coli (data not shown). To compare the levels of antimicrobial activities of longer-peptide McC-like compounds, each adenylate was purified, concentrations of each compound were matched, and their activities were semiquantitatively determined by depositing drops containing serial dilutions of each compound on lawns of wild-type laboratory E. coli tester cells and two different E. coli clinical isolates. The results are shown in Table 2. One clinical strain, E. coli 0256, demonstrated low sensitivity to all compounds tested. In contrast, the other two strains were sensitive. Adenylates of extended MccA peptides were more active than wild-type MccA adenylate on the laboratory strain BL21(DE3). The activities of adenylated GMRTGNAN, G2MRTGNAN, and G3MRTGNAN were increased 4-fold, while G6MRTGNAN was twice as active as MccA adenylate. The clinical isolate E. coli 0193 showed similar trends. Thus, increasing the length of the peptide moiety beyond the natural seven amino acids improves the bioactivity of McC-like compounds.
TABLE 2.
Bioactivity of elongated MccA peptide adenylates
| Compound | MIC (μM) for E. coli straina: |
||
|---|---|---|---|
| BL21(DE3) | 0256 | 0193 | |
| MRTGNAD-AMP | 10 | >60 | 10 |
| MRTGNAD-AMP(ap)b | 2.5 | >60 | 1.25 |
| GMRTGNAD-AMP | 2.5 | >60 | 5 |
| GMRTGNAD-AMP(ap) | 0.3 | >60 | 0.3 |
| GGMRTGNAD-AMP | 2.5 | >60 | 5 |
| GGMRTGNAD-AMP(ap) | 0.3 | 60 | 0.3 |
| GGGMRTGNAD-AMP | 2.5 | >60 | 5 |
| GGGMRTGNAD-AMP(ap) | 0.3 | 60 | 0.3 |
| GGGGGGMRTGNAD-AMP | 5 | >60 | 5 |
| GGGGGGMRTGNAD-AMP(ap) | 1.3 | 60 | 0.6 |
Bioactivity was measured with serial 2-fold dilutions of each compound in the diffusion test on a lawn of the indicated E. coli cells. MIC values presented are the minimal concentration at which the inhibition zone was visible around a 10-μl aliquot of compound.
ap, aminopropyl.
The activity of the product of MccB-catalyzed adenylation of natural MccA is increased 4-fold by aminopropyl decoration on the phosphate (14). Using the general conditions described in reference 14, we prepared aminopropylated variants of MccB adenylation products of peptides of various lengths (Fig. 5) and determined their bioactivities. The results are presented in Table 2. As can be seen, aminopropylated derivatives of longer peptide adenylates were more active than corresponding compounds without this decoration. For E. coli 0256, which was practically resistant to compounds without aminopropyl, growth inhibition zones around modified 9-, 10-, and 13-amino-acid-long peptides were observed. For the more susceptible BL21(DE3) and 0193 strains, an 8- to 16-fold stimulatory effect of aminopropyl was observed. We conclude that increased peptide length and aminopropylation have an additive effect and together can increase the bioactivity of adenylated MccA heptapeptide by as much as 30-fold, changing the MIC from 10 to 0.3 μM.
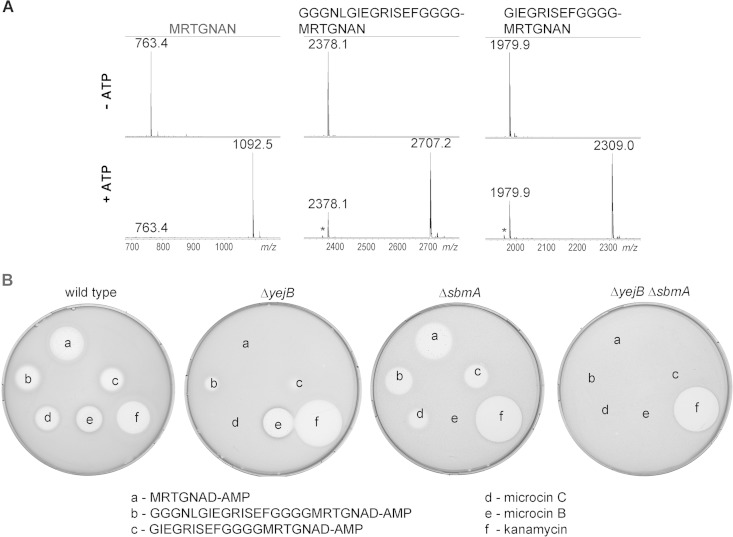
It could be argued that extending the MccA peptide with homogeneous contiguous glycines represents a special case that does not report on the MccB ability to recognize substrates with extra heterogeneous sequences containing bulky amino acids. We tested two MccA-based peptides extended to a total length of 20 and 25 amino acids with a randomly chosen sequence. Both peptides were readily adenylated by MccB, and the resulting peptide adenylates inhibited growth of wild-type E. coli (Fig. 6). Interestingly, when the activities of these peptide adenylates were tested on the McC-resistant yejB mutant strain, clear growth inhibition zones (which were nevertheless much smaller than those on wild-type cell lawns) were observed, suggesting that an additional transporter is involved in the uptake. When cells lacking SbmA, a transporter responsible for microcin B transport (19), were tested, growth inhibition zones around adenylated 20- and 25-amino-acid peptides were diminished marginally, while full resistance to McB was observed, as expected. However, when activity was tested on lawns of double mutant cells (ΔyejB ΔsbmA) lacking both transporters, no growth inhibition was detected with adenylated 20- and 25-amino-acid peptides, McC, or McB. The double mutant cells were as sensitive to kanamycin, which was used as control, as single mutants or wild-type cells (Fig. 6). The result thus suggests that McC analogs with longer peptide chains are transported inside the cells through a joint function of the YejABEF and SbmA inner membrane transporters.
FIG 6.
In vitro-adenylated extended-length MccA variants are active and enter cells through both YejABEF and SbmA. (A) Wild-type MccA or 20- and 25-amino-acid-long peptides containing C-terminal MccA sequences were adenylated by MccB in vitro and products analyzed by MALDI-MS. (B) Adenylation products, in 10-μl reaction aliquots, were deposited onto cell lawns formed by wild-type E. coli or the indicated mutants along with several control antibiotics. The results of overnight growth at 37°C are shown. The plate shown is representative of one of three independent experiments.
Use of MccA/MccB for terminal protein labeling.
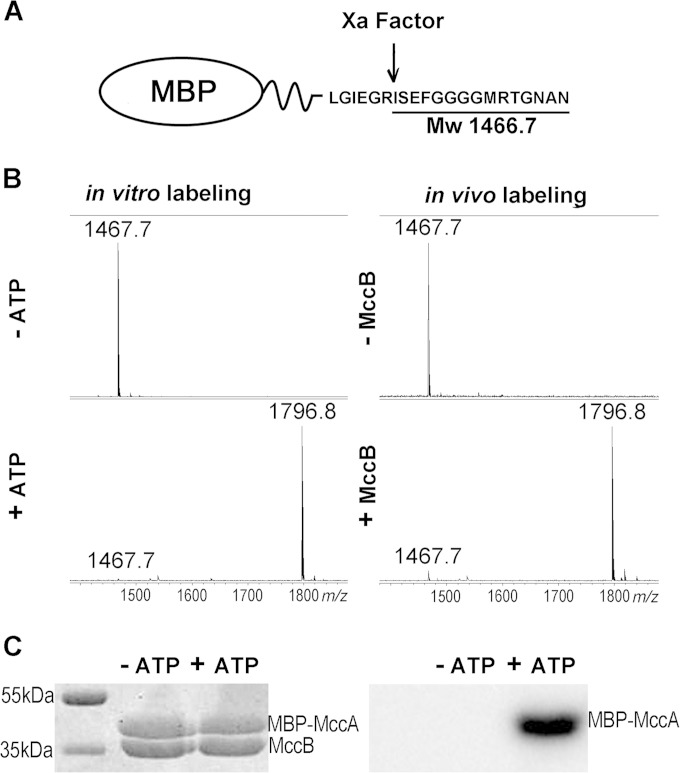
The results presented above clearly show that E. coli MccB can adenylate MccA-based peptides with substantial N-terminal extensions. This observation raised the question whether MccA can function as a C-terminal tag specifically recognized and adenylated by MccB. To answer this question, a plasmid expressing MBP C-terminally fused to MccA was created. A linker between MBP and MccA contained a site of recognition by factor Xa protease (Fig. 7A). Two types of experiments were performed. First the fusion protein was purified and combined with MccB in the presence or in the absence of ATP under conditions of MccA adenylation, and the products were treated with factor Xa, followed by mass spectrometric analysis. The results are shown in Fig. 7B (left). As can be seen, a mass peak with m/z 1,467.5 was seen in reactions without ATP. This mass peak corresponds to expected peptide ISEFGGGGMRTGNAN that should be generated upon factor Xa cleavage (Fig. 7A). In reaction mixtures containing ATP, this mass peak was not present. Instead, a mass peak with m/z 1,796.8, matching adenylated ISEFGGGGMRTGNAD, was observed. The in vitro adenylation experiment was repeated using [α-32P]ATP, and reaction products were separated by SDS-PAGE. The gel was stained and then subjected to autoradiography. As can be seen, in reaction mixtures containing the MBP-MccA fusion protein, MccB, and [α-32P]ATP, the MBP-MccA became radioactively labeled. No labeling was observed in lanes where one of the reaction components was missing (Fig. 7C).
FIG 7.
MccA can be used as a C-terminal tag for adenylation of proteins in vivo and in vitro. (A) The structure of the fusion protein containing MBP fused to MccA through a linker containing a factor Xa cleavage site is schematically shown. Factor Xa cleavage results in generation of an ISEFGGGGMRTGNAN peptide with an Mw of 1,466.7. (B) Left, the MBP-MccA fusion protein was purified and combined with MccB in the presence or in the absence of ATP under conditions favoring adenylation. Reaction mixtures were treated with factor Xa and subjected to MALDI-MS. Right, MBP-MccA protein was expressed in cells with or without MccB, purified, and subjected to factor Xa treatment, and products were analyzed by MALDI-MS. (C) Purified MBP-MccA was incubated with MccB in the presence or in the absence of [α-32P]ATP. Reaction products were resolved by SDS-PAGE and stained with Coomassie blue (left) (bands corresponding to MBP-MccA and MccB are indicated). The leftmost lane contains molecular mass markers. An autoradiograph of the gel is shown on the right.
To show that labeling of proteins tagged by the MccA peptide tag can also occur in vivo, E. coli cells coexpressing MBP-MccA and MccB were obtained. Upon induction, MccB- and MBP-MccA-cooverproducing cell cultures continued growth for several hours but stopped growing afterwards. No such effect was seen in cultures cooverexpressing MccB and MBP. The cessation of growth of cultures cooverproducing MccB and MBP-MccA could have been due to degradation of the adenylated MBP-MccA fusion, which eventually should lead to accumulation of processed McC. Indeed, when extracts of cells cooverproducing MccB and MBP-MccA were loaded on an amylose resin column followed by factor Xa treatment of affinity-purified material, a mass peak with m/z 1,796.8, matching the adenylated ISEFGGGGMRTGNAD terminal peptide of MBP-MccA, was detected (Fig. 7B, right). Only a mass peak with m/z 1,487.5, corresponding to unmodified ISEFGGGGMRTGNAN, was present in cells expressing MBP-MccA alone. We conclude that the MccA peptide can serve as a terminal tag, which is adenylated by MccB either in vivo or in vitro with high specificity and efficiency.
DISCUSSION
In this work, we extended the structure-activity analysis of microcin C by preparing a panel of modified McCs in an in vitro adenylation reaction catalyzed by the McC synthase MccB. Earlier, structure-activity relationships of McC were studied by introducing mutations in a cloned mccA gene or by chemical synthesis of McC analogs. Introducing substitutions in the mccA gene did not allow the role of MccA peptide Met1 to be assessed. The chemical approach has proven to be impossible or very complicated when attempts to synthesize McC-like compounds with longer peptide parts were made. Further, since a sulfonamide rather than a phosphoamide bond was used to connect adenosine to the peptide during chemical synthesis, decoration with aminopropyl or other modifications, which could increase bioactivity, became impossible. The enzymatic synthetic approach is free from these limitations. The MccB enzyme is highly active in vitro, and large amounts (tens of milligrams) of peptide adenylates can be routinely prepared using chemically synthesized peptides as adenylation substrates. Using this strategy, we show that the N-terminal amino acid of MccA plays a critical role in adenylation by the MccB enzyme. Only peptides with bulky hydrophobic residues (which includes the natural methionine) are accepted by the enzyme.
McC variants with substituted N-terminal methionine residues are taken up through the YejABEF inner membrane transporter. While the physiological function of YejABEF is not known, it has been proposed that in Salmonella it is involved in the uptake of peptides containing N-terminal formyl-methionine, contributing to avoidance of the host immune response (9, 20). Our data show that mutant MccA peptide with replacement of the N-terminal methionine by glutamine does not compete with McC, confirming the importance of the terminal Met residue for YejABEF recognition. However, just like in the case of MccB binding, the situation appears to be more complex, since MRTGNAD also poorly competes with McC, indicating that the presence of the N-terminal methionine cannot be the only determinant of recognition by YejABEF. While MRTGNAD is unable to compete with McC for entry through YejABEF, its sequence is identical to that of the peptide moiety of McC. This observation suggests that YejABEF also recognizes the nucleotide part of McC and may thus be functionally similar to its close relative NppABCD from Pseudomonas aeruginosa, which contributes to resistance to peptidyl-nucleoside antibiotics (21).
The wild-type McC processing pathway requires deformylation by methionine deformylase followed by the action of methionine aminopeptidase (MAP) and degradation by nonspecific aminopeptidase A, N, or B of the resulting hexapeptide adenylate (2). Compounds with substituted Met1 must be processed without the involvement of methionine deformylase or MAP. The bioactivities of some McC-like compounds produced by bacteria other than E. coli have been shown to be limited by the rate of proteolytic processing (13). Combining peptide stability/degradation rate analysis with MccB-catalyzed in vitro adenylation could yield adenylation-competent peptides with faster processing, which may result in peptide adenylates with increased bioactivity.
Enzymatic synthesis allowed us to probe longer peptides as adenylation substrates and determine the bioactivities of the resulting adenylates. Using a series of N-terminally extended MccA variants, we showed that extension of the McC peptide length by a single amino acid increases bioactivity. Further increase has no additional effect, followed by an eventual decline in activity. The transport of longer peptide adenylates remains YejABEF dependent up to a certain point. However, McC variants with peptides lengths of 20 and 25 amino acids are transported jointly by YejABEF and SbmA. Earlier, we showed that a peptide adenylate prepared by adenylation of a 25-amino-acid-long MccA-like peptide from a Synechococcus sp. by cognate MccB enzyme enters E. coli cells exclusively through SbmA (13). The difference between the entry pathways of E. coli and Synechococcus sp. peptide-based adenylates indicates that both YejABEF and SbmA exhibit sequence or structural specificity toward the peptides they transport. The fact that certain peptide adenylates are taken up by two transport systems is potentially very attractive, as it could allow limitation of the appearance of spontaneously resistant mutants.
The biological activity of longer peptide adenylates can be further increased by aminopropylation, providing an additional way of obtaining compounds with higher activity levels. Since multiple MccA-MccB pairs have been validated in in vitro adenylation reactions, the opportunities for obtaining modified peptide adenylates with increased activity levels explored here using the E. coli system are also fully open for other McC-like compounds, some of which are produced by (and presumably target) important pathogens.
We finally showed that MccA fusions to full-size proteins such as MBP are also subject to adenylation by MccB in vivo and in vitro. This finding opens several interesting possibilities. First, a C-terminal MccA tag offers an opportunity to selectively label proteins in a way that is complementary and orthogonal to popular methods that rely on phosphorylation by a kinase (22). Second, and perhaps more interestingly, introducing an adenylated tag into a protein in vivo should allow one to study protein stability, for upon degradation of a tagged protein, processed toxic McC should accumulate inside the cell.
ACKNOWLEDGMENTS
This work was supported by the Skoltech Translational Research and Innovation Program, Russian Foundation for Basic Research grant 15-04-07598, Russian Academy of Sciences Program Grants in Molecular and Cell Biology and Bionanotechnology, and Ministry of Education and Science of Russian Federation project 14.B25.31.0004 (to K.S.) and NIAID grant R56AI117210 (to Satish A. Nair and K.S.). The MS facility was available to us in the framework of the Moscow State University Development Program PNG 5.13.
REFERENCES
- 1.Guijarro JI, González-Pastor JE, Baleux F, San Millán JL, Castilla MA, Rico M, Moreno F, Delepierre M. 1995. Chemical structure and translation inhibition studies of the antibiotic microcin C7. J Biol Chem 270:23520–23532. doi: 10.1074/jbc.270.40.23520. [DOI] [PubMed] [Google Scholar]
- 2.Kazakov T, Vondenhoff GH, Datsenko KA, Novikova M, Metlitskaya A, Wanner BL, Severinov K. 2008. Escherichia coli peptidase A, B, or N can process translation inhibitor microcin C. J Bacteriol 190:2607–2610. doi: 10.1128/JB.01956-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Metlitskaya A, Kazakov T, Kommer A, Pavlova O, Praetorius-Ibba M, Ibba M, Krasheninnikov I, Kolb V, Khmel I, Severinov K. 2006. Aspartyl-tRNA synthetase is the target of peptide nucleotide antibiotic Microcin C. J Biol Chem 281:18033–18042. doi: 10.1074/jbc.M513174200. [DOI] [PubMed] [Google Scholar]
- 4.Novikova M, Metlitskaya A, Datsenko K, Kazakov T, Kazakov A, Wanner B, Severinov K. 2007. The Escherichia coli Yej transporter is required for the uptake of translation inhibitor microcin C. J Bacteriol 189:8361–8365. doi: 10.1128/JB.01028-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garcia-Bustos JF, Pezzi N, Mendez E. 1985. Structure and mode of action of microcin 7, an antibacterial peptide produced by Escherichia coli. Antimicrob Agents Chemother 27:791–797. doi: 10.1128/AAC.27.5.791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Metlitskaya A, Kazakov T, Vondenhoff GH, Novikova M, Shashkov A, Zatsepin T, Semenova E, Zaitseva N, Ramensky V, Van Aerschot A, Severinov K. 2009. Maturation of the translation inhibitor microcin C. J Bacteriol 191:2380–2387. doi: 10.1128/JB.00999-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vondenhoff GH, Blanchaert B, Geboers S, Kazakov T, Datsenko KA, Wanner BL, Rozenski J, Severinov K, Van Aerschot A. 2011. Characterization of peptide chain length and constituency requirements for YejABEF-mediated uptake of microcin C analogues. J Bacteriol 193:3618–3623. doi: 10.1128/JB.00172-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eswarappa SM, Panguluri KK, Hensel M, Chakravortty D. 2008. The yejABEF operon of Salmonella confers resistance to antimicrobial peptides and contributes to its virulence. Microbiology 154:666–678. doi: 10.1099/mic.0.2007/011114-0. [DOI] [PubMed] [Google Scholar]
- 9.Qimron U, Madar N, Mittrücker HW, Zilka A, Yosef I, Bloushtain N, Kaufmann SH, Rosenshine I, Apte RN, Porgador A. 2004. Identification of Salmonella typhimurium genes responsible for interference with peptide presentation on MHC class I molecules: Deltayej Salmonella mutants induce superior CD8+ T-cell responses. Cell Microbiol 6:1057–1070. doi: 10.1111/j.1462-5822.2004.00418.x. [DOI] [PubMed] [Google Scholar]
- 10.González-Pastor JE, San Millán JL, Moreno F. 1994. The smallest known gene. Nature 369:281. doi: 10.1038/369281a0. [DOI] [PubMed] [Google Scholar]
- 11.Roush RF, Nolan EM, Löhr F, Walsh CT. 2008. Maturation of an Escherichia coli ribosomal peptide antibiotic by ATP-consuming N-P bond formation in microcin C7. J Am Chem Soc 130:3603–3609. doi: 10.1021/ja7101949. [DOI] [PubMed] [Google Scholar]
- 12.Severinov K, Semenova E, Kazakov A, Kazakov T, Gelfand MS. 2007. Low-molecular-weight post-translationally modified microcins. Mol Microbiol 65:1380–1394. doi: 10.1111/j.1365-2958.2007.05874.x. [DOI] [PubMed] [Google Scholar]
- 13.Bantysh O, Serebryakova M, Makarova KS, Dubiley S, Datsenko KA, Severinov K. 2014. Enzymatic synthesis of bioinformatically predicted microcin C-like compounds encoded by diverse bacteria. mBio 5(3):e01059-14. doi: 10.1128/mBio.01059-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kulikovsky A, Serebryakova M, Bantysh O, Metlitskaya A, Borukhov S, Severinov K, Dubiley S. 2014. The molecular mechanism of aminopropylation of peptide-nucleotide antibiotic microcin C. J Am Chem Soc 136:11168–11175. doi: 10.1021/ja505982c. [DOI] [PubMed] [Google Scholar]
- 15.Kazakov T, Metlitskaya A, Severinov K. 2007. Amino acid residues required for maturation, cell uptake, and processing of translation inhibitor microcin C. J Bacteriol 189:2114–2118. doi: 10.1128/JB.01609-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Van de Vijver P, Vondenhoff GH, Kazakov TS, Semenova E, Kuznedelov K, Metlitskaya A, Van Aerschot A, Severinov K. 2009. Synthetic microcin C analogs targeting different aminoacyl-tRNA synthetases. J Bacteriol 191:6273–6280. doi: 10.1128/JB.00829-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thomason LC, Costantino N, Court DL. 2007. E. coli genome manipulation by P1 transduction. Curr Protoc Mol Biol Chapter 1:Unit 1.17. doi: 10.1002/0471142727.mb0117s79. [DOI] [PubMed] [Google Scholar]
- 18.Regni CA, Roush RF, Miller DJ, Nourse A, Walsh CT, Schulman BA. 2009. How the MccB bacterial ancestor of ubiquitin E1 initiates biosynthesis of the microcin C7 antibiotic. EMBO J 28:1953–1964. doi: 10.1038/emboj.2009.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laviña M, Pugsley AP, Moreno F. 1986. Identification, mapping, cloning and characterization of a gene (sbmA) required for microcin B17 action on Escherichia coli K12. J Gen Microbiol 132:1685–1693. [DOI] [PubMed] [Google Scholar]
- 20.Shawar SM, Cook RG, Rodgers JR, Rich RR. 1990. Specialized functions of MHC class I molecules. I. An N-formyl peptide receptor is required for construction of the class I antigen Mta. J Exp Med 171:897–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pletzer D, Braun Y, Dubiley S, Lafon C, Köhler T, Page MGP, Mourez M, Severinov K, Weingart H. 2015. The Pseudomonas aeruginosa ABC transporter NppA1A2BCD is required for uptake of peptidyl nucleoside antibiotics. J Bacteriol 197:2217–2228. doi: 10.1128/JB.00234-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen BP, Hai T. 1994. Expression vectors for affinity purification and radiolabeling of proteins using Escherichia coli as host. Gene 139:73–75. doi: 10.1016/0378-1119(94)90525-8. [DOI] [PubMed] [Google Scholar]