ABSTRACT
Pseudomonas aeruginosa is a ubiquitously occurring environmental bacterium and opportunistic pathogen responsible for various acute and chronic infections. Obviously, anaerobic energy generation via denitrification contributes to its ecological success. To investigate the structural basis for the interconnection of the denitrification machinery to other essential cellular processes, we have sought to identify the protein interaction partners of the denitrification enzyme nitrite reductase NirS in the periplasm. We employed NirS as an affinity-purifiable bait to identify interacting proteins in vivo. Results obtained revealed that both the flagellar structural protein FliC and the protein chaperone DnaK form a complex with NirS in the periplasm. The interacting domains of NirS and FliC were tentatively identified. The NirS-interacting stretch of amino acids lies within its cytochrome c domain. Motility assays and ultrastructure analyses revealed that a nirS mutant was defective in the formation of flagella and correspondingly in swimming motility. In contrast, the fliC mutant revealed an intact denitrification pathway. However, deletion of the nirF gene, coding for a heme d1 biosynthetic enzyme, which leads to catalytically inactive NirS, did not abolish swimming ability. This pointed to a structural function for the NirS protein. FliC and NirS were found colocalized with DnaK at the cell surface of P. aeruginosa. A function of the detected periplasmic NirS-DnaK-FliC complex in flagellum formation and motility was concluded and discussed.
IMPORTANCE Physiological functions in Gram-negative bacteria are connected with the cellular compartment of the periplasm and its membranes. Central enzymatic steps of anaerobic energy generation and the motility mediated by flagellar activity use these cellular structures in addition to multiple other processes. Almost nothing is known about the protein network functionally connecting these processes in the periplasm. Here, we demonstrate the existence of a ternary complex consisting of the denitrifying enzyme NirS, the chaperone DnaK, and the flagellar protein FliC in the periplasm of the pathogenic bacterium P. aeruginosa. The dependence of flagellum formation and motility on the presence of an intact NirS was shown, structurally connecting both cellular processes, which are important for biofilm formation and pathogenicity of the bacterium.
INTRODUCTION
Pseudomonas aeruginosa is a metabolically versatile bacterium inhabiting multiple environmental niches (1). It is known for its highly efficient growth in the absence of oxygen. Fast anaerobic growth is mediated via the utilization of different N-oxides as electron acceptors in the respiratory chains of denitrification (2). Anaerobic respiratory growth via denitrification also sustains biofilm formation on environmental surfaces, in the mucus in the lung of cystic fibrosis patients, on the epithelium of the urinary tract infected individuals, and in burn wounds (3).
The periplasm, the cellular compartment bounded by the cytoplasmic membrane and the outer membrane of Gram-negative bacteria, is a unique compartment, rich in signal transduction and transport systems and other connections between the outer cell surface and the cytoplasm. It selectively links and buffers external and internal environments and connects systems that monitor external parameters with internal systems that respond to such parameters. It is the “telephone exchange” and logistics center of the cell envelope. It also orchestrates cellular actions directed at the external environment, including the attachment to and attack of host cells by infecting pathogens. Present knowledge of periplasm structure and function is patchy, and a deeper understanding of bacterial behavior in environmental settings, in particular of bacterial activities related to infections, will inter alia depend upon new advances in periplasm biology.
In the process of denitrification, nitrate is used as a terminal electron acceptor and is reduced to N2 in four steps catalyzed by nitrate (Nar), nitrite (Nir), nitric oxide (Nor), and nitrous oxide (Nos) reductases, respectively (1, 4–6). Except for the first reduction catalyzed by the Nar enzyme, all of these reactions are carried out in the bacterial periplasm. The periplasmic denitrification pathway is intimately connected with other central cellular processes, including respiratory energy generation, transmembrane transport, protein translocation, environmentally controlled gene regulation, disulfide bond formation, flagellum assembly and function, the biogenesis of cytochrome c and heme d1 (2), and various processes important to pathogenesis. Of special interest is the P. aeruginosa cytochrome cd1 nitrite reductase (NirS), a homodimer composed of two distinct domains. The N-terminal domain is a c-type cytochrome accommodating a covalently bound heme c, while the C-terminal domain harbors the catalytic heme d1 molecule (7, 8). This enzyme catalyzes the reduction of nitrite to nitric oxide. Interestingly, a nirS knockout mutant of P. aeruginosa PA14 showed deficiency in swarming motility (9) which, in P. aeruginosa, involves both flagella and pili, unlike swimming motility, which is accomplished exclusively by flagella (10). The flagellum, in combination with the chemotaxis system, mediates directional movement of the bacterium in response to attractants and repellents (10) and plays a crucial role in initial cell-cell and cell-surface interactions, including biofilm formation (11). A nirS mutant produces poorly dispersing biofilms, which partially regain dispersal ability upon addition of exogenous nitric oxide (12, 13). These observations suggest a coupling between denitrification and motility, although direct evidence for this was lacking.
Although small signaling molecules are involved in the coupling of many cellular processes, physical protein-protein interactions are equally important in orchestrating metabolic and regulatory networks. They are expected to be particularly relevant for the control of biochemical processes in the periplasm.
In this study, we have sought the interaction partners of NirS through application of interactomic methods, phenotypic characterizations, and electron microscopy-based imaging. This has revealed the existence of a periplasmic protein interaction triad composed of NirS, the flagellar protein FliC, and, surprisingly for a protein previously thought to be cytoplasmic, the molecular chaperone DnaK. This complex connecting the denitrification machinery with motility via flagellum assembly might be only the beginning for the understanding of complex dynamic protein-protein interactions in the periplasmic compartment.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Bacterial strains and plasmids used in this study are listed in Table 1 (also see Table SA1 in the supplemental material). Escherichia coli DH10b (ThermoFisher Scientific, Waltham, MA) was employed as the host propagator of the constructs obtained in this study. E. coli BL21 was used to overproduce DnaK for polyclonal antibody generation. The P. aeruginosa PA14 wild-type strain (14) and several PA14 MAR2xT7 library mutants (15) were used throughout this study. The pAS40 vector was utilized to express nirS, nosZ, and fliC by their natural promoters in P. aeruginosa. The pJET1.2 plasmid (ThermoFisher Scientific, Waltham, MA) served as an intermediate cloning vector carrying the promoter-gene insert prior to its final cloning into pAS40 for the fusion of the target proteins with Strep tag II. The pET14b plasmid was used to produce DnaK-His6 protein for polyclonal antibody production (Metabion AG, Planegg/Steinkirchen, Germany). E. coli was transformed by the heat shock method (16) and P. aeruginosa by electroporation (17). Plasmid preparations were carried out according to the manufacturer's guidelines (Qiagen Miniprep kit). Molecular DNA techniques were performed as formerly described (18). DNA sequencing of the constructs was performed by GATC Biotech AG (Constance, Germany). All enzymes used during the study were obtained from New England BioLabs GmbH (Frankfurt am Main, Germany).
TABLE 1.
Bacterial strains used in this study
| Strain | Relevant feature(s)a | Source or referenceb |
|---|---|---|
| E. coli | ||
| DH10b | F− endA1 recA1 galE15 galK16 nupG rpsL ΔlacX74 ϕ80lacZΔM15 araD139 Δ(ara,leu)7697 mcrA Δ(mrr-hsdRMS-mcrBC) λ− | ThermoFisher Scientific, Waltham, MA |
| BL21 | B F− dcm ompT hsdS(rB − mB−) gal (DE3) | Stratagene, Santa Clara, CA |
| P. aeruginosa | ||
| PA14 wild type | Parental strain | 14 |
| PA14 narH mutant | PA14 narH transposon mutant strain (MAR2 × T7) | 15 |
| PA14 nirS mutant | PA14 nirS transposon mutant strain (MAR2 × T7) | 15 |
| PA14 nirF mutant | PA14 nirF transposon mutant strain (MAR2 × T7) | 15 |
| PA14 nosZ mutant | PA14 nosZ transposon mutant strain (MAR2 × T7) | 15 |
| PA14 fliC mutant | PA14 fliC transposon mutant strain (MAR2 × T7) | 15 |
| PA14 pAS40 | PA14 harboring the plasmid without cloned gene | This study |
| PA14 nirS mutant, nirS+ | PA14 nirS mutant complemented with nirS-Strep tag II expression plasmid pAS40 | This study |
| PA14 nirS mutant, fliC+ | PA14 nirS mutant carrying the fliC-Strep tag II expression plasmid pAS40 | This study |
| PA14 fliC mutant, fliC+ | PA14 fliC mutant complemented with fliC-Strep tag II expression plasmid pAS40 | This study |
| PA14 fliC mutant, nirS+ | PA14 fliC mutant carrying nirS-Strep tag II expression plasmid pAS40 | This study |
| PA14 nosZ mutant, nosZ+ | PA14 nosZ mutant complemented with nosZ-Strep tag II expression plasmid pAS40 | This study |
Significant characteristic(s) of the strain.
Source from which the strain was obtained.
For aerobic growth, all strains were grown in Luria-Bertani (LB) medium in shake flasks aerated by rotation at 180 rpm. For anaerobic growth, strains were grown until late exponential phase in LB supplemented with 50 mM nitrate in rubber-stoppered serum flasks rotated at 100 rpm (19). Where specified, P. aeruginosa strains were cultivated in swimming medium (20) supplemented with 20 mM arginine aerobically until an optical density (OD) of 1.0 at 578 nm was reached, immediately shifted into anaerobic flasks, and further incubated for 5 days. The incubation temperature was 37°C. All experiments were done in triplicate. Required antibiotics were added at the following concentrations (in micrograms per milliliter): ampicillin (Ap), 100; carbenicillin (Cb), 100 for E. coli DH10b and 250 for P. aeruginosa.
Construction of P. aeruginosa strains for tagged protein complex formation.
Construction of the plasmid carrying the target genes (nirS, fliC, and nosZ) under the control of their natural promoters but fused to the DNA encoding Strep tag II was achieved using a slightly modified Golden Gate cloning protocol. This cloning strategy is shown in Fig. SA1 in the supplemental material (also see Fig. SA2). During the procedure, restriction endonuclease BsaI recognizes the DNA sequence GGTCTC and cleaves 1 nucleotide upstream in the same strand and 5 nucleotides in the complementary strand, leaving a sticky end of 4 nucleotides, which was customized. Two single-strand DNA sequences (primers; 5′ to 3′ orientation) were annealed. The forward primer consisted of an EcoRI restriction site, the first 4 nucleotides of the cloning region (∼400 to 500 bp upstream of the gene of interest to cover the promoter region), one random nucleotide, the restriction sites for BsaI (GAGACC; inverse orientation), BglII, EcoRV, BsaI (GGTCTC), one random nucleotide, the last 4 nucleotides of the gene of interest (stop codon omitted), the DNA encoding Strep tag II, and a stop codon (TCA). The reverse primer was the forward complementary strand lacking the EcoRI restriction site at the 5′ end and instead including a HindIII restriction site at its 3′ end. After the fragment was annealed, it was ligated into the EcoRI-HindIII-digested pAS40 plasmid. The fragments cloned were nirSStreptagIIFw-nirSStreptagIIRv, fliCStreptagIIFw-fliCStreptagIIRv, and nosZStreptagIIFw-nosZStreptagIIRv (see Table SA2 in the supplemental material). The BglII and EcoRV digestion sites were employed to confirm proper cloning. In parallel, nirS and fliC genes, along with approximately 400 to 500 bp upstream (including promoter regions) (21, 22), were amplified and further cloned into the plasmid pJET1.2. Both genes included one BsaI (GGTCTC) restriction site at the 5′ and 3′ extremes. The nosZ gene and corresponding nos promoter (approximately 400 to 500 bp upstream of nosR) (23) were amplified separately and cloned into pJET1.2. Afterwards, both constructed plasmids pAS40 and pJET1.2 were mixed and subjected to 50 cycles of 5 min of BsaI digestion followed by 5 min of ligation. This way, the pJET1.2-constructed vectors were continuously subjected to digestion with a release of the genes of interest, whereas once the genes were inserted into pAS40, resulting constructs were stable due to the loss of BsaI restriction sites. This cloning approach resulted in a high yield of pAS40 plasmids harboring the genes of interest fused to the Strep tag II at the C terminus. Five strains were obtained by following this strategy, expressing nirS, fliC, and nosZ in three different mutant backgrounds. They are listed in Table 1.
Isolation of the periplasmic fraction.
P. aeruginosa strains grown anaerobically to the exponential phase were harvested by centrifugation at 4,000 × g at 4°C for 20 min. The resulting pellets were washed twice in phosphate-buffered saline (PBS) buffer (137 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4 × 7 H2O, and 1.4 mM KH2PO4), and the isolation of the periplasmic contents was performed by following a previously described method with minor changes (24). Washed cells were resuspended in PE buffer (20% [wt/vol] sucrose, 50 mM Tris, pH 8.0, and 1 mg/ml polymyxin B) supplemented with 1 tablet/10 ml of protease inhibitor cocktail (Roche Diagnostics, Berlin, Germany). Cell suspensions were incubated overnight at 4°C with continuous mixing, followed by centrifugation at 100,000 × g for 1 h and 4°C in an Optima L-90K ultracentrifuge (Beckman Coulter, Krefeld, Germany) with a 70.1 Ti rotor (Beckman Coulter, Krefeld, Germany). The resulting supernatant fluid constituted the periplasmic fraction.
In vivo cross-linking.
Where indicated, native protein assemblies were stabilized prior to periplasmic extraction by in vivo cross-linking, performed as formerly described (25). The introduction of covalent bonds between interacting proteins enabled more stringent washing reactions and reduces background by largely conserving the native protein ratios in a multiprotein assembly. Formaldehyde, which mediates very specific cross-linking between proteins in close proximity (26), was injected by syringe into the anaerobic cultures to a final concentration of 0.125% (wt/vol). The cross-linking reaction was carried out by incubation at 150 rpm for 20 min at 37°C to enable permeation through cell membranes and was terminated by injecting glycine to a final concentration of 130 mM, followed by incubation for a further 5 min with shaking. Protein samples then were heated to 95°C for 20 to 30 min prior to analysis by SDS-gel electrophoresis (27).
Affinity copurification of the bait proteins and their interaction partners.
Periplasmic fractions were subjected to affinity purification by applying the samples to a gravity-flow column of a Strep-Tactin superflow high-capacity device (IBA GmbH, Göttingen, Germany). Washing and elution were carried out according to the manufacturer's guidelines. A periplasmic extract of the P. aeruginosa PA14 wild-type strain harboring the parental plasmid was used as a background control. Protein concentrations of the eluate fractions were determined by means of the Bradford assay (28), and eluates between 0.05 and 0.2 mg/ml were analyzed by SDS-PAGE or liquid chromatography-tandem mass spectrometry (LC-MS/MS). For SDS-PAGE analyses, eluted fractions were precipitated by adding 50% (vol/vol) trichloroacetic acid (TCA), 0.1% (wt/vol) sodium desoxycholate to the protein sample (ratio of 2.5:9:1) and subsequently incubated on ice for 30 min. Precipitates were collected by centrifugation at 13,000 rpm for 20 min and washed twice with cold acetone, followed by centrifugation for 10 min at 13,000 rpm and 4°C. Precipitates were resuspended in 20 μl SDS loading buffer (15% [vol/vol] glycerol, 5% [vol/vol] β-mercaptoethanol, 2.4% [wt/vol] SDS, 1% [wt/vol] bromophenol blue, 0.8% [wt/vol] Tris, pH 6.8), heated at 95°C for 20 to 30 min, and subsequently loaded on a 12.5% SDS gel (29).
Antibody generation.
Specific polyclonal antibodies were obtained against overproduced DnaK protein and synthetic peptides from NirS and FliC. For this purpose, DnaK-His6 was produced in E. coli BL21 using the pET14b plasmid. Purification of approximately 2 mg/ml recombinant protein was achieved by following the manufacturer's guidelines (Protino, Macherey-Nagel, Düren, Germany). Polyclonal rabbit antibodies were generated by Metabion International AG (Planegg/Steinkirchen, Germany). NirS peptides covering the regions from amino acid 379 to 392, 526 to 540, and 541 to 555 were synthesized and the mixture was employed for raising polyclonal rabbit antibodies. For the generation of anti-FliC antibodies, a peptide consisting of the first 174 amino acids of FliC was used for rabbit immunization. IgGs were purified on a Sepharose-protein A column.
Colocalization experiments using transmission electron microscopy.
P. aeruginosa PA14 was grown anaerobically in LB supplemented with 50 mM nitrate until late exponential phase (8 h) and fixed with 1% formaldehyde in growth medium overnight at 5°C. After centrifugation at 4,000 × g for 20 min at 5°C, harvested bacteria were resuspended in Tris-EDTA (TE) buffer containing 10 mM glycine to quench free aldehyde groups. Bacteria then were dehydrated according to the progressive-lowering-of-temperature protocol (PLT embedding) using ethanol and embedded in Lowicryl K4M resin. After polymerization with UV light, ultrathin sections were cut with a diamond knife, collected onto butvar-coated nickel grids, and incubated with the specific antibodies. First, sections were incubated with a 1:25 dilution of the purified DnaK IgG antibodies overnight at 5°C. After washing with PBS, bound antibodies were made visible by incubating with protein A/G conjugated with gold nanoparticles (PAG) 15 nm in size (1:75 dilution of the stock solution) for 30 min at room temperature. After washing with PBS containing 0.1% Tween, 100 sections were incubated with a protein A solution (0.1 mg/ml) for 15 min at room temperature. Sections were incubated either with anti-NirS or anti-FliC antibodies (1:20 dilution of the purified IgG) for 3 h at room temperature. After PBS washing, sections were incubated with PAG (10 nm in size, 1:200 dilution of the stock solution) for 30 min at room temperature. Subsequently, sections were washed with PBS containing 0.1% Tween 100, TE buffer, and distilled water. Sections were counterstained with 4% aqueous uranyl acetate for 1 min. Samples then were examined in a TEM910 transmission electron microscope (Carl Zeiss, Oberkochen, Germany) at an acceleration voltage of 80 kV. Images were recorded digitally at calibrated magnifications with a slow-scan charge-coupled-device (CCD) camera (1024 by 1024 pixels; ProScan, Scheuring, Germany) with ITEM Software (Olympus Soft Imaging Solutions, Münster, Germany). Contrast and brightness were adjusted with Adobe Photoshop CS4.
Immunofluorescence microscopy.
For immunofluorescence staining of flagella, bacteria were grown aerobically in swimming medium or LB supplemented with 20 mM arginine to an OD of 1.0 and then switched to anaerobic growth (as described before) for a period of 5 days. Samples were taken and fixed with 3% paraformaldehyde in PBS for 30 min at room temperature. No washing and centrifugation steps were performed to avoid possible damage of the flagellar structures. A drop (25 μl) of bacterial culture was placed on a glass poly-l-lysine-coated coverslip. Fixed coverslips were rinsed twice after each immunostaining step with PBS. Samples were treated with blocking buffer (10% fetal calf serum in PBS) for 1 h at room temperature. Coverslips were incubated with the anti-FliC primary rabbit polyclonal antibody in blocking buffer for 1 h at room temperature. After washing, samples were incubated with secondary Alexa Fluor 488-conjugated goat anti-rabbit antibody (Molecular Probes, Eugene, OR). Coverslips were mounted using ProLong Gold containing nuclear staining (4′,6-diamidino-2-phenylindole [DAPI]; Molecular Probes). Fluorescence images were obtained with an Axio Imager A1, AxioCam MRm camera (Zeiss, Oberkochen, Germany), and AxioVision Rel. 4.6 software using AxioVision 4 module multichannel fluorescence and filters 49 and 44.
SDS-PAGE and Western blot detection.
Bait proteins and interaction partners were separated by SDS-PAGE, stained in a solution of 0.05% (wt/vol) Coomassie brilliant blue GS-250 in 25% (vol/vol) isopropanol and 10% (vol/vol) acetic acid, and visualized under a GS-800 calibrated densitometer (Bio-Rad, Munich, Germany). Interactions involving proteins NirS and FliC were corroborated by Western blotting. Proteins were transferred to a polyvinylidene difluoride (PVDF) membrane and detected with Strep-Tactin-conjugated alkaline phosphatase according to the procedure recommended by the supplier (IBA GmbH, Göttingen, Germany).
LC-MS/MS and data analyses.
Prominent proteins were excised from the polyacrylamide gels and prepared for LC-MS/MS analysis (30). Protein samples were supplemented with 1 μl GlycoBlue (Ambion, Darmstadt, Germany), 80 μl 2.5 M Na acetate, pH 5.0, and 1.5 ml ethanol and incubated overnight at room temperature to precipitate the solved proteins. The precipitated proteins were pelleted (16,000 rpm, 30 min, 4°C) and air dried. Air-dried proteins were resolubilized in 40 μl of 50 mM trimethylammonium bicarbonate buffer (TEAB), reduced by addition of 4 μl of 20 mM Tris-(2-carboxyethyl)-phosphine-hydrochloride (TCEP), alkylated with 2 μl of 200 mM methyl methane thiosulfonate (MMTS), digested by the addition trypsin to a final ratio of 50:1 (protein to protease), and incubated at 37°C overnight. After evaporation of all fluid in a SpeedVac (Eppendorf, Hamburg, Germany), the peptides were desalted using stage tips with an additional layer of 5 μl of 10-μm LiChrosorb RP-18 (Merck Millipore, Darmstadt, Germany) material as previously described (31). Peptides were resolubilized in 20 μl of 3% acetonitrile (ACN) containing 0.2% TFA and adsorbed to the RP-18 material. After washing with resolubilization solution, the peptides were eluted with 60% ACN containing 0.2% TFA. To remove the organic phase, the peptides were dried in a SpeedVac and resolubilized in 12 μl of 3% ACN containing 0.2% TFA.
LC-MS/MS analyses were performed on a Dionex UltiMate 3000 n-RSLC (rapid-separation LC) system connected to an LTQ Orbitrap Velos mass spectrometer (ThermoFisher Scientific, Waltham, MA). Peptides were loaded onto a C18 precolumn (3 μm; 75 μm by 20 mm; Acclaim; Dionex, ThermoFisher Scientific, Waltham, MA) in 3% ACN containing 0.1% TFA, washed for 3 min in 3% ACN containing 0.1% formic acid (FA) at a flow rate of 6 μl/min, and subsequently fractionated on a C18 analytical column (3 μm; 75 μm by 25 cm; Acclaim PepMap RSLC; Dionex) at 350 nl/min with a linear 120-min gradient of 0 to 80% ACN in 0.1% FA. The LC system was operated with Chromeleon software (version 6.8; Dionex), which was embedded in the Xcalibur software (version 2.1; ThermoFisher Scientific, Waltham, MA). The effluent from the column was electrosprayed (Picotip emitter needles; New Objectives, Woburn, MA) into the mass spectrometer controlled by Xcalibur software and operated in the data-dependent mode, allowing the automatic selection of a maximum of 10 doubly and triply charged peptides and their subsequent fragmentation. Peptide fragmentation was carried out using the CID mode in the ion trap. MS/MS raw data files were processed via Proteome Discoverer 1.3.0.339-mediated searches against a P. aeruginosa PA14 database, extracted from Swiss-Prot on a Mascot server (v. 2.4; Matrix Science, Boston, MA). The following search parameters were used: enzyme, trypsin; maximum missed cleavages, 1; fixed modification, carbamidomethylation (C); variable modification, oxidation (M); peptide tolerance, 5 ppm; MS/MS tolerance, 0.5 Da.
Analysis of the peptides involved in the NirS-FliC interaction.
To investigate the peptides involved in the interaction between NirS and FliC, eluates obtained from the affinity chromatography of all free extracts from PA14 complemented strains (a nirS mutant complemented with nirS, an fliC mutant complemented with nirS, and a nirS mutant complemented with fliC) were digested by trypsin and analyzed by LC-MS/MS for complete peptide composition analysis. A schematic representation of the strategy followed is shown in Fig. SA3 in the supplemental material. The NirS peptides involved in the interaction with FliC were superimposed on an existing protein model (PDB code 1NIR) (32) using the PyMOL Molecular Graphics System, version 1.5.04 (Schrödinger LLC) (33).
Swimming assay.
Swimming motility was tested on swimming medium as described before (20). Petri dishes were prepared with 1% (wt/vol) tryptone, 0.5% NaCl, 0.3% (wt/vol) agarose medium containing 20 mM l-arginine as the energy source. The plates were point inoculated in the middle by stabbing with a toothpick that had been soaked in overnight culture and were placed in an Anaerocult anaerobic incubation container with an Anaerocult A sachet (Merck Millipore, Darmstadt, Germany). Oxygen depletion was confirmed colorimetrically by an Anaerotest indicator strip (Merck Millipore). The plates were incubated bottom up at 37°C for 5 days. Swimming motility was determined by measuring the colony halo diameter.
RESULTS
NirS, FliC, and DnaK form a complex in the periplasm of P. aeruginosa grown under denitrifying conditions.
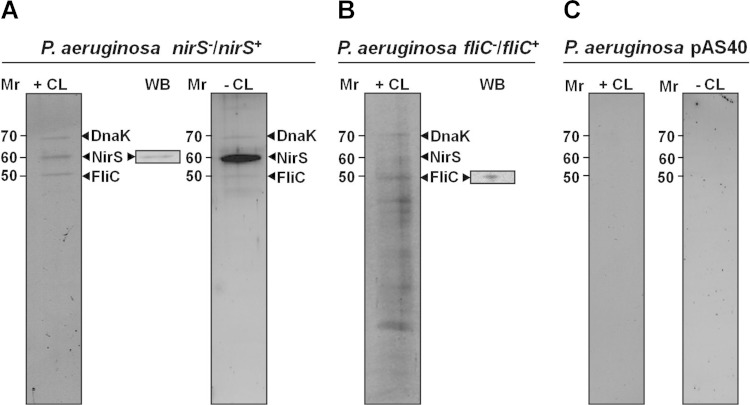
In order to investigate anaerobic energy generation via the periplasmic nitrite reductase NirS- and FliC-containing flagellum-mediated motility, physical protein-protein interaction studies were performed under anaerobic denitrifying conditions. First, for this purpose the interaction partners of the periplasmic cytochrome cd1 nitrite reductase NirS were investigated in a Strep-protein interaction experiment (SPINE) (34), with NirS as a bait protein. The nirS gene and approximately 500 bp of its upstream sequence were cloned into the P. aeruginosa plasmid pAS40. Thus, nirS expression was controlled by the native transcriptional regulation machinery under denitrifying conditions (35). The plasmid was introduced into the PA14 nirS mutant strain to yield the PA14 strain complemented with nirS. Samples of growing bacteria were taken from cultures incubated under anaerobic denitrifying conditions, left untreated (control), or treated with cross-linker. Periplasmic extracts were prepared and subjected to affinity purification of the resulting NirS-protein complexes. SDS-PAGE analyses of these NirS-protein complexes eluted from the column revealed two distinct protein species in addition to the NirS bait protein. The identity of the purified bait Strep-tagged NirS was confirmed by Western blotting (Fig. 1A). The other two proteins were always present in the experiments, which were performed in triplicates (Fig. 1A). These copurifying proteins were identified as FliC (flagellin type B) and the chaperone DnaK by LC-MS/MS analyses. A reciprocal experiment with FliC as the bait protein produced in the PA14 fliC mutant background (PA14 fliC mutant complemented with fliC) confirmed the initial observation. In this case, NirS and DnaK were found bound and copurifying with FliC (Fig. 1B). Other coeluting proteins were identified as multiple subunits of the flagellum (data not shown). The NirS-FliC-DnaK triad was detected in samples from both in vivo cross-linked cultures and untreated cultures, indicating that the observed interactions were rather stable, although the yields of copurifying proteins were substantially lower from cultures that had not been treated with cross-linker (Fig. 1A). No column-binding proteins were observed with periplasmic samples from control cells of the wild-type PA14 strain bearing the cloning vector pAS40 alone (Fig. 1C). In a further control involving another periplasmic denitrification pathway enzyme, the nitrous oxide reductase NosZ, as a bait protein, FliC and DnaK were not found bound to the enzyme, which underlines the specificity of the NirS-FliC-DnaK interactions (data not shown).
FIG 1.
Protein interaction partners of P. aeruginosa NirS and FliC. SDS-PAGE analysis of periplasmic proteins copurifying with affinity-purified bait proteins NirS (A) and FliC (B). Protein extracts from P. aeruginosa without bait protein chromatographed through the same affinity material served as a negative control (C). +CL and −CL indicate whether or not the cell samples were treated with cross-linker prior to protein isolation. Proteins obtained from cross-linked samples were heated to 95°C in SDS sample buffer to reverse cross-linking prior to loading the gel. The identity of the proteins, indicated by arrowheads, was determined via their excision from the gels. The proteins were eluted and tryptic digests were analyzed by LC-MS/MS, as described in Materials and Methods. WB (inset) indicates Western blot confirmation of the identity of the bait protein. The relative molecular weights (in thousands) are indicated. nirS−/nirS+ indicates a nirS mutant strain that has been complemented with nirS, and fliC−/fliC+ indicates an fliC mutant strain that has been complemented with fliC.
A P. aeruginosa nirS mutant is deficient in swimming motility.
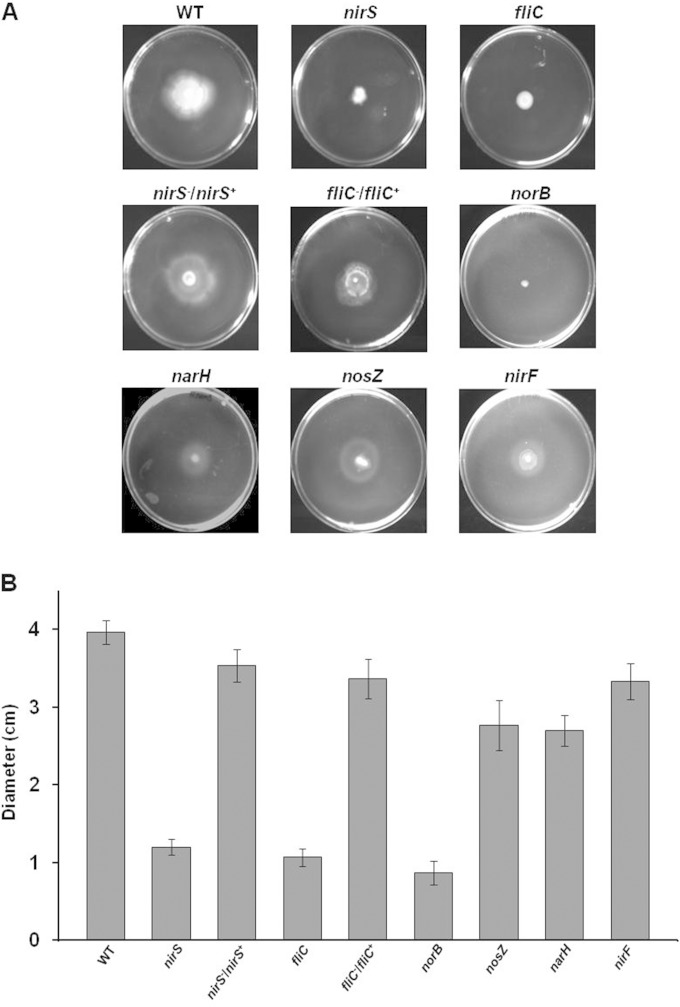
To investigate the relevance of enzymes of the denitrification pathway for P. aeruginosa, the swimming motility of mutant strains defective in different denitrification reductases was tested in appropriate motility assays. Wild-type P. aeruginosa PA14 and a corresponding fliC mutant strain served as positive and negative controls, respectively. Because denitrification mutants fail to grow anaerobically with nitrate as an electron acceptor, the fermentable substrate arginine was provided in the cultures to enable anaerobic growth of the mutants. Under these conditions, any swimming defect should be related to a malfunction of the flagellum rather than to impaired denitrification. As shown in Fig. 2, the P. aeruginosa PA14 nirS mutant was clearly impaired in its swimming ability. The degree was comparable to that of the fliC mutant strain. The P. aeruginosa PA14 nirS mutant strain complemented with nirS and the P. aeruginosa PA14 fliC mutant strain complemented with fliC both exhibited normal motility, i.e., the characteristic large swimming halo of the wild-type parental strain (Fig. 2). Most importantly, the P. aeruginosa PA14 narH and nosZ mutant strains also were able to swim, which demonstrated that denitrification per se is not crucial for P. aeruginosa motility. Thus, the involvement of the denitrification machinery in motility appears to be NirS specific. In order to distinguish between a poorly structural and a catalytic, i.e., energy-delivering, function of NirS, a catalytically inactive NirS was tested. For this purpose, a P. aeruginosa nirF mutant, defective in the biosynthesis of the essential heme d1 cofactor of nitrite reductase, was analyzed for its swimming behavior (25). nirF was described to produce catalytically inactive but stable NirS in the periplasm (36). Clearly, the swimming activity observed for the nirF mutant, carrying a catalytically inactive NirS, pointed toward a structural function of the NirS protein in the triple complex. In agreement, the functional consequences of the NirS-FliC interaction were not reciprocal, i.e., the nirS mutant did not swim, whereas the denitrifying growth of the fliC mutant was not affected (see Fig. 4A). The anaerobic growth and motility defect of the P. aeruginosa PA14 norB mutant most likely is caused by the previously described essential role of NorB in the formation of catalytically active NirS (37) (Fig. 2). Finally, testing of the nirS mutant for swimming under aerobic growth conditions revealed a partial restoration of swimming ability. Approximately 40% of the analyzed strains showed the restored phenotype. We observed a mixture of strains in their aerobic and anaerobic modes. However, de la Fuente-Nunez et al. showed the swarming motility defect in P. aeruginosa for a nirS mutant under aerobic conditions (9). Obviously, further research is needed to clarify the structural basis of aerobic versus anaerobic flagellum assembly.
FIG 2.
Swimming motility assays of the P. aeruginosa wild type (WT) and strains carrying inactivated genes for various enzymes of denitrification. (A) Soft agar plates containing arginine as energy source were point inoculated with overnight cultures of the strains to be tested, incubated anaerobically at 37°C for 5 days, as described in Materials and Methods. (B) The diameters of the halos of bacterial growth, representing the distance migrated by the strain via swimming, were measured. The plots show the mean halo diameters from triplicate measurements and their standard deviations.
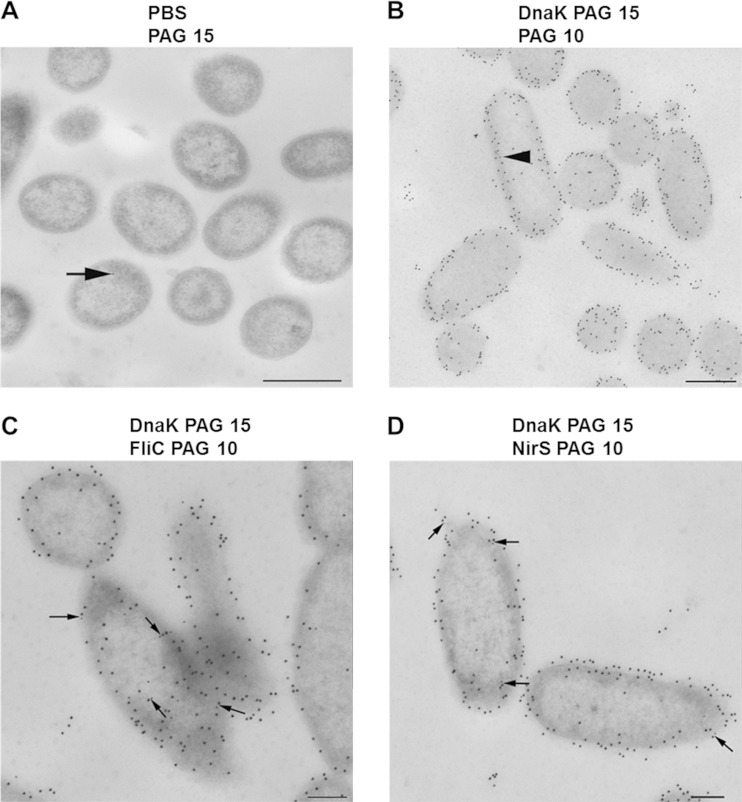
FIG 4.
Cellular localization and colocalization of DnaK, FliC, and NirS in anaerobically grown, denitrifying cells of P. aeruginosa PA14. Cells were prepared for transmission electron microscopy, treated with specific antibody, and protein A/G conjugated with 10- or 15-nm-diameter gold nanoparticles (PAG 10 and PAG 15, respectively), counterstained, and examined in a TEM910 transmission electron microscope, as described in Materials and Methods. (A) Antibody-negative control with PAG 15; (B) anti-DnaK antibodies with PAG 15, followed by protein A blocking, and then PAG 10; (C) anti-DnaK antibodies with PAG 15, followed by protein A blocking, and then anti-FliC antibodies with PAG 10; (D) anti-DnaK antibodies with PAG 15, followed by protein A blocking, and then anti-NirS antibodies with PAG 10. DnaK is seen to be distributed mostly in the extracytoplasmic region; several cocomplexes between DnaK and FliC as well as DnaK and NirS are observed and are indicated by arrows. Scale bars are 500 nm (A and B) and 200 nm (C and D).
P. aeruginosa PA14 nirS mutant was impaired in flagellum assembly.
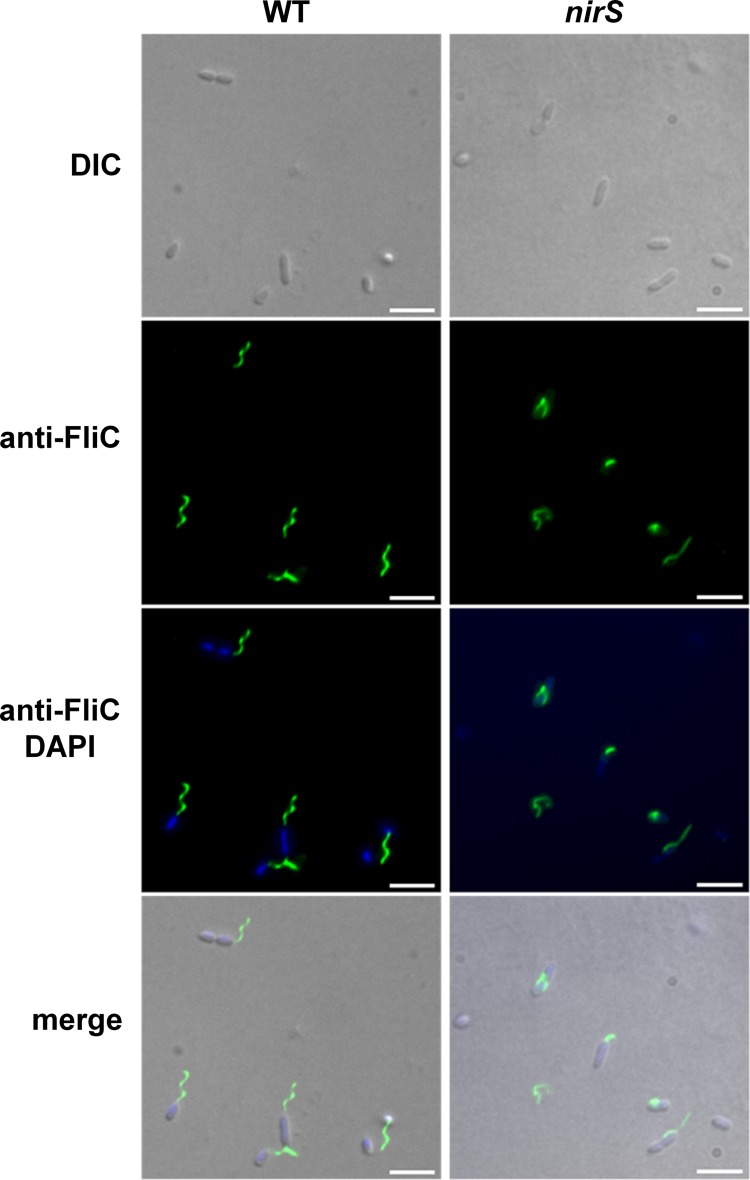
In order to investigate the functional basis for the observed motility defect of the P. aeruginosa PA14 nirS mutant, the flagellation of this mutant strain was tested by immunofluorescence microscopy using specific antibodies against FliC. The flagellar abundance and morphology of wild-type P. aeruginosa and the nirS mutant were compared. The wild-type parental strain produced flagella under aerobic and anaerobic growth conditions when grown in LB, although flagellar abundance varied from cell to cell (Fig. 3).
FIG 3.
Flagellar morphology in the wild type and nirS mutant visualized by immunofluorescence microscopy. For visualization of extracellular FliC protein, samples of bacteria cultured anaerobically in LB containing arginine were labeled with primary rabbit anti-FliC antibody followed by secondary Alexa Fluor 488-conjugated goat anti-rabbit antibody (green), and the DNA was labeled with DAPI (blue). Arrowheads point to nonfunctional flagella in the NirS mutant strain. Fluorescence images were taken and processed as described in Materials and Methods. Scale bars, 10 μm.
The number of flagellated P. aeruginosa PA14 nirS mutant bacteria was significantly lower than that for the parental strain. Many short, incomplete, and abnormal nonspiral (nonrotating) flagella were observed (Fig. 3, arrowheads). Similar results were obtained with bacteria cultured in so-called swimming medium. These results demonstrated that the NirS protein is required for correct flagellum functionality and, as a consequence, for motility in P. aeruginosa.
Immunolocalization of NirS-FliC-DnaK.
In order to directly visualize the spatial distribution of NirS, FliC, and DnaK in cells, purified specific IgG antibodies followed by protein A/G-coated gold nanoparticles (PAG) of different sizes (10 and 15 nm in diameter) were used for labeling of ultrathin sections of P. aeruginosa PA14. The anti-DnaK antibodies revealed the predominant localization of DnaK at the cell periphery and in the extracytoplasmic region (Fig. 4B to D). Almost no labeling of P. aeruginosa PA14 was observed using PAG nanoparticles alone in the absence of specific antibody (Fig. 4A). We then performed a second control, which is depicted in Fig. 4B, where after labeling DnaK with specific antibodies, possibly free binding sites on the first bound antibody were blocked by incubation with protein A alone. Only a few single PAG 10 particles could be detected (arrow in Fig. 4B), validating the colocalization studies we performed. This method detects proteins localizing less than 25 nm from each other, since each of the two applied antibodies can maximally span a distance of approximately 12 nm due to the hinge in the Fab region. Double labeling with anti-DnaK and anti-FliC as well as anti-NirS antibodies first revealed that both FliC and NirS proteins were much less abundant than DnaK (Fig. 4C and D). Performing the experiments in reverse, e.g., first incubating with anti-NirS and subsequently with anti-DnaK antibodies as well as for the other possible pairings (FliC and DnaK), we observed the same results as those for the original experiment. However, when they were detected, they were mostly colocalized with DnaK, providing ultrastructural support for direct DnaK-NirS and DnaK-FliC interaction in the bacterial periplasm.
The NirS is directly interacting via its cytochrome c domain with FliC.
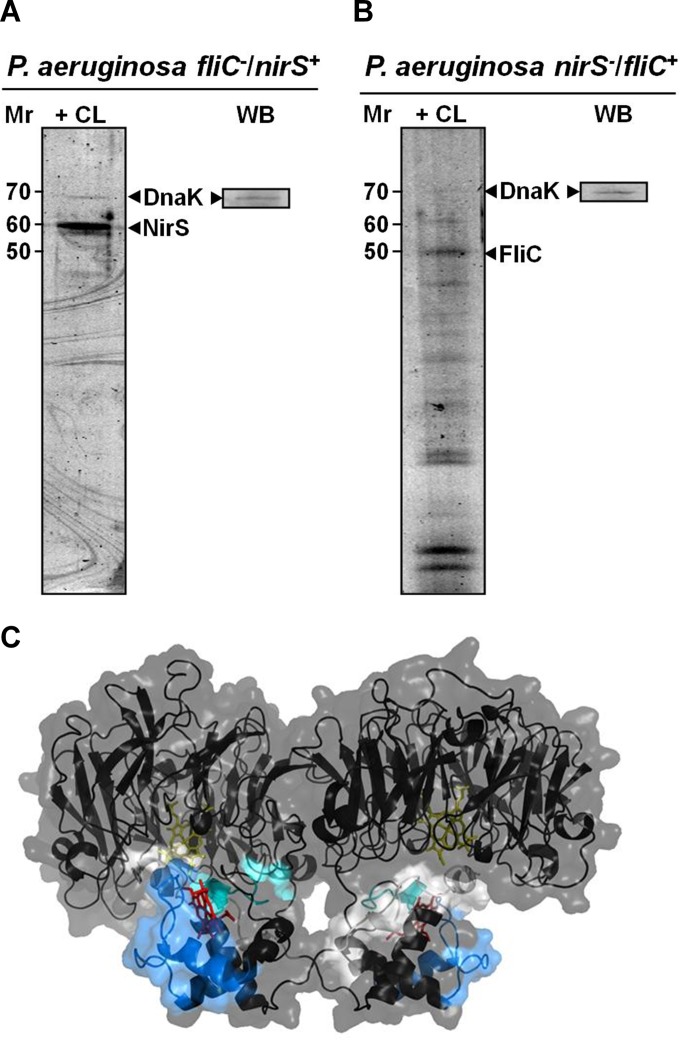
In order to obtain further information on the protein-protein interactions in the detected NirS-DnaK-FliC triad, we repeated the bait-prey binding studies as shown in Fig. 1 in fliC (Fig. 5A) and nirS (Fig. 5B) mutant backgrounds. This approach tested for sole DnaK-FliC and DnaK-NirS interaction. As can be seen in Fig. 5, FliC is not required for interaction of NirS with DnaK (Fig. 5A), and NirS is not required for FliC interaction with DnaK (Fig. 5B). Obviously, both NirS and FliC are directly linked to DnaK. To specify the interaction of FliC and NirS, we analyzed the complete peptide composition from the samples obtained from the tagged FliC and NirS. Isolated protein complexes were subjected to trypsinization and peptide analysis by LC-MS/MS (see Fig. SA3 in the supplemental material). In these experiments, NirS-FliC cross-linked peptides were identified, showing a direct interaction. The NirS-FliC peptides from the cross-linked complex were AAEQYQGAASAVDPTHVVR, CAGCHGVLRK, and GQQYLEALITYGTPLGMPNWGSSGELSK from NirS and NQVLQQAGTAILAQANQLPQAVLSLLR and LGITASINDK from FliC. Since interaction surfaces of the complex potentially are contained within these peptides, we wanted to map them onto available structure models. As can be seen in Fig. 5C, which shows the NirS peptides that cross-link to FliC highlighted in the NirS structure model (obtained from the PDB database under code 1NIR [32]), these peptides are located in the cytochrome c domain of NirS (2, 32). Unfortunately, the entire structure, especially the detected peptide-containing regions, of the P. aeruginosa FliC was not available. Consequently, it was not possible to deduce a model of the possible NirS-FliC docking surfaces.
FIG 5.
Structural basis of NirS-FliC-DnaK interactions. (A) NirS copurification with DnaK as bait in the PA14 strain mutated in fliC but complemented with nirS strain. (B) FliC copurification in the P. aeruginosa nirS mutant strain complemented with fliC. (C) Structural model of dimeric P. aeruginosa NirS showing the peptides hypothesized to participate in the interaction with FliC. These peptides were determined as outlined in the legend to Fig. SA3 in the supplemental material. NirS structure is displayed as a cartoon model with transparent surface as inferred from PDB entry 1NIR (32). The stick models indicate the heme c (red) and heme d molecules (yellow). Interacting peptides are shown in light gray (AAEQYQGAASAVDPTHVVR), cyan (CAGCHGVLRK), and light blue (GQQYLEALITYGTPLGMPNWGSSGELSK).
DISCUSSION
In this study, a novel triple complex consisting of the denitrification enzyme nitrite reductase (NirS), the flagellar filament protein FliC, and the chaperone DnaK was identified in the periplasm of P. aeruginosa PA14. The current understanding of FliC function localizes the protein in the flagellum outside the cell, while the ATP-dependent DnaK chaperone is considered to be cytoplasmic. However, E. coli FliC and several other FliC proteins, including P. aeruginosa FliC, were detected in the inner membrane fraction and the periplasm during various proteomic analyses (38–43). In multiple approaches, the FliC protein was detected together with NirS (41) or even with NirS and DnaK (43). Similarly, DnaK was found in the cell envelope of Aggregatibacter actinomycetemcomitans (44) and Vibrio cholerae (14) and cross-linked to E. coli respiratory formate dehydrogenase during mapping of cell envelope and periplasmic protein interaction partners (45). Even a first mechanism for the transport of DnaK into the periplasm of E. coli during osmostress was described (46, 47). In this case, the mechano-sensitive channel protein McsL was involved in the export of DnaK. However, other examples for the involvement of DnaK in the export of periplasmic and outer membrane proteins even in the presence of a defective SEC system pointed toward alternative protein export systems involving DnaK (48). All of these observations suggest a periplasmic function of FliC and DnaK. Such a periplasmic FliC function was described in the context of the E. coli LeoA protein activity during enterotoxin secretion (49). Deletion of leoA led to reduced toxin export, accumulation of FliC in the periplasm, and a nonmotile phenotype (49). Moreover, FliC function in adhesion and virulence has been described in detail before (50, 51). In this context, the existence of various differentially N-glycosylated forms of FliC offer the basis for multiple cellular functions in various cellular compartments (52). Species-specific differences in FliC function, indicated by the failure to exchange FliC proteins between, for example, E. coli and P. aeruginosa, point to additional FliC function in these bacteria (53). In contrast, FliD can be exchanged among a variety of bacteria without further problems (53). Finally, the detected direct interaction of the cytochrome c domain of NirS with both the proposed long N- and C-terminal helices of FliC might give insights into the structural interconnection of denitrification and motility (54). Overall, we assume that the periplasmic compartment represents a highly ordered and structured cellular organelle in Gram-negative bacteria. Most likely, the membrane-associated anaerobic energy generation machinery of denitrification forms a megacomplex, as observed for the electron transport chain in mammalian, yeast, and plant mitochondria (55–61). Therefore, since several protein-protein interactions of the involved proteins have been shown already, one can assume the presence of a large denitrification megacomplex in bacteria. The dynamic but mostly stable complex of denitrification provides an ideal assembly and mobility platform for the multiple other periplasmic proteins involved in processes like ATP generation, disulfide bond formation, protein and metabolite transport, communication to the outer membrane, environmental signal perception, and transduction. One of the major challenges of motility is the trans-cytoplasmic, periplasmic, and outside formation of the flagellum. Here, highly ordered molecular transport and assembly processes are required. Obviously, the already determined multiple protein-protein contact-forming nitrite reductase most likely serves as a major driving force (25). The NirS-FliC interaction would attach FliC to the proposed megacomplex and consequently stabilize the overall flagellum assembly process. The periplasmic NirS is dedicated via the lipoprotein NirF to the outer membrane, opening the flagellum assembly pathway toward extracellular localization. Association with DnaK, which is usually stabilizing partly denatured, not completely soluble, and unstable proteins, might provide the opportunity to stabilize involved, structurally unstable forms of FliC prior to cellular export and assembly. Therefore, DnaK might serve as an FliC stabilizer and NirS as a guide from the cytoplasmic to the outer membrane during the process of flagellum assembly. Future investigation will determine the exact molecular mechanism underlying the NirS-DnaK-FliC triad function.
Supplementary Material
ACKNOWLEDGMENTS
J.M.B.-D.A. gratefully acknowledges funding from the DFG grant SCHO 888/4-1 and from an ERC grant (IPBSL) awarded to Ricardo Amils (Centro de Astrobiología-CSIC, Spain) and K.N.T. T.D., L.J., M.J., and D.J. acknowledge funding from the Deutsche Forschungsgemeinschaft (Forschergruppe PROTRAIN).
We thank Katharina Riedel (Greifswald University, Germany) and Susanne Häussler (Twincore, Hannover, Germany) for helpful discussions and Ina Schleicher for excellent help in the electron microscopic studies and the Helmholtz International Graduate School for Infection Research for continuous support. We are grateful for technical assistance from Simone Virus.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00415-15.
REFERENCES
- 1.Williams HD, Zlosnik JE, Ryall B. 2007. Oxygen, cyanide and energy generation in the cystic fibrosis pathogen Pseudomonas aeruginosa. Adv Microb Physiol 52:1–71. [DOI] [PubMed] [Google Scholar]
- 2.Zumft WG. 1997. Cell biology and molecular basis of denitrification. Microbiol Mol Biol Rev 61:533–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lyczak JB, Cannon CL, Pier GB. 2000. Establishment of Pseudomonas aeruginosa infection: lessons from a versatile opportunist. Microbes Infect 2:1051–1060. doi: 10.1016/S1286-4579(00)01259-4. [DOI] [PubMed] [Google Scholar]
- 4.Toyofuku M, Uchiyama H, Nomura N. 2012. Social behaviours under anaerobic conditions in Pseudomonas aeruginosa. Int J Microbiol 2012:405191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hassett DJ, Cuppoletti J, Trapnell B, Lymar SV, Rowe JJ, Yoon SS, Hilliard GM, Parvatiyar K, Kamani MC, Wozniak DJ, Hwang SH, McDermott TR, Ochsner UA. 2002. Anaerobic metabolism and quorum sensing by Pseudomonas aeruginosa biofilms in chronically infected cystic fibrosis airways: rethinking antibiotic treatment strategies and drug targets. Adv Drug Deliv Rev 54:1425–1443. doi: 10.1016/S0169-409X(02)00152-7. [DOI] [PubMed] [Google Scholar]
- 6.Arai H. 2011. Regulation and function of versatile aerobic and anaerobic respiratory metabolism in Pseudomonas aeruginosa. Front Microbiol 2:103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nurizzo D, Cutruzzola F, Arese M, Bourgeois D, Brunori M, Cambillau C, Tegoni M. 1998. Conformational changes occurring upon reduction and NO binding in nitrite reductase from Pseudomonas aeruginosa. Biochemistry 37:13987–13996. doi: 10.1021/bi981348y. [DOI] [PubMed] [Google Scholar]
- 8.Arese M, Zumft WG, Cutruzzola F. 2003. Expression of a fully functional cd1 nitrite reductase from Pseudomonas aeruginosa in Pseudomonas stutzeri. Protein Expr Purif 27:42–48. doi: 10.1016/S1046-5928(02)00600-9. [DOI] [PubMed] [Google Scholar]
- 9.de la Fuente-Nunez C, Reffuveille F, Fairfull-Smith KE, Hancock RE. 2013. Effect of nitroxides on swarming motility and biofilm formation, multicellular behaviors in Pseudomonas aeruginosa. Antimicrob Agents Chemother 57:4877–4881. doi: 10.1128/AAC.01381-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kohler T, Curty LK, Barja F, van Delden C, Pechere JC. 2000. Swarming of Pseudomonas aeruginosa is dependent on cell-to-cell signaling and requires flagella and pili. J Bacteriol 182:5990–5996. doi: 10.1128/JB.182.21.5990-5996.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O'Toole GA, Kolter R. 1998. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol Microbiol 30:295–304. doi: 10.1046/j.1365-2958.1998.01062.x. [DOI] [PubMed] [Google Scholar]
- 12.Barraud N, Hassett DJ, Hwang SH, Rice SA, Kjelleberg S, Webb JS. 2006. Involvement of nitric oxide in biofilm dispersal of Pseudomonas aeruginosa. J Bacteriol 188:7344–7353. doi: 10.1128/JB.00779-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barraud N, Schleheck D, Klebensberger J, Webb JS, Hassett DJ, Rice SA, Kjelleberg S. 2009. Nitric oxide signaling in Pseudomonas aeruginosa biofilms mediates phosphodiesterase activity, decreased cyclic di-GMP levels, and enhanced dispersal. J Bacteriol 191:7333–7342. doi: 10.1128/JB.00975-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rahme LG, Stevens EJ, Wolfort SF, Shao J, Tompkins RG, Ausubel FM. 1995. Common virulence factors for bacterial pathogenicity in plants and animals. Science 268:1899–1902. doi: 10.1126/science.7604262. [DOI] [PubMed] [Google Scholar]
- 15.Liberati NT, Urbach JM, Miyata S, Lee DG, Drenkard E, Wu G, Villanueva J, Wei T, Ausubel FM. 2006. An ordered, nonredundant library of Pseudomonas aeruginosa strain PA14 transposon insertion mutants. Proc Natl Acad Sci U S A 103:2833–2838. doi: 10.1073/pnas.0511100103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chan WT, Verma CS, Lane DP, Gan SK. 2013. A comparison and optimization of methods and factors affecting the transformation of Escherichia coli. Biosci Rep 33:e00086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Enderle PJ, Farwell MA. 1998. Electroporation of freshly plated Escherichia coli and Pseudomonas aeruginosa cells. Biotechniques 25:954–958. [DOI] [PubMed] [Google Scholar]
- 18.Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, New York, NY. [Google Scholar]
- 19.Benkert B, Quack N, Schreiber K, Jaensch L, Jahn D, Schobert M. 2008. Nitrate-responsive NarX-NarL represses arginine-mediated induction of the Pseudomonas aeruginosa arginine fermentation arcDABC operon. Microbiology 154:3053–3060. doi: 10.1099/mic.0.2008/018929-0. [DOI] [PubMed] [Google Scholar]
- 20.Rashid MH, Kornberg A. 2000. Inorganic polyphosphate is needed for swimming, swarming, and twitching motilities of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 97:4885–4890. doi: 10.1073/pnas.060030097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arai H, Igarashi Y, Kodama T. 1994. Structure and ANR-dependent transcription of the nir genes for denitrification from Pseudomonas aeruginosa. Biosci Biotechnol Biochem 58:1286–1291. doi: 10.1271/bbb.58.1286. [DOI] [PubMed] [Google Scholar]
- 22.Frisk A, Jyot J, Arora SK, Ramphal R. 2002. Identification and functional characterization of flgM, a gene encoding the anti-sigma 28 factor in Pseudomonas aeruginosa. J Bacteriol 184:1514–1521. doi: 10.1128/JB.184.6.1514-1521.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arai H, Mizutani M, Igarashi Y. 2003. Transcriptional regulation of the nos genes for nitrous oxide reductase in Pseudomonas aeruginosa. Microbiology 149:29–36. doi: 10.1099/mic.0.25936-0. [DOI] [PubMed] [Google Scholar]
- 24.Dammeyer T, Timmis KN, Tinnefeld P. 2013. Broad host range vectors for expression of proteins with (Twin-) Strep-tag, His-tag and engineered, export optimized yellow fluorescent protein. Microb Cell Fact 12:49. doi: 10.1186/1475-2859-12-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nicke T, Schnitzer T, Munch K, Adamczack J, Haufschildt K, Buchmeier S, Kucklick M, Felgentrager U, Jansch L, Riedel K, Layer G. 2013. Maturation of the cytochrome cd1 nitrite reductase NirS from Pseudomonas aeruginosa requires transient interactions between the three proteins NirS, NirN and NirF. Biosci Rep 33:e00048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sutherland BW, Toews J, Kast J. 2008. Utility of formaldehyde cross-linking and mass spectrometry in the study of protein-protein interactions. J Mass Spectrom 43:699–715. doi: 10.1002/jms.1415. [DOI] [PubMed] [Google Scholar]
- 27.Dammeyer T, Schobert M. 2010. Interactomics, p 4407–4428. In Timmis K. (ed), Handbook of hydrocarbon and lipid microbiology. Springer Verlag, Berlin, Germany. [Google Scholar]
- 28.Noble JE, Bailey MJ. 2009. Quantitation of protein. Methods Enzymol 463:73–95. doi: 10.1016/S0076-6879(09)63008-1. [DOI] [PubMed] [Google Scholar]
- 29.Laemmli UK. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 30.Duvel J, Bertinetti D, Moller S, Schwede F, Morr M, Wissing J, Radamm L, Zimmermann B, Genieser HG, Jansch L, Herberg FW, Haussler S. 2012. A chemical proteomics approach to identify c-di-GMP binding proteins in Pseudomonas aeruginosa. J Microbiol Methods 88:229–236. doi: 10.1016/j.mimet.2011.11.015. [DOI] [PubMed] [Google Scholar]
- 31.Rappsilber J, Ishihama Y, Mann M. 2003. Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal Chem 75:663–670. doi: 10.1021/ac026117i. [DOI] [PubMed] [Google Scholar]
- 32.Nurizzo D, Silvestrini MC, Mathieu M, Cutruzzola F, Bourgeois D, Fulop V, Hajdu J, Brunori M, Tegoni M, Cambillau C. 1997. N-terminal arm exchange is observed in the 2.15 A crystal structure of oxidized nitrite reductase from Pseudomonas aeruginosa. Structure 5:1157–1171. doi: 10.1016/S0969-2126(97)00267-0. [DOI] [PubMed] [Google Scholar]
- 33.DeLano WL. 2002. The PyMOL molecular graphics system. Schrödinger, Mannheim, Germany. [Google Scholar]
- 34.Herzberg C, Weidinger LA, Dorrbecker B, Hübner S, Stülke J, Commichau FM. 2007. SPINE: a method for the rapid detection and analysis of protein-protein interactions in vivo. Proteomics 7:4032–4035. doi: 10.1002/pmic.200700491. [DOI] [PubMed] [Google Scholar]
- 35.Trunk K, Benkert B, Quack N, Munch R, Scheer M, Garbe J, Jansch L, Trost M, Wehland J, Buer J, Jahn M, Schobert M, Jahn D. 2010. Anaerobic adaptation in Pseudomonas aeruginosa: definition of the Anr and Dnr regulons. Environ Microbiol 12:1719–1733. [DOI] [PubMed] [Google Scholar]
- 36.Kawasaki S, Arai H, Igarashi Y, Kodama T. 1995. Sequencing and characterization of the downstream region of the genes encoding nitrite reductase and cytochrome c551 (nirSM) from Pseudomonas aeruginosa: identification of the gene necessary for biosynthesis of heme d1. Gene 167:87–91. doi: 10.1016/0378-1119(95)00641-9. [DOI] [PubMed] [Google Scholar]
- 37.Zumft WG, Braun C, Cuypers H. 1994. Nitric oxide reductase from Pseudomonas stutzeri. Primary structure and gene organization of a novel bacterial cytochrome bc complex. Eur J Biochem 219:481–490. [DOI] [PubMed] [Google Scholar]
- 38.Han MJ, Kim JY, Kim JA. 2014. Comparison of the large-scale periplasmic proteomes of the Escherichia coli K-12 and B strains. J Biosci Bioeng 117:437–442. doi: 10.1016/j.jbiosc.2013.09.008. [DOI] [PubMed] [Google Scholar]
- 39.Imperi F, Ciccosanti F, Perdomo AB, Tiburzi F, Mancone C, Alonzi T, Ascenzi P, Piacentini M, Visca P, Fimia GM. 2009. Analysis of the periplasmic proteome of Pseudomonas aeruginosa, a metabolically versatile opportunistic pathogen. Proteomics 9:1901–1915. doi: 10.1002/pmic.200800618. [DOI] [PubMed] [Google Scholar]
- 40.Choi DS, Kim DK, Choi SJ, Lee J, Choi JP, Rho S, Park SH, Kim YK, Hwang D, Gho YS. 2011. Proteomic analysis of outer membrane vesicles derived from Pseudomonas aeruginosa. Proteomics 11:3424–3429. doi: 10.1002/pmic.201000212. [DOI] [PubMed] [Google Scholar]
- 41.Arts IS, Ball G, Leverrier P, Garvis S, Nicolaes V, Vertommen D, Ize B, Tamu Dufe V, Messens J, Voulhoux R, Collet JF. 2013. Dissecting the machinery that introduces disulfide bonds in Pseudomonas aeruginosa. mBio 4:e00912–00913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nouwens AS, Willcox MD, Walsh BJ, Cordwell SJ. 2002. Proteomic comparison of membrane and extracellular proteins from invasive (PAO1) and cytotoxic (6206) strains of Pseudomonas aeruginosa. Proteomics 2:1325–1346. doi:. [DOI] [PubMed] [Google Scholar]
- 43.Casabona MG, Vandenbrouck Y, Attree I, Coute Y. 2013. Proteomic characterization of Pseudomonas aeruginosa PAO1 inner membrane. Proteomics 13:2419–2423. doi: 10.1002/pmic.201200565. [DOI] [PubMed] [Google Scholar]
- 44.Smith KP, Fields JG, Voogt RD, Deng B, Lam YW, Mintz KP. 2015. The cell envelope proteome of Aggregatibacter actinomycetemcomitans. Mol Oral Microbiol 30:97–110. doi: 10.1111/omi.12074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zorn M, Ihling CH, Golbik R, Sawers RG, Sinz A. 2014. Mapping cell envelope and periplasm protein interactions of Escherichia coli respiratory formate dehydrogenases by chemical cross-linking and mass spectrometry. J Proteome Res 13:5524–5535. doi: 10.1021/pr5004906. [DOI] [PubMed] [Google Scholar]
- 46.el Yaagoubi A, Kohiyama M, Richarme G. 1994. Localization of DnaK (chaperone 70) from Escherichia coli in an osmotic-shock-sensitive compartment of the cytoplasm. J Bacteriol 176:7074–7078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Berrier C, Garrigues A, Richarme G, Ghazi A. 2000. Elongation factor Tu and DnaK are transferred from the cytoplasm to the periplasm of Escherichia coli during osmotic downshock presumably via the mechanosensitive channel mscL. J Bacteriol 182:248–251. doi: 10.1128/JB.182.1.248-251.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Qi HY, Hyndman JB, Bernstein HD. 2002. DnaK promotes the selective export of outer membrane protein precursors in SecA-deficient Escherichia coli. J Biol Chem 277:51077–51083. doi: 10.1074/jbc.M209238200. [DOI] [PubMed] [Google Scholar]
- 49.Brown EA, Hardwidge PR. 2007. Biochemical characterization of the enterotoxigenic Escherichia coli LeoA protein. Microbiology 153:3776–3784. doi: 10.1099/mic.0.2007/009084-0. [DOI] [PubMed] [Google Scholar]
- 50.Haiko J, Westerlund-Wikstrom B. 2013. The role of the bacterial flagellum in adhesion and virulence. Biology 2:1242–1267. doi: 10.3390/biology2041242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cohen TS, Prince AS. 2013. Activation of inflammasome signaling mediates pathology of acute Pseudomonas aeruginosa pneumonia. J Clin Investig 123:1630–1637. doi: 10.1172/JCI66142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Khemiri A, Naudin B, Franck X, Song PC, Jouenne T, Cosette P. 2013. N-glycosidase treatment with 18O labeling and de novo sequencing argues for flagellin FliC glycopolymorphism in Pseudomonas aeruginosa. Anal Bioanal Chem 405:9835–9842. doi: 10.1007/s00216-013-7424-x. [DOI] [PubMed] [Google Scholar]
- 53.Inaba S, Hashimoto M, Jyot J, Aizawa S. 2013. Exchangeability of the flagellin (FliC) and the cap protein (FliD) among different species in flagellar assembly. Biopolymers 99:63–72. doi: 10.1002/bip.22141. [DOI] [PubMed] [Google Scholar]
- 54.Song WS, Yoon SI. 2014. Crystal structure of FliC flagellin from Pseudomonas aeruginosa and its implication in TLR5 binding and formation of the flagellar filament. Biochem Biophys Res Commun 444:109–115. doi: 10.1016/j.bbrc.2014.01.008. [DOI] [PubMed] [Google Scholar]
- 55.Krause F, Reifschneider NH, Vocke D, Seelert H, Rexroth S, Dencher NA. 2004. “Respirasome”-like supercomplexes in green leaf mitochondria of spinach. J Biol Chem 279:48369–48375. doi: 10.1074/jbc.M406085200. [DOI] [PubMed] [Google Scholar]
- 56.Bultema JB, Braun HP, Boekema EJ, Kouril R. 2009. Megacomplex organization of the oxidative phosphorylation system by structural analysis of respiratory supercomplexes from potato. Biochim Biophys Acta 1787:60–67. doi: 10.1016/j.bbabio.2008.10.010. [DOI] [PubMed] [Google Scholar]
- 57.Marques I, Dencher NA, Videira A, Krause F. 2007. Supramolecular organization of the respiratory chain in Neurospora crassa mitochondria. Eukaryot Cell 6:2391–2405. doi: 10.1128/EC.00149-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schagger H, Pfeiffer K. 2000. Supercomplexes in the respiratory chains of yeast and mammalian mitochondria. EMBO J 19:1777–1783. doi: 10.1093/emboj/19.8.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vonck J, Schafer E. 2009. Supramolecular organization of protein complexes in the mitochondrial inner membrane. Biochim Biophys Acta 1793:117–124. doi: 10.1016/j.bbamcr.2008.05.019. [DOI] [PubMed] [Google Scholar]
- 60.Dudkina NV, Kudryashev M, Stahlberg H, Boekema EJ. 2011. Interaction of complexes I, III, and IV within the bovine respirasome by single particle cryoelectron tomography. Proc Natl Acad Sci U S A 108:15196–15200. doi: 10.1073/pnas.1107819108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lapuente-Brun E, Moreno-Loshuertos R, Acin-Perez R, Latorre-Pellicer A, Colas C, Balsa E, Perales-Clemente E, Quiros PM, Calvo E, Rodriguez-Hernandez MA, Navas P, Cruz R, Carracedo A, Lopez-Otin C, Perez-Martos A, Fernandez-Silva P, Fernandez-Vizarra E, Enriquez JA. 2013. Supercomplex assembly determines electron flux in the mitochondrial electron transport chain. Science 340:1567–1570. doi: 10.1126/science.1230381. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.