ABSTRACT
Motility is a beneficial attribute that enables cells to access and explore new environments and to escape detrimental ones. The organelle of motility in Escherichia coli is the flagellum, and its production is initiated by the activating transcription factors FlhD and FlhC. The expression of these factors by the flhDC operon is highly regulated and influenced by environmental conditions. The flhDC promoter is recognized by σ70 and is dependent on the transcriptional activator cyclic AMP (cAMP)-cAMP receptor protein complex (cAMP-CRP). A number of K-12 strains exhibit limited motility due to low expression levels of flhDC. We report here a large number of mutations that stimulate flhDC expression in such strains. They include single nucleotide changes in the −10 element of the promoter, in the promoter spacer, and in the cAMP-CRP binding region. In addition, we show that insertion sequence (IS) elements or a kanamycin gene located hundreds of base pairs upstream of the promoter can effectively enhance transcription, suggesting that the topology of a large upstream region plays a significant role in the regulation of flhDC expression. None of the mutations eliminated the requirement for cAMP-CRP for activation. However, several mutations allowed expression in the absence of the nucleoid organizing protein, H-NS, which is normally required for flhDC expression.
IMPORTANCE The flhDC operon of Escherichia coli encodes transcription factors that initiate flagellar synthesis, an energetically costly process that is highly regulated. Few deregulating mutations have been reported thus far. This paper describes new single nucleotide mutations that stimulate flhDC expression, including a number that map to the promoter spacer region. In addition, this work shows that insertion sequence elements or a kanamycin gene located far upstream from the promoter or repressor binding sites also stimulate transcription, indicating a role of regional topology in the regulation of flhDC expression.
INTRODUCTION
Flagellar synthesis in Escherichia coli is an intricately programmed process that begins with the expression of the master operon, flhDC, which encodes the transcription factors FlhD and FlhC. These factors form homodimers that combine to form a heterohexameric FlhD4C2 complex (1) that activates transcription of the numerous σ70-dependent genes required for construction and regulation of the basic flagellar apparatus, including the FliA sigma factor (σ28), which is required for the expression of other downstream flagellar and sensory genes involved in motility and chemotaxis. More than 50 genes comprise the FlhDC-dependent flagellar regulon (2, 3). FlhD2 and FlhD4C2 are also implicated in the control of expression of other genes that are not directly related to flagellar synthesis (4–8).
The flhDC operon itself is a σ70-dependent operon that is regulated on multiple levels. It is controlled by both transcriptional and posttranscriptional regulators that are responsive to environmental factors, such as nutrient type and availability, osmotic pressure, oxygen tension, pH, and temperature (9). It has a 198-nucleotide (nt) mRNA leader sequence that interacts with the regulator of carbohydrate metabolism, CsrA, resulting in increased translation (10). Also, flhDC mRNA interacts with several environmentally responsive small regulatory RNAs (sRNAs) (11, 12). Several of these sRNAs are negative regulators (ArcZ, OmrA, OmrB, and OxyS), while one (McsA) is a positive regulator.
A number of transcriptional regulators acting as direct repressors of flhDC have been identified. The phosphorylated form of the osmotic response regulator, OmpR, inhibits flhDC expression under conditions of increased osmotic pressure, as well as increased cytoplasmic concentrations of acetyl-phosphate (13). Mutations in the type I fimbrial regulator, LrhA, whose target site overlaps one of the two OmpR binding regions, result in derepressed flagellar synthesis and increased biofilm formation (14, 15). A mutation in HdfR results in increased transcription from the flhDC promoter in a strain with hns deleted, and overexpression of HdfR decreases flhDC transcription (16). Similarly, overexpression of MatA, an activator of MatA/ECP fimbrial expression, inhibits flhDC transcription (17). RcsB, a component of a complex phosphorelay system that is involved in the positive regulation of capsular exopolysaccharide synthesis, biofilm formation, and acid resistance, both activates MatA and, in complex with RcsA, represses flagellar synthesis (18, 19); its binding site has been identified just downstream of the promoter. Another transcriptional repressor with a binding site just downstream of the promoter is FliZ (20). Requiring FlhDC for expression, this factor accumulates during post-exponential-phase growth and exerts weak negative-feedback control on flagellar synthesis. The binding regions of FliZ, RcsAB, and one of the two OmpR binding sites overlap.
Only two transcription factors that directly activate flhDC expression have been found thus far in E. coli. One that appears to be absolutely required for expression in vivo is the global regulator cyclic AMP (cAMP)-cAMP receptor protein complex (cAMP-CRP), which binds a region upstream of the flhDC promoter (21, 22). The second factor is the phosphorylated form of the quorum-sensing response component, QseB, which enhances flhDC transcription in the presence of autoinducer 2 (23). This activator has been shown to function in a non-K-12 strain in conjunction with σ28 at a promoter downstream of the σ70-dependent promoter (24).
Two nucleoid-associated proteins, H-NS and HU, are implicated in positive regulation of flhDC transcription, as both hns and hupA hupB cells are immotile (22, 25, 26). H-NS is a small, abundant dimeric protein involved in shaping chromosomal architecture through extensive oligomerization. It preferentially binds curved, AT-rich DNA (25), common in promoter regions (26, 27), and has been shown to bind the regulatory region of flhDC (22). This suggests that the interaction of H-NS with the regulatory region accounts at least in part for its positive influence on flhDC transcription. In addition, H-NS reduces the expression of the HdfR repressor (16). HU, a heterodimer of HupA and HupB that constrains supercoils, tends to bind DNA that is bent or of noncanonical form (29). While HU has not been demonstrated to bind the flhDC regulatory region, Salmonella enterica serovar Typhimurium cells mutant for both hupA and hupB exhibit reduced flhC expression and reduced motility (30), and E. coli hupA hupB cells are nonmotile (31). The involvement of H-NS and possibly HU in flhDC expression suggests that DNA topology plays a role in the control of flhDC expression. A role of DNA topology in flhDC regulation by H-NS in response to low pH has been suggested previously by Soutourina et al. (32).
Relatively few stimulating cis-acting mutations of the flhDC regulatory region have been reported in E. coli. Several cfs (constitutive for flagellar synthesis) strains that do not require cAMP-CRP for flagellar synthesis have been described (33); one of these was shown to contain a mutation in the −10 promoter element that improves its match to the σ70 consensus sequence (22). It has been recognized that insertion sequence (IS) elements in the flhDC regulatory region result in activation of flhDC transcription, likely due, in part, to disruption of repressor binding sites (34–36). The two motile sequenced K-12 strains MG1655 [CGSC 7740, referred to here as MG1655(Seq)] and W3110 have an IS1 [MG1655(Seq)] or IS5 (W3110) element just upstream of the cAMP-CRP binding site, both of which disrupt the LrhA and one of two OmpR binding sites. In addition, an IS1 and an IS5 element several hundred base pairs from the flhDC promoter that each lead to increased transcription of the operon have been identified (36).
We report here the identification of a large number of cis-acting mutations that stimulate flhDC expression. The bulk of these are IS elements, many of which are located far upstream of the cAMP-CRP binding site, suggesting that the flhDC regulatory region can be considered much more extensive than previously thought. Also identified were single nucleotide mutations located in either the promoter or cAMP-CRP binding region. The mechanism of stimulation by these mutations is discussed. In addition, we show that some mutations can override the requirement for H-NS.
MATERIALS AND METHODS
Strains and plasmids.
The strains and plasmids used in this study are shown in Table 1. flhDC, including 50-bp upstream and 21-bp downstream flanking sequences, was cloned into the EcoRI and HindIII sites of pBAD24 to give pVN15. l-Arabinose was added to 0.2% as an inducer where indicated.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Genotype/description | Reference | Source |
|---|---|---|---|
| Strains | |||
| JW1225-2 | Δ(araD-araB)567 ΔlacZ4787(::rrnB-3) Δhns-746::kan rph-1 Δ(rhaD-rhaB)568 hsdR514 | 85 | CGSC 9111 |
| JW5702-4 | Δ(araD-araB)567 ΔlacZ4787(::rrnB-3) Δcrp-765::kan rph-1 Δ(rhaD-rhaB)568 hsdR514 | 85 | CGSC 11596 |
| MG1655 | rph-1 | 86 | CGSC 6300 |
| MG1655(Seq) | MG1655 with IS1 insertion upstream of flhD | 87 | CGSC 7740 |
| Y10 | λ thr-1 leuB6(Am) glnV44(AS) rfbC1 thiE1 | 38 | CGSC 5037 |
| Plasmids | |||
| pBAD24 | araBp araC bla | 88 | |
| pKD45 | aphA-2 ccdB | J. S. Parkinson | |
| pVN15 | pBAD24 carrying flhDC | This study |
Motility studies.
Motility plates contained TB (1% tryptone and 0.5% NaCl) in 0.26% Bacto agar (Becton, Dickinson & Co., Sparks, MD) and were incubated at 30°C unless otherwise indicated. The plates were inoculated with 2.5 μl from a saturated LB (1% tryptone, 0.5% NaCl, 0.5% yeast extract) culture grown at 37°C. Microscopic motility studies were performed on cells grown in TB to an optical density at 600 nm (OD600) of ∼0.6. To count motile cells, fields of cells were recorded and analyzed using ImageJ (http://imagej.nih.gov/ij/). A minimum of 500 cells were counted, and all counts were repeated twice.
Sequencing.
The primers for sequencing the intergenic region between flhDC and uspC were 5′ GTCGCCATTTCTTCATTTATGCCGAG 3′ and 5′ CCGGGTCGGAAGCGAGAGTAATTAAA 3′. The primers for sequencing crp were 5′ ATTCGCCCAGAAAAGTTAACCCTTCGACC 3′ and 5′ TGTCGAAGTGCATAGTTGATATCGGGGTG 3′. Sequencing was done by Eurofins MGWOperon (Alabama) or GeneWiz, Inc. (New Jersey).
Generation of mutations.
DNA fragments containing single nucleotide mutations were amplified using Herculase II Fusion DNA polymerase (Agilent Technologies, California) and transferred to MG1655 by the λ Red method of Datsenko and Wanner (37). All mutations were checked by sequencing. A crp deletion of the second through the penultimate codon was also introduced by λ Red, as was the kanamycin resistance gene aphA-2 from plasmid pKD45. crp and hns deletions were also transduced into recipient strains by P1 transduction.
Polyacrylamide gel electrophoresis.
Nondenaturing 15% gels containing acrylamide–bis-acrylamide (29:1) in TBE (89 mM Tris, 89 mM boric acid, 1 mM EDTA, pH 8.4) were run at 4 V/cm at room temperature.
Curvature analysis.
Analysis of curvature was performed using available online programs, Bend.it (http://pongor.itk.ppke.hu/dna/bend_it.html#/bendit_intro) and DNA Curvature Analysis (http://www.lfd.uci.edu/∼gohlke/dnacurve/), with windows of 21 bp.
RESULTS
Expression of flhDC is low in strain Y10.
Y10 is a strain three steps removed from the original K-12 strain isolated in 1922 (38) and is ancestral to strain RP437. RP437 has been used extensively for studies of chemotaxis and motility and has been shown to have an activating IS element in the regulatory region of the flhDC operon (34). In contrast to RP437, Y10 does not have an activating IS element and exhibits poor motility under conditions that promote flagellar synthesis. Its motility was found to be temperature sensitive: while about 25 to 50% of cells were motile at room temperature, less than 0.2% were motile at 30°C and none at 37°C. When a plasmid expressing flhDC was introduced, cells rapidly spread through soft-agar plates (Fig. 1), and >99% of the cells were motile in liquid, even at 37°C. (See Fig. S1 in the supplemental material for the correspondence between spreading on plates and motility in liquid.) The intergenic region between flhDC and the upstream universal stress protein gene, uspC, was found to be identical to that in MG1655, another strain with low flhDC expression (36) that was determined here to be similarly temperature sensitive for flagellar synthesis. Expression of flhD in MG1655 has been shown to be repressed 3- to 4-fold by LrhA (14, 36). LrhA likely represses flagellation of Y10, as well.
FIG 1.
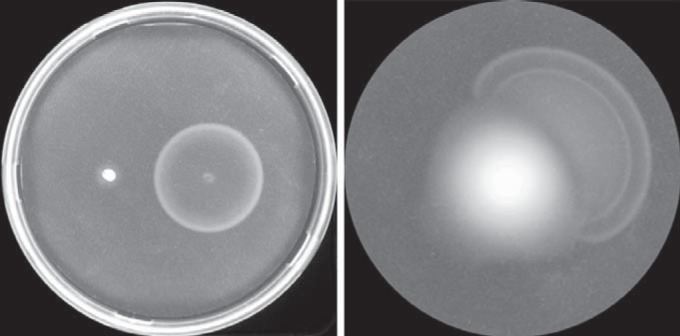
Migration through motility plates. (Left) Spreading of Y10 and Y10 expressing flhDC from plasmid pVN15 after 6 h. The plate contained 0.2% arabinose. (Right) Formation of a rapidly spreading offshoot of Y10 after 24 h. The plates were incubated at 30°C.
Isolation of fast-spreading Y10 mutants.
As has been found for MG1655 and a number of other strains with limited motility (34–36), prolonged incubation (12 to 36 h) of Y10 cells on soft-agar plates frequently gave rise to highly motile offshoots from the main population (Fig. 1). Fifty such offshoots were isolated, and all but four were found to contain an IS element at various sites upstream of the flhDC cAMP-CRP recognition site (Fig. 2). The remaining four mutants were found to have single nucleotide changes in the regulatory region. Two of these were in the promoter region and the other two near or in the cAMP-CRP binding region. These four mutations and all other single nucleotide mutations described below were generated in MG1655 for analysis, since Y10 was found to exhibit a high degree of phenotypic variation, including heterogeneous colony morphology, and tendencies to form small clumps when grown in liquid or to adhere to a coverslip. Spreading of the four single nucleotide mutants on a soft-agar plate is shown in Fig. S2 in the supplemental material.
FIG 2.
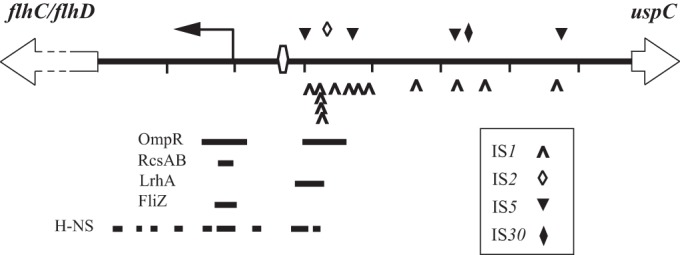
Distribution of IS elements in the flhDC-uspC intergenic region (shown in genomic orientation). The cAMP-CRP binding site is shown as an open hexagon, and the transcriptional start site is shown by a bent arrow. The tick marks are at 100-bp intervals from the start of flhD. The locations of binding regions for H-NS and four transcriptional repressors are shown as solid bars (13, 14, 18, 20, 22).
IS elements stimulate flhDC transcription independently of orientation and have a long-range effect.
Four types of IS elements were recovered, IS1, IS5, IS2, and IS30, the last two of which have not been isolated in this region before. The most proximal element was an IS5 at position −100 relative to the transcription site, and the most distal was an IS5 at position −476. IS element types, insertion sites, orientations of insertion, and duplicated sequences are shown in Table S1 in the supplemental material. Of 22 IS5 elements, 5 inserted at −100, 5 at −170, 10 at −319, and 2 at −476. These sites correspond to IS5 preferred target sequences (5′-YTAR-3′, where Y is C/T and R is A/G) (39). One of the IS5 elements (at −319) appeared to have originated from the IS5Y element embedded in the rac prophage lom pseudogene, based on sequence identity. All five IS30 insertions were found solely at −340. Eighteen IS1 elements inserted at 13 distinct AT-rich sites between −108 and −469, with eight clustered between −125 and −128. These elements showed a strong bias of divergent orientation (14 of 18) with respect to the flhDC promoter. Duplicated sequences ranged from 8 to 10 bp, one of which was an imperfect duplication. A single IS2 element was recovered, at −135.
Stimulation of flhDC expression was found to be independent of the IS element orientation, as there was no correlation between orientation and migration rates on soft-agar plates (data not shown). Independence of orientation also has been found for activating IS elements of the bgl and ade operons (40, 41). Consistent with this are the results of Barker et al., who demonstrated that the transcription start site of flhDC was not altered in strains with differently oriented upstream IS1 or IS5 elements, including the vigorously motile strain MG1655(Seq), which has an IS1 element at −108 (34). Certainly, if any outwardly directed promoters within IS elements were responsible for flhDC expression, cAMP-CRP would not be required for activation of transcription. Deletions of crp were transduced into a subset of IS5-containing variants at all five insertion sites, including those with insertions in both orientations. All such crp derivatives were nonmotile. We note, however, that an example of internal transcriptional initiation has been demonstrated for the IS1 element in the flhDC-uspC intergenic region of MG1655(Seq) upon overexpression of rpoN (σ54) (42).
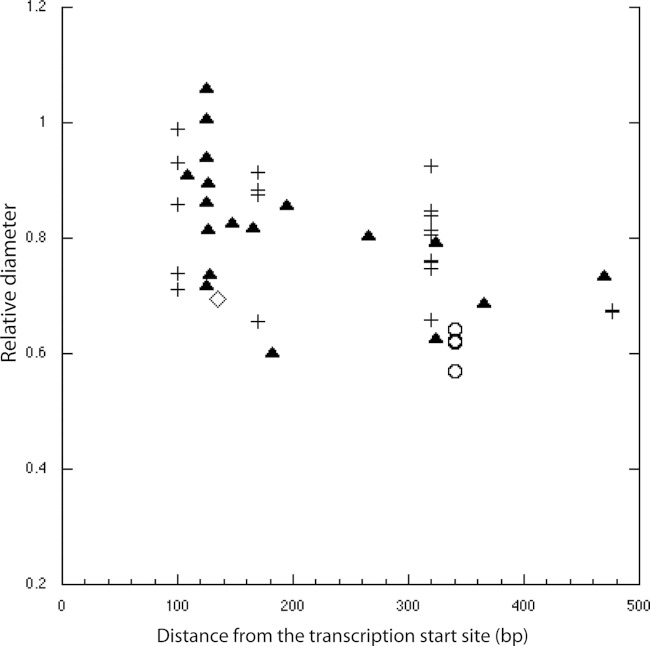
The stimulatory effect of IS elements declined somewhat with distance from the promoter, based on migration on soft-agar plates (Fig. 3). This is consistent with a previous observation of somewhat weaker flhD expression for strains with IS elements located at −303 and −313 than for strains with IS elements located at −166 (36). The migration of variants with IS30 insertions was noticeably slower than the migration of those with other types of IS elements located in the same region. Interference from the phenotypic variation of Y10 mentioned above may account for the spread of migration rates at given insertion sites (different strains had different tendencies to clump or form heterogeneous colony types, etc.).
FIG 3.
Spreading of Y10 variants versus IS element insertion sites relative to MG1655(Seq) (IS1 element at −108) on motility plates at 7 h at 30°C. The sites are given in base pairs from the start of transcription. Shown are the averages from two experiments. The standard deviations were all under 12%. IS1, IS2, IS5, and IS30 elements are represented by triangles, diamonds, pluses, and circles, respectively.
The AT content of the flhDC-uspC intergenic region of Y10 is high (65%), with a number of poly(A) tracts and long mixed AT tracts (up to 12 nt), sequence features tending to form curved DNA, commonly found in regulatory regions that provide target sites for transcription factors and nucleoid-associated proteins (27, 43). In contrast, IS elements, coding for transposases, have an AT content not atypical of E. coli coding sequences that are about 1 kb in length (average, ∼48%) (44). IS1 (768 bp), IS2 (1,331 bp), and IS5 (1,195 bp) have an overall AT content of ∼47%. IS30 (1,221 bp), which was less effective in stimulating flhDC expression, has a higher AT content, 57%. It seemed likely that insertion of any typical coding sequence in the intergenic region would also stimulate flhDC expression. Indeed, a divergently oriented kanamycin resistance gene (aphA-2 of Tn5) having ∼40% AT content inserted into MG1655 at −319 resulted in rapid spreading on soft-agar plates [90% of the rate of MG1655(Seq)] and greater than 90% motility in liquid. The long-range effect of insertions of coding sequences suggests that the intergenic region participates in setting the topology of the regulatory region, which, when disrupted, leads to unregulated transcription of the flhDC operon.
Whereas many strains of E. coli are motile at 30°C, a loss in motility at 37°C has been noted for some (45, 46), and as mentioned above, Y10 flagellation is highly temperature sensitive. Most variants were quite motile at 37°C, especially those with IS elements within 200 bp of the transcription start site (see Fig. S3 in the supplemental material). Strains with more distal elements began to show some temperature sensitivity. It may be that the thermal sensitivity of flhDC expression resides with an inherent temperature-sensitive conformation of the regulatory region, a conformation that is altered by insertion of coding sequences.
Single nucleotide mutations in the promoter region that stimulate flhDC expression include one in the −10 element and one in the spacer.
The promoter contains two key recognition elements for sigma factors, a −10 hexamer and a −35 hexamer. One of the single nucleotide mutations found to stimulate expression was a C-to-T transition at −10 with respect to the start of transcription within the −10 element. Cells with this mutation spread through soft agar 10 to 20% faster than the MG1655(Seq) strain, and >99% were motile in liquid, even at 37°C. The −10T mutation improves the match to the σ70 consensus recognition sequence (47) (Fig. 4). Interestingly, this site of the −10 element is the least conserved (48).
FIG 4.
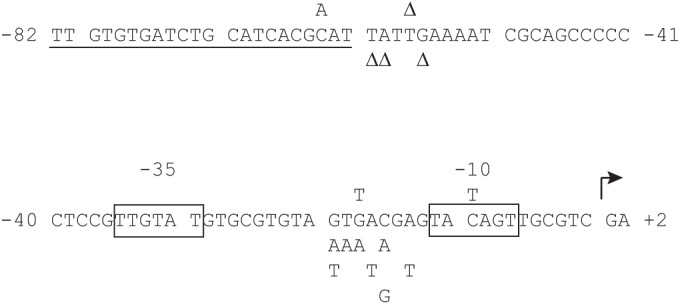
Part of the regulatory region of flhDC showing sites of single nucleotide substitutions and deletions that result in highly motile cells. The four mutations isolated in the screen are shown above the sequence; the stimulatory mutations generated subsequently are below. The cAMP-CRP recognition site is underlined, the −10 and −35 elements are boxed, and the start of transcription is shown by a bent arrow. The consensus sequence for cAMP-CRP binding is AAATGTGATCTAGATCACATTT (52), the −35 element σ70 consensus sequence is TTGACA, and the −10 element σ70 consensus is TATAAT (47).
The same mutation is present in a strain reported to be constitutive for flagellar synthesis (cfs) that does not require cAMP-CRP for activation of flhDC expression (22, 33). The regulatory region of this cfs strain was found to be otherwise identical to the regulatory region of Y10 and MG1655. However, when a crp deletion was transduced into the −10T mutant, or generated by lambda Red recombination, all cells were nonmotile. We were unable to obtain the original cfs strain for direct comparison.
The second promoter mutation recovered was a −18 G-to-T change in the 17-nt spacer region between the −10 and −35 promoter elements, resulting in a >90% motile strain that migrated at 80% the rate of the −10T mutant (see Fig. S2 in the supplemental material). This strain was motile at 37°C, and it required an intact crp gene. No consensus sequence exists among σ70-dependent promoter spacers, so it seemed likely that the −18T mutation might in some way affect local DNA conformation and hence spacer interaction with σ70. Indeed, altered curvature due to this substitution was predicted by curvature analyses, as well as for a number of other substitutions at various sites in the spacer. However, different programs gave different results (data not shown). Nevertheless, to test for possible correspondence between altered curvature and sequence, all possible single nucleotide mutations from −13 to −23 were made. Eight other significantly activating substitutions (>50% motility) were found that gave rise to rapid spreading with well-formed chemotaxis rings on motility plates, mostly changes to A or T: −14AT, −16CA, −16CG, −17AT, −18GA, −19TA, −20GA, and −20GT (Fig. 5). However, these particular activating mutations did not necessarily correspond to predictions of changes in curvature. Tests for altered curvature were conducted by running DNA fragments containing mutations near or at their centers on native acrylamide gels. PCR-amplified fragments of 48 bp were run for every mutation, and annealed 48- or 29-bp oligonucleotide fragments were run for a subset. No differences in migration rates were detected among any of them. Examples are shown in Fig. S4 in the supplemental material. Given the potential limitation of demonstrating conformational changes of DNA fragments with single substitutions on polyacrylamide gels, it remains possible that altered curvature plays a role in the effect of the stimulating spacer mutations.
FIG 5.
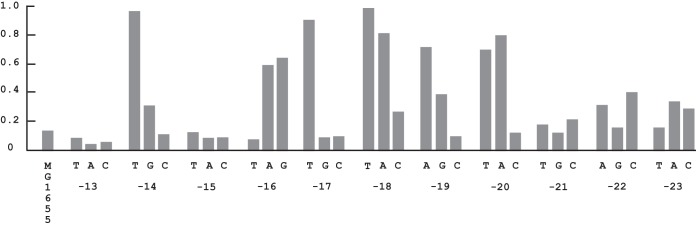
Diameters of strains with single nucleotide substitutions in the promoter spacer region on motility plates relative to MG1655(Seq) after 7 h at 30°C. Locations are given relative to the start of transcription. Shown are the averages from two separate assays; the standard deviations were under 8%.
In addition to single nucleotide mutations, a double mutation generating an extended −10 element (TG at positions −15 and −14) was constructed, which resulted in a strain that was >99% motile. This is consistent with findings that an extended −10 element can compensate for poor matches to the σ70 consensus sequence (49–51).
Two mutations map to the CAP-binding region.
One of the two mutations found near the cAMP-CRP binding site was a deletion of a T 4 bp downstream, at −57 (Fig. 4). This single mutation resulted in cell populations that were >96% motile. Two other deletions were made in the region, at −59 and −60 (the nucleotide at position −58 is also a T), that resulted in cell populations that were also >96% motile. A deletion at −56 gave ∼75% motility. The −57, −59, and −60 deletion mutants were highly motile at 37°C (>90%); the one at −56 was less so (<50%). Spreading on soft-agar plates is shown in Fig. S2 in the supplemental material. The proximity to the cAMP-CRP binding site suggests that these deletions affect the interaction between cAMP-CRP and RNA polymerase.
The other mutation in this region was a C-to-A mutation at −63, near the proximal end of the cAMP-CRP binding region. This was the least robust mutant recovered, with only about 40% of cells motile at 30°C and none at 37°C. The −63A mutation does not improve the match to the consensus sequence for binding by cAMP-CRP (Fig. 4) (52). We note that the nucleotide at this site was also reported to be an A in a strain (FB8) considered to be wild type for motility (22).
Some mutations can stimulate flhDC expression in the absence of H-NS.
Strains mutant for hns have been reported to be nonmotile and have greatly diminished expression of flhDC (22, 28, 53). Consistent with these reports, when hns deletions were introduced into Y10 and MG1655, cells were completely nonmotile (assayed at room temperature). To test whether IS elements or single nucleotide mutations could suffice to stimulate flhDC expression in the absence of H-NS, deletions of hns were transduced into a subset of mutants (for Y10 variant hns strains, see Table S1 in the supplemental material). All such hns strains, including the donor strain, grew much more slowly than the parental strains at room temperature and at 30°C and less so at 37°C (∼34- versus 31-min doubling times in TB). Growth defects of hns strains at room temperature have been reported previously (54). In addition, hns cells formed clumps at lower temperatures, so motility was tested at 37°C.
The −10T hns mutant was highly motile when grown in liquid. The same was true for hns mutants with an IS5 element at −100, as well as for MG1655(Seq) hns with its IS1 element at −108. Thus, it appears that these particular mutations were able to substantially overcome the requirement for H-NS. Other hns strains carrying IS elements or other single nucleotide mutations were less or not at all motile in liquid and on motility plates. It was not possible to determine the extent of motility because, while cells grown at 37°C tended not to form clumps, motile cells rapidly stuck to coverslips. In spite of their high motility, these strains migrated more slowly on motility plates than the parental strains. Induced expression of plasmid-based flhDC in the highly motile hns strains improved migration, although spreading rates were also lower than those of their parental strains (Fig. 6). Possible contributing factors that could account for this are as follows: (i) the growth rate is slightly lower at 37°C in hns strains; (ii) the stickiness of hns cells could interfere with migration; (iii) ability to perform chemotaxis could be affected by altered metabolism of chemoattractants or expression of chemoreceptors; (iv) since H-NS is a component of wild-type flagellar motors thought to provide motor stability (55, 56), motors devoid of H-NS could be subfunctional when migrating through media such as soft agar. Regardless, it appears that H-NS is still required for full expression of flhDC in these strains.
FIG 6.

Comparison of the −10T mutant with its derivative hns strains on motility plates. (Left) −10T. (Middle) −10T hns. (Right) −10T hns expressing flhDC from pVN15. The plates contained 0.2% l-arabinose and were incubated at 37°C for 6 h.
DISCUSSION
Expression of flhDC is too low to significantly initiate flagellar synthesis in strains Y10 and MG1655 under growth conditions typical for studies of motility (references 14 and 34 and this study). In this study, 50 highly motile mutants of Y10 were isolated, all of which were altered in the flhDC-uspC intergenic region. Forty-six of these contained IS elements upstream of the cAMP-CRP binding site, nearly half at sites between 240 and 476 bp distant from the transcriptional start site. Four stimulating single nucleotide mutations were also recovered, two in the promoter region and two near or in the cAMP-CRP binding region. These differing mutations would be expected to stimulate flhDC expression in distinct ways while remaining dependent on the cAMP-CRP transcriptional activator.
Promoter region mutations.
The −10 C-to-T mutation in the −10 hexamer element of the flhDC promoter results in a match of 5 out of 6 bp to the σ70 consensus (Fig. 4). This element is recognized initially as a duplex by domain 2 of σ70 and subsequently melts (beginning at approximately −11) during open-complex formation with RNA polymerase holoenzyme (RNAP). The −10T mutation may improve binding recognition by subdomain 2.4 of σ70, and/or it may help stabilize the open complex preceding elongation via interaction with subdomain 2.3 (for reviews on σ70 domain interactions with promoters and RNAP transcription initiation, see references 57–60). Contrary to what has been reported previously for a cfs strain with the same mutation (22, 33), the presence of cAMP-CRP was required for activation of flhDC transcription. We cannot account for this discrepancy.
The length, composition, and sequence of the promoter spacer greatly influence promoter strength. A length of 17 bp is generally optimal for productive positioning of σ70 recognition domains 2 and 4 with respect to the −10 and −35 elements of the promoter (61–63). Spacers with higher GC content can result in weaker activity (64). A −15TG−14 motif (called, when it includes the −10 element, the extended −10 element) that is present in about 20% of E. coli promoters (65) can compensate for weak −35 or −10 elements through its specific interaction with σ70 domain 3.0, stabilizing RNAP-DNA interactions (49–51). The flhDC spacer is 17 bp but has a high GC content (59%). It does not contain the −15TG−14 motif, but when the motif was introduced, it compensated for mismatches to the −10 and/or −35 hexamer consensuses (the latter flhDC hexamer matches only 3 out of 6 bp).
While there is no consensus sequence for spacers, studies of specific promoters with significantly altered spacer sequences and compositions have led to the proposal that RNAP senses overall spacer structure and that spacer flexibility is required to maintain contact with the promoter DNA during transcription initiation (64, 66–69). How RNAP might sense spacer conformation is not clear. There may be contact between the Zn2+-binding domain of the β′ subunit and the distal portion of the spacer, as indicated from structural studies of Thermus aquaticus RNAP (70). Residue R451 in the linker between domains 2 and 3 of σ70 interacts with spacer position −18 in a manner that is affected by local curvature (69), and σ70 region 1.1, while not contacting DNA, affects the transition from the closed to the open complex in a way that is influenced by spacer structure (68).
The isolation of the stimulating −18T spacer mutation and the discovery of eight other significantly stimulating single nucleotide substitutions (at positions −14, −16, −17, −18, −19, and −20) highlight the importance of the spacer in determining promoter strength. Relatively few examples of single stimulating substitutions in the spacer have been reported in E. coli. One example is a substitution at position −23 of the gal P1 promoter, a cAMP-CRP-dependent class II promoter in which the cAMP-CRP binding site replaces the −35 element (71). This substitution is thought to increase the twist angle between the −10 element and the cAMP-CRP binding site, presumably affecting the interaction between cAMP-CRP and RNAP. Other examples of stimulation are single substitutions at positions −14, −15, −17, and −18 at the cbpA promoter and at −18 at the LEE1 promoter (69). The −18 substitutions are associated with increased curvature. In the case of the flhDC spacer substitutions, it was not possible to show that altered curvature was responsible for their stimulatory effects. Mobility shifts on polyacrylamide gels were not observed for fragments containing these mutations. However, small conformational changes may not always be detectable by this method. Also, analyses predicting altered curvature did not consistently match the effects of various single nucleotide substitutions. It remains possible that single nucleotide substitutions in the flhDC spacer do affect local conformation. However, there may also be some sequence specificity to the interactions between specific sites in the spacer and RNAP. The mechanism by which single nucleotide substitutions and specific sequences in the spacer region can affect RNAP function will require further study. In any case, these mutations demonstrate that subtle contextual changes in spacer sequences can have profound effects.
We consider it unlikely that any of these spacer mutations generate a new −10 element and hence a new transcriptional start site, as none of the permutations would create a strong −10 element. In addition, a new −10 element would result in a displaced, weaker or nonexistent −35 element (Fig. 4) and would alter the spacing between the promoter and the cAMP-CRP binding site (see below).
cAMP-CRP binding region mutations.
The flhDC promoter does not have an UP element, an ∼22-bp AT-rich region to which the C-terminal domains of one or both α subunits (α-CTDs) of RNAP preferentially bind, stabilizing the interaction of the enzyme complex with the promoter (see reference 72 for a review). Rather, it has a very GC-rich region immediately upstream of the −35 element, which likely contributes to the promoter's dependency on cAMP-CRP for activation of transcription. At class I promoters, cAMP-CRP interacts with one of the α-CTDs, facilitating its binding to DNA and thus recruitment of RNAP to the core promoter elements (see reference 73 for a review). Because the interface between cAMP-CRP and the α-CTD is small (74) and cAMP-CRP must be bound on the same side of the helix as RNAP for interaction, the orientation and spacing between this activator and RNAP are critical for its function (75, 76). For σ70-dependent class I promoters, the cAMP-CRP binding site is often 61.5 bp from the transcription start site (75). The cAMP-CRP binding site of the class I flhDC promoter is centered one helical turn farther away, 71.5 bp upstream of the transcription start site. This distance is similar to that for the malT promoter, centered at −70.5 (77). At the malT promoter, it has been shown that a spacing shift of the cAMP-CRP binding site by as little as 1 or 2 bp can have a large effect on transcriptional activation by cAMP-CRP (76). The stimulating −57ΔT deletion mutation isolated in this study, and the three others that were generated, all adjacent to the cAMP-CRP binding site, may well optimize the alignment between CAP and the α-CTD.
The final single nucleotide mutation isolated, a C-to-A change at −63, is within the CAP-binding consensus sequence, but it does not improve the match (Fig. 4). This is the weakest of the stimulating single nucleotide mutations isolated in this study, and the strain containing it is nonmotile at 37°C. Presumably, this mutation also affects the strength of interaction between cAMP-CRP and the α-CTD, but this remains to be determined.
Long-range effects of IS elements and the role of H-NS.
Transcriptional activation by IS elements is a well-established phenomenon that can occur via various mechanisms. Initiation of transcription can occur from within an IS element (42, 78). In fortuitous cases, when inserted into a weak promoter, an IS element can form a “hybrid promoter” due to an internal −35 hexamer in the inverted-repeat region of some IS elements (39). Insertion into a repressor binding site can relieve repression, and this could explain, in part, the activating effect on the flhDC operon by those IS elements that are inserted in or near the LrhA repressor binding site (34)or binding sites of other repressors such as HdfR and MatA that have yet to be mapped. [The IS element at −108 of MG1655(Seq) does not appear to interfere with the repressing effect of HdfR, since a deletion in hdfR partially restores motility in an hns background (79).] In fact, insertions of the thyA gene at sites near the LrhA repressor binding region also activate flhDC expression (7). However, these mechanisms cannot account for orientation-independent activation by IS elements hundreds of base pairs from the promoter or for that by a divergently transcribed kanamycin gene inserted at −319. The long-range effects of IS elements found in this study suggest the involvement of a higher-order regulating structure encompassing much of the AT-rich flhDC-uspC intergenic region and that insertion of coding sequences of typical length and composition (more GC rich) disrupts the structure. Such a structure could result from the inherent DNA topology of this AT-rich region, or it could result from a nucleoprotein complex, perhaps organized by H-NS. H-NS is known to bind to the flhDC regulatory region (22) and likely binds throughout the intergenic region, given its preference for interacting with AT-rich DNA (79, 80). It is possible that a higher-order structure renders the region in a topologically unfavorable state for σ70 binding at the relatively weak flhDC promoter, especially at higher temperatures. Alternatively, it may enhance the interaction of repressors with their binding sites. Disruption of repressing local DNA topology based in part on AT-rich composition has been proposed for the activating effects of IS elements or an inserted kanamycin gene for the cryptic bgl operon (40, 81). IS elements have also been shown to relieve H-NS-mediated repression of the cryptic ade operon (41). In these cases, the activating IS elements were located within ∼110 bp of the promoters.
H-NS generally acts as a repressor of transcription. If the architecture of the intergenic region is organized by H-NS, one might expect H-NS to act as a repressor for the flhDC operon, given that disruption of the region by IS elements stimulates expression. Wang and Wood found that overexpression of H-NS repressed motility (35). However, Paul et al. found that overexpression of H-NS did not repress motility but rather improved it under conditions of high salt or high agar concentrations (56). Genetic data have shown that H-NS is required for flhDC expression (22, 25, 53) and that it mitigates the effect of the HdfR repressor. Interestingly, in vitro and in vivo experiments with a plasmid containing the flhDC promoter and the upstream regulatory region showed that transcription was inhibited in the presence of H-NS (22). If the region corresponding to the mRNA leader sequence was included, however, H-NS stimulated expression in vivo. Similar results were found when the regulatory region was located on the chromosome. In the current study, H-NS was not required for stimulation of flagellar synthesis in the presence of the strong −10 promoter element generated by the −10T mutation or for strains with IS elements closest to the cAMP-CRP binding site (at −100 and −108). A similar result has been reported for an hns strain with an IS5 at −100 (35). Other mutant strains were less motile or nonmotile in the absence of H-NS, consistent with the general positive role of H-NS in flhDC expression. At this time, the mechanism by which H-NS influences flhDC expression remains unclear.
The flagellum is an energetically costly organelle, and the shift to or from motility is regulated by internal and external cues that directly or indirectly control flhDC expression. Thus, the flhDC regulatory region must be poised to respond appropriately. The activating mutations identified in this study would presumably lead to compromised control of flhDC expression and may not be beneficial in the natural environment. For example, for some species, biofilm formation and dispersal involve differentiable states of both motile and nonmotile cells. One study reported that the highly motile MG1655(Seq) strain with its activating IS1 element was outcompeted in the mouse intestine by nonmotile mutants that occurred over time during colonization (82). These mutants had developed deletions downstream of the IS1 element in the flhDC operon. Similar findings have been reported by Leatham et al. (83) and Fabich et al. (8). These studies suggest that FlhDC represses the expression of certain carbon-metabolizing genes, slowing growth, and imply that the inability to turn off flhDC expression would likely reduce fitness. We note that the closely related Shigella spp., which also form biofilms, are nonmotile, mostly due to various deletions and insertions related to the action of IS elements in the flhDC region (84). In contrast, another study with a K-12 strain found that an activating upstream IS5 element was associated with increased biofilm formation (adherence to polystyrene and glass wool) (35). The relative fitness of cells expressing high levels of flhDC deserves further exploration, as do the mechanisms involved in flhDC regulation.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grant AI016478 from the U.S. National Institutes of Health.
We thank Vedavalli S. J. Nathan for technical assistance.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00455-15.
REFERENCES
- 1.Wang S, Fleming RT, Westbrook EM, Matsumura P, McKay DB. 2006. Structure of the Escherichia coli FlhDC complex, a prokaryotic heteromeric regulator of transcription. J Mol Biol 355:798–808. doi: 10.1016/j.jmb.2005.11.020. [DOI] [PubMed] [Google Scholar]
- 2.Chilcott GS, Hughes KT. 2000. Coupling of flagellar gene expression to flagellar assembly in Salmonella enterica serovar Typhimurium and Escherichia coli. Microbiol Mol Biol Rev 64:694–708. doi: 10.1128/MMBR.64.4.694-708.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chevance FF, Hughes KT. 2008. Coordinating assembly of a bacterial macromolecular machine. Nat Rev Microbiol 6:455–465. doi: 10.1038/nrmicro1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prüss BM, Liu X, Hendrickson W, Matsumura P. 2001. FlhD/FlhC-regulated promoters analyzed by gene array and lacZ gene fusions. FEMS Microbiol Lett 197:91–97. doi: 10.1111/j.1574-6968.2001.tb10588.x. [DOI] [PubMed] [Google Scholar]
- 5.Stafford GP, Ogi T, Hughes C. 2005. Binding and transcriptional activation of non-flagellar genes by the Escherichia coli flagellar master regulator FlhD2C2. Microbiology 151:1779–1788. doi: 10.1099/mic.0.27879-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhao K, Liu M, Burgess RR. 2007. Adaptation in bacterial flagellar and motility systems: from regulon members to ‘foraging’-like behavior in E. coli. Nucleic Acids Res 35:4441–4452. doi: 10.1093/nar/gkm456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fitzgerald DM, Bonocora RP, Wade JT. 2014. Comprehensive mapping of the Escherichia coli flagellar regulatory network. PLoS Genet 10:e1004649. doi: 10.1371/journal.pgen.1004649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fabich AJ, Leatham MP, Grissom JE, Wiley G, Lai H, Najar F, Roe BA, Cohen PS, Conway T. 2011. Genotype and phenotypes of an intestine-adapted Escherichia coli K-12 mutant selected by animal passage for superior colonization. Infect Immun 79:2430–2439. doi: 10.1128/IAI.01199-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Soutourina OA, Bertin PN. 2003. Regulation cascade of flagellar expression in Gram-negative bacteria. FEMS Microbiol Rev 27:505–523. doi: 10.1016/S0168-6445(03)00064-0. [DOI] [PubMed] [Google Scholar]
- 10.Wei BL, Brun-Zinkernagel AM, Simecka JW, Prüss BM, Babitzke P, Romeo T. 2001. Positive regulation of motility and flhDC expression by the RNA-binding protein CsrA of Escherichia coli. Mol Microbiol 40:245–256. doi: 10.1046/j.1365-2958.2001.02380.x. [DOI] [PubMed] [Google Scholar]
- 11.De Lay N, Gottesman S. 2012. A complex network of small non-coding RNAs regulate motility in Escherichia coli. Mol Microbiol 86:524–538. doi: 10.1111/j.1365-2958.2012.08209.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thomason MK, Fontaine F, De Lay N, Storz G. 2012. A small RNA that regulates motility and biofilm formation in response to changes in nutrient availability in Escherichia coli. Mol Microbiol 84:17–35. doi: 10.1111/j.1365-2958.2012.07965.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shin S, Park C. 1995. Modulation of flagellar expression in Escherichia coli by acetyl phosphate and the osmoregulator OmpR. J Bacteriol 177:4696–4702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lehnen D, Blumer C, Polen T, Wackwitz B, Wendisch VF, Unden G. 2002. LrhA as a new transcriptional key regulator of flagella, motility and chemotaxis genes in Escherichia coli. Mol Microbiol 45:521–532. doi: 10.1046/j.1365-2958.2002.03032.x. [DOI] [PubMed] [Google Scholar]
- 15.Blumer C, Kleefeld A, Lehnen D, Heintz M, Dobrindt U, Nagy G, Michaelis K, Emödy L, Polen T, Rachel R, Wendisch VF, Unden G. 2005. Regulation of type 1 fimbriae synthesis and biofilm formation by the transcriptional regulator LrhA of Escherichia coli. Microbiology 151:3287–3298. doi: 10.1099/mic.0.28098-0. [DOI] [PubMed] [Google Scholar]
- 16.Ko M, Park C. 2000. H-NS-dependent regulation of flagellar synthesis is mediated by a LysR family protein. J Bacteriol 182:4670–4672. doi: 10.1128/JB.182.16.4670-4672.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lehti TA, Bauchart P, Dobrindt U, Korhonen TK, Westerlund-Wikström B. 2012. The fimbriae activator MatA switches off motility in Escherichia coli by repression of the flagellar master operon flhDC. Microbiology 158:1444–1455. doi: 10.1099/mic.0.056499-0. [DOI] [PubMed] [Google Scholar]
- 18.Francez-Charlot A, Laugel B, Van Gemert A, Dubarry N, Wiorowski F, Castanié-Cornet MP, Gutierrez C, Cam K. 2003. RcsCDB His-Asp phosphorelay system negatively regulates the flhDC operon in Escherichia coli. Mol Microbiol 49:823–832. [DOI] [PubMed] [Google Scholar]
- 19.Lehti TA, Heikkinen J, Korhonen TK, Westerlund-Wikström B. 2012. The response regulator RcsB activates expression of Mat fimbriae in meningitic Escherichia coli. J Bacteriol 194:3475–3485. doi: 10.1128/JB.06596-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pesavento C, Hengge R. 2012. The global repressor FliZ antagonizes gene expression by σS-containing RNA polymerase due to overlapping DNA binding specificity. Nucleic Acids Res 40:4783–4793. doi: 10.1093/nar/gks055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yokota T, Gots JS. 1970. Requirement of adenosine 3′, 5′-cyclic phosphate for flagella formation in Escherichia coli and Salmonella typhimurium. J Bacteriol 103:513–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Soutourina O, Kolb A, Krin E, Laurent-Winter C, Rimsky S, Danchin A, Bertin P. 1999. Multiple control of flagellum biosynthesis in Escherichia coli: role of H-NS protein and the cyclic AMP-catabolite activator protein complex in transcription of the flhDC master operon. J Bacteriol 181:7500–7508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sperandio V, Torres AG, Kaper JB. 2002. Quorum sensing Escherichia coli regulators B and C (QseBC): a novel two-component regulatory system involved in the regulation of flagella and motility by quorum sensing in E. coli. Mol Microbiol 43:809–821. doi: 10.1046/j.1365-2958.2002.02803.x. [DOI] [PubMed] [Google Scholar]
- 24.Clarke MB, Sperandio V. 2005. Transcriptional regulation of flhDC by QseBC and σ28 (FliA) in enterohaemorrhagic Escherichia coli. Mol Microbiol 57:1734–1749. doi: 10.1111/j.1365-2958.2005.04792.x. [DOI] [PubMed] [Google Scholar]
- 25.Yamada H, Muramatsu S, Mizuno T. 1990. An Escherichia coli protein that preferentially binds to sharply curved DNA. J Biochem 108:420–425. [DOI] [PubMed] [Google Scholar]
- 26.Pérez-Martín J, Rojo F, de Lorenzo V. 1994. Promoters responsive to DNA bending: a common theme in prokaryotic gene expression. Microbiol Rev 58:268–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gabrielian AE, Landsman D, Bolshoy A. 1999. Curved DNA in promoter sequences. In Silico Biol 1:183–196. [PubMed] [Google Scholar]
- 28.Bertin P, Terao E, Lee EH, Lejeune P, Colson C, Danchin A, Collatz E. 1994. The H-NS protein is involved in the biogenesis of flagella in Escherichia coli. J Bacteriol 176:5537–5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pontiggia A, Negri A, Beltrame M, Bianchi ME. 1993. Protein HU binds specifically to kinked DNA. Mol Microbiol 7:343–350. doi: 10.1111/j.1365-2958.1993.tb01126.x. [DOI] [PubMed] [Google Scholar]
- 30.Mangan MW, Lucchini S, Ó Cróinín T, Fitzgerald S, Hinton JC, Dorman CJ. 2011. Nucleoid-associated protein HU controls three regulons that coordinate virulence, response to stress and general physiology in Salmonella enterica serovar Typhimurium. Microbiology 157:1075–1087. doi: 10.1099/mic.0.046359-0. [DOI] [PubMed] [Google Scholar]
- 31.Nishida S, Mizushima T, Miki T, Sekimizu K. 1997. Immotile phenotype of an Escherichia coli mutant lacking the histone-like protein HU. FEMS Microbiol Lett 150:297–301. doi: 10.1016/S0378-1097(97)00134-1. [DOI] [PubMed] [Google Scholar]
- 32.Soutourina OA, Krin E, Laurent-Winter C, Hommais F, Danchin A, Bertin PN. 2002. Regulation of bacterial motility in response to low pH in Escherichia coli: the role of H-NS protein. Microbiology 148:1543–1551. [DOI] [PubMed] [Google Scholar]
- 33.Silverman M, Simon M. 1974. Characterization of Escherichia coli flagellar mutants that are insensitive to catabolite repression. J Bacteriol 120:1196–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barker CS, Prüss BM, Matsumura P. 2004. Increased motility of Escherichia coli by insertion sequence element integration into the regulatory region of the flhD operon. J Bacteriol 186:7529–7537. doi: 10.1128/JB.186.22.7529-7537.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang X, Wood TK. 2011. IS5 inserts upstream of the master motility operon flhDC in a quasi-Lamarckian way. ISME J 5:1517–1525. doi: 10.1038/ismej.2011.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee C, Park C. 2013. Mutations upregulating the flhDC operon of Escherichia coli K-12. J Microbiol 51:140–144. doi: 10.1007/s12275-013-2212-z. [DOI] [PubMed] [Google Scholar]
- 37.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bachmann BJ. 1972. Pedigrees of some mutant strains of Escherichia coli K-12. Bacteriol Rev 36:525–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mahillon J, Chandler M. 1998. Insertion sequences. Microbiol Mol Biol Rev 62:725–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schnetz K, Rak B. 1992. IS5: a mobile enhancer of transcription in Escherichia coli. Proc Natl Acad Sci U S A 89:1244–1248. doi: 10.1073/pnas.89.4.1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Petersen C, Møller LB, Valentin-Hansen P. 2002. The cryptic adenine deaminase gene of Escherichia coli. Silencing by the nucleoid-associated DNA-binding protein, H-NS, and activation by insertion elements. J Biol Chem 277:31373–31380. [DOI] [PubMed] [Google Scholar]
- 42.Zhao K, Liu M, Burgess RR. 2010. Promoter and regulon analysis of nitrogen assimilation factor, σ54, reveal alternative strategy for E. coli MG1655 flagellar biosynthesis. Nucleic Acids Res 38:1273–1283. doi: 10.1093/nar/gkp1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jáuregui R, Abreu-Goodger C, Moreno-Hagelsieb G, Collado-Vides J, Merino E. 2003. Conservation of DNA curvature signals in regulatory regions of prokaryotic genes. Nucleic Acids Res 31:6770–6777. doi: 10.1093/nar/gkg882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oliver JL, Marín A. 1996. A relationship between GC content and coding-sequence length. J Mol Evol 43:216–223. doi: 10.1007/BF02338829. [DOI] [PubMed] [Google Scholar]
- 45.Adler J, Templeton B. 1967. The effect of environmental conditions on the motility of Escherichia coli. J Gen Microbiol 46:175–184. doi: 10.1099/00221287-46-2-175. [DOI] [PubMed] [Google Scholar]
- 46.Morrison RB, McCapra J. 1961. Flagellar changes in Escherichia coli induced by temperature of the environment. Nature 192:774–776. doi: 10.1038/192774a0. [DOI] [Google Scholar]
- 47.Hawley DK, McClure WR. 1983. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res 11:2237–2255. doi: 10.1093/nar/11.8.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Djordjevic M. 2011. Redefining Escherichia coli σ70 promoter elements: −15 motif as a complement of the −10 motif. J Bacteriol 193:6305–6314. doi: 10.1128/JB.05947-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Keilty S, Rosenberg M. 1987. Constitutive function of a positively regulated promoter reveals new sequences essential for activity. J Biol Chem 262:6389–6395. [PubMed] [Google Scholar]
- 50.Kumar A, Malloch RA, Fujita N, Smillie DA, Ishihama A, Hayward RS. 1993. The minus 35-recognition region of Escherichia coli sigma 70 is inessential for initiation of transcription at an “extended minus 10” promoter. J Mol Biol 232:406–418. doi: 10.1006/jmbi.1993.1400. [DOI] [PubMed] [Google Scholar]
- 51.Hook-Barnard I, Johnson XB, Hinton DM. 2006. Escherichia coli RNA polymerase recognition of a σ70-dependent promoter requiring a −35 DNA element and an extended −10 TGn motif. J Bacteriol 188:8352–8359. doi: 10.1128/JB.00853-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lawson CL, Swigon D, Murakami KS, Darst SA, Berman HM, Ebright RH. 2004. Catabolite activator protein: DNA binding and transcription activation. Curr Opin Struct Biol 14:10–20. doi: 10.1016/j.sbi.2004.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ko M, Park C. 2000. Two novel flagellar components and H-NS are involved in the motor function of Escherichia coli. J Mol Biol 303:371–382. doi: 10.1006/jmbi.2000.4147. [DOI] [PubMed] [Google Scholar]
- 54.Dersch P, Kneip S, Bremer E. 1994. The nucleoid-associated DNA-binding protein H-NS is required for the efficient adaptation of Escherichia coli K-12 to a cold environment. Mol Gen Genet 245:255–259. [DOI] [PubMed] [Google Scholar]
- 55.Donato GM, Kawula TH. 1998. Enhanced binding of altered H-NS protein to flagellar rotor protein FliG causes increased flagellar rotational speed and hypermotility in Escherichia coli. J Biol Chem 273:24030–24036. doi: 10.1074/jbc.273.37.24030. [DOI] [PubMed] [Google Scholar]
- 56.Paul K, Carlquist WC, Blair DF. 2011. Adjusting the spokes of the flagellar motor with the DNA-binding protein H-NS. J Bacteriol 193:5914–5922. doi: 10.1128/JB.05458-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Murakami KS, Darst SA. 2003. Bacterial RNA polymerases: the wholo story. Curr Opin Struct Biol 13:31–39. doi: 10.1016/S0959-440X(02)00005-2. [DOI] [PubMed] [Google Scholar]
- 58.Hook-Barnard IG, Hinton DM. 2007. Transcription initiation by mix and match elements: flexibility for polymerase binding to bacterial promoters. Gene Regul Syst Bio 1:275–293. [PMC free article] [PubMed] [Google Scholar]
- 59.Haugen SP, Ross W, Gourse RL. 2008. Advances in bacterial promoter recognition and its control by factors that do not bind DNA. Nat Rev Microbiol 6:507–519. doi: 10.1038/nrmicro1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee DJ, Minchin SD, Busby SJ. 2012. Activating transcription in bacteria. Annu Rev Microbiol 66:125–152. doi: 10.1146/annurev-micro-092611-150012. [DOI] [PubMed] [Google Scholar]
- 61.Stefano JE, Gralla JD. 1982. Spacer mutations in the lac ps promoter. Proc Natl Acad Sci U S A 79:1069–1072. doi: 10.1073/pnas.79.4.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mulligan ME, Brosius J, McClure WR. 1985. Characterization in vitro of the effect of spacer length on the activity of Escherichia coli RNA polymerase at the TAC promoter. J Biol Chem 260:3529–3538. [PubMed] [Google Scholar]
- 63.Dombroski AJ, Johnson BD, Lonetto M, Gross CA. 1996. The sigma subunit of Escherichia coli RNA polymerase senses promoter spacing. Proc Natl Acad Sci U S A 93:8858–8862. doi: 10.1073/pnas.93.17.8858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu M, Tolstorukov M, Zhurkin V, Garges S, Adhya S. 2004. A mutant spacer sequence between −35 and −10 elements makes the Plac promoter hyperactive and cAMP receptor protein-independent. Proc Natl Acad Sci U S A 101:6911–6916. doi: 10.1073/pnas.0401929101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Burr T, Mitchell J, Kolb A, Minchin S, Busby S. 2000. DNA sequence elements located immediately upstream of the −10 hexamer in Escherichia coli promoters: a systematic study. Nucleic Acids Res 28:1864–1870. doi: 10.1093/nar/28.9.1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Auble DT, Allen TL, deHaseth PL. 1986. Promoter recognition by Escherichia coli RNA polymerase. Effects of substitutions in the spacer DNA separating the −10 and −35 regions. J Biol Chem 261:11202–11206. [PubMed] [Google Scholar]
- 67.Jensen PR, Hammer K. 1998. The sequence of spacers between the consensus sequences modulates the strength of prokaryotic promoters. Appl Environ Microbiol 64:82–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hook-Barnard IG, Hinton DM. 2009. The promoter spacer influences transcription initiation via σ70 region 1.1 of Escherichia coli RNA polymerase. Proc Natl Acad Sci U S A 106:737–742. doi: 10.1073/pnas.0808133106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Singh SS, Typas A, Hengge R, Grainger DC. 2011. Escherichia coli σ70 senses sequence and conformation of the promoter spacer region. Nucleic Acids Res 39:5109–5118. doi: 10.1093/nar/gkr080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Murakami KS, Masuda S, Campbell EA, Muzzin O, Darst SA. 2002. Structural basis of transcription initiation: an RNA polymerase holoenzyme-DNA complex. Science 296:1285–1290. doi: 10.1126/science.1069595. [DOI] [PubMed] [Google Scholar]
- 71.Busby S, Truelle N, Spassky A, Dreyfus M, Buc H. 1984. The selection and characterisation of two novel mutations in the overlapping promoters of the Escherichia coli galactose operon. Gene 28:201–209. doi: 10.1016/0378-1119(84)90257-9. [DOI] [PubMed] [Google Scholar]
- 72.Gourse RL, Ross W, Gaal T. 2000. UPs and downs in bacterial transcription initiation: the role of the alpha subunit of RNA polymerase in promoter recognition. Mol Microbiol 37:687–695. doi: 10.1046/j.1365-2958.2000.01972.x. [DOI] [PubMed] [Google Scholar]
- 73.Busby S, Ebright RH. 1999. Transcription activation by catabolite activator protein (CAP). J Mol Biol 293:199–213. doi: 10.1006/jmbi.1999.3161. [DOI] [PubMed] [Google Scholar]
- 74.Benoff B, Yang H, Lawson CL, Parkinson G, Liu J, Blatter E, Ebright YW, Berman HM, Ebright RH. 2002. Structural basis of transcription activation: the CAP-αCTD-DNA complex. Science 297:1562–1566. doi: 10.1126/science.1076376. [DOI] [PubMed] [Google Scholar]
- 75.Gaston K, Bell A, Kolb A, Buc H, Busby S. 1990. Stringent spacing requirements for transcription activation by CRP. Cell 62:733–743. doi: 10.1016/0092-8674(90)90118-X. [DOI] [PubMed] [Google Scholar]
- 76.Déthiollaz S, Eichenberger P, Geiselmann J. 1996. Influence of DNA geometry on transcriptional activation in Escherichia coli. EMBO J 15:5449–5458. [PMC free article] [PubMed] [Google Scholar]
- 77.Chapon C, Kolb A. 1983. Action of CAP on the malT promoter in vitro. J Bacteriol 156:1135–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Charlier D, Piette J, Glansdorff N. 1982. IS3 can function as a mobile promoter in E. coli. Nucleic Acids Res 10:5935–5948. doi: 10.1093/nar/10.19.5935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yamada H, Yoshida T, Tanaka K, Sasakawa C, Mizuno T. 1991. Molecular analysis of the Escherichia coli hns gene encoding a DNA-binding protein, which preferentially recognizes curved DNA sequences. Mol Gen Genet 230:332–336. doi: 10.1007/BF00290685. [DOI] [PubMed] [Google Scholar]
- 80.Atlung T, Ingmer H. 1997. H-NS: a modulator of environmentally regulated gene expression. Mol Microbiol 24:7–17. doi: 10.1046/j.1365-2958.1997.3151679.x. [DOI] [PubMed] [Google Scholar]
- 81.Singh J, Mukerji M, Mahadevan S. 1995. Transcriptional activation of the Escherichia coli bgl operon: negative regulation by DNA structural elements near the promoter. Mol Microbiol 17:1085–1092. doi: 10.1111/j.1365-2958.1995.mmi_17061085.x. [DOI] [PubMed] [Google Scholar]
- 82.Gauger EJ, Leatham MP, Mercado-Lubo R, Laux DC, Conway T, Cohen PS. 2007. Role of motility and the flhDC operon in Escherichia coli MG1655 colonization of the mouse intestine. Infect Immun 75:3315–3324. doi: 10.1128/IAI.00052-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Leatham MP, Stevenson SJ, Gauger EJ, Krogfelt KA, Lins JJ, Haddock TL, Autieri SM, Conway T, Cohen PS. 2005. Mouse intestine selects nonmotile flhDC mutants of Escherichia coli MG1655 with increased colonizing ability and better utilization of carbon sources. Infect Immun 73:8039–8049. doi: 10.1128/IAI.73.12.8039-8049.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tominaga A, Lan R, Reeves PR. 2005. Evolutionary changes of the flhDC flagellar master operon in Shigella strains. J Bacteriol 187:4295–4302. doi: 10.1128/JB.187.12.4295-4302.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2:2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Guyer MS, Reed RR, Steitz JA, Low KB. 1981. Identification of a sex-factor-affinity site in E. coli as gamma delta. Cold Spring Harbor Symp Quant Biol 45:135–140. doi: 10.1101/SQB.1981.045.01.022. [DOI] [PubMed] [Google Scholar]
- 87.Blattner FR, Plunkett G III, Bloch CA, Perna NT, Burland V, Riley M, Collado-Vides J, Glasner JD, Rode CK, Mayhew GF, Gregor J, Davis NW, Kirkpatrick HA, Goeden MA, Rose DJ, Mau B, Shao Y. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 88.Guzman LM, Belin D, Carson MJ, Beckwith J. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol 177:4121–4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.