ABSTRACT
A number of investigations of Escherichia coli have suggested that the DNA-binding protein H-NS, in addition to its well-known functions in chromosome organization and gene regulation, interacts directly with the flagellar motor to modulate its function. Here, in a study initially aimed at characterizing the H-NS/motor interaction further, we identify problems and limitations in the previous work that substantially weaken the case for a direct H-NS/motor interaction. Null hns mutants are immotile, largely owing to the downregulation of the flagellar master regulators FlhD and FlhC. We, and others, previously reported that an hns mutant remains poorly motile even when FlhDC are expressed constitutively. In the present work, we use better-engineered strains to show that the motility defect in a Δhns, FlhDC-constitutive strain is milder than that reported previously and does not point to a direct action of H-NS at the motor. H-NS regulates numerous genes and might influence motility via a number of regulatory molecules besides FlhDC. To examine the sources of the motility defect that persists in an FlhDC-constitutive Δhns mutant, we measured transcript levels and overexpression effects of a number of genes in candidate regulatory pathways. The results indicate that H-NS influences motility via multiple regulatory linkages that include, minimally, the messenger molecule cyclic di-GMP, the biofilm regulatory protein CsgD, and the sigma factors σS and σF. The results are in accordance with the more standard view of H-NS as a regulator of gene expression rather than a direct modulator of flagellar motor performance.
IMPORTANCE Data from a number of previous studies have been taken to indicate that the nucleoid-organizing protein H-NS influences motility not only by its well-known DNA-based mechanisms but also by binding directly to the flagellar motor to alter function. In this study, H-NS is shown to influence motility through diverse regulatory pathways, but a direct interaction with the motor is not supported. Previous indications of a direct action at the motor appear to be related to the use of nonnull strains and, in some cases, a failure to effectively bypass the requirement for H-NS in the expression of the flagellar regulon. These findings call for a substantially revised interpretation of the literature concerning H-NS and flagellar motility and highlight the importance of H-NS in diverse regulatory processes involved in the motile-sessile transition.
INTRODUCTION
H-NS is a small DNA-binding protein that occurs widely in Gram-negative bacteria, where it functions to organize chromosomal DNA and regulate hundreds of genes (1–3). Binding of H-NS to the chromosome is not strictly sequence specific but exhibits a preference for certain motifs (4) and for intrinsically curved regions of DNA (5). While the logic underlying the very wide regulatory action of H-NS is not fully understood, generally speaking, it appears to modulate the expression of genes involved in sensing and responding to changes in the environment (6), including genes involved in pathogenesis (7).
A number of studies have given indications that H-NS might interact directly with the flagellar motor. The first of these studies used the yeast two-hybrid method (8) to identify a possible interaction with FliG, a protein of the rotor that functions closely in the generation of torque. A number of studies followed up on this observation to provide what appeared to be a more detailed picture of the H-NS/FliG interaction and its possible contribution to motor function. Certain mutations in H-NS were reported to confer a hypermotile (better-than-wild-type) phenotype on motility plates while at the same time increasing its affinity for FliG (9). H-NS acts indirectly through the transcription factors HdfR and RcsB to stimulate the expression of the flagellar master regulators FlhD and FlhC (10–12), so investigations of the putative motor-binding action of H-NS were carried out by using hns strains in which FlhDC were expressed constitutively from plasmids. The first such study found that an FlhDC-constitutive hns mutant was normally flagellated yet still very poorly motile (13), and on this basis, it was proposed that H-NS might engage in functionally important interactions with the motor. Another study, from our laboratory, described physiological properties of an hns mutant that appeared to indicate that H-NS acts on FliG to stabilize the rotor and enhance motor performance in the face of challenging conditions of increased ionic strength or load (14).
We sought to extend this picture through further characterization of the H-NS/FliG interaction and its influences on motor function. In initial experiments, we noted behaviors indicating that the critical regulatory proteins FlhDC were not expressed at sufficient levels in our previous study (14) and that other studies (9, 13) were likely complicated by the use of nonnull backgrounds. New strains were constructed to enable more reliable characterization of the Δhns, FlhDC-constitutive phenotype. The results differ from those of our previous report and the other literature on the Δhns motility phenotype and the H-NS/motor interaction. We find a motility defect milder than that reported previously (13, 14); mutant variants of H-NS that were believed to confer a hypermotile phenotype (9) appear to function similarly to the wild type. The proposal that H-NS carries out a direct, functionally important function at the flagellar motor appears not to be supported by data from current experiments.
Although less severe than was thought, the motility defect in an FlhDC-constitutive Δhns mutant is still significant, particularly at a relatively low temperature (23°C). To identify the sources of this motility defect, we measured transcript levels and overexpression effects of a number of genes in regulatory pathways that might be expected to connect H-NS with motility. Functionally significant linkages were found to involve the second messenger cyclic di-GMP (c-di-GMP), the biofilm regulator CsgD, the flagellar sigma factor σF, and the stress- and stationary-phase-associated sigma factor σS. The results thus highlight the modulating role of H-NS in the regulatory web orchestrating the motile-sessile transition. The motility phenotype of a Δhns mutant appears to be due to altered gene expression, in accordance with the standard view of H-NS as a DNA-binding regulatory protein.
MATERIALS AND METHODS
Strains and media.
Escherichia coli strains and plasmids are listed in Table 1. Most strains were derivates of wild-type strain MS296, described previously by Ko and Park (13). Deletions and deletion replacements were made by using the lambda red method (15). The tetRA genes were amplified by using primers that included ∼40 bp of homology to regions upstream and downstream of the target. Amplified tetRA DNA was introduced by electroporation into target strains previously transformed with plasmid pKD46, which encodes the lambda recombinase under the control of the ara promoter. Integrants were selected on tetracycline (15 μg/ml) plates and recultured on tetracycline-containing LB plates at 42°C to drive the loss of the pKD46 plasmid, which has a temperature-sensitive origin of replication.
TABLE 1.
Escherichia coli strains and plasmids used in this study
| Strain or plasmid | Relevant genotype and/or property(ies) | Source or reference |
|---|---|---|
| Strains | ||
| DB9 (RP437) | Wild type for motility and chemotaxis | J. S. Parkinson |
| MS365 | Wild-type E. coli strain from C. Park; also named MS296a | 11 |
| MS531 | hns::neo in the background of MS296 from C. Park; also named MS299a | 11 |
| MS1162 | Δhns (hns::FRT-FRT) in MS365 | 14 |
| MS1277 | ΔfliG (fliG::FRT-FRT) in MS365 | 14 |
| MS1278 | Δhns ΔfliG (fliG::FRT-FRT) in MS1162 | 14 |
| MS1517 | ΔycgR::tetRA in MS1162 | This study |
| EK1 | ΔflhDC in MS365 | This study |
| EK2 | ΔflhDC in MS1162 | This study |
| EK3 | ΔflhDC in MS1277 | This study |
| EK4 | ΔflhDC in MS1278 | This study |
| Plasmids | ||
| pRR48 | Ptac expression vector; Ampr | J. S. Parkinson |
| pKG116 | Psalicylate expression vector; Cmr | J. S. Parkinson |
| pKP147 | flhDC in pKG116; Cmr | K. Paul |
| pKP148 | flhDC in pRR48; Ampr | K. Paul |
| pKP94 | hns in pET19B His-FLAG tag; Ampr | K. Paul |
| pKP133 | hns with mutation T108I in pKG116; Cmr | K. Paul |
| pKP179 | hns with mutation A18E in pKG116; Cmr | K. Paul |
| pKP648 | fliG with mutations S117C and G166C in pRR48; Ampr | K. Paul |
| pTBM30 | Ptac expression vector; Ampr | 59 |
| pKP58 | hns in pKG116 | 14 |
| pKP85 | flhDC in pTM30; Ampr | 14 |
| pKP141 | flhDC in pTRC99A; Ampr | 13 |
| pKD46 | Lambda red recombinase gene (arabinose inducible); Ampr | 15 |
| pEK1 | fliG wild type in pET19B His; Ampr | This study |
| pEK4 | fliG H126W in pET19B His; Ampr | This study |
| pEK5 | fliG R160W in pET19B His; Ampr | This study |
| pEK8 | rssB in pKG116; Cmr | This study |
| pEK9 | ydeH in pKG116; Cmr | This study |
| pEK18 | ycdT in pKG116; Cmr | This study |
| pEK21 | fliA in pKG116; Cmr | This study |
| pEK16 | fliF in pKG116; Cmr | This study |
| pEK30 | fliE in pKG116; Cmr | This study |
| pEK29 | yhjH in pKG116; Cmr | This study |
| pEK23 | flhD with mutation L19D in pKP148; Ampr | This study |
| pEK25 | yeaJ in pKG116; Cmr | This study |
| pEK27 | yeaI in pKG116; Cmr | This study |
| pEK28 | yeaI in pRR48; Ampr | This study |
See reference 11.
The Δhns strain, described previously (14), retains 5 starting and 5 ending codons of hns, separated by an FLP recombination target (FRT)-FRT sequence. The flhDC deletion made for the present work retains the 4 starting codons of flhD and 27 presumably untranslated 3′ codons of flhC, flanking a tetRA insert. Deletions extending further into flhC were found to interfere with the expression of the downstream Mocha operon. Plasmid isolation and transformation were performed according to standard procedures (16). TB medium contained (per liter) 10 g tryptone and 5 g NaCl; LB additionally contained 5 g yeast extract. Ampicillin (Amp) was used at 125 μg/ml in liquid media, 100 μg/ml in selective plates, and 50 μg/ml in soft-agar motility plates. Chloramphenicol (Cm) was used at 50 μg/ml in liquid media and selective plates and at 12.5 μg/ml in motility plates. Isopropyl-β-d-thiogalactopyranoside (IPTG) and sodium salicylate were prepared as aqueous 0.1 M and 10 mM stocks, respectively, and used at the concentrations indicated. DNA sequencing and oligonucleotide synthesis were carried out by core facilities at the University of Utah.
Motility assay.
Rates of migration in soft-agar plates were measured as described previously (17). Plates contained TB medium, 0.26% Bacto agar, antibiotics as needed, and inducers (IPTG and salicylate) at the concentrations given in the figures and figure legends. To examine the effects of hns mutations, mutant H-NS proteins were expressed from variants of the salicylate-inducible plasmid pKP58, with wild-type hns on pKP58 serving as a positive control. pKG116, the parent of pKP58, was used as a negative control. Experiments were performed by using either a Δhns strain or, when H-NS-dependent regulation of FlhDC was to be bypassed, a Δhns ΔflhDC strain containing the flhDC-expressing plasmid pKP148, induced at the levels indicated in the text and figures. In experiments examining agar and salt effects, the composition of the motility plates was changed as indicated in the text and figures. Standard plates contained 0.5% (∼86 mM) NaCl.
Protein purification.
Wild-type or mutant (H126W and R160W) fliG genes were cloned into plasmid pET-19B, which attaches an N-terminal His10 tag and uses the T7 promoter, and expressed in the T7 polymerase-containing strain BL21(DE3). Wild-type hns was likewise cloned into pET-19B and expressed in BL21(DE). Single colonies of protein-expressing strains were inoculated into 20 ml LB-Amp broth and grown overnight at 37°C. The culture grown overnight was used to inoculate 250 ml of LB-Amp medium (1:100 dilution) and grown to an optical density at 600 nm (OD600) of 0.6. IPTG was added to 0.5 mM, and cultures were shaken overnight at 37°C. Cells were collected by centrifugation (5,000 × g for 20 min at 4°C) and resuspended in 4 ml lysis buffer (50 mM Na-phosphate [pH 8.0], 1 M NaCl, 10 mM imidazole). Following the addition of lysozyme (final concentration, 0.3 mg/ml) and phenylmethylsulfonyl fluoride (PMSF) (1 mM final concentration from a 200 mM stock in ethanol), cells were incubated on ice for 1 h. Cells were lysed by three freeze-thaw cycles followed by sonication (five times for 60 s each with a Branson model 450 at power 4 and 50% duty cycle).
Lysates were centrifuged (10,000 × g for 25 min at 4°C) to remove cellular debris, and the supernatant was applied to a Ni-nitrilotriacetic acid (NTA)-agarose column (Qiagen). The column was washed three times with 5 bed volumes of wash buffer (50 mM Na-phosphate [pH 8.0], 1 M NaCl, 20 mM imidazole) and a fourth time with the same buffer except containing just 0.6 M NaCl. Bound protein was released from the column with elution buffer (50 mM Na-phosphate [pH 8.0], 300 mM NaCl, 250 mM imidazole) and concentrated by ultrafiltration (molecular weight [MW] cutoffs of 12,000 for FliG and 3,000 for H-NS) (Centriprep; Millipore). H-NS protein samples were supplemented with 1% sodium deoxycholate at this step to reduce sticking to the membrane.
Cross-linking.
For experiments examining cross-linking of H-NS and FliG with DMS (dimethyl suberimidate), DTBP (dimethyl 3,3′-dithiobispropionimidate), sulfo-EGS [ethylene glycol bis(sulfosuccinimidyl succinate)], or BS3 [bis(sulfosuccinimidyl)suberate] (Pierce), the buffer contained 25 mM HEPES (pH 8.0) and 10 mM NaCl. Proteins were mixed, and a cross-linking agent was added to a final concentration of 3 or 5 mM (from 50 mM stocks in cross-linking buffer). After 1 h of gentle rotation at room temperature, reactions were stopped by adding Tris-HCl (pH 7.5) (final concentration of 100 mM from a 1 M stock) and incubating the mixture for 15 min. Samples were prepared for electrophoresis by the addition of 2× reducing (for DMS, sulfo-EGS, and BS3 samples) or nonreducing (for DTBP samples) dye, followed by heating at 95°C for 10 min. Samples were electrophoresed and immunoblotted as described below.
For experiments using ANB-NOS (N-5-azido-2-nitrobenzoyloxysuccinimide; Pierce) or ATFB,SE (4-azido-2,3,5,6-tetrafluorobenzoic acid; Life Technologies), the cross-linking buffer contained 25 mM HEPES (pH 8.0) and 10 mM NaCl, and cross-linking reagents were prepared as 50 mM stocks in dimethyl sulfoxide (DMSO), stored in the dark. Cross-linkers were added in the dark to a final concentration of 2 mM or 5 mM and then activated by irradiation for 5 min (Fisher Biotech transilluminator with a maximum of 312 nm), and the reaction then was stopped by the addition of nonreducing dye to the mixture, followed by heating at 95°C for 10 min. Electrophoresis and blotting were performed as described below.
Experiments with EMCS (N-maleimidocaproyl-oxysulfosuccinimide ester; Pierce) were performed by using a solution containing 25 mM HEPES (pH 8.0) and 10 mM NaCl, and the cross-linking reagent was prepared as a 20 mM stock in DMSO. EMCS was added to the proteins to a final concentration of 1 mM EMCS, and the mixture was incubated with gentle rotation for 1 h at room temperature. The reaction was quenched by using Tris-HCl (final concentration, 100 mM), added from a 1 M stock. After 15 min at room temperature, samples were prepared for electrophoresis by adding 2× nonreducing dye and heating the mixture to 95°C for 10 min.
In vivo cross-linking.
Experiments to measure H-NS cross-linking in cells were performed by using the ΔflhDC Δhns strain containing plasmid pKP148 to express FlhDC and plasmid pKP58 to express H-NS (or pKG116 as a negative control). Cells were grown overnight at 37°C in LB containing Amp and Cm and used to inoculate 20 ml of TB medium containing the antibiotics and the inducers salicylate (2.5 μM) and IPTG (0.1 mM), using dilutions (∼100-fold) adjusted to give similar cell densities based on the OD600 of the cultures grown overnight. Cells were grown at 32°C to mid-log phase (∼4 h), and DMS was added to 2 mM, 4 mM, or 5 mM from a 50 mM stock in cross-linking buffer. Following gentle rotation for 1 h at room temperature (ca. 23°C), the reaction was quenched with 100 mM Tris-HCl (pH 7.5) for 15 min at room temperature. Reducing dye (2×) was added, and samples were heated at 95°C for 10 min. Samples were resolved on SDS-PAGE gels and immunoblotted as described below.
Studies of FliG-FliG cross-linking in cells were performed by using the ΔflhDC ΔfliG and ΔflhDC ΔfliG Δhns strains, with FlhDC being expressed from the salicylate-regulated plasmid pKP147 and FliG (either the wild type or with the S117C G166C double-Cys replacement) being expressed from derivatives of the IPTG-regulated plasmid pRR48. Cells were cultured overnight at 32°C in LB containing Amp and Cm, diluted 100-fold into TB medium containing the antibiotics and inducers (50 μM IPTG and 2.5 μM Na-salicylate), and grown at 32°C to mid-log phase (OD600 = 0.5 to 0.6). Equal amounts of cells (as determined by the OD600) were pelleted and resuspended in XL buffer (20 mM Na-phosphate [pH 7.4], 150 mM NaCl). For cross-linking with 1,6-bis-maleimidohexane (BMH; Pierce), 0.1 ml of cells was mixed with 2 μl BMH stock (50 mM in DMSO; final concentration, ∼1 mM). Following incubation on ice for 30 min, the reaction was quenched by adding 2.5 μl 2-mercaptoethanol to the mixture. Cells were mixed with 2× reducing dye and heated to 95°C for 10 min. Proteins were resolved on SDS-PAGE gels and immunoblotted as described below. For experiments using iodine, cells were prepared similarly but with 0.25 mM I2 (from a 25 mM stock in ethanol) for 3 min at room temperature, followed by quenching with 1 mM N-ethyl maleimide (from a 1 M stock in ethanol). Cells were mixed with 2× nonreducing loading dye and heated at 95°C for 10 min in preparation for electrophoresis.
SDS-PAGE and immunoblotting.
Proteins were resolved on 10% or 12% SDS-polyacrylamide gels (Bio-Rad Mini) and transferred onto nitrocellulose by using a semidry transfer apparatus (Bio-Rad). Rabbit polyclonal antibodies against FliG, FliM, or FliN were prepared in a solution containing 1× phosphate-buffered saline (PBS) (pH 7.4), 0.1% gelatin, and 0.01% Na-azide, at dilutions of 1:2,000, 1:5,000, and 1:2,000, respectively. His-tagged FliG and H-NS were detected by using mouse anti-His antibody (Qiagen) at a 1:1,000 dilution. Immunoblots were visualized and analyzed by using the Odyssey infrared imaging system.
RNA extraction and preparation of cDNA.
Cultures grown overnight in TB-Amp-Cm medium, each grown from three colonies, were used to inoculate 20-ml cultures in TB-Amp-Cm containing inducers (0.1 mM IPTG and 10 μM Na-salicylate). Cultures were grown at 32°C to mid-log phase (OD600 of 0.5 to 0.6). Prior to RNA extraction, cells were incubated overnight in ice-cold phenol-ethanol (5:95, vol/vol) to stabilize the RNA (18). Samples were centrifuged at 6,000 × g for 10 min, and the supernatant was discarded. Cells were resuspended in 1 ml Tris-EDTA (TE) to remove residual phenol and then pelleted once more. Cells were resuspended in 100 μl TE and then incubated with lysozyme (10 mg/ml from a 100-mg/ml stock) for 8 min at room temperature, and RNA was then extracted by using the GenCatch total RNA miniprep kit (Epoch Life Sciences) according to the manufacturer's protocol. RNA concentration and quality were determined by using a NanoDrop spectrophotometer (ND-1000; NanoDrop Technologies) and agarose (1.5%) gels.
Prior to reverse transcription, RNA was subjected to on-column DNase I treatment (DNA-free RNA kit; Zymo Research), according to the manufacturer's protocol, and a second treatment with RQ1 RNase-free DNase (Promega), again according to the manufacturer's instructions. Following treatment, 1 μg of RNA was reverse transcribed to cDNA by using the Maxima first-strand cDNA synthesis kit (Thermo Scientific), using random-hexamer and oligo(dT) primers provided with the kit.
Real-time quantitative PCR (RT-qPCR).
Real-time PCR (RT-PCR) was carried out by using the CFX96 Touch real-time PCR detection system (Bio-Rad) with Sso Fast Evagreen supermix (Bio-Rad) in 96-well plates. Two microliters of cDNA (previously diluted to 10 ng/μl) was added to 8 μl of the PCR mixture (5 μl Evagreen mix and 1.5 μl of each target gene primer at 3 μM). Primers were designed by using the PrimerQuest tool (Integrated DNA Technologies). The following thermal cycling program was used: an initial denaturation step at 98°C for 30 s followed by 40 cycles of 98°C for 5 s and 63°C for 10 s (annealing/synthesis step), followed by a melting curve from 65°C to 95°C in 0.5°C and 5-s increments. The rpoD, cysG, and hisG genes were used as internal controls. All experiments were performed as three biological replicates, with three technical replicates each. Results were analyzed by using the comparative critical threshold (ΔΔCT) method, in which the relative quantities of the genes of interest are normalized to the relative quantities of the reference genes across samples.
RESULTS
FlhDC-constitutive Δhns mutants show moderate motility defects.
The immotility of hns mutants of E. coli is due mainly to the essential involvement of H-NS in the transcription of the flhDC operon encoding the top-level transcription factors of the flagellar regulatory hierarchy (10). In two previous studies aimed at characterizing other possible functions of H-NS in motility, this requirement was bypassed by expressing the FlhDC proteins from plasmids in hns mutant backgrounds. Ko and Park (13) found that an FlhDC-constitutive hns mutant was well flagellated yet still very poorly motile. A similarly conceived strain made in our laboratory was also essentially immotile (14). Ko and Park made the further, important observation that the motility of the FlhDC-constitutive hns mutant is significantly improved upon disruption of ycgR (13), which has since been shown to encode a motor-regulating protein responsive to cyclic di-GMP (20–22). A previous study from our laboratory (14) examined various aspects of flagellar motor performance in the putatively FlhDC-constitutive hns strain but did not examine the role of ycgR. In experiments first intended to characterize the H-NS/motor interaction further, we found that the strain did not show the expected motility improvement upon deletion of ycgR, prompting closer examination.
A concern with both previous studies using plasmid-expressed flhDC (13, 14) in hns mutant backgrounds is the retention of the flhDC genes in the chromosome, where their expression could be influenced by the reintroduction of wild-type or mutant H-NS or by other factors that modulate the flhD promoter. To allow better separation of the flhDC-regulating effects of H-NS from any direct effects that it might have at the motor, the chromosomal flhDC genes were replaced with a tetRA cassette (leaving just 4 codons at the 5′ end of flhD) in both the hns deletion strain and the wild-type parent strain. The ΔflhDC (hns+) strain was used to test complementation by various flhDC plasmids and to determine the levels of induction sufficient to give wild-type migration rates in soft agar. At 32°C, plasmid pKP148 gave migration rates similar to those of the wild type when induced with 80 μM IPTG (see Fig. S1 in the supplemental material). At 23°C, lower-level induction (20 to 30 μM IPTG) sufficed (data not shown). The pTrc-flhDC plasmid used previously (13) also complemented the ΔflhDC mutant although somewhat less effectively than pKP148 (see Fig. S1 in the supplemental material). Plasmid pKP85 used in our previous study (14) did not complement the flhDC-null mutant (see Fig. S1 in the supplemental material). Sequencing showed errors in its design that prevented effective expression. It follows that in the previous experiments on the Δhns strain, the main role of H-NS would have been to stimulate the expression of the flhDC genes still present in the chromosome (14).
Plasmid pKP148 was then introduced into the Δhns ΔflhDC background, and motility was tested at induction levels slightly above those giving wild-type motility in the ΔflhDC background (100 μM IPTG at 32°C or 40 μM IPTG at 23°C). In contrast to data from both previous reports (13, 14), the strain displayed fair motility, moving in soft-agar plates at ∼50% of the wild-type rate at 32°C (Fig. 1). The motility defect was somewhat more severe at a lower temperature (23°C), but substantial motility persisted (Fig. 1). Microscopic examination of cells in late-log-phase tryptone broth cultures showed that most cells were motile at both temperatures.
FIG 1.
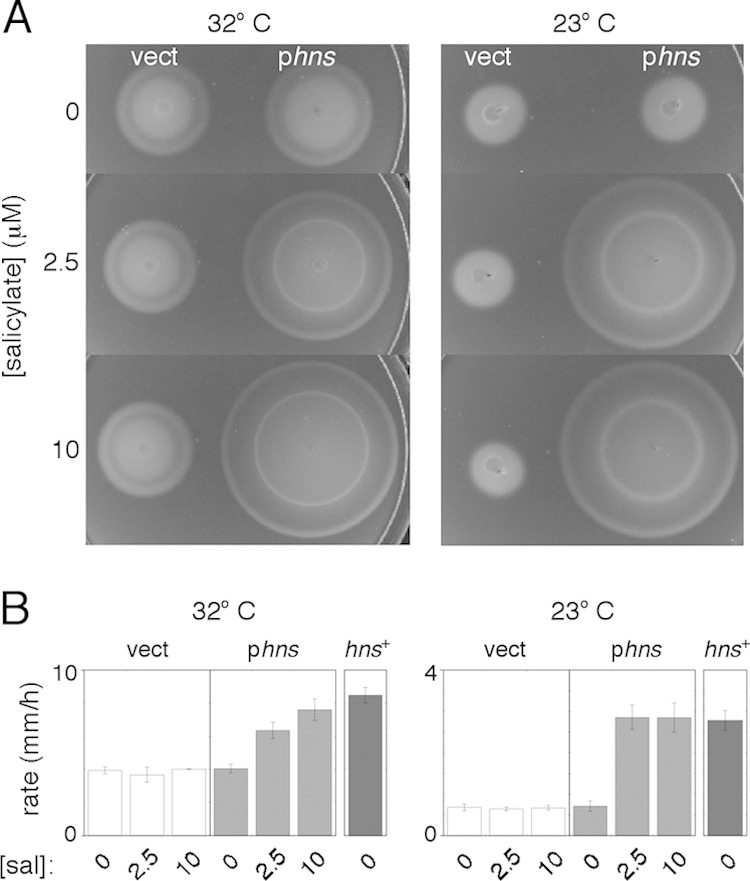
Motility of a Δhns, FlhDC-constitutive strain. (A) Motility phenotype on soft (0.26% agar) tryptone plates at two temperatures. The background was strain EK2 deleted for chromosomal hns and flhDC genes. To ensure constitutive expression of FlhDC, strains contained plasmid pKP148 induced with 100 μM IPTG at 32°C or 40 μM IPTG at 23°C, levels just above those that matched the motility of the wild type in calibration experiments. Cells contained either the salicylate-regulated hns plasmid pKP58 (phns) or the vector-only control plasmid pKG116 (vect). Results are shown for three levels of induction by salicylate. Plates were inoculated with 3-μl drops from cultures grown overnight and incubated for 7.3 h (32°C) or 18.5 h (23°C). (B) Migration rates of the strains, determined from linear fits to measurements of colony diameter versus time. Measurements are averages of data from three (32°C) or four (23°C) determinations (±standard deviations) and are shown for various levels of induction by salicylate (sal). For comparison, results are also shown for an hns+ strain (EK1 transformed with plasmid pKP148) for the same levels of IPTG induction.
The poor motility observed previously by Paul et al. (14) can be understood as the result of poor expression of FlhDC, but the same should not be true in the study by Ko and Park (13), where the better-complementing plasmid pTrc-flhDC was used. The nature of the hns mutation appears to be responsible instead; poor motility was reproduced when plasmid pTrc-flhDC was present in the hns::neo background used by Ko and Park (13), but motility was better with the same plasmid in the Δhns background (Fig. 2A). The hns::neo strain was also less effectively complemented by hns on a plasmid and grew in cultures overnight to a lower density (and with greater clonal variation) than the Δhns strain (Fig. 2B and C). The hns::neo strain retains 37 hns codons (the segment before the HpaI restriction site used for construction). The nonnull behavior of hns alleles with insertions after codon 37 was noted by others previously (23, 24); experiments demonstrating the nonnull behavior of a similar strain (with a Kanr cassette at the same position) were reported previously by Battesti et al. (23). The structure of H-NS shows that these amino-terminal residues, together with the same segment from another subunit, would fold into a discrete domain, important for holding subunits together (25, 26). Residues at the intersubunit interface in this domain are conserved in the H-NS homolog StpA (26), so the H-NS fragment encoded in the hns::neo strain could form heterodimers with StpA. Structural and mutational evidence points to the involvement of the amino-terminal domain not only in intersubunit associations but also in DNA binding (26). The H-NS fragment in the hns::neo strain could then affect cellular physiology either by binding to StpA or through direct binding to the chromosome.
FIG 2.
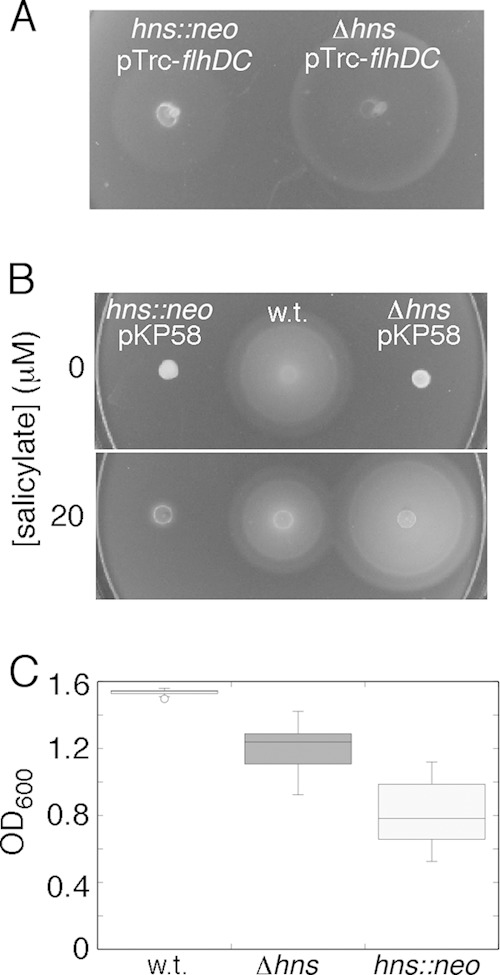
Comparison of strains with different hns disruptions. (A) Motility plate phenotypes of hns mutant strains with either a deletion retaining the first 5 codons (strain MS1162) or a neomycin resistance cassette preserving the first 37 codons (strain MS531). Both strains were transformed with plasmid pTrc-flhDC (13) as a source of FlhDC, with no induction by IPTG (the conditions described in reference 13). Motility became worse upon induction (see Fig. S1 in the supplemental material). (B) Complementation of the two strains by the hns-expressing plasmid pKP58, induced at various levels. (C) Densities (OD600) of cultures of the two hns mutants and of a wild-type (w.t.) strain (strain MS365, identical to MS296 [13]) grown overnight. Cultures were in tryptone broth, inoculated from single colonies and grown for 14 h at 32°C (n = 24, 23, and 28 for the wild-type, Δhns, and hns::neo strains, respectively). Standard box plots are shown.
H-NS phenotypes do not support a direct action at the motor.
The motility defect that persists in the FlhDC-constitutive Δhns strain, though milder than previously thought, might yet indicate a direct action of H-NS at the motor. To characterize this motility impairment further, we measured the effects of increasing salt and agar concentrations, as was done previously (14). In contrast to results described previously, migration rates of the Δhns-deficient strain remained the same relative to those of the wild type as salt and agar concentrations were changed (see Fig. S2A and B in the supplemental material). Early studies had implicated the rotor protein FliG as a site for binding of H-NS (8, 9), which motivated cross-linking experiments by Paul et al. (14) that were purported to show a significant loss of FliG subunit organization in hns-deficient cells. Given the issues described above, this disorganization likely stemmed from an underexpression of FlhDC and consequent defects in flagellar assembly. Experiments with the new strain (see Fig. S2C in the supplemental material) showed very similar patterns of FliG-FliG cross-linking in the presence and in the absence of H-NS, indicating little or no effect of H-NS on FliG organization.
Key evidence for the functional relevance of an H-NS/FliG interaction came from a study by Donato and Kawula (9), who found that certain H-NS mutations conferred a hypermotile phenotype, enabling cells to migrate at more than twice the wild-type rate in soft-agar plates. Their strain carried an insertional disruption of hns similar to that reported previously (13) (also after codon 37 but with Tetr instead of Neor [27]). The flhDC genes were retained in the chromosome, and no additional source of FlhDC was provided. In this background, the major role of H-NS would presumably have been to enable the expression of the flhDC genes in the chromosome, and hypermotility would appear most likely to result from altered DNA-binding properties. When tested in the Δhns strain, the putatively hypermotile H-NS variants did not appear to confer motility better than that of the wild type (see Fig. S3 in the supplemental material). The hns::neo strain was complemented more effectively by a hypermotile variant (A18E) than by wild-type H-NS on the plasmid, but the motility was not better than that of a true wild-type strain (i.e., having hns+ in the chromosome) (not shown).
Involvement of c-di-GMP and YcgR.
Ko and Park's (13) findings that an hns-deficient strain is partially rescued by deletion of ycgR or overexpression of yhjH led to the discovery of motility regulation by the second messenger c-di-GMP, acting through the motor-binding protein YcgR (20–22). To test for the involvement of c-di-GMP in the motility defect of the FlhDC-constitutive Δhns strain, a plasmid expressing the c-di-GMP phosphodiesterase YhjH was introduced into the cells. Motility was improved significantly at both temperatures (by ∼50% at 32°C and 60% at 23°C) (Fig. 3A). To examine the role of YcgR, motilities of the Δhns and Δhns ΔycgR strains were compared, with FlhDC being expressed from plasmid pKP148 as described above. Migration rates were significantly increased in the double-deletion strain, as expected if c-di-GMP acts primarily through YcgR (Fig. 3B). Overexpression of the phosphodiesterase YhjH did not exhibit a measurable rescue effect in the Δhns ΔycgR background, also as expected if the action of c-di-GMP is mediated chiefly through YcgR.
FIG 3.
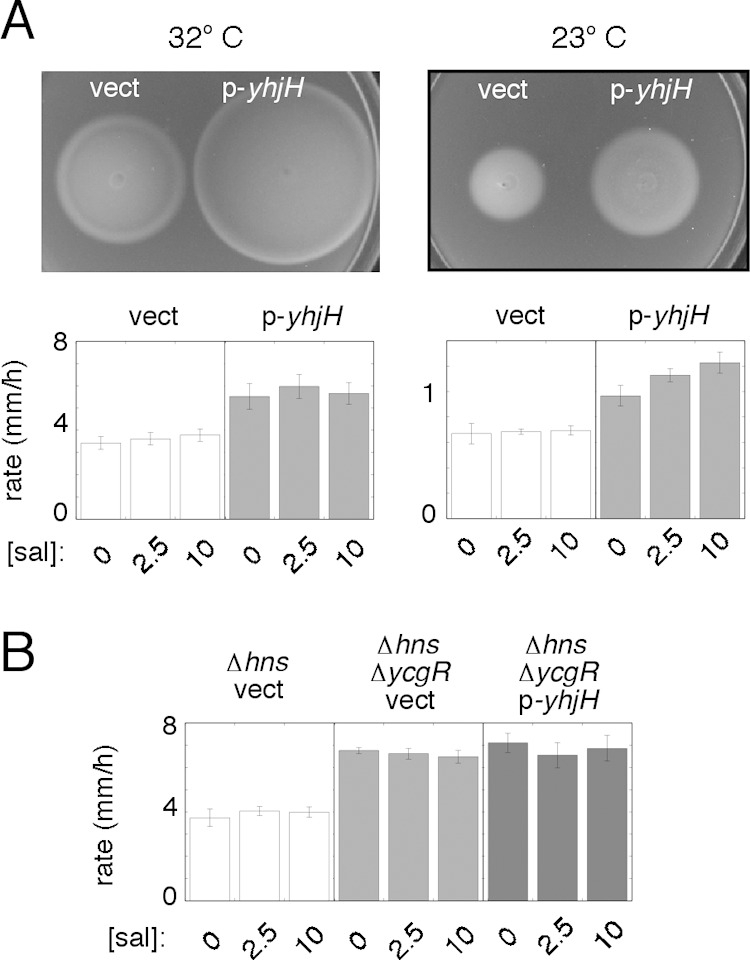
Involvement of c-di-GMP and YcgR in the motility defect of the FlhDC-constitutive Δhns mutant (strain EK2 transformed with plasmid pKP148 as a source of FlhDC and with other plasmids as indicated). (A) Effect of expression of the c-di-GMP phosphodiesterase YhjH in the mutant strain at two temperatures. Cells contained either the yhj-expressing plasmid pEK29 (p-yhjH) or the vector control pKG116 (vect), as indicated. Plates were inoculated with 3-μl spots from cultures grown overnight and incubated at 32°C (for 9 h) or 23°C (28.2 h). Plates contained 2.5 mM salicylate to induce yhjH. Induction of flhDC was done with 100 mM IPTG at 32°C or 40 μM IPTG at 23°C. Migration rates for three levels of yhjH induction are shown in the graphs at the bottom. Values are averages of data from 5 determinations (±standard errors of the means) at 32°C or 3 determinations (±standard deviations) at 23°C. (B) Motility improvement of a Δhns strain upon deletion of ycgR and lack of any additional motility rescue by expression of YhjH in the Δhns ΔycgR background. Experiments were done at 32°C. The Δhns ΔycgR strain used was MS1517; cells were transformed with plasmid pKP148 as a source of FlhDC, with induction with 100 μM IPTG. Strains expressing YhjH contained the salicylate-regulated plasmid pEK29, induced at the levels indicated; the “vect” control contained the parent vector pKG116. Values are averages of data from 3 determinations (±standard deviations).
Altered expression of diguanylate cyclase and c-di-GMP hydrolase genes in an hns mutant.
The motility improvement conferred by a c-di-GMP-hydrolyzing enzyme might indicate increased activity of diguanylate cyclase (c-di-GMP-forming) enzymes in the Δhns mutant. We measured transcript levels of several candidate diguanylate cyclase genes and found significantly increased levels of ydeH (150-fold), ycdT (60-fold), yeaI (24-fold), yeaJ (19-fold), and yaiC (3-fold) in the FlhDC-constitutive Δhns strain relative to the hns+-complemented strain (Fig. 4A). Levels of yeaP were not increased (not shown). To determine which of the strongly upregulated cyclase genes encode enzymes that might be active under the conditions of our assays, each gene (ycdT, yeaI, yeaJ, or ydeH) was expressed from an inducible plasmid in the FlhDC-constitutive Δhns mutant, and motility was tested. Motility was strongly inhibited upon induction of yeaJ (Fig. 4B).
FIG 4.
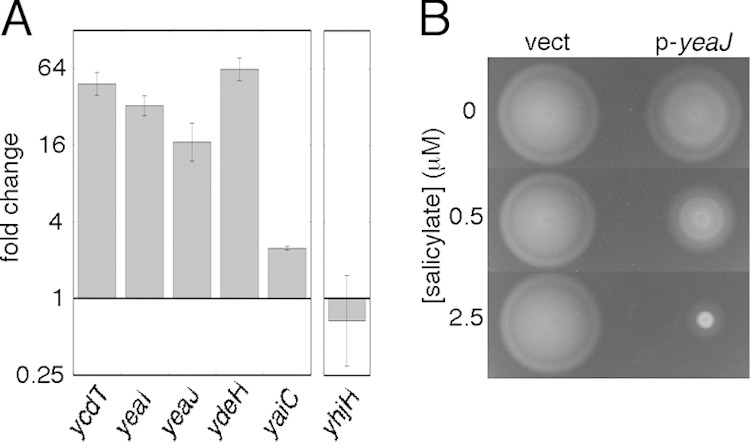
Altered expression of several diguanylate cyclase genes, and of yhjH, in the hns mutant. (A) Upregulation of diguanylate cyclase genes, and downregulation of yhjH, in the FlhDC-constitutive Δhns ΔflhDC mutant. The strain used was EK2 transformed with plasmid pKP148 as a source of FlhDC, with induction with 100 μM IPTG. Transcript levels were determined by the 2−ΔΔCT method and are relative to the levels in the strain complemented with wild-type hns on a plasmid (pKP58 induced with 10 μM salicylate). Values are the means of data from 3 independent experiments (±standard deviations), each representing an average of results based on three reference genes (rpoD, hisG, and cysG), as described in Materials and Methods. (B) Motility inhibition upon overexpression of the diguanylate cyclase YeaJ. The strain used was EK2 transformed with pKP148 to provide FlhDC (with induction with 100 μM IPTG) and with either plasmid pEK25 expressing yeaJ under salicylate control (p-yeaJ) or the parent vector pKG116 (vect). Results are shown for three levels of induction by salicylate. Plates were inoculated with 3-μl drops from cultures grown overnight and incubated at 32°C for 8.5 h.
Levels of cyclic di-GMP will also reflect the activity of the enzymes catalyzing its hydrolysis, and the phosphodiesterase YhjH appears to be of particular importance, judging from the poor motility of ΔyhjH mutants (20, 22). We measured levels of the yhjH transcript in the FlhDC-constitutive Δhns strain and found a possible, but modest, decrease (Fig. 4A). yhjH is a class 3 flagellar gene and thus is under the control of the flagellar sigma factor σF, which is expressed at a decreased level in the mutant (see below).
Master biofilm regulator CsgD.
The transcription factor CsgD is a key regulator in the motile-sessile transition (28) and in promoting tolerance to drying (29), acting to promote the production of curli fimbriae and cellulose while downregulating motility. CsgD is itself subject to multiple regulatory inputs (28, 30–32), including positive regulation by c-di-GMP (33). Measurements by RT-qPCR showed that levels of the csgD transcript were increased ∼50-fold in the Δhns strain relative to the strain complemented with plasmid-borne hns (Fig. 5A). This finding contrasts with the previously reported (33) decrease in the csgD transcript level in a strain that used a different hns disruption (hns-205::Tn10) and that also retained the chromosomal flhDC genes. To allow more direct comparison with the findings reported previously (33), we compared csgD transcript levels in a simple Δhns strain (i.e., retaining chromosomal flhDC) and its wild-type parent. Levels of the csgD transcript were elevated significantly (by ∼8-fold) in the Δhns strain (Fig. 5B).
FIG 5.
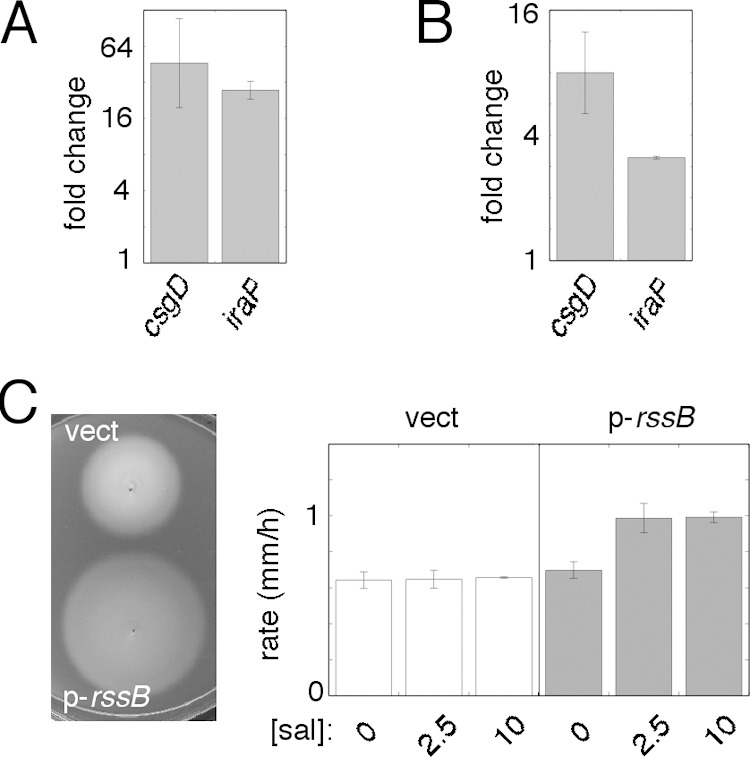
Upregulation of the csgD and iraP genes in hns mutants and involvement of the stress response sigma factor σS in motility impairment. (A) Upregulation of csgD and iraP in the FlhDC-constitutive Δhns strain (EK2 transformed with pKP148, induced with 100 μM IPTG) relative to the strain complemented with wild-type hns (using plasmid pKP58, induced with 10 μM salicylate). Values shown are means ± standard deviations from 3 biological replicates, each based on the use of 3 reference genes. (B) Upregulation of csgD and iraP in a Δhns strain (MS1162) relative to the wild-type strain (MS365). Values are means ± standard deviations from 2 biological replicates, each based on the use of 3 reference genes. (C) Effect of overexpression of the σS degradation factor RssB on the motility defect of the FlhDC-constitutive Δhns strain in an experiment at 23°C. The strain used was EK2 transformed with pKP148 to provide FlhDC (induction with 40 μM IPTG) and pEK8 to express RssB (p-rssB) or the vector control pKG116 (vect). (Left) Motility plate. The plate was inoculated with 3-μl drops from cultures grown overnight and incubated for 36 h; induction was done with 2.5 μΜ salicylate. (Right) Migration rates at three levels of RssB induction (means ± standard deviations; n = 3).
CsgD might affect motility by activating the transcription of iraP (34), encoding the “antiadaptor” protein IraP that inhibits the ability of RssB (the “adaptor”) to target the sigma factor σS for degradation by ClpXP protease (35, 36). Levels of iraP transcript measured by RT-qPCR were increased >20-fold in the FlhDC-constitutive Δhns mutant (Fig. 5A) and also, though more modestly, in the simple Δhns strain (Fig. 5B). The iraM and iraD genes (encoding two other antiadaptors) were previously shown to be more highly expressed in an hns mutant (23); thus, all three Ira proteins could contribute to the stabilization of σS in a Δhns strain. Increased levels of σS could stimulate the transcription of csgD in a positive-feedback loop and might also decrease transcription directed by the flagellar sigma factor σF through competition for RNA polymerase (and possibly other regulatory connections). To assess the possible role of σS in the motility defect in the FlhDC-constitutive Δhns mutant, the σS degradation factor RssB was overexpressed from a regulatable plasmid in the cells. The migration rate was not measurably affected at 32°C (not shown) but was increased by ∼50% at 23°C, a temperature at which σS is expected to be more highly expressed (37) (Fig. 5C).
Flagellar sigma factor σF.
Dudin et al. (19) recently found that CsgD can directly inhibit the transcription of fliA, which encodes the sigma factor (σF) that directs the expression of the late (class 3) flagellar genes. This might provide a more direct regulatory link between CsgD and motility. Elevated levels of CsgD should, through repression of fliA, cause the underexpression of proteins needed to build the flagellar filament and to energize and regulate its rotation. Motility might also be decreased as a result of the underexpression of yhjH, which is a class 3 flagellar gene. To determine whether a limitation in σF contributes to the motility defect, we measured the effects of expressing additional σF from a plasmid in the FlhDC-constitutive Δhns strain. At 32°C, the σF level appears to be a factor limiting motility; the migration rate was increased ∼50% upon modest induction of FliA (using 1 μM salicylate) (Fig. 6A). RT-qPCR measurements showed a rather modest (1.5-fold) but apparently reproducible decrease in fliA transcript levels in the Δhns mutant (Fig. 6B). The effects were smaller at 23°C: the migration rate was increased by ∼25% upon low-level FliA overexpression (with no induction by salicylate; stronger induction led to impaired growth) (Fig. 6C).
FIG 6.
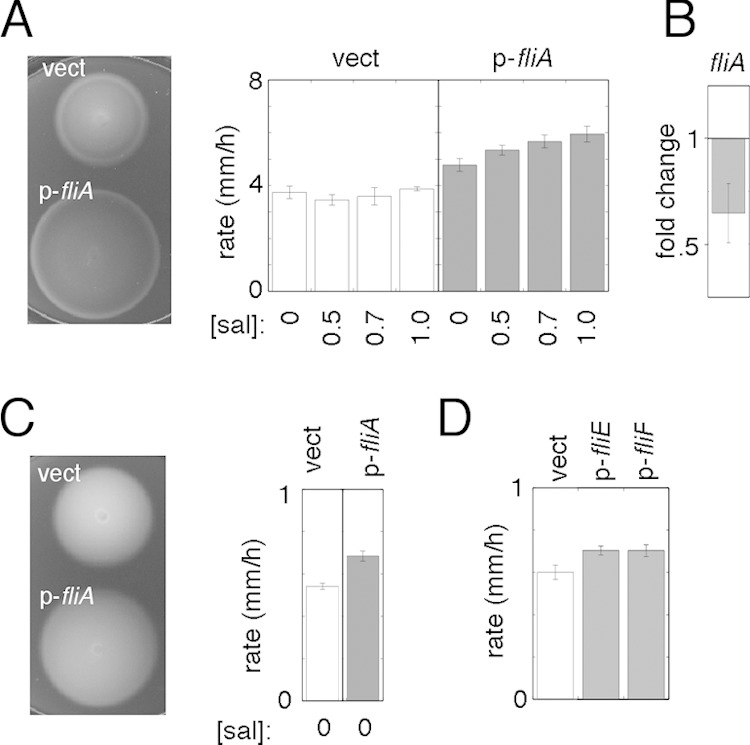
Involvement of FliA, FliE, and FliF in the motility defect of the hns mutant. (A) Significant motility enhancement upon expression of FliA in the FlhDC-constitutive Δhns strain at 32°C. The strain used was EK2 transformed with pKP148 (vect) to provide FlhDC, induced with 100 μM IPTG. FliA was expressed from the salicylate-regulated plasmid pEK21 (p-fliA). (Left) Motility plate containing 1 μM salicylate. (Right) Migration rates at various levels of induction (means ± standard deviations; n = 3). (B) Modestly decreased level of the fliA transcript in the FlhDC-constitutive hns mutant [EK2(pKP158) induced with 100 μM IPTG, also containing the control plasmid pKG116] relative to the strain complemented with wild-type hns (having pKP58 instead of pKG116). Induction of H-NS was done with 10 μM salicylate. Values are means ± standard errors of the means from 4 biological replicates. (C) Effects of modest overexpression of FliA in the FlhDC-constitutive Δhns strain at 23°C. The strains used were as described above for panel A. Salicylate was not added, because even low-level induction of fliA caused growth impairment at this temperature. (D) Effects of modest overexpression of the basal body protein FliE or FliF at 23°C. The strain used was EK2 transformed with pKP148 to provide FlhDC induced with 40 μM IPTG. Expression of FliE and FliF was obtained with plasmids pEK30 and pEK16, respectively, with no induction with salicylate; higher-level expression impaired motility (FliE) or growth (FliF). Values are the averages of data from 8 measurements (±standard errors of the means); P values determined by Student's t test are 0.025 for FliE versus the control and 0.036 for FliF versus the control.
Flagellar fliE and fliF operons.
In a recent study to identify regulatory targets of CsgD, Ogasawara et al. (32) found that CsgD can directly inhibit the transcription of the fliF and fliE operons, which encode proteins used at early stages of basal body assembly (38, 39). At 32°C, the motility of the FlhDC-constitutive Δhns strain was not measurably improved by plasmid-directed expression of fliF (the first of six genes in its operon) or fliE (data not shown). At 23°C, the migration rate of the strain was increased slightly (by 15 to 20%) by low-level expression (using no induction by salicylate) of either fliF or fliE (Fig. 6D). Higher-level expression led to an inhibition of either growth (FliF) or motility (FliE) (data not shown). Decreased levels of FliE and FliF may therefore contribute, but in a relatively minor way, to the motility impairment of the Δhns mutant.
Anti-FlhDC factor YdiV.
In the experiments described above, the plasmid-borne flhDC genes were induced at levels sufficient to give wild-type or slightly greater motility in a ΔflhDC background. FlhDC activity could nevertheless be subnormal in the (nominally) FlhDC-constitutive Δhns strain if the absence of H-NS enables processes that destabilize or otherwise oppose FlhDC. The YdiV protein functions as an anti-FlhD factor, targeting FlhD for proteolysis in a manner similar to the action of RssB on σS (40). Measurements by RT-qPCR showed a substantial (∼30-fold) increase in the ydiV transcript level in the Δhns mutant (see Fig. S4 in the supplemental material). Levels of the YdiV protein might not be correspondingly increased, because the ydiV transcript has been shown to be inefficiently translated in E. coli (41). To test for the involvement of YdiV in the motility impairment of the Δhns strain, we used a point mutant of FlhD in Salmonella that was found to make the protein resistant to the action of YdiV (40). If YdiV-directed FlhD degradation contributes to the motility defect in the Δhns strain, then this FlhD mutation (L19H in the protein of E. coli) should confer better motility. With flhDC induced at normal levels (100 μM IPTG at 32°C or 40 μM IPTG at 23°C), the L19H mutation yielded no increase in motility (see Fig. S4B, right, in the supplemental material). In an experiment where the FlhDC level was intentionally made more limiting by induction with just 50 μM IPTG (at 32°C), the mutant form of FlhD enabled an ∼40% increase in the migration rate (see Fig. S4B, left, in the supplemental material), indicating that FlhDC levels are in fact limiting under these conditions and that the L19H mutation has the expected stabilizing effect. Together, the results indicate that while ydiV transcript levels are significantly increased in the Δhns mutant, destabilization of FlhD by YdiV is not a substantial contributor to its motility defect.
DISCUSSION
Available evidence does not point to a direct action of H-NS at the motor.
Results from three studies (9, 13, 14) have been taken to indicate a functionally important interaction between H-NS and the flagellar motor. The observations reported here cast some doubt on this proposition. The reportedly severe motility defect in FlhDC-constitutive Δhns cells appears to have resulted from the use of either a faulty plasmid (14) or a background that retained an interfering fragment of H-NS (13). The apparent hypermotility of certain mutant variants of H-NS (9), important for establishing the functional relevance of the interaction, was measured in an experiment where the main role of H-NS would have been to stimulate the expression of chromosomal flhDC; also, the strain carried an allele with the potential to encode an interfering H-NS fragment, and we find that in a cleaner Δhns background, the putatively hypermotile variants display motility no better than that of the wild type.
Indications of an H-NS/motor interaction have also come from experiments not involving measurements of motility. The first such study used the yeast two-hybrid system, primarily providing the earliest documentation of interactions among the so-called “switch complex” proteins (FliG, FliM, and FliN) but also giving evidence of an interaction between H-NS and FliG (8). The H-NS/FliG interaction gave a relatively weak signal in the two-hybrid assay (0.5 units of β-galactosidase activity, compared to values of between 39 and 651 units for the FliG-FliM, FliM-FliN, and FliM-FliM interactions). A somewhat stronger interaction (7 units) was observed when the experiment was performed by using a FliG variant lacking residues 1 to 45. The subsequently solved structure of FliG (42) shows that this truncation would remove about half of the N-terminal domain of the protein, leaving a fragment that would be expected to fold poorly and show an increased propensity for nonspecific binding (owing to the exposure of about a dozen hydrophobic side chains). A more recent, wide-ranging screen of the E. coli protein interaction landscape by the yeast two-hybrid method did not give evidence of a FliG/H-NS interaction (43), instead identifying several candidate FliG partners without obvious connections to the flagellum (PabA, ProS, RpsS, YcbY, YcfM, and AscG). The same broad screen identified a few candidate H-NS partners, including its homolog StpA but not FliG. We suggest that FliG, which has a multidomain structure and engages in multiple interactions in its native situation within the flagellar motor, might be prone to nonspecific interactions.
In addition to their motility measurements of H-NS variants, Donato and Kawula (9) reported evidence of physical interactions between H-NS and FliG, using two assays. Cross-linking experiments using purified FliG and H-NS gave a FliG/H-NS heterodimer, in a yield that was low but appeared to be somewhat increased by the H-NS T108I mutation, a putatively hypermotile variant. The low yield could be due to a nonoptimal arrangement of the reacting chemical groups or alternatively might indicate the absence of a specific binding interaction. We carried out similar experiments with isolated His-tagged FliG and H-NS using a number of cross-linking reagents, including a photoactivated one with broad chemical reactivity, but did not observe a FliG/H-NS-cross-linked product in a significant yield (data not shown). Experiments with whole cells using a membrane-permeant cross-linker (DMS) likewise failed to show clear indications of a FliG/H-NS interaction (see Fig. S5 in the supplemental material). Support for an H-NS/FliG interaction also came from measurements of fluorescence anisotropy in mixtures of fluorescein-labeled H-NS with various amounts of FliG (9). Those measurements might reflect the formation of an H-NS/FliG complex; we have not undertaken comparable measurements but note that they might be complicated by the demonstrated ability of H-NS to form dimers and larger multimers (44).
More recently, evidence of both an H-NS/FliG interaction and an H-NS/FliN interaction came from experiments using fluorescence resonance energy transfer (FRET) (45). In both cases, FRET signals were in a fairly weak range where false-positive results are often a concern (46). Thus, while the FRET results are consistent with an H-NS/FliG interaction, they might not, by themselves, constitute firm support. Finally, in our previous study, pulldown experiments with a glutathione S-transferase (GST)–H-NS fusion protein implicated the middle domain of FliG as a site of interaction for H-NS (14). In follow-up experiments, we obtained various results suggesting that while the proteins have some affinity for one another (possibly reflecting the “stickiness” of FliG alluded to above), their interaction is not strong enough to persist through the stringent washing steps needed to give convincingly clear negative controls (data not shown).
Multiple regulatory pathways connect H-NS to flagellar motility.
While arguing against a direct action at the motor, the present findings highlight the modulating influence of H-NS over several regulatory pathways that govern motility. A scheme that organizes these results is presented in Fig. 7. The regulatory web pictured is based on existing literature on the motile-sessile (planktonic-biofilm) transition in E. coli (19–22, 28, 32–35, 47–49), together with the new findings here on the effects of hns deletion, which include (i) upregulation of several diguanylate cyclase genes, (ii) upregulation of csgD, (iii) upregulation of iraP, (iv) mild downregulation of fliA, and (v) upregulation of ydiV. H-NS acts through multiple, interconnected pathways, each having the net effect of promoting motility. We suggest that these several regulatory linkages, together with the observed changes in gene expression, are likely enough to account for the observed motility defect in a Δhns mutant. Factors that appear most responsible for the motility impairment are increased levels of the biofilm regulator CsgD and the intracellular messenger molecule c-di-GMP.
FIG 7.
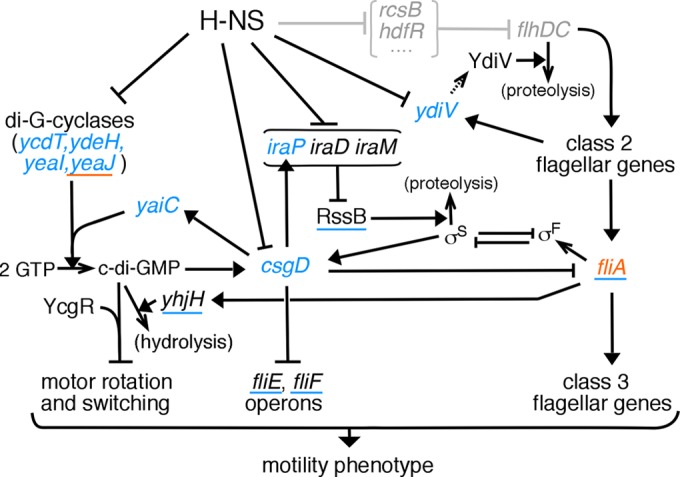
Summary of regulatory actions of H-NS on motility. Filled arrowheads signify stimulatory actions, and T bars indicate inhibition. Simple arrowheads denote chemical processes, including protein synthesis. Note that translation of ydiV (dashed arrow) has been shown to be inefficient in E. coli (41). The scheme is based on data from literature concerning the motile-sessile transition and associated processes in E. coli (11, 12, 19–22, 28, 32–35, 47–49), together with the present findings on transcript levels and overexpression effects in hns mutants, shown in color. Genes in blue letters showed increased expression in Δhns mutants, and that in red showed decreased expression. Blue underlining indicates genes that, when overexpressed, enhanced the motility of an hns mutant. Red underlining indicates a gene that, when overexpressed, aggravated the motility defect of an hns mutant. Regulatory actions shown in gray at the top are those which should be bypassed by the constitutive expression of FlhDC.
We note some instances where our measurements of hns effects on gene expression differ significantly from those reported in previous work. In contrast to the substantial increase in csgD expression in a Δhns strain observed here, two previous studies found csgD expression that was either moderately decreased (33) or not greatly affected (decreasing or increasing slightly depending on medium osmolarity) (50). Differences might be due, at least in part, to the use of different hns alleles. The hns-205 allele used by Weber et al. (33) has a Tn10 insertion after codon 93, while the Δhns-118 mutation used by Jubelin et al. (50) is a large deletion that removes a number of adjacent genes along with hns (51). In a third study (31) that used an engineered hns deletion retaining only a single codon, H-NS was found to repress csgD, in better agreement with data from the present experiments. Expression of iraP in an hns-disrupted strain was also measured previously and was found to be only modestly increased (by <2-fold) (23), whereas we observed a more significant (∼30-fold) increase. Differences in this case could reflect the use of cultures grown overnight rather than mid-log-phase cultures or possibly, again, the hns disruption used (in this case a Kanr insertion after codon 37, the nonnull behavior of which has been well discussed [23]). The additional presence of the flhDC deletion in our strain might also be a factor in this case, because increases in iraP expression were more modest in the simple Δhns strain (compare Fig. 5A and B). Whatever the precise magnitude of iraP upregulation, it should join with the upregulation of iraD and iraM (23) to increase the level of σS and thus decrease motility (Fig. 7).
The strong involvement of c-di-GMP (and YcgR) in the motility defect of the hns mutant presumably accounts for the identification of yhjH and ycgR in the suppressor screen of Ko and Park (13). We found strong modulation of several diguanylate cyclase genes by H-NS, at least one of which (yeaJ) encodes an enzyme that is active under the conditions of the motility assays, as judged by the ability to impair motility in a YcgR-dependent fashion. The apparent inactivity of several other cyclases (as judged by the absence of a motility impairment upon overexpression) is also of interest and might indicate that these enzymes require specific stimuli, such as environmental stresses, to become active. The strong upregulation of these genes in Δhns cells would then fit with the proposal that H-NS modulates genes involved in responses to the environment.
Other sources of the motility defect in the Δhns strain involve the biofilm regulator CsgD, which is strongly upregulated in the Δhns mutant and which was recently shown to inhibit transcription of the fliA (19) as well as of the fliE and fliF (32) operons. Each of these repressing actions appears significant under some conditions. Motility of the hns mutant was improved significantly when cells were provided extra FliA, particularly at 32°C. Extra FliEF proteins were also beneficial, although to a lesser extent, measurable only at a lower temperature (23°C). Through these repressing actions, CsgD could act both to decrease the material investment in existing flagella (by reducing the transcription of class 3 genes, especially fliC) and to retard the construction of new basal bodies (by reducing the availability of proteins used at early stages of assembly).
The elevated expression of csgD in the Δhns mutant might largely reflect a direct repression of its promoter by H-NS but also appears to be due at least in part to an increased level of σS, a known property of hns mutants (52) that is supported in the present study by the motility increase seen upon overexpression of the σS degradation factor RssB. Regulation of the rpoS promoter is complex (53, 54), and the increased activity of σS could have a number of causes, but among the likely causes are increased levels of the antiadaptor genes iraP, iraD, and iraM (23) (Fig. 5).
Although YdiV-mediated degradation of FlhD does not account for a significant part of the motility impairment that we observed in the Δhns mutant, ydiV transcription was greatly increased in the Δhns strain. This again appears to agree with the notion of H-NS as a modulator of the cellular response to the environment; in Salmonella, YdiV mediates responses to changes in nutritional status (48). Control at the level of translation would ensure that the ydiV transcript is present in a standby mode that would allow a rapid response to environmental cues, not yet identified in E. coli, that signal the advisability of conserving resources rather than committing to the synthesis of new flagella.
Correlation with ChIP measurements of H-NS localization.
In the course of testing candidate genes to identify those with altered expression in the hns mutant, we found that data from a chromosome-wide chromatin immunoprecipitation (ChIP) study by Uyar et al. (55) showed a clear correspondence with our measurements by RT-qPCR. Data in that ChIP study (55) are organized to allow ready identification of chromosomal regions highly enriched for H-NS interactions. The strongly upregulated genes found in the RT-qPCR experiments (ycdT, ydeH, yeaJ, yeaI, csgD, iraP, and ydiV) all display very prominent peaks indicative of strong overrepresentation in the ChIP data; less-affected genes (yeaP, yhjH, and fliA) do not display such peaks. Further study should determine if this correlation holds more widely, in which case the ChIP data appear to provide a useful predictive tool.
Regulation by small noncoding RNA.
Noncoding RNA molecules have key roles in regulating the expression of FlhDC, CsgD, and RpoS (56, 57). We have not attempted here to address the question of whether H-NS modulates the action of these regulatory RNAs. Although regulatory RNAs are not included in the regulatory scheme presented, we note an aspect of their function that might be especially important in connection with the present study. Some of these regulatory RNA molecules interact with mRNAs transcribed from both the flhD and csgD promoters, thus constituting regulatory cross-connections between the master regulators of motility and biofilm (30, 57, 58). Deletion of the chromosomal flhDC genes (as was required for most experiments reported here) might then influence expression from the csgD operon through mechanisms unrelated to FlhDC protein levels, and deletion of H-NS could have different consequences for gene expression in a ΔflhDC context (even with FlhDC proteins maintained at nominal levels through plasmid expression) than in cells retaining chromosomal flhDC. A similar pattern of gene expression effects was seen in the RT-qPCR measurements on a simple Δhns (flhDC+) strain, however (Fig. 5B; see also Fig. S6 in the supplemental material), arguing that regulatory interconnections of this kind do not contribute in a very major way to the changes in gene expression reported here.
In conclusion, although the case for a direct action of H-NS at the flagellar motor seemed persuasive and was supported by results from multiple laboratories, new experiments and a reexamination of the existing data substantially undercut this idea. The motility defect in cells lacking H-NS is most likely due to several, largely already characterized, regulatory pathways. It is difficult to prove the absence of any functionally relevant H-NS/motor interaction, and in this sense, the question is not fully settled. Persuasive evidence that H-NS interacts with the motor may yet emerge but must come from experiments that take full account of its complex regulatory actions.
Supplementary Material
ACKNOWLEDGMENTS
We thank Kelly Hughes, Fabienne Chevance, and Sandy Parkinson for helpful discussions.
This work was supported by grant R01-GM64664 from the National Institutes of Health.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00309-15.
REFERENCES
- 1.Dorman CJ. 2004. H-NS: a universal regulator for a dynamic genome. Nat Rev Microbiol 2:391–400. doi: 10.1038/nrmicro883. [DOI] [PubMed] [Google Scholar]
- 2.Rimsky S. 2004. Structure of the histone-like protein H-NS and its role in regulation and genome superstructure. Curr Opin Microbiol 7:109–114. doi: 10.1016/j.mib.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 3.Tendeng C, Bertin PN. 2003. H-NS in Gram-negative bacteria: a family of multifaceted proteins. Trends Microbiol 11:511–518. doi: 10.1016/j.tim.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 4.Lang B, Blot N, Bouffartigues E, Buckle M, Geertz M, Gualerzi CO, Mavathur R, Muskhelishvili G, Pon CL, Rimsky S, Stella S, Babu MM, Travers A. 2007. High-affinity DNA binding sites for H-NS provide a molecular basis for selective silencing within proteobacterial genomes. Nucleic Acids Res 35:6330–6337. doi: 10.1093/nar/gkm712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dame RT, Wyman C, Goosen N. 2001. Structural basis for preferential binding of H-NS to curved DNA. Biochimie 83:231–234. doi: 10.1016/S0300-9084(00)01213-X. [DOI] [PubMed] [Google Scholar]
- 6.Atlung T, Ingmer H. 1997. H-NS: a modulator of environmentally regulated gene expression. Mol Microbiol 24:7–17. doi: 10.1046/j.1365-2958.1997.3151679.x. [DOI] [PubMed] [Google Scholar]
- 7.Müller CM, Dobrindt U, Nagy G, Emödy L, Uhlin BE, Hacker J. 2006. Role of histone-like proteins H-NS and StpA in expression of virulence determinants of uropathogenic Escherichia coli. J Bacteriol 188:5428–5438. doi: 10.1128/JB.01956-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marykwas DL, Schmidt SA, Berg HC. 1996. Interacting components of the flagellar motor of Escherichia coli revealed by the two-hybrid system in yeast. J Mol Biol 256:564–576. doi: 10.1006/jmbi.1996.0109. [DOI] [PubMed] [Google Scholar]
- 9.Donato GM, Kawula TH. 1998. Enhanced binding of altered H-NS protein to flagellar rotor protein FliG causes increased flagellar rotation speed and hypermotility in Escherichia coli. J Biol Chem 273:24030–24036. doi: 10.1074/jbc.273.37.24030. [DOI] [PubMed] [Google Scholar]
- 10.Bertin P, Terao E, Lee EH, Lejeune P, Colson C, Danchin A, Collatz E. 1994. The H-NS protein is involved in the biogenesis of flagella in Escherichia coli. J Bacteriol 176:5537–5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ko M, Park C. 2000. H-NS-dependent regulation of flagellar synthesis is mediated by a LysR family protein. J Bacteriol 182:4670–4672. doi: 10.1128/JB.182.16.4670-4672.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krin E, Danchin A, Soutourina O. 2010. RcsB plays a central role in H-NS-dependent regulation of motility and acid stress resistance in Escherichia coli. Res Microbiol 161:363–371. doi: 10.1016/j.resmic.2010.04.002. [DOI] [PubMed] [Google Scholar]
- 13.Ko M, Park C. 2000. Two novel flagellar components and H-NS are involved in the motor function of Escherichia coli. J Mol Biol 303:371–382. doi: 10.1006/jmbi.2000.4147. [DOI] [PubMed] [Google Scholar]
- 14.Paul K, Carlquist WC, Blair DF. 2011. Adjusting the spokes of the flagellar motor with the DNA-binding protein H-NS. J Bacteriol 193:5914–5922. doi: 10.1128/JB.05458-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 17.Tang H, Blair DF. 1995. Regulated underexpression of the FliM protein of Escherichia coli and evidence for a location in the flagellar motor distinct from the MotA/MotB torque generators. J Bacteriol 177:3485–3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Simm R, Remminghorst U, Ahmad I, Zakikhany K, Römling U. 2009. A role for the EAL-like protein STM1344 in regulation of CsgD expression and motility in Salmonella enterica serovar Typhimurium. J Bacteriol 191:3928–3937. doi: 10.1128/JB.00290-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dudin O, Geiselmann J, Ogasawara H, Ishihama A, Lacour S. 2014. Repression of flagellar genes in exponential phase by CsgD and CpxR, two crucial modulators of Escherichia coli biofilm formation. J Bacteriol 196:707–715. doi: 10.1128/JB.00938-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boehm A, Kaiser M, Li H, Spangler C, Kasper CA, Ackermann M, Kaever V, Sourjik V, Roth V, Jenal U. 2010. Second messenger-mediated adjustment of bacterial swimming velocity. Cell 141:107–116. doi: 10.1016/j.cell.2010.01.018. [DOI] [PubMed] [Google Scholar]
- 21.Fang X, Gomelsky M. 2010. A post-translational, c-di-GMP dependent mechanism regulating flagellar motility. Mol Microbiol 76:1295–1305. doi: 10.1111/j.1365-2958.2010.07179.x. [DOI] [PubMed] [Google Scholar]
- 22.Paul K, Nieto V, Carlquist WC, Blair DF, Harshey RM. 2010. The c-di-GMP binding protein YcgR controls flagellar motor direction and speed to affect chemotaxis by a “backstop brake” mechanism. Mol Cell 38:128–139. doi: 10.1016/j.molcel.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Battesti A, Tesgaye YM, Gottesman S. 2012. H-NS regulation of IraD and IraM antiadaptors for control of RpoS degradation. J Bacteriol 194:2470–2478. doi: 10.1128/JB.00132-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wolf T, Janzen W, Blum C, Schnetz K. 2006. Differential dependence of StpA on H-NS in autoregulation of stpA and in regulation of bgl. J Bacteriol 188:6728–6738. doi: 10.1128/JB.00586-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arold ST, Leonard PG, Parkinson GN, Ladbury JE. 2010. H-NS forms a superhelical protein scaffold for DNA condensation. Proc Natl Acad Sci U S A 107:15728–15732. doi: 10.1073/pnas.1006966107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bloch V, Yang Y, Margeat E, Chaanieu A, Auge MT, Robert B, Arold ST, Rimsky S, Kochoyan M. 2003. The H-NS dimerization domain defines a new fold contributing to DNA recognition. Nat Struct Biol 10:212–218. doi: 10.1038/nsb904. [DOI] [PubMed] [Google Scholar]
- 27.Kawula TH, Orndorff PE. 1991. Rapid site-specific DNA inversion in Escherichia coli mutants lacking the histonelike protein H-NS. J Bacteriol 173:4116–4123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pesavento C, Becker G, Sommerfeldt N, Possling A, Tschowri N, Mehlis A, Hengge R. 2008. Inverse regulatory coordination of motility and curli-mediated adhesion in Escherichia coli. Genes Dev 22:2434–2446. doi: 10.1101/gad.475808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gualdi L, Tagliabue L, Bergtagnoli S, Ierano T, DeCastro C, Landini P. 2008. Cellulose modulates biofilm formation by counteracting curli-mediated colonization of solid surfaces in Escherichia coli. Microbiology 154:2017–2024. doi: 10.1099/mic.0.2008/018093-0. [DOI] [PubMed] [Google Scholar]
- 30.Boehm A, Vogel J. 2012. The csgD mRNA as a hub for signal integration via multiple small RNAs. Mol Microbiol 84:1–5. doi: 10.1111/j.1365-2958.2012.08033.x. [DOI] [PubMed] [Google Scholar]
- 31.Ogasawara H, Yamada K, Kori A, Yamamoto K, Ishihama A. 2010. Regulation of the Escherichia coli csgD promoter: interplay between five transcription factors. Microbiology 156:2470–2483. doi: 10.1099/mic.0.039131-0. [DOI] [PubMed] [Google Scholar]
- 32.Ogasawara H, Yamamoto K, Ishihama A. 2011. Role of the biofilm master regulator CsgD in cross-regulation between biofilm formation and flagellar synthesis. J Bacteriol 193:2587–2597. doi: 10.1128/JB.01468-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weber H, Pesavento C, Possling A, Tischendorf G, Hengge R. 2006. Cyclic-di-GMP-mediated signalling within the σS network of Escherichia coli. Mol Microbiol 62:1014–1034. doi: 10.1111/j.1365-2958.2006.05440.x. [DOI] [PubMed] [Google Scholar]
- 34.Gualdi L, Tagliabue L, Landini P. 2007. Biofilm formation-gene expression relay system in Escherichia coli: modulation of σS-dependent gene expression by the CsgD regulatory protein via σS protein stabilization. J Bacteriol 189:8034–8043. doi: 10.1128/JB.00900-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bougdour A, Cunning C, Baptiste PJ, Elliott T, Gottesman S. 2008. Multiple pathways for regulation of sigmaS (RpoS) stability in Escherichia coli via the action of multiple anti-adaptors. Mol Microbiol 68:298–313. doi: 10.1111/j.1365-2958.2008.06146.x. [DOI] [PubMed] [Google Scholar]
- 36.Bougdour A, Wickner S, Gottesman S. 2006. Modulating RssB activity: IraP, a novel regulator of sigma(S) stability in Escherichia coli. Genes Dev 20:884–897. doi: 10.1101/gad.1400306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sledjeski DD, Gupta A, Gottesman S. 1996. The small RNA, DsrA, is essential for the low temperature expression of RpoS during exponential growth of Escherichia coli. EMBO J 15:3993–4000. [PMC free article] [PubMed] [Google Scholar]
- 38.Minamino T, Yamaguchi S, Macnab RM. 2000. Interaction between FliE and FlgB, a proximal rod component of the flagellar basal body of Salmonella. J Bacteriol 182:3029–3036. doi: 10.1128/JB.182.11.3029-3036.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ueno T, Oosawa K, Aizawa S-I. 1992. M ring, S ring and proximal rod of the flagellar basal body of Salmonella typhimurium are composed of subunits of a single protein, FliF. J Mol Biol 227:672–677. doi: 10.1016/0022-2836(92)90216-7. [DOI] [PubMed] [Google Scholar]
- 40.Takaya A, Erhardt M, Karata K, Winterberg K, Yamamoto T, Hughes KT. 2012. YdiV: a dual function protein that targets FlhDC for ClpXP-dependent degradation by promoting release of DNA-bound FlhDC complex. Mol Microbiol 83:1268–1284. doi: 10.1111/j.1365-2958.2012.08007.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wada T, Hatamoto Y, Kutsukake K. 2012. Functional and expressional analyses of the anti-FlhD4C2 factor gene ydiV in Escherichia coli. Microbiology 158:1533–1542. doi: 10.1099/mic.0.056036-0. [DOI] [PubMed] [Google Scholar]
- 42.Lee LK, Ginsburg MA, Crovace C, Donohoe M, Stock D. 2010. Structure of the torque ring of the flagellar motor and the molecular basis for rotational switching. Nature 466:996–1000. doi: 10.1038/nature09300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rajagopala SV, Sikorski P, Kumar A, Mosca R, Vlasblom J, Arnold R, Franca-Koh J, Pakala SB, Phanse S, Ceol A, Hauser R, Siszler G, Wuchty S, Emili A, Babu M, Aloy P, Pieper R, Uetz P. 2014. The binary protein-protein interaction landscape of Escherichia coli. Nat Biotechnol 32:285–290. doi: 10.1038/nbt.2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ceschini S, Lupidi G, Coletta M, Pon CL, Fioretti E, Angeletti M. 2000. Multimeric self-assembly equilibria involving the histone-like protein H-NS. A thermodynamic study. J Biol Chem 275:729–734. [DOI] [PubMed] [Google Scholar]
- 45.Li H, Sourjik V. 2011. Assembly and stability of flagellar motor in Escherichia coli. Mol Microbiol 80:886–899. doi: 10.1111/j.1365-2958.2011.07557.x. [DOI] [PubMed] [Google Scholar]
- 46.Karpova TS, Baumann CT, He L, Wu X, Grammer A, Lipsky P, Hager GL, McNally JG. 2003. Fluorescence resonance energy transfer from cyan to yellow fluorescent protein detected by acceptor photobleaching using confocal microscopy and a single laser. J Microsc 209:56–70. doi: 10.1046/j.1365-2818.2003.01100.x. [DOI] [PubMed] [Google Scholar]
- 47.Brombacher E, Dorel C, Zehnder AJ, Landini P. 2003. The curli biosynthesis regulator CsgD co-ordinates the expression of both positive and negative determinants for biofilm formation in Escherichia coli. Microbiology 149:2847–2857. doi: 10.1099/mic.0.26306-0. [DOI] [PubMed] [Google Scholar]
- 48.Wada T, Morizane T, Abo T, Tominaga A, Inoue-Tanaka K, Kutsukake K. 2011. EAL domain protein YdiV acts as an anti-FlhD4C2 factor responsible for nutritional control of the flagellar regulon in Salmonella enterica serovar Typhimurium. J Bacteriol 193:1600–1611. doi: 10.1128/JB.01494-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou Y, Gottesman S, Hoskins JR, Maurizi MR, Wickner S. 2001. The RssB response regulator directly targets sigma(S) for degradation by ClpXP. Genes Dev 15:627–637. doi: 10.1101/gad.864401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jubelin G, Vianney A, Beloin C, Ghigo J-M, Lazzaroni J-C, Lejeune P, Dorel C. 2005. CpxR/OmpR interplay regulates curli gene expression in response to osmolarity in Escherichia coli. J Bacteriol 187:2038–2049. doi: 10.1128/JB.187.6.2038-2049.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dersch P, Kneip S, Bremer E. 1994. The nucleoid-associated DNA-binding protein H-NS is required for the efficient adaptation of Escherichia coli K-12 to a cold environment. Mol Gen Genet 245:255–259. [DOI] [PubMed] [Google Scholar]
- 52.Zhou Y, Gottesman S. 2006. Modes of regulation of RpoS by H-NS. J Bacteriol 188:7022–7025. doi: 10.1128/JB.00687-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hengge-Aronis R. 2002. Signal transduction and regulatory mechanisms involved in control of the σS (RpoS) subunit of RNA polymerase. Microbiol Mol Biol Rev 66:373–395. doi: 10.1128/MMBR.66.3.373-395.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Majdalani N, Vanderpool CK, Gottesman S. 2005. Bacterial small RNA regulators. Crit Rev Biochem Mol Biol 40:93–113. doi: 10.1080/10409230590918702. [DOI] [PubMed] [Google Scholar]
- 55.Uyar E, Kurokawa K, Oshima T. 2009. Differential binding profiles of StpA in wild-type and hns mutant cells: a comparative analysis of cooperative partners by chromatin immunoprecipitation-microarray analysis. J Bacteriol 191:2388–2391. doi: 10.1128/JB.01594-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.De Lay N, Gottesman S. 2012. A complex network of small noncoding RNAs regulate motility in Escherichia coli. Mol Microbiol 86:524–538. doi: 10.1111/j.1365-2958.2012.08209.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mika F, Hengge R. 2014. Small RNAs in the control of RpoS, CsgD, and biofilm architecture of Escherichia coli. RNA Biol 11:494–507. doi: 10.4161/rna.28867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mika F, Hengge R. 2013. Small regulatory RNAs in the control of motility and biofilm formation in E. coli and Salmonella. Int J Mol Sci 14:4560–4579. doi: 10.3390/ijms14034560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tang H, Billings S, Wang X, Sharp L, Blair DF. 1995. Regulated underexpression and overexpression of the FliN protein of Escherichia coli and evidence for an interaction between FliN and FliM in the flagellar motor. J Bacteriol 177:3496–3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


