Abstract
Shox2 is expressed in several developing organs in a tissue specific manner in both mice and humans, including the heart, palate, limb, and nervous system. To better understand the spatial and temporal expression patterns of Shox2 and to systematically dissect the genetic cascade regulated by Shox2, we created Shox2-LacZ and Shox2-Cre knock-in mouse lines. We show that the Shox2-LacZ allele expresses beta-galactosidase reporter gene in a fashion that recapitulates the endogenous Shox2 expression pattern in developing organs, including the sinoatrial node (SAN), the anterior portion of the palate, and the proximal region of the limb bud. Conditional deletion of Shox2 in mice carrying the Shox2-Cre allele yielded SAN phenotypes that resemble conventional Shox2 knockout mice. Our results indicate that the Shox2-Cre allele offer a useful tool for tissue specific manipulation of genes in a number of developing organs, particularly in the developing SAN.
Keywords: Shox2, Cre, gene deletion, heart, palate, limb
During development, the vertebrate heart develops from a tubular structure with a single circulation to a four chambered structure with a dual circulation. This correlates with change of the heart beating pattern from a peristaltic motion to a sophisticated synchronous contraction due to the maturation of the cardiac conduction system (CCS). The CCS initiates and conducts electric signals to stimulate the contraction of atrial and ventricular working myocardium in a coordinated manner. This conduction system is comprised of the following components: the sino-atrial node (SAN) the atrio-ventricular node (AVN), the Bachmann’s bundle (in the atrium), and ventricular conduction sites including the His bundle, right bundle branch, left bundle branch and Purkinje fibers.
The SAN is the first site of CCS components to become functional and the origin of myocardial stimulating electrical current. The SAN is located at the junction between the vena cava and the right atrium. At E10.5, the SAN is fully functional and can be identified histologically (van Mierop and Gessner, 1970, Christoffels et al., 2010). During embryogenesis from E11.5 onward, the SAN appears to be a bulging structure at the dorsal side of the right atrium wall. Previous studies revealed several SAN specific molecular markers including Tbx3 and the hyperpolarization-activated cyclic nucleotide-gated channel 4 (Hcn4) (Ludwig et al., 1999; Hoogaars et al., 2004). These markers are expressed distinctively in CCS and are important to maintain SAN identity and functions during cardiogenesis.
Heart disease is the leading cause of the deaths in the developed countries. (Xu et al., 2010; Kochanek et al., 2011). Various types of heart diseases from the most common atrial fibrillation to the lethal sudden cardiac death have been associated with pacemaker defects (Tsai et al., 2000; Marsman et al., 2011). Recent studies have established a casual relationship between certain genetic defects and SAN abnormalities, though the genetic regulatory network is still not well understood (Puskaric et al., 2010; Aanhaanen et al., 2011). Understanding such genetic regulatory mechanism may shed light on therapeutic approaches for both congenital and acquired CCS defects.
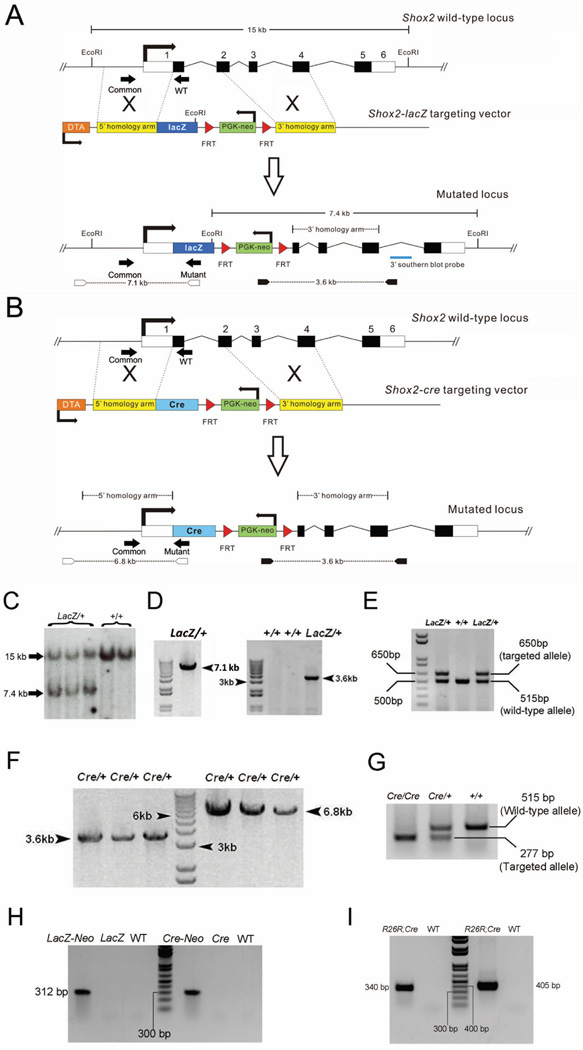
The Short stature homeobox 2 (Shox2) gene has been shown to be expressed in a number of developing organs in a tissue specific manner (Yu et al., 2005; Blaschke et al., 2007). In the developing palate, Shox2 expression is restricted in the anterior portion of the mesenchyme; and in the developing limb, its expression is only found in the proximal region of the limb (Yu et al., 2005; 2007; Cobb et al., 2006). In the developing heart, Shox2 is expressed specifically in the inflow tract region where the SAN derives and later in the developing SAN (Blaschke et al., 2007; Espinoza-Lewis et al., 2009). Targeted inactivation of Shox2 leads to severe defects in multiple organs including anterior clefting of the palate, elimination of the stylopod, and defective differentiation of the SAN (Yu et al., 2007; 2007; Cobb et al., 2006; Blaschke et al., 2007; Gu et al., 2008a; Espinoza-Lewis et al., 2009). In order to reveal a fine and real time expression pattern of Shox2 in developing mouse embryos, we generated a Shox2-LacZ knock-in allele by gene targeting in ES cells. In this case, the beta-galactosidase protein coding sequence and a FRT flanked PGK-neo cassette were introduced to replace Shox2’s exon1, intron 1 and a small proportion of exon 2 (Fig. 1A and B). Southern blotting and a PCR screen confirmed the successful targeting of the knock-in alleles in ES cells (Fig. 1C and D). The PCR screening strategy and probe used for Southern blotting are indicated in Fig. 1A. The numbers of positive ES cell clones were 9 out of 180 for Shox2-LacZ allele. Germline transmitted heterozygous mice were genotyped using a PCR based method as described in Materials and Methods. 10 agouti (indicates SV129 CJ-7 cell lineage) pups out of 13 (76.9%) F1 mice were observed in two litters from Shox2-LacZ chimeras.
Figure 1.
Targeting strategy of generating Shox2-LacZ and Shox2-Cre mice. (A and B) Targeted Cre or LacZ replacement of the Shox2 gene in mice. The mouse Shox2 genomic structure comprises an 8.3 kb region, and contains 6 exons, as indicated by numbered blocks. Black blocks indicate coding regions. The targeting vector contains genomic fragments flanking the lacZ/Cre-FRT-neo-PGK-FRT cassette which replaces exon 1 and part of exon 2 of Shox2 when correct homologous recombination occurs in ES cells. (C) Southern blot analysis for Shox2-LacZ targeted allele (digested with EcoRI and probed with 3’southern blot probe). (D) PCR product for Shox2-LacZ knock-in allele in ES cells. 7.1 kb and 3.6 kb PCR products indicate the correct homologous recombination.at 5' and 3' of the Shox2 locus respectively. Primer sets are labeled by arrow heads in A. (E) PCR analysis of DNA extracted from yolk sac of E10.5 embryos reveals the wild-type and LacZ targeted alleles. Primer sets are labeled by arrow heads in B. (F) PCR screen showing correct homologous recombination of Shox2-Cre targeted vector in ES cells. A set of common, WT and Mutant primers are used and labeled by black arrows in A. (G) PCR analysis of DNA extracted from yolk sac of E10.5 embryos reveals the Shox2-Cre alleles. The primer sets are shown by black arrowheads in B. (H) Verification of neo-PGK deletion by PCR analysis. The neo-PGK cassette is detected in F1 mice (LacZ-Neo and Cre-Neo) and undetectable in WT mice and offspring from F1 mice and FLP deleter mice (LacZ and Cre). (I) Identification of R26R;Shox2Cre/+ double heterozygous mice by PCR assay. A 340 bp PCR product indicates the R26R allele in double heterozygote. The 405 bp product indicates Cre allele.
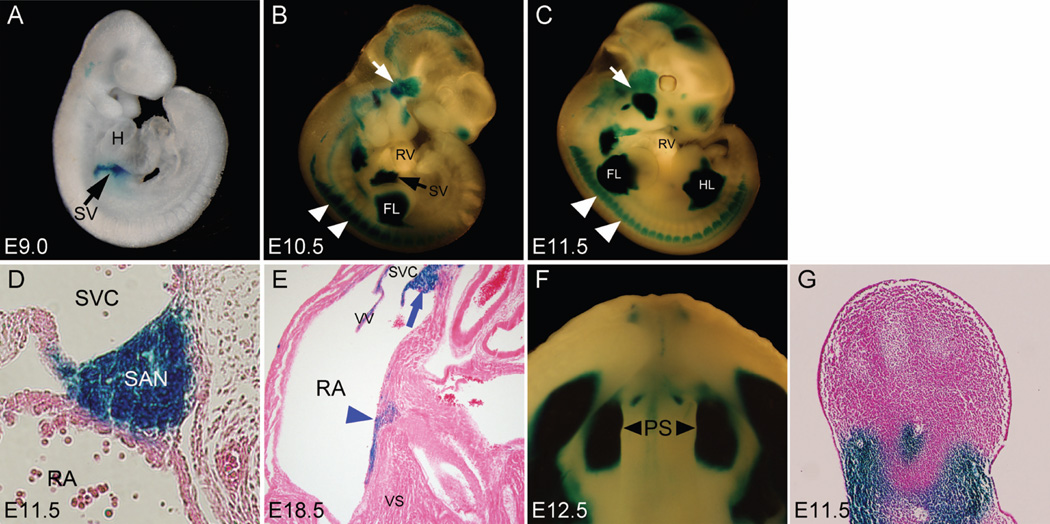
After removal of the PGK-Neo cassette by crossing F1 heterozygous mice to FLP deleter mice, Shox2LacZ/+ mice were used to examine Shox2 expression patterns by X-gal staining in developing embryos at selected stages. As shown in Fig. 2, X-gal staining indeed revealed Shox2 expression patterns identical to that was reported previously by in situ hybridization in developing embryos at E9.0, E10.5, and E11.5 (Yu et al., 2005; 2007; Blaschke et al., 2007; Espinoza-Lewis et al., 2009). At E9.0, beta-galactosidase activity was detected in the sinus venosus of the developing heart (Fig. 2A). At 10.5 and E11.5, Robust expression of beta-galactosidase activity persisted in the SAN and surrounding superior vena cava tissue (Fig. 2B–D). We observed beta-galactosidase activity in the SAN region and tissue on top of the ventricular septum in the atrioventricular junction at E18.5 (Fig. 2E) as we predicted from previous report (Blaschke et al., 2007; Hahurij, 2011). We identified that this group of cells contribute to the developing atrioventricular conduction system (Data not shown). Shox2 expression was restricted in the atrium and was not found in the ventricular proportion of the heart. In addition, beta-galactosidase activity was observed in the proximal region of the limb, the anterior portion of the palatal shelves, and the temporomandibular junction in E10.5 and E11.5 embryos (Fig. 2B, C, F and G), similar to the Shox2 mRNA expression patterns reported previously (Yu et al., 2005, 2007; Gu et al., 2008b). We also observed beta-galactosidase activity in the dorsal root ganglia and brain region (Fig. 2B and C), consistent with previous reports (Scott et al., 2011). These observations indicate that the knock-in strategy we used does not affect the regulatory elements required for the tissue specific expression of Shox2, allowing recapitulation of endogenous Shox2 expression.
Figure 2.
Expression of beta-galactosidase in Shox2lacZ/+ embryos revealed by X-gal staining. (A) beta-galactosidase expression in the sinus venosus (black arrow) was detectable as early as E9.0. (B) At E10.5, beta-galactosidase expression was seen, besides the sinus venosus (SV), in other developing organs and tissues including the proximal parts of the limb buds, the dorsal root ganglion (white arrowheads), several craniofacial structures including the temporomandibular junction (white arrow), and the brain region. (C) At E11.5, beta-galactosidase was continuously expressed in the developing organs and tissues observed at E10.5. (D) X-gal staining replicated Shox2 expression pattern in the sinoatrial node (SAN) at E11.5. (E) beta-galactosidase expression was detected the SAN and on top of the ventricular septum at E18.5. (F) At E12.5, beta-galactosidase was strongly expressed in the anterior portion (black arrow heads) of the palatal shelves (PS). (G) Section of an E11.5 Shox2LacZ/+ limb showing beta-galactosidase expression in the mesenchymal cells of the proximal limb bud. H, heart; SV, sinus venosus; RV, right ventricle; RA, right atrium; FL, forelimb bud; HL, hindlimb bud; SAN, sinoatrial node; SVC, superior vena cava; PS, palatal shelves; VS, ventricular septum.
The unique expression pattern of Shox2 in the developing SAN, palatal shelves, and the limb in Shox-LacZ knock-in mice prompted us to generate Shox2-Cre knock-in mice, which would provide a valuable research tool for studying gene function in the developing SAN, the palate and limb. Using the same targeting strategy for the creation of Shox2-LacZ allele (Fig. 1B), we generated the Shox2-Cre knock-in allele in ES cells and Shox2-Cre mice, as confirmed by PCR genotyping (Fig. 1F–G). The Shox2-Cre construct was targeted into G4 ES cell line (George et al. 2007). The numbers of positive ES cell clones were 7 out of 144 for Shox2-Cre allele. 8 Cre positive F1 mice out of 14 were identified in two litters from chimeras.
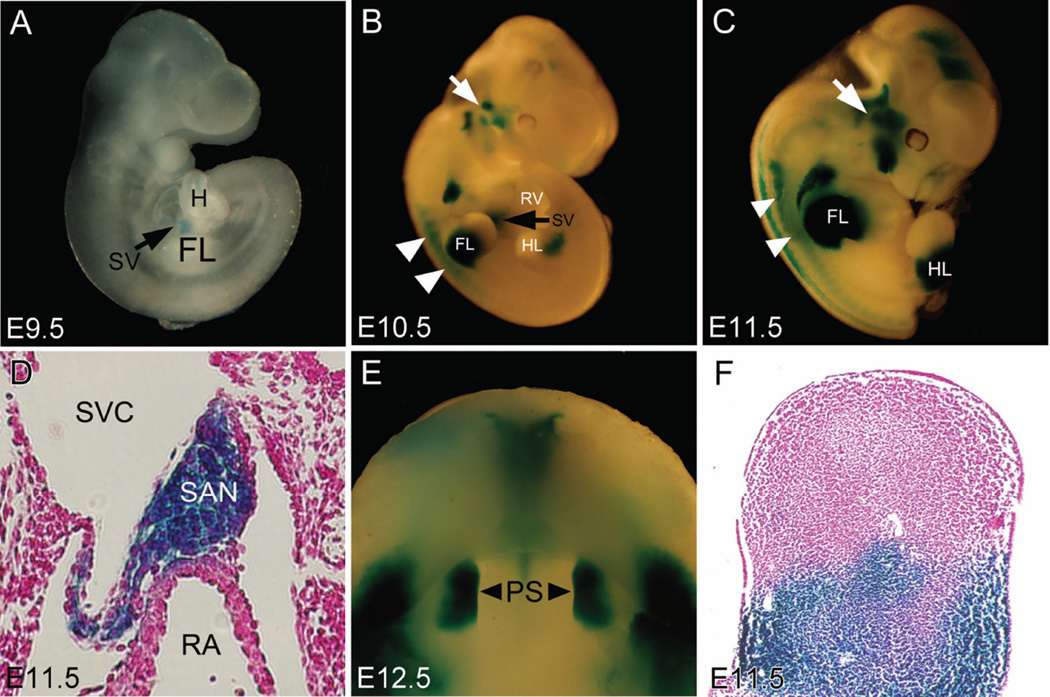
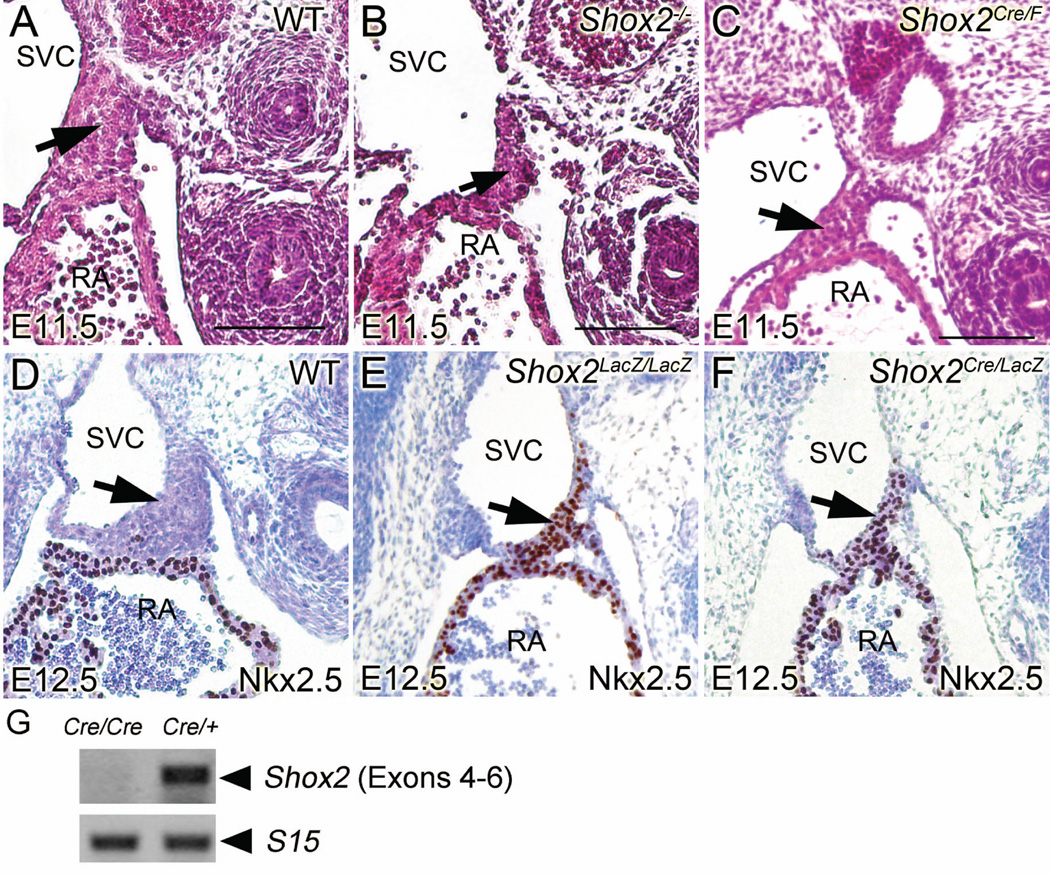
To evaluate the Cre activity in Shox2-Cre knock-in mice, R26R reporter mice were crossed to Shox2-Cre knock-in mice to generate Shox2Cre/+;R26R double heterozygous embryos for X-gal staining (Fig. 1I). At E9.5, beta-galactosidase activity was first detected in the developing sinus venosus (Fig. 3A). Compared to the earlier expression of beta-galactosidase in Shox2-LacZ embryos, such delay of about a half day in Shox2Cre/+;R26R double heterozygous embryos is likely attributed by the time needed for accumulation of Cre recombinase protein and transcription/translation of the activated R26R allele. At E10.5 and E11.5, in whole mount stained embryos, beta-galactosidase was robustly expressed in all early Shox2-expressing tissues including the SAN, the proximal domain of the limb, the anterior region of the palatal shelves, the temporomandibular junction, as well as the dorsal root ganglia and brain (Fig. 3B–F). These expression domains correlate with that was observed in Shox2-LacZ mice. To determine the efficiency of Cre recombinase in inactivating floxed gene, we crossed Shox2-Cre mice to Shox2flox mice (Cobb et al., 2006) to generate Shox2Cre/F embryos. In Shox2Cre/F embryos, we observed hypoplastic SAN phenotype similar to that found in Shox2−/− embryos at E11.5 (Fig. 4A–C). At E12.5, We also found that both Shox2Cre/LacZ and Shox2Cre/F exhibited similar phenotypes in the SAN as seen in Shox2-null mice (Fig. 4D–F). Similar to that was found in Shox2-null mutants (Yu et al., 2005; Blaschke et al., 2007), Shox2LacZ/LacZ, Shox2Cre/LacZ and Shox2Cre/F embryos began to exhibit embryonic lethality at E11.5 (Supplementary data). To confirm that the Shox2-Cre line is a null allele, reverse transcription PCR was performed using methods described previously (Yu et al., 2005), Shox2 mRNA was undetectable in Shox2Cre/Cre homozygous mice (Fig. 4G). This proved that Shox2 transcription was successfully inactivated in Shox2-Cre knock-in alleles demonstrating that the Shox2-Cre allele can be used as an efficient tool for tissue specific inactivation of targeted genes.
Figure. 3.
Detection of Cre activity in Shox2cre/+;R26R double heterozygous embryos. (A) Cre-mediated beta-galactosidase expression (blue) in the sinus venosus (black arrow) was detectable at E9.5. (B) At E10.5, beta-galactosidase expression in Shox2cre/+;R26R double heterozygous embryos persisted in the sinus venosus (SV), and was also found in the proximal domain of the limb buds, the dorsal root ganglion (white arrowheads), and the temporomandibular junction (white arrow). (C) An E11.5 Shox2cre/+;R26R embryo showing beta-galactosidase activity in the same organs and tissues as described in E10.5 embryo. (D) X-gal staining was seen in the SAN of an E11.5 Shox2cre/+;R26R embryo. (E) Beta-galactosidase expression was detected in the anterior domain (black arrowheads) of the palatal shelves (PS). (F) Beta-galactosidase expression was found restrictedly in the proximal region of the limb bud at E11.5.
Figure. 4.
Similar hypoplasia phenotype was observed in the SAN of Shox2Cre/F, Shox2LacZ/LacZ and Shox2Cre/LacZ mice as seen in Shox2-null mice. (A–C) representative H&E staining pictures of the developing SAN in E11.5 WT (A), Shox2 conventional knockout (B) and Shox2Cre/F embryos(C). Arrows denote SAN region. (D–F) representative IHC pictures of the developing SAN in E11.5 WT (D), Shox2LacZ/LacZ (E) and Shox2Cre/LacZ embryos (F). Arrows denote SAN region. Note the ectopic expression of Nkx2.5 in the SAN region of Shox2LacZ/LacZ and Shox2Cre/LacZ embryos. (G) RT-PCR assays demonstrating the absence of Shox2 transcript in E11.5 Shox2Cre/Cre mice. Scale bar: 100 µm.
While several Cre lines have been created previously for manipulating gene expression in the CCS including the SAN (Hoesl et al., 2011; Arnolds et al., 2011), our Shox2-Cre allele provide another unique Cre line to manipulate gene function in SAN development. It has been demonstrated that heterogeneity, at both the cellular and molecular levels, exists in the developing palatal shelves along the anteroposterior (A-P) axis (Hilliard et al., 2005; Okano et al., 2006; Gritli-Linde, 2007). A number of genes including Shox2 exhibit differential expression and play different role in regulating development of different portion of the palate along the A-P axis (Zhang et al., 2002; Yu et al., 2005; He et al., 2008; Liu et al., 2008; Xiong et al., 2009; Li et al., 2011). The vertebrate limb grows and lengthens along the proximodistal axis, and is patterned into the stylopod, zeugopod, and autopod by differential expression of genes such as Hox genes (Davis et al., 1995; Wellik and Capecchi, 2003; Boulet and Capecchi, 2004). The unique Cre expression in the anterior portion of the developing palatal shelves and the proximal domain of the limb bud in the Shox2-Cre mice suggest that Shox2-Cre mice could be a valuable tool for gene manipulation in the specific domain of the developing palate and limb to study gene function in these two organs.
MATERIALS AND METHODS
Mouse Strains
The FLP deleter and R26R mice (Soriano, 1999) were obtained from the Jackson laboratory (Bar Harbor, ME). The Shox2flox mice have been described previously (Cobb et al., 2006). A set of primers (5’-AAAGTCGCTCTGAGTTGTTAT-3’ and 5’-GCGAAGAGTTTGTCCTCAACC-3’) is used to identify the R26R allele.
Generation of Shox2-lacZ and Shox2-Cre knock-in mice
A 15 kb EcorI genomic DNA fragment comprising all six exons of the Shox2 gene was cloned and used in generation of Shox2-lacZ and Shox2-Cre allele, respectively. To insert the LacZ or Cre expression cassette into the Shox2 locus, we constructed a targeting vector containing a 5.6 kb homologous arm and a 2.9 kb homologous arm flank the lacZ/Cre-FRT-neo-PGK-FRT cassette (Fig. 1A and B). Additionally, a diphtheria toxin expression cassette was placed in the 5’ flanking region of the 5’ homologous arm to rule out random insertion. The targeting vector was digested by AscI restriction endonucleases for linearization and subsequently electroporated into mouse ES cells. The Shox2-LacZ construct was targeted in CJ7 ES cells (Swiatek and Gridley, 1993), while the Shox2-Cre construct was targeted in G4 ES cells (George et al. 2007). To obtain ES cell clones with correct homologous recombination, Neomycin (G418) resistant colonies were selected and screened by Southern blotting assay (Fig. 1C), or by a long range PCR screening strategy (Fig. 1D). For Shox2-LacZ targeted allele, the 5’ PCR primer located in the neo-PGK cassette (5’-CGAAACGATCCTCATCCTGT-3’) and the 3’ PCR primer located in 3’ flaking region of 3’ homologous arm (5’-GGACCAGCCTCTGTATTGGA-3’) (Fig. 1A) amplified a 3.6 kb product from the genuine Shox2-lacZ allele (Fig. 1D). As to the identification of 5’ homologous recombination, a forward primer (5’-AGTTTGGGAGATGAGGGAAAACCATCATCAGAATAT-ATTGTATACAG-3’) located in 5’ flaking region of 5’ homologous arm and a reverse primer (5’-GACAGTATCGGCCTCAGGAA-3’) located in the LacZ cassette were used and a PCR product of 7.1 kb indicated targeted Shox2 locus (Fig. 1D). For Shox2-Cre allele, long range PCR on both 5’ end and 3’ end of the targeted Shox2 locus was performed to identify correctly targeted allele using two pairs of primers. The first set of primers is identical to that used to identify 3’ homologous recombination of Shox2-lacZ allele, and the second set of primers, with the same forward primer used in Shox2-LacZ 5’ homologous recombination screening and the reverse primer (5’-GCAAACGGACAGAAGCATTT-3’) located in the Cre cassette, identified 5’ homologous recombination by amplifying a product of 6.8 kb (Fig1. F). For both lines, ES cells from two different positive clones were injected in to blastocysts with C57BL/6 background. Chimeras were crossed with C57BL/6 females (Charles River) to generate F1 mice and they were genotyped by long ranged PCR of tail DNA. Mice and embryos, maintained on C57BL/6 and CD-1 background separately, were routinely genotyped by PCR analysis of tail DNA or yolk sacs of embryos. CD-1 mice with knock-in alleles are used for subsequent analysis in this study. For Shox2-LacZ allele, a set of three primers (Common: 5’-GCCCCATTGATGTGTTATTA-3’; Mutant: 5’-GACAGTATCGGCCTCAGGAA-3’; WT: 5’-GATAAGGGAAGGCAGTAAGG-3’) produced a product of 650bp from the targeted allele and a product of 515 bp from the wild type allele (Fig.1A and E). PCR genotyping of Shox2-Cre allele was conducted using a set of three primers (Common: 5’-GCCCCATTGATGTGTTATTA-3’; Mutant: 5’-GCAGTTTCCAGGTATGCCCAG-3’; WT: 5’-GATAAGGGAAGGCAGTAAGG-3’) produced a product of 277bp from the targeted allele and a product of 515 bp from the wild type allele (Fig. 1B and G). After crossing F1 mice with FLP deleter mice, to confirm the removal of neo-PGK cassette, a set of primers (5’-AGAGGCTATTCGGCTAT-GACTG-3’ and 5’-ATACTTTCTCGGCAGGAGCA-3’) was used to amplify a 312 bp product from neo-PGK fragment. F1 mice are positive for neo-PGK detection, in contrast, the neo-PGK cassette is not detectable in WT mice and offspring form F1 mice and FLP deleter mice (Fig. 1H). The Shox2-LacZ and Shox2-Cre alleles will be available upon request to the research community.
Long Range PCR
DNA sample was prepared following DNA extraction procedure for southern blot assay described previously (Alexandra, 2000). TaKaRa LA Taq enzyme was used for long range PCR. Annealing temperature was set to 58°C for 45 seconds and the time for elongation was set to 8 minutes for 5’ homologous recombination screening and 5 minutes for 3’ homologous recombination screening. Cycle number was set to 35.
X-Gal Staining
X-gal staining on cryosections and whole embryo was carried out following the procedures described previously (Hogan et al., 1994). After X-gal staining, sections were counterstained with nuclear fast red subsequently.
Immunohistochemistry Assay
Immunohistochemistry assay using antibody against Nkx2.5 (abcam ab35842) on paraffinsections was carried out following the procedures available online (Invitrogen). After diaminobenzidine color development, sections were counterstained with methyl green subsequently.
Reverse Transcription PCR
E11.5 mouse heart with sinus venosus were used to isolate mRNA by TRIzol reagent (Invitrogen). Subsequent first-strand cDNA synthesis was performed using a SuperScript kit (Invitrogen). A set of primers (5-ACCAGCAAGAACTCCAGCAT-3 and 5-GCCACACTCCTTTGTCCAGT-3) that cover exon 4 to exon 6 of Shox2 amplified a 371 bp cDNA product to detect Shox2 transcript in the Shox2Cre/Cre mice. As the positive control, a pair of primers (5-TTCCGCAAGTTCACCTACC-3 and 5-CGGGCCGGCCATGCTTTACG-3) that amplify a 361 bp sequence of S15 RNA was included.
Table 1. Embryonic lethality in Shox2LacZ/LacZ, Shox2Cre/LacZ and Shox2Cre/F mice.
Embryonic lethality was observed at different developmental stages. This table shows the number of dead and survived mutant embryos. Total embryo numbers include WT embryos.
| Mutant Allele | Stage | Dead Mutants | Survived Mutants | Total Embryos |
|---|---|---|---|---|
| Shox2LacZ/LacZ | E11.5 | 3 | 6 | 33 |
| E12.5 | 7 | 3 | 42 | |
| E14.5 | 12 | 1 | 58 | |
| Shox2Cre/LacZ | E11.5 | 2 | 3 | 21 |
| E12.5 | 6 | 3 | 35 | |
| E14.5 | 1 | 0 | 10 | |
| Shox2Cre/F | E11.5 | 2 | 4 | 13 |
| E12.5 | 1 | 1 | 6 | |
| E14.5 | 2 | 0 | 7 | |
Acknowledgements
This work was supported by NIH grant R01 DE17792 to Y.C.
Literature Cited
- Aanhaanen WTJ, Boukens BJD, Sizarov A, Wakker V, de Gier-de Vries C, van Ginneken A, Moorman AFM, Coronel R, Christoffels VM. Defective Tbx2-dependent patterning of the atrioventricular canal myocardium causes accessory pathway formation in mice. The Journal of Clinical Investigation. 2011;121:534–544. doi: 10.1172/JCI44350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnolds DE, Moskowitz IP. Inducible recombination in the cardiac conduction system of minK: CreERT2 BAC transgenic mice. Genesis. 2011;49(11):878–884. doi: 10.1002/dvg.20759. [DOI] [PubMed] [Google Scholar]
- Alexandra JL. Gene Targeting: A Practical Approach. Oxford University Press; 2000. pp. 122–123. [Google Scholar]
- Blaschke RJ, Hahurij ND, Kuijper S, Just S, Wisse LJ, Deissler K, Maxelon T, Anastassiadis K, Spitzer J, Hardt SE, Schöler H, Feitsma H, Rottbauer W, Blum M, Meijlink F, Rappold G, Gittenberger-de Groot AC. Targeted Mutation Reveals Essential Functions of the Homeodomain Transcription Factor Shox2 in Sinoatrial and Pacemaking Development. Circulation. 2007;115:1830–1838. doi: 10.1161/CIRCULATIONAHA.106.637819. [DOI] [PubMed] [Google Scholar]
- Boulet AM, Capecchi MR. Multiple roles of Hoxa11 and Hoxd11 in the formation of the mammalian forelimb zeugopod. Development. 2004;131:299–309. doi: 10.1242/dev.00936. [DOI] [PubMed] [Google Scholar]
- Christoffels VM, Smits GJ, Kispert A, Moorman AFM. Development of the pacemaker tissues of the heart. Circ Res. 2010;106:240–254. doi: 10.1161/CIRCRESAHA.109.205419. [DOI] [PubMed] [Google Scholar]
- Davis AP, Witte DP, Hsieh-Li HM, Potter SS, Capecchi MR. Absence of radius and ulna in mice lacking hoxa-11 and hoxd-11. Nature. 1995;375:791–795. doi: 10.1038/375791a0. [DOI] [PubMed] [Google Scholar]
- Espinoza-Lewis RA, Yu L, He F, Liu H, Tang R, Shi J, Sun X, Martin J, Wang D, Yang J, Chen YP. Shox2 is essential for the differentiation of cardiac pacemaker cells by repressing Nkx2-5. Developmental Biology. 2009;327:376–385. doi: 10.1016/j.ydbio.2008.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gritli-Linde A. Molecular control of secondary palate development. Developmental Biology. 2007;301:309–326. doi: 10.1016/j.ydbio.2006.07.042. [DOI] [PubMed] [Google Scholar]
- Gu S, Wei N, Yu X, Jiang Y, Fei J, Chen YP. Mice with an anterior cleft of the palate survive neonatal lethality. Developmental Dynamics. 2008;237:1509–1516. doi: 10.1002/dvdy.21534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahurij ND. Cardiac Development The Posterior Heart Field and Atrioventricular Reentry Tachycardia. Vrije Universiteit; 2011. p. 78. [Google Scholar]
- He F, Xiong W, Espinoza-Lewis R, Liu C, Gu S, Nishita M, Suzuki K, Yamada G, Minami Y, Chen YP. Wnt5a regulates directional cell migration and cell proliferation via Ror2-mediated noncanonical pathway in mammalian palate development. Development. 2008;135:3871–3879. doi: 10.1242/dev.025767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilliard SA, Yu L, Gu S, Zhang Z, Chen YP. Regional regulation of palatal growth and patterning along the anterior–posterior axis in mice. Journal of Anatomy. 2005;207:655–667. doi: 10.1111/j.1469-7580.2005.00474.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoesl E, Stieber J, Herrmann S, Feil S, Tybl E, Hofmann F, Feil R, Ludwig A. Tamoxifen-inducible gene deletion in the cardiac conduction system. Journal of Molecular and Cellular Cardiology. 2008;45:62–69. doi: 10.1016/j.yjmcc.2008.04.008. [DOI] [PubMed] [Google Scholar]
- Hoogaars WMH, Hoogaars WMH, Tessari A, Moorman AFM, de Boer PAJ, Hagoort J, Soufan AT, Campione M, Christoffels VM. The transcriptional repressor Tbx3 delineates the developing central conduction system of the heart. Cardiovascular Research. 2004;62:489–499. doi: 10.1016/j.cardiores.2004.01.030. [DOI] [PubMed] [Google Scholar]
- Kochanek K, Xu J, Murphy S, Miniño A, Kung H. Deaths: Preliminary Data for 2009. National Vital Statistics Reports. 2011;59(4) [PubMed] [Google Scholar]
- Li L, Lin M, Wang Y, Cserjesi P, Chen Z, Chen YP. BmprIa is required in mesenchymal tissue and has limited redundant function with BmprIb in tooth and palate development. Developmental Biology. 2011;349:451–461. doi: 10.1016/j.ydbio.2010.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Lan Y, Pauws E, Meester-Smoor MA, Stanier P, Zwarthoff EC, Jiang R. The Mn1 transcription factor acts upstream of Tbx22 and preferentially regulates posterior palate growth in mice. Development. 2008;135:3959–3968. doi: 10.1242/dev.025304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig A, Ludwig A, Zong X, Stieber J, Hullin R, Hofmann F, Biel M. Two pacemaker channels from human heart with profoundly different activation kinetics. EMBO J. 1999;18:323–2329. doi: 10.1093/emboj/18.9.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano J, Suzuki S, Shiota K. Regional heterogeneity in the developing palate: morphological and molecular evidence for normal and abnormal palatogenesis. Congenital Anomalies. 2006;46:49–54. doi: 10.1111/j.1741-4520.2006.00103.x. [DOI] [PubMed] [Google Scholar]
- Puskaric S, Schmitteckert S, Mori AD, Glaser A, Schneider KU, Bruneau BG, Blaschke RJ, Steinbeisser H, Rappold G. Shox2 mediates Tbx5 activity by regulating Bmp4 in the pacemaker region of the developing heart. Human Molecular Genetics. 2010;19:4625–4633. doi: 10.1093/hmg/ddq393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott A, Hasegawa H, Sakurai K, Yaron A, Cobb J, Wang F. Transcription factor short stature homeobox 2 is required for proper development of tropomyosin-related kinase B-expressing mechanosensory neurons. J Neurosci. 2011;31(18):6741–6749. doi: 10.1523/JNEUROSCI.5883-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21(1):70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Tsai C-F, Tai C-T, Hsieh M-H, Lin W-S, Yu W-C, Ueng K-C, Ding Y-A, Chang M-S, Chen S-A. Initiation of Atrial Fibrillation by Ectopic Beats Originating From the Superior Vena Cava : Electrophysiological Characteristics and Results of Radiofrequency Ablation. Circulation. 2000;102:67–74. doi: 10.1161/01.cir.102.1.67. [DOI] [PubMed] [Google Scholar]
- van Mierop LHS, Gessner IH. The morphologic development of the sinoatrial node in the mouse. Am J Cardiol. 1970;25:204–212. doi: 10.1016/0002-9149(70)90580-1. [DOI] [PubMed] [Google Scholar]
- Wellik DM, Capecchi MR. Hox10 and Hox11 Genes Are Required to Globally Pattern the Mammalian Skeleton. Science. 2003;301:363–367. doi: 10.1126/science.1085672. [DOI] [PubMed] [Google Scholar]
- Xiong W, He F, Morikawa Y, Yu X, Zhang Z, Lan Y, Jiang R, Cserjesi P, Chen YP. Hand2 is required in the epithelium for palatogenesis in mice. Developmental Biology. 2009;330:131–141. doi: 10.1016/j.ydbio.2009.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Kochanek KD, Murphy SL, Tejada-Vera B. Deaths: Final Data for 2007. National Vital Statistics Reports. 2010;58(19) [PubMed] [Google Scholar]
- Yu L, Gu S, Alappat S, Song Y, Yan M, Zhang X, Zhang G, Jiang Y, Zhang Z, Zhang Y, Chen YP. Shox2-deficient mice exhibit a rare type of incomplete clefting of the secondary palate. Development. 2005;132:4397–4406. doi: 10.1242/dev.02013. [DOI] [PubMed] [Google Scholar]
- Yu L, Liu H, Yan M, Yang J, Long F, Muneoka K, Chen YP. Shox2 is required for chondrocyte proliferation and maturation in proximal limb skeleton. Developmental Biology. 2007;306:549–559. doi: 10.1016/j.ydbio.2007.03.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Song Y, Zhao X, Zhang X, Fermin C, Chen YP. Rescue of cleft palate in Msx1-deficient mice by transgenic Bmp4 reveals a network of BMP and Shh signaling in the regulation of mammalian palatogenesis. Development. 2002;129:4135–4146. doi: 10.1242/dev.129.17.4135. [DOI] [PubMed] [Google Scholar]