Abstract
Background
Hyper-IgE Syndrome (HIES) is a rare, autosomal dominant (AD) immunodeficiency characterized by eczema, Staphylococcus aureus skin abscesses, pneumonia with pneumatocele formation, Candida infections, and skeletal/connective tissue abnormalities. Recently it was shown that heterozygous STAT3 mutations cause AD-HIES.
Objective
To determine the spectrum and functional consequences of heterozygous STAT3 mutations in a cohort of HIES patients.
Methods
We sequenced the STAT3 gene in 38 HIES patients (NIH-score >40 points) from 35 families, quantified TH17 cells in peripheral blood, and evaluated tyrosine phosphorylation of STAT3.
Results
Most STAT3 mutations in our cohort were in the DNA-binding domain (DBD) (22/35 families) or SH2 domain (10/35), and were missense mutations. We identified two intronic mutations resulting in exon skipping and in-frame deletions within the DBD. In addition, we identified two mutations located in the transactivation domain downstream of the SH2 domain: A ten amino acid deletion and an amino acid substitution. In one patient, we were unable to identify a STAT3 mutation. TH17 cells were absent or low in the peripheral blood of all patients who were evaluated (n=17). IL-6 induced STAT3-phosphorylation was consistently reduced in patients with SH2 domain mutations, but comparable to normal controls in patients with mutations in the DBD.
Conclusion
Heterozygous STAT3 mutations were identified in 34/35 unrelated HIES families. Patients had impaired TH17 cell development, and those with SH2 domain mutations had reduced STAT3 phosphorylation.
Clinical implication
Mutations in STAT3 and decreased TH17 cells identify individuals with AD-HIES, thereby allowing timely diagnosis and early treatment of these patients.
Capsule summary
Results from this patient cohort expand the spectrum of heterozygous STAT3 mutations in AD-HIES, and demonstrate impaired development of TH17 cells in all and reduced STAT3-phosphorylation in patients with SH2-domain mutations.
Keywords: Hyper-IgE syndrome, HIES, Autosomal-dominant Hyper-IgE Syndrome, Job syndrome, IgE, STAT3 mutations, STAT3 phosphorylation, TH17 cells
INTRODUCTION
Hyper-IgE Syndrome (HIES) is a rare autosomal dominant or sporadic multisystem immunodeficiency characterized by eczema, Staphylococcus aureus skin abscesses, pneumonia with abscess and pneumatocele formation, Candida infections, and skeletal and connective tissue abnormalities.1
Two patients with eczema, recurrent respiratory tract infections, and “cold” Staphylococcus aureus skin abscesses were reported in 1966 as suffering from “Job’s syndrome” because of the phenotypic similarity to the Biblical figure Job who had been “smitten with sore boils from the soles of his feet unto his crown” (Job 2:7).2 Subsequently, patients with similar clinical findings were reported3 and additional characteristic abnormalities recognized including distinct facial features, hyperextensible joints, pathologic bone fractures, scoliosis, craniosynostosis, and retained primary dentition.1,4 Among the immunologic abnormalities reported, patients had markedly elevated serum IgE which led to the disorder being named “Hyper-IgE Syndrome”.3
Most patients with this syndrome were noted to arise sporadically from unaffected healthy parents. The first observation of a possible familial occurrence was made in 1975.5 With the advent of improved antibiotic therapy, patients survived into adulthood, had children of their own, and an autosomal dominant pattern of inheritance became evident.6,7
Recently, a homozygous mutation of Tyk2, a tyrosine kinase involved in type I IFN and IL-12 induced phosphorylation of STAT4, was identified in a single patient with eczema, moderately elevated serum IgE, and a mild T-cell deficiency.8 This observation suggested that other defects in JAK/STAT signaling may cause HIES. Indeed, heterozygous mutations of the gene encoding the transcription factor STAT3 (Signal Transducer and Activator of Transcription 3) have recently been identified as the cause of autosomal dominant HIES (AD-HIES).9–11
In this report, we analyzed the spectrum of STAT3 mutations in a large cohort of patients with the HIES phenotype and examined the functional effect of selected mutations on TH17 cell development and STAT3 tyrosine phosphorylation.
METHODS
Subjects
We enrolled 38 patients (28 males and 10 females, age range 15 months to 50 years) from 35 unrelated families with diverse ethnic backgrounds living in the US and central Europe. All patients had the characteristic clinical symptoms of HIES, defined as presenting with a NIH-score above 40 points7 with the exception of the previously reported 15-month-old affected grandson of the index patient in family 711 (Table 1). In addition, we sequenced DNA from 44 asymptomatic members of those families in which a STAT3 mutation was identified and analyzed 100 normal chromosomes (50 control individuals of diverse ethnic backgrounds). The study was approved by the local Institutional Review Boards. Written informed consent was obtained.
Table 1.
Clinical and laboratory findings and STAT3 mutations in Hyper-IgE syndrome
| ID | Age, year |
Sex | IgE, IU/ml |
Eczema | Abscesses1 | Pneumonia | Pneumatoceles | Candida infection2 |
Associated findings3 |
NIH- score4 |
Heterozygous STAT3 mutation |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1.1 | 12 | f | 10460 | yes | no | yes | yes | no | no | 42 | 1003 C>T, R335W |
| 1.2 | 43 | m | 11689 | yes | yes | yes | yes | no | no | 43 | 1003 C>T, R335W |
| 2 | †35 | f | 2890 | yes | yes | yes | yes | yes | yes | 48 | 1020delGAC, K340N/T341del |
| 3 | 34 | m | 3310 | yes | yes | no | no | yes | yes | 42 | 1027 G>C, V343L |
| 4 | 34 | m | 11860 | yes | yes | yes | yes | yes | yes | 63 | IVS11+1g>t |
| 58 | 25 | m | 2831 | yes | yes | no | no | yes | yes | 46 | IVS11+1g>t |
| 6 | 48 | m | 45900 | yes | yes | yes | no | yes | yes | 66 | IVS11–2a>g |
| 7.15 | 50 | f | 4560 | yes | yes | yes | yes | yes | yes | 83 | 1144 C>T, R382W6,7 |
| 7.25 | †29 | m | 10400 | yes | yes | yes | yes | yes | yes | 66 | 1144 C>T, R382W6,7 |
| 7.35 | 1.25 | m | 491 | yes | yes | yes | yes | no | yes | 34 | 1144 C>T, R382W6,7 |
| 88 | 8 | m | 3489 | yes | no | yes | yes | yes | yes | 55 | 1144 C>T, R382W6,7 |
| 98 | 25 | m | 32500 | yes | yes | yes | yes | yes | yes | 68 | 1144 C>T, R382W6,7 |
| 108 | 41 | m | 106320 | yes | yes | yes | no | yes | yes | 51 | 1144 C>T, R382W6,7 |
| 118 | 23 | m | 10700 | yes | yes | yes | no | yes | yes | 42 | 1144 C>T, R382W6,7 |
| 12 | 31 | m | 7100 | yes | yes | yes | no | yes | yes | 67 | 1144 C>T, R382W6,7 |
| 13 | 50 | m | >2000 | yes | yes | yes | yes | yes | yes | 65 | 1144 C>T, R382W6,7 |
| 14 | 38 | m | 5958 | yes | yes | yes | yes | yes | yes | 53 | 1144 C>T, R382W6,7 |
| 15 | 24 | m | 8000 | yes | yes | yes | yes | yes | yes | 76 | 1144 C>T, R382W6,7 |
| 16 | 30 | m | 29899 | yes | yes | no | yes | yes | yes | 62 | 1144 C>T, R382W6,7 |
| 17 | 40 | f | 21700 | yes | yes | yes | yes | no | yes | 69 | 1144 C>T,R382W6,7 |
| 18 | 25 | m | 811 | yes | yes | yes | yes | no | yes | 55 | 1144 C>T,R382W6,7 |
| 198 | 9 | m | 3529 | yes | yes | yes | yes | yes | no | 51 | 1145 G>A, R382Q6,7 |
| 208 | 19 | f | 15641 | yes | yes | yes | yes | yes | yes | 57 | 1145 G>A, R382Q6,7 |
| 218 | 25 | m | 52887 | yes | yes | yes | yes | yes | yes | 48 | 1152 T>A, F384L7 |
| 228 | 16 | m | 2483 | yes | yes | yes | no | yes | yes | 56 | 1407 G>T, Q469H |
| 238 | †18 | m | 18110 | yes | yes | yes | no | yes | yes | 70 | 1858A>G, T620A |
| 24 | 19 | f | 17407 | yes | yes | yes | no | yes | yes | 57 | 190C>T, S636F |
| 258 | 31 | m | 4840 | yes | yes | yes | yes | no | yes | 63 | 1909 G>A,V637M7 |
| 26 | 18 | m | 31160 | yes | yes | yes | yes | yes | yes | 70 | 1909 G>A, V637M7 |
| 278 | 5 | f | 14060 | yes | no | yes | yes | no | no | 42 | 1909 G>A, V637M7 |
| 288 | 10 | m | 39917 | yes | yes | yes | yes | no | yes | 72 | 1909G>A, V637M7 |
| 29 | 24 | m | 5080 | yes | yes | yes | yes | no | yes | 75 | 1909 G>A, V637M7 |
| 30 | 21 | f | >2000 | yes | yes | yes | no | yes | yes | 74 | 1909 G>A, V637M7 |
| 318 | 13 | m | 3189 | yes | yes | yes | yes | yes | yes | 56 | 1909 G>A, V637M7 |
| 32 | 25 | m | 2600 | yes | yes | yes | yes | yes | yes | 70 | 1913 A>G, E638G |
| 33 | †46 | m | 8127 | yes | yes | yes | yes | yes | yes | 64 | 2069del30bp, E690_P699del |
| 34 | 20 | f | >2000 | yes | yes | yes | yes | yes | yes | 74 | 2137 G>C, V713L |
| 35 | 7 | f | 13550 | yes | yes | yes | no | yes | yes | 72 | No mutation identified |
|
Percent of patients symptom Present |
100% | 92% | 92% | 72% | 81% | 89% | |||||
Abscess formation of skin, lymph nodes, and organs other than lung
History of hyperkeratoric fingernails and/or oral thrush
Including retained primary teeth, hyperextensible joints, pathologic fractures, scoliosis, and/or characteristic facial features
scoring system described in reference (7)
family has been reported previously (11)
mutation previous reported by Minegishi et al. (9)
mutation previous reported by Holland et al. (10)
confirmed de-novo mutation because both parents have two wild type alleles
age patient died
Mutation analysis
Genomic DNA (gDNA) was prepared from heparinized venous blood using the QIAamp DNA Blood Mini Kit (QIAGEN, Valencia, CA) according to the manufacturer's protocol. Total mRNA was extracted from 5×106 fresh peripheral blood mononuclear cells (PBMCs) with Trizol (Invitrogen, Carlsbad, CA) and subjected to first strand cDNA synthesis using the Omniscript RT Kit (QIAGEN). The STAT3 gene was amplified from gDNA and cDNA using specific oligonucleotide primers (available upon request) and polymerase chain reaction. Where available, cDNA was sequenced first followed by confirmatory sequencing of gDNA if a STAT3 mutation was identified. The amplified gene fragments were sequenced using the ABI Big Dye Terminator mix (Applied Biosystem, Foster City, CA), and analyzed with a 3730×1 DNA Analyzer (Applied Biosystem). Mutations are reported using the nomenclature of den Dunnen and Antonarakis.12 PolyPhen (=Polymorphism Phenotyping),13 a bioinformatics method to predict the possible impact of an amino acid substitution on the structure and function of a human protein using physical and comparative considerations, was used to estimate the effect of the identified STAT3 missense mutations. PolyPhen scores the resulting protein damage as “putative benign” (score 0.00 to 1.50), “possibly damaging” (score 1.51 to 2.00), and “probably damaging” (score > 2.01). Structural modeling to visualize the dimerized STAT3 protein was performed using Pymol (DeLano Scientific LLC, San Carlos, CA) and structural coordinates from the RCSB (1BG1.pdb)14 database.
Evaluation of TH17 cells
PBMCs were isolated using Ficoll-Paque™ plus (Bioscience AB, Uppsala). TH17 cells were identified by intracellular staining of CD4+ T cells for IL-17 production. Briefly, 2.5×106 PBMCs from patients and controls were stimulated overnight with 10 ng/ml phorbol myristate acetate (PMA) and 1 µg/ml lonomycin (Sigma-Aldrich, St. Louis, MO) in the presence of GolgiPlug® (BD Biosciences, San Jose, CA). After cell surface staining with PE conjugated anti-CD4 (eBioscience, San Diego, CA), cells were fixed and permeabilized (Cytofix/Cytoperm, BD Biosciences) and stained with Alexa Fluor 647 conjugated anti-IL17A (eBioscience). As a control for cellular activation and intracellular staining, CD4+ T cells were also evaluated for IFNγ production (FITC conjugated; eBioscience). All flow cytometry studies were performed on an LSRII instrument (BD Biosciences) and analyzed using FlowJo software (TreeStar, Ashland, OR).
Evaluation of STAT3 phosphorylation
Tyrosine phosphorylation of STAT3 was assessed in patient and control PBMCs by flow cytometry using the BD™ Phosflow reagents per the manufacturer’s instruction (BD Biosciences). Briefly, 1×106 cells were stimulated with various concentrations of IL-6 (R&D Systems, Inc., Minneapolis, MN) for 20 min at 37°C (5% C02). Cells were fixed in buffered paraformaldehyde (4%). After washing with staining buffer, cells were permeabilized with Perm Buffer III (BD Biosciences) and stained with Alexa Fluor 647 conjugated mouse anti-STAT3 (pY705) monoclonal antibody (BD Biosciences).
Phosphorylation was also evaluated in Cos-7 cells (ATCC, Manassas, VA) transiently transfected with wild-type or mutant STAT3. The human STAT3α cDNA (Open Biosystems, Huntsville, AL) was cloned into the pAcGFP1-N1 vector (Clontech, Mountain View, CA) using the In-Fusion Dry-Down PCR cloning kit (Clontech). Mutations were introduced into the STAT3 cDNA using specific oligonucleotide primers and the QuickChange II XL site directed mutagenesis kit per the manufacturer’s recommendations (Stratagene, La Jolla, CA). Cells were transfected with plasmids encoding either wild-type or mutant STAT3 using Fugene HD (Roche, Indianapolis, IN). Twenty-four hours after transfection, cells were stimulated with Epidermal Growth Factor (EGF) (100 ng/ml) for 20 min and whole-cell extracts prepared using M-PER extraction buffer (Pierce, Rockford, IL). Following standard SDS-PAGE and protein transfer, blots were probed with antibodies to total STAT3, tyrosine phosphorylated (pY705) STAT3 (Cell Signaling, Danvers, MA), and actin (Sigma-Aldrich). Blots were developed using secondary horseradish peroxidase conjugated polyclonal antibodies (Biosource-Invitrogen, Carlsbad, CA) and Supersignal West Pico chemiluminescent substrate (Pierce, Rockford, IL).
Statistics
Differences in the percentage of CD4+IL-17+ T cells in patients and healthy controls were tested for significance using the Student t Test.
RESULTS
Patient characteristics
All 38 patients had classic findings of HIES with high NIH-scores (range 42 to 83 points) except for patient #7.3 who at 15 months of age had a score of 34 points; serum IgE levels were at least 2SD above normal for age (Table 1). Most developed chronic eczema during the first six months of life. Every patient had a history of recurrent Staphylococcus aureus skin and/or visceral abscesses. Recurrent episodes of pneumonia were documented in all but three patients and 27 developed pneumatoceles following the formation of lung abscesses. Twenty-nine patients had recurrent oral thrush beyond infancy or a history of chronic hyperkeratotic fingernails due to Candida albicans. At least one of the associated findings such as characteristic facial features, retained primary teeth, hyperextensible joints, pathologic fractures, and/or scoliosis was present in all but four patients. Two patients (#2 and #7.2, the latter reported previously11) died of respiratory complications after multiple pulmonary infections and pneumatocele formation; one patient (#23) died of liver failure caused by progressive Echinococcus multilocularis infection; patient #33 died of lung fibrosis six months after bone marrow transplantation for lymphoma.15
Mutations of STAT3
Following the recent discovery that heterozygous STAT3 mutations are responsible for AD-HIES9–11 we sequenced the STAT3 gene in 38 HIES patients and their asymptomatic family members. Fourteen different STAT3 mutations were identified in 37 patients from 34 unrelated families (Table 1, Figure 1). Despite repeated evaluation of cDNA and gDNA samples including intron/exon boundaries, we failed to identify a STAT3 mutation in the remaining patient (#35). None of the 44 available asymptomatic parents, siblings or close relatives from twenty affected families carried a STAT3 mutation. In fourteen informative families, both parents were shown to have two wild-type alleles demonstrating that the STAT3 mutation identified in the index patient arose de novo.
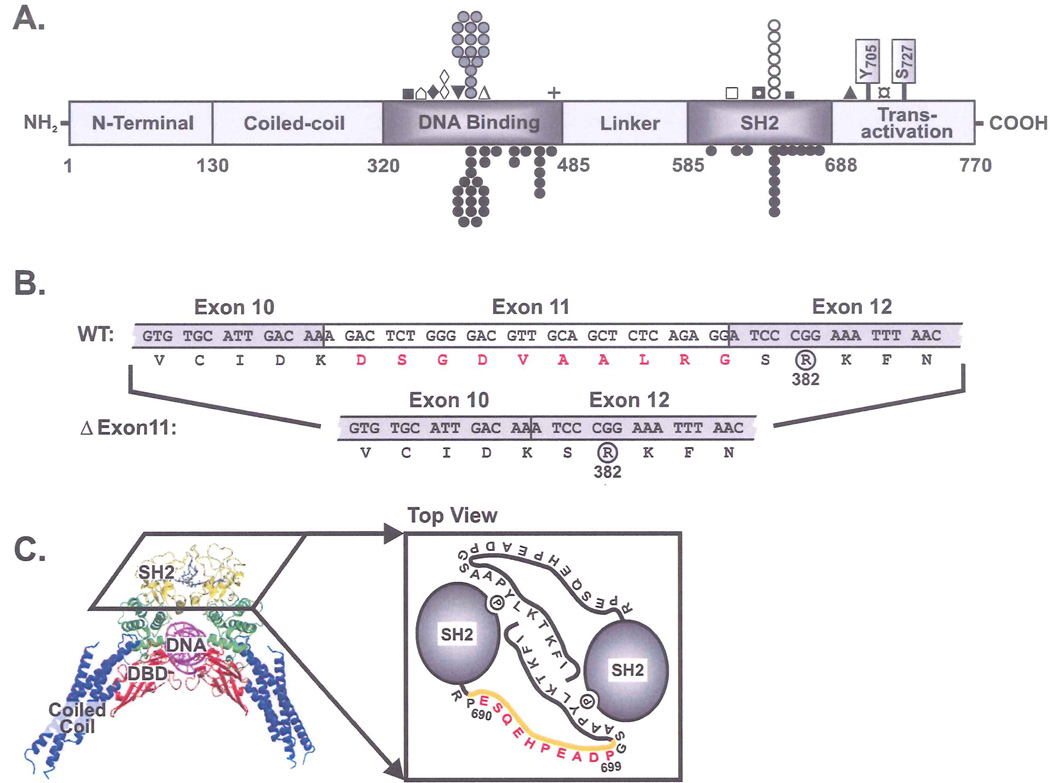
Figure 1.
A. Protein structure of STAT3 showing the identified mutations (one symbol per family). Our cohort (above): DNA binding domain (n=22: ■R335W, ⌂K340N/T341del, ♦V343L, ◊IVS11+1g>t, ▼IVS11-2a>g, ( )R382W/Q, ΔF384L, +Q469H), SH2 domain (n=10: □T620A, ◘S636F, ○V637M, ■E638G), Transactivation (n=2: ▲E690_P699del, ¤V713L). Other published mutations (below) 9,10.
)R382W/Q, ΔF384L, +Q469H), SH2 domain (n=10: □T620A, ◘S636F, ○V637M, ■E638G), Transactivation (n=2: ▲E690_P699del, ¤V713L). Other published mutations (below) 9,10.
B. Skipping of Exon 11 due to the identified splicing mutation IVS11+1g>t causes an in-frame deletion of 10 amino acids (red). Proximity to the frequently mutated R382 residue is shown.
C. Crystal structure of STAT3β dimer bound to DNA with SH2 domains at top (yellow)14. Cartoon view of SH2 domains from top showing phosphotyrosine-dependent dimerization. Deleted residues (E690_P699del) (red) remove most of the identified flexible loop (orange).
Similar to previous reports9,10 the R382W/Q mutation in the DNA-binding domain (DBD) was the most frequent STAT3 mutation observed in our cohort accounting for 41% of the 34 families with a STAT3 mutation. The second most frequent hotspot, V637M, located in the SH2 domain, accounted for 21% of our families. While the third hotspot mutation F384L was present in one family, the fourth reported hotspot (V463del) was not observed in our cohort (Figure 1A).
We identified three novel mutations that result in larger in-frame amino acid deletions than any previously reported. Two of these mutations affect the invariant splice donor and splice acceptor sites flanking exon 11 and are predicted to cause aberrant mRNA splicing. Analysis of cDNA from patient #4 confirmed that the IVS11+1 g>t mutation results in skipping of exon 11 and consequently in-frame deletion of 10 amino acids (deletion shown in red, Figure 1B). The IVS11-2a>g mutation (patient #6, no cDNA available) is also expected to result in skipping of exon 11 thus causing the same truncated protein product. The other in-frame deletion (E690_P699del) and the amino acid substitution V713L are the first reported mutations outside the DNA-binding or SH2 domains of STAT3. The 10 amino acid in-frame deletion (E690_P699del) removes most of a flexible, “disordered” loop (residues 689–701) shown in the STAT3 crystal structure to form a spacer between the SH2 domain and the critical phosphotyrosine residue, Y705 (Figure 1C).14
The remaining eight novel mutations observed include a complex mutation in the DNA-binding domain consisting of an amino acid substitution with an in-frame deletion of an adjacent amino acid (K340N/T341del) and seven missense mutations, three located in the DNA-binding domain (R335W, V343L, Q469H), three in the SH2 domain (T620A, S636F, E638G), and one in the transactivation domain (V713L) which is downstream of the SH2 domain (Figure 1A). None of the amino acid substitutions identified in our patient cohort were reported as SNPs in the NCBI dbSNP database; nor did we observe any of these alterations in 100 normal chromosomes from 50 control individuals of diverse ethnic backgrounds or in the 88 chromosomes from 44 unaffected close relatives. The PolyPhen analysis which predicts the possible impact of an amino acid substitution on the structure and function of a protein13, suggests that 5 of the 7 novel amino acid substitutions identified in our cohort have the highest probability of altering local protein structure (score > 2.01) and the remaining two (both Valine to Leucine substitutions) had intermediate probabilities (1.53 and 1.54, respectively). None were predicted to be conservative/benign changes.
Based on the clinical features (Table 1), no significant genotype/phenotype correlation was evident between patients with mutations in the DNA-binding, the SH2, or the transactivation domains. Similarly, there was no difference in the clinical presentation between the cohort of patients with STAT3 mutations and the one patient without a demonstrable mutation.
Decreased number of circulating TH17 cells
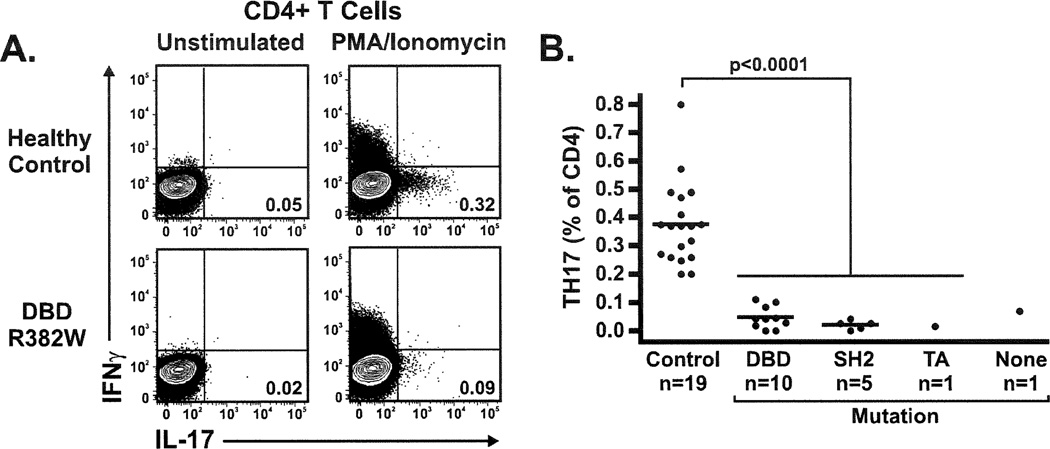
To determine whether the heterozygous STAT3 mutations identified in our cohort are associated with deficits in STAT3 function, we enumerated IL-17 producing CD4+ T cells (TH17 cells) in peripheral blood of available patients and normal controls. This subset of effector T cells has been shown to be largely dependent on STAT3 function.16,17 As recently reported by Milner, et al.18 all 17 HIES patients tested had significantly reduced or absent TH17 cells in the peripheral blood compared to healthy controls. We observed a marked reduction in TH17 cells that was similar in all patients irrespective of the type or location of the mutation. A similar reduction was found in the one patient lacking a STAT3 mutation (Figure 2).
Figure 2.
A. Intracellular staining for IL-17 production in PMA/lonomycin activated CD4+ T cells. The HIES patient with a DBD(R382W) mutation has fewer circulating TH17 cells compared to the healthy control. B. Percentage of TH17 cells in PBMCs is similarly low in all HIES patients tested: DBD (R335W, IVS11+1g>t, R382W, R382Q), SH2 (S636F, V637M), transactivation (V713L), and one patient without a detectable STAT3 mutation.
Defective STAT3-phosphorylation
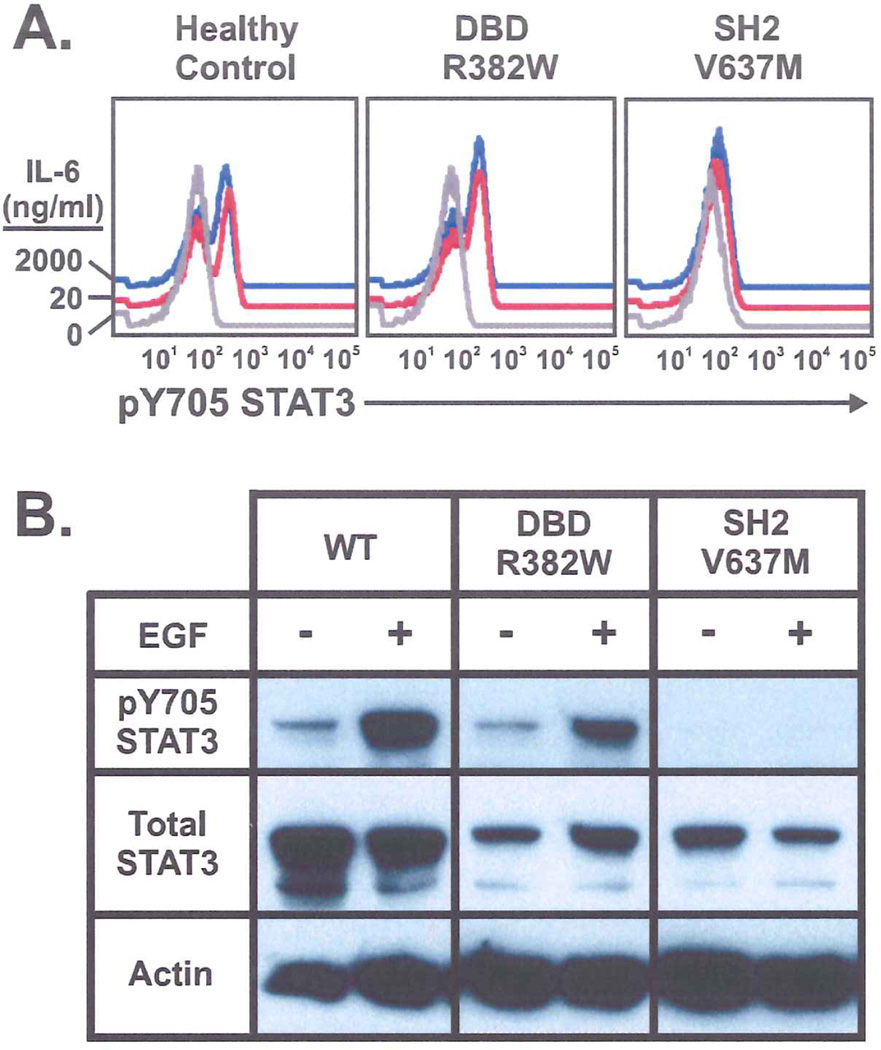
Because of the critical role played by the SH2 domain in receptor docking and kinase interactions, we hypothesized that mutations in this domain would result in defective STAT3 phosphorylation. To explore this possibility, we evaluated STAT3 tyrosine phosphorylation (pY705-STAT3) in IL-6 stimulated PBMCs by flow cytometry (Figure 3A). PBMCs from patients with mutations in the SH2 domain (cells available from four patients with the V637M and one patient with the S636F mutation) showed decreased pY705-STAT3 when compared to healthy controls. Increasing the strength of the IL-6 stimulus 100 fold failed to overcome the observed phosphorylation defect (shown for the V637M mutation in the right panel, Figure 3A). In contrast, PBMCs from seven patients with DBD mutations exhibited normal STAT3 phosphorylation (shown for the R382W mutation, middle panel), comparable in magnitude to that observed in a healthy control (left panel).
Figure 3.
A. Phospho-flow cytometry showing IL-6 induced tyrosine phosphorylation of STAT3 is similar in control and DBD (R382W) mutant PBMCs but decreased in SH2 (V637M) mutant PBMCs. The defect is not overcome with 100x IL-6 concentration. B. EGF induced tyrosine phosphorylation of GFP-STAT3 in transiently transfected Cos-7 cells. Lower band in total STAT3 lanes is a degradation product of transfected STAT3 (data not shown).
The defect in STAT3 phosphorylation observed in PBMCs from patients with SH2 domain mutations was confirmed by western blotting of STAT3 in transiently transfected Cos-7 cells stimulated with Epidermal Growth Factor (EGF). Under these conditions, wild-type STAT3 is phosphorylated on Y705 in response to EGF as previously shown.19. STAT3 bearing the most common DBD mutation (R382W) was phosphorylated normally whereas the most common SH2 domain mutant (V637M) was not significantly phosphorylated (Figure 3B). Similar results were obtained using transiently transfected HepG2 cells stimulated with IL-6 (data not shown).
DISCUSSION
This study of a large cohort of patients with HIES and high NIH clinical scores (> 40 points) expands the spectrum of heterozygous STAT3 mutations observed in AD-HIES.9–11 In addition to confirming three of four previously reported mutation hotspots,9,10 we have identified eleven novel mutations; nine of which were located in the DNA-binding and SH2 domains and two in the transactivation domain, including a 30 base pair deletion found in a flexible loop connecting the critical phosphotyrosine residue (Y705) to the SH2 domain. Models based on the crystal structure of STAT3β14 reveal that this flexible loop positions the Y705 residue adjacent to the opposite SH2 domain and its deletion may affect activation-induced STAT3 dimerization (Figure 1C).
Most STAT3 mutations identified to date are missense mutations9–11. The seven novel missense mutations reported here result in amino acid changes that are most likely pathogenic because the affected residues are highly conserved among various species, none were reported as SNPs, and none were identified in normal controls. Using the PolyPhen algorithm13, five scored within the highest and two within intermediate probability. Two of these missense mutations (S636F and E638G) flank the hotspot mutation (V637M) in the SH2 domain and are thought to cause a similar functional defect. For the patients with the Q469H and T620A mutations, both parents were available for analysis and had only wild-type STAT3 suggesting that the de novo STAT3 mutation in the index case is pathogenic. Finally, patients with the R335W, S636F, and V713L mutations showed low TH17 cell numbers and the S636F and V713L mutations resulted in decreased STAT3 phosphorylation.
Although more than 90% of our HIES patients had STAT3 mutations, we were unable to identify a mutation in patient #35 who has characteristic symptoms of HIES, an NIH-score of 74 points, and very low numbers of TH17 cells. Despite extensive investigation, it is unclear whether this patient has a STAT3 mutation that was missed or if this indicates heterogeneity of the molecular etiology. The latter possibility is supported by the observation of Minegishi et al. who identified STAT3 mutations in only eight of fifteen unrelated families with a diagnosis of HIES.9
Like others10, we did not observe a significant genotype/phenotype correlation. The total number of patients described to-date is, however, limited and there may be a selection bias resulting from the inclusion criteria of a NIH score of >40 points. Thus, subtle differences may not become apparent until more HIES patients with STAT3 mutations have been characterized.
The mechanism by which heterozygous mutations in STAT3 cause HIES remains unclear. The autosomal dominant inheritance suggests either haploinsuffiency or a dominant negative mechanism by which mutant STAT3 fully or partly abrogates the function of wild-type STAT3 expressed by the normal allele. The fact that complete knockout of Stat3 in mice leads to an embryonic lethal phenotype whereas Stat3+/− heterozygote mice are phenotypically normal suggests that haploinsufficiency is not sufficient to cause the symptoms of HIES. The finding that all STAT3 mutations identified in HIES patients to-date are either amino acid substitutions or in-frame deletions strongly supports the concept that co-expression of wild-type and mutant STAT3 protein is required to cause HIES. The notable lack of nonsense or frameshift mutations strengthens the notion that AD-HIES is the result of a dominant negative effect as suggested by Minegishi et al.9 and similar to that observed with the well characterized Y705F mutant of Stat320.
The fact that STAT3 mutations associated with HIES cluster in three areas of the protein known to have distinct functions (DNA-binding, SH2, and transactivation) suggests that more than one molecular mechanism causes a similar clinical presentation. This is supported by our observation that mutations in the SH2 domain but not those in the DBD affect activation-induced tyrosine phosphorylation of STAT3, while DBD mutants were shown by Minigeshi et al.9 to be defective in DNA binding. Further studies will be required to fully elucidate these divergent mechanisms.
Since STAT3 plays a key role in the development of IL-17 producing TH17 cells,16,17 we have quantified this T cell subset in PMA/lonomycin stimulated PBMCs from seventeen HIES patients. Similar to a recent report18, we found that every HIES patient tested had markedly decreased or absent circulating TH17 cells including the patient without a detectable STAT3 mutation. Given that IL-17 plays an important role in host defense against extracellular bacteria and fungi (particularly Candida) and upregulates production of β-defensins and S100 proteins by neutrophils,21 absence of TH17 cells may directly affect susceptibility to Staphylococcus aureus and Candida.22 Mice with conditional Stat3 ablation in hematopoietic cells demonstrate eosinophilia and accumulation, in the bone marrow, of mature granulocytes with relative paucity of immature granulocytes.23 Neutrophils from these animals show defective regulation of chemoattractant-induced actin polymerization and fail to mobilize during acute inflammatory response due to impaired neutrophil chemotactic responses to natural ligands such as CXCR2.23 Interestingly, neutrophil chemotaxis has been reported to be variably impaired in HIES patients.5,6,24,25 In addition, hematopoetic cell specific Stat3 knockout mice develop osteoporosis and decreased bone volume due to enhanced osteoclastogenesis.26 This finding may explain the pathologic fractures and retention of primary teeth in HIES.
There is no clear experimental explanation for the striking elevation of serum IgE and pronounced eosinophilia consistently observed in AD-HIES. The altered cytokine pattern observed27–30 suggests that a decreased production of IFN-γ and overproduction of IL-4 and IL-5 may lead to elevated serum IgE levels and eosinophilia.
STAT3 is ubiquitously expressed and not limited to immune cells31, thus defective wound healing and extracellular matrix remodeling observed in several murine models with tissue specific Stat3-disruption may explain some of the characteristic bone and connective tissue abnormalities observed in HIES.32–35 For example, mice conditionally lacking Stat3 in respiratory epithelial cells demonstrate increased epithelial cell damage with airspace enlargement following oxygen-induced lung injury.33 Similar processes may explain the tendency of HIES patients to develop pneumatoceles following pneumonias. Mice with Stat3-disruption in keratinocytes show a profound wound healing defect with delayed keratinocyte migration and spontaneous development of skin ulcers32, processes that may contribute to the development of cutaneous infections and “cold” Staph abscesses seen in HIES.
In conclusion, HIES is a monogenic defect caused by heterozygous STAT3 mutations affecting the DNA-binding domain, SH2 domain, or the transactivation domain downstream of the SH2 domain. Mutations within the SH2 domain interfere with STAT3 phosphorylation and mutations in the DNA-binding, SH2, and transactivation domains deleteriously affect the generation of TH17 cells. A better understanding of the biochemical defects resulting from heterozygous STAT3 mutations will predictably lead to new treatment modalities for HIES.
ACKNOWLEDGMENTS
We acknowledge the efforts of Elizabeth Ocheltree and Taylor LaFlam in creating the STAT3 expression vectors used in the transient transfection assays, Karin Golob for assisting with sequencing, and Julie Sawalle for help with patient recruitment. We thank all patients, families, and primary care physicians for generously providing samples for analysis.
Funding: This work was supported by grants from the Fritz Thyssen Foundation (Az. 10.07.1.159) and the American Academy of Allergy, Asthma and Immunology’s Strategic Training in Allergy Research (ST*AR) Award 2007 (EDR), NIH grant AI063267-01 and USIDNet grant N01A130070 (TRT), NIH grant HD017427-41 and grants from the Jeffrey Modell and Immune Deficiency Foundations (HDO).
Abbreviations
- AD-HIES
Autosomal-dominant Hyper-IgE Syndrome
- DBD
DNA binding domain
- EGF
Epidermal Growth Factor
- HIES
Hyper-IgE syndrome
- IL-17
Interleukin 17
- IFN-α
Interferon-alpha
- IFN-γ
Interferon-gamma
- PBMCs
Peripheral blood mononuclear cells
- pY705-STAT3
Tyrosine 705 phosphorylated STAT3
- MFI
Mean fluorescence index
- SH2
Src homology 2
- SNP
Single nucleotide polymorphism
- STAT3
Signal transducer and activator of transcription 3
- Tyk2
Tyrosine kinase 2
REFERENCES
- 1.Grimbacher B, Holland SM, Gallin JI, Greenberg F, Hill SC, Malech HL, et al. Hyper-IgE syndrome with recurrent infections--an autosomal dominant multisystem disorder. N Engl J Med. 1999;340(9):692–702. doi: 10.1056/NEJM199903043400904. [DOI] [PubMed] [Google Scholar]
- 2.Davis SD, Schaller J, Wedgwood RJ. Job's Syndrome. Recurrent, “cold”, staphylococcal abscesses. Lancet. 1966;1(7445):1013–1015. doi: 10.1016/s0140-6736(66)90119-x. [DOI] [PubMed] [Google Scholar]
- 3.Buckley RH, Wray BB, Belmaker EZ. Extreme hyperimmunoglobulinemia E and undue susceptibility to infection. Pediatrics. 1972;49(1):59–70. [PubMed] [Google Scholar]
- 4.Borges WG, Hensley T, Carey JC, Petrak BA, Hill HR. The face of Job. J Pediatr. 1998;133(2):303–305. doi: 10.1016/s0022-3476(98)70243-4. [DOI] [PubMed] [Google Scholar]
- 5.Van Scoy RE, Hill HR, Ritts RE, Quie PG. Familial neutrophil chemotaxis defect, recurrent bacterial infections, mucocutaneous candidiasis, and hyperimmunoglobulinemia E. Ann Intern Med. 1975;82(6):766–771. doi: 10.7326/0003-4819-82-6-766. [DOI] [PubMed] [Google Scholar]
- 6.Blum R, Geller G, Fish LA. Recurrent severe staphylococcal infections, eczematoid rash, extreme elevations of IgE, eosinophilia, and divergent chemotactic responses in two generations. J Pediatr. 1977;90(4):607–609. doi: 10.1016/s0022-3476(77)80380-6. [DOI] [PubMed] [Google Scholar]
- 7.Grimbacher B, Schaffer AA, Holland SM, Davis J, Gallin JI, Malech HL, et al. Genetic linkage of hyper-IgE syndrome to chromosome 4. Am J Hum Genet. 1999;65(3):735–744. doi: 10.1086/302547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Minegishi Y, Saito M, Morio T, Watanabe K, Agematsu K, Tsuchiya S, et al. Human tyrosine kinase 2 deficiency reveals its requisite roles in multiple cytokine signals involved in innate and acquired immunity. Immunity. 2006;25(5):745–755. doi: 10.1016/j.immuni.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 9.Minegishi Y, Saito M, Tsuchiya S, Tsuge I, Takada H, Hara T, et al. Dominant-negative mutations in the DNA-binding domain of STAT3 cause hyper-IgE syndrome. Nature. 2007;448(7157):1058–1062. doi: 10.1038/nature06096. [DOI] [PubMed] [Google Scholar]
- 10.Holland SM, DeLeo FR, Elloumi HZ, Hsu AP, Uzel G, Brodsky N, et al. STAT3 mutations in the hyper-IgE syndrome. N Engl J Med. 2007;357(16):1608–1619. doi: 10.1056/NEJMoa073687. [DOI] [PubMed] [Google Scholar]
- 11.Renner ED, Torgerson TR, Rylaarsdam S, Anover-Sombke S, Golob K, LaFlam T, et al. STAT3 mutation in the original patient with Job's syndrome. N Engl J Med. 2007;357(16):1667–1668. doi: 10.1056/NEJMc076367. [DOI] [PubMed] [Google Scholar]
- 12.den Dunnen JT, Antonarakis SE. Mutation nomenclature extensions and suggestions to describe complex mutations: a discussion. Hum Mutat. 2000;15(1):7–12. doi: 10.1002/(SICI)1098-1004(200001)15:1<7::AID-HUMU4>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 13.Ng PC, Henikoff S. Accounting for human polymorphisms predicted to affect protein function. Genome Res. 2002;12(3):436–446. doi: 10.1101/gr.212802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Becker S, Groner B, Muller CW. Three-dimensional structure of the Stat3beta homodimer bound to DNA. Nature. 1998;394(6689):145–151. doi: 10.1038/28101. [DOI] [PubMed] [Google Scholar]
- 15.Nester TA, Wagnon AH, Reilly WF, Spitzer G, Kjeldsberg CR, Hill HR. Effects of allogeneic peripheral stem cell transplantation in a patient with job syndrome of hyperimmunoglobulinemia E and recurrent infections. Am J Med. 1998;105(2):162–144. doi: 10.1016/s0002-9343(98)00200-9. [DOI] [PubMed] [Google Scholar]
- 16.Yang XO, Panopoulos AD, Nurieva R, Chang SH, Wang D, Watowich SS, et al. STAT3 regulates cytokine-mediated generation of inflammatory helper T cells. J Biol Chem. 2007;282(13):9358–9363. doi: 10.1074/jbc.C600321200. [DOI] [PubMed] [Google Scholar]
- 17.Nishihara M, Ogura H, Ueda N, Tsuruoka M, Kitabayashi C, Tsuji F, et al. IL-6-gp130-STAT3 in T cells directs the development of IL-17+ Th with a minimum effect on that of Treg in the steady state. Int Immunol. 2007;19(6):695–702. doi: 10.1093/intimm/dxm045. [DOI] [PubMed] [Google Scholar]
- 18.Milner JD, Brenchley JM, Laurence A, Freeman AF, Hill BJ, Elias KM, et al. Impaired T(H)17 cell differentiation in subjects with autosomal dominant hyper-IgE syndrome. Nature. 2008 doi: 10.1038/nature06764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barboza JA, Wang S, Schaefer TS. Generation and characterization of a constitutively active Stat3 protein. Mol Biol Rep. 2004;31(1):13–21. doi: 10.1023/b:mole.0000013503.16301.82. [DOI] [PubMed] [Google Scholar]
- 20.Kaptein A, Paillard V, Saunders M. Dominant negative stat3 mutant inhibits interleukin-6-induced Jak-STAT signal transduction. J Biol Chem. 1996;271(11):5961–5964. doi: 10.1074/jbc.271.11.5961. [DOI] [PubMed] [Google Scholar]
- 21.Huang W, Na L, Fidel PL, Schwarzenberger P. Requirement of interleukin-17A for systemic anti-Candida albicans host defense in mice. J Infect Dis. 2004;190(3):624–631. doi: 10.1086/422329. [DOI] [PubMed] [Google Scholar]
- 22.Acosta-Rodriguez EV, Rivino L, Geginat J, Jarrossay D, Gattorno M, Lanzavecchia A, et al. Surface phenotype and antigenic specificity of human interleukin 17-producing T helper memory cells. Nat Immunol. 2007;8(6):639–646. doi: 10.1038/ni1467. [DOI] [PubMed] [Google Scholar]
- 23.Panopoulos AD, Zhang L, Snow JW, Jones DM, Smith AM, El Kasmi KC, et al. STAT3 governs distinct pathways in emergency granulopoiesis and mature neutrophils. Blood. 2006;108(12):3682–3690. doi: 10.1182/blood-2006-02-003012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hill HR, Ochs HD, Quie PG, Clark RA, Pabst HF, Klebanoff SJ, et al. Defect in neutrophil granulocyte chemotaxis in Job's syndrome of recurrent “cold” staphylococcal abscesses. Lancet. 1974;2(7881):617–619. doi: 10.1016/s0140-6736(74)91942-4. [DOI] [PubMed] [Google Scholar]
- 25.Donabedian H, Gallin JI. Two inhibitors of neutrophil chemotaxis are produced by hyperimmunoglobulin E recurrent infection syndrome mononuclear cells exposed to heat-killed staphylococci. Infect Immun. 1983;40(3):1030–1037. doi: 10.1128/iai.40.3.1030-1037.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Z, Welte T, Troiano N, Maher SE, Fu XY, Bothwell AL. Osteoporosis with increased osteoclastogenesis in hematopoietic cell-specific STAT3-deficient mice. Biochem Biophys Res Commun. 2005;328(3):800–807. doi: 10.1016/j.bbrc.2005.01.019. [DOI] [PubMed] [Google Scholar]
- 27.Borges WG, Augustine NH, Hill HR. Defective interleukin-12/interferon-gamma pathway in patients with hyperimmunoglobulinemia E syndrome. J Pediatr. 2000;136(2):176–180. doi: 10.1016/s0022-3476(00)70098-9. [DOI] [PubMed] [Google Scholar]
- 28.Vercelli D, Jabara HH, Cunningham-Rundles C, Abrams JS, Lewis DB, Meyer J, et al. Regulation of immunoglobulin (Ig)E synthesis in the hyper-IgE syndrome. J Clin Invest. 1990;85(5):1666–1671. doi: 10.1172/JCI114618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chehimi J, Elder M, Greene J, Noroski L, Stiehm ER, Winkelstein JA, et al. Cytokine and chemokine dysregulation in hyper-IgE syndrome. Clin Immunol. 2001;100(1):49–56. doi: 10.1006/clim.2001.5039. [DOI] [PubMed] [Google Scholar]
- 30.Renner ED, Pawlita I, Hoffmann F, Hornung V, Hartl D, Albert M, et al. No indication for a defect in toll-like receptor signaling in patients with hyper-IgE syndrome. J Clin Immunol. 2005;25(4):321–328. doi: 10.1007/s10875-005-4183-2. [DOI] [PubMed] [Google Scholar]
- 31.Levy DE, Lee CK. What does Stat3 do? J Clin Invest. 2002;109(9):1143–1148. doi: 10.1172/JCI15650. [DOI] [PMC free article] [PubMed] [Google Scholar]