Abstract
One-bead-one-compound (OBOC) combinatorial library screening has been broadly utilized for the last two decades to identify small molecules, peptides or peptidomimetics targeting variable screening probes such as cell surface receptors, bacteria, protein kinases, phosphatases, proteases etc. In previous screening methods, library beads were suspended in solution and screened against one single probe. Only the positive beads were tracked and isolated for additional screens and finally selected for chemical decoding. During this process, the remaining negative beads were not tracked and discarded. Here we report a novel bead immobilization method such that a bead library array can be conveniently prepared and screened in its entirety, sequentially many times with a series of distinct probes. This method not only allows us to increase the screening efficiency but also permits us to determine the binding profile of each and every library bead against a large number of target receptors. As proof of concept, we serially screened a random OBOC disulfide containing cyclic heptapeptide library with three water soluble dyes as model probes: malachite green, bromocresol purple and indigo carmine. This multiplicative screening approach resulted in a rapid determination of the binding profile of each and every bead respective to each of the three dyes. Beads that interacted with malachite green only, bromocresol purple only, or both indigo carmine and bromocresol purple were isolated, and their peptide sequences were determined with microsequencer. Ultimately, the novel OBOC multiplicative screening approach could play a key role in the enhancement of existing on-bead assays such as whole cell binding, bacteria binding, protein binding, post-translational modifications etc. with increased efficiency, capacity, and specificity.
Keywords: One-bead-one-compound combinatorial chemistry, PDMS affixed bead array, High throughput screening, Multiplicative screening, Water soluble organic dye
INTRODUCTION
The one-bead-one-compound combinatorial (OBOC) library chemistry was invented two decades ago.[1] In this method, OBOC libraries are prepared using a “split-mix” approach which creates individual 90 μm beads[2] each displaying up to 1013 copies of only one chemical entity.[1] These chemical compounds can include small molecules, peptides, glycopeptides and peptidomimetics. Since it was established, OBOC has been used broadly in many high throughput screening assays to discover ligands against a variety of biological targets such as protein kinases,[2, 3] proteases,[4–6] cell surface receptors,[7, 8] G-coupled protein receptor inhibitors,[9] mRNA precursors [10], etc.
In the previous traditional bead screening methodologies, libraries were screened against specific targets such as proteins, proteases or live cells in suspension with no means of reliably immobilizing the beads without fouling the bead surface for molecular interactions. The positive beads were then selected for identification or for next round of screening. Any subsequent screening analyses performed on the positive or negative beads identified in the initial screen could not be conveniently tracked and compared to the result from previous screens. Within each library, some beads may be negative for one target but positive for another, or vice versa. If we can interrogate each individual bead with various targets sequentially and be able to track the bead-target interactions, we will be able to greatly increase the screening efficiency and specificity of the OBOC method. To achieve this, the library of microbeads will need to be affixed on a planar surface to form a bead array without fouling the bead surface using adhesives. The library will then be probed with related or different probes sequentially and the interactions recorded. This approach would provide a much more informative and complete binding profile for each compound bead to the binding targets. Furthermore, because the same batch of beads from a given library could be used multiple times against numerous targets, library synthesis time could be conserved and usage and waste of materials (minimal as they may be) would be significantly minimized.
In previous work, a two-step subtraction method was used to eliminate false positives from a library prior to screening with the desired target.[11] In this method a preliminary screening step is performed to colorimetrically mark nonspecific interactions in the presence of background proteins or protein extracts. The bead library is then incubated with the protein target of interest. Prior to the secondary colorimetric labeling step, the bead library is immobilized in agarose and scanned on a flat-bed transparency scanner. The beads are then developed a second time and a second scanned image is obtained. Comparing the two images allows the identification of beads that demonstrate non-specific interaction during the first colorimetric labeling step. While this method permits the screening of two probes sequentially, no further probe binding is feasible once the beads are immobilized and embedded with the agarose. In our present immobilization method, many more probes can be screened against the immobilized library because the surface of the securely fixed beads are readily accessible to the free aqueous environment, and the bead array can be recycled after each round of screening.
Here we describe a method that securely affixes library micro-beads to the bottom of a standard tissue culture dish or microscope slide spin-coated with a thin layer of PDMS (polydimethylsiloxane), leaving approximately 95% of the bead surface freely exposed to the surrounding environment. We previously reported a bead immobilization strategy in which beads were loaded into fabricated PDMS micro-well cassettes and surrounded by a gel cell growth matrix (Matrigel™ or agarose) facilitating a high-throughput releasable compound assay.[12] In the PDMS affixed bead array format described here, the beads are immobilized more securely with little chance of becoming dislodged even with vigorous shaking steps in incubation or washing protocols. Furthermore, unlike the releasable assay approach where a micro-well partitioned gel matrix environment is desirable to achieve maximum compound concentrations in distinct regions[12], the PDMS affixed bead array described here results in beads which are freely exposed to surrounding aqueous environment. This method of bead immobilization is more suitable for screens in which the surface of beads need to be readily exposed to surrounding aqueous environment to permit free interaction with target probes in solution such as proteins in buffer, cells in media, etc. thereby maximizing screening versatility and efficiency in a given assay.
The OBOC combinatorial method can be used as a useful high throughput strategy to identify sequence specific colorimetric dye interactions within a randomized peptide library.[13],[14] The ease and simplicity of this approach are made possible by taking advantage of the dye's optical properties, permitting direct visualization of dye binding to individual beads under a bench top light microscope in a manner analogous to other commonly screened binding targets in combinatorial methodology. Here we use a relatively straightforward and simplistic approach to demonstrate the potentials and utility of the PDMS affixed bead assay.
In this report, water soluble dyes such as indigo carmine, malachite green oxalate and bromocresol purple were chosen as the screening probes to validate the affixed bead library sequential binding assay. The peptide binding properties of each dye were characterized upon sequential screening of the same affixed bead peptide library against each of these dyes. Using this screening approach, the feasibility and utility of the PDMS affixed bead multiplicative library screening method and the adaptability of this approach to other screening methodologies is demonstrated.
RESULTS AND DISCUSSION
Dishes prepared with OBOC combinatorial library beads immobilized on the bottom
Using a relatively simple method outlined in the Experimental Section, we have succeeded in generating a reproducible array of library beads affixed to the bottom of the polystyrene tissue culture dish via a thin layer of PDMS. The dish and bead arrays were found to be optically clear and the beads remained attached to the bottom of the dish even after repeated washes with water and buffer solutions.
Color Dye Binding Profile for CX1X2X3X4X5C
In this work, an OBOC combinatorial cyclic heptamer peptide library, CX1X2X3X4X5C, was prepared and screened for target binding. Nineteen L-amino acids, excluding L-Cys, were included on each position from X1 to X5 and the fixed cysteines on both ends were subsequently cyclized through a disulfide bond. The library beads were fixed on the PDMS layer in a tissue culture dish. Three water soluble dyes (indigo carmine, malachite green and bromocresol purple) with different chemical structures (Figure 1) were selected as probes for sequential screening of the library. After incubation with each dye, the immobilized bead library was washed with PBS and inspected under a dissecting microscope and image recorded serially over time. Prior to the next dye screening, the immobilized bead library was washed thoroughly with 70% ethanol.
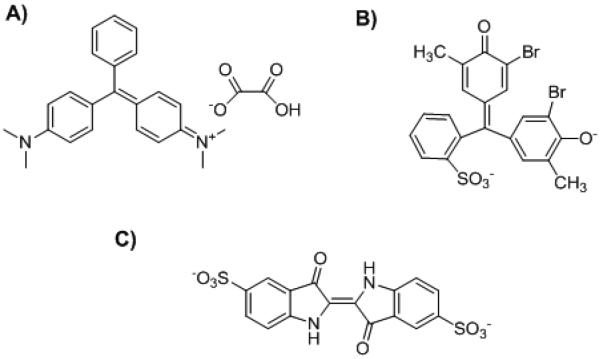
Figure 1.
Structure of the color dyes. A) Malachite green oxalate; B) Bromocresol purple; C) Indigo carmine
The binding profile of the bead library to each color dye is shown in Figure 2.
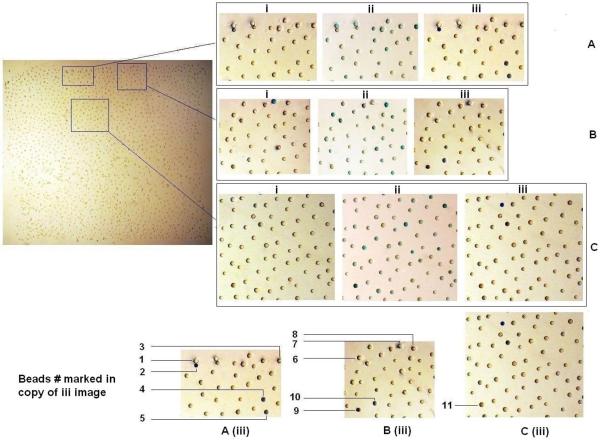
Figure 2.
PDMS affixed bead array was incubated with three different dyes sequentially. Beads were incubated with indigo carmine (i) till the color was saturated, and then the dye was washed away completely and incubated with the second dye malachite green (ii). The third dye bromocresol purple (iii) was last incubated after malachite green was removed. A, B, and C three different areas were selected to zoom in for more clear visualization. Among total 11 beads with characteristic binding profile, #1, #3, #6, #8 and #11 beads only bind to malachite green. #4, #5 and #9 beads only bind to bromocresol purple. #2, #7 and #10 beads bind to both indigo carmine and bromocresol purple.
For beads #1, #3, #6, #8 and #11, binding was only observed with the malachite green oxalate dye. Similarly, beads #4, #5 and #9 only bound to the bromocresol purple dye. Beads #2, #7 and #10, however, displayed some level of binding interaction with both indigo carmine and bromocresol purple dyes. These eleven beads were picked and the sequences were decoded (Table 1). For the sequences binding to malachite green oxalate only, no strong consensus motif was apparent although four of the five peptides has one acidic residue at the 4th or 5th position and 1 peptide has acidic residues in both 4th and 5th position. In addition, every peptide has either one Ser or one Thr residue. One or two hydrophobic residues are present in four of the five peptides. Beads #1 and #3 both contained a Pro followed by Asp. Malachite green is a triphenylmethane basic dye (positive charge, see Figure 1) often used in the aquaculture industry to control fungus and protozoa.[15] This dye has also been used extensively for dyeing anionic fabrics such as silk, wool, jute, cotton, nylon and acrylics.[16] Therefore it is not surprising that all the five binding peptides contain one or two acidic residues. In response to concerns regarding the health risks associated with the use of the dye, adsorption is, by far, the most versatile and widely used technique for its removal from contaminated aqueous solutions.[15]'[16] The OBOC may therefore be used to identify high affinity adsorbent agents for decontamination of water with malachite green dye.
Table 1.
The peptide sequences of the selected beads in Figure 2.
| Bead number | Sequence | Color dye binding |
|---|---|---|
| # 1 | CNSPDIC | Malachite green |
| # 3 | CQPDLTC | |
| # 6 | CGNTEPC | |
| # 8 | CMTQEAC | |
| # 11 | CLSDEFC | |
| # 4 | CHTHILC | Bromocresol purple |
| # 5 | CHPLLPC | |
| # 9 | CEIHRIC | |
| # 2 | CLMNKWC | Indigo carmine + Bromocresol purple |
| # 7 | CYKWWVC | |
| # 10 | CKWILPC |
For the three peptides that only bind bromocresol purple alone, every one has one or two His and two hydrophobic residues (Leu and Ile). In the sequences which were shown to bind both bromocresol purple and indigo carmine (5,5'-indigodisulfonic acid sodium salt), there is a highly consistent appearance of the motif Lys-Trp and additional hydrophobic residues. Interesting, in this limited screening experiment, we did not find any beads that only bind to indigo carmine but not bromocresol purple.
A key advantage of multiplicative beads screening strategy is that differences and/or similarities of conformation and structure among different targets can be directly assessed and considered for each bead/target interaction. For example, there were two different groups of beads which only bound to malachite green or bromocresol purple, indicating a separate and distinct structure/binding property between these two dyes. Accordingly, the sequence results demonstrate a completely different preference such that peptides binding to malachite green include more acidic amino acids such as Asp or Glu, while more basic amino acids such as His or Lys were contained in peptides binding to bromocresol purple. The marked difference in sequences between these two targets likely results from the dyes' noticeable differences in structure and properties. Malachite green tends to be more basic in nature while bromocresol purple is predisposed to be more acidic. By contrast, all of the beads shown to bind Indigo carmine also bound to bromocresol purple, suggesting a structure/property similarity between these two dyes. Indeed, the sequences obtained from the beads that interacted with both of these dyes shared a noticeable motif perhaps indicating a common interaction with sulfonic group characteristic to both dyes.
Using these results, the design and synthesis of focused libraries could facilitate the elucidation of more stringent and specific peptide-dye interactions.
Binding and Dissociation Profile Based on Color Quantification
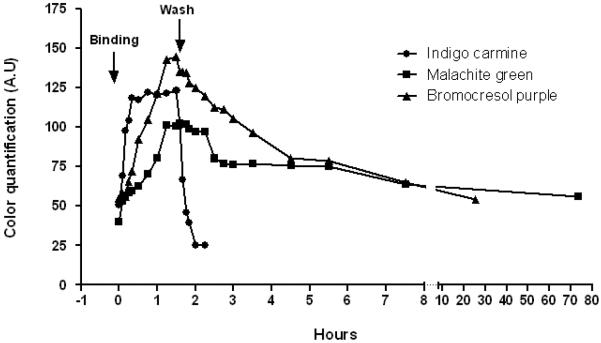
In the PDMS affixed bead approach, it is possible to dynamically observe the peptide-dye interaction on each individual immobilized bead in real time. To further characterize the binding profile for each of the target/sequence interactions, the representative positive beads #1, #4 and #10 were selected for binding and dissociation study with each respective target dye, by measuring the intensity of the resulting color accumulation on the bead surface at different time points. The values obtained for color intensity across progressing time points were plotted and the profiles for the three dye/bead interactions were compared. In this way the association and dissociation rate of each organic dye to its respective binding peptide could be evaluated.
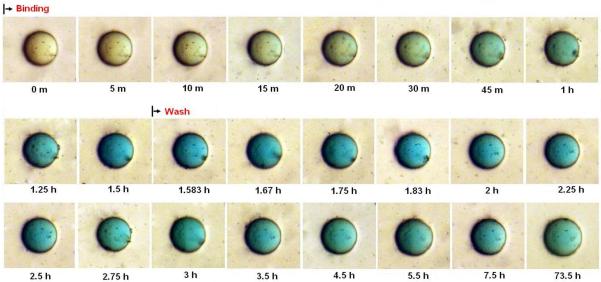
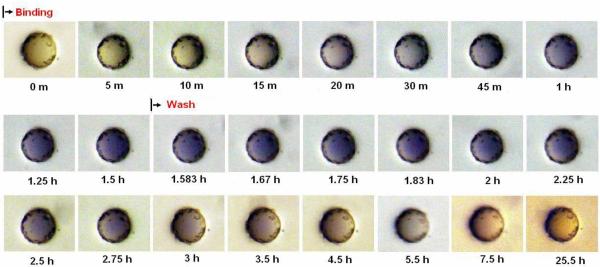
Upon incubation of bead #10 with indigo carmine dye, colorization can be seen after just five minutes and peaks after 30 to 45 minutes. The dissociation process also occurred relatively fast as the color began to fade immediately upon washing with PBS and was completely gone after 20 minutes (Figure 3A and Figure 3D). Incubation of bead #1 with malachite green oxalate dye resulted in a colorization rate that was markedly slower than the indigo carmine to bead #10 interaction. Measurable colorization could only be seen after 20 to 30 minutes incubation. Conversely, after washing, the color did not recede completely even after 72 hours. Only after thorough washing with 70% ethanol was complete dissociation observed (Figure 3B and Figure 3D).
Figure 3A.
Real time association and dissociation of indigo carmine dye to #10 bead.
Figure 3D.
The real time association and dissociation curve of malachite green to #1 bead, bromocresol purple to #4 bead and indigo carmine to #10 after color quantitative analysis.
Figure 3B.
Real time association and dissociation of the malachite green dye to #1 bead.
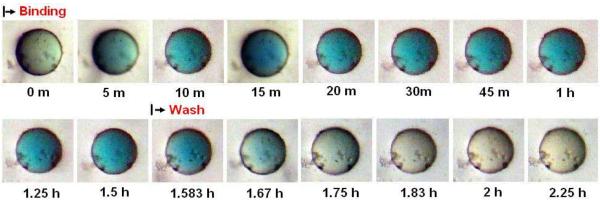
Incubating bead #4 with bromocresol purple dye also resulted in a slower color binding by comparison to the indigo carmine/bead #10 incubation, reaching its maximum around 75 minutes. The dissociation of the color also occurred slowly disappearing by 72 hours (Figure 3C and Figure 3D). Interestingly, #10 bead was shown to also bind bromocresol purple, but the binding and dissociation rates appeared much slower than was observed with indigo carmine, suggesting a difference in binding profile between each of these two dyes and the cyclic peptide in spite of share similarities discussed previously. It is important to note that the color intensity of the bead is uniform throughout the entire study, thus eliminating the possibility that dye diffusion through the bead is the rate-limiting step of staining. Furthermore, the majority of the beads in the library remain colorless indicating that non-specific binding by the polystyrene matrix is totally absent.
Figure 3C.
Real time association and dissociation of the bromocresol purple dye to #4 bead.
In summary, we have developed a simple and reliable method to immobilize microbeads on planar surface without fouling the bead surface necessary for many different biological assays. This PDMS affixed bead library screening approach allows us to not only identify positive bead/target binding as permitted by existing bead screening assays, but can also track beads through successive target binding assays, resulting in a more informative and complete binding profile to multiple strategic targets of interest. Furthermore, this approach facilitates real time binding analysis for each individual bead within a library or sample, delivering more characteristic information from bead compounds and their targets. In addition to the colorimetric binding analysis of simple dye targets, this methodology can be expanded to study the interaction/activity displayed by a bead library/sample with live cells, bacteria, protein or mixture of proteins of interest and more, with direct observation under microscope (live cells, bacteria) or readout in fluorescence or radio image (protein). PDMS is optically transparent and fully compatible with essentially all biochemical or cell-based screening assays that have been described for OBOC libraries.
EXPERIMENTAL SECTION
Materials
The Silicone Elastomer Base and Silicone Elastomer Curing Agent kit (Sylgard® 184) were purchased from Dow Corning Corporation (Midland, MI). The color dyes Malachite green oxalate (Figure 1A), Bromocresol purple (Figure 1B) and Indigo carmine (Figure 1C) were obtained from Sigma-Aldrich (Saint Louis, MO). TentaGel S NH2 resin (90 μm, 0.26 mmol/g) was purchased from Rapp Polymere GmbH (Tübingen, Germany). Fmoc-Amino acids, 1-hydroxybenzotriazole (HOBt), and N,N'-diisopropylcarbodiimide (DIC) were purchased from GL Biochem (Shanghai, China). All solvents and other chemical reagents were purchased from Aldrich (Milwaukee, WI) and were analytical grade.
Library Synthesis
The OBOC libraries were synthesized on TentaGel S NH2 resin using Fmoc-chemistry and “split-and-mix” method. [1] The library used in this experiment is the CX1X2X3X4X5C peptide library. The synthesis procedure is described briefly as following: TentaGel beads (2.0 g, loading 0.26 mmol/g) were swollen in DMF (30 mL) in a column for 3 h. After filtration and washing with DMF, a mixture of Fmoc-L-Cys(Trt)-OH (3 equiv), HOBt (3 equiv) and DIC (3 equiv) was added to the beads. The column was rotated until a Kaiser test [17] was negative. The resins were washed and subjected to Fmoc deprotection with 20% piperidine (5min, 15min). After washing with DMF, methanol (MeOH), DCM, and DMF, respectively, the beads were split into 19 equal portions in 19 tubes (5 mL). Nineteen different Fmoc-L-amino acids except L-cystine (3 equiv.), HOBt (3 equiv) and DIC (3 equiv) were dissolved in DMF, and separately added to the 19 tubes. The coupling was carried out at room temperature for 2 h. Four random tubes were chosen for a Kaiser test. After the Kaiser test was negative, the beads were pooled to a column, drained and washed with DMF three times. The same coupling procedure was repeated for additional 4 cycles with 19 Fmoc-L-amino acids. After last cycle of coupling, the beads were combined, and Fmoc was deprotected, and coupled with Fmoc-L-Cys(Trt)-OH as described above. After Fmoc deprotection, the beads were washed with DMF, MeOH, and dichloromethane (DCM), respectively, three times. The beads were then dried under vacuum. Side-chain deprotection was achieved using a mixture of 82.5% trifluoroacetic acid: 5% phenol: 5% thioanisole: 5% water: 2.5% triisopropylsilane. After neutralization with 3% N,N-diisopropylethylamine (DIEA)/DMF (twice), the beads were washed sequentially with DMF, MeOH, DCM, DMF, DMF/water (60%, 30%) and water. The beads were transferred to a 1 liter bottle, to which was added 500 mL mixture of water, acetic acid and dimethyl sulfoxide (DMSO) (75:5:20) with the pH adjusted with ammonium hydroxide to 6.0. The bottle was shaken for two days until the Ellman test was negative. After filtration, the beads were thoroughly washed with H2O. Finally, the bead library was stored in 70% ethanol/water and was ready for screening.
Beads affixation and Screening
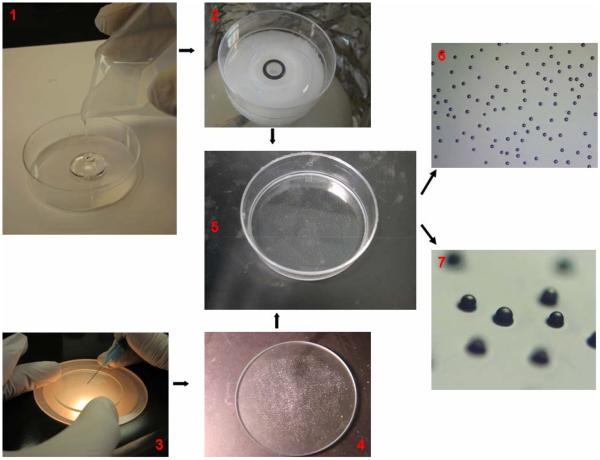
Silicon Elastomer Base was mixed completely with Silicone Elastomer Curing Agent at ratio 10:1 in volume and the solution was kept in vacuum container for 15 to 20 minutes to eliminate the gas bubbles formed during the mixing process. After the removal of all residual bubbles, approximately 0.5 mL solution was added into the bottom of a 60 mm culture dish. The dish was then spun on a single wafer spin processor (Laurell Technologies Corporation®, model WS-650S-6NPP/Lite) to evenly distribute the solution over the plates, at a speed of 4000 rpm for 30 s. A 5 μm layer of PDMS was formed on the bottom of dish and was cured at 80 °C for 5 min or at room temperature for 24 hrs to allow for beads immobilization. Knowing that the PDMS coating was about 5 μm thick and the radius of bead was around 45 μm, the percentage of free bead surface area was estimated by the formula of [1–2πrh/4πr2]×100% (h= thickness of PDMS layer; r= radius of bead) to be approximately 95%. The library beads resuspended in 70% ethanol solution were first randomly placed on the surface of a glass plate that was 10% smaller than the dish used in the experiment. After the ethanol evaporated, a syringe needle was used to adjust the dry beads to evenly redistribute beads on the glass surface. Then the dish was inverted and the PDMS layer was pressed down onto the glass plate with beads. The library beads would stick and embed partly inside the PDMS layer (Figure 4).
Figure 4.
Photographs illustrating how library beads are affixed on the bottom of dish. Around 1 mL PDMS solution was poured into the 60 mm cell culture dish (1). The dish was placed on the top of the rotator of spinning machine and a 5 μm thin layer of solution would form after spinning. The PDMS layer was partially cured overnight prior to bead affixation (2). A mono-layer of library beads was distributed evenly on a glass plate (50 mm in diameter) with the help of a hyperdermic needle (3). The dish was inverted and the PDMS layer on the bottom of the dish was pressed down onto the glass plate supporting the beads (4). The beads would be affixed strongly to the bottom of the dish (5) and the glass plate removed. The beads were observed from top view (10x) (6) and at an angle (40x) under the dissecting microscope (7).
Dye solutions were prepared in PBS to a 100 μM final concentration and 5 mL of each dye solution were added into the 60 mm dish with beads. Three dyes were tested sequentially. The dish was kept swirling at 60 rpm on the rotator. The dye binding was monitored for 90 minutes, and then the dye solution was removed from the dishes and PBS was added to rinse the beads on the bottom to remove the color dye. The PBS solution was changed every 30 min and the dish was always swirled as above. The beads color change was also observed and recorded at certain time interval during the whole wash process to collect data of color dissociation. Before adding the next dye solution, beads were washed with 70% ethanol to completely remove residual dye and were rinsed with PBS solution three times afterwards.
Beads Color Quantification
A representative positive bead for each color was selected for the color binding and dissociation study. A series of images were taken at different time points during the binding and washing process. From the images taken, regions of interest (ROI) with the same size were picked up from the color beads and colorless control beads. The value from the control bead was subtracted from the color bead to get quantitated color value for each single color bead. The measure of the color intensity for each color dye was performed using the Kodak Image Station 2000MM.
Beads Sequencing
Beads with different binding profiles were removed from the plates and washed sequentially with 8 M Guanidine HCl solution and water. Beads were then decoded using the Perkin-Elmer/ Applied Biosystems Protein Sequencer (ABI Procise 494). This system works by sequentially cleaving N-terminal amino acids from a protein or peptide chains and then analyzing the formed phenylthiohydantoin (PTH)-amino acid derivatives.
ACKNOWLEDGMENT
We thanks to Ms. Diana Lac for her editing work.
Funding Sources
This study is supported by funds from NIH grants 1R21HL108300-01A1, 1R33CA160132-01A1.
Footnotes
ASSOCIATED CONTENT None
REFERENCES
- [1].Lam KS, Salmon SE, Hersh EM, Hruby VJ, Kazmierski WM, Knapp RJ. A new type of synthetic peptide library for identifying ligand-binding activity. Nature. 1991;354(6348):82–84. doi: 10.1038/354082a0. [DOI] [PubMed] [Google Scholar]
- [2].Lam KS, Lebl M, Krchnak V. The “One-Bead-One-Compound” Combinatorial Library Method. Chem Rev. 1997;97(2):411–448. doi: 10.1021/cr9600114. [DOI] [PubMed] [Google Scholar]
- [3].Lam KS, Liu R, Miyamoto S, Lehman AL, Tuscano JM. Applications of one-bead one-compound combinatorial libraries and chemical microarrays in signal transduction research. Acc Chem Res. 2003;36(6):370–377. doi: 10.1021/ar0201299. [DOI] [PubMed] [Google Scholar]
- [4].Meldal M, Svendsen I, Breddam K, Auzanneau FI. Portion-mixing peptide libraries of quenched fluorogenic substrates for complete subsite mapping of endoprotease specificity. Proc Natl Acad Sci U S A. 1994;91(8):3314–3318. doi: 10.1073/pnas.91.8.3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Olsen JA, Jensen KJ, Nielsen J. Combinatorial solid-phase synthesis of apalosin mimetics. J Comb Chem. 2000;2(2):143–150. doi: 10.1021/cc990062p. [DOI] [PubMed] [Google Scholar]
- [6].Groth T, Renil M, Meinjohanns E. PEG based resins for protease drug discovery synthesis, screening and analysis of combinatorial on-bead libraries. Comb Chem High Throughput Screen. 2003;6(7):589–610. doi: 10.2174/138620703771981188. [DOI] [PubMed] [Google Scholar]
- [7].Peng L, Liu R, Marik J, Wang X, Takada Y, Lam KS. Combinatorial chemistry identifies high-affinity peptidomimetics against alpha4beta1 integrin for in vivo tumor imaging. Nat Chem Biol. 2006;2(7):381–389. doi: 10.1038/nchembio798. [DOI] [PubMed] [Google Scholar]
- [8].Yao N, Xiao W, Wang X, Marik J, Park SH, Takada Y, Lam KS. Discovery of targeting ligands for breast cancer cells using the one-bead one-compound combinatorial method. J Med Chem. 2009;52(1):126–133. doi: 10.1021/jm801062d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Appell KC, Chung TDY, Ohlmeyer MJH, Sigal NH, Baldwin JJ, Chelsky D. Biological screening of a large combinatorial library. J Biomol Screening. 1996;1:5. [Google Scholar]
- [10].Chirayil S, Chirayil R, Luebke KJ. Discovering ligands for a microRNA precursor with peptoid microarrays. Nucleic Acids Res. 2009;37(16):5486–5497. doi: 10.1093/nar/gkp549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Lehman A, Gholami S, Hahn M, Lam KS. Image subtraction approach to screening one-bead-one-compound combinatorial libraries with complex protein mixtures. J Comb Chem. 2006;8(4):562–70. doi: 10.1021/cc0600268. [DOI] [PubMed] [Google Scholar]
- [12].Townsend JB, Shaheen F, Liu R, Lam KS. Jeffamine derivatized TentaGel beads and poly(dimethylsiloxane) microbead cassettes for ultrahigh-throughput in situ releasable solution-phase cell-based screening of one-bead-one-compound combinatorial small molecule libraries. J Comb Chem. 2010;12(5):700–712. doi: 10.1021/cc100083f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Wennemers H, Still WC. Peptide complexation in water : Sequence-Selective binding with simple dye molecules. Tetrahedron Letters. 1994;35(3):4. [Google Scholar]
- [14].Lam KS, Zhao ZG, Wade S, Krchnak V, Lebl M. Identification of small peptides that interact specifically with a small organic dye. Drug Develop Res. 1994;33:4. [Google Scholar]
- [15].Alderman DJ. Malachite green: a review. J Fish Dis. 1985;8:10. [Google Scholar]
- [16].Culp SJ, Beland FA. Malachite green: a toxicological review. J Am Coll Toxicol. 1996;15:20. [Google Scholar]
- [17].Kaiser E, Colescott RL, Bossinger CD, Cook PI. Color test for detection of free terminal amino groups in the solid-phase synthesis of peptides. Anal Biochem. 1970;34(2):595–598. doi: 10.1016/0003-2697(70)90146-6. [DOI] [PubMed] [Google Scholar]