Abstract
Black tooth stain is a characteristic extrinsic discoloration commonly seen on the cervical enamel following the contour of the gingiva. To investigate the relationship between black tooth stain and the oral microbiota, we used 16S rRNA gene sequencing to compare the microbial composition of dental plaque and saliva among caries-free children with and without black stain. Dental plaque and saliva, as well as black stain, were sampled from 10 children with and 15 children without black stain. Data were analyzed using the pipeline tool MOTHUR. Student’s t-test was used to compare alpha diversities and the Mann-Whitney U test to compare the relative abundances of the microbial taxa. A total of 10 phyla, 19 classes, 32 orders, 61 families and 102 genera were detected in these samples. Shannon and Simpson diversity were found to be significantly lower in saliva samples of children with black stain. Microbial diversity was reduced in the black stain compared to the plaque samples. Actinomyces, Cardiobacterium, Haemophilus, Corynebacterium, Tannerella and Treponema were more abundant and Campylobacter less abundant in plaque samples of children with black stain. Principal component analysis demonstrated clustering among the dental plaque samples from the control group, while the plaque samples from the black stain group were not and appeared to cluster into two subgroups. Alterations in oral microbiota may be associated with the formation of black stain.
Introduction
Black tooth stain is a type of extrinsic discoloration of the tooth. It may be clinically diagnosed as pigmented, dark lines parallel to the gingival margin or as an incomplete coalescence of dark dots rarely extending beyond the cervical third of the crown[1]. Both primary and permanent teeth can be affected, with a reported prevalence of 1–20%[2].
Black stain (BS) is hard to be wiped off by daily cleaning via tooth brushing, and tends to re-form after professional scaling. BS can cause an aesthetic problem for individuals, but no associated impairment of dental health has been reported. Most epidemiological studies worldwide found that children with black-stained teeth had lower caries prevalence or experiences[3–7]. A study of a Brazilian population-based birth cohort also suggested that BS might be a protective factor for dental caries development[8].
Analysis of the black material in black-stained teeth indicates that it contains insoluble ferric salt, which is likely to be ferric sulfide, and a high content of calcium and phosphate[9, 10]. These studies suggested that the ferric sulfide might be formed by the reaction between hydrogen sulfide produced by bacterial action and iron in the saliva or gingival exudates. In one study, a group of 6-year-old Spanish children with black stain were found to have consumed a significantly higher proportion of iron supplements than children without black stain; an increased rate of iron supplementation was also found among the mothers of these children during pregnancy[11]. This study also reported higher consumption of specific foods rich in iron, such as vegetables, dairy products and eggs, in children with black stain. Furthermore, another study revealed a positive correlation between black stain and the concentration of iron in water sources[12]. These findings suggested that the formation of black tooth stain may be associated with ferric sulfide.
Chromogenic bacteria were proposed as an etiological factor in the production of black pigment. Periodontal pathogens such as Porphyromonas gingivalis, Prevotella intermedia, and Prevotella nigrescens are reported to be black-pigmented anaerobes in oral cavity[13]. Former studies assumed Prevotella melaninogenica was closely related to black tooth stain [14]. Traditional bacteriological examinations have implicated Actinomycetes as the predominant cultivable microorganisms found in black stain. However, almost 50% of oral bacteria are non-culturable. Saba et al. detected the presence of four distinct subspecies in dental plaque samples of 100 BS-affected children between 6 and 12 years old and 100 stain-free control subjects by PCR and electrophoresis. Significantly higher prevalences of Actinomyces and Aggregatibacter actinomycetemcomitans were observed in black stain compared to the plaque of the control group. Porphyromonas gingivalis and Prevotella melaninogenica were absent from the samples of both black stain and control subjects[15]. Furthermore, real-time PCR was performed to determine the microbial compositions of samples of black stain from 46 children aged from 3 to 10 years and non-discolored dental plaque from 47 counterparts without black stain. Plaque samples of the black stain group were found to contain higher numbers of Actinomyces naeslundii and lower numbers of Fusobacterium nucleatum and Lactobacillus spp. No significant differences were observed for Aggregatibacter actinomycetemcomitans and Prevotella intermedia[6]. The above studies indicate a unique microbiota in dental plaque with black stain. However, as the results were not unanimous and only a few selected species were evaluated, the relationship between microorganisms and black stain remains uncertain.
The traditional method of assessing oral microorganisms is bacterial culture. The disadvantage of this method is that almost 50% of oral bacteria are non-culturable. In addition, conventional culture methods are time-consuming. By contrast, molecular techniques have the advantage of detecting non-culturable bacteria as well as those that are difficult to cultivate. Besides, number of target bacteria for polymerase chain reaction (PCR) techniques is restricted, only expected species can be detected. In this study, we utilized next-generation sequencing of the bacterial 16S rRNA gene to detect oral microorganisms in children with or without BS. Compared to automated Sanger sequencing, which is considered as the ‘first-generation’ technology, NGS allows sequencing whole genomes without cloning in E. coli or any host cell, and can provides a larger number of reads and greater depth of coverage[16]. This is for the first time that NGS was applied to study black tooth stain. BS-relative bacteria were supposed to be found, which may offer more information for the research of the mechanism of black stain formation.
Materials and Methods
Subject Selection
A total of 25 caries-free children aged 4–5 years were recruited from the third kindergarten of the Chinese Academy of Sciences. 10 children with at least six black-stained teeth were included in group A, while children free of BS were in group B. No significant difference was found between groups involving age and gender. From each child in group A, samples of saliva (SS), plaque (PS) and black stain (BS) were obtained. Samples of supra-gingival plaque (PS) were collected from dental surface where there was no black material. Black stain (BS) was exactly the black material on tooth surface and was scraped off the teeth using dental curette. (Fig 1). From children in group B samples of saliva (SC) and plaque (PC) were obtained. Children in both groups were free of dental caries (dmfs = 0). None had systemic or infectious diseases, enamel hypoplasia or dentinogenesis imperfecta, or had used antibiotic drugs within the 2-week period before sample collection. The study design, protocol, and informed consent were approved by the Ethics Committee of Stomatological Hospital of Peking University (PKUSSIRB- 201311085). And written informed consent was obtained from the parents of all children enrolled in this study.
Fig 1. Primary dentition with more than six black-stained teeth.
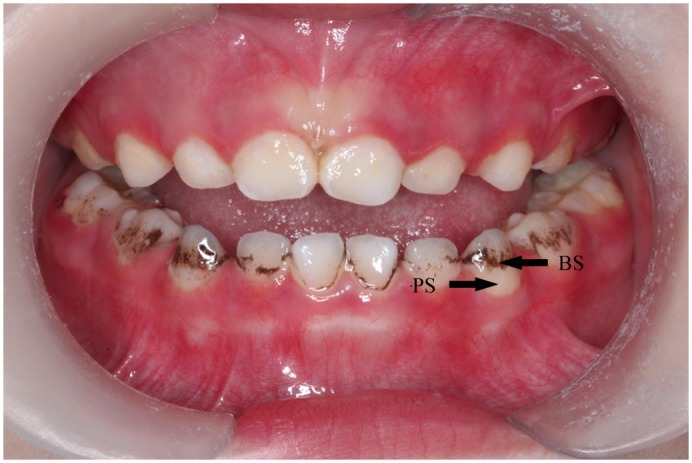
Black stain (BS) was found as dark pigmented lines or incomplete coalescence of dark dots on the cervical enamel following the contour of the gingiva. Samples of both black stain (BS) and dental plaque (PS) were collected from children with black stain.
Sample Collection
Samples were collected from both groups at approximately 8:40–11:20 in the morning at the third kindergarten. Informed consent was obtained from parents of participating children.
Subjects were asked to rinse their mouths with water before sample collection. Samples of dental plaque and black stain were collected using new metal curettes. An expectation of 1-2mg dental plaque and black stain were obtained. For children with black stain, we first collected their dental plaque on non-black stained surface, and then scraped their black stain into Eppendorf tubes. These children then had their teeth dry-brushed to collect stimulated whole saliva into a 50-ml centrifuge tube, for an expected amount of 1–2 ml. Saliva collection was performed within 5 min.
The plaque and black stain samples were immediately transferred to an Eppendorf tube containing 400 μl of sterile water. All samples were placed on ice before being transported to the laboratory.
DNA Extraction
Saliva samples were centrifuged at 13,400g for 15 min and the precipitates were collected. Bacterial DNA was extracted from all samples using the QIAamp DNA Mini Kit (Qiagen, Hilden, Germany), following the manufacturer’s instructions, after initial treatment with lysozyme (20 mg/ml, 37°C for 1 h). The final quality of DNA was evaluated by Nanodrop2000 (Thermo, USA). A standard concentration of 10 ng/μl was prepared for each individual sample for all PCR assays.
PCR amplification of the V3-V4 region of the bacterial 16S rRNA gene was performed using universal primers (338F 5’-ACTCCTACGGGAGGCAGCA-3’, 806R 5’-GGACTACHVGGGTWTCTAAT-3’) incorporating a sample barcode sequence. The cycling parameters were as follows: initial denaturation at 94°C for 10 min; 30 cycles of denaturation at 94°C (30s), annealing at 65°C (30s), elongation at 72°C (30s); and final extension at 72°C for 10 min. PCR products were separated by 2% agarose gel electrophoresis and the fragments of ~500 bp were purified. Equal qualities of PCR products from each sample were pooled for further sequencing.
Bioinformatic Analysis
Sixty samples were sequenced, and the raw data generated were analyzed using the pipeline tool MOTHUR[17, 18]. The multiplexed samples were deconvoluted based on the unique barcode assigned to each sample. The barcodes and primers were then trimmed off and sequences with a quality value of <20 and low-quality sequences were removed[19]. High-quality reads were clustered into operational taxonomic units (OTUs) at 97% similarity. Microbial diversity was estimated and the relative abundance of microbial taxa were calculated and compared. Student’s t-test was used to compare alpha diversities and the Mann-Whitney U test to compare the relative abundances of microbial taxa between two groups. Principal component analysis (PCA) was conducted according to the matrix of distance.
Results
Community distribution
The total number of samples was 60, from which 2 samples of PS were excluded due to the lack of sufficient sequences, resulting in 58 samples that were analyzed further. Totals of 10 phyla, 19 classes, 32 orders, 61 families and 102 genera were detected in these samples.
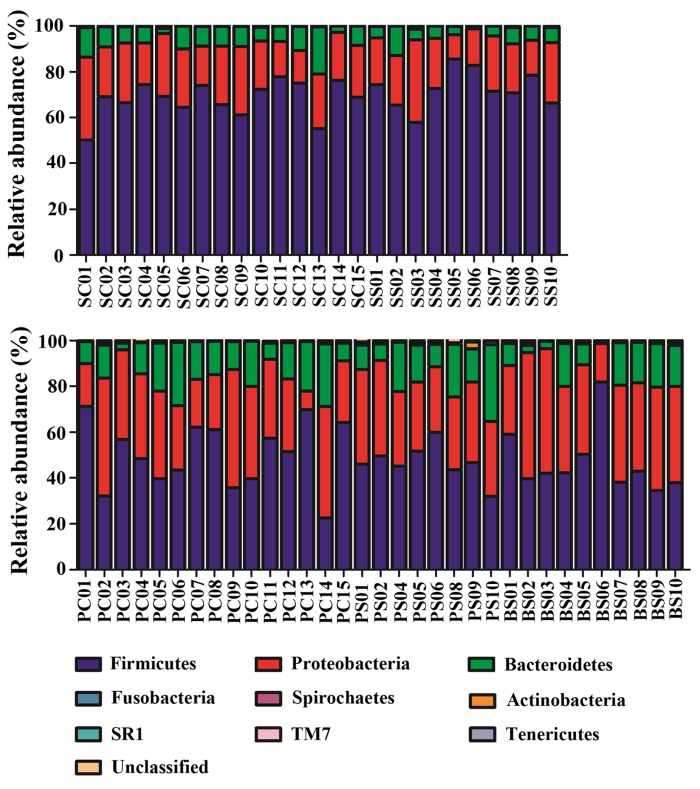
In the PC, PS and BS samples, the 3 most abundant of the 10 phyla were Firmicutes, Proteobacteria and Bacteroidetes, which represented more than 95% of the total sequences. In samples of saliva, SC and SS, the aforementioned three phyla also constituted the majority of the sequences. Firmicutes represented more than 50% of the total sequences (Fig 2).
Fig 2. Relative abundance of bacterial composition at phylum level in saliva and plaque as well as black stain (BS) samples.
At the genus level, among the 102 genera detected, the five most abundant in plaque and BS samples were Streptococcus, Neisseria, Burkholderiales unclassified, Capnocytophaga and Neisseriaceae unclassified. These constituted more than 60% of the total sequences. In saliva, Streptococcus, Neisseria and Capnocytophaga represented more than 60% of the total sequences (S1 Fig).
OTU diversity
In total, 3475 OTUs were obtained at a 97% similarity level. The richness of the total amount of bacteria was estimated by ACE and Chao. The diversity of the microbiota was estimated by Shannon and Simpson indices[20]. Student’s t-test was used to compare alpha diversities between groups. The Shannon and Simpson index showed a significant difference between SC and SS samples. The saliva microbial diversity in children with BS was lower than that in children without BS. Though there was no significant difference among the PC, PS and BS groups, the Shannon and Simpson index suggested a tendency towards a reduction in the BS group (Fig 3).
Fig 3. Microbiota diversity as calculated by Shannon and Simpson index.
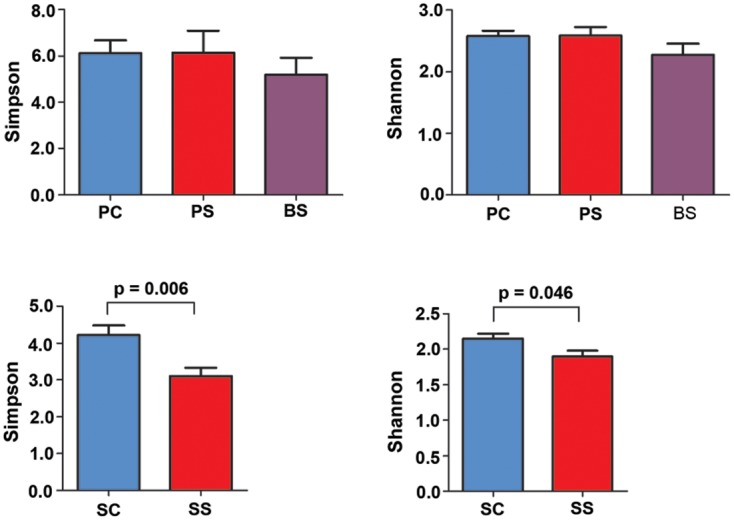
The Shannon and Simpson index values were significantly higher in salivary samples of the control group (SC) than the black stain group (SS). A tendency towards a reduction in the Shannon and Simpson index was observed in samples of black stain (BS) compared to dental plaque from the control (PC) and BS groups (PS); however, the difference was not significant. The error bars indicate mean with standard error.
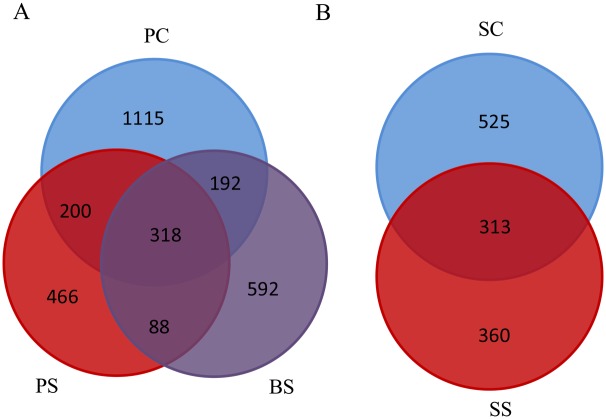
After removing the duplicate sequences, 2971 OTUs were measured in the PC, PS and BS groups, with 318, 200, 88,192 OTUs shared by samples PC/PS/BS, PC/PS, PS/BS, PC/BS, respectively. Unique measurements of OTUs in the PC, PS, and BS groups were 1115, 466 and 592, respectively. The number of OTUs in saliva samples was 1198, with 313 shared by SC and SS, and 525 and 360 OTUs unique to the respective groups (Fig 4).
Fig 4. Venn diagram of the number of OTUs common/unique to the black stain and control groups.
The overlapping areas represent the number of OTUs shared by the counterpart samples. A: A total of 2971 OTUs were detected in black stain (BS) and the dental plaque of the black stain group (PS) and control group (PC). Only 318 OTUs were shared by PS, PC and BS, while 1115, 466, and 592 OTUs were unique to the respective groups. B: A total of 1198 OTUs were detected in the salivary samples of the black stain group (SS) and control group (SC). Only 313 OTUs were shared by the SC and SS groups, and 525 and 360 OTUs were unique to the respective groups.
Taxonomic analysis
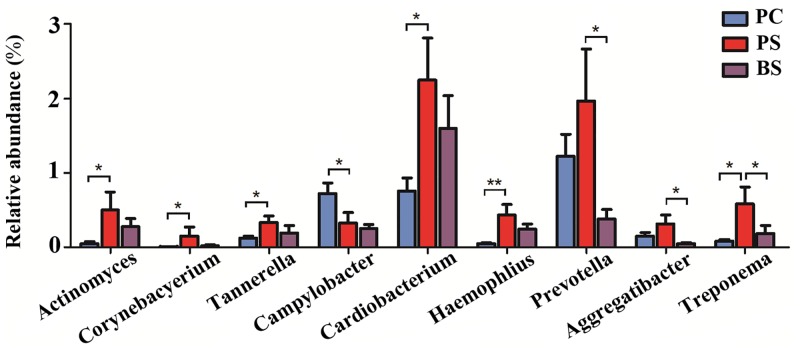
After removing the 2 samples of poor quality, we compared microbiota between groups at various levels using the Mann-Whitney U test. Each of the 10 phyla, 19 classes, 32 orders, 61 families and 102 genera was compared between PC and PS, PS and BS, SC and SS. There were significant differences between the PC and PS groups at the following levels: Actinobacteria (phylum), Actinobacteria, Gammaproteobacteria, and Epsilonproteobacteria (class), Actinomycetales, Cardiobacteriales, Pasteurellales, and Campylobacterales (order), Actinomycetaceae, Cardiobacteriaceae, Pasteurellaceae, Campylobacteraceae, Corynebacteriaceae, and Porphyromonadaceae (family) (S3 Fig), Actinomyces, Cardiobacterium, Haemophilus, Campylobacter, Corynebacterium, and Tannerella (genus) (Fig 5).
Fig 5. Comparisons of microbiota that presented significantly different contents in dental plaque of the black stain group (PS) and control group (PC) and black stain (BS) at genus level.
*P<0.05; **P<0.01.
There were significant differences between the PS and BS groups at the following levels: Spirochaetes (phylum), Spirochaetes (class), Spirochaetales and Pasteurellales (order), Spirochaetaceae, Pasteurellaceae and Prevotellaceae (family) (S3 Fig), and Treponema, Aggregatibacter and Prevotella (genus) (Fig 5). Bacterial genera that were more abundant in children with BS are listed in Table 1.
Table 1. Bacterial phylotype that were more abundant in children with black stain.
| Bacterial phylotype | Characteristics | References |
|---|---|---|
| Actinomyces | Produce H2S | [21] |
| Pigmentation | [22] | |
| Normal oral flora | ||
| Tannerella | Periodontitis(Tannerella forsythia) | [27] |
| Treponema | Periodontal diseases | [26] |
| Corynebacterium | Dental calculi (C. matruchotii) | [25] |
| Opportunistic pathogen | ||
| Cardiobacterium | Endocarditis | [23] |
| Normal flora | ||
| Haemophilus | Respiratory tract infection | [24] |
| Normal flora |
We also compared the salivary microbiota composition of the two groups and found differences in the following microorganisms: Bacteroidetes (phylum), Flavobacteria (class), Flavobacteriales (order), Flavobacteriaceae (family) and Capnocytophaga (genus) (S4 Fig).
Principal Component Analysis (PCA)
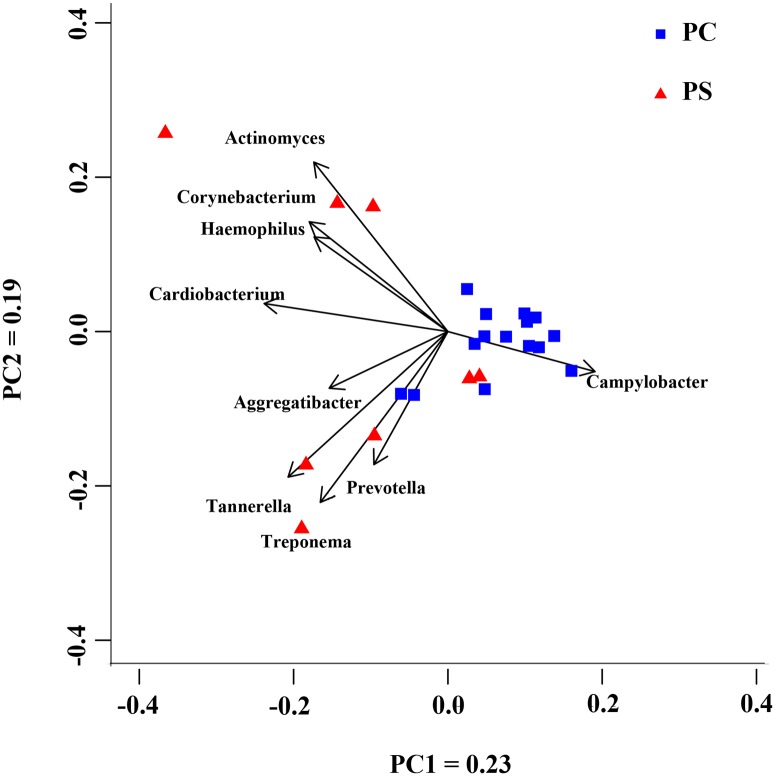
To classify the bacteria into two groups, a PCA was implemented (Fig 6) based on genus information. The PC samples appeared to cluster together, with two samples mixed in the PS cluster, while the PS samples were more dispersive and seemed to cluster into two subgroups. This implied there may be complex mechanisms for black stain formation, but a study involving a larger sample size would be required to confirm this.
Fig 6. Principal component analysis of the bacterial genera detected in the dental plaque of the control group (PC) and black stain group (PS).
The PC samples clustered together, with two samples mixed into the PS cluster, while the PS samples were more dispersive and seemed to cluster into two subgroups.
Discussion
This study used next-generation sequencing of the bacterial 16S rRNA gene to evaluate the oral microbiota in children with and without black stain. The salivary microbiota diversity in children with black stain was significantly lower than in children without black stain. Regarding the plaque microbiota, although the Shannon and Simpson index showed no significant differences among PC, PS and BS groups, the microbial diversity tended to be lower in BS samples compared with PC and PB samples. This suggested that microbial diversity is reduced in children with black stain, which might be associated with black stain formation.
Former studies had found the relative abundance of Actinomyces was higher in children with black stain compared to children without black stain[6, 15]. Our study confirmed these findings. Hence it is possible that Actinomycetes is involved in the deposition of BS. Actinomyces belong to the resident oral microbiota of supra-gingival plaque. Some Actinomyces strains produce hydrogen sulfide[10, 21], which can result in ferric sulfide formation in the presence of iron in saliva or gingival exudates. Sarkonen et al. found that A. odontolyticus, A. graevenitzii and A. radicidentis produce pigments, with colors ranging from brown to black, when cultured on rabbit slaked blood agar[22]. However, this pigmentation may not be true pigment production, but rather a result of medium decomposition. Nevertheless, the proportions of Actinomyces could account for different susceptibilities to extrinsic black stain formation.
Cardiobacterium, Haemophilus, Corynebacterium, Tannerella and Treponema were also more abundant in plaque samples from children with black stain. These bacteria may not produce pigments or hydrogen sulfide. However, the overall plaque genome suggests that Actinomyces may not be the only microorganism involved in black stain formation. Alterations in the abundance of several other species could provide an environment that facilitates black stain deposition on the enamel surface. Corynebacterium, Cardiobacterium[23] and Haemophilus[24] were not previously reported to be associated with extrinsic black stain. Corynebacterium matruchotii was supposed to be the main Corynebacterium species in the oral cavity of humans, and is associated with the formation of dental calculi[25]. On the other hand, Campylobacter was detected in this study to be more abundant in PC than in PS. These genus are commonly found in oral cavity. As a more sensitive and culture-independent method, 16S rRNA gene sequencing makes it possible to detect previously undetected species, but whether these microorganisms are related to black stain formation remains to be determined.
Saba et al. detected the periodontal pathogen A. actinomycetemcomitans at a higher frequency in the black stain group and discussed the potential future risk of developing periodontitis[15]. Using real-time PCR, Heinrich-Weltzien et al. found no significant difference in the prevalence of A. actinomycetemcomitans and P. intermedia between black stain group and control group[6], which is consistent with our results. In addition, we detected Tannerella and Treponema at a higher prevalence in the BS group (Table 1). Both Tannerella and Treponema are related to periodontal disease[26, 27]. Sakai et al. evaluated the prevalence of putative periodontal pathogens (Aggregatibacter actinomycetemcomitans, Porphyromonas gingivalis, Prevotella nigrescens, Treponema denticola) in the saliva of children with mixed dentition, and found a high percentage of children harbored at least one of these four pathogens without signs of periodontal disease[28]. Further studies should analyze the role of periodontopathogenic bacteria in BS and the susceptibility of children with black stain to the development of periodontal disease.
The presence of black stain has been reported to be associated with low frequency of caries, for as-yet-unknown reasons. Slots proposed that the low caries occurrence might be due to lower numbers of Streptococcus, one of the main pathogens of dental caries[14]. According to Heinrich-Weltzien, counts of S. mutans and S. sobrinus were not significantly different between samples of BS and non-discolored plaque in both caries-free and -affected children, but counts of Lactobacillus spp. were higher in non-discolored plaque samples[6]. In our study, no significant difference of Streptococcus among groups was found. Reid and Beeley suggested that the reduction in the incidence of caries in individuals with black extrinsic tooth stains is due to the calcium and phosphate content of the biofilm of the stain[10]. Considering the multifactorial nature of caries disease, the hypothesis that the microbial composition of BS might be associated with the lower caries experience of children is questionable.
In conclusion, black stain in the primary dentition might be more correlated with differences in the microbiota in dental plaque than in saliva. Children with black stain exhibit a reduced microbial diversity and black stain formation may be related to alteration of the plaque microbiota. Actinomyces was more abundant in plaque samples of children with black stain and thus might be a causative agent. Alterations in the abundance of other species might provide an environment that enables black stain formation and deposition onto the surface of enamel. Furthermore, the role of these microorganisms in black stain formation and the association of black stain with periodontitis and caries require further research.
This is the first time that next generation sequencing has been applied to assess the microbiology of black tooth stain. It provided us a broader view of the microbiota in black stain from the aspect of bacterial composition compared to former studies using PCR or rt-PCR. However, the descriptive results cannot confirm the mechanisms of black stain formation. We need further studies to analyze functions of these bacteria. On the other hand, our sample size is relatively small, we will collect more samples in our clinic and kindergarten for further study to verify our conclusion.
The English in this document has been checked by at least two professional editors, both native speakers of English. For a certificate, please see: http://www.textcheck.com/certificate/ZAboUD
Supporting Information
(TIF)
No significant difference was found (P>0.05).
(TIF)
*P<0.05; **P<0.01.
(TIF)
(TIF)
An average of 4638 reads was randomly drawing out from each sample for subsequent OTU analysis.
(TIF)
(DOCX)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was funded by the National Natural Science Foundation of China (81200762 to FC) and Peking University School of Stomatology (PKUSS20110301 to FC). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Koch MJ, Bove M, Schroff J, Perlea P, Garcia-Godoy F, Staehle HJ. Black stain and dental caries in schoolchildren in Potenza, Italy. ASDC J Dent Child. 2001;68(5–6):353–5, 02 . [PubMed] [Google Scholar]
- 2. Ronay V, Attin T. Black stain—a review. Oral Health Prev Dent. 2011;9(1):37–45. . [PubMed] [Google Scholar]
- 3. Chen X, Zhan JY, Lu HX, Ye W, Zhang W, Yang WJ, et al. Factors associated with black tooth stain in Chinese preschool children. Clin Oral Investig. 2014;18(9):2059–66. 10.1007/s00784-013-1184-z . [DOI] [PubMed] [Google Scholar]
- 4. Gasparetto A, Conrado CA, Maciel SM, Miyamoto EY, Chicarelli M, Zanata RL. Prevalence of black tooth stains and dental caries in Brazilian schoolchildren. Braz Dent J. 2003;14(3):157–61. . [DOI] [PubMed] [Google Scholar]
- 5. Heinrich-Weltzien R, Monse B, van Palenstein Helderman W. Black stain and dental caries in Filipino schoolchildren. Community Dent Oral Epidemiol. 2009;37(2):182–7. 10.1111/j.1600-0528.2008.00458.x . [DOI] [PubMed] [Google Scholar]
- 6. Heinrich-Weltzien R, Bartsch B, Eick S. Dental caries and microbiota in children with black stain and non-discoloured dental plaque. Caries Res. 2014;48(2):118–25. 10.1159/000353469 . [DOI] [PubMed] [Google Scholar]
- 7. Mellanby M, Coumoulos H, Kelley M. Teeth of 5-year-old London schoolchildren (1955) with a comparison of results obtained from 1929 to 1955. Br Med J. 1957;2(5040):318–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Franca-Pinto CC, Cenci MS, Correa MB, Romano AR, Peres MA, Peres KG, et al. Association between black stains and dental caries in primary teeth: findings from a Brazilian population-based birth cohort. Caries Res. 2012;46(2):170–6. 10.1159/000337280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Reid JS, Beeley JA, MacFarlane TW. A study of the pigment produced by Bacteroides melaninogenicus. J Dent Res. 1976;55(6):1130 . [DOI] [PubMed] [Google Scholar]
- 10. Reid JS, Beeley JA, MacDonald DG. Investigations into black extrinsic tooth stain. J Dent Res. 1977;56(8):895–9. [DOI] [PubMed] [Google Scholar]
- 11. Garcia Martin JM, Gonzalez Garcia M, Seoane Leston J, Llorente Pendas S, Diaz Martin JJ, Garcia-Pola MJ. Prevalence of black stain and associated risk factors in preschool Spanish children. Pediatr Int. 2013;55(3):355–9. 10.1111/ped.12066 . [DOI] [PubMed] [Google Scholar]
- 12. Pushpanjali K, Khanal SS, Niraula SR. The relationship of dental extrinsic stains with the concentration of trace elements in water sources in a district of Nepal. Oral Health Prev Dent. 2004;2(1):33–7. . [PubMed] [Google Scholar]
- 13. Soukos NS, Som S, Abernethy AD, Ruggiero K, Dunham J, Lee C, et al. Phototargeting oral black-pigmented bacteria. Antimicrob Agents Chemother. 2005;49(4):1391–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Slots J. The microflora of black stain on human primary teeth. Scand J Dent Res. 1974;82(7):484–90. . [DOI] [PubMed] [Google Scholar]
- 15. Saba C, Solidani M, Berlutti F, Vestri A, Ottolenghi L, Polimeni A. Black stains in the mixed dentition: a PCR microbiological study of the etiopathogenic bacteria. J Clin Pediatr Dent. 2006;30(3):219–24. . [DOI] [PubMed] [Google Scholar]
- 16. Hutchison CA 3rd. DNA sequencing: bench to bedside and beyond. Nucleic Acids Res. 2007;35(18):6227–37. 10.1093/nar/gkm688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75(23):7537–41. 10.1128/AEM.01541-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Unno T. Bioinformatic Suggestions on MiSeq-based Microbial Community Analysis. J Microbiol Biotechnol. 2015. . [DOI] [PubMed] [Google Scholar]
- 19. Li Y, He J, He Z, Zhou Y, Yuan M, Xu X, et al. Phylogenetic and functional gene structure shifts of the oral microbiomes in periodontitis patients. ISME J. 2014;8(9):1879–91. 10.1038/ismej.2014.28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kemp PF, Aller JY. Bacterial diversity in aquatic and other environments: what 16S rDNA libraries can tell us. FEMS Microbiol Ecol. 2004;47(2):161–77. 10.1016/S0168-6496(03)00257-5 [DOI] [PubMed] [Google Scholar]
- 21. Tek M, Metin M, Sener I, Bereket C, Tokac M, Kazancioglu HO, et al. The predominant bacteria isolated from radicular cysts. Head Face Med. 2013;9:25 10.1186/1746-160X-9-25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sarkonen N, Kononen E, Summanen P, Kononen M, Jousimies-Somer H. Phenotypic identification of Actinomyces and related species isolated from human sources. J Clin Microbiol. 2001;39(11):3955–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Han XY, Falsen E. Characterization of oral strains of Cardiobacterium valvarum and emended description of the organism. J Clin Microbiol. 2005;43(5):2370–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Murphy TF. Respiratory infections caused by non-typeable Haemophilus influenzae. Curr Opin Infect Dis. 2003;16(2):129–34. . [DOI] [PubMed] [Google Scholar]
- 25. Tsuzukibashi O, Uchibori S, Shinozaki-Kuwahara N, Kobayashi T, Takada K, Hirasawa M. A selective medium for the isolation of Corynebacterium species in oral cavities. J Microbiol Methods. 2014;104:67–71. 10.1016/j.mimet.2014.06.005 . [DOI] [PubMed] [Google Scholar]
- 26. Sela MN. Role of Treponema denticola in periodontal diseases. Crit Rev Oral Biol Med. 2001;12(5):399–413. . [DOI] [PubMed] [Google Scholar]
- 27. Tomita S, Komiya-Ito A, Imamura K, Kita D, Ota K, Takayama S, et al. Prevalence of Aggregatibacter actinomycetemcomitans, Porphyromonas gingivalis and Tannerella forsythia in Japanese patients with generalized chronic and aggressive periodontitis. Microb Pathog. 2013;61–62:11–5. 10.1016/j.micpath.2013.04.006 . [DOI] [PubMed] [Google Scholar]
- 28. Sakai VT, Campos MR, Machado MA, Lauris JR, Greene AS, Santos CF. Prevalence of four putative periodontopathic bacteria in saliva of a group of Brazilian children with mixed dentition: 1-year longitudinal study. Int J Paediatr Dent. 2007;17(3):192–9. 10.1111/j.1365-263X.2006.00813.x . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIF)
No significant difference was found (P>0.05).
(TIF)
*P<0.05; **P<0.01.
(TIF)
(TIF)
An average of 4638 reads was randomly drawing out from each sample for subsequent OTU analysis.
(TIF)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.