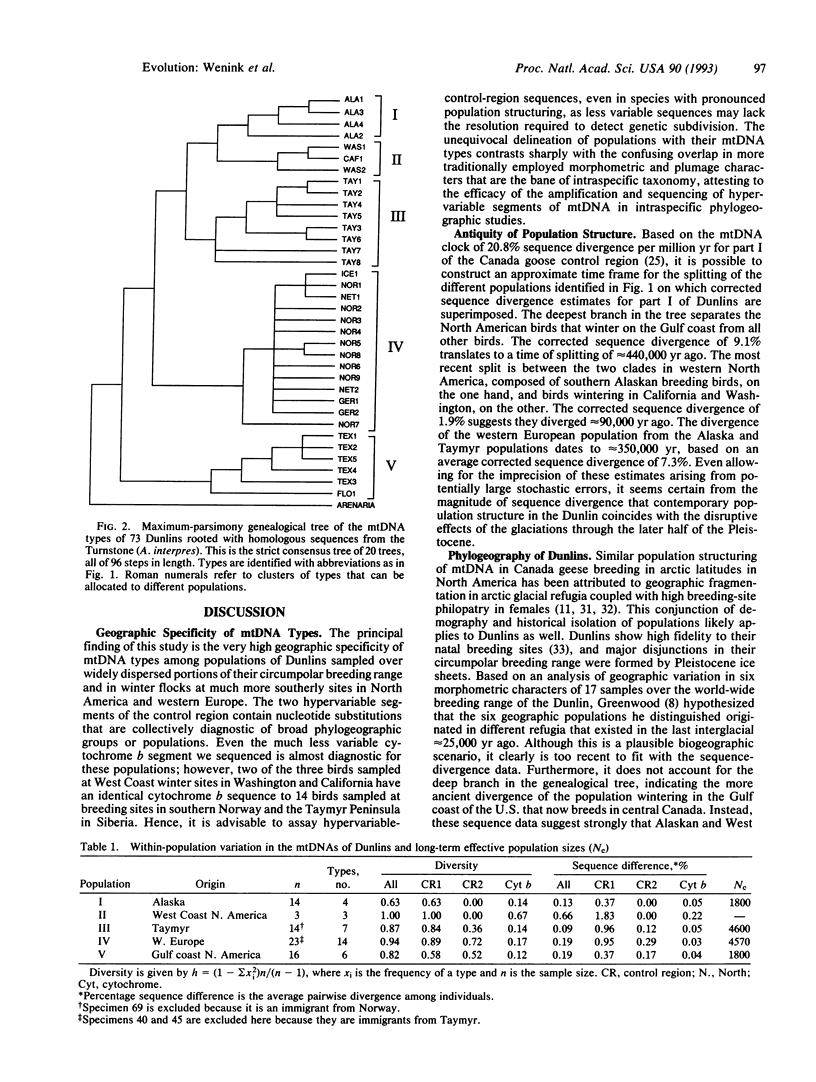
Abstract
Hypervariable segments of the control region of mtDNA as well as part of the cytochrome b gene of Dunlins were amplified with PCR and sequenced directly. The 910 base pairs (bp) obtained for each of 73 individuals complete another of the few sequencing studies that examine the global range of a vertebrate species. A total of 35 types of mtDNA were detected, 33 of which were defined by the hypervariable-control-region segments. Thirty of the latter were specific to populations of different geographic origin in the circumpolar breeding range of the species. The remaining three types indicate dispersal between populations in southern Norway and Siberia, but female-mediated flow of mtDNA apparently is too low to overcome the effects of high mutation rates of the control-region sequences, as well as population subdivision associated with historical range disjunctions. A genealogical tree relating the types grouped them into five populations: Alaska, West Coast of North America, Gulf of Mexico, western Europe, and the Taymyr Peninsula. The Dunlin is thus highly structured geographically, with measures of mutational divergence approaching 1.0 for fixation of alternative types in different populations. High diversity of types within populations as well as moderate long-term effective population sizes argue against severe population bottlenecks in promoting this differentiation. Instead, population fragmentation in Pleistocene refuges is the most plausible mechanism of mtDNA differentiation but at a much earlier time scale than suggested previously with morphometric data.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aquadro C. F., Greenberg B. D. Human mitochondrial DNA variation and evolution: analysis of nucleotide sequences from seven individuals. Genetics. 1983 Feb;103(2):287–312. doi: 10.1093/genetics/103.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball R. M., Freeman S., James F. C., Bermingham E., Avise J. C. Phylogeographic population structure of Red-winged Blackbirds assessed by mitochondrial DNA. Proc Natl Acad Sci U S A. 1988 Mar;85(5):1558–1562. doi: 10.1073/pnas.85.5.1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cann R. L., Brown W. M., Wilson A. C. Polymorphic sites and the mechanism of evolution in human mitochondrial DNA. Genetics. 1984 Mar;106(3):479–499. doi: 10.1093/genetics/106.3.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cann R. L., Stoneking M., Wilson A. C. Mitochondrial DNA and human evolution. Nature. 1987 Jan 1;325(6099):31–36. doi: 10.1038/325031a0. [DOI] [PubMed] [Google Scholar]
- Desjardins P., Morais R. Sequence and gene organization of the chicken mitochondrial genome. A novel gene order in higher vertebrates. J Mol Biol. 1990 Apr 20;212(4):599–634. doi: 10.1016/0022-2836(90)90225-B. [DOI] [PubMed] [Google Scholar]
- Di Rienzo A., Wilson A. C. Branching pattern in the evolutionary tree for human mitochondrial DNA. Proc Natl Acad Sci U S A. 1991 Mar 1;88(5):1597–1601. doi: 10.1073/pnas.88.5.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyllensten U. B., Erlich H. A. Generation of single-stranded DNA by the polymerase chain reaction and its application to direct sequencing of the HLA-DQA locus. Proc Natl Acad Sci U S A. 1988 Oct;85(20):7652–7656. doi: 10.1073/pnas.85.20.7652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyllensten U., Wharton D., Josefsson A., Wilson A. C. Paternal inheritance of mitochondrial DNA in mice. Nature. 1991 Jul 18;352(6332):255–257. doi: 10.1038/352255a0. [DOI] [PubMed] [Google Scholar]
- Hedges S. B., Kumar S., Tamura K., Stoneking M. Human origins and analysis of mitochondrial DNA sequences. Science. 1992 Feb 7;255(5045):737–739. doi: 10.1126/science.1738849. [DOI] [PubMed] [Google Scholar]
- Horai S., Hayasaka K. Intraspecific nucleotide sequence differences in the major noncoding region of human mitochondrial DNA. Am J Hum Genet. 1990 Apr;46(4):828–842. [PMC free article] [PubMed] [Google Scholar]
- Kimura M., Maruyama T. Pattern of neutral polymorphism in a geographically structured population. Genet Res. 1971 Oct;18(2):125–131. doi: 10.1017/s0016672300012520. [DOI] [PubMed] [Google Scholar]
- Kocher T. D., Thomas W. K., Meyer A., Edwards S. V., Päbo S., Villablanca F. X., Wilson A. C. Dynamics of mitochondrial DNA evolution in animals: amplification and sequencing with conserved primers. Proc Natl Acad Sci U S A. 1989 Aug;86(16):6196–6200. doi: 10.1073/pnas.86.16.6196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latter B. D. The island model of population differentiation: a general solution. Genetics. 1973 Jan;73(1):147–157. doi: 10.1093/genetics/73.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn T. W. The genetic legacy of Mother Goose--phylogeographic patterns of lesser snow goose Chen caerulescens caerulescens maternal lineages. Mol Ecol. 1992 Aug;1(2):105–117. doi: 10.1111/j.1365-294x.1992.tb00162.x. [DOI] [PubMed] [Google Scholar]
- Stoneking M., Hedgecock D., Higuchi R. G., Vigilant L., Erlich H. A. Population variation of human mtDNA control region sequences detected by enzymatic amplification and sequence-specific oligonucleotide probes. Am J Hum Genet. 1991 Feb;48(2):370–382. [PMC free article] [PubMed] [Google Scholar]
- Templeton A. R. Human origins and analysis of mitochondrial DNA sequences. Science. 1992 Feb 7;255(5045):737–737. doi: 10.1126/science.1590849. [DOI] [PubMed] [Google Scholar]
- Vigilant L., Pennington R., Harpending H., Kocher T. D., Wilson A. C. Mitochondrial DNA sequences in single hairs from a southern African population. Proc Natl Acad Sci U S A. 1989 Dec;86(23):9350–9354. doi: 10.1073/pnas.86.23.9350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson G. S., Chapman A. M. Length and sequence variation in evening bat D-loop mtDNA. Genetics. 1991 Jul;128(3):607–617. doi: 10.1093/genetics/128.3.607. [DOI] [PMC free article] [PubMed] [Google Scholar]




