Abstract
Ongoing DNA damage is a common feature of epithelial cancers. Here we show that tumor cells derived from multiple myeloma (MM), a disease of clonal plasma cells, demonstrate DNA replicative stress leading to DNA damage. We identified a poor prognosis subset of MM with extensive chromosomal instability and replicative stress which rely on ATR to compensate for DNA replicative stress; conversely, silencing of ATR or treatment with a specific ATR inhibitor triggers MM cell apoptosis. We show that oncogenes such as MYC induce DNA damage in MM cells not only by increased replicative stress, but also via increased oxidative stress, and that ROS-inducer piperlongumine triggers further DNA damage and apoptosis. Importantly, ATR inhibition combined with piperlongumine triggers synergistic MM cytotoxicity. This synthetic lethal approach, enhancing oxidative stress while concomitantly blocking replicative stress response, provides a novel combination targeted therapy to address an unmet medical need in this subset of MM.
Introduction
MM is a clonal proliferation of malignant plasma cells. The genome of MM cells is remarkably complex with profoundly altered karyotypes including aneuploidies, chromosomal translocations, and frequent DNA copy-number variations (1–3). The causes underlying these extensive genomic rearrangements in MM remain largely unknown. We and others have recently shown that hematological cancers including MM have constitutive ongoing DNA damage, evidenced by high number of γ-H2A.X foci in their nuclei (4, 5). As a result, the DNA damage response (DDR) is activated, with phosphorylation of ATM and CHK2, as well as ATR and CHK1. These findings suggest that the complex karyotypes in MM cells may result from ongoing intrinsic DNA damage in MM cells.
The mechanisms responsible for DNA damage in hematological cancers remain unclear. In epithelial cancers, activated oncogenes elicit double-strand breaks (DSB) and ultimately genomic instability (6–10). Classical studies on instability arising from overexpression of mutated HRAS (11) and MYC (12) support this notion. A recent model in epithelial cancers proposed that activated oncogenes trigger inordinate DNA replication, thereby leading to replicative stress (6, 8, 9, 13) which results in DNA double strand breaks (DSBs). For example, MYC interacts with the pre-replicative complex and localizes in proximity of replication origins early in S phase; when overexpressed, it increases replicons due to unscheduled origin activation (14). Through CDC45, MYC overexpression reduces inter-origin distances, independent of its transcriptional activity (15). Additionally, oncogenes can upregulate components of the pre-replicative complex (pre-RC) and the replication machinery, including CDC6 and members of the ORC and MCM families. These proteins are frequently overexpressed and amplified in various cancers, associated with a poor prognosis, and behave as oncogenes when overexpressed both in vitro and in vivo (7, 16).
Oncogene-induced replicative stress also triggers a DNA damage response and senescence (7), and cancer cells overcome these suppressive responses via several compensatory mechanisms. For example, tumors frequently inactivate the phosphoinositide 3-kinase (PI3K)-related protein kinase (PIKK) ATM and its downstream target p53, thereby abrogating apoptosis after DSBs. However, the relevance of the inactivation of the p53 pathway in hematological cancers is unclear, since we have recently shown that hematological cancers preferentially activate an alternative pathway in response to DNA damage. Specifically, after ATM activation the serine-threonine ABL1 relocalizes to the nucleus, where it interacts with the Hippo co-factor YAP1 and the tumor suppressor TP73 to induce apoptosis. Importantly, a subset of hematologic malignancies genetically or functionally disables YAP1, thereby preventing apoptosis (4).
Surprisingly, ATR, the other major PIKK, has an opposite effect than ATM, since it is required for survival of cancer cells under conditions of increased DNA damage (17). Unlike ATM, ATR is activated during S phase and regulates firing of replication origins and repair of damaged replication forks. Indeed, during replicative stress, single stranded DNA (ssDNA) increases, which is coated by the single-stranded-(ss) DNA-binding replication protein A (RPA), thereby activating ATR and its main downstream target, CHK1. As a result, replication forks are stabilized and stalled forks resolved, in order to assure completion of replication. If the resolution of the stalled forks does not succeed, ssDNAs evolve into double stranded breaks (DSBs), followed by ATM and H2A.X phosphorylation and apoptosis.
An intact ATR/CHK1 pathway is crucial for the survival of tumor cells in vivo (18), especially in the presence of activated oncogenes. For instance, Eμ-myc transgenic mice develop B-cell lymphomas with ATM activation (19) and intense replicative stress, as well as ATR and CHK1 phosphorylation (20). Remarkably, crossing the Eμ-myc transgenic mice with a hypomorphic Atr mouse strain (Atr-Seckel; AtrS/S) (21) prevents this development of lymphomas (18). ATR also suppresses MYC-induced replicative stress and apoptosis. Accordingly, treatment with CHK1 inhibitors (UCN-01 or SB-218078) was highly effective in human cancer cell lines and tumors from mice overexpressing MYC, especially in p53-null background. Similar results can be observed in tumors with replicative stress due to dysregulation of other oncogenes including members of the RAS family and MLL-ENL (22), although the effect of mutated RAS might be context-dependent (18). Importantly, ongoing replicative stress may allow specific targeting of cancer cells in a synthetic lethal approach, since reduction of ATR levels induces apoptosis only in tumor cells, with no apparent effects on normal cells. These studies have provided the rationale for the development of ATR inhibitors for the treatment of cancers with enhanced replicative stress (22–24).
Additionally, oxidative stress, an imbalance between the production and elimination of free radicals and reactive metabolites (so-called reactive oxygen species, ROS) of mitochondrial origin, can lead to DNA damage and promote neoplastic transformation. ROS can also activate cellular proliferation via AKT activation or modulate the expression and enzymatic activity of the DNA mismatch repair genes (25). Interestingly mutated RAS (26), MYC (27), and cyclin E have been reported to increase ROS (28) without eliciting replicative stress (29, 30), and therefore represent an independent manner to induce DNA damage in cancer.
Several oncogenes have been associated with MM pathogenesis. MYC in particular is overexpressed in MM, and it has been proposed that low-level overexpression of MYC drives the transition from precursor stages to active MM. Additionally, genomic rearrangements directly targeting the MYC locus occur during disease progression, resulting in further upregulation of MYC expression levels (31). In this study, we delineated mechanisms underlying constitutive ongoing DNA damage in MM and identify a novel synthetic lethal approach to target these cells and induce their apoptosis.
Results
Intense replicative stress in MM cell lines with constitutive DNA damage
To obtain insights into the mechanisms underlying widespread DNA damage in MM cells, we first compared gene expression profiles of MM cell lines with ongoing DNA damage to those without this phenotype (4) using Gene Set Enrichment Analysis (GSEA) (32). Remarkably, examination of the Kegg repository revealed that the DNA replication pathway was the most significantly enriched in MM cells with enhanced DNA damage (NES=1.89, P<0.0001, FDR=0.014) (Fig. 1A) (Supplementary Table 1). Indeed, oncogene-induced replicative stress is often associated with enhanced expression of components of the replication machinery (7). Cell cycle regulatory genes were also significantly altered in these cells (NES=1.92, P<0.0001, FDR=0.047), suggesting an association between enhanced proliferation and replicative stress in MM cells.
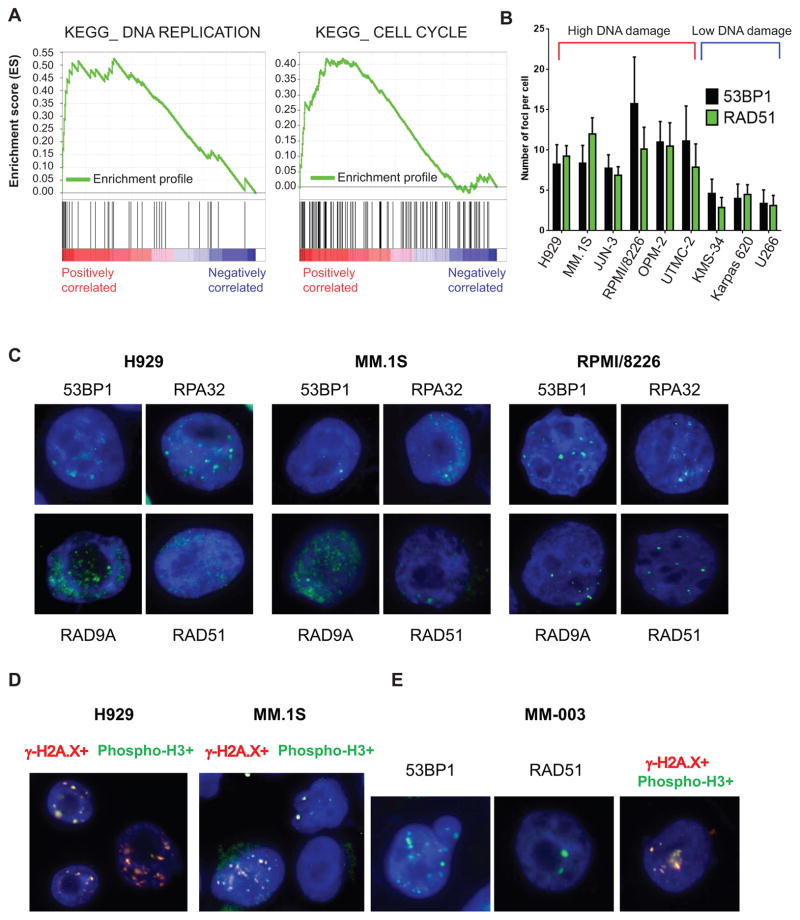
Figure 1. Replicative stress in MM cells.
A, GSEA enrichment profiles of KEGG DNA replication (left) and cell cycle (right) pathways as annotated in MSigDB, comparing MM cell lines with or without ongoing DNA damage. B, number of foci in a panel of MM cells presenting with high DNA damage (red bracket) and low DNA damage (blue bracket) C, MM.1S, H929, and RPMI/8226 MM cell lines were stained by immunofluorescence for known replicative stress markers, including 53BP1, RAD51, RPA32, and RAD9A. D, γ-H2A.X/phospho-histone H3 double staining in MM cell lines. E, replicative stress markers in one representative MM patient.
To explore whether ongoing DNA damage in MM cells led to increased replicative stress, we next assessed a large panel of replicative stress markers including 53BP1, RPA32, RAD9A, and RAD51. MM cell lines (H929, MM.1S, and RPMI/8226) all demonstrated markers of replicative stress (Fig. 1B and 1C, Supplementary Fig. S1A–B and S2A-B) that were absent in peripheral blood mononuclear cells (PBMCs) (Supplementary Fig. S3) or only minimally expressed in MM cell lines without DNA damage.
Moreover, we found a significant colocalization of γ-H2A.X with phospho-histone H3 (Fig. 1D and Supplementary Fig. S4), a marker of proliferation phosphorylated only during mitosis (33), further indicating that cells with DNA damage have activated replicative capacity. We then explored whether replicative stress was also evident in patient MM cells. As shown in Fig. 1E and Supplementary Fig. S5, patient MM cells showed signs of replicative stress, evidenced by 53BP1 and RAD51 positivity, as well as by co-localization of γ-H2A.X and phospho-histone H3. Taken together, these results indicate that MM cells with DNA damage also have increased replicative stress.
A signature of chromosomal instability defines a subset of MM patients with increased expression of replicative genes and poor prognosis
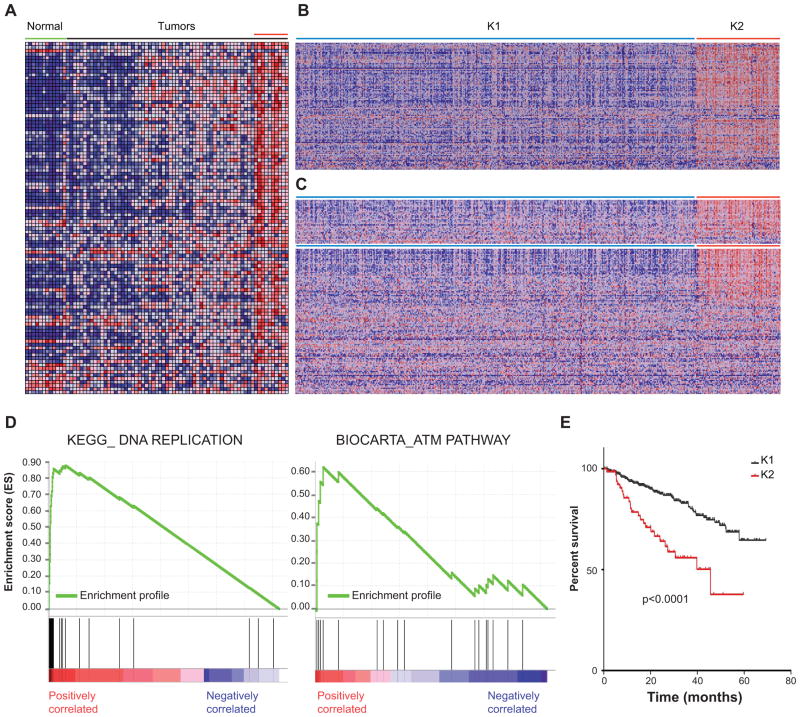
We next sought to determine whether patient MM cells also show DNA damage associated with replicative stress. To this end, we examined a widely-used gene expression signature (34) to identify patients characterized by increased chromosomal instability and DNA damage, and then applied this signature to our published MM gene expression profiling (GEP) data, which includes healthy as well as patient-derived plasma cells (1). Remarkably, a subset of approximately 20% MM patients demonstrated overexpression of the probe sets belonging to the chromosomal instability signature (Fig. 2A). In contrast, plasma cells derived from healthy individuals consistently showed the lowest level of expression of these probe sets. We next extended our analysis to a GEP set that includes 559 MM patients (35). K means clustering sharply divided these patients into two groups, including 96 patients (17%) with overexpression of the instability signature (K2; Fig. 2B). We then compared and contrasted this subset with the remaining K1 patients using GSEA. As in MM cell lines with DNA damage, GSEA in the Kegg repository revealed that DNA replication was the most significantly altered pathway in the K2 subgroup, compared to the remaining patients (NES=2.49, P<0.0001, FDR<0.0001) (Fig. 2C–D and Supplementary Table 2); the cell cycle pathway also significantly differed in these patient subgroups (NES=2.20, P<0.0001, FDR=0.003) (Fig. 2C). We similarly explored the Biocarta data set and identified the pathway centered on ATM to be among the most significantly dysregulated in the K2 patient subgroup (NES=1.84, P<0.006, FDR=0.05), again with significant differences in the cell cycle pathway as well (Fig. 2C, Supplementary Table 3). We next sought to identify any potential association with prognosis, and found that patients in the K2 group demonstrated a poor prognosis when compared with the other patients (p<0.0001) (Fig. 2E and Supplementary Table 4). Importantly, in a multivariate analysis, this instability signature was independent from other poor prognostic criteria, including the proliferation signature and the presence of MMSET/FGFR3 or MAF translocations (35) (Supplementary Table 5). These results therefore indicate that there is a subset of MM with genomic instability associated with replicative stress, ATM activation, and active proliferation, and that patient in this subgroup have a poor prognosis.
Figure 2. Definition of a MM patient subset with increased genomic instability, enhanced expression of DNA replication and cell cycle genes, and poor prognosis.
A, expression levels in plasma cells derived from healthy individuals and MM patients for the probe sets corresponding to the chromosomal instability signature described in (34). B, heat map showing the K-means clustering (clusters=2) of 559 MM patient GEP data using the probe sets corresponding to the chromosomal instability signature described in (34). C, heat map in the K1 and K2 groups showing the expression levels corresponding to probe sets included in the DNA replication (Top) and cell cycle (Bottom) KEGG pathways as annotated in MSigDB. D, GSEA ES enrichment profiles for 559 patients divided in K1 and K2 groups of KEGG DNA replication (Left panel) and BIOCARTA ATM (Right panel) pathways as annotated in MSigDB. E, Kaplan-Meyer survival curves of K1 and K2 patients (414 patients analyzed).
MYC triggers replicative stress and DNA damage in MM cells
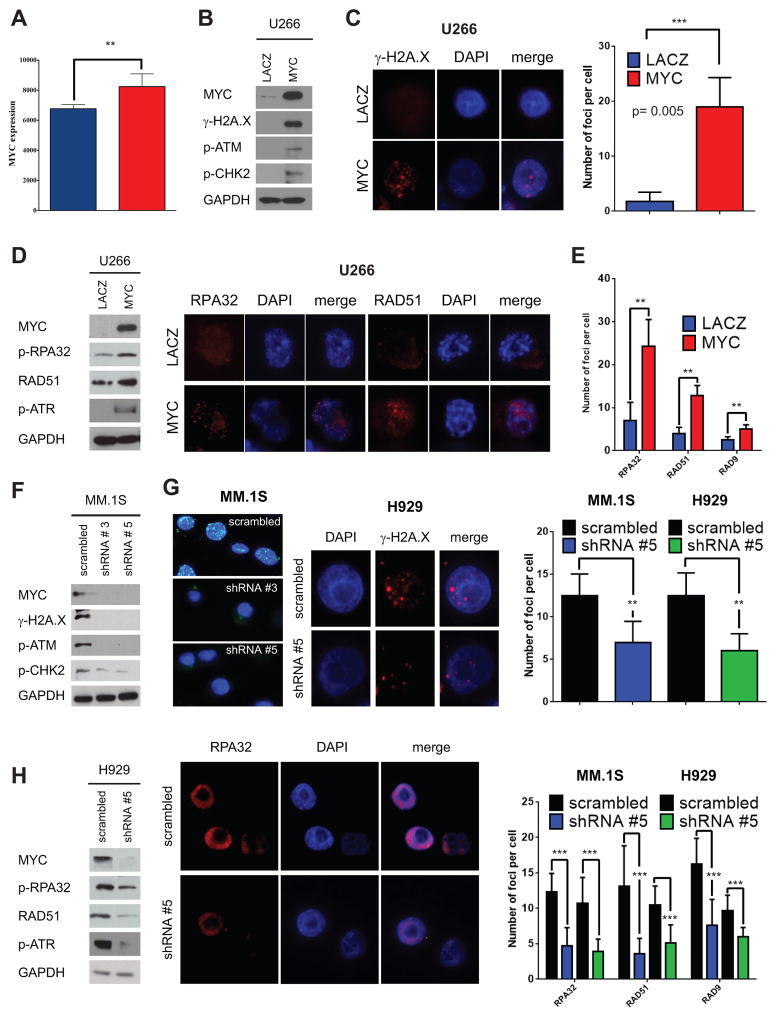
One of the mechanisms by which mutated and dysregulated oncogenes induce DNA damage is via induction of replicative stress (6, 8, 9, 13). Since the oncogene MYC can induce replicative stress (14, 15) and plays a prominent role in MM pathogenesis (31), we next explored its expression in the K2 patient subgroup overexpressing genes included in the chromosomal instability signature. MYC was significantly upregulated in the K2 subgroup (Fig. 3A), suggesting its potential role in replication stress and DNA damage in this MM subset. We therefore next determined whether MYC overexpression triggers replicative stress in MM cells. As a gain-of-function model, we choose the U266 MM cell line, which expresses L-MYC and does not have either genomic rearrangements affecting the MYC locus nor MYC overexpression (36). Despite a reported role for L-MYC in inducing DNA damage (14), we were not able to detect ongoing DNA damage in U266 cells, evidenced by the lack of γ-H2A.X foci or of markers of DNA damage response activation including p-ATM and p-ATR and their downstream effectors p-CHK2 and p-CHK1, respectively (4) (Fig. 3 and data not shown). The overexpression of MYC in U266 cells triggered an increase of γ-H2A.X, as assessed by western blotting (Fig. 3B). γ-H2A.X foci also appeared after MYC overexpression, detected by immunofluorescence (from 1.75 ± 1.7 to 19.0 ± 5.2, on average, P= 0.0052) (Fig. 3C). These changes were associated with activation of the DNA damage response, since both p-ATM and p-CHK2 were increased after MYC overexpression (Fig. 3B).
Figure 3. MYC is involved in DNA damage and replicative stress in MM.
A, MYC expression levels in K1 (blue) and K2 (red) patient groups. B, U266 MM cells were transiently transfected with MYC-EGFP or LACZ-EGFP. Lysates were obtained at 48 hours after transfection and assayed for western blot analysis using antibodies against c-MYC, γ-H2A.X, p-CHK2, and p-ATM. GAPDH is used as loading control. C, immunofluorescence staining for γ-H2A.X at 48 hours from transfection. Right panel: Number of γ-H2A.X foci in U266 MYC-EGFP cells in comparison with LACZ-EGFP cells at 48 hours. D, replicative stress markers by western blot analysis and immunofluorescence evaluated at 48 hours. Left panel: Western blot analysis for p-RPA32, RAD51, p-ATR, MYC, and GAPDH in U266 MYC-EGFP and U266 LACZ-EGFP transfected cells. Right panel: RPA32 and RAD51 immunofluorescence staining in U266 MYC-EGFP cells versus U266-LACZ-EGFP cells. E, number of RPA32, RAD51, and RAD9A foci in U266 MYC-EGFP cells in comparison with LACZ-EGFP cells. F, MM.1S cells were silenced for MYC using shRNAs #3 and #5 and a vector including a scrambled sequence. Western blot analysis was performed on lysates obtained at 48 hours after transfection using antibodies against c-MYC, γ-H2A.X, p-ATM, p-CHK2, and GAPDH. G, immunofluorescence to detect γ-H2A.X foci was performed in MM.1S and H929 48 hours after transfection with scrambled or shRNAs against c-MYC. Number of γ-H2A.X foci is shown on the right, from 2 representative experiments and 10 different cells. H, replicative stress markers in MM.1S and H929 silenced for c-MYC by western blot analysis (left panel) in H929 cells and by immunofluorescence with foci count (right panel).
We next explored whether the increased DNA damage triggered by MYC overexpression was associated with enhanced replicative stress. We first assayed for single-stranded DNA, typically associated with stalled DNA replication forks, both by IF using antibodies specific for RPA32 and RAD51, and by western blotting (Fig. 3D–E). RPA32, RAD51, and RAD9 all increased after MYC overexpression. Phosphorylation of ATR also increased, further confirming the notion that overexpression of MYC in MM cells induces DNA replicative stress that in turn leads to enhanced DNA damage.
To further demonstrate the link between MYC overexpression and replicative stress with DNA damage, loss-of-function studies were next undertaken. In this case, we choose two MM cell lines, MM.1S and H929, with robust MYC expression and γ-H2A.X staining. To reduce MYC levels, we chose two shRNA constructs targeting MYC, based on their ability to decrease MYC expression (Fig. 3F). Upon MYC knockdown a reduction of γH2A.X staining, assessed by western blot and immunofluorescence, was evident, indicating a concurrent reduction in DNA damage upon MYC downregulation (Fig. 3F–G). We further explored the effects of MYC down regulation on markers of DNA damage activation, and found that p-ATM and p-CHK2 levels were greatly reduced. To assess the effect of MYC downregulation on markers of replicative stress, we carried out both immunofluorescence as well as western blotting: a marked reduction in replicative stress markers including RPA32, RAD51, and p-ATR was observed in both MM1.S and H929 MM cells (Fig. 3H). Taken together, these results suggest that MYC overexpression in MM cells drives replicative stress, associated with the appearance of DNA damage and activation of DNA damage response.
ATR is required for survival of MYC-overexpressing cells
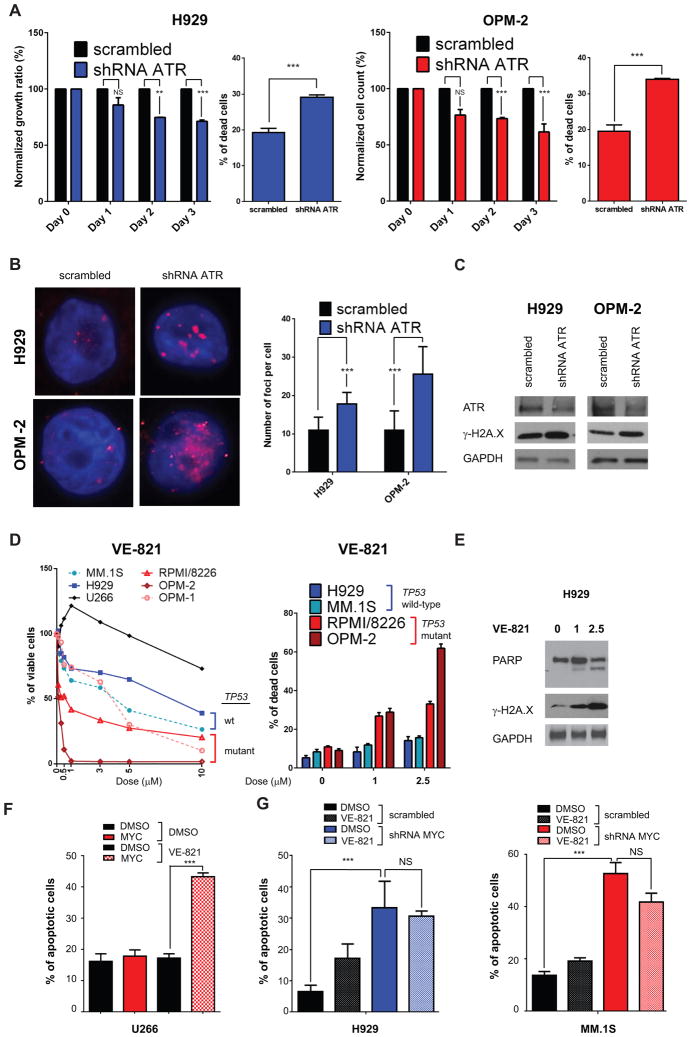
To mend stalled forks and prevent the apoptotic response triggered by replicative stress, epithelial cancer cells require ATR and its downstream target CHK1. Recent evidence suggests that ATR may exert a similar role in hematological cancers. Indeed ATR inactivation prevents MYC-induced lymphomas (18). Notably, our K2 MM patient subset demonstrated significant upregulation of genes belonging to the ATR pathway, suggesting that these tumors rely on activated ATR for survival (Supplementary Fig. S6A–B, Supplementary Table 3). We therefore next assessed whether ATR is required for survival of MM cells. To this end, we downregulated ATR in H929 and OPM-2 MM cells using a specific shRNA. ATR knockdown significantly reduced viable cell count and increased apoptosis in both cell lines, assessed with Annexin V-PI staining (Fig. 4A). ATR knockdown was also associated with enhanced DNA damage, evidenced by increased γ-H2A.X levels both by immunofluorescence (Fig. 4B) and western blotting (Fig. 4C). A potent and selective small molecule ATR inhibitor VE-821 has been recently described (37), which we next used to determine whether pharmacological inhibition of ATR could trigger apoptosis in MM cells. As anticipated, U266 cells were only modestly sensitive to the drug (Fig. 4D); however, other MM cells with ongoing DNA damage responded to VE-821 treatment, as evidenced by reduced viability and increased apoptosis (Fig. 4D). Consistent with previous results showing that cells without p53 are more sensitive to ATR inhibitors (18, 24), p53-mutant MM cell lines (OPM-1, OPM-2, and RPMI/8266) showed greater response to VE-821 than p53 wild-type MM cell lines (MM.1S and H929). Moreover, treatment with VE-821 induced PARP cleavage and increased γ-H2A.X (Fig. 4E). To determine the potential role of MYC-induced replicative stress in this phenotype, we re-expressed MYC in U266 cells. Remarkably, U266 cells became more sensitive to VE-821 after MYC re-expression (Fig. 4F and Supplementary Fig. S6C). In contrast, silencing of MYC in MM.1S and H929 MM cells blocks the effect of VE-821 treatment (Fig. 4G). These results suggest that inhibition of ATR can selectively target the subset of MM with increased replicative stress and ongoing DNA damage.
Figure 4. ATR modulation by shRNAs or using a specific inhibitor (VE-821) reduces MM cell growth.
A, H929 MM cells (left panels, blue) and OPM-2 MM cells (right panels, red) were transiently transfected with a shRNA against ATR or with a scrambled shRNA. Data derived from two independent experiments in triplicates are reported. For each cell line, cellular growth is shown on the left and apoptosis on the right. Cellular growth is measured by cell count with trypan blue exclusion at day 0–3, while apoptosis is evaluated by Annexin V-PE/7-AAD on GFP-gated cells at 48 hours. B, Left panel: Immunofluorescence staining for γ-H2A.X foci was performed in H929 and OPM-2 cells silenced with scrambled and ATR shRNAs at 48 hours. Right panel: Number of γ-H2A.X foci with average ± SD is reported. C, Lysates were obtained at 48 hours after transfection. Western blot analysis using antibodies against ATR, γ-H2A.X, and GAPDH (as loading control) was used. D, Left panel: MTT viability assay was used to evaluate growth inhibitory effects upon incubation for 72 hours with VE-821, with doses ranging from 0 to 10μM. Right panel: Apoptosis was measured with Annexin V-FITC/PI staining, upon incubation with DMSO, 1 μM, and 2.5 μM for 48 hours. Data are obtained from two independent experiments, performed in triplicate. E, western blot analysis for total PARP, γ-H2A.X, and GAPDH after treatment with DMSO, 1 μM, and 2.5 μM for 48 hours. F, U266 cells were transfected with MYC-EGFP and LACZ-EGFP and then treated with 5μM VE-821 for 48 h, starting from Day 0 of transfection. Annexin V-PE/7-AAD staining was used to measure the percentage of dead cells. G, H929 and MM.1S myeloma cells were silenced with shRNAs targeting c-MYC and scrambled and then treated with 2.5 μM VE-821 for 48 hours. Apoptosis was evaluated as in (F).
Genes associated with reactive oxygen species are dysregulated in the K2 MM patient subgroup
In a few cellular settings, oncogenes such as MYC have been shown to enhance DNA damage through enhanced production of ROS (27, 30). Although oncogene induced-DNA damage in epithelial cancer cells has been primarily attributed to increased replicative stress (29), a recent report in glioblastoma multiforme showed that MYC and RAS can elicit DNA damage through both enhanced replicative stress and increased ROS production, depending on the cellular context (38). High levels of ROS in the setting of oxidative stress reflect an imbalance between production and termination of reactive species.
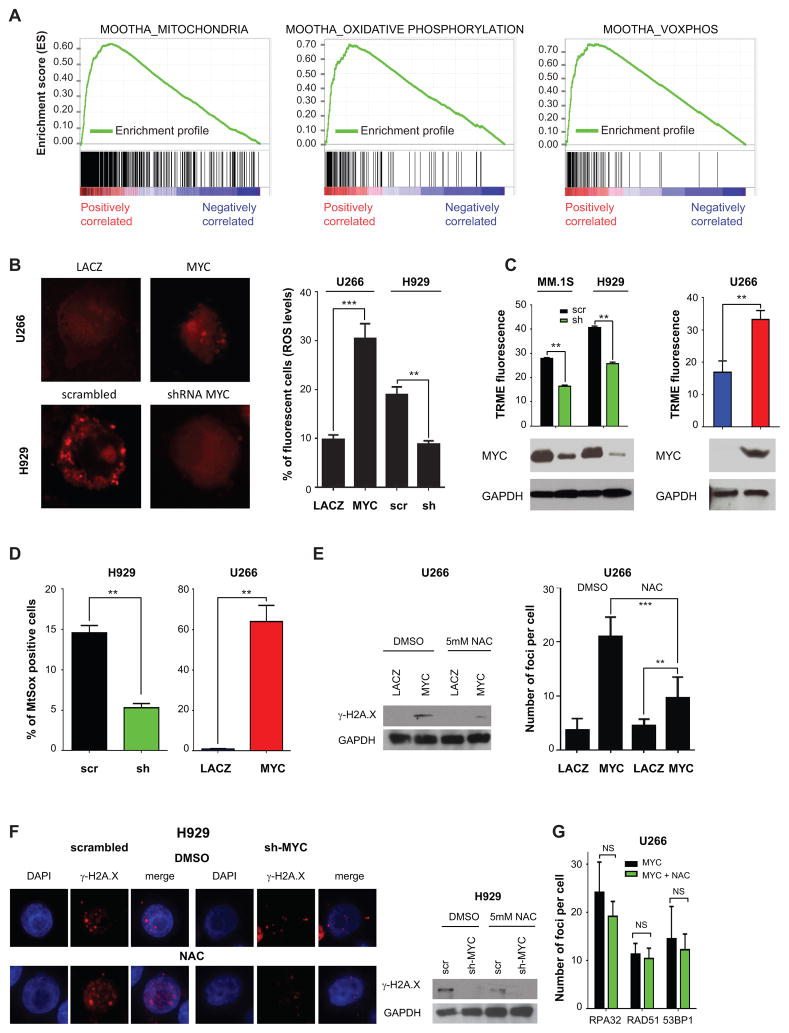
To determine whether pathways involved in ROS metabolism are dysregulated in MM patients, especially in the subset with enhanced DNA damage, we first determined whether the K2 patient group demonstrated dysregulated expression of genes implicated in oxidative stress. To this end, we compiled a list of pathways implicated in ROS and NOS metabolism collected from the Molecular Signature Database MSigDB (32) (Supplementary Table 6). Remarkably, in the K2 group there was a highly significant enrichment for pathways related to ROS metabolism, including Mootha_Mitochondria, Kegg_Oxidative_Phosphorylation, and Mootha_Voxphos (NES=2.23, 2.04 and 1.90, P<0.0001, P=0.002 and 0.006, and FDR<0.0001, FDR=0.002 and 0.014, respectively) (Fig. 5A and data not shown). In fact, most genes within these pathways were upregulated in the K2 compared with the K1 MM subgroups (Supplementary Fig. S7A–C). Notably, at the gene expression level, NOS signaling was not enriched in this patient subset, and inducible NOS was barely detectable in the tested MM cell lines (data not shown). Taken together, these results indicate that the K2 MM subgroup demonstrates replicative stress, as well as activation of pathways implicated in generation and metabolism of ROS.
Figure 5. MYC induces oxidative stress mediating DNA damage in MM cells.
A, GSEA ES enrichment profiles for 559 patients divided in K1 and K2 groups of MOOTHA_MITOCHONDRIA (Left panel), MOOTHA_OXIDATIVE PHOSPHORYLATION (Middle panel), and MOOTHA VOXPHOS (Right panel) pathways as annotated in MSigDB. B, U266 cells were transfected with MYC-EGFP and LACZ-EGFP; superoxide levels were detected by immunofluorescence (Left panel), while ROS levels were evaluated by flow cytometry at 48 hours. C, TRME staining to detect active mitochondria upon MYC silencing in MM.1S and H929 or overexpression of MYC in U266 cells. Cells were gated to exclude Annexin V-FITC positive cells. Western blot analysis using antibodies against MYC and GAPDH levels is shown and is representative of one replicate experiment. The same cells were also used for experiments shown in panel B and D. D, MitoSOX staining upon MYC silencing in H929 cells or MYC overexpression in U266 cells. E, MYC-EGFP and LACZ-EGFP U266 transfected cells were treated with DMSO or 5mM N-Acetyl L-Cysteine (NAC) for 48 hours. Left panel: Western blot analysis for γ-H2A.X and GAPDH on U266 MYC-EGFP and LACZ-EGFP at 48 hours after transfection. Right panel: Immunofluorescence was performed. Number of γ-H2A.X foci was counted in up to 10 fields of cells per condition. F, H929 cells were silenced for MYC using a specific shRNA (#5) and scrambled vectors and incubated with DMSO or 5mM N-Acetyl Cysteine (NAC) for 48 hours. Immunofluorescence for γ-H2A.X foci (left panel) and western blot analysis using antibodies against γ-H2A.X and GAPDH (right panel). G, replicative stress markers in U266 overexpressing MYC after treatment with DMSO or NAC for 48 hours. The number of RPA32, RAD51, and RAD9A foci is shown.
MYC induces oxidative stress in MM cells
We next sought to test whether the oncogene MYC can elicit ROS in MM cells. To this end, we first tested the levels of both free-radicals ROS (superoxide) and non radical ROS (hydrogen peroxide). Overexpression of MYC in U266 cells increased both superoxide and hydrogen peroxide levels (Fig. 5B), suggesting that MYC regulates ROS levels in MM cells. Conversely, downregulation of MYC in MM.1S and H929 MM cell lines reduced these ROS levels (Fig. 5B and data not shown). Notably, MYC overexpression did not increase levels of nitrogen oxygen (NO) species, unlike the NO inducer L-arginine, which served as a positive control (Supplementary Fig. S8A).
Cellular ROS originates from peroxisomes, endoplasmic reticulum, and mitochondria, with mitochondria representing the predominant source. We therefore next sought to determine whether MYC triggered ROS via mitochondria. Indeed, MYC downregulation increased mitochondrial depolarization in both MM.1S and H929 MM cells (Fig. 5C). In contrast, MYC overexpression in U266 MM cells significantly decreased their depolarization, suggesting a prominent role for mitochondria as a source of the increased ROS induced by MYC (Fig. 5C). Indeed, the production of superoxide by mitochondria was significantly affected by MYC, as demonstrated by the reduction of mitochondria superoxide levels after MYC downregulation in MM.1S and H929 cells; and conversely, by the increase in mitochondrial superoxide levels after MYC overexpression in U266 cell line (Fig. 5D). These results indicate that MYC regulates ROS levels by modulating the activity of mitochondria.
MYC-induced oxidative stress triggers DNA damage in MM cells
To determine whether ROS elicited by MYC overexpression was causally linked to DNA damage, cells overexpressing MYC were next treated with an antioxidant reagent N-Acetyl-L-cysteine (NAC), which scavenges ROS by replenishing glutathione stores. NAC reduced DNA double strand breaks, assessed by γ-H2A.X staining (Fig. 5E and Supplementary Figure S8B). Interestingly, NAC did not completely block the formation of DNA double strand breaks in the presence of MYC, and a significant increase in γ-H2A.X foci remained after MYC overexpression even in the presence of NAC. We next explored the effects of NAC treatment on MM cell lines with replicative stress. NAC markedly reduced γ-H2A.X in MM cells (Supplementary Fig. S8C), and concomitant downregulation of MYC further decreased its levels (Fig. 5F). Notably, NAC treatment did not affect replicative stress, assessed by the number of RPA32, RAD51 and 53BP1 foci (Fig. 5G), and minimally reduced the efficacy of VE-821 treatment (Supplementary Fig. S8D). Additionally, neither VE-821 nor an ATM inhibitor Ku55933 increased reactive oxygen species (Supplementary Fig. S8E). These data indicate that DNA damage in MM cells induced by MYC through ROS is not mediated by and is independent of replicative stress (Fig. 5G). Since RNS can also be important in inducing DNA DSBs, we treated U266 and H929 cells with a RNS inducer L-arginine and a NOS inhibitor, L-NAME (L-NG-Nitroarginine Methyl Ester), respectively. Although we observed modulation of γ-H2A.X levels (Supplementary Fig. S8F), the effects of RNS in inducing DNA damage are likely independent of MYC, since MYC overexpression was not modulating RNS levels (Supplementary Fig. S8A). Altogether, these results demonstrate that MYC elicits DNA damage in MM cells both by inducing DNA replicative stress and by increasing ROS levels.
MYC modulates antioxidant pathways
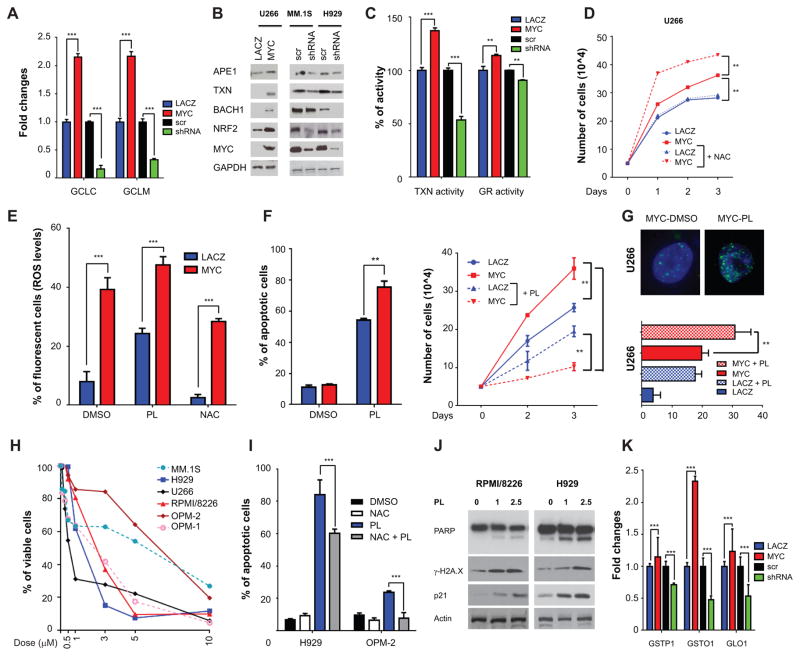
Elevated oxidative stress is often present in cancer, and strong evidence links ROS to carcinogenesis. Conversely, other data indicates that ROS increase in cancer cells as a byproduct of the tumorigenic process, and that excessive ROS may be detrimental to cancer growth (39). As such, cancer cells tightly regulate ROS levels via various antioxidant pathways (40). Glutathione (GSH) and thioredoxin (TXN) are among the main mechanisms involved in ROS detoxification. We therefore next tested whether MYC overexpression was able to modulate the levels of the enzymes involved in GSH production including glutamate-cysteine ligase, catalytic subunit (GCLC), and glutamate–cysteine ligase complex modifier subunit (GCLM). MYC overexpression increased mRNA levels of both enzymes; conversely, MYC downregulation reduced their mRNA levels (Fig. 6A). MYC similarly modulated the levels of TXN, APE1 (important in hydrogen peroxide-induced oxidative stress in the mitochondria), and BACH1, which blocks oxidative stress-induced senescence either directly or through nuclear factor (erythroid-derived 2)-like 2 (NRF2) (Fig. 6B). We therefore next tested GSH and TXN activity after MYC modulation. MYC overexpression increased, while MYC downregulation decreased GSH and TXN activities, indicating that MYC regulates ROS levels through the modulation of these proteins (Fig. 6C). To test the hypothesis that MM cells tightly control ROS levels and that unrestrained ROS may be detrimental to MM cells, we overexpressed MYC in U266 cells, in the presence of NAC. Remarkably, the addition of NAC increased MYC-induced proliferation (Fig. 6D), which was not rescued by L-NAME (Supplementary Fig. S9A). These results suggest that MYC both increases and at the same time stringently controls ROS levels, to prevent ROS-mediated apoptosis in MM cells.
Figure 6. PL treatment causes MM cell death through ROS.
A, mRNA level evaluation by qPCR of genes important in GSH metabolism (GCLC and GCLM) after MYC overexpression in U266 cells or downregulation in H929 cells at 48 hours. B, Western blot analysis using antibodies against APE1, TXN, BACH1, NRF2, MYC, and GAPDH after MYC overexpression in U266 cells or MYC downregulation in MM.1S and H929 cells. Lysates were obtained at 48 hours. C, Thioredoxin (TXN) and glutathione reductase (GR) activity in the same cell settings of panel B. D, Cellular growth by cell count with trypan blue exclusion in U266 cells transfected with EGFP-MYC or EGFP-LACZ after treatment with DMSO or 5mM NAC. NAC was added at Day 0 of transfection. E, ROS levels were evaluated in U266 cells transfected with EGFP-MYC and EGFP-LACZ upon incubation with DMSO, 5mM NAC, and 1μM piperlongumine (PL) for 48 hours. F, U266 cells were transfected with EGFP-MYC and EGFP-LACZ and incubated with DMSO or 1μM PL. Apoptosis at 48 hours by Annexin V-PE/7AAD staining (left panel) and cell growth by cell count (right panel) are shown. G, Evaluation of number and size of γ-H2A.X foci in U266 cells overexpressing MYC in the presence or absence of 1 μM PL by immunofluorescence (top panel). In the lower panel are compared the number of foci in the same cellular settings as F. H, MTT viability assay was used to evaluate growth inhibitory effects upon incubation for 72 hours with piperlongumine (PL), with doses ranging from 0 to 10μM. I, H929 and OPM-2 MM cells were treated with 1μM piperlongumine (PL) and 5mM NAC, alone or in combination. Apoptosis with Annexin V-FITC/PI staining was measured at 72 hours. J, H929 and OPM-2 myeloma cells were treated with 1μM piperlongumine and lysates were obtained after 48 hour treatment. Western blot analysis for PARP, γ-H2A.X, p21, and GAPDH was performed. K, mRNA level evaluation by qPCR of PL-interacting protein genes (GSTP1, GSTO1, and GLO1) upon MYC overexpression in U266 cells or downregulation in H929 cells.
Enhanced oxidative stress promotes MM cell toxicity
Prompted by these data, we reasoned that increasing ROS levels in MM cells could further promote DNA damage and induce apoptosis. Indeed novel compounds have recently been described which exploit induction of ROS for treatment of cancer (41). Specifically, a recent cell-based small molecule screening identified Piperlongumine (PL) as a compound that increases ROS levels and thereby selectively inducing apoptosis in cancer cells, while sparing normal cells (42). We therefore next tested whether further increasing ROS levels in MM cells could enhance DNA damage and induce cell death. Indeed treatment of U266 cells for a short period (30 minutes) with PL triggered a surge in ROS levels (Fig. 6E), which was enhanced by MYC overexpression and abrogated by treatment with NAC. PL also dramatically increased apoptosis and decreased proliferation, both of which were surprisingly enhanced by MYC overexpression (Fig. 6F). Indeed PL induced DNA double strand breaks, which were further increased by MYC overexpression (Fig. 6G). As expected, PL induced a robust increase of both ROS and superoxide levels in MM cell lines (Supplementary Fig. S9B). Most MM cell lines were sensitive to PL; OPM2 cells were least sensitive, while U266 were the most sensitive to PL (Fig. 6H). To confirm that the activity of PL was mediated by an increase in ROS, H929 and OPM2 cells were treated with PL, with or without NAC. Indeed, concomitant treatment of MM cells with PL and NAC significantly reduced the apoptotic effects of PL (Fig. 6I and Supplementary Fig. S9C), whereas L-NAME did not impact the activity of PL (Supplementary Fig. S9D). Finally, we assessed the overall effects of PL on apoptosis and DNA damage. PL significantly induced PARP cleavage, as well as increased γ-H2A.X and p21 levels, in both H929 and OPM-2 MM cells (Fig. 6J). We then tested the effect of arsenic trioxide (ATO), a well-known inducer of ROS. Indeed ATO increased ROS/superoxide levels and γ-H2A.X (Supplementary Fig. S10A–B). Importantly, in our study, PL was more potent in terms of growth inhibition than ATO, despite inducing lower levels of ROS and SOD (Supplementary Fig. S10C). PL acts predominantly by reducing GSH levels, without affecting glutathione reductase activity (Supplementary Fig. S11A) (42). On the other hand, PL directly interacts with several proteins including glutathione S-transferase pi 1 (GSTP1), glutathione S-transferase omega 1 (GSTO1), and glyoxalase I (GLO1), which are important in catalyzing the conjugation of reduced glutathione to electrophilic substances (43). Interestingly, we found that these proteins are significantly upregulated in MM in comparison to plasma cells from healthy individuals (Supplementary Fig. S11B). We also evaluated whether their expression was also modulated by MYC, and found that MYC overexpression increased, whereas MYC silencing decreased, the levels of GSTP1, GSTO1, and GLO1 (Fig. 6K). Of note, PL did not exhibit any cytotoxic effect on PBMCs (Supplementary Fig. S12), suggesting a favorable therapeutic index. Altogether, these data suggest that increasing ROS levels in MM induces apoptosis and may represent a novel therapeutic option.
ATR inhibition combined with PL treatment induces synergistic MM cell death
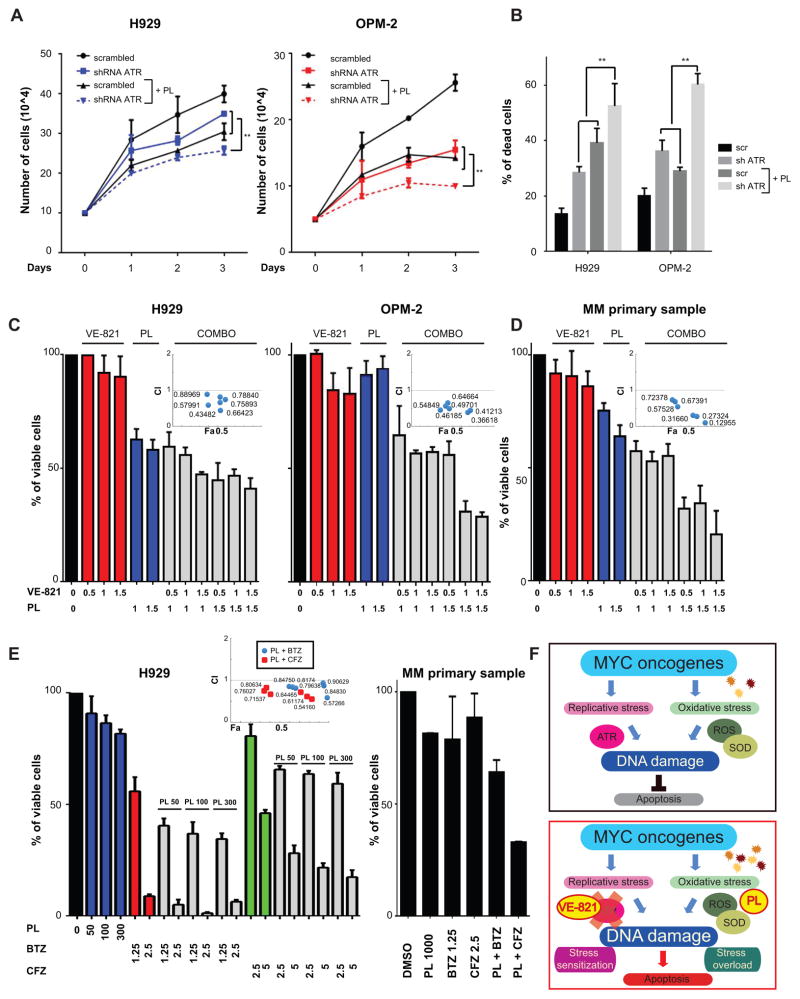
To assess whether combining therapies that elicit oxidative stress, like PL, with ATR inhibition may have higher cytotoxic effects on MM cells, we next examined the effect of combined genetic knockdown of ATR with PL treatment. In H929 and OPM-2 cells transfected with ATR shRNAs, we observed decreased proliferation (Fig. 7A) and enhanced apoptosis (Fig. 7B), when we combined ATR inactivation and PL treatment. Similar results were obtained with VE-821 (1–1.5μM) in combination with PL (0.5–1.5μM): the combination had synergistic effects, with a combination index (C.I.) below 1 for both H929 and OPM-2 cells (Fig. 7C). Similar synergistic effects were observed in MM cell lines MM.1S and RPMI/8226 cell lines, but no synergism was detectable in U266 cells (Supplementary Fig. S13A–B). MM cells isolated from patients also showed a similar pattern, with synergistic/additive effects of combination treatment (Fig. 7D). Interestingly, combination treatment of ATO with VE-821 was less effective than combination of PL with VE-821 (Supplementary Fig. S14). Indeed, a recent study reported that ROS are not essential for the induction MM cell death by ATO (44). Interestingly, the proteasome cascade was one of the pathways most prominently associated with the K2 subgroup, and bortezomib can induce a “BRCAness” phenotype by abrogating ubiquitination of H2A.X, thereby reducing recruitment of repair proteins such as RAD51 and BRCA1 and acting as stress sensitizer (45). We therefore next assessed whether the combination of PL and VE-821 demonstrated synergistic effects with proteasome inhibitors. To this end, we have tested both MM cell lines and patient MM cells with bortezomib and carfilzomib. Indeed, we detected a robust synergy between proteasome inhibitors and PL (Fig. 7E and Supplementary Fig. S15A–B). On western blot analysis, we observed that combination treatment with bortezomib and PL induced higher levels of γ-H2A.X than either single agent alone (Supplementary Fig. 15C). Similar results were obtained combining proteasome and ATR inhibitors (Supplementary Fig. 16). Taken together, these results indicate that combined inhibition of ATR with pharmacological induction of ROS may trigger synergistic cytotoxicity and apoptosis in MM cells with ongoing DNA damage (Fig. 7F).
Figure 7. ROS induction and ATR inhibition synergize in inducing MM cell death.
A, H929 MM cells (left panel) and OPM-2 MM cells (right panel) were transiently transfected with ATR- or scrambled-shRNAs and incubated with DMSO or 1μM piperlongumine starting from Day 0 of transfection. Cell growth by cell counting with trypan blue exclusion was performed in triplicate. B, Apoptosis by Annexin V-PI staining was evaluated in the same conditions as A at 48 hours from transfection. Summary of two independent experiments is shown. C, combination studies by MTT viability assay using 0.5–1.5 μM VE-821 and 1–1.5 μM PL were performed in H929 and OPM-2 cells after 48 hour treatment. Combination Index (CI) plot with CI values are shown as insets. D, combination studies by MTT viability assay in MM patient cells as in C. E, combination studies by MTT viability assay using 50–300 nM PL, 1.25–2.5 nM bortezomib (BTZ), and 2.5–5 nM carfilzomib (CFZ) in H929 and one representative patient. Combination Index (CI) plot with CI values are shown as insets. F, schematic model. Top panel: Oncogene activation (as for example MYC) increases replicative stress and ROS, thus triggering DNA damage in MM cells that is maintained below a critical threshold by ATR-mediated resolution of DNA replicative stress and increase of enzymes reducing ROS levels. Lower panel: Combined inhibition of ATR (stress sensitization) and increase in ROS (stress overload) elicits an increase in unresolved DNA damage that leads to apoptosis.
Discussion
Ongoing DNA damage is a hallmark of epithelial cancers that leads to genomic instability and ultimately to more aggressive tumors, often resistant to current therapies. Dysregulated oncogene expression triggers replicative stress in these cancers, leading to DSBs. We have recently demonstrated constitutive ongoing DNA damage in hematological malignancies (4). In this study, we show that replicative stress causes DNA damage in blood cancers, specifically in MM. Indeed, we identified a subset of MM characterized by chromosomal instability, replicative stress, and poor prognosis. These patients had increased expression of the oncogene MYC, suggesting a prominent role for MYC in conferring this phenotype. We show that MYC is able to trigger replicative stress in MM cells. Besides replicative stress, oncogenes, including MYC, can induce DNA damage by increasing production of ROS (27, 46). We here provide evidence that MYC also triggers DNA damage by increasing ROS levels in MM cells. Furthermore, a recently described ROS inducer piperlongumine, enhances DNA damage via increased ROS. These data indicate that both replicative stress and ROS induction trigger DNA damage in MM cells, providing the basis for novel combination treatment strategies.
A model has been recently proposed in which a step-wise increase in MYC levels is associated with disease progression. For example, a relatively modest, yet critical, increase in MYC levels is implicated in driving the evolution from MGUS toward MM (47). As the disease progresses, genomic rearrangements affecting the MYC locus occur, resulting in much higher levels of MYC expression (31). Indeed mouse models support the notion that distinct expression thresholds may direct the role of MYC in oncogenesis (48, 49). Although our data demonstrate that MYC contributes to the replicative stress and enhanced proliferation in the K2 MM subgroup, we would argue that oncogenes other than MYC might also play a role. Further studies are needed to comprehensively evaluate the genetic or epigenetic anatomy of this patient subset.
Oncogenesis causes cellular stresses in cancer cells which are not evident in normal cells (50). As a consequence, tumor cells become dependent on stress response pathways for their survival. These stresses include DNA damage/replication, proteotoxic, mitotic, metabolic, and oxidative stresses. This so-called non-oncogene addiction represents a vulnerability of cancer cells that can be targeted therapeutically. For example, PARP inhibitors are under clinical evaluation in breast and ovarian cancer patients with hereditary mutations of BRCA1 or BRCA2 (51) to exploit one of these liabilities. This approach offers several advantages over the current model of targeting proteins arising from mutated genes. First, the development of compounds against members of a pathway, rather than against single mutated proteins, is more feasible from a medicinal chemistry perspective. Indeed recent large sequencing efforts have revealed a panoply of mutations in various, unrelated genes. As a consequence, the development of compounds targeting a large series of mutated proteins remains a formidable challenge. Second, drug resistance develops more promptly against compounds targeting single mutated amino acids, as is often the case in conventional targeted therapies, than against compounds targeting an entire pathway. Third, recent reports in MM (52, 53) have demonstrated exceedingly high levels of genetic and clonal heterogeneity. Therefore targeting one clone endowed with a specific set of mutations may not be effective, since additional clones may quickly overcome the targeted one. Hence it may be more effective to target a pathway to which a cancer in its entirety has become addicted, irrespective of the mutations present in each single clone. In this regard, MM represents a particularly apt example. MM plasma cells are characterized by increased protein synthesis, well in excess of the levels present in healthy cells, irrespective of their genetic makeup. MM cells cope with this stress by increasing the activity of the proteasome to facilitate the degradation of unfolded proteins. Importantly, the introduction of proteasome inhibitors such as bortezomib for the treatment of MM is a landmark achievement, targeting a vulnerability which is based on non-oncogene addiction. Additional such approaches, such as the one described here, may similarly provide an important advance in the treatment of this disease.
To therapeutically target these hallmarks of stress, one could envision two approaches, stress overload or stress sensitization (50). We have exploited both paths, with the goal of maximizing the effects on tumor cells (Fig. 7F). Blocking ATR (stress sensitization) prevents the repair of stalled forks during proliferation induced by unconstrained oncogenic stimulus, thereby leading to DNA damage and cell death. In parallel, PL treatment increases ROS levels and exacerbates the oncogene-induced DNA damage (stress overload). Although drugs targeting MYC or MYC-interactors have recently been proposed (54–56), in our approach interfering with MYC activity would be detrimental, since it would reduce the stress overload on MM cells.
Based on gene expression profiling, MM patients have been subdivided into 7 subgroups, each endowed with specific features, plus an additional subgroup characterized by a “myeloid” signature (35). Among these 8 subtypes, one subgroup of patients, the proliferation (PR) subgroup, specifically overexpresses cell cycle- and proliferation-related genes, and has a significantly higher gene expression-defined proliferation index (PI) than the other groups. Importantly, patients in the PR group have the worst prognosis. Remarkably, however, a multivariate analysis demonstrated that the chromosomal instability classification that we propose herein was nevertheless able to identify patients with a poor prognosis who are not captured by other genomic features, including the proliferation group, as outlined in Supplemental Table 5. Specifically, 33 patients belonged to both the K2 and high proliferation groups. However, 34 patients in the low-proliferation group and were also included within the K2 group, and these patients did show a poor prognosis, despite being included in the low-proliferation group (p<0.013). Importantly, a likelihood ratio test comparing the multivariate model with and without the K1/K2 clusters was significant (p=0.023). Therefore, the K1/K2 classification provides additional information and is independent of other established prognostic factors derived from gene expression profiling, such as proliferation and the presence of MMSET/FGFR3 translocations.
We demonstrate here the rationale for a novel therapeutic approach specifically targeting this patient group with aggressive disease, a poor prognosis, and lack of effective therapeutic options. In the context of their endogenous rapid tumor cell proliferation and associated DNA damage, ATR inhibition coupled with the induction of further oxidative stress could both inhibit proliferation and trigger apoptosis. Our study provides the framework for derived combination clinical trials to address and unmet medical need and improve outcome in this patient subgroup.
Methods
Reagents
An ATR inhibitor VE-821, bortezomib, and carfilzomib were purchased from Selleck Chemicals LLC (Houston, TX, United States); piperlongumine, N-acetyl-L-cysteine from Sigma–Aldrich (St. Louis, MO, Unites States); and L-NG-Nitroarginine Methyl Ester (L-NAME) from Santa Cruz (Dallas, TX, United States).
Cell lines and culture
The MM cell lines MM1.S, U266, RPMI-8226 and H929 MM cells were purchased from American Type Culture Collection (ATCC, Manassas, VA); plasma cell leukemia (PCL) cells OPM-1 and OPM-2 were provided by Dr. Edward Thompson (University of Texas Medical Branch, Galveston, TX). Cell lines have been tested and authenticated by STR DNA fingerprinting analysis (Molecular Diagnostic Laboratory, DFCI), and used within three months after thawing. All MM cell lines were cultured in RPMI–1640 media containing 10% fetal bovine serum (FBS, GIBCO, Life technologies, Carlsbad, CA, United States), 2 μM L-1 glutamine, 100 U mL-1 penicillin, 100 μg mL-1 streptomycin (GIBCO, GIBCO, Life technologies, Carlsbad, CA, United States).
Primary cells
Blood samples collected from healthy volunteers were processed by Ficoll–Paque (GE Healthcare, Boston, MA, United States) gradient to obtain peripheral blood mononuclear cells (PBMCs). MM cells from individuals affected by MM were obtained from bone marrow samples after informed consent was obtained in accordance with the Declaration of Helsinki and approval by the Institutional Review Board of the Dana–Farber Cancer Institute. Mononuclear cells were separated using Ficoll–Paque density sedimentation, and plasma cells were purified (>95% CD138+) by positive selection with anti–CD138 magnetic activated cell separation micro beads (Miltenyi Biotec, Cambridge, MA, United States).
Western blotting, RNA extraction and reverse transcription polymerase chain reaction, immunofluorescence staining, transient transfection of MM cell lines, foci number quantification, viability and cellular growth assays, apoptotic assays and ROS assays
Detailed protocols are included in the Supplementary Methods
Gene expression analysis
The Affymetrix H133A gene expression dataset for MM cell lines (57) was downloaded from the Multiple Myeloma Research Consortium Genomics Portal at (58). The cell lines used for the analysis were KMS-34, U266, and Karpas-620 (DNA damage negative (4)), and INA-6, JJN-3, KMS-11, KMS-12PE, KMS-18, NCI-H929, OCI-MY5, RPMI-8226, and UTMC-2 (DNA damage positive cell lines (4)). The Affymetrix U133 Plus 2.0 GEPs were derived from (1) (GEO Accession number GSE4452) and from (35) (GEO GSE2658). Gene set enrichment analysis (GSEA) was performed as previously described (2) (GSEA v2.0 at (59)) using gene set as permutation type and 1,000 permutations and signal to noise as metric for ranking genes. Both absolute and real data pre-processing and K-means clustering was performed with GenePattern (60).
Statistical analysis
Statistical significance was determined by Student’s t test on average values ± SD. Survival analysis was assessed with the “survival” package in R. For the survival analysis, a subset of 414 patients was analyzed (patients with the myeloid signature were excluded), in line with (34, 35).
Supplementary Material
Statement of significance.
MM remains an incurable disease. We have identified a subset of MM patients with poor prognosis, whose tumors present chromosomal instability, replicative and oxidative stress, and DNA damage. We define a synthetic lethal approach enhancing oxidative stress while targeting replicative stress response, inducing tumor cell apoptosis in this patient subset.
Acknowledgments
We thank W. Hahn (Dana-Farber Cancer Institute) for pLKO.1 shRNA lentiviral vectors, the Dana-Farber Cancer Institute Flow cytometry facility and members of the Anderson and Tonon lab for sharing reagents and critical reading of the manuscript. This work was supported by the Associazione Italiana per la Ricerca sul Cancro (AIRC Investigator Grants and Special Program Molecular Clinical Oncology, 5 per mille no. 9965 to G.T.) (G.T.). K.C.A. is an American Cancer Society Clinical Research Professor and is supported by NIH grants NIH SPORE P50 100707, PO-1 78378 and RO-1 50947.
Footnotes
The Authors declare that they have no conflict of interest to disclose.
References
- 1.Carrasco DR, Tonon G, Huang Y, Zhang Y, Sinha R, Feng B, et al. High-resolution genomic profiles define distinct clinico-pathogenetic subgroups of multiple myeloma patients. Cancer cell. 2006;9(4):313–25. doi: 10.1016/j.ccr.2006.03.019. [DOI] [PubMed] [Google Scholar]
- 2.Chapman MA, Lawrence MS, Keats JJ, Cibulskis K, Sougnez C, Schinzel AC, et al. Initial genome sequencing and analysis of multiple myeloma. Nature. 2011;471(7339):467–72. doi: 10.1038/nature09837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Egan JB, Shi CX, Tembe W, Christoforides A, Kurdoglu A, Sinari S, et al. Whole-genome sequencing of multiple myeloma from diagnosis to plasma cell leukemia reveals genomic initiating events, evolution, and clonal tides. Blood. 2012;120(5):1060–6. doi: 10.1182/blood-2012-01-405977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cottini F, Hideshima T, Xu C, Sattler M, Dori M, Agnelli L, et al. Rescue of Hippo coactivator YAP1 triggers DNA damage-induced apoptosis in hematological cancers. Nature medicine. 2014;20(6):599–606. doi: 10.1038/nm.3562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Walters DK, Wu X, Tschumper RC, Arendt BK, Huddleston PM, Henderson KJ, et al. Evidence for ongoing DNA damage in multiple myeloma cells as revealed by constitutive phosphorylation of H2AX. Leukemia. 2011;25(8):1344–53. doi: 10.1038/leu.2011.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartkova J, Rezaei N, Liontos M, Karakaidos P, Kletsas D, Issaeva N, et al. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature. 2006;444(7119):633–7. doi: 10.1038/nature05268. [DOI] [PubMed] [Google Scholar]
- 7.Di Micco R, Fumagalli M, Cicalese A, Piccinin S, Gasparini P, Luise C, et al. Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature. 2006;444(7119):638–42. doi: 10.1038/nature05327. [DOI] [PubMed] [Google Scholar]
- 8.Gorgoulis VG, Vassiliou LV, Karakaidos P, Zacharatos P, Kotsinas A, Liloglou T, et al. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature. 2005;434(7035):907–13. doi: 10.1038/nature03485. [DOI] [PubMed] [Google Scholar]
- 9.Halazonetis TD, Gorgoulis VG, Bartek J. An oncogene-induced DNA damage model for cancer development. Science. 2008;319(5868):1352–5. doi: 10.1126/science.1140735. [DOI] [PubMed] [Google Scholar]
- 10.Negrini S, Gorgoulis VG, Halazonetis TD. Genomic instability--an evolving hallmark of cancer. Nature reviews Molecular cell biology. 2010;11(3):220–8. doi: 10.1038/nrm2858. [DOI] [PubMed] [Google Scholar]
- 11.Denko NC, Giaccia AJ, Stringer JR, Stambrook PJ. The human Ha-ras oncogene induces genomic instability in murine fibroblasts within one cell cycle. Proceedings of the National Academy of Sciences of the United States of America. 1994;91(11):5124–8. doi: 10.1073/pnas.91.11.5124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Felsher DW, Bishop JM. Transient excess of MYC activity can elicit genomic instability and tumorigenesis. Proceedings of the National Academy of Sciences of the United States of America. 1999;96(7):3940–4. doi: 10.1073/pnas.96.7.3940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ekholm-Reed S, Mendez J, Tedesco D, Zetterberg A, Stillman B, Reed SI. Deregulation of cyclin E in human cells interferes with prereplication complex assembly. The Journal of cell biology. 2004;165(6):789–800. doi: 10.1083/jcb.200404092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dominguez-Sola D, Ying CY, Grandori C, Ruggiero L, Chen B, Li M, et al. Non-transcriptional control of DNA replication by c-Myc. Nature. 2007;448(7152):445–51. doi: 10.1038/nature05953. [DOI] [PubMed] [Google Scholar]
- 15.Srinivasan SV, Dominguez-Sola D, Wang LC, Hyrien O, Gautier J. Cdc45 is a critical effector of myc-dependent DNA replication stress. Cell reports. 2013;3(5):1629–39. doi: 10.1016/j.celrep.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blow JJ, Gillespie PJ. Replication licensing and cancer--a fatal entanglement? Nature reviews Cancer. 2008;8(10):799–806. doi: 10.1038/nrc2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cimprich KA, Cortez D. ATR: an essential regulator of genome integrity. Nature reviews Molecular cell biology. 2008;9(8):616–27. doi: 10.1038/nrm2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murga M, Campaner S, Lopez-Contreras AJ, Toledo LI, Soria R, Montana MF, et al. Exploiting oncogene-induced replicative stress for the selective killing of Myc-driven tumors. Nature structural & molecular biology. 2011;18(12):1331–5. doi: 10.1038/nsmb.2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shreeram S, Hee WK, Demidov ON, Kek C, Yamaguchi H, Fornace AJ, Jr, et al. Regulation of ATM/p53-dependent suppression of myc-induced lymphomas by Wip1 phosphatase. The Journal of experimental medicine. 2006;203(13):2793–9. doi: 10.1084/jem.20061563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Campaner S, Doni M, Hydbring P, Verrecchia A, Bianchi L, Sardella D, et al. Cdk2 suppresses cellular senescence induced by the c-myc oncogene. Nature cell biology. 2010;12(1):54–9. doi: 10.1038/ncb2004. sup pp 1–14. [DOI] [PubMed] [Google Scholar]
- 21.Murga M, Bunting S, Montana MF, Soria R, Mulero F, Canamero M, et al. A mouse model of ATR-Seckel shows embryonic replicative stress and accelerated aging. Nature genetics. 2009;41(8):891–8. doi: 10.1038/ng.420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schoppy DW, Ragland RL, Gilad O, Shastri N, Peters AA, Murga M, et al. Oncogenic stress sensitizes murine cancers to hypomorphic suppression of ATR. The Journal of clinical investigation. 2012;122(1):241–52. doi: 10.1172/JCI58928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nghiem P, Park PK, Kim Y, Vaziri C, Schreiber SL. ATR inhibition selectively sensitizes G1 checkpoint-deficient cells to lethal premature chromatin condensation. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(16):9092–7. doi: 10.1073/pnas.161281798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Toledo LI, Murga M, Zur R, Soria R, Rodriguez A, Martinez S, et al. A cell-based screen identifies ATR inhibitors with synthetic lethal properties for cancer-associated mutations. Nature structural & molecular biology. 2011;18(6):721–7. doi: 10.1038/nsmb.2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schetter AJ, Heegaard NH, Harris CC. Inflammation and cancer: interweaving microRNA, free radical, cytokine and p53 pathways. Carcinogenesis. 2010;31(1):37–49. doi: 10.1093/carcin/bgp272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee AC, Fenster BE, Ito H, Takeda K, Bae NS, Hirai T, et al. Ras proteins induce senescence by altering the intracellular levels of reactive oxygen species. The Journal of biological chemistry. 1999;274(12):7936–40. doi: 10.1074/jbc.274.12.7936. [DOI] [PubMed] [Google Scholar]
- 27.Vafa O, Wade M, Kern S, Beeche M, Pandita TK, Hampton GM, et al. c-Myc can induce DNA damage, increase reactive oxygen species, and mitigate p53 function: a mechanism for oncogene-induced genetic instability. Molecular cell. 2002;9(5):1031–44. doi: 10.1016/s1097-2765(02)00520-8. [DOI] [PubMed] [Google Scholar]
- 28.Maya-Mendoza A, Ostrakova J, Kosar M, Hall A, Duskova P, Mistrik M, et al. Myc and Ras oncogenes engage different energy metabolism programs and evoke distinct patterns of oxidative and DNA replication stress. Molecular oncology. 2015;9(3):601–16. doi: 10.1016/j.molonc.2014.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bartkova J, Horejsi Z, Koed K, Kramer A, Tort F, Zieger K, et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005;434(7035):864–70. doi: 10.1038/nature03482. [DOI] [PubMed] [Google Scholar]
- 30.Ray S, Atkuri KR, Deb-Basu D, Adler AS, Chang HY, Herzenberg LA, et al. MYC can induce DNA breaks in vivo and in vitro independent of reactive oxygen species. Cancer research. 2006;66(13):6598–605. doi: 10.1158/0008-5472.CAN-05-3115. [DOI] [PubMed] [Google Scholar]
- 31.Kuehl WM, Bergsagel PL. MYC addiction: a potential therapeutic target in MM. Blood. 2012;120(12):2351–2. doi: 10.1182/blood-2012-08-445262. [DOI] [PubMed] [Google Scholar]
- 32.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(43):15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burrell RA, McClelland SE, Endesfelder D, Groth P, Weller MC, Shaikh N, et al. Replication stress links structural and numerical cancer chromosomal instability. Nature. 2013;494(7438):492–6. doi: 10.1038/nature11935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carter SL, Eklund AC, Kohane IS, Harris LN, Szallasi Z. A signature of chromosomal instability inferred from gene expression profiles predicts clinical outcome in multiple human cancers. Nature genetics. 2006;38(9):1043–8. doi: 10.1038/ng1861. [DOI] [PubMed] [Google Scholar]
- 35.Zhan F, Huang Y, Colla S, Stewart JP, Hanamura I, Gupta S, et al. The molecular classification of multiple myeloma. Blood. 2006;108(6):2020–8. doi: 10.1182/blood-2005-11-013458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dib A, Gabrea A, Glebov OK, Bergsagel PL, Kuehl WM. Characterization of MYC translocations in multiple myeloma cell lines. Journal of the National Cancer Institute Monographs. 2008;(39):25–31. doi: 10.1093/jncimonographs/lgn011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reaper PM, Griffiths MR, Long JM, Charrier JD, Maccormick S, Charlton PA, et al. Selective killing of ATM- or p53-deficient cancer cells through inhibition of ATR. Nature chemical biology. 2011;7(7):428–30. doi: 10.1038/nchembio.573. [DOI] [PubMed] [Google Scholar]
- 38.Bartkova J, Hamerlik P, Stockhausen MT, Ehrmann J, Hlobilkova A, Laursen H, et al. Replication stress and oxidative damage contribute to aberrant constitutive activation of DNA damage signalling in human gliomas. Oncogene. 2010;29(36):5095–102. doi: 10.1038/onc.2010.249. [DOI] [PubMed] [Google Scholar]
- 39.Schumacker PT. Reactive oxygen species in cancer cells: live by the sword, die by the sword. Cancer cell. 2006;10(3):175–6. doi: 10.1016/j.ccr.2006.08.015. [DOI] [PubMed] [Google Scholar]
- 40.Diehn M, Cho RW, Lobo NA, Kalisky T, Dorie MJ, Kulp AN, et al. Association of reactive oxygen species levels and radioresistance in cancer stem cells. Nature. 2009;458(7239):780–3. doi: 10.1038/nature07733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Trachootham D, Zhou Y, Zhang H, Demizu Y, Chen Z, Pelicano H, et al. Selective killing of oncogenically transformed cells through a ROS-mediated mechanism by beta-phenylethyl isothiocyanate. Cancer cell. 2006;10(3):241–52. doi: 10.1016/j.ccr.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 42.Raj L, Ide T, Gurkar AU, Foley M, Schenone M, Li X, et al. Selective killing of cancer cells by a small molecule targeting the stress response to ROS. Nature. 2011;475(7355):231–4. doi: 10.1038/nature10167. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 43.Deponte M. Glutathione catalysis and the reaction mechanisms of glutathione-dependent enzymes. Biochimica et biophysica acta. 2013;1830(5):3217–66. doi: 10.1016/j.bbagen.2012.09.018. [DOI] [PubMed] [Google Scholar]
- 44.Morales AA, Gutman D, Cejas PJ, Lee KP, Boise LH. Reactive oxygen species are not required for an arsenic trioxide-induced antioxidant response or apoptosis. The Journal of biological chemistry. 2009;284(19):12886–95. doi: 10.1074/jbc.M806546200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Neri P, Ren L, Gratton K, Stebner E, Johnson J, Klimowicz A, et al. Bortezomib-induced “BRCAness” sensitizes multiple myeloma cells to PARP inhibitors. Blood. 2011;118(24):6368–79. doi: 10.1182/blood-2011-06-363911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Campaner S, Amati B. Two sides of the Myc-induced DNA damage response: from tumor suppression to tumor maintenance. Cell division. 2012;7(1):6. doi: 10.1186/1747-1028-7-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chesi M, Robbiani DF, Sebag M, Chng WJ, Affer M, Tiedemann R, et al. AID-dependent activation of a MYC transgene induces multiple myeloma in a conditional mouse model of post-germinal center malignancies. Cancer cell. 2008;13(2):167–80. doi: 10.1016/j.ccr.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen D, Kon N, Zhong J, Zhang P, Yu L, Gu W. Differential effects on ARF stability by normal versus oncogenic levels of c-Myc expression. Molecular cell. 2013;51(1):46–56. doi: 10.1016/j.molcel.2013.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Murphy DJ, Junttila MR, Pouyet L, Karnezis A, Shchors K, Bui DA, et al. Distinct thresholds govern Myc’s biological output in vivo. Cancer cell. 2008;14(6):447–57. doi: 10.1016/j.ccr.2008.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Luo J, Solimini NL, Elledge SJ. Principles of cancer therapy: oncogene and non-oncogene addiction. Cell. 2009;136(5):823–37. doi: 10.1016/j.cell.2009.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Garber K. PARP inhibitors bounce back. Nature reviews Drug discovery. 2013;12(10):725–7. doi: 10.1038/nrd4147. [DOI] [PubMed] [Google Scholar]
- 52.Bolli N, Avet-Loiseau H, Wedge DC, Van Loo P, Alexandrov LB, Martincorena I, et al. Heterogeneity of genomic evolution and mutational profiles in multiple myeloma. Nature communications. 2014;5:2997. doi: 10.1038/ncomms3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lohr JG, Stojanov P, Carter SL, Cruz-Gordillo P, Lawrence MS, Auclair D, et al. Widespread genetic heterogeneity in multiple myeloma: implications for targeted therapy. Cancer cell. 2014;25(1):91–101. doi: 10.1016/j.ccr.2013.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146(6):904–17. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Holien T, Vatsveen TK, Hella H, Waage A, Sundan A. Addiction to c-MYC in multiple myeloma. Blood. 2012;120(12):2450–3. doi: 10.1182/blood-2011-08-371567. [DOI] [PubMed] [Google Scholar]
- 56.Mertz JA, Conery AR, Bryant BM, Sandy P, Balasubramanian S, Mele DA, et al. Targeting MYC dependence in cancer by inhibiting BET bromodomains. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(40):16669–74. doi: 10.1073/pnas.1108190108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Keats JJ, Fonseca R, Chesi M, Schop R, Baker A, Chng WJ, et al. Promiscuous mutations activate the noncanonical NF-kappaB pathway in multiple myeloma. Cancer cell. 2007;12(2):131–44. doi: 10.1016/j.ccr.2007.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.mit.edu/mmgphwb. Available from: http://www.broad.mit.edu/mmgp.
- 59.http://www.broad.mit.edu/gsea.
- 60.http://www.genepattern.broadinstitute.org/gp.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.