Abstract
Why breast cancers become resistant to tamoxifen despite continued expression of the estrogen receptor alpha (ERα) and what factors are responsible for high HER2 expression in these tumors remains an enigma. HOXB7 ChIP analysis followed by validation showed that HOXB7 physically interacts with ERα, and that the HOXB7-ERα complex enhances transcription of many ERα target genes including HER2. Investigating strategies for controlling HOXB7, our studies revealed that MYC, stabilized via phosphorylation mediated by EGFR-HER2 signaling, inhibits transcription of miRNA-196a, a HOXB7 repressor. This leads to increased expression of HOXB7, ER-target genes and HER2. Repressing MYC using small molecule inhibitors reverses these events, and causes regression of breast cancer xenografts. The MYC-HOXB7-HER2 signaling pathway is eminently targetable in endocrine-resistant breast cancer.
Keywords: HOXB7, HER2, endocrine resistance, MYC, miR-196a
INTRODUCTION
In order to antagonize estrogen receptor-α (ER) function in ER-positive breast cancer, various endocrine therapies have been employed such as selective estrogen receptor modulation via SERMs, ligand deprivation using aromatase inhibitors, as well as ER downregulation with fulvestrant (1–4). In spite of the world-wide use of tamoxifen as adjuvant treatment for postmenopausal women with ER-positive breast cancer, cancer recurs in about one-third of tamoxifen treated patients (5). Thus far, two major pathways of endocrine resistance- through ER itself or receptor tyrosine kinases (RTKs) have played pivotal roles in endocrine therapy. In acquired tamoxifen resistance, ER expression is maintained at detectable levels in the majority of tumors and ER continues to promote tumor proliferation (6). With regard to RTKs, upregulation of ErbB/HER family members such as EGFR and HER2 was observed in endocrine resistant tumors, but the mechanism underlying increased expression of ErbB/HERs is not clear (6–8).
Several studies on HOX genes including HOXB7, HOXB13, HOXC10, HOXC11, and a cofactor for the homeobox gene, PBX1, have been reported in endocrine-resistant breast cancer (9–13). We recently reported that HOXB7 overexpression confers TAM-resistance through upregulation of EGFR signaling (9). Here, we have provided evidence for the function of HOXB7 as an ER-cofactor in the activation of ER-target genes and HER2. Further, we have identified the upstream regulators of HOXB7 which are amenable to therapeutic targeting directed at overcoming endocrine resistance in breast cancer.
RESULTS
HOXB7 promotes ER target gene expression
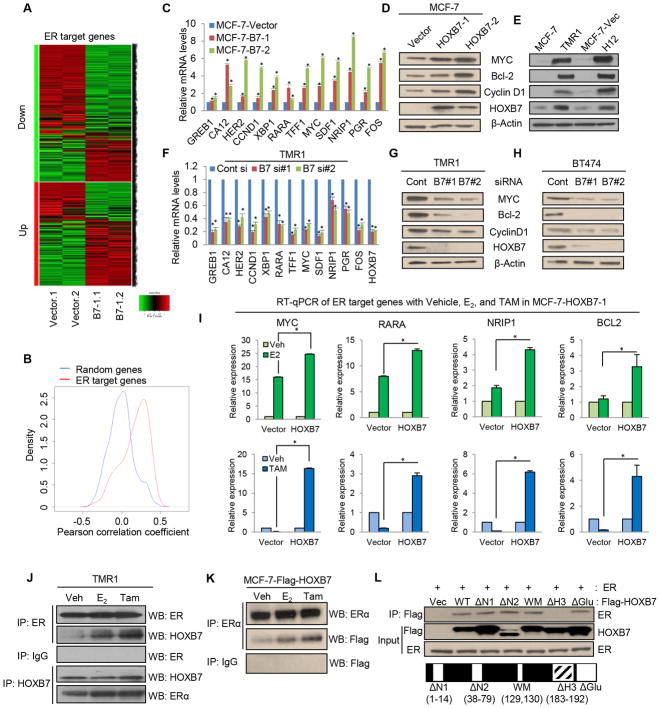
It is known that crosstalk between ER and other transcription factors can promote ER target gene expression (14–16). We had previously observed that estrogen supplementation of mice caused an increase in size of xenografts of ER-positive MCF-7 cells overexpressing HOXB7, but not of vector control MCF-7 cells (9). This finding motivated the hypothesis that HOXB7 may participate in the ER-signaling pathway as an ER-co-activator. Microarray expression analysis was performed with MCF-7-HOXB7 cells and MCF-7-Vector cells. Mean rank gene set enrichment test (17) using moderated t-statistics (Supplementary Methods) showed a significant number of genes induced by HOXB7 overlapping genes in the same direction altered in MCF-7 in response to estrogen (18). Specifically, we observed a FWER of < 0.001 (Fig 1A, Supplementary Fig S1A, S1B, Table S1). This finding is also corroborated by a Gene Ontology (GO) analysis (Supplementary Fig. S1C). A large scale cross-sample gene expression correlation analysis based on the ChIP-PED dataset (n=13,182) (19) (which uses the combined strength of ChIP/ChIP and Chip-Seq data along with the large amounts of publicly available gene expression data to discover new biological contexts with potential TF regulatory activities), allowed us to confirm that HOXB7 expression significantly correlated with ER target genes (Fig. 1B; Supplementary Fig. S1D–S1K and Table S2). Real time q-PCR and immunoblot analysis showed that HOXB7 enhances endogenous ER-target gene expression in cells stably overexpressing MCF-7-HOXB7 cells (two clones: MCF-7-B7-1 and -2) (Fig. 1C and 1D), T47D-HOXB7 and ERIN-HOXB7 (MCF10A-ER) (Supplementary Fig. S2A–C), tamoxifen-resistant MCF-7 cells (TMR) and in MCF-7 cells overexpressing both EGFR and HER2 (H12) (Fig. 1E). Conversely, siRNA-mediated depletion of HOXB7 expression in cells with high endogenous HOXB-7 expression, TMR (Fig. 1F and 1G) and BT-474 cells (Fig. 1H) resulted in reduced expression of ER-target genes. Interestingly, we observed that when cells were treated with estrogen (E2), the mRNA expression of the ER-target genes was significantly increased in MCF-7-HOXB7 (Fig. 1I; Supplementary Fig. S2D), T47D-HOXB7 (Supplementary Fig. S2E), and ERIN-HOXB7 cells (Supplementary Fig. S2F), when compared to vector control cells which suggested a cooperative action of both HOXB7 and ER on target gene expression.
Figure 1. HOXB7 interacts with ERα and enhances expression of ER-target genes.
A, Heatmap representing relative expression of ER target genes (identified in Dutertre et al., 2010) in MCF-7-HOXB7 cells compared to MCF-7-Vector determined by microarray analysis. B, Density curves for cross-sample (n=13,182) gene expression correlations between HOXB7 and ER target genes versus randomly selected genes. Correlation between HOXB7 and ER target genes is significantly (P< 10−15) different from correlation between HOXB7 and random genes. C, Real time RT-qPCR analysis of ER target genes in stable HOXB7-overexpressing MCF-7 cells. Immunoblot analysis of HOXB7 and ER target genes in D, MCF-7-HOXB7 and E, MCF-7-TMR1 and H12 (MCF-7-EGFR/HER2) cells compared to vector control cells. F, RT-qPCR analysis of ER target genes in HOXB7 depleted MCF-7-TMR1 cells using siRNAs. Immunoblot analysis of HOXB7 and ER target genes in HOXB7-depleted G, TMR1 and H, BT474 cells compared to vector control cells. I, RT-qPCR analysis of ER target genes in MCF-7-HOXB7 stable cells incubated in estrogen deprived medium (5 % charcoal stripped serum in phenol red free DMEM) for 48 hours before treatment with vehicle, 10 nM E2, and 1 uM TAM for 24 hours. Co-immunoprecipitation (Co-IP) analysis performed J, in TMR cells using anti-ER or HOXB7 antibody and western blot analysis using anti-HOXB7 or ER antibody, or K, co-IP analysis in MCF-7-Flag-tagged-HOXB7 cells using anti-ER antibody and western blot analysis using anti-Flag antibody. L, Co-IP analysis performed with anti-flag antibodies in MCF-7 cells transfected with-Flag-tagged-full length and -deletion mutants of HOXB7 constructs. (WT: full length HOXB7, N1: N-terminal deletion 1 (1–14), N2: N-terminal deletion (38–79), WM: W129F and M130I, ΔH3: deletion of Helix domain 3 of homeodomain (183–192), ΔGlu: deletion of glutamic acid tail. Mean ± s.d. for three independent replicates. (*P<0.001).
Previously, we proposed that as a result of HOXB7 overexpression, tamoxifen might change from antagonist to an agonist for ER in TMR cells (9). In fact, in contrast to MCF-7-Vector cells, ER target gene expression was upregulated upon tamoxifen (TAM) treatment of MCF-7-HOXB7, T47D-HOXB7, and ERIN-HOXB7 cells (Fig. 1I; Supplementary Fig. S2D–I). This raised the possibility that HOXB7 can interact with ER bound to either estrogen or tamoxifen. To address this, we performed co-immunoprecipitation and GST pulldown analysis. This revealed a direct interaction between HOXB7 and ER, which was enhanced upon estrogen and TAM treatment as well (Fig. 1J and 1K; Supplementary Fig. S2J). Further definition was sought for the genomic regions of HOXB7-ER interaction with ER using co-IP analysis with multiple deletion mutants of HOXB7. Helix 3 of the homeodomain was identified as the key region of physical interaction for HOXB7-ER (Fig. 1L). Together, these results suggest that upon estrogen and TAM treatment, HOXB7 physically interacts with ER and that the resulting HOXB7-ER complex could promote ER transcriptional activity at promoters of multiple ER-target genes.
Identification of novel HOXB7 binding sites in ER target genes
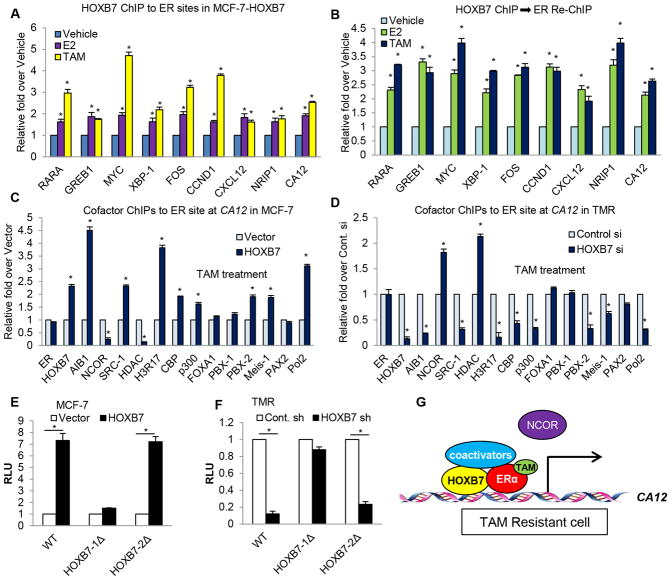
Given the robust upregulation of ER-target genes by HOXB7, we explored the role of HOXB7 in regulating the interaction of ER with chromatin at the promoters of ER-target genes. ER-binding sites are frequently located further upstream of the genes, such as in enhancer regions (20–23). Using ER binding sites identified by ER-ChIP analysis in published data profiles (24), present in the proximal promoter or intron regions in ER-target gene loci (Supplementary Fig. S3A), we performed HOXB7 chromatin immunoprecipitation (ChIP) analysis of known ER binding regions in the ER target genes loci, RARA, GREB1, MYC, XBP1, FOS, CCND1, CXCL12, NRIP1, and CA12. ER ChIP analysis was also performed as a positive control. As predicted, following estrogen or tamoxifen treatment, ER was recruited to its binding sites. Interestingly, HOXB7 was also recruited significantly to the ER binding site. In addition, the occupancy of HOXB7 to the site was enhanced with TAM treatment in ER target genes loci (Fig. 2A; Supplementary Fig. S3B and S3C). A Re-ChIP assay confirmed concurrent presence of both HOXB7 and ER proteins at the same ER binding region within the target genes (Fig. 2B; Supplementary Fig. 3D–F). To investigate the mechanism by which the HOXB7-ER complex may enhance CA12 gene transcription, ChIP assays were performed with pioneer factors FOXA1 (24) and PBX1 (25), ER cofactors (AIB1, SRC-1, CBP, p300, NCOR, and PAX2), and HOXB7 cofactors (PBX2 and Meis1) to measure their occupancy at ER binding site within the CA12 gene in MCF-7-HOXB7 cells compared to MCF-7-Vector cells. Although the recruitment of ER itself and pioneer factors was not significantly altered in ER binding regions of the CA12 gene in MCF-7-HOXB7 cells, ER coactivators were strikingly enriched at levels higher than the HOXB7 coactivators. In contrast, the recruitment of NCOR, an ER corepressor, was decreased at the ER binding sites (Supplementary Fig. S3G). Following knockdown of HOXB7 expression, recruitment of both ER and HOXB7 co-activators was significantly reduced, whereas NCOR recruitment was increased (Supplementary Fig. S3H). When TAM binds ER in TAM-sensitive cells, it induces a conformational change in ER, and recruits co-repressors to inhibit ER-target gene transcription. However, when TAM binds ER in TAM-resistant cells, coactivators are recruited to ER-binding sites instead, resulting in select ER-target gene transcription. However, the detailed mechanism remains unclear (26). To shed light on this question, we investigated whether HOXB7 functions as a major recruiter of ER-coactivators in TAM-resistant cells. The same ChIP assays were performed as in Supplementary Fig. S3G and 3H, now, after TAM treatment. As predicted, in the presence of TAM, recruitment of coactivators at the ER-binding site in the CA12 gene locus was higher in MCF-7-HOXB7 cells compared to vector controls. Depletion of HOXB7, on the other hand, resulted in enrichment of corepressors at the site (Fig. 2C and 2D). These events were also confirmed by HOXB7 ChIP analysis to ER binding sites at a second ER-target gene, MYC (Supplementary Fig. S3I–L). In addition, we found that the recruitment of ER-coactivator or repressor to ER binding site in CA12 or MYC loci was regulated by HOXB7 expression in a dose-dependent manner (Supplementary Fig. S4A and S4B). To investigate the detailed mechanism of how the ER-HOXB7 complex promotes CA12 transcription, we created CA12-luciferase constructs where we subcloned the genomic 1.7 kb CA12-promoter sequence, included the ER-binding sites and two putative HOXB7 binding sites, into the pGL3 promoter vector (Supplementary Fig. S4C and S4D). We found that overexpression of HOXB7 enhanced luciferase activity, and HOXB7 depletion (using shRNA) resulted in decreased luciferase activity for CA12-WT and CA12-HOXB7-2Δ constructs, but not of CA12-HOXB7-1Δ construct. This data suggested that HOXB7 was recruited to the HOXB7-1 site in the CA12 gene locus (Fig. 2E and 2F), and that the ER-HOXB7 complex is critical for activating CA12 transcription (Supplementary Fig. S4E and S4F). To further confirm these findings in a second ER-target gene, we created MYC-luciferase constructs containing an ER enhancer region (27) tagged to three different putative HOXB7 binding regions in MYC. These were MYC-B7-1, MYC-B7-2, and MYC-B7-3, which were selected through analysis of DNase-seq data detailed in Materials and Methods (Supplementary Fig. S4G). We found that overexpression of HOXB7 enhanced luciferase activity and HOXB7 depletion (using shRNA) resulted in decreased luciferase activity for constructs containing MYC-B7-1 or -2 but not MYC-B7-3 (Supplementary Fig. S4H and S4I). These results suggested that HOXB7 was recruited to the binding sites 1 and 2 in the MYC gene. Furthermore, we verified the formation of a chromatin loop between the ER-binding site and HOXB7 binding sites by using the chromosome conformation capture (3C) assay for MYC gene in MCF-7-HOXB7 cells after treatment with estrogen and TAM (Supplementary Fig. S4J–L). This finding confirmed the occurrence of dynamic long-range chromatin interaction (~65 kb) between ER and HOXB7 bound to their cognate sites, in order to promote MYC transcription. Collectively, these results suggest that when overexpressed HOXB7 binds to TAM-bound ER, the HOXB7-ER complex tethers coactivators, resulting in ER-target gene transcription in TAM-resistant cells. Both HOXB7 and ER cooperate to upregulate CA12 and MYC expression and that HOXB7 augments ER genomic functions as an important co-activator (Fig. 2G; Supplementary Fig. S4M and S4N).
Figure 2. The ER-HOXB7 complex directly enhances transcriptional activity of ER target genes.
A, HOXB7 ChIP assay of known ER-binding sites in ER target genes was performed in MCF-7-HOXB7 cells incubated in estrogen depleted medium (5 % charcoal stripped serum in phenol red free DMEM) for 48 hours before treatment with vehicle, 10 nM E2, and 1 uM TAM for 45 minutes. B, ER-re-ChIP analysis following HOXB7 ChIP was performed as in (A). ChIP analysis of each factor to the ER binding site in the CA12 gene locus in C, MCF-7-HOXB7 cells compared to MCF-7-vector cells and in D, HOXB7 depleted TMR cells treated with tamoxifen. RNA Polymerase II ChIP analysis was performed for CA12 promoter region as a positive control for CA12 transcription. E, Luciferase assay was performed in MCF-7 cells transiently co-transfected with HOXB7 or vector control plasmid along with wildtype (WT) CA12 promoter-luciferase constructs, or those with deleted HOXB7 binding sites- HOXB7-1Δ or HOXB7-2Δ. F, Same assay was performed as (E) in HOXB7 depleted TMR cells. G, An activation model of CA12 transcription through interaction between HOXB7-ER complex and coactivators in TAM-resistant cells. Mean ± s.d. for at least three independent replicates (*P<0.001).
HOXB7 enhances HER2 expression
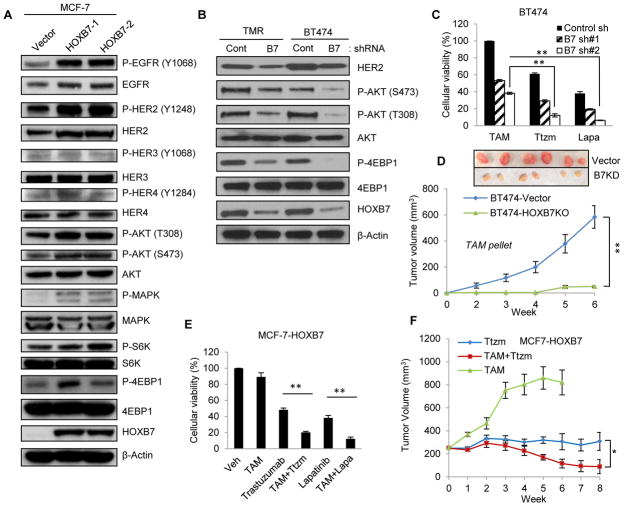
Upregulation of HER2 leads to the poor prognosis in ER positive breast cancer (28). As a consequence, the nature of the crosstalk between HER2 and ER has been studied for more than a decade (29) with a recent study concluding that ER and its cofactors directly regulate HER2 transcription (24). Our previous studies had also shown that HER2 expression is upregulated in HOXB7 overexpressing breast cancer (30). ChIP-PED analysis (19) showed a strong correlation (r=0.4670, P<10−15) between HOXB7 and HER2 expression (Supplementary Fig. S5A). In the absence of evidence of direct transcriptional regulation, and based on the finding that HOXB7/ER interactions occur at many ER-target gene promoters, we hypothesized that HOXB7 can regulate HER2 transcriptional activity via direct binding to its enhancer region along with ER, in a manner similar to that observed in regulation of other ER target genes. Western blot analysis showed that overexpression of HOXB7 in MCF-7 cells resulted in enhanced HER2 expression and phosphorylation at the Y1248 residue in the kinase domain, and activation of downstream effectors of HER2 signaling such AKT, p44/42 MAPK, S6K, and 4EBP1 (Fig. 3A). Similar overexpression of EGFR and HER2, but not HER3 and HER4, was observed in TMR1 and TMR2 cells with endogenous high HOXB7 expression (Supplementary Fig. S5B); and similar HER signaling responses were seen in TMR and H12 cells (Supplementary Fig. S5C) and in two additional ER+ breast cancer cells, T47D-HOXB7 and ERIN-HOXB7 cells (Supplementary Fig. S5D). To define the precise contribution of HOXB7 in HER2 overexpression and TAM sensitivity, we used tamoxifen-resistant TMR and BT474 cells. Stable depletion of HOXB7 in both cell lines using HOXB7 shRNAs decreased HER2 expression and the downstream effectors of the HER2 signaling pathway (Fig. 3B). Knockdown of HOXB7 in MCF7-TMR (Supplementary Fig. S5E) and in tamoxifen resistant BT474 cells (Fig. 3C; Supplementary Fig. S5F) restored TAM sensitivity. TMR and BT474 cells, are HER2-dependent for growth. Treatment of TMR and BT474 cells with anti-HER2 antibody-trastuzumab (31) or dual EGFR/HER2 inhibitor, lapatinib (32) (Fig. 3C; Supplementary Fig. S5G) resulted in enhanced cell death and this effect was even more significant upon depletion of HOXB7 (Fig. 3C; Supplementary Fig. S5H–J). Xenografts of MCF-7-TMR-shHOXB7 regained sensitivity to tamoxifen and showed significant tumor regression (Fig. 3D; P<0.001). Growth of MCF7-HOXB7 cells, that also overexpress HER2, in culture (Fig. 3E) and as xenografts (IHC; Supplementary Fig. S5K), remained unaffected by TAM alone, but was suppressed by HER2 inhibitor, trastuzumab. Addition of TAM to trastuzumab caused significantly greater inhibition of tumor growth (Fig. 3F). These results suggest that depletion of HOXB7 may be a potential strategy for reversing TAM-resistance in breast cancer showing concurrent overexpression of HER2.
Figure 3. HOXB7 promotes HER2 expression.
A, Immunoblotting analysis of P-EGFR (Y1068), EGFR, P-HER2 (Y1248), HER2, P-HER3 (Y1068), HER2, P-HER4 (Y1248), HER4, P-AKT (T308), P-AKT (S473), AKT, P-MAPK, MAPK, P-S6K, S6K, P-4EBP1, 4EBP1 and HOXB7 in MCF-7-HOXB7 (two clones: MCF-7-B7-1 and -2) and B, HOXB7 depleted TMR-shHOXB7 and BT474-shHOXB7 cells. C, Cellular viability assay was performed in BT474-shHOXB7 cells after treatment of 2 uM tamoxifen (TAM), 100 ug/ml trastuzumab (Ttzm) or 1 uM Lapatinib (Lapa). D, Tumor growth of BT474-vector and BT474-shHOXB7 as m.f.p. xenografts in NSG mice treated with tamoxifen (slow release pellets). E, Cellular viability assay of MCF-7-HOXB7 cells treated with TAM, trastuzumab (Ttzm), and lapatinib (Lapa). F, Tumor growth of MCF-7-HOXB7 cells as S.C. xenografts in athymic mice with E2 supplementation until tumors reached an approximate volume of 200 mm3, then treated with tamoxifen pellets (TAM) implanted S.C, 20 mg/kg trastuzumab (Ttzm) twice a week, and a combination of Ttzm and TAM. Mean ± s.d. for three independent replicates (A–C). Mean ± s.e.m. n=6 (D), n=10 (F) (*P<0.02, **P<0.001).
To evaluate if the HOXB7-ER complex could directly regulate HER2 transcription, we performed ChIP assays with HOXB7, ER, and their cofactors to the previously described ER binding site in the enhancer present in the first exon of the HER2 gene (24). Consistent with previous studies (24, 33), and our prior results with other ER target genes, we found that HOXB7, ER, and their cofactors were recruited to ER binding site within the HER2 gene (Supplementary Fig. S6A–C). Together, these results supported our hypothesis that the HOXB7-ER complex regulates HER2 transcriptional activity in tamoxifen resistant breast cancer (Supplementary Fig. S6D).
To investigate the clinical relevance of HOXB7-mediated regulation of HER2 expression, we performed real time q-PCR analysis of HOXB7 and HER2 from 48 breast cancer cell lines which were divided into three groups according to their relative HER2 gene expression levels. We found that high HOXB7 expression correlated significantly (p<0.0001) with high expression of HER2 (Supplementary Fig. S6E). Further, upon analysis of the Cancer Genome Atlas (TCGA) data set, HOXB7 mRNA levels were found to be significantly increased (p<0.0001) in HER2-positive tumors (Supplementary Fig. S6F).
miR-196a regulates HOXB7 expression in tamoxifen resistance
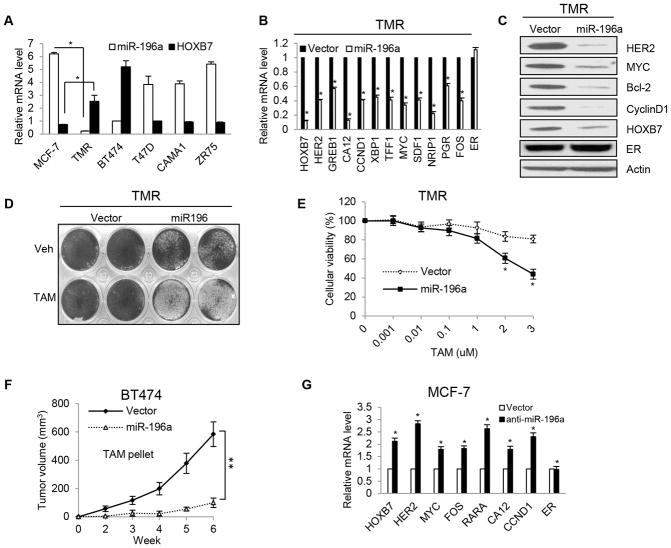
In malignant melanoma cells, microRNA-196a binds directly to the HOXB7 3′UTR and inhibits HOXB7 expression (34). MicroRNA array analysis showed that miR-196a expression is downregulated in endocrine-resistant breast cancer cells (35). From these observations, we posited that the upregulation of HOXB7 in MCF-7-TMR cells was likely caused by reduced miR-196a expression. Consistent with this notion, we found that miR-196a expression level was lower in TMR cells than in the parental MCF-7 cells, as well as in BT474 cells, in comparison to other ER-positive TAM-sensitive cells such as T47D, CAMA1, and ZR75-1, where HOXB7 expression was relatively low (Fig. 4A). Overexpression of miR-196a inhibited HOXB7 expression, leading to the decrease of ER target gene and HER2 expression in TMR cells as shown by RT-qPCR (Fig. 4B) and western blot analysis for prototypic molecules (Fig. 4C). Enforced miR-196a expression re-sensitized TMR-miR-196a cells to TAM as shown by colony formation (Fig. 4D) and MTT assays (Fig. 4E), and caused regression of stably transfected BT474-miR-196a tumors upon treatment with TAM (Fig. 4F). Introducing the miR-196a-inhibitor (36) into MCF-7 cells increased ER target gene expression (Fig. 4G). Using HOXB7-3′UTR luciferase constructs (WT and mutant MT-Δ10 bp miR-196a target sequence), we confirmed the inhibitory effect of miR-196a on HOXB7 transcription in MCF7-miR-196a (Supplementary Fig. S7A), H12 (MCF7-EGFR-HER2) (Supplementary Fig. S7B), TMR (Supplementary Fig. S7C) and MCF7-miR-196a-inhibitor expressing cells (Supplementary Fig. S7D). We also confirmed that ER transcriptional activity is not modulated by miR-196a, by conducting ERE-Luciferase assays in TMR cells and in MCF-7 cells. ERE-luciferase activity was not significantly altered in both miR-196a overexpressing TMR, and miR-196a inhibitor overexpressing MCF-7 cells (Supplementary Fig. S7E). These findings suggest that downregulation of miR-196a induced HOXB7 overexpression in TMR cells, and conversely, reconstituting miR-196a expression in cells should elicit therapeutic effects. Also, as demonstrated by others (37), the success in restoring miRNA expression with nanoparticle preparations to achieve therapeutic advantage supports the rationale of this approach.
Figure 4. miRNA-196a regulates HOXB7 expression.
A, Real time RT-qPCR analysis of miR-196a and HOXB7 in TMR cells compared to MCF-7-parental cells and in four ER-positive breast cancer cell lines. B, Real time RT-qPCR analysis and C, Immunoblot analysis of ER target genes in TMR cells transiently transfected with miR-196a compared to vector control cells. D, Crystal violet staining of TMR cells with enforced miR-196a expression after 7 days in culture with and without TAM treatment. E, Cellular viability assay in TMR-miR-196a cells compared to vector control with TAM treatment at different doses. F, Tumor growth of BT474-vector cells (same as in Figure 3D) and BT474-miR-196a cells xenografts (m.f.p.) in NSG mice treated with tamoxifen (pellets). G, Real time RT-qPCR analysis of ER target genes and HOXB7 expression in MCF-7 cells transiently transfected with two different miR-196a inhibitors compared to vector control cells. Mean ± s.d. for three independent replicates. Mean ± s.e.m. n=10 (*P<0.0001 and **P<0.00001).
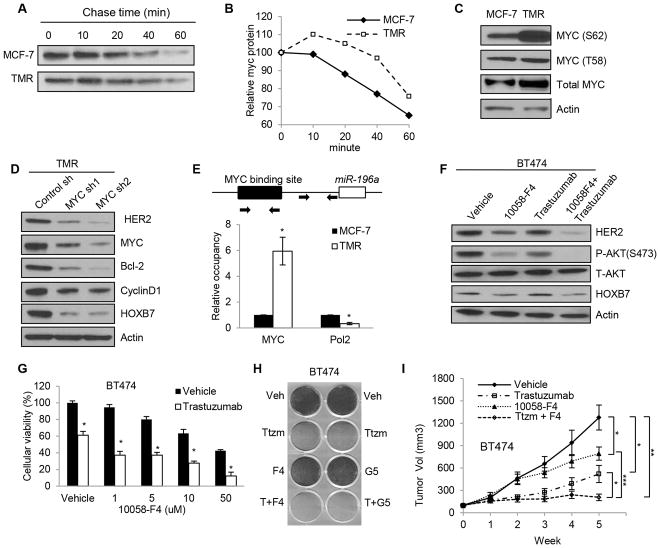
Stability of MYC controls miR-196a-HOXB7 signaling
The relationship between miRNAs and their target genes has been studied in depth; however, the regulation of miRNA expression itself remains largely unclear (38). So we considered alternate approaches to resolving this problem. It was established early on that EGFR/HER2 signaling regulates MYC stability (39–41). Recent reports suggest that MYC activity may be a key pathway in endocrine resistance (42) and that MYC has the ability to repress miRNA expression (43). These studies led us to propose that a result of MYC-mediated repression of miR-196a expression would be a subsequent upregulation of HOXB7 expression. To determine how MYC functions in tamoxifen resistance, we first examined MYC protein stability following treatment with cycloheximide. We found that MYC protein was more stable in TMR cells (Fig. 5A and 5B), H12 (MCF-7-EGFR+HER2), and MCF-7-HOXB7 cells (Supplementary Fig. S8A–D) when compared to control cells. MAPK and AKT signaling pathways phosphorylate the MYC protein at the serine 62 residue, which stabilizes MYC protein (41, 44). Interestingly, we found that the hyperphosphorylation of MYC at serine (Ser 62), but not at threonine (Thr 58) through EGFR/HER2 signaling in MCF7-HOXB7, TMR, and H12 cells (Fig. 5C; Supplementary Fig. S8E and F). MYC phosphorylation levels are responsive to HOXB7 levels; stable depletion of HOXB7 in TMR or BT474 cells results in significantly lower levels of MYC phosphorylation at Ser62 (Supplementary Fig. S8G). As predicted, depletion of MYC in TMR cells by MYC shRNAs resulted in decreased HOXB7, HER2, and ER target gene expression as measured by Western blot analysis (Fig. 5D); here, a higher level of miR-196a gene expression was also observed (Supplementary Fig. S8H). Similarly, inhibiting MYC using MYC-MAX dimerization inhibitor, 10058-F4 (45), also results in higher levels of miR-196a expression in TMR cells (Supplementary Fig. S8I). In addition, depletion of MYC or overexpression of a dominant negative MYC (S62A) in MCF-7 cells caused a dramatic decrease in the 3′UTR-HOXB7 luciferase activity (Supplementary Fig. S8J and S8K). This allowed us to conclude that depletion of MYC decreased HOXB7 expression, in all probability, via upregulation of miR-196a. Thus far, the results support our hypothesis that MYC inhibits miR-196a expression, which results in HOXB7 expression. However, MYC does not directly upregulate HOXB7 and HER2 function as shown by lack of change in HOXB7-promoter-luciferase and HER2 promoter-luciferase assays in scrambled or sh-MYC transfected cells (Supplementary Fig. S8L and S8M). miR-196a was identified as a putative MYC target gene through global mapping of MYC binding sites (46). We performed ChIP analysis to investigate if MYC can bind directly to putative binding sites in the miR-196a gene locus using both TMR-shMYC cells, and TMR cells treated with MYC inhibitors. We found that MYC binds directly to the MYC binding site (chr17:46707861-46708471) and recruits corepressors to the miR-196a gene locus to inhibit miR-196a transcription (Fig. 5E; Supplementary Fig. S8N–P). Using a miR-196a luciferase construct that contained a putative MYC binding site, we showed that the overexpression of dominant-negative MYC (S62A) in MCF-7 cells, or MYC depletion in TMR cells significantly increased luciferase activity (Supplementary Fig. S8Q). These results suggested that the recruitment of MYC to its putative binding site in the miR-196a promoter contributes to the repression of miR-196a expression in TMR cells.
Figure 5. MYC-miR-196a-HOXB7 signaling controls TAM-resistance.
A, Cycloheximide chase examining MYC protein stability between MCF-7 parental cells and TMR cells by immunoblot analysis. B, Quantification of MYC decay following CHX treatment based on densitometric scanning of the immunohybridization signals. C, Immunoblot analysis of phosphorylation of MYC at serine 62 and threonine 58 residues in TMR cells compared to MCF-7 cells. D, Immunoblot analysis of HER2, HOXB7, and ER target gene in MYC-depleted TMR cells using two different MYC shRNAs. E, MYC ChIP analysis for a putative MYC binding site in the miR-196a gene locus, and RNA Polymerase II ChIP analysis for miR-196a promoter region in TMR cells. F, Immunoblot analysis of HER2, P-AKT, and HOXB7 in BT474 cells after treatment with 10 uM 10058-F4, 100 ug/ml of trastuzumab, or a combination of 10058-F4 and trastuzumab. G, Cellular viability of BT474 cells after treatment with 1, 5, 10, or 50 uM of 10058-F4 plus 100 ug/ml of trastuzumab for 48 hours. H, Crystal violet staining of BT474 cells treated with 5 uM 10058-F4, 5 uM 10074-G5, or 100 ug/ml trastuzumab alone, or a combination of 10058-F4 or 10074-G5 with trastuzumab for a week. I, Tumor growth of BT474 cells m.f.p. xenografts in NSG mice. When the tumors reached an approximate volume of 100 mm3, trastuzumab (20 mg/kg, twice weekly), 10058-F4 (30 mg/kg/day), or a combination of trastuzumab and 10058-F4 were administered by i.p. injection. Mean ± s.d. for three independent replicates. Mean ± s.e.m. n=10; (*P<0.03, **P<0.001, and ***P<0.0001).
To determine if inactivation of MYC can re-sensitize TMR cells to TAM, we first depleted MYC using shRNA in TMR cells. Decrease in MYC levels significantly attenuated TAM resistance in TMR cells (Supplementary Fig. S8R and S8S). Although targeted drug development against MYC has been a challenge, we projected that MYC inhibitors could alter tamoxifen resistance via HOXB7 reduction. Using two MYC inhibitors of MYC-MAX dimerization, 10058-F4 and 10074-G5 (47), not only did we observe a decrease of HOXB7 expression, but also a reduction of HER2 and ER target gene expression (Fig. 5F; Supplementary Fig. S8T and S8U). In addition, combined treatment with 10058-F4 and trastuzumab synergistically reduced cellular viability of BT474 cells (Fig. 5G; Supplementary Fig. S8V) and colony formation (Fig. 5H), effects that could be rescued by exogenous expression of HOXB7 in these cells (Supplementary Fig. S8W). Further, the drug combination caused significant reduction of BT474 tumor growth in immunodeficient NSG mice compared to single agents (Fig. 5I; Supplementary Fig. S9A–S9C). In addition, fulvestrant reduced cellular viability of TMR and BT474 cells and caused regression of MCF-7-HOXB7 xenografts (Supplementary Fig. S9D and S9E). Collectively, the data strongly support the notion that MYC is a critical molecule in regulating HOXB7 expression.
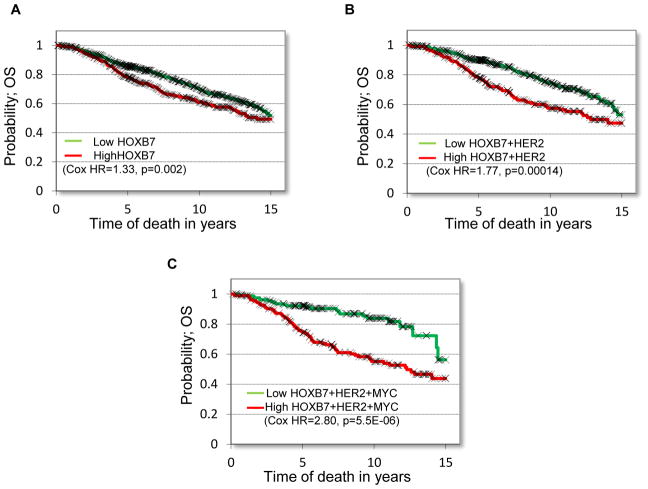
MYC-HOXB7-HER2 predicts clinical outcome in tamoxifen resistant breast cancer patients
In order to examine if the MYC-HOXB7-HER2 pathway can be predictive of endocrine resistance in breast cancer, we first examined HOXB7 expression in endocrine therapy-treated ER-positive breast cancer patients (n=1208) from the Metabric database. We found a statistically significant association between high HOXB7 expression and poor overall survival (OS, HR=1.37, p=0.002). In addition, co-expression of HOXB7, HER2, and MYC was significantly prognostic of overall survival (HR=2.80, p=5.5E-06) and more significant than HOXB7 alone. To determine commonality across various datasets, we investigated three independent ER-positive breast cancer patient data sets and found that a combination of HOXB7 and MYC or HOXB7 and HER2 expression was significantly prognostic of OS or relapse-free survival (RFS) in each data set (Fig. 6A–C; Supplementary Fig. S10A–F). To further confirm the results of these analyses, we performed IHC for HOXB7, HER2 and MYC in a tissue microarray containing 72 tumor samples of ER+ breast cancer from patients treated with TAM monotherapy. We found that a combination of HOXB7 and MYC or HOXB7 and HER2 expression was significantly correlated with OS or relapse-free survival (RFS) (Fig. S10G–K). These results provide further support to our postulate that the MYC-HOXB7-HER2 signaling pathway is associated with endocrine resistance in patients diagnosed with breast cancer.
Figure 6. MYC-HOXB7-HER2 predicts clinical outcome in breast cancer patients treated with tamoxifen.
Kaplan-Meier plots of overall survival probability in endocrine therapy treated breast cancer patients with ER+ tumors (n=1208) expressing A, HOXB7 B, HOXB7 and HER2 and C, HOXB7, HER2, and MYC.
DISCUSSION
Estrogen receptor targeted therapy by using selective ER modulator (SERM); tamoxifen (1, 2, 48) or aromatase inhibitors (AIs) such as anastrozole, exemestane, letrozole (3) or the selective ER downregulator (SERD), Fulvestrant (4) is effective in treating breast cancer in postmenopausal women (49). However, long term endocrine treatment often leads to development of de novo resistance in ERα-positive breast cancer. Many underlying molecular events that confer resistance are known, but a unifying theme is yet to be revealed. Here, we have described findings that support a key role of HOXB7 in endocrine resistance and identify novel potential therapeutic targets among the upstream regulators of HOXB7.
Despite the fact that in time many ER+ breast tumors lose estrogen dependence for growth, nearly 30% retain ER expression and can therefore respond to a different endocrine therapy (6). This implies that activated ER can still serve as a therapeutic target. One of the thought provoking findings of our previous study was that xenograft growth of ER-positive breast cancer cells overexpressing HOXB7 was more robust upon supplementation of the mice with exogenous estrogen (9), suggesting a cross talk between HOXB7 and ER. If so, the two transcription factors, acting together, could confer TAM resistance in breast cancer cells. In line with this prediction, we found that a large number of genes induced by HOXB7 were common to those induced by ER. This overlap prompted the hypothesis that HOXB7 may act as an ER cofactor to activate the ER signaling pathway. This hypothesis gained strength through observations of upregulation of ER target genes in conjunction with HOXB7 overexpression and downregulation of ER target genes upon HOXB7 depletion, aided by co-IP studies that supported their physical interaction (Fig. 1). ChIP analyses strengthened the finding that HOXB7-ER complex transcriptionally activates several ER-target genes. For the first time, both CA12 and MYC were identified as putative HOXB7 targets (Fig. 2). Our data implicated HOXB7 as a key ER cofactor that promotes ER target gene expression; which also could serve as yet another target in endocrine therapy.
Receptor tyrosine kinases (RTKs) regulate cellular differentiation and proliferation through transmitting extracellular signals into cells. In endocrine resistance, upregulation of ErbB/HER signaling plays an important role in altering cellular response to tamoxifen (6–8). Thus far, the current strategy for overcoming endocrine resistant breast cancer is i) to use a EGFR inhibitor (Gefitinib), ii) HER2 inhibitor (Trastuzumab), iii) EGFR/HER2 dual tyrosine kinase inhibitor (Lapatinib), or iv) mTOR inhibitor (everolimus) combined with tamoxifen or aromatase inhibitors (50–55). Although these inhibitors have shown some clinical benefits for treating tamoxifen or AI-resistant breast cancer, toxicity and development of drug resistance remain major obstacles (56). Furthermore, it highlights the need to debilitate multiple RTK-pathways in order to achieve efficient cytotoxic effects. Is it possible to destroy most, if not all, of the receptor tyrosine kinase-mediated pathways leading to endocrine resistance by targeting just one upstream gene? Our previous data provided evidence that HOXB7 is a direct, upstream regulator of the EGFR signaling pathway, and that long-term exposure to TAM can lead to activation of the HER2 signaling pathway (9). Through a series of studies deciphering the complexes formed at the HER2 enhancer between ER and HOXB7, we were able to provide a common mechanistic basis to the upregulation of HER2 seen in TMR and in HOXB7 overexpressing cells. Functionally, not only did knockdown of HOXB7 correlate with a decrease in HER2 expression, it also restored TAM sensitivity to TAM-resistant cell lines both in vitro and in vivo (Fig. 3). These observations identify HOXB7 as a critical molecule in the control of HER2 expression in TAM-resistance.
Searching for clinically feasible strategies for inhibiting HOXB7, we investigated potential upstream regulators of HOXB7, and found miR-196a, a potential negative regulator of HOXB7 expression. We showed that miR-196a binds directly to the HOXB7 3′UTR, resulting in downregulation of HOXB7 expression. Loss of miR-196a in TMR cells permits over expression of HOXB7 in TMR breast cancer models both in vitro and in vivo. Through several lines of experimentation, we have provided convincing evidence that HOXB7 expression is regulated by miR-196a (Fig. 4). While miRNAs constitute a highly attractive target for therapy, the field is in its infancy since there are yet many aspects of targeted delivery that need to be addressed before they become commonly used (57). The first microRNA currently in Phase 1 clinical trials is MRX34, a synthetic miRNA34 (a tumor suppressor downstream of p53) incorporated into liposomal nanoparticles, for liver cancer (58), while the most advanced is the anti-miR-122, complementary in sequence to miR-122, with a modified locked-nucleic acid structure (Miraversen), which is directed against the hepatitis C virus (59).
Probing this question further, we investigated the regulation of miR-196a expression and literature searches identified MYC as its upstream transcriptional regulator (46). This finding was particularly relevant because other studies have also suggested that MYC signaling could be a potential target in endocrine therapy (42, 60, 61). In this study, we show that increased stability of MYC protein, conferred by phosphorylation at Serine 62 residue in TAM-resistant cells, results in suppression of miR-196a expression and consequent promotion of HOXB7 expression. In support of this finding, through global mapping in silico of binding sites for MYC and additional ChIP analysis, we confirmed that MYC directly regulates miR-196a expression (Fig. 5). These findings provide a mechanistic explanation of how MYC can contribute to the increase of HOXB7 expression in models of TAM resistance.
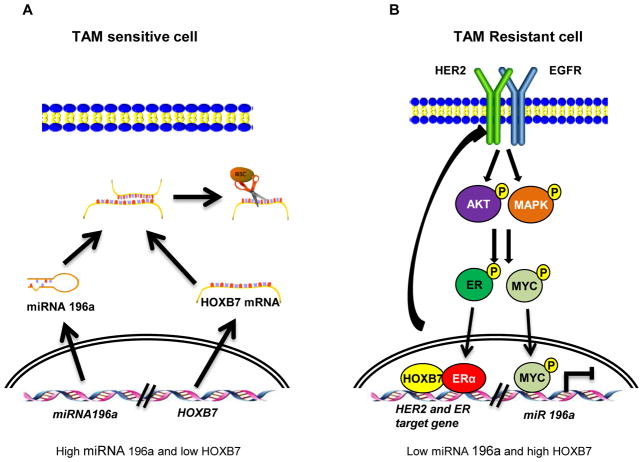
In summary, our mechanistic studies present evidence for amplification of a positive feedback loop where the interaction between HOXB7 and ER promotes activation of the HER2 signaling pathway and ER target gene expression including MYC. Activated HER2 signaling results in phosphorylation and greater stability of MYC, which suppresses miR-196a, permitting HOXB7 expression in tamoxifen-resistant breast cancer (Fig. 7A and 7B).
Figure 7. Model of HOXB7 interactions in tamoxifen resistance.
A, In TAM-sensitive ER-positive breast cancer, HOXB7 expression is suppressed by miR-196a. Conversely, in tamoxifen resistant cells B, increased MYC protein stability mediated by HER-signaling inhibits miR-196a expression, thus relieving its control of HOXB7 expression. Consequently, HOXB7 is upregulated, which promotes RTKs and ERα-induced gene expression in TAM-resistant breast cancer.
Through preclinical testing performed in the present study, we propose two potential therapeutic strategies to overcome hormone therapy resistance: First, downregulation of MYC activity by utilizing a MYC inhibitor. Several pharmacological approaches to inhibit MYC, drugs that interfere with MYC-MAX binding- 10058-F4; chromatin modification- JQ1; cell cycle-Dinaciclib; SUMOylation- Ginkgolic acid; and metabolism- BPTES, have been reported, but MYC-targeted therapeutic strategy has not been clinically successful in treating high-MYC expressing tumors (62). In the present study a combination of 10058-F4 and trastuzumab reduced TAM-resistant tumor burden by downregulating HER2 expression. Whether other MYC inhibitors have the same effects combined with trastuzumab in TAM-resistant breast cancer deserves attention. Second, enforced miR-196a expression could repress HOXB7 expression. Recently, it was reported that nanoparticle-mediated delivery of a dicer product (miRNA duplex) or mature miRNA (single stranded) as plasmid DNA was effective in silencing target gene expression. To carry miRNA, three different nanoparticles are used: i) Inorganic nanoparticle such as gold (Au), quantum dots, silicon oxide and iron oxide. ii) as liposomes such as DOTAP and DOTMA or iii) as polymers such PEI and PLGA. The advantage of the nanoparticle/miRNA complex is stability, resistance to degradation, and protection from its niche compared to naked miRNA (63). Although miRNAs have multiple targets, we have demonstrated that overexpressed miR-196a re-sensitizes TAM-resistant cells to TAM by inhibition of HOXB7 expression without much effect of cell proliferation. This implies that increasing miR-196a could be a specific therapeutic intervention for TAM-resistant breast cancer, suggesting that future studies could investigate effects of nanoparticle/miR-196a complexes in HER2-MYC-HOXB7 positive tumors. Finally, our studies showed that destruction of ER using fulvestrant (ICI 182,780) (64) reduced CA12 and MYC luciferase activity in MCF-7-HOXB7 cells (Supplementary Fig. S4F). In addition, fulvestrant inhibited growth of TMR cells and caused regression of MCF-7-HOXB7 xenografts (Supplementary Fig. S9D and S9E). Thus, fulvestrant, a well-established selective estrogen response modifier in the clinic, could antagonize these functions and provides an attractive and feasible alternative to targeting MYC or miRNAs.
Taken together, these studies contribute significantly to our understanding of the role of HOXB7 in TAM resistance in ER-positive breast cancer and provide critical insights as to how HOXB7 overexpression may be targeted therapeutically to prevent TAM-resistance.
METHODS
Additional details are provided in Supplementary Information online.
Cell Lines
MCF-7, MCF-7-HOXB7, MCF-7-TMR, MCF-7-EGFR, MCF-7-HER2, MCF-7-EGFR+HER2 (H12), BT474, BT474-HOXB7KO, T47D, T47D-HOXB7, ERIN, and ERIN-HOXB7 cells were used. MCF-7, BT474 and T47D cells were purchased from the ATCC (authenticated using STR profile analysis). Stable cell lines were not authenticated independently. Cells were cultured for a maximum of 4 weeks before thawing fresh, early passage cells. HOXB7, EGFR and HER2 status were confirmed by Western blot analysis and Real-Time RT-qPCR. All cells were confirmed to be Mycoplasma negative (Hoechst stain and PCR; tested in 2014). For deriving vector control and HOXB7-overexpressing cell lines, the pcDNA3 vector or pcDNA3-Flag-HOXB7 was stably transfected into MCF-7, T47D, and ERIN cells using Lipofectamine 2000 (Invitrogen, Life Technologies, Carlsbad, California, United States). Cell lines were maintained at 37°C in a 5% CO2 incubator. MCF-7 cells and derivatives were cultured in DMEM medium (Mediatech, Manassas, Virginia) supplemented with 10% heat-inactivated FBS (Mediatech), 100 IU/mL penicillin, and 100 μg/mL streptomycin (GIBCO, Life Technologies, Carlsbad, California). BT474 and T47D cells were cultured in RPMI medium 1640 supplemented with 10% heat-inactivated FBS, 100 IU/mL penicillin, and 100 μg/mL streptomycin. ERIN (MCF10A cells stably expressing ER) cells (kindly provided by Ben H. Park, JH) and derivatives were cultured in DMEM/F-12 medium supplemented with 5% horse serum (Invitrogen), 2 mM glutamine, 100 μg/mL streptomycin, 100 IU/mL penicillin, 0.25 μg/mL ampicillin B, 100 ng/mL cholera toxin, 20 ng/mL epidermal growth factor (EMD Millipore, Darmstadt, Germany), 0.5 μg/mL hydrocortisone (EMD Millipore), and 10 μg/mL insulin. Cell lines were maintained at 37°C in a 5% CO2 as described in the supplemental information.
Antibodies and Reagents
The antibodies used in this study were from the following sources: EGFR (Santa Cruz Biotechnology, Dallas, Texas), HER2 (Cell Signaling Technology, Danvers, Massachusetts), HER3 (Cell Signaling Technology), HER4 (Cell Signaling Technology), HOXB7 (Invitrogen and Abcam, Cambridge, Massachusetts), ERα (Santa Cruz Biotechnology), MYC (Abcam), phosphor-MYC-Threonine 58 (Applied Biological Materials Richmond, Canada), phosphor-MYC-Serine 62 (Abcam), BCL-2 (Cell Signaling Technology), CyclinD1 (Cell Signaling Technology), Actin (Sigma-Aldrich, St. Louis, Missouri), phospho-p44/42 MAPK (Cell Signaling Technology), phospho-AKT-serine and threonine (Cell Signaling Technology), phospho-p70 (Cell Signaling Technology), phosphor-4EBP1 (Cell Signaling Technology), Flag tag (Sigma), AIB1 (Abcam), NCOR (Abcam), SRC-1 (Abcam), HDAC (Abcam), H3R17-di-methyl (Abcam), p300 (Invitrogen), FOXA1 (Abcam), PBX-1(Santa Cruz Biotechnology), PBX-2 (Abcam), Meis-1 (Abcam), PAX-2 (Abcam), and RNA polymerase II (EMD Millipore). For in vitro assays, 4-hydroxytamoxifen (5 mg), β-estradiol (1 g), 10058-F4, and 10074-G5 (5 mg) were used and purchased from Sigma.
Co-immunoprecipitation
For co-immunoprecipitation from cell cultures, MCF-7 cells were transfected with 3 μg of each plasmid. After 48 hours, cells were washed with cold PBS and harvested in immunoprecipitation buffer (10 mM Tris-HCl, pH 7.4, 150 mM NaCl, 5 mM EDTA, 0.5% Nonidet P-40, Phospho-stop (Roche Diagnostics, Indianapolis, Indiana), and Complete Protease Inhibitor Mixture (Roche). The lysate was then rotated at 4°C for 1 h, followed by centrifugation at 14,000 rpm for 20 min. The supernatants were then combined with 50 μl of protein G Sepharose (GE Healthcare, Boston, Massachusetts) preincubated with antibodies against FLAG or ER or HOXB7, followed by rotating at 4°C for 2 h. The protein G Sepharose was pelleted and washed three times using immunoprecipitation buffer. The precipitates were resolved on SDS-PAGE gel and subjected to immunoblot analysis. Immunoblot signals were visualized with chemiluminescence (GE Healthcare). For mapping of the binding region between ER and HOXB7, Flag-tagged HOXB7 deletion constructs were co-transfected with full-length ER plasmid.
Luciferase Assay
MCF-7 or MCF-7-TAMR cells were transiently co-transfected with CA12 or 3′UTR region of HOXB7-luciferase construct plus pcDNA3.1-empty vector or pcDNA3.1-HOXB7 or HOXB7 shRNAs or miR-196a plus β-galactosidase (β-GAL) plasmid for normalization. Cells were harvested 24 hours post-transfection and lysates were assayed sequentially for luciferase and β-galactosidase activity, which were measured by following a protocol (Promega, Madison, Wisconsin). Assays were conducted in triplicate in a single experiment, and then as three independent experiments.
Chromatin Immunoprecipitation
Chromatin immunoprecipitation was carried out according to manufacturer’s instruction (EMD Millipore) with modifications. Briefly, cells were fixed with 1% formaldehyde for 10 min at room temperature. Glycerol quenched samples were lysed in 1 ml of SDS buffer containing protease inhibitors. The lysates were incubated for 10 min on ice and sonicated with a Covaris S220 (5% duty cycle, 4 intensity, 200 burst per cycle, 3 cycles of 60 sec) for 30 min on ice. The samples were centrifuged at 10,000 × g at 4°C for 10 min and supernatant was taken. With pre-cleared samples, Immunoprecipitation was performed by using antibodies and Dynabead-protein G. Eluates were subjected to reverse cross-linking and DNA was recovered by phenol-chloroform-ethanol purification. Q-PCR was performed using following primers described (24, 65).
Real-Time RT-qPCR
Total RNA was extracted with Trizol reagent (Invitrogen), and cDNA was synthesized from total RNA (2 μg) using an M-MLV Reverse Transcriptase (Promega). Aliquots of cDNA were used as templates for real-time RT-qPCR procedure. Relative quantitation of mRNA expression was achieved using real-time PCR (Applied Biosystem 7500 Real-Time PCR system, Applied Biosystems, Grand Island, New York). The Maxima SYBR Green/ROX Master Mix (Fermentas, Pittsburgh, Pennsylvania) was used according to the manufacturer’s instruction.
Immunoblotting
Cells were rinsed with a cold 1xPBS for three times and lysed with RIPA buffer. 40 μg of extracted lysate were vertically electrophoresed on 4–12% Bis-Tris NuPage Novex Gel in MOPS SDS running buffer (Invitrogen), then transferred to Hybond C Extra membrane (GE Healthcare). Membranes were stained with Ponceau stain to confirm protein transfer, then blocked with 4% powdered milk in PBS with 0.2% Tween-20 (PBST) for one hour at room temperature. Membranes were probed with primary antibody in 4% milk/PBST overnight, rinsed three times with 1X PBST for 5 minutes, then probed with secondary antibody (either anti-rabbit-HRP or anti-mouse-HRP (GE Healthcare) at 1:2000 dilution in 5% milk/PBST for 1 hour. Membranes were rinsed three times with 1X PBST for 5 minutes, and then treated with ECL Detection Reagent (GE Healthcare) for 1 minute. Membranes were exposed to Hyblot CL autoradiography film to determine protein expression. The quantification of protein level was performed by densitometric scanning and normalizing to intensity of actin.
Cell viability assay
Cells were plated at 2.5×103 cells per well in 96-well plates, in triplicate, with 200μL media, with drug treatments at stated concentrations, a combination, or vehicle for three days. 30 μL of MTT solution (5mg/mL in PBS) were added to each well and cells incubated for 2 hours. Media was then aspirated and cells resuspended in 200 μL DMSO. Absorbance at 560 nm was measured, with background at 670 nm subtracted. Triplicates were averaged for a mean absorbance, and then a percentage calculated of survival of drug-treated cells versus time-matched vehicle-treated cells. Experiments were performed in triplicate.
Microarray data analysis
Data was preprocessed using Illumina GenomeStudio where background level intensities were subtracted and quantile normalized. Data was exported into R Statistical Software where probes with p-detection value <0.05 were filtered out and further analysis was performed using the limma package from Bioconductor and custom functions where necessary. An unsupervised clustering heatmap was generated using the 1000 most variable probes across all samples. Differentially expressed probes, defined as probes with 2-fold change in expression and an FDR<0.05, were converted into Entrez gene identifiers and exported into Cytoscape where Gene Ontology analysis was performed using ClueGO with default settings. To understand the relationship between HOXB7 and ER-target genes, an established ER-target gene set was obtained from Dutertre et al. (18) and mapped to their respective probe IDs through Entrez Gene identifiers. Genes that were not represented by the array were discarded and the remaining genes were plotted on a supervised heatmap organized by ER-regulation status (up or down-regulated). The limma implementation of mean-rank gene set enrichment (MR-GSE) (17, 66) was used to identify the significance of enrichment of gene sets. MR-GSE uses the average ranks of t-statistics which reduces individual effects of individual genes and gives additional weight to gene sets with large number of active genes. Such implementation also allows for tests against individual gene sets. Moderated t-statistics obtained from limma comparing HOXB7 and vector controls were used as the input into MR-GSE and were tested for enrichment by up-regulated or down-regulated genes. The gene sets that were tested were the two estradiol response gene sets identified by Dutertre et al. (18) and a housekeeping gene set identified by Hsiao et al. (67) as a negative control. P-values were corrected using Bonferroni correction to obtain family-wise error rate (FWER) (68). The GEO accession number for the microarray data reported in this paper is GSE63607.
Mouse Xenograft Studies
All animal studies were approved by the Institutional Animal Care and Use Committee at Johns Hopkins University. For experiments described in Figure 3F, 6–8 week old female athymic nude mice were used, and study approved by Johns Hopkins Institutional Animal Care and Use Committee. MCF-7-HOXB7 cells were grown to 90% confluence, trypsinized, resuspended in serum free medium, and mixed 1:1 with Matrigel (BD Biosciences, Franklin Lakes, New Jersey). Estrogen pellets (60-d slow release pellet containing 0.72 mg) (Innovative Research of America, Sarasota, Florida) were implanted s.c. at the nape of the neck two days before 2 ×106 cancer cells were injected subcutaneously (s.c.) with 50 ul of 1:1 PBS/Matrigel (BD Biosciences) of nude or NSG mice. When the tumor size reached about 200 mm3, six mice in each group were treated with either 1) tamoxifen pellet implants s.c. (60-d slow release pellet containing 5 mg), 2) 20 mg/kg trastuzumab twice weekly, i.p., or 3) combination of tamoxifen and trastuzumab. For Figure 3D and 4F, tamoxifen pellet was implanted one week before 2 ×106 BT474-HOXB7KO or BT474-miR-196a or BT474-Vector control cells were injected into two mammary fat pads (mfp) of immunodeficient NSG mice with 50 ul of 1:1 PBS/Matrigel (BD Biosciences) using 3 mice per group. For Figure 5I, 2 ×106 BT474 cells were injected into the NSG mouse mammary fat pads (mfp) with 50 ul of 1:1 PBS/Matrigel (BD Biosciences). When the tumor size reached about 100 mm3, six mice in each group were treated with either 1) Vehicle, 2) 20 mg/kg trastuzumab twice weekly, i.p., 3) 30 mg/kg 10058-F4 (Selleckchem, Houston, Texas) i.p., daily, or 4) combination of trastuzumab and 10058-F4. Mice were measured weekly for tumor growth. After 6–8 weeks, mice were euthanized and tumors were sectioned and fast-frozen, or formalin-fixed and paraffin-embedded and H&E stained slides were made. Tumor volume was estimated by the calculation V=(length × width × height × 0.5236) mm3. The 10058-F4 was a kind gift of Selleckchem (Houston, Texas).
Statistical Analysis
HOXB7 expression levels determined by RT-PCR were dichotomized into low and high groups using the median as cutoff. Kaplan-Meier analysis was performed as described previously (69). Kaplan-Meier survival plot, and the hazard ratio and logrank P value were calculated and plotted using Winstat 2013 (R. Fitch Software, Germany). All statistical tests were two sided, and differences were considered statistically significant at P<0.05.
Supplementary Material
SIGNIFICANCE.
HOXB7 acts as an ERα cofactor regulating a myriad of ER-target genes, including HER2, in tamoxifen resistant breast cancer. HOXB7 expression is controlled by MYC via transcriptional regulation of the HOXB7 repressor, miRNA-196a; consequently, antagonists of MYC cause reversal of SERM-resistance both in vitro and in vivo.
Acknowledgments
Grant support: This work was supported by, Susan G. Komen Foundation Leadership Grant # SAC110050 (S.S.), the Department of Defense Center of Excellence-W81XWH-04-1-0595 (S.S.), the SKCCC Core grant P30 CA006973 (S.S.), Susan G. Komen Foundation Postdoctoral Fellowship KG101506 (K.J.), The OTKA K108655 Grant (B.G.), SAC110025 Komen Grant (H.N.) and the NIH R01HG006282 (H.J.).
We thank Dr. Ben H. Park for providing the ERIN cell line, Dr. Daniel Leahy for the full-length human HER2 plasmid, Dr. Care for the HOXB7 promoter luciferase plasmid, and Selleckchem for the MYC inhibitor, 10058-F4. We are immensely grateful to Dr. Zaver Bhujwalla for her constant support.
Footnotes
Disclosure of Potential Conflicts of Interest
All the authors declare no potential conflicts of interest
Authors’ Contributions
Conception and design: K. Jin, S. Sukumar
Development of methodology: K. Jin, S. Park, W.W. Teo, P. Korangath, T. Yoshida, L. Cruz, S.Sukumar
Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): K. Jin, S. Park, P. Korangath, T. Yoshida, Z. Wu, Z. Tao, S. Sukumar
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): W. Zhou, H. Ji, B. Gyorffy, C.P. Goswami, H. Nakshatri, S.S. Cho
Writing, review, and/or revision of the manuscript: K. Jin, S. Sukumar
Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): K. Jin, S. Sukumar, Y. Su, M. Ekram, K. Polyak
Study supervision: S. Sukumar
References
- 1.Frasor J, Stossi F, Danes JM, Komm B, Lyttle CR, Katzenellenbogen BS. Selective estrogen receptor modulators: discrimination of agonistic versus antagonistic activities by gene expression profiling in breast cancer cells. Cancer research. 2004;64:1522–33. doi: 10.1158/0008-5472.can-03-3326. [DOI] [PubMed] [Google Scholar]
- 2.Jordan VC, O’Malley BW. Selective estrogen-receptor modulators and antihormonal resistance in breast cancer. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2007;25:5815–24. doi: 10.1200/JCO.2007.11.3886. [DOI] [PubMed] [Google Scholar]
- 3.Jordan VC, Brodie AM. Development and evolution of therapies targeted to the estrogen receptor for the treatment and prevention of breast cancer. Steroids. 2007;72:7–25. doi: 10.1016/j.steroids.2006.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dowsett M, Nicholson RI, Pietras RJ. Biological characteristics of the pure antiestrogen fulvestrant: overcoming endocrine resistance. Breast cancer research and treatment. 2005;93 (Suppl 1):S11–8. doi: 10.1007/s10549-005-9037-3. [DOI] [PubMed] [Google Scholar]
- 5.Brown RJ, Davidson NE. Adjuvant hormonal therapy for premenopausal women with breast cancer. Seminars in oncology. 2006;33:657–63. doi: 10.1053/j.seminoncol.2006.08.014. [DOI] [PubMed] [Google Scholar]
- 6.Gutierrez MC, Detre S, Johnston S, Mohsin SK, Shou J, Allred DC, et al. Molecular changes in tamoxifen-resistant breast cancer: relationship between estrogen receptor, HER-2, and p38 mitogen-activated protein kinase. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2005;23:2469–76. doi: 10.1200/JCO.2005.01.172. [DOI] [PubMed] [Google Scholar]
- 7.Lipton A, Leitzel K, Ali SM, Demers L, Harvey HA, Chaudri-Ross HA, et al. Serum HER-2/neu conversion to positive at the time of disease progression in patients with breast carcinoma on hormone therapy. Cancer. 2005;104:257–63. doi: 10.1002/cncr.21202. [DOI] [PubMed] [Google Scholar]
- 8.Meng S, Tripathy D, Shete S, Ashfaq R, Haley B, Perkins S, et al. HER-2 gene amplification can be acquired as breast cancer progresses. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:9393–8. doi: 10.1073/pnas.0402993101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jin K, Kong X, Shah T, Penet MF, Wildes F, Sgroi DC, et al. The HOXB7 protein renders breast cancer cells resistant to tamoxifen through activation of the EGFR pathway. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:2736–41. doi: 10.1073/pnas.1018859108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McIlroy M, McCartan D, Early S, POG, Pennington S, Hill AD, et al. Interaction of developmental transcription factor HOXC11 with steroid receptor coactivator SRC-1 mediates resistance to endocrine therapy in breast cancer [corrected] Cancer research. 2010;70:1585–94. doi: 10.1158/0008-5472.CAN-09-3713. [DOI] [PubMed] [Google Scholar]
- 11.Shah N, Jin K, Cruz LA, Park S, Sadik H, Cho S, et al. HOXB13 mediates tamoxifen resistance and invasiveness in human breast cancer by suppressing ERalpha and inducing IL-6 expression. Cancer research. 2013;73:5449–58. doi: 10.1158/0008-5472.CAN-13-1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pathiraja TN, Nayak SR, Xi Y, Jiang S, Garee JP, Edwards DP, et al. Epigenetic reprogramming of HOXC10 in endocrine-resistant breast cancer. Sci Transl Med. 2014;6:229ra41. doi: 10.1126/scitranslmed.3008326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Magnani L, Stoeck A, Zhang X, Lanczky A, Mirabella AC, Wang TL, et al. Genome-wide reprogramming of the chromatin landscape underlies endocrine therapy resistance in breast cancer. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:E1490–9. doi: 10.1073/pnas.1219992110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnston SR, Lu B, Scott GK, Kushner PJ, Smith IE, Dowsett M, et al. Increased activator protein-1 DNA binding and c-Jun NH2-terminal kinase activity in human breast tumors with acquired tamoxifen resistance. Clinical cancer research : an official journal of the American Association for Cancer Research. 1999;5:251–6. [PubMed] [Google Scholar]
- 15.Schiff R, Reddy P, Ahotupa M, Coronado-Heinsohn E, Grim M, Hilsenbeck SG, et al. Oxidative stress and AP-1 activity in tamoxifen-resistant breast tumors in vivo. Journal of the National Cancer Institute. 2000;92:1926–34. doi: 10.1093/jnci/92.23.1926. [DOI] [PubMed] [Google Scholar]
- 16.Zhou Y, Yau C, Gray JW, Chew K, Dairkee SH, Moore DH, et al. Enhanced NF kappa B and AP-1 transcriptional activity associated with antiestrogen resistant breast cancer. BMC Cancer. 2007;7:59. doi: 10.1186/1471-2407-7-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Michaud J, Simpson KM, Escher R, Buchet-Poyau K, Beissbarth T, Carmichael C, et al. Integrative analysis of RUNX1 downstream pathways and target genes. BMC Genomics. 2008;9:363. doi: 10.1186/1471-2164-9-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dutertre M, Gratadou L, Dardenne E, Germann S, Samaan S, Lidereau R, et al. Estrogen regulation and physiopathologic significance of alternative promoters in breast cancer. Cancer research. 2010;70:3760–70. doi: 10.1158/0008-5472.CAN-09-3988. [DOI] [PubMed] [Google Scholar]
- 19.Wu G, Yustein JT, McCall MN, Zilliox M, Irizarry RA, Zeller K, et al. ChIP-PED enhances the analysis of ChIP-seq and ChIP-chip data. Bioinformatics (Oxford, England) 2013;29:1182–9. doi: 10.1093/bioinformatics/btt108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carroll JS, Meyer CA, Song J, Li W, Geistlinger TR, Eeckhoute J, et al. Genome-wide analysis of estrogen receptor binding sites. Nature genetics. 2006;38:1289–97. doi: 10.1038/ng1901. [DOI] [PubMed] [Google Scholar]
- 21.Fullwood MJ, Liu MH, Pan YF, Liu J, Xu H, Mohamed YB, et al. An oestrogen-receptor-alpha-bound human chromatin interactome. Nature. 2009;462:58–64. doi: 10.1038/nature08497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hsu PY, Hsu HK, Lan X, Juan L, Yan PS, Labanowska J, et al. Amplification of distant estrogen response elements deregulates target genes associated with tamoxifen resistance in breast cancer. Cancer cell. 2013;24:197–212. doi: 10.1016/j.ccr.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sanyal A, Lajoie BR, Jain G, Dekker J. The long-range interaction landscape of gene promoters. Nature. 2012;489:109–13. doi: 10.1038/nature11279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hurtado A, Holmes KA, Geistlinger TR, Hutcheson IR, Nicholson RI, Brown M, et al. Regulation of ERBB2 by oestrogen receptor-PAX2 determines response to tamoxifen. Nature. 2008;456:663–6. doi: 10.1038/nature07483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Magnani L, Ballantyne EB, Zhang X, Lupien M. PBX1 genomic pioneer function drives ERalpha signaling underlying progression in breast cancer. PLoS Genet. 2011;7:e1002368. doi: 10.1371/journal.pgen.1002368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stanya KJ, Kao HY. New insights into the functions and regulation of the transcriptional corepressors SMRT and N-CoR. Cell Div. 2009;4:7. doi: 10.1186/1747-1028-4-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang C, Mayer JA, Mazumdar A, Fertuck K, Kim H, Brown M, et al. Estrogen induces c-myc gene expression via an upstream enhancer activated by the estrogen receptor and the AP-1 transcription factor. Molecular endocrinology. 2011;25:1527–38. doi: 10.1210/me.2011-1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kun Y, How LC, Hoon TP, Bajic VB, Lam TS, Aggarwal A, et al. Classifying the estrogen receptor status of breast cancers by expression profiles reveals a poor prognosis subpopulation exhibiting high expression of the ERBB2 receptor. Hum Mol Genet. 2003;12:3245–58. doi: 10.1093/hmg/ddg347. [DOI] [PubMed] [Google Scholar]
- 29.Shou J, Massarweh S, Osborne CK, Wakeling AE, Ali S, Weiss H, et al. Mechanisms of tamoxifen resistance: increased estrogen receptor-HER2/neu cross-talk in ER/HER2-positive breast cancer. Journal of the National Cancer Institute. 2004;96:926–35. doi: 10.1093/jnci/djh166. [DOI] [PubMed] [Google Scholar]
- 30.Chen H, Lee JS, Liang X, Zhang H, Zhu T, Zhang Z, et al. Hoxb7 inhibits transgenic HER-2/neu-induced mouse mammary tumor onset but promotes progression and lung metastasis. Cancer research. 2008;68:3637–44. doi: 10.1158/0008-5472.CAN-07-2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang YC, Morrison G, Gillihan R, Guo J, Ward RM, Fu X, et al. Different mechanisms for resistance to trastuzumab versus lapatinib in HER2-positive breast cancers--role of estrogen receptor and HER2 reactivation. Breast Cancer Res. 2011;13:R121. doi: 10.1186/bcr3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rimawi MF, Wiechmann LS, Wang YC, Huang C, Migliaccio I, Wu MF, et al. Reduced dose and intermittent treatment with lapatinib and trastuzumab for potent blockade of the HER pathway in HER2/neu-overexpressing breast tumor xenografts. Clin Cancer Res. 2011;17:1351–61. doi: 10.1158/1078-0432.CCR-10-1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Deblois G, Hall JA, Perry MC, Laganiere J, Ghahremani M, Park M, et al. Genome-wide identification of direct target genes implicates estrogen-related receptor alpha as a determinant of breast cancer heterogeneity. Cancer research. 2009;69:6149–57. doi: 10.1158/0008-5472.CAN-09-1251. [DOI] [PubMed] [Google Scholar]
- 34.Braig S, Mueller DW, Rothhammer T, Bosserhoff AK. MicroRNA miR-196a is a central regulator of HOX-B7 and BMP4 expression in malignant melanoma. Cellular and molecular life sciences : CMLS. 2010;67:3535–48. doi: 10.1007/s00018-010-0394-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Manavalan TT, Teng Y, Appana SN, Datta S, Kalbfleisch TS, Li Y, et al. Differential expression of microRNA expression in tamoxifen-sensitive MCF-7 versus tamoxifen-resistant LY2 human breast cancer cells. Cancer letters. 2011;313:26–43. doi: 10.1016/j.canlet.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sun M, Liu XH, Li JH, Yang JS, Zhang EB, Yin DD, et al. MiR-196a is upregulated in gastric cancer and promotes cell proliferation by downregulating p27(kip1) Mol Cancer Ther. 2012;11:842–52. doi: 10.1158/1535-7163.MCT-11-1015. [DOI] [PubMed] [Google Scholar]
- 37.Piao L, Zhang M, Datta J, Xie X, Su T, Li H, et al. Lipid-based nanoparticle delivery of Pre-miR-107 inhibits the tumorigenicity of head and neck squamous cell carcinoma. Molecular therapy : the journal of the American Society of Gene Therapy. 2012;20:1261–9. doi: 10.1038/mt.2012.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Adams BD, Kasinski AL, Slack FJ. Aberrant Regulation and Function of MicroRNAs in Cancer. Current biology : CB. 2014;24:R762–R76. doi: 10.1016/j.cub.2014.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gregory MA, Qi Y, Hann SR. Phosphorylation by glycogen synthase kinase-3 controls c-myc proteolysis and subnuclear localization. The Journal of biological chemistry. 2003;278:51606–12. doi: 10.1074/jbc.M310722200. [DOI] [PubMed] [Google Scholar]
- 40.Sears R, Nuckolls F, Haura E, Taya Y, Tamai K, Nevins JR. Multiple Ras-dependent phosphorylation pathways regulate Myc protein stability. Genes & development. 2000;14:2501–14. doi: 10.1101/gad.836800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yeh E, Cunningham M, Arnold H, Chasse D, Monteith T, Ivaldi G, et al. A signalling pathway controlling c-Myc degradation that impacts oncogenic transformation of human cells. Nature cell biology. 2004;6:308–18. doi: 10.1038/ncb1110. [DOI] [PubMed] [Google Scholar]
- 42.Miller TW, Balko JM, Ghazoui Z, Dunbier A, Anderson H, Dowsett M, et al. A gene expression signature from human breast cancer cells with acquired hormone independence identifies MYC as a mediator of antiestrogen resistance. Clinical cancer research : an official journal of the American Association for Cancer Research. 2011;17:2024–34. doi: 10.1158/1078-0432.CCR-10-2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang Z, Lin S, Li JJ, Xu Z, Yao H, Zhu X, et al. MYC protein inhibits transcription of the microRNA cluster MC-let-7a-1~let-7d via noncanonical E-box. The Journal of biological chemistry. 2011;286:39703–14. doi: 10.1074/jbc.M111.293126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Welcker M, Orian A, Jin J, Grim JE, Harper JW, Eisenman RN, et al. The Fbw7 tumor suppressor regulates glycogen synthase kinase 3 phosphorylation-dependent c-Myc protein degradation. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:9085–90. doi: 10.1073/pnas.0402770101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guo J, Parise RA, Joseph E, Egorin MJ, Lazo JS, Prochownik EV, et al. Efficacy, pharmacokinetics, tisssue distribution, and metabolism of the Myc-Max disruptor, 10058-F4 [Z,E]-5-[4-ethylbenzylidine]-2-thioxothiazolidin-4-one, in mice. Cancer Chemother Pharmacol. 2009;63:615–25. doi: 10.1007/s00280-008-0774-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zeller KI, Zhao X, Lee CW, Chiu KP, Yao F, Yustein JT, et al. Global mapping of c-Myc binding sites and target gene networks in human B cells. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:17834–9. doi: 10.1073/pnas.0604129103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Muller I, Larsson K, Frenzel A, Oliynyk G, Zirath H, Prochownik EV, et al. Targeting of the MYCN protein with small molecule c-MYC inhibitors. PLoS One. 2014;9:e97285. doi: 10.1371/journal.pone.0097285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jordan VC. The science of selective estrogen receptor modulators: concept to clinical practice. Clinical cancer research : an official journal of the American Association for Cancer Research. 2006;12:5010–3. doi: 10.1158/1078-0432.CCR-06-1136. [DOI] [PubMed] [Google Scholar]
- 49.Osborne CK, Schiff R. Mechanisms of endocrine resistance in breast cancer. Annual review of medicine. 2011;62:233–47. doi: 10.1146/annurev-med-070909-182917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Osborne CK, Neven P, Dirix LY, Mackey JR, Robert J, Underhill C, et al. Gefitinib or placebo in combination with tamoxifen in patients with hormone receptor-positive metastatic breast cancer: a randomized phase II study. Clinical cancer research : an official journal of the American Association for Cancer Research. 2011;17:1147–59. doi: 10.1158/1078-0432.CCR-10-1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Carter P, Presta L, Gorman CM, Ridgway JB, Henner D, Wong WL, et al. Humanization of an anti-p185HER2 antibody for human cancer therapy. Proceedings of the National Academy of Sciences of the United States of America. 1992;89:4285–9. doi: 10.1073/pnas.89.10.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hudis CA. Trastuzumab--mechanism of action and use in clinical practice. The New England journal of medicine. 2007;357:39–51. doi: 10.1056/NEJMra043186. [DOI] [PubMed] [Google Scholar]
- 53.Xia W, Gerard CM, Liu L, Baudson NM, Ory TL, Spector NL. Combining lapatinib (GW572016), a small molecule inhibitor of ErbB1 and ErbB2 tyrosine kinases, with therapeutic anti-ErbB2 antibodies enhances apoptosis of ErbB2-overexpressing breast cancer cells. Oncogene. 2005;24:6213–21. doi: 10.1038/sj.onc.1208774. [DOI] [PubMed] [Google Scholar]
- 54.Xia W, Mullin RJ, Keith BR, Liu LH, Ma H, Rusnak DW, et al. Anti-tumor activity of GW572016: a dual tyrosine kinase inhibitor blocks EGF activation of EGFR/erbB2 and downstream Erk1/2 and AKT pathways. Oncogene. 2002;21:6255–63. doi: 10.1038/sj.onc.1205794. [DOI] [PubMed] [Google Scholar]
- 55.Baselga J, Campone M, Piccart M, Burris HA, 3rd, Rugo HS, Sahmoud T, et al. Everolimus in postmenopausal hormone-receptor-positive advanced breast cancer. The New England journal of medicine. 2012;366:520–9. doi: 10.1056/NEJMoa1109653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Potente M, Gerhardt H, Carmeliet P. Basic and therapeutic aspects of angiogenesis. Cell. 2011;146:873–87. doi: 10.1016/j.cell.2011.08.039. [DOI] [PubMed] [Google Scholar]
- 57.Hayes J, Peruzzi PP, Lawler S. MicroRNAs in cancer: biomarkers, functions and therapy. Trends in molecular medicine. 2014;20:460–9. doi: 10.1016/j.molmed.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 58.Bouchie A. First microRNA mimic enters clinic. Nat Biotechnol. 2013;31:577. doi: 10.1038/nbt0713-577. [DOI] [PubMed] [Google Scholar]
- 59.Gebert LF, Rebhan MA, Crivelli SE, Denzler R, Stoffel M, Hall J. Miravirsen (SPC3649) can inhibit the biogenesis of miR-122. Nucleic acids research. 2014;42:609–21. doi: 10.1093/nar/gkt852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mukherjee S, Conrad SE. c-Myc suppresses p21WAF1/CIP1 expression during estrogen signaling and antiestrogen resistance in human breast cancer cells. The Journal of biological chemistry. 2005;280:17617–25. doi: 10.1074/jbc.M502278200. [DOI] [PubMed] [Google Scholar]
- 61.Feng Q, Zhang Z, Shea MJ, Creighton CJ, Coarfa C, Hilsenbeck SG, et al. An epigenomic approach to therapy for tamoxifen-resistant breast cancer. Cell Res. 2014;24:809–19. doi: 10.1038/cr.2014.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Horiuchi D, Anderton B, Goga A. Taking on challenging targets: making MYC druggable. Am Soc Clin Oncol Educ Book. 2014:e497–502. doi: 10.14694/EdBook_AM.2014.34.e497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Muthiah M, Park IK, Cho CS. Nanoparticle-mediated delivery of therapeutic genes: focus on miRNA therapeutics. Expert opinion on drug delivery. 2013;10:1259–73. doi: 10.1517/17425247.2013.798640. [DOI] [PubMed] [Google Scholar]
- 64.Macedo LF, Sabnis GJ, Goloubeva OG, Brodie A. Combination of anastrozole with fulvestrant in the intratumoral aromatase xenograft model. Cancer research. 2008;68:3516–22. doi: 10.1158/0008-5472.CAN-07-6807. [DOI] [PubMed] [Google Scholar]
- 65.Hurtado A, Holmes KA, Ross-Innes CS, Schmidt D, Carroll JS. FOXA1 is a key determinant of estrogen receptor function and endocrine response. Nature genetics. 2011;43:27–33. doi: 10.1038/ng.730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hsiao LL, Dangond F, Yoshida T, Hong R, Jensen RV, Misra J, et al. A compendium of gene expression in normal human tissues. Physiological genomics. 2001;7:97–104. doi: 10.1152/physiolgenomics.00040.2001. [DOI] [PubMed] [Google Scholar]
- 67.Hsiao LL, Dangond F, Yoshida T, Hong R, Jensen RV, Misra J, et al. A compendium of gene expression in normal human tissues. Physiol Genomics. 2001;7:97–104. doi: 10.1152/physiolgenomics.00040.2001. [DOI] [PubMed] [Google Scholar]
- 68.Bland JM, Altman DG. Multiple significance tests: the Bonferroni method. BMJ. 1995;310:170. doi: 10.1136/bmj.310.6973.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gyorffy B, Lanczky A, Szallasi Z. Implementing an online tool for genome-wide validation of survival-associated biomarkers in ovarian-cancer using microarray data from 1287 patients. Endocrine-related cancer. 2012;19:197–208. doi: 10.1530/ERC-11-0329. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.