Abstract
Mycobacterium tuberculosis (Mtb) induces necrosis of infected cells to evade immune responses. Recently, we found that Mtb utilizes the protein CpnT to kill human macrophages by secreting its C-terminal domain, named tuberculosis necrotizing toxin (TNT) that induces necrosis by an unknown mechanism. Here we show that TNT gains access to the cytosol of Mtb-infected macrophages, where it hydrolyzes the essential co-enzyme nicotinamide adenine dinucleotide (NAD+). Expression or injection of a non-catalytic TNT mutant showed no cytotoxicity in macrophages or zebrafish zygotes, respectively, demonstrating that the NAD+-glycohydrolase activity is required for TNT-induced cell death. To prevent self-poisoning, Mtb produces an immunity factor for TNT (IFT) that binds TNT and inhibits its activity. The crystal structure of the TNT-IFT complex revealed a novel NAD+-glycohydrolase fold of TNT, which constitutes the founding member of a toxin family wide-spread in pathogenic microorganisms.
INTRODUCTION
Tuberculosis is a highly infectious disease and a global health threat. Survival within macrophages is a key feature of Mycobacterium tuberculosis (Mtb) pathogenesis and is crucial to a persistent infection in the human host1. The battle between Mtb and the human immune system to control the fate of infected macrophages is critical in determining the outcome of the infection2. The ability to control the timing and mode of host cell death plays a pivotal role in many microbial infections3. Virulent Mtb induces necrosis of infected cells and suppresses host cell apoptosis to evade immune responses and disseminate4,5. Killing of infected macrophages by necrosis is dependent on the ESX-1 secretion system and has been attributed to the membrane activity of ESAT-66. In contrast, apoptosis of infected macrophages is induced by the host immune system to control the bacterial infection. Subsequently, apoptotic bodies containing Mtb are scavenged by other activated macrophages, or taken up by dendritic cells, to facilitate the priming of antigen-specific T cells to stimulate adaptive immunity4. Thus the fate of infected macrophages profoundly affects host resistance to Mtb2.
The molecular mechanisms by which Mtb induces macrophage necrosis are largely unknown7,8. Many bacterial pathogens utilize toxic proteins to kill host cells9. The lack of any protein in the Mtb genome with homologs of known bacterial toxins and the failure to isolate secreted proteins with strong toxicity against host cells led to the widespread belief that Mtb does not encode these classical virulence factors10-12. However, this paradigm was challenged by our discovery of the outer membrane protein CpnT as the main cytotoxicity factor of Mtb in macrophages13. CpnT is utilized by Mtb to secrete its toxic C-terminal domain (residues 651 – 846), which is sufficient to induce necrotic death in host cells by an unknown mechanism13. Hence, we named the secreted C-terminal domain of CpnT as TNT (tuberculosis necrotizing toxin).
In this study, we set out to identify the mechanism of TNT-induced necrotic cell death at the functional and structural level. We demonstrate that TNT possesses robust NAD+-glycohydrolase activity, which depletes cellular NAD+ pools resulting in host cell death. TNT mutants with lower or abrogated NAD+-glycohydrolase activity, show reduced or no cytotoxicity, respectively, in macrophages and in zebrafish establishing a link between the enzymatic activity and toxicity of TNT. We further identify an endogenous Mtb protein which acts as an anti-toxin for TNT to prevent self-poisoning. The crystal structure of TNT in complex with its anti-toxin shows a novel NAD+ binding and hydrolysis module distinct from that of known NAD+-utilizing toxins.
RESULTS
TNT hydrolyzes NAD+
Recombinant expression of tnt (3′ end of rv3903c encoding the secreted C-terminal domain) is toxic in all tested prokaryotic and eukaryotic cells, suggesting a common cellular target. Deep sequencing of E. coli RNA revealed that tnt expression induced nadA and nadB transcription by 16- and 44-fold, respectively (Supplementary Fig. 1a). These genes encode key enzymes in nicotinamide adenine dinucleotide (NAD+) biosynthesis suggesting a link between TNT and NAD+. Indeed, tnt expression completely depleted NAD+ in E. coli and Jurkat T-cells (Supplementary Fig. 1b), indicating an involvement of TNT in degradation of cellular NAD+. Purified recombinant TNT (Supplementary Fig. 2) hydrolyzed NAD+ in vitro and was inhibited by a TNT-specific antibody demonstrating that TNT degrades NAD+ (Fig. 1a). NAD+ is an essential coenzyme in many redox reactions as well as a substrate for NAD+-consuming enzymes14 that play important roles in transcriptional regulation and longevity15. Depletion of cytosolic NAD+ compromises ATP generation by glycolysis and leads to necrotic cell death16,17. Thus, NAD+ hydrolysis might explain TNT’s cytotoxicity.
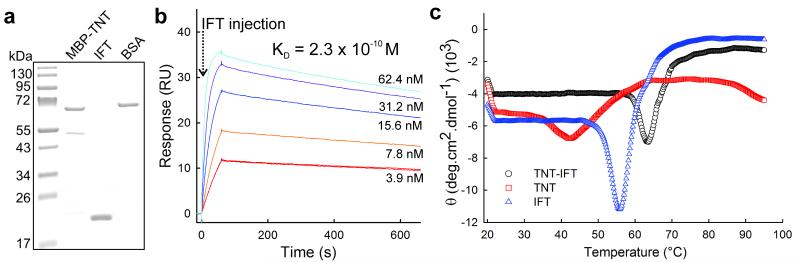
Figure 1. TNT is a novel β-NAD+-glycohydrolase of M. tuberculosis.
(a) NAD+-glycohydrolase assay for wt TNT in the absence and presence of TNT antibody (400 ng) or IFT (100 ng).(b) The kinetics of NAD+ hydrolysis by TNT (10 nM) was determined using a fluorometric assay at pH 7.4. The values of Km and Vmax were determined by non-linear regression analysis using the Michaelis-Menten equation. (c) NAD+-glycohydrolase activity of TNT (10 nM) after heating to 65°C or 95°C for 5 minutes. The initial velocity v0 of NAD+ hydrolysis was determined at a substrate concentration of 200 μM. All data in this figure represent the means ± s.e.m. of three independent experiments.
All known NAD+-degrading enzymes release nicotinamide and a second reaction product that varies with the specific hydrolytic mechanism. NAD+-glycohydrolases and ADP-ribosyl-cyclases produce ADP-ribose and cyclic ADP-ribose, respectively, while ADP-ribosyltransferases attach ADP-ribose to a specific target protein. Thin layer chromatography revealed that TNT is a glycohydrolase that efficiently cleaves NAD+ into nicotinamide and ADP-ribose in vitro in the absence of any other protein (Supplementary Fig. 1c). A kinetic analysis of NAD+ hydrolysis by TNT (Fig. 1b, Supplementary Fig. 3a, b) yielded a Michaelis constant Km of 614 ± 43 μM, near the physiological NAD+ concentration of ~500-600 μM in human cells18, a turnover number kcat of 52 s−1 and a catalytic efficiency kcat/Km of 8.4 × 104 M−1 s−1 (Fig. 1b, Supplementary Table 1). TNT has maximal activity at pH 6.5, but is only marginally active at pH 5.5 (Supplementary Fig. 3c) suggesting that TNT is not active in a mature phagolysosome19. TNT is also remarkably heat-stable retaining approximately 50% of its activity after prolonged heating at 95°C (Fig. 1c). The pH dependency and heat stability of TNT match the uncharacterized NAD+-glycohydrolase activity identified in Mtb cell extracts half a century ago20,21.
The Mtb protein Rv3902c binds to and inactivates TNT
The only other known bacterial NAD+-glycohydrolase is the secreted toxin SPN of Streptococcus pyogenes22. S. pyogenes protects itself against the toxicity of SPN by producing the immunity factor IFS which is encoded in the spn operon, and tightly binds to and inactivates SPN23. However, SPN and IFS have no sequence similarities to any Mtb protein. Since tnt expression is toxic in Mtb (not shown), we reasoned that rv3902c, the last gene in the cpnT operon, might encode an antitoxin to TNT. Indeed, equimolar amounts of purified recombinant Rv3902c protein completely inhibited TNT NAD+-glycohydrolase activity (Fig. 1a). Surface plasmon resonance (SPR) experiments using purified MBP-TNT and Rv3902c proteins (Fig. 2a) revealed that the two proteins form a stable complex with a dissociation constant KD of 2.3 × 10−10 M (Fig. 2b). Remarkably, an uncharacterized heat-labile “inhibitor” of the Mtb NAD+-glycohydrolase activity was also described in the 1960’s24,25. In agreement with this report, folding studies revealed that a complex of Rv3902c with TNT denatured irreversibly with an apparent melting temperature of ~ 65.4 °C, leading to dramatic conformational changes of the TNT-IFT complex and liberating TNT (Fig. 2c and Supplementary Fig. 2c). Taken together, these experiments demonstrate that Rv3902c inhibits the NAD+-glycohydrolase activity of TNT by complex formation and constitutes the first inhibitor of a secreted toxin in Mtb. Hence, we named Rv3902c as Immunity Factor for TNT (IFT).
Figure 2. M. tuberculosis produces the endogenous immunity factor IFT that binds to and inactivates TNT.
(a) Coomassie-stained SDS-polyacrylamide gel with purified MBP-TNT and IFT used for SPR interaction assays. (b) Surface plasmon resonance analysis of the interaction between TNT and IFT. Sensorgrams show the concentration-dependent kinetic analysis of two-fold serial dilutions of IFT (0 - 62.5 nM). (c) Secondary structure changes of TNT, IFT and TNT-IFT monitored by the ellipticity θ at 222 nm as a function of temperature. The apparent transition temperatures were ~53, 58 and 65.4°C, respectively.
The atomic structure of the TNT-IFT complex
The discovery of IFT enabled us to purify milligram quantities of the TNT-IFT complex from E. coli after co-expression of the corresponding genes (Supplementary Fig. 2). We crystallized the TNT-IFT complex at physiological pH and determined its structure to a Rwork/Rfree of 12.8/14.3%, at 1.1 Å resolution (Table 1). The TNT-IFT complex adopts a blocky conformation of 50×50×35 Å3 that resembles a ‘ball-in-a-hand’ (Fig. 3a). IFT (‘ball’) fills a deep cavity in TNT (‘hand’) that buries ~20% of the complex solvent-accessible area (~3,490 Å2). Consistent with the picomolar KD measured by SPR, the extensive binding interface between TNT and IFT involves approximately one third of the residues in each protein and results in over 50, mainly electrostatic, contacts. Strikingly, both TNT and IFT have minimal internal hydrophobic cores and as much as 97% of their residues are exposed on the structure surface, resulting in huge predicted solvation energies (ΔGf = −157 and −163 kcal/mol for TNT and IFT, respectively).
Table 1.
Crystallographic data collection and refinement statistics.
| TNT-IFT Crystal form 1 |
TNT-IFT Crystal form 2 |
|
|---|---|---|
| Data collection | ||
| Space group | P212121 | P212121 |
| Cell dimensions | ||
| a, b, c (Å) | 70.6, 86.2, 62.6 | 75.3, 85.3, 63.2 |
| α, β, γ (°) | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 |
| Wavelength (Å) | 0.97 | 0.97 |
| Resolution (Å) | 20-1.10 (1.14-1.10) | 20-1.9 (1.97-1.90) |
| R sym | 5.7 (50.2) | 7.2 (33.5) |
| I / σI | 29.5 (4.5) | 66.6 (10.8) |
| Completeness (%) | 90.8 (55.5) | 99.5 (93.3) |
| Redundancy | 12.6 (8.8) | 13.2 (9.7) |
| Refinement | ||
| Resolution (Å) | 1.0-1.1 | 20-1.9 |
| No. reflections | 133,725 | 32,610 |
| Rwork / Rfree | 12.8 / 14.3 | 17.0 / 20.8 |
| No. atoms | ||
| Protein | 2,990 | 2,889 |
| Water | 506 | 461 |
| B-factors (Å2) | ||
| Protein (TNT/IFT) | 13.6 / 15.2 | 25.5 / 26.4 |
| Water | 29.9 | 36.1 |
| R.m.s deviations | ||
| Bond lengths (Å) | 0.006 | 0.008 |
| Bond angles (°) | 1.09 | 1.19 |
Values in parentheses are for highest-resolution shells.
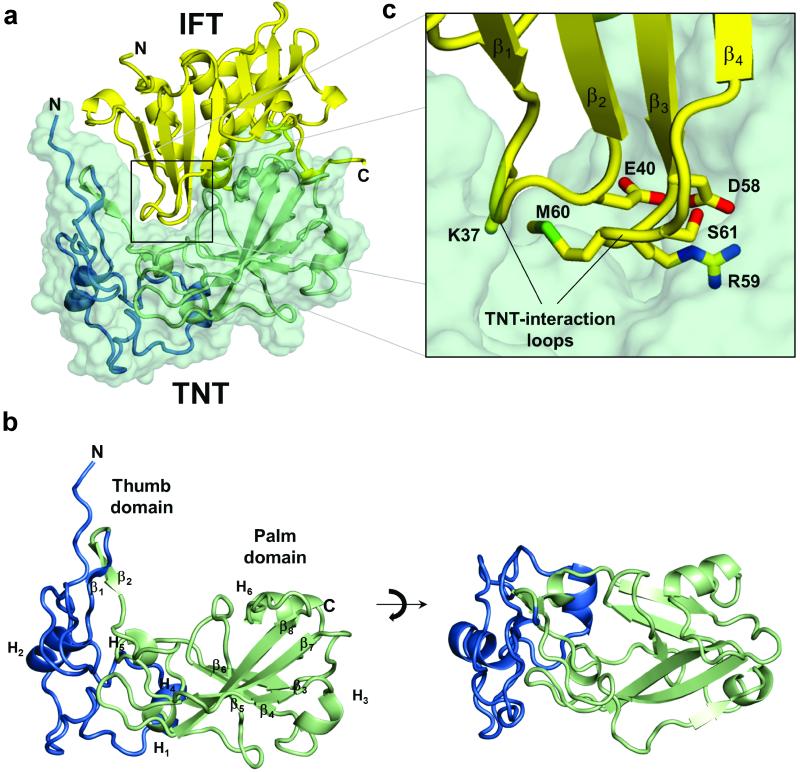
Figure 3. Atomic structure of the TNT-IFT complex.
(a) Ribbon representation of TNT (overlaid to a green semi-transparent solvent/protein contact surface) bound to IFT (in yellow). (b) Ribbon representation of free TNT with the thumb-domain (residues 648-719) and the palm-domain (residues 720-846), colored in blue and green, respectively. (c) Magnified view of TNT-IFT binding interface. IFT side chains projecting from TNT-interaction loops inside TNT crevice are shown as sticks.
The TNT structure consists of two regions: a ‘thumb’ (residues 648-736) and a ‘palm-domain’ (residues 747-846) (Fig. 3b). Three-quarters of the residues in the thumb domain adopt a random coiled conformation interrupted by two shorts α-helices (α1, α2) and two antiparallel β-strands (β3, β4). The conformations of both main and side chains in the thumb domain between two crystal forms of the TNT-IFT complex (Table 1) revealed subtle structural differences between residues 669-679, possibly caused by the lack of crystal contacts in this region and underscoring the structural plasticity of this domain. The thumb wraps around the palm domain, which is formed by a six-stranded β-sheet surrounded by two short α-helices and two 3/10 helices. Four strands, arranged into two nearly orthogonal antiparallel β-hairpins (β12-β13 and β7-β11), are substantially longer and resemble the four fingers of a hand (Fig. 3b). IFT adopts a globular structure that consists of a β-rich domain containing two antiparallel β-sheets (β1–β5 and β7–β9) and a helical domain, exposed on the opposite surface, formed by 4 α-helices. Between the TNT palm and the thumb domain is a ~15 Å deep crevice occupied by two IFT β-turns named TNT-interaction loops 1 and 2 (Fig. 3c, 4a) that penetrate deeply inside the TNT core and make extensive electrostatic contacts. Although the structure of IFT in complex with TNT superimposes well to the structure of free IFT26 (rmsd ~0.52 Å), the side chain conformations in the TNT-interaction loops (N36, K37, E40, and D58, R59, M60, S61) projecting inside the TNT crevice are different (Fig. 3c).
Figure 4. Identification of TNT residues involved in NAD+ hydrolysis and cytotoxicity in macrophages.
(a) A cut-through view of the TNT molecular surface calculated using Pymol reveals a ~15 Å deep crevice involved in NAD+-binding. (b) Ribbon diagram of TNT showing residues lining the crevice and involved in NAD+-binding and/or hydrolysis. (c) in vitro NAD+-glycohydrolase activity assay for wt and mutant TNT proteins. (d) Cytotoxicity of egfp fused wt and mutant TNT expression in RAW 264.7 macrophages were assessed by flow cytometry to measure of the percentage of 7AAD-positive cells. (e) The intracellular NAD+ content of Mtb-infected THP-1 macrophages was determined by an enzyme coupling assay in comparison to non-infected cells. (f) Cytotoxicity of Mtb-infected THP-1 macrophages (MOI 20) was determined by flow cytometry 48 h post infection after staining with the Live/Dead fixable green cell stain. Data in (c-f) represent the means ± s.d. of three independent experiments.
Identification of putative NAD+-binding site of TNT
TNT has no sequence similarity to any protein of known function13 and an exhaustive search for similar structures failed to yield significant hits. However, a restraint-based structural alignment of TNT with the SPN glycohydrolase domain27 and the ribosyltransferase domains of the diphtheria and cholera toxins28,29 revealed a similar basic architecture of the NAD+-binding core. Noticeable differences are that the TNT core contains only six β-strands as opposed to seven found in all known NAD+-utilizing toxins, and is significantly smaller, with only two short α-helices and two 3/10 helices. In silico docking readily placed NAD+ in the deep crevice of TNT (Supplementary Fig. 4a) with a predicted free-energy of binding of −8.7 kcal/mol. All solutions share a similar position of the nicotinamide group that inserts deeply inside the TNT crevice. Inspection of the amino acids surrounding the putative NAD+-binding pocket identified Q822, Y765 and R757 as residues possibly involved in NAD+-binding and hydrolysis (Supplementary Fig. 4b) based on similar positions of catalytic amino acids of ADP-ribosylating toxins30. Out of these residues, the glutamine 822 is highly conserved among TNT homologs. To determine the role of Q822 in NAD+ hydrolysis, we produced the TNTQ822A protein in E. coli in the presence of the anti-toxin IFT to prevent secondary mutations and separated the complex by selective thermal denaturation (as shown for wt TNT in Supplementary Fig. 2c). Purified TNTQ822A maintained structural integrity, as determined by IFT binding (Supplementary Fig. 5a), but its NAD+-glycohydrolase activity was reduced by two-fold compared to wild-type TNT (Fig. 4c), suggesting that glutamine 822 is important, but not essential, for binding and/or hydrolysis of NAD+. Similarly, the TNTY765A mutant protein also had reduced NAD+-glycohydrolase activity in vitro (Fig. 4c) and cytotoxicity in macrophages (Fig. 4d). By contrast, mutation of the corresponding glutamine 148 and tyrosine 65 residues in diphtheria toxin resulted in the drastic loss of its ADP-ribosyltransferase and NAD+-glycohydrolase activities31,32, suggesting that TNT uses a different mechanism of NAD+ binding and possibly hydrolysis. The highly conserved G818 residue also lines the putative NAD+ binding pocket of TNT (Fig. 4b). However, the purified TNTG818V protein was unfolded as indicated by circular dichroism and lack of IFT binding (not shown) and hence did not hydrolyze NAD+ (Fig. 4c). Since several single mutants including TNTQ822A did not completely eliminate the NAD+-glycohydrolase activity of TNT, except for the unfolded TNTQ818V mutant (Fig. 4c), we exploited the inadvertent selection of an additional mutation, H792N, in E. coli carrying a tntQ822K expression vector in the absence of IFT. Indeed, the purified TNTH792N Q822K protein had no detectable NAD+-glycohydrolase activity in a sensitive assay with a detection limit of 0.1 μM NAD+ (Fig. 4c), but efficiently bound to IFT in pull-down experiments (Supplementary Fig. 5b) demonstrating that the loss of NAD+-glycohydrolase activity was not caused by impaired protein folding. Taken together, this mutational analysis identifies the putative NAD+ binding site of TNT, but also reveals substantial differences compared to SPN and ADP-ribosylating toxins30.
TNT-mediated NAD+ depletion causes macrophage cell death
To assess whether NAD+ depletion by TNT leads to cell death, RAW 264.7 macrophages were transiently transfected with plasmids expressing egfp fusions with wt tnt or mutant tnt genes. Thereafter, macrophage viability was assessed by 7AAD staining and flow cytometry. While wt TNT killed more than 50% of the transfected macrophages, the non-catalytic TNTH792N Q822K mutant completely lost toxicity as did the unfolded TNT G818V mutant (Fig. 4d) indicating that macrophage cell death is indeed dependent on the NAD+-glycohydrolase activity of TNT. The TNTQ822A mutant showed intermediate cytotoxicity (Fig. 4d), which is consistent with its partial enzymatic activity (Fig. 4c). Expression of all TNT mutants was much higher than wt TNT as shown by fluorescence of the GFP fusion proteins thus demonstrating that the lower toxicity was not due to lower expression levels.
To examine the role of the NAD+-glycohydrolase activity of TNT in the context of Mtb infection, we infected THP-1 macrophages with wt Mtb, the ΔcpnT mutant, and the ΔcpnT mutant complemented with cpnT encoding the wt TNT or the non-catalytic TNTH792N Q822K domain. Macrophages infected with wt Mtb showed a drastic decline in NAD+ levels 24 h post infection in contrast to macrophages infected with the Mtb ΔcpnT deletion mutant where NAD+ levels remained high (Fig. 4e). Importantly, the non-catalytic TNTH792N Q822K mutant was expressed to similar protein levels as wt TNT and showed a similar subcellular localization pattern (Fig. 5), but did not reduce NAD+ levels compared to that of the Mtb ΔcpnT deletion mutant (Fig. 4e). This result is consistent with the complete loss of NAD+-glycohydrolase activity of the TNTH792N Q822K mutant (Fig. 4c) and demonstrates that NAD+-depletion in macrophages infected with Mtb mainly depends on the enzymatic activity of TNT. It should be noted that NAD+ levels are also reduced in Mtb-infected macrophages by 40% through TNT-independent mechanisms compared to uninfected macrophages (Fig. 4e).
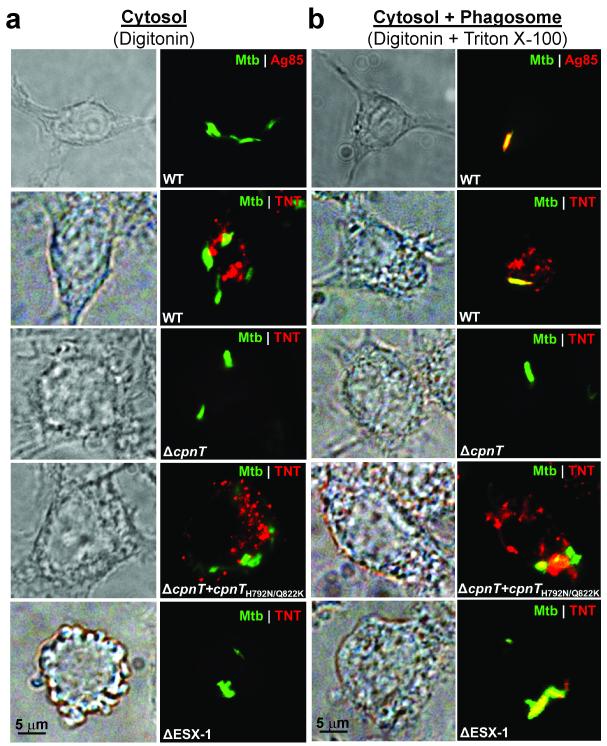
Figure 5. M. tuberculosis secretes TNT into the macrophage cytosol.
(a, b) THP-1 macrophages were infected with gfp expressing strains of wt Mtb, Mtb ΔcpnT, Mtb ΔcpnT::cpnTH792N Q822K or Mtb ΔESX-1 (mc26230) at an MOI of 10:1. Representative images are shown 48 h post infection when macrophages were fixed, selectively or completely permeabilized with digitonin (a) or digitonin plus Triton X-100 (b), respectively, and stained with α-TNT or α-Ag85 antibody (red). Magnification: 100x; Scale bar: 5 μm. N = 50 cells from three independent experiments.
To correlate the observed TNT-dependent NAD+ depletion during infection with the known cytotoxicity of Mtb7, we measured the viability of macrophages infected with Mtb strains secreting wt TNT or the catalytically inactive TNTH792N Q822K. Importantly, the cytotoxicity of Mtb secreting the non-catalytic TNTH792N Q822K was reduced to the same level as that of the Mtb cpnT deletion mutant (Fig. 4f). The interpretation of the infection experiments is complicated by the attenuation of the Mtb cpnT deletion mutant in macrophages13, because the reduced bacillary load may also reduce cytotoxicity by mechanisms other than NAD+ hydrolysis. However, the bacterial load of the Mtb strain expressing only the N-terminal domain of CpnT was increased by 12-fold, but showed the same cytotoxicity as the cpnT mutant indicating that the observed difference in cytotoxicity is likely not a consequence of fewer bacteria, but is dependent on TNT activity13. Furthermore, these results link NAD+ levels to cytotoxicity and are also consistent with our observations that no NAD+ is detectable in Jurkat T cells 24 hours after induction of tnt expression (Supplementary Fig. 1b) and that more than 90% of these cells died during this time13. Supplementation of this Jurkat T cell line with nicotinamide or nicotinic acid, both NAD+ precursors that elevate intracellular NAD+ levels (Supplementary Fig. 6a)18,33, reduced TNT-induced toxicity (Supplementary Fig. 6c). This effect is not due to inhibition of the enzymatic activity of TNT caused by addition of nicotinamide or nicotinic acid (Supplementary Fig. 6b). These NAD+ precursor supplementation experiments support the direct role of NAD+ hydrolysis in the toxicity of TNT. Taken together, these experiments provide compelling evidence that NAD+ hydrolysis by TNT is the main toxicity mechanism of Mtb in macrophages.
TNT gains access to the macrophage cytoplasm
Uptake of many bacterial toxins into target cells occurs by receptor-mediated endocytosis34-36. To examine putative cellular uptake pathways for TNT, purified TNT protein was added to THP-1 macrophages. However, extracellular TNT toxin did not induce macrophage cell death (Supplementary Fig. 7a) indicating that TNT lacks a membrane translocation domain. Hence, secreted TNT needs to exit the Mtb-containing phagosome in order to gain access to the NAD+ pools in the macrophage cytosol and induce necrotic cell death of infected macrophages as we proposed previously13. Furthermore, internalized latex beads coated with purified TNT protein did not cause macrophage death despite showing glycohydrolase activity in vitro (Supplementary Fig. 7b, c) suggesting that TNT itself is not capable of phagosomal exit. To examine the cellular localization of TNT, we selectively permeabilized the plasma- but not the phagosomal-membrane by digitonin to visualize proteins within the cytosol of macrophages infected with Mtb37. To validate this method, we stained Mtb-infected macrophages using an antibody against the antigen 85 complex (Ag85), a major secreted protein complex of Mtb38. Ag85 was only detected when macrophage membranes were solubilized by Triton X-100 but not in the presence of digitonin alone (Fig. 5), indicating that the phagosomal membrane is not permeabilized by digitonin. Then, we showed that TNT is secreted by Mtb into the cytosol of infected macrophages, while the bacteria are still confined to the phagosome (Fig. 5a, b). The fraction of macrophages with cytosolic localization of TNT 48 h after infection was ~ 50%, 0%, and 10% for wt Mtb, Mtb ΔcpnT, and Mtb Δesx-1, respectively. Interestingly, TNT is not evenly distributed in the cytosol of macrophages, but rather shows punctate localization, possibly from association with cellular vesicles and/or organelles. A similar phenotype was observed for the catalytically inactive TNTH792N Q822K mutant (Fig. 5a, b), indicating that the punctate distribution of TNT in macrophages is not associated with its enzymatic activity. Access to the macrophage cytosol, but not secretion of TNT by Mtb, completely depends on the ESX-1 secretion system (Fig. 5a, b) consistent with previous findings that the phagosomal membrane is perforated in an ESX-1 dependent manner39 and that the ESX-1 system is required for Mtb cytotoxicity7.
Direct injection of TNT protein kills zebrafish zygotes
Zebrafish zygotes are a well-established model to study higher order vertebrates. Fundamental cellular processes are highly conserved and the short, 24 hour, developmental period from a single-celled zygote to an embryo enables rapid phenotype detection40. Further, defined quantities of protein and protein complexes such as TNT-IFT can be easily injected directly into the cytoplasm eliminating potential confounding factors of transfection and infection experiments such as varying gene expression, potentially altered properties of GFP fusion proteins and the co-secretion of other proteins and/or lipids. To examine the toxicity of purified TNT protein in vivo, zebrafish zygotes were injected with buffer, BSA, wt TNT, catalytically inactive TNTH792N Q822K or wt TNT-IFT complex proteins at the 1-2 cell state. While zygotes injected with buffer and BSA developed into morphologically normal larvae with a heartbeat 24 hours after injection, injection of wt TNT dramatically compromised embryonic development (Fig. 6). This result demonstrates that the TNT protein itself is toxic when directly introduced into the host cell cytosol. In contrast, injection of the non-catalytic TNTH792N Q822K mutant was not lethal to embryonic development demonstrating that NAD+ hydrolysis is required for TNT toxicity (Fig. 6 and Supplementary Table 2). The TNT-IFT complex was not toxic when injected into zebrafish zygotes (Supplementary Table 2) indicating that this protein complex is not dissociated under physiological conditions. Furthermore, TNT protein injected in the tail vein of zebrafish larvae 24 hours after fertilization was not toxic (not shown) in contrast to cholera toxin41 indicating that TNT lacks a cellular signal to access the host cell cytoplasm unlike the cholera toxin which contains a cell receptor-binding subunit42. Taken together, the results of the zebrafish protein injection experiments are consistent with the macrophage transfection (Fig. 4d) and infection experiments (Fig. 4e, f), and show that TNT toxicity in eukaryotic cells is completely dependent on its cytosolic localization and NAD+ glycohydrolase activity. These results are consistent with the lack of toxicity of TNT externally added to macrophages as well as the macrophage transfection and infection experiments.
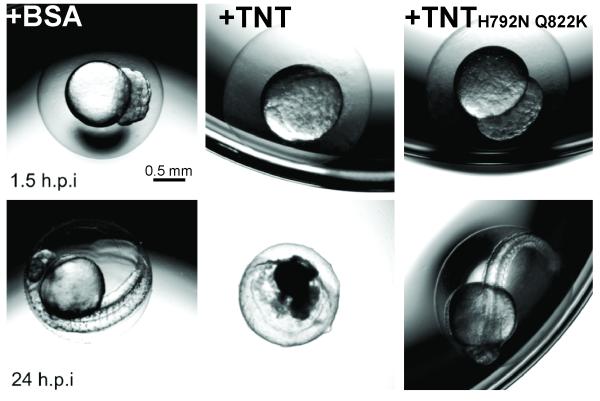
Figure 6. TNT is toxic in Zebrafish.
Zebrafish zygotes were injected at the 1-2 cell stage with 80 pg of purified BSA, wt TNT or the non-catalytic TNTH792N Q822K mutant or with 160 pg of the purified TNT-IFT complex. Representative images from two separate experiments are shown for embryos at 1.5 and 24 hours post injection (h.p.i.). Survival was determined by the presence of a heartbeat. Scale bar: 0.5 mm. Quantification of zebrafish zygote survival is shown in Supplementary Table 2.
DISCUSSION
The control of host cell death is of utmost importance for the survival, escape, and dissemination of M. tuberculosis. It has been clearly shown that Mtb induces a necrotic-like cell death7, but the components and molecular mechanism of this process were unclear43. Our recent discovery of TNT as a novel Mtb toxin led us to hypothesize that TNT is secreted by Mtb to induce cell death of infected macrophages13. In this study, we found that TNT (i) hydrolyzes the essential cellular co-factor NAD+ in vitro, (ii) translocates to the macrophage cytosol in an ESX-1-dependent manner, while Mtb is still trapped in phagosomes, and (iii) kills macrophages infected with Mtb by depleting NAD+. Nonetheless, it remains unclear whether TNT-induced cell death results directly from metabolic collapse after NAD+ depletion or from other molecular events triggered by low NAD+ levels, or from a combination of different mechanisms. For example, low NAD+ levels have been shown to impair glycolysis, which prevents cells from rapidly generating ATP. Metabolic collapse leading to necrotic cell death can be prevented by NAD+ replenishment16,17,44,45 as also shown for TNT in this study (Supplementary Fig. 8). On the other hand, depletion of cytoplasmic NAD also deprives the cell from pyruvate blocking the tricarboxylic acid (TCA) cycle in mitochondria46. Mitochondrial dysfunction is known to increase production of reactive oxygen species (ROS) and to activate calcium-dependent proteases (calpains) that disrupt the cytoskeletal network47. Moreover, ROS and calpains permeabilize lysosomes and thereby release lipases which degrade phospholipids and disrupt mitochondrial and cellular membranes as observed during Mtb infection of macrophages7. Further, the observation that a TNT deletion mutant has a residual cytotoxicity of about 20-30% compared to wt Mtb in macrophage infection experiments (Fig. 4f) indicates the involvement of other factors in Mtb-mediated cell death13. These factors may include the ESX-1 substrate ESAT-6, which damages lipid membranes6. In addition, Mtb activates the cytosolic “danger” sensor NLRP3 by Mtb in an ESAT-6 dependent manner that initiates inflammasome formation48 and impairs the membrane repair machinery by lipoxin A4, which down-regulates the pro-apoptotic and necrosis-protecting prostaglandin E2 (PGE2)4,5. Taken together, this study shows that Mtb-induced necrosis of infected macrophages is mainly dependent on NAD+ hydrolysis by TNT, although the exact sequence of downstream events that lead to cell death remain to be elucidated.
Since necrosis of infected cells promotes bacterial dissemination, the utilization of TNT’s NAD+-glycohydrolase activity is highly advantageous for Mtb compared to ADP-ribosylating toxins of other bacterial pathogens that trigger apoptosis49-51. TNT may also enable Mtb to control the timing and mode of cell death in other cell types such as neutrophils and dendritic cells, which Mtb is known to invade52,53. However, the toxicity of TNT is also detrimental to Mtb13. To protect itself from from self-poisoning, Mtb produces the immunity factor IFT (Fig. 2). The atomic structure of the TNT-IFT complex presented in this study reveals a fully electrostatic binding interface, which ensures picomolar binding affinity between these two proteins and may enable disassembly of IFT in the phagosome at acidic pH. TNT has similar catalytic activity as the SPN glycohydrolase of group A streptococci54 and a similar protective mechanism23, although TNT and IFT lack identifiable sequence and structural similarities with proteins of known function. Although functionally equivalent, TNT and SPN differ in at least three ways. First, TNT is significantly smaller (~150 versus 450 residues) and represents one of the smallest NAD+-binding domain identified to date. Second, TNT has ~160-fold slower catalytic turnover of NAD+, but similar substrate affinity (Supplementary Table 1), possibly resulting in slower toxicity kinetics in vivo. Finally, the secretion mechanisms of TNT and SPN differ greatly. While a signal peptide is sufficient to secrete SPN across the membrane of S. pyogenes, TNT constitutes the C-terminal domain of the outer membrane protein CpnT which resembles autotranporters of Gram-negative bacteria13. In the Pfam database, TNT is annotated as a member of the Domain of Unknown Function 4237, which comprises over 600 homologs encompassing approximately 300 bacterial and fungal species55. Thus, our results identify a class of novel toxins present in a large group of microorganisms including many mycobacteria, Listeria monocytogenes and Yersinia pestis, which are known to induce necrosis of infected host cells56,57. In conclusion, our study revealed the structure and enzymatic activity of TNT as an NAD+ glycohydrolase that affects host cell fate on multiple levels. These findings have broad implications in microbial pathogenesis.
ONLINE METHODS
Bacterial strains and reagents
E. coli strains DH5α or DH5αZ1 and BL21 (DE3) were used for cloning and expression experiments respectively, and was routinely grown in Luria-Bertani broth at 37°C. The M. tuberculosis H37Rv derivative avirulent strains mc26206 or mc26230 strains were grown in Middlebrook 7H9 medium (Difco) supplemented with 0.2% glycerol, 0.02% Tyloxapol and 10% OADC (Remel) or on Middlebrook 7H10 plates supplemented with 0.5% glycerol and 10% OADC (Remel). All bacteria strains used are shown in Supplementary Table 3. Growth media of the auxotrophic M. tuberculosis strains were supplemented with 24 μg/ml pantothenate and 50 μg/ml L-leucine. Hygromycin B was purchased from Calbiochem. All other chemicals were purchased from Merck or Sigma at the highest purity available. Enzymes for DNA restriction and modification were from New England Biolabs and Invitrogen. Oligonucleotides were obtained from IDT. β-NAD+ and doxycycline was purchased from Sigma. Antibiotics were used when required at the following concentrations: hygromycin (200 μg/ml for E. coli, 50 μg/ml for mycobacteria), kanamycin (30 μg/ml), carbenicillin (100 μg/ml), and chloramphenicol (25 μg/ml). Live/Dead Fixable Green Cell Stain was purchased from Invitrogen. Polyclonal rabbit anti-TNT antibody was generated previously58. Monoclonal Anti-Ag85 complex (Clone IT-44 (HBT7)) was obtained from BEI Resources (NR-13826). Validation is provided on the manufacturer’s website.
Plasmid construction
Construction of plasmids are described in a flow-chart that includes the oligonucleotide pairs (Supplementary Table 4) used for PCR amplification and restriction enzymes utilized for cloning (Supplementary Fig. 8). All plasmids used are shown in Supplementary Table 5.
Purification of the TNT-IFT complex
E. coli BL21 (DE3) cells harboring both plasmids pML1995 and pML1999 were grown in 2 liters of the auto induction liquid medium ZYP-505259 supplemented with 100 μg/mL carbenicillin and 25 μg/mL chloramphenicol until the culture reached saturation (OD600 ~ 10) for 3 hours. The cells were harvested by centrifugation (7,000 × g, 4°C, 20 min) and washed with a buffer containing 20 mM Tris (pH 7.6) and200 mM NaCl. The cell pellet (24 g) was resuspended with 130 mL of the same buffer supplemented with 100 μg/mL lysozyme, 1 mM PMSF, 1 mM EDTA and 2.5 units benzonase per mL of suspension and was submitted to lysis using a French press (five cycles at 1,000, 5,000, 10,000 and 15,000 psi each). The resulting lysate was cleared by centrifugation (60,000 × g, 4°C, 45 min). The complex MBP-TNT-IFT was first purified using Ni-NTA affinity and then the elution was further purified using amylose resin, both according to manufactures’ protocols. The protein concentration of the fusion complex was determined with the Bradford assay and the corresponding amount of TEV protease NIa and 1 mM TCEP was added. The digestion was performed at 4°C and followed by SDS-PAGE analysis over a period of 96 h when > 90% of the fusion protein was cleaved. The resulting protein mixture was passed through a Talon-Co column to retain MBP, the uncleaved fusion protein and the NIa protease, with the complex TNT-IFT being recovered in the flow-through. The fractions containing the toxin/immunity factor complex were pooled, concentrated by ultrafiltration using an Amicon Ultracel unit (3 kDa cut-off) and submitted to size-exclusion chromatography using a Superdex sd75pg 16/60 column, bathed with 20 mM Tris pH7.6, 300 mM NaCl at a 1 mL/min flow-rate and kept at 15°C. Absorbance at 280 nm was recorded on a paper chart and the chromatograms were digitized using a scanner then converted using the XYscan program (available at http://rhig.physics.yale.edu/~ullrich/xyscanDistributionPage/). The column was calibrated using albumin from bovine serum (66 kDa), carbonic anhydrase from bovine erythrocytes (29 kDa) and cytochrome C from horse heart (12.4 kDa). The fractions containing the complex TNT-IFT were pooled together and the concentration measured using Bradford assay.
Crystallization of the TNT-IFT complex and structure determination
Crystals of the TNT-IFT complex were obtained using the hanging-drop vapor diffusion method by mixing together 2 μl of TNT-IFT complex at 25 mg/ml with 2 μl of 0.2 M ammonium formate, 24% (w/v) PEG 3,350 at pH 6.2 and the resultant drop equilibrated against 600 μl of crystallization solution, at 18 °C. Diffraction quality, large rod-like crystals grew in 2-7 days to exceed ~1 mm in length. Crystals were harvested in nylon cryo-loops, cryo-protected by quick passage through a solution containing mother liquor solution supplemented with 27% ethylene glycol and flash-frozen in liquid nitrogen. Diffraction data were collected at beamlines X6A and X29 at the National Synchrotron Light Source (NSLS) on a ADSC Quantum Q270 and a Quantum-315r CCD detector, respectively. Data indexing, integration and scaling were carried out with the HKL2000 software package60 (Table 1). Initial phases were obtained by molecular replacement with Phaser61 using the deposited structure of Rv3902c (PDB entry 4O6G) as search model and diffraction intensities from Crystal form 2, that extends to 1.9 Å resolution (Table 1). An initial solution, which accounts for only ~40% of atoms in the asymmetric unit, was rebuilt automatically with Buccaneer62 and phenix.autobuild63 and manually with Coot64, and refined with phenix.refine63, yielding an initial complete model of the TNT-IFT complex. This model was then replaced in the high resolution crystal form I (Table 1) and refined to 1.10 Å resolution using phenix.refine63. The final model, which includes residues 1-175 of IFT, residues 648-846 of TNT, riding hydrogens and 798 water molecules, was refined to a Rwork/Rfree of 12.8/14.3%, at 1.10 Å resolution (the Rfree was calculated using 2,000 randomly selected reflections). The final model has excellent geometry (r.m.s.d. bond = 0.010 Å, r.m.s.d. angle = 0.14 °) and the Ramachandran plot shows over 95% of residues in the most favored regions with no disallowed residues, as assessed by MolProbity65 (Table 1). The structure was analyzed using the PISA server66, PDBsum67 and all structural illustrations in the paper were done with Pymol.
Docking studies
Docking of NAD+ inside the refined structure of TNT (stripped of IFT) was performed using AutoDock Vina68. Atomic coordinates for NAD+ were downloaded from Coot64 and idealized using phenix.elbow69. Only polar hydrogens in TNT were used for docking simulation. To define an appropriate search space to dock NAD+ onto TNT, a generous grid box of 44 × 32 × 26 Å3 (along x, y and z) was generated around TNT crevice. 10 solutions were generated by Vina with calculated free binding energies (ΔGB) between −8.6 and −7.7 kcal/mol (Supplementary Fig 6). All solutions share a similar position of the nicotinamide group that inserts deeply inside the TNT crevice placing the hydrolysable bond between nicotinamide and ribose in close proximity to Q822 and Y765. In contrast, the position of the NAD+ adenosine-dinucleotide group varies greatly among different solutions as a result of the greater degree of freedom of this group inside TNT. The phosphate at position β is always positioned close (~4 Å) to R757.
Purification of TNT for functional assays and SPR analysis
E. coli DH5αZ1 harboring the plasmid pML1993 (TNTwt) was grown in LB medium supplemented with 30 μg/mL kanamycin to an OD600 of 0.6. The culture was then induced with 200 ng/mL doxycycline for 1 h. Cells were harvested by centrifugation (7,000 × g, 4°C, 20 min.) and washed with 20 mM Tris pH 7.6, 200 mM NaCl. The cell pellet was resuspended with 20 mL (per 500 mL of original culture) of the same buffer supplemented with 1 mg/mL lysozyme, 1 mM PMSF, and 2.5 units benzonase per mL of suspension and was submitted to lysis by sonication. The resulting lysate was cleared by centrifugation (10,000 × g, 4°C, 45 min). The cleared supernatant was used for purification by Ni-NTA resin (Pierce) following manufacturer’s protocol. Subsequently, the TNT containing elution was used for a second affinity chromatography step using Amylose resin (NEB) according to manufacturer’s protocol. The corresponding amount of TEV protease NIa and 1 mM TCEP was added to cleave MBP from TNT for 72 h at 4°C. The resulting protein mixture was passed through a Talon-Co column to retain MBP, the uncleaved fusion protein and the NIa protease, with the TNT being recovered in the flow-through. E. coli BL21 (DE3) cells harboring the plasmid pML3339 was used to express and purify TNTQ822K by auto induction as described for the TNT-IFT complex. To obtain enough fusion protein for SPR experiments, E. coli BL21 (DE3) cells harboring the plasmid pML1997 was used to target expression of TNT into the periplasmic space. TNT was extracted by osmotic shock as previously described70, and purified by Ni-NTA resin followed by Amylose resin as above.
NAD+ glycohydrolase assays
Recombinant TNT (100 ng) was incubated with 1 μM β-NAD+ for 30 min at 37°C in reaction buffer (20 mM Tris, 200 mM NaCl, pH 7.4). The amount of NAD+ remaining in each sample was then measured using the EnzyFluo NAD/NADH Assay Kit (Bioassay Systems) according to manufacturer’s protocol. The EnzyChrom NAD/NADH Assay Kit was used to measure the amount of NAD+ in E. coli or Jurkat T-cells. Protocols were carried out according to manufacturer’s directions with the minor modification of an additional 5 min heat extraction at 90°C for E. coli samples.
TLC assays
NAD+ glycohydrolase assays were performed by incubating TNT (20 nM) with 1 mM NAD+ for 30 min at 37°C. 2 μL of this reaction were spotted onto silica-gel glass TLC plates (Sigma). The same amount of standard molecules NAD, ADP-R, and cADP-R were spotted. The TLC was developed in the ascending direction for 20 min at room temperature with a mixture of 30% water, 70% ethanol and 0.2 M ammonium bicarbonate. After drying, the TLC plates were sprayed uniformly with 10% cupric sulfate in 8% aqueous phosphoric acid, allowed to dry, and then placed into a 145 °C oven for 10 - 20 min.
Enzyme kinetics of TNT
Hydrolysis of β-NAD+ by TNT was monitored by detecting fluorescence as previously described71 with minor modifications. Briefly, initial rates of hydrolysis were measured by incubation of various concentrations of NAD+ (50 μM to 1.6 mM) with 10 nM TNT in assay buffer (20 mM Tris, pH 7.4 and 200 mM NaCl) at 37°C. 100 μL aliquots of the reaction were taken out at various time points (1 – 10 min) and quenched by the addition of 70 μL of 5 M NaOH in a 96 well plate. The plate was then incubated at room temperature for 1 h in the dark to develop the alkaline treatment generated fluorescence signal of NAD. Subsequently, the relative fluorescence at 360/460 nm (excitation/emission filter set) was determined on a BioTek Synergy HT plate reader and compared to a standard NAD+ concentration curve. The initial rate of NAD+ hydrolysis was determined by linear regression of plots of substrate depletion versus time. Thereafter, the rate was plotted as a function of NAD+ concentration and was fit using the Michaelis-Menten equation to determine the Km, Vmax, and Kcat for the enzyme. Non-linear regression analysis was performed with SigmaPlot 11.
Purification of IFT for surface plasmon resonance experiments
E. coli BL21 (DE3) cells harboring pML1974 were grown in liquid LB medium supplemented with 100 μg/mL carbenicillin to an OD600 of 0.8. The culture was then induced with 1 mM IPTG for 3 h at 37°C. The bacterial pellet was resuspended in 20 mM Tris pH 7.4, 200 mM NaCl (5 mL/g cells) supplemented with 1 mg/mL lysozyme, 1 mM PMSF, and 2.5 units benzonase per mL of suspension. The lysate was centrifuged at 10,000 × g, 4°C for 45 min to obtain the soluble fraction. Purification of polyhistidine tagged MBP-IFT from this fraction was completed using Talon-Co affinity chromatography according to manufacturer’s protocol. The eluted fractions containing the MBP-IFT fusion protein were pooled and dialyzed against 20 mM Tris pH 7.4, 200 mM NaCl to decrease the imidazole concentration prior to the addition of TEV protease NIa and 1 mM TCEP. The digestion was performed at 4°C over a period of 72 hours and was followed by SDS-PAGE analysis until > 90% of the fusion protein was cleaved. The resulting protein mixture was passed through Amylose resin to retain MBP and the uncleaved fusion protein, the IFT being recovered in the flow-through. IFT was further purified by anion exchange (Hi-Trap Q FF) in 20 mM Tris pH 8 using a 10 to 500 mM NaCl gradient.
Surface plasmon resonance binding assays and kinetic analysis
Binding interactions between TNT and IFT were examined using a Biacore T200 molecular interaction analysis system (GE Healthcare). HBS-EP (0.01 M HEPES, 150 mM NaCl, 3 mM EDTA, 0.005% polysorbate 20; pH 7.4) was used as the running buffer for the immobilization and kinetic studies. A Series S Sensor Chip NTA (Biacore) was used to immobilize polyhistidine tagged MBP-TNT (160 nM) onto its flow cell at a flow rate of 5 μl/min for 120 s to reach a ligand density of approximately 500 RU. A control reference flow cell was used where the Ni-loading step onto the NTA chip was omitted prior to ligand capture, but otherwise contained all the same steps as the active flow cell. Different concentrations of the analyte IFT (0 – 62.5 nM) were diluted in running buffer and injection was carried out for 60 s at a flow rate of 20 μL/min to observe association, and then dissociation was followed for 10 min. A constant temperature of 25°C was used throughout the entire kinetic study. In order to minimize nonspecific binding and instrumental drift on the generated binding sensorgrams, a double referencing of the data was performed. The binding responses over the reference flow cell were subtracted from the active surface to correct for non-specific binding. Furthermore, an analyte run using running buffer alone was also subtracted from the binding sensorgrams to minimize noise. Biacore evaluation software was used to analyze the binding kinetics. A 1:1 binding model was used to fit the binding response curves over five analyte concentrations which resulted in residuals well within quality control standards as indicated by the chi-square value of 0.183.
In vitro pull down assay
Purified recombinant TNT (5μg) was incubated with equimolar amounts of MBP-IFT overnight at 4°C with 25 μL packed Amylose resin (NEB) in a final volume of 75 μL reaction buffer (20 mM Tris, 200 mM NaCl, pH 7.4). Equivalent amounts of TNT and MBP-IFT were incubated independently with Amylose resin as controls. Post incubation, the resin was washed thoroughly with reaction buffer. The washed resin was combined with an equivalent volume of protein loading buffer and heated at 95°C for 10 minutes to dissociate resin-bound protein. Captured proteins were analyzed by SDS-PAGE stained with Coomassie blue.
Cell culture, transfection and flow cytometry
Human THP-1 monocytic cell line (ATCC TIB-202) and mouse RAW 264.7 macrophage cell line (ATCC TIB-71) were propagated in RPMI-1640 or Dulbecco’s modified Eagle’s medium (DMEM), respectively, supplemented with 10% fetal bovine serum (FBS), 10 mM HEPES, 2 mM L-glutamine (Thermo), 100 IU/mL penicillin, and 100 μg/mL streptomycin. Jurkat-derived cell lines58 were maintained in RPMI-1640 medium supplemented with the same reagents. All cells were grown in a 37°C humidified incubator at 5% CO2. Cell lines were routinely tested negative for mycoplasma contamination every month. For transfections, 5 × 105 RAW macrophages were allowed to adhere onto glass coverslips in 24-well plates overnight. Transfection was achieved using FuGENE HD (Promega). In brief, 0.5 μg of purified plasmid DNA (pML3035, pML3036, and pML3059) were mixed with 1 uL of FuGENE reagent in a final volume of 50 uL with DMEM. This mixture was incubated at room temperature for 15 min and then added to the macrophages contained in 450 uL of media. Thirty hour post-transfection, cells were washed and detached by trypsin-EDTA. Cells were harvested by low-speed centrifugation and resuspended in PBS containing 1% FBS, and stained with 7-amino-actinomycin D (7AAD, eBioscience) according to the manufacturer’s recommendation. Analysis of cell viability was quantified by flow cytometry using the Guava easyCyte. Positively transfected cells were gated according to green fluorescence (FL-1; GFP), and the percentage of non-viable cells out of the transfected population was gated according to red fluorescence (FL-2; 7AAD).
NAD+ content in macrophages infected with Mtb
THP-1 monocytes were differentiated with 100 ng/mL PMA for 24 h, and subsequently infected with Mtb strains at a MOI of 20:1. 24 h post infection, macrophages were lysed in 0.025% SDS to release all NAD+ from the macrophages, but not mycobacteria. The EnzyFluo NAD/NADH kit was then used to quantify the amount of NAD+ in each sample.
Intracellular staining
Mtb infected RAW 264.7 macrophages on glass coverslips were fixed with 4% paraformaldehyde for 1 h at room temperature and subsequently washed multiple times with PBS prior to staining. Cells were selectively permeabilized as described72. In brief, cells were permeabilized with 25 μg/mL digitonin in PBS alone for 5 min or in combination with 0.2% Triton-X100 in PBS for an additional 5 min. Then, cells were washed and blocked with PBS containing 1% BSA (S-Buffer) for 20 min. Thereafter, polyclonal α-TNT antibody (1:75) or monoclonal α-Ag85 antibody (1:50) was used in S-Buffer and incubated with the cells for 60 min. After 3 washes with PBS, secondary anti-rabbit or anti-mouse Alexa Fluor 555 (Invitrogen) was used at a dilution of 1:1000 and incubated with the cells for 30 min. Cells were then washed 3 times with PBS and coverslips were mounted onto microscope slides with FluorSave (Calbiochem) for fluorescence imaging. Imaging was performed using an Axiovert 200 microscope (Carl Zeiss) equipped with a 100x/1.4 Plan-Apochromat (Carl Zeiss). Images were recorded using an AxioCam MRc camera (Zeiss) coupled to Axiovision v4.5 software (Carl Zeiss).
TNT toxicity in Jurkat cells and rescue by nicotinamide
Jurkat cells expressing tnt under an inducible system58 were pre-treated with either 5 mM nicotinamide, 10 μM nicotinic acid, or in combination for 2 h. At this point, expression of tnt was induced by addition of 100 ng/mL doxycycline for 2 h. Cells were subsequently washed free of doxycycline and incubated in regular media for 24 h. Cell viability was then assessed using the 7AAD dye followed by flow cytometry.
Supplementary Material
ACKNOWLEDGMENTS
We thank L. DeLucas for providing the Rv3902c structure before its publication, J. Pascal for stimulating discussions, Y. Wang for initial purification of IFT, the scientific staff at National Synchrotron Light Source beamlines X6A and X29 for beamtime, the University of Alabama at Birmingham Zebrafish Core for generous support of the zebrafish experiments, F. Wolschendorf for use of his microscope and P. Patel and U. Tak for technical assistance. Some work was carried out at the Sidney Kimmel Cancer Center X-ray Crystallography and Molecular Interaction Facility, which is supported in part by the US National Cancer Institute Cancer Center Support Grant P30 CA56036. This work was supported by the US National Institutes of Health grants GM100888 (G.C.), AI63432 (M.N.), AI083632 (M.N.) and AI074805 (M.N.).
Footnotes
AUTHOR CONTRIBUTIONS
J.S., A.Si., A.Sp., K.S.D., G.C. and M.N conceived and designed experiments and analysed data; J.S, A.Si., R.K.L., K.S.D., A.Sp. and G.C. performed experiments; J.S., G.C. and M.N. wrote and edited the paper.
REFERENCES
- 1.Russell DG, Barry CE, 3rd, Flynn JL. Tuberculosis: what we don’t know can, and does, hurt us. Science. 2010;328:852–6. doi: 10.1126/science.1184784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Behar SM, Divangahi M, Remold HG. Evasion of innate immunity by Mycobacterium tuberculosis: is death an exit strategy? Nat Rev Microbiol. 2010;8:668–74. doi: 10.1038/nrmicro2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Butler RE, et al. The balance of apoptotic and necrotic cell death in Mycobacterium tuberculosis infected macrophages is not dependent on bacterial virulence. PLoS One. 2012;7:e47573. doi: 10.1371/journal.pone.0047573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Behar SM, et al. Apoptosis is an innate defense function of macrophages against Mycobacterium tuberculosis. Mucosal Immunol. 2011;4:279–87. doi: 10.1038/mi.2011.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Divangahi M, Behar SM, Remold H. Dying to live: how the death modality of the infected macrophage affects immunity to tuberculosis. Adv Exp Med Biol. 2013;783:103–20. doi: 10.1007/978-1-4614-6111-1_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith J, et al. Evidence for pore formation in host cell membranes by ESX-1-secreted ESAT-6 and its role in Mycobacterium marinum escape from the vacuole. Infect Immun. 2008;76:5478–87. doi: 10.1128/IAI.00614-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee J, Repasy T, Papavinasasundaram K, Sassetti C, Kornfeld H. Mycobacterium tuberculosis induces an atypical cell death mode to escape from infected macrophages. PLoS One. 2011;6:e18367. doi: 10.1371/journal.pone.0018367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abdallah AM, et al. Mycobacterial secretion systems ESX-1 and ESX-5 play distinct roles in host cell death and inflammasome activation. J Immunol. 2011;187:4744–53. doi: 10.4049/jimmunol.1101457. [DOI] [PubMed] [Google Scholar]
- 9.Henkel JS, Baldwin MR, Barbieri JT. Toxins from bacteria. EXS. 2010;100:1–29. doi: 10.1007/978-3-7643-8338-1_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gordon SV, Bottai D, Simeone R, Stinear TP, Brosch R. Pathogenicity in the tubercle bacillus: molecular and evolutionary determinants. Bioessays. 2009;31:378–88. doi: 10.1002/bies.200800191. [DOI] [PubMed] [Google Scholar]
- 11.Mukhopadhyay S, Nair S, Ghosh S. Pathogenesis in tuberculosis: transcriptomic approaches to unraveling virulence mechanisms and finding new drug targets. FEMS Microbiol Rev. 2012;36:463–85. doi: 10.1111/j.1574-6976.2011.00302.x. [DOI] [PubMed] [Google Scholar]
- 12.Forrellad MA, et al. Virulence factors of the Mycobacterium tuberculosis complex. Virulence. 2013;4:3–66. doi: 10.4161/viru.22329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Danilchanka O, et al. An outer membrane channel protein of Mycobacterium tuberculosis with exotoxin activity. Proc Natl Acad Sci U S A. 2014;111:6750–5. doi: 10.1073/pnas.1400136111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Belenky P, Bogan KL, Brenner C. NAD+ metabolism in health and disease. Trends Biochem Sci. 2007;32:12–9. doi: 10.1016/j.tibs.2006.11.006. [DOI] [PubMed] [Google Scholar]
- 15.Lin SJ, Guarente L. Nicotinamide adenine dinucleotide, a metabolic regulator of transcription, longevity and disease. Curr Opin Cell Biol. 2003;15:241–6. doi: 10.1016/s0955-0674(03)00006-1. [DOI] [PubMed] [Google Scholar]
- 16.Du L, et al. Intra-mitochondrial poly(ADP-ribosylation) contributes to NAD+ depletion and cell death induced by oxidative stress. J Biol Chem. 2003;278:18426–33. doi: 10.1074/jbc.M301295200. [DOI] [PubMed] [Google Scholar]
- 17.Zong WX, Thompson CB. Necrotic death as a cell fate. Genes Dev. 2006;20:1–15. doi: 10.1101/gad.1376506. [DOI] [PubMed] [Google Scholar]
- 18.Hara N, et al. Elevation of cellular NAD levels by nicotinic acid and involvement of nicotinic acid phosphoribosyltransferase in human cells. J Biol Chem. 2007;282:24574–82. doi: 10.1074/jbc.M610357200. [DOI] [PubMed] [Google Scholar]
- 19.Russell DG. New ways to arrest phagosome maturation. Nat Cell Biol. 2007;9:357–9. doi: 10.1038/ncb0407-357. [DOI] [PubMed] [Google Scholar]
- 20.Artman M, Bekierkunst A. Mycobacterium tuberculosis H37Rv grown in vivo: nature of the inhibitor of lactic dehydrogenase of Mycobacterium phlei. Proc Soc Exp Biol Med. 1961;106:610–4. doi: 10.3181/00379727-106-26419. [DOI] [PubMed] [Google Scholar]
- 21.Gopinathan KP, Sirsi M, Vaidyanathan CS. Nicotinamide-adenine dinucleotide glycohydrolase of Mycobacterium tuberculosis H37Rv. Biochem J. 1964;91:277–82. doi: 10.1042/bj0910277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stevens DL, Salmi DB, McIndoo ER, Bryant AE. Molecular epidemiology of nga and NAD glycohydrolase/ADP-ribosyltransferase activity among Streptococcus pyogenes causing streptococcal toxic shock syndrome. J Infect Dis. 2000;182:1117–28. doi: 10.1086/315850. [DOI] [PubMed] [Google Scholar]
- 23.Meehl MA, Pinkner JS, Anderson PJ, Hultgren SJ, Caparon MG. A novel endogenous inhibitor of the secreted streptococcal NAD-glycohydrolase. PLoS Pathog. 2005;1:e35. doi: 10.1371/journal.ppat.0010035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gopinathan KP, Sirsi M, Ramakrishnan T. Nicotin-amide-adenine nucleotides of Mycobacterium tuberculosis H37Rv. Biochem J. 1963;87:444–8. doi: 10.1042/bj0870444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gopinathan KP, Ramakrishnan T, Vaidyanathan CS. Purification and properties of an inhibitor for nicotinamide-adenine dinucleotidase from Mycobacterium tuberculosis H-37-Rv. Arch Biochem Biophys. 1966;113:376–82. doi: 10.1016/0003-9861(66)90201-3. [DOI] [PubMed] [Google Scholar]
- 26.Reddy BG, et al. 1.55 A resolution X-ray crystal structure of Rv3902c from Mycobacterium tuberculosis. Acta Crystallogr F Struct Biol Commun. 2014;70:414–7. doi: 10.1107/S2053230X14003793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith CL, et al. Structural basis of Streptococcus pyogenes immunity to its NAD+ glycohydrolase toxin. Structure. 2011;19:192–202. doi: 10.1016/j.str.2010.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Choe S, et al. The crystal structure of diphtheria toxin. Nature. 1992;357:216–22. doi: 10.1038/357216a0. [DOI] [PubMed] [Google Scholar]
- 29.Zhang RG, et al. The three-dimensional crystal structure of cholera toxin. J Mol Biol. 1995;251:563–73. doi: 10.1006/jmbi.1995.0456. [DOI] [PubMed] [Google Scholar]
- 30.Fieldhouse RJ, Turgeon Z, White D, Merrill AR. Cholera- and anthrax-like toxins are among several new ADP-ribosyltransferases. PLoS Comput Biol. 2010;6:e1001029. doi: 10.1371/journal.pcbi.1001029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tweten RK, Barbieri JT, Collier RJ. Diphtheria toxin. Effect of substituting aspartic acid for glutamic acid 148 on ADP-ribosyltransferase activity. J Biol Chem. 1985;260:10392–4. [PubMed] [Google Scholar]
- 32.Blanke SR, Huang K, Collier RJ. Active-site mutations of diphtheria toxin: role of tyrosine-65 in NAD binding and ADP-ribosylation. Biochemistry. 1994;33:15494–500. doi: 10.1021/bi00255a031. [DOI] [PubMed] [Google Scholar]
- 33.Van Gool F, et al. Intracellular NAD levels regulate tumor necrosis factor protein synthesis in a sirtuin-dependent manner. Nat Med. 2009;15:206–10. doi: 10.1038/nm.1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lord JM, Smith DC, Roberts LM. Toxin entry: how bacterial proteins get into mammalian cells. Cell Microbiol. 1999;1:85–91. doi: 10.1046/j.1462-5822.1999.00015.x. [DOI] [PubMed] [Google Scholar]
- 35.Torgersen ML, Skretting G, van Deurs B, Sandvig K. Internalization of cholera toxin by different endocytic mechanisms. J Cell Sci. 2001;114:3737–47. doi: 10.1242/jcs.114.20.3737. [DOI] [PubMed] [Google Scholar]
- 36.Barth H, Aktories K. New insights into the mode of action of the actin ADP-ribosylating virulence factors Salmonella enterica SpvB and Clostridium botulinum C2 toxin. Eur J Cell Biol. 2011;90:944–50. doi: 10.1016/j.ejcb.2010.11.007. [DOI] [PubMed] [Google Scholar]
- 37.Manzanillo PS, et al. The ubiquitin ligase parkin mediates resistance to intracellular pathogens. Nature. 2013;501:512–6. doi: 10.1038/nature12566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kremer L, Maughan WN, Wilson RA, Dover LG, Besra GS. The M. tuberculosis antigen 85 complex and mycolyltransferase activity. Lett Appl Microbiol. 2002;34:233–7. doi: 10.1046/j.1472-765x.2002.01091.x. [DOI] [PubMed] [Google Scholar]
- 39.Manzanillo PS, Shiloh MU, Portnoy DA, Cox JS. Mycobacterium tuberculosis activates the DNA-dependent cytosolic surveillance pathway within macrophages. Cell Host Microbe. 2012;11:469–80. doi: 10.1016/j.chom.2012.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Spitsbergen JM, Kent ML. The state of the art of the zebrafish model for toxicology and toxicologic pathology research--advantages and current limitations. Toxicol Pathol. 2003;31(Suppl):62–87. doi: 10.1080/01926230390174959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saslowsky DE, et al. Intoxication of zebrafish and mammalian cells by cholera toxin depends on the flotillin/reggie proteins but not Derlin-1 or -2. J Clin Invest. 2010;120:4399–4409. doi: 10.1172/JCI42958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vanden Broeck D, Horvath C, De Wolf MJ. Vibrio cholerae: cholera toxin. Int J Biochem Cell Biol. 2007;39:1771–5. doi: 10.1016/j.biocel.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 43.Moraco AH, Kornfeld H. Cell death and autophagy in TB. Semin Immunol. 2014;26:497–511. doi: 10.1016/j.smim.2014.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ha HC, Snyder SH. Poly(ADP-ribose) polymerase is a mediator of necrotic cell death by ATP depletion. Proc Natl Acad Sci U S A. 1999;96:13978–82. doi: 10.1073/pnas.96.24.13978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Herceg Z, Wang ZQ. Failure of poly(ADP-ribose) polymerase cleavage by caspases leads to induction of necrosis and enhanced apoptosis. Mol Cell Biol. 1999;19:5124–33. doi: 10.1128/mcb.19.7.5124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McCommis KS, Finck BN. Mitochondrial pyruvate transport: a historical perspective and future research directions. Biochem J. 2015;466:443–54. doi: 10.1042/BJ20141171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kennedy CL, Smith DJ, Lyras D, Chakravorty A, Rood JI. Programmed cellular necrosis mediated by the pore-forming alpha-toxin from Clostridium septicum. PLoS Pathog. 2009;5:e1000516. doi: 10.1371/journal.ppat.1000516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wong KW, Jacobs WR., Jr. Critical role for NLRP3 in necrotic death triggered by Mycobacterium tuberculosis. Cell Microbiol. 2011;13:1371–84. doi: 10.1111/j.1462-5822.2011.01625.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Heine K, Pust S, Enzenmuller S, Barth H. ADP-ribosylation of actin by the Clostridium botulinum C2 toxin in mammalian cells results in delayed caspase-dependent apoptotic cell death. Infect Immun. 2008;76:4600–8. doi: 10.1128/IAI.00651-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Morimoto H, Bonavida B. Diphtheria toxin- and Pseudomonas A toxin-mediated apoptosis. ADP ribosylation of elongation factor-2 is required for DNA fragmentation and cell lysis and synergy with tumor necrosis factor-alpha. J Immunol. 1992;149:2089–94. [PubMed] [Google Scholar]
- 51.Pessina A, et al. Bcl-2 down modulation in WEHI-3B/CTRES cells resistant to Cholera Toxin (CT)-induced apoptosis. Cell Res. 2006;16:306–12. doi: 10.1038/sj.cr.7310038. [DOI] [PubMed] [Google Scholar]
- 52.Wolf AJ, et al. Mycobacterium tuberculosis infects dendritic cells with high frequency and impairs their function in vivo. J Immunol. 2007;179:2509–19. doi: 10.4049/jimmunol.179.4.2509. [DOI] [PubMed] [Google Scholar]
- 53.Eum SY, et al. Neutrophils are the predominant infected phagocytic cells in the airways of patients with active pulmonary TB. Chest. 2010;137:122–8. doi: 10.1378/chest.09-0903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ajdic D, McShan WM, Savic DJ, Gerlach D, Ferretti JJ. The NAD-glycohydrolase (nga) gene of Streptococcus pyogenes. FEMS Microbiol Lett. 2000;191:235–41. doi: 10.1111/j.1574-6968.2000.tb09345.x. [DOI] [PubMed] [Google Scholar]
- 55.Finn RD, et al. Pfam: the protein families database. Nucleic Acids Res. 2014;42:D222–30. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Florido M, Cooper AM, Appelberg R. Immunological basis of the development of necrotic lesions following Mycobacterium avium infection. Immunology. 2002;106:590–601. doi: 10.1046/j.1365-2567.2002.01459.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sridharan H, Upton JW. Programmed necrosis in microbial pathogenesis. Trends Microbiol. 2014;22:199–207. doi: 10.1016/j.tim.2014.01.005. [DOI] [PubMed] [Google Scholar]
- 58.Danilchanka O, et al. An outer membrane channel protein of Mycobacterium tuberculosis with exotoxin activity. Proc Natl Acad Sci U S A. 2014;111:6750–5. doi: 10.1073/pnas.1400136111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Studier FW. Protein production by auto-induction in high density shaking cultures. Protein Expr Purif. 2005;41:207–34. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 60.Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 61.McCoy AJ. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr D Biol Crystallogr. 2007;63:32–41. doi: 10.1107/S0907444906045975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cowtan K. The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr D Biol Crystallogr. 2006;62:1002–11. doi: 10.1107/S0907444906022116. [DOI] [PubMed] [Google Scholar]
- 63.Adams PD, et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–54. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 64.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–32. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 65.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–97. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 67.Laskowski RA. PDBsum new things. Nucleic Acids Res. 2009;37:D355–9. doi: 10.1093/nar/gkn860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31:455–61. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Moriarty NW, Grosse-Kunstleve RW, Adams PD. electronic Ligand Builder and Optimization Workbench (eLBOW): a tool for ligand coordinate and restraint generation. Acta Crystallogr D Biol Crystallogr. 2009;65:1074–80. doi: 10.1107/S0907444909029436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ghosh J, Anderson PJ, Chandrasekaran S, Caparon MG. Characterization of Streptococcus pyogenes beta-NAD+ glycohydrolase: re-evaluation of enzymatic properties associated with pathogenesis. J Biol Chem. 2010;285:5683–94. doi: 10.1074/jbc.M109.070300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Meehl MA, Pinkner JS, Anderson PJ, Hultgren SJ, Caparon MG. A novel endogenous inhibitor of the secreted streptococcal NAD-glycohydrolase. PLoS Pathog. 2005;1:e35. doi: 10.1371/journal.ppat.0010035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Manzanillo PS, et al. The ubiquitin ligase parkin mediates resistance to intracellular pathogens. Nature. 2013;501:512–6. doi: 10.1038/nature12566. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.