Summary
The pluripotent epiblast (EPI) is the founder tissue of almost all somatic cells. EPI and primitive endoderm (PrE) progenitors arise from the inner cell mass (ICM) of the blastocyst stage embryo. The EPI lineage is distinctly identified by its expression of pluripotency-associated factors. Many of these factors have been reported to exhibit dynamic fluctuations of expression in embryonic stem cell cultures. Whether these fluctuations correlating with ICM fate choice occur in vivo remains an open question. Using single-cell resolution quantitative imaging of a Nanog transcriptional reporter, we noted an irreversible commitment to EPI/PrE lineages in vivo. A period of apoptosis occurred concomitantly with ICM cell fate choice, followed by a burst of EPI-specific cell proliferation. Transitions were occasionally observed from PrE-to-EPI, but not vice versa, suggesting that they might be regulated and not stochastic. We propose that the rapid timescale of early embryonic development prevents frequent fluctuations in cell fate.
Keywords: pluripotency, blastocyst, epiblast, mouse embryo, embryonic stem cells, dynamic gene expression, live imaging, nuclear segmentation, cell tracking, NANOG, GFP
Introduction
Pluripotency is defined as the ability of a cell to differentiate and give rise to all somatic and germ cells (Nichols and Smith, 2012). Although pluripotency can be induced in differentiated cells (Takahashi and Yamanaka, 2006), how a pluripotent population emerges in its native context, within the early mammalian embryo, remains an open question. Insight will come from elucidating the dynamic cell behaviors and molecular mechanisms underlying the development of the mammalian blastocyst, the embryonic stage at which a bona fide pluripotent population – the epiblast (EPI) – is established.
The EPI is molecularly-distinct and spatially segregated from the two extra-embryonic lineages, the primitive endoderm (PrE) and trophectoderm (TE) of the mouse blastocyst. The specification of these lineages occurs as two sequential binary cell fate decisions. The first involves specification and segregation of TE from ICM, while the second occurs within the ICM, and involves the specification of EPI and PrE precursors, and their eventual segregation into adjacent tissue layers [reviewed in (Schrode et al., 2013)]. By late blastocyst stage, the EPI and PrE lineages are defined both by their position within the embryo and expression of lineage-specific transcription factors, such as NANOG in the EPI, and GATA6 and GATA4 in the PrE (Xenopoulos et al., 2012). Recent studies have illustrated that EPI/PrE allocation occurs in at least three successive steps (Chazaud et al., 2006; Frankenberg et al., 2011; Plusa et al., 2008). Initially, lineage-specific transcription factors, such as NANOG and GATA6 are co-expressed by all ICM cells, suggesting a multi-lineage priming state. Thereafter, NANOG and PrE lineage-specific transcription factors exhibit mutually-exclusive expression, as lineage progenitors emerge in a salt-and-pepper distribution within the ICM. At this stage GATA4 becomes activated in PrE progenitors, concomitant with NANOG downregulation. Finally, lineage segregation is achieved with the localization of PrE cells to the surface of the ICM. At this time, other pluripotency-associated factors become restricted to EPI cells, which have become positioned internally within the ICM. Notably, NANOG is one of the first markers to be restricted within the EPI, while OCT4 and SOX2, become subsequently dowregulated in PrE progenitors, and restricted to EPI progenitors.
The initial specification of EPI and PrE progenitors appears to occur in a spatially random manner (Schrode et al., 2014), and could be achieved if a stochastic process were to underlie this second fate decision. Indeed, an analysis of transcriptomes of single ICM cells revealed that gene expression is highly heterogeneous at earlier stages, exhibiting no apparent lineage-specificity, and a hierarchical relationship of marker expression only appearing in the late blastocyst (Guo et al., 2010; Kurimoto et al., 2006; Ohnishi et al., 2014).
A degree of heterogeneity has been observed at both protein and mRNA level for various pluripotency-associated factors in embryonic stem cell (ESC) cultures. Many studies have focused on Nanog, a central component of the core pluripotency transcriptional network (Chambers et al., 2007; Kalmar et al., 2009). Experiments in ESCs have suggested that Nanog expression displays dynamic fluctuations that may correlate with a cell's fate choice between self-renewal and differentiation. However, it is unclear whether fluctuations in gene expression take place in vivo in embryos where cell differentiation occurs on a shorter time-scale, nor whether they predict fate choice or fate reversion. Notably, understanding how pluripotent cells behave in embryos may provide information that can be reconciled with observations made in ESCs (Smith, 2013).
To determine how the EPI emerges within the mouse blastocyst we generated a reporter of Nanog transcription (Nanog:H2B-GFP). Derivation of ESCs from reporter-expressing embryos revealed heterogeneous gene expression as an adaptation to ESC propagation. Using live imaging, we quantified the dynamics of Nanog expression in individual cells of live blastocysts, establishing how Nanog expression influences the fate of ICM cells. By contrast to ESCs maintained in culture, fluctuations in Nanog expression between distinct developmental states did not, generally, occur in vivo. However, we noted rare cases of cells unidirectionally switching their fate, from a PrE to EPI identity. Since ICM fate change was only observed towards the EPI, and not towards PrE, we concluded that this change was not stochastic. Our analyses also revealed events of selective apoptosis at the onset of ICM lineage differentiation, followed by a burst of cell proliferation in EPI-committed cells. Collectively, these data suggest that although it may be dynamic in ESCs, the emergence of a pluripotent identity is sequential and linear in vivo, not accommodating reversibility in fate. In this way, after its allocation, the pluripotent population might be protected ensuring the development of somatic lineages.
Results
BAC-based Nanog transcriptional reporters mark the pluripotent state in ESCs and embryos
To probe the dynamics of the pluripotent state, we developed a Nanog-based BAC transgenic transcriptional reporter, based on a previous design used as a readout of cellular reprogramming during iPS cell generation (Okita et al., 2007) (Figure S1H). For single-cell resolution readouts of Nanog expression we generated nuclear-localized human histone H2B fusion versions of the reporter (Figure 1A and C and S1E).
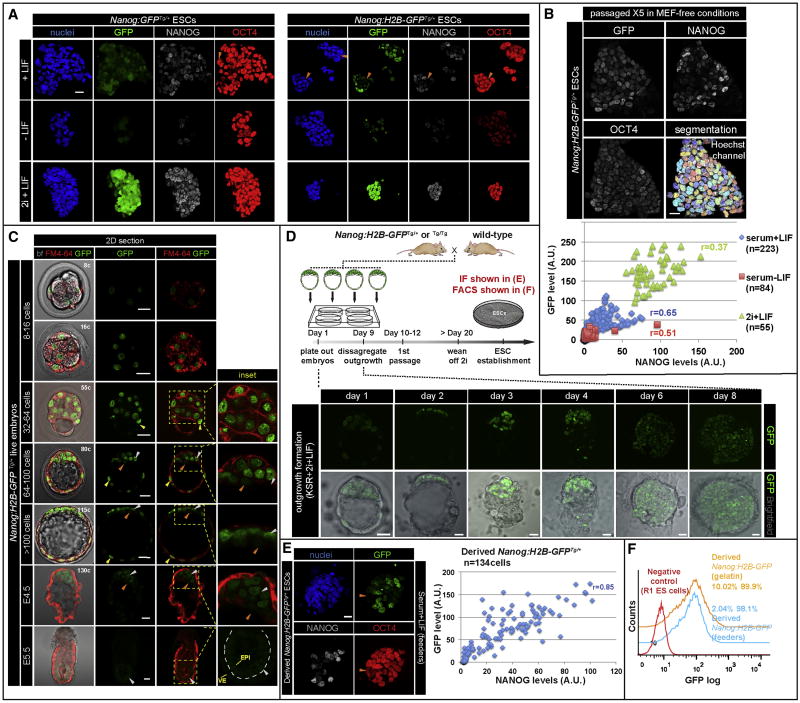
Figure 1. BAC-based Nanog transcriptional reporters faithfully mark the pluripotent state in ESCs and embryos.
(A) Immunofluorescence images of Nanog:GFPTg/+ and Nanog:H2B-GFPTg/+ ESCs grown in MEF-free conditions including serum+LIF, 2i+LIF and serum-LIF for three passages. Orange arrowheads identify GFP-low and NANOG-low cells within ESC colonies. (B) Quantitative immunofluorescence analysis after nuclear segmentation of Nanog:H2B-GFPTg/+ ESCs. GFP (y-axis) and NANOG (x-axis) fluorescence values plotted for individual ESCs propagated for five passages in MEF-free serum+LIF conditions then grown for 4 days in various culture conditions. (C) Reporter expression in live embryos stained with membrane marker FM4-64. GFP-hi cells, white arrowheads; GFP-low cells, orange arrowheads; TE cells expressing GFP, yellow arrowheads. Dashed line depicts boundary between epiblast (EPI) and visceral endoderm (VE) layers of an E5.5 embryo. Cell number was determined by staining with Hoechst. (D) Schematic of ESC derivation. After 20 days, Nanog:H2B-GFPTg/+ ESCs were established in the presence of MEF feeders in serum+LIF conditions, then propagated in the presence or absence of MEFs. (E) Immunostaining and analysis derived Nanog:H2B-GFPTg/+ ESCs. Orange arrowheads mark GFP-low/NANOG-low/OCT4+ cells. (F) FACS analysis of derived Nanog:H2B-GFPTg/+ ESCs grown in serum+LIF conditions in the presence or absence of MEFs. Numbers in FACS histograms indicate percentage of GFP- (left) and GFP+ (right) populations. r = Pearson correlation coefficient. Scale bar: 20μm.
To validate transgene activity, we analyzed reporter expression in transgenic ESCs under various culture conditions. These conditions included the presence or absence of LIF, and 2i+LIF, which promote the self-renewal of ESCs, induce differentiation or ground state pluripotency, respectively (Ying et al., 2008). Immunostaining of ESCs in 2i+LIF or serum-LIF conditions revealed markedly increased or decreased expression, respectively, of both reporter and NANOG protein. Heterogeneous but correlated GFP and NANOG expression was observed in Nanog:GFPTg/+ and Nanog:H2B-GFPTg/+ ESCs maintained in serum+LIF conditions (Figure 1A-B and S1D-E). Single-cell quantitative image analyses of immunostained Nanog:H2B-GFPTg/+ ESCs maintained under different conditions further validated reporter efficacy (Figure 1B and S1E-F). Moreover, we observed an increased correlation of reporter activity with NANOG protein for the Nanog:H2B-GFP transgene, compared to the targeted Nanog transcriptional reporter in the heterozygous TNGA ESCs (Chambers et al., 2007) (Figure S1E-F). These data suggest that the BAC transgenic reporters we constructed faithfully marked the pluripotent state in ESC cultures, and could be used to probe Nanog expression dynamics at single-cell resolution.
We next generated Nanog:H2B-GFP transgenic mice for single-cell resolution quantitative visualization of the EPI lineage in vivo. We used live imaging to analyze the distribution and validate the reporter in embryos (Figure 1C). We first observed reporter activity in embryos at the 8-16 cell stage (Plusa et al., 2008). Cells displaying high and low GFP levels were first observed at the mid-blastocyst stage (70-100cells), as a salt-and-pepper distribution of EPI and PrE precursors was established within the ICM (Chazaud et al., 2006; Plusa et al., 2008). In late blastocysts (>100cells), where the EPI and PrE lineages have sorted into distinct layers, cells within the ICM forming the EPI exhibited significantly elevated levels of GFP compared to PrE cells located on the surface of the ICM. Thereafter, GFP was detected within the EPI, albeit at reduced levels in implanting E4.5 and post-implantation E5.5 embryos consistent with the downregulation of NANOG observed at periimplantation (Chambers et al., 2003). Of note, the reporter was strongly expressed in TE cells of early and mid-blastocysts (corresponding to ∼32-90 cell stage), possibly resulting from robust NANOG localization in ICM and TE cells at these stages (Figure S1I) (Dietrich and Hiiragi, 2007; Messerschmidt and Kemler, 2010; Morgani et al., 2013). However, GFP expression in the TE was significantly reduced in late (>100cells) and in implanting blastocysts (Figure 1C and S1G). TE localization was also noted for other Nanog-based reporters analyzed at comparable embryonic stages (Figure S1G). Collectively, these data lead us to conclude that the Nanog:H2B-GFP reporter faithfully marks the pluripotent state both in vitro in ESC cultures, and in vivo in the emerging EPI lineage of the mouse blastocyst.
Derivation of ESCs from mouse blastocysts results in highly variable pluripotency-associated gene expression
The EPI lineage of the blastocyst represents the in vivo counterpart to ESC cultures propagated in vitro (Boroviak et al., 2014). We therefore sought to investigate the profile of Nanog:H2B-GFP reporter activity in transgenic embryo-derived ESCs. We used an ESC derivation protocol in which an ICM outgrowth emerges in conditions promoting ground state pluripotency in serum-free medium containing 2i (Czechanski et al., 2014). Indeed, under these conditions GFP was strongly expressed by all cells of outgrowths from Nanog:H2B-GFPTg/+ blastocysts (Figure 1D). However, both reporter and NANOG expression became heterogeneous when the derived ESCs were propagated under standard serum+LIF conditions, either in the presence or absence of mouse embryonic feeders (MEFs). Even in the presence of MEFs, GFP-low/NANOG-low cells were observed within ESC colonies (Figure 1E). FACS and quantitative immunofluorescence analyses confirmed the presence of a highly heterogeneous population, where both reporter and protein expression where highly correlated (r = 0.85) (Figure 1E-F). From these observations we conclude that gene expression heterogeneities of the pluripotent state likely arise in ESCs as a result of their in vitro propagation.
Quantitative single-cell analysis of Nanog:H2B-GFP expression in the emerging pluripotent cells of the blastocyst
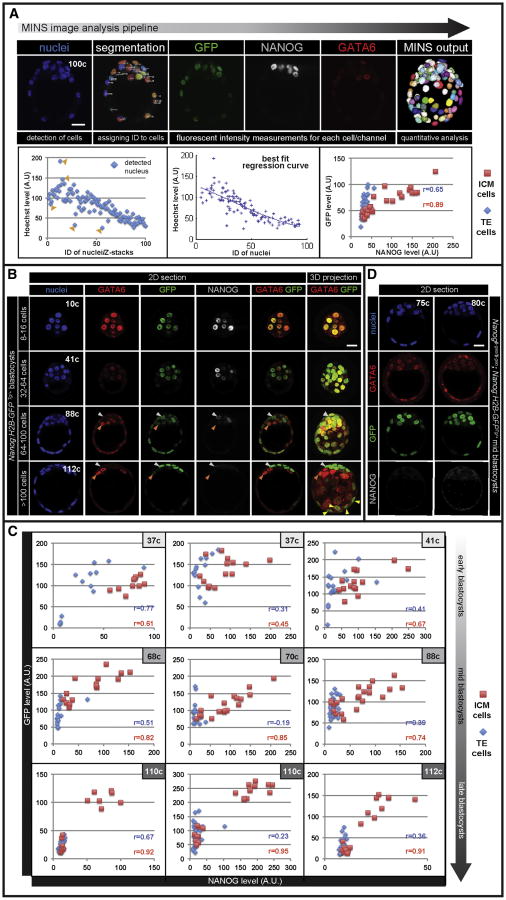
To determine whether the transgenic reporter allowed a quantitative evaluation of Nanog expression in vivo, we analyzed the pattern of reporter expression in transgenic embryos that had been fixed and stained for lineage-specific markers such as GATA6/PrE and NANOG/EPI using single-cell quantitative image analysis (Lou et al., 2014)(Figure 2A). In morulae (8-16cells) and early blastocyst (32-64cells) stages, GFP was observed throughout the embryo, reflecting a double (GATA6+ NANOG+) positive state. The differential levels of GFP expression were evident in mid-blastocysts (∼70-100cells), as embryos established a mutually-exclusive distribution of GATA6+ PrE and NANOG+ EPI progenitors (Figure 2B and S2A). In late blastocysts (>100cells), where the EPI/PrE sorting had occurred, reporter expression was markedly elevated in NANOG+ EPI progenitor cells and diminished in GATA6+ PrE progenitor cells (Figure 2B). Quantitative fluorescence analysis of immunostained Nanog:H2B-GFPTg/+ blastocysts at different stages confirmed a strong correlation between GFP levels and NANOG levels in ICM cells, as seen in ESCs, thus indicating that the reporter could be used to quantitatively infer levels of expression of Nanog (Figure 2C and S2B). Of note, reporter expression in TE cells was prominent in early and mid-blastocysts, but displayed reduced correlation with NANOG protein compared to the ICM, and hence was not investigated further (Figure 2C and S2B). The H2B-GFP reporter responds to changes in Nanog expression with a characteristic time that is determined by the stability of the H2B-GFP fusion protein. Our control experiments and computational analysis indicate that we can measure changes in Nanog expression higher than 2-fold with about 1 hour resolution (see Supplemental Experimental Procedures). Such temporal resolution is much smaller than the timescales of cell differentiation. Finally, by analyzing Nanog mutant embryos carrying the reporter, we confirmed that no functional NANOG protein was produced from the BAC transgene (Figure 2D). We therefore conclude that the Nanog:H2B-GFP reporter faithfully marks pluripotent cells emerging within the ICM, and provides an accurate quantitative readout of Nanog expression.
Figure 2. Differential expression of Nanog:H2B-GFP reporter in segregating EPI and PrE cells.
(A) MINS image analysis pipeline (see Experimental Procedures). GFP and NANOG expression plotted for individual cells after automated nuclear segmentation and fluorescence intensity normalization (best fit regression curve). (B) Immunofluorescence images of fixed Nanog:H2B-GFPTg/+ embryos. EPI cells, white arrowheads; PrE cells, orange arrowheads; TE cells expressing GFP, yellow arrowheads. (C) Quantitative immunofluorescence analyses. (D) Reporter and GATA6 and NANOG expression in Nanog®-geo/®-geo blastocysts carrying Nanog:H2B-GFP reporter. NANOG staining was absent in Nanog mutant transgenic embryos. r = Pearson correlation coefficient. Scale bar: 20μm.
Distribution of NANOG expression in the blastocyst is altered by modulation of FGF signaling
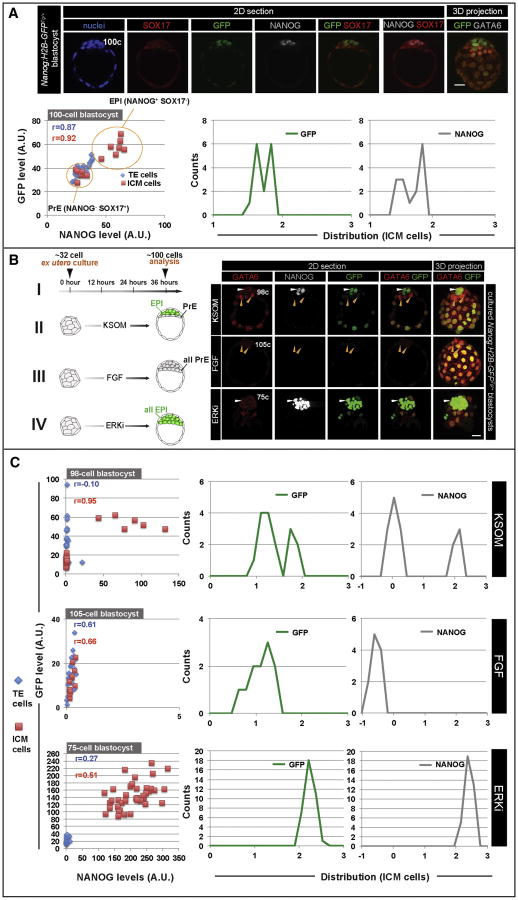
We next investigated the distribution of the Nanog:H2B-GFP transcriptional reporter, as well as NANOG protein, as the EPI compartment emerges within the ICM of late blastocysts. We performed quantitative immunofluorescence analysis on mid-to-late blastocysts (90-110cells), stained with Hoechst, NANOG and the PrE marker SOX17 (Figure 3A). We noted a bimodal distribution of reporter expression within the ICM, which corresponded to bimodality in NANOG distribution within prospective EPI and PrE cells (Figure 3A and S3A-D).
Figure 3. Nanog expression in the ICM is altered by modulation of FGF signaling.
(A) Immunofluorescence images of late blastocyst stage Nanog:H2B-GFPTg/+ embryo. Fluorescence values for GFP and NANOG plotted for individual cells. Values for GFP and NANOG of ICM cells also plotted as frequency distributions obtained by binning fluorescence values in 20 logarithmically spaced categories, as described previously (Munoz Descalzo et al., 2012). (B) Regime used for exogenous FGF and ERK1/2 inhibitor (ERKi) treatment experiments. Nanog:H2B-GFPTg/+ blastocysts recovered at E2.75 and cultured for 36 hours in KSOM medium (I), KSOM + FGF2 (II) and ERKi (III), followed by staining for Hoechst, NANOG and SOX17. White arrowheads identify NANOG+;SOX17- ICM cells exhibiting high levels of GFP. Orange arrowheads identify SOX17+;NANOG-;GFP-low ICM cells. (C) Reporter and NANOG distribution analysis in cells of cultured embryos shown in (B). Note that x- and y-axis scales for scatter plots vary due to changes in reporter and NANOG expression between embryos cultured under different conditions. r = Pearson correlation coefficient. Scale bar: 20μm.
We investigated how these distributions could be modulated by perturbation of FGF signaling, which plays a critical role in ICM lineage specification. Pathway inhibition results in an ICM comprised exclusively of EPI precursors, while incubation of embryos in exogenous FGF results in an all-PrE ICM (Chazaud et al., 2006; Kang et al., 2013; Nichols et al., 2009; Yamanaka et al., 2010). Consistent with these studies we observed that the bimodal distribution of GFP and NANOG levels within the ICM was lost when FGF signaling activity was modulated (Figure 3B-C and S3E-J). We therefore conclude that a signature heterogeneous distribution of NANOG expression distribution exists in vivo. This is represented as a stable bimodal distribution within the ICM, and reflects cell fate specification towards the PrE and EPI lineages, and that this distribution is altered by modulation of FGF signaling.
Highly variable Nanog dynamics are initially observed followed by the establishment of differential expression at the onset of ICM lineage specification
A bimodal distribution of Nanog expression is established as the pluripotent EPI emerges within the ICM. However, we currently do not know whether fluctuating expression of pluripotency-associated factors, such as Nanog, occurs during EPI specification, nor, if they do exist, whether such fluctuations correlate or predict state reversions (Smith, 2013). To address this question, we investigated the behavior of individual ICM cells in Nanog:H2B-GFPTg/+ blastocysts using time-lapse imaging coupled with quantitative image analyses performed in an accurate and automated fashion (Supplemental Experimental Procedures).
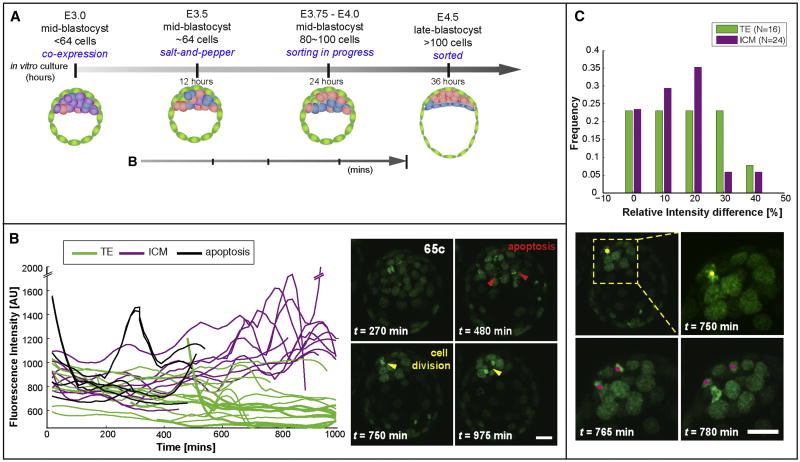
Using this methodology, we first observed that in early blastocysts (until ∼60 cell stage) all cells (ICM and TE) displayed highly heterogeneous reporter activity (0-400min in Figure 4B), consistent with previous observations (Dietrich and Hiiragi, 2007; Ohnishi et al., 2014; Plusa et al., 2008). Following this phase, TE cells downregulated reporter expression, whereas ICM cells retained or increased their expression, presumably coinciding with the establishment of a lineage bias (400min-end of movie, Figure 4B). We also noted apoptotic events, occurring during the process of lineage specification in randomly positioned GFP-hi or GFP-low ICM cells (red arrowheads; Figure 4B). As described previously, apoptotic events occur during blastocyst development and are not a consequence of in vitro culture or phototoxicity (Artus et al., 2013; Plusa et al., 2008). Furthermore, we noted several cell divisions occurring in GFP-hi ICM cells (yellow arrowheads; Figure 4B). Heritability of Nanog:H2B-GFP levels, and likely cell fate, was observed in all ICM cells after division (Figure 4C). Of note, an increase in GFP fluorescence was routinely observed during mitosis, due to chromosome condensation; thus any resulting rapid oscillations (of the order of 150 mins) in reporter activity were excluded from the analysis, as they were not considered as reflecting changes in gene expression. By using a computational approach we confirmed that the GFP expression was not diluted in dividing cells, suggesting that daughter cells inherited the lineage identity of their parental cell (Figure S4). Finally, cells within the TE displayed low, somewhat variable and continually decreasing levels of reporter activity (Figure 4B).
Figure 4. EPI cells emerge in vivo with an accompanying increase in Nanog expression.
(A) Schematic of embryo development. (B) Quantification of GFP intensity in single nuclei of a living Nanog:H2B-GFPTg/+ blastocyst recovered at E3.5 (65-cells) and corresponding images of single time-points. Single-cell intensity traces were obtained by tracking single cells in time-lapse experiments. Mitotic and apoptotic cells were also tracked, resulting in abrupt peaks in fluorescence intensity. Cells belonging to the ICM are depicted in purple, while TE cells are depicted in green; cells having undergone apoptosis are depicted as black lines. The developmental timing and the time spanned by tracks analysed in this panel are illustrated by an arrow (A). (C) Top panel, quantification of relative GFP intensity in daughters/sisters versus mother cells upon division in cells of TE (green bars) or ICM (purple bars) origin. Daughter cells retained expression levels after mitosis, with most changes within 20%. Data were obtained from the analysis of time-lapse movies of 5 or more embryos. Bottom panel, cell divisions of EPI-progenitors occurring at late blastocyst stages shown in (B). Red arrowheads identify apoptotic events, and yellow arrowheads mark cell divisions. Scale bar: 20 μm.
We next focused our analyses on subsequent phases of development, initiating from early-to-mid blastocyst stages (at around 50-60 cell) until late blastocyst stages (at around 100 cells), where the EPI and PrE cells have sorted to their final positions (Figure 5A-B and Movie S1). A subpopulation of ICM cells is specified to the EPI-lineage, with GFP expression maintained or increased. By contrast, cells biased toward the PrE lineage extinguished the reporter during differentiation (Figure 5A).
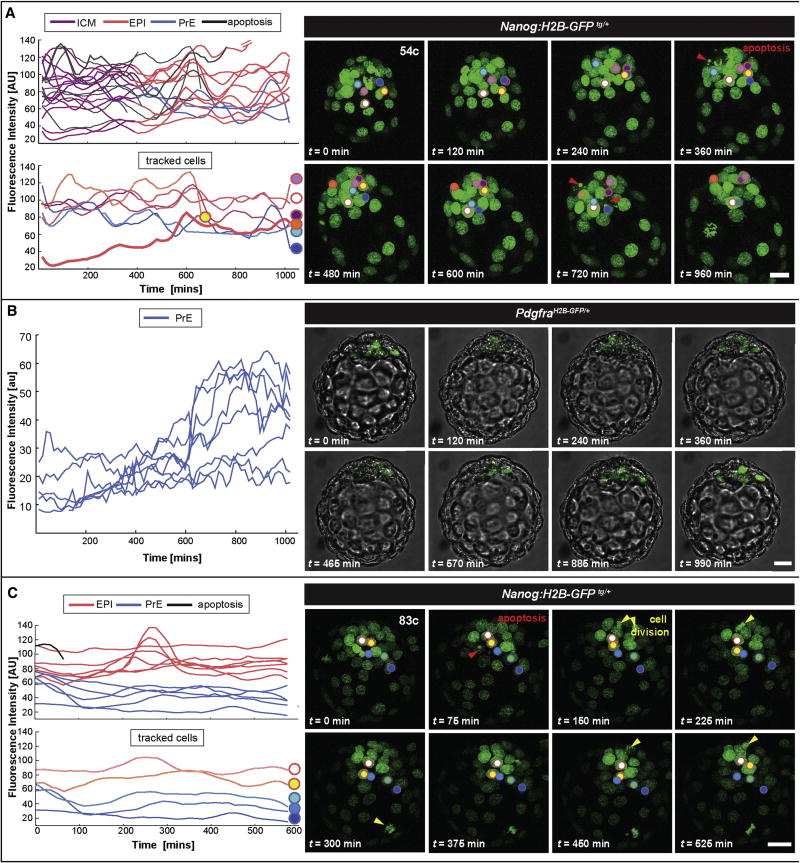
Figure 5. State reversals can occur towards a pluripotent identity during lineage specification in mid-blastocysts, but cells do not change fate in late embryos.
(A) Quantification of GFP intensity in single nuclei of a living Nanog:H2B-GFPTg/+ at E3.5 (54 cells) and corresponding snapshots from a time-lapse movie. A subset of tracks are detailed in the lower plot, corresponding to cells highlighted in the images in the panel on the right. Cell highlighted with an orange dot and red outline represents a GFP-low cell that upregulated reporter expression and contributed to EPI. (B) Quantification of GFP intensity in single nuclei of a living PdgfraH2B-GFP/+ at E3.5 (∼60 cells) and corresponding snapshot of fluorescence channel overlapped with bright field images from a time-lapse movie. Only PrE-biased cells express H2B-GFP. (C) The same analysis applied in panel (A) was repeated in a later Nanog:H2B-GFPTg/+ embryo at E3.75 (85 cells). ICM cells (purple) segregated towards EPI (red) and PrE (blue) lineages. Cells that underwent apoptosis depicted with black lines. Red arrowheads mark apoptotic events and yellow arrowheads mark cell divisions. Tracked nuclei are highlighted by dots; the outline of each dot depicts the lineage choice; red-outline depicts EPI progenitor cells, blue-outline depicts PrE progenitor cells. Scale bar: 20 μm.
Infrequent cell state reversals occur towards, not away from, a pluripotent identity
Notably, we observed few instances where cells in mid-stage blastocysts, exhibiting low levels of reporter activity, would rapidly increase reporter activity, and concomitantly become segregated with the EPI (Figure 5A). These could, in principle, represent rare PrE-to-EPI conversions. Importantly, we never observed events of EPI-to-PrE progenitor switching associated with downregulation of Nanog expression. To obtain an independent confirmation of the absence of EPI-to-PrE transitions, we analyzed time-lapse data from a PrE-specific single-cell resolution reporter (PdgfraH2B-GFP/+, Plusa et al., 2008). In contrast to Nanog:H2B-GFP, expression of the Pdgfra reporter is activated only in PrE-biased cells after cell fate specification. Therefore, PrE-to-EPI conversion would be detected as downregulation of the reporter. On the other hand, EPI-to-PrE conversion would give rise to significantly delayed activation of the reporter in cells lacking expression. We imaged embryos expressing the Pdgfra reporter starting at the time of cell differentiation, i.e. the time when expression of the reporter becomes reliably detectable. We found that all positive cells at the onset of differentiation retained their expression and no new cells initiated expression with a significant delay, corresponding to an EPI-to-PrE conversion (Figure 5B). Collectively, our results with the Nanog:H2B-GFP and Pdgfra reporters imply that cell fate reversals are extremely rare and preferentially happen from the PrE to the EPI state.
The PrE-to-EPI unidirectionality would suggest that fate transitions are regulated, and do not result from purely stochastic fluctuations in gene expression. To rule out that the inability to detect EPI-to-PrE transition in Nanog:H2B-GFP embryos was due to perdurance of GFP reporter, we performed the following analysis. First, we quantified the dynamics of downregulation of Nanog:H2B-GFP in PrE cells (Figure S6) and estimated the lifetime of H2B-GFP to be shorter than 4 hours, which is short enough to detect cell fate reversal events (details in Supplemental Experimental Procedures). Second, we analyzed time-lapse movies of the Nanog:H2B-GFP reporter in embryos treated with exogenous FGF (all ICM cells would downregulate NANOG) and showed that a clear downregulation can be observed (Figure S5). In addition, we calculated the half-life of H2B-GFP from time-lapse movies of Nanog:H2B-GFP in exogenous FGF or cycloheximide (CHX). The decay rates of H2B-GFP in ICM cells in the embryos under CHX or FGF treatments were around 6.5 and 5.5 hours, respectively (Figure S6). Collectively, these experiments allow us to conclude that the half-life of H2B-GFP is shorter than 6 hours and thus not dissimilar to the half-life of NANOG, which has been calculated to be approximately 4 hours (Abranches et al., 2013). Altogether, these results show that the Nanog:H2B-GFP reporter allows measuring Nanog transcriptional dynamics on timescales longer than 1 hour (see Supplemental Experimental Procedures).
At late blastocyst stages, EPI and PrE do not exhibit fluctuations
We next examined whether fluctuations in Nanog:H2B-GFP, and thus Nanog, levels were observed after ICM lineage specification. At these late blastocyst stages (>90-100cells), the active sorting of EPI and PrE populations to adjacent tissue layers is evident. Our analyses revealed that the EPI cells exhibited increasing levels of reporter activity, whereas a decrease in GFP levels was observed in cells forming the emergent PrE epithelial layer (0min-end of movie; Figure 5C and Movie S1). At the end of the specification period, apoptotic events were observed (red arrowhead; 0-100min; Figure 5C). Thereafter, we did not observe any apoptosis. Instead, after ICM lineage specification, several cell divisions were observed in EPI-progenitors (yellow arrowheads; Figure 5C). Collectively these observations suggest a wave of temporarily restricted apoptotic events, occurring around the period of lineage specification, followed by a burst of EPI lineage-specific cell proliferation. Nanog levels remained stable after ICM cell fate divergence suggesting that fluctuations between EPI and PrE states do not occur. Based on these data, we conclude that within the ICM the majority of pluripotent EPI and extra-embryonic PrE progenitor cells do not change their fate after specification. Furthermore, our data lead us to propose that a burst of cell proliferation following cell differentiation ensures EPI lineage-specific expansion.
Quantitative analyses of Nanog:H2B-GFP time-lapse movies revealed a correlation between cell behaviors and cell fate choice within the ICM
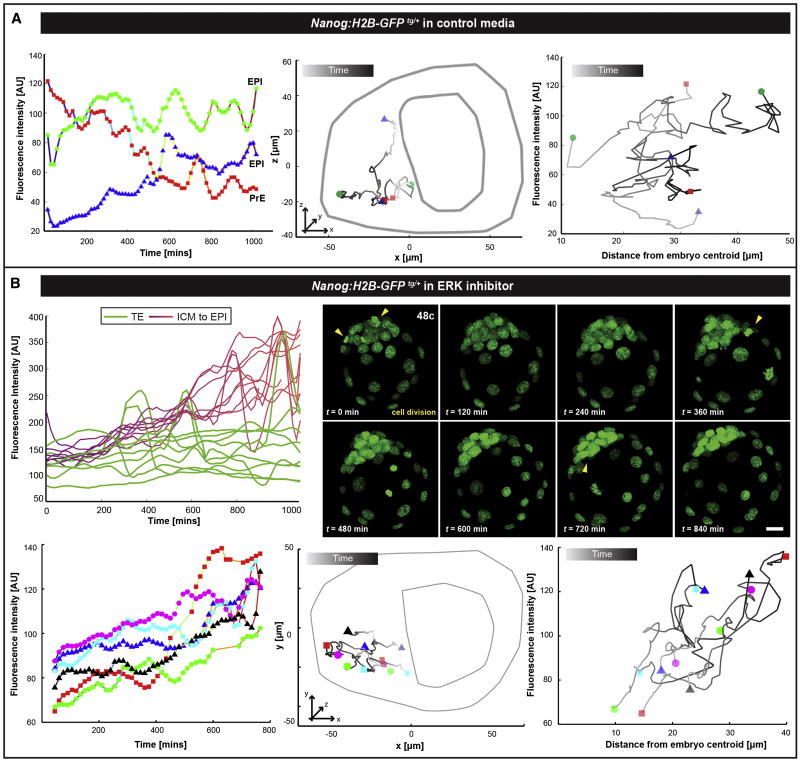
Time-lapse imaging in combination with cell tracking and quantitative analysis allowed us to determine whether fate reversals correlate with the spatial position of individual cells within the ICM. We simultaneously analyzed the position of a cell relative to the blastocyst cavity, and its Nanog reporter expression levels as a function of time. Analysis of the spatial distribution of cells converting from PrE-to-EPI indicated that the cells migrated towards the inner region of the ICM (Figure 6A, blue cell). The cell undergoing fate switching exhibited similar rate of GFP increase as in cells of the embryo under conditions of ERK inhibition. This observation suggests that the fate switching might be a result of PrE-biased cells that stop responding to ERK signaling (due to their localization and fates of their neighbors) and, as a consequence, increase Nanog expression and change fate (Figure 6B). Consistently, under conditions of ERK inhibition, all ICM cells committed to an EPI fate and displayed increasing reporter activity, whereas GFP expression in TE cells remained unaffected, consistent with our previous observations in fixed embryos (Figure 3B and C and FigureS3E-J). Notably, we did not observe any apoptotic events during ERK inhibition, whereas a burst of proliferation was evident in the all-EPI ICM (yellow arrowheads in Figure 6B and Movie S2). These findings support our previous observations, suggesting that apoptosis serves as a selection mechanism within the ICM, while the proliferation burst in EPI-committed cells ensures the rapid expansion of the pluripotent lineage after its specification. In addition, we observed that there was no abrupt spatial position change of EPI cells within the ICM of ERK inhibitor-treated embryos; instead cell motility was restrained and only passive movement of cells was observed as these embryos developed (Figure 6B).
Figure 6. Cell state reversals towards a pluripotent identity are associated with changes in position within ICM.
Fluorescence levels of single ICM cells (A, left and B lower left panels) and two-dimensional projections of representative space trajectories (A central and B lower central panels) are plotted for the duration of time-lapse movies of Nanog:H2B-GFPTg/+ (A) corresponding to embryo in Figure 5A and for a Nanog:H2B-GFPTg/+ embryo grown in the presence of ERKi (B). The distance of each cell from the barycenter of the embryo is plotted vs. GFP intensity (A right and B lower right panels). (A) EPI cells: green and blue; PrE cell: red trajectories. Blue trajectory represents a cell, which switched state acquiring an EPI identity; moving from an initial position on the surface of ICM inwards while increasing Nanog expression. (B) Top panels, quantification of GFP fluorescence intensities and corresponding snapshots. Bottom panels, all six trajectories depict the behavior of ICM cells that acquire an EPI fate increasing reporter activity. All cells change their absolute position due to embryo growth, but there are no events of abrupt spatial change in position of individual cells within the cohort. Lines represent cell trajectories. Displayed trajectories for mitotic cells were excluded. Unique colors identify individual cells. The outline of the embryo is drawn for plotting relative position of nuclei. Shades of gray in the trajectory depict time; earlier (light) to later (darker).
Discussion
Pluripotency-associated factors such as Nanog have been reported to exhibit dynamic fluctuations of expression in ESC cultures. Whether fluctuations in gene expression correlating with cell state transitions occur in vivo remains an open question. Here we have investigated Nanog expression heterogeneities using single-cell resolution Nanog transcriptional reporters coupled with 3D time-lapse imaging and high-resolution automated quantitative image analyses. Our data suggest that BAC-based Nanog:H2B-GFP transgenic reporters are expressed at physiological levels, exhibit minimal reporter perdurance, and serve as a faithful readouts of Nanog expression both in vitro in ESCs, and in vivo in mouse embryos.
For both ESCs rendered transgenic through introduction of a Nanog:H2B-GFP construct, as well as transgenic embryo-derived ESCs, heterogeneities in the levels of reporter, as well as NANOG protein were noted. A GFP-low cell population, which also displayed low levels of NANOG protein, was evident in cells that were propagated in MEF-free conditions, which promote a differentiation bias; this population displayed OCT4 expression (Figure 1A-B and S1E), suggesting that it might consist of pluripotent cells that were actively differentiating and/or primed for differentiation. Importantly, this variability, and the presence of a GFP-low/NANOG-low population, in ESC cultures have also been recently reported with another Nanog transcriptional reporter (Abranches et al., 2013). These observations therefore call for caution in the choice of culture conditions for ESC propagation; hence, it was recently shown that random monoallelic gene expression could occur stochastically as ESCs differentiated, resulting in the acquisition of heterogeneities during the adaptation of cells to in vitro culture (Eckersley-Maslin et al., 2014; Gendrel et al., 2014).
Furthermore, it has been suggested that reporters targeted to the Nanog locus which concomitantly ablate Nanog activity, might exhibit behaviors resulting from Nanog heterozygosity (Faddah et al., 2013; Filipczyk et al., 2013). We noted that GFP expression from the knock-in/knock-out TNGA reporter was elevated compared to a BAC-based Nanog:GFPTg/+ reporter (Figure S1A-C). We therefore investigated whether Nanog allele heterozygosity per se might result in increased NANOG expression, consistent with its reported auto-repressive activity (MacArthur et al., 2012; Navarro et al., 2012). However, we failed to observe any noticeable difference in reporter activity between Nanog:H2B-GFPTg/+ ESCs that harbored a two (wild-type) or one functional Nanog alleles (Figure S1J-K). Thus, our data suggest that, the elevated GFP expression observed in the TNGA ESCs is not the result of Nanog heterozygosity, and could result from allele design. The development of a faithful EPI lineage-specific reporter providing a single-cell resolution quantitative readout of Nanog expression, coupled with high-resolution image data analyses, allowed us to address a central open question pertaining the behavior of EPI cells in vivo. Critical for this type of analysis was a single-cell resolution live imaging reporter that was sufficiently bright for time-lapse image acquisition, but expressed at physiological levels, so that reporter perdurance would not mask downregulation in cells. Of note, our experience with destabilized fluorescent protein reporters reveals reduced levels of fluorescence, not amenable to the image analyses methodologies used in this study.
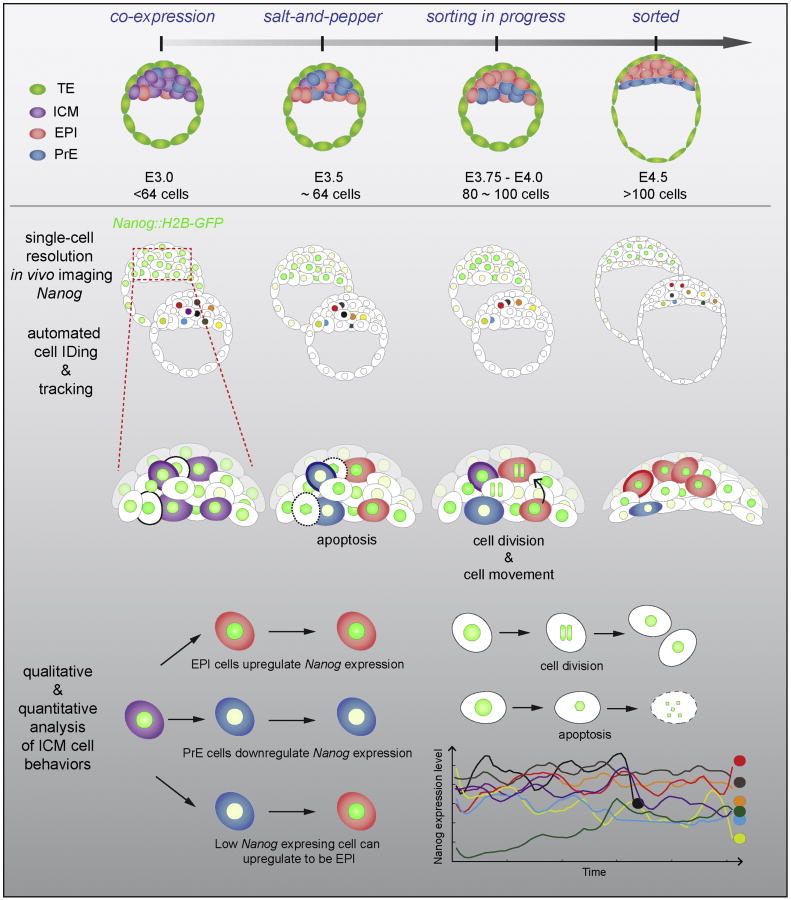
Our data reveal a range of cell behaviors before and after the pluripotent EPI population has been specified in vivo within the ICM (Figure 7). Prior to EPI vs. PrE specification has occurred, apoptosis serves as a selective mechanism to ensure proper segregation of lineage progenitors. Rare fate reversal events likely occur whereby GFP-low/PrE-progenitor cells convert to a GFP-hi/EPI-progenitor state as cells migrate inwards in the ICM. At this time, cell migration correlates with fate choice and is linked to the presence of a heterogeneous population of EPI and PrE progenitors. Progenitors could be sorting towards a niche comprising cells with a similar lineage bias. By contrast, in the presence of ERK inhibition, lineage specification is forced towards one direction (all-EPI) resulting in a homogenous ICM population, and is accompanied by a lack of cell movement (Figure 6B). Finally, after lineage specification has occurred within the ICM, fluctuations between EPI and PrE progenitors were not observed. However, a burst of cell proliferation was observed in EPI progenitors, as they sorted to the interior of the ICM.
Figure 7. Cell behaviors during the emergence of the pluripotent EPI lineage.
Schematic representation of embryo development from mid-to-late blastocyst. Prior to lineage specification (around 64-80 cells): (1) apoptosis occurs randomly in EPI or PrE-progenitors as they segregate to their appropriate layers, (2) segregation is linked to spatial movements within the ICM, and (3) few GFP-low cells might acquire a pluripotent identity and migrate inwards the ICM. After lineage specification (>80-90 cells): (1) cells do not fluctuate between EPI and PrE states, (2) cell divisions occur in EPI-specified population while relocating to the interior of the ICM.
Rarely cells changed their state towards, but never away from, a pluripotent identity. By using a single-cell resolution reporter for the PrE lineage, we previously showed that PrE-progenitor cells could downregulate reporter expression, but could not confirm the fate reversal to EPI, as cells could not be followed after the downregulation (Plusa et al., 2008). Here, using a single-cell resolution reporter of the EPI lineage, we directly visualized these transitions. Perhaps cells converting from PrE-to-EPI might cease responding to an FGF signal, as in the presence of an ERK-inhibitor, resulting in Nanog upregulation and establishment of pluripotency (Figure 6A, blue cell). These data agree with recent studies suggesting that pulsatile FGF signaling induces differential Nanog expression within ICM cells, and drives Nanog mRNA degradation for rapid post-transcriptional control of pluripotency (Torres-Padilla and Chambers, 2014, Tan and Elowitz, 2014).
Importantly, we observed no fate reversals between EPI and PrE subsequent to their specification. This is in agreement with a recent mathematical model accounting for the dynamics of the regulatory network that controls ICM differentiation; simulations indicated that after specification, cells do not change identity, and thus EPI and PrE states are not interchangeable (Bessonnard et al., 2014). In addition, we observed that after specification, a burst of cell proliferation in the pluripotent compartment was evident. This might ensure adequate numbers of EPI progenitors available for subsequent development. Furthermore, our observations suggest that PrE progenitors exhibit increased plasticity, compared to EPI progenitors, consistent with recent studies suggesting that PrE progenitors have a broader developmental potential than their EPI counterparts (Grabarek et al., 2012), and observations reporting that ESCs primed towards endoderm co-express embryonic and extra-embryonic markers (Morgani et al., 2013).
Our data suggest that once specified in wild-type embryos, pluripotent cells do not, in general, change their fate, but a very limited number of state reversals may occur, but only towards a pluripotent identity. During development, sufficient numbers of lineage progenitors must be generated, and there is a time window during which cell fate reversals can occur. Moreover, mechanisms involving apoptotic events and symmetric cell divisions may ensure that a pluripotent identity is protected and maintained in vivo. These observations within the embryonic environment appear contrasting with studies in ESCs. A possible explanation for this disparity could be that ESCs in culture do not receive the appropriate inputs from a niche, namely neighboring cells and extra-cellular components. Another could be due to the differential time-scales. ICM cells commit to PrE and EPI fates in less than 24 hours, and since development proceeds in a unidirectionally, even though there may be non-differentiating (or symmetric) cell divisions, neither cell population arising within the ICM self-renews. By contrast, ESCs can be maintained indefinitely in vitro under conditions of self-renewal; a time-scale that presents an unrestricted period for conversion between alternative states.
Experimental Procedures
Mouse husbandry
Mice were maintained under a 12-hour light-dark cycle. Mouse lines used in this study were Nanog:H2B-GFPTg/+, Nanog®-geo/+ (Mitsui et al., 2003) and NanogGFP/+ (Hatano et al., 2005). Alleles are schematized in Figure S1H.
Live embryo imaging
For live imaging, embryos were cultured in glass-bottomed dishes (MatTek) in an environmental chamber as previously (Kang et al., 2013). Live imaging conditions used were compatible with normal development as shown previously (Plusa et al., 2008). For incubation experiments an ERK1/2 inhibitor, 1 μM PD0325901 (StemGent) was added to medium 2-3 hours prior to initiation of 3D time-lapse imaging. Green fluorescent protein (GFP) was excited using a 488-nm Argon laser. Live image data were acquired using three laser scanning confocal imaging systems: Zeiss LSM510META, LSM710, LSM780 and Leica SP8. Images were acquired using 20×/0.75, 40×/1.3 or 63×/1.4 objectives. 20-30 xy planes separated by 2 μm were acquired per z-stack, every 15 minutes. Movies of time-lapse sequences were compiled and annotated using QuickTime Pro (Apple Inc.).
Immunostaining of ESCs and embryos
Immunostaining of ESCs and embryos was performed as previously (Kalmar et al., 2009; Kang et al., 2013; Munoz Descalzo et al., 2012). Primary antibodies used were CDX2 (1:100, Biogenex), GATA6 (1:100, R&D systems), NANOG (1:500, Cosmobio), OCT4 (1:100, Santa Cruz) and SOX17 (1:100, R&D systems). Secondary AlexaFluor (Invitrogen) conjugated antibodies were used at 1:500. DNA was visualized with Hoechst 33342 (5 μg/ml; Invitrogen).
Quantitative fluorescence image analysis
Quantitative fluorescence measurements from images of fixed ESCs, and fixed or live embryos were performed using an automated image processing workflow comprising the segmentation of multiple nuclei in 3D data (Figure 2A). The frontend software, MINS, is a MATLAB (Mathworks) based Graphic User Interface described previously (Lou et al., 2014, http://katlab-tools.org). We noted that for normalizing fluorescence values of each channel the Hoechst channel per cell was not optimal to compensate for loss of fluorescence intensity throughout the sample depth (differences in Hoechst fluorescence intensity throughout z-stack indicated by orange arrowheads in Figure 2A). We thus developed an algorithm to generate a regression curve across values of the Hoechst channel for individual cells (details in Supplemental Experimental Procedures). Fluorescence values for other channels were normalized using this curve (Figure 2A). To compare fluorescence values between ICM and TE cells, TE cells being analyzed needed to be in the same focal plane as ICM cells. Thus, TE cells positioned at the beginning and end of the z-stack were excluded from the analysis. Only accurately segmented nuclei were included in analyses.
Nuclear segmentation and cell tracking
Using the segmentation output of the MINS software we developed an algorithm for cell tracking. To determine the position of any given cell at a subsequent time-point, the algorithm first seeks to identify a clear, well-defined minimum in the distance between the cell centroid and the centroids of all cells in the subsequent frame. If such minimum does not exist, the algorithm uses cell segmentation to look for maximum overlap between cells at the subsequent time-point and the cell being tracked. As additional criteria, changes in nuclear shape and overall distance are required to lie within control ranges. Such criteria generate unambiguous cell assignments and provide reliable tracking of a large fraction of cells (60-70% in this study). The algorithm was validated by visual inspection, and each track was obtained by matching forward (prospective) and reverse (retrospective) tracking data (details provided in Supplemental Experimental Procedures).
Supplementary Material
Highlights.
Single-cell resolution live imaging of the pluripotent EPI lineage in vivo
Fluctuations between distinct developmental states do not occur in vivo
Occasionally cells can change their fate towards a pluripotent identity
Rapid expansion of the pluripotent lineage once specified
Acknowledgments
We thank X. Lou for developing the algorithm used to calculate regression curves for fluorescence intensity normalizations; J. Nichols for the Nanog®geo/+ mouse strain, A. Martinez-Arias for TNGA ESCs; S. Nowotschin, N. Saiz and N. Schrode for discussions and comments on the manuscript; the Memorial Sloan Kettering Molecular Cytology and Rockefeller University Bio-Imaging Core Facilities for use of their instruments for live embryo imaging. Work in A.-KH's. laboratory is supported by the National Institutes of Health (R01-HD052115 and R01-DK084391) and NYSTEM (N13G-236). S.D. is supported by National Institutes of Health (R00-HD074670). A.P. is supported by Finalized Research and Founding for Investments in Basic Research (RBAP11BYNP-Newton).
Footnotes
Author Contributions: P.X. and A.-K.H. conceived the project and designed the experiments. P.X. generated the Nanog-H2B-GFP mouse line and performed ESC and fixed embryo imaging experiments. M.K. performed time-lapse embryo imaging experiments. A.P. and S.D.T. carried out quantitative analyses of experimental datasets. P.X., M.K. and A.-K.H. wrote the manuscript with input from A.P. and S.D.T.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abranches E, Bekman E, Henrique D. Generation and characterization of a novel mouse embryonic stem cell line with a dynamic reporter of Nanog expression. PLoS One. 2013;8:e59928. doi: 10.1371/journal.pone.0059928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artus J, Kang M, Cohen-Tannoudji M, Hadjantonakis AK. PDGF signaling is required for primitive endoderm cell survival in the inner cell mass of the mouse blastocyst. Stem cells. 2013;31:1932–1941. doi: 10.1002/stem.1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessonnard S, De Mot L, Gonze D, Barriol M, Dennis C, Goldbeter A, Dupont G, Chazaud C. Gata6, Nanog and Erk signaling control cell fate in the inner cell mass through a tristable regulatory network. Development. 2014 doi: 10.1242/dev.109678. [DOI] [PubMed] [Google Scholar]
- Boroviak T, Loos R, Bertone P, Smith A, Nichols J. The ability of inner-cell-mass cells to self-renew as embryonic stem cells is acquired following epiblast specification. Nature cell biology. 2014;16:516–528. doi: 10.1038/ncb2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers I, Colby D, Robertson M, Nichols J, Lee S, Tweedie S, Smith A. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell. 2003;113:643–655. doi: 10.1016/s0092-8674(03)00392-1. [DOI] [PubMed] [Google Scholar]
- Chambers I, Silva J, Colby D, Nichols J, Nijmeijer B, Robertson M, Vrana J, Jones K, Grotewold L, Smith A. Nanog safeguards pluripotency and mediates germline development. Nature. 2007;450:1230–1234. doi: 10.1038/nature06403. [DOI] [PubMed] [Google Scholar]
- Chazaud C, Yamanaka Y, Pawson T, Rossant J. Early lineage segregation between epiblast and primitive endoderm in mouse blastocysts through the Grb2-MAPK pathway. Dev Cell. 2006;10:615–624. doi: 10.1016/j.devcel.2006.02.020. [DOI] [PubMed] [Google Scholar]
- Czechanski A, Byers C, Greenstein I, Schrode N, Donahue LR, Hadjantonakis AK, Reinholdt LG. Derivation and characterization of mouse embryonic stem cells from permissive and nonpermissive strains. Nat Protoc. 2014;9:559–574. doi: 10.1038/nprot.2014.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich JE, Hiiragi T. Stochastic patterning in the mouse pre-implantation embryo. Development. 2007;134:4219–4231. doi: 10.1242/dev.003798. [DOI] [PubMed] [Google Scholar]
- Eckersley-Maslin MA, Thybert D, Bergmann JH, Marioni JC, Flicek P, Spector DL. Random Monoallelic Gene Expression Increases upon Embryonic Stem Cell Differentiation. Developmental cell. 2014;28:351–365. doi: 10.1016/j.devcel.2014.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faddah DA, Wang H, Cheng AW, Katz Y, Buganim Y, Jaenisch R. Single-cell analysis reveals that expression of nanog is biallelic and equally variable as that of other pluripotency factors in mouse ESCs. Cell Stem Cell. 2013;13:23–29. doi: 10.1016/j.stem.2013.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipczyk A, Gkatzis K, Fu J, Hoppe PS, Lickert H, Anastassiadis K, Schroeder T. Biallelic expression of nanog protein in mouse embryonic stem cells. Cell Stem Cell. 2013;13:12–13. doi: 10.1016/j.stem.2013.04.025. [DOI] [PubMed] [Google Scholar]
- Frankenberg S, Gerbe F, Bessonnard S, Belville C, Pouchin P, Bardot O, Chazaud C. Primitive endoderm differentiates via a three-step mechanism involving Nanog and RTK signaling. Developmental cell. 2011;21:1005–1013. doi: 10.1016/j.devcel.2011.10.019. [DOI] [PubMed] [Google Scholar]
- Gendrel AV, Attia M, Chen CJ, Diabangouaya P, Servant N, Barillot E, Heard E. Developmental dynamics and disease potential of random monoallelic gene expression. Developmental cell. 2014;28:366–380. doi: 10.1016/j.devcel.2014.01.016. [DOI] [PubMed] [Google Scholar]
- Grabarek JB, Zyzynska K, Saiz N, Piliszek A, Frankenberg S, Nichols J, Hadjantonakis AK, Plusa B. Differential plasticity of epiblast and primitive endoderm precursors within the ICM of the early mouse embryo. Development. 2012;139:129–139. doi: 10.1242/dev.067702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo G, Huss M, Tong GQ, Wang C, Li Sun L, Clarke ND, Robson P. Resolution of cell fate decisions revealed by single-cell gene expression analysis from zygote to blastocyst. Developmental cell. 2010;18:675–685. doi: 10.1016/j.devcel.2010.02.012. [DOI] [PubMed] [Google Scholar]
- Hatano SY, Tada M, Kimura H, Yamaguchi S, Kono T, Nakano T, Suemori H, Nakatsuji N, Tada T. Pluripotential competence of cells associated with Nanog activity. Mech Dev. 2005;122:67–79. doi: 10.1016/j.mod.2004.08.008. [DOI] [PubMed] [Google Scholar]
- Kalmar T, Lim C, Hayward P, Munoz-Descalzo S, Nichols J, Garcia-Ojalvo J, Martinez Arias A. Regulated fluctuations in nanog expression mediate cell fate decisions in embryonic stem cells. PLoS Biol. 2009;7:e1000149. doi: 10.1371/journal.pbio.1000149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang M, Piliszek A, Artus J, Hadjantonakis AK. FGF4 is required for lineage restriction and salt-and-pepper distribution of primitive endoderm factors but not their initial expression in the mouse. Development. 2013;140:267–279. doi: 10.1242/dev.084996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurimoto K, Yabuta Y, Ohinata Y, Ono Y, Uno KD, Yamada RG, Ueda HR, Saitou M. An improved single-cell cDNA amplification method for efficient high-density oligonucleotide microarray analysis. Nucleic acids research. 2006;34:e42. doi: 10.1093/nar/gkl050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou X, Kang M, Xenopoulos P, Munoz-Descalzo S, Hadjantonakis AK. A Rapid and Efficient 2D/3D Nuclear Segmentation Method for Analysis of Early Mouse Embryo and Stem Cell Image Data. Stem cell reports. 2014;2:382–397. doi: 10.1016/j.stemcr.2014.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacArthur BD, Sevilla A, Lenz M, Muller FJ, Schuldt BM, Schuppert AA, Ridden SJ, Stumpf PS, Fidalgo M, Ma'ayan A, et al. Nanog-dependent feedback loops regulate murine embryonic stem cell heterogeneity. Nature cell biology. 2012;14:1139–1147. doi: 10.1038/ncb2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messerschmidt DM, Kemler R. Nanog is required for primitive endoderm formation through a non-cell autonomous mechanism. Dev Biol. 2010;344:129–137. doi: 10.1016/j.ydbio.2010.04.020. [DOI] [PubMed] [Google Scholar]
- Mitsui K, Tokuzawa Y, Itoh H, Segawa K, Murakami M, Takahashi K, Maruyama M, Maeda M, Yamanaka S. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell. 2003;113:631–642. doi: 10.1016/s0092-8674(03)00393-3. [DOI] [PubMed] [Google Scholar]
- Morgani SM, Canham MA, Nichols J, Sharov AA, Migueles RP, Ko MS, Brickman JM. Totipotent embryonic stem cells arise in ground-state culture conditions. Cell reports. 2013;3:1945–1957. doi: 10.1016/j.celrep.2013.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz Descalzo S, Rue P, Garcia-Ojalvo J, Martinez Arias A. Correlations between the levels of Oct4 and Nanog as a signature for naive pluripotency in mouse embryonic stem cells. Stem cells. 2012;30:2683–2691. doi: 10.1002/stem.1230. [DOI] [PubMed] [Google Scholar]
- Navarro P, Festuccia N, Colby D, Gagliardi A, Mullin NP, Zhang W, Karwacki-Neisius V, Osorno R, Kelly D, Robertson M, et al. OCT4/SOX2-independent Nanog autorepression modulates heterogeneous Nanog gene expression in mouse ES cells. Embo J. 2012;31:4547–4562. doi: 10.1038/emboj.2012.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols J, Silva J, Roode M, Smith A. Suppression of Erk signalling promotes ground state pluripotency in the mouse embryo. Development. 2009;136:3215–3222. doi: 10.1242/dev.038893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols J, Smith A. Pluripotency in the embryo and in culture. Cold Spring Harb Perspect Biol. 2012;4:a008128. doi: 10.1101/cshperspect.a008128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi Y, Huber W, Tsumura A, Kang M, Xenopoulos P, Kurimoto K, Oles AK, Arauzo-Bravo MJ, Saitou M, Hadjantonakis AK, et al. Cell-to-cell expression variability followed by signal reinforcement progressively segregates early mouse lineages. Nature cell biology. 2014;16:27–37. doi: 10.1038/ncb2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313–317. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- Plusa B, Piliszek A, Frankenberg S, Artus J, Hadjantonakis AK. Distinct sequential cell behaviours direct primitive endoderm formation in the mouse blastocyst. Development. 2008;135:3081–3091. doi: 10.1242/dev.021519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrode N, Saiz N, Di Talia S, Hadjantonakis AK. GATA6 levels modulate primitive endoderm cell fate choice and timing in the mouse blastocyst. Developmental cell. 2014;29:454–467. doi: 10.1016/j.devcel.2014.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrode N, Xenopoulos P, Piliszek A, Frankenberg S, Plusa B, Hadjantonakis AK. Anatomy of a blastocyst: cell behaviors driving cell fate choice and morphogenesis in the early mouse embryo. Genesis. 2013;51:219–233. doi: 10.1002/dvg.22368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A. Nanog heterogeneity: tilting at windmills? Cell Stem Cell. 2013;13:6–7. doi: 10.1016/j.stem.2013.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Xenopoulos P, Kang M, Hadjantonakis AK. Cell lineage allocation within the inner cell mass of the mouse blastocyst. Results Probl Cell Differ. 2012;55:185–202. doi: 10.1007/978-3-642-30406-4_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka Y, Lanner F, Rossant J. FGF signal-dependent segregation of primitive endoderm and epiblast in the mouse blastocyst. Development. 2010;137:715–724. doi: 10.1242/dev.043471. [DOI] [PubMed] [Google Scholar]
- Ying QL, Wray J, Nichols J, Batlle-Morera L, Doble B, Woodgett J, Cohen P, Smith A. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.