Abstract
Tick selenoproteins have been associated with antioxidant activity in ticks. Thioredoxin reductase (TrxR), also a selenoprotein, belongs to the pyridine nucleotide-disulfide oxidoreductase family of proteins and is an important antioxidant protein. Molecular interaction between native microbiota and tick hosts are barely investigated. In this study, we have determined the functional role of TrxR in tick feeding, and maintenance of native microbial community. TrxR transcript levels remained high and microbial load was reduced throughout tick attachment to the vertebrate host. Results of RNA interference (RNAi) show that depletion of TrxR activity did not interfere with tick hematophagy or phenotype but did reduce the viability of the microbiome within the tick tissues, presumably by perturbing redox homeostasis. The transcriptional activity of various antioxidant genes remained unaffected while antioxidant genes MnSOD, Cu/ZnSOD and SelM were significantly downregulated in salivary glands of the ticks subjected to RNAi. The perturbed TrxR enzymatic activity in the knocked down tick tissues negatively affected the bacterial load as well. Furthermore, the bacterial profiles in all the tissues dominated by Rickettisiaceae family decreased in TrxR silenced tissues. Taken together, these results indicate an essential functional role for TrxR in maintaining the bacterial community associated with ticks.
Keywords: Thioredoxin reductase, Selenoprotein, Ixodidae Ticks, Amblyomma maculatum, RNA interference, microbiota, Caspases, Superoxide dismutase, transcriptional expression
Background
Thioredoxin reductase (TrxR) belongs to the pyridine nucleotide-disulfide oxidoreductase family. This family also includes glutathione reductase (GSHR), lipoamide dehydrogenase, and mercuric ion reductase. Members of this homodimeric flavoprotein family contain one redox-active disulfide bond and one FAD per subunit; these reduce the active site disulfide in oxidized thioredoxin (Trx) (Holmgren 1989). TrxR is the only enzyme known to reduce thioredoxin; it accomplishes this by transferring an electron from bound NADPH to the thioredoxin active site, thereby allowing thioredoxin to reduce its disulfide bonds. The reduced thioredoxin reductase is a powerful protein disulfide reductase catalyzing electron transport to ribonucleotide reductase and other reductive enzymes and transcription factors (Nordberg et al. 1998). TrxR is a unique selenoprotein, having a selenocysteine (Sec) incorporated at the opal (UGA) stop codon through a complex process utilizing a Sec incorporation sequence element (SECIS) in the 3’ untranslated region (Adamson et al. 2013).
Blood feeding arthropods like ticks face a variety of oxidative stress conditions both on and off the vertebrate host. Stress in ticks can be caused by external stimuli, starvation or during blood meal acquisition when imbalances in oxidants and antioxidants can manifest themselves. An imbalance between reactive oxygen species (ROS) generation and detoxification of their reactive intermediates is known as oxidative stress. Living organisms experience a variety of oxidative stress conditions while facing abiotic or biotic stressors and must initiate a counter-response before highly reactive biological molecules can damage their cellular structures. Severe or prolonged oxidative stress can cause cellular necrosis and the induction of apoptosis. To counteract the deleterious effects of ROS and achieve redox hemostasis, organisms utilize a battery of antioxidant molecules such as catalases, peroxidases, superoxide dismutase (SOD) metallo-enzymes, glutathione reductases and thioredoxin reductases.
As a ferocious blood feeder, the Gulf Coast tick, Amblyomma maculatum, is a serious threat to livestock production and public health in the USA. This tick is a vector of the rickettsial pathogen Rickettsia parkeri, which causes a disease similar to Rocky Mountain spotted fever (Paddock et al. 2008). The sialotranscriptome of this tick reveals the presence of various antioxidants and selenoproteins including TrxR, which is a major contributor to the tick antioxidant system (Karim et al. 2011). Tick selenoproteins have been shown to play important roles in mitigating oxidative stress and pathogen colonization (Adamson et al. 2014; Adamson et al. 2013). The functional role of TrxR has been implicated in various metabolic pathways including control of organismal growth, immune functions, and anti-apoptosis via the thioredoxin-thioredoxin reductase system (Muller 1991; Salz et al. 1994; Yodoi and Uchiyama 1992). In Drosophila, TrxR has been shown to be involved in longevity and oxidative stress tolerance (Svensson and Larsson 2007). Interestingly, two isoforms of TrxR have been reported in Drosophila, neither of which can substitution for the other, as a mutation in either isoform results in a lethal phenotype (Missirlis et al. 2002).
Microbes in the tick midgut facilitate colonization of infectious agents, such as Borrelia burgdorferi, as evidenced by the reduction in Borrelia colonization following perturbation of gut microbiota (Narasimhan et al., 2014). Understanding the mechanisms by which microbiota colonize ticks would enhance our fundamental knowledge of the processes governing vector competence. Therefore, better understanding of the molecular mechanisms involved in the regulation and control of tick microbiota could benefit the development of new strategies to control and prevent tick-borne diseases. Blood meal processing involves a complex molecular interplay between the vector and microbiota residing within the vector cells. However, many of the mechanisms underlying blood meal processing, particularly its regulation, have not been elucidated.
Here, we report a link between tick TrxR and its association with the microbial community residing in A. maculatum. To the best of our knowledge, the role of TrxR has not been described previously in ticks. Hence, our findings extend current knowledge of this important enzyme in maintaining hemostasis of bacterial community in blood-sucking ticks.
RESULTS
Bioinformatics analysis
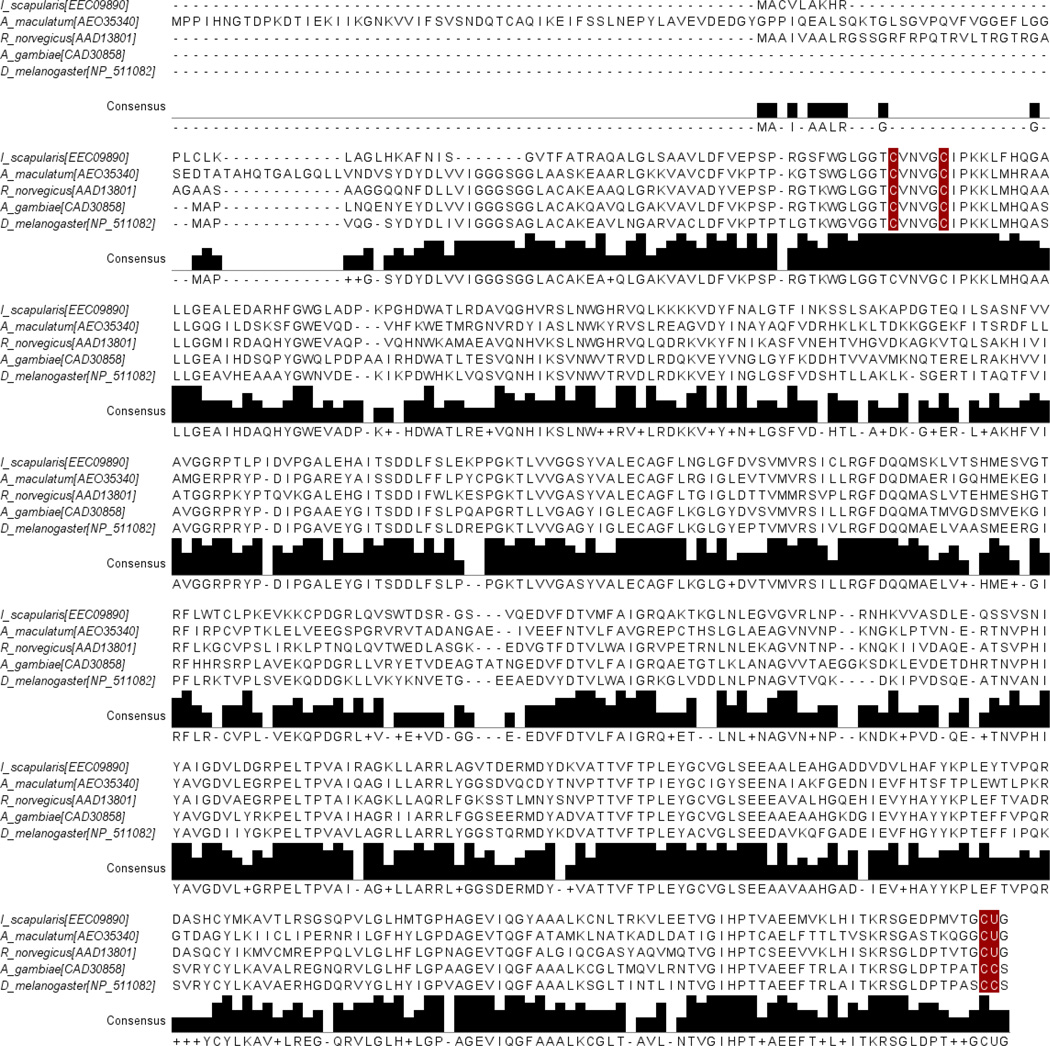
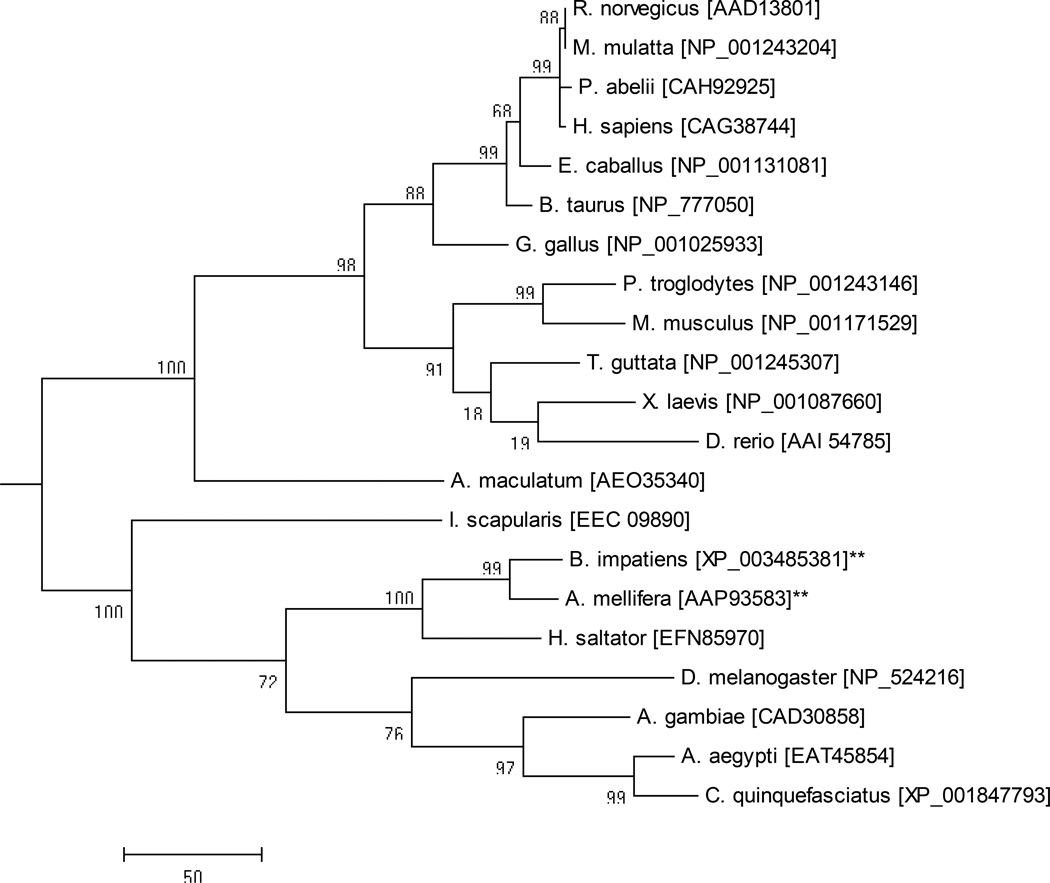
A full-length sequence (GenBank Accession: JO843723) with significant amino acid homology to TrxR was found in the transcriptional shotgun assembly of the A. maculatum sialotranscriptome (Karim et al. 2011). The deduced TrxR amino acid sequence was aligned against the deduced amino acid sequences of other invertebrates and vertebrates species to observe the similarity levels between them (Fig. 1). The A. maculatum TrxR amino acid sequence was also aligned against other TrxR orthologs previously identified from Ixodes scapularis, Rattus norvegicus, Drosophila melanogaster, and Anopheles gambiae. The residues highlighted in the alignment correspond to the conserved cysteine/Sec residues (Fig. 1). The A. maculatum TrxR protein sequence shared 47% identity with TrxR from I. scapularis, 51% with R. norvegicus, 52% with D. melanogaster, and 53% with A. gambiae (Tusnady and Simon 1998; Tusnady and Simon 2001). The phylogenetic relatedness of TrxR orthologs from vertebrates, invertebrates and A. maculatum was investigated (Fig. 2) using Mega 6 software (Tamura et al. 2013). The results showed the expected pattern of speciation of eukaryotic organisms, with A. maculatum TrxR grouping between prokaryotes and eukaryotes (Fig. 2). The A. maculatum TrxR protein lacks a secretory signal peptide suggesting it has an intracellular localization (Petersen et al. 2011). A possible intracellular location for TrxR is further supported by the dense alignment surface method transmembrane prediction server; this software predicts that TrxR contains three transmembrane helices, possibly indicating that it is localized in the mitochondrial membrane
Figure 1. Multiple sequence alignment of TrxR.
The Clustal-X alignment was imported into Jalview. Amino acids highlighted in red are key cysteine/ or selenocysteine residues within TrxR. At the C-terminus, the opal codon (UGA or TGA in case of DNA) translated to selenocysteine (Sec, represented by U). The Gly-Cys-Sec-Gly at C-terminus is essential for catalytic activity. The selenocysteine containing thioredoxin reductase from I. scapularis, A. maculatum and R. norvegicus were aligned with cysteine homologs containing thioredoxin reductases from A. gambiae and D. melanogaster. I_scapularis (Ixodes scapularis); A_maculatum (Amblyomma maculatum); R_norvegicus (Rattus norvegicus); A_gambiae (Anopheles gambiae); D_melanogaster (Drosophila melanogaster).
Figure 2. Phylogenetic analysis of TrxR from various eukaryotes using the neighbor joining method.
The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) is shown next to the branch. The sequences denoted with asterisks are derived from species that lack the capacity to synthesize selenoprotein and represent cysteine-containing homologues. The scale bar represents the degree of amino acid substitutions per position. The following vertebrate and invertebrate species were used for the analysis: Brown rat (Rattus norvegicus), Rhesus macaque (Macaca mulatta), Sumatran orangutan (Pongo abelli), Human (Homo sapiens), Horse (Equus caballus), Cattle (Bos taurus), Chicken (Gallus gallus), Chimpanzee (Pan troglodytes), House mouse (Mus musculus), Zebra Finch (Taeniopygia guttata), African clawed frog (Xenopus laevis), Zebra fish (Danio rerio), Gulf Coast tick (Amblyomma maculatum), Black legged tick (Ixodes scapularis), Bumble bee (Bombus impatiens), Honey bee (Apis mellifera), Jumping ant (Harpegnathos saltator), Fruit fly (Drosophila melanogaster), malaria-transmitting mosquito (Anopheles gambiae), Yellow fever mosquito (Aedes aegypti), Southern house mosquito (Culex quinquefasciatus).
TrxR transcriptional expression and immunolocalization
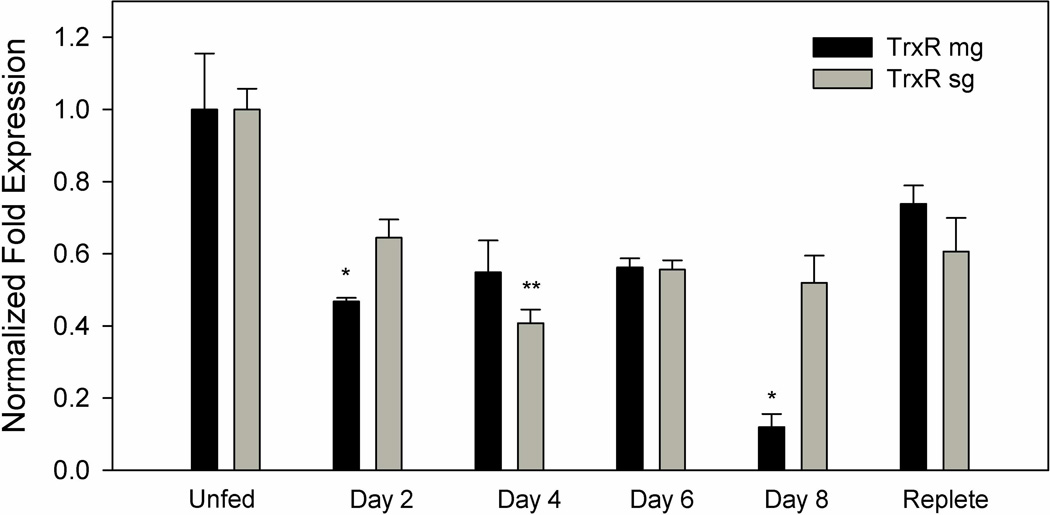
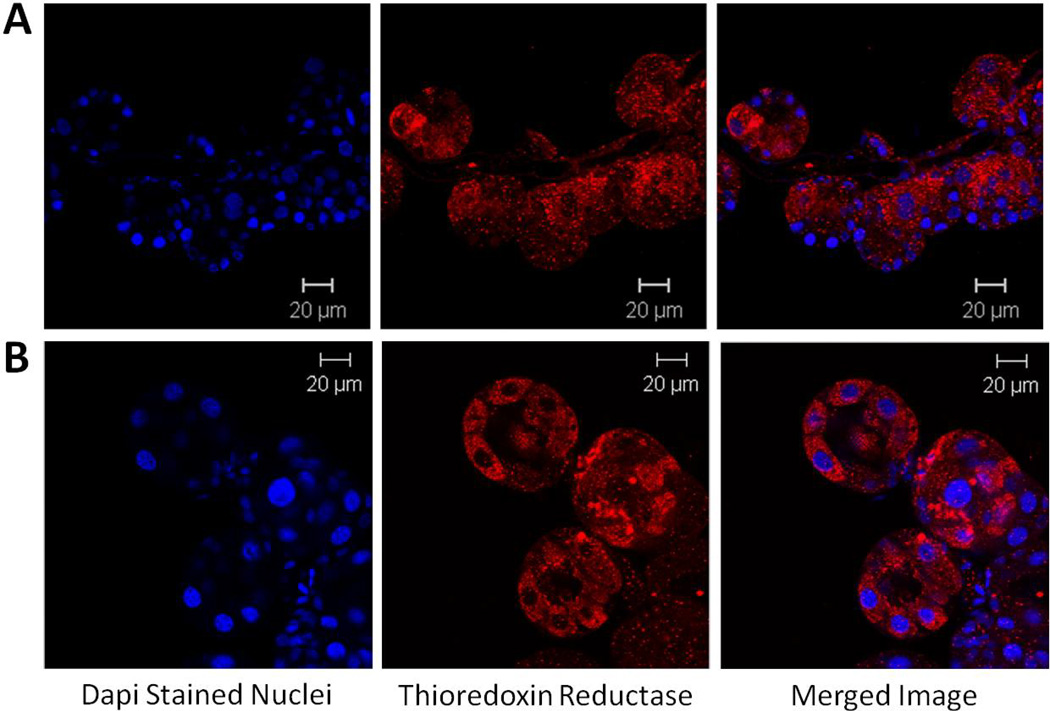
TrxR transcript expression in the midgut and salivary gland tissues of A. maculatum had similar patterns of expression across the normal blood meal cycle (Fig. 3). The relatively high level of transcriptional gene activity throughout the blood meal in both tissues underscores their potential role in blood feeding. The transcriptional expression of TrxR in salivary glands are significantly down regulated on day 4 (P-value <0.05) and remain similar across other time points but significant depletion was reported on days 2 and 8 (P-value <0.01) in midgut tissues compared to unfed tissues. Immunolocalization studies of TrxR were performed in unfed and partially blood-fed A. maculatum salivary glands using a rabbit TrxR antibody (Sigma-Aldrich, St. Louis, MO, USA). TrxR was localized in cells of the salivary gland acini in unfed (Figure 4A) and partially fed ticks (Fig. 4B). An increased level of staining was apparent in all cells of acini II and III upon blood feeding.
Figure 3. Transcriptional gene expression of TrxR in untreated midguts and salivary glands throughout the blood meal of A. maculatum adult females.
Transcriptional expression fluctuates throughout the blood meal, but never rises above the unfed stage in both the salivary glands and midguts. The transcriptional expression in salivary glands significantly down regulated on day 4 (P-value <0.05) and remained similar across other time points. But transcriptional expression in midgut significantly depleted on days 2 and 8 (P-value <0.01). TrxR gene expression was normalized against the unfed developmental stage using tick β-actin as the reference gene.
Figure 4. Immunolocalization of thioredoxin reductase in salivary glands from unfed (A) and 4 day partially fed (B) A. maculatum.
In the unfed salivary glands (A), thioredoxin reductase was localized in the cells most closely associated with the salivary duct and punctate staining is evident in the remaining cells. In the partially fed salivary glands (B), thioredoxin reductase was seen throughout the acini cells.
TrxR gene silencing in vivo
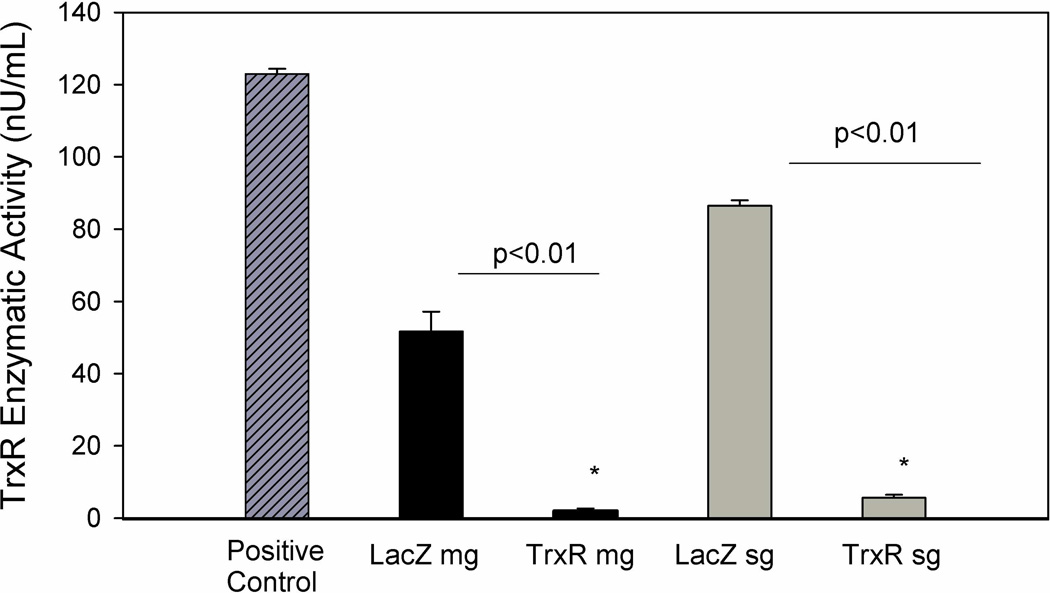
TrxR transcripts were silenced using RNAi in order to assess their likely role in tick blood feeding. The qRT-PCR results showed significant depletion of the TrxR transcript levels, with a 66% reduction in transcripts from the midguts compared with 99% depletion in the salivary gland tissues (Fig. 5). Interestingly, no statistically significant differences were noted for mean engorged body weight, tick lethality, oviposition, and egg conversion (mass of egg/engorged tick weight) between the dsTrxR- and dsLacZ-injected groups (Table 2). However, TrxR enzymatic assay revealed a decreased level of TrxR enzyme activity in the knocked down ticks, a finding supported by the level of transcript depletion. Enzymatic activity was reduced by 96.0% in the midguts and 93% in the salivary glands of the knocked down ticks compared with the dsLacZ tissue controls (Fig. 6).
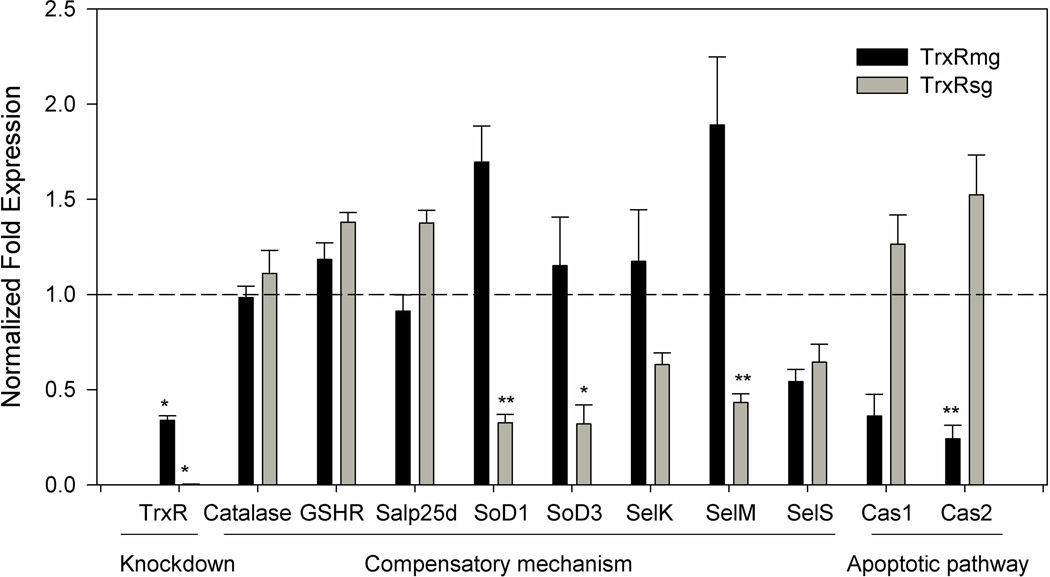
Figure 5. Knockdown of thioredoxin reductase and evaluation gene expressions of select antioxidants in knocked down tick tissues.
AmTrxR transcript depletion was seen in the midguts (66.15%) and salivary glands (99.7%) indicating successful knockdown. The normalized fold change in transcriptional expression of selected antioxidant genes in thioredoxin reductase depleted midgut and salivary gland tissues are shown. The transcript level of each candidate gene in the tissues injected with dsLacZ was set to 1.0 as a reference point. Gene expression was normalized against the tick β-actin gene (* p<0.01; ** p<0.05).
Table 2.
Phenotype data for dsLacZ and dsTrxR treated A. maculatum ticks (Mean ± SD), the weights are in miligram scale.
| Group | # of ticks | Tick weight (Range) |
Egg mass (Range) |
Egg conversion (Range) |
|---|---|---|---|---|
| dsLacZ | 7 | 712 ± 145 (475–875) |
343 ± 188 (80–620) |
0.464±0.224 (0.168–0.839) |
| dsTrxR | 21 | 786±130 (515–987) |
437± 164 (160–710) |
0.542±0.148 (0.207–0.771) |
Figure 6. Enzymatic activity of TrxR in midguts and salivary glands from partially fed A. maculatum injected with dsLacZ (control) or dsRNA-TrxR.
The enzymatic activity of thioredoxin reductase was reduced by 96.0% in midguts and 93.5% in the salivary glands of dsTrxR-injected ticks, when compared to the control dsLacZ group. The depletion of the enzymatic activity in dsTrxR knockdown tick tissues were statistically significant (P-value < 0.01).
Compensatory mechanism for TrxR inhibition
The in vivo silencing of the TrxR gene confirmed the successful knockdown of transcript and protein levels in the tick tissues, although no obvious phenotypic changes were observed. To examine the potential compensatory mechanism(s) associated with the depletion of the A. maculatum TrxR gene, the transcriptional expression levels of various antioxidant encoding genes, specifically catalase, glutathione reductase (GSHR), glutathione peroxidase (Salp25d), SODs (MnSOD and Cu/ZnSOD), and selenoprotein genes (SelK, SelM, SelS) were assessed. The transcript levels of catalase, GSHR, and Salp25d genes remained unchanged in both tissues after TrxR gene silencing. In contrast, the transcript expression levels of MnSOD and Cu/ZnSOD decreased significantly in the salivary glands of the knocked down ticks (Fig. 5). Interestingly, SelK and SelM transcriptional expression appeared to be increased in the midgut tissues although not significantly. SelM, and SelS were depleted in salivary glands and SelS was also depleted in the midguts (Fig. 5). Overall, TrxR knockdown triggered various transcriptional activities in the antioxidant factors tested herein, and may have contributed to normal tick feeding on the host (Table 2). Importantly, the significant depletion of SoDs and SelM in tick TrxR depleted tick salivary glands suggested differential impact of the TrxR depletion inside the tick. The A. maculatum tick sialotranscriptome also contained sequences related to caspases (Karim et al. 2011) such as caspase1 and 2 (Table 1); these were selected to test for apoptotic signaling in the TrxR knockdown ticks. Low levels of ROS are mitogenic and facilitate cell growth, while increased levels of ROS stimulate pro-apoptotic kinases, leading to cell death (Halliwell and Gutteridge 1984). Apoptosis in tick tissues was assessed by monitoring the transcript levels of selected caspases genes in the TrxR deficient tissues. Both caspases (Caspase 1 and 2) had increased expression in tick salivary glands though not statistical significantly while in tick midgut tissues capapase 2 was significantly reduced (Fig. 5).
Table 1.
Gene-specific primers used in this study
| Gene | Accession Number |
Forward Reverse |
Size (bp) |
|---|---|---|---|
| AmTrxR | JO843723 (dsRNA and qRT PCR) | 5’-TCGACACAAGCTGAAGCTGACTGA-3’ 5’-CAATGCGTTCTGCCATGTCTTGGT-3’ |
320 |
| AmTrxR | 5’-TGTGACTACACCAACGTGCCTACA-3’ 5’-AGTAGCCTGCATCCGTTCCTCTTT-3’ |
175 | |
| AmCatalase | JO843741 | 5’-AAAGGACGTCGACATGTTCTGGGA-3’ 5’-ACTTGCAGTAGACTGCCTCGTTGT-3’ |
173 |
| AmGSHR | JO844062 | 5’-ACCTGACCAAGAGCAACGTTGAGA-3’ 5’-ATCGCTTGTGATGCCAAACTCTGC-3’ |
170 |
| AmSalp25d (GPx) | JO843645 | 5’-TGCCGCGCTGTCTTTATTATTGGC-3’ 5’-AGTTGCACGGAGAACCTCATCGAA-3’ |
102 |
| Am MnSoD (SOD3) | JO843979 | 5’-GCATCTACTGGAC AAACCTCTC-3’ 5’-GCAGACATCAGGCCTTTGA-3’ |
115 |
| Am Cu/ZnSoD (SOD1) | JO844140 | 5’-GGAACCGAAGACAGCAAGAA-3’ 5’-GAGAAGAGGCCGATGACAAA-3’ |
143 |
| AmSelK | JO843326 | 5’-AGTTCCAGCAGGTCATCAGTGTCA-3’ 5’-TCCAGGAATAGGGCAGTCCATTGT-3’ |
132 |
| AmSelM | JO842653 | 5’-ATGATACCTGAATGGCCATCCGCA-3’ 5’-TGATCGCGGGTCATCTTCTCCAAA-3’ |
171 |
| AmSelS | JO842687 | 5’-AGAACAAGTGCACCACAACAGCAG-3’ 5’-ATTTCTTGCATCCTTCGACGTGCC-3’ |
107 |
| AmActin | JO842238 | 5’-TGGCTCCTTCCACCATGAAGATCA-3’ 5’-TAGAAGCACTTGCGGTGCACAATG-3’ |
177 |
| Amcaspase1 | JO842755 | Am2600-F(qrtpcr) : GAGGAGTCTAGCAGGATGTTTC Am2600-R(qrtpcr): ACTGTCATGCTCCGTGTAATC |
127 |
| Amcaspase2 | JO845022 | Am29082-F(qrtpcr): GGTGATCGTGATGTCCTGTATG Am29082-R(qrtpcr): CGACAGGCCTGAATGAAGAA |
128 |
| 16srRNA | microbial-F(qrt): AGAGTTTGATCCTGGCTCAG microbial- R(qrt): CATGCTGCCTCCCGTAGGAGT |
Total bacterial load and bacterial profile in TrxR depleted tick tissues
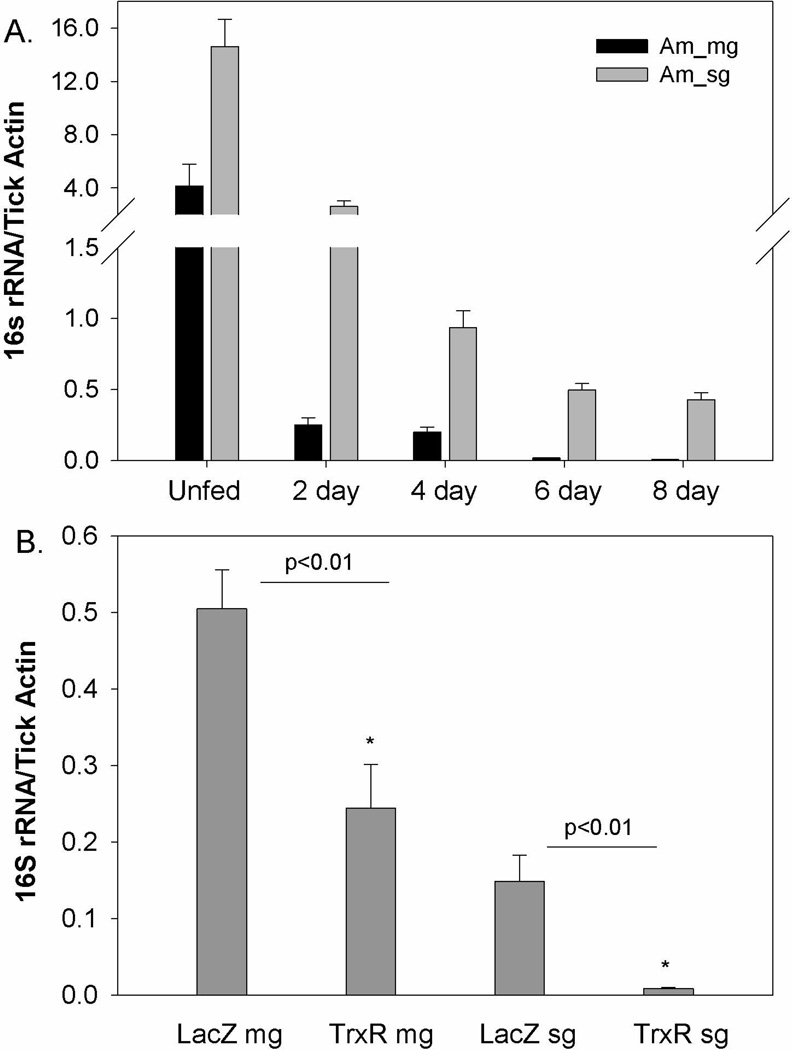
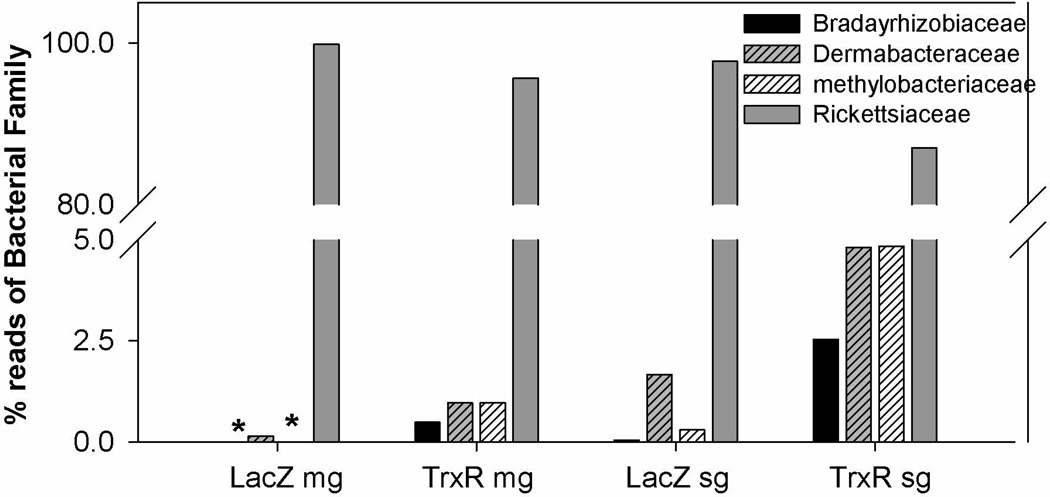
The normal bacterial load was estimated in naïve ticks tissues across blood meal cycle (Fig. 7A) and the impact of TrxR depletion (Fig. 7B) on the native microbes residing inside the ticks was examined by comparing the overall levels of bacterial 16s rRNA genes quantified and normalized against tick β-actin copies (see Methods). The bacterial load in normal ticks in both midgut and salivary glands tissues was observed to be significantly reduced along the blood meal (Fig. 7A). The reactive oxidative stress upon blood meal digestion might have made bacteria unfavorable in tick tissues. TrxR gene silencing caused a significant decrease in the native bacterial loads of the ticks (p<0.01) in both the midgut and salivary glands (Fig. 7B). A 454-pyrosequencer was used to determine the variation in the bacterial community in the knocked down tick tissues (Budachetri et al. 2014). In total, 9948 reads were obtained after bioinformatic quality control, as described in the methods section. The results revealed the presence of the following three bacterial phyla: Proteobacteria, Actinobacteria and Firmicutes. Protobacteria were dominantly prevalent (>98%) in the samples. In total, 11 bacterial families were found and dominant bacterial families included Bradyrhizobiaceae, Methylobacteriaceae and Rickettsiaceae. Rickettsiaceae was the most prevalent bacterial family residing in the tick tissues and the prevalence of which were decreased in TrxR silenced tick tissues (Fig. 8). Interestingly, in the TrxR depleted tissues, additional bacterial sequence reads representing families Bradyrhizobiaceae, Dermabacteraceae, and Methylobacterium were observed (Fig. 8).
Figure 7. Native microbial load estimation in tick tissues.
(A) The bacterial load estimation across the normal blood meal cycle in A. maculatum tick tissues. (B) The bacterial load in dsLacZ and dsTrxR injected tick tissues. The bacterial loads were calculated as number of bacterial 16S rRNA gene per tick β-actin (mean ± SD). The copy number of each gene was derived from the standard equations obtained from Ct values of known concentration of 16s rRNA and tick β-actin as described in method. The depletion of bacterial load/100 tick actin were significant in tick tissues depleted with thioredoxin reductase (P-value <0.01).
Figure 8. Bacterial profile changes after the TrxR silencing in tick tissues.
Bacterial profiles were assessed from 16S rRNA amplicon sequencing from representative tick tissues cDNAs with 454-pyrosequening approach, the sequences representing bacterial families were used in estimation of percent in tick tissues.
DISCUSSION
In this study, we investigated the role of tick thioredoxin reductase by computational analysis and characterized its function via an RNAi assay. Our data indicate a role for this gene in preserving tick microbiota. The Sec-encoded motif is found at the C-terminus in I. scapularis, A. maculatum and R. norvegicus whereas cysteine residues are present in other species, A. gambiae and D. melanogaster (Fig. 1), and has been described in other tick selenoproteins, specifically SelK and SelM (Adamson et al. 2014). At the C-terminus the Gly-Cys-Sec-Gly motif is essential for catalytic activity of enzyme thioredoxin reductase as determined by mutational analysis, and compared to the C-terminus of glutathione reductase, the presence of selenocysteine is the only change (Zhong and Holmgren 2000). The Sec-containing TrxR, along with other cysteine-containing homologs (Fig. 2, asterisk) in invertebrate and vertebrate TrxR sequences were used to identify phylogenetic relatedness among these proteins (Fig. 2). The invertebrate and vertebrate clades were distinct, while I. scapularis TrxR formed its own outgroup from the invertebrates, and A. maculatum formed an outgroup from the vertebrates. The divergence of A. maculatum TrxR from both the invertebrate and I. scapularis groupings suggests an evolutionarily bridge between the arthropods and higher vertebrates. The sequences identified in Bombus impatiens, Apis mellifera, and Harpegnathos saltator encode Cys containing TrxR homologs which do not have the capacity to be synthesized as selenoproteins, and, therefore, form a distinct clade.
In A. maculatum TrxR is transcriptionally active during the prolonged period the tick is attached to its host, but it remains high in the tissues of unfed questing ticks (Fig. 3).. Transcriptional expression of TrxR in midgut tissues was similar throughout early feeding until day 8 post-attachment when a marked decrease of 86% was detected (Fig. 3). The 24–48 h time period before a tick drops off of its host represents a fast feeding stage where more than two-thirds of the total blood engorgement is ingested by the tick (Horn et al. 2009). The marked decrease in TrxR levels at the onset of the fast feeding stage suggests that the tick prepares itself for the assimilation of a large blood meal. Oxidative stress in unfed or feeding ticks is possibly responsible for the relatively high transcriptional expression level of TrxR. This stress could be related to starvation, and/or desiccation, osmoregulation, or acquisition of a blood meal and its subsequent processing (heme degradation). Blood meal induced oxidative stress might have a role in significantly reducing bacterial loads in both tissues compared to unfed tissues (Fig. 7A). The immunolocalization of TrxR in unfed and 4 day partially blood-fed salivary glands appears to increase (Fig. 4). These cells are primed with synthesized proteins, such as host immunomodulatory proteins and cement cone proteins, for secretion via the saliva prior to tick attachment. It is reasonable to speculate that that these acini cells are sites of high oxidative stress and deployment of antioxidant molecules, such as thioredoxin reductase. As a tick feeds, more cellular machinery is activated throughout the cells of the acini, accounting for the widespread protein localization seen in its partially fed tissues (Fig. 4B). Significant structural, cellular and biochemical changes are known to occur in tick salivary glands during blood feeding (Binnington 1978).
A significant depletion of TrxR enzymatic activity occurs in the midgut tissues at a level greater than that expected from the depleted level of transcripts (Figs. 5,6); this can potentially be attributed to the natural degradation of TrxR transcripts within tick tissue coupled with reduction from induced degradation via RNAi (Low and Berry 1996; Xu et al. 1997). Day-to-day variation in gene depletion among the attached ticks cannot be ruled out either. Because most central antioxidant enzyme reactions can be catalyzed by at least one or more proteins of the antioxidant defense network, inhibition of a single target antioxidant protein is unlikely to be efficient at impairing tick feeding (Table 2) through induction of oxidative stress. The normal tick phenotype (i.e., tick attachment duration, weight gain/loss, fecundity and vitellogenesis) in TrxR-deficient ticks was not impacted, as has been shown in our previous studies (Adamson et al. 2014; Adamson et al. 2013).
Other antioxidants (catalase, GSHR, and Salp25d) with important roles in redox hemostasis within the tick vector showed no changes in transcript levels (Fig. 5). Catalase and glutathione peroxidase catalyze the reduction of hydrogen peroxide to form water (Aebi et al. 1974; Epp et al. 1983). GSHR is an important enzyme for catalysis of glutathione disulfides and generates a reduced environment within the cells. SODs catalyze two molecules of superoxide to form hydrogen peroxide and molecular oxygen, which is a source of cellular hydrogen peroxide (McCord and Fridovich 1969), but somewhat surprisingly, tissue-specific SODs gene expression was downregulated in the TrxR knocked down salivary glands (Fig. 7). This suggests a direct link between the function of TrxR and SODs and further amelioration of the elevated stress conditions in the TrxR knocked down salivary glands when compared with midgut tissues (Weisiger and Fridovich 1973). Similarly, selK and selM transcript levels were differentially regulated in the midguts and salivary glands, while selS transcript levels remained decreased in both of the tissues tested (Fig. 7). Both selK and selM have been shown to be involved in maintaining the antioxidant capacity of A. maculatum tissues and saliva (Adamson et al. 2014), and SelS is an important endoplasmic and plasma membrane protein involved in alleviating oxidative injury in human endothelial cells by increasing SOD activity (Zhao et al. 2013).
Midgut tissues are getting compensation by increased expression of SoDs, SelK and SelM. However, the transcript levels of SODs and SelM significantly downregulated in the tick salivary glands deprived of thioredoxin reductase. Low level of transcriptional expression of SelK and SelS compared to control tissues might have aggravated the situation (Fig. 5). Hypothetically, elevated oxidative stress levels in the knocked down salivary glands may triggered caspase expression leading to apoptosis signaling (Fig. 5); elevated oxidative stress and apoptotic cell death are a closely related phenomena (Kannan and Jain 2000). Increased transcript level of caspases in salivary glands indicates the onset of apoptotic processes (Fig. 5), however the salivary gland is a highly secretory tissue and undergoes incredible biochemical and physiological changes during the fast blood-feeding phase in ticks and caspase activation cannot be ruled out (Binnington 1978).
Depleted TrxR activity (Figs. 5, 6) possibly altered the redox status of the tick tissues. The possible imbalance in the redox status of the tick tissues might have altered the native microbiota in the ticks (Fig. 7B). In contrast to invertebrates, microbes, and mammals (Kanzok et al. 2001), thiol-based intracellular antioxidants in ticks and possibly other blood-sucking invertebrates are generated by a single enzyme system in which TrxR plays a key role in maintaining the intracellular redox homeostasis (Kanzok et al. 2001). The results presented here show that depleting TrxR activity does not affect the tick phenotype or blood feeding, rather, it reduces the viability of the microbiome residing inside the tick tissues, seemingly caused by a perturbed redox homeostasis balance within the tick host. TrxR system-related genes are also transcribed in the salivary glands of A. gambiae. It is assumed that the corresponding proteins are especially important for protecting glands from heme-driven free radical attack (Francischetti et al. 2002).
Previously, we have examined the microbial diversity in A. maculatum and observed each tissue harbored distinct bacterial species (Budachetri et al. 2014), although potential nonpathogenic interactions inside the tick tissues remain unknown. Microbial diversity in arthropods has been implicated in innate immunity and nutrition (Dillon and Dillon 2004), and alteration of these native microbial species has been shown to impair pathogen colonization or transmission success (Weiss and Aksoy 2011). Though, we did not observe significant depletion of reads from Rickettsiaceae but decreased level of percent sequence reads and occurrence of new bacterial families in TrxR depleted tick tissues is interesting. Our findings open new avenues for further investigating the functional role of TrxR in the colonization of pathogenic bacteria such as Rickettsia parkeri within the tick host.
In conclusion, the work presented here suggests that A. maculatum TrxR is important for neutralizing elevated ROS levels and maintaining redox hemostasis, and thereby provides new insights into the metabolic needs of blood-sucking arthropods. Silencing TrxR activity does not interfere with hematophagy but diminishes the viability of the microbiome residing inside the tick. This effect is possibly the result of a perturbed redox homeostasis balance within the tick host. Understanding how we can disturb tick redox balance during infections with these pathogens could lead to novel strategies to prevent tick-borne diseases.
Methods
Tick rearing
Gulf Coast ticks (Amblyomma maculatum) were purchased from the tick rearing facility at Oklahoma State University, and maintained according to standard methods (Patrick and Hair 1975). Prior to infestation of the sheep, all unfed adult ticks were kept at ~ 24–26°C in 90% relative humidity for a 14h light/10h dark photoperiod. All animal protocols were approved by the Institutional Animal Care and Use Committee (IACUC) at the University of Southern Mississippi, USA (protocol # 10042001).
Tick tissue dissection
Naive adult ticks were allowed to infest the sheep and were then removed routinely by pulling them off their hosts. The ticks (5–10) removed at each time point from a host were dissected to isolate their midguts and salivary glands tissues, which were transferred to ice-cold 100 mM MOPS buffer containing 20 mM EGTA, pH 6.8. Once removed, the tick tissues were gently washed in the same ice-cold buffer and the pooled dissected tissues were used either immediately or stored at −80°C in RNAlater (Invitrogen, Carlsbad, CA, USA).
Bioinformatics analyses
The TrxR coding sequence from A. maculatum (GenBank Accession JO843723) was obtained by pyrosequencing an A. maculatum salivary gland cDNA library(Karim et al. 2011). Nucleotide sequences were conceptually translated, initially aligned using ClustalX2 (Thompson et al. 2002), refined by eye, and graphically presented using Jalview 2.7 (Larkin et al. 2007; Waterhouse et al. 2009). Phylogenetic relationships were inferred by MEGA 6 using the neighbor-joining method (Tamura et al. 2013).
RNA Isolation, cDNA synthesis and qRT PCR
Total RNA extraction, conversion into complementary DNA and qRT-PCR were performed as previously described (Browning and Karim 2013). Briefly, the tick midguts and salivary gland tissues stored in RNAlater were used for total RNA extraction using Illustra™ RNAspin Mini Isolation Kit (GE Healthcare, Piscataway, NJ, USA) according to manufacturer’s instructions. Total RNA (~ 1 µg) was reverse transcribed into cDNA using Moloney murine leukemia virus reverse transcriptase according to the manufacturer’s protocol (Invitrogen). Gene-specific primers were designed to amplify the cDNA fragments from A. maculatum tissues. All primer sequences used in this study are listed in Table 1. First-strand cDNA was used to measure mRNA levels using qRT-PCR. Maxima™ SYBR Green qPCR Master Mix (2×) (Fermentas Life Sciences, Waltham, MA, USA). cDNA (25 ng) and 150 nM of gene-specific primers (Table 1) were used in each reaction mixture (Browning et al. 2013). Reaction mixtures were subjected to 95°C for 10 min, followed by 35 cycles of 95°C for 15 s, 60°C for 30 s, and 72°C for 30 s using the CFX96 Real Time System (BIORAD Inc.). All samples were run in triplicate and Bio-Rad Software was used for data analysis with the 2^ddCt method.
Bacterial load quantitation
We followed the bacterial load estimation protocol described previously (Narasimhan et al. 2014). 16S rRNA primers were used to amplify the Staphylococcus aureus 16S rRNA bacterial gene. Similarly, we amplified the A. maculatum tick β-actin gene using tick actin primers (Table 1). The amplified PCR products were serially diluted 10-fold (108 to 101 copies) and used to generate a standard curve. The qRT PCR reactions consists of 200nM of the primers, Maxima™ SYBR Green Master Mix (2×) (Fermentas Life Sciences, Waltham, MA, USA) and the serially diluted PCR products prepared for each standard curve. The reaction mixture was subjected to thermal cycling at 94°C for 5 min followed by 35 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 30 s in a CFX96 Real Time System (Bio Rad Inc.) The standard curve generated was used to calculate the copy number of bacterial 16s rRNA genes and the tick β-actin gene. Using the same reaction conditions and thermal cycle parameters, 25ng of A. maculatum cDNA for each control and dsTrxR knocked down tissues were amplified and the standard curves produced were used to estimate the copy numbers of each gene in a given sample. The bacterial 16s gene copy numbers were normalized against the A. maculatum β-actin gene. As with the other qRT PCR reactions, all the samples were run in triplicate.
Tick cDNA and 454 pyrosequencing
Tick salivary gland cDNAs were individually processed for the bacterial tag-encoded titanium amplicon pyrosequencing (bTETAP) approach (Dowd et al. 2008a). In a modified version of this process, 16S universal eubacterial primers 530F (5'-GTG CCA GCM GCN GCG G) and 1100R (5'-GGG TTN CGN TCG TTG) were used to amplify the 600 bp region of the 16S rRNA genes. A single-step 30 cycle PCR using a HotStarTaq plus master mix kit (Qiagen, Valencia, CA, USA) was used with the following conditions: 94°C for 3 min followed by 32 cycles of 94°C for 30 s, 60°C for 40 s and 72°C for 1 min, and a final elongation step at 72°C for 5 min. Following PCR, all amplicons from the different samples were mixed in equal concentrations. The mixtures were purified using Agecourt Ampure beads (Agencourt Bioscience Corporation, Danvers, MA, USA). Samples were sequenced with Roche 454 FLX titanium instruments and reagents (Roche, Branford, CT, USA), following the manufacturer’s guidelines. The amplicons obtained were curated on a proprietary analysis pipeline (www.mrdnalab.com, MR DNA, Shallowater, TX, USA). Sequences were depleted of barcodes and primers. Next, short sequences <200 bp were removed, as were those with ambiguous base calls and homopolymer runs exceeding 6-bp. Sequences were then denoised and the chimeras removed. Operational taxonomic units were defined after removal of singleton sequences, clustering at 3% divergence (97% similarity) (Dowd et al. 2008a; Dowd et al. 2008b; Edgar 2010; Eren et al. 2011; Swanson et al. 2011). Thereafter, the sequences obtained were subjected to BLASTn analysis against the GreenGenes database (DeSantis et al. 2006). The homologies obtained were assigned to taxonomic classification for bacteria and operational taxonomic units.
dsRNA synthesis, tick injections and feeding
Synthesis of dsRNA for the TrxR gene and tick manipulations were performed according to the methods described previously (Browning and Karim 2013). Briefly, TrxR PCR products were joined to the Block-iT™ T7 TOPO® linker using a BLOCK-iT™ RNAi TOPO® Transcription Kit (Invitrogen, Carlsbad, CA, USA). The TOPO® linking reaction was used to produce sense and antisense linear DNA templates using gene-specific and BLOCK-iT™ T7 primers in two separate PCR reactions. The sense and antisense DNA templates were used to transcribe sense and antisense ssRNA, and were then heated to facilitate dsRNA formation. The dsRNA generated was analyzed to verify its size using agarose gel electrophoresis. Subsequently, unfed adult females were injected with 1–1.5 µL of TrxR dsRNA or LacZ dsRNA using a 31-gauge needle. After injection, the ticks were maintained at 37°C overnight under high humidity to promote their survival. The surviving ticks were placed in designated, contained cells on a naive sheep and allowed to take blood feeds. The ticks (5–10) were removed on days six and eleven post-infestation, but the remaining ticks were allowed to remain attached and ingest blood until repletion. Ticks feeding success was determined by attachment duration, replete weight, and oviposition (Karim and Adamson 2012). Ticks removed from the dsLacZ and dsTrxR groups were dissected individually and their tissues were pooled in assigned groups. Some dissected salivary glands and midgut tissues were kept for total RNA extraction for investigating the knockdown effect of the dsTrxR RNAi assay.
Protein extraction and enzyme assays
Total soluble proteins from pooled tick midguts and salivary glands tissues from 5–7 ticks were extracted using a phosphate-buffered saline (1X PBS) cocktail containing protease inhibitors with brief sonication (Branson Sonifier Model 250, 35% output, two 3 s pulses, Branson, Danbury, CT, USA). The tissue homogenates were centrifuged at 5000 × g for 10 min at −4°C and the supernatant was collected. The protein concentration of each supernatant was estimated (Bradford 1976). TrxR enzymatic activity assays were performed on tick midgut and salivary glands protein homogenates using a commercial kit (BioVision, Milpitas, CA, USA). Briefly, a comparative standard curve was generated using standard TNB concentrations of 0, 10, 20, 30, 40, and 50 nmol. All samples were run in duplicate, both with and without the inhibitor, and in accordance with the manufacturer’s recommendations. Approximately 12 µg of total soluble protein was used for each sample. The total reaction mixture containing 40 µL of assay buffer, 5,5’-dithio-bis-(2-nitrobenzoic acid (DTNB) solution, and NADPH was added to each sample, and a final volume of 100 µL was reached by adding an appropriate additional amount of assay buffer. The optical density (OD) was measured at 412 nm to generate a T1 reading (before incubation) and T2 reading (after incubation) at 25°C for 20 min. TrxR activity was calculated using the formula provided by the manufacturer according to the standard curve.
TrxR immunolocalization
Immunolocalization studies of tick TrxR were performed on unfed and partially fed salivary glands from untreated controls and TrxR knocked down ticks. The tick salivary glands were fixed in 1X PBS containing 4% formaldehyde and kept at −4°C until further manipulation, as described elsewhere (Villarreal et al. 2013). Briefly, the salivary glands were permeabilized using 0.5% Triton X-100 for 30 min and then blocked in 1X PBS containing 3% bovine serum albumin (BSA) for 1 h at room temperature. Salivary glands were incubated overnight at −4°C with a rabbit TrxR antibody (1:100) (Sigma-Aldrich, St. Louis, MO, USA) in 1X PBS containing 3% BSA, after which they were incubated with an anti-rabbit Alexa Fluor® 555 secondary antibody (1:100) in 1X PBS containing 3% BSA for 1 h in the dark. All incubations were maintained on a rocking plate at room temperature unless otherwise indicated. Salivary glands were mounted on glass slides using VECTASHIELD® mounting medium with 4’,6-diamidino-2-phenylindole (DAPI) stain. Tissues prepared in this manner were mounted and viewed under a Zeiss LSM 510 META confocal microscope (40× objective) using 555 nm and 220 nm lasers.
Statistical analysis
All data are expressed as the mean ± SEM. Statistical significance between the two groups was determined by a Student’s t-test and comparative differences among multiple experimental groups were determined by ANOVA (SigmaPlot ver. 11, San Jose, CA, USA). Transcription expression levels considered by Bio-Rad Software (Bio-Rad CFX manager V 3.1) to be significantly different between samples for two-fold differences with p-values of <0.01.
Acknowledgements
We thank Rebecca Browning, Leshia Hobbard (AGEM fellow) and Baobin Kang for technical support. Funding was provided by the National Institute of Allergy and Infectious Diseases award # AI099919, United States Department of State award # PGA-P21049 (Pakistan-United States Science and Technology Cooperation Program) to SK. The MS-INBRE core facility is supported by the National Institute of General Medical Sciences, National Institutes of Health award (P20RR016476). We also thank the company EdanZ for providing editorial assistance.
List of Abbreviations
- Trx
Thioredoxin
- TrxR
Thioredoxin reductase
- SOD
Superoxide dismutase
- GR
Glutathione reductase
- ROS
Reactive Oxygen Species
- RNAi
RNA interference
Footnotes
Competing interests
The authors declare that they have no competing interests.
References
- Adamson S, Browning R, Singh P, Nobles S, Villarreal A, Karim S. Transcriptional activation of antioxidants may compensate for selenoprotein deficiencies in Amblyomma maculatum (Acari: Ixodidae) injected with selK- or selM-dsRNA. Insect Mol Biol. 2014;23(4):497–510. doi: 10.1111/imb.12098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamson SW, Browning RE, Budachetri K, Ribeiro JMC, Karim S. Knockdown of Selenocysteine-Specific Elongation Factor in Amblyomma maculatum Alters the Pathogen Burden of Rickettsia parkeri with Epigenetic Control by the Sin3 Histone Deacetylase Corepressor Complex. Plos One. 2013;8(11):e82012. doi: 10.1371/journal.pone.0082012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aebi H, Wyss SR, Scherz B, Skvaril F. Heterogeneity of erythrocyte catalase II. Isolation and characterization of normal and variant erythrocyte catalase and their subunits. Eur J Biochem. 1974;48:137–145. doi: 10.1111/j.1432-1033.1974.tb03751.x. [DOI] [PubMed] [Google Scholar]
- Binnington KC. Sequential changes in salivary gland structure during attachment and feeding of the cattle tick, Boophilus microplus. Int J Parasitol. 1978;8:97–115. doi: 10.1016/0020-7519(78)90004-8. [DOI] [PubMed] [Google Scholar]
- Bradford MM. Rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Browning R, Adamson SW, Karim S. Choice of a Stable Set of Reference Genes for qRT-PCR Analysis in Amblyomma maculatum (Acari: Ixodidae) Journal of Medical Entomology. 2013;49:1339–1346. doi: 10.1603/me12123. [DOI] [PubMed] [Google Scholar]
- Browning R, Karim S. RNA interference-mediated depletion of N-ethylmaleimide Sensitive Fusion Protein and Synaptosomal Associated Protein of 25 kDa results in the inhibition of blood feeding of the Gulf Coast tick, Amblyomma maculatum. Insect Mol Biol. 2013;22(3):245–257. doi: 10.1111/imb.12017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budachetri K, Browning RE, Adamson SW, Dowd SE, Chao CC, Ching WM, Karim S. An Insight Into the Microbiome of the Amblyomma maculatum (Acari: Ixodidae) Journal of Medical Entomology. 2014;51:119–129. doi: 10.1603/me12223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillon RJ, Dillon VM. The gut bacteria of insects: nonpathogenic interactions. Annu Rev Entomol. 2004;49:71–92. doi: 10.1146/annurev.ento.49.061802.123416. [DOI] [PubMed] [Google Scholar]
- Dowd SE, Callaway TR, Wolcott RD, Sun Y, McKeehan T, Hagevoort RG, Edrington TS. Evaluation of the bacterial diversity in the feces of cattle using 16S rDNA bacterial tag-encoded FLX amplicon pyrosequencing (bTEFAP) BMC Microbiol. 2008a;8:125. doi: 10.1186/1471-2180-8-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowd SE, Sun Y, Wolcott RD, Domingo A, Carroll JA. Bacterial tag-encoded FLX amplicon pyrosequencing (bTEFAP) for microbiome studies: bacterial diversity in the ileum of newly weaned Salmonella-infected pigs. Foodborne Pathog Dis. 2008b;5:459–472. doi: 10.1089/fpd.2008.0107. [DOI] [PubMed] [Google Scholar]
- Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- Epp O, Ladenstein R, Wendel A. The refined structure of the selenoenzyme glutathione peroxidase at 0.2-nm resolution. Eur J Biochem. 1983;133:51–69. doi: 10.1111/j.1432-1033.1983.tb07429.x. [DOI] [PubMed] [Google Scholar]
- Eren AM, Zozaya M, Taylor CM, Dowd SE, Martin DH, Ferris MJ. Exploring the diversity of Gardnerella vaginalis in the genitourinary tract microbiota of monogamous couples through subtle nucleotide variation. PLoS One. 2011;6:e26732. doi: 10.1371/journal.pone.0026732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francischetti IM, Valenzuela JG, Pham VM, Garfield MK, Ribeiro JM. Toward a catalog for the transcripts and proteins (sialome) from the salivary gland of the malaria vector Anopheles gambiae. J Exp Biol. 2002;205:2429–2451. doi: 10.1242/jeb.205.16.2429. [DOI] [PubMed] [Google Scholar]
- Halliwell B, Gutteridge JM. Free radicals, lipid peroxidation, and cell damage. Lancet. 1984;2:1095. doi: 10.1016/s0140-6736(84)91530-7. [DOI] [PubMed] [Google Scholar]
- Holmgren A. Thioredoxin and glutaredoxin systems. J Biol Chem. 1989;264:13963–13966. [PubMed] [Google Scholar]
- Horn M, Nussbaumerova M, Sanda M, Kovarova Z, Srba J, Franta Z, Sojka D, Bogyo M, Caffrey CR, Kopacek P, Mares M. Hemoglobin digestion in blood-feeding ticks: mapping a multipeptidase pathway by functional proteomics. Chem Biol. 2009;16:1053–1063. doi: 10.1016/j.chembiol.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannan K, Jain SK. Oxidative stress and apoptosis. Pathophysiology. 2000;7:153–163. doi: 10.1016/s0928-4680(00)00053-5. [DOI] [PubMed] [Google Scholar]
- Kanzok SM, Fechner A, Bauer H, Ulschmid JK, Muller HM, Botella-Munoz J, Schneuwly S, Schirmer RH, Becker K. Substitution of the thioredoxin system for glutathione reductase in Drosophila melanogaster. Science. 2001;291:643–646. doi: 10.1126/science.291.5504.643. [DOI] [PubMed] [Google Scholar]
- Karim S, Adamson SW. RNA Interference in Ticks: A Functional Genomics Tool for the Study of Physiology. In: Jockusch E, editor. Advances in Insect Physiology Small RNAs: Their diversity, Roles and Practical uses. Vol. 42. San Diego, CA: Academic Press; 2012. pp. 119–154. [Google Scholar]
- Karim S, Singh P, Ribeiro JM. A deep insight into the sialotranscriptome of the gulf coast tick, Amblyomma maculatum. PLoS One. 2011;6:e28525. doi: 10.1371/journal.pone.0028525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Low SC, Berry MJ. Knowing when not to stop: selenocysteine incorporation in eukaryotes. Trends Biochem Sci. 1996;21:203–208. [PubMed] [Google Scholar]
- McCord JM, Fridovich I. Superoxide dismutase. An enzymic function for erythrocuprein (hemocuprein) J Biol Chem. 1969;244:6049–6055. [PubMed] [Google Scholar]
- Missirlis F, Ulschmid JK, Hirosawa-Takamori M, Gronke S, Schafer U, Becker K, Phillips JP, Jackle H. Mitochondrial and cytoplasmic thioredoxin reductase variants encoded by a single Drosophila gene are both essential for viability. J Biol Chem. 2002;277:11521–11526. doi: 10.1074/jbc.M111692200. [DOI] [PubMed] [Google Scholar]
- Muller EG. Thioredoxin deficiency in yeast prolongs S phase and shortens the G1 interval of the cell cycle. J Biol Chem. 1991;266:9194–9202. [PubMed] [Google Scholar]
- Narasimhan S, Rajeevan N, Liu L, Zhao YO, Heisig J, Pan J, Eppler-Epstein R, Deponte K, Fish D, Fikrig E. Gut microbiota of the tick vector Ixodes scapularis modulate colonization of the lyme disease spirochete. Cell Host Microbe. 2014;15:58–71. doi: 10.1016/j.chom.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordberg J, Zhong L, Holmgren A, Arner ES. Mammalian thioredoxin reductase is irreversibly inhibited by dinitrohalobenzenes by alkylation of both the redox active selenocysteine and its neighboring cysteine residue. J Biol Chem. 1998;273:10835–10842. doi: 10.1074/jbc.273.18.10835. [DOI] [PubMed] [Google Scholar]
- Paddock CD, Finley RW, Wright CS, Robinson HN, Schrodt BJ, Lane CC, Ekenna O, Blass MA, Tamminga CL, Ohl CA, McLellan SL, Goddard J, Holman RC, Openshaw JJ, Sumner JW, Zaki SR, Eremeeva ME. Rickettsia parkeri rickettsiosis and its clinical distinction from Rocky Mountain spotted fever. Clin Infect Dis. 2008;47:1188–1196. doi: 10.1086/592254. [DOI] [PubMed] [Google Scholar]
- Patrick CD, Hair JA. Laboratory rearing procedures and equipment for multi-host ticks (Acarina: Ixodidae) J Med Entomol. 1975;12:389–390. doi: 10.1093/jmedent/12.3.389. [DOI] [PubMed] [Google Scholar]
- Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- Salz HK, Flickinger TW, Mittendorf E, Pellicena-Palle A, Petschek JP, Albrecht EB. The Drosophila maternal effect locus deadhead encodes a thioredoxin homolog required for female meiosis and early embryonic development. Genetics. 1994;136:1075–1086. doi: 10.1093/genetics/136.3.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svensson MJ, Larsson J. Thioredoxin-2 affects lifespan and oxidative stress in Drosophila. Hereditas. 2007;144:25–32. doi: 10.1111/j.2007.0018-0661.01990.x. [DOI] [PubMed] [Google Scholar]
- Swanson KS, Dowd SE, Suchodolski JS, Middelbos IS, Vester BM, Barry KA, Nelson KE, Torralba M, Henrissat B, Coutinho PM, Cann IK, White BA, Fahey GC., Jr Phylogenetic and gene-centric metagenomics of the canine intestinal microbiome reveals similarities with humans and mice. ISME J. 2011;5:639–649. doi: 10.1038/ismej.2010.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Higgins DG. Multiple sequence alignment using ClustalW and ClustalX. Curr Protoc Bioinformatics. 2002;Chapter 2(Unit 2):3. doi: 10.1002/0471250953.bi0203s00. [DOI] [PubMed] [Google Scholar]
- Tusnady GE, Simon I. Principles governing amino acid composition of integral membrane proteins: application to topology prediction. J Mol Biol. 1998;283:489–506. doi: 10.1006/jmbi.1998.2107. [DOI] [PubMed] [Google Scholar]
- Tusnady GE, Simon I. The HMMTOP transmembrane topology prediction server. Bioinformatics. 2001;17:849–850. doi: 10.1093/bioinformatics/17.9.849. [DOI] [PubMed] [Google Scholar]
- Villarreal AM, Adamson SW, Browning RE, Budachetri K, Sajid MS, Karim S. Molecular characterization and functional significance of the Vti family of SNARE proteins in tick salivary glands. Insect Biochem Mol Biol. 2013;43:483–493. doi: 10.1016/j.ibmb.2013.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview Version 2--a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisiger RA, Fridovich I. Mitochondrial superoxide simutase. Site of synthesis and intramitochondrial localization. J Biol Chem. 1973;248:4793–4796. [PubMed] [Google Scholar]
- Weiss B, Aksoy S. Microbiome influences on insect host vector competence. Trends Parasitol. 2011;27:514–522. doi: 10.1016/j.pt.2011.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu N, Chen CY, Shyu AB. Modulation of the fate of cytoplasmic mRNA by AU-rich elements: key sequence features controlling mRNA deadenylation and decay. Mol Cell Biol. 1997;17:4611–4621. doi: 10.1128/mcb.17.8.4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yodoi J, Uchiyama T. Diseases associated with HTLV-I: virus, IL-2 receptor dysregulation and redox regulation. Immunol Today. 1992;13:405–411. doi: 10.1016/0167-5699(92)90091-K. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Li H, Men LL, Huang RC, Zhou HC, Xing Q, Yao JJ, Shi CH, Du JL. Effects of selenoprotein S on oxidative injury in human endothelial cells. J Transl Med. 2013;11:287. doi: 10.1186/1479-5876-11-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong L, Holmgren A. Essential role of selenium in the catalytic activities of mammalian thioredoxin reductase revealed by characterization of recombinant enzymes with selenocysteine mutations. J Biol Chem. 2000;275:18121–18128. doi: 10.1074/jbc.M000690200. [DOI] [PubMed] [Google Scholar]