Abstract
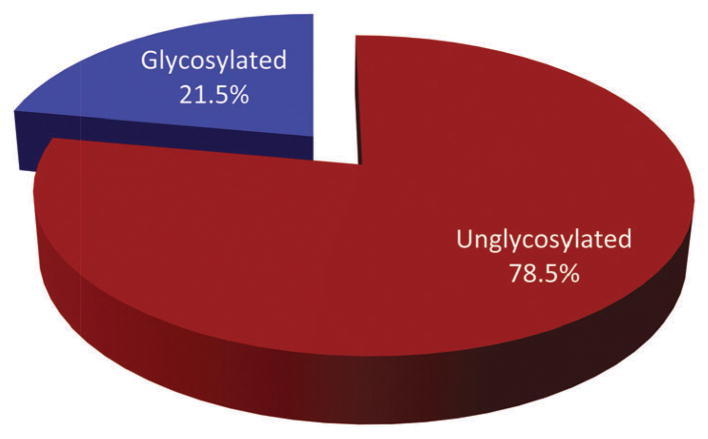
A systematic analysis of all naturally-occurring glycosylated bacterial secondary metabolites reported in the scientific literature up through early 2013 is presented. This comprehensive analysis of 15 940 bacterial natural products revealed 3426 glycosides containing 344 distinct appended carbohydrates and highlights a range of unique opportunities for future biosynthetic study and glycodiversification efforts.
1. Introduction
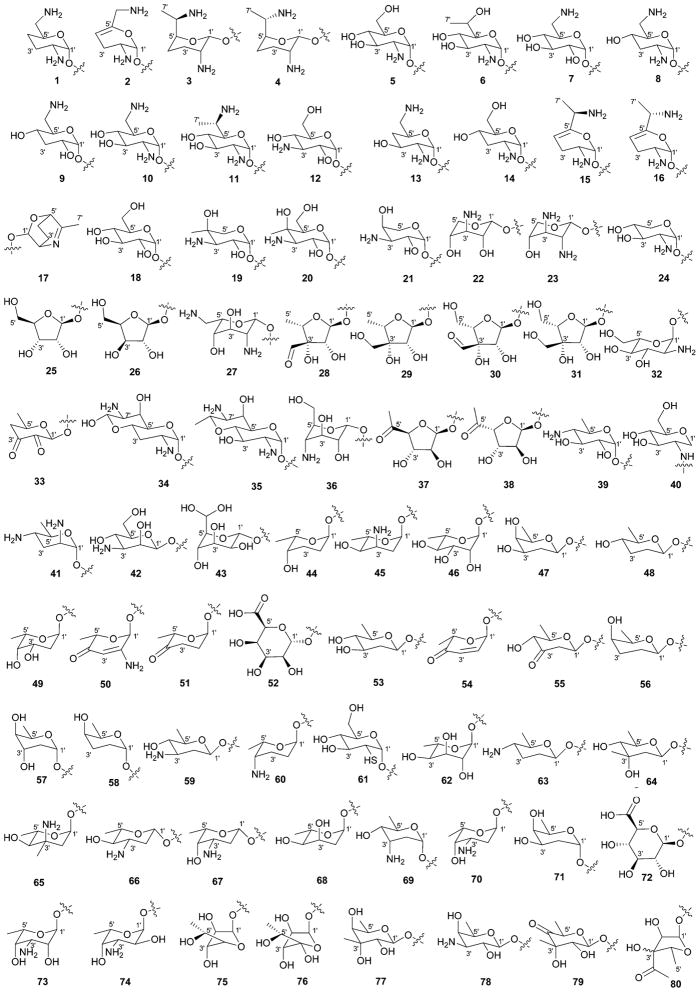
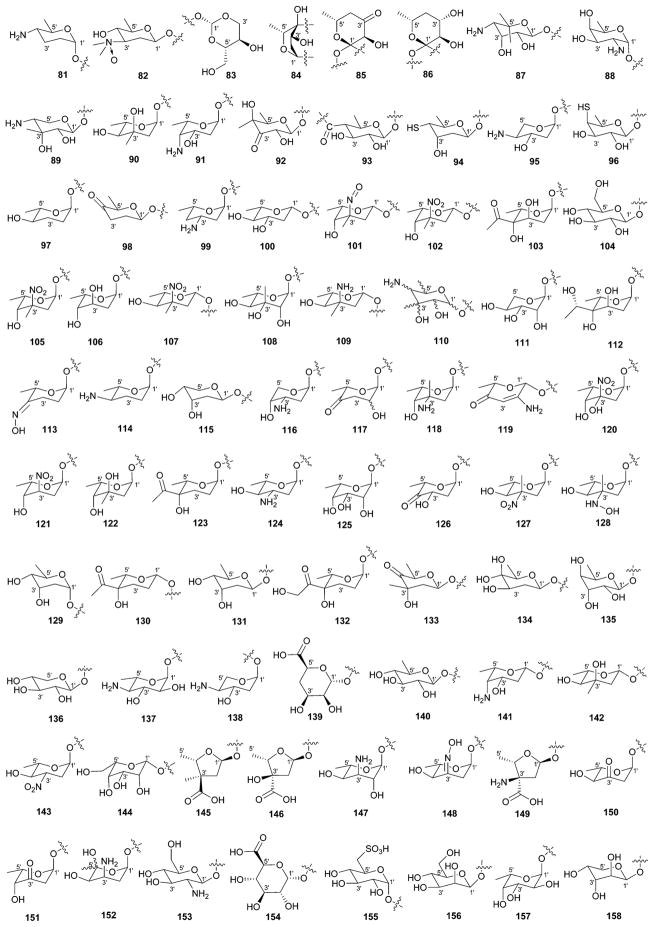
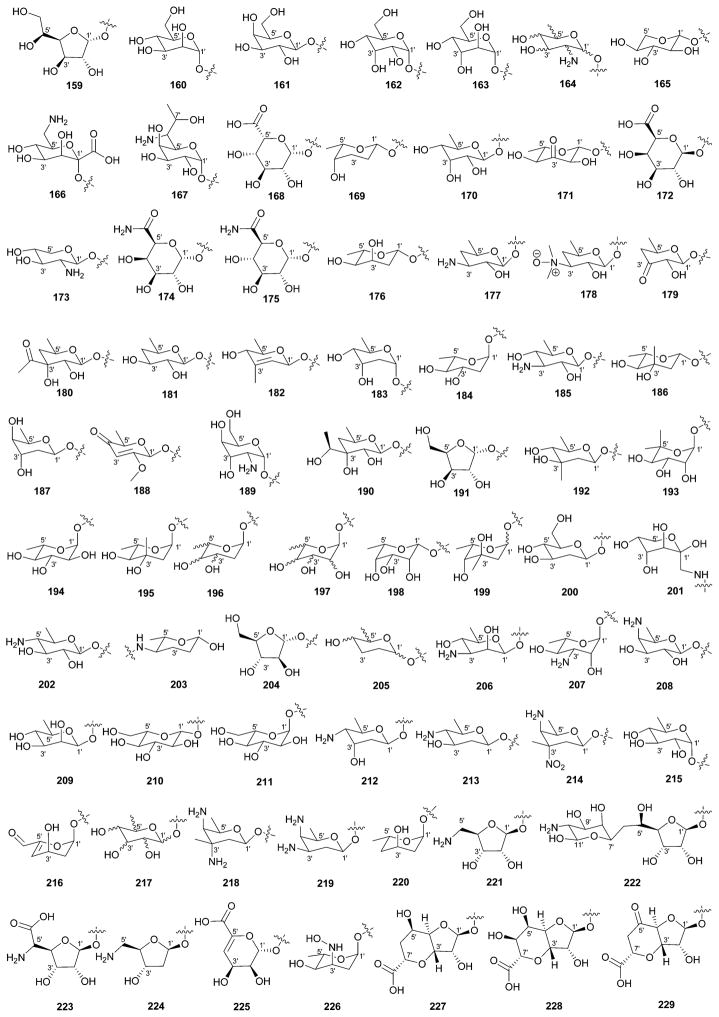
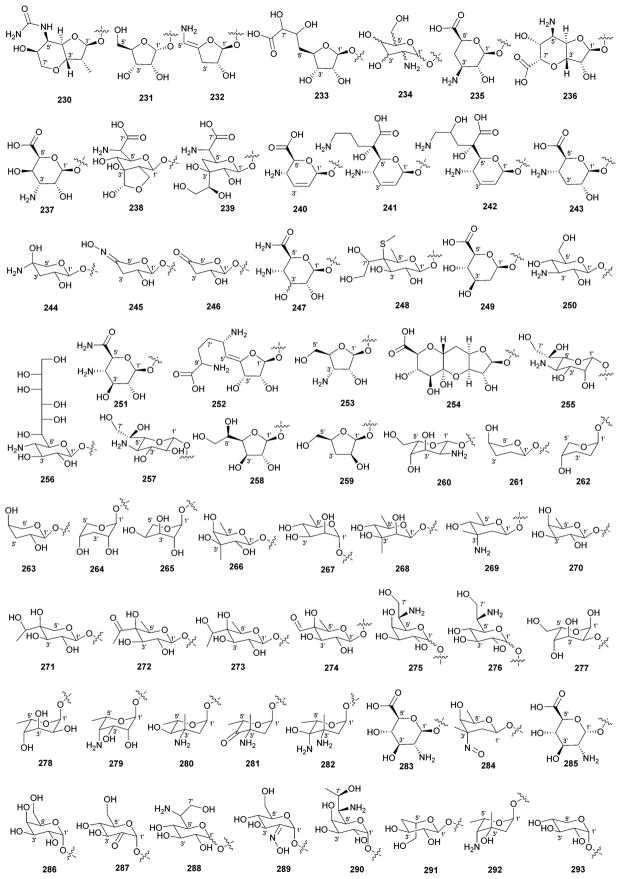
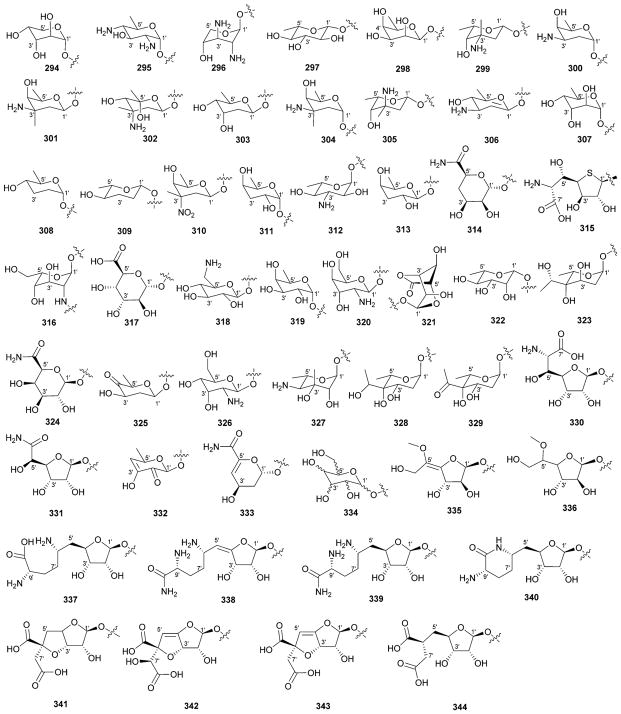
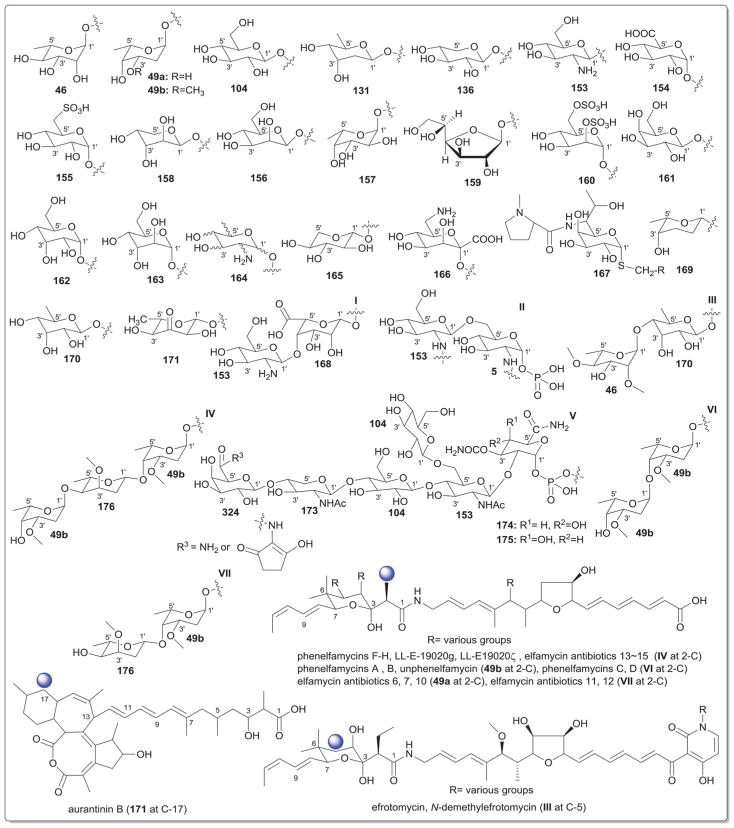
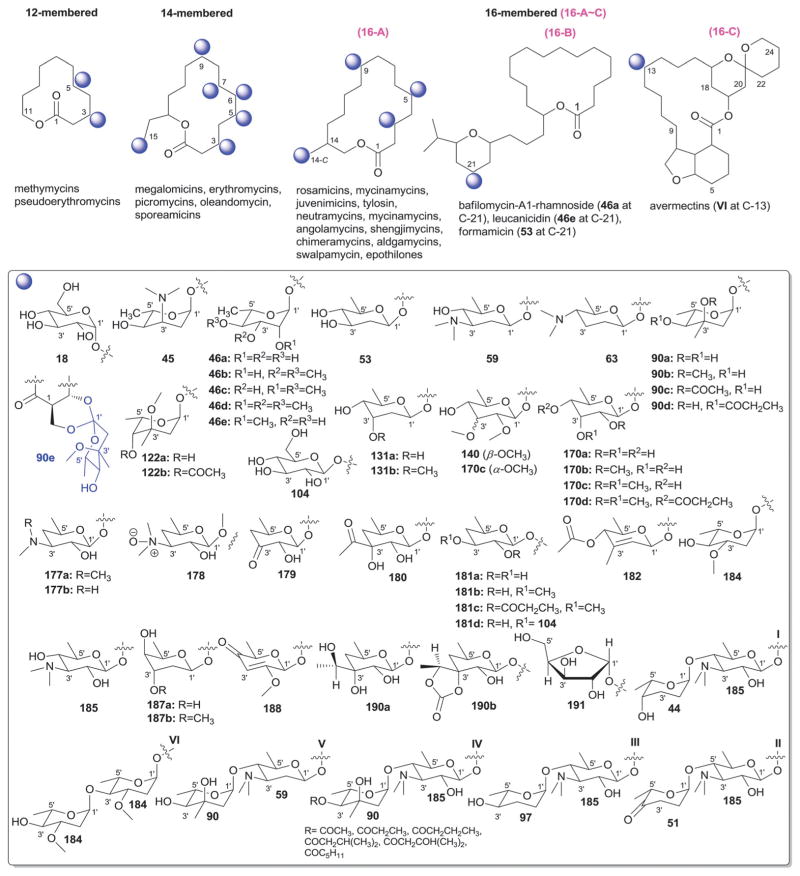
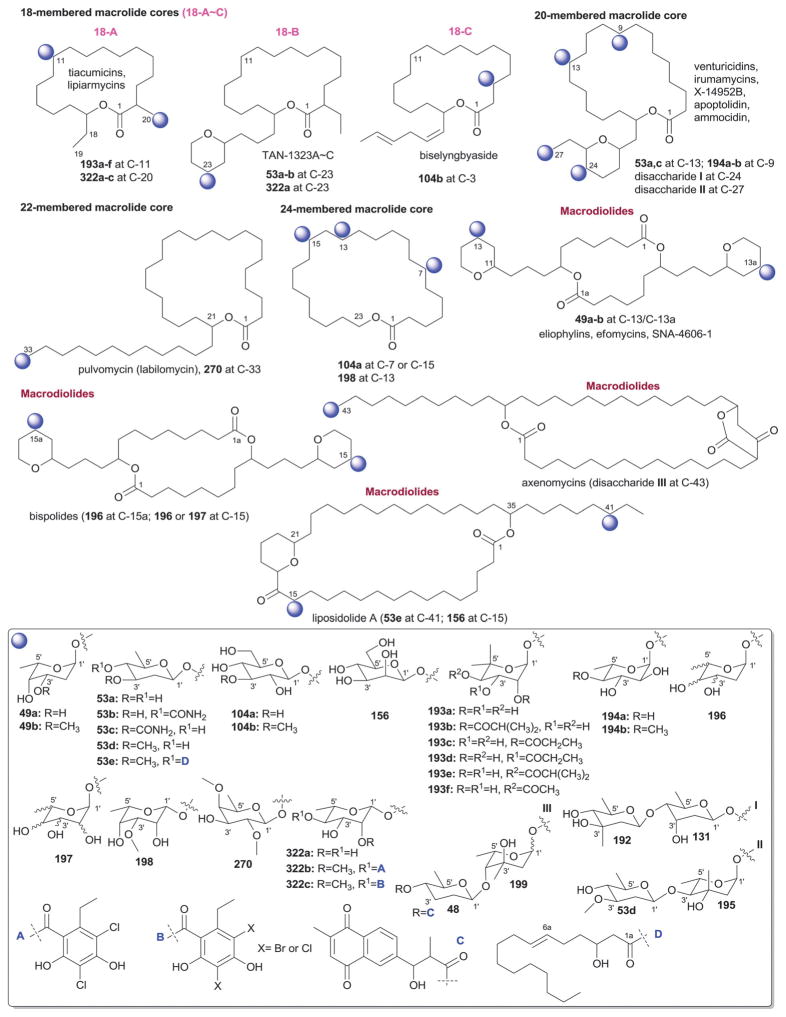
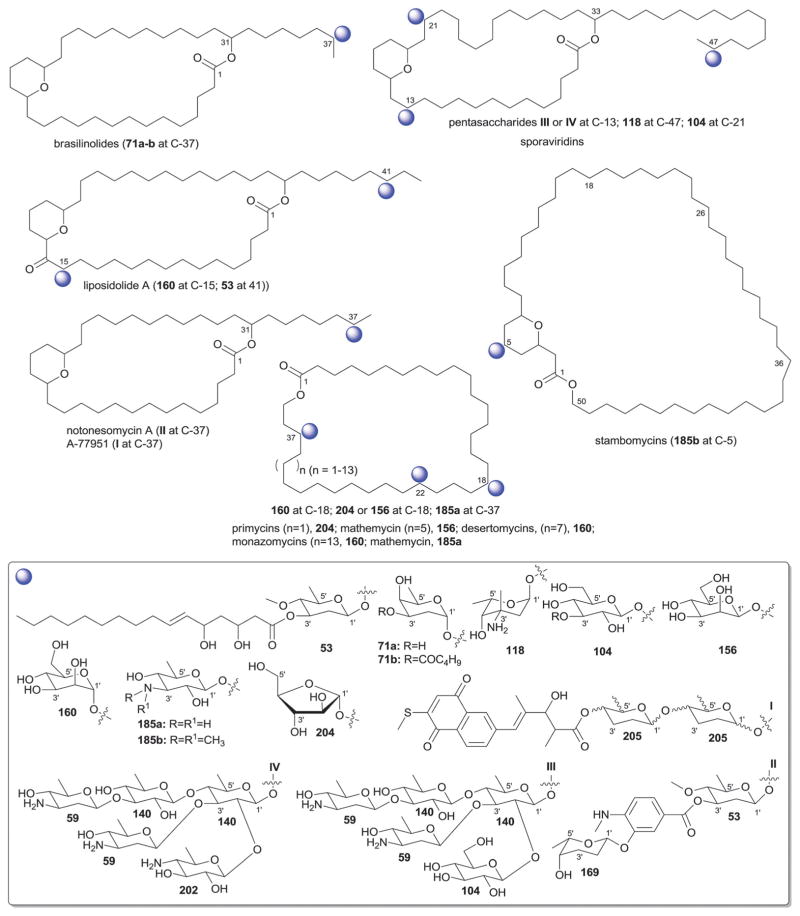
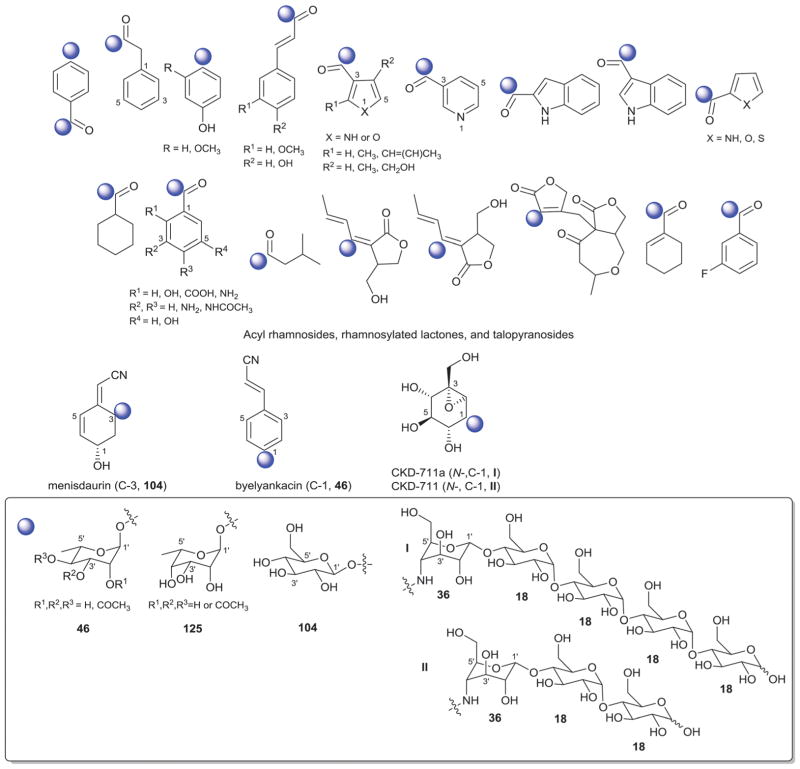
While it is well-established that the glycosylation of naturally-occurring and/or synthetic small molecule-based drugs can dramatically influence the pharmacological properties of the parent scaffold,1–17 there remains a lack of accuracy regarding the prevalence and/or extent of glycosidic diversity in the context of naturally-occuring glycosides. The current review attempts to address this gap of knowledge by providing a systematic analysis of all naturally-occurring glycosylated bacterial secondary metabolites reported in the scientific literature. AntiBase 201218 served as a key resource for glycoside identification and all glycosides were structurally validated via an analysis of the primary literature and corrected where necessary prior to integration into this compilation. Based upon an analysis of 15 940 bacterial natural products, over one fifth (3426 compounds, 21.5%, Fig. 1) are glycosides wherein glycosylated macrolides and macrolactams represent the largest allocation (738 compounds, 21.5% of all bacterial glycosides, Fig. 2). Further analysis of the range of saccharides represented across all 3426 bacterial glycosides revealed 344 distinct carbohydrates (Fig. 3 and Table 1). For this latter consideration, carbohydrates were only designated as ‘distinct’ based upon differences within the fundamental monsaccharide core (specifically, notable stereochemical and/or functional group variation, including anomeric configuration) whereas simple modifications of a given common sugar core (e.g., O/N/S-alkyl/acyl substitutions) were designated as identical to the parental core saccharide. The content of this comprehensive review has been organized based upon aglycon class/structure as indicated in Fig. 2 wherein each section provides additional relevant information pertaining to the glycosides represented within the selected metabolite classification. Illustrations throughout this review employ two standard conventions. First, regiospecificity of glycosylation is represented as a colored ball within the context of a representative aglycon, the latter of which are presented in many cases in generic form in an effort to emphasize glycosylation. Second, for simplicity, D-pyranoses are represented in the 4C1 conformation while L-pyranoses are illustrated as 1C4 conformers. For furanoses, and/or in cases where the chair conformation may obstruct field of view, a standard planar pentose or hexose ring is used. In addition, all pseudosugars found as part of the microbial glycosylated natural products discussed within this review are summarized in Fig. 4.
Fig. 1.
Bacterial glycosylated (21.5%; 3426 compounds) and unglycosylated (78.5%; 12 514 compounds) natural products.
Fig. 2.
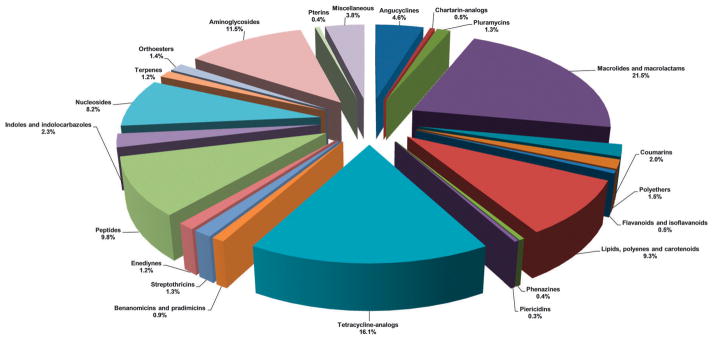
Chemical classes of glycosylated bacterial natural products (total 3426 compounds).
Fig. 3.
Summary of all sugars present in bacterial natural products (1–344). Only sugars that displayed differences within the fundamental monosaccharide core (specifically, notable stereochemical and/or functional group variation, including anomeric configuration) were considered as distinct. Modifications of a common sugar core (e.g., O/N/S-alkyl/acyl substitutions) were designated as identical to the parental core saccharide.
Table 1.
Names of sugars present in bacterial natural products. Numbers correspond to sugars in Fig. 3. For simplicity and convenience for the readers, common names were preferred than IUPAC nomenclature
| Sugar no. | Sugar name |
|---|---|
| 1 | 2′,6′-Diamino-2′,3′,4′,6′-tetradeoxy-α-D-glucose |
| 2 | 2′,6′-Diamino-2′,3′,4′,6′-tetradeoxy-4′,5′-unsaturated-α-D-glucose |
| 3 | (6′R)-2′,6′-Diamino-2′,3′,4′,6′,7′-pentadeoxy-β-L-glucoheptose |
| 4 | (6′S)-2′,6′-Diamino-2′,3′,4′,6′,7′-pentadeoxy-β-L-glucoheptose |
| 5 | 2′-Amino-2′-deoxy-α-D-glucose |
| 6 | 2′-Amino-2′,7′-dideoxy-α-D-glucoheptose |
| 7 | 6′-Amino-6′-deoxy-α-D-glucose |
| 8 | 2′,6′-Diamino-2′,3′,6′-trideoxy-α-D-glucose |
| 9 | 6′-Amino-3′,6′-dideoxy-α-D-glucose |
| 10 | 2′,6′-Diamino-2′,6′-dideoxy-α-D-glucose |
| 11 | 2′,6′-Diamino-2′,6′,7′-trideoxy-α-D-glucoheptose |
| 12 | 3′-Amino-3′-deoxy-α-D-glucose |
| 13 | 2′,6′-Diamino-2′,4′,6′-trideoxy-α-D-glucose |
| 14 | 2′-Amino-2′,3′-dideoxy-α-D-glucose |
| 15 | (6′R)-2′,6′-Diamino-2′,3′,4′,6′,7′-pentadeoxy-4′,5′-unsaturated-α-D-glucoheptose |
| 16 | (6′S)-2′,6′-Diamino-2′,3′,4′,6′,7′-pentadeoxy-4′,5′-unsaturated-α-D-glucoheptose |
| 17 | Cyclic unsaturated analog of 3/4 |
| 18 | α-D-Glucose |
| 19 | 3′-Amino-3′-deoxy-4′-C-methyl-4′-epi-α-D-xylose |
| 20 | 3′-Amino-3′-deoxy-4′-C-methyl-α-D-galactose |
| 21 | 3′-Amino-3′-deoxy-4′-epi-α-D-xylose |
| 22 | 3′-Amino-3′-deoxy-β-L-xylose |
| 23 | 2′,3′-Diamino-2′,3′-dideoxy-β-L-xylose |
| 24 | 2′-Amino-2′-deoxy-α-D-xylose |
| 25 | β-D-Ribofuranose |
| 26 | β-D-Xylofuranose |
| 27 | 2′,6′-Diamino-2′,6′-dideoxy-β-L-idose |
| 28 | 3′-C-Carbaldehyde-5′-deoxy-α-L-lyxofuranose |
| 29 | 5′-Deoxy-3′-C-hydroxymethyl-α-L-lyxofuranose |
| 30 | 3′-C-Carbaldehyde-α-L-lyxofuranose |
| 31 | 3′-C-Hydroxymethyl-α-L-lyxofuranose |
| 32 | α-L-Glucosamine |
| 33 | α-D-Spectinose |
| 34 | Unusual cyclized analog of 3′-deoxyglucosamine |
| 35 | Unusual cyclized analog of glucosamine |
| 36 | 4′-Deoxy-4′-amino-β-L-glucose |
| 37 | 5′-Keto-5′-C-methyl-β-D-arabinofuranose |
| 38 | 5′-Keto-5′-C-methyl-α-L-xylofuranose |
| 39 | 4′-Amino-4′,6′-dideoxy-α-D-glucose |
| 40 | 2′-Amino-1′,2′-dideoxy-α-D-glucose |
| 41 | 2′,4′-Diamino-2′,3′,4′,6′-tetradeoxy-α-D-mannose |
| 42 | 3′-Amino-3′-deoxy-β-D-mannose |
| 43 | 6′-Hydroxy-β-L-mannose |
| 44 | α-L-Rhodinose |
| 45 | α-L-Ristosamine |
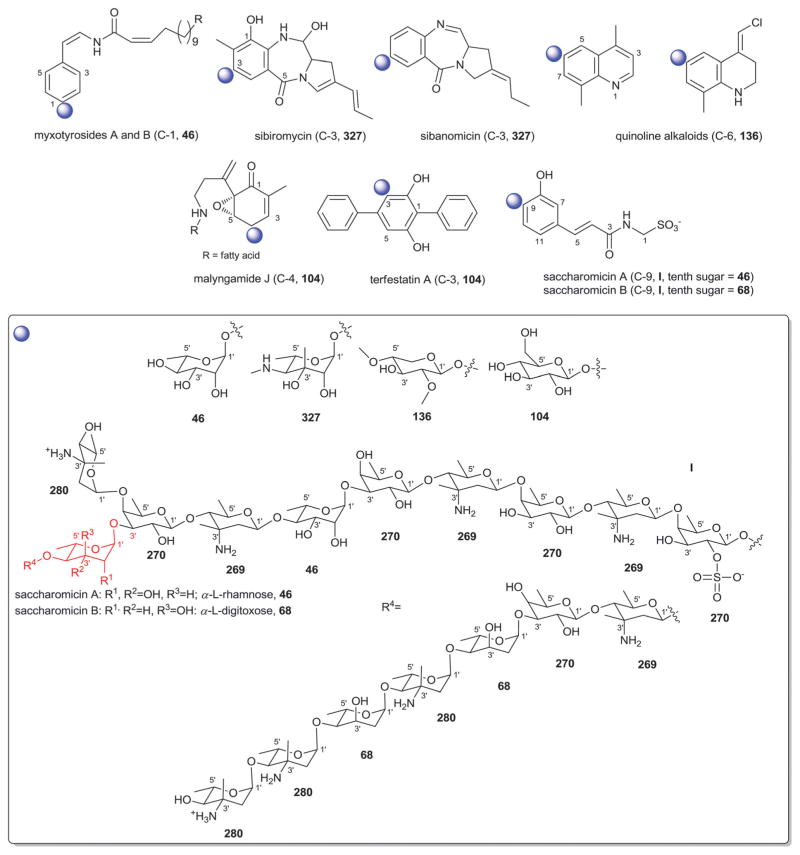
| 46 | α-L-Rhamnose |
| 47 | β-D-Oliose |
| 48 | β-D-Amicetose |
| 49 | α-L-Oliose |
| 50 | α-L-Rednose |
| 51 | α-L-Cinerulose |
| 52 | α-D-Taluronic acid |
| 53 | β-D-Olivose |
| 54 | α-L-Aculose |
| 55 | β-D-Kerriose |
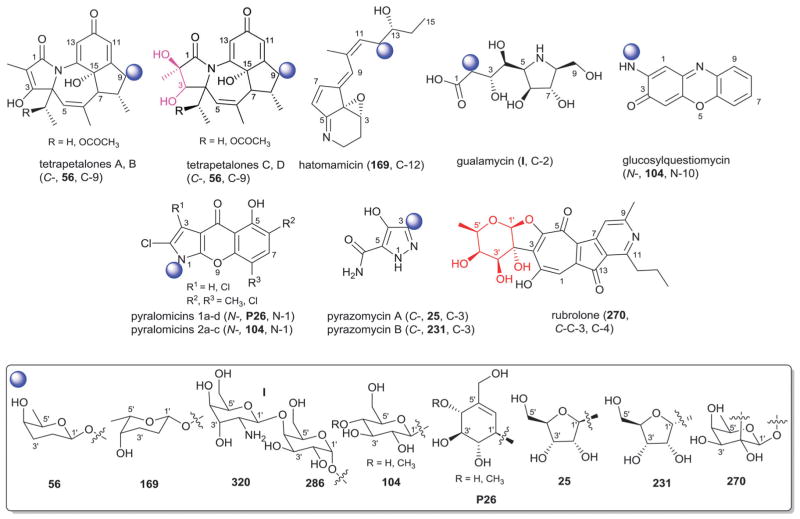
| 56 | β-D-Rhodinose |
| 57 | α-D-Boivinose |
| 58 | α-D-Rhodinose |
| 59 | 3′-Amino-2′,3′,6′-trideoxy-β-D-glucose |
| 60 | 4′-epi-α-L-Tolyposamine |
| 61 | 2′-Thio-α-D-glucose |
| 62 | 6′-Deoxy-α-L-altrose |
| 63 | 4′-Amino-4′-deoxy-β-D-amicetose |
| 64 | β-D-Mycarose |
| 65 | 3′-epi-4′-epi-α-L-Vancosamine |
| 66 | 3′-Amino-2′,3′,6′-trideoxy-β-L-glucose |
| 67 | 3′-Amino-2′,3′-dideoxy-β-L-fucose |
| 68 | α-L-Digitoxose |
| 69 | α-D-Ristosamine |
| 70 | 3′-Amino-2′,3′-dideoxy-α-L-fucose |
| 71 | α-D-Oliose |
| 72 | β-D-Glucuronic acid |
| 73 | 3′-Amino-3′,6′-dideoxy-α-L-talose |
| 74 | 3′-Amino-3′,6′-dideoxy-α-L-galactose |
| 75 | β-D-Fucofuranose |
| 76 | 4′-Hydroxy-β-D-fucofuranose |
| 77 | β-D-Virenose |
| 78 | 3′-Deoxy-3′-amino-β-D-fucose (β-D-ravidosamine) |
| 79 | 4′-Keto-β-D-virenose |
| 80 | 3′-C-Acetylpentofuranose |
| 81 | 4′-Amino-4′-deoxy-α-D-amicetose (α-D-forasmine) |
| 82 | 3′-Amino-N′,N′-dimethyl-N-oxido-2′,3′,6′-trideoxy-β-D-glucose |
| 83 | Unusual cyclized sugar I |
| 84 | Unusual cyclized sugar II |
| 85 | Potential sugar I |
| 86 | Potential sugar II |
| 87 | 4′-Amino-4′,6′-dideoxy-β-D-allose |
| 88 | α-D-Fucosamine; α-D-elsaminose |
| 89 | 4′-Amino-4′-deoxy-3′-C-methyl-β-D-ribose |
| 90 | α-L-Mycarose |
| 91 | 4′-Amino-2′,4′-dideoxy-β-L-fucose |
| 92 | 3′-Keto-4′-C-methyl-β-D-fucose |
| 93 | 4′,6′-Dideoxy-4′-carbonyl-β-D-glucose |
| 94 | 4′-Thio-2′,4′,6′-trideoxy-β-D-altrose |
| 95 | 4′-Amino-2′,4′-dideoxy-α-L-xylose |
| 96 | 4′-Deoxy-4′-thio-β-D-fucose |
| 97 | α-L-Amicetose |
| 98 | β-D-Cinerulose |
| 99 | 4′-Deoxy-α-L-daunosamine |
| 100 | β-L-Olivose |
| 101 | 3′-C-Methyl-3′-nitrose-2′,3′,6′-trideoxy-β-L-gulose |
| 102 | β-L-Decilonitrose |
| 103 | 4′-C-Acetyl-2′,6′-dideoxy-α-L-gulose (α-L-trioxacarcinose B) |
| 104 | β-D-Glucose |
| 105 | α-L-Decilonitrose |
| 106 | 2′,6′-Dideoxy-α-L-gulose |
| 107 | 4′-epi-β-L-Decilonitrose |
| 108 | 3′-C-Methyl-α-L-rhamnose |
| 109 | β-L-Avidinosamine |
| 110 | 4′-Amino-4′,6′-dideoxy-3′-C-methylhexose |
| 111 | α-L-Lyxose |
| 112 | 2′,6′-Dideoxy-4′-C-hydroxyethyl-α-L-gulose |
| 113 | 4′-Oximo-2′,3′,4′-trideoxy-α-L-fucose |
| 114 | 4′-Amino-2′,3′,4′,6′-tetradeoxy-α-L-glucose |
| 115 | 2′-Deoxy-β-D-ribose |
| 116 | 3′-Amino-2′,3′-dideoxy-4′-epi-α-L-xylose |
| 117 | 3′,6′-Dideoxy-4′-keto-α-L-hexose |
| 118 | α-L-Vancosamine |
| 119 | β-L-Rednose |
| 120 | 3′-Hydroxy-3′-C-nitro-2′,6′-dideoxy-α-L-talose |
| 121 | 3′-Nitro-2′,3′,6′-trideoxy-α-L-gulose (3′-desmethyl-α-L-decilonitrose) |
| 122 | 4′-epi-α-L-Mycarose |
| 123 | 4′-C-Acetyl-2′,3′,6′-trideoxy-α-L-gulose |
| 124 | α-L-Actinosamine |
| 125 | 6′-Deoxy-α-L-talose |
| 126 | 4′-Keto-α-L-olivose |
| 127 | 3′-epi-4′-epi-α-L-Decilonitrose (4′-desmethyl-α-L-evernitrose) |
| 128 | 3′-Denitro-3′-hydroxylamine-3′-epi-4′-epi-α-L-decilonitrose |
| 129 | α-D-Digitoxose |
| 130 | 4′-C-Acetyl-2′,3′,6′-trideoxy-β-L-gulose |
| 131 | β-D-Digitoxose |
| 132 | 4′-C-Hydroxyethanone-2′,3′,6′-trideoxy-α-L-gulose |
| 133 | 4′-Keto-β-D-mycarose |
| 134 | 4′-Hydroxy-β-D-olivose |
| 135 | 6′-Deoxy-β-D-gulose |
| 136 | β-D-Xylose |
| 137 | 4′-Amino-4′,6′-dideoxy-α-L-glucose |
| 138 | 4′-Amino-2′,4′-dideoxy-α-L-xylose |
| 139 | 4′-Deoxy-α-D-taluronic acid |
| 140 | β-D-Quinovose |
| 141 | 4′-Amino-2′,4′,6′-trideoxy-β-L-galactose |
| 142 | β-L-Mycarose |
| 143 | 3′-Nitro-2′,3′,6′-trideoxy-α-L-glucose |
| 144 | β-L-Talose |
| 145 | 3′-C-Carboxy-3′-C-methyl-2′,3′,5′-trideoxy-α-L-xylofuranose |
| 146 | 3′-C-Carboxy-2′,5′-dideoxy-α-L-ribofuranose |
| 147 | 3′-Amino-3′,6′-dideoxy-α-L-altrose |
| 148 | 2′,3′,6′-Trideoxy-3′-oximo-α-L-altrose |
| 149 | 3′-Amino-3′-C-carboxy-2′,3′,5′-trideoxy-α-L-ribofuranose |
| 150 | 3′-Keto-2′,3′,6′-trideoxy-α-L-glucose |
| 151 | 2′-Deoxy-3′-keto-α-L-fucose |
| 152 | 3′-Amino-5′-hydroxy-2′,3′,6′-trideoxyhexose |
| 153 | β-D-Glucosamine |
| 154 | α-D-Glucuronic acid |
| 155 | α-D-Quinovose-6′-sulfonic acid |
| 156 | β-D-Mannose |
| 157 | α-L-Fucose |
| 158 | β-D-Arabinose |
| 159 | α-D-Glucofuranose |
| 160 | α-D-Mannose |
| 161 | β-D-Galactose |
| 162 | α-D-Allose |
| 163 | α-D-Altrose |
| 164 | 6′-Deoxyhexosamine |
| 165 | β-L-Xylose |
| 166 | 6′-Amino-1′-carboxy-6′-deoxy-β-D-mannose |
| 167 | 6′-Amino-6′,8′-dideoxy-α-D-galactoctose |
| 168 | β-L-Iduronic acid |
| 169 | β-L-Rhodinose |
| 170 | 6′-Deoxy-β-D-allose |
| 171 | 6′-Deoxy-3′-keto-β-L-glucose |
| 172 | β-D-Galacturonic acid |
| 173 | 2′-Amino-2′-deoxy-β-D-xylose |
| 174 | α-D-Galacturonamide |
| 175 | α-D-Glucuronamide |
| 176 | β-L-Digitoxose |
| 177 | 3′-Amino-3′,4′-dideoxy-β-D-fucose |
| 178 | 3′-N-Oxido-β-D-desosamine |
| 179 | 4′,6′-Dideoxy-3′-keto-β-D-fucose |
| 180 | 3′-C-Acetyl-4′,6′-dideoxy-β-D-allose |
| 181 | 4′-Deoxy-β-D-fucose |
| 182 | 3′-C-Methyl-2′,3′,6′-trideoxy-2′,3′-unsaturated-β-D-glucose |
| 183 | 2′,6′-Dideoxy-α-D-altrose |
| 184 | α-L-Olivose |
| 185 | 3′-Amino-3′,6′-dideoxy-β-D-glucose |
| 186 | β-L-Chromose |
| 187 | β-D-Boivinose |
| 188 | 3′,6′-Dideoxy-4′-keto-2′-O-methyl-2′,3′-unsaturated-β-D-glucose |
| 189 | α-D-Gulosamine |
| 190 | 4′,6′-Dideoxy-3′-C-hydroxyethyl-β-D-allose |
| 191 | α-D-Xylofuranose |
| 192 | β-D-Olivomicose |
| 193 | 5′-C-Methyl-α-L-rhamnose (4′-O-desmethyl-α-L-noviose) |
| 194 | α-L-Quinovose |
| 195 | α-L-Chromose |
| 196 | 2′,6′-Dideoxyhexose |
| 197 | 6′-Deoxyhexose |
| 198 | 6′-Deoxy-β-L-talose |
| 199 | L-Axenose |
| 200 | 2′-Deoxy-β-D-glucose |
| 201 | 1′-C-Methylamine-β-D-arabinose |
| 202 | 4′-Amino-4′,6′-dideoxy-β-D-glucose |
| 203 | 4′-Amino-2′,3′,4′,6′-tetradeoxyhexose |
| 204 | α-D-Arabinofuranose |
| 205 | 2′,3′,6′-Trideoxyhexose |
| 206 | β-D-Mycosamine |
| 207 | α-L-Mycosamine |
| 208 | 4′-Amino-4′-deoxy-β-D-fucose |
| 209 | 6′-Deoxy-β-D-mannose |
| 210 | β-L-Glucose |
| 211 | α-L-Glucose |
| 212 | N-Desmethyl-β-D-vicenisamine |
| 213 | β-D-Pyrrolosamine |
| 214 | N-Desmethylcarbamate-β-D-tetronitrose |
| 215 | α-D-Quinovose |
| 216 | 5′-C-Carbaldehyde-4′,5′-unsaturated-β-D-digitoxose |
| 217 | 6′-Deoxyhexose |
| 218 | 3′,4′-Diamino-3′-C-methyl-2′,3′,4′,6′-tetradeoxy-β-D-gulose |
| 219 | 3′,4′-Diamino-2′,3′,4′,6′-tetradeoxy-β-D-galactose |
| 220 | 4′-Deoxy-α-L-digitoxose |
| 221 | 5′-Amino-5′-deoxy-β-D-ribofuranose |
| 222 | Tunicamine |
| 223 | 5′-Amino-5′-C-carboxy-5′-deoxy-β-D-ribofuranose |
| 224 | 2′,5′-Dideoxy-5′-amino-β-D-ribofuranose |
| 225 | 4′,5′-Unsaturated-α-D-mannuronic acid |
| 226 | 3′-Hydroxylamine-2′,3′,6′-trideoxy-α-L-allose |
| 227 | Unusual bicyclic sugar I |
| 228 | Unusual bicyclic sugar II |
| 229 | Unusual bicyclic sugar III |
| 230 | Unusual bicyclic sugar IV |
| 231 | α-D-Ribofuranose |
| 232 | 5′-Amino-5′-deoxy-4′,5′-unsaturated-β-D-ribofuranose |
| 233 | 5′-Deoxy-β-D-ribo-octofuranuronic acid |
| 234 | 2′-Deoxy-2′-aminohexose |
| 235 | 3′-Amino-3′,4′-dideoxy-β-D-glucuronic acid |
| 236 | Unusual dicyclic sugar |
| 237 | 2′-Amino-2′-deoxy-β-D-guluronic acid |
| 238 | Unusual β-D-sugar |
| 239 | Unusual β-D-sugar analog of 238 |
| 240 | 4′-Amino-4′-deoxy-2′,3′-unsaturated-β-D-glucuronic acid |
| 241 | 4′-Amino-6′-C-carboxy-4′-deoxy-6′-C-propylamine-2′,3′-unsaturated-β-D-glucose |
| 242 | Hydroxy analog of 241 |
| 243 | 4′-Amino-3′-4′-dideoxy-β-D-glucuronic acid |
| 244 | 4′-C-Amino-3′-deoxypentose |
| 245 | 4′-Oximo-3′,4′,5′-trideoxy-β-D-xylose |
| 246 | 4′-Keto-3′,4′,5′-trideoxy-β-D-xylose |
| 247 | 4′-Amino-4′-deoxy-β-D-hexuronamide |
| 248 | 6′-Deoxy-4′-thio-4′-S-methyl-4′-C-(1,2-dihydroxyethyl)-β-D-galactose |
| 249 | 2′-Deoxy-β-D-glucuronic acid |
| 250 | 3′-Amino-3′-deoxy-β-D-glucose |
| 251 | 4′-Amino-4′-deoxy-β-D-glucuronamide |
| 252 | 6′,9′-Diamino-5′,6′,7′,8′,9′-pentadexoy-4′,5′-unsaturated-α-L-ribo-decofuranuronic acid |
| 253 | 3′-Amino-3′-deoxy-β-D-ribofuranose |
| 254 | Unusual tricyclic sugar |
| 255 | 4′-Amino-4′-deoxy-β-L-mannoheptose |
| 256 | Unusual C–C sugar |
| 257 | 4′-Amino-4′-deoxy-β-L-glucoheptose |
| 258 | β-D-Glucofuranose |
| 259 | 3′-Deoxy-β-D-arabinofuranose |
| 260 | β-L-Gulosamine |
| 261 | 2′,3′-Dideoxy-4′-epi-β-D-xylose |
| 262 | 2′,3′-Dideoxy-4′-epi-α-L-xylose |
| 263 | 3′-Deoxy-4′-epi-β-D-xylose |
| 264 | 3′-Deoxy-4′-epi-α-L-arabinose |
| 265 | 4′-α-L-Arabinose |
| 266 | β-D-Elsarose |
| 267 | 6′-Deoxy-α-D-mannose |
| 268 | 6′-Deoxy-3′-C-methyl-β-D-mannose |
| 269 | β-D-Saccharosamine |
| 270 | β-D-Fucose |
| 271 | β-D-Eurekanate I |
| 272 | β-D-Eurekanate II |
| 273 | β-D-Eurekanate III |
| 274 | β-D-Eurekanate IV |
| 275 | D-Destomic acid |
| 276 | 4′-epi-Destomic acid |
| 277 | α-L-Gulose |
| 278 | 6′-Deoxy-α-L-gulose |
| 279 | 4′-Amino-4′,6′-dideoxy-α-L-talose |
| 280 | 4′-epi-α-L-Vancosamine |
| 281 | 4′-Keto-α-L-vancosamine |
| 282 | 4′-C-Amino-4′-epi-α-L-vancosamine |
| 283 | 2′-Amino-2′-deoxy-β-D-glucuronic acid |
| 284 | 3′-C-Methyl-3′-nitroso-2′,3′,6′-trideoxy-β-D-gulose |
| 285 | 2′-Amino-2′-deoxy-α-D-glucuronic acid |
| 286 | α-D-galactose |
| 287 | 2′-Keto-α-D-glucose |
| 288 | 6′-Amino-6′-deoxy-D-glucoheptose |
| 289 | 2′-Oximo-α-D-glucose |
| 290 | 7′-C-Methyl-destomic acid |
| 291 | 4′-Deoxy-β-D-glucose |
| 292 | 4′-Amino-4′-deoxy-3′-C-methyl-α-L-fucose |
| 293 | α-D-Xylose |
| 294 | α-D-Arabinose |
| 295 | 2′,4′-Diamino-2′,4′,6′-trideoxy-α-D-glucose |
| 296 | 2′,3′-Diamino-2′,3′-dideoxy-α-L-pentose |
| 297 | β-L-Quinovose |
| 298 | 6′-Deoxy-β-D-talose |
| 299 | β-L-Vancosamine |
| 300 | 3′-Amino-2′,3′-dideoxy-α-D-fucose |
| 301 | β-D-Vancosamine |
| 302 | 3′-epi-4′-epi-5′-Hydroxy-β-D-vancosamine |
| 303 | 2′,6′-Dideoxy-β-D-altrose |
| 304 | α-D-Vancosamine |
| 305 | 3′-epi-β-L-Vancosamine |
| 306 | Δ1,2-D-Anglosamine |
| 307 | 6′-Deoxy-α-D-altrose |
| 308 | α-D-Amicetose |
| 309 | β-L-Amicetose |
| 310 | 3′-O-desmethyl-β-D-rubranitrose |
| 311 | 3′-Deoxy-α-D-fucose |
| 312 | 3′-Amino-3′,6′-dideoxy-α-L-glucose |
| 313 | 3′-Deoxy-β-D-fucose |
| 314 | 4′-Deoxy-α-D-taluronamide |
| 315 | 6′-Amino-6′-deoxy-4′-thio-heptofuranuronic acid |
| 316 | 2′-Amino-2′-deoxy-α-L-idose |
| 317 | α-D-Altruronic acid |
| 318 | 6′-Amino-6′-deoxy-β-D-glucose |
| 319 | α-D-Fucose; 3-O-desmthyl-α-D-digitalose |
| 320 | 2′-Deoxy-2′-amino-β-D-gulose |
| 321 | 3′,5′-Unsaturated galactouronic acid |
| 322 | β-L-Rhamnose |
| 323 | 2′,6′-Dideoxy-3′-hydroxyethyl-α-L-gulose (analog of 324) |
| 324 | β-D-Galacturonamide |
| 325 | 4′-Keto-β-D-olivose |
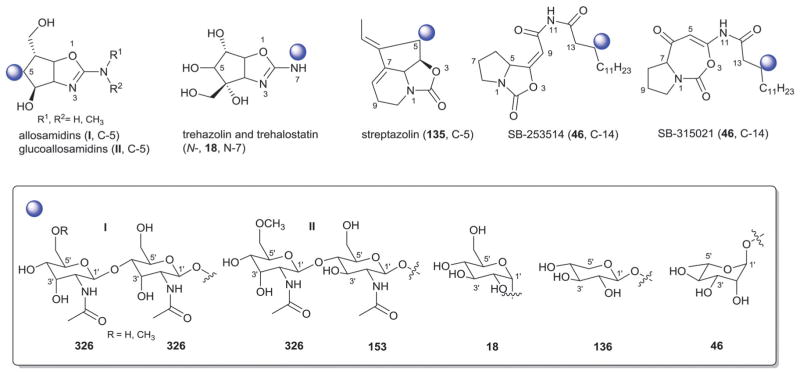
| 326 | β-D-Allosamine |
| 327 | α-L-Sibirosamine |
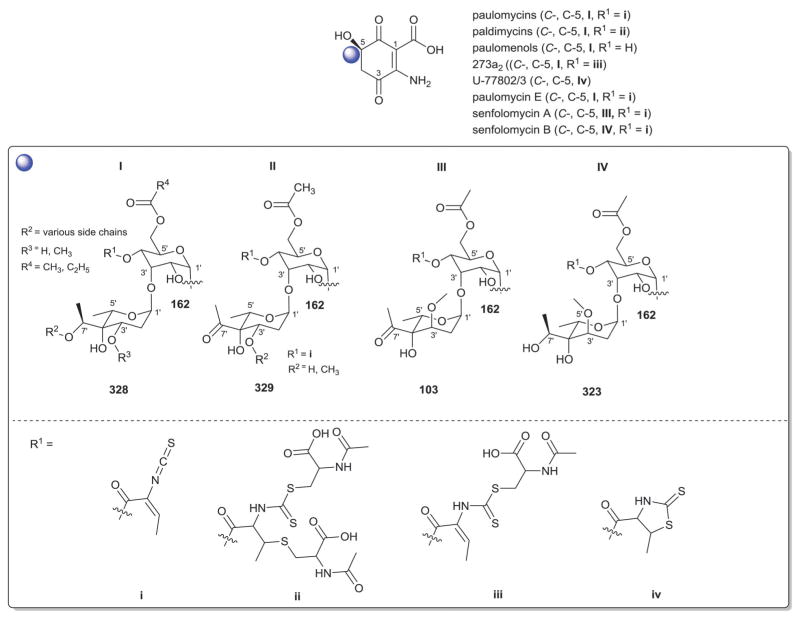
| 328 | α-L-Paulomycose |
| 329 | α-L-Keto analog of 328 |
| 330 | 6′-Amino-6′-deoxy-β-D-ribofuranhepturonic acid |
| 331 | β-D-Ribofuranhexuronamide |
| 332 | enol form of spectinose (33) |
| 333 | 2′,4′-Dideoxy-4′,5′-unsaturated-α-D-glucuronamide |
| 334 | Hexose |
| 335 | 5′-O-Methyl-4′,5′-unsaturated-β-D-arabinofuranohexose |
| 336 | 5′-O-Methyl-β-D-arabinofuranohexose |
| 337 | 6′,9′-Diamino-5′,6′,7′,8′,9′-pentadeoxy-β-D-ribofuranodecuronic acid |
| 338 | 6′,9′-Diamino-5′,6′,7′,8′,9′-pentadeoxy-4′,5′-unsaturated-β-D-ribofuranodecuronamide |
| 339 | 6′,9′-Diamino-5′,6′,7′,8′,9′-pentadeoxy-β-D-ribofuranodecuronamide |
| 340 | Cyclic analog of 339 |
| 341 | Unusual nine carbon bicyclic sugar I (cyclic form of 344) |
| 342 | Unusual nine carbon bicyclic sugar II |
| 343 | Unusual nine carbon bicyclic sugar III |
| 344 | 6′-C-Carboxy-5′,6′,7′-trideoxy-β-D-ribofuranoocturonic acid |
Fig. 4.
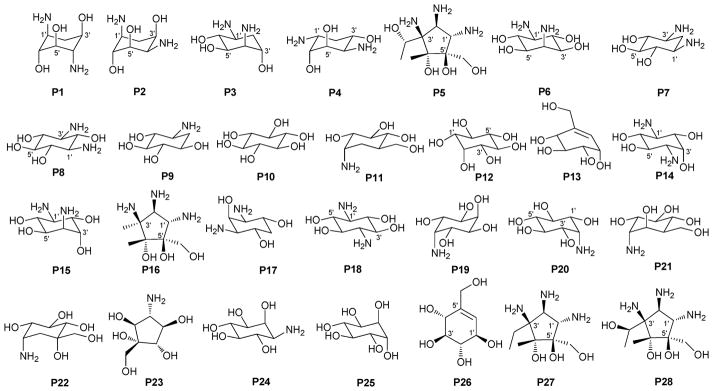
Summary of pseudosugars P1–P28 present in bacterial natural products. This list represents only those pseudosugar found within the context of bacterial glycosides and does not reflect an exhaustive list of naturally-occurring pseudosugars.
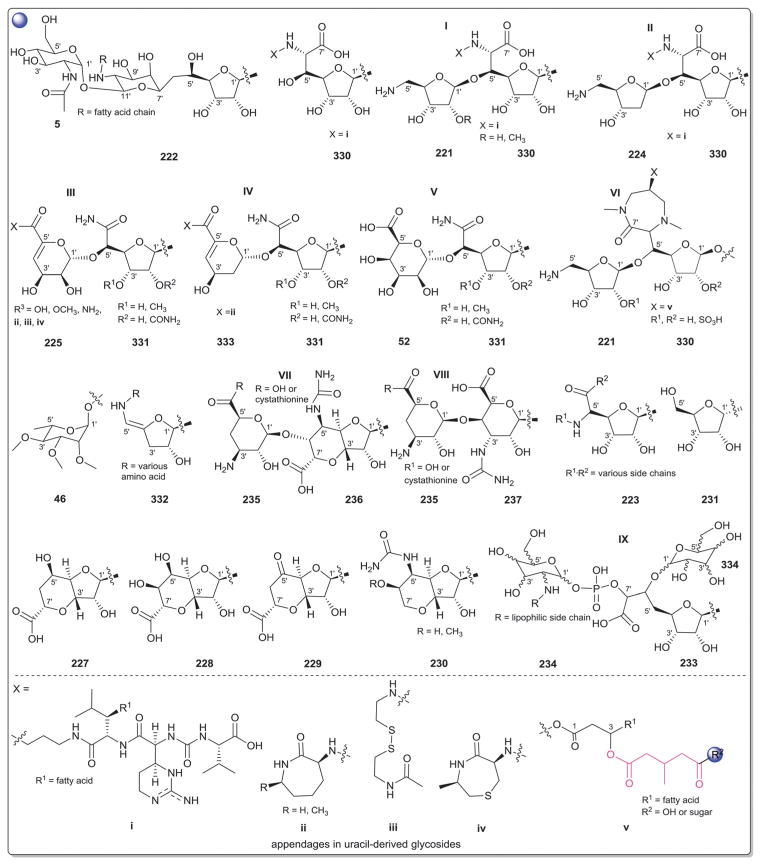
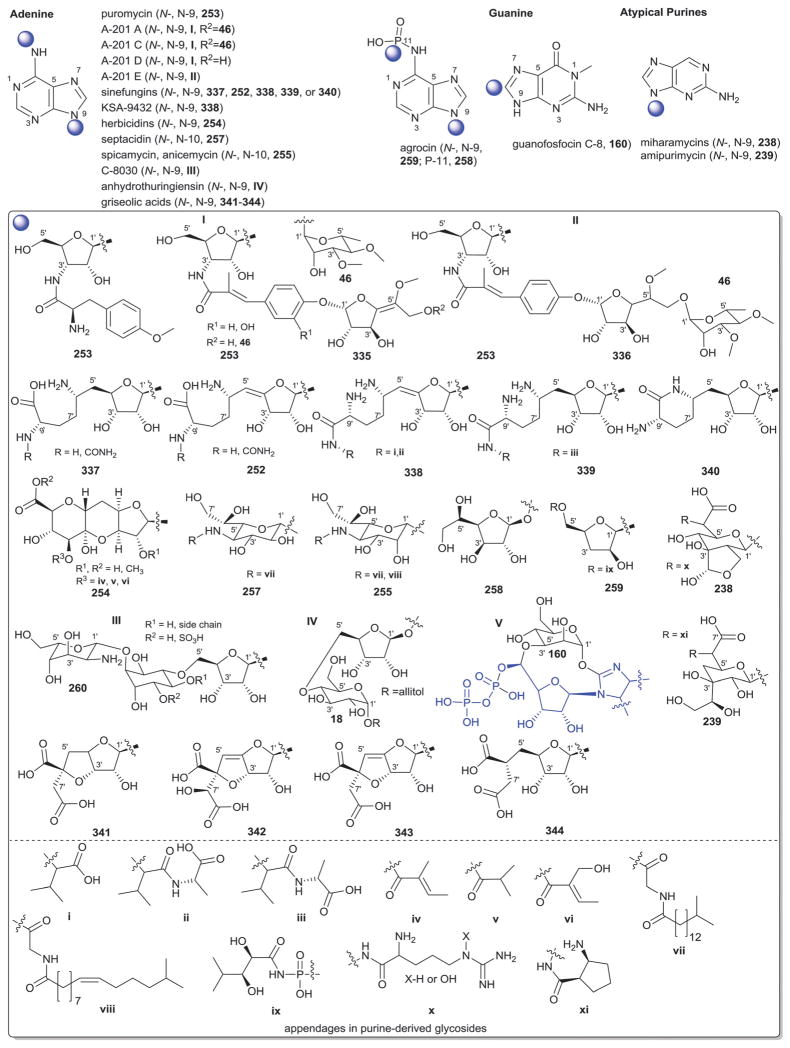
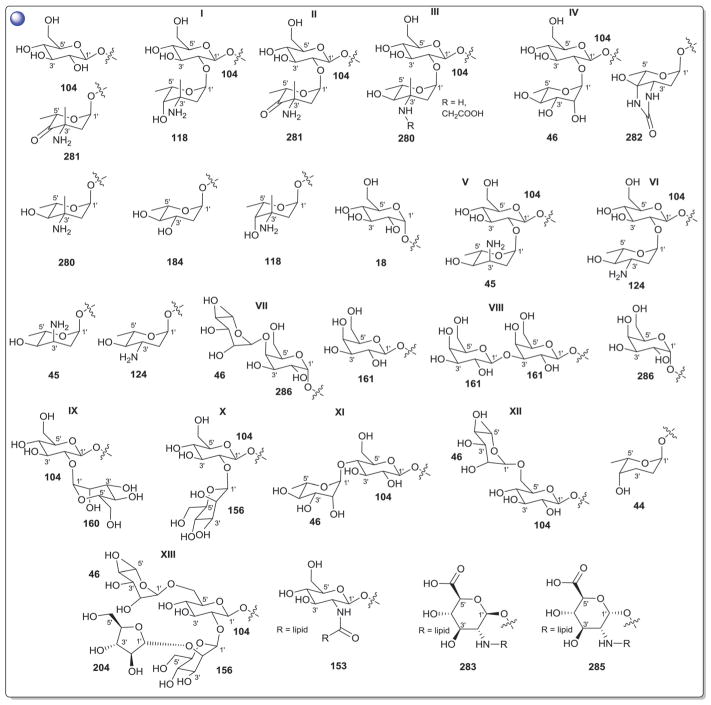
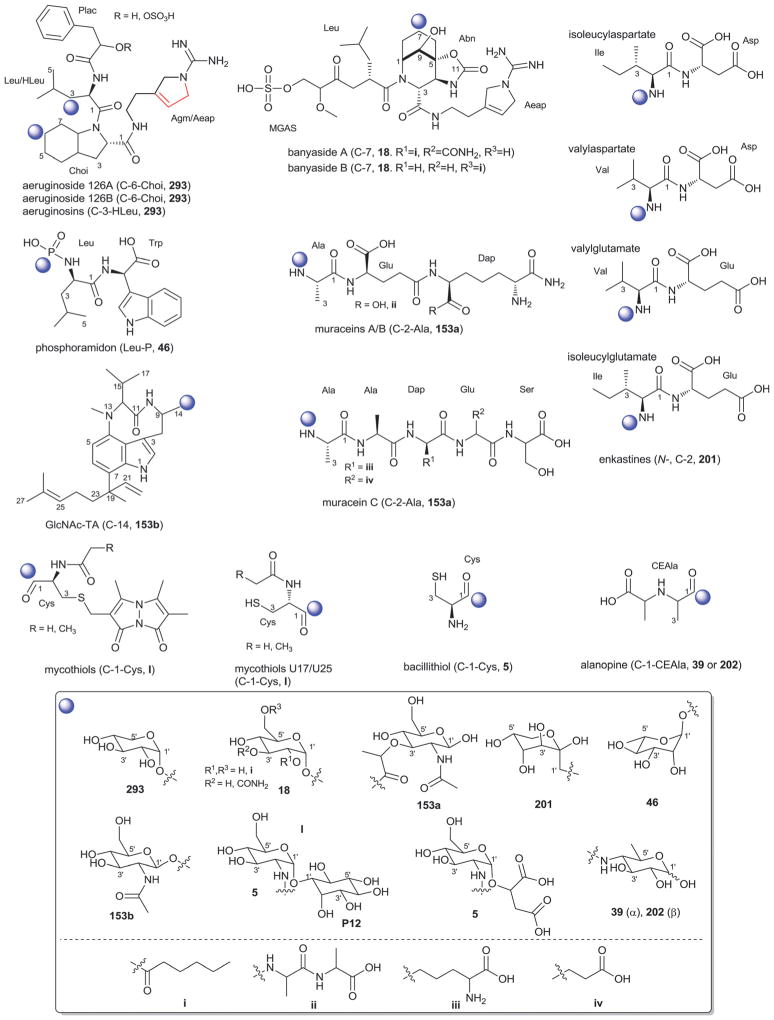
2. Aminoglycosides and related secondary metabolites
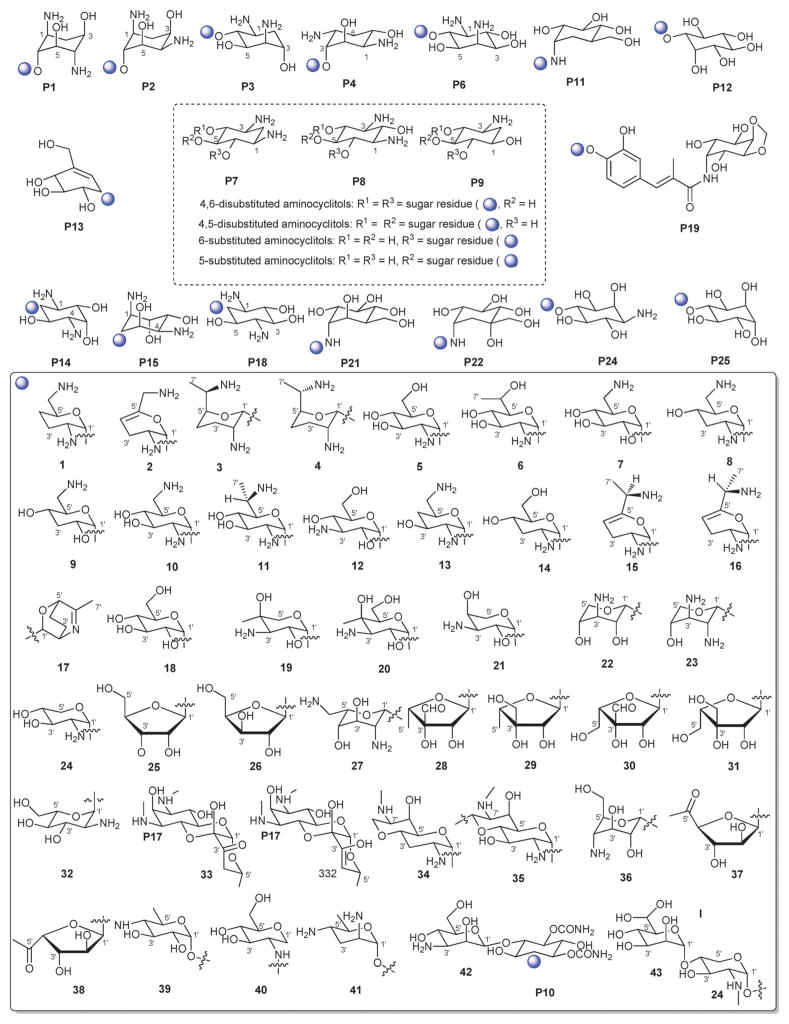
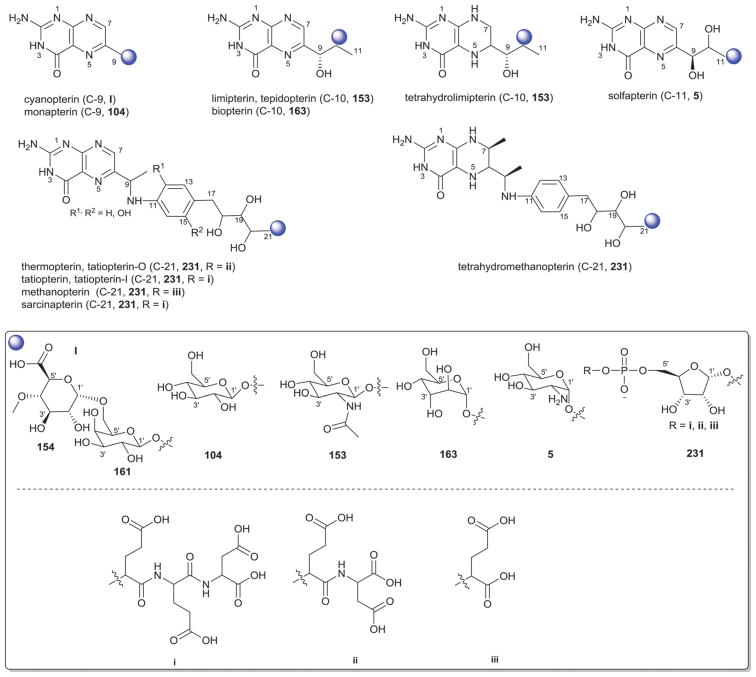
Aminoglycosides are structurally and functionally diverse and are noted for a range of biological activities including inhibition of bacterial protein synthesis (e.g., anti-infectives such as gentamicin, kanamycin, streptomycin), glucosidase inhibition (e.g., the antidiabetic acarbose), trehalase inhibition (e.g., the crop protectant validamycin) and inhibition of bacterial/eukaryotic protein synthesis (e.g. the cytotoxin pactamycin).19 The core sugar-derived carbocycle is the key structural signature of aminocylitols and this core is further diversified via variant functionalization (such as various degrees of deoxygenation and amination) where glycosylation at one or more positions is critical to the wide range of structural and functional diversity of this important natural product family.
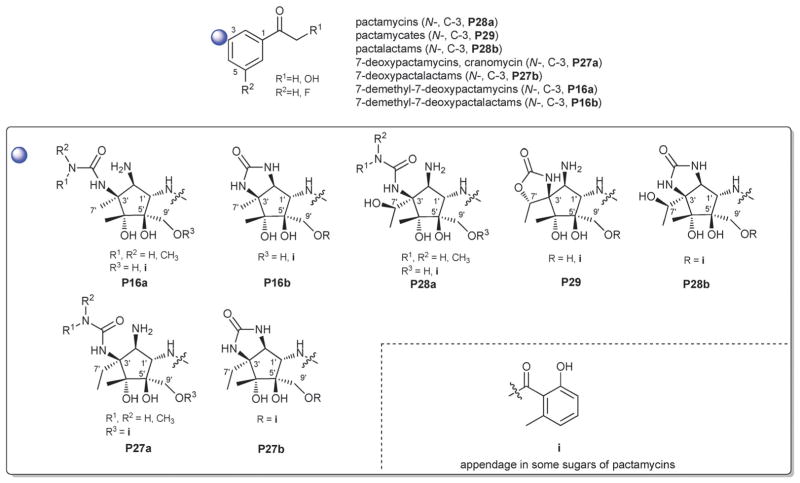
2.1. Pactamycins
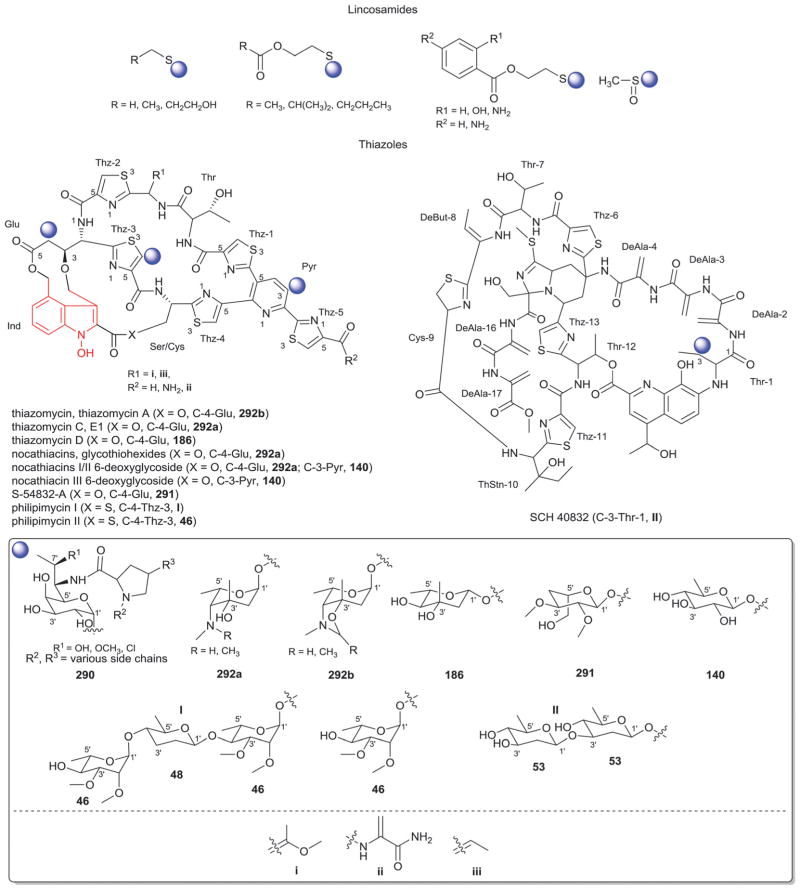
Members of this class include pactamycins, pactalactams, pactamycates, and cranomycin.19–21 They all contain pseudosugars and are characterized by the presence of an acetophenone unit which is N-linked at the C-3 position to an aminocyclopentinol pseudosugar. Twenty compounds have been found to be produced naturally, through feeding studies, or through mutasynthesis.22–27 Pseudosugar features common to all members include C-3′, C-4′ and C-5′-disubstitution (‘branching’) and conserved stereochemistry at all positions where pseudosugar divergence stems from variation of the C-2′ and C-3′-amine and/or C-9′-hydroxy substituents (Fig. 5). This family inhibits protein synthesis by binding the 30S subunit and displays broad activities including antibacterial, antiviral, antimalarial, and general cytotoxicity. This broad range of activities is due to the inhibition of protein synthesis in most organisms through binding with the 30S ribosomal subunit. The C-3′-1,3-dimethyl urea moiety of pactamycins at C-3′ (Fig. 5) has been noted to be important for activity while the C-9′ 6-methyl salicylic ester is considered dispensible.25 Variation of the pseudosugar C-3′-branching (P27a and P16a) favored antimalarial acitivity with lowered overall mammalian cell line cytotoxicity.26
Fig. 5.
Pactamycin aglycons and associated pseudosugars.
2.2. Other members
While a diverse array of carbocyles serves as the core aglycon of aminoglycosides, diamino carbocycles are among the most common. Metabolites that contain a 1,4-diamino carbocycle core include both the 2-deoxy variants (P1–P4, Fig. 6) such as that found within the istamycins and related metabolites sporaricins and sannamycins,28–31 as well as 2-hydroxy-containing cores found within metabolites like lysinomicin (P6)32 and the fortimicins (P6, P14, P15, and P18).33–36 The corresponding 2-deoxy-1,3-diamino carbocyclic aglycon 2-deoxystreptamine (2-DOS, P7) was found in majority of clinically used aminocyclitols including gentamicins,37 kanamycins,38 ribostamycin39 and tobramycins while the related 2-hydroxy aglycon streptamine (P8) serves as the core of several aminoglycosides including streptomycins40 and ashimycins.41 In other aminoglycosides, including gentamicins and sagamycins,42 a 1-deamino-1-hydroxy-2-DOS (P9) was reported as the aglycon. Aminoglycosides hygromycin A and epi-hygromycin contain the methylene-bridged aminocyclitol neo-inosamine (P19)43 while a handful of secondary metabolites including acarbose,44 amylostatins45 and validamycins46 utilize an unsaturated C7 cyclitol aglycon (P13). The D-myo-inositol (P12) and its aminated derivative P24 were recently discovered in mycothiol and minosaminomycin, respectively.47,48 The epimer of myo-inositol (P25) found as the aminocyclitol core of kasugamycin49 and carbamoylated inositol (P10) anchors boholmycin.50
Fig. 6.
Aminoglycoside pseudosugars and associated sugars.
While a diverse range of aglycon glycosylation patterns exist among aminoglycosides, glycosylation at C-4 and C-6 are among the most prevalent. Sugars employed the context of C-4 glycosylation of core aglycons include: the highly deoxygenated diaminosugar 1 and its C-5′ branched 3 and 4 (gentamicins)39,51 as well as the corresponding C-4′/C-5′-unsaturated variants 2 (sisomycin),52 15 and 16 (verdamicins);53 2′-amino-2′-deoxy-α-D-glucose (5), its C-5′-methyl branched analog 6 (gentamicins)37 or its 2′-deoxy 14 (nebramycin);54 6′-amino-6′-deoxy-α-D-glucose (7, gentamicins and combimicins)37,55 its corresponding 3′-deoxy 9 (combimicins)56 or its C-5′-methyl branched 11 (gentamicins);37 2′,6′-diamino-2′,6′-dideoxy-α-D-glucose 10 (combimicins)55 and its corresponding 3′-deoxy 8 (nebramine and tobramycin);57 3′-amino-3′-deoxy-α-D-glucose (12, nebramycins)54 and 2′,6′-diamino-4′-deoxy 13 (seldomycins).58 The novel 2′,4′-diamino-2′,3′,4′,6′-tetradeoxyhexose 41 comprised the C-4 attachment in minosaminomycin and C-1 in kasugamycin (Fig. 6). The bicyclic iminosugar 17 (gentamicins)37 stands out as particularly unique among those found within the C4 glycoside series. Also noteworthy are the C-4′ octadiose moieties (34 and 35) observed among saccharomycins and apramycin.59 The C-8′ of this unusual eight carbon carbohydrate was found to be further modified by 4′-amino-4′-deoxy-β-L-glucose (36) or α-D-glucose (18) in apramycin. C-4 glycosylation using select members of the sugars described above is also represented among other aminoglycosides including butirosins,60 xylostatins and ribostamycins.61
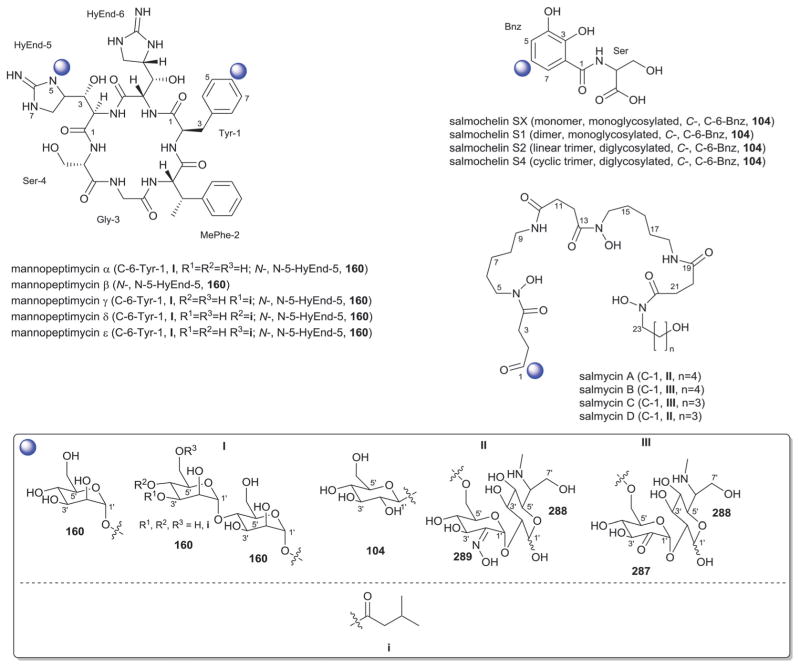
In addition to a few sugars highlighted in the previous paragraph found appended at C-6 of aminoglycoside core aglycons (2, 4, 8, 10, 12),54,62,63 additional sugar diversity has been found at C-6 including: α-D-glucose (18);64 3′-amino-3′-deoxy-α-D-xylose 21 (gentamicins and sisomicins)37,65 and its C-4′-methyl branched 19 (prevalent in a range of aminoglycosides);51,66,67 C-4′-methyl branched 3′-amino-3′-deoxy-α-D-galactose 20 (combimicins);56 3′-amino-3′-deoxy-β-L-arabinose (22, gentamicins and related compounds);37,65 2′-amino-2′-deoxy-α-D-xylose 24 and its corresponding 2′,3′-diamino-2′,3′-dideoxy analog 23 (seldomycins);58,68 as well as 3′-amino-3′-deoxy-β-D-mannose (42) and the diasaccharide I [comprised of the uniquely C-6′ oxidized mannose 43 and 2′-amino-2′-deoxy-α-D-xylose (24) in boholmycin].50 Other C-6′-appended sugars include L-streptose (28), 5′-hydroxy-L-streptose (30) and the corresponding reduced forms 29 and 31 have been found among streptomycins69–71 wherein further C-2′ glycosylation of the streptose moiety with 2′-amino-2′-deoxy-α-L-glucose (32) has been observed.
In addition to the previously noted 1 and 2, D-xylose (26, xylostatin and butirosin A)61 and D-ribose (25) are also found among the core C5 glycosides with the latter being most prevalent (as exemplified by butirosin B,60 lividomycins,72 paromomycins72 and ribostamycin61). In addition, glycosides of the C-3 position of the appended ribose with 5 (neomycin),73 27 (neomycin B,73 paromomycin and lividomycin A74) and 10 (neomycin C)75 have been characterized. A C-4′/C-5′-diether bridged glycosidic connection to 3′-keto-sugar (33) and its enol form (332) has been observed in the context of spectinomycin and spenolimycin, respectively.
Additional glycosides that fall outside the scope of those described in the preceding paragraphs include the aromatic hexofuranose glycosides (37 and 38) of hygromycins,43 various N-glycosides (4′,6′-dideoxy-α-D-glucose 39 in acarbose44 and the unique 1′,2′-dideoxy-2′-aminohexose 40 in salbostatin76). It is important to note that minor modifications of functional groups such as N- and O-methylation, acetylation, carbamoylation, and formylation of sugar monomers, not specificied herein, are also prevalent.
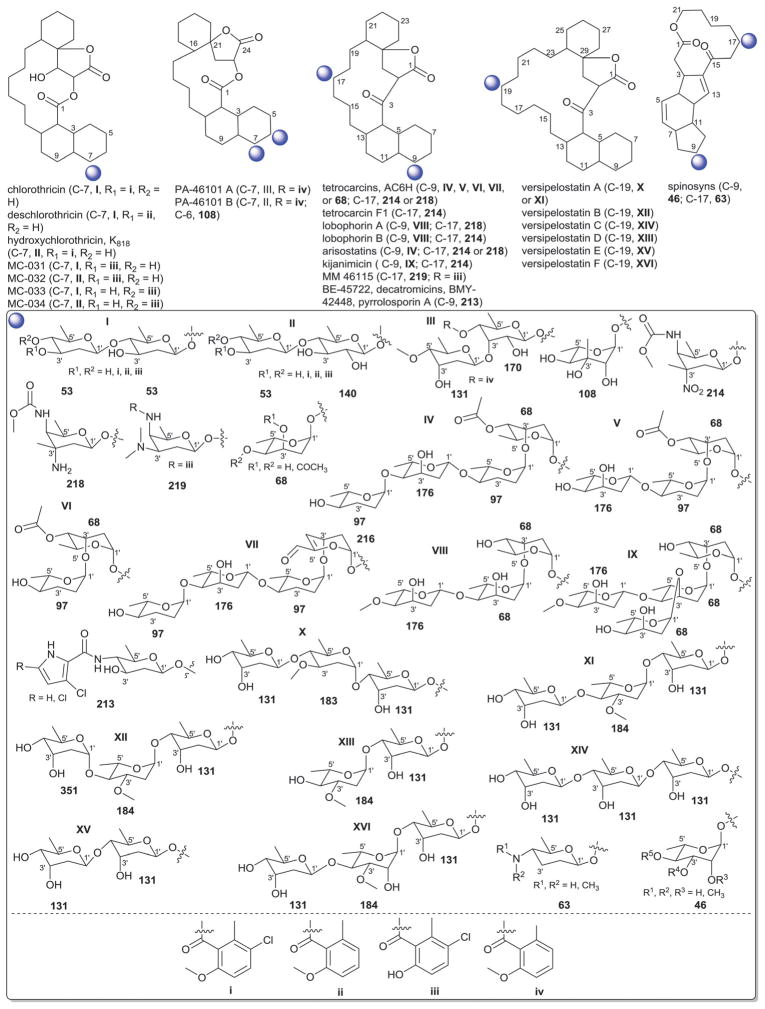
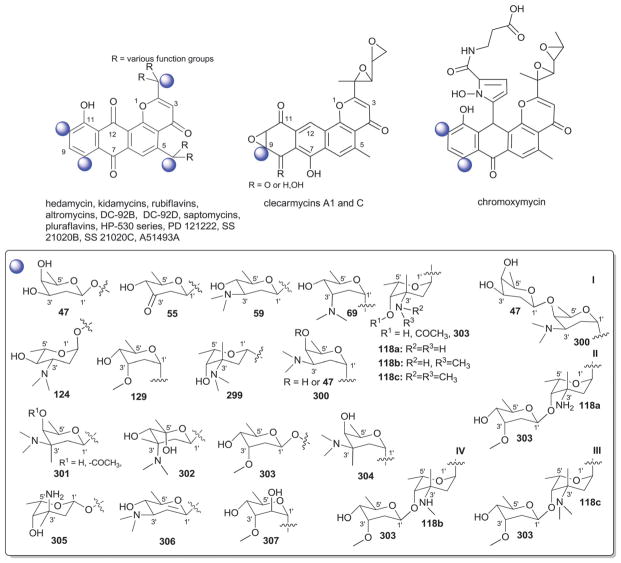
3. Angucyclines
With more than 200 known bacterial-derived members,18 angucyclines comprise one of the largest groups of polycyclic aromatic polyketides. Angucyclines are classified based upon their signature tetracyclic benz[a]anthracene system into five major classes: tetrangomycin-type (e.g., landomycins); aquayamycin-type (e.g., saquayamycins and urdamycins); benzanthrin-type (e.g., benzanthrins); gilvocarcin-type and jadomycin-type (Fig. 7).77–79 Despite their remarkable chemical diversity and divergent biological activties, angucyclines have failed to advance in clinical development due to off-target toxicities and/or poor drug-like properties.79
Fig. 7.
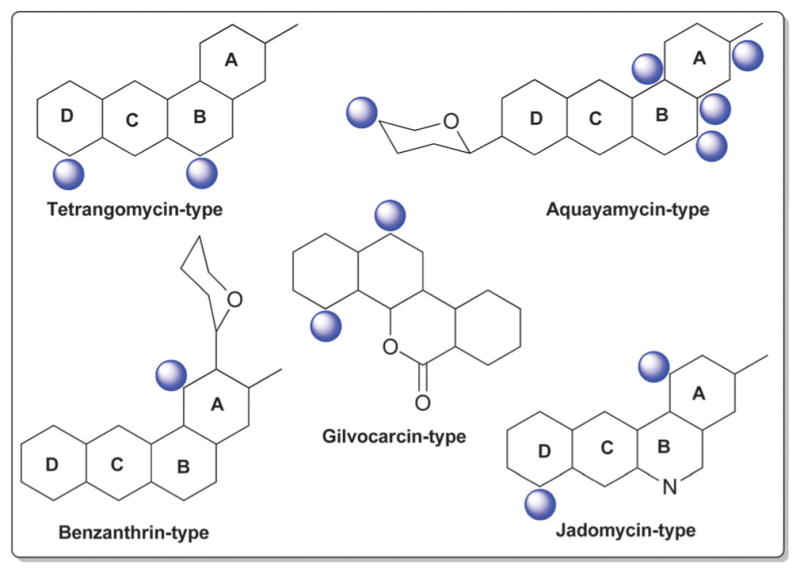
The five major types in angucyclines and glycosylation positions.
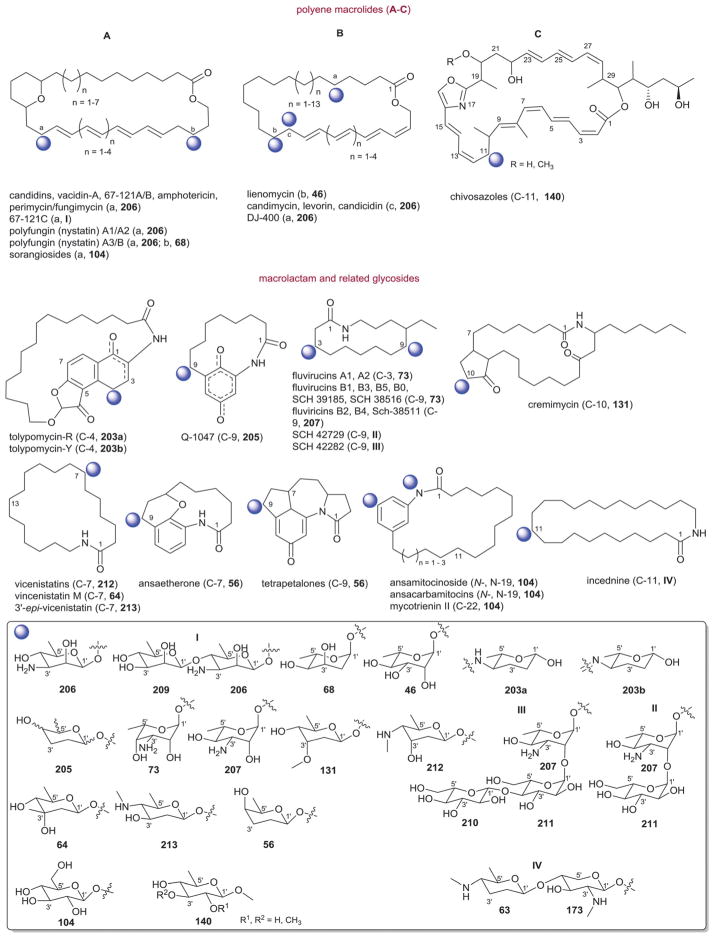
3.1. Tetrangomycin-type (landomycins)
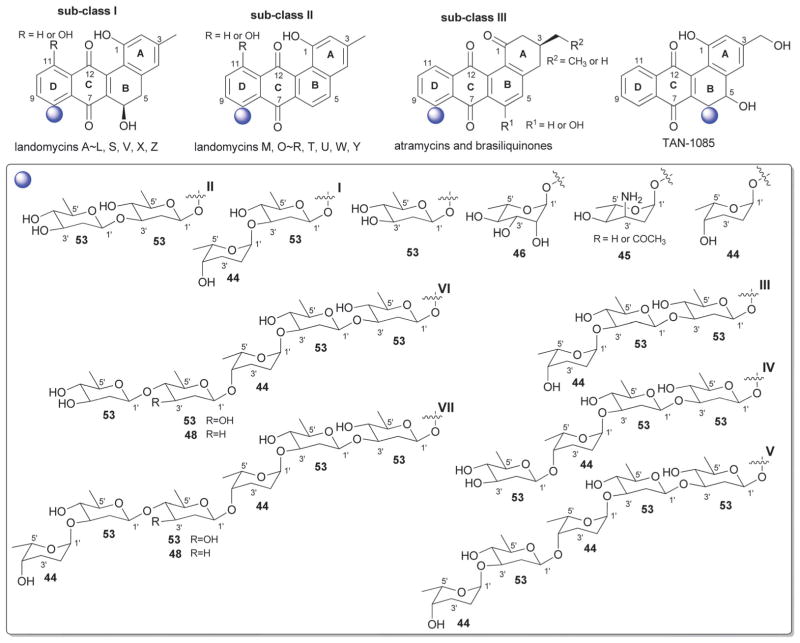
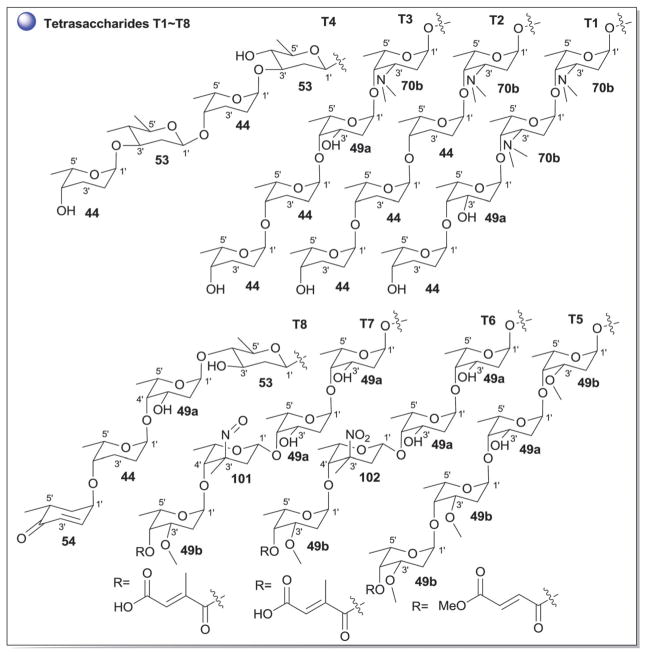
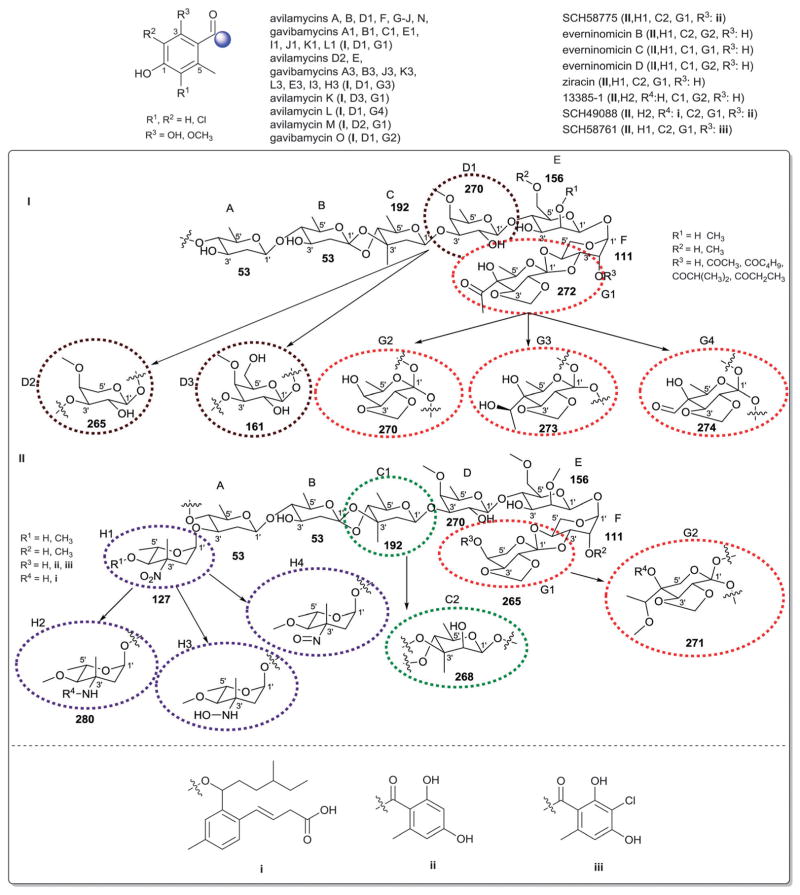
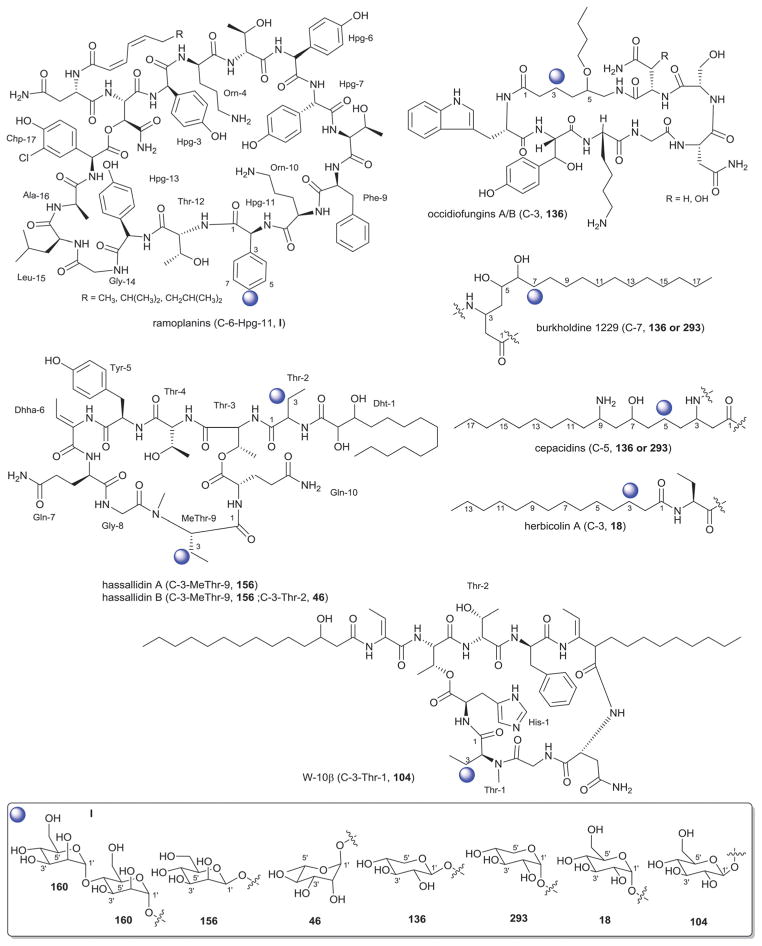
The landomycins (Fig. 8) are one of the largest angucycline subclasses and all members contain a C-8 oligosaccharide of varying lengths.77,79 Landomycins have been further subdivided based upon the polycyclic aromatic aglycon into three sub-types: landomycinone-based (sub-class I; landomycins A–L, S, V, X and Z); tetrangulol-based (sub-class II; landomycins M, O–R, T, U, W, and Y) and tetrangomycin-based (sub-class III; atramycins, brasiliquinones and TAN-1085). Despite their notable diversity, only three core sugars (β-D-olivose, 53; β-D-amicetose, 48 and α-L-rhodinose, 44) comprise the glycosyl components of landomycins.
Fig. 8.
Landomycin aglycons and associated sugars.
Natural variants within sub-class I contain disaccharide to hexasaccharide glycosyl substitutions and include landomycins A–E, K, L and S–U and members of this group display potent in vitro cytotoxicity against cancer cell lines.80–84 A common theme is the prevalence of the α-L-rhodinose-(1→3)-β-D-olivose and β-D-olivose-(1→4)-β-D-olivose repeat units within the oligosaccharide chains followed by chain termination with β-D-olivose (53). A notable distinction of landomycins K and L is the alternative termination of the saccharide chain with α-L-rhodinose (44). Additional members of sub-class I (landomycins F–J) have been generated via strain engineering where H and J stand out as among the only monosaccharide and trisaccharide-substituted members, respectively.85–87
For subclass II, landomycins M–Z have been isolated from a combination of native and engineered bacterial hosts.88–90 Similar to sub-class I, a common glycosyl theme is the prevalence of β-D-olivose-3-1-α-L-rhodinose and β-D-olivose-4-1-β-D-olivose repeat units followed by chain termination with β-D-olivose (53). In contrast, in certain members of sub-class II (such as landomycins C, X–Z), β-D-olivose has been replaced with its corresponding 3-deoxy analog β-D-amicetose (48). A comparison of the in vitro cancer cell line cytotoxicities displayed by members within sub-class I and II revealed the aglycon C6 and C11 hydroxyls as important for activity and the saccharide length to also modulate potency. Specifically for the latter, the most active landomycins were those lacking sugars (e.g., landomycinone, tetrangomycin, or tetrangulol) and those appended by penta- or hexasaccharide chains (e.g., landomycins A and B). Based upon this comparison, it was proposed that the oligosaccharide-substituted landomycins and their sugar-free congeners may function via distinct mechanisms.89,90
Members of sub-class III also display notable in vitro cytotoxitity against representative cancer cell lines and include the atramycins,91 brasiliquinones92–94 and TAN-1085.95–97 While the regiospecificity of glycosylation (aglycon C-8-glycosylation) of atramycins and brasiliquinones is reminiscent of that described for sub-classes I and II, the C-6 glycosylation of TAN-1085 stands out as an exception among tetrangomycin-type angucyclines. The brasiliquinones are unique among tetrangomycin-type angucyclines as they are the only members isolated from a non-Streptomyces strain (Nocardia brasiliensis) and the only members with glycosides reported to contain α-L-ristosamine-based sugars (45). These latter metabolites were active against Gram-positive bacteria (including Mycobacterium) and multiple drug-resistant P388/ADR tumor cell lines.
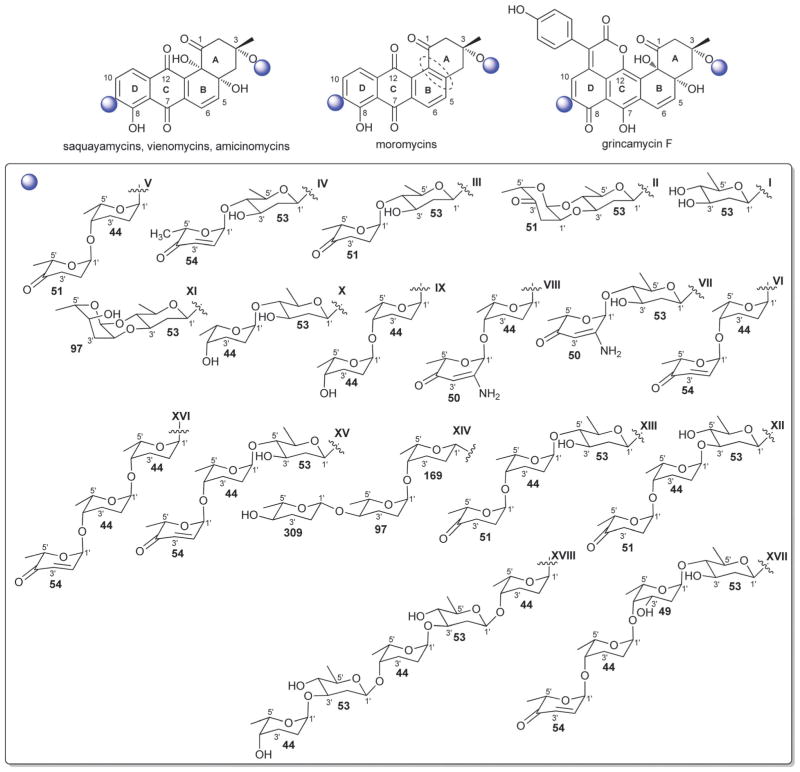
3.2. Aquayamycin-type (saquayamycin and urdamycin analogs)
C-9 C-glycosylation is a common signature among both the saquayamycin-type and urdamycin-type angucyclines and the main structural divergence stems from C-3 (saquayamycins) or C-12b (urdamycins) O-glycosylation. In addition to saquayamycins,98–104 other structurally related saquayamycin-type bacterial metabolites include moromycins,105 vieneomycins,104,106,107 PI-080/083/085/087,108,109 grincamycins,110,111 Sch 47554/47555,112,113 and amicenomycins (Fig. 9).114 In addition to potent antibacterial activity and in vitro cytotoxicity against cancer cell lines, other activities noted among this broad group of metabolites include antifungal activity, and inhibition of platelet aggregation, UDP-GlcNAc enolpyruvyl transferase (EPTase) and inducible nitric oxide synthase (iNOS). Many saquayamycin-type members have also been demonstrated to be efficacious in standard murine xenografts and, at least in some cases, to be reasonably well-tolerated (e.g., the acute IP LD50 of the potent antibacterial amicenomycin A = 100.0 mg kg−1).114
Fig. 9.
Saquayamycin aglycons and associated sugars.
Only ten distinct monosaccharides (β-D-olivose, 53; α-L-rhodinose, 44; β-L-rhodinose, 169; α-L-aculose, 54; α-L-cinerulose, 51; α-L-rednose, 50; α-L-oliose, 49; α-L-amicetose, 97; β-D-amicetose, 48; β-D-oliose, 47; β-L-amicetose, 309), have been observed among the saccharide constituents of saquayamycin-type bacterial natural products. Of these, only four (β-D-olivose, 53; β-D-amicetose, 48; α-L-rhodinose, 44 and β-L-rhodinose, 169) have been found as C-glycosides where β-D-olivose (53), α-L-rhodinose, (44) and the β-D-amicetose (48) enantiomer α-L-amicetose (97) were also noted among O-glycosides. With respect to general trends, β-D-olivose (53), α-L-rhodinose, (44) and, to a lesser extent, α-L-amicetose (97) and α-L-oliose (49), predominate as the ‘internal’ sugars of appended oligosaccharides within this series wherein uniquely oxidized sugars (α-L-aculose, 54; α-L-cinerulose, 51; α-L-rednose, 50) predominate among the terminal ‘capping’ sugars. Glycosidic bonds within the corresponding oligosaccharides extend from either C-3 or C-4 in β-D-olivose (53), C-4 of β-D-oliose (49), and the single C-4 hydroxyl group within rhodinose, (44/169) and amicetose (48/97/309). Among the ‘capping’ sugars, the terminal α-L-aculoside (54) was reported to convert to the corresponding α-L-cineruloside (51) during silica gel chromatography raising the question of whether this latter sugar is artifactual rather than a biosynthetic product. The aminated α-β-unsaturated ketosugar α-L-rednose (50) in saquayamycins H and I stands out as particularly unique and this sugar was found to contribute to enhanced in vitro cytotoxicity against certain cancer cell lines.103
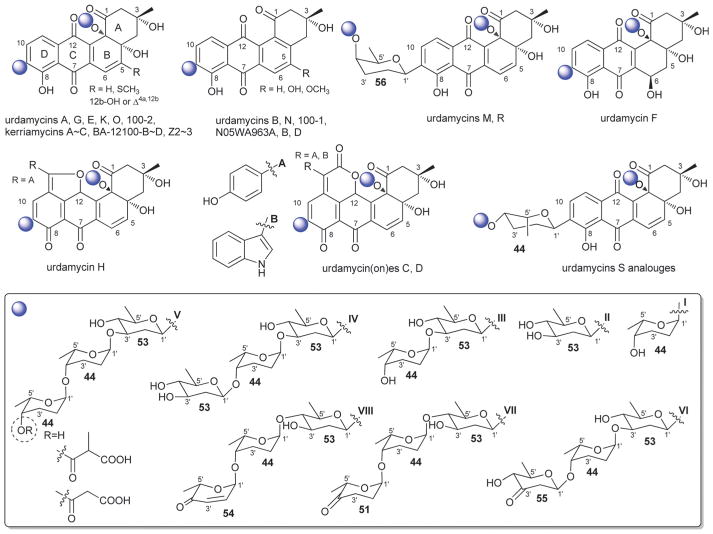
Urdamycins differ from saquayamycins based upon their distinct regiospecificity of glycosylation (C-9 and C-12b of the angucycline core; Fig. 10). In addition to the numerous natural115–119 and engineered urdamycins,120–125 members of this subclass include the kerriamycins,126 the N05WA963 series127 and the urdamycin BA-12100 set of metabolites,77,128 all of which derive from Streptomyces. Members of this sub-type are best known for their anti-proliferative properties against various cancers where C-9 C-trisaccharyl-substituted analogs are generally more potent. Six distinct monosaccharides (β-D-olivose, 53; α-L-rhodinose, 44; α-L-aculose, 54; α-L-cinerulose, 51; β-D-rhodinose, 56; and β-D-kerriose, 55) have been observed among the saccharides of urdamycin-type bacterial natural products with latter (β-D-kerriose, 55) unique to this sub-type of aquayamycin-type angucyclines. Of these, only three (β-D-olivose, 53; α-L-rhodinose, 44; and β-D-rhodinose, 56) have been found as C-9 C-glycosides where the predominant substitution at C-9 is comprised of α-L-rhodinosyl-(1→3)-β-D-olivose disaccharide (III, Fig. 10). In most members, this disaccharide is further capped by one of four monosaccharides (β-D-olivose, 53; α-L-aculose, 54; α-L-cinerulose, 51; or β-D-kerriose, 55). Urdamycins M, R, S and 12b,4′-di-urdaderhodinosyl-urdamycin S serve as the exception in this regard wherein each bear distinct C-9 C-rhodinosyl-based disaccharides.
Fig. 10.
Urdamycin aglycons and associated sugars.
3.3. Other related aquayamycin-type angucyclines
In addition to the above described angucyclines, a number of additional miscellaneous angucyclines have been reported and are organized herein based upon the regiospecificity of glycosylation. Angucyline C-1 or C-2 glycosylation is relatively rare with the benzanthrins from Nocardia,129,130 pseudonocardones from Pseudonocardia (associated with the fungus-growing ant Apterostigma dentigerum)131 and P371A1/A2 from Streptomyces132,133 as the only representative bacterial metabolites within this subset. Members display variant activities including antibacterial, antiplasmodial, and/or cancer cell line cytotoxicity as well as inhibition of gastric acid secretion. Sugars employed in C-1-O-glycosides within this series include α-D-boivinose (57), β-L-rhodosamine (67), β-L-angolosamine (66), and a β-hexuronic acid moiety (72, absolute stereochemistry not determined). In contrast, glycosylation at C-2 is in the form of C-glycosylation with β-D-angolosamine (59b) as the sole sugar represented. P371A1/A2 also contain a C-9 C-trisaccharyl moiety (V, Fig. 11) initiating with the typical angucycline C-9 C-β-D-olivose (53) modified at C-3′ with a dideoxy branched β-D-mycarose (64) which is capped at C-4′ with a notably unique 4′-ureido tetradeoxysugar 4′-amino-4′-deoxy-β-D-amicetose (63).
Fig. 11.
Other related angucycline aglycons and associated sugars.
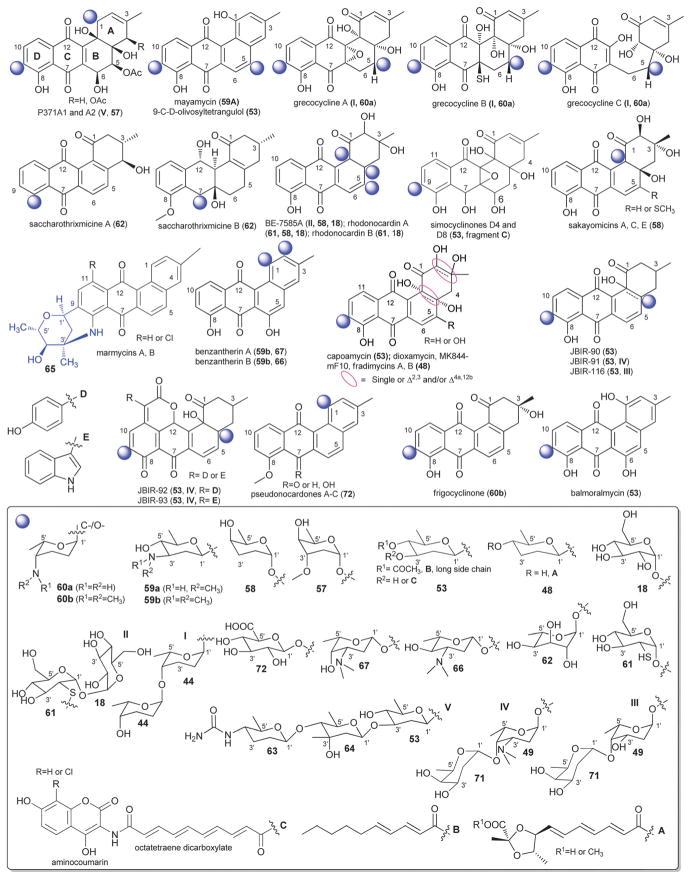
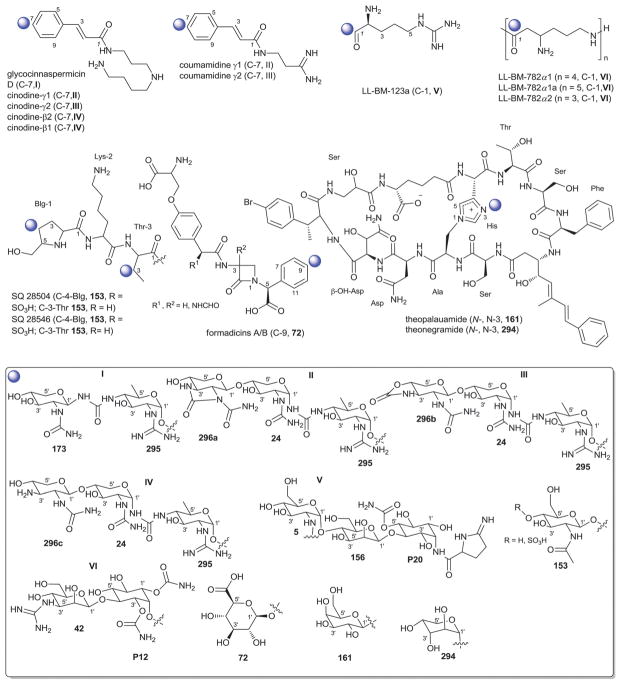
Angucycline glycosylation at C-4a, C-5 or C-12b is also relatively rare. Bacterial metabolites representing C-4a-O-glycosides include the JBIR series of metabolites from Streptomyces,134 and the rhodonocardins from Nocardia135 where the sugars employed include α-D-glucose (18), α-L-rhodosamine (70) and α-L-oliose (49) and, in the case of the JBIR series, the C-4a-appended sugar is further substituted at C-4′ by α-D-oliose (71). This latter set of metabolites also contains the typical angucycline C-9 C-glycosidic β-D-olivose (53). The rhodonocardins, and the structurally-related BE-7585A from Amycolatopsis,136 also carry the rare sugar 2′-thio-α-D-glucose (61) at C-5, connected as a typical O-glycoside in rhodonocardins or via an unsual thioether bond at C-2′ of the thiosugar (which is part of a ‘head-to-head’ α-D-glucose-containing disaccharide, 18) in BE-7585A. In addition, rhodonocardin A and BE-7585A contain the C-12b-O-glycosidic α-D-rhodinose (58). Mayamycin from a marine Streptomyces137 (unique among angucyclines as the only C-5 C-glycosyl member) contains N-desmethyl-β-D-angolosamine (59a) as the C-5 sugar. Grecocyclines from Streptomyces,138 the other C-5-O-glycoside member within this subset, contains a C-5-appended O-4-epi-α-L-tolyposamine (60a) and a C-9 C-disaccharyl α-L-rhodinosyl-(1→4)-α-L-rhodinose moiety (disaccharide I, Fig. 11). The remaining C-12b-O-glycosides within this subset, sakyomicins from Nocardia,139 contain α-D-rhodinose (58) at the C-12b-position reminiscent of rhodonocardin A and BE-7585A. Members within this cumulative subset have been reported as cytotoxic against representative cancer cell lines, to display antibacterial activity (including for antibiotic resistant strains) and as protein tyrosine phosphatase 1B and thymidylate synthase inhibitors.
Saccharothrixmicines A and B from a marine Saccharothrix,140 represent the only angucycline C-8 and C-7 O-glycosides, respectively. Both metabolites contain the same sugar, α-L-6-deoxyaltrose (62), and were reported to display antifungal activities in vitro.
As with angucyclines discussed within preceding sections, C-9 C-glycosylation is a predominate modification among members described herein and include naturally-occurring simocyclinones,141–144 capoamycin,145 dioxamycin,146 MK844-mF10,147 fradimycins,148 frigocyclinone,149 balmoralmycin,150 and marmycins151 as well as the engineered landomycin–urdamycin hybrid metabolite 9-C-D-olivosyl-tetrangulol.152 With the exception of marmycin (produced by a marine actinomycete), all other metabolites within this grouping are Streptomyces metabolites. In addition to the the typical β-D-olivose (53), three additional sugars are represented as C-9-C-glycosides among members within this subgroup including β-D-amicetose (48), 3′/4′-epi-α-L-vancosamine (65) and the rare aminodeoxysugar α-L-ossamine (60b). In contrast to many previously discussed C-9-modified angucyclines (which contain C-9 C-oligosaccharyl modifications), the C-9 C-glycosylation patterns within this subgroup are limited to monosaccharides. Typically within this subgroup, β-D-olivose (53) and β-D-amicetose (48) is further acylated at C-3′/4′ with, in many cases, uniquely functionalized lipids which, in some cases (e.g., simocyclinones), is further conjugated to an aminocoumarin. A notable standout within this series is the unique C-8/C-9 ring fusion that occurs from an additional bond between the angucycline C-8 and C-3′-amine 3′/4′-epi-α-L-vancosamine (65) within the marmycins. In addition to reported cancer cell line cytotoxicity and antibacterial activities, other reported activities of members of this subgroup include differentiation-inducing activity on myeloid leukemia cells (M1) and inhibition of protein kinase C-α (PKC-α).
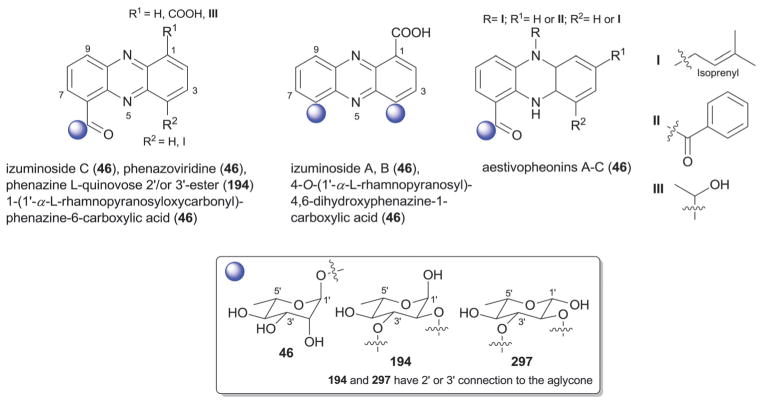
3.4. Jadomycin- and gilvocarcin-types (jadomycins, ravidomycins, chrysomycins and gilvocarcins)
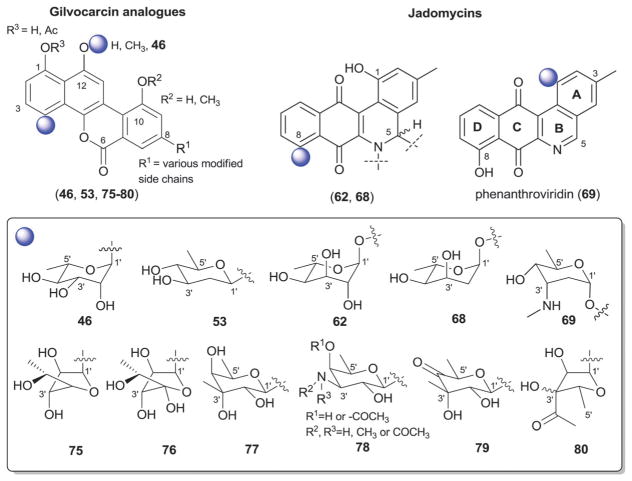
The gilvocarcin-type aryl-C-glycosides79 contain a number of naturally-occurring (gilvocarcins,153–162 polycarcins, BE-12406A/B,163–165 ravidomycins166–171 chrysomycins172 and virenomycins173,174) and engineered (gilvocarcins and polycarcins)172,175–177 Streptomyces metabolites wherein the signature C-glycosylation occurs at C-4 (Fig. 12). The monosaccharides found among naturally-occurring C-4-C-glycosides of this class include β-D-fucofuranose (75), α-L-rhamnose (46), β-D-ravidosamine (78), β-D-virenose (77), the 4′-keto analog of β-D-virenose (79) and a C-3′-branched analog of β-D-fucofuranose (80), wherein the presence of β-D-fucofuranose stands out as relatively rare among bacterial secondary metabolites. This set has been expanded to also include 4′-hydroxy-β-D-fucofuranose (76) and β-D-olivose (53) via metabolic engineering.172,175,176 Some members of this subclass also contain a C-12-O-α-L-rhamnose (46) substituent. These metabolites are generally known as anticancer antibiotics where subtle differences within the appended sugar were found to improve the perceived therapeutic index (potency versus general toxicity).
Fig. 12.
Gilvocarcin and jadomycin aglycons and associated sugars.
The jadomycins from Streptomyces are angucyclines that contain a nitrogen at position 6 within ring B (Fig. 12).79,178–181 Naturally-occurring jadomycins are C-8-O-glycosides of α-L-digitoxose (68) and an engineered variant that led to a replacement of this sugar with 6′-deoxy-α-L-altrose (62) has been reported.180 The naturally-occurring B ring Streptomyces metabolite phenanthroviridin and is alternatively conjugated at C-1 to α-D-ristosamine (69).182,183 Jadomycins display a range of bioactivities, including anticancer cytotoxicity, antimicrobial, anti-viral, and inhibition aurora-B kinase.
4. Anthracyclines, tetracyclines, quinones and tricyclines
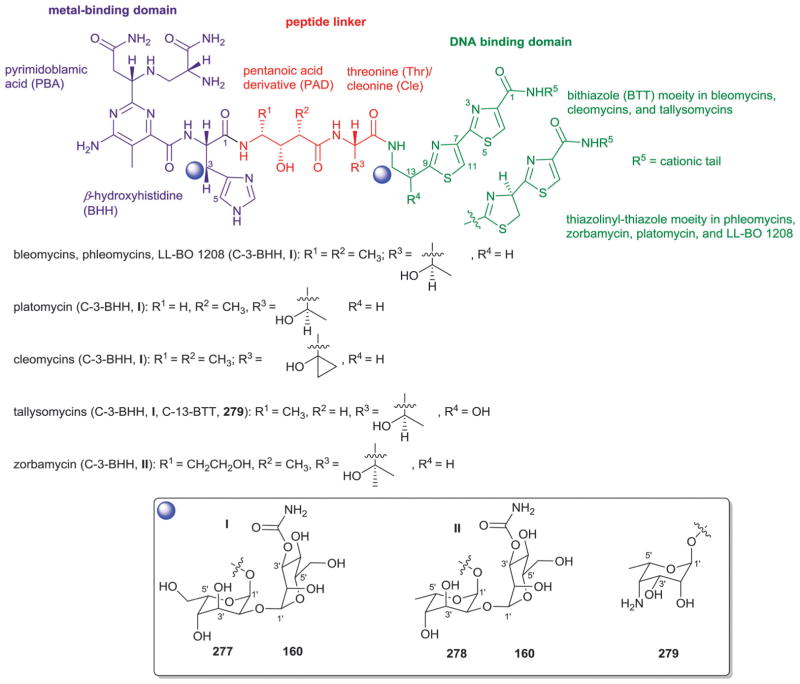
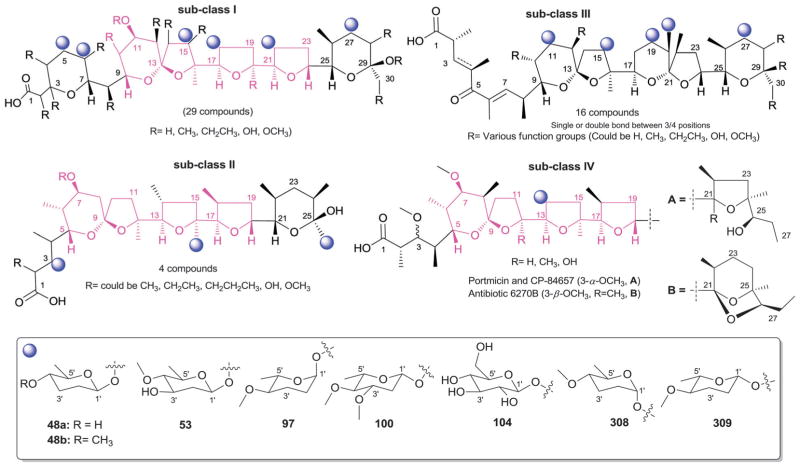
4.1. Anthracyclines
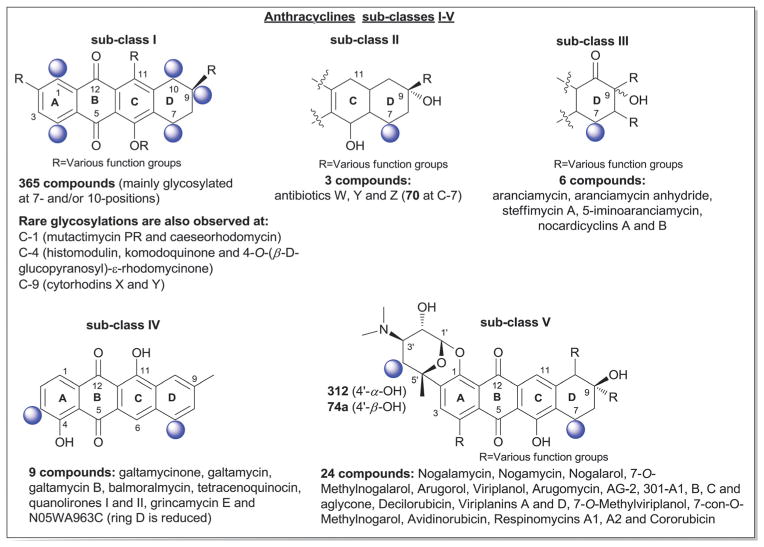
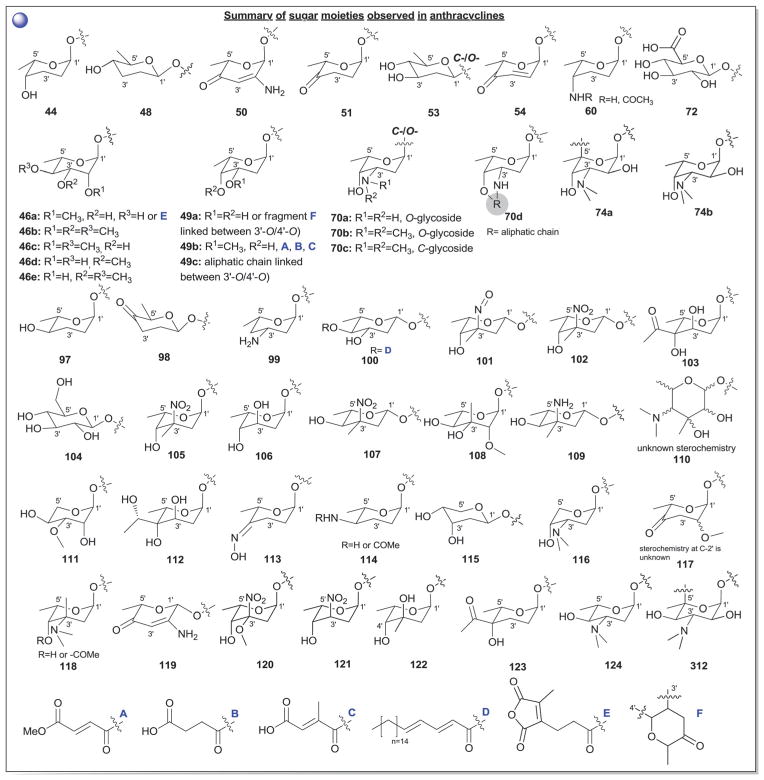
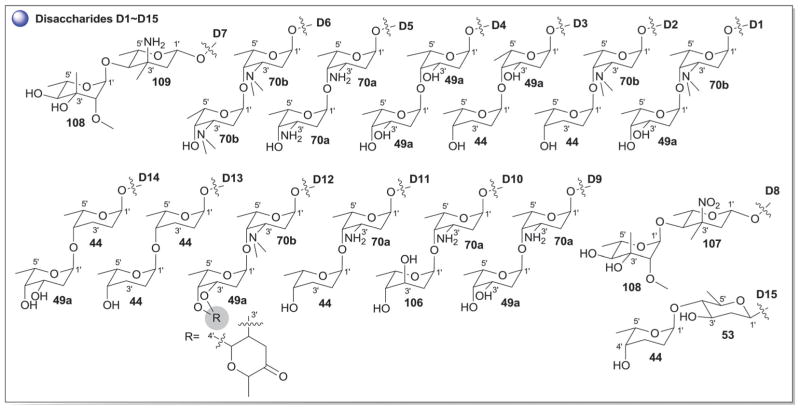
According to 1963 Hans Brockman definition, anthracyclines are defined as red to orange natural/synthetic dyes with a skeleton of 7,8,9,10-tetrahydro-tetracene-5,12-quinone decorated with mono-, di-, tri-, tetra- and/or pentasaccharide side chains (Fig. 13, subclass I).184 As exemplified by the early protypical members daunomycin (discovered in 1964)185 and doxorubicin (adriamycin, discovered in 1969)186,187 used to successfully treat cancer for nearly four decades, anthracyclines display a range of bioactivities of pharmaceutical utility.188 To date,18 407 bacterial-derived anthracycline glycosides have been reported from a range of bacteria including, but not limited to Streptomyces, Micromonospora, Actinomadura, Nomadura Actinosporangium, Chaetomium, Actinoplanes, Ampullariella and Nocardia. For the scope of this discussion, members are classified into five sub-classes I (365 compounds), II (3 compounds), III (6 compounds), IV (9 compounds), and V (24 compounds), based upon aglycon structural divergence (Fig. 13). Cumulatively, anthracycline glycosides integrate 51 structurally distinct monosaccharide units (Fig. 14) incorporated within the anthracycline-appended mono-, di-(15 variations, D1–D15; Fig. 15), tri- (35 variations, trisccharides I–XXXV; Fig. 16), tetra- (eight variations, T1–T8; Fig. 17), and pentasaccharides (two variations, P1 and P2; Fig. 18). While the predominant regiospecificity of anthracycline glycosylation occurs at C-7 and/or C-10, C-1-, C-3-, C-4-, and/or C-9-glycosylation (as well as C-4′-glycosylation in subclass V) has been observed (Fig. 13). Also, both O- and C-glycosides have been reported, the latter of which is less prevalent and mainly occurs at C-3 in sub-class IV (reminiscent of angucyclines and/or pluramycins).
Fig. 13.
The five anthracycline sub-classes I–V and glycosylation positions.
Fig. 14.
Sugars associated with anthracyclines.
Fig. 15.
Disaccharide variations D1–D15 observed in anthracyclines.
Fig. 16.
Trisaccharide variations (I–XXXV) observed in anthracyclines.
Fig. 17.
Tetrasaccharide variations (T1–T8) observed in anthracyclines.
Fig. 18.
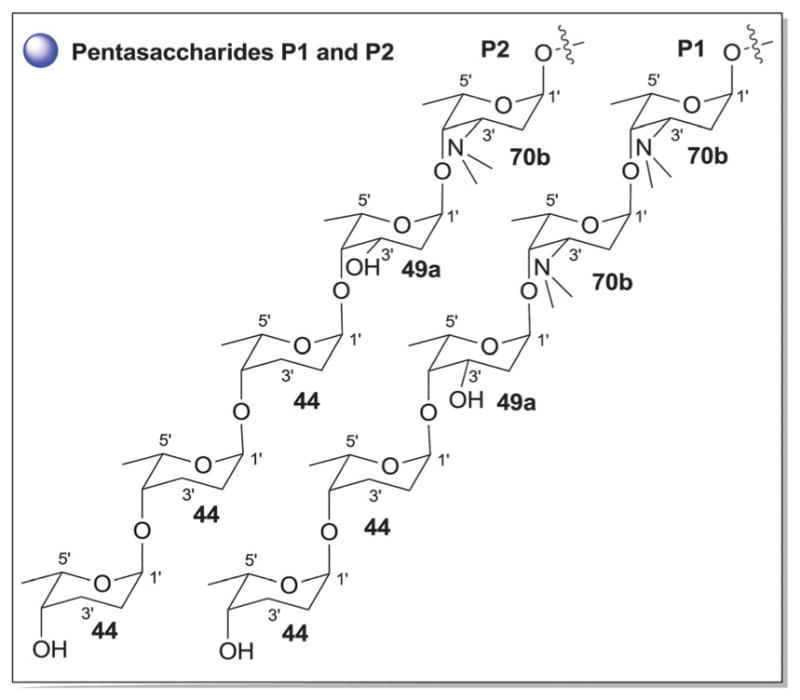
Pentasaccharide variations P1–P2 observed in anthracyclines.
Sub-class I contains 365 anthracycline O-glycosides (and a few C-glycosides) bearing variant carbohydrate substitution ranging from mono- to pentasaccharides. C7- and/or C10-O-glycosylation predominates (358 compounds) within sub-class I anthracyclines including D-788,189–193 oxaunomycins,193 doxorubicins,194,195 spartanamicins,196 daunorubicins,194,195 daunomycins,185,197,198 aclacinomycins (aklavins),199–205 rhodomycins,184,206–210 betaclamycins (and CG-21-C, CG1-C, CG21-B analogs),211–213 auramycins,214–219 MA-144-U and G analogs,220–222 steffimycins,223–226 baumycins,227,228 pyrrocyclines,222 rubeomycins,229,230 pyrromycins,231 oblemycins (cytorhodins),232,233 sulfurmycins,214–219 cinerubins,188,234–236 mutactimycins,237–239 alldimycins,233,240 nocardicyclins,241 violamycins,242 ditrisarubicins,243 aranciamycins,244–247 A447A~D1,248 ciclamycins,249 isoquinocycline A,188,250 kosinostatin (quinocycline B),251,252 isoquinocycline B,252,253 micromonomycin254 and other representative anthracyclines.100,188,201,202,211,229,255–263 Less common sub-class I O-glycoside regioselectivity was observed at C-1 (mutactimycin PR237,238), C-4 [histomodulin,264 komodoquinone A265,266 and 4-O-(β-D-glucopyranosyl)-ε-rhodomycinone267], and C-9 (cytorhodins X and Y).188,268 In addition to the mono- to tetrasccharides observed, two pentasaccharide-containing members were reported [roseorubicin A (pentasaccharide P1 at C-10; Fig. 18)229 and β-rhodomycin-Roa2.deofuc.rod3 (pentasaccharide P2 at C-7, and 70b at C-10; Fig. 14 and 18)269] where roseorubicin SAR revealed improved potency to correlate with increase oligosaccharide length.229 Caeseorhodomycin256 is distinguished among this series as a C-1-C-glycoside (70c; α-L-rhodosamine).
A reduced ring-C is the distinguishing feature of sub-class II. Only 3 metabolites (antibiotics W, Y and Z) fall within this sub-class, all of which are C-7-O-glycosides bearing α-L-daunosamine (70a; Fig. 14).270 Sub-class III is unique by virtue of the C-10-keto ring D and contains only 6 metabolites, all of which are C-7-O-glycosides bearing α-L-sugars [2′-OMe-α-L-rhamnose (46a) or α-L-vancosamine (118)]. Sub-class III members include steffimycin A (46a at C-7),223 5-iminoaranciamycin (46a at C-7),247 aranciamycin (46a at C-7),244 aranciamycin anhydride (46a, R3 = fragment E; at C-7),245 and nocardicyclins A (118 at C-7) and B (118 at C-7).241 The 7 glycosidic sub-class IV members contain a fully aromatized tetracycline backbone with, typically, C-3-C-glycosylation, and include galtamycin(one)s,100,271,272 balmoralmycin (β-L-olivose, 100; at C-3),272 tetracenoquinocin (46a at C-7),247 quanolirones I (trisaccharide XXXII, at C-3; Fig. 16) and II (disaccharide D15, at C-3; Fig. 15),273 and the two D-ring-reduced metabolites N05WA963C127 and grincamycin E111 (both bearing the C-3-C-trisaccharide XXXIII). Finally, sub-class V members are differentiated by the presence of a fused ring A-sugar (connected 1-/1′-O- and 2/5′-C-) where the carbohydrate unit is comprised of α-L-mycaminose (74a) or 4′-epi-α-L-mycaminose (312). The 24 metabolites that fall within this sub-class include nogalamycin,274–276 nogamycin,229 nogalarol,276,277 7-O-methylnogalarol,276 7-con-O-methylnogarol,229 arugorol,277,278 arugomycin,277–279 decilorubi-cin,280–283 avidinorubicin,284 respinomycins,285–287 cororubicin,288 and viriplanin and viriplanol analogs.289,290 Additional C-4′ and/or C-7-O-glycosylation is observed with sugars 74a and 312 and/or various lengths of mono- to tetra-saccharide side chains.
From a global perspective (Fig. 14), anthracyclines present several unique carbohydrates. For example, C-3′- and C-4′-branching is prevalent as exemplified by 101–103, 105, 107–110, 112, 118, 120, 122, and 123. Additionally, nitroso (101), hydroxylimino (113) and nitro (102, 105, 107, 120, 121) sugars are quite common. Unsaturated (50, 54, 119) and ketosugars (50, 51, 54, 98, 117, 119) are also observed and the C-3′-N/C-4′-O-macrocylic substitution of 70d is uncommon. The C-3′-disubstitution pattern of 120 3′-methoxy-3′-nitro-2′,6′-dideoxy-α-L-talose in trisaccharide XXXV of 301-C291 is of particular note as unprecedented carbohydrate functionality. In general, glycosylation is considered to be important to bioactivity in most cases studied. For example, Matsuzawa and Oki reported SAR on ~100 anthracyclines where they found the presence of an aminosugar (or amine-substituted aglycon) to be important and increased potency to also correlate with increased saccharide chain length.229
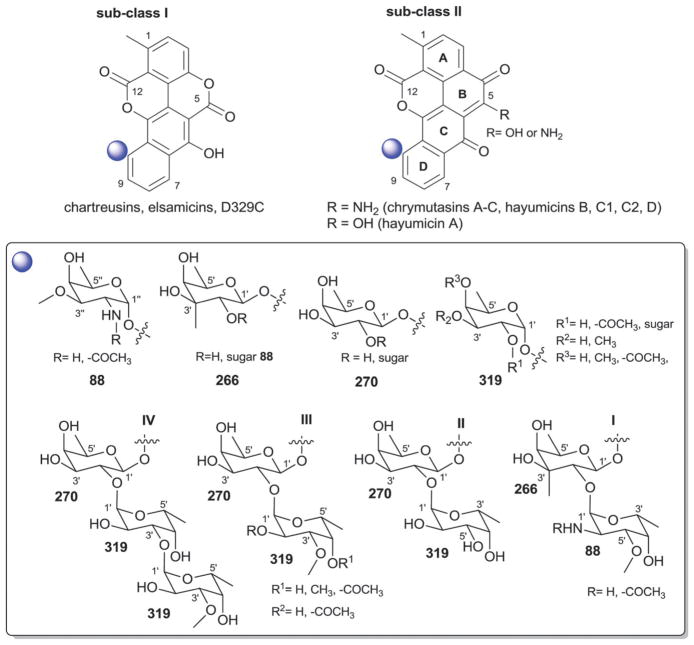
4.2. Hibarimicins
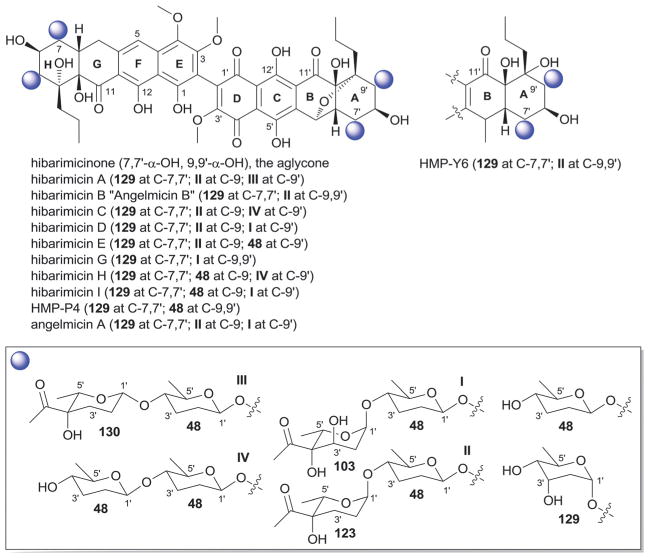
Hibarimicins are distinguished by their highly oxidized naphthyl-naphthoquinone aglycon which derives biosynthetically from dimerization of undecaketide-based precursors (Fig. 19).292 To date, 11 herbimycin O-glycosides have been reported from Microbiospora where the regiospecificity of glycosylation is restricted to C-7/7′ and C-9/9′ (Fig. 19).18,293–299 Glycosylation at these positions is composed of mono- and/or disaccharide side chains (Fig. 19) where five distinct sugar residues were observed including α-L-digitoxose (129), β-D-amicetose (48), 4′-C-acetyl-2′,3′,6′-trideoxy-α-L-gulose (123), 4′-C-acetyl-2′,3′,6′-trideoxy-β-L-gulose (130) and α-L-4-C-acetyl-2,6-dideoxy-xylo-hexopyranosyl (103). In all cases, the C-7/C-7′-sugar was α-L-digitoxose (129) and C-9-/9′-sugar was β-D-amicetose (48), the latter of which in certain analogs was further 4′-O-glycosylated (48, 103, 123 or 130 to present disaccharides I–IV, Fig. 19). HMP-P4 and HMB-Y6 are metabolites produced by Microbiospora engineered mutants.295 Hibarimcins are selective src tyrosine kinase inhibitors and do not inhibit protein kinase C.296 They display potent in vitro cytotoxicity against cancer cell lines and moderate Gram-positive antibacterial activity.296
Fig. 19.
Hibarimicin aglycons and associated sugars.
4.3. Tetracycline-type antibiotics
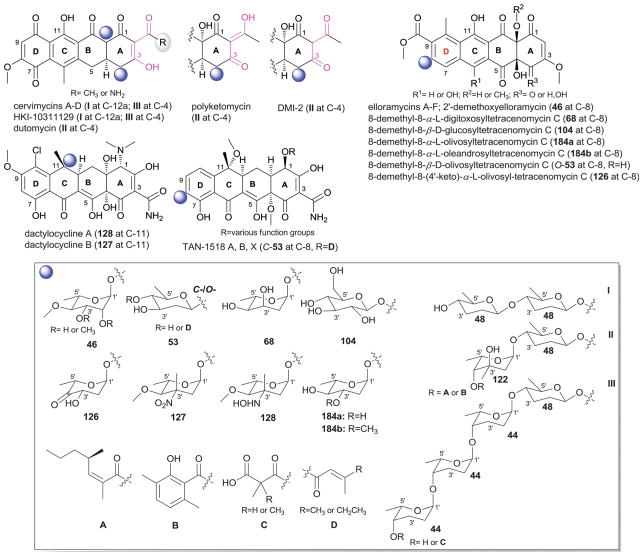
Tetracyclines are tetracyclic polyketide-based broad spectrum antibiotics with a rich history that includes the discovery and implementation of a number of clinically important tetracycline-based drugs, extensive litigation beginning in the 1950’s over tetracycline intellectual property and price fixing among major pharmaceutical companies of the time (Pfizer, American Cyanamid, Bristol-Myers) and a more recent discovery suggesting tetracycline ‘use’ dates back possibly to before 400–500 A.D. There are 26 glycosylated tetracyclines reported to date including the cervimycins,300–302 HKI10311129303 dutomycin,304,305 polyketomycin,306–308 elloramycins,176,309,310 tetracenomycins,176,311,312 TAN-1518 A, B and X (also known as SF-2575),313–315 and dactylocyclines,316–319 the latter of which being the only member of non-Streptomyces origin (Dactylosporangium). Regiospecificity of tetracycline glycosylation is limited to C-4, C-8, C-11 and C-12a where all members with the exception of one subgroup (the C-8-C-glycosidic TAN-1518 analogs) are O-glycosides.
Cervimycins A–D are glycosylated at C-4 and C-12a with tetra-[consisting of one β-D-amicetose (48) and three α-L-rhodinoses (44), tetrasaccharide III] and disaccharide (β-D-amicetosyl-(1→4)-β-D-amicetose, disaccharide I) moieties, respectively (Fig. 20).300–302 The terminal rhodinose (44) of tetrasaccharide III is further modified with a 4′-O-dimethylmalonyl/or monomethylmalonyl ester. The structurally similar HKI10311129303 displays an identical glycosylation pattern but lacks the terminal rhodinose ester modification while the related dutomycin and polyketomycin are C-4-O-glycosides bearing an a α-L-4′-epi-mycarosyl-(1′→4′)-β-D-amicetose (disaccharide II) but lacking C-12a glycosylation.304–308 In these latter metabolites, the terminating mycarose is further modified via 4″-O-esterification and both have been noted as anticancer cytotoxins, antibiotics while dutomycin was also reported as a DNA methyltransferase inhibitor. Elloramycins are cytotoxic C-8-O-glycosides bearing per-methyl-α-L-rhamnose (46) where cytotoxicity, in this case, is attenuated via the presence of the sugar.176,309,310,320 The corresponding tetracenomycin C-8-O-glycosides from strain engineering include substitution with α-L-digitoxose (68), β-D-glucose (104), 4′-keto-α-L-olivose (126), α-L-olivose (184a) and α-L-oleandrose (184b).176,311,312 Dactylocyclines A and B from Dactylosporangium sp. (ATCC 53693)316–319 are C-11-O-glycosides substituted with α-L-evernitrose (127) and its hydroxyl-amino congener 128 (monosaccharides more commonly associated with the orthosomycin everninomicin, Section 14.1).321 Intriguingly, removal of the dactylocycline sugar improves Gram-negative antibacterial activity.318 Finally, the naphthacenecarboxamides TAN-1518 A, B and X (also known as SF-2575) are C-8-C-glycosides of β-D-olivose (C-53).313–315 TAN-1518 A and B have been noted as topoismerase I inhibitors while TAN-1518 X displayed both Gram-positive antibacterial activity and in vitro and in vivo anticancer activity.314 The structural elucidation of the TAN-X C-glycosyltransferase SsfS6 was recently reported as one among only a few C-glycosyltransferases to be structurally characterized to date.322
Fig. 20.
Tetracycline aglycons and associated sugars.
4.4. Aureolic acids and related tetracyclines
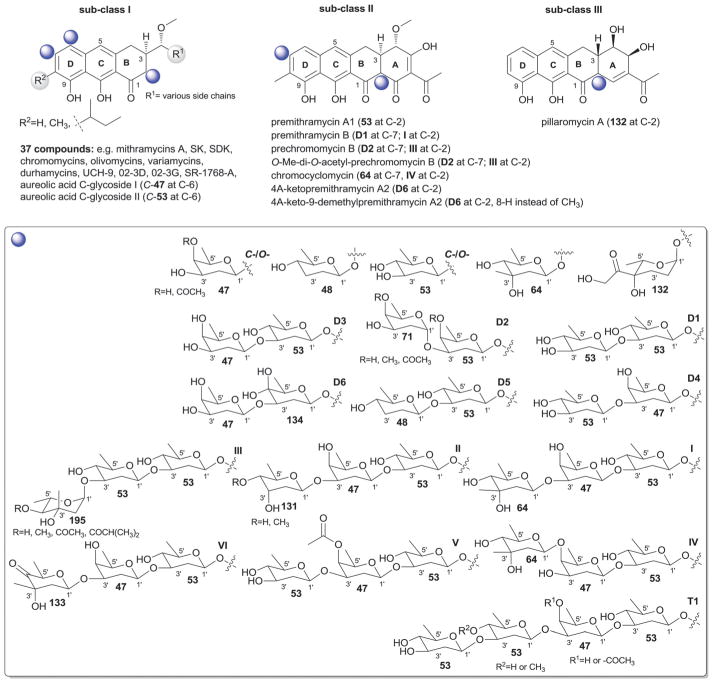
Mithramycin314 is the prototypical member of the aureolic acid family of antitumor antibiotics that also includes chromomycins,323 olivomycins,324–326 durhamycins,327 SR1768A,328 UCH9,329,330 chromocyclomycin,325,331 02-3D and 02-3G,332 and variamycins333,334 from Streptomyces and Actinoplanes as well as several other analogs (e.g., ketopremithramycins and ketomithramycins)335 generated by pathway engineering.323–326,331,333,336–340 The defining structural feature of aureolic acids is their tricyclic polyketide architecture and, to date, 45 glycosylated analogs have been reported from bacteria. The predominant regiospecificity of glycosylation among naturally-occurring aureolic acids is C-2-and/or C-7-O-glycosylation with two family members [aureolic acid C-glycoside I (C-47 at C-6) and aureolic acid C-glycoside II (C-53 at C-6); Fig. 21] displaying atypical C-6-C-glycosylation. For the scope of this discussion, members have been divided into subclasses I–III based upon aglycon distinctions where the tetracyclic subclass II are primarily considered as biosynthetic precursors and subclass III (pillaromycin A)341 also displays similarity to II.
Fig. 21.
Aureolic acid analogs and associated sugars.
Family members are glycosylated with mono-, di-, tri-, or tetrasaccharide side chains (Fig. 21). Sugars found as monosaccharide substitutions include the uncommon C-6-C-glycosidic β-D-oliose (C-47) and β-D-olivose (C-53)-substituted aureolic acid C-glycosides I and II342 and the unusual C4′-branched sugar 4′-C-hydroxyethanone-2′,3′,6′-trideoxy-α-L-gulose (132, pillaromycin A).341 Six different disaccharide (D1–D6) and trisccharide (I–VI) variations are represented among family members, which are comprised of 10 different distinct monosaccharide units where β-D-oliose (47) and β-D-olivose (53), are the most predominant. Other saccharides employed include β-D-amicetose (48), β-D-mycarose (64), α-D-oliose (71, R = H; α-D-chromose, R = CH3), β-D-digitoxose (131), 4′-C-hydroxyethanone-2′,3′,6′-trideoxy-α-L-gulose (132), 4′-keto-β-D-mycarose (133), 4′-hydroxy-β-D-olivose (134), and 3′-epi-αL-mycarose (195, α-L-chromose B). Tetrasaccharide substitution is uncommon with UCH-9 and durhamycin A as the only C2-tetrasaccharide-bearing examples [β-D-olivosyl-(1→3)-β-D-olivosyl-(1→3)-β-D-oliosyl-(1→3)-β-D-olivose, tetrasaccharide T1].
Aureolic acids display potent Gram-positive antibacterial and anticancer activities. Mithramycin (plicamycin) was originally approved for the treatment of cancer however, off-target toxicities, potentially deriving from the ability of mithramycin to target the ubiquitous transcription factor Sp1, limited clinical use. Recent studies revealed mithramycin to selectively target the EWS/FLI1 transcription factor fusion found within Ewing’s sarcoma, renewing interest in mithramycin and development of less toxic, more selective analogs.343,344 Other activities noted among aureolic acids include inhibition of mdr1 gene expression and viral Tat transactivation inhibition. Intriguingly, pillaromycin A, a potent anticancer cytotoxin from this family, has been noted to display lower overall toxicity.341,345
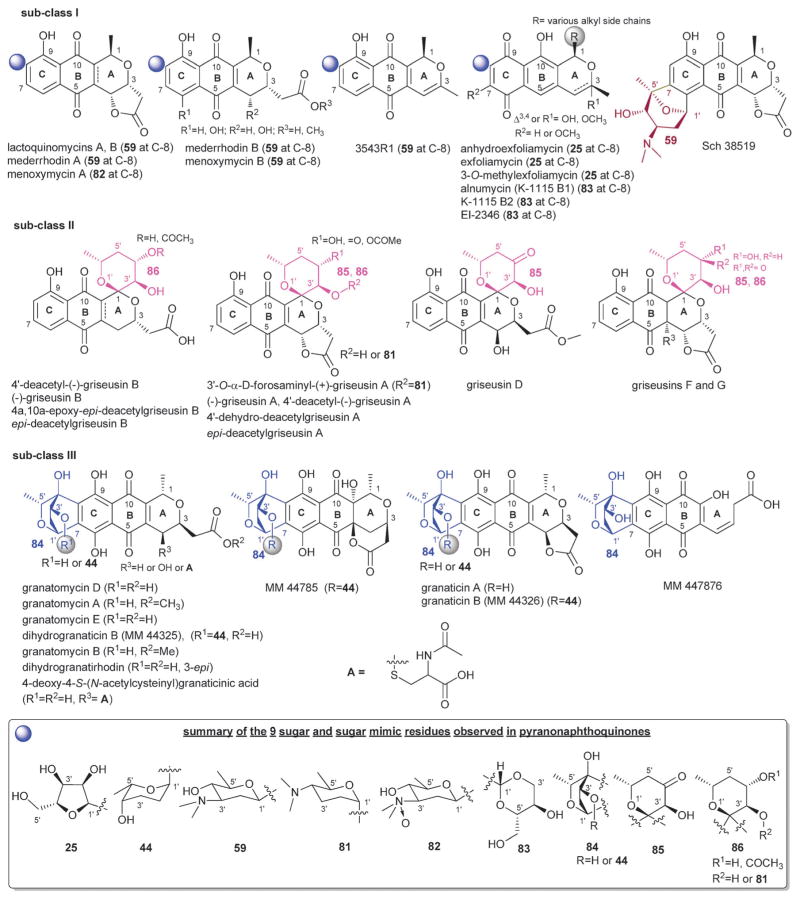
4.5. Pyranonaphthoquinones
Pyranonaphthoquinones contain a naphtho[2,3-c]pyran-5,10-dione core where some members present an additional γ-lactone ring, or corresponding open ring carboxylic acid, fused to the dihydropyran ring (Fig. 22). Pyranonaphthoquinone antibiotics isolated from various bacterial strains (including Streptomyces, Actinomycete, and alkaphilic Nocardiopsis) exhibited antibacterial, antifungal, antiviral, as well as cytotoxic activities.346 To date, 37 bacterial pyranonaphthoquinone glycosides have been reported, all of which contain single C-glycosidic sugars consisting of α-L-rhodinose (44), β-D-ribofuranose (25), β-D-angolosamine (59), α-D-forosamine (81), 3′-amino-N′,N′-dimethyl-N-oxido-2′,3′,6′-trideoxy-β-D-glucose (82), unusual cyclized sugars I (83) and II (84) or sugar mimetics I (85) and II (86) (Fig. 22). For the scope of this discussion, this family is further divided into subclasses I–III as further described below where the C6-C1′/C7-C5′ glycoside fusion found in Sch38519 from Thermomonospora is a notable outlier. This latter metabolite was reported to inhibit thrombin-induced aggregation of human platelets.347,348
Fig. 22.
Pyranonaphthoquinone related aglycons and associated sugars.
Sub-class I is distinguished by a single C-8-C-glycosidic substitution and includes lactoquinomycins,349–352 menoxymycins353 exfoliamycins,354,355 mederrhodins,356 alnumycin (also known as K1115 B1 and K1115 A),357,358 K 1115 B2,355,356 and EI-2346.359,360 Of the four sugars found among sub-class I members (59 or 25 or 82 or 83), N-oxide 82 (menoxymycin A) is unique.353 The C-ribosyl 25 (exfoliamycins) was also observed to rearrange to unusual dioxane 83 (alnumycin,361 K 1115 B2,355,356 and EI-2346331,332).362 Sub-class II members are distinguished by the 1,7-dioxaspiro-[5.5]undecane ring system (85 or 86), the C-1-spiroketal fused moiety of which, while likely polyketide-based, serves as a carbohydrate mimic.363,364 Of the 12 griseusins mainly produced by Nocardiopsis,337–343,365–371 the C-3′-O-α-D-forosaminyl-(+)-griseusin A is the only O-glycosylated member. Finally, sub-class III contains a C-7-C-glycosidic linkage where the corresponding sugar 84 is also fused to the pyranonapthoquinone core via a C-8-C-4′ bond to provide a bicyclic structure (Fig. 22). Eleven sub-class III members have been reported including granatomycins, granaticins, dihydrogranatirhodins, 4-deoxy-4-S-(N-acetylcysteinyl)granaticinic acid, MM 44785 and MM 447876.256,346,372,373 Sugar 84 in three granaticin analogs [dihydrogranaticin B (MM 44325), MM 44785 and granaticin B (MM 44326)] is further C-3′-O-modified with α-L-rhodinose (44).
4.6. Benzoquinones related antibiotics
Napthoquinones represent a large class of bacterial natural products, the structural signature of which is a bicyclic aromatic-1,4-benzoquinone fused core structure 1,4-napthoquinone. Also included within this section are metabolites related to this core structure where the central benzoquinone has been reduced and/or further modified. The predominant regiospecificity of glycosylation across this series is glycosylation of the fused aromatic moiety either at the α- or β-carbon in relation to the ring junction or glycosylation of the quinone core (the precise numbering of which, in all cases, varies depending upon the specific aglycon, Fig. 23.
Fig. 23.
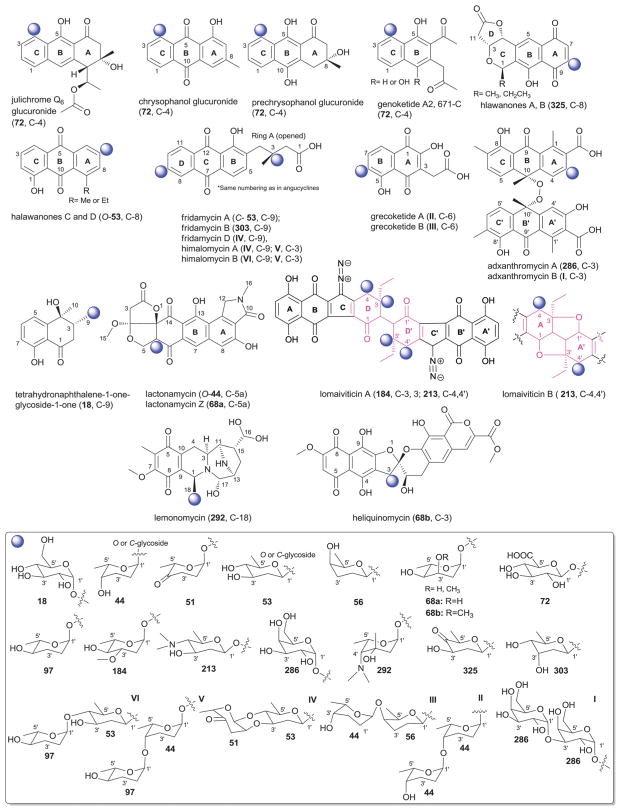
Benzoquinone-related aglycons and associated sugars.
A series of α-aromatic O-glucuronides including julichrome Q6 glucuronide374 671-C, genoketide A2, chrysophanol glucuronide and prechrysophanol glucuronide have been reported as metabolites of Streptomyces.375 All share a common C-4-β-D-glucuronic acid (72) substitution. Julichrome Q6 glucuronide was reported as the first monomeric member of the julimycin-B analogs, and displayed moderate unselective cytotoxicity against human tumor cell lines.374 Genoketide A2 and prechrysophanol glucuronide were reported to inhibit the lymphoma cell proliferation in vitro.375
The β-aromatic glycosides include halawanones,376 fridamycins/himalomycins,77,377 adxanthromycins378–382 and grecoketides383 from various Streptomyces. Halawanones C and D, two tricyclic quinone-containing metabolites, are C-7-O-glycosides (β-D-olivose, 53).376 Fridamycins A (also known as vineomycinone B2), B, and D, and the structurally related himalomycins A and B,377 are angucycline-related antitumor antibiotics that likely arise from a cleavage of the corresponding C-12b/C-l bond to afford the substituted tricyclic angucyclinone lacking ring-A.77,377 Similar to the saquayamycins (Section 3.2), they are C-3- or C-9-O-glycosides bearing mono- or disaccharides (IV–VI) comprised of β-D-olivose (53), 2′,6′-dideoxy-β-D-altrose (303), α-L-rhodinose (44), α-L-cinerulose (51) and α-L-amicetose (97).77,377,384 Himalomycins A and B along with fridamycin D exhibited strong antibacterial activity against Gram-positive and Gram-negative bacteria.377 The two naphthoquinones grecoketides A and B are both disaccharyl-containing C-6-C-glycosides of the grecoketidone aglycon that differ in disaccharide composition.383 Specifically, the C-6-C-glycosyl moiety of grecoketide A is α-L-rhodinose (44) while that in grecoketide B is β-D-rhodinose (56). In both, the rhodinose is further C-4′-O-glycosylated with α-L-rhodinose (44).383 Finally, adxanthromycins A and B are unique dimeric peroxo-anthrone C-3-O-glycosides bearing α-D-galactose (286) and α-D-galactosyl-(1′→3′)-α-D-galactose (disaccharide I), respectively.379–381 These compounds are reported as inhibitors of ICAM-1/LFA-1-mediated cell adhesion.378–381
Glycosides of the core quinone include halawanones A and B,376 substituted naphthalene-1-ones,382 lactonamycins385,386 and lomaiviticins387 from Streptomyces and Micromonospora. Halawanones A and B are structurally related to other isochromane quinone antibiotics from Streptomyces (including exfoliamycins, granaticins and griseusins; see the lactoquinomycins, Section 4.5) and, as C-8-C-glycosides (2′-keto-β-D-oliose, 325) of the quinone core are unique among this class.376 A product of a mutant Streptomyces, 4β,8-dihydroxy-3α-O-(α-glucopyranosyl)hydroxymetyl-4α-methyl-1,2,3,4-tetrahydron-naphthalene-1-one is a simple side chain C-9-O-α-D-glucoside(44).382 Lactonamycin, an antibiotic active against Gram-positive bacteria including MRSA and VRE, consists of a hexacyclic C-5a-O-glycoside bearing α-L-rhodinose (44).385,386 In the related lactonamycin Z, rhodinose is replaced by α-L-digitoxose (68a).388 This latter compound displayed potent antiproliferative activity against gastric adenocarcinoma cell lines. Finally, lomaiviticins A and B are two unique diazobenzofluorene O-glycosides produced by Micromonospora.387 Both are C4/4′-O-glycosides of β-D-pyrrolosamine (213), a sugar previously identified in pyrrolosporin A.389 Lomaiviticin A is further C-3/3′-O-glycosylated with an α-L-oleandrose (184), presumably preventing the tetrahydrofuran ring fusion observed in lomaiviticin B. Diazo-containing natural products like lomaiviticins and the structurally-related kinamycins are rare.390,391 Lomaiviticins function as DNA-damaging agents where lomaiviticin A cleaves double stranded DNA under reducing conditions.387,392 Both were also reported as potent antibiotics against Gram-positive bacteria Staphylococcus aureus and Enterococcus faecium.387
Two metabolites display unique glycosylation patterns that fall outside the scope of those described in the preceding paragraphs namely, lemonomycin393 and heliquinomycin394–396 from Streptomyces. Lemonomycin is C-18-O-glycoside (4′-amino-4′-deoxy-3′-C-methyl-6′-deoxy-α-L-fucose; 292) of a uniquely fused pyrrolidine-tetrahydroisoquinoline aglycon while heliquinomycin is a C-3-O-glycosyl rubromycin member bearing α-L-cymarose (68b).394–396 A glycoside of the rubromycins/griseorhodins,395 this metabolite was found to selectively inhibit cellular DNA replication without affecting level of chromatin-bound MCM4 or activation of the DNA replication stress checkpoint system.394–397
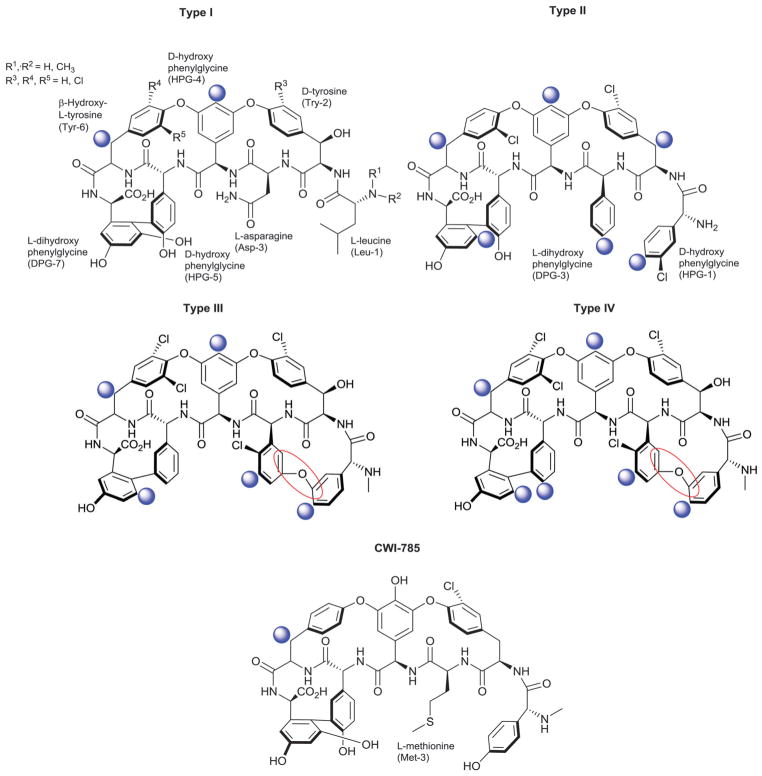
5. Benanomicins and pradimicins
Benanomicin and pradimicin analogs consist of a benzo[a]-naphthacene quinone conjugated to various modified aminoacids at C-15. Glycosylation within this subclass consists of mono or disaccharide side chains at C-5 and, in some cases, also C-11 of the benzoquinone nucleus (rings B and E; Fig. 24).398–404 These compounds have excellent in vitro and in vivo activities against a wide range of fungal strains including Candida, Cryptococcus, Aspergillus and Trichophyton and also are effective inhibitors of HIV syncytium formation.405,406 The pradamicins uniquely function as small molecule ‘lectins’ wherein antifungal activity derives from specific binding to terminal D-mannosides of the fungal cell wall while specific interactions with the N-glycosylation patterns of HIV-1 gp120 contribute to antiviral activity via inhibition of viral entry. The monomers β-D-glucose (104), β-D-xylose (136), 3′-amino-β-D-fucose (208), and β-D-fucose (270), or modifications thereof, encompass the sugars represented and SAR studies to date point the importance of both the C-15 amino acid substitution and C-5 glycosylation for antifungal and anti-HIV activities.357 C-5-glycosidic examples include pradimicins A–E,400,406–409 FA-1,410 FA-2,410 FB,411 FH, L, FL,412 S,413–415 FS411 and other analogs.17,18,399,416–419 C-11 glycosylation occurs less frequently and is restricted to β-D-xylose (136) as illustrated by pradimicin T2,420,421 11-O-L-xylosyl-pradimicin H422 and 11-O-L-xylosyl-pradimicin FH,422 all of which contain C-5 mono- or disaccharide substitutions.
Fig. 24.
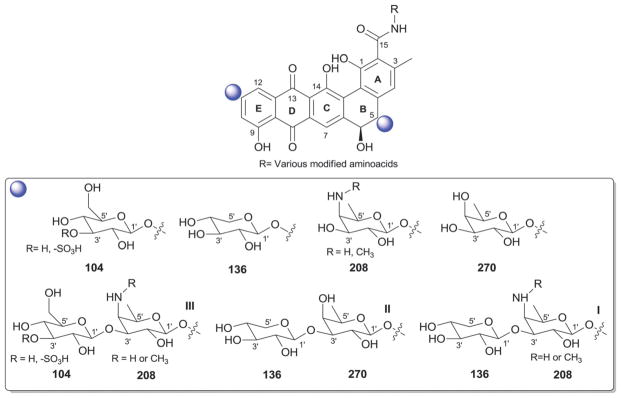
Benanomicin and pradimicin aglycons and associated sugars.
6. Chartarin-analogs
Chartarin analogs are divided into two main classes distinguished by the B ring oxidation state and C-5 substitution of the fused pentacyclic ring aglycon (sub-classes I and II; Fig. 25). Sixteen naturally-occurring chartarin glycosides have been reported to date as bacterial metabolites (mainly from Streptomyces, Fig. 25) including the chartreusins (lambdamycins),423–433 chrymutasins,434–436 D329C,437 elsamicins,438–440 and hayumicins.441 These compounds display notable antimicrobial and antitumor activities where glyco-sylation plays a key role in improving potency, formulation and in vivo properties wherein aminosugar-containing variants are among the most advantagous.423,438–440,442–445 In all cases reported to date, O-glycosylation is restricted to C-10 mono-, di- or trisaccharide substitution. Four different appended modified sugars are found among existing charterin analogs [β-D-fucose (270), β-D-elsarose (266), α-D-fucosamine (also known as α-D-elsaminose, 88), and α-D-fucose (319)]. Of these four sugars, D-fucose (270) is the most predominant with a range of C-2′-, C-3′-, and/or C-4′-O-methyl/acetyl-substituted, as well as the C-3′-C-methyl-branched (β-D-elsarose, 266) or the corresponding α-D-fucosamine (also known as α-D-elsaminose, 88) analogs observed.
Fig. 25.
Chartarin aglycons and associated sugars.
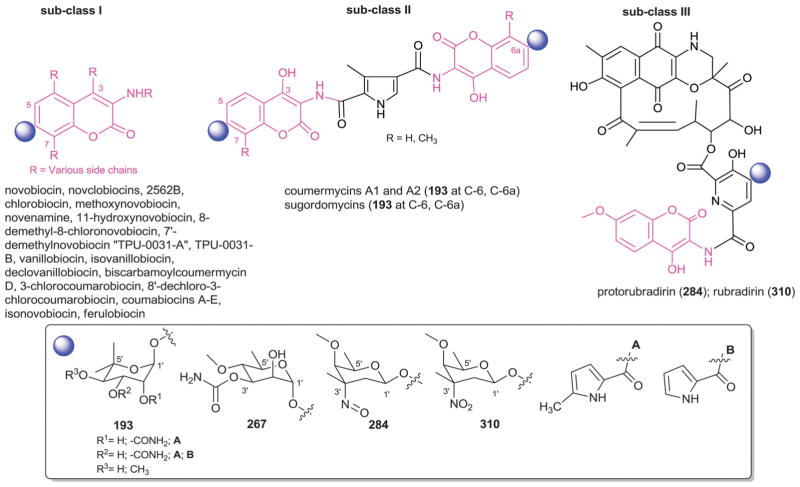
7. Coumarins
The aminocoumarin group of antibiotics are characterized by their 3-amino-4,7-dihydroxycoumarin moiety, the microbial source for which is currently restricted to Streptomyces. This family of antibiotics [which includes novobiocin, isonovobiocin,446 2562B, clorobiocin (2562A; RP 18 631),447,448 coumermycin A1,449,450 coumermycin A2,451 vanillobiocin,452 novenamine,453,454 11-hydroxynovobiocin,455 isovanillobiocin,455 declovanillobiocin,455 biscarbamoylcoumermycin D,456 3-chlorocoumarobiocin,457 and 8′-dechloro-3-chlorocoumarobiocin457] function as potent bacterial DNA gyrase inhibitors and were more recently discovered to inhibit Hsp90.458–462 To date, 84 naturally-occurring aminocoumarins have been reported from bacteria, 65 of which are glycosylated representatives.463–467 Importantly, the 3′-carbamoylation of the appended sugar in these metabolites is critical for DNA gyrase inhibitory activity but detrimental to Hsp90 inhibitory function.460,468,469 The scope of glycosylated aminocoumarin derivatives has been notably expanded by Heide et al. via metabolic engineering, mutasynthesis and chemoenzymatic synthetic methods.456,467,470–473 It is also important to note that there are currently no examples of native bacterial coumarin/isocoumarin glycoside production.
The glycosylated aminocoumarins are classified into subclasses I–III based upon subtle substitution/conjugation distinctions within the core aminocoumarin (Fig. 26). From the perspective of glycosylation, subclasses I and II are markedly similar in both the regiospecificity of glycosylation (C-6 of the aminocoumarin core) and the predominate sugar employed (4′-O-methyl-5′-C-methyl-L-rhamnose, more commonly referred to as α-L-noviose, 193). In both subclasses I and II, the core sugar moiety is further modified via differential acylation of the 2′-OH and/or 3′-OH with a carbamoyl, 5-methyl-pyrrole-2-carboxyl or pyrrole-2-carboxyl moiety (Fig. 26, 193). In addition, 4′-OH methylation was observed among some members. A notable departure from the structural/biosynthetic conservation among subclass I members is TPU-0031-B (5′-demethyl-novobiocin) which reportedly bears the distinct sugar α-D-5′-demethyl-noviose (267) and, given the glycosidic conservation among the metabolites in this family, may raise questions regarding the structural assignment in this case.474
Fig. 26.
Coumarin aglycons and associated sugars.
Subclass III members are structurally distinguished by their conjugated system comprised of a novobiocin-type amino-coumarin connected to an ansamycin-like moiety via a 3,4-dihydroxydipioolinic acid central bridge, the latter of which serves as the site of glycosylation (specifically, C-4 of the 3,4-dihydroxydipioolinate). This subclass contains two antibiotics (rubradirin475,476 and protorubradirin477 from Streptomyces) distinguished by their corresponding uniquely functionalized appended sugars [the C-3′-nitro sugar β-D-rubranitrose (310) and its C-3′-nitroso-analog (284), Fig. 26]. As in the case of the nitro sugar-containing orthosomycins (where aminosugars with differing levels of amine oxidation have been observed, Section 14), it is expected that NDP-rubranitrose biosynthesis proceeds via the C-3′-nitroso-analog. Interestingly, these two sugars have also been identified in anthracycline antibiotics (viriplanins A and D, Section 4).289,290,477
8. Enediynes
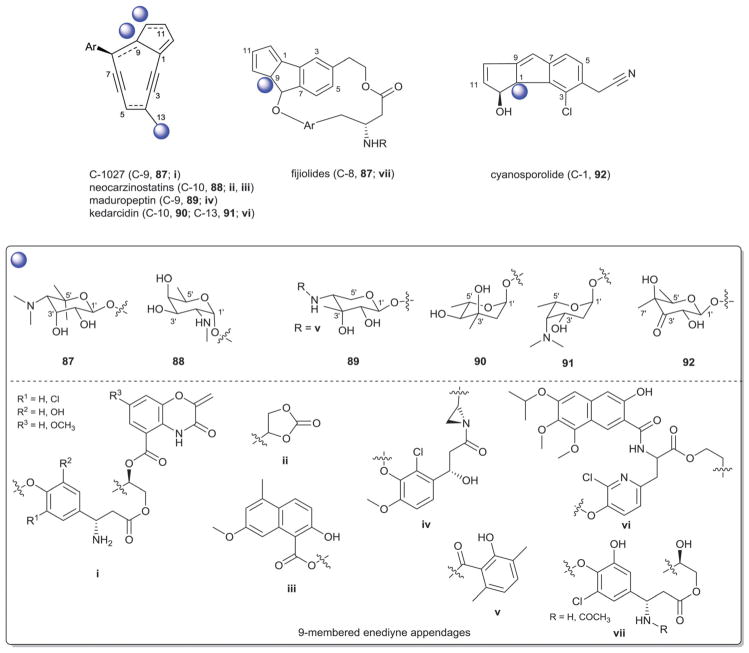
Enediynes are a group of antibiotic anticancer compounds characterized by the presence of bicyclo[7.3.0]dodecadienediyne or bicyclo[7.3.1]tridecenediyne and this structural distinction serves as a basis for classification of family members within 9- or 10-membered subclasses, respectively.478 As an additional signature, 9-membered enediynes are commonly associated with a stabilizing apoprotein and 10-membered enediynes have been more recently been associdated with a novel self-sacrifice resistance mechansism.479–481 Enediynes bind the minor groove of dsDNA in a sequence/context-specific fashion which is often mediated via their appended carbohydrates. Subsequent reductive/nucleophilic activation initiates a Bergman or Myers-Saito cyclization, the reactive diradical intermediate of which abstracts hydrogen from the DNA backbone to ultimately afford oxidative DNA strand cleavage.482 Out of 46 enediynes and enediyne derived compounds isolated from bacteria, 38 glycosides have been characterized (Fig. 27). Members of this family are treated separately herein based upon enediyne ring size.
Fig. 27.
9-Membered enediyne aglycons and associated sugars. Ar, aromatic moiety.
8.1. 9-Membered enediynes
The glycosylated 9-membered enediynes (Fig. 27) include C-1027,483–485 neocarzinostatin,486–488 maduropeptin,489 kedar-cidin,490–492 and two putative cycloaromatized analogs (NFκB inhibiting fijiolides493 and cyanosporasides494) from Streptomyces, Actinomadura, Streptoalloteichus, Salinospora, and Nocardiopsis. Nine-membered enediyne glycosylation regiospecificity is restricted to C-9 or C-10 of the enediyne core. With one exception (cyanosporoside), all 9-membered enediyne glycosides contain an amino sugar (C-1027 and fijiolides, 4′-deoxy-4′-dimethylamino-5′,5′-dimethyl-β-D-ribose 87; neocarzinostatin, N-methyl-α-D-fucosamine 88; maduropeptin, 4′-amino-4′-deoxy-3′-methyl-β-D-ribose 89; kedarcidin, α-L-kedarosamine 91) where kedarcidin is the only member containing more than one carbohydrate (an additional α-L-mycarose 90). Interestingly, the sugar appended to cyanosporoside is a rare oxo-β-D-fucose (92), reflecting a potential lack of a functional keto-sugar nucleotide aminotransferase common to members of this subclass. C-3′-, C-4′- or C-5′-branched sugars are also a predominate feature among 9-membered enediynes with N-methyl-α-D-fucosamine 88 (neocarzinostatin) and α-L-kedarosamine 91 (kedarcidin) as the only exceptions.
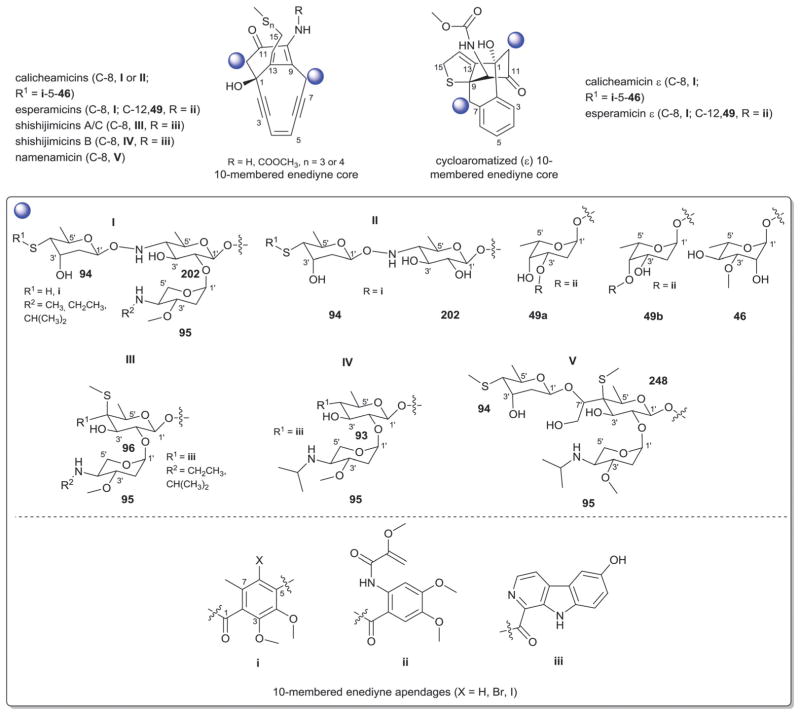
8.2. 10-Membered enediynes
Glycosylated 10-membered enediyne glycosides (Fig. 28) include the calicheamicins,495–498 esperamicins,499–505 shishijimicins,506 and namenamicin507 from Micromonospora, Actinomadura and potentially unidentified bacterial symbionts. With respect to the latter, shishijimicins and namenamicin were isolated from marine ascidians but based upon their structural resemblance to bacterial counterparts they are assumed to be of bacterial origin and therefore included herein. Among the glycosylated 10-membered enediynes, C-8 and C-12 of the enediyne core are the primary points of glycosylation (Fig. 28). Both the calicheamicins and esperamicins contain a conserved C-8 trisaccharide moeity I comprised of 4′,6′-dideoxy-4′-hydroxyamino-β-D-glucose (202), 4′-thio-2′,4′,6′-trideoxy-β-D-altrose (94) and 4′-amino-2′,4′-dideoxy-α-L-xylose (95) where, in some calicheamicins (calicheamicin , LL-E33288 B, and LL-E33288 ), the aminopentose is lacking.508–511 The corresponding terminal thiosugar is 4′-S-methylated (esperamicins) and 4′-S-acylated with a modified orsellinate (calicheamicins), the latter of which is typically O-glycosylated (3′-O-methyl-α-L-rhamnose, 46; exceptions being calicheamicin , LL-E33288 B and LL-E33288 ). Esperamicin contains an additional C-12-O-glycoside (α-L-oliose, 49) which is further esterified at C-3′ (esperamicin A1 series, 49a) or C-4′ (esperamicin A2 series, 49b) with a modified anthranilate (Fig. 28). Shishijimicins A and C contain an alternative C-8 disaccharide III comprised of a branched thiosugar (6′-deoxy-4′-pyridoindolylcarbonyl-4′-S-methyl-4′-thio-β-D-galactose, 96) and 4′-amino-2′,4′-dideoxy-4′-O-methyl-α-L-xylose (95)506 while shishijimicin B contains disaccharide IV where the branched thio sugar 96 present in A and C derivatives is replaced with a 4′,6′-dideoxy-4′-pyridoindolylcarbonyl-β-D-glucose (93). Namenamicin507 contains the alternative C-8 trisaccharide V comprised of a non-branched thiosugar [6′-deoxy-4′-thio-4′-S-methyl-4′-C-(1,2-dihydroxyethyl)-β-D-galactose, 248], and 4′-amino-2′,4′-dideoxy-4′-O-methyl-α-L-xylose (95), the thiosugar branch of which is C-7′-O-glycosylated with the esperamicin 4′-S-methylthiosugar 94 (Fig. 28).506 It is also important to note that aminopentose 95 was also found appended to indolocarbazoles AT2433 (Section 10) and, as expected, common biosynthetic elements have been noted (Fig. 28).512 In the calicheamicins and esperamicins, the C-8-saccharide substitution is known to be important for DNA binding. While there remains some controversy regarding whether the metabolite or DNA alters configuration upon complex formation,513–515 the unique conformation of the calicheamicin/esperamicin hydroxyaminosugar glycoside bond is believed to be a key contributor. Interesting, this hydroxylaminoglycosidic bond also was recently demonstrated to serve as a chemo-selective handle for neoglycosylation.10,516 It should also be noted that the biochemical study of the glycosyltransferases involved in calicheamicin biosynthesis revealed these reactions to be much more reversible, a phenomenon that has since to be found as relatively universal among glycosyltransferase-catalyzed reactions.509
Fig. 28.
10-Membered enediyne aglycons and associated sugars. Ar, aromatic moiety.
9. Flavonoids and isoflavonoids
(Iso)flavonoids are a prevalent group of plant secondary metabolites that share a common three ring architecture with subtle ring regiospecificity distinctions as a defining feature. Specifically, the term ‘flavonoid’ generally encompasses scaffolds containing a 2-phenylchromen-4-one core while ‘isoflavonoid’ typically corresponds to compounds containing the corresponding 3-phenylchromen-4-one backbone. While the core scaffolds are not of bacterial origin, several novel analogs deriving from microbial bioconversion of soy-based media components have been reported in the context of bacterial natural products discovery.517 Within this context, it is important to note recent strain engineering efforts to enable microbial (iso)flavonoid scaffold production518,519 as well as engineered microbial strains recently developed to enable differential glycosylation of (iso)-flavonoids.520–523 Many diverse activities have been attributed to the vast repertoire of naturally-occurring (iso)flavonoids including antioxidative properties and/or inhibition of lytic/digestive enzymes as well as anticancer, antifungal and/or estrogenic action.524 To date, 16 glycosylated (iso)flavonoid analogs have been reported from native bacterial media bioconversion by Streptomyces,525–530 Kitasatospora,531,532 and Sorangium18,533 to afford the installation of five distinct sugars [α-L-rhamnose, 46; α-D-ristosamine, 69; 6′-deoxy-α-L-talose 125; α-L-chinovose (6′-deoxy-4′-epi-α-L-glucose), 157; and β-L-cymarose, 176], the most predominate of which being α-L-rhamnose (46) (Fig. 29).
Fig. 29.
Flavonoid and isoflavonoid aglycons and associated sugars.
The dominate glycoside regiospecificity within this class is C-4a- and/or C7-O-glycosylation as exemplified by the various daidzein and genistein rhamnosides from Streptomyces,525,526,530 the corresponding genistein-derived talosins A and B from Kitasatospora kifunensis MJM341 [which contain the unusual sugar 6′-deoxy-α-L-talose (125)531,532 better known as a monosaccharide integrated within cell wall oligosaccharides of certain Gram negative bacteria]534–536 and the 4′,7-bis-(β-cymaropyranosyl)-genistein from Streptomyces.528 In addition, the Streptomyces-derived actinoflavoside carries the rare 3′-amino-2′,3′,6′-trideoxy-ribopyranoside (α-D-ristosamine, 69) at C-7.527 Intriguingly, while genistein rhamnosides lack antifungal activity, the corresponding talosides (talosins A and B)532 were reported as potent antifungals, implicating the inversion of a single stereo-center (C-3′) within the appended sugar as the distinguishing feature that invokes antifungal activity.
C-3a and C-6 represent less common points of O-glycosylation with N-99-596 A (7,4a-dihydroxyisoflavone-3a-O-α-L-rhamno-pyranoside) and N-99-596 B (5,7,4a-trihydroxyisoflavone-3a-O-αL-rhamnopyranoside)529 from Streptomyces as the only examples of the former. For the latter, the α-L-chinovose (6′-deoxy-4′-epi-αL-glucose, 157) containing luteolin-6-chinovoside from Sorangium represents the only C-6-glycoside within this series.18,533
10. Indoles and indolocarbazoles
For the scope of this discussion, indole-derived metabolites have been divided into two simple classes – those that contain a single indole as part of the aglycon core (indoles, kahakamides, neosidomycin, SF-2140, oxopropalines, pyridindolols, pyrroin-domycins and tryptamines), and aglycons comprised of fused bis-indole systems (indolocarbazoles, tjipanazoles, and akashins). Cumulatively, 79 glycosylated members are represented among these two classes where an intriguing bifurcation exists. Namely, with a few exceptions (kahakamides, neosidomycin, and SF-2140), indolocarbazole N-glycosylation is restricted to the fused bis-indole-containing members.
10.1. Fused indoles
Indolocarbazoles, the predominate metabolites within the fused indole series, are actinomycete-dervived alkaloids which inhibit topoisomerase I and kinases relevant to anticancer, antitubercular, antimalarial and antiviral lead development.537–540 While many indolocarbazole analogs have been discovered or synthesized,537,538 it is noteworthy that those that have advanced furthest clinically are either a known natural product (CT327, a pegylated formulation of the natural product K252a currently in phase IIb for treating psoriasis)541 or subtle variations thereof including N-acylated staurosporine [midostaurin/PKC412, which recently completed a successful phase II trial for acute myeloid leukemia (AML) and is currently in phase II evaluation for metastatic melanoma],542 the N-alkylated rebeccamycin (becatecarin)543,544 and a reduced K252a (lestauritinib/CEP-701 currently in clinical phase II/III evaluation for treating FLT3-ITD AML).545
Indolocarbazoles are characterized by their indolo[2,3-a]pyrrolo[3,4-c]carbazole core and generally divided into two separate sub-classes exemplified by rebeccamycin546 and staurosporine547 with 64 glycosides cumulatively reported to date (Fig. 30). The distinguishing features of the rebeccamycin-like indolocarbazoles (including both AT2433 and rebeccamycin analogs) include a 1H-pyrrole-2,5-dione, indole N-glycosylation and often indole halogenation. In contrast, staurosporine-like indolocarbazoles (including, indocarbazostatins, K-252, MLR-52, RK-286, RK-1409, staurosporine, UCN and ZHD-0501 variants) contain a reduced 1H-pyrrol-2(5H)-one, a signature fused N,N′-12,13-glycoside, and typically lack indole halogenation (Fig. 30). In general, mechanism of action coincides with these structural distinctions where staurosporine-like indolocarbazoles typically function as kinase inhibitors548 while rebeccamycin-like indolocarbazoles inhibit topoisomerase I549 and, in either case, glycosylation generally contributes to improved bioactivity.549 Staurosporine-like indolocarbazoles have been reported from Micromonospora, Nocardiopsis, and Streptomyces while rebeccamycin-like analogs have been reported from Saccharothrix (later reclassified as Lechevalieria) and Actinomadura. In addition to the native natural products, indolocarbazole analogs have been generated via strain engineering,540,548,549 feeding native producing strains with 6-fluoro-tryptophan,550 bioconversion of aglycon mimetics by E. coli strains expressing genes encoding tailoring enzymes (N-glycosyltransferases and N-methyltransferases),551 and exploiting the permissive nature of corresponding methyltransferases for differential alkylation.552,553 In addition, the heterologous production of rebeccamycin in E. coli has been achieved.554
Fig. 30.
Fused indole-related aglycons and associated sugars where different colors distinguish multiple points of attachment.
The staurosporine-like members can be further divided into N,N′-12,13-fused pyranosides and N,N′-12,13 furanosides. Of the former, a biosynthetic progression can be observed among isolates (suggesting notable sugar nucleotide tolerance within the corresponding N,N′-12,13 glycosyl-forming enzymes) beginning with the simple 6′-deoxy sugar α-L-rhamnose (46b) found in MLR-52 (Fig. 30).555 Continuing, sugar C-2′-deoxygenation and C-3′-epimerization presents the methylated α-L-digitoxose 68b of RK-286C556,557 while corresponding sugar C-3′oxidation contributes to a staurosporine analog bearing the C-3′-keto sugar, 3′-keto-2′,3′,6′-trideoxy-α-L-glucose (150).558 Subsequent C-3′ sugar transamination and C-3′ sugar amino modification provides a range of analogs including the flagship 3′-N-methyl-4′-O-methyl-α-L-ristosamine (45b, staurosporine,547,559–561 TAN-999,562 UCNs,563,564 and RK-1409565) as well as N,N-dimethyl,566 N-desmethyl,567 and N-amide derivatives568 in staurosporines; N-formyl and the corresponding C-3′/C-4′-cyclic carboxamide 45c of ZHD-0501;569 and N-oxidation to afford the hydroxyl-amino analog (226, associated with two N-hydroxystaurosporine derivatives and 4′-demethyl-N-formyl-N-hydroxystaurosporine) and hydroxylimino analog (148) of the TAN-1030A series (Fig. 30).558 The same strain afforded metabolites containing the 3′-keto corresponding sugars 150 and 151.558 Three divergent N,N′-12,13-pyranosides have also been observed including: 4′-O-methyl-α-L-olivose (106, RK-1409B);570 3′-nitro-2′,3′,6′-trideoxy-α-L-glucose (143, 4′-demethylamino-4′-nitro-staurosporine);568 and 3′-amino-3′,6′-dideoxy-4′-O-methyl-α-L-altrose (147, 5′-hydroxystaurosporine and 4′-N-methyl-5′-hydroxystaurosporine from Micromonospora).571 The corresponding N,N′-12,13-fused furanoside-containing members of this group include indocarbazostatins A–D,572–574 all of which contain a novel branched α-L pentose 145 [differing via a C-3′-side chain ethyl (A/B) or methyl (C/D) ester], and K-252a-b.566,575,576 K-252a and K-252b contain a novel branched α-L pentose 146 [differing via the presence of a C-3′-side chain free acid (b) or methyl ester (a)] while, in N-methyl-3′-amino-3′-deoxy K-252a, the 146 C-3′-hydroxyl has been replaced with a N-methyl-amino group to afford 149 (Fig. 30). From a biosynthetic perspective, general formation of the indole-N-sugar-C-5′ fusion, sugar branching (145, 146 and 149), sugar N-oxidation (143, 148, 335) and formation of the C-3′/C-4′-cyclic carboxamide 45c are expected to offer potential new chemistries.
In contrast to the N,N′-12,13-fused pyranosides and furanosides discussed in the preceding paragraph, rebeccamycins (from Lechevalieria),546,577–579 AT2433 analogs (from Actinomadura),580,581 holyrines (from a marine actinomycete strain N96C-47),582 RK-286D (from Streptomyces),557 tjipanazoles (from Tolypothrix),583 akashins (from Streptomyces),584,585 and K-252d (from a marine Streptomyces and Nocardiopsis)566,575,576 represent related fused tryptophan ring-based systems that contain a single N-glycosidic attachment. A range of monosaccharides have been observed among the appended sugars within this subclass including: β-D-glucose (104) and its 4′-O-methyl-analog (rebeccamycins); α-L-digitoxose (68a) (RK-286D); α-L-ristosamine (45a) and the 2′,3′,6′-trideoxyaminohexose (152) (holyrines A and B, respectively); α-L-rhamnose 46a (K-252d); 6′-deoxy-β-D-gulose (135), α-L-rhamnose (46a), β-D-glucose (104), or β-D-xylose (136) (tjipanazoles); 4′-amino-4′,6′-dideoxy-α-L-glucose (137a), 4′-acetamido-4′,6′-dideoxy-α-L-glucose (137a) and the corresponding C-3′/C-4′-oxazole sugar 137b (akashins A, B and C, respectively), the latter of which is perhaps the most uniquely functionalized sugar among this series. The four AT2433 variants stand out as the only disaccharide-containing metabolites within the fused indole series where the deoxyaminopentose 4′-amino-2′,4′-dideoxy-α-L-xylose (138) or its 4′-N-methyl derivative is attached at 6′-position of indole 4′-O-methyl-β-D-N-glucoside (104) (reminiscent of rebeccamycin core structure) (Fig. 30). AT2433 and rebeccamycin share many common biosynthetic features and a biosynthetic relationship between AT2433 and the enediyne calicheamicin (Section 8.2, by virtue of their shared aminopentose) has also been noted.512
10.2. Simple indoles
Unlike members discussed in the previous subsection, indole N-glycosylation is uncommon among the simple indoles metabolites. Specifically, kahakamides (a marine Nocardiopsis),586 neosidomycin (Streptomyces),587 and SF-2140 (Actinomadura)588 are the only members that contain indole N-glycosides, all of which contain either 4-deoxy-α-D-taluronamide (314; kahakamide A) or 4′-deoxy-α-D-taluronate (139; kahakamide B, neosidomycin, and SF-2140) (Fig. 31). Of these, SF-2140 displayed notable antiviral activity and a higher survival rate than amantadine in viral-challenged mice. Among other simple indole/tryptophan analogs, two C-5-glycosylated N-acetyl tryptamine derivatives [β-D-quinovose (140) or α-L-rhamnose (46)], were discovered in the context of metabolic feeding studies using the staurosporine producer.589 Sugar 46 was also present, as an unusual anomeric glycosyl ester, in an indolyl-3-carbonyl derivative isolated from Streptomyces.590
Fig. 31.
Simple indole-related aglycons and associated sugars.
The pyrroindomycins from Streptomyces represent the only trisaccharyl-containing members of the simple indole group. Unlike the indolyl-glycosyl esters described above, the trisaccharide of pyrroindomycins is connected via an ester bond to the C-3′ terminus of a 4′-amino-2′,4′,6′-trideoxy-β-L-galactosyl-(1′→4′)-β-L-mycarosyl-(1′→4′)-β-D-rhodinosyl trisaccharide appended to a polyketide tetramic acid-containing macro-ring system (trisaccharide I, Fig. 31). These unique metabolites were noted for their antibacterial activity against the drug resistant pathogens including MRSA and VRSA.
The β-carbolines represent the last group of simple indole-containing metabolites and are signified by a fused pyridine-indole skeleton (Fig. 31). Members include the oxopropalines591,592 and pyridindolols593–596 from Streptomyces and have been noted for herbicidal and anti-fungal activities and as benzodiazepine-receptor ligands.597,598 To date, only six glycosylated β-carbolines have been reported from bacteria. Variant side-chain regiospecificity of O-glycosylation was observed among this group with C-10, C-11, and C-12 mono-glycosylation [β-D-glucose (104)] observed in pyridindolols and C-11 and C-12 mono-glycosylation [α-L-rhamnose (46)] found among oxopropalines (Fig. 31). In the latter case, rhamnosylation was found to be detrimental to the observed anticancer activity of the parental aglycon.592
11. Lipids, polyenes and carotenoids
Microbial glycosylated lipids are ubiquitous and include both primary metabolites common to cell wall/membrane architecture as well as a range of bioactive secondary metabolites. As aglycon structure and glycosyl regioselectivity is highly diverse, this section simply highlights the diverse sugar structures observed among bacterial glycolipids.
Not surprisingly, O-β-D-glucosides (104) are among the most prevalent glycosyl substitutions represented as exemplified by fattiviracins,599 moenomycins,600 arthrobacillins,601 glucolipsins,602 and carotenoids.603 The α-anomer 18 was also observed in a small set of glycosylated lipids604,605 along with the corresponding common aminosugar variant (2′-amino-2′-deoxy-β-D-glucose, 153), α-D-anomeric phosphate [2-amino-2-deoxy-α-D-glucose-1-phosphate (5) of lipid A]606 α-D-glucuronic acid (154), and C-5′ sulfonates (155).607,608 The common 2′-epimer β-D-mannose (156) is also represented among many bacterial carotenoids609 and Flavobacterium glycerolipids610 while the corresponding C-2′, C-6′-disulfate (160),611 as well as α-L-fucose (157),612 β-D-arabinose (158),613 α-L-rhamnose (46),614 β-D-xylose (136),615 β-D-glucofuranose (159),604 β-D-galactose (161),607 α-D-allose (162),616 α-D-altrose (163),617 Gram-negative phosphosphingolipid heptose sugar 166,618 and the unique 8-carbon thio sugar 167 (desalicetin 2′-butyrate)619 are additional sugars represented among bacterial glycolipids, carotenoids and related metabolites. Di-saccharides I and II (Fig. 32), consisting of β-L-hexanoic acid (168, β-L-iduronic acid) and β-D-glucosamine (153), or glucose-1-phosphate (18) and β-D-glucosamine (153), are also components of some glycolipids.620,621
Fig. 32.
Glycolipids, polyenes and carotenoids and associated sugars.
Monosaccharides represented among secondary metabolites include the undefined 2′-amino-2′,6′-dideoxy hexose 164 (vancoresmycin),622 β-L-xylose (165, malyngamide J),623 β-L-rhodinose (169, streptolydigin)624 β-D-digitoxose (131, α-lipomycin)625 and α-D-altrose (163, moritoside).617 While glucosamine (153) is commonly found as a lipid O-glycoside, it exists as an N-glycoside among racemomycins.626 Alternatively, some secondary metabolites within this family contain O-oligosaccharyl substitutions as exemplified by moenomycin [Fig. 32; hexasaccharide chain V comprised of β-D-galacturonamide (324), 2′-N-acetyl-β-D-xylose (173), β-D-glucose (104), N-acetyl-β-D-2′-glucosamine (153) and phosphosugars 174 and 175].600
Elfamycins and phenelfamycins represent a structurally unique group of metabolites from Streptomyces.627–629 The polyene moieties found within these molecules are glycosylated with mono-, di-, or trisaccharides (Fig. 32) where α-L-oliose (49a/49b) serves as the predominate monomer and typically as the point of contact to the aglycon. Exceptions to this include disaccharide III of efrotomycin and N-demethylefrotomycin, comprised of the 6′-deoxy-β-D-allose (170) and α-L-rhamnose (46),630–632 and the inclusion of β-L-digitoxose (176) in elfamycin analogs (disaccharide VII and trisaccharide IV). Aurantinin B from Bacillus, while also structurally related to elfamycins, is also unique by virtue of its rare 3′-keto-β-L-sugar (171).633
12. Macrolides and macrolactams
Macrolides and macrolactams are highly functionalized macro-cyclic polyketide-derived metabolites, the ring structures of which are formed via an intramolecular ester (macrolides) or amide (macrolactams) bond. This section summarizes the vast array of glycosylated macrolides (categorized by ring size and architecture in Sections 12.1–12.7), macrodiolides (Section 12.8), super macrolactones (Section 12.9), polyene macrolides Section 12.10), macrolactams (Section 12.11), spirotetronates (Section 12.12) and spinosyns (Section 12.13).
12.1. 12-Membered macrolides
Methymycin and pseudoerythromycin derivatives constitute the known 12-membered bacterial glycosylated macrolides from Streptomyces, where O-glycosylation is restricted to C-3 and/or C-5 of the macrolide core (Fig. 33). The most predominate sugar found appended to this class is β-D-desosamine (177a), at either C-3 or C-5,634,635 but other sugars observed include the corresponding desosamine analogs N-desmethyl 177b or N-oxide 178. In addition, C-3-appended α-L-cladinose (90b) is present pseudo-erythromycins634 while β-D-olivosyl (53) substituted methymycin has been generated via pathway engineering636 and a range of non-native desosamine replacements (in the context of 12-, 14- and 16-membered macrolides) have been accomplished via chemoenzymatic methods.637 Classical 12-, 14- and 16-membered macrolides function as antibacterials in a mechanistically similar manner as described in Section 12.3.
Fig. 33.
12-, 14- and 16-membered macrolactones and associated sugars.
12.2. 14-Member macrolides
As exemplified by the naturally-occurring erythromycin or semi-synthetic clarithromycin, 14-membered macrolides are a mainstay in the clinical treatment of Gram-positive/negative infections.638,639 There are 96 naturally-occurring 14-membered glycosylated macrolides reported to date from bacterial sources including Streptomyces, Nocardia, Saccharopolyspora and Amycolatopsis. C-3- and C-5-O-glycosylation is the most predominate among 14-membered macrolides, but C-6-, C-7-, C-9- and, in one case (CP 64537, containing either 140 or 170c),640 C15-O-glycosylation has also been observed. Reminiscent of classical 12- and 16-membered macrolides, 14-membered macrolides (including erythromycins,634,641–643 megalomycin,644,645 picromycin,646 oleandomycins,647 sporeamicins;648–651 Sections 12.1 and 12.3) typically contain the common C-5 β-D-desosamine (177a). The corresponding N-desmethyl-β-D-desosamine (177b)634 is also found at this position as exemplified by the cineromycins [which also contain a C7 α-D-glucose (18)]652 C-3-O-glycosylation is also prevalent, albeit less important for bioactivity, and includes 3-O-methyl-α-L-mycarose (also known as α-L-cladinose, 90b),634,648,653,654 α-L-mycarose (90a; as observed in CP64593,640 sporeamicin-C650 or the 2-norerythromycins deriving from pathway engineering),655 or the C-3′/C-4′-modified form (90c/90d, Fig. 33) in megalomycin analogs.656
Additional C-3-appended sugars observed include 4′-O-acetyl-αL-arcanose (122b) [and corresponding engineered 2′,3′-anhydro-arcanose (182) and 3′-acetylated 180657,658] of the lankamycins659 and the kujimycins [which contain a C-3 α-L-arcanose (122a) or 4′-O-acetyl-α-L-arcanose (122b) and a C-5 β-D-lankavose (181b)]660 and α-L-oleandrose (184); found in oleandomycin647,661 and the 3-O-oleandrosyl-5-O-desosaminyl-(8S)-8-hydroxyerythronolide B662 from bioconversion. Additional sugars attached to C-5 include 3-keto-4,6-dideoxy-β-D-hexopyranose 179 (CP-63693640), β-D-chalcose (181b; lankamycins659 and kujimycins654) and β-D-mycaminose (185; narbolide,663 via biotransformation). C-6 substitution is restricted to α-L-megosamine (45; megalomycin656 and SF2748664) while that of C-9 is limited to variants of α-L-rhamnose (14–18; lyngbyaloside and lyngbuilloside665,666). Arguably the most unique glycosylation among 14-membered macrolides is the α-L-cladinose (90e) orthoester of erythromycin E originally produced via bio-conversion667 and later discovered as a metabolite of a pathogenic Nocardia.668 Such orthoester glycosides are more typically associated with orthoester natural products (see Section 14). Classical 12-, 14- and 16-membered macrolides function as antibacterials in a mechanistically similar manner as described in Section 12.3.
12.3. 16-Membered macrolides
Bacterial 16-membered macrolide glycosides represent the largest subgroup with a total of 284 glycosylated members reported thus far primarly from actinomycetes. This group is further divided into two subgroups based upon macrolactone ring architecture of as depicted in Fig. 33, where the predominant subgroup 16-A contains classical antibacterial macrolides reminiscent of their smaller counterparts described in the preceding sections. As with other classical macrolides, the prominent observed glycosyl regioselectivity within subgroup 16-A is C-3 and/or C-5 where some members also display C-9 and/or 14-C O-glycosylation. A total of 21 sugar monomers are represented among 16-membered macrolides including β-D-desosamine (177a, typically C-5), β-D-mycaminose (185, typically C-5) and α-L-mycarose (90a, typically C-5/C-3) commonly found in 12- and/or 14-membered counterparts as illustrated by rosamicins,669 mycinamicins,670–678 juvenimicins.679 Similar to the 12-membered comparators, C-3 3′-N-oxo-β-D-desosamine (also called desosamine N-oxide, 178) is observed among rosaramicins from Micromonospora680 while the only pentose (α-D-arabinofuranose, 191) is found at this position in epothilones.681 C-5-O-glycosides of 4′,6′-dideoxy-β-D-glucose (181a), or subtle variations thereof (such as β-D-chalcose, 181b, or 2-propionyl 181c) are also observed as exemplified by chalcomycins,682 GERI-155683 and neutramycin A.684 Angolamycins, in contrast, carry the 2′-deoxy-β-D-mycaminose (β-D-angolosamine, 59)685 while β-D-aldgarose (190b) and its acyclic precursor (190a) are found at the same position of aldgamycins686–691 and swalpamycin.692,693 C-4′-O-glycosylation of the C-5 β-D-mycaminose (185) is also a fairly common occurrence within this subgroup (disaccharides I–IV, Fig. 33)694–696 where four L-sugars [α-L-amicetose (97), α-L-cinerulose (51), α-L-rhodinose (44) and α-L-mycarose (90a)], or subtle modifications thereof, make up the terminal sugar.696–706 The angolamycins are distinguished within this context by their C-5-initiating sugar as described above (α-L-mycarose, 90; disaccharide V). The highly deoxygenated sugar β-D-forosamine (63) is restricted to C-9-O-glycosylation as found in the spiramycins (also known as shengjimycins)707–711 and chimeramycins.699 Finally, C-14-O-glycosides have also been reported where the sugars utilized include: β-D-mycinose (170c in tylosin,703 neutramycins,684 mycinamycins678 and GERI-155683); 6′-deoxy-β-D-allose, 6′-deoxy-2′-O-methyl-β-D-allose, or 4′-O-propionyl-β-D-mycinose (170a, 170b or 170d, respectively, in mycinamycin677 and tylosin analogs712); β-D-boivinose and β-D-digitoxose variants (187a/b and 131a/b, respectively in engineered tylosins703) as well as 3′,6′-dideoxy-4′-keto-2′-O-methyl-2′,3′-unsaturated-β-D-glucose (188) also via pathway engineering.713
A small set of 16-membered glycosylated macrolides are distinguished by the subgroup 16-B branched macrocyclic lactone architecture, where O-glycosylation occurs at C-21 (Fig. 33). Examples include bafilomycin-A1-rhamnoside (α-L-rhamnose, 46a)714 leucanicidin (2′-methoxy-α-L-rhamnose, 46e)715 and formamicin (β-D-olivose, 53).716,717
The final sub-group of the 16-membered macrolides are the avermectins (sub-class 16-C, Fig. 33).718–720 They are 16-membered macrolides produced by Streptomyces, Amycolatopsis and Nocardia. Avermectins typically contain a C-13 disaccharyl substitution (disaccharide VI). More recently, a variety of sugar analogs of this sub-class have been made via chemoenzymatic methods.721
Classical 12-, 14- and 16-membered macrolides inhibit bacterial protein synthesis by blocking the 50S ribosomal peptide exit tunnel.722,723 Core contributions to the specific macrolide-ribosome interaction derive from the C-3 sugar (e.g., desosamine, 177a) 2′-hydroxyl and 3′-dimethylamino groups, the latter of which has been deemed most important, based upon both macrolide-ribosome complex structure elucidation724 and bioactivity assessment of analogs.725–727 Glucosylation of 2′-hydroxyl group of this aminosugar is an established mechanism of macrolide resistance,728–730 where the corresponding O-glucosyltransferases have been recognized as highly permissive.731,732 C-5-disaccharyl-substituted macrolides further perturb the relative positioning of the 3′-end of P-site bound tRNA and 23 S rRNA in the ribosome where the terminal sugar of the disaccharide moiety extends into the peptidyl transferase center.684,702,733–735 Some classical macrolides (e.g., megalomicins) have been found to inhibit protein trafficking in the golgi and, while the megalomicin C-6 α-L-megosamine (45) has been put forth as a contributor to this activity,736,737 this contention has not been experimentally validated. A number of 14-membered macrolides have also been noted for their immunomodulatory activities,738–740 the mechanism for which remains poorly understood. Exceptions to the classical mode of action, the epothiolones are potent tubulin-targeted anticancer agents that advanced to clinical evaluation,741,742 subgroup 16-B members such as bafilomycins are potent inhibitors of vacuolar type H+-ATPase,743,744 and the avermectins are potent antihelmenthics.745
12.4. 18-Membered macrolides
There are only 23 reported bacterial 18-membered glycosylated macrolides the majority of which are tiacumycins and lipiarmycins746,747 from Micromonospora, Dactylosporangium, Actinoplanes and Catellatospora. All analogs share a common macrolactone ring O-glycosylated at C-11 with variants of the branched 5′-C-methyl-β-L-rhamnose (193a–f) or at C-20 with variants of β-L-rhamnose (322a) (Fig. 34, 18-A). TAN-1323 A-C (18-B)748 and the recently isolated biselyngbyaside (18-C)749 are distinguished from the tiacumicins by their C-17 side chains and corresponding glycosylation patterns. TAN-1323 A-C are C-23-O-glycosides of β-D-olivose (53a), 4-carbamyol-β-D-olivose (53b) and β-L-rhamnose (322a), respectively, while biselyngbyaside is a C-3-O-glucoside (2′-O-methyl-β-D-glucose, 104b).
Fig. 34.
18-, 20-, 22-, 24-Membered macrolactones and macrodiolides and associated sugars.
12.5. 20-Membered macrolides
Venturicidins, irumamycins, ammodicin and apoptolidins are the only known glycosylated 20-membered macrolides from Nocardiopsis, Saccharotrix, and Streptomyces (Fig. 34). Glycosylation regiospecificity is variant across this series with C-9-O-glycosides represented by ammocidin (6-deoxy-α-L-glucose, 194a) and apoptolidin (4′-O-methyl-6-deoxy-α-L-glucose, 194b). C-13-O-glycosides include the venturicidins (β-D-olivose, 53a) and irumamycin (3′-carbamoyl-β-D-olivose, 53c).750–752 C-24-O-glycosides are disaccharide substitutions as exemplified by ammocidin [disaccharide I comprised of β-D-olivomicose (192) and β-D-digitoxose (131)].753 In contrast, apoptolidin contains a C-27-O-disaccharide [disaccharide II comprised of 3′-O-methyl-β-D-olivose (53d) and α-L-chromose (195)].754 Apoptolidin specifically induces apoptosis in E1A-transformed glial cells with little or no detectable normal cell toxicity and is believed to function via inhibition of mitochondrial F1F0-ATPase where the disaccharide appears to be dispensable.754–756 Venturicidins and irumamycins are potent antifungals believed to inhibit ATP-driven proton transport and hydrolytic processes.757,758 Interestingly, X-14952B which differs from irumamycin in the C-19 alkyl side chain possesses antibacterial activities.759
12.6. 22-Membered macrolactones
Pulvomycin from Streptouerticillium represents the only bacterial 22-membered macrolide (Fig. 34).760 This metabolite is a C-33-O-glycoside of 2′,4′-di-O-methyl-β-D-fucose (270). Pulvomycin inhibits protein synthesis by preventing the formation of the elongation factor Tu (EF-Tu)/GTP/aa-tRNA ternary complex, the pulvomycin sugar contribution to which remains unknown.761
12.7. 24-Membered macrolactones
Macrolactins,762 archazolids763 and maduralides764 represent 24-membered glycosylated macrolactones from Bacillus, Cystobacter, Archangium and Maduramycetes (Fig. 34). Macrolactins and archazolids are C-7 and/or C-15-O-β-D-glucosides (104a). Glycosylation in these molecules has been demonstrated to suppress their cytotoxic effects. Maduralide, a weak Gram-positive antibacterial, is a C-13-O-glycoside of 6-deoxy-3′-O-methyl-β-L-talose (198).
12.8. Macrodiolides
A small set of glycosylated macrocyclic dilactones have been reported from Streptomyces and Microbiospora. Elaiophylin, SNA-4606-1 and efomycins are C-13/13a-di-O-α-L-oliosides (49a),765 where the sugars are 3′-O-methylated in (49b) efomycin A.766 Bispolides from Microbiospora are comprised of larger dilactone rings containing 2′,6′-dideoxyhexose 196 at C15a and the same sugar or 6′-deoxyhexose 197 at C-15.767 Axenomycins from Streptomyces are C-43-O-disaccharyl-substituted metabolites [disaccharide III comprised of β-D-amicetose (48) and L-axenose (199, anomeric stereocenter undefined)].768 Liposidolide A encompasses a C-15-O-β-D-mannoside (156) and C-41-O-β-D-olivoside (53e), the latter of which is modified as depicted (Fig. 34).769 Efomycins are non-toxic inhibitors of selectin-mediated leukocyte adhesion and have been considered as candidates for further development to treat autoimmune disorders.770 Elaiophylin, efomycin G and SNA-4606 inhibit testosterone 5α-reductase and display growth-promoting, antiviral and antibacterial activities. Glycosylation does not impact upon efomycin activity and the glycosyl contribution to the latter metabolites has not been studied.
12.9. Super macrolactones (ring size 32–48)
There are 25 glycosylated super macrolactones (defined as macrolides with ≥32-membered ring systems) from bacterial sources including Streptosporangium, Streptomyces and Nocardia (Fig. 35). Brasilinolides are C-37-O-α-D-oliosyl (71a) bearing 32-membered macrolides where brasilinolide C contains unmodified α-D-oliose (71a),771 brasilinolide A carries 3′-O-acyl-α-D-oliose (71b),772 and bracilinolide B has the permethylated counterpart.773 Liposidolide A has two appended sugars, a C-15-O-α-D-mannose (160) and a C-41-O-β-D-olivose (53) derivative modified at C-3′.769 The 34-membered macrocyclic sporaviridins from Streptosporangium are glycosylated to a greater extent with a C-13-O-pentasaccharide (III and IV), a C-21-O-β-D-glucose (104) and a C-47-O-α-L-vancosamine (118).774 The core of the sporaviridin pentasaccharide (III and IV) is a tri-substituted β-D-quinovose (140) which contains a C-2′-O-β-D-glucose (104) or 4′-amino-4,6-dideoxy-β-D-glucose (202), a C-3′-O-3′-amino-3,6-dideoxy-β-D-glucose (59) and a C-4′-O-disccharide comprised of β-D-quinovose and 3-amino-3,6-dideoxy-β-D-glucose. The 32-membered macrolides notonesomycin775 and A-77951776 both possess C-37–O-glycosides. The former contains both 3-O-methyl-β-D-olivose (53) and β-L-rhodinose (169), connected via a modified para-aminobenzoic acid (fragment II) while the latter contains an undefined modified disaccharide (205, disaccharide I). The recently discovered 51-membered stambomycins represent the largest glycosylated bacterial macrolide and share a common C-5-O-N,N-dimethyl-3′,6′-dideoxy-β-D-glucoside (185b).777 Primycins (C-18-O-α-D-arabinofuranose, 204) and mathemycins (C-18-O-β-D-mannose, 156) both carry an O-glycoside three carbons from the macro-cyclic ring forming oxygen. In addition, mathemycin contains a C-37-O-3′-amino-2,6-dideoxy-β-D-glucose (185a).778 Finally, the 42-membered desertomycins and 48-membered monazomycins both share a C-22-O-α-D-mannose (160).779–783
Fig. 35.
Super macrolactone (ring size 32–48) aglycons and associated sugars.
In addition to antifungal activities, brasilinolide A and A-77951 were noted for their immunosuppressive effects.772,776 Stambomycins, moderatively active against Gram-positive and Gram-negative bacteria, also were reportedly cytotoxic against a number of human cancer cell lines.777 It should also be noted that acetylation of the sporaviridin aminosugar abolished antimicrobial activity.774
12.10. Polyene macrolide
A total of 49 glycosylated polyene macrolides have been reported from Streptomyces, Actinoplanes, and Sorangium. Members within this group share a common macrocyclic lactone of various sizes (26 to 38-membered rings) with 4–7 conjugated double bonds where the classical antifungals amphotericin and nystatin serve as prototypical examples. With the exception of chivosazoles784–786 aglycons can be further distinguished by the presence or lack of an endocyclic pyran ring (core structures A and B, respectively; Fig. 36).
Fig. 36.
Polyene and macrolactam aglycons and associated sugars.
The aminosugar β-D-mycosamine (206) is the predominate O-glycoside among polyene macrolides as exemplified by amphotericins,787 candicidins,788 candihexins,789 vacidin-A,790 SCH 16656 complex (67–121 analogs)791 and polyfungins.792 With the exception of sorangiosides (glycosylated with β-D-glucose, 104),793 regioselectivity of glycosylation as β to the pyran ring is strictly conserved among prototypical polyene members (Fig. 36, position a, β-D-mycosamine 206). Some polyfungins carry an additional C-35-O-α-L-digitoxose (68) and the mycosamine C-4′ is further modified with 6′-deoxy-β-D-mannose (209; disaccharide I) in 67-121C (Fig. 36). The 44-membered polyene antibiotic lienomycin is distinguished by its C-25-O-α-L-rhamnose (46),794 while DJ-400-B and B2 are the only members with a C-5 β-D-mycosamine (206) (Fig. 36, position a).795 The appended mycosamine is important to the antifungal activity of these metabolites where subtle changes dramatically influence potency.796 While systemic toxicity presents a challenge to the clinical use of amphotericin, recent work with analogs bearing 2-deoxy mycosamine suggest a notable improvement of selectivity for ergosterol-containing fungal membranes.797 As with other bacterial systems, reactions catalyzed by polyene glycosyltransferases have been found to be reversible and used to generate differentially-glycosylated polyene analogs.798
Distinct from the prototypical antifungal polyenes, chivosazoles are cytotoxins that contain an integrated aglycon oxazole ring and are C-11 glycosides (β-D-quinovose, 140).784,785
12.11. Macrolactams and related glycosylated metabolites
Similar to the macrolactones discussed in the previous section, macrolactams, from Streptomyces, Actinosynnema, Amicolatopsis, Actinomadura, Maduromycetes, Microtetraspora, Nonomuraea and Pseudonocardia are large macrocyclic structures but are distinguished by a ring-closing amide bond. Compared to macrolactones, glycosylated macrolactams are fewer in number (53 total) and display variant glycosyl regiospecificity as summarized in Fig. 36.18 Many members contain an integrated aromatic ring as part of the overall macrolactam aglycon, the exceptions being fluvirubicins, SCH 39185, SCH 38511, SCH 38516, SCH 42729, SCH 42282, cremimycins, vicenistatin, and incednine. Tolypomycins from Streptomyces stand out by virtue of their conjugation to the C-4′-amine of tolyposamine (203a) or via a corresponding C-4′-imine in tolypomycin-Y (203b).18 Ansamitocinosides from Actinosynnema are unique as N-β-D-glucosides (104),799 the sugar of which can be further modified as in ansacarbamitocins from Amicolatopsis.800 In contrast, the phenolic hydroxyl is the glycosyl acceptor in the related C-22-Oβ-D-glucosyl-mycotrienin II.801
Fluvirucins,802,803 SCH 39185, SCH 42729, and SCH 42282 are 14-membered macrolactams bearing either a C-3-O-3′,6′-dideoxy-3′-amino-α-L-talose (73) or C-9-O-glycoside comprised of the same sugar or α-L-mycosamine (207) (Fig. 36).804 The C-2′-OH of α-L-mycosamine (207) is further glucosylated with α-L-glucose (211, disaccharide II, SCH-42729) which can also be C-4′-O-glucosylated with β-L-glucose (210, trisaccharide III, SCH-42282).805 Modified macrolactams, ansaetherone,806 and tetrapetalones (described in details in Section 23.5.3.),806 Q-1047-A807 and its hydroquinone analog Q-1047-R-A all are believed to contain C-9-O-β-D-rhodinose (56), although in some cases, the nature of the sugar has not been completely defined. The vicenistatins also contain a single C-7-O-glycoside comprised of β-D-vicenisamine (212), 3′-epi-β-D-vicenisamine (213), or β-D-mycarose (64)808 while cremimycin contains a C-10-O-3′-O-methyl-β-D-digitoxose (131).809 Finally, incedine from Streptomyces bears a C-11-O-disaccharyl moiety constructed from the two amino sugars 4′-N-methyl-β-D-forosamine (63) and 2′-deoxy-2′-methyl-amino-β-D-xylose (173).810
Glycosylated macrolactams are known to possess a variety of biological activities including antifungal, antibacterial, antihelmintic, antiviral, antitumor, radical scavenger and phospholipase C inhibition.800,801,803,806,809–812 While the specific impact of the sugar upon bioactivity has not been delineated in most cases, recent studies in which the vicenistatin D-vicenisamine was replaced with the D-mycarose led to a complete loss of activity, implicating the aminosugar as important to metabolite cytotoxicity.808 It should be noted that the glycosyltransferase involved in vicenistatin biosynthesis displays broad substrate tolerance and also was among the first to be studied in the context of glycosyltransferase reversibility.813,814
12.12. Spirotetronates
Spirotetronates are pentacyclic polyketides that contain a signature fused 6-membered/γ-butyrolactone spiro-bicyclic ring substructure and are produced by Amycolatopsis, Streptomyces, Actinomadura and Micromonospora. Several spirotetronate O-glycosides have been reported (Fig. 37) where dominant glycosyl regioselectivity observed is α- to the bicyclo[4.4.0]decene ring fusion (as exemplified by C-7 of chlorothricins or C-9 of tetrocarcins). Additional O-glycosylation is observed at C-17/19 of the largest macrocyclic ring in some members (as exemplified by tetrocarcins and versipelostatins, respectively) and, in one case, C-6 (PA-46101 B).
Fig. 37.
Spirotetronates and spinosyn aglycons and associated sugars.
As noted above, C-7/C-9 O-glycosylation is a predominate feature. Examples of C-7 glycosylation include disaccharide I comprised of two β-D-olivose (53) units (chlorothricins,815 MC-031 and MC-033816), disaccharide II comprised of a β-D-olivose (53) and β-D-quinovose (140) (hydroxychlorothricins,815 K818,817 MC-032, and MC-034),816,817 or disaccharide III continaing 6′-deoxy-β-D-allose (170) and 5′-O-methyl-β-D-digitoxose (131) (PA-46101A and B).818 The terminal olivose sugar (53) of these metabolites is often further modified as observed in chlorothricin and related analogs (Fig. 37, appendages i–iii) and, as previously noted, PA-46101 B also contains an additional C-6-appended 3′-C-methyl-α-L-rhamnose (108). C-9 glycosylation also predominates with monosaccharyl substitution observed on occasion (e.g., the acylated trideoxy amino sugar 213 in BE-45722,819 BMY-42448,819 decatromicins,820 pyrrolosporin A389) and oligosaccharyl substitutions units typically containing α- and/or β-L-digitoxose (68 and 176, respectively) and α-L-amicetose (97) as more prevalent (e.g., oligosaccharides IV–VII, tetrocarcins821 and AC6H);822 disaccharide VI, arisostatins;823 trisaccharide VIII, lobophorins;824,825 tetrasaccharide IX, kijanimicin.826,827 The α,β-unsaturated aldehyde-containing deoxysugar (216) of tetrocarcin K stands out as a unique glycoside among this series (Fig. 37, VII).828
C-17 and C-19 glycosylation is less common. Examples of C-17 glycosylation include the unique nitro sugar β-D-kijanose (also known as β-D-tetronitrose, 214; tetrocarcins,821,822 lobophorins B,824,825 kijanimicin,826,827 arisostatin A823), the corresponding amino analog 218 (AC6H, lobophorin A,824,825 arisostatin B823), and variations of 3′,4′-diamino-2′,3′,4′,6′-tetradeoxy-β-D-galactose (219, MM 46115).829 C-19 glycosylation is restricted to the larger versipelostatin aglycon and employs a series of oligosaccharides (X–XVI, Fig. 37) comprised of β–D-digitoxose (131), 3′-O-methyl-α-D-olivose (183) and/or α-L-oleandrose (184) monomers.830–832
Spirotetronates possess antitumor, antiviral and antimicrobial activities where metabolite glycosylation, in most cases, is critical to activity.829 For example, a reduction in tetrocarcin glycosylation correlates to diminished antibacterial activity,833 while the sugar variation among lobophorin analogs contributes to antibacterial activity modulation.825 Sugar variation of MC-031 congeners also contributes to potency modulation in the context of cholesterol biosynthesis inhibition.816 Versipelostatins were reported to down-regulate the GRP78 molecular chaperone which has potential application in cancer, Alzheimer’s and Parkinson’s.832
12.13. Spinosyns
Spinosyns are a structurally unique group of reduced polyketide macrolactams from Saccharopolyspora. A total of 23 spinosyn O-glycosides have been reported thus far.18,834 Spinosyns contain two O-glycosides critical to their insecticidal activities, a C-9 α-L-rhamnose (46) and a C-17 β-D-forosamine (63) (Fig. 37). Spinosyn variation is based, in part, upon differential methylation of the corresponding glycosides and SAR studies revealed both sugars to contribute to bioactivity.835
13. Nucleosides and nucleoside-derived compounds
Compounds in this class are divided into pyrimidines (uracil- or cytosine-derived) and purines (adenine- or guanine- derived) metabolites. While metabolites with a modified ribose core are included herein, classical ribose-based nucleosides are considered outside the scope of this discussion. A total of 280 glycosylated bacterial metabolites are encompassed within this broad class where pyrimidine nucleoside antibiotics comprise the predominate group.836
13.1. Pyrimidine-derived nucleosides
13.1.1 Uracil-derived nucleosides
The first major subclass of uracil-derived nucleosides includes streptovirudins, tunicamycins, and corynetoxin isolated from Streptomyces, Bacillus, and Coryne-bacterium-infected Lolium.837–840 This group includes 23 metabolites and structural signatures include a uracil or dihydrouracil (Fig. 38), an amide-linked fatty acid, a dialdose sugar attached to the N-1 of the uracil moeity, and N-acetylglucosamine (5). The corresponding dialdose sugar, a unique 11-carbon amino sugar (tunicamine, 222) comprised of a carbon-carbon bond fusion between C-6 of galactosamine and the uridine ribose C-5, is connected head-to-head to N-acetyl-α-D-glucosamine (5) (Fig. 39).841 While potent antibacterial cell wall inhibitors, these agents lack clinical utility due to their inhibition of N-linked protein glycosylation in mammalian cells.842
Fig. 38.
Uracil-derived aglycons.
Fig. 39.
Sugars associated with uracil-derived aglycons. Pink fragment is absent in some derivatives.
The next subclass, the uridylpeptide antibiotics, includes mureidomycins,843,844 napsamycins,845 pacidamycins,846,847 and sansanmycins from Streptomyces and Pseudomonas.848–850 The 25 members of this group share a common skeleton that consists of a uracil or dihydrouracil attached to a 4′,5′-unsaturated-3′,5′-dideoxyribose (332) at the uracil N-1. This modified ribose is connected to a 2,3-diaminobutyric acid (DABA) via a 4′,5′-enamide linkage to which are appended various amino acids (Fig. 39). The uridylpeptides selectively inhibit bacterial translocase (MraY) and, while both the modified ribose and uracil are critical to activity, analogs in which the enamide has been reduced retain activity.851
Twenty-two glycosylated muraymycins and related metabolites from Streptomyces comprise the next subclass.852 The core structure is composed of a peptide, uracil, uronic acid (330), and a modified ribose which terminates with a hexahydro-2-imino-4-pyrimidylglycyl (epicapreomycidine)–urea–valine moiety (Fig. 39, X = i). Members of this family differ via either the terminal amino sugar [5′-amino-5′-deoxy-β-D-ribose (221), 2′-methoxy-221, or 5′-amino-2′,5′-dideoxy-β-D-ribose (224)] and/or the fatty acid (Fig. 39). Three related epicapreomycidine-containing analogs [AA-896-A6, C5 and D4 (Fig. 39, X = i)] contain sugar 221 attached to C-5′ of the ribose moiety of uridine.853 Muraymycins also effectively inhibit bacterial translocase (MraY). Interestingly, while removal of the aminoribose primary amine abolishes activity,854 derivatives lacking aminoribose remain active.855
Eighteen glycosylated capuramycins (A-500359) and related metabolites produced by Streptomyces and Amycolatopsis comprise the next subclass. In addition to the structural aminocaprolactam signature (Fig. 39, X = ii), the capuramycin skeleton consists of a uracil nucleoside with a unique 4′,5′-unsaturated α-D-mannuronic acid (225) connected to the C-5′ of a modified ribose (331 forming disaccharide III, Fig. 39). Exceptions include A-500359 D, which contains the 2′-deoxy analog of 225 (333), and A500359 J which bears the C-4′-C-5′-hydrated form of 225 (α-D-taluronic acid, 52).856 Inhibition of lysine biosynthesis, via feeding the capuramycin production strain 2-aminoethyl-L-cysteine, led to additional analogs where the signature aminocaprolactam moiety was lacking (A-500359 E, F, F-amide, H), or replaced with acetylcystamine (M-1) (Fig. 39, X = iii) or a thiazepanone moeity (M-2) (Fig. 39, X = iv).857–859 A-503083 A, B, E, and F also contain disaccharide III composed of 225 and 331.860 A-102395, which has a benzene group in place of the caprolactam moiety, is the only analog isolated from a non-Streptomyces species.861 Additional modifications of capuramycin-type metabolites include ribose 2′-O-acylation and 3′-O-methylation, the former of which has been noted to improve cell permeability.862 Members of this class also function as translocase (MraY) inhibitors and display notable anti-tubercular activity but suffer from poor bioavailability. Based upon existing SAR, a core structure containing the modified ribose, an unsaturated hexuronic acid and an aminocaprolactam offers the best activity.
The liposidomycins and caprazamycins from Streptomyces comprise the next subclass and the 38 glycosylated members within this group share a common structural skeleton that is composed of uracil, a modified ribose (330), a corresponding ribose C-5′-appended aminoribose (5′-amino-5′-deoxy-β-D-ribose, 221) and an acylated diazepanone (Fig. 39, X = v). Liposidomycins are subclassified into four types based on the presence or absence of the aminoribose (221) 2′-sulfate and/or the diazepanone side chain C-3 modifications (Fig. 39, X = v).863–865 Type I contains both groups while type IV lacks both. Type II and III lack the diazepanone glutaryl modification and sulfation, respectively. Caprazamycins resemble type III, and to lesser extent type I, liposidomycins but contain an additional 2′,3′,4′-tri-O-methyl-α-L-rhamnose (46) attached to C-5 of diazepanone-appended methyl-glutaryl group (Fig. 39, appendage v). In A-90289 A and B, C-2′ sulfation of the modified ribose 330 rather than aminoribose 221 is observed.866,867 Liposidomycins, caprazamycins, and A-90289 also inhibit translocase (MraY) where the type I analogs were found to be most active.836,868 Synthetic derivatives modified at the ribose C-5′ display improved potency.836
Ezomycins are a group of pyrimidine-based N- and C- glycosidic antifungal compounds isolated from Streptomyces.869–877 Glycosylation within this subclass occurs at either the N-1 (ezomycins A1 and A2), C-5 (ezomycins B1, B2, C1, C2), or C-8 (ezomycins D1/D2) (Fig. 38) where the predominate glycosylation signature is comprised of a trans-fused furopyranoside 3′,7′-anhydrooctose (236) which is further glycosylated at C-6′ with 3′-amino-3′,4′-dideoxy-β-D-glucuronic acid (235, also called ezoaminuroic acid).875 This octosyl nucleoside-ezoaminuroic acid (235) ezomycin core is sometimes referred to as ezomycin disaccharide (VII). Exceptions include ezomycins D1 and D2 which carry a 3′-amino-3′,4′-dideoxy-β-D-glucuronic acid-(1→4)-3′-deoxy-3′-ureal-β-D-guluronic acid disaccharide (VIII). Some ezomycins are further substituted via a hexuronic acid C-6′-L-cystathionine amide, a modification important to antifungal activity.
Nikkomycins (also referred to as neopolyoxins)878–883 and polyoxins884,885 are peptidyl nucleosides isolated from Streptomyces that contain a pyrimidine aglycon typically comprised of uracil, thymine or, in some cases, 4-formyl-4-imidazolin-2-one. For simplicity, all glycosylated nikkomycins and polyoxins regardless of the base are included herein. Nucleoside N-1-glycosylation with 5′-amino-5′-carboxy-5′-deoxy-β-D-ribose (223) is a predominate feature (Fig. 39) where this hexuronic acid is typically further modified in most nikkomycins by a rare amino acid hydroxypyridylhomo-threonine via a C-6′-amide linkage. Exceptions include nikkomycins Sx/Sz and Sox/Soz, which carry the bicyclic sugars 227 and 228, respectively,882 and the uracil C-5-C-glycosides nikkomycins pseudo J/Z. Octosyl acids are also structurally related to nikkomycins and polyoxins but lack the aminohexuronic acid.886 Derivatives A and B contain modified pentose 227 while C contains the keto derivative 229. While nikkomycins and polyoxins are potent inhibitors of chitin biosynthesis in fungi and display notable antifungal activity,887 their unfavorable physicochemical properties has hampered their clinical development.
Other uracil-based metabolites include the C-5-C-nucleosides malayamycin A and de-O-methylmalayamycin A from Streptomyces, which are inhibitors of fungal sporulation and contain the bicyclic sugar 230 (Fig. 39).888–890 A bacterial translocase inhibitor, A-94964A contains two unidentified sugars, 2′-deoxy-2′-aminohexosyl-1-phosphate (234) and a hexose (334), attached to a riboocturonic acid (233) appended to N-1 of uracil.891,892
13.1.2 Cytosine-derived nucleosides
The first main group in this subclass includes amicetins, cytosaminomycins, and SF-2457. This group is comprised of eleven metabolites from Streptomyces, Arthrobacter, and Nocardia,893–899 most of which contain a N-1-β-disaccharide (I or II) and p-aminobenzamide-modification of the cytosine 4-NH2. The dissaccharide is comprised of either β-D-amicetose (48) or β-D-olivose (53) attached α-(1′→4′) to either α-D-amosamine (39) or N-demethyl-α-D-amosamine (39). Amicetin inhibits the bacterial peptidyl transferase by binding 23S-like rRNA.900 Amicetins were also found to be active against herpes virus 1 and poliovirus901 cytosaminomycins have been noted for anticoccidial activity.898,902
Mildiomycins from Streptoverticillium are a group of cytosine N-1-glycosidic peptidyl nucleosides bearing C-2′-C-3′ unsaturated C-4′-N-acyl-β-D-hepturonic acid derivatives (241 and 242) (Fig. 40).903–905 These molecules have activity against powdery mildews and function as protein synthesis inhibitors via binding the ribosomal large subunit.906 The related sugar 240a and corresponding esters are present in the antibacterial arginomycin and related 10381 series from Streptomyces,907,908 while 240b or its hydrated congener 243 exists as part of the antifungal blasticidins from Streptomyces. Cytosylglucuronic acid bearing a N1-2′-deoxy-β-D-glucuronic acid (249) was isolated from the same strain while cytosyl-N-1-glycosides bearing unique 4′-amino-, keto-, or oxime-functionality (244a, 244b, 245, and 246), were also isolated from bioconversion experiments with this same blasticidin producer.909–913 Additional studies, in conjunction with enzyme inhibitors, yielded an even extended glycoside diversity to include β-D-glucose (104), 261–265, 136, 72 and 243 attached to pentopyranines.910,913–915 Hikizimycin, also referred to as anthelmycin, from Streptomyces is a cytosyl-N-1-β-D-dissacharide (III), the latter of which is comprised of 3′-amino-3′-deoxy-β-D-glucose (kanosamine, 250) linked β-(1′→6′) to an unusual 4′-amino-undecose (hikosamine, 256) (Fig. 40).916–918 Hikizimycin displays antiparasitic activities and inhibits protein synthesis.919 The Bacillus metabolites bagougeramines A and B both contain modified cytosyl-N-1-4′-amino-4′-deoxy-β-D-glucuronamides (251, appendage iv) and function as antimicrobials.920 Sugar 251 was also found to be present in mitaimeisu (appendage v),921 gougerotin (appendage vi),922 and possibly ningnanmycin;923 although in the latter case the C-3′-stereochemistry remains undefined (247). Albomycins from Streptomyces are siderophore antibiotics similar to other sideromycins (Section 15.4) are composed of iron chelating portion in addition to a 6′-amino-6′-deoxy-4′-thio-heptofuranose (315) uronic acid moiety (Fig. 40).924,925 Dapiramicins A and B from Micromonospora are N-glycosidic pyrrolopyrimidines bearing 6′-deoxy-α-D-glucose (215) or β-D-glucose (104), respectively, each capped (β1→4) with 4′-methyl-β-D-glucose (104).926,927 The α-configured dapiramicin A with disaccharide IV possessed significantly higher activity against the plant disease, sheath blight than the analog bearing V.
Fig. 40.
Cytosine-derived aglycons and associated sugars.
13.2 Purine-derived nucleosides
13.2.1 Adenine-derived nucleosides
There are 32 adenine-based glycosides reported where N-glycosylation is restricted to the adenosyl N-9 and C-6-amine. The adenosyl N-9-amino-pentose 3′-amino-3′-deoxy-α-L-ribose (253) was found in puromycin928 and A-201-A, C, D, and E,929,930 from Streptomyces (Fig. 41) where the 3′-N-tyrosyl moiety contains additional glycosylation in the A-201 series (335 in A, C, and D; 336 in E – capped with 3′,4′-di-O-methylated α-L-rhamnose 46 in A, C, and E).
Fig. 41.
Purine-derived aglycons and associated sugars.
Sinefungins from Streptomyces are a group of adenosyl-N-9-glycosidic antileshmanial and antitrypanosomal metabolites signified by an unusual ribose-derived 10-carbon sugar, likely formed via a carbon–carbon bond-forming reaction between adenosine and an amino acid.931–933 Sugar 337 and its amide derivative 339 are present in sinefungin and ureidosinefungin, respectively. Sugar 339 is also present in sinefungin VA with appendage iii attached to the terminal amino group (Fig. 41). Sugar 340 is a cyclic version of 339 and is present in cyclosinefungin. Sugars 252 and 338 are 4′,5′-unsaturated derivatives of 337 and 339, respectively, and are present in other sinefungin derivatives. Dehydrosinefungin V and KSA-9432 contain the same sugar 338 with appendages i and ii, respectively (Fig. 41). These metabolites function as S-adenosylmethionine (AdoMet) mimetics and methyltransferase inhibitors where derivatives with unsaturated sugars were found to generally be less potent.934
Herbicidins are a group of herbicidal compounds isolated from Streptomyces that contain an interesting tricyclic furanopyranoside (254) attached to the adenosyl N-9 (Fig. 41).935–939 The antitumor compound septacidin from Streptomyces contains a C-4′-amino-modified 4′-amino-4′-deoxy-β-L-glucoheptose (257) connected to C-6 amino group of adenine (Fig. 41, appendage vii). Replacement of the natural sugar with 4′-amino-4′-deoxy- or 4′-amino-4′,6′-dideoxy-L-glucose did not affect activity.940 C-4′-amino-modified analogs of the C-2′-epimer (4′-amino-4′-deoxy-β-L-mannoheptose, 255) are found in the antitumor compounds spicamycin and anicemycin from Streptomyces.941,942 Agrocin 84 is a diglycoside structurally related to sugar phosphate opines used to control crown gall disease in plants.943 Agrocin 84 contains a C-5′-modifed adenosyl-N-3′-deoxy-β-D-arabinofuranose (259) and a corresponding β-D-glucofuranoside (258) linked to the adenosyl C-6-amine via a phosphoramidate bond (Fig. 41 appendage ix). The β-L-gulosamine (260) in the pseudodisaccharide III, present in mitocides C-8030 C, D, and E from Streptomyces, is connected to the C-5′ of the ribose through a cyclohexane ring.944 Anhydrothuriniensin isolated from Bacillus contains a disaccharide IV composed of α-D-glucose (18) linked to the β-D-ribose sugar of adenosine.945,946 Griseolic acids isolated from a Streptomyces species, contain four unique nine carbon bicyclic sugars 341–344 (Fig. 41).947,948 They were found to inhibit cyclic nucleotide phosphodiesterase.949
13.2.2. Guanine and other miscellaneous nucleosides
The sugar α-D-mannose (160) is present in the chitin synthase inhibitor guanofosfocin A from Streptomyces, the only guanine-based glycoside reported.950 In this unique metabolite, the mannose serves to bridge the base C-8 and ribose C-5 of guanosine diphosphate (via connections at C-1′ and C-3′, respectively) (Fig. 41).
Miharamycins A and B,951 amipurimycin952,953 from Streptomyces belong to atypical purines. All are aminopurine N-9-glycosides where miharamycins contain modified bicyclic D-sugars (238) and amipurimycins the unusual modified D-sugar 239 (Fig. 41).
14. Orthoester glycosides
Orthoester glycosides are a group of metabolites that are characterized by the presence of one or more of acid sensitive orthoester glycosides. A total of 50 orthoester glycoside-containing bacterial metabolites have been reported, 47 of which fall within two major subclasses – orsellinic acids (subclass I) and aminocyclitols (subclass II). Three additional orthoester glycoside-containing outliers can be found within the macrolide (kanglemycin A and swalpamycin, Section 12) and the polyene (polyene SE-73, Section 11) sections.
14.1. Orsellinic acid derivatives
Members of the orsellinic acid subclass of the orthoester glycosides have attracted attention for both their exciting bioactivity and unprecedented oligosaccharide-based structural diversity. Members of this class possess potent activity against antibiotic-resistant Staphylococcus and Enterococci and include the everninomicins, avilamycins, and flambamycin.954 These unique metabolites function via inhibiting bacterial protein synthesis through binding at a unique site of the bacterial 50S ribosomal subunit.955,956 One member of this family, ziracin, advanced to phase III clinical studies but was ultimately discontinued due to side effects and poor pharmacokinetic properties.957
Sixteen avilamycins from Streptomyces contain a signature chloroisoeverninic C-4-heptasaccharide ester.958–960 Additional avilamycin-derived gavibamycins were generated through pathway engineering.961–964 The heptasaccharide moiety I of avilamycin A (Fig. 42), is composed of two units of β-D-olivose (A and B, 53), 2′-deoxy-β-D-evalose (C, 192), 4′-O-methyl-β-D-fucose (D1, 270), 2′,6′-di-O-methyl-β-D-mannose (E, 156), 2′-O-isobutyryl-α-L-lyxose (F, 111), and the C-4′-branched sugar methyleurekanate (G1, 272). Subtle variants where 272 has been substituted with 270 (β-D-fucose, avilamycin A1 and gavibamycin O), 273 (gavibamycins A3, B3, J3, K3, L3, E3, H3, I3) or 274 (avilamycin L) have also been reported. In addition, analogs where β-D-fucose (D1, 270) has been replaced by 4′-O-methyl-β-D-arabinose (D2, 265; avilamycin M) or β-D-galactose (D3, 161; avilamycin K) are also known (Fig. 42).
Fig. 42.
Orsellinic acid aglycons and associated sugars.
The closely related everninomicins965–968 from Micromonospora include both ziracin (evernimicin) and a series of SCH-compounds identified by Schering-Plough.969–971 Everninomicins are distinguished by an additional sugar (C-3′-nitrosugar α-L-evernitrose, H1, 127 or aminosugar H2, 280) attached to C-3′ of sugar A to provide octasaccharide II. Analogs that contain reduced nitro sugar such as the hydroxylamino (H3) or nitroso (H4) derivatives have also been obtained by reduction during structure elucidation or derivatization and have shown comparable antibacterial activity.967 A second delineation stems from the replacement of the avilamycin sugar G (β-D-methyleurekanate) by β-D-eurekanate 265 (everninomicin C, ziracin, SCH49088, SCH58775) (Fig. 42) or the C-4′-branched sugar 271 (everninomicins B and D, 13385-1).972 In some everninomicins, 2′-deoxy-β-D-evalose (C1, 192) is replaced by 6′-deoxy-3′-C-methyl-β-D-mannose (C2, 268). The hydrolytic products (sporacuracins and Sch-58777 which contain only sugars E, F, and G)973 lack antibacterial activity970 while removal of the nitro sugar has no major effect.958,974
14.2. Aminocyclitol derivatives
Members in this class are also considered aminoglycosides (Section 1) and include destomycins,975–977 hygromycin B,978 RH5012 C,979 and SS-56-C980 from different bacteria including Streptomyces, Streptoverticillium, and Saccaropolyspora (Fig. 43). The aglycon in this subclass contains an aminocyclitol moiety derived from D-streptamine (hyosamine, P7) bearing a C-5 glycoside comprised of either β-L-talose (144) or β-D-mannose (156). The signature orthoester glycoside linkage occurs between the C-2′/C-3′ and the anomeric carbon of the 7-membered amino sugar, destomic acid 275 (Fig. 43). It is worth noting that analogs which lack destomic acid (and thus, the orthoester linkage) are inactive.
Fig. 43.
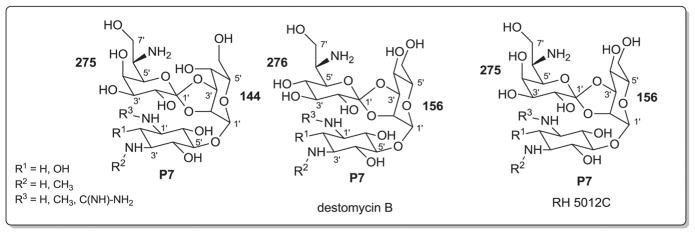
Sugars associated with orthoester aminocyclitols.
15. Peptides
Glycosylated peptides represent a large and clinically-important group of bacterial metabolites and its members invoke a broad range of activities particularly relevant to anti-cancer and anti-infective development. For the scope of this discussion, glycosylated peptides have divided into distinct sections covering bleomycins, vancomycins, mannopeptimycins, salmochelins and salmycins, linocosamides, thiazoles, lipoglycopeptides, miscellaneous antiinfective peptides and miscellaneous (other) members.
15.1. Bleomycins
Bleomycins are a group of cytotoxic glycopeptides from Streptomyces that bind both DNA and a metal [typically Cu(I) or Fe(II)] and invoke oxidative DNA strand cleavage.981–986 While members of this family (Blenoxane®) have been used with great success clinically to treat malignant lymphomas,478,986 dose-dependent pulmonary fibrosis and drug resistance restrict broader application. In conjunction with the structurally-related metabolites phleomycins,987,988 tallysomycins,989,990 zorbamycin,991,992 cleomycins,993,994 LL-BO 1208,995 and platomycin,996 the total number of glycopeptides in this class is 69.985 Structure divergence among members derive primary from subtle variations within the peptide linker, oxidative state of the bithiazole (BTT), the terminal substituted amide58,990,997,998 and, the presence (as in tallysomycins) or absence of a second point of glycosylation (Fig. 44). All members contain a conserved O-disaccharyl-substituted β-hydroxyhistidine, the disaccharide composition of which is 3′-O-carbamoyl-α-D-mannopyranosyl-(1′→2′)-α-L-gulose (I) in most [the exception being 3′-O-carbamoyl-α-D-mannopyranosyl-(1′→2′)-6′-deoxy-α-L-gulose in zorbamycin, II].999 Tallysomycins differ from other members via an additional O-glycosylcarbinolamide bearing 4′-amino-4′,6′-dideoxy-α-L-talose (279) at C-13 of the bithiazole moiety (Fig. 44). Zorbamycin-bleomycin disaccharyl hybrids have also been constructed via strain engineering.1000 The precise role of the disaccharide remains controversial and hypotheses include improving DNA affinity and cleavage985,1000,1001 as well as extracellular recognition and uptake.1002,1003 In support of the latter, the disaccharide moiety was demonstrated to target tumor cells possibly by a specific ATP-dependent uptake mechanism.1002,1003
Fig. 44.
Bleomycin aglycons and associated sugars. Platomycin structure is based on partial structure reported.
15.2. Vancomycins
Vancomycins are cyclic glycopeptides comprised of seven amino acids, the heptapeptide core of which presents two 16-membered and one 12-membered ring. Members are classified into four different subclasses based on the heptapeptide core and cumulatively, there are 133 corresponding bacterial glycopeptide members isolated from different bacteria including Streptomyces, Nocardia, Pseudonocardia, Amycolatopsis, Actinoplanes, Kibdelosporangium, Micropolyspora, Saccharothrix, Actinomadura.1004 Type I glycopeptides contain two aliphatic (residues 1 and 3) and five aromatic amino acids, while types II–IV contain seven aromatic amino acids (Fig. 45). Type III differs from type II by an additional ether-bridged ring system between residues 1 and 3 while type IV (generally referred as lipoglycopeptides) differs from type III via N-acyl-substitution of appended sugars. Other compounds that are structurally related to vancomycins but lack the sugar include the complestatins,1005–1007 chloropeptins,1008 and A-47934.1009 Each of the previous types, with a particular focus upon glycosylation, is further elaborated herein.
Fig. 45.
Aglycons of the five types of vancomycin. The ether bridge in types III and IV is highlighted in red. Locataions of attachment to most sugars in CWI-785 series is not determined.
Vancomycin from Amycolatopsis is the prototypical member of the type I sublcass. First discovered in the mid-fifties, the structure of this metabolite was not elucidated until 30 years later.1010–1012 Vancomycin is a potent antibiotic to treat life-threatening Gram-positive infections and inhibits bacterial cell wall biosynthesis by binding peptidoglycan L-Lys-D-Ala-D-Ala.1013–1015 In addition to vancomycin, there are 37 glycosylated type I metabolites. The predominate site of O-glycosylation within type I–IV members is the phenolic hydroxyl of residue 4 (hydroxyphenylglycine, HPG-4, Fig. 45). To a lesser extent, O-glycosylation of the substituted β-hydroxy-L-tyrosine (Tyr-6) is also observed among type I–IV members. HPG-4 O-glycosylation is comprised of a β-D-glucose (104, devancosamine vancomycin, M43C, most balhimycins, and chloro-orienticins B and E) or an α-L-sugar-(1′→2′)-β-D-glucose disaccharide (I–IV) where the L-sugar varies among members and includes: α-L-vancosamine (118, vancomycins)1016–1018 4′-oxo-α-L-vancosamine (281, balhimycin V, A-83850);1019–1021 4′-epi-α-L-vancosamine (280, eremomycin, orienticins, and chloro-orienticins A and D)1022 and its N-carboxymethyl derivative (280, eremomycin B);1023 or α-L-rhamnose (46, decaplanin, A-42867, MM 47761, and MM 49721) (Fig. 46).1024 Sugars employed for residue 6 tyrosine (Tyr-6) β-hydroxy O-glycosylation within type I members include: ureido-α-L-vancosamine (282, ureidobalhimycin);1019–1021 4′-oxo-α-L-vancosamine (281, balhimycins);1021 4′-epi-α-L-vancosamine (280; eremomycin,1022 orienticins, A, C, D, chloro-orienticins, decaplanin, MM 47761, MM 497211025); α-L-olivose (184, orienticin B);1026,1027 or α-L-vancosamine (118, MM 49727, A-42867) (Fig. 46).1024 OA-76531028,1029 from S. hygroscopicus is unique among type I members as it contains α-D-glucose (18) attached to the residue 6 tyrosine-β-hydroxyl but, similar to chloro-orienticins C and F, lacks residue 4 phenolic hydroxyl glycosylation. Importantly, the broad substrate specificity of the residue 4 phenolic glucosyltransferase GtfE has been exploited to generate a wide range of differentially glycosylated analogs.1030–1032
Fig. 46.
Sugars associated with vancomycins.
There are 21 metabolites classified as type II including avoparcins,1033,1034 galacardins,1035 helvecardins,1036 chloropolysporins,1037 and actinoidins1038–1041 from Streptomyces, Nocardia, Pseudonocardia, Micropolyspora, Saccharothrix, and Nonomuraea. Variant glycosylation is observed among type II metabolites with heavily glycosylated members containing maximally seven sugars at five glycosylation positions (galacardins) while sparsely glycosylated members contain just a single monosaccharide (demannosyl-A-40926). Of these, only avoparcins, galacardins, and actinoidins contain the residue 4 phenolic β-D-glucose (104, chloropolysporins and deristosaminyl-avoparcins) or α-L-sugar-(1′→2′)-β-D-glucose disaccharyl signature (IV–VI) typically observed among type I members where the terminating sugar can be α-L-rhamnose (46; actinoidin A2), α-L-ristosamine (45; avoparcins, galacardins and helvecardins), or α-L-actinosamine (124; actinoidins A and B, MM 47766, MM 47767, MM 55256, MM 55260) (Fig. 46). Like type I members, residue 6 tyrosine-β-hydroxyl O-glycosylation is also prevalent among type II members and includes α-L-ristosamine (45; avoparcins, galacardins, helvecardins, chloropolysporins) or 4′-O-methyl-α-L-actinosamine (124, actinoidins, MM 47766, MM 47767, MM 55256, MM 55260, MM 55261). Distinct from type I members, O-glycosylation of the residue 2 tyrosine-β-hydroxyl and/or phenolic hydroxyl of residues 1, 3 and/or 7 is also observed among type II members. Type II member residue 1 phenolic hydroxyl O-glycosylation employs two sugars α-L-rhamnose (46, chloropolysporins, helvecardin B, avoparcins) or a disaccharide α-L-rhamnosyl-(1′→4′)-α-D-galactose (VII, galacardins), while a single sugar, α-D-mannose (160), is observed for residue 2 tyrosine-β-hydroxyl O-glycosylation (avoparcins, galacardins, chloropolysporins, helvecardin A) (Fig. 46). An α-D-mannose (160) or β D-mannose (156) are observed at residue 7 phenolic O-glycosylation (actinoidins, MM 47766, MM 47767, MM 55260). Finally, residue 3 phenolic O-glycosides are comprised of α-D-galactose (286, avoparcins and galacardin A). With respect to activity/use, the use of avoparcins as animal growth promoters was banned due to reports of increased vancomycin resistance1042 while helvecardins were demonstrated to inhibit Neisseria gonorrhoeae in addition to Gram positive bacteria.1036
There are 23 metabolites classified as type III including ristocetins,1043–1046 A41030 C, F, and G,1047 actaplanins,1048–1051 UK-69542, and UK-68,5971052 from Actinoplanes, Nocardia, and Streptomyces species. Variant glycosylation is observed among type III metabolites with heavily glycosylated members containing maximally six sugars at three glycosylation positions (ristocetin A) and sparsely glycosylated members containing just single monosaccharides (A41030C). O-glycosylation of the residues 1, 3, 4, and/or 7 phenolic hydroxyls and/or the residue 6 tyrosine β-hydroxyl is observed among type II metabolites. Residue 1 phenolic hydroxyl O-glycosylation is only observed among A41030 metabolites with β-D-galactose (161) or a β-D-galactosyl-(1′→4′)-β-D-galactose disaccharide (VIII). Likewise, only a single sugar is observed [D-mannose, α-160, β-156] as residue 3 (actaplanins) and/or residue 7 (actaplanins, ristocetin A) phenolic hydroxyl O-glycosyl substituents. Wider variation is observed among the residue 4 phenolic hydroxyl O-glycosylation patterns. For example, actaplanins B2, C3, G, O, and brominated actaplanins contain the single sugar β-D-glucose (104) while other type III metabolites contain D-hexosyl-(1′→2′)-β-D-glucose (IX, X, I) disaccharyl substitution reminiscent of type I/II metabolites where the terminal sugar is either D-mannose (α-160, β-156) or α-L-vancosamine (118, UK-68597). Other observed disaccharides at this position include α-L-rhamnosyl-(1′→4′)-β-D-glucose (XI, actaplanins) and α-L-rhamnosyl-(1′→6′)-β-D-glucose (XII, actaplanins) (Fig. 46). Alternatively, in ristocetin A the α-D-mannosyl-(1′→2′)-β-D-glucose disaccharide is further modified at the glucose C-6′ by α-L-rhamnose (46) and at the mannose C-2′ by α-D-arabinose (204), forming tetrasaccharide XIII. Like type I/II metabolites, residue 6 tyrosine β-hydroxyl O-glycosylation in type III is observed to engage uniquely functionalized sugars including α-L-ristosamine (45; ristocetins and actaplanins) and the 2′,3′,6′-trideoxy-α-L-hexose 44 of UK-69542 (Fig. 46). Regarding activity/use, ristocetins were approved for clinical use but were subsequently removed due to thrombocytopenia and platelet agglutination.1043–1045 While the corresponding mechanism remains unclear, ristocetin causes von Willebrand factor to bind the platelet receptor glycoprotein Ib (GpIb), and is now used solely in the context of in vitro assays for the diagnosis of conditions such as von Willebrand disease (vWD) and Bernard–Soulier syndrome. Semisynthetic analogs, including those deriving from differential glycosylation, have revealed derivatives with improved antibacterial/antiviral potency, and/or less toxicity.
Type IV is the largest subclass with 46 members including teicoplanins,1053–1056 RS1-4,1054 kibdelins (AAD-609),1057 aridicins,1058 A-40926 analogs, parvodicins,1059–1061 and MM 49728, MM 55266, and MM 552681062 from Actinoplanes, Actinomadura, Kibdelosporangium and Amycolatopsis. CWI-785 A-C (symonicins) are comprised of a similar aglycon where methionine has replaced dihydroxyphenylglycine (residue 3).1063 O-glycosylation of the residues 4, 5, and/or 7 phenolic hydroxyls and/or the residue 6 tyrosine β-hydroxyl is observed among type IV metabolites. Residue 4 phenolic O-glycosylation has been observed with various monosaccharides including N-acyl-2′-amino-2′-deoxy-β-D-glucosamine (153; teicoplanins, RS1-4, kibdelins, AAD-609), N-acyl-2′-amino-2′-deoxy-β-D-glucuronic acid (283, aridicins, parvodicins, A-40926, A-84575A), β-D-mannose (156; MM 49728, MM 55266, MM 55268, MM 56597), or a disaccharide β-D-mannosyl-(1′→2′)-β-D-glucose (X, A-39893) (Fig. 46). It is noteworthy that teicoplanins (approved an an antibacterial, Targocid®) and RS1-4 isolated from the same strain differ mainly in their sugar 2′-N-acyl substitution and this served as inspiration for the clinically approved semi-synthetic analogs such as dalbavancin (Dalvance™). MM 49728, MM 56597, MM 56598, MM 55266 and MM 55268 are the only type IV members that contain residue 5 phenolic O-glycosylation (β-D-glucose, 104) while some variation in the sugars employed for residue 6 tyrosine β-hydroxyl O-glycosylation is observed [N-acyl-β-D-glucosamine (153), teicoplanins; 2′-amino-2′-deoxy-α-D-glucuronic acid (285), MM 49728, MM 55266, and MM 55268; α-L-ristosamine (45), CWI-785 A-C (symonicins), A-39893]. Teicoplanins, kibdelins (with the exception of AAD-609D), and aridicins also carry an α-D-mannose (160) as a residue 7 phenolic hydroxyl modification. CWI-785 A-C (symonicins) also contain β-D-glucose (104), α and β-D-mannose (160 and 156, respectively), and α-L-rhamnose (46), the position of attachment for which has not been confirmed.1063
15.3. Mannopeptimycins
Mannopeptimycins α-ε are hexapeptides made from D-tyrosine(Tyr-1), β-methylphenylalanine (MePhe-2), glycine (Gly-3), L-serine (Ser-4), and 2 units of β-hydroxyenduracididines (HyEnd-5 and HyEnd-6) (Fig. 47).1064 All members are N-glycosylated with α-D-mannose (160) at the N-5-terminus of HyEnd-5. In addition, all members except mannopeptimycin β contain a common α-D-mannosyl-(1′→4′)-α-D-mannose (disaccharide I) appended to Tyr-1 phenolic hydroxyl group (C-6). Analogs γ, δ, and ε contain a C-2′, C-3′, C-4′-isovaleryl (Fig. 47, appendage i) modification of the terminal mannose (160), respectively. Like vancomycins, mannopeptins inhibit bacterial cell wall biosynthesis and display potent Gram positive antibacterial activity. The presence of the isovaleryl moiety (i) is important for antibacterial potency possibly by virtue of improving cell membrane penetration.
Fig. 47.
Mannopeptimycin, salmochelin, and salmomycin aglycons and associated sugars.
15.4. Salmochelins and salmycins
Salmochelins are glycosides of the catecholate-type enterobactins from Salmonella which function as siderophores for metal sequestration.1065,1066 Four salmochelins have been characterized including SX (monomer), S1 (dimer), S2 (linear trimer) and S4 (cyclic trimer) where each monomeric unit is comprised of a L-serine (Ser) attached to a dihydroxybenzamide (Bnz). Salmochelins exist as C-glucosides (β-D-glucose, 104) of the dihydroxybenzamide core structure (Fig. 47). Salmycins, hydroxamate-containing siderophores from Streptomyces violaceus, are glycosylated danoxamins at the C-1 position with disaccharides II or III.1067,1068 Disaccharide II present in salmycins A and D consists of a unique 6′-methylamino-D-heptopyranose (288) attached at the C-2′ position to the rare 2′-hydroxyimino-α-D-glucose (289). Disaccharide III present in salmycins B and C contains the α-D-keto sugar 2′-oxo-α-D-glucose (287) in place of the hydroxylimino sugar (289) (Fig. 47). This disaccharide is attached to the aglycon via an ester bond to C-6′ of the modified glucosyl unit. Salmycins were found to be strong inhibitors of the growth of Staphylococci and Streptococci and are therefore considered siderophore antibiotics (sideromycins).
15.5. Lincosamides
Lincosamides from Streptomyces are a group of clinically important antibacterials that target Gram positive and anaerobic bacteria as well as some protozoa through inhibition of protein synthesis. Including the lincomycins first discovered in 1963 (which serve as the basis for the semi-synthetic drug clindamycin, Dalacin®), 29 naturally-occurring members have been reported from Streptomyces species including celesticetins and U-57930 analogs.1069–1074 Their core structure is composed of a unique thioglycoside with variable substitution of the anomeric thioether and the 6′-prolylamide (Fig. 48). The anomeric stereochemistry of the central sugar, 6′-amino-6′,8′-dideoxy-1′-thio-D-erythro-α-D-galactose (methylthio-lincosamide, 290) (Fig. 48) is critical for antibacterial activity and oxidation of the thioether (to a sulfoxide) is also detrimental to function.1075,1076 Additional derivatives have been obtained via precursor-directed feeding1077–1081 including ribonucleotide derivatives where the proline has been replaced with a 4-ethyl-piperidine moiety.1082
Fig. 48.
Lincosamide and thiazolyl peptide aglycons and associated sugars. Indole moiety in red is absent or replaced by quinoline in different members.
15.6. Thiazoles
Thiazolyl peptides contain one or more signature thiazoles embedded within their macrocyclic architecture. In spite of their potent antibacterial activity members of this family have not advanced to the clinic due to poor biodistribution and pharmacokinetic properties. Currently 21 thiazole glycosides have been reported from different Actinomycetes including Micromonospora, Amycolatopsis, Nocardia, Actinoplanes, and Amycolata. Regiospecificity of glycosylation among the thiazomycin-type members (which includes thiazomycins,1083,1084 nocathiacins,1085–1087 glycothiohexides,1088,1089 philipimycins1090 and S-54832-A series1091) is restricted to the hydroxyl group of a modified glutamate residue (Glu), the C-4 of a thiazole ring (Thz-3) or a C-3 phenolic substitution of the unique 2,5,6-trithiazolyl-3-hydroxypyridine (Pyr) (Fig. 48). In contrast, SCH 40832 glycosylation occurs at the Thr-1 side chain hydroxyl (disaccharide II composed of two units of β-D-olivose 53 connected 1′→3′)1092 (Fig. 48). The sugars found within thiazomycins, nocathiacins, and glycothiohexides include 2′,4′-dideoxy-4′-dimethylamino-3′-C-methyl-α-L-fucose (292a), oxazolidine-substituted 2′,4′-dideoxy-4′-amino-3′-C-methyl-α-L-fucose (292b), β-L-chromose 186, 4′-deoxy-2′,3′-di-O-methyl-β-D-glucose 291 or β-D-quinovose (140) (Fig. 48). The related philipimycin I contains a trisaccharide [trisaccharide I comprised of 2′,3′-dimethoxy-α-L-rhamnosyl-(1′→4′)-β-D-amicetosyl-(1′→4′)-2′,3′-dimethoxy-α-L-rhamnose].1090 The degradation product of philipimycin I (philipimycin II) contains only the monosaccharide 2′,3′-dimethoxy-α-L-rhamnose (46). In general, thiazole glycosylation was found to improve both solubility and antibacterial potency.
15.7. Lipoglycopeptides
The lipoglycopeptides presented herein are macrocyclic peptides bearing one or more signature lipophilic side chains and are separated for discussion by glycosides of the peptide core backbone and glycosides of lipid side chains. While most members in this group display antifungal activity (hassillidins from Hassallia,1093,1094 occidiofungins1095 and burkholdine1096 from Burkholderia, cepacidines from Pseudomonas,1097,1098 herbicolins from Erwinia1099,1100 and W-10β from Aeromonas1101), the ramoplanins from Actinoplanes stand out as agents recently approved by the FDA to treat C. difficile (Fig. 49).1102,1103 Ramoplanins are phenolic O-disaccharyl-substituted metabolites, the disaccharide I of which is comprised of two α-D-mannose (160) monomers. Ramoplanins display broad antibacterial activity via inhibition of cell wall biosynthesis however; serum instability restricts current use to oral applications. The cyclic lipopeptide hassallidins are also peptide core O-mannosides, both containing N-methylthreonine (MeThr-9) C-3-O-β-D-mannose (156) and hassallidin B having an additional threonine (Thr-2) C-3-O-α-L-rhamnose (46) (Fig. 49).1093,1094 Both the mono and diglycosylated analogs of hassallidins presented comparable antifungal activity against Aspergillus and Candida. Finally, W-10β isolated from Aeromonas is a threonine residue (Thr-1) C-3-O-β-D-glucoside (104) with antifungal activity (Fig. 49).1101
Fig. 49.
Lipoglycopeptide aglycons and associated sugars. Only partial structures of burkholdine, cepacidins, and herbicolin are shown.
The octapeptides occidiofungins1095 and burkholdine1096 also possess broad antifungal properties and display both core peptide and side chain O-glycosylation. Occidiofungins are C-3-O-β-D-xylosides (136) of a deaminated lysyl monomer (Fig. 49) while burkholdines are C-7-O-D-xylosides (136 or 293, anomeric stereochemistry undetermined) of the appended lipid side chain. Importantly, the corresponding glycoside displayed 4–8 fold increased antifungal potency compared to the aglycon burkholdine.1097 Lipid side chain O-D-xylosyl modification (136 or 293, anomeric stereochemistry undetermined) was also observed in the antifungal metabolites cepacidines A1 and A2 (Fig. 49), where glycosylation occurs at C-5 of the 5,7-dihydroxy-3,9-diaminooctadecanoic acid.1097,1098 Finally, the antifungal lipodepsinonapeptide herbicolin A also displays lipid side chain O-glycosylation with α-D-glucose (18) found at the C-3 position of the hydroxytetradecanoic acid residue.1099,1100
15.8. Miscellaneous anti-infective peptides
Miscellaneous glycopeptides with reported antibacterial and antifungal activities that fall outside the scope of those previously discussed are combined within this subsection, the predominant representation of which are polyamine peptides. The first group of compounds are comprised of an acylated spermidine (glycocinnaspermicin D and cinodines from Nocardia)1104,1105 or 3-aminopropanamidine (coumamidines from Saccharopolyspora)846,1105 conjugated to a cinnamoic acid moiety, the latter of which is glycosylated at C-7 with unique di- or trisaccharides (Fig. 50). The corresponding disaccharide I and trisaccharides II–IV are comprised of the core sugars 2′,4′-diamino-2′,4′,6′-trideoxy-α-D-glucose (295), 2′-amino-2′-deoxy-β-D-xylose (173), 2′-amino-2′-deoxy-α-D-xylose (24) and 2′,3′-diamino-2′,3′-dideoxy-α-L-pentose (296a–296c) (Fig. 50). Unique features among this series include the high degree of C-2′, C-3′ and/or C-4′ carbamidyl/guanidinyl modification of the corresponding sugar monomers and the carbamide-glycosyl linkage exclusive to this family of metabolites. These compounds exhibited broad spectrum antibacterial activity. Other polyamine antibacterial compounds include the cyclitol antibiotics LL-BM-123α and LL-BM-782 series from Nocardia (Fig. 50). They are Gram negative bacterial growth inhibitors that contain a pseudotri- and pseudodisaccharide, respectively (V and VI).1106,1107 The pseudotrisaccharide V in LL-BM-123α consists of an arginine N-linked to the C-2′ of α-D-glucosaminyl-(1′→4′)-β-D-mannose conjugated to a modified myoinosamine (P20). The corresponding sugar of pseudodisaccharide VI in LL-BM-782 series is 3′-guanidino-β-mannose (42) which is conjugated to the aglycon via a modified myoinosamine (P12). Both LL-BM-123α and LL-BM-782 compounds displayed in vivo efficacy against Gram negative bacteria where LL-BM-782 was more potent but also more cytotoxic.
Fig. 50.
Miscellaneous antiinfective peptide aglycons and associated sugars.
Non-polyamine metabolites include SQ 28504 and SQ 28546 from Chromobacterium,964,965 formadicins from Flexibacter,1108,1109 and theopalauamide and theonegramide from bacterial symbionts of Theonella.1110–1112 SQ 28504 and SQ 28546 are C-4 bulgecinine (Blg-1) and C-3 threonine (Thr-3) di-O-glycosides [N-acetyl-β-D-glucosamine (153)] where the Blg-1-appended sugar is C-4′-O-sulfated (Fig. 50). Both compounds potentiate the activity of β-lactam antibiotics. Formadicins are β-lactam antibiotics, two of which (formadicins A and B) are C-9-O-β-D-glucuronidates (72) (Fig. 50). Glycosylation is notably rare among naturally occurring β-lactams and these unique metabolites displayed antibacterial activity againts Pseudomonas and Proteus species where glyco-sylation contributed to reduced potency. Finally, the anti-fungals theopalauamide and theonegramide are histidine (His) N-3 N-glycosides of β-D-galactose (161) and a β-D-arabinose (294), respectively.1110–1112
15.9. Other peptides
Miscellaneous peptides that lack anti-infective related activity are summarized herein. These include aeruginosides,1113 aeruginosins,1114 muraceins,1115 enkastines,1116 banyasides,1117 phosphoramidon,1118 GlcNAc-TA,1119 mycothiols,47,1120 bacillithiols,1121 and alanopine1122 from Streptomyces, Nostoc, Nocardia, Planktothrix, Bacillus, Staphylococcus Providencia, Proteus and Oscillatoria (Fig. 51). Aeruginosides and aeruginosins share a common peptide skeleton [composed of 2-carboxy-6-hydroxyoctahydroindole (Choi), leucine (Leu) or hydroxyleucine (HLeu), phenyl lactic acid (Plac), and agmatin (Agm) or 1-amino-2-(N-amidino-Δ3-pyrrolinyl)ethyl (Aeap)], a common sugar (α-D-xylose, 293) but distinct regioselectivity of glycosylation (aeruginosides, Choi C-6; aeruginosins, HLeu C-3; Fig. 51).1113,1114 Banyaside A and B are comprised of 1-amidino-3-(2-aminoethyl)-3-pyrroline (Aaep), an azabicyclononane (Abn) C-7-O-α-D-glucoside (18), D-leucine (Leu), and a 2-O-methylglyceric acid-3-O-sulfate (MGAS).1117 They differ by subtle modifications of the appended glucose and, like aeruginosins, are potent inhibitors of trypsin and thrombin.
Fig. 51.
Miscellaneous peptide aglycons and associated sugars.
Muraceins are a group of N-acetylmuramyl peptides composed of alanine (Ala), glutamic acid (Glu), diaminopimelic acid (Dap), and, in the case of muracein C, serine (Ser).1115 Glycosylation of muraceins is afforded via an amide bond between the N-terminus Ala α-amine and the C3′-side chain carboxylate of N-acetyl muramic acid (153a, Fig. 51). They inhibit angiotensin-converting enzyme (ACE) and have been put forth as potential antihypertensives.
Enkastines I–IV are dipeptides composed of an N-terminal hydrophobic amino acid [isoleucine (Ile) or valine (Val)] and an acid amino acid [aspartic acid (Asp) or glutamic acid (Glu)]. All exist as N-terminal N-glycosides of 1′-deoxyfructopyranose (201) and inhibit enkephalinase endopeptidase.1116 Phosphoramidon, another dipeptide endopeptidase inhibitor, consists of a C-terminal tryptophan (Trp) and and N-terminal leucine (Leu), the N-terminal amine of which is uniquely connected to α-L-rhamnose (46) via a phosphoramidate bridge.1118 GlcNAc-TA is a prenylated indolactam C-14-O-glycoside (N-acetyl-β-D-glucosamine, 153).1119 This is the only naturally-occurring teleocidin glycoside and is a small molecule activator of protein kinase C and induces the release of excitatory neuropeptides.
Examples of amino acid glycosides include mycothiols,47,1120 bacillithiol,1121 and alanopine. Mycothiols are constructed of N-acetyl or N-propionyl cysteine (Cys) modified at C-1 with pseudodisaccharide I (composed of α-D-glucosamine 5 and D-myoinositol P12). In a similar manner, bacillithiol is a 1′-(2-butanedioic acid)-α-D-glucosamine (5)-conjugated L-cysteine. Both mycothiols and bacillithiol protect against oxidative stress.1123 Alanopine, a component of bacterial lipopolysaccharide, is an N-carboxyethylalanine (CEAla) attached to 4′-amino-4′-deoxy-D-quinovose (α, 39; β, 202), the anomeric configuration for which was not established.1122
16. Phenazines
Phenazines comprise a large group of nitrogen-containing heterocyclic bacterial metabolites that display a range of activities including antibacterial, antitumor, antimalaria, and antiparasitic functions.1124 Over 140 of different phenazine metabolites have been reported from bacteria, only 13 of which are glycosylated (Fig. 52).18 Regiospecificity of O-glycosylation among these metabolites is restricted to the 4- and 6-positions of the phenazine ring as well as the less common glycosyl esters which occur at a phenazine 6-carboxylic acid. Only three sugars were observed among the corresponding glycosides where α-L-quinovose (194) and β-L-quinovose (297), a sugar common to saponin glycosides,1125 were found as sugars with free reducing termini attached as sugar 2′/or 3′-esters of the phenazine C6-carboxylic acid [phenazine α-L-quinovose 2′-ester (194, C-6), phenazine α-L-quinovose 3′-ester (194, C-6), phenazine β-L-quinovose 2′-ester (297, C-6) and phenazine β-L-quinovose 3′-ester (297, C-6)] in metabolites of a marine Streptomyces.1126 The remaining glycosylated phenazines (aestivophoenins A-C,1127,1128 izuminosides A-C,1129 and phenazoviridin1130–1132 from Streptomyces) are mono-glycosylated with α-L-rhamnose (46) at the phenazine C-4- or C-6-position, or as anomeric glycosyl esters of the phenazine 6-carboxylic acid similar to that described above (Fig. 52). Izuminosides B and C were reported to have a synergistic activity in sensitizing tumor necrosis factor-related apoptosis-inducing ligand “TNF-RAIL”-resistance AGS cells.1129
Fig. 52.
Phenazine aglycons and associated sugars.
17. Piericidins and pyranones
Piericidins are prenylated polyoxypyridines with variant functions that include general cytotoxicity and insecticidal activity. More than 38 naturally occurring piericidins and closely related pyranones have been reported from Streptomyces, 11 of which are piericidin C-3a or C-10 O-monoglycosides (Fig. 53).18 Three different monosaccharides have been observed among glycoconjugates of this class including β-D-glucose (104), α-L-rhamnose (46) and 2′-epi-β-D-fucose (also known as 6′-deoxy-β-D-talose, 298). Of these, β-D-glucose (104) is the most predominant, found in 6 of the 11 naturally-occurring members of this class including glucopiericidins A-C,236,1133–1135 13-hydroxyglucopiericidin A,1136 and glucopiericidinols A1 and A2.1134 In contrast, the C-3a-appended α-L-rhamnose (46) was only observed in 3a-rhamnopiericidin A1 (SN-198-C),1137,1138 and 7-demethyl-3a-rhamnopiericidin A1 (SN-l98-B),1139 while the C-3a attached 2′-epi-β-D-fucose (298) was observed only in 3-deoxytalopiericidin A1.1140 Additionally, the two glucopyranone (PM-050511 and PM-060431) metabolites of a marine Streptomyces display C-10-O-β-D-glucosyl (104) regiospecificity reminiscent of corresponding glucopiericidin.1141 Importantly, glycosylation generally correlates with improved antimicrobial properties and reduced general in vivo toxicity.1133–1135,1137,1139–1141
Fig. 53.
Piericidin and pyranone aglycons and associated sugars.
18. Pluramycins
The 4H-anthra[1,2-b]pyran-4,7,12-trione core is a characteristic feature of the pluramycin antitumor antibiotics produced by Streptomyces, Actinomyces, Actinomadura and Saccharotrix.1142,1143 To date, 45 pluramycin glycosides have been reported (Fig. 54) and a unique signature of most pluramycin members is the presence of C-8- and/or C-10-C-glycosides. C-2- and/or C-5-side chain O- and/or C-glycosylation has also been observed and two metabolites (clecarmycins)1144 distinctly contain C-9-C-glycoside substitution. Those members that present the prototypical C-8-/C-10-C-glycosyl archetype include hedamycin,1145–1148 DC92-B and DC92-D,1149,1150 kidamycin and epoxy-kidamycin (largomycin FII chromophore component 4),1151 PD 121222,1152 ankinomycin,1145,1146,1153–1155 altromycins,1156–1159 saptomycins,1160–1162 neopluramcin and pluramycin A,1163–1165 pluraflavins,1166 A51493A,1167 rubiflavins1168 and chromoxymycin,1169–1171 the latter of which has a uniquely modified aglycon core at C-12. Eighteen monosaccharide units are represent among pluramycin glycosides where N-alkyl aminosugars are a predominate feature known to be critical for pluramycin metabolite DNA interaction and cytotoxicity.940,1153,1168,1172–1177
Fig. 54.
Pluramycin aglycons and associated sugars.
Among the sugars found as C-8-C-glycosides, β-D-anglosamine (59) is most common with 3′-epi-α-D-anglosamine (69; SS 21020B) and Δ1,2-D-anglosamine (306; DC92-D) also observed. Of these, the C-1′/C-2′-unsaturated Δ1,2-D-anglosamine (306; DC92-D) is perhaps the most unique. For C-10-C-glycosides, N,N-dimethyl-α-L vancosamine (118c) is the chief sugar where X-ray analyses revealed this sugar to adopt a boat confirmation.1178–1180 Other monosaccharyl C-10-C-glycosides include N,N-dimethyl-β-L-vancosamine (299; SS 21020B, SS 21020C, isokidamycin), N,N-dimethyl-α-D-vancosamine (304; chromoxymycin), N,N-dimethyl-3′-epi-5′-hydroxy-β-D-vancosamine (302; DC-92B, DC-92D), α-L-vancosamine (118a; altromycin G) and N,N-dimethyl-β-D-vancosamine (301; saptomycin B) and corresponding disaccharides have also been reported (altromycins, disaccharides II, III, IV; pluraflavins, disaccharide I). It is important to note that the C-10 N,N-dimethyl-α-D-vancosamine (304; chromoxymycin) was not observed in any other pluramycin member, potentially calling into question this assignment. Finally, altromycins A-G also present C-glycosylation of the C-5 side chain (2′,6′-didexoy-3′-methoxy-α-D-altrose, 129; 3′-methoxy-6′-deoxy-α-D-altrose, 307) while clecarmycins contain C9-C-glycosides of β-D-kerriose (55). While the genes encoding for the unique C-8-/C-10-C-glycosyltransferases have been identified,1181 they remain resistant to biochemical study.
19. Polyethers
Polyether ionophores represent a large group of natural occurring biologically active compounds produced mainly by Streptomyces.18,1182 This family of secondary metabolite contributes to the transport of metal cations across cell membranes and members also display a broad spectrum of activities including antibacterial, antifungal, antiviral, antiparasitic, and cancer cell line cytotoxicity. To date, 52 glycosylated bacterial polyethers have reported, subdivided herein into four sub-classes I–IV according to their aglycon core structural conservation (Fig. 55). Despite the rather large group of glycosylated analogs observed, the glycosides found are restricted to the use only eight distinct sugars (Fig. 55). The sugar 4′-O-methyl-β-D-amicetose (48b) represents the most predominant sugar, found in 34 of the 52 glycosylated bacterial polyethers. Exceptions to the use of 48b are noted herein.
Fig. 55.
Polyethers aglycons and associated sugars.
In sub-class I, 29 O-glycosylated polyethers were found with variant glycosyl regiospecificity (C5, C6, C15, C18, C22 and C27) including; antibiotics 6016,1183–1186 A204-A [α-D-amicetose (308)],1187 carriomycin,1188,1189 K-41-A,1190,1191 K-41-B,1192 X-14868A (maduramicin α),1193–1196 X-14868C~D,1193 etheromycin,1189,1197 >UK-58852, nanhumycin, RP-37454, A80789, CP-120509 [α-D-amicetose (308)],1198 CP-91243,1199 CP-91244,1199 CP-96797,1200 octacyclomycin,1201 CP-82009,1202 semduramicin [α-D-amicetose (308)],1203 UK-58,852-methylketal,1204 A-13001-C1196 and SF-2361,1205 along with the septamycin analogs.1206,1207 Of these, β-D-amicetose (48a) was observed in CP-91243 and CP-91244.1199 In addition, 4′-O-methyl-α-L-amicetose (97) was observed only in etheromycin (CP-38295) while X-14868A through D1193 contain β-L-olivose (100) connected at the same position (C-22) of the aglycon. Sub-class II, contains the four glycosylated polyethers 26-O-β-D-glucopyranosylmonensin (β-D-glucose, 104) and the α-D-amicetose (308)-bearing CP-47433 and CP-47434, and CP-82996.1208 Alternatively, sub-class III members (16 compounds) contain 4′-O-methyl-β-D-amicetose (48b) connected at five different positions of the aglycon (C-11, C-15, 20-CH2, and C-27) as exemplified by A-130-B,1209 A-130-C,1209 X-14934A, moyukamycin,1210 lenoremycin (A-130),1211,1212 leuseramycin,1213 endusamycin,1214 CP-60993,1215 CP-80219,1216 nangchangmycin1217–1219 and dianemycin analogs.1212,1220,1221 All subclass VI analogs are C-14-O-glycosides of 4′-O-methyl-β-D-amicetose (48b), including, 6270B,1222 portmicin1223 and CP-84657.1224
20. Pterins
Pterins are characterized by the presence of a pteridine or a tetrahydropterine ring and have been proposed to prevent the bleaching of photosynthetic pigments due to irradiation in bacteria.1225 Fifteen glycosylated metabolites of this class were reported predominately from cyanobacteria, green sulfur bacteria or methanogens where, in all cases, regioselectivity of glycosylation was observed to occur at variable positions upon side chains appended at C-6 of the pterin ring structure (Fig. 56). Cyanopterin1226,1227 and monapterin, representing members that display O-glycosylation closest to the ring, contains a C-9 4′-O-methyl-α-D-glucuronic acid-(1′→6′)-β-D-galactose disaccharide (I) and β-D-glucose (104), respectively. Limipterin, tetrahydrolimipterin, tepidopterin and biopterin glucoside represent C-10-O-glycosides, all which contain N-acetyl-β-D-glucosamine (153) with one exception (biopterin).1225,1228,1229 The latter, incorrectly labeled 2′-O-(α-D-glucopyranosyl)biopterin, actually contains α-D-altrose (163). Solfapterin, the only C-11-O-glycoside within this series, contains α-D-glucosamine (5).1230 Finally, tatiopterins, thermopterin, methanopterin, sarcinapterin, and tetrahydromethanopterin all contain uniquely functionalized anilines at C-9 that terminate with an O-glycosidic 5′-phospho-α-D-ribose (231) (Fig. 56).1231–1233 In some cases, the corresponding 5′-phosphate is further modified with α-hydroxyglutaric moiety via a phosphodiester bond (Fig. 56, appendages i, ii, or iii).
Fig. 56.
Pterin aglycons and associated sugars.
21. Streptothricins
Streptothricins are N-glycosides composed of streptolidine or streptolidine lactam and a sugar (typically, β-D-gulosamine 320) attached to the C-2 exocyclic amino group, the latter of which (320) is C-2′-amino modified via a β-lysine derived moiety (Fig. 57). The streptolidine lactam is typically trans but a few cis conformers were recently reported.1234,1235 Including the first member discovered (streptothricin F in 1942),1236–1238 45 analogs have been reported as metabolites of Streptomyces and Bacillus including streptothricins, racemomycins, albothricin, glycinothricin, lavenothricins, boseimycin, and sclerothricin.1234,1235,1239–1251 These metabolites act as broad spectrum antibiotics through inhibition of protein synthesis and lavendothricins B and C have also been reported as calcium channel blockers.1252,1253 However, apparent renal toxicity has hampered further development.1254,1255 As mentioned, the core sugar employed among these metabolites is β-D-gulosamine (320) where 43 out of the 45 derivatives contain subtle modifications of the C-2′ amino group and/or C-4′ and/or C-6′-carbamoylation and are contributors to activity modulation.1246,1250,1251 The two exceptions include streptothricin F (which carries a corresponding C-2′, C-6′-modified α-D-gulosamine, 189)1235 and racemomycin O (which alternatively contains a corresponding C-2′,C-4′,C-5′-modifed β-D-glucosamine 153).1256 Sugar C-4′ carbomylation favors activity while cleavage of streptolidine lactam amide bond reduces activity and toxicity.1251,1257
Fig. 57.
Streptothricin aglycons and associated sugars.
22. Terpenes and sterols
While prominent among plant metabolites, terpenes are relatively rare in the context of bacterial natural products with only 41 glycosylated terpenoids reported from bacteria to date. These are discussed below in the context of standard terpene classification.
22.1. Sesquiterpenes and diterpenes
Sesquiterpenes are comprised of three isoprene units and only one, the mildly cytotoxic deoxypentalenylglucuron (containing an aglycon exocyclic C-6 acid anomeric ester of β-D-glucuronic acid 72), has been reported from various Streptomyces.1258
Diterpenes are comprised of four isoprene units and 16 glycosylated diterpenoids have been reported as metabolites of Strepomyces and Nocardia including the phenalinolactones,1259 lucensimycins,1260 brasillicardins,1261,1262 platensimycins,1263 and platencins.1264,1265 Phenalinolactones, C-4 glycosides of a methylated α-L-amicetose (97) (Fig. 58), display moderate activity against Gram positive bacteria.1259 In contrast, brasillicardins are C-2 (α-L-rhamnose 46) glycosides which, in brasillicardins A and B, are further modified at the rhamnose C-3′ with N-acetyl-β-D-glucosamine (153).1261,1262 The latter sugar is believed to be essential for the immunosuppressive activity of brasillicardin A. Two of the seven lucensimycins from Streptomyces are glycosides,1260 attached to the modified cysteine side chain via an amide bond between the cysteine C-1′ carboxylate and an aminosugar (2′-amino-2′-deoxy-α-L-idose, 316) amine. The anomeric position of the latter forms a α-glycosidic bond with the C-1′ of myo-inositol (P12) to form pseudodisaccharide II (Fig. 58) and it is important to point out that some confusion exists in the literature regarding the structure of pseudodisaccharide II.1131 Platensimycin B4 and its methyl ester1263 and platencins A9, A10 and A12–A151264,1265 are the known glycosylated derivatives of the platensimycins and platencins. Members of this class were found to be potent inhibitors of mammalian and bacterial fatty acid synthases with potential relevance for the treatment of metabolic disease and bacterial infections.1266 All glycosylated analogs exist as 4′-β-D-glucosides (104) of the corresponding 3-amino-2,4-dihydroxybenzoic acid moiety (Fig. 58) and display attenuated inhibition of FAS.1263
Fig. 58.
Sesquiterpene and diterpene aglycons and associated sugars.
22.2. Triterpenes
Triterpenes consist of six isoprene units and are hypothesized to function as cell membrane constituents in bacteria. Cholesteryl glucosides with hemolytic activity have been reported as metabolites of Helicobacter and Streptomyces (Fig. 59).1267,1268 The former are C-3-α-glycosides bearing C-6′-modified α-D-glucose (18) while corresponding metabolites from Streptomyces are β-glucosides (104).1268 Hopanoids are pentacyclic triterpenoids thought to function as membrane stabilizers in bacteria to enable adaptation to extreme environments and are occasionally used as biomarkers for bacterial chemical taxonomy.1269–1276 Bacteriohopanepolyols are hopanes characterized by a signature C-21-polyol side chain where C-36-glycosylated members have been characterized from different bacterial species including Zymomonas, Synechocystis, Synechococcus, Prochlorothrix, Acetobacter, Pseudomonas, Ralstonia, and Burkholderia. Nine different monosaccharides have been observed among this group including 2′-N-methyl-β-D-glucosamine (153), 6′-amino-β-D-glucose (318), β-D-glucose (104), 2′-amino-β-L-glucose (260), β-D-galacturonic acid (172), α-D-glucuronic acid (154), α-D-altropyranouronic acid (317), pseudosugar P23, and the unusual sugar 3,5-anhydrogalactouronic acid (321) (Fig. 59). MK800-62F1, a gammacerane and inhibitor of apotosis isolated from Streptomyces, is a C-22-glycoside of α-L-arabinose (265).1277 It should also be noted that glycosylated bacterial carotenoids are briefly discussed in Section 11.
Fig. 59.
Triterpenes aglycons and associated sugars.
23. Miscellaneous
23.1. Trioxacarcin analogs
Trioxacarcins and LL-D49194 analogs are metabolites from Streptomyces that broadly inhibit the growth of cultured bacterial and eukaryotic cells.1278–1281 To date, 16 glycosylated trioxacarcin metabolites have been reported, glycosylated at C-4-, C-13- and/or C-16 of the aglycon (Fig. 60). Three branched-chain sugar monomers are represented among this group including α-L-trioxacarcinose A (4′-epi-α-L-mycarose; 122), α-L-trioxacarcinose B (103), and the corresponding keto-reduced trioxacarcinose B (323).18 The C-4-O-α-L-trioxacarcinose A moiety (122) is common to all trioxacarcin glycosides, including the monoglycosidic trioxacarcins A1 (DC-45-A1) and E, the diglycosidic trioxacarcins (A, B, D, F), gutingimycin,1282 and the triglycosidic LL-D49194 (α1, β1, β2, e and w1).1283 Additional C16-O-glycosylation with a mono-(122) or disaccharide (fragment I) side chain was observed in the LL-D49194 analogs while additional C-13-O-glycosylation with α-L-trioxacarcinose B (103) or its corresponding keto-reduced trioxacarcinose B (323), was observed in trioxacarcins A–D, F and gutingimycin. Sugar 323 of trioxacarcin C and its keto-form 103 have also been found appended to the anthracycline antibiotics isoquinocycline A and kosinostatin (Section 4.1), respectively.188,251
Fig. 60.
Trioxacarcins and LL-D49194 aglycons and associated sugars.
23.2. Polycyclic xanthones and related analogs
Polycyclic xanthone natural products are a family of diverse polyketides characterized by highly oxygenated, angular hexacyclic frameworks, many of which are glycosylated. As described herein, C-11-O-glycosylation predominates but select members representing C-4-, C-6-, C-9- and C-10-O-glycosylation and C-11-N-glycosylation have also been characterized (Fig. 61). Among the C-11-glycosyl series, kibdelones A-C rhamnosides and isokibdelone A rhamnoside from Kibdelosporangium all exist as C-11-O-α-L-rhamnosides (46).1284,1285 Interestingly, kibdelones display potent anticancer, antibacterial and nematocidal activity while isokibdelones lack activity.1284,1285 Neocitreamicin II from Nocardia is a C-11-O-glycoside bearing 4′-acetylated-β-D-oliose (47).1286 Both neocitreamicin II and its aglycon neocitreamicin I display equal in vitro potency against Gram-positive bacteria including methicillin-resistant Staphylococcus aureus and vancomycin-resistant Enterococcus faecalis.1286 FDA-594,1287,1288 MSO 9018091289 and BE-13793X,1290 from Streptomyces are three O-trisaccharyl-appended antitumor antibiotics. MSO 901809 and FD-594 contain the same trisaccharide [β-D-oleandrosyl-(1′→4′)-O-β-D-olivosyl-(1′→4′)-O-β-D-olivose, trisaccharide V] connected at C-9 and C-11 of the xanthone backbone, respectively. A similar C-11-trisaccharide is found in BE-13793X which contains a distinct internal β-D-olivosyl-(1′→3′)-O-β-D-olivose connection [β-D-oleandrosyl-(1′→4′)-O-β-D-olivosyl-(1′→3′)-O-β-D-olivose, trisaccharide VI]. Antibacterials/antimycoplasmodials SF2446A1, A2, B1 and B2 from Streptomyces along with the non-selective cytotoxin arenimycin from Salinospora also reflect C-11 glycosyl regiospecificity reminiscent of the kibdelones but are the only N-glycosidic members of this group, all bearing 2′,4′-di-O-methyl-β-L-rhamnose (322).1291–1293
Fig. 61.
Polycyclic xanthone aglycons and associated sugars.
Distinct from the C-11 glycosyl series, kigamicins A–E are antitumor antibiotics from Amicolatopsis that possess a fused octcyclic ring system comprised of seven six membered rings and an oxazolidine.1294–1296 Kigamicins are C-10-O-glycosides bearing a mono- to tetrasaccharide comprised of β-D-amicetose (48) and β-D-oleandrose (53b).1294–1296 The C-6-O-glycosyl regiospecificity of benaphthamycin-β-D-glucoside (104) from Streptomyces is unique from other members in this class. Likewise, the C4-O-glycosyl regiospecificity of IB-00208 (β-D-quinovose, 140) from Actinomadura is unique from other members in this class.236,1297,1298 IB-00208 displays both in vitro cytotoxicity against various tumor cell lines (P-388, A-549, HT-29 and SK-MEL-28) and activity against Gram-positive bacteria.236,1297,1298
23.3. Paulomycins related compounds
This class of compounds share a 2-amino-5-hydroxy-3,6-dioxocyclohex-1-enecarboxylate-based aglycon. Twenty analogs belong to this class and include ten paulomycins, two paldimycins, two 273a2 analogs, two paulomenols, U-77,802 and U-77,803, and two senfolomycins from Streptomyces.1299–1303 These metabolites carry a C-5-C-glycoside which, in most cases, is α-L-paulomycosyl-(1′→3′)-α-D-allose (disaccharide I, Fig. 62). Exceptions to this include paulomycin E and senfolomycins which differ via subtle variations of the disaccharide C-4′-branched α-L-paulomycose (328) monomer (ketosugar 329 in disaccharide II, paulomycin E; ketosugar 103 in disaccharide III, senfolomycin A; epimer 323, disaccharide IV, senfolomycin A). Modification of the disaccharide core [C-6′ of α-D-allose (162), C-3′ (328 and 329) and/or C-7′ (328) of the terminal branched sugar] further differentiates members as illustrated in Fig. 62. Members of this family display potent Gram positive antibacterial activity.
Fig. 62.
Paulomycins and associated sugars.
23.4. Phenyls and related compounds
Metabolites that contain a phenyl ring and do not belong to one of the previous classes are consolidated within this section which contains 10 structurally/functionally diverse glycosides. Myxotyrasoides, moderately cytotoxic N-acyl tyrosine derived metabolites from Myxococcus,614 contain a phenolic-O-α-L-rhamnose (46) (Fig. 63). Myxotyrasoides, along with chivosazoles and sorangiosids (Section 12), are among the only glycosylated secondary metabolites reported from myxobacteria.
Fig. 63.
Phenyl glycosides and associated sugars.
Sibiromycin and sibanomicin from Streptosporangium and Micromonospora, respectively, are C3-O-glycosides (α-L-sibirosamine, 327) of modified pyrrolo[1,4]benzodiazepines.1304–1306 Both are cytotoxic and sibiromycin binds the minor groove of DNA.1307 Two C-6-O-2′,4′-di-O-methyl-β-D-xylosyl (136) quinoline alkaloids were isolated from the cyanobacterium Lyngbya1308,1309 and the same sugar was also found appended at C-4 of malyngamide J cyclohexanone core (a cytotoxin).1310 Terfestatin A, a specific inhibitor of auxin signaling in Arabidopsis, is a C-3-O-β-D-glucoside (104).1311
Saccharomicins are O-heptadecaglycosides of N-(m,p-dihydroxycinnamoyl) taurine from Saccharothrix.1312 Common monosaccharide units of the saccharomicin oligosaccharide include β-D-fucose (270), 2′-sulfate-β-D-fucose (270), the C-3′-branched β-D-saccharosamine (269), α-L-rhamnose (46), 4′-epi-α-L-vancosamine (280), and α-L-digitoxose (68). Saccharomicins A and B differ via a single tenth sugar residue (α-L-rhamnose 46 or α-L-digitoxose 68, respectively). Both compounds display potent Gram-positive and Gram-negative antibacterial activity where the primary cellular target put forth was the bacterial membrane. While a new potential antibacterial class, their narrow therapeutic index has prohibited further development.
23.5. Heterocycles
23.5.1. Oxazoles
Fourteen oxazole glycosides have been reported from Streptomyces. Of these, seven are allosamidins (chitinase inhibitors) distinguished by the fused bicyclic pentanoidoxazolidine aglycon.1313–1315 Members contain a C-5-O-β-D-N-acetyl-β-D-allosaminyl-β-(1′→4′)-N-acetyl-β-D-allosamine (disaccharide I, monomer 326) or the C-6′-O-methyl variant (Fig. 64). Two related glucoallosamidins exist in which the proximal monomer (326) has been replaced by N-acetyl-β-D-glucosamine (153). N-glucosides trehazolin and trehalostatin share a similar bicyclic aglycon (trehazolamine) but carry a N-7-α-D-glucose (18).1316,1317 Both function as specific irreversible inhibitors of trehalase, with trehazolin as the most potent.1316,1318 One tricyclic streptazolin C-5-O-β-D-xyloside (136) was reported where the glycoside displayed notably higher in vitro anticancer cytotoxicity than non-glycosylated comparators.1319 Finally, two glycosylated cyclocarbamides (SB-253514 and SB-315021 from Pseudomonas) both exist as C-14-O-α-L-rhamnosides (46).1320 Both metabolites inhibit lipoprotein-associated phospholipase A2 and exhibit anti-inflammatory properties.
Fig. 64.
Oxazole aglycons and associated sugars.
23.5.2. Porphyrins
Porphyrins are heterocyclic tetrapyrrole-based macrocyclic metabolites (Fig. 65). Of the 27 porphyrins reported from bacteria, seven tolyporphins from Tolypothrix exist as C17-C-glycosides or C-7,C-17-di-C-glycosides. While 3′,6′-dideoxy-β-D-xylohexopyranose (313) was initially reported as the corresponding monosaccharide, a tolyporphin A structure revision by Moore and coworkers implicated 3′,6′-dideoxy-α-D-xylo-hexopyranose (311)1321 and may call into question the anomeric configuration of other reported members. Tolyporphins inhibit P-glycoprotein1321–1324 and can photosensitize tumor cells.1325
Fig. 65.
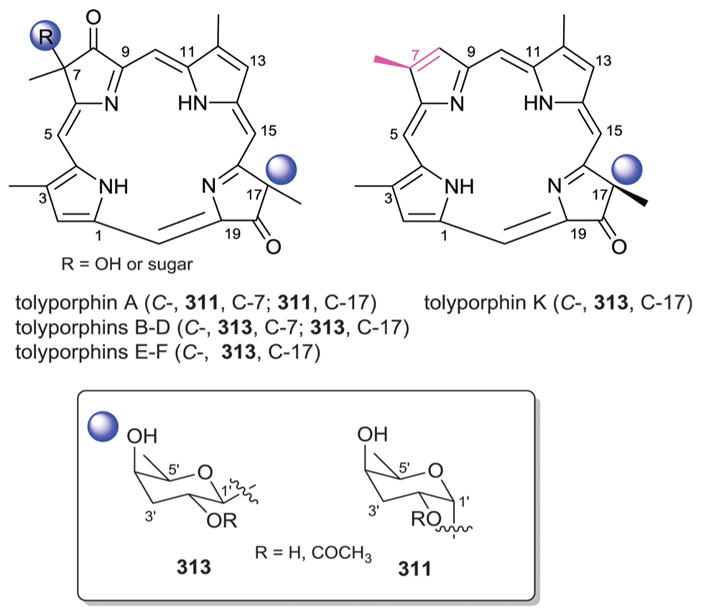
Porphyrin aglycons and associated sugars.
23.5.3. Other heterocycles
Eighteen other bacterial heterocyclic glycosides have been reported. Of these, the macrolactam tetracyclic tetrapetalones A-D from Streptomyces were discovered via a lipooxygenase inhibition guided-assay.1326,1327 They are C-9-β-D-rhodinosides (56) (Fig. 66). The corresponding enantiomer, β-L-rhodinose (169), is appended to C12 of the C2-heptenylidene side chain in hatomamicin, a cytotoxic Saccharopolyspora metabolite.1328 Gualaymycin from Streptomyces is a pyrrolidinyl butanoate metabolite bearing a C2-C-disaccharyl moiety [disaccharide I; 2′-deoxy-2′-amino-β-D-gulosyl-(1′→4′)-α-D-galactose]. These metabolites are superior to dicofol as acaricidals.1329 A phenoxazine N-β-D-glucosidic (104), questiomycin from Microbispora, was reported to have both antibacterial and cytotoxic activity where N-glucosylation was found to suppress bioactivity.1330 Pyralomicins are chlorinated benzopyranopyrrole N-(pseudo)-glycosides produced by Nonomuraea.1331–1333 Four derivatives 1a–d contain the pseudosugar 1′-epi-valienol (P26) proposed to arise from D-seduheptulose-7-phosphate1334 while three others (2a–c) are N-β-D-glucosides (104). Pyralomicins display Gram-positive antibacterial activity where pseudosugar-based derivatives are superior to the corresponding glucosides.1331 Pyrazomycins A and B from Streptomyces are pyrazole C-3-C-glycosides bearing β-D-ribose (25) and α-D-ribose (231), respectively. Two rings of the pentacyclic red pigment rubrolone from Streptomyces derive from a unique C-4-O-C1′/C-3-C-2′ fusion with β-D-fucose (270) (Fig. 66).1335
Fig. 66.
Other heterocycle aglycons and associated sugars. Pink fragment highlights differences between tetrapetalones. Red fragment highlights sugar in rubrolone.
23.6. Additional simple aglycons
Compounds combined with this collection include 44 metabolites. While some are bacterial metabolites, most derive from precursor directed biosynthesis by feeding Streptomyces GT61150 a range of putative aglycons including benzoyls, pyrrolycarbonyls, furanylcarbonyls, phenylacetyls, thiophenylcarbonyls, indolylcarbonyls, pyridylcarbonyls, phenyls, and isovaleryls (Fig. 67) to afford the corresponding α-L-rhamnosides (46) or 6′-deoxy-α-L-talosides (125).590,1336–1339 All corresponding products were monosaccharide-substituted with one exception, benzoyl dirhamnoside bearing both a phenolic hydroxyl- and ester-linked α-L-rhamnose (46) (Fig. 67). Rhamnosylated lactones analogs were found to inhibit the 3α-hydroxysteroid dehydrogenase.590
Fig. 67.
Aglycons with simple moieties and associated sugars.
Additional metabolites include menisdaurin,1340 byelyankacin,1341 and two CKD-711 derivatives1341–1343 from Streptomyces or Enterobacter. Menisdaurin is a C-3-β-D-glucoside (104) while byelyankacin is a C-1-α-L-rhamnoside (46) and potent tyrosinase inhibitor.1341 CKD-711 and CKD-711a are epoxycyclohexanol C-1-N-glycosides bearing either a pentasaccharide I or a trisaccharide II (Fig. 67). Both oligosaccharides are comprised of one or two α-D-glucosyl-(1′→4′)-α-D-glucose units “18-(1′→4′)-18” terminating with a C-4-O-4′-deoxy-4′-amino-β-L-glucose (36) and inhibited α-glucosidase with the trisaccharyl derivative as the most potent. Both compounds also selectively inhibited the growth of the Gram negative bacteria Comamonas terrigena but displayed no other antibacterial/antifungal effects.1342
24. Conclusions
This cumulative analysis reveals a relatively high degree of natural product family-specific conservation in both glycosyl regiospecificity and the specific sugars employed as well as a moderate degree of crossover among natural product families in the use of less functionalized ‘common’ sugars (e.g., standard metabolic sugars and simple corresponding deoxysugars). This likely parallels a higher prevalence (and therefore a higher likelihood of lateral gene transfer) of the core genes encoding some of the common sugar functionalization chemistry in both primary (cell wall) and secondary metabolism where the more complex natural product family-specific sugar functionalization may have derived via divergent evolution. No clear overall general trends relating glycosylation to biological activity were observed (i.e., glycosylation does not always improve or suppress biological activity) however, within a number of natural product subfamilies such trends were apparent. While this points to a lack of a predictive model to apply glycosylation as a medicinal chemistry tool for natural product optimization, it suggests efficient glycodiversification strategies (chemoenzymatic,9,1344 chemoselective10 and/or pathway engineering1345) to offer unique opportunities for improving and/or broadening natural product therapeutic potential. While notable advances have been made toward understanding the fundamental mechanisms and structural biology of enzymes involved in natural product glycosylation,1346–1348 this review also exposes a number of uniquely functionalized sugars and glycosidic connections as a basis for the future discovery of unprecedented enzymatic chemistry.
Acknowledgments
JST would like to specifically acknowledge the heroic effort and commitment of the two lead contributing authors (Drs Elshahawi and Shaaban) and note that, while listed in alphabetical order, these authors contributed equally to this substantial body of work. This work was also supported in part by the University of Kentucky College of Pharmacy, the University of Kentucky Markey Cancer Center, the National Center for Advancing Translational Sciences (UL1TR000117), and National Institutes of Health (NIH) grants R37 AI52218 and R01 CA84374.
Biographies

Sherif I. Elshahawi
Sherif received his BS degree in Pharmaceutical Sciences (2000) from Cairo University, a Master’s degree in Pharmacognosy (2006) from the University of Mississippi and a PhD degree in Biochemistry and Molecular Biology (2012) from the School of Medicine at Oregon Health & Science University with Professor Margo Haygood. He is currently a postdoctoral scholar in the Center for Pharmaceutical Research and Innovation at University of Kentucky College of Pharmacy. Dr Elshahawi’s research interests include the isolation and phylogenetic analyses of microorganisms from unusual environments and the biosynthesis, isolation, bioassay and resistance of bioactive secondary metabolites.

Khaled A. Shaaban
Dr Khaled A. Shaaban obtained his BS degree in Chemistry (2000) from Mansoura University (Egypt) and MS (2005) and PhD degrees in Organic Chemistry (2009) from the University of Göttingen (Germany) with Professor Hartmut Laatsch. He subsequently held postdoctoral appointments at the University of Kentucky (Professor Jürgen Rohr, 2009–2011) and The Scripps Research Institute (TSRI Florida, Professor Ben Shen, 2011–2012) prior to joining the Center for Pharmaceutical Research and Innovation at University of Kentucky College of Pharmacy as a Research Scientist. His major research interests include isolation of bacterial strains, screening, scale-up fermentation, isolation and structure elucidation of novel bioactive microbial natural products.

Madan K. Kharel
Dr Kharel obtained his BS (1998) and MS (2000) from the Tribhuvan University (Nepal), a PhD in biochemistry from the Sun Moon University (South Korea, 2007) and a PhD in pharmaceutical sciences from the University of Kentucky (2010, Professor Jürgen Rohr). Dr Kharel served as a research faculty in the University of Kentucky Center for Pharmaceutical Research and Innovation (2012–2013) and currently is an Assistant Professor of Medicinal Chemistry, Department of Pharmaceutical Sciences, University of Maryland Eastern Shore. His research interests include elucidation of function of novel enzymes, enzymatic synthesis of natural products, and isolation of new bioactive microbial natural products.

Jon S. Thorson
Jon received his BA from Augsburg College (1986), a PhD from the University of Minnesota (1993, Professor Hung-wen Liu) and was a Helen Hay Whitney postdoctoral fellow at the University of California, Berkeley (1993–1996, Professor Peter Schultz). He has served on the faculty of the Memorial Sloan-Kettering Cancer Center and the Sloan-Kettering Division, Joan and Sanford I. Weill Graduate School of Medical Sciences, Cornell University (1996–2001), the University of Wisconsin –Madison School of Pharmacy (2001–2011), and the University of Kentucky College of Pharmacy (2011–present) where he is also the director of the Center for Pharmaceutical Research and Innovation. His research interests include bioprospecting, biocatalysis and biosynthesis.
References
- 1.Kapoor VK, Kaur A. Mini-Rev Med Chem. 2013;13:584–596. doi: 10.2174/1389557511313040010. [DOI] [PubMed] [Google Scholar]
- 2.Desmet T, Soetaert W, Bojarova P, Kren V, Dijkhuizen L, Eastwick-Field V, Schiller A. Chem – Eur J. 2012;18:10786–10801. doi: 10.1002/chem.201103069. [DOI] [PubMed] [Google Scholar]
- 3.La Ferla B, Airoldi C, Zona C, Orsato A, Cardona F, Merlo S, Sironi E, D’Orazio G, Nicotra F. Nat Prod Rep. 2011;28:630–648. doi: 10.1039/c0np00055h. [DOI] [PubMed] [Google Scholar]
- 4.Calvaresi EC, Hergenrother PJ. Chem Sci. 2013;4:2319–2333. doi: 10.1039/C3SC22205E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thuan NH, Sohng JK. J Ind Microbiol Biotechnol. 2013;40:1329–1356. doi: 10.1007/s10295-013-1332-0. [DOI] [PubMed] [Google Scholar]
- 6.Thibodeaux CJ, Melancon CE, Liu H-w. Angew Chem, Int Ed. 2008;47:9814–9859. doi: 10.1002/anie.200801204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hancock SM, Vaughan MD, Withers SG. Curr Opin Chem Biol. 2006;10:509–519. doi: 10.1016/j.cbpa.2006.07.015. [DOI] [PubMed] [Google Scholar]
- 8.Ostash B, Yan X, Fedorenko V, Bechthold A. Top Curr Chem. 2010;297:105–148. doi: 10.1007/128_2010_78. [DOI] [PubMed] [Google Scholar]
- 9.Gantt RW, Peltier-Pain P, Thorson JS. Nat Prod Rep. 2011;28:1811–1853. doi: 10.1039/c1np00045d. [DOI] [PubMed] [Google Scholar]
- 10.Goff RD, Thorson JS. MedChemComm. 2014;5:1036–1047. doi: 10.1039/C4MD00117F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cipolla L, Peri F. Mini-Rev Med Chem. 2011;11:39–54. doi: 10.2174/138955711793564060. [DOI] [PubMed] [Google Scholar]
- 12.Olano C, Méndez C, Salas JA. Nat Prod Rep. 2010;27:571–616. doi: 10.1039/b911956f. [DOI] [PubMed] [Google Scholar]
- 13.Kolarich D, Lepenies B, Seeberger PH. Curr Opin Chem Biol. 2012;16:214–220. doi: 10.1016/j.cbpa.2011.12.006. [DOI] [PubMed] [Google Scholar]
- 14.Chang PV, Bertozzi CR. Chem Commun. 2012;48:8864–8879. doi: 10.1039/c2cc31845h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wilkinson B, Bachmann BO. Curr Opin Chem Biol. 2006;10:169–176. doi: 10.1016/j.cbpa.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 16.Doores KJ, Gamblin DP, Davis BG. Chem – Eur J. 2006;12:656–665. doi: 10.1002/chem.200500557. [DOI] [PubMed] [Google Scholar]
- 17.Schmaltz RM, Hanson SR, Wong CH. Chem Rev. 2011;111:4259–4307. doi: 10.1021/cr200113w. [DOI] [PubMed] [Google Scholar]
- 18.Laatsch H. AntiBase 2012: The natural compound identifier. Wiley-VCH; Weinheim: 2012. [Google Scholar]
- 19.Flatt PM, Mahmud T. Nat Prod Rep. 2007;24:358–392. doi: 10.1039/b603816f. [DOI] [PubMed] [Google Scholar]
- 20.Mahmud T, Flatt PM, Wu X. J Nat Prod. 2007;70:1384–1391. doi: 10.1021/np070210q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mahmud T. Curr Opin Chem Biol. 2009;13:161–170. doi: 10.1016/j.cbpa.2009.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dobashi K, Isshiki K, Sawa T, Obata T, Hamada M, Naganawa H, Takita T, Takeuchi T, Umezawa H, Bei HS. J Antibiot. 1986;39:1779–1783. doi: 10.7164/antibiotics.39.1779. [DOI] [PubMed] [Google Scholar]
- 23.Hurley TR, Smitka TA, Wilton JH, Bunge RH, Hokanson GC, French JC. J Antibiot. 1986;39:1086–1091. doi: 10.7164/antibiotics.39.1086. [DOI] [PubMed] [Google Scholar]
- 24.Adams ES, Rinehart KL. J Antibiot. 1994;47:1456–1465. doi: 10.7164/antibiotics.47.1456. [DOI] [PubMed] [Google Scholar]
- 25.Ito T, Roongsawang N, Shirasaka N, Lu W, Flatt PM, Kasanah N, Miranda C, Mahmud T. ChemBioChem. 2009;10:2253–2265. doi: 10.1002/cbic.200900339. [DOI] [PubMed] [Google Scholar]
- 26.Lu W, Roongsawang N, Mahmud T. Chem Biol. 2011;18:425–431. doi: 10.1016/j.chembiol.2011.01.016. [DOI] [PubMed] [Google Scholar]
- 27.Mahmud T. US20130231377 A1. US Pat. 2013
- 28.Okami Y, Hotta K, Yoshida M, Ikeda D, Kondo S, Umezawa H. J Antibiot. 1979;32:964–966. doi: 10.7164/antibiotics.32.964. [DOI] [PubMed] [Google Scholar]
- 29.Umezawa H, Kondo S, Okami Y. EP 0048549A1. European Pat. 1981
- 30.Collins PM. Dictionary of carbohydrates. 1992:S-00012. and reference cited therein. [Google Scholar]
- 31.Watanabe I, Deushi T, Yamaguchi T, Kamiya K, Nakayama M, Mori T. J Antibiot. 1979;32:1066–1068. doi: 10.7164/antibiotics.32.1066. [DOI] [PubMed] [Google Scholar]
- 32.Kurath P, Rosenbrook W, Dunnigan DA, Mcalpine JB, Egan RS, Stanaszek RS, Cirovic M, Mueller SL, Washburn WH. J Antibiot. 1984;37:1130–1143. doi: 10.7164/antibiotics.37.1130. [DOI] [PubMed] [Google Scholar]
- 33.Tadanier J, Hallas R. J Antibiot. 1983;36:256–266. doi: 10.7164/antibiotics.36.256. [DOI] [PubMed] [Google Scholar]
- 34.Iida T, Sato M, Matsubara I, Mori Y, Shirahata K. J Antibiot. 1979;32:1273–1279. doi: 10.7164/antibiotics.32.1273. [DOI] [PubMed] [Google Scholar]
- 35.Martin JR. US4213972. US Pat. 1980
- 36.Carney RE. US4214080. US Pat. 1980
- 37.Berdy J, Pauncz JK, Vajna ZM, Horvath G, Gyimesi J, Koczka I. J Antibiot. 1977;30:945–954. doi: 10.7164/antibiotics.30.945. [DOI] [PubMed] [Google Scholar]
- 38.Ogawa H, Ito T. J Antibiot. 1957;10:267. [PubMed] [Google Scholar]
- 39.Baud H, Betencourt A, Peyre M, Penasse L. J Antibiot. 1977;30:720–723. doi: 10.7164/antibiotics.30.720. [DOI] [PubMed] [Google Scholar]
- 40.McGilveray IJ, Rinehart KL. J Am Chem Soc. 1965;87:4003–4004. doi: 10.1021/ja01095a054. [DOI] [PubMed] [Google Scholar]
- 41.Tohma S, Kondo H, Yokotsuka J, Iwamoto J, Matsuhashi G, Ito T, Seto H. J Antibiot. 1989;42:1205–1212. doi: 10.7164/antibiotics.42.1205. [DOI] [PubMed] [Google Scholar]
- 42.Shirahata K, Kase H, Kitamura S, Iida T. J Antibiot. 1982;35:520–523. doi: 10.7164/antibiotics.35.520. [DOI] [PubMed] [Google Scholar]
- 43.Wakisaka Y, Koizumi K, Nishimoto Y, Kobayashi M, Tsuji N. J Antibiot. 1980;33:695–704. doi: 10.7164/antibiotics.33.695. [DOI] [PubMed] [Google Scholar]
- 44.Truscheit E, Frommer W, Junge B, Muller L, Schmidt DD, Wingender W. Angew Chem, Int Ed Engl. 1981;20:744–761. [Google Scholar]
- 45.Fukuhara K, Murai H, Murao S. Agric Biol Chem. 1982;46:1941–1945. [Google Scholar]
- 46.Kameda Y, Asano N, Yamaguchi T, Matsui K, Horii S, Fukase H. J Antibiot. 1986;39:1491–1494. doi: 10.7164/antibiotics.39.1491. [DOI] [PubMed] [Google Scholar]
- 47.Newton GL, Jensen PR, Macmillan JB, Fenical W, Fahey RC. Arch Microbiol. 2008;190:547–557. doi: 10.1007/s00203-008-0405-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Iinuma K, Kondo S, Maeda K, Umezawa H. J Antibiot. 1975;28:613–615. doi: 10.7164/antibiotics.28.613. [DOI] [PubMed] [Google Scholar]
- 49.Umezawa H, Hamada M, Suhara Y, Hashimoto T, Ikekawa T. Antimicrob Agents Chemother. 1965;5:753–757. [PubMed] [Google Scholar]
- 50.Saitoh K, Tsunakawa M, Kawasaki K, Miyaki T. US4972545. US Pat. 1988
- 51.Nagabhushan TL, Wright JJ, Cooper AB, Turner WN, Miller GH. J Antibiot. 1978;31:43–54. doi: 10.7164/antibiotics.31.43. [DOI] [PubMed] [Google Scholar]
- 52.Reimann H, Cooper DJ, Mallams AK, Jaret RS, Yehaskel A, Kugelman M, Vernay HF, Schumach D. J Org Chem. 1974;39:1451–1457. doi: 10.1021/jo00924a001. [DOI] [PubMed] [Google Scholar]
- 53.Hanessian S, Szychowski J, Maianti JP. Org Lett. 2009;11:429–432. doi: 10.1021/ol802421d. [DOI] [PubMed] [Google Scholar]
- 54.Koch KF, Merkel KE, Oconnor SC, Occolowitz JL, Paschal JW, Dorman DE. J Org Chem. 1978;43:1430–1434. [Google Scholar]
- 55.Lee BK, Wagman GH, Rane DF, Marquez JA, Daniels PJL. US4234685. US Pat. 1979
- 56.Oka Y, Ishida H, Morioka M, Numasaki Y, Yamafuji T, Osono T, Umezawa H. J Antibiot. 1981;34:777–781. doi: 10.7164/antibiotics.34.777. [DOI] [PubMed] [Google Scholar]
- 57.Dorman DE, Paschal JW, Merkel KE. J Am Chem Soc. 1976;98:6885–6888. doi: 10.1021/ja00438a020. [DOI] [PubMed] [Google Scholar]
- 58.Egan RS, Sinclair AC, De Vault RL, McAlpine JB, Mueller SL, Goodley PC, Stanaszek RS, Cirovic M, Mauritz RJ, Mitscher LA, Shirahata K, Sato S, Iida T. J Antibiot. 1977;30:31–38. doi: 10.7164/antibiotics.30.31. [DOI] [PubMed] [Google Scholar]
- 59.Tatsuta K, Akimoto K, Takahashi H, Hamatsu T, Annaka M, Kinoshita M. Bull Chem Soc Jpn. 1984;57:529–538. [Google Scholar]
- 60.Dion HW, Woo PW, Willmer NE, Kern DL, Onaga J, Fusari SA. Antimicrob Agents Chemother. 1972;2:84–88. doi: 10.1128/aac.2.2.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Horii S, Nogami I, Mizokami N, Arai Y, Yoneda M. Antimicrob Agents Chemother. 1974;5:578–581. doi: 10.1128/aac.5.6.578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang WX, Chen Y, Liang QZ, Li H, Jin HW, Zhang LR, Meng XB, Li ZJ. J Org Chem. 2013;78:400–409. doi: 10.1021/jo302247x. [DOI] [PubMed] [Google Scholar]
- 63.Claes PJ, Comperno F, Vanderha H. J Antibiot. 1974;27:931–942. doi: 10.7164/antibiotics.27.931. [DOI] [PubMed] [Google Scholar]
- 64.Nishimur Y, Tsuchiya T, Umezawa S. Bull Chem Soc Jpn. 1971;44:2521–2527. [Google Scholar]
- 65.Davies DH, Greeves D, Mallams AK, Morton JB, Tkach RW. J Chem Soc, Perkin Trans 1. 1975:814–818. doi: 10.1002/chin.197529439. [DOI] [PubMed] [Google Scholar]
- 66.Odakura Y, Kase H, Nakayama K. J Antibiot. 1983;36:125–130. doi: 10.7164/antibiotics.36.125. [DOI] [PubMed] [Google Scholar]
- 67.Daniels PJL, Jaret RS, Nagabhushan TL, Turner WN. J Antibiot. 1976;29:488–491. doi: 10.7164/antibiotics.29.488. [DOI] [PubMed] [Google Scholar]
- 68.Mcalpine JB, Sinclair AC, Egan RS, Devault RL, Stanaszek RS, Cirovic M, Mueller SL, Goodley PC, Mauritz RJ, Wideburg NE, Mitscher LA, Shirahata K, Matsushima H, Sato S, Iida T. J Antibiot. 1977;30:39–49. doi: 10.7164/antibiotics.30.39. [DOI] [PubMed] [Google Scholar]
- 69.Tatsuoka S, Kusaka A, Miyake M, Inoue M, Hitomi H, Shiraishi H, Iwasaki H, Imanishi M. Chem Pharm Bull. 1957;5:343–349. doi: 10.1248/cpb1953.5.343. [DOI] [PubMed] [Google Scholar]
- 70.Benedict RG, Lindenfelser LA, Stodola FH, Traufler DH. J Bacteriol. 1951;62:487–497. doi: 10.1128/jb.62.4.487-497.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kondo S, Ikeda Y, Ikeda T, Gomi S, Naganawa H, Hamada M, Umezawa H. J Antibiot. 1985;38:433–435. doi: 10.7164/antibiotics.38.433. [DOI] [PubMed] [Google Scholar]
- 72.Mori T, Kyotani Y, Watanabe I, Oda T. J Antibiot. 1972;25:317–319. doi: 10.7164/antibiotics.25.317. [DOI] [PubMed] [Google Scholar]
- 73.Autissier D, Barthelemy P, Mazieres N, Peyre M, Penasse L. J Antibiot. 1981;34:536–543. doi: 10.7164/antibiotics.34.536. [DOI] [PubMed] [Google Scholar]
- 74.Oda T, Mori T, Kyotani Y, Nakayama M. J Antibiot. 1971;24:511–518. doi: 10.7164/antibiotics.24.511. [DOI] [PubMed] [Google Scholar]
- 75.Umezawa S, Nishimura Y. J Antibiot. 1977;30:189–191. doi: 10.7164/antibiotics.30.189. [DOI] [PubMed] [Google Scholar]
- 76.Vertesy L, Fehlhaber HW, Schulz A. Angew Chem, Int Ed Engl. 1994;33:1844–1846. [Google Scholar]
- 77.Rohr J, Thiericke R. Nat Prod Rep. 1992;9:103–137. doi: 10.1039/np9920900103. [DOI] [PubMed] [Google Scholar]
- 78.Krohn K, Rohr J. Top Curr Chem. 1997;188:127–195. [Google Scholar]
- 79.Kharel MK, Pahari P, Shepherd MD, Tibrewal N, Nybo SE, Shaaban KA, Rohr J. Nat Prod Rep. 2012;29:264–325. doi: 10.1039/c1np00068c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Henkel T, Rohr J, Beale JM, Schwenen L. J Antibiot. 1990;43:492–503. doi: 10.7164/antibiotics.43.492. [DOI] [PubMed] [Google Scholar]
- 81.Weber S, Zolke C, Rohr J, Beale JM. J Org Chem. 1994;59:4211–4214. [Google Scholar]
- 82.Luzhetskii AN, Ostash BE, Fedorenko VA. Genetika. 2001;37:1340–1347. [PubMed] [Google Scholar]
- 83.Matseliukh BP, Lavrinchuk V. Mikrobiol Zh. 1999;61:22–27. [PubMed] [Google Scholar]
- 84.Erb A, Krauth C, Luzhetskyy A, Bechthold A. Appl Microbiol Biotechnol. 2009;83:1067–1076. doi: 10.1007/s00253-009-1993-9. [DOI] [PubMed] [Google Scholar]
- 85.Ostash B, Rix U, Rix LL, Liu T, Lombo F, Luzhetskyy A, Gromyko O, Wang C, Brana AF, Méndez C, Salas JA, Fedorenko V, Rohr J. Chem Biol. 2004;11:547–555. doi: 10.1016/j.chembiol.2004.03.011. [DOI] [PubMed] [Google Scholar]
- 86.Luzhetskyy A, Fedoryshyn M, Durr C, Taguchi T, Novikov V, Bechthold A. Chem Biol. 2005;12:725–729. doi: 10.1016/j.chembiol.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 87.Zhu L, Luzhetskyy A, Luzhetska M, Mattingly C, Adams V, Bechthold A, Rohr J. ChemBioChem. 2007;8:83–88. doi: 10.1002/cbic.200600360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Luzhetskyy A, Zhu L, Gibson M, Fedoryshyn M, Durr C, Hofmann C, Hoffmeister D, Ostash B, Mattingly C, Adams V, Fedorenko V, Rohr J, Bechthold A. ChemBioChem. 2005;6:675–678. doi: 10.1002/cbic.200400316. [DOI] [PubMed] [Google Scholar]
- 89.Shaaban KA, Stamatkin C, Damodaran C, Rohr J. J Antibiot. 2011;64:141–150. doi: 10.1038/ja.2010.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Shaaban KA, Srinivasan S, Kumar R, Damodaran C, Rohr J. J Nat Prod. 2011;74:2–11. doi: 10.1021/np100469y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Fujioka K, Furihata K, Shimazu A, Hayakawa Y, Seto H. J Antibiot. 1991;44:1025–1028. doi: 10.7164/antibiotics.44.1025. [DOI] [PubMed] [Google Scholar]
- 92.Nemoto A, Tanaka Y, Karasaki Y, Komaki H, Yazawa K, Mikami Y, Tojo T, Kadowaki K, Tsuda M, Kobayashi J. J Antibiot. 1997;50:18–21. doi: 10.7164/antibiotics.50.18. [DOI] [PubMed] [Google Scholar]
- 93.Tsuda M, Sato H, Tanaka Y, Yazawa G, Mikami Y, Sasaki T, Kobayashi J. J Chem Soc, Perkin Trans 1. 1996:1773–1775. [Google Scholar]
- 94.Tsuda M, Nemoto A, Komaki H, Tanaka Y, Yazawa K, Mikami Y, Kobayashi J. J Nat Prod. 1999;62:1640–1642. doi: 10.1021/np970132e. [DOI] [PubMed] [Google Scholar]
- 95.Ohmori K, Mori K, Ishikawa Y, Tsuruta H, Kuwahara S, Harada N, Suzuki K. Angew Chem, Int Ed. 2004;43:3167–3171. doi: 10.1002/anie.200453801. [DOI] [PubMed] [Google Scholar]
- 96.Akerling ZR, Norton JE, Houk KN. Org Lett. 2004;6:4273–4275. doi: 10.1021/ol048277h. [DOI] [PubMed] [Google Scholar]
- 97.Kanamaru T, Nozaki Y, Muroi M. JP 02 289-532/1990. Japanese Pat. 1991
- 98.Uchida T, Imoto M, Watanabe Y, Miura K, Dobashi T, Matsuda N, Sawa T, Naganawa H, Hamada M, Takeuchi T. J Antibiot. 1985;38:1171–1181. doi: 10.7164/antibiotics.38.1171. [DOI] [PubMed] [Google Scholar]
- 99.Sekizawa R, Iinuma H, Naganawa H, Hamada M, Takeuchi T, Yamaizumi J, Umezawa K. J Antibiot. 1996;49:487–490. doi: 10.7164/antibiotics.49.487. [DOI] [PubMed] [Google Scholar]
- 100.Stroch K, Zeeck A, Antal N, Fiedler HP. J Antibiot. 2005;58:103–110. doi: 10.1038/ja.2005.13. [DOI] [PubMed] [Google Scholar]
- 101.Antal N, Fiedler HP, Stackebrandt E, Beil W, Stroch K, Zeeck A. J Antibiot. 2005;58:95–102. doi: 10.1038/ja.2005.12. [DOI] [PubMed] [Google Scholar]
- 102.Erb A, Luzhetskyy A, Hardter U, Bechthold A. ChemBioChem. 2009;10:1392–1401. doi: 10.1002/cbic.200900054. [DOI] [PubMed] [Google Scholar]
- 103.Shaaban KA, Ahmed TA, Leggas M, Rohr J. J Nat Prod. 2012;75:1383–1392. doi: 10.1021/np300316b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Alvi KA, Baker DD, Stienecker V, Hosken M, Nair BG. J Antibiot. 2000;53:496–501. [PubMed] [Google Scholar]
- 105.Abdelfattah MS, Kharel MK, Hitron JA, Baig I, Rohr J. J Nat Prod. 2008;71:1569–1573. doi: 10.1021/np800281f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Imamura N, Kakinuma K, Ikekawa N, Tanaka H, Omura S. J Antibiot. 1981;34:1517–1518. doi: 10.7164/antibiotics.34.1517. [DOI] [PubMed] [Google Scholar]
- 107.Imamura N, Kakinuma K, Ikekawa N, Tanaka H, Omura S. J Antibiot. 1982;35:602–608. doi: 10.7164/antibiotics.35.602. [DOI] [PubMed] [Google Scholar]
- 108.Kawashima A, Kishimura Y, Tamai M, Hanada K. Chem Pharm Bull. 1989;37:3429–3431. doi: 10.1248/cpb.37.3429. [DOI] [PubMed] [Google Scholar]
- 109.Kawashima A, Yoshimura Y, Goto J, Nakaike S, Mizutani T, Hanada K, Omura S. J Antibiot. 1988;41:1913–1914. doi: 10.7164/antibiotics.41.1913. [DOI] [PubMed] [Google Scholar]
- 110.Hayakawa Y, Iwakiri T, Imamura K, Seto H, Otake N. J Antibiot. 1987;40:1785–1787. doi: 10.7164/antibiotics.40.1785. [DOI] [PubMed] [Google Scholar]
- 111.Huang H, Yang T, Ren X, Liu J, Song Y, Sun A, Ma J, Wang B, Zhang Y, Huang C, Zhang C, Ju J. J Nat Prod. 2012;75:202–208. doi: 10.1021/np2008335. [DOI] [PubMed] [Google Scholar]
- 112.Basnet DB, Oh TJ, Vu TTH, Sthapit B, Liou K, Lee HC, Yoo JC, Sohng JK. Mol Cells. 2006;22:154–162. [PubMed] [Google Scholar]
- 113.Chu M, Yarborough R, Schwartz J, Patel MG, Horan AC, Gullo VP, Das PR, Puar MS. J Antibiot. 1993;46:861–865. doi: 10.7164/antibiotics.46.861. [DOI] [PubMed] [Google Scholar]
- 114.Kawamura N, Sawa B, Takahashi Y, Sawa T, Kinoshita N, Naganawa H, Hamada M, Takeuchi T. J Antibiot. 1995;48:1521–1524. doi: 10.7164/antibiotics.48.1521. [DOI] [PubMed] [Google Scholar]
- 115.Rohr J, Beale JM, Floss HG. J Antibiot. 1989;42:1151–1157. doi: 10.7164/antibiotics.42.1151. [DOI] [PubMed] [Google Scholar]
- 116.Rohr J. J Antibiot. 1989;42:1482–1488. doi: 10.7164/antibiotics.42.1482. [DOI] [PubMed] [Google Scholar]
- 117.Rohr J, Zeeck A. J Antibiot. 1987;40:459–467. doi: 10.7164/antibiotics.40.459. [DOI] [PubMed] [Google Scholar]
- 118.Drautz H, Zahner H, Rohr J, Zeeck A. J Antibiot. 1986;39:1657–1669. doi: 10.7164/antibiotics.39.1657. [DOI] [PubMed] [Google Scholar]
- 119.Zeeck A, Rohr J, Sheldrick GM, Jones PG, Paulus EF. J Chem Res, Synop. 1986:104–105. [Google Scholar]
- 120.Kunzel E, Faust B, Oelkers C, Weissbach U, Bearden DW, Weitnauer G, Westrich L, Bechthold A, Rohr J. J Am Chem Soc. 1999;121:11058–11062. [Google Scholar]
- 121.Faust B, Hoffmeister D, Weitnauer G, Westrich L, Haag S, Schneider P, Decker H, Kunzel E, Rohr J, Bechthold A. Microbiology. 2000;146:147–154. doi: 10.1099/00221287-146-1-147. [DOI] [PubMed] [Google Scholar]
- 122.Hoffmeister D, Ichinose K, Domann S, Faust B, Trefzer A, Drager G, Kirschning A, Fischer C, Kunzel E, Bearden D, Rohr J, Bechthold A. Chem Biol. 2000;7:821–831. doi: 10.1016/s1074-5521(00)00029-6. [DOI] [PubMed] [Google Scholar]
- 123.Hoffmeister D, Drager G, Ichinose K, Rohr J, Bechthold A. J Am Chem Soc. 2003;125:4678–4679. doi: 10.1021/ja029645k. [DOI] [PubMed] [Google Scholar]
- 124.Rohr J, Schonewolf M, Udvarnoki G, Eckardt K, Schumann G, Wagner C, Beale JM, Sorey SD. J Org Chem. 1993;58:2547–2551. [Google Scholar]
- 125.Trefzer A, Hoffmeister D, Kunzel E, Stockert S, Weitnauer G, Westrich L, Rix U, Fuchser J, Bindseil KU, Rohr J, Bechthold A. Chem Biol. 2000;7:133–142. doi: 10.1016/s1074-5521(00)00079-x. [DOI] [PubMed] [Google Scholar]
- 126.Hayakawa Y, Iwakiri T, Imamura K, Seto H, Otake N. J Antibiot. 1985;38:960–963. doi: 10.7164/antibiotics.38.960. [DOI] [PubMed] [Google Scholar]
- 127.Ren X, Lu X, Ke A, Zheng Z, Lin J, Hao W, Zhu J, Fan Y, Ding Y, Jiang Q, Zhang H. J Antibiot. 2011;64:339–343. doi: 10.1038/ja.2011.4. [DOI] [PubMed] [Google Scholar]
- 128.Okabe T, Funaishi K, Hagiwara M, Kawamura K, Suda H, Sato F. JP 02 291 287. Japanese Pat. 1990
- 129.Theriault RJ, Rasmussen RR, Kohl WL, Prokop JF, Hutch TB, Barlow GJ. J Antibiot. 1986;39:1509–1514. doi: 10.7164/antibiotics.39.1509. [DOI] [PubMed] [Google Scholar]
- 130.Rasmussen RR, Nuss ME, Scherr MH, Mueller SL, McAlpine JB, Mitscher LA. J Antibiot. 1986;39:1515–1526. doi: 10.7164/antibiotics.39.1515. [DOI] [PubMed] [Google Scholar]
- 131.Carr G, Derbyshire ER, Caldera E, Currie CR, Clardy J. J Nat Prod. 2012;75:1806–1809. doi: 10.1021/np300380t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Uesato S, Tokunaga T, Takeuchi K. Bioorg Med Chem Lett. 1998;8:1969–1972. doi: 10.1016/s0960-894x(98)00340-0. [DOI] [PubMed] [Google Scholar]
- 133.Uesato S, Tokunaga T, Mizuno Y, Fujioka H, Kada S, Kuwajima H. J Nat Prod. 2000;63:787–792. doi: 10.1021/np990533p. [DOI] [PubMed] [Google Scholar]
- 134.Ueda JY, Izumikawa M, Mukai A, Nagai A, Hwang JH, Takagi M, Shin-ya K. J Antibiot. 2011;64:367–372. doi: 10.1038/ja.2011.8. [DOI] [PubMed] [Google Scholar]
- 135.Etoh H, Iguchi M, Nagasawa T, Tani Y, Yamada H, Fukami H. Agric Biol Chem. 1987;51:1819–1824. [Google Scholar]
- 136.Sasaki E, Ogasawara Y, Liu H-w. J Am Chem Soc. 2010;132:7405–7417. doi: 10.1021/ja1014037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Schneemann I, Kajahn I, Ohlendorf B, Zinecker H, Erhard A, Nagel K, Wiese J, Imhoff JF. J Nat Prod. 2010;73:1309–1312. doi: 10.1021/np100135b. [DOI] [PubMed] [Google Scholar]
- 138.Paululat T, Kulik A, Hausmann H, Karagouni AD, Zinecker H, Imhoff JF, Fiedler HP. Eur J Org Chem. 2010:2344–2350. [Google Scholar]
- 139.Nagasawa T, Fukao H, Irie H, Yamada H. J Antibiot. 1984;37:693–699. doi: 10.7164/antibiotics.37.693. [DOI] [PubMed] [Google Scholar]
- 140.Kalinovskaya NI, Kalinovsky AI, Romanenko LA, Dmitrenok PS, Kuznetsova TA. Nat Prod Commun. 2010;5:597–602. [PubMed] [Google Scholar]
- 141.Holzenkampfer M, Walker M, Zeeck A, Schimana J, Fiedler HP. J Antibiot. 2002;55:301–307. doi: 10.7164/antibiotics.55.301. [DOI] [PubMed] [Google Scholar]
- 142.Schimana J, Fiedler HP, Groth I, Sussmuth R, Beil W, Walker M, Zeeck A. J Antibiot. 2000;53:779–787. doi: 10.7164/antibiotics.53.779. [DOI] [PubMed] [Google Scholar]
- 143.Schimana J, Walker M, Zeeck A, Fiedler HP. J Ind Microbiol Biotechnol. 2001;27:144–148. doi: 10.1038/sj.jim.7000024. [DOI] [PubMed] [Google Scholar]
- 144.Galm U, Schimana J, Fiedler HP, Schmidt J, Li SM, Heide L. Arch Microbiol. 2002;178:102–114. doi: 10.1007/s00203-002-0429-z. [DOI] [PubMed] [Google Scholar]
- 145.Hayakawa Y, Iwakiri T, Imamura K, Seto H, Otake N. J Antibiot. 1985;38:957–959. doi: 10.7164/antibiotics.38.957. [DOI] [PubMed] [Google Scholar]
- 146.Sawa R, Matsuda N, Uchida T, Ikeda T, Sawa T, Naganawa H, Hamada M, Takeuchi T. J Antibiot. 1991;44:396–402. doi: 10.7164/antibiotics.44.396. [DOI] [PubMed] [Google Scholar]
- 147.Igarashi M, Utsumi R, Watanabe T. JP 2011201843A. Japanese Pat. 2011
- 148.Xin W, Ye X, Yu S, Lian XY, Zhang Z. Mar Drugs. 2012;10:2388–2402. doi: 10.3390/md10112388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Bruntner C, Binder T, Pathomaree W, Goodfellow M, Bull AT, Potterat O, Puder C, Horer S, Schmid A, Bolek W, Wagner K, Mihm G, Fiedler HP. J Antibiot. 2005;58:346–349. doi: 10.1038/ja.2005.43. [DOI] [PubMed] [Google Scholar]
- 150.Bindseil KU, Hug P, Peter HH, Petersen F, Roggo BE. J Antibiot. 1995;48:457–461. doi: 10.7164/antibiotics.48.457. [DOI] [PubMed] [Google Scholar]
- 151.Martin GDA, Tan LT, Jensen PR, Dimayuga RE, Fairchild CR, Raventos-Suarez C, Fenical W. J Nat Prod. 2007;70:1406–1409. doi: 10.1021/np060621r. [DOI] [PubMed] [Google Scholar]
- 152.Luzhetskyy A, Taguchi T, Fedoryshyn M, Durr C, Wohlert SE, Novikov V, Bechthold A. ChemBioChem. 2005;6:1406–1410. doi: 10.1002/cbic.200500018. [DOI] [PubMed] [Google Scholar]
- 153.Balitz DM, O’Herron FA, Bush J, Vyas DM, Nettleton DE, Grulich RE, Bradner WT, Doyle TW, Arnold E, Clardy J. J Antibiot. 1981;34:1544–1555. doi: 10.7164/antibiotics.34.1544. [DOI] [PubMed] [Google Scholar]
- 154.Nakano H, Matsuda Y, Ito K, Ohkubo S, Morimoto M, Tomita F. J Antibiot. 1981;34:266–270. doi: 10.7164/antibiotics.34.266. [DOI] [PubMed] [Google Scholar]
- 155.Takahashi KY, Yoshida M, Tomita F, Shirahata K. J Antibiot. 1981;34:271–275. doi: 10.7164/antibiotics.34.271. [DOI] [PubMed] [Google Scholar]
- 156.Morimoto MO, Okubo S, Tomita F, Marumo H. J Antibiot. 1981;34:701–707. doi: 10.7164/antibiotics.34.701. [DOI] [PubMed] [Google Scholar]
- 157.Tomita F, Takahashi K, Tamaoki T. J Antibiot. 1982;35:1038–1041. doi: 10.7164/antibiotics.35.1038. [DOI] [PubMed] [Google Scholar]
- 158.Takahashi K, Tomita F. J Antibiot. 1983;36:1531–1535. doi: 10.7164/antibiotics.36.1531. [DOI] [PubMed] [Google Scholar]
- 159.Horii S, Fukase H, Mizuta E, Hatano K, Mizuno K. Chem Pharm Bull. 1980;28:3601–3611. [Google Scholar]
- 160.Hatano K, Higashide E, Shibata M, Kameda Y, Horii S, Mizuno K. Agric Biol Chem. 1980;44:1157–1163. [Google Scholar]
- 161.Jain TC, Simolike GC, Jackman LM. Tetrahedron. 1983;39:599–605. [Google Scholar]
- 162.Hou J, Liu P, Qu H, Fu P, Wang Y, Wang Z, Li Y, Teng X, Zhu W. J Antibiot. 2012;65:523–526. doi: 10.1038/ja.2012.61. [DOI] [PubMed] [Google Scholar]
- 163.Li YQ, Huang XS, Ishida K, Maier A, Kelter G, Jiang Y, Peschel G, Menzel KD, Li MG, Wen ML, Xu LH, Grabley S, Fiebig HH, Jiang CL, Hertweck C, Sattler I. Org Biomol Chem. 2008;6:3601–3605. doi: 10.1039/b808633h. [DOI] [PubMed] [Google Scholar]
- 164.Nakajima S, Kojiri K, Suda H, Okanishi M. J Antibiot. 1991;44:1061–1064. doi: 10.7164/antibiotics.44.1061. [DOI] [PubMed] [Google Scholar]
- 165.Kojiri K, Arakawa H, Satoh F, Kawamura K, Okura A, Suda H, Okanishi M. J Antibiot. 1991;44:1054–1060. doi: 10.7164/antibiotics.44.1054. [DOI] [PubMed] [Google Scholar]
- 166.Narita T, Matsumoto M, Mogi K, Kukita K, Kawahara R, Nakashima T. J Antibiot. 1989;42:347–356. doi: 10.7164/antibiotics.42.347. [DOI] [PubMed] [Google Scholar]
- 167.Yamashita N, Shin-Ya K, Furihata K, Hayakawa Y, Seto H. J Antibiot. 1998;51:1105–1108. doi: 10.7164/antibiotics.51.1105. [DOI] [PubMed] [Google Scholar]
- 168.Arai M, Tomoda H, Matsumoto A, Takahashi Y, Woodruff BH, Ishiguro N, Kobayashi S, Omura S. J Antibiot. 2001;54:554–561. doi: 10.7164/antibiotics.54.554. [DOI] [PubMed] [Google Scholar]
- 169.Arai M, Tomoda H, Tabata N, Ishiguro N, Kobayashi S, Omura S. J Antibiot. 2001;54:562–566. doi: 10.7164/antibiotics.54.562. [DOI] [PubMed] [Google Scholar]
- 170.Carter GT, Fantini AA, James JC, Borders DB, White RJ. Tetrahedron Lett. 1984;25:255–258. [Google Scholar]
- 171.Kharel MK, Lian H, Rohr J. Org Biomol Chem. 2011;9:1799–1808. doi: 10.1039/c0ob00854k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Fujii T, Kumagai H, Nakashima T, Sakai K, Sameshima T, Yoshioka T. US6030951 A. US Pat. 2000
- 173.Weiss U, Yoshihira K, Highet RJ, White RJ, Wei TT. J Antibiot. 1982;35:1194–1201. doi: 10.7164/antibiotics.35.1194. [DOI] [PubMed] [Google Scholar]
- 174.Wei TT, Byrne KM, Warnick-Pickle D, Greenstein M. J Antibiot. 1982;35:545–548. doi: 10.7164/antibiotics.35.545. [DOI] [PubMed] [Google Scholar]
- 175.Shepherd MD, Liu T, Méndez C, Salas JA, Rohr J. Appl Environ Microbiol. 2011;77:435–441. doi: 10.1128/AEM.01774-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Rodriguez L, Oelkers C, Aguirrezabalaga I, Brana AF, Rohr J, Méndez C, Salas JA. J Mol Microbiol Biotechnol. 2000;2:271–276. [PubMed] [Google Scholar]
- 177.Liu T, Kharel MK, Zhu L, Bright S, Rohr J. ChemBioChem. 2009;10:278–286. doi: 10.1002/cbic.200800348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Rix U, Zheng JT, Rix LLR, Greenwell L, Yang KQ, Rohr J. J Am Chem Soc. 2004;126:4496–4497. doi: 10.1021/ja031724o. [DOI] [PubMed] [Google Scholar]
- 179.Liu T, Fischer C, Beninga C, Rohr J. J Am Chem Soc. 2004;126:12262–12263. doi: 10.1021/ja0467521. [DOI] [PubMed] [Google Scholar]
- 180.Jakeman DL, Borissow CN, Graham CL, Timmons SC, Reid TR, Syvitski RT. Chem Commun. 2006:3738–3740. doi: 10.1039/b608847c. [DOI] [PubMed] [Google Scholar]
- 181.Dupuis SN, Veinot T, Monro SM, Douglas SE, Syvitski RT, Goralski KB, McFarland SA, Jakeman DL. J Nat Prod. 2011;74:2420–2424. doi: 10.1021/np200689w. [DOI] [PubMed] [Google Scholar]
- 182.Mohri S, Stefinovic M, Sneickus V. J Org Chem. 1998;63:1746. doi: 10.1021/jo971123d. [DOI] [PubMed] [Google Scholar]
- 183.Mohri SI, Stefinovic M, Snieckus V. J Org Chem. 1997;62:7072–7073. doi: 10.1021/jo971123d. [DOI] [PubMed] [Google Scholar]
- 184.Brockmann H. Fortschr Chem Org Naturst. 1963;21:121–182. [PubMed] [Google Scholar]
- 185.Dimarco A, Gaetani M, Orezzi P, Scarpinato BM, Silvestrini R, Soldati M, Dasdia T, Valentini L. Nature. 1964;201:706–707. doi: 10.1038/201706a0. [DOI] [PubMed] [Google Scholar]
- 186.Arcamone F, Cassinelli G, Fantini G, Grein A, Orezzi P, Pol C, Spalla C. Biotechnol Bioeng. 2000;67:704–713. doi: 10.1002/(sici)1097-0290(20000320)67:6<704::aid-bit8>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 187.Arcamone F, Cassinelli G, Fantini G, Grein A, Orezzi P, Pol C, Spalla C. Biotechnol Bioeng. 1969;11:1101–1110. doi: 10.1002/bit.260110607. [DOI] [PubMed] [Google Scholar]
- 188.Laatsch H, Fotso S. Top Curr Chem. 2008;282:3–74. [Google Scholar]
- 189.Yoshimoto A, Fujii S, Johdo O, Kubo K, Nishida H, Okamoto R, Takeuchi T. J Antibiot. 1993;46:56–64. doi: 10.7164/antibiotics.46.56. [DOI] [PubMed] [Google Scholar]
- 190.Yoshimoto A, Johdo O, Fujii S, Kubo K, Nishida H, Okamoto R, Takeuchi T. J Antibiot. 1992;45:1255–1267. doi: 10.7164/antibiotics.45.1255. [DOI] [PubMed] [Google Scholar]
- 191.Yoshimoto A, Johdo O, Nishida H, Okamoto R, Takeuchi T. J Antibiot. 1993;46:1758–1761. doi: 10.7164/antibiotics.46.1758. [DOI] [PubMed] [Google Scholar]
- 192.Fujii S, Kubo K, Johdo O, Yoshimoto A, Ishikura T, Naganawa H, Sawa T, Takeuchi T, Umezawa H. J Antibiot. 1986;39:473–475. doi: 10.7164/antibiotics.39.473. [DOI] [PubMed] [Google Scholar]
- 193.Yoshimoto A, Johdo O, Tone H, Okamoto R, Naganawa H, Sawa T, Takeuchi T. J Antibiot. 1992;45:1609–1617. doi: 10.7164/antibiotics.45.1609. [DOI] [PubMed] [Google Scholar]
- 194.Cassinelli G, Di Matteo F, Forenza S, Ripamonti MC, Rivola G, Arcamone F, Di Marco A, Casazza AM, Soranzo C, Pratesi G. J Antibiot. 1980;33:1468–1473. doi: 10.7164/antibiotics.33.1468. [DOI] [PubMed] [Google Scholar]
- 195.Cassinelli G, Rivola G, Ruggieri D, Arcamone F, Grein A, Merli S, Spalla C, Casazza AM, Di Marco A, Pratesi G. J Antibiot. 1982;35:176–183. doi: 10.7164/antibiotics.35.176. [DOI] [PubMed] [Google Scholar]
- 196.Nair MG, Mishra SK, Putnam AR, Pandey RC. J Antibiot. 1992;45:1738–1745. doi: 10.7164/antibiotics.45.1738. [DOI] [PubMed] [Google Scholar]
- 197.Yoshimoto A, Matsuzawa Y, Oki T, Naganawa H, Takeuchi T, Umezawa H. J Antibiot. 1980;33:1150–1157. doi: 10.7164/antibiotics.33.1150. [DOI] [PubMed] [Google Scholar]
- 198.Yoshimoto A, Oki T, Takeuchi T, Umezawa H. J Antibiot. 1980;33:1158–1166. doi: 10.7164/antibiotics.33.1158. [DOI] [PubMed] [Google Scholar]
- 199.Oki T, Matsuzawa Y, Yoshimoto A, Numata K, Kitamura I. J Antibiot. 1975;28:830–834. doi: 10.7164/antibiotics.28.830. [DOI] [PubMed] [Google Scholar]
- 200.Oki T, Kitamura I, Matsuzawa Y, Shibamoto N, Ogasawara T, Yoshimoto A, Inui T, Naganawa H, Takeuchi T, Umezawa H. J Antibiot. 1979;32:801–819. doi: 10.7164/antibiotics.32.801. [DOI] [PubMed] [Google Scholar]
- 201.Oki T, Yoshimoto A, Matsuzawa Y, Takeuchi T, Umezawa H. J Antibiot. 1980;33:1331–1340. doi: 10.7164/antibiotics.33.1331. [DOI] [PubMed] [Google Scholar]
- 202.Matsuzawa Y, Yoshimoto A, Oki T, Naganawa H, Takeuchi T, Umezawa H. J Antibiot. 1980;33:1341–1347. doi: 10.7164/antibiotics.33.1341. [DOI] [PubMed] [Google Scholar]
- 203.Kumar V, Remers WA, Grulich R. J Antibiot. 1977;30:881–882. doi: 10.7164/antibiotics.30.881. [DOI] [PubMed] [Google Scholar]
- 204.Kim HS, Hong YS, Kim YH, Yoo OJ, Lee JJ. J Antibiot. 1996;49:355–360. doi: 10.7164/antibiotics.49.355. [DOI] [PubMed] [Google Scholar]
- 205.Hoshino T, Sekine Y, Fujiwara A. J Antibiot. 1983;36:1458–1462. doi: 10.7164/antibiotics.36.1458. [DOI] [PubMed] [Google Scholar]
- 206.Brockman H, Scheffer B, Stein C. Tetrahedron Lett. 1973;14:3699–3702. [Google Scholar]
- 207.Niemi J, Ylihonko K, Hakala J, Parssinen R, Kopio A, Mantsala P. Microbiology. 1994;140:1351–1358. doi: 10.1099/00221287-140-6-1351. [DOI] [PubMed] [Google Scholar]
- 208.Saito S, Katsuda Y, Johdo O, Yoshimoto A. Biosci, Biotechnol, Biochem. 1995;59:135–137. doi: 10.1271/bbb.59.135. [DOI] [PubMed] [Google Scholar]
- 209.Reddy GC, Sahai R, Fehlhaber HW, Ganguli BN. J Antibiot. 1985;38:1423–1425. doi: 10.7164/antibiotics.38.1423. [DOI] [PubMed] [Google Scholar]
- 210.Johdo O, Yoshioka T, Takeuchi T, Yoshimoto A. J Antibiot. 1997;50:522–525. doi: 10.7164/antibiotics.50.522. [DOI] [PubMed] [Google Scholar]
- 211.Yoshimoto A, Matsuzawa Y, Ishikura T, Sawa T, Takeuchi T, Umezawa H. J Antibiot. 1984;37:920–922. doi: 10.7164/antibiotics.37.920. [DOI] [PubMed] [Google Scholar]
- 212.Yoshimoto A, Johdo O, Watanabe Y, Nishida H, Okamoto R, Takeuchi T. J Antibiot. 1992;45:1005–1007. doi: 10.7164/antibiotics.45.1005. [DOI] [PubMed] [Google Scholar]
- 213.Johdo O, Yoshioka T, Naganawa H, Takeuchi T, Yoshimoto A. J Antibiot. 1996;49:669–675. doi: 10.7164/antibiotics.49.669. [DOI] [PubMed] [Google Scholar]
- 214.Fujiwara A, Hoshino T, Tazoe M, Fujiwara M. J Antibiot. 1981;34:608–610. doi: 10.7164/antibiotics.34.608. [DOI] [PubMed] [Google Scholar]
- 215.Fujiwara A, Tazoe M, Hoshino T, Sekine Y, Masuda S, Nomura S. J Antibiot. 1981;34:912–915. doi: 10.7164/antibiotics.34.912. [DOI] [PubMed] [Google Scholar]
- 216.Hoshino T, Tazoe M, Nomura S, Fujiwara A. J Antibiot. 1982;35:1271–1279. doi: 10.7164/antibiotics.35.1271. [DOI] [PubMed] [Google Scholar]
- 217.Fujiwara A, Hoshino T, Tazoe M, Fujiwara M. J Antibiot. 1982;35:164–175. doi: 10.7164/antibiotics.35.164. [DOI] [PubMed] [Google Scholar]
- 218.Hoshino T, Fujiwara A. J Antibiot. 1984;37:1473–1474. doi: 10.7164/antibiotics.37.1473. [DOI] [PubMed] [Google Scholar]
- 219.Hoshino T, Setoguchi Y, Fujiwara A. J Antibiot. 1984;37:1469–1472. doi: 10.7164/antibiotics.37.1469. [DOI] [PubMed] [Google Scholar]
- 220.Matsuzawa Y, Yoshimoto A, Shibamoto N, Tobe H, Oki T, Naganawa H, Takeuchi T, Umezawa H. J Antibiot. 1981;34:959–964. doi: 10.7164/antibiotics.34.959. [DOI] [PubMed] [Google Scholar]
- 221.Yoshimoto A, Matsuzawa Y, Oki T, Takeuchi T, Umezawa H. J Antibiot. 1981;34:951–958. doi: 10.7164/antibiotics.34.951. [DOI] [PubMed] [Google Scholar]
- 222.Oki T, Shibamoto N, Matsuzawa Y, Ogasawara T, Yoshimoto A, Kitamura I, Inui T, Naganawa H, Takeuchi T, Umezawa H. J Antibiot. 1977;30:683–687. doi: 10.7164/antibiotics.30.683. [DOI] [PubMed] [Google Scholar]
- 223.Brodasky TF, Mizsak S, Hoffstetter JR. J Antibiot. 1985;38:849–855. doi: 10.7164/antibiotics.38.849. [DOI] [PubMed] [Google Scholar]
- 224.Suzukake-Tsuchiya K, Moriya Y, Yamazaki K, Hori M, Hosokawa N, Sawa T, Iinuma H, Naganawa H, Imada C, Hamada M. J Antibiot. 1990;43:1489–1496. doi: 10.7164/antibiotics.43.1489. [DOI] [PubMed] [Google Scholar]
- 225.Kunnari T, Tuikkanen J, Hautala A, Hakala J, Ylihonko K, Mantsala P. J Antibiot. 1997;50:496–501. doi: 10.7164/antibiotics.50.496. [DOI] [PubMed] [Google Scholar]
- 226.Olano C, Abdelfattah MS, Gullon S, Brana AF, Rohr J, Méndez C, Salas JA. ChemBioChem. 2008;9:624–633. doi: 10.1002/cbic.200700610. [DOI] [PubMed] [Google Scholar]
- 227.Komiyana T, Matsuzawa Y, Oki T, Inui T, Takahashi Y, Naganawa H, Takeuchi T, Umezawa H. J Antibiot. 1977;30:619–621. doi: 10.7164/antibiotics.30.619. [DOI] [PubMed] [Google Scholar]
- 228.Takahashi Y, Naganawa H, Takeuchi T, Umezawa H, Komiyana T, Oki T, Iui T. J Antibiot. 1977;30:622–624. doi: 10.7164/antibiotics.30.622. [DOI] [PubMed] [Google Scholar]
- 229.Matsuzawa Y, Oki T, Takeuchi T, Umezawa H. J Antibiot. 1981;34:1596–1607. doi: 10.7164/antibiotics.34.1596. [DOI] [PubMed] [Google Scholar]
- 230.Fomichova EV, Fedorova GB, Potapova NP, Katrukha GS. J Antibiot. 1992;45:1185–1186. doi: 10.7164/antibiotics.45.1185. [DOI] [PubMed] [Google Scholar]
- 231.Phipps RK, Blunt JW, Cole ALJ, Munro MHG. ARKIVOC. 2004;10:94–100. [Google Scholar]
- 232.Johdo O, Watanabe Y, Ishikura T, Yoshimoto A, Naganawa H, Sawa T, Takeuchi T. J Antibiot. 1991;44:1130–1140. doi: 10.7164/antibiotics.44.1130. [DOI] [PubMed] [Google Scholar]
- 233.Johdo O, Ishikura T, Yoshimoto A, Takeuchi T. J Antibiot. 1991;44:1110–1120. doi: 10.7164/antibiotics.44.1110. [DOI] [PubMed] [Google Scholar]
- 234.Nakata M, Saito M, Inouye Y, Nakamura S, Hayakawa Y, Seto H. J Antibiot. 1992;45:1599–1608. doi: 10.7164/antibiotics.45.1599. [DOI] [PubMed] [Google Scholar]
- 235.Nakagawa M, Furihata K, Furihata K, Adachi K, Seto H, Otake N. J Antibiot. 1986;39:1178–1179. doi: 10.7164/antibiotics.39.1178. [DOI] [PubMed] [Google Scholar]
- 236.Shaaban KA. PhD thesis. University of Göttingen; 2009. [Google Scholar]
- 237.Zitouni A, Mathieu F, Coppel Y, Pont F, Sabaou N, Lebrihi A. J Antibiot. 2004;57:373–378. doi: 10.7164/antibiotics.57.373. [DOI] [PubMed] [Google Scholar]
- 238.Zitouni A, Boudjella H, Mathieu F, Sabaou N, Lebrihi A. J Antibiot. 2004;57:367–372. doi: 10.7164/antibiotics.57.367. [DOI] [PubMed] [Google Scholar]
- 239.Hopp DC, Rabenstein J, Rhea J, Smith C, Romari K, Clarke M, Francis L, Irigoyen M, Milanowski D, Luche M, Carr GJ, Mocek U. J Antibiot. 2008;61:675–679. doi: 10.1038/ja.2008.95. [DOI] [PubMed] [Google Scholar]
- 240.Johdo O, Tone H, Okamoto R, Yoshimoto A, Naganawa H, Sawa T, Takeuchi T. J Antibiot. 1991;44:1160–1164. doi: 10.7164/antibiotics.44.1160. [DOI] [PubMed] [Google Scholar]
- 241.Tanaka Y, Grafe U, Yazawa K, Mikami Y, Ritzau M. J Antibiot. 1997;50:822–827. doi: 10.7164/antibiotics.50.822. [DOI] [PubMed] [Google Scholar]
- 242.Fleck W, Strauss D, Koch W, Prauser H. Z Allg Mikrobiol. 1974;14:551–558. doi: 10.1002/jobm.3630140702. [DOI] [PubMed] [Google Scholar]
- 243.Uchida T, Imoto M, Masuda T, Imamura K, Hatori Y, Sawa T, Naganawa H, Hamada M, Takeuchi T, Umezawa H. J Antibiot. 1983;36:1080–1083. doi: 10.7164/antibiotics.36.1080. [DOI] [PubMed] [Google Scholar]
- 244.Luzhetskyy A, Hoffmann J, Pelzer S, Wohlert SE, Vente A, Bechthold A. Appl Microbiol Biotechnol. 2008;80:15–19. doi: 10.1007/s00253-008-1515-1. [DOI] [PubMed] [Google Scholar]
- 245.Nachtigall J, Schulz D, Beil W, Sussmuth RD, Fiedler HP. J Antibiot. 2010;63:397–399. doi: 10.1038/ja.2010.59. [DOI] [PubMed] [Google Scholar]
- 246.Luzhetskyy A, Mayer A, Hoffmann J, Pelzer S, Holzenkamper M, Schmitt B, Wohlert SE, Vente A, Bechthold A. ChemBioChem. 2007;8:599–602. doi: 10.1002/cbic.200600529. [DOI] [PubMed] [Google Scholar]
- 247.Motohashi K, Takagi M, Shin-Ya K. J Nat Prod. 2010;73:755–758. doi: 10.1021/np9007409. [DOI] [PubMed] [Google Scholar]
- 248.Shimosaka A, Hayakawa Y, Nakagawa M, Furihata K, Seto H, Otake N. J Antibiot. 1987;40:116–121. doi: 10.7164/antibiotics.40.116. [DOI] [PubMed] [Google Scholar]
- 249.Bieber LW, da Silva Filho AA, de Mello JF, de Lima OG, do Nascimento MS, Veith HJ, von der Saal W. J Antibiot. 1987;40:1335–1338. doi: 10.7164/antibiotics.40.1335. [DOI] [PubMed] [Google Scholar]
- 250.Cordes J, Harms K, Koert U. Org Lett. 2010;12:3808–3811. doi: 10.1021/ol101500k. [DOI] [PubMed] [Google Scholar]
- 251.Furumai T, Igarashi Y, Higuchi H, Saito N, Oki T. J Antibiot. 2002;55:128–133. doi: 10.7164/antibiotics.55.128. [DOI] [PubMed] [Google Scholar]
- 252.Igarashi Y, Higuchi H, Oki T, Furumai T. J Antibiot. 2002;55:134–140. doi: 10.7164/antibiotics.55.134. [DOI] [PubMed] [Google Scholar]
- 253.Matern U, Grisebach H, Karl W, Achenbach H. Eur J Biochem. 1972;29:1–4. doi: 10.1111/j.1432-1033.1972.tb01949.x. [DOI] [PubMed] [Google Scholar]
- 254.Yang SW, Chan TM, Terracciano J, Patel R, Loebenberg D, Chen G, Patel M, Gullo V, Pramanik B, Chu M. J Antibiot. 2004;57:601–604. doi: 10.7164/antibiotics.57.601. [DOI] [PubMed] [Google Scholar]
- 255.Fuoco D. Antibiotics. 2012;1:1–13. doi: 10.3390/antibiotics1010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Hultin PG. Curr Top Med Chem. 2005;5:1299–1331. doi: 10.2174/156802605774643015. [DOI] [PubMed] [Google Scholar]
- 257.Johdo O, Nishida H, Okamoto R, Yoshimoto A, Takeuchi T. J Antibiot. 1995;48:1153–1158. doi: 10.7164/antibiotics.48.1153. [DOI] [PubMed] [Google Scholar]
- 258.Yoshimoto A, Johdo O, Takatsuki Y, Ishikura T, Sawa T, Takeuchi T, Umezawa H. J Antibiot. 1984;37:935–938. doi: 10.7164/antibiotics.37.935. [DOI] [PubMed] [Google Scholar]
- 259.Kimura K, Nakayama S, Miyata N, Takeshita Y, Kawanishi G. J Antibiot. 1988;41:411–414. doi: 10.7164/antibiotics.41.411. [DOI] [PubMed] [Google Scholar]
- 260.Kimura K, Morinaga T, Miyata N, Kawanishi G. J Antibiot. 1989;42:1838–1843. doi: 10.7164/antibiotics.42.1838. [DOI] [PubMed] [Google Scholar]
- 261.Kimura K, Nakayama S, Miyata N, Kawanishi G. J Antibiot. 1989;42:127–131. doi: 10.7164/antibiotics.42.127. [DOI] [PubMed] [Google Scholar]
- 262.Cassinelli G, Arlandini E, Ballabio M, Bordoni T, Geroni C, Giuliani F, Grein A, Merli S, Rivola G. J Antibiot. 1990;43:19–28. doi: 10.7164/antibiotics.43.19. [DOI] [PubMed] [Google Scholar]
- 263.Miyamoto Y, Ohta S, Johdo O, Nagamatsu Y, Yoshimoto A. J Antibiot. 2000;53:828–836. doi: 10.7164/antibiotics.53.828. [DOI] [PubMed] [Google Scholar]
- 264.Komatsu Y, Takahashi O, Hayashi H. J Antibiot. 1998;51:85–88. doi: 10.7164/antibiotics.51.85. [DOI] [PubMed] [Google Scholar]
- 265.Itoh T, Kinoshita M, Wei H, Kobayashi M. Chem Pharm Bull. 2003;51:1402–1404. doi: 10.1248/cpb.51.1402. [DOI] [PubMed] [Google Scholar]
- 266.Itoh T, Kinoshita M, Aoki S, Kobayashi M. J Nat Prod. 2003;66:1373–1377. doi: 10.1021/np030212k. [DOI] [PubMed] [Google Scholar]
- 267.Nakagawa M, Kawai H, Hayakawa Y, Seto H, Otake N. J Antibiot. 1985;38:1622–1624. doi: 10.7164/antibiotics.38.1622. [DOI] [PubMed] [Google Scholar]
- 268.Hedtmann U, Fehlhaber HW, Sukatsch DA, Weber M, Hoffmann D, Kraemer HP. J Antibiot. 1992;45:1373–1375. doi: 10.7164/antibiotics.45.1373. [DOI] [PubMed] [Google Scholar]
- 269.Brockmann H, Greve H. Tetrahedron Lett. 1975:831–834. [Google Scholar]
- 270.Cassinelli G, Grein A, Merli S, Rivola G. US4421851 A. US Pat. 1983
- 271.Murenets NV, Kudinova MK, Kliuev NA, Chernyshev AI, Shorshnev SV. Antibiot Med Biotekhnol. 1986;31:431–434. [PubMed] [Google Scholar]
- 272.Arnone A, Cardillo R, Nasini G, Depava OV, Quaroni S. Phytochemistry. 1988;27:3611–3617. [Google Scholar]
- 273.Qian-Cutrone J, Kolb JM, McBrien K, Huang S, Gustavson D, Lowe SE, Manly SP. J Nat Prod. 1998;61:1379–1382. doi: 10.1021/np980234k. [DOI] [PubMed] [Google Scholar]
- 274.Kunnari TJ, Ylihonko KP, Klika KD, Mantsala PI, Hakala JM. J Org Chem. 2000;65:2851–2855. doi: 10.1021/jo980345o. [DOI] [PubMed] [Google Scholar]
- 275.Wiley PF, Johnson JL, Houser DJ. J Antibiot. 1977;30:628–629. doi: 10.7164/antibiotics.30.628. [DOI] [PubMed] [Google Scholar]
- 276.Wiley PF, Kelly RB, Caron EL, Wiley VH, Johnson JH, MacKellar FA, Mizsak SA. J Am Chem Soc. 1977;99:542–549. doi: 10.1021/ja00444a038. [DOI] [PubMed] [Google Scholar]
- 277.Kawai H, Hayakawa Y, Nakagawa M, Furihata K, Furihata K, Shimazu A, Seto H, Otake N. J Antibiot. 1987;40:1266–1272. doi: 10.7164/antibiotics.40.1266. [DOI] [PubMed] [Google Scholar]
- 278.Kawai H, Hayakawa Y, Nakagawa M, Furihata K, Seto H, Otake N. J Antibiot. 1987;40:1273–1282. doi: 10.7164/antibiotics.40.1273. [DOI] [PubMed] [Google Scholar]
- 279.Shimosaka A, Kawai H, Hayakawa Y, Komeshima N, Nakagawa M, Seto H, Otake N. J Antibiot. 1987;40:1283–1291. doi: 10.7164/antibiotics.40.1283. [DOI] [PubMed] [Google Scholar]
- 280.Ishii K, Nishimura Y, Kondo S, Umezawa H. J Antibiot. 1983;36:454–456. doi: 10.7164/antibiotics.36.454. [DOI] [PubMed] [Google Scholar]
- 281.Ishii K, Kondo S, Nishimura Y, Hamada M, Takeuchi T, Umezawa H. J Antibiot. 1983;36:451–453. doi: 10.7164/antibiotics.36.451. [DOI] [PubMed] [Google Scholar]
- 282.Ishii K, Nishimura Y, Naganawa H, Kondo S, Umezawa H. J Antibiot. 1984;37:344–353. doi: 10.7164/antibiotics.37.344. [DOI] [PubMed] [Google Scholar]
- 283.Nishimura Y, Ishii K, Kondo S. J Antibiot. 1990;43:54–61. doi: 10.7164/antibiotics.43.54. [DOI] [PubMed] [Google Scholar]
- 284.Aoki M, Shirai H, Nakayama N, Itezono Y, Mori M, Satoh T, Ohshima S, Watanabe J, Yokose K, Seto H. J Antibiot. 1991;44:635–645. doi: 10.7164/antibiotics.44.635. [DOI] [PubMed] [Google Scholar]
- 285.Ubukata M, Uzawa J, Osada H, Isono K. J Antibiot. 1993;46:942–951. doi: 10.7164/antibiotics.46.942. [DOI] [PubMed] [Google Scholar]
- 286.Ubukata M, Osada H, Kudo T, Isono K. J Antibiot. 1993;46:936–941. doi: 10.7164/antibiotics.46.936. [DOI] [PubMed] [Google Scholar]
- 287.Ubukata M, Tanaka C, Osada H, Isono K. J Antibiot. 1991;44:1274–1276. doi: 10.7164/antibiotics.44.1274. [DOI] [PubMed] [Google Scholar]
- 288.Ishigami K, Hayakawa Y, Seto H. J Antibiot. 1994;47:1219–1225. doi: 10.7164/antibiotics.47.1219. [DOI] [PubMed] [Google Scholar]
- 289.Hutter K, Baader E, Frobel K, Zeeck A, Bauer K, Gau W, Kurz J, Schroder T, Wunsche C, Karl W, et al. J Antibiot. 1986;39:1193–1204. doi: 10.7164/antibiotics.39.1193. [DOI] [PubMed] [Google Scholar]
- 290.Kind R, Hutter K, Zeeck A, Schmidt-Base K, Egert E. J Antibiot. 1989;42:7–13. doi: 10.7164/antibiotics.42.7. [DOI] [PubMed] [Google Scholar]
- 291.Kiyo K, Sho S, Marunaka T, Miyake Y, Minami Y. JP 62 030 793. Japanese Pat. 1987
- 292.Hori H, Kajiura T, Igarashi Y, Furumai T, Higashi K, Ishiyama T, Uramoto M, Uehara Y, Oki T. J Antibiot. 2002;55:46–52. doi: 10.7164/antibiotics.55.46. [DOI] [PubMed] [Google Scholar]
- 293.Cho SI, Fukazawa H, Honma Y, Kajiura T, Hori H, Igarashi Y, Furumai T, Oki T, Uehara Y. J Antibiot. 2002;55:270–278. doi: 10.7164/antibiotics.55.270. [DOI] [PubMed] [Google Scholar]
- 294.Hori H, Igarashi Y, Kajiura T, Furumai T, Higashi K, Ishiyama T, Uramota M, Uehara Y, Oki T. J Antibiot. 1998;51:402–417. doi: 10.7164/antibiotics.51.402. [DOI] [PubMed] [Google Scholar]
- 295.Igarashi Y, Kajiura T, Furumai T, Hori H, Higashi K, Ishiyama T, Uramoto M, Uehara Y, Oki T. J Antibiot. 2002;55:61–70. doi: 10.7164/antibiotics.55.61. [DOI] [PubMed] [Google Scholar]
- 296.Kajiura T, Furumai T, Igarashi Y, Hori H, Higashi K, Ishiyama T, Uramoto M, Uehara Y, Oki T. J Antibiot. 1998;51:394–401. doi: 10.7164/antibiotics.51.394. [DOI] [PubMed] [Google Scholar]
- 297.Kajiura T, Furumai T, Igarashi Y, Hori H, Higashi K, Ishiyama T, Uramoto M, Uehara Y, Oki T. J Antibiot. 2002;55:53–60. doi: 10.7164/antibiotics.55.53. [DOI] [PubMed] [Google Scholar]
- 298.Uehara Y, Li PM, Fukazawa H, Mizuno S, Nihei Y, Nishio M, Hanada M, Yamamoto C, Furumai T, Oki T. J Antibiot. 1993;46:1306–1308. doi: 10.7164/antibiotics.46.1306. [DOI] [PubMed] [Google Scholar]
- 299.Yokoyama A, Okabe-Kado J, Uehara Y, Oki T, Tomoyasu S, Tsuruoka N, Honma Y. Leuk Res. 1996;20:491–497. doi: 10.1016/0145-2126(96)00014-8. [DOI] [PubMed] [Google Scholar]
- 300.Herold K, Gollmick FA, Groth I, Roth M, Menzel KD, Mollmann U, Grafe U, Hertweck C. Chem – Eur J. 2005;11:5523–5530. doi: 10.1002/chem.200500320. [DOI] [PubMed] [Google Scholar]
- 301.Herold K, Xu Z, Gollmick FA, Grafe U, Hertweck C. Org Biomol Chem. 2004;2:2411–2414. doi: 10.1039/B409221J. [DOI] [PubMed] [Google Scholar]
- 302.Krugel H, Licht A, Biedermann G, Petzold A, Lassak J, Hupfer Y, Schlott B, Hertweck C, Platzer M, Brantl S, Saluz HP. FEMS Microbiol Lett. 2010;313:155–163. doi: 10.1111/j.1574-6968.2010.02143.x. [DOI] [PubMed] [Google Scholar]
- 303.Moellmann U, Herold K, Roth M, Groth I, Gollmick F, Graefe U. US7557091 B2. US Pat. 2009
- 304.Xuan LJ, Xu SH, Zhang HL, Xu YM, Chen MQ. J Antibiot. 1992;45:1974–1976. doi: 10.7164/antibiotics.45.1974. [DOI] [PubMed] [Google Scholar]
- 305.Nagao K, Suzuki K, Tokunaga J, Miyazaki H, Katayama N, Mitsuyama R, Uyeda M. J Enzyme Inhib. 1996;10:115–124. doi: 10.3109/14756369609020164. [DOI] [PubMed] [Google Scholar]
- 306.Momose I, Chen W, Kinoshita N, Iinuma H, Hamada M, Takeuchi T. J Antibiot. 1998;51:21–25. doi: 10.7164/antibiotics.51.21. [DOI] [PubMed] [Google Scholar]
- 307.Momose I, Chen W, Nakamura H, Naganawa H, Iinuma H, Takeuchi T. J Antibiot. 1998;51:26–32. doi: 10.7164/antibiotics.51.26. [DOI] [PubMed] [Google Scholar]
- 308.Paululat T, Zeeck A, Gutterer JM, Fiedler HP. J Antibiot. 1999;52:96–101. doi: 10.7164/antibiotics.52.96. [DOI] [PubMed] [Google Scholar]
- 309.Drautz H, Reuschenbach P, Zahner H, Rohr J, Zeeck A. J Antibiot. 1985;38:1291–1301. doi: 10.7164/antibiotics.38.1291. [DOI] [PubMed] [Google Scholar]
- 310.Fiedler HP, Rohr J, Zeeck A. J Antibiot. 1986;39:856–859. doi: 10.7164/antibiotics.39.856. [DOI] [PubMed] [Google Scholar]
- 311.Fischer C, Rodriguez L, Patallo EP, Lipata F, Brana AF, Méndez C, Salas JA, Rohr J. J Nat Prod. 2002;65:1685–1689. doi: 10.1021/np020112z. [DOI] [PubMed] [Google Scholar]
- 312.Nybo SE, Shaaban KA, Kharel MK, Sutardjo H, Salas JA, Méndez C, Rohr J. Bioorg Med Chem Lett. 2012;22:2247–2250. doi: 10.1016/j.bmcl.2012.01.094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Hatsu M, Sasaki T, Gomi S, Kodama Y, Sezaki M, Inouye S, Kondo S. J Antibiot. 1992;45:325–330. doi: 10.7164/antibiotics.45.325. [DOI] [PubMed] [Google Scholar]
- 314.Hatsu M, Sasaki T, Watabe H, Miyadoh S, Nagasawa M, Shomura T, Sezaki M, Inouye S, Kondo S. J Antibiot. 1992;45:320–324. doi: 10.7164/antibiotics.45.320. [DOI] [PubMed] [Google Scholar]
- 315.Horiguchi T, Hayashi K, Tsubotani S, Iinuma S, Harada S, Tanida S. J Antibiot. 1994;47:545–556. doi: 10.7164/antibiotics.47.545. [DOI] [PubMed] [Google Scholar]
- 316.Devasthale PV, Mitscher LA, Telikepalli H, Vander Velde D, Zou JY, Ax HA, Tymiak AA. J Antibiot. 1992;45:1907–1913. doi: 10.7164/antibiotics.45.1907. [DOI] [PubMed] [Google Scholar]
- 317.Tymiak AA, Ax HA, Bolgar MS, Kahle AD, Porubcan MA, Andersen NH. J Antibiot. 1992;45:1899–1906. doi: 10.7164/antibiotics.45.1899. [DOI] [PubMed] [Google Scholar]
- 318.Wells JS, O’Sullivan J, Aklonis C, Ax HA, Tymiak AA, Kirsch DR, Trejo WH, Principe P. J Antibiot. 1992;45:1892–1898. doi: 10.7164/antibiotics.45.1892. [DOI] [PubMed] [Google Scholar]
- 319.Wang P, Kim W, Pickens LB, Gao X, Tang Y. Angew Chem, Int Ed. 2012;51:11136–11140. doi: 10.1002/anie.201205426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Rohr J, Zeeck A. J Antibiot. 1990;43:1169–1178. doi: 10.7164/antibiotics.43.1169. [DOI] [PubMed] [Google Scholar]
- 321.Ganguly AK, Sarre OZ, Reimann H. J Am Chem Soc. 1968;90:7129–7130. doi: 10.1021/ja01027a047. [DOI] [PubMed] [Google Scholar]
- 322.Wang F, Zhou M, Singh S, Yennamalli RM, Bingman CA, Thorson JS, Phillips GN., Jr Proteins. 2013;81:1277–1282. doi: 10.1002/prot.24289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Bosserman MA, Florez AB, Shaaban KA, Brana AF, Salas JA, Méndez C, Rohr J. Biochemistry. 2011;50:1421–1428. doi: 10.1021/bi1016205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Berlin YA, Esipov SE, Kolosov MN, Shemyakin MM. Tetrahedron Lett. 1966;15:1643–1647. doi: 10.1016/s0040-4039(01)99770-9. [DOI] [PubMed] [Google Scholar]
- 325.Berlin YA, Kolosov MN, Yartseva IV. Chem Nat Compd. 1973;9:504–508. [Google Scholar]
- 326.Yoshimura Y, Koenuma M, Matsumoto K, Tori K, Terui Y. J Antibiot. 1988;41:53–67. doi: 10.7164/antibiotics.41.53. [DOI] [PubMed] [Google Scholar]
- 327.Jayasuriya H, Lingham RB, Graham P, Quamina D, Herranz L, Genilloud O, Gagliardi M, Danzeisen R, Tomassini JE, Zink DL, Guan Z, Singh SB. J Nat Prod. 2002;65:1091–1095. doi: 10.1021/np010642f. [DOI] [PubMed] [Google Scholar]
- 328.Oki T, Matsuzawa Y, Numata K, Takamatsu A. J Antibiot. 1973;26:701–704. doi: 10.7164/antibiotics.26.701. [DOI] [PubMed] [Google Scholar]
- 329.Katahira R, Uosaki Y, Ogawa H, Yamashita Y, Nakano H, Yoshida M. J Antibiot. 1998;51:267–274. doi: 10.7164/antibiotics.51.267. [DOI] [PubMed] [Google Scholar]
- 330.Ogawa H, Yamashita Y, Katahira R, Chiba S, Iwasaki T, Ashizawa T, Nakano H. J Antibiot. 1998;51:261–266. doi: 10.7164/antibiotics.51.261. [DOI] [PubMed] [Google Scholar]
- 331.Berlin YA, Kolosov MN, Vasina IV, Yartseva IV. Chem Commun. 1968:762–763. [Google Scholar]
- 332.Kawano T, Hidaka T, Furihata K, Mochizuki J, Nakayama H, Hayakawa Y, Seto H. J Antibiot. 1990;43:110–113. doi: 10.7164/antibiotics.43.110. [DOI] [PubMed] [Google Scholar]
- 333.Zhdanovich Iu V, Kuzovkov AD, Veselova SI, Vagina IM, Sapozhnikov Iu M. Antibiotiki. 1982;27:83–87. [PubMed] [Google Scholar]
- 334.Shashkov AS, Iashunskii DV, Zhdanovich Iu V, Lokshin GB. Bioorg Khim. 1991;17:410–416. [PubMed] [Google Scholar]
- 335.Remsing LL, Garcia-Bernardo J, Gonzalez A, Kunzel E, Rix U, Brana AF, Bearden DW, Méndez C, Salas JA, Rohr J. J Am Chem Soc. 2002;124:1606–1614. doi: 10.1021/ja0105156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Nunez LE, Nybo SE, Gonzalez-Sabin J, Perez M, Menendez N, Brana AF, Shaaban KA, He M, Moris F, Salas JA, Rohr J, Méndez C. J Med Chem. 2012;55:5813–5825. doi: 10.1021/jm300234t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Remsing LL, Gonzalez AM, Nur-e-Alam M, Fernandez-Lozano MJ, Brana AF, Rix U, Oliveira MA, Méndez C, Salas JA, Rohr J. J Am Chem Soc. 2003;125:5745–5753. doi: 10.1021/ja034162h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Koenuma M, Uchida N, Yamaguchi K, Kawamura Y, Matsumoto K. J Antibiot. 1988;41:45–52. doi: 10.7164/antibiotics.41.45. [DOI] [PubMed] [Google Scholar]
- 339.Koenuma M, Yoshimura Y, Matsumoto K, Terui Y. J Antibiot. 1988;41:68–72. doi: 10.7164/antibiotics.41.68. [DOI] [PubMed] [Google Scholar]
- 340.Miyamoto M, Kawamatsu Y, Kawashima K, Shinohara M, Nakanishi K. Tetrahedron Lett. 1966;6:545–552. doi: 10.1016/s0040-4039(01)99663-7. [DOI] [PubMed] [Google Scholar]
- 341.Pezzanite JO, Clardy J, Lau PY, Wood G, Walker DL, Fraser-Reid B. J Am Chem Soc. 1975;97:6250–6251. doi: 10.1021/ja00854a054. [DOI] [PubMed] [Google Scholar]
- 342.Tagashira M, Kitagawa T, Nozato N, Isonishi S, Okamoto A, Ochiai K, Ohtake Y. Chem Pharm Bull. 2000;48:575–578. doi: 10.1248/cpb.48.575. [DOI] [PubMed] [Google Scholar]
- 343.Grohar PJ, Woldemichael GM, Griffin LB, Mendoza A, Chen QR, Yeung C, Currier DG, Davis S, Khanna C, Khan J, McMahon JB, Helman LJ. J Natl Cancer Inst. 2011;103:962–978. doi: 10.1093/jnci/djr156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Subbiah V, Kurzrock R. Discov Med. 2012;13:405–415. [PMC free article] [PubMed] [Google Scholar]
- 345.Asai M. Chem Pharm Bull. 1970;18:1699–1705. [Google Scholar]
- 346.Brimble MA, Duncalf LJ, Nairn MR. Nat Prod Rep. 1999;16:267–281. doi: 10.1039/a804287j. [DOI] [PubMed] [Google Scholar]
- 347.Hegde VR, King AH, Patel MG, Puar MS, Mcphail AT. Tetrahedron Lett. 1987;28:4485–4488. [Google Scholar]
- 348.Patel M, Hegde V, Horan A, Barrett T, Bishop R, King A, Marquez J, Hare R, Gullo V. J Antibiot. 1989;42:1063–1069. doi: 10.7164/antibiotics.42.1063. [DOI] [PubMed] [Google Scholar]
- 349.Okabe T, Nomoto K, Funabashi H, Okuda S, Suzuki H, Tanaka N. J Antibiot. 1985;38:1333–1336. doi: 10.7164/antibiotics.38.1333. [DOI] [PubMed] [Google Scholar]
- 350.Tanaka N, Okabe T, Isono F, Kashiwagi M, Nomoto K, Takahashi M, Shimazu A, Nishimura T. J Antibiot. 1985;38:1327–1332. doi: 10.7164/antibiotics.38.1327. [DOI] [PubMed] [Google Scholar]
- 351.Toral-Barza L, Zhang WG, Huang X, McDonald LA, Salaski EJ, Barbieri LR, Ding WD, Krishnamurthy G, Hu YB, Lucas J, Bernan VS, Cai P, Levin JI, Mansour TS, Gibbons JJ, Abraham RT, Yu K. Mol Cancer Ther. 2007;6:3028–3038. doi: 10.1158/1535-7163.MCT-07-0211. [DOI] [PubMed] [Google Scholar]
- 352.Okabe T, Nomoto K, Tanaka N. J Antibiot. 1986;39:1–5. doi: 10.7164/antibiotics.39.1. [DOI] [PubMed] [Google Scholar]
- 353.Hayakawa Y, Ishigami K, Shin-Ya K, Seto H. J Antibiot. 1994;47:1344–1347. doi: 10.7164/antibiotics.47.1344. [DOI] [PubMed] [Google Scholar]
- 354.Potterat O, Zahner H, Volkmann C, Zeeck A. J Antibiot. 1993;46:346–349. doi: 10.7164/antibiotics.46.346. [DOI] [PubMed] [Google Scholar]
- 355.Volkmann C, Zeeck A, Potterat O, Zahner H, Bohnen FM, Herbst-Irmer R. J Antibiot. 1995;48:431–432. doi: 10.7164/antibiotics.48.431. [DOI] [PubMed] [Google Scholar]
- 356.Omura S, Ikeda H, Malpartida F, Kieser HM, Hopwood DA. Antimicrob Agents Chemother. 1986;29:13–19. doi: 10.1128/aac.29.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357.Naruse N, Goto M, Watanabe Y, Terasawa T, Dobashi K. J Antibiot. 1998;51:545–552. doi: 10.7164/antibiotics.51.545. [DOI] [PubMed] [Google Scholar]
- 358.Goto M, Masegi M, Yamauchi T, Chiba K, Kuboi Y, Harada K, Naruse N. J Antibiot. 1998;51:539–544. doi: 10.7164/antibiotics.51.539. [DOI] [PubMed] [Google Scholar]
- 359.Koizumi F, Hasegawa A, Ochiai K, Ando K, Kondo H, Yoshida M, Matsuda Y, Nakanishi S. J Antibiot. 2003;56:985–992. doi: 10.7164/antibiotics.56.985. [DOI] [PubMed] [Google Scholar]
- 360.Koizumi F, Hasegawa A, Yoshida M, Matsuda Y, Nakanishi S. J Antibiot. 2004;57:289–290. doi: 10.7164/antibiotics.57.289. [DOI] [PubMed] [Google Scholar]
- 361.Bieber B, Nuske J, Ritzau M, Grafe U. J Antibiot. 1998;51:381–382. doi: 10.7164/antibiotics.51.381. [DOI] [PubMed] [Google Scholar]
- 362.Oja T, Niiranen L, Sandalova T, Klika KD, Niemi J, Mantsala P, Schneider G, Metsa-Ketela M. Proc Natl Acad Sci U S A. 2013;110:1291–1296. doi: 10.1073/pnas.1207407110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 363.Swigelaar WP. Master thesis. Murdoch University; 2005. [Google Scholar]
- 364.Naysmith BJ, Brimble MA. Org Lett. 2013;15:2006–2009. doi: 10.1021/ol400686f. [DOI] [PubMed] [Google Scholar]
- 365.Maruyama M, Nishida C, Takahashi Y, Naganawa H, Hamada M, Takeuchi T. J Antibiot. 1994;47:952–954. doi: 10.7164/antibiotics.47.952. [DOI] [PubMed] [Google Scholar]
- 366.Igarashi M, Chen W, Tsuchida T, Umekita M, Sawa T, Naganawa H, Hamada M, Takeuchi T. J Antibiot. 1995;48:1502–1505. doi: 10.7164/antibiotics.48.1502. [DOI] [PubMed] [Google Scholar]
- 367.Tsuji N, Kobayashi M, Wakisaka Y, Kawamura Y, Mayama M. J Antibiot. 1976;29:7–9. doi: 10.7164/antibiotics.29.7. [DOI] [PubMed] [Google Scholar]
- 368.Tsuji N, Kobayashi M, Terui Y, Tori K. Tetrahedron. 1976;32:2207–2210. [Google Scholar]
- 369.Li YQ, Li MG, Li W, Zhao JY, Ding ZG, Cui XL, Wen ML. J Antibiot. 2007;60:757–761. doi: 10.1038/ja.2007.100. [DOI] [PubMed] [Google Scholar]
- 370.Ding ZG, Zhao JY, Li MG, Huang R, Li QM, Cui XL, Zhu HJ, Wen ML. J Nat Prod. 2012;75:1994–1998. doi: 10.1021/np3004936. [DOI] [PubMed] [Google Scholar]
- 371.He J, Roemer E, Lange C, Huang X, Maier A, Kelter G, Jiang Y, Xu LH, Menzel KD, Grabley S, Fiebig HH, Jiang CL, Sattler I. J Med Chem. 2007;50:5168–5175. doi: 10.1021/jm070170n. [DOI] [PubMed] [Google Scholar]
- 372.Elson AL, Box SJ, Gilpin ML. J Antibiot. 1988;41:570–572. doi: 10.7164/antibiotics.41.570. [DOI] [PubMed] [Google Scholar]
- 373.Gilpin ML, Box SJ, Elson AL. J Antibiot. 1988;41:512–518. doi: 10.7164/antibiotics.41.512. [DOI] [PubMed] [Google Scholar]
- 374.Shaaban KA, Shaaban M, Grun-Wollny I, Maier A, Fiebig HH, Laatsch H. J Nat Prod. 2007;70:1545–1550. doi: 10.1021/np070196h. [DOI] [PubMed] [Google Scholar]
- 375.Fiedler HP, Dieter A, Gulder TA, Kajahn I, Hamm A, Brown R, Jones AL, Goodfellow M, Muller WE, Bringmann G. J Antibiot. 2008;61:464–473. doi: 10.1038/ja.2008.63. [DOI] [PubMed] [Google Scholar]
- 376.Ford PW, Gadepalli M, Davidson BS. J Nat Prod. 1998;61:1232–1236. doi: 10.1021/np980126y. [DOI] [PubMed] [Google Scholar]
- 377.Maskey RP, Helmke E, Laatsch H. J Antibiot. 2003;56:942–949. doi: 10.7164/antibiotics.56.942. [DOI] [PubMed] [Google Scholar]
- 378.Nakagawa A, Koshi T. JP 2000-103798. Japanese Pat. 2000
- 379.Koiwa T, Nakano T, Takahashi S, Koshino H, Yamazaki M, Takashi T, Nakagawa A. J Antibiot. 1999;52:198–200. doi: 10.7164/antibiotics.52.198. [DOI] [PubMed] [Google Scholar]
- 380.Takahashi S, Nakano T, Koiwa T, Noshita T, Funayama S, Koshino H, Nakagawa A. J Antibiot. 2000;53:163–170. doi: 10.7164/antibiotics.53.163. [DOI] [PubMed] [Google Scholar]
- 381.Nakano T, Koiwa T, Takahashi S, Nakagawa A. J Antibiot. 2000;53:12–18. doi: 10.7164/antibiotics.53.12. [DOI] [PubMed] [Google Scholar]
- 382.Deseo MA, Hunter IS, Waterman PG. J Antibiot. 2005;58:822–827. doi: 10.1038/ja.2005.110. [DOI] [PubMed] [Google Scholar]
- 383.Paululat T, Katsifas EA, Karagouni AD, Fiedler HP. Eur J Org Chem. 2008:5283–5288. [Google Scholar]
- 384.Krohn K, Baltus W. Tetrahedron. 1988;44:49–54. [Google Scholar]
- 385.Matsumoto N, Tsuchida T, Nakamura H, Sawa R, Takahashi Y, Naganawa H, Iinuma H, Sawa T, Takeuchi T, Shiro M. J Antibiot. 1999;52:276–280. doi: 10.7164/antibiotics.52.276. [DOI] [PubMed] [Google Scholar]
- 386.Matsumoto N, Tsuchida T, Maruyama M, Kinoshita N, Homma Y, Iinuma H, Sawa T, Hamada M, Takeuchi T, Heida N, Yoshioka T. J Antibiot. 1999;52:269–275. doi: 10.7164/antibiotics.52.269. [DOI] [PubMed] [Google Scholar]
- 387.He H, Ding WD, Bernan VS, Richardson AD, Ireland CM, Greenstein M, Ellestad GA, Carter GT. J Am Chem Soc. 2001;123:5362–5363. doi: 10.1021/ja010129o. [DOI] [PubMed] [Google Scholar]
- 388.Holtzel A, Dieter A, Schmid DG, Brown R, Goodfellow M, Beil W, Jung G, Fiedler HP. J Antibiot. 2003;56:1058–1061. doi: 10.7164/antibiotics.56.1058. [DOI] [PubMed] [Google Scholar]
- 389.Schroeder DR, Colson KL, Klohr SE, Lee MS, Matson JA, Brinen LS, Clardy J. J Antibiot. 1996;49:865–872. doi: 10.7164/antibiotics.49.865. [DOI] [PubMed] [Google Scholar]
- 390.Blair LM, Sperry J. J Nat Prod. 2013;76:794–812. doi: 10.1021/np400124n. [DOI] [PubMed] [Google Scholar]
- 391.Herzon SB, Woo CM. Nat Prod Rep. 2012;29:87–118. doi: 10.1039/c1np00052g. [DOI] [PubMed] [Google Scholar]
- 392.Lee HG, Ahn JY, Lee AS, Shair MD. Chem – Eur J. 2010;16:13058–13062. doi: 10.1002/chem.201002157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 393.He HY, Shen B, Carter GT. Tetrahedron Lett. 2000;41:2067–2071. [Google Scholar]
- 394.Chino M, Nishikawa K, Umekita M, Hayashi C, Yamazaki T, Tsuchida T, Sawa T, Hamada M, Takeuchi T. J Antibiot. 1996;49:752–757. doi: 10.7164/antibiotics.49.752. [DOI] [PubMed] [Google Scholar]
- 395.Chino M, Nishikawa K, Sawa R, Hamada M, Naganawa H, Sawa T, Takeuchi T. J Antibiot. 1997;50:781–784. doi: 10.7164/antibiotics.50.781. [DOI] [PubMed] [Google Scholar]
- 396.Chino M, Nishikawa K, Tsuchida T, Sawa R, Nakamura H, Nakamura KT, Muraoka Y, Ikeda D, Naganawa H, Sawa T, Takeuchi T. J Antibiot. 1997;50:143–146. doi: 10.7164/antibiotics.50.143. [DOI] [PubMed] [Google Scholar]
- 397.Sugiyama T, Chino M, Tsurimoto T, Nozaki N, Ishimi Y. J Biochem. 2012;151:129–137. doi: 10.1093/jb/mvr130. [DOI] [PubMed] [Google Scholar]
- 398.Takeuchi T, Hara T, Naganawa H, Okada M, Hamada M, Umezawa H, Gomi S, Sezaki M, Kondo S. J Antibiot. 1988;41:807–811. doi: 10.7164/antibiotics.41.807. [DOI] [PubMed] [Google Scholar]
- 399.Gomi S, Sezaki M, Kondo S, Hara T, Naganawa H, Takeuchi T. J Antibiot. 1988;41:1019–1028. doi: 10.7164/antibiotics.41.1019. [DOI] [PubMed] [Google Scholar]
- 400.Oki T, Saitoh K, Tomatsu K, Tomita K, Konishi M, Kawaguchi H. Ann N Y Acad Sci. 1988;544:184–187. doi: 10.1111/j.1749-6632.1988.tb40402.x. [DOI] [PubMed] [Google Scholar]
- 401.Gomi S, Sezaki M, Hamada M, Kondo S, Takeuchi T. J Antibiot. 1989;42:1145–1150. doi: 10.7164/antibiotics.42.1145. [DOI] [PubMed] [Google Scholar]
- 402.Furumai T, Kakinuma S, Yamamoto H, Komiyama N, Suzuki K, Saitoh K, Oki T. J Antibiot. 1993;46:412–419. doi: 10.7164/antibiotics.46.412. [DOI] [PubMed] [Google Scholar]
- 403.Tsuno T, Yamamoto H, Narita Y, Suzuki K, Hasegawa T, Kakinuma S, Saitoh K, Furumai T, Oki T. J Antibiot. 1993;46:420–429. doi: 10.7164/antibiotics.46.420. [DOI] [PubMed] [Google Scholar]
- 404.Kakinuma S, Suzuki K, Hatori M, Saitoh K, Hasegawa T, Furumai T, Oki T. J Antibiot. 1993;46:430–440. doi: 10.7164/antibiotics.46.430. [DOI] [PubMed] [Google Scholar]
- 405.Hoshino H, Seki J, Takeuchi T. J Antibiot. 1989;42:344–346. doi: 10.7164/antibiotics.42.344. [DOI] [PubMed] [Google Scholar]
- 406.Oki T, Tenmyo O, Hirano M, Tomatsu K, Kamei H. J Antibiot. 1990;43:763–770. doi: 10.7164/antibiotics.43.763. [DOI] [PubMed] [Google Scholar]
- 407.Tomita K, Nishio M, Saitoh K, Yamamoto H, Hoshino Y, Ohkuma H, Konishi M, Miyaki T, Oki T. J Antibiot. 1990;43:755–762. doi: 10.7164/antibiotics.43.755. [DOI] [PubMed] [Google Scholar]
- 408.Tsunakawa M, Nishio M, Ohkuma H, Tsuno T, Konishi M, Naito T, Oki T, Kawaguchi H. J Org Chem. 1989;54:2532–2536. [Google Scholar]
- 409.Sawada Y, Nishio M, Yamamoto H, Hatori M, Miyaki T, Konishi M, Oki T. J Antibiot. 1990;43:771–777. doi: 10.7164/antibiotics.43.771. [DOI] [PubMed] [Google Scholar]
- 410.Sawada Y, Hatori M, Yamamoto H, Nishio M, Miyaki T, Oki T. J Antibiot. 1990;43:1223–1229. doi: 10.7164/antibiotics.43.1223. [DOI] [PubMed] [Google Scholar]
- 411.Saitoh K, Suzuki K, Hirano M, Furumai T, Oki T. J Antibiot. 1993;46:398–405. doi: 10.7164/antibiotics.46.398. [DOI] [PubMed] [Google Scholar]
- 412.Saitoh K, Sawada Y, Tomita K, Tsuno T, Hatori M, Oki T. J Antibiot. 1993;46:387–397. doi: 10.7164/antibiotics.46.387. [DOI] [PubMed] [Google Scholar]
- 413.Saitoh K, Furumai T, Oki T, Nishida F, Harada K, Suzuki M. J Antibiot. 1995;48:162–168. doi: 10.7164/antibiotics.48.162. [DOI] [PubMed] [Google Scholar]
- 414.Saitoh K, Tsuno T, Kakushima M, Hatori M, Furumai T, Oki T. J Antibiot. 1993;46:406–411. doi: 10.7164/antibiotics.46.406. [DOI] [PubMed] [Google Scholar]
- 415.Saitoh K, Tenmyo O, Yamamoto S, Furumai T, Oki T. J Antibiot. 1993;46:580–588. doi: 10.7164/antibiotics.46.580. [DOI] [PubMed] [Google Scholar]
- 416.Kondo S, Gomi S, Uotani K, Inouye S, Takeuchi T. J Antibiot. 1991;44:123–129. doi: 10.7164/antibiotics.44.123. [DOI] [PubMed] [Google Scholar]
- 417.Sawada Y, Murakami T, Ueki T, Fukagawa Y, Oki T. J Antibiot. 1991;44:119–121. doi: 10.7164/antibiotics.44.119. [DOI] [PubMed] [Google Scholar]
- 418.Furumai T, Saitoh K, Kakushima M, Yamamoto S, Suzuki K, Ikeda C, Kobaru S, Hatori M, Oki T. J Antibiot. 1993;46:265–274. doi: 10.7164/antibiotics.46.265. [DOI] [PubMed] [Google Scholar]
- 419.Oki T, Kakushima M, Hirano M, Takahashi A, Ohta A, Masuyoshi S, Hatori M, Kamei H. J Antibiot. 1992;45:1512–1517. doi: 10.7164/antibiotics.45.1512. [DOI] [PubMed] [Google Scholar]
- 420.Hasegawa T, Kakushima M, Hatori M, Aburaki S, Kakinuma S, Furumai T, Oki T. J Antibiot. 1993;46:598–605. doi: 10.7164/antibiotics.46.598. [DOI] [PubMed] [Google Scholar]
- 421.Furumai T, Hasegawa T, Kakushima M, Suzuki K, Yamamoto H, Yamamoto S, Hirano M, Oki T. J Antibiot. 1993;46:589–597. doi: 10.7164/antibiotics.46.589. [DOI] [PubMed] [Google Scholar]
- 422.Furumai T, Yamamoto H, Narita Y, Hasegawa T, Aburaki S, Kakushima M, Oki T. J Antibiot. 1993;46:1589–1597. doi: 10.7164/antibiotics.46.1589. [DOI] [PubMed] [Google Scholar]
- 423.Hanka LJ, Gerpheide SA. Antimicrob Agents Chemother. 1977;12:571–572. doi: 10.1128/aac.12.5.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 424.Leach BE, Calhoun KM, Johnson LE, Teeters CM, Jackson WG. J Am Chem Soc. 1953;75:4011–4012. [Google Scholar]
- 425.Strauss DG, Fleck WF. Z Allg Mikrobiol. 1977;17:227–234. doi: 10.1002/jobm.3630170308. [DOI] [PubMed] [Google Scholar]
- 426.Fleck W, Strauss D, Prauser H, Jungstand W, Heinecke H, Gutsche W, Wohlrabe K. Z Allg Mikrobiol. 1976;16:521–528. doi: 10.1002/jobm.3630160704. [DOI] [PubMed] [Google Scholar]
- 427.Beisler JA. Prog Med Chem. 1982;19:247–268. doi: 10.1016/s0079-6468(08)70331-x. [DOI] [PubMed] [Google Scholar]
- 428.Uramoto M, Kusano T, Nishio T, Isono K, Shishido K, Ando T. FEBS Lett. 1983;153:325–328. doi: 10.1016/0014-5793(83)80634-6. [DOI] [PubMed] [Google Scholar]
- 429.Kamitori S, Tanaka M, Akita Y, Yamamoto K. Carbohydr Res. 2003;338:1523–1525. doi: 10.1016/s0008-6215(03)00200-3. [DOI] [PubMed] [Google Scholar]
- 430.Weissman K. Chem Biol. 2005;12:512–514. doi: 10.1016/j.chembiol.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 431.Xu Z, Jakobi K, Welzel K, Hertweck C. Chem Biol. 2005;12:579–588. doi: 10.1016/j.chembiol.2005.04.017. [DOI] [PubMed] [Google Scholar]
- 432.Aoyama Y, Katayama T, Yamamoto M, Tanaka H, Kon K. J Antibiot. 1992;45:875–878. doi: 10.7164/antibiotics.45.875. [DOI] [PubMed] [Google Scholar]
- 433.Maskey RP, Pusecker K, Speitling M, Monecke P, Helmke E, Laatsch H. Z Naturforsch, B: J Chem Sci. 2002;57:823–829. [Google Scholar]
- 434.Uchida H, Nakakita Y, Enoki N, Abe N, Nakamura T, Munekata M. J Antibiot. 1994;47:655–667. doi: 10.7164/antibiotics.47.655. [DOI] [PubMed] [Google Scholar]
- 435.Uchida H, Nakakita Y, Enoki N, Abe N, Nakamura T, Munekata M. J Antibiot. 1994;47:648–654. doi: 10.7164/antibiotics.47.648. [DOI] [PubMed] [Google Scholar]
- 436.Uchida H, Nakakita Y, Enoki N, Abe N, Nakamura T, Munekata M. J Chem Soc, Chem Commun. 1994:323–324. [Google Scholar]
- 437.Uchida H, Nakakita Y, Enoki N, Abe N, Nakamura T, Munekata M. J Antibiot. 1993;46:1611–1615. doi: 10.7164/antibiotics.46.1611. [DOI] [PubMed] [Google Scholar]
- 438.Konishi M, Sugawara K, Kofu F, Nishiyama Y, Tomita K, Miyaki T, Kawaguchi H. J Antibiot. 1986;39:784–791. doi: 10.7164/antibiotics.39.784. [DOI] [PubMed] [Google Scholar]
- 439.Sugawara K, Tsunakawa M, Konishi M, Kawaguchi H, Krishnan B, He CH, Clardy J. J Org Chem. 1987;52:996–1001. [Google Scholar]
- 440.Lorico A, Long BH. Eur J Cancer. 1993;29A:1985–1991. doi: 10.1016/0959-8049(93)90459-s. [DOI] [PubMed] [Google Scholar]
- 441.Furumai T, Menzel R, Numata K, Taylor ST, Tsunakawa M. US5639735 A. US Pat. 1997
- 442.McGovren JP, Neil GL, Crampton SL, Robinson MI, Douros JD. Cancer Res. 1977;37:1666–1672. [PubMed] [Google Scholar]
- 443.Matsumae A, Shimosato Y, Igarashi Y, Hoshino M, Sano Y, Hata T. Kitasato Arch Exp Med. 1964;37:75–83. [PubMed] [Google Scholar]
- 444.Matsumae A. Kitasato Arch Exp Med. 1965;38:73–78. [PubMed] [Google Scholar]
- 445.Sano Y, Kanda N, Hata T. Kitasato Arch Exp Med. 1967;40:69–75. [PubMed] [Google Scholar]
- 446.Cheenpracha S, Vidor NB, Yoshida WY, Davies J, Chang LC. J Nat Prod. 2010;73:880–884. doi: 10.1021/np900843b. [DOI] [PubMed] [Google Scholar]
- 447.Heide L. Methods Enzymol. 2009;459:437–455. doi: 10.1016/S0076-6879(09)04618-7. [DOI] [PubMed] [Google Scholar]
- 448.Dolak L. J Antibiot. 1973;26:121–125. doi: 10.7164/antibiotics.26.121. [DOI] [PubMed] [Google Scholar]
- 449.Kawaguchi H, Naito T, Tsukiura H. J Antibiot. 1965;18:11–25. [PubMed] [Google Scholar]
- 450.Kawaguchi H, Tsukiura H, Okanishi M, Miyaki T, Ohmori T, Fujisawa K, Koshiyama H. J Antibiot. 1965;18:1–10. [PubMed] [Google Scholar]
- 451.Kawaguchi H, Miyaki T, Tsukiura H. J Antibiot. 1965;18:220–222. [PubMed] [Google Scholar]
- 452.Freitag A, Galm U, Li SM, Heide L. J Antibiot. 2004;57:205–209. doi: 10.7164/antibiotics.57.205. [DOI] [PubMed] [Google Scholar]
- 453.Reusser F, Dolak LA. J Antibiot. 1986;39:272–274. [PubMed] [Google Scholar]
- 454.Sebek OK, Hoeksema H. J Antibiot. 1972;25:434–436. doi: 10.7164/antibiotics.25.434. [DOI] [PubMed] [Google Scholar]
- 455.Sebek OK, Dolak LA. J Antibiot. 1984;37:136–142. doi: 10.7164/antibiotics.37.136. [DOI] [PubMed] [Google Scholar]
- 456.Li SM, Heide L. Curr Med Chem. 2005;12:419–427. doi: 10.2174/0929867053363063. [DOI] [PubMed] [Google Scholar]
- 457.Anderle C, Li SM, Kammerer B, Gust B, Heide L. J Antibiot. 2007;60:504–510. doi: 10.1038/ja.2007.64. [DOI] [PubMed] [Google Scholar]
- 458.Hooper DC, Wolfson JS, McHugh GL, Winters MB, Swartz MN. Antimicrob Agents Chemother. 1982;22:662–671. doi: 10.1128/aac.22.4.662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 459.Steffensky M, Muhlenweg A, Wang ZX, Li SM, Heide L. Antimicrob Agents Chemother. 2000;44:1214–1222. doi: 10.1128/aac.44.5.1214-1222.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 460.Galm U, Heller S, Shapiro S, Page M, Li SM, Heide L. Antimicrob Agents Chemother. 2004;48:1307–1312. doi: 10.1128/AAC.48.4.1307-1312.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 461.Neidle S. Cancer Drug Design and Discovery. 2. 2014. p. 260. [Google Scholar]
- 462.Zhao H, Yan B, Peterson LB, Blagg BS. ACS Med Chem Lett. 2012;3:327–331. doi: 10.1021/ml300018e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 463.Dolak L. J Antibiot. 1976;29:710–713. doi: 10.7164/antibiotics.29.710. [DOI] [PubMed] [Google Scholar]
- 464.Shane SJ. Can Med Assoc J. 1956;75:51–52. [PMC free article] [PubMed] [Google Scholar]
- 465.Crow FW, Duholke WK, Farley KA, Hadden CE, Hahn DA, Kaluzny BD, Mallory CS, Martin GE, Smith RF, Thamann TJ. J Heterocycl Chem. 1999;36:365–370. [Google Scholar]
- 466.Kaczka EA, Wolf FJ, Rathe FP, Folkers K. J Am Chem Soc. 1955;77:6404–6405. [Google Scholar]
- 467.Heide L, Gust B, Anderle C, Li SM. Curr Top Med Chem. 2008;8:667–679. doi: 10.2174/156802608784221505. [DOI] [PubMed] [Google Scholar]
- 468.Hooper DC, Wolfson JS, Mchugh GL, Winters MB, Swartz MN. Antimicrob Agents Chemother. 1982;22:662–671. doi: 10.1128/aac.22.4.662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 469.Burlison JA, Neckers L, Smith AB, Maxwell A, Blagg BS. J Am Chem Soc. 2006;128:15529–15536. doi: 10.1021/ja065793p. [DOI] [PubMed] [Google Scholar]
- 470.Heide L. Biotechnol Adv. 2009;27:1006–1014. doi: 10.1016/j.biotechadv.2009.05.017. [DOI] [PubMed] [Google Scholar]
- 471.Heide L. Nat Prod Rep. 2009;26:1241–1250. doi: 10.1039/b808333a. [DOI] [PubMed] [Google Scholar]
- 472.Xu H, Heide L, Li SM. Chem Biol. 2004;11:655–662. doi: 10.1016/j.chembiol.2004.02.028. [DOI] [PubMed] [Google Scholar]
- 473.Alt S. PhD thesis. University of Tübingen; 2010. [Google Scholar]
- 474.Sasaki T, Igarashi Y, Saito N, Furumai T. J Antibiot. 2001;54:441–447. doi: 10.7164/antibiotics.54.441. [DOI] [PubMed] [Google Scholar]
- 475.Hoeksema H, Mizsak SA, Baczynskyj L. J Antibiot. 1979;32:773–776. doi: 10.7164/antibiotics.32.773. [DOI] [PubMed] [Google Scholar]
- 476.Hoeksema H, Mizsak SA, Baczynskyj L, Pschigoda LM. J Am Chem Soc. 1982;104:5173–5181. [Google Scholar]
- 477.Bannister B, Zapotocky BA. J Antibiot. 1992;45:1313–1324. doi: 10.7164/antibiotics.45.1313. [DOI] [PubMed] [Google Scholar]
- 478.Galm U, Hager MH, Van Lanen SG, Ju J, Thorson JS, Shen B. Chem Rev. 2005;105:739–758. doi: 10.1021/cr030117g. [DOI] [PubMed] [Google Scholar]
- 479.Elshahawi SI, Ramelot TA, Seetharaman J, Chen J, Singh S, Yang Y, Pederson K, Kharel MK, Xiao R, Lew S, Yennamalli RM, Miller MD, Wang F, Tong L, Montelione GT, Kennedy MA, Bingman CA, Zhu H, Phillips GN, Jr, Thorson JS. ACS Chem Biol. 2014;9:2347–2358. doi: 10.1021/cb500327m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 480.Singh S, Hager MH, Zhang C, Griffith BR, Lee MS, Hallenga K, Markley JL, Thorson JS. ACS Chem Biol. 2006;1:451–460. doi: 10.1021/cb6002898. [DOI] [PubMed] [Google Scholar]
- 481.Biggins JB, Onwueme KC, Thorson JS. Science. 2003;301:1537–1541. doi: 10.1126/science.1086695. [DOI] [PubMed] [Google Scholar]
- 482.Liu C, Smith BM, Ajito K, Komatsu H, Gomez-Paloma L, Li T, Theodorakis EA, Nicolaou KC, Vogt PK. Proc Natl Acad Sci U S A. 1996;93:940–944. doi: 10.1073/pnas.93.2.940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 483.Yoshida K-i, Yoshinori M, Ryotaro A, Mayuko S, Toshio O. Tetrahedron Lett. 1993;34:2637–2640. [Google Scholar]
- 484.Minami Y, Yoshida K-I, Azuma R, Saeki M, Otani T. Tetrahedron Lett. 1993;34:2633–2636. [Google Scholar]
- 485.Otani T, Yoshida K, Sasaki T, Minami Y. J Antibiot. 1999;52:415–421. doi: 10.7164/antibiotics.52.415. [DOI] [PubMed] [Google Scholar]
- 486.Albers-Schönberg G, Dewey RS, Hensens OD, Liesch JM, Napier MA, Goldberg IH. Biochem Biophys Res Commun. 1980;95:1351–1356. doi: 10.1016/0006-291x(80)91622-8. [DOI] [PubMed] [Google Scholar]
- 487.Edo K, Mizugaki M, Koide Y, Seto H, Furihata K, Ôtake N, Ishida N. Tetrahedron Lett. 1985;26:331–334. [Google Scholar]
- 488.Edo K, Akiyama Y, Saito K, Mizugaki M, Koide Y, Ishida N. J Antibiot. 1986;39:1615–1619. doi: 10.7164/antibiotics.39.1615. [DOI] [PubMed] [Google Scholar]
- 489.Hanada M, Ohkuma H, Yonemoto T, Tomita K, Ohbayashi M, Kamei H, Miyaki T, Konishi M, Kawaguchi H, Forenza S. J Antibiot. 1991;44:403–414. doi: 10.7164/antibiotics.44.403. [DOI] [PubMed] [Google Scholar]
- 490.Leet JE, Schroeder DR, Hofstead SJ, Golik J, Colson KL, Huang S, Klohr SE, Doyle TW, Matson JA. J Am Chem Soc. 1992;114:7946–7948. [Google Scholar]
- 491.Leet JE, Golik J, Hofstead SJ, Matson JA, Lee AY, Clardy J. Tetrahedron Lett. 1992;33:6107–6110. [Google Scholar]
- 492.Ren F, Hogan PC, Anderson AJ, Myers AG. J Am Chem Soc. 2007;129:5381–5383. doi: 10.1021/ja071205b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 493.Nam S-J, Gaudêncio SP, Kauffman CA, Jensen PR, Kondratyuk TP, Marler LE, Pezzuto JM, Fenical W. J Nat Prod. 2010;73:1080–1086. doi: 10.1021/np100087c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 494.Oh D-C, Williams PG, Kauffman CA, Jensen PR, Fenical W. Org Lett. 2006;8:1021–1024. doi: 10.1021/ol052686b. [DOI] [PubMed] [Google Scholar]
- 495.Lee MD, Manning JK, Williams DR, Kuck NA, Testa RT, Borders DB. J Antibiot. 1989;42:1070–1087. doi: 10.7164/antibiotics.42.1070. [DOI] [PubMed] [Google Scholar]
- 496.Lee MD, Dunne TS, Chang CC, Siegel MM, Morton GO, Ellestad GA, McGahren WJ, Borders DB. J Am Chem Soc. 1992;114:985–997. [Google Scholar]
- 497.Ellestad GA, McGahren WJ. EP0330874 A3. European Pat. 1989
- 498.Lee MD-M, Greenstein M, Labeda DP, Fantini AA. EP0276485 (A2) European Pat. 1988
- 499.Konishi M, Ohkuma H, Saitoh K, Kawaguchi H, Golik J, Dubay G, Groenewold G, Krishnan B, Doyle TW. J Antibiot. 1985;38:1605–1609. doi: 10.7164/antibiotics.38.1605. [DOI] [PubMed] [Google Scholar]
- 500.Golik J, Dubay G, Groenewold G, Kawaguchi H, Konishi M, Krishnan B, Ohkuma H, Saitoh K, Doyle TW. J Am Chem Soc. 1987;109:3462–3464. doi: 10.7164/antibiotics.38.1605. [DOI] [PubMed] [Google Scholar]
- 501.Golik J, Clardy J, Dubay G, Groenewold G, Kawaguchi H, Konishi M, Krishnan B, Ohkuma H, Saitoh K, Doyle TW. J Am Chem Soc. 1987;109:3461–3462. [Google Scholar]
- 502.Kiessling LL, Schreiber SL. Chemtracts: Org Chem. 1988;1:4–6. [Google Scholar]
- 503.Golik J, Wong H, Vyas DM, Doyle TW. Tetrahedron Lett. 1989;30:2497–2500. [Google Scholar]
- 504.Golik J, Doyle TW, Van Duyne G, Clardy J. Tetrahedron Lett. 1990;31:6149–6150. [Google Scholar]
- 505.Beutler JA, Clark P, Alvarado AB, Golik J. J Nat Prod. 1994;57:629–633. doi: 10.1021/np50107a010. [DOI] [PubMed] [Google Scholar]
- 506.Oku N, Matsunaga S, Fusetani N. J Am Chem Soc. 2003;125:2044–2045. doi: 10.1021/ja0296780. [DOI] [PubMed] [Google Scholar]
- 507.McDonald LA, Capson TL, Krishnamurthy G, Ding W-D, Ellestad GA, Bernan VS, Maiese WM, Lassota P, Kramer RA, Ireland CM. J Am Chem Soc. 1996;118:10898–10899. [Google Scholar]
- 508.Ahlert J, Shepard E, Lomovskaya N, Zazopoulos E, Staffa A, Bachmann BO, Huang K, Fonstein L, Czisny A, Whitwam RE, Farnet CM, Thorson JS. Science. 2002;297:1173–1176. doi: 10.1126/science.1072105. [DOI] [PubMed] [Google Scholar]
- 509.Zhang C, Griffith BR, Fu Q, Albermann C, Fu X, Lee I-K, Li L, Thorson JS. Science. 2006;313:1291–1294. doi: 10.1126/science.1130028. [DOI] [PubMed] [Google Scholar]
- 510.Zhang C, Bitto E, Goff RD, Singh S, Bingman CA, Griffith BR, Albermann C, Phillips GN, Jr, Thorson JS. Chem Biol. 2008;15:842–853. doi: 10.1016/j.chembiol.2008.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 511.Chang A, Singh S, Helmich KE, Goff RD, Bingman CA, Thorson JS, Phillips GN., Jr Proc Natl Acad Sci U S A. 2011;108:17649–17654. doi: 10.1073/pnas.1108484108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 512.Gao Q, Zhang C, Blanchard S, Thorson JS. Chem Biol. 2006;13:733–743. doi: 10.1016/j.chembiol.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 513.Walker S, Valentine KG, Kahne D. J Am Chem Soc. 1990;112:6428–6429. [Google Scholar]
- 514.Kumar RA, Ikemoto N, Patel DJ. J Mol Biol. 1997;265:173–186. doi: 10.1006/jmbi.1996.0719. [DOI] [PubMed] [Google Scholar]
- 515.Kumar RA, Ikemoto N, Patel DJ. J Mol Biol. 1997;265:187–201. doi: 10.1006/jmbi.1996.0718. [DOI] [PubMed] [Google Scholar]
- 516.Goff RD, Singh S, Thorson JS. ChemMedChem. 2011;6:774–776. doi: 10.1002/cmdc.201100028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 517.Ndejouong Ble S, Sattler I, Dahse HM, Kothe E, Hertweck C. Bioorg Med Chem Lett. 2009;19:6473–6476. doi: 10.1016/j.bmcl.2009.08.084. [DOI] [PubMed] [Google Scholar]
- 518.Cress BF, Linhardt RJ, Koffas MAG. Natural Products, Isoflavonoid Production by Genetically Engineered Microorganisms. 2013:1647–1681. [Google Scholar]
- 519.Du J, Shao Z, Zhao H. J Ind Microbiol Biotechnol. 2011;38:873–890. doi: 10.1007/s10295-011-0970-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 520.Williams GJ, Yang J, Zhang C, Thorson JS. ACS Chem Biol. 2011;6:95–100. doi: 10.1021/cb100267k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 521.Pandey RP, Li TF, Kim EH, Yamaguchi T, Park YI, Kim JS, Sohng JK. Appl Environ Microbiol. 2013;79:3516–3521. doi: 10.1128/AEM.00409-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 522.Pandey RP, Malla S, Simkhada D, Kim BG, Sohng JK. Appl Microbiol Biotechnol. 2013;97:1889–1901. doi: 10.1007/s00253-012-4438-9. [DOI] [PubMed] [Google Scholar]
- 523.Thuan NH, Pandey RP, Thuy TT, Park JW, Sohng JK. Appl Biochem Biotechnol. 2013;171:1956–1967. doi: 10.1007/s12010-013-0459-9. [DOI] [PubMed] [Google Scholar]
- 524.Dixon RA, Ferreira D. Phytochemistry. 2002;60:205–211. doi: 10.1016/s0031-9422(02)00116-4. [DOI] [PubMed] [Google Scholar]
- 525.Hazato T, Naganawa H, Kumagai M, Aoyagi T, Umezawa H. J Antibiot. 1979;32:217–222. doi: 10.7164/antibiotics.32.217. [DOI] [PubMed] [Google Scholar]
- 526.Aoyagi T, Hazato T, Kumagai M, Hamada M, Takeuchi T. J Antibiot. 1975;28:1006–1008. doi: 10.7164/antibiotics.28.1006. [DOI] [PubMed] [Google Scholar]
- 527.Jiang ZD, Jensen PR, Fenical W. Tetrahedron Lett. 1997;38:5065–5068. [Google Scholar]
- 528.Maskey RP, Asolkar RN, Speitling M, Hoffmann V, Grun-Wollny I, Felck WF, Laatsch H. Z Naturforsch, B: J Chem Sci. 2003;58:686–691. [Google Scholar]
- 529.Dong Y, Yang J, Ren X, Zhang H, He J. J Antibiot. 2005;58:737–739. doi: 10.1038/ja.2005.100. [DOI] [PubMed] [Google Scholar]
- 530.Hu JF, Wunderlich D, Sattler I, Thiericke R, Grabley S, Feng XZ. Nat Prod Res. 2003;17:451–458. doi: 10.1080/1478641031000120534. [DOI] [PubMed] [Google Scholar]
- 531.Kim WG, Yoon TM, Kwon HJ, Suh JW. J Antibiot. 2006;59:640–645. doi: 10.1038/ja.2006.85. [DOI] [PubMed] [Google Scholar]
- 532.Yoon TM, Kim JW, Kim JG, Kim WG, Suh JW. J Antibiot. 2006;59:633–639. doi: 10.1038/ja.2006.84. [DOI] [PubMed] [Google Scholar]
- 533.Jansen R. DECHEMA Biotechnology Conferences, Part B. 1992;5:729–733. [Google Scholar]
- 534.Knirel YA, Shashkov AS, Senchenkova SN, Merino S, Tomas JM. Carbohydr Res. 2002;337:1381–1386. doi: 10.1016/s0008-6215(02)00136-2. [DOI] [PubMed] [Google Scholar]
- 535.Shibuya N, Amano K, Azuma J, Nishihara T, Kitamura Y, Noguchi T, Koga T. J Biol Chem. 1991;266:16318–16323. [PubMed] [Google Scholar]
- 536.Torgov VI, Shashkov AS, Kochanowski H, Jann B, Jann K. Carbohydr Res. 1996;283:223–227. doi: 10.1016/0008-6215(95)00411-4. [DOI] [PubMed] [Google Scholar]
- 537.Nakano H, Omura S. J Antibiot. 2009;62:17–26. doi: 10.1038/ja.2008.4. [DOI] [PubMed] [Google Scholar]
- 538.Speck K, Magauer T. Beilstein J Org Chem. 2013;9:2048–2078. doi: 10.3762/bjoc.9.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 539.Méndez C, Moris F, Salas JA. Drug Discovery from Natural Products: Biosynthesis of indolocarbazole alkaloids and generation of novel derivatives by combinatorial biosynthesis. 2012 [Google Scholar]
- 540.Sanchez C, Zhu LL, Brana AF, Salas AP, Rohr J, Méndez C, Salas JA. Proc Natl Acad Sci U S A. 2005;102:461–466. doi: 10.1073/pnas.0407809102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 541.Ortiz-Ibanez K, Alsina MM, Munoz-Santos C. Actas Dermo-Sifiliogr. 2013;104:304–310. doi: 10.1016/j.adengl.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 542.Fischer T, Stone RM, Deangelo DJ, Galinsky I, Estey E, Lanza C, Fox E, Ehninger G, Feldman EJ, Schiller GJ, Klimek VM, Nimer SD, Gilliland DG, Dutreix C, Huntsman-Labed A, Virkus J, Giles FJ. J Clin Oncol. 2010;28:4339–4345. doi: 10.1200/JCO.2010.28.9678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 543.Robey RW, Obrzut T, Shukla S, Polgar O, Macalou S, Bahr JC, Di Pietro A, Ambudkar SV, Bates SE. Cancer Chemother Pharmacol. 2009;64:575–583. doi: 10.1007/s00280-008-0908-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 544.Marinelli F. Methods Enzymol. 2009;458:29–58. doi: 10.1016/S0076-6879(09)04802-2. [DOI] [PubMed] [Google Scholar]
- 545.Shabbir M, Stuart R. Expert Opin Invest Drugs. 2010;19:427–436. doi: 10.1517/13543781003598862. [DOI] [PubMed] [Google Scholar]
- 546.Nettleton DE, Doyle TW, Krishnan B, Matsumoto GK, Clardy J. Tetrahedron Lett. 1985;26:4011–4014. [Google Scholar]
- 547.Funato N, Takayanagi H, Konda Y, Toda Y, Harigaya Y, Iwai Y, Ômura S. Tetrahedron Lett. 1994;35:1251–1254. [Google Scholar]
- 548.Sánchez C, Butovich IA, Braña AF, Rohr J, Méndez C, Salas JA. Chem Biol. 2002;9:519–531. doi: 10.1016/s1074-5521(02)00126-6. [DOI] [PubMed] [Google Scholar]
- 549.Sánchez C, Méndez C, Salas JA. Nat Prod Rep. 2006;23:1007–1045. doi: 10.1039/b601930g. [DOI] [PubMed] [Google Scholar]
- 550.Lam KS, Schroeder DR, Veitch JM, Colson KL, Matson JA, Rose WC, Doyle TW, Forenza S. J Antibiot. 2001;54:1–9. doi: 10.7164/antibiotics.54.1. [DOI] [PubMed] [Google Scholar]
- 551.Zhang C, Albermann C, Fu X, Peters NR, Chisholm JD, Zhang G, Gilbert EJ, Wang PG, Van Vranken DL, Thorson JS. ChemBioChem. 2006;7:795–804. doi: 10.1002/cbic.200500504. [DOI] [PubMed] [Google Scholar]
- 552.Zhang C, Weller RL, Thorson JS, Rajski SR. J Am Chem Soc. 2006;128:2760–2761. doi: 10.1021/ja056231t. [DOI] [PubMed] [Google Scholar]
- 553.Singh S, Zhang J, Huber TD, Sunkara M, Hurley K, Goff RD, Wang G, Zhang W, Liu C, Rohr J, Van Lanen SG, Morris AJ, Thorson JS. Angew Chem, Int Ed. 2014;53:3965–3969. doi: 10.1002/anie.201308272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 554.Hyun CG, Bililign T, Liao J, Thorson JS. ChemBioChem. 2003;4:114–117. doi: 10.1002/cbic.200390004. [DOI] [PubMed] [Google Scholar]
- 555.McAlpine JB, Karwowski JP, Jackson M, Mullally MM, Hochlowski JE, Premachandran U, Burres NS. J Antibiot. 1994;47:281–288. doi: 10.7164/antibiotics.47.281. [DOI] [PubMed] [Google Scholar]
- 556.Takahashi H, Osada H, Uramoto M, Isono K. J Antibiot. 1990;43:168–173. doi: 10.7164/antibiotics.43.168. [DOI] [PubMed] [Google Scholar]
- 557.Osada H, Satake M, Koshino H, Onose R, Isono K. J Antibiot. 1992;45:278–279. doi: 10.7164/antibiotics.45.278. [DOI] [PubMed] [Google Scholar]
- 558.Cai Y, Fredenhagen A, Hug P, Meyer T, Peter HH. J Antibiot. 1996;49:519–526. doi: 10.7164/antibiotics.49.519. [DOI] [PubMed] [Google Scholar]
- 559.Omura S, Iwai Y, Hirano A, Nakagawa A, Awaya J, Tsuchya H, Takahashi Y, Masuma R. J Antibiot. 1977;30:275–282. doi: 10.7164/antibiotics.30.275. [DOI] [PubMed] [Google Scholar]
- 560.Xie Q, Wang Y, Huang Y, Wu Y, Ba F, Liu Z. Int J Syst Evol Microbiol. 2002;52:1815–1820. doi: 10.1099/00207713-52-5-1815. [DOI] [PubMed] [Google Scholar]
- 561.Rüegg UT, Gillian B. Trends Pharmacol Sci. 1989;10:218–220. doi: 10.1016/0165-6147(89)90263-0. [DOI] [PubMed] [Google Scholar]
- 562.Tanida S, Takizawa M, Takahashi T, Tsubotani S, Harada S. J Antibiot. 1989;42:1619–1630. doi: 10.7164/antibiotics.42.1619. [DOI] [PubMed] [Google Scholar]
- 563.Takahashi I, Saitoh Y, Yoshida M, Sano H, Nakano H, Morimoto M, Tamaoki T. J Antibiot. 1989;42:571–576. doi: 10.7164/antibiotics.42.571. [DOI] [PubMed] [Google Scholar]
- 564.Omura S, Sasaki Y, Iwai Y, Takeshima H. J Antibiot. 1995;48:535–548. doi: 10.7164/antibiotics.48.535. [DOI] [PubMed] [Google Scholar]
- 565.Koshino H, Osada H, Isono K. J Antibiot. 1992;45:195–198. doi: 10.7164/antibiotics.45.195. [DOI] [PubMed] [Google Scholar]
- 566.Cai Y, Fredenhagen A, Hug P, Peter HH. J Antibiot. 1996;49:1060–1062. doi: 10.7164/antibiotics.49.1060. [DOI] [PubMed] [Google Scholar]
- 567.Hoehn P, Ghisalba O, Moerker T, Peter HH. J Antibiot. 1995;48:300–305. doi: 10.7164/antibiotics.48.300. [DOI] [PubMed] [Google Scholar]
- 568.Cai Y, Fredenhagen A, Hug P, Peter HH. J Antibiot. 1995;48:143–148. doi: 10.7164/antibiotics.48.143. [DOI] [PubMed] [Google Scholar]
- 569.Han X-X, Cui C-B, Gu Q-Q, Zhu W-M, Liu H-B, Gu J-Y, Osada H. Tetrahedron Lett. 2005;46:6137–6140. [Google Scholar]
- 570.Koshino H, Osada H, Amano S, Onose R, Isono K. J Antibiot. 1992;45:1428–1432. doi: 10.7164/antibiotics.45.1428. [DOI] [PubMed] [Google Scholar]
- 571.Hernández LM, Blanco JA, Baz JP, Puentes JL, Millán FR, Vázquez FE, Fernández-Chimeno RI, Grávalos DG. J Antibiot. 2000;53:895–902. doi: 10.7164/antibiotics.53.895. [DOI] [PubMed] [Google Scholar]
- 572.Ubukata M, Tamehiro N, Matsuura N, Nakajima N. J Antibiot. 1999;52:921–924. doi: 10.7164/antibiotics.52.921. [DOI] [PubMed] [Google Scholar]
- 573.Tamehiro N, Matsuura N, Feng Y, Nakajima N, Ubukata M. J Antibiot. 2002;55:363–370. doi: 10.7164/antibiotics.55.363. [DOI] [PubMed] [Google Scholar]
- 574.Feng Y, Matsuura N, Ubukata M. J Antibiot. 2004;57:627–633. doi: 10.7164/antibiotics.57.627. [DOI] [PubMed] [Google Scholar]
- 575.Nakanishi S, Matsuda Y, Iwahashi K, Kase H. J Antibiot. 1986;39:1066–1071. doi: 10.7164/antibiotics.39.1066. [DOI] [PubMed] [Google Scholar]
- 576.Yasuzawa T, Iida T, Yoshida M, Hirayama N, Takahashi M, Shirahata K, Sano H. J Antibiot. 1986;39:1072–1078. doi: 10.7164/antibiotics.39.1072. [DOI] [PubMed] [Google Scholar]
- 577.Anizon F, Belin L, Moreau P, Sancelme M, Voldoire A, Prudhomme M, Ollier M, Sevère D, Riou JF, Bailly C, Fabbro D, Meyer T. J Med Chem. 1997;40:3456–3465. doi: 10.1021/jm9702084. [DOI] [PubMed] [Google Scholar]
- 578.Anizon F, Moreau P, Sancelme M, Laine W, Bailly C, Prudhomme M. Bioorg Med Chem. 2003;11:3709–3722. doi: 10.1016/s0968-0896(03)00343-2. [DOI] [PubMed] [Google Scholar]
- 579.Labeda DP, Hatano K, Kroppenstedt RM, Tamura T. Int J Syst Evol Microbiol. 2001;51:1045–1050. doi: 10.1099/00207713-51-3-1045. [DOI] [PubMed] [Google Scholar]
- 580.Golik J, Doyle TW, Krishnan B, Dubay G, Matson JA. J Antibiot. 1989;42:1784–1789. doi: 10.7164/antibiotics.42.1784. [DOI] [PubMed] [Google Scholar]
- 581.Chisholm JD, Golik J, Krishnan B, Matson JA, Van Vranken DL. J Am Chem Soc. 1999;121:3801–3802. [Google Scholar]
- 582.Williams DE, Bernan VS, Ritacco FV, Maiese WM, Greenstein M, Andersen RJ. Tetrahedron Lett. 1999;40:7171–7174. [Google Scholar]
- 583.Bonjouklian R, Smitka TA, Doolin LE, Molloy RM, Debono M, Shaffer SA, Moore RE, Stewart JB, Patterson GML. Tetrahedron. 1991;47:7739–7750. [Google Scholar]
- 584.Maskey RP, Grün-Wollny I, Fiebig HH, Laatsch H. Angew Chem, Int Ed. 2002;41:597–599. [Google Scholar]
- 585.Maskey RP, Grün-Wollny I, Laatsch H. Nat Prod Res. 2005;19:137–142. doi: 10.1080/14786410410001704741. [DOI] [PubMed] [Google Scholar]
- 586.Schumacher RW, Harrigan BL, Davidson BS. Tetrahedron Lett. 2001;42:5133–5135. [Google Scholar]
- 587.Furuta R, Naruto S, Tamura A, Yokogawa K. Tetrahedron Lett. 1979;20:1701–1704. [Google Scholar]
- 588.Ito T, Ohba K, Koyama M, Sezaki M, Tohyama H, Shomura T, Fukuyasu H, Kazuno Y, Niwa T, Kojima M. J Antibiot. 1984;37:931–934. doi: 10.7164/antibiotics.37.931. [DOI] [PubMed] [Google Scholar]
- 589.Yang SW, Cordell GA. J Nat Prod. 1997;60:230–235. doi: 10.1021/np960674g. [DOI] [PubMed] [Google Scholar]
- 590.Hu JF, Wunderlich D, Sattler I, Härtl A, Papastavrou I, Grond S, Grabley S, Feng XZ, Thiericke R. J Antibiot. 2000;53:944–953. doi: 10.7164/antibiotics.53.944. [DOI] [PubMed] [Google Scholar]
- 591.Abe N, Enoki N, Nakakita Y, Uchida H, Nakamura T, Munekata M. J Antibiot. 1993;46:1678–1686. doi: 10.7164/antibiotics.46.1678. [DOI] [PubMed] [Google Scholar]
- 592.Abe N, Nakakita Y, Nakamura T, Enoki N, Uchida H, Takeo S, Munekata M. J Antibiot. 1993;46:1672–1677. doi: 10.7164/antibiotics.46.1672. [DOI] [PubMed] [Google Scholar]
- 593.Hagmann L, Keller-Schierlein W, Wahl B, Zahner H. J Antibiot. 1988;41:289–295. doi: 10.7164/antibiotics.41.289. [DOI] [PubMed] [Google Scholar]
- 594.Kumagai M, Aoyagi T, Umezawa H. J Antibiot. 1976;29:696–703. doi: 10.7164/antibiotics.29.696. [DOI] [PubMed] [Google Scholar]
- 595.Kumagai M, Naganawa H, Aoyagi T, Umezawa H. J Antibiot. 1975;28:876–880. doi: 10.7164/antibiotics.28.876. [DOI] [PubMed] [Google Scholar]
- 596.Aoyagi T, Kumagai M, Hazato T, Hamada M, Takeuchi T. J Antibiot. 1975;28:555–557. doi: 10.7164/antibiotics.28.555. [DOI] [PubMed] [Google Scholar]
- 597.De Sarro G, Carotti A, Campagna F, McKernan R, Rizzo M, Falconi U, Palluotto F, Giusti P, Rettore C, De Sarro A. Pharmacol, Biochem Behav. 2000;65:475–487. doi: 10.1016/s0091-3057(99)00230-0. [DOI] [PubMed] [Google Scholar]
- 598.Shaaban M, Schroder D, Shaaban KA, Helmke E, Grun-Wollny I, Wagner-Dobler I, Laatsch H. Rev Latinoam Quím. 2007;35:58–67. [Google Scholar]
- 599.Habib ES, Yokomizo K, Murata K, Uyeda M. J Antibiot. 2000;53:1420–1423. doi: 10.7164/antibiotics.53.1420. [DOI] [PubMed] [Google Scholar]
- 600.Donnerstag A, Marzian S, Muller D, Welzel P, Bottger D, Stark A, Fehlhaber HW, Markus A, Vanheijenoort Y, Vanheijenoort J. Tetrahedron. 1995;51:1931–1940. [Google Scholar]
- 601.Ohtsuka T, Itezono Y, Nakayama N, Kurano M, Nakada N, Tanaka H, Sawairi S, Yokose K. Tetrahedron Lett. 1992;33:2705–2708. [Google Scholar]
- 602.Qian-Cutrone J, Ueki T, Huang S, Mookhtiar KA, Ezekiel R, Kalinowski SS, Brown KS, Golik J, Lowe S, Pirnik DM, Hugill R, Veitch JA, Klohr SE, Whitney JL, Manly SP. J Antibiot. 1999;52:245–255. doi: 10.7164/antibiotics.52.245. [DOI] [PubMed] [Google Scholar]
- 603.Burgess ML, Barrow KD, Gao C, Heard GM, Glenn D. J Nat Prod. 1999;62:859–863. doi: 10.1021/np980573d. [DOI] [PubMed] [Google Scholar]
- 604.Wicke C, Huners M, Wray V, Nimtz M, Bilitewski U, Lang S. J Nat Prod. 2000;63:621–626. doi: 10.1021/np990313b. [DOI] [PubMed] [Google Scholar]
- 605.Batrakov SG, Nikitin DI, Pitryuk IA. Biochim Biophys Acta. 1996;1303:39–46. doi: 10.1016/0005-2760(96)00072-0. [DOI] [PubMed] [Google Scholar]
- 606.Kumada H, Haishima Y, Umemoto T, Tanamoto K. J Bacteriol. 1995;177:2098–2106. doi: 10.1128/jb.177.8.2098-2106.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 607.Reshef V, Mizrachi E, Maretzki T, Silberstein C, Loya S, Hizi A, Carmeli S. J Nat Prod. 1997;60:1251–1260. doi: 10.1021/np970327m. [DOI] [PubMed] [Google Scholar]
- 608.Kawahara K, Moll H, Knirel YA, Seydel U, Zahringer U. Eur J Biochem. 2000;267:1837–1846. doi: 10.1046/j.1432-1327.2000.01189.x. [DOI] [PubMed] [Google Scholar]
- 609.Taylor RF, Davies BH. Can J Biochem Cell Biol. 1983;61:892–905. doi: 10.1139/o83-114. [DOI] [PubMed] [Google Scholar]
- 610.Yagi H, Maruyama A. J Nat Prod. 1999;62:631–632. doi: 10.1021/np9804280. [DOI] [PubMed] [Google Scholar]
- 611.Matsubara T, Iida-Tanaka N, Kamekura M, Moldoveanu N, Ishizuka I, Onishi H, Hayashi A, Kates M. Biochim Biophys Acta. 1994;1214:97–108. doi: 10.1016/0005-2760(94)90014-0. [DOI] [PubMed] [Google Scholar]
- 612.Becker DJ, Lowe JB. Glycobiology. 2003;13:41R–53R. doi: 10.1093/glycob/cwg054. [DOI] [PubMed] [Google Scholar]
- 613.Anjaneyulu V, Rao PS, Radhika P, Laatsch H, Asolkar RN. Ind J Chem. 2001;40:405–409. [Google Scholar]
- 614.Ohlendorf B, Lorenzen W, Kehraus S, Krick A, Bode HB, Konig GM. J Nat Prod. 2009;72:82–86. doi: 10.1021/np8005875. [DOI] [PubMed] [Google Scholar]
- 615.Shindo K, Mikami K, Tamesada E, Takaichi S, Adachi K, Misawa N, Maoka T. Tetrahedron Lett. 2007;48:2725–2727. [Google Scholar]
- 616.Antonopoulou S, Karantonis HC, Nomikos T, Oikonomou A, Fragopoulou E, Pantazidou A. Comp Biochem Physiol, Part B: Biochem Mol Biol. 2005;142:269–282. doi: 10.1016/j.cbpc.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 617.Fusetani N, Yasukawa K, Matsunaga S, Hashimoto K. Tetrahedron Lett. 1985;26:6449–6452. [Google Scholar]
- 618.Batrakov SG, Sheichenko VI, Nikitin DI. Biochim Biophys Acta. 1999;1440:163–175. doi: 10.1016/s1388-1981(99)00119-5. [DOI] [PubMed] [Google Scholar]
- 619.Argoudelis AD, Coats JH, Johnson LE. US3907774. US Pat. 1975
- 620.Yagi H, Maruyama A. Biochim Biophys Acta. 1998;1393:161–165. doi: 10.1016/s0005-2760(98)00070-8. [DOI] [PubMed] [Google Scholar]
- 621.Vorob’eva EV, Dmitrenok AS, Dmitrenok PS, Isakov VV, Krasikova IN, Solov’eva TF. Bioorg Khim. 2005;31:404–413. [PubMed] [Google Scholar]
- 622.Hopmann C, Kurz M, Bronstrup M, Wink J, LeBeller D. Tetrahedron Lett. 2002;43:435–438. [Google Scholar]
- 623.Gross H, McPhail K, Goeger DE, Valeriote FA, Gerwick WH. Phytochemistry. 2010;71:1729–1735. doi: 10.1016/j.phytochem.2010.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 624.Pronin SV, Kozmin SA. J Am Chem Soc. 2010;132:14394–14396. doi: 10.1021/ja107190w. [DOI] [PubMed] [Google Scholar]
- 625.Schabacher K, Zeeck A. Tetrahedron Lett. 1973;29:2691–2694. [Google Scholar]
- 626.Taniyama H, Sawada Y, Kitagawa T. J Antibiot. 1971;24:390–392. doi: 10.7164/antibiotics.24.390. [DOI] [PubMed] [Google Scholar]
- 627.Hochlowski JE, Buytendorp MH, Whittern DN, Buko AM, Chen RH, Mcalpine JB. J Antibiot. 1988;41:1300–1315. doi: 10.7164/antibiotics.41.1300. [DOI] [PubMed] [Google Scholar]
- 628.Brotz E, Kulik A, Vikineswary S, Lim CT, Tan GY, Zinecker H, Imhoff JF, Paululat T, Fiedler HP. J Antibiot. 2011;64:257–266. doi: 10.1038/ja.2010.170. [DOI] [PubMed] [Google Scholar]
- 629.McDonald LA, Lotvin JA, Bailey AE, Carter GT. J Nat Prod. 1998;61:217–226. doi: 10.1021/np970408i. [DOI] [PubMed] [Google Scholar]
- 630.Dolle RE, Nicolaou KC. J Am Chem Soc. 1985;107:1695–1698. [Google Scholar]
- 631.Jefson MR, Kaneda K, Nishiyama S, Tone J. US4937184 A. US Pat. 1990
- 632.Dewey RS, Arison BH, Hannah J, Shih DH, Albers-Schonberg G. J Antibiot. 1985;38:1691–1698. doi: 10.7164/antibiotics.38.1691. [DOI] [PubMed] [Google Scholar]
- 633.Konda Y, Nakagawa A, Harigaya Y, Onda M, Masuma R, Omura S. J Antibiot. 1988;41:268–270. doi: 10.7164/antibiotics.41.268. [DOI] [PubMed] [Google Scholar]
- 634.Kibwage IO, Janssen G, Busson R, Hoogmartens J, Vanderhaeghe H, Verbist L. J Antibiot. 1987;40:1–6. doi: 10.7164/antibiotics.40.1. [DOI] [PubMed] [Google Scholar]
- 635.Park JW, Oh HS, Jung WS, Park SR, Han AR, Ban YH, Kim EJ, Kang HY, Yoon YJ. Chem Commun. 2008:5782–5784. doi: 10.1039/b814603a. [DOI] [PubMed] [Google Scholar]
- 636.Hong JS, Park SH, Choi CY, Sohng JK, Yoon YJ. FEMS Microbiol Lett. 2004;238:391–399. doi: 10.1016/j.femsle.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 637.Borisova SA, Zhang C, Takahashi H, Zhang H, Wong AW, Thorson JS, Liu H-w. Angew Chem, Int Ed. 2006;45:2748–2753. doi: 10.1002/anie.200503195. [DOI] [PubMed] [Google Scholar]
- 638.Zuckerman JM. Infect Dis Clin North Am. 2004;18:621–649. doi: 10.1016/j.idc.2004.04.010. [DOI] [PubMed] [Google Scholar]
- 639.Zuckerman JM, Qamar F, Bono BR. Med Clin North Am. 2011;95:761–791. doi: 10.1016/j.mcna.2011.03.012. [DOI] [PubMed] [Google Scholar]
- 640.Celmer WD, Cullen WP, Maeda H, Tone J. US 4543334 A. US Pat. 1985
- 641.Martin JR, DeVault RL, Sinclair AC, Stanaszek RS, Johnson P. J Antibiot. 1982;35:426–430. doi: 10.7164/antibiotics.35.426. [DOI] [PubMed] [Google Scholar]
- 642.El-Bondkly AM, Abd-Alla HI, Shaaban M, Shaaban KA. Indian J Pharm Sci. 2008;70:310–319. doi: 10.4103/0250-474X.42979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 643.Paterson I, Mansuri MM. Tetrahedron. 1985;41:3569–3624. [Google Scholar]
- 644.Mallams AK, Jaret RS, Reimann H. J Am Chem Soc. 1969;91:7506–7508. doi: 10.1021/ja01054a047. [DOI] [PubMed] [Google Scholar]
- 645.Weinstei Mj, Wagman GH, Marquez JA, Testa RT, Oden E, Waitz JA. J Antibiot. 1969;22:253–258. doi: 10.7164/antibiotics.22.253. [DOI] [PubMed] [Google Scholar]
- 646.Rickards RW, Smith RM, Majer J. Chem Commun. 1968:1049–1051. [Google Scholar]
- 647.Belyakova YG, Viktorovsky IV, Vyunov KA, Vorontsov VM, Kanev AS, Komarov EV. Antibiot Khimioter. 1989;34:199–202. [PubMed] [Google Scholar]
- 648.Yaginuma S, Morishita A, Ishizawa K, Murofushi S, Hayashi M, Mutoh N. J Antibiot. 1992;45:599–606. doi: 10.7164/antibiotics.45.599. [DOI] [PubMed] [Google Scholar]
- 649.Morishita A, Ishizawa K, Mutoh N, Yamamoto T, Hayashi M, Yaginuma S. J Antibiot. 1992;45:607–612. doi: 10.7164/antibiotics.45.607. [DOI] [PubMed] [Google Scholar]
- 650.Morishita A, Murofushi S, Ishizawa K, Mutoh N, Yaginuma S. J Antibiot. 1992;45:1011–1015. doi: 10.7164/antibiotics.45.1011. [DOI] [PubMed] [Google Scholar]
- 651.Morishita A, Murofushi S, Ishizawa K, Mutoh N, Yaginuma S. J Antibiot. 1992;45:809–812. doi: 10.7164/antibiotics.45.809. [DOI] [PubMed] [Google Scholar]
- 652.Schiewe HJ, Zeeck A. J Antibiot. 1999;52:635–642. doi: 10.7164/antibiotics.52.635. [DOI] [PubMed] [Google Scholar]
- 653.Cevallos A, Guerriero A. J Antibiot. 2003;56:280–288. doi: 10.7164/antibiotics.56.280. [DOI] [PubMed] [Google Scholar]
- 654.Sawada J, Namiki S, Onodera M, Omura S, Tanaka I. J Antibiot. 1974;27:639–641. doi: 10.7164/antibiotics.27.639. [DOI] [PubMed] [Google Scholar]
- 655.McAlpine JB, Tuan JS, Brown DP, Grebner KD, Whittern DN, Buko A, Katz L. J Antibiot. 1987;40:1115–1122. doi: 10.7164/antibiotics.40.1115. [DOI] [PubMed] [Google Scholar]
- 656.Bartner P, Boxler DL, Brambilla R, Mallams AK, Morton JB, Reichert P, Sancilio FD, Surprenant H, Tomalesky G, Lukacs G, Olesker A, Thang TT, Valente L, Omura S. J Chem Soc, Perkin Trans 1. 1979:1600–1624. [Google Scholar]
- 657.Martin JR, Egan RS, Goldstein AW, Mueller SL, Kellerschierlein W, Mitscher LA, Foltz RL. Helv Chim Acta. 1976;59:1886–1894. doi: 10.1002/hlca.19760590545. [DOI] [PubMed] [Google Scholar]
- 658.Arakawa K, Cao ZS, Suzuki N, Kinashi H. Tetrahedron. 2011;67:5199–5205. [Google Scholar]
- 659.Egan RS, Martin JR. J Am Chem Soc. 1970;92:4129–4130. doi: 10.1021/ja00716a062. [DOI] [PubMed] [Google Scholar]
- 660.Omura S, Muro T, Namiki S, Shibata M, Sawada J. J Antibiot. 1969;22:629–634. doi: 10.7164/antibiotics.22.629. [DOI] [PubMed] [Google Scholar]
- 661.Hernandez C, Olano C, Méndez C, Salas JA. Gene. 1993;134:139–140. doi: 10.1016/0378-1119(93)90189-a. [DOI] [PubMed] [Google Scholar]
- 662.Spagnoli R, Cappelletti L, Toscano L. J Antibiot. 1983;36:365–375. doi: 10.7164/antibiotics.36.365. [DOI] [PubMed] [Google Scholar]
- 663.Maezawa I, Kinumaki A, Suzuki M. J Antibiot. 1976;29:1203–1208. doi: 10.7164/antibiotics.29.1203. [DOI] [PubMed] [Google Scholar]
- 664.Sato E, Gomi S, Amano S, Hara O, Imai S, Koyama M, Nagaoka K. JP 05178881. Japanese Pat. 1993
- 665.Luesch H, Yoshida WY, Harrigan GG, Doom JP, Moore RE, Paul VJ. J Nat Prod. 2002;65:1945–1948. doi: 10.1021/np0202879. [DOI] [PubMed] [Google Scholar]
- 666.Tan LT, Marquez BL, Gerwick WH. J Nat Prod. 2002;65:925–928. doi: 10.1021/np010526c. [DOI] [PubMed] [Google Scholar]
- 667.Martin JR, Egan RS, Goldstein AW, Collum P. Tetrahedron. 1975;31:1985–1989. [Google Scholar]
- 668.Mikami Y, Yazawa K, Nemoto A, Komaki H, Tanaka Y, Grafe U. J Antibiot. 1999;52:201–202. doi: 10.7164/antibiotics.52.201. [DOI] [PubMed] [Google Scholar]
- 669.Anzai Y, Sakai A, Li W, Iizaka Y, Koike K, Kinoshita K, Kato F. J Antibiot. 2010;63:325–328. doi: 10.1038/ja.2010.38. [DOI] [PubMed] [Google Scholar]
- 670.Hayashi M, Kinoshita K, Sudate Y, Satoi S, Sakakibara H, Harada K, Suzuki M. J Antibiot. 1983;36:175–178. doi: 10.7164/antibiotics.36.175. [DOI] [PubMed] [Google Scholar]
- 671.Hayashi M, Ohno M, Katsumata S, Satoi S, Harada K, Takeda M, Suzuki M. J Antibiot. 1981;34:276–281. doi: 10.7164/antibiotics.34.276. [DOI] [PubMed] [Google Scholar]
- 672.Hayashi M, Ohno M, Kinoshita K, Satoi S, Suzuki M, Harada K. J Antibiot. 1981;34:346–349. doi: 10.7164/antibiotics.34.346. [DOI] [PubMed] [Google Scholar]
- 673.Kinoshita K, Imura Y, Takenaka S, Hayashi M. J Antibiot. 1989;42:1869–1872. doi: 10.7164/antibiotics.42.1869. [DOI] [PubMed] [Google Scholar]
- 674.Kinoshita K, Satoi S, Hayashi M, Harada K, Suzuki M, Nakatsu K. J Antibiot. 1985;38:522–526. doi: 10.7164/antibiotics.38.522. [DOI] [PubMed] [Google Scholar]
- 675.Kinoshita K, Satoi S, Hayashi M, Nakatsu K. J Antibiot. 1989;42:1003–1005. doi: 10.7164/antibiotics.42.1003. [DOI] [PubMed] [Google Scholar]
- 676.Kinoshita K, Takenaka S, Hayashi M. J Antibiot. 1991;44:1270–1273. doi: 10.7164/antibiotics.44.1270. [DOI] [PubMed] [Google Scholar]
- 677.Kinoshita K, Takenaka S, Suzuki H, Morohoshi T, Hayashi M. J Antibiot. 1992;45:1–9. doi: 10.7164/antibiotics.45.1. [DOI] [PubMed] [Google Scholar]
- 678.Satoi S, Muto N, Hayashi M, Fujii T, Otani M. J Antibiot. 1980;33:364–376. doi: 10.7164/antibiotics.33.364. [DOI] [PubMed] [Google Scholar]
- 679.Kishi T, Harada S, Yamana H, Miyake A. J Antibiot. 1976;29:1171–1181. doi: 10.7164/antibiotics.29.1171. [DOI] [PubMed] [Google Scholar]
- 680.Puar MS, Schumacher D. J Antibiot. 1990;43:1497–1501. doi: 10.7164/antibiotics.43.1497. [DOI] [PubMed] [Google Scholar]
- 681.Wang J, Zhang H, Ying L, Wang C, Jiang N, Zhou Y, Wang H, Bai H. J Antibiot. 2009;62:483–487. doi: 10.1038/ja.2009.55. [DOI] [PubMed] [Google Scholar]
- 682.Asolkar RN, Maskey RP, Helmke E, Laatsch H. J Antibiot. 2002;55:893–898. doi: 10.7164/antibiotics.55.893. [DOI] [PubMed] [Google Scholar]
- 683.Kim SD, Ryoo IJ, Kim CJ, Kim WG, Kim JP, Kong JY, Koshino H, Uramoto M, Yoo ID. J Antibiot. 1996;49:955–957. doi: 10.7164/antibiotics.49.955. [DOI] [PubMed] [Google Scholar]
- 684.Graziani EI, Overk CR, Carter GT. J Nat Prod. 2003;66:1149–1153. doi: 10.1021/np0301691. [DOI] [PubMed] [Google Scholar]
- 685.Chan YA, Boyne MT, Podevels AM, Klimowicz AK, Handelsman J, Kelleher NL, Thomas MG. Proc Natl Acad Sci U S A. 2006;103:14349–14354. doi: 10.1073/pnas.0603748103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 686.Brimacombe JS, Minshall J, Smith CW. J Chem Soc, Perkin Trans 1. 1975:682–686. [PubMed] [Google Scholar]
- 687.Park JS, Yang HO, Kwon HC. J Antibiot. 2009;62:171–175. doi: 10.1038/ja.2009.6. [DOI] [PubMed] [Google Scholar]
- 688.Achenbach H, Karl W. Chem Ber/Recl. 1975;108:759–771. [Google Scholar]
- 689.Achenbach H, Karl W. Chem Ber/Recl. 1975;108:780–789. [Google Scholar]
- 690.Ellestad GA, Kunstman MP, Lancaste JE, Mitscher LA, Morton G. Tetrahedron. 1967;23:3893–3902. doi: 10.1016/s0040-4020(01)97899-8. [DOI] [PubMed] [Google Scholar]
- 691.Mizobuchi S, Mochizuki J, Soga H, Tanba H, Inoue H. J Antibiot. 1986;39:1776–1778. doi: 10.7164/antibiotics.39.1776. [DOI] [PubMed] [Google Scholar]
- 692.Chatterjee S, Reddy GC, Franco CM, Rupp RH, Ganguli BN, Fehlhaber HW, Kogler H. J Antibiot. 1987;40:1368–1374. doi: 10.7164/antibiotics.40.1368. [DOI] [PubMed] [Google Scholar]
- 693.Franco CM, Gandhi JN, Chatterjee S, Ganguli BN. J Antibiot. 1987;40:1361–1367. doi: 10.7164/antibiotics.40.1361. [DOI] [PubMed] [Google Scholar]
- 694.Kirst HA, Wild GM, Baltz RH, Seno ET, Hamill RL, Paschal JW, Dorman DE. J Antibiot. 1983;36:376–382. doi: 10.7164/antibiotics.36.376. [DOI] [PubMed] [Google Scholar]
- 695.Lotvin J, Puar MS, Patel M, Lee BK, Schumacher D, Waitz JA. J Antibiot. 1982;35:1407–1408. doi: 10.7164/antibiotics.35.1407. [DOI] [PubMed] [Google Scholar]
- 696.Furumai T, Takeda K, Suzuki M. J Antibiot. 1975;28:789–797. doi: 10.7164/antibiotics.28.789. [DOI] [PubMed] [Google Scholar]
- 697.Sawada Y, Tsuno T, Miyaki T, Naito T, Oki T. J Antibiot. 1989;42:242–253. doi: 10.7164/antibiotics.42.242. [DOI] [PubMed] [Google Scholar]
- 698.Nash SM, Koch KF, Hoehn MM. US4252898 A. US Pat. 1981
- 699.Omura S, Sadakane N, Tanaka Y, Matsubara H. J Antibiot. 1983;36:927–930. doi: 10.7164/antibiotics.36.927. [DOI] [PubMed] [Google Scholar]
- 700.Nakahama K, Kishi T, Igarasi S. J Antibiot. 1974;27:433–441. doi: 10.7164/antibiotics.27.433. [DOI] [PubMed] [Google Scholar]
- 701.Vezina C, Bolduc C, Kudelski A, Audet P. Antimicrob Agents Chemother. 1979;15:738–746. doi: 10.1128/aac.15.5.738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 702.Okamoto R, Nomura H, Tsuchiya M, Tsunekawa H, Fukumoto T, Inui T, Sawa T, Takeuchi T, Umezawa H. J Antibiot. 1979;32:542–544. doi: 10.7164/antibiotics.32.542. [DOI] [PubMed] [Google Scholar]
- 703.Kiyoshima K, Takada K, Yamamoto M, Kubo K, Okamoto R, Fukagawa Y, Ishikura T, Naganawa H, Sawa T, Takeuchi T, et al. J Antibiot. 1987;40:1123–1130. doi: 10.7164/antibiotics.40.1123. [DOI] [PubMed] [Google Scholar]
- 704.Shimauchi Y, Kubo K, Osumi K, Okamura K, Fukagawa Y, Ishikura T, Lein J. J Antibiot. 1978;31:270–275. doi: 10.7164/antibiotics.31.270. [DOI] [PubMed] [Google Scholar]
- 705.Shimauchi Y, Okamura K, Koki A, Hasegawa M, Date M, Fukagawa Y, Kouno K, Ishikura T. J Antibiot. 1978;31:261–269. doi: 10.7164/antibiotics.31.261. [DOI] [PubMed] [Google Scholar]
- 706.Shimauchi Y, Kubo K, Osumi K, Okamura K, Fukagawa Y, Ishikura T, Lein J. J Antibiot. 1979;32:878–883. doi: 10.7164/antibiotics.32.878. [DOI] [PubMed] [Google Scholar]
- 707.Chenhang S, Wei J, Wenzao J, Yiguang W. Chin J Antibiot. 2000;25:1–4. [Google Scholar]
- 708.Wei J, Cheng-hang S, Wen-zao J. Chin J Antibiot. 2003;28:385–387. [Google Scholar]
- 709.Wei J, Cheng-hang S, Wen-zao J. Chin J Antibiot. 2002;27:209–213. [Google Scholar]
- 710.Yafei L, Chenhang S, Wenzao J. Chin J Antibiot. 1997;22:328–333. [Google Scholar]
- 711.Omura S, Nakagawa A. J Antibiot. 1975;28:401–433. doi: 10.7164/antibiotics.28.401. [DOI] [PubMed] [Google Scholar]
- 712.Baltz RH, Seno ET, Stonesifer J, Wild GM. J Antibiot. 1983;36:131–141. doi: 10.7164/antibiotics.36.131. [DOI] [PubMed] [Google Scholar]
- 713.Okamoto R, Kiyoshima K, Yamamoto M, Takada K, Ohnuki T, Ishikura T, Naganawa H, Tatsuta K, Takeuchi T, Umezawa H. J Antibiot. 1982;35:921–924. doi: 10.7164/antibiotics.35.921. [DOI] [PubMed] [Google Scholar]
- 714.Meyer M, Kellerschierlein W, Drautz H, Blank W, Zahner H. Helv Chim Acta. 1985;68:83–94. [Google Scholar]
- 715.Isogai A, Sakuda S, Matsumoto S, Ogura M, Furihata K, Seto H, Suzuki A. Agric Biol Chem. 1984;48:1379–1381. [Google Scholar]
- 716.Igarashi M, Kinoshita N, Ikeda T, Nakagawa E, Hamada M, Takeuchi T. J Antibiot. 1997;50:926–931. doi: 10.7164/antibiotics.50.926. [DOI] [PubMed] [Google Scholar]
- 717.Igarashi M, Nakamura H, Naganawa H, Takeuchi T. J Antibiot. 1997;50:932–936. doi: 10.7164/antibiotics.50.932. [DOI] [PubMed] [Google Scholar]
- 718.Burg RW, Miller BM, Baker EE, Birnbaum J, Currie SA, Hartman R, Kong YL, Monaghan RL, Olson G, Putter I, Tunac JB, Wallick H, Stapley EO, Oiwa R, Omura S. Antimicrob Agents Chemother. 1979;15:361–367. doi: 10.1128/aac.15.3.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 719.Chabala JC, Mrozik H, Tolman RL, Eskola P, Lusi A, Peterson LH, Woods MF, Fisher MH, Campbell WC, Egerton JR, Ostlind DA. J Med Chem. 1980;23:1134–1136. doi: 10.1021/jm00184a014. [DOI] [PubMed] [Google Scholar]
- 720.Siddique S, Syed Q, Adnan A, Qureshi FA. Jundishapur J Microbiol. 2014;7:e8626. doi: 10.5812/jjm.8626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 721.Zhang CS, Albermann C, Fu X, Thorson JS. J Am Chem Soc. 2006;128:16420–16421. doi: 10.1021/ja065950k. [DOI] [PubMed] [Google Scholar]
- 722.Poehlsgaard J, Douthwaite S. Nat Rev Microbiol. 2005;3:870–881. doi: 10.1038/nrmicro1265. [DOI] [PubMed] [Google Scholar]
- 723.Kannan K, Mankin AS. Ann N Y Acad Sci. 2011;1241:33–47. doi: 10.1111/j.1749-6632.2011.06315.x. [DOI] [PubMed] [Google Scholar]
- 724.Schlunzen F, Zarivach R, Harms J, Bashan A, Tocilj A, Albrecht R, Yonath A, Franceschi F. Nature. 2001;413:814–821. doi: 10.1038/35101544. [DOI] [PubMed] [Google Scholar]
- 725.LeTourneau N, Vimal P, Klepacki D, Mankin A, Melman A. Bioorg Med Chem Lett. 2012;22:4575–4578. doi: 10.1016/j.bmcl.2012.05.110. [DOI] [PubMed] [Google Scholar]
- 726.Chen X, Xu P, Xu Y, Liu L, Liu Y, Zhu D, Lei P. Bioorg Med Chem Lett. 2012;22:7402–7405. doi: 10.1016/j.bmcl.2012.10.064. [DOI] [PubMed] [Google Scholar]
- 727.Omura S, Namiki S, Shibata M, Muro T, Machida S. J Antibiot. 1971;24:717–718. doi: 10.7164/antibiotics.24.717. [DOI] [PubMed] [Google Scholar]
- 728.Morisaki N, Hashimoto Y, Furihata K, Yazawa K, Tamura M, Mikami Y. J Antibiot. 2001;54:157–165. doi: 10.7164/antibiotics.54.157. [DOI] [PubMed] [Google Scholar]
- 729.Jenkins G, Cundliffe E. Gene. 1991;108:55–62. doi: 10.1016/0378-1119(91)90487-v. [DOI] [PubMed] [Google Scholar]
- 730.Quiros LM, Carbajo RJ, Brana AF, Salas JA. J Biol Chem. 2000;275:11713–11720. doi: 10.1074/jbc.275.16.11713. [DOI] [PubMed] [Google Scholar]
- 731.Bolam DN, Roberts S, Proctor MR, Turkenburg JP, Dodson EJ, Martinez-Fleites C, Yang M, Davis BG, Davies GJ, Gilbert HJ. Proc Natl Acad Sci U S A. 2007;104:5336–5341. doi: 10.1073/pnas.0607897104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 732.Gantt RW, Goff RD, Williams GJ, Thorson JS. Angew Chem, Int Ed. 2008;47:8889–8892. doi: 10.1002/anie.200803508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 733.Kirillov SV, Porse BT, Garrett RA. RNA. 1999;5:1003–1013. doi: 10.1017/s1355838299990568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 734.Petropoulos AD, Kouvela EC, Dinos GP, Kalpaxis DL. J Biol Chem. 2008;283:4756–4765. doi: 10.1074/jbc.M708371200. [DOI] [PubMed] [Google Scholar]
- 735.Hansen JL, Ippolito JA, Ban N, Nissen P, Moore PB, Steitz TA. Mol Cell. 2002;10:117–128. doi: 10.1016/s1097-2765(02)00570-1. [DOI] [PubMed] [Google Scholar]
- 736.Alarcon B, Gonzalez ME, Carrasco L. FEBS Lett. 1988;231:207–211. doi: 10.1016/0014-5793(88)80732-4. [DOI] [PubMed] [Google Scholar]
- 737.Blanchard S, Thorson JS. Curr Opin Chem Biol. 2006;10:263–271. doi: 10.1016/j.cbpa.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 738.Cory TJ, Birket SE, Murphy BS, Hayes D, Jr, Anstead MI, Kanga JF, Kuhn RJ, Bush HM, Feola DJ. J Cystic Fibrosis. 2014;13:164–171. doi: 10.1016/j.jcf.2013.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 739.Kovaleva A, Remmelts HH, Rijkers GT, Hoepelman AI, Biesma DH, Oosterheert JJ. J Antimicrob Chemother. 2012;67:530–540. doi: 10.1093/jac/dkr520. [DOI] [PubMed] [Google Scholar]
- 740.Amsden GW. J Antimicrob Chemother. 2005;55:10–21. doi: 10.1093/jac/dkh519. [DOI] [PubMed] [Google Scholar]
- 741.Hofle GH, Bedorf N, Steinmetz H, Schomburg D, Gerth K, Reichenbach H. Angew Chem, Int Ed. 1996;35:1567–1569. [Google Scholar]
- 742.Altmann K-H, Prantz K, Mulzer J, Müller R, Höfle G. The Epothilones: An Outstanding Family of Anti-Tumor Agents: From Soil to the Clinic, Fortschr. Chem Org Naturst, Progress In The Chemistry Of Organic Natural Products. 2009 [PubMed] [Google Scholar]
- 743.Yoshimori T, Yamamoto A, Moriyama Y, Futai M, Tashiro Y. J Biol Chem. 1991;266:17707–17712. [PubMed] [Google Scholar]
- 744.Harada M, Shakado S, Sakisaka S, Tamaki S, Ohishi M, Sasatomi K, Koga H, Sata M, Tanikawa K. Liver. 1997;17:244–250. doi: 10.1111/j.1600-0676.1997.tb01025.x. [DOI] [PubMed] [Google Scholar]
- 745.Egerton JR, Ostlind DA, Blair LS, Eary CH, Suhayda D, Cifelli S, Riek RF, Campbell WC. Antimicrob Agents Chemother. 1979;15:372–378. doi: 10.1128/aac.15.3.372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 746.Cavalleri B, Arnone A, Di Modugno E, Nasini G, Goldstein BP. J Antibiot. 1988;41:308–315. doi: 10.7164/antibiotics.41.308. [DOI] [PubMed] [Google Scholar]
- 747.Hochlowski JE, Swanson SJ, Ranfranz LM, Whittern DN, Buko AM, McAlpine JB. J Antibiot. 1987;40:575–588. doi: 10.7164/antibiotics.40.575. [DOI] [PubMed] [Google Scholar]
- 748.Ishii T, Hida T, Iinuma S, Muroi M, Nozaki Y. J Antibiot. 1995;48:12–20. doi: 10.7164/antibiotics.48.12. [DOI] [PubMed] [Google Scholar]
- 749.Teruya T, Sasaki H, Kitamura K, Nakayama T, Suenaga K. Org Lett. 2009;11:2421–2424. doi: 10.1021/ol900579k. [DOI] [PubMed] [Google Scholar]
- 750.Brufani M, Cellai L, Musu C, Keller-Schierlein W. Helv Chim Acta. 1972;55:2329–2346. doi: 10.1002/hlca.19720550706. [DOI] [PubMed] [Google Scholar]
- 751.Omura S, Tanaka Y, Nakagawa A, Iwai Y, Inoue M, Tanaka H. J Antibiot. 1982;35:256–257. doi: 10.7164/antibiotics.35.256. [DOI] [PubMed] [Google Scholar]
- 752.Shaaban KA, Singh S, Elshahawi SI, Wang X, Ponomareva LV, Sunkara M, Copley GC, Hower JC, Morris AJ, Kharel MK, Thorson JS. J Antibiot. 2014;67:223–230. doi: 10.1038/ja.2013.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 753.Murakami R, Tomikawa T, Shin-ya K, Shinozaki J, Kajiura T, Seto H, Hayakawa Y. J Antibiot. 2001;54:714–717. doi: 10.7164/antibiotics.54.714. [DOI] [PubMed] [Google Scholar]
- 754.Kim JW, Adachi H, Shin-ya K, Hayakawa Y, Seto H. J Antibiot. 1997;50:628–630. doi: 10.7164/antibiotics.50.628. [DOI] [PubMed] [Google Scholar]
- 755.Salomon AR, Voehringer DW, Herzenberg LA, Khosla C. Proc Natl Acad Sci U S A. 2000;97:14766–14771. doi: 10.1073/pnas.97.26.14766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 756.Salomon AR, Zhang YB, Seto H, Khosla C. Org Lett. 2001;3:57–59. doi: 10.1021/ol006767d. [DOI] [PubMed] [Google Scholar]
- 757.Fourati-Ben Fguira L, Smaoui S, Karray-Rebai I, Bejar S, Mellouli L. Biotechnol J. 2008;3:1058–1066. doi: 10.1002/biot.200700155. [DOI] [PubMed] [Google Scholar]
- 758.Perlin DS, Latchney LR, Senior AE. Biochim Biophys Acta. 1985;807:238–244. doi: 10.1016/0005-2728(85)90254-3. [DOI] [PubMed] [Google Scholar]
- 759.Omura S, Nakagawa A, Imamura N, Kushida K, Liu CM, Sello LH, Westley JW. J Antibiot. 1985;38:674–676. doi: 10.7164/antibiotics.38.674. [DOI] [PubMed] [Google Scholar]
- 760.Smith RJ, Williams DH, Barna JCJ, Mcdermott IR, Haegele KD, Piriou F, Wagner J, Higgins W. J Am Chem Soc. 1985;107:2849–2857. [Google Scholar]
- 761.Parmeggiani A, Krab IM, Okamura S, Nielsen RC, Nyborg J, Nissen P. Biochemistry. 2006;45:6846–6857. doi: 10.1021/bi0525122. [DOI] [PubMed] [Google Scholar]
- 762.Zheng CJ, Lee S, Lee CH, Kim WG. J Nat Prod. 2007;70:1632–1635. doi: 10.1021/np0701327. [DOI] [PubMed] [Google Scholar]
- 763.Horstmann N, Essig S, Bockelmann S, Wieczorek H, Huss M, Sasse F, Menche D. J Nat Prod. 2011;74:1100–1105. doi: 10.1021/np200036v. [DOI] [PubMed] [Google Scholar]
- 764.Pathirana C, Tapiolas D, Jensen PR, Dwight R, Fenical W. Tetrahedron Lett. 1991;32:2323–2326. [Google Scholar]
- 765.Nakakoshi M, Kimura K, Nakajima N, Yoshihama M, Uramoto M. J Antibiot. 1999;52:175–177. doi: 10.7164/antibiotics.52.175. [DOI] [PubMed] [Google Scholar]
- 766.Antonicek H-P, Gladbach B, Bischoff E, Gondol D, Gutbrod O, Krahn T, Rodriguez M-L, Schütz H. US 6291515 B1. US Pat. 2001
- 767.Okujo N, Iinuma H, George A, Eim KS, Li TL, Ting NS, Jye TC, Hotta K, Hatsu M, Fukagawa Y, Shibahara S, Numata K, Kondo S. J Antibiot. 2007;60:216–219. doi: 10.1038/ja.2007.26. [DOI] [PubMed] [Google Scholar]
- 768.Arcamone F, Barbieri W, Francesc G, Penco S, Vigevani A. J Am Chem Soc. 1973;95:2008–2009. doi: 10.1021/ja00787a048. [DOI] [PubMed] [Google Scholar]
- 769.Kihara T, Koshino H, Shin YC, Yamaguchi I, Isono K. J Antibiot. 1995;48:1385–1387. doi: 10.7164/antibiotics.48.1385. [DOI] [PubMed] [Google Scholar]
- 770.Wienrich BG, Krahn T, Schon M, Rodriguez ML, Kramer B, Busemann M, Boehncke WH, Schon MP. J Invest Dermatol. 2006;126:882–889. doi: 10.1038/sj.jid.5700159. [DOI] [PubMed] [Google Scholar]
- 771.Komatsu K, Tsuda M, Tanaka Y, Mikami Y, Kobayashi J. J Org Chem. 2004;69:1535–1541. doi: 10.1021/jo035773v. [DOI] [PubMed] [Google Scholar]
- 772.Tanaka Y, Komaki H, Yazawa K, Mikami Y, Nemoto A, Tojyo T, Kadowaki K, Shigemori H, Kobayashi J. J Antibiot. 1997;50:1036–1041. doi: 10.7164/antibiotics.50.1036. [DOI] [PubMed] [Google Scholar]
- 773.Mikami Y, Komaki H, Imai T, Yazawa K, Nemoto A, Tanaka Y, Graefe U. J Antibiot. 2000;53:70–74. doi: 10.7164/antibiotics.53.70. [DOI] [PubMed] [Google Scholar]
- 774.Harada K, Kimura I, Sakazaki T, Murata H, Suzuki M. J Antibiot. 1989;42:1056–1062. doi: 10.7164/antibiotics.42.1056. [DOI] [PubMed] [Google Scholar]
- 775.Sasaki T, Furihata K, Shimazu A, Seto H, Iwata M, Watanabe T, Otake N. J Antibiot. 1986;39:502–509. doi: 10.7164/antibiotics.39.502. [DOI] [PubMed] [Google Scholar]
- 776.Serizawa N, Suga S, Nakajima M, Nishizaki T, Kizuka M. WO 2000032604. Japenese Pat. 2000
- 777.Laureti L, Song L, Huang S, Corre C, Leblond P, Challis GL, Aigle B. Proc Natl Acad Sci U S A. 2011;108:6258–6263. doi: 10.1073/pnas.1019077108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 778.Mukhopadhyay T, Vijayakumar EK, Nadkarni SR, Fehlhaber HW, Kogler H, Petry S. J Antibiot. 1998;51:582–585. doi: 10.7164/antibiotics.51.582. [DOI] [PubMed] [Google Scholar]
- 779.Horvath G, Batta G, Bognar R, Sztaricskai F. Acta Chim Hung. 1988;125:767–770. [Google Scholar]
- 780.Dinya Z, Sztaricskai F, Horvath G. Rapid Commun Mass Spectrom. 1991;5:534–537. doi: 10.1002/rcm.1290051110. [DOI] [PubMed] [Google Scholar]
- 781.Mayer M, Thiericke R. J Chem Soc, Perkin Trans 1. 1993:2525–2531. [Google Scholar]
- 782.Ivanova V. Prep Biochem Biotechnol. 1997;27:19–38. doi: 10.1080/10826069708001275. [DOI] [PubMed] [Google Scholar]
- 783.Kuo MS, Yurek DA, Laborde AL, Truesdell SE, Nielsen JW, Argoudelis AD, Baczynskyj L. J Antibiot. 1990;43:438–440. doi: 10.7164/antibiotics.43.438. [DOI] [PubMed] [Google Scholar]
- 784.Jansen R, Irschik H, Reichenbach H, Hofle G. Liebigs Ann/Recl. 1997:1725–1732. [Google Scholar]
- 785.Janssen D, Albert D, Jansen R, Muller R, Kalesse M. Angew Chem, Int Ed. 2007;46:4898–4901. doi: 10.1002/anie.200605198. [DOI] [PubMed] [Google Scholar]
- 786.Diestel R, Irschik H, Jansen R, Khalil MW, Reichenbach H, Sasse F. ChemBioChem. 2009;10:2900–2903. doi: 10.1002/cbic.200900562. [DOI] [PubMed] [Google Scholar]
- 787.Wang YH, Zhang JP, Chang Y, Hu CQ. J Antibiot. 2010;63:553–557. doi: 10.1038/ja.2010.80. [DOI] [PubMed] [Google Scholar]
- 788.Pawlak J, Sowinski P, Borowski E, Gariboldi P. J Antibiot. 1993;46:1598–1604. doi: 10.7164/antibiotics.46.1598. [DOI] [PubMed] [Google Scholar]
- 789.Martin JF, McDaniel LE. J Antibiot. 1974;27:610–619. doi: 10.7164/antibiotics.27.610. [DOI] [PubMed] [Google Scholar]
- 790.Cybulska B, Ziminski T, Borowski E, Garybobo CM. Mol Pharmacol. 1983;24:270–276. [PubMed] [Google Scholar]
- 791.Wagman GH, Testa RT, Patel M, Marquez JA, Oden EM, Waitz JA, Weinstein MJ. Antimicrob Agents Chemother. 1975;7:457–461. doi: 10.1128/aac.7.4.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 792.Zielinski J, Jereczek E, Sowinski P, Falkowski L, Rudowski A, Borowski E. J Antibiot. 1979;32:565–568. doi: 10.7164/antibiotics.32.565. [DOI] [PubMed] [Google Scholar]
- 793.Jansen R, Irschik H, Reichenbach H, Wray V, Hofle G. Liebigs Ann Chem. 1989:213–222. [Google Scholar]
- 794.Pawlak J, Nakanishi K, Iwashita T, Borowski E. J Org Chem. 1987;52:2896–2901. [Google Scholar]
- 795.Mechlinski W, Schaffner CP. J Antibiot. 1980;33:591–599. doi: 10.7164/antibiotics.33.591. [DOI] [PubMed] [Google Scholar]
- 796.Croatt MP, Carreira EM. Org Lett. 2011;13:1390–1393. doi: 10.1021/ol2000765. [DOI] [PubMed] [Google Scholar]
- 797.Wilcock BC, Endo MM, Uno BE, Burke MD. J Am Chem Soc. 2013;135:8488–8491. doi: 10.1021/ja403255s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 798.Zhang CS, Moretti R, Jiang JQ, Thorson JS. ChemBioChem. 2008;9:2506–2514. doi: 10.1002/cbic.200800349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 799.Lu CH, Bai LQ, Shen YM. J Antibiot. 2004;57:348–350. doi: 10.7164/antibiotics.57.348. [DOI] [PubMed] [Google Scholar]
- 800.Snipes CE, Duebelbeis DO, Olson M, Hahn DR, Dent WH, Gilbert JR, Werk TL, Davis GE, Lee-Lu R, Graupner PR. J Nat Prod. 2007;70:1578–1581. doi: 10.1021/np070275t. [DOI] [PubMed] [Google Scholar]
- 801.Sugita M, Hiramoto S, Ando C, Sasaki T, Furihata K, Seto H, Otake N. J Antibiot. 1985;38:799–802. doi: 10.7164/antibiotics.38.799. [DOI] [PubMed] [Google Scholar]
- 802.Ayers S, Zink DL, Powell JS, Brown CM, Grund A, Genilloud O, Salazar O, Thompson D, Singh SB. J Antibiot. 2008;61:59–62. doi: 10.1038/ja.2008.110. [DOI] [PubMed] [Google Scholar]
- 803.Naruse N, Konishi M, Oki T, Inouye Y, Kakisawa H. J Antibiot. 1991;44:756–761. doi: 10.7164/antibiotics.44.756. [DOI] [PubMed] [Google Scholar]
- 804.Cooper R, Truumees I, Yarborough R, Loebenberg D, Marquez J, Horan A, Patel M, Gullo V, Puar M, Pramanik B. J Antibiot. 1992;45:633–638. doi: 10.7164/antibiotics.45.633. [DOI] [PubMed] [Google Scholar]
- 805.Hegde VR, Patel MG, Horan AC, King AH, Gentile F, Puar MS, Loebenberg D. J Antibiot. 1998;51:464–470. doi: 10.7164/antibiotics.51.464. [DOI] [PubMed] [Google Scholar]
- 806.Komoda T, Akasaka K, Hirota A. Biosci, Biotechnol, Biochem. 2008;72:2392–2397. doi: 10.1271/bbb.80282. [DOI] [PubMed] [Google Scholar]
- 807.Yazawa H, Imai H, Suzuki K, Kadota S, Saito T. US 4912215. US Pat. 1990
- 808.Matsushima Y, Nakayama T, Fujita M, Bhandari R, Eguchi T, Shindo K, Kakinuma K. J Antibiot. 2001;54:211–219. doi: 10.7164/antibiotics.54.211. [DOI] [PubMed] [Google Scholar]
- 809.Igarashi M, Tsuchida T, Kinoshita N, Kamijima M, Sawa R, Sawa T, Naganawa H, Hamada M, Takeuchi T, Yamazaki K, Ishizuka M. J Antibiot. 1998;51:123–129. doi: 10.7164/antibiotics.51.123. [DOI] [PubMed] [Google Scholar]
- 810.Takaishi M, Kudo F, Eguchi T. Tetrahedron. 2008;64:6651–6656. [Google Scholar]
- 811.Naruse N, Tenmyo O, Kawano K, Tomita K, Ohgusa N, Miyaki T, Konishi M, Oki T. J Antibiot. 1991;44:733–740. doi: 10.7164/antibiotics.44.733. [DOI] [PubMed] [Google Scholar]
- 812.Futamura Y, Sawa R, Umezawa Y, Igarashi M, Nakamura H, Hasegawa K, Yarnasaki M, Tashiro E, Takahashi Y, Akarnatsu Y, Imoto M. J Am Chem Soc. 2008;130:1822–1823. doi: 10.1021/ja710124p. [DOI] [PubMed] [Google Scholar]
- 813.Minami A, Eguchi T. J Am Chem Soc. 2007;129:5102–5107. doi: 10.1021/ja0685250. [DOI] [PubMed] [Google Scholar]
- 814.Minami A, Kakinuma K, Eguchi T. Tetrahedron Lett. 2005;46:6187–6190. [Google Scholar]
- 815.Yamamoto I, Nakagawa M, Hayakawa Y, Adachi K, Kobayashi E. J Antibiot. 1987;40:1452–1454. doi: 10.7164/antibiotics.40.1452. [DOI] [PubMed] [Google Scholar]
- 816.Kawashima A, Nakamura Y, Ohta Y, Akama T, Yamagishi M, Hanada K. J Antibiot. 1992;45:207–212. doi: 10.7164/antibiotics.45.207. [DOI] [PubMed] [Google Scholar]
- 817.Yamamoto K, Hayakawa Y. JP 63203695 A. Japanese Pat. 1988
- 818.Matsumoto M, Kawamura Y, Yoshimura Y, Terui Y, Nakai H, Yoshida T, Shoji J. J Antibiot. 1990;43:739–747. doi: 10.7164/antibiotics.43.739. [DOI] [PubMed] [Google Scholar]
- 819.Schroeder DR, Lam KS, Hesler GA, Gustavson DR, Tomita K, Berry RL. US 5082933 A. US Pat. 1992
- 820.Momose I, Hirosawa S, Nakamura H, Naganawa H, Iinuma H, Ikeda D, Takeuchi T. J Antibiot. 1999;52:787–796. doi: 10.7164/antibiotics.52.787. [DOI] [PubMed] [Google Scholar]
- 821.Tomita F, Tamaoki T, Shirahata K, Kasai M, Morimoto M, Ohkubo S, Mineura K, Ishii S. J Antibiot. 1980;33:668–670. doi: 10.7164/antibiotics.33.668. [DOI] [PubMed] [Google Scholar]
- 822.Shimotohno KW, Endo T, Furihata K. J Antibiot. 1993;46:682–686. doi: 10.7164/antibiotics.46.682. [DOI] [PubMed] [Google Scholar]
- 823.Igarashi Y, Takagi K, Kan Y, Fujii K, Harada K, Furumai T, Oki T. J Antibiot. 2000;53:233–240. doi: 10.7164/antibiotics.53.233. [DOI] [PubMed] [Google Scholar]
- 824.Wei RB, Xi T, Li J, Wang P, Li FC, Lin YC, Qin S. Mar Drugs. 2011;9:359–368. doi: 10.3390/md9030359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 825.Niu S, Li S, Chen Y, Tian X, Zhang H, Zhang G, Zhang W, Yang X, Zhang S, Ju J, Zhang C. J Antibiot. 2011;64:711–716. doi: 10.1038/ja.2011.78. [DOI] [PubMed] [Google Scholar]
- 826.Mallams AK, Puar MS, Rossman RR, Mcphail AT, Macfarlane RD. J Am Chem Soc. 1981;103:3940–3943. [Google Scholar]
- 827.Waitz JA, Horan AC, Kalyanpur M, Lee BK, Loebenberg D, Marquez JA, Miller G, Patel MG. J Antibiot. 1981;34:1101–1106. doi: 10.7164/antibiotics.34.1101. [DOI] [PubMed] [Google Scholar]
- 828.Fusao T, Tatsuya T, Kimikatsu S, Masaji K, Takemitsu A, Shinzou I. JP 57171997 A. Japanese Pat. 1982
- 829.Ashton RJ, Kenig MD, Luk K, Planterose DN, Scottwood G. J Antibiot. 1990;43:1387–1393. doi: 10.7164/antibiotics.43.1387. [DOI] [PubMed] [Google Scholar]
- 830.Park HR, Chijiwa S, Furihata K, Hayakawa Y, Shin-Ya K. Org Lett. 2007;9:1457–1460. doi: 10.1021/ol070042t. [DOI] [PubMed] [Google Scholar]
- 831.Ueda JY, Chijiwa S, Takagi M, Shin-Ya K. J Antibiot. 2008;61:752–755. doi: 10.1038/ja.2008.89. [DOI] [PubMed] [Google Scholar]
- 832.Zhao P, Ueda JY, Kozone I, Chijiwa S, Takagi M, Kudo F, Nishiyama M, Shin-ya K, Kuzuyama T. Org Biomol Chem. 2009;7:1454–1460. doi: 10.1039/b817312e. [DOI] [PubMed] [Google Scholar]
- 833.Tamaoki T, Kasai M, Shirahata K, Tomita F. J Antibiot. 1982;35:979–984. doi: 10.7164/antibiotics.35.979. [DOI] [PubMed] [Google Scholar]
- 834.Kirst HA, Michel KH, Martin JW, Creemer LC, Chio EH, Yao RC, Nakatsukasa WM, Boeck L, Occolowitz JL, Paschal JW, Deeter JB, Jones ND, Thompson GD. Tetrahedron Lett. 1991;32:4839–4842. [Google Scholar]
- 835.Sparks TC, Thompson GD, Kirst HA, Hertlein MB, Larson LL, Worden TV, Thibault ST. J Econ Entomol. 1998;91:1277–1283. [Google Scholar]
- 836.Winn M, Goss RJM, Kimura K-i, Bugg TDH. Nat Prod Rep. 2010;27:279–304. doi: 10.1039/b816215h. [DOI] [PubMed] [Google Scholar]
- 837.Eckardt K, Thrum H, Bradler G, Tonew E, Tonew M. J Antibiot. 1975;28:274–279. doi: 10.7164/antibiotics.28.274. [DOI] [PubMed] [Google Scholar]
- 838.Eckardt K. J Nat Prod. 1983;46:544–550. doi: 10.1021/np50028a020. [DOI] [PubMed] [Google Scholar]
- 839.Takatsuki A, Arima K, Tamura G. J Antibiot. 1971;24:215–223. doi: 10.7164/antibiotics.24.215. [DOI] [PubMed] [Google Scholar]
- 840.Vogel P, Petterson DS, Berry PH, Frahn JL, Anderton N, Cockrum PA, Edgar JA, Jago MV, Lanigan GW, Payne AL, Culvenor CC. Aust J Exp Biol Med Sci. 1981;59:455–467. doi: 10.1038/icb.1981.39. [DOI] [PubMed] [Google Scholar]
- 841.Wyszynski FJ, Lee SS, Yabe T, Wang H, Gomez-Escribano JP, Bibb MJ, Lee SJ, Davies GJ, Davis BG. Nat Chem. 2012;4:539–546. doi: 10.1038/nchem.1351. [DOI] [PubMed] [Google Scholar]
- 842.Heifetz A, Keenan RW, Elbein AD. Biochemistry. 1979;18:2186–2192. doi: 10.1021/bi00578a008. [DOI] [PubMed] [Google Scholar]
- 843.Isono F, Sakaida Y, Takahashi S, Kinoshita T, Nakamura T, Inukai M. J Antibiot. 1993;46:1203–1207. doi: 10.7164/antibiotics.46.1203. [DOI] [PubMed] [Google Scholar]
- 844.Isono F, Inukai M, Takahashi S, Haneishi T, Kinoshita T, Kuwano H. J Antibiot. 1989;42:667–673. doi: 10.7164/antibiotics.42.667. [DOI] [PubMed] [Google Scholar]
- 845.Chatterjee S, Nadkarni SR, Vijayakumar EK, Patel MV, Ganguli BN, Fehlhaber HW, Vertesy L. J Antibiot. 1994;47:595–598. doi: 10.7164/antibiotics.47.595. [DOI] [PubMed] [Google Scholar]
- 846.Chen RH, Buko AM, Whittern DN, McAlpine JB. J Antibiot. 1989;42:512–520. doi: 10.7164/antibiotics.42.512. [DOI] [PubMed] [Google Scholar]
- 847.Fronko RM, Lee JC, Galazzo JG, Chamberland S, Malouin F, Lee MD. J Antibiot. 2000;53:1405–1410. doi: 10.7164/antibiotics.53.1405. [DOI] [PubMed] [Google Scholar]
- 848.Xie Y, Chen R, Si S, Sun C, Xu H. J Antibiot. 2007;60:158–161. doi: 10.1038/ja.2007.16. [DOI] [PubMed] [Google Scholar]
- 849.Xie Y, Xu H, Si S, Sun C, Chen R. J Antibiot. 2008;61:237–240. doi: 10.1038/ja.2008.34. [DOI] [PubMed] [Google Scholar]
- 850.Xie Y, Xu H, Sun C, Yu Y, Chen R. J Antibiot. 2010;63:143–146. doi: 10.1038/ja.2010.6. [DOI] [PubMed] [Google Scholar]
- 851.Lemoine RC, Magon A, Hecker SJ. Bioorg Med Chem Lett. 2002;12:1121–1123. doi: 10.1016/s0960-894x(02)00100-2. [DOI] [PubMed] [Google Scholar]
- 852.McDonald LA, Barbieri LR, Carter GT, Lenoy E, Lotvin J, Petersen PJ, Siegel MM, Singh G, Williamson RT. J Am Chem Soc. 2002;124:10260–10261. doi: 10.1021/ja017748h. [DOI] [PubMed] [Google Scholar]
- 853.Norton EB, Yamashita A. WO2002086139 A1. Japenese Pat. 2002
- 854.Lin Y-I, Li Z, Francisco GD, McDonald LA, Davis RA, Singh G, Yang Y, Mansour TS. Bioorg Med Chem Lett. 2002;12:2341–2344. doi: 10.1016/s0960-894x(02)00469-9. [DOI] [PubMed] [Google Scholar]
- 855.Yamashita A, Norton E, Petersen PJ, Rasmussen BA, Singh G, Yang Y, Mansour TS, Ho DM. Bioorg Med Chem Lett. 2003;13:3345–3350. doi: 10.1016/s0960-894x(03)00671-1. [DOI] [PubMed] [Google Scholar]
- 856.Ohnuki T, Muramatsu Y, Miyakoshi S, Takatsu T, Inukai M. J Antibiot. 2003;56:268–279. doi: 10.7164/antibiotics.56.268. [DOI] [PubMed] [Google Scholar]
- 857.Yamaguchi H, Sato S, Yoshida S, Takada K, Itoh M, Seto H, Otake N. J Antibiot. 1986;39:1047–1053. doi: 10.7164/antibiotics.39.1047. [DOI] [PubMed] [Google Scholar]
- 858.Muramatsu Y, Muramatsu A, Ohnuki T, Ishii MM, Kizuka M, Enokita R, Tsutsumi S, Arai M, Ogawa Y, Suzuki T, Takatsu T, Inukai M. J Antibiot. 2003;56:243–252. doi: 10.7164/antibiotics.56.243. [DOI] [PubMed] [Google Scholar]
- 859.Muramatsu Y, Miyakoshi S, Ogawa Y, Ohnuki T, Ishii MM, Arai M, Takatsu T, Inukai M. J Antibiot. 2003;56:259–267. doi: 10.7164/antibiotics.56.259. [DOI] [PubMed] [Google Scholar]
- 860.Muramatsu Y, Ohnuki T, Ishii MM, Kizuka M, Enokita R, Miyakoshi S, Takatsu T, Inukai M. J Antibiot. 2004;57:639–646. doi: 10.7164/antibiotics.57.639. [DOI] [PubMed] [Google Scholar]
- 861.Murakami R, Fujita Y, Kizuka M, Kagawa T, Muramatsu Y, Miyakoshi S, Takatsu T, Inukai M. J Antibiot. 2007;60:690–695. doi: 10.1038/ja.2007.88. [DOI] [PubMed] [Google Scholar]
- 862.Hotoda H, Daigo M, Furukawa M, Murayama K, Hasegawa CA, Kaneko M, Muramatsu Y, Ishii MM, Miyakoshi S-i, Takatsu T, Inukai M, Kakuta M, Abe T, Fukuoka T, Utsui Y, Ohya S. Bioorg Med Chem Lett. 2003;13:2833–2836. doi: 10.1016/s0960-894x(03)00597-3. [DOI] [PubMed] [Google Scholar]
- 863.Isono K, Uramoto M, Kusakabe H, Kimura K, Isaki K, Nelson CC, McCloskey JA. J Antibiot. 1985;38:1617–1621. doi: 10.7164/antibiotics.38.1617. [DOI] [PubMed] [Google Scholar]
- 864.Kimura K, Ikeda Y, Kagami S, Yoshihama M, Ubukata M, Esumi Y, Osada H, Isono K. J Antibiot. 1998;51:647–654. doi: 10.7164/antibiotics.51.647. [DOI] [PubMed] [Google Scholar]
- 865.Ubukata M, Isono K, Kimura K, Nelson CC, McCloskey JA. J Am Chem Soc. 1988;110:4416–4417. [Google Scholar]
- 866.Funabashi M, Baba S, Nonaka K, Hosobuchi M, Fujita Y, Shibata T, Van Lanen SG. ChemBioChem. 2010;11:184–190. doi: 10.1002/cbic.200900665. [DOI] [PubMed] [Google Scholar]
- 867.Fujita Y, Kizuka M, Funabashi M, Ogawa Y, Ishikawa T, Nonaka K, Takatsu T. J Antibiot. 2011;64:495–501. doi: 10.1038/ja.2011.38. [DOI] [PubMed] [Google Scholar]
- 868.Kimura K, Ikeda Y, Kagami S, Yoshihama M, Suzuki K, Osada H, Isono K. J Antibiot. 1998;51:1099–1104. doi: 10.7164/antibiotics.51.1099. [DOI] [PubMed] [Google Scholar]
- 869.Sakata K, Sakurai A, Tamura S. Agric Biol Chem. 1973;37:697–699. [Google Scholar]
- 870.Sakata K, Sakurai A, Tamura S. Tetrahedron Lett. 1974;15:1533–1536. [Google Scholar]
- 871.Sakata K, Sakurai A, Tamura S. Agric Biol Chem. 1974;38:1883–1890. [Google Scholar]
- 872.Sakata K, Uzawa J. Agric Biol Chem. 1977;41:413–415. [Google Scholar]
- 873.Sakata K, Sakurai A, Tamura S. Agric Biol Chem. 1977;41:2027–2032. [Google Scholar]
- 874.Sakata K, Uzawa J, Sakurai A. Org Magn Reson. 1977;10:230–234. [Google Scholar]
- 875.Sakata K, Sakurai A, Tamura S. Agric Biol Chem. 1977;41:2033–2039. [Google Scholar]
- 876.Sakata K, Sakurai A, Tamura S. Agric Biol Chem. 1976;40:1993–1999. [Google Scholar]
- 877.Sakata K, Sakurai A, Tamura S. Tetrahedron Lett. 1975;16:3191–3194. [Google Scholar]
- 878.Delzer J, Fiedler HP, Müller H, Zähner H, Rathmann R, Ernst K, König WA. J Antibiot. 1984;37:80–82. doi: 10.7164/antibiotics.37.80. [DOI] [PubMed] [Google Scholar]
- 879.Bormann C, Huhn W, Zähner H, Rathmann R, Hahn H, König WA. J Antibiot. 1985;38:9–16. doi: 10.7164/antibiotics.38.9. [DOI] [PubMed] [Google Scholar]
- 880.Decker H, Bormann C, Fiedler HP, Zähner H, Heitsch H, König WA. J Antibiot. 1989;42:230–235. doi: 10.7164/antibiotics.42.230. [DOI] [PubMed] [Google Scholar]
- 881.Decker H, Walz F, Bormann C, Zähner H, Fiedler HP, Heitsch H, König WA. J Antibiot. 1990;43:43–48. doi: 10.7164/antibiotics.43.43. [DOI] [PubMed] [Google Scholar]
- 882.Schüz TC, Fiedler HP, Zähner H, Rieck M, Konig WA. J Antibiot. 1992;45:199–206. doi: 10.7164/antibiotics.45.199. [DOI] [PubMed] [Google Scholar]
- 883.Obi K, Uda J, Iwase K, Sugimoto O, Ebisu H, Matsuda A. Bioorg Med Chem Lett. 2000;10:1451–1454. doi: 10.1016/s0960-894x(00)00256-0. [DOI] [PubMed] [Google Scholar]
- 884.Isono K, Suzuki S. Heterocycles. 1979;13:333–351. [Google Scholar]
- 885.Uramoto M, Kobinata K, Isono K, Jenkins EE, McCloskey JA, Higashijima T, Miyazawa T. Nucleic Acids Symp Ser. 1980:s69–s71. [PubMed] [Google Scholar]
- 886.Isono K, Crain PF, McCloskey JA. J Am Chem Soc. 1975;97:943–945. [Google Scholar]
- 887.Endo A, Kakiki K, Misato T. J Bacteriol. 1970;104:189–196. doi: 10.1128/jb.104.1.189-196.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 888.Hanessian S, Marcotte S, Machaalani R, Huang G. Org Lett. 2003;5:4277–4280. doi: 10.1021/ol030095k. [DOI] [PubMed] [Google Scholar]
- 889.Benner JP, Boehlendorf BGH, Kipps MR, Lambert NEP, Luck R, Molleyres L-P, Neff S, Schuez TC, Stanley PD. WO2003062242 A1. WIPO Pat. 2003
- 890.Li W, Csukai M, Corran A, Crowley P, Solomon PS, Oliver RP. Pest Manage Sci. 2008;64:1294–1302. doi: 10.1002/ps.1632. [DOI] [PubMed] [Google Scholar]
- 891.Fujita Y, Murakami R, Muramatsu Y, Miyakoshi S, Takatsu T. J Antibiot. 2008;61:545–549. doi: 10.1038/ja.2008.72. [DOI] [PubMed] [Google Scholar]
- 892.Murakami R, Fujita Y, Kizuka M, Kagawa T, Muramatsu Y, Miyakoshi S, Takatsu T, Inukai M. J Antibiot. 2008;61:537–544. doi: 10.1038/ja.2008.71. [DOI] [PubMed] [Google Scholar]
- 893.Hinuma Y, Kuroya M, Yajima T, Ishihara K, Hamada S, Watanabe K, Kikuchi K. J Antibiot. 1955;8:148–152. [PubMed] [Google Scholar]
- 894.Konishi M, Naruishi M, Tsuno T, Tsukiura H, Kawaguchi H. J Antibiot. 1973;26:757–764. doi: 10.7164/antibiotics.26.757. [DOI] [PubMed] [Google Scholar]
- 895.Konstantinova NV, Fedorova GB, Borisova VN, Shapovalova SP, Malkova IV. Antibiotiki. 1982;27:403–409. [PubMed] [Google Scholar]
- 896.Evans JR, Weare G. J Antibiot. 1977;30:604–606. doi: 10.7164/antibiotics.30.604. [DOI] [PubMed] [Google Scholar]
- 897.Itoh J, Miyadoh S. J Antibiot. 1992;45:846–853. doi: 10.7164/antibiotics.45.846. [DOI] [PubMed] [Google Scholar]
- 898.Shiomi K, Haneda K, Tomoda H, Iwai Y, Omura S. J Antibiot. 1994;47:782–786. doi: 10.7164/antibiotics.47.782. [DOI] [PubMed] [Google Scholar]
- 899.Smith JL, Sundaralingam M. Acta Crystallogr, Sect B: Struct Crystallogr Cryst Chem. 1981:1095–1101. [Google Scholar]
- 900.Leviev IG, Rodriguez-Fonseca C, Phan H, Garrett RA, Heilek G, Noller HF, Mankin AS. EMBO J. 1994;13:1682–1686. doi: 10.1002/j.1460-2075.1994.tb06432.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 901.Alarcón B, Lacal JC, Fernández-Sousa J, Carrasco L. Antiviral Res. 1984;4:231–244. doi: 10.1016/0166-3542(84)90029-9. [DOI] [PubMed] [Google Scholar]
- 902.Haneda K, Shinose M, Seino A, Tabata N, Tomoda H, Iwai Y, Omura S. J Antibiot. 1994;47:774–781. doi: 10.7164/antibiotics.47.774. [DOI] [PubMed] [Google Scholar]
- 903.Harada S, Kishi T. J Antibiot. 1978;31:519–524. doi: 10.7164/antibiotics.31.519. [DOI] [PubMed] [Google Scholar]
- 904.Takashi S, Hidekazu S, Kazuyoshi K. EP0024866 A2. Eur Pat. 1981
- 905.Harada S, Mizuta E, Kishi T. Tetrahedron. 1981;37:1317–1327. [Google Scholar]
- 906.Feduchi E, Cosín M, Carrasco L. J Antibiot. 1985;38:415–419. doi: 10.7164/antibiotics.38.415. [DOI] [PubMed] [Google Scholar]
- 907.Argoudelis AD, Baczynskyj L, Kuo MT, Laborde AL, Sebek OK, Truesdell SE, Shilliday FB. J Antibiot. 1987;40:750–760. doi: 10.7164/antibiotics.40.750. [DOI] [PubMed] [Google Scholar]
- 908.Argoudelis AD, Laborde AL, Sebek OK, Truesdell SE. WO 8605785 A1. US Pat. 1986
- 909.Takeuchi S, Hirayama K, Ueda K, Sakai H, Yonehara H. J Antibiot. 1958;11:1–5. [PubMed] [Google Scholar]
- 910.Seto H, Furihata K, Yonehara H. J Antibiot. 1976;29:595–596. doi: 10.7164/antibiotics.29.595. [DOI] [PubMed] [Google Scholar]
- 911.Seto H, Yonehara H. J Antibiot. 1977;30:1019–1021. doi: 10.7164/antibiotics.30.1019. [DOI] [PubMed] [Google Scholar]
- 912.Larsen SH, Berry DM, Paschal JW, Gilliam JM. J Antibiot. 1989;42:470–471. doi: 10.7164/antibiotics.42.470. [DOI] [PubMed] [Google Scholar]
- 913.Gould SJ, Guo J, Geitmann A, Dejesus K. Can J Chem. 1994;72:6–11. [Google Scholar]
- 914.Seto H. Agric Biol Chem. 1973;37:2415–2419. [Google Scholar]
- 915.Seto H, Otake N, Yonehara H. Tetrahedron Lett. 1972;13:3991–3994. [Google Scholar]
- 916.Das BC, Defaye J, Uchida K. Carbohydr Res. 1972;22:293–299. doi: 10.1016/s0008-6215(00)81279-3. [DOI] [PubMed] [Google Scholar]
- 917.Uchida K, Das BC. Biochimie. 1973;55:635–636. doi: 10.1016/s0300-9084(73)80425-0. [DOI] [PubMed] [Google Scholar]
- 918.Vuilhorgne M, Ennifar S, Das BC, Paschal JW, Nagarajan R, Hagaman EW, Wenkert E. J Org Chem. 1977;42:3289–3291. doi: 10.1021/jo00440a019. [DOI] [PubMed] [Google Scholar]
- 919.Uchida K, Wolf H. J Antibiot. 1974;27:783–787. doi: 10.7164/antibiotics.27.783. [DOI] [PubMed] [Google Scholar]
- 920.Takahashi A, Ikeda D, Naganawa H, Okami Y, Umezawa H. J Antibiot. 1986;39:1041–1046. doi: 10.7164/antibiotics.39.1041. [DOI] [PubMed] [Google Scholar]
- 921.Bei M, Wu Y, Du C, Liu Q. CN 1428429A. Chinese Pat. 2003
- 922.Iwasaki H. Yakugaku Zasshi. 1962;82:1393–1395. doi: 10.1248/yakushi1947.82.10_1387. [DOI] [PubMed] [Google Scholar]
- 923.Xiang G, Houzhi H, Chen J, Chen W. Weishengwu Xuebao. 1995;35:368–374. [Google Scholar]
- 924.Turkova J, Mikes O, Sorum F. Experientia. 1963;19:633–634. doi: 10.1007/BF02151289. [DOI] [PubMed] [Google Scholar]
- 925.Benz G, Schroder T, Kurz J, Wunsche C, Karl W, Steffens G, Pfitzner J, Schmidt D. Angew Chem, Int Ed. 1982;21:527–528. [Google Scholar]
- 926.Seto H, Otake N, Koyama M, Ogino H, Kodama Y, Nishizawa N, Tsuruoka T, Inouye S. Tetrahedron Lett. 1983;24:495–498. [Google Scholar]
- 927.Nishizawa N, Kondo Y, Koyama M, Omoto S, Iwata M, Tsuruoka T, Inouye S. J Antibiot. 1984;37:1–5. doi: 10.7164/antibiotics.37.1. [DOI] [PubMed] [Google Scholar]
- 928.Fryth PW, Waller CW, Hutchings BL, Williams JH. J Am Chem Soc. 1958;80:2736–2740. [Google Scholar]
- 929.Kirst HA, Dorman DE, Occolowitz JL, Jones ND, Paschal JW, Hamill RL, Szymanski EF. J Antibiot. 1985;38:575–586. doi: 10.7164/antibiotics.38.575. [DOI] [PubMed] [Google Scholar]
- 930.Kirst HA. US4205164 A. US Pat. 1980
- 931.Hamil RL, Hoehn MM. J Antibiot. 1973;26:463–465. doi: 10.7164/antibiotics.26.463. [DOI] [PubMed] [Google Scholar]
- 932.Berry DR, Abbott BJ. J Antibiot. 1978;31:185–191. doi: 10.7164/antibiotics.31.185. [DOI] [PubMed] [Google Scholar]
- 933.Niitsuma M, Hashida J, Iwatsuki M, Mori M, Ishiyama A, Namatame M, Nishihara-Tsukashima A, Matsumoto A, Takahashi Y, Yamada H, Otoguro K, Shiomi K, Omura S. J Antibiot. 2010;63:673–679. doi: 10.1038/ja.2010.102. [DOI] [PubMed] [Google Scholar]
- 934.Vedel M, Lawrence F, Robert-Gero M, Lederer E. Biochem Biophys Res Commun. 1978;85:371–376. doi: 10.1016/s0006-291x(78)80052-7. [DOI] [PubMed] [Google Scholar]
- 935.Haneishi T, Terahara A, Kayamori H, Yabe J, Arai M. J Antibiot. 1976;29:870–875. doi: 10.7164/antibiotics.29.870. [DOI] [PubMed] [Google Scholar]
- 936.Takiguchi Y, Yoshikawa H, Terahara A, Torikata A, Terao M. J Antibiot. 1979;32:857–861. doi: 10.7164/antibiotics.32.857. [DOI] [PubMed] [Google Scholar]
- 937.Takiguchi Y, Yoshikawa H, Terahara A, Torikata A, Terao M. J Antibiot. 1979;32:862–867. doi: 10.7164/antibiotics.32.862. [DOI] [PubMed] [Google Scholar]
- 938.Terahara A, Haneishi T, Arai M, Hata T, Kuwano H, Tamura C. J Antibiot. 1982;35:1711–1714. doi: 10.7164/antibiotics.35.1711. [DOI] [PubMed] [Google Scholar]
- 939.Kizuka M, Enokita R, Takahashi K, Okamoto Y, Otsuka T, Shigematsu Y, Inoue Y, Okazaki T. Actinomycetologica. 1998;12:89–91. [Google Scholar]
- 940.Acton EM, Ryan KJ, Luetzow AE. J Med Chem. 1977;20:1362–1371. doi: 10.1021/jm00221a002. [DOI] [PubMed] [Google Scholar]
- 941.Sakai T, Shindo K, Odagawa A, Suzuki A, Kawai H, Kobayashi K, Hayakawa Y, Seto H, Otake N. J Antibiot. 1995;48:899–900. doi: 10.7164/antibiotics.48.899. [DOI] [PubMed] [Google Scholar]
- 942.Igarashi Y, Ootsu K, Onaka H, Fujita T, Uehara Y, Furumai T. J Antibiot. 2005;58:322–326. doi: 10.1038/ja.2005.40. [DOI] [PubMed] [Google Scholar]
- 943.Roberts WP, Tate ME, Kerr A. Nature. 1977;265:379–381. doi: 10.1038/265379a0. [DOI] [PubMed] [Google Scholar]
- 944.Iwasa T, Harada S, Sato Y. Takeda Kenkyushoho. 1981;40:12–26. [Google Scholar]
- 945.Prystaš M, Kalvoda L, Šorm F. Collect Czech Chem Commun. 1975;40:1775–1785. [Google Scholar]
- 946.Farkaš J, Šebesta K, Horská K, Samek Z, Dolejš L, Šorm F. Collect Czech Chem Commun. 1977;42:909–929. [Google Scholar]
- 947.Takahashi S, Nakagawa F, Kawazoe K, Furukawa Y, Sato S, Tamura C, Naito A. J Antibiot. 1985;38:830–834. doi: 10.7164/antibiotics.38.830. [DOI] [PubMed] [Google Scholar]
- 948.Takahashi S, Nakagawa F, Sato S. J Antibiot. 1988;41:705–706. doi: 10.7164/antibiotics.41.705. [DOI] [PubMed] [Google Scholar]
- 949.Iijima Y, Nakagawa F, Handa S, Oda T, Naito A, Yamazaki M. FEBS Lett. 1985;192:179–183. doi: 10.1016/0014-5793(85)80103-4. [DOI] [PubMed] [Google Scholar]
- 950.Katoh H, Yamada M, Lida K, Aoki M, Itezono Y, Nakayama N, Suzuki Y, Watanabe M, Shimada H. Tennen Yuki Kagobutsu Toronkai Koen Yoshishu. 1996;38:115–120. [Google Scholar]
- 951.Seto H, Koyama M, Ogino H, Tsuruoka T, Inouye S, Otake N. Tetrahedron Lett. 1983;24:1805–1808. [Google Scholar]
- 952.Harada S, Kishi T. J Antibiot. 1977;30:11–16. doi: 10.7164/antibiotics.30.11. [DOI] [PubMed] [Google Scholar]
- 953.Goto T, Toya Y, Kondo T. Nucleic Acids Symp Ser. 1980:s73–s74. [PubMed] [Google Scholar]
- 954.McCranie EK, Bachmann BO. Nat Prod Rep. 2014;31:1026–1042. doi: 10.1039/c3np70128j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 955.Belova L, Tenson T, Xiong L, McNicholas PM, Mankin AS. Proc Natl Acad Sci U S A. 2001;98:3726–3731. doi: 10.1073/pnas.071527498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 956.McNicholas PM, Najarian DJ, Mann PA, Hesk D, Hare RS, Shaw KJ, Black TA. Antimicrob Agents Chemother. 2000;44:1121–1126. doi: 10.1128/aac.44.5.1121-1126.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 957.Belanger AE, Shryock TR. J Mass Spectrom. 2005;40:1109. doi: 10.1002/jms.889. [DOI] [PubMed] [Google Scholar]
- 958.Wright DE. Tetrahedron. 1979;35:1207–1237. [Google Scholar]
- 959.Mertz JL, Peloso JS, Barker BJ, Babbitt GE, Occolowitz JL, Simson VL, Kline RM. J Antibiot. 1986;39:877–887. doi: 10.7164/antibiotics.39.877. [DOI] [PubMed] [Google Scholar]
- 960.Weitnauer G, Muhlenweg A, Trefzer A, Bechthold A. US20040006026 A1. US Pat. 2004
- 961.Weitnauer G, Muhlenweg A, Trefzer A, Hoffmeister D, Sussmuth RD, Jung G, Welzel K, Vente A, Girreser U, Bechthold A. Chem Biol. 2001;8:569–581. doi: 10.1016/s1074-5521(01)00040-0. [DOI] [PubMed] [Google Scholar]
- 962.Weitnauer G, Hauser G, Hofmann C, Linder U, Boll R, Pelz K, Glaser SJ, Bechthold A. Chem Biol. 2004;11:1403–1411. doi: 10.1016/j.chembiol.2004.08.016. [DOI] [PubMed] [Google Scholar]
- 963.Treede I, Hauser G, Mühlenweg A, Hofmann C, Schmidt M, Weitnauer G, Glaser S, Bechthold A. Appl Environ Microbiol. 2005;71:400–406. doi: 10.1128/AEM.71.1.400-406.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 964.Hofmann C, Boll R, Heitmann B, Hauser G, Durr C, Frerich A, Weitnauer G, Glaser SJ, Bechthold A. Chem Biol. 2005;12:1137–1143. doi: 10.1016/j.chembiol.2005.08.016. [DOI] [PubMed] [Google Scholar]
- 965.Bartner P, Pramanik BN, Saksena AK, Liu YH, Das PR, Sarre O, Ganguly AK. J Am Soc Mass Spectr. 1997;8:1134–1140. [Google Scholar]
- 966.Ganguly AK, Sarre OZ, Greeves D, Morton J. J Am Chem Soc. 1975;97:1982–1985. doi: 10.1021/ja00840a078. [DOI] [PubMed] [Google Scholar]
- 967.Ganguly AK, Szmulewicz S, Sarre OZ, Girijavallabhan VM. J Chem Soc, Chem Commun. 1976:609–611. [Google Scholar]
- 968.Sanders WE, Jr, Sanders CC. Antimicrob Agents Chemother. 1974;6:232–238. doi: 10.1128/aac.6.3.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 969.Ganguly AK. J Antibiot. 2000;53:1038–1044. doi: 10.7164/antibiotics.53.1038. [DOI] [PubMed] [Google Scholar]
- 970.Chu M, Mierzwa R, Jenkins J, Chan T-M, Das P, Pramanik B, Patel M, Gullo V. J Nat Prod. 2002;65:1588–1593. doi: 10.1021/np020093t. [DOI] [PubMed] [Google Scholar]
- 971.Chu M, Mierzwa R, Patel M, Jenkins J, Das P, Pramanik B, Chan T-M. Tetrahedron Lett. 2000;41:6689–6693. [Google Scholar]
- 972.Ganguly AK, McCormick JL, Chan T-M, Saksena AK, Das PR. Tetrahedron Lett. 1997;38:7989–7992. [Google Scholar]
- 973.Tamura A, Furuta R, Kotani H. JP 50125094. Japanese Pat. 1975
- 974.Timmons SC, Thorson JS. Curr Opin Chem Biol. 2008;12:297–305. doi: 10.1016/j.cbpa.2008.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 975.Kondo SI, Sezaki M, Koike M, Shimura M, Akita E, Satoh K, Hara T. J Antibiot. 1965;18:38–42. [PubMed] [Google Scholar]
- 976.Kondo S, Iinuma K, Naganawa H, Shimura M, Sekizawa Y. J Antibiot. 1975;28:79–82. doi: 10.7164/antibiotics.28.79. [DOI] [PubMed] [Google Scholar]
- 977.Shimura M, Sekizawa Y, Iinuma K, Naganawa H, Kondo S. J Antibiot. 1975;28:83–84. doi: 10.7164/antibiotics.28.83. [DOI] [PubMed] [Google Scholar]
- 978.Ikeda Y, Kondo S, Kanai F, Sawa T, Hamada M, Takeuchi T, Umezawa H. J Antibiot. 1985;38:436–438. doi: 10.7164/antibiotics.38.436. [DOI] [PubMed] [Google Scholar]
- 979.Yoshihisa U, Manabu I, Hiroyuki K, Teruya N, Shingo M. JP 60002317. Japanese Pat. 1985
- 980.Inouye S, Shomura T, Watanabe H, Totsugawa K, Niida T. J Antibiot. 1973;26:374–385. doi: 10.7164/antibiotics.26.374. [DOI] [PubMed] [Google Scholar]
- 981.Umezawa H, Maeda K, Takeuchi T, Okami Y. J Antibiot. 1966;19:200–209. [PubMed] [Google Scholar]
- 982.Umezawa H, Suhara Y, Takita T, Maeda K. J Antibiot. 1966;19:210–215. [PubMed] [Google Scholar]
- 983.Omoto S, Takita T, Maeda K, Umezawa H, Umezawa S. J Antibiot. 1972;25:752–754. doi: 10.7164/antibiotics.25.752. [DOI] [PubMed] [Google Scholar]
- 984.Shomura T, Omoto S, Ohba K, Ogino H, Kojima M, Inouye S. J Antibiot. 1980;33:1243–1248. doi: 10.7164/antibiotics.33.1243. [DOI] [PubMed] [Google Scholar]
- 985.Chen J, Stubbe J. Nat Rev Cancer. 2005;5:102–112. doi: 10.1038/nrc1547. [DOI] [PubMed] [Google Scholar]
- 986.Hecht SM. J Nat Prod. 2000;63:158–168. doi: 10.1021/np990549f. [DOI] [PubMed] [Google Scholar]
- 987.Maeda K, Kosaka H, Yagishita K, Umezawa H. J Antibiot. 1956;9:82–85. [PubMed] [Google Scholar]
- 988.Ikekawa T, Iwami F, Hiranaka H, Umezawa H. J Antibiot. 1964;17:194–199. [PubMed] [Google Scholar]
- 989.Konishi M, Saito K, Numata K, Tsuno T, Asama K. J Antibiot. 1977;30:789–805. doi: 10.7164/antibiotics.30.789. [DOI] [PubMed] [Google Scholar]
- 990.Kawaguchi H, Konishi M, Miyaki T, Tenmyo O. US4246400 A. US Pat. 1981
- 991.Argoudelis AD, Bergy ME, Pyke TR. J Antibiot. 1971;24:543–557. doi: 10.7164/antibiotics.24.543. [DOI] [PubMed] [Google Scholar]
- 992.Wang L, Yun B-S, George NP, Wendt-Pienkowski E, Galm U, Oh T-J, Coughlin JM, Zhang G, Tao M, Shen B. J Nat Prod. 2007;70:402–406. doi: 10.1021/np060592k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 993.Umezawa H, Muraoka Y, Fujii A, Naganawa H, Takita T. J Antibiot. 1980;33:1079–1082. doi: 10.7164/antibiotics.33.1079. [DOI] [PubMed] [Google Scholar]
- 994.Fujii A, Kunishima M, Muraoka Y, Takita T, Umezawa H. US4326054 A. US Pat. 1982
- 995.Kirby JP, Fantini AA, Borders DB, Testa RT, Martin J. US4650765 A. US Pat. 1987
- 996.Takasawa S, Kawamoto I, Sato S, Yahashi R, Okachi R. J Antibiot. 1975;28:662–667. doi: 10.7164/antibiotics.28.662. [DOI] [PubMed] [Google Scholar]
- 997.Fujii A, Takita T, Maeda K, Umezawa H. J Antibiot. 1973;26:396–397. doi: 10.7164/antibiotics.26.396. [DOI] [PubMed] [Google Scholar]
- 998.Fujii A, Takita T, Shimada N, Umezawa H. J Antibiot. 1974;27:73–77. doi: 10.7164/antibiotics.27.73. [DOI] [PubMed] [Google Scholar]
- 999.Galm U, Wendt-Pienkowski E, Wang L, Huang S-X, Unsin C, Tao M, Coughlin JM, Shen B. J Nat Prod. 2011;74:526–536. doi: 10.1021/np1008152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1000.Huang S-X, Feng Z, Wang L, Galm U, Wendt-Pienkowski E, Yang D, Tao M, Coughlin JM, Duan Y, Shen B. J Am Chem Soc. 2012;134:13501–13509. doi: 10.1021/ja3056535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1001.Tounekti O, Kenani A, Foray N, Orlowski S, Mir LM. Br J Cancer. 2001;84:1272–1279. doi: 10.1054/bjoc.2001.1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1002.Chapuis J-C, Schmaltz RM, Tsosie KS, Belohlavek M, Hecht SM. J Am Chem Soc. 2009;131:2438–2439. doi: 10.1021/ja8091104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1003.Yu Z, Schmaltz RM, Bozeman TC, Paul R, Rishel MJ, Tsosie KS, Hecht SM. J Am Chem Soc. 2013;135:2883–2886. doi: 10.1021/ja311090e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1004.Wolter F, Schoof S, Süssmuth RD. In: Glycopeptides and Glycoproteins. Wittmann V, editor. Springer; Berlin Heidelberg: 2007. pp. 143–185. [Google Scholar]
- 1005.Kaneko I, Fearon DT, Austen KF. J Immunol. 1980;124:1194–1198. [PubMed] [Google Scholar]
- 1006.Kaneko I, Kamoshida K, Takahashi S. J Antibiot. 1989;42:236–241. doi: 10.7164/antibiotics.42.236. [DOI] [PubMed] [Google Scholar]
- 1007.Singh SB, Jayasuriya H, Salituro GM, Zink DL, Shafiee A, Heimbuch B, Silverman KC, Lingham RB, Genilloud O, Teran A, Vilella D, Felock P, Hazuda D. J Nat Prod. 2001;64:874–882. doi: 10.1021/np000632z. [DOI] [PubMed] [Google Scholar]
- 1008.Tanaka H, Matsuzaki K, Nakashima H, Ogino T, Matsumoto A, Ikeda H, Woodruff HB, Omura S. J Antibiot. 1997;50:58–65. doi: 10.7164/antibiotics.50.58. [DOI] [PubMed] [Google Scholar]
- 1009.Boeck LD, Mertz FP. J Antibiot. 1986;39:1533–1540. doi: 10.7164/antibiotics.39.1533. [DOI] [PubMed] [Google Scholar]
- 1010.McCormick MH, McGuire JM, Pittenger GE, Pittenger RC, Stark WM. Antibiot Annu. 1955;3:606–611. [PubMed] [Google Scholar]
- 1011.Harris CM, Harris TM. J Am Chem Soc. 1982;104:4293–4295. [Google Scholar]
- 1012.Schäfer M, Schneider TR, Sheldrick GM. Structure. 1996;4:509–1515. doi: 10.1016/s0969-2126(96)00156-6. [DOI] [PubMed] [Google Scholar]
- 1013.Kahne D, Leimkuhler C, Lu W, Walsh C. Chem Rev. 2005;105:425–448. doi: 10.1021/cr030103a. [DOI] [PubMed] [Google Scholar]
- 1014.Mackay JP, Gerhard U, Beauregard DA, Maplestone RA, Williams DH. J Am Chem Soc. 1994;116:4573–4580. [Google Scholar]
- 1015.Ashford P-A, Bew SP. Chem Soc Rev. 2012;41:957–978. doi: 10.1039/c1cs15125h. [DOI] [PubMed] [Google Scholar]
- 1016.Nagarajan R, Merkel KE, Michel KH, Higgins HM, Hoehn MM, Hunt AH, Jones ND, Occolowitz JL, Schabel AA, Swartzendruber JK. J Am Chem Soc. 1988;110:7896–7897. [Google Scholar]
- 1017.Boeck LD, Hoehn MM, Marconi GG. US4558008 A. US Pat. 1985
- 1018.Li S, Lindbeck AC, Nilius AM, Towne TB, Zhou CC, Paulus TJ. J Antibiot. 2004;57:691–694. doi: 10.7164/antibiotics.57.691. [DOI] [PubMed] [Google Scholar]
- 1019.Nadkarni SR, Patel MV, Chatterjee S, Vijayakumar EK, Desikan KR, Blumbach J, Ganguli BN, Limbert M. J Antibiot. 1994;47:334–341. doi: 10.7164/antibiotics.47.334. [DOI] [PubMed] [Google Scholar]
- 1020.Chatterjee S, Vijayakumar EKS, Nadkarni SR, Patel MV, Blumbach J, Ganguli BN, Fehlhaber HW, Kogler H, Vertesy L. J Org Chem. 1994;59:3480–3484. [Google Scholar]
- 1021.Vértesy L, Fehlhaber HW, Kogler H, Limbert M. J Antibiot. 1996;49:115–118. doi: 10.7164/antibiotics.49.115. [DOI] [PubMed] [Google Scholar]
- 1022.Gause GF, Brazhnikova MG, Lomakina NN, Berdnikova TF, Fedorova GB, Tokareva NL, Borisova VN, Batta GY. J Antibiot. 1989;42:1790–1797. doi: 10.7164/antibiotics.42.1790. [DOI] [PubMed] [Google Scholar]
- 1023.Berdnikova TF, Shashkov AS, Katrukha GS, Lapchinskaya OA, Yurkevich NV, Grachev AA, Nifant’ev NE. Bioorg Khim. 2009;35:497–503. doi: 10.1134/s1068162009040128. [DOI] [PubMed] [Google Scholar]
- 1024.Riva E, Gastaldo L, Beretta MG, Ferrari P, Zerilli LF, Cassani G, Selva E, Goldstein BP, Berti M, Parenti F. J Antibiot. 1989;42:497–505. doi: 10.7164/antibiotics.42.497. [DOI] [PubMed] [Google Scholar]
- 1025.Box SJ, Elson AL, Gilpin ML, Winstanley DJ. J Antibiot. 1990;43:931–937. doi: 10.7164/antibiotics.43.931. [DOI] [PubMed] [Google Scholar]
- 1026.Tsuji N, Kobayashi M, Kamigauchi T, Yoshimura Y, Terui Y. J Antibiot. 1988;41:819–822. doi: 10.7164/antibiotics.41.819. [DOI] [PubMed] [Google Scholar]
- 1027.Tsuji N, Kamigauchi T, Kobayashi M, Terui Y. J Antibiot. 1988;41:1506–1510. doi: 10.7164/antibiotics.41.1506. [DOI] [PubMed] [Google Scholar]
- 1028.Ang S-G, Williamson MP, Williams DH. J Chem Soc, Perkin Trans 1. 1988;7:1949–1956. [Google Scholar]
- 1029.Jeffs PW, Yellin B, Mueller L, Heald SL. J Org Chem. 1988;53:471–477. [Google Scholar]
- 1030.Fu X, Albermann C, Jiang J, Liao J, Zhang C, Thorson JS. Nat Biotechnol. 2003;21:1467–1469. doi: 10.1038/nbt909. [DOI] [PubMed] [Google Scholar]
- 1031.Fu X, Albermann C, Zhang C, Thorson JS. Org Lett. 2005;7:1513–1515. doi: 10.1021/ol0501626. [DOI] [PubMed] [Google Scholar]
- 1032.Peltier-Pain P, Marchillo K, Zhou M, Andes DR, Thorson JS. Org Lett. 2012;14:5086–5089. doi: 10.1021/ol3023374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1033.McGahren WJ, Leese RA, Barbatschi F, Morton GO, Kuck NA, Ellestad GA. J Antibiot. 1983;36:1671–1682. doi: 10.7164/antibiotics.36.1671. [DOI] [PubMed] [Google Scholar]
- 1034.Ellestad GA, Swenson W, McGahren WJ. J Antibiot. 1983;36:1683–1690. doi: 10.7164/antibiotics.36.1683. [DOI] [PubMed] [Google Scholar]
- 1035.Takeuchi M, Takahashi S, Enokita R, Sakaida Y, Haruyama H, Nakamura T, Katayama T, Inukai M. J Antibiot. 1992;45:297–305. doi: 10.7164/antibiotics.45.297. [DOI] [PubMed] [Google Scholar]
- 1036.Takeuchi M, Takahashi S, Inukai M, Nakamura T, Kinoshita T. J Antibiot. 1991;44:271–277. doi: 10.7164/antibiotics.44.271. [DOI] [PubMed] [Google Scholar]
- 1037.Takatsu T, Takahashi S, Nakajima M, Haneishi T, Nakamura T, Kuwano H, Kinoshita T. J Antibiot. 1987;40:933–940. doi: 10.7164/antibiotics.40.933. [DOI] [PubMed] [Google Scholar]
- 1038.Berdnikova TF, Lomakina NN, Potapova NP. Antibiotiki. 1982;27:252–258. [PubMed] [Google Scholar]
- 1039.Batta G, Sztaricskai F, Csanádi J, Komáromi I, Bognár R. J Antibiot. 1986;39:910–913. doi: 10.7164/antibiotics.39.910. [DOI] [PubMed] [Google Scholar]
- 1040.Dingerdissen JJ, Sitrin RD, DePhillips PA, Giovenella AJ, Grappel SF, Mehta RJ, Oh YK, Pan CH, Roberts GD, Shearer MC. J Antibiot. 1987;40:165–172. doi: 10.7164/antibiotics.40.165. [DOI] [PubMed] [Google Scholar]
- 1041.Heald SL, Mueller L, Jeffs PW. J Antibiot. 1987;40:630–645. doi: 10.7164/antibiotics.40.630. [DOI] [PubMed] [Google Scholar]
- 1042.Bager F, Madsen M, Christensen J, Aarestrup FM. Prev Vet Med. 1997;31:95–112. doi: 10.1016/s0167-5877(96)01119-1. [DOI] [PubMed] [Google Scholar]
- 1043.Fehlner JR, Hutchinson REJ, Tarbell DS, Schenck JR. Proc Natl Acad Sci U S A. 1972;69:2420–2421. doi: 10.1073/pnas.69.9.2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1044.Kalman JR, Williams DH. J Am Chem Soc. 1980;102:897–905. [Google Scholar]
- 1045.Harris CM, Harris TM. J Am Chem Soc. 1982;104:363–365. [Google Scholar]
- 1046.Pintér G, Bereczki I, Batta G, Otvös R, Sztaricskai F, Roth E, Ostorházi E, Rozgonyi F, Naesens L, Szarvas M, Boda Z, Herczegh P. Bioorg Med Chem Lett. 2010;20:2713–2717. doi: 10.1016/j.bmcl.2010.03.080. [DOI] [PubMed] [Google Scholar]
- 1047.Eggert JH, Michel KH. J Antibiot. 1986;39:792–799. doi: 10.7164/antibiotics.39.792. [DOI] [PubMed] [Google Scholar]
- 1048.Debono M, Merkel KE, Molloy RM, Barnhart M, Presti E, Hunt AH, Hamill RL. J Antibiot. 1984;37:85–95. doi: 10.7164/antibiotics.37.85. [DOI] [PubMed] [Google Scholar]
- 1049.Merkel KE. US4558036 A. US Pat. 1985
- 1050.Debono M. US4497802 A. US Pat. 1985
- 1051.Huber FM, Michel KH, Hunt AH, Martin JW, Molloy RM. J Antibiot. 1988;41:798–801. doi: 10.7164/antibiotics.41.798. [DOI] [PubMed] [Google Scholar]
- 1052.Skelton NJ, Williams DH, Monday RA, Ruddock JC. J Org Chem. 1990;55:3718–3723. [Google Scholar]
- 1053.Borghi A, Coronelli C, Faniuolo L, Allievi G, Pallanza R, Gallo GG. J Antibiot. 1984;37:615–620. doi: 10.7164/antibiotics.37.615. [DOI] [PubMed] [Google Scholar]
- 1054.Borghi A, Antonini P, Zanol M, Ferrari P, Zerilli LF, Lancini GC. J Antibiot. 1989;42:361–366. doi: 10.7164/antibiotics.42.361. [DOI] [PubMed] [Google Scholar]
- 1055.Quarta C, Borghi A, Zerilli LF, De Pietro MT, Ferrari P, Trani A, Lancini GC. J Antibiot. 1996;49:644–650. doi: 10.7164/antibiotics.49.644. [DOI] [PubMed] [Google Scholar]
- 1056.Lazzarini A, Borghi A, Zerilli LF, Ferrari P, Colombo L, Lancini GC. J Antibiot. 1997;50:180–183. doi: 10.7164/antibiotics.50.180. [DOI] [PubMed] [Google Scholar]
- 1057.Folena-Wasserman G, Poehland BL, Yeung EW, Staiger D, Killmer LB, Snader K, Dingerdissen JJ, Jeffs PW. J Antibiot. 1986;39:1395–1406. doi: 10.7164/antibiotics.39.1395. [DOI] [PubMed] [Google Scholar]
- 1058.Sitrin RD, Chan GW, Dingerdissen JJ, Holl W, Hoover JR, Valenta JR, Webb L, Snader KM. J Antibiot. 1985;38:561–571. doi: 10.7164/antibiotics.38.561. [DOI] [PubMed] [Google Scholar]
- 1059.Selva E, Gastaldo L, Beretta G, Borghi A, Goldstein BP, Arioli V, Cassani G, Parenti F. EP0177882 (A2) European Pat. 1986
- 1060.Beltrametti F, Lazzarini A, Brunati C, Selva E, Marinelli F. J Antibiot. 2003;56:310–313. doi: 10.7164/antibiotics.56.310. [DOI] [PubMed] [Google Scholar]
- 1061.Beltrametti F, Lazzarini A, Brunati C, Marazzi A, Jovetic S, Selva E, Marinelli F. J Antibiot. 2003;56:773–782. doi: 10.7164/antibiotics.56.773. [DOI] [PubMed] [Google Scholar]
- 1062.Box SJ, Coates NJ, Davis CJ, Gilpin ML, Houge-Frydrych CS, Milner PH. J Antibiot. 1991;44:807–813. doi: 10.7164/antibiotics.44.807. [DOI] [PubMed] [Google Scholar]
- 1063.Verma AK, Goel AK, Rao VA, Venkateswarlu A, Sitrin RD, Christensen SB. EP0255299 (A2) European Pat. 1988
- 1064.He H, Williamson RT, Shen B, Graziani EI, Yang HY, Sakya SM, Petersen PJ, Carter GT. J Am Chem Soc. 2002;124:9729–9736. doi: 10.1021/ja020257s. [DOI] [PubMed] [Google Scholar]
- 1065.Hantke K, Nicholson G, Rabsch W, Winkelmann G. Proc Natl Acad Sci U S A. 2003;100:3677–3682. doi: 10.1073/pnas.0737682100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1066.Bister B, Bischoff D, Nicholson GJ, Valdebenito M, Schneider K, Winkelmann G, Hantke K, Süssmuth RD. BioMetals. 2004;17:471–481. doi: 10.1023/b:biom.0000029432.69418.6a. [DOI] [PubMed] [Google Scholar]
- 1067.Vértesy L, Aretz W, Fehlhaber H-W, Kogler H. Helv Chim Acta. 1995;78:46–60. [Google Scholar]
- 1068.Braun V, Pramanik A, Gwinner T, Köberle M, Bohn E. BioMetals. 2009;22:3–13. doi: 10.1007/s10534-008-9199-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1069.Hoeksema H. J Am Chem Soc. 1968;90:755–757. doi: 10.1021/ja01005a036. [DOI] [PubMed] [Google Scholar]
- 1070.Toda S, Nakagawa S, Naito T, Kawaguchi H. J Antibiot. 1981;34:596–599. doi: 10.7164/antibiotics.34.596. [DOI] [PubMed] [Google Scholar]
- 1071.Ulanova D, Novotná J, Smutná Y, Kameník Z, Gazák R, Sulc M, Sedmera P, Kadlcík S, Plhácková K, Janata J. Antimicrob Agents Chemother. 2010;54:927–930. doi: 10.1128/AAC.00918-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1072.Mason DJ, Dietz A, DeBoer C. Antimicrob Agents Chemother. 1963;1962:554–559. [Google Scholar]
- 1073.Herr RR, Bergy ME. Antimicrob Agents Chemother. 1963;1962:560–564. [Google Scholar]
- 1074.Hoeksema H, Bannister B, Birkenmeyer RD, Kagan F, Magerlein BJ, MacKellar FA, Schroeder W, Slomp G, Herr RR. J Am Chem Soc. 1964;86:4223–4224. [Google Scholar]
- 1075.Magerlein BJ. Adv Appl Microbiol. 1971;14:185–229. doi: 10.1016/s0065-2164(08)70544-6. [DOI] [PubMed] [Google Scholar]
- 1076.Pospísil S, Sedmera P, Halada P, Spizek J. Folia Microbiol. 2001;46:376–378. doi: 10.1007/BF02814424. [DOI] [PubMed] [Google Scholar]
- 1077.Argoudelis AD, Mason DJ. Biochemistry. 1965;4:704–709. doi: 10.1021/bi00880a015. [DOI] [PubMed] [Google Scholar]
- 1078.Argoudelis AD, Mason DJ. J Antibiot. 1969;22:289–291. [PubMed] [Google Scholar]
- 1079.Argoudelis AD, Eble TE, Mason DJ. J Antibiot. 1970;23:1–8. doi: 10.7164/antibiotics.23.1. [DOI] [PubMed] [Google Scholar]
- 1080.Argoudelis AD, Coats JH, Magerlein BJ. J Antibiot. 1972;25:191–193. doi: 10.7164/antibiotics.25.191. [DOI] [PubMed] [Google Scholar]
- 1081.Argoudelis AD, Johnson LE, Pyke TR. J Antibiot. 1973;26:429–436. doi: 10.7164/antibiotics.26.429. [DOI] [PubMed] [Google Scholar]
- 1082.Womack SD, Demetrios AA. FR2504142 (A1) France Pat. 1982
- 1083.Jayasuriya H, Herath K, Ondeyka JG, Zhang C, Zink DL, Brower M, Gailliot FP, Greene J, Birdsall G, Venugopal J, Ushio M, Burgess B, Russotti G, Walker A, Hesse M, Seeley A, Junker B, Connors N, Salazar O, Genilloud O, Liu K, Masurekar P, Barrett JF, Singh SB. J Antibiot. 2007;60:554–564. doi: 10.1038/ja.2007.70. [DOI] [PubMed] [Google Scholar]
- 1084.Zhang C, Herath K, Jayasuriya H, Ondeyka JG, Zink DL, Occi J, Birdsall G, Venugopal J, Ushio M, Burgess B, Masurekar P, Barrett JF, Singh SB. J Nat Prod. 2009;72:841–847. doi: 10.1021/np800783b. [DOI] [PubMed] [Google Scholar]
- 1085.Li W, Leet JE, Ax HA, Gustavson DR, Brown DM, Turner L, Brown K, Clark J, Yang H, Fung-Tomc J, Lam KS. J Antibiot. 2003;56:226–231. doi: 10.7164/antibiotics.56.226. [DOI] [PubMed] [Google Scholar]
- 1086.Leet JE, Li W, Ax HA, Matson JA, Huang S, Huang R, Cantone JL, Drexler D, Dalterio RA, Lam KS. J Antibiot. 2003;56:232–242. doi: 10.7164/antibiotics.56.232. [DOI] [PubMed] [Google Scholar]
- 1087.Lam KS, Leet JE, Li W. EP1109820 A1. Eur Pat. 2001
- 1088.Steinberg DA, Bernan VS, Montenegro DA, Abbanat DR, Pearce CJ, Korshalla JD, Jacobus NV, Petersen PJ, Mroczenski-Wildey MJ, Maiese WM. J Antibiot. 1994;47:887–893. doi: 10.7164/antibiotics.47.887. [DOI] [PubMed] [Google Scholar]
- 1089.Northcote PT, Siegel M, Borders DB, Lee MD. J Antibiot. 1994;47:901–908. doi: 10.7164/antibiotics.47.901. [DOI] [PubMed] [Google Scholar]
- 1090.Zhang C, Occi J, Masurekar P, Barrett JF, Zink DL, Smith S, Onishi R, Ha S, Salazar O, Genilloud O, Basilio A, Vicente F, Gill C, Hickey EJ, Dorso K, Motyl M, Singh SB. J Am Chem Soc. 2008;130:12102–12110. doi: 10.1021/ja803183u. [DOI] [PubMed] [Google Scholar]
- 1091.Keller-Juslen C, Kuhn M, King HD. US4478831 (A) US Pat. 1984
- 1092.Puar MS, Chan TM, Hegde V, Patel M, Bartner P, Ng KJ, Pramanik BN, MacFarlane RD. J Antibiot. 1998;51:221–224. doi: 10.7164/antibiotics.51.221. [DOI] [PubMed] [Google Scholar]
- 1093.Neuhof T, Schmieder P, Preussel K, Dieckmann R, Pham H, Bartl F, von Döhren H. J Nat Prod. 2005;68:695–700. doi: 10.1021/np049671r. [DOI] [PubMed] [Google Scholar]
- 1094.Neuhof T, Schmieder P, Seibold M, Preussel K, von Döhren H. Bioorg Med Chem Lett. 2006;16:4220–4222. doi: 10.1016/j.bmcl.2006.05.094. [DOI] [PubMed] [Google Scholar]
- 1095.Lu S-E, Novak J, Austin FW, Gu G, Ellis D, Kirk M, Wilson-Stanford S, Tonelli M, Smith L. Biochemistry. 2009;48:8312–8321. doi: 10.1021/bi900814c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1096.Tawfik KA, Jeffs P, Bray B, Dubay G, Falkinham JO, Mesbah M, Youssef D, Khalifa S, Schmidt EW. Org Lett. 2010;12:664–666. doi: 10.1021/ol9029269. [DOI] [PubMed] [Google Scholar]
- 1097.Lee CH, Kim S, Hyun B, Suh JW, Yon C, Kim C, Lim Y, Kim C. J Antibiot. 1994;47:1402–1405. doi: 10.7164/antibiotics.47.1402. [DOI] [PubMed] [Google Scholar]
- 1098.Lim Y, Suh JW, Kim S, Hyun B, Kim C, Lee CH. J Antibiot. 1994;47:1406–1416. doi: 10.7164/antibiotics.47.1406. [DOI] [PubMed] [Google Scholar]
- 1099.Winkelmann G, Lupp R, Jung G. J Antibiot. 1980;33:353–358. doi: 10.7164/antibiotics.33.353. [DOI] [PubMed] [Google Scholar]
- 1100.Aydin M, Lucht N, König WA, Lupp R, Jung G, Winkelmann G. Liebigs Ann Chem. 1985;1985:2285–2300. [Google Scholar]
- 1101.Taplin D, Weinstein MJ, Testa RT, Marque JA, Patel MG. US 4137224 A. US Pat. 1979
- 1102.Ciabatti R, Kettenring JK, Winters G, Tuan G, Zerilli L, Cavalleri B. J Antibiot. 1989;42:254–267. doi: 10.7164/antibiotics.42.254. [DOI] [PubMed] [Google Scholar]
- 1103.Freeman J, Baines SD, Jabes D, Wilcox MH. J Antimicrob Chemother. 2005;56:717–725. doi: 10.1093/jac/dki321. [DOI] [PubMed] [Google Scholar]
- 1104.Dobashi K, Nagaoka K, Watanabe Y, Nishida M, Hamada M, Naganawa H, Takita T, Takeuchi T, Umezawa H. J Antibiot. 1985;38:1166–1170. doi: 10.7164/antibiotics.38.1166. [DOI] [PubMed] [Google Scholar]
- 1105.Martin JH, Tresner HD, Porter JN. US4007167 (A) US Pat. 1977
- 1106.Ellestad GA, Martin JH, Morton GO, Sassiver ML, Lancaster JE. J Antibiot. 1977;30:678–680. doi: 10.7164/antibiotics.30.678. [DOI] [PubMed] [Google Scholar]
- 1107.Ellestad GA, Morton GO, McGahren WJ. J Antibiot. 1982;35:1418–1421. doi: 10.7164/antibiotics.35.1418. [DOI] [PubMed] [Google Scholar]
- 1108.Katayama N, Nozaki Y, Okonogi K, Ono H, Harada S, Okazaki H. J Antibiot. 1985;38:1117–1127. doi: 10.7164/antibiotics.38.1117. [DOI] [PubMed] [Google Scholar]
- 1109.Hida T, Tsubotani S, Katayama N, Okazaki H, Harada S. J Antibiot. 1985;38:1128–1140. doi: 10.7164/antibiotics.38.1128. [DOI] [PubMed] [Google Scholar]
- 1110.Schmidt EW, Bewley CA, Faulkner DJ. J Org Chem. 1998;63:1254–1258. [Google Scholar]
- 1111.Bewley CA, Faulkner DJ. J Org Chem. 1994;59:4849–4852. [Google Scholar]
- 1112.Bewley CA, Faulkner DJ. J Org Chem. 1995;60:2644. [Google Scholar]
- 1113.Ishida K, Christiansen G, Yoshida WY, Kurmayer R, Welker M, Valls N, Bonjoch J, Hertweck C, Börner T, Hemscheidt T, Dittmann E. Chem Biol. 2007;14:565–576. doi: 10.1016/j.chembiol.2007.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1114.Shin HJ, Matsuda H, Murakami M, Yamaguchi K. J Org Chem. 1997;62:1810–1813. [Google Scholar]
- 1115.Singh PD, Johnson JH. J Antibiot. 1984;37:336–343. doi: 10.7164/antibiotics.37.336. [DOI] [PubMed] [Google Scholar]
- 1116.Vertesy L, Fehlhaber H-W, Kogler H, Schindler PW. Liebigs Ann. 1996:121–126. [Google Scholar]
- 1117.Pluotno A, Carmeli S. Tetrahedron. 2005;61:575–583. [Google Scholar]
- 1118.Umezawa S, Tatsuta K, Izawa O, Tsuchiya T, Umezawa H. Tetrahedron Lett. 1972;13:97–100. [Google Scholar]
- 1119.Nakae K, Hosokawa N, Sawa R, Kubota Y, Masuda T, Ohba S, Igarashi M, Nakagawa N, Nishimura Y, Akamatsu Y. J Antibiot. 2006;59:11–17. doi: 10.1038/ja.2006.3. [DOI] [PubMed] [Google Scholar]
- 1120.Spies HS, Steenkamp DJ. Eur J Biochem. 1994;224:203–213. doi: 10.1111/j.1432-1033.1994.tb20013.x. [DOI] [PubMed] [Google Scholar]
- 1121.Newton GL, Rawat M, La Clair JJ, Jothivasan VK, Budiarto T, Hamilton CJ, Claiborne A, Helmann JD, Fahey RC. Nat Chem Biol. 2009;5:625–627. doi: 10.1038/nchembio.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1122.Kocharova NA, Kondakova AN, Ovchinnikova OG, Perepelov AV, Shashkov AS, Knirel YA. Carbohydr Res. 2009;344:2060–2062. doi: 10.1016/j.carres.2009.07.003. [DOI] [PubMed] [Google Scholar]
- 1123.Sakuda S, Zhou ZY, Yamada Y. Biosci, Biotechnol, Biochem. 1994;58:1347–1348. doi: 10.1271/bbb.58.1347. [DOI] [PubMed] [Google Scholar]
- 1124.Laursen JB, Nielsen J. Chem Rev. 2004;104:1663–1686. doi: 10.1021/cr020473j. [DOI] [PubMed] [Google Scholar]
- 1125.Caulier G, Van Dyck S, Gerbaux P, Eeckhaut I, Flammang P. SPC Beche-de-mer Inf Bull. 2011;31:48–54. [Google Scholar]
- 1126.Pathirana C, Jensen PR, Dwight R, Fenical W. J Org Chem. 1992;57:740–742. [Google Scholar]
- 1127.Shin-ya K, Shimizu S, Kunigami T, Furihata K, Hayakawa Y, Seto H. J Antibiot. 1995;48:1378–1381. doi: 10.7164/antibiotics.48.1378. [DOI] [PubMed] [Google Scholar]
- 1128.Kunigami T, Shin-Ya K, Furihata K, Furihata K, Hayakawa Y, Seto H. J Antibiot. 1998;51:880–882. doi: 10.7164/antibiotics.51.880. [DOI] [PubMed] [Google Scholar]
- 1129.Abdelfattah MS, Toume K, Ishibashi M. J Antibiot. 2011;64:271–275. doi: 10.1038/ja.2010.172. [DOI] [PubMed] [Google Scholar]
- 1130.Kato S, Shindo K, Yamagishi Y, Matsuoka M, Kawai H, Mochizuki J. J Antibiot. 1993;46:1485–1493. doi: 10.7164/antibiotics.46.1485. [DOI] [PubMed] [Google Scholar]
- 1131.Krastel P, Zeeck A, Gebhardt K, Fiedler HP, Rheinheimer J. J Antibiot. 2002;55:801–806. doi: 10.7164/antibiotics.55.801. [DOI] [PubMed] [Google Scholar]
- 1132.Gebhardt K, Schimana J, Krastel P, Dettner K, Rheinheimer J, Zeeck A, Fiedler HP. J Antibiot. 2002;55:794–800. doi: 10.7164/antibiotics.55.794. [DOI] [PubMed] [Google Scholar]
- 1133.Matsumoto M, Mogi K, Nagaoka K, Ishizeki S, Kawahara R, Nakashima T. J Antibiot. 1987;40:149–156. doi: 10.7164/antibiotics.40.149. [DOI] [PubMed] [Google Scholar]
- 1134.Funayama S, Ishibashi M, Anraku Y, Miyauchi M, Mori H, Komiyama K, Omura S. J Antibiot. 1989;42:1734–1740. doi: 10.7164/antibiotics.42.1734. [DOI] [PubMed] [Google Scholar]
- 1135.Shaaban KA, Helmke E, Kelter G, Fiebig HH, Laatsch H. J Antibiot. 2011;64:205–209. doi: 10.1038/ja.2010.125. [DOI] [PubMed] [Google Scholar]
- 1136.Mori H, Funayama S, Sudo Y, Komiyama K, Omura S. J Antibiot. 1990;43:1329–1331. doi: 10.7164/antibiotics.43.1329. [DOI] [PubMed] [Google Scholar]
- 1137.Kimura K, Nakayama S, Nakajima N, Yoshihama M, Miyata N, Kawanishi G. J Antibiot. 1990;43:1341–1343. doi: 10.7164/antibiotics.43.1341. [DOI] [PubMed] [Google Scholar]
- 1138.Kimura K, Miyata N, Nakayama S. EP 399505 A1. European Pat. 1990
- 1139.Kimura K, Takahashi H, Miyata N, Yoshihama M, Uramoto M. J Antibiot. 1996;49:697–699. doi: 10.7164/antibiotics.49.697. [DOI] [PubMed] [Google Scholar]
- 1140.Iwasaki H, Kamisango K, Kuboniwa H, Sasaki H, Matsubara S. J Antibiot. 1991;44:451–452. doi: 10.7164/antibiotics.44.451. [DOI] [PubMed] [Google Scholar]
- 1141.Schleissner C, Perez M, Losada A, Rodriguez P, Crespo C, Zuniga P, Fernandez R, Reyes F, de la Calle F. J Nat Prod. 2011;74:1590–1596. doi: 10.1021/np200196j. [DOI] [PubMed] [Google Scholar]
- 1142.Berdy J. Adv Appl Microbiol. 1974;18:309–406. [PubMed] [Google Scholar]
- 1143.Bililign T, Griffith BR, Thorson JS. Nat Prod Rep. 2005;22:742–760. doi: 10.1039/b407364a. [DOI] [PubMed] [Google Scholar]
- 1144.Fujii N, Katsuyama T, Kobayashi E, Hara M, Nakano H. J Antibiot. 1995;48:768–772. doi: 10.7164/antibiotics.48.768. [DOI] [PubMed] [Google Scholar]
- 1145.Sequin U. Tetrahedron. 1978;34:761–767. [Google Scholar]
- 1146.Sequin U, Furukawa M. Tetrahedron. 1978;34:3623–3629. [Google Scholar]
- 1147.Sequin U, Bedford CT, Chung SK, Scott AI. Helv Chim Acta. 1977;60:896–906. doi: 10.1002/hlca.19770600320. [DOI] [PubMed] [Google Scholar]
- 1148.Hansen M, Yun S, Hurley L. Chem Biol. 1995;2:229–240. doi: 10.1016/1074-5521(95)90273-2. [DOI] [PubMed] [Google Scholar]
- 1149.Takahashi I, Takahashi K, Asano K, Kawamoto I, Yasuzawa T, Ashizawa T, Tomita F, Nakano H. J Antibiot. 1988;41:1151–1153. doi: 10.7164/antibiotics.41.1151. [DOI] [PubMed] [Google Scholar]
- 1150.Yasuzawa T, Saitoh Y, Sano H. J Antibiot. 1990;43:485–491. doi: 10.7164/antibiotics.43.485. [DOI] [PubMed] [Google Scholar]
- 1151.Byrne KM, Gonda SK, Hilton BD. J Antibiot. 1985;38:1040–1049. doi: 10.7164/antibiotics.38.1040. [DOI] [PubMed] [Google Scholar]
- 1152.Nadig H, Sequin U, Bunge RH, Hurley TR, Murphey DB, French JC. Helv Chim Acta. 1985;68:953–957. [Google Scholar]
- 1153.Ishii S, Satoh Y, Tsuruo T. Cancer Chemother Pharmacol. 1993;32:173–178. doi: 10.1007/BF00685831. [DOI] [PubMed] [Google Scholar]
- 1154.Ishii S, Nagasawa M, Kariya Y, Itoh O, Yamamoto H, Inouye S, Kondo S. J Antibiot. 1989;42:1518–1519. doi: 10.7164/antibiotics.42.1518. [DOI] [PubMed] [Google Scholar]
- 1155.Sato Y, Watabe H, Nakazawa T, Shomura T, Yamamoto H, Sezaki M, Kondo S. J Antibiot. 1989;42:149–152. doi: 10.7164/antibiotics.42.149. [DOI] [PubMed] [Google Scholar]
- 1156.Sun D, Hansen M, Clement JJ, Hurley LH. Biochemistry. 1993;32:8068–8074. doi: 10.1021/bi00083a003. [DOI] [PubMed] [Google Scholar]
- 1157.Brill GM, Jackson M, Whittern DN, Buko AM, Hill P, Chen RH, McAlpine JB. J Antibiot. 1994;47:1160–1164. doi: 10.7164/antibiotics.47.1160. [DOI] [PubMed] [Google Scholar]
- 1158.Brill GM, McAlpine JB, Whittern DN, Buko AM. J Antibiot. 1990;43:229–237. doi: 10.7164/antibiotics.43.229. [DOI] [PubMed] [Google Scholar]
- 1159.Jackson M, Karwowski JP, Theriault RJ, Hardy DJ, Swanson SJ, Barlow GJ, Tillis PM, McAlpine JB. J Antibiot. 1990;43:223–228. doi: 10.7164/antibiotics.43.223. [DOI] [PubMed] [Google Scholar]
- 1160.Abe N, Enoki N, Nakakita Y, Uchida H, Nakamura T, Munekata M. J Antibiot. 1993;46:1536–1549. doi: 10.7164/antibiotics.46.1536. [DOI] [PubMed] [Google Scholar]
- 1161.Abe N, Nakakita Y, Nakamura T, Enoki N, Uchida H, Munekata M. J Antibiot. 1993;46:1530–1535. doi: 10.7164/antibiotics.46.1530. [DOI] [PubMed] [Google Scholar]
- 1162.Abe N, Enoki N, Nakakita Y, Uchida H, Sato R, Watanabe N. J Antibiot. 1991;44:908–911. doi: 10.7164/antibiotics.44.908. [DOI] [PubMed] [Google Scholar]
- 1163.Kondo S, Miyamoto M, Naganawa H, Takeuchi T, Umezawa H. J Antibiot. 1977;30:1143–1145. doi: 10.7164/antibiotics.30.1143. [DOI] [PubMed] [Google Scholar]
- 1164.Kondo S, Wakashiro T, Hamada M, Maeda K, Takeuchi T. J Antibiot. 1970;23:354–359. doi: 10.7164/antibiotics.23.354. [DOI] [PubMed] [Google Scholar]
- 1165.Sequin U, Ceroni M. Helv Chim Acta. 1978;61:2241–2242. [Google Scholar]
- 1166.Vertesy L, Barbone FP, Cashmen E, Decker H, Ehrlich K, Jordan B, Knauf M, Schummer D, Segeth MP, Wink J, Seibert G. J Antibiot. 2001;54:718–729. doi: 10.7164/antibiotics.54.718. [DOI] [PubMed] [Google Scholar]
- 1167.Paschal JW, Occolowitz JL, Larsen SH, Boeck LD, Mertz FP. J Antibiot. 1989;42:623–626. doi: 10.7164/antibiotics.42.623. [DOI] [PubMed] [Google Scholar]
- 1168.Nadig H, Sequin U. Helv Chim Acta. 1987;70:1217–1228. [Google Scholar]
- 1169.Iwami M, Kawai Y, Kiyoto S, Terano H, Kohsaka M, Aoki H, Imanaka H. J Antibiot. 1986;39:6–11. doi: 10.7164/antibiotics.39.6. [DOI] [PubMed] [Google Scholar]
- 1170.Hori Y, Hino M, Kawai Y, Kiyoto S, Terano H, Kohsaka M, Aoki H, Hashimoto M, Imanaka H. J Antibiot. 1986;39:12–16. doi: 10.7164/antibiotics.39.12. [DOI] [PubMed] [Google Scholar]
- 1171.Kawai Y, Furihata K, Seto H, Otake N. Tetrahedron Lett. 1985;26:3273–3276. [Google Scholar]
- 1172.Umezawa I, Komiyama K, Takeshima H, Hata T, Kono M. J Antibiot. 1973;26:669–675. doi: 10.7164/antibiotics.26.669. [DOI] [PubMed] [Google Scholar]
- 1173.Kanda N. J Antibiot. 1972;25:557–560. doi: 10.7164/antibiotics.25.557. [DOI] [PubMed] [Google Scholar]
- 1174.Kanda N, Kono M, Asano K. J Antibiot. 1972;25:553–556. doi: 10.7164/antibiotics.25.553. [DOI] [PubMed] [Google Scholar]
- 1175.Kanda N. J Antibiot. 1971;24:599–606. doi: 10.7164/antibiotics.24.599. [DOI] [PubMed] [Google Scholar]
- 1176.White HL, White JR. Biochemistry. 1969;8:1020–1042. doi: 10.1021/bi00831a034. [DOI] [PubMed] [Google Scholar]
- 1177.Aszalos A, Lemanski P, Berk B, Dutcher JD. Antimicrob Agents Chemother. 1965;5:845–849. [PubMed] [Google Scholar]
- 1178.Furukawa M, Itai A, Iitaka Y. Tetrahedron Lett. 1973:1065–1068. [Google Scholar]
- 1179.Furukawa M, Iitaka Y. Tetrahedron Lett. 1974:3287–3290. [Google Scholar]
- 1180.Furukawa M, Hayakawa I, Ohta G, Iitaka Y. Tetrahedron. 1975;31:2989–2995. [Google Scholar]
- 1181.Bililign T, Hyun CG, Williams JS, Czisny AM, Thorson JS. Chem Biol. 2004;11:959–969. doi: 10.1016/j.chembiol.2004.04.016. [DOI] [PubMed] [Google Scholar]
- 1182.Rutkowski J, Brzezinski B. BioMed Res Int. 2013;2013:162513. doi: 10.1155/2013/162513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1183.Ogita T, Nakayama H, Otake N, Miyamae H, Sato S, Saito Y. Agric Biol Chem. 1979;43:1537–1542. [Google Scholar]
- 1184.Kusakabe Y, Mitsuoka S, Omuro Y, Seino A. J Antibiot. 1980;33:1437–1442. doi: 10.7164/antibiotics.33.1437. [DOI] [PubMed] [Google Scholar]
- 1185.Otake N, Ogita T, Nakayama H, Miyamae H, Sato S, Saito Y. J Chem Soc, Chem Comm. 1978:875–876. [Google Scholar]
- 1186.Seto H, Nakayama H, Ogita T, Furihata K, Mizove K, Otake N. J Antibiot. 1979;32:244–246. doi: 10.7164/antibiotics.32.244. [DOI] [PubMed] [Google Scholar]
- 1187.Jones ND, Chaney MO, Chamberlin JW, Hamill RL, Chen S. J Am Chem Soc. 1973;95:3399–3400. doi: 10.1021/ja00791a062. [DOI] [PubMed] [Google Scholar]
- 1188.Imada A, Nozaki Y, Hasegawa T, Mizuta E, Igarasi S, Yoneda M. J Antibiot. 1978;31:7–14. doi: 10.7164/antibiotics.31.7. [DOI] [PubMed] [Google Scholar]
- 1189.Mitani M, Otake N. J Antibiot. 1978;31:750–755. doi: 10.7164/antibiotics.31.750. [DOI] [PubMed] [Google Scholar]
- 1190.Otoguro K, Ishiyama A, Ui H, Kobayashi M, Manabe C, Yan G, Takahashi Y, Tanaka H, Yamada H, Omura S. J Antibiot. 2002;55:832–834. doi: 10.7164/antibiotics.55.832. [DOI] [PubMed] [Google Scholar]
- 1191.Tsuji N, Nagashima K, Kobayashi M, Wakisaka Y, Kawamura Y. J Antibiot. 1976;29:10–14. doi: 10.7164/antibiotics.29.10. [DOI] [PubMed] [Google Scholar]
- 1192.Tsuji N, Nagashima K, Terui Y, Tori K. J Antibiot. 1979;32:169–172. doi: 10.7164/antibiotics.32.169. [DOI] [PubMed] [Google Scholar]
- 1193.Liu CM, Hermann TE, Downey A, Prosser BL, Schildknecht E, Palleroni NJ, Westley JW, Miller PA. J Antibiot. 1983;36:343–350. doi: 10.7164/antibiotics.36.343. [DOI] [PubMed] [Google Scholar]
- 1194.Tsou H, Rajan S, Fiala R, Mowery PC, Bullock MW, Borders DB, James JC, Martin JH, Morton GO. J Antibiot. 1984;37:1651–1663. doi: 10.7164/antibiotics.37.1651. [DOI] [PubMed] [Google Scholar]
- 1195.Rajan S, Tsou HR, Mowery PC, Bullock MW, Stockton GW. J Antibiot. 1984;37:1495–1500. doi: 10.7164/antibiotics.37.1495. [DOI] [PubMed] [Google Scholar]
- 1196.Schneider RP, Lynch MJ, Ericson JF, Fouda HG. Anal Chem. 1991;63:1789–1794. doi: 10.1021/ac00017a024. [DOI] [PubMed] [Google Scholar]
- 1197.Mitani M, Umetsu T, Yamanishi T. J Antibiot. 1977;30:239–243. doi: 10.7164/antibiotics.30.239. [DOI] [PubMed] [Google Scholar]
- 1198.Dirlam JP, Bordner J, Chang SP, Grizzuti A, Nelson TH, Tynan EJ, Whipple EB. J Antibiot. 1992;45:1544–1548. doi: 10.7164/antibiotics.45.1544. [DOI] [PubMed] [Google Scholar]
- 1199.Dirlam JP, Cullen WP, Huang LH, Nelson TH, Oscarson JR, Presseau-Linabury L, Tynan EJ, Whipple EB. J Antibiot. 1991;44:1262–1266. doi: 10.7164/antibiotics.44.1262. [DOI] [PubMed] [Google Scholar]
- 1200.Dirlam JP, Bordner J, Cullen WP, Jefferson MT, Presseau-Linabury L. J Antibiot. 1992;45:1187–1189. doi: 10.7164/antibiotics.45.1187. [DOI] [PubMed] [Google Scholar]
- 1201.Funayama S, Nozoe S, Tronquet C, Anraku Y, Komiyama K, Omura S. J Antibiot. 1992;45:1686–1691. doi: 10.7164/antibiotics.45.1686. [DOI] [PubMed] [Google Scholar]
- 1202.Dirlam JP, Belton AM, Bordner J, Cullen WP, Huang LH, Kojima Y, Maeda H, Nishiyama S, Oscarson JR, Ricketts AP, et al. J Antibiot. 1992;45:331–340. doi: 10.7164/antibiotics.45.331. [DOI] [PubMed] [Google Scholar]
- 1203.Tynan EJ, Nelson TH, Davies RA, Wernau WC. J Antibiot. 1992;45:813–815. doi: 10.7164/antibiotics.45.813. [DOI] [PubMed] [Google Scholar]
- 1204.Cullen WP, Maeda H, Ruddock JC, Tone J. US 4746650 A. US Pat. 1988
- 1205.Nakamura M, Kunimoto S, Takahashi Y, Naganawa H, Sakaue M, Inoue S, Ohno T, Takeuchi T. Antimicrob Agents Chemother. 1992;36:492–494. doi: 10.1128/aac.36.2.492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1206.Keller-Juslen C, King HD, Kis ZL, von Wartburg A. J Antibiot. 1975;28:854–859. doi: 10.7164/antibiotics.28.854. [DOI] [PubMed] [Google Scholar]
- 1207.Dorman DE, Hamill RL, Occolowitz JL, Terui Y, Tori K, Tsuji N. J Antibiot. 1980;33:252–255. doi: 10.7164/antibiotics.33.252. [DOI] [PubMed] [Google Scholar]
- 1208.Dirlam JP, Belton AM, Bordner J, Cullen WP, Huang LH, Kojima Y, Maeda H, Nishiyama S, Oscarson JR, Ricketts AP, et al. J Ind Microbiol. 1990;6:135–142. doi: 10.1007/BF01576433. [DOI] [PubMed] [Google Scholar]
- 1209.Tsuji N, Terui Y, Nagashima K, Tori K, Johnson LF. J Antibiot. 1980;33:94–97. doi: 10.7164/antibiotics.33.94. [DOI] [PubMed] [Google Scholar]
- 1210.Nakayama H, Seto H, Otake N, Yamagishi M, Kawashima A, Mizutani T, Omura S. J Antibiot. 1985;38:1433–1436. doi: 10.7164/antibiotics.38.1433. [DOI] [PubMed] [Google Scholar]
- 1211.Kubota T, Hinoh G, Mayama M, Motokawa K, Yasuda Y. J Antibiot. 1975;28:931–934. doi: 10.7164/antibiotics.28.931. [DOI] [PubMed] [Google Scholar]
- 1212.Mizoue K, Seto H, Mizutani T, Yamagishi M, Kawashima A, Omura S, Ozeki M, Otake N. J Antibiot. 1980;33:144–156. doi: 10.7164/antibiotics.33.144. [DOI] [PubMed] [Google Scholar]
- 1213.Mizutani T, Yamagishi M, Hara H, Kawashima A, Omura S, Ozeki M. J Antibiot. 1980;33:137–143. doi: 10.7164/antibiotics.33.137. [DOI] [PubMed] [Google Scholar]
- 1214.Oscarson JR, Bordner J, Celmer WD, Cullen WP, Huang LH, Maeda H, Moshier PM, Nishiyama S, Presseau L, Shibakawa R, et al. J Antibiot. 1989;42:37–48. doi: 10.7164/antibiotics.42.37. [DOI] [PubMed] [Google Scholar]
- 1215.Cullen WP, Bordner J, Huang LH, Moshier PM, Oscarson JR, Presseau LA, Ware RS, Whipple EB, Kojima Y, Maeda H, et al. J Ind Microbiol. 1990;5:365–374. doi: 10.1007/BF01578095. [DOI] [PubMed] [Google Scholar]
- 1216.Dirlam JP, Presseau-Linabury L, Koss DA. J Antibiot. 1990;43:727–730. doi: 10.7164/antibiotics.43.727. [DOI] [PubMed] [Google Scholar]
- 1217.Wang XJ, Guo SL, Guo WQ, Xi D, Xiang WS. J Antibiot. 2009;62:309–313. doi: 10.1038/ja.2009.33. [DOI] [PubMed] [Google Scholar]
- 1218.Sun Y, Zhou X, Dong H, Tu G, Wang M, Wang B, Deng Z. Chem Biol. 2003;10:431–441. doi: 10.1016/s1074-5521(03)00092-9. [DOI] [PubMed] [Google Scholar]
- 1219.Harvey BM, Mironenko T, Sun Y, Hong H, Deng Z, Leadlay PF, Weissman KJ, Haydock SF. Chem Biol. 2007;14:703–714. doi: 10.1016/j.chembiol.2007.05.011. [DOI] [PubMed] [Google Scholar]
- 1220.Hamill RL, Hoehn MM, Pittenger GE, Chamberlin J, Gorman M. J Antibiot. 1969;22:161–164. doi: 10.7164/antibiotics.22.161. [DOI] [PubMed] [Google Scholar]
- 1221.Czerwinski EW, Steinrauf LK. Biochem Biophys Res Commun. 1971;45:1284–1287. doi: 10.1016/0006-291x(71)90157-4. [DOI] [PubMed] [Google Scholar]
- 1222.Kusakabe Y, Suzuki H, Kudo H. EP 293787 A1. European Pat. 1988
- 1223.Kusakabe Y, Takahashi N, Iwagaya Y, Seino A. J Antibiot. 1987;40:237–238. doi: 10.7164/antibiotics.40.237. [DOI] [PubMed] [Google Scholar]
- 1224.Dirlam JP, Belton AM, Bordner J, Cullen WP, Huang LH, Kojima Y, Maeda H, Nishida H, Nishiyama S, Oscarson JR, et al. J Antibiot. 1990;43:668–679. doi: 10.7164/antibiotics.43.668. [DOI] [PubMed] [Google Scholar]
- 1225.Noguchi Y, Ishii A, Matsushima A, Haishi D, Yasumuro K-i, Moriguchi T, Wada T, Kodera Y, Hiroto M, Nishimura H, Sekine M, Inada Y. Mar Biotechnol. 1999;1:207–210. doi: 10.1007/pl00011769. [DOI] [PubMed] [Google Scholar]
- 1226.Lee HW, Oh CH, Geyer A, Pfleiderer W, Park YS. Biochim Biophys Acta. 1999;1410:61–70. doi: 10.1016/s0005-2728(98)00175-3. [DOI] [PubMed] [Google Scholar]
- 1227.Moon YJ, Kim SJ, Park YM, Chung YH. Plant Signaling Behav. 2010;5:1127–1130. doi: 10.4161/psb.5.9.12567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1228.Choi YK, Hwang YK, Kang YH, Park YS. Pteridines. 2001;12:121–125. [Google Scholar]
- 1229.Cha KW, Pfleiderer W, Yim JJ. Helv Chim Acta. 1995;78:600–614. [Google Scholar]
- 1230.Lin XL, White RH. J Bacteriol. 1988;170:1396–1398. doi: 10.1128/jb.170.3.1396-1398.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1231.Raemakers-Franken PC, Voncken FG, Korteland J, Keltjens JT, van der Drift C, Vogels GD. BioFactors. 1989;2:117–122. [PubMed] [Google Scholar]
- 1232.Raemakers-Franken PC, van Elderen CH, van der Drift C, Vogels GD. BioFactors. 1991;3:127–130. [PubMed] [Google Scholar]
- 1233.Raemakers-Franken PC, Bongaerts R, Fokkens R, van der Drift C, Vogels GD. Eur J Biochem. 1991;200:783–787. doi: 10.1111/j.1432-1033.1991.tb16245.x. [DOI] [PubMed] [Google Scholar]
- 1234.Gan M, Guan Y, Zheng X, Yang Y, Hao X, Liu Y, Yu L, Xiao C. J Antibiot. 2012;65:513–516. doi: 10.1038/ja.2012.55. [DOI] [PubMed] [Google Scholar]
- 1235.Gan M, Zheng X, Gan L, Guan Y, Hao X, Liu Y, Si S, Zhang Y, Yu L, Xiao C. J Nat Prod. 2011;74:1142–1147. doi: 10.1021/np2000733. [DOI] [PubMed] [Google Scholar]
- 1236.Waksman SA, Woodruff HB. Exp Biol Med. 1942;49:207–210. [Google Scholar]
- 1237.Khokhlov AS, Shutova KI. J Antibiot. 1972;25:501–508. doi: 10.7164/antibiotics.25.501. [DOI] [PubMed] [Google Scholar]
- 1238.Kusumoto S, Kambayashi Y, Imaoka S, Shima K, Shiba T. J Antibiot. 1982;35:925–927. doi: 10.7164/antibiotics.35.925. [DOI] [PubMed] [Google Scholar]
- 1239.Kono Y, Makino S, Takeuchi S, Yonehara H. J Antibiot. 1969;22:583–589. doi: 10.7164/antibiotics.22.583. [DOI] [PubMed] [Google Scholar]
- 1240.Tsuruoka T, Shomura T, Ezaki N, Niwa T, Niida T. J Antibiot. 1968;21:237–238. doi: 10.7164/antibiotics.21.237. [DOI] [PubMed] [Google Scholar]
- 1241.Voronina OI, Shutova KI, Tovarova II, Khokhlov AS. Antibiotiki. 1969;14:1063–1069. [PubMed] [Google Scholar]
- 1242.Borders DB, Sax KJ, Lancaster JE, Hausmann WK, Mitscher LA, Wetzel ER, Patterson EL. Tetrahedron. 1970;26:3123–3133. doi: 10.1016/s0040-4020(01)92895-9. [DOI] [PubMed] [Google Scholar]
- 1243.Sinha RK. J Antibiot. 1970;23:360–364. doi: 10.7164/antibiotics.23.360. [DOI] [PubMed] [Google Scholar]
- 1244.Sawada Y, Kawakami S, Taniyama H. J Antibiot. 1977;30:460–467. doi: 10.7164/antibiotics.30.460. [DOI] [PubMed] [Google Scholar]
- 1245.Kawakami Y, Yamasaki K, Nakamura S. J Antibiot. 1981;34:921–922. doi: 10.7164/antibiotics.34.921. [DOI] [PubMed] [Google Scholar]
- 1246.Miyashiro S, Ando T, Hirayama K, Kida T, Shibai H, Murai A, Shiio T, Udaka S. J Antibiot. 1983;36:1638–1643. doi: 10.7164/antibiotics.36.1638. [DOI] [PubMed] [Google Scholar]
- 1247.Hunt AH, Hamill RL, DeBoer JR, Presti EA. J Antibiot. 1985;38:987–992. doi: 10.7164/antibiotics.38.987. [DOI] [PubMed] [Google Scholar]
- 1248.Ohba K, Nakayama H, Furihata K, Furihata K, Shimazu A, Seto H, Otake N, Yang ZZ, Xu LS, Xu WS. J Antibiot. 1986;39:872–875. doi: 10.7164/antibiotics.39.872. [DOI] [PubMed] [Google Scholar]
- 1249.Ando T, Miyashiro S, Hirayama K, Kida T, Shibai H, Murai A, Udaka S. J Antibiot. 1987;40:1140–1145. doi: 10.7164/antibiotics.40.1140. [DOI] [PubMed] [Google Scholar]
- 1250.Ji Z, Wang M, Zhang J, Wei S, Wu W. J Antibiot. 2007;60:739–744. doi: 10.1038/ja.2007.96. [DOI] [PubMed] [Google Scholar]
- 1251.Ji Z, Wang M, Wei S, Zhang J, Wu W. J Antibiot. 2009;62:233–237. doi: 10.1038/ja.2009.16. [DOI] [PubMed] [Google Scholar]
- 1252.Haupt I, Hübener R, Thrum H. J Antibiot. 1978;31:1137–1142. doi: 10.7164/antibiotics.31.1137. [DOI] [PubMed] [Google Scholar]
- 1253.Hisamoto M, Inaoka Y, Sakaida Y, Kagazaki T, Enokida R, Okazaki T, Haruyama H, Kinoshita T, Matsuda K. J Antibiot. 1998;51:607–617. doi: 10.7164/antibiotics.51.607. [DOI] [PubMed] [Google Scholar]
- 1254.Härtl A, Güttner J, Stöckel U, Hoffmann H. Arch Exp Veterinarmed. 1986;40:727–735. [PubMed] [Google Scholar]
- 1255.Bräunlich H, Hoffmann H, Bocker H. Pharmazie. 1988;43:200–202. [PubMed] [Google Scholar]
- 1256.Takemura S. Chem Pharm Bull. 1960;8:578–582. [Google Scholar]
- 1257.Hamano Y, Matsuura N, Kitamura M, Takagi H. J Biol Chem. 2006;281:16842–16848. doi: 10.1074/jbc.M602294200. [DOI] [PubMed] [Google Scholar]
- 1258.Takahashi S, Takeuchi M, Arai M, Seto H, Otake N. J Antibiot. 1983;36:226–228. doi: 10.7164/antibiotics.36.226. [DOI] [PubMed] [Google Scholar]
- 1259.Gebhardt K, Meyer SW, Schinko J, Bringmann G, Zeeck A, Fiedler H-P. J Antibiot. 2011;64:229–232. doi: 10.1038/ja.2010.165. [DOI] [PubMed] [Google Scholar]
- 1260.Singh SB, Zink DL, Dorso K, Motyl M, Salazar O, Basilio A, Vicente F, Byrne KM, Ha S, Genilloud O. J Nat Prod. 2009;72:345–352. doi: 10.1021/np8005106. [DOI] [PubMed] [Google Scholar]
- 1261.Shigemori H, Komaki H, Yazawa K, Mikami Y, Nemoto A, Tanaka Y, Sasaki T, In Y, Ishida T, Kobayashi JI. J Org Chem. 1998;63:6900–6904. doi: 10.1021/jo9807114. [DOI] [PubMed] [Google Scholar]
- 1262.Komatsu K, Tsuda M, Shiro M, Tanaka Y, Mikami Y, Kobayashi JI. Bioorg Med Chem. 2004;12:5545–5551. doi: 10.1016/j.bmc.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 1263.Zhang C, Ondeyka J, Guan Z, Dietrich L, Burgess B, Wang J, Singh SB. J Antibiot. 2009;62:699–702. doi: 10.1038/ja.2009.106. [DOI] [PubMed] [Google Scholar]
- 1264.Yu Z, Smanski MJ, Peterson RM, Marchillo K, Andes D, Rajski SR, Shen B. Org Lett. 2010;12:1744–1747. doi: 10.1021/ol100342m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1265.Yu Z, Rateb ME, Smanski MJ, Peterson RM, Shen B. J Antibiot. 2013;66:291–294. doi: 10.1038/ja.2013.1. [DOI] [PubMed] [Google Scholar]
- 1266.Wu M, Singh SB, Wang J, Chung CC, Salituro G, Karanam BV, Lee SH, Powles M, Ellsworth KP, Lassman ME, Miller C, Myers RW, Tota MR, Zhang BB, Li C. Proc Natl Acad Sci U S A. 2011;108:5378–5383. doi: 10.1073/pnas.1002588108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1267.Hirai Y, Haque M, Yoshida T, Yokota K, Yasuda T, Oguma K. J Bacteriol. 1995;177:5327–5333. doi: 10.1128/jb.177.18.5327-5333.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1268.Nagao K, Suzuki K, Hamada S, Yahara S, Yamamura R, Uyeda M. J Enzyme Inhib. 1998;13:135–146. doi: 10.3109/14756369809035832. [DOI] [PubMed] [Google Scholar]
- 1269.Flesch G, Rohmer M. Biochem J. 1989;262:673–675. doi: 10.1042/bj2620673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1270.Llopiz P, Jürgens UJ, Rohmer M. FEMS Microbiol Lett. 1996;140:199–202. [Google Scholar]
- 1271.Herrmann D, Bisseret P, Connan J, Rohmer M. Tetrahedron Lett. 1996;37:1791–1794. [Google Scholar]
- 1272.Neunlist S, Bisseret P, Rohmer M. Eur J Biochem. 1988;171:245–252. doi: 10.1111/j.1432-1033.1988.tb13783.x. [DOI] [PubMed] [Google Scholar]
- 1273.Rezanka T, Siristova L, Melzoch K, Sigler K. Lipids. 2011;46:249–261. doi: 10.1007/s11745-010-3482-4. [DOI] [PubMed] [Google Scholar]
- 1274.Simonin P, Jürgens UJ, Rohmer M. Tetrahedron Lett. 1992;33:3629–3632. [Google Scholar]
- 1275.Simonin P, Jürgens UJ, Rohmer M. Eur J Biochem. 1996;241:865–871. doi: 10.1111/j.1432-1033.1996.00865.x. [DOI] [PubMed] [Google Scholar]
- 1276.Cvejic JH, Putra SR, El-Beltagy A, Hattori R, Hattori T, Rohmer M. FEMS Microbiol Lett. 2000;183:295–299. doi: 10.1111/j.1574-6968.2000.tb08974.x. [DOI] [PubMed] [Google Scholar]
- 1277.Yoshimoto Y, Sawa T, Naganawa H, Sugai T, Takeuchi T, Imoto M. J Antibiot. 2000;53:575–578. doi: 10.7164/antibiotics.53.575. [DOI] [PubMed] [Google Scholar]
- 1278.Magauer T, Smaltz DJ, Myers AG. Nat Chem. 2013;5:886–893. doi: 10.1038/nchem.1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1279.Maskey RP, Helmke E, Kayser O, Fiebig HH, Maier A, Busche A, Laatsch H. J Antibiot. 2004;57:771–779. doi: 10.7164/antibiotics.57.771. [DOI] [PubMed] [Google Scholar]
- 1280.Tomita F, Tamaoki T, Morimoto M, Fujimoto K. J Antibiot. 1981;34:1519–1524. doi: 10.7164/antibiotics.34.1519. [DOI] [PubMed] [Google Scholar]
- 1281.Tamaoki T, Shirahata K, Iida T, Tomita F. J Antibiot. 1981;34:1525–1530. doi: 10.7164/antibiotics.34.1525. [DOI] [PubMed] [Google Scholar]
- 1282.Maskey RP, Sevvana M, Uson I, Helmke E, Laatsch H. Angew Chem, Int Ed. 2004;43:1281–1283. doi: 10.1002/anie.200352312. [DOI] [PubMed] [Google Scholar]
- 1283.Maiese WM, Labeda DP, Korshalla J, Kuck N, Fantini AA, Wildey MJ, Thomas J, Greenstein M. J Antibiot. 1990;43:253–258. doi: 10.7164/antibiotics.43.253. [DOI] [PubMed] [Google Scholar]
- 1284.Ratnayake R, Lacey E, Tennant S, Gill JH, Capon RJ. Chemistry. 2007;13:1610–1619. doi: 10.1002/chem.200601236. [DOI] [PubMed] [Google Scholar]
- 1285.Ratnayake R, Lacey E, Tennant S, Gill JH, Capon RJ. Org Lett. 2006;8:5267–5270. doi: 10.1021/ol062113e. [DOI] [PubMed] [Google Scholar]
- 1286.Peoples AJ, Zhang Q, Millett WP, Rothfeder MT, Pescatore BC, Madden AA, Ling LL, Moore CM. J Antibiot. 2008;61:457–463. doi: 10.1038/ja.2008.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1287.Kondo K, Eguchi T, Kakinuma K, Mizoue K, Qiao YF. J Antibiot. 1998;51:288–295. doi: 10.7164/antibiotics.51.288. [DOI] [PubMed] [Google Scholar]
- 1288.Qiao YF, Okazaki T, Ando T, Mizoue K. J Antibiot. 1998;51:282–287. doi: 10.7164/antibiotics.51.282. [DOI] [PubMed] [Google Scholar]
- 1289.Franco CM, Upadhyay DJ, Coutinho LL, Ganguli BN, Blumbach J, Fehlhaber H, Kogler H. US5270182. US Pat. 1993
- 1290.Ojiri K, Nishioka H, Torigoe K, Nakajima S, Kawamura K, Suda H. JP 07025893. Japenese Pat. 1995
- 1291.Takeda U, Okada T, Takagi M, Gomi S, Itoh J, Sezaki M, Ito M, Miyadoh S, Shomura T. J Antibiot. 1988;41:417–424. doi: 10.7164/antibiotics.41.417. [DOI] [PubMed] [Google Scholar]
- 1292.Gomi S, Sasaki T, Itoh J, Sezaki M. J Antibiot. 1988;41:425–432. doi: 10.7164/antibiotics.41.425. [DOI] [PubMed] [Google Scholar]
- 1293.Asolkar RN, Kirkland TN, Jensen PR, Fenical W. J Antibiot. 2010;63:37–39. doi: 10.1038/ja.2009.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1294.Kunimoto S, Someno T, Yamazaki Y, Lu J, Esumi H, Naganawa H. J Antibiot. 2003;56:1012–1017. doi: 10.7164/antibiotics.56.1012. [DOI] [PubMed] [Google Scholar]
- 1295.Kunimoto S, Lu J, Esumi H, Yamazaki Y, Kinoshita N, Honma Y, Hamada M, Ohsono M, Ishizuka M, Takeuchi T. J Antibiot. 2003;56:1004–1011. doi: 10.7164/antibiotics.56.1004. [DOI] [PubMed] [Google Scholar]
- 1296.Someno T, Kunimoto S, Nakamura H, Naganawa H, Ikeda D. J Antibiot. 2005;58:56–60. doi: 10.1038/ja.2005.6. [DOI] [PubMed] [Google Scholar]
- 1297.Rodriguez JC, Fernandez Puentes JL, Baz JP, Canedo LM. J Antibiot. 2003;56:318–321. doi: 10.7164/antibiotics.56.318. [DOI] [PubMed] [Google Scholar]
- 1298.Malet-Cascon L, Romero F, Espliego-Vazquez F, Gravalos D, Fernandez-Puentes JL. J Antibiot. 2003;56:219–225. doi: 10.7164/antibiotics.56.219. [DOI] [PubMed] [Google Scholar]
- 1299.Argoudelis AD, Brinkley TA, Brodasky TF, Buege JA, Meyer HF, Mizsak SA. J Antibiot. 1982;35:285–294. doi: 10.7164/antibiotics.35.285. [DOI] [PubMed] [Google Scholar]
- 1300.Argoudelis AD, Baczynskyj L, Buege JA, Marshall VP, Mizsak SA, Wiley PF. J Antibiot. 1987;40:408–418. doi: 10.7164/antibiotics.40.408. [DOI] [PubMed] [Google Scholar]
- 1301.Argoudelis AD, Baczynskyj L, Haak WJ, Knoll WM, Mizsak SA, Shilliday FB. J Antibiot. 1988;41:157–169. doi: 10.7164/antibiotics.41.157. [DOI] [PubMed] [Google Scholar]
- 1302.Argoudelis AD, Baczynskyj L, Mizsak SA, Shilliday FB. J Antibiot. 1988;41:1316–1330. doi: 10.7164/antibiotics.41.1316. [DOI] [PubMed] [Google Scholar]
- 1303.Argoudelis AD, Baczynskyj L, Mizsak SA, Shilliday FB, Wiley PF. J Antibiot. 1988;41:1212–1222. doi: 10.7164/antibiotics.41.1212. [DOI] [PubMed] [Google Scholar]
- 1304.Brazhnikova MG, Konstantinova NV, Mesentsev AS. J Antibiot. 1972;25:668–673. doi: 10.7164/antibiotics.25.668. [DOI] [PubMed] [Google Scholar]
- 1305.Leber JD, Hoover JRE, Holden KG, Johnson RK, Hecht SM. J Am Chem Soc. 1988;110:2992–2993. [Google Scholar]
- 1306.Itoh J, Watabe H, Ishii S, Gomi S, Nagasawa M, Yamamoto H, Shomura T, Sezaki M, Kondo S. J Antibiot. 1988;41:1281–1284. doi: 10.7164/antibiotics.41.1281. [DOI] [PubMed] [Google Scholar]
- 1307.Hertzberg RP, Hecht SM, Reynolds VL, Molineux IJ, Hurley LH. Biochemistry. 1986;25:1249–1258. doi: 10.1021/bi00354a009. [DOI] [PubMed] [Google Scholar]
- 1308.Orjala J, Gerwick WH. Phytochemistry. 1997;45:1087–1090. doi: 10.1016/s0031-9422(97)00084-8. [DOI] [PubMed] [Google Scholar]
- 1309.Nogle LM, Gerwick WH. J Nat Prod. 2003;66:217–220. doi: 10.1021/np020332c. [DOI] [PubMed] [Google Scholar]
- 1310.Wu M, Milligan KE, Gerwick WH. Tetrahedron. 1997;53:15983–15990. [Google Scholar]
- 1311.Yamazoe A, Hayashi K-i, Kepinski S, Leyser O, Nozaki H. Plant Physiol. 2005;139:779–789. doi: 10.1104/pp.105.068924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1312.Kong F, Zhao N, Siegel MM, Janota K, Ashcroft JS, Koehn FE, Borders DB, Carter GT. J Am Chem Soc. 1998;120:13301–13311. [Google Scholar]
- 1313.Sakuda S, Isogai A, Matsumoto S, Suzuki A, Koseki K. Tetrahedron Lett. 1986;27:2475–2478. [Google Scholar]
- 1314.Nishimoto Y, Sakuda S, Takayama S, Yamada Y. J Antibiot. 1991;44:716–722. doi: 10.7164/antibiotics.44.716. [DOI] [PubMed] [Google Scholar]
- 1315.Zhou ZY, Sakuda S, Kinoshita M, Yamada Y. J Antibiot. 1993;46:1582–1588. doi: 10.7164/antibiotics.46.1582. [DOI] [PubMed] [Google Scholar]
- 1316.Ando O, Satake H, Itoi K, Sato A, Nakajima M, Takahashi S, Haruyama H, Ohkuma Y, Kinoshita T, Enokita R. J Antibiot. 1991;44:1165–1168. doi: 10.7164/antibiotics.44.1165. [DOI] [PubMed] [Google Scholar]
- 1317.Nakayama T, Amachi T, Murao S, Sakai T, Shin T, Kenny PTM, Iwashita T, Zagorski M, Komura H, Nomoto K. J Chem Soc, Chem Commun. 1991;14:919–921. [Google Scholar]
- 1318.Kobayashi Y, Miyazaki H, Shiozaki M, Haruyama H. J Antibiot. 1994;47:932–938. doi: 10.7164/antibiotics.47.932. [DOI] [PubMed] [Google Scholar]
- 1319.Puder C, Loya S, Hizi A, Zeeck A. J Nat Prod. 2001;64:42–45. doi: 10.1021/np000377i. [DOI] [PubMed] [Google Scholar]
- 1320.Busby DJ, Copley RC, Hueso JA, Readshaw SA, Rivera A. J Antibiot. 2000;53:670–676. [PubMed] [Google Scholar]
- 1321.Minehan TG, Cook-Blumberg L, Kishi Y, Prinsep MR, Moore RE. Angew Chem, Int Ed. 1999;38:926–928. doi: 10.1002/(SICI)1521-3773(19990401)38:7<926::AID-ANIE926>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 1322.Prinsep MR, Caplan FR, Moore RE, Patterson GML, Smith CD. J Am Chem Soc. 1992;114:385–387. [Google Scholar]
- 1323.Prinsep MR, Patterson GML, Larsen LK, Smith CD. Tetrahedron. 1995;51:10523–10530. [Google Scholar]
- 1324.Prinsep MR, Patterson GM, Larsen LK, Smith CD. J Nat Prod. 1998;61:1133–1136. doi: 10.1021/np970566+. [DOI] [PubMed] [Google Scholar]
- 1325.Morliere P, Maziere JC, Santus R, Smith CD, Prinsep MR, Stobbe CC, Fenning MC, Golberg JL, Chapman JD. Cancer Res. 1998;58:3571–3578. [PubMed] [Google Scholar]
- 1326.Komoda T, Yoshida K, Abe N, Sugiyama Y, Imachi M, Hirota H, Koshino H, Hirota A. Biosci, Biotechnol, Biochem. 2004;68:104–111. doi: 10.1271/bbb.68.104. [DOI] [PubMed] [Google Scholar]
- 1327.Komoda T, Kishi M, Abe N, Sugiyama Y, Hirota A. Biosci, Biotechnol, Biochem. 2004;68:903–908. doi: 10.1271/bbb.68.903. [DOI] [PubMed] [Google Scholar]
- 1328.Imai H, Fujita S, Suzuki K, Morioka M, Tokunaga T, Shimizu M, Kadota S, Furuya T, Saito T. J Antibiot. 1989;42:1043–1048. doi: 10.7164/antibiotics.42.1043. [DOI] [PubMed] [Google Scholar]
- 1329.Tsuchiya K, Kobayashi S, Harada T, Kurokawa T, Nakagawa T, Shimada N, Kobayashi K. J Antibiot. 1995;48:626–629. doi: 10.7164/antibiotics.48.626. [DOI] [PubMed] [Google Scholar]
- 1330.Igarashi Y, Takagi K, Kajiura T, Furumai T, Oki T. J Antibiot. 1998;51:915–920. doi: 10.7164/antibiotics.51.915. [DOI] [PubMed] [Google Scholar]
- 1331.Kawamura N, Sawa R, Takahashi Y, Issiki K, Sawa T, Kinoshita N, Naganawa H, Hamada M, Takeuchi T. J Antibiot. 1995;48:435–437. doi: 10.7164/antibiotics.48.435. [DOI] [PubMed] [Google Scholar]
- 1332.Kawamura N, Nakamura H, Sawa R, Takahashi Y, Sawa T, Naganawa H, Takeuchi T. J Antibiot. 1997;50:147–149. doi: 10.7164/antibiotics.50.147. [DOI] [PubMed] [Google Scholar]
- 1333.Zhang Z, Wang Y, Ruan J. Int J Syst Bacteriol. 1998;48(pt 2):411–422. doi: 10.1099/00207713-48-2-411. [DOI] [PubMed] [Google Scholar]
- 1334.Flatt PM, Wu X, Perry S, Mahmud T. J Nat Prod. 2013;76:939–946. doi: 10.1021/np400159a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1335.Schüep W, Blount JF, Williams TH, Stempel A. J Antibiot. 1978;31:1226–1232. doi: 10.7164/antibiotics.31.1226. [DOI] [PubMed] [Google Scholar]
- 1336.Grond S, Langer H-J, Henne P, Sattler I, Thiericke R, Grabley S, Zähner H, Zeeck A. Eur J Org Chem. 2000:929–937. [Google Scholar]
- 1337.Grond S, Papastavrou I, Zeeck A. Eur J Org Chem. 2000:1875–1881. [Google Scholar]
- 1338.Bitzer J, Zeeck A. Eur J Org Chem. 2006:3661–3666. [Google Scholar]
- 1339.Ben Ameur Mehdi R, Shaaban KA, Rebai IK, Smaoui S, Bejar S, Mellouli L. Nat Prod Res. 2009;23:1095–1107. doi: 10.1080/14786410802362352. [DOI] [PubMed] [Google Scholar]
- 1340.Abdalla MA. PhD thesis. Georg-August University of Göttingen; 2010. [Google Scholar]
- 1341.Takahashi S, Iwai H, Kosaka K, Miyazaki T, Osanai Y, Arao N, Tanaka K, Nagai K, Nakagawa A. J Antibiot. 2007;60:717–720. doi: 10.1038/ja.2007.93. [DOI] [PubMed] [Google Scholar]
- 1342.Kwon Y-I, Son H-J, Moon KS, Kim JK, Kim J-G, Chun H-S, Ahn SK, Hong CI. J Antibiot. 2002;55:462–466. doi: 10.7164/antibiotics.55.462. [DOI] [PubMed] [Google Scholar]
- 1343.Chang H-B, Kim S-H, Kwon Y-I, Choung D-H, Choi W-K, Kang TW, Lee S, Kim J-G, Chun H-S, Ahn SK, Hong CI, Han K-H. J Antibiot. 2002;55:467–471. doi: 10.7164/antibiotics.55.467. [DOI] [PubMed] [Google Scholar]
- 1344.Song MC, Kim E, Ban YH, Yoo YJ, Kim EJ, Park SR, Pandey RP, Sohng JK, Yoon YJ. Appl Microbiol Biotechnol. 2013;97:5691–5704. doi: 10.1007/s00253-013-4978-7. [DOI] [PubMed] [Google Scholar]
- 1345.Salas JA, Méndez C. Trends Microbiol. 2007;15:219–232. doi: 10.1016/j.tim.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 1346.Singh S, Phillips GN, Jr, Thorson JS. Nat Prod Rep. 2012;29:1201–1237. doi: 10.1039/c2np20039b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1347.Lin CI, McCarty RM, Liu H-W. Chem Soc Rev. 2013;42:4377–4407. doi: 10.1039/c2cs35438a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1348.Thibodeaux CJ, Melancon CE, Liu HW. Nature. 2007;446:1008–1016. doi: 10.1038/nature05814. [DOI] [PubMed] [Google Scholar]