Abstract
The flow of λ-DNA solutions in a gradual micro-contraction was investigated using direct measurement techniques. The effects on DNA transport in microscale flows are significant because the flow behavior is influenced by macromolecular conformations, both viscous and elastic forces dominate inertial forces at this length scale, and the fully extended length of the molecule approaches the characteristic channel length wc (L/wc ∼ 0.13). This study examines the flow of semi-dilute and entangled DNA solutions in a gradual planar micro-contraction for low Reynolds numbers (3.7 × 10−6 < Re < 3.1 × 10−1) and high Weissenberg numbers (0.4 < Wi < 446). The semi-dilute DNA solutions have modest elasticity number, El = Wi/Re = 55, and do not exhibit viscoelastic behavior. For the entangled DNA solutions, we access high elasticity numbers (7.9 × 103 < El < 6.0 × 105). Video microscopy and streak images of entangled DNA solution flow reveal highly elastic behavior evidenced by the presence of large, stable vortices symmetric about the centerline and upstream of the channel entrance. Micro-particle image velocimetry measurements are used to obtain high resolution, quantitative velocity measurements of the vortex growth in this micro-contraction flow. These direct measurements provide a deeper understanding of the underlying physics of macromolecular transport in microfluidic flow, which will enable the realization of enhanced designs of lab-on-a-chip systems.
I. INTRODUCTION
Lab-on-a-chip systems for biological applications such as DNA sequencing, hybridization, and pathogen detection will incorporate numerous microfluidic components and require the flow of macromolecules such as DNA, which may give the bulk fluid viscoelastic behavior. As compared to macroscale flows, the fully extended lengths of macromolecules approach the width of the fluid channels in microfluidic flows, and the small characteristic length scale of these devices leads to increasing influence of elastic and viscous forces. This is a unique flow environment that is not well understood. For viscoelastic flows, not only does the fluid flow influence the conformation of the macromolecule but also the presence of macromolecules affects the flow behavior. A deeper understanding of the viscoelastic flow behavior in micro-environments will enable optimized lab-on-a-chip system designs for enhanced sensing or rapid diagnostic applications.
Because complex fluids may display very different behavior in shear-dominated and extension-dominated flows, it is important to consider the variety of typical geometries that comprise lab-on-a-chip devices. Flow studies in microfluidic elements where the fluid experiences rapid changes to the flow area or flow direction are particularly valuable, since the spatial variation reveals the viscoelastic behavior.
This investigation examines viscoelastic flows in a microscale planar gradual contraction, a canonical microfluidic component. In this flow structure, as the fluid approaches the channel from the upstream reservoir, it is spatially accelerated. The reservoir is concave with a large radius of curvature. There is nominal convex rounding at the channel entrance (radius of curvature of the rounding is much smaller than the channel width) due to the difficulty of obtaining sharp corners by microfabrication techniques.
Viscoelastic flows have been explored in other common microfluidic elements at negligible to moderate Reynolds (Re) number (10−7 < Re < 102). Recent attention has been given to flows through microscale abrupt planar contraction geometries, since it is the microfluidic analog of the viscoelastic benchmark flow. In this geometry, an upstream channel of larger width suddenly contracts to a downstream channel of smaller width, resulting in sharp corners in the region of the contraction. A global characteristic of viscoelastic flow in abrupt micro-contraction geometries is vortex growth and dynamics in the upstream channel with increasing flow rate. Studies have been conducted for flows of synthetic polymer solutions in abrupt micro-contraction-expansions (Rodd et al., 2005, 2007; Lanzaro and Yuan, 2011, 2014; Li et al., 2011a, 2011b) and DNA solutions in an abrupt micro-contraction (Gulati et al., 2008). Additionally, there have been studies characterizing viscoelastic vortex dynamics in micro-bifurcations (Balan et al., 2012) and in micro-bends (Gulati et al., 2010), as well as utilizing viscoelastic instabilities in serpentine channels for micro-mixing (Burghelea et al., 2004) and for micro-rectifier performance (Groisman and Quake, 2004; Jensen et al., 2012).
Experimental and numerical studies in a related geometry to the gradual micro-contraction, a microfluidic planar hyperbolic contraction-abrupt expansion, have been reported recently. The hyperbolic contraction-abrupt expansion has a smoothly converging channel entrance but with convex curvature. It has been evaluated as an extensional micro-rheometer, since it enables nearly uniform strain rate along the channel centerline for geometries with high Hencky strains under Hele–Shaw flow conditions (Oliveira et al., 2007). This geometry has also been used to characterize low viscosity, non-shear thinning (Boger) PAA solutions (Campo-Deano et al., 2011) and for characterization of blood simulants (Sousa et al., 2011) at small Re (1 < Re < 102).
To our knowledge, experimental studies of viscoelastic flows in microscale planar gradual contractions have not been reported in the literature. Laminar flows through macroscale planar gradual contractions at small to moderate Re (1 < Re < 104), however, have received some attention. Poole et al. (2005) explored flows of a moderately shear-thinning polyacrylamide (PAA) solution in a macroscale planar sudden expansion preceded by a gradual contraction. This geometry had a concave shape followed by significant convex curvature at the connection to the expansion; the radius of curvature of the concave shape was two times that of the convex. In the region of the gradual contraction, large velocities with large velocity gradients were observed near the sidewalls as compared with the smaller magnitude centerline velocity, leading to a velocity profile which they termed “cat's ears” due to the profile shape. The magnitude of the velocity overshoot (i.e., the difference between the velocity at the sidewalls compared with the centerline) in the plane with the flat sidewalls was found to be larger than the profiles in the plane with curved sidewalls of the contraction (Poole et al., 2005). Symmetric recirculation regions are only observed in the expansion portion of this device for viscoelastic flows. Three-dimensional numerical simulations of viscoelastic flows in gradual planar contraction-abrupt expansion geometries modelled using the Phan-Thien–Tanner (PTT) model (Poole et al., 2007) and modelled by both the upper-convected Maxwell (UCM) and PTT model (Poole and Alves, 2009) qualitatively agreed with experimental observations of the “cat's ears” velocity effect.
In this investigation, λ-DNA serves as the biopolymer model because of its direct use in lab-on-a-chip applications and the extensive rheological characterization (LeDuc et al., 1999; Perkins et al., 1997). The molecular contour length L approaches the device length scale wc; L/wc ∼ 0.13 for these flows. We explore flows of semi-dilute DNA solutions, where intermolecular interactions become important, and entangled DNA solutions and compare with the flows of a Newtonian fluid. While the flow field affects the conformation of the DNA at vanishing concentrations of DNA (Shrewsbury et al., 2001; 2002), we show that the presence of the DNA affects the flow field only at sufficiently high concentrations and flow rates. Streak images were obtained to characterize the upstream flow kinematics to determine these critical conditions. Pressure drops were measured to quantify the pressure drop due to the strong elastic flow behavior. Finally, micro-particle image velocimetry (μPIV) was used to quantify the velocity fields of the recirculation region with high-resolution. It is hoped that these measurements will be used for validation of numerical simulations as well as to determine when viscoelastic effects are important in microflows of DNA.
II. EXPERIMENT
A. Device design and fabrication
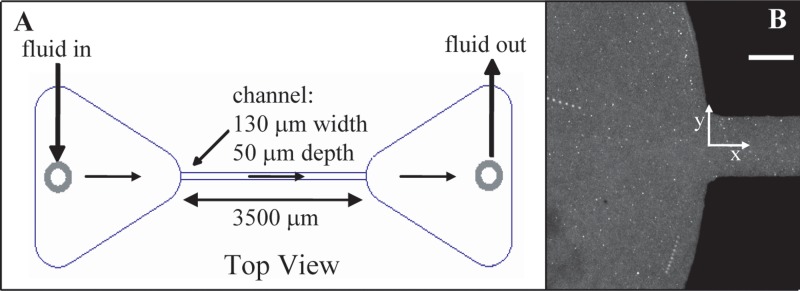
Figure 1 contains a schematic and an image of the gradual contraction device. The device consists of two reservoirs connected by a 3.5 mm channel with rectangular cross-section, 130 μm wide and 50 μm deep. Arrows in Figure 1(a) indicate the fluidic path. Fluid enters through a through-hole located in the inlet reservoir approximately 0.5 cm upstream of the contraction. The fluid experiences contraction flow when accelerating from the converging reservoir into the straight channel. In the region nearest to the contraction, the converging shape is circular with radius of curvature of ∼1800 μm (Figure 1(b)). After traveling the length of the channel, the fluid exits the device through a through-hole in the outlet reservoir. The gradual contraction device was pre-loaded with distilled water under vacuum to remove all air bubbles from the system. Subsequently, the test fluid was delivered to the device.
FIG. 1.
(a) Top view of gradual contraction device. (b) Image of contraction region with a DNA solution (40 μg/ml, Re = 0.314, Wi = 17.5, and El = 55) flowing through the device. Scale bar represents 100 μm.
The gradual contraction device was fabricated using conventional silicon microfabrication methods. A detailed description of the fabrication and bonding process flow are given by Shrewsbury et al. (2001); however, we provide a brief summary here. The device substrate was an n-type, 4 in. ⟨100⟩, single surface polished, silicon wafer. A 1 μm layer of silicon dioxide was formed by wet oxidation of the wafer at 1100 °C for 3 h. Two photolithography sequences using a positive resist were performed to pattern first the contraction geometry and subsequently through-holes for external fluidic connection. In the first sequence, a thin layer of photoresist (∼1.3 μm) was spun on, exposed, and developed to create the device pattern and then the exposed silicon dioxide layer was etched using oxygen plasma. In the second photolithography step, a thicker layer of photoresist of ∼10 μm was spun on in order to protect the regions not to be etched during the subsequent etching of through-holes. The through-holes (1 mm diameter) that serve as the inlet and outlet located in the device reservoirs were patterned, developed, and then formed by a deep reactive ion etch (DRIE) through the thickness of the wafer. The photoresist mask was then removed, exposing the underlying silicon dioxide masking layer, and subsequent RIE created the 50 μm deep device reservoirs and channels. Finally, the silicon dioxide layer was removed by buffered hydrofluoric acid.
A 170 μm thick Pyrex® coverslip was bonded to the silicon wafer using epoxy, which enabled optical access for flow visualization. A 5–10 μm thick layer of Epo-tek 301 epoxy (Epoxy Technology, Billerica, MA) was uniformly spun onto the coverslip, placed in contact with the etched silicon wafer, and cured. Oxygen plasma was directed into the device via the through-holes to ash away any unbonded epoxy. Finally, Tygon® 1/16 in. inner diameter flexible tubing (Cole-Parmer) was affixed to the through-holes of both reservoirs on the backside of the device with a quick setting two part epoxy (J-B Kwik and J-B Weld) to provide the external fluidic connection to the device. A syringe pump is used to drive the flow during experiments.
B. Fluid rheology
Two viscoelastic DNA solutions of different concentrations were prepared as test fluids for this study using λ-DNA (New England Biolabs). λ-DNA has a molecular weight of 31.5 × 106 Da or 48 502 base pairs, a fully extended length of 17 μm, and an equilibrium radius of gyration of 0.5 μm (Bustamante et al., 1994; Sanger et al., 1982; Verma et al., 1998). Additionally, the dynamics of this molecule in pure shear and pure extensional flow has been carefully documented (LeDuc et al., 1999; Perkins et al., 1997). Further, the conformational changes of λ-DNA at dilute concentrations in a similar microfluidic gradual contraction geometry (Shrewsbury et al. 2001, 2002) and the vortex growth of the higher concentration λ-DNA solutions in 2:1 abrupt contraction geometries (Gulati et al., 2008) have been characterized. The DNA solutions were diluted with 1× TAE buffer (40 mM Tris acetate and 1 mM EDTA, pH 8.3) to 40 μg/ml and 400 μg/ml DNA, respectively. In the absence of added monovalent salt, the polyelectrolyte nature of DNA will likely influence the dynamics and it is not anticipated that the solutions will behave as neutral synthetic polymer solutions. A similar influence is expected for our DNA solutions to those prepared at vanishing salt concentration conditions, resulting in an increased persistence length and hydrodynamic radius as compared with DNA in the fully screened high salt limit (Pan et al., 2014). Distilled water was tested as a Newtonian comparison.
In the previous work, we determined the overlap concentration c* of our DNA solution, the concentration where equilibrium coils begin to overlap each other, from the intrinsic viscosity by c* = 1/[η]. The intrinsic viscosity can be estimated by the y-intercept of the plot of the reduced viscosity against the concentration in the low concentration limit (Shrewsbury et al., 2001). The reduced viscosity is the ratio of the specific viscosity and concentration or ηsp/c and the specific viscosity is given by ηsp = (η − ηsol)/ ηsol, where ηsol is the solution viscosity. This method gives an intrinsic viscosity [η] of 0.01 ml/μg for DNA in TAE buffer and the overlap concentration c* = 1/[η] = 100 μg/ml (Gulati et al., 2008). Other studies have defined overlap concentration by the expression, , where M is the molecular weight, Rg is the radius of gyration, and NA is Avogadro's number. Using this definition, c* is approximately 40 μg/ml for λ-DNA solutions under good solvent conditions (Hur et al., 2001; Pan et al., 2014). Hence, from the intrinsic viscosity and the direct estimate of c*, the 40 μg/ml DNA solution is found to be 0.4c* and 1c*, respectively, and the 400 μg/ml DNA solution is 4c* and 10c*, respectively. As a result, the 40 μg/ml DNA solution is classified in the semi-dilute (unentangled) regime (0.5 < c/c* < 3) and 400 μg/ml DNA in the entangled regime (c/c* > 3) (Pan et al., 2014). Additionally, as elaborated above, the equilibrium description of c* is used to describe the DNA solutions; however, it should be noted that under the high shear conditions explored in this study the dynamic chain overlap concentration cs* is expected to be a lower concentration than c* (Salamone, 1996).
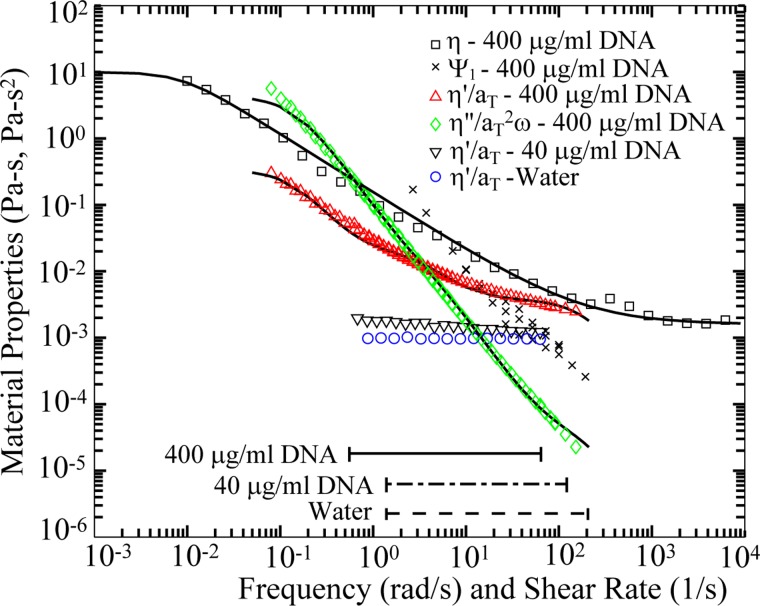
The steady shear viscosity η and the first normal stress coefficient Ψ1 for the 400 μg/ml DNA solution were characterized at 20 °C using a Malvern Bohlin Gemini rheometer and a Rheometrics RMS-800 mechanical spectrometer over the achievable shear rate range, 10−2 s−1 < < 103 s−1. These material properties for the 400 μg/ml DNA solution are given in Figure 2. Both the viscosity η and the first normal stress coefficient Ψ1 are highly shear-thinning. Steady shear viscosity for the 400 μg/ml DNA solution is fit to a Carreau model, , where η0 is the zero-shear rate viscosity, η∞ is the infinite shear rate viscosity, n is the power law index, and Λ is a time constant. The fitted model parameters are η0 = 10 Pa s, η∞ = 0.016 Pa s, Λ = 115 s, and n = 0.121.
FIG. 2.
Steady shear viscosity η and first normal stress coefficient Ψ1 as a function of shear rate for 400 μg/ml DNA solution. Master curves of dynamic viscosity η′/aT and dynamic rigidity η′′/aT2ω at Tref = 20 °C as a function of reduced frequency aTω for 400 μg/ml DNA solution. Also shown is the dynamic viscosity of the 40 μg/ml DNA solution and water as a function of frequency ω. The shear rate ranges ( = V/(wc/2)) probed for these fluids through the gradual contraction device are indicated by the horizontal lines. The dynamic shear data are fit to a 3-mode Maxwell model; the parameters are given in Table I. The steady shear viscosity is fit to a Carreau model with parameters η0 = 10 Pa-s, η∞ = 0.016 Pa-s, Λ = 115 s, n = 0.121.
Dynamic rheological properties were determined by small amplitude oscillatory shear (SAOS) measurements using a Vilastic-3 Viscoelasticity Analyzer (Vilastic Scientific, Inc.), an oscillatory flow capillary viscometer. SAOS measurements were taken at three temperatures T = 5, 10, and 20 °C, and combined into master curves at Tref = 20 °C by time-temperature superposition using shift factors aT(5 °C) = 1.45 and aT(10 °C) = 1.30. The master curves of dynamic viscosity η′/aT and dynamic rigidity 2η′′/aT2ω at Tref = 20 °C as a function of reduced frequency aTω for the 400 μg/ml DNA solution are shown in Figure 2. The dynamic viscosity of 40 μg/ml DNA and water at Tref = 20 °C are also given in Figure 2. The 40 μg/ml and 400 μg/ml DNA solutions were found to be mildly and highly shear-thinning, respectively, at high frequencies. The 40 μg/ml DNA solution viscosity is taken to be the maximum value of viscosity η = 1.965 × 10−3 Pa s, since it is only modestly dependent on shear rate.
The 400 μg/ml DNA master curves of dynamic viscosity η′/aT and dynamic rigidity 2η′′/aT2ω were fit to a multi-mode generalized Maxwell model describing the storage and loss moduli G′ and G′′
| (1) |
The relaxation spectrum λk and modulus parameters gk are given in Table I for the three-mode best fit of the data. The relaxation time of the 400 μg/ml DNA solution is taken as the longest relaxation time λ1. The relaxation time of the 40 μg/ml DNA solution is calculated using the Rouse model given by
| (2) |
where [η] is the intrinsic viscosity of the polymer solution, ηsol is the solution viscosity, M is the molecular weight of the polymer, R is the gas constant, and T is the temperature. The relaxation times for both DNA solutions are reported in Table II.
TABLE I.
Relaxation times and moduli for 400 μg/ml DNA.
| k | λk (s) | gk (Pa) |
|---|---|---|
| 1 | 6.79 | 4.72 × 10−2 |
| 2 | 0.259 | 5.89 × 10−2 |
| 3 | 0.005 | 7.54 × 10−1 |
TABLE II.
Rheological properties at 20 °C.
| Relaxation time, λ (s) | Density, ρ (g/cm3) | c/c* | Concentration regime | |
|---|---|---|---|---|
| 40 μg/ml DNA | 0.133 | 1.007 | 0.4,a 1b | Semi-dilute |
| 400 μg/ml DNA | 6.79 | 1.000 | 4,a 10b | Entangled |
c* = 100 μg/ml calculated from the intrinsic viscosity (Gulati et al., 2008).
c* = 40 μg/ml from Pan et al. (2014).
For flow visualization and μPIV experiments of flows of 400 μg/ml DNA solution and water, the test fluids are seeded with neutrally buoyant 1.0 μm orange fluorescent polystyrene tracer particles (FluoSpheres®, Molecular Probes). For flow visualization experiments, the water and 400 μg/ml DNA were seeded at concentrations of 0.0125% by volume and 0.4% by volume, respectively. For μPIV experiments, both solutions are seeded at concentrations of 0.4% by volume. At these volume fractions, 0.0125 = 1.25 × 10−4 and 0.4 = 4 × 10−3, by the Einstein expression, there is only a negligible increase in viscosity, η = η0{1 + 2.5 0.0125} = 1.0003η0 and η = η0{1 + 2.5 0.4} = 1.01η0. Preliminary flow visualization of 40 μg/ml DNA flow through the gradual contraction appears indistinguishable from water (cf. Figures 1(b), 6(c), and 6(b)) and hence, detailed pressure drop and μPIV studies were not conducted for this fluid.
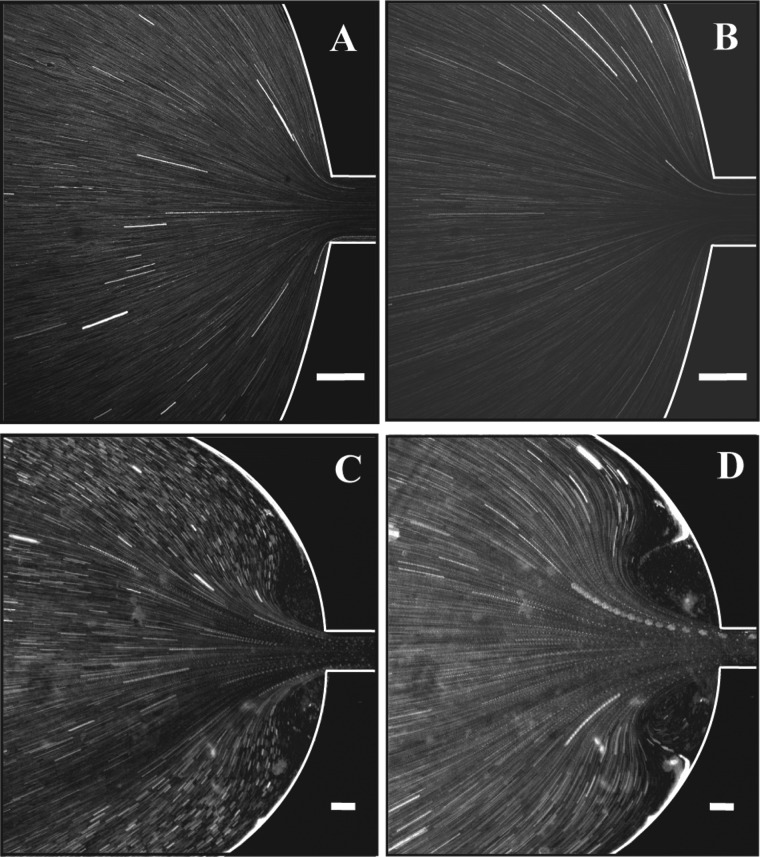
FIG. 6.
Streak images of a microscale gradual contraction device for flows of water at (a) Re = 0.08 and (b) Re = 0.31 and 400 μg/ml DNA at (c) Re = 5.2 × 10 − 3, Wi = 112, and El = 2.1 × 104 and (d) Re = 1.8 × 10−2, Wi = 223, and El = 1.3 × 104. Each scale bar represents 100 μm.
C. Dimensionless parameters
The microfluidic contraction flows in this study were characterized by three dimensionless parameters. The Reynolds (Re) number is defined as
| (3) |
where η() is the shear rate dependent viscosity, ρ is the solution density, Dh = 2wch/(wc + h) is the hydraulic diameter, V = Q/(hwc) is the average velocity, Q is the flow rate, = V/(wc/2) is the characteristic shear rate, wc is the channel width, and h is the channel depth. As with most microfluidic flows, the Re numbers accessed in this study are less than 1.
The Weissenberg (Wi) number is used to describe viscoelastic flows and is defined as
| (4) |
where λ is the relaxation time of the solution, taken as the longest Maxwell relaxation time for the 400 μg/ml DNA solution and the Rouse relaxation time for the 40 μg/ml DNA solution. Finally, the elasticity number (El) is the ratio of the dimensionless Wi to the Re
| (5) |
Since the viscosity of the 40 μg/ml DNA solution is only modestly dependent on shear rate, the maximum value of viscosity is used to compute the dimensionless parameters for all shear rates probed. Testing was conducted for flows of water, 40 μg/ml DNA, and 400 μg/ml DNA over the range of operating conditions given in Table III and the shear rate ranges probed are indicated by the horizontal lines in Figure 2.
TABLE III.
Experimental operating space.
| Re | Wi | El | |
|---|---|---|---|
| Water | 0.015–0.926 | 0 | 0 |
| 40 μg/ml DNA | 0.008–0.314 | 0.4–17.5 | 55 |
| 400 μg/ml DNA | 3.7 × 10−6–5.6 × 10−2 | 2.2–446 | 7.9 × 103–6.0 × 105 |
Experimental measurements in this study are also reported as dimensionless values. The non-dimensional pressure drop Δp is computed for water and 400 μg/ml DNA flows. At flow rate Q, Δp(Q) is given by
| (6) |
where ΔP(Q) is the measured pressure drop for water or the 400 μg/ml DNA solution at flow rate Q and ΔPsim(Q) is the simulated pressure drop for a Newtonian fluid or for an inelastic, shear-thinning fluid at flow rate Q, respectively. The simulated pressure drops are calculated from three-dimensional finite element simulations of a Newtonian fluid with viscosity of water and a fluid with identical Carreau model parameters describing the steady shear viscosity of the 400 μg/ml DNA solution and provided the viscous contribution to the pressure drop. Hence, Δp uncovers the significance, if any, of the elastic contribution to the total pressure drop for 400 μg/ml DNA over the range of flow rates.
Additionally, the dimensionless vortex reattachment length χ is given as follows:
| (7) |
where the vortex length LV is the arc length along the upstream channel wall between the boundary of the secondary flow vortex and the contraction plane and wc is the channel width.
Spatial positions are reported using x as the axial coordinate and y as the transverse coordinate. The origin is set at the x-position at the contraction plane, where the converging, upstream reservoir meets the downstream rectangular channel, and the y-position at the centerline of the device. The normalized x- and y-positions are given by x′ = x/wc and y′ = y/wc. The normalized x- and y-velocities are given by vx/V and vy/V, respectively.
D. Pressure drop measurements
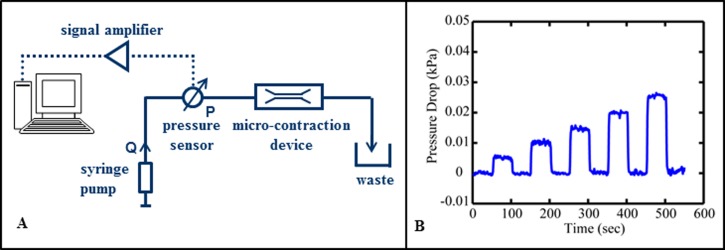
A schematic of the pressure measurement set-up is shown in Figure 3(a). The pressure drop is measured across the entire micro-contraction device using a flow-through pressure sensor (Honeywell Micro Switch 26PCA) with a nominal pressure range of 1 psi. The voltage signal from the pressure transducer is amplified by an instrumentation amplifier and subsequently recorded to a 12 bit I/O board on a computer. The sensor is placed in the tubing just upstream of the device. Upstream of the sensor, a syringe pump delivers the fluid to the micro-contraction device at controlled volumetric flow rates. The outlet flow from the micro-contraction is directed to a waste reservoir at atmospheric pressure. The measured pressure drops are dominated by the pressure drop through the straight channel but contain contributions from the gradual contraction and expansion regions and a minuscule contribution from the connected tubing.
FIG. 3.
(A) Schematic of the pressure measurement setup. (b) Pressure drop for flow of water through gradual contraction demonstrating the transient response of the system as the flow rate is stepped from 5 to 25 μl/h in five equal increments of 5 μl/h and in between dropped to 0 μl/h.
Pressure drops were measured for flows of water and 400 μg/ml DNA. For water flow through the gradual contraction, a commercial syringe pump (Cole-Parmer 40900) with a 1000 μl volume glass syringe is used to mechanically control the flow rate over the testing range, 5 < Q < 300 μl/h. For 400 μg/ml DNA flow through the gradual contraction, a custom syringe pump with a 10 μl volume glass syringe is used to deliver flows over the range, 0.5 < Q < 40 μl/h. The syringe sizes were chosen in order to maximize the plunger velocity and glass syringes were chosen so that the fluids could be delivered as smoothly as possible. Data were acquired at 300 Hz and the average value was reported each second. The pressure drop is computed as the difference between the mean steady-state values at zero flow (over a minimum of 60 s) and at the flow rate of interest (over a minimum of 100 s). The difference was taken to account for any slight drift in the transducer signal that may have occurred over the duration of testing. The transient response was recorded but not included in the differential pressure drop calculation. Between runs, the system was brought to rest and pressure measured for a minimum of 60 s in order to check that the device was operating properly (i.e., no clogs or leaks were present). The transient response of the pressure system for flows of water in the micro-contraction device is shown in Figure 3(b). The system response time to steady-state is rapid and on the order of seconds.
E. Visualization techniques
The steady flow kinematics in the region of the contraction (in the reservoir upstream of the straight channel) for flows of water and 400 μg/ml DNA are investigated using video microscopy, streak imaging, and μPIV. An inverted epifluorescence microscope equipped with either a 20×/0.5 NA Plan objective lens, a 10×/0.3 NA Universal Plan objective lens, or a 4×/0.13 NA Universal Plan objective lens was used for flow visualization measurements. For water flows, a mercury burner served as a constant illumination source for streak imaging and two alternating Nd:YAG lasers, with a wavelength of 532 nm and each firing at 15 Hz for a pulse duration of ∼6 ns, illuminated the flow for μPIV imaging. For 400 μg/ml DNA flows, the two alternating Nd:YAG lasers were illumination sources for both streak imaging and μPIV. Lasers were used as the illumination source for 400 μg/ml DNA flows because the images obtained were sufficient to produce high quality streak images. Images with 1013 × 1000 pixels were captured using a progressive scanning full-frame shutter camera (Pulnix TM-9701) and recorded to a computer for post-processing. All images were obtained at the midplane of the channel (25 μm from the top and bottom of the device) at 20×, 10×, or 4× magnifications depending on the size of the vortex. Streak imaging and μPIV analyses were not conducted for 40 μg/ml DNA flows because initial visualization studies using the two alternating Nd:YAG lasers across the range of operating conditions given in Table III did not reveal kinematic differences from water.
The external connections to the micro-contraction were identical to the pressure measurement setup except there is no pressure sensor placed in the upstream tubing. A commercial syringe pump (Cole-Parmer 40900) was used to control the volumetric flow rate over the range, 2 < Q < 100 μl/h, for visualization experiments. A 1000 μl volume glass syringe was used with the syringe pump. As with the pressure measurements, in between runs, the flow was turned off for a minimum of 60 s and then allowed to flow for a minimum of 60 s to ensure steady-state behavior prior to image acquisition.
Streak images were constructed by overlaying 4 s of sequential images taken at either 4× or 10× magnification of the steady flow taken at 30 fps using Scion Image Beta 4.0.2 image processing software. For μPIV measurements, the lasers are controlled independently to fire at given time intervals, ranging from 0.2 ms to 33 ms, in order to optimize the particle displacements for high quality cross-correlation μPIV algorithms. For each flow rate tested, 300 image pairs were collected for μPIV analysis at either 10× or 20× magnification, depending on the size of the vortex. The size of interrogation regions were chosen such that they contained a minimum of 8 particles per region and the displacements were chosen to be less than 33% the length of the interrogation region in the axial and transverse directions. For the flows of water, this corresponded to the regions closest to the contraction where the fluid was rapidly accelerating. For 400 μg/ml DNA flows, the focus was on the kinematics within the vortex region and the timing intervals were chosen to optimize the displacements in fast flow region towards the outer edge of the vortex. The primary flow was significantly faster than the secondary flow, and consequently the timing and interrogation region sizes optimized for the vortex region resulted in spurious cross-correlation vectors in the primary flow and were subsequently removed. Additionally, spurious vectors in the regions past the wall where there was no flow (for the silicon substrate) were removed for both water and 400 μg/ml DNA velocity fields. Individual velocity fields constructed from single image pairs were ensemble averaged over all 300 pairs of images to construct an averaged velocity vector field.
Numerous factors in the visualization system setup contribute to accuracy and resolution of post-processed images. For our measurement system, we have previously reported measurement depth δzm (Meinhart et al., 2000) because it more accurately reflects the depth of the image plane as compared to the depth of field. The measurement depth is given by
| (8) |
where λ0 is the wavelength of the imaged light, n is the refractive index, NA is the numerical aperture, dp is the tracer particle diameter, and θ = sin−1(NA/n). For this imaging system, λ0 = 560 nm, n = 1.33, dp = 1 μm, and NA = 0.5, 0.3, and 0.13 for the 20×, 10×, and 4× magnifications, respectively. Hence, for this study, δzm is 15.3 μm or 28% of the channel depth for the 20× objective lens, δzm is 35.2 μm or 64% of the channel depth for the 10× objective lens, and δzm is 155 μm or over 100% of the channel depth for the 4× objective lens. Additionally, uncertainty also arises in applying the cross-correlation algorithm over parts of the flow field where particle movement within interrogation regions is not spatially uniform, such as at the interface between the primary and secondary flow regions and nearby walls. Ensemble averaging was used to minimize uncertainty due to poor correlations.
F. Computational fluid dynamics simulations
Three-dimensional numerical simulations were conducted using Comsol Multiphysics 3.2 in order to model the flow of a Newtonian fluid with the viscosity of water and a Carreau fluid with the same shear-thinning behavior as the 400 μg/ml DNA solution. The non-dimensional momentum equations were used to simulate the fluid flow in the channel downstream of the gradual micro-contraction. Simulations were performed at the same flow rates as experimental pressure drops across a portion of the total channel length where the flow is fully developed. For the simulations, the walls were specified as no-slip boundaries, the average velocity was specified as the inlet boundary condition for each flow rate, and the pressure was set to zero at the outlet boundary. The simulated pressure drop was then scaled for the total channel length of 3500 μm in order to determine the total channel pressure drop experienced by water and the Carreau fluid. Finally, the non-dimensional pressure drop was computed (cf. Eq. (6)).
III. RESULTS AND DISCUSSION
A. Pressure drop measurements
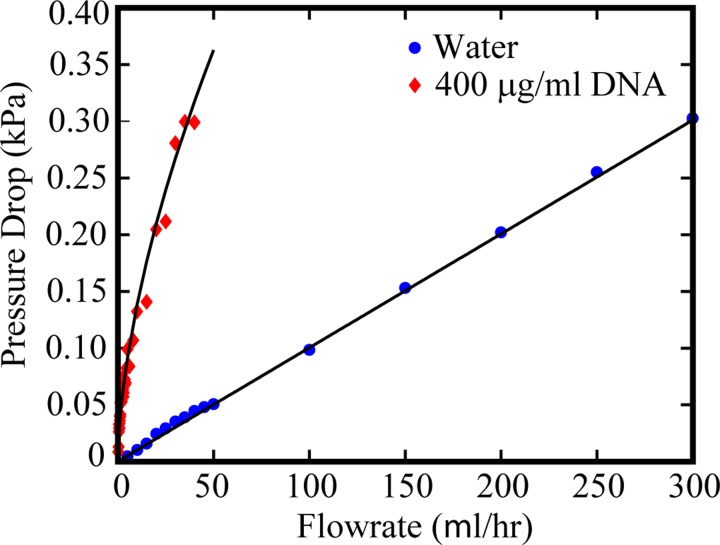
The measured steady-state pressure drops for water and 400 μg/ml DNA flows in the gradual contraction device are given in Figure 4. The pressure drop relationships for water and 400 μg/ml DNA are given by the functional forms, ΔP = 0.001Q and ΔP = 0.035Q0.6, respectively. The dimensionless pressure drop Δp of water is computed as the ratio of the measured pressure drop and the simulated pressure drop over the channel region for water as given in Eq. (6). The Δp for water as a function of flow rate is given in Figure 5(a). The error bars indicate the uncertainty in Δp measurements, which are propagated from the uncertainty in the differential pressure drop, estimated as 0.017 kPa or 0.25% of the range of the pressure sensor.
FIG. 4.
Pressure drop for flow of water and 400 μg/ml DNA through gradual contraction device as a function of flow rate Q. The pressure drop for water follows the functional form, ΔP = 0.001Q, and the pressure drop for 400 μg/ml DNA follows the functional form, ΔP = 0.035Q0.6.
FIG. 5.
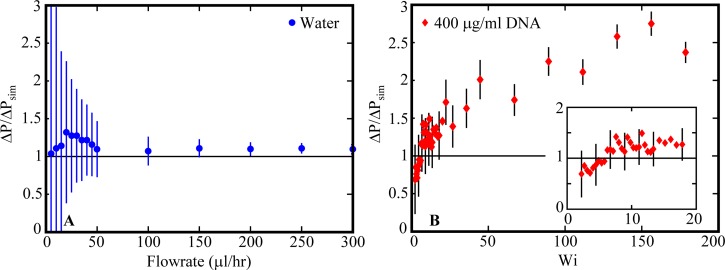
Dimensionless pressure drop ΔP/ΔPsim for flow of (a) water as a function of flowrate and (b) 400 μg/ml DNA through gradual contraction device as a function of Wi. Error bars indicate the uncertainty in ΔP/ΔPsim propagated from the uncertainty in the differential pressure drop measurements. Inset in (b) is the dimensionless pressure drop of 400 μg/ml DNA at low Wi with some representative error bars included.
For water flow, Δp ∼ 1.1 for the higher flow rates, Q > 50 μl/h (Figure 5(a)). The portion of Δp above unity (∼10% of the simulated channel pressured drop ΔPsim) is expected to contain the pressure contributions from the turn and expansion flow at the tubing entrance to the upstream inlet reservoir, the gradually converging upstream reservoir that joins the channel at the contraction plane, and the gradually expanding outlet reservoir. A range of Δp from 1.05 to 1.3 are obtained for the slower flows with notably high error uncertainty due to the measured pressure drops falling below or being close in magnitude to the uncertainty from the pressure sensor. Additionally, the nature of the drive mechanism of the syringe pump and the short duration of the test may have produced slightly elevated flow rates than expected, resulting in higher Δp.
In a similar manner, the Δp for 400 μg/ml DNA is computed as the ratio of the measured pressure drop of 400 μg/ml DNA flow and the simulated pressure drop over the channel region for a fluid with an identical Carreau model viscosity relationship to the 400 μg/ml DNA solution (Eq. (6)). Recasting pressure drops as Δp isolates the viscoelastic contributions from the contraction and expansion regions (i.e., the tubing entrance to the inlet, the inlet reservoir, and outlet reservoir) of the microdevice. The dimensionless pressure drop as a function of Wi is given in Figure 5(b). As Wi approaches zero, we expect Δp for 400 μg/ml DNA to approach 1.1 as was observed for the Newtonian case. Additionally, viscous behavior would dominate the DNA solution flow at this Wi limit. For Wi < 5.8, Δp falls below this asymptote. The measured pressure drops in this Wi range are close in magnitude to the uncertainty from the pressure sensor, yielding large error uncertainty. Moreover, the flows are very slow (0.5 μl/h < Q < 1.3 μl/h) and smooth, consistent flow may have been difficult to achieve with the syringe pump and throughout the device. For 6 < Wi < 18, Δp remains between 1.1 and 1.3, and for Wi > 20, Δp increases with Wi until reaching a plateau value of Δp ∼ 2.75 at high Wi. The saturation of Δp in our work, as was previously suggested by Rodd et al. (2005), is likely a consequence of polymer chains reaching their finite extensibility limit.
The growth and asymptotic approach of Δp with increasing Wi can be compared with the previously reported micro-contraction flows. The growth mostly closely resembles the results of Rodd et al. (2005), in which a plateau of slightly higher magnitude (Δp ∼ 3.5) was observed for flows of semi-dilute, mildly shear thinning 3.5c* PEO solutions in a 16:1:16 abrupt planar contraction-expansion over a wider Wi range. Additionally, the plateau magnitude is slightly lower than the maximum value reported in our past studies in a 2:1 abrupt micro-contraction of Δp ∼ 3.5 for flow of the same DNA solution over a wider Wi range, although there is no plateau observed for the 2:1 abrupt contraction flow (Gulati et al., 2008). The Δp maximum in the gradual micro-contraction is larger than the reported peak values of Δp ∼ 2.15 and ∼2.0 for flows of mildly shear-thinning semi-dilute 15c* and 20c* PEO solutions, respectively, in 8:1:8 contraction-expansion over a similar Wi range (Li et al., 2011a) and peaks of Δp ∼ 1.8 for weakly shear-thinning 8.3c* PAA solution flows in an 8:1:8 contraction-expansion and 3.3c* PAA solution flows in a 16:1:16 contraction-expansion over a similar Wi range (Lanzaro and Yuan, 2011).
B. Flow visualization
Streak imaging techniques were used to elucidate fluid pathlines in the reservoir upstream of the channel (Figure 6). These were taken at the lowest magnification (4×) in order to view the regions on both sides of the channel centerline. For flows of distilled water, no vortices are present across the range of Re probed. Examples of the streak images obtained for water are given in Figures 6(a) and 6(b) for flow at Re = 0.08 and Re = 0.31, respectively. However, symmetric, stable vortices are observed in streak images for flows of 400 μg/ml DNA at Re = 5.2 × 10−3, Wi = 112, and El = 2.1 × 104 and Re = 1.8 × 10−2, Wi = 223, and El = 1.3 × 104 (Figures 6(c) and 6(d)) illustrating the strong elastic flow behavior. These vortices grow dramatically with increasing Re and Wi and do not exhibit time-dependence across the Wi range probed.
Although detailed streak images are not reported here, it should be noted that video microscopy revealed no recirculation across the Re and Wi range for 40 μg/ml flows in the gradual contraction, indicating non-elastic behavior (cf. Figure 1(b) and Figures 6(a) and 6(b)). This is consistent with the observations of non-shear thinning fluids in other planar geometries. Vortices are not present for experimental flows of Boger (non-shear thinning) fluids in macroscale planar abrupt contractions, whereas vortex growth occurs for shear-thinning fluids (Nigen and Walters, 2002; Evans and Walters, 1986). Similarly, flows of non-shear thinning dilute PEO solutions in a microscale abrupt planar bend show the absence of vortices, while concentrated semi-dilute solutions of the same PEO, which have shear thinning behavior, exhibit elastic vortex growth (Gulati et al., 2010). Additionally, the chain rigidity of the DNA molecule may play a role in the lack of vortex dynamics. Arratia et al. (2006) do not observe an elastic instability for dilute solutions of semirigid xanthum gum in cross-channel flow. Under the same low Re conditions, dilute solutions of flexible PAA reveal flow instability (Arratia et al., 2006).
C. Evolution in vortex length
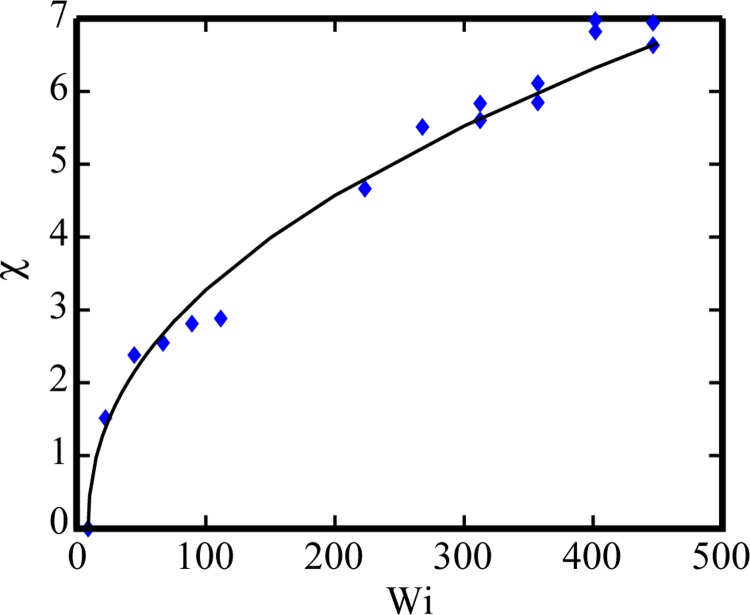
The vortex length LV was obtained by measuring the arc length along the wall from the edge of the vortex boundary to the corner of the contraction in pixels. The size of the vortex above the centerline was determined from digital microscopy videos taken at the higher magnifications. The length in pixels was then converted to a dimensional length scale (1 pixel = 2.26 μm for 20× magnification and 1 pixel = 1.13 μm for 10× magnification). Dimensionless vortex length χ was computed using Eq. (7). Figure 7 shows the relationship between χ and Wi for flows of 400 μg/ml DNA. The increase in the dimensionless vortex length is well described by χ = 0.43(Wi – Wicrit)0.45 where Wicrit is 8.9 over the Wi range 8.9 < Wi < 446. At low Wi, no vortex was present, consistent with expected behavior for Newtonian flow (cf. streak image of water flow in Figure 9(a)). The onset and growth of vortices occur in the range 8.9 < Wi < 22.
FIG. 7.
Average dimensionless vortex length for flows of 400 μg/ml DNA through gradual micro-contraction device as a function of Wi. The vortex length follows the functional form, χ = 0.43(Wi − Wicrit)0.45 where Wicrit is 8.9.
FIG. 9.
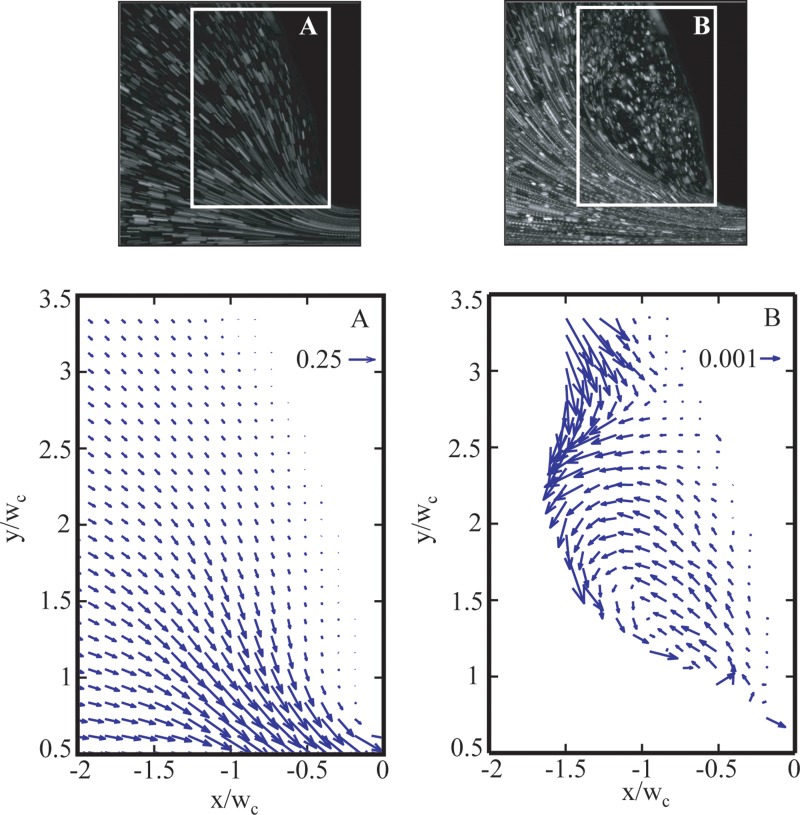
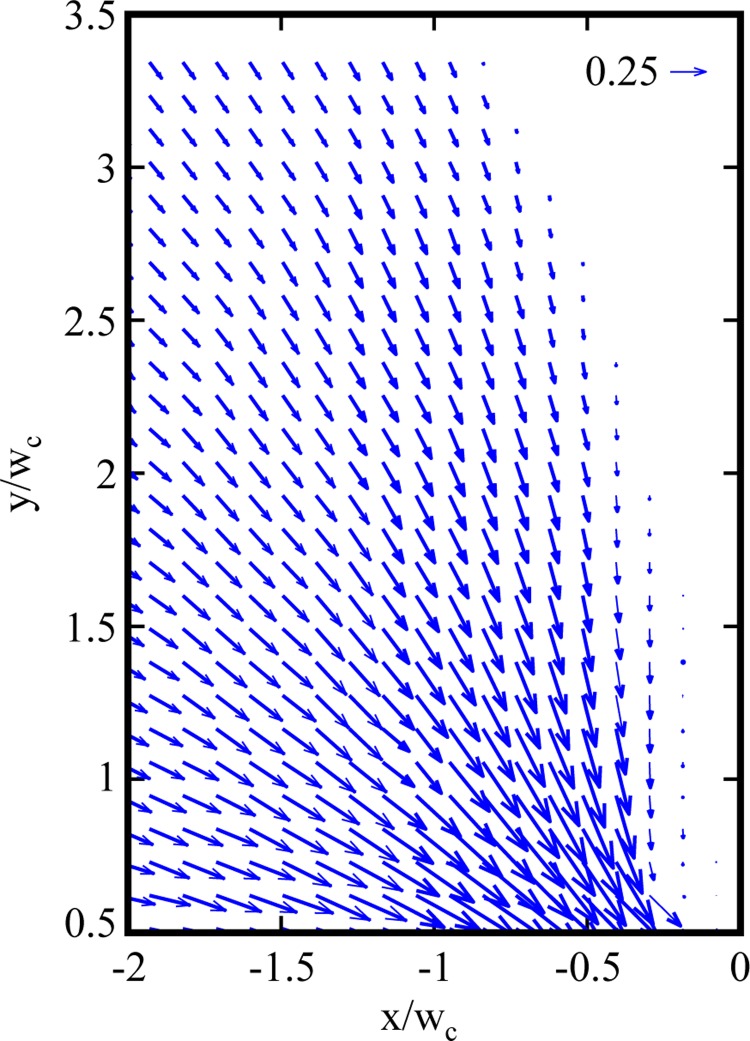
Velocity vector fields for 400 μg/ml DNA flow at (a) Re = 5.0 × 10−5, Wi = 8.9, and El = 1.8 × 105 and (b) Re = 2.1 × 10−3, Wi = 67, and El = 3.2 × 104 through gradual contraction. The corresponding streak images and region of interest are shown at top.
It is difficult to make comparisons of vortex growth with the previous studies of viscoelastic flows in micro-contractions due to the unique continuously converging shape of the gradual micro-contraction. Hyperbolic micro-contractions provide the closest comparison given their converging shape though the contraction is preceded by a straight channel section where the vortex develops. The non-dimensional vortex length χu is calculated as the ratio of the vortex length (measured along the straight section) and the upstream width wu. If we recast the dimensionless vortex length (the arc length of vortex) for our 400 μg/ml DNA flows in the gradual micro-contraction by an estimate of the upstream width of the reservoir of 2000 μm (the width of the gradual contraction that is visible in the images at the lowest magnification), the vortex grows to a maximum χu = 0.45. The size and growth of the vortex are similar to flows of moderately shear-thinning xanthan gum solutions in hyperbolic contractions (Hencky strains, εH = ln(wu/wc) = 2 and 3) across a similar Wi range (Sousa et al., 2011).
D. Micro-particle image velocimetry
Velocity vector fields for flows of water and 400 μg/ml DNA through the gradual micro-contraction were obtained by μPIV techniques. A series of 300 image pairs were taken and flow fields for all pairs of images are averaged to construct the velocity vector field. Images are obtained at 20× magnification for water flow and 400 μg/ml DNA at low Wi, where no or small vortices are present. For 400 μg/ml DNA flows at higher Wi where larger vortices are present, images are obtained at 10× magnification. The interrogation regions at 10× and 20× magnification are ∼28 × 28 μm and ∼14 × 14 μm, respectively, with a 50% overlap.
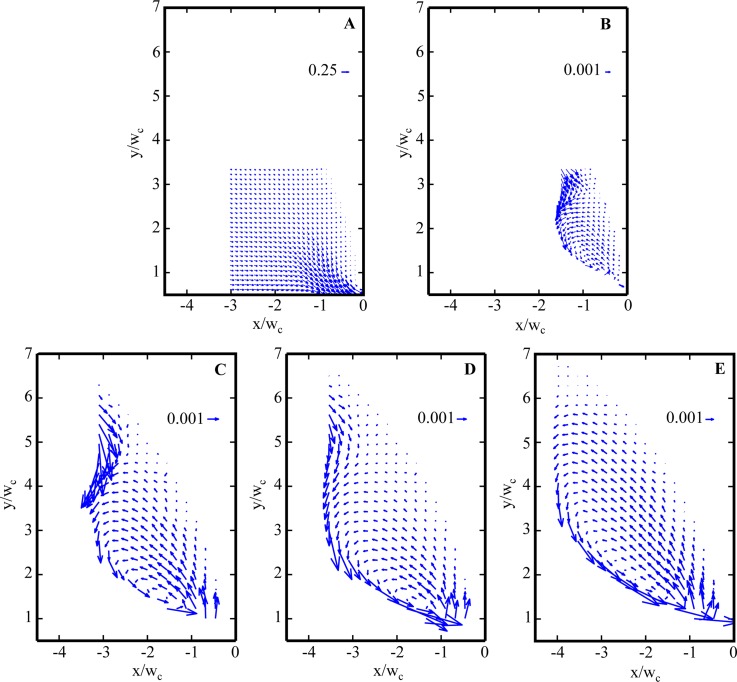
Figure 8 shows the velocity field for water flow at Re = 0.31 above the centerline of the gradual micro-contraction. Figure 9 contains velocity fields for flows of 400 μg/ml DNA in the recirculation region at low Wi (Wi = 8.9 and 67). The contraction is located at x′ = 0 and the centerline is located at y′ = 0. For 400 μg/ml DNA flows in the gradual micro-contraction, no recirculation is observed for the low Wi case and the transition to vortices (i.e., to elastic flow behavior) occurs at Wi > 8.9. Figure 10 indicates the dramatic vortex growth with increasing Wi for flows of 400 μg/ml DNA in the gradual micro-contraction. The flow conditions shown in Figure 10 are (a) Wi = 8.9, (b) Wi = 67, (c) Wi = 223, (d) = 268, and (e) Wi = 312.
FIG. 8.
Velocity vector field for water flow at Re = 0.31 through gradual contraction. The contraction is located at x′ = 0 and the centerline is located at y′ = 0.
FIG. 10.
Velocity vector fields for 400 μg/ml DNA flow at (a) Re = 5.0 × 10−5, Wi = 8.9, and El = 1.8 × 105 and (b) Re = 2.1 × 10−3, Wi = 67, and El = 3.2 × 104; (c) Re = 1.8 × 10−2, Wi = 223, and El = 1.3 × 104; (d) Re = 2.4 × 10−2, Wi = 268, and El = 1.1 × 104; and (e) Re = 3.1 × 10−2, Wi = 312, and El = 1.0 × 104 through gradual contraction.
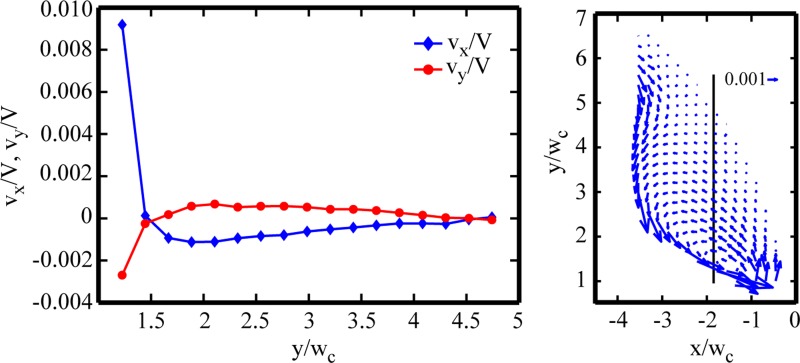
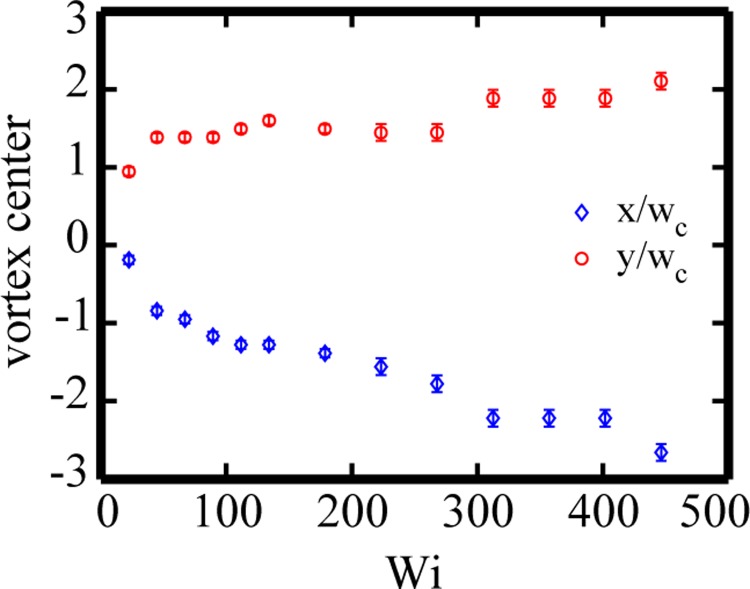
The normalized x- and y-velocity profiles for 400 μg/ml DNA flow (Wi = 268) at the x-position of the vortex center (x′ = −1.77) is given in Figure 11. The y-position of the vortex center is y′ = 1.45, where the normalized x- and y-velocities, vx/V and vy/V, both experience a zero velocity within the μPIV spatial resolution. Figure 12 shows the evolution of the normalized x- and y-positions of the vortex center with increasing Wi. The error bars correspond to the spatial resolution of the interrogation regions (normalized by the channel width) used to construct the velocity fields. With increasing Wi, the vortex center moves farther into the upstream reservoir, with shifts of greater magnitude in the axial direction compared with the transverse direction.
FIG. 11.
Normalized velocity profiles for 400 μg/ml DNA flow (Re = 2.4 × 10 − 2, Wi = 268, and El = 1.1 × 104) through the gradual contraction at the x-position of the vortex center (x′ = x/wc = −1.77, corresponding velocity field at right). Velocities are normalized by the average velocity in the downstream channel. The connected lines are to guide the eye.
FIG. 12.
Evolution of the vortex center for flows of 400 μg/ml DNA through the gradual contraction as a function of Wi. The error bars correspond to the spatial resolution of the interrogation regions (normalized by the channel width) used to construct the velocity fields.
It is hoped that the experimental velocity fields constructed in the vortex region demonstrate the use of μPIV for quantitative validation of computational design tools. Preliminary comparisons to simulations have been presented elsewhere (Nonaka et al., 2005; Trebotich et al., 2005).
IV. CONCLUSION
The small length scales and high shear rates characteristic of polymer flow in microfluidic structures enable access to very high elasticity numbers, which are unable to be explored at the macroscale. In this flow environment, high Weissenberg number and very low Reynolds number flows are simultaneously attained. DNA flows in a gradual micro-contraction geometry were characterized via experimental measurements of pressure drops, visualization of the flow kinematics, and velocity fields using μPIV.
The flow of semi-dilute (unentangled) and entangled DNA solutions of modest and high El respectively were investigated over a range of low Re (3.7 × 10−6 < Re < 3.1 × 10−1) and high Wi (0.4 < Wi < 446). The semi-dilute DNA flows are constant El = 55 and the entangled DNA solution flows probe the El range, 7.9 × 103 < El < 6.0 × 105. Preliminary flow visualization experiments indicated no vortex behavior for flows of the non-shear-thinning semi-dilute DNA solutions over the entire parameter range probed. For flows of the entangled DNA solutions, strong, steady, symmetric vortex growth is observed for increasing Re and Wi. The dimensionless vortex length χ is quantified as a function of Weissenberg number; it is well described by a power law of the form χ = 0.43(Wi − Wicrit)0.45 where Wicrit is 8.9. Dimensionless pressure drops Δp are also determined for the entangled DNA flows which reach a peak value of 2.75 at around Wi ∼ 155. Finally, velocity fields constructed using μPIV quantify the vortex dynamics from Newtonian-like flow at low Wi to the onset of vortex activity and dramatic growth with increasing Wi.
It is hoped that these direct measurements will be useful for forming comparisons with numerical simulation for optimization of lab-on-a-chip designs incorporating macromolecular flows. Additionally, these measurements of macromolecular flow in microfluidic geometries reveal the fundamental physics of this unique flow environment where the molecular lengths approach the critical device lengths (L/wc ∼ 0.13). The issue of molecular confinement and the influence of molecular concentration on flow are particularly relevant concerns for realistic design and operation of lab-on-a-chip applications incorporating macromolecular flows.
ACKNOWLEDGMENTS
The authors would like to acknowledge financial support from Laboratory Directed Research and Development (LDRD) Grant No. 03-ERI-003, of the U.S. Department of Energy, through Lawrence Livermore National Laboratory, and useful discussions with the Principal Investigator of the grant Dr. David Trebotich. The authors are also grateful to Dr. Polly Shrewsbury for fabrication of test devices. All fabrication work was completed at the Microfabrication Laboratory at the University of California, Berkeley.
References
- 1. Arratia, P. E. , Thomas, C. C. , Diorio, J. , and Gollub, J. P. , “ Elastic instabilities of polymer solutions in cross-channel flow,” Phys. Rev. Lett. 96, 144502 (2006). 10.1103/PhysRevLett.96.144502 [DOI] [PubMed] [Google Scholar]
- 2. Balan, C. M. , Broboana, D. , and Balan, C. , “ Investigations of vortex formation in microbirfurcations,” Microfluidics Nanofluidics 13, 819–833 (2012). 10.1007/s10404-012-1005-8 [DOI] [Google Scholar]
- 3. Burghelea, T. , Segre, E. , Bar-Joseph, I. , Groisman, A. , and Steinberg, V. , “ Chaotic flow and efficient mixing in a microchannel with a polymer solution,” Phys. Rev. E 69, 066305 (2004). 10.1103/PhysRevE.69.066305 [DOI] [PubMed] [Google Scholar]
- 4. Bustamante, C. , Marko, J. F. , Siggia, E. D. , and Smith, S. , “ Entropic elasticity of lambda-phage DNA,” Science 265, 1599–1601 (1994). 10.1126/science.8079175 [DOI] [PubMed] [Google Scholar]
- 5. Campo-Deano, L. , Galindo-Rosales, F. J. , Pinho, F. T. , Alves, M. A. , and Oliveira, M. S. N. , “ Flow of low viscosity Boger fluids through a microfluidic hyperbolic contraction,” J. Non-Newtonian Fluid Mech. 166, 1286–1296 (2011). 10.1016/j.jnnfm.2011.08.006 [DOI] [Google Scholar]
- 6. Evans, R. E. and Walters, K. , “ Flow characteristics associated with abrupt changes in geometry in the case of highly elastic liquids,” J. Non-Newtonian Fluid Mech. 20, 11–29 (1986). 10.1016/0377-0257(86)80013-1 [DOI] [Google Scholar]
- 7. Groisman, A. and Quake, S. R. , “ A microfluidics rectifier: Anisotropic flow resistance at low Reynolds numbers,” Phys. Rev. Lett. 92(9), 094501 (2004). 10.1103/PhysRevLett.92.094501 [DOI] [PubMed] [Google Scholar]
- 8. Gulati, S. , Dutcher, C. S. , Liepmann, D. , and Muller, S. J. , “ Elastic secondary flows in sharp 90 degree micro-bends: A comparison of PEO and DNA solutions,” J. Rheol. 54(2), 375–392 (2010). 10.1122/1.3308643 [DOI] [Google Scholar]
- 9. Gulati, S. , Muller, S. J. , and Liepmann, D. , “ Direct measurements of viscoelastic flows of DNA in a 2:1 abrupt planar micro-contraction,” J. Non-Newtonian Fluid Mech. 155, 51–66 (2008). 10.1016/j.jnnfm.2008.05.005 [DOI] [Google Scholar]
- 10. Hur, J. S. , Shaqfeh, E. S. G. , Babcock, H. P. , Smith, D. E. , and Chu, S. , “ Dynamics of dilute and semidilute DNA solutions in the start-up of shear flow,” J. Rheol. 45(2), 421–450 (2001). 10.1122/1.1339246 [DOI] [Google Scholar]
- 11. Jensen, K. E. , Szabo, P. , Okkels, F. , and Alves, M. A. , “ Experimental characterisation of a novel viscoelastic rectifier design,” Biomicrofluidics 6, 044112 (2012). 10.1063/1.4769781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lanzaro, A. and Yuan, X.-F. , “ Effects of contraction ratio on non-linear dynamics of semi-dilute, highly polydisperse PAAm solutions in microfluidics,” J. Non-Newtonian Fluid Mech. 166, 1064–1075 (2011). 10.1016/j.jnnfm.2011.06.004 [DOI] [Google Scholar]
- 13. Lanzaro, A. and Yuan, X.-F. , “ A quantitative analysis of spatial extensional rate distribution in nonlinear viscoelastic flows,” J. Non-Newtonian Fluid Mech. 207, 32–41 (2014). 10.1016/j.jnnfm.2014.03.005 [DOI] [Google Scholar]
- 14. LeDuc, P. , Haber, C. , Bao, G. , and Wirtz, D. , “ Dynamics of individual flexible polymers in a shear flow,” Nature 399, 564–566 (1999). 10.1038/21148 [DOI] [PubMed] [Google Scholar]
- 15. Li, Z. , Yuan, X.-F. , Haward, S. J. , Odell, J. A. , and Yeates, S. , “ Non-linear dynamics of semi-dilute polydisperse polymer solutions in microfluidics: A study of a benchmark flow problem,” J. Non-Newtonian Fluid Mech. 166, 951–963 (2011a). 10.1016/j.jnnfm.2011.04.010 [DOI] [Google Scholar]
- 16. Li, Z. , Yuan, X.-F. , Haward, S. J. , Odell, J. A. , and Yeates, S. , “ Non-linear dynamics of semi-dilute polydisperse polymer solutions in microfluidics: Effects of flow geometry,” Rheol. Acta 50, 277–290 (2011b). 10.1007/s00397-011-0539-0 [DOI] [Google Scholar]
- 17. Meinhart, C. D. , Wereley, S. T. , and Gray, M. H. B. , “ Volume illumination for two-dimensional particle image velocimetry,” Meas. Sci. Technol. 11(6), 809–814 (2000). 10.1088/0957-0233/11/6/326 [DOI] [Google Scholar]
- 18. Nigen, S. and Walters, K. , “ Viscoelastic contraction flows: Comparison of axisymmetric and planar configurations,” J. Non-Newtonian Fluid Mech. 102(2), 343–359 (2002). 10.1016/S0377-0257(01)00186-0 [DOI] [Google Scholar]
- 19. Nonaka, A. , Gulati, S. , Trebotich, D. , Miller, G. H. , Muller, S. J. , and Liepmann, D. , “ A computational model with experimental validation for DNA flow in microchannels,” in Proceedings of the 2005 NSTI Nanotechnology Conference and Trade Show (2005). [Google Scholar]
- 20. Oliveira, M. S. N. , Alves, M. A. , Pinho, F. T. , and McKinley, G. H. , “ Viscous flow through microfabricated hyperbolic contractions,” Exp. Fluids 43, 437–451 (2007). 10.1007/s00348-007-0306-2 [DOI] [Google Scholar]
- 21. Pan, S. , Nguyen, D. A. , Sridhar, T. , Sunthar, P. , and Prakash, J. R. , “ Universal solvent quality crossover of the zero shear rate viscosity of semidilute DNA solutions,” J. Rheol. 58(2), 339–368 (2014). 10.1122/1.4861072 [DOI] [Google Scholar]
- 22. Perkins, T. T. , Smith, D. E. , and Chu, S. , “ Single polymer dynamics in an elongational flow,” Science 276(5321), 2016–2021 (1997). 10.1126/science.276.5321.2016 [DOI] [PubMed] [Google Scholar]
- 23. Poole, R. J. and Alves, M. A. , “ Velocity overshoots in gradual contraction flows,” J. Non-Newtonian Fluid Mech. 160, 47–54 (2009). 10.1016/j.jnnfm.2009.03.005 [DOI] [Google Scholar]
- 24. Poole, R. J. , Escudier, M. P. , Afonso, A. , and Pinho, F. T. , “ Laminar flow of a viscoelastic shear-thinning liquid over a backward-facing step preceded by a gradual contraction,” Phys. Fluids 19, 093101 (2007). 10.1063/1.2769380 [DOI] [Google Scholar]
- 25. Poole, R. J. , Escudier, M. P. , and Oliveira, P. J. , “ Laminar flow of a viscoelastic shear-thinning liquid through a plane sudden expansion preceded by a gradual contraction,” Proc. R. Soc., London, Ser. A 461, 3827–3845 (2005). [Google Scholar]
- 26. Rodd, L. E. , Cooper-White, J. J. , Boger, D. V. , and McKinley, G. H. , “ Role of the elasticity number in the entry flow of dilute polymer solutions in micro-fabricated contraction geometries,” J. Non-Newtonian Fluid Mech. 143, 170–191 (2007). 10.1016/j.jnnfm.2007.02.006 [DOI] [Google Scholar]
- 27. Rodd, L. E. , Scott, T. P. , Boger, D. V. , Cooper-White, J. J. , and McKinley, G. H. , “ The inertio-elastic planar entry flow of low-viscosity elastic fluids in micro-fabricated geometries,” J. Non-Newtonian Fluid Mech. 129, 1–22 (2005). 10.1016/j.jnnfm.2005.04.006 [DOI] [Google Scholar]
- 28. Salamone, J. C. , Polymeric Materials Encyclopedia ( CRC Press, 1996), Vol. 3, p. 1941. [Google Scholar]
- 29. Sanger, F. , Coulson, A. R. , Hong, G. F. , Hill, D. F. , and Petersen, G. B. , “ Nucleotide sequence of bacteriophage λ-DNA,” J. Mol. Biol. 162(4), 729–773 (1982). 10.1016/0022-2836(82)90546-0 [DOI] [PubMed] [Google Scholar]
- 30. Shrewsbury, P. J. , Liepmann, D. , and Muller, S. J. , “ Concentration effects of a biopolymer in a microfluidic device,” Biomed. Microdevices 4(1), 17–26 (2002). 10.1023/A:1014263611236 [DOI] [Google Scholar]
- 31. Shrewsbury, P. J. , Muller, S. J. , and Liepmann, D. , “ Effect of flow on complex biological macromolecules in microfluidic devices,” Biomed. Microdevices 3(3), 225–238 (2001). 10.1023/A:1011415414667 [DOI] [Google Scholar]
- 32. Sousa, P. C. , Pinho, F. T. , Oliveira, M. S. N. , and Alves, M. A. , “ Extensional flow of blood analog solutions in microfluidic devices,” Biomicrofluidics 5, 014108 (2011). 10.1063/1.3567888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Trebotich, D. , Colella, P. , and Miller, G. , “ A stable and convergent scheme for viscoelastic flow in contraction channels,” J. Comput. Phys. 205(1), 315–342 (2005). 10.1016/j.jcp.2004.11.007 [DOI] [Google Scholar]
- 34. Verma, R. , Crocker, J. C. , Lubensky, T. C. , and Yodh, A. G. , “ Entropic colloidal interactions in concentrated DNA solutions,” Phys. Rev. Lett. 81(18), 4004–4007 (1998). 10.1103/PhysRevLett.81.4004 [DOI] [Google Scholar]