Abstract
Vascular endothelial growth factors (VEGFs) and their receptors (VEGF-R) are central regulators of vasculogenesis, angiogenesis, and lymphangiogenesis. They contribute to many vascular-related pathologies, and hence VEGF-targeted therapies have been widely sought after. In this study, the authors investigated the ability of promoter-targeted small hairpin RNAs (shRNAs) to regulate VEGF-A, VEGF-C and VEGF-R1 in different cell lines. The authors identified shRNAs that can upregulate hVEGF-C at both the mRNA and protein levels, and differentially regulate hVEGF-A depending on the cell type. Likewise, the authors identified shRNA that downregulated VEGF-R1 gene expression. Hence, promoter-targeted shRNAs can affect endogenous gene expression not only bimodally, but also differentially in a cell-type specific manner. Importantly, all three genes tested were regulated by at least one shRNA, supporting the idea that nuclear RNA interference is a widespread phenomenon. The level of regulation across the panel of shRNAs varied maximally from a 2.2-fold increase to a 4-fold decrease. This level of change should be useful in fine-tuning and modulating target gene expression, which for potent molecules, such as VEGF-A and VEGF-C, can be very beneficial. These promoter-targeted shRNAs may facilitate the design and development of targeted, context-dependent strategies for both pro- and antiangiogenic therapies for the treatment of vascular-related pathologies.
Keywords: lentiviral vector, promoter-targeted shRNA, RNA interference, RNA activation, RNAa, vasculogenesis, vascular endothelial growth factors
Introduction
Small RNAs (sRNA) targeting promoters and subsequently regulating gene expression were observed in plants1 and subsequently in mammalian cells.2 Although originally, inhibition of gene expression by the promoter-targeted sRNA was observed, soon after gene activation by these sRNA was demonstrated.3 Importantly, these sRNA have since then been shown to be active not only in vitro,3,4,5,6,7,8,9,10,11 but also in vivo in mouse models of hind-limb ischemia10 and myocardial infarction.12 In addition, a functional outcome of promoter-targeted regulation of tumor-related genes has been demonstrated in cancer models.13,14,15
The sequence requirements4,5,16 of activating RNAs have been characterized and their mechanisms of action have been determined to some extent,17 but these still need to be clearly defined. To date, downstream modifications of epigenetic marks have been observed, in particular histone modifications characteristic of the active marks.3,4,10,18 However, although DNA methylation of the targeted promoters was observed with silencing sRNA,7,19,20 changes in DNA methylation were not observed for activating sRNA.3 These sRNA have demonstrated some dependence on RNAi factors. In particular, they have been associated with a requirement for Argonaut (Ago), especially Ago23,5,6,10,21 and GW182.22 Furthermore, in some cases they have induced enrichment of RNA pol II at the activated promoters23,24 or have been associated with specific heterogeneous nuclear ribonucleoproteins.25 In other instances binding to an antisense transcript overlapping the activated promoter has been observed6,23,24 and proposed as a putative regulatory mechanism.
Although the mechanisms of promoter-targeted sRNA still need to be elucidated, the potential of these as therapeutic agents is obvious. Of particular interest is the activating sRNA, since only a few systems exist that can upregulate endogenous gene expression, in contrast to gene silencing which can be efficiently achieved by traditional RNAi and antisense technologies. Importantly, promoter-targeted sRNA can simultaneously regulate the expression of multiple isoforms generated from the targeted gene12 and thereby can maintain a better physiological balance of the isoforms. One of the genetic targets to which this type of regulation would be very useful is the vascular endothelial growth factor (VEGF) family of proteins.
The VEGF family consists of proteins which are critical for vascular growth and homeostasis. In particular, VEGF-A mediates angiogenesis and vasculogenesis,26 whereas VEGF-C regulates lymphangiogenesis.27 The biological effects of these factors are mediated via their receptors VEGF-R1, 2, and 3.28 Because of their important roles in vasculature, VEGFs are putative therapeutic targets for many different pathologies, such as ischemic diseases and vulnerable atherosclerotic plaques29 as well as cancer.30 However, the therapeutic requirements are different depending on the targeted disease, i.e., there is a need for both pro- and antiangiogenic therapies.31 The dichotomy of required therapies subsequently makes promoter-targeted sRNA an appealing strategy. In this study the authors' intention was to test the effectiveness of promoter-targeted short hairpin (sh)RNAs in the regulation of endogenous genes of the VEGF family, including VEGF-A, VEGF-C, and VEGF-R1. The authors' second intention was to test these shRNAs in a variety of cell types to address potential issues of cell specificity.3,10
Results
To study the regulation of promoter-targeted shRNAs on VEGF-A, VEGF-C or VEGF-R1 cells were transduced with viral-like particles encapsulating lentiviral vectors (LVs) encoding for specific shRNAs under the hU6 promoter and coexpressing enhanced green fluorescent protein (eGFP) under the human phosphoglycerate kinase-1 promoter. The transduction efficiency of the cells as indicated by eGFP expression was monitored by flow cytometry. The authors used MOI of 10–20 to achieve transduction efficiencies of ≥80%. There were no significant differences in the transduction efficiencies of different viral-like particles as compared with the control transductions in any of the cell types tested (data not shown).
Regulation of the VEGF-C gene
The regulation of the VEGF-C gene by promoter-targeted shRNAs was investigated and the authors identified one shRNA that upregulated VEGF-C (Figure 1). Specifically, shRNA516 upregulated hVEGF-C mRNA and protein levels in PC3 following 4 days of transduction and the secreted protein further accumulated up to day 7 (Figure 1a). Likewise, shRNA516, which has 100% homology with both the mouse and the human VEGF-C promoters, significantly increased VEGF-C mRNA expression in the human primary cells, human umbilical vein endothelial cells, and in the mouse endothelial cell line, C166 (Figure 1b). In addition, a mouse VEGF-C promoter sequence specific shRNA, shRNA348, significantly increased VEGF-C mRNA in C166 cells. Of note, expression of a scrambled shRNA (shCTRL) did not affect hVEGF-C mRNA and protein levels, as there were no differences between LV-GFP and LV-shCTRL (Figure 1a). In addition, hVEGF-A was measured in the 7-day media of PC3 samples, and no change in hVEGF-A was observed following hVEGF-C promoter-targeted shRNA treatment (data not shown), suggesting that the changes are specific for hVEGF-C.
Figure 1.
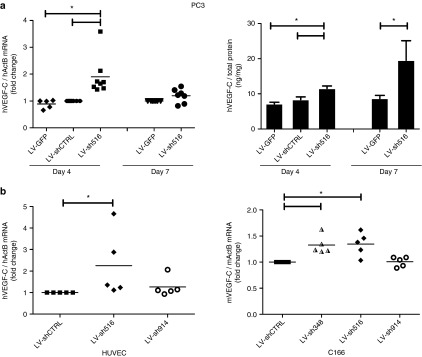
Promoter-targeted shRNA regulation of vascular endothelial growth factor (VEGF)-C. (a) Human PC3 were transduced with LV coding for shRNA targeting the VEGF-C promoter (LV-sh516, LV-sh914), or scrambled shRNA (LV-shCTRL) or LV not expressing a shRNA (LV-GFP). Cells and media were harvested following 4 and 7 days of transduction. The VEGF-C mRNA expression was determined by RT-qPCR (dotted diagrams) and the secreted VEGF-C protein was quantified by ELISA (bar diagrams). (b) Human umbilical vein endothelial cells (HUVECs) and mouse C166 were transduced with LV-shCTRL, LV-348, LV-sh516, or LV-sh914, and VEGF-C mRNA expression was determined by RT-qPCR following 7 days of transduction. Data in bar diagrams are presented as the mean ± SEM (n = 5–7) and were analyzed by analysis of variance (ANOVA) and Tukey's multiple comparison tests. Nonhomogenous data (mRNA) were analyzed by Kruskal–Wallis and Dunn's multiple comparison tests. Statistical significance (P < 0.05) between groups is indicated by (*). LV, lentiviral vector; shRNA, small hairpin RNA; VEGF, vascular endothelial growth factor.
Regulation of the VEGF-A gene
The regulation of VEGF-A gene by promoter-targeted shRNAs was investigated and in both PC3 and EAhy.926 cells activity of shRNAs targeting the VEGF-A promoter was observed. In EA.hy926 cells, hVEGF-A mRNA was significantly increased by shRNA1212 (Figure 2). In addition, there was a corresponding increase in the frequency and median of detectable secreted hVEGF-A protein in EA.hy926-conditioned media with shRNA1212. In PC3 cells, shRNA674 resulted in significantly reduced expression of hVEGF-A mRNA and protein (Figure 2).
Figure 2.
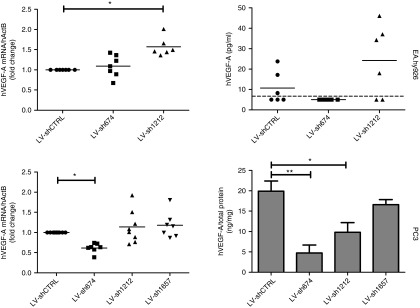
Promoter-targeted shRNA regulation of hVEGF-A. EA.hy926 and PC3 cells were transduced with LV coding for shRNA targeting the VEGF-A promoter (LV-sh674, LV-sh1212, and LV-sh1657), or scrambled shRNA (LV-shCTRL). Cells and media were harvested following 7 days of transduction. The hVEGF-A mRNA expression was determined by RT-qPCR (dotted diagrams) and the secreted hVEGF-A protein was quantified by ELISA (diagrams in the right panel). The dotted line across the graph for secreted hVEGF-A protein from EA.hy926 data indicates the sensitivity of the assay, points below this line indicate samples with undetectable hVEGF-A. In the bar diagram, the data are presented as the mean ± SEM (n = 6–8) and were analyzed by analysis of variance (ANOVA) and Tukey's multiple comparison tests. Nonhomogenous data (mRNA) were analyzed by Kruskal–Wallis and Dunn's multiple comparison tests. Statistical significance (P < 0.05) between groups is indicated by (*). LV, lentiviral vector; shRNA, small hairpin RNA; VEGF, vascular endothelial growth factor.
Regulation of the VEGF-R1 gene
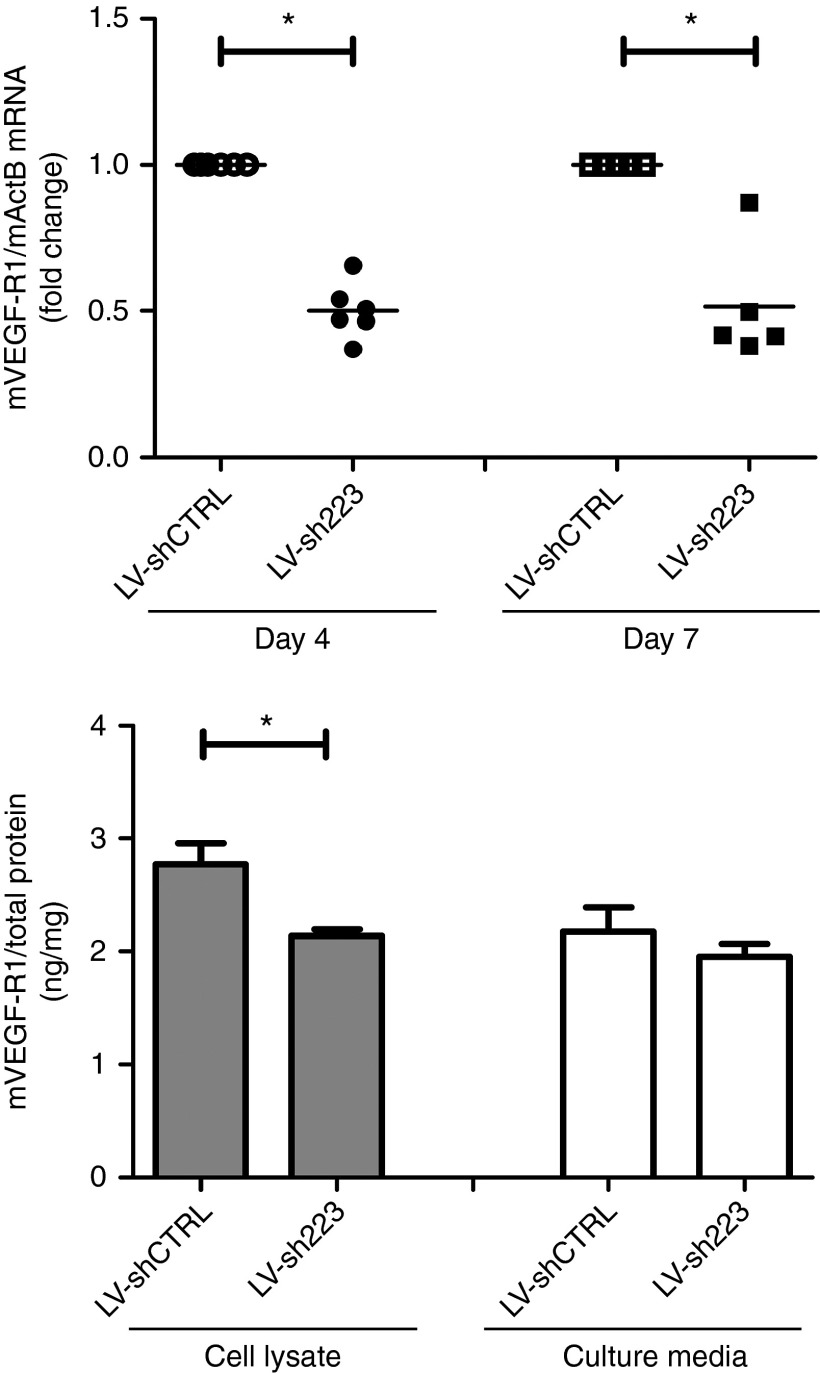
The regulation of VEGF-R1 expression was investigated, and the authors observed that shRNA223 downregulated VEGF-R1 mRNA expression in C166 (Figure 3). The VEGF-R1 mRNA was reduced by shRNA223 as early as 4 days after transduction and was maintained for at least 7 days in C166. The reduced VEGF-R1 mRNA corresponded to significantly lower cellular VEGF-R1 protein expression (Figure 3). In addition, shRNA223 downregulated hVEGF-R1 mRNA expression (0.6785 ± 0.044-fold change, n = 5) in EAhy.926 cells.
Figure 3.
Promoter-targeted shRNA regulation of VEGF-R1. Mouse C166 cells were transduced with LV coding for shRNA targeting the VEGF-R1 promoter (LV-sh223), or scrambled shRNA (LV-shCTRL). Cells and media were harvested following 4 and/or 7 days of transduction. The VEGF-R1 mRNA expression was determined by RT-qPCR (dotted diagrams) and the secreted or cellular VEGF-R1 protein (day 7) was quantified by ELISA (bar diagrams). Data in bar diagrams are presented as the mean ± SEM (n = 5–6) and were analyzed by analysis of variance (ANOVA) and Tukey's multiple comparison tests. Nonhomogenous data (mRNA) were analyzed by Kruskal–Wallis and Dunn's multiple comparison tests. Statistical significance (P < 0.05) between groups is indicated by (*).
Effect of combined treatment of activating and repressing shRNAs
To address the effects of coexpression of both activating and repressing shRNAs in the same cells, the authors tested previously published LV-shRNAs targeting mouse VEGF-A promoters, LV-451 and LV-856, which upregulate and downregulate, respectively, mouse VEGF-A mRNA in mouse C166 cells.10 C166 cells were cotransduced with both LV-451 and LV-856 (MOI 10), and mouse VEGF-A mRNA was measured after 4 days. Combined expression of both shRNA-451 and -856 resulted overall in an unchanged expression of mouse VEGF-A (fold change 1.22 ± 0.094, n = 3, P = 0.25) as compared to LV-shCTRL transduced cells. These data demonstrate that coexpression of activating and suppressing shRNAs can balance each other's effects on gene expression and suggest that shared cellular factors are not limiting, as both shRNAs were functional.
Effects of active shRNAs on histone methylation and acetylation of targeted promoters
Previously, promoter-targeted shRNA have been shown to affect histone modifications at the target site, and hence to delineate the mechanisms of regulation by promoter-targeted shRNA, the authors investigated the histone methylation and acetylation status of the genomic DNA at the targeted promoters. Figure 4 represents the chromatin immunoprecipitation (ChIP) analysis of these cells at the specific site of the targeting shRNA as well as an additional promoter site. Antibodies recognizing both active (H3K27ac, H4K5ac) and inactive (H3K27me3) histone markers were used in the ChIP analysis. The authors observed a consistent trend for a higher association of the activating markers in C166 and PC3 with activating shRNA at the shRNA target site but not at other promoter areas (Figure 4). In contrast, shRNA674 which suppressed hVEGF-A expression in PC3 cells (Figure 2) resulted in a reduced association of active histone mark H4K5ac at the targeted promoter site when compared with cells expressing control shRNA (Figure 4.). In addition, shRNA674 resulted in an increased association of H3K27me3 suppressing mark both at the targeted site and at other sites within the hVEGF-A promoter (Figure 4).
Figure 4.
ChIP analysis of VEGF-C and hVEGF-A promoters following activating shRNA treatment. Cells were transduced with LV-shRNAs either scrambled shRNA (shCTRL) or active shRNA (LVsh348, LVsh516, LVsh674, or LVsh1212) for 4 days, after which the chromatin was extracted, sheared by sonication, and immunoprecipitated using rabbit antibodies against H3K27me3, H3K27ac, and H4K5ac, or rabbit IgG. Quantitative PCR was performed using promoter site specific Taqman-based assays. The results were normalized with respect to input (% input). The data is presented as the mean ± SEM (n = 4–10). ChIP, chromatin immunoprecipitation; LV, lentiviral vector; shRNA, small hairpin RNA; VEGF, vascular endothelial growth factor.
Discussion
This study demonstrates differential regulation of multiple genes of the VEGF family by promoter-targeted shRNAs in various cell types. The authors demonstrated novel upregulation of VEGF-C and downregulation of VEGF-R1, in addition to dual regulation of hVEGF-A. Importantly, all three genes tested were regulated by at least one shRNA that had been designed to target their promoters. This supports the idea that nuclear RNA interference, as defined by promoter targeting, is a more general phenomenon associated with both upregulation and downregulation of the target genes and may be a more widespread concept than classical RNAi in the cytoplasm. Evidence is accumulating that factors involved in RNAi can be found in the nucleus,21,32,33 and hence nuclear generated shRNAs may be processed in the nucleus.
Importantly, in this study the authors demonstrated upregulation of gene expression from their endogenous promoters by the shRNAs. These promoter-targeted shRNAs may become useful tools for cell-type specific gene regulation in multicellular organisms. Although the mechanisms underlying the cell specificity of the shRNA activity remain to be elucidated, these may be multifactorial and may include variation in promoter methylation status, differential expression of noncoding RNAs or promoter miRNAs, or modulation of transcriptional regulatory complexes. In particular, DNA methylation has been observed to play a role in the activity of promoter-targeted sRNA.2,3 However, this effect was limited to shRNA inducing transcriptional gene silencing and not activation.12 In addition, differential expression of an antisense RNA or promoter RNA of the gene of interest may affect the shRNAs activity. Earlier, promoter-targeted sRNA modulation of Progesterone receptor,6,34 low-density lipoprotein,24 and cyclooxegenase-222 were linked to promoter-targeted sRNA binding of an antisense transcript or long noncoding RNA overlapping the promoters of these genes. These interactions were proposed as a putative mechanism of transcriptional regulation either by degradation of the promoter RNA or by interference with a putative scaffolding function.22 Alternatively, differential expression of promoter-targeting miRNAs may affect promoter-targeted shRNA activity.23 Recently, Ago-bound sRNA from proximal promoters (named TSS-miRNA) were found globally.35,36 Thus, promoter-targeted shRNAs may interfere with these promoter-specific miRNA targets to induce regulation.22,23 Furthermore, it was also shown that sRNA targeting cyclooxygenase-2 promoter could upregulate not only cyclooxygenase-2, but also phospholipase A2 potentially because of interrupted looping of chromosomal DNA by which these two genes are coregulated.22 This raises a possibility whether shRNA can in a similar fashion interfere with or augment enhancer activity by affecting its looping to the gene promoter(s).22,37,38
Overall, our study demonstrates regulation of members of the VEGF family by promoter-targeted shRNAs. The level of regulation was often moderate which in the case of hVEGF-A can be beneficial, because earlier hVEGF-A therapies tested in the clinics have been associated with unwanted edema because of the strong potency of this molecule.29 Hence, these promoter-targeted shRNAs can be used for fine-tuning or modulating target gene expression. Careful assessment of any particular promoter-targeted shRNA has to take place to define their cell specificity. However, they have a clear potential for cell-type specific, context-dependent therapeutic applications.
Materials and methods
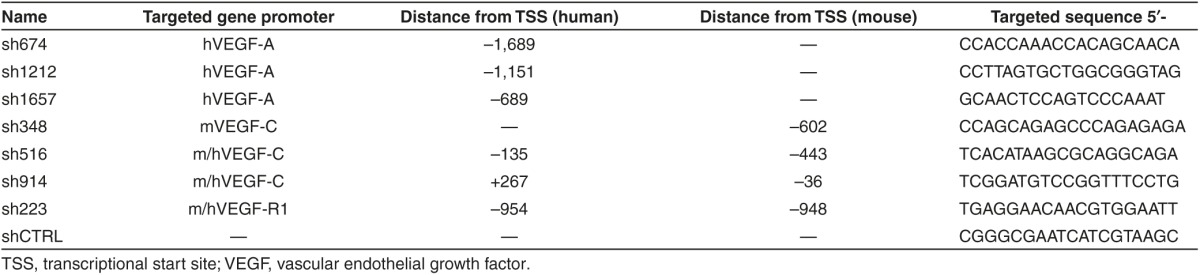
Vectors and lentiviral production. The promoter targeted shRNA sequences were designed using the Dharmacon siDESIGN Center algorithm (http://www.dharmacon.gelifesciences.com/design-center/). Specifically, of the algorithm-derived siRNA sequences, the authors chose those ones that did not overlap GC rich areas or known transcription factor binding sites in the promoter. The selected shRNA sequences with reference to the genes TSS and a mismatched control sequence are listed in Table 1. The shRNAs were cloned under the hU6 promoter in the context of the third generation HIV-1 derived LVs coexpressing eGFP under the human phosphoglycerate kinase-1 promoter [LV-hU6shRNAXXX-hPGK-GFP].39 The viral-like particles encapsulating the LV-hU6shRNAXXX-hPGK-GFP vectors were prepared using standard calcium phosphate transfection in 293T-cells as described earlier.39
Table 1. List of promoter-targeted small hairpin RNA (shRNAs) specified with their gene promoters, distance from TSS and sequence.
Cell culture and transductions. The following cell lines were used in this study: C166 (ATCC: CRL-2583), EA.hy926,39 and PC3. All cell lines were cultured in their specified media (RPMI for PC3 and DMEM for remaining cell lines; Sigma) supplemented with 10% fetal bovine serum, 100 units/ml penicillin and 100µg/ml streptomycin. Primary human umbilical vein endothelial cells were either isolated from umbilical cords obtained from the maternity ward of Kuopio University Hospital, Kuopio, Finland, with the approval of local Ethics Committee or purchased from Gibco (C-003-5C; Grand Island, NY). The cells were used at early passages and grown on 0.05% gelatine/10 μg/ml fibronectin (Sigma) coated plastic plates, in EBM endothelial cell basal medium supplemented with endothelial growth medium SingleQuots (Lonza).
Cells were seeded onto six-well plates the day before the experiment to achieve ~30% confluence. The cells were transduced with vectors encoding for the shRNAs at an MOI of 10–20 to achieve ≥80% transduction efficiency. The transduction efficiency was determined by quantification of eGFP expression by flow cytometry using FACSCalibre (Becton Dickinson, Franklin Lakes, NJ) with a 488-nm laser. The transduced cells were washed and split 1 : 3, 1 day after the transduction and maintained in culture till harvested either on day 4 or day 7. In each independent experiment (n = 5–14), the transductions for each LV-shRNA were performed in duplicate wells.
Reverse transcription and quantitative PCR. Total cellular RNA was extracted from cells with Tri-reagent (Molecular Research Center Inc, Cincinnati, OH) according to the manufacturer's instructions. First-strand cDNA synthesis, from 1 µg RNA, was performed using RevertAid reverse transcriptase (RT; 200 U; Fermentas, Vantaa, Finland) and random primers (0.5 µg; Promega, Madison, WI) in the presence of RevertAid RT buffer, RiboLock (20 U RNase inhibitor; Fermentas), and deoxynucleoside triphosphates (1 mM each; Fermentas). The RT reaction was carried out at 25°C for 10 minutes, followed by 42°C for 1 hour.
Real-time qPCR was used to quantify mRNA expression and chromosomal promoter DNA content (following immunoprecipiation; see ChIP) using Taqman gene expression assays (Table 2; Applied Biosystems, Grand Island, NY) and Taqman 2× PCR Master mix (Applied Biosystems). β-Actin mRNA levels were used as an endogenous control for normalization (mouse or human ACTB Endogenous Control FAM-MGB Probe, 4352933 and 4352935, respectively; Applied Biosystems). Real-time qPCR was carried out in an ABI STEP One Plus Prism 7700, and the data were analyzed with the corresponding software (Applied Biosystems).
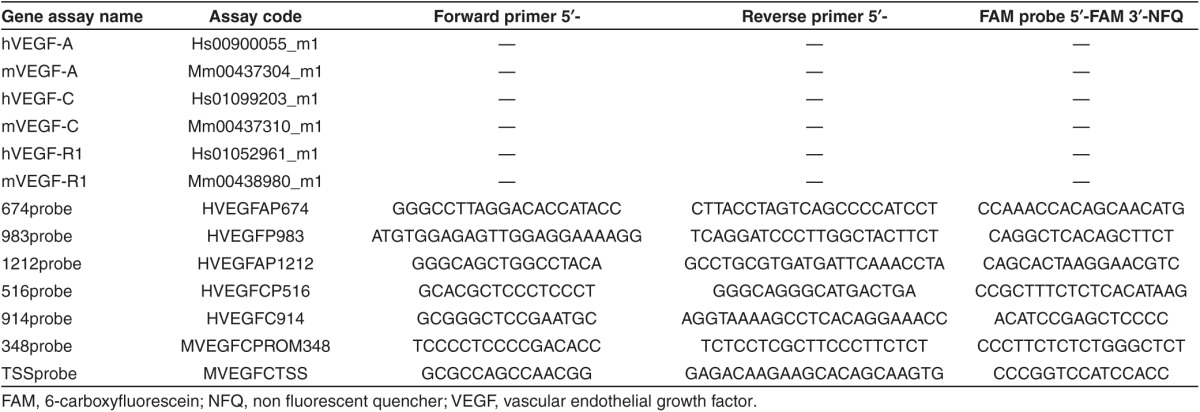
Table 2. List of Taqman assays for gene expression and chromatin immunoprecipitation analysis.
Protein analysis. Fresh cells or frozen cell pellets were lysed as described earlier40 in the presence of 1× complete protease inhibitor (Roche Diagnostics Oy, Espoo, Finland). The total protein content of the precleared cell lysates were quantified using the BCA Protein Assay kit (Pierce Biotechnology, Rockford, IL) according to the manufacturer's instructions.
The VEGF-A, VEGF-C, or VEGF-R1 content in cell lysates or conditioned media was quantified using enzyme-linked immunosorbent assays (ELISA) according to the manufacturer's instructions. Specifically, human VEGF-A and VEGF-C were assayed using relevant Quantikine ELISA kits (R&D Systems), the human and mouse VEGF-R1 were measured using DuoSet ELISA development kits (R&D Systems), and Mouse VEGF-C using mVEGF-C ELISA kit (MyBioSource, San Diego, CA).
ChIP assay. ChIP assays were performed as described earlier.41 The chromatin was sheared by sonication with a 30 second on/30 second off cycle for 15 minutes with Bioruptor resulting in 500–1000 bp chromatin fragments. Antibodies against trimethylated histone H3K27 (H3K27me3, 07-449), acetylated histone H3K27 (H3K27ac, 07-360) and acetylated histone H4K5 (H4K5ac, 07-327) as well as rabbit IgG serum (12–370) and Magna ChIP Protein A Magnetic beads were from Merck Millipore.
ChIP DNA samples were assayed by qPCR as described earlier. Results were normalized with respect to inputs as follows: derived ΔCt, which is a subtraction of the threshold cycles input from the IP samples [Ct(immunoprecipitated DNA) – Ct(input)] and then calculated fold change using the formula [2–(ΔCt)] and subsequently % input.
Statistics. Statistical analysis of the data was performed using GraphPad Prism (5.03; GraphPad Software, San Diego, CA). The data are presented as mean ± SEM unless otherwise stated. When the data conformed to homogeneity of variance, then either one-way analysis of variance (ANOVA) or two-tailed Student's t-test was performed; otherwise, nonparametric tests were used, including Kruskal–Wallis with Dunns post hoc test or Wilcoxon matched-pair signed rank test. The specific tests used for each data set are indicated in the figure legends. Statistical significance was ascribed at P < 0.05 and is indicated by (*).
Acknowledgments
The authors wish to thank the following people at (UEF): Joonas Malinen, Anne Martikainen, Maarit Mähönen, Anneli Miettinen, and Mervi Nieminen for technical support, as well as Mikko Turunen and Minna Kaikkonen for design of shRNAs and helpful discussions. This work was supported by a Finnish Cultural Foundation award (NLK) and grants from the Finnish Academy, Sigrid Juselius Foundation, ERC Advanced grant (SYH) and Kuopio University Hospital. The authors declare no conflict of interest.
References
- Mette MF, Aufsatz W, van der Winden J, Matzke MA, Matzke AJ. Transcriptional silencing and promoter methylation triggered by double-stranded RNA. EMBO J. 2000;19:5194–5201. doi: 10.1093/emboj/19.19.5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris KV, Chan SW, Jacobsen SE, Looney DJ. Small interfering RNA-induced transcriptional gene silencing in human cells. Science. 2004;305:1289–1292. doi: 10.1126/science.1101372. [DOI] [PubMed] [Google Scholar]
- Li LC, Okino ST, Zhao H, Pookot D, Place RF, Urakami S, et al. Small dsRNAs induce transcriptional activation in human cells. Proc Natl Acad Sci USA. 2006;103:17337–17342. doi: 10.1073/pnas.0607015103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janowski BA, Younger ST, Hardy DB, Ram R, Huffman KE, Corey DR. Activating gene expression in mammalian cells with promoter-targeted duplex RNAs. Nat Chem Biol. 2007;3:166–173. doi: 10.1038/nchembio860. [DOI] [PubMed] [Google Scholar]
- Place RF, Noonan EJ, Földes-Papp Z, Li LC. Defining features and exploring chemical modifications to manipulate RNAa activity. Curr Pharm Biotechnol. 2010;11:518–526. doi: 10.2174/138920110791591463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz JC, Younger ST, Nguyen NB, Hardy DB, Monia BP, Corey DR, et al. Antisense transcripts are targets for activating small RNAs. Nat Struct Mol Biol. 2008;15:842–848. doi: 10.1038/nsmb.1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins PG, Santoso S, Adams C, Anest V, Morris KV. Promoter targeted small RNAs induce long-term transcriptional gene silencing in human cells. Nucleic Acids Res. 2009;37:2984–2995. doi: 10.1093/nar/gkp127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang V, Qin Y, Wang J, Wang X, Place RF, Lin G, et al. RNAa is conserved in mammalian cells. PLoS One. 2010;5:e8848. doi: 10.1371/journal.pone.0008848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, Kelleher AD. Transcriptional regulation by promoter targeted RNAs. Curr Top Med Chem. 2009;9:1079–1087. doi: 10.2174/156802609789630875. [DOI] [PubMed] [Google Scholar]
- Turunen MP, Lehtola T, Heinonen SE, Assefa GS, Korpisalo P, Girnary R, et al. Efficient regulation of VEGF expression by promoter-targeted lentiviral shRNAs based on epigenetic mechanism: a novel example of epigenetherapy. Circ Res. 2009;105:604–609. doi: 10.1161/CIRCRESAHA.109.200774. [DOI] [PubMed] [Google Scholar]
- Voutila J, Sætrom P, Mintz P, Sun G, Alluin J, Rossi JJ, et al. Gene expression profile changes after short-activating RNA-mediated induction of endogenous pluripotency factors in human mesenchymal stem cells. Mol Ther Nucleic Acids. 2012;1:e35. doi: 10.1038/mtna.2012.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turunen MP, Husso T, Musthafa H, Laidinen S, Dragneva G, Laham-Karam N, et al. Epigenetic upregulation of endogenous VEGF-A reduces myocardial infarct size in mice. PLoS One. 2014;9:e89979. doi: 10.1371/journal.pone.0089979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Place RF, Wang J, Noonan EJ, Meyers R, Manoharan M, Charisse K, et al. Formulation of small activating RNA into lipidoid nanoparticles inhibits xenograft prostate tumor growth by inducing p21 expression. Mol Ther Nucleic Acids. 2012;1:e15. doi: 10.1038/mtna.2012.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang MR, Yang G, Charisse K, Epstein-Barash H, Manoharan M, Li LC. An orthotopic bladder tumor model and the evaluation of intravesical saRNA treatment. J Vis Exp. 2012;65:4207. doi: 10.3791/4207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren S, Kang MR, Wang J, Huang V, Place RF, Sun Y, et al. Targeted induction of endogenous NKX3-1 by small activating RNA inhibits prostate tumor growth. Prostate. 2013;73:1591–1601. doi: 10.1002/pros.22709. [DOI] [PubMed] [Google Scholar]
- Watts JK, Yu D, Charisse K, Montaillier C, Potier P, Manoharan M, et al. Effect of chemical modifications on modulation of gene expression by duplex antigene RNAs that are complementary to non-coding transcripts at gene promoters. Nucleic Acids Res. 2010;38:5242–5259. doi: 10.1093/nar/gkq258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris KV, Santoso S, Turner AM, Pastori C, Hawkins PG. Bidirectional transcription directs both transcriptional gene activation and suppression in human cells. PLoS Genet. 2008;4:e1000258. doi: 10.1371/journal.pgen.1000258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang K, Shen J, Xie YQ, Lin YW, Qin J, Mao QQ, et al. Promoter-targeted double-stranded small RNAs activate PAWR gene expression in human cancer cells. Int J Biochem Cell Biol. 2013;45:1338–1346. doi: 10.1016/j.biocel.2013.03.022. [DOI] [PubMed] [Google Scholar]
- Mette MF, Aufsatz W, Kanno T, Daxinger L, Rovina P, Matzke M, et al. Analysis of double-stranded RNA and small RNAs involved in RNA-mediated transcriptional gene silencing. Methods Mol Biol. 2005;309:61–82. doi: 10.1385/1-59259-935-4:061. [DOI] [PubMed] [Google Scholar]
- Janowski BA, Huffman KE, Schwartz JC, Ram R, Nordsell R, Shames DS, et al. Involvement of AGO1 and AGO2 in mammalian transcriptional silencing. Nat Struct Mol Biol. 2006;13:787–792. doi: 10.1038/nsmb1140. [DOI] [PubMed] [Google Scholar]
- Chu Y, Yue X, Younger ST, Janowski BA, Corey DR. Involvement of argonaute proteins in gene silencing and activation by RNAs complementary to a non-coding transcript at the progesterone receptor promoter. Nucleic Acids Res. 2010;38:7736–7748. doi: 10.1093/nar/gkq648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui M, Chu Y, Zhang H, Gagnon KT, Shaikh S, Kuchimanchi S, et al. Promoter RNA links transcriptional regulation of inflammatory pathway genes. Nucleic Acids Res. 2013;41:10086–10109. doi: 10.1093/nar/gkt777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Place RF, Li LC, Pookot D, Noonan EJ, Dahiya R. MicroRNA-373 induces expression of genes with complementary promoter sequences. Proc Natl Acad Sci USA. 2008;105:1608–1613. doi: 10.1073/pnas.0707594105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui M, Sakurai F, Elbashir S, Foster DJ, Manoharan M, Corey DR. Activation of LDL receptor expression by small RNAs complementary to a noncoding transcript that overlaps the LDLR promoter. Chem Biol. 2010;17:1344–1355. doi: 10.1016/j.chembiol.2010.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Chen Z, Xia D, Wu J, Xu H, Ye ZQ. Promoter-associated small double-stranded RNA interacts with heterogeneous nuclear ribonucleoprotein A2/B1 to induce transcriptional activation. Biochem J. 2012;447:407–416. doi: 10.1042/BJ20120256. [DOI] [PubMed] [Google Scholar]
- Ylä-Herttuala S, Rissanen TT, Vajanto I, Hartikainen J. Vascular endothelial growth factors: biology and current status of clinical applications in cardiovascular medicine. J Am Coll Cardiol. 2007;49:1015–1026. doi: 10.1016/j.jacc.2006.09.053. [DOI] [PubMed] [Google Scholar]
- Lohela M, Bry M, Tammela T, Alitalo K. VEGFs and receptors involved in angiogenesis versus lymphangiogenesis. Curr Opin Cell Biol. 2009;21:154–165. doi: 10.1016/j.ceb.2008.12.012. [DOI] [PubMed] [Google Scholar]
- Koch S, Tugues S, Li X, Gualandi L, Claesson-Welsh L. Signal transduction by vascular endothelial growth factor receptors. Biochem J. 2011;437:169–183. doi: 10.1042/BJ20110301. [DOI] [PubMed] [Google Scholar]
- Ylä-Herttuala S. Cardiovascular gene therapy with vascular endothelial growth factors. Gene. 2013;525:217–219. doi: 10.1016/j.gene.2013.03.051. [DOI] [PubMed] [Google Scholar]
- Samaranayake H, Määttä AM, Pikkarainen J, Ylä-Herttuala S. Future prospects and challenges of antiangiogenic cancer gene therapy. Hum Gene Ther. 2010;21:381–396. doi: 10.1089/hum.2010.017. [DOI] [PubMed] [Google Scholar]
- Vuorio T, Jauhiainen S, Ylä-Herttuala S. Pro- and anti-angiogenic therapy and atherosclerosis with special emphasis on vascular endothelial growth factors. Expert Opin Biol Ther. 2012;12:79–92. doi: 10.1517/14712598.2012.641011. [DOI] [PubMed] [Google Scholar]
- Ahlenstiel CL, Lim HG, Cooper DA, Ishida T, Kelleher AD, Suzuki K. Direct evidence of nuclear Argonaute distribution during transcriptional silencing links the actin cytoskeleton to nuclear RNAi machinery in human cells. Nucleic Acids Res. 2012;40:1579–1595. doi: 10.1093/nar/gkr891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagnon KT, Li L, Chu Y, Janowski BA, Corey DR. RNAi factors are present and active in human cell nuclei. Cell Rep. 2014;6:211–221. doi: 10.1016/j.celrep.2013.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui M, Prakash TP, Corey DR. Transcriptional silencing by single-stranded RNAs targeting a noncoding RNA that overlaps a gene promoter. ACS Chem Biol. 2013;8:122–126. doi: 10.1021/cb300490j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burroughs AM, Ando Y, de Hoon MJ, Tomaru Y, Suzuki H, Hayashizaki Y, et al. Deep-sequencing of human Argonaute-associated small RNAs provides insight into miRNA sorting and reveals Argonaute association with RNA fragments of diverse origin. RNA Biol. 2011;8:158–177. doi: 10.4161/rna.8.1.14300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamudio JR, Kelly TJ, Sharp PA. Argonaute-bound small RNAs from promoter-proximal RNA polymerase II. Cell. 2014;156:920–934. doi: 10.1016/j.cell.2014.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Laat W, Duboule D. Topology of mammalian developmental enhancers and their regulatory landscapes. Nature. 2013;502:499–506. doi: 10.1038/nature12753. [DOI] [PubMed] [Google Scholar]
- Ghavi-Helm Y, Klein FA, Pakozdi T, Ciglar L, Noordermeer D, Huber W, et al. Enhancer loops appear stable during development and are associated with paused polymerase. Nature. 2014;512:96–100. doi: 10.1038/nature13417. [DOI] [PubMed] [Google Scholar]
- Mäkinen PI, Koponen JK, Kärkkäinen AM, Malm TM, Pulkkinen KH, Koistinaho J, et al. Stable RNA interference: comparison of U6 and H1 promoters in endothelial cells and in mouse brain. J Gene Med. 2006;8:433–441. doi: 10.1002/jgm.860. [DOI] [PubMed] [Google Scholar]
- Melamed D, Mark-Danieli M, Kenan-Eichler M, Kraus O, Castiel A, Laham N, et al. The conserved carboxy terminus of the capsid domain of human immunodeficiency virus type 1 gag protein is important for virion assembly and release. J Virol. 2004;78:9675–9688. doi: 10.1128/JVI.78.18.9675-9688.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey MF, Peterson CL, Smale ST. Chromatin Immunoprecipitation (ChIP) CSHL Press; Cold Spring Harbor, NY; 2009. [DOI] [PubMed] [Google Scholar]