Abstract
Although interferon (IFN)-α is known to exert immunomodulatory and antiproliferative effects on dendritic cells (DCs) through induction of protein-coding IFN-stimulated genes (ISGs), little is known about IFN-α-regulated miRNAs in DCs. Since several miRNAs are involved in regulating DC functions, it is important to investigate whether IFN-α's effects on DCs are mediated through miRNAs as well. In this study, we examined miRNA expression patterns in myeloid DCs (mDCs) and plasmacytoid DCs after exposing them to IFN-α. We report that IFN-α downregulates miR-221 in both DC subsets via inhibition of STAT3. We validated proapoptotic proteins BCL2L11 and CDKN1C as miR-221 targets suggesting that IFN-α can induce DC apoptosis via miR-221 downregulation. In addition, we validated another miR-221 target, SOCS1, which is known to be a negative regulator of JAK/STAT signaling. Consistent with this, miR-221 overexpression in mDCs enhanced the secretion of proinflammatory cytokines IL-6 and TNF-α. In peripheral blood mononuclear cells (PBMCs) of HIV-1/HCV co-infected individuals undergoing IFN-α-based treatment the baseline miR-221 expression was lower in non-responders compared with responders; and miR-221 expression directly correlated with DC frequency and IL-6/TNF-α secretion. In addition to PBMCs, we isolated total liver cells and kupffer cells from HCV-infected individuals and individuals with alcoholic cirrhosis. We found that both total liver cells and kupffer cells from HCV-infected individuals had significantly higher miR-221 levels compared with individuals with cirrhosis. Overall, we demonstrate that IFN-α exerts both antiproliferative and immunomodulatory effects on mDCs via miR-221 downregulation; and IFN-miR-221 axis can play important role in HCV pathogenesis and treatment.
Introduction
Mammalian cells are equipped with several classes of pattern recognition receptors (PRRs) whose function is to sense the presence of different pathogens (Gilliet and others 2008). Upon engagement with pathogen-associated molecular patterns, PRRs trigger intracellular signaling cascades resulting in production of type I interferons (IFN-I), predominantly IFN-α/β . Most cell types produce IFN-β upon encountering pathogens, whereas hematopoietic cells, particularly plasmacytoid dendritic cells (pDCs) are the major producers of IFN-α (Jarrossay and others 2001; Liu 2005; Ivashkiv and Donlin 2014). IFN-α binds to a ubiquitously expressed heterodimeric transmembrane receptor, known as IFN-α receptor (IFNAR), and activates Janus kinase (JAK)-signal transducer and activator of transcription (STAT) pathway ultimately leading to transcription of IFN-stimulated genes (ISGs) that exhibit antiviral, immunomodulatory and antiproliferative activities (Isaacs and Lindenmann 1957; Perry and others 2005; Bieniasz and others 2011). In addition to protein coding ISGs, IFN-α can modulate the expression of cellular microRNAs (miRNAs) in various cell types (Pedersen and others 2007; Ohno and others 2009; Yang and others 2010; Tanaka and others 2012; Cheng and others 2013; Hao and others 2013; Parlato and others 2013; Zhang and others 2013) including those associated with the innate and adaptive immune system (Lodish and others 2008; O'Neill and others 2011; O'Connell and others 2012). Mature miRNAs act as important post-transcriptional regulators of gene expression by binding to and degrading/translationally repressing their target mRNAs (Nilsen 2007). It is becoming increasingly clear that miRNAs have profound effects on DC functions such as activation, maturation, cytokine secretion, and antigen presentation (Turner and others 2011; Busch and Zernecke 2012; Zhan and Wu 2012; Brain and others 2013; Karrich and others 2013; Kim and others 2013; Montagner and others 2013; Parlato and others 2013; Riepsaame and others 2013). Optimal DC functionality is a crucial requirement for successful resolution of persistent viral infections such as HIV-1 and HCV (Lambotin and others 2010; Sehgal and others 2013), and it is important to understand the molecular mechanisms that shape DC functions. Since IFN-α is known to regulate the maturation, migratory potential, and immunostimulatory capacity of DCs (Luft and others 1998; Ito and others 2001; Longhi and others 2009; Pantel and others 2014) we explored the possibility that these effects are mediated through modulation of miRNAs. Specifically, we examined changes in expression of 113 miRNAs in myeloid DCs (mDCs) and pDCs after in vitro treatment with IFN-α. These 113 miRNAs belonged to one or more of these 4 categories: (1) miRNAs known to directly regulate DC functions, (2) miRNAs known to regulate innate immune response and inflammatory signaling, (3) miRNAs whose expression is modulated by IFN-α in any cell type, (4) 84 most abundantly expressed and best characterized miRNAs in miRBase. We report that IFN-α downregulates miR-221 in both DC subsets. With the help of JAK1/2 inhibitor Ruxolitinib we confirmed that IFN-α-induced miR-221 downregulation in mDCs is mediated by JAK/STAT pathway. Since STAT3 has been shown to upregulate miR-221 expression in various cancer cells, we investigated its role in miR-221 regulation in mDCs. We observed that BP-1-102 (a STAT3 inhibitor) downregulates miR-221. Exposure of mDCs to IFN-α led to STAT1 activation but STAT3 inhibition suggesting that IFN-α-induced miR-221 downregulation is a result of STAT3 inhibition. Next, we validated proapoptotic proteins BCL2L11 and CDKN1C as miR-221 targets in mDCs, which interestingly suggested that IFN-α can induce mDC apoptosis via miR-221 downregulation. We also validated another miR-221 target, SOCS1, which is known to be a negative regulator of JAK/STAT signaling. Consistent with SOCS1 as miR-221 target, transfection of miR-221 mimic in mDCs enhanced the secretion of proinflammatory cytokines IL-6, and TNF-α. In peripheral blood mononuclear cells (PBMCs) isolated from HIV-1/HCV co-infected individuals on IFN-α-based therapy, pretreatment expression of miR-221 was significantly lower in non-responders (NRs) compared with responders and healthy controls. Also, miR-221 expression directly correlated with DC frequency but indirectly correlated with IL-6/TNF-α secretion. Finally, we demonstrate that total liver cells and kupffer cells (antigen-presenting cells in liver) isolated from HCV-infected individuals and individuals with alcoholic cirrhosis had significantly higher miR-221 levels.
Materials and Methods
Isolation of primary human immune cells
mDCs, pDCs, and B cells were isolated from PBMCs of anonymized healthy donors (Biological Specialty Corporation). PBMCs were isolated by density gradient centrifugation using Ficoll Paque Plus (GE Healthcare Life Sciences), washed in cell isolation buffer (DPBS without calcium and magnesium containing 0.5% BSA and 2 mM EDTA) and cleared of contaminating red blood cells using ACK lysis buffer (Quality Biological). PBMCs were washed 2× before purification of specific cell types. mDCs and pDCs were isolated using CD1c+ DC isolation kit (positive selection) and pDC isolation kit II (negative selection) respectively (Miltenyi Biotech). B cells were isolated in one of the intermittent steps (involving depletion of B cells) during the isolation of mDCs. After isolation, 5×105 cells were resuspended in 1 mL AIM-V cell culture media (Life Technologies) and cultured in 12-well cell culture plates. Cells were always rested for 1 h before proceeding with any experiment.
miRNA expression profiling of IFN-α-treated cells
Cells (mDCs, pDCs, and B cells) were treated with IFN-α (2,000 U/mL) for 24 h. After treatment, total RNA was isolated using the miRNeasy Mini kit (Qiagen) and miRNA expression profiling was performed using qPCR. cDNA required for qPCR was generated using miScript II RT Kit (SA Biosciences, Qiagen). Human miFinder miScript miRNA PCR Array (MIHS-001Z; SA Biosciences) consisting of predesigned forward primers (corresponding to each miRNA) was used to profile the expression of 84 most abundantly expressed and best characterized miRNAs in miRBase (listed in Supplementary Table S1; Supplementary Data are available online at www.liebertonline.com/jir). miScript SYBR Green PCR Kit (SA Biosciences) was used for amplifying cDNA during qPCR. Profiling of remaining miRNAs investigated in our study (listed in Supplementary Table S2) was performed using the same procedure except that we designed our own forward primers (containing the exact sequence as its corresponding miRNA except that the “U” in mature miRNA sequence is replaced with “T” in forward primer sequence). SNORD68 and SNORD95 were used as normalization controls to calculate fold change between untreated and treated cells. Roche LightCycler 480 Instrument II was used for all qPCR reactions.
Inhibition of JAK1/2 and STAT3 activity using small molecule inhibitors
To demonstrate that IFN-α downregulates miR-221 through JAK/STAT signaling, mDCs were pretreated with a selective small molecule JAK1/2 inhibitor, Ruxolitinib (1 μM; Selleckchem) for 3 h before IFN-α treatment. Similarly, dependency of IFN-α-induced miR-221 downregulation on STAT3 was demonstrated by treating cells with small molecule STAT3 inhibitor, BP-1-102 (10 μM; EMD Millipore) for 2 h before IFN-α treatment. BP-1-102 blocks STAT3 phosphorylation, dimerization, and DNA-binding activity. Percentage of DMSO (vehicle used to dissolve the inhibitors) in cell culture media was 0.2% and was confirmed to be nontoxic to cells.
Detection of IFN-α induced changes in pSTAT1 and pSTAT3 levels in mDCs
mDCs were treated with IFN-α for 15 min. Immediately after treatment, cells were fixed with BD Cytofix™ buffer for 10 min at 37°C followed by permeabilization with BD™ Phosflow Perm Buffer III for 30 min on ice. Cells were then washed twice in staining buffer (PBS with 2% heat inactivated FBS, 0.09% sodium azide, and 2 mM EDTA) and stained separately with PE mouse anti-STAT1 (pY701) antibody (Affymetrix eBioscience) and PE mouse anti-STAT3 (pY705) antibody (BD Biosciences) for 30 min at room temperature. After staining, cells were washed with staining buffer, resuspended in PBS, and analyzed on a BD FACSCalibur™ flow cytometery instrument.
Validation of miR-221-3p targets in mDCs
mDCs were transfected with mirVana miRNA miR-221-3p mimic (Ambion) using Lipofectamine RNAiMAX transfection reagent (Life Technologies). Briefly, 106 cells were plated in a 6-well cell culture treated plate at a density of 5×105 cells/mL and transfected with 30 nM of miR-221 mimic (Mature miRNA sequence: AGCUACAUUGUCUGCUGGGUUUC; miRBase Accession No. MIMAT0000278) or miRNA mimic negative control according to the manufacturer's protocol. cDNA required for qPCR-based mRNA quantitation was generated using QuantiTect Reverse Transcription kit (Qiagen). qPCR was performed using QuantiTect SYBR Green PCR kit (Qiagen). GAPDH was used as normalization control to calculate fold change between untreated and treated cells. To further validate miR-221 targets, we generated luciferase-target mRNA 3′UTR reporter constructs corresponding to each of the 6 mRNA targets and transfected mDCs with these constructs. Forty-eight hours post-transfection, we transfected mDCs with miR-221 mimic and measured the luciferase activity using dual Luciferase Assay (Promega) after 24 h.
Annexin V/7-AAD staining of mDCs
About 1×106 mDCs at the concentration of 5×105 cells/mL were treated with IFN-α for 48 h and then washed with DPBS (without calcium and magnesium) supplemented with 2% FBS, 2 mM EDTA, and 0.09% sodium azide. After washing, cells were stained with Annexin V-FITC and 7-AAD using Annexin V-FITC/7-AAD Apoptosis Detection Kit (BioLegend) according to manufacturer's directions. Briefly, mDCs were pelleted (300 g, 10 min) and resuspended in 100 μL Annexin V binding buffer. Next, 5 μL Annexin V-FITC and μL 7-AAD was added and cells were incubated for 15 min at room temperature. After incubation, 400 μL Annexin V binding buffer was added and cells were analyzed on a BD FACSCalibur flow cytometery instrument.
Th1/Th2/Th17 cytokines secreted by mDCs transfected with miR-221 mimic
Culture supernatants collected from IFN-treated cells were used to measure the concentration of a panel of 12 cytokines (IL-2, IL-4, IL-5, IL-6, IL-10, IL-12, IL-13, IL-17A, IFN-γ, TNF-α, G-CSF, and TGF-β1) using Human Th1/Th2/Th17 Cytokines Multi-Analyte ELISArray Kit (SA Biosciences, Qiagen) according to manufacturer's protocol. Briefly, antigen standards (500 ng/mL) corresponding to each of the 12 cytokines were prepared. Fifty microliters of assay buffer (supplied with the kit) was pipetted into each well (every well has a capture antibody specific to a cytokine) followed by addition of 50 μL antigen standards or undiluted culture supernatant in the appropriate wells. Plate was gently shaken and incubated for 2 h at room temperature. After 2 h, plates were decanted and washed thrice with the washing buffer (supplied with the kit) followed by incubation with biotin-conjugated detection antibodies for 1 h at room temperature and washed thrice thereafter. In the end, cytokines were detected colorimetrically by addition of Avidin-HRP solution followed by addition of substrate solution.
Ethics statement
A total of 18 HIV-1/HCV co-infected, IFN/Ribavirin treatment naïve individuals were recruited from the hepatitis clinic of Weill Cornell Medical College in a cohort study. The study was approved by the Institutional Review Board of Weill Medical College of Cornell University and conforms to the 1975 Helsinki guidelines. All study participants were adults and written informed consent was obtained from all of them. Inclusion criteria required that patients have detectable HCV RNA, be on a stable course of antiretroviral therapy or no antiretroviral agents for at least 4 weeks before treatment and to have a CD4+ T-cell count ≥100 cells/mm3. Patients were excluded if they had severe depression, immunodeficiency-related opportunistic infections, on active substance abuse, or were pregnant or lactating. Subjects were treated with 1.5 μg/kg of PEG-INTRON (Schering Plough) once weekly and a 1,000–1,200 mg total daily dose of RBV (Rebitrol; Schering Plough), taken every 12 h for up to 48 weeks. Individuals with HCV RNA below detection limit (<29 IU/mL) 24 weeks after treatment are considered sustained virological responders (SVRs), whereas those with detectable HCV RNA levels 24 weeks after treatment are considered as NRs.
Isolation of Kupffer cells from HCV-infected and alcoholic cirrhotic patients
We obtained total liver cells from HCV-infected and alcoholic cirrhotic patients from Emory University liver transplant center. For isolation of kupffer cells, we first isolated the mononuclear cell fraction using percoll density gradient centrifugation. From the mononuclear cells, CD14+ kupffer cells were isolated using positive selection with CD14+ positive isolation kit (StemCell).
Results
IFN-α downregulates miR-221-3p in mDCs and pDCs
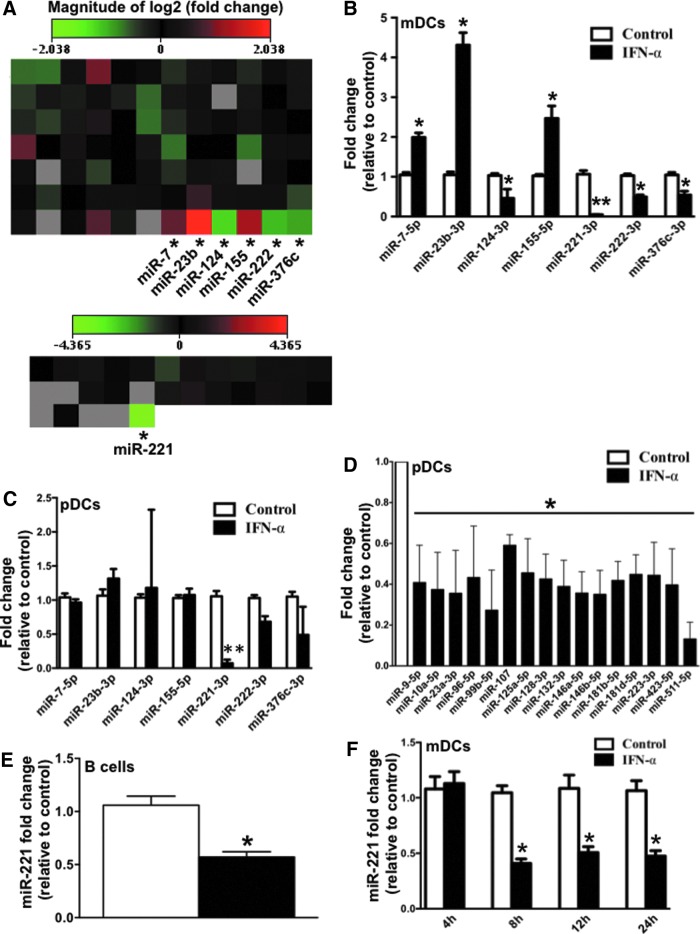
To determine whether IFN-α can regulate the expression of miRNAs in DCs we screened a library of 113 miRNAs consisting of most abundantly expressed and best-characterized miRNAs in miRBase (Supplementary Table S1) as well as miRNAs associated with innate immunity and DC functions (Supplementary Table S2). DCs were treated with IFN-α-2a (2,000 U/mL) for 24 h, following which, the IFN-induced changes in expression of 113 miRNAs were measured using miScript miRNA qPCR assay (SA Biosciences). Fold-change was calculated using 2 small nucleolar RNAs SNORD68 and SNORD95 as normalization controls. Figure 1A shows heat map representing changes in expression of 113 miRNAs in mDCs (n=2) upon IFN-α treatment. A cutoff fold change of 1.5 was used to screen potential IFN-α-regulated miRNAs and 7 miRNAs: miR-7-5p, miR-23b-3p, miR-155-5p, miR-124-3p, miR-221-3p, miR-222-3p, and miR-376c-3p were identified as potential IFN-α-regulated miRNAs. IFN-α-induced changes in these 7 miRNAs were reconfirmed in mDCs (n=3) (Fig. 1B). We found that miR-7-5p, miR-23b-3p and miR-155-5p were upregulated, whereas miR-124-3p, miR-221-3p, miR-222-3p, and miR-376c-3p were downregulated in response to IFN-α (Fig. 1B). Exact strategy was employed to investigate IFN-α-regulated miRNAs in another DC subset, pDCs. Out of the 7 miRNAs that were significantly modulated in mDCs, only miR-221 downregulation was observed in pDCs as well (Fig. 1C). Other than miR-221, 16 more miRNAs were downregulated in pDCs in response to IFN-α (Fig. 1D). This shows that pDCs, major producers of IFN-α, are highly responsive to IFN-α-induced changes in miRNAs. It is possible that IFN-α induces global downregulation of miRNAs in pDCs by inhibiting the miRNA biogenesis pathway. In fact there is evidence that IFN-α can repress the expression and activity of Dicer, RNAse III enzyme that plays an important role in miRNA biogenesis. IFN-α-induced miR-221 downregulation was also confirmed in B cells, another important professional antigen-presenting cell (Fig. 1E). We also studied the kinetics of IFN-induced miR-221 downregulation and found that miR-221 downregulation began as early as 8 h and persisted until 24 h (Fig. 1F). These studies establish that IFN-α downregulates miR-221 expression in 3 of the most important antigen-presenting cells: mDCs, pDCs, and B cells. In subsequent experiments, we decided to focus on the mechanism of miR-221 downregulation and its impact on mDCs.
FIG. 1.
Interferon (IFN)-α induces downregulation of miR-221-3p in dendritic cells (DCs). (A) Heat map representing mean fold change in expression of 113 miRNAs (84 miRNAs profiled using Human miFinder miScript miRNA qPCR Array, SA Biosciences plus 29 miRNAs whose qPCR primers were designed according to similar strategy as miFinder array) in myeloid dendritic cells (mDCs) (n=2) upon 24 h IFN-α treatment. Asterisk (*) indicates miRNA with greater than 1.5-fold upregulation/downregulation in both biological replicates. (B) Bar graph indicating mean fold change in expression of 7 miRNAs (which were marked with asterisk in the previous graph) in mDCs (n=3). (C) Bar graph indicating mean fold change in expression of same 7 miRNAs in plasmacytoid dendritic cells (pDCs) (n=3) upon 24 h IFN-α treatment. (D) Out of the remaining 116 miRNAs profiled in pDCs, 16 miRNAs showed significant downregulation upon 24 h IFN-α treatment. Bar graph indicates the mean fold-downregulation of those 16 miRNAs. (E) Bar graph indicating mean fold change in expression of miR-221 in B cells upon 24 h IFN-α treatment. (F) Bar graph indicating the kinetics of IFN-α-induced downregulation of miR-221 in mDCs. P values were calculated using Student's t test (*P<0.05; **P<0.01). Error bars represent standard deviation (n=3). Color images available online at www.liebertpub.com/jir
IFN-α-induced miR-221 downregulation is mediated by JAK/STAT pathway
IFN-α activates multiple pathways, including the canonical JAK/STAT signaling pathway, and non-canonical signaling cascades such as those involving p38 mitogen-activated protein kinase and phosphatidylinositol 3-kinase. To determine whether the JAK/STAT pathway is involved in the IFN-α-induced downregulation of miR-221, we pretreated mDCs with Ruxolitinib, a small molecule JAK1/2 inhibitor (Heine and others 2013), before IFN-α treatment. JAK1/2 inhibition prevented miR-221 downregulation in response to IFN-α, whereas Ruxolitinib treatment alone had no significant effect on miR-221 levels (Fig. 2A). It is worth noting that patterns of ISGs can vary from one cell type to another. This is because IFN-α can differentially activate various STAT family members, each having distinct target genes (ISGs) and biological functions. Various studies have suggested that relative expression of various STAT family members (STAT1, 2, 3, 4, 5A, 5B, and 6) can vary from one cell type to another and can dictate which STAT family member gets predominantly activated in response to IFN-α (Ho and Ivashkiv 2006; Kallal and Biron 2013; Ivashkiv and Donlin 2014). Therefore, relative expression of STATs can be an important determinant of the pattern of ISGs induced upon IFN-α signaling. In light of this, we profiled the basal mRNA levels of all STAT family members in mDCs to determine whether the expression of one or more STATs was significantly different from the rest. Our data show that the mRNA expression of all STATs was comparable in mDCs (Fig. 2B). This suggests that relative expression of STATs may not be playing an important role in activation of one STAT member versus another. Although there are studies that have shown that type I IFNs can induce the expression of STAT1 in PBMCs and macrophages (Lehtonen and others 1997), little is known about IFN-induced STATs in human mDCs. Through our profiling of mRNA expression levels of all STATs in IFN-α-treated mDCs, we were able to confirm that IFN-α can induce STAT1 in mDCs as well. In addition, we were able to show that IFN-α induces STAT4 in mDCs. In future, it will be important to investigate the functional consequences of STAT1 and STAT4 induction in mDCs in response to IFN-α. In summary, we were able to establish that IFN-α-induced miR-221 downregulation is dependent on activation of the JAK/STAT pathway; and IFN-α can induce STAT1 and STAT4 expression in mDCs.
FIG. 2.
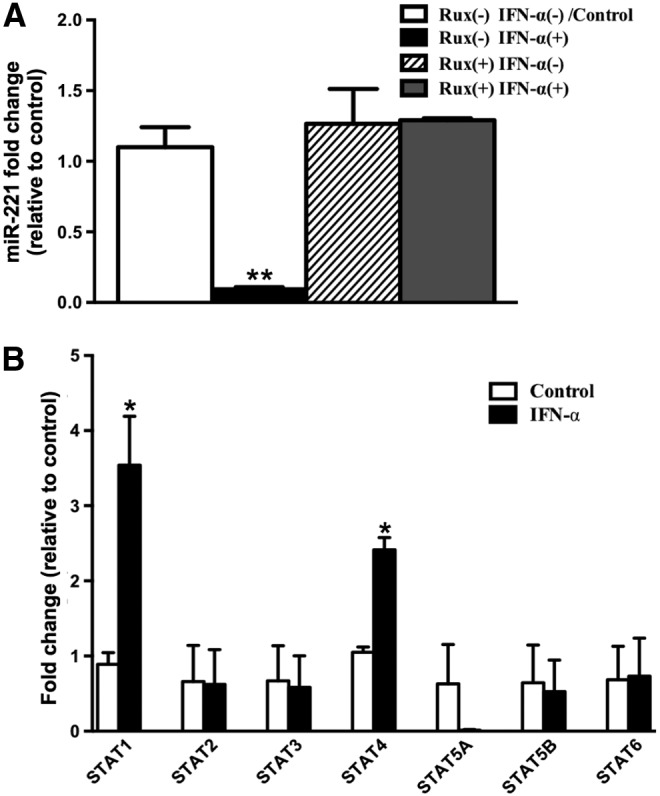
IFN-α-induced miR-221 downregulation is mediated by JAK/STAT pathway. (A) Bar graph indicating mean miR-221-fold change in mDCs upon 3 h pre-treatment with JAK1/2 inhibitor, Ruxolitinib (Selleckchem) followed by 24 h IFN-α treatment. The graph shows 4 bars (white: untreated with Ruxolitinib, untreated with IFN-α; black: untreated with Ruxolitinib, but treated with IFN-α; stripes: treated with Ruxolitinib, but untreated with IFN-α; gray: treated with Ruxolitinib, treated with IFN-α). (B) Bar graph indicating mean fold change in expression of 7 STAT family members (STAT1, 2, 3, 4, 5A, 5B, and 6) in mDCs upon 24 h IFN-α treatment. P values were calculated using Student's t test (*P<0.05; **P<0.01). Error bars represent standard deviation (n=3).
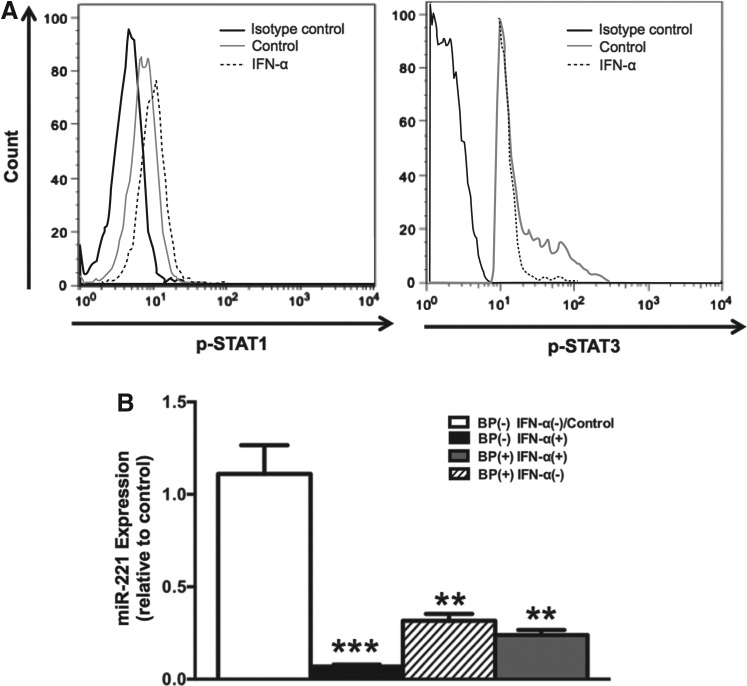
Role of STAT3 in IFN-α-induced miR-221 downregulation
STAT3 has been shown to upregulate miR-221 expression in various cancer cells (Kneitz and others 2014; Liu and others 2014). To determine whether IFN-α-induced miR-221 downregulation is mediated by STAT3, we first investigated the effect of IFN-α on STAT3 phosphorylation. As expected, we observed that IFN-α upregulated pSTAT1 levels (Fig. 3A); however, it downregulated pSTAT3 levels (Fig. 3A). We next wanted to determine whether IFN-α-induced downregulation of miR-221 is STAT3 dependent. For this, we pretreated mDCs with BP-1-102 (a small molecule inhibitor that blocks STAT3 phosphorylation, dimerization, and DNA-binding activity) before IFN-α treatment. Interestingly, we found that BP-1-102 treatment alone (without subsequent IFN-α treatment) downregulated miR-221 (Fig. 3B) indicating that STAT3 is essential for maintaining steady state levels of miR-221. We saw a similar downregulation when we pretreated mDCs with BP-1-102 before IFN-α treatment (Fig. 3B). Together, our data indicate that IFN-α-induced miR-221 downregulation is mediated via STAT3 inhibition.
FIG. 3.
Role of STAT3 in IFN-α-induced miR-221 downregulation. (A) Freshly isolated mDCs were rested for 1 h followed by 15-min IFN-α treatment. After treatment, pSTAT1 and pSTAT3 levels were measured by flow cytometry. The graph shows control mDCs (no IFN-α treatment) in dark gray histogram and IFN-α-treated mDCs in dashed lines histogram. Black histogram shows mDCs stained with isotype (IgG-2a κ) control antibody. (B) Bar graph indicating mean fold change in miR-221 expression in mDCs pre-treated with STAT3 inhibitor, BP-1-102 (2 h) before exposing them to IFN-α (24 h). The graph shows 4 bars, white: untreated with BP-1-102, untreated with IFN-α; black: untreated with BP-1-102, but treated with IFN-α; stripes: treated with BP-1-102, but untreated with IFN-α; gray: treated with BP-1-102, treated with IFN-α. P values were calculated using Student's t test (**P<0.01; ***P<0.001). Error bars represent standard deviation (n=3).
Validation of miR-221 targets in mDCs
To more clearly define the role of IFN-α-induced miR-221 downregulation in mDCs, it is important to identify miR-221 targets. BBC3, BCL2L11, BNIP3, CDKN1B, CDKN1C, DDIT4, ESR1, FOS, FOXO3, ICAM1, PTEN, TBK1, and SOCS1 are miR-221 targets experimentally validated in other cell types (Galardi and others 2007; Fornari and others 2008; Zhao and others 2008; Garofalo and others 2009; Gramantieri and others 2009; Chun-Zhi and others 2010; Hu and others 2010; Pineau and others 2010; Zhang and others 2010; Lu and others 2011) [except CDKN1C, which is validated in monocyte-derived DCs (Lu and others 2011)] and we decided to validate all the above genes plus few bioinformatically predicted genes as miR-221 targets in mDCs. Cells were transfected with mirVana miR-221 mimic or a negative control mimic followed by quantitation of target mRNAs 24 h post-transfection (Supplementary Fig. S1 shows the extent of miR-221 overexpression in our system). Introduction of miR-221 mimic resulted in downregulation of all but 4 of the selected mRNAs, including a novel target RIG-I that was identified using in silico approaches (Fig. 4A). Next, we further validated BBC3, BCL2L11, CDKN1C, SOCS1, ICAM1, and RIG-I (mRNAs whose expression was reduced the most after transfection of miR-221 mimic) as miR-221 targets using luciferase-3′UTR reporter plasmid constructs. Out of the 6 mRNAs, BCL2L11, CDKN1C, and SOCS1 were finally validated as miR-221 targets (Fig. 4B).
FIG. 4.
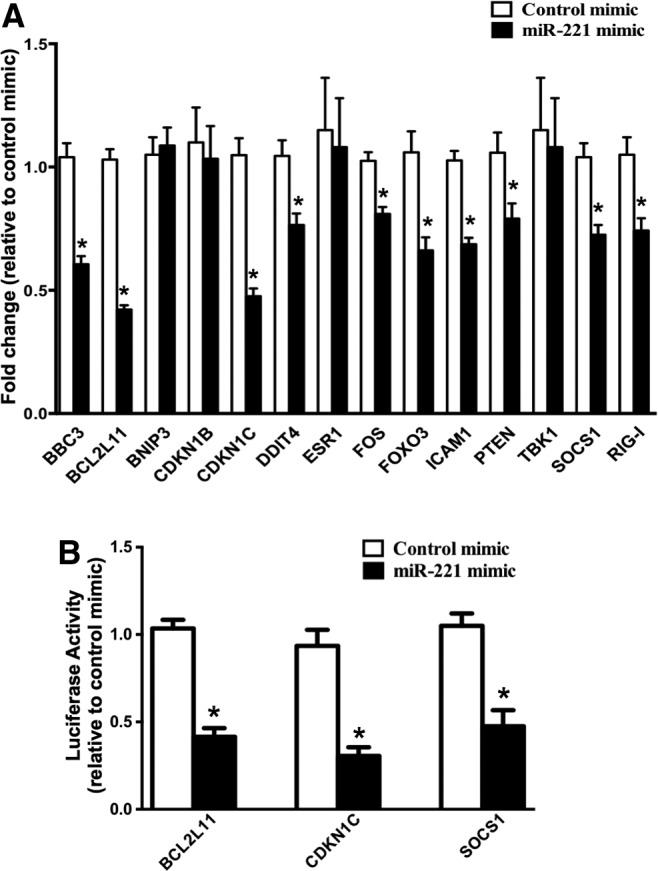
Validation of miR-221 targets in mDCs. (A) mDCs were transfected with mirVana miR-221 mimic or negative control mimic. Bar graph indicates mean fold change in mRNA levels of potential miR-221 targets in mDCs transfected with miR-221 mimic relative to mDCs transfected with negative control mimic. (B) Luciferase-target mRNA 3′UTR reporter constructs corresponding to each potential miR-221 target were generated and mDCs were transfected with these constructs (using Lipofectamine LTX). Forty-eight hours post-transfection, mDCs were transfected with miR-221 mimic and luciferase activity was measured after 24 h. Bar graph indicates the mean luciferase activity relative to controls. (*P<0.05). Error bars represent standard deviation (n=3).
Effect of miR-221 on apoptosis and cytokine secretion profile of mDCs
Since BCL2L11 and CDKN1C are proapoptotic proteins, we hypothesized that IFN-α-induced miR-221 downregulation leads to accumulation of BCL2L11 and CDKN1C in mDCs resulting in their apoptosis. We further explored the verity of this hypothesis by investigating the effect of miR-221 mimic on the expression of various apoptosis-associated ISGs. Out of 35 apoptosis-associated ISGs that we investigated, we observed that miR-221 mimic downregulated CAV1, MX1, SHB, and TNFSF10 (Fig. 5A) suggesting that IFN-α can induce apoptosis in mDCs by downregulating miR-221. We verified the proapoptotic effect of IFN-α on mDCs by performing Annexin V/7-AAD staining of mDCs following IFN-α treatment. Our results (Fig. 5B) show that IFN-α treatment of mDCs results in increased apoptosis compared with control (5.55% versus 2.15% Annexin V+/7-AAD+ cells, as well as 15.4% versus 6.96% Annexin V+ cells). In addition, because SOCS1 (another miR-221 target validated in our study, Fig. 4B) is capable of inhibiting JAK/STAT pathway thereby regulating the expression of various cytokines, we evaluated the Th1/Th2/Th17 cytokine secretion profile of mDCs after transfecting them with miR-221 mimic. We found that transfection of mDCs with miR-221 mimic enhanced their IL-6 and TNF-α secretion (Fig. 5C).
FIG. 5.
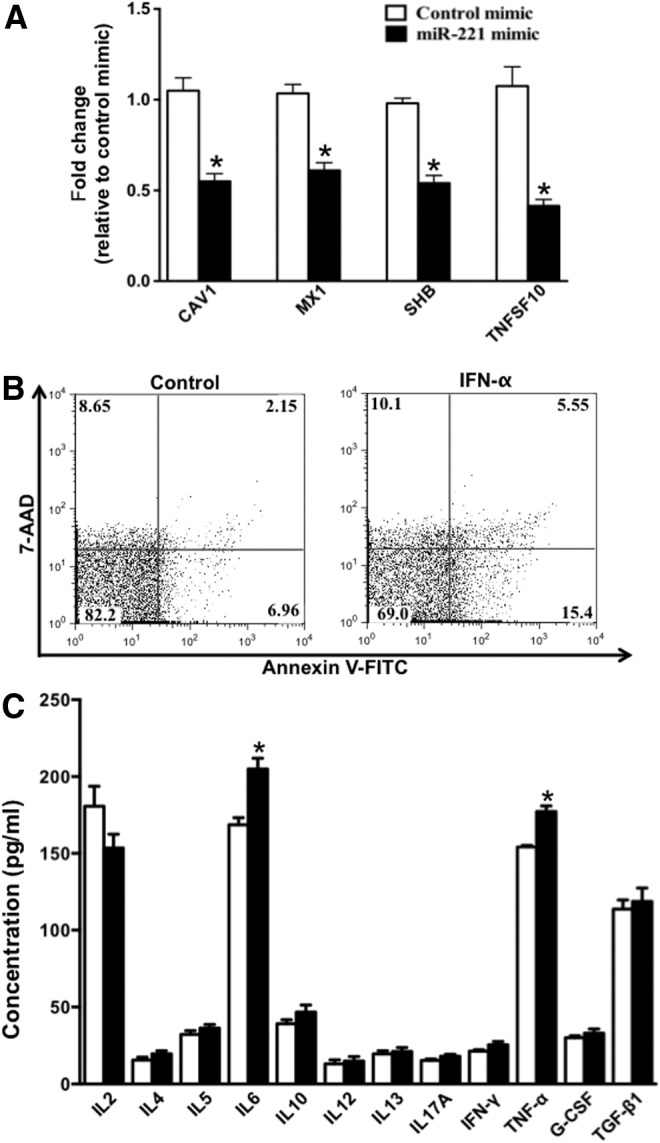
Effect of miR-221 on apoptosis and cytokine secretion profile of mDCs. (A) Bar graph indicating the mean fold change in expression of 4 proapoptotic ISGs in mDCs transfected with miR-221 mimic. (B) mDCs were treated with IFN-α for 48 h and then stained with Annexin V and 7-AAD. After staining, cells were analyzed using flow cytometry. Figure shows the dot plot indicating the fluorescent intensity of Annexin V-FITC and 7-AAD. (C) Bar graph indicating the mean concentration (pg/mL) of a panel of 12 Th1/Th2/Th17 cytokines secreted by mDCs transfected with miR-221 mimic. P values were calculated using Student's t test (*P<0.05). Error bars represent standard deviation (n=3).
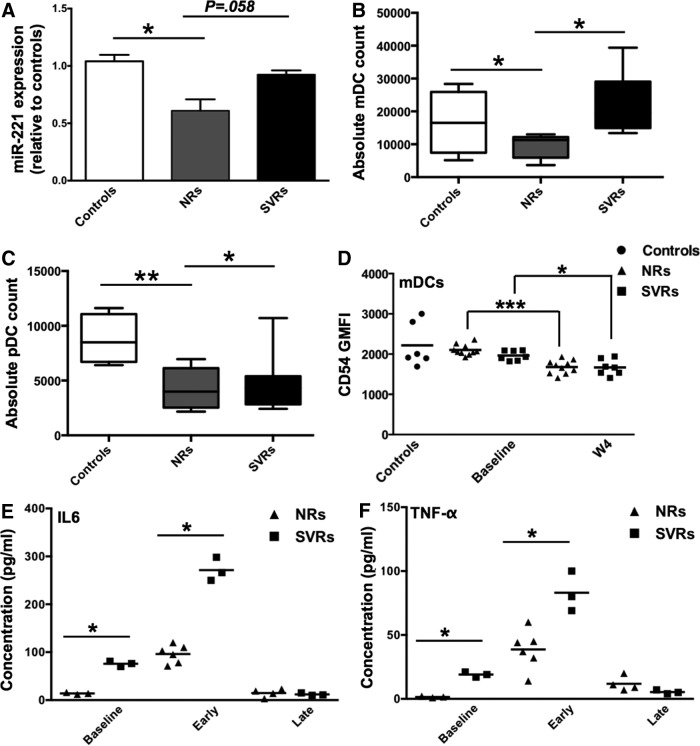
Role of miR-221 in IFN/Ribavirin treatment response against HCV
Thus far, we have established that IFN-α is capable of modulating miR-221 expression in mDCs through a STAT3-dependent mechanism. In addition, we have validated miR-221 targets: BCL2L11, CDKN1C, and SOCS-1 in mDCs and demonstrated that miR-221 overexpression in mDCs leads to enhanced secretion of IL-6 and TNF-α. Next, we were interested in evaluating how IFN-miRNA axis in DCs is affected during chronic infections such as HIV-1 and HCV. miRNAs are being increasingly recognized to play an important role in anti-HCV response and clinical outcome of standard IFN-based HCV treatment. In light of these observations, we utilized a well-characterized cohort of HIV-1/HCV co-infected patients (Supplementary Table S3) to investigate whether miR-221 is differentially regulated in SVRs and NRs of standard IFN/Ribavirin treatment. First, we determined baseline (pre-IFN/Ribavirin treatment) miR-221 expression in PBMCs of seronegative controls, NRs and SVRs. We found that NRs had lower miR-221 baseline levels compared with controls (Fig. 6A). We also profiled the expression of 84 genes associated with IFN response (using SA Biosciences Type I IFN response qPCR array) and identified certain signatures associated with treatment outcome (Supplementary Figs S2 and 3). Next, since NRs were characterized by lower miR-221 expression compared with controls and SVRs, we investigated how it affected their frequency of mDCs and pDCs. We observed that NRs had reduced numbers of mDCs and pDCs compared with controls and SVRs (Fig. 6B, C). We also monitored the expression of CD54 (miR-221 target that showed only moderate degradation/translational repression by miR-221) on mDCs of seronegative controls, NRs and SVRs. CD54 is a DC adhesion molecule that plays an important role in DC-T cells interaction. CD54 expression can thus be an important predictor of DC functionality. Although there were no significant differences in CD54 expression between NRs and SVRs, both at baseline and week 4 of treatment, it is important to note that by week 4 of IFN/RBV treatment, CD54 expression was significantly downregulated (although the extent of downregulation was greater in NRs) in both NRs and SVRs (Fig. 6D). This necessitates the need to profile miR-221 expression in mDCs (instead of PBMCs) of NRs and SVRs and directly correlate that to CD54 expression. Finally, we isolated PBMCs from NRs and SVRs at week 0, early (first 4 weeks of the 48-week treatment), and late (last 2 weeks of the 48-week treatment) and evaluated their Th1/Th2/Th17 cytokine secretion profile in response to in vitro IFN-α treatment. We found that SVRs secrete higher amounts of IL-6 and TNF-α, which is consistent with their higher miR-221 levels. These results show that lower miR-221 expression in PBMCs of NRs directly correlates with their mDC and pDC frequency in addition to IL-6 and TNF-α secretion.
FIG. 6.
Role of miR-221 in IFN/Ribavirin treatment response against HCV. (A) Peripheral blood mononuclear cell (PBMCs) were isolated from seronegative controls (n=6) and non-responders (NRs) (n=10) and sustained virological responders (SVRs) (n=7) at week 0 (baseline) of IFN/Ribavirin treatment. Bar graph indicates mean fold change in miR-221 levels in NRs (gray bar) and SVRs (black bar) relative to seronegative controls (white bar). P values were calculated using Student's t test (*P<0.05). Error bars represent standard deviation. (B) Box and Whiskers plot indicating mDC count in seronegative controls, NRs and SVRs (C) Box and Whiskers plot indicating pDC count in seronegative controls, NRs and SVRs (D) PBMCs from seronegative controls, NRs and SVRs were stained with polychromatic flow cytometry Ab cocktail consisting of Lin-1, CD11c, CD123, and CD54 Abs. Lin-1 was used to gate on DCs and CD11c and CD123 were used to gate on mDCs (CD11c+ CD123−). Scatter plot indicates CD54 GMFI in controls, NRs, and SVRs at week 0 and week 4 of treatment. (E) PBMCs isolated from NRs and SVRs at week 0 (baseline), early weeks (within first 4 weeks) and last weeks (last 4 weeks) of treatment were stimulated in vitro with IFN-α-2a/Ribavirin for 24 h, and concentrations of a panel of 12 Th1/Th2/Th17 cytokines were determined in culture supernatant using ELISA. Scatter plot indicates the concentration of IL-6, and (F) TNF-α in NRs and SVRs. P values were calculated using Mann–Whitney's test (*P<0.05; **P<0.01). Horizontal bars represent mean concentration.
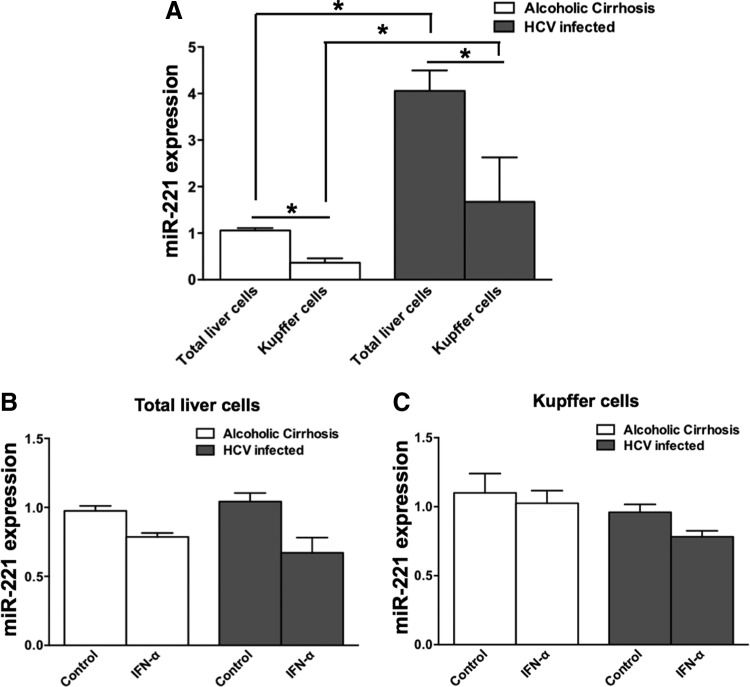
Higher miR-221 expression in total liver cells and kupffer cells of HCV-infected individuals compared to individuals with alcoholic cirrhosis
Since liver is the primary site of HCV infection, we investigated miR-221 expression in total liver cells and kupffer cells, which are major liver-resident antigen-presenting cells. We found that miR-221 expression in total liver cells and kupffer cells was higher in individuals with chronic HCV infection compared with individuals with alcoholic cirrhosis (Fig. 7A). Another important thing to note is that within both the groups, miR-221 expression is lower in kupffer cells relative to total liver cells. We also investigated whether IFN-α treatment of total liver cells and kupffer cells from both the groups led to miR-221 downregulation, but we did not observe miR-221 downregulation in any group (Fig. 7B, C).
FIG. 7.
Higher miR-221 expression in total liver cells and kupffer cells of HCV-infected individuals compared to individuals with alcoholic cirrhosis. (A) Bar graph indicating mean miR-221 expression in total liver cells and kupffer cells of HCV-infected individuals and individuals with alcoholic cirrhosis. (B) Bar graph indicating mean miR-221 expression in total liver cells of HCV-infected individuals and individuals with alcoholic cirrhosis upon 24 h IFN-α treatment. (C) Bar graph indicating mean miR-221 expression in kupffer cells of HCV-infected individuals and individuals with alcoholic cirrhosis upon 24 h IFN-α treatment. *P<0.05.
Discussion
IFN-α is known for its antiviral, antiproliferative, and immunomodulatory effects. IFN-α exerts these pleiotropic effects on target cells by regulating the expression of protein-coding genes; however, recent studies have strongly suggested that IFN-α can regulate the expression of cellular miRNAs as well. Little is known about IFN-α-regulated miRNAs and their role in controlling DC functions. Here, we report that IFN-α downregulates miR-221 and upregulates miR-155 in mDCs (Fig. 1). Lu and others recently reported that miR-221 is downregulated upon DC maturation. miR-221 downregulation leads to accumulation of its target, CDKN1C (a proapoptotic protein) and hence apoptosis of mature DCs. In the same study, miR-155 was shown to be upregulated during DC maturation. Increased miR-155 led to downregulation of KPC1 (protein that is responsible for degrading CDKN1C). Overall, Lu and others have showed that DC maturation results in accumulation of CDKN1C due to a direct effect of miR-221 downregulation and an indirect effect of miR-155 upregulation, and this can ultimately increase the rate of DC apoptosis. In fact our study also demonstrates the pro-apoptotic effect of IFN-α on DCs (Fig. 5B). These observations suggest that DC apoptosis plays a specific pathophysiological role in response to activation/maturation stimuli including IFN-I (Chen and Wang 2010). The most compelling role of DC apoptosis is in maintaining self-tolerance (Chen and others 2006). Consistent with this, individuals with autoimmune lymphoproliferative syndrome are characterized by DCs that are defective in undergoing apoptosis (Wang and others 1999). Although type I IFNs are known to induce DC apoptosis, the underlying molecular mechanisms are not completely understood. Here, we show that IFN-α downregulates miR-221 in mDCs. We also validate proapoptotic proteins BCL2L11 and CDKN1C as miR-221 targets in mDCs for the first time (Fig. 4). Therefore, we propose that IFN-α-induced miR-221 downregulation leads to upregulation of CDKN1C and BCL2L11, which results in mDC apoptosis. Our study also shows that miR-221 overexpression can downregulate the expression of proapoptotic ISGs CAV1, MX1, SHB, and TNFSF10 (TRAIL) as well (Fig. 5A), some of which are already implicated in DC apoptosis (Blum and others 2006). It is very important to note that reduced frequency of DCs as a result of HIV-1 and HCV infection is known to impair the antiviral immune response (Grassi and others 1999; Donaghy and others 2001; Pacanowski and others 2001; Soumelis and others 2001; Chehimi and others 2002; Kanto and others 2004; Cavaleiro and others 2009; Sehgal and others 2013). Therefore, the proapoptotic effect of PEG-IFN on DCs can further exacerbate this situation. We also validated SOCS1 as an miR-221 target in mDCs (Fig. 4). SOCS1 has been previously shown to negatively regulate IL-6 and TNF-α production by inhibiting the JAK/STAT pathway (Kimura and others 2005; Prêle and others 2008) and our study shows that miR-221 can control IL-6 and TNF-α levels by knocking down SOCS1 (Fig. 5).
In addition to the JAK-STAT pathway, IFN-I can activate several other signaling pathways such as C3G/Rap1 pathway, p38 mitogen-activated protein kinase and phosphatidylinositol 3-kinase signaling pathway to induce gene transcription (Platanias 2005). Here, we ascertained that IFN-α-induced miR-221 downregulation is dependent on activation of JAK/STAT pathway (Fig. 2A).
It is worth noting that IFN-α can have distinct biological outcomes despite a common proximal signal. This is possible because IFN-α can differentially activate various STAT family members (STAT1, 2, 3, 4, 5, and 6) each having distinct target genes and biological functions. STAT1, 2, and 3 are most commonly studied STATs. While STAT1 and STAT2 are associated with antiviral and proinflammatory response, STAT3 has been shown to balance and restrain the proinflammatory pathways induced by IFN-α (Ho and Ivashkiv 2006; Wang and others 2011). Higher pSTAT3 levels, observed in several cancers, have been demonstrated to upregulate miR-221 expression (Kneitz and others 2014; Liu and others 2014), which promotes proliferation of cancer cells (Nassirpour and others 2013; Yang and others 2014). STAT3 is known to play a critical role in normal DC differentiation (Miller and others 2012). Interestingly, inhibition of JAK2/STAT3 signaling leads to activation of immature DCs (Nefedova and others 2005). In light of these observations, it is possible that both antiproliferative and immunomodulatory effects of IFN-α on DCs are mediated through inhibition of STAT3 and resulting miR-221 downregulation; however, there is no clear consensus on this (Kirkwood and others 1999; Humpoliková-Adámková and others 2009). In this study we have shown that IFN-α inhibits STAT3, which results in miR-221 downregulation (Fig. 3). Our findings are summarized in the form of a model (Supplementary Fig. S4).
Role of miR-221 in HIV-1 and HCV replication is also beginning to be clear. Recently, Xu and others showed that miR-221 is upregulated in serum of chronic HCV individuals and Huh7.5.1 cells infected with HCVcc (Xu and others 2014). The authors also showed that miR-221 mimic could enhance anti-HCV effect of IFN-α in HCVcc model, whereas miR-221 inhibitor had reverse effects. miR-221 expression in PBMCs of HIV-1 elite controllers is higher than viremic progressors (Egaña-Gorroño and others 2014). Overall, this suggests that miR-221 can play an important role in antiviral defense. Our study shows that PBMCs of NRs have lower baseline expression of miR-221 compared with SVRs and seronegative controls (Fig. 6A). Also, miR-221 expression directly correlated with mDC/pDC frequency (Fig. 6B, C) and IL-6/TNF-α secretion (Fig. 6E, F). Since our study underscores the role of STAT3 in upregulating miR-221, it will be interesting to investigate whether lower miR-221 expression in NRs is a result of lower STAT3 expression/activity. We also found that miR-221 expression is higher in total liver cells and kupffer cells of individuals with chronic HCV infection relative to individuals with alcoholic cirrhosis (Fig. 7A). Since our data show that miR-221 can positively regulate proinflammatory cytokines, IL-6 and TNF-α, it will be very interesting to study whether high miR-221 levels are a cause of chronic liver inflammation associated with HCV infection.
Overall, this study significantly advances our understanding of molecular mechanisms by which IFN-α exerts its immunomodulatory and antiproliferative effects. This will greatly help us address why it effectively controls the development of some immunopathologies while exacerbate the severity of others.
Supplementary Material
Acknowledgments
We wish to acknowledge U.S. Public Health Service/National Institutes of Health Grants 1R01AI077414-06A1 to P.J. We also wish to acknowledge Mr. Matt Chomo (Flowmetric, Inc.) for helping us in designing and optimization of the antibody cocktail.
Author Disclosure Statement
No competing financial interests exist.
References
- Bieniasz P, Jones CT, Murphy MY, Panis M, Rice CM, et al. 2011. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature 472:481–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum A, Chaperot L, Molens JP, Foissaud V, Plantaz D, et al. 2006. Mechanisms of TRAIL-induced apoptosis in leukemic plasmacytoid dendritic cells. Exp Hematol 34:1655–1662 [DOI] [PubMed] [Google Scholar]
- Brain O, Owens BM, Pichulik T, Allan P, Khatamzas E, et al. 2013. The intracellular sensor NOD2 induces microRNA-29 expression in human dendritic cells to limit IL-23 release. Immunity 39:521–536 [DOI] [PubMed] [Google Scholar]
- Busch M, Zernecke A. 2012. microRNAs in the regulation of dendritic cell functions in inflammation and atherosclerosis. J Mol Med (Berl) 90:877–885 [DOI] [PubMed] [Google Scholar]
- Cavaleiro R, Baptista AP, Soares RS, Tendeiro R, Foxall RB, et al. 2009. Major depletion of plasmacytoid dendritic cells in HIV-2 infection, an attenuated form of HIV disease. PLoS Pathog 5:e1000667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chehimi J, Campbell DE, Azzoni L, Bacheller D, Papasavvas E, et al. 2002. Persistent decreases in blood plasmacytoid dendritic cell number and function despite effective highly active antiretroviral therapy and increased blood myeloid dendritic cells in HIV-infected individuals. J Immunol 168:4796–4801 [DOI] [PubMed] [Google Scholar]
- Chen M, Wang J. 2010. Programmed cell death of dendritic cells in immune regulation. Immunol Rev 236:11–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Wang YH, Wang Y, Huang L, Sandoval H, et al. 2006. Dendritic cell apoptosis in the maintenance of immune tolerance. Science 311:1160–1164 [DOI] [PubMed] [Google Scholar]
- Cheng M, Si Y, Niu Y, Liu X, Li X, et al. 2013. High-throughput profiling of alpha interferon- and interleukin-28B-regulated microRNAs and identification of let-7s with anti-hepatitis C virus activity by targeting IGF2BP1. J Virol 87:9707–9718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun-Zhi Z, Lei H, An-Ling Z, Yan-Chao F, Xiao Y, et al. 2010. MicroRNA-221 and microRNA-222 regulate gastric carcinoma cell proliferation and radioresistance by targeting PTEN. BMC Cancer 10:367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaghy H, Pozniak A, Gazzard B, Qazi N, Gilmour J, et al. 2001. Loss of blood CD11c(+) myeloid and CD11c(-) plasmacytoid dendritic cells in patients with HIV-1 infection correlates with HIV-1 RNA virus load. Blood 98:2574–2576 [DOI] [PubMed] [Google Scholar]
- Egaña-Gorroño L, Escribà T, Boulanger N, Guardo AC, León A, et al. 2014. Differential MicroRNA expression profile between stimulated PBMCs from HIV-1 infected elite controllers and viremic progressors. PLoS One 9:e106360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornari F, Gramantieri L, Ferracin M, Veronese A, Sabbioni S, et al. 2008. MiR-221 controls CDKN1C/p57 and CDKN1B/p27 expression in human hepatocellular carcinoma. Oncogene 27:5651–5661 [DOI] [PubMed] [Google Scholar]
- Galardi S, Mercatelli N, Giorda E, Massalini S, Frajese GV, et al. 2007. miR-221 and miR-222 expression affects the proliferation potential of human prostate carcinoma cell lines by targeting p27Kip1. J Biol Chem 282:23716–23724 [DOI] [PubMed] [Google Scholar]
- Garofalo M, Di Leva G, Romano G, Nuovo G, Suh SS, et al. 2009. miR-221&222 regulate TRAIL resistance and enhance tumorigenicity through PTEN and TIMP3 downregulation. Cancer Cell 16:498–509 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Gilliet M, Cao W, Liu YJ. 2008. Plasmacytoid dendritic cells: sensing nucleic acids in viral infection and autoimmune diseases. Nat Rev Immunol 8:594–606 [DOI] [PubMed] [Google Scholar]
- Gramantieri L, Fornari F, Ferracin M, Veronese A, Sabbioni S, et al. 2009. MicroRNA-221 targets Bmf in hepatocellular carcinoma and correlates with tumor multifocality. Clin Cancer Res 15:5073–5081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grassi F, Hosmalin A, McIlroy D, Calvez V, Debre P, et al. 1999. Depletion in blood CD11c-positive dendritic cells from HIV-infected patients. AIDS 13:759–766 [DOI] [PubMed] [Google Scholar]
- Hao J, Jin W, Li X, Wang S, Zhang X, et al. 2013. Inhibition of alpha interferon (IFN-alpha)-induced microRNA-122 negatively affects the anti-hepatitis B virus efficiency of IFN-alpha. J Virol 87:137–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heine A, Held SA, Daecke SN, Wallner S, Yajnanarayana SP, et al. 2013. The JAK-inhibitor ruxolitinib impairs dendritic cell function in vitro and in vivo. Blood 122:1192–1202 [DOI] [PubMed] [Google Scholar]
- Ho HH, Ivashkiv LB. 2006. Role of STAT3 in type I interferon responses. Negative regulation of STAT1-dependent inflammatory gene activation. J Biol Chem 281:14111–14118 [DOI] [PubMed] [Google Scholar]
- Hu G, Gong AY, Liu J, Zhou R, Deng C, et al. 2010. miR-221 suppresses ICAM-1 translation and regulates interferon-gamma-induced ICAM-1 expression in human cholangiocytes. Am J Physiol Gastrointest Liver Physiol 298:G542–G550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humpoliková-Adámková L, Kovařík J, Dušek L, Lauerová L, Boudný V, et al. 2009. Interferon-alpha treatment may negatively influence disease progression in melanoma patients by hyperactivation of STAT3 protein. Eur J Cancer 45:1315–1323 [DOI] [PubMed] [Google Scholar]
- Isaacs A, Lindenmann J. 1957. Virus interference. I. The interferon. Proc R Soc Lond B Biol Sci 147:258–267 [PubMed] [Google Scholar]
- Ito T, Amakawa R, Inaba M, Ikehara S, Inaba K, et al. 2001. Differential regulation of human blood dendritic cell subsets by IFNs. J Immunol 166:2961–2969 [DOI] [PubMed] [Google Scholar]
- Ivashkiv LB, Donlin LT. 2014. Regulation of type I interferon responses. Nat Rev Immunol 14:36–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarrossay D, Napolitani G, Colonna M, Sallusto F, Lanzavecchia A. 2001. Specialization and complementarity in microbial molecule recognition by human myeloid and plasmacytoid dendritic cells. Eur J Immunol 31:3388–3393 [DOI] [PubMed] [Google Scholar]
- Kallal LE, Biron CA. 2013. Changing partners at the dance. JAKSTAT 2:e23504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanto T, Inoue M, Miyatake H, Sato A, Sakakibara M, et al. 2004. Reduced numbers and impaired ability of myeloid and plasmacytoid dendritic cells to polarize T helper cells in chronic hepatitis C virus infection. J Infect Dis 190:1919–1926 [DOI] [PubMed] [Google Scholar]
- Karrich JJ, Jachimowski LC, Libouban M, Iyer A, Brandwijk K, et al. 2013. MicroRNA-146a regulates survival and maturation of human plasmacytoid dendritic cells. Blood 122:3001–3009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SJ, Gregersen PK, Diamond B. 2013. Regulation of dendritic cell activation by microRNA let-7c and BLIMP1. J Clin Invest 123:823–833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura A, Naka T, Muta T, Takeuchi O, Akira S, et al. 2005. Suppressor of cytokine signaling-1 selectively inhibits LPS-induced IL-6 production by regulating JAK–STAT. Proc Natl Acad Sci U S A 102:17089–17094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkwood JM, Farkas DL, Chakraborty A, Dyer KF, Tweardy DJ, et al. 1999. Systemic interferon-alpha (IFN-alpha) treatment leads to Stat3 inactivation in melanoma precursor lesions. Mol Med 5:11–20 [PMC free article] [PubMed] [Google Scholar]
- Kneitz B, Krebs M, Kalogirou C, Schubert M, Joniau S, et al. 2014. Survival in patients with high-risk prostate cancer is predicted by miR-221, which regulates proliferation, apoptosis, and invasion of prostate cancer cells by inhibiting IRF2 and SOCS3. Cancer Res 74:2591–2603 [DOI] [PubMed] [Google Scholar]
- Lambotin M, Raghuraman S, Stoll-Keller F, Baumert TF, Barth H. 2010. A look behind closed doors: interaction of persistent viruses with dendritic cells. Nat Rev Microbiol 8:350–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehtonen A, Matikainen S, Julkunen I. 1997. Interferons up-regulate STAT1, STAT2, and IRF family transcription factor gene expression in human peripheral blood mononuclear cells and macrophages. J Immunol 159:794–803 [PubMed] [Google Scholar]
- Liu S, Sun X, Wang M, Hou Y, Zhan Y, et al. 2014. A microRNA 221- and 222-mediated feedback loop, via PDLIM2, maintains constitutive activation of NFkappaB and STAT3 in colorectal cancer cells. Gastroenterology 147:847–859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YJ. 2005. IPC: professional type 1 interferon-producing cells and plasmacytoid dendritic cell precursors. Annu Rev Immunol 23:275–306 [DOI] [PubMed] [Google Scholar]
- Lodish HF, Zhou B, Liu G, Chen CZ. 2008. Micromanagement of the immune system by microRNAs. Nat Rev Immunol 8:120–130 [DOI] [PubMed] [Google Scholar]
- Longhi MP, Trumpfheller C, Idoyaga J, Caskey M, Matos I, et al. 2009. Dendritic cells require a systemic type I interferon response to mature and induce CD4+ Th1 immunity with poly IC as adjuvant. J Exp Med 206:1589–1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Huang X, Zhang X, Roensch K, Cao Q, et al. 2011. miR-221 and miR-155 regulate human dendritic cell development, apoptosis, and IL-12 production through targeting of p27kip1, KPC1, and SOCS-1. Blood 117:4293–4303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luft T, Pang KC, Thomas E, Hertzog P, Hart DN, et al. 1998. Type I IFNs enhance the terminal differentiation of dendritic cells. J Immunol 161:1947–1953 [PubMed] [Google Scholar]
- Miller JC, Brown BD, Shay T, Gautier EL, Jojic V, et al. 2012. Deciphering the transcriptional network of the dendritic cell lineage. Nat Immunol 13:888–899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montagner S, Orlandi EM, Merante S, Monticelli S. 2013. The role of miRNAs in mast cells and other innate immune cells. Immunol Rev 253:12–24 [DOI] [PubMed] [Google Scholar]
- Nassirpour R, Mehta PP, Baxi SM, Yin MJ. 2013. miR-221 promotes tumorigenesis in human triple negative breast cancer cells. PLoS One 8:e62170. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Nefedova Y, Cheng P, Gilkes D, Blaskovich M, Beg AA, et al. 2005. Activation of dendritic cells via inhibition of Jak2/STAT3 signaling. J Immunol 175:4338–4346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsen TW. 2007. Mechanisms of microRNA-mediated gene regulation in animal cells. Trends Genet 23:243–249 [DOI] [PubMed] [Google Scholar]
- O'Connell RM, Rao DS, Baltimore D. 2012. microRNA regulation of inflammatory responses. Annu Rev Immunol 30:295–312 [DOI] [PubMed] [Google Scholar]
- O'Neill LA, Sheedy FJ, McCoy CE. 2011. MicroRNAs: the fine-tuners of Toll-like receptor signalling. Nat Rev Immunol 11:163–175 [DOI] [PubMed] [Google Scholar]
- Ohno M, Natsume A, Kondo Y, Iwamizu H, Motomura K, et al. 2009. The modulation of microRNAs by type I IFN through the activation of signal transducers and activators of transcription 3 in human glioma. Mol Cancer Res 7:2022–2030 [DOI] [PubMed] [Google Scholar]
- Pacanowski J, Kahi S, Baillet M, Lebon P, Deveau C, et al. 2001. Reduced blood CD123+ (lymphoid) and CD11c+ (myeloid) dendritic cell numbers in primary HIV-1 infection. Blood 98:3016–3021 [DOI] [PubMed] [Google Scholar]
- Pantel A, Teixeira A, Haddad E, Wood EG, Steinman RM, et al. 2014. Direct type I IFN but not MDA5/TLR3 activation of dendritic cells is required for maturation and metabolic shift to glycolysis after poly IC stimulation. PLoS Biol 12: e1001759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parlato S, Bruni R, Fragapane P, Salerno D, Marcantonio C, et al. 2013. IFN-α regulates Blimp-1 expression via miR-23a and miR-125b in both monocytes-derived DC and pDC. PLoS One 8:e72833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen IM, Cheng G, Wieland S, Volinia S, Croce CM, et al. 2007. Interferon modulation of cellular microRNAs as an antiviral mechanism. Nature 449:919–922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry AK, Chen G, Zheng D, Tang H, Cheng G. 2005. The host type I interferon response to viral and bacterial infections. Cell Res 15:407–422 [DOI] [PubMed] [Google Scholar]
- Pineau P, Volinia S, McJunkin K, Marchio A, Battiston C, et al. 2010. miR-221 overexpression contributes to liver tumorigenesis. Proc Natl Acad Sci U S A 107:264–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platanias LC. 2005. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat Rev Immunol 5:375–386 [DOI] [PubMed] [Google Scholar]
- Prêle CM, Woodward EA, Bisley J, Keith-Magee A, Nicholson SE, et al. 2008. SOCS1 Regulates the IFN but not NFκB pathway in TLR-stimulated human monocytes and macrophages. J Immunol 181:8018–8026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riepsaame J, van Oudenaren A, den Broeder BJ, van Ijcken WF, Pothof J, et al. 2013. MicroRNA-mediated down-regulation of M-CSF receptor contributes to maturation of mouse monocyte-derived dendritic cells. Front Immunol 4:353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehgal M, Khan ZK, Talal AH, Jain P. 2013. Dendritic cells in HIV-1 and HCV infection: can they help win the battle? Virology 4:1–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soumelis V, Scott I, Gheyas F, Bouhour D, Cozon G, et al. 2001. Depletion of circulating natural type 1 interferon-producing cells in HIV-infected AIDS patients. Blood 98:906–912 [DOI] [PubMed] [Google Scholar]
- Tanaka , Sugaya , Kita , Arai , Kanda , et al. , 2012. Inhibition of cell viability by human IFN-β is mediated by microRNA-431. Int J Oncol 40:1470–1476 [DOI] [PubMed] [Google Scholar]
- Turner ML, Schnorfeil FM, Brocker T. 2011. MicroRNAs regulate dendritic cell differentiation and function. J Immunol 187:3911–3917 [DOI] [PubMed] [Google Scholar]
- Wang J, Zheng L, Lobito A, Chan FK, Dale J, et al. 1999. Inherited human Caspase 10 mutations underlie defective lymphocyte and dendritic cell apoptosis in autoimmune lymphoproliferative syndrome type II. Cell 98:47–58 [DOI] [PubMed] [Google Scholar]
- Wang W-B, Levy DE, Lee C-K. 2011. STAT3 negatively regulates type I IFN-mediated antiviral response. J Immunol 187:2578–2585 [DOI] [PubMed] [Google Scholar]
- Xu G, Yang F, Ding CL, Wang J, Zhao P, et al. 2014. MiR-221 accentuates IFNs anti-HCV effect by downregulating SOCS1 and SOCS3. Virology 462–463:343–350 [DOI] [PubMed] [Google Scholar]
- Yang CH, Yue J, Fan M, Pfeffer LM. 2010. IFN induces miR-21 through a signal transducer and activator of transcription 3-dependent pathway as a suppressive negative feedback on IFN-induced apoptosis. Cancer Res 70:8108–8116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Yang Y, Gan R, Zhao L, Li W, et al. 2014. Down-regulation of mir-221 and mir-222 restrain prostate cancer cell proliferation and migration that is partly mediated by activation of SIRT1. PLoS One 9:e98833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan Y, Wu L. 2012. Functional regulation of monocyte-derived dendritic cells by microRNAs. Protein Cell 3:497–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang CZ, Zhang JX, Zhang AL, Shi ZD, Han L, et al. 2010. MiR-221 and miR-222 target PUMA to induce cell survival in glioblastoma. Mol Cancer 9:229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Li S, Yan Q, Chen X, Yang Y, et al. 2013. Interferon-beta induced microRNA-129-5p down-regulates HPV-18 E6 and E7 viral gene expression by targeting SP1 in cervical cancer cells. PLoS One 8:e81366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao JJ, Lin J, Yang H, Kong W, He L, et al. 2008. MicroRNA-221/222 negatively regulates estrogen receptor alpha and is associated with tamoxifen resistance in breast cancer. J Biol Chem 283:31079–31086 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.