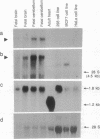
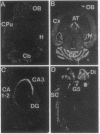
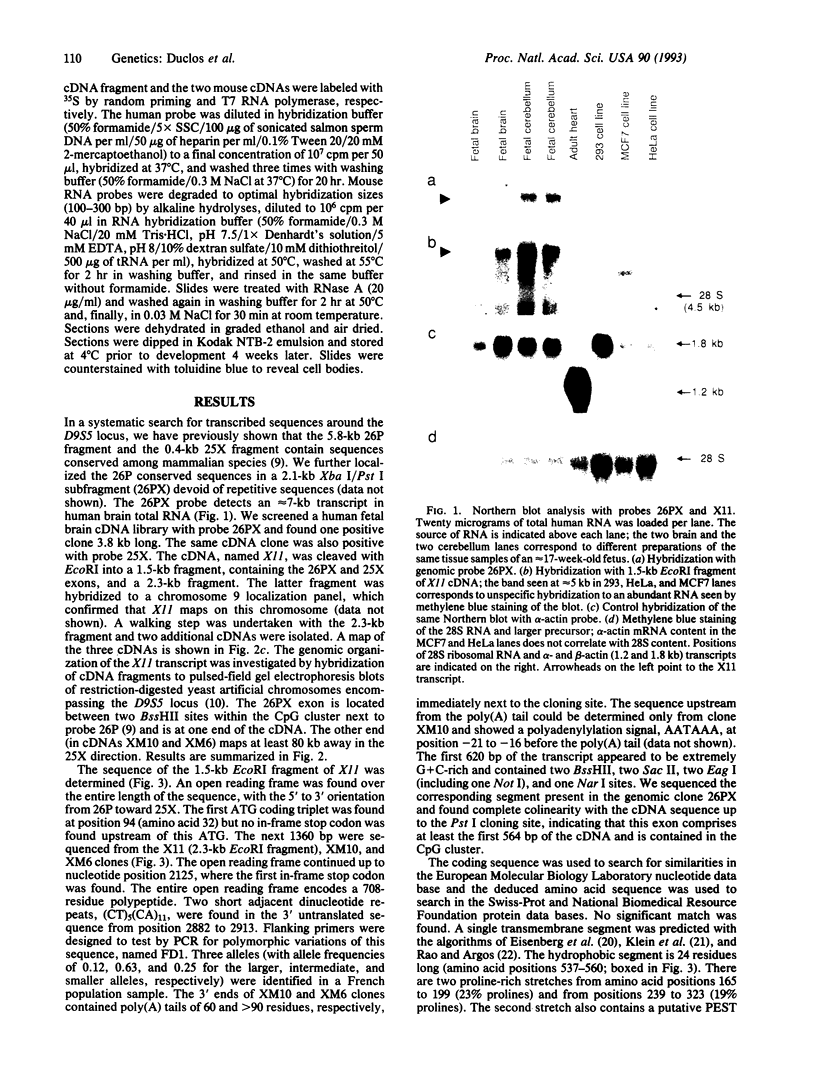
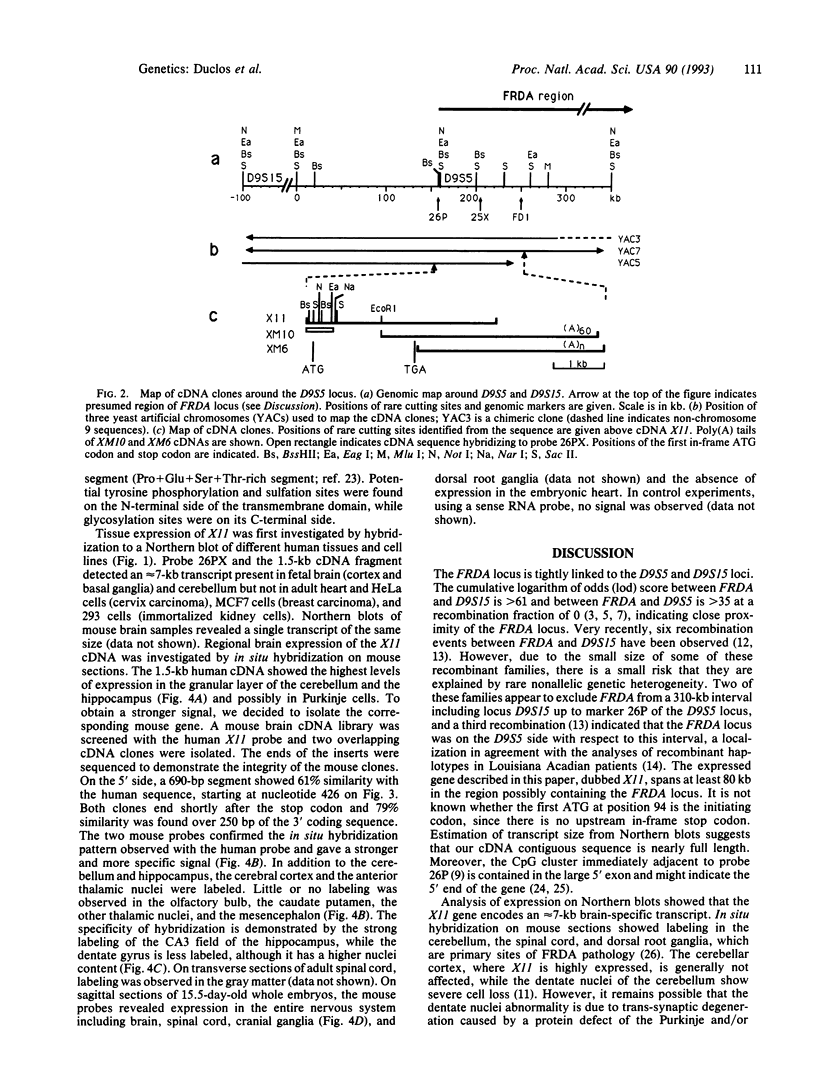
Abstract
Friedreich ataxia (FRDA) is an autosomal recessive degenerative disorder that affects the cerebellum, spinal cord, and peripheral nerves. The FRDA gene was localized in 9q13-q21 within 0.7 centimorgan of the D9S5 and D9S15 loci. One recently reported recombination event and haplotype analysis in a population with a founder effect suggested that the FRDA locus is on the D9S5 side. Using a conserved probe from the D9S5 locus, we have now identified an approximately 7-kilobase (kb) transcript and report cloning of its cDNA. The corresponding gene, X11, extends at least 80 kb in a direction opposite D9S15. The gene is expressed in the brain, including the cerebellum, but is not detectable in several nonneuronal tissues and cell lines. In situ hybridization of adult mouse brain sections showed prominant expression in the granular layer of the cerebellum. Expression was also found in the spinal cord. The cDNA contains an open reading frame encoding a 708-amino acid sequence that shows no significant similarity to other known proteins but contains a unique, 24-residue-long, putative transmembrane segment. On the basis of its genomic localization and its neuronal site of expression, particularly in the cerebellum, this "pioneer" gene represents a candidate for FRDA. Direct evidence of its involvement in FRDA will require a search for causative point mutations in patients.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. Basic local alignment search tool. J Mol Biol. 1990 Oct 5;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Bird A. P. CpG-rich islands and the function of DNA methylation. Nature. 1986 May 15;321(6067):209–213. doi: 10.1038/321209a0. [DOI] [PubMed] [Google Scholar]
- Chamberlain S., Shaw J., Rowland A., Wallis J., South S., Nakamura Y., von Gabain A., Farrall M., Williamson R. Mapping of mutation causing Friedreich's ataxia to human chromosome 9. Nature. 1988 Jul 21;334(6179):248–250. doi: 10.1038/334248a0. [DOI] [PubMed] [Google Scholar]
- Chamberlain S., Shaw J., Wallis J., Rowland A., Chow L., Farrall M., Keats B., Richter A., Roy M., Melancon S. Genetic homogeneity at the Friedreich ataxia locus on chromosome 9. Am J Hum Genet. 1989 Apr;44(4):518–521. [PMC free article] [PubMed] [Google Scholar]
- Eisenberg D., Schwarz E., Komaromy M., Wall R. Analysis of membrane and surface protein sequences with the hydrophobic moment plot. J Mol Biol. 1984 Oct 15;179(1):125–142. doi: 10.1016/0022-2836(84)90309-7. [DOI] [PubMed] [Google Scholar]
- Feener C. A., Koenig M., Kunkel L. M. Alternative splicing of human dystrophin mRNA generates isoforms at the carboxy terminus. Nature. 1989 Apr 6;338(6215):509–511. doi: 10.1038/338509a0. [DOI] [PubMed] [Google Scholar]
- Fujita R., Agid Y., Trouillas P., Seck A., Tommasi-Davenas C., Driesel A. J., Olek K., Grzeschik K. H., Nakamura Y., Mandel J. L. Confirmation of linkage of Friedreich ataxia to chromosome 9 and identification of a new closely linked marker. Genomics. 1989 Jan;4(1):110–111. doi: 10.1016/0888-7543(89)90323-6. [DOI] [PubMed] [Google Scholar]
- Fujita R., Hanauer A., Sirugo G., Heilig R., Mandel J. L. Additional polymorphisms at marker loci D9S5 and D9S15 generate extended haplotypes in linkage disequilibrium with Friedreich ataxia. Proc Natl Acad Sci U S A. 1990 Mar;87(5):1796–1800. doi: 10.1073/pnas.87.5.1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita R., Hanauer A., Vincent A., Mandel J. L., Koenig M. Physical mapping of two loci (D9S5 and D9S15) tightly linked to Friedreich ataxia locus (FRDA) and identification of nearby CpG islands by pulse-field gel electrophoresis. Genomics. 1991 Aug;10(4):915–920. doi: 10.1016/0888-7543(91)90179-i. [DOI] [PubMed] [Google Scholar]
- Fujita R., Sirugo G., Duclos F., Abderrahim H., Le Paslier D., Cohen D., Brownstein B. H., Schlessinger D., Mandel J. L., Koenig M. A 530kb YAC contig tightly linked to the Friedreich ataxia locus contains five CpG clusters and a new highly polymorphic microsatellite. Hum Genet. 1992 Jul;89(5):531–538. doi: 10.1007/BF00219179. [DOI] [PubMed] [Google Scholar]
- Hafen E., Levine M., Garber R. L., Gehring W. J. An improved in situ hybridization method for the detection of cellular RNAs in Drosophila tissue sections and its application for localizing transcripts of the homeotic Antennapedia gene complex. EMBO J. 1983;2(4):617–623. doi: 10.1002/j.1460-2075.1983.tb01472.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanauer A., Chery M., Fujita R., Driesel A. J., Gilgenkrantz S., Mandel J. L. The Friedreich ataxia gene is assigned to chromosome 9q13-q21 by mapping of tightly linked markers and shows linkage disequilibrium with D9S15. Am J Hum Genet. 1990 Jan;46(1):133–137. [PMC free article] [PubMed] [Google Scholar]
- Klein P., Kanehisa M., DeLisi C. The detection and classification of membrane-spanning proteins. Biochim Biophys Acta. 1985 May 28;815(3):468–476. doi: 10.1016/0005-2736(85)90375-x. [DOI] [PubMed] [Google Scholar]
- Lindsay S., Bird A. P. Use of restriction enzymes to detect potential gene sequences in mammalian DNA. 1987 May 28-Jun 3Nature. 327(6120):336–338. doi: 10.1038/327336a0. [DOI] [PubMed] [Google Scholar]
- Lipman D. J., Pearson W. R. Rapid and sensitive protein similarity searches. Science. 1985 Mar 22;227(4693):1435–1441. doi: 10.1126/science.2983426. [DOI] [PubMed] [Google Scholar]
- Mohana Rao J. K., Argos P. A conformational preference parameter to predict helices in integral membrane proteins. Biochim Biophys Acta. 1986 Jan 30;869(2):197–214. doi: 10.1016/0167-4838(86)90295-5. [DOI] [PubMed] [Google Scholar]
- Oppenheimer D. R. Brain lesions in Friedreich's ataxia. Can J Neurol Sci. 1979 May;6(2):173–176. doi: 10.1017/s0317167100119596. [DOI] [PubMed] [Google Scholar]
- Pandolfo M., Sirugo G., Antonelli A., Weitnauer L., Ferretti L., Leone M., Dones I., Cerino A., Fujita R., Hanauer A. Friedreich ataxia in Italian families: genetic homogeneity and linkage disequilibrium with the marker loci D9S5 and D9S15. Am J Hum Genet. 1990 Aug;47(2):228–235. [PMC free article] [PubMed] [Google Scholar]
- Rogers S., Wells R., Rechsteiner M. Amino acid sequences common to rapidly degraded proteins: the PEST hypothesis. Science. 1986 Oct 17;234(4774):364–368. doi: 10.1126/science.2876518. [DOI] [PubMed] [Google Scholar]
- Sirugo G., Keats B., Fujita R., Duclos F., Purohit K., Koenig M., Mandel J. L. Friedreich ataxia in Louisiana Acadians: demonstration of a founder effect by analysis of microsatellite-generated extended haplotypes. Am J Hum Genet. 1992 Mar;50(3):559–566. [PMC free article] [PubMed] [Google Scholar]
- Wilkes D., Shaw J., Anand R., Riley J., Winter P., Wallis J., Driesel A. G., Williamson R., Chamberlain S. Identification of CpG islands in a physical map encompassing the Friedreich's ataxia locus. Genomics. 1991 Jan;9(1):90–95. doi: 10.1016/0888-7543(91)90224-3. [DOI] [PubMed] [Google Scholar]