Introductory Paragraph
An increase in amyloid β (Aβ) production is a major pathogenic mechanism associated with Alzheimer's Disease (AD)1,2, but little is known about possible homeostatic control of the amyloidogenic pathway. Here we report that the amyloid precursor protein (APP) intracellular domain (AICD) downregulates WASP (Wiskott-Aldrich syndrome protein) family verprolin homologous protein 1 (WAVE1 or WASF1) as part of a negative feedback mechanism to limit Aβ production. AICD binds to the Wasf1 promoter, negatively regulates its transcription and down-regulates Wasf1 mRNA and protein in Neuro 2a (N2a) cells. WAVE1 interacts with and colocalizes with APP in the Golgi apparatus. Experimentally reducing WAVE1 in N2a cells decreases the budding of APP-containing vesicles, and reduces cell surface APP, thereby reducing the production of Aβ. WAVE1 downregulation is observed in mouse models of AD. Reduction of Wasf1 gene dosage dramatically reduces Aβ levels and restores memory deficits in a mouse model of AD. A decrease in WASF1 mRNA is also observed in human AD brains, suggesting clinical relevance of the negative feedback circuit involved in homeostatic regulation of Aβ production.
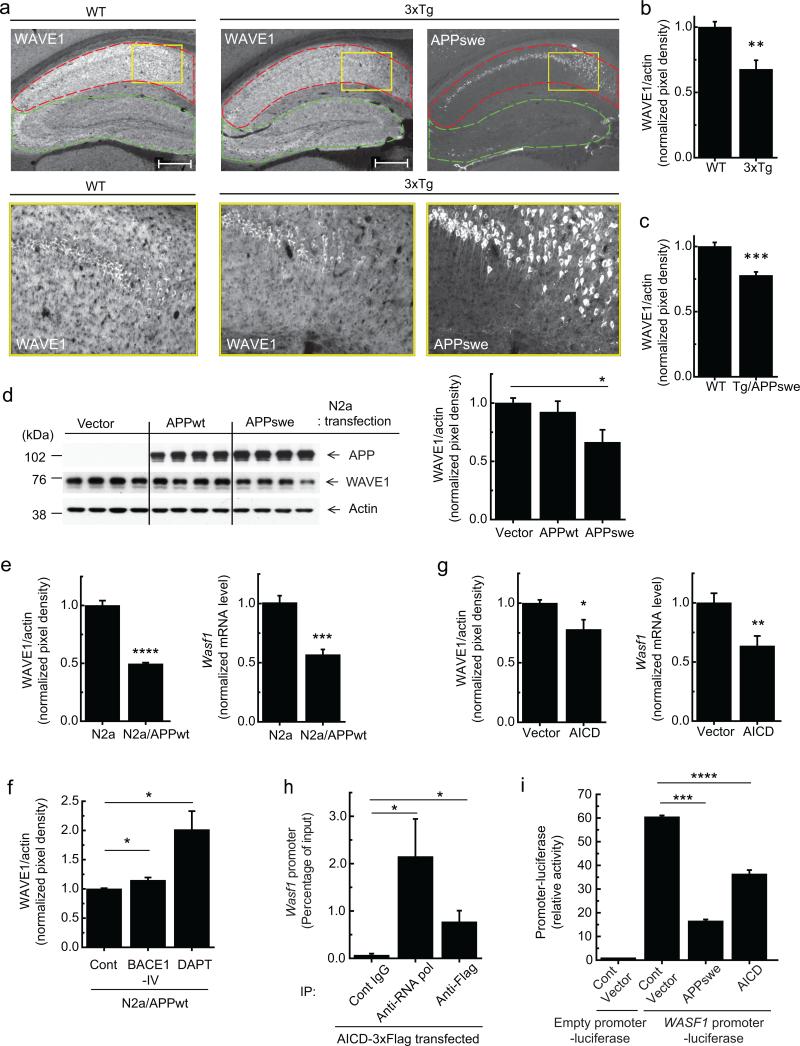
WAVE1, as a member of the WASP/WAVE family proteins, activates the actin-related protein 2/3 (Arp2/3) complex and initiates actin polymerization3. WAVE1 is highly expressed in the brain4, where it exists as a heteropentameric complex together with PIR121, Nap1, Abi2 and HSPC30005,6. Previously, NAPAP1 (human Nap1) expression was found to be markedly decreased in AD-affected brains7. Since WAVE1 is a core component of the complex required for actin polymerization, we measured the level of WAVE1 protein in mouse models of AD. WAVE1 expression was examined by immunohistochemistry in the hippocampus of a triple-transgenic AD model (3xTg) harboring presenilin 1 (PS1 M146V), Swedish mutant of APP (APPswe), and tau (P301L) transgenes8. Compared to wild-type (WT) mouse brain, 3xTg mice showed decreased WAVE1-immunoreactivity (WAVE1-IR) in the CA1 region where APPswe was highly expressed, but showed a comparable WAVE1-IR in the CA2-3 region and dentate gyrus, where there was little expression of APPswe (Fig. 1a). In separate experiments, a lower level of WAVE1 in the entire hippocampus of 3xTg compared to WT mice was confirmed by immunoblotting (Fig. 1b). WAVE1 downregulation was also observed in the hippocampus of a transgenic AD mouse model overexpressing only APPswe9 (Tg/APPswe) (Fig. 1c), indicating a critical role for APP in the downregulation of WAVE1. WAVE1 was also significantly down-regulated by transient overexpression of the APPswe, which strongly facilitates β-secretase-mediated processing of APP10, but not by WT APP695 (APPwt) in neuroblastoma N2a cells (Fig. 1d and Supplementary Fig. 1a). Overexpression of APPwt or APPswe did not change the level of actin (Supplementary Fig. 1b) and was not associated with cell toxicity (Supplementary Fig. 1c).
Figure 1.
Downregulation of WAVE1 expression by overexpression of APP or AICD. (a) Immunohistochemistry of WAVE1 and APPswe in 12 month-old WT and 3xTg male mice. High magnification images (bottom) of the rectangular regions (yellow) in top images. CA1 (red), and CA2, CA3 and dentate gyrus (green). Scale bars, 500 μm. (b, c) Immunoblotting of WAVE1 and actin in the hippocampus from 12 month-old WT (n = 4) and 3xTg (n = 8) (b) or 8 month-old WT (n = 10) and Tg/APPswe (n = 10) (c) male mice. The quantified protein level of WAVE1 was normalized to the level of actin. (d) N2a cells were transiently transfected as indicated. Representative immunoblotting images (left), and quantification (right, n = 5). (e) WAVE1 protein (left, n = 6) and mRNA (right, n = 6) levels in normal N2a and N2a/APPwt cells. (f) Effect of the β-secretase (BACE1-IV) or γ-secretase (DAPT) inhibitors on WAVE1 protein level in N2a/APPwt cells (Cont and BACE1-IV, n = 6; DAPT, n = 8). (g) WAVE1 protein (left, n = 4) and mRNA (right, n = 6) levels in N2a cells transiently transfected with AICD. (h) ChIP analysis of N2a cells transiently transfected with 3xFlag-tagged AICD. Immunoprecipitation (IP) was performed with preimmune (Cont) IgG, anti-RNA polymerase antibody (anti-RNA pol) as a positive control, or anti-Flag antibody. A fragment of the Wasf1 gene promoter in the immune complex was amplified by PCR and quantified (n = 9). (i) N2a cells were transiently co-transfected as indicated. Luciferase activity was measured (n = 6). Means ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001 and ****P < 0.0001, two-tailed t-test.
To further study the regulation of WAVE1, we used a stable N2a cell line expressing WT APP (N2a/APPwt), where all cells express APPwt, unlike the fractional expression of APPwt in transiently transfected cells. WAVE1 was down-regulated in N2a/APPwt cells compared to its level in non-transfected N2a cells (Fig. 1e, left). Treatment with a specific γ-secretase inhibitor markedly increased the level of WAVE1 (Fig. 1f). APP processing by β- and γ-secretases not only generates Aβ but also produces AICD11,12. A higher level of AICD was generated in cells expressing APPswe compared to the cells expressing APPwt (Supplementary Fig. 1d). Notably, the effect of APP overexpression on WAVE1 was mimicked by overexpression of AICD (left graph in Fig. 1g and Supplementary Fig. 1e). Downregulation of WAVE1 by overexpresssion of APP or AICD takes place at the level of mRNA (right graphs in Fig. 1e, g). AICD is known to be a transcription factor13 and transcriptionally active AICD is preferentially generated by the amyloidogenic pathway14-16. Previously, direct interactions of AICD with several target genes including an Aβ-degrading enzyme, neprilysin, were characterized by chromatin immunoprecipitation (ChIP) assay17,18. We performed ChIP assays to examine possible interaction of AICD with the Wasf1 promoter. 3xflag-tagged AICD was transiently expressed in N2a cells. Immunoprecipitation with anti-RNA polymerase (a positive control) or anti-flag antibody, but not with preimmune IgG, co-precipitated the Wasf1 promoter region (Fig. 1h). A WASF1 promoter fused-luciferase assay showed suppression of WASF1 promoter activity by overexpression of APPswe or AICD (Fig. 1i). As a positive control, AICD increased expression of neprilysin in a MME (human neprilysin)-promoter fused luciferase reporter assay (Supplementary Fig. 1f) as expected18. The potency of suppression by APPswe or AICD overexpression was well correlated with the protein levels of AICD in the transfected cells (Supplementary Fig. 1g). WAVE1 expression was also regulated by the intracellular domains (ICDs) of APP like protein (APLP) 1 and 2, but not by notch intracellular domain (NICD) (Supplementary Fig. 2a–e). Overexpression of APLP1-ICD reduced the level of Wasf11 mRNA and WASF1 promoter-luciferase activity (Supplementary Fig. 2c, d), but did not significantly alter the level of WAVE1 protein (Supplementary Fig. 2e). This may be due to a long half-life of WAVE1 protein (~24 h) (Supplementary Fig. 2f, g) and a relatively weaker inhibitory activity of APLP1-ICD compared to AICD and APLP2-ICD in the regulation of the WASF1 promoter (Supplementary Fig. 2d). Together these data suggest a critical role for AICD and ICDs of APLPs in the regulation of WAVE1 expression.
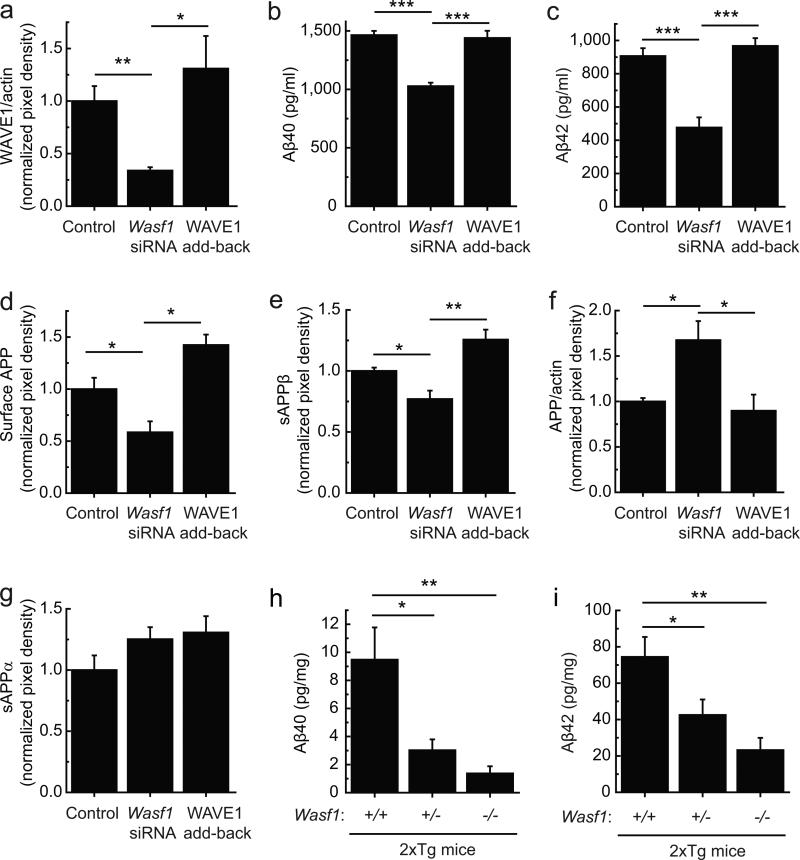
We next investigated the possibility that WAVE1 regulates the amyloidogenic pathway. Lowering WAVE1 by a synthetic duplex of small interfering RNA (siRNA) (34% of WAVE1 level compared to control; Fig. 2a) reduced the levels of Aβ40 (70% of control) and Aβ42 (53% of control) in a double transgenic N2a cell line overexpressing APPswe and familial Alzheimer's Disease (FAD) presenilin1 mutant ΔE9 (N2a/APPswe.PS1ΔE9) (Fig. 2b, c). We also observed that lowering WAVE1 was associated with a lower level of surface APP (Fig. 2d), a lower level of the soluble ectodomain of APP (sAPPβ) produced by β-secretase (Fig. 2e), a higher level of total APP (Fig. 2f) and an unchanged level of the soluble ectodomain of APP (sAPPα) produced by α-secretase (Fig. 2g). Restoration of WAVE1 level by expressing siRNA-resistant WAVE1 in conjunction with siRNA (Fig. 2a) reversed these effects (Fig. 2b–g). To address the physiological relevance of the regulation of Aβ formation by WAVE1, double transgenic AD mice (2xTg) were bred with Wasf1 knockout (KO) mice. We chose 2xTg mice harboring APPswe and PS1ΔE919 because the pathological phenotype appears earlier than Tg/APPswe but is not influenced by tau as with 3xTg mice. We generated constitutive Wasf1 KO mice by crossing floxed Wasf1 with Cre-deleter mice (Supplementary Fig. 3). The brains of 2xTg mice harboring lower Wasf1 gene dosage, compared to the brains with Wasf1+/+, showed dramatically reduced levels of Aβ40 (Wasf11+/–, 32% and Wasf1–/–, 14% of Wasf1+/+) and Aβ42 (Wasf1+/–, 57% and Wasf1–/–, 31% of Wasf1+/+) (Fig. 2h, i).
Figure 2.
Lowering WAVE1 expression reduces Aβ. (a–g) N2a/APPswe.PS1ΔE9 cells were transfected with control siRNA plus control plasmid (Control), Wasf1 siRNA plus control plasmid (Wasf1 siRNA), or Wasf1 siRNA plus siRNA-resistant plasmid for WAVE1 (WAVE1 add-back). WAVE1 (a, n = 9), Aβ40 (b, n = 6), Aβ42 (c, n = 6), total APP (f, n = 4) and actin were measured in cell lysates. Surface APP was measured by a biotinylation assay (d, n = 6). Soluble ectodomain of APP (sAPPβ) produced by β-secretase (e, n = 4), and sAPPα, a product of α-secretase (g, n = 4), were measured in the medium. Data represent mean ± SEM. *P < 0.05, **P < 0.01 and ***P < 0.001, two-tailed t-test. (h, i) The levels of Aβ40 (h) and Aβ42 (i) were measured in mouse brains of 4–5 month-old 2xTg AD male mice (tg/+) harboring Wasf1+/+ (n = 13), Wasf1+/– (n = 13) or Wasf1–/– (n = 6). Data represent means ± SEM. *P < 0.05 and **P < 0.01, one-way analysis of variance (ANOVA), Dunnett's post hoc test.
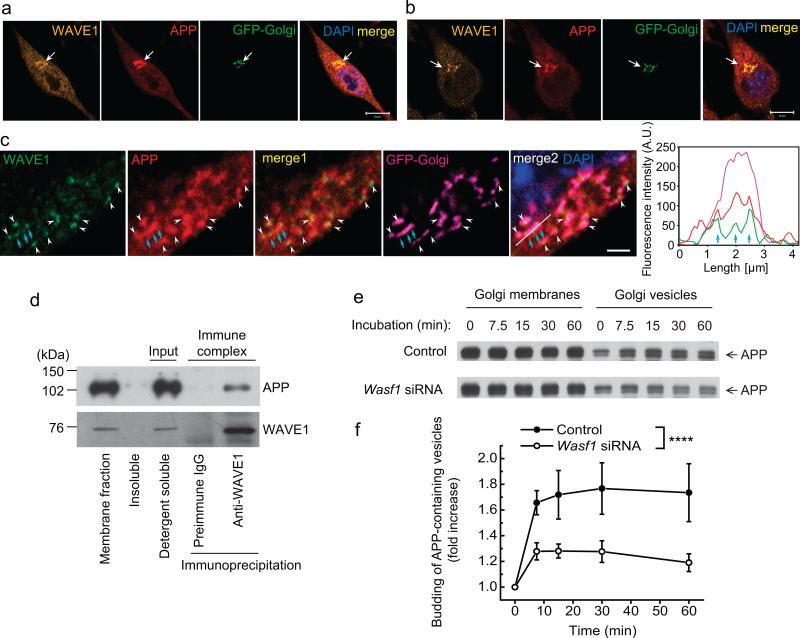
APP is trafficked to and is processed within various cellular organelles. After translation of APP in the endoplasmic reticulum (ER), APP is transported to the plasma membrane via the Golgi apparatus and trans-Golgi network12,20. Within this constitutive secretory pathway, immature APP molecules mature through post-translational modifications including glycosylation, phosphorylation and tyrosine sulphation12,20. The surface APP is then endocytosed to endosomes, and a fraction in endosomes is recycled to the trans-Golgi network or to the cell surface. During this trafficking cycle, β-secretase cleaves APP mainly in endosomes, and γ-secretase cleaves APP carboxy-terminal fragment (APP-CTF) mainly at the plasma membrane and in endosomes/lysosomes12,20. Immunocytochemical analysis of N2a/APPswe.PS1ΔE9 cells and N2a/APPwt cells showed that WAVE1 and APP were enriched and co-localized in the Golgi apparatus (Fig. 3a, b and Supplementary Fig. 4a). High magnification of Golgi area and line scan data show coinciding fluorescence signal for WAVE1, APP and Golgi, indicating co-localization of WAVE1 and APP in the Golgi apparatus (Fig. 3c). However, WAVE1 was rarely found to be localized in early-endosomes, late-endosomes, lysosomes or ER (Supplementary Fig. 4b–e). Biochemical studies showed that APP and WAVE1 were co-immunoprecipitated from a detergent-soluble membrane fraction of N2a/APPwt cells (Fig. 3d and Supplementary Fig. 5a, b). Furthermore, in vitro budding of APP-containing vesicles from Golgi membranes was significantly reduced following suppression of WAVE1 expression (Fig. 3e, f and Supplementary Fig. 5c, d). Lowering WAVE1 expression by siRNA also significantly decreased the ratio of mature APP to immature APP, supporting a role for WAVE1 in APP trafficking (Supplementary Fig. 6a–e).
Figure 3.
WAVE1 facilitates budding of APP-containing vesicles from the Golgi apparatus. (a–c) Immunocytochemistry of WAVE1 and APP in N2a/APPswe.PS1ΔE9 cells (a, low magnification; c, high magnification; a and c are different cells) and in N2a/APPwt cells (b). Cells were infected with a viral vector expressing a fluorescent protein fused-Golgi targeting sequence (GFP-Golgi). DAPI counterstaining was used to show the nucleus. Line scan (white line in merge 2 of c) shows coinciding fluorescence signal (cyan arrows) for WAVE1, APP and GFP-Golgi (right panel of c). Scale bars, 10 μm (a, b) and 2 μm (c). Arrows (a, b) and arrow heads (c) indicate the co-localization of WAVE1 and APP in the Golgi apparatus. (d) A detergent soluble membrane fraction from N2a/APPwt cells was used for immunoprecipitation with preimmune IgG or anti-WAVE1 antibody. WAVE1 co-precipitated with APP. (e, f) Golgi membrane (from N2a/APPwt cells) and cytosol (from N2a cells) were prepared from cells transfected with control siRNA or Wasf1 siRNA for in vitro budding assay. Reconstituted Golgi membrane and cytosol were incubated at 37 °C for the indicated times, and released vesicles were separated from Golgi membrane by centrifugation. Representative images showing time course of APP-containing vesicle formation (e) and time-dependent fold increase of APP-containing vesicle formation from six independent experiments (f, n = 6). Data represent mean ± SEM. ****P < 0.0001, Control versus Wasf1 siRNA, two-way ANOVA.
In the case of APP overexpressing cell lines, the majority of APP is known to localize to the Golgi apparatus and trans-Golgi network, and only a small fraction of nascent APP molecules (~10%) reach the plasma membrane12. We also observed an enrichment of APP in the Golgi apparatus in N2a/APPwt cells (ratio of Golgi/cytoplasm for APP mean density is 12) but we did not observe further accumulation of APP in the Golgi as a result of WAVE1 reduction by siRNA (Supplementary Fig. 7a). Reducing WAVE1 by siRNA also did not alter the turnover rate of Aβ peptides (Supplementary Fig. 7b, c) or alter protein levels of the γ-secretase complex, BACE1 or NICD (as a readout of γ-secretase activity) (Supplementary Fig. 7d–i). Altogether these results suggest that downregulation of WAVE1 results in a lower surface level of APP and lower production of Aβ by the amyloidogenic pathway through reducing vesicular trafficking of APP-containing vesicles from the Golgi. This conclusion is consistent with a growing appreciation for the role of actin polymerization mediated by WASP and WAVE family proteins in facilitation of transport carrier biogenesis21,22.
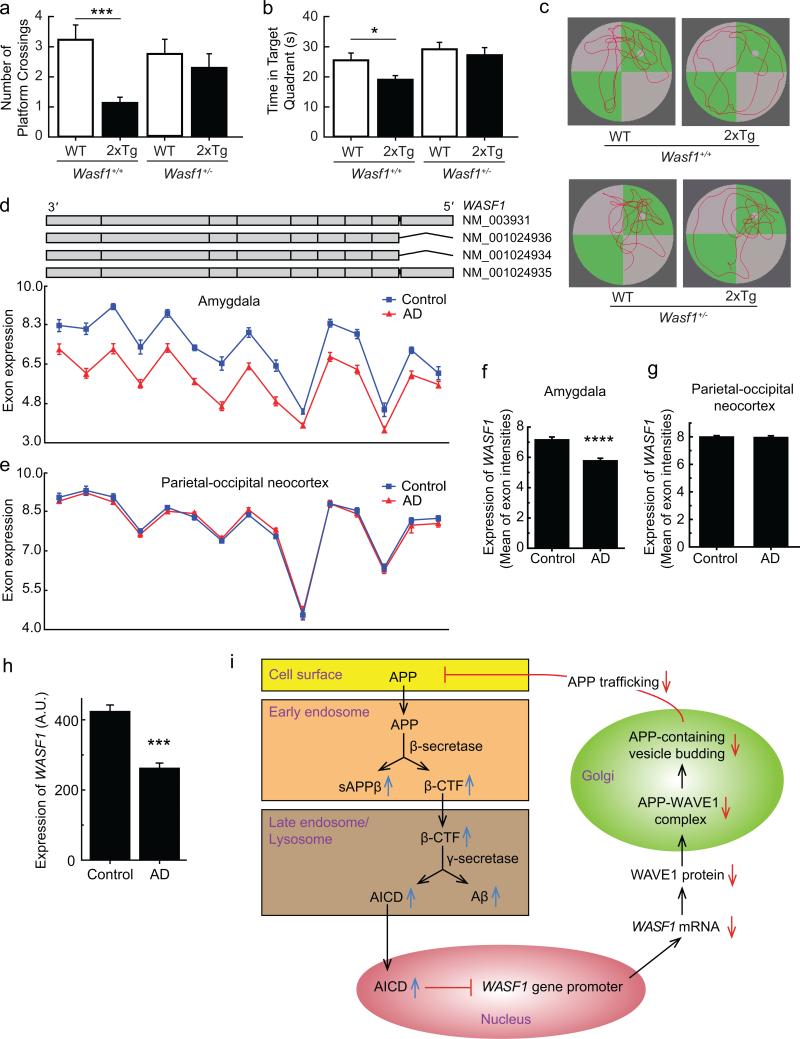
To assess the behavioral consequences of the reduced level of WAVE1, we tested WT mice or 2xTg mice harboring Wasf1+/+ or Wasf1+/– in the Morris water maze task. We did not test Wasf1–/– KO mice in this task because of a previously characterized sensorimotor retardation phenotype4. There were no significant differences between the groups for swim time (Supplementary Fig. 8a) or speed (Supplementary Fig. 8b) during the spatial acquisition sessions (Sessions 1–4; hidden platform), indicating that there was no difference in learning. There were also no significant differences between the groups during the cued session when the platform was visible (Supplementary Fig. 8a, b). During the probe trial, the platform was removed, and the number of platform crossings made where the platform had been located (Fig. 4a) and the amount of time spent swimming in the target quadrant (Fig. 4b) were measured. The 2xTg mice with Wasf1+/+ alleles made significantly fewer platform crossings and spent significantly less time in the target quadrant compared to their non-2xTg (WT) littermates, indicating a memory deficit. These deficits were not observed in the 2xTg mice heterozygous for WAVE1 KO, suggesting an ameliorated memory deficit (Fig. 4a–c). There was no statistically significant difference in the APP level between 2xTg mice groups harboring Wasf1+/+ and Wasf1+/– alleles (Supplementary Fig. 8c). Thus, the beneficial effect of lower Wasf1 gene dosage on the behavioral phenotype is consistent with its role in lowering Aβ production.
Figure 4.
Behavioral consequence and clinical relevance of WAVE1 downregulation, and a model for negative feedback regulation of Aβ production. (a–c) WT (non-Tg) (Wasf1+/+, n = 17; Wasf1+/–, n = 15) and 2xTg (tg/+) (Wasf1+/+, n = 15; Wasf1+/–, n = 10) AD mice (7 month-old, males) were analyzed in the Morris water maze. The number of platform crossings (a) and time spent swimming in the target quadrant (b) were measured (mean ± SEM). ***P < 0.001, *P < 0.05, two-tailed t-test. Representative swim paths recorded during the probe trial (c). (d–g) WASF1 exon expression profiles (d, e) and the mean value of the exon intensities (f, g) from the amygdala (d, f) or the parietal-occipital neocortex (e, g) in 19 AD cases and 10 controls. Each red triangle or blue square represents least squares mean expression of an exon in AD or control tissues, respectively. The upper part in d shows the structure of the four WASF1 isoforms retrieved from the UCSC browser. Mean ± SEM. ****P < 0.00001, one-way ANOVA. (h) Differences in WASF1 transcript expression between 173 AD cases and 187 controls in an independent publicly available dataset23. Mean ± SEM. ***P < 0.001, one-way ANOVA. (i)Proposed model of negative feedback circuit. The processing of APP in the amyloidogenic pathway increases Aβ and AICD. AICD negatively regulates WASF1 gene promoter activity. Downregulation of WAVE1 inhibits the trafficking of APP from the Golgi apparatus to the cell surface and endosomal membranes, resulting in reduced production of Aβ. Colored backgrounds indicate different subcellular compartments.
In support of the clinical relevance of WAVE1 downregulation by the amyloidogenic pathway, lower WASF1 expression level was observed in human AD brains compared to healthy control brains. Multiple probes were used to quantify the WASF1 exon expression profile in the amygdala in 19 AD cases and 10 controls (Fig. 4d). Since neuronal loss in the amygdala is less excessive compared to the hippocampus, the use of amygdala tissue as an affected brain region minimized the impact of AD-related neuronal loss on expression levels. The least squares mean of the gene expression intensity of AD versus controls was 5.8 ± 0.14 versus 7.2 ± 0.16 (p = 4.5E–06, One-way ANOVA) across all exons (Fig. 4f). However, comparable levels of ACTB and GAPDH mRNA in AD and control samples were measured in the same amygdala samples (Supplementary Fig. 9a–d). Such WAVE1 downregulation in AD cases was not observed in unaffected brain regions such as the parietal-occipital neocortex (Fig. 4e, g) and cerebellum (Supplementary Fig. 9e, f). The consistent differences in WASF1 transcript expression between 173 AD cases and 187 controls was also observed in an independent publicly available dataset23 (Fig. 4h).
According to our model (Fig. 4i), when the amyloidogenic pathway is augmented, increased AICD would bind to the promoter of WASF1 in the nucleus and negatively regulate transcription of the WASF1 gene. Decreased WAVE1 would then retard APP trafficking from the Golgi to the plasma membrane and endosomes, resulting in lower Aβ production. Thus, the lower expression of WASF1 observed in AD patients may reflect a negative feedback and homeostatic control for Aβ during disease progression. Previously we characterized WAVE1 as an activity-regulated phosphoprotein in neurons6,24. Recent studies showed that APP processing is regulated by neuronal activity25-27. Whether WAVE1 signaling pathways are involved in neuronal activity-regulated APP processing remains to be addressed. The role of AICD in AD has remained obscure, probably because of complex regulation of various downstream targets11,17,28,29. Our current study elucidates a signaling mechanism for homeostatic regulation of the amyloidogenic pathway by AICD, and suggests a novel pathway for the development of therapeutic agents. Particularly, WAVE1 interaction with the cytoplasmic domain of APP might be targeted to achieve specific inhibition of APP vesicular trafficking.
ONLINE METHODS
Animals
All procedures involving animals were approved by the Rockefeller University Institutional Animal Care and Use Committee and were in accordance with the National Institutes of Health guidelines. 3xTg and 2xTg AD mice were purchased from the Jackson Laboratory (Jax MMRRC Stock Number 034832). Tg/APPswe AD mice were purchased from Taconic (Model 2789). The constitutive Wasf1 KO mice were generated by Taconic-Artemis (Germany) and maintained at The Rockefeller University. We produced the progeny using in vitro fertilization (IVF) and embryo transfer (ET) techniques, to generate the number of animals needed for the biochemical and behavioral tests. Mice were assigned to experimental groups based on their genotype. Selection of animal samples out of different experimental groups for biochemical analyses was performed randomly and in a blinded fashion. The entire water maze test was run by an experimenter who was blinded to the genotype group allocation. No blinding was done for the measurements of WAVE1 (Fig. 1a–c) and Aβ (Fig. 2h, i).
Cell culture and transfection
Mouse N2a neuroblastoma WT cells, an established N2a cell line expressing APP695 (N2a/APPwt), and an established N2a cell line overexpressing APPswe and familial AD presenilin1 mutant ΔE9 (N2a/APPswe.PS1ΔE9) were maintained in medium containing 50% DMEM and 50% Opti-MEM, supplemented with 5% FBS, antibiotics and 200 μg/ml G418 (only for the established cell lines, Invitrogen)30-33. We confirmed that all cell lines used in this study were not contaminated with mycoplasma (LookOut Mycoplasma PCR Detection Kit, Sigma-Aldrich). Transient transfections of plasmids encoding APP695, APP695swe, AICD, AICD-3xFlag, APLP1-ICD-3xFlag, APLP2-ICD-3xFlag, NICD-3xFlag, mNotchΔE, MME promoter-luciferase and WASF1 promoter-luciferase used Fugene 6 (Roche) or Lipofectamine 2000 (Invitrogen). N2a, N2a/APPwt or N2a/APPswe.PS1ΔE9 cells were transfected with control siRNA or siRNA for the mouse Wasf1 gene using Dharmafect II (Thermo Scientific) or Lipofectamine 2000 (Invitrogen). The On-TARGET sequence was AACGATGAGAAAGGCTTTCCG6,34, and On-TARGETplus Non-targeting siRNA#1 was used as a negative control (Thermo Scientific). siRNA-resistant WAVE1 plasmid was generated previously6. β-secretase 1 inhibitor IV (10 μM, EMD Millipore) and γ-secretase inhibitor (DAPT, 5 μM, BD Biosciences) were incubated overnight (18 h). Staurosporine (100 nM for 24 h, EMD Millipore) and cycloheximide (25 μg/ml for various times as indicated, Sigma-Aldrich ) were used. Lactate dehydrogenase (LDH) assay was performed with LDH Cytotoxicity Detection Kit (Clontech Laboratories, Inc.) according to the manufacturer's instructions.
Plasmids
pCB6-APP695wt and pCB6-APP695swe were reported previously10. The nucleotides of AICD48 were synthesized (Genewiz) and subcloned into pCMV-Script. The cDNA sequence of human AICD50 was amplified from pCB6-APP695. The full length sequence of AICD50 was cloned into pGEM-T Easy vector (Promega). Successful clones were confirmed by sequencing. The full length human AICD50 was also subcloned into the Not I/EcoR I site of pCMV-Tag4A (Stratagene). 3xFlag-tagged (at C-terminus) AICD50 was generated using a 3xFlag-fusion mammalian expression vector system. The cDNA of AICD50 was subcloned into the Hind III/BamH I site of 3xFlag-CMV 14 (Sigma-Aldrich) for 3xFlag tagging the C-terminus. The cDNA sequence of mouse NICD was amplified from p3xFlag-CMV7-NICD (Addgene No. 20183). The cDNA sequence of human APLP1-ICD was amplified from pCAX-APLP1 (Addgene No. 30141). The cDNA sequence of human APLP2-ICD was amplified from pcDNATO-myc-His A-APLP2 (Abgent No. DC00166). 3xFlag-tagged (at C-terminus) NICD, APLP1-ICD and APLP2-ICD were generated by subcloning into the EcoR I/BamH I site of 3xFlag-CMV-14.
SDS-PAGE and immunoblot analysis
Cultured cells or mouse hippocampal tissues were lysed in buffer A (50 mM Tris-HCl, pH 7.5, 150 mM NaCl and 2 mM MgCl2) supplemented with 1% Triton X-100, a protease inhibitor cocktail (Complete, EDTA-free, Roche) and phosphatase inhibitors (30 mM NaF, 1 mM orthovanadate and 30 mM pyrophosphate). The cell or tissue lysates were sonicated twice with a probe-type sonicator (Branson) for 10–20 s, centrifuged and the protein level in the supernatant measured by the BCA method. For sAPPα and sAPPβ, conditioned cell culture media were analyzed. The samples were boiled in standard protein sample buffer, and subjected to 4–20% SDS-PAGE with Tris-Glycine gels (Life Technologies) followed by protein transfer onto a nitrocellulose membrane. For the detection of N-terminal fragment of presenilin1, the samples were not boiled and Tricine gels (Life Technologies) were used. Pre-stained protein size markers were used (Amersham full-range rainbow molecular weight markers, GE Healthcare; SeeBlue Plus2 Pre-stained Standard, Invitrogen). Immunoblotting was performed with a standard protocol using the following antibodies: anti-C-terminal WAVE1 (rabbit polyclonal, 1:5,000)6, anti-WAVE1 (mouse monoclonal, 1:1,000, EMD Millipore), anti-APP (6E10, mouse monoclonal, 1:1,000, Covance) for total APP and sAPPα, anti-giantin (rabbit polyclonal, 1:5,000, Abcam), anti-GM130 (mouse monoclonal, 1:1,000, BD Transduction Laboratories), anti-BiP (mouse monoclonal, 1:1,000, BD Transduction Laboratories), anti-actin (rabbit polyclonal, 1:1,000, Cytoskeleton), anti-sAPPβ (unpublished, a generous gift from Dr. William J. Netzer), anti-N-terminal (PS1 Ab14, rabbit polyclonal, 1:100)35, anti-Pen2 (mouse monoclonal, 1:1,000, Abcam), anti-Notch (Cleaved Notch1 (Val 1744) (rabbit polyclonal, 1:1,000, Cell Signaling), anti-Aph1(anti-Aph1a, rabbit polyclonal, 1:100, Invitrogen), anti-Nicastrin (mouse monoclonal, 1:1,000, BD Transduction Lab), and anti-BACE1 (B690 C-Terminal, rabbit polyclonal antibody, 1:500, BioLegend). Flag-tagged ICDs were detected by immunoblotting with anti-Flag antibody (rabbit polyclonal, 1:1,000, Sigma-Aldrich) following immunoprecipitation with anti-Flag antibody (M2, mouse monoclonal, Sigma-Aldrich).
Immunoprecipitation
Cultured cells were lysed by sonication in buffer A (plus supplements of inhibitors but without Triton X-100). After centrifugation at 100,000 × g for 10 min, the supernatant was discarded and the pellet (membrane fraction) was solubilized with buffer A (plus supplements of inhibitors and 1% Triton X-100). The supernatant obtained after centrifugation at 100,000 × g for 10 min was used as the detergent soluble fraction. The detergent soluble fraction was incubated with anti-C-terminal WAVE1 antibody or control antibody-coupled A/G agarose beads (Pierce) overnight at 4 °C with constant rotation. After washing three times with buffer A containing 1% Triton X-100, bound proteins were eluted by boiling in the SDS sample buffer for 3 min. Samples were subjected to SDS-PAGE with 4–20% Tris-Glycine gels and immunoblot analysis with anti-WAVE1 and anti-APP antibodies.
RNA isolation and quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA was extracted using the RNeasy Plus Mini Kit (Qiagen) according to the manufacturer's protocol. One microgram of total RNA was converted to complementary DNA using Superscript II reverse transcriptase (Invitrogen) and oligo-(dT)12-18 primers (Bioline) according to the manufacturer's instructions. qRT-PCR was performed in a 20 μl reaction mixture containing 1 μl complementary DNA, 10 μl Taqman Gene Expression master mix (Applied Biosystems), and 1 μl Taqman primers for each gene. The mouse Gapdh primer was Mm99999915_g1 and the mouse Wasf1 primer was Mm01189596_m1 (Life technologies). Reactions were run at 50 °C for 2 min and 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 1 min on an ABI PRISM 7500 Fast sequence detection system (Applied BioSystems). Comparative quantitation of each target gene was performed based on the cycle threshold (CT), which was normalized to Gapdh.
Enzyme-linked immunosorbent assay (ELISA) for Aβ
The analysis of Aβ40 or Aβ42 was performed by ELISA kit according to the manufacturer's instructions (Life Technologies). Briefly, supernatants of conditioned media from N2a/APPswe.PS1ΔE9 cells were diluted in a buffer provide by the kit and incubated in a 96 well plate with Aβ40 or Aβ42 antibody for 3 h at room temperature. After washes of unbound material, a secondary antibody conjugated to horse-radish peroxidase (HRP) was added for 30 min. Unbound secondary antibody was washed, and subsequently samples were incubated with a developing reagent for 30 min. A stop solution was added to block further reaction between horse-radish peroxidase (HRP) and the colorimetric substrate. An absorbance multiplate reader was used to quantitate the colorimetric reaction at 450 nm. For the analysis of mouse brains, 4–5 month-old male mice were euthanized by CO2 followed by decapitation. Brains were removed and frozen on dry ice and stored at –80 °C until use. Half brains were homogenized in 5 volumes of Tris Buffered Saline (Sigma-Aldrich) containing protease inhibitors (Roche Diagnostics, Mannheim, Germany), and then centrifuged at 50,000 × g, at 4 °C for 20 min. The resulting pellet was resuspended in 70% formic acid and sonicated for 30 s. The samples were neutralized in 10 volumes of neutralization buffer (1 M Tris-base, 0.5 M Na2HPO4, 0.05% NaN3 pH 9.0) and Aβ40 or Aβ42 were measured by ELISA. A preliminary study showed a water soluble fraction gave a negligible amount of Aβ40 and 42 at 4–5 months of age. Although we analyzed a formic acid soluble fraction in this study, the size of plaques at this age is not big enough to be detectable by immunohistochemistry.
Chromatin immunoprecipitation (ChIP)
ChIP assay was performed using the EZ ChIP Kit (Millipore) according to the manufacturers’ instructions. DNA was cross-linked with formaldehyde (1% final concentration for 10 min). N2a cells transfected with 3xflag-tagged AICD were lysed in the presence of protease inhibitors. Lysates were sonicated using a Bioruptor (Diagenode) to shear DNA (average size 200–600 bp). Immunoprecipitation was performed with 1 μg of mouse anti-Flag M2 (Sigma-Aldrich), or 1 μg of anti-RNA polymerase II as a positive control or 1 μg normal mouse IgG as a negative control. Immune complexes collected with protein A/G agarose were subjected to decrosslinking by adding 200 mM NaCl followed by incubation at 65 °C for 4–5 h. Uncrosslinked samples were treated with RNase and Proteinase K and were loaded onto polypropylene spin columns. After washing the column, DNA was eluted in a low-salt buffer (the columns, washing and eluting buffers are included in the kit). Conventional PCR was performed with primers 5′-GGGGCACAGCTGGTATATGT-3′ and 5′-CTTTTCACCAACGGCATCTT-3′. The primers were designed with a software Primer3 WWW primer tool, available at http://biotools.umassmed.edu/bioapps/primer3_www.cgi. Real-time PCR data are represented as percentage of input.
Cell surface biotinylation
N2a/APPswe.PS1ΔE9 cells were cultured on a 60 mm dish and transfected with siRNA targeting Wasf1 or control siRNA. 48 h after transfection, cells were washed three times with ice-cold 1 × phosphate buffered saline (PBS) containing 2.5 mM CaCl2 and 1 mM MgCl2 (pH 8.0). Cells were then incubated with either ice-cold PBS (negative control) or ice-cold PBS plus 10 mM Sulfo-NHS-SS-biotin at 4 °C for 30 min. Cells were washed one time with ice-cold 50 mM Tris (pH 8.0) and twice with ice-cold PBS (pH 8.0) to quench the reaction before being lysed in 1 × RIPA buffer containing protease inhibitors (EDM Millipore). Cell lysate from each condition was incubated with 50 μl streptavidin-coupled magnetic beads (Invitrogen) rotating on a tumbling wheel for 1 h at 4 °C. The streptavidin-coupled beads were then washed four times with ice-cold 1 × PBS (pH 7.0) containing 0.01% Tween 20 and protease inhibitors. Samples were centrifuged at 5,000 rpm at 4 °C for 5 min and resuspended in Laemmli buffer. Finally, beads were removed using a magnet and samples were incubated at 55 °C for 15 min before analysis by SDS-PAGE and immunodetection with RU-369 antibody (rabbit polyclonal, 1:1,000) which recognizes the C-terminus of APP69536.
WASF1 or MME promoter-luciferase assay
N2a cells were co-transfected with a plasmid vector expressing luciferase under the human WASF1 gene promoter, MME (human neprilysin) promoter or a control vector with an empty promoter (a plasmid expressing only luciferase without any gene promoter) (SWITCHGEAR Genomics) with a plasmid expressing APPswe, AICD, APLP1-ICD, APLP2-ICD or NICD. The sequence of WASF1 promoter is:
CAGATTGCAATGGAACACATCATTAACTTTCTTTGCTAAGAAATAATGTCTTACATTT TGAAACAATAAGCTAATTATTATTAGAATAACTACTGTCGAGCTGAGAGCTTAAAAA AGTTTATCTAGGGAAAAAGGGAGAATTATAATAGTTTATAACATTTAGGACCTAGTT GACTAAAGAATATGAGTTCCAAAATACCTTGAGGCCTCGCGGTCCTGCACTCCCAGC GTGCAGCGCGGCAGGAAGCGCGAGAGGGCACAATGGGAAGTGTAGTTTGCAACGA GGGGGCGGAACGGGGGCGTGTCCCCCTTTGACCCCGCCCCCGAATGGATGTTCCATT CAATTGGCTATCATCTTCCCTCATTCCCTAGCCAAGCAGTGTTCTCTTCGTCCCCCTC CCCCAAACTGAGGATTGGGCAATACCACAGAACCTCAGGAAAGGGGGGAAGAGCG AGCTTCGGCCCCACTAATGGGGGAGTGGGCGGAGGCTGGATTTCCCACCTCGGCTG CACCTGGGCACTGGAGGCTGAAGAGGAAAGTGAGAATCTGAAGTTTTGAGACCTCT GACTGGCCAGGAATAGCTCCTGGGGCGGGGGGCAAGGATGGGACCATAGGCGGAA AGAGTCTCGCGGTCCCCCCTGCTTCTGGCGCGGGTCCCTGCGCCCGGTTGTGGAGCG TCTCGCGCGGGGAGGGGGCGGGGGGAACGGCAGCTCGCGGTGTTGTTCACTCGCGC GTCGAGCACACGGTGGGTCCGGCGGCGGGTTGGCGCCCCAGGCGGCGTTCCCTGTG GCCTGGCGCCTGGGCCGCTGCCCTGAGCGGGTTCCGCCCCAGAGCCCGACCCTCCTG GGGGCTCTAGGCGGAGTCCCGCGAGCCGAGGGGGACCGGCGACCGCTGCCGAAGC ATGAAGAAGGGGTAAGGCGTGAGCCCCCAAGATTTCACGGTGAGCGCGAGTCCCTG AGGAATTGCTTTTT.
The sequence of MME promoter is:
ATGAAGTGACTGGATTTGGGAGACCTAGCGGAAACCTTCAGGTCACCCAGGGAACT GCTCCTCTCTCTGGTCCTGCAGCGGCGAGGCGGGGTAGGGGGTGGGGGGGGTGGGC CGTGAGAGCGCCGAGACGCGCGGGGCGCGGAGATGTGCAAGTGGCGAAGCTTGAC CGAGAGCAGGCTGGAGCAGCCGCCCAACTCCTGGCGCGGGATCTGCTGAGGGGTCA CGGTGAGTCTCTGTGGGTGCGGGTCGGAGGGATGCCCAGGTGCCTCGTCGCCCCAC GGCCCGGGCGGAGGCGGGTCCGGAACCTCCCCCAAGTCCGGCCTCAGCCTGGAGCC CGCGCGCCTTGGGCGGATGCACGGACTGAGAGGCGCTTGGCTGGGCTCTCAGCCCG CCACTGCCACCCGGCCCCGTGCGCTCATTGGTCGGGATGTGTCGGCTCAGCAGCCTC CAACTTCTCCCGAATCCCACTGGTGAGTCCCAGGAGAGCGAGCTGAGGGAGAAAGG TCCAAAGGGCGCGAGCGCCCAGGGCGCTGGGAGCCCGTGGGACGGGCACGGGGAG GGCAGAGCCAGCCGAGGGGAGCGGCTGCCGGGAGGTGCGCTGAGCAGGGTCCGCA GCTAAGGTCCAGCGCGATCCGAGCGCCCAGGACCCTGCGGCCGTGGAGGAGGCGTG CCCTGGGAAGGAGCCGCTACTGGGACCTGAAGAGTGAGGGGAGGGAGAGGGGCCC AGAGGGTGACCTGGAGGAGGGCTCTGGAAGTCACGTCAGGTTGGCTCTTCAGGTTC ATTTCCATAGTTCCCTGCGGCCTCTGCCTTGGGGAGTTATGTTTTGTTACCGAGATCC GCGCTACCAGATTGCACCGGGGCTGATTTGGGGGCTGGGAATTTGCCATTCTGCTGT ACAGACACTGATTTTTTTTTCTTCTTTTTAAAAAGCAAGGTTTGTTTTCATTTTGGTTT CTGTCGGATTTTCTCAT.
The intensity of luminescence was measured at 48 h post transfection by the MISSON LightSwitch Luciferase Assay system (Sigma-Aldrich) according to the manufacturer's protocol using a Veritas microplate luminometer (Turner Biosystems, Sunnyvale, CA). The data represent the ratio of luminescence intensity from the experimental cell extracts to intensity from the extracts of the cells transfected with the control vector with empty promoter.
Immunocytochemistry
Cells were grown on poly-L-lysine coated coverslips (12 mm) in 12 well dishes and were infected with viral vector expressing a fluorescent protein fused-Golgi targeting sequence from a marker (human Golgi-resident enzyme N-acetylgalactosaminyltransferase 2) (GFP-Golgi) (Invitrogen) 16 h prior to fixation. Cells were fixed with 1 mL of 4% paraformaldehyde for 20 min. After fixation, coverslips were washed three times with DPBS (1 × Dulbecco's phosphate-buffered saline, Invitrogen). They were then transferred into 24 well dishes. Triton X-100 (3% in DPBS) was added to each well for 15 min to permeabilize cell membranes. For blocking, 3 ml of 10% normal goat serum (NGS) (Thermo Scientific) or 10% bovine serum albumin (BSA) was added to each well for at least 1 h. After blocking, coverslips underwent a single wash with DPBS. Primary antibodies were diluted in 1% NGS or BSA and added to each well for overnight incubation at 4 °C. The primary antibodies were: anti-WAVE1 antibody (rabbit polyclonal, 1:1,000, V010137), anti-APP (mouse monoclonal, 1:500, Covance), anti-GM130 (mouse monoclonal, 1:500, BD Transduction Laboratories) and anti-Giantin (rabbit polyclonal, 1:1,000, Abcam). The next day, the coverslips were washed three times with DPBS. Secondary antibodies (Alexa Fluor 488, 568, and 633/647 dyes, 1:500 dilutions, Invitrogen) were diluted in 1% NGS or BSA and added to each well for 1 h incubation at room temperature. Coverslips were then washed three times with DPBS, removed from their wells, and mounted on microscope slides using ProLong Gold Antifade reagent with or without DAPI mounting medium. After drying overnight, each coverslip was secured to its slide using nail polish.
Brain perfusion and immunohistochemistry
12 month-old 3xTg and age-matched control male mice were deeply anesthetized with Nembutal and transcardially perfused with 10 ml of PBS followed by 40 ml of 4% paraformaldehyde (PFA) in PBS. Brains were dissected, postfixed for 1 h in PFA and cryoprotected in different percentage weight/volume (w/v) sucrose in PBS at 4 °C (5% for 1 h, 15% overnight, and 30% overnight, at 4 °C). Brains were placed in an embedding mold filled with Neg-50 embedding medium (Richard Allan Scientific, Kalamazoo, MI) for 1 h at room temperature, and were subsequently incubated on dry ice for 1 h to freeze the embedding medium. Brains were then stored at –80 °C until being sectioned. Sagittal sections (12 μm) were cut, mounted on glass slides, and kept overnight at –20 °C, then transferred to – 80 °C until used for immunohistochemistry. Slides were thawed and dried at room temperature for 20 min and washed with PBS. Then, sections were blocked in 2% normal serum (vol/vol) at room temperature for 1 h and incubated overnight at 4 °C with primary antibody against β-amyloid (6E10, mouse monoclonal, 1:1,000, Covance) or against WAVE1 (rabbit polyclonal, 1:1,000, V010137). The specificity of anti-WAVE1 antibody was previously demonstrated by immunohistochemistry with the Wasf1–/– brain4 and also confirmed by us. The next day, sections were washed and incubated with Alexa-fluor-conjugated secondary antibodies (1:500, Invitrogen) at room temperature for 1 h. Slides were mounted with Prolong Gold Antifade contained DAPI staining (Life Technologies), dried overnight, and fluorescence was visualized on a Zeiss confocal microscope.
Image acquisition and quantification
Fluorescent images were taken using a confocal microscope (Zeiss LSM 710) with an oil immersion lens (Plan-Apochromat 100 ×/1.40 N.A. Oil DIC) and a 2 × digital zoom. DAPI was excited using the 405 nm emitting diode laser; Alexa Fluor 488 or organelle markers labeled with GFP (Cell Light fluorescent proteins, Invitrogen) using an argon laser (488 nm); Alexa Fluor 568 dye using a 561 nm DPSS laser; Alexa Fluor 633 and 647 dyes using a HeNe laser (633 nm). To obtain line scans to test for co-localization, the fluorescence intensity signals for WAVE1, APP and Golgi-GFP were plotted against distance in μm for all three colors (Fig. 3c, indicated by the white line). An average of 5 pixels was used to obtain line scans for the data shown in Supplemental Fig. 4 and plotted against distance in μm. For the quantification of APP in the Golgi apparatus, the z slice with the strongest Golgi signal was chosen. The images were analyzed with the MetaMorph software (ver. 7.7.8). The immunostaining of a Golgi marker protein, giantin, was used to define a mask for the Golgi area to quantify the mean intensity of APP signal in this area. One cytoplasmic area of the same dimension as the Golgi area was randomly chosen. The ratio of the average intensity of APP signal on Golgi over the average intensity of APP signal in the cytoplasm was determined as APP enrichment.
Measurement of AICD expression
N2a cells transiently transfected with plasmids were gently scrapped in cold PBS and collected by centrifugation at 12,000 rpm for 1 min at 4 °C. Cell pellet was lysed in RIPA buffer (150 mM NaCl, 1.0% IGEPAL CA-630, 0.5% sodium deoxycholate, 0.1% SDS, and 50 mM Tris, pH 8.0) (Sigma-Aldrich) containing protease inhibitors (Complete mini, EDTA-free, Roche) and centrifuged at 12,000 rpm for 20 min at 4 °C. Protein concentration of the supernatant was measured by the BCA method (Thermo Scientific). Supernatant (50 μg) was loaded onto a Tris-HCl precast gel (Bio-Rad) and run with MOPS buffer. The gel was transferred to a PVDF membrane and incubated with RU-369 rabbit antibody (1:1,000 dilution) developed in house to detect the C-terminus of APP36.
Metabolic pulse labeling and chase experiments
N2a/APPswe.PS1ΔE9 cells were transfected with control siRNA or Wasf1 siRNA. 2 days after transfection, cells were washed with serum-free and methionine-free DMEM medium (high glucose, no glutamine, no methionine and no cysteine; Invitrogen) supplemented with 4 mM L-Glutamine (Invitrogen) and incubated with the medium at 37 °C to starve for 30 min. Cells were then pulse-labeled with [35S]methionine (250 μCi/ml) in the medium at 37 °C for 15 min. After washing with pre-warmed PBS, cells were chased with serum-containing normal medium for indicated times. Cells were lysed in buffer A supplemented with 1% Triton X-100 and a protease inhibitor cocktail (Complete, EDTA-free, Roche). To measure Aβ levels, soluble lysates were subjected to immunoprecipitation with 4G8 antibody, followed by SDS-PAGE with 10–20% Tricine gels and protein transferring to PVDF membrane.
Previous studies examined immature and mature APP using metabolic pulse labeling and chase38,39. To measure immature and mature APP, N2a/APPswe.PS1ΔE9 cells were pulse-labeled and chased for 20 min as described above. The immunoprecipitation of APP was performed with 6E10 antibody, followed by SDS-PAGE with 8% Tris-Glycine gels and proteins were transferred to nitrocellulose membrane. Membranes were exposed to autoradiography film (LABSCIENTIFIC, inc) and also scanned using a Typhoon 9400 (GE Healthcare) followed by quantification with Image Quant (Ver. 5.2, Molecular Dynamics) or line scanning with ImageQuant TL Toolbox (Ver. 7.0, GE Healthcare).
In vitro budding assay
An in vitro budding assay was performed with non-radiolabeled Golgi membranes prepared from N2a/APPwt cells transiently transfected with control siRNA or Wasf1 siRNA, and cytosol prepared from N2a cells transiently transfected with control siRNA or Wasf1 siRNA.
Preparation of Golgi membranes
All procedures were performed at 4 °C. N2a/APPwt cells were homogenized using a Dounce homogenizer (20 strokes with a tight pestle, Wheaton) in sucrose buffer (0.25 M sucrose in 10 mM Tris-HCl, pH 7.4, 2 mM MgAc2, 0.5 mM EDTA) containing a protease inhibitor cocktail (Complete, EDTA-free, Roche). The homogenate was loaded on the top of a step gradient composed of 1.5 ml of 2 M sucrose, 2.5 ml of 1.3 M sucrose, 2.5 ml of 1.16 M sucrose, 2.0 ml of 0.8 M sucrose and 2.0 ml of 0.5 M sucrose. All sucrose solutions contained 10 mM Tris-HCl (pH 7.4) and 1 mM MgAc2. The gradients were centrifuged for 2.5 h at 39,000 rpm in a Beckman SW41 Ti rotor. Twelve fractions (each 1 ml) were collected from the top of each gradient. The same volume of each fraction (15 μl) was subjected to 4–12% SDS/PAGE, followed by protein transfer onto nitrocellulose membrane and immunoblotting. Fraction #6 was used for Golgi membranes (Supplementary Fig. 5c). The 1-ml Golgi fraction was mixed with 2 ml of breaking buffer (10 mM Hepes-KOH, pH 7.2, 90 mM KCl) and centrifuged at 100,000 × g for 6 min. The pellet was resuspended in 300 μl of breaking buffer, and then centrifuged at 11,000 × g for 1 min. The pellet was resuspended in 300 μl of breaking buffer. The total protein concentration was determined using the Bradford method. Fresh Golgi membranes were used for each in vitro budding assay.
Preparation of cytosol
All procedures were performed at 4 °C. N2a cells were homogenized using a Dounce homogenizer (20 strokes with a tight pestle) in homogenization buffer (25 mM Hepes-KOH, pH 7.2, 125 mM KCl). The homogenates were centrifuged at 100,000 × g for 1 h. The supernatant was passed through a Sephadex G-25 column (PD-10, GE Healthcare) equilibrated with the homogenization buffer. The cytosol fraction was pooled, and total protein concentration was determined using the by Bradford method. Aliquots were stored at –80 °C.
Reconstitution
Golgi membranes (10 μg) was reconstituted with 15 μg of cytosol in a final volume of 150 μl of reaction buffer (12 mM Hepes-KOH, pH7.2, 110 mM KCl, 2.5 mM MgCl2, 0.1 mM CaCl2) containing an energy-regenerating system consisting of 1 mM ATP, 0.2 mM GTP, 10 mM creatine phosphate, 80 μg/ml of creatine phosphokinase. The reaction mixture was incubated at 37 °C for the indicated times. Golgi membranes and vesicles were separated by centrifugation at 4 °C at 11,000 × g for 1 min. The pellet (Golgi membranes) was resuspended with 45 μl of Laemmli buffer and boiled for 3 min. The supernatant (vesicles) was concentrated using Microcon-30 kDa centrifugal filter (EMD Millipore), and completely drained. The protein that remained on the filter membrane was recovered in 45 μl of Laemmli buffer with vigorous vortexing (the yield of APP protein recovery was over 95%) and boiled for 3 min. Golgi membranes (5 μl) and vesicles (5 μl) were subjected to 8% Tris-Glycine SDS-PAGE followed by protein transfer onto nitrocellulose membrane and immunoblotting with anti-APP antibody (6E10, mouse monoclonal, 1:1,000, Covance). APP-containing vesicles (inferred from APP level in vesicles) was quantified as % of total APP (APP in Golgi membranes plus vesicles) and the time-dependent fold increase of APP-containing vesicles was calculated.
Analysis of WASF1 gene expression in human brain tissue
All procedures involving human brain tissue were approved by institutional review board of the Columbia University Medical Center. Expression profiling was performed separately for amygdala (as affected brain region), and the cerebellum and parietal-occipital neocortex (as unaffected brain regions) from 19 AD and 10 control brains from the New York Brain Bank (www.nybb.hs.columbia.edu). All the brains were from AD or healthy control subjects without clinical or pathological comorbidity, and high quality RNA samples were available in the brain bank. This 3-region approach allowed us to determine whether the changes in expression patterns of candidate genes such as WASF1 are specific for late-onset AD and consistent with distribution of AD pathology. For instance, WASF1 downregulation in AD cases was observed only in the amygdala but not in the cerebellum and parietal-occipital neocortex. This selection of clean brain samples with internal brain control tissues largely increases a priori power. Based on power analysis, this sample size is adequate to detect small differences in mean expression level.
For the expression profiling of AD and control brains, Affymetrix GeneChip® Human Exon 1.0 ST Arrays were used. Frozen brain tissue was ground over liquid nitrogen and stored at –80 °C until use. Total RNA was extracted and purified using TRIzol Plus RNA purification kit (Invitrogen). Quantification and qualification of all RNA preparations was performed using an Agilent Bioanalyzer (RNA 6000 nano-kit) and only samples with RNA integrity number (RIN) > 8 were used in the subsequent RNA amplification and hybridization steps. The GeneChip expression 2-cycle target labeling kit (Affymetrix) was used for all samples according to Affymetrix protocols. Briefly, the procedure consists of an initial ribosomal RNA (rRNA) reduction step and 2 cycles of reverse transcription followed by in vitro transcription (IVT). For each sample, 1 μg of total RNA was initially subjected to removal of rRNA using the RiboMinus™ Transcriptome Isolation Kit (Invitrogen) and spiked with Eukaryotic PolyA RNA controls (Affymetrix). The rRNA-depleted fraction was used for complementary DNA (cDNA) synthesis by reverse-transcription primed with T7-random hexamer primers, followed by second-strand synthesis. This cDNA served as template for in vitro transcription to obtain amplified antisense complementary RNA (cRNA). Subsequently cRNA from the first round was reverse-transcribed using random primers to obtain single-stranded sense DNA. In this second reverse-transcription, dUTP was incorporated into the DNA to allow for subsequent enzymatic fragmentation using a combination of UDG and APE1. All reverse and in vitro transcription steps were performed using the GeneChip WT cDNA synthesis and amplification Kit (Affymetrix). The resulting fragmented DNA was labeled with Affymetrix DNA Labeling Reagent. Labeled fragmented DNA was hybridized to Affymetrix Human Exon 1.0 ST arrays, then washed and stained using the GeneChip Hybridization, Wash and Stain Kit. Fluorescent images were recorded on a GeneChip scanner 3000 and analyzed with the GeneChip operating software.
Morris Water Maze
The Morris water maze procedure was adapted from previous reports40,41. The entire water maze test was run by an experimenter who was blinded to the genotype group allocation. Testing occurred in a white circular tank (135 cm diameter) filled with water (24 ± 1 °C) made opaque with white non-toxic tempera paint. Various extra-maze cues (e.g., black and white abstract designs) surrounded the tank. Data were collected using a Noldus Ethovision (Leesburg, VA) automated video tracking system. A five-trial shaping procedure occurred one day prior to testing, during which a smaller tank (54 cm) was inserted into the center of the larger tank to decrease the swimming area. In the first shaping trial, mice were placed on a visible circular escape platform (covered in red tape; 10 cm diameter) for 15 s. The mice were then placed in the water at 4 distances progressively further from the platform and given 30 s to locate the platform. If the mouse did not find the platform within 30 s, then it was led to the platform by the experimenter. No data were collected during shaping.
Spatial acquisition and memory in the water maze were evaluated as described below. A transparent acrylic escape platform (10 cm) was submerged 0.8 cm below the water surface in the northeast quadrant of the tank where it remained for all trials during the acquisition sessions. One session occurred on four consecutive days, and each session consisted of five trials. The mouse's start position varied for each trial and each mouse was given 60 s to locate the escape platform, upon which it sat for 15 s before being removed. If the mouse did not locate the platform within 60 s, then the experimenter led the mouse to the platform where it sat for 15 s. The mouse was then dried and returned to the home cage for an inter-trial interval of approximately 20 min. Swim time (s) and swim speed (cm/s) were recorded during each trial.
Spatial memory was evaluated in a probe trial, which occurred one day after the fourth session. During the probe trial (60 s), the platform was removed from the tank. The number of times that the mouse crossed the platform location (i.e. platform crossings) and the amount of time that the mouse spent swimming in the target quadrant (i.e. the quadrant which contained the platform during Sessions 1–4) were measured. Higher numbers during the probe trial indicated better performance.
A non-spatial cued water maze task was conducted to evaluate potential sources of non-mnemonic contributions (for example, swimming ability, visual acuity, motivation) to task performance. Cued testing occurred one day after the probe trial and consisted of 5 trials in one session. During the cued trials, the platform was made visible by raising it above the water surface (0.5 cm), covering it with red tape, and attaching a white circular flag to the side of the platform. The extra-maze cues surrounding the tank were removed. The platform was moved to a different location in the tank for each trial. Mice were given 60 s to locate the platform and the inter-trial interval was approximately 20 min. Swim time (s) and swim speed (cm/s) were recorded during each trial.
Mice included in the data analysis were in good health throughout behavioral testing. Spatial acquisition and cued water maze measurements were averaged within each group for each session. To evaluate the rate of task acquisition, we used ANOVA to analyze the data set from multiple sessions. We used a three-way ANOVA with factors of 2xTg mouse genotype (tg/+, +/+), Wasf1 KO mouse genotype (+/+, +/–), and a repeated measure of Session (StatView). The data passed normality tests. For the probe trial, the number of platform crossings and the time spent in the target quadrant during the probe were analyzed using an unpaired t-test (two-tailed) to compare behavioral performance between two groups, namely WT (non-Tg) mice versus 2xTg mice harboring Wasf1+/+ or Wasf1+/– (GraphPad Prism). Outliers were defined as scores being 2 standard deviations above or below the group mean in the spatial acquisition, cued task, or probe task were excluded prior to statistical data analysis. Data from one WT mouse with Wasf1+/+, two WT mice with Wasf1+/–, and three 2xtg mice with Wasf1+/+ were excluded based on this criteria.
Statistics
For all graphic data, n indicates number of biological replicates. All quantitative data for N2a cells are representative of two or three independent experiments and have been replicated at least two times. To determine the numbers of animal to use per group (for biochemical and behavioral experiments), we based our calculations on empirical data accumulated and also on power analysis. The number of animals for each experiment was appropriate to detect biochemical and behavioral differences. Differences between groups were assessed using unpaired two-tailed t-test or analysis of variance (ANOVA) as indicated in figure legends. The data met the assumptions of the tests. When the variances in two groups were significantly different, we performed unequal variance t-test (Welch's correction).
Supplementary Material
Acknowledgements
This work was supported by National Institutes of Health grant AG09464 and Fisher Center for Alzheimer's Research Foundation to P.G.. Y.K. was supported by US Department of Defense/USAMRAA grants W81XWH-09-1-0392 and P.G. by W81XWH-09-1-0402. A.C.N. was supported by National Institutes of Health grant AG047270. J.-H. A. was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT & Future Planning (2010-0027945). We would like to thank Dr. W. Luo for metabolic pulse labeling and chase protocol, and organelle fractionation protocol; Dr. W. J. Netzer for sAPPβ antibody and mNotchΔE plasmid; Dr. D. Cai for the budding assay protocol; Dr. S. S. Sisodia for the pCB6-APP695wt and pCB6-APP695swe plasmids. We also would like to thank Drs. A. Schaeffer, P. Kurup, P. Lombroso, D. Tampellini, G. Gouras and R. Moir for discussion or sharing materials. We acknowledge R. Norinsky and the Rockefeller University Transgenics Services Laboratory for their excellent IVF services, Z. Dong for research assistance and E. Griggs for graphics.
Footnotes
Author Contributions
Y.K., I.C., A.C.N. and P.G. designed experiments and wrote manuscript. I.C. and Y.K. analyzed WAVE1 in N2a cells and mice. I.C. did ChIP assay and immunohistochemistry of WAVE1. J.-H.A. constructed plasmids including AICD-3xFlag and performed real-time PCR. I.C. and J.-H.A. performed WAVE1-luciferase assays. V.B. and I.C. analyzed AICD and Aβ levels. Y.K. and I.C. performed metabolic pulse label and chase experiments, and in vitro budding assay. C.R. analyzed WAVE1 expression in human samples. X.Z. prepared mice. J.G. performed behavioral experiments. G.M., M.B., S.M.S. and Y.K performed immunocytochemical experiments and data analysis.
Competing Financial Interests
The authors declare no competing financial interests.
References
- 1.Tanzi RE, Bertram L. Twenty years of the Alzheimer's disease amyloid hypothesis: a genetic perspective. Cell. 2005;120:545–555. doi: 10.1016/j.cell.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 2.Ballard C, et al. Alzheimer's disease. Lancet. 2011;377:1019–1031. doi: 10.1016/S0140-6736(10)61349-9. [DOI] [PubMed] [Google Scholar]
- 3.Takenawa T, Suetsugu S. The WASP-WAVE protein network: connecting the membrane to the cytoskeleton. Nat. Rev. Mol. Cell Biol. 2007;8:37–48. doi: 10.1038/nrm2069. [DOI] [PubMed] [Google Scholar]
- 4.Soderling SH, et al. Loss of WAVE-1 causes sensorimotor retardation and reduced learning and memory in mice. Proc. Natl. Acad. Sci. USA. 2003;100:1723–1728. doi: 10.1073/pnas.0438033100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eden S, Rohatgi R, Podtelejnikov AV, Mann M, Kirschner MW. Mechanism of regulation of WAVE1-induced actin nucleation by Rac1 and Nck. Nature. 2002;418:790–793. doi: 10.1038/nature00859. [DOI] [PubMed] [Google Scholar]
- 6.Kim Y, et al. Phosphorylation of WAVE1 regulates actin polymerization and dendritic spine morphology. Nature. 2006;442:814–817. doi: 10.1038/nature04976. [DOI] [PubMed] [Google Scholar]
- 7.Suzuki T, et al. Molecular cloning of a novel apoptosis-related gene, human Nap1 (NCKAP1), and its possible relation to Alzheimer disease. Genomics. 2000;63:246–254. doi: 10.1006/geno.1999.6053. [DOI] [PubMed] [Google Scholar]
- 8.Oddo S, et al. Triple-transgenic model of Alzheimer's disease with plaques and tangles: intracellular Abeta and synaptic dysfunction. Neuron. 2003;39:409–421. doi: 10.1016/s0896-6273(03)00434-3. [DOI] [PubMed] [Google Scholar]
- 9.Hsiao K, et al. Correlative memory deficits, Abeta elevation, and amyloid plaques in transgenic mice. Science. 1996;274:99–102. doi: 10.1126/science.274.5284.99. [DOI] [PubMed] [Google Scholar]
- 10.Lo AC, Haass C, Wagner SL, Teplow DB, Sisodia SS. Metabolism of the “Swedish” amyloid precursor protein variant in Madin-Darby canine kidney cells. J. Biol. Chem. 1994;269:30966–30973. [PubMed] [Google Scholar]
- 11.Muller T, Meyer HE, Egensperger R, Marcus K. The amyloid precursor protein intracellular domain (AICD) as modulator of gene expression, apoptosis, and cytoskeletal dynamics-relevance for Alzheimer's disease. Prog. Neurobiol. 2008;85:393–406. doi: 10.1016/j.pneurobio.2008.05.002. [DOI] [PubMed] [Google Scholar]
- 12.Haass C, Kaether C, Thinakaran G, Sisodia S. Trafficking and proteolytic processing of APP. Cold Spring Harb. Perspect. Med. 2012;2:a006270. doi: 10.1101/cshperspect.a006270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cao X, Sudhof TC. A transcriptionally [correction of transcriptively] active complex of APP with Fe65 and histone acetyltransferase Tip60. Science. 2001;293:115–120. doi: 10.1126/science.1058783. [DOI] [PubMed] [Google Scholar]
- 14.Goodger ZV, et al. Nuclear signaling by the APP intracellular domain occurs predominantly through the amyloidogenic processing pathway. J. Cell Sci. 2009;122:3703–3714. doi: 10.1242/jcs.048090. [DOI] [PubMed] [Google Scholar]
- 15.Belyaev ND, et al. The transcriptionally active amyloid precursor protein (APP) intracellular domain is preferentially produced from the 695 isoform of APP in a {beta}-secretase-dependent pathway. J. Biol. Chem. 2010;285:41443–41454. doi: 10.1074/jbc.M110.141390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Flammang B, et al. Evidence that the amyloid-beta protein precursor intracellular domain, AICD, derives from beta-secretase-generated C-terminal fragment. J. Alzheimers Dis. 2012;30:145–153. doi: 10.3233/JAD-2012-112186. [DOI] [PubMed] [Google Scholar]
- 17.Pardossi-Piquard R, Checler F. The physiology of the beta-amyloid precursor protein intracellular domain AICD. J. Neurochem. 2012;120(Suppl 1):109–124. doi: 10.1111/j.1471-4159.2011.07475.x. [DOI] [PubMed] [Google Scholar]
- 18.Belyaev ND, Nalivaeva NN, Makova NZ, Turner AJ. Neprilysin gene expression requires binding of the amyloid precursor protein intracellular domain to its promoter: implications for Alzheimer disease. EMBO Rep. 2009;10:94–100. doi: 10.1038/embor.2008.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jankowsky JL, et al. Mutant presenilins specifically elevate the levels of the 42 residue beta-amyloid peptide in vivo: evidence for augmentation of a 42-specific gamma secretase. Hum. Mol. Genet. 2004;13:159–170. doi: 10.1093/hmg/ddh019. [DOI] [PubMed] [Google Scholar]
- 20.Rajendran L, Annaert W. Membrane trafficking pathways in Alzheimer's disease. Traffic. 2012;13:759–770. doi: 10.1111/j.1600-0854.2012.01332.x. [DOI] [PubMed] [Google Scholar]
- 21.Suetsugu S, Gautreau A. Synergistic BAR-NPF interactions in actin-driven membrane remodeling. Trends Cell. Biol. 2012;22:141–150. doi: 10.1016/j.tcb.2012.01.001. [DOI] [PubMed] [Google Scholar]
- 22.Anitei M, Hoflack B. Bridging membrane and cytoskeleton dynamics in the secretory and endocytic pathways. Nat. Cell Biol. 2012;14:11–19. doi: 10.1038/ncb2409. [DOI] [PubMed] [Google Scholar]
- 23.Webster JA, et al. Genetic control of human brain transcript expression in Alzheimer disease. Am. J. Hum. Genet. 2009;84:445–458. doi: 10.1016/j.ajhg.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ceglia I, Kim Y, Nairn AC, Greengard P. Signaling pathways controlling the phosphorylation state of WAVE1, a regulator of actin polymerization. J. Neurochem. 2010;114:182–190. doi: 10.1111/j.1471-4159.2010.06743.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kamenetz F, et al. APP processing and synaptic function. Neuron. 2003;37:925–937. doi: 10.1016/s0896-6273(03)00124-7. [DOI] [PubMed] [Google Scholar]
- 26.Lesne S, et al. NMDA receptor activation inhibits alpha-secretase and promotes neuronal amyloid-beta production. J. Neurosci. 2005;25:9367–9377. doi: 10.1523/JNEUROSCI.0849-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cirrito JR, et al. Synaptic activity regulates interstitial fluid amyloid-beta levels in vivo. Neuron. 2005;48:913–922. doi: 10.1016/j.neuron.2005.10.028. [DOI] [PubMed] [Google Scholar]
- 28.Konietzko U. AICD nuclear signaling and its possible contribution to Alzheimer's disease. Curr. Alzheimer Res. 2012;9:200–216. doi: 10.2174/156720512799361673. [DOI] [PubMed] [Google Scholar]
- 29.Nalivaeva NN, Turner AJ. The amyloid precursor protein: a biochemical enigma in brain development, function and disease. FEBS Lett. 2013;587:2046–2054. doi: 10.1016/j.febslet.2013.05.010. [DOI] [PubMed] [Google Scholar]
- 30.Gasparini L, et al. Stimulation of beta-amyloid precursor protein trafficking by insulin reduces intraneuronal beta-amyloid and requires mitogen-activated protein kinase signaling. J Neurosci. 2001;21:2561–2570. doi: 10.1523/JNEUROSCI.21-08-02561.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.He G, et al. Gamma-secretase activating protein is a therapeutic target for Alzheimer's disease. Nature. 2010;467:95–98. doi: 10.1038/nature09325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cai D, et al. Presenilin-1 uses phospholipase D1 as a negative regulator of beta-amyloid formation. Proc Natl Acad Sci U S A. 2006;103:1941–1946. doi: 10.1073/pnas.0510708103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cai D, et al. Presenilin-1 regulates intracellular trafficking and cell surface delivery of beta-amyloid precursor protein. J Biol Chem. 2003;278:3446–3454. doi: 10.1074/jbc.M209065200. [DOI] [PubMed] [Google Scholar]
- 34.Suetsugu S, Yamazaki D, Kurisu S, Takenawa T. Differential roles of WAVE1 and WAVE2 in dorsal and peripheral ruffle formation for fibroblast cell migration. Dev Cell. 2003;5:595–609. doi: 10.1016/s1534-5807(03)00297-1. [DOI] [PubMed] [Google Scholar]
- 35.Thinakaran G, et al. Endoproteolysis of presenilin 1 and accumulation of processed derivatives in vivo. Neuron. 1996;17:181–190. doi: 10.1016/s0896-6273(00)80291-3. [DOI] [PubMed] [Google Scholar]
- 36.Buxbaum JD, et al. Processing of Alzheimer beta/A4 amyloid precursor protein: modulation by agents that regulate protein phosphorylation. Proc Natl Acad Sci U S A. 1990;87:6003–6006. doi: 10.1073/pnas.87.15.6003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Soderling SH, et al. The WRP component of the WAVE-1 complex attenuates Rac-mediated signalling. Nat Cell Biol. 2002;4:970–975. doi: 10.1038/ncb886. [DOI] [PubMed] [Google Scholar]
- 38.Thinakaran G, Teplow DB, Siman R, Greenberg B, Sisodia SS. Metabolism of the “Swedish” amyloid precursor protein variant in neuro2a (N2a) cells. Evidence that cleavage at the “beta-secretase” site occurs in the golgi apparatus. J Biol Chem. 1996;271:9390–9397. doi: 10.1074/jbc.271.16.9390. [DOI] [PubMed] [Google Scholar]
- 39.Urmoneit B, Turner J, Dyrks T. Pulse-chase experiments revealed beta-secretase cleavage from immature full-length amyloid precursor protein harboring the Swedish mutation. Implications for distinct pathways. J Mol Neurosci. 1998;11:141–150. doi: 10.1385/JMN:11:2:141. [DOI] [PubMed] [Google Scholar]
- 40.Gresack JE, Kerr KM, Frick KM. Life-long environmental enrichment differentially affects the mnemonic response to estrogen in young, middle-aged, and aged female mice. Neurobiol Learn Mem. 2007;88:393–408. doi: 10.1016/j.nlm.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Harburger LL, Lambert TJ, Frick KM. Age-dependent effects of environmental enrichment on spatial reference memory in male mice. Behav Brain Res. 2007;185:43–48. doi: 10.1016/j.bbr.2007.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.