Abstract
Heterozygous mutations of the NRXN1 gene, which encodes the presynaptic cell-adhesion molecule neurexin-1, were repeatedly associated with autism and schizophrenia. However, diverse clinical presentations of NRXN1 mutations in patients raise the question whether heterozygous NRXN1 mutations alone directly impair synaptic function. To address this question under conditions that precisely control for genetic background, we generated human ES cells with different heterozygous conditional NRXN1 mutations, and analyzed two different types of isogenic control and NRXN1-mutant neurons derived from these ES cells. Both heterozygous NRXN1 mutations selectively impaired neurotransmitter release in human neurons without changing neuronal differentiation or synapse formation. Moreover, NRXN1-mutant human neurons exhibited increased levels of CASK, a critical synaptic scaffolding protein that binds to neurexin-1. Our results show that, unexpectedly, heterozygous inactivation of NRXN1 directly impairs synaptic function in human neurons, and illustrate the value of this conditional deletion approach for studying the functional effects of disease-associated mutations.
Keywords: Schizophrenia, autism, synapse, human neurons, neurexin, synaptic cell adhesion, iN cells
INTRODUCTION
Autism Spectrum Disorders (ASDs) and schizophrenia impose major disease burdens. Recent advances revealed a complex genetic landscape in neuropsychiatric disorders (reviewed in Murdoch and State, 2013; Giusti-Rodríguez and Sullivan, 2013). Mutations in hundreds of genes with high effect sizes were identified (O’Roak et al., 2012; Sanders et al., 2012; Neale et al., 2012; Purcell et al., 2014; Fromer et al., 2014). However, many of these mutations are so rare that their association with a particular disease was not statistically significant. Moreover, it often remained unclear whether a particular mutation was related to a specific disorder, and whether haploinsufficiency of a gene actually produces a direct functional impairment.
Several candidate ASD genes encode proteins involved in synapse development and/or function. Among these, NRXN1 was significantly associated with both ASDs and schizophrenia in many studies (reviewed in Südhof, 2008; Doherty et al., 2012; Clarke and Eapen, 2014). The NRXN1 gene encodes neurexin-1, a presynaptic cell-adhesion molecule that binds to postsynaptic cell-adhesion molecules such as neuroligins and LRRTMs, which were also associated with ASDs or schizophrenia. Most NRXN1 mutations are heterozygous copy number variations (CNVs) that delete only NRXN1 due to the large size of the NRXN1 gene, while missense and truncation mutations are less frequent (for recent papers, see Enggaard Hoeffding et al., 2014; Gauthier et al., 2011; Todarello et al., 2014; Béna et al., 2014). Although NRXN1 gene mutations are rare (~0.18% of schizophrenia patients; Rees et al., 2014), this frequency translates into thousands of patients. As a result, NRXN1 mutations at present represent the most frequent single-gene mutation in schizophrenia. Moreover, NRXN1 gene polymorphisms have been associated with differential responses to antipsychotic medication in schizophrenia (Souza et al., 2010; Lett et al., 2011), further buttressing the link between NRXN1 and schizophrenia. However, the effects of heterozygous NRXN1 mutations on synaptic transmission in human neurons are unknown.
Neurexin-1 is an evolutionarily conserved presynaptic cell-adhesion molecule (Ushkaryov et al., 1992; Tabuchi and Südhof, 2002). Vertebrates express three neurexin genes (NRXN1, NRXN2 and NRXN3 in humans), each of which includes separate promoters for longer α- and shorter β-neurexins (Ushkaryov et al., 1992 and 1993; Tabuchi and Südhof, 2002; Rowen et al., 2002). Moreover, neurexins are alternatively spliced at six canonical sites, generating >1000 protein isoforms (Ullrich et al., 1995; Treutlein et al., 2014). Triple Nrxn1α/2α/3α knockout (KO) mice exhibit severe impairments in synaptic transmission and die at birth (Missler et al., 2003). Recordings in acute hippocampal slices revealed discrete changes in synaptic transmission but not neurotransmitter release in homozygous Nrxn1α KO mice (Etherton et al., 2009). No studies analyzing cultured neurons from either homo- or heterozygous Nrxn1α KO mice are available, and the role of NRXN1 in synaptic transmission remains unclear.
With the advent of induced pluripotent stem (iPS) cells, it has become feasible to analyze neurons from patient-derived iPS cells. Multiple studies have proved the utility of this approach (reviewed in Hanna et al., 2010; Han et al., 2011; Bellin et al., 2012; Imaizumi and Okano, 2013). Moreover, recent studies showed that mutations in patient-derived iPS cells can be corrected by genomic manipulations (Raya et al., 2009; Zou et al., 2009; Howden et al., 2011). However, many ASD- and schizophrenia-associated mutations exhibit incomplete penetrance, suggesting that genetic background effects significantly influence the clinical presentation and could contribute to the observed phenotypes. This issue is particularly relevant for NRXN1 mutations which have been associated with several different neuropsychiatric disorders (Ching et al., 2010; reviewed in Südhof, 2008; Doherty et al., 2012; Clarke and Eapen, 2014).
Here we assessed the functional significance of a heterozygous disease-associated mutation on human neurons by introducing heterozygous conditional NRXN1 mutations into human embryonic stem (ES) cells. This approach allowed us to analyze the functional effects of NRXN1 mutations on synaptic transmission in neurons derived from the mutant ES cells. We found that heterozygous loss-of-function NRXN1 mutations had no effect on neuronal differentiation and synaptogenesis, but severely impaired neurotransmitter release in a stimulus-dependent pattern. This phenotype was specific to human neurons as mouse Nrxn1α mutations exhibited no phenotype in comparable experiments. Moreover, NRXN1 mutations increased the levels of CASK, a synaptic membrane associated guanylate kinase (MAGUK) that binds to neurexin-1 (Hata et al., 1996). Our findings demonstrate that heterozygous NRXN1 mutations have profound but selective effects on synaptic transmission, demonstrating a haploinsufficiency phenotype in human cells and suggesting a potential mechanism for the role of NRXN1 mutations in ASD and schizophrenia pathogenesis.
RESULTS
Generation of human ES cells with heterozygous conditional NRXN1 mutations
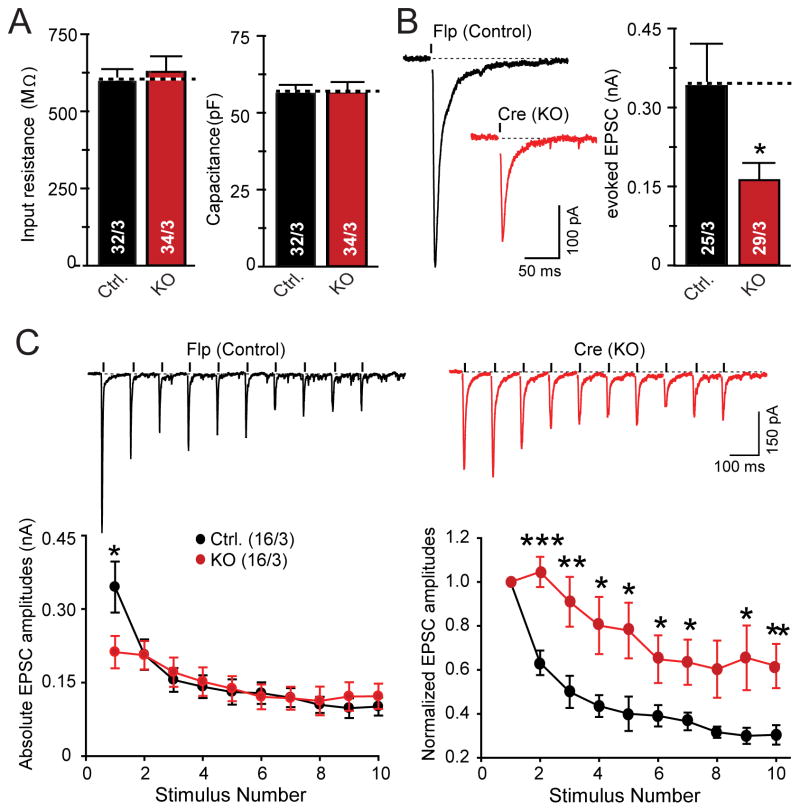
Although hundreds of ASD and schizophrenia cases with heterozygous NRXN1 mutations have been described and constitutive mouse mutants for Nrxn1α (which encodes the major transcript of the Nrxn1 locus) were reported, no functional analyses of cultured neurons from hetero- or homozygous Nrxn1α KO mice are available, and no mouse mutants of the entire Nrxn1 locus were described. The only available data are from slice physiology experiments suggesting a small impairment in synaptic transmission in homozygous Nrxn1α KO mice (Etherton et al., 2009). Thus, it is unknown whether heterozygous or homozygous Nrxn1 mutations in mice produce a synaptic or non-synaptic neuronal phenotype.
To address this gap in our understanding, we analyzed cortical neurons cultured from littermate wild-type and heterozygous and homozygous Nrxn1α KO mice. However, we detected no change in spontaneous miniature excitatory postsynaptic currents (mEPSCs) or action-potential evoked excitatory postsynaptic currents (EPSCs) in hetero- or homozygous Nrxn1α KO neurons (Fig. S1). These results suggest that Nrxn1α is not essential for excitatory synaptic transmission in mouse cortical neurons, but do not rule out the possibility of a more severe human-specific effect.
Thus, to test whether NRXN1 mutations may potentially have a more severe phenotypic effect in human neurons than in mouse neurons, we used recombinant adeno-associated viruses (AAVs) to introduce conditional mutations into the NRXN1 gene in human H1 embryonic stem (ES) cells (Kohli et al., 2004). We separately introduced two different NRXN1 mutations and obtained correctly targeted clones with recombination frequencies of 5–10% (correct clones/total clones), verified by PCR screening and Southern blot analysis (Figs. 1A, S2). Both mutations produce loss-of-function alleles (see below), and control for each other.
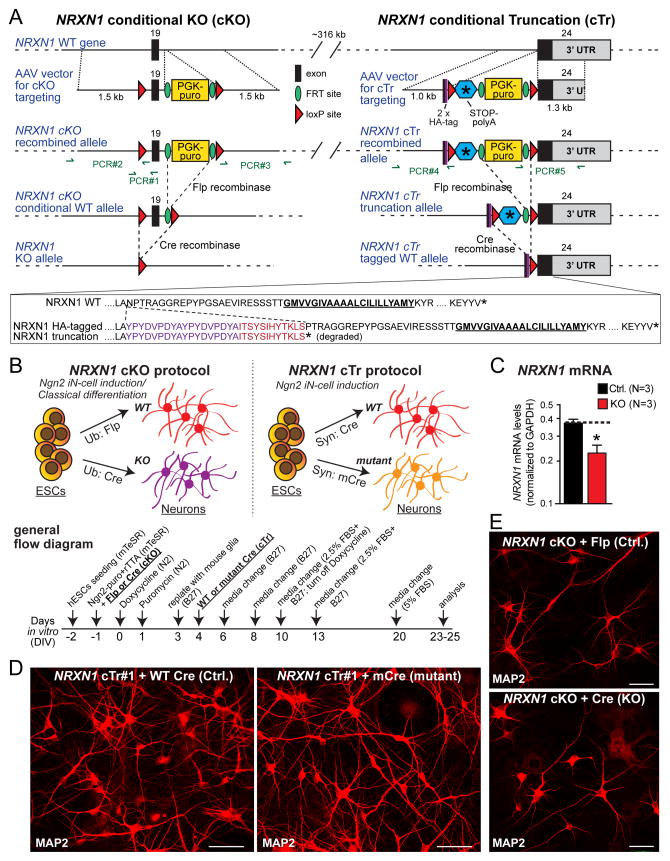
Figure 1. Gene targeting in human ES cells and generation of NRXN1-mutant induced neuronal (iN) cells.
(A) NRXN1 targeting strategy. In the conditional knockout (cKO) strategy (left), we flanked the first exon shared by all neurexin-1 isoforms (exon 19) with LoxP sites and inserted an FRT-flanked PGK-puromycin resistance cassette (PGK-puro). In the conditional truncation (cTr) strategy (right), we inserted a cassette containing two HA epitopes, a loxP site, a stop codon and a polyadenylation signal followed by an FRT-flanked PGK-puro cassette and another loxP site in-frame into the beginning of exon 24, the last NRXN1 coding exon NRXN1 (see sequences below the diagram). All gene modifications were introduced by AAV-mediated homologous recombination in H1 ES cells and validated by PCR and Southern blotting (Fig. S2).
(B) Procedure for generating precisely matching control and NRXN1-mutant iN cells from targeted conditionally mutant ES cells (top; schematic of the overall procedure and the recombinase applications used for the two different mutations; bottom, flow diagram of the overall procedure).
(C) NRXN1 mRNA measurements show that the heterozygous NRXN1 cKO decreases total NRXN1 mRNA levels nearly 2-fold as measured by qRT-PCR of mRNAs spanning exon 19. Data are means ± SEM (n=3). Statistical analyses were performed by Student’s t-test comparing test samples to control (*p<0.05).
(D & E) Representative images of iN cells produced from targeted ES cells and immunolabeled with MAP2 (D, NRXN1 cTr#1 line; E, cKO line; scale bars = 50 μm (D), 40 μm (E)). For analysis of HA-tagged NRXN1 protein produced by cTr iN cells and additional immunolabeled images of cKO iN cells, see Figs. S3, S4.
For the first of the two NRXN1 mutations (the conditional knockout [cKO]), we flanked exon 19 with loxP sites. Exon 19 is the first exon that is shared by all NRXN1 transcripts (Tabuchi and Südhof, 2002). After homologous recombination, the mutated NRXN1 allele carries a selectable marker surrounded by FRT sites, and loxP sites that flank both exon 19 and the selectable marker (Fig. 1A). Flp-recombinase deletes the selectable marker, allowing wild-type expression of neurexin-1 (control), while Cre-recombinase deletes exon 19, disrupting the reading frame of the NRXN1 mRNA and blocking NRXN1 expression (KO; Fig. 1B).
Because there are no working antibodies that specifically detect neurexin-1 protein in human cells, we pursued a second NRXN1 mutagenesis strategy that introduces an epitope tag into neurexin-1 and additionally generates a conditional loss-of-function mutation. In this second mutation (referred to as conditional truncation [cTr]), we inserted into exon 24 (Tabuchi and Südhof, 2002) a cassette containing tandem HA epitopes, a loxP site, a stop codon, and a polyadenylation signal, followed by an FRT-flanked selectable marker (a puromycin resistance cassette) and another loxP site (Fig. 1A). Flp-recombinase deletes the selectable marker but retains the stop codon, producing a truncated HA-tagged neurexin-1 protein. Cre-recombinase, conversely, generates a control NRXN1 allele encoding HA-tagged full-length neurexin-1 (Fig. 1B). The inserted stop codon in the cTr mutation mimics a de novo mutation (G1402fs) found in a schizophrenia patient that truncates neurexin-1 protein 25 residues C-terminal to the cTr mutation (Gauthier et al., 2011).
Generation of NRXN1-mutant human induced neurons (iN cells)
We converted ES cells with conditional NRXN1 mutations into induced neurons using forced expression of the transcription factor Ngn2 (Fig. 1B). This method produces a homogenous well characterized population of excitatory cortical neurons (Zhang et al., 2013). During iN cell differentiation, we co-expressed the appropriate recombinases with Ngn2 in the ES cells to produce precisely matching sets of control and heterozygous NRXN1-mutant iN cells (Fig. 1B). We analyzed iN cells derived from one cKO-mutant and two cTr-mutant ES cell clones. We observed efficient conversion of all genetically altered ES cells into iN cells, with expression of recombinases in virtually 100% of iN cells (Fig. S3). mRNA measurements demonstrated that the cKO reduces NRXN1 mRNA levels approximately 2-fold (Fig. 1C). No effect of NRXN1 mutations on neuronal differentiation was detected, as indicated by unchanged dendritic and cellular morphological properties (Figs. 1D, 1E, S4).
Immunocytochemistry and immunoblotting showed that in control iN cells from cTr-mutant ES cells (i.e., Cre-recombinase treated cTr iN cells), full-length HA-tagged neurexin-1 was abundantly expressed and displayed on the neuronal surface (Fig. S3). Without Cre-recombinase treatment (i.e., in cTr iN cells treated with inactive Cre-recombinase), HA-tagged full-length or truncated neurexin-1 was neither detectable by immunocytochemistry on the neuronal surface nor observed by immunoblotting in the cell lysate or the media. Thus, full-length HA-tagged neurexin-1 appears to be trafficked normally to the neuronal cell surface in control cTr neurons, whereas truncated HA-tagged neurexin-1 is unstable and degraded in mutant cTr neurons. Similarly, mRNA measurements in control and mutant neurons from NRXN1 cKO ES cells failed to detect the mutant mRNA despite robust expression of wild-type mRNA, suggesting that the mutant mRNA is rapidly degraded by nonsense-mediated decay (Fig. S3).
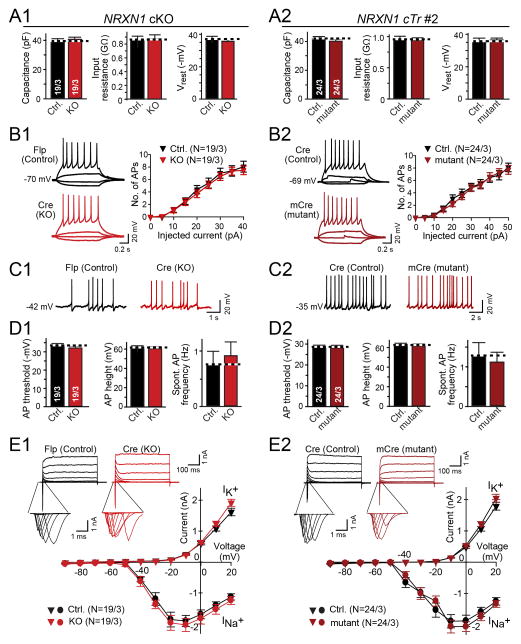
Next, we performed electrophysiological recordings in matched control and NRXN1-mutant iN cells to measure fundamental parameters, such as, membrane capacitance, input resistance, resting membrane potential, neuronal excitability, and voltage-dependent Na+ and K+ currents responsible for action potential generation. We detected no differences between corresponding NRXN1-mutant and control neurons in either cKO- or cTr-mutant NRXN1 iN cells (Fig. 2 and S5). These data suggest that loss of one NRXN1 allele does not lead to major changes in non-synaptic properties of human neurons.
Figure 2. Intrinsic membrane properties are not changed in NRXN1-mutant iN cells.
(A) Summary graphs of the passive membrane properties (left, capacitance; middle, input resistance; right, resting membrane potential) of control and NRXN1-mutant iN cells derived from the NRXN1 cKO (A1) or the cTr#2 line (A2).
(B) Neuronal excitability in control and NRXN1-mutant iN cells is unchanged. Left, representative traces of action potentials induced by step current injections (from −5 pA to +50 pA); right, intensity-frequency plots in the number of action potentials as a function of step current injections (B1, cKO line; B2, cTr#2 line).
(C) Representative traces of spontaneous action potentials recorded from control and NRXN1 mutant iN cells.
(D) Summary graphs of action potential properties in control and NRXN1-mutant iN cells (left, firing threshold; middle, action potential amplitude; right, spontaneous firing frequency).
(E) Voltage-dependent Na+ (INa+) and K+ (IK+) currents are unchanged. Representative traces are shown on top (insets = Na+ currents on an expanded time scale), and current/voltage plots at the bottom. Currents were recorded in voltage-clamp mode; cells were held at −70 mV; stepwise depolarization from −90 mV to +20 mV at 10 mV intervals was delivered.
‘1’ and ‘2’ panels depict data for the NRXN1 cKO and the cTr#2 line, respectively. For data from the cTr#1 line, see Fig. S5. All summary data are means ± SEM (number in bars or brackets show number of cells/cultures analyzed).
Heterozygous NRXN1 mutations decrease the frequency of spontaneous mEPSCs in human neurons without affecting synapse density
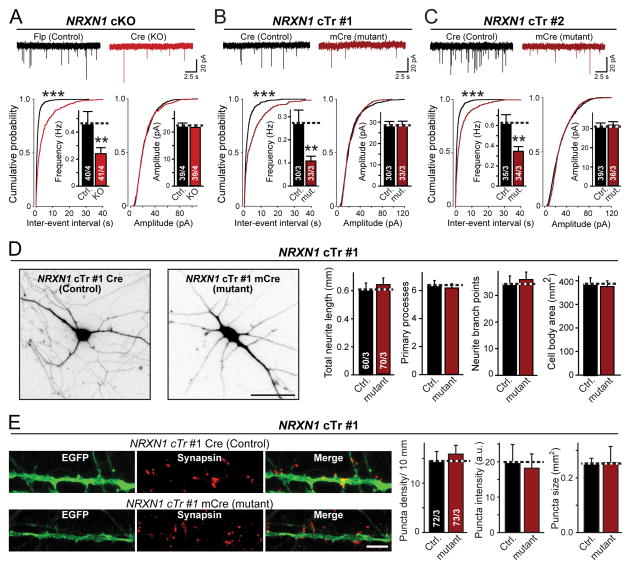
We next asked whether heterozygous NRXN1 mutations alter synapse formation or function, again monitoring the effects of both mutations under precisely controlled conditions. We first measured mEPSCs. Strikingly, we found that both the cTr and the cKO NRXN1 mutation caused an ~2-fold decrease in mEPSC frequency without altering the mEPSC amplitude (Figs. 3A–3C).
Figure 3. Heterozygous NRXN1 mutation decreases the frequency of spontaneous mEPSCs without altering neuronal morphology or synapse numbers.
(A–C) Heterozygous NRXN1 mutations uniformly decrease the mEPSC frequency approximately 2-fold. Top; representative traces recorded from iN cells derived from the NRXN1 cKO line (A) or the two different cTr lines (B and C); bottom, summary plots and graphs of the frequency (left) and amplitudes (right) of mEPSCs. Quantitations are shown both as cumulative probability plots and as bar diagrams (inserts).
(D) Morphological properties are unchanged in NRXN1-mutant human neurons (left, representative images of control and NRXN1–mutant cTr#1 iN cells that were sparsely infected with lentiviruses expressing tdTomato to label individual neurons [scale bar, 40 μm]; right, quantifications of the total neurite length, number of primary processes, neurite branch points, and cell size in control and NRXN1-mutant iN cells).
(E) Synapse density is unchanged in NRXN1-mutant human neurons (left, representative images of isolated control and mutant cTr#1 iN cells that were co-infected with GFP and stained for the synaptic marker synapsin-1 [scale bar, 5 μm]; right, quantification of the density, staining intensity, and size of synapses in control and mutant iN cells).
Data are means ± SEM; numbers of cells/cultures analyzed are shown in the bars. Statistical significance (**, p<0.01; ***, p<0.001) was evaluated with the Kolmogorov-Smirnov-test (cumulative probability plots) and Student’s t-test (bar graphs). Fig. S6 shows a similar characterization of the other lines of NRXN1-mutant ES cells.
A change in mEPSC frequency suggests a presynaptic alteration in the number of synapses and/or their release probability. To differentiate between these two possibilities, we performed a morphological analysis of synapse numbers and dendritic arborization in NRXN1-mutant iN cells.
We sparsely labeled control and NRXN1-mutant iN cells with lentiviruses expressing fluorescent markers (EGFP or TdTomato), immunostained the neurons for synapsin-1, and examined the size and arborization of dendrites as well as the density, size, and labeling intensity of synapses (Figs. 3D, 3E, S6). Neither NRXN1 mutation altered any of these parameters, suggesting that the NRXN1 mutations did not impair neuronal development or synaptogenesis. Thus, the decreased mEPSC frequency observed in NRXN1-mutant neurons is not due to a decrease in synapse numbers.
Heterozygous NRXN1 mutations impair evoked neurotransmitter release but not the readily-releasable pool (RRP) of vesicles
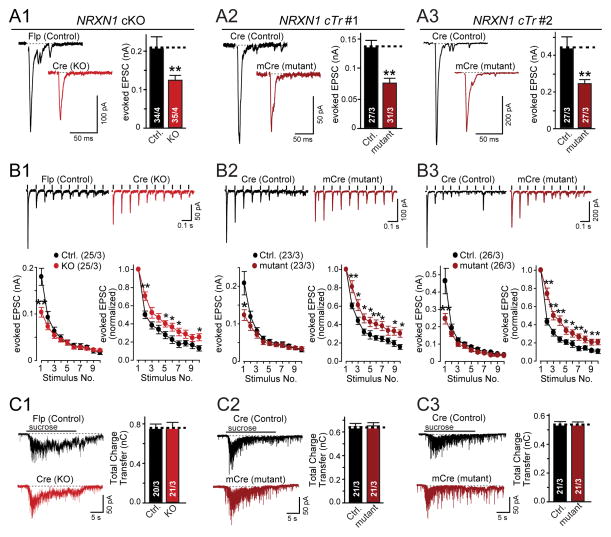
To analyze synaptic function in heterozygous NRXN1-mutant neurons in more detail, we measured evoked EPSCs mediated by AMPA-type glutamate receptors (AMPARs). Consistent with the decrease in mEPSC frequency, we observed an ~2-fold decrease in EPSC amplitude in cTr and cKO mutant neurons compared to corresponding control neurons (Fig. 4A). This surprisingly large effect was not due to the viral expression of recombinases because the two different NRXN1 mutations depend on different recombinases (Fig. 1A) and expression of various recombinases in naïve human neurons had no effect on EPSC amplitudes (Fig. S7).
Figure 4. Heterozygous NRXN1 mutations decrease the initial probability of neurotransmitter release but not the size of the readily releasable pool.
(A) Evoked EPSC amplitudes are decreased approximately 2-fold by heterozygous NRXN1 mutations (left, representative traces; right, summary graphs of the amplitudes of EPSCs evoked by isolated action potentials in control and NRXN1-mutant iN cells as indicated).
(B) Heterozygous NRXN1 mutations selectively impair only the first EPSC in response to a 10 Hz stimulus train (top, representative traces; bottom, summary plots of the absolute (left) and relative EPSC amplitudes normalized to the first response (right)).
(C) Heterozygous NRXN1 mutations do not alter the size of the readily-releasable pool as measured by the amount of release induced by an application of 0.5 M hypertonic sucrose for 20 s (left, representative traces; right, summary graphs of the synaptic charge transfer; numbers in bars indicate the number of cells/cultures analyzed).
The ‘1’, ‘2’, and ‘3’ panels show results separately performed with control and mutant iN cells from the cKO and the two different cTr lines to demonstrate reproducibility as indicated. Data are means ± SEM. Numbers in bars or brackets show number of cells/independent cultures analyzed. Statistical significance (*, p<0.05; **, p<0.01) was determined by Student’s t-test (A) or the Mann-Whitney rank sum t-test (B).
The decreased mEPSC frequency, normal synapse numbers, and reduced evoked EPSC amplitudes suggest that the NRXN1 mutations decrease neurotransmitter release probability. To further explore this possibility, we analyzed short-term synaptic plasticity which represents an indirect measure of release probability (Regehr, 2012).
We found that both heterozygous NRXN1 mutations significantly decreased only the first EPSC response during a 10 Hz stimulus train (Fig. 4B). As a result, normalized plots of EPSCs as a function of stimulus number showed that mutant neurons exhibited significantly less synaptic depression. In accordance with the decreases in mEPSC frequency and EPSC amplitude, this finding strongly indicates that conditional NRXN1 mutations cause a selective large decrease in the initial neurotransmitter release probability in human neurons, but not in the release probability overall. This decrease is not due to changes in the size of the RRP of synaptic vesicles, which can be measured as the cumulative EPSC elicited by a hypertonic sucrose stimulus (Rosenmund and Stevens, 1996), because we detected no change in the RRP size between control and heterozygous NRXN1-mutant neurons (Fig. 4C). Importantly, the two different mutations (the ‘cTr’ and ‘cKO’ mutations; Fig. 1A) exhibited indistinguishable phenotypes.
Heterozygous NRXN1 mutation phenotype is independent of the neuronal differentiation protocol
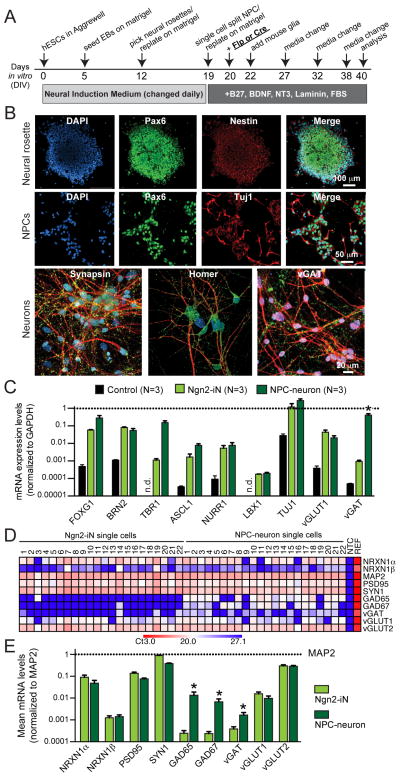
Our analyses of NRXN1 mutations used the iN cell protocol for generating neurons (Zhang et al., 2013), which differs from traditional methods of converting ES cells into neurons via neural precursor cell (NPC) intermediates (e.g., see Zhang et al., 2001, Wu et al., 2007; Chambers et al., 2009, Koch et al., 2009). Because the phenotype of heterozygous NRXN1 mutations in iN cells was surprisingly severe when compared to the lack of a phenotype in similar mouse mutants (Fig. S1), we asked whether this phenotype may be specific to neurons produced by the iN cell method.
We differentiated NRXN1 cKO ES cells into a mixed population of excitatory and inhibitory neurons using a traditional protocol (Figs. 5A, 5B; Zhang et al., 2001, Wu et. al, 2007; Koch et al., 2009). Comparative gene expression analyses of NPC-derived neurons and iN cells using quantitative RT-PCR of select genes both for bulk total mRNA (Fig. 5C) and for single-cell mRNA analyzed on a fluidigm chip (Figs. 5D, 5E; see Pang et al., 2011) demonstrated that gene expression in the two types of human neurons was overall rather similar. Both expressed comparable levels of the pan-neuronal marker Tuj1 and of the telencephalic progenitor marker FOXG1 and cortical layer 2/3 marker BRN2, whereas the layer 6 marker TBR1 was more strongly induced in NPC-derived neurons than in iN cells (Fig. 5C). Expression of other regional markers, such as, ASCL1 and LBX1 was comparably low in both neuronal types. The excitatory presynaptic markers vGlut1 and vGlut2 were similarly induced in both types of neurons, whereas the inhibitory presynaptic marker vGAT was expressed ~1,000-fold more strongly in NPC-derived neurons than in iN cells, consistent with the production of inhibitory neurons only by the traditional protocol (Figs. 5C–5E). Strikingly, NRXN1α was expressed at ~100-fold higher levels than NRXN1β in both types of neurons (Figs. 5D, 5E). Thus, neurons produced from NPC intermediates were cortical in nature similar to iN cells and primarily expressed neurexin-1α, motivating us to analyze the NRXN1-mutant phenotype in these neurons.
Figure 5. Generation of control and heterozygous NRXN1-mutant human neurons using a standard differentiation protocol involving neuronal precursor cell (NPC) intermediates.
(A) Flow diagram of the differentiation protocol (modified from Zhang et al., 2001). Abbreviations: hESCs, human embryonic stem cells; EBs, embryoid bodies.
(B) Representative images of neural rosettes (day 12), NPCs (day 20), and neurons (day 40) obtained as described in A. Cells were immunostained for the indicated markers (scale bars as marked). In neurons, synaptic markers (synapsin-1, homer, and vGAT in green) were co-stained with MAP2 (red). DAPI is indicated in blue.
(C) Comparative analysis reveals similar gene expression patterns in human neurons generated by iN cell and NPC-intermediate based protocols. Data show mRNA levels of the indicated genes (normalized for those of GAPDH [dotted line]) measured by quantitative RT-PCR in total RNA isolated from control cells (an unrelated iPS cell line) and the two different kinds of neurons (day 23 from Ngn2-iN cells and day 40 from NPC-neurons). Data are means ± SEM (* = p<0.05 by Student’s t-test).
(D & E) Analysis of gene expression in single neurons using quantitative RT-PCR. Total cytosol was aspirated from patched iN cells or NPC-derived neurons and mRNA levels were measured on a fluidigm chip for the indicated genes, including separate assays for NRXN1α and NRXN1β. Data are shown for individual cells as a heat map (D) or in a bar diagram as means ± SEM for all cells (E; normalized to MAP2 levels [dotted line] and shown on a logarithmic scale; *p<0.05 by Student’s t-test).
We found that the phenotype of NPC-derived NRXN1-mutant human neurons was indistinguishable from that of NRXN1-mutant iN cells, with no changes in intrinsic electrical properties but a decrease in EPSC amplitude and a selective loss of EPSC size during the first response in a 10 Hz stimulus train (Figs. 6A–6C). Thus, the heterozygous NRXN1 deletion causes the same phenotype in NPC-derived human neurons as in human iN cells, validating the accuracy of that phenotype and demonstrating that iN cells can be used as a useful preparation for the analysis of neuronal phenotypes.
Figure 6. Heterozygous NRXN1 cKO impairs evoked excitatory synaptic responses in human neurons derived by a standard differentiation protocol.
All data are from human neurons generated from NRXN1 cKO ES cells as shown in Fig. 5A, with lentiviral expression of Flp- or Cre-recombinases at day 20.
(A) Input resistance and capacitance values are identical in control and NRXN1 cKO neurons.
(B) ESPCs evoked by isolated action potentials exhibit a ~2-fold decrease in amplitude in NRXN1 cKO neurons (left, representative traces; right, summary graphs).
(C) EPSCs evoked by a 10 Hz stimulus train exhibit a decreased initial amplitude, but subsequent normal amplitudes in NRXN1 cKO neurons (top, representative traces; bottom left, summary plot of absolute EPSC amplitudes; bottom right, summary plot of normalized amplitudes to illustrate the apparent decrease in synaptic depression).
Data are means ± SEM; numbers in graphs represent number of total cells/independent cultures analyzed. Statistical analyses were performed by Student’s t-test (A,B) or the Mann-Whitney rank sum t-test (C) comparing test samples to control (*p<0.05, **p<0.01, ***p<0.001).
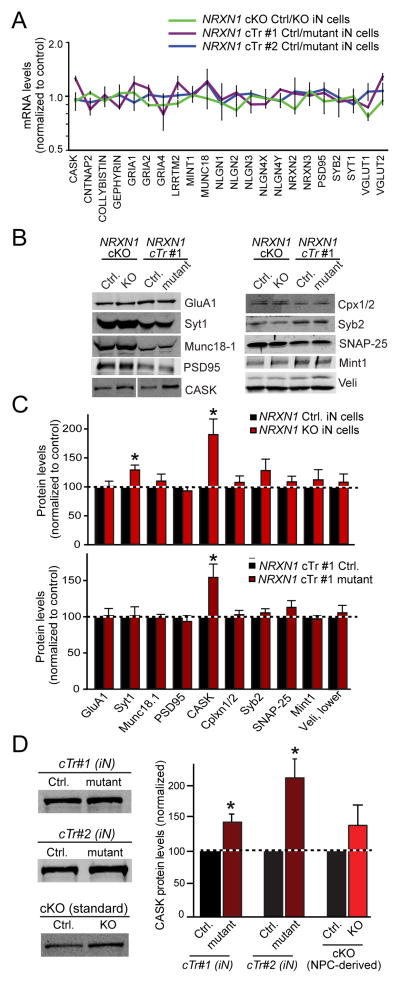
Heterozygous NRXN1 mutations increase CASK protein levels in human neurons
To explore the molecular basis for the decreased initial release probability in NRXN1-mutant human neurons, we tested for gene expression changes in mRNAs and proteins that are critical for synaptic function, adhesion, and signaling. We found no significant change in mRNA levels as far analyzed, including CASK mRNAs (Fig. 7A). For the protein analyses, we were limited to human synaptic marker proteins with validated antibodies. Strikingly, we observed a 60–80% increase in the levels of CASK protein in both cKO and cTr NRXN1-mutant iN cells (Figs. 7B–7D). This increase was observed with two different antibodies and in both cKO and cTr mutant neurons.
Figure 7. Heterozygous NRXN1 inactivation stabilizes CASK protein levels in human neurons.
(A) Quantifications of selected total mRNA levels in matched control vs. NRXN1-mutant iN cells as measured by quantitative RT-PCR. Average ΔCt values (normalized to synapsin-1 mRNA) were converted to a ratio of control/mutant and plotted on a logarithmic scale.
(B & C) Quantifications of protein levels in control iN cells and in iN cells with heterozygous NRXN1 mutations (Syt1, synaptotagmin-1; Cpx 1/2, complexin-1 and -2; Syb2, synaptobrevin-2). Protein levels were determined by quantitative immunoblotting using fluorescently labeled secondary antibodies (B, representative immunoblots; C, summary graphs). NeuN was used as a loading control.
(D) Quantifications of CASK protein levels in control and NRXN1-mutant human neurons using a different antibody to CASK that was used from panels B & C. iN cells from both clones with heterozygous cTr NRXN1 mutations and neurons obtained by a standard protocol (Fig. 5) were analyzed.
All quantitative data are normalized for the controls analyzed in parallel, and expressed as means ± SEM (n=3–5 independent cultures). Statistical analyses were performed by Student’s t-test comparing test samples to control (*p<0.05).
CASK is a cytoplasmic scaffolding protein that interacts with neurexins (Hata et al., 1996). Mutations in the CASK gene are associated with ASDs, X-linked mental retardation, and brain malformations (Najm et al., 2008; Hackett et al., 2010; Sanders et al., 2012), suggesting that regulation of CASK levels is critical for normal brain function. Since we found that CASK mRNA levels were normal in NRXN1-mutant neurons (Fig. 7A), the increased CASK protein levels in NRXN1-mutant neurons suggest that the synthesis or stability of CASK protein is enhanced by the decrease in neurexin-1 levels. The magnitude of the effect is surprising, given that only a single allele of the three biallelic human NRXN genes was mutated. No other protein tested was reproducibly altered, consistent with the observed lack of a change in neuronal morphology and synapse density (Fig. 3).
DISCUSSION
Heterozygous NRXN1 mutations are among the more frequently observed single-gene mutations predisposing to schizophrenia and ASDs, and are additionally associated with other neuropsychiatric disorders (Südhof, 2008; Doherty et al., 2012; Clarke and Eapen, 2014; Béna et al., 2013; Ching et al., 2010; Rees et al., 2014). Despite their importance, however, it is unclear how heterozygous NRXN1 mutations predispose human patients to neuropsychiatric diseases. This question is particularly pungent because in mouse, homozygous Nrxn1α mutations cause only a minor phenotype in acute hippocampal slices (Etherton et al., 2009) and no significant phenotype in cultured cortical neurons (Fig. S1).
To address this question, we introduced two different conditional loss-of-function mutations into human ES cells: A conditional exon deletion that causes a frame shift and disrupts both neurexin-1α and neurexin-1β (the cKO mutation), and a conditional truncation of neurexin-1α and -1β that introduces a stop codon into the protein N-terminal to the transmembrane region and that results in a secreted protein which is rapidly degraded (the cTr mutation; Figs. 1A, S3). Although most clinical NRXN1 mutations affect only NRXN1α, many alter both NRXN1α and NRXN1β, and our cTr mutation closely mimics a schizophrenia patient-associated mutation (Enggaard Hoeffding et al., 2014; Gauthier et al., 2011).
The two heterozygous conditional NRXN1 mutations produced a severe and selective, indistinguishable synaptic phenotype in human neurons. Specifically, we detected a striking decrease in presynaptic neurotransmitter release in NRXN1 mutant neurons that manifested as a reduction in spontaneous mEPSC frequency but not amplitude and as a parallel decrease in evoked EPSC amplitude (Figs. 3, 4, 6). At the same time, the mutations did not alter the electrical properties of human neurons (Fig. 2 and Fig. S5), or their synapse numbers and dendritic arborization (Fig. 3 and Fig. S6). The key features of the NRXN1 heterozygous mutant phenotype were observed in two different types of human neurons, iN cells that are composed of a homogeneous population of excitatory neurons (Zhang et al., 2013), and a more heterogeneous population of neurons generated by a standard method involving an NPC intermediate (Zhang et al., 2001).
The phenotype of the human heterozygous NRXN1 mutations is broadly consistent with previous work in mouse neurons (Missler et al., 2003; Etherton et al., 2009) but exhibits two unexpected features. First, it is surprisingly severe. In human neurons which express ~100-fold more NRXN1α than NRXN1β (Fig. 5), the heterozygous NRXN1 mutations caused an almost 2-fold decrease in EPSC amplitude (Figs. 4, 6) whereas in mouse neurons the homozygous Nrxn1α KO caused no change in EPSC amplitude (Fig. S1). Second, the decrease in EPSC amplitude produced by heterozygous NRXN1 mutations in human neurons was rapidly alleviated during a stimulus train, i.e. the phenotype did not consist of a general decrease of the release probability, but exhibit a specific decrease in release probability for only the first stimulus (Figs. 4, 6). The observation of the same phenotype in two types of human neurons obtained by different methods strengthens the notion that heterozygous loss of NRXN1 gene leads to selective impairments in synaptic transmission.
What mechanism underlies this unusual phenotype of heterozygous NRXN1-mutant human neurons? The most straightforward hypothesis for this phenotype is an impairment in presynaptic Ca2+-influx during an action potential. Such an impairment would impair release only initially in a high-frequency stimulus train because of the accumulation of residual Ca2+ later in the train. Although this phenotype differs from that observed previously in triple α-neurexin KO mice, it is consistent with long-standing ideas about neurexin function derived from these mice (Missler et al., 2003). Our results thus suggest that heterozygous NRXN1 mutations may predispose to schizophrenia and other neuropsychiatric disorders by impairing synapse function in a highly specific manner that may be amenable to pharmacological intervention.
In addition to the synaptic phenotype in heterozygous NRXN1-mutant neurons, we observed a dramatic increase in CASK protein levels (Fig. 7), validating the interaction of neurexin-1 with CASK (Hata et al., 1996). It seems likely that neurexin-1 normally regulates CASK levels which might be critical for controlling neurotransmitter release; indeed, constitutive CASK deletion in mice results in an increased mESPC frequency in cortical neurons (Atasoy et al., 2007). Interestingly, mutations in the human CASK gene are also associated with ASDs, mental retardation, and brain malformations similar to NRXN1 mutations (Najm et al., 2008; Hackett et al., 2010; Sanders et al., 2012).
We believe that our results may provide a significant advance not only in our understanding of how NRXN1 mutations predispose to schizophrenia and ASDs, but also in developing technologies for analyzing the biological significance of gene mutations observed in neuropsychiatric diseases. It is puzzling that many independent gene mutations were detected in schizophrenia and ASDs; nearly all of these mutations are so rare that their significance is difficult to evaluate (O’Roak et al., 2012; Sanders et al., 2012; Purcell et al., 2014; Fromer et al., 2014). The approach used here allows direct testing of whether or not a given heterozygous mutation has a significant functional effect on at least a subset of neurons, with the underlying notion that any mutation with a functional effect in human neurons is likely pathogenic. Thus, our approach may be generally applicable to the large-scale analysis of mutations observed in human patients, especially those mutations that are rare and patient derived materials are limited.
Several potential concerns about our study may be raised. First, given the location of the cKO mutation, is it possible that the NRXN1 cKO we use produces a truncated protein that may have residual functions, or exert a dominant negative activity? We believe that this is highly unlikely because the mutant cKO mRNA is not detectable, suggesting that it is degraded by nonsense-mediated decay (Fig. S3), and because the cTr mutation did not produce a stable truncated protein, probably because that protein is also unstable (Fig. S3). Thus, the cKO and the cTr mutations both are effectively loss-of-function mutations. Second, is it possible that the Cre- and Flp-recombinase manipulations we perform to produce NRXN1-mutant and control iN cells from the same ES cell clone induce toxicity, and create part of the observed phenotype? We believe this is improbable because the Cre- and Flp-manipulations do not create a phenotype when instituted with wild-type ES cells (Fig. S7), and because Cre induces the loss-of-function state in the NRXN1 cKO but prevents the loss-of-function state in the NRXN1 cTr (Fig. 1B). Third, is it possible that the genomic condition of the ES cells we use or that clonal variations among ES cells may contribute to the phenotype? Although the particular genetic background of the ES cells we used (H1 cells) could have significantly contributed to the phenotype we observed, we observed the same phenotype analyzed in different clones and different mutations of the same gene. This consistency documents that the phenotype is specifically induced by the NRXN1 mutation and rules out clonal variation, but does not exclude a possible enabling contribution by genetic background effects in H1 cells. It will be interesting in future expansions of the current iN cell/mutagenesis approach to compare the phenotype produced by NRXN1 mutations in different types of ES and iPS cells, and to systematically use this approach to dissect genetic background effects in patient-derived iPS cells.
The experiments described here thus suggest new avenues to analyze the biological significance of rare disease-associated mutations. Moreover, our approach provides insight into the similarity of disease pathways produced by different types of mutations. In addition, our data show in human neurons that heterozygous NRXN1 mutations produce a functional impairment in synaptic transmission, raising the possibility of therapeutic interventions at later, post-developmental stages in life even though ASDs and schizophrenia are considered neurodevelopmental disorders. Finally, the experiments described here show that neurons produced by the iN cell method and by traditional NPC-dependent methods exhibit similar NRXN1-mutant phenotypes, suggesting that iN cells are reasonable experimental substrates that can in principle be generated in copious amounts for large-scale screening experiments.
EXPERIMENTAL PROCEDURES
General procedures
All experiments were performed under anonymized sample conditions, and independently repeated multiple times. All animal experiments were approved by the Stanford animal use committee.
ES cells
H1 ES cells were obtained from WiCell Research Resources (Wicell, WI) and used at intermediate (50) passage numbers. ES cells were maintained as feeder-free cells in mTeSR™1 medium (Stem Cell Technologies).
Viruses
Cloning of AAV targeting constructs and of lentiviral vectors is described in detail in the supplementary Methods. Lentiviruses and AAVs were produced as described (Pang et al., 2010; Xu et al., 2012); for details and vector sequences, see Supplemental Methods.
Transduction of ES cells using AAVs was performed essentially as described (Kohli et al., 2004). Cells were then selected in puromycin (1 mg/ml) containing mTeSR1 medium until independent clones emerged. Clones were manually picked under sterile conditions and transferred to 24-well or 12-well dishes in mTeSR1+puromycin, and further grown until confluence. Cells were harvested and prepared for frozen stocks or genomic DNA preps for genotyping.
Generation of iN cells from naïve and genetically engineered human ES cells
iN cells were generated from ES cells according to Zhang et al. (2013) as described in detail in the Supplementary Methods. From day 6, 50% of the medium in each well was exchanged every 2–3 days. FBS (2.5%) was added to the culture medium on day 10 to support astrocyte viability, and iN cells were assayed on day 23 in most experiments.
Primary cultures of neurons from neurexin-1α KO mice (Missler et al., 2003) were performed and analyzed electrophysiologically as described (Maximov and Südhof, 2005).
Neural differentiation via NPC intermediates was performed as described (Fig. 5A; Zhang et al., 2001, Wu et. al, 2007, Koch et al., 2009,) with the modifications described in the Supplementary Methods. Differentiated neurons were maintained with half medium change every five days until day 40 for analysis.
Morphological analyses
Immunofluorescence labeling experiments were performed and analyzed as described (Pang et al., 2011, Anderson et al., 2012). For analysis of dendritic arborizations, iN cells were sparsely infected with lentiviruses expressing tdTomato to obtain fluorescent images of individual neurons. Image acquisition and analysis were performed as described (Anderson et al., 2012). See Supplemental Methods for details.
Electrophysiological recordings in cultured iN cells were performed in the patch-clamp whole-cell configuration as described (Maximov and Südhof, 2005; Zhang et al., 2013; for details, see Supplemental Methods).
Gene expression analyses
Qualitative and quantitative RT–PCR analyses of cultured cells were performed in bulk on total RNA using Superscript-III Reverse Transcriptase reaction (Invitrogen; RT-PCR) and a one-step reaction (Roche) and an Applied Biosystems 7900HT apparatus (qRT-PCR). To ensure the specificity of the amplification, titrations of total human brain RNA were used to eliminate primers without a linear amplification relation. Single-cell quantitative RT-PCR was performed using fluidigm methods (Pang et al., 2011). For details and PCR primer sequences, see Supplemental Methods.
Immunoblotting and protein quantifications were performed with fluorescently labeled secondary antibodies and an Odyssey Infrared Imager and Odyssey software (LI-COR Biosciences). Signals were normalized for NeuN probed on the same blots as loading controls. For antibody details, see Supplemental Methods.
Supplementary Material
Acknowledgments
We thank Drs. Yan Han, Nan Yang, Samuele Marro, Csaba Földy, Justin Trotter, Patrick Rothwell, and Daniel Haag for advice. This paper was supported by grants from the NIMH (MH-092931 to MW), NINDS (NS077906 to TCS), and CIRM (RT2 02061 to MW) and by a fellowship from the NICHD (F32 HD078051-02 to CP), and helped by the Stanford Schizophrenia Genetics Research Fund established by an anonymous donor.
Footnotes
AUTHOR CONTRIBUTIONS
CP performed the cell culture, targeting, morphological, and gene and protein expression analyses; TD performed all electrophysiological experiments and mouse cortical cultures; YZ performed the standard differentiation experiments; JA, GA, and FY performed ancillary supporting experiments; SM, MW, and TCS provided critical advice, and CP, TD, and TCS analyzed the data and wrote the paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anderson GR, Galfin T, Xu W, Aoto J, Malenka RC, Südhof TC. Candidate autism gene screen identifies critical role for cell-adhesion molecule CASPR2 in dendritic arborization and spine development. Proc Natl Acad Sci USA. 2012;109:18120–18125. doi: 10.1073/pnas.1216398109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atasoy D, Schoch S, Ho A, Nadasy KA, Liu X, Zhang W, Mukherjee K, Nosyreva ED, Fernandez-Chacon R, Missler M, et al. Deletion of CASK in mice is lethal and impairs synaptic function. Proc Natl Acad Sci USA. 2007;104:2525–2530. doi: 10.1073/pnas.0611003104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellin M, Marchetto MC, Gage FH, Mummery CL. Induced pluripotent stem cells: the new patient? Nat Rev Mol Cell Biol. 2012;13:713–726. doi: 10.1038/nrm3448. [DOI] [PubMed] [Google Scholar]
- Béna F, Bruno DL, Eriksson M, van Ravenswaaji-Arts C, Stark Z, Dijkhuizen T, Gerkes E, Gimelli S, Ganesamoorthy D, Thuresson AC, et al. Molecular and clinical characterization of 25 individuals with exonic deletions of NRXN1 and comprehensive review of the literature. Am J Med Genet B Neuropsychiatr Genet. 2013;162B:388–403. doi: 10.1002/ajmg.b.32148. [DOI] [PubMed] [Google Scholar]
- Chambers SM, Fasano CA, Papapetrou EP, Tomishima M, Sadelain M, Studer L. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat Biotechnol. 2009;27:275–280. doi: 10.1038/nbt.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ching MS, Shen Y, Tan WH, Jeste SS, Morrow EM, Chen X, Mukaddes NM, Yoo SY, Hanson E, Hundley R, et al. Deletions of NRXN1 (neurexin-1) predispose to a wide spectrum of developmental disorders. Am J Med Genet B Neuropsychiatr Genet. 2010;153B:937–947. doi: 10.1002/ajmg.b.31063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke RA, Eapen V. Balance within the Neurexin Trans-Synaptic Connexus Stabilizes Behavioral Control. Front Hum Neurosci. 2014;8:52. doi: 10.3389/fnhum.2014.00052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty JL, O’Donovan MC, Owen MJ. Recent genomic advances in schizophrenia. Clin Genet. 2012;81:103–109. doi: 10.1111/j.1399-0004.2011.01773.x. [DOI] [PubMed] [Google Scholar]
- Enggaard Hoeffding LK, Hansen T, Ingason A, Doung L, Thygesen JH, Møller RS, Tommerup N, Kirov G, Rujescu D, Larsen LA, et al. Sequence analysis of 17 NRXN1 deletions. Am J Med Genet B Neuropsychiatr Genet. 2014;165:52–61. doi: 10.1002/ajmg.b.32204. [DOI] [PubMed] [Google Scholar]
- Etherton MR, Blaiss C, Powell CM, Südhof TC. Mouse neurexin-1α deletion causes correlated electrophysiological and behavioral changes consistent with cognitive impairments. Proc Natl Acad Sci USA. 2009;106:17998–18003. doi: 10.1073/pnas.0910297106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromer M, Pocklington AJ, Kavanagh DH, Williams HJ, Dwyer S, Gormley P, Georgieva L, Rees E, Palta P, Ruderfer DM, et al. De novo mutations in schizophrenia implicate synaptic networks. Nature. 2014;506:179–184. doi: 10.1038/nature12929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauthier J, Siddiqui TJ, Huashan P, Yokomaku D, Hamdan FF, Champagne N, Lapointe M, Spiegelman D, Noreau A, Lafrenière RG, et al. Truncating mutations in NRXN2 and NRXN1 in autism spectrum disorders and schizophrenia. Hum Genet. 2011;130:563–573. doi: 10.1007/s00439-011-0975-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giusti-Rodríguez P, Sullivan PF. The genomics of schizophrenia: update and implications. J Clin Invest. 2013;123:4557–4563. doi: 10.1172/JCI66031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackett A, Tarpey PS, Licata A, Cox J, Whibley A, Boyle J, Rogers C, Grigg J, Partington M, Stevenson RE, et al. CASK mutations are frequent in males and cause X-linked nystagmus and variable XLMR phenotypes. European J Human Genetics. 2010;18:544–552. doi: 10.1038/ejhg.2009.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han SS, Williams LA, Eggan KC. Constructing and deconstructing stem cell models of neurological disease. Neuron. 2011;70:626–644. doi: 10.1016/j.neuron.2011.05.003. [DOI] [PubMed] [Google Scholar]
- Hanna JH, Saha K, Jaenisch R. Pluripotency and cellular reprogramming: facts, hypotheses, unresolved issues. Cell. 2010;143:508–525. doi: 10.1016/j.cell.2010.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hata Y, Butz S, Südhof TC. CASK: A novel dlg/PSD95 homologue with an n-terminal CaM kinase domain identified by interaction with neurexins. J Neurosci. 1996;16:2488–2494. doi: 10.1523/JNEUROSCI.16-08-02488.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howden SE, Gore A, Li Z, Fung HL, Nisler BS, Nie J, Chen G, McIntosh BE, Gulbranson DR, Diol NR, et al. Genetic correction and analysis of induced pluripotent stem cells from a patient with gyrate atrophy. Proc Natl Acad Sci USA. 2011;108:6537–42. doi: 10.1073/pnas.1103388108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imaizumi Y, Okano H. Modeling human neurological disorders with induced pluripotent stem cells. J Neurochem. 2013;129:388–399. doi: 10.1111/jnc.12625. [DOI] [PubMed] [Google Scholar]
- Koch P, Opitz T, Steinbeck JA, Ladewig J, Brustle O. A rosette-type, self-renewing human ES cell-derived neural stem cell with potential for in vitro instruction and synaptic integration. Proc Natl Acad Sci USA. 2009;106:3225–3230. doi: 10.1073/pnas.0808387106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohli M, Rago C, Lengauer C, Kinzler KW, Vogelstein B. Facile methods for generating human somatic cell gene knockouts using recombinant adeno-associated viruses. Nucleic Acids Res. 2004;32:e3. doi: 10.1093/nar/gnh009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lett TA, Tiwari AK, Meltzer HY, Lieberman JA, Potkin SG, Voineskos AN, Kennedy JL, Müller DJ. The putative functional rs1045881 marker of neurexin-1 in schizophrenia and clozapine response. Schizophr Res. 2011;132:121–124. doi: 10.1016/j.schres.2011.08.007. [DOI] [PubMed] [Google Scholar]
- Maximov A, Südhof TC. Autonomous function of synaptotagmin-1 in triggering synchronous release independent of asynchronous release. Neuron. 2005;48:547–554. doi: 10.1016/j.neuron.2005.09.006. [DOI] [PubMed] [Google Scholar]
- Missler M, Zhang W, Rohlmann A, Kattenstroth G, Hammer RE, Gottmann K, Südhof TC. α-Neurexins couple Ca2+ channels to synaptic vesicle exocytosis. Nature. 2003;423:939–948. doi: 10.1038/nature01755. [DOI] [PubMed] [Google Scholar]
- Murdoch JD, State MW. Recent developments in the genetics of autism spectrum disorders. Curr Opin Genet Dev. 2013;23:310–315. doi: 10.1016/j.gde.2013.02.003. [DOI] [PubMed] [Google Scholar]
- Najm J, Horn D, Wimplinger I, Golden JA, Chizhikov VV, Sudi J, Christian SL, Ullmann R, Kueschler A, Haas CA, et al. Mutations of CASK cause an X-linked brain malformation phenotype with microcephaly and hypoplasia of the brainstem and cerebellum. Nat Genet. 2008;40:1065–1067. doi: 10.1038/ng.194. [DOI] [PubMed] [Google Scholar]
- Neale BM, Kou Y, Liu L, Ma’ayan A, Samocha KE, Sabo A, Lin CF, Stevens C, Wang LS, Makarov V, et al. Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature. 2012;485:242–245. doi: 10.1038/nature11011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Roak BJ, Vives L, Girirajan S, Karakoc E, Krumm N, Coe BP, Levy R, Ko A, Lee C, Smith JD, et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature. 2012;485:246–50. doi: 10.1038/nature10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang ZP, Xu W, Cao P, Südhof TC. Calmodulin suppresses synaptotagmin-2 transcription in cortical neurons. J Biol Chem. 2010;285:33930–33939. doi: 10.1074/jbc.M110.150151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang ZP, Yang N, Vierbuchen T, Ostermeier A, Fuentes DR, Yang TQ, Citri A, Sebastiano V, Marro S, Südhof TC, et al. Induction of human neuronal cells by defined transcription factors. Nature. 2011;476:220–223. doi: 10.1038/nature10202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raya A, Rodríguez-Pizà I, Guenechea G, Vassena R, Navarro S, Barrero MJ, Consiglio A, Castellà M, Rio P, Sleep E, et al. Disease-corrected haematopoietic progenitors from Fanconi anaemia induced pluripotent stem cells. Nature. 2009;460:53–59. doi: 10.1038/nature08129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees E, Walters JT, Georgieva L, Isles AR, Chambert KD, Richards AL, Mahoney-Davies G, Legge SE, Moran JL, McCarroll SA, et al. Analysis of copy number variations at 15 schizophrenia-associated loci. Br J Psychiatry. 2014;204:108–14. doi: 10.1192/bjp.bp.113.131052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regehr WG. Short-term presynaptic plasticity. Cold Spring Harb Perspect Biol. 2012;4:a005702. doi: 10.1101/cshperspect.a005702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenmund C, Stevens CF. Definition of the readily releasable pool of vesicles at hippocampal synapses. Neuron. 1996;16:1197–207. doi: 10.1016/s0896-6273(00)80146-4. [DOI] [PubMed] [Google Scholar]
- Rowen L, Young J, Birditt B, Kaur A, Madan A, Philipps DL, Qin S, Minx P, Wilson RK, Hood L, et al. Analysis of the human neurexin genes: alternative splicing and the generation of protein diversity. Genomics. 2002;79:587–597. doi: 10.1006/geno.2002.6734. [DOI] [PubMed] [Google Scholar]
- Purcell SM, Moran JL, Fromer M, Ruderfer D, Solovieff N, Roussos P, O’Dushlaine C, Chambert K, Bergen SE, Kähler A, et al. A polygenic burden of rare disruptive mutations in schizophrenia. Nature. 2014;506:185–190. doi: 10.1038/nature12975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders SJ, Murtha MT, Gupta AR, Murdoch JD, Raubeson MJ, Wilsey AJ, Ercan-Sencicek AG, DiLullo NM, Parikshak NN, Stein JL, et al. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature. 2012;485:237–241. doi: 10.1038/nature10945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souza RP, Meltzer HY, Lieberman JA, Le Foll B, Kennedy JL. Influence of neurexin 1 (NRXN1) polymorphisms in clozapine response. Hum Psychopharmacol. 2010;25:582–585. doi: 10.1002/hup.1146. [DOI] [PubMed] [Google Scholar]
- Südhof TC. Neuroligins and Neurexins Link Synaptic Function to Cognitive Disease. Nature. 2008;455:903–911. doi: 10.1038/nature07456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabuchi K, Südhof TC. Structure and evolution of neurexin genes: insight into the mechanism of alternative splicing. Genomics. 2002;79:849–859. doi: 10.1006/geno.2002.6780. [DOI] [PubMed] [Google Scholar]
- Todarello G, Feng N, Kolachana BS, Li C, Vakkalanka R, Bertolino A, Weinberger DR, Straub RE. Incomplete penetrance of NRXN1 deletions in families with schizophrenia. Schizophr Res. 2014;155:1–7. doi: 10.1016/j.schres.2014.02.023. [DOI] [PubMed] [Google Scholar]
- Treutlein B, Gokce O, Quake SR, Südhof TC. Cartography of Neurexin Alternative Splicing Mapped by Single-Molecule Long-Read mRNA Sequencing. Proc Natl Acad Sci USA. 2014;111:E1291–1299. doi: 10.1073/pnas.1403244111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullrich B, Ushkaryov YA, Südhof TC. Cartography of neurexins: More than 1000 isoforms generated by alternative splicing and expressed in distinct subsets of neurons. Neuron. 1995;14:497–507. doi: 10.1016/0896-6273(95)90306-2. [DOI] [PubMed] [Google Scholar]
- Ushkaryov YA, Petrenko AG, Geppert M, Südhof TC. Neurexins: Synaptic cell surface proteins related to the α-latrotoxin receptor and laminin. Science. 1992;257:40–56. doi: 10.1126/science.1621094. [DOI] [PubMed] [Google Scholar]
- Ushkaryov YA, Südhof TC. Neurexin IIIα: Extensive alternative splicing generates membrane-bound and soluble forms in a novel neurexin. Proc Natl Acad Sci USA. 1993;90:6410–6414. doi: 10.1073/pnas.90.14.6410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Xu J, Pang ZP, Ge W, Kim KJ, Blanchi B, Chen C, Südhof TC, Sun YE. Integrative genomic and functional analyses reveal neuronal subtype differentiation bias in human embryonic stem cell lines. Proc Natl Acad Sci USA. 2007;104:13821–13826. doi: 10.1073/pnas.0706199104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Morishita W, Buckmaster PS, Pang ZP, Malenka RC, Südhof TC. Distinct neuronal coding schemes in memory revealed by selective erasure of fast synchronous synaptic transmission. Neuron. 2012;73:990–1001. doi: 10.1016/j.neuron.2011.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang SC, Wernig M, Duncan ID, Brustle O, Thomson JA. In vitro differentiation of transplantable neural precursors from human embry- onic stem cells. Nat Biotechnol. 2001;19:1129–1133. doi: 10.1038/nbt1201-1129. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Pak C, Han Y, Ahlenius H, Zhang Z, Chanda S, Marro S, Patzke C, Acuna C, Covy J, et al. Rapid Single-Step Induction of Functional Neurons from Human Pluripotent Stem Cells. Neuron. 2013;78:785–798. doi: 10.1016/j.neuron.2013.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J, Maeder ML, Mali P, Pruett-Miller SM, Thibodeau-Beganny S, Chou BK, Chen G, Ye Z, Park IH, Daley GQ, et al. Gene targeting of a disease-related gene in human induced pluripotent stem and embryonic stem cells. Cell Stem Cell. 2009;5:97–110. doi: 10.1016/j.stem.2009.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.