Abstract
Purpose
A simplified acquisition and analysis approach for spin- and gradient-echo (SAGE) based DSC-MRI data that is free of contrast agent T1 leakage effects is proposed.
Methods
A five-echo SAGE sequence was used to acquire DSC-MRI data in rat C6 tumors (n=7). Non-linear fitting of all echoes was performed to obtain T1-insensitive ΔR2* and ΔR2 time series. The simplified approach, which includes two gradient echoes and one spin echo, was also used to analytically compute T1-insensitive ΔR2*, using the two gradient echoes, and ΔR2, using all three echoes. The blood flow, blood volume and vessel size values derived from each method were compared.
Results
In all cases, the five-echo and simplified SAGE ΔR2* and ΔR2 were in excellent agreement and demonstrated significant T1-leakage correction compared to the uncorrected single-echo data. The derived hemodynamic parameters for blood volume, blood flow and vessel size were not significantly different between the two methods.
Conclusions
The proposed simplified SAGE technique enables the acquisition of gradient and spin echo DSC-MRI data corrected for T1 leakage effects, yields parameters that are in agreement with the five echo SAGE, and does not require non-linear fitting to extract ΔR2* and ΔR2 time series.
Keywords: spin-echo EPI, contrast agent leakage, multi-echo sequence, dynamic susceptibility-contrast MRI, perfusion imaging, SAGE
Introduction
Unlike other tracer-based perfusion imaging modalities, dynamic susceptibility-contrast magnetic resonance imaging (DSC-MRI) is unique in its acquisition-dependent vessel size sensitivity. When acquired with spin echo (SE) sequences, the derived DSC parameters, including cerebral blood volume (CBV), cerebral blood flow (CBF), and mean transit time (MTT), are maximally sensitive to capillary-sized vascular structures, while gradient echo (GE) derived hemodynamic parameters are sensitive to vessels of all sizes (1). Although GE imaging is preferred for clinical practice due to its higher SNR, the addition of SE imaging may provide complementary information due to its microvascular sensitivity (2–5). Moreover, the combination of GE and SE imaging permits analysis of mean vessel diameter (mVD) (1,4,6–9). However, quantification of these parameters relies on the assumption that the contrast agent (CA) remains confined to the intravascular space, which may not be the case in tumors (10) or stroke (11).
In tumors, a compromised blood-brain barrier (BBB) leads to extravasation of small molecular weight Gd-based contrast agents (CA) and can severely reduce the reliability of the derived perfusion measures due to competing T1 effects (10,12–14). The use of a preload CA dose can reduce, but not eliminate, the magnitude of T1 leakage effects on DSC-MRI signals (15). In contrast, dual gradient echo sequences (16) provide a simple analytical method to obtain both T1-insensitive ΔR2* measures and T1-weighted signals for Dynamic Contrast Enhanced (DCE) MRI analysis (17). In a comparison with other leakage correction methods, dual-echo sequences were found to provide the most robust T1-insensitive gradient-echo hemodynamic measures (14). However, no analogous method exists to obtain T1-insensitive spin-echo hemodynamic measures. Towards this end, a combined spin- and gradient-echo (SAGE) EPI method was recently proposed to simultaneously obtain T1-insensitive ΔR2 and ΔR2* dynamic time-courses (4,18). This method relies upon the acquisition of multiple echoes (typically between 5 and 7 echoes) and non-linear fitting of each dynamic in order to compute the ΔR2 and ΔR2* time courses. Here, we propose a simplified SAGE approach that utilizes a combined dual gradient-echo and spin-echo pulse sequence and an analytical solution for computing T1-insensitive ΔR2* and ΔR2 time series. As this approach only requires the acquisition and storage of three echoes and does not rely upon computationally demanding non-linear fitting algorithms, it could facilitate the more rapid clinical translation and adoption of SAGE-based DSC-MRI.
Theory
As described above, the simplified SAGE approach, which we will henceforth term sSAGE to delineate it from the original SAGE technique, relies upon the acquisition of two gradient echoes followed by a spin echo. To remove contrast-agent induced T1 leakage effects from the GE and SE data, the MRI signal is expressed in terms of the combined dynamic T1 and T2 (=1/R2) or T2* (=1/R2*) changes. In the case of the spin echo, the MRI signal at a given echo time, STE(t), and resulting ΔR2, assuming exponential decay, are given by:
| [1] |
| [2] |
where f(T1) describes the dynamic changes in the tissue T1 due to contrast agent leakage, α is the flip angle, TR is the pulse sequence repetition time, TE is the echo time, Mo is the initial magnetization, and “pre” designates the mean signal prior to contrast arrival. The GE signal can be similarly derived using Eqs. [1] and [2], with T2 and ΔR2 replaced by T2* and ΔR2*.
While f(T1) is typically assumed to be negligible, this assumption is not valid in cases of compromised BBB. As previously shown (16), T1 effects can be completely removed from ΔR2* through the use of a dual gradient echo sequence:
| [3] |
where STE1 and STE2 are the gradient echo signals at each echo time. A dual-echo acquisition also has the advantage of providing a T1-weighted signal extrapolated to TE=0 (16):
| [4] |
The main purpose of this study is to provide a similar analytical solution to eliminate T1 effects from ΔR2 time series, thus enabling simultaneous extraction of T1-insensitive GE and SE DSC-MRI data. Using the signal extrapolated to TE=0 as f(T1) in Eqs. [1] and [2], the T1 leakage effects can be removed from spin echo signals using:
| [5] |
Methods
Animals Studies
All animal studies were performed in accordance with Vanderbilt University’s Institutional Animal Care and Use Committee (IACUC) protocols. For all procedures and imaging, the animals were immobilized in a stereotactic head holder. Anesthesia was induced using 3–5% isoflurane in air and maintained with 1–2.5% isoflurane in air. Body temperature was maintained at 38°C using forced warm air. The sSAGE and SAGE signal and SNR were compared in normal male Wistar rats (n = 3). For the tumor studies, male Wistar rats (Harlan Laboratories, Indianapolis, IN) were inoculated with 1×105 C6 glioma cells (American Type Culture Collection, Manassas VA), respectively, at 1 mm anterior and 3 mm lateral to the bregma and 4 mm depth from the dural surface. Imaging was performed after 16 days (n = 7).
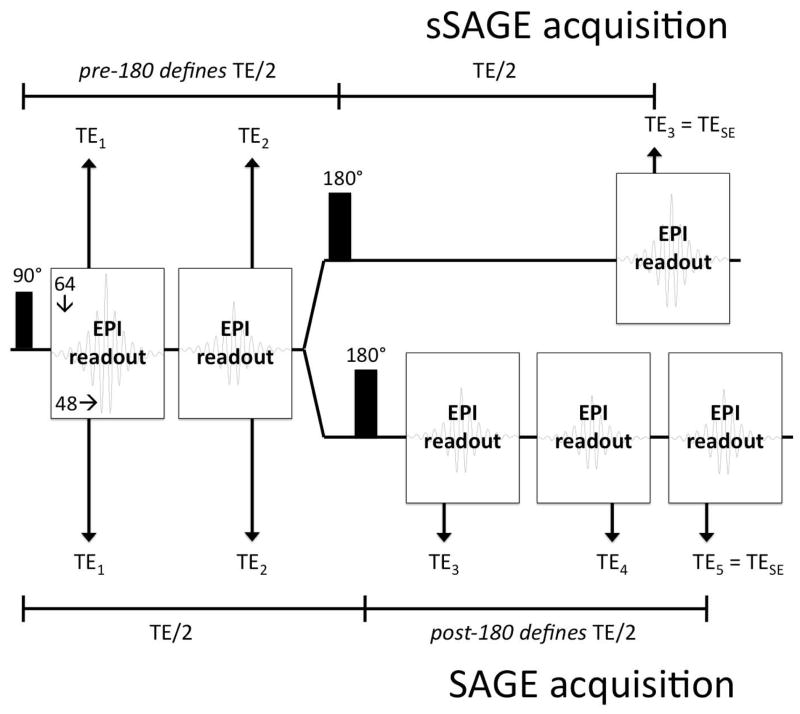
MRI was performed at 4.7T (Agilent, Santa Clara, CA). The sSAGE- and SAGE-EPI sequences, shown in Figure 1, were used to obtain three and five echoes, respectively. Both sequences incorporate two gradient echoes before a 180° pulse; the sSAGE sequence includes a single spin echo, while the SAGE sequence includes two asymmetric spin echoes and a final spin echo. For the sSAGE acquisition, the 180° pulse follows immediately after the 2nd gradient echo, as the first TE/2 period determines the where the SE (TE3) occurs. In the original SAGE acquisition, the SE (TE5) is determined by the second TE/2 period, which depends on the number and length of the acquired asymmetric spin echoes. Partial Fourier encoding (48 of 64 lines acquired) was used to obtain acceptable echo times (Table 1; TE1 – TE3 = 8.6/35/86 ms for sSAGE; TE1 – TE5 = 8.6/35/56/82/96 ms for SAGE). As a result of the shorter final TE, the sSAGE sequence provided 10 slices in a 1 s TR, while the SAGE sequence provided 8 slices in the same TR. The partially sampled k-space data were reconstructed to full Fourier space using an iterative homodyne reconstruction algorithm (19,20) in Matlab (Mathworks, Inc.). Standard slice-selective sinc pulses were used for excitation and refocusing, with crusher gradients surrounding the refocusing pulse. A 64×64 acquisition matrix within a 36×36 mm2 FOV was acquired with 1-mm thick slices. To obtain adequate temporal resolution for dynamic studies, a TR of 1 s was used for at least 5 minutes. After 80 s of baseline images, 0.4 mmol/kg gadopentetate dimeglumine (Gd-DTPA) was injected via jugular catheter.
Figure 1.
Pulse sequence diagram for the sSAGE (top) and SAGE (bottom) acquisitions with three and five echoes, respectively.
Table 1.
Comparison of TE, signal, and SNR for sSAGE and SAGE acquisitions in normal rat brain (n=3).
| TE1 | TE2 | TE3 | TE4 | TESE | ||
|---|---|---|---|---|---|---|
| TE (ms) | sSAGE | 8.6 | 35 | -- | -- | 86 |
| SAGE | 8.6 | 35 | 54 | 80 | 96 | |
| Signal (SD) | sSAGE | 1.73 (0.04) | 1.01 (0.05) | -- | -- | 0.40 (0.01) |
| SAGE | 1.69 (0.05) | 0.99 (0.05) | 0.58 (0.02) | 0.42 (0.01) | 0.33 (0.01) | |
| SNR (SD) | sSAGE | 39.2 (0.6) | 23.2 (0.4) | -- | -- | 10.1 (0.2) |
| SAGE | 38.7 (0.4) | 23.1 (0.7) | 14.4 (0.4) | 10.5 (0.2) | 8.5 (0.2) |
Post-processing and Analysis
The SAGE-derived ΔR2 and ΔR2* time-courses were obtained using nonlinear least squares fits to a piecewise function (Eq. [5]) as previously described (18). Due to slice profile imperfections between the excitation and refocusing pulses (18), the signal intensities SI0 and SII0 were permitted to differ in the fitting.
| [6] |
The baseline signals were averaged to obtain the pre-bolus signal, and the voxel-wise R2, R2*, SI0, and SII0 were determined from Eq. [6]. The full four-parameter fit for ΔR2* and ΔR2 was compared to a reduced fit with the ratio SI0/SII0 – a measure of the slice profile mismatch – held constant for the dynamic time course (4). Due to temporal inconsistencies observed with the full four-parameter fit (Supporting Figure S1), the reduced 3-parameter fit was used for all remaining data to obtain R2(t), R2*(t) and SI0(t) (with SII0(t) replaced with SI0(t)/(SI0/SII0) in the fit function).
To avoid differences that may occur between multiple injections, the sSAGE data in tumor-bearing rats were obtained from the full SAGE datasets using only TE1, TE2, and TE5. The 2nd (gradient-echo) and 5th echo (spin-echo) signals were used to determine the single-echo-based ΔR2* and ΔR2 time series. The first two gradient echoes and the 5th echo were used, along with Eqns. [3] and [5], to compute the T1-insensitive sSAGE ΔR2* and ΔR2 time series. The SAGE fits for R2*(t) and R2(t) were used to determine the T1-insensitive ΔR2* and ΔR2 time series. The arterial input function (AIF) was selected from the T1-insensitive ΔR2* time courses using an automated method (21) specifically adapted for use with multi-echo acquisitions (22). CBV was determined from the ratio of the scaled integrals of the tissue ΔR2* and ΔR2 curves and the arterial input function curve. To avoid artifactually low CBV values that are often observed in single echo brain tumor data, negative ΔR2* and ΔR2 values were not included in the integration. CBF was taken as the maximum of the tissue impulse response function determined from the circular singular value decomposition (SVD) of the AIF and tissue ΔR2* and ΔR2 (23). For display purposes, CBV and CBF were normalized to 4% and 60 ml/100g/min in gray matter. Relative mVD maps were calculated from the ratio of the integrals of the single-echo, sSAGE and SAGE GE ΔR2* and SE ΔR2 curves during bolus passage (1,6,7). Regions of interest (ROIs) were initially drawn from a fast spin echo (FSE) image at a long TE (80ms), and the tumor ROIs were further refined using a 15% enhancement threshold compared to the normal tissue ROIs. Group means were compared using a paired t-test with 5% and 1% significance levels.
Results
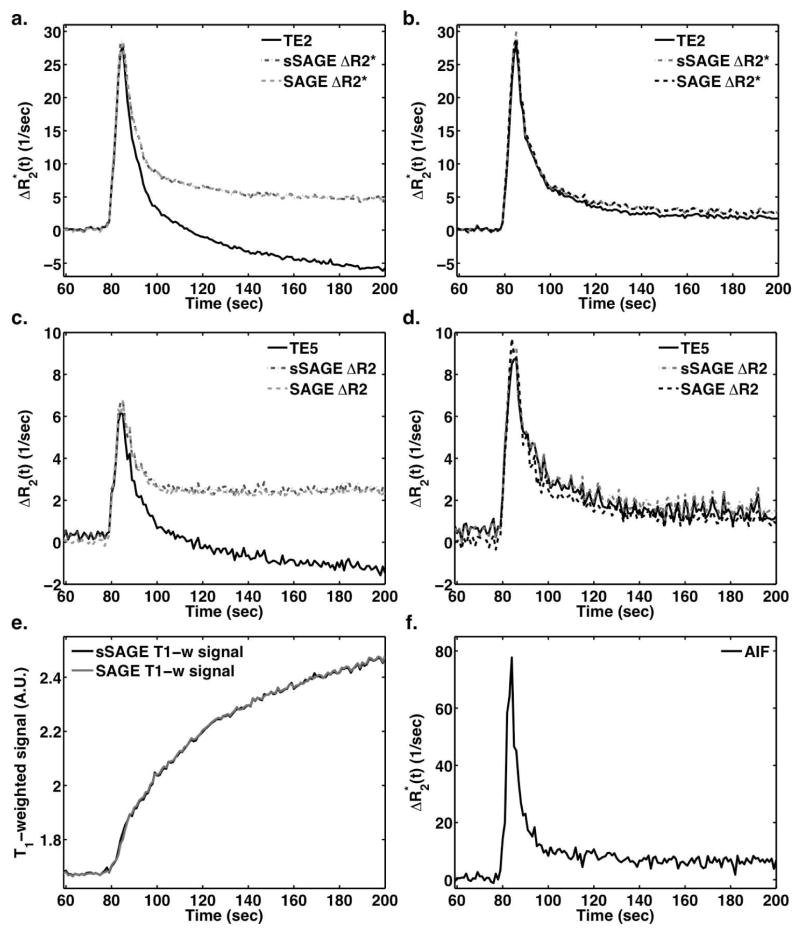
Figure 2 demonstrates representative examples of dynamic DSC data following Gd-DTPA injection in a C6 rat brain tumor ROI (a,c) and normal brain ROI (b,d). In tumor, Gd-DTPA extravasates out of the vasculature, leading to T1-shortening effects that manifest as lower post-bolus ΔR2* and ΔR2 for single echo data (TE2 and TE5, respectively). The SAGE (three-parameter fit) and sSAGE ΔR2* curves, both corrected for T1 leakage effects, do not exhibit reduced post-bolus ΔR2* and are in close agreement. Similarly, the T1-corrected sSAGE derived ΔR2 curve matches well with the T1-insensitive SAGE ΔR2. In normal tissue (b,d), where CA does not typically extravasate, the various ΔR2* and ΔR2 measures are similar. In the bottom left panel, the T1-weighted signals in tumor derived from the two SAGE techniques are in good agreement.
Figure 2.
Dynamic ΔR2* (a,b) and ΔR2 (c,d) curves for a tumor ROI (a,c) and normal ROI (b,d) following bolus injection of 0.4 mmol/kg Gd-DTPA. The sSAGE and SAGE-based T1-weighted signals in a tumor ROI are also shown (e), along with the AIF used for DSC-MRI analysis (f).
Supporting Figure S1 compares dynamic SAGE and sSAGE ΔR2* and ΔR2 obtained with the full four-parameter fit, a reduced three-parameter fit, and the sSAGE method. The four-parameter fit results in identical ΔR2* curves to sSAGE in both C6 tumor (a) and normal ROIs (b). However, ΔR2 from the four-parameter fit is dramatically narrowed and reduced during bolus passage compared to sSAGE. Further analysis shows that this can be attributed to the large, incorrect change in the fitted SII parameter during bolus passage (e,f). The T1-weighted signals for the fitted SI parameter are nearly identical to the T1-weighted signal from sSAGE. To improve temporal stability, a reduced three-parameter fit without dynamic compensation for slice profile mismatch was compared to sSAGE in tumor (c,g) and normal brain (d,h), where the curves were in close agreement for ΔR2*, ΔR2, and T1-weighted signals.
The reduced SE TE (86 ms) of sSAGE provides higher SE signal and SNR compared to SAGE (SE TE = 96 ms) (Table 1). The two GE acquisitions for both sSAGE and SAGE have the same TEs and similar signals and SNR.
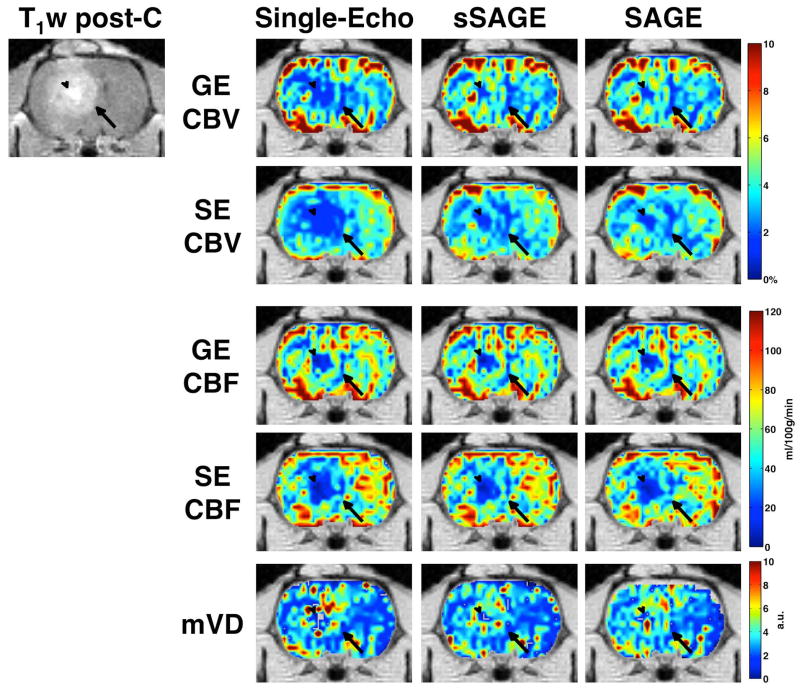
The CBV and CBF maps for gradient-echo and spin-echo are shown in Figure 3 for the single-echo (TE2 and TE5), sSAGE and SAGE. Both the gradient-echo and spin-echo CBV maps for single-echo are substantially underestimated in the tumor region, while the sSAGE- and SAGE-derived maps both exhibit similarly higher CBV. For CBF, the single echo, sSAGE, and SAGE-derived maps for gradient-echo and spin-echo are similar.
Figure 3.
GE (TE2, sSAGE, and SAGE) CBV and CBF, SE (TE5, sSAGE, and SAGE) CBV and CBF, and mVD in a tumor-bearing rat (T1-weighted post-contrast image shows tumor edge, indicated by arrow, and necrotic core, indicated by arrowhead).
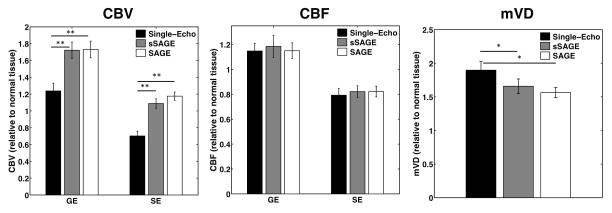
The bar plots in Figure 4 show the mean CBF, CBV, and mVD in tumor relative to normal tissue using the single-echo, sSAGE, and SAGE ΔR2* and ΔR2 (n=7). The gradient-echo CBF in tumor was slightly higher than normal tissue, while the spin-echo CBF was slightly lower than normal tissue. None of the gradient-echo or spin-echo CBF measures were significantly different (p>0.05). T1-leakage effects led to significantly reduced single-echo CBV for both gradient-echo and spin-echo compared to the sSAGE and SAGE measures. The single-echo CBV values were significantly different from the sSAGE and SAGE CBV (p<0.0005 for both GE and SE values), while the sSAGE and SAGE CBV were not significantly different from each other (p>0.05). All three mVD measures were similarly increased in tumors, and the sSAGE and SAGE measures were not significantly different (p>0.05). The single-echo measures were significantly different from the sSAGE and SAGE measures (p=0.0429 and p=0.0222, respectively).
Figure 4.
Bar plots showing mean GE and SE CBV and CBF and mVD relative to normal tissue for the single-echo, sSAGE, and SAGE methods (n=7). **p<0.01 and *p<0.05.
Discussion
The proposed simplified SAGE technique leverages the known insensitivity of dual-gradient echo DSC-MRI data to T1 leakage effects and provides a simple, computationally efficient analytical solution for T1-correction of SE data, thereby yielding T1-insensitive GE and SE hemodynamic parameters, plus measures of vessel size. On a computer with a 2.4 GHz dual-core processor with 8 GB of RAM, the computation time for deriving the ΔR2* and ΔR2 curves for a typical whole rat brain (approximately 1800 voxels with 200 repetitions) was 2 hours for SAGE and 16 seconds for sSAGE. Consequently, the proposed analytic approach is 450 times faster than the non-linear fitting procedure used for full SAGE data. In reality, all voxels can be analytically calculated simultaneously, and thus, sSAGE for the entire rat brain and all repetitions can be calculated in less than 1 second. Furthermore, the sSAGE method will prove especially advantageous for human DSC-MRI that has substantially more voxels.
A limitation of the proposed approach is its inability to correct for slice profile mismatch that can occur with spin echo sequences (18). While this is important for absolute quantification of T2, it is less important for DSC measures that rely on assessing changes in R2 to obtain CBF, CBV, and MTT. Moreover, current SAGE DSC implementations quantify slice profile mismatch during baseline, which is held constant during the dynamic time-course (4,24,25). Thus, the effects of slice profile mismatch are effectively subtracted when ΔR2 and ΔR2* are obtained for DSC purposes. In addition, dynamically correcting for slice profile mismatch using a four-parameter fit can incorrectly assign temporal ΔR2 changes during bolus passage to changes in the post-180 signal intensity SII. This is avoided in the sSAGE method or can be effectively rectified by using a reduced three-parameter fit with constant slice profile mismatch SI/SII. Because the sSAGE sequence requires only three echoes, shorter SE TEs may be feasible, which improves the SE SNR and may increase slice coverage (or yield shorter TRs). In addition, the later echoes (such as the 3rd and 4th asymmetric spin-echoes in SAGE) tend to be more sensitive to signal voids due to high CA concentration (26) or susceptibility-induced edge artifacts, which can yield inaccurate fits (25). The method proposed herein does not depend on asymmetric spin-echoes, although this also makes the method more sensitive to potential errors in any of the three echoes. Of the hemodynamic parameters, CBF is especially sensitive to noise due to the deconvolution step, and thus, the higher SE SNR of the simplified approach may be advantageous to improve the overall reliability of the derived hemodynamic parameters.
Many methods exist to mitigate or correct leakage effects (historically focusing on T1 effects) in single gradient echo data, including preload dose, sequence modifications, and model-based corrections (12–14). With the advent of dual-echo acquisitions, the T1 effects can be eliminated from the DSC ΔR2* data, and the derived T1-weighted signal can subsequently be used for DCE analysis (17). This study presents an analogous solution for spin-echo data by utilizing the dual gradient-echoes to quantifying the dynamic T1 changes and then removing these effects from ΔR2. With the exception of the previously published SAGE papers (4,24), no studies have considered the effects of CA leakage on spin echo data. In this study, SE CBF was not significantly different for single-echo, sSAGE, or SAGE, consistent with previous studies showing minor leakage effects on GE CBF. On the other hand, GE and SE CBV values were significantly underestimated by the single echoes compared to the T1-insensitive measures (sSAGE and SAGE), further demonstrating the importance of leakage correction for CBV assessment. Interestingly, single echo mVD was significantly higher than the sSAGE and SAGE measures, which were not significantly different, indicating that T1 leakage effects do not simply cancel out when computing the ratio of ΔR2* and ΔR2. In most vessel-size sensitive measurement methods (8), CA leakage effects are mitigated by γ-variate fitting, and this is likely sufficient for T1-predominant leakage effects (9).
For the purposes of this study, tumors that exhibited predominantly T1-leakage effects were deliberately chosen, as the primary focus was removing T1 leakage effects from spin- and gradient echo DSC-MRI data. However, other tumor types (in both rat and human brains) that exhibit a range of T1 and T2*/T2 leakage effects should yield similar curves between SAGE and sSAGE, as these techniques should exhibit similar sensitivity to T2 and T2* leakage effects. As T2 leakage effects would undoubtedly affect the ΔR2 curves and derived hemodynamic parameters, obtaining quantitative measures of SE hemodynamics will require both removal of T1 leakage effects and corrections for T2 leakage effects. While multi-echo acquisitions remove T1 leakage effects, unfortunately, no consensus currently exists on the best method to correct T2* and T2 leakage effects (12–14,24). While this is outside the scope of this study, such topics will be the focus of future investigations.
In conclusion, T1-insensitive GE and SE hemodynamic parameters can be obtained using a simplified spin-and gradient-echo sequence with three total echoes (two gradient-echoes and one spin-echo). The T1-insensitive ΔR2* and ΔR2 time courses can be calculated using the previously proposed dual-echo equation and the spin-echo correction presented here. As this method does not require time-consuming nonlinear fitting, it is an efficient and clinically feasible method. Moreover, the ΔR2 curves from the sSAGE method match well with the reduced three-parameter SAGE fit, both of which are more accurate than the originally proposed four-parameter SAGE fit. In addition to T1-insensitive CBF, CBV, and MTT with both GE (total vasculature) and SE (microvasculature) contrast, this sequence provides measures of mVD. Aside from the addition of the T1-insensitive spin-echo hemodynamic parameters, the proposed approach may still be used to obtain ΔR1 curves for DCE analysis, thereby providing simultaneous measures of perfusion and permeability (16,17).
Supplementary Material
Dynamic ΔR2* and ΔR2 (c,d) curves comparing the four-parameter SAGE fit (a,b) and the three-parameter SAGE fit (c,d) to sSAGE in tumor (a,c) and normal (b,d) ROIs. The four-parameter signals SI and SII (e,f) and the three-parameter signal SI (g,h) are also compared to the sSAGE-based signal STE=0 in tumor (e,g) and normal (f,h) ROIs.
Acknowledgments
We would like to thank Jack Skinner and Allen Newton for many helpful discussions and Zou Yue for help with animal prep. This work was performed at the Vanderbilt University Institute of Imaging Science, with support from NCI P30 CA68485 and 1R01CA158079.
References
- 1.Dennie J, Mandeville JB, Boxerman JL, Packard SD, Rosen BR, Weisskoff RM. NMR imaging of changes in vascular morphology due to tumor angiogenesis. Magn Reson Med. 1998;40(6):793–799. doi: 10.1002/mrm.1910400602. [DOI] [PubMed] [Google Scholar]
- 2.Boxerman JL, Hamberg LM, Rosen BR, Weisskoff RM. MR contrast due to intravascular magnetic susceptibility perturbations. Magn Reson Med. 1995;34(4):555–566. doi: 10.1002/mrm.1910340412. [DOI] [PubMed] [Google Scholar]
- 3.Kiselev VG, Strecker R, Ziyeh S, Speck O, Hennig J. Vessel size imaging in humans. Magn Reson Med. 2005;53(3):553–563. doi: 10.1002/mrm.20383. [DOI] [PubMed] [Google Scholar]
- 4.Schmiedeskamp H, Straka M, Newbould RD, Zaharchuk G, Andre JB, Olivot JM, Moseley ME, Albers GW, Bammer R. Combined spin- and gradient-echo perfusion-weighted imaging. Magn Reson Med. 2012;68(1):30–40. doi: 10.1002/mrm.23195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Troprès I, Grimault S, Vaeth A, Grillon E, Julien C, Payen J-F, Lamalle L, Décorps M. Vessel size imaging. Magn Reson Med. 2001;45(3):397–408. doi: 10.1002/1522-2594(200103)45:3<397::aid-mrm1052>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 6.Donahue KM, Krouwer HGJ, Rand SD, Pathak AP, Marszalkowski CS, Censky SC, Prost RW. Utility of simultaneously acquired gradient-echo and spin-echo cerebral blood volume and morphology maps in brain tumor patients. Magn Reson Med. 2000;43(6):845–853. doi: 10.1002/1522-2594(200006)43:6<845::aid-mrm10>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 7.Schmainda KM, Rand SD, Joseph AM, Lund R, Ward BD, Pathak AP, Ulmer JL, Baddrudoja MA, Krouwer HGJ. Characterization of a first-pass gradient-echo spin-echo method to predict brain tumor grade and angiogenesis. Am J Neuroradiol. 2004;25(9):1524–1532. [PMC free article] [PubMed] [Google Scholar]
- 8.Lemasson B, Valable S, Farion R, Krainik A, Remy C, Barbier EL. In vivo imaging of vessel diameter, size, and density: a comparative study between MRI and histology. Magn Reson Med. 2013;69(1):18–26. doi: 10.1002/mrm.24218. [DOI] [PubMed] [Google Scholar]
- 9.Pannetier N, Lemasson B, Christen T, Tachrount M, Tropres I, Farion R, Segebarth C, Remy C, Barbier EL. Vessel size index measurements in a rat model of glioma: comparison of the dynamic (Gd) and steady-state (iron-oxide) susceptibility contrast MRI approaches. Nmr Biomed. 2012;25(2):218–226. doi: 10.1002/nbm.1734. [DOI] [PubMed] [Google Scholar]
- 10.Boxerman JL, Schmainda KM, Weisskoff RM. Relative Cerebral Blood Volume Maps Corrected for Contrast Agent Extravasation Significantly Correlate with Glioma Tumor Grade, Whereas Uncorrected Maps Do Not. Am J Neuroradiol. 2006;27(4):859–867. [PMC free article] [PubMed] [Google Scholar]
- 11.Kassner A, Roberts T, Taylor K, Silver F, Mikulis D. Prediction of Hemorrhage in Acute Ischemic Stroke Using Permeability MR Imaging. Am J Neuroradiol. 2005;26(9):2213–2217. [PMC free article] [PubMed] [Google Scholar]
- 12.Bjornerud A, Sorensen AG, Mouridsen K, Emblem KE. T1- and T2*-dominant extravasation correction in DSC-MRI: part I--theoretical considerations and implications for assessment of tumor hemodynamic properties. Journal of cerebral blood flow and metabolism : official journal of the International Society of Cerebral Blood Flow and Metabolism. 2011;31(10):2041–2053. doi: 10.1038/jcbfm.2011.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boxerman JL, Prah DE, Paulson ES, Machan JT, Bedekar D, Schmainda KM. The Role of preload and leakage correction in gadolinium-based cerebral blood volume estimation determined by comparison with MION as a criterion standard. AJNR American journal of neuroradiology. 2012;33(6):1081–1087. doi: 10.3174/ajnr.A2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paulson ES, Schmainda KM. Comparison of Dynamic Susceptibility-weighted Contrast-enhanced MR Methods: Recommendations for Measuring Relative Cerebral Blood Volume in Brain Tumors. Radiology. 2008;249(2):601–613. doi: 10.1148/radiol.2492071659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Quarles CC, Ward BD, Schmainda KM. Improving the reliability of obtaining tumor hemodynamic parameters in the presence of contrast agent extravasation. Magn Reson Med. 2005;53(6):1307–1316. doi: 10.1002/mrm.20497. [DOI] [PubMed] [Google Scholar]
- 16.Vonken E-jPA, van Osch MJP, Bakker CJG, Viergever MA. Simultaneous quantitative cerebral perfusion and Gd-DTPA extravasation measurement with dual-echo dynamic susceptibility contrast MRI. Magn Reson Med. 2000;43(6):820–827. doi: 10.1002/1522-2594(200006)43:6<820::aid-mrm7>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 17.Quarles CC, Gore JC, Xu L, Yankeelov TE. Comparison of dual-echo DSC-MRI- and DCE-MRI-derived contrast agent kinetic parameters. Magn Reson Imaging. 2012;30(7):944–953. doi: 10.1016/j.mri.2012.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schmiedeskamp H, Straka M, Bammer R. Compensation of slice profile mismatch in combined spin- and gradient-echo echo-planar imaging pulse sequences. Magn Reson Med. 2012;67(2):378–388. doi: 10.1002/mrm.23012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McGibney G, Smith MR, Nichols ST, Crawley A. Quantitative evaluation of several partial fourier reconstruction algorithms used in mri. Magn Reson Med. 1993;30(1):51–59. doi: 10.1002/mrm.1910300109. [DOI] [PubMed] [Google Scholar]
- 20.Noll DC, Nishimura DG, Macovski A. Homodyne detection in magnetic resonance imaging. Medical Imaging, IEEE Transactions on. 1991;10(2):154–163. doi: 10.1109/42.79473. [DOI] [PubMed] [Google Scholar]
- 21.Carroll TJ, Rowley HA, Haughton VM. Automatic Calculation of the Arterial Input Function for Cerebral Perfusion Imaging with MR Imaging1. Radiology. 2003;227(2):593–600. doi: 10.1148/radiol.2272020092. [DOI] [PubMed] [Google Scholar]
- 22.Newton AT, Skinner JT, Quarles CC. Automatic AIF Estimation in Multi-Echo DSC-MRI of Pediatric Patients - Avoiding the Noise Floor. Proceedings of the 21st Annual Meeting of ISMRM; Salt Lake City, UT, USA. 2013; p. 3064. [Google Scholar]
- 23.Willats L, Calamante F. The 39 steps: evading error and deciphering the secrets for accurate dynamic susceptibility contrast MRI. Nmr Biomed. 2013;26(8):913–931. doi: 10.1002/nbm.2833. [DOI] [PubMed] [Google Scholar]
- 24.Schmiedeskamp H, Andre JB, Straka M, Christen T, Nagpal S, Recht L, Thomas RP, Zaharchuk G, Bammer R. Simultaneous perfusion and permeability measurements using combined spin- and gradient-echo MRI. Journal of cerebral blood flow and metabolism : official journal of the International Society of Cerebral Blood Flow and Metabolism. 2013;33(5):732–743. doi: 10.1038/jcbfm.2013.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stokes AM, Skinner JT, Quarles CC. Assessment of a combined spin- and gradient-echo (SAGE) DSC-MRI method for preclinical neuroimaging. Magn Reson Imaging. 2014;32(10):1181–1190. doi: 10.1016/j.mri.2014.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Newbould RD, Skare ST, Jochimsen TH, Alley MT, Moseley ME, Albers GW, Bammer R. Perfusion mapping with multiecho multishot parallel imaging EPI. Magn Reson Med. 2007;58(1):70–81. doi: 10.1002/mrm.21255. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Dynamic ΔR2* and ΔR2 (c,d) curves comparing the four-parameter SAGE fit (a,b) and the three-parameter SAGE fit (c,d) to sSAGE in tumor (a,c) and normal (b,d) ROIs. The four-parameter signals SI and SII (e,f) and the three-parameter signal SI (g,h) are also compared to the sSAGE-based signal STE=0 in tumor (e,g) and normal (f,h) ROIs.