Abstract
Background. Gene expression profiles of 181 breast cancer samples were analyzed to identify prognostic features of nuclear receptors NR5A1 and NR5A2 based upon their associated transcriptional networks. Methods. A supervised network analysis approach was used to build the NR5A-mediated transcriptional regulatory network. Other bioinformatic tools and statistical methods were utilized to confirm and extend results from the network analysis methodology. Results. NR5A2 expression is a negative factor in breast cancer prognosis in both ER(−) and ER(−)/ER(+) mixed cohorts. The clinical and cohort significance of NR5A2-mediated transcriptional activities indicates that it may have a significant role in attenuating grade development and cancer related signal transduction pathways. NR5A2 signature that conditions poor prognosis was identified based upon results from 15 distinct probes. Alternatively, the expression of NR5A1 predicts favorable prognosis when concurrent NR5A2 expression is low. A favorable signature of eight transcription factors mediated by NR5A1 was also identified. Conclusions. Correlation of poor prognosis and NR5A2 activity is identified by NR5A2-mediated 15-gene signature. NR5A2 may be a potential drug target for treating a subset of breast cancer tumors across breast cancer subtypes, especially ER(−) breast tumors. The favorable prognostic feature of NR5A1 is predicted by NR5A1-mediated 8-gene signature.
1. Introduction
Breast cancer (BC) is the second most commonly diagnosed cancer but ranks 5th as cause of death worldwide in 2012 [1]. Incidence and mortality rates vary among populations in the tested areas worldwide by more than 5-fold [2]. In general, the more developed areas have higher ratio of age-standardized incidence rate/mortality rate (ratio = 4.34) than less developed areas (ratio = 2.53) [3]. Despite significant advances in treatment options being available for BC, the heterogeneous nature of breast cancers demands a personalized medicine approach [4].
Differentiating the signal transduction pathways governing development and prognosis of breast cancer subtypes is vital to design optimal intervention strategies especially for the ER(−) subtype that has fewer treatment options than the ER(+) subtype [5]. Research suggests that dissection of integrated transcriptional data and identification of relevant signal transduction pathways can be statistically correlated with tumor subtype, thus predicting clinical disease progression and informing treatment options [6, 7]. We have recently developed a supervised network analysis approach to effectively predict functional regulatory networks mediated by a target transcription factor within a given population [8–10]. The target transcription factor involved in controlling disease progression can be initially characterized using the supervised network prediction. We further demonstrate the utility of this approach in predicting the prognosis relevant signature in a subset of tumor samples [11]. Several aberrantly expressed transcription factors including estrogen receptor alpha (ERα) and progesterone receptor (PR) are known as the clinical biomarkers in breast cancers. Zheng et al. suggested that there may be 1,850 to 4,105 putative human transcription factors (TFs) [12]. We suspect that a significant subset of those TFs may be involved in key regulatory events governing BC development. Dissecting the complex interplay between TFs and their gene partners on a system-wide basis in breast cancer subtypes is critical for better management and outcomes for breast cancer patients.
Human NR5A2 was first cloned and characterized as a novel human hepatocyte transcription factor, hB1F [13]. Computer analysis of the NR5A2 promoter region indicates several transcription factor binding sites including SREBP, RORA1, GATA, TBP, C/EBP, HNF1, and HNF3 [14]. Liver receptor homologue-1 (LRH-1) or NR5A2 belongs to one of six subfamilies of nuclear receptors (NRs 1–6) [15]. Fayard et al. [16] described the biological evidence supporting LRH-1(NR5A2) as an orphan nuclear receptor involved in development, metabolism, and steroidogenesis. The phosphatidyl inositols are ligands of both NR5A2 and NR5A1 [17]. However, NR5A1 and NR5A2 are still considered to be orphan receptors [15]. There is also a positive correlation between immunohistochemistry (IHC) and NR5A2(LRH-1) mRNA levels [18]. Based upon NR5A2(LRH-1) immunolocalization in human breast carcinomas, Miki et al. suggested NR5A2 as a regulator of in situ steroidogenesis.
The physiological and pathophysiological activities of NR5A2 can be due to both estrogen dependent and independent activities. Annicotte et al. [19] directly implicated NR5A2 in estrogen dependent breast cancer development. It is expressed via ERα binding to the estrogen response element (ERE) within its promoter region. The immunohistochemistry (IHC) stain of NR5A2 is elevated in breast tumors and is preferentially coexpressed with ERα. In addition, Thiruchelvam et al. [20] reported that ERα is NR5A2(LRH-1) target gene product and Chand and colleagues [21] implicated NR5A2 in promotion of migration and invasion in breast cancer independent of estrogen sensitivity.
In this study, we demonstrate the clinical importance of NR5A2 relative to its prognostic role in breast cancers. To identify specific prognostic features of NR5A2, we analyzed the network of NR5A2 using the supervised approach. We conclude that NR5A2 is an indicator of poor clinical outcomes in ER(+)/ER(−) tumor (1 : 1 ratio) cohort and its ER(−) subcohort.
2. Materials and Methods
2.1. Features of Surgical Specimens for Generating the Dataset of Gene Expression Profiles
We analyzed 181 tumor samples from primary infiltrating ductal breast carcinomas (IDCs) that have eight molecular subtypes based on the immunohistochemical analysis of estrogen receptor (ER) alpha, progesterone receptor (PR) A, and Her-2/neu (HER) biomarkers. Determination of Her-2/neu gene copy number for HER (IHC protein intensity (IHC score): 2+) was established by chromogenic in situ hybridization (CISH), and IHC/CISH status was used for determining HER status.
Ninety IDC specimens (90/181) were in subgroups IE (ER(+)PR(+)) (n = 61) and IIE (ER(+)PR(−)) (n = 29). Ninety-one of the 181 IDC samples were in subgroup “triple negative” (TN) (ER(−)PR(−)HER(−)) (n = 48), ERBB2+(ER(−)PR(−)HER(+)) (n = 29), ER(−) PR(+)HER(−) (n = 5), ER(−)PR(+)HER(+) (n = 6), and ER(−)HER(?) (n = 3). All samples were obtained from patients who underwent surgery at the National Taiwan University Hospital (NTUH) between 1995 and 2007. All patients provided informed consent according to the guidelines approved by the Institutional Review Board at NTUH (200706039R, Research Ethics Committee at National Taiwan University Hospital, Taipei, Taiwan). The microarray data from this study have been submitted to the NCBI Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE24124. The abbreviation for each gene expression array dataset was “A.” In this study, we designated 90A as the gene expression microarray dataset for 90 ER(+) breast tumors. It consists of two subsets that are group IE (61A) and group IIE (29A). The definition for 91A is the gene expression microarray dataset of 91 ER(−) breast tumors. It consists of subsets for TN (48A), ERBB2+(29A), ER(−) PR(+)HER(−)(5A), ER(−)PR(+)HER(+)(6A), and ER(−)HER(?)(3A). The 181A cohort includes datasets from the 90A and 91A cohorts.
2.2. Microarray Data Analyses
The global gene expression profile per breast tumor specimen was analyzed using a Human 1A (version 2) oligonucleotide microarray (half a genome size: 22 k) (Agilent Technologies, USA). Heatmaps of gene expression data were displayed after unsupervised hierarchical clustering [10]. For unsupervised hierarchical clustering, the log2 ratio of mean expression data for each gene was first centered by subtracting the median across all samples to discriminate the subclass of the dataset. Then, the selected gene expression profiles were analyzed by R2.15.1 software for displaying the gene list (y-axis) derived from hierarchical clustering analysis on the gene profiles of selected arrays (x-axis) to generate the heatmaps. In addition, Gene Spring GX7.3.1 (Agilent Technologies, USA) was used for generating Venn diagrams and for retrieving updated gene annotations. ANOVA tests for the relationship between mRNA level of NR5A2 and a clinical index of interest in a given population as well as the statistical methods for establishing NR5A2 transcriptional regulatory network were described previously [8, 10, 22]. The same data analyses described above were used for analyzing other transcription factors of interest.
Kaplan-Meier (K-M) survival analyses [23] using the “survival” package in R (version 2.15.1) were performed using the gene profiles from the 90A cohort, 91A cohort, and 181A cohort or the extracted gene pools of interest in the assigned cohorts. The weight of hazard ratios associated with the prognostic gene signature and the traditional prognostic factors in a given cohort of interest was quantified using both the univariate and multivariate COX proportional hazard (COXPH) regression model in the R package.
2.3. Experimental Design
Network analysis only predicts the TF-target regulatory associations in a given population transcriptome but not in a given population proteome. Moreover, this association is measured using the data from the sample population prepared for systems biochemistry study. Therefore, the expression of mRNA levels in the microarray setting represents the sum expression of mRNAs due to transcriptional activities that may occur across multiple organelles, cells, and tissues of the tumor or nontumor samples. Network analysis predicts the interactions of the TF and its target gene at the systems transcriptome scale. Further localization of the network gene expression for the given TF using the cell model or other appropriate models is required for subsequent validation of the network prediction.
The association between NR5A2 and the identified target gene was measured by both the coefficient of intrinsic dependence (CID) and Galton Pearson's correlation coefficient (GPCC). CID measures nonlinear association and GPCC measures linear association. Such combined statistical analyses (CIDUGPCC) provide predictive power in measuring gene-gene expression relationship particularly for the regulatory relationship between TF and its target gene. Multivariate CID measures the association between TFs and their shared target gene.
Colocalization of the transcription factors or of other inferred target genes with the given TF conditions multiple outcomes. TF-target regulatory interaction predicted by CIDUGPCC is one such outcome. However, the supervised network analysis specifically enriches and efficiently dissects the inferred functional regulatory association that can also be partially validated by in vitro biological evidence [8–11, 24]. The positive regulatory associations at the transcriptome level support but do not confirm similar associations at the proteome level.
Network analysis is a highly sensitive measurement of both known and novel TF-target gene expression relationships in a population of interest. Here, we employ a strategy to couple network analysis with K-M survival analysis to identify the prognostic relevant transcriptional regulatory subnetwork of NR5A2. Further, we integrate steps to control confounders and quickly locate major NR5A2 prognostic features. First, three populations of interest (i.e., 90A cohort, 91A cohort, and 181A cohort) were subcategorized based upon prognostic potentials into four types. Second, the subpool of genes was identified which shared prognostic predictors of a given type with the putative network of the NR5A2. Third, genes in a given feature type were proposed to be major prognostic features of NR5A2 based on cumulative analysis of the NR5A2 transcriptional regulatory network in relation to biochemical profiles, malignant phenotypes, and supporting evidence from other studies that are described in the Results and Discussion. Fourth, the overlapping gene set in the 91A and 181A cohorts was selected as the consensus prognostic signature of NR5A2 and the overlapping gene set between feature type IV (the overlapping gene pool between type IV and 181A relevant network; see Table S5.8 in Supplementary Material available online at http://dx.doi.org/10.1155/2015/403576) of NR5A2 and NR5A1 in the 181A cohort was selected. This effectively distinguished the prognostic features of NR5A2 from those of NR5A1. Finally, the prognostic signatures relevant to clinicopathological parameter(s), subtype(s), and treatment response(s) were predicted and their prognostic relevance in a subset of breast cancers was identified using K-M and COXPH survival analysis.
3. Results and Discussion
3.1. The Clinical and Prognostic Relevance of NR5A2 in a Breast Cancer Population Is ERα Independent
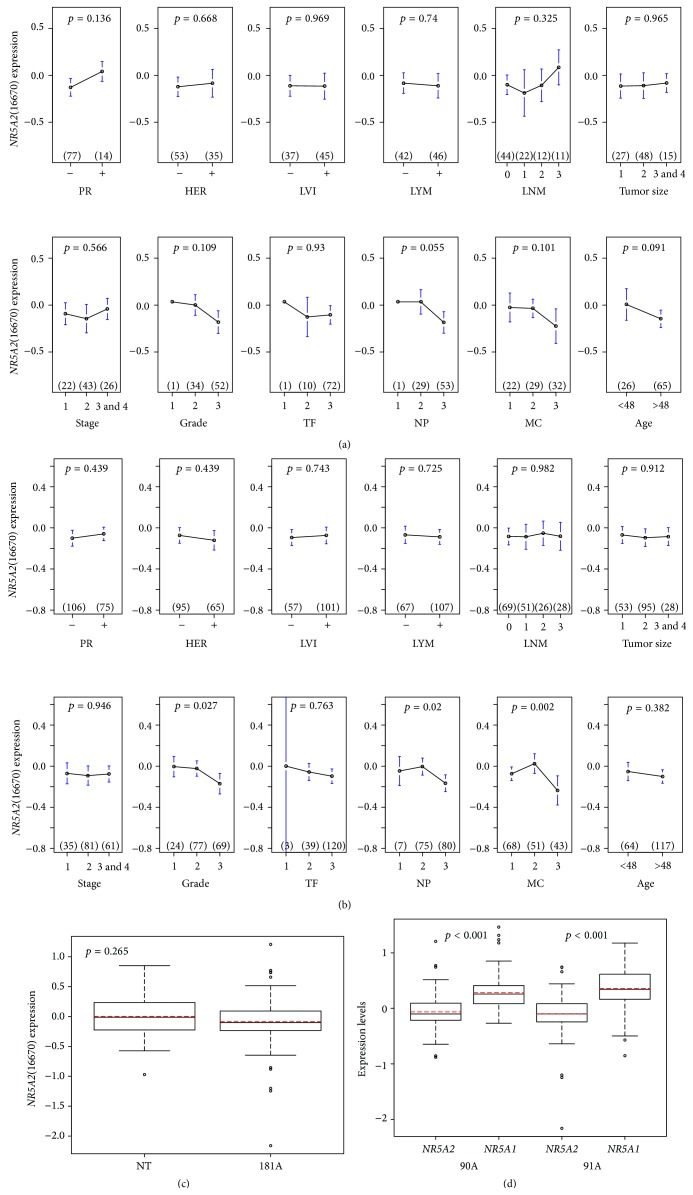
Preliminary data suggests the prognostic value of NR5A2 in 91A and 181A cohorts, supporting the concept that these NR5A2 features may be clinically relevant in both cohorts (Figure 1). We found no significant clinical impact of NR5A2 in the 91A cohort (Figure 2(a)). However, ANOVA tests indicate that histological grade, mitotic counts, and nuclear pleomorphism are positively associated with NR5A2 in the 181A cohort (Figure 2(b)).
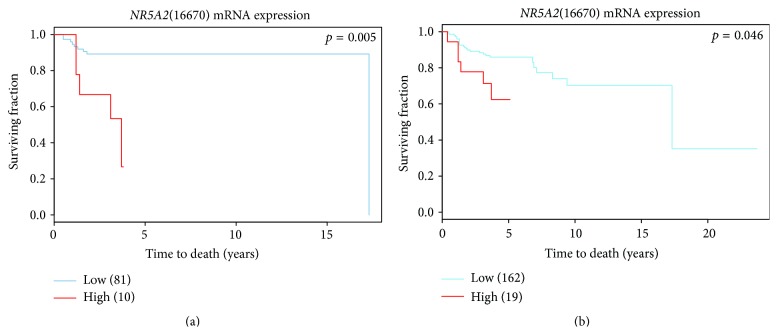
Figure 1.
Kaplan-Meier survival analysis on NR5A2 in 91A and 181A cohorts. The survival curves of a breast tumor group with top 10% high NR5A2 mRNA levels versus the group with low NR5A2 mRNA levels in 91A cohort and 181A cohort, respectively.
Figure 2.
Clinical impact of NR5A2 in two cohorts of infiltrating ductal breast carcinomas. ANOVA test results of NR5A2(16670) mRNA levels in eight clinical indices: progesterone receptor (PR), HER-2/neu (HER), lymphovascular invasion (LVI), lymph nodal category (lymph node metastasis status (LYM), number of lymph node metastases (LNM)), age, tumor size (size), histological grade (grade), nuclear pleomorphism (NP), mitotic count (MC), tubule formation (TF), and cancer stage in 91A cohort and 181A cohort, respectively (a, b). (c, d): box plot analysis of NR5A2(16670) mRNA levels in two cohorts (i.e., nontumor component (NT) and 181A cohort) (c). The number on the y-axis inside the box plot C stands for the expression level of NR5A2. The x-axis for each box stands for 25 nontumor (NT) and 181A cohorts, respectively. Box plot analysis for pairwise comparison of NR5A2(16670) mRNA levels and NR5A1(652) mRNA levels in three cohorts (i.e., 90A cohort, 91A cohort, and 181A cohort) (d). The number on the y-axis inside the box plot D stands for the expression level of the given TF which is listed in the x-axis, respectively. The red dot line within the box is the mean value for each subgroup in the plot. The black line within the box is the median value for each subgroup in the plot. The Agilent feature number for NR5A2 is 16670. 652 is the Agilent feature number for NR5A1.
Both NR5A1 and NR5A2 are members of the same transcription factor family. However, NR5A1 is a prognostic predictor for favorable clinical outcomes in the 181A cohorts (Figure S9.2 in Supplementary Material) and is a positive determinant of lymphovascular invasion (LVI) in both the 91A and 181A cohorts (Figure S9.3 in Supplementary Material). It is also a positive determinant of HER(+) in the 181A and 90A cohorts (Figure S9.3 in Supplementary Material), while it is a negative determinant of LYM in the 90A cohort.
We investigated the expression patterns of NR5A family members in different cohorts. The median expression level of NR5A2 in the breast tumor component is not significantly different from that in the nontumor component (Figure 2(c)). In contrast, Zhou et al. [25] showed elevated NR5A2 in microdissected tumor versus nontumor tissues using real-time PCR that quantified NR5A2 mRNA levels. This discrepancy may be due to cohort dependent factors and methodology differences in tumor sample preparation.
Figure 2(d) demonstrates a significant difference in NR5A2 and NR5A1 expression levels in the 90A, 91A, and 181A cohorts. NR5A1 levels also are significantly higher than NR5A2 levels and independent of ER status (Figure 2(d)).
NR5A2 and ESR1 may mutually upregulate each other in the 90A cohort but differences are not significant based on network analysis in our model. The median expression level of NR5A2 in ER(+) breast tumor samples (90A cohort) is similar to that in ER(−) ones (91A cohort) (Figure 2(d)). Moreover, the top 10% high level or 90th percentile of NR5A2 in ER(−) as well as in 181 breast cancer samples predicts poor prognosis (Figures 1(a) and 1(b)). This suggests that the prognostic value of NR5A2 may be mainly due to ERα independent transcriptional events of NR5A2.
3.2. NR5A2 Tumor Suppressive and Tumor Promoting Features
3.2.1. Clinically Significant Transcriptional Profiling of NR5A2
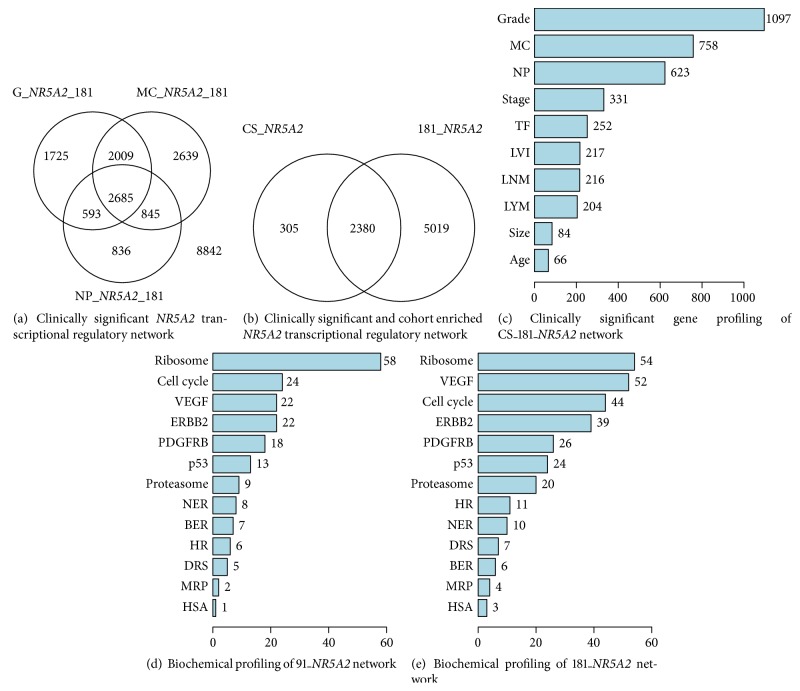
We identified the clinically significant NR5A2 cluster including 39 TFs (39/2299) as NR5A2 partners during early tumor development in the 181A cohort (Table S1.1). This was obtained by overlapping the significant gene pools of each clinical parameter (i.e., histological grade, mitotic count, and nuclear polymorphism), which includes NR5A2 as one of the clinical relevant determinants, derived from ANOVA tests. Forty-six TFs (46/2685) with clinical relevance are identified in the putative NR5A2 transcriptional regulatory network (Figure 3(a), Table S1.2). Only six TFs (6/400) are shared by the NR5A2 cluster and the network (Table S1.3). Based on the clinically significant gene profiling, NR5A2 may be involved in regulating SOX15, SALL2, NR4A2, GTF2i, and TFEC. In addition, both clinically significant and cohort relevant networks of NR5A2 share 39 TFs (39/2380) as TF partners of NR5A2 (Figure 3(b), Table S1.4). The putative network of clinically significant (CS) and 181A cohort relevant NR5A2 appears to preferentially regulate genes which are determinants of histological grade, mitotic counts, and nuclear pleomorphism (Figure 3(c)). The gene expression pattern of these putative NR5A2 target genes suggests a role in early tumor development that may predominantly suppress tumor progression (Figures S10.1–10.3). Network analysis that identified components that overlap with the gene determinants of 10 clinicopathological parameters predicts an array of NR5A2-regulated target genes that, in part, may contribute to the tumor suppressive activities. The heatmaps of these overlapping gene pools align tumor suppressive gene expression patterns with early onset of select clinical parameters. No further statistical evaluation was possible because the partial gene pool regulating the clinical parameters was also a component of NR5A2 transcriptional regulatory network. In addition, the consensus expression pattern had only a relatively small n number of samples that limits value for further statistical analysis. The tumor suppressors, SALL2, SOX15, and FOXJ1, are upregulated by NR5A2 (Figure S10.1) [26–28] and the malignant activities of ESRRA and MYBL2 are suppressed by NR5A2 (Figure S10.1) [29–32]. Oncogenic activities also may be stimulated by NR5A2 due to upregulation of NR4A2 and HOXC6 gene expression (Figure S10.1). For example, expression of NR4A2 has been reported to increase proliferation [33] and HOXC6 expression conditions tumorigenesis and drug resistance [34].
Figure 3.
Clinical and/or cohort significance of transcriptional activities of NR5A2 in given population(s). (a) Clinically significant (CS) NR5A2 transcriptional regulatory network (CS_NR5A2 network). (b) Clinically significant and cohort enriched NR5A2 transcriptional regulatory network (CS_181_NR5A2 network). (c) The bar chart for overlapping gene pools (x-axis) between the CS_181_NR5A2 network and clinical relevant genes in 10 clinical parameters (y-axis). (d) The bar chart for overlapping gene pools (x-axis) between 91_NR5A2 network and genes in 13 signal transduction pathways (y-axis). (e) The bar chart for overlapping gene pools (x-axis) between the 181_NR5A2 network and genes in 13 signal transduction pathways (y-axis). The signal transduction pathways are derived from Kyoto Encyclopedia of Genes and Genomes (KEGG) database and National Center for Biotechnology Information (NCBI) pathway interaction database.
3.2.2. Pathway Analysis of NR5A2
Cancer related profiling of NR5A2 was evaluated in 13 signal transduction pathways. The most relevant activities of NR5A2 in 91A and 181A cohorts are characterized by similar target preference but with a different preferential order (Figures 3(d) and 3(e)). Importantly, these data indicate that the pathways are suppressed by NR5A2 (see partial results in Figures S7.1–7.12). NR5A2 preferentially suppresses ribosome, VEGF, cell cycle, ERBB2, and PDGFRB signal transduction pathways. Moreover, suppression of ribosomal protein mRNA levels in the histological grade category (grade, NP, and MC) (Figures S7.13–7.15) indicates NR5A2 tumor suppressive role due to its regulation of common genes in both clinically significant and cancer related pathway profiling.
3.3. The Prognostic Relevant Gene Profiles in Two Feature Types Are Differentially Regulated by NR5A2 and NR5A1
NR5A1 and NR5A2 recognize the same promoter regions of their shared target genes [35]. Network analysis predicts that NR5A2 and NR5A1 significantly downregulate each other in 90A and 181A cohorts but not in the 91A cohort. Such differential regulatory patterns among cohorts may affect the most relevant prognostic features of both. In addition, the clinical relevance of NR5A2, as predicted by the ANOVA test, is only significant in the 181A cohort. Therefore, we selected the 181A cohort population to further identify significant prognostic features of NR5A2.
In this study, the 91A, 90A, and 181A cohorts were subclassified according to prognostic predictors into four types. Type 1 is defined as the gene pool significant for prognosis in the 90A, 91A, and 181A cohorts based on K-M analysis. Type 2 is the gene pool significant for prognosis in the 91A and 181A cohorts but less significant for prognosis in the 90A cohort. Type 3 is characterized by the gene pool significant for prognosis in the 90A and 181A cohorts but less significant for prognosis in the 91A cohort. Type 4 is the gene pool significant for prognosis in the 181A cohort but less significant for prognosis in the 91A and 90A cohorts. We also identified a subcategory of genes that contain prognostic predictors in each cohort and components of the putative NR5A2 transcriptional regulatory network. These were classified as genes within a given feature type. Four gene subcategories have been derived from this classification strategy and have been designated as feature types I–IV.
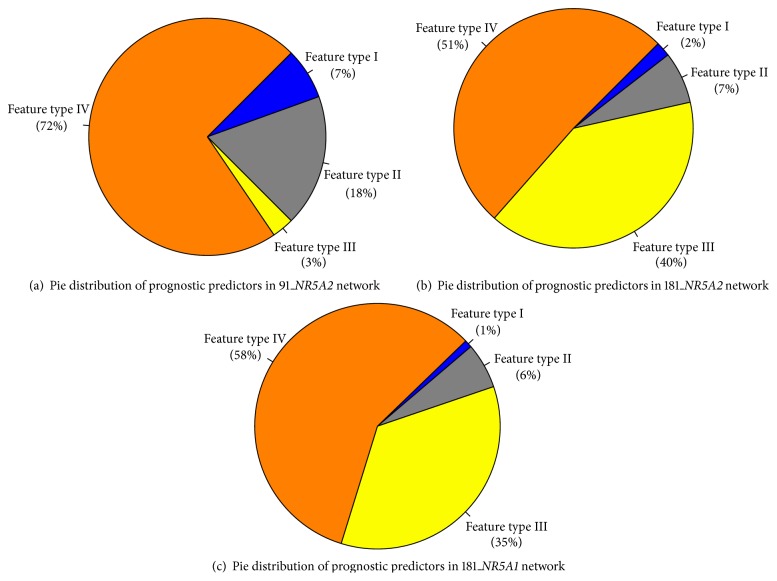
Figure 4 shows the pie distribution of prognostic relevant gene pools identified in the NR5A2 transcriptional regulatory network of the 91A (Figure 4(a)) and 181A cohorts (Figure 4(b)) and the NR5A1 transcriptional regulatory network of the 181A cohort (Figure 4(c)). The percentage of feature types I, II, III, and IV in the 91A cohort relevant NR5A2 network is 7, 18, 3, and 72, respectively. The percentage of feature types I, II, III, and IV in the 181A cohort relevant NR5A2 network is 2, 7, 40, and 51, respectively. The percentage of feature types I, II, III, and IV in the 181A cohort relevant NR5A1 network is 1, 6, 35, and 58, respectively.
Figure 4.
Pie distribution for four subpools of prognostic predictors from the networks of NR5A2 and NR5A1. (a, b) Pie distribution for four subpools of classified prognostic predictors derived from the networks of NR5A2 in two selected populations. Two cohort relevant networks of NR5A2 (91A cohort and 181A cohort) identify 131 probes and 302 probes to be the potential prognostic factors in the 91A and 181A cohort, respectively. The pie distribution illustrates four classified prognostic predictor subpools derived from overlapping between the cohort relevant network of NR5A2 and four types of prognostic indicators in three selected populations (91A cohort, 90A cohort, and 181A cohort). (a) The four feature types follow an order of type IV > type II > type III > type I. (b) Four feature types follow an order of type IV > type III > type II > type I. (c) Four subpools of classified prognostic predictors derived from the networks of NR5A1 in 181A cohort; the 181A cohort relevant network of NR5A1 predicts 1,334 probes to be the potential prognostic factors in 181A cohort. The feature type distribution follows an order of type IV > type III > type II > type I.
3.3.1. Clinical Outcomes due to Feature Type II Gene Activity Modulated by NR5A2
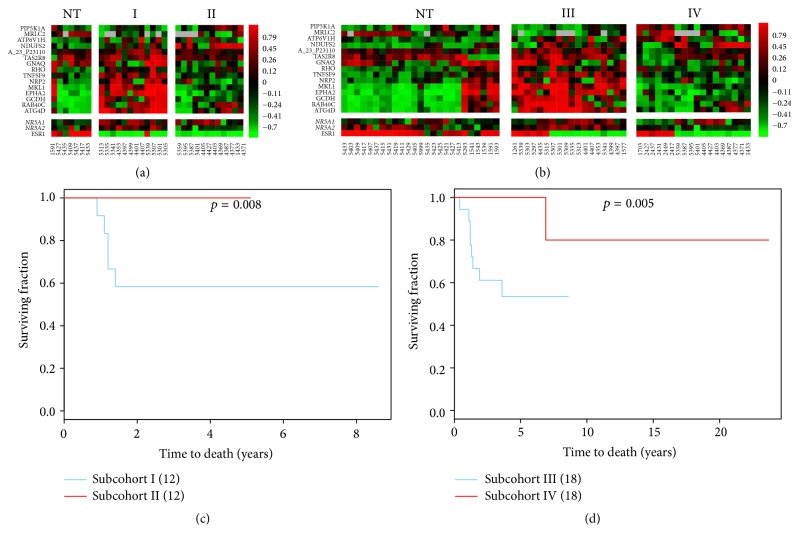
Feature types II and IV were selected as the most relevant indicators of clinical outcomes for NR5A1 and NR5A2 because NR5A1 and NR5A2 recognize the same promoter regions of their shared target genes [35]. This suggests that the unique prognostic roles of NR5A1 and NR5A2 are due to their differential interactions in a cohort dependent manner. We found NR5A1 to be a favorable prognostic indicator in 181A cohort (Figure S9.2), and NR5A2 is a factor conditioning poor prognosis in the 91A and 181A cohorts (Figure 1). Thus, the major prognostic features of NR5A2 in relation to the regulatory interaction with NR5A1 are classified in feature type II. The feature type II predicts a signature of 16 probes (Table S5.9 in Supplementary Material) that condition poor clinical outcomes with NR5A2 activation. As a result, the transcriptional dynamic of NR5A2 shows a differential gene expression pattern in Figure 5. In this case, the expression levels of NR5A1 are not suppressed by NR5A2 in the subset of tumor samples as predicted by network analysis. Moreover, NR5A1 and NR5A2 do not compete for gene regulation of the 16 probes that are putative shared target genes. Instead, they are differentially coexpressed at mRNA level and NR5A2 may override the regulatory activities of NR5A1. Based upon these analyses, we identified a 15-gene signature as a poor prognostic predictor in subcohorts I/II and subcohorts III/IV (Figure 5 and Table S10.1). It is also an independent prognostic factor in subcohorts III/non-III (Table S10.1). This signature is shared across eight molecular subtypes but enriched in ER(−) breast tumors. The prognosis related activities of the 15-gene signature are listed in Table 1. Only five (5/16) have been documented in support of the network prediction including chemoresistant enhancers, ATG4D and ATP6V1H [36, 37], EPHA2, which is a resistance marker for anti-HER therapy [38], and two new drug targets NR5A2 and NRP2 [39, 40].
Figure 5.
Validation of a prognostic signature conditioning poor outcomes in subsets of ER(−) IDCs and 181 IDCs. Poor clinical outcome in a subset of ER(−) IDCs and a subset of 181 IDCs is determined by 15 probes that are predicted to be regulated by a transcription factor, NR5A2. Panel (a, b) presents the heatmap displaying 15 prognostic relevant probes, which are predicted to show the consensus expression pattern in both the 91A and 181A cohorts. These 15 probes are part of the most relevant transcriptional activities of NR5A2 in a subset of ER(−) IDCs (a) or in the subset of 181 IDCs predominantly containing ER(−) subtype (b). Panel (c, d) demonstrates significant differences in clinical outcomes based on Kaplan-Meier survival curves when comparing subcohorts I (12A)/II (12A) (c) and subcohorts III (18A)/IV (18A) (d). “NT” stands for nontumor component.
Table 1.
Literature based validation of 16 prognostic predictors predicted in the NR5A2 transcriptional regulatory network.
| Feature number | Gene symbol | Elevated in 91A and 181A cohorts | Prognostic features listed in the literature |
|---|---|---|---|
| 2551 | TAS2R8 | Poor prognosis | Undefined |
| 16670 | NR5A2 | Poor prognosis | Poor prognosis in PR(−) breast cancer [15] |
| 1761 | ATG4D | Poor prognosis | Enhancing chemoresistance [36] |
| 2967 | ATP6V1H | Poor prognosis | Early relapse in ER(+) BCs treated with tamoxifen [37] |
| 11875 | EPHA2 | Poor prognosis | Poor prognosis (resistance to anti-HER (trastuzumab) therapy) [38] |
| 4013 | A_23_P23110 | Poor prognosis | Undefined |
| 10235 | RAB40C | Poor prognosis | Undefined |
| 4332 | MKL1 | Poor prognosis | Undefined |
| 8615 | GNAQ | Poor prognosis | Undefined |
| 7774 | NRP2 | Poor prognosis | Poor prognosis in lung cancer and other cancers but not in breast cancer [40] |
| 7864 | TNFSF9 | Poor prognosis | Undefined |
| 14025 | RHO | Poor prognosis | Undefined |
| 4337 | GCDH | Poor prognosis | Undefined |
| 17978 | MRLC2 | Poor prognosis | Undefined |
| 22368 | NDUFS2 | Good prognosis | Undefined |
| 15037 | PIP5K1A | Good prognosis | Undefined |
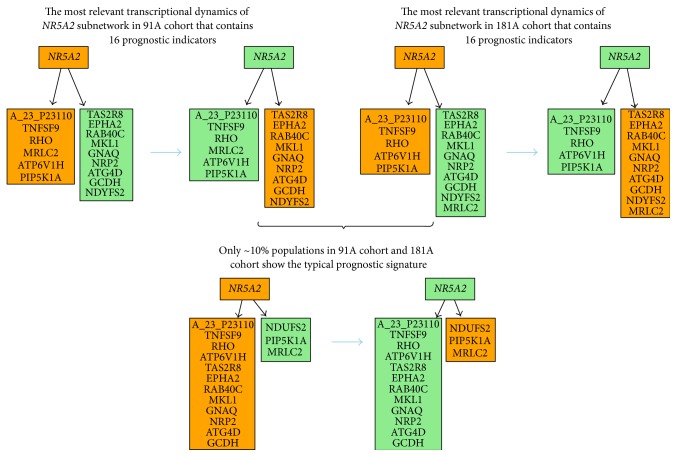
Figure 6 illustrates differences in the transcriptional dynamics of 15 probes in the 91A and 181A cohorts. The regulatory mode of MRLC2 in cohort relevant NR5A2 networks (91A cohort versus 181A cohort) is switched. However, approximately 10% of breast tumors in both cohorts share the same gene expression pattern that was previously proven to be a poor prognostic signature in subcohorts I/non-I, subcohorts III/IV, and subcohorts III/non-III based on univariate COXPH analysis (Table S10.1). The 15-gene signature is a prognostic indicator for poor outcomes in subcohort III of the 181A cohort based on results from multivariate COXPH analysis. This functional prognostic signature does not fully match the most relevant subnetwork of NR5A2 in 91A cohort. This misalignment occurs because the MRLC2 expression distribution in subcohort I of 91A cohort is an exception for cohort relevant NR5A2 network but normal for the 181A cohort.
Figure 6.
Inferred subnetwork of NR5A2 in regulating prognostic predictors during tumor progression. Tumor progression is predicted to be controlled by an ERα independent NR5A2-mediated mechanism. The deviation between prognostic relevant subnetwork and the inferred subnetwork of NR5A2 is due to differential data distribution among tumor samples. This diagram demonstrates that the supervised network analysis can identify signatures conditioning poor prognosis possibly driven by NR5A2 in ~10% studied cohort.
Interestingly, the prognostic relevant subcohorts I and III have 7 probes (EPHA2, RAB40C, MKL1, GNAQ, NRP2, ATG4D, and GCDH) that show a shifted expression pattern when compared to the most relevant networks of NR5A2 in both 91A and 181A cohorts (Figure 6). Additionally, no transcriptional dynamics are observed for ATP6V1H in subcohorts I/II and III/IV (Figure 5). This is an atypical case [11] derived from the dual coupling of the supervised network and 90th percentile K-M survival analyses. The latter method typically enriches the prognostic relevant subgroup with relatively high sub-CID values for a given gene signature [24].
3.3.2. Clinical Outcomes due to Feature Type IV Gene Activity Conditioned by NR5A1
We found NR5A1 to be a prognostic indicator for favorable outcomes in the 181A cohort (Figure S9.2). NR5A1 downregulates NR5A2 in the 90A and 181A cohorts but not in the 91A cohort as shown by network analysis. However, Figure 2(d) shows that NR5A2 levels are lower than NR5A1 levels in the 90A, 91A, and 181A cohorts. We suspect that the competitive interaction between NR5A1 and A2, or NR5A1 predominant transcriptional regulatory pattern, may determine the favorable prognosis across molecular subtypes. Therefore, we proposed feature type IV, which includes genes regulated via mixed regulatory patterns, to be the most relevant prognostic event driven by NR5A1.
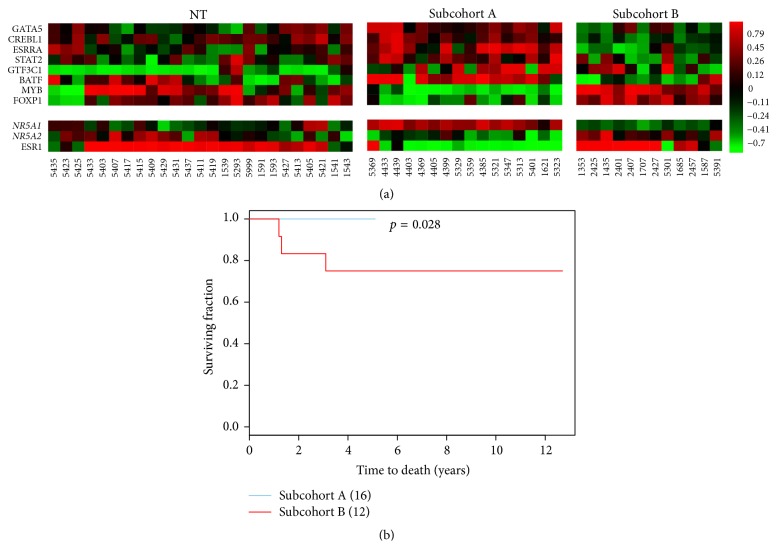
Both NR5A2 and NR5A1 share common target genes (292 probes) in feature type IV (Table S6.6). We found 9 TFs (9/14) within 292 probes that showed a distinct expression pattern due to the opposite regulatory modes of NR5A1 and NR5A2. Figure 7 shows heatmaps demonstrating differential gene expression patterns of 8 common components of subnetworks for NR5A1 and NR5A2 in subcohorts A and B. Kaplan-Meier survival analyses indicate their significance in prognosis. A favorable prognostic signature regulated by NR5A1 has been identified and its prognostic relevant activities have been partially validated (Table 2). For instance, GTF3C1 is a chemoresistance marker in breast cancer [41] and MYB is a favorable prognostic factor [42]. Interestingly, BATF is a poor prognostic factor in B-cell lymphoma [43] and ESRRA is a poor prognostic factor in breast cancer [44] and ovarian cancer [45]. FOXP1 predicts a favorable prognosis in breast cancer [46] STAT2 may predict favorable prognosis in carcinoid tumors [47] and COXPH analysis suggests this 8-gene signature is not a favorable prognostic signature in our model system. Further evaluations of more comprehensive populations are needed to fully establish the utility of these interactions.
Figure 7.
In vivo validation of a favorable prognostic signature in a subset of 181 IDCs. Panel (a) shows the heatmaps of a signature (8 probes) for favorable prognosis to be regulated by NR5A1 and NR5A2 in two subcohorts (subcohort A and subcohort B), respectively. NR5A1 has an opposite regulatory mode as compared to NR5A2 in a subset of 181 IDCs for this inferred subnetwork. Panel (b) shows the survival curves of two subcohorts based on Kaplan-Meier survival analysis.
Table 2.
Summary of eight transcription factors regulated by NR5A1 predicting favorable prognostic values.
| Feature number | Gene symbol | High level and prognosis | Prognostic features listed in the literature | Regulation by NR5A1 |
|---|---|---|---|---|
| 17400 | GTF3C1 | Poor | Adriamycin/cytoxan resistance [41] | Up |
| 5586 | MYB | Good | Good prognosis [42] | Down |
| 6776 | BATF | Good | Poor prognostic predictor in B-cell lymphoma [43] | Up |
| 5480 | ESRRA | Good | Poor prognosis [44, 45] | Up |
| 14511 | FOXP1 | Poor | Good prognosis [46] | Down |
| 878 | ATF6B | Good | Undefined | Up |
| 3113 | GATA5 | Good | Undefined | Up |
| 2002 | STAT2 | Poor | Good prognosis in carcinoid tumors [47] | Up |
3.4. Partially Antagonistic Interactions between NR5A1 and A2 in the Regulation of Pathophysiological and Prognostic Relevant Activities
NR5A1 and A2 show an inverse regulatory mode in six biochemical activities: cell cycle regulation, tumor progression and carcinogenesis, steroidogenesis [48], sustained angiogenesis, the Warburg effect, and epithelial mesenchymal transition (EMT) (Figure S10.5). Network analysis predicts that these tumor promoting activities are partially regulated by NR5A1 and A2. NR5A1 has relatively higher activities than NR5A2 in cell cycle regulation, sustained angiogenesis, the Warburg effect, and EMT. However, NR5A2 may impact steroidogenesis, tumor progression, and carcinogenesis to a greater degree than NR5A1. Typically, CYP19A1 coding for aromatase is upregulated by NR5A2 in the 181A cohort (Figures S10.5B and S10.5C). CYP19A1 also indicates poor prognosis in ER(−) breast cancers comprised of TN and ERBB2 (Figure S9.1_77A). Our data support the concept that NR5A2 is a regulator of steroidogenesis in breast cancers. Also, this suggests the need for further investigation of TN and/or ERBB2 subtypes to evaluate whether an aromatase inhibitor may be a treatment option for a subset of ER(−) breast cancers.
3.5. Functional Subtyping of Breast Cancers via the Gene Signatures Potentially Driven by a Transcription Factor or Multiple Transcription Factors
The final goal of functional subtyping of breast cancers via the TF(s) mediated gene signature(s) is to develop the clinical biomarker(s) that can be located by IHC stain and be reproducible in other gene expression datasets. Functional subtyping of breast cancer via gene signature may facilitate decision making for cocktail treatments in clinical experiments to optimize personalized medicine strategies. Although using meta-analysis may add some clarity to the interpretation of our results, this has not been done for this study, in part, due to the absence of appropriate comparator data or groups.
The tumor section contains ~1 : 1 ratio of tumor cell and nontumor components in our model. Microdissection was not performed as a part of the sample preparation because both tumor initiation and development involve the interactions between tumor cells and nontumor cells [25, 49]. As a result, there is a low probability for accurately validating an inferred gene signature containing the coexpressed TFs with NR5A2 derived from the supervised network analysis and functional clustering (e.g., 90th percentile K-M survival analysis) to be colocalized in the nucleus of the same cell. Similarly, there is a low probability for validating the 15 target genes of NR5A2 localization in the same cell. For instance, some limitations described below suggest the validation of the 8-gene signature driven by NR5A2 using IHC staining of clinical tumor samples and the reproducibility of this gene signature in other datasets may not be a viable option for our model.
Multiple biological and/or genetic effects impacting pathophysiological outcomes can alter the intrinsic TF mediated transcriptional pathways during tumor initiation and development. As a result, biological outcomes mediated by altered TF transcriptional activities may have changed. Statistical measures (CIDUGPCC and multivariate version of CID) facilitate predicting the most relevant transcriptional pathways in a given sample population. However, several limiting factors impact model validation. These include the following: (1) due to tumor sample preparation without microdissection, the microarray gene expression data used for the network analysis cannot dissect cell type specific networks of NR5A2. (2) There are a suboptimal number of paraffin-embedded tumor samples (~10% population) for IHC staining. This is because NR5A2 is expressed in both stromal and epithelial cells in the breast cancer tumor section [25] and the tumor sections are collected at a single time point. This limiting factor is important because of the large number of gene products for the network components needed to be stained. (3) The candidate cohort, which may reproduce the key signature of NR5A2, needs to align with the specific defining characteristics of the 181A cohort to conclude the same prognostic relevance. The public datasets have unique cohort characters that do not conclusively reproduce the gene signatures from the181A cohort study based on our limited meta-analysis experience.
Alternatively, the cell models (e.g., human breast cancer cell lines) or other appropriate model systems (e.g., the mouse model organism and others) can be used to evaluate the unique or common activities of TFs in tumor cells in a time dependent manner, which are predicted by the supervised network analysis. Furthermore, results from time course studies in the cell models and other model systems inform validation of the predicted NR5A2 activities in a cell specific manner or between different cell types via a paracrine mechanism. Both IHC staining in cell models and collecting an appropriate sample of each testing cohort will be necessary to reproduce the gene signature in the testing cohort for the identification of clinical biomarkers.
4. Conclusions
The prognostic value of NR5A2 is established by identifying the most relevant prognostic indicators regulated by NR5A2, which are expressed in both ER(−) (91A cohort) and ER(+)/ER(−) (181A cohort) tumor cells. We analyzed two cohort relevant networks to identify a common prognostic signature which is relevant in 91A and 181A cohorts but less so in the 90A cohort (i.e., feature type II). A 15-gene signature was identified which is an independent prognostic factor across eight molecular subtypes of 181 IDCs especially enriched in ER(−) IDCs. This is the first report where NR5A2-mediated signature is a poor prognostic indicator in a subset of breast cancers. Relative to therapeutic potential, NR5A2 may be a new target for the effective treatment of a subset of breast cancers with the 15-gene signature.
In this study, we also found NR5A1 to be a favorable prognostic indicator in the breast cancer population with feature type IV and we identified an 8-gene signature to be significant in a subset of breast cancers (subcohort A) based on Kaplan-Meier survival analysis but not significant by COXPH analysis.
Supplementary Material
There are ten supplemental files gathered to be the supplementary information. We collected the Venn diagram figures and their corresponding gene pools to form the unique table. It consists of supplemental files 1-6. We have gathered the key heatmaps results of the network analyses in supplemental files 7-8. It includes the clinical relevant gene profiling, the pathway analyses and the partially validated results of the network analysis. The major results of survival analyses are listed in supplemental file 9. In addition, we put the ANOVA test results of a few important transcription factors for showing their clinical impacts in supplemental file 9. The supplemental file 10 includes (1) the gene expression patterns of the gene signature (292 probes), the gene components of NR5A2 network overlapping with clinical relevant genes; (2) the potential pathophysiological activities driven by NR5A1 and NR5A2 in 181A cohort, respectively; and (3) univariate and multivariate analyses for survival on prognostic factors in 91A cohort and 181A cohort.
Acknowledgments
The financial support of this work was mainly from Grants NSC95-2314-B-002-255-MY3 and NSC98-2314-B-002-093-MY2 (to Dr. Fon-Jou Hsieh). The authors appreciate the technical support from Welgene Biotechnology company in Taiwan.
Abbreviations
- ATF6B:
Activating transcription factor 6 beta. Its old symbol is CREBL1 (see Figure 7)
- ATG4D:
Autophagy related 4D, cysteine peptidase
- ATP6V1H:
ATPase, H+ transporting, lysosomal 50/57 kDa, V1 subunit H
- BATF:
Basic leucine zipper transcription factor, ATF-like
- BER:
Signal transduction pathway of base excision repair
- Cell cycle:
Signal transduction pathway of cell cycle
- CID:
Coefficient of intrinsic dependence
- CYP19A1:
Cytochrome P450, family 19, subfamily A, polypeptide 1
- DRS:
Signal transduction pathway of DNA replication
- EPHA2:
EPH receptor A2
- ER(+):
Estrogen receptor positive
- ERBB2+:
ER(−), PR(−) and HER(+)
- ERBB2:
Signal transduction pathway of erb-b2 receptor tyrosine kinase 2
- ESR1:
Estrogen receptor 1
- ESRRA:
Estrogen-related receptor alpha
- FOXJ1:
Forkhead box J1
- FOXP1:
Forkhead box P1
- GPCC:
Galton Pearson's Correlation Coefficient
- GTF2i:
General transcription factor IIi
- GTF3C1:
General transcription factor IIIC, polypeptide 1, alpha
- HER(−):
v-erb-b2 erythroblastic leukemia viral oncogene homolog 2 negative
- HOXC6:
Homeobox C6
- HR:
Signal transduction pathway of homologous recombination (HR)
- HSΑ:
Signal transduction pathway of $hsa03450$ (HSΑ)
- MRP:
Signal transduction pathway of mismatch repair pathway (MRP)
- MYB:
v-myb avian myeloblastosis viral oncogene homolog
- MYBL2:
v-myb avian myeloblastosis viral oncogene homolog-like 2
- NER:
Signal transduction pathway of nucleotide excision repair
- NR4A2:
Nuclear receptor subfamily 4, group A, member 2
- NR5A1:
Nuclear receptor subfamily 5, group A, member 1
- NR5A2:
Nuclear receptor subfamily 5, group A, member 2
- NRP2:
Neuropilin 2
- p53:
Signal transduction pathway of p53
- PDGFRB:
Platelet-derived growth factor receptor, beta polypeptide or signal transduction pathway of PDGFRB
- PR(−):
Progesterone receptor negative
- Proteasome:
Signal transduction pathway of proteasome
- Ribosome:
Signal transduction pathway of ribosome
- SALL2:
Sal-like 2 (Drosophila)
- SOX15:
SRY (sex determining region Y)-box 15
- TAM:
Tamoxifen
- TFEC:
Transcription factor EC
- VEGF:
Vascular endothelial growth factor or signal transduction pathway of VEGF.
Conflict of Interests
The authors declare that there is no conflict of interests regarding the publication of this paper.
References
- 1.Ferlay J., Soerjomataram I., Dikshit R., et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. International Journal of Cancer. 2015;136(5):E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A., Center M. M., DeSantis C., Ward E. M. Global patterns of cancer incidence and mortality rates and trends. Cancer Epidemiology Biomarkers and Prevention. 2010;19(8):1893–1907. doi: 10.1158/1055-9965.epi-10-0437. [DOI] [PubMed] [Google Scholar]
- 3.Jemal A., Bray F., Center M. M., Ferlay J., Ward E., Forman D. Global cancer statistics. CA: Cancer Journal for Clinicians. 2011;61(2):69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 4.Patani N., Martin L.-A., Dowsett M. Biomarkers for the clinical management of breast cancer: international perspective. International Journal of Cancer. 2013;133(1):1–13. doi: 10.1002/ijc.27997. [DOI] [PubMed] [Google Scholar]
- 5.Crown J., O'Shaughnessy J., Gullo G. Emerging targeted therapies in triple-negative breast cancer. Annals of Oncology. 2012;23(6):vi56–vi65. doi: 10.1093/annonc/mds196. [DOI] [PubMed] [Google Scholar]
- 6.Gonzalez-Angulo A. M., Hennessy B. T. J., Mills G. B. Future of personalized medicine in oncology: a systems biology approach. Journal of Clinical Oncology. 2010;28(16):2777–2783. doi: 10.1200/jco.2009.27.0777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang H.-H., Dreyfuss J. M., Ramoni M. F. A transcriptional network signature characterizes lung cancer subtypes. Cancer. 2011;117(2):353–360. doi: 10.1002/cncr.25592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu L.-Y. D., Chen C.-Y., Chen M.-J. M., et al. Statistical identification of gene association by CID in application of constructing ER regulatory network. BMC Bioinformatics. 2009;10, article 85 doi: 10.1186/1471-2105-10-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu L.-Y. D., Chang L.-Y., Kuo W.-H., et al. Major functional transcriptome of an inferred center regulator of an ER(-) breast cancer model system. Cancer Informatics. 2012;11:87–111. doi: 10.4137/cin.s8633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu L.-Y. D., Chang L.-Y., Kuo W.-H., et al. In Silico prediction for regulation of transcription factors on their shared target genes indicates relevant clinical implications in a breast cancer population. Cancer Informatics. 2012;11:113–137. doi: 10.4137/cin.s8470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu L.-Y. D., Chang L.-Y., Kuo W.-H., et al. Prognostic features of signal transducer and activator of transcription 3 in an ER(+) breast cancer model system. Cancer Informatics. 2014;13:21–45. doi: 10.4137/cin.s12493. correction in: Cancer Informatics, vol. 13, pp. 125–129, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zheng G., Tu K., Yang Q., et al. ITFP: an integrated platform of mammalian transcription factors. Bioinformatics. 2008;24(20):2416–2417. doi: 10.1093/bioinformatics/btn439. [DOI] [PubMed] [Google Scholar]
- 13.Li M., Xie Y.-H., Kong Y.-Y., Wu X., Zhu L., Wang Y. Cloning and characterization of a novel human hepatocyte transcription factor, hB1F, which binds and activates enhancer II of hepatitis B virus. The Journal of Biological Chemistry. 1998;273(44):29022–29031. doi: 10.1074/jbc.273.44.29022. [DOI] [PubMed] [Google Scholar]
- 14.Zhang C.-K., Lin W., Cai Y.-N., et al. Characterization of the genomic structure and tissue-specific promoter of the human nuclear receptor NR5A2 (hB1F) gene. Gene. 2001;273(2):239–249. doi: 10.1016/S0378-1119(01)00586-8. [DOI] [PubMed] [Google Scholar]
- 15.Riggins R. B., Mazzotta M. M., Maniya O. Z., Clarke R. Orphan nuclear receptors in breast cancer pathogenesis and therapeutic response. Endocrine-Related Cancer. 2010;17(3):R213–R231. doi: 10.1677/erc-10-0058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fayard E., Auwerx J., Schoonjans K. LRH-1: an orphan nuclear receptor involved in development, metabolism and steroidogenesis. Trends in Cell Biology. 2004;14(5):250–260. doi: 10.1016/j.tcb.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 17.Krylova I. N., Sablin E. P., Moore J., et al. Structural analyses reveal phosphatidyl inositols as ligands for the NR5 orphan receptors SF-1 and LRH-1. Cell. 2005;120(3):343–355. doi: 10.1016/j.cell.2005.01.024. [DOI] [PubMed] [Google Scholar]
- 18.Miki Y., Clyne C. D., Suzuki T., et al. Immunolocalization of liver receptor homologue-1 (LRH-1) in human breast carcinoma: possible regulator of in situ steroidogenesis. Cancer Letters. 2006;244(1):24–33. doi: 10.1016/j.canlet.2005.11.038. [DOI] [PubMed] [Google Scholar]
- 19.Annicotte J.-S., Chavey C., Servant N., et al. The nuclear receptor liver receptor homolog-1 is an estrogen receptor target gene. Oncogene. 2005;24(55):8167–8175. doi: 10.1038/sj.onc.1208950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thiruchelvam P. T. R., Lai C.-F., Hua H., et al. The liver receptor homolog-1 regulates estrogen receptor expression in breast cancer cells. Breast Cancer Research and Treatment. 2011;127(2):385–396. doi: 10.1007/s10549-010-0994-9. [DOI] [PubMed] [Google Scholar]
- 21.Chand A. L., Herridge K. A., Thompson E. W., Clyne C. D. The orphan nuclear receptor LRH-1 promotes breast cancer motility and invasion. Endocrine-Related Cancer. 2010;17(4):965–975. doi: 10.1677/ERC-10-0179. [DOI] [PubMed] [Google Scholar]
- 22.Kuo W. H., Chang L. Y., Liu D. L., et al. The interactions between GPR30 and the major biomarkers in infiltrating ductal carcinoma of the breast in an Asian population. Taiwanese Journal of Obstetics and Gynecology. 2007;46(2):135–145. doi: 10.1016/S1028-4559(07)60007-2. [DOI] [PubMed] [Google Scholar]
- 23.Ray P. S., Wang J., Qu Y., et al. FOXC1 is a potential prognostic biomarker with functional significance in basal-like breast cancer. Cancer Research. 2010;70(10):3870–3876. doi: 10.1158/0008-5472.can-09-4120. [DOI] [PubMed] [Google Scholar]
- 24.Liu L.-Y. D., Chang L.-Y., Kuo W.-H., Hwa H.-L., Chang K.-J., Hsieh F.-J. A supervised network analysis on gene expression profiles of breast tumors predicts a 41-gene prognostic signature of the transcription factor MYB across molecular subtypes. Computational and Mathematical Methods in Medicine. 2014;2014:19. doi: 10.1155/2014/813067.813067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou J., Suzuki T., Kovacic A., et al. Interactions between prostaglandin E2, liver receptor homologue-1, and aromatase in breast cancer. Cancer Research. 2005;65(2):657–663. [PubMed] [Google Scholar]
- 26.Liu H., Adler A. S., Segal E., Chang H. Y. A transcriptional program mediating entry into cellular quiescence. PLoS Genetics. 2007;3(6, article e91) doi: 10.1371/journal.pgen.0030091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thu K. L., Radulovich N., Becker-Santos D. D., et al. SOX15 is a candidate tumor suppressor in pancreatic cancer with a potential role in Wnt/β-catenin signaling. Oncogene. 2014;33(3):279–288. doi: 10.1038/onc.2012.595. [DOI] [PubMed] [Google Scholar]
- 28.Demircan B., Dyer L. M., Gerace M., Lobenhofer E. K., Robertson K. D., Brown K. D. Comparative epigenomics of human and mouse mammary tumors. Genes Chromosomes and Cancer. 2009;48(1):83–97. doi: 10.1002/gcc.20620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chisamore M. J., Cunningham M. E., Flores O., Wilkinson H. A., Chen J. D. Characterization of a novel small molecule subtype specific estrogen-related receptor α antagonist in MCF-7 breast cancer cells. PLoS ONE. 2009;4(5) doi: 10.1371/journal.pone.0005624.e5624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thorner A. R., Hoadley K. A., Parker J. S., Winkel S., Millikan R. C., Perou C. M. In vitro and in vivo analysis of B-Myb in basal-like breast cancer. Oncogene. 2009;28(5):742–751. doi: 10.1038/onc.2008.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sala A. B-MYB, a transcription factor implicated in regulating cell cycle, apoptosis and cancer. European Journal of Cancer. 2005;41(16):2479–2484. doi: 10.1016/j.ejca.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 32.Kalashnikova E. V., Revenko A. S., Gemo A. T., et al. ANCCA/ATAD2 overexpression identifies breast cancer patients with poor prognosis, acting to drive proliferation and survival of triple-negative cells through control of B-Myb and EZH2. Cancer Research. 2010;70(22):9402–9412. doi: 10.1158/0008-5472.can-10-1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Han Y.-F., Cao G.-W. Role of nuclear receptor NR4A2 in gastrointestinal inflammation and cancers. World Journal of Gastroenterology. 2012;18(47):6865–6873. doi: 10.3748/wjg.v18.i47.6865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim K.-J., Moon S.-M., Kim S.-A., Kang K.-W., Yoon J.-H., Ahn S.-G. Transcriptional regulation of MDR-1 by HOXC6 in multidrug-resistant cells. Oncogene. 2013;32(28):3339–3349. doi: 10.1038/onc.2012.354. [DOI] [PubMed] [Google Scholar]
- 35.Martin L. J., Taniguchi H., Robert N. M., Simard J., Tremblay J. J., Viger R. S. GATA factors and the nuclear receptors, steroidogenic factor 1/liver receptor homolog 1, are key mutual partners in the regulation of the human 3 beta-hydroxysteroid dehydrogenase type 2 promoter. Molecular Endocrinology. 2005;19(9):2358–2370. doi: 10.1210/me.2004-0257. [DOI] [PubMed] [Google Scholar]
- 36.Frankel L. B., Wen J., Lees M., et al. microRNA-101 is a potent inhibitor of autophagy. The EMBO Journal. 2011;30(22):4628–4641. doi: 10.1038/emboj.2011.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bilal E., Vassallo K., Toppmeyer D., et al. Amplified Loci on chromosomes 8 and 17 predict early relapse in ER-positive breast cancers. PLoS ONE. 2012;7(6) doi: 10.1371/journal.pone.0038575.e38575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rexer B. N., Arteaga C. L. Intrinsic and acquired resistance to HER2-targeted therapies in HER2 gene-amplified breast cancer: mechanisms and clinical implications. Critical Reviews in Oncogenesis. 2012;17(1):1–16. doi: 10.1615/critrevoncog.v17.i1.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Busby S., Nuhant P., Cameron M., et al. Probe Reports from the NIH Molecular Libraries Program. Bethesda, Md, USA: National Center for Biotechnology Information; 2010. Discovery of inverse agonists for the liver receptor homologue-1 (LRH1; NR5A2) [PubMed] [Google Scholar]
- 40.Yasuoka H., Kodama R., Tsujimoto M., et al. Neuropilin-2 expression in breast cancer: correlation with lymph node metastasis, poor prognosis, and regulation of CXCR4 expression. BMC cancer. 2009;9, article 220 doi: 10.1186/1471-2407-9-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cleator S., Tsimelzon A., Ashworth A., et al. Gene expression patterns for doxorubicin (Adriamycin) and cyclophosphamide (Cytoxan) (AC) response and resistance. Breast Cancer Research and Treatment. 2006;95(3):229–233. doi: 10.1007/s10549-005-9009-7. [DOI] [PubMed] [Google Scholar]
- 42.Guérin M., Sheng Z.-M., Andrieu N., Riou G. Strong association between c-myb and oestrogen-receptor expression in human breast cancer. Oncogene. 1990;5(1):131–135. [PubMed] [Google Scholar]
- 43.Cai Y.-D., Huang T., Feng K.-Y., Hu L., Xie L. A unified 35-gene signature for both subtype classification and survival prediction in diffuse large B-cell Lymphomas. PLoS ONE. 2010;5(9) doi: 10.1371/journal.pone.0012726.e12726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ariazi E. A., Clark G. M., Mertz J. E. Estrogen-related receptor α and estrogen-related receptor γ associate with unfavorable and favorable biomarkers, respectively, in human breast cancer. Cancer Research. 2002;62(22):6510–6518. [PubMed] [Google Scholar]
- 45.Fujimoto J., Alam S. M., Jahan I., Sato E., Sakaguchi H., Tamaya T. Clinical implication of estrogen-related receptor (ERR) expression in ovarian cancers. Journal of Steroid Biochemistry and Molecular Biology. 2007;104(3-5):301–304. doi: 10.1016/j.jsbmb.2007.03.016. [DOI] [PubMed] [Google Scholar]
- 46.Fox S. B., Brown P., Han C., et al. Expression of the forkhead transcription factor FOXP1 is associated with estrogen receptor α and improved survival in primary human breast carcinomas. Clinical Cancer Research. 2004;10(10):3521–3527. doi: 10.1158/1078-0432.CCR-03-0461. [DOI] [PubMed] [Google Scholar]
- 47.Zhou Y., Wang S., Gobl A., Oberg K. Interferon alpha induction of Stat1 and Stat2 and their prognostic significance in carcinoid tumors. Oncology. 2001;60(4):330–338. doi: 10.1159/000058529. [DOI] [PubMed] [Google Scholar]
- 48.Lazarus K. A., Wijayakumara D., Chand A. L., Simpson E. R., Clyne C. D. Therapeutic potential of liver receptor homolog-1 modulators. The Journal of Steroid Biochemistry and Molecular Biology. 2012;130(3–5):138–146. doi: 10.1016/j.jsbmb.2011.12.017. [DOI] [PubMed] [Google Scholar]
- 49.Liu C.-C., Shen Z., Kung H.-F., Lin M. C. M. Cancer gene therapy targeting angiogenesis: an updated review. World Journal of Gastroenterology. 2006;12(43):6941–6948. doi: 10.3748/wjg.v12.i43.6941. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
There are ten supplemental files gathered to be the supplementary information. We collected the Venn diagram figures and their corresponding gene pools to form the unique table. It consists of supplemental files 1-6. We have gathered the key heatmaps results of the network analyses in supplemental files 7-8. It includes the clinical relevant gene profiling, the pathway analyses and the partially validated results of the network analysis. The major results of survival analyses are listed in supplemental file 9. In addition, we put the ANOVA test results of a few important transcription factors for showing their clinical impacts in supplemental file 9. The supplemental file 10 includes (1) the gene expression patterns of the gene signature (292 probes), the gene components of NR5A2 network overlapping with clinical relevant genes; (2) the potential pathophysiological activities driven by NR5A1 and NR5A2 in 181A cohort, respectively; and (3) univariate and multivariate analyses for survival on prognostic factors in 91A cohort and 181A cohort.