Abstract
Meiotic recombination has two key functions: the faithful assortment of chromosomes into gametes and the creation of genetic diversity. Both processes require that meiotic recombination occurs between homologous chromosomes, rather than sister chromatids. Accordingly, a host of regulatory factors are activated during meiosis to distinguish sisters from homologues, suppress recombination between sister chromatids and promote the chromatids of the homologous chromosome as the preferred recombination partners. Here, we discuss the recent advances in our understanding of the mechanistic basis of meiotic recombination template choice, focusing primarily on developments in the budding yeast, Saccharomyces cerevisiae, where the regulation is currently best understood.
Meiosis
Sexually reproducing organisms are faced with the challenge of promoting genetic diversity in offspring whilst maintaining genome stability. The controlled reshuffling of genetic information is achieved by meiosis, a specialized cell division program that generates haploid gametes from a diploid progenitor. The unique division pattern of meiosis features one round of DNA replication followed by two successive nuclear divisions. The first division segregates homologous chromosomes inherited from different parents, whereas the second division separates sister chromatids. Prior to homologue disjunction in meiosis I, self-inflicted DNA double-strand breaks (DSBs) trigger recombination between homologous chromosomes. In addition to providing a source of sequence diversity in offspring, the immediate purpose of this genetic exchange is to create physical links between homologues in the form of crossovers (Figure 1A). These links resist the pulling force of the meiosis I spindle and help properly orient homologous chromosome pairs in the division plane, thereby ensuring accurate homologue segregation. Errors in in this process, in particular failure to form a crossover and inappropriate placement of crossovers, are a major cause of infertility and birth defects, such as trisomy 21 (Down Syndrome) [1]. As a result, there is constant regulatory pressure throughout meiotic recombination to reach a suitable crossover outcome.
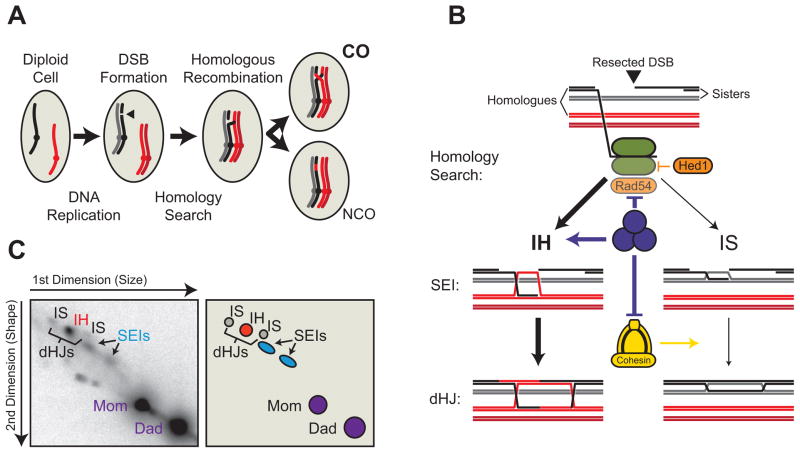
Figure 1. Meiotic Template Choice Overview.
A. Inter-homologue (IH) recombination by homologous recombination (HR) can be repaired as the preferred crossover (CO) or alternative non-crossover (NCO) products. B. Initial meiotic template choice. IH or inter-sister (IS) strand invasion results in single-end invasions (SEIs), which progress to double Holliday junctions (dHJs). MRH represents Mek1, Red1, Hop1 axis proteins. C. Example Southern blot of 2D-gel analysis at the HIS4-LEU2 hotspot in a wild-type strain in meiosis, showing positions of parental DNA and recombination intermediates – SEIs and IH/IS dHJs.
The Mechanism of Meiotic Recombination
Meiotic recombination occurs through highly controlled DSB repair. DSBs are created by the Spo11 endonuclease and form predominantly at DSB hotspots. Following breakage, DSB ends get rapidly resected to produce 3′ single-stranded tails, which become coated with replication protein A and stimulate a DNA damage signaling cascade related to the DNA damage response in mitotic cells. DSB repair in meiosis occurs exclusively by homologous recombination (HR), which requires a homologous DNA sequence (the homologous chromosome or the sister chromatid) to act as a template molecule for repair-associated DNA synthesis. Locating the correct repair template is aided by a variety of pairing mechanisms and relies on sequence- or chromosome-structure-related homology [2]. Individual 3′ single-stranded tails, coated with recombinase enzymes, are thought to be the primary effectors of sequence-dependent homology search, and have been suggested to act as a ‘homology-searching tentacle’ extending into the nucleus [3]. Strand invasion occurs by insertion of the resected end into the template duplex, followed by limited synthesis-dependent extension of the invading end and displacement of the non-template strand. If stabilized, this structure matures into a single-end invasion (SEI) intermediate [4]. The other resected end is then incorporated into this structure, forming a stable double Holliday junction (dHJ), a major recombination intermediate in many organisms [5]. Under normal circumstances, dHJs are almost exclusively resolved as crossovers, leading to the reciprocal exchange of DNA sequences between homologues. If the invading strand fails to be stabilized, it is expunged and processed by synthesis-dependent strand annealing (SDSA) to form a non-crossover product. Unlike crossovers, non-crossovers do not contribute to accurate homologue segregation (Figure 1A & B).
Distinguishing Repair Templates
To ensure productive crossover linkage, meiotic recombination is strongly biased toward the homologous chromosome, rather than the sister chromatid, which is the preferred repair template of DNA damage in mitotic cells. The need to differentiate between sister chromatids and homologous (non-sister) chromatids represents an intriguing mechanistic problem, as there is no intrinsic difference at the DNA level that reliably distinguishes the two. Although homologous chromosomes often encode abundant sequence polymorphisms (in particular in outcrossing populations), repair template bias is also observed in isogenic laboratory organisms, whose homologous chromosome pairs are for all intents and purposes identical at the sequence level. Any mechanisms controlling repair template choice must therefore be acting at the level of epigenetics, chromosome structure or spatial arrangement of chromosomes in the nucleus. Available evidence suggests that the mitotic cells’ inter-sister (IS) repair bias results to a large extent from the spatial proximity of sister chromatids. This proximity is an inherent consequence of DNA replication and is stabilized by deposition of cohesin complexes, and cohesin-independent associations including sister chromatid intertwining [3, 6], making the sister chromatids more readily available for repair interactions. Meiotic cells must overcome this inherent sister bias to promote inter-homologue (IH) recombination. In recent years, a number of molecular mechanisms, involving specialized recombinase pathways and profound modifications of chromosome structure, have been uncovered that play fundamental roles in mediating meiotic repair template choice. Here, we give a brief overview of the molecular assays of template choice and then describe our current understanding of the molecular mechanisms driving repair template bias in meiosis.
Experimental Determination of Template Choice
The quantitative measurement of recombination partner choice is technically challenging because of the difficulty in measuring recombination between sister chromatids. IH recombination often can be readily detected by the genetic transfer of sequence information between parental homologues. The occurrence of such ‘molecular scars’ of recombination has been used successfully in S. cerevisiae to capture the majority of crossover and non-crossover events genome-wide and obtain a global view of IH recombination [7–9]. By contrast, sister chromatids are genetically identical, rendering observation of IS recombination outcome impossible. As a result, IS recombination is frequently inferred indirectly by comparing IH recombination outcome to the levels of DSB formation in the corresponding genomic region [10, 11]. Regions that exhibit high DSB formation, but low IH recombination are then assumed to be repaired by IS recombination. For example, IS recombination was inferred to be increased near centromeres based on the observation that DSB formation was repressed 2-fold in centromere-proximal regions compared to the genome average [12], whereas IH recombination was shown to be 6-fold less [7]. This type of inference can also be made by comparing DSB levels to binding of known markers of IH-crossovers, for example Zip3 in S. cerevisiae [13] or COSA-1 in C. elegans [14], but is inherently limited by the lack of direct evidence of IS recombination.
Direct analysis of IS recombination is possible as long as the two recombining chromatids are in physical contact during repair, because the recombination intermediates form branched DNA species that can be analyzed using 2D gel separation and Southern blotting (Figure 1B & C). If the two homologous chromosomes carry restriction site polymorphisms surrounding the site of repair, this assay allows quantitative analysis of both IH and IS intermediates. The HIS4-LEU2 hotspot in S. cerevisiae is the most widely used for this purpose and has greatly advanced our understanding of the role of meiotic factors involved in template choice and progression of recombination [4]. 2D gel quantification of dHJs in wild-type cells reveals a 5:1 IH:IS ratio at this hotspot in meiotic cells (Figure 2A, red bars) [3], and a 1:4 IH:IS ratio following DSB induction in mitotic cells, showing the expected strong IS bias to preserve genome integrity during vegetative growth (Figure 2A) [15]. Although HIS4-LEU2 is not an endogenous hotspot and contains bacterially derived DNA sequences, a similar meiotic 4.5:1 ratio has also been observed at the naturally ‘DSB-hot’ ERG1 hotspot, suggesting that HIS4-LEU2 is a good model for other ‘DSB-hot’ hotspots [4, 16]. It is currently not possible to determine whether a similar bias also applies to weaker hotspots in S. cerevisiae. This is particularly relevant because weak and strong DSB hotspots in the fission yeast S. pombe have been inferred to be subject to dramatically different levels of template bias. Direct physical analysis of template choice at the mbs1 ‘DSB-hot’ hotspot, revealed a strong repair bias toward the sister chromatid of ~1:4 IH:IS [17], whereas a DSB-cold region was found by genetic analysis to exhibit a high level of IH recombination [18, 19], suggesting that weak S. pombe hotspots have a strong IH bias. However, confirmation of these inferences will have to await the development of unbiased genomic tools for direct analysis of meiotic template bias. For the purpose of this review, we will focus predominantly on studies using the HIS4-LEU2 hotspot, which has been the workhorse for the mechanistic analysis of meiotic template choice.
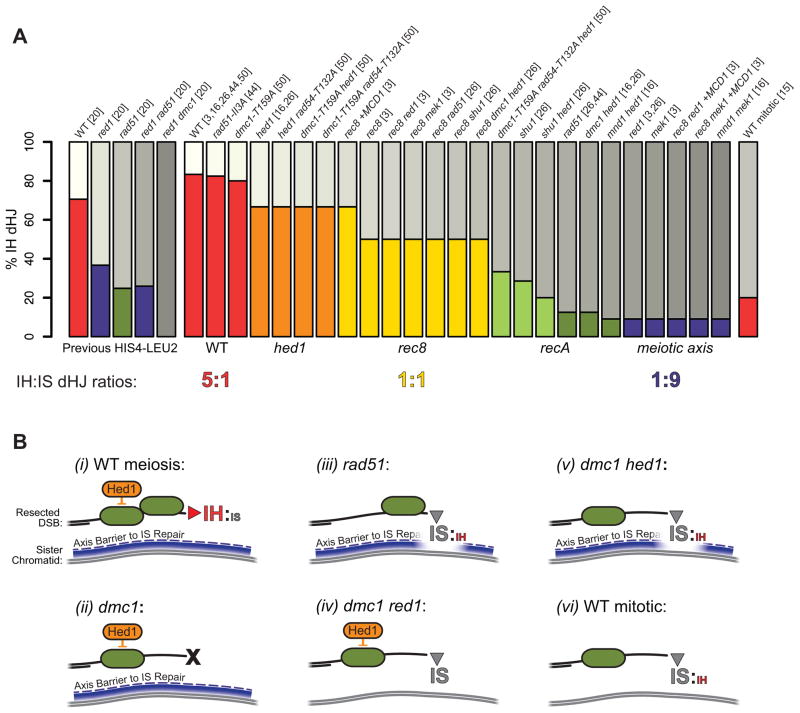
Figure 2. Template Choice Bias in Meiotic Mutants.
A. IH:IS dHJ ratios at the HIS4-LEU2 hotspot. Two versions of the HIS4-LEU2 hotspot are shown: a previous hotspot is shown on the left [20], and the currently used hotspot is shown on the right [4]. Five main mutant groups have been classified (left to right), wild type (WT; red), hed1-like (orange), rec8/cohesin (yellow), recA-associated (green), and axis-defective (blue). B. Schematics of altered responses to template choice regulatory signals in meiotic mutants.
Mechanisms Controlling Meiotic Template Choice
Studies in various organisms have demonstrated that multiple cooperative groups of factors are involved in regulating meiotic template bias (summarized in Figure 1B). These factors can be grouped roughly into instructive signals and interpreters. The instructive signals comprise factors (mostly chromosomal proteins) that differentiate sister chromatids from homologous chromosomes and create a barrier to IS repair. The interpreters (essentially the recombinases and their accessory factors) in turn read these instructive signals to avoid the sister and thus invade the homologous chromosome. Furthermore, it appears that the initial determination of template choice occurs at a very early stage of meiosis, and must be maintained until resolution of recombination intermediates to obtain suitable outcomes, a process that requires the meiotic cohesin complex [3].
Instructive Signals of Meiotic Inter-Homologue Bias
Available evidence suggests that meiotic cells take advantage of the inherent spatial proximity of sister chromatids and the assembly of a highly specialized chromosome architecture to establish a barrier to IS repair. Meiotic chromosomes assemble into characteristic structures featuring associated sister chromatids compacted along a dense axis, from which loops of chromatin protrude. This architecture plays a central role in the control of multiple steps of meiotic recombination, including the establishment of IH bias [20–24]. In S. cerevisiae, the meiotic chromosome axis comprises the meiosis-specific phospho-proteins Red1 and Hop1, as well as the meiotic cohesin complex, which is partially responsible for recruiting Red1 and Hop1 to chromosomes [25]. Consistent with a central function of the axis in mediating meiotic template choice, deletion of Red1 dramatically alters the IH:IS dHJ ratio at HIS4-LEU2 from 5:1 in wild type to 1:9, producing a strong IS bias (Figure 2A- blue bars) [20, 26]. The strong IS bias caused by defective axis factors reflects a likely reversion to mitotic-like HR, based on the associated rapid resection rates, a feature of mitotic HR [3, 27], the independence from meiosis-specific repair factors [20], and the fact that the axis-less chromatin probably highly resembles mitotic chromosomes. Analysis of SEIs (Figure 1C) suggests that axis proteins create template choice bias at an early stage of recombination. IH SEIs appear to dominate in wild-type cells, whereas IS SEIs are the major species in axis-defective mutants [3], suggesting that the meiotic axis functions at the strand-invasion stage to promote IH bias, which appears intuitive as the inhibition of recombinase function is a major output of axis-dependent signaling (see below).
The major effector of axis-mediated IH bias is the meiotic kinase Mek1. Mek1 is recruited to chromosomes in a Red1 and Hop1-dependent manner and phosphorylates a number of targets to locally suppress the activity of homologous recombination factors. Accordingly, an inhibitor-sensitive allele of Mek1 (mek1-as), which allows inhibition of kinase activity when meiotic recombination is due to commence, exhibits the same strong IS bias as deletion of Red1 [3, 21, 28]. Notably, constitutive activation of Mek1 delays not only IS repair but also IH repair, suggesting that Mek1 activity is generally counteracting the HR machinery [29]. Thus, Mek1 cannot be active chromosome-wide, as it would then likely inhibit repair from all possible repair templates. Instead, Mek1 activation is coupled to sites of DSB formation through the action of the DNA damage-responsive kinases Mec1 and Tel1. These two kinases phosphorylate Hop1 on threonine 318, which along with Red1 is required for the recruitment, dimerization and activation and subsequent autophosphorylation of Mek1 kinase [21, 30–32]. In this manner, Mek1 activity is limited to the sequences in immediate spatial proximity of DSBs (i.e. the sister), and leaves the allelic sequences of the homologous chromosomes available for repair.
Signal Interpreters: the Role of RecA-like Recombinases
The regulation of RecA-like recombinases is one of the central mechanisms for establishing meiotic template choice. RecA is a bacterial recombinase that binds single-stranded DNA to form a nucleoprotein filament that facilitates recombination by catalyzing strand invasion [33]. Many organisms, including yeasts, plants and mammals, possess two RecA homologues; Rad51, which functions in both mitotic and meiotic cells, and a meiosis-specific protein, Dmc1 [34]. The interplay between these two recombinases to promote homologue bias in meiosis presents an intriguingly complex situation. The theoretical simplicity of having one recombinase for each of the two break ends could suggest an asymmetrical loading model, whereby one resected end would contain Dmc1 and be capable of IH invasion, and the other would be bound by Rad51 to keep the second break end dormant until the first end is engaged for repair. Some evidence for this binding pattern was found in certain contexts in yeast [35], and it was an accepted mechanism in A. thaliana, where each recombinase bound to a different side of the DSB was observed [36]. However, recent evidence discussed below does not support the presence of exclusive Dmc1 filaments in S. cerevisiae or A. thaliana.
Rad51 and Dmc1 appear biochemically almost identical, able to functionally substitute for one another in vitro, and it is thought to be mostly the plethora of accessory proteins that gives them their distinct roles in vivo. The DNA translocases Rad54 and Rdh54, which are thought to function with Rad51 and Dmc1 respectively, are chromatin remodelers of the Swi/Snf superfamily and are required for strand invasion, filament stability, and removal of recombinases from DNA to allow DNA synthesis to occur. In vitro experiments show some evidence of recombinase-translocase promiscuity [37], although other in vitro experiments and extensive genetic studies would suggest a more monogamous relationship [38], probably facilitated by additional accessory factors. Rad54 is a target of Mek1 kinase, which phosphorylates Rad54 on threonine 132 to destabilize interaction with Rad51, thus downregulating Rad51 activity [39]. Rad51 is also regulated by the Rad55-Rad57 heterodimer to promote strand exchange by stabilizing Rad51 filaments [40], and a recently discovered, non-essential Rad51 accessory complex formed of Psy3-Csm2-Shu1-Shu2 was found to promote Rad51 filament assembly and stability [41]. Dmc1 activity, in turn, is specifically stimulated by Hop2-Mnd1 [42] and Sae3-Mei5 [43].
The involvement of these recombinases in template choice is paramount. Deletion of Rad51 (or Rad55 or Rad57) creates a strong IS bias (1:4 IH:IS at HIS4-LEU2, Figure 2A- green bars), which, importantly, also demonstrates the competency of Dmc1 for IS repair and indicates that Rad51 is required for IH bias [20]. Deletion of the non-essential accessory factors Shu1 and/or Psy3 results in an intermediate ratio of 1:2.5 IH:IS (Figure 2A- light green bars) [26, 41]. By contrast, in the absence of Dmc1, very little recombination is observed [20]. Rad51 foci are present on chromatin in dmc1 strains, but DSBs remain unrepaired, thus putting into question whether Rad51 functions as a recombinase in meiosis. In fact, meiotic recombination occurs normally with a Rad51 allele (rad51-II3A) that is solely deficient in strand exchange activity [44]. This finding suggests that the function of Rad51 in Dmc1-mediated repair is predominantly structural (Figure 2A- red and green bars), whereas this is not the case with the equivalent dmc1-II3A allele, which phenocopies the dmc1 null mutant [44]. This finding was confirmed in A. thaliana, where a similarly repair-defective Rad51 allele was sufficient to promote normal Dmc1-mediated IH recombination [45].
The failure of Rad51 to promote DSB repair in the absence of Dmc1 suggests that Rad51 recombinase activity is not active during meiosis. Indeed, Rad51 recombinase activity appears to be specifically downregulated, which occurs in part by its meiosis-specific inhibitor, Hed1. Hed1 binds to Rad51 on ssDNA filaments and prevents interaction between Rad51 and Rad54, thus inhibiting Rad51 activity [46–48]. Deletion of Hed1 in an otherwise wild-type strain causes an intermediate decrease in homologue bias (2:1 IH:IS, Figure 2A- orange bars). However, deletion of Hed1 in a dmc1 (or mnd1) strain alleviates the repair defect of these strains by allowing IS recombination to occur, thus producing a mitotic-like 1:7 IH:IS ratio (Figure 2A- green bars) [16, 26]. DSB repair can also be restored in dmc1 strains by over-expressing RAD51 or RAD54 during meiosis [49]. These observations suggest that Rad51 recombinase activity is inhibited in meiosis to prevent IS recombination.
A recent study [50] set out to challenge these interpretations, and questioned whether Rad51 downregulation was a feature of normal meiosis, or a phenomenon seen only in meiotically arrested cells lacking Dmc1. In this study, constitutive activation of Rad51 during meiosis was found to cause a modest decrease in IH recombination (Figure 2A- orange bars). This was achieved by deleting Hed1 to prevent inhibition of Rad51 and/or preventing phosphorylation of Rad54 by the Mek1 kinase. In order to avoid using a dmc1 null strain, a functionally compromised Dmc1 allele (dmc1-T159A) was used that did not trigger checkpoint-mediated arrest. When combined with a partially activated Rad51 background (hed1 or unphosphorylatable Rad54) no further IS bias was observed, however an additive effect was seen when Rad51 activity was doubly uninhibited (Figure 2A- light green bars). These data confirm that although the presence of Rad51 protein is required for effective meiotic recombination [44], Rad51 recombinase activity is detrimental to effective IH-biased repair.
The data outlined above suggest that a combined RecA filament, including Hed1-bound Rad51 and active Dmc1, is required to interpret the regulatory signals of template choice and establish IH bias (Figure 2B(i)). Dmc1 activity appears to provide the recombinase activity of this filament, as removal of Dmc1 prevents any recombination from occurring at all (Figure 2B(ii)). However, the presence of Rad51 is necessary to allow the filament to respond to signals of IH bias, as loss of Rad51 causes IS bias (Figure 2B(iii)). Conversely, Rad51 alone can also not respond to signals of IH bias, because removal of constraints of Rad51 activity in dmc1 cells, namely deletion of Hed1 or removal of the meiotic axis permits mitotic-like IS-biased repair (Figure 2B(iv-vi)). The meiotic axis is known to inhibit Rad51 activity via Mek1-mediated phosphorylation of Rad54 [39], however a broader positive effect on IH-bias appears to occur for the combined RecA filament, possibly due to the specialized chromatin structure creating a barrier to IS repair. These data support the notion that the combined RecA filament is necessary to correctly interpret the axis-mediated IS barrier.
Maintenance of IH Bias: the Role of Sister Chromatid Cohesion
Although homologue bias appears to be established at the strand-invasion step, the resulting repair intermediates must be stabilized for this bias to be reflected at the dHJ stage. Recent work indicates a role for the cohesin complex in this process. S. cerevisiae mutants lacking the meiosis-specific cohesin subunit Rec8 exhibit a normal level of homologue bias for SEI formation, but a 1:1 IH:IS dHJ ratio (Figure 2A- yellow bars), suggesting that cohesin helps maintain IH bias beyond the SEI stage. Given that cohesin/Rec8 froms part of the meiotic chromosome axis, a wild-type level IH bias at the SEI level is somewhat surprising. However, Hop1 (and presumably Red1) does not depend on Rec8 for its association in the vicinity of HIS4-LEU2 [51]. Moreover, a certain degree of spatial proximity and intertwining of sister chromatids is likely maintained in the early stages of prophase even in the absence of cohesin, thus allowing axis-dependent establishment of IH bias. At later stages of meiotic prophase, chromosomes undergo profound chromosomal movements [52], which may necessitate cohesin-dependent linkages to stabilize chromosome axes and maintain repair intermediates. The 1:1 IH:IS dHJ ratio observed in rec8 is close to the ratio expected from random repair template choice. The slight bias toward the sister (due to the presence of two homologues and one sister) may be explained by persistent proximity of sisters in the absence of cohesin. Deletion of Rec8 in axis-defective mutants also exhibited the same 1:1 IH:IS dHJ ratio during meiosis, consistent with a role of cohesin in stabilizing repair intermediates. However, intermediates are overall significantly depleted and SEI levels are not measurable in these mutants. Both phenotypes can be suppressed to a degree by meiotically expressing the cohesin Scc1/Mcd1 (Figure 2A- yellow/blue bars), Rec8’s mitotic counterpart, which suggests that it is mostly sister cohesion but also additional regulatory functions of Rec8 that affect the bias, as Rec8 is known to perform regulatory roles in meiotic recombination independently of sister chromatid cohesion [53]. The rec8 mutant exhibits variable defects in axis distribution across the genome, which suggests that different loci could resemble more or less wild-type axis conditions [51]. Collectively, these analyses suggest an antagonistic relationship between meiotic cohesin and axis proteins, whereby cohesin promotes IS recombination, but the axis counteracts this bias to a greater extent to allow IH recombination to dominate in wild-type cells (Figure 1B). In addition to effects of Mek1 kinase on combined RecA filament activity, this could be due to axis-mediated localized depletion of cohesin proteins to facilitate IH recombination [26].
Conclusion and Future Directions
The study of template choice in meiosis presents a fascinating field, with recent advances in our ability to observe recombination intermediates on an increasingly global scale occurring in parallel with elegant in vitro and in vivo investigations dissecting the mechanistic functions of the host of regulatory factors involved in meiotic recombination. Many fundamental questions still remain, particularly concerning the spatial and temporal regulation of template choice. Template choice bias across the genome in S. pombe appears to effectively compensate for local differences in DSB formation in order to maintain a constant rate of crossover formation [19]. The presence of high IH bias at DSB-hot hotspots in S. cerevisiae suggests an opposite trend in the spatial regulation of bias. Evidence of a DSB-colder hotspot in S. cerevisiae exists from quantification at a previous version of the HIS4-LEU2 hotspot, which contained 2 DSB sites and a lower overall frequency of DSB formation. This version exhibits a lower wild-type IH:IS dHJ ratio of 2.5:1, compared to 5:1 at the currently used HIS4-LEU2 hotspot. The red1 and rad51 mutants also exhibited less extreme dHJ ratios of 1:1.72 and 1:3 respectively (Figure 2A- left side), compared to the 1:9 and 1:7 IH:IS ratios obtained using the current hotspot (Figure 2A- right side). This could be due to improved methods of quantification but could also suggest a reduction in bias using the less ‘hot’ hotspot. The inter-species differences in bias could be a consequence of the higher chromosome number of 16 in S. cerevisiae, compared to 3 in S. pombe, whereby initial homologue engagement may require strong IH bias at ‘hot’ hotspots in S. cerevisiae, and a more passive approach in S. pombe can achieve the same ultimate goal of sufficient IH-crossover formation. Emergence of other hotspots suitable for analysis, and the advancement of genome-wide technologies are expected to further improve our understanding of template choice bias across the genome.
Temporal regulation of template choice is another important aspect in promoting correct crossover numbers. Although IH recombination is strongly upregulated during meiosis, there is a vast excess of DSBs compared to crossover numbers (~15 DSBs per CO in mouse, 25 in A. thaliana and 2 in S. cerevisiae [54]), which requires alternative repair pathways, including IS repair, to be employed accordingly. The wild-type 5:1 IH:IS dHJ ratio demonstrates that IS repair remains active in meiosis, and IS repair has been shown to function efficiently when a homologue is not available [55]. This suggests that in addition to promoting IH-noncrossover products, IS repair could also be used to repair excess DSBs in order to maintain correct crossover number. Indeed, a mechanism involving meiotic chromosome restructuring that appears to target DSBs formed late in meiotic prophase into the sister was recently reported for C. elegans [56]. Thus, spatial and temporal regulation of template choice bias is likely a pervasive feature of meiosis in many organisms and presents an exciting frontier in trying to integrate template bias with the meiotic program and the formation of healthy gametes.
Acknowledgments
This work was supported in part supported by NIH grant GM088248 and Research Grant 6-FY13-105 from the March of Dimes Foundation to A.H.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hassold T, Hall H, Hunt P. The origin of human aneuploidy: where we have been, where we are going. Hum Mol Genet. 2007;16(Spec No. 2):R203–208. doi: 10.1093/hmg/ddm243. [DOI] [PubMed] [Google Scholar]
- 2.Klutstein M, Cooper JP. The chromosomal courtship dance-homolog pairing in early meiosis. Curr Op Cell Biol. 2014;26:123–131. doi: 10.1016/j.ceb.2013.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim KP, Weiner BM, Zhang L, Jordan A, Dekker J, Kleckner N. Sister cohesion and structural axis components mediate homolog bias of meiotic recombination. Cell. 2010;143:924–937. doi: 10.1016/j.cell.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hunter N, Kleckner N. The single-end invasion: an asymmetric intermediate at the double-strand break to double-holliday junction transition of meiotic recombination. Cell. 2001;106:59–70. doi: 10.1016/s0092-8674(01)00430-5. [DOI] [PubMed] [Google Scholar]
- 5.Schwacha A, Kleckner N. Identification of double Holliday junctions as intermediates in meiotic recombination. Cell. 1995;83:783–791. doi: 10.1016/0092-8674(95)90191-4. [DOI] [PubMed] [Google Scholar]
- 6.Nasmyth K, Haering CH. Cohesin: its roles and mechanisms. Annu Rev Genet. 2009;43:525–558. doi: 10.1146/annurev-genet-102108-134233. [DOI] [PubMed] [Google Scholar]
- 7.Chen SY, Tsubouchi T, Rockmill B, Sandler JS, Richards DR, Vader G, Hochwagen A, Roeder GS, Fung JC. Global analysis of the meiotic crossover landscape. Dev Cell. 2008;15:401–415. doi: 10.1016/j.devcel.2008.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mancera E, Bourgon R, Brozzi A, Huber W, Steinmetz LM. High-resolution mapping of meiotic crossovers and non-crossovers in yeast. Nature. 2008;454:479–485. doi: 10.1038/nature07135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martini E, Borde V, Legendre M, Audic S, Regnault B, Soubigou G, Dujon B, Llorente B. Genome-wide analysis of heteroduplex DNA in mismatch repair-deficient yeast cells reveals novel properties of meiotic recombination pathways. PLoS Genet. 2011;7:e1002305. doi: 10.1371/journal.pgen.1002305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pan J, Sasaki M, Kniewel R, Murakami H, Blitzblau HG, Tischfield SE, Zhu X, Neale MJ, Jasin M, Socci ND, Hochwagen A, Keeney S. A hierarchical combination of factors shapes the genome-wide topography of yeast meiotic recombination initiation. Cell. 2011;144:719–731. doi: 10.1016/j.cell.2011.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blitzblau HG, Bell GW, Rodriguez J, Bell SP, Hochwagen A. Mapping of meiotic single-stranded DNA reveals double-stranded-break hotspots near centromeres and telomeres. Curr Biol. 2007;17:2003–2012. doi: 10.1016/j.cub.2007.10.066. [DOI] [PubMed] [Google Scholar]
- 12.Buhler C, Borde V, Lichten M. Mapping meiotic single-strand DNA reveals a new landscape of DNA double-strand breaks in Saccharomyces cerevisiae. PLoS Biol. 2007;5:e324. doi: 10.1371/journal.pbio.0050324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Serrentino ME, Chaplais E, Sommermeyer V, Borde V. Differential association of the conserved SUMO ligase Zip3 with meiotic double-strand break sites reveals regional variations in the outcome of meiotic recombination. PLoS Genet. 2013;9:e1003416. doi: 10.1371/journal.pgen.1003416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yokoo R, Zawadzki KA, Nabeshima K, Drake M, Arur S, Villeneuve AM. COSA-1 reveals robust homeostasis and separable licensing and reinforcement steps governing meiotic crossovers. Cell. 2012;149:75–87. doi: 10.1016/j.cell.2012.01.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bzymek M, Thayer NH, Oh SD, Kleckner N, Hunter N. Double Holliday junctions are intermediates of DNA break repair. Nature. 2010;464:937–941. doi: 10.1038/nature08868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lao JP, Cloud V, Huang CC, Grubb J, Thacker D, Lee CY, Dresser ME, Hunter N, Bishop DK. Meiotic crossover control by concerted action of Rad51-Dmc1 in homolog template bias and robust homeostatic regulation. PLoS Genet. 2013;9:e1003978. doi: 10.1371/journal.pgen.1003978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hyppa RW, Smith GR. Using Schizosaccharomyces pombe meiosis to analyze DNA recombination intermediates. Methods Mol Biol. 2009;557:235–252. doi: 10.1007/978-1-59745-527-5_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cromie GA, Hyppa RW, Taylor AF, Zakharyevich K, Hunter N, Smith GR. Single Holliday junctions are intermediates of meiotic recombination. Cell. 2006;127:1167–1178. doi: 10.1016/j.cell.2006.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hyppa RW, Smith GR. Crossover invariance determined by partner choice for meiotic DNA break repair. Cell. 2010;142:243–255. doi: 10.1016/j.cell.2010.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schwacha A, Kleckner N. Interhomolog bias during meiotic recombination: meiotic functions promote a highly differentiated interhomolog-only pathway. Cell. 1997;90:1123–1135. doi: 10.1016/s0092-8674(00)80378-5. [DOI] [PubMed] [Google Scholar]
- 21.Niu H, Wan L, Baumgartner B, Schaefer D, Loidl J, Hollingsworth NM. Partner choice during meiosis is regulated by Hop1-promoted dimerization of Mek1. Mol Biol Cell. 2005;16:5804–5818. doi: 10.1091/mbc.E05-05-0465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Latypov V, Rothenberg M, Lorenz A, Octobre G, Csutak O, Lehmann E, Loidl J, Kohli J. Roles of Hop1 and Mek1 in meiotic chromosome pairing and recombination partner choice in Schizosaccharomyces pombe. Mol Cell Biol. 2010;30:1570–1581. doi: 10.1128/MCB.00919-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sanchez-Moran E, Santos JL, Jones GH, Franklin FC. ASY1 mediates AtDMC1-dependent interhomolog recombination during meiosis in Arabidopsis. Genes Dev. 2007;21:2220–2233. doi: 10.1101/gad.439007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ferdous M, Higgins JD, Osman K, Lambing C, Roitinger E, Mechtler K, Armstrong SJ, Perry R, Pradillo M, Cunado N, Franklin FC. Inter-homolog crossing-over and synapsis in Arabidopsis meiosis are dependent on the chromosome axis protein AtASY3. PLoS Genet. 2012;8:e1002507. doi: 10.1371/journal.pgen.1002507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klein F, Mahr P, Galova M, Buonomo SB, Michaelis C, Nairz K, Nasmyth K. A central role for cohesins in sister chromatid cohesion, formation of axial elements, and recombination during yeast meiosis. Cell. 1999;98:91–103. doi: 10.1016/S0092-8674(00)80609-1. [DOI] [PubMed] [Google Scholar]
- 26.Hong S, Sung Y, Yu M, Lee M, Kleckner N, Kim KP. The logic and mechanism of homologous recombination partner choice. Mol Cell. 2013;51:440–453. doi: 10.1016/j.molcel.2013.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chung WH, Zhu Z, Papusha A, Malkova A, Ira G. Defective resection at DNA double-strand breaks leads to de novo telomere formation and enhances gene targeting. PLoS Genet. 2010;6:e1000948. doi: 10.1371/journal.pgen.1000948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Terentyev Y, Johnson R, Neale MJ, Khisroon M, Bishop-Bailey A, Goldman AS. Evidence that MEK1 positively promotes interhomologue double-strand break repair. Nucleic Acids Res. 2010;38:4349–4360. doi: 10.1093/nar/gkq137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu HY, Ho HC, Burgess SM. Mek1 kinase governs outcomes of meiotic recombination and the checkpoint response. Curr Biol. 2010;20:1707–1716. doi: 10.1016/j.cub.2010.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carballo JA, Johnson AL, Sedgwick SG, Cha RS. Phosphorylation of the axial element protein Hop1 by Mec1/Tel1 ensures meiotic interhomolog recombination. Cell. 2008;132:758–770. doi: 10.1016/j.cell.2008.01.035. [DOI] [PubMed] [Google Scholar]
- 31.Niu H, Li X, Job E, Park C, Moazed D, Gygi SP, Hollingsworth NM. Mek1 kinase is regulated to suppress double-strand break repair between sister chromatids during budding yeast meiosis. Mol Cell Biol. 2007;27:5456–5467. doi: 10.1128/MCB.00416-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wan L, de los Santos T, Zhang C, Shokat K, Hollingsworth NM. Mek1 kinase activity functions downstream of RED1 in the regulation of meiotic double strand break repair in budding yeast. Mol Biol Cell. 2004;15:11–23. doi: 10.1091/mbc.E03-07-0499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Neale MJ, Keeney S. Clarifying the mechanics of DNA strand exchange in meiotic recombination. Nature. 2006;442:153–158. doi: 10.1038/nature04885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bishop DK, Park D, Xu L, Kleckner N. DMC1: a meiosis-specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell. 1992;69:439–456. doi: 10.1016/0092-8674(92)90446-j. [DOI] [PubMed] [Google Scholar]
- 35.Shinohara M, Gasior SL, Bishop DK, Shinohara A. Tid1/Rdh54 promotes colocalization of Rad51 and Dmc1 during meiotic recombination. PNAS. 2000;97:10814–10819. doi: 10.1073/pnas.97.20.10814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kurzbauer MT, Uanschou C, Chen D, Schlogelhofer P. The recombinases DMC1 and RAD51 are functionally and spatially separated during meiosis in Arabidopsis. Plant Cell. 2012;24:2058–2070. doi: 10.1105/tpc.112.098459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Busygina V, Gaines WA, Xu Y, Kwon Y, Williams GJ, Lin SW, Chang HY, Chi P, Wang HW, Sung P. Functional attributes of the Saccharomyces cerevisiae meiotic recombinase Dmc1. DNA Repair. 2013;12:707–712. doi: 10.1016/j.dnarep.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nimonkar AV, Dombrowski CC, Siino JS, Stasiak AZ, Stasiak A, Kowalczykowski SC. Saccharomyces cerevisiae Dmc1 and Rad51 proteins preferentially function with Tid1 and Rad54 proteins, respectively, to promote DNA strand invasion during genetic recombination. J Biol Chem. 2012;287:28727–28737. doi: 10.1074/jbc.M112.373290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Niu H, Wan L, Busygina V, Kwon Y, Allen JA, Li X, Kunz RC, Kubota K, Wang B, Sung P, Shokat KM, Gygi SP, Hollingsworth NM. Regulation of meiotic recombination via Mek1-mediated Rad54 phosphorylation. Mol Cell. 2009;36:393–404. doi: 10.1016/j.molcel.2009.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu J, Renault L, Veaute X, Fabre F, Stahlberg H, Heyer WD. Rad51 paralogues Rad55-Rad57 balance the antirecombinase Srs2 in Rad51 filament formation. Nature. 2011;479:245–248. doi: 10.1038/nature10522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sasanuma H, Tawaramoto MS, Lao JP, Hosaka H, Sanda E, Suzuki M, Yamashita E, Hunter N, Shinohara M, Nakagawa A, Shinohara A. A new protein complex promoting the assembly of Rad51 filaments. Nat Commun. 2013;4:1676. doi: 10.1038/ncomms2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chan YL, Brown MS, Qin D, Handa N, Bishop DK. The 3rd exon of the budding yeast meiotic recombination gene HOP2 is required for calcium-dependent and recombinase Dmc1-specific stimulation of homologous strand assimilation. J Biol Chem. 2014 doi: 10.1074/jbc.M114.558601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ferrari SR, Grubb J, Bishop DK. The Mei5-Sae3 protein complex mediates Dmc1 activity in Saccharomyces cerevisiae. J Biol Chem. 2009;284:11766–11770. doi: 10.1074/jbc.C900023200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cloud V, Chan YL, Grubb J, Budke B, Bishop DK. Rad51 is an accessory factor for Dmc1-mediated joint molecule formation during meiosis. Science. 2012;337:1222–1225. doi: 10.1126/science.1219379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Da Ines O, Degroote F, Goubely C, Amiard S, Gallego ME, White CI. Meiotic recombination in Arabidopsis is catalysed by DMC1, with RAD51 playing a supporting role. PLoS Genet. 2013;9:e1003787. doi: 10.1371/journal.pgen.1003787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Busygina V, Saro D, Williams G, Leung WK, Say AF, Sehorn MG, Sung P, Tsubouchi H. Novel attributes of Hed1 affect dynamics and activity of the Rad51 presynaptic filament during meiotic recombination. J Biol Chem. 2012;287:1566–1575. doi: 10.1074/jbc.M111.297309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Busygina V, Sehorn MG, Shi IY, Tsubouchi H, Roeder GS, Sung P. Hed1 regulates Rad51-mediated recombination via a novel mechanism. Genes Dev. 2008;22:786–795. doi: 10.1101/gad.1638708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tsubouchi H, Roeder GS. Budding yeast Hed1 down-regulates the mitotic recombination machinery when meiotic recombination is impaired. Genes Dev. 2006;20:1766–1775. doi: 10.1101/gad.1422506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bishop DK, Nikolski Y, Oshiro J, Chon J, Shinohara M, Chen X. High copy number suppression of the meiotic arrest caused by a dmc1 mutation: REC114 imposes an early recombination block and RAD54 promotes a DMC1-independent DSB repair pathway. Genes Cells. 1999;4:425–444. doi: 10.1046/j.1365-2443.1999.00273.x. [DOI] [PubMed] [Google Scholar]
- 50.Liu Y, Gaines WA, Callender T, Busygina V, Oke A, Sung P, Fung JC, Hollingsworth NM. Down-regulation of Rad51 activity during meiosis in yeast prevents competition with Dmc1 for repair of double-strand breaks. PLoS Genet. 2014;10:e1004005. doi: 10.1371/journal.pgen.1004005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Panizza S, Mendoza MA, Berlinger M, Huang L, Nicolas A, Shirahige K, Klein F. Spo11-accessory proteins link double-strand break sites to the chromosome axis in early meiotic recombination. Cell. 2011;146:372–383. doi: 10.1016/j.cell.2011.07.003. [DOI] [PubMed] [Google Scholar]
- 52.Conrad MN, Lee CY, Chao G, Shinohara M, Kosaka H, Shinohara A, Conchello JA, Dresser ME. Rapid telomere movement in meiotic prophase is promoted by NDJ1, MPS3, and CSM4 and is modulated by recombination. Cell. 2008;133:1175–1187. doi: 10.1016/j.cell.2008.04.047. [DOI] [PubMed] [Google Scholar]
- 53.Brar GA, Hochwagen A, Ee LS, Amon A. The multiple roles of cohesin in meiotic chromosome morphogenesis and pairing. Mol Biol Cell. 2009;20:1030–1047. doi: 10.1091/mbc.E08-06-0637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Serrentino ME, Borde V. The spatial regulation of meiotic recombination hotspots: are all DSB hotspots crossover hotspots? Exp Cell Res. 2012;318:1347–1352. doi: 10.1016/j.yexcr.2012.03.025. [DOI] [PubMed] [Google Scholar]
- 55.Goldfarb T, Lichten M. Frequent and efficient use of the sister chromatid for DNA double-strand break repair during budding yeast meiosis. PLoS Biol. 2010;8:e1000520. doi: 10.1371/journal.pbio.1000520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Libuda DE, Uzawa S, Meyer BJ, Villeneuve AM. Meiotic chromosome structures constrain and respond to designation of crossover sites. Nature. 2013;502:703–706. doi: 10.1038/nature12577. [DOI] [PMC free article] [PubMed] [Google Scholar]