Abstract
Immunoregulatory functions of vitamin D have been well documented in various immunological disorders including multiple sclerosis, arthritis, and asthma. IL-10 has been considered as a chief effector molecule that promotes vitamin D-induced immunosuppressive states of T cells and accessory cells. Here we demonstrate that active from of vitamin D, 1,25-dihydroxyvitamin D3 [Calcitiriol] has a profound inhibitory effect on the development of human Th9, a CD4 T cell subset that is highly associated with asthma, in an IL-10 independent manner. Our data show that Calcitriol represses expression of BATF, a transcription factor essential for Th9, via suppressing aryl hydrocarbon receptor (AhR) expression, without an increase of IL-10. The data show a novel link between vitamin D and two key transcription factors involved in T cell differentiation.
Introduction
Vitamin D plays a significant role in immunity and its deficiency has been associated with multiple inflammatory disorders (1). An active metabolite of vitamin D, Calcitriol (2), exerts its influence through the vitamin D receptor (VDR), which regulates over 200 genes. VDR is present in circulating mononuclear cells, dendritic cells, thymus and peripheral T and B cells. Calcitriol decreases the maturation of antigen presenting cells and their ability to activate T lymphocytes (3). IL-10 has been shown to mediate Calcitriol effects on immune modulation (4). Other than IL-10, factors that are targeted by Calcitriol in primary T cells are yet to be delineated.
In this study, we determined the effect of Calcitriol on Th9 cells. Th9 cells are defined as a subset of CD4 T cells that produce IL-9, a pleiotropic cytokine which supports mast cell growth, enhances production of IgE, promotes eosinophil maturation, and stimulates mucin transcription in respiratory epithelial cells (5). Th9 cells also play pivotal roles in anti-tumor immunity (6, 7). Immunosuppressive effect of Calcitriol on mouse Th9 responses required IL-10 (8). However, the effect of Calcitriol on human Th9 cells was unknown. Here, we demonstrate that Calcitriol suppresses human Th9 differentiation in vitro in a manner independent from IL-10 and show the link between Calcitriol with two transcription factors essential for Th9, AhR and BATF.
Materials and Methods
Cell preparation and culture
Naïve CD4 T cells were sorted from PBMCs using EasySep™ Human naïve CD4+ T cell isolation kit (STEMCELL™ TECHNOLOGIES, Vancouver, BC). For cell stimulation, CytoOne24/48-well-non-treated-plates (USA SCIENTIFIC, Ocala, FL) coated with anti-CD3 (OKT3, 5μg/ml, Biolegend, San Diego, CA) and anti-CD28 (28.2, 5μg/ml, Biolegend) were used. For induction of Th9 cells, naïve CD4 T cells were stimulated in the presence of IL-4 (20ng/ml, PeproTech, Rocky Hill, NJ) and IL-1β (20ng/ml, PeproTech) for 3 days (RNA isolation and western blot) or 5 days (ELISA). Anti-IL-10R Ab (10μg/ml, Biolegend) was used for inhibition of IL-10. Calcitriol, AhR antagonist (CH223191 ) (both from Sigma, St. Louis, MO), or FICZ (Enzo Life Sciences, Farmingdale, NY) was added where indicated.
siRNA/DNA transfection
Predesigned siRNA for ahr and its negative control siRNA were obtained from Life technologies (Grand Island, NY). Human BATF cDNA (Genescript, Piscatway, NJ) was inserted to pMAX plasmid (Lonza, Allendale, NJ). Transfection was performed by electroporation using human T cell Nucleofector Kit (Lonza, Allendale, NJ).
Enzyme-linked immunosorbent assay (ELISA) and cytokine staining
Culture supernatant (day 5) was used to determine IL-9 or IL-10 concentrations using Human IL-9/IL-10 ELISA MAX set (Biolegend, San Diego, CA). For cytokine staining, cells harvested at day 5 were restimulated with 50ng/ml PMA and 1μM Ionomycin in the presence of 2μM monensin for 4 hours, then stained with anti-IL-4 and anti-IL-9 Abs (Biolegend). Data were analyzed using FlowJo software (Tree Star, Ashland, OR).
RNA isolation and Real time PCR
Total RNA was isolated using ReliaPrep™ RNA cell MiniPrep system (Promega, Madison, WI) followed by cDNA synthesis using oligo (dT) and superscriptIII First-strand synthesis system (Life technology, Grand Island, NY). PCR was performed using SsoAdvanced™ SYBR Green super mix (BioRAD, Hercules, CA). The following primers were used. IL-9 forward 5′-TCTGACAACTGCACCAGACC-3′; IL-9 reverse 5′-TTGCATGGCTGTTCACAGGA-3′; Ahr forward 5′-GACTGGACCCAAGTCCATCG-3′; Ahr reverse 5′-TTGGTTGTGATGCCAAAGGA-3′; Gapdh forward 5′-TGTTGCCATCAATGACCCCTT-3′, Batf forward 5′-AAATCGTATTGCCGCCCAGA-3′; Batf reverse 5′-TAGAGCCGCGTTCTGTTTCT-3′, Gapdh reverse 5′-CTCCACGACGTACTCAGCG-3′. The fold change was calculated based on ΔΔCt. GAPDH was used to normalize the quantity of RNA in each sample.
Western blot
Equal numbers of cells (1.0×106 cells/ 100μl) were lysed in SDS sample buffer (2% SDS, 125mM DTT, 10% glycerol, 62.5mM Tris-HCl, pH6.8) and proteins were subjected to western blot analysis using following Abs. anti-AhR Ab and anti-GATA3 Ab: Santacruz Biotechnology (Santa Cruz, CA); anti-PU.1 Ab and anti-IRF-4 Ab: Cell Signaling technology (Danvers, MA); anti-BATF Ab: Biolegend (San Diego, CA); anti-β actin Ab: Sigma-Aldrich (St. Louis, MO); anti-Erg Ab: Thermo Fisher Scientific (West Palm Beach, FL). Signals were detected with the ECL system (GE Healthcare, Piscataway, NJ). Relative intensity of each band (shown below each lane) was determined by Image J software (NIH) after normalization using β-actin or HDAC as the control.
Network analysis of transcription factors and genes highly expressed by Th9 cells
Genes specifically enriched more than twofold in Th9 cells compared to Th2 or iTreg cells (9) were analyzed using a list of transcription factors with their potential target genes based on databases and published articles (10-18). Transcription factors and their predicted target genes were combined together to create integrated transcription factors target dataset without allowing redundancy. Total of 1503 transcription factors and its 18523 putative target genes have been obtained from above data integration. The network analysis on this integrated dataset and 560 Th9 enriched genes was performed using a software package Gephi (19). Data preparation was conducted by programming language python and python package NetworkX (20). The shape of the network was determined by Force Atlas layout in Gephi.
Statistical analysis
Statistical analysis was performed using two-tailed Student T test.
Study approval
This study was approved by Loyola University Chicago IRB.
Results and Discussion
To address whether vitamin D inhibits human Th9 cell development, we cultured naïve CD4 T cells under Th9-inducing conditions in the presence or absence of Calcitriol. In a dose dependent manner, Calcitriol abrogated IL-9 protein and mRNA production by human T cells (Fig. 1A). Although a high dose of Calcitriol treatment is known to have an effect on T cell proliferation (21), we did not observe any significant inhibition of T cell growth by Calcitriol (data not shown).
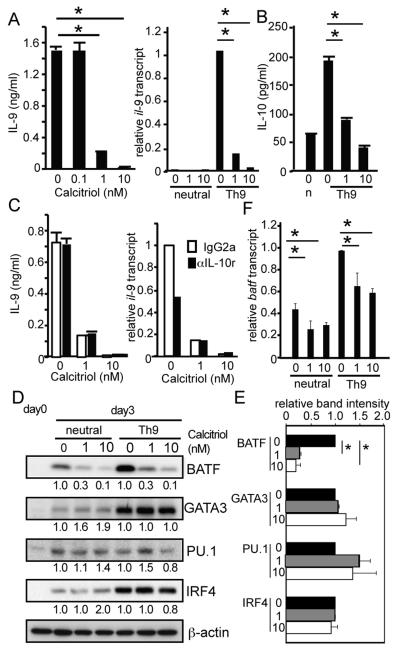
Fig. 1. Calcitriol represses BATF expression and inhibits IL-9 production.
(A) Effect of Calcitriol on IL-9 expression. Human naïve CD4 T cells from adult PBMCs were stimulated under neutral or Th9-inducing conditions in the presence or absence of Calcitriol [Concentration (nM) is shown below each lane]. (left) Secreted IL-9 concentrations determined by ELISA. Data represent one of 5 independent experiments. *p<0.001 (right) Relative il-9 mRNA level analyzed by real time PCR. Normalization was performed using gapdh transcripts as a control. Data represent one of 3 independent experiments. Error bars represent standard deviation. (B) Secreted IL-10 concentrations in the presence or absence of Calcitriol. Data represent one of 3 independent experiments. *P<0.001 (C) Effect of IL-10 inhibition on IL-9. Naïve CD4 T cells were stimulated in the presence or absence of Calcitriol, anti-IL-10R Ab (closed bars, 10μg/ml) or anti-rat IgG2a Ab (open bars, 10μg/ml). (left) IL-9 concentrations determined by ELISA. Data represent one of 3 independent experiments. (right) il-9 mRNA level determined by real time PCR. Data represent one of 2 independent experiments. (D, E, and F) Expression of transcription factors by T cells treated with Calcitriol. Cells cultured as in (A) were subjected to western blot analysis with Abs specific for proteins listed on the right. A set of data from 3 independent samples is shown. Relative intensity of each band (top band for ERG) is shown below. Data from Calcitriol untreated conditions were arbitrary set as 1. (E) shows average from three samples cultured under the Th9-polarizing conditions. *p<0.001. (F) Effect of Calcitriol on batf mRNA expression. Relative batf mRNA level analyzed by real time PCR as in (A). Data represent average of 3 donor samples.
A previous study on murine CD4 T cells showed that Calcitriol abrogated IL-9 production in an IL-10 dependent manner (8). Contrary to the mouse T cell observations, Calcitriol decreased IL-10 production by human T cells cultured under Th9 conditions (Fig. 1B). Moreover, blockade of IL-10 signaling by anti-IL-10 receptor Ab didn’t enhance IL-9 production (Fig. 1C). These data suggest that Calcitriol suppresses human Th9 differentiation in an IL-10 independent manner. The difference between human and mouse Th9 cells may be due to the species specific requirement for factors involved in differentiation. Alternatively this may be caused by the differences in culture conditions. While our study used purified T cells only, mouse T cell study (8) was performed in the presence of antigen presenting cells that could produce IL-10 in response to Calcitriol.
GATA3, PU.1, IRF4, and BATF are known transcription factors involved in Th9 differentiation (5, 8). BATF was shown to be a critical transcription factor both for mouse and human Th9 development (9). Under the Th9-inducing conditions, BATF expression was highly upregulated compared to T cells stimulated without exogenous cytokines (termed ‘neutral conditions’) (Fig. 1D, E, and F). Calcitriol significantly reduced the level of BATF protein and mRNA expression but did not cause a significant change in the expression of GATA3, IRF4, and PU.1. These finding suggested that inhibition of Th9 differentiation by Calcitriol, at least in part, is due to a reduction in BATF expression.
To decipher detailed mechanisms by which Calcitriol controls human Th9 development, we carried out transcription factor network analysis on genes highly expressed by Th9, based on previously published data (9). We focused on 560 genes expressed more than two fold in Th9 cells compared to Th2 or inducible Tregs (iTregs) (termed DEG herein) (10-13) (supplemental Table 1). Total of 1503 transcription factors and their 18523 putative target genes have been obtained from above data integration. Among the 560 Th9 DEGs, 458 genes (81.8 %) were found in the integrated transcription factor-target gene dataset, and we analyzed these 458 genes for their possible transcription factor-target relationship (Fig. 2A). The results showed that a significant majority of DEGs maybe controlled by two transcription factors, AhR and ERG. AhR is required in the differentiation pathways of various T cell subsets, including Foxp3+ regulatory T cells, IL-10 secreting Type 1 regulatory T cells (Tr1), Th17 cells, and Th22 cells (22). ERG is an ETS family transcription factor known for its role in thymocyte development but its function in effector T cell differentiation was not clearly identified (23).
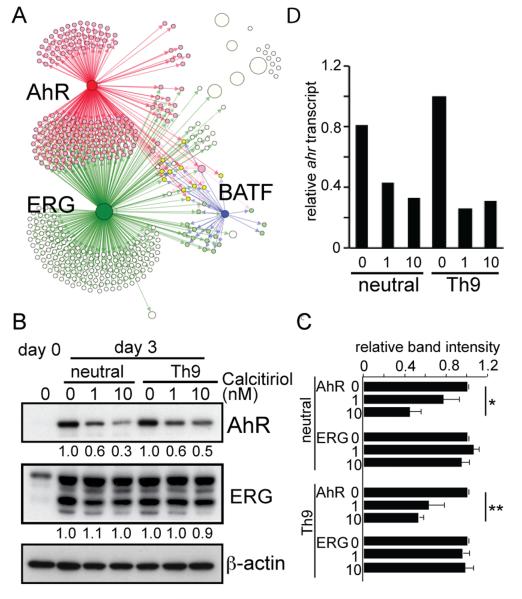
Fig. 2. AhR expression is suppressed by Calcitriol.
(A) Potential regulatory network of transcription factors and their possible targets among the Th9 enriched genes. Each node represents a Th9 enriched gene, and nodes are connected by directional edges if there are regulator-target relationships predicted. The sizes of the nodes reflect eccentricity of the genes in this network. Colored circles represent AhR(RED), and ERG(Green) related genes. (B and C) Suppression of AhR expression by Calcitriol. Naïve CD4 T cells were stimulated under neutral or Th9 inducing conditions in the presence or absence of Calcitriol. (B) Total cell lysates were subjected to western blot analysis with Abs specific for proteins listed on the right. Data represent one of 3 independent experiments. Relative intensity of each band was determined as in Fig.1 and average from 3 samples is shown in (C). * p<0.005, **p<0.001 (D) mRNA was subjected to analysis by real time PCR for ahr expression. Samples were normalized using gapdh transcripts as a control. Data represent one of 3 independent experiments.
Upon stimulation under both Th9 and neutral conditions, AhR and ERG expression was substantially increased (Fig.2B). When Calcitriol was present, AhR, but not ERG expression, was significantly reduced (Fig.2B and C). The reduction of AhR by Calcitriol was also observed at the transcriptional level under both neutral and Th9-polarizing conditions (Fig. 2D).
Suppression of AhR may be mediated by direct binding of vitamin D receptor (VDR) in the ahr gene. Indeed, we have identified at least10 potential binding sites of VDR in the ahr gene based on the consensus binding motif (24). Reduced AhR expression maybe a cause of Calcitriol-induced decrease of IL-10 since AhR is required for Tr1 differentiation (25).
Based on these data, we hypothesized that Calcitriol inhibits Th9 development by reduction of AhR expression and tested if inhibition of AhR function by an antagonist (CH223191) blocks Th9 development. When AhR antagonist was added to the culture, IL-9 production and IL-9+ cell frequency were decreased significantly (Fig. 3A and 3B). We also used siRNA-based knock down against ahr (Fig. 3C). As observed with the antagonist, siRNA against ahr markedly reduced il-9 mRNA and IL-9 protein expression (Fig. 3D). Together, the data demonstrate that AhR is essential for Th9 development.
Fig. 3. AhR is required for BATF expression and Th9 differentiation.
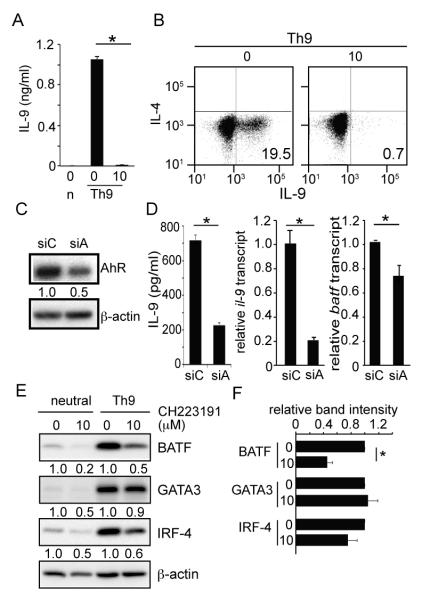
(A and B) IL-9 expression by AhR inhibited T cells. Naïve CD4 T cells were stimulated under neutral (n) or Th9 culture condition in the presence of AhR antagonist (10μM) or career control. (A) IL-9 concentrations determined by ELISA. Data represent one of 6 independent experiments. *P<0.001 (B) Cytokine staining for IL-9 and IL-4. (C and D) Expression of IL-9 or BATF by naïve CD4 T cells transfected with siRNAs against ahr. (C) shows western blot analysis of AhR protein expression by T cells transfected with siRNA against ahr (siA) or non-specific target control (siC). Data represent one of 3 independent experiments. (D) shows secreted IL-9 concentrations (left panel), relative IL-9 (middle ) or batf (right) mRNA level. Normalization of real time PCR was performed using gapdh transcripts as a control. Data represent one of 3 independent experiments. *P<0.001 (E and F) Western blot analysis of cell lysates cultured with or without AhR antagonist. Data represent one of 3 independent experiments (E). Average band intensity from 3 independent experiments under Th9 polarizing conditions is shown in (F). *p<0.001
To determine how AhR functions in Th9 development, we hypothesized that AhR promotes BATF expression and tested if inhibition of AhR signaling reduces BATF expression. Indeed, siRNA against ahr mRNA reduced batf mRNA expression (Fig.3D). Moreover, AhR antagonist caused a significant reduction of BATF protein expression by T cells cultured under Th9 conditions (Fig. 3E). These data demonstrate that AhR signaling is required for BATF expression during Th9 differentiation and that Calcitriol abrogates Th9 differentiation in part by inhibiting the AhR-BATF transcriptional network since BATF is essential for Th9 differentiation (9). .
Next we tested if Ahr is the only functional target of Calcitriol required for Th9 differentiation. If it is, then activation of AhR could restore expression of BATF and IL-9 in the presence of Calcitriol. To activate AhR, we added a ligand (FICZ) along with Calcitriol (supplemental Fig.1A and B). Under these conditions, FICZ enhanced expression of BATF and IL-9, but the restoration was not complete. The data suggest that, in addition to AhR, there are additional Th9-related molecules controlled by Calcitriol in T cells. In support of this, overexpression of BATF in the presence of AhR antagonist also did not restore IL-9 expression (supplemental Fig.1C and D).
This is the first time that Calcitriol is shown to suppress AhR or BATF expression in human T cells. AhR is required in development of multiple CD4 T cell subsets (26) and may serve as a potential molecular link between vitamin D and immune homeostasis (4). The data also reveal that perturbation of AhR inhibits BATF. Previously, the link between AhR and BATF has not been demonstrated. The data indicate the potential mechanism by which exposure to AhR ligands could increase expression of BATF and promotes effector T cell differentiation including Th9.
Since Calcitriol suppresses AhR and Th9 differentiation, vitamin D deficiency could lead to enhanced differentiation of Th9 cells and promote activation and expansion of mast cells. Th9-associated diseases like asthma maybe one of such conditions whereby vitamin D plays a protective role via reduction of Th9 cells while AhR ligand could exacerbate the disease (27). Vitamin D deficiency has also been linked to atopic dermatitis (1). Vitamin D and AhR ligands are both generated from UV light exposure and have effects on lymphoid and non-lymphoid tissues (28). A recent study showed that human Th9 cells preferentially localize in skin (29). Although the role Th9 plays in skin inflammations are not clearly defined, it is conceivable that the vitamin D and/or AhR local ligand levels would alter the differentiation process of antigen-activated T cells in the skin. In addition, AhR is also involved in differentiation of pro-inflammatory Th17 cells and anti-inflammatory Tregs and Tr1 cells (22, 25). Together, these data implicate that the balance between vitamin D and AhR ligand maybe a critical factor that determines the overall outcomes of CD4 T cell differentiations under pathophysiological conditions.
Supplementary Material
Acknowledgements
The authors thank Dr. Julie Swartzendruber for critical reading of the manuscript.
This work was supported by NIH PO1 CA154778 (M.N) and the Van Kampen Cardiopulmonary Research Fund (M.I).
Abbreviations used in this article
- AhR
aryl hydrocarbon receptor
- BATF
Basic leucine zipper transcription factor, ATF-like
- ERG
E26 transforming sequence related gene
- IRF4
interferon regulatory factor 4
References
- 1.Yin K, Agrawal DK. Vitamin D and inflammatory diseases. Journal of inflammation research. 2014;7:69–87. doi: 10.2147/JIR.S63898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Holick MF, Schnoes HK, DeLuca HF, Suda T, Cousins RJ. Isolation and identification of 1,25-dihydroxycholecalciferol. A metabolite of vitamin D active in intestine. Biochemistry. 1971;10:2799–2804. doi: 10.1021/bi00790a023. [DOI] [PubMed] [Google Scholar]
- 3.Baeke F, Takiishi T, Korf H, Gysemans C, Mathieu C. Vitamin D: modulator of the immune system. Current opinion in pharmacology. 2010;10:482–496. doi: 10.1016/j.coph.2010.04.001. [DOI] [PubMed] [Google Scholar]
- 4.Sandhu MS, Casale TB. The role of vitamin D in asthma. Annals of allergy, asthma & immunology : official publication of the American College of Allergy, Asthma, & Immunology. 2010;105:191–199. doi: 10.1016/j.anai.2010.01.013. quiz 200-192, 217. [DOI] [PubMed] [Google Scholar]
- 5.Kaplan MH. Th9 cells: differentiation and disease. Immunol Rev. 2013;252:104–115. doi: 10.1111/imr.12028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Purwar R, Schlapbach C, Xiao S, Kang HS, Elyaman W, Jiang X, Jetten AM, Khoury SJ, Fuhlbrigge RC, Kuchroo VK, Clark RA, Kupper TS. Robust tumor immunity to melanoma mediated by interleukin-9-producing T cells. Nature medicine. 2012;18:1248–1253. doi: 10.1038/nm.2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu Y, Hong S, Li H, Park J, Hong B, Wang L, Zheng Y, Liu Z, Xu J, He J, Yang J, Qian J, Yi Q. Th9 cells promote antitumor immune responses in vivo. The Journal of clinical investigation. 2012;122:4160–4171. doi: 10.1172/JCI65459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Palmer MT, Lee YK, Maynard CL, Oliver JR, Bikle DD, Jetten AM, Weaver CT. Lineage-specific effects of 1,25-dihydroxyvitamin D(3) on the development of effector CD4 T cells. J Biol Chem. 2011;286:997–1004. doi: 10.1074/jbc.M110.163790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jabeen R, Goswami R, Awe O, Kulkarni A, Nguyen ET, Attenasio A, Walsh D, Olson MR, Kim MH, Tepper RS, Sun J, Kim CH, Taparowsky EJ, Zhou B, Kaplan MH. Th9 cell development requires a BATF-regulated transcriptional network. The Journal of clinical investigation. 2013;123:4641–4653. doi: 10.1172/JCI69489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dere E, Lo R, Celius T, Matthews J, Zacharewski TR. Integration of genome-wide computation DRE search, AhR ChIP-chip and gene expression analyses of TCDD-elicited responses in the mouse liver. BMC genomics. 2011;12:365. doi: 10.1186/1471-2164-12-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zheng G, Tu K, Yang Q, Xiong Y, Wei C, Xie L, Zhu Y, Li Y. ITFP: an integrated platform of mammalian transcription factors. Bioinformatics. 2008;24:2416–2417. doi: 10.1093/bioinformatics/btn439. [DOI] [PubMed] [Google Scholar]
- 12.Wilson NK, Foster SD, Wang X, Knezevic K, Schutte J, Kaimakis P, Chilarska PM, Kinston S, Ouwehand WH, Dzierzak E, Pimanda JE, de Bruijn MF, Gottgens B. Combinatorial transcriptional control in blood stem/progenitor cells: genome-wide analysis of ten major transcriptional regulators. Cell Stem Cell. 2010;7:532–544. doi: 10.1016/j.stem.2010.07.016. [DOI] [PubMed] [Google Scholar]
- 13.Heng TS, Painter MW, C. Immunological Genome Project The Immunological Genome Project: networks of gene expression in immune cells. Nat Immunol. 2008;9:1091–1094. doi: 10.1038/ni1008-1091. [DOI] [PubMed] [Google Scholar]
- 14.Ciofani M, Madar A, Galan C, Sellars M, Mace K, Pauli F, Agarwal A, Huang W, Parkurst CN, Muratet M, Newberry KM, Meadows S, Greenfield A, Yang Y, Jain P, Kirigin FK, Birchmeier C, Wagner EF, Murphy KM, Myers RM, Bonneau R, Littman DR. A validated regulatory network for Th17 cell specification. Cell. 2012;151:289–303. doi: 10.1016/j.cell.2012.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wei G, Abraham BJ, Yagi R, Jothi R, Cui K, Sharma S, Narlikar L, Northrup DL, Tang Q, Paul WE, Zhu J, Zhao K. Genome-wide analyses of transcription factor GATA3-mediated gene regulation in distinct T cell types. Immunity. 2011;35:299–311. doi: 10.1016/j.immuni.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wei L, Vahedi G, Sun HW, Watford WT, Takatori H, Ramos HL, Takahashi H, Liang J, Gutierrez-Cruz G, Zang C, Peng W, O’Shea JJ, Kanno Y. Discrete roles of STAT4 and STAT6 transcription factors in tuning epigenetic modifications and transcription during T helper cell differentiation. Immunity. 2010;32:840–851. doi: 10.1016/j.immuni.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu JQ, Seay M, Schulz VP, Hariharan M, Tuck D, Lian J, Du J, Shi M, Ye Z, Gerstein M, Snyder MP, Weissman S. Tcf7 is an important regulator of the switch of self-renewal and differentiation in a multipotential hematopoietic cell line. PLoS genetics. 2012;8:e1002565. doi: 10.1371/journal.pgen.1002565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhu J, Jankovic D, Oler AJ, Wei G, Sharma S, Hu G, Guo L, Yagi R, Yamane H, Punkosdy G, Feigenbaum L, Zhao K, Paul WE. The transcription factor T-bet is induced by multiple pathways and prevents an endogenous Th2 cell program during Th1 cell responses. Immunity. 2012;37:660–673. doi: 10.1016/j.immuni.2012.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bastian MH, S., Jacomy M. Gephi: An Open Source Software for Exploring and Manipulating Networks. Proceedings of the Third International ICWSM Conference.2009. pp. 361–362. [Google Scholar]
- 20.Hagberg AS, D.A., Swart PJ. Exploring Network Structure, Dynamics, and Function using NetworkX. Proceedings of the 7th Python in Science Conference.2008. pp. 11–15. [Google Scholar]
- 21.Khoo AL, Joosten I, Michels M, Woestenenk R, Preijers F, He XH, Netea MG, van der Ven AJ, Koenen HJ. 1,25-Dihydroxyvitamin D3 inhibits proliferation but not the suppressive function of regulatory T cells in the absence of antigen-presenting cells. Immunology. 2011;134:459–468. doi: 10.1111/j.1365-2567.2011.03507.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Esser C, Rannug A, Stockinger B. The aryl hydrocarbon receptor in immunity. Trends Immunol. 2009;30:447–454. doi: 10.1016/j.it.2009.06.005. [DOI] [PubMed] [Google Scholar]
- 23.Loughran SJ, Kruse EA, Hacking DF, de Graaf CA, Hyland CD, Willson TA, Henley KJ, Ellis S, Voss AK, Metcalf D, Hilton DJ, Alexander WS, Kile BT. The transcription factor Erg is essential for definitive hematopoiesis and the function of adult hematopoietic stem cells. Nat Immunol. 2008;9:810–819. doi: 10.1038/ni.1617. [DOI] [PubMed] [Google Scholar]
- 24.Ramagopalan SV, Heger A, Berlanga AJ, Maugeri NJ, Lincoln MR, Burrell A, Handunnetthi L, Handel AE, Disanto G, Orton SM, Watson CT, Morahan JM, Giovannoni G, Ponting CP, Ebers GC, Knight JC. A ChIP-seq defined genome-wide map of vitamin D receptor binding: associations with disease and evolution. Genome research. 2010;20:1352–1360. doi: 10.1101/gr.107920.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mascanfroni ID, Takenaka MC, Yeste A, Patel B, Wu Y, Kenison JE, Siddiqui S, Basso AS, Otterbein LE, Pardoll DM, Pan F, Priel A, Clish CB, Robson SC, Quintana FJ. Metabolic control of type 1 regulatory T cell differentiation by AHR and HIF1-alpha. Nature medicine. 2015;21:638–646. doi: 10.1038/nm.3868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee JS, Cella M, Colonna M. AHR and the Transcriptional Regulation of Type-17/22 ILC. Frontiers in immunology. 2012;3:10. doi: 10.3389/fimmu.2012.00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chiba T, Chihara J, Furue M. Role of the Arylhydrocarbon Receptor (AhR) in the Pathology of Asthma and COPD. Journal of allergy. 2012;2012:372384. doi: 10.1155/2012/372384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Esser C, Bargen I, Weighardt H, Haarmann-Stemmann T, Krutmann J. Functions of the aryl hydrocarbon receptor in the skin. Seminars in immunopathology. 2013;35:677–691. doi: 10.1007/s00281-013-0394-4. [DOI] [PubMed] [Google Scholar]
- 29.Schlapbach C, Gehad A, Yang C, Watanabe R, Guenova E, Teague JE, Campbell L, Yawalkar N, Kupper TS, Clark RA. Human TH9 cells are skin-tropic and have autocrine and paracrine proinflammatory capacity. Science translational medicine. 2014;6:219ra218. doi: 10.1126/scitranslmed.3007828. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.