Abstract
Quantitative assays are of great importance for point-of-care (POC) diagnostics because they can offer accurate information on the analytes. However, current POC devices often require an accessory instrument to give quantitative readouts for protein biomarkers, especially for those at very low concentration levels. Here, we report a microfluidic platform, the digital volumetric bar-chart chip (DV-chip), for quantitative POC diagnostics with ultra-low detection limits that are readable with naked eyes. Requiring no calibration, the DV-chip presents a digital ink bar chart (representing multiple bits composed of 0 and 1) for the target biomarker based on direct competition between O2 generated by the experimental and control samples. The bar chart clearly and accurately defines target concentration, allowing identification of disease status. For the standard PtNP solutions, the detection limit of the platform is approximately 0.1 pM and the dynamic range covers four orders of magnitude from 0.1 to 1000 pM. CEA samples with concentrations of 1 ng/mL and 1.5 ng/mL could be differentiated on the device. We also performed ELISA assay for B-type natriuretic peptide (BNP) in 20 plasma samples from heart failure patients and the obtained on-chip data were in agreement with the clinical results. In addition, BNP was detectable at concentrations of less than 5 pM, which is three orders of magnitude lower than the detection limit of the previously reported readerless digital methods. By the integration of gas competition, volumetric bar chart, and digital readout, the DV-chip possesses merits of portability, visible readout, and ultra-low detection limit, which should offer a powerful platform for quantitative POC diagnostics in clinical settings and personalized detection.
Graphic abstract
the dv-chip presents a digital bar chart (representing multiple bits composed of 0 and 1) for the biomarker detection based on direct competition between o2 generated by the experimental and control samples. the digital readout clearly and accurately defines target concentration and further indicates the disease status.
Introduction
Accurate and rapid detection of biomarkers is extremely useful in the screening, diagnosis, or management of many diseases,1-7 such as breast cancer,8 ovarian cancer9, and heart failure (HF).10, 11 Typically, biomarkers are present at very low levels in human blood, and their concentrations change at different stages of the diseases.12, 13 For instance, B-type natriuretic peptide (BNP) is an important biomarker for the diagnosis of HF14, 15 in clinical settings, and two cutoff values in the picomolar range (i.e., 29 pM and 116 pM) are established to distinguish different stages of the disease.5, 16, 17 Quantitative monitoring of changes in levels of biomarkers like BNP facilitates the assessment of disease severity and determination of further treatment.10, 18 Many existing POC devices display quantitative results in a readerless manner, such as change in color or color intensity, 4, 19, 20 but the change in readout is discriminable only if the corresponding variations are of one order of magnitude or greater. To quantify smaller changes at low concentration (e.g., picomolar) ranges, the signal must be detected based on UV absorbance, fluorescence, chemiluminescence, electrochemistry or others.21-24 For example, Mok et al. reported a microfluidics platform for protein measurement with a detection limit of 50 pM, based on an electrical impedance sensor.25
Unlike the conventional readerless method based on color change, a digital readout shows a quantitative result as a series of positive and negative signals and is much more accurate and easily read with the naked eye.26, 27 However, very few papers reported equipment-free digital platforms for POC diagnostics because it is difficult to accurately control the transit between positive and negative. By employing a hydrophobic-to-hydrophilic reaction, Phillips' group developed a novel digital assay carried out on paper-based devices to detect hydrogen peroxide in the millimolar range.26 Another promising method reported by Ismagilov's group had the ability to quantify a 1.5-fold change in cystatin C in a digital manner by using threshold chemistry based on enzymatic inhibition.27 Still, this system can only detect changes in target concentration within the nanomolar range, and the method can potentially be interfered by confounding factors (e.g., changes in enzymatic activity in the presence of proteins in serum or competition between the inhibitor and the substrate used for the readout of enzymatic activity).27 Consequently, it remains an important challenge to develop a rapid and sensitive POC platform based on detection by naked eyes for accurate measurement of disease biomarkers, such as BNP at picomolar levels.
Here, we describe a microfluidic platform, the DV-chip, for quantitative POC diagnostics with digital readout and an ultra-low detection limit. The DV-chip utilizes platinum nanoparticles (PtNPs) as the ELISA probe to catalyze release of O2 from H2O2 and presents digital ink bar-chart readouts based on direct competition between O2 produced by control samples (representing cutoff values for disease activity levels) and experimental samples. The concentration range of the target biomarker and, therefore, the stage of the disease of interest, can be obtained by reading the array of bar charts formed by pairing of the experimental sample of unknown biomarker concentration with each of a range of controls of known concentration. The basic mechanism of the DV-chip was adapted from and improves upon our previous volumetric bar-chart chip (V-chip) studies,28, 29 which demonstrated quantitative readouts in an analog manner based on calibrations. Compared to the earlier design, the DV-chip integrates gas-based competition and both volumetric bar-chart and digital readouts in a single device; it requires no calibration, and the results are more accurate, easier to interpret, and better facilitate monitoring of disease stage via efficient measurement of biomarker levels. Accurate detection of carcinoembryonic antigen (CEA) in phosphate-buffered saline (PBS) and BNP in patient plasma clearly demonstrate the reliability and low detection limit of this microfluidics-based digital platform for POC diagnostics.
Materials and methods
Reagents and materials
Glass slides (75×50×1 mm) were obtained from Corning (Corning, NY, USA). Hydrogen peroxide solution (H2O2, 35 % w/v in H2O), silicone oil, CEA, and BNP were purchased from Sigma-Aldrich (St. Louis, MO, USA). The capture antibody and detection antibody for CEA and BNP were purchased from Abcam (Cambridge, UK).
Device operation
The DV-chip is based on slipchip technology.28, 30 The top plate and bottom plate of the device were fabricated from two 75×50×1-mm glass slides using standard photolithography and glass wet-etching methods (Figure S1, Supporting Information).28, 31 To operate the device, 5 μL silicone oil was added to the top plate with the patterns facing up, followed by assembly of the device from top and bottom plates. The two plates were slid back and forth repeatedly to evenly distribute the silicone oil, which seals the plates together and prevents solution leakage. The two plates were then aligned so that the relevant wells overlapped and formed an N-shaped fluidic path in the horizontal direction. Solutions of sample or reagent were dispensed from a 10-μL pipette tip inserted into the right inlet on the top plate. To obtain readout, the top plate was slid obliquely so that wells were connected in a Z-shaped path in the vertical direction.
Biomarker assay on the DV-chip
Assay of biomarker concentration on the DV-chip is based on sandwich ELISA. After the silane treatment (see Supporting Information for more details), the ELISA wells (the first lanes counted either from the top or the bottom end of DV-chips) on the bottom plate are modified with epoxy groups.28 Then 2 μL of the capture antibody (∼ 10 μg/mL) was added to the wells, followed by incubation overnight at 4°C. The antibodies are covalently immobilized on the well surface by the reaction between their amino groups and the epoxy groups.28 Then the wells were washed with PBS (containing 0.05% v/v Tween) four times and blocked with 5% w/v BSA for 1 h, followed by assembly of the chip from top and bottom plates. Ten microliters of control and experimental samples were loaded into the device and incubated for 1 h at room temperature. Then, 10 μL of the detection antibody-conjugated PtNP (∼ 10 μg/mL) was added and incubated for 1 h followed by washing. After loading of H2O2 and red ink, the top plate was carefully slid at an oblique angle, forming a Z-shaped fluidic path in the vertical direction and allowing reaction between the PtNP probe and H2O2. The red ink was propelled into the small channels due to the difference in quantity of oxygen gas generated by the experimental and control samples. Readout was conducted at 5 min. The concentration range of the tested sample is determined by the combination of upward and downward bars or the digital readouts.
Results and discussion
Working principle of the platform
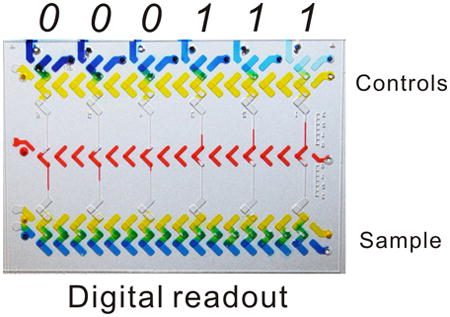
The original V-chip presents the analog readout in ink bar charts and quantitative results are obtained based on the distance traveled by the ink (i.e., the length of the bars; Figure 1a); thus, its use requires calibration before sampling. In addition, we found that the distances advanced by the ink may be hard to discriminate when two samples are as concentrated as ng/ml in concentration (e.g., 1.2 and 2.4 ng/mL CEA).28 We also reported a competitive V-chip for binary detection (positive and negative) of biomarkers that is not suitable for quantitative assay.32 In contrast, the DV-chip presents the quantitative, visually detected result in a series of downward and upward ink bars (Figure 1a). Each ink bar represents one ‘bit’, with downward bars (resulting from relatively greater O2 production by control sample)representing ‘0’ and upward bars (resulting from greater O2 production by experimental sample) representing ‘1’. Multiple bits combine to produce the digital result for samples. Such a digital readout is clear and accurate and can distinguish small differences between samples based on the transition point of ink bars. Figure 1b illustrates the principle of detection by the DV-chip using a three-plex device as an example. Biomarker detection is based on ELISA. Three control samples (C1, C2, and C3; C1<C2<C3) are loaded at the top end and the experimental sample (S) is loaded at the bottom end. PtNPs are conjugated to the biomarker detection antibody and serve as the ELISA probe because they have been reported to be efficient catalysts of O2 production from H2O233, 34 The biomarker concentration is related to the quantity of PtNPs and, therefore, to the quantity of oxygen gas produced. Readouts can be obtained based on the competition of oxygen gas generated from controls, which can be set as the biomarker cutoff values for certain disease stages, and experimental samples (Figure 1b). Because a lower concentration of a certain biomarker produces less oxygen, the unit with less concentrated sample will generate a downward ink bar (‘0’) in the corresponding channel after the gas competition. Conversely, an upward ink bar (‘1’) will be generated if the sample concentration is higher than the relevant control. Thus, two downward ink bars and one upward ink bar (‘001’) will be generated if C1<S<C2 and suggest ‘stage A’ of the disease. Otherwise, the bar charts will demonstrate a larger digital readout of ‘011’ for ‘stage B’ (if C2<S<C3) and ‘111’ for ‘stage C’ (if S>C3). Therefore, the concentration range of a biomarker and the stage of the related disease can be determined easily and accurately based on the combination of upward and downward ink bars, without any calibration. Since the concentration of the controls is prepared in advance of the assay, we can modify the concentration of each control, the dynamic coverage of all the controls, and the interval between controls. The controls are selected based on the cutoff value and dynamic range of a specific biomarker and the requirement of the resolution. In addition, the digital readouts are mainly based on the direction (up or down) of the ink bars, not the length of the ink bars (though we can take the length of the ink bars to determine the accuracy of the measurements), thus the result will be much readable.
Figure 1.
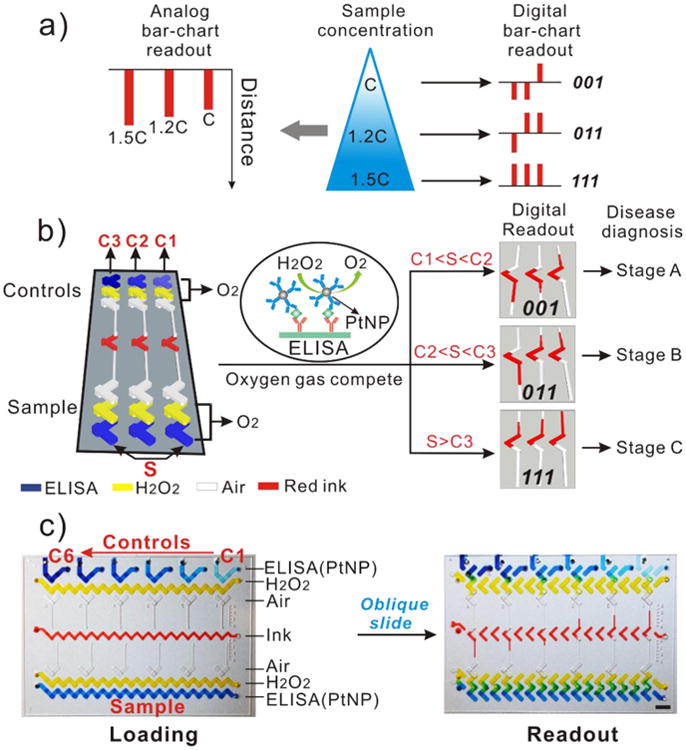
Working principle of the DV-chip. (a) Schematic diagram of analog and digital bar-chart readouts. The analog readout of the original V-chip is based on the distance travelled by the ink, whereas the digital readout of the DV-chip consists of a series of ink bars, with the downward bar representing ‘0’ and the upward bar representing ‘1’. (b) Detection mechanism of the DV-chip. Experimental samples are loaded at the bottom end of the DV-chip and controls at the top.‘S’ represents ‘sample’ and ‘C1’,‘C2’ and ‘C3’ represents ‘control 1’, ‘control 2’ and ‘control 3’, respectively. The colour rectangle boxes represent various reagents. ELISA probe PtNP catalyzes release of O2 from H2O2 at both ends of the chip. The two ends compete with each other on gas production and generate downward or upward ink bars, which together indicate the concentration of the biomarker of interest and, therefore, the stage of the disease of interest. (c) Operation of the DV-chip. In the loading phase, six control samples with known concentrations (Ci<Ci+1, 1≤i≤5) are loaded at the top end of the device. The experimental sample of unknown concentration is loaded at the bottom end. Food dyes were mixed with reagents to give a clear view. After the oblique sliding of the top plate, the wells are connected in the vertical direction to initiate the reaction and digital readout can be obtained based on the competition between O2 pressure generated by the control and experimental samples.
The fabricated DV-chips were shown in Figure 1c and Figure S2. The two plates of the chip are placed against each other and can be shifted between loading phase and reaction phase (Figure 1c). The bottom end of the device is for loading sample, and the top end is for loading six controls. Typically, ELISA takes place in the topmost and bottommost lanes, which can also be filled directly with PtNP solution as a test of O2 production33; the second lanes from the top and bottom are loaded with H2O2, which reacts with PtNP to generate O2; the third lanes are kept blank to serve as air spacers, preventing direct contact between the sample and the ink; the central lane is loaded with red ink. After oblique sliding of the top plate, the horizontally connected fluid flow paths will be separated and re-connected into independent parallel vertical units, bringing H2O2 into contact with the PtNP probe to generate oxygen gas (Figure 1b). Then, an array of ink bars will form based on the competition of oxygen gas produced at the opposite ends of each vertical unit to propel ink away from the site of the respective control or experimental reaction (Figure 1b, c).
Evaluation of the DV-chip
To validate the performance of the DV-chip, a series of evaluation experiments was conducted using PtNPs (average diameter, ∼ 56.2 nm; Figure S3, Supporting Information). First, a series of standard PtNP solutions (5, 8, 10, 20, 30, 60 pM) was loaded at the top end as controls and 3.5 pM PtNP was loaded at the bottom end (Figure 2a) as the experimental sample. Bar-chart results were recorded at different time points. The first ink bar appeared at 0.3 min in the bottom half of the unit on the leftmost side of the device, where the greatest difference in concentration of control (60 pM) and experimental samples occurred. Over a period of 4.2 min, ink bars of decreasing length formed in each vertical unit from left to right (Figure 2a, b and Figure S5). Based on these results, we maintained a consistent readout time of 5 min in the following experiments. Second, a series of standard PtNP solutions (0, 5, 10, 20, 30, 60 pM) was loaded at the top end as multiplex threshold controls, and solutions ranging from 3.5 to 35 pM were loaded at the bottom end as experimental samples (Figure 2c and Figure S6). A PtNP solution of higher concentration forms a bar chart with more upward bars and hence, a larger digital value (Figure 2c, d). Third, the device was loaded with an experimental sample (1 pM PtNP) and two controls (0.8 and 1.2 pM PtNP) to test the sensitivity of the device. The results showed that such small differences (approximately 1.2 fold) can be discerned (Figure 2e). Last, the dynamic range of the device was characterized. Figure 2f shows that ink bars could be discerned for the pair of 0.07 and 0.14 pM PtNP solutions. Hence, we determined that the detection limit of the device is approximately 0.1 pM for the standard PtNP solutions. Figure 2g shows formation of bars uniform in length for the pairs 0.07 vs. 0.14 pM and 1000 vs. 2000 pM. Actually, we also tested the concentrated PtNPs (∼ 120 nM)) from the stocked PtNP solution, the minimal resolution is around 1.2 folds for the control and experimental samples (data not shown). However, the reaction rate is too fast to show a stable result. Taking these data together with the results shown in Figure S7, we concluded that the DV-chip spans a dynamic range of four orders of magnitude from 0.1 to 1000 pM. Consequently, for a biomarker in concentrations spanning a wide dynamic range, the controls could be set to cover the entire concentration range, whereas for a biomarker requiring high-resolution detection (e.g., discrimination of two samples with concentrations of 1 pM and 1.5 pM), a small value could be chosen for the interval between the controls (e.g., C1=1.3 pM, C2=2 pM).
Figure 2.
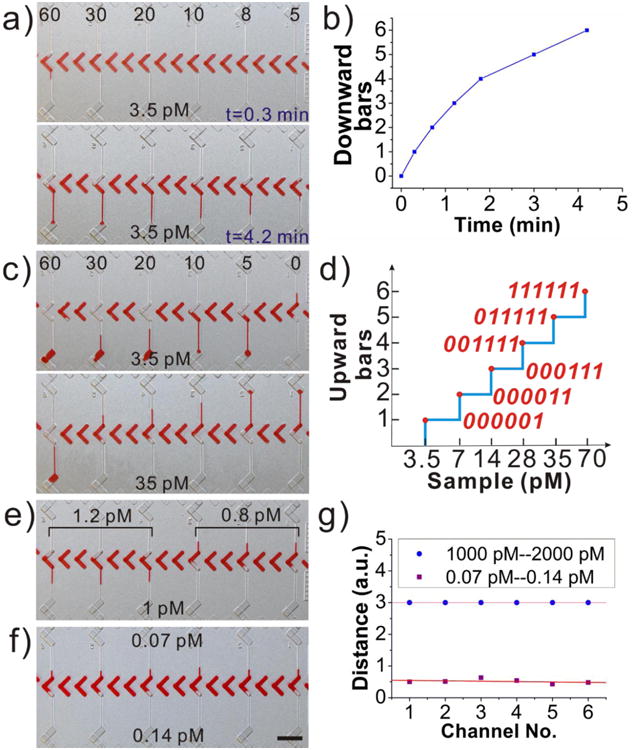
Evaluation of the DV-chip performance with standard PtNP solutions. (a) Test of bar-chart formation with time at 0.3 and 4.2 min. The concentration of the experimental sample (3.5 pM) is lower than that of all controls (5–60 pM). (b) Quantitation of downward bar formation vs. time for samples shown in (a). (c) Bar-chart formation with time for controls of 0–60 pM and experimental samples of 3.5 and 35 pM PtNPs. (d) Quantitation of the upward bar formation for experimental samples of indicated PtNP concentrations after competition with controls of 0–60 pM. Numbers in red font indicate digital results for each sample. (e) Test of the DV-chip sensitivity. (f) Test of the DV-chip detection resolution. Samples of control (upper) and experimental (lower) PtNP solutions with minimal difference in concentration still generate discernable bar charts, indicating a detection limit of ∼0.1 pM. (g) Quantitation of constant bar-chart length for samples shown in (f) and for the 1000 pM vs. 2000 pM pair. Distances traveled by ink bars exceeding the channel length were designated ‘3’. a.u., arbitrary units. Scale bar, 0.5 cm for (a), (c), (e) and (f).
Detection of carcinoembryonic antigen (CEA)
After evaluating the DV-chip, we applied the device to quantitatively detect CEA. CEA has become one of the most reliable serum biomarkers for clinical diagnosis, such as for the screening of colon cancer35, 36 or monitoring the response of breast cancer patients to therapy. 37 CEA assay is based on sandwich ELISA, as shown in Figure 3a. The wells of the first lane were coated with capture antibody after silane treatment (Supporting Information). The rough surface of the wells after wet etching increased the surface area, which contributed to the effective antibody coating (Figure S8). The assay wells were then washed with PBS and blocked with BSA. The DV-chip was then assembled and sealed with silicon oil. Subsequently, control samples of 0, 1.3, 2, 4, 6, and 10 ng/mL CEA in PBS were loaded into the assay lane at the top end. Samples of 1 ng/mL or 13 pM (sample 1) and 1.5 ng/mL or 19.5 pM (sample 2) were loaded at the bottom end and tested individually. To detect the quantity of CEA, detection antibody conjugated PtNP (Figure S4, Supporting Information) served as probe to catalyze generation of O2 from H2O2. Then red ink and H2O2 were loaded. After the initiation of the reaction by oblique sliding of the top plate, the readouts were conducted at 5 min. The digital results in Figure 3b (000001 for sample 1 and 000011 for sample 2) showed that the two samples with minimal differences could be discerned on the device. Then, sample 3 (85 ng/mL or 1105 pM) and sample 4 (450 ng/mL or 5850 pM) were tested by changing the controls to 0, 10, 50, 100, 200 and 400 ng/mL and the digital readouts of 000111 and 111111 were obtained, as shown in Figure 3b. Three parallel experiments were conducted and the quantitative plots are shown in Figure 3c. These results showed that the DV-chip has high sensitivity and wide dynamic range for assaying picomolar levels of biomarkers with visual digital readouts.
Figure 3.
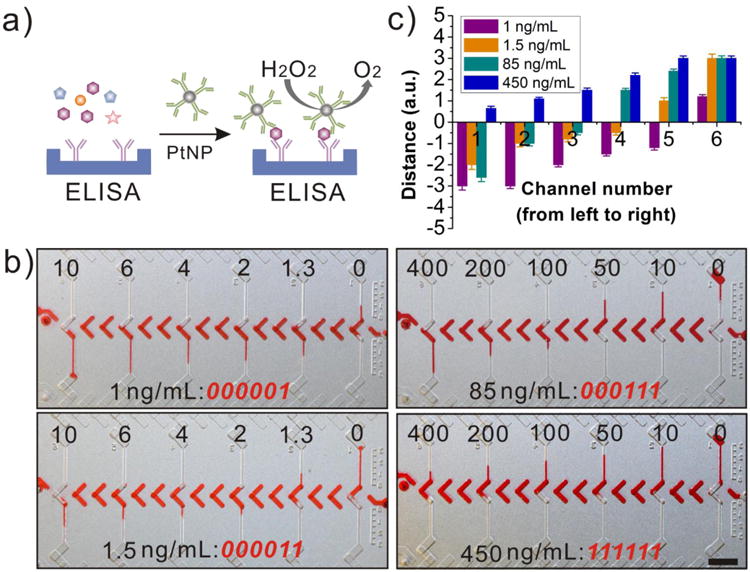
CEA assay on the DV-chip. (a) Schematic diagram of oxygen generation from the sandwiched ELISA. (b) Bar charts and corresponding digital readouts (red font) for CEA samples with indicated concentrations (lower labels). The concentrations of control CEA samples are indicated by upper labels. (c) Quantitation of bar-chart lengths for samples shown in (b). Distances travelled by ink bars exceeding the channel length were designated ‘3’. Values are the mean and standard deviation (SD) of three independent experiments. a.u., arbitrary units. Scale bar, 0.5 cm for (b).
BNP assay on the DV-chip
Furthermore, to demonstrate the ability of the DV-chip to facilitate assessment of disease status by biomarker quantification, we employed the device to analyze BNP, which has been reported to be a highly specific and sensitive marker of HF.11, 16 HF is a major cause of morbidity and mortality worldwide.38 POC detection of BNP has the potential to significantly accelerate diagnosis and decision-making for patients in the emergency department and thus, greatly increase efficiency of evaluation.39, 40
Two concentrations of BNP, 100 pg/mL (29 pM) and 400 pg/mL (116 pM), are recognized as cutoffs for different stages of HF.5, 16, 17 Samples of BNP in PBS or human serum were loaded at the bottom end. BNP solutions of 0, 50, 100, 200, 400, and 600 pg/mL were loaded at the top end and served as controls. In principle, oxygen gas produced by experimental and control samples compete to generate the bar charts (digital readout), and samples in different concentration ranges generate different bar charts, as shown in Figure 4a. Based on the digital readouts, we can easily determine the BNP concentration range in a sample and determine the stage of HF. We first tested samples of 60 and 120 pg/mL BNP in PBS (Figure S9a) and obtained the digital results 000011 and 000111, respectively. The ability to distinguish the small differences in concentration between experimental sample and control (60 vs. 50 pg/mL; 120 vs. 100 pg/mL) confirmed the high sensitivity of the assay. We further tested two BNP samples spiked in serum and compared the results with those obtained in PBS (Figure S9b). The digital readouts were the same for these two groups of samples, but the length of ink bar in each relevant channel for BNP in serum is a little shorter (within 10%) than that for BNP in PBS. We also tested samples of 250 and 500 pg/mL BNP in neat serum and the results were in accordance with the assay principles (Figure S9b). These results suggested that the complex components in human serum provide little or no interference with the performance of our device, demonstrating the stability of our method.
Figure 4.
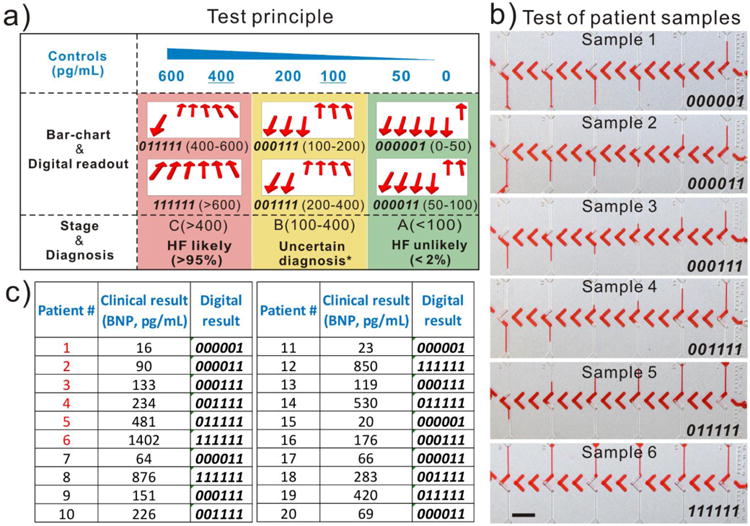
DV-chip assay of BNP in patient samples. (a) Principle of the BNP assay. Control samples of BNP of the indicated concentrations (blue) were loaded at the top; 100 and 400 pg/mL are the cutoff values for different stages of HF. Bar charts and digital readouts (middle) indicate the concentration range of BNP and thus, the stage of the HF disease (bottom). *Several other diagnostic possibilities have to be considered for these patients. [2] (b) Bar charts for patient samples 1–6 shown in red font in (c). (c) Clinical information and digital readouts for all 20 patient samples. Scale bar, 0.5 cm for (b).
In addition, we tested whether this approach performed well with patient samples. Twenty samples of plasma from HF patients were collected from the clinical department of The Houston Methodist Hospital. Control solutions of 0, 50, 100, 200, 400, and 600 pg/mL BNP were prepared in human plasma. The collected patient plasma samples were individually assayed directly (no dilution) on the DV-chip. Figure 4b shows the bar charts and digital readouts for samples 1–6, with BNP levels ranging from 16 pg/mL (∼ 4.6 pM) to 1402 pg/mL (∼ 404.7 pM). The results of clinical laboratory analysis of BNP levels (measured by Siemens Centaur Immunoassay System) and the on-chip digital readouts for the 20 patient samples are shown in Figure 4c and indicate good correlation between the two methods. Notably, the DV-chip exhibited a readable digital signal for BNP in sample 1, which is only 4.6 pM in concentration. The result demonstrates that the DV-chip possesses a detection limit of less than 5 pM for BNP in patient plasma samples, which is three orders of magnitude lower than that of the previously described readerless digital method.27 The data in Figure 4b and c also show that BNP level is correlated with clinical outcome (Table S1), proving that this biomarker is of great value for the diagnosis of HF.5, 16 In addition, the sensitivity and specificity of BNP for HF at a cutoff of 100 pg/ml was 62.5% and 83.3%, respectively; if taking 200 pg/mL as the cutoff value, the sensitivity and specificity would be 87.5% and 66.6%, respectively.
Conclusions
In summary, a digital microfluidic platform with ultra-low detection limit was developed for quantitative POC diagnostics based on detection by the naked eye. The DV-chip employs the competition between oxygen gas produced by control and experimental samples to propel red ink through channels, generating visible digital ink bar charts. The results are clear, accurate, and require no calibration for the definition of biomarker concentration and, therefore, disease state. The DV-chip platform was characterized with the standard PtNP solutions and the detection limit is approximately 0.1 pM due to the high efficiency of PtNPs in catalyzing generation of O2 from H2O2. The detection dynamic range was found to cover four orders of magnitude from 0.1 to 1000 pM. Minimal differences between CEA samples (1 ng/mL and 1.5 ng/mL) could be discerned on the device. In addition, BNP in 20 plasma samples from the heart failure patients was assayed with an ultra-low detection limit of less than 5 pM. With the successful detection of CEA and BNP, the methodology can potentially be adapted to detect any analyte with available antibodies by simple modification of the controls. Based on the advantages of low cost, ultra-low detection limit, and absence of a requirement for accessory equipment, the DV-chip could have great potential as a quantitative POC diagnostics tool.
Supplementary Material
Acknowledgments
We are grateful for the funding support from NIH/NIDA 1R01DA035868-01, the Cancer Prevention and Research Institute of Texas (CPRIT-R1007), and the Golfers Against Cancer Foundation.
Footnotes
Electronic Supplementary Information (ESI) available. See DOI: 10.1039/x0xx00000x
Contributor Information
Ping Wang, Email: pwang@houstonmethodist.org.
Lidong Qin, Email: lqin@houstonmethodist.org.
Notes and references
- 1.Giljohann DA, Mirkin CA. Nature. 2009;462:461–464. doi: 10.1038/nature08605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stoeva SI, Lee JS, Smith JE, Rosen ST, Mirkin CA. J Am Chem Soc. 2006;128:8378–8379. doi: 10.1021/ja0613106. [DOI] [PubMed] [Google Scholar]
- 3.Zhang B, Kumar RB, Dai H, Feldman BJ. Nat Med. 2014;20:948–953. doi: 10.1038/nm.3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de la Rica R, Stevens MM. Nat Nanotechnol. 2012;7:821–824. doi: 10.1038/nnano.2012.186. [DOI] [PubMed] [Google Scholar]
- 5.Chan CPY, Mak WC, Cheung KY, Sin KK, Yu CM, Rainer TH, Renneberg R. Annu Rev Anal Chem. 2013;6:191–211. doi: 10.1146/annurev-anchem-062012-092641. [DOI] [PubMed] [Google Scholar]
- 6.Richards-Kortum R, Oden M. Science. 2013;342:1055–1057. doi: 10.1126/science.1243473. [DOI] [PubMed] [Google Scholar]
- 7.Sackmann EK, Fulton AL, Beebe DJ. Nature. 2014;507:181–189. doi: 10.1038/nature13118. [DOI] [PubMed] [Google Scholar]
- 8.Rakha EA, Ellis IO. Nat Rev Clin Oncol. 2014;11:8–9. doi: 10.1038/nrclinonc.2013.230. [DOI] [PubMed] [Google Scholar]
- 9.Kulasingam V, Pavlou MP, Diamandis EP. Nat Rev Cancer. 2010;10:371–378. doi: 10.1038/nrc2831. [DOI] [PubMed] [Google Scholar]
- 10.Maisel AS, Choudhary R. Nat Rev Cardiol. 2012;9:478–490. doi: 10.1038/nrcardio.2012.60. [DOI] [PubMed] [Google Scholar]
- 11.Ahmad T, Fiuzat M, Felker GM, O'Connor C. Nat Rev Cardiol. 2012;9:347–359. doi: 10.1038/nrcardio.2012.37. [DOI] [PubMed] [Google Scholar]
- 12.Polanski M, Anderson NL. Biomarker insights. 2007;1:1–48. [PMC free article] [PubMed] [Google Scholar]
- 13.Das PM, Bast RC., Jr Biomark Med. 2008;2:291–303. doi: 10.2217/17520363.2.3.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Januzzi JL, Troughton R. Circulation. 2013;127:500–507. doi: 10.1161/CIRCULATIONAHA.112.120485. [DOI] [PubMed] [Google Scholar]
- 15.O'Donoghue M, Braunwald E. Nat Rev Cardiol. 2010;7:13–20. doi: 10.1038/nrcardio.2009.197. [DOI] [PubMed] [Google Scholar]
- 16.Maisel A. Rev Cardiovasc Med. 2002;3 Suppl 4:S10–S17. [PubMed] [Google Scholar]
- 17.Dickstein K, Cohen-Solal A, Filippatos G, McMurray JJ, Ponikowski P, Poole-Wilson PA, Stromberg A, van Veldhuisen DJ, Atar D, Hoes AW, Keren A, Mebazaa A, Nieminen M, Priori SG, Swedberg K E. S. C. C. f. P. Guidelines. Eur Heart J. 2008;29:2388–2442. doi: 10.1093/eurheartj/ehn309. [DOI] [PubMed] [Google Scholar]
- 18.Eroglu S, Bozbas H, Muderrisoglu H. Biochem Medica. 2008;18:183–192. [Google Scholar]
- 19.Hsu CK, Huang HY, Chen WR, Nishie W, Ujiie H, Natsuga K, Fan ST, Wang HK, Lee JY, Tsai WL, Shimizu H, Cheng CM. Anal Chem. 2014;86:4605–4610. doi: 10.1021/ac500835k. [DOI] [PubMed] [Google Scholar]
- 20.Pollock NR, Rolland JP, Kumar S, Beattie PD, Jain S, Noubary F, Wong VL, Pohlmann RA, Ryan US, Whitesides GM. Sci Transl Med. 2012;4:152ra12901–12152ra12910. doi: 10.1126/scitranslmed.3003981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu H, Isikman SO, Mudanyali O, Greenbaum A, Ozcan A. Lab Chip. 2013;13:51–67. doi: 10.1039/c2lc40864c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boppart SA, Richards-Kortum R. Sci Transl Med. 2014;6:253rv201–253Rrv212. doi: 10.1126/scitranslmed.3009725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sousa CP, Coutinho-Neto MD, Liberato MS, Kubota LT, Alves WA. J Phys Chem C. 2015;119:1038–1046. [Google Scholar]
- 24.Gervais L, de Rooij N, Delamarche E. Adv Mater. 2011;23:H151–H176. doi: 10.1002/adma.201100464. [DOI] [PubMed] [Google Scholar]
- 25.Mok J, Mindrinos MN, Davis RW, Javanmard M. Proc Natl Acad Sci USA. 2014;111:2110–2115. doi: 10.1073/pnas.1323998111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lewis GG, DiTucci MJ, Phillips ST. Angew Chem Int Ed. 2012;51:12707–12710. doi: 10.1002/anie.201207239. [DOI] [PubMed] [Google Scholar]
- 27.Huynh T, Sun B, Li L, Nichols KP, Koyner JL, Ismagilov RF. J Am Chem Soc. 2013;135:14775–14783. doi: 10.1021/ja4062882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Song Y, Zhang Y, Bernard PE, Reuben JM, Ueno NT, Arlinghaus RB, Zu Y, Qin L. Nat Commun. 2012;3:1283–1291. doi: 10.1038/ncomms2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Song YJ, Wang YC, Qin LD. J Am Chem Soc. 2013;135:16785–16788. doi: 10.1021/ja4085397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Du W, Li L, Nichols KP, Ismagilov RF. Lab Chip. 2009;9:2286–2292. doi: 10.1039/b908978k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wheeler AR, Chah S, Whelan RJ, Zare RN. Sensors and Actuators B: Chemical. 2004;98:208–214. [Google Scholar]
- 32.Li Y, Xuan J, Xia T, Han X, Song Y, Cao Z, Jiang X, Guo Y, Wang P, Qin L. Anal Chem. 2015;87:3771–3777. doi: 10.1021/ac504301y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Song Y, Xia X, Wu X, Wang P, Qin L. Angew Chem Int Ed. 2014;53:1–6. doi: 10.1002/anie.201404349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhu Z, Guan Z, Jia S, Lei Z, Lin S, Zhang H, Ma Y, Tian ZQ, Yang CJ. Angew Chem Int Ed. 2014;53:12503–12507. doi: 10.1002/anie.201405995. [DOI] [PubMed] [Google Scholar]
- 35.Shibutani M, Maeda K, Nagahara H, Ohtani H, Sakurai K, Toyokawa T, Kubo N, Tanaka H, Muguruma K, Ohira M, Hirakawa K. Anticancer Res. 2014;34:3753–3758. [PubMed] [Google Scholar]
- 36.Fakih MG, Padmanabhan A. Oncology-Ny. 2006;20:579–587. [PubMed] [Google Scholar]
- 37.Duffy MJ. Clin Chem. 2006;52:345–351. doi: 10.1373/clinchem.2005.059832. [DOI] [PubMed] [Google Scholar]
- 38.Go AS, Mozaffarian D, Roger VL, Benjamin EJ, Berry JD, Borden WB, Bravata DM, Dai SF, Ford ES, Fox CS, Franco S, Fullerton HJ, Gillespie C, Hailpern SM, Heit JA, Howard VJ, Huffman MD, Kissela BM, Kittner SJ, Lackland DT, Lichtman JH, Lisabeth LD, Magid D, Marcus GM, Marelli A, Matchar DB, McGuire DK, Mohler ER, Moy CS, Mussolino ME, Nichol G, Paynter NP, Schreiner PJ, Sorlie PD, Stein J, Turan TN, Virani SS, Wong ND, Woo D, Turner MB, Assoc AH, Subcomm SS. Circulation. 2013;127:E6–E245. doi: 10.1161/CIR.0b013e31828124ad. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hill SA, Booth RA, Santaguida PL, Don-Wauchope A, Brown JA, Oremus M, Ali U, Bustamam A, Sohel N, McKelvie R, Balion C, Raina P. Heart Fail Rev. 2014;19:421–438. doi: 10.1007/s10741-014-9447-6. [DOI] [PubMed] [Google Scholar]
- 40.Schwam E. Acad Emerg Med. 2004;11:686–691. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


