Abstract
Background
Immunohistochemical staining has been widely used in distinguishing lung adenocarcinoma (LUAD) from lung squamous cell carcinoma (LUSC), which is of vital importance for the diagnosis and treatment of lung cancer. Due to the lack of a comprehensive analysis of different lung cancer subtypes, there may still be undiscovered markers with higher diagnostic accuracy.
Methods
Herein first, we systematically analyzed high-throughput data obtained from The Cancer Genome Atlas (TCGA) database. Combining differently expressed gene screening and receiver operating characteristic (ROC) curve analysis, we attempted to identify the genes which might be suitable as immunohistochemical markers in distinguishing LUAD from LUSC. Then we detected the expression of six of these genes (MLPH, TMC5, SFTA3, DSG3, DSC3 and CALML3) in lung cancer sections using immunohistochemical staining.
Results
A number of genes were identified as candidate immunohistochemical markers with high sensitivity and specificity in distinguishing LUAD from LUSC. Then the staining results confirmed the potentials of the six genes (MLPH, TMC5, SFTA3, DSG3, DSC3 and CALML3) in distinguishing LUAD from LUSC, and their sensitivity and specificity were not less than many commonly used markers.
Conclusions
The results revealed that the six genes (MLPH, TMC5, SFTA3, DSG3, DSC3 and CALML3) might be suitable markers in distinguishing LUAD from LUSC, and also validated the feasibility of our methods for identification of candidate markers from high-throughput data.
Keywords: Lung cancer, immunohistochemical marker, receiver operating characteristic (ROC) curve analysis, The Cancer Genome Atlas (TCGA)
Introduction
As the most frequently diagnosed cancer and the leading cause of tumor death, lung cancer was estimated to account for more than 1.8 million new cases and nearly 1.6 million deaths worldwide in 2012, with a sharp rising from 2008 (1,2). Lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC) are the two major pathologic subtypes of lung cancer, constituting the vast majority of diagnosed lung cancers, but there are a lot of differences in their molecular profiling and characteristics, as well as therapeutic methods (3-5). Therefore, to accurately distinguish these two subtypes is important for the diagnosis and treatment of lung cancer.
Recently the main method used to distinguish LUAD and LUSC is hematoxylin-eosin (HE) staining of the tumor tissue sections observed under a light microscope. But in tumors with unclear structures caused by low differentiation, necrosis, or serious extrusion, small biopsies or cytologies with a limited number of tumor cells, it is difficult to make a precise diagnosis relying on HE staining alone. At this time, combining immunohistochemical results can refine the diagnosis, thus immunohistochemical staining is now recommended and widely applied in clinical practices (4-6).
At present, there are a number of reliable immunohistochemical markers that have been adopted to distinguish LUAD from LUSC, including thyroid transcription factor-1 (TTF-1, also called NKX2-1), napsin-A (NAPSA), tumor protein p63 (TP63), and cytokeratin (CK) 5/6 (3-5,7-10). These markers are highly sensitive, specific, and can be easily detected, the expression is significantly different between LUAD and LUSC. However, due to the lack of a comprehensive analysis of different lung cancer subtypes, there may still be undiscovered markers with higher sensitivity, specificity and application value. In the current study, we systematically analyzed high-throughput data obtained from The Cancer Genome Atlas (TCGA) database. Combining differently expressed gene screening and receiver operating characteristic (ROC) curve analysis, we identified and validated a number of genes which can be used as candidate immunohistochemical markers in distinguishing LUAD from LUSC.
Materials and methods
Ethics statement
This study was approved by the Ethics Committee of Zhongshan Hospital, Fudan University, Shanghai, China (Approval No. 2014-101). All work conformed to the provisions of the Declaration of Helsinki. Written informed consent was obtained from all patients participating in this research at the time of hospitalization.
Data acquisition and differently expressed gene screening
Level 3 RNA sequencing (RNA-Seq) V2 data of human LUAD and LUSC samples, which was released by TCGA before April 15, 2014, were obtained from the TCGA data portal (https://tcga-data.nci.nih.gov/tcga/tcgaHome2.jsp), including 490 LUAD samples and 490 LUSC samples. RNA-Seq by expectation maximization (RSEM) values were used to represent the levels of expression of these genes. The data are presented as means and standard deviations (SD).
All genes recorded in the TCGA data were filtered using the following criteria:
mean (LUAD) ≥1,000 and mean (LUAD)/mean (LUSC) ≥4;
mean (LUSC) ≥1,000 and mean (LUSC)/mean (LUAD) ≥4.
Here, mean (LUAD) and mean (LUSC) denote the mean of the RSEM value of the gene in the LUAD and LUSC samples, respectively. When a gene met one of the two conditions above, it was then entered in the subsequent analyses. Through these criteria, we attempted to identify those genes which were highly elevated and could be easily detected, with tremendous differences between the LUAD and LUSC samples.
Patient selection
Fifty patients with LUAD who underwent curative surgery between Jan 1 and Feb 19, 2014, and 50 other patients with LUSC who underwent curative surgery between Jan 1 and Apr 25, 2014, in the Department of Thoracic Surgery, Zhongshan Hospital, Fudan University, were included in this research. All of the cases were clearly confirmed by pathologic evaluation. Immunohistochemistry results of TTF1, CK7, NAPSA, surfactant protein A (SPA), TP63, HCK proto-oncogene, Src family tyrosine kinase (HCK) and P40 in the specimens were obtained from the pathologists’ original reports. Sections of paraffinembedded tumor tissues were obtained from all cases involved.
Immunohistochemistry
Immunohistochemical staining was performed using an EnVisionTM HRP-polymer anti-mouse/rabbit IHC Kit (KeyGEN BioTECH, Nanjing, Jiangsu, China) according to the manufacturer’s guidelines. Briefly, the primary antibodies specific for melanophilin (MLPH, 1:100 dilution), transmembrane channel-like 5 (TMC5, 1:100 dilution), surfactant associated 3 (SFTA3, 1:100 dilution), desmoglein 3 (DSG3, 1:100 dilution), desmocollin 3 (DSC3, 1:100 dilution) and calmodulin-like 3 (CALML3, 1:100 dilution) were applied to detect the expressions of these genes. Stained specimens were then viewed independently at 100× independently by two investigators. Expression of these genes was determined by semiquantitatively assessing the percentage of marked tumor cells and the staining intensity as previously reported (11,12). Finally, we separated the specimens according to expression in four groups (negative, weak, moderate, and strong).
The primary antibodies [anti-MLPH (HPA014685), anti-TMC5 (HPA042037), anti-SFTA3 (HPA059427), anti-DSC3 (HPA049265) and anti-CALML3 (HPA044999)] were obtained from Sigma-Aldrich (St. Louis, MO, USA). Anti-DSG3 (ab183743) was obtained from Abcam (Cambridge, MA, USA).
Statistical analysis
Data were analyzed using IBM SPSS for Windows, version 20 (Armonk, NY, USA). ROC curve analysis was used to identify the candidate genes for distinguishing LUAD from LUSC. The Mann-Whitney U test was used to evaluate the differences in genes and markers between LUAD and LUSC samples.
Results
After differently expressed gene screening, 228 genes were filtered out for the next analysis. One hundred and ten genes were elevated in LUAD compared with LUSC, the other 118 genes were upregulated in LUSC (Tables S1 and S2).
Then, ROC curve analysis was used to evaluate the effectiveness of these 228 genes when applied to distinguish LUAD from LUSC based on the TCGA data (Tables S1 and S2). Part of the genes with the highest area under curve (AUC) values in LUAD and LUSC can be found in Tables 1 and 2, respectively. The higher AUC value is indicative of greater sensitivity and specificity. MLPH, SFTA2, TMC5, SFTA3, DSG3, KRT5, DSC3 and CALML3 rank highest in these two tables.
Table 1. Fifteen genes greatly elevated in LUAD with highest AUC values.
| Gene | LUAD | LUSC | Fold-change (LUAD/LUSC) | AUC value |
|---|---|---|---|---|
| MLPH | 3,961±3,315 | 521±769 | 7.60 | 0.953 |
| SFTA2 | 2,833±3,115 | 161±327 | 17.59 | 0.946 |
| TMC5 | 3,045±2,381 | 428±646 | 7.11 | 0.943 |
| SFTA3 | 3,073±2,704 | 271±761 | 11.33 | 0.937 |
| DDAH1 | 2,446±1,405 | 544±462 | 4.50 | 0.934 |
| RORC | 1,213±952 | 130±232 | 9.31 | 0.933 |
| TMEM125 | 1,873±1,362 | 297±351 | 6.29 | 0.931 |
| SMPDL3B | 1,482±1,421 | 238±284 | 6.22 | 0.930 |
| ALDH3B1 | 2,509±2,619 | 378±646 | 6.62 | 0.930 |
| ACSL5 | 4,050±3,178 | 604±775 | 6.70 | 0.926 |
| NKX2-1 | 3,246±2,233 | 309±940 | 10.50 | 0.926 |
| ATP11A | 7,025±5,571 | 1,356±1,261 | 5.18 | 0.924 |
| CGN | 3,626±2,448 | 796±777 | 4.55 | 0.922 |
| FMO5 | 1,174±1,575 | 86±136 | 13.51 | 0.921 |
| MUC1 | 22,301±16,816 | 3,137±3,945 | 7.11 | 0.921 |
LUAD, lung adenocarcinoma; AUC: area under curve; LUSC: lung squamous cell carcinoma.
Table 2. Fifteen genes greatly elevated in LUSC with highest AUC values.
| Gene | LUAD | LUSC | Fold-change (LUSC/LUAD) | AUC value |
|---|---|---|---|---|
| DSG3 | 88±777 | 8,728±8,556 | 98.77 | 0.973 |
| KRT5 | 1,227±10,342 | 116,689±96,742 | 95.03 | 0.972 |
| DSC3 | 128±789 | 7,515±6,291 | 58.62 | 0.970 |
| CALML3 | 141±1,096 | 10,039±11,031 | 71.17 | 0.964 |
| SERPINB13 | 22±191 | 2,166±3,217 | 95.70 | 0.956 |
| KRT6B | 310±1,208 | 17,808±27,334 | 57.45 | 0.954 |
| KRT6C | 136±529 | 7,372±12,063 | 54.13 | 0.954 |
| KRT6A | 2,297±8,724 | 87,096±81,359 | 37.91 | 0.951 |
| PVRL1 | 1,204±1,177 | 11,200±7,063 | 9.30 | 0.950 |
| LOC642587 | 59±213 | 1,247±1,247 | 20.99 | 0.949 |
| PERP | 6,258±4,951 | 31,500±21,939 | 5.03 | 0.947 |
| TP63 | 325±914 | 10,976±9,139 | 33.72 | 0.946 |
| TRIM29 | 861±1,930 | 11,291±7,291 | 13.10 | 0.945 |
| ATP1B3 | 1,866±1,138 | 9,231±6,592 | 4.94 | 0.945 |
| FAT2 | 125±383 | 3,737±3,587 | 29.82 | 0.943 |
LUSC: lung squamous cell carcinoma; AUC: area under curve; LUAD, lung adenocarcinoma.
Because the appropriate primary antibody of human SFTA2 could not be obtained when we performed this study, and KRT5 is one part of CK5/6 which has been frequently used to distinguish the subtypes of lung cancer, we selected MLPH, TMC5, SFTA3, DSG3, DSC3, and CALML3 for the next immunohistochemical staining. As Figure 1 and Figure 2 show, the expression distribution profiles of these six genes were quite different in LUAD and LUSC, and the sensitivity and specificity for distinguishing between the two types of lung cancer was high.
Figure 1.
The distribution of expression of the six genes in LUAD and LUSC. LUAD, lung adenocarcinoma; LUSC: lung squamous cell carcinoma.
Figure 2.
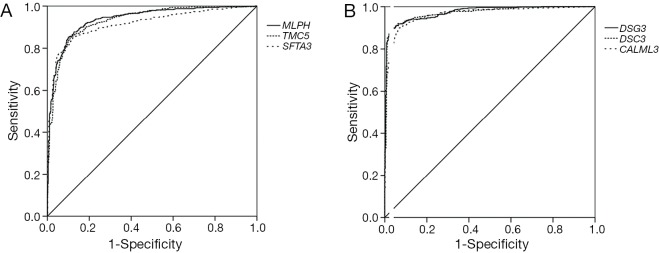
The ROC curves of the six genes when they were used in distinguishing LUAD from LUSC. (A) The ROC curves of MLPH, TMC5, and SFTA3; (B) the ROC curves of DSG3, DSC3, and CALML3. ROC, receiver operating characteristic; LUAD, lung adenocarcinoma; LUSC: lung squamous cell carcinoma.
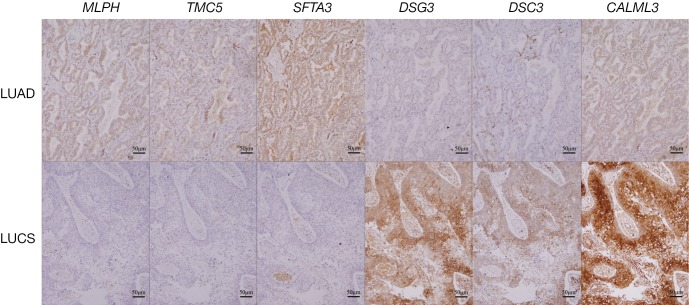
As Figure 3 and Table 3 show, the results of immunohistochemical staining further confirmed the elevation of MLPH, TMC5, and SFTA3 in LUAD, and DSG3, DSC3, and CALML3 in LUSC. Then the immunohistochemical results were compared to the markers used in our hospital clinic; the staining scores were obtained from the pathologists’ original reports. As Table 3 shows, the sensitivity and specificity of the six genes could be more than 80% and higher than some markers frequently used.
Figure 3.
The immunohistochemical staining results of the six genes in LUAD and LUSC. Scale bar: 50 µm. LUAD, lung adenocarcinoma; LUSC: lung squamous cell carcinoma.
Table 3. The immunohistochemical staining results.
| Gene and markers | LUAD |
LUSC |
P value | Threshold (LUAD/LUSC) |
Sensitivity (%) | Specificity (%) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Negative | Weak | Moderate | Strong | Negative | Weak | Moderate | Strong | ||||||
| LUAD | |||||||||||||
| MLPH | 1 | 20 | 23 | 6 | 44 | 5 | 1 | 0 | <0.001 | weak/negative | 98 | 88 | |
| TMC5 | 2 | 17 | 31 | 0 | 43 | 7 | 0 | 0 | <0.001 | weak/negative | 96 | 86 | |
| SFTA3 | 0 | 6 | 39 | 5 | 38 | 12 | 0 | 0 | <0.001 | weak/negative | 88 | 100 | |
| TTF1 | 0 | 24 | 21 | 5 | 44 | 6 | 0 | 0 | <0.001 | weak/negative | 100 | 88 | |
| CK7 | 0 | 11 | 28 | 11 | 42 | 5 | 3 | 0 | <0.001 | weak/negative | 100 | 84 | |
| NAPSA | 3 | 39 | 5 | 3 | 47 | 3 | 0 | 0 | <0.001 | weak/negative | 94 | 94 | |
| SPA | 24 | 26 | 0 | 0 | 47 | 3 | 0 | 0 | <0.001 | weak/negative | 52 | 94 | |
| LUSC | |||||||||||||
| DSG3 | 40 | 10 | 0 | 0 | 5 | 11 | 29 | 5 | <0.001 | negative/weak | 90 | 98 | |
| DSC3 | 35 | 12 | 3 | 0 | 5 | 9 | 24 | 12 | <0.001 | negative/weak | 90 | 97 | |
| CALML3 | 38 | 11 | 1 | 0 | 0 | 5 | 17 | 28 | <0.001 | weak/moderate | 90 | 98 | |
| TP63 | 41 | 9 | 0 | 0 | 3 | 24 | 20 | 3 | <0.001 | negative/weak | 94 | 86 | |
| HCK | 3 | 37 | 10 | 0 | 0 | 7 | 13 | 30 | <0.001 | weak/moderate | 86 | 80 | |
| P40 | 50 | 0 | 0 | 0 | 17 | 33 | 0 | 0 | <0.001 | negative/weak | 66 | 100 | |
The staining scores of TTF1, CK7, NAPSA, SPA, TP63, HCK and P40 were obtained from the pathologists’ original reports. The threshold indicates the criteria to distinguish LUAD from LUSC when the sum of the sensitivity and specificity reaches a peak. e.g., “weak/negative” means if the sample’s staining score ranks from weak to strong it will be identified as LUAD, and negative as LUSC. LUAD, lung adenocarcinoma; LUSC: lung squamous cell carcinoma.
Discussion
Combining differently expressed gene screening and ROC curve analysis, we identified the differently expressed genes with the highest AUC values based on TCGA data, which might be suitable to be applied as markers in distinguishing LUAD from LUSC. To validate our analyses, the expression of six candidate genes was detected in lung cancer samples by immunohistochemical staining. The staining results confirmed the potentials of these six genes in distinguishing LUAD from LUSC, and also validated the feasibility of our methods for identification of candidate markers from high-throughput data.
Our analyses revealed that the expression distribution profiles of MLPH, TMC5, SFTA3, DSG3, DSC3, and CALML3 were markedly different between LUAD and LUSC, and their sensitivity and specificity were not less than many commonly used markers. And we believed that the sensitivity and specificity would be improved after wide use in clinical practices. DSG3 and DSC3 are both transmembrane glycoproteins that belong to calcium-dependent cell adhesion molecules, and their diagnostic values in distinguishing LUSC from LUSC have been frequently reported (13-18). DSG3 and DSC3 are also greatly elevated in other squamous tumors and reduced in many other adenocarcinomas (19-21). The downregulation of DSG3 and DSC3 is in part due to DNA methylation and associated with poor prognosis in tumors (13,15,22-24). Although our results showed the potential diagnostic abilities of MLPH, TMC5, SFTA3, and CALML3, their expressions and functions in lung cancer have received little attention and remain unclear.
Most of the genes recommended as markers in distinguishing LUAD from LUSC also ranked tops in our tables according to the order of the AUC values, such as TTF-1 (NKX2-1), NAPSA, TP63 and S100 calcium binding protein A7 (S100A7) (Tables 1, 2, S1, and S2) (4-6). Another commonly used marker, CK5/6, detects the proteins coded by keratin (KRT) 5, KRT6A, and KRT6B, all three genes ranked high in Table 2 (4-6). Many other genes ranked high in our tables such as mucin 1 (MUC1), carcinoembryonic antigen-related cell adhesion molecule 6 (CEACAM6), tripartite motif containing 29 (TRIM29) and S100 calcium binding protein A2 (S100A2), were also reported that they could be used in distinguishing LUAD from LUSC (17,25,26).
With the rapid development of microarrays and RNA-Seq in recent years, more and more high-throughput data have been accumulated. How to effectively identify suitable biomarkers from these data for disease diagnosis and sub-classification is now receiving a lot of attention. Therefore, we hope our method to investigate candidate markers by combing differently expressed gene screening and ROC curve analysis, will be widely applied and further improved in the future.
Acknowledgements
The results published here are based upon data generated by the TCGA Research Network (http://cancergenome.nih.gov/).
Funding: This analysis is supported by the National Natural Science Foundation of China (Grant Nos. 81401875, 81472225) (http://www.nsfc.gov.cn/) and the Natural Science Foundation of Shanghai, China (Grant No. 14ZR1406000) (http://www.stcsm.gov.cn/).
Supplementary
Table S1. The ROC curve analyze results of genes greatly elevated in LUAD.
| Gene | LUAD | LUSC | Fold-change (LUAD/LUSC) | AUC value |
|---|---|---|---|---|
| MLPH | 3,961±3,315 | 521±769 | 7.60 | 0.953 |
| SFTA2 | 2,833±3,115 | 161±327 | 17.59 | 0.946 |
| TMC5 | 3,045±2,381 | 428±646 | 7.11 | 0.943 |
| SFTA3 | 3,073±2,704 | 271±761 | 11.33 | 0.937 |
| DDAH1 | 2,446±1,405 | 544±462 | 4.50 | 0.934 |
| RORC | 1,213±952 | 130±232 | 9.31 | 0.933 |
| TMEM125 | 1,873±1,362 | 297±351 | 6.29 | 0.931 |
| SMPDL3B | 1,482±1,421 | 238±284 | 6.22 | 0.930 |
| ALDH3B1 | 2,509±2,619 | 378±646 | 6.62 | 0.930 |
| ACSL5 | 4,050±3,178 | 604±775 | 6.70 | 0.926 |
| NKX2-1 | 3,246±2,233 | 309±940 | 10.50 | 0.926 |
| ATP11A | 7,025±5,571 | 1,356±1,261 | 5.18 | 0.924 |
| CGN | 3,626±2,448 | 796±777 | 4.55 | 0.922 |
| FMO5 | 1,174±1,575 | 86±136 | 13.51 | 0.921 |
| MUC1 | 22,301±16,816 | 3,137±3,945 | 7.11 | 0.921 |
| KCNK5 | 1,458±1,260 | 212±262 | 6.86 | 0.921 |
| PRR15L | 1,306±1,207 | 187±334 | 6.96 | 0.915 |
| SLC44A4 | 2,905±2,552 | 387±636 | 7.50 | 0.907 |
| CLDN3 | 2,127±2,016 | 356±930 | 5.97 | 0.907 |
| ST3GAL5 | 1,751±1,535 | 318±304 | 5.49 | 0.906 |
| CD55 | 9,112±9,307 | 2,068±2,001 | 4.41 | 0.898 |
| LPCAT1 | 17,427±17,015 | 3,703±5,206 | 4.71 | 0.895 |
| CEACAM6 | 41,068±39,526 | 4,992±11,717 | 8.23 | 0.889 |
| SELENBP1 | 4,213±4,536 | 697±820 | 6.04 | 0.889 |
| GPR116 | 5,436±5,921 | 842±1,175 | 6.46 | 0.887 |
| SLC34A2 | 42,409±40,305 | 5,358±10,219 | 7.91 | 0.886 |
| HPN | 1,351±1,788 | 219±406 | 6.16 | 0.885 |
| TESC | 1,759±3,143 | 126±754 | 13.92 | 0.882 |
| PLEKHA6 | 1,199±943 | 269±402 | 4.45 | 0.882 |
| FOLR1 | 3,586±4,963 | 305±641 | 11.76 | 0.881 |
| NAPSA | 35,629±37,838 | 3,240±6,098 | 11.00 | 0.879 |
| LMO3 | 2,516±2,520 | 318±722 | 7.91 | 0.878 |
| STEAP4 | 4,339±4,707 | 753±1,528 | 5.76 | 0.877 |
| B3GNT7 | 2,440±3,524 | 421±761 | 5.79 | 0.875 |
| VSTM2L | 1,714±2,342 | 213±496 | 8.03 | 0.874 |
| MUC21 | 2,461±4,873 | 103±613 | 23.87 | 0.873 |
| RHOBTB2 | 3,058±3,121 | 731±806 | 4.18 | 0.873 |
| DPP4 | 3,010±3,391 | 389±1,004 | 7.74 | 0.872 |
| MACC1 | 1,519±1,287 | 369±402 | 4.12 | 0.872 |
| ABCC3 | 5,208±3,908 | 1,169±1,428 | 4.45 | 0.869 |
| FGL1 | 1,227±4,239 | 50±553 | 24.17 | 0.868 |
| SPINK1 | 3,748±10,070 | 134±1,321 | 27.86 | 0.868 |
| C16orf89 | 5,412±8,524 | 326±626 | 16.60 | 0.866 |
| ATP8A1 | 1,186±1,289 | 289±329 | 4.10 | 0.863 |
| AHCYL2 | 3,891±4,065 | 782±626 | 4.97 | 0.861 |
| CYP2B7P1 | 3,261±9,555 | 259±714 | 12.58 | 0.856 |
| PON3 | 1,042±1,294 | 235±662 | 4.43 | 0.855 |
| TMPRSS2 | 2,486±2,505 | 565±827 | 4.40 | 0.853 |
| AGR2 | 11,318±15,822 | 1,998±3,064 | 5.66 | 0.852 |
| C1orf116 | 5,471±5,568 | 931±814 | 5.88 | 0.850 |
| C4orf31 | 1,549±1,809 | 301±725 | 5.13 | 0.850 |
| RNASE1 | 13,190±15,196 | 2,749±2,810 | 4.80 | 0.846 |
| ALPK3 | 1,139±1,068 | 224±372 | 5.08 | 0.846 |
| HOPX | 7,935±12,980 | 1,136±1,974 | 6.98 | 0.845 |
| DPCR1 | 1,687±14,092 | 17±40 | 99.16 | 0.835 |
| C5orf4 | 1,037±1,551 | 230±450 | 4.51 | 0.834 |
| XAGE1D | 3,375±4,395 | 413±1,514 | 8.16 | 0.817 |
| SLC26A9 | 1,281±2,386 | 116±229 | 10.99 | 0.816 |
| TREM1 | 1,139±1,735 | 248±357 | 4.58 | 0.807 |
| C4BPA | 5,525±10,596 | 733±1,371 | 7.53 | 0.807 |
| CLIC6 | 3,400±3,554 | 658±1,120 | 5.16 | 0.806 |
| RASD1 | 2,210±3,304 | 393±728 | 5.62 | 0.800 |
| SFTPB | 195,735±252,122 | 29,275±45,424 | 6.69 | 0.799 |
| TSPAN8 | 2,050±5,256 | 220±659 | 9.32 | 0.799 |
| AGR3 | 1,328±1,793 | 205±376 | 6.47 | 0.799 |
| SUSD2 | 4,164±7,302 | 600±1,568 | 6.93 | 0.790 |
| MFSD4 | 1,158±1,461 | 172±214 | 6.72 | 0.790 |
| PIGR | 20,188±41,363 | 1,719±3,039 | 11.74 | 0.788 |
| HPGD | 2,926±6,201 | 489±1,115 | 5.98 | 0.788 |
| FGB | 5,412±24,204 | 312±3,894 | 17.32 | 0.788 |
| MSLN | 10,685±21,563 | 1,039±6,275 | 10.28 | 0.785 |
| SERPINA1 | 24,209±47,249 | 5,747±8,054 | 4.21 | 0.781 |
| GCNT3 | 1,071±2,121 | 195±496 | 5.47 | 0.777 |
| MUC5B | 22,738±53,189 | 1,754±8,646 | 12.96 | 0.775 |
| FGA | 8,319±35,185 | 500±4,168 | 16.61 | 0.772 |
| TFPI2 | 3,447±14,530 | 525±4,287 | 6.56 | 0.764 |
| ALOX15B | 1,444±2,164 | 327±623 | 4.41 | 0.763 |
| AMY1A | 1,596±6,215 | 220±473 | 7.25 | 0.754 |
| HLA-DQB2 | 1,216±3,432 | 259±462 | 4.70 | 0.751 |
| CLDN2 | 1,224±4,183 | 63±287 | 19.17 | 0.748 |
| PGC | 33,835±138,066 | 389±1,462 | 86.86 | 0.748 |
| PPP1R1B | 1,452±2,143 | 299±789 | 4.85 | 0.747 |
| CACNA2D2 | 1,313±2,012 | 221±376 | 5.93 | 0.746 |
| AQP5 | 1,562±3,262 | 125±322 | 12.49 | 0.745 |
| FGG | 10,438±37,227 | 1,092±7,279 | 9.55 | 0.739 |
| PAEP | 1,822±6,186 | 78±975 | 23.27 | 0.738 |
| CTSE | 6,809±12,689 | 1,058±1,650 | 6.44 | 0.735 |
| MUC13 | 1,434±3,740 | 175±955 | 8.16 | 0.731 |
| AZGP1 | 2,531±6,377 | 583±3891 | 4.34 | 0.730 |
| CEACAM5 | 20,407±34,340 | 4,095±12,219 | 4.98 | 0.723 |
| SLC7A2 | 2,658±4,735 | 515±909 | 5.16 | 0.723 |
| CYP4B1 | 2,242±4,144 | 444±875 | 5.05 | 0.721 |
| LGALS4 | 1,133±4,373 | 17±96 | 64.50 | 0.715 |
| TFF3 | 3,040±8,131 | 457±1,565 | 6.65 | 0.713 |
| VSIG1 | 1,259±4,284 | 73±352 | 17.04 | 0.712 |
| SCGB3A1 | 10,328±58,644 | 585±1,433 | 17.63 | 0.711 |
| CRLF1 | 2,809±6,631 | 319±1,329 | 8.80 | 0.695 |
| S100P | 5,442±10,667 | 1,111±3,795 | 4.90 | 0.693 |
| GPR110 | 1,332±1,797 | 306±564 | 4.34 | 0.688 |
| PLUNC | 10,603±42,374 | 851±3,069 | 12.46 | 0.683 |
| MUC6 | 1,217±8,355 | 75±611 | 16.22 | 0.681 |
| CALCA | 3,578±19,341 | 224±3,022 | 15.96 | 0.679 |
| SCGB3A2 | 8,546±23,575 | 1,224±2,096 | 6.98 | 0.670 |
| CLDN18 | 2,013±7,033 | 307±823 | 6.55 | 0.653 |
| TFF1 | 1,249±5,541 | 34±230 | 36.46 | 0.647 |
| CPS1 | 5,079±15,544 | 436±3515 | 11.63 | 0.593 |
| HP | 4,502±22,250 | 1,056±2,141 | 4.26 | 0.591 |
| PCSK2 | 1,817±10,039 | 100±397 | 18.01 | 0.568 |
| MSMB | 1,343±7,980 | 175±874 | 7.67 | 0.560 |
| PCSK1 | 1,049±6,553 | 142±1,047 | 7.36 | 0.340 |
ROC, receiver operating characteristic; LUAD, lung adenocarcinoma; LUSC: lung squamous cell carcinoma; AUC: area under curve.
Table S2. The ROC curve analyze results of genes greatly elevated in LUSC.
| Gene | LUAD | LUSC | Fold-change (LUSC/LUAD) | AUC value |
|---|---|---|---|---|
| DSG3 | 88±777 | 8,728±8,556 | 98.77 | 0.973 |
| KRT5 | 1,227±10,342 | 116,689±96,742 | 95.03 | 0.972 |
| DSC3 | 128±789 | 7,515±6,291 | 58.62 | 0.970 |
| CALML3 | 141±1,096 | 10,039±11,031 | 71.17 | 0.964 |
| SERPINB13 | 22±191 | 2,166±3,217 | 95.70 | 0.956 |
| KRT6B | 310±1,208 | 17,808±27,334 | 57.45 | 0.954 |
| KRT6C | 136±529 | 7,372±12,063 | 54.13 | 0.954 |
| KRT6A | 2,297±8,724 | 87,096±81,359 | 37.91 | 0.951 |
| PVRL1 | 1,204±1,177 | 11,200±7,063 | 9.30 | 0.950 |
| LOC642587 | 59±213 | 1,247±1,247 | 20.99 | 0.949 |
| PERP | 6,258±4,951 | 31,500±21,939 | 5.03 | 0.947 |
| TP63 | 325±914 | 10,976±9,139 | 33.72 | 0.946 |
| TRIM29 | 861±1,930 | 11,291±7,291 | 13.10 | 0.945 |
| ATP1B3 | 1,866±1,138 | 9,231±6,592 | 4.94 | 0.945 |
| FAT2 | 125±383 | 3,737±3,587 | 29.82 | 0.943 |
| CLCA2 | 87±691 | 6,787±7,536 | 77.23 | 0.943 |
| SPRR2A | 43±546 | 4,036±8,211 | 93.51 | 0.940 |
| JAG1 | 1,118±1,157 | 7,365±7,830 | 6.58 | 0.939 |
| KRT14 | 315±3,191 | 26,428±57,383 | 83.77 | 0.939 |
| SERPINB5 | 358±904 | 4,421±3,570 | 12.32 | 0.937 |
| KRT13 | 225±2,423 | 18,866±41,338 | 83.76 | 0.934 |
| CSTA | 190±403 | 4,222±5,543 | 22.20 | 0.934 |
| PKP1 | 882±2,176 | 19,788±16,151 | 22.42 | 0.934 |
| DAPL1 | 15±102 | 1,098±1,932 | 69.02 | 0.933 |
| IRF6 | 647±369 | 3,108±1,757 | 4.80 | 0.932 |
| KRT16 | 310±1,070 | 17,386±35,463 | 56.03 | 0.932 |
| SLC6A8 | 965±1,028 | 7,254±5,830 | 7.52 | 0.929 |
| SPRR2E | 13±179 | 1,158±3,196 | 84.41 | 0.929 |
| A2ML1 | 106±1,345 | 1,717±3,166 | 16.10 | 0.929 |
| GPC1 | 1,375±1,171 | 9,223±8,003 | 6.71 | 0.926 |
| HR | 60±115 | 1,104±1,530 | 18.30 | 0.923 |
| KRT17 | 2,926±8,839 | 62,551±69,399 | 21.37 | 0.921 |
| COL7A1 | 442±945 | 5,390±5,665 | 12.17 | 0.919 |
| SLC2A1 | 4,007±4,652 | 23,021±18,217 | 5.74 | 0.918 |
| ANXA8 | 240±740 | 3,194±3,237 | 13.30 | 0.916 |
| PTHLH | 149±307 | 3,642±5,287 | 24.41 | 0.914 |
| GBP6 | 71±203 | 2,247±2,528 | 31.33 | 0.913 |
| ABCC5 | 1,037±1,012 | 7,355±7,806 | 7.09 | 0.912 |
| SPRR1A | 36±250 | 2,333±4,852 | 63.44 | 0.912 |
| SNAI2 | 255±444 | 1,149±731 | 4.49 | 0.911 |
| SLC16A1 | 597±1,019 | 2,486±1,753 | 4.16 | 0.910 |
| TFRC | 3,415±3,639 | 18,175±19,185 | 5.32 | 0.910 |
| FOXE1 | 80±276 | 1,593±1,939 | 19.72 | 0.908 |
| BMP7 | 172±530 | 1,843±1,470 | 10.70 | 0.907 |
| ITGA6 | 1,937±3,063 | 8,650±7,228 | 4.46 | 0.906 |
| NTRK2 | 173±794 | 7,764±9,701 | 44.79 | 0.905 |
| ST6GALNAC2 | 287±316 | 1,438±978 | 5.00 | 0.904 |
| CELSR2 | 487±386 | 2,204±1,814 | 4.53 | 0.904 |
| ODZ2 | 29±146 | 1,147±1,729 | 38.99 | 0.904 |
| ADAM23 | 26±90 | 1,535±2,091 | 57.10 | 0.902 |
| GJB6 | 96±265 | 2,657±4,069 | 27.65 | 0.899 |
| ANXA8L2 | 133±347 | 1,201±1,194 | 8.99 | 0.897 |
| LGALS7 | 33±147 | 1,397±3,297 | 41.66 | 0.897 |
| S100A7 | 79±824 | 2,320±11,972 | 29.29 | 0.896 |
| RHCG | 62±554 | 2,294±5,834 | 36.71 | 0.894 |
| NRARP | 217±196 | 1,068±1,082 | 4.92 | 0.894 |
| S100A2 | 1,037±4,073 | 14,533±20,550 | 14.01 | 0.890 |
| ADH7 | 71±513 | 2,704±3,930 | 37.83 | 0.887 |
| LYPD3 | 428±839 | 3,478±4,530 | 8.12 | 0.886 |
| SPRR3 | 75±497 | 4,179±9,702 | 55.54 | 0.884 |
| COL4A5 | 312±414 | 1,956±2,391 | 6.26 | 0.884 |
| CXCR7 | 609±1,045 | 4,107±4,471 | 6.74 | 0.883 |
| C3orf58 | 458±333 | 1,881±1,718 | 4.10 | 0.883 |
| PTPRZ1 | 222±538 | 2,422±2,239 | 10.88 | 0.882 |
| GPR87 | 239±399 | 1,358±1,159 | 5.68 | 0.881 |
| RAPGEFL1 | 302±456 | 1,882±1,782 | 6.22 | 0.880 |
| UGT1A7 | 8±77 | 1,054±2,247 | 128.92 | 0.880 |
| SPRR2D | 87±428 | 2,165±4,477 | 24.63 | 0.878 |
| SPRR1B | 178±777 | 3,747±6,231 | 20.96 | 0.878 |
| KRT15 | 1,280±4,508 | 20,918±28,994 | 16.33 | 0.878 |
| PI3 | 352±4431 | 5,523±12,731 | 15.67 | 0.876 |
| SFN | 3,844±3,146 | 17,013±14,551 | 4.43 | 0.876 |
| FABP5 | 157±305 | 1,443±2,707 | 9.15 | 0.876 |
| RBP1 | 360±732 | 2,217±3,706 | 6.15 | 0.873 |
| DST | 2,550±2,332 | 10,378±8,529 | 4.07 | 0.873 |
| PITX1 | 329±586 | 2,003±2,523 | 6.08 | 0.870 |
| FAM84A | 302±428 | 1,341±1,198 | 4.44 | 0.865 |
| UPK1B | 266±1,452 | 2,995±5,424 | 11.24 | 0.864 |
| ADM | 503±728 | 2,123±2,249 | 4.22 | 0.862 |
| SOX2 | 479±830 | 43,21±4,483 | 9.02 | 0.862 |
| CLDN1 | 2,085±3,554 | 15,300±19,672 | 7.34 | 0.861 |
| MAGEA4 | 323±2,589 | 2,327±4,114 | 7.19 | 0.860 |
| NDUFA4L2 | 632±1,412 | 4,587±5,094 | 7.25 | 0.860 |
| SERPINB4 | 78±380 | 1,223±3,002 | 15.63 | 0.853 |
| FGFBP1 | 236±491 | 2,053±2,791 | 8.70 | 0.851 |
| SERPINB3 | 344±1,591 | 3,359±6,296 | 9.75 | 0.848 |
| NTS | 1,909±15,405 | 8,452±21,005 | 4.43 | 0.846 |
| FGFR2 | 547±653 | 2,244±2,092 | 4.10 | 0.845 |
| RGMA | 233±383 | 1,250±1,463 | 5.35 | 0.841 |
| ALDH3B2 | 288±450 | 1,176±1,362 | 4.08 | 0.838 |
| CYP2S1 | 568±775 | 3,034±2,938 | 5.33 | 0.833 |
| GPNMB | 6,752±7,084 | 30,334±47,047 | 4.49 | 0.831 |
| NDRG4 | 172±226 | 1,102±1,372 | 6.39 | 0.825 |
| GJB2 | 862±1,422 | 6,171±10,796 | 7.15 | 0.820 |
| ABCA13 | 257±471 | 1,296±1,327 | 5.04 | 0.812 |
| FBN2 | 154±1,446 | 1,750±3,324 | 11.34 | 0.812 |
| CRYAB | 187±291 | 1,272±4,611 | 6.80 | 0.811 |
| MMP10 | 194±1,193 | 3,002±7,273 | 15.47 | 0.808 |
| NRCAM | 221±609 | 1,241±1,578 | 5.61 | 0.806 |
| HAS3 | 1,028±1,839 | 4,158±4,225 | 4.04 | 0.804 |
| IL1RN | 449±537 | 2,017±2,468 | 4.49 | 0.804 |
| S100A8 | 1,344±8,937 | 1,1440±28,668 | 8.51 | 0.802 |
| CNTNAP2 | 164±561 | 1,116±1,722 | 6.78 | 0.798 |
| COL17A1 | 1,339±3,023 | 6,832±10,661 | 5.10 | 0.797 |
| AKR1B10 | 2,145±7,972 | 9,111±13,901 | 4.25 | 0.794 |
| WNT5A | 633±563 | 2,606±2,816 | 4.12 | 0.789 |
| CYP4F3 | 141±485 | 1,153±1,964 | 8.14 | 0.773 |
| LY6D | 214±729 | 3,033±6,896 | 14.13 | 0.765 |
| ALDH3A1 | 1,848±7,693 | 8,124±17,776 | 4.40 | 0.759 |
| IVL | 207±501 | 1,093±2,097 | 5.26 | 0.758 |
| CYP4F11 | 271±579 | 2,195±3,752 | 8.09 | 0.725 |
| GSTM2 | 458±496 | 2,044±3,044 | 4.46 | 0.703 |
| GSTM3 | 609±941 | 2,866±4,641 | 4.70 | 0.696 |
| GPC3 | 540±1,255 | 2,291±3,642 | 4.24 | 0.684 |
| KRT4 | 228±1,156 | 2,160±9,487 | 9.45 | 0.644 |
| OLFM1 | 248±296 | 1,325±2,310 | 5.33 | 0.642 |
| GSTM1 | 257±559 | 1,626±4,391 | 6.32 | 0.557 |
| C4orf7 | 87±314 | 1,896±12,269 | 21.63 | 0.530 |
ROC, receiver operating characteristic; LUSC: lung squamous cell carcinoma; LUAD, lung adenocarcinoma; AUC: area under curve.
Footnotes
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- 1.Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin 2015;65:87-108. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Bray F, Center MM, et al. Global cancer statistics. CA Cancer J Clin 2011;61:69-90. [DOI] [PubMed] [Google Scholar]
- 3.Herbst RS, Heymach JV, Lippman SM. Lung cancer. N Engl J Med 2008;359:1367-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Travis WD, Brambilla E, Noguchi M, et al. International association for the study of lung cancer/american thoracic society/european respiratory society international multidisciplinary classification of lung adenocarcinoma. J Thorac Oncol 2011;6:244-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Travis WD, Rekhtman N, Riley GJ, et al. Pathologic diagnosis of advanced lung cancer based on small biopsies and cytology: a paradigm shift. J Thorac Oncol 2010;5:411-4. [DOI] [PubMed] [Google Scholar]
- 6.Schwartz AM, Rezaei MK. Diagnostic surgical pathology in lung cancer: Diagnosis and management of lung cancer, 3rd ed: American College of Chest Physicians evidence-based clinical practice guidelines. Chest 2013;143:e251S-62S. [DOI] [PubMed] [Google Scholar]
- 7.Nagashio R, Ueda J, Ryuge S, et al. Diagnostic and prognostic significances of MUC5B and TTF-1 expressions in resected non-small cell lung cancer. Sci Rep 2015;5:8649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Khayyata S, Yun S, Pasha T, et al. Value of P63 and CK5/6 in distinguishing squamous cell carcinoma from adenocarcinoma in lung fine-needle aspiration specimens. Diagn Cytopathol 2009;37:178-83. [DOI] [PubMed] [Google Scholar]
- 9.Simsir A, Wei XJ, Yee H, et al. Differential expression of cytokeratins 7 and 20 and thyroid transcription factor-1 in bronchioloalveolar carcinoma: an immunohistochemical study in fine-needle aspiration biopsy specimens. Am J Clin Pathol 2004;121:350-7. [DOI] [PubMed] [Google Scholar]
- 10.Ao MH, Zhang H, Sakowski L, et al. The utility of a novel triple marker (combination of TTF1, napsin A, and p40) in the subclassification of non-small cell lung cancer. Hum Pathol 2014;45:926-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhan C, Shi Y, Lu C, et al. Pyruvate kinase M2 is highly correlated with the differentiation and the prognosis of esophageal squamous cell cancer. Dis Esophagus 2013;26:746-53. [DOI] [PubMed] [Google Scholar]
- 12.Remmele W, Stegner HE. Recommendation for uniform definition of an immunoreactive score (IRS) for immunohistochemical estrogen receptor detection (ER-ICA) in breast cancer tissue. Pathologe 1987;8:138-40. [PubMed] [Google Scholar]
- 13.Saaber F, Chen Y, Cui T, et al. Expression of desmogleins 1-3 and their clinical impacts on human lung cancer. Pathol Res Pract 2015;211:208-13. [DOI] [PubMed] [Google Scholar]
- 14.Gómez-Morales M, Cámara-Pulido M, Miranda-León MT, et al. Differential immunohistochemical localization of desmosomal plaque-related proteins in non-small-cell lung cancer. Histopathology 2013;63:103-13. [DOI] [PubMed] [Google Scholar]
- 15.Cui T, Chen Y, Yang L, et al. Diagnostic and prognostic impact of desmocollins in human lung cancer. J Clin Pathol 2012;65:1100-6. [DOI] [PubMed] [Google Scholar]
- 16.Cui T, Chen Y, Yang L, et al. The p53 target gene desmocollin 3 acts as a novel tumor suppressor through inhibiting EGFR/ERK pathway in human lung cancer. Carcinogenesis 2012;33:2326-33. [DOI] [PubMed] [Google Scholar]
- 17.Tsuta K, Tanabe Y, Yoshida A, et al. Utility of 10 immunohistochemical markers including novel markers (desmocollin-3, glypican 3, S100A2, S100A7, and Sox-2) for differential diagnosis of squamous cell carcinoma from adenocarcinoma of the Lung. J Thorac Oncol 2011;6:1190-9. [DOI] [PubMed] [Google Scholar]
- 18.Brown AF, Sirohi D, Fukuoka J, et al. Tissue-preserving antibody cocktails to differentiate primary squamous cell carcinoma, adenocarcinoma, and small cell carcinoma of lung. Arch Pathol Lab Med 2013;137:1274-81. [DOI] [PubMed] [Google Scholar]
- 19.Aizawa S, Ochiai T, Ara T, et al. Heterogeneous and abnormal localization of desmosomal proteins in oral intraepithelial neoplasms. J Oral Sci 2014;56:209-14. [DOI] [PubMed] [Google Scholar]
- 20.Wang L, Liu T, Wang Y, et al. Altered expression of desmocollin 3, desmoglein 3, and beta-catenin in oral squamous cell carcinoma: correlation with lymph node metastasis and cell proliferation. Virchows Arch 2007;451:959-66. [DOI] [PubMed] [Google Scholar]
- 21.Hamidov Z, Altendorf-Hofmann A, Chen Y, et al. Reduced expression of desmocollin 2 is an independent prognostic biomarker for shorter patients survival in pancreatic ductal adenocarcinoma. J Clin Pathol 2011;64:990-4. [DOI] [PubMed] [Google Scholar]
- 22.Wang Q, Peng D, Zhu S, et al. Regulation of Desmocollin3 Expression by Promoter Hypermethylation is Associated with Advanced Esophageal Adenocarcinomas. J Cancer 2014;5:457-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pan J, Chen Y, Mo C, et al. Association of DSC3 mRNA down-regulation in prostate cancer with promoter hypermethylation and poor prognosis. PLoS One 2014;9:e92815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cui T, Chen Y, Yang L, et al. DSC3 expression is regulated by p53, and methylation of DSC3 DNA is a prognostic marker in human colorectal cancer. Br J Cancer 2011;104:1013-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ring BZ, Seitz RS, Beck RA, et al. A novel five-antibody immunohistochemical test for subclassification of lung carcinoma. Mod Pathol 2009;22:1032-43. [DOI] [PubMed] [Google Scholar]
- 26.Mai KT, Perkins DG, Zhang J, et al. ES1, a new lung carcinoma antibody--an immunohistochemical study. Histopathology 2006;49:515-22. [DOI] [PubMed] [Google Scholar]