Abstract
Microalgae accumulate triacylglycerols (TAGs) under conditions of nutrient stress. Phosphorus (P) starvation induces the accumulation of TAGs, and the cells under P starvation maintain growth through photosynthesis. We recently reported that P starvation–dependent overexpression of type-2 diacylglycerol acyl-CoA acyltransferase (CrDGTT4) from Chlamydomonas reinhardtii using a sulfoquinovosyldiacylglycerol synthase 2 (SQD2) promoter, which has increased activity during P starvation, enhances TAG accumulation in C. reinhardtii cells. As a result, the content of C18:1 fatty acid, a preferred substrate of CrDGTT4, is increased in TAGs. Here we isolated genes encoding SQD2 from strain NIES-2145 of the eustigmatophyte Nannochloropsis and showed that their expression, like that in C. reinhardtii, was up-regulated during P starvation. To enhance oil accumulation under P starvation, we transformed pCrSQD2-CrDGTT4 into Nannochloropsis strain NIES-2145. The transformants had a fatty acid composition that was more similar to that of C. reinhardtii, which resulted in enhanced TAG accumulation and higher 18:1(9) content. The results indicated that the P starvation–inducible promoter of C. reinhardtii was able to drive expression of the CrDGTT4 gene in Nannochloropsis strain NIES-2145 under P starvation. We conclude that the heterologous CrSQD2 promoter is effective in manipulating TAG synthesis in Nannochloropsis during P starvation.
Keywords: algae, Nannochloropsis, phosphorus starvation, inducible promoter, triacylglycerol
Introduction
Algal biofuel technology exploits algal photosynthesis and biosynthesis processes to produce oils using only sunlight, CO2, water and limited nutrients. Many microalgae accumulate triacylglycerols (TAGs) during nutrient stress (Giroud et al., 1988; Guschina and Harwood, 2006; Hu et al., 2008). It is estimated that the annual oil production from algae is in the range of 40,700–53,200 L ha−1 year−1 (Weyer et al., 2010). Therefore, the potential use of microalgae to provide biofuel feedstock is receiving significant attention.
A eukaryotic microalga, Chlamydomonas reinhardtii, is a model organism for studying algal biodiesel production because of its available whole-genome sequence and the ability to manipulate gene expression within this organism (Harris, 2009). Under stress conditions, such as nitrogen (N) starvation, Chlamydomonas nearly stops its growth and accumulates large amounts of TAGs (Grossman, 2000; Zhang et al., 2004). We reported that C. reinhardtii cells in logarithmic growth phase that were diluted into fresh medium showed substantial TAG accumulation under N or P deprivation (Iwai et al., 2014). P deprivation substantially induced the accumulation of oil droplets in the cytosol but allowed the maintenance of thylakoid membranes in C. reinhardtii (Iwai et al., 2014).
Nannochloropsis species are unicellular photosynthetic microalgae in the class Eustigmatophyceae. Previously known as “marine Chlorella,” this class was identified on the basis of its ultrastructure and named Nannochloropsis by Maruyama et al. (1986). Its cells are spherical to slightly ovoid, are 2–4 μm in diameter and contain ovoid or cup-shaped chloroplasts (Maruyama et al., 1986). Nannochloropsis are of interest because of their rapid growth and their ability to produce large quantities of TAGs and be cultured on an industrial scale (Hodgson et al., 1991; Rodolfi et al., 2009; Huerlimann et al., 2010) In recent years, Nannochloropsis strains have been studied for their biomass production and their lipid composition and content under different growth conditions (Hu and Gao, 2006; Converti et al., 2009; Rodolfi et al., 2009; Simionato et al., 2011; Arudchelvam and Nirmalakhandan, 2012; Vieler et al., 2012b; Wang et al., 2014). In Nannochloropsis strains, N deprivation induces lipid accumulation and lipid droplet formation (Rodolfi et al., 2009; Vieler et al., 2012a,b; Martin et al., 2014), and their lipid content also increases with decreasing P concentrations (Hu and Gao, 2006; Rodolfi et al., 2009; Bondioli et al., 2012).
Diacylglycerol acyltransferase (DGAT) catalyzes the last step of TAG synthesis involving sn-1,2 diacylglycerol and acyl-CoA (Lung and Weselake, 2006; Rajakumari et al., 2008) and includes two major types, type 1 DGAT (DGAT1) and type 2 DGAT (DGAT2) (Cases et al., 1998; Lardizabal et al., 2001; Shockey et al., 2006). There are six DGAT genes in C. reinhardtii (encoding one DGAT1 and five DGAT2s) (Miller et al., 2010; Boyle et al., 2012). In contrast, 12 or 13 DGAT genes (encoding one or two DGAT1s and 11 DGAT2s) are present in Nannochloropsis strains (Radakovits et al., 2012; Vieler et al., 2012b; Wang et al., 2014). During N starvation in Nannochloropsis oceanica IMET1, seven DGAT transcripts are up-regulated and six other DGAT transcripts are down-regulated (Li et al., 2014).
Endogenous promoters are mainly used for the nuclear transformation of algae such as C. reinhardtii (Eichler-Stahlberg et al., 2009; Harris, 2009; Brueggeman et al., 2014), although stable nuclear transformation of algae is feasible with promoters from heterologous sources that are categorized in the same class (Hirata et al., 2011; Lerche and Hallmann, 2013, 2014). Transformation experiments using Nannochloropsis with endogenous promoters have recently been reported (Kilian et al., 2011; Vieler et al., 2012b). Additionally, a construct with the C. reinhardtii α-tubulin promoter was used for transforming Nannochloropsis (Vieler et al., 2012b). However, the stable nuclear transformation of Nannochloropsis using an inducible promoter from a heterologous source under specific conditions, in particular an oil-accumulating condition such as nutrient starvation, has not been reported.
In this study, we found that Nannochloropsis strain NIES-2145 has a homolog of the Arabidopsis SQD2 gene. Our results suggest that there is a common expression control system in a wide range of algal species, including primary and secondary microalgae, for adaptation to low P. We used the promoter of sulfoquinovosyldiacylglycerol (SQDG) synthase 2 (SQD2), which encodes the sulfoquinovosyl transferase that catalyzes the second step of sulfolipid biosynthesis (Yu et al., 2002). The transcript levels of the SQD2 gene homologs are increased in C. reinhardtii and Arabidopsis by P starvation concomitant with increases in the SQDG content (Yu et al., 2002; Chang et al., 2005; Okazaki et al., 2009; Iwai et al., 2014). The promoter of SQD2 induced expression of the down stream gene during P starvation, to successfully overexpress C. reinhardtii type-2 diacylglycerol acyl-CoA acyltransferase (CrDGTT4) in Nannochloropsis strain NIES-2145 under P starvation. The total lipid content, neutral lipid content and fatty acid profiles were determined. CrDGTT4 enhanced TAG accumulation under P starvation by changing the fatty acid composition to be more similar to that of C. reinhardtii. Therefore, the heterologous CrSQD2 promoter is effective in manipulating oil synthesis in Nannochloropsis during P starvation.
Materials and methods
Materials and culture conditions
Nannochloropsis strain NIES-2145 was obtained from the Microbial Culture Collection of the National Institute for Environmental Studies, Japan. Nannochloropsis strain NIES-2145 was grown photoautotrophically in f/2 medium (Guillard and Ryther, 1962) or F2N50%SW medium (standard medium), which is F2N medium (Kilian et al., 2011) made with 50% artificial seawater (Wako Pure Chemical Industries, Ltd., Japan). Liquid cultures were grown in continuous white light (20–40 μmol photons m−2 s−1) at room temperature. Nutrient deficiency was induced by centrifuging the cells for 10 min at 2000 × g, washing them twice with the respective medium and subsequently resuspending them in the standard medium without NaNO3 and NH4Cl (–N) or phosphate (–P) solutions. Agar plates were prepared using 0.8% Bacto agar (Difco, USA) in standard medium. The cells were maintained on these plates at the same light intensity at room temperature.
Analysis of differential gene expression levels
The differential expression levels of the CrDGAT gene and SQD2 genes in Nannochloropsis strain NIES-2145 cultured in standard, -P and -N media were determined using quantitative real-time PCR (qPCR). RNA was extracted from standard, -P and -N cultures using the phenol/chloroform method. Total RNA (500 ng) was used for the synthesis of cDNA with an oligo(dT)18 primer, random hexamers and Superscript II reverse transcriptase (Invitrogen, Carlsbad, CA). cDNA was amplified using SYBR Premix Ex Taq II (Takara, Japan). The Thermal Cycler Dice Real Time System and Multiplate RQ software (Takara) were used for the analysis. Expression levels of the CrDGAT gene and SQD2 genes were normalized to TUB mRNA expression. The CrDGTT4 expression levels in Nannochloropsis cultured in -P medium were compared with those of Nannochloropsis cultured in the standard medium (the values of which were set to 1). Primers are listed in Supplemental Table 1.
Construction and identification of CrDGTT4-overexpressing NIES-2145 cell lines
Transformation experiments were performed using a construct developed for the nuclear transformation of Nannochloropsis, pMD20 with a NT7 cassette (Kilian et al., 2011), except that a truncated bidirectional violaxanthin–chlorophyll a binding protein 2 promoter and the violaxanthin–chlorophyll a binding protein 1 3′ UTR from Nannochloropsis strain NIES-2145 were used and that the EcoRV restriction site was inserted between the ble gene and the violaxanthin–chlorophyll a binding protein 1 3′ UTR. To construct the expression plasmid carrying the CrDGTT4 (XM_001693137.1) gene, pCrSQD2aDGTT4, which contains the C. reinhardtii SQD2 promoter region and the coding sequence of the DGTT4 gene (Iwai et al., 2014), was digested with restriction endonucleases EcoRV and SpeI and was treated using a DNA Blunting Kit (Takara). The resulting blunt-ended DNA was cloned into pMD-NT7, which was digested with restriction endonuclease EcoRV. Electroporation with a supplier was then used to transform the nuclear genome of Nannochloropsis strain NIES-2145 cells with 2–10 μg vector construct (Vieler et al., 2012b). The selection of transformants was performed on standard medium supplemented with 2 μg mL−1 Zeocin. Approximately 100 positive transformants were identified by growth on the selective medium, and overexpression levels in each resulting line were determined using qPCR.
Extraction and separation of lipids
Nannochloropsis cells were harvested by centrifugation (10 min at 3000 × g), and total lipids were extracted as described (Bligh and Dyer, 1959). The lipids were dissolved in chloroform/methanol (2:1, v/v) and stored at −20°C. Lipid classes were separated using two-dimensional thin-layer chromatography (TLC). The first dimension was developed using chloroform/methanol/7 N ammonia water (115:80:8, v/v/v). After the plates were dried for 1 h, the second dimension was developed using chloroform/methanol/acetic acid/water (170:25:15:3, v/v/v/v). TAGs were separated by TLC using the solvent system hexane/diethyl ether/acetic acid (160:40:4, v/v/v). Lipids were visualized under UV light after spraying the TLC plates with 0.001% (w/v) primuline in 80% (v/v) acetone.
Preparation of fatty acid methyl esters (FAMEs)
Each lipid was scraped off of the TLC plates. FAMEs were obtained by incubating lipids for 1 h at 85°C in the presence of 5% (v/v) hydrogen chloride–methanol solution (Wako Pure Chemical Industries) (Iwai et al., 2014). FAMEs were extracted using hexane and were determined by gas chromatography (GC). Fatty acid quantification was performed by comparing samples with the FAMEs derived from the internal standard, pentadecanoic acid (15:0).
GC analysis
FAMEs were analyzed using a GC-2014 (Shimadzu Corporation, Kyoto, Japan) with a HR-SS-10 capillary column (length, 25 m; internal diameter, 0.25 mm; Shinwa Chemical Industries, Ltd., Japan). The fatty acid profiles were determined by comparison with a standard reference mix composed of FAMEs (Supelco 18917-1AMP, 18913-1AMP, CRM47885; Sigma-Aldrich, GLC411, GLC462; Funakoshi, Japan).
GC-mass spectrometry (MS)
The qualitative composition of FAMEs was studied using a GS-MS (model GCMS-TQ8030; Shimadzu Corporation). High-grade pure helium was used as the carrier gas. The ionization voltage was 70 eV, and the ionization temperature was 200°C. Mass spectra were scanned every 0.2 s. For the analysis of the FAMEs, a DB-5 ms column (length, 30 m; internal diameter, 0.25 mm; Agilent Technologies, Inc., CA, USA) was used. For each sample, 1 μL was injected on the column into a helium gas flow held constant at 1.4 mL min−1. The column temperature was elevated from 40°C to 320°C at a rate of 6°C min−1 and then kept at 320°C for 1.15 min.
SQD2 cDNA
RNA was extracted from standard and -P cultures using the phenol/chloroform method. Total RNA (500 ng) was used for the synthesis of cDNA with an oligo(dT)18 primer, random hexamers and Superscript II reverse transcriptase (Invitrogen, Carlsbad, CA). The primers used are listed in Supplemental Table 1. The gene sequences were then cloned into the pMD20 vector (Takara) for DNA sequence.
Phylogenetic analyses
Amino acid sequences were aligned using MAFFT v7.220 (Katoh and Standley, 2013). Gblocks 0.91b (Talavera and Castresana, 2007) was used to remove any poorly conserved regions. The phylogenetic analyses were performed using the maximum likelihood and the neighbor-joining methods in MEGA6 (Tamura et al., 2013), and a Bayesian analysis in MrBayes 3.2.3 (Ronquist et al., 2012). The amino acid substitution model for the maximum likelihood and Bayesian inference methods was selected using Aminosan (Tanabe, 2011). The maximum likelihood and neighbor-joining methods were performed based on the LG model + Gamma (eight categories) with 1000 bootstraps and the JTT model with 1000 bootstraps, respectively. The Bayesian analysis was performed based on the LG model + Gamma (eight categories) for 1,000,000 generations. Every 500th generation was sampled, and the first 200 trees were discarded as burn-in.
Motifs analysis
The analysis of the motifs was performed in silico using New PLACE (https://sogo.dna.affrc.go.jp/cgi-bin/sogo.cgi?sid=&lang=en&pj=640&action=page&page=newplace), a database of cis-acting regulatory DNA elements from plants, to find the binding motifs of transcription factors.
Results
Cell growth and tag accumulation in cells under P deprivation
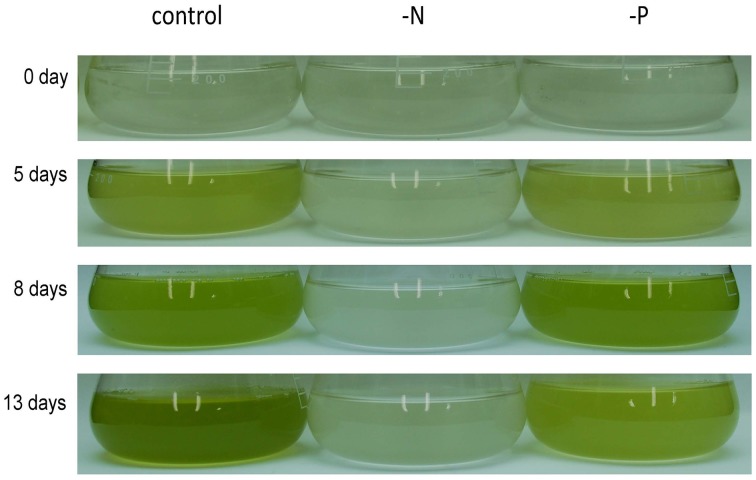
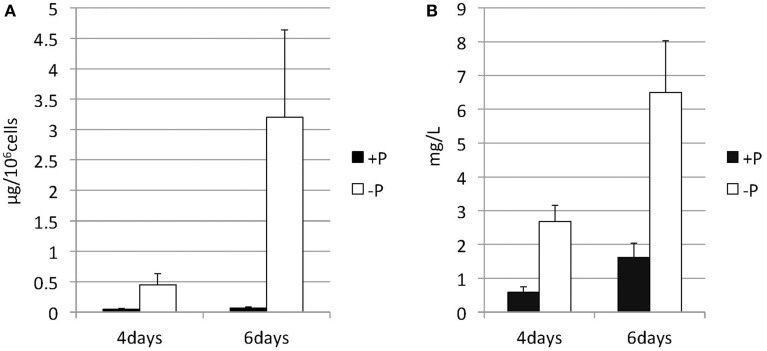
Nannochloropsis strain NIES-2145 was cultured to the logarithmic phase in f/2 medium. Cultures were then centrifuged and resuspended in fresh f/2 or in N-depleted (-N) or P-depleted (-P) f/2 medium. The cultures derived from logarithmic-phase cultures were inoculated at low cell densities (1 × 106 cells mL−1). Under N-deprived conditions, the Nannochloropsis cells were nearly white, but the cells exposed to P starvation were still green (Figure 1). Because C. reinhardtii cells diluted into P-depleted medium show substantial TAG accumulation (Iwai et al., 2014), we investigated whether TAG was accumulated in these nutrient-starved cells by quantifying lipid-derived FAMEs using GC. Cells had substantially increased TAG levels under both N and P deprivation (Supplemental Figure 1). Compared with cells grown in complete medium, the N-starved cells contained ~11-fold and the P-starved cells contained ~6-fold more TAG per cell after 13 days. The TAG accumulation in the N-starved cells is consistent with that in recent reports (Bondioli et al., 2012; Vieler et al., 2012b; Simionato et al., 2013). When the cultures were inoculated at 5 × 106 cells mL−1 in standard medium or –P medium, P-starved cells contained more TAG per cell than the corresponding cells maintained in the standard medium (~7-fold more at 4 days and ~47-fold more at 6 days; Figure 2A). P-starved cultures contained ~4.6-fold more TAG per liter at 4 days and ~4-fold more TAG per liter at 6 days than the corresponding culture maintained in the standard medium (Figure 2B).
Figure 1.
Growth of Nannochloropsis NIES-2145 under standard growth conditions, N starvation and P starvation. Nannochloropsis cells cultured to the logarithmic phase were inoculated into standard (control), -N and -P media. Cells were cultured for the indicated times after transfer before being photographed.
Figure 2.
Changes in the TAG content of Nannochloropsis in response to P starvation. Cells cultured to a logarithmic phase under standard conditions were then inoculated into standard (+P) or -P medium and cultured for 4 or 6 days. The y-axis is (A) Total TAG per 106 cells or (B) total TAG per liter of culture. Values represent the mean from three independent experiments ± SD.
Increase in SQDG and SQD2 homolog promoter activity during P starvation
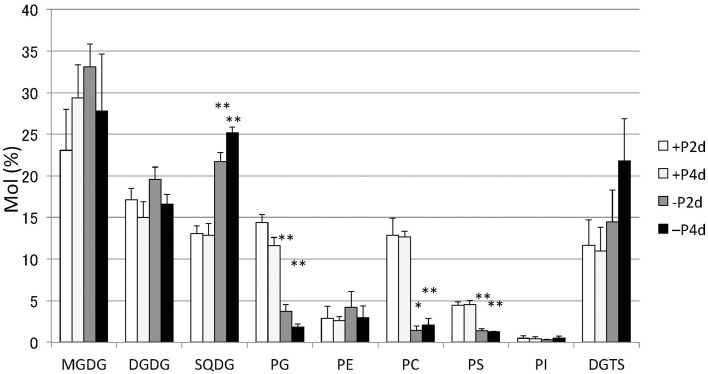
Lipids extracted from Nannochloropsis strain NIES-2145 were separated by two-dimensional TLC, followed by GC-flame ionization detection and GC-MS. P starvation caused significant changes in lipid composition. The glycerolipid composition of Nannochloropsis strain NIES-2145 resembles that of the photosynthetic organism Arabidopsis thaliana (Li-Beisson et al., 2010), being comprised mostly of the prevalent glycoglycerolipids monogalactosyldiacylglycerol (MGDG), DGDG and SQDG, as well as the common phospholipids phosphatidylcholine (PC), phosphatidylethanolamine (PE), phosphatidylserine (PS), phosphatidylinositol (PI), and phosphatidylglycerol (PG). In addition, a betaine lipid diacylglycerol-N,N,N-trimethylhomoserine (DGTS) is present, as in C. reinhardtii (Sato and Furuya, 1985; Giroud et al., 1988). P deprivation resulted in elevated levels of SQDG and DGTS. PG, PC, and PS which contain phosphates in their head groups, decreased after P deprivation (Figure 3). This result corroborates previous reports on P deprivation in several species (Benning et al., 1993; Essigmann et al., 1998; Sato et al., 2000; Khozin-Goldberg and Cohen, 2006).
Figure 3.
Changes in major lipid classes in P-starved cells. Cells cultured to a logarithmic phase under standard conditions were then inoculated into standard (+P) or -P medium and cultured for 2 days (2 d) or 4 days (4 d). Lipid levels were determined by GC. Values represent the mean from three independent experiments ± SD. Asterisks indicate a statistically significant difference compared with the empty vector control based on a two-tailed Student's t-test (*P < 0.05 and **P < 0.01).
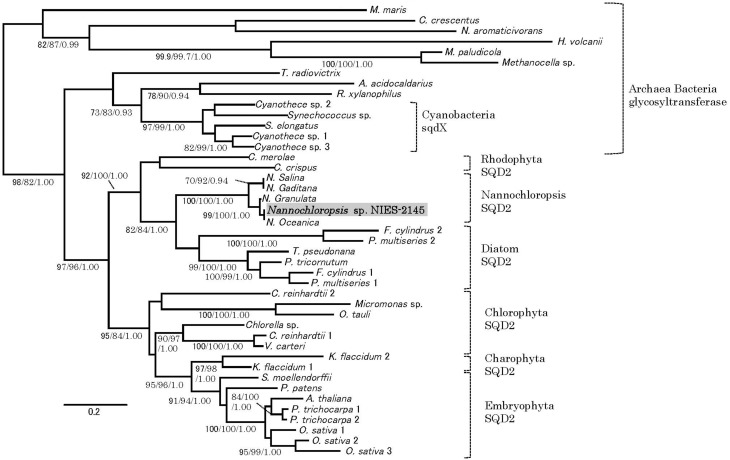
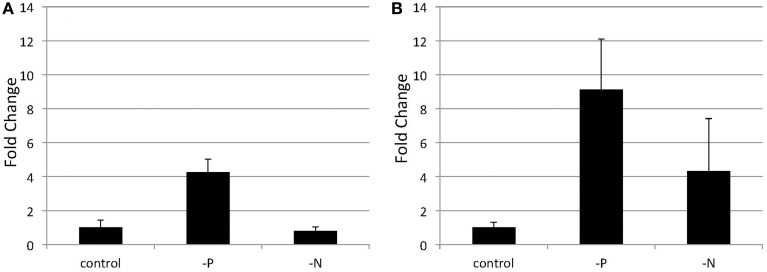
Orthologs of the SQD2 encoding the sulfoquinovosyl transferase (Yu et al., 2002) and a cyanobacterial counterpart, the sqdX, are highly conserved across different groups of algae and plants (Figure 4). We expected that the expression of homologs of the SQD2 gene would be induced during P starvation. To determine the expression of SQD2 homologs in Nannochloropsis strain NIES-2145, we searched the N. oceanica genome (Vieler et al., 2012b) using a BLAST algorithm to identify amino acid sequences with similarity to C. reinhardtii and Arabidopsis SQD2 genes. Using consensus sequences of the SQD2 genes, we amplified the corresponding cDNA from Nannochloropsis strain NIES-2145. We sequenced the cDNA and found that Nannochloropsis strain NIES-2145 has a homolog of the Arabidopsis SQD2 gene (Figure 4). Figure 4 is based on protein comparison. Expression of the SQD2 gene homolog, as measured by qPCR, was induced after P deprivation in Nannochloropsis strain NIES-2145 (Figure 5). The Chlamydomonas SQD2 promoter is useful for improving transgene expression in C. reinhardtii when P is depleted (Iwai et al., 2014). Therefore, we expected that the Chlamydomonas SQD2 promoter would be available for inducing transgene expression to increase TAG production in Nannochloropsis under P-depleted conditions.
Figure 4.
Phylogenetic tree analysis of SQD2. A phylogeny incorporating eukaryotic SQD2, cyanobacterial sqdX and the glycosyl transferase proteins of other organisms was constructed using Bayesian inference, maximum likelihood and neighbor-joining algorithms. It is based on a protein comparison. The topologies and branch lengths were calculated using maximum likelihood. Bootstrap values (maximum likelihood and neighbor-joining) higher than 70 and Bayesian posterior probabilities (Bayesian inference) higher than 0.9 are indicated under each branch (maximum likelihood/neighbor-joining/Bayesian inference). The scale bar represents 0.2 amino acid substitutions per site. Sequence accession numbers or sequence resources for the phylogenetic tree are as follows: Archaea glycosyl transferase: Methanocella paludicola (YP_003355097), Methanocella sp. (YP_685710) and Haloferax volcanii (YP_003535220); Alpha-proteobacteria glycosyl transferase: Caulobacter crescentus (YP_002516166), Novosphingobium aromaticivorans (WP_011446186), and Maricaulis maris (WP_011642337); Cyanobacteria sqdX: Cyanothece sp. 1 (WP_015784192), Cyanothece sp. 2 (WP_012629991), Synechococcus elongatus (WP_011243256), Synechococcus sp. (WP_011429929), and Cyanothece sp. 3 (WP_015956855); Other bacterial glycosyl transferases: Truepera radiovictrix (WP_013178429), Rubrobacter xylanophilus (WP_011564279), and Alicyclobacillus acidocaldarius (WP_012810695); Rhodophyta SQD2: Chondrus crispus (XP_005715110), and Cyanidioschyzon merolae (XP_005538341); Stramenopile SQD2: Nannochloropsis sp. NIES-2145 (DDBJ Accession LC061442), Nannochloropsis oceanica IMET1 (BioProject: PRJNA202418, scaffold00247.g6721), Nannochloropsis granulata CCMP529 (BioProject: PRJNA65111, evm.model.NODE_3998_length_11613_cov_18.861534.4), Nannochloropsis salina CCMP537 (BioProject: PRJNA62503, evm.model.NODE_9973_length_179428_cov_24.782236.3), Nannochloropsis gaditana (EWM27152), Phaeodactylum tricornutum (JGI Protein ID: 50356), Fragilariopsis cylindrus 1 (JGI Protein ID: 207999), Fragilariopsis cylindrus 2 (JGI Protein ID: 158007), Pseudo-nitzschia multiseries 1 (JGI Protein ID: 252457), Pseudo-nitzschia multiseries 2 (JGI Protein ID: 145536), Thalassiosira pseudonana (JGI Protein ID: 38775), [Chlorophyta] Ostreococcus tauri (JGI Protein ID: 3203), Micromonas sp. (JGI Protein ID: 58169), Chlorella sp. (JGI Protein ID: 33086), Volvox carteri (Phytozome Transcript Name: Vocar20009459m), Chlamydomonas reinhardtii 1 (Phytozome Transcript Name: Cre01.g038550.t1.3), and Chlamydomonas reinhardtii 2 (Phytozome Transcript Name: Cre16.g689150.t1.2); Charophyta: Klebsormidium flaccidum 1 (BioProject: PRJDB718, kfl00392_0070) and Klebsormidium flaccidum 2 (BioProject: PRJDB718, kfl00041_0140), and Embryophyta (Phytozome Transcript Name): Arabidopsis thaliana (AT5G01220.1), Populus trichocarpa 1 (Potri.006G097600.1), Populus trichocarpa 2 (Potri.016G112600.1), Oryza sativa 1 (Os07g01030.1), Oryza sativa 2 (Os01g04920.1), Oryza sativa 3 (Os03g15840.1), Selaginella moellendorffii (170091), and Physcomitrella patens (Pp1s24_194V6.1).
Figure 5.
Quantitative real-time PCR showing the induction of SQD2 gene expression in Nannochloropsis after P or N starvation. Logarithmic-phase cells were transferred to standard (control), -P and -N media and cultured for 4 days (A) or 6 days (B). The values are normalized to the expression of TUB and the SQD2/TUB ratio under standard conditions was set as 1. Values represent the mean ± SD from three independent replicates.
DGAT enzymes are important for TAG accumulation. During N starvation in N. oceanica IMET1, seven DGAT transcripts are up-regulated and six other DGAT transcripts are down-regulated (Li et al., 2014). However, it is unclear how DGAT transcripts are regulated during P starvation in Nannochloropsis strain NIES-2145. We therefore hypothesized that the overexpression of C. reinhardtii DGTT4 in Nannochloropsis would be a convenient way to increase the lipid content or alter the lipid composition of these cells.
Enhanced tag accumulation and changes in fatty acid profiles in CrDGTT4-overexpressing lines under P deprivation
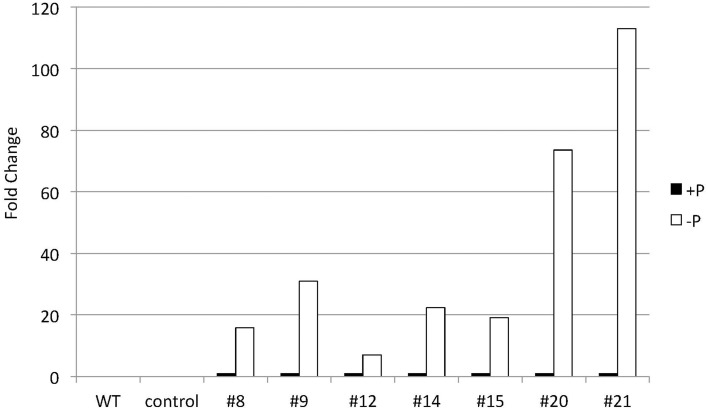
To explore CrDGTT4 as a tool to produce TAGs in Nannochloropsis cells, we expressed the CrDGTT4 coding sequence in Nannochloropsis under the control of the CrSQD2 promoter, which increases transcription levels during P starvation in C. reinhardtii (Iwai et al., 2014). The relative level of overexpression of CrDGTT4 was measured using qPCR. CrDGTT4 mRNA levels in the overexpressing lines under P starvation were determined to be between 7- and 112-fold higher than in those under standard conditions (Figure 6), whereas no transcripts were detected in the wild-type and the empty vector controls. Thus, the SQD2 promoter from C. reinhardtii is quite useful as a heterologous promoter in Nannochloropsis strain NIES-2145. Three overexpressing lines, #8, 9, and 21, were then selected for the subsequent experiments. To confirm the effect of CrDGAT gene overexpression, we compared the lipid content and fatty acid composition of the wild-type and overexpressing lines. All lines were cultured to the logarithmic phase prior to P depletion, as was done in the previous experiments.
Figure 6.
Quantitative real-time PCR showing the induction of CrDGTT4 gene expression in CrDGTT4-overexpressing cells after P deprivation. Cells were cultured for 5 days. The relative levels of CrDGTT4 mRNA overexpression in seven cell lines were assessed under standard (+P) and -P growth conditions. The values were normalized to the expression level of TUB. The CrDGTT4/TUB ratio under standard conditions was set as 1. The levels of the wild type (WT) and empty vector control (control) are below the detection threshold.
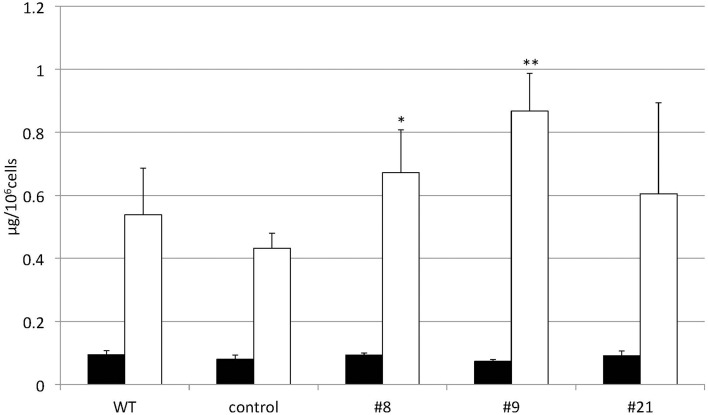
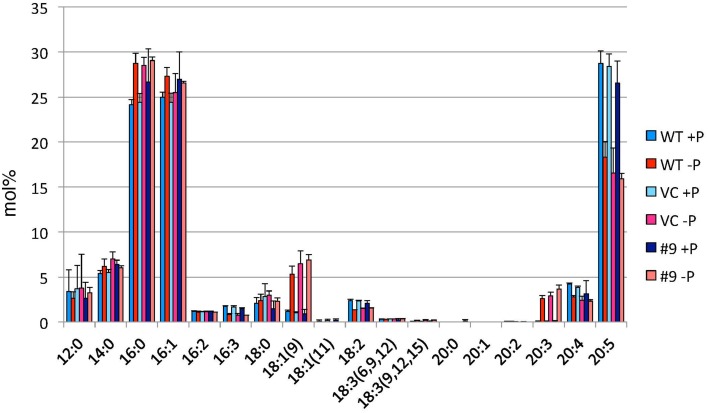
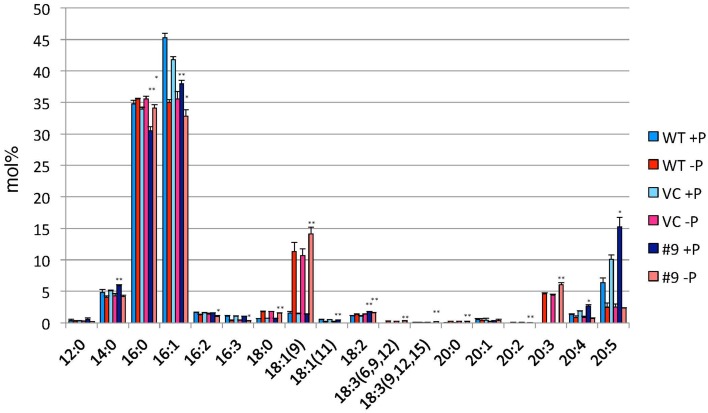
TAG accumulation increased in CrDGTT4-overexpressing lines compared with the control and wild-type lines in P-depleted medium. CrDGTT4-overexpressing lines contained ~1.7-fold and ~1.3-fold more TAG per cell than the empty vector control cells at 4 and 6 days, respectively (Figure 7, Supplemental Figure 2). We next investigated if the fatty acid composition of total lipids and TAGs was altered in the CrDGTT4-overexpressing cell line. One of the overexpressing lines, line #9, was analyzed because of TAG accumulation most increased in this line. The major total fatty acids in either control or P-depleted medium were eicosapentaenoic (20:5 ω-3), palmitoleic (16:1 ω-7), and palmitic (16:0) acids (Figure 8). This agreed with previous analyses (Maruyama et al., 1986; Patil et al., 2007). The levels of 16:0, 16:1, oleic (18:1 ω-9), and dihomo-gamma-linolenic (20:3 ω-6) fatty acids increased, whereas 16:3, linoleic (18:2 ω-6), arachidonic (20:4 ω-6) and 20:5 fatty acids decreased under P starvation conditions (Figure 8). The degree of unsaturation for each fatty acid decreased as the unsaturated fatty acids were replaced with saturated fatty acids. As for the total fatty acid composition, no statistically significant differences could be observed among the wild-type, control and overexpressing lines (Figure 8). In the case of TAG fatty acids in the overexpressing line, the levels of 18:1 and 20:3 fatty acids were higher, whereas 16:0, 16:1, 16:2, and 16:3 fatty acids were lower than those in the wild-type or control lines during P starvation (Figure 9). The levels of 20:4 and 20:5 bound to TAGs (~0.8% and ~2.3%, respectively) were remarkably reduced under P starvation in comparison with the levels of these fatty acids in total fatty acids (~1–3% and ~15%, respectively; Figure 9). Under P starvation, the increase in the 20:3 fatty acid in the total lipids was a reflection of its increase in TAG. It is interesting to note that the level of 18:1 fatty acids was higher in the CrDGTT4-overexpressing cell line than in the control line under P starvation conditions (Figure 9). This is consistent with our previous report on CrDGTT4-overexpressing C. reinhardtii cell lines during P starvation (Iwai et al., 2014). These results showed that the C. reinhardtii SQD2 promoter is useful as a heterologous promoter in the secondary endosymbiotic alga Nannochloropsis strain NIES-2145.
Figure 7.
Changes in the TAG content of CrDGTT4-overexpressing cells in response to P starvation. Nannochloropsis cells were transformed with pCrSQD2-CrDGTT4 to overexpress CrDGTT4. Cells cultured to a logarithmic phase under standard conditions were then inoculated into P-starved medium and cultured for 4 days. Three transformants, the empty vector control (control) and the wild type (WT) are shown. Values represent the mean ± SD from four independent replicates. Asterisks indicate a statistically significant difference compared with the empty vector control based on a two-tailed Student's t-test (*P < 0.05 and **P < 0.01).
Figure 8.
Analysis of the fatty acid composition of total lipids in pCrSQD2-CrDGTT4 (#9), vector control (VC) and wild-type (WT) lines. Cells were cultured in control (+P) or -P medium for 4 days. Values are the mean ± SD from three independent experiments.
Figure 9.
Analysis of the fatty acid composition of the TAG fraction in pCrSQD2-CrDGTT4 (#9), the vector control (VC) and wild-type (WT) lines. Cells were cultured in control (+P) or -P medium for 4 days. Values are the mean ± SD from three independent experiments. Asterisks indicate a statistically significant difference compared with VC based on a two-tailed Student's t-test (*P < 0.05 and **P < 0.01).
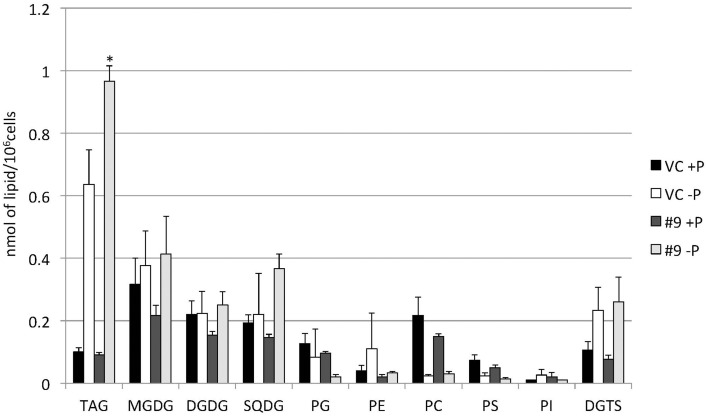
P starvation did not cause significant changes in membrane lipid composition between the control line and overexpressing line
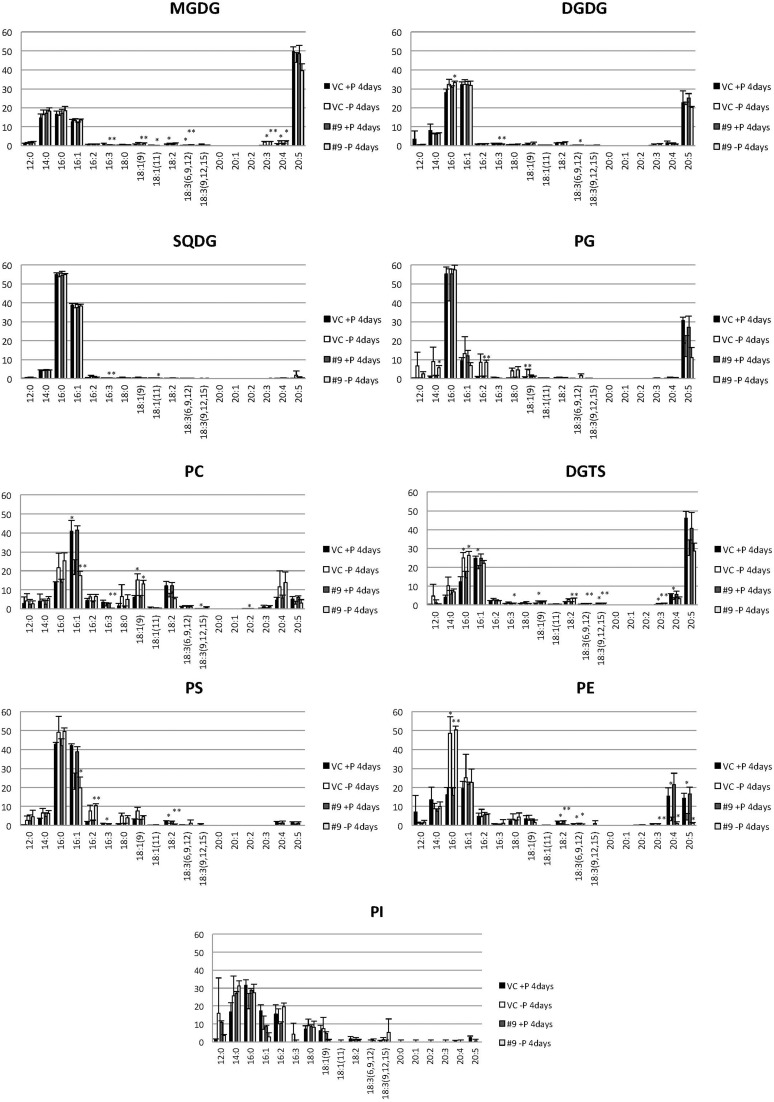
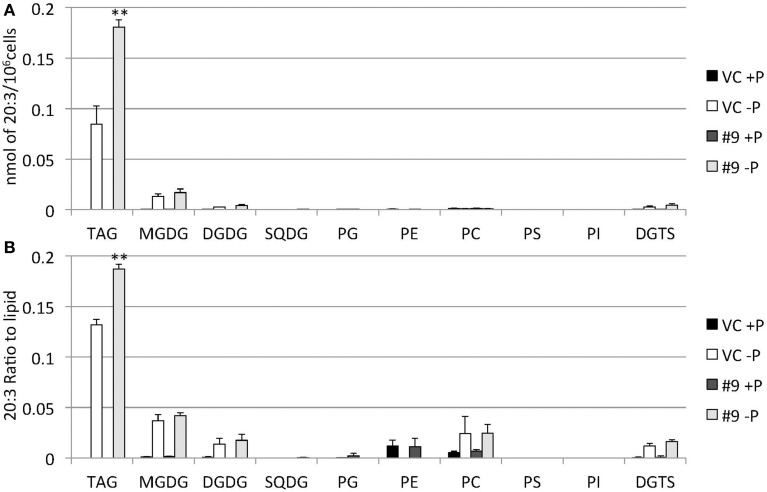
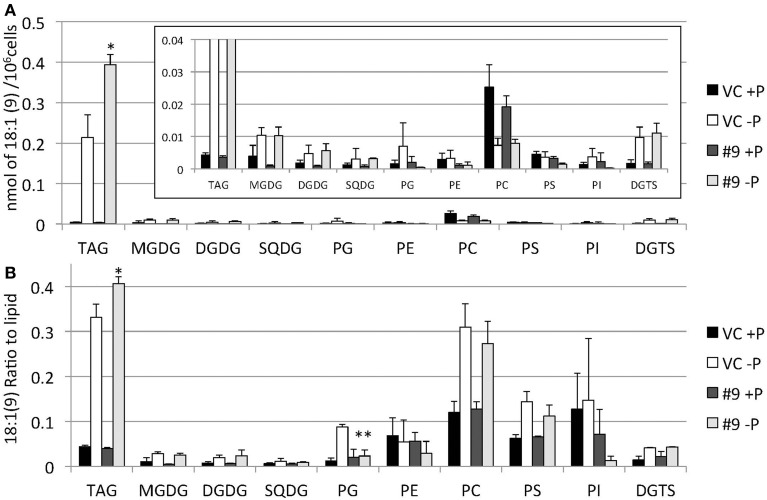
The overexpressing line showed alteration in TAG, whereas no serious alterations in the major membrane lipids compared with the control line under standard growth conditions or conditions of P stress, except for PS level (Figure 10, Supplemental Figure 3). PS decreased more in the overexpressing line than in the control line under P starvation (Supplemental Figure 3). We investigated whether the fatty acid composition of the major membrane lipids was altered in the CrDGTT4-overexpressing cell line (Figures 11–13, Supplemental Figures 4–7). During P-depleted conditions, the enhanced incorporation of 18:1(9) fatty acid was observed in the major plastidic membrane lipids MGDG, DGDG and PG and the extra-plastidic membrane lipids PC, PS, and DGTS (Figures 11, 12), whereas the enhanced incorporation of 20:3 fatty acid was observed in MGDG, DGDG, PC, and DGTS (Figures 11, 13). In the contrast, there were decreases of 18:1(9) and 20:3 fatty acids in PE. There was a global decrease in the long polyunsaturated fatty acid, 20:5 (Figure 11 and Supplemental Figure 4), and an increase in saturated fatty acids, for example 16:0 (Figure 11 and Supplemental Figure 5), during P-depleted conditions. Although large changes occur during P depravation, no significant differences could be observed in the major membrane lipids between the control line and overexpressing line, except for the decrease of 18:1 (9) fatty acid in the major plastidic membrane lipid PG.
Figure 10.
Quantitative analysis of the various lipids. Cells were cultured in control (+P) or -P medium for 4 days. Each lipid is expressed in nmol per 106 cells. Values are the mean ± SD from three independent experiments. Asterisks indicate a statistically significant difference compared with VC based on a two-tailed Student's t-test (*P < 0.05).
Figure 11.
Analysis of the fatty acid composition of the major lipids in pCrSQD2-CrDGTT4 (#9) and the vector control (VC) lines. Cells were cultured in control (+P) or -P medium for 4 days. Black bars, the vector control (VC) 4 days in control medium; white bars, VC 4 days in -P; dark gray bars, pCrSQD2-CrDGTT4 (#9) 4 days in control medium; and light gray bars, pCrSQD2-CrDGTT4 (#9) 4 days in -P. Values are the mean ± SD from three independent experiments. Asterisks indicate a statistically significant difference compared with wild-type based on a two-tailed Student's t-test (*P < 0.05 and **P < 0.01).
Figure 13.
Quantitative analysis and ratio of 20:3 in the various lipids. Cells were cultured in control (+P) or -P medium for 4 days. (A) Each 20:3 in the various lipids is expressed in nmol per 106 cells. (B) The y-axis is ratio of 20:3 in the various lipids per 106 cells. Values are the mean ± SD from three independent experiments. Asterisks indicate a statistically significant difference compared with VC based on a two-tailed Student's t-test (**P < 0.01).
Figure 12.
Quantitative analysis and ratio of 18:1(9) in the various lipids. Cells were cultured in control (+P) or -P medium for 4 days. (A) Each 18:1(9) in the various lipids is expressed in nmol per 106 cells. (B) The y-axis is ratio of 18:1(9) in each lipid per 106 cells. Values are the mean ± SD from three independent experiments. Asterisks indicate a statistically significant difference compared with VC based on a two-tailed Student's t-test (*P < 0.05 and **P < 0.01).
Discussion
Whereas TAG amounts increase under N starvation, MGDG, DGDG, SQDG, PG, and PI decrease in C. reinhardtii cells (Siaut et al., 2011; Sakurai et al., 2014) and the polar glycerolipid contents, represented mainly by MGDG, DGDG, and SQDG and phospholipids PE and PC, are reduced in Nannochloropsis strains (Simionato et al., 2013; Li et al., 2014; Martin et al., 2014). However, we found that SQDG and DGTS accumulated, replacing PG and PC, respectively, in Nannochloropsis cells under P starvation conditions (Figures 3, 10, Supplemental Figure 3). This result corroborates previous reports on P deprivation in several species (Benning et al., 1993, 1995; Essigmann et al., 1998; Sato et al., 2000; Khozin-Goldberg and Cohen, 2006). In Monodus subterraneus, which is a freshwater microalga in the class Eustigmatophyceae, DGTS and SQDG increased under P starvation conditions (Khozin-Goldberg and Cohen, 2006). In A. thaliana, SQDG is synthesized to replace PG in chloroplasts during P starvation so that the amount of anionic thylakoid lipid is maintained (Essigmann et al., 1998; Yu and Benning, 2003). In C. reinhardtii, mechanisms exist that maintain a constant total amount of SQDG and PG under sulfur- or P-limiting conditions (Sato et al., 2000). Therefore, our results suggest that SQDG substitutes for PG, to some extent, to sustain the functional activity of the decreased chloroplast membranes in Nannochloropsis, as in C. reinhardtii.
When the SQD2 promoter is applied, total TAG levels per cell after 4 days of P starvation were ~1.7-fold greater in CrDGTT4-overexpressing lines compared with the control line (Figure 7). However, the difference between CrDGTT4-overexpressing lines and the control line after 6 days of P starvation was less than after 4 days (Supplemental Figure 2). During N starvation in N. oceanica IMET1, seven DGAT transcripts are up-regulated and six other DGAT transcripts are down-regulated (Li et al., 2014). Our results in Nannochloropsis might be explained by the up-regulation of Nannochloropsis's own DGATs, particularly in later stages of P depletion. The levels of 18:1(9) and 20:3 fatty acids bound to TAGs increased during P starvation in comparison with the levels of these fatty acids under standard conditions (Figure 9). The levels of these two fatty acids in TAG molecular species were higher in the CrDGTT4-overexpressing cell line than in the control line during P starvation. This may reflect the specificity of CrDGTT4. In contrast to the TAG fraction, the 18:1(9) molecular ratio of PG decreased in the CrDGTT4-overexpressing cell line than in the control line during P starvation (Figures 11, 12). This fatty acid change may be associated with the preferential incorporation of the 18:1(9) molecular species into the TAG fraction.
It was unexpected that 20:3 fatty acid also increased in the CrDGTT4-overexpressing line (Figures 11, 13). This may have been due to the suppression of desaturation in very long chain fatty acids, like 20:4 and 20:5 (Figure 11 and Supplemental Figures 6, 7), because of accelerated TAG accumulation. Although the exact reason is unclear, 20:3 fatty acid tends to accumulate under P starvation conditions, but not under N starvation conditions (Simionato et al., 2013; Martin et al., 2014). This fact suggests that a specific pathway for fatty acid incorporation is activated during P starvation.
The SQD2 is highly conserved from red algae to plants, as are its primary structures (Figure 4), and the algal SQD2 is also conserved in secondary algae, including Nannochloropsis. This may be due to the importance of the acidic membrane lipid SQDG, which is common to various algal species. The importance of SQDG for photosystem II was demonstrated clearly for C. reinhardtii and the cyanobacterium Synechocystis sp. PCC6803 through the characterization of mutants from each species that are deficient in the ability to synthesize SQDG (Minoda et al., 2002; Sato et al., 2003; Aoki et al., 2004; Sato, 2004). Recent work on higher plant SQD2 genes indicated that SQD2 is also involved with another acidic lipid, glucuronosyldiacylglycerol (Okazaki et al., 2013). It is possible that algal SQD2 also produces glucuronosyldiacylglycerol, which would thus be more important under P starvation conditions.
Our results showed that the SQD2 promoter from the green alga C. reinhardtii is useful as heterologous promoter in Nannochloropsis strain NIES-2145 cells. The results of this study suggest that there is a common expression control system in a wide range of algal species, including primary and secondary microalgae, for adaptation to low P. This result, together with the occurrence of SQDG synthesis genes throughout algal species, suggests that a membrane remodeling system in chloroplasts under low-P stress is itself common among various microalgae and, thus, related genes and cis- and trans-acting elements are highly conserved. This also suggests that the SQD2 promoter from C. reinhardtii could be applied in various algae.
Arabidopsis SQD2 contains three PHR1-binding sequence (P1BS) motifs in its promoter. The PHR1 transcription factor binds to P1BS elements containing the consensus sequence “GNATATNC” in the promoters of a large number of P starvation–responsive genes (Franco-Zorrilla et al., 2004; Bustos et al., 2010; Pant et al., 2015). PHR1 is a well-described transcriptional regulator of P starvation in the MYB family. Most of the genes and their promoters that are involved in phospholipid degradation and glycolipid biosynthesis contain P1BS motifs in Arabidopsis (Pant et al., 2015). As there are no significant effects of a PHR1 null allele on DGDG synthesis during P limitation in Arabidopsis, a role for PHR1 in lipid remodeling was thus excluded (Gaude et al., 2008). However, glycerolipids, including MGDG, DGDG and SQDG, their composition and the expression of most lipid-remodeling gene transcripts analyzed were all altered in the phr1 mutant under P starvation when compared with the wild type (Pant et al., 2015). We used the C. reinhardtii SQD2 gene (Cre01.g038550) promoter for CrDGTT4 overexpression. Another SQD2 gene homolog (Cre16.g689150) exists in C. reinhardtii. Both C. reinhardtii SQD2 genes were up-regulated under P starvation (data not shown). We searched the promoter regions of these genes, and they do not contain P1BS motifs in their promoters but have binding motifs for many other transcription factors. C. reinhardtii SQD2 has 14 MYB or MYC binding domains in its promoter region, whereas Nannochloropsis oceanica SQD2 has 12 MYB or MYC binding domains in its promoter region (Supplemental Figure 8). There is no reliable evidence revealing which motif offers the most effective transcriptional regulation under P starvation. Further work is necessary to understand the regulation of lipid remodeling during P stress in algae.
Author contributions
MI prepared samples of Nannochloropsis strain NIES-2145 for each experiment. KH performed in silico analysis. YS performed GC-MS analysis. MI, KH, YS and HO wrote the manuscript. MS and HO planned the project.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was partially supported by the KAO Corporation and JST CREST from the Ministry of Education, Sports, Science and Culture in Japan.
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2015.00912
Changes in the TAG content of Nannochloropsis in response to N or P starvation. Cells cultured to the logarithmic phase under f/2 medium were then inoculated into f/2, -N or -P medium and cultured for 13 d.
Changes in the TAG content of CrDGTT4-overexpressing cells in response to P starvation. Nannochloropsis cells were transformed with pCrSQD2-CrDGTT4 to overexpress CrDGTT4. Cells cultured to the logarithmic phase under standard conditions were then inoculated into control (+P) or -P medium and cultured for 6 days (6 d). The transformants (#8, #9, and #21), the empty vector control (VC) and the wild type (WT) are shown. Values represent the mean ± SD from three independent replicates.
Changes in major lipid classes in P-starved cells. Black bars, the vector control (VC) 4 days in control medium (+P); white bars, VC 4 days in -P; dark gray bars, pCrSQD2-CrDGTT4 (#9) 4 days in control medium (+P); and light gray bars, pCrSQD2-CrDGTT4 (#9) 4 days in -P. Values are the mean ± SD from three independent experiments. Asterisks indicate a statistically significant difference compared with VC based on a two-tailed Student's t-test (*P < 0.05). Small letters indicate statistically significant difference compared with the control medium based on a two-tailed Student's t-test (aP < 0.05 and bP < 0.01).
Quantitative analysis and ratio of 20:5 in the various lipids. Cells were cultured in control (+P) or -P medium for 4 days. (A) Each 20:5 in the various lipids is expressed in nmol per 106 cells. (B) The y-axis is ratio of 20:5 in each lipid per 106 cells. Values are the mean ± SD from three independent experiments. Asterisks indicate a statistically significant difference compared with VC based on a two-tailed Student's t-test (*P < 0.05).
Quantitative analysis and ratio of 16:0 in the various lipids. Cells were cultured in control (+P) or -P medium for 4 days. (A) Each 16:0 in the various lipids is expressed in nmol per 106 cells. (B) The y-axis is ratio of 16:0 in each lipid per 106 cells. Values are the mean ± SD from three independent experiments. Asterisks indicate a statistically significant difference compared with VC based on a two-tailed Student's t-test (*P < 0.05).
Quantitative analysis and ratio of 16:1 in the various lipids. Cells were cultured in control (+P) or –P medium for 4 days. (A) Each 16:1 in the various lipids is expressed in nmol per 106 cells. (B) The y-axis is ratio of 16:1 in each lipid per 106 cells. Values are the mean ± SD from three independent experiments. Asterisks indicate a statistically significant difference compared with VC based on a two-tailed Student's t-test (*P < 0.05).
Quantitative analysis and ratio of 20:4 in the various lipids. Cells were cultured in control (+P) or -P medium for 4 days. (A) Each 20:4 in the various lipids is expressed in nmol per 106 cells. (B) The y-axis is ratio of 20:4 in each lipid per 106 cells. Values are the mean ± SD from three independent experiments. Asterisks indicate a statistically significant difference compared with VC based on a two-tailed Student's t-test (*P < 0.05).
MYB/MYC binding motif in the SQD2 promoter region of Nannochloropsis oceanica and Chlamydomonas reinhardtii. Yellow character, CTGTTA (+) strand MYB/MYC binding motif; red character, (+) and (−) strand MYB/MYC binding motif; blue character, (−) strand MYB/MYC binding motif; green character, CDS; pale green character, UTR region.
Primer sequences.
References
- Aoki M., Sato N., Meguro A., Tsuzuki M. (2004). Differing involvement of sulfoquinovosyl diacylglycerol in photosystem II in two species of unicellular cyanobacteria. Eur. J. Biochem. 271, 685–693. 10.1111/j.1432-1033.2003.03970.x [DOI] [PubMed] [Google Scholar]
- Arudchelvam Y., Nirmalakhandan N. (2012). Optimizing net energy gain in algal cultivation for biodiesel production. Bioresour. Technol. 114, 294–302. 10.1016/j.biortech.2012.02.126 [DOI] [PubMed] [Google Scholar]
- Benning C., Beatty J. T., Prince R. C., Somerville C. R. (1993). The sulfolipid sulfoquinovosyldiacylglycerol is not required for photosynthetic electron transport in Rhodobacter sphaeroides but enhances growth under phosphate limitation. Proc. Natl. Acad. Sci. U.S.A. 90, 1561–1565. 10.1073/pnas.90.4.1561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benning C., Huang Z. H., Gage D. A. (1995). Accumulation of a novel glycolipid and a betaine lipid in cells of Rhodobacter sphaeroides grown under phosphate limitation. Arch. Biochem. Biophys 317, 103–111. 10.1006/abbi.1995.1141 [DOI] [PubMed] [Google Scholar]
- Bligh E. G., Dyer W. J. (1959). A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37, 911–917. 10.1139/o59-099 [DOI] [PubMed] [Google Scholar]
- Bondioli P., Della Bella L., Rivolta G., Chini Zittelli G., Bassi N., Rodolfi L., et al. (2012). Oil production by the marine microalgae Nannochloropsis sp. F&M-M24 and Tetraselmis suecica F&M-M33. Bioresour. Technol. 114, 567–572. 10.1016/j.biortech.2012.02.123 [DOI] [PubMed] [Google Scholar]
- Boyle N. R., Page M. D., Liu B., Blaby I. K., Casero D., Kropat J., et al. (2012). Three acyltransferases and nitrogen-responsive regulator are implicated in nitrogen starvation-induced triacylglycerol accumulation in Chlamydomonas. J. Biol. Chem. 287, 15811–15825. 10.1074/jbc.M111.334052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brueggeman A. J., Kuehler D., Weeks D. P. (2014). Evaluation of three herbicide resistance genes for use in genetic transformations and for potential crop protection in algae production. Plant Biotechnol. J. 12, 894–902. 10.1111/pbi.12192 [DOI] [PubMed] [Google Scholar]
- Bustos R., Castrillo G., Linhares F., Puga M. I., Rubio V., Pérez-Pérez J., et al. (2010). A central regulatory system largely controls transcriptional activation and repression responses to phosphate starvation in Arabidopsis. PLoS Genet. 6:e1001102. 10.1371/journal.pgen.1001102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cases S., Smith S. J., Zheng Y. W., Myers H. M., Lear S. R., Sande E., et al. (1998). Identification of a gene encoding an acyl CoA:diacylglycerol acyltransferase, a key enzyme in triacylglycerol synthesis. Proc. Natl. Acad. Sci. U.S.A. 95, 13018–13023. 10.1073/pnas.95.22.13018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C. W., Moseley J. L., Wykoff D., Grossman A. R. (2005). The LPB1 gene is important for acclimation of Chlamydomonas reinhardtii to phosphorus and sulfur deprivation. Plant Physiol. 138, 319–329. 10.1104/pp.105.059550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Converti A., Casazza A. A., Ortiz E. Y., Perego P., Borghi M. D. (2009). Effect of temperature and nitrogen concentration on the growth and lipid content of Nannochloropsis oculata and Chlorella vulgaris for biodiesel production. Chem. Eng. Process. 48, 1146–1151. 10.1016/j.cep.2009.03.006 [DOI] [Google Scholar]
- Eichler-Stahlberg A., Weisheit W., Ruecker O., Heitzer M. (2009). Strategies to facilitate transgene expression in Chlamydomonas reinhardtii. Planta 229, 873–883. 10.1007/s00425-008-0879-x [DOI] [PubMed] [Google Scholar]
- Essigmann B., Güler S., Narang R. A., Linke D., Benning C. (1998). Phosphate availability affects the thylakoid lipid composition and the expression of SQD1, a gene required for sulfolipid biosynthesis in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 95, 1950–1955. 10.1073/pnas.95.4.1950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco-Zorrilla J. M., González E., Bustos R., Linhares F., Leyva A., Paz-Ares J. (2004). The transcriptional control of plant responses to phosphate limitation. J. Exp. Bot. 55, 285–293. 10.1093/jxb/erh009 [DOI] [PubMed] [Google Scholar]
- Gaude N., Nakamura Y., Scheible W. R., Ohta H., Dörmann P. (2008). Phospholipase C5 (NPC5) is involved in galactolipid accumulation during phosphate limitation in leaves of Arabidopsis. Plant J. 56, 28–39. 10.1111/j.1365-313X.2008.03582.x [DOI] [PubMed] [Google Scholar]
- Giroud C., Gerber A., Eichenberger W. (1988). Lipids of Chlamydomonas reinhardtii. Analysis of molecular species and intracellular site(s) of biosynthesis. Plant Cell Physiol. 29, 587–595. [Google Scholar]
- Grossman A. (2000). Acclimation of Chlamydomonas reinhardtii to its nutrient environment. Protist 151, 201–224. 10.1078/1434-4610-00020 [DOI] [PubMed] [Google Scholar]
- Guillard R. R., Ryther J. H. (1962). Studies of marine planktonic diatoms. I. Cyclotella nana Hustedt, and Detonula confervacea (cleve) Gran. Can. J. Microbiol. 8, 229–239. 10.1139/m62-029 [DOI] [PubMed] [Google Scholar]
- Guschina I. A., Harwood J. L. (2006). Lipids and lipid metabolism in eukaryotic algae. Prog. Lipid. Res. 45, 160–186. 10.1016/j.plipres.2006.01.001 [DOI] [PubMed] [Google Scholar]
- Harris E. H. (2009). The Chlamydomonas Sourcebook: Introduction to Chlamydomonas and its Laboratory Use, 2nd Edn. Oxford, UK: AcademicPress. [Google Scholar]
- Hirata R., Jeong W. J., Saga N., Mikami K. (2011). Heterologous activation of the Porphyra tenera HSP70 promoter in Bangiophycean algal cells. Bioeng. Bugs 2, 271–274. 10.4161/bbug.2.5.16938 [DOI] [PubMed] [Google Scholar]
- Hodgson P. A., Henderson R. J., Sargent J. R., Leftley J. W. (1991). Patterns of variation in the lipid class and fatty acid composition of Nannochloropsis oculata (Eustigmatophyceae) during batch culture. J. Appl. Phycol. 3, 169–181. 10.1007/BF00003699 [DOI] [Google Scholar]
- Hu H., Gao K. (2006). Response of growth and fatty acid compositions of Nannochloropsis sp. to environmental factors under elevated CO2 concentration. Biotechnol. Lett. 28, 987–992. 10.1007/s10529-006-9026-6 [DOI] [PubMed] [Google Scholar]
- Hu Q., Sommerfeld M., Jarvis E., Ghirardi M., Posewitz M., Seibert M., et al. (2008). Microalgal triacylglycerols as feedstocks for biofuel production: perspectives and advances. Plant J. 54, 621–639. 10.1111/j.1365-313X.2008.03492.x [DOI] [PubMed] [Google Scholar]
- Huerlimann R., de Nys R., Heimann K. (2010). Growth, lipid content, productivity, and fatty acid composition of tropical microalgae for scale-up production. Biotechnol. Bioeng. 107, 245–257. 10.1002/bit.22809 [DOI] [PubMed] [Google Scholar]
- Iwai M., Ikeda K., Shimojima M., Ohta H. (2014). Enhancement of extraplastidic oil synthesis in Chlamydomonas reinhardtii using a type-2 diacylglycerol acyltransferase with a phosphorus starvation-inducible promoter. Plant Biotechnol. J. 12, 808–819. 10.1111/pbi.12210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Standley D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khozin-Goldberg I., Cohen Z. (2006). The effect of phosphate starvation on the lipid and fatty acid composition of the fresh water eustigmatophyte Monodus subterraneus. Phytochemistry 67, 696–701. 10.1016/j.phytochem.2006.01.010 [DOI] [PubMed] [Google Scholar]
- Kilian O., Benemann C. S., Niyogi K. K., Vick B. (2011). High-efficiency homologous recombination in the oil-producing alga Nannochloropsis sp. Proc. Natl. Acad. Sci. U.S.A. 108, 21265–21269. 10.1073/pnas.1105861108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lardizabal K. D., Mai J. T., Wagner N. W., Wyrick A., Voelker T., Hawkins D. J. (2001). DGAT2 is a new diacylglycerol acyltransferase gene family: purification, cloning, and expression in insect cells of two polypeptides from Mortierella ramanniana with diacylglycerol acyltransferase activity. J. Biol. Chem. 276, 38862–38869. 10.1074/jbc.M106168200 [DOI] [PubMed] [Google Scholar]
- Lerche K., Hallmann A. (2013). Stable nuclear transformation of Eudorina elegans. BMC Biotechnol. 13:11. 10.1186/1472-6750-13-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerche K., Hallmann A. (2014). Stable nuclear transformation of Pandorina morum. BMC Biotechnol. 14:65. 10.1186/1472-6750-14-65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Han D., Wang D., Ning K., Jia J., Wei L., et al. (2014). Choreography of transcriptomes and lipidomes of Nannochloropsis reveals the mechanisms of oil synthesis in microalgae. Plant Cell 26, 1645–1665. 10.1105/tpc.113.121418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li-Beisson Y., Shorrosh B., Beisson F., Andersson M. X., Arondel V., Bates P. D., et al. (2010). Acyl-lipid metabolism. Arabidopsis Book 8:e0133. 10.1199/tab.0133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lung S. C., Weselake R. J. (2006). Diacylglycerol acyltransferase: a key mediator of plant triacylglycerol synthesis. Lipids 41, 1073–1088. 10.1007/s11745-006-5057-y [DOI] [PubMed] [Google Scholar]
- Martin G. J., Hill D. R., Olmstead I. L., Bergamin A., Shears M. J., Dias D. A., et al. (2014). Lipid profile remodeling in response to nitrogen deprivation in the microalgae Chlorella sp. (Trebouxiophyceae) and Nannochloropsis sp. (Eustigmatophyceae). PLoS ONE 9:e103389. 10.1371/journal.pone.0103389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama I., Nakamura T., Matubayashi T., Ando Y., Maeda T. (1986). Identification of the alga known as “marine Chlorella” as a member of the Eustigmatophyceae. Jpn. J. Phycol. 34, 319–325. [Google Scholar]
- Miller R., Wu G., Deshpande R. R., Vieler A., Gärtner K., Li X., et al. (2010). Changes in transcript abundance in Chlamydomonas reinhardtii following nitrogen deprivation predict diversion of metabolism. Plant Physiol. 154, 1737–1752. 10.1104/pp.110.165159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minoda A., Sato N., Nozaki H., Okada K., Takahashi H., Sonoike K., et al. (2002). Role of sulfoquinovosyl diacylglycerol for the maintenance of photosystem II in Chlamydomonas reinhardtii. Eur. J. Biochem. 269, 2353–2358. 10.1046/j.1432-1033.2002.02896.x [DOI] [PubMed] [Google Scholar]
- Okazaki Y., Otsuki H., Narisawa T., Kobayashi M., Sawai S., Kamide Y., et al. (2013). A new class of plant lipid is essential for protection against phosphorus depletion. Nat. Commun. 4, 1510. 10.1038/ncomms2512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki Y., Shimojima M., Sawada Y., Toyooka K., Narisawa T., Mochida K., et al. (2009). A chloroplastic UDP-glucose pyrophosphorylase from Arabidopsis is the committed enzyme for the first step of sulfolipid biosynthesis. Plant Cell 21, 892–909. 10.1105/tpc.108.063925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pant B. D., Burgos A., Pant P., Cuadros-Inostroza A., Willmitzer L., Scheible W. R. (2015). The transcription factor PHR1 regulates lipid remodeling and triacylglycerol accumulation in Arabidopsis thaliana during phosphorus starvation. J. Exp. Bot. 66, 1907–1918. 10.1093/jxb/eru535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil V., Källqvist T., Olsen E., Vogt G., Gislerød H. R. (2007). Fatty acid composition of 12 microalgae for possible use in aquaculture feed. Aquacult Int. 15, 1–9. 10.1007/s10499-006-9060-3 [DOI] [Google Scholar]
- Radakovits R., Jinkerson R. E., Fuerstenberg S. I., Tae H., Settlage R. E., Boore J. L., et al. (2012). Draft genome sequence and genetic transformation of the oleaginous alga Nannochloropis gaditana. Nat. Commun. 3, 686 10.1038/ncomms1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajakumari S., Grillitsch K., Daum G. (2008). Synthesis and turnover of non-polar lipids in yeast. Prog. Lipid. Res. 47, 157–171. 10.1016/j.plipres.2008.01.001 [DOI] [PubMed] [Google Scholar]
- Rodolfi L., Chini Zittelli G., Bassi N., Padovani G., Biondi N., Bonini G., et al. (2009). Microalgae for oil: strain selection, induction of lipid synthesis and outdoor mass cultivation in a low-cost photobioreactor. Biotechnol. Bioeng. 102, 100–112. 10.1002/bit.22033 [DOI] [PubMed] [Google Scholar]
- Ronquist F., Teslenko M., Van der Mark P., Ayres D. L., Darling A., Höhna S., et al. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. 10.1093/sysbio/sys029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai K., Moriyama T., Sato N. (2014). Detailed identification of fatty acid isomers sheds light on the probable precursors of triacylglycerol accumulation in photoautotrophically grown Chlamydomonas reinhardtii. Eukaryot. Cell 13, 256–266. 10.1128/EC.00280-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato N. (2004). Roles of the acidic lipids sulfoquinovosyl diacylglycerol and phosphatidylglycerol in photosynthesis: their specificity and evolution. J. Plant Res. 117, 495–505. 10.1007/s10265-004-0183-1 [DOI] [PubMed] [Google Scholar]
- Sato N., Aoki M., Maru Y., Sonoike K., Minoda A., Tsuzuki M. (2003). Involvement of sulfoquinovosyl diacylglycerol in the structural integrity and heat-tolerance of photosystem II. Planta 217, 245–251. 10.1007/s00425-003-0992-9 [DOI] [PubMed] [Google Scholar]
- Sato N., Furuya M. (1985). Distribution of diacylglyceryltrimethylhomoserine and phosphatidylcholine in non-vascular green plants. Plant Sci. 38, 81–85. 10.1016/0168-9452(85)90134-7 [DOI] [Google Scholar]
- Sato N., Hagio M., Wada H., Tsuzuki M. (2000). Environmental effects on acidic lipids of thylakoid membranes. Biochem. Soc. Trans. 28, 912–914. 10.1042/bst0280912 [DOI] [PubMed] [Google Scholar]
- Shockey J. M., Gidda S. K., Chapital D. C., Kuan J. C., Dhanoa P. K., Bland J. M., et al. (2006). Tung tree DGAT1 and DGAT2 have nonredundant functions in triacylglycerol biosynthesis and are localized to different subdomains of the endoplasmic reticulum. Plant Cell 18, 2294–2313. 10.1105/tpc.106.043695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siaut M., Cuiné S., Cagnon C., Fessler B., Nguyen M., Carrier P., et al. (2011). Oil accumulation in the model green alga Chlamydomonas reinhardtii: characterization, variability between common laboratory strains and relationship with starch reserves. BMC Biotechnol. 11:7. 10.1186/1472-6750-11-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simionato D., Block M. A., La Rocca N., Jouhet J., Maréchal E., Finazzi G., et al. (2013). The response of Nannochloropsis gaditana to nitrogen starvation includes de novo biosynthesis of triacylglycerols, a decrease of chloroplast galactolipids, and reorganization of the photosynthetic apparatus. Eukaryot. Cell 12, 665–676. 10.1128/EC.00363-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simionato D., Sforza E., Corteggiani Carpinelli E., Bertucco A., Giacometti G. M., Morosinotto T. (2011). Acclimation of Nannochloropsis gaditana to different illumination regimes: effects on lipids accumulation. Bioresour. Technol. 102, 6026–6032. 10.1016/j.biortech.2011.02.100 [DOI] [PubMed] [Google Scholar]
- Talavera G., Castresana J. (2007). Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 56, 564–577. 10.1080/10635150701472164 [DOI] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. (2013). MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanabe A. S. (2011). Kakusan4 and Aminosan: two programs for comparing nonpartitioned, proportional and separate models for combined molecular phylogenetic analyses of multilocus sequence data. Mol. Ecol. Resour. 11, 914–921. 10.1111/j.1755-0998.2011.03021.x [DOI] [PubMed] [Google Scholar]
- Vieler A., Brubaker S. B., Vick B., Benning C. (2012a). A lipid droplet protein of Nannochloropsis with functions partially analogous to plant oleosins. Plant Physiol. 158, 1562–1569. 10.1104/pp.111.193029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieler A., Wu G., Tsai C. H., Bullard B., Cornish A. J., Harvey C., et al. (2012b). Genome, functional gene annotation, and nuclear transformation of the heterokont oleaginous alga Nannochloropsis oceanica CCMP1779. PLoS Genet. 8:e1003064. 10.1371/journal.pgen.1003064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Ning K., Li J., Hu J., Han D., Wang H., et al. (2014). Nannochloropsis genomes reveal evolution of microalgal oleaginous traits. PLoS Genet. 10:e1004094. 10.1371/journal.pgen.1004094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weyer K. M., Bush D. R., Darzins A., Wilso B. D. (2010). Theoretical maximum algal oil production. Bioenergy Res. 3, 204–213. 10.1007/s12155-009-9046-x [DOI] [Google Scholar]
- Yu B., Benning C. (2003). Anionic lipids are required for chloroplast structure and function in Arabidopsis. Plant J. 36, 762–770. 10.1046/j.1365-313X.2003.01918.x [DOI] [PubMed] [Google Scholar]
- Yu B., Xu C., Benning C. (2002). Arabidopsis disrupted in SQD2 encoding sulfolipid synthase is impaired in phosphate-limited growth. Proc. Natl. Acad. Sci. U.S.A. 99, 5732–5737. 10.1073/pnas.082696499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Shrager J., Jain M., Chang C. W., Vallon O., Grossman A. R. (2004). Insights into the survival of Chlamydomonas reinhardtii during sulfur starvation based on microarray analysis of gene expression. Eukaryot. Cell 3, 1331–1348. 10.1128/EC.3.5.1331-1348.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Changes in the TAG content of Nannochloropsis in response to N or P starvation. Cells cultured to the logarithmic phase under f/2 medium were then inoculated into f/2, -N or -P medium and cultured for 13 d.
Changes in the TAG content of CrDGTT4-overexpressing cells in response to P starvation. Nannochloropsis cells were transformed with pCrSQD2-CrDGTT4 to overexpress CrDGTT4. Cells cultured to the logarithmic phase under standard conditions were then inoculated into control (+P) or -P medium and cultured for 6 days (6 d). The transformants (#8, #9, and #21), the empty vector control (VC) and the wild type (WT) are shown. Values represent the mean ± SD from three independent replicates.
Changes in major lipid classes in P-starved cells. Black bars, the vector control (VC) 4 days in control medium (+P); white bars, VC 4 days in -P; dark gray bars, pCrSQD2-CrDGTT4 (#9) 4 days in control medium (+P); and light gray bars, pCrSQD2-CrDGTT4 (#9) 4 days in -P. Values are the mean ± SD from three independent experiments. Asterisks indicate a statistically significant difference compared with VC based on a two-tailed Student's t-test (*P < 0.05). Small letters indicate statistically significant difference compared with the control medium based on a two-tailed Student's t-test (aP < 0.05 and bP < 0.01).
Quantitative analysis and ratio of 20:5 in the various lipids. Cells were cultured in control (+P) or -P medium for 4 days. (A) Each 20:5 in the various lipids is expressed in nmol per 106 cells. (B) The y-axis is ratio of 20:5 in each lipid per 106 cells. Values are the mean ± SD from three independent experiments. Asterisks indicate a statistically significant difference compared with VC based on a two-tailed Student's t-test (*P < 0.05).
Quantitative analysis and ratio of 16:0 in the various lipids. Cells were cultured in control (+P) or -P medium for 4 days. (A) Each 16:0 in the various lipids is expressed in nmol per 106 cells. (B) The y-axis is ratio of 16:0 in each lipid per 106 cells. Values are the mean ± SD from three independent experiments. Asterisks indicate a statistically significant difference compared with VC based on a two-tailed Student's t-test (*P < 0.05).
Quantitative analysis and ratio of 16:1 in the various lipids. Cells were cultured in control (+P) or –P medium for 4 days. (A) Each 16:1 in the various lipids is expressed in nmol per 106 cells. (B) The y-axis is ratio of 16:1 in each lipid per 106 cells. Values are the mean ± SD from three independent experiments. Asterisks indicate a statistically significant difference compared with VC based on a two-tailed Student's t-test (*P < 0.05).
Quantitative analysis and ratio of 20:4 in the various lipids. Cells were cultured in control (+P) or -P medium for 4 days. (A) Each 20:4 in the various lipids is expressed in nmol per 106 cells. (B) The y-axis is ratio of 20:4 in each lipid per 106 cells. Values are the mean ± SD from three independent experiments. Asterisks indicate a statistically significant difference compared with VC based on a two-tailed Student's t-test (*P < 0.05).
MYB/MYC binding motif in the SQD2 promoter region of Nannochloropsis oceanica and Chlamydomonas reinhardtii. Yellow character, CTGTTA (+) strand MYB/MYC binding motif; red character, (+) and (−) strand MYB/MYC binding motif; blue character, (−) strand MYB/MYC binding motif; green character, CDS; pale green character, UTR region.
Primer sequences.