Abstract
The identification of cell-free fetal DNA (cffDNA) in maternal plasma in 1997 heralded the most significant change in obstetric care for decades, with the advent of safer screening and diagnosis based on analysis of maternal blood. Here, we describe how the technological advances offered by next-generation sequencing have allowed for the development of a highly sensitive screening test for aneuploidies as well as definitive prenatal molecular diagnosis for some monogenic disorders.
Next-generation sequencing technologies have facilitated the development of sensitive noninvasive prenatal diagnostic tests for aneuploidies and some monogenic disorders. Ethical and economic barriers may complicate their clinical application.
Since the identification of cell-free fetal DNA (cffDNA) in maternal plasma in the late 1990s (Lo et al. 1997), there has been considerable progress in developing safer methods for noninvasive prenatal testing (NIPT) based on analysis of nucleic acids circulating in maternal blood. Analysis of cffDNA was rapidly incorporated into clinical practice, albeit largely on a research basis for the first few years, for the identification of alleles not present in the mother but present in her blood because they had been inherited from the father or arisen de novo at conception. The first clinical applications included fetal sex determination in pregnancies at risk of sex-linked conditions to direct invasive testing and in utero therapy (Costa et al. 2002; Hyett et al. 2005), and fetal RHD typing in pregnancies of RhD-negative mothers to direct the monitoring of pregnancies at high-risk of hemolytic disease of the newborn (Faas et al. 1998; Lo et al. 1998a; Finning et al. 2002). This was followed shortly by reports of NIPT for selected single gene disorders (Saito et al. 2000; Chiu et al. 2002). These early noninvasive prenatal tests were predominantly based on applying various polymerase chain reaction (PCR) approaches to detect or exclude mutations in maternal plasma (Lench et al. 2013). With the advent of massively parallel sequencing (MPS) and digital PCR, applications have rapidly expanded to include a highly sensitive screening test for aneuploidies (Chiu et al. 2008; Fan et al. 2008; Boon and Faas 2013), the ability to determine both the paternal and maternal origins of fetal mutations in monogenic conditions (Lun et al. 2008a; Lam et al. 2012; New et al. 2014), and the use of multiple gene panels to screen for multiple disease-causing mutations (Lench et al. 2013). Progress has been rapid (Table 1) (Chitty and Bianchi 2013). Here, we discuss the technological developments that have enabled this progress, and the challenges that this progress brings when implementing advances in technology into a public healthcare system.
Table 1.
Chronology of development of cell-free fetal testing (1997–2014)
| Year | Milestone | Technology | Reference | Clinical availability |
|---|---|---|---|---|
| 1997 | Discovery of cffDNA in the maternal plasma | Lo et al. 1997 | ||
| 1997 | Fetal sex determination using cffDNA | PCR | Lo et al.1997 | |
| 1998 | RHD genotyping in RhD-negative mothers | PCR | Faas et al. 1998; Lo et al.1998a | |
| 2000 | NIPD test for single-gene disorders | PCR | Amicucci et al. 2000; Saito et al. 2000 | |
| 2001 | Fetal sex determination using cffDNA available in clinical practice | PCR | Research basis | |
| 2001 | Fetal RHD genotyping using cffDNA available in clinical practice in high-risk pregnancies | PCR | Finning et al. 2002 | Public Health Service |
| 2002 | Prenatal exclusion for autosomal recessive conditions (cystic fibrosis, β-thalassemia, and CAH) | PCR | Chiu et al. 2002; Gonzalez-Gonzalez et al. 2002 | |
| 2006 | Methylation-based approach for trisomy 18 | Methylation-specific PCR and mass spectrometry | Tong et al. 2006 | |
| 2007 | RhC, RhE, and Kell geno-typing service | PCR | Finning et al. 2007 | Public Health Service |
| 2007 | RNA-based approach for trisomy 21 | Digital PCR | Lo et al. 2007a | |
| 2007 | Molecular counting for trisomy 21 | Digital PCR | Lo et al. 2007b; Fan et al. 2008 | |
| 2008 | Detection of trisomies 21, 18, and 13 | NGS | Chiu et al. 2008; Fan et al. 2008 | |
| 2008 | NIPD for a monogenic disorder in which parents carry the same mutation (male fetuses only) | Digital PCR and RMD | Lun et al. 2008a | |
| 2010 | NIPD for selected single-gene disorders offered clinically | PCR-RED | Chitty et al. 2011 | Research basis |
| 2010 | Whole-fetal genome mapping from maternal plasma | NGS | Lo et al. 2010 | |
| 2010 | NIPD for fetal sex determination approved for use in the NHS | PCR | UKGTN1; Hill et al. 2011 | Public Health Service |
| 2011 | First large validation studies of NIPT for trisomies 21, 18, and 13 in high-risk pregnancies reported | NGS | Chiu et al. 2011; Ehrich et al. 2011; Sehnert et al. 2011 | |
| 2011 | NIPT for trisomies 21, 18, and 13, offered by commercial companies in Hong Kong/China and the United States | NGS | Commercial sector | |
| 2011 | Routine 3rd trimester RHD genotyping in all RhD-women offered to direct anti-RhD immunoglobulin administration | PCR | Clausen et al. 2012; de Haas et al. 2012 | Public Health Service |
| 2012 | NIPD for a monogenic disorder in which parents carry the same mutation reported (male and female fetuses) | NGS | Lam et al. 2012 | |
| 2012 | NIPT for sex chromosome aneuploidies launched as a commercial service | Mazloom et al. 2013 | Commercial sector | |
| 2012 | NIPD for selected single gene disorders approved for use in United Kingdom NHS | PCR-RED | Lench et al. 2013; UKGTN2 | Public Health Service |
| 2013 | NIPD for using gene panels approved for use in United Kingdom NHS | NGS | Chitty et al. 2015; UKGTN3 | Public Health Service |
| 2013 | NIPT for aneuploidy reimbursed by some insurance companies in the United States | NGS | Genomeweb3 | State funded private service |
| 2013 | Evaluation of NIPT for aneuploidy in a public health service begins | NGS | Hill et al. 2014 | |
| 2013 | Molecular karyotyping of subchromosomal abnormalities | NGS | Srinivasan et al. 2013; Yu et al. 2013 | |
| 2013 | NIPT for subchromosomal abnormalities launched as a service | NGS | Sequenom4; Natera5 | Commercial sector |
| 2013/4 | Fetal methylome and transcriptome sequencing | NGS | Koh et al. 2014; Lun et al. 2013; Tsui et al. 2014 |
NIPD, noninvasive prenatal diagnosis; NIPT, noninvasive prenatal testing; CAH, congenital adrenal hyperplasia; NGS, next-generation sequencing, RMD, relative mutation dosage.
BRIEF HISTORICAL SURVEY OF THE APPROACHES FOR ANALYZING cffDNA
Following the discovery of cffDNA in 1997, Lo et al. developed a methodology for the measurement of cffDNA using real-time PCR (Lo et al. 1998b). This study revealed the gestational variations in the absolute and fractional concentrations of cffDNA. In particular, it was shown that fetal DNA represents a mean of some 3%–6% of all DNA in maternal plasma. These preliminary figures have played a critical role in the subsequent development of NIPT, in which the designers of such tests needed to develop assays could work precisely at very low concentrations. This study also showed that the fractional concentrations of cffDNA in maternal plasma are higher than those in maternal serum and established plasma as the preferred sample type for carrying out NIPT. cffDNA is cleared very rapidly from the maternal circulation following parturition, with a half-life of 16 min (Lo et al. 1999b). This is a crucial finding, as it means that cffDNA, unlike some fetal cells (Bianchi et al. 1996), will not persist from one pregnancy to the next, thereby removing one potential source of false results.
These pioneering studies were followed rapidly by those focusing on pathological pregnancies, which showed that the absolute concentrations of cffDNA in maternal plasma/serum in women carrying fetuses with Down syndrome were higher than in those carrying euploid fetuses (Lo et al. 1999a). These results showed that it might be possible to carry out noninvasive prenatal screening for Down syndrome using cffDNA. However, there is considerable overlap in the cffDNA concentrations in the aneuploid and euploid groups and further technological developments were necessary to improve the clinical sensitivity and specificity of such an approach. One development was the demonstration of cell-free fetal RNA in maternal plasma in 2000 (Poon et al. 2000). Another was the demonstration that DNA methylation differences between the mother and fetus could be exploited for the development of unique fetal epigenetic markers that could be detected in maternal plasma (Poon et al. 2002). These two developments were important because they offered the possibility of identifying markers in maternal plasma that are virtually entirely fetal-specific. Using such markers for aneuploidy detection would potentially be more robust, as the fractional contribution of the fetal component for some of such markers could be close to 100%, allowing more reliably detection of a quantitative aberration, as in Down syndrome when the fetus has an extra dose of chromosome 21.
This possibility was first realized for methylation markers with the demonstration that unmethylated SERPINB5, a gene present on chromosome 18, could serve as a fetal-specific marker in maternal plasma (Chim et al. 2005). This was promptly followed by the demonstration that allelic ratio analysis of a single nucleotide polymorphism (SNP) present on the SERPINB5 gene could be used to detect a trisomy 18 fetus noninvasively by analysis of maternal plasma, as long as the fetus was heterozygous for the SNP (Tong et al. 2006). An analogous development took place for fetal RNA markers when it was shown that PLAC4 mRNA, derived from a gene on chromosome 21, could serve as a fetal-specific mRNA marker in maternal plasma (Lo et al. 2007b). It was also shown that allelic ratio analysis of a SNP on the PLAC4 gene could be used for detecting Down syndrome, as long as the fetus was heterozygous for the SNP analyzed. Although these two lines of development further confirm the utility of cell-free nucleic acid markers in maternal plasma for the noninvasive prenatal detection of fetal aneuploidies, they have relatively limited population coverage, as they can only be used for fetuses heterozygous for the SNPs analyzed. This limitation prompted the development of methods that were not dependent on genetic polymorphisms.
In 2007, Lo et al. (2007a) reported that digital PCR, in which multiple PCRs are performed, with each amplification typically containing either a single, or no, target DNA molecule, could be used to detect a fetus with Down syndrome from maternal plasma. At the same time, Fan and Quake (2007) showed a similar concept using artificial DNA mixtures. The core concept of this approach is that a Down syndrome fetus would release an increased amount of chromosome 21 sequences into maternal plasma compared with other chromosomal sequences. Thus, by measuring the precise ratio of a target present on chromosome 21 relative to one present on another chromosome, a diagnosis of trisomy 21 could be mathematically derived. Digital PCR allows measurement of such a ratio relatively precisely, provided that a sufficient number of reactions are performed. The disadvantage of this method is that a very large number of digital PCRs are needed to construct a system that would deliver clinically useful diagnostic sensitivity and specificity. Furthermore, as digital PCR requires that the identity of the detected target is decided ahead of time, diagnostic information contained in plasma DNA molecules not targeted by the digital PCR primers is “wasted.”
MASSIVELY PARALLEL DNA SEQUENCING OF MATERNAL PLASMA
In 2008, two groups reported on the use of random MPS of maternal plasma for detecting fetal chromosomal aneuploidies (Chiu et al. 2008; Fan et al. 2008). The principle of this method is that a fetus with Down syndrome should release a higher amount of chromosome 21 sequences into maternal plasma relative to sequences from the other chromosomes. Thus, by sequencing millions of plasma DNA molecules from maternal plasma and then aligning each sequenced molecule back to its chromosome of origin, a proportional representation of each chromosome in maternal plasma can be calculated. To determine if a fetus has Down syndrome, the proportional representation of chromosome 21 can be compared with a group of healthy pregnant women carrying euploid fetuses. A statistically significant overrepresentation in such a comparison suggests the presence of an aneuploid fetus in the test case. Such an approach has been found to be highly robust and has been replicated in many clinical trials (Ehrich et al. 2011; Palomaki et al. 2011; Sehnert et al. 2011; Bianchi et al. 2012, 2014; Palomaki et al. 2012). As this approach involves MPS in a genome-wide fashion, it can also be applied to detection of other chromosomal aneuploidies, including trisomies 18 and 13 (Chen et al. 2011; Bianchi et al. 2012, 2014; Palomaki et al. 2012), and the sex chromosome aneuploidies (Mazloom et al. 2013). Furthermore, with sufficient depth of DNA sequencing, this approach can also be used for detecting subchromosomal deletions and duplications (Peters et al. 2011; Jensen et al. 2012; Srinivasan et al. 2013; Yu et al. 2013).
The advantage of the random genome-wide sequencing approach is that the same methodology can be used for performing NIPT on a spectrum of chromosomal and subchromosomal aberrations. The disadvantage, however, is that a proportion of the sequencing has been expended on genomic regions that might not be of clinical value and thus could be considered to be not optimally cost-effective. This has prompted the development of approaches based on targeted sequencing, in which the sequencing is focused on selected genomic regions. One method is based on the hybridization-based targeting of selected genomic regions, followed by sequencing of these regions (Norton et al. 2012; Sparks et al. 2012a,b; Ashoor et al. 2013). A second method is based on highly multiplexed PCR-based amplification of many SNPs across chromosomal regions of interest, followed by sequencing of the amplicons (Zimmermann et al. 2012). The allelic ratios of such SNPs are then analyzed and copy number aberrations at the targeted chromosomal regions are deduced from the allelic ratios. This method can be used for detecting subchromosomal microdeletions (Rabinowitz et al. 2014) and a proportion of triploidy cases (Nicolaides et al. 2014). The advantage of these targeted sequencing approaches is that, as the sequencing power is focused on genomic regions of clinical interest, it is possible to analyze more cases per multiplexed sequencing run. The main disadvantage is that the methodology is the need for specific design and optimization for different targets. It is also of interest to note that one group of investigators has recently reported a version of the targeted assay that has substituted sequencing by microarray analysis (Juneau et al. 2014). It will be useful to see independent validation of this work and a comparison of its performance with the sequencing-based approaches.
SIZE-, METHYLATION-, AND TRANSCRIPTOME-BASED APPROACHES
In 2004, it was shown that the size profile of cffDNA was shorter than that of the maternally derived DNA in maternal plasma (Chan et al. 2004). With the advent of paired-end MPS, differences in size profile at single-nucleotide resolution can be elucidated. It has subsequently been found that maternally derived DNA in maternal plasma has a size peak at 166 bp, whereas the cffDNA has a size peak of 143 bp (Lo et al. 2010). Below 143 bp, there are a series of peaks at 10 bp intervals. In 2014, Yu et al. showed that a fetus with Down syndrome would release an extra dose of chromosome 21 sequences into maternal plasma resulting in a reduction in the size distribution of DNA molecules derived from chromosome 21 in maternal plasma (Yu et al. 2014). Such a strategy has also been shown to work for trisomies 18 and 13. Conversely, for a fetus with Turner syndrome, the lack of one copy of fetal chromosome X would result in a lengthening in the size distribution of DNA molecules derived from that chromosome, when compared with that of normal pregnancies with a female fetus. This size-based diagnostic strategy can potentially be used in a synergistic manner with the count-based approaches mentioned in the previous paragraph, and potentially enhance the specificity of plasma DNA-based NIPT.
Earlier in this article, we discussed the use of fetal epigenetic markers for NIPT (Poon et al. 2002; Chim et al. 2005; Tong et al. 2006). With the advent of MPS, it is now possible to sequence the entire plasma methylome through the use of bisulfite conversion of plasma DNA, followed by MPS (Chan et al. 2013; Lun et al. 2013). When DNA is treated with sodium bisulfite, unmethylated cytosine residues are converted into uracil, whereas methylated cytosine residues will remain unchanged. In this way, bisulfite conversion converts an epigenetic change into a genetic one that can be read using DNA sequencing. Lun et al. (2013) performed genome-wide bisulfite MPS on maternal plasma DNA. They were able to deduce the methylome of cffDNA from the sequencing data in two ways. The first involved the elucidation of the methylation status of plasma DNA molecules carrying a paternally inherited fetal allele. The second involved the assumption that maternal plasma DNA was a mixture of fetal and maternal DNA, with the latter being mainly of hematopoietic origin and could be approximated to the methylome of the maternal buffy coat. Hence, provided the fractional concentration of fetal DNA in maternal plasma could be estimated (e.g., using SNP allelic ratios), the fetal methylation level at a particular genomic region using the total methylation level and the methylation status of the maternal buffy coat at that region could be deduced. Lun et al. also studied matched placental samples at the same gestational age. They showed that cffDNA in maternal plasma was hypomethylated and was similar to that of the placenta, thereby providing further powerful evidence that cffDNA is derived from the placenta.
This development has a number of implications. First, it allows one to identify from maternal plasma, genomic regions that are differentially methylated for the fetally derived and maternally derived DNA in maternal plasma. Second, it offers a new approach for the NIPT of chromosomal aneuploidies. A fetus with Down syndrome would release an extra dose of hypomethylated DNA from chromosome 21 into maternal plasma, thereby reducing the methylation level of plasma DNA sequences originating from chromosome 21 compared with samples obtained from euploid pregnancies (Lun et al. 2013). Future work is needed to explore the synergistic utility of this methylation-based approach with the count-based method (Chiu et al. 2008; Fan et al. 2008) and size-based (Yu et al. 2014) methods described above. Theoretically, combining all three methods would allow development of an extremely specific method for NIPT.
Very recently, two groups have reported the profiling of the fetal transcriptome by massively parallel RNA sequencing from maternal plasma (Koh et al. 2014; Tsui et al. 2014). It would be of interest to see the potential application of such an approach for the detection and monitoring pathological pregnancies, such as those affected by preeclampsia and preterm labor.
MONOGENIC DISEASES
In 2010, it was shown that by using MPS of maternal plasma DNA together with knowledge of the genetic maps of the paternal and maternal DNA, the fetal genome could be deduced from the maternal plasma DNA sequencing data (Lo et al. 2010). These results have been reproduced by a number of independent investigators (Fan et al. 2012; Kitzman et al. 2012; Chen et al. 2013) and have potential to be applied to enable NIPT of a broad spectrum of monogenic diseases. To make this approach more cost-effective, Lam et al. (2012) and New et al. (2014) applied targeted versions of this approach to the genomic regions flanking the HBB gene and the CYP21A2 gene, for the NIPT of β-thalassemia and congenital adrenal hyperplasia (CAH) (Ma et al. 2014), respectively. Using this approach for NIPT of a monogenic autosomal recessive disease, the method is typically applied to a family with a previously affected child. In this situation, genomic DNA samples from the affected child and parents are analyzed to determine the haplotype structure of SNPs flanking the disease gene and to deduce the linkage of the parental haplotypes to the mutant and normal genes. This is followed by targeted sequencing of SNPs using maternal plasma DNA obtained from the pregnant woman. In one application of this approach, this targeted sequencing assay can also be used to determine the haplotype structure using the genomic DNA (New et al. 2014). The analysis of the maternal plasma DNA sequencing data is performed in two phases (Fig. 1). The first is to find which SNP alleles in maternal plasma the fetus has inherited from its father, but also which are absent in the mother’s genome. This determines which of the two paternal haplotypes the fetus has inherited in the disease gene region. The second is to determine the relative dosage of the two maternal haplotypes in maternal plasma. The maternal haplotype that is in excess in maternal plasma is the one inherited by the fetus. This strategy is known as the relative haplotype dosage (RHDO) approach (Lam et al. 2012; Lo et al. 2010; New et al. 2014). Because of the statistical power conferred by the multiple SNPs targeted (typically in the range of thousands), the diagnostic sensitivity and specificity of this approach seems to be very high. However, the number of cases published in the literature is still relatively small and future large-scale validation is needed to confirm these results. Furthermore, it is labor intensive, expensive, and requires knowledge of both parental genomes. For applications in which one wishes to directly target the disease-causing mutation, instead of the SNPs linked to the mutation, the digital PCR-based approach can be used to measure the relatively dosage of the mutant and normal alleles in maternal plasma. This approach is known as the relative mutation dosage (RMD) approach (Lun et al. 2008a; Tsui et al. 2011; Barrett et al. 2012).
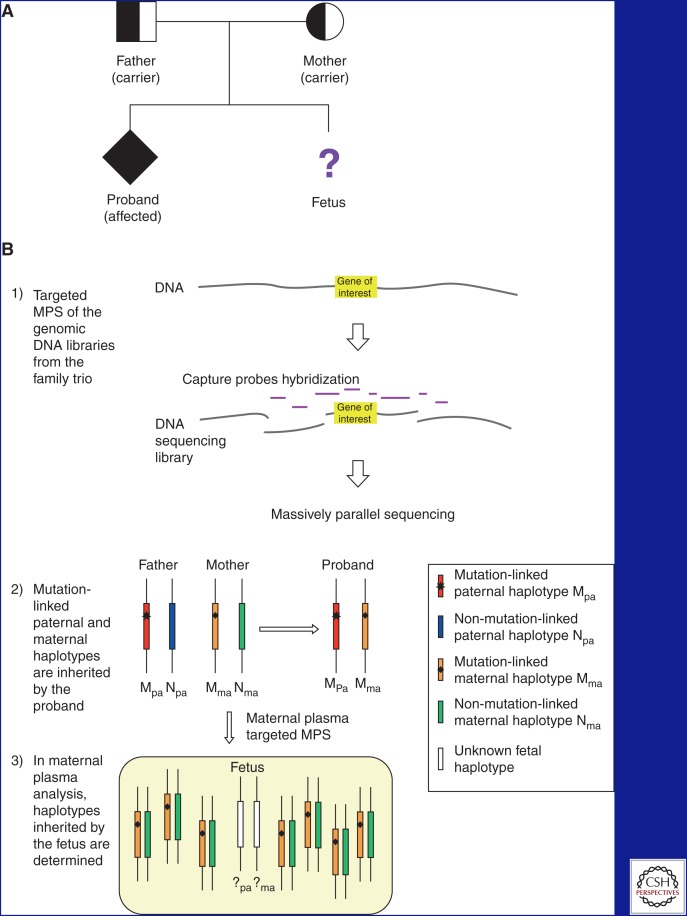
Figure 1.
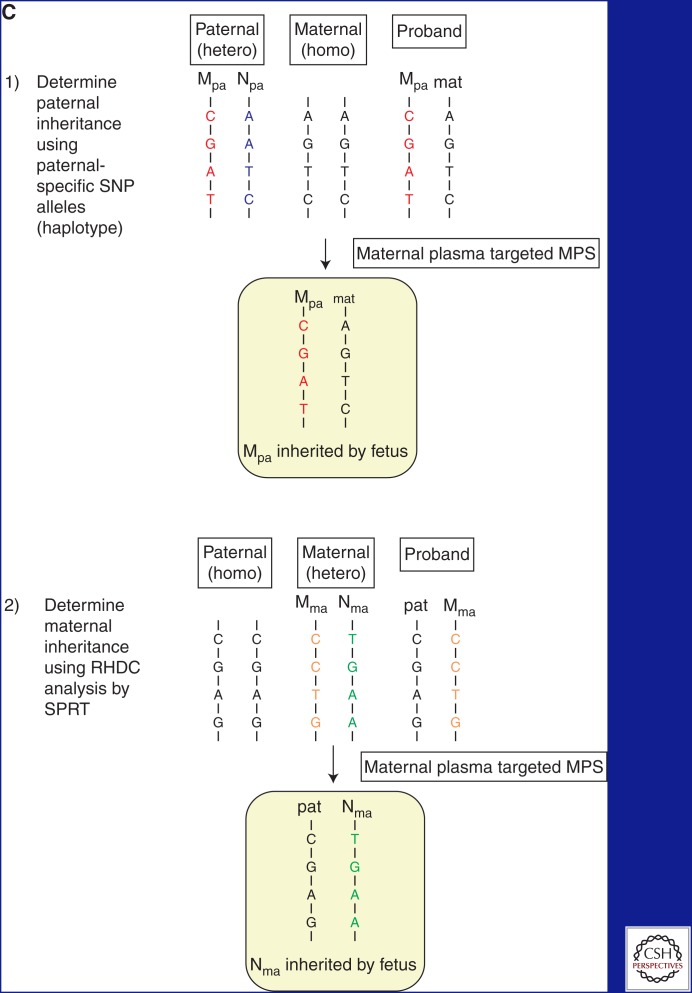
Approach for NIPD of monogenic autosomal recessive diseases. (A) Typical pedigree of a family affected by monogenic autosomal recessive diseases seeking prenatal diagnosis. A proband is present and has inherited both mutations from the carrier parents. (B) Overview of the targeted massively parallel sequencing (MPS) approach. (1) Genomic DNA samples are obtained from the father, mother, and proband. DNA sequencing libraries are constructed and hybridized with target capture probes spanning the gene of interest (not drawn to scale). The target-enriched DNA libraries are subjected to MPS. Target capture increases the sequencing depth of the region of interest with the same sequencing efforts. The enrichment fold depends on the extent of the target region. (2) Mutation-linked haplotypes of the father and mother are deduced from the genotyping results from trio genomic DNA. The paternal and maternal haplotypes present in the proband are linked to the mutations, whereas the haplotypes absent from the proband are nonmutation-linked. (3) Maternal plasma samples are subjected to targeted MPS analysis. Two and 14 DNA copies are shown from the fetus and the mother, respectively. Inheritance of the paternal haplotype is determined by the presence of paternal-specific SNP alleles in maternal plasma. Inheritance of the maternal haplotype is determined by the over-representation of the maternal SNP alleles. (C) Determination of fetal inheritance. (1) To determine the paternal inheritance, SNPs that are heterozygous (hetero) in the father and homozygous (homo) in the mother are used. The paternal haplotype present in the proband is mutation-linked. The mutation-linked paternal haplotype is present in the maternal plasma, thus the fetus has inherited the mutation from the father. (2) To determine the maternal inheritance, SNPs that are homozygous in the father and heterozygous in the mother are used. The maternal haplotype present in the proband is mutation-linked. In maternal plasma targeted MPS, an over-representation of the nonmutation-linked maternal haplotype was determined using the relative haplotype dosage (RHDO) analysis by sequential probability ratio test (SPRT) (New et al. 2014). Thus, the fetus has inherited the nonmutation-linked maternal haplotype.
IMPLICATIONS FOR IMPLEMENTATION INTO CLINICAL PRACTICE
NIPT for Aneuploidy
Shortly after the publication of the first clinical validation studies showing that whole-genome sequencing of cell-free DNA (cfDNA) in maternal plasma could reliably detect fetal trisomy 21 (Chiu et al. 2011; Palomaki et al. 2011), the first commercial test was launched by Sequenom Inc., in the United States, although testing through another company had been available commercially in Asia for a few months before this. The commercial launch in the United States was rapidly followed by other companies reporting clinical validation studies in high-risk populations using the whole-genome (Bianchi et al. 2012) or targeted approaches (Sparks et al. 2012a,b; Zimmermann et al. 2012). Initially, recommendations from professional bodies were for use only in high-risk singleton pregnancies (Benn et al. 2011) for the detection of the three major autosomal trisomies. In a recent meta-analysis, performance for these autosomal aneuploidies was shown to be very good, with sensitivities of ∼99% (95% CI 98.2–99.6), 96.8% (95% CI 94.5–98.4), and 92.1% (95% CI 85.9–96.7) for trisomies 21, 18, and 13, respectively (Gil et al. 2014). However, the appeal of a safer test for fetal aneuploidies, which is readily accessible, has led to a more widespread uptake and concerns have been raised about its performance in low-risk pregnancies. However, this concern has recently largely been laid to rest following the publication of clinical validation studies in lower risk populations that have confirmed good performance regardless of earlier risk (Bianchi et al. 2014; Hudecova et al. 2014; Norton et al. 2014), although only one study prospectively compares NIPT and Down syndrome screening performed at the same time in pregnancy (Norton et al. 2014). For the majority of studies, NIPT is performed variably from the first to the third trimester when the levels of cffDNA may be higher and performance of NIPT improved. There are fewer data available in multiple pregnancies, with reports largely being based on stored plasma samples (del Mar Gil et al. 2014) or small prospective studies (Canick et al. 2012; Lau et al. 2012; Huang et al. 2014). However, a meta-analysis of studies determined a detection rate of 94.4% for trisomy 21 (95% CI 74.2–99.0) in twin pregnancies (Gil et al. 2014) and most commercial companies now offer testing in multiple pregnancies. Recently, testing for sex chromosome anomalies has been offered, although, as with multiple pregnancies, there are fewer data available on performance than are available for aneuploidy testing. Sensitivities of ∼88.6% (95% CI 83.0–93.1) for Turner syndrome and 93.8% (95% CI 85.9–98.7) for other sex chromosome aneuploidies were reported in a meta-analysis of 12 studies reporting a total of 139 Turner cases and 44 other sex chromosome abnormalities (Gil et al. 2014). Finally, following the reports of the detection of other chromosomal aberrations using MPS (Chen et al. 2013; Srinivasan et al. 2013; Yu et al. 2013), several companies have now launched supplementary testing for a limited number of microdeletion syndromes (22q-, 5p-, 4p-, 1p36-, and Prader Willi or Angelman syndrome) and other aneuploidies (trisomies 9, 12, 16, and 22). To date, there are no published large-scale validation data to support implementation of screening for these very rare conditions that are often not associated with any particular risk factors. An exception is 22q-, which may underlie conotruncal cardiac anomalies. Given the very low prevalence of these conditions (1:4000 to 1:50,000) the prior risk will be low and false-positive results will be inevitable. The clinical utility of such testing in the routine population is likely to be small and, if applied widely, will lead to an increase in the invasive testing rate.
Following the launch of NIPT for autosomal aneuploidies, in late 2011, there has been an unprecedented uptake of testing in the private sector, largely driven by the commercial sector that wishes to capitalize on a potential market thought to be worth around $1.3 billion in the United States alone (Hayden 2012). Verinata Health Inc., an Illumina company, launched the Verifi test in March 2012, Ariosa Diagnostics Inc. the Harmony test in May 2012, and Natera the Panorama test in February 2013. Initially, these tests were only available in the United States or Asia, but access to testing has rapidly spread to Europe and other parts of the world (Chandrasekharan et al. 2014). Initially, samples were sent to companies in China or the United States, but more recently there has been technology transfer and licensing of local organizations to increasingly allow NIPT to be offered regionally. It is estimated that by the end of 2014 more than 1,000,000 tests will have been performed with availability in more than 60 countries. A dramatic decrease in the uptake of invasive diagnostic testing (such as CVS or amniocentesis) was reported just a year after the first NIPT was available clinically (Chetty et al. 2013; Musci et al. 2013; Larion et al. 2014a,b).
Discordant Results
Most of the early validation studies were relatively small, with only one study reporting more than 1000 cases (Palomaki et al. 2011) and it is only with widespread uptake in the private sector that some of the drawbacks associated with NIPT are more clearly appreciated. Regardless of the approach taken (genome-wide or targeted), NIPT for aneuploidy depends on sequencing a combination of both fetal and maternal cfDNA in maternal plasma, the majority of which emanates from the mother herself (Lo et al. 1998b), with the fetal fraction being placental in origin (Alberry et al. 2007; Lun et al. 2013). This means that there is a risk of detecting chromosomal aberrations present in the mother but not necessarily in the fetus as well as those confined to the placenta. In most validation studies the false-positive, or discordant, rates reported were low and of the order of 1%–2% (Palomaki et al. 2011, 2012; Bianchi et al. 2012). Discordant results are now well documented to be a result of confined placental mosaicism (Choi et al. 2013; Pan et al. 2013), early demise of an aneuploid cotwin (Futch et al. 2013), maternal chromosomal abnormalities (Sehnert et al. 2011; Lau et al. 2013) or maternal mosaicism (Wang et al. 2014a), which is a particular problem when reporting sex chromosome aneuploidies, and also maternal malignancy (Osborne et al. 2013). As the reasons for discordant results are biological and thus essentially unavoidable, most professional bodies endorse NIPT for aneuploidy as an advanced screening test, rather than a diagnostic test, and recommend that all positive results should be confirmed by invasive testing (Benn et al. 2011; ACOG 2012; Devers et al. 2013; Gregg et al. 2013; Michaelson-Cohen et al. 2014).
Reports from several companies describing large-scale experience of NIPT for aneuploidies in clinical practice in the United States and China have confirmed the high sensitivities defined in the earlier validation studies, but these studies do not have complete outcome data (Dan et al. 2012; Futch et al. 2013; McCullough et al. 2014) and thus the incidence of less common outcomes such as discordant results, may remain underestimated. Increasing numbers of anecdotal reports as well as published series (Wang et al. 2014b) of discordant results indicate that the positive predictive value of NIPT in clinical practice may not be as high as previously thought and the discordant rate may be >1%–2%. This may reflect increasing use in the low-risk population, as well as the recent addition of testing for sex-chromosome anomalies, in which, particularly in populations with an older average maternal age, it is to be expected that the incidence of maternal mosaicism will rise. Furthermore, as many positive NIPT results are confirmed using CVS, and confined placental mosaicism is a well-documented cause of discordant results, the use of CVS as a confirmatory test may underestimate the true incidence of discordance by confirming a result that was confined to the placenta. In a recent report describing a series of follow-up of positive NIPT results in a single center, results were confirmed as positive by invasive diagnostic cytogenetic testing in 38/41 fetuses with trisomy 21, 16/25 for trisomy 18, 7/16 for trisomy 13, and only 6/16 for fetuses with sex chromosome aneuploidies (Wang et al. 2014b). These investigators then combined their data with that from two other series to estimate the positive predictive value for the detection or trisomy 21 by NIPT to be 94.4%, with values of 59.5% for trisomy 18, 44.4% for trisomy 13, and 37.9% for sex chromosome aneuploidy. A few of these discordant results were shown to result from confined placental mosaicism or an abnormal maternal cell line, but many remained unexplained. These findings lead one to question the etiology of these anomalies and confirm the need for invasive testing, preferably by amniocentesis, to confirm any abnormal NIPT result. One must also ponder what the true impact of NIPT for aneuploidy will be on the invasive testing rate for the delivery of a safe and highly accurate aneuploidy screening and diagnosis service. While we may not yet know the full impact of NIPT, it is clear that its performance as a highly accurate screening test is significantly better than other screening tests such as the combined test (Bianchi et al. 2014; Norton et al. 2014). However, there has been no direct comparison of the targeted versus whole-genome approaches for NIPT for aneuploidy. It is clear that targeted approaches will not detect anomalies, fetal or maternal, that lie in areas of the genome not covered by the assay. In addition, there are reports that suggest that detection of sex chromosome abnormalities reveal differences in performance of the targeted and whole-genome approaches. Using the targeted approach, the Ariosa group found a sensitivity of 97% (33/34 cases) and a positive predictive value of 89% (34/38) in a retrospective study of 432 frozen plasma samples (Hooks et al. 2014), whereas a group in China using the BGI-Shenzhen whole-genome approach, reported a positive predictive value of 54.2% in 24 cases in which the karyotype has been confirmed to verify an NIPT result positive for a sex chromosome abnormality. This latter group reports a sensitivity of 100% (13/13 cases) and a false-positive rate of 0.18% (11/5937) but acknowledges that these figures may not be a true reflection of performance, as negative findings were not confirmed by karyotyping (Yao et al. 2014).
Implementation of NIPT for Aneuploidy into Public Sector Health Care
As yet, NIPT for aneuploidy is not routinely offered as part of a public sector maternity care pathway. Implementation into state funded healthcare systems requires further information to determine where it fits in the care pathway, what the uptake might be in clinical practice, whether the rate of invasive testing will change and if so how this might impact on service provision and training, the costs for the public health service and for service users, and how best to educate women and health professionals to ensure that informed choice is maintained. There is information available to address some of these issues, but it is largely based on studies assessing patient and health care views based on hypothetical scenarios in which indications are that uptake will be high (Sayres et al. 2011; Tischler et al. 2011; Hill et al. 2012a; Lewis et al. 2013, 2014a) or modeled data to inform discussions on potential economic impact (Chitty et al. 2012b; Beulen et al. 2014; Morris et al. 2014). However, it is well documented that what happens in clinical practice does not always reflect what people say/believe they will do hypothetically. Furthermore, any data currently reported from experience in clinical practice (Chitty et al. 2013) will relate to a different healthcare setting that will not be publicly funded with some costs still attributable to the patient, and so may not be relevant to implementation in any one particular individual country’s public health care service. Research is essential to explore the implementation of NIPT into public sector healthcare and studies and there are currently three studies evaluating all aspects of implementing NIPT for aneuploidy in publicly run health services, the RAPID evaluation of NIPT in the NHS study (Hill et al. 2014; http://www.rapid.nhs.uk), the TRIDENT study in the Netherlands (http://www.emgo.nl/news-and-events/news/486/non-invasive-prenatal-test-nipt-now-allowed-in-the-netherlands-within-trident-study/) and the Pegasus study in Canada (http://pegasus-pegase.ca/).
Economic Aspects
Cost is a major factor when deciding how NIPT might fit into existing care pathways, particularly for state-funded healthcare systems. All three studies addressing the implementation of NIPT in the public sector are evaluating test performance in local laboratories, as the current cost of provision through most commercial partners is significant. Furthermore, as yet, there are no FDA approved or CE marked tests available from commercial providers. This standard is increasingly being required by government organizations providing commercial tests on a national basis, and indeed may be required for tests provided on a large scale by accredited public sector laboratories in the near future (http://conventions.coe.int/Treaty/en/Treaties/html/203.htm). The models used for offering NIPT in these studies and in practice will vary. While there are many advocates suggesting that as NIPT is superior to any of the current Down syndrome screening programs (Bianchi et al. 2014; Norton et al. 2014), it should be offered to all pregnant women requesting Down syndrome screening; there are others, on both sides of the Atlantic, who suggest that the cost is too great at the current time and that NIPT should be offered as a contingent test using traditional screening to triage for a cutoff above which NIPT should be offered (Chitty et al. 2012b; Cuckle et al. 2013; Ohno and Caughey 2013; Beulen et al. 2014; Morris et al. 2014). Thus, in one study modeled around care in the United Kingdom National Health Service in which a screen cutoff of 1:150 is currently used to identify women who will be offered invasive testing, NIPT as a contingent test at a cost of £500 would be cost-neutral or cost-saving compared with the current Down syndrome screening pathway, the economic savings arising from a decrease in invasive testing, which would result in fewer miscarriages (Morris et al. 2014). Using the same model, but with NIPT as first line testing, more Down syndrome cases would be detected; but, even at a cost per test of £50, this would cost more than the current Down syndrome screening and diagnosis program (Morris et al. 2014). This group has estimated that if the Down syndrome screen positive level were changed from 1:150 to 1:1000 and women were offered NIPT, with invasive testing only offered to those who have a positive NIPT, ∼15% of women would be screen positive, compared with 2%–3% currently, and ∼95% of Down syndrome cases would be identified compared with 85%–90% currently (Chitty et al. 2012b). This implies that to identify the remaining 5%–6% of Down syndrome cases would require offering NIPT to the remaining 85% of the population. However, this assumed 100% uptake of testing and did not take account of women declining the offer of NIPT or leaving the screening pathway and opting for invasive testing following detection of an increased nuchal translucency or other fetal abnormality. Evaluation in clinical practice, albeit as part of a trial (Hill et al. 2014), is showing that a small but significant proportion of women with a very high-risk (>1:30) or an ultrasound abnormality are being advised to opt for invasive testing, and that some of the women in the lower risk category (1:150–1: 1000) are content with their risk and decline NIPT (LS Chitty, unpubl.). One other factor influencing health economic aspects is that studies based on hypothetical scenarios of offering NIPT have indicated that an additional group of women, who would decline current screening and diagnosis because of the risk of miscarriage, will undergo NIPT to plan and prepare for the birth of an affected child (Sayres et al. 2011). Both uptake and clinical utility will clearly influence the economic aspects of implementation. Furthermore, as current screening methods also identify pregnancies at risk of other adverse pregnancy outcomes, IUGR, preeclampsia, etc. (Goetzinger and Odibo 2014; Poon and Nicolaides 2014), further evaluation of what might be lost by abandoning current screening practices is required before changing policy and implementing new care pathways.
Implications of Failed Tests
In addition to cost, one key aspect requiring consideration when implementing NIPT into public sector care is the rate of failed tests. In recent reports this has varied between providers from <1% (Futch et al. 2013), through 1.3% (McCullough et al. 2014), 2.6% (Norton et al. 2014) to 6.3% (Dar et al. 2014). If offered as a contingent screen, when NIPT fails, the traditional Down syndrome screening risk is available on which to base an offer of invasive testing. If offered as the primary screen and NIPT fails, the only recourse would be maternal age, which is a poor screen. To offer invasive testing to everyone with a failed result could potentially increase the invasive rate beyond current levels thereby increasing costs and iatrogenic miscarriages.
Stakeholder Opinions and Education
It is clear from many studies conducted across the globe that NIPT for aneuploidy is welcomed by women and health professionals alike and uptake is high in most societies. Women value the improved safety secondary to the reduction in invasive testing as well as early, easy access to a highly sensitive test (Hill et al. 2012a; Allyse et al. 2014; Lewis et al. 2014a; Sayres et al. 2011) and health professionals value the improved accuracy of screening as well as the safety (Hill et al. 2012b). Furthermore, studies based on hypothetical scenarios of offering NIPT have indicated that an additional group of women, who would decline current screening and diagnosis because of the risk of miscarriage, will undergo NIPT to plan and prepare for the birth of an affected child (Sayres et al. 2011). Both uptake and clinical utility will clearly influence the economic aspects of implementation. Furthermore, studies of uptake in the United States indicate that cost is an important factor, as insurance coverage is variable; some people had out-of-pocket expenses for NIPT but not other types of testing (Chetty et al. 2013; Allyse et al. 2014; Vahanian et al. 2014). Other factors thought to contribute to NIPT uptake include having certainty and additional information from invasive testing (e.g., pathogenic chromosomal rearrangements, microdeletions, etc.).
However, despite the positive attitudes, concerns have also been raised predominantly because of the ease of access, NIPT requiring “just a blood test” and the fact that women undergo many blood tests in pregnancy and so may not realize they have had NIPT unless there are very high standards for pretest counseling. Further, because the risk associated with testing is removed for many women undergoing NIPT, there may be societal and family pressure applied to undergo testing (Lewis et al. 2013). Widespread availability through the private sector may also compromise informed consent unless health care providers are well informed and offer expert, detailed counseling as women may opt for NIPT having been made aware of it through a variety of public media which tend to report the positive, rather than negative features of NIPT (Lewis et al. 2014b). Given these caveats and the other features described above such as discordant results, detection of maternal chromosomal rearrangements, and failed tests, although NIPT still offers improved sensitivity and positive predictive value over current screening tests, it is clear that there is a huge need for health professional education and time for both pre- and post-test counseling. This applies to both the private and public health sectors but can only realistically be mandated in the public sector in which a detailed and widespread educational program must precede implementation, as is being trialed in the RAPID study (Hill et al. 2014; www.rapid.nhs.uk).
Another implication for implementation arises from the profound fall in invasive testing we are experiencing in many countries (Larion et al. 2014b). In the public sector, this will impact on service provision and training as there are guidelines defining the minimum number of invasive tests a provider should perform to maintain competency (https://www.rcog.org.uk/globalassets/documents/guidelines/gt8amniocentesis0111.pdf). Furthermore, evidence suggests that a trainee should perform ∼50–100 tests (Nizard et al. 2002) to achieve competency. The fall in invasive testing will adversely influence both on training and maintenance of skills and may require service reconfiguration to centralize services in fewer centers (Rose et al. 2013).
Intellectual Property Issues
Intellectual property (IP) issues may impact on the implementation of NIPT in the public sector as there are a number of companies claiming intellectual property rights in this area (Agarwal et al. 2013) with a number of ongoing legal battles, particularly in the United States. Such IP issues might ultimately have an impact to the cost of test provision. The recent settlement between two of the major commercial players in NIPT (http://www.utsandiego.com/news/2014/dec/03/Illumina-sequenom-verinata-nipt/) signals the first evidence of resolution of some of these disputes.
Implementation of NIPD for Monogenic Disorders
While noninvasive prenatal diagnosis (NIPD) for fetal sex determination, to triage invasive testing for sex-linked disorders and maternal dexamethasone treatment in pregnancies at risk of CAH (Chitty et al. 2012a; Hill et al. 2012b) based on PCR-based methodologies has been widely implemented in Europe, largely through accredited public sector laboratories, there has been very little development and implementation of sequencing applications for NIPD of monogenic disorders, despite proof-of-principle studies clearly showing these tests are possible (Lun et al. 2008b; Lam et al. 2012; Lench et al. 2013). This is because there is less demand for testing for monogenic disorders, it being largely confined to families at known high-risk with tests being provided on a bespoke, patient- or disease-specific basis and, until recently, the methods and workflows were labor intensive and not readily scalable. All of these factors suggest that the market opportunity may be small and not commercially attractive, leaving development to academics using increasingly scarce research funding.
The sequencing approaches described above for the NIPD of recessive disorders are labor intensive and costly in terms of both manpower and sequencing as they require detailed knowledge of both parental genotypes (Lam et al. 2012; New et al. 2014). As such, these have not been applied in clinical practice as yet. In the United Kingdom there are clear guidelines for the validation of any molecular genetic test that must occur before commissioning and subsequent funding by the National Health Service (NHS). This process is underpinned by the ACCE framework (analytic validity, clinical validity, clinical utility, and associated ethical, legal, and social implications) (Sanderson et al. 2005). Other aspects required by the United Kingdom Genetic Testing Network (http://ukgtn.nhs.uk/) before issuing approval for use include a detailed health economic evaluation, definition of care pathways and counseling approaches, patient information, health professional education, and guidance for policy makers. Fetal sex determination using a PCR-based method was the first noninvasive prenatal diagnostic test approved by the UKGTN in 2011 (Hill et al. 2012b). Since then, NIPD for achondroplasia and thanatophoric dysplasia using a polymerase chain reaction–restriction enzyme digest (PCR–RED) approach was approved by the UKGTN for use in the NHS in 2012 (Chitty et al. 2011; Chitty et al. 2013). While these approaches are very good to detect or exclude a paternal mutation in dominant conditions or in recessive conditions in which both parents carry different mutations, they have limited clinical applicability, particularly for the diagnosis of conditions with multiple causal mutations arising de novo, as each mutation has to be assayed individually.
With the advent of desktop massively parallel DNA sequencers, it has been possible to develop gene panels that can be used to screen for multiple mutations in a single assay. At the moment, these are used in clinical practice for the detection or exclusion of the paternally inherited in recessive conditions such as cystic fibrosis in which the parents carry different mutations (Hill et al. 2015) or for the detection of dominantly inherited conditions in which the father carries the mutation or it arises de novo. A number of these panels have now been approved for use in the NHS by the UKGTN, including an FGFR3 panel for the diagnosis of achondroplasia, thanatophoric dysplasia and other FGFR3-related skeletal dysplasias (http://ukgtn.nhs.uk/find-a-test/search-by-disorder-gene/test-service/fgfr3-related-skeletal-dysplasias-panel-test-nipd-654/; Chitty et al. 2015), an FGFR2 panel for Apert syndrome (http://ukgtn.nhs.uk/find-a-test/search-by-disorder-gene/test-service/apert-syndrome-nipd-697/) and a panel covering the 10 most common cystic fibrosis mutations (http://ukgtn.nhs.uk/find-a-test/search-by-disorder-gene/details/5062/). The sequencing approach offers flexibility, the ability to screen a case for multiple mutations at one time, and several cases can be multiplexed either when testing for the same condition or multiple conditions. These features are very advantageous in a busy public sector genetics laboratory and facilitate a rapid turnaround time or 3–6 d required for a timely prenatal diagnostic service. Sequencing approaches also offer the potential for the estimation of fetal fraction, opening the way for determining allelic ratios and inheritance of the maternal mutant allele in either X-linked or recessive conditions.
CONCLUSIONS
The technological advances that have enabled the vision of NIPT and diagnosis based on cfDNA in maternal plasma to become a reality have transformed prenatal screening and diagnosis for many women. In a technology that has advanced as rapidly as NIPT, one has to carefully assess the applications that are best supported by scientific evidence and those that would provide the biggest benefits to the tested subjects. It is likely that the main challenges, as we go forward, will be ethical and economical rather than technological ones.
ACKNOWLEDGMENTS
Y.M.D.L. has support from University Grants Committee of the Government of the Hong Kong Special Administration Region, China, under the Areas of Excellence Scheme (AoE/M-04/06), the Li Ka Shing Foundation and the S.K. Yee Foundation. Y.M.D.L. also has support from, holds equities in, and is a consultant to Sequenom and is a founder of Xcelom Limited. Y.M.D.L. has licensed technologies to Sequenom, Xcelom, and Illumina.
L.S.C. is partially funded by the Great Ormond Street Hospital Children’s Charity and the National Institute for Health Research (NIHR) Biomedical Research Centre at Great Ormond Street Hospital. The views expressed are those of the authors and not necessarily those of the National Health Service (NHS), the NIHR or the Department of Health.
Footnotes
Editors: Diana W. Bianchi and Errol R. Norwitz
Additional Perspectives on Molecular Approaches to Reproductive and Newborn Medicine available at www.perspectivesinmedicine.org
REFERENCES
- ACOG Committee Opinion 545. 2012. Noninvasive prenatal testing for fetal aneuploidy. Obstet Gynecol 120: 1532–1534. [DOI] [PubMed] [Google Scholar]
- Agarwal A, Sayres LC, Cho MK, Cook-Deegan R, Chandrasekharan S. 2013. Commercial landscape of non-invasive prenatal testing in the United States. Prenat Diagn 33: 521–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberry M, Maddocks D, Jones M, Abdel Hadi M, Abdel-Fattah S, Avent N, Soothill P. 2007. Free fetal DNA in maternal plasma in anembryonic pregnancies: Confirmation that the origin is the trophoblast. Prenat Diagn 27: 415–418. [DOI] [PubMed] [Google Scholar]
- Allyse M, Sayres LC, Goodspeed TA, Cho MK. 2014. Attitudes towards non-invasive prenatal testing for aneuploidy among US adults of reproductive age. J Perinatol 34: 429–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amicucci P, Gennarelli M, Novelli G, Dallapiccola B. 2000. Prenatal diagnosis of myotonic dystrophy using fetal DNA obtained from maternal plasma. Clin Chem 46: 301–302. [PubMed] [Google Scholar]
- Ashoor G, Syngelaki A, Wang E, Struble C, Oliphant A, Song K, Nicolaides KH. 2013. Trisomy 13 detection in the first trimester of pregnancy using a chromosome-selective cell-free DNA analysis method. Ultrasound Obstet Gynecol 41: 21–25. [DOI] [PubMed] [Google Scholar]
- Barrett AN, McDonnell TC, Chan KC, Chitty LS. 2012. Digital PCR analysis of maternal plasma for non-invasive detection of sickle cell anemia. Clin Chem 58: 1026–1032. [DOI] [PubMed] [Google Scholar]
- Benn P, Borrell A, Crossley J, Cuckle H, Dugoff L, Gross S, Johnson JA, Maymon R, Odibo A, Schielen P, et al. 2011. Aneuploidy screening: A position statement from a committee on behalf of the Board of the International Society for Prenatal Diagnosis, January 2011. Prenat Diagn 31: 519–522. [DOI] [PubMed] [Google Scholar]
- Beulen L, Grutters JP, Faas BH, Feenstra I, van Vugt JM, Bekker MN. 2014. The consequences of implementing non-invasive prenatal testing in Dutch national health care: A cost-effectiveness analysis. Eur J Obstet Gynecol Reprod Biol 182: 53–61. [DOI] [PubMed] [Google Scholar]
- Bianchi DW, Zickwolf GK, Weil GJ, Sylvester S, DeMaria MA. 1996. Male fetal progenitor cells persist in maternal blood for as long as 27 years postpartum. Proc Natl Acad Sci 93: 705–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi DW, Platt LD, Goldberg JD, Abuhamad AZ, Sehnert AJ, Rava RP; MatErnal Blood IS Source to Accurately diagnose fetal aneuploidy (MELISSA Study Group). 2012. Genome-wide fetal aneuploidy detection by maternal plasma DNA sequencing. Obstet Gynecol 119: 890–901. [DOI] [PubMed] [Google Scholar]
- Bianchi DW, Parker RL, Wentworth J, Madankumar R, Saffer C, Das AF, Craig JA, Chudova DI, Devers PL, Jones KW, et al. 2014. DNA sequencing versus standard prenatal aneuploidy screening. N Engl J Med 370: 799–808. [DOI] [PubMed] [Google Scholar]
- Boon EM, Faas BH. 2013. Benefits and limitations of whole genome versus targeted approaches for non-invasive prenatal testing for fetal aneuploidies. Prenat Diagn 33: 563–568. [DOI] [PubMed] [Google Scholar]
- Canick JA, Kloza EM, Lambert-Messerlian GM, Haddow JE, Ehrich M, van den Boom D, Bombard AT, Deciu C, Palomaki GE. 2012. DNA sequencing of maternal plasma to identify Down syndrome and other trisomies in multiple gestations. Prenat Diagn 32: 730–734. [DOI] [PubMed] [Google Scholar]
- Chan KCA, Zhang J, Hui AB, Wong N, Lau TK, Leung TN, Lo KW, Huang DW, Lo YMD. 2004. Size distributions of maternal and fetal DNA in maternal plasma. Clin Chem 50: 88–92. [DOI] [PubMed] [Google Scholar]
- Chan KC, Jiang P, Chan CW, Sun K, Wong J, Hui EP, Chan SL, Chan WC, Hui DS, Ng SS, et al. 2013. Non-invasive detection of cancer-associated genome-wide hypomethylation and copy number aberrations by plasma DNA bisulfite sequencing. Proc Natl Acad Sci 110: 18761–18768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrasekharan S, Minear MA, Hung A, Allyse M. 2014. Non-invasive prenatal testing goes global. Sci Transl Med 6: 231–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen EZ, Chiu RW, Sun H, Akolekar R, Chan KC, Leung TY, Jiang P, Zheng YW, Lun FM, Chan LY, et al. 2011. Non-invasive prenatal diagnosis of fetal trisomy 18 and trisomy 13 by maternal plasma DNA sequencing. PLoS One 6: e21791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Ge H, Wang X, Pan X, Yao X, Li X, Zhang C, Chen F, Jiang F, Li P, et al. 2013. Haplotype-assisted accurate non-invasive fetal whole genome recovery through maternal plasma sequencing. Genome Med 5: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chetty S, Garabedian MJ, Norton ME. 2013. Uptake of non-invasive prenatal testing (NIPT) in women following positive aneuploidy screening. Prenat Diagn 33: 542–546. [DOI] [PubMed] [Google Scholar]
- Chim SSC, Tong YK, Chiu RWK, Lau TK, Leung TN, Chan LYS, Oudejans CBM, Ding C, Lo YMD. 2005. Detection of the placental epigenetic signature of the maspin gene in maternal plasma. Proc Natl Acad Sci 102: 14753–14758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitty LS, Bianchi DW. 2013. Non-invasive prenatal testing: The paradigm is shifting rapidly. Prenat Diagn 33: 511–513. [DOI] [PubMed] [Google Scholar]
- Chitty LS, Griffin DR, Meaney C, Barrett A, Khalil A, Pajkrt E, Cole TJ. 2011. New aids for the non-invasive prenatal diagnosis of achondroplasia: Dysmorphic features, charts of fetal size and molecular confirmation using cell-free fetal DNA in maternal plasma. Ultrasound Obstet Gynecol 37: 283–289. [DOI] [PubMed] [Google Scholar]
- Chitty LS, Chatelain P, Wolffenbuttel KP, Aigrain Y. 2012a. Prenatal management of disorders of sex development. J Pediat Urol 8: 576–584. [DOI] [PubMed] [Google Scholar]
- Chitty LS, Hill M, White H, Wright D, Morris S. 2012b. Non-invasive prenatal testing for aneuploidy-ready for prime time? Am J Obstet Gynecol 206: 269–275. [DOI] [PubMed] [Google Scholar]
- Chitty LS, Khalil A, Barrett AN, Pajkrt E, Griffin DR, Cole T. 2013. Safer, accurate prenatal diagnosis of thanatophoric dysplasia using ultrasound and cell free fetal DNA. Prenat Diagn 33: 416–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitty LS, Mason S, Barrett AB, McKay F, Lench N, Daley R, Jenkins LA. 2015. Non-invasive prenatal diagnosis of achondroplasia and thanatophoric dysplasia: Next generation sequencing allows for a safer, more accurate and comprehensive approach. Prenat Diagn 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu RW, Lau TK, Leung TN, Chow KC, Chui DH, Lo YM. 2002. Prenatal exclusion of β thalassaemia major by examination of maternal plasma. Lancet 360: 998–1000. [DOI] [PubMed] [Google Scholar]
- Chiu RW, Chan KC, Gao Y, Lau VY, Zheng W, Leung TY, Foo CH, Xie B, Tsui NB, Lun FM, et al. 2008. Non-invasive prenatal diagnosis of fetal chromosomal aneuploidy by massively parallel genomic sequencing of DNA in maternal plasma. Proc Natl Acad Sci 105: 20458–20463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu RW, Akolekar R, Zheng YW, Leung TY, Sun H, Chan KC, Lun FM, Go AT, Lau ET, To WW, et al. 2011. Non-invasive prenatal assessment of trisomy 21 by multiplexed maternal plasma DNA sequencing: Large scale validity study. BMJ 342: c7401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi H, Lau TK, Jiang FM, Chan MK, Zhang HY, Lo PS, Chen F, Zhang L, Wang W. 2013. Fetal aneuploidy screening by maternal plasma DNA sequencing: ‘false positive’ due to confined placental mosaicism. Prenat Diagn 33: 198–200. [DOI] [PubMed] [Google Scholar]
- Clausen FB, Christiansen M, Steffensen R, Jørgensen S, Nielsen C, Jakobsen MA, Madsen RD, Jensen K, Krog GR, Rieneck K, et al. 2012. Report of the first nationally implemented clinical routine screening for fetal RHD in D- pregnant women to ascertain the requirement for antenatal RhD prophylaxis. Transfusion 52: 752–758. [DOI] [PubMed] [Google Scholar]
- Costa JM, Benachi A, Gautier E. 2002. New strategy for prenatal diagnosis of X-linked disorders. N Engl J Med 346: 1502. [DOI] [PubMed] [Google Scholar]
- Cuckle H, Benn P, Pergament E. 2013. Maternal cfDNA screening for Down syndrome—A cost sensitivity analysis. Prenat Diagn 33: 636–642. [DOI] [PubMed] [Google Scholar]
- Dan S, Wang W, Ren J, Li Y, Hu H, Xu Z, Lau TK, Xie J, Zhao W, Huang H, et al. 2012. Clinical application of massively parallel sequencing-based prenatal non-invasive fetal trisomy test for trisomies 21 and 18 in 11,105 pregnancies with mixed risk factors. Prenat Diagn 32: 1225–1232. [DOI] [PubMed] [Google Scholar]
- Dar P, Curnow KJ, Gross SJ, Hall MP, Stosic M, Demko Z, Zimmermann B, Hill M, Sigurjonsson S, Ryan A, et al. 2014. Clinical experience and follow-up with large scale single-nucleotide polymorphism-based non-invasive prenatal aneuploidy testing. Am J Obstet Gynecol 211: 527.e1–527.e17. [DOI] [PubMed] [Google Scholar]
- de Haas M, van der Ploeg CPB, Scheffer PG, Verlinden DA, Hirschberg H, Abbink F, van der Schoot CE. 2012. A nationwide fetal RHD screening programme for targeted antenatal and postnatal anti-D. ISBT Science Series 7: 164–167. [Google Scholar]
- del Mar Gil M, Quezada MS, Bregant B, Syngelaki A, Nicolaides KH. 2014. Cell-free DNA analysis for trisomy risk assessment in first-trimester twin pregnancies. Fetal Diagn Ther 35: 204–211. [DOI] [PubMed] [Google Scholar]
- Devers PL, Cronister A, Ormond KE, Facio F, Brasington CK, Flodman P. 2013. Non-invasive prenatal testing/non-invasive prenatal diagnosis: The position of the National Society of Genetic Counselors. J Genet Couns 22: 291–295. [DOI] [PubMed] [Google Scholar]
- Ehrich M, Deciu C, Zwiefelhofer T, Tynan JA, Cagasan L, Tim R, Lu V, McCullough R, McCarthy E, Nygren AO, Dean J, et al. 2011. Non-invasive detection of fetal trisomy 21 by sequencing of DNA in maternal blood: A study in a clinical setting. Am J Obstet Gynecol 204: 205.e201–211. [DOI] [PubMed] [Google Scholar]
- Faas BH, Beuling EA, Christiaens GC, von dem Borne AE, van der Schoot CE. 1998. Detection of fetal RHD-specific sequences in maternal plasma. Lancet 352: 1196. [DOI] [PubMed] [Google Scholar]
- Fan HC, Quake SR. 2007. Detection of aneuploidy with digital polymerase chain reaction. Anal Chem 79: 7576–7579. [DOI] [PubMed] [Google Scholar]
- Fan HC, Blumenfeld YJ, Chitkara U, Hudgins L, Quake SR. 2008. Non-invasive diagnosis of fetal aneuploidy by shotgun sequencing DNA from maternal blood. Proc Natl Acad Sci 105: 16266–16271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan HC, Gu W, Wang J, Blumenfeld YJ, El-Sayed YY, Quake SR. 2012. Non-invasive prenatal measurement of the fetal genome. Nature 487: 320–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finning KM, Martin PG, Soothill PW, Avent ND. 2002. Prediction of fetal D status from maternal plasma: Introduction of a new non-invasive fetal RHD genotyping service. Transfusion 42: 1079–1085. [DOI] [PubMed] [Google Scholar]
- Finning K, Martin P, Summers J, Daniels G. 2007. Fetal genotyping for the K (Kell) and Rh C, c, and E blood groups on cell-free fetal DNA in maternal plasma. Transfusion 47: 2126–2133. [DOI] [PubMed] [Google Scholar]
- Futch T, Spinosa J, Bhatt S, de Feo E, Rava RP, Sehnert AJ. 2013. Initial clinical laboratory experience in non-invasive prenatal testing for fetal aneuploidy from maternal plasma DNA samples. Prenat Diagn 33: 569–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil MM, Akolekar R, Quezada MS, Bregant B, Nicolaides KH. 2014. Analysis of cell-free DNA in maternal blood in screening for aneuploidies: Meta-analysis. Fetal Diagn Ther 35: 156–173. [DOI] [PubMed] [Google Scholar]
- Goetzinger KR, Odibo AO. 2014. Screening for abnormal placentation and adverse pregnancy outcomes with maternal serum biomarkers in the second trimester. Prenat Diagn 34: 635–641. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Gonzalez MC, Garcia-Hoyos M, Trujillo MJ, Rodriguez de Alba M, Lorda-Sanchez I, Diaz-Recasens J, Gallardo E, Ayuso C, Ramos C. 2002. Prenatal detection of a cystic fibrosis mutation in fetal DNA from maternal plasma. Prenat Diagn 22: 946–948. [DOI] [PubMed] [Google Scholar]
- Gregg AR, Gross SJ, Best RG, Monaghan KG, Bajaj K, Skotko BG, Thompson BH, Watson MS. 2013. ACMG statement on non-invasive prenatal screening for fetal aneuploidy. Genet Med 15: 395–398. [DOI] [PubMed] [Google Scholar]
- Hayden EC. 2012. Fetal tests spur legal battle: A newborn industry based on non-invasive genetic testing turns combative. Nature 486: 454. [DOI] [PubMed] [Google Scholar]
- Hill M, Finning K, Martin P, Hogg J, Meaney C, Norbury G, Daniels G, Chitty LS. 2011. Non-invasive prenatal determination of fetal sex: Translating research into clinical practice. Clin Genet 80: 68–75. [DOI] [PubMed] [Google Scholar]
- Hill M, Fisher J, Chitty LS, Morris S. 2012a. Women’s and health professionals’ preferences for prenatal tests for Down syndrome: A discrete choice experiment to contrast non-invasive prenatal diagnosis with current invasive tests. Genet Med 14: 905–913. [DOI] [PubMed] [Google Scholar]
- Hill M, Lewis C, Jenkins L, Allen S, Elles RG, Chitty LS. 2012b. Implementing non-invasive prenatal fetal sex determination using cell-free fetal DNA in the United Kingdom. Expert Opin Biol Ther 12: S119–S126. [DOI] [PubMed] [Google Scholar]
- Hill M, Wright D, Daley R, Lewis C, McKay F, Mason S, Lench N, Howarth A, Boustred C, Lo K, et al. 2014. Evaluation of non-invasive prenatal testing (NIPT) for aneuploidy in an NHS setting: A reliable accurate prenatal non-invasive diagnosis (RAPID) protocol. BMC Pregnancy Childbirth 14: 229–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill M, Twiss P, Verhoef T, Drury S, McKay F, Mason S, Jenkins L, Morris S, Chitty LS. 2015. Non-invasive prenatal diagnosis for cystic fibrosis: Detection of paternal mutations, exploration of patient preferences and cost analysis. Prenat Diagn 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooks J, Wolfberg AJ, Wang ET, Struble CA, Zahn J, Juneau K, Mohseni M, Huang S, Bogard P, Song K, et al. 2014. Non-invasive risk assessment of fetal sex chromosome aneuploidy through directed analysis and incorporation of fetal fraction. Prenat Diagn 34: 496–499. [DOI] [PubMed] [Google Scholar]
- Huang X, Zheng J, Chen M, Zhao Y, Zhang C, Liu L, Xie W, Shi S, Wei Y, Lei D, et al. 2014. Non-invasive prenatal testing of trisomies 21 and 18 by massively parallel sequencing of maternal plasma DNA in twin pregnancies. Prenat Diagn 34: 335–340. [DOI] [PubMed] [Google Scholar]
- Hudecova I, Sahota D, Heung MM, Jin Y, Lee WS, Leung TY, Lo YM, Chiu RW. 2014. Maternal plasma fetal DNA fractions in pregnancies with low and high risks for fetal chromosomal aneuploidies. PLoS ONE 28: e88484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyett JA, Gardener G, Stojilkovic-Mikic T, Finning KM, Martin PG, Rodeck CH, Chitty LS. 2005. Reduction in diagnostic and therapeutic interventions by non-invasive determination of fetal sex in early pregnancy. Prenat Diagn 25: 1111–1116. [DOI] [PubMed] [Google Scholar]
- Jensen TJ, Dzakula Z, Deciu C, van den Boom D, Ehrich M. 2012. Detection of microdeletion 22q11.2 in a fetus by next-generation sequencing of maternal plasma. Clin Chem 58: 1148–1151. [DOI] [PubMed] [Google Scholar]
- Juneau K, Bogard PE, Huang S, Mohseni M, Wang ET, Ryvkin P, Kingsley C, Struble CA, Oliphant A, Zahn JM. 2014. Microarray-based cell-free DNA analysis improves non-invasive prenatal testing. Fetal Diagn Ther 36: 282–286. [DOI] [PubMed] [Google Scholar]
- Kitzman JO, Snyder MW, Ventura M, Lewis AP, Qiu R, Simmons LE, Gammill HS, Rubens CE, Santillan DA, Murray JC, et al. 2012. Non-invasive whole-genome sequencing of a human fetus. Sci Transl Med 4: 137ra176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh W, Pan W, Gawad C, Fan HC, Kerchner GA, Wyss-Coray T, Blumenfeld YJ, El-Sayed YY, Quake SR. 2014. Non-invasive in vivo monitoring of tissue-specific global gene expression in humans. Proc Natl Acad Sci 111: 7361–7366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam KW, Jiang P, Liao GJ, Chan KC, Leung TY, Chiu RW, Lo YMD. 2012. Non-invasive prenatal diagnosis of monogenic diseases by targeted massively parallel sequencing of maternal plasma: Application to β-thalassemia. Clin Chem 58: 1467–1475. [DOI] [PubMed] [Google Scholar]
- Larion S, Warsof SL, Romary L, Mlynarczyk M, Peleg D, Abuhamad AZ. 2014a. Association of combined first-trimester screen and non-invasive prenatal testing on diagnostic procedures. Obstet Gynecol 123: 1303–1310. [DOI] [PubMed] [Google Scholar]
- Larion S, Warsof SL, Romary L, Mlynarczyk M, Peleg D, Abuhamad AZ. 2014b. Uptake of non-invasive prenatal testing at a large academic referral center. Am J Obstet Gynecol 211: 651.e1–651.e7. [DOI] [PubMed] [Google Scholar]
- Lau TK, Jiang F, Chan MK, Zhang H, Lo PS, Wang W. 2012. Non-invasive prenatal screening of fetal Down syndrome by maternal plasma DNA sequencing in twin pregnancies. J Matern Fetal Neonatal Med 26: 434–437. [DOI] [PubMed] [Google Scholar]
- Lau TK, Jiang FM, Stevenson RJ, Lo TK, Chan LW, Chan MK, Lo PS, Wang W, Zhang HY, Chen F, et al. 2013. Secondary findings from non-invasive prenatal testing for common fetal aneuploidies by whole genome sequencing as a clinical service. Prenat Diagn 33: 602–608. [DOI] [PubMed] [Google Scholar]
- Lench N, Barrett A, Fielding S, McKay F, Hill M, Jenkins L, White H, Chitty LS. 2013. The clinical implementation of non-invasive prenatal diagnosis for single-gene disorders: Challenges and progress made. Prenat Diagn 33: 555–562. [DOI] [PubMed] [Google Scholar]
- Lewis C, Silcock C, Chitty LS. 2013. Non-invasive prenatal testing for Down’s syndrome: Pregnant women’s views and likely uptake. Public Health Genomics 16: 223–232. [DOI] [PubMed] [Google Scholar]
- Lewis C, Hill M, Silcock C, Daley R, Chitty LS. 2014a. Non-invasive prenatal testing for trisomy 21: A cross-sectional survey of service users’ views and likely uptake. BJOG 121: 582–594. [DOI] [PubMed] [Google Scholar]
- Lewis C, Choudhury M, Chitty LS. 2014b. ‘Hope for safe prenatal gene tests’. A content analysis of how the UK press media are reporting advances in non-invasive prenatal testing. Prenat Diagn 10.1002/pd.4488. [DOI] [PubMed] [Google Scholar]
- Lo YMD, Corbetta N, Chamberlain PF, Rai V, Sargent IL, Redman CWG. 1997. Presence of fetal DNA in maternal plasma and serum. Lancet 350: 485–487. [DOI] [PubMed] [Google Scholar]
- Lo YMD, Hjelm NM, Fidler C, Sargent IL, Murphy MF, Chamberlain PF, Poon PM, Redman CW, Wainscoat JS. 1998a. Prenatal diagnosis of fetal RhD status by molecular analysis of maternal plasma. N Engl J Med 339: 1734–1738. [DOI] [PubMed] [Google Scholar]
- Lo YMD, Tein MS, Lau TK, Haines CJ, Leung TN, Poon PM, Wainscoat JS, Johnson PJ, Chang AM, Hjelm NM. 1998b. Quantitative analysis of fetal DNA in maternal plasma and serum: Implications for non-invasive prenatal diagnosis. Am J Hum Genet 62: 768–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo YMD, Lau TK, Zhang J, Leung TN, Chang AM, Hjelm NM, Elmes RS, Bianchi DW. 1999a. Increased fetal DNA concentrations in the plasma of pregnant women carrying fetuses with trisomy 21. Clin Chem 45: 1747–1751. [PubMed] [Google Scholar]
- Lo YMD, Zhang J, Leung TN, Lau TK, Chang AM, Hjelm NM. 1999b. Rapid clearance of fetal DNA from maternal plasma. Am J Hum Genet 64: 218–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo YMD, Tsui NB, Chiu RW, Lau TK, Leung TN, Heung MM, Gerovassili A, Jin Y, Nicolaides KH, Cantor CR, et al. 2007a. Plasma placental RNA allelic ratio permits non-invasive prenatal chromosomal aneuploidy detection. Nat Med 13: 218–223. [DOI] [PubMed] [Google Scholar]
- Lo YMD, Lun FMF, Chan KCA, Tsui NBY, Chong KC, Lau TK, Leung TY, Zee BCY, Cantor CR, Chiu RWK. 2007b. Digital PCR for the molecular detection of fetal chromosomal aneuploidy. Proc Natl Acad Sci 104: 13116–13121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo YMD, Chan KC, Sun H, Chen EZ, Jiang P, Lun FM, Zheng YW, Leung TY, Lau TK, Cantor CR, et al. 2010. Maternal plasma DNA sequencing reveals the genome-wide genetic and mutational profile of the fetus. Sci Transl Med 2: 61ra91. [DOI] [PubMed] [Google Scholar]
- Lun FM, Tsui NB, Chan KC, Leung TY, Lau TK, Charoenkwan P, Chow KC, Lo WY, Wanapirak C, Sanguansermsri T, et al. 2008a. Non-invasive prenatal diagnosis of monogenic diseases by digital size selection and relative mutation dosage on DNA in maternal plasma. Proc Natl Acad Sci 105: 19920–19925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lun FM, Chiu RW, Allen Chan KC, Yeung Leung T, Kin Lau T, Lo YMD. 2008b. Microfluidics digital PCR reveals a higher than expected fraction of fetal DNA in maternal plasma. Clin Chem 54: 1664–1672. [DOI] [PubMed] [Google Scholar]
- Lun FM, Chiu RW, Sun K, Leung TY, Jiang P, Chan KC, Sun H, Lo YMD. 2013. Non-invasive prenatal methylomic analysis by genome-wide bisulfite sequencing of maternal plasma DNA. Clin Chem 59: 1583–1594. [DOI] [PubMed] [Google Scholar]
- Ma D, Ge H, Li X, Jiang T, Chen F, Zhang Y, Hu P, Chen S, Zhang J, Ji X, et al. 2014. Haplotype-based approach for non-invasive prenatal diagnosis of congenital adrenal hyperplasia by maternal plasma DNA sequencing. Gene 544: 252–258. [DOI] [PubMed] [Google Scholar]
- Mazloom AR, Dzakula Z, Oeth P, Wang H, Jensen T, Tynan J, McCullough R, Saldivar JS, Ehrich M, van den Boom D, et al. 2013. Non-invasive prenatal detection of sex chromosomal aneuploidies by sequencing circulating cell-free DNA from maternal plasma. Prenat Diagn 33: 591–597. [DOI] [PubMed] [Google Scholar]
- McCullough RM, Almasri EA, Guan X, Geis JA, Hicks SC, Mazloom AR, Deciu C, Oeth P, Bombard AT, Paxton B, et al. 2014. Non-invasive prenatal chromosomal aneuploidy testing - clinical experience: 100,000 clinical samples. PLoS ONE 9: e109173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelson-Cohen R, Gershoni-Baruch R, Sharoni R, Shochat M, Yaron Y, Singer A. 2014. Israeli Society of Medical Genetics NIPT Committee Opinion 072013: Non-invasive prenatal testing of cell-free DNA in maternal plasma for detection of fetal aneuploidy. Fetal Diagn Ther 36: 242–244. [DOI] [PubMed] [Google Scholar]
- Morris S, Karlsen S, Chung N, Hill M, Chitty LS. 2014. Model-based analysis of costs and outcomes of non-invasive prenatal testing for Down’s syndrome using cell free fetal DNA in the UK National Health Service. PLoS ONE 9: e93559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musci TJ, Fairbrother G, Batey A, Bruursema J, Struble C, Song K. 2013. Non-invasive prenatal testing with cell-free DNA: US physician attitudes toward implementation in clinical practice. Prenat Diagn 33: 424–428. [DOI] [PubMed] [Google Scholar]
- New MI, Tong YK, Yuen T, Jiang P, Pina C, Chan KC, Khattab A, Liao GJ, Yau M, Kim SM, et al. 2014. Non-invasive prenatal diagnosis of congenital adrenal hyperplasia using cell-free fetal DNA in maternal plasma. J Clin Endocrinol Metab 99: E1022–E1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolaides KH, Syngelaki A, del Mar Gil M, Quezada MS, Zinevich Y. 2014. Prenatal detection of fetal triploidy from cell-free DNA testing in maternal blood. Fetal Diagn Ther 35: 212–217. [DOI] [PubMed] [Google Scholar]
- Nizard J, Duyme M, Ville Y. 2002. Teaching ultrasound-guided invasive procedures in fetal medicine: Learning curves with and without an electronic guidance system. Ultrasound Obstet Gynecol 19: 274–277. [DOI] [PubMed] [Google Scholar]
- Norton ME, Brar H, Weiss J, Karimi A, Laurent LC, Caughey AB, Rodriguez MH, Williams J III, Mitchell ME, Adair CD, et al. 2012. Non-Invasive Chromosomal Evaluation (NICE) Study: Results of a multicenter prospective cohort study for detection of fetal trisomy 21 and trisomy 18. Am J Obstet Gynecol 207: e131–e138. [DOI] [PubMed] [Google Scholar]
- Norton ME, Jacobsson B, Swamy G, Laurent LC, Ranzini A, Brar H, Tomlinson M, Pereira L, Spitz J, Holleman D, et al. 2014. Non-invasive examination of trisomy using directed cell-free DNA analysis. The NEXT study. Prenat Diagn 34: e2. [DOI] [PubMed] [Google Scholar]
- Ohno M, Caughey A. 2013. The role of non-invasive prenatal testing as a diagnostic versus a screening tool—A cost-effectiveness analysis. Prenat Diagn 33: 630–635. [DOI] [PubMed] [Google Scholar]
- Osborne CM, Hardisty E, Devers P, Kaiser-Rogers K, Hayden MA, Goodnight W, Vora NL. 2013. Discordant non-invasive prenatal testing results in a patient subsequently diagnosed with metastatic disease. Prenat Diagn 33: 609–611. [DOI] [PubMed] [Google Scholar]
- Palomaki GE, Kloza EM, Lambert-Messerlian GM, Haddow JE, Neveux LM, Ehrich M, van den Boom D, Bombard AT, Deciu C, Grody WW, et al. 2011. DNA sequencing of maternal plasma to detect Down syndrome: An international clinical validation study. Genet Med 13: 913–920. [DOI] [PubMed] [Google Scholar]
- Palomaki GE, Deciu C, Kloza EM, Lambert-Messerlian GM, Haddow JE, Neveux LM, Ehrich M, van den Boom D, Bombard AT, Grody WW, et al. 2012. DNA sequencing of maternal plasma reliably identifies trisomy 18 and trisomy 13 as well as Down syndrome: An international collaborative study. Genet Med 14: 296–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan M, Li FT, Li Y, Jiang FM, Li DZ, Lau TK, Liao C. 2013. Discordant results between fetal karyotyping and non-invasive prenatal testing by maternal plasma sequencing in a case of uniparental disomy 21 due to trisomic rescue. Prenat Diagn 33: 598–601. [DOI] [PubMed] [Google Scholar]
- Peters D, Chu T, Yatsenko SA, Hendrix N, Hogge WA, Surti U, Bunce K, Dunkel M, Shaw P, Rajkovic A. 2011. Non-invasive prenatal diagnosis of a fetal microdeletion syndrome. N Engl J Med 365: 1847–1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon LC, Nicolaides K. 2014. First-trimester maternal factors and biomarker screening for pre-eclampsia. Prenat Diagn 34: 618–627. [DOI] [PubMed] [Google Scholar]
- Poon LLM, Leung TN, Lau TK, Lo YMD. 2000. Presence of fetal RNA in maternal plasma. Clin Chem 46: 1832–1834. [PubMed] [Google Scholar]
- Poon LL, Leung TN, Lau TK, Chow KC, Lo YM. 2002. Differential DNA methylation between fetus and mother as a strategy for detecting fetal DNA in maternal plasma. Clin Chem 48: 35–41. [PubMed] [Google Scholar]
- Rabinowitz M, Savage M, Pettersen B, Sigurjonsson S, Hill M, Zimmermann B. 2014. Non-invasive cell-free DNA-based prenatal detection of microdeletions using single nucleotide polymorphism-targeted sequencing. Obstet Gynecol 123: 167S. [Google Scholar]
- Rose NC, Lagrave D, Hafen B, Jackson M. 2013. The impact of utilization of early aneuploidy screening on amniocenteses available for training in obstetrics and fetal medicine. Prenat Diagn 33: 242–244. [DOI] [PubMed] [Google Scholar]
- Saito H, Sekizawa A, Morimoto T, Suzuki M, Yanaihara T. 2000. Prenatal DNA diagnosis of a single-gene disorder from maternal plasma. Lancet 356: 1170. [DOI] [PubMed] [Google Scholar]
- Sanderson S, Zimmern R, Kroese M, Higgins J, Patch C, Emery J. 2005. How can the evaluation of genetic tests be enhanced? Lessons learned from the ACCE framework and evaluating genetic tests in the United Kingdom. Genet Med 7: 495–500. [DOI] [PubMed] [Google Scholar]
- Sayres LC, Allyse M, Norton ME, Cho MK. 2011. Cell-free fetal DNA testing: A pilot study of obstetric healthcare provider attitudes toward clinical implementation. Prenat Diagn 31: 1070–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehnert AJ, Rhees B, Comstock D, de Feo E, Heilek G, Burke J, Rava RP. 2011. Optimal detection of fetal chromosomal abnormalities by massively parallel DNA sequencing of cell-free fetal DNA from maternal blood. Clin Chem 57: 1042–1049. [DOI] [PubMed] [Google Scholar]
- Sparks AB, Struble CA, Wang ET, Song K, Oliphant A. 2012a. Non-invasive prenatal detection and selective analysis of cell-free DNA obtained from maternal blood: Evaluation for trisomy 21 and trisomy 18. Am J Obstet Gynecol 206: e311–e319. [DOI] [PubMed] [Google Scholar]
- Sparks AB, Wang ET, Struble CA, Barrett W, Stokowski R, McBride C, Zahn J, Lee K, Shen N, Doshi J, et al. 2012b. Selective analysis of cell-free DNA in maternal blood for evaluation of fetal trisomy. Prenat Diagn 32: 3–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan A, Bianchi DW, Huang H, Sehnert AJ, Rava RP. 2013. Non-invasive detection of fetal subchromosome abnormalities via deep sequencing of maternal plasma. Am J Hum Genet 92: 167–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tischler R, Hudgins L, Blumenfeld YJ, Greely HT, Ormond KE. 2011. Non-invasive prenatal diagnosis: Pregnant women’s interest and expected uptake. Prenat Diagn 31: 1292–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong YK, Ding C, Chiu RWK, Gerovassili A, Chim SSC, Leung TY, Leung TN, Lau TK, Nicolaides KH, Lo YMD. 2006. Non-invasive prenatal detection of fetal trisomy 18 by epigenetic allelic ratio analysis in maternal plasma: Theoretical and empirical considerations. Clin Chem 52: 2194–2202. [DOI] [PubMed] [Google Scholar]
- Tsui NB, Kadir RA, Chan KC, Chi C, Mellars G, Tuddenham EG, Leung TY, Lau TK, Chiu RW, Lo YMD. 2011. Non-invasive prenatal diagnosis of hemophilia by microfluidics digital PCR analysis of maternal plasma DNA. Blood 117: 3684–3691. [DOI] [PubMed] [Google Scholar]
- Tsui NBY, Jiang P, Wong YF, Leung TY, Chan KC, Chiu RW, Sun H, Lo YMD. 2014. Maternal plasma RNA sequencing for genome-wide transcriptomic profiling and identification of pregnancy-associated transcripts. Clin Chem 60: 954–962. [DOI] [PubMed] [Google Scholar]
- Vahanian SA, Baraa Allaf M, Yeh C, Chavez MR, Kinzler WL, Vintzileos Am. 2014. Patient acceptance of non-invasive testing for fetal aneuploidy via cell-free fetal DNA. J Matern Fetal Neonatal Med 27: 106–109. [DOI] [PubMed] [Google Scholar]
- Verweij EJ, Jacobsson B, van Scheltema PA, de Boer MA, Hoffer MJ, Hollemon D, Westren M, Song K, Oepkes D. 2013. European non-invasive trisomy evaluation (EU-NITE) study: A multicenter prospective cohort study for non-invasive fetal trisomy 21 testing. Prenat Diagn 33: 996–1001. [DOI] [PubMed] [Google Scholar]
- Wang Y, Chen Y, Tian F, Zhang J, Song Z, Wu Y, Han X, Hu W, Ma D, Cram D, et al. 2014a. Maternal mosaicism is a significant contributor to discordant sex chromosomal aneuploidies associated with non-invasive prenatal testing. Clin Chem 60: 251–259. [DOI] [PubMed] [Google Scholar]
- Wang JC, Sahoo T, Schonberg S, Kopita KA, Ross L, Patek K, Strom CM. 2014b. Discordant non-invasive prenatal testing and cytogenetic results: A study of 109 consecutive cases. Genet Med 10.1038/gim.2014.92. [DOI] [PubMed] [Google Scholar]
- Yao H, Jiang F, Hu H, Gao Y, Zhu Z, Zhang H, Wang Y, Guo Y, Liu L, Yuan Y, et al. 2014. Detection of fetal sex chromosome aneuploidy by massively parallel sequencing of maternal plasma DNA: Initial experience in a Chinese hospital. Ultrasound Obstet Gynecol 44: 17–24. [DOI] [PubMed] [Google Scholar]
- Yu SCY, Jiang P, Choy KW, Chan KC, Won HS, Leung WC, Lau ET, Tang MH, Leung TY, Lo YMD, et al. 2013. Non-invasive prenatal molecular karyotyping from maternal plasma. PLoS ONE 8: e60968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu SCY, Chan KC, Zheng YW, Jiang P, Liao GJ, Sun H, Akolekar R, Leung TY, Go AT, van Vugt JM, et al. 2014. Size-based molecular diagnostics using plasma DNA for non-invasive prenatal testing. Proc Natl Acad Sci 111: 8583–8588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann B, Hill M, Gemelos G, Demko Z, Banjevic M, Baner J, Ryan A, Sigurjonsson S, Chopra N, Dodd M, et al. 2012. Non-invasive prenatal aneuploidy testing of chromosomes 13, 18, 21, X, and Y, using targeted sequencing of polymorphic loci. Prenat Diagn 32: 1233–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]