Abstract
The sheer volume and complexity of publications in the biological sciences are straining traditional approaches to research planning. Nowhere is this problem more serious than in molecular and cellular cognition, since in this neuroscience field, researchers routinely use approaches and information from a variety of areas in neuroscience and other biology fields. Additionally, the multilevel integration process characteristic of this field involves the establishment of experimental connections between molecular, electrophysiological, behavioral, and even cognitive data. This multidisciplinary integration process requires strategies and approaches that originate in several different fields, which greatly increases the complexity and demands of this process. Although causal assertions, where phenomenon A is thought to contribute or relate to B, are at the center of this integration process and key to research in biology, there are currently no tools to help scientists keep track of the increasingly more complex network of causal connections they use when making research decisions. Here, we propose the development of semiautomated graphical and interactive tools to help neuroscientists and other biologists, including those working in molecular and cellular cognition, to track, map, and weight causal evidence in research papers. There is a great need for a concerted effort by biologists, computer scientists, and funding institutions to develop maps of causal information that would aid in integration of research findings and in experiment planning.
Information in biology, including neuroscience, is growing at an unprecedented pace that demands new tools and new approaches (Lok 2010; Silva et al. 2014). Because of the ever-growing number, complexity, and interconnectedness of research publications and biological concepts, it is simply no longer possible for individual biologists to be aware of even a fraction of the published findings potentially pertinent to their work. The library of medicine, for example, now includes more than 25 million research papers reporting the results of at least 100 million experiments. Even a young field like Molecular and Cellular Cognition includes tens of thousands of research papers reporting millions of experiments. When the implications of what has already been published remain buried in the never-ending avalanche of published information, how can scientists reasonably optimize future research plans? Although there is a great deal of work on-going to tackle different components of this problem, from annotation of the literature, to curation of databases and automated reasoning, much remains to be done. Here, we address the need for graphical and interactive tools that track and map causal evidence in research papers (i.e., research maps). Although causal assertions are the very fabric of biology, there are currently no tools to help biologists keep track of the increasingly more complex network of causal connections derived from published findings. A causal connection is defined by evidence (see below) that phenomenon A contributes to the occurrence or state of phenomenon B. Maps of causal information derived from research papers would help biologists plan experiments, as well as track and gage the success (both prospectively and retrospectively) of different experiment planning strategies. Maps of causal information could also contribute to how science is reviewed and funded by providing an objective and inclusive tool for helping to evaluate the content of research papers and grant proposals.
Molecular and cellular cognition in the age of information
In the last two decades, information technology (Akil et al. 2011) has slowly transformed biological fields as diverse as molecular genetics and cognitive neuroscience. Using powerful computer science approaches, biologists have developed methods to both organize and explore the content of complex petabyte-scale data sets (Dai et al. 2012), such as those generated by whole-genome sequencing, brain imaging, and drug effects. For example, network analyses based on the US Food and Drug Administration's Adverse Event Reporting System allowed investigators to find beneficial drug combinations that were then tested in animal models (Zhao et al. 2013). Innovative natural language processing methods have been developed to automatically find and abstract specific content from large bodies of text, as well as identify latent relations among biological entities as diverse as genes, signaling pathways and anatomical structures. Increasingly more complex and sophisticated hierarchical ontologies have considerably facilitated these automated processes. With the rising complexity of biological assertions, researchers have also sought to derive meta-analytical approaches to mine and synthesize large bodies of published information on specific topics (Akil et al. 2011).
These and other efforts to tackle the problem of information overload in the biological sciences, helped to highlight another problem in this discipline: the need for developing new approaches to generate theoretical frameworks that accommodate the dizzying growth and diversity of discovery in biology. Historically, these frameworks have emerged from causal insights, such as those inspired by evolution and the structure of DNA. The resulting frameworks were essential from an information perspective, since they allowed biologists to synthesize and, more important, simplify large bodies of otherwise complex information. The current immensity of the published record provides an unprecedented challenge for this integrative process, since potentially pertinent causal information could be spread through millions (not tens or even hundreds!) of published research papers.
This problem is especially acute in molecular and cellular cognition because in this field researchers routinely integrate findings and use methods from widely different areas of neuroscience and biology, from molecular and cellular biology, to neurophysiology, systems neuroscience, behavioral neuroscience, and cognitive neuroscience. The sheer breadth of concepts and literature base that span these different disciplines is a true obstacle for optimal progress in highly integrative and multidisciplinary fields like molecular and cellular cognition. Until we develop methods to automatically abstract, integrate and easily interact with this buried treasure throve of causal information, we can only wonder about the number of potentially transforming insights that will remain just beyond our reach. The research maps we introduce next, provide a conceptual solution to this problem, one that is built on principles used by experimentalists in evaluating the strength and reliability of causal information in biology. But, how exactly could causal information be represented in a map?
Research maps for molecular and cellular cognition
It is important to note that although research maps share a superficial resemblance to Concept Maps (Novak 1990), a tool used for science education (Novak and Musonda 1991), we will show below that the two types of maps are very different conceptually, structurally, and in organization. Like concept maps, research maps have nodes and edges, but the resemblance stops there. Concept maps are hierarchical structures that represent information that addresses a specific question or topic, with the most general information at the top of the hierarchy, and the increasingly specific information below. Nodes are linked by edges (signal directionality) with words that define a proposition involving both nodes.
Knowledge Engineering from Experimental Design (KEfED) provides a powerful computational framework for representing, and publishing experiments (Russ et al. 2011). KEfED elements can be derived from a flow diagram of an experimental protocol. Although KEfED and research maps are both based on scientific evidence rather than interpretations (Russ et al. 2011), the goals, structure, and conceptual framework of KEfED are distinct from the research maps proposed here.
At the highest zoom levels, maps of causal information (research maps) would be simply networks where biological phenomena (their identity and properties; the nodes in the map) are linked by weighted causal connections (the edges in this network; Fig. 1). These edges would represent one of three possible types of causal connections between two phenomena: excitatory, where one phenomenon promotes the other, inhibitory where one inhibits another, or simply that one phenomenon has no measurable effect on the other. A score assigned to each edge would give users a sense for the strength and consistency of evidence represented by each connection among the phenomena represented. Additionally, symbols would inform users of the types of experiments represented in each edge.
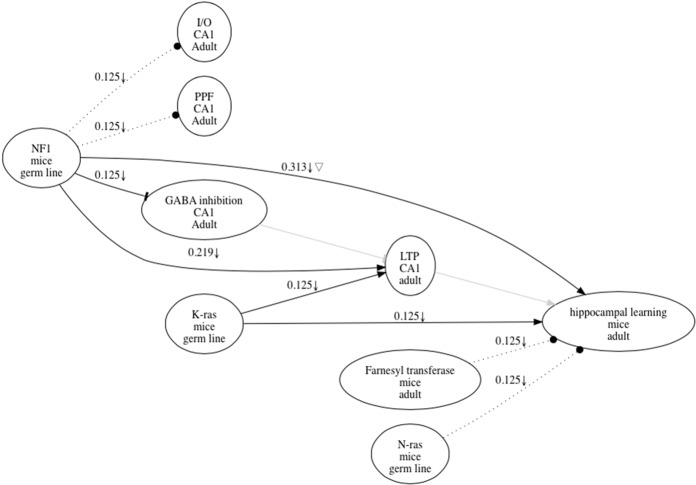
Figure 1.
Research map representing results in a published paper (Costa et al. 2002). Each node in the graph has three items that describe the name of the item (top), as well as spatial (middle) and temporal (bottom) information that defines it. Nodes are connected by edges that characterize the nature of the causal relations represented, including excitatory (sharp edges), inhibitory (dull edge), and no relation (dotted line). Each edge also has a score that reflects the amount of evidence represented, and symbols that reflect the types of experiments carried out, including upward arrow for Positive Manipulations, downward arrow for Negative Manipulations, and triangle for Mediation Experiments (see text for definitions). Edges representing key hypothetical information mentioned in the article are represented as thick gray lines; since these edges are hypothetical, they have no weights or experimental symbols. The weights or scores of the edges in the map were determined according to the following simple rules; any one of the four types of experiments described in the text was given a score of 0.125. Additional experiments of the same type were scored according to a geometric progression with a start term of 0.125 and r factor of 0.5. For example, the first negative manipulation and mediation experiments supporting a causal connection between NF1 and LTP (long-term potentiation) in the graph contributed each 0.125 weight. The second negative manipulation contributed 0.125 × 0.51 or 0.6025. Thus, adding the scores of these three experiments, we derived the rounded up score of 0.313 shown above the NF1-LTP edge. Contradictory evidence, when available, would detract from the score of that edge.
Although there are tens of millions of experiments testing causal relations in biology, they fall into a small number of classes (Landreth and Silva 2013). For example, in molecular and cellular cognition, researchers commonly use at least four major types of experiments to test a possible causal connection between two entities A and B, where A is manipulated or simply tracked and the results are measured in B: positive Manipulations where A's levels or activity are increased, Negative Manipulations when A's levels or activity are decreased, Non-Interventions whose goal is to track how A covaries with B, and Mediation experiments, designed to determine whether C is part of the mechanism by which A contributes to B. B's levels or probability increases or decreases with changes in A will determine whether the possible causal connection is excitatory, inhibitory or absent. Any one of these four types of experiments can contribute to testing whether A causes B. For example, if the levels of A and B are both increased, this would be evidence for an excitatory connection; If B's levels are decreased instead, the connection would likely be inhibitory, and if B's levels do not change, this would be evidence that there is no causal connection between A and B.
Accordingly, no one single experiment can establish causality. Instead, biologists use convergency (different experiments with a single interpretation) and consistency (similar experiments yielding similar results) among these different types of experiments to judge the strength of any causal assertion. Therefore, in research maps convergency and consistency among results increases the score (from 0 to 1) assigned to each edge, while contradictions have the opposite effect. By selecting any one edge in the map, users could be directed to the exact research papers and experiments represented by that edge, and therefore explore how that score was computed. In addition, hypothetical connections (those representing causal hypothetical statements) could help keep track of key postulates in hypotheses or ideas that organize sets of research information. Hypothesis guide experimentation and in research maps hypothetical connections (connections that were not tested by experiments) help to structure research maps representing those experiments. At the lowest zoom levels, the maps would guide users to different biological sub-disciplines, while intermediate-zoom levels would reveal topics and domains represented in the maps.
It is unlikely that maps of causal information, for any major biological subject such as learning and memory, will be complete any time in the foreseeable future. Indeed, the very goal of the research maps proposed here is to provide a tool to help individual researchers evaluate available information so that they can decide what do next (e.g., how to best complement existing information). In this respect, it is important to stress that the concepts used to build research maps should reflect principles and practices routinely used in many areas of biology, such as in molecular and cellular cognition. Otherwise, the maps would be little more than intellectual curiosities, destined to be ignored by the very people that they were designed for.
It is important to note that the research map shown in Figure 1 represents a relatively small set of experiments published in a single research paper. We chose to use a relatively simple research map so that we could more effectively illustrate the purpose and content of these maps. In practice, users of research maps will be able to interact dynamically with far more complex maps representing thousands if not millions of experiments. Just as a reader opens a specific section of a textbook, users of research maps will be able to direct their searches to specific sets and combinations of nodes in the map (e.g., specific molecules, types of physiology, particular behaviors, etc.), thus reducing the complexity of the maps and focusing their analyses on the components that interest them.
How would these maps be assembled?
Graduate students, post-doctoral fellows, principal investigators, and other biologists would initially extract from published research papers findings that describe the identity of biological phenomena (the nodes in the map), as well as those experiments that test causal connections between these phenomena (the edges in the maps). This would be routinely done as part of reading and analyzing research papers critical for their work. With a little practice, the process of entering these data into a suitable interface should take only a little longer than simply reading the paper. In our experience this adds only another 20%–30% of the time it takes to carefully read a research paper. Part of the additional time required by the manual curation of a research map is not simply due to the process of entering the required data. Instead, the additional time is needed to understand the research well enough to derive a map. In our experience, the rigor required to derive a research map adds to the understanding of the research being mapped.
Using manually entered examples, eventually, machine-learning routines (Bishop 2006; Mohri et al. 2012) could systematically populate the maps with similar and related experiments by trawling multiple resources of published information, such as the Library of Medicine. Even though at this point only some research papers in biology are available for data mining, this could change in the near future, as journals recognize the importance of changing current business models. We implore the science publishing houses to open their resources to big data projects, such as the one proposed here. In the future, authors could also include a research map of their findings with their manuscript submissions, thus facilitating the incorporation of new results into an overall research map.
The process of generating manual maps of causal information provides scientists with the opportunity to analyze in detail published experiments important to their work. This process can be generative because it requires close attention to the experiments being mapped. In an age of information overload, it is all too easy to gloss over content, and miss crucial details that may have otherwise led to important insights. This is especially true in areas of biology, such as molecular and cellular cognition, where interdisciplinary studies have become the norm, and where biologists struggle to master knowledge and approaches from several very different disciplines.
In the immediate future, manual entry will be key to accommodate the needs of individual scientists, and the considerable complexity of mapping causal information that involves new concepts and experimental paradigms. Thus, we expect that initially research maps will be a personalized tool that individual investigators use to track published work and plan future experiments. Therefore, individual investigators will be able to control the quality and standards of the experiments represented in the maps they use for planning their own work. However, as machine-learning routines get better, with more experience and feedback from biologists, manual entry could become an increasingly smaller component of updating research maps. In the distant future, we imagine that these maps will be updated automatically every time a new article or any other research resource is made public.
With such a map, scientists could instantly evaluate the amount and type of evidence available for any one causal connection of interest. Research maps would be machine readable, and therefore users would be able to interact with these maps dynamically: for example, they could query them for possible connections between any two phenomena in the maps, mine them for hitherto unsuspected relations and for micro and macro trends. Moreover, to facilitate research planning, users could also generate personalized private maps with their own unpublished results. These private maps of unpublished information are a real help in deciding how to best complement existing experiments before submitting a manuscript for publication. Although there is a freely available web-driven interface for assembling and interacting with research maps (www.researchmaps.org), users can draw their own maps with nothing more than pen and paper (Landreth and Silva 2013; Silva et al. 2014).
Beyond representations of published experiments, research maps could also be linked to the growing number of biological resources that curate large bodies of information about genes, proteins, cells, biological systems, clinical resources, etc. These resources would be not only accessible from research maps, they would also be appropriately integrated with the nodes and edges of these maps. For example, by selecting any one node in the map, a user would get instant access to the resources that reference phenomena related to that node, whether this information was originally reported in a research paper or in one of the numerous databases that curate the ever growing and increasingly sophisticated collections of biological phenomena (Wren and Bateman 2008).
With maps of causal research statements, the size and complexity of the biological corpus would be less overwhelming during research planning. These maps would provide versatile visual interactive depictions of critical causal findings that could potentially reveal links between experiments that would otherwise be lost within the immensity of the published record. While planning the next series of experiments, biologists could use the maps to scan relevant findings (even in unfamiliar areas), get an immediate sense for the amount of evidence and types of experiments already published in that topic, and quickly judge whether further work, if any, should be carried out next. Scientists could also use such an interactive system to add to their intuitions, and thus better estimate the relative merits of alternative experimental plans. Sophisticated machine-learning routines could conceivably learn from how scientists use this resource, and then aid in its use as research maps grow in size and complexity. The ease and clarity afforded by such causal maps would facilitate creative exploration of ideas that may otherwise lie buried by the crashing size of the literature.
Research maps at work
Figure 1 shows a research map for a molecular and cellular cognition article that includes 14 different causal experiments exploring the role of the NF1 gene on key physiological (e.g., paired pulse facilitation or PPF; long-term potentiation or LTP) and behavioral phenomena (e.g., hippocampal learning) (Costa et al. 2002). Reading and understanding the causal content of this article would take a considerable amount of time. However, even a brief inspection of the map of the article (Fig. 1), gives trained neuroscientists a fairly comprehensible view of the results in that paper (Costa et al. 2002). More important, since the results in that paper would be part of a larger database with other related information, it would be possible to quickly explore connections between those findings and other findings. It is easy to see how this same approach could be scaled and used to integrate vast amounts of causal information while allowing users to interact with chosen subsets of these data without being overwhelmed by the cheer quantity of the overall information. The problem with information is not quantity but usability. Research maps get around the quantity problem by placing causal information in a format that biologist can easily use.
A science of experiment planning
Every scientist works hard to optimize experiment planning. Beyond methods that advise scientists on sample size and other statistical parameters (Fisher 1935; Winer et al. 1962; Quinn and Keough 2002) experiment planning is mostly intuitive and depends on insights and wisdom gained from experience, as well as advice from colleagues and mentors. These factors are important and will undoubtedly continue to be key to how scientists make research choices. However, we imagine a future where we would more easily and efficiently learn from the collective countless research choices made by scientists world-wide. An accurate and objective record of how scientists interact with research maps and the research choices they make (recorded in the experiments they publish), could lead to studies designed to both learn from this collective experience and eventually optimize experiment planning. Machine learning could also potentially have a role in this optimization process by finding hidden patterns among successful cases. We caution, however, that principles of experiment planning derived from these studies should never play a restrictive role in research planning: we are strong believers in serendipity and the extraordinary power of human creativity and intuition! Instead, insights gained from studying our collective scientific choices would simply help scientists use their intuition and creativity to address the unprecedented complexity of the ever-growing published record. In another 20 yr the library of medicine will have an estimated half a billion published experiments. How will we address this complexity without new tools and new methods?
Next steps
The semiautomated research maps of causal information that we propose could be built in the next 5 yr. However, meanwhile biologists could use the concepts and tools we mentioned here to keep track of research findings and resources critical for their work. One of the ironies of the problems biologists face is that the vast majority of the informatics resources that have been developed to address specific needs in this community are under used (Akil et al. 2011). There are many reasons for this, but there is no doubt that the current training divide between informatics and biology is a key contributor (Akil et al. 2011). Beyond the urgent need for a greater effort to provide students with basic training in informatics, biologists and computer scientists need to form collaborations to continue to find solutions to the information problem. Additionally, funding sources both public (e.g., National Institutes of Health in the USA) and private (e.g., foundations) should expand their efforts to promote and support these collaborations so that we stimulate much needed growth in this area. Publishers also need to remove the restrictions that currently hamper access to published research, so that data mining efforts can flourish (Neylon 2012). It will also be critical that experimental findings, including causal statements, are reported in a machine-readable format (e.g., nano-publication; Groth et al. 2010) to facilitate access and mining of this resource. The process of submitting an article to publication could easily include a summary of experimental findings in a machine-readable format. This will require a little training and some extra work for biologists, but it is not an unreasonable demand considering the potential benefits.
Biology and computer science students should be made aware of the challenges discussed here, because we need their energy, imagination and creativity to solve this growing problem. What we need is nothing short of reinventing how we integrate, plan, and report research findings. It is an enormous but incredibly exciting challenge with far reaching impact that is clearly not restricted to the biological sciences, but that will affect sister disciplines, such as chemistry and the social sciences, and potentially all of science. One thing, we are certain: with the current vertiginous growth of the scientific literature, the status quo is no longer tenable. We need to dramatically expand current computation efforts, and we need to do this now!
Acknowledgments
We would like to thank Anthony Landreth, Pranay Doshi, Nicholas Matiasz, Justin Wood, Darin Nee, Tawnie Silva, William Hsu, Wei Wang, Alan Smith, and Peter Whybrow for discussions that shaped the ideas presented here.
Footnotes
Article is online at http://www.learnmem.org/cgi/doi/10.1101/lm.029355.112.
References
- Akil H, Martone ME, Van Essen DC. 2011. Challenges and opportunities in mining neuroscience data. Science 331: 708–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop CM. 2006. Pattern recognition and machine learning. Springer, New York. [Google Scholar]
- Costa RM, Federov NB, Kogan JH, Murphy GG, Stern J, Ohno M, Kucherlapati R, Jacks T, Silva AJ. 2002. Mechanism for the learning deficits in a mouse model of neurofibromatosis type 1. Nature 415: 526–530. [DOI] [PubMed] [Google Scholar]
- Dai L, Gao X, Guo Y, Xiao J, Zhang Z. 2012. Bioinformatics clouds for big data manipulation. Biol Direct 7: 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher RA. 1935. The design of experiments. Oliver and Boyde, Edinburgh, London. [Google Scholar]
- Groth P, Gibson A, Velterop J. 2010. The anatomy of a nanopublication. Inf Serv Use 30: 51–56. [Google Scholar]
- Landreth A, Silva AJ. 2013. The need for research maps to navigate published work and inform experiment planning. Neuron 79: 411–415. [DOI] [PubMed] [Google Scholar]
- Lok C. 2010. Literature mining: speed reading. Nature 463: 416–418. [DOI] [PubMed] [Google Scholar]
- Mohri M, Rostamizadeh A, Talwalkar A. 2012. Foundations of machine learning. MIT Press, Boston. [Google Scholar]
- Neylon C. 2012. Science publishing: open access must enable open use. Nature 492: 348–349. [DOI] [PubMed] [Google Scholar]
- Novak JD. 1990. Concept maps and Vee diagrams: two metacognitive tools for science and mathematics education. Instr Sci 19: 29–52. [Google Scholar]
- Novak JD, Musonda D. 1991. A twelve-year longitudinal study of science concept learning. Am Educ Res J 28: 117–153. [Google Scholar]
- Quinn GP, Keough MJ. 2002. Experimental design and data analysis for biologists. Cambridge University Press, Cambridge. [Google Scholar]
- Russ T, Ramakrishnan C, Hovy EH, Bota M, Burns G. 2011. Knowledge engineering tools for reasoning with scientific observations and interpretations: a neural connectivity use case. BMC Bioinformatics 12: 351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva AJ, Landreth A, Bickle J. 2014. Engineering the next revolution in neuroscience: the new science of experiment planning. Oxford Press, New York, New York. [Google Scholar]
- Winer B, Brown D, Michels K. 1962. Statistical principles in experimental design. McGraw-Hill, New York. [Google Scholar]
- Wren JD, Bateman A. 2008. Databases, data tombs and dust in the wind. Bioinformatics 24: 2127–2128. [DOI] [PubMed] [Google Scholar]
- Zhao S, Nishimura T, Chen Y, Azeloglu EU, Gottesman O, Giannarelli C, Zafar MU, Benard L, Badimon JJ, Hajjar RJ, et al. 2013. Systems pharmacology of adverse event mitigation by drug combinations. Sci Transl Med 5: 206ra140. [DOI] [PMC free article] [PubMed] [Google Scholar]