Abstract
The sh3bgr (SH3 domain binding glutamate-rich) gene encodes a small protein containing a thioredoxin-like fold, SH3 binding domain, and glutamate-rich domain. Originally, it was suggested that increased expression of Sh3bgr may cause the cardiac phenotypes in Down's syndrome. However, it was recently reported that the overexpression of Sh3bgr did not cause any disease phenotypes in mice. In this study, we have discovered that Sh3bgr is critical for sarcomere formation in striated muscle tissues and also for heart development. Sh3bgr is strongly expressed in the developing somites and heart in Xenopus. Morpholino mediated-knockdown of sh3bgr caused severe malformation of heart tissue and disrupted segmentation of the somites. Further analysis revealed that Sh3bgr specifically localized to the Z-line in mature sarcomeres and that knockdown of Sh3bgr completely disrupted sarcomere formation in the somites. Moreover, overexpression of Sh3bgr resulted in abnormally discontinues thick firmaments in the somitic sarcomeres. We suggest that Sh3bgr does its function at least partly by regulating localization of Enah for the sarcomere formation. In addition, we provide the data supporting Sh3bgr is also necessary for proper heart development in part by affecting the Enah protein level.
Introduction
The sh3bgr gene was originally identified in an effort to discover the causative genes for Down's syndrome (Scartezzini et al., 1997). The sh3bgr gene lies in the 21q22.3 region and is known to be expressed in fetal heart and skeletal muscle tissues in mammal (Egeo et al., 2000). Three additional sh3bgr homologues, sh3bgrl, sh3bgrl2, and sh3bgrl3, define the sh3bgr family (Mazzocco et al., 2001; Mazzocco et al., 2002). Among them, Sh3bgr contains an extra proline rich and a glutamic acid rich domain in the C-terminal region adjacent to the N-terminal thioredoxin fold domain. Although the sh3bgr gene lies in the Down's syndrome critical region, overexpression by inserting one extra copy of the gene did not show any scorable phenotypes (Sandri et al., 2004). Recent data on sh3bgrl suggest that the sh3bgr family may have a distinct function from other thioredoxin fold proteins which generally control redox potentials. Sh3bgrl seems to possess a tumor suppressor function in the v-Rel-mediated tumor transformation process (Majid et al., 2006). However, the biological functions of Sh3bgr are yet to be determined.
The sarcomere is a structural and functional building unit of striated cardiac and skeletal muscle fibers. The Z-line is the structural border of each sarcomere, and the thin actin filaments are tightly associated with the Z-line at each end of the sarcomere (Luther, 2009). The thick myosin filaments are also compartmentalized with the distinct M-band containing several proteins which are critical for the contractile function of the muscle fibers (Agarkova and Perriard, 2005).
During sarcomere assembly, the Enah protein localizes to the Z-line and is required for the α-II-spectrin and α-actin assembly at the Z-line of cardiac muscles and intercalated disc (ICD) in cardiomyocytes (Benz et al., 2013). Also, the Enah protein level is dramatically increased in the ICD and Z-line upon calsequestrin overexpression in mice (Aguilar et al., 2011). Loss of both Enah and Vasp in cardiomyocytes caused dilated cardiomyopathy which is due to the defective ICD and Z-line formation (Benz et al., 2013) and overexpression of Enah resulted in the exacerbation of heart failure after cardiac injury in mice (Belmonte et al., 2013).
In this report we show that Sh3bgr localizes to the Z-line in the somitic sarcomeres and is critical for proper development of the somites and heart. Furthermore, we show the evidence supporting the idea that Sh3bgr is required for the proper localization and expression level of the Enah protein. Our data also suggest that overexpression of Sh3bgr may cause defects in muscle function by changing the architecture of thick filaments of the skeletal muscles. Further, overexpression of Sh3bgr resulted in an enlarged heart during development. These are the first experimental data suggesting a possible mode of action whereby Sh3gbr may contribute to cardiac and skeletal muscle development.
Results and discussion
Sh3bgr is expressed in the somite and myocardium
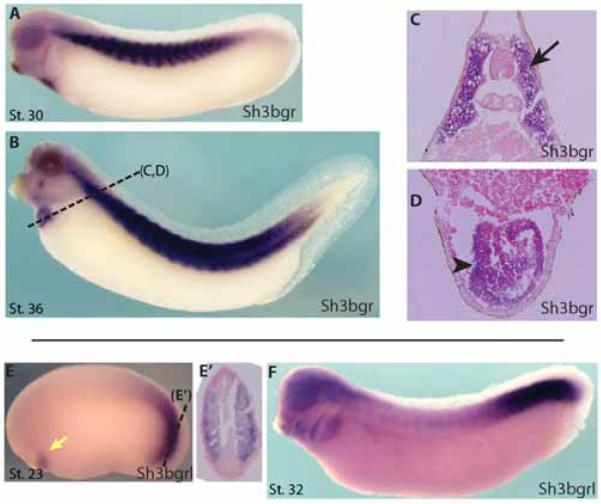
In an effort to elucidate the biological functions of the Sh3bgr family genes in developing vertebrate embryos, we cloned Xenopus homologues of Sh3bgr family genes according to the sequences from the Xenbase (James-Zorn et al., 2013). Next, we analyzed expression of the Sh3bgr family by whole-mount in situ hybridization assay in Xenopus embryos. In gastrulating embryos, sh3bgr is expressed in the presumptive somitic mesoderm. At the tailbud stage, sh3bgr is strongly expressed in developing somite and cardiac tissues as previously reported for mammals (Fig.1A-D). At a later stage, sh3bgr expression was also observed in facial muscles (Fig.1B). In situ hybridization of tissue sections revealed that sh3bgr is expressed in the myocardial precursor tissues and the entire somitic tissues (Fig. 1C-D). These data suggest that Sh3bgr may have roles in striated muscle development. The sh3bgrl gene is also expressed in the somite and heart tissues but the expression pattern was restricted to the presomitic tissues and early cardiac tissues in contrast to Sh3bgr expression (Fig. 1E-F). Two other homologues are expressed in trigeminal neurons and will be discussed elsewhere. Interestingly, the sh3bgr gene seems to be expressed as several splicing variants during development (Fig. S1D). We cloned the two major splicing variants and found that the long form of sh3bgr transcript (Sh3bgr-a) contains an additional exon which is 57 nucleotides long compared to the shorter version of the transcript (Sh3bgr-b) (Fig. S1E).
Figure 1. sh3bgr is expressed in the skeletal muscles and heart tissues.
Whole mount in situ hybridization showing that sh3bgr is expressed in the somites and heart at Stage 30 (A), Stage 36 (B). C, D. Sectioned samples shown in (B). E. sh3bgrl is expressed in the presomitic tissue and early cardiac tissue at Stage 23. F. At Stage 32, sh3bgrl is expressed in the pharyngeal arches and the presomitic expression was maintained.
Knockdown of Sh3bgr causes severe defects in somite and heart development
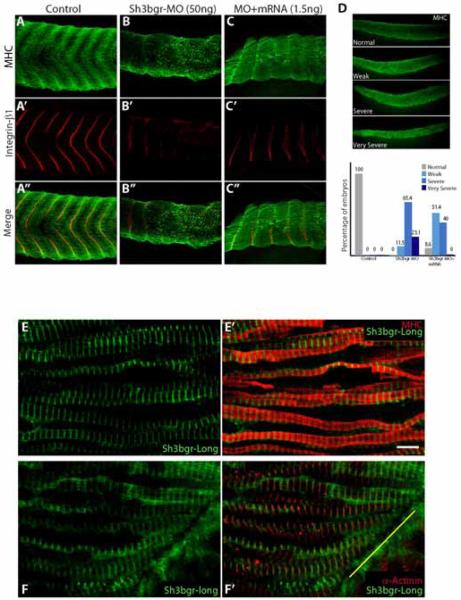
In order to study the functions of sh3bgr in vertebrate development, we assessed the loss-of-function phenotypes by specifically repressing the expression of sh3bgr or sh3bgrl using the antisense morpholino (Sh3bgr-MO, Sh3bgrl-MO). Sh3bgr-MO and Sh3bgrl-MO were both designed to block the splicing of the transcript. Microinjection of either Sh3bgr-MO or Sh3bgrl-MO at 2 cell stage embryos efficiently inhibited the mature mRNA formation (Fig. S2B, data not shown). Knockdown of sh3bgr expression caused severe defects in developing somite and heart, revealed by myosin heavy chain (MHC) staining (Fig. 2A-D and Fig. S2C). Also, integrin-β1 staining showed that the boundaries of somites were severely disrupted in the morphants (Fig. 2A-C). However, Sh3bgrl-MO did not cause any noticeable phenotype although it efficiently inhibited the mature RNA formation (data not shown). So we focused on studying sh3bgr thereafter. The ectopic expression of the in vitro transcribed mRNA reduced the severity of the somite loss of function phenotype, suggesting that the striated muscle phenotypes are due to the knockdown of sh3bgr expression (Fig. 2D). Muscle differentiation during development is achieved by sequential expression of muscle regulatory transcription factors such as myoD, myf5 and myogenin (Buckingham and Rigby, 2014). We have examined the expression patterns of several muscle regulatory factors in Sh3bgr morphants. The unique chevron pattern of somite tissues seemed to be severely distorted in the morphants. However, we did not observe any significant differences in the expression levels of MRF genes between control and Sh3bgr morphants (Fig. S2A-B). It seems the differentiation processes of striated muscles are not significantly affected by sh3bgr knockdown. Rather, Sh3bgr seems to regulate structural aspects of skeletal muscle formation.
Figure 2. Knockdown of sh3bgr caused severe defects in somite development.
A-C. Microinjection of sh3bgr antisense morpholino (Sh3bgr-MO; 50ng per embryo) disrupted normal somite development. The somite boundaries were labeled with anti-integrin-β1 antibody (Red) and the muscle fibers were stained with the myosin heavy chain (MHC) antibody (Green) at Stage 33. D. The muscle phenotypes were categorized according to the severity. Weak; the chevron pattern is partially disrupted, Severe; the chevron pattern is completely lost. Very severe; the somatic tissue is severely hypomorphic. The muscle defects in the morphants were recovered by co-injection with 1.5ng of sh3bgr mRNA. The numbers in the graph are the percentage of the embryos. The numbers of embryos analyzed: Control-13, sh3bgr morphants-26, Rescue-35. E-E’. Sh3bgr localizes to the muscle fibers. 150ng of Sh3bgr-flag mRNA was injected into Stage 2 embryos. Stage 33 embryos were fixed and the coronal sections of somitic tissues were stained for MHC (Red) and Sh3bgr-flag (Green). F-F’. Sh3bgr-flag fusion protein localized to the Z-line in sarcomeres. Sh3bgr-flag (Green) was co-immunostained with α-actinin (Red). The yellow line indicates a somitic boundary. Scale bar = 5μm
Sh3bgr localizes to the somitic boundaries and the z-lines
Because the knockdown of sh3bgr expression caused severe defects in somite and cardiac muscle development without affecting muscle differentiation status, we reasoned that sh3bgr may be required to organize muscle fibers. Although the thioredoxin fold of Sh3bgr family proteins is very similar to that of glutaredoxin, the molecular functions of Sh3bgr have not yet been elucidated. To address the biochemical functions of Sh3bgr in muscle development, we have injected very low dose of Sh3bgr-flag fusion construct (150 ng of mRNA) targeting somitic tissues at 2 cell stage Xenopus embryos and visualized the fusion protein using confocal microscopy. The Sh3bgr-flag fusion protein was observed in a repeating pattern along with the muscle fibers (Fig. S2D). Co-immunostaining with myosin heavy chain (MHC) revealed the alternating patterns of Sh3bgr fusion protein with MHC signals, indicating that Sh3bgr protein localizes to the Z-lines of sarcomeres(Fig. 2E-E’). Indeed, co-immunostaining with a Z-line marker α-actinin revealed that the Sh3bgr signal overlaps with the α-actinin signals (Fig. 2F-F’). We have also examined localization of the shorter version of Sh3bgr splicing variants and found that the splicing variant is also localized to the Z-line (data not shown). The Z-line is not only known to function as a structural unit for sarcomere formation but is also critical for the contractile ability of mature striated muscles (Luther, 2009). Given the specific localization of Sh3bgr protein to the Z-line, we reasoned that the gross defects of developing somite and myocardium might be due to defective muscle fiber formation.
Sh3bgr is necessary for sarcomere formation
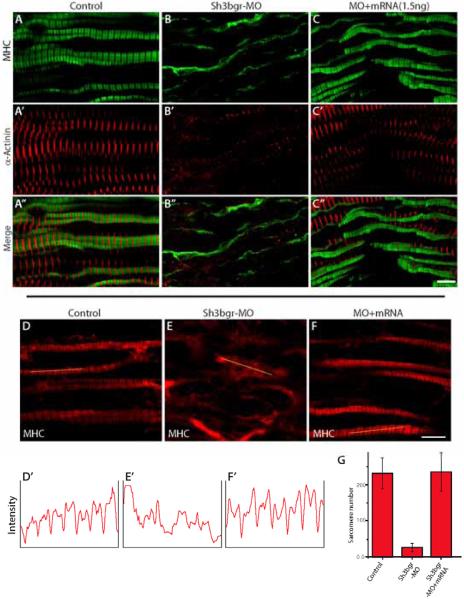
In order to examine the function of Sh3bgr in muscle fiber formation, we observed somitic muscle fibers using confocal microscopy. In Sh3bgr morphant embryos, the sarcomeres in somites were completely disrupted and the regular spacing of MHC was not observed (Fig. 3A-B). The co-injection of mature sh3bgr mRNA with Sh3bgr-MO can effectively reverse the sarcomere defects (Fig. 3C, F, G). It seemed the fluorescent intensity of myosin heavy chain in the morphants’ tissue was comparable to that of the control. Also, the western blot analysis revealed that the level of myosin heavy chain was not significantly affected by the knockdown of Sh3bgr (Fig. S3D). We have also analyzed the localization of α-actinin and myomesin in sarcomeres for Z-line and M-band markers respectively upon knockdown of Sh3bgr (Fig. 3A’-C’ and Fig. 4A-B). Indeed, both Z-line and M-band were not observed in the Sh3bgr knockdowned somites. When we counted the intact sarcomeres, we observed a significantly reduced number of sarcomeres in the morphants (Fig. 3D-G). Next, we examined the expression level of α-actinin and myomesin in Sh3bgr compromised embryos, however we did not observe any significant differences compared to control (Fig. S3D). These data indicate that Sh3bgr may not be necessary for the specification of the striated muscles but is critical for the sarcomere formation in the striated muscles.
Figure 3. sh3bgr is necessary for sarcomere formation.
A-C. Stage 33 embryos were fixed and the coronal sections of somitic tissues were stained for α-actinin (Red) and MHC (Green). Knockdown of sh3bgr caused severe defects in the sarcomeres. Sh3bgr knockdown caused almost compete loss of sarcomere structures (B). Co-injection of Sh3bgr mRNA (1.5ng) with Sh3bgr-MO rescued sarcomere defects (C). D-F. Representative images of thick filaments. The fluorescent intensity plots of indicated muscle fibers are shown in (D’-F’). The average numbers of intact sarcomeres in each sample are shown in (G). Scale bar = 5μm
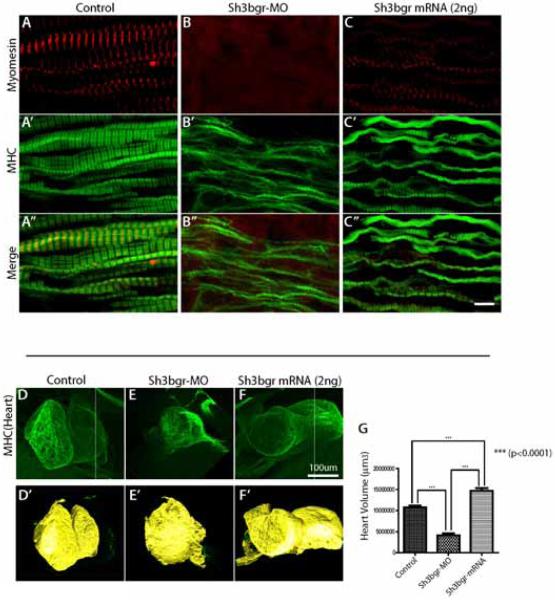
Figure 4. Overexpression of Sh3bgr caused abnormal sarcomere formation and increased heart volume.
A-C. Stage 33 embryos were fixed and the coronal sections of somitic tissues were stained for myomesin (Red) and MHC (Green). Overexpression of sh3bgr (2ng of Sh3bgr-flag mRNA) caused abnormally discontinuous thick filaments and mislocalizaiton of myomesin. D-F. Overexpression of Sh3bgr increased the heart volumes. The embryonic hearts were stained with anti-MHC antibody and the volumes of Stage 45 embryos were measured using the Imaris program. D’-F’. The surface views of 3D confocal images in (D-F) rendered using the Imaris program. The heart volume was plotted in (G). The numbers of embryos analyzed: Control-13, sh3bgr morphants-13, Rescue-13. Scale bar = 5μm
Overexpression of sh3bgr alters sarcomere morphology and heart development
Although one extra copy of sh3bgr did not seem to cause critical defects in striated muscles in mice, sh3bgr was originally identified as a putative cause of heart defects in Down's syndrome. Since the data suggest that Sh3bgr is a critical component for the structural integrity of sarcomeres, we reasoned that the strong overexpression of sh3bgr might also cause abnormal muscle formation. Consistent with the transgenic mouse data, the overexpression of the gene did not cause noticeable embryonic phenotypes. Also, the overall morphology of the somite muscle looked comparable to that of control embryos (data not shown). However, when we observed the sarcomere structures of the somite, we found the thick filaments of Sh3bgr overexpressing embryos were noticeably more discontinuous than those of the control (Fig. 4C, S3B-C). The overall length of the thick filament is about 0.7-8 μm in contrast to the 1.5 μm of the control thick filaments. This suggested us that the M-band of the thick filaments may not form properly in Sh3bgr overexpressing somitic cells. Then, we observed if the M-band was properly formed in Sh3bgr overexpressed embryos by analyzing the M-band marker myomesin. Compared to control, myomesin localization on the M-band was significantly disorganized (Fig. 4A, C). In addition, we examined the morphology of the thick filaments using super-resolution structured illumination microscopy (SIM) (Fig. 5A-C). Indeed, the SIM images of the sarcomeres showed that the thick filaments in Sh3bgr overexpressing cells were significantly discontinuous.
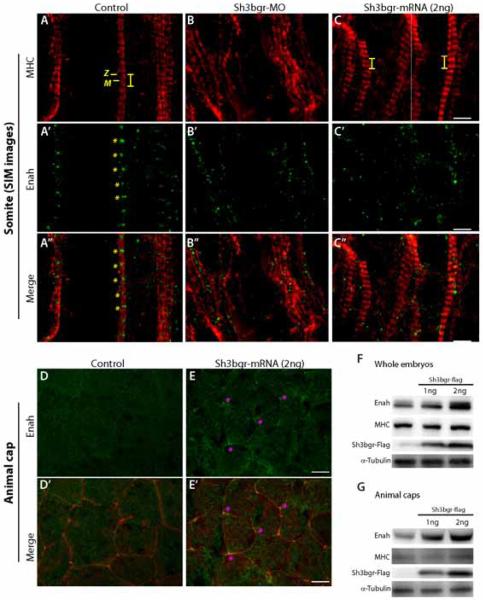
Figure 5. Sh3bgr is an upstream regulator of Enah.
A-C. Super resolution SIM images of sarcomeres in control (A) Sh3bgr knockdown (B), and Sh3bgr overexpressing embryos (C). Enah protein was labeled in green and MHC was labeled in red. The Z-line and M-band are indicated within a sarcomere. Both knockdown and overexpression of Sh3bgr delocalize Enah from the Z-lines. D-E. Enah staining in control (D) and Sh3bgr overexpressing (E) animal cap cells. In Sh3bgr overexpressing animal cap cells, the Enah protein is highly enriched in tri-junctional foci (asterisk). F-G. The western blot analysis showed that the endogenous Enah protein level was sharply increased in whole embryos (F) or animal cap tissues (G) upon overexpression of Sh3bgr.
Scale bar (A-C) = 2μm Scale bar (D, E) = 10μm
Given the strong expression of Sh3gbr in the heart tissue and the consistent overexpression phenotypes in somitic muscles, we turned our attention to the heart development in Sh3bgr compromised embryos. To this end, we imaged whole heart tissues upon Sh3bgr knockdown or overexpression by performing MHC immunostaining and analyzed the heart development. While knockdown of Sh3bgr drastically disrupted the overall heart development (Fig. 4E, G), the heart development in sh3bgr overexpressing embryos did not seem to be noticeably disrupted. Due to the heterogeneity of the heart morphology at the early embryonic stages, we were not able to directly compare the morphogenetic differences in Sh3bgr overexpressed embryos. However, to our surprise, when we measured the volume of the embryonic hearts, Sh3bgr overexpressing embryos developed with consistently and significantly enlarged hearts (Fig. 4 F, G).
These data suggest that the precise level of Sh3bgr is critical for both skeletal muscle and heart development.
Sh3bgr regulates Enah protein for striated muscle development
Although the X-ray crystal structure of the thioredoxin fold in Sh3bgr is solved, no data on the biochemical functions of Sh3bgr have been published yet. Since Sh3bgr localizes to the Z-lines and is necessary for sarcomere formation, we hypothesized that Sh3bgr may be involved in actomyosin organization. The primary sequences of sh3bgr suggest the protein contains a SH3 binding domain and an EVH1 binding domain (Mazzocco et al., 2002). We especially focused on the EVH1 domain containing proteins because the EVH1 proteins are known to be necessary for proper actin dynamics and muscle formation (Bear and Gertler, 2009). There are three known proteins with an EVH1 domain in vertebrates: Vasp, Enah, and Evl. Among them, the Enah protein was previously reported to be involved in the morphogenetic process for somite development (Kragtorp and Miller, 2006). Also, the Enah protein is critical for the Z-line and intercalated disc (ICD) formation in cardiac muscles (Aguilar et al., 2011; Belmonte et al., 2013; Eigenthaler et al., 2003). Moreover, Enah overexpression caused a mild increase of heart mass and exacerbated heart failure after cardiac injury in mice (Belmonte et al., 2013). We therefore examined the interactions between Sh3bgr and Enah in the formation of striated muscles.
In order to examine possible molecular interactions between Sh3bgr and the Enah/Vasp protein, we observed the localization and expression level of the Enah protein in Sh3bgr compromised embryos. As previously reported for the cardiac muscles (Benz et al., 2013), both confocal and super resolution SIM images clearly showed the Enah protein is also localized to the Z-line of somitic sarcomeres (Fig. S3E, 5A’). In contrast, the Enah protein in the morphants was completely disorganized (Fig. S3F, 5B’), but this may be secondary to defective sarcomere formation. Next, we observed the Enah protein in Sh3bgr overexpressing somite tissue. Excess Sh3bgr did not severely disrupt the sarcomere structure. However, the localization of the Enah protein was completely disrupted upon Sh3bgr overexpression (Fig. S3G, 5C’). The Enah protein was no longer associated to the Z-line; instead, it was broadly distributed along with the muscle fibers. In some somitic cells, the Enah protein was observed in the cytosol associated with the non-sarcomeric MHC signals (Fig. 5C’).
Interestingly, the level of the Enah protein was sharply increased by overexpression of Sh3bgr in the whole embryo lysate (Fig. 5F). However, in contrast to the increased Enah protein level in the embryo lysate, the fluorescent intensity of the Enah protein in the Sh3bgr overexpressing somitic cells was not significantly different from the control. This may be because the Enah gene is already strongly expressed in the somite and not very sensitive to the excess Sh3bgr protein. Indeed, developing somites are the major Enah expressing tissues in tail bud stage embryos (Xanthos et al., 2005). Therefore, we speculated that the increased Enah signals on the western blot of whole embryo lysate may due to the increase of Enah protein in non-muscle cells which minimally express Enah.
To analyze how Enah is affected by Sh3bgr in non-muscle cells, we used Xenopus animal cap tissue. In control animal cap cells, a weak Enah signal was observed at cell boundaries, whereas in Sh3bgr overexpressing animal cap cells, the strong Enah signal was frequently observed around the cell-cell junctions (Fig 5D, E). These tri-junctional foci of the Enah protein were not observed in control animal cap tissues at all. In addition to that, we normalized the fluorescent intensity of the Enah protein with auto-fluorescence in the DAPI channel and compared the normalized intensity between control and Sh3bgr overexpressing samples. The mean fluorescent intensity was significantly increased in Sh3bgr overexpressing cells (Fig. S3H). Likewise, the western blot analysis showed that the overexpression of Sh3bgr in animal cap tissues strongly increased the Enah protein level compared the control (Fig. 5G).
Our data demonstrate for the first time that Sh3bgr is necessary for both heart development and sarcomere formation in the somites. In this study, we show that upon knockdown of sh3bgr, the Z-line proteins are no longer recruited to the Z-line, suggesting that Sh3bgr is required for the early steps of Z-line assembly. One of the surprising aspects of Sh3bgr is that it regulates not only the localization, but also the amount of Enah, which is known to function as an actin anti-capping and bundling protein. Furthermore, we present the first evidence that overexpression of Sh3bgr may cause abnormal assembly of sarcomeres in the somites and also give rise to abnormal heart development. Interestingly, increased Enah expression was shown to be correlated with heart failure in mice and humans (Blaxall et al., 2003a; Blaxall et al., 2003b) . Also, Enah overexpression exacerbated cardiac injury after transverse aortic constriction in mice (Belmonte et al., 2013). Taken altogether, our data revealed that the precise level of Sh3bgr is critical for striated muscle development in the somites. Although we were not able to thoroughly examine the heart muscle phenotypes in sh3bgr knockdown embryos due to the technical difficulties, we observed that the embryonic heart was severely hypomorphic in the Sh3bgr morphants (Fig. S2C) and the overexpression of Sh3bgr resulted in increased heart volume (Fig. 4D-G). Overall these results illustrate the importance of the Sh3bgr protein in muscle development, through its interactions with EVH1 domain-containing proteins such as Enah.
Materials and Methods
Xenopus Embryo manipulations
Adult female Xenopus laevis were ovulated by injection of human chorionic gonadotropin (HCG), and eggs were fertilized in vitro and dejellied by pH 7.9, 3% cysteine in 1/3X MMR (Marc's Modified Ringers) solution. Fertilized eggs were reared in 1/3X MMR.
Injection procedure
For microinjections, embryos were placed in 3% ficoll in 1/3X MMR solution, injected in the ventral-vegetal site for somite targeting at stage 2 embryos (staged according to Nieuwkoop and Faber (1967)) by using a Picospritzer III microinjector (Parker), and grown in 1/3X MMR with gentamycin. For experiments, we injected morpholinos at 40-80ng per blastomere and mRNAs in various amounts.
Morpholino
We designed splicing-blocking antisense morpholinos for Sh3bgr on the basis of the sequences from the Xenbase database. We obtained morpholinos from Gene Tools; (Sh3bgr Int MO, TAAAAACCCTCAAACTTACCGCAGT)
Fixation
To analyze embryonic muscle development, especially striated somite muscle, we fixed embryos between stages 33 using MEMFA (MEM salts and 4% formaldehyde) or Dent's fixative solution (Methanol and 20% DMSO).
Sectioning
Fixed embryos were washed with 1X PBS, embedded in 3% low-melted agarose in 1X PBS and sectioned coronally in 100μm thickness using vibratome (Leica, VT1000S).
For cryo-sectioning, fixed embryos were washed with 5, 10, 15% sucrose, embedded in Optimal Cutting Temperature compound (Sakura Finetek) in the direction orthogonal to the embryonic anterior-posterior axis, and sectioned in 15μm thickness using a cryostat (Thermo Scientific, HM56OM).
In situ hybridization
The whole-mount in situ hybridization was performed as described previously (Harland, 1991).
Cloning and plasmid information
The full-length sequence of the Xenopus laevis Sh3bgr gene (942bp) was obtained from the Xenbase database.
We designed the primer pairs with a restriction enzyme site (SalI-forward, 5’- aattgtcgacGA TCTGTGCTCCCTGTTCCAAG-3’; NotI-backward, 5’-aattgcggccgcATTCTTGCTCTACTTCTTCTTCTACTTC TTC-3’). Total RNA was purified from stage 30-33 embryos and reverse-transcribed with random hexamer (Life technologies) and Reverse Transcriptase (Promega). Amplified PCR product was cloned into the CS107 and 3X Flag tag sequences were added to the C-terminus of the ORF.
We have cloned two splicing variants of Sh3bgr. Two forms of Sh3bgr mRNAs were transcribed with SP6 RNA polymerase (NEB) from cloned plasmid.
Immunofluoresence, Immunoblotting and microscopy
Immunostaining
Embryos were fixed with MEMFA or Dent's fixative overnight at 4°C. Fixed embryos were rehydrated and all embryos or sectioned embryo slices were incubated in blocking solution (10% FBS + 2% DMSO in TBS + 0.1% Triton X-100) at room temperature for 30 minutes to block non-specific binding. Immunostaining was performed with the following antibodies: anti-DDDD-K (abcam), anti-Xenopus Enah (obtained from Dr. Jeffrey Miller), anti-GFP (abcam), anti-MyHC (DSHB), anti-actin (Thermo), anti-α-actinin (Sigma-aldrich), anti-myomesin (DSHB), anti-α-tubulin (abcam), and anti-integrin β1 (DSHB) at a 1:300 dilution for 3 hours at room temperature. Fluorescent labeling was performed using Alexa Fluore 405-, Alexa Fluore 488-, Alexa Fluore 555-, Alexa Fluore 633-conjugated (all 1:300; invitrogen), Phalloidin 633 (1:20; Life technologies), and DAPI (1:1000;) for secondary antibodies. All sectioned slices were run through washes in 100% methanol before clearing in BA:BB (benzyl alcohol/benzyl benzoate, 1:2) and mounting on Sylgard slides for imaging. Images were captured using a confocal microscope (LSM700) and SR-SIM (super resolution-structured illumination microscope; ELYRA S1) (both Zeiss).
Immunoblotting
For western blot analysis, control and injected embryos were collected at stage 29, homogenized in lysis buffer (50mM Tris pH 7.4, 105mM NaCl, 0.1% Triton X-100, 5% Glycerol), and supplemented with protease inhibitor. Homogenates were cleared by 13,200rpm centrifugation for 15 minutes at 4°C. Proteins were blotted to 6-12% polyacrylamide gel. Blots were blocked in TBS + 0.05% Tween 20 with skimmed milk and immunoblots were performed with anti-DDDD-K (abcam), anti-Xena (obtained from Dr. Jeffrey Miller), anti-GFP (abcam), anti-MyHC (DSHB), anti-HA (Roche), anti-α-actinin (Sigma-aldrich) and anti-myomesin (DSHB), for experiments, and anti-α-tubulin (abcam) for loading control antibodies at a 1:4000 dilution for 1 hour at room temperature. Visualization was performed using HRP-conjugated antibodies (1:4000) with SuperSignal West Pico Chemiluminescent Substrate or SuperSignal West Dura Extended Duration Substrate (both Thermo) and the blots were exposed to ChemiDoc MP (Bio-Rad).
Image analysis
Image analysis was performed using the ZEN program for merging and 3D images. Quantitative analysis of average length and the number of sarcomeres were performed using ImageJ software. The average volume of hearts was measured using Imaris software. P values were calculated using Origin9 two-simple t-test and Prism5 two-tail t-test.
Supplementary Material
Acknowledgement
We appreciate Dr. JB Wallingford, RM Harland and CY Park for critical reading and discussion, and JH Hur at UNIST-Olympus Biomedical imaging Center (UOBC) for technical support. This work was supported by grants from the Korea National Research Foundation (2012R1A1A1011604) and the research fund from UNIST (1.140034.01). This work was initiated in Richard Harland's lab, with support from NIH GM42341, and a Miller Fellowship to TJP.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author contributions
DG Jang and TJ Park performed the experiments and wrote the manuscript. HJ Sim, EK Song and S Medina-Ruiz performed the experiments. TJ Park designed the experiments
Conflict of Interest
The authors declare no conflict of interest.
References
- Agarkova I, Perriard JC. The M-band: an elastic web that crosslinks thick filaments in the center of the sarcomere. Trends Cell Biol. 2005;15:477–485. doi: 10.1016/j.tcb.2005.07.001. [DOI] [PubMed] [Google Scholar]
- Aguilar F, Belmonte SL, Ram R, Noujaim SF, Dunaevsky O, Protack TL, Jalife J, Todd Massey H, Gertler FB, Blaxall BC. Mammalian enabled (Mena) is a critical regulator of cardiac function. American journal of physiology. Heart and circulatory physiology. 2011;300:H1841–1852. doi: 10.1152/ajpheart.01127.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bear JE, Gertler FB. Ena/VASP: towards resolving a pointed controversy at the barbed end. J Cell Sci. 2009;122:1947–1953. doi: 10.1242/jcs.038125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belmonte SL, Ram R, Mickelsen DM, Gertler FB, Blaxall BC. Cardiac overexpression of Mammalian enabled (Mena) exacerbates heart failure in mice. American journal of physiology. Heart and circulatory physiology. 2013;305:H875–884. doi: 10.1152/ajpheart.00342.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benz PM, Merkel CJ, Offner K, Abesser M, Ullrich M, Fischer T, Bayer B, Wagner H, Gambaryan S, Ursitti JA, Adham IM, Linke WA, Feller SM, Fleming I, Renne T, Frantz S, Unger A, Schuh K. Mena/VASP and alphaII-Spectrin complexes regulate cytoplasmic actin networks in cardiomyocytes and protect from conduction abnormalities and dilated cardiomyopathy. Cell Commun Signal. 2013;11:56. doi: 10.1186/1478-811X-11-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaxall BC, Spang R, Rockman HA, Koch WJ. Differential myocardial gene expression in the development and rescue of murine heart failure. Physiol Genomics. 2003a;15:105–114. doi: 10.1152/physiolgenomics.00087.2003. [DOI] [PubMed] [Google Scholar]
- Blaxall BC, Tschannen-Moran BM, Milano CA, Koch WJ. Differential gene expression and genomic patient stratification following left ventricular assist device support. J Am Coll Cardiol. 2003b;41:1096–1106. doi: 10.1016/s0735-1097(03)00043-3. [DOI] [PubMed] [Google Scholar]
- Buckingham M, Rigby PW. Gene regulatory networks and transcriptional mechanisms that control myogenesis. Dev Cell. 2014;28:225–238. doi: 10.1016/j.devcel.2013.12.020. [DOI] [PubMed] [Google Scholar]
- Egeo A, Di Lisi R, Sandri C, Mazzocco M, Lapide M, Schiaffino S, Scartezzini P. Developmental expression of the SH3BGR gene, mapping to the Down syndrome heart critical region. Mech Dev. 2000;90:313–316. doi: 10.1016/s0925-4773(99)00253-1. [DOI] [PubMed] [Google Scholar]
- Eigenthaler M, Engelhardt S, Schinke B, Kobsar A, Schmitteckert E, Gambaryan S, Engelhardt CM, Krenn V, Eliava M, Jarchau T, Lohse MJ, Walter U, Hein L. Disruption of cardiac Ena-VASP protein localization in intercalated disks causes dilated cardiomyopathy. American journal of physiology. Heart and circulatory physiology. 2003;285:H2471–2481. doi: 10.1152/ajpheart.00362.2003. [DOI] [PubMed] [Google Scholar]
- James-Zorn C, Ponferrada VG, Jarabek CJ, Burns KA, Segerdell EJ, Lee J, Snyder K, Bhattacharyya B, Karpinka JB, Fortriede J, Bowes JB, Zorn AM, Vize PD. Xenbase: expansion and updates of the Xenopus model organism database. Nucleic Acids Res. 2013;41:D865–870. doi: 10.1093/nar/gks1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kragtorp KA, Miller JR. Regulation of somitogenesis by Ena/VASP proteins and FAK during Xenopus development. Development. 2006;133:685–695. doi: 10.1242/dev.02230. [DOI] [PubMed] [Google Scholar]
- Luther PK. The vertebrate muscle Z-disc: sarcomere anchor for structure and signalling. J Muscle Res Cell Motil. 2009;30:171–185. doi: 10.1007/s10974-009-9189-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majid SM, Liss AS, You M, Bose HR. The suppression of SH3BGRL is important for v-Rel-mediated transformation. Oncogene. 2006;25:756–768. doi: 10.1038/sj.onc.1209107. [DOI] [PubMed] [Google Scholar]
- Mazzocco M, Arrigo P, Egeo A, Maffei M, Vergano A, Di Lisi R, Ghiotto F, Ciccone E, Cinti R, Ravazzolo R, Scartezzini P. A novel human homologue of the SH3BGR gene encodes a small protein similar to Glutaredoxin 1 of Escherichia coli. Biochem Biophys Res Commun. 2001;285:540–545. doi: 10.1006/bbrc.2001.5169. [DOI] [PubMed] [Google Scholar]
- Mazzocco M, Maffei M, Egeo A, Vergano A, Arrigo P, Di Lisi R, Ghiotto F, Scartezzini P. The identification of a novel human homologue of the SH3 binding glutamic acid-rich (SH3BGR) gene establishes a new family of highly conserved small proteins related to Thioredoxin Superfamily. Gene. 2002;291:233–239. doi: 10.1016/s0378-1119(02)00602-9. [DOI] [PubMed] [Google Scholar]
- Sandri C, Di Lisi R, Picard A, Argentini C, Calabria E, Myklak K, Scartezzini P, Schiaffino S. Heart morphogenesis is not affected by overexpression of the Sh3bgr gene mapping to the Down syndrome heart critical region. Hum Genet. 2004;114:517–519. doi: 10.1007/s00439-004-1088-8. [DOI] [PubMed] [Google Scholar]
- Scartezzini P, Egeo A, Colella S, Fumagalli P, Arrigo P, Nizetic D, Taramelli R, Rasore-Quartino A. Cloning a new human gene from chromosome 21q22.3 encoding a glutamic acid-rich protein expressed in heart and skeletal muscle. Hum Genet. 1997;99:387–392. doi: 10.1007/s004390050377. [DOI] [PubMed] [Google Scholar]
- Xanthos JB, Wanner SJ, Miller JR. Cloning and developmental expression of Xenopus Enabled (Xena). Dev Dyn. 2005;233:631–637. doi: 10.1002/dvdy.20358. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.