Abstract
DNase I hypersensitive sites (DHSs) are a hallmark of chromatin regions containing regulatory DNA such as enhancers and promoters; however, the factors affecting the establishment and maintenance of these sites are not fully understood. We now show that HMGN1 and HMGN2, nucleosome-binding proteins that are ubiquitously expressed in vertebrate cells, maintain the DHS landscape of mouse embryonic fibroblasts (MEFs) synergistically. Loss of one of these HMGN variants led to a compensatory increase of binding of the remaining variant. Genome-wide mapping of the DHSs in Hmgn1−/−, Hmgn2−/−, and Hmgn1−/−n2−/− MEFs reveals that loss of both, but not a single HMGN variant, leads to significant remodeling of the DHS landscape, especially at enhancer regions marked by H3K4me1 and H3K27ac. Loss of HMGN variants affects the induced expression of stress-responsive genes in MEFs, the transcription profiles of several mouse tissues, and leads to altered phenotypes that are not seen in mice lacking only one variant. We conclude that the compensatory binding of HMGN variants to chromatin maintains the DHS landscape, and the transcription fidelity and is necessary to retain wild-type phenotypes. Our study provides insight into mechanisms that maintain regulatory sites in chromatin and into functional compensation among nucleosome binding architectural proteins.
Tissue- and developmental-specific gene expression is facilitated by the binding of transcription factors to unique DNA sequences at regulatory sites in chromatin, such as enhancers and promoters. At these sites, the organization of the nucleosomes is altered, rendering the DNA more accessible to regulatory factors and hypersensitive to digestion by various nucleases including DNase I. DNase I hypersensitive sites (DHSs) are a hallmark of chromatin regions containing regulatory DNA and are used to identify regulatory sites in chromatin. The DHSs chromatin landscape is cell-type specific and dynamic, reflecting changes in the cellular transcription profile occurring during development, or as a consequence of genomic changes leading to altered cellular phenotypes (Gross and Garrard 1988; Boyle et al. 2008; Thurman et al. 2012; Calo and Wysocka 2013; Stergachis et al. 2013).
The molecular mechanisms that establish and maintain DHSs and their role in gene expression are key questions in chromatin biology. DHSs are established by the combined action of transcription factors that bind dynamically to specific DNA sequences and of chromatin remodeling complexes that alter the organization of the nucleosomes at these sites. Chromatin dynamics play an important role in the plasticity of the DHS landscape since it affects the ability of transcription factors to access potential new regulatory sites and facilitate the loss of unused sites. Although DHSs are stable and perpetuated to daughter cells, they are not necessarily static; they are maintained by a steady state, rapid, and continuous turnover of regulatory factors at specific sites (Narlikar et al. 2002; Erdel and Rippe 2011; Wippo et al. 2011; Calo and Wysocka 2013; Mueller-Planitz et al. 2013; Swygert and Peterson 2014; Voss and Hager 2014).
Given the dynamic nature of the DHSs, it could be expected that nucleosome binding proteins known to affect chromatin compaction and accessibility, such as linker histone H1 variants or members of the high mobility group (HMG) families, could play a role in establishing or maintaining the DHS landscape, a possibility that has not yet been examined. The three distinct families of HMG proteins named HMGA, HMGB, and HMGN, are each characterized by a specific functional domain (Bustin 1999; Bianchi and Agresti 2005). The HMGN family is unique in its ability to bind specifically to the 147 base pair nucleosome core particle, but not to purified DNA (Bustin 1999; Bianchi and Agresti 2005). HMGNs have been shown to bind to chromatin with a short residence time, efficiently counteract the chromatin condensing activity of histone H1, and induce changes in chromatin structure (Bustin 2001; Rochman et al. 2009). Chromatin immunoprecipitation studies revealed that HMGN1, one of the two major HMGN variants, colocalizes genome-wide with DHSs (Cuddapah et al. 2011; Deng et al. 2013); however, it is not clear whether HMGNs affect the organization of DHSs in chromatin.
Here, we examine the role of HMGN1 and HMGN2, the predominant variants of the HMGN nucleosome binding protein family, in modulating the DHS landscape of mouse embryonic fibroblasts (MEFs). HMGN1 and HMGN2 are widely expressed in vertebrate cells and compete for nucleosome binding sites (Bustin 2001; Catez et al. 2002; Ueda et al. 2008), raising the possibility of functional redundancy among these variants. Therefore we generated Hmgn2−/− mice and mated them with Hmgn1−/− mice to obtain Hmgn1−/−n2−/− mice, lacking both variants. We prepared MEFs from the three mouse strains and used genome-wide approaches and transcriptional analyses to examine whether the highly conserved HMGN nucleosome binding architectural proteins impact chromatin organization and gene expression. To gain insights into the biological function of the proteins in the contexts of an entire organism, we subjected the mice to a battery of phenotypic tests.
Our studies provide new insights into the biological function and mechanism of action of a ubiquitous family of chromatin binding architectural proteins.
Results
Compensatory binding of HMGN1 and HMGN2 to chromatin
HMGN1 and HMGN2 are the major variants of the HMGN protein family that is ubiquitously expressed in vertebrate cells (Bustin 2001). Since the HMGN1 variant localizes with DHSs in both human T cells and mouse neuroprogenitors (NPs), we examined the genome-wide organization of the HMGN2 variant in NPs. We identified 21,600 HMGN2 binding sites; 15,000 (74%) of these sites colocalized with DHSs, showing a Pearson correlation coefficient of 0.91 (Fig. 1A). The HMGN2 distribution across the genome was similar to that of HMGN1; we find that 16,500 (76%) of the HMGN2 sites overlapped with HMGN1 sites (Fig. 1B), showing a Pearson correlation coefficient of 0.66 (Fig. 1C).
Figure 1.
Compensatory binding of HMGN variants to NP chromatin. (A) Scatter plot showing correlation between HMGN2 binding and DNase I hypersensitivity at all DHSs. Normalized coverage depths of both HMGN2 and hypersensitivity were calculated at all DHSs, sorted by DHS coverage depths, grouped into 50 data point bins, and averaged. (R) Pearson correlation coefficient. (B) Venn diagram showing the overlap in the chromatin occupancy between HMGN1 and HMGN2 in wild-type (WT) NPs and the HMGN2 occupancy in Hmgn1−/− NPs. (C) Scatter plot showing colocalization of HMGN1 and HMGN2 in WT NPs. ChIP-seq signals are normalized over each site in reads per million (RPM). (D–F) Redistribution of HMGN2 occupancy upon loss of HMGN1. Shown is the occupancy of HMGN2 in WT and Hmgn1−/− NPs at sites occupied by either HMGN1, HMGN2, or both of the variants in WT cells. Note the shift in HMGN2 occupancy from HMGN2 unique sites to HMGN1 unique sites. (G) Genome browser view of HMGN occupancy in WT and Hmgn1−/− NPs. All the y-axes span from 0 to 0.5 as indicated.
Given the extensive colocalization between the HMGN variants we tested for possible interplay between these variants by examining the genome-wide organization of HMGN2 in NPs lacking HMGN1 (Hmgn1−/− NPs). In Hmgn1−/− NPs, we detect 39,700 HMGN2 binding sites, an 80% increase compared with the number of binding sites in wild-type NPs (Fig. 1B). Compared to WT cells, in Hmgn1−/− NPs, the occupancy of HMGN2 did not change significantly at sites that were shared with HMGN1 (Fig. 1D), but it significantly increased at sites that were exclusively occupied by HMGN1 (Fig. 1E) and decreased at sites that were exclusively occupied only by HMGN2 (Fig. 1F). Thus, loss of the HMGN1 variant increases the chromatin occupancy of HMGN2 (Fig. 1G), raising the possibility of functional compensation among these two major HMGN variants.
To test for possible functional compensation between the HMGN variants, we generated Hmgn2−/− mice (Supplemental Fig. S1A–D) and mated them with Hmgn1−/− mice (Birger et al. 2003) to obtain Hmgn1−/−n2−/− mice, lacking both variants (Supplemental Fig. S1E). We derived mouse embryonic fibroblasts (MEFs) from 13.5-d-old embryos of Hmgn1−/−, Hmgn2−/−, and Hmgn1−/−n2−/− mice and used these cells to examine whether HMGN variants affect the organization of regulatory sites in chromatin.
Chromatin immunoprecipitation analyses indicated that the antibodies to either HMGN1 or HMGN2 specifically recognize the chromatin-bound HMGNs since the amount of chromatin immunoprecipitated by anti-HMGN1 from WT MEFs was 18 times higher than the amount of chromatin precipitated from Hmgn1−/− MEFs. Similarly, the anti-HMGN2 antibodies immunoprecipitated 22-fold more chromatin from WT MEFs compared with Hmgn2−/− MEFs (Fig. 2A). Significantly, compared with WT MEFs, anti-HMGN2 antibody immunoprecipitated twice as much chromatin from Hmgn1−/− MEFs, and anti-HMGN1 immunoprecipitated twice as much chromatin from Hmgn2−/− cells (Fig. 2A). Thus, loss of HMGN1 increases the amount of chromatin-bound HMGN2, whereas loss of HMGN2 increases the amount of chromatin-bound HMGN1—a finding fully compatible with the analysis of HMGN2 organization in NPs and with previous FRAP analyses of living cells, which demonstrated competition among HMGN variants for chromatin binding sites (Catez et al. 2002). Taken together, the results suggest that loss of an HMGN variant is compensated by increased chromatin binding of the remaining variant.
Figure 2.
Altered DHS landscape in Hmgn1−/−n2−/− MEFs. (A) Quantitative analyses of the chromatin immunoprecipitated from either WT, Hmgn1−/−, or Hmgn2−/− MEFs with antibodies to either HMGN1 or HMGN2. Note that loss of either HMGN1 or HMGN2 variant increased the amount of chromatin precipitated by the remaining variant. (*) P < 0.001. (B–D) Venn diagrams showing the overlap in the DHSs between WT and either Hmgn1−/−, Hmgn2−/−, or Hmgn1−/−n2−/− MEFs. (E–G) Scatter plot showing similar intensity of DHSs in WT and Hmgn1−/− or Hmgn2−/−, but not in Hmgn1−/−n2−/− MEFs. DHS signals are normalized by library size and averaged over two biological replicates over each site in reads per million (RPM). (H–J) Reduced DHS width in Hmgn1−/−n2−/− MEFs. In Hmgn1−/−n2−/− MEFs, the mean width of all DHSs is narrower (H), the width of DHSs lost is larger than that of DHSs gained (I), and the shared DHSs are narrower (J) compared with WT MEFs. Bars show frequency, and curves show density. (K) Genome browser snapshot visualizes the lost or gained DHSs in Hmgn1−/−n2−/− MEFs. Two biological replicates for each genotype are shown. Numbers in parentheses indicate the scales of the y-axes.
Synergistic effects of HMGN variants on the DHS landscape of MEFs
DNase I hypersensitivity mapping identified 102,034 DHSs in wild-type (WT) MEFs, 101,159 sites in Hmgn1−/− MEFs, and 107,150 sites in Hmgn2−/− MEFs, with an 80% of overlap between WT and each of the Hmgn−/− cells (Fig. 2B,C). The DHS distribution and intensity across the genome was not significantly affected by loss of a single HMGN variant; the Pearson correlation coefficient between WT and either Hmgn1−/− or Hmgn2−/−MEFs was 0.86 and 0.93, respectively (Fig. 2E,F), similar to that of two biological replicates of the same genotype (Supplemental Fig. S2).
Although loss of a single HMGN variant did not affect significantly the DHS landscape, loss of both HMGN variants did alter the DHS landscape. We identified only 67,363 DHSs in Hmgn1−/−n2−/− MEFs; of these, 57% overlapped with DHSs seen in WT MEFs and 43% were newly formed. Thus, >60% of the DHSs identified in WT cells were lost in Hmgn1−/−n2−/− (Fig. 2D). The correlation plot of the overall DHS intensity between WT and Hmgn1−/−n2−/− MEFs shows a high scatter with a Pearson correlation coefficient of 0.65 (Fig. 2G). The genomic snapshot shown in Figure 2K visualizes the significant loss and gain of DHSs in Hmgn1−/−n2−/− compared with WT, Hmgn1−/−, or Hmgn2−/− MEFs. Thus, the combined loss of HMGN1 and HMGN2 led to both loss and gain of DHSs, suggesting that the proteins synergistically remodeled the DHS landscape of MEFs.
Loss of both HMGN variants affected not only the number of sites detected but also significantly decreased the width of the remaining DHSs, as measured by the amount of base pairs encompassed in a site. Thus, in Hmgn1−/−n2−/− MEFs, the median DHS width is only half of that in WT cells (Fig. 2H), and the total genomic region covered by DHSs in Hmgn1−/−n2−/− MEFs is less than half of that covered in WT cells (Supplemental Fig. S3A, all DHSs). This trend was even more pronounced when the width of the sites that were present in WT but absent in Hmgn1−/−n2−/− MEFs (black in Fig. 2D) is compared to the sites that were absent in WT but present in Hmgn1−/−n2−/− MEFs (red in Fig. 2D): The median width of lost DHSs was 2.5 times larger than the newly formed, gained sites in Hmgn1−/−n2−/− cells (Fig. 2I); and the total genomic region covered by unique peaks in WT cells was four times larger than in Hmgn1−/−n2−/− MEFs (Supplemental Fig. S3A, unique DHSs). Likewise, the shared sites, although present both in WT and Hmgn1−/−n2−/− MEFs (white in Fig. 2D), showed a similar effect; the median width of these sites decreased from 2000 bp in WT to 800 bp in Hmgn1−/−n2−/− MEFs (Fig. 2J; Supplemental Fig. S3A, shared DHSs). Furthermore, to avoid the potential issue that the peak calling software may not accurately assess the width of a peak, we also plotted the aggregate DHS accessibility around all the peaks in WT and Hmgn1−/−n2−/− MEFs. The results indicate that indeed, the average half-height width of the DNase I peaks in Hmgn1−/−n2−/−cells (510 bp) is significantly narrower than that in WT cells (920 bp). Thus, the combined loss of HMGN1 and HMGN2 significantly altered the DNase I sensitivity of the MEFs. The newly formed sites in Hmgn1−/−n2−/− MEFs were significantly narrower than the average sites seen in WT cells, suggesting a general decrease in DNA accessibility at chromatin regulatory regions.
Preferential loss of DHSs at enhancers in Hmgn1−/−n2−/− MEFs
Chromatin regulatory regions are marked not only by increased DNase I hypersensitivity but also by specific histone modifications, such as H3K4me1, H3K27ac and H3K4me3, which mark enhancers, active enhancers, and promoters, respectively (Thurman et al. 2012; Calo and Wysocka 2013). Genome-wide mapping of these histone marks in WT and Hmgn1−/−n2−/− MEFs indicated that loss of both HMGN1 and HMGN2 did not affect the level of these three histone modifications (Supplemental Fig. S4). However, alignment of the DHSs with these histone modifications reveals that loss of both HMGN1 and HMGN2 primarily affected the DHSs located in enhancer regions. We detected 60,000 DHSs at enhancer regions of WT MEFs but only 23,000 DHSs at the Hmgn1−/−n2−/− enhancers; thus, >50% of the DHSs detected at the WT enhancers were lost or significantly diminished in Hmgn1−/−n2−/− MEFs (Fig. 3A; Supplemental Fig. S5). Indeed, only 3% of the lost and 1% of the gained DHSs overlapped with H3K4me3, a marker for promoters; the great majority of DHSs that were either lost or gained overlapped with H3K4me1 and H3K27ac, histone marks for enhancer regions (Fig. 3B). The loss of DHSs at enhancer regions marked by H3K4me1 in Hmgn1−/−n2−/− MEFs, but not in Hmgn1−/−, or Hmgn2−/− MEFs lacking a single HMGN variant, is visualized by the heat map in Figure 3C and by the decreased normalized DHS intensity overlapping the H3K4me1 or H3K27ac marks (Fig. 3D,E). The genomic snapshots of Hmgn1−/−n2−/− cells shown in Figure 3G exemplify the unaltered profiles of H3K4me1, H3K4me3, and H3K27ac and the preferential loss of DHSs at sites overlapping with H3K4me1 and H3K27ac, but not at H3K4me3.
Figure 3.
Altered DHSs at Hmgn1−/−n2−/− enhancers. (A) Loss of DHSs at enhancers of Hmgn1−/−n2−/− MEFs. Shown is the DHS distribution among annotated sequences in genome of WT, Hmgn1−/−, Hmgn2−/−, and Hmgn1−/−n2−/− MEFs. (B) Changes in DHSs at regions overlapping with enhancers (H3K4me1 or H3K27ac) but not promoters (H3K4me3). (C) Heat maps of DHS intensities across all H3K4me1, not overlapping H3K4me3, peak regions (center aligned), visualizes the loss of DHSs at H3K4me1 regions in Hmgn1−/−n2−/− MEFs. (D–F) Normalized average intensity of DHSs in WT and mutant MEFs, at indicated histone marks. (G) Genomic browser snapshot visualizing the preferential loss of DHSs overlapping H3K4me1 and H3K27ac in Hmgn1−/−n2−/− MEFs. Two biological replicates are shown for the DHS maps. Numbers in parenthesis indicate the scales of the y-axis.
As a consequence of the significant loss of DHSs at enhancer regions, the DNA fragments at promoter regions, which are marked by H3K4me3, were disproportionally overrepresented in the sequenced libraries, resulting in a significant increase in the normalized intensity (reads per million) at H3K4me3 sites (Fig. 3F), thereby giving the false impression that in Hmgn1−/−n2−/− MEFs the DNase I accessibility at these sites is increased. In fact, quantitative DNase I digestion analyses of regions overlapping with either H3K4me1 or H3K4me3 from the same gene indicates that both the promoter and the enhancer region in Hmgn1−/−n2−/− MEFs were less digested than the same regions in WT MEFs (Fig. 4A–C). In agreement with the genome-wide analyses, the effects at H3K4me1 sites are significantly bigger than at H3K4me3 sites. The differences are most obvious when comparing the percentage of digested chromatin in WT to that in Hmgn1−/−n2−/− MEFs at the same DNase I concentration. For example, at the H3K4me1 site in the Wrb locus, 2000 units/mL DNase I digested 74.6% of the site in WT cells but only 13.3% of the sites in Hmgn1−/−n2−/− cells, a 5.6-fold difference. However, at the H3K4me3 site, loss of HMGN1 and HMGN2 reduced the percentage of chromatin digested by only 1.5-fold, from 87% to 60.2% (Fig. 4A).
Figure 4.
Altered nuclease sensitivity at regulatory chromatin sites of Hmgn1−/−n2−/− MEFs. (A–C) Quantitative analysis of DNase I sensitivity at promoters and enhancers of three selected genes. The extent of DNase I digestion at regions overlapping either H3K4me1 or H3K4me3 (pink or blue in genome browser), quantified by PCR amplification. Loss of HMGNs affected regions overlapping with H3K4me1 more significantly than regions overlapping H3K4me3. Numbers in parentheses indicate the scales of the y-axes. (D) Loss of HMGN does not affect the kinetics of MNase digestion. (E) Deep sequencing of nucleosome positions reveals enhanced occupancy of unstable nucleosomes at the TSS of Hmgn1−/−n2−/− MEFs. Arrows point to the presence of a nucleosome detectable at the TSS in limited, but not extensive digests.
Taken together, our results reveal that loss of both HMGN1 and HMGN2 alters the DNase I hypersensitivity in chromatin globally, and these affects are more pronounced at enhancer regions marked by H3K4me1-modified histones than at promoter regions marked by H3K4me3 histones.
Altered nucleosome organization at promoters of Hmgn1−/−n2−/− MEFs
DNase I hypersensitivity at regulatory sites results from perturbed nucleosome occupancy. The nucleosome ladders originating during the MNase digestion of WT and Hmgn1−/−n2−/− cells were indistinguishable, indicating that loss of HMGNs does not have significant effects on global chromatin compaction (Fig. 4D). To test the effects of HMGN variants on nucleosome occupancy at regulatory sites, we mapped the genomic nucleosome positions in WT and Hmgn1−/−n2−/− MEFs by deep sequencing of mononucleosomal DNA purified from either limited or extensive micrococcal nuclease (MNase) digestions, when either only ∼5% or more than ∼80% of the chromatin is converted to mononucleosomes, respectively (Fig. 4D). The limited digests are enriched in DNA fragments originating from the nucleosomes located in the decondensed, regulatory regions of chromatin. The linker regions of these nucleosomes are more accessible to the enzymes; and therefore, during the digestions, these nucleosomes are released first and then further digested. Consequently, the DNA from regulatory regions is disproportionally underrepresented in extensive MNase digests.
Deep sequencing of DNA recovered from extensive digests of WT and Hmgn1−/−n2−/− MEFs revealed a “nucleosome-free region” at the TSS (Fig. 4E, top), as was seen in other cells (Jiang and Pugh 2009). In contrast, the DNA of the nucleosomes from the limited MNase digestions clearly reveals the presence of a nucleosome positioned at the TSS, in the “nucleosome-free regions” seen in extensive digests. Significantly, in the limited digests, the occupancy of nucleosomes at the TSS of Hmgn1−/−n2−/− MEFs was higher than in WT MEFs (Fig. 4E, bottom) indicating that loss of HMGNs enhances the stability of the nucleosomes that are located at regulatory sites, a finding that is in full agreement with the reduced DNase I sensitivity of these cells.
Altered stress-induced gene expression in Hmgn1−/−n2−/− MEFs
Since changes in DNase I hypersensitivity could affect gene expression, we sequenced the RNA isolated from exponentially growing MEFs and tested whether loss of HMGNs changed the cellular transcriptome. We find that loss of HMGN1, HMGN2, or both these variants, altered the expression of 22, 49, and 77 genes, respectively (Supplemental Fig. S6; Supplemental Table S3). GO analysis failed to reveal specific pathways affected by these genes, reinforcing previous findings that unlike transcription factors, HMGNs do not target specific genes and are not major regulators of specific gene expression.
As an additional test of the consequences of the altered DHS landscape on gene expression, we measured the kinetics of the transcriptional responses known to involve rapid changes in chromatin. To this end, we measured the transcriptional response to stress induced either by treatment with the protein synthesis inhibitor anisomycin (Mahadevan et al. 1991) or by exposure to heat shock (Corey et al. 2003), treatments that induce the expression of specific genes. We exposed WT and Hmgn1−/−n2−/− MEFs to anisomycin and quantified the transcriptional response of 44 “immediate-early” genes (Fig. 5A; Supplemental Table S4) known to be induced by stress (Tullai et al. 2007). Of the 44 genes tested, 22 showed significant differences in the induction kinetics with 18 of these genes induced faster and four genes induced slower in Hmgn1−/−n2−/− MEFs, as exemplified by the induction kinetics of Ccl2 and Il6 genes (Fig. 5B,C). We also calculated the difference in induction ratio between the two biological replicates of WT and Hmgn1−/−n2−/− MEFs and compared them with the differences seen between Hmgn1−/−n2−/− and WT MEFs (Supplemental Fig. S7). The observed significant differences in the induction kinetics between Hmgn1−/−n2−/− and WT cells were eliminated between the replicates of the same genotype, indicating that the differences are due to the loss of HMGNs and not to variance between biological replicates. The genes with altered expression showed loss of DHSs in Hmgn1−/−n2−/− MEFs, especially at enhancer sites marked by H3K4me1, as shown by the example in Figure 5D. Likewise, the heat shock-induced transcriptional activation of Hspa1a and Hspa1b, known to be accompanied by a rapid chromatin restructuring, was slower in Hmgn1−/−n2−/− MEFs (Fig. 5E). Loss of HMGN1 and HMGN2 leads to significant changes in the DHSs at the promoters of both Hspa1a and Hspa1b genes (Fig. 5F).
Figure 5.
Altered immediate early (IE) gene expression in Hmgn1−/−n2−/− MEFs. (A) Heat map showing the induction fold ratios of IE genes in WT and Hmgn1−/−n2−/− MEFs. Boxes separate the genes that were up-regulated and down-regulated from the unaffected genes. (*) P < 0.05; two biological replicates were analyzed. (B,C) Kinetics of anisomycin-induced expression of Ccl2 and Il6. (D) Genome browser view of changes in DHSs at H3K4me1 and H3K4me3 sites 5′ to the Ccl2 gene. Numbers in parentheses indicate the scales of the y-axes. (E) Altered induction of heat shock genes in Hmgn1−/−n2−/− MEFs. (F) Genome browser view of changes in DHSs at H3K4me1 and H3K4me3 sites near the Hspa1a and Hspa1b genes. Numbers in parentheses indicate the scales of the y-axes.
Altered phenotypes in Hmgn1−/−n2−/− mice
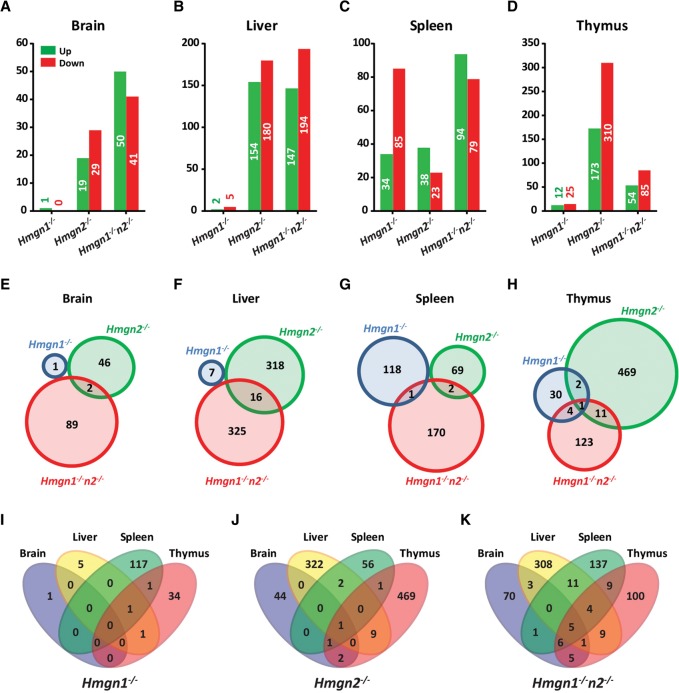
In the biological context of an entire organism, we compared the transcription profile in brain, liver, spleen, and thymus in each of the three Hmgn−/− lines to the corresponding wild-type controls. Loss of HMGN1, HMGN2, or both of these variants affected the expression of a limited number of genes in each tissue, with a similar number of genes up- and down-regulated. The number of genes changed in Hmgn2−/− tissues was comparable to the changes in tissues from Hmgn1−/−n2−/− mice, both of which were higher than the number of genes changed in Hmgn1−/− tissues (Fig. 6A–D). There was very little overlap between the genes affected in the various mouse lines (Fig. 6E–H) or between the four tissues taken from a mouse line (Fig. 6I–K), reinforcing previous findings indicating tissue-specific and HMGN variant–specific effects on transcription (Kugler et al. 2013).
Figure 6.
Effect of HMGN loss on global gene expression in mouse tissues. (A–D) Number of genes up- and down-regulated in the four tissues of the three Hmgn−/− mice lines compared to WT littermates. (E–H) Venn diagrams showing the overlap in the number of genes with altered expression in the tissues of each of the three genotypes examined. (I–K) Venn diagrams showing the overlap between the genes altered in the different tissues of each genotype.
In addition to examining the genes with significant changes in gene expression, we also analyzed the effect of HMGN loss on cellular transcription pathways, disregarding the magnitude of change in the expression levels of individual genes. In this analysis, we averaged the expression level of the four WT mice and compared this average to each of their four genetically altered littermates. Running a one-sample t-test on fold changes of genes, we identified several pathways in which the expression of a significant number of genes was affected in each of the four mice in the three mouse lines examined (Supplemental Table S5). The significant number of genes affected in a particular pathway raises the possibility that the cumulative effects of small changes in the transcription of several genes may be functionally relevant and lead to altered phenotypes.
Phenotypic screening of 15 male and 15 female Hmgn mutant mice and their WT controls, a total of 30 mice for each genotype, was performed at the German Mouse Clinic using a standard battery of tests (Fuchs et al. 2009; Gailus-Durner et al. 2009). In these tests, Hmgn1−/−n2−/− mice showed significantly more altered phenotypes than either Hmgn1−/− or Hmgn2−/− mice (Table 1; Supplemental Table S1). Metabolic screening of Hmgn1−/−n2−/− mice indicated increased food intake but decreased body mass, perhaps linked to increased energy expenditure. Likewise, clinical chemistry analyses of these mice revealed changes in the levels of albumin and alpha amylase activity, as well as changes in triglyceride, cholesterol, and insulin levels of mice lacking both HMGN variants that were not detected in mice lacking a single variant. Additional noticeable differences were detected as changes in the distribution of leukocyte subpopulations in the blood of Hmgn1−/−n2−/− mice, which showed higher frequencies of T, NK, and NKT cells, altered CD4+ to CD8+ ratios in T cells, and changes in the distribution of the cells in the NK and B cell compartment. Furthermore, Hmgn1−/−n2−/− mice showed not only a reduced corneal thickness, which was also seen in Hmgn1−/− mice (Birger et al. 2006), but also significantly enlarged eye axial lengths and increased number of the fundic main blood vessels, phenotypes which were not observed in either Hmgn1−/− or Hmgn2−/− eyes. Thus, Hmgn1−/−n2−/− mice show changes in eye morphology that were not seen in mice lacking a single HMGN variant, most likely because both variants are strongly expressed in the eye (Lucey et al. 2008). Thus, the phenotypic analyses indicate that loss of both HMGN1 and HMGN2 lead to more detectable phenotypes than loss of only HMGN1 or HMGN2, a finding that is in general agreement the compensatory binding of HMGN variants to chromatin and with the more severe changes in the DHS landscape of Hmgn1−/−n2−/− mice.
Table 1.
Phenotypes detected in Hmgn1−/−, Hmgn2−/−, and Hmgn1−/− n2−/− mice

Discussion
Elucidation of the factors that affect the establishment and maintenance of the DHSs may provide insight into the mechanisms whereby chromatin dynamics affect the cellular phenotype. We now show that HMGN variants modulate the DHS landscape in the chromatin of MEFs. The combined loss of both HMGN1 and HMGN2 remodels the DHS landscape of MEFs in three measurable ways: Part of the DHSs are lost, new DHSs are formed, and all the DHSs are less prominent, as measured by the number of nucleotides encompassed across a site. The overall effect of loss of the HMGN variants is a decrease in the DNase I hypersensitivity. Given the ubiquitous presence of HMGN proteins in vertebrate cells, it is likely that HMGN variants will similarly affect the organization of chromatin regulatory sites in other cells.
Most of the prominent DHSs are located at promoters and enhancers, regulatory chromatin sites that are also marked by specific modifications in the N-terminal tail of histone H3. Although loss of HMGNs did not affect the levels of these histone modifications, it affected the DNase I hypersensitivity at enhancers to a significantly larger extent than at promoters. Over 70% of the DHSs that changed in Hmgn1−/−n2−/− MEFs localized to enhancer regions; there were twice as many DHSs lost as newly formed. Furthermore, all the DHSs of Hmgn1−/−n2−/− MEFs, regardless whether lost, gained, or shared, were narrower than the sites of WT MEFs. For these experiments, the quality and depth of all our sequencing samples are sufficiently high (Supplemental Table S1). Although there are known limitations in peak calling algorithms, we think that it is an indispensable initial screening in identifying potential regions of interest. Additionally, we included an aggregate DHS accessibility plot (Supplemental Fig. S5).
DNase I digestion is a continuous enzymatic process in which most of the chromatin will eventually be reduced to very small fragments; the DHS landscape provides a snapshot of an early kinetic point in the digestion. Therefore, the disappearance of a site does not necessarily mean that the site is fully lost; it means that the site is less accessible to the enzyme, as we demonstrate in the quantitative DNase I digestion of promoter and enhancer regions of the same gene (Fig. 4A). Significantly, although loss of HMGNs leads to a global reduction in DNase I hypersensitivity, we also find that Hmgn1−/−n2−/− MEFs contain new DHSs that were not detected in WT cells, an indication that these regions became more accessible to the enzyme. Thus, loss of HMGNs remodels the DHS landscape of the cells: While most of the regulatory sites become less sensitive to DNase I, new sites that are more accessible to digestion are formed.
Enhancer regions are the most variable and rapidly evolving class of chromatin regulatory sites and show a cell-type–specific pattern that is altered during differentiation and development (Heintzman et al. 2009; Thurman et al. 2012; Calo and Wysocka 2013; Villar et al. 2015). Our finding that HMGN variants preferentially remodel DHSs at enhancers, the most malleable and dynamic part of the genome, raises the possibility that these proteins help optimize chromatin remodeling during evolution, differentiation, or in response to stimuli that evoke changes in gene expression.
HMGNs could remodel the DHS landscape by altering the action of factors known to establish and modulate DNase I hypersensitivity. The levels of H3K4me1, H3K4me3, and H3K27ac modifications were not altered in Hmgn1−/−n2−/− MEFS, indicating that the HMGN-mediated chromatin remodeling is not due to changes in these histone modifications. HMGNs could modulate the ability of regulatory factors to interact with chromatin, as was shown for PCNA (Postnikov et al. 2012), or the action of ATP-dependent nucleosome remodeling factors (Rattner et al. 2009), which play a role in the organization of local chromatin structures. An additional important mechanism potentially contributing to the HMGN-mediated changes in DHSs could involve alterations in the interaction of the linker histone H1 with chromatin. Linker H1 variants, the most abundant chromatin binding proteins, have been shown to stabilize chromatin compaction and decrease chromatin accessibility (Bustin et al. 2005; Woodcock et al. 2006; Li et al. 2012). The interaction of HMGNs with the nucleosome is known to reduce the binding of H1 to chromatin, both in vivo and in vitro (Ding et al. 1997; Catez et al. 2002, 2006; Rochman et al. 2009). Loss of HMGNs could enhance H1 binding, thereby reducing the accessibility of the DNA at chromatin regulatory sites to DNase I digestion. Most likely, the cumulative effects of changes in several factors that affect chromatin accessibility and remodeling lead to changes in nucleosome occupancy at regulatory sites, as we detected in the promoters of Hmgn1−/−n2−/− MEFs (Fig. 4D). Thus, we suggest that HMGNs play a role in maintaining the DHS landscape by modulating the interaction of regulatory factors and architectural proteins with chromatin.
Global changes in gene expression occurring during development and differentiation, or in response to alteration in transcription factor availability, are often associated with changes in the DHS landscape (Voss and Hager 2014). Surprisingly, in Hmgn1−/−n2−/− MEFs growing at optimal conditions, the changes in DHSs were accompanied with only minor changes in gene expression, perhaps because loss of the HMGN variants did not alter the histone marks at regulatory sites and did not detectably change the level of chromatin regulators. Thus, in the absence of additional changes or stress, an altered DHS landscape does not necessarily lead to significant changes in transcription. Indeed, the stress-induced transcriptional response in Hmgn1−/−n2−/− MEFs was different from that of WT MEFs, supporting the suggestion that the presence of HMGNs facilitates chromatin remodeling. We note however that these changes were not very large; and in most cases, the loss of HMGNs increased the rate of induction of the immediate early genes, an unexpected result. An obvious possible explanation is that loss of DHSs decreased the binding of a negative regulator to a site that affects the induction of immediate gene expression. In addition, it is known that the induction of immediate early gene expression is associated with, and affected by, phosphorylation of H3S10, and that loss of HMGN1 enhances the rate of this phosphorylation (Lim et al. 2005). The results reinforce the general notion that by itself, the presence of DNase I sites cannot predict the gene expression levels.
Transcriptional profiling of tissues from genetically altered HMGN mice showed few significant differences between the four tissues taken from mice lacking HMGN variants and their wild-type littermates. Yet, compared to WT littermates, the phenotype of Hmgn1−/−n2−/− mice was more significantly affected than that of mice lacking a single variant. Most likely, the phenotypes result from the accumulated changes in the transcription level of numerous genes that at the individual gene are not highly significant, as suggested by the small but numerous changes in the expression of genes in several distinct pathways.
Considering the interaction of HMGN variants with chromatin, our ChIP analyses demonstrating that loss of one variant increases the nucleosome binding of the remaining variant provide direct evidence for an interplay among HMGNs, as was suggested by fluorescence analyses of living cells (Catez et al. 2002). The interactions among HMGN variants are functionally relevant since distinct changes in the DHS landscape are seen when both variants are missing. However, the biological functions of the various HMGNs are not necessarily identical; each HMGN variant forms unique complexes with nucleosomes (Postnikov et al. 1995), raising the possibility of variant-specific effects on the function of chromatin regulators thereby leading to variant-specific contributions to an observed phenotype.
Methods
Generation of Hmgn1−/−, Hmgn2−/−, and Hmgn1−/−n2−/− knockout mice
Hmgn1−/− mice, lacking exons 2–4 of the gene, were previously described (Birger et al. 2003; Kugler et al. 2013). The targeting vector for generating the Hmgn2−/−, lacking the entire gene, was constructed by a recombinogenic cloning strategy (Liu et al. 2003) with a murine bacterial artificial chromosome clone, RP23-31L13. The vector was constructed to remove the entire gene, encompassing exons 1–6 (Supplemental Fig. S1A). A 16-kb fragment containing the Hmgn2 gene was retrieved from the bacterial artificial chromosome clone and inserted into the targeting vector PL253 by recombination in bacterial strain DY380. The neo gene with the phosphoglycerate kinase 1 promoter (pGK-neo) was used as a positive selectable marker, and the pGK-thymidine kinase cassette was used as a negative selectable marker. The FRT-flanked pGK-neo and the LoxP sites for conditional deletion of Hmgn2 were inserted as described in Supplemental Figure S1A. Electroporation and selection were performed by using the v6.4 embryonic stem (ES) cell line. DNAs derived by G418/1,2′-deoxy-2′-fluoro-1-β-D-arabinofuranosyl-5-iodo-uracil-resistant ES cell clones were screened by diagnostic BamHI restriction enzyme digestion using the 5′ probes external to the targeting vector sequence. Two independent ES cell clones targeted to Hmgn2 were injected into C57BL/6 blastocysts and generated chimeras that transmitted the mutated allele to the progeny. The neomycin resistance (neo) and the Hmgn2 gene were removed by crossing with beta actin Cre mice. The mice containing the targeted allele were backcrossed into the C57BL/6 background for at least five generations. To generate Hmgn1−/−n2−/− mice, the Hmgn2−/− mice were crossed with Hmgn1−/−. All mice were bred in a specific pathogen-free facility with food and water ad libitium. At the GMC, mice were maintained according to the GMC housing conditions and German laws. All tests performed at the GMC were approved by the responsible authority of district government of Upper Bavaria, Germany.
Phenotype screening of 15 male and 15 female Hmgn mutant mice and their age-matched WT littermates or corresponding WT controls for the Hmgn1−/−n2−/− knockout mice, a total of 30 mutant and 30 control mice for each mouse line, were performed at the German Mouse Clinic using a standard battery of tests (Gailus-Durner et al. 2005, 2009; Fuchs et al. 2009, 2011) The detailed phenotypic analysis of the mice is described in Supplemental Data. A complete description of the methods can be found at the German Mouse Clinic website (http://www.mouseclinic.de).
Preparation of primary mouse embryonic fibroblasts (MEFs) and neuroprogenitors
MEFs were prepared from E13.5 embryos. After removal of the head and viscera, the embryos were digested in 0.25% trypsin at 37°C, with gentle pipetting for 30 min. The dissociated fibroblasts were allowed to settle and then cultured in 150-cm plates in DMEM (Corning) with 10% FBS (Life Technologies) under 5% CO2 at 37°C. All experiments were done with MEFs at passage 3. For each genotype, two independent MEF clones (biological replicates) were used in every experiment, including DNase-seq and MNase-seq. Neuroprogenitors were prepared from embryonic stem cells differentiated along the neural lineage using a mouse dopaminergic neuron differentiation kit (R&D Systems) as described (Deng et al. 2013).
Western blotting and immunostaining
Rabbit affinity pure polyclonal anti-mouse HMGN1, anti-human HMGN2, and anti-calf Histone H1 antibodies were generated in our laboratory (Birger et al. 2003). HRP-conjugated secondary antibodies for Western blots were from Pierce. Fluorescence secondary antibody was AlexaFluor488-donkey-anti-rabbit IgG (Invitrogen). For Western blots, whole cell lysates were prepared in 1× SDS PAGE sample buffer (Bio-Rad) supplemented with protease inhibitors (Roche). The samples were fractionated on 15% precast Criterion gels, transferred by semidry method to PVDF membrane, blocked with nonfat milk in TBST, and probed with antibodies. Chemiluminiscent detection using ECL Plus has been performed according to Amersham (GE Healthcare) recommendations.
For immunostaining, 5 μm paraffin-embedded liver sections were prepared from 8-wk-old Hmgn2+/+ and Hmgn2−/− mice. Immunofluorescence staining was performed using anti-human HMGN2 antibody as previously described (Cherukuri et al. 2008). The images were taken using Nikon Eclipse E800 fluorescence microscope and processed using NIS-Elements software (Nikon).
ChIP-seq
MEFs were crosslinked directly in culture medium with 1% formaldehyde solution for 10 min at room temperature. Chromatin was fragmented to 200–300 bp by 15 cycles of sonication (30 sec on/30 sec off) (Bioruptor). Chromatin from 2 × 107 cells and 10 µg antibodies against HMGN1, HMGN2, H3K4me1 (Abcam, ab8895), H3K4me3 (Abcam, ab8580), and H3K27ac (Abcam, ab4729) were used for each ChIP experiment. Sequencing libraries were prepared using Illumina TruSeq ChIP Sample Prep Kit with compatible indexed adaptors according to the manufacturer's instruction. Equal amounts of 4–6 indexed library samples were pooled for cluster generation and sequenced on Illumina HiSeq 2000 system with 2 × 100-bp paired-end reads.
Genome-wide mapping of DNase I hypersensitive sites
For preparation of nuclei, MEFs were incubated on ice for 10 min in Buffer A (0.15 M Tris-HCl pH8.0, 15 mM NaCl, 60 mM KCl, 1 mMEDTA, 0.5 mMEGTA) containing 0.03% IGEPAL (Sigma), 1 mM EDTA, 0.5 mM EGTA, 0.5 mM spermidine, and protease inhibitor (Roche). DNase I digestion was carried out by incubation of 2 × 107 nuclei with 2000 units/mL DNase I (Roche) for 3 min at 37°C. After treatment by RNase A and proteinase K, DNA was recovered by phenol/chloroform extraction. DNA fragments with size 150–500 bp were isolated by 9% sucrose gradient centrifugation. Library construction and sequencing were performed as described above for ChIP-seq samples. DNase I accessibility assays were performed to validate the sequencing results. Kinetic DNase I digestion assays were done using 1000, 2000, or 3000 units/mL DNase I. The digested DNA was precipitated, and an equal amount of DNA was analyzed by quantitative real-time PCR. Primer information is presented in Supplemental Table S2.
Genome-wide nucleosome positioning assay
Mononucleosomal DNA from MEFs was isolated by micrococcal nuclease digestion using EZ Nucleosomal DNA Prep Kit (Zymo Research) according to the manufacturer's instruction. In brief, nuclei were isolated from trypsinized cells by incubation on ice for 5 min in Nuclei Prep Buffer, 1.5 × 106 nuclei were then digested by 0.05–0.5 units of micrococcal nuclease in 100 μL MN Digestion Buffer at room temperature for 10 min, and DNA was purified from the nuclear digests using the columns supplied with the kit and fractionated on 2% agarose gels. Mononucleosomal DNA was purified from the gels using a Qiagen gel extraction kit and processed for library construction and deep sequencing as described above for ChIP-seq.
Gene expression analysis
Transcription analysis in MEFs was performed by RNA sequencing. Total RNA was isolated by RNeasy Mini Kit (Qiagen) followed by “on-column” DNase I treatment. mRNA-seq libraries were prepared from 1 µg total RNA using the Illumina TruSeq RNA Sample Preparation Kit, following the manufacturer's instructions. Libraries were sequenced on the Illumina HiSeq 2000 with 2 × 100-bp paired-end reads. The data analysis was performed using the RNA-seq pipeline in Partek Genome Suite. A gene was considered expressed if reads per kilobase of transcript model per million mapped reads (RPKM) was ≥1.0. |Fold Change| ≥ 1.5 with P-value <0.05 was considered significantly differentially expressed between two genotypes.
For analysis of gene expression in mouse tissues and identification of differentially expressed genes between wild-type and mutant mice, amplified cRNA was hybridized to MouseRef-8 Version 2 Expression BeadChips (Illumina); and after a 16-h incubation, staining and scanning were done according to the Illumina expression protocol. To identify differentially regulated genes, we used the mattest function of the MATLAB package. A permutation method (Storey and Tibshirani 2003) has been used to address the small sample size, and 5000 permutations have been performed using the mattest function. Differently expressed genes were selected using a uniform cutoff—P-value < 0.05 and fold change > 1.5—in all cases.
Bioinformatic DNase-seq, ChIP-seq, and MNase-seq analyses
Unfiltered sequencing reads (from DNase-seq, ChIP-seq, or MNase-seq) were aligned to the mouse reference genome (NCBI build 37, mm9) using Bowtie (Langmead et al. 2009). Up to two mismatches was allowed for each aligned read. Only uniquely aligned reads were collected for further analysis. The information of the quality and depth of all the sequencing samples are in Supplemental Table S1.
DNase-seq and ChIP-seq peaks were identified using SICER (Zang et al. 2009) with the following parameters: effective genome size, 0.875 (87.5% of the mouse genome is mapped by 100-bp reads); window size, 50 bp; gap size, 50 bp. Calculation of coverage and identification of overlapping regions were performed with the “chipseq” and “GenomicRanges” packages in BioConductor (Gentleman et al. 2004).
For normalization, the calculation of coverage at any region and the comparisons between different data sets were preceded by library size normalization. Control subtraction was carried out in the following way: coverage (exp)/N1 − coverage (control)/N2, in which “exp” is the data set (in .bam format) to be examined, N1 is the library size of the experimental data (“exp”), and N2 is the library size of the control. In this study, input sequences (DNA sequences after sonication only without immunoprecipitation) were used as a control for all histone modification ChIP-seq data. The function coverage that calculates genome coverage from .bam files is from the “chipseq” package in BioConductor.
Specifically, in analyzing MNase-seq data, since the reads are from the ends of the mononucleosome particle, to best visualize the nucleosome positions, all the reads were shifted to the middle of the corresponding nucleosome core. The average sizes of mononucleosome fragments purified from a 2% agarose gel were determined by Bioanalyzer (Agilent).
Immediate early gene induction
WT and Hmgn1−/−n2−/− MEFs were grown in 1× DMEM (#10-013-CV, Corning) supplemented with 10% fetal calf serum (#16000-044, Life Technologies). After reaching 60% confluence, the cells were stimulated for the indicated times with 50 ng/mL anisomycin (#A9789, Sigma-Aldrich). Next, the cells were scraped and washed with saline. Total RNA was extracted with TRIzol reagent (#15596-026, Invitrogen), followed by purification using RNeasy Plus Mini kit (#74134, Qiagen). Transcription levels of 42 selected genes (Supplemental Table S4) were determined by NanoString Expression Analysis, using custom-made nCounter Gene Expression CodeSets (NanoString Technologies). The hybridization mix containing 10 µL Reporter CodeSet, 10 µL hybridization buffer, 5 µL (100 ng) RNA and 5 µL Capture ProbeSet were incubated for at least 12 h at 65°C and immediately post-hybridized in an nCounter Prep-station (NanoString Technologies). The data were analyzed with nSlover Analysis Software version 1.1. The expression levels of the 42-immediate early genes were normalized to Gapdh, Gusb, and Pgk1. The induction ratio for each gene at any time was calculated by dividing the fold induction of the gene in WT by that of Hmgn1−/−n2−/−. The Log2 of the ratios were subjected to a hierarchical clustering algorithm of self-organizing heat maps (GeneSpring GX version 12.6, Agilent Technologies). All analyses were done on two biological replicates.
Heat shock
Primary mouse WT and Hmgn1−/− n2−/− embryonic fibroblasts were subjected to heat shock at 42°C for the times indicated. RNA was isolated immediately with TRIzol reagent (#15596-026, Invitrogen), followed by purification using RNeasy Plus Mini kit (#74134, Qiagen). Quantitative real-time RT-PCR analysis was done using two primer pairs per gene and two biological replicates using SYBR Green RT-PCR reagent kits (#4309155, Applied Biosystems). ΔCt values were determined by subtracting the Ct value of the PCR reaction obtained using Hspa1a and Hspa1b genes from that of control sets of primers. Expression of Hspa1a and Hspa1b was calculated as value for each time point and each genotype, assigning the expression of Hspa1a and Hspa1b at 0 min as 1 and calculating fold induction as a ratio to the expression at 0 min.
Functional analysis of expression profiles
To predict the impact of HMGN knockouts on gene expression and alteration of biological pathways (each being comprised of a group of the genes), we also adopted a computation approach based on genome-wide expression profiles as described before (Kugler et al. 2013).
Data access
The data from this study have been submitted to the NCBI Sequence Read Archive (SRA; http://www.ncbi.nlm.nih.gov/sra/) under accession number SRS779760.
Supplementary Material
Acknowledgments
This research was supported by the Intramural Research Programs of the CCR, NCI, NIH (grant ZIABC011154); by the NCBI, NLM, NIH; by the German Federal Ministry of Education and Research (Infrafrontier grant 01KX1012); by the Helmholtz Portfolio Theme “Metabolic Dysfunction and Common Disease”; by the Helmholtz Alliance “ICEMED”; and by the German Center for Diabetes Research (DZD).
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.192229.115.
References
- Bianchi ME, Agresti A. 2005. HMG proteins: dynamic players in gene regulation and differentiation. Curr Opin Genet Dev 15: 496–506. [DOI] [PubMed] [Google Scholar]
- Birger Y, West KL, Postnikov YV, Lim JH, Furusawa T, Wagner JP, Laufer CS, Kraemer KH, Bustin M. 2003. Chromosomal protein HMGN1 enhances the rate of DNA repair in chromatin. EMBO J 22: 1665–1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birger Y, Davis J, Furusawa T, Rand E, Piatigorsky J, Bustin M. 2006. A role for chromosomal protein HMGN1 in corneal maturation. Differentiation 74: 19–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle AP, Davis S, Shulha HP, Meltzer P, Margulies EH, Weng Z, Furey TS, Crawford GE. 2008. High-resolution mapping and characterization of open chromatin across the genome. Cell 132: 311–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin M. 1999. Regulation of DNA-dependent activities by the functional motifs of the high-mobility-group chromosomal proteins. Mol Cell Biol 19: 5237–5246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin M. 2001. Chromatin unfolding and activation by HMGN(*) chromosomal proteins. Trends Biochem Sci 26: 431–437. [DOI] [PubMed] [Google Scholar]
- Bustin M, Catez F, Lim JH. 2005. The dynamics of histone H1 function in chromatin. Mol Cell 17: 617–620. [DOI] [PubMed] [Google Scholar]
- Calo E, Wysocka J. 2013. Modification of enhancer chromatin: what, how, and why? Mol Cell 49: 825–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catez F, Brown DT, Misteli T, Bustin M. 2002. Competition between histone H1 and HMGN proteins for chromatin binding sites. EMBO Rep 3: 760–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catez F, Ueda T, Bustin M. 2006. Determinants of histone H1 mobility and chromatin binding in living cells. Nat Struct Mol Biol 13: 305–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherukuri S, Hock R, Ueda T, Catez F, Rochman M, Bustin M. 2008. Cell cycle-dependent binding of HMGN proteins to chromatin. Mol Biol Cell 19: 1816–1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corey LL, Weirich CS, Benjamin IJ, Kingston RE. 2003. Localized recruitment of a chromatin-remodeling activity by an activator in vivo drives transcriptional elongation. Genes Dev 17: 1392–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuddapah S, Schones DE, Cui K, Roh TY, Barski A, Wei G, Rochman M, Bustin M, Zhao K. 2011. Genomic profiling of HMGN1 reveals an association with chromatin at regulatory regions. Mol Cell Biol 31: 700–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng T, Zhu ZI, Zhang S, Leng F, Cherukuri S, Hansen L, Mariño-Ramírez L, Meshorer E, Landsman D, Bustin M. 2013. HMGN1 modulates nucleosome occupancy and DNase I hypersensitivity at the CpG island promoters of embryonic stem cells. Mol Cell Biol 33: 3377–3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding HF, Bustin M, Hansen U. 1997. Alleviation of histone H1-mediated transcriptional repression and chromatin compaction by the acidic activation region in chromosomal protein HMG-14. Mol Cell Biol 17: 5843–5855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdel F, Rippe K. 2011. Chromatin remodelling in mammalian cells by ISWI-type complexes—where, when and why? FEBS J 278: 3608–3618. [DOI] [PubMed] [Google Scholar]
- Fuchs H, Gailus-Durner V, Adler T, Pimentel JA, Becker L, Bolle I, Brielmeier M, Calzada-Wack J, Dalke C, Ehrhardt N, et al. 2009. The German Mouse Clinic: a platform for systemic phenotype analysis of mouse models. Curr Pharm Biotechnol 10: 236–243. [DOI] [PubMed] [Google Scholar]
- Fuchs H, Gailus-Durner V, Adler T, Aguilar-Pimentel JA, Becker L, Calzada-Wack J, Da Silva-Buttkus P, Neff F, Götz A, Hans W, et al. 2011. Mouse phenotyping. Methods 53: 120–135. [DOI] [PubMed] [Google Scholar]
- Gailus-Durner V, Fuchs H, Becker L, Bolle I, Brielmeier M, Calzada-Wack J, Elvert R, Ehrhardt N, Dalke C, Franz TJ, et al. 2005. Introducing the German Mouse Clinic: open access platform for standardized phenotyping. Nat Methods 2: 403–404. [DOI] [PubMed] [Google Scholar]
- Gailus-Durner V, Fuchs H, Adler T, Aguilar Pimentel A, Becker L, Bolle I, Calzada-Wack J, Dalke C, Ehrhardt N, Ferwagner B, et al. 2009. Systemic first-line phenotyping. Methods Mol Biol 530: 463–509. [DOI] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. 2004. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5: R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross DS, Garrard WT. 1988. Nuclease hypersensitive sites in chromatin. Annu Rev Biochem 57: 159–197. [DOI] [PubMed] [Google Scholar]
- Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, et al. 2009. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 459: 108–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang C, Pugh BF. 2009. A compiled and systematic reference map of nucleosome positions across the Saccharomyces cerevisiae genome. Genome Biol 10: R109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kugler JE, Horsch M, Huang D, Furusawa T, Rochman M, Garrett L, Becker L, Bohla A, Hölter SM, Prehn C, et al. 2013. High mobility group N proteins modulate the fidelity of the cellular transcriptional profile in a tissue- and variant-specific manner. J Biol Chem 288: 16690–16703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JY, Patterson M, Mikkola HK, Lowry WE, Kurdistani SK. 2012. Dynamic distribution of linker histone H1.5 in cellular differentiation. PLoS Genet 8: e1002879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim JH, West KL, Rubinstein Y, Bergel M, Postnikov YV, Bustin M. 2005. Chromosomal protein HMGN1 enhances the acetylation of lysine 14 in histone H3. EMBO J 24: 3038–3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Jenkins NA, Copeland NG. 2003. A highly efficient recombineering-based method for generating conditional knockout mutations. Genome Res 13: 476–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucey MM, Wang Y, Bustin M, Duncan MK. 2008. Differential expression of the HMGN family of chromatin proteins during ocular development. Gene Expr Patterns 8: 433–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahadevan LC, Willis AC, Barratt MJ. 1991. Rapid histone H3 phosphorylation in response to growth factors, phorbol esters, okadaic acid, and protein synthesis inhibitors. Cell 65: 775–783. [DOI] [PubMed] [Google Scholar]
- Mueller-Planitz F, Klinker H, Becker PB. 2013. Nucleosome sliding mechanisms: new twists in a looped history. Nat Struct Mol Biol 20: 1026–1032. [DOI] [PubMed] [Google Scholar]
- Narlikar GJ, Fan HY, Kingston RE. 2002. Cooperation between complexes that regulate chromatin structure and transcription. Cell 108: 475–487. [DOI] [PubMed] [Google Scholar]
- Postnikov YV, Trieschmann L, Rickers A, Bustin M. 1995. Homodimers of chromosomal proteins HMG-14 and HMG-17 in nucleosome cores. J Mol Biol 252: 423–432. [DOI] [PubMed] [Google Scholar]
- Postnikov YV, Kurahashi T, Zhou M, Bustin M. 2012. The nucleosome binding protein HMGN1 interacts with PCNA and facilitates its binding to chromatin. Mol Cell Biol 32: 1844–1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rattner BP, Yusufzai T, Kadonaga JT. 2009. HMGN proteins act in opposition to ATP-dependent chromatin remodeling factors to restrict nucleosome mobility. Mol Cell 34: 620–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochman M, Postnikov Y, Correll S, Malicet C, Wincovitch S, Karpova TS, McNally JG, Wu X, Bubunenko NA, Grigoryev S, et al. 2009. The interaction of NSBP1/HMGN5 with nucleosomes in euchromatin counteracts linker histone-mediated chromatin compaction and modulates transcription. Mol Cell 35: 642–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stergachis AB, Neph S, Reynolds A, Humbert R, Miller B, Paige SL, Vernot B, Cheng JB, Thurman RE, Sandstrom R, et al. 2013. Developmental fate and cellular maturity encoded in human regulatory DNA landscapes. Cell 154: 888–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R. 2003. Statistical significance for genomewide studies. Proc Natl Acad Sci 100: 9440–9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swygert SG, Peterson CL. 2014. Chromatin dynamics: interplay between remodeling enzymes and histone modifications. Biochim Biophys Acta 1839: 728–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurman RE, Rynes E, Humbert R, Vierstra J, Maurano MT, Haugen E, Sheffield NC, Stergachis AB, Wang H, Vernot B, et al. 2012. The accessible chromatin landscape of the human genome. Nature 489: 75–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tullai JW, Schaffer ME, Mullenbrock S, Sholder G, Kasif S, Cooper GM. 2007. Immediate-early and delayed primary response genes are distinct in function and genomic architecture. J Biol Chem 282: 23981–23995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda T, Catez F, Gerlitz G, Bustin M. 2008. Delineation of the protein module that anchors HMGN proteins to nucleosomes in the chromatin of living cells. Mol Cell Biol 28: 2872–2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villar D, Berthelot C, Aldridge S, Rayner TF, Lukk M, Pignatelli M, Park TJ, Deaville R, Erichsen JT, Jasinska AJ, et al. 2015. Enhancer evolution across 20 mammalian species. Cell 160: 554–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voss TC, Hager GL. 2014. Dynamic regulation of transcriptional states by chromatin and transcription factors. Nat Rev Genet 15: 69–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wippo CJ, Israel L, Watanabe S, Hochheimer A, Peterson CL, Korber P. 2011. The RSC chromatin remodelling enzyme has a unique role in directing the accurate positioning of nucleosomes. EMBO J 30: 1277–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodcock CL, Skoultchi AI, Fan Y. 2006. Role of linker histone in chromatin structure and function: H1 stoichiometry and nucleosome repeat length. Chromosome Res 14: 17–25. [DOI] [PubMed] [Google Scholar]
- Zang C, Schones DE, Zeng C, Cui K, Zhao K, Peng W. 2009. A clustering approach for identification of enriched domains from histone modification ChIP-seq data. Bioinformatics 25: 1952–1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.