Abstract
SETDB1, a histone methyltransferase responsible for methylation of histone H3 lysine 9 (H3K9), is involved in maintenance of embryonic stem (ES) cells and early embryonic development of the mouse. However, how SETDB1 regulates gene expression during development is largely unknown. Here, we characterized genome-wide SETDB1 binding and H3K9 trimethylation (H3K9me3) profiles in mouse ES cells and uncovered two distinct classes of SETDB1 binding sites, termed solo and ensemble peaks. The solo peaks were devoid of H3K9me3 and enriched near developmental regulators while the ensemble peaks were associated with H3K9me3. A subset of the SETDB1 solo peaks, particularly those near neural development–related genes, was found to be associated with Polycomb Repressive Complex 2 (PRC2) as well as PRC2-interacting proteins JARID2 and MTF2. Genetic deletion of Setdb1 reduced EZH2 binding as well as histone 3 lysine 27 (H3K27) trimethylation level at SETDB1 solo peaks and facilitated neural differentiation. Furthermore, we found that H3K27me3 inhibits SETDB1 methyltransferase activity. The currently identified reciprocal action between SETDB1 and PRC2 reveals a novel mechanism underlying ES cell pluripotency and differentiation regulation.
The self-renewal and pluripotency of ES cells are regulated by both genetic and epigenetic mechanisms (Boyer et al. 2005; Surani et al. 2007; Chen and Daley 2008; Chen et al. 2008; Kim et al. 2008). Epigenetic silencing represses developmental programs, guiding ES cell differentiation into distinct lineages (Surani et al. 2007; Hong et al. 2011). SETDB1, also known as ESET or KMT1E, was identified as a key chromatin modifier required for ES cell maintenance through the suppression of developmental regulators (Bilodeau et al. 2009). Consistently, we have shown previously that Setdb1 is essential for embryogenesis (Dodge et al. 2004). In coordination with POU5F1, SETDB1 suppresses the trophectoderm cell lineage (Yuan et al. 2009; Lohmann et al. 2010). SETDB1 was also shown to be involved in various developmental processes (Wang et al. 2011; Mysliwiec et al. 2012) as well as silencing of endogenous retrovirus (Matsui et al. 2010; Karimi et al. 2011).
Although SETDB1 is a major histone methyltransferase (HMT) responsible for transcription repression mainly via H3K9me3 (Schultz et al. 2002), deletion of Setdb1 has little or no effect on global H3K9me3 level in ES cells (Dodge et al. 2004). Brain-specific deletion of Setdb1 leads to impaired brain development accompanying the suppressed expression of neuronal genes (Tan et al. 2012). Since SETDB1 is a transcription suppressor, it is unlikely that the suppression of neuronal genes upon Setdb1 deletion is directly resulted from the change of H3K9me3 mediated by SETDB1. These observations indicated that there may be H3K9me3 independent roles of SETDB1 in gene expression and development regulation.
Consistent with the roles of SETDB1 in early development, searching genome-wide SETDB1 binding sites in mouse ES cells has indicated that SETDB1 is associated with developmental regulators which are often poised as bivalent genes that are manifested by the co-occupancy of the repressive H3K27me3 and the active H3K4me3 methyl marks (Bilodeau et al. 2009; Yuan et al. 2009). Moreover, SETDB1 bindings are found to overlap with the Polycomb proteins (Bilodeau et al. 2009). There is also evidence indicating that SETDB1 could interact with JARID2, an interacting protein of the PRC2 complex (Mysliwiec et al. 2012). Since PRC2 is thought to maintain ES cell pluripotency by silencing developmental regulators (Margueron and Reinberg 2011), it is of importance to determine whether SETDB1 can act in coordination with the PRC complex to regulate stem cell pluripotency and differentiation; and if so, whether such coordination will require H3K9 trimethylation mediated by SETDB1.
Results
SETDB1 targets demonstrate distinct H3K9 trimethylation status
To better understand how SETDB1 regulates gene expression and cell differentiation in ES cells, we analyzed global H3K9me3 in inducible Setdb1 knockout (iKO) mouse ES cells using an H3K9me3-specific antibody (Supplemental Fig. S1). By comparing H3K9me3 ChIP (chromatin immunoprecipitation)-seq (deep sequencing) signals of four biological replicates from our experiment with previous SETDB1 ChIP-seq data performed under similar conditions (Yuan et al. 2009), we found, on average, only 4.7% of the H3K9me3 signal peaks overlapping with SETDB1 binding sites, indicating that a large number of H3K9me3 peaks may be attributed to other H3K9 HMTs, such as SUV39H1/H2. For each SETDB1 binding site, we calculated the average H3K9me3 signal within its center 2-kb window. Then we drew the distribution of the H3K9me3 signals derived from all SETDB1 binding sites, and to our surprise, we observed a clear bimodal distribution for each H3K9me3 ChIP-seq biological replicate (Supplemental Fig. S2A–D), suggesting there are two kinds of SETDB1 peaks with distinct H3K9me3 enrichment status. We further checked the observation using previously published H3K9me3 data sets, and three data sets from two studies (Yuan et al. 2009; Karimi et al. 2011) also displayed a clear bimodal distribution (Supplemental Fig. S2E,G,H), while one data set (Bilodeau et al. 2009) showed a unimodal pattern (Supplemental Fig. S2F).
Next we examined the H3K9me3 signal within 10 kb of the center of SETDB1 binding sites. Unsurprisingly, a significant portion (2689 out of 5882; 45.7%) of SETDB1 binding sites did not have nearby H3K9me3 signal peaks in any of four H3K9me3 ChIP-seq replicates, and those peaks were termed SETDB1 solo peaks. Accordingly, SETDB1 binding sites with nearby H3K9me3 signal peaks in all of four H3K9me3 ChIP-seq replicates were termed ensemble peaks (1522 out of 5882, 25.9%). Binding profiles of representative SETDB1 solo and ensemble peaks are shown in Figure 1A,B. The average profile of H3K9me3 signal around SETDB1 solo peaks indeed presented a much lower level than that around SETDB1 ensemble peaks (Supplemental Fig. S2I–M,O,P). We further confirmed H3K9me3 signal and SETDB1 binding at those solo and ensemble loci by ChIP-qPCR analysis (Fig. 1C,D). Setdb1 deletion diminished SETDB1 binding at those loci, indicating that the lack of H3K9me3 at the SETDB1 solo peaks is not due to artifacts of SETDB1 antibody (Supplemental Fig. S3). Further analysis of H3K9me1 and H3K9me2 status at SETDB1 solo and ensemble peaks by ChIP-qPCR revealed that SETDB1 ensemble peaks often had H3K9me1 and H3K9me2 signals all five SETDB1 solo peaks tested were devoid of H3K9me2 or H3K9me1 (Supplemental Fig. S4).
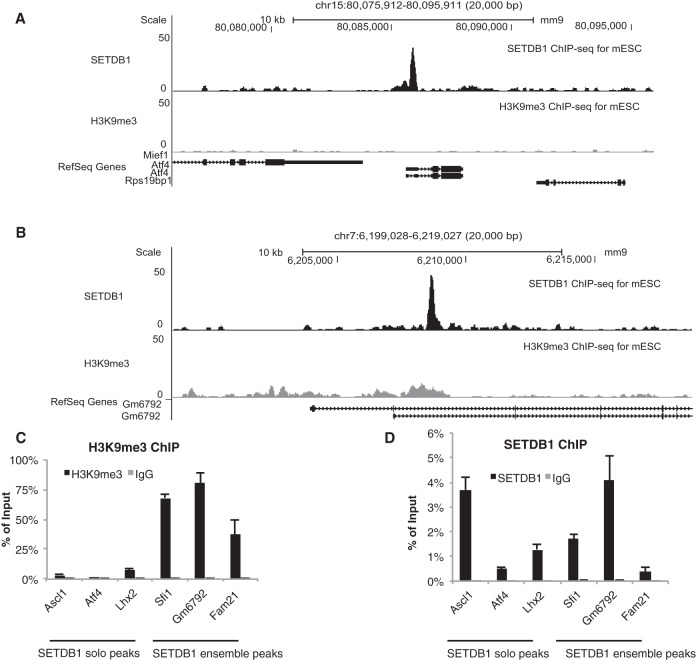
Figure 1.
Classification of SETDB1 targets to solo and ensemble peaks. (A) Examples for SETDB1 solo (around Atf4 promoter) peaks. (Top) Track is the ChIP-seq profile of SETDB1 in wild-type mouse ES cells (Yuan et al. 2009). (Bottom) Track is the H3K9me3 ChIP-seq (biological replicate 1) signal in Setdb1 iKO cells (without Tam treatment). (B) Examples for SETDB1 ensemble (around Gm6792 promoter) peaks. (Top) Track is the ChIP-seq profile of SETDB1 in wild-type mouse ES cells (Yuan et al. 2009). (Bottom) Track is the H3K9me3 ChIP-seq (biological replicate 1) signal in Setdb1 iKO cells (without Tam treatment). (C) ChIP-qPCR of H3K9me3 in selected SETDB1 peak loci. Setdb1 iKO mouse ES cells underwent ChIP with H3K9me3 antibody. Anti-rabbit normal IgG was used as a negative control. The enrichment level was normalized to input. The selected SETDB1 peak loci were marked by their nearby gene symbols (Atf4, Lhx2, Ascl1 for SETDB1 solo peaks; Fam21, Gm6792, Sfi1 for SETDB1 ensemble peaks). (D) ChIP-qPCR of SETDB1 in selected SETDB1 peak loci. Setdb1 iKO mouse ES cells underwent ChIP with SETDB1 antibody. Anti-rabbit normal IgG was used as a negative control. The enrichment level was normalized to input. The selected SETDB1 peak loci were marked by their nearby gene symbols (Atf4, Lhx2, Ascl1 for SETDB1 solo peaks; Fam21, Gm6792, Sfi1 for SETDB1 ensemble peaks).
SETDB1 solo peak targets enriched for developmental regulators
To investigate the potential functional difference between SETDB1 solo and ensemble peaks, we examined their genomic distributions. SETDB1 solo peaks were highly enriched in promoter regions (9.6-fold; P-value <1.0 × 10−15, Fisher's exact test), while the enrichment for ensemble peaks (3.0-fold; P-value: 6.0 × 10−7, Fisher's exact test) in promoter regions was not as strong (Fig. 2A). We further performed Gene Ontology (GO) analysis on all SETDB1 target genes, defined as genes having transcription start sites (TSSs) within 5 kb of a SETDB1 binding site. Consistent with one previous study (Yuan et al. 2009), several GO categories in neural development–related processes were significantly overrepresented. Next, we divided SETDB1 target genes into solo and ensemble peak target genes, and we observed that biological processes related to neural development were only specifically enriched for SETDB1 solo peak target genes (Fig. 2B). Consistently, among 2073 SETDB1 target genes, 150 (7.2%) were neural development related, and 126 of them were SETDB1 solo peak target genes. Furthermore, among all SETDB1 peak target genes, 12 were key ES regulators (Nishiyama et al. 2009); 11 of them were SETDB1 solo peak target genes, suggesting that SETDB1 solo peaks represent major roles of SETDB1 in ES regulation.
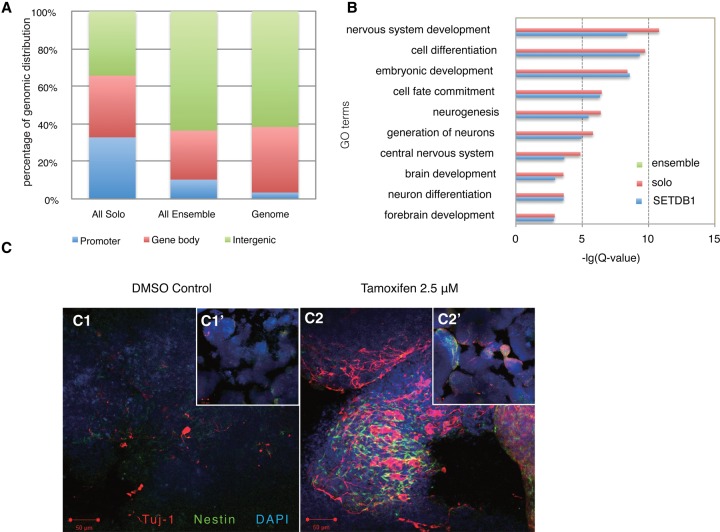
Figure 2.
SETDB1 solo and ensemble peaks display distinct biological functions. (A) Genomic distribution of SETDB1 solo and ensemble peaks. We defined regions from 2 kb upstream of the TSS to 2 kb downstream from the TSS as promoters (blue bars), from 2 kb downstream from the TSS to the transcription termination sites (TTS) as gene bodies (red bars), and all other genomic regions as intergenic regions (green bars). (B) GO analysis on all SETDB1 target genes (blue bars), SETDB1 solo target genes (red bars), and SETDB1 ensemble target genes (green bars). Fisher's exact test and Benjamini correction were performed. (C) Setdb1 was deleted in iKO mouse ES cells by Tam treatment, and ES cells were induced for neural differentiation in suspension cultures for 8 d. Immunostaining for TUJ1 (red) was used to assess neuronal cell formation. Cultures were costained with Nestin (green) and DAPI (blue). Note that more neural differentiation, as manifested by TUJ1-positive cells, was seen in Setdb1-deleted iKO cells compared with the DMSO control. (Insets) Small-magnitude images for a complete view of the embryoid bodies (EBs). Bars, 50 μm. (Insets) Bars, 100 μm.
To determine whether SETDB1 regulates neural differentiation in ES cells, we conditionally deleted Setdb1 in adherent cultures for 3 d before differentiating them into the neuronal lineage using suspension cultures. Setdb1 deletion efficiency was confirmed by Western blot analysis (Supplemental Fig. S5). Deletion of Setdb1 by tamoxifen (Tam) dramatically increased neuronal differentiation, as visualized by TUJ1 staining (Fig. 2C). Wild-type mouse ES cells used as controls did not show any significant effect on neuronal differentiation (Supplemental Fig. S6), suggesting that enhanced neuronal differentiation observed in Setdb1 iKO cells results from the loss of Setdb1. As GO categories in neural development were specifically enriched for SETDB1 solo peak target genes, we hypothesized that SETDB1 may regulate neural developmental genes through a mechanism other than H3K9 methylation.
A subset of SETDB1 solo peaks overlap with PRC binding sites
To explore the potential regulatory mechanism of SETDB1 in mouse ES cells, we examined the colocalization between SETDB1 and other transcription factors (TFs). In total, 43 TFs with public ChIP-seq data sets were used (Supplemental Table S1), including key stem cell factors (POU5F1, NANOG, SOX2) and histone or DNA modifying enzymes (EZH2, SUZ12, TET1). At SETDB1 ensemble peaks and a subset of solo peaks (not associated with neural development genes), binding levels for the 43 factors were slightly above or below average (signal ratio = 1.0) (for details, see Methods; Supplemental Fig. S7A,B). However, at those solo peaks near genes related to neural development, two core PRC2 members (EZH2 and SUZ12), one PRC1 core member (RNF2), and two PRC2-interacting factors (JARID2 and MTF2) displayed distinctly high binding intensity (Fig. 3A), indicating that a fraction of SETDB1 solo peaks were closely associated with PRC binding. Moreover, k-means clustering of the five PRC components or interacting factors showed SETDB1 solo peaks that overlapped with PRC2 (termed as SETDB1 solo/PRC2 peaks) were accompanied by abundant proximal H3K27me3 modification (Fig. 3B,C).
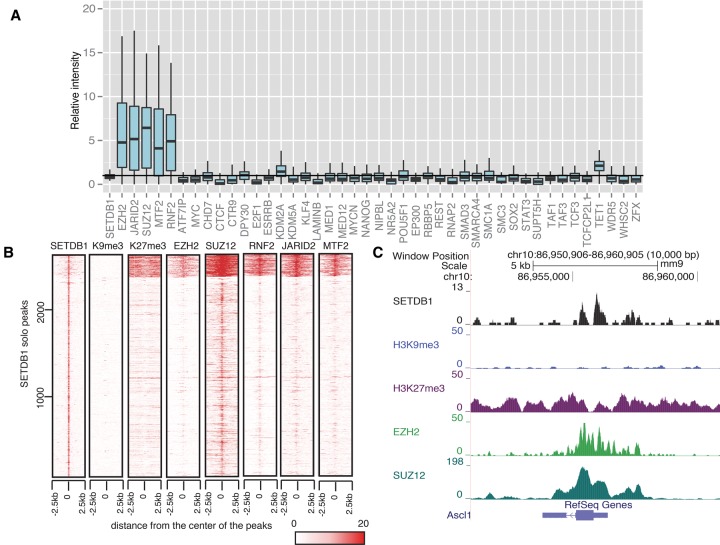
Figure 3.
SETDB1 solo peaks overlap with PRC complex. (A) Relative binding intensity of 43 TFs at all neural development–related SETDB1 solo peak loci. All 43 TFs have public ChIP-seq data in mouse ES cells (Supplemental Table S1). The value of base pair count/peak length for all neural development–related solo peaks was further divided by the corresponding average level of all SETDB1 peaks for each TF. These ratios were used to depict the deviation from the average binding intensity across all SETDB1 peaks, which was marked with the horizontal line. (B) In the panel, 2689 SETDB1 solo peak loci were clustered to two groups by k-means according to binding intensities of SETDB1, H3K9me3 (biological replicate 1), H3K27me3 (biological replicate 1), EZH2, SUZ12, RNF2, JARID2, and MTF2. In the clustering, each line represents a genomic location around a SETDB1 binding site (±2.5 kb). The color scale indicates normalized ChIP-seq enrichment level from bigWig files generated by MACS2. (C) An example of SETDB1 solo/PRC2 peak locus (around Ascl1 promoter). SETDB1 (black track) (Yuan et al. 2009), H3K9me3 (blue track; biological replicate 1), H3K27me3 (purple track; biological replicate 1), EZH2 (light green track) (Peng et al. 2009), and SUZ12 (dark green track) (Peng et al. 2009) ChIP-seq profiles.
SETDB1 target genes are believed to partially overlap with SUZ12 target genes among development-related genes (Bilodeau et al. 2009); our data further indicated that the co-occupancy of SETDB1 and PRC2 in mouse ES cells occurs at loci without detectable H3K9me3 signals. To avoid the potential biases from analysis parameters, different peak calling cutoffs for H3K9me3 (Q-value 0.001, 0.01; P-value 0.001) were applied to validate the rationality. The looser the cutoff for H3K9me3 peak calling was, the more stringent the standard for SETDB1 solo peaks would be. SETDB1 solo peaks were divided into three groups based on the peak calling cutoffs of H3K9me3 data (Supplemental Fig. S8A). GO analysis demonstrated that neural development–related biological processes became gradually enriched in solo peak targets when the standard was tightened up (Supplemental Fig. S8B). In addition, the ChIP-seq signals for H3K27me3 and PRC components at SETDB1 solo peaks increased steadily with more stringent cutoffs (Supplemental Fig. S9). Therefore, the existence of SETDB1 solo peaks related to neural development–related genes and co-occupied with PRC2 is reliable.
H3K27 methylation inhibits SETDB1-mediated H3K9 trimethylation
Given that a subset of SETDB1 solo peaks are associated with H3K27me3 while having an undetectable level of H3K9me3 and that SETDB1 solo and ensemble peaks display distinct H3K9me3 and H3K27me3 patterns (Supplemental Fig. S10), we hypothesized that H3K27 methylation may inhibit SETDB1-mediated H3K9 trimethylation. To test this, we performed in vitro biochemical assays using recombinant nucleosomes as the substrates. Prior to the H3K9 methylation by SETDB1, the nucleosomes were first methylated by a five-member complex consisting of recombinant EED, SUZ12, RBBP4, AEBP2, and a wild-type or a Y641F mutant form of EZH2, which is known to facilitate H3K27me2-to-H3K27me3 conversion (Yap et al. 2011). We found that H3K27 methylation by EZH2 did inhibit SETDB1 activity on H3K9 methylation (Fig. 4A); the inhibition was particularly apparent on H3K9me3, but not on H3K9me1 or H3K9me2. Prereacted nucleosomes with an HMT activity-dead mutant of EZH2, however, showed no inhibition on H3K9me3 methylation by SETDB1. These data indicate that EZH2-mediated H3K27 trimethylation inhibits SETDB1 activity, particularly the H3K9me2-to-H3K9me3 conversion.
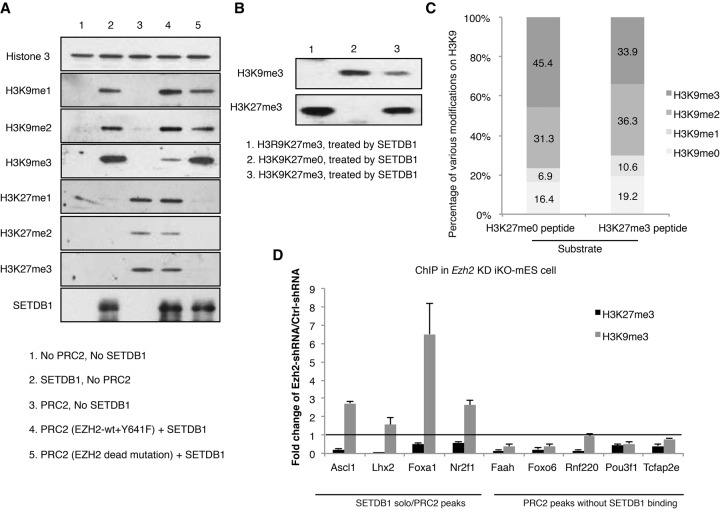
Figure 4.
H3K27 methylation inhibits SETDB1-mediated H3K9 methylation. (A) In vitro methylation assay was performed using recombinant nucleosomes. The recombinant nucleosome was first reacted with EZH2 (1:1 mixture of wild-type and the Y641F mutant EZH2 protein together with EED, RBBP4, AEBP2, and SUZ12) to induce H3K27 methylation. Recombinant human SETDB1 protein was then added to the reaction for 4 h at room temperature. The reaction was measured by Western blot analysis using antibodies against various H3K9 methyl marks, as well as H3K27 methyl marks. SETDB1 Western blot analysis confirmed the equal amount of the SETDB1 protein in the reaction. (B) Chemically modified H3 (1-84aa) peptides were used as the substrates in the in vitro methylation assay with SETDB1. Peptides with H3K27me3 modification (lane 3) showed reduced H3K9 trimethylation by SETDB1 in vitro compared to H3K27me0 peptides (lane 2). A peptide with K9 mutated to R was used as the control (lane 1). (C) The end products of the peptide-based, in vitro methylation assay were quantified by LC/MS analysis. The percentage of the various modifications was shown in the stack bar graphs. The left and right plots correspond to the product from the peptide substrate with (lane 3 in B) or without H3K27me3 (lane 2 in B) modification, respectively. Note that the H3K9me3 production is dramatically reduced when the H3K27me3 peptide was used as the substrate. (D) Setdb1 iKO cells were infected with a lentiviral shRNA against Ezh2 or control scramble shRNA. The chromatin was immunoprecipitated with antibodies against H3K27me3 or H3K9me3 histone marks. The data are normalized to the control shRNA samples. Four representative SETDB1 solo/PRC2 loci were analyzed by qPCR using the immunoprecipitated DNA. The selected SETDB1 solo/PRC2 peak loci were marked by their nearby gene symbols (Ascl1, Lhx2, Foxa1, and Nr2f1). Five PRC2 peaks without SETDB1 binding were used as the control. The selected PRC2 peak loci without SETDB1 binding were marked by their nearby gene symbols (Faah, Pou3f1, Tcfap2e, Rnf220, and Foxo6).
To rule out the possibility that the observed inhibition of H3K9 trimethylation was due to the depletion of S-adenosyl methionine (SAM), the methyl donor for HMTs in the reaction, we used synthetic histone H3 peptides with chemically modified K27me3 as the substrates in the in vitro methylation assay. We found that K27 trimethylated peptides had a significantly lower efficiency for K9 trimethylation by SETDB1 (Fig. 4B). The reduction of H3K9me3 in the K27 methylated peptide was also confirmed and quantified by using LC/MS analysis (Fig. 4C).
To extend our in vitro findings to in vivo, we next asked if reduced H3K27me3 in mouse ES cells would release suppression of H3K9me3 at the SETDB1 solo/PRC2 peaks. We thus attenuated EZH2 expression in the iKO mouse ES cells using an shRNA against EZH2. The knockdown efficiency was shown in Supplemental Figure S11. We tested four SETDB1 solo/PRC2 peaks and five PRC2 peaks without SETDB1 binding as the control. We found that attenuation of EZH2 reduced H3K27me3 in all of the loci tested. The SETDB1 solo/PRC2 peaks indeed regained H3K9me3, but not the PRC2 peaks without SETDB1 binding (Fig. 4D). These data suggest that nearby inhibition of SETDB1-mediated H3K9 methylation by H3K27me3 may also take place in vivo.
SETDB1 complexes with and regulates PRC2 activity at a subset of SETDB1 solo peaks
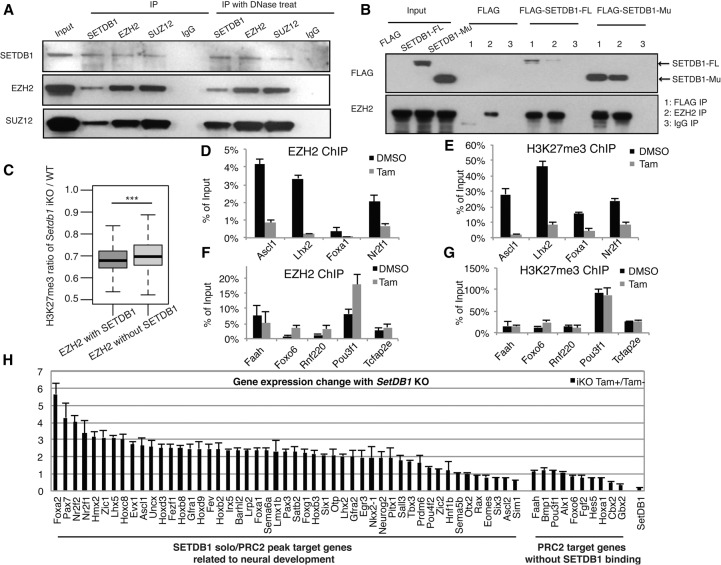
Next, we performed coimmunoprecipitation (co-IP) to pull down either SETDB1 or EZH2/SUZ12 to determine if SETDB1 and PRC2 can coexist in a complex. Indeed, SETDB1 and EZH2/SUZ12 reciprocally pulled down one another (Fig. 5A), suggesting that SETDB1 may physically interact with the PRC2 complex at least in mouse ES cells. As a control, in the Setdb1-deleted ES cells, we did not detect the cocomplex (data not shown). To rule out the DNA- or nucleosome-dependent interaction, we further performed DNase I treatment prior to the co-IP experiment. We found that DNase treatment did not affect the co-IP efficiency (Fig. 5A). To further understand the SETDB1 and PRC2 interaction, we generated a SET domain-deleted SETDB1 mutant that lost the HMT enzymatic activity. We overexpressed this mutant SETDB1 in the iKO mouse ES cells and performed the co-IP experiments. We found that such a SETDB1 mutant was still able to pull down the endogenous PRC2 complex (Fig. 5B). This observation suggested that the interaction between SETDB1 and PRC2 does not require the SET domain. Identification of the protein domain(s) of SETDB1 necessary for this interaction warrants further investigation.
Figure 5.
SETDB1 modulates H3K27 methylation and EZH2 binding. (A) Coimmunoprecipitation (co-IP) was performed to validate SETDB1 and PRC2 correlation. Nuclear proteins of SETDB1 iKO-ES cells were extracted for IP with SETDB1, EZH2, and SUZ12 antibodies. IgG was used as a negative control. Western blot analysis was performed with SETDB1, EZH2, and SUZ12 antibodies. Cell cytoplasmic and nuclear fractions were used as the input controls. (Left) co-IP results without DNase treatment; (right) those with DNase treatment prior to the IP procedure. (B) 3xFLAG-tagged, full-length wild-type or SET domain-deleted SETDB1 was transfected into iKO mouse ES cells. Nuclear proteins were extracted for IP using the antibodies for SETDB1 or EZH2. IgG was used as a negative control. (C) H3K27me3 level variation at all ±10-kb regions of EZH2 peak loci before and after Setdb1 iKO (biological replicate pair 1). Only EZH2 peaks with enriched H3K27me3 signal before Tam treatment were included in this analysis. Those EZH2 peaks were divided into two categories based on the overlap status with SETDB1 binding: with SETDB1 binding (431 peaks) and without SETDB1 binding (13,138 peaks). Two-sided Welch's t-test was performed for the change of H3K27me3 between the two groups; (***) P < 0.001. Supplemental Figure S12 showed the results using biological replicate pair 2. (D) ChIP-qPCR of EZH2 in selected SETDB1 solo/PRC2 peak loci upon Setdb1 knockout. Chromatin of Setdb1 iKO ES with or without Tam treatment was treated with anti-EZH2 antibody. The selected SETDB1 solo/PRC2 peak loci were marked by their nearby gene symbols (Ascl1, Lhx2, Foxa1, and Nr2f1). (E) ChIP-qPCR of H3K27me3 in selected SETDB1 solo/PRC2 peak loci upon Setdb1 knockout. Chromatin of Setdb1 iKO ES with or without Tam treatment was treated with anti-H3K27me3 antibody. (F) ChIP-qPCR of EZH2 in selected PRC2 peak loci without SETDB1 binding upon Setdb1 knockout. Chromatin of Setdb1 iKO ES with or without Tam treatment was treated with anti-EZH2 antibody. The selected PRC2 peak loci without SETDB1 binding were marked by their nearby gene symbols (Faah, Foxo6, Rnf220, Pou3f1, and Tcfap2e). No reduction of EZH2 binding was observed. (G) ChIP-qPCR of H3K27me3 in selected PRC2 peak loci without SETDB1 binding upon Setdb1 knockout. Chromatin of Setdb1 iKO ES with or without Tam treatment was treated with anti-H3K27me3 antibody. No reduction of H3K27me3 enrichment was observed. (H) Setdb1 iKO ES cells were treated with or without Tam for 3 d. Setdb1 expression level was detected by qPCR for knockout efficiency confirmation. The expression level of the 49 SETDB1 solo/PRC2 peak target neural development–related genes was examined by RT-qPCR analysis. Ten PRC2 target genes without SETDB1 binding were used as the negative control. Please note that while there is significant reactivation of the SETDB1 solo/PRC2 peak target genes related to neural development (P-value <0.05, paired t-test) as a group, there is no consistent change of the PRC2 target genes without SETDB1 binding.
We next hypothesized that SETDB1 may regulate solo/PRC2 peak targets by modulating PRC2 activity. We first compared genome-wide H3K27me3 levels upon Setdb1 deletion. We found that loss of Setdb1 caused a moderate decrease in H3K27me3 level at SETDB1 solo/PRC2 peaks compared with PRC2 peaks without SETDB1 binding (P-value: 2.79 × 10−7, two-sample Welch's t-test) (Fig. 5C; Supplemental Fig. S12). We next examined four SETDB1 solo/PRC2 peaks in Setdb1 iKO mouse ES cells using ChIP-qPCR. Setdb1 deletion resulted in significantly reduced EZH2 binding at these loci (Fig. 5D), along with significant loss of H3K27me3 (Fig. 5E). However, in wild-type D3 control mouse ES cells, the addition of Tam did not reduce EZH2 binding or H3K7me3 level at these loci (data not shown). To further confirm that this reduction is specific to the SETDB1 binding, we did in parallel similar EZH2 and H3K27me3 ChIP-qPCR analyses on five randomly selected PRC2 peak loci without SETDB1 binding. We found that Setdb1 deletion did not reduce the EZH2 binding (Fig. 5F) or H3K27me3 (Fig. 5G) level at those loci. To test if loss of SETDB1 would reactivate the expression of the SETDB1 solo/PRC2 genes, we carried out a large-scale RT-qPCR analysis. Out of 96 neural development–related genes that were SETDB1 solo/PRC2 peak targets, we randomly selected 49 (51.0%) SETDB1 solo/PRC2 genes and examined their expression upon Tam treatment by RT-qPCR. We found that Setdb1 deletion reactivated the expression of 44 out of 49 genes. As a control, we did not see consistent changes for randomly selected PRC2 target genes without SETDB1 binding (Fig. 5H).
To determine if the reduced EZH2 binding and H3K27me3 following Setdb1 deletion did not result from general protein destabilization caused by Setdb1 depletion, we examined the total EZH2 protein amount upon Setdb1 deletion. We found that Setdb1 deletion did not affect the total EZH2 or SUZ12 protein level (Supplemental Fig. S13); global H3K27me1, me2, and me3 levels remained unaffected as well. Together, these data suggest that SETDB1 regulates a subset of SETDB1 solo/PRC2 peaks by modulating PRC2 activity.
Discussion
In the present study, we found that substantial SETDB1 targets indeed have very low H3K9me3. To corroborate our findings, we also reanalyzed the genome-wide distribution of SETDB1 and H3K9me3 signals in mouse ES cells using data reported in three previous studies(Bilodeau et al. 2009; Yuan et al. 2009; Karimi et al. 2011). Data sets from two studies also showed a general bimodal distribution of H3K9me3 signals in all SETDB1 peaks, indicating there was a subset of SETDB1 peaks with low H3K9me3 signal, but the concept and mechanism of the SETDB1 solo peaks were not addressed in those studies (Yuan et al. 2009; Karimi et al. 2011). Another study revealed the coexistence of SETDB1 binding and H3K9me3 enrichment as an overall pattern (Bilodeau et al. 2009). In that study, most euchromatic genes with H3K9me3 signal peaks also showed strong H3K4me3 signals (Bilodeau et al. 2009; Lohse et al. 2013). As H3K4me3 and H3K9me3 marks were considered to be mutually exclusive with each other (Wang et al. 2001), this might imply the possible lack of antibody specificity in that exceptive data set. It was recently reported that the non-PRC2 functions of EZH2 may play important roles for its oncogenic functions (Xu et al. 2012), suggesting that the disparity between HMTs and their respective canonical histone methylations may represent a novel and general epigenetic mechanism.
Based on the H3K9me3 profiles generated by our ChIP-seq experiment, we categorized SETDB1 binding sites into ensemble and solo peaks. The average profile of H3K9me3 signal around SETDB1 solo peaks presented a much lower level than that around SETDB1 ensemble peaks. SETDB1 peak targets are enriched for development-related genes (Bilodeau et al. 2009; Yuan et al. 2009). An estimated 10% of genes in ES cells are occupied by PRC2, including many developmental regulators. Consistent with this, we found that a subset of SETDB1 solo peaks overlap with PRC2 targets with abundant H3K27me3. That subset of SETDB1 peaks contains a higher percentage and longer length of CpG islands than other SETDB1 peaks (data not shown), suggesting more and longer CpG islands may afford a greater opportunity for cobinding of other TFs. We further showed that SETDB1 complexes with PRC2. Our data indicate that SETDB1 solo/PRC2 peak targets are the most significantly enriched group for neural development genes, which supports the observation that inactivation of Setdb1 leads to enhanced neurogenesis competence in mouse ES cells.
Long noncoding RNAs, active TF binding, and DNA-binding proteins have been implicated in the poorly understood process of PRC2 recruitment to specific loci (Plath et al. 2003; Rinn et al. 2007; Mendenhall et al. 2010; Rapicavoli et al. 2011). JARID2, a substoichiometric subunit of the PRC2 complex that can directly bind to DNA, is thought to be involved in PRC2 recruitment (Peng et al. 2009; Herz and Shilatifard 2010). We showed obvious enrichments of two PRC2 core members, EZH2 and SUZ12, as well as JARID2 and MTF2, at the loci of a fraction of the SETDB1 solo peaks but not ensemble peaks. Deletion of Setdb1 reduced both the binding of EZH2 to SETDB1 solo/PRC2 peaks and the presence of H3K27me3 marks at those loci. Therefore, SETDB1 may help target the PRC2 complex to developmental regulators through stoichiometric regulation. However, such speculation needs further experimental validation. The co-occupancy of SETDB1 solo peaks with JARID2, which modulates H3K9 methylation at the Notch1 (Mysliwiec et al. 2012), and with the PRC2 complex suggests an important role for JARID2 in coordinating gene silencing by H3K9 and H3K27 methylation mediated by SETDB1 and PRC2, respectively. This mechanism may contribute to the fine-tuning of gene expression that is critical for ES differentiation.
Histone modifications are often combinatory context dependent; cross-talk between different forms of epigenetic modulations adds another layer to the complexity of epigenetic regulation (Lee et al. 2010). Histone methylation itself may be affected by methylation status of nearby residues (Binda et al. 2010; Schmitges et al. 2011; Yuan et al. 2011). Although the general mechanism of SETDB1 solo peak generation remains elusive, our findings that H3K27me3 inhibits H3K9 trimethylation by SETDB1 suggest that, at least, a subset of the SETDB1 solo peaks may result from nearby inhibition by H3K27me3 at SETDB1 solo/PRC2 loci. Moreover, there is a general dip of the average H3K27me3 profile at the peak of SETDB1 solo/PRC2 targets (data not shown). The H3K9me3/H3K27me3 cross-talk of histone methylation at nearby residues adds to the previously identified antagonism between H3K4me3 and H3K9me3 (Binda et al. 2010), as well as H3K27me3 and H3K4me3 (Schmitges et al. 2011) / H3K36me2/3 (Yuan et al. 2011). Further investigation is needed to elucidate the origination of the SETDB1 solo peaks without PRC2 binding, as well as the recruitment mechanism of SETDB1 to the ensemble and solo peaks.
Both SETDB1 and PRC2 are thought to maintain ES cell pluripotency by silencing developmental regulators (Boyer et al. 2006; Lee et al. 2006). ES cells with PRC2 core subunits deleted can remain undifferentiated, suggesting other factors may act in coordination with the PRC2 complex to maintain ES cell pluripotency (Pasini et al. 2007; Chamberlain et al. 2008; Margueron and Reinberg 2011). H3K27me3 was found to be associated with facultative heterochromatin, which is highly developmentally regulated, while H3K27me1 is associated with constitutive heterochromatin (Peters et al. 2003). It was recently reported that SETDB1 could play an important role in establishing and/or maintaining H3K9me3, H3K27me3, and DNA methylation at some retrotransposon loci and also function as an essential regulator in controlling proviral gene expression prior to the onset of de novo DNA methylation in the prenatal germline (Liu et al. 2014). Our results indicated the existence of PRC2-dependent deposition of H3K27me3 at SETDB1 solo peak regions, and we propose that SETDB1 acts in coordination with PRC2 to suppress developmental regulators, which may also contribute to the transition of the facultative heterochromatin to the constitutive heterochromatin, a process important for the cells to become terminally differentiated during development. Our current findings may shed light on a novel mechanism underlying stem cell pluripotency and differentiation regulation.
Methods
Cell culture
Conditional Setdb1 ablation mouse embryonic stem cells (iKO ES cells) were generated as previously described (Lohmann et al. 2010). Both the iKO and the wild-type D3 ES cells (purchased from ATCC) were cultured feeder-free in 60-mm or 100-mm tissue culture dishes (Corning) at 37°C with 5% CO2. The culture surface was precoated with 0.1% (w/v) gelatin (Sigma) in PBS. The ES cell culture medium consists of Dulbecco's modified Eagle's medium (Gibco) supplemented with 15% fetal bovine serum (Invitrogen), 1 mM glutamine, 0.1 mM β-mercaptoethanol, 0.1 mM nonessential amino acid, 50 U/mL penicillin plus 50 µg/mL streptomycin, and 1000 units ESGRO (LIF; Millipore). To induce the deletion of Setdb1, cells were treated with 2.5–5 μM Tam (Sigma), with DMSO used as the solvent control.
Mouse ES cell–derived neural differentiation
iKO ES cells were pretreated with 2.5 μM Tam (Sigma) for 3 d to induce Setdb1 deletion. Pretreated ES cells were trypsinized and dispersed into single-cell suspension for embryoid body (EB) culture in 60- or 100-mm ultralow adhesion dishes (Corning). Cells were plated at a concentration of 2.5 × 104 cells/mL in mouse ES cells medium without ESGRO (LIF). For in vitro induced neural differentiation, EB cultures were switched to neural precursor cell culture medium (NP medium) on day 3. Serum-free NP medium contains DMEM/F12 (Invitrogen), N2 (1%) and B27 (2%; Invitrogen), bFGF (20 ng/mL; Invitrogen), and EGF (20 ng/mL; R&D). After 5 more days in serum-free culture, differentiated EBs were fixed in 4% paraformaldehyde for 15 min at room temperature and processed for immunofluorescence.
Immunofluorescence
Immunofluorescence was performed following standard procedures. Briefly, cells were permeabilized for 10 min in 0.2% Triton X-100 in PBS after fixation. Then cells were blocked with 2.5% BSA in PBS for 30 min at room temperature and incubated with primary antibody overnight at 4°C. Primary antibodies used in the studies were NESTIN (1:100; Millipore, no. IHCR1006-6), TUJ1 (1:200; Sigma, no. T2200), and H3K9me3 (1:500; Active Motif, no. 39161). After washing, cells were incubated with the appropriate secondary antibodies conjugated with Alexa546 (1:1000; Invitrogen, no. A11035) or Alexa488 (1:1000; Invitrogen, no. A21202) for 1 h at room temperature. Cell nuclei were labeled by DAPI staining (0.5 μg/mL; Sigma). Cells were then washed in PBS and mounted for examination under a fluorescence microscope.
Coimmunoprecipitation
Co-IP experimental procedures were performed following the protocol of nuclear complex co-IP kit (Active Motif, no. 54001). The nuclear complex was collected for IP using antibodies against SETDB1 (Santa Cruz, no. 66884), EZH2 (Cell Signaling, no. 3147), and SUZ12 (Cell Signaling, no. 3737). Post-IP protein G beads were washed three times with 1× wash buffer, and proteins were eluted with 2× SDS-loading buffer. Samples were then incubated for 10 min at 99°C before being loaded for SDS-PAGE.
In vitro methylation assay
SAM was purchased from USB. The mononucleosome was prepared essentially according to the protocol described by Luger et al. (1999). The H3K27 methylated modified nucleosome was produced according to the protocol described by Simon et al. (2007). Briefly, histone H2A, H2B, H3, and H4 was expressed, refolded, and purified, respectively. Core histone was then reconstituted and purified to homogeneity by size-exclusion chromatography. The 147-bp wisdom 601 DNA was prepared and purified by standard protocol. The nucleosome was then reconstituted and further purified by gel filtration to homogeneity. The quality of the nucleosomes was assured by LC/MS analysis, as well as histone methylation assessment using several histone HMTs on specific histone marks. For the in vitro methylation assay, 10 µM SAM and 0.4 µM recombinant nucleosome were first mixed with 2 µM EZH2 (in a five-member complex containing EZH2, EED, SUZ12, RBBP4, and AEBP2) in the assay buffer containing 20 mM Tris (pH 8), 0.01% Tween-20, 10 mM MgCl2, and 1 mM DTT. The reactions were allowed to proceed at room temperature for 4 h before 0.2 µM SETDB1 was added to the reaction for H3K9 methylation. After 4 more hours of incubation at room temperature, the reaction was stopped for analysis by Western blot. For the peptide-based assay, synthetic peptides (1 µM, Histone H3, 1-84aa) with or without K27 trimethyated chemically were incubated with 2 µM SETDB1 only under the same reaction conditions as the nucleosome assay. The quality of the peptide with or without chemically modified histone methylation was confirmed by LC/MS analysis prior to the reaction.
Western blot analysis
Samples were collected directly in 1× NuPAGE LDS sample buffer with 1× sample reducing buffer (Invitrogen) and denatured for 5 min at 95°C before being centrifuged for 5 min at 13,200 rpm. Supernatants were electrophoresed on a 4%–12% Tris-HCl gel and transferred to nitrocellulose membrane (Invitrogen). After blocking with superblock T20 blocking buffer (Thermo Scientific), the membrane was incubated with primary antibody overnight at 4°C and, subsequently, with HRP-conjugated secondary antibody (1 h at room temperature). The signal was detected using ECL (Pierce). The following primary antibodies were used: anti-SETDB1 (Santa Cruz, no. 66884; 1:1000), anti-Histone 3 (Cell Signaling, no. 9715; 1:1000), anti-H3K9me3 (Active Motif, no. 39161; 1:1000), anti-H3K27me2 (Cell Signaling, no. 9728; 1:1000), anti-H3K27me3 (Millipore, 07-449; 1:1000), anti-EZH2 (Cell Signaling, no. 3147; 1:1000), anti-SUZ12 (Cell Signaling, no. 3737; 1:1000), and anti-GAPDH (Santa Cruz, no. 137179; 1:10000).
RNA isolation and real-time RT-PCR
RNA was extracted using RNeasy mini kit (Qiagen). RNA quality was confirmed by Nanodrop. Real-time RT-PCR analysis was performed on an ABI prism 7900 sequence detection system using the SYBR green PCR master mix (Applied Biosystems). The relative expression for each gene was normalized against Gapdh. The primer sequences are listed in Supplemental Table S2.
Chromatin immunoprecipitation
Mouse Setdb1 iKO-ES cells were treated with Tam for 3 d. Cells were then treated with fresh culture medium containing 1% formaldehyde for 10 min and washed twice with ice-cold PBS containing protease inhibitors. Cell pellets were resuspended in SDS lysis buffer also containing protease inhibitors (200 µL lysis buffer for every 1 × 106 cells) and incubated on ice for 10 min. Cell lysate was sonicated (15 W for 10 sec, six times) to shear DNA to lengths between 100 and 500 bp. Subsequently, ChIP was performed according to the ChIP assay kit (Millipore17-295) instructions using antibodies against SETDB1 (Santa Cruz, no. 66884), EZH2 (Millipore, no. 17-662), H3K27me3 (Millipore, no. 07-449), and H3K9me3 (Active Motif, no. 39161). Eluted DNA was used for PCR, qPCR, or deep sequencing. For ChIP-qPCR analysis, the relative binding level of each gene was normalized against input. Primer sequences are listed in Supplemental Table S2. For ChIP-seq libraries, 10 ng of input chromatin DNA or ChIP DNA was processed using the ChIP-seq sample prep kit (Illumina). Gel-purified ChIP-seq library DNA was further purified by phenol-chloroform extraction and ethanol precipitation and was processed for cluster generation, 15-cycle sequencing, and sequence analysis using Illumina HiSeq. The summary of generated ChIP-seq data sets is listed in Supplemental Table S3.
Bioinformatics analyses
Analysis of ChIP-seq data sets
ChIP-seq reads were aligned to mouse genome (mm9) using Bowtie (version 0.12.7; parameter -m 1) (Langmead et al. 2009), and only uniquely mapped reads (or read pairs) were kept. Peak calling was performed using MACS2, a new version of MACS (Zhang et al. 2008), with a q-value cutoff of 0.01. If there were more than one read mapped to the same location in the Bowtie-generated BAM files, only one was kept in the peak calling process. An additional parameter, “broad,” was set for wide peak detection of histone marks H3K9me3 and H3K27me3. SETDB1 ChIP-seq data were derived from GEO (GSE17642) (Yuan et al. 2009), and 5882 peaks were detected. A cis-regulatory element annotation system (CEAS) was used to get genome-wide peak distribution. The overlap ratio was defined by BEDTools (Quinlan and Hall 2010), and two binding sites were considered to overlap when they had at least 1 bp in common. For each ChIP-seq data set, a bigWig file was generated by MACS2 (with parameter -B; the values in the bigWig file were the extended reads pileup at every bp). In this study, the bigWig file was used to indicate the signals of a given histone mark (or TF) across the genome.
In this study, four types of metrics were applied for ChIP-seq data set quality control. First, as both H3K9me3 and H3K27me3 are repressive marks, their ChIP-seq signals should be anti-correlated with gene expression level. One public RNA-seq data set in mouse ES cells (GSM1499144) was used to divide the genes into three categories based on their FPKM values: low (0–10), moderate (10–100), and high (>100). For each H3K9me3 or H3K27me3 ChIP-seq biological replicate generated in this study, the average signal profiles across gene bodies for each gene category were drawn, as shown in Supplemental Figure S14, A through H. Second, the distribution of fold enrichment scores for each ChIP-seq data set was displayed in updated Supplemental Figure S14, I through J. Third, the library complexity for each ChIP-seq data set was calculated using the definition in the study by Landt et al. (2012), and the values were shown in Supplemental Table S3. Last, the signal correlations between our ChIP-seq data sets and public H3K9me3 or H3K27me3 ChIP-seq data sets in mouse ES cells were calculated, as shown in Supplemental Figure S15.
Measurement of the level of cobinding between SETDB1 and other factors
In total, 43 TFs (Supplemental Table S1) were selected for assaying their cobinding with SETDB1 loci. For each factor, we defined the signal ratios at every SETDB1 peak as their total reads counts first divided by the corresponding lengths of peak region and then by the average value for all SETDB1 peaks.
H3K27me3 variation for EZH2 before and after Setdb1 iKO
EZH2 peaks (Supplemental Table S1) were also detected by MACS2 with the same parameters mentioned above. Those EZH2 peaks with enriched H3K27me3 signal before Tam treatment were then included in this analysis. The resulting peaks were further divided into two categories based on the status of whether SETDB1 overlapped or not. Every single site was extended to 10 kb, and the ratio of per base sequence coverage for mapped H3K27me3 reads was calculated based on H3K27me3 profiles upon Setdb1 deletion and then plotted against the respective peak groups. Two-sided Welch's t-test was performed for the change of H3K27me3 between the two groups.
GO enrichment analysis
SETDB1 target genes were defined as genes (RefSeq ID) having TSSs within 5 kb of a SETDB1 peak. All GO enrichment analyses were performed by the functional annotation tool from DAVID (http://david.abcc.ncifcrf.gov/), and all P-values were generated by a modified Fisher's exact test and then corrected by Benjamini-Hochberg correction.
Data access
The ChIP-seq data from this study have been submitted to the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE62666.
Supplementary Material
Acknowledgments
We thank Thomas Jenuwein for the H3K9me3 antibody for our initial study. We also thank Drs. Song Shi, Shannon Chuai, and Haiyan Xu for insightful discussion and suggestions. We thank Dr. Tengfei Zhang for critical reading of the paper. We thank Drs. Teddy Yang and Kehao Zhao for help with making the nucleosomes and Drs. Ling Li and Justin Gu for the help with the in vitro methylation assay. This project is sponsored by China Novartis Institutes for BioMedical Research. The fourteenth author (Y.Z.) is partially supported by the National Natural Science Foundation of China (31371288, 31322031) and the Shanghai Rising-Star Program (13QH1402200).
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.177576.114.
References
- Bilodeau S, Kagey MH, Frampton GM, Rahl PB, Young RA. 2009. SetDB1 contributes to repression of genes encoding developmental regulators and maintenance of ES cell state. Genes Dev 23: 2484–2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binda O, LeRoy G, Bua DJ, Garcia BA, Gozani O, Richard S. 2010. Trimethylation of histone H3 lysine 4 impairs methylation of histone H3 lysine 9: regulation of lysine methyltransferases by physical interaction with their substrates. Epigenetics 5: 767–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, et al. 2005. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 122: 947–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al. 2006. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 441: 349–353. [DOI] [PubMed] [Google Scholar]
- Chamberlain SJ, Yee D, Magnuson T. 2008. Polycomb repressive complex 2 is dispensable for maintenance of embryonic stem cell pluripotency. Stem Cells 26: 1496–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Daley GQ. 2008. Molecular basis of pluripotency. Hum Mol Genet 17: R23–R27. [DOI] [PubMed] [Google Scholar]
- Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, et al. 2008. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell 133: 1106–1117. [DOI] [PubMed] [Google Scholar]
- Dodge JE, Kang YK, Beppu H, Lei H, Li E. 2004. Histone H3-K9 methyltransferase ESET is essential for early development. Mol Cell Biol 24: 2478–2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herz HM, Shilatifard A. 2010. The JARID2–PRC2 duality. Genes Dev 24: 857–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong SH, Rampalli S, Lee JB, McNicol J, Collins T, Draper JS, Bhatia M. 2011. Cell fate potential of human pluripotent stem cells is encoded by histone modifications. Cell Stem Cell 9: 24–36. [DOI] [PubMed] [Google Scholar]
- Karimi MM, Goyal P, Maksakova IA, Bilenky M, Leung D, Tang JX, Shinkai Y, Mager DL, Jones S, Hirst M, et al. 2011. DNA methylation and SETDB1/H3K9me3 regulate predominantly distinct sets of genes, retroelements, and chimeric transcripts in mESCs. Cell Stem Cell 8: 676–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Chu J, Shen X, Wang J, Orkin SH. 2008. An extended transcriptional network for pluripotency of embryonic stem cells. Cell 132: 1049–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landt SG, Marinov GK, Kundaje A, Kheradpour P, Pauli F, Batzoglou S, Bernstein BE, Bickel P, Brown JB, Cayting P, et al. 2012. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res 22: 1813–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg S. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, et al. 2006. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell 125: 301–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JS, Smith E, Shilatifard A. 2010. The language of histone crosstalk. Cell 142: 682–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Brind'Amour J, Karimi MM, Shirane K, Bogutz A, Lefebvre L, Sasaki H, Shinkai Y, Lorincz MC. 2014. Setdb1 is required for germline development and silencing of H3K9me3-marked endogenous retroviruses in primordial germ cells. Genes Dev 28: 2041–2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohmann F, Loureiro J, Su H, Fang Q, Lei H, Lewis T, Yang Y, Labow M, Li E, Chen T, et al. 2010. KMT1E mediated H3K9 methylation is required for the maintenance of embryonic stem cells by repressing trophectoderm differentiation. Stem Cells 28: 201–212. [DOI] [PubMed] [Google Scholar]
- Lohse B, Helgstrand C, Kristensen JBL, Leurs U, Cloos PAC, Kristensen JL, Clausen RP. 2013. Posttranslational modifications of the histone 3 tail and their impact on the activity of histone lysine demethylases in vitro. PLoS One 8: e67653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K, Rechsteiner TJ, Richmond TJ. 1999. Expression and purification of recombinant histones and nucleosome reconstitution. Methods Mol Biol 119: 1–16. [DOI] [PubMed] [Google Scholar]
- Margueron R, Reinberg D. 2011. The Polycomb complex PRC2 and its mark in life. Nature 469: 343–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui T, Leung D, Miyashita H, Maksakova IA, Miyachi H, Kimura H, Tachibana M, Lorincz MC, Shinkai Y. 2010. Proviral silencing in embryonic stem cells requires the histone methyltransferase ESET. Nature 464: 927–931. [DOI] [PubMed] [Google Scholar]
- Mendenhall EM, Koche RP, Truong T, Zhou VW, Issac B, Chi AS, Ku M, Bernstein BE. 2010. GC-rich sequence elements recruit PRC2 in mammalian ES cells. PLoS Genet 6: e1001244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mysliwiec MR, Carlson CD, Tietjen J, Hung H, Ansari AZ, Lee Y. 2012. Jarid2 (Jumonji, AT rich interactive domain 2) regulates NOTCH1 expression via histone modification in the developing heart. J Biol Chem 287: 1235–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama A, Xin L, Sharov AA, Thomas M, Mowrer G, Meyers E, Piao Y, Mehta S, Yee S, Nakatake Y, et al. 2009. Uncovering early response of gene regulatory networks in ESCs by systematic induction of transcription factors. Cell Stem Cell 5: 420–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini D, Bracken AP, Hansen JB, Capillo M, Helin K. 2007. The polycomb group protein Suz12 is required for embryonic stem cell differentiation. Mol Cell Biol 27: 3769–3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng JC, Valouev A, Swigut T, Zhang J, Zhao Y, Sidow A, Wysocka J. 2009. Jarid2/Jumonji coordinates control of PRC2 enzymatic activity and target gene occupancy in pluripotent cells. Cell 139: 1290–1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters AH, Kubicek S, Mechtler K, O'Sullivan RJ, Derijck AA, Perez-Burgos L, Kohlmaier A, Opravil S, Tachibana M, Shinkai Y, et al. 2003. Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol Cell 12: 1577–1589. [DOI] [PubMed] [Google Scholar]
- Plath K, Fang J, Mlynarczyk-Evans SK, Cao R, Worringer KA, Wang H, de la Cruz CC, Otte AP, Panning B, Zhang Y. 2003. Role of histone H3 lysine 27 methylation in X inactivation. Science 300: 131–135. [DOI] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapicavoli NA, Poth EM, Zhu H, Blackshaw S. 2011. The long noncoding RNA Six3OS acts in trans to regulate retinal development by modulating Six3 activity. Neural Dev 6: 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, et al. 2007. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129: 1311–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitges FW, Prusty AB, Faty M, Stutzer A, Lingaraju GM, Aiwazian J, Sack R, Hess D, Li L, Zhou S, et al. 2011. Histone methylation by PRC2 is inhibited by active chromatin marks. Mol Cell 42: 330–341. [DOI] [PubMed] [Google Scholar]
- Schultz DC, Ayyanathan K, Negorev D, Maul GG, Rauscher FJ III. 2002. SETDB1: a novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev 16: 919–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon MD, Chu F, Racki LR, de la Cruz CC, Burlingame AL, Panning B, Narlikar GJ, Shokat KM. 2007. The site-specific installation of methyl-lysine analogs into recombinant histones. Cell 128: 1003–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surani MA, Hayashi K, Hajkova P. 2007. Genetic and epigenetic regulators of pluripotency. Cell 128: 747–762. [DOI] [PubMed] [Google Scholar]
- Tan SL, Nishi M, Ohtsuka T, Matsui T, Takemoto K, Kamio-Miura A, Aburatani H, Shinkai Y, Kageyama R. 2012. Essential roles of the histone methyltransferase ESET in the epigenetic control of neural progenitor cells during development. Development 139: 3806–3816. [DOI] [PubMed] [Google Scholar]
- Wang H, Cao R, Xia L, Erdjument-Bromage H, Borchers C, Tempst P, Zhang Y. 2001. Purification and functional characterization of a histone H3-lysine 4-specific methyltransferase. Mol Cell 8: 1207–1217. [DOI] [PubMed] [Google Scholar]
- Wang X, Pan L, Wang S, Zhou J, McDowell W, Park J, Haug J, Staehling K, Tang H, Xie T. 2011. Histone H3K9 trimethylase Eggless controls germline stem cell maintenance and differentiation. PLoS Genet 7: e1002426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K, Wu ZJ, Groner AC, He HH, Cai C, Lis RT, Wu X, Stack EC, Loda M, Liu T, et al. 2012. EZH2 oncogenic activity in castration-resistant prostate cancer cells is Polycomb-independent. Science 338: 1465–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap DB, Chu J, Berg T, Schapira M, Cheng SW, Moradian A, Morin RD, Mungall AJ, Meissner B, Boyle M, et al. 2011. Somatic mutations at EZH2 Y641 act dominantly through a mechanism of selectively altered PRC2 catalytic activity, to increase H3K27 trimethylation. Blood 117: 2451–2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan P, Han J, Guo G, Orlov YL, Huss M, Loh YH, Yaw LP, Robson P, Lim B, Ng HH. 2009. Eset partners with Oct4 to restrict extraembryonic trophoblast lineage potential in embryonic stem cells. Genes Dev 23: 2507–2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan W, Xu M, Huang C, Liu N, Chen S, Zhu B. 2011. H3K36 methylation antagonizes PRC2-mediated H3K27 methylation. J Biol Chem 286: 7983–7989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. 2008. Model-based analysis of ChIP-Seq (MACS). Genome Biol 9: R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.