Abstract
Long noncoding (lnc)RNAs have recently emerged as key regulators of gene expression. Here, we performed high-depth poly(A)+ RNA sequencing across multiple clonal populations of mouse embryonic stem cells (ESCs) and neural progenitor cells (NPCs) to comprehensively identify differentially regulated lncRNAs. We establish a biologically robust profile of lncRNA expression in these two cell types and further confirm that the majority of these lncRNAs are enriched in the nucleus. Applying weighted gene coexpression network analysis, we define a group of lncRNAs that are tightly associated with the pluripotent state of ESCs. Among these, we show that acute depletion of Platr14 using antisense oligonucleotides impacts the differentiation- and development-associated gene expression program of ESCs. Furthermore, we demonstrate that Firre, a lncRNA highly enriched in the nucleoplasm and previously reported to mediate chromosomal contacts in ESCs, controls a network of genes related to RNA processing. Together, we provide a comprehensive, up-to-date, and high resolution compilation of lncRNA expression in ESCs and NPCs and show that nuclear lncRNAs are tightly integrated into the regulation of ESC gene expression.
Landmark cDNA cloning efforts and transcriptional interrogation using tiling arrays, together with more recent genome-wide RNA sequencing and chromatin state analyses, have led to the discovery that the majority of the mammalian genome is transcribed to yield tens of thousands of RNA products (Kapranov et al. 2002, 2007; Okazaki et al. 2002; Bertone et al. 2004; Carninci et al. 2005; Cheng et al. 2005; Katayama et al. 2005; Guttman et al. 2009, 2010; Khalil et al. 2009; Kim et al. 2010; Djebali et al. 2012; The ENCODE Project Consortium 2012; Hangauer et al. 2013). A significant subset of these RNAs are spliced, yielding transcripts >200 nt in length, but without apparent protein coding potential, and are hence referred to as long noncoding RNAs (lncRNAs). A small, but steadily increasing fraction of these lncRNAs has been studied on the molecular level (Quek et al. 2015). These lncRNAs have been reported to act through a staggering variety of mechanisms mediating a large number of biological processes (for review, see Wilusz et al. 2009; Rinn and Chang 2012; Fatica and Bozzoni 2013; Bergmann and Spector 2014). These examples also indicate that a large fraction of lncRNAs function in the nucleus, regulating local or global chromatin state, gene expression, and nuclear structures (for review, see Lee 2012; Bergmann and Spector 2014).
Many lncRNAs are developmentally regulated and expressed in a cell-type–specific manner (Dinger et al. 2008; Mercer et al. 2010; Derrien et al. 2012; Hu et al. 2013). Furthermore, a growing number of individual lncRNA candidates have been shown to be required for cellular differentiation and tissue development (Ulitsky et al. 2011; Grote et al. 2013; Kretz et al. 2013; Ng et al. 2013; Lin et al. 2014; Zhao et al. 2014). Conversely, several lncRNAs have been implicated in maintaining the pluripotent state of embryonic stem cells (ESCs) (Sheik Mohamed et al. 2010; Guttman et al. 2011; Lin et al. 2014). In light of the large number of lncRNAs yet to be characterized, it is to be expected that many more of these transcripts will be linked to critical differentiation processes and will ultimately help to refine our understanding of normal development and disease.
Here, we set out to provide a comprehensive analysis of the (lncRNA) transcriptome of mouse ESCs and neural progenitor cells (NPCs) in the context of the latest GENCODE M3 annotation (including more than 5000 mouse lncRNAs). We identify a large number of presently uncharacterized lncRNAs tightly associated with the pluripotency gene expression profile of ESCs and address the immediate functional consequences on ESC gene expression following independent depletion of two abundant, nuclear-enriched lncRNAs. These analyses uncover a broad but gene set-specific requirement of lncRNAs for the maintenance of the ESC transcriptome.
Results
Transcriptome analysis of mouse ESCs and NPCs
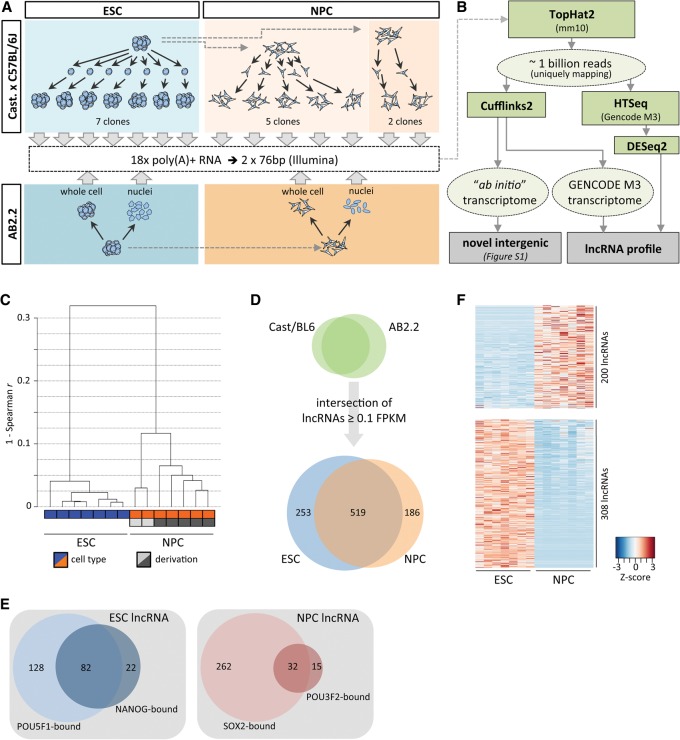
As a basis for our analysis of mouse ESC and NPC transcriptomes, we utilized high quality, poly(A)-selected RNA isolated from seven single cell-derived ESC clones in the Castaneous/C57BL/6J hybrid background (hereafter referred to as “Cast/BL6,” clones 1–7). Similarly, we included poly(A)+ RNA obtained from seven single-cell-derived mouse NPC clones that were differentiated from Cast/BL6 ESCs in two separate derivations (clones 1–5 and clones 6–7, respectively) (Fig. 1A; Eckersley-Maslin et al. 2014). We also derived NPCs from AB2.2 ESCs (129S5/SvEvBrd) and isolated RNA both from whole-cell and nuclear fractions to estimate nuclear enrichment of a given gene product (see below). Together, these biological replicates control for gene expression differences arising during routine cell culture and in vitro differentiation and capture differences based on the genetic background of each cell type. We obtained a total of approximately one billion uniquely mapping paired-end reads (Supplemental Table S1), representing to our knowledge the deepest RNA sequencing data set for ESCs and NPCs available to date.
Figure 1.
Transcriptome characterization in ESCs and NPCs. (A) Schematic of RNA sources for high-throughput RNA sequencing. (B) Pipeline for ESC and NPC transcriptome analysis. (C) Average linkage clustering dendrogram based on Spearman rank correlation of mRNA and lncRNA expression levels. (D) LncRNAs in the intersection of AB2.2 and Cast/BL6 backgrounds detected in ESCs and NPCs. (E) Analysis of POU5F1, NANOG, SOX2, and POU3F2 binding within 2.5 kb of the transcription start site of expressed lncRNAs (ChIP-seq data reanalyzed from Marson et al. 2008; Lodato et al. 2013). (F) Heatmap (gene-wise Z-score) of lncRNAs in Cast/BL6 clones determined as differentially expressed between ESCs and NPCs (FDR < 0.01).
We subsequently used Cufflinks2 (Trapnell et al. 2010) using the recently released GENCODE M3 annotation (April 2014, Ensembl v.76; www.ensembl.org) to provide a standardized and up-to-date analysis of gene expression (Fig. 1B). As expected, correlation of gene expression between ESCs and NPCs was considerably weaker than the correlation within each cell type. The strongest correlations were found between ESCs (Supplemental Fig. S1A,B), resulting in a tight clustering of all seven clones (Fig. 1C) (Spearman r: minimum about 0.88 for NPCs versus about 0.96 for ESCs). Notably, the two groups of NPCs obtained from separate derivations showed distinct differences in correlation of their RNA levels, resulting in a marked separation of the cluster comprising clones 1–5 (derivation 1) from clones 6 and 7 (derivation 2) (Fig. 1C). Somewhat surprisingly, the pair-wise correlation coefficients of the five NPC clones obtained from the same parent population were on average lower than those of ESCs (Supplemental Fig. S1B). These data suggest that gene expression or post-transcriptional regulation of steady-state RNA levels may be more tightly controlled in ESCs than in NPCs.
Profile of long noncoding RNA expression in ESCs and NPCs
Next, we characterized the expression of putative “intergenic” lncRNAs within the ESC to NPC differentiation paradigm. Of the 3506 genes annotated in GENCODE M3 as “lincRNA” or “processed transcript” (referred to as “lncRNA” for the remainder of this study), we detected 1433 expressed at ≥0.1 FPKM in the union of our ESC or NPC data sets. Of these, 111 genes overlapped annotated small RNAs (miRNA, snRNA, or snoRNA) on the same strand and were thus removed as putative small RNA hosts. To increase confidence in the potential relevance of a lncRNA within the given cell type, we only considered those common to both genetic backgrounds (Fig. 1D). This set of 958 lncRNAs contained 74 of the 226 putative noncoding genes previously compiled for characterization in ESCs by Guttman et al. (2011) (Supplemental Table S2; Supplemental Methods) and represents a collection of well over 800 lncRNAs largely uncharacterized on the molecular level. We note that although the majority of the remaining genes described by Guttman et al. (2011) are also detected here, these are not included due to differing biotype annotations (including small RNA host genes, pseudogenes, and antisense transcripts).
We also used ab initio transcript assembly using Cufflinks (Fig. 1B) to assess the completeness of the GENCODE annotation with respect to intergenic lncRNAs expressed in ESCs and NPCs. This analysis revealed only 34 (ESC) and 18 (NPC) additional novel high-confidence intergenic transcription units producing spliced transcripts (Supplemental Fig. S2A,B; Supplemental Information). The majority of these RNAs are detected at low levels only, with ∼32% lacking apparent protein-coding potential (Supplemental Fig. S2C,D), demonstrating that our analysis essentially provides a comprehensive resource with respect to gene-level expression of intergenic lncRNAs in ESCs and NPCs.
As reported previously (Guttman et al. 2010; Derrien et al. 2012), transcript levels of lncRNAs were considerably lower than those of mRNAs (Supplemental Fig. S3A,B). In contrast to low- or nonexpressed lncRNAs, a large fraction of the transcription start sites of more abundantly transcribed (>1 FPKM) lncRNAs were enriched for trimethylated histone H3 lysine 4 (H3K4me3), a modification associated with active RNA Pol II promoters (Supplemental Fig. S3C,D; Supplemental Table S2; Mikkelsen et al. 2007). We also reanalyzed transcription factor ChIP-seq data from ESCs and NPCs (Marson et al. 2008; Lodato et al. 2013). We detected binding peaks of the pluripotency master-regulators POU5F1 (also known as OCT4) or NANOG within 2.5 kb of the transcription start sites of 232 of 772 (30%) lncRNAs expressed in ESCs. Similarly, in NPCs, 294 of 705 (42%) detected lncRNAs were associated with SOX2. Expression of these lncRNAs may thus be regulated by these core transcription factors. Of the SOX2-occupied fraction, 32 NPC lncRNAs were also bound by POU3F2 (previously known as BRN2), a neurogenic Pou-family member recently suggested to associate with distal enhancer elements in NPCs (Fig. 1E; Supplemental Table S2; Lodato et al. 2013).
We further note that the transcription start site of ∼8.2% of lncRNAs expressed in ESCs fell within long terminal repeat (LTR) elements, a proportion significantly larger than the ∼4.4% in NPCs (P < 0.002; one-sided χ2) (Supplemental Fig. S3E). In contrast, no difference was observed for the small fractions of lncRNAs originating from within long interspersed elements (LINE; ∼2.4% and ∼1.8% in ESCs and NPCs, respectively).
Of the 958 lncRNAs identified, 508 (53%) displayed a significant change in expression level upon differentiation of ESCs to NPCs (DESeq2, FDR < 0.01), with 439 lncRNAs being exclusively detected in either cell type (Fig. 1B,D,F; Supplemental Table S2). The fraction of lncRNAs expressed exclusively in one cell type was considerably larger than that of mRNAs detected at the same expression cutoff (Supplemental Fig. S3F), consistent with the notion that lncRNAs are more cell- and tissue-specific in expression than protein-coding genes (Cabili et al. 2011; Derrien et al. 2012). To independently assess and validate expression changes determined by RNA-seq, we selected 38 genes (35 noncoding, three protein-coding) (Supplemental Table S3) with a wide range of expression levels and fold changes and performed real-time RT-PCR in ESCs and NPCs of both genetic backgrounds. Count-based and FPKM-calculated log2-fold changes (DESeq2 and Cufflinks, respectively) correlated very well with one another (Pearson r 0.99, data not shown). Both RNA-seq-based methods also showed strong correlation with the log2-fold changes determined by real-time RT-PCR (Pearson r between 0.86 and 0.95) (Supplemental Fig. S4A–C).
Global identification of nuclear-enriched lncRNAs
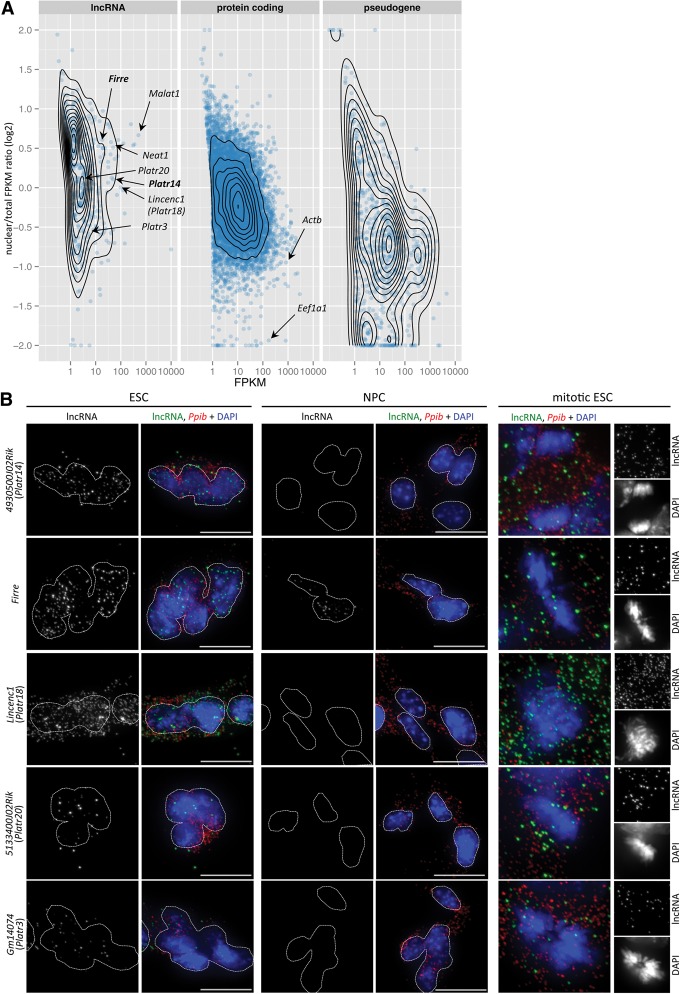
Many of the lncRNAs for which molecular data has been acquired to date are found to exert their function within the nucleus (for review, see Bergmann and Spector 2014; Rinn and Guttman 2014), and recent data suggest that a large fraction of human lncRNAs are enriched in the nuclear compartment (Derrien et al. 2012; Djebali et al. 2012). We thus used nuclear fractions from ESCs and NPCs to globally assess the relative distribution of gene products in these cell types. As expected, the majority of mRNAs were enriched in the cytoplasm (Fig. 2A; Supplemental Fig. S5A). In contrast, the majority of lncRNAs show preferential enrichment in the nucleus, consistent with potential roles of these transcripts in this cellular compartment. Nuclear-enriched lncRNAs include transcripts known to be retained in the nucleus, such as Malat1 and Neat1 (Clemson et al. 2009). We also found the lncRNA Firre to display moderately high expression and pronounced nuclear localization in both ESCs and NPCs. Firre, which escapes X inactivation in female mouse ESCs (Yang et al. 2010), was recently reported to show exclusive focal enrichment at its transcription start site (Hacisuleyman et al. 2014). Compared to mRNAs and lncRNAs, pseudogene transcripts displayed the strongest shift toward cytoplasmic localization (Fig. 2A; Supplemental Fig. S5A), consistent with proposed roles of some pseudogene transcripts acting as miRNA “sponges” (Poliseno et al. 2010).
Figure 2.
Identification of nuclear-enriched lncRNAs. (A) Nuclear to total log2 FPKM ratio by annotated gene biotype (Ensembl v.76) in AB2.2 ESCs plotted against FPKM values obtained from total cell RNA-seq. Only genes with expression >1 FPKM in either nuclear or total fraction are plotted. (B) Single-molecule RNA FISH in AB2.2 ESCs and NPCs using probe pairs specific for Platr14, Firre, and other indicated lncRNAs (green). Housekeeping Ppib mRNA (red) is used as a control. Images represent maximum intensity projections. Dotted lines demarcate the nuclear outline of individual cells or ESC colonies. (Scale bar) 10 µm. Mitotic images are enlarged twofold.
To confirm nuclear localization and to investigate expression of lncRNAs at the single cell level, we selected 11 lncRNAs, including Firre, Tug1 (Young et al. 2005) and lncRNAs Platr2, -14, -15, -19 and -20 (see below) and performed single-molecule RNA FISH in ESCs and NPCs (see Methods). Qualitative and quantitative analysis of RNA FISH hybridization strongly supported both nuclear enrichment and cell-type expression patterns inferred from RNA-seq analysis (Fig. 2B; Supplemental Fig. S5B,C). When transcript levels determined by RNA-seq were compared to the average number of RNA FISH hybridization foci per cell, the Pearson correlation coefficient was approximately 0.58 (Supplemental Fig. S5B). Notably, we detected on average 139 and 62 Gm11974 lncRNA hybridization foci in ESCs and NPCs, respectively (Supplemental Fig. S5C), corresponding to an approximately twofold difference that is tightly reflected by the corresponding FPKM values (44 and 19, approximately a twofold difference). Similarly, an approximate fourfold difference in Firre hybridization foci between ESCs and NPCs (40 and 10 foci, respectively) (Fig. 2B) was closely reflected by an approximate threefold change in FPKM values (24 and 9 in ESCs and NPCs, respectively). All the lncRNAs examined were also detectable in mitotic cells (Fig. 2B; Supplemental Fig. S5C), indicating stability of these transcripts following breakdown of the nuclear envelope. Transcripts with high enrichment in the interphase nucleus may thus be subject to an active process restricting their localization to the nucleus via the association with nuclear proteins or chromatin.
Identification of lncRNAs associated with the ESC state
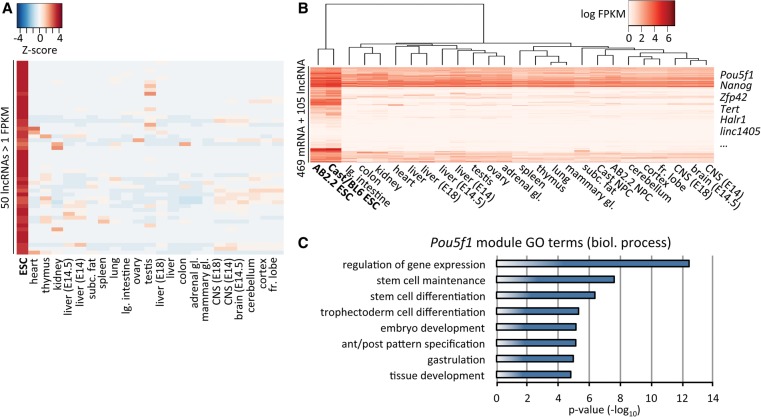
We were interested in identifying lncRNAs whose expression was strongly associated with the ESC state, thus potentially implicating them in the coregulation of self renewal or pluripotency-related networks. To this end, we analyzed raw poly(A)+ RNA-seq data from 22 mouse tissues (including fetal liver and neural tissues of embryonic origin) released by the ENCODE Project. Following identical processing, we created an FPKM-based expression matrix of the ENCODE data and our ESC and NPC data sets. Gene-level analysis of this matrix demonstrated that the majority of the 50 lncRNAs most specifically expressed in ESCs displayed exclusive expression in this cell type, with few also being appreciably detected in a specific tissue, rendering them useful candidates as developmental biomarkers (Fig. 3A).
Figure 3.
Identification of ESC-associated lncRNAs. (A) Heatmap (GeneWise Z-score) of the 50 lncRNAs most specifically (calculated based on Z-score) expressed in ESCs at ≥1 FPKM. ENCODE mouse tissue poly(A)+ RNA-seq data represent the average expression of two biological replicates. (B) Expression heatmap of the gene module containing Pou5f1 as determined by weighted gene coexpression analysis. (C) Gene ontology (GO) terms for biological process selected from the top 20 nonredundant terms enriched in the Pou5f1 gene module. Enrichment was ranked based on Fisher's exact test.
To more formally identify lncRNAs correlated with the expression of key pluripotency factors, we performed weighted gene coexpression network analysis (Langfelder and Horvath 2008). We collated modules of highly inter-connected genes using hierarchical clustering of a topological overlap matrix from a signed coexpression network (Methods). As a marker for undifferentiated pluripotent cells, we subsequently selected the module containing Pou5f1 for further investigation. As expected, the module's gene expression profile specifically clustered ESCs away from more differentiated tissues (Fig. 3B). Some of the most strongly enriched gene ontology terms revolved around stem cell maintenance and embryonic differentiation-associated biological processes (Fig. 3C). The Pou5f1 module included other prominent factors associated with ESC maintenance and early development, such as Nanog, Zfp42 (also known as Rex1), Dnmt3b, and Tert (Rogers et al. 1991; Armstrong et al. 2000; Watanabe et al. 2002; Chambers et al. 2003). Among the module's 574 genes were 105 lncRNAs (Supplemental Table S4). Of these, Halr1 (also known as linc-Hoxa1) was recently shown to act in cis to suppress Hoxa1 expression in mouse ESCs (Maamar et al. 2013). Another 10 lncRNAs were also found to be required for normal gene expression in ESCs (Guttman et al. 2011). To our knowledge, the remaining 93 lncRNAs are presently uncharacterized on the molecular level, highlighting the potential for identifying novel regulators of ESC biology within this set of genes (see below).
Platr14 is associated with maintenance of the ESC gene expression profile
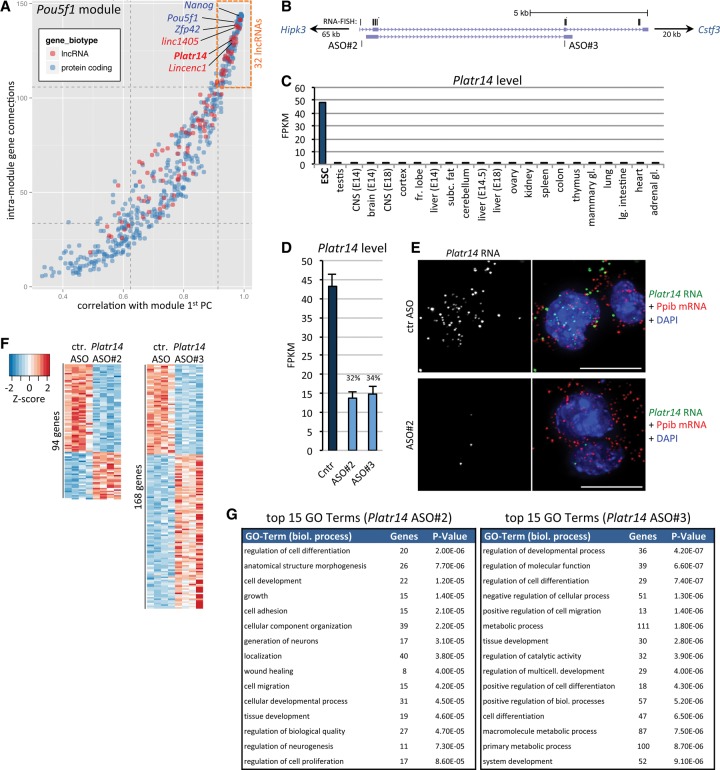
We went on to identify “high-profile” lncRNA candidates within the Pou5f1 module described above. We took into consideration a gene's number of connections with other genes in the module and further determined the correlation of a given gene's expression with the overall module gene expression (determined as the first principal component of the module). We reasoned that a gene would be relatively more important with increasing connectivity and module correlation. Consistently, Pou5f1, Nanog, and Zfp42 were among the highest-ranking genes (Fig. 4A).
Figure 4.
Platr14 lncRNA is functionally integrated in ESC gene expression regulation. (A) Connectivity between genes within the Pou5f1 module (see Fig. 3B) is plotted against the module's first principal component (module gene expression). Dotted lines correspond to first and third quartiles. (Red) lncRNA genes; (blue) mRNA genes. (B) Schematic of annotated Platr14 isoforms. Targeting sites of ASO#2 and ASO#3 are indicated. Black lines above the transcript model represent targeting sites of RNA FISH probes. For greater detail and cloned Platr14 cDNA, see Supplemental Figure S7. (C) Expression of Platr14 in AB2.2 ESCs and ENCODE tissues. (D) Expression level of Platr14 in ESCs 24 h after transfection with control ASO (Cntr) or two independent ASOs specific against Platr14 lncRNA. Data represent the mean and standard deviation of four biological replicates. (E) Single-molecule RNA FISH for Platr14 lncRNA (green) and Ppib housekeeping mRNA (red) in AB2.2 ESCs transfected with control ASO or Platr14 specific ASO. (Scale bar) 10 µm. (F) Heatmap (gene-wise Z-score) of genes significantly affected within 24 h of transfection of Platr14 specific ASOs (FDR < 0.05). (G) Top 15 nonredundant GO terms enriched in the set of differentially expressed genes for each ASO. Enrichment was ranked based on Fisher's exact test.
We identified 32 lncRNAs clustering tightly among the known pluripotency factors in the top quartiles of importance (Fig. 4A; Supplemental Table S5), suggesting their functional integration into the ESC gene expression program. We refer to these lncRNAs as Platr1 to -32 (pluripotency-associated transcript; ranked by relative module importance). Importantly, two of these lncRNAs were previously implicated in the maintenance of the ESC state: Platr11 (linc1405; Gm26975/ENSMUSG00000098161) was found to be required for Zfp42 expression (Guttman et al. 2011). Platr18 (Lincenc1; ENSMUSG00000078952) (Fig. 2A,B) was part of an early knockdown screen and is critically necessary for ESC colony formation (Ivanova et al. 2006). From the other 30 presently uncharacterized Platr lncRNAs, we selected Platr14 to further assess its potential role in ESC state. Platr14 (4930500J02Rik/ENSMUSG00000086454) (Fig. 4B) is specifically expressed in ESCs at a comparatively high expression level (49 and 101 FPKM in AB2.2 and Cast/BL6, respectively) (Fig. 4C; Supplemental Table S2). Localization of Platr14 lncRNA displayed pronounced nuclear enrichment (Figs. 2A,B, 4E), further supporting a regulatory role within ESC nuclei. Although we did not detect POU5F1 or NANOG binding sites in the vicinity of Platr14, we found its TSS to fall within an LTR. Using rapid amplification of cDNA ends (RACE) and RT-PCR, we confirmed the overall structure of the main Platr14 isoforms (ranging between ∼600 and ∼900 nt), splicing of which is markedly diverse both at the 5′ and 3′ end of the transcript (Supplemental Fig. S7A–C).
To determine if loss of Platr14 impacts global ESC gene expression, we treated AB2.2 ESCs with a control 2′-O-methoxyethyl gapmer antisense oligonucleotide (ASO) or two independent ASOs complementary to Platr14. These chemically modified ASOs act through RNase H-mediated degradation of their target and are extremely well suited for the knockdown of nuclear RNAs, including primary, unprocessed transcripts (Wheeler et al. 2012; Meng et al. 2015). We decided to analyze the effect of lncRNA depletion by RNA-seq 24 h after ASO transfection, for technical and biological reasons: (1) The majority of lncRNAs characterized to date regulate transcriptional or post-transcriptional RNA levels, rendering RNA-seq a first-choice method to investigate lncRNA function globally at high resolution and sensitivity; (2) in contrast to mRNAs, for which a knockdown phenotype is delayed due to a critical dependence on the half-life of the encoded protein, lncRNAs should exert an immediate phenotypic effect upon knockdown; (3) similarly, observed phenotypic changes should be a direct consequence of lncRNA loss, rather than secondary effects and thus allow one to more narrowly pinpoint potential target genes; and (4) extended culture following ASO introduction may cause a passive dilution of ASO molecules that results in the partial recovery of lncRNA levels, thus potentially “rescuing” earlier transient effects (JH Bergmann and DL Spector, unpubl.).
To enable a statistically robust analysis of differential expression at this early time point, in particular in light of the expected biological variability in gene expression between independent replicates (Supplemental Fig. S6), we performed ASO transfections in four biological replicates, sequenced high-quality poly(A)+ RNA at high depth, and considered only uniquely mapping reads (on average 240 million mapped reads per condition) (Supplemental Table S1). Each specific ASO resulted in a ∼70% reduction of Platr14 levels compared to a control ASO or mock-transfected cells (Fig. 4D,E). Within 24 h of Platr14 knockdown by either ASO#2 or ASO#3, we observed a significant impact on the expression of 94 and 168 genes, respectively (DESeq2, FDR < 0.05) (Fig. 4F). We note that these two ASOs have different preferences for individual Platr14 transcript isoforms (Fig. 4B; Supplemental Fig. S7) and may thus exert different biological activity. Comparison of RNA-seq knockdown data with potential off-targets based on ASO sequence complementary to all expressed primary and spliced ESC transcripts confirmed the high specificity of ASO action toward their cognate targets in vivo (Supplemental Fig. S8). Strikingly, a large fraction of the genes affected by loss of Platr14 in either knockdown data set were directly related to differentiation and tissue development (Fig. 4G). Importantly, the enrichment for these terms does not constitute a “default” gene response preference within ESCs, as ASOs targeting other transcripts show target-dependent gene expression changes and enrichment for different functional GO terms (see Firre knockdown below).
Together, these data confirm that Platr14 tightly associates with expression of key pluripotency genes and has a functional impact with respect to maintenance of the ESC gene expression program. Although beyond the scope of the present study, it will be extremely interesting to investigate the mechanistic function of Platr14 on the molecular level.
lncRNA Firre is integrated into the control of a modular gene expression program
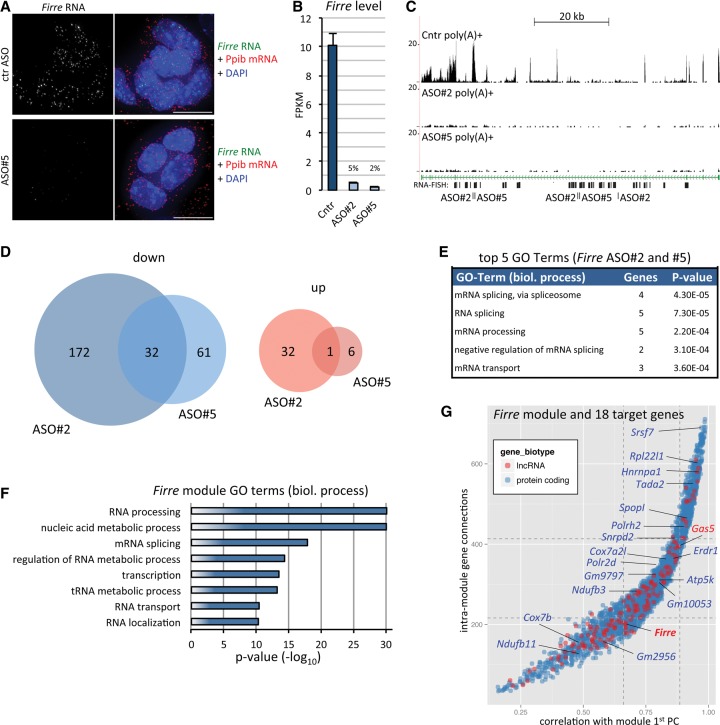
Firre was previously proposed to act as a molecular scaffold required for the spatial clustering of five genic regions in ESCs and other cell lines (Hacisuleyman et al. 2014). Contrary to the focal nuclear localization of Firre transcripts described previously (Hacisuleyman et al. 2014), we instead detected a disperse distribution of Firre transcripts within the nucleoplasm of ESCs and NPCs (Figs. 2B, 5A). A similar nuclear distribution was observed in female PGK12.1 ESCs (Supplemental Fig. S9A). In addition to single-molecule RNA FISH, we also performed “standard” RNA FISH using a nick-translated Firre cDNA probe as an independent technical control. This alternate protocol also confirmed the distribution of Firre transcripts (Supplemental Fig. S9B). In addition to the dispersed Firre signal, some cells also displayed a brighter hybridization focus likely corresponding to the Firre transcription site on the single X Chromosome in male cells, as typically seen with this approach (Supplemental Fig. S9B). As expected for the highly specific nature of our single-molecule RNA FISH approach, hybridization signals were essentially abolished in ESCs depleted of Firre transcripts by ASO knockdown (Fig. 5A, see below).
Figure 5.
Firre lncRNA is functionally integrated into an RNA processing gene expression module. (A) Single-molecule RNA FISH for Firre (green) and Ppib housekeeping mRNA (red) 24 h after transfection of AB2.2 ESCs with control (ctr) ASO or a Firre-specific ASO. (B) Firre expression level in ESCs transfected as in A with control or two independent Firre-specific ASOs. Data represent mean and standard deviation of four biological replicates. (C) Normalized poly(A)+ read coverage across the Firre locus in ESCs transfected as in A and B. RNA FISH target sites and ASO target sites are indicated. Due to the repeat motif content, ASOs are complementary to more than one site of the Firre transcript. (D) Overview of differentially expressed genes 24 h after Firre knockdown with two ASOs in four biological replicates each (FDR < 0.05; see Supplemental Fig. S9 for the full heatmap). (E) GO enrichment for the genes affected by both ASO treatments. Top five biological process terms are shown based on Fisher's exact test and after redundancy trimming. (F) GO terms as in Figure 3C obtained from the weighted gene coexpression analysis-derived gene module containing Firre. (G) Firre module genes (red: lncRNA; blue: mRNA) plotted as in Figure 4A. Labeled genes correspond to 18 of the 31 genes (indicated in D, excluding Firre) significantly down-regulated in both Firre ASO treatments.
Our data thus suggest that Firre transcripts may act more globally within the nucleus. The previous deletion of its >70 kb genomic locus (Hacisuleyman et al. 2014) does not allow one to distinguish between RNA-dependent effects and those mediated by the loss of the chromosomal region, including potential regulatory sites (Bassett et al. 2014), and long-term culture further precludes the discrimination between direct and indirect changes. We thus decided to investigate the immediate impact of Firre depletion on the ESC gene expression profile. To this end, we transfected AB2.2 ESCs with control or two independent Firre-specific ASOs for 24 h after which poly(A)+ RNA-seq was performed. As for Platr14 above, we integrated approximately 240 million uniquely mapping reads from four biological replicates per condition (Supplemental Table S1). Each of the two Firre-specific ASOs achieved substantial reduction of Firre RNA (>95% and 90% based on RNA-seq and qRT-PCT, respectively) (Fig. 5A–C; Supplemental Fig. S10A). Depletion of Firre was paralleled by a significant change (DESeq2, FDR < 0.05) of 100 and 238 genes for ASO#5 and ASO#2, respectively. The majority of affected genes were down-regulated, suggesting that Firre RNA positively mediates the levels of these transcripts (Fig. 5D; Supplemental Fig. S10B). Thirty-three of these genes (including Firre) were detected in both ASO sets. Notably, Eef1a1, one of the five genes found to associate with the Firre locus in ESCs (Hacisuleyman et al. 2014), displayed a reproducible decrease in transcript levels in both treatment conditions, although only ASO#2 resulted in a significant call at the FDR threshold used (Supplemental Fig. S10C). The other four previously identified interacting targets (Hacisuleyman et al. 2014) did not respond to Firre knockdown, with Ypel4 not being detectably expressed in our data set (Supplemental Fig. S10C).
Gene ontology analysis of the 32 differentially expressed genes detected in both treatments indicated that depletion of Firre RNA impacted expression of genes encoding factors involved in RNA processing, including splicing regulators (Fig. 5E). Intriguingly, when we assessed the module membership of Firre using our coexpression analysis above, we found that genes in the Firre-containing module were strongly enriched for gene ontology terms related to RNA metabolism, splicing, and processing (Fig. 5F). Most strikingly, 18 of 32 genes affected by Firre knockdown were members of the same coexpression module as Firre (P < 10−9, hypergeometric test) (Fig. 5G). Together, these data provide compelling evidence for a tight positive relationship between Firre RNA levels and expression of genes encoding for factors regulating RNA fate.
Discussion
LncRNAs have been implicated in playing key roles in the regulation of a multitude of cellular processes, by acting through a large variety of transcript-specific molecular functions (for review, see Lee 2012; Rinn and Chang 2012; Fatica and Bozzoni 2013; Bergmann and Spector 2014). The small fraction of lncRNAs functionally characterized to date suggests that the plethora of presently unstudied molecules may hold the potential to uncover additional layers in the regulation of basic cellular functions as well as roles in differentiation and development. ESCs and their differentiation in culture is a widely used model system to study regulatory function in the context of development and disease (Keller 2005; Evans 2011). We thus set out to provide a comprehensive analysis of the ESC and NPC poly(A)+ lncRNA transcriptome at an unprecedented high depth. Drawing from a panel of biologically meaningful replicate samples, we detect a large number of lncRNAs expressed in the two genetic backgrounds used, generating confidence for their relevance in the given cell type. The use of the latest GENCODE M3 annotation assures that our data set is readily assessable and directly comparable to external research based on this annotation. Importantly, in light of the relatively small number of unannotated “high confidence” intergenic transcription units found in both genetic backgrounds, our analysis represents a valuable near-complete expression profile for the “intergenic” class of lncRNAs studied here.
In line with the work of others (Cabili et al. 2011; Derrien et al. 2012; Djebali et al. 2012), we find that a large fraction of these lncRNAs are highly cell-type specific. We further show that lncRNAs as a class are enriched in the nucleus, consistent with potential roles in this compartment. The finding that many of the nuclear-enriched transcripts remain detectable throughout mitosis may suggest that, rather than being the target of rapid turnover, passive or active mechanisms retain these transcripts within the nucleoplasm of interphase cells. These could include the association of lncRNAs with nuclear protein complexes or interphase chromatin, or the presence of specific motifs causing nuclear import or retention, for example as recently identified for lncRNA BORG (GenBank sequence ID AB010885) (Zhang et al. 2014).
As a central key to our lncRNA characterization and classification, we used robust gene coexpression analysis and, in an unprecedented manner, implemented a stringent importance filter to identify lncRNAs tightly associated with the gene expression state of ESCs. This allowed us to short list a number of presently uncharacterized lncRNAs that can serve as novel developmental biomarkers for the pluripotent state and may be functionally integrated into the ESC gene expression program. As proof of principle, we identify Platr14, a nuclear-enriched lncRNA required to maintain typical expression of a wider range of genes associated with differentiation and tissue development. Interestingly, in contrast to many other pluripotency-related genes, Platr14 is not bound by POU5F1 or NANOG, but instead appears to be expressed from within an endogenous LTR. We generally observed a greater propensity for the expression of LTR-associated lncRNAs in ESCs over more differentiated cells, consistent with the work of others (Kelley and Rinn 2012). Indeed, expression of endogenous retroviral elements is common in mouse and human ESCs and was postulated to contribute to species-specific regulation of pluripotency and differentiation (Macfarlan et al. 2012; Lu et al. 2014). It will be interesting to investigate loss of additional LTR-associated lncRNAs with respect to their contribution to the ESC gene expression program.
Our global cellular fractionation analysis and RNA FISH confirm the recent finding of Hacisuleyman et al. (2014) that the X Chromosome encoded lncRNA Firre is highly enriched in the nucleus (Hacisuleyman et al. 2014). However, both our single-molecule as well as standard RNA FISH approaches indicate that Firre transcripts are widely dispersed throughout the nucleoplasm rather than showing the exclusive focal aggregation reported previously. We can exclude the possibility that these disperse signals observed here are a result of unspecific hybridization, because the method applied critically depends on multiple pairs of independent primary oligos to stably anneal at the expected distance from one another, which drastically increases specificity over conventional RNA FISH techniques. More importantly, ASO-mediated depletion of Firre transcripts to barely detectable levels causes a complete loss of Firre hybridization signal. One possibility for the observed disparities in Firre localization could stem from subtle differences in its basal expression. Indeed, ESC culture conditions differ between the previous study (Hacisuleyman et al. 2014) and our study, although we also do not observe focal aggregation of Firre in other cell types and backgrounds.
It is worth noting that the reported trans-interaction of the Firre locus with the indicated target genes does not appear to be required for their steady-state expression (Hacisuleyman et al. 2014). We also conclude that, with the possible exception of Eef1a1, loss of Firre RNA had no immediate effect on expression of these loci in our ESC system. Instead, Firre down-modulation appears to have a broader impact on the ESC gene expression program, consistent with the ubiquitous distribution of Firre lncRNA in the nucleus. We identify a number of genes encoding proteins related to RNA processing and transport to be negatively impacted by Firre depletion. Notably, gene sets corresponding to such biological processes were also down-regulated in ESCs with a deleted Firre locus (Hacisuleyman et al. 2014). Our data thus markedly extend these previous findings by identifying these genes as primary Firre-dependent targets and further by directly implicating Firre lncRNA molecules in the upkeep of their expression. Our findings that Firre is part of a gene module related to RNA processing and highly connected with 18 of its immediate target genes based on coexpression network analysis further suggests an intimate and positive regulatory connection.
Together, we confirm that both, Platr14 and Firre lncRNAs exert a molecular function that is directly related to the functional associations of the genes within their respective coexpression modules. These data thus strongly support the value of coexpression analysis for predictive “guilt-by-association” inferences (Hughes et al. 2000; Stuart et al. 2003; Basso et al. 2005; Guttman et al. 2009). In particular, integration of the topological overlap measure as used here was shown to aid in the identification of biologically meaningful networks (Li and Horvath 2007; Yip and Horvath 2007). This type of expression profile-based analysis is particularly relevant for the study of lncRNAs, for which transcript levels and changes therein have a direct impact on the cell state. Beyond supporting the generation of functional hypotheses for a given lncRNA, the assessment of module membership and “hub gene”-analogous features, such as intra-module connectivity, enables a robust classification of groups of uncharacterized lncRNAs, as demonstrated here for lncRNAs associated with pluripotency. With increasing numbers of cell type, tissue as well as clinical expression data becoming available, this approach may ultimately provide a highly refined classification of lncRNA associations.
In summary, we globally identified and characterized lncRNAs enriched in the nucleus of pluripotent ESCs and NPCs. We provide a robust and stringent classification of lncRNAs within the pluripotency-associated gene module. Our investigation of two specific lncRNAs on the molecular level demonstrates their requirement for and functional integration into the ESC gene expression program.
Methods
RNA isolation and quality control, nucleofection procedure, ChIP-seq data analysis, ab initio transcript assembly, qRT-PCR, and RACE analysis as well as additional details are provided in the Supplemental Methods.
Cell culture
All cell culture reagents were obtained from Gibco (Life Technologies), unless stated otherwise. ESC colonies were maintained on gelatinized cell culture dishes (Corning), on a feeder layer of irradiated mouse embryonic fibroblasts (GlobalStem) in knockout DMEM supplemented with 15% fetal bovine serum, nonessential amino acids, and 1000 units/mL leukemia inhibiting factor (Millipore). Prior to RNA isolation, feeder cells were removed during a 1-h soaking period. AB2.2 NPCs were differentiated from ESCs via neurospheres as described previously (Conti et al. 2005; Eckersley-Maslin et al. 2014). NPCs were subsequently maintained on gelatinized cell culture dishes in N2 expansion medium composed of 50:50 DMEM/F12: neurobasal medium, supplemented with 1× N2, 0.05× B-27, 50 µg/mL BSA (fraction V), 1 µg/mL laminin, and 10 ng/mL each murine basic fibroblast growth factor and epidermal growth factor (FGF and EGF, PeproTech).
Read mapping and transcriptome analysis
Reads were mapped to the mouse mm10 reference assembly (GRCm38, patch 3) using TopHat2 (version 2.0.9) (Kim et al. 2013). The GENCODE M3 GTF was provided as reference. Default parameters were used, except that only uniquely mapping, “no mixed” and “no discordant” reads were retained. Furthermore, maximum insertion and deletion length was reduced to 2 bp. For transcriptome analysis, Cufflinks2 (version 2.1.1; http://cufflinks.cbcb.umd.edu/) was run by providing transcript models from GENCODE M3 (“-G”). For expression level analysis, the following parameters were set: Effective length correction was suppressed, resulting in improved expression correlation within replicates (data not shown); “max-bundle-length” was increased to 6 Mbp; reads were normalized to those compatible with the reference annotation only. Reads mapping to ribosomal RNAs and small RNA species were masked (“-M”).
Expression analysis was performed on the gene level using a semiconservative cutoff at ≥0.1 FPKM (AB2.2, or the mean expression across seven Cast/BL6 clones). At this threshold, >95% of filtered genes had a nonzero FPKM at the lower bound of the 95% confidence interval. For differential gene expression analysis, raw read count tables for GENCODE M3 gene models were compiled using HTSeq (Anders et al. 2015). Differential expression statistics were obtained using DESeq2, which also handles library size normalization, independent filtering, and dispersion shrinkage (Love et al. 2014). Default settings were used, except for reducing the FDR threshold to a more stringent 0.01 or 0.05 for ESC versus NPC characterization and ASO knockdown studies, respectively.
For mouse tissue expression data, the indicated poly(A)-selected ENCODE data sets (CSHL long RNA sequencing) were downloaded and processed as above. For each tissue, Cufflinks’ FPKM values were averaged from two replicate samples.
Gene ontology analysis was performed using TopGO (version 2.16) considering differentially expressed genes over a background gene universe comprised of all genes assessed in DESeq2. Fisher's exact test was used to calculate P-values and to rank terms. GO terms were subsequently filtered for redundancy using a trimming algorithm at a soft threshold of 0.4 (Jantzen et al. 2011).
Weighted gene coexpression network analysis
Weighted gene coexpression network analysis was performed based on Langfelder and Horvath (2008). Log-transformed FPKM for ESCs and NPCs (AB2.2 and mean values of Cast/BL6 clones) were combined with those of the ENCODE tissue data. Only mRNA and lncRNA biotypes expressed at ≥0.1 FPKM in any one sample were retained. The adjacency matrix was calculated based on pair-wise Pearson correlation coefficients for a signed network (considering only positive expression correlation). A value of β = 15 was empirically chosen as soft-threshold to maximize the number of modules with at least 30 genes while minimizing the number of genes not assigned to any module. Modules were formed based on the adjacency matrix's topological overlap, subsequent average linkage hierarchical clustering, and by using the dynamic tree cut function implemented in the WGCNA R package (Langfelder and Horvath 2008). This analysis yielded 37 modules with a median of about 350 genes. GO analysis was performed using TopGO and followed by term redundancy filtering as above.
RNA Fluorescence In Situ Hybridization (FISH)
For single-molecule RNA FISH, custom Type-6 primary probes targeting Platr14, Firre, and other lncRNAs were designed and synthesized by Affymetrix (Supplemental Table S6). Affymetrix's QuantiGene ViewRNA Cell ISH reagents (Affymetrix) were used to perform dual-color multiplex hybridization on PFA-fixed ESCs with a mouse Ppib-specific Type-1 probe set as endogenous control. Hybridization was performed according to the manufacturer's instructions with some modifications to improve the detection of nuclear transcripts. See Supplemental Methods for full details as well as standard RNA FISH and imaging.
Data access
RNA-seq data from this study have been submitted to the EMBL-EBI ArrayExpress database (http://www.ebi.ac.uk/arrayexpress/) under accession number E-MTAB-3198.
Competing interest statement
D.L.S. is a consultant to Isis Pharmaceuticals (Carlsbad, CA).
Supplementary Material
Acknowledgments
J.H.B. was supported by a DAAD postdoctoral fellowship. M.A.E.-M. was supported by a Genentech Foundation Fellowship and the George A. and Marjorie H. Anderson Fellowship. Research in the Spector laboratory is supported by grants from the National Institute of General Medical Sciences (NIGMS) 42694 and the National Cancer Institute (NCI) 5PO1CA013106-Project 3. The CSHL Microscopy Shared Resource and Next-Generation Sequencing Shared Resource are supported by NCI 2P30CA45508.
Author contributions: J.H.B. and D.L.S. conceived the study, designed the experiments, and wrote the manuscript. J.H.B. performed the experiments and analyzed the data. J.L. and M.A.E.-M. prepared RNA-seq libraries. F.R. and S.M.F. designed and provided ASOs.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.189027.114.
References
- Anders S, Pyl PT, Huber W. 2015. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong L, Lako M, Lincoln J, Cairns PM, Hole N. 2000. mTert expression correlates with telomerase activity during the differentiation of murine embryonic stem cells. Mech Dev 97: 109–116. [DOI] [PubMed] [Google Scholar]
- Bassett AR, Akhtar A, Barlow DP, Bird AP, Brockdorff N, Duboule D, Ephrussi A, Ferguson-Smith AC, Gingeras TR, Haerty W, et al. 2014. Considerations when investigating lncRNA function in vivo. Elife 3: e03058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basso K, Margolin AA, Stolovitzky G, Klein U, Dalla-Favera R, Califano A. 2005. Reverse engineering of regulatory networks in human B cells. Nat Genet 37: 382–390. [DOI] [PubMed] [Google Scholar]
- Bergmann JH, Spector DL. 2014. Long non-coding RNAs: modulators of nuclear structure and function. Curr Opin Cell Biol 26: 10–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertone P, Stolc V, Royce TE, Rozowsky JS, Urban AE, Zhu X, Rinn JL, Tongprasit W, Samanta M, Weissman S, et al. 2004. Global identification of human transcribed sequences with genome tiling arrays. Science 306: 2242–2246. [DOI] [PubMed] [Google Scholar]
- Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, Rinn JL. 2011. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev 25: 1915–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, et al. 2005. The transcriptional landscape of the mammalian genome. Science 309: 1559–1563. [DOI] [PubMed] [Google Scholar]
- Chambers I, Colby D, Robertson M, Nichols J, Lee S, Tweedie S, Smith A. 2003. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell 113: 643–655. [DOI] [PubMed] [Google Scholar]
- Cheng J, Kapranov P, Drenkow J, Dike S, Brubaker S, Patel S, Long J, Stern D, Tammana H, Helt G, et al. 2005. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science 308: 1149–1154. [DOI] [PubMed] [Google Scholar]
- Clemson CM, Hutchinson JN, Sara SA, Ensminger AW, Fox AH, Chess A, Lawrence JB. 2009. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell 33: 717–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti L, Pollard SM, Gorba T, Reitano E, Toselli M, Biella G, Sun Y, Sanzone S, Ying QL, Cattaneo E, et al. 2005. Niche-independent symmetrical self-renewal of a mammalian tissue stem cell. PLoS Biol 3: e283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, et al. 2012. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res 22: 1775–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinger ME, Amaral PP, Mercer TR, Pang KC, Bruce SJ, Gardiner BB, Askarian-Amiri ME, Ru K, Soldà G, Simons C, et al. 2008. Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome Res 18: 1433–1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, et al. 2012. Landscape of transcription in human cells. Nature 489: 101–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckersley-Maslin MA, Thybert D, Bergmann JH, Marioni JC, Flicek P, Spector DL. 2014. Random monoallelic gene expression increases upon embryonic stem cell differentiation. Dev Cell 28: 351–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The ENCODE Project Consortium. 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489: 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans M. 2011. Discovering pluripotency: 30 years of mouse embryonic stem cells. Nat Rev Mol Cell Biol 12: 680–686. [DOI] [PubMed] [Google Scholar]
- Fatica A, Bozzoni I. 2013. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet 15: 7–21. [DOI] [PubMed] [Google Scholar]
- Grote P, Wittler L, Hendrix D, Koch F, Währisch S, Beisaw A, Macura K, Bläss G, Kellis M, Werber M, et al. 2013. The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev Cell 24: 206–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al. 2009. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 458: 223–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Garber M, Levin JZ, Donaghey J, Robinson J, Adiconis X, Fan L, Koziol MJ, Gnirke A, Nusbaum C, et al. 2010. Ab initio reconstruction of cell type–specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat Biotechnol 28: 503–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L, et al. 2011. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature 477: 295–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacisuleyman E, Goff LA, Trapnell C, Williams A, Henao-Mejia J, Sun L, McClanahan P, Hendrickson DG, Sauvageau M, Kelley DR, et al. 2014. Topological organization of multichromosomal regions by the long intergenic noncoding RNA Firre. Nat Struct Mol Biol 21: 198–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hangauer MJ, Vaughn IW, McManus MT. 2013. Pervasive transcription of the human genome produces thousands of previously unidentified long intergenic noncoding RNAs. PLoS Genet 9: e1003569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G, Tang Q, Sharma S, Yu F, Escobar TM, Muljo SA, Zhu J, Zhao K. 2013. Expression and regulation of intergenic long noncoding RNAs during T cell development and differentiation. Nat Immunol 14: 1190–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes TR, Marton MJ, Jones AR, Roberts CJ, Stoughton R, Armour CD, Bennett HA, Coffey E, Dai H, He YD, et al. 2000. Functional discovery via a compendium of expression profiles. Cell 102: 109–126. [DOI] [PubMed] [Google Scholar]
- Ivanova N, Dobrin R, Lu R, Kotenko I, Levorse J, DeCoste C, Schafer X, Lun Y, Lemischka IR. 2006. Dissecting self-renewal in stem cells with RNA interference. Nature 442: 533–538. [DOI] [PubMed] [Google Scholar]
- Jantzen SG, Sutherland BJG, Minkley DR, Koop BF. 2011. GO Trimming: systematically reducing redundancy in large Gene Ontology datasets. BMC Res Notes 4: 267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapranov P, Cawley SE, Drenkow J, Bekiranov S, Strausberg RL, Fodor SPA, Gingeras TR. 2002. Large-scale transcriptional activity in chromosomes 21 and 22. Science 296: 916–919. [DOI] [PubMed] [Google Scholar]
- Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermüller J, Hofacker IL, et al. 2007. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science 316: 1484–1488. [DOI] [PubMed] [Google Scholar]
- Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, Nishida H, Yap CC, Suzuki M, Kawai J, et al. 2005. Antisense transcription in the mammalian transcriptome. Science 309: 1564–1566. [DOI] [PubMed] [Google Scholar]
- Keller G. 2005. Embryonic stem cell differentiation: emergence of a new era in biology and medicine. Genes Dev 19: 1129–1155. [DOI] [PubMed] [Google Scholar]
- Kelley D, Rinn J. 2012. Transposable elements reveal a stem cell-specific class of long noncoding RNAs. Genome Biol 13: R107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, et al. 2009. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci 106: 11667–11672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, et al. 2010. Widespread transcription at neuronal activity-regulated enhancers. Nature 465: 182–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretz M, Siprashvili Z, Chu C, Webster DE, Zehnder A, Qu K, Lee CS, Flockhart RJ, Groff AF, Chow J, et al. 2013. Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature 493: 231–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P, Horvath S. 2008. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9: 559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JT. 2012. Epigenetic regulation by long noncoding RNAs. Science 338: 1435–1439. [DOI] [PubMed] [Google Scholar]
- Li A, Horvath S. 2007. Network neighborhood analysis with the multi-node topological overlap measure. Bioinformatics 23: 222–231. [DOI] [PubMed] [Google Scholar]
- Lin N, Chang KY, Li Z, Gates K, Rana ZA, Dang J, Zhang D, Han T, Yang CS, Cunningham TJ, et al. 2014. An evolutionarily conserved long noncoding RNA TUNA controls pluripotency and neural lineage commitment. Mol Cell 53: 1005–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodato MA, Ng CW, Wamstad JA, Cheng AW, Thai KK, Fraenkel E, Jaenisch R, Boyer LA. 2013. SOX2 co-occupies distal enhancer elements with distinct POU factors in ESCs and NPCs to specify cell state. PLoS Genet 9: e1003288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Sachs F, Ramsay L, Jacques PE, Göke J, Bourque G, Ng HH. 2014. The retrovirus HERVH is a long noncoding RNA required for human embryonic stem cell identity. Nat Struct Mol Biol 21: 423–425. [DOI] [PubMed] [Google Scholar]
- Maamar H, Cabili MN, Rinn J, Raj A. 2013. linc-HOXA1 is a noncoding RNA that represses Hoxa1 transcription in cis. Genes Dev 27: 1260–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macfarlan TS, Gifford WD, Driscoll S, Lettieri K, Rowe HM, Bonanomi D, Firth A, Singer O, Trono D, Pfaff SL. 2012. Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nature 487: 57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marson A, Levine SS, Cole MF, Frampton GM, Brambrink T, Johnstone S, Guenther MG, Johnston WK, Wernig M, Newman J, et al. 2008. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell 134: 521–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng L, Ward AJ, Chun S, Bennett CF, Beaudet AL, Rigo F. 2015. Towards a therapy for Angelman syndrome by targeting a long non-coding RNA. Nature 518: 409–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer TR, Qureshi IA, Gokhan S, Dinger ME, Li G, Mattick JS, Mehler MF. 2010. Long noncoding RNAs in neuronal-glial fate specification and oligodendrocyte lineage maturation. BMC Neurosci 11: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. 2007. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448: 553–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng SY, Bogu GK, Soh BS, Stanton LW. 2013. The long noncoding RNA RMST interacts with SOX2 to regulate neurogenesis. Mol Cell 51: 349–359. [DOI] [PubMed] [Google Scholar]
- Okazaki Y, Furuno M, Kasukawa T, Adachi J, Bono H, Kondo S, Nikaido I, Osato N, Saito R, Suzuki H, et al. 2002. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature 420: 563–573. [DOI] [PubMed] [Google Scholar]
- Poliseno L, Salmena L, Zhang J, Carver B, Haveman WJ, Pandolfi PP. 2010. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature 465: 1033–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quek XC, Thomson DW, Maag JLV, Bartonicek N, Signal B, Clark MB, Gloss BS, Dinger ME. 2015. lncRNAdb v2.0: expanding the reference database for functional long noncoding RNAs. Nucleic Acids Res 43: D168–D173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Chang HY. 2012. Genome regulation by long noncoding RNAs. Annu Rev Biochem 81: 145–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn J, Guttman M. 2014. RNA function. RNA and dynamic nuclear organization. Science 345: 1240–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers MB, Hosler BA, Gudas LJ. 1991. Specific expression of a retinoic acid-regulated, zinc-finger gene, Rex-1, in preimplantation embryos, trophoblast and spermatocytes. Development 113: 815–824. [DOI] [PubMed] [Google Scholar]
- Sheik Mohamed J, Gaughwin PM, Lim B, Robson P, Lipovich L. 2010. Conserved long noncoding RNAs transcriptionally regulated by Oct4 and Nanog modulate pluripotency in mouse embryonic stem cells. RNA 16: 324–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart JM, Segal E, Koller D, Kim SK. 2003. A gene-coexpression network for global discovery of conserved genetic modules. Science 302: 249–255. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. 2010. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28: 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulitsky I, Shkumatava A, Jan CH, Sive H, Bartel DP. 2011. Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell 147: 1537–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe D, Suetake I, Tada T, Tajima S. 2002. Stage- and cell-specific expression of Dnmt3a and Dnmt3b during embryogenesis. Mech Dev 118: 187–190. [DOI] [PubMed] [Google Scholar]
- Wheeler TM, Leger AJ, Pandey SK, MacLeod AR, Nakamori M, Cheng SH, Wentworth BM, Bennett CF, Thornton CA. 2012. Targeting nuclear RNA for in vivo correction of myotonic dystrophy. Nature 488: 111–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz JE, Sunwoo H, Spector DL. 2009. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev 23: 1494–1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, Babak T, Shendure J, Disteche CM. 2010. Global survey of escape from X inactivation by RNA-sequencing in mouse. Genome Res 20: 614–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yip AM, Horvath S. 2007. Gene network interconnectedness and the generalized topological overlap measure. BMC Bioinformatics 8: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young TL, Matsuda T, Cepko CL. 2005. The noncoding RNA taurine upregulated gene 1 is required for differentiation of the murine retina. Curr Biol 15: 501–512. [DOI] [PubMed] [Google Scholar]
- Zhang B, Gunawardane L, Niazi F, Jahanbani F, Chen X, Valadkhan S. 2014. A novel RNA motif mediates the strict nuclear localization of a long non-coding RNA. Mol Cell Biol 34: 2318–2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao XY, Li S, Wang GX, Yu Q, Lin JD. 2014. A long noncoding RNA transcriptional regulatory circuit drives thermogenic adipocyte differentiation. Mol Cell 55: 372–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.