Abstract
Precise regulation of microRNA (miRNA) expression is critical for diverse physiologic and pathophysiologic processes. Nevertheless, elucidation of the mechanisms through which miRNA expression is regulated has been greatly hindered by the incomplete annotation of primary miRNA (pri-miRNA) transcripts. While a subset of miRNAs are hosted in protein-coding genes, the majority of pri-miRNAs are transcribed as poorly characterized noncoding RNAs that are 10's to 100's of kilobases in length and low in abundance due to efficient processing by the endoribonuclease DROSHA, which initiates miRNA biogenesis. Accordingly, these transcripts are poorly represented in existing RNA-seq data sets and exhibit limited and inaccurate annotation in current transcriptome assemblies. To overcome these challenges, we developed an experimental and computational approach that allows genome-wide detection and mapping of pri-miRNA structures. Deep RNA-seq in cells expressing dominant-negative DROSHA resulted in much greater coverage of pri-miRNA transcripts compared with standard RNA-seq. A computational pipeline was developed that produces highly accurate pri-miRNA assemblies, as confirmed by extensive validation. This approach was applied to a panel of human and mouse cell lines, providing pri-miRNA transcript structures for 1291/1871 human and 888/1181 mouse miRNAs, including 594 human and 425 mouse miRNAs that fall outside protein-coding genes. These new assemblies uncovered unanticipated features and new potential regulatory mechanisms, including links between pri-miRNAs and distant protein-coding genes, alternative pri-miRNA splicing, and transcripts carrying subsets of miRNAs encoded by polycistronic clusters. These results dramatically expand our understanding of the organization of miRNA-encoding genes and provide a valuable resource for the study of mammalian miRNA regulation.
MicroRNAs (miRNAs) are a broad class of ∼18–24 nt RNA molecules that play a critical role in regulating gene expression in diverse physiologic settings and diseases by negatively regulating the translation and stability of target messenger RNAs (mRNAs) (Bartel 2009). Over the past decade, significant progress has been made in identifying miRNA targets and dissecting the mechanisms through which they are regulated by miRNA-directed protein complexes (Pasquinelli 2012; Gurtan and Sharp 2013). However, much less is known about how miRNA expression is regulated (Winter et al. 2009; Schanen and Li 2011). Through examination of mature miRNA levels, it is well established that miRNA abundance is tightly controlled during development and across tissues (Landgraf et al. 2007; Chiang et al. 2010). Moreover, dysregulated expression of specific miRNAs plays a causative role in a number of human diseases, including cancer and cardiovascular disease (Di Leva et al. 2014; Olson 2014). Indeed, key transcription factors and signaling pathways have been shown to strongly regulate miRNA expression under diverse physiologic and pathophysiologic conditions (Lotterman et al. 2008). Nevertheless, a major bottleneck in the dissection of the mechanisms through which these pathways control miRNA levels has been our incomplete understanding of miRNA gene structures.
miRNAs are initially transcribed by RNA polymerase II as long primary transcripts (pri-miRNAs) that can extend hundreds of kilobases in length (Cai et al. 2004; Lee et al. 2004). The mature miRNA sequences are located in introns or exons of pri-miRNAs, within regions that fold into imperfect hairpin structures (Rodriguez et al. 2004). The RNA-binding protein DGCR8 and the RNase III enzyme DROSHA together recognize and cleave the hairpins, generating ∼60–80 nt precursors (pre-miRNAs) that are subsequently exported to the cytoplasm where they are processed into mature miRNAs by DICER. Once loaded into the Argonaute family of RNA-binding proteins, miRNAs select mRNA targets for repression (Ha and Kim 2014). While a subset of miRNAs are hosted in well-characterized protein-coding genes, the majority of pri-miRNAs are transcribed as poorly characterized noncoding transcripts (Rodriguez et al. 2004). Due to the nature of rapid and efficient DROSHA/DGCR8 processing, the abundance of pri-miRNAs is very low at steady state. Therefore, elucidation of pri-miRNA structure has remained a significant challenge. A further understanding of the organization of miRNA transcription units will likely reveal new transcriptional and post-transcriptional regulatory mechanisms that influence miRNA biogenesis and potentially uncover new opportunities to manipulate miRNA expression for experimental or therapeutic applications.
Previous studies have systematically identified genomic locations of the promoters and transcription start sites (TSSs) of miRNAs by integrating chromatin signatures such as H3K4me3 histone modifications, nucleosome position, cap analysis of gene expression (CAGE) tags, and high-throughput TSS sequencing (TSS-Seq) (Marson et al. 2008; Ozsolak et al. 2008; Megraw et al. 2009; Chien et al. 2011; Marsico et al. 2013; Georgakilas et al. 2014; Xiao et al. 2014). Nevertheless, while providing valuable information regarding the boundaries of miRNA transcription units, these approaches do not provide annotation of the often complex splicing patterns of miRNA primary transcripts, and thus provide an incomplete picture of miRNA gene structure. Moreover, miRNA promoters that are located at great distances from the mature miRNA sequence are not easily associated with a given miRNA transcription unit and alternative promoter usage can be difficult to discern. Finally, without an understanding of the structure of the pri-miRNA itself, it is impossible to determine whether miRNAs encoded by polycistronic clusters are always cotranscribed or whether transcripts carrying subsets of the clustered miRNAs are produced through use of alternative promoters, polyadenylation sites, or even through alternative splicing.
In recent years, high-throughput RNA sequencing (RNA-seq) has emerged as a powerful tool for transcriptome reconstruction (Martin and Wang 2011; McGettigan 2013). Unfortunately, due to their low abundance, pri-miRNAs are poorly represented in standard RNA-seq data sets, thus preventing comprehensive annotation of their structures using existing methodologies. To overcome this limitation, we developed a highly effective experimental and computational approach that allows genome-wide mapping of miRNA primary transcript structures. By performing deep RNA-seq in cells expressing a dominant-negative DROSHA mutant protein, we demonstrated dramatic enrichment of intact pri-miRNAs, resulting in much greater coverage of these transcripts compared with standard RNA-seq. This strategy permitted the reconstruction of pri-miRNA structures in a high-throughput manner. We applied this approach to human and mouse cell lines of diverse origins, thereby significantly improving the existing annotation of mammalian miRNA genes. These new assemblies revealed new regulatory mechanisms for many miRNAs, including previously unknown connections between pri-miRNAs and distant protein-coding genes, alternative pri-miRNA splicing, and pri-miRNA transcripts that produce subsets of miRNAs encoded by polycistronic clusters. This new genome-wide map of pri-miRNA structure provides a valuable resource for investigating the mechanisms that control miRNA expression in normal physiology and disease.
Results
Pri-miRNAs are poorly represented in standard RNA-seq data sets
In order to globally reconstruct pri-miRNA structures, we first examined existing RNA-seq data sets to determine whether they could be used for this purpose. The Illumina BodyMap 2.0 represents a collection of RNA-seq data sets generated from 16 human tissues, each sequenced very deeply (∼80 million 50-bp paired-end reads per sample) (www.ebi.ac.uk/arrayexpress; ArrayExpress ID: E-MTAB-513). As described in greater detail below, we determined that StringTie, a transcriptome assembler that we recently described (Pertea et al. 2015), outperforms other existing assembly algorithms for pri-miRNA reconstruction. We therefore utilized StringTie to assess pri-miRNA assembly using Illumina BodyMap data.
Although assemblies were attempted for all human pri-miRNAs, the quality and extent of pri-miRNA reconstruction was assessed by examining a well-annotated set of miRNAs that are highly conserved among mammals (Chiang et al. 2010). Nonconserved human miRNAs were excluded from this performance analysis since these are frequently expressed at low levels, and there is no current consensus regarding which of these represent bona fide miRNAs as opposed to nonfunctional RNAs that spuriously enter the miRNA processing pathway (Chiang et al. 2010; Kozomara and Griffiths-Jones 2014). A total of 295 human miRNAs, produced from 183 transcription units, are classified as conserved among mammals (Supplemental Fig. S1). Of these 183 transcription units, 80 represent well-annotated protein-coding genes, whereas the remaining 103 are intergenic. While the structures of 29 of these intergenic pri-miRNAs are annotated in RefSeq, the majority (74 of 103) have no existing annotation. Assembly of all 16 BodyMap data sets using StringTie, which comprised the analysis of over 1.2 × 109 reads, resulted in the assembly of only 11 additional novel pri-miRNA structures covering the set of conserved miRNAs (Supplemental Table S1). These results indicate that standard RNA-seq libraries are inadequate for transcriptome-wide reconstruction of pri-miRNA structures.
DROSHA inhibition facilitates pri-miRNA assembly
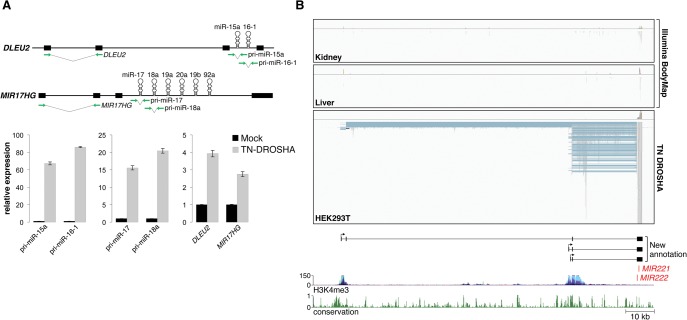
During miRNA biogenesis, pri-miRNAs are first processed in the nucleus by the microprocessor complex composed of DROSHA and DGCR8. We reasoned that the low steady-state abundance of pri-miRNAs, and their poor representation in standard RNA-seq libraries, is most likely due to their rapid degradation following microprocessor-mediated cleavage. Therefore, we hypothesized that slowed or disrupted DROSHA/DGCR8 activity may result in an enrichment of pri-miRNAs in RNA-seq libraries and thereby facilitate pri-miRNA assembly. To test this concept, a trans-dominant-negative DROSHA mutant protein (TN-DROSHA) containing inactivating mutations in critical residues in the catalytic RNase IIIa and IIIb domains (Heo et al. 2008) was ectopically expressed in HEK293T cells, and nuclear RNA was analyzed by quantitative real time PCR (qRT-PCR). Amplicons spanning pre-miRNA hairpins in the primary transcripts that encode the miR-15a/16-1 and miR-17-92 clusters (DLEU2 and MIR17HG, respectively) were strongly enriched following TN-DROSHA expression, indicating efficient inhibition of microprocessor activity (Fig. 1A). Importantly, distant regions of these pri-miRNAs that do not span the pre-miRNA hairpins also showed significant enrichment, suggesting that the entire pri-miRNA was stabilized.
Figure 1.
DROSHA inhibition facilitates pri-miRNA assembly. (A) qPCR analysis of pri-miRNA abundance in HEK293T cells with or without expression of TN-DROSHA. The assayed transcripts DLEU2 and MIR17HG are depicted in the top panel with green arrows indicating the location of primers. qPCR results are shown in the bottom panel with error bars representing standard deviations derived from three independent measurements. (B) Visualization of RNA-seq data from Illumina Human BodyMap 2.0 (kidney and liver) and TN-DROSHA-transfected HEK293T cells. The Integrative Genomics Viewer (IGV) was used to visualize mapped read alignments. Segments of reads that are aligned to the genome are shown in gray, while blue lines represent spliced sequences. StringTie assembled transcripts produced from this locus are shown at the bottom of the panel. Plots representing H3K4me3 histone marks and evolutionary conservation were generated using the UCSC Genome Browser (human genome GRCh37/hg19 assembly). The y-axes for UCSC Genome Browser tracks shown in this and all other figures represent the default vertical viewing range settings.
Next, we subjected the same nuclear RNA from TN-DROSHA expressing HEK293T cells to Illumina RNA sequencing to test its suitability for transcriptome-wide pri-miRNA assembly. After generating a very deep RNA-seq data set (193,346,087 100-bp paired-end reads), we evaluated several transcriptome assemblers, such as StringTie, Cufflinks (Trapnell et al. 2010), IsoLasso (Li et al. 2011), and Scripture (Guttman et al. 2010), to assess their performance for this application (Supplemental Table S2). By evaluating the assembly of pri-miRNAs that are annotated in RefSeq, we found that StringTie correctly assembled the highest number of pri-miRNA transcripts in considerably less time than the other assemblers. We therefore used StringTie for all subsequent pri-miRNA assembly experiments.
When RNA-seq data from TN-DROSHA expressing HEK293T cells were used, pri-miRNA assembly was dramatically improved compared with results obtained using the Illumina BodyMap. From this single cell line, 24/74 conserved intergenic pri-miRNAs that lack existing annotation were assembled. When combined with RefSeq annotation, 53/103 conserved intergenic pri-miRNAs in total were defined, essentially doubling the available annotation of conserved non-protein-coding pri-miRNAs. Reads mapping to miRNA loci were highly enriched for those that span splice sites, allowing reconstruction of multi-exonic pri-miRNA structures. Illustrative of these improved assemblies, three multi-exonic transcripts that encode miR-221 and miR-222 were reconstructed using RNA-seq data generated from TN-DROSHA-expressing HEK293T cells, while few reads mapping to these transcripts were present in Illumina BodyMap data (Fig. 1B). These transcript assemblies were validated by confirming the predicted exon–exon junctions using reverse-transcriptase PCR (RT-PCR) with primers near the 5′ and 3′ ends of the transcripts (Supplemental Fig. S2). Notably, although the 5′ ends of these transcripts are ∼25- to 100-kb upstream of the MIR221 and MIR222 sequences, analysis of ENCODE chromatin immunoprecipitation sequencing (ChIP-seq) data (Ernst et al. 2011) revealed precise colocalization with H3K4me3 promoter marks (Fig. 1B), supporting the correct identification of these transcription start sites. These results demonstrate that inhibition of microprocessor activity by expression of TN-DROSHA greatly improves pri-miRNA assembly in RNA-seq data.
Genome-wide annotation of pri-miRNAs
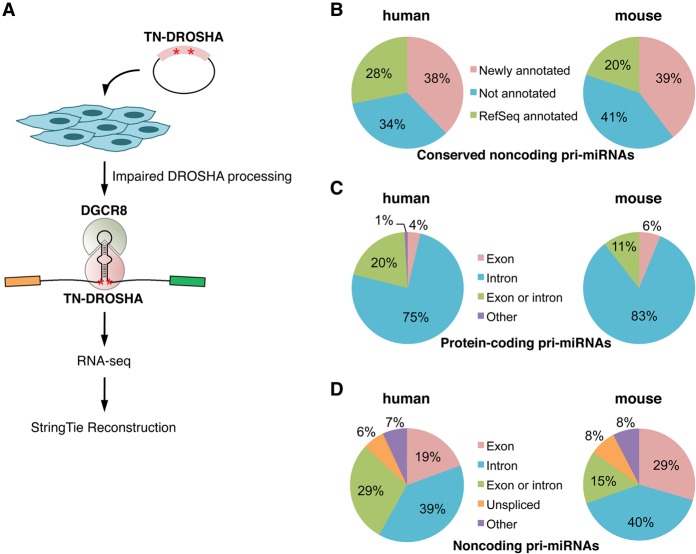
Having established an experimental and computational strategy suitable for pri-miRNA reconstruction, we next sought to apply this approach to generate a genome-wide map of human and mouse pri-miRNA structures. Since miRNA expression is often cell-type- and tissue-specific (Olive et al. 2015), we selected for analysis a panel of eight human cell lines (A-172, A-673, HCT116, HEK293T, HepG2, MCF-7, NCCIT, and primary fibroblasts) and six mouse cell lines (C2C12, CT-26, Hepa1-6, Neuro-2a, mouse embryonic fibroblasts [MEF], and E14TG2a embryonic stem cells) derived from a diverse array of cell types. Transfection conditions were optimized for each cell line and TN-DROSHA was introduced, followed by RNA-seq and StringTie transcriptome reconstruction (Fig. 2A). On average, ∼180 million 100-bp paired-end reads were generated per sample (Supplemental Table S3).
Figure 2.
General characteristics of human and mouse pri-miRNAs. (A) Overview of the experimental workflow used to generate pri-miRNA assemblies. (B) Proportion of conserved non-protein-coding human and mouse pri-miRNAs annotated in this study or in RefSeq in at least one cell type. (C,D) Intronic or exonic locations of conserved miRNAs transcribed within protein-coding (C) or non-protein-coding genes (D).
Using these data, pri-miRNA assemblies were provided for 1291/1871 (69%) of human miRNAs and 888/1181 (75%) of mouse miRNAs that are annotated in miRBase version 20. This includes assemblies for 594 human and 425 mouse miRNAs that are not hosted by annotated protein-coding genes. As mentioned above, nonconserved intergenic miRNAs are generally very low in abundance and consensus is lacking regarding which of these represent true miRNA genes. Therefore, to more accurately assess the quality of these pri-miRNA assemblies, we focused on the pri-miRNA transcripts that encode the set of 295 human and 297 mouse miRNAs that are conserved among mammals (Chiang et al. 2010), which represents a more reliable set of bona fide miRNAs. A total of 38% (39 of 103) of human and 39% (41 of 104) of mouse conserved non-protein-coding pri-miRNAs were successfully reconstructed in at least one cell line (Fig. 2B). When combined with existing RefSeq data, annotation for 66% and 59% of conserved intergenic miRNA genes was provided in total for human and mouse, respectively. These new assemblies are supplied as Gene Transfer Files (GTF), which can be directly visualized in standard genome browsers (Supplemental Data), and can also be viewed in the UCSC Genome Browser (see Data access).
General characteristics and conservation of pri-miRNAs
Using these improved pri-miRNA maps, we examined the characteristics that typify miRNA-encoding genes. As expected, of the conserved miRNAs that are hosted within protein-coding genes, a large majority of pre-miRNA hairpins are located in introns (75% in human and 83% in mouse) (Fig. 2C). For conserved intergenic miRNAs, the frequency of intronic miRNAs drops to ∼40%, with the remainder in exons or regions that may be intronic or exonic due to alternative splicing (Fig. 2D). In some cases, intergenic miRNAs are hosted in unspliced noncoding RNAs (6% in human and 8% in mouse).
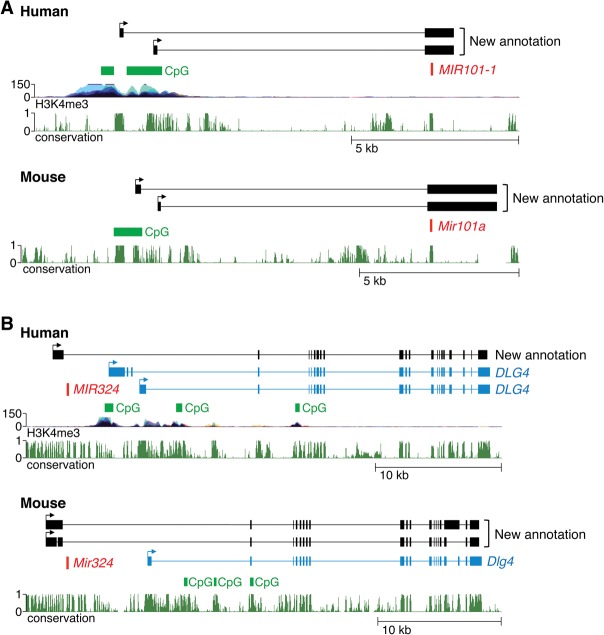
In cases where orthologous human and mouse intergenic pri-miRNAs were assembled, we frequently observed conservation of the organization of these miRNA-encoding loci. The locations of pri-miRNA promoters were particularly highly conserved, with the 5′ ends of these transcripts almost always mapping to orthologous regions in the human and mouse genomes when pri-miRNA assemblies were available for both species. Representative examples of conserved pri-miRNAs are shown in Figure 3. For instance, we identified two distinct pri-miRNAs that encode human miR-101-1 that each utilized different transcription start sites located ∼9 kb upstream of the miRNA (Fig. 3A). The presence of CpG islands and H3K4me3 histone marks near the transcript 5′ ends support these assemblies. Likewise, two transcription start sites were also mapped to a GC-rich region 9 kb upstream of the sequence that encodes mouse miR-101a (Fig. 3A). Both the human and mouse pri-miRNA transcripts are composed of two exons, with the miRNA located in exon 2. These transcript structures were confirmed by RT-PCR (Supplemental Figs. S3,S4). Human and mouse miR-324 are also representative of miRNAs encoded by transcription units with conserved organization, and, as discussed in greater detail below, represent a class of pri-miRNAs that are transcribed as 5′ extensions of annotated protein-coding genes (Fig. 3B).
Figure 3.
Examples of evolutionarily conserved pri-miRNAs. (A) Genomic loci encoding human and mouse miR-101-1. StringTie assembled transcripts, as well as H3K4me3 marks, CpG islands, and conservation tracks from the UCSC Genomic Browser (hg19 and mm10) are shown. (B) Genomic loci encoding human and mouse miR-324 as in A. The RefSeq protein-coding transcript DLG4 is shown in blue.
Classification of miRNA gene structures
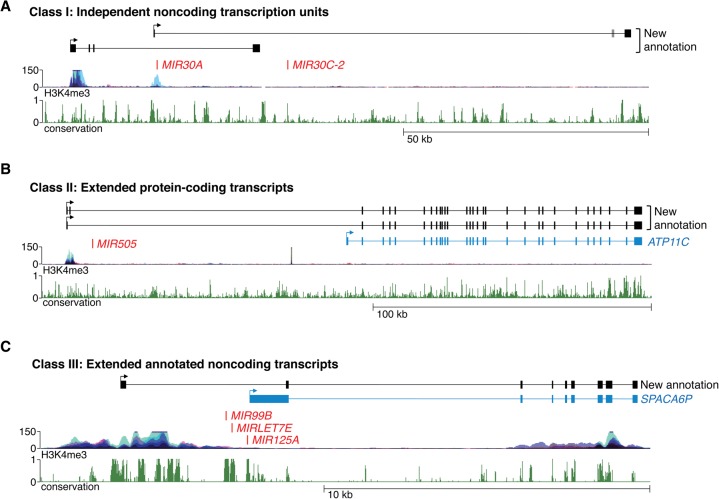
Examination of miRNAs that are not hosted within protein-coding genes revealed that their primary transcripts could be cataloged into three broad classes (Supplemental Table S1), each described below and illustrated in Figure 4.
Figure 4.
Classification of newly annotated miRNA genes. (A) Class I pri-miRNAs, represented by the transcripts that encode miR-30a and miR-30c-2, are independent noncoding transcription units with no existing annotation. (B) Class II pri-miRNAs, represented by the transcript that encodes miR-505, are extensions of annotated protein-coding transcripts. The RefSeq protein-coding transcript ATP11C is shown in blue. (C) Class III pri-miRNAs, represented by the transcript that encodes miR-99b, let-7e, and miR-125a, are extensions of annotated noncoding transcripts. The RefSeq noncoding transcript SPACA6P is shown in blue.
Class I: independent noncoding transcription units
Approximately 60%–70% of newly defined noncoding pri-miRNAs that host conserved miRNAs do not overlap any existing annotated genes and likely represent independent transcription units (Supplemental Table S1). For example, MIR30A and MIR30C-2 are intergenic miRNA genes with no existing annotation of their primary transcripts (Fig. 4A). Our assemblies revealed two putative overlapping pri-miRNAs that initiate and terminate at distinct sites. The 5′ ends of both transcripts colocalize with ENCODE H3K4me3 ChIP-seq signals and were validated using 5′ rapid amplification of cDNA ends (RACE) (Supplemental Fig. S5). 3′ RACE was used to confirm the distal termini of the transcripts while RT-PCR verified their exonic structure (Supplemental Figs. S5,S6). Although it is generally assumed that clustered miRNAs such as these are always cotranscribed, it is noteworthy that use of the upstream promoter produces a transcript that encodes miR-30a but not miR-30c-2. These results suggest that production of miR-30a is uncoupled from miR-30c-2 in some settings. As discussed further below, we found additional examples of pri-miRNA transcripts that produce subsets of clustered miRNAs.
Class II: extended protein-coding transcripts
In addition to completely independent transcription units, we unexpectedly observed that several pri-miRNAs are produced as extended isoforms of annotated protein-coding genes (Fig. 4B; Supplemental Table S1). This configuration is illustrated by MIR505, which is located ∼100 kb upstream of the gene that encodes the ATP11C protein. Remarkably, we observed that the predominant promoter that drives ATP11C transcription is located upstream of MIR505, with the miRNA hairpin located within intron 1 of the extended transcript. Indeed, ENCODE H3K4me3 ChIP-seq signal is significantly higher at the extended transcript 5′ end compared with the RefSeq annotated ATP11C promoter. RT-PCR confirmed the existence of the extended miRNA-hosting transcript (Supplemental Fig. S7). Additional examples of similarly organized pri-miRNAs encoding miR-181c/181d and miR-219-1 are provided in Supplemental Figure S8.
Class III: extended annotated noncoding transcripts
The third class of pri-miRNAs that we observed were a set that overlap annotated RefSeq noncoding RNAs. This type of transcript is exemplified by the pri-miRNA that encodes miR-99b, let-7e, and miR-125a (Fig. 4C). These miRNAs are located immediately upstream of an annotated noncoding RNA, SPACA6P. In our assemblies, a longer transcript that encompasses both the miRNAs and SPACA6P was detected. RT-PCR confirmed the transcript structure predicted by our data (Supplemental Fig. S9). It is likely that the existing annotation of SPACA6P actually represents the 3′ cleavage product of the MIR99B/MIRLET7E/MIR125A pri-miRNA that is produced by DROSHA processing, since the 5′ end of SPACA6P is immediately adjacent to the 3′ end of the pre-miR-125a hairpin. We speculate that this class of pri-miRNAs is largely composed of transcripts that are incompletely annotated in RefSeq.
Pri-miRNA structures reveal novel regulatory mechanisms
Inspection of pri-miRNA gene structure using our assemblies uncovered new potential regulatory mechanisms that likely influence the production of specific miRNAs. These mechanisms include alternative promoters, partially transcribed miRNA clusters, and alternative splicing, each discussed in turn below and summarized in Supplemental Tables S4 and S5.
Alternative promoters
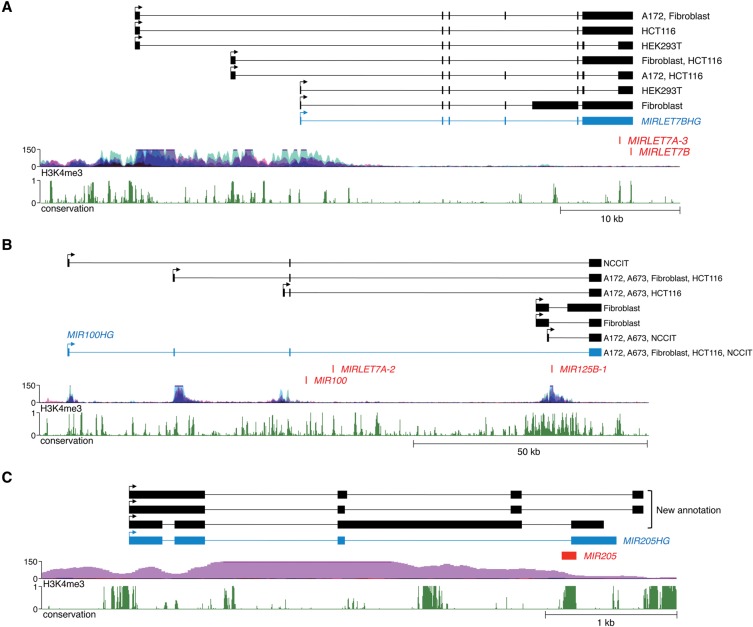
Perhaps unsurprisingly given the incomplete existing annotation of pri-miRNA genes, our assemblies frequently identified alternative promoters that drive miRNA expression in different cell types. This phenomenon is exemplified by the gene that encodes let-7a-3 and let-7b. This pri-miRNA, annotated in RefSeq as MIRLET7BHG, initiates 27 kb upstream of the miRNA sequences, in a region rich in H3K4me3-modified histones (Fig. 5A). We observed two additional transcription start sites further upstream, also associated with H3K4me3. These transcript structures and 5′ ends were validated by RT-PCR and RACE (Supplemental Figs. S10 and S11). While all cell lines tested used the most upstream promoter, the alternative downstream transcription start sites were differentially utilized in a cell-line-specific manner. These results suggest that these distinct promoters may be differentially regulated. Of the 103 human intergenic conserved miRNA transcription units, we documented that at least 25 have multiple alternative promoters (Supplemental Table S4), indicating that this is a very common mode of miRNA regulation.
Figure 5.
Representative examples of newly identified miRNA regulatory mechanisms. (A) Pri-miRNA genes frequently utilize multiple alternative promoters, as exemplified by the transcript that encodes let-7a-3 and let-7b. The RefSeq noncoding transcript MIRLET7BHG is shown in blue. (B) Pri-miRNAs may host subsets of clustered miRNAs, as illustrated by transcripts that encode miR-100, let-7a-2, and miR-125b-1. The RefSeq noncoding transcript MIR100HG is shown in blue. (C) miRNAs may span splice sites and thereby may be regulated by alternative splicing. The pri-miRNA that encodes miR-205 is shown as a representative example of this configuration. The RefSeq noncoding transcript MIR205HG is shown in blue.
Transcription of subsets of clustered miRNAs
Many miRNA sequences are clustered in the genome and it is generally assumed that miRNAs that are located within ∼50 kb of one another are cotranscribed as polycistronic transcripts (Baskerville and Bartel 2005; Liang et al. 2007). Unexpectedly, we observed multiple examples of pri-miRNA transcripts that encode subsets of clustered miRNAs (Supplemental Tables S4,S5). The transcripts that host miR-30a and miR-30c-2, described above (Fig. 4), represent examples of this phenomenon. Another interesting example is the miRNA cluster that encodes miR-100, let-7a-2, and miR-125b-1. Notably, the clustering of these miRNAs and even their order in the cluster is conserved between mammals and Drosophila, suggesting that their coordinated regulation has been subject to strong evolutionary selection (Roush and Slack 2008). Our assemblies confirmed the existence of a previously annotated RefSeq transcript, MIR100HG, which encompasses all three human miRNAs in the cluster (Fig. 5B). The 5′ end of this pri-miRNA is supported by H3K4me3 data. In addition, we identified three additional alternative transcription start sites also corroborated by H3K4me3 histone modifications. Use of the most downstream promoter produces a transcript that encodes only miR-125b-1. RT-PCR and 5′ RACE confirmed the accuracy of all of these pri-miRNA transcript assemblies (Supplemental Figs. S12, S13). These findings demonstrate that production of individual miRNAs in polycistronic clusters can be uncoupled through the use of alternative promoters.
Alternative splicing
A previous analysis of existing expressed sequence tags (ESTs) and mRNAs revealed a class of pre-miRNA sequences that span intron–exon junctions such that splicing prevents processing of these miRNA hairpins by the microprocessor complex (Melamed et al. 2013). We were able to confirm the existence of pri-miRNAs with this configuration using our assemblies (Supplemental Table S4). For example, the pre-miR-205 hairpin spans the splice donor site immediately upstream of the final exon of an annotated pri-miRNA, MIR205HG (Fig. 5C). Use of this splice site disrupts the pre-miR-205 sequence and is thus mutually exclusive with production of the mature miRNA. Interestingly, we found alternatively spliced isoforms that utilize a distinct 3′ terminal exon, placing the pre-miRNA hairpin within an intron, a location permissive for miRNA processing. RT-PCR confirmed the use of both alternative terminal exons (Supplemental Fig. S14). These observations lend further support for the regulation of miRNA biogenesis by alternative splicing.
Discussion
Investigation of miRNA functions in numerous biological settings has advanced our understanding of the roles of miRNAs in development and disease and the downstream targets that they regulate (Vidigal and Ventura 2015). On the other hand, considerably less is known about the pathways that govern miRNA biogenesis at transcriptional and post-transcriptional levels. Elucidation of such miRNA regulatory mechanisms has been hindered by the poor annotation of pri-miRNA gene structures. Indeed, a frequent misperception is that miRNA promoters are located in the genomic sequence immediately adjacent to pre-miRNA hairpins when, in fact, these promoters are often located 10's to 100's of kilobases upstream (Cai et al. 2004; Chang et al. 2007). Clearly, dissection of cis- and trans-regulation of miRNA transcription requires an accurate description of the relevant transcription units. Putative post-transcriptional regulatory mechanisms may also be overlooked without an understanding of the splicing patterns or polyadenylation sites of pri-miRNA transcripts. In light of these limitations, we set out to establish a resource of miRNA gene structures that could be easily accessed by investigators in the field in order to improve the study of miRNA regulation. Herein, we describe a novel experimental and computational approach that we developed to achieve this goal.
Having demonstrated that comprehensive pri-miRNA annotation cannot be easily accomplished using existing RNA-seq data, we devised a multi-step strategy to enable genome-wide pri-miRNA reconstruction. First, a dominant-negative DROSHA protein that globally impairs pri-miRNA processing is expressed, thereby stabilizing pri-miRNA transcripts and dramatically improving their coverage in RNA-seq libraries. Next, StringTie, an advanced transcriptome assembler that is capable of accurately reconstructing pri-miRNAs, is used. Since miRNA expression is often cell-type-specific, we applied this strategy to a panel of human and mouse cell lines of diverse origins, thereby successfully annotating ∼70% of pri-miRNAs in these species. We anticipate that near complete assembly of annotated miRNAs is possible by applying this approach to additional cell types.
Multiple lines of evidence support the accuracy of the new pri-miRNA annotations provided here. First, the 5′ ends of the assembled transcripts are frequently located within regions enriched in H3K4me3 histone marks and CpG islands, features that are associated with RNA polymerase II promoters (Mikkelsen et al. 2007). Moreover, we extensively validated new pri-miRNA assemblies using 5′ and 3′ RACE as well as RT-PCR, demonstrating strong concordance between predicted and actual pri-miRNA structures. Additionally, mature miRNAs are highly conserved and we reasoned that their gene structures would tend to be conserved as well. Indeed, in cases where orthologous pri-miRNAs were annotated in human and mouse, we frequently found similar gene structures and promoter locations. Overall, these findings support the reliability of these new pri-miRNA assemblies.
This new map of pri-miRNA structure has revealed previously unrecognized potential regulatory mechanisms for many miRNAs. In particular, we found that alternative promoter usage is a frequent feature of miRNA genes, underscoring the need for a thorough understanding of a given miRNA transcription unit to fully dissect its cis- and trans-regulation. Unexpectedly, we also found several examples of pri-miRNAs that are contiguous with downstream protein-coding genes, suggesting possible coordinated expression. In light of these findings, it will be interesting to investigate whether the miRNAs and proteins encoded by these linked transcripts function within or control common cellular or developmental pathways. In addition, analysis of pri-miRNAs spanning polycistronic clusters revealed that these miRNAs are not always cotranscribed, even in cases where the clustered organization is deeply conserved, such as the miRNA cluster that encodes miR-100, let-7a-2, and miR-125b-1. These results indicate that expression of these apparently linked miRNAs may be uncoupled in some settings. Finally, our data confirm previous analyses that identified miRNAs that span splice sites (Melamed et al. 2013), supporting a role for alternative splicing in regulating the expression of specific miRNAs.
In summary, our results highlight the importance of precise annotation of miRNA gene structures, provide assemblies for a large majority of human and mouse pri-miRNAs, and offer an experimental framework for further reconstruction of the remaining pri-miRNAs yet-to-be described. We anticipate that these annotations will be highly valuable for ongoing efforts to dissect mechanisms of miRNA regulation in diverse biological settings.
Methods
Cell culture
E14TG2a embryonic stem cells were cultured in GMEM with 1% nonessential amino acids, β-mercaptoethanol, and leukocyte inhibitory factor. A-172, A-673, C2C12, HEK293T, Hepa1-6, MCF-7, and MEF cell lines were cultured in DMEM. CT-26 and NCCIT cells were cultured in RPMI 1640. HCT116 cells were cultured in McCoy's 5A. HepG2, human primary fibroblasts, and Neuro-2a were cultured in EMEM. All media was supplemented with 10% fetal bovine serum (FBS) and Antibiotic-Antimycotic.
Plasmids
To generate pcDNA5/FLAG-HA-DGCR8, FLAG-HA-DGCR8 was amplified from pFLAG/HA-DGCR8 (Landthaler et al. 2004) and cloned into the HindIII site of pcDNA5/FRT (Life Technologies). To construct the TN-DROSHA expression plasmid, E1045Q and E1222Q mutations were introduced into pcDNA3.1/V5-His-DROSHA (Rakheja et al. 2014) using the QuikChange Lightning Site-Directed Mutagenesis kit (Stratagene). This plasmid also carries synonymous mutations at codons T438-L444 that render it resistant to commonly used siRNAs. Primer sequences for mutagenesis are provided in Supplemental Table S6.
RNA preparation
Cells were cotransfected with pcDNA3.1/V5-His-TN-DROSHA and pcDNA5/FLAG-HA-DGCR8 under optimized conditions (Supplemental Table S7), and harvested 48 h after transfection. To isolate nuclear RNA, cells were lysed on ice for 5 min in 10 mM Tris-HCl (pH 7.5), 10 mM NaCl, 0.2 mM EDTA, and 0.05% NP-40; and nuclei were spun at 2500g for 3 min and then resuspended in QIAzol for RNA isolation using the miRNeasy kit with DNase I digestion according to the manufacturer's instructions (Qiagen).
RT-PCR, qPCR, and RACE
RNA was reverse-transcribed using the QuantiTect Reverse Transcription Kit (Qiagen) prior to PCR amplification. qPCR was performed using an ABI 7900HT Sequence Detection System with the SYBR Green PCR core reagent kit (Life Technologies). Eukaryotic 18S rRNA endogenous control (Life Technologies) was used as an internal standard. RACE was performed using the GeneRacer kit (Life Technologies). Primer sequences are provided in Supplemental Table S6.
RNA-seq library preparation and sequencing
RNA-seq libraries were generated using the Illumina TruSeq RNA Sample Preparation Kit v2 according to the manufacturer's protocol, and sequenced in one lane of a HiSeq 2000 using the 100-bp paired-end protocol.
Alignment of reads and transcriptome assembly
Reads with a length shorter than 25 nt were first filtered and discarded using fqtrim (http://ccb.jhu.edu/software/fqtrim/index.shtml). The remaining reads were aligned to the human (hg19) or mouse (mm10) reference genome using TopHat2 (Kim et al. 2013). The alignments were assembled using StringTie-v0.97 (Pertea et al. 2015).
fqtrim command line
fqtrim -A -p 5 -l 25 -o trimmed.fq.gz R1.fastq.gz,R2.fastq.gz.
tophat command line
tophat2 -p 10 -o tophat -G known_genes.gff3 --transcriptome-index=./tindex --library-type fr-firststrand hg19 R1.trimmed.fq.gz R2.trimmed.fq.gz >& run.tophat.
stringtie command line
stringtie accepted_hits.bam -p 10 -S -g 0 -f 0.1 -o accepted_hits.gtf.
Data access
The RNA-seq data sets from this study have been submitted to the NCBI Sequence Read Archive (SRA; http://www.ncbi.nlm.nih.gov/sra) under accession number SRP057660. Human and mouse assemblies are available in the Supplemental Data, and can also be viewed in the UCSC Genome Browser using the following link: http://www4.utsouthwestern.edu/mendell-lab/resources.html.
Supplementary Material
Acknowledgments
We thank Vanessa Schmid, Ashley Guzman, and Rachel Bruce in the McDermott Center Next Generation Sequencing Core for assistance with library preparation and high-throughput sequencing, Stephen Johnson for software implementation and computational support, and Kathryn O'Donnell for critical reading of the manuscript. This work was supported by grants from the Cancer Prevention and Research Institute of Texas (CPRIT) (R1008 to J.T.M.) and the National Institutes of Health (NIH) (R01CA120185 and P01CA134292 to J.T.M.; R01HG006677 to S.L.S.). J.T.M. is a CPRIT Scholar in Cancer Research.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.193607.115.
References
- Bartel DP. 2009. MicroRNAs: target recognition and regulatory functions. Cell 136: 215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskerville S, Bartel DP. 2005. Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA 11: 241–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X, Hagedorn CH, Cullen BR. 2004. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA 10: 1957–1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang TC, Wentzel EA, Kent OA, Ramachandran K, Mullendore M, Lee KH, Feldmann G, Yamakuchi M, Ferlito M, Lowenstein CJ, et al. 2007. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell 26: 745–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang HR, Schoenfeld LW, Ruby JG, Auyeung VC, Spies N, Baek D, Johnston WK, Russ C, Luo S, Babiarz JE, et al. 2010. Mammalian microRNAs: experimental evaluation of novel and previously annotated genes. Genes Dev 24: 992–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien CH, Sun YM, Chang WC, Chiang-Hsieh PY, Lee TY, Tsai WC, Horng JT, Tsou AP, Huang HD. 2011. Identifying transcriptional start sites of human microRNAs based on high-throughput sequencing data. Nucleic Acids Res 39: 9345–9356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Leva G, Garofalo M, Croce CM. 2014. MicroRNAs in cancer. Annu Rev Pathol 9: 287–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, Zhang X, Wang L, Issner R, Coyne M, et al. 2011. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature 473: 43–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgakilas G, Vlachos IS, Paraskevopoulou MD, Yang P, Zhang Y, Economides AN, Hatzigeorgiou AG. 2014. microTSS: Accurate microRNA transcription start site identification reveals a significant number of divergent pri-miRNAs. Nat Commun 5: 5700. [DOI] [PubMed] [Google Scholar]
- Gurtan AM, Sharp PA. 2013. The role of miRNAs in regulating gene expression networks. J Mol Biol 425: 3582–3600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Garber M, Levin JZ, Donaghey J, Robinson J, Adiconis X, Fan L, Koziol MJ, Gnirke A, Nusbaum C, et al. 2010. Ab initio reconstruction of cell type–specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat Biotechnol 28: 503–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha M, Kim VN. 2014. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol 15: 509–524. [DOI] [PubMed] [Google Scholar]
- Heo I, Joo C, Cho J, Ha M, Han J, Kim VN. 2008. Lin28 mediates the terminal uridylation of let-7 precursor microRNA. Mol Cell 32: 276–284. [DOI] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozomara A, Griffiths-Jones S. 2014. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res 42: D68–D73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, et al. 2007. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 129: 1401–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landthaler M, Yalcin A, Tuschl T. 2004. The human DiGeorge syndrome critical region gene 8 and its D. melanogaster homolog are required for miRNA biogenesis. Curr Biol 14: 2162–2167. [DOI] [PubMed] [Google Scholar]
- Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN. 2004. MicroRNA genes are transcribed by RNA polymerase II. EMBO J 23: 4051–4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Feng J, Jiang T. 2011. IsoLasso: a LASSO regression approach to RNA-Seq based transcriptome assembly. J Comput Biol 18: 1693–1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y, Ridzon D, Wong L, Chen C. 2007. Characterization of microRNA expression profiles in normal human tissues. BMC Genomics 8: 166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotterman CD, Kent OA, Mendell JT. 2008. Functional integration of microRNAs into oncogenic and tumor suppressor pathways. Cell Cycle 7: 2493–2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsico A, Huska MR, Lasserre J, Hu H, Vucicevic D, Musahl A, Orom U, Vingron M. 2013. PROmiRNA: A new miRNA promoter recognition method uncovers the complex regulation of intronic miRNAs. Genome Biol 14: R84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marson A, Levine SS, Cole MF, Frampton GM, Brambrink T, Johnstone S, Guenther MG, Johnston WK, Wernig M, Newman J, et al. 2008. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell 134: 521–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin JA, Wang Z. 2011. Next-generation transcriptome assembly. Nat Rev Genet 12: 671–682. [DOI] [PubMed] [Google Scholar]
- McGettigan PA. 2013. Transcriptomics in the RNA-seq era. Curr Opin Chem Biol 17: 4–11. [DOI] [PubMed] [Google Scholar]
- Megraw M, Pereira F, Jensen ST, Ohler U, Hatzigeorgiou AG. 2009. A transcription factor affinity-based code for mammalian transcription initiation. Genome Res 19: 644–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melamed Z, Levy A, Ashwal-Fluss R, Lev-Maor G, Mekahel K, Atias N, Gilad S, Sharan R, Levy C, Kadener S, et al. 2013. Alternative splicing regulates biogenesis of miRNAs located across exon-intron junctions. Mol Cell 50: 869–881. [DOI] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. 2007. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448: 553–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olive V, Minella AC, He L. 2015. Outside the coding genome, mammalian microRNAs confer structural and functional complexity. Sci Signal 8: re2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson EN. 2014. MicroRNAs as therapeutic targets and biomarkers of cardiovascular disease. Sci Transl Med 6: 239ps3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozsolak F, Poling LL, Wang Z, Liu H, Liu XS, Roeder RG, Zhang X, Song JS, Fisher DE. 2008. Chromatin structure analyses identify miRNA promoters. Genes Dev 22: 3172–3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquinelli AE. 2012. MicroRNAs and their targets: recognition, regulation and an emerging reciprocal relationship. Nat Rev Genet 13: 271–282. [DOI] [PubMed] [Google Scholar]
- Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, Salzberg SL. 2015. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol 33: 290–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakheja D, Chen KS, Liu Y, Shukla AA, Schmid V, Chang TC, Khokhar S, Wickiser JE, Karandikar NJ, Malter JS, et al. 2014. Somatic mutations in DROSHA and DICER1 impair microRNA biogenesis through distinct mechanisms in Wilms tumours. Nat Commun 2: 4802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez A, Griffiths-Jones S, Ashurst JL, Bradley A. 2004. Identification of mammalian microRNA host genes and transcription units. Genome Res 14: 1902–1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roush S, Slack FJ. 2008. The let-7 family of microRNAs. Trends Cell Biol 18: 505–516. [DOI] [PubMed] [Google Scholar]
- Schanen BC, Li X. 2011. Transcriptional regulation of mammalian miRNA genes. Genomics 97: 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. 2010. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28: 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidigal JA, Ventura A. 2015. The biological functions of miRNAs: lessons from in vivo studies. Trends Cell Biol 25: 137–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter J, Jung S, Keller S, Gregory RI, Diederichs S. 2009. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol 11: 228–234. [DOI] [PubMed] [Google Scholar]
- Xiao Y, Liu T, Zhao H, Li X, Guan J, Xu C, Ping Y, Fan H, Wang L, Zhao T, et al. 2014. Integrating epigenetic marks for identification of transcriptionally active miRNAs. Genomics 104: 70–78. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.