Summary
Microbial inhabitants of the bovine rumen fulfil the majority of the normal caloric requirements of the animal by fermenting lignocellulosic plant polysaccharides and releasing short chain fatty acids that are then metabolized by the host. This process also occurs within the human colon, although the fermentation products contribute less to the overall energy requirements of the host. Mounting evidence, however, indicates that the community structure of the distal gut microbiota is a critical factor that influences the inflammatory potential of the immune system thereby impacting the progression of inflammatory bowel diseases. Non-digestible dietary fibre derived from plant material is highly enriched in the lignocellulosic polysaccharides, cellulose and xylan. Members of the Bacteroidetes constitute a dominant phylum in both the human colonic microbiome and the rumen microbial ecosystem. In the current article, we review recent insights into the molecular mechanisms for xylan degradation by rumen and human commensal members of the Bacteroidetes phylum, and place this information in the context of the physiological and metabolic processes that occur within these complex microbial environments.
Xylan degradation by gut-associated Bacteroidetes
The plant cell wall polysaccharide, xylan, is an abundant substrate supporting microbial fermentation in the digestive tracts of ruminants as well as humans. Xylan is the second most abundant plant cell wall polysaccharide next to cellulose in monocotyledonous grasses, which comprise the primary feedstuff of grazing animals. Cereal grains are a primary component of dietary fibre consumed by humans and possess a very high xylan content (Selvendran, 1984). Xylan consists of a β-1,4-linked xylose backbone which may be substituted at the 2′-OH or 3′-OH with other molecules such as acetyl groups, 4-O-methyl glucuronyl groups, or arabinose. Furthermore, ferulic acid or p-coumaric acid esterified to arabinose moieties may facilitate cross-linking of this polysaccharide to other hemicellulosic polymers or to the structural polymer, lignin. Xylan therefore contains a variety of chemical linkages, and thus its degradation requires a number of different enzymatic activities. These enzymes include endoxylanases, β-xylosidases, α-l-arabinofuranosidases, α-glucuronidases, ferulic acid esterases and acetyl xylan esterases, which have all been recently reviewed in the context of xylan degradation (Dodd and Cann, 2009).
In the bovine rumen approximately 36–79% of the ingested xylan is degraded (Van Soest, 1994). In humans the extent of xylan degradation has not been directly measured, but from studies of total hemicellulose degradation, it is estimated to be between 51% and 72% (Slavin et al., 1981). These studies suggest that xylan degradation is an important process that contributes to the maintenance of microbial communities in the rumen and human colonic ecosystems. Despite this importance, the detailed biochemical processes employed by the resident xylanolytic microbes to acquire energy from recalcitrant xylan polymers are yet to be completely elucidated. Two genera of the phylum Bacteroidetes, the Bacteroides spp. (Chassard et al., 2007) and Prevotella spp. (Bryant et al., 1958; Dehority, 1966) are among the most frequently isolated xylanolytic organisms from human fecal samples and the bovine rumen respectively (Table 1). The rumen Prevotella spp. were first classified as two species, Bacteroides ruminicola ssp. ruminicola and B. ruminicola ssp. brevis; however, as more strains were isolated, it became clear that these organisms differed considerably from the human colonic Bacteroides and thus they were re-classified into the new genus Prevotella (Shah and Collins, 1990). Prevotella ruminicola was later subdivided into four species, P. ruminicola, Prevotella bryantii, Prevotella albensis and Prevotella brevis on the basis of carbohydrate utilization patterns and genotypic characteristics (Avgustin et al., 1997). Recently, the analysis of xylanolytic bacteria in the human colon has been revisited (Chassard et al., 2007) and xylanolytic Bacteroides spp. previously thought to be dominant xylan utilizers, such as B. ovatus and B. fragilis ssp. A (Salyers et al., 1977; Hespell and Whitehead, 1990), were not isolated. Additional xylanolytic members of this genus were, however, identified and these included B. intestinalis DSM 17393 (Bakir et al., 2006), B. cellulosilyticus DSM 14838 (Robert et al., 2007) and B. xylanisolvens XB1A (Chassard et al., 2008). These studies revealed that the xylan degrading community within the human colon is more complex than previously envisioned and although these studies might be biased by the culture-based techniques employed, the results clearly indicate that the Bacteroidetes represent a significant xylanolytic group within the human colonic microbial ecosystem.
Table 1.
Xylanolytic gut Bacteroidetes.
| Organism | Gut environment | Type of xylan fermenteda | Genome (GenBank Accession No.) |
|---|---|---|---|
| B. cellulosilyticus DSM 14838 | Colon | OSX (Robert et al., 2007) | Draft (NZ_ACCH00000000) |
| B. eggerthii DSM 20697 | Colon | N.S. (Salyers et al., 1977) | Draft (NZ_ABVO00000000) |
| B. fragilis subsp. A b | Colon | N.S. (Salyers et al., 1977) | No |
| B. intestinalis DSM 17393 | Colon | OSX (Robert et al., 2007) | Draft (NZ_ABJL00000000) |
| B. ovatus ATCC 8483 | Colon | OSX (Weaver et al., 1992) | Draft (NZ_AAXF00000000) |
| B. fragilis ANH 11543 | Colon | RAX (Crittenden et al., 2002) | No |
| B. fragilis RHI 3001 | Colon | XOS, RAX (Crittenden et al., 2002) | No |
| B. fragilis AHN 2981 | Colon | RAX (Crittenden et al., 2002) | No |
| B. fragilis AHN 2898 | Colon | RAX (Crittenden et al., 2002) | No |
| B. thetaiotaomicron RHI 4171 | Colon | XOS, RAX (Crittenden et al., 2002) | No |
| B. thetaiotaomicron AHN 1368 | Colon | RAX (Crittenden et al., 2002) | No |
| B. vulgatus RHI 3621 | Colon | RAX (Crittenden et al., 2002) | No |
| B. vulgatus AHN 413 | Colon | RAX (Crittenden et al., 2002) | No |
| B. xylanisolvens XB1A | Colon | OSX, BWX (Chassard et al., 2008) | Draft (FP929033) |
| P. albensis M384 | Rumen | OSX (Matsui et al., 2000) | No |
| P. bryantii B14 | Rumen | OSX (Miyazaki et al., 1997, Matsui et al., 2000), BWX (Miyazaki et al., 1997), WAX (Dodd et al., 2010b) | Draft (NZ_ADWO00000000) |
| P. ruminicola 23 | Rumen | N.S. (Bryant et al., 1958), OSX (Matsui et al., 2000) | Complete (NC_014033) |
| P. ruminicola D31d | Rumen | OSX (Hespell and Whitehead, 1990) | No |
Abbreviations are: N.S., not specified; OSX, oat spelt xylan; RAX, rye arabinoxylan; WAX, wheat arabinoxylan; BWX, birchwood xylan; XOS, xylo-oligosaccharides.
Only certain strains of these species which were tested ferment xylan.
Genes encoding enzymes involved in xylan degradation have been identified previously in Bacteroidetes members from the rumen, including P. ruminicola 23, P. ruminicola D31d and P. bryantii B14, as well as from the human colon such as Bacteroides ovatus V975 and Bacteroides xylanisolvens XB1A (Table 2). Of these xylanolytic bacteria, P. bryantii B14 and B. ovatus 0038 (ATCC 8483) are the best studied organisms. Flint and colleagues identified a gene cluster in the genome of P. bryantii B14, which contained β-xylosidase and endoxylanase genes as well as other genes predicted to encode proteins with roles in xylan degradation, including a putative xylose/sodium symporter, a putative acetylxylan esterase and a putative α-glucuronidase (Gasparic et al., 1995a). The expression of these genes is controlled at the transcriptional level by a hybrid two-component system (HTCS) regulator (XynR) (Miyazaki et al., 2003).
Table 2.
Summary of xylanolytic enzymes characterized from gut-associated Bacteroidetes.
| Gene | Organism | CAZy familya,b | Enzymatic activity (reference) | Notesc |
|---|---|---|---|---|
| xyn5A | B. eggerthii DSM 20697 | GH5 | endoxylanase (Dodd et al., 2010a) | Part of a xylan utilization cluster |
| xyn5A | B. intestinalis DSM 17393 | GH5 | endoxylanase (Dodd et al., 2010a) | Part of a xylan utilization cluster |
| xyn5B | B. intestinalis DSM 17393 | GH5/GH43 | endoxylanase (Dodd et al., 2010a) | Part of a xylan utilization cluster |
| xyll | B. ovatus V975 | GH10 | endoxylanase (Whitehead, 1995) | Part of a xylan utilization cluster |
| xsa | B. ovatus V975 | GH43 | β-xylosidase/α-L-arabinofuranosidase (Whitehead, 1995) | Part of a xylan utilization cluster |
| xyn10A | B. xylanisolvens XB1A | GH10 | endoxylanase (Mirande et al., 2010) | |
| xyl3A | P. bryantii B14 | GH3 | β-xylosidase (Dodd et al., 2010b) | Possesses a PA14 domain |
| xyl3B | P. bryantii B14 | GH3 | β-xylosidase (Dodd et al., 2010b) | |
| xyl3C | P. bryantii B14 | GH3 | β-xylosidase (Dodd et al., 2010b) | Possesses a PA14 domain |
| xyn5A | P. bryantii B14 | GH5 | endoxylanase (Dodd et al., 2010a) | Part of a xylan utilization cluster |
| xyn5B | P. bryantii B14 | GH5 | endoxylanase (Dodd et al., 2010a) | Part of a xylan utilization cluster |
| xynA | P. bryantii B14 | GH10 | endoxylanase (Gasparic et al., 1995a) | Part of a xylan utilization cluster |
| xynC | P. bryantii B14 | GH10 | endoxylanase (Flint et al., 1997) | Possesses a putative CBM 4 domain |
| xynB | P. bryantii B14 | GH43 | β-xylosidase/α-L-arabinofuranosidase (Gasparic et al., 1995a) | Part of a xylan utilization cluster |
| agu67A | P. bryantii B14 | GH67 | α-glucuronidase (Dodd et al., 2010b) | |
| xyl3A | P. ruminicola 23 | GH3 | β-xylosidase (Dodd et al., 2009) | Possesses a PA14 domain |
| xyn10D-fae1A | P. ruminicola 23 | GH10/CE1 | endoxylanase/ferulic acid esterase (Dodd et al., 2009) | Two domain bifunctional xylanase-esterase protein |
| xyn10A | P. ruminicola D31d | GH10 | endoxylanase (Flint et al., 1997) | Possesses a putative CBM 4 |
Family designations were made using the Carbohydrate Active Enzymes (CAZy) database (http://www.cazy.org/).
GH, glycoside hydrolase; CE, carbohydrate esterase.
PA14 domains are β-sheet protein domains involved in carbohydrate binding (Yoshida et al., 2010).
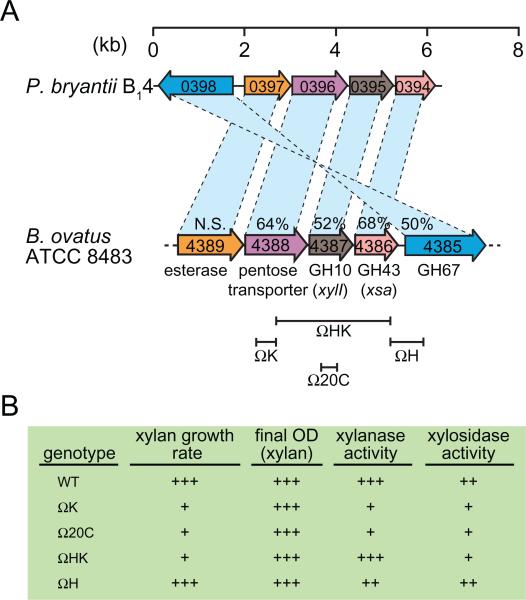
The arrangement of this gene cluster in P. bryantii B14 exhibits similarities to a gene cluster that was previously cloned from the B. ovatus ATCC 8483 chromosome (Fig. 1A) (Whitehead and Hespell, 1990). To analyse the importance of this gene cluster for xylan utilization, four different segments of the DNA fragment containing the gene cluster were subsequently cloned into pBT-2, a suicide vector, and used to construct disruptions in the B. ovatus ATCC 8483 chromosome (Fig. 1A) (Weaver et al., 1992). Insertion of this plasmid results in a duplication of the integrated region, which then flanks the vector sequence, which has an approximate length of 14 kb. Insertion of pBT-2 in the gene immediately upstream of xylI (mutant ΩK, Fig. 1A) led to a reduction in the growth rate with hot-water extracted xylans from oat spelts and a decrease in xylanase, xylosidase and arabinosidase activities in cell extracts (Fig. 1B). Insertion of pBT-2 within xylI (mutant Ω20C, Fig. 1A) also resulted in a reduction in growth rate with xylan and decreases in xylanase and xylosidase activities; however the arabinosidase activity was comparable to that of the wild-type strain. The observation that the ΩK mutant exhibited reduced arabinosidase activity whereas the Ω20C activity was comparable to the wild-type indicated both that xsa does not encode the only arabinosidase enzyme in B. ovatus ATCC 8483 and that the gene immediately upstream of xylI (a putative pentose transporter) may influence arabinosidase expression.
Fig. 1.
A. Conservation of xylan hydrolase gene clusters in B. ovatus ATCC 8483 and P. bryantii B14. The corresponding genes in the two bacteria are connected by dashed lines, and the per cent amino acid identity between the gene products is indicated. Open reading frame numbers are indicated within each of the genes. N.S., not significant amino acid identity. A map of insertional mutants constructed in the chromosome of B. ovatus ATCC 8483, as described in the text, is indicated below the corresponding genes.
B. Phenotypes associated with the insertional mutants indicated in (A) and described in the text. Data are derived from Weaver et al. (1992). OD, optical density.
The Ω20C mutant exhibited a growth rate defect, thus to evaluate whether xylI or xsa was responsible for this phenotype, a mutant was generated that resulted in a disruption of the distal 300 nucleotides of the xsa gene (mutant ΩHK, Fig. 1A). Mutant ΩHK exhibited xylanase and arabinosidase activities comparable to the wild-type strain, however the xylosidase activity was lower and a growth rate defect with xylan was observed. Since disruption of the gene downstream of xsa did not yield a discernable phenotype (mutant ΩH, Fig. 1A), the authors concluded that disruption of xsa alone was responsible for the growth rate defect with xylan as a carbon source. Thus, the degradation of xylo-oligosaccharides to xylose by Xsa appears to be the rate-limiting step for xylan utilization by B. ovatus ATCC 8483.
Although several of these mutants exhibited growth rate defects with xylan, they eventually reached the same cell density as the wild-type and the overall extent of xylan degradation in mutant ΩK was similar to the wild-type. These observations indicate that compensatory mechanisms exist within B. ovatus ATCC 8483 to permit complete utilization of xylan in the absence of xylI, xsa, as well as the putative pentose transporter located upstream of xylI. Indeed, the recent genome sequence for B. ovatus ATCC 8483 has revealed the presence of five putative GH10 endo-xylanase genes and ~13 putative β-xylosidase genes, which indicates that the organism possesses additional enzymes that participate in xylan utilization. Furthermore, it is noteworthy that a second gene cluster that contains homologues of xylI, xsa and the pentose transporter (BACOVA_00487, BACOVA_00488, BACOVA_00489) is present in the genome in the same orientation (data not shown).
Sus-like genes are involved in xylan utilization by gut Bacteroidetes
Xylanase activity is cell-associated in the rumen bacterium P. bryantii B14 (Miyazaki et al., 1997) as well as the human colonic bacteria, B. eggerthii and B. ovatus (Salyers et al., 1982; Weaver et al., 1992). This raises an important question of how these bacteria efficiently utilize large polysaccharides in the apparent absence of an extensive extracellular xylanase system. The answer to this question may lie in the Bacteroides thetaiotaomicron starch utilization system (Sus), which was discovered and characterized by Salyers and colleagues and has recently been reviewed (Martens et al., 2009a). The Sus system includes four proteins, which are anchored on the outer leaflet of the outer membrane (SusDEFG) (Shipman et al., 2000), and a TonB-dependent receptor protein (SusC) (Reeves et al., 1997), which spans the outer membrane and couples oligosaccharide transport to the proton motive force. Four of these proteins (SusCDEF) are involved in the cellular attachment to starch (Shipman et al., 2000), and SusG exhibits α-amylase activity but is not essential for starch binding (Shipman et al., 1999). Despite the observation that SusG is not essential for starch binding by B. thetaiotaomicron, recent crystallo-graphic analysis of this enzyme has revealed the presence of two previously undetected carbohydrate binding sites, including a CBM 58 and a secondary surface starch binding site (Koropatkin and Smith, 2010). Two additional proteins, SusA and SusB, are located within the periplasm (Shipman et al., 2000) and have neopullulanase and α-glucosidase activities (D'Elia and Salyers, 1996a), which are involved in the degradation of starch oligosaccharides. Expression of the sus genes is controlled at the transcriptional level by SusR (D'Elia and Salyers, 1996b) and MalR (Cho et al., 2001), and deletion of both of these genes completely attenuates the expression of starch utilization genes (Cho et al., 2001). The Sus system therefore requires only a single extracellular endo-acting enzyme (SusG) that is closely associated with the outer membrane transport apparatus (SusCD). Through this close interaction, oligosaccharides that are released from long starch polymers can be immediately sequestered into the periplasm, where they are further degraded into monosaccharides by SusA and SusB for subsequent transport into the cytoplasm, the site for fermentation. This mechanism minimizes the loss of monosaccharides to other bacteria within the colon and represents an evolutionary adaptation that permits the Bacteroides spp. to effectively compete in the human colonic ecosystem.
Recently, a number of genome sequences for Prevotella spp. and Bacteroides spp. have been made available through the Human Microbiome Project (HMP) (Turn-baugh et al., 2007) as well as the North American Consortium for Genomics of Fibrolytic Rumen Bacteria (Morrison et al., 2009; Purushe et al., 2010). These genome sequences have revealed that gut associated Bacteroidetes possess an expansive repertoire of genes predicted to encode carbohydrate active enzymes (Nelson et al., 2010). Furthermore, both the human colonic Bacteroides spp. and the rumen Prevotella spp. exhibit a greatly expanded set of starch utilization system homologues, specifically homologues for SusC and SusD (Xu et al., 2003; 2007; Martens et al., 2009a). Many of these SusC- and SusD-like genes cluster in the genome with genes that are predicted to be involved in the metabolism of various polysaccharides. This observation suggests that the SusC- and SusD-like genes have expanded through gene duplication and then diverged to participate in the utilization of various dietary and host-derived polysaccharides (Martens et al., 2009a).
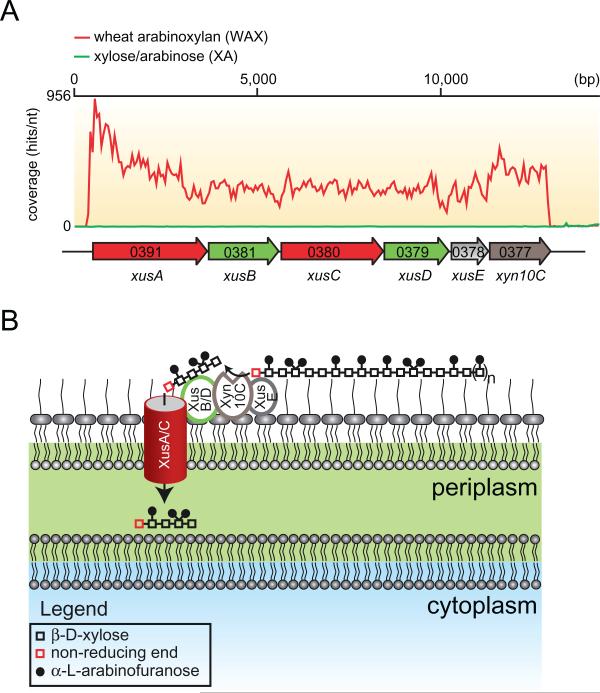
In B. thetaiotaomicron VPI-5482, approximately 88 polysaccharide utilization loci (PULs) (Bjursell et al., 2006) that contain genes encoding SusC/SusD homologues as well as putative carbohydrate active enzymes are present. The proteins encoded by the genes at these loci are predicted to play roles in the metabolism of host-derived as well as dietary polysaccharides. Deletion of the five regulatory genes, which control the expression of PULs associated with mucin O-glycan utilization in B. thetaiotaomicron, resulted in impairment of persistence in mice as well as mother to pup transmission of these bacteria (Martens et al., 2008). These studies revealed that degradation of host-derived saccharides by these PULs, which contain one SusC/SusD gene pair per locus, is important for the persistence of these bacteria in vivo. A recent functional genomics study of the rumen bacterium, P. bryantii B14 revealed that an operon of six genes including two tandem repeats of SusC/SusD homologues were the most highly induced genes during growth with soluble wheat arabinoxylan relative to the component monosaccharides, xylose and arabinose (Dodd et al., 2010a) (Fig. 2A). The xylan-specific induction coupled with the high conservation of this cluster among other xylanolytic Bacteroidetes indicates that these genes are important for xylan utilization, thus they are termed members of the xylan utilization system (Xus). The two repeats of SusC and SusD homologues (xusA, xusB, xusC, xusD) are followed by a hypothetical protein encoding gene (xusE), and then followed by an endoxylanase encoding gene (xyn10C). The function of XusE is unknown; however the presence of an N-terminal signal peptidase II cleavage site supports the prediction that this protein is anchored onto the outside leaflet of the outer membrane. The gene xyn10C encodes a GH family 10 endoxylanase (Flint et al., 1997), which is interrupted by a carbohydrate binding module (CBM) family 4 that is homologous to the CBM 4 domain in Xyn10A from Rhodothermus marinus (Simpson et al., 2002). Furthermore, this protein possesses an N-terminal signal peptidase II cleavage site similar to XusE, which indicates that this protein may also be anchored on the outer leaflet of the outer membrane. Thus, Xyn10C may represent a functional homologue of the B. thetaiotaomicron SusG protein, catalysing the extracellular cleavage of xylan polymers to oligosaccha-rides that may be subsequently transported into the cell (Fig. 2B).
Fig. 2.
Model for the xylan utilization system in xylanolytic Bacteroidetes.
A. RNAseq coverage map of the major xylan utilization system in P. bryantii B14 during growth on soluble wheat arabinoxylan (WAX, red line) or a mixture of xylose and arabinose (XA, green line). Total RNA was extracted from P. bryantii B14 cultured on either growth substrate (WAX or XA), and then rRNA was subtracted and the enriched mRNA was converted to cDNA. The cDNA was directly sequenced by Illumina technology and individual sequence reads were assembled onto the genome sequence of P. bryantii B14. These data are derived from Dodd et al. (2010a).
B. Predicted model for binding of xylan, cleavage and transport of xylan fragments across the outer membrane by components of the Xus cluster in P. bryantii B14. The proteins, XusA and XusC, are homologues of the SusC TonB-dependent receptor, which is involved in oligosaccharide transport across the outer membrane in B. thetaiotaomicron VPI-5482. XusB and XusD are homologues of B. thetaiotaomicron VPI-5482 SusD, which binds polysaccharides on the outer leaflet of the outer membrane. XusE has no homology to other characterized proteins, but the presence of an N-terminal signal peptidase II cleavage site in this protein suggests that it is tethered to the outer leaflet of the outermembrane. Xyn10C is an endoxylanase gene that also possesses a putative N-terminal signal peptidase cleavage II site. In the predicted model, XusE, Xyn10C and the XusB/D proteins bind to extracellular xylan polymers. Xyn10C then catalyses the endo-cleavage of these polymers and XusB/D facilitate transport of these fragments across the outer membrane and into the periplasm through the TonB-dependent receptors XusA and XusC.
The presence of Sus homologues in this gene cluster and the fact that these genes are highly induced during growth on xylan indicates that this gene cluster is adapted to the acquisition of energy from xylan polysaccharides. This gene cluster represents a specific example of the Sus paradigm for nutrient acquisition, which is common among many environmental as well as gut-associated organisms within the phylum Bacteroidetes (Martens et al., 2009a). Analysis of the genome sequences for several Bacteroides spp. and Prevotella spp. revealed that this cluster of genes is highly conserved among Bacteroidetes members that are known to be xylanolytic (Dodd et al., 2010a). Moreover, this gene cluster is present in several members of the phylum Bacteroidetes that have not been assessed for the ability to degrade xylan (data not shown), and this may be predictive of their potential to degrade this polymer. These observations are highly suggestive that xylan degradation is relatively wide-spread among the human-associated Bacteroides and Prevotella spp.
Multiplicity of xylanolytic enzymes in gut Bacteroidetes
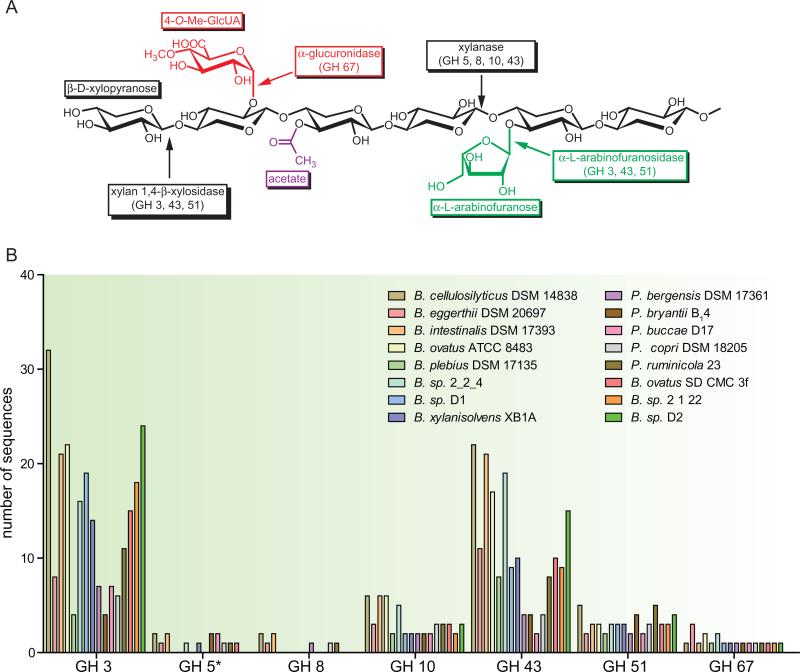
The Xus genes described above may comprise the core, conserved set of genes required for extracellular outer membrane-associated binding of xylan and transport of xylan fragments by gut-associated Bacteroidetes. However, due to the structural complexity of xylans, additional enzymes are required to breakdown this substrate into fermentable monosaccharides (Fig. 3A). The availability of genomic sequences in the publicly available databases provides us with resources to examine how well-equipped members of the phylum Bacteroidetes are to completely release the fermentable sugars from complex hemicelluloses. The molecular scissors required for degradation of these complex polysaccharides likely derive from glycoside hydrolase (GH) enzyme families. The GH enzymes are grouped into different families based on their three-dimensional folds and their catalytic mechanisms (Cantarel et al., 2009). Within many GH families, the protein domain represents a scaffold in which different substrate specificities may evolve, provided that the correct positioning of catalytic residues is maintained. Although bioinformatic approaches are largely successful at assigning GH genes into appropriate families, attempting to infer substrate specificity and thus physiological function by sequence alone represents a significant challenge.
Fig. 3.
Presence of carbohydrate active enzymes associated with xylan degradation in Bacteroidetes genomes that possess a Xus gene cluster.
A. The general structure for xylan is shown and the relevant glycoside hydrolase (GH) enzymes are indicated. Only the GH families that are present within the genome sequences of Xus-containing Bacteroidetes are shown.
B. Frequency distribution plot for the GH families from panel A in xylanolytic Bacteroidetes. For GH 5, only those members with high homology [expect (E) value < 1 × 10−15] to the Bacteroidetes-specific endoxylanase enzymes identified by Dodd et al. (2010a) are included. With the exception of Prevotella ruminicola 23, which has a complete genome, all numbers of genes are estimates based on the analysis of data from partially sequenced genomes. Analyses were performed by identifying a biochemically verified member from each GH family on the CAZy website, and using it as a query for a BLASTp search of the listed bacterial genome. Only those results that exhibited E-values < 1 × 10−5 were included.
This is particularly relevant to the saccharolytic gut Bacteroidetes whose genomes exhibit a highly expanded repertoire of glycolytic enzymes. Many of these bacterial genomes contain multiple copies of members of a particular GH family and this raises the important question of whether these genes code for enzymes with redundant activities or whether each of these genes encodes an enzyme with a unique functional role within the cell. GH enzymes with similar biochemical activities may derive from different families and the major xylanolytic enzymes are classified as follows: endoxylanases (GH 5, 7, 8, 10, 11, 43), β-xylosidases (GH 3, 39, 43, 52, 54), α-l-arabinofuranosidases (GH 3, 43, 51, 54, 62) and α-glucuronidases (GH 67, 115). Bacteroidetes members that harbour a Xus gene cluster also possess a large number of genes that derive from these families (Fig. 3B). The most highly represented families include GH family 3 and GH family 43, with Bacteroides intestinalis DSM 17393 possessing 32 GH family 3 genes and 22 GH family 43 genes. Metagenomic studies have also confirmed that these two families are among the most abundant microbiota-specific GH families (Gill et al., 2006).
The question then arises; are any of these GH3 or GH43 genes involved with xylan degradation, and if so, how many? A recent study addressed this question by evaluating the role of four GH family 3 genes in P. bryantii B14. Each of these genes was cloned, expressed as a recombinant protein in E. coli, and the biochemical properties were evaluated with a range of oligo- and polysaccharides. These studies revealed that one of the proteins (CdxA) has β-glucosidase and cellodextrinase activities, whereas the three remaining GH3 enzymes (Xyl3A, Xyl3B and Xyl3C) each possesses β-xylosidase activity (Dodd et al., 2010b). Furthermore, transcriptional analysis of P. bryantii B14 revealed that expression of xyl3A was induced during growth on arabinoxylan relative to glucose, whereas the expression of xyl3B and xyl3C was constitutive. Analysis of enzymatic activity with substituted xylo-oligosaccharides revealed that Xyl3C exhibits a different cleavage pattern during the hydrolysis of 4-O-methyl glucuronyl substituted xylo-oligosaccharides relative to Xyl3B, suggesting a fundamental difference in substrate specificity between these enzymes. These results provide evidence that these three β-xylosidases possess different transcriptional and biochemical profiles and support the prediction that each enzyme fulfils a unique role in xylan degradation within the cell.
The findings above have important consequences for analysing the metabolic potential of the gut microbiota and suggest that many glycolytic enzymes have evolved not only to cleave a specific chemical linkage, but also to recognize the structural context in which the linkage occurs. This added layer of complexity represents a considerable limitation to our ability to correctly assign functions of GH enzymes using bioinformatics alone. Emphasis should therefore be placed on studies that incorporate functional genomics as well as detailed biochemical approaches for assigning functions to glycolytic enzymes.
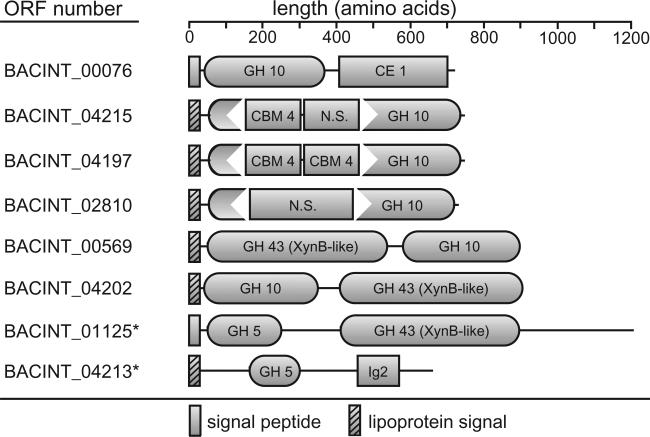
The genome sequence of B. intestinalis DSM 17393 harbours eight different putative endoxylanase genes, six from GH family 10 and two biochemically verified GH family 5 endoxylanases (Dodd et al., 2010a) (Fig. 4). All of these genes exhibit unique domain architecture, which indicates that these proteins may serve different functions. This is the first evidence of a human gut bacterium with such an impressive arsenal of endoxylanase enzymes that are linked to such a variety of catalytic domains and carbohydrate binding modules. Furthermore, six of these eight proteins possess putative signal peptidase II cleavage sites, suggesting that the cell surface of this bacterium is decorated with a number of different endoxylanase enzymes during xylan metabolism. Several of these endoxylanase genes encode GH10 catalytic modules that are interrupted with CBM4 domains and this may influence the substrate specificity of these enzymes. Indeed, Flint and colleagues identified different xylo-oligosaccharide product distribution profiles for Xyn10A (no CBM domain) and Xyn10C (putative CBM4 domain) cloned from P. bryantii B14 when incubated with intact xylans. Xyn10A produced shorter xylo-oligosaccharide products (Gasparic et al., 1995b), whereas Xyn10C produced longer xylo-oligosaccharides (Flint et al., 1997), indicating that the domain architecture for these proteins may influence product distribution profiles. Taken together, these observations suggest that each of the B. intestinalis DSM 17393 endoxylanases possess distinct substrate specificities or product distribution profiles and collectively, this large number of enzymes may improve the digestibility of a wide variety of xylans from different dietary sources.
Fig. 4.
Expansion of xylanase enzymes in the genome of Bacteroides intestinalis DSM 17393. Putative GH family 10 xylanase enzymes were identified in the B. intestinalis genome using P. bryantii B14 Xyn10A (Gasparic et al., 1995b) as a query in a BLASTp search of the genome. Domain architectures were predicted using the Conserved Domains Database (CDD) on the NCBI website (Marchler-Bauer et al., 2007). Domains were included if the E-value was less than 1 × 10−5. The two biochemically verified (Dodd et al., 2010a) GH 5 endoxylanases from B. intestinalis DSM 17393 are also included and indicated by asterisks (BACINT_01125 and BACINT_04213). Signal peptides and lipoprotein signal sequences were predicted using SignalP v3.0 (Emanuelsson et al., 2007) and LipoP v1.0 (Juncker et al., 2003) respectively. N.S., no significant match to any domain within the CD database.
Co-ordinated regulation of xylanolytic genes by XynR
Of the xylan degrading Bacteroidetes described to date, the xylanolytic system of the rumen bacterium P. bryantii B14 is the best studied. This bacterium grows efficiently on soluble xylan fragments (Miyazaki et al., 1997; 2005) and responds at the transcriptional level to medium to large-sized xylan fragments via the HTCS regulator, XynR (Miyazaki et al., 2003). This HTCS regulator is also conserved in the genomes of all other xylanolytic Bacteroidetes that contain Xus clusters.
The XynR of P. bryantii B14 is a large protein of about 1369 amino acids and contains multiple domains, including a putative periplasmic sensing region as well as all of the signal transduction machinery required to initiate a transcriptional response to an environmental signal. XynR possesses three putative transmembrane helices, which predict that the N-terminal residues 21–750 are periplasmic and the C-terminal residues 770–1336 are cytoplasmic, while the final 12 amino acids are either membrane associated or periplasmic. There are many protein sequences, which exhibit homology to the N-terminal periplasmic region of XynR in the GenBank database. Most of the homologous proteins derive from gut-associated Bacteroidetes. However, none of these proteins has demonstrated function. The N-terminal periplasmic region is grouped within the Cluster of Orthologous Groups (COG) of proteins number 3292, which according to the NCBI Conserved Domains Database (CDD) contains approximately 3854 members. No domain hits exist within the Pfam database, and a BLASTp search of the RCSB Protein Data Bank (PDB) retrieved no significant hits.
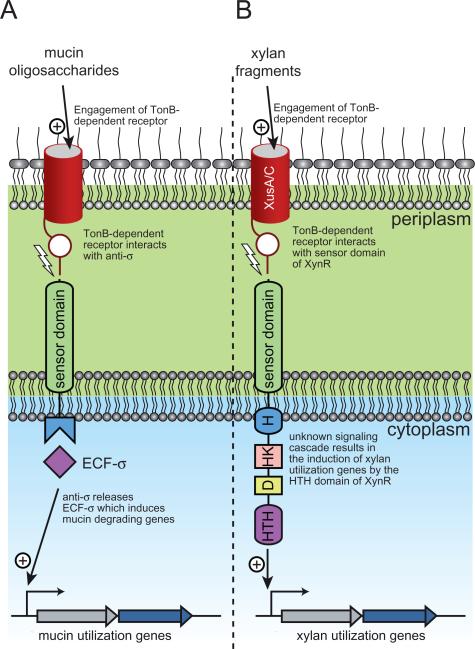
Recently, the periplasmic sensing domain for an HTCS that controls expression of a fructan utilization locus in B. thetaiotaomicron (BT1754) was crystallized and shown to bind to the monosaccharide, fructose, which is the end-product of fructan degradation (Sonnenburg et al., 2010). While the C-terminal region of XynR, which contains the histidine kinase and response regulator elements, shares high amino acid sequence identity with the C-terminal region of BT1754, the N-terminal periplasmic regions of these proteins are not related. This observation suggests a fundamental difference in the sensing mechanism between these two proteins. It is unlikely that XynR binds to monomeric xylan degradation products, as previous studies revealed that induction of xylanase activity is unaffected by xylose, arabinose or glucuronic acid. Xylanase activity was rather specifically induced in the presence of medium to large xylan fragments (Miyazaki et al., 2005). It is possible that the periplasmic region of XynR binds directly to xylan fragments that are transported into the periplasmic space. However, previous data indicate that xylanase induction is low even in the presence of xylo-oligosaccharides with degree of polymerization (DP) up to five, and that substitutions on the xylan chain are not critical for xylanase induction (Miyazaki et al., 1997; 2005). Therefore to initiate signalling, the transported xylan fragments would have to be rather large. It is currently unknown whether xylan fragments of sufficient size can be transported across the outer membrane via XusA or XusC, and if transported, whether periplasmic degradation by constitutively expressed β-xylosidase enzymes might precede the recognition of these fragments by XynR. Another possibility is that extracellular engagement of xylan fragments by the outer membrane associated Xus apparatus may initiate a signalling event that is transmitted to the periplasmic region of XynR through a protein–protein interaction. This could potentially be mediated through one of the two TonB-dependent receptors, XusA or XusC, which are predicted to possess periplasmic N-terminal amino acid extensions. Indeed, it has been shown that TonB-dependent receptors in the related bacterium, B. thetaiotaomicron VPI-5482, transduce signals from the outside of the cell into the cytoplasm via protein–protein interactions between the N-terminal region of TonB-dependent receptor domain and an anti-σ factor (Fig. 5A) (Martens et al., 2009b). For the prototypical E. coli Fec system, this trans-membrane signalling leads to a conformational change in the cytoplasmic region of the inner membrane spanning anti-σ factor causing release of the ECF-σ factor, which then potentiates an appropriate transcriptional response (Braun and Mahren, 2005). While a similar protein–protein interaction between the periplasmic region of XynR and the N-terminal region of XusA or XusC would be a very interesting paradigm for transcriptional control of xylan degrading genes in the Bacteroidetes (Fig. 5B), future studies will need to be performed to evaluate this hypothesis.
Fig. 5.
Model for signal transduction in gut Bacteroidetes.
A. Mechanism revealed by Martens et al., (2009b) for trans-envelope signalling in response to mucin oligosaccharide degradation by Bacteroides thetaiotaomicron VPI-5482. Mucin oligosaccharides engage a TonB-dependent receptor, which releases the N-terminal plug domain of the receptor. The N-terminal extension of the TonB receptor is then able to interact with the sensor domain of an anti-σ factor. This interaction causes the anti-σ to release the ECF-σ that was being sequestered in the cytoplasm and is now free to induce the transcription of mucin utilization genes.
B. Potential model for xylan signal recognition involving XusC/D and XynR. Extracellular xylan fragments engage the TonB-dependent receptors XusC and/or XusD and this triggers release of the N-terminal plug domain of the receptor. The N-terminal extension of XusC/D is then able to interact with the sensor domain of XynR. An unknown signalling cascade, probably involving the histidine kinase (H and HK) and response regulator (D and HTH) domains, activates the C-terminal helix-turn-helix (HTH) domain of XynR, allowing it to bind to regulatory elements and thus inducing transcription of xylan utilization genes.
Of particular note, the transcription of several genes involved in the utilization of chondroitin sulfate by B. thetaiotaomicron VPI5482 is induced by oligosaccharides with DP of eight or higher (Salyers and Kotarski, 1980). This system also contains an HTCS that is homologous to XynR (27% amino acid identity over entire polypeptide sequence). Therefore, there may be parallels between these two systems in their mode of regulation. Irrespective of how the signal is sensed, these data clearly suggest that P. bryantii B14 (and by extension other XynR-containing Bacteroidetes) has evolved an elaborate mechanism for specifically recognizing the intact structure of xylan. This is of particular physiological significance in the gut environment because polysaccharides from various plant sources or tissues, such as arabinan and xyloglucan, contain arabinose or xylose in linkages that are inaccessible to xylanolytic enzyme systems. Thus sensing intact β-1,4-linked xylan fragments ensures that the ensuing transcriptional response is appropriate and only those enzymes required for efficient utilization of xylan are produced.
Unresolved issues in xylan utilization by gut Bacteroidetes
Recent genome sequencing of xylanolytic gut bacteria has provided unprecedented insight into the mechanisms that members of the Bacteroidetes phylum might employ to harvest energy from xylan. Although yet to be fully investigated, the xylan degrading systems of these gut bacteria promise to be much more complex than previously envisaged. Furthermore, the molecular machinery used to capture energy from xylan is analogous to the Sus-like paradigm, which is common to other glycan assimilating pathways in these organisms (Martens et al., 2009a). In addition to providing an immense wealth of information to the scientific community, it is also clear that the genome sequences of the gut Bacteroidetes have raised many more questions than answers. Thus, there are several unresolved issues, especially with respect to xylan utilization and an elaboration is provided below.
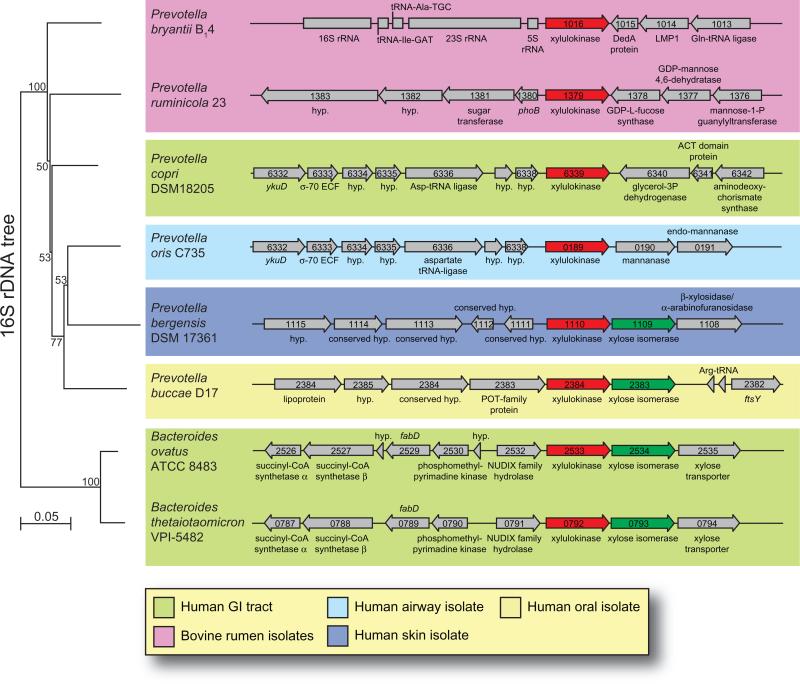
How do certain Prevotella spp. metabolize xylose in the apparent absence of a xylose isomerase gene? This is an important question that is fundamental to our basic understanding of microbial fermentation by these gut bacteria. There are two major pathways for xylose utilization, the xylose isomerase pathway, which is common among bacteria, and the redox pathway, which is common among eukaryotes (Jeffries, 2006). In the bacterial pathway, xylose is first converted to xylulose by the action of xylose isomerase (E.C. 5.3.1.5) and then xylulokinase (E.C. 2.7.1.17) phosphorylates xylulose to produce xylulose-5-phosphate, which may enter the pentose phosphate pathway. Of the xylanolytic gut Bacteroidetes sequenced to date, most possess a conserved gene cluster containing a xylulokinase gene followed by a xylose isomerase gene (Fig. 6, lower half of figure). In contrast, four recently sequenced Bacteroidetes members are apparently missing the xylose isomerase gene from this cluster (Fig. 6, upper half of figure). These organisms include the rumen bacteria, P. bryantii B14, P. ruminicola 23 and the human-associated bacteria, Prevotella copri DSM 18205 and Prevotella oris C735. The genome sequence for P. ruminicola 23 is complete or closed, whereas the genomes for P. bryantii, P. copri and P. oris are only partially sequenced. Analysis of the region immediately downstream of the xylulokinase gene for these three partially sequenced organisms revealed no large gaps in the DNA sequence, indicating the absence of a xylose isomerase gene. This observation suggests that these four Prevotella spp. exhibit a fundamental difference in the genomic organization of xylose-metabolizing genes compared with other Bacteroidetes (Fig. 5). BLASTp searches of the genome sequences for these four bacteria using the xylose isomerase from B. thetaiotaomicron VPI-5482 (BT_0793) as a query returned no significant matches. Further analyses of the genome sequence of these bacteria did not yield any candidate xylose reductase or xylitol dehydrogenase genes, indicating that the eukaryotic redox pathway is not at play in these bacteria.
Fig. 6.
Genomic organization of xylulokinase genes from selected human or bovine-associated Bacteroidetes. The Prevotella bryantii B14 xylulokinase gene (GenBank accession no. EFI71375.1) was used as the query sequence in a BLASTp search of the GenBank database. Next, the Bacteroides thetaiotaomicron VPI-5482 xylose isomerase gene (BT_0793) was used as a query sequence in a BLASTp search of the GenBank database. The four organisms that possess only a xylulose kinase gene and no xylose isomerase gene are grouped in the upper half of the figure, and representative organisms that contain both genes are grouped in the lower half. The genomic context is shown for the region surrounding the xylulose kinase genes in these organisms. ORF numbers are indicated within each of the genes as derived from the genome project for each organism in the GenBank database. The small subunit (16S) ribosomal RNA genes from the organisms were aligned and a neighbour-joining tree was constructed using the CLC Genomics Workbench v3.0 software. Each alignment was re-sampled 100 times and the bootstrap values are indicated on the internal branches. The branch length is reported as the expected number of substitutions per nucleotide position. The rumen isolates are in pink and the human isolates are colour-coded as indicated in the legend based upon the reported body isolation site as listed in the Human Microbiome Projects Catalog (http://www.hmpdacc.org/).
Xylose isomerase activity, although detected in cell extracts of xylose-grown P. bryantii B14 and P. ruminicola 23 cultures, was very low as compared with cell extracts of E. coli strain LE 392, whereas xylulokinase activity was robust and inducible in these two Prevotella strains during growth with xylose as compared with glucose (Matte et al., 1992). Do these bacteria employ a unique mechanism for the conversion of xylose to xylulose? Perhaps there is another isomerase enzyme that has evolved to moonlight as a xylose isomerase enzyme in these bacteria. Whichever the case, the genomic data provide support that these Prevotella spp. possess a fundamental difference in their xylose utilization system as compared with related bacteria within the Bacteroidetes phylum.
Another fundamental question, which arises from these genome sequences, is whether or not xylan degradation in the human colon is a physiologically important process that contributes to the maintenance and population dynamics of the microbial community in this ecosystem. The presence of numerous bacteria, which possess extensive xylan degrading systems as well as an even larger presence of xylo-oligosaccharide fermenting bacteria, would suggest that xylan utilization is important. Our increased understanding of the Xus system coupled with the genetic tools that are available for xylanolytic Bacteroides spp. will allow this question to be answered by constructing mutant bacteria, which have genetic lesions in their xylan utilizing systems and then to test whether these mutations affect their fitness and transmissibility in vivo.
Acknowledgements
This research was supported by the Energy Biosciences Institute. D.D. was partially supported by an NIH NRSA fellowship (fellowship no. 1F30DK084726). We thank Shosuke Yoshida, Young-Hwan Moon, Michael Iakiviak, Yejun Han and Xiaoyun Su of the Energy Biosciences Institute, UIUC and Eric C. Martens of the Department of Microbiology and Immunology, University of Michigan for valuable scientific discussions.
References
- Avgustin G, Wallace RJ, Flint HJ. Phenotypic diversity among ruminal isolates of Prevotella ruminicola: proposal of Prevotella brevis sp. nov., Prevotella bryantii sp. nov., and Prevotella albensis sp. nov. and redefinition of Prevotella ruminicola. Int J Syst Bacteriol. 1997;47:284–288. doi: 10.1099/00207713-47-2-284. [DOI] [PubMed] [Google Scholar]
- Bakir MA, Kitahara M, Sakamoto M, Matsumoto M, Benno Y. Bacteroides intestinalis sp. nov., isolated from human faeces. Int J Syst Evol Microbiol. 2006;56:151–154. doi: 10.1099/ijs.0.63914-0. [DOI] [PubMed] [Google Scholar]
- Bjursell MK, Martens EC, Gordon JI. Functional genomic and metabolic studies of the adaptations of a prominent adult human gut symbiont, Bacteroides thetaiotaomicron, to the suckling period. J Biol Chem. 2006;281:36269–36279. doi: 10.1074/jbc.M606509200. [DOI] [PubMed] [Google Scholar]
- Braun V, Mahren S. Transmembrane transcriptional control (surface signalling) of the Escherichia coli. Fec type. FEMS Microbiol Rev. 2005;29:673–684. doi: 10.1016/j.femsre.2004.10.001. [DOI] [PubMed] [Google Scholar]
- Bryant MP, Small N, Bouma C, Chu H. Bacteroides ruminicola n. sp. and Succinimonas amylolytica; the new genus and species; species of succinic acid-producing anaerobic bacteria of the bovine rumen. J Bacteriol. 1958;76:15–23. doi: 10.1128/jb.76.1.15-23.1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for Glycogenomics. Nucleic Acids Res. 2009;37:D233–D238. doi: 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chassard C, Goumy V, Leclerc M, Del'homme C, Bernalier-Donadille A. Characterization of the xylan-degrading microbial community from human faeces. FEMS Microbiol Ecol. 2007;61:121–131. doi: 10.1111/j.1574-6941.2007.00314.x. [DOI] [PubMed] [Google Scholar]
- Chassard C, Delmas E, Lawson PA, Bernalier-Donadille A. Bacteroides xylanisolvens sp. nov., a xylan-degrading bacterium isolated from human faeces. Int J Syst Evol Microbiol. 2008;58:1008–1013. doi: 10.1099/ijs.0.65504-0. [DOI] [PubMed] [Google Scholar]
- Cho KH, Cho D, Wang GR, Salyers AA. New regulatory gene that contributes to control of Bacteroides thetaiotaomicron starch utilization genes. J Bacteriol. 2001;183:7198–7205. doi: 10.1128/JB.183.24.7198-7205.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crittenden R, Karppinen S, Ojanen S, Tenkanen M, Fagerstrom R, Matto J, et al. In vitro fermentation of cereal dietary fibre carbohydrates by probiotic and intestinal bacteria. J Sci Food Agric. 2002;82:781–789. [Google Scholar]
- D'Elia JN, Salyers AA. Contribution of a neopullulanase, a pullulanase, and an alpha-glucosidase to growth of Bacteroides thetaiotaomicron on starch. J Bacteriol. 1996a;178:7173–7179. doi: 10.1128/jb.178.24.7173-7179.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Elia JN, Salyers AA. Effect of regulatory protein levels on utilization of starch by Bacteroides thetaiotaomicron. J Bacteriol. 1996b;178:7180–7186. doi: 10.1128/jb.178.24.7180-7186.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehority BA. Characterization of several bovine rumen bacteria isolated with a xylan medium. J Bacteriol. 1966;91:1724–1729. doi: 10.1128/jb.91.5.1724-1729.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd D, Cann IKO. Enzymatic deconstruction of xylan for biofuel production. GCB Bioenergy. 2009;1:2–17. doi: 10.1111/j.1757-1707.2009.01004.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd D, Kocherginskaya SA, Spies MA, Beery KE, Abbas CA, Mackie RI, Cann IK. Biochemi cal analysis of a beta-D-xylosidase and a bifunctional xylanase-ferulic acid esterase from a xylanolytic gene cluster in Prevotella ruminicola 23. J Bacteriol. 2009;191:3328–3338. doi: 10.1128/JB.01628-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd D, Moon Y-H, Mackie RI, Cann IK. Transcriptomic analyses of xylan degradation by Prevotella bryantii and insights into energy acquisition by xylanolytic Bacteroidetes. J Biol Chem. 2010a;285:30261–30273. doi: 10.1074/jbc.M110.141788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd D, Kiyonari S, Mackie RI, Cann IK. Functional diversity of four glycoside hydrolase family 3 enzymes from the rumen bacterium, Prevotella bryantii B14. J Bacteriol. 2010b;192:2335–2345. doi: 10.1128/JB.01654-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O, Brunak S, von Heijne G, Nielsen H. Locating proteins in the cell using TargetP, SignalP and related tools. Nat Protoc. 2007;2:953–971. doi: 10.1038/nprot.2007.131. [DOI] [PubMed] [Google Scholar]
- Flint HJ, Whitehead TR, Martin JC, Gasparic A. Interrupted catalytic domain structures in xylanases from two distantly related strains of Prevotella ruminicola. Biochim Biophys Acta. 1997;1337:161–165. doi: 10.1016/s0167-4838(96)00213-0. [DOI] [PubMed] [Google Scholar]
- Gasparic A, Martin J, Daniel AS, Flint HJ. A xylan hydrolase gene cluster in Prevotella ruminicola B(1)4: sequence relationships, synergistic interactions, and oxygen sensitivity of a novel enzyme with exoxylanase and beta-(1,4)-xylosidase activities. Appl Environ Microbiol. 1995a;61:2958–2964. doi: 10.1128/aem.61.8.2958-2964.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasparic A, Marinsek-Logar R, Martin J, Wallace RJ, Nekrep FV, Flint HJ. Isolation of genes encoding beta-D-xylanase, beta-D-xylosidase and alpha-L-arabinofuranosidase activities from the rumen bacterium Prevotella ruminicola B(1)4. FEMS Microbiol Lett. 1995b;125:135–141. doi: 10.1111/j.1574-6968.1995.tb07349.x. [DOI] [PubMed] [Google Scholar]
- Gill SR, Pop M, Deboy RT, Eckburg PB, Turnbaugh PJ, Samuel BS, et al. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312:1355–1359. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hespell RB, Whitehead TR. Physiology and genetics of xylan degradation by gastrointestinal tract bacteria. J Dairy Sci. 1990;73:3013–3022. doi: 10.3168/jds.S0022-0302(90)78988-6. [DOI] [PubMed] [Google Scholar]
- Jeffries TW. Engineering yeasts for xylose metabolism. Curr Opin Biotechnol. 2006;17:320–326. doi: 10.1016/j.copbio.2006.05.008. [DOI] [PubMed] [Google Scholar]
- Juncker AS, Willenbrock H, Heijne GV, Brunak S, Nielsen H, Krogh A. Prediction of lipoprotein signal peptides in Gram-negative bacteria. Protein Sci. 2003;12:1652–1662. doi: 10.1110/ps.0303703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koropatkin NM, Smith TJ. SusG: a unique cell-membrane-associated alpha-amylase from a prominent human gut symbiont targets complex starch molecules. Structure. 2010;18:200–215. doi: 10.1016/j.str.2009.12.010. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A, Anderson JB, Derbyshire MK, DeWeese-Scott C, Gonzales NR, Gwadz M, et al. CDD: a conserved domain database for interactive domain family analysis. Nucleic Acids Res. 2007;35:D237–D240. doi: 10.1093/nar/gkl951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens EC, Chiang HC, Gordon JI. Mucosal glycan foraging enhances fitness and transmission of a saccharolytic human gut bacterial symbiont. Cell Host Microbe. 2008;4:447–457. doi: 10.1016/j.chom.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens EC, Koropatkin NM, Smith TJ, Gordon JI. Complex glycan catabolism by the human gut microbiota: the Bacteroidetes Sus-like paradigm. J Biol Chem. 2009a;284:24673–24677. doi: 10.1074/jbc.R109.022848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens EC, Roth R, Heuser JE, Gordon JI. Coordinate regulation of glycan degradation and polysaccharide capsule biosynthesis by a prominent human gut symbiont. J Biol Chem. 2009b;284:18445–18457. doi: 10.1074/jbc.M109.008094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui H, Ogata K, Tajima K, Nakamura M, Nagamine T, Aminov RI, Benno Y. Phenotypic characterization of polysaccharidases produced by four Prevotella type strains. Curr Microbiol. 2000;41:45–49. doi: 10.1007/s002840010089. [DOI] [PubMed] [Google Scholar]
- Matte A, Forsberg CW, Verrinder AM. Enzymes associated with metabolism of xylose and other pentoses by Prevotella (Bacteroides) ruminicola strains, Selenomonas ruminantium D, and Fibrobacter succino-genes S85. Can J Microbiol. 1992;38:370–376. doi: 10.1139/m92-063. [DOI] [PubMed] [Google Scholar]
- Mirande C, Mosoni P, Bera-Maillet C, Bernalier-Donadille A, Forano E. Characterization of Xyn10A, a highly active xylanase from the human gut bacterium Bacteroides xylanisolvens XB1A. Appl Microbiol Biotechnol. 2010;87:2097–2105. doi: 10.1007/s00253-010-2694-0. [DOI] [PubMed] [Google Scholar]
- Miyazaki K, Martin JC, Marinsek-Logar R, Flint HJ. Degradation and utilization of xylans by the rumen anaerobe Prevotella bryantii (formerly P. ruminicola subsp. brevis) B(1)4. Anaerobe. 1997;3:373–381. doi: 10.1006/anae.1997.0125. [DOI] [PubMed] [Google Scholar]
- Miyazaki K, Miyamoto H, Mercer DK, Hirase T, Martin JC, Kojima Y, Flint HJ. Involvement of the multidomain regulatory protein XynR in positive control of xylanase gene expression in the ruminal anaerobe Prevotella bryantii B(1)4. J Bacteriol. 2003;185:2219–2226. doi: 10.1128/JB.185.7.2219-2226.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki K, Hirase T, Kojima Y, Flint HJ. Medium- to large-sized xylo-oligosaccharides are responsible for xylanase induction in Prevotella bryantii B(1)4. Microbiology. 2005;151:4121–4125. doi: 10.1099/mic.0.28270-0. [DOI] [PubMed] [Google Scholar]
- Morrison M, Daugherty SC, Nelson WC, Davidsen T, Nelson KE. The FibRumBa database: a resource for biologists with interests in gastrointestinal microbial ecology, plant biomass degradation, and anaerobic microbiology. Microb Ecol. 2009;59:212–213. doi: 10.1007/s00248-009-9562-4. [DOI] [PubMed] [Google Scholar]
- Nelson KE, Weinstock GM, Highlander SK, Worley KC, Creasy HH, Wortman JR, et al. A catalog of reference genomes from the human microbiome. Science. 2010;328:994–999. doi: 10.1126/science.1183605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purushe J, Fouts DE, Morrison M, White BA, Mackie RI, Coutinho PM, et al. Comparative genome analysis of Prevotella ruminicola and Prevotella bryantii: Insights into their environmental niche. Microb Ecol. 2010;60:721–729. doi: 10.1007/s00248-010-9692-8. [DOI] [PubMed] [Google Scholar]
- Reeves AR, Wang GR, Salyers AA. Characterization of four outer membrane proteins that play a role in utilization of starch by Bacteroides thetaiotaomicron. J Bacteriol. 1997;179:643–649. doi: 10.1128/jb.179.3.643-649.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert C, Chassard C, Lawson PA, Bernalier-Donadille A. Bacteroides cellulosilyticus sp. nov., a cellulolytic bacterium from the human gut microbial community. Int J Syst Evol Microbiol. 2007;57:1516–1520. doi: 10.1099/ijs.0.64998-0. [DOI] [PubMed] [Google Scholar]
- Salyers AA, Kotarski SF. Induction of chondroitin sulfate lyase activity in Bacteroides thetaiotaomicron. J Bacteriol. 1980;143:781–788. doi: 10.1128/jb.143.2.781-788.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salyers AA, Vercellotti JR, West SE, Wilkins TD. Fermentation of mucin and plant polysaccharides by strains of Bacteroides from the human colon. Appl Environ Microbiol. 1977;33:319–322. doi: 10.1128/aem.33.2.319-322.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salyers AA, Balascio JR, Palmer JK. Break down of xylan by enzymes from human colonic bacteria. J Food Biochem. 1982;6:39–56. [Google Scholar]
- Selvendran RR. The plant cell wall as a source of dietary fiber: chemistry and structure. Am J Clin Nutr. 1984;39:320–337. doi: 10.1093/ajcn/39.2.320. [DOI] [PubMed] [Google Scholar]
- Shah HN, Collins DM. Prevotella, a new genus to include Bacteroides melaninogenicus and related species formerly classified in the genus Bacteroides. Int J Syst Bacteriol. 1990;40:205–208. doi: 10.1099/00207713-40-2-205. [DOI] [PubMed] [Google Scholar]
- Shipman JA, Cho KH, Siegel HA, Salyers AA. Physiological characterization of SusG, an outer membrane protein essential for starch utilization by Bacteroides thetaiotaomicron. J Bacteriol. 1999;181:7206–7211. doi: 10.1128/jb.181.23.7206-7211.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shipman JA, Berleman JE, Salyers AA. Characterization of four outer membrane proteins involved in binding starch to the cell surface of Bacteroides thetaiotaomicron. J Bacteriol. 2000;182:5365–5372. doi: 10.1128/jb.182.19.5365-5372.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson PJ, Jamieson SJ, Abou-Hachem M, Karlsson EN, Gilbert HJ, Holst O, Williamson MP. The solution structure of the CBM4-2 carbohydrate binding module from a thermostable Rhodothermus marinus xylanase. Biochemistry. 2002;41:5712–5719. doi: 10.1021/bi012093i. [DOI] [PubMed] [Google Scholar]
- Slavin JL, Brauer PM, Marlett JA. Neutral detergent fiber, hemicellulose and cellulose digestibility in human subjects. J Nutr. 1981;111:287–297. doi: 10.1093/jn/111.2.287. [DOI] [PubMed] [Google Scholar]
- Sonnenburg ED, Zheng H, Joglekar P, Higginbottom SK, Firbank SJ, Bolam DN, Sonnenburg JL. Specificity of polysaccharide use in intestinal Bacteroides species determines diet-induced microbiota alterations. Cell. 2010;141:1241–1252. doi: 10.1016/j.cell.2010.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI. The human microbiome project. Nature. 2007;449:804–810. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Soest PJ. Nutritional Ecology of the Ruminant. Cornell University Press; Ithaca, NY: 1994. [Google Scholar]
- Weaver J, Whitehead TR, Cotta MA, Valentine PC, Salyers AA. Genetic analysis of a locus on the Bacteroides ovatus chromosome which contains xylan utilization genes. Appl Environ Microbiol. 1992;58:2764–2770. doi: 10.1128/aem.58.9.2764-2770.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead TR. Nucleotide sequences of xylaninducible xylanase and xylosidase/arabinosidase genes from Bacteroides ovatus V975. Biochim Biophys Acta. 1995;1244:239–241. doi: 10.1016/0304-4165(95)00051-c. [DOI] [PubMed] [Google Scholar]
- Whitehead TR, Hespell RB. The genes for three xylan-degrading activities from Bacteroides ovatus are clustered in a 3.8-kilobase region. J Bacteriol. 1990;172:2408–2412. doi: 10.1128/jb.172.5.2408-2412.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Bjursell MK, Himrod J, Deng S, Carmichael LK, Chiang HC, et al. A genomic view of the human-Bacteroides thetaiotaomicron symbiosis. Science. 2003;299:2074–2076. doi: 10.1126/science.1080029. [DOI] [PubMed] [Google Scholar]
- Xu J, Mahowald MA, Ley RE, Lozupone CA, Hamady M, Martens EC, et al. Evolution of symbiotic bacteria in the distal human intestine. PLoS Biol. 2007;5:e156. doi: 10.1371/journal.pbio.0050156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida E, Hidaka M, Fushinobu S, Koyanagi T, Minami H, Tamaki H, et al. Role of a PA14 domain in determining substrate specificity of a glycoside hydrolase family 3 beta-glucosidase from Kluyveromyces marxianus. Biochem J. 2010;431:39–49. doi: 10.1042/BJ20100351. [DOI] [PubMed] [Google Scholar]