Abstract
The mammalian immune system has been traditionally subdivided into two compartments known as the innate and the adaptive. T cells and B cells, which rearrange their antigen receptor genes using the RAG recombinase, comprise the adaptive arm of immunity. Meanwhile, every other white blood cell has been grouped together under the broad umbrella of innate immunity, including natural killer (NK) cells. NK cells are considered innate lymphocytes because of their rapid responses to stressed cells and their ability to develop without receptor gene rearrangement (i.e. in RAG-deficient mice). However, new findings implicate a critical function for RAG proteins during NK cell ontogeny, and suggest a novel mechanism by which controlled DNA breaks during NK cell development dictate the fitness, function, and longevity of these cells. This review highlights recent work describing how DNA break events can impact cellular differentiation and fitness in a variety of cell types and settings.
INTRODUCTION
The appearance of the recombination activating genes (RAG) in jawed vertebrates during evolution endowed T cells and B cells with the ability to mediate V(D)J gene rearrangement at their antigen receptor loci, providing these lymphocytes with a molecular mechanism for diversifying their antigen receptor repertoire. In contrast to these adaptive immune cells, natural killer (NK) cells classically represent the third lineage of lymphocytes (i.e. innate lymphocytes), which possess germline-encoded antigen receptors and do not require RAG for their development [1]. As innate lymphocytes, these cells are the body’s first line of defense against pathogen invasion and are thought to be short-lived effector cells that do not need prior sensitization for their activation. Thus, since their discovery 40 years ago, NK cells have been placed in immunology textbook chapters devoted to the innate immune system [2].
However, this classical view of NK cells has been rapidly changing in past decade. Recent evidence suggests that this cell type possesses traits attributable to adaptive immunity [3, 4]. These characteristics include education mechanisms involving NK receptor-MHC interactions to ensure self-tolerance during NK cell development (reviewed in [5]), and clonal-like expansion of antigen-specific NK cells during viral infection, followed by the ability to generate long-lived progeny known as “memory” NK cells [6–9]. NK cell memory has also been described in a plethora of non-pathogen settings (reviewed in [10, 11]). For instance, several groups have described the ability of NK cells to mediate anamnestic responses against chemical haptens, demonstrating a surprising degree of specificity [10, 12, 13]. Another study found that activation by cytokines, including interleukin (IL)-12, IL-15, and IL-18, can result in the generation of NK cells with memory-like properties [14] However, few distinct surface markers discriminate cytokine-induced memory-like NK cells from their naïve or activated counterparts.
NK cells specifically share similarities with CD8+ T cells, including their development from the common lymphoid progenitor (CLP), their requirement for the IL-2 common gamma chain receptor for their survival, their expression of similar activation/maturation markers, and their use of identical cytolytic machinery (perforin and granzymes) to destroy transformed or virally-infected target cells (reviewed in [3]). Because we are beginning to understand the diversity and heterogeneity within the effector and memory CD8+ T cell response during infection, this body of literature can serve as a guide to investigating the “adaptive” NK cell response against pathogens.
Some of the underlying molecular mechanisms that control NK cell function and longevity, resulting in effector and memory NK cells subsets during pathogen challenge, have only recently come to light. DNA damage is generally assumed to be a detrimental event, frequently associated with impaired cell survival or cellular transformation [15, 16]. However, a growing body of evidence suggests that focal DNA breaks are a mechanism for normal cell development, differentiation, and function. The most well-studied examples are homologous recombination during cellular meiosis [17], and programmed DNA double strand break (DSB) and repair during V(D)J recombination in developing lymphocytes. DNA cleavage in V(D)J recombination is mediated by specifically-timed expression of the evolutionarily conserved and lymphocyte specific RAG recombinase and repaired by the ubiquitous non homologous end joining (NHEJ) DNA damage response (DDR) pathway. Since adaptive immune cells require RAG for the development of their antigenic receptors, genetic ablation of RAG in mice results in a loss of T cells and B cells [18, 19]. However, innate lymphocyte lineages such as NK cells express germ-line encoded antigen receptors and are not thought to require RAG for the development and function.
Our recent study [20] demonstrates that RAG expression in CLPs and NK cell precursors that eventually develop into mature NK cells marks functionally distinct subsets of NK cells in the periphery. Those NK cells that have had a history of RAG expression during development are more “fit”, and are capable of greater survival following bursts of robust proliferation, suggesting a role for the RAG recombinase and following DDR repair pathway beyond their canonical pathways. This review will attempt to integrate our prior understanding of RAG activity in adaptive lymphocytes with our recent discovery of a role for RAG in innate lymphocytes. We will also discuss the implications of these advances in understanding programmed induction of DNA damage/repair and their role in maintaining the genomic integrity of cells and determining cell fate.
CANONICAL ROLE OF RAG PROTEINS IN ADAPTIVE LYMPHOCYTES
The RAG recombinase mediates V(D)J recombination, an intricate and tightly regulated process that requires the programmed induction and subsequent repair of DSBs at the antigen receptor gene loci of developing T cells and B cells during the G1 phase of the cell cycle [21]. Diversity in the antigen receptor repertoire is generated by shuffling of variable (V), diversity (D), and joining (J) gene segments that are found within the T-cell receptor and B-cell receptor loci. RAG1 and RAG2 (collectively known as RAG) function as ‘Y-shaped’ heterotetramer consisting of two subunits each of RAG1 and RAG2 [22], where the RAG1 subunits contain the endonuclease catalytic center (necessary for cleavage) [23, 24], and is only active in the presence of its binding partner RAG2 [25]. RAG proteins bind to highly conserved target recombination signal sequences (RSS) flanking the V, D, and J gene segments (Figure 1A). These RSS reside next to conserved segments containing a palindromic heptamer (CACAGTG) and an A/T-rich nonamer (ACAAAAACC) [26–29] separated by non-conserved spaces of either 12 or 23 bases. During V(D)J recombination, only coding segments flanked by RSS with nonidentical spacer lengths can be combined, known as the “12/23 rule” [30]. This spatial restriction helps prevent non-productive rearrangements. RAG initiates recombination by nicking DNA at the border between the heptamer of RSS and the coding segment [31], leading to programmed induction of DSBs within the antigen receptor loci.
Figure 1. Canonical and revised view of RAG binding.
(A) Traditionally, the RAG recombinase (RAG1/2) was thought to only bind to the antigen receptor locus to facilitate rearrangement of the V, D, and J gene segments of the antigen receptor, and generate receptor diversity in T cells and B cells. However, recent studies have led to a revision of this canonical view of RAG binding. (B) In this revised view, RAG2 binds at points of ‘active’ chromatin throughout the genome (at thousands of sites), marked by the histone modification H3K4me3. Whether RAG1 also binds outside the recombination center remains to be determined.
Recent advances have been made in determining the mechanisms of how the RAG proteins are guided to appropriate target sites. It is now known that RAG2 contains a plant homeodomain (PHD) finger which targets the RAG1/2 complex to activated or “open” chromatin through binding of histone 3 trimethylated at lysine 4 (H3K4me3) [32–34]. Therefore, recognition of the RSS and H3K4me3 chromatin mark by RAG1/2 facilitates the recombination reaction [33, 34]. Although RAG is depicted in all immunology textbooks to exclusively bind at RSS within V, D, and J genes, there was never direct evidence to suggest RAG binding was restricted to H3K4me3 at the antigen receptor loci. Recent genome wide ChIP-seq analysis of lymphocytes in mice indicates that RAG2 recruitment mirrors the footprint of the H3K4me3 activation mark, binding to thousands of sites throughout the genome (Figure 1B), whereas RAG1 binding is more directed and occurs predominantly at conserved RSSs [32]. It remains to be determined if RAG1 may also bind outside the recombination center. Further studies have linked RAG1/2 to DNA breaks and chromosomal rearrangements (including translocations) at RSS-like “cryptic” sites (sites that mimic RSSs outside of canonical antigen-receptor loci) and non-RSS sequences that are scattered throughout the genome [35–38]. In previously documented oncogenic rearrangements, many translocation breakpoints on the non-antigen receptor gene partner contain RSS-like sequences at or near the breakpoint [39]. However, the function of RAG proteins at these “cryptic” RSS remains to be determined.
Following DNA cleavage, RAG stabilizes the four broken DNA ends in a post-cleavage complex that directs repair by the ubiquitous classical NHEJ pathway [40, 41]. In response to DNA DSBs, PI3K-like Ser/Thr kinases ataxia-telangiectasia mutated (ATM) kinase and DNA-dependent protein kinase (DNA-PK), which phosphorylate hundreds of targets necessary for the downstream activation of DDR pathways and the initiation of apoptosis in cells that fail to repair DNA breaks [16, 42–44]. Specifically, ATM phosphorylates the CHK2 kinase leading to the phosphorylation of Cdc25a promoting cell cycle arrest and apoptosis [45]. Furthermore, these kinases activate a multitude of downstream effector molecules that regulate transcriptional programs to pro-survival pathways (e.g. downregulate Bcl-2) within successfully rearranging cells [16].
During this catalytic reaction, ATM and DNA-PK also phosphorylate the histone protein H2AX in chromatin flanking DNA DSBs [46, 47]. Phosphorylated H2AX (γH2AX) at the site of DSB recruits DNA damage response proteins to the site of broken DNA [46]. Clearance of phosphorylation signifies repair of the DSB. To further increase receptor diversity, several insertions and deletions occur at the ligated junction by specialized proteins terminal deoxynucleotidyl transferase (TdT) and DNA Pol μ[48]. This leads to a final antigen receptor count of approximately 1011 possible BCRs/antibodies for B cells and 1015 possible TCRs for T cells [49, 50]. High efficiency is required in this system, as aberrant rearrangement events can lead to apoptosis, genomic instability, and cellular transformation [36, 37, 51, 52].
RAG EXPRESSION IN INNATE LYMPHOCYTES
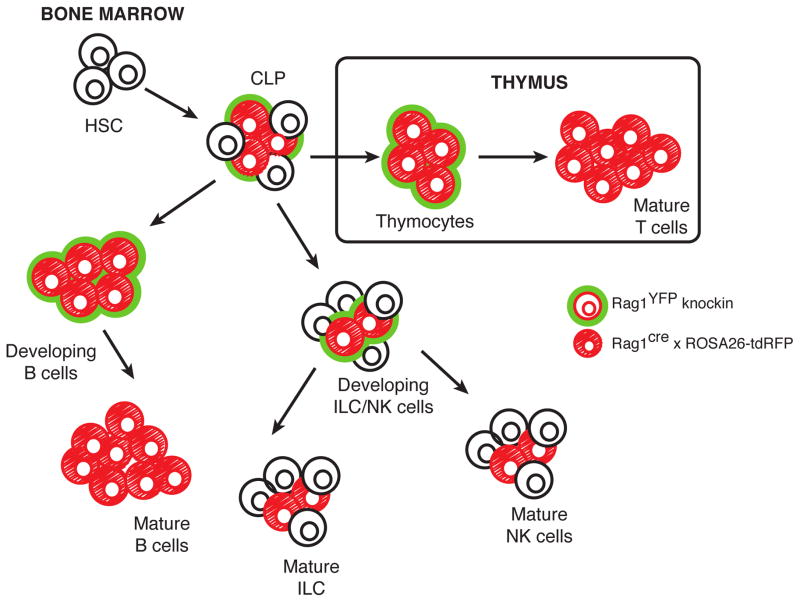
Although the generation of the lymphoid compartment has been studied extensively, unifying models of lymphocyte development have been difficult to assemble, and the ontogeny of NK cells is not yet fully understood. Progress in understanding stem and progenitor cell populations is aided by experimental mouse models that report current RAG expression (RAGGFP knockin mice [53]) or a history of RAG expression (‘fate mapping’ mice: Rag1Cre x Rosa26-floxed STOP-tdRFP) (Figure 2) [54, 55]. Within the bone marrow, CLPs are destined to become B cells or commit to the NK cell lineage through the upregulation of the IL-2/IL-15 receptor (CD122) [56, 57]. All developing B cells and T cells in the bone marrow and thymus respectively have been shown to exhibit RAG expression (using the RAG reporter mice), and all peripheral B cells and T cells have been shown to exhibit a history of RAG expression (using the RAG ‘fate mapping’ mice); however, a surprisingly large fraction of developing NK cells (~40%) were shown to derive from RAG-expressing CLPs [20, 58] (Figure 2). Prior work utilized a transgenic mouse recombination reporter system [59], whereby recombinase activity is indicated by the permanent expression of violet light-excited (VEX) fluorescence [60], to mark NK cells that have undergone recombination events. Analysis of recombination in NK cells revealed that approximately 50% of CLP in bone marrow are VEX+ and 20–30% of peripheral NK cells express the reporter, in agreement with our observations in RAG fate-mapping mice [20].
Figure 2. Innate and adaptive lymphocytes demonstrating a history of RAG expression.
Lymphocytes from RAG reporter mice (green-outlined cells) and RAG ‘fate-mapping’ mice (red-filled cells) demonstrate that a subset of developing and mature innate lymphoid cells (ILCs) and natural killer (NK) cells have a history of RAG gene expression, whereas all mature B cells and T cells have previously expressed RAG.
Through analysis of RAG fate mapping mice, 20–30% of peripheral NK cells were shown to have had, at one point, expressed RAG (i.e. are permanently marked by RFP expression) [20, 55, 59, 61–63] (Figure 2). Analysis of NK cell development in RAG reporter mice by flow cytometry [64, 65] revealed that the highest levels of RAG expression occurs in CLPs from the bone marrow, with GFP expression decreasing as developing NK cells undergo maturation and migration from the bone marrow. In support of these observations, previous independent studies in both human and mouse cells have shown that NK cells can possess rearrangements within the immunoglobulin (Ig) and TCR loci, but these rearrangements do not result in a functional antigen receptor [55, 59, 62, 66]. Whether a physiological consequence of these unproductive rearrangements exists remains unknown.
Innate lymphoid cells (ILCs) have recently between described to maintain tissue homeostasis and provide protection against pathogens at mucosal surfaces [67, 68]. Similar to T cells, B cells, and NK cells, ILCs develop from the CLP and require the common gamma chain for their survival. ILCs have been divided into three classes based on their transcription factor requirements, response to cytokines, cytokine production, and anatomical location. Group 1 ILCs (ILC1s) produce IFN-γ in response to IL-12 and IL-18, and are broadly found throughout lymphoid and non-lymphoid organs (e.g. liver, intestine, spleen, peritoneal cavity). ILC1s can be distinguished from NK cells by requiring the transcription factor T-bet but not Eomes for their development, and ILC1s have been shown to be important for immunity against viruses, intracellular bacteria and parasites [67, 68]. Unlike NK cells, ILC1s are not thought to mediate cytotoxicity [67, 68]. Group 2 ILCs (ILC2s) require GATA3, RORα, and GFI1 expression for their differentiation/development, produce type 2 cytokines (IL-4, IL-5, and IL-13) and amphiregulin in response to IL-25 and IL-33, and are predominantly found in the lung and adipose tissue to protect against helminth infection [67, 68]. Group 3 ILCs (ILC3s) require RORγt and T-bet for their differentiation, and are important for immunity against extracellular bacteria. ILC3s are found primarily in the gut, and produce a variety of cytokines, including IL-17 and IL-22, in response to IL-1β and IL-23 [67, 68]. Although ILCs are not thought to require RAG for their development [69], we showed that nearly half of all ILCs among the three subsets have a history of RAG expression during ontogeny [20] (Figure 2). The functional consequence of this subset heterogeneity is currently being investigated. These novel findings suggest that RAG proteins may possess more ubiquitous and genome-wide functions in developing lymphocytes, including NK cells and ILCs, than previously thought.
A NOVEL ROLE FOR RAG IN INNATE LYMPHOCYTES
A. HETEROGENEOUS SUBSETS DISTINGUISHED BY RAG EXPRESSION
In our recent findings, RAG expression in NK cells clearly defined subset heterogeneity in the periphery [20]. Using the RAG fate mapping mice, NK cells that never expressed RAG during development (RFP−) were more terminally differentiated (as defined by greater KLRG1 and CD11b expression), and demonstrated a higher degree of cytotoxicity in ex vivo and in vivo kill assays [20]. We investigated whether RAG influenced in vivo NK cell responses by incorporating a well-established viral model of antigen-specific NK cell expansion [8]. NK cells expressing the mouse cytomegalovirus (MCMV)-specific activating receptor Ly49H have been shown to undergo a robust antigen-driven proliferation (100–1000 fold expansion) following MCMV infection, and following viral clearance, a population of long-lived memory NK cells persists in both lymphoid and non-lymphoid organs [8]. In this system, donor NK cells from RAG1- or RAG2-deficient mice were outcompeted by WT NK cells following adoptive transfer and MCMV infection. NK cells that lacked either RAG or a history of RAG expression were defective in virus-driven expansion and failed to persist due to an increase in NK-cell apoptosis [20] (Figure 3). These findings suggest a novel role for RAG outside of V(D)J recombination, where RAG expression during NK cell development results in the enhanced cellular “fitness” of peripheral NK cells.
Figure 3. RAG expression during ontogeny delineates functional heterogeneity and cellular fitness in NK cell subsets.
NK cells from B6 mice expressing the activating Ly49H receptor will recognize MCMV-encoded m157 on infected cells, leading to activation and clonal proliferation of antigen-specific NK cells. NK cell with a history of RAG expression (red) were shown to have enhanced DNA damage response (DDR) and survival following rapid proliferation. After the NK cell expansion phase and viral control, effector Ly49H+ NK cells undergo a contraction phase resulting in long-lived memory NK cells, where only the NK cells that previously expressed RAG remain. These memory NK cells are able to mount a recall response when virus is re-encountered.
A recent study [70] demonstrated that KLRG1- NK cells, which are considered less terminally differentiated, are more likely to represent memory progenitor cells, retaining the ability to generate long-lived memory cells. Several cell extrinsic factors were shown to govern the development of these KLRG1- NK-cell memory precursor populations, including presence of T cells and the microbiota. During MCMV infection, the authors implicate T cells competing for IL-15 (critical for NK cell development and survival [71–73]) as responsible for diminished survival of KLRG1+ NK cells [70]. On the other hand, reduction of the microbiota (using antibiotics) increased the memory potential of KLRG1+ NK cells [70]. Thus, the lack of T cells in RAG-deficient mice may also contribute to the defect in memory NK cell generation in these animals, and represents an extrinsic mechanism that complements the cell-intrinsic role for RAG described in our study. Additional cell-intrinsic and extrinsic factors that dictate memory-precursor NK cell populations remain to be determined.
Interestingly, a paradigm of subset heterogeneity has recently been ascribed to CD8+ T cells following antigen encounter. Following LCMV infection, responding CD8+ T cells were shown to generate a heterogeneous effector population consisting of KLRG1hi short-lived effector cells (SLECs) which are more terminally differentiated, and KLRG1lo memory precursor effector cells (MPECs) which can give rise to long-lived memory cells [74]. Given that NK cells are more similar in function and phenotype to effector/memory CD8+ T cells than naïve CD8+ T cells [3], it will be of interest to further investigate whether a parallel SLEC/MPEC paradigm can explain the heterogeneous peripheral NK cell pool in wildtype mice, where certain subsets of NK cells (those that haven’t expressed RAG during development) are more lethal killers in the face of immediate pathogen insult, whereas other subsets (those that have expressed RAG) contribute to the long-lived memory population which can respond to repeated pathogen exposure.
B. DNA DAMAGE RESPONSE & CELLULAR FITNESS
Our recent findings suggest RAG expression during NK cell ontogeny establishes functional heterogeneity in the periphery, likely through transcriptional or epigenetic changes. To this end, NK cells that lacked RAG expression have a decreased expression in essential components of the DDR including DNA-PKcs (Prkdc), Ku80 (Xrcc5), Chk2 (Chek2), and ATM (Atm) in mature cells [20]. These perturbations in gene expression strongly correlated with inefficient DNA repair, as measured by γ-H2AX foci appearance and removal, following introduction of exogenous DNA damaging agents such as radiation [20]. Interestingly, this phenomenon is not restricted to innate lymphocytes. Using TCR transgenic CD8+ T cells from OT-1 (RAG-competent) versus OT-1 x Rag2−/− mice [75], it was also shown that RAG-deficient OT-1 cells possess higher levels of genomic instability and decreased levels of essential DNA damage repair proteins as in NK cells [20]. Radiation exposure resulted in delayed γ-H2AX foci resolution in RAG-deficient T cells, and virus infection revealed a reduced capacity for survival, compared to OT-1 cells [20]. Thus, RAG expression during ontogeny endows both innate and adaptive lymphocytes with enhanced cellular fitness, suggesting this may be a general mechanism for producing immune memory in lymphocytes.
Consistent with the finding that poorly surviving RAG-deficient NK cells possess diminished levels of DNA-PK, NK cells from SCID mice (which possess a genetic mutation in DNA-PK) have also been shown to have an expansion defect following viral infection [20]. Furthermore, NK cells from RAG1 endonuclease-dead mice [32] exhibited the same defects as RAG1-deficient mice in response to viral infection, suggesting the RAG-mediated DNA cleavage is critical in endowing cellular fitness. Altogether, these data suggest that RAG-mediated DNA damage plus repair during NK cell ontogeny “educates” the developing NK cells to better respond to subsequent DNA damage, possibly through the manipulation of the transcriptional landscape in these cells (further discussed below). Future studies will determine whether NK cells that have previously expressed RAG may become functionally specialized through upregulation of lymphoid-related transcription factors, resulting in the cellular “fitness” observed following periods of robust proliferation and cellular stress.
Recent studies support the idea that controlled DNA damage during cellular development acts as a feed-forward mechanism that propagates the transcription and translation of proteins and factors that have important consequences for normal development and differentiation [76–78]. In pre-B cells, RAG-mediated DSBs induce the expression of homing receptors important for lymphocyte egress from the bone marrow and promote homing to secondary lymphoid organs (e.g. CD62L, CD69) [76, 79]. The lack of these homing receptors prior to RAG expression prevent pre-B cells from prematurely exiting the bone marrow before complete rearrangement of B cell receptors (BCR). In another study [80], caspase-3-mediated DNA damage was shown to lead to muscle cell differentiation by inducing expression of non-apoptotic genetic pathways. These studies begin to highlight the potential functional consequence of DNA DSBs beyond their detrimental effect on cell survival. In fact, downstream DSB-mediated transcriptional events may be necessary for the productive maturation of specific cell subsets, and support the idea that DDR by programmed DSBs may “imprint” lymphocyte functions long after the breakpoint has been repaired.
RAG-mediated DSBs have been shown to activate the canonical NF-kB pathway [81, 82], leading to the expression of several pro-survival factors in response to genotoxic DSBs [82–84]. Approximately half of the ATM-dependent gene expression changes that occur in response to RAG DSBs require NF-kB [76]. For example, activation of NF-kB by RAG DSBs leads to the upregulation of genes such as Pim2, a constitutively active anti-apoptotic serine-threonine kinase, shown to be important for lymphocyte proliferation and survival [76, 85–87]. Pim2 expression is increased in an ATM-dependent manner in response to RAG DSBs [76, 78]. Upregulation of Pim2 maintains phosphorylation and inactivation of BAD, which antagonizes the pro-apoptotic function of BAX, promoting increased levels of Bcl-2 and associated cell survival [78, 88]. In these studies, upregulation of Pim2 by RAG DSBs results in reduced proliferation of IL-7 stimulated pre-B cells [78] by enforcing the G1 to S checkpoint and inhibiting the proliferation of pre-B cells with unrepaired DSBs [16, 89, 90]. Pim2 is not sufficient to block proliferation, but maintains cell cycle arrest due to coordination with other DSB-dependent signals [78]. In these cells, expression of Pim2 promotes genomic stability by preventing unrepaired breaks to enter cell cycle resulting in further chromosomal aberrations during replication. Therefore, analysis of Pim2 levels and cell cycle checkpoints in NK cells may provide insight to a mechanism whereby RAG-mediated breaks suppress cell cycle progression during development, leading less mature but more fit peripheral cells.
Interestingly, RAG2 is known to possess highly conserved domains that are important for maintaining genome integrity [91]. Specifically, the RAG2 C-terminus domain is required to escort the post-cleavage RAG1/2:DNA complex into the canonical NHEJ pathway for proper DNA repair [91, 92]. Genetic ablation of only the C-terminal “non-core” domain in Rag2c/c mice [93] (which still contains a functional cleavage activity) has been shown to induce genome-wide chromosomal aberrations and lymphomagenesis when crossed to highly proliferative p53−/− mice [94]. Furthermore, threonine 490 (T490) within the C-terminal domain is a target for the cyclin A-Cdk2 kinase and is phosphorylated at the G1 to S cell cycle transition, leading to proteasomal destruction [95–97]. Defined RAG2(T490A) knockin mice show RAG2 does not undergo cell-cycle mediated degradation and V(D)J recombination persists throughout the cell cycle leading to aberrant repair of RAG-medicate DSBs [98]. When crossed onto a p53−/− background these mice also developed T lymphomas, further reiterating that erroneous V(D)J recombination can promote lymphomagenesis and genomic instability [98]. Thus, whether the additional “non-core” functions of RAG activity (beyond the catalytic cleavage of DNA) further endows innate lymphocytes such as NK cells with greater genomic integrity remains to be elucidated.
Although a global reduction in DNA damage repair enzymes poses an attractive mechanism for the decreased cellular fitness of RAG-deficient NK cells, a close correlation has been reported between RAG2 binding and the activating H3K4me4 histone mark at the promoters of many tissue-specific genes throughout the entire genome [32–34]. Therefore, epigenetic analysis of regulatory regions surrounding DNA damage repair genes in WT compared to RAG-deficient NK cells might provide mechanistic insights into how damage response protein levels are modulated in any given individual resting lymphocyte. Furthermore, ATM-mediated changes in the nuclear chromatin structure of the post-cleavage complex limits the number of potential substrates for recombination, protecting against aberrant translocations [99]. Therefore, lower levels of ATM in RAG-deficient NK cells may further lead to the increase in genomic stability seen by disrupting this chromatin organization. It is possible the RAG1/2 complex plays a regulatory function during ontogeny at sites lacking RSS sequences, resulting in changes in transcriptional activity, histone modifications, or chromatin structure of non-antigen receptor loci.
The ability of RAG2 (and potentially RAG1) to bind to transcriptionally active sites outside of antigen receptor loci raises questions about specificity and fidelity of V(D)J recombination. There exists evidence that suggests RAG-mediated cleavage may be occurring at these non-antigen receptor sites, contributing to lymphoma-associated genome alterations [36, 37, 51]. The data from our group and others suggest that these ectopic breaks by RAG might be evolutionarily conserved in order to create heterogeneity among cell types, rather than be purely detrimental to the genome. Many questions remain unanswered. Are these RAG-mediated DNA break/repair events controlled or stochastic? How and when do the RAG proteins create DSBs at these non-antigen receptor sites in the genome? Finally, what additional mechanisms or other molecular chaperones exist to suppress (or promote) this ectopic DNA damage leading to differential cellular fates?
NK cells that have previously expressed RAG proteins during their ontogeny are less terminally differentiated, suggesting RAG-mediated DSBs may inhibit rapid cell maturation and promote a genetic program that is important for processes beyond canonical DDRs. This hypothesis is supported in other cell types where the DDR may be affecting cell differentiation, which has been observed in myoblasts, melanocyte stem cells (MSCs), and hematopoietic stem cells (HSCs) [100–102]. For example, C2C12 myoblasts undergo cell-cycle arrest and a block in differentiation in the presence of genotoxic stress, due to a DDR-regulated differentiation checkpoint during muscle differentiation [100]. This block can be overcome following the repair of the damaged DNA. Furthermore, DNA DSB-initiated ATM signaling has been shown to maintain MSCs within the stem cell niche and blocks differentiation into mature melanocytes [101]. It is also thought that DNA DSB accumulation over time leads to the functional decline of HSCs. HSCs from DDR-deficient mice show significantly higher apoptosis and decreased proliferation, suggested that proper repair of DNA damage is required for efficient HSC renewal and function [102]. Overall, these studies suggest a protective function for DDR in stem and progenitor cells, similar to that observed in NK cells that have expressed RAG activity, in order to promote normal maturation and prevent the propagation of genetic errors.
Irrespective of the precise molecular mechanism behind the RAG-endowed cellular fitness of lymphocytes, these examples suggest a paradigm where controlled DNA damage is able to alter the functional properties of a developmental lineage during ontogeny, leading to alterations that condition subsequent cellular behavior.
CONCLUSION
Adaptive lymphocytes have long been known to require RAG-mediated DNA DSBs for development of functional antigen receptor genes. Recent findings demonstrate that controlled DNA breaks are also required for the development and long-lived “fitness” of innate lymphocytes, and that RAG expression is a major player in these events. The sum of these results indicate that RAG-mediated DSBs outside of canonical V(D)J recombination in developing lymphocytes may be regulating processes that impact their ultimate survival as mature cells. From these studies a more general paradigm is emerging where activation of the DDR by DSBs generated during timed cell processes (such as transient RAG expression and activity in lymphocyte progenitors) regulate a multitude of cell-type specific programs. This endonuclease-mediated DDR mechanism may be dictating the genomic integrity and fitness of select cells within the total adaptive and innate lymphocyte populations, and ensuring the longevity of specific lymphocyte subsets during periods of rapid proliferation or stress during pathogen invasion.
Acknowledgments
The authors thank David Schatz for insightful discussions and critical reading of the manuscript. J.M.K. was supported by a Geoffrey Beene Fellowship and a Cancer Research Institute (CRI) STaRT grant. J.C.S. is supported by the Searle Scholars Program, the CRI, and NIH grant AI100874.
Footnotes
The authors declare no financial conflicts of interest.
References
- 1.Kondo M, Weissman IL, Akashi K. Identification of clonogenic common lymphoid progenitors in mouse bone marrow. Cell. 1997;91:661–672. doi: 10.1016/s0092-8674(00)80453-5. [DOI] [PubMed] [Google Scholar]
- 2.Murphy K, Travers P, Walport M, Janeway C. Janeway’s Immunobiology. New York: Garland Science; 2012. [Google Scholar]
- 3.Sun JC, Lanier LL. NK cell development, homeostasis and function: parallels with CD8(+) T cells. Nature reviews. Immunology. 2011;11:645–657. doi: 10.1038/nri3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vivier E, Raulet DH, Moretta A, Caligiuri MA, Zitvogel L, Lanier LL, Yokoyama WM, Ugolini S. Innate or adaptive immunity? The example of natural killer cells. Science. 2011;331:44–49. doi: 10.1126/science.1198687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Orr MT, Lanier LL. Natural killer cell education and tolerance. Cell. 2010;142:847–856. doi: 10.1016/j.cell.2010.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dokun AO, Kim S, Smith HR, Kang HS, Chu DT, Yokoyama WM. Specific and nonspecific NK cell activation during virus infection. Nat Immunol. 2001;2:951–956. doi: 10.1038/ni714. [DOI] [PubMed] [Google Scholar]
- 7.Daniels KA, Devora G, Lai WC, O’Donnell CL, Bennett M, Welsh RM. Murine cytomegalovirus is regulated by a discrete subset of natural killer cells reactive with monoclonal antibody to Ly49H. J Exp Med. 2001;194:29–44. doi: 10.1084/jem.194.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun JC, Beilke JN, Lanier LL. Adaptive immune features of natural killer cells. Nature. 2009;457:557–561. doi: 10.1038/nature07665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun JC, Beilke JN, Lanier LL. Immune memory redefined: characterizing the longevity of natural killer cells. Immunol Rev. 2010;236:83–94. doi: 10.1111/j.1600-065X.2010.00900.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paust S, von Andrian UH. Natural killer cell memory. Nat Immunol. 2011;12:500–508. doi: 10.1038/ni.2032. [DOI] [PubMed] [Google Scholar]
- 11.Cooper MA, Yokoyama WM. Memory-like responses of natural killer cells. Immunol Rev. 2010;235:297–305. doi: 10.1111/j.0105-2896.2010.00891.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O’Leary JG, Goodarzi M, Drayton DL, von Andrian UH. T cell- and B cell-independent adaptive immunity mediated by natural killer cells. Nat Immunol. 2006;7:507–516. doi: 10.1038/ni1332. [DOI] [PubMed] [Google Scholar]
- 13.Paust S, Gill HS, Wang BZ, Flynn MP, Moseman EA, Senman B, Szczepanik M, Telenti A, Askenase PW, Compans RW, von Andrian UH. Critical role for the chemokine receptor CXCR6 in NK cell-mediated antigen-specific memory of haptens and viruses. Nat Immunol. 2010;11:1127–1135. doi: 10.1038/ni.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cooper MA, Elliott JM, Keyel PA, Yang L, Carrero JA, Yokoyama WM. Cytokine-induced memory-like natural killer cells. Proc Natl Acad Sci U S A. 2009;106:1915–1919. doi: 10.1073/pnas.0813192106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sherman MH, Bassing CH, Teitell MA. Regulation of cell differentiation by the DNA damage response. Trends Cell Biol. 2011;21:312–319. doi: 10.1016/j.tcb.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bednarski JJ, Sleckman BP. Integrated signaling in developing lymphocytes: the role of DNA damage responses. Cell Cycle. 2012;11:4129–4134. doi: 10.4161/cc.22021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li W, Ma H. Double-stranded DNA breaks and gene functions in recombination and meiosis. Cell Res. 2006;16:402–412. doi: 10.1038/sj.cr.7310052. [DOI] [PubMed] [Google Scholar]
- 18.Mombaerts P, Iacomini J, Johnson RS, Herrup K, Tonegawa S, Papaioannou VE. RAG-1-deficient mice have no mature B and T lymphocytes. Cell. 1992;68:869–877. doi: 10.1016/0092-8674(92)90030-g. [DOI] [PubMed] [Google Scholar]
- 19.Shinkai Y, Rathbun G, Lam KP, Oltz EM, Stewart V, Mendelsohn M, Charron J, Datta M, Young F, Stall AM, et al. RAG-2-deficient mice lack mature lymphocytes owing to inability to initiate V(D)J rearrangement. Cell. 1992;68:855–867. doi: 10.1016/0092-8674(92)90029-c. [DOI] [PubMed] [Google Scholar]
- 20.Karo JM, Schatz DG, Sun JC. The RAG recombinase dictates functional heterogeneity and cellular fitness in natural killer cells. Cell. 2014;159:94–107. doi: 10.1016/j.cell.2014.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Swanson PC. The bounty of RAGs: recombination signal complexes and reaction outcomes. Immunological reviews. 2004;200:90–114. doi: 10.1111/j.0105-2896.2004.00159.x. [DOI] [PubMed] [Google Scholar]
- 22.Kim MS, Lapkouski M, Yang W, Gellert M. Crystal structure of the V(D)J recombinase RAG1-RAG2. Nature. 2015;518:507–511. doi: 10.1038/nature14174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim DR, Dai Y, Mundy CL, Yang W, Oettinger MA. Mutations of acidic residues in RAG1 define the active site of the V(D)J recombinase. Genes Dev. 1999;13:3070–3080. doi: 10.1101/gad.13.23.3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Landree MA, Wibbenmeyer JA, Roth DB. Mutational analysis of RAG1 and RAG2 identifies three catalytic amino acids in RAG1 critical for both cleavage steps of V(D)J recombination. Genes Dev. 1999;13:3059–3069. doi: 10.1101/gad.13.23.3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oettinger MA, Schatz DG, Gorka C, Baltimore D. RAG-1 and RAG-2, adjacent genes that synergistically activate V(D)J recombination. Science. 1990;248:1517–1523. doi: 10.1126/science.2360047. [DOI] [PubMed] [Google Scholar]
- 26.Schatz DG, Spanopoulou E. Biochemistry of V(D)J recombination. Curr Top Microbiol Immunol. 2005;290:49–85. doi: 10.1007/3-540-26363-2_4. [DOI] [PubMed] [Google Scholar]
- 27.Sakano H, Huppi K, Heinrich G, Tonegawa S. Sequences at the somatic recombination sites of immunoglobulin light-chain genes. Nature. 1979;280:288–294. doi: 10.1038/280288a0. [DOI] [PubMed] [Google Scholar]
- 28.Max EE, Seidman JG, Leder P. Sequences of five potential recombination sites encoded close to an immunoglobulin kappa constant region gene. Proc Natl Acad Sci U S A. 1979;76:3450–3454. doi: 10.1073/pnas.76.7.3450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Early P, Huang H, Davis M, Calame K, Hood L. An immunoglobulin heavy chain variable region gene is generated from three segments of DNA: VH, D and JH. Cell. 1980;19:981–992. doi: 10.1016/0092-8674(80)90089-6. [DOI] [PubMed] [Google Scholar]
- 30.van Gent DC, Ramsden DA, Gellert M. The RAG1 and RAG2 proteins establish the 12/23 rule in V(D)J recombination. Cell. 1996;85:107–113. doi: 10.1016/s0092-8674(00)81086-7. [DOI] [PubMed] [Google Scholar]
- 31.Schlissel M, Constantinescu A, Morrow T, Baxter M, Peng A. Double-strand signal sequence breaks in V(D)J recombination are blunt, 5′-phosphorylated, RAG-dependent, and cell cycle regulated. Genes Dev. 1993;7:2520–2532. doi: 10.1101/gad.7.12b.2520. [DOI] [PubMed] [Google Scholar]
- 32.Ji Y, Resch W, Corbett E, Yamane A, Casellas R, Schatz DG. The in vivo pattern of binding of RAG1 and RAG2 to antigen receptor loci. Cell. 2010;141:419–431. doi: 10.1016/j.cell.2010.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu Y, Subrahmanyam R, Chakraborty T, Sen R, Desiderio S. A plant homeodomain in RAG-2 that binds Hypermethylated lysine 4 of histone H3 is necessary for efficient antigen-receptor-gene rearrangement. Immunity. 2007;27:561–571. doi: 10.1016/j.immuni.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matthews AG, Kuo AJ, Ramon-Maiques S, Han S, Champagne KS, Ivanov D, Gallardo M, Carney D, Cheung P, Ciccone DN, Walter KL, Utz PJ, Shi Y, Kutateladze TG, Yang W, Gozani O, Oettinger MA. RAG2 PHD finger couples histone H3 lysine 4 trimethylation with V(D)J recombination. Nature. 2007;450:1106–1110. doi: 10.1038/nature06431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gostissa M, Alt FW, Chiarle R. Mechanisms that promote and suppress chromosomal translocations in lymphocytes. Annu Rev Immunol. 2011;29:319–350. doi: 10.1146/annurev-immunol-031210-101329. [DOI] [PubMed] [Google Scholar]
- 36.Lieber MR, Yu K, Raghavan SC. Roles of nonhomologous DNA end joining, V(D)J recombination, and class switch recombination in chromosomal translocations. DNA Repair (Amst) 2006;5:1234–1245. doi: 10.1016/j.dnarep.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 37.Mills KD, Ferguson DO, Alt FW. The role of DNA breaks in genomic instability and tumorigenesis. Immunological reviews. 2003;194:77–95. doi: 10.1034/j.1600-065x.2003.00060.x. [DOI] [PubMed] [Google Scholar]
- 38.Papaemmanuil E, Rapado I, Li Y, Potter NE, Wedge DC, Tubio J, Alexandrov LB, Van Loo P, Cooke SL, Marshall J, Martincorena I, Hinton J, Gundem G, van Delft FW, Nik-Zainal S, Jones DR, Ramakrishna M, Titley I, Stebbings L, Leroy C, Menzies A, Gamble J, Robinson B, Mudie L, Raine K, O’Meara S, Teague JW, Butler AP, Cazzaniga G, Biondi A, Zuna J, Kempski H, Muschen M, Ford AM, Stratton MR, Greaves M, Campbell PJ. RAG-mediated recombination is the predominant driver of oncogenic rearrangement in ETV6-RUNX1 acute lymphoblastic leukemia. Nat Genet. 2014;46:116–125. doi: 10.1038/ng.2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tycko B, Sklar J. Chromosomal translocations in lymphoid neoplasia: a reappraisal of the recombinase model. Cancer Cells. 1990;2:1–8. [PubMed] [Google Scholar]
- 40.Agrawal A, Schatz DG. RAG1 and RAG2 form a stable postcleavage synaptic complex with DNA containing signal ends in V(D)J recombination. Cell. 1997;89:43–53. doi: 10.1016/s0092-8674(00)80181-6. [DOI] [PubMed] [Google Scholar]
- 41.Gellert M. V(D)J recombination: RAG proteins, repair factors, and regulation. Annu Rev Biochem. 2002;71:101–132. doi: 10.1146/annurev.biochem.71.090501.150203. [DOI] [PubMed] [Google Scholar]
- 42.Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, 3rd, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, Shiloh Y, Gygi SP, Elledge SJ. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 43.Rouse J, Jackson SP. Interfaces between the detection, signaling, and repair of DNA damage. Science. 2002;297:547–551. doi: 10.1126/science.1074740. [DOI] [PubMed] [Google Scholar]
- 44.Zhou BB, Elledge SJ. The DNA damage response: putting checkpoints in perspective. Nature. 2000;408:433–439. doi: 10.1038/35044005. [DOI] [PubMed] [Google Scholar]
- 45.Reinhardt HC, Yaffe MB. Kinases that control the cell cycle in response to DNA damage: Chk1, Chk2, and MK2. Curr Opin Cell Biol. 2009;21:245–255. doi: 10.1016/j.ceb.2009.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fernandez-Capetillo O, Lee A, Nussenzweig M, Nussenzweig A. H2AX: the histone guardian of the genome. DNA Repair (Amst) 2004;3:959–967. doi: 10.1016/j.dnarep.2004.03.024. [DOI] [PubMed] [Google Scholar]
- 47.Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 48.Desiderio SV, Yancopoulos GD, Paskind M, Thomas E, Boss MA, Landau N, Alt FW, Baltimore D. Insertion of N regions into heavy-chain genes is correlated with expression of terminal deoxytransferase in B cells. Nature. 1984;311:752–755. doi: 10.1038/311752a0. [DOI] [PubMed] [Google Scholar]
- 49.Davis MM, Bjorkman PJ. T-cell antigen receptor genes and T-cell recognition. Nature. 1988;334:395–402. doi: 10.1038/334395a0. [DOI] [PubMed] [Google Scholar]
- 50.Nishana M, Raghavan SC. Role of recombination activating genes in the generation of antigen receptor diversity and beyond. Immunology. 2012;137:271–281. doi: 10.1111/imm.12009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gladdy RA, Taylor MD, Williams CJ, Grandal I, Karaskova J, Squire JA, Rutka JT, Guidos CJ, Danska JS. The RAG-1/2 endonuclease causes genomic instability and controls CNS complications of lymphoblastic leukemia in p53/Prkdc-deficient mice. Cancer Cell. 2003;3:37–50. doi: 10.1016/s1535-6108(02)00236-2. [DOI] [PubMed] [Google Scholar]
- 52.Nussenzweig A, Nussenzweig MC. Origin of chromosomal translocations in lymphoid cancer. Cell. 2010;141:27–38. doi: 10.1016/j.cell.2010.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yu W, Nagaoka H, Jankovic M, Misulovin Z, Suh H, Rolink A, Melchers F, Meffre E, Nussenzweig MC. Continued RAG expression in late stages of B cell development and no apparent re-induction after immunization. Nature. 1999;400:682–687. doi: 10.1038/23287. [DOI] [PubMed] [Google Scholar]
- 54.Welner RS, Esplin BL, Garrett KP, Pelayo R, Luche H, Fehling HJ, Kincade PW. Asynchronous RAG-1 expression during B lymphopoiesis. Journal of immunology. 2009;183:7768–7777. doi: 10.4049/jimmunol.0902333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Borghesi L, Hsu LY, Miller JP, Anderson M, Herzenberg L, Schlissel MS, Allman D, Gerstein RM. B lineage-specific regulation of V(D)J recombinase activity is established in common lymphoid progenitors. J Exp Med. 2004;199:491–502. doi: 10.1084/jem.20031800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kouro T, Kumar V, Kincade PW. Relationships between early B- and NK-lineage lymphocyte precursors in bone marrow. Blood. 2002;100:3672–3680. doi: 10.1182/blood-2002-02-0653. [DOI] [PubMed] [Google Scholar]
- 57.Fathman JW, Bhattacharya D, Inlay MA, Seita J, Karsunky H, Weissman IL. Identification of the earliest natural killer cell-committed progenitor in murine bone marrow. Blood. 2011;118:5439–5447. doi: 10.1182/blood-2011-04-348912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ichii M, Shimazu T, Welner RS, Garrett KP, Zhang Q, Esplin BL, Kincade PW. Functional diversity of stem and progenitor cells with B-lymphopoietic potential. Immunol Rev. 2010;237:10–21. doi: 10.1111/j.1600-065X.2010.00933.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pilbeam K, Basse P, Brossay L, Vujanovic N, Gerstein R, Vallejo AN, Borghesi L. The ontogeny and fate of NK cells marked by permanent DNA rearrangements. J Immunol. 2008;180:1432–1441. doi: 10.4049/jimmunol.180.3.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Borghesi L, Gerstein RM. Developmental separation of V(D)J recombinase expression and initiation of IgH recombination in B lineage progenitors in vivo. J Exp Med. 2004;199:483–489. doi: 10.1084/jem.20031802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ichii M, Shimazu T, Welner RS, Garrett KP, Zhang Q, Esplin BL, Kincade PW. Functional diversity of stem and progenitor cells with B-lymphopoietic potential. Immunological reviews. 2010;237:10–21. doi: 10.1111/j.1600-065X.2010.00933.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lanier LL, Chang C, Spits H, Phillips JH. Expression of cytoplasmic CD3 epsilon proteins in activated human adult natural killer (NK) cells and CD3 gamma, delta, epsilon complexes in fetal NK cells. Implications for the relationship of NK and T lymphocytes. Journal of immunology. 1992;149:1876–1880. [PubMed] [Google Scholar]
- 63.Welner RS, Esplin BL, Garrett KP, Pelayo R, Luche H, Fehling HJ, Kincade PW. Asynchronous RAG-1 expression during B lymphopoiesis. J Immunol. 2009;183:7768–7777. doi: 10.4049/jimmunol.0902333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Igarashi H, Kuwata N, Kiyota K, Sumita K, Suda T, Ono S, Bauer SR, Sakaguchi N. Localization of recombination activating gene 1/green fluorescent protein (RAG1/GFP) expression in secondary lymphoid organs after immunization with T-dependent antigens in rag1/gfp knockin mice. Blood. 2001;97:2680–2687. doi: 10.1182/blood.v97.9.2680. [DOI] [PubMed] [Google Scholar]
- 65.Kuwata N, Igarashi H, Ohmura T, Aizawa S, Sakaguchi N. Cutting edge: absence of expression of RAG1 in peritoneal B-1 cells detected by knocking into RAG1 locus with green fluorescent protein gene. Journal of immunology. 1999;163:6355–6359. [PubMed] [Google Scholar]
- 66.Fronkova E, Krejci O, Kalina T, Horvath O, Trka J, Hrusak O. Lymphoid differentiation pathways can be traced by TCR delta rearrangements. Journal of immunology. 2005;175:2495–2500. doi: 10.4049/jimmunol.175.4.2495. [DOI] [PubMed] [Google Scholar]
- 67.Cortez VS, Robinette ML, Colonna M. Innate lymphoid cells: new insights into function and development. Curr Opin Immunol. 2015;32C:71–77. doi: 10.1016/j.coi.2015.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Artis D, Spits H. The biology of innate lymphoid cells. Nature. 2015;517:293–301. doi: 10.1038/nature14189. [DOI] [PubMed] [Google Scholar]
- 69.Spits H, Cupedo T. Innate lymphoid cells: emerging insights in development, lineage relationships, and function. Annu Rev Immunol. 2012;30:647–675. doi: 10.1146/annurev-immunol-020711-075053. [DOI] [PubMed] [Google Scholar]
- 70.Kamimura Y, Lanier LL. Homeostatic Control of Memory Cell Progenitors in the Natural Killer Cell Lineage. Cell Rep. 2015 doi: 10.1016/j.celrep.2014.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Di Santo JP. Natural killer cell developmental pathways: a question of balance. Annu Rev Immunol. 2006;24:257–286. doi: 10.1146/annurev.immunol.24.021605.090700. [DOI] [PubMed] [Google Scholar]
- 72.Prlic M, Blazar BR, Farrar MA, Jameson SC. In vivo survival and homeostatic proliferation of natural killer cells. J Exp Med. 2003;197:967–976. doi: 10.1084/jem.20021847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cooper MA, Bush JE, Fehniger TA, VanDeusen JB, Waite RE, Liu Y, Aguila HL, Caligiuri MA. In vivo evidence for a dependence on interleukin 15 for survival of natural killer cells. Blood. 2002;100:3633–3638. doi: 10.1182/blood-2001-12-0293. [DOI] [PubMed] [Google Scholar]
- 74.Kaech SM, Wherry EJ. Heterogeneity and cell-fate decisions in effector and memory CD8+ T cell differentiation during viral infection. Immunity. 2007;27:393–405. doi: 10.1016/j.immuni.2007.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hogquist KA, Jameson SC, Heath WR, Howard JL, Bevan MJ, Carbone FR. T cell receptor antagonist peptides induce positive selection. Cell. 1994;76:17–27. doi: 10.1016/0092-8674(94)90169-4. [DOI] [PubMed] [Google Scholar]
- 76.Bredemeyer AL, Helmink BA, Innes CL, Calderon B, McGinnis LM, Mahowald GK, Gapud EJ, Walker LM, Collins JB, Weaver BK, Mandik-Nayak L, Schreiber RD, Allen PM, May MJ, Paules RS, Bassing CH, Sleckman BP. DNA double-strand breaks activate a multi-functional genetic program in developing lymphocytes. Nature. 2008;456:819–823. doi: 10.1038/nature07392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sherman MH, Kuraishy AI, Deshpande C, Hong JS, Cacalano NA, Gatti RA, Manis JP, Damore MA, Pellegrini M, Teitell MA. AID-induced genotoxic stress promotes B cell differentiation in the germinal center via ATM and LKB1 signaling. Mol Cell. 2010;39:873–885. doi: 10.1016/j.molcel.2010.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bednarski JJ, Nickless A, Bhattacharya D, Amin RH, Schlissel MS, Sleckman BP. RAG-induced DNA double-strand breaks signal through Pim2 to promote pre-B cell survival and limit proliferation. J Exp Med. 2012;209:11–17. doi: 10.1084/jem.20112078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Schwab SR, Cyster JG. Finding a way out: lymphocyte egress from lymphoid organs. Nat Immunol. 2007;8:1295–1301. doi: 10.1038/ni1545. [DOI] [PubMed] [Google Scholar]
- 80.Larsen BD, Rampalli S, Burns LE, Brunette S, Dilworth FJ, Megeney LA. Caspase 3/caspase-activated DNase promote cell differentiation by inducing DNA strand breaks. Proc Natl Acad Sci U S A. 2010;107:4230–4235. doi: 10.1073/pnas.0913089107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Miyamoto S. Nuclear initiated NF-kappaB signaling: NEMO and ATM take center stage. Cell Res. 2011;21:116–130. doi: 10.1038/cr.2010.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wu ZH, Shi Y, Tibbetts RS, Miyamoto S. Molecular linkage between the kinase ATM and NF-kappaB signaling in response to genotoxic stimuli. Science. 2006;311:1141–1146. doi: 10.1126/science.1121513. [DOI] [PubMed] [Google Scholar]
- 83.Rashi-Elkeles S, Elkon R, Weizman N, Linhart C, Amariglio N, Sternberg G, Rechavi G, Barzilai A, Shamir R, Shiloh Y. Parallel induction of ATM-dependent pro- and antiapoptotic signals in response to ionizing radiation in murine lymphoid tissue. Oncogene. 2006;25:1584–1592. doi: 10.1038/sj.onc.1209189. [DOI] [PubMed] [Google Scholar]
- 84.Elkon R, Rashi-Elkeles S, Lerenthal Y, Linhart C, Tenne T, Amariglio N, Rechavi G, Shamir R, Shiloh Y. Dissection of a DNA-damage-induced transcriptional network using a combination of microarrays, RNA interference and computational promoter analysis. Genome Biol. 2005;6:R43. doi: 10.1186/gb-2005-6-5-r43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Amaravadi R, Thompson CB. The survival kinases Akt and Pim as potential pharmacological targets. J Clin Invest. 2005;115:2618–2624. doi: 10.1172/JCI26273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Nawijn MC, Alendar A, Berns A. For better or for worse: the role of Pim oncogenes in tumorigenesis. Nat Rev Cancer. 2011;11:23–34. doi: 10.1038/nrc2986. [DOI] [PubMed] [Google Scholar]
- 87.Malin S, McManus S, Cobaleda C, Novatchkova M, Delogu A, Bouillet P, Strasser A, Busslinger M. Role of STAT5 in controlling cell survival and immunoglobulin gene recombination during pro-B cell development. Nat Immunol. 2010;11:171–179. doi: 10.1038/ni.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Youle RJ, Strasser A. The BCL-2 protein family: opposing activities that mediate cell death. Nat Rev Mol Cell Biol. 2008;9:47–59. doi: 10.1038/nrm2308. [DOI] [PubMed] [Google Scholar]
- 89.Domen J, van der Lugt NM, Acton D, Laird PW, Linders K, Berns A. Pim-1 levels determine the size of early B lymphoid compartments in bone marrow. J Exp Med. 1993;178:1665–1673. doi: 10.1084/jem.178.5.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mikkers H, Nawijn M, Allen J, Brouwers C, Verhoeven E, Jonkers J, Berns A. Mice deficient for all PIM kinases display reduced body size and impaired responses to hematopoietic growth factors. Mol Cell Biol. 2004;24:6104–6115. doi: 10.1128/MCB.24.13.6104-6115.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Fugmann SD, Lee AI, Shockett PE, Villey IJ, Schatz DG. The RAG proteins and V(D)J recombination: complexes, ends, and transposition. Annual review of immunology. 2000;18:495–527. doi: 10.1146/annurev.immunol.18.1.495. [DOI] [PubMed] [Google Scholar]
- 92.Lee GS, Neiditch MB, Salus SS, Roth DB. RAG proteins shepherd double-strand breaks to a specific pathway, suppressing error-prone repair, but RAG nicking initiates homologous recombination. Cell. 2004;117:171–184. doi: 10.1016/s0092-8674(04)00301-0. [DOI] [PubMed] [Google Scholar]
- 93.Liang HE, Hsu LY, Cado D, Cowell LG, Kelsoe G, Schlissel MS. The “dispensable” portion of RAG2 is necessary for efficient V-to-DJ rearrangement during B and T cell development. Immunity. 2002;17:639–651. doi: 10.1016/s1074-7613(02)00448-x. [DOI] [PubMed] [Google Scholar]
- 94.Deriano L, Chaumeil J, Coussens M, Multani A, Chou Y, Alekseyenko AV, Chang S, Skok JA, Roth DB. The RAG2 C terminus suppresses genomic instability and lymphomagenesis. Nature. 2011;471:119–123. doi: 10.1038/nature09755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Desiderio S. Temporal and spatial regulatory functions of the V(D)J recombinase. Semin Immunol. 2010;22:362–369. doi: 10.1016/j.smim.2010.09.001. [DOI] [PubMed] [Google Scholar]
- 96.Jiang H, Chang FC, Ross AE, Lee J, Nakayama K, Nakayama K, Desiderio S. Ubiquitylation of RAG-2 by Skp2-SCF links destruction of the V(D)J recombinase to the cell cycle. Mol Cell. 2005;18:699–709. doi: 10.1016/j.molcel.2005.05.011. [DOI] [PubMed] [Google Scholar]
- 97.Li Z, Dordai DI, Lee J, Desiderio S. A conserved degradation signal regulates RAG-2 accumulation during cell division and links V(D)J recombination to the cell cycle. Immunity. 1996;5:575–589. doi: 10.1016/s1074-7613(00)80272-1. [DOI] [PubMed] [Google Scholar]
- 98.Zhang L, Reynolds TL, Shan X, Desiderio S. Coupling of V(D)J recombination to the cell cycle suppresses genomic instability and lymphoid tumorigenesis. Immunity. 2011;34:163–174. doi: 10.1016/j.immuni.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chaumeil J, Skok JA. A new take on v(d)j recombination: transcription driven nuclear and chromatin reorganization in rag-mediated cleavage. Front Immunol. 2013;4:423. doi: 10.3389/fimmu.2013.00423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Puri PL, Bhakta K, Wood LD, Costanzo A, Zhu J, Wang JY. A myogenic differentiation checkpoint activated by genotoxic stress. Nat Genet. 2002;32:585–593. doi: 10.1038/ng1023. [DOI] [PubMed] [Google Scholar]
- 101.Inomata K, Aoto T, Binh NT, Okamoto N, Tanimura S, Wakayama T, Iseki S, Hara E, Masunaga T, Shimizu H, Nishimura EK. Genotoxic stress abrogates renewal of melanocyte stem cells by triggering their differentiation. Cell. 2009;137:1088–1099. doi: 10.1016/j.cell.2009.03.037. [DOI] [PubMed] [Google Scholar]
- 102.Rossi DJ, Bryder D, Seita J, Nussenzweig A, Hoeijmakers J, Weissman IL. Deficiencies in DNA damage repair limit the function of haematopoietic stem cells with age. Nature. 2007;447:725–729. doi: 10.1038/nature05862. [DOI] [PubMed] [Google Scholar]