Abstract
Carcinogenesis is a complex process during which cells undergo genetic and epigenetic alterations. These changes can lead tumor cells to acquire characteristics that enable movement from the primary site of origin when conditions become unfavorable. Such characteristics include gain of front-rear polarity, increased migration/invasion, and resistance to anoikis, which facilitate tumor survival during metastasis. An epithelial to mesenchymal transition (EMT) constitutes one way that cancer cells can gain traits that promote tumor progression and metastasis. Two microRNA (miRNA) families, the miR-200 and miR-221 families, play crucial opposing roles that affect the differentiation state of breast cancers. These two families are differentially expressed between the luminal A subtype of breast cancer as compared to the less well-differentiated triple negative breast cancers (TNBCs) that exhibit markers indicative of an EMT. The miR-200 family promotes a well-differentiated epithelial phenotype, while high miR-221/222 results in a poorly differentiated, mesenchymal-like phenotype. This review focuses on the mechanisms (specific proven targets) by which these two miRNA families exert opposing effects on cellular plasticity during breast tumorigenesis and metastasis.
Keywords: miR-200, miR-221, miR-222, EMT, MET, breast cancer
Introduction
miRNAs are small (18–25 nucleotide) non-coding RNAs that regulate gene expression post-transcriptionally by binding to the 3’ untranslated region (UTR) of target messenger RNAs (mRNAs) (1), and inhibiting translation or targeting the mRNA for degradation (2). The extent to which miRNAs regulate the human transcriptome is still under investigation; however, miRNAs can target hundreds of genes, suggesting that their regulatory role may be as significant as that of transcription factors. miRNAs are differentially regulated during development (3–5). Controlled epithelial to mesenchymal transition (EMT) is a normal process in development, required for processes such as gastrulation, mammary gland branching, and neural crest formation (reviewed in (6)). However, EMT is a pathological event in cancer that contributes to the gain of aggressive characteristics that facilitate metastasis (7–10). In cancer EMT, carcinoma cells do not become mesenchymal cells, although there can be a marked loss of epithelial hallmarks and a shift toward mesenchymal and even neuronal gene expression. It is widely believed that acquisition of these characteristics can allow tumor cells to become motile, invasive, and able to intravasate into the blood and lymph vessels and survive the metastatic journey. Transcription factors, such as Twist, Snai1, and ZEB1/2 (Reviewed in (11)) regulate both normal and oncogenic EMT. ZEB1 (zinc finger E-box binding homeobox 1) and ZEB2 (also known as SIP1) directly repress the adherens junction protein E-cadherin (12–14) and other genes involved in polarity and epithelial identity (15,16).
ZEB1/2 are post-transcriptionally controlled by the miR-200 family of miRNAs (17–19), and ZEB2 is indirectly controlled by the miR-221 family (20). Indeed, recent studies have identified the miR-200 and miR-221 families as differentially expressed in carcinomas, particularly in breast cancer (20,21). Specifically, the miR-200 family is high in the luminal breast cancer subtypes, while miR-221/222 is overexpressed in triple negative breast cancers (TNBCs), particularly those that have undergone EMT. These miRNAs control expression of many genes that define the EMT-like phenotype and likely affect tumor behavior and clinical outcome by influencing metastatic potential. Thus, in this review we focus on the opposing roles of these two miRNA families in controlling differentiation state or epithelial identity in breast cancer.
miR-200 protection of the epithelial phenotype
miR-200 family regulation of EMT in breast cancer
The miR-200 family of miRNAs is comprised of two polycistronic clusters – miR-200c and miR-141 on chromosome 12 and miR-200b, miR-200a and miR-429 on chromosome 1. miR-200a and miR-141 share a seed sequence, while miR-200b, miR-200c and miR-429 also share a seed sequence, which differs from that of miR-200a/141 by one nucleotide. Because of their sequence similarity, the miRNAs are predicted to share gene targets; however, there is evidence that the two clusters control different regulatory networks even in the same model. In MDA-231 cells the miR-200bc/429 cluster induces G2/M arrest, while miR-200a/141 induces G0/1 arrest (22). Additionally, miR-200c directly targets and down-regulates the transcription factor ZEB1, while miR-200a does not (23).
The miR-200 family was first discovered to directly target and down-regulate the E-cadherin transcriptional repressors ZEB1 and ZEB2, leading to restoration of an epithelial phenotype in breast cancer cell lines, characterized by an increase in E-cadherin expression, and decreased migration and invasion (17–19). Expression of the miR-200 family correlates with an epithelial-like phenotype in the National Cancer Institute (NCI) panel of 60 cancer cells lines (19), and suppresses EMT in several additional cancer models, including bladder (24), colorectal (25,26), and lung (27–30). Although genes encoding ZEB1/2 are the best-studied targets of the miR-200 family, the small consensus binding sequence of miRNAs results in many bioinformatically predicted targets. The miR-200 family has now been confirmed to directly target other genes involved in various aspects of EMT. One aspect of EMT that has been particularly well studied is the increase in migratory and invasive capacity. Targeting and repression of the genes encoding ZEB1/2 by miR-200c and the resultant increase in E-cadherin decreases migration and invasion; however, direct targeting of genes encoding the actin cytoskeleton associated proteins WAVE3 (31) and MSN (32), and the extracellular matrix component FN1 (32) also contribute to suppression of motility and invasion. The miR-200 family also targets two genes involved in cell cycle control, RND3 (33) and FOG2 (34).
The power of miRNAs lies in their ability to target multiple genes that contribute to a pathway or phenotype. For instance, normal well-differentiated mammary epithelial cells exhibit hallmarks such as E-cadherin and hormone receptor expression, while poorly differentiated breast carcinoma cells loose these characteristics. When carcinoma cells revert towards a less-differentiated state, in addition to loosing expression of epithelial hallmarks, they also inappropriately gain expression of proteins that confer the ability to move away from the primary tumor when conditions are harsh (hypoxia, lack of nutrients, and build-up of waste products). The tumor cells must also be able to resist anoikis in order to survive detachment from the basement membrane.
Anoikis resistance is a relatively poorly understood and understudied aspect of EMT. Anoikis is apoptosis induced when cells lose attachment to their native extracellular matrix (ECM), and resistance to anoikis is required for cancer cells to survive as they move away from the primary tumor, and travel through the vasculature or lymphatics to metastatic sites. Data from our lab demonstrate that miR-200c suppresses anoikis resistance through direct targeting of NTRK2, the gene encoding TrkB (32), a receptor tyrosine kinase involved in neuronal development and differentiation. TrkB was first associated with anoikis resistance when it was isolated from a cDNA library screen designed to identify genes capable of conferring anoikis resistance to normal intestinal epithelial cells (35). TrkB is involved in anoikis resistance in breast cancer (32,35–38) and is specifically expressed in TNBCs that have undergone EMT, but not luminal A lines (32).
Resistance to chemotherapy is a critical aspect of tumorigenesis also associated with acquisition of an EMT phenotype. The miR-200 family has been found to be involved in maintaining sensitivity to two classes of chemotherapeutics to date, microtubule targeting agents, and DNA damaging drugs. In aggressive cancer cells resistant to taxanes, restoration of miR-200c increases sensitivity due to its direct targeting of TUBB3, the gene encoding class III beta tubulin (39,40). TUBB3 is a tubulin isoform aberrantly expressed in several types of carcinomas (41–43), including breast (44,45), that leads to resistance to taxanes (Reviewed in (46)). Additionally, the miR-200 family is down-regulated in MCF7 cells selected for resistance to cisplatin (47), or doxorubicin (48). Indeed, miR-200 expression correlates with sensitivity to EGFR blocking agents in bladder cancer, and restoration of miR-200 family members increased sensitivity to EGFR inhibitors in mesenchymal-like cell lines (49). Additionally, lower expression of miR-200c was observed in a panel of 39 breast cancer patients resistant to chemotherapy (48). The authors speculate that these effects may be due to the predicted targeting of the multidrug resistance gene 1 by miR-200c, but this remains to be proven. Finally, miR-200c directly targets FAP-1, leading to restoration of sensitivity to CD-95 (Fas) -mediated apoptosis (50). Thus, the miR-200 family exerts multi-level control over apoptosis in epithelial cells. The family promotes sensitivity to natural apoptotic stimuli, including loss of adhesion and Fas signaling, while also preventing resistance to several classes of therapeutic agents.
While not classically thought of as a characteristic of EMT, an overall decrease in miRNA abundance is found in aggressive cancer cells (51,52). Dicer, an enzyme involved in the maturation of miRNAs, is often low in cancers that have undergone EMT (53). While the mechanism remains to be elucidated, we demonstrated that restoration of miR-200c to TNBC cell lines causes an increase in Dicer protein (21). Since relatively high levels of Dicer and overall miRNA abundance are characteristic of normal epithelial cells, this is a unique mechanism through which the miR-200 family promotes an epithelial phenotype.
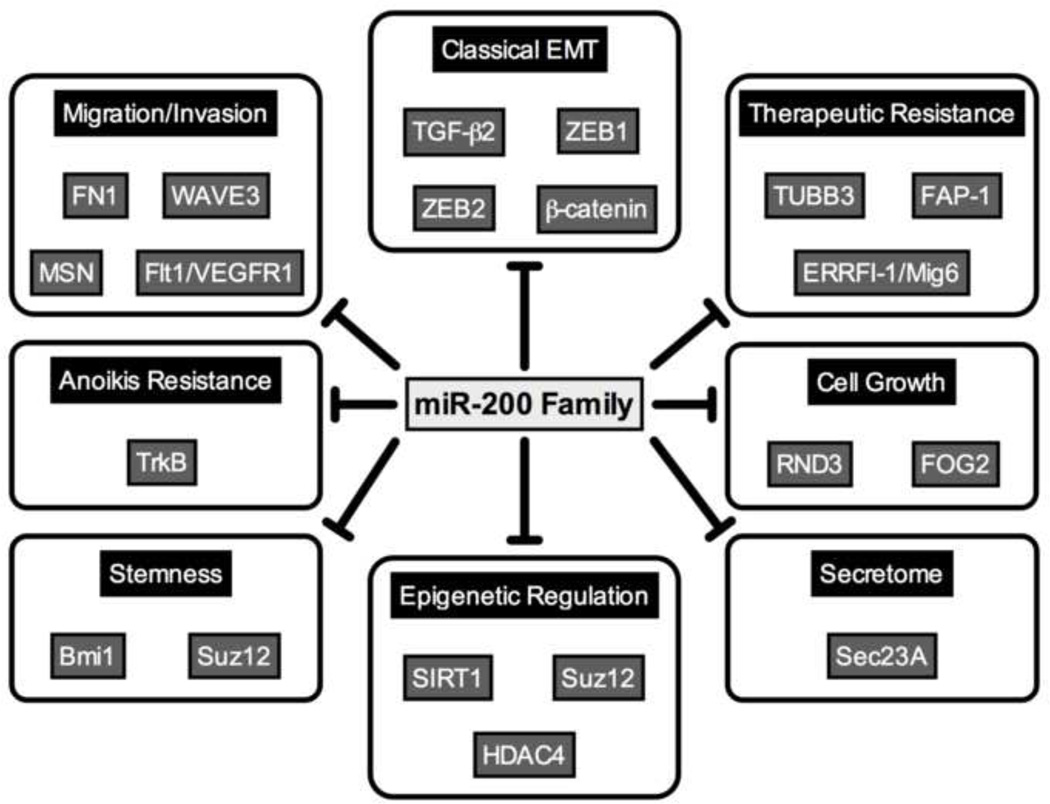
In addition to regulation of EMT, there is emerging evidence that the miR-200 family plays a role in epigenetic regulation and inhibition of stem cell-like qualities in breast, prostate (54,55), and colorectal cancer cells (26). Expression of both miR-200 family clusters is down-regulated in stem cells isolated from normal human breast, and murine mammary glands, as well as in stem cells isolated from breast cancer patients (56). Inhibition of miR-200 leads to an enrichment of the stem cell population, and up-regulation of the miR-200b direct target Suz12, a subunit of the polycomb repressor complex. Increased Suz12 leads to trimethylation and polycomb-mediated repression of the E-cadherin promoter (57). Another direct target, the gene encoding class III histone deacetylase, SIRT1, deacetylates histone H3 at the E-cadherin promoter, and miR-200 mediated repression of SIRT further relieves repression of E-cadherin (58). The miR-200 family also directly targets and represses Bmi1, allowing further repression of stemness (26). Additionally, expression of miR-200c inhibits clonal expansion of stem cells, and prevents tumor formation from patient-derived breast cancer stem cells transplanted into mice (56). Finally, two important stem cell factors, Sox2 and KLF4 have been found to be down-regulated following restoration of miR-200c (26). Thus, the miR-200 family controls multiple genes that repress cancer stem cells, leading to restoration of an epithelial phenotype and decreased aggressiveness. The genes and aggressive phenotypes repressed by the miR-200 family are detailed in Figure 1.
Figure 1.
Direct targets of the miR-200 family. Members of the miR-200 family directly target and down-regulate genes involved in a variety of processes that contribute to tumorigenesis and metastasis. References are included in the text.
The miR-200 family is highly expressed in luminal A breast cancer cell lines and lost in TNBC lines (21); however, data from primary and metastatic breast cancer samples are not as clear. Based on the cell line data, it was expected that the miR-200 family would be down-regulated in aggressive tumors and metastases. While this is true in some models, and restoration of miR-200 to a TNBC cell line prevents metastases (59), in other models the miR-200 family positively correlates with metastases (60,61). Consistent with the theory that miR-200c positively correlates with a well-differentiated phenotype, the miR-200 family is very low in the poorly differentiated claudin-low subtype of breast cancer , while expression of ZEB1/2, vimentin, and Twist are high and these tumors are enriched for tumor initiating cells, suggesting that the miR-200 family must be down-regulated for formation of an aggressive subpopulation of tumor cells (62). However, while several profiling studies found that expression of the miR-200 family is lost between normal breast tissue and malignant breast cancers (18,63) one profiling experiment (64), comparing luminal A, luminal B, basal-like and malignant myoepithelioma, revealed that while the miR-200 family is highly expressed in luminal tumors, it is also highly expressed in basal-like tumors. Only malignant myoepitheliomas showed down-regulation of the miR-200 family, which is consistent with a strong EMT phenotype (64).
Expression of the miR-200 family in metastatic disease has been even more contested. While one group found the miR-200 family to be down-regulated between matched primary versus metastatic breast, colon, lung and bladder cancers (65), another showed that the miR-200 family is over-expressed in matched metastases, and that higher than median expression of several family members correlates with decreased progression free survival in estrogen receptor (ER) positive breast tumors (61). In contrast, high expression of miR-200b, and low expression of Suz12 can distinguish primary breast tumors from metastases, which express low miR-200b and high Suz12 (57). Further complicating the matter are two studies performed in syngeneic mouse mammary carcinoma models. In one study, using the 4T1 panel of cells lines, expression of miR-200 in a non-metastatic cell line increased metastasis (60). Forced expression of miR-200c and miR-141, or all members of the miR-200 family led to increased metastasis in a similar model, the 4TO7 cell line (61). These studies suggest that expression of the miR-200 family may induce mesenchymal to epithelial transition (MET) during the metastatic cascade. Induction of MET may be necessary for colonization of cells at the metastatic site, which would be consistent with increased expression of the miR-200 family. It is also possible that EMT is not required for metastasis in these models. Another possible explanation is that there are differences in the rate limiting steps of the metastatic cascade across models, which could affect the necessity of MET in colonization. Finally, regulated expression of miR-200 may be important for phenotypic plasticity, and may allow cells to transition between epithelial and mesenchymal states as needed.
miR-200 family in plasticity
There is mounting evidence that both EMT and MET are important in the progression of carcinomas, and that carcinoma cells exhibit increased plasticity, allowing them to transition as necessary. Both EMT and MET are required for proper development, and the role of the miR-200 family in transitions between the epithelial and mesenchymal states is becoming clear. During embryonic stem cell differentiation, the miR-200 family is down-regulated by Snai1 and Wnt signaling, and forced expression of miR-200 leads to cells stalling at the epiblast-like stem cell stage of differentiation (66). The miR-200 family is also regulated by c-Myc in differentiating embryonic stem cells (67).
Forced expression of miR-200c in epithelial cells of the developing mammary gland suppresses ductal growth (56), suggesting that plasticity is required for proper formation of the ducts. Similarly, forced expression of miR-200 in plastic, metastatic lung adenocarcinoma cells reversed plasticity, preventing the cells from undergoing EMT or metastasizing (68). Manipulation of ZEB1/2 and the miR-200 family in Madin-Darby canine kidney (MDCK) cells leads to EMT and MET, respectively, but the states remain plastic and can be reversed (69). miRNA profiling of embryonic stem cells, induced pluripotent stem (iPSC) cells, differentiated cells and cancer cells revealed that the pluripotent stem cells formed two clusters, irrespective of the origin of the cells (embryonic versus induced). The miRNAs that distinguished these groups also differentiated normal cells from cancer cells. Expression of miR-92 or miR-200 family members in iPSCs changed their classification status, leading the authors to suggest that the subdivision in pluripotent stem cell states does not reflect their origin, but rather miRNA and gene expression network (70). Similarly, the miR-200 family is regulated during reprogramming of somatic cells into iPSCs (71). Thus, the miR-200 family, as well as EMT-inducing transcription factors, must be expressed in the proper order to allow differentiation of embryonic stem cells.
Regulation of the miR-200 family
The most potent regulators of the miR-200 family are ZEB1 and ZEB2, which have been demonstrated to target E-boxes in the miR-200 cluster promoters (72,73). Another well recognized EMT inducer, transforming growth factor beta (TGF-β), has also been shown to reduce expression of the miR-200 family in transformed human breast epithelial cells (74), murine mammary epithelial cells (75), prostate cancer cells (76), and canine renal MDCK cells, a model of the epithelial phenotype (18,77). Indeed, treatment with TGF-β leads to hypermethylation of the miR-200 promoters, potentially through miR-200a-mediated direct targeting of the histone deacetylase SIRT1 (74). Further study of the role of epigenetic regulation of the family revealed that the promoters are unmethylated in epithelial cells, and in cancer cells that express the family, but heavily methylated in fibroblasts and tumors that do not express the miR-200 family (78,79). Furthermore, the permissive epigenetic mark, histone H3 acetylation, is decreased at the miR-200 promoter in cancer cells lacking expression of the family (80), an epigenetic mark potentially influenced by miR-200a direct targeting of HDAC4. Together, this data indicates that while classical EMT-inducers control expression of the miR-200 family in tumorigenesis, epigenetic control is also important, and potentially forms feedback loops through miR-200 control of epigenetic regulators, including SIRT1, HDAC4, and Suz12.
Several other EMT inducers down-regulate the miR-200 family, including platelet derived growth factor (PDGF) (81), long-term treatment with the epidermal growth factor receptor (EGFR) inhibitor gemcitabine (82), and carcinogen induced tumorigenesis (83). Interestingly, treatment of pancreatic cancer cells with curcumin, or the analog CDF, along with gemcitabine lead to increased miR-200 family expression (81,84). Additionally, Akt isoforms leads to differential miRNA expression profiles. Expression of only Akt2 dramatically decreases expression of the miR-200 family, while knockdown of Akt1 induced EMT by reducing expression of the miR-200 family. The authors suggest that the expression of miR-200 family members depends on the ratio of Akt1/Akt2, rather than the overall activity of Akt (85). To date, the only known activators of miR-200 expression are the tumor suppressors p53 (86,87), p63, and p73 (88), and ERalpha (89). However, there are likely other positive-regulators of the miR-200 family.
miR-221/222 suppression of the epithelial phenotype
miR-221/222 expression in breast cancer and other carcinomas
miR-221 and miR-222 are found on the X chromosome and are expressed from a single transcript. For many cancer types, miR-221/222 are considered oncomiRs, and are overexpressed in tumor compared to normal tissue of origin. This expression pattern holds true in breast (63), prostate (90), gastric (91), bladder (92), papillary thyroid carcinoma (93), colorectal cancer (94), melanoma (95), and acute myeloid leukemia (96). High miR-221/222 expression is associated with increased tumor grade (97,98) and poor prognosis (99). High miR-221 is found in prostate cancer cell lines, where it is associated with aggressive phenotypes, such as androgen-independence and neuroendocrine differentiation (90).
Several studies have demonstrated that miR-221/222 directly target ERα (21,100,101). In breast cancer, miR-221/222 negatively correlate with ER status, and are more highly expressed in triple negative cell lines as compared to luminal (20,21,100) and the same holds true in clinical samples (21,102). Additionally, in the murine mammary tumor virus (MMTV)-c-myc mouse model of mammary carcinoma, miR-222 is increased during tumorigenesis (103). However, some controversy exists, since one study observed that although miR-221 is overexpressed in TNBCs and is associated with poor disease-free and overall survival, there was no difference in miR-222 expression between breast cancer and normal epithelial tissue (99). Additionally, another study found that miR-221 expression positively correlated with ER status in breast cancer patient samples, while miR-222 expression did not change between ER positive and ER negative samples (104). Thus, as with the miR-200 family, although expression of miR-221/222 correlates strongly with specific phenotypes in vitro in breast cancer cell lines, more work is required to fully elucidate the role of the family in human tumors.
miR-221/222 in EMT and metastasis
Since miR-221/222 are often overexpressed in poorly differentiated, aggressive cancers, it stands to reason that these miRNAs play an active role in promoting EMT. Increasing miR-221 or 222 can affect various characteristics associated with EMT, including increased invasive capacity (90,105), and anoikis resistance (106). Low Dicer is characteristic of poorly differentiated cells and cells that have undergone EMT. In TNBC lines, miR-221/222 directly target and repress Dicer1 (21), leading to the possibility that aberrant expression of miR-221/222 leads to decreased Dicer, which in turn leads to a decrease in overall miRNA abundance.
Long term mammosphere culture of MCF7 cells induces EMT, with the resulting cells displaying a basal B phenotype (107). The cells also exhibit increased expression of stem cell markers (CD44 + /CD24 − /low), and exhibited stem cell-like characteristics, including chemoresistance. qRT-PCR miRNA profiling demonstrates that miR-200c, −203 and 205 are decreased, while miR-221/222 are increased in the mammosphere cultured cells, with miR-222 increased 20-fold (107). Thus, although further more exhaustive and rigorous genetic analysis of necessity and sufficiency remains to be performed, it appears that induction of EMT in luminal breast cancer cells involves decreased expression of the miR-200 family and increased expression of miR-221/222. Although miR-221/222 are high in both basal A and B breast cancer, their expression is higher in the basal B subtype, which has a more mesenchymal phenotype (20), consistent with the role of miR-221/222 in EMT. Forced expression of miR-221/222 in luminal breast cancer cells causes a decrease in E-cadherin and an increase in the mesenchymal marker vimentin (20). Luminal cells expressing miR-221/222 gained a more mesenchymal morphology and had increased migratory and invasive capacity. Conversely, inhibition of miR-221/222 in basal-like cells promoted MET (108). miR-221/222 promote a mesenchymal phenotype in part by directly targeting trichorhinophalangeal 1 (TRPS1), and keeping its levels low (20). TRPS1 is a transcriptional repressor that binds to GATA sites that can promote MET (20), and is underexpressed in breast cancers with poor clinical outcome (109). TRPS1 represses the mesenchymal transcription factor ZEB2 through a GATA site in its promoter. As ZEB2 is a repressor of E-cadherin, this provides a functional link between expression of miR-221/222 and repression of E-cadherin in basal breast cancers (20,110).
miR-221/222 control of proliferation
miR-221/222 positively influence cellular proliferation in many types of cancers. While there are several mechanisms through which increased growth rate is achieved, the best studied is direct targeting of p27KIP1 (98,111), and p57KIP2 (112,113). In patient samples, miR-221 or miR-222 levels are often inversely correlated with p27KIP1 (111,114–116) or p57KIP2 (94,112). Increasing the expression of miR-221 or miR-222 causes increased proliferation in vitro (111,114), and increased tumor growth in xenograft tumor models (117). Conversely, antagonizing miR-221/222 results in decreased proliferation both in vitro (94) and in vivo (118). In one study, decreased tumor growth was achieved through in vivo administration of cholesterol modified anti-miR-221, which suggests that miR-221 can be a viable therapeutic target for the treatment of aggressive cancers (119).
Direct targets other than p27KIP1 and p57KIP2 can also mediate the proliferative effects of miR-221/222. In gastric cancer cells, the proliferative effects of miR-221/222 are partially due to their ability to directly target PTEN (105), and targeting of PTEN is also likely to play an important role in breast carcinomas. Additionally, miR-221/222 directly target ARH1 (120), a tumor suppressor protein decreased in many types of cancers (121–123). Loss of ARH1 results in increased proliferation, colony formation and invasion (120). Thus, miR-221/222 promote proliferation by suppressing targets that normally serve to repress proliferative pathways.
miR-221/222 in resistance to apoptotic stimuli
Overexpression of miR-221/222 serves to protect cancer cells against various forms of apoptotic stimuli, including chemotherapeutics, endocrine therapies, radiotherapy and detached growth conditions. MCF7 cells resistant to cisplatin have increased miR-221/222 expression compared to the wild type cells (47). Antagonizing miR-221 in pancreatic cell lines causes increased apoptosis and sensitized the cells to gemcitabine (124). miR-221 and miR-222 are increased in taxol resistant cells, and addition of miR-221 to breast cancer cells results in increased survival in response to paclitaxel treatment (125). One of the mechanisms through which miR-221/222 repress apoptosis is through direct targeting of pro-apototic genes, such as PUMA (126) and BMF (106).
Her2/neu amplified breast cancers tend to be resistant to endocrine therapy (127,128). miR-221/222 are high in breast cancers that are positive for Her2/neu, compared to Her2/neu negative breast cancers, and overexpression of miR-221/222 causes MCF7 cells to become tamoxifen resistant (129). miR-221/222 directly target p27KIP1 (114) and this is one of the mechanisms through which the cells become tamoxifen-resistant. In xenograft tumors that are resistant to tamoxifen, antagonizing miR-222 sensitizes tumors to tamoxifen (130). miR-221/222 directly target TIMP3, a tissue metalloproteinase inhibitor that normally inhibits tamoxifen resistant tumor growth. In breast cancer cells that have become resistant to tamoxifen through increased miR-221/222 expression, TIMP3 is repressed, and there is a resultant increase in the expression of metalloproteases ADAM17 and ADAM 10, as well as increased growth factor signaling (130).
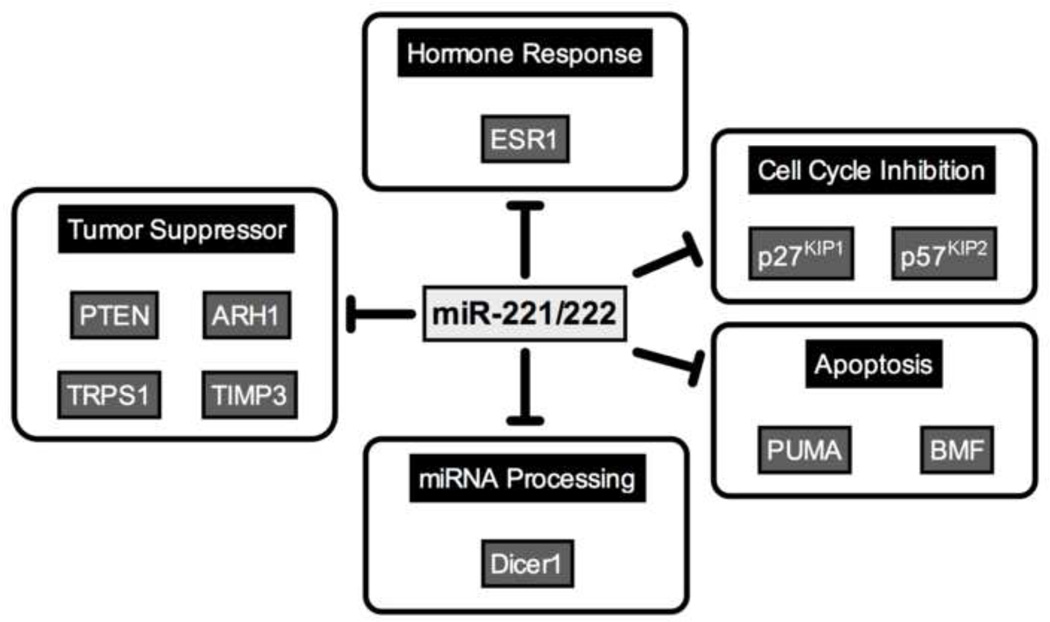
While MCF7 cells treated with tamoxifen have slightly decreased levels of miR-221/222, cells treated with fulvestrant, either alone or in combination with E2, have increased miR-221/222 expression (131), likely because ER represses miR-221/222 (101), so degradation of ER after fulvestrant binding could relieve repression of miR-221/222. Inhibition of miR-221/222 activity causes decreased proliferation. Fulvestrant resistance is explained in part by the downregulation of p27KIP1 and p57KIP2 (111,112), and ER (100,101). Increased p-catenin contributes to fulvestrant resistance and E2 independent growth (132). Cells overexpressing miR-221/222 have increased nuclear β-catenin, corresponding to increased β-catenin-mediated transcriptional activity. TGF-β1 blocks proliferation in wild type MCF7s, but not the fulvestrant resistant cells (133,134). However, overexpression of miR-221 or miR-222 in wild type cells increases survival in response to TGF-β1, and antagonizing these miRNAs in resistant cells increases sensitivity (131). Therefore, it is possible that miR-221/222 are involved in switching the effect of TGF-β signaling from tumor suppressive to tumor promotional. The genes and phenotypes regulated by miR-221/222 are depicted in Figure 2.
Figure 2.
Direct targets of miR-221/222. miR-221/222 directly target and down-regulate genes associated with differentiation or tumor suppression. References are included in the text.
Regulation of miR-221/222
There is a negative feedback loop between miR-221/222 and ERα. miR-221/222 directly bind to and down-regulate ERα, while ERα binds to estrogen response elements in the promoter of miR-221/222 and represses transcription (101). Other transcriptional repressors of miR-221/222 function in a cell-type specific manner. For example, in AML cells, the AML1 protein binds to the promoter of miR-221/222 and represses transcription (135). In melanoma cells, a transcriptional repressor, PLZF (promyelocytic leukemia zinc finger) binds to the promoter of miR-221/222 (136).
FOSL1 (Fra-1) is part of the AP-1 transcription complex and promotes invasiveness and metastatic potential of breast cancers (137–139). FOSL1 binds an AP-1 site upstream of miR-221/222 and promotes transcription (20). Activation of the RAS/RAF/MEK pathway increased expression of miR-221/222 in basal breast cancer cells via FOSL1 (20), and activation of the MAPK pathway also increases miR-221/222 expression [D. El-Ashry, Personal Communication].
Interplay between the miR-200 and miR-221 families
Perhaps the most convincing evidence that these two families play an important role in epithelial plasticity in breast cancer comes from the White lab, in a study where breast cancer cells were forced to undergo EMT by being grown in mammosphere conditions. The resulting cells had decreased miR-200, and increased miR-221/222 (107). Collectively, as described above, these two families clearly exert opposing effects on polarity, migration and invasion, proliferation, apoptosis, and differentiation.
ZEB1/2 transcription factors promote a mesenchymal phenotype by repressing genes involved in polarity. Therefore, ZEB1/2 is detrimental to an epithelial phenotype, and it is essential that these genes remain suppressed in differentiated epithelial cells. While they are most definitely repressed at the promoter level, epithelial cells have evolved an additional layer of protection against their expression, which is miR-200 mediated repression at the post-transcriptional level. Conversely, miR-221/222 promote expression of ZEB2 indirectly through TRPS1, and therefore these miRNAs tend to only be expressed in cells that have undergone EMT (20).
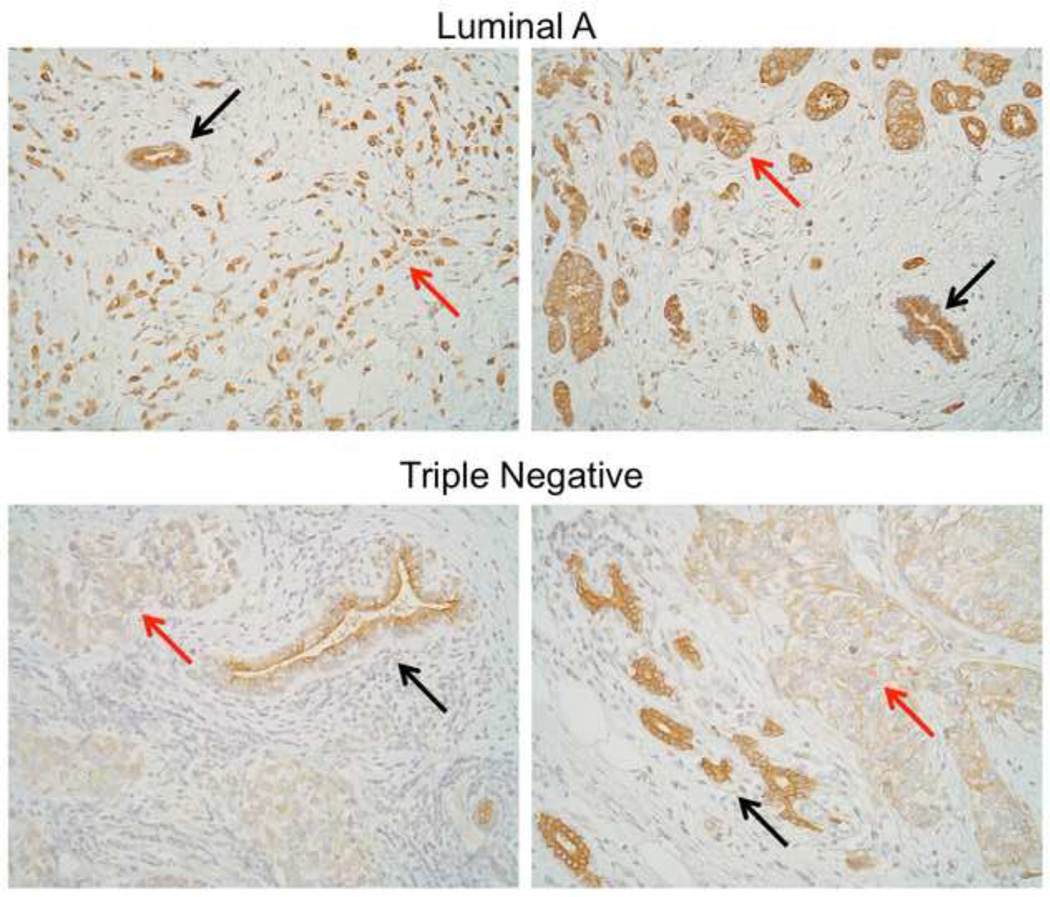
miR-221/222 directly target and repress Dicer, while miR-200c increases Dicer by a yet to be identified mechanism (21). miR-221/222 are more highly expressed in TNBC (21,100). miR-103/107 have also been demonstrated to directly target Dicer (140); however, an inverse correlation between these miRNA and Dicer has not been as well documented as it has for miR-221/222 which are high in tumors in which Dicer levels are low (TNBC). Thus, miR-221/222 may keep Dicer levels low in poorly differentiated breast cancers (21). Since Dicer is required for the maturation of most miRNAs, this may explain why overall miRNA expression is lower in TNBC than luminal. Dicer is often low in cancers that have undergone EMT (53). Dicer is clearly lower in TNBC than adjacent normal breast epithelial cells, while in luminal A breast cancers the difference between tumor and normal is much less dramatic (Figure 3). Interestingly, TAp63 was recently discovered to suppress metastasis by positively regulating Dicer (141). It is possible that miR-200c increases Dicer through its ability to repress ZEB1, which upregulates deltaNp63 (142), a dominant negative inhibitor of TAp63.Consequently, the miR-221 and miR-200 families may control the global miRNA landscape in normal and cancerous cells by dueling for control of Dicer. Much remains to be explored to fully determine how the influence of these miRNA families over Dicer might control motility and metastasis in normal development and cancer.
Figure 3.
Dicer protein expression in luminal A and triple negative breast cancer. Formalin-fixed paraffin embedded sections of human breast cancers were stained for Dicer using ab5818 polyclonal antibody (Abcam, Cambridge, MA). Two representative cases each of luminal and triple negative are shown in which adjacent normal glands are present in the same field of vision (top = luminal, bottom = triple negative) with adjacent normal tissue. Red arrows = tumor, black arrows = normal, 200X.
Conclusions
The role of miRNAs in tumorigenesis and the power they wield with respect to phenotypic control and tumor behavior is just beginning to be understood. In this review we focus on two of the most dysregulated miRNA families in breast cancer, the miR-200 and miR-221 families. The miR-200 family serves to protect the epithelial phenotype, while simultaneously suppressing EMT and tumorigenesis. The miR-200 family protects against migration/invasion, anoikis- and therapeutic resistance, and stem cell-like properties. Conversely, miR-221/222 promote a mesenchymal-like phenotype, and support tumorigenesis. Expression of miR-221/222 inhibits tumor suppressors and genes involved in apoptosis, cell cycle inhibition, and miRNA processing. Both miRNA families impinge on two important pathways: EMT through ZEB1/2, and miRNA processing through Dicer.
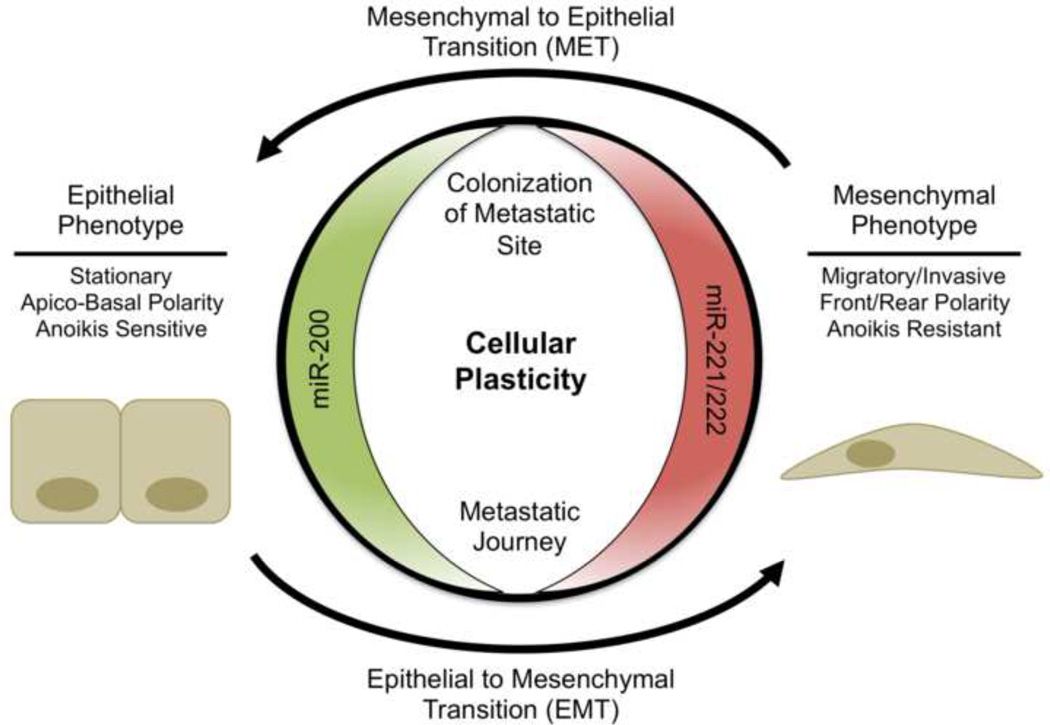
These two miRNA families promote dueling phenotypes, thus they are coordinately regulated during cellular transformations such as EMT and MET (Figure 4). During oncogenic EMT the miR-200 family is strongly down-regulated, while miR-221/222 are highly up-regulated and the reverse is true during MET. This suggests that not only is each miRNA family important for induction of their respective phenotypes, but that the coordinated inverse regulation of these families is required to fully achieve an epithelial or mesenchymal phenotype and associated functional properties. In contrast to their now quite evident role in breast cancer, to date, these miRNA families have not been specifically examined in the normal human breast or mouse mammary gland, although some of their identified targets are clearly relevant in the normal gland.
Figure 4.
Phenotypic consequences of miR-200 or miR-221/222 expression. In addition to the roles of miR-200 and miR-221/222 in protecting the epithelial or mesenchymal phenotype, respectively, they are also actively regulated during EMT and MET. Green indicates expression of the miRNA is associated with a less aggressive, epithelial phenotype, while red indicates the miRNA is associated with aggressive behavior.
Abbreviations
- EMT
Epithelial to mesenchymal transition
- ZEB1/2
Zinc finger E-box binding homeobox 1/2
- UTR
Untranslated Region
- MET
Mesenchymal to epithelial transition
- MDCK
Madin-Darby Canine Kidney
- iPSC
Induced pluripotent stem cell
- TGF-β
Transforming growth factor beta
- PDGF
Platelet derived growth factor
- EGFR
Epidermal growth factor receptor
- NCI
National Cancer Institute
- VEGF
Vascular endothelial growth factor
- ER
Estrogen receptor
- MMTV
Murine mammary tumor virus
Footnotes
Financial Disclosure: Nothing to disclose.
References
- 1.Pillai RS, Bhattacharyya SN, Filipowicz W. Repression of protein synthesis by miRNAs: how many mechanisms? Trends in Cell Biology. 2007 Mar;17(3):118–126. doi: 10.1016/j.tcb.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 2.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009 Jan 23;136(2):215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Williams AE, Moschos SA, Perry MM, Barnes PJ, Lindsay MA. Maternally imprinted microRNAs are differentially expressed during mouse and human lung development. Developmental Dynamics: An Official Publication of the American Association of Anatomists. 2007 Feb;236(2):572–580. doi: 10.1002/dvdy.21047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blakaj A, Lin H. Piecing together the mosaic of early mammalian development through microRNAs. The Journal of Biological Chemistry. 2008 Apr 11;283(15):9505–9508. doi: 10.1074/jbc.R800002200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stefani G, Slack FJ. Small non-coding RNAs in animal development. Nature Reviews. Molecular Cell Biology. 2008 Mar;9(3):219–230. doi: 10.1038/nrm2347. [DOI] [PubMed] [Google Scholar]
- 6.Micalizzi DS, Farabaugh SM, Ford HL. Epithelial-mesenchymal transition in cancer: parallels between normal development and tumor progression. J Mammary Gland Biol Neoplasia. 2010 Jun;15(2):117–134. doi: 10.1007/s10911-010-9178-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huber MA, Kraut N, Beug H. Molecular requirements for epithelial-mesenchymal transition during tumor progression. Curr. Opin. Cell Biol. 2005 Oct;17(5):548–558. doi: 10.1016/j.ceb.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 8.Thiery JP. Epithelial-mesenchymal transitions in tumour progression. Nature Reviews. Cancer. 2002 Jun;2(6):442–454. doi: 10.1038/nrc822. [DOI] [PubMed] [Google Scholar]
- 9.Thiery JP, Sleeman JP. Complex networks orchestrate epithelial-mesenchymal transitions. Nature Reviews. Molecular Cell Biology. 2006 Feb;7(2):131–142. doi: 10.1038/nrm1835. [DOI] [PubMed] [Google Scholar]
- 10.Yang J, Weinberg RA. Epithelial-mesenchymal transition: at the crossroads of development and tumor metastasis. Developmental Cell. 2008 Jun;14(6):818–829. doi: 10.1016/j.devcel.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 11.Peinado H, Olmeda D, Cano A. Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat. Rev. Cancer. 2007 Jun;7(6):415–428. doi: 10.1038/nrc2131. [DOI] [PubMed] [Google Scholar]
- 12.Remacle JE, Kraft H, Lerchner W, Wuytens G, Collart C, Verschueren K, et al. New mode of DNA binding of multi-zinc finger transcription factors: deltaEF1 family members bind with two hands to two target sites. EMBO J. 1999 Sep 15;18(18):5073–5084. doi: 10.1093/emboj/18.18.5073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guaita S, Puig I, Franci C, Garrido M, Dominguez D, Batlle E, et al. Snail induction of epithelial to mesenchymal transition in tumor cells is accompanied by MUC1 repression and ZEB1 expression. J. Biol. Chem. 2002 Oct 18;277(42):39209–39216. doi: 10.1074/jbc.M206400200. [DOI] [PubMed] [Google Scholar]
- 14.Eger A, Aigner K, Sonderegger S, Dampier B, Oehler S, Schreiber M, et al. DeltaEF1 is a transcriptional repressor of E-cadherin and regulates epithelial plasticity in breast cancer cells. Oncogene. 2005 Mar 31;24(14):2375–2385. doi: 10.1038/sj.onc.1208429. [DOI] [PubMed] [Google Scholar]
- 15.Aigner K, Dampier B, Descovich L, Mikula M, Sultan A, Schreiber M, et al. The transcription factor ZEB1 (deltaEF1) promotes tumour cell dedifferentiation by repressing master regulators of epithelial polarity. Oncogene. 2007 Oct 25;26(49):6979–6988. doi: 10.1038/sj.onc.1210508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shirakihara T, Saitoh M, Miyazono K. Differential regulation of epithelial and mesenchymal markers by deltaEF1 proteins in epithelial mesenchymal transition induced by TGF-beta. Mol. Biol. Cell. 2007 Sep;18(9):3533–3544. doi: 10.1091/mbc.E07-03-0249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hurteau GJ, Carlson JA, Spivack SD, Brock GJ. Overexpression of the microRNA hsa-miR-200c leads to reduced expression of transcription factor 8 and increased expression of E-cadherin. Cancer Res. 2007 Sep 1;67(17):7972–7976. doi: 10.1158/0008-5472.CAN-07-1058. [DOI] [PubMed] [Google Scholar]
- 18.Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat. Cell Biol. 2008 May;10(5):593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 19.Park S-M, Gaur AB, Lengyel E, Peter ME. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008 Apr 1;22(7):894–907. doi: 10.1101/gad.1640608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stinson S, Lackner MR, Adai AT, Yu N, Kim H-J, O’Brien C, et al. TRPS1 targeting by miR-221/222 promotes the epithelial-to-mesenchymal transition in breast cancer. Sci Signal. 2011;4(177):ra41. doi: 10.1126/scisignal.2001538. [DOI] [PubMed] [Google Scholar]
- 21.Cochrane DR, Cittelly DM, Howe EN, Spoelstra NS, McKinsey EL, LaPara K, et al. MicroRNAs link estrogen receptor alpha status and Dicer levels in breast cancer. Horm Cancer. 2010 Dec;1(6):306–319. doi: 10.1007/s12672-010-0043-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Uhlmann S, Zhang JD, Schwäger A, Mannsperger H, Riazalhosseini Y, Burmester S, et al. miR-200bc/429 cluster targets PLCgamma1 and differentially regulates proliferation and EGF-driven invasion than miR-200a/141 in breast cancer. Oncogene. 2010 Jul 29;29(30):4297–4306. doi: 10.1038/onc.2010.201. [DOI] [PubMed] [Google Scholar]
- 23.Hurteau GJ, Carlson JA, Roos E, Brock GJ. Stable expression of miR-200c alone is sufficient to regulate TCF8 (ZEB1) and restore E-cadherin expression. Cell Cycle. 2009 Jul 1;8(13):2064–2069. doi: 10.4161/cc.8.13.8883. [DOI] [PubMed] [Google Scholar]
- 24.Kenney PA, Wszolek MF, Rieger-Christ KM, Neto BS, Gould JJ, Harty NJ, et al. Novel ZEB1 expression in bladder tumorigenesis. BJU Int. 2011 Feb;107(4):656–663. doi: 10.1111/j.1464-410X.2010.09489.x. [DOI] [PubMed] [Google Scholar]
- 25.Hu M, Xia M, Chen X, Lin Z, Xu Y, Ma Y, et al. MicroRNA-141 regulates Smad interacting protein 1 (SIP1) and inhibits migration and invasion of colorectal cancer cells. Dig. Dis. Sci. 2010 Aug;55(8):2365–2372. doi: 10.1007/s10620-009-1008-9. [DOI] [PubMed] [Google Scholar]
- 26.Wellner U, Schubert J, Burk UC, Schmalhofer O, Zhu F, Sonntag A, et al. The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nat. Cell Biol. 2009 Dec;11(12):1487–1495. doi: 10.1038/ncb1998. [DOI] [PubMed] [Google Scholar]
- 27.Yang Y, Ahn Y-H, Gibbons DL, Zang Y, Lin W, Thilaganathan N, et al. The Notch ligand Jagged2 promotes lung adenocarcinoma metastasis through a miR-200-dependent pathway in mice. J. Clin. Invest. 2011 Apr 1;121(4):1373–1385. doi: 10.1172/JCI42579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Z, Zhao Y, Smith E, Goodall GJ, Drew PA, Brabletz T, et al. Reversal and prevention of arsenic-induced human bronchial epithelial cell malignant transformation by microRNA-200b. Toxicol. Sci. 2011 May;121(1):110–122. doi: 10.1093/toxsci/kfr029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rui W, Bing F, Hai-Zhu S, Wei D, Long-Bang C. Identification of microRNA profiles in docetaxel-resistant human non-small cell lung carcinoma cells (SPC-A1) J. Cell. Mol. Med. 2010 Jan;14(1–2):206–214. doi: 10.1111/j.1582-4934.2009.00964.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ceppi P, Mudduluru G, Kumarswamy R, Rapa I, Scagliotti GV, Papotti M, et al. Loss of miR-200c expression induces an aggressive, invasive, and chemoresistant phenotype in non-small cell lung cancer. Mol. Cancer Res. 2010 Sep;8(9):1207–1216. doi: 10.1158/1541-7786.MCR-10-0052. [DOI] [PubMed] [Google Scholar]
- 31.Sossey-Alaoui K, Bialkowska K, Plow EF. The miR200 family of microRNAs regulates WAVE3-dependent cancer cell invasion. J. Biol. Chem. 2009 Nov 27;284(48):33019–33029. doi: 10.1074/jbc.M109.034553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Howe EN, Cochrane DR, Richer JK. Targets of miR-200c mediate suppression of cell motility and anoikis resistance. Breast Cancer Res. 2011;13(2):R45. doi: 10.1186/bcr2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xia W, Li J, Chen L, Huang B, Li S, Yang G, et al. MicroRNA-200b regulates cyclin D1 expression and promotes S-phase entry by targeting RND3 in HeLa cells. Mol. Cell. Biochem. 2010 Nov;344(1–2):261–266. doi: 10.1007/s11010-010-0550-2. [DOI] [PubMed] [Google Scholar]
- 34.Hyun S, Lee JH, Jin H, Nam J, Namkoong B, Lee G, et al. Conserved MicroRNA miR-8/miR-200 and its target USH/FOG2 control growth by regulating PI3K. Cell. 2009 Dec 11;139(6):1096–1108. doi: 10.1016/j.cell.2009.11.020. [DOI] [PubMed] [Google Scholar]
- 35.Douma S, Van Laar T, Zevenhoven J, Meuwissen R, Van Garderen E, Peeper DS. Suppression of anoikis and induction of metastasis by the neurotrophic receptor TrkB. Nature. 2004 Aug 26;430(7003):1034–1039. doi: 10.1038/nature02765. [DOI] [PubMed] [Google Scholar]
- 36.Geiger TR, Peeper DS. Critical role for TrkB kinase function in anoikis suppression, tumorigenesis, and metastasis. Cancer Res. 2007 Jul 1;67(13):6221–6229. doi: 10.1158/0008-5472.CAN-07-0121. [DOI] [PubMed] [Google Scholar]
- 37.Cameron HL, Foster WG. Dieldrin promotes resistance to anoikis in breast cancer cells in vitro. Reprod. Toxicol. 2008 Feb;25(2):256–262. doi: 10.1016/j.reprotox.2007.11.013. [DOI] [PubMed] [Google Scholar]
- 38.Smit MA, Geiger TR, Song J-Y, Gitelman I, Peeper DS. A Twist-Snail axis critical for TrkB-induced epithelial-mesenchymal transition-like transformation, anoikis resistance, and metastasis. Mol. Cell. Biol. 2009 Jul;29(13):3722–3737. doi: 10.1128/MCB.01164-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cochrane DR, Spoelstra NS, Howe EN, Nordeen SK, Richer JK. MicroRNA-200c mitigates invasiveness and restores sensitivity to microtubule-targeting chemotherapeutic agents. Mol. Cancer Ther. 2009 May;8(5):1055–1066. doi: 10.1158/1535-7163.MCT-08-1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cochrane DR, Howe EN, Spoelstra NS, Richer JK. Loss of miR-200c: A Marker of Aggressiveness and Chemoresistance in Female Reproductive Cancers. J Oncol. 2010;2010:821717. doi: 10.1155/2010/821717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mozzetti S, Ferlini C, Concolino P, Filippetti F, Raspaglio G, Prislei S, et al. Class III beta-tubulin overexpression is a prominent mechanism of paclitaxel resistance in ovarian cancer patients. Clin. Cancer Res. 2005 Jan 1;11(1):298–305. [PubMed] [Google Scholar]
- 42.Umezu T, Shibata K, Kajiyama H, Terauchi M, Ino K, Nawa A, et al. Taxol resistance among the different histological subtypes of ovarian cancer may be associated with the expression of class III beta-tubulin. Int. J. Gynecol. Pathol. 2008 Apr;27(2):207–212. doi: 10.1097/PGP.0b013e318156c838. [DOI] [PubMed] [Google Scholar]
- 43.Stengel C, Newman SP, Leese MP, Potter BVL, Reed MJ, Purohit A. Class III beta-tubulin expression and in vitro resistance to microtubule targeting agents. Br. J. Cancer. 2010 Jan 19;102(2):316–324. doi: 10.1038/sj.bjc.6605489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tommasi S, Mangia A, Lacalamita R, Bellizzi A, Fedele V, Chiriatti A, et al. Cytoskeleton and paclitaxel sensitivity in breast cancer: the role of beta-tubulins. Int. J. Cancer. 2007 May 15;120(10):2078–2085. doi: 10.1002/ijc.22557. [DOI] [PubMed] [Google Scholar]
- 45.Paradiso A, Mangia A, Chiriatti A, Tommasi S, Zito A, Latorre A, et al. Biomarkers predictive for clinical efficacy of taxol-based chemotherapy in advanced breast cancer. Ann. Oncol. 2005 May;16(Suppl 4):14–19. doi: 10.1093/annonc/mdi902. [DOI] [PubMed] [Google Scholar]
- 46.Kavallaris M. Microtubules and resistance to tubulin-binding agents. Nat. Rev. Cancer. 2010 Mar;10(3):194–204. doi: 10.1038/nrc2803. [DOI] [PubMed] [Google Scholar]
- 47.Pogribny IP, Filkowski JN, Tryndyak VP, Golubov A, Shpyleva SI, Kovalchuk O. Alterations of microRNAs and their targets are associated with acquired resistance of MCF-7 breast cancer cells to cisplatin. Int. J. Cancer. 2010 Oct 15;127(8):1785–1794. doi: 10.1002/ijc.25191. [DOI] [PubMed] [Google Scholar]
- 48.Chen J, Tian W, Cai H, He H, Deng Y. Medical Oncology. Northwood, London, England: 2011. Nov 19, [[cited 2011 Dec 12]]. Down-regulation of microRNA-200c is associated with drug resistance in human breast cancer. [Internet] Available from: http://www.ncbi.nlm.nih.gov/pubmed/22101791. [DOI] [PubMed] [Google Scholar]
- 49.Adam L, Zhong M, Choi W, Qi W, Nicoloso M, Arora A, et al. miR-200 expression regulates epithelial-to-mesenchymal transition in bladder cancer cells and reverses resistance to epidermal growth factor receptor therapy. Clin. Cancer Res. 2009 Aug 15;15(16):5060–5072. doi: 10.1158/1078-0432.CCR-08-2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schickel R, Park S-M, Murmann AE, Peter ME. miR-200c regulates induction of apoptosis through CD95 by targeting FAP-1. Mol. Cell. 2010 Jun 25;38(6):908–915. doi: 10.1016/j.molcel.2010.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, et al. MicroRNA expression profiles classify human cancers. Nature. 2005 Jun 9;435(7043):834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 52.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat. Rev. Cancer. 2006 Nov;6(11):857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 53.Grelier G, Voirin N, Ay A-S, Cox DG, Chabaud S, Treilleux I, et al. Prognostic value of Dicer expression in human breast cancers and association with the mesenchymal phenotype. Br. J. Cancer. 2009 Aug 18;101(4):673–683. doi: 10.1038/sj.bjc.6605193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kong D, Banerjee S, Ahmad A, Li Y, Wang Z, Sethi S, et al. Epithelial to mesenchymal transition is mechanistically linked with stem cell signatures in prostate cancer cells. PLoS ONE. 2010;5(8):e12445. doi: 10.1371/journal.pone.0012445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vallejo DM, Caparros E, Dominguez M. Targeting Notch signalling by the conserved miR-8/200 microRNA family in development and cancer cells. EMBO J. 2011 Feb 16;30(4):756–769. doi: 10.1038/emboj.2010.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shimono Y, Zabala M, Cho RW, Lobo N, Dalerba P, Qian D, et al. Downregulation of miRNA-200c links breast cancer stem cells with normal stem cells. Cell. 2009 Aug 7;138(3):592–603. doi: 10.1016/j.cell.2009.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Iliopoulos D, Lindahl-Allen M, Polytarchou C, Hirsch HA, Tsichlis PN, Struhl K. Loss of miR-200 inhibition of Suz12 leads to polycomb-mediated repression required for the formation and maintenance of cancer stem cells. Mol. Cell. 2010 Sep 10;39(5):761–772. doi: 10.1016/j.molcel.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tryndyak VP, Beland FA, Pogribny IP. E-cadherin transcriptional down-regulation by epigenetic and microRNA-200 family alterations is related to mesenchymal and drug-resistant phenotypes in human breast cancer cells. Int. J. Cancer. 2010 Jun 1;126(11):2575–2583. doi: 10.1002/ijc.24972. [DOI] [PubMed] [Google Scholar]
- 59.Ahmad A, Aboukameel A, Kong D, Wang Z, Sethi S, Chen W, et al. Phosphoglucose isomerase/autocrine motility factor mediates epithelial-mesenchymal transition regulated by miR-200 in breast cancer cells. Cancer Res. 2011 May 1;71(9):3400–3409. doi: 10.1158/0008-5472.CAN-10-0965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dykxhoorn DM, Wu Y, Xie H, Yu F, Lal A, Petrocca F, et al. miR-200 enhances mouse breast cancer cell colonization to form distant metastases. PloS One. 2009;4(9):e7181. doi: 10.1371/journal.pone.0007181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Korpal M, Ell BJ, Buffa FM, Ibrahim T, Blanco MA, Celià-Terrassa T, et al. Direct targeting of Sec23a by miR-200s influences cancer cell secretome and promotes metastatic colonization. Nat. Med. 2011 Sep;17(9):1101–1108. doi: 10.1038/nm.2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Herschkowitz JI, Zhao W, Zhang M, Usary J, Murrow G, Edwards D, et al. Comparative oncogenomics identifies breast tumors enriched in functional tumor-initiating cells. [[cited 2011 Dec 12]];Proceedings of the National Academy of Sciences of the United States of America [Internet] 2011 Jun 1; doi: 10.1073/pnas.1018862108. Available from: http://www.ncbi.nlm.nih.gov/pubmed/21633010. [DOI] [PMC free article] [PubMed]
- 63.Hui ABY, Shi W, Boutros PC, Miller N, Pintilie M, Fyles T, et al. Robust global micro-RNA profiling with formalin-fixed paraffin-embedded breast cancer tissues. Lab. Invest. 2009 May;89(5):597–606. doi: 10.1038/labinvest.2009.12. [DOI] [PubMed] [Google Scholar]
- 64.Bockmeyer CL, Christgen M, Müller M, Fischer S, Ahrens P, Länger F, et al. MicroRNA profiles of healthy basal and luminal mammary epithelial cells are distinct and reflected in different breast cancer subtypes. Breast Cancer Res. Treat. 2011 Dec;130(3):735–745. doi: 10.1007/s10549-010-1303-3. [DOI] [PubMed] [Google Scholar]
- 65.Baffa R, Fassan M, Volinia S, O’Hara B, Liu C-G, Palazzo JP, et al. MicroRNA expression profiling of human metastatic cancers identifies cancer gene targets. J. Pathol. 2009 Oct;219(2):214–221. doi: 10.1002/path.2586. [DOI] [PubMed] [Google Scholar]
- 66.Gill JG, Langer EM, Lindsley RC, Cai M, Murphy TL, Kyba M, et al. Snail and the microRNA-200 family act in opposition to regulate epithelial-to-mesenchymal transition and germ layer fate restriction in differentiating ESCs. Stem Cells. 2011 May;29(5):764–776. doi: 10.1002/stem.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lin C-H, Jackson AL, Guo J, Linsley PS, Eisenman RN. Myc-regulated microRNAs attenuate embryonic stem cell differentiation. EMBO J. 2009 Oct 21;28(20):3157–3170. doi: 10.1038/emboj.2009.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gibbons DL, Lin W, Creighton CJ, Rizvi ZH, Gregory PA, Goodall GJ, et al. Contextual extracellular cues promote tumor cell EMT and metastasis by regulating miR-200 family expression. Genes Dev. 2009 Sep 15;23(18):2140–2151. doi: 10.1101/gad.1820209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gregory PA, Bracken CP, Smith E, Bert AG, Wright JA, Roslan S, et al. An autocrine TGF-beta/ZEB/miR-200 signaling network regulates establishment and maintenance of epithelial-mesenchymal transition. Mol. Biol. Cell. 2011 May 15;22(10):1686–1698. doi: 10.1091/mbc.E11-02-0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Neveu P, Kye MJ, Qi S, Buchholz DE, Clegg DO, Sahin M, et al. MicroRNA profiling reveals two distinct p53-related human pluripotent stem cell states. Cell Stem Cell. 2010 Dec 3;7(6):671–681. doi: 10.1016/j.stem.2010.11.012. [DOI] [PubMed] [Google Scholar]
- 71.Samavarchi-Tehrani P, Golipour A, David L, Sung H-K, Beyer TA, Datti A, et al. Functional genomics reveals a BMP-driven mesenchymal-to-epithelial transition in the initiation of somatic cell reprogramming. Cell Stem Cell. 2010 Jul 2;7(1):64–77. doi: 10.1016/j.stem.2010.04.015. [DOI] [PubMed] [Google Scholar]
- 72.Bracken CP, Gregory PA, Kolesnikoff N, Bert AG, Wang J, Shannon MF, et al. A double-negative feedback loop between ZEB1-SIP1 and the microRNA-200 family regulates epithelial-mesenchymal transition. Cancer Res. 2008 Oct 1;68(19):7846–7854. doi: 10.1158/0008-5472.CAN-08-1942. [DOI] [PubMed] [Google Scholar]
- 73.Burk U, Schubert J, Wellner U, Schmalhofer O, Vincan E, Spaderna S, et al. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep. 2008 Jun;9(6):582–589. doi: 10.1038/embor.2008.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Eades G, Yao Y, Yang M, Zhang Y, Chumsri S, Zhou Q. miR-200a regulates SIRT1 expression and epithelial to mesenchymal transition (EMT)-like transformation in mammary epithelial cells. J. Biol. Chem. 2011 Jul 22;286(29):25992–26002. doi: 10.1074/jbc.M111.229401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Korpal M, Lee ES, Hu G, Kang Y. The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J. Biol. Chem. 2008 May 30;283(22):14910–14914. doi: 10.1074/jbc.C800074200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Slabáková E, Pernicová Z, Slavíčková E, Staršíchová A, Kozubík A, Souček K. TGF-β1-induced EMT of non-transformed prostate hyperplasia cells is characterized by early induction of SNAI2/Slug. Prostate. 2011 Sep;71(12):1332–1343. doi: 10.1002/pros.21350. [DOI] [PubMed] [Google Scholar]
- 77.Davalos V, Moutinho C, Villanueva A, Boque R, Silva P, Carneiro F, et al. Dynamic epigenetic regulation of the microRNA-200 family mediates epithelial and mesenchymal transitions in human tumorigenesis. [[cited 2011 Dec 13]];Oncogene [Internet] 2011 Aug 29; doi: 10.1038/onc.2011.383. Available from: http://www.ncbi.nlm.nih.gov/pubmed/21874049. [DOI] [PMC free article] [PubMed]
- 78.Vrba L, Jensen TJ, Garbe JC, Heimark RL, Cress AE, Dickinson S, et al. Role for DNA methylation in the regulation of miR-200c and miR-141 expression in normal and cancer cells. PLoS ONE. 2010;5(1):e8697. doi: 10.1371/journal.pone.0008697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wiklund ED, Bramsen JB, Hulf T, Dyrskjøt L, Ramanathan R, Hansen TB, et al. Coordinated epigenetic repression of the miR-200 family and miR-205 in invasive bladder cancer. Int. J. Cancer. 2011 Mar 15;128(6):1327–1334. doi: 10.1002/ijc.25461. [DOI] [PubMed] [Google Scholar]
- 80.Yuan J-H, Yang F, Chen B-F, Lu Z, Huo X-S, Zhou W-P, et al. The histone deacetylase 4/SP1/microrna-200a regulatory network contributes to aberrant histone acetylation in hepatocellular carcinoma. Hepatology. 2011 Dec;54(6):2025–2035. doi: 10.1002/hep.24606. [DOI] [PubMed] [Google Scholar]
- 81.Kong D, Li Y, Wang Z, Banerjee S, Ahmad A, Kim H-RC, et al. miR-200 regulates PDGF-D-mediated epithelial-mesenchymal transition, adhesion, and invasion of prostate cancer cells. Stem Cells. 2009 Aug;27(8):1712–1721. doi: 10.1002/stem.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Li Y, VandenBoom TG2nd, Kong D, Wang Z, Ali S, Philip PA, et al. Up-regulation of miR-200 and let-7 by natural agents leads to the reversal of epithelial-to-mesenchymal transition in gemcitabine-resistant pancreatic cancer cells. Cancer Res. 2009 Aug 15;69(16):6704–6712. doi: 10.1158/0008-5472.CAN-09-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tellez CS, Juri DE, Do K, Bernauer AM, Thomas CL, Damiani LA, et al. EMT and stem cell-like properties associated with miR-205 and miR-200 epigenetic silencing are early manifestations during carcinogen-induced transformation of human lung epithelial cells. Cancer Res. 2011 Apr 15;71(8):3087–3097. doi: 10.1158/0008-5472.CAN-10-3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bao B, Ali S, Kong D, Sarkar SH, Wang Z, Banerjee S, et al. Anti-tumor activity of a novel compound-CDF is mediated by regulating miR-21, miR-200, and PTEN in pancreatic cancer. PLoS ONE. 2011;6(3):e17850. doi: 10.1371/journal.pone.0017850. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 85.Iliopoulos D, Polytarchou C, Hatziapostolou M, Kottakis F, Maroulakou IG, Struhl K, et al. MicroRNAs differentially regulated by Akt isoforms control EMT and stem cell renewal in cancer cells. Sci Signal. 2009;2(92):ra62. doi: 10.1126/scisignal.2000356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chang C-J, Chao C-H, Xia W, Yang J-Y, Xiong Y, Li C-W, et al. p53 regulates epithelial-mesenchymal transition and stem cell properties through modulating miRNAs. Nat. Cell Biol. 2011 Mar;13(3):317–323. doi: 10.1038/ncb2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kim T, Veronese A, Pichiorri F, Lee TJ, Jeon Y-J, Volinia S, et al. p53 regulates epithelial-mesenchymal transition through microRNAs targeting ZEB1 and ZEB2. J. Exp. Med. 2011 May 9;208(5):875–883. doi: 10.1084/jem.20110235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Knouf EC, Garg K, Arroyo JD, Correa Y, Sarkar D, Parkin RK, et al. An integrative genomic approach identifies p73 and p63 as activators of miR-200 microRNA family transcription. [[cited 2011 Dec 13]];Nucleic Acids Research [Internet] 2011 Sep 14; doi: 10.1093/nar/gkr731. Available from: http://www.ncbi.nlm.nih.gov/pubmed/21917857. [DOI] [PMC free article] [PubMed]
- 89.Bhat-Nakshatri P, Wang G, Collins NR, Thomson MJ, Geistlinger TR, Carroll JS, et al. Estradiol-regulated microRNAs control estradiol response in breast cancer cells. Nucleic Acids Res. 2009 Aug;37(14):4850–4861. doi: 10.1093/nar/gkp500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zheng C, Yinghao S, Li J. Medical Oncology. Northwood, London, England: 2011. Apr 13, [[cited 2011 Dec 13]]. MiR-221 expression affects invasion potential of human prostate carcinoma cell lines by targeting DVL2. [Internet] Available from: http://www.ncbi.nlm.nih.gov/pubmed/21487968. [DOI] [PubMed] [Google Scholar]
- 91.Li X, Zhang Y, Zhang H, Liu X, Gong T, Li M, et al. miRNA-223 promotes gastric cancer invasion and metastasis by targeting tumor suppressor EPB41L3. Mol. Cancer Res. 2011 Jul;9(7):824–833. doi: 10.1158/1541-7786.MCR-10-0529. [DOI] [PubMed] [Google Scholar]
- 92.Gottardo F, Liu CG, Ferracin M, Calin GA, Fassan M, Bassi P, et al. Micro-RNA profiling in kidney and bladder cancers. Urol. Oncol. 2007 Oct;25(5):387–392. doi: 10.1016/j.urolonc.2007.01.019. [DOI] [PubMed] [Google Scholar]
- 93.Chou C-K, Chen R-F, Chou F-F, Chang H-W, Chen Y-J, Lee Y-F, et al. miR-146b is highly expressed in adult papillary thyroid carcinomas with high risk features including extrathyroidal invasion and the BRAF(V600E) mutation. Thyroid. 2010 May;20(5):489–494. doi: 10.1089/thy.2009.0027. [DOI] [PubMed] [Google Scholar]
- 94.Sun K, Wang W, Zeng J-jie, Wu C-tang, Lei S-tong, Li G-xin. MicroRNA-221 inhibits CDKN1C/p57 expression in human colorectal carcinoma. Acta Pharmacol. Sin. 2011 Mar;32(3):375–384. doi: 10.1038/aps.2010.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Felicetti F, Errico MC, Bottero L, Segnalini P, Stoppacciaro A, Biffoni M, et al. The promyelocytic leukemia zinc finger-microRNA-221/−222 pathway controls melanoma progression through multiple oncogenic mechanisms. Cancer Res. 2008 Apr 15;68(8):2745–2754. doi: 10.1158/0008-5472.CAN-07-2538. [DOI] [PubMed] [Google Scholar]
- 96.Cammarata G, Augugliaro L, Salemi D, Agueli C, La Rosa M, Dagnino L, et al. Differential expression of specific microRNA and their targets in acute myeloid leukemia. Am. J. Hematol. 2010 May;85(5):331–339. doi: 10.1002/ajh.21667. [DOI] [PubMed] [Google Scholar]
- 97.Veerla S, Lindgren D, Kvist A, Frigyesi A, Staaf J, Persson H, et al. MiRNA expression in urothelial carcinomas: important roles of miR-10a, miR-222, miR-125b, miR-7 and miR-452 for tumor stage and metastasis, and frequent homozygous losses of miR-31. Int. J. Cancer. 2009 May 1;124(9):2236–2242. doi: 10.1002/ijc.24183. [DOI] [PubMed] [Google Scholar]
- 98.Lu X, Zhao P, Zhang C, Fu Z, Chen Y, Lu A, et al. Analysis of miR-221 and p27 expression in human gliomas. Mol Med Report. 2009 Aug;2(4):651–656. doi: 10.3892/mmr_00000152. [DOI] [PubMed] [Google Scholar]
- 99.Radojicic J, Zaravinos A, Vrekoussis T, Kafousi M, Spandidos DA, Stathopoulos EN. MicroRNA expression analysis in triple-negative (ER, PR and Her2/neu) breast cancer. Cell Cycle. 2011 Feb 1;10(3):507–517. doi: 10.4161/cc.10.3.14754. [DOI] [PubMed] [Google Scholar]
- 100.Zhao J-J, Lin J, Yang H, Kong W, He L, Ma X, et al. MicroRNA-221/222 negatively regulates estrogen receptor alpha and is associated with tamoxifen resistance in breast cancer. J. Biol. Chem. 2008 Nov 7;283(45):31079–31086. doi: 10.1074/jbc.M806041200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 101.Di Leva G, Gasparini P, Piovan C, Ngankeu A, Garofalo M, Taccioli C, et al. MicroRNA cluster 221–222 and estrogen receptor alpha interactions in breast cancer. J. Natl. Cancer Inst. 2010 May 19;102(10):706–721. doi: 10.1093/jnci/djq102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Buffa FM, Camps C, Winchester L, Snell CE, Gee HE, Sheldon H, et al. microRNA-associated progression pathways and potential therapeutic targets identified by integrated mRNA and microRNA expression profiling in breast cancer. Cancer Res. 2011 Sep 1;71(17):5635–5645. doi: 10.1158/0008-5472.CAN-11-0489. [DOI] [PubMed] [Google Scholar]
- 103.Sun Y, Wu J, Wu S-hung, Thakur A, Bollig A, Huang Y, et al. Expression profile of microRNAs in c-Myc induced mouse mammary tumors. Breast Cancer Res. Treat. 2009 Nov;118(1):185–196. doi: 10.1007/s10549-008-0171-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yoshimoto N, Toyama T, Takahashi S, Sugiura H, Endo Y, Iwasa M, et al. Distinct expressions of microRNAs that directly target estrogen receptor α in human breast cancer. Breast Cancer Res. Treat. 2011 Nov;130(1):331–339. doi: 10.1007/s10549-011-1672-2. [DOI] [PubMed] [Google Scholar]
- 105.Chun-Zhi Z, Lei H, An-Ling Z, Yan-Chao F, Xiao Y, Guang-Xiu W, et al. MicroRNA-221 and microRNA-222 regulate gastric carcinoma cell proliferation and radioresistance by targeting PTEN. BMC Cancer. 2010;10:367. doi: 10.1186/1471-2407-10-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Gramantieri L, Fornari F, Ferracin M, Veronese A, Sabbioni S, Calin GA, et al. MicroRNA-221 targets Bmf in hepatocellular carcinoma and correlates with tumor multifocality. Clin. Cancer Res. 2009 Aug 15;15(16):5073–5081. doi: 10.1158/1078-0432.CCR-09-0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Guttilla IK, Phoenix KN, Hong X, Tirnauer JS, Claffey KP, White BA. Prolonged mammosphere culture of MCF-7 cells induces an EMT and repression of the estrogen receptor by microRNAs. [[cited 2011 Dec 14]];Breast Cancer Res. Treat. [Internet] 2011 May 7; doi: 10.1007/s10549-011-1534-y. Available from: http://www.ncbi.nlm.nih.gov/pubmed/21553120. [DOI] [PubMed]
- 108.Gai Z, Zhou G, Itoh S, Morimoto Y, Tanishima H, Hatamura I, et al. Trps1 functions downstream of Bmp7 in kidney development. J. Am. Soc. Nephrol. 2009 Nov;20(11):2403–2411. doi: 10.1681/ASN.2008091020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Chen JQ, Litton J, Xiao L, Zhang H-Z, Warneke CL, Wu Y, et al. Quantitative immunohistochemical analysis and prognostic significance of TRPS-1, a new GATA transcription factor family member, in breast cancer. Horm Cancer. 2010 Feb;1(1):21–33. doi: 10.1007/s12672-010-0008-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Shah MY, Calin GA. MicroRNAs miR-221 and miR-222: a new level of regulation in aggressive breast cancer. Genome Med. 2011;3(8):56. doi: 10.1186/gm272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Galardi S, Mercatelli N, Giorda E, Massalini S, Frajese GV, Ciafrè SA, et al. miR-221 and miR-222 expression affects the proliferation potential of human prostate carcinoma cell lines by targeting p27Kip1. J. Biol. Chem. 2007 Aug 10;282(32):23716–23724. doi: 10.1074/jbc.M701805200. [DOI] [PubMed] [Google Scholar]
- 112.Fornari F, Gramantieri L, Ferracin M, Veronese A, Sabbioni S, Calin GA, et al. MiR-221 controls CDKN1C/p57 and CDKN1B/p27 expression in human hepatocellular carcinoma. Oncogene. 2008 Sep 25;27(43):5651–5661. doi: 10.1038/onc.2008.178. [DOI] [PubMed] [Google Scholar]
- 113.Medina R, Zaidi SK, Liu C-G, Stein JL, van Wijnen AJ, Croce CM, et al. MicroRNAs 221 and 222 bypass quiescence and compromise cell survival. Cancer Res. 2008 Apr 15;68(8):2773–2780. doi: 10.1158/0008-5472.CAN-07-6754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.le Sage C, Nagel R, Egan DA, Schrier M, Mesman E, Mangiola A, et al. Regulation of the p27(Kip1) tumor suppressor by miR-221 and miR-222 promotes cancer cell proliferation. EMBO J. 2007 Aug 8;26(15):3699–3708. doi: 10.1038/sj.emboj.7601790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Fu X, Wang Q, Chen J, Huang X, Chen X, Cao L, et al. Clinical significance of miR-221 and its inverse correlation with p27Kip1 in hepatocellular carcinoma. Mol. Biol. Rep. 2011 Jun;38(5):3029–3035. doi: 10.1007/s11033-010-9969-5. [DOI] [PubMed] [Google Scholar]
- 116.Frenquelli M, Muzio M, Scielzo C, Fazi C, Scarfò L, Rossi C, et al. MicroRNA and proliferation control in chronic lymphocytic leukemia: functional relationship between miR-221/222 cluster and p27. Blood. 2010 May 13;115(19):3949–3959. doi: 10.1182/blood-2009-11-254656. [DOI] [PubMed] [Google Scholar]
- 117.Mercatelli N, Coppola V, Bonci D, Miele F, Costantini A, Guadagnoli M, et al. The inhibition of the highly expressed miR-221 and miR-222 impairs the growth of prostate carcinoma xenografts in mice. PLoS ONE. 2008;3(12):e4029. doi: 10.1371/journal.pone.0004029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zhang C, Kang C, You Y, Pu P, Yang W, Zhao P, et al. Co-suppression of miR-221/222 cluster suppresses human glioma cell growth by targeting p27kip1 in vitro and in vivo. Int. J. Oncol. 2009 Jun;34(6):1653–1660. doi: 10.3892/ijo_00000296. [DOI] [PubMed] [Google Scholar]
- 119.Park J-K, Kogure T, Nuovo GJ, Jiang J, He L, Kim JH, et al. miR-221 Silencing Blocks Hepatocellular Carcinoma and Promotes Survival. Cancer Res. 2011 Dec 15;71(24):7608–7616. doi: 10.1158/0008-5472.CAN-11-1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Chen Y, Zaman MS, Deng G, Majid S, Saini S, Liu J, et al. MicroRNAs 221/222 and genistein-mediated regulation of ARHI tumor suppressor gene in prostate cancer. Cancer Prev Res (Phila) 2011 Jan;4(1):76–86. doi: 10.1158/1940-6207.CAPR-10-0167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Janssen EAM, Øvestad IT, Skaland I, Søiland H, Gudlaugsson E, Kjellevold KH, et al. LOH at 1p31 (ARHI) and proliferation in lymph node-negative breast cancer. Cell. Oncol. 2009;31(5):335–343. doi: 10.3233/CLO-2009-0479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Yuan J, Luo RZ, Fujii S, Wang L, Hu W, Andreeff M, et al. Aberrant methylation and silencing of ARHI, an imprinted tumor suppressor gene in which the function is lost in breast cancers. Cancer Res. 2003 Jul 15;63(14):4174–4180. [PubMed] [Google Scholar]
- 123.Bao J-J, Le X-F, Wang R-Y, Yuan J, Wang L, Atkinson EN, et al. Reexpression of the tumor suppressor gene ARHI induces apoptosis in ovarian and breast cancer cells through a caspase-independent calpain-dependent pathway. Cancer Res. 2002 Dec 15;62(24):7264–7272. [PubMed] [Google Scholar]
- 124.Park J-K, Lee EJ, Esau C, Schmittgen TD. Antisense inhibition of microRNA-21 or −221 arrests cell cycle, induces apoptosis, and sensitizes the effects of gemcitabine in pancreatic adenocarcinoma. Pancreas. 2009 Oct;38(7):e190–e199. doi: 10.1097/MPA.0b013e3181ba82e1. [DOI] [PubMed] [Google Scholar]
- 125.Zhou M, Liu Z, Zhao Y, Ding Y, Liu H, Xi Y, et al. MicroRNA-125b confers the resistance of breast cancer cells to paclitaxel through suppression of pro-apoptotic Bcl-2 antagonist killer 1 (Bak1) expression. J. Biol. Chem. 2010 Jul 9;285(28):21496–21507. doi: 10.1074/jbc.M109.083337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Zhang J, Han L, Ge Y, Zhou X, Zhang A, Zhang C, et al. miR-221/222 promote malignant progression of glioma through activation of the Akt pathway. Int. J. Oncol. 2010 Apr;36(4):913–920. doi: 10.3892/ijo_00000570. [DOI] [PubMed] [Google Scholar]
- 127.Zhu L, Chow LWC, Loo WTY, Guan X-Y, Toi M. Her2/neu expression predicts the response to antiaromatase neoadjuvant therapy in primary breast cancer: subgroup analysis from celecoxib antiaromatase neoadjuvant trial. Clin. Cancer Res. 2004 Jul 15;10(14):4639–4644. doi: 10.1158/1078-0432.CCR-04-0057. [DOI] [PubMed] [Google Scholar]
- 128.Piccart M, Lohrisch C, Di Leo A, Larsimont D. The predictive value of HER2 in breast cancer. Oncology. 2001;61(Suppl 2):73–82. doi: 10.1159/000055405. [DOI] [PubMed] [Google Scholar]
- 129.Miller TE, Ghoshal K, Ramaswamy B, Roy S, Datta J, Shapiro CL, et al. MicroRNA-221/222 confers tamoxifen resistance in breast cancer by targeting p27Kip1. J. Biol. Chem. 2008 Oct 31;283(44):29897–29903. doi: 10.1074/jbc.M804612200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lu Y, Roy S, Nuovo G, Ramaswamy B, Miller T, Shapiro C, et al. Anti-microRNA-222 (Anti-miR-222) and −181B Suppress Growth of Tamoxifen-resistant Xenografts in Mouse by Targeting TIMP3 Protein and Modulating Mitogenic Signal. J. Biol. Chem. 2011 Dec 9;286(49):42292–42302. doi: 10.1074/jbc.M111.270926. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 131.Rao X, Di Leva G, Li M, Fang F, Devlin C, Hartman-Frey C, et al. MicroRNA-221/222 confers breast cancer fulvestrant resistance by regulating multiple signaling pathways. Oncogene. 2011 Mar 3;30(9):1082–1097. doi: 10.1038/onc.2010.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Fan M, Yan PS, Hartman-Frey C, Chen L, Paik H, Oyer SL, et al. Diverse gene expression and DNA methylation profiles correlate with differential adaptation of breast cancer cells to the antiestrogens tamoxifen and fulvestrant. Cancer Res. 2006 Dec 15;66(24):11954–11966. doi: 10.1158/0008-5472.CAN-06-1666. [DOI] [PubMed] [Google Scholar]
- 133.Kalkhoven E, Kwakkenbos-Isbrücker L, Mummery CL, de Laat SW, van den Eijnden-van Raaij AJ, van der Saag PT, et al. The role of TGF-beta production in growth inhibition of breast-tumor cells by progestins. Int. J. Cancer. 1995 Mar 29;61(1):80–86. doi: 10.1002/ijc.2910610114. [DOI] [PubMed] [Google Scholar]
- 134.Sovak MA, Arsura M, Zanieski G, Kavanagh KT, Sonenshein GE. The inhibitory effects of transforming growth factor beta1 on breast cancer cell proliferation are mediated through regulation of aberrant nuclear factor-kappaB/Rel expression. Cell Growth Differ. 1999 Aug;10(8):537–544. [PubMed] [Google Scholar]
- 135.Brioschi M, Fischer J, Cairoli R, Rossetti S, Pezzetti L, Nichelatti M, et al. Down-regulation of microRNAs 222/221 in acute myelogenous leukemia with deranged core-binding factor subunits. Neoplasia. 2010 Nov;12(11):866–876. doi: 10.1593/neo.10482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Felicetti F, Bottero L, Felli N, Mattia G, Labbaye C, Alvino E, et al. Role of PLZF in melanoma progression. Oncogene. 2004 Jun 3;23(26):4567–4576. doi: 10.1038/sj.onc.1207597. [DOI] [PubMed] [Google Scholar]
- 137.Belguise K, Kersual N, Galtier F, Chalbos D. FRA-1 expression level regulates proliferation and invasiveness of breast cancer cells. Oncogene. 2005 Feb 17;24(8):1434–1444. doi: 10.1038/sj.onc.1208312. [DOI] [PubMed] [Google Scholar]
- 138.Luo YP, Zhou H, Krueger J, Kaplan C, Liao D, Markowitz D, et al. The role of proto-oncogene Fra-1 in remodeling the tumor microenvironment in support of breast tumor cell invasion and progression. Oncogene. 2010 Feb 4;29(5):662–673. doi: 10.1038/onc.2009.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Ndlovu ‘Matladi N, Van Lint C, Van Wesemael K, Callebert P, Chalbos D, Haegeman G, et al. Hyperactivated NF-{kappa}B and AP-1 transcription factors promote highly accessible chromatin and constitutive transcription across the interleukin-6 gene promoter in metastatic breast cancer cells. Mol. Cell. Biol. 2009 Oct;29(20):5488–5504. doi: 10.1128/MCB.01657-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Martello G, Rosato A, Ferrari F, Manfrin A, Cordenonsi M, Dupont S, et al. A MicroRNA targeting dicer for metastasis control. Cell. 2010 Jun 25;141(7):1195–1207. doi: 10.1016/j.cell.2010.05.017. [DOI] [PubMed] [Google Scholar]
- 141.Su X, Chakravarti D, Cho MS, Liu L, Gi YJ, Lin Y-L, et al. TAp63 suppresses metastasis through coordinate regulation of Dicer and miRNAs. Nature. 2010 Oct 21;467(7318):986–990. doi: 10.1038/nature09459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Fontemaggi G, Gurtner A, Damalas A, Costanzo A, Higashi Y, Sacchi A, et al. deltaEF1 repressor controls selectively p53 family members during differentiation. Oncogene. 2005 Nov 10;24(49):7273–7280. doi: 10.1038/sj.onc.1208891. [DOI] [PubMed] [Google Scholar]