Abstract
MicroRNAs regulate eukaryotic gene expression upon pairing onto target mRNAs. This targeting is influenced by the complementarity between the microRNA “seed” sequence at its 5′ end and the seed-matching sequences in the mRNA. Here, we assess the efficiency and specificity of 8-mer locked nucleic acid (LNA)-modified oligonucleotides raised against the seeds of miR-372 and miR-373, two embryonic stem cell-specific microRNAs prominently expressed in the human gastric adenocarcinoma AGS cell line. Provided that the pairing is perfect over all the eight nucleotides of the seed and starts at nucleotide 2 or 1 at the microRNA 5′ end, these short LNAs inhibit miR-372/373 functions and derepress their common target, the cell cycle regulator LATS2. They decrease cell proliferation in vitro upon either transfection at nanomolar concentrations or unassisted delivery at micromolar concentrations. Subcutaneously delivered LNAs reduce tumor growth of AGS xenografts in mice, upon formation of a stable, specific heteroduplex with the targeted miR-372 and -373 and LATS2 upregulation. Their therapeutic potential is confirmed in fast-growing, miR-372-positive, primary human gastric adenocarcinoma xenografts in mice. Thus, microRNA silencing by 8-mer seed-targeting LNAs appears a valuable approach for both loss-of-function studies aimed at elucidating microRNA functions and for microRNA-based therapeutic strategies.
Keywords: gastric adenocarcinoma, LATS2 tumor suppressor, miR-372, miR-373, oncomirs, short LNA oligonucleotides, therapeutic strategy, xenografts in mice
Introduction
MicroRNAs (miRNAs) are small, evolutionarily conserved, noncoding RNAs of approximately 22 nucleotides in length, which are prominently involved in gene regulation in nearly every biological process. Thirty thousands of mature miRNA sequences have been identified in multiple organisms and deposited into the miRBase database.1 MiRNA bind to partly complementary sequences preferentially located in the 3′-untranslated region (3′ UTR) of mRNAs of target genes. For most miRNAs, the targeting of cognate mRNAs is strongly influenced by the complementarity between the short “seed” sequence at nucleotides 2–8 at the 5′ end of the miRNA and the seed matching sequences contained in the mRNA. The pairing of a miRNA onto target mRNAs most often decreases the mRNA translation and stability, subsequently leading to decreased production of the target protein.2 MiRNAs sharing the same seed sequence are grouped into families and are theorized to target overlapping sets of genes.
Perturbations in miRNA biogenesis or expression contribute to diseases, including cancer, immune pathologies or infection. The widespread alterations in the miRNA profiles are ubiquitous features of cancer and affect all steps of the malignant phenotype from cellular transformation to tumor progression. In the same way as protein-coding genes, some miRNA behave as cancer “drivers” (the so-called oncomirs) or tumor suppressors depending on the cellular context in which they are expressed.3
The involvement of deregulated miRNA expression in the malignant phenotype prompts the exploitation of miRNAs as anticancer therapeutic targets. Numerous studies have demonstrated the utility of inhibiting miRNAs using complementary anti-miRNA oligonucleotides. Antisense anti-miRNAs sequester the miRNA in competition with target mRNAs, leading to functional inhibition of the miRNA and derepression of the direct targets. Antisense-mediated miRNA inhibition requires optimization of the oligonucleotides for increased binding affinity, improved nuclease resistance and efficient in vivo delivery. This can be achieved using a variety of chemical modifications, which show particular promise in vivo. In most studies, fully complementary 22-mer antimiRs have been used to target the mature miRNA. Introduction of locked nucleic acids (LNA), in which the furanose ring in the sugar-phosphate backbone is chemically locked in a RNA mimicking N-type (C3′-endo) conformation by a 2′-O,4′-C methylene bridge, into an antisense sequence dramatically enhances its affinity for the target.4 Thus, an efficient antagonism of several miRNAs has been reported using either high-affinity 15-16-mer LNA/DNA antimiRs targeting the 5′ region of the mature miRNA, or even 12-14-mer seed-directed LNA oligonucleotides with a phosphodiester backbone.5 Furthermore, the outstanding binding affinity of LNAs has been exploited in an approach using only 8-mer LNA-phosphorothioate oligonucleotides complementary to the miRNA seed sequence.6 These 8-mer oligos, designated as tiny LNAs (TL), enable specific inhibition of miRNA function with concomitant derepression of direct targets in cultured cells. In an orthotopic mouse model of breast tumor xenografts, unconjugated anti-miR-21 TL21 efficiently reached the miR-21 oncomir within the tumors upon systemic injection and derepressed one of its target, the neoplasic transformation inhibitor programmed cell death 4 (PDCD4).6 This strategy has been thereafter successfully applied to other miRNA-associated diseases. TL21 are able to reduce autoimmune splenomegaly in lupus mice.7 TL155 targeting the miR-155 oncomir inhibit the tumor growth in a mouse xenograft model of a B-cell lymphoproliferative disease.8 TL17 and TL19 targeting the miRNA seed family expressed by the oncogenic miR-17~92 and miR-106b~25 clusters inhibit medulloblastoma growth in vivo and prolong mice survival.9 TL33 targeting the miR-33 family, which regulates lipid/cholesterol homeostasis, increase circulating high-density lipoprotein cholesterol at long-term in obese and insulin-resistant nonhuman primates.10 Altogether, these data stress the potential of these tiny oligonucleotides in the therapeutic targeting of aberrantly expressed miRNAs.
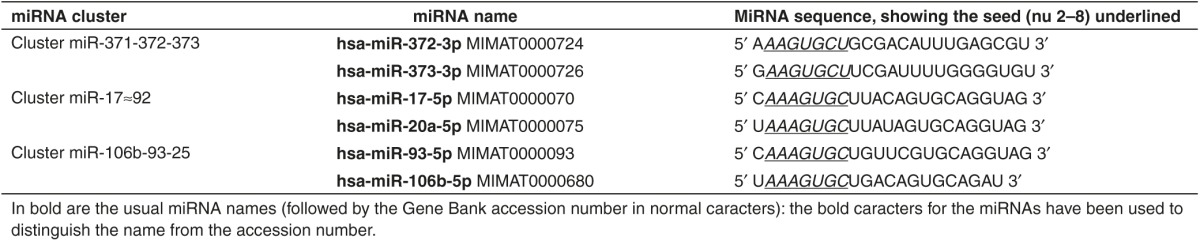
We previously reported that the human gastric adenocarcinoma-derived AGS cell line express high levels of miR-372, which confer an active proliferation rate to these cells.11 MiR-372 belongs to the miRNA-371-372-373 (miR-371–373) cluster, originally found to be specifically expressed in human embryonic stem cells and involved in both stem cell pluripotency and cell cycle regulation.12,13,14 In this cluster, miR-372 and miR-373 are homologous and likely target the same protein-coding genes. The miR-371–373 cluster is frequently deregulated in some human tumors , leading to high expression of mir-372 and -373 in testicular germ cell tumors, esophageal carcinoma, thyroid adenoma, hepatoblastoma, colorectal carcinoma, and gastric carcinoma.15,16,17,18,19,20,21 The oncogenic role of this miRNA cluster has been defined through the identification of some of the miR-372 and -373 targets, which are silenced in tumor cells expressing high levels of these miRNAs: the LArge Tumor Suppressor 2 (LATS2), a protein kinase regulating cell cycle progression,11,15,17 the hyaluronic acid receptor CD44 regulating tumor invasion and metastasis,22 the transforming growth factor β receptor 2 (TGFβR2), which upon TGFβ binding controls cell growth and promotes epithelial to mesenchymal transition,23 or else the tumor necrosis factor α-induced protein 1 (TNFAIP1) involved in the regulation of the Nuclear Factor κB (NFκB) signaling pathway.21 miR-372 and miR-373 belong to the miR-17 family along with miR-17-5p, miR-20a, miR-106b, and miR-93.24 All these miRNAs harbor similar seed sequence (Table 1).
Table 1. Sequences of the miRNAs of the miR-17 family.
Based on both the efficient silencing of miRNA families by seed-targeting tiny LNAs and our previous results on the high miR-372 and -373 expression in the human gastric adenocarcinoma AGS cells,6,11 the present work aims at adapting this anti-miRNA approach to inhibit gastric adenocarcinoma cell growth both in vitro and in vivo. We report here that 8-mer LNAs targeting miR-372 and -373 seeds are as efficient as full-length anti-miR-372/373 oligonucleotides upon transfection to antagonize the function of these oncomirs in cultured AGS cells, derepress LATS2 and subsequently inhibit their growth rate. They also enter without any transfection agent into cultured cells at micromolar concentrations to achieve cell growth inhibition. Moreover, they are able to specifically sequester their target miRNAs within tumor cells in AGS xenografts in mice and impair tumor growth. Their antitumor effect is particularly noticeable in fast-growing primary human gastric adenocarcinoma xenografts in mice.
Results
Specificity of 8-mer LNAs targeting miR-372 and miR-373 seeds
We previously reported the efficiency of full-length antisense oligonucleotides to miR-372 and miR-373 (AS372, AS373) to inhibit the functions of these miRNAs in AGS cells.11 To assess whether miR-372 seed-targeting LNAs (TL372) are as efficient as AS372 in antagonizing miR-372, 8-mer TL372, 22-mer AS372 or their respective controls TLCo and SC372 were transfected into growing AGS cells at final concentration of 10 nmol/l and miR-372 expression was evaluated by RT-qPCR and nondenaturating northern blot analyses. Upon AS372 transfection, the miR-372 level is drastically reduced as determined by RT-qPCR, and becomes undetectable by northern blot analyses, compared to untransfected cells or SC372-transfected ones (Figure 1a,b, respectively), in agreement with our previous data.11 Similar results were obtained by determining miR-373 content upon AS373 transfection (data not shown). However, the RT-qPCR data suggest that TL372 does not reduce the miR-372 content as efficiently as AS372 (Figure 1a). Northern blot analysis of miR-372 reveals a slower-migrating band in TL372-transfected cells, as compared to TLCo-, SC-transfected or untransfected cells (Figure 1b), likely being the miR-372/TL372 heteroduplex: indeed, anti-miRNA LNAs may sequester their targeted miRNA without degrading it.6 There is still room left in the miR-372/TL372 heteroduplex for the anti-miR-372 probe or the RT-qPCR primers to pair onto the miR-372 sequence and to make it detectable both by northern blot or RT-qPCR analyses (lower part of Figure 1b). On the contrary, in AS372-transfected cells, miR-372 remains sequestered on its full length by the specific 22-mer antisense oligonucleotide, and hence unable to bind either the probe or the primers: this makes it undetectable either on the northern blot or by RT-qPCR. Thus, using anti-miRNA 8-mer LNAs, quantification of residual miRNA levels by RT-qPCR may be misleading.
Figure 1.

Specificity of miRNA targeting by 22-mer full-length antisense or 8-mer seed-directed oligonucleotides in cultured AGS cells. AGS cells were transfected with either the 22-mer full-length anti-miR-372 LNA/DNA antisense (AS372) or its scrambled sequence (SC372), or with the 8-mer LNA oligonucleotide targeting the nu 2–9 of miR-372 including the seed (TL372), or its negative control sequence (TLCo), at a final concentration of 10 nmol/l in the presence of Lipofectamine 2000. Cells were grown for 5 days. (a) RT-qPCR analysis of miR-372 levels; the bars represent the miR-372 levels normalized with snor25 and U6 and compared to mock treated cells (mean ± SD, n = 4). (b) Northern blot analysis of miR-372 (medium panel) and U6 (upper panel) in nondenaturing conditions; the lower panel schematizes miR-372 paired or not with the specific antimiR for each lane. (c) Nondenaturing northern blot analyses of miR-372, miR-373, miR-17-5p, and miR-93 in AGS cells transfected with either TL372, TL373, TL17, TL21, or TLCo at 10 nmol/l.
The TL372 ability to sequester miR-372 is likely related to the high stability of LNAs in a duplex.4 Indeed, duplex formation between the miR-372/TL372 (RNA/full LNA) or the miR-372/TDNA372 (RNA/full DNA) was followed by thermal denaturation monitored by UV-spectroscopy. A shift is observed in the melting curves (Supplementary Figure S1), allowing the determination of melting temperature (Tm) of 79.7 ± 5 °C and 38.9 ± 0.7 °C for miR-372/TL372 and miR-372/TDNA372 respectively (n = 5; P value = 1.2 × 10−7). This important and significant increase in the thermal stability of the 8-mer LNA oligonucleotide as compared to the 8-mer DNA in duplex with the targeted miRNA, corresponding to a gain of 5 °C per LNA modification, confers to TL372 outstanding antisense properties despite its tiny size.
MiR-372 and miR-373 share similar seed sequences, which resemble those of miR-17-5p, miR-20a miR-106b, and miR-93 (Table 1). We synthesized 8-mer TL372, TL373, and TL17 LNAs perfectly matching the two to nine nucleotides of miR-372, miR-373, or miR-17-5, respectively (Supplementary Table S1). In order to assess their respective ability to target the different miRNAs of this miRNA family, we transfected them in AGS cells at 10 nmol/l and, 4 days later, we analyzed their ability to form a stable TL/miRNA heteroduplex in the cells. TLCo and the miR-21-targeting TL21 were used as negative controls. Figure 1c shows the northern blot analyses of miR-372 (Figure 1c-A), miR-373 (Figure 1c-B), miR-17-5p (Figure 1c-C) and miR-93 (Figure 1c-D) in nondenaturating conditions, and Table 2 summarizes the results, along with the likely pairing of the 8-mer LNAs to the miRNA sequences. As expected, TL372 is able to sequester miR-372, which appears as a shifted band as compared to mock, TLCo, and TL21-treated cells. TL372 also partly retains miR-93, with which it pairs beyond the seed from nucleotides 3 to 10. TL373 does not form a heteroduplex with any miRNA but miR-373, and the 10 nmol/l concentration seems insufficient to sequester the whole amount of miR-373 of the AGS cells. TL17 forms stable heteroduplexes with miR-17-5p and miR-93, as expected, as well as with miR-372, to which it pairs from nucleotides 1 to 8. It forms a smeary duplex with miR-373, to which it pairs from nucleotides 2 to 8. TL21 does not form a heteroduplex with any of those miRNAs but miR-21, as shown in Supplementary Figure S2. All combined, these data suggest that the ability of a 8-mer LNAs to efficiently target miRNAs of a same family resides in the perfect pairing over all the eight nucleotides and starting at nucleotide 2 or 1 at the 5′ end of the miRNA. Moving forward at nucleotide 3 of the miRNA and/or mispairing at nucleotide 9 lead to substantial or complete loss of their efficacy to sequester this miRNA. Therefore, although they harbor similar seeds, but differ on nucleotide 9, miR-372 and miR-373 cannot be targeted efficiently by a unique 8-mer seed-targeting sequence.
Table 2. Theoretical 8-mer LNA pairing with the miRNAs of the miR-17 family.

The 8-mer LNAs against miR-372 and miR-373 derepress LATS2 and inhibit AGS cell growth in vitro
LATS2 is one of the major targets of both miR-372 and miR-373, and contains two miR-372 and -373 pairing sites in its 3′UTR11. It has been validated in other cancer cells.15,17 To compare the efficiency of the 8-mer LNA and full-length AS against miR-372 and miR-373 to derepress LATS2, the luciferase reporter plasmid containing the 3′UTR of LATS2, named pGL3-3'UTR-LATS2wt, was transiently transfected into AGS cells along with 8-mer LNAs or 22-mer AS. The resulting luciferase activity was compared to that of either the control pGL3 vector or the pGL3-3'UTR-LATS2mut vector harboring mutated miR-372 and -373 pairing sites. In the absence of oligonucleotides, the luciferase activity generated by pGL3-3′UTR-LATS2wt, which senses the endogenous miR-372 and miR-373 levels, represented 30% of that of the control pGL3 reporter and 37% of that of the pGL3-3′UTR-LATS2mut vector (Figure 2a). In the presence of an equimolar mix of TL372+TL373, but not TLCo or SC372, the luciferase activity of the pGL3-3'UTR-LATS2wt increased in the same extend that in the presence of an equimolar mix of AS372+AS373 (Figure 2a). The TL372+373-derepressed luciferase activity reached the level of that generated by the pGL3-3′UTR-LATS2mut. This indicates that the 8-mer LNAs are as efficient as the AS372+373 to inhibit miR-372 and -373 functions and derepress LATS2. In consequence, LATS2 protein accumulates in cells treated by either 8-mer or full-length antisenses against miR-372 and miR-373, but not in cells treated by the respective control oligonucleotides, in which LATS2 expression pattern remained as faint as in untreated cells (Figure 2b,d). The derepressed LATS2 mainly accumulates in the cell nucleus upon treatment with either 22-mer AS or 8-mer LNAs (Figure 2d). TL372+373-transfected AGS cells exhibit a noticeably reduced proliferation rate, which is slowed down by 50 or 40% compared to those of untransfected or TLCo-transfected cells, respectively (Figure 2c). TLCo-transfected cells grow slightly slower as compared to untransfected cells: this effect could be attributed either to the transfection procedure using Lipofactamin or to some noxious properties of the LNA/phosphorothiate chemistry of oligonucleotides, which may have reached unspecific targets as well through their strong binding capacities, leading to slight off-target harmfulness. Of note, TL21, which totally sequesters miR-21, another oncogenic miRNA that is also highly expressed in AGS cells (Supplementary Figure S2),11 has no more effect on AGS proliferation rate that TLCo, suggesting that this cell line may not be particularly dependent on miR-21 for its growth. All together, these results show that 8-mer LNAs, inhibiting miR-372 and -373 functions and derepressing LATS2, inhibit AGS cell growth. Their effects are similar as those of the 22-mer AS, as reported in our previous data.11
Figure 2.
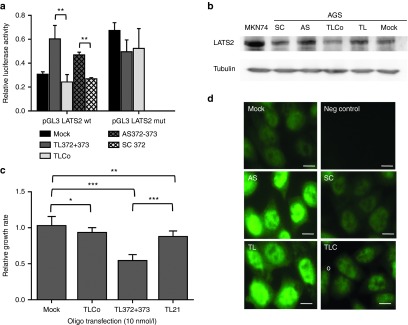
8-mer LNAs against miR-372 and miR-373 de-repress LATS2 and inhibit the growth of cultured AGS cells. (a) Relative luciferase activity (mean ± SD, n = 3) of the LATS2 reporter system pGL3-LATS2wt containing the wt LATS2 3'UTR, or of the mutated LATS2 reporter system pGL3-LATS2mut, compared to that of the control vector pGL3. The vectors have been cotransfected with pRL-SV40 and 10 nmol/l of the indicated oligonucleotides using lipofectamin, and their expression was analyzed 48 hours later. (b) Western blot analysis of LATS2 (upper panel) and α-tubulin (lower panel) protein expression in untreated MKN74 cells (positive control) or in AGS cells treated with an equimolar mix of either 22-mer anti-miR-372+miR-373 antisenses (AS) or their scrambled sequence (SC), or with 8-mer LNA against miR-372 and miR-373 (TL) or their negative control (TLCo). (c) Relative growth rate (mean ± SD, n = 6) of tiny LNA-transfected cultured AGS cells between day 2 and day 5 post-treatment, compared to TLCo-treated cells. (d) LATS2 immunofluorescent staining in AGS cells in the same conditions than in (b); scale bars, 10 μm.
The experiments shown in Figures 1 and 2 have been performed by transfecting the 8-mer LNAs with lipofectamin, which allows using concentrations in nanomolar range. As tiny LNA have been proven to enter cells unassisted,6 we verified this property by treating AGS cell cultures with them without any transfection agent. We established a stable AGS cell line containing the pGL4-PM372 luciferase reporter system, which contains a single site perfectly matching miR-372 downstream of the luc coding sequence. This AGS-luc-PM372 line was compared to the AGS-luc-Mut372, a stable cell line constitutively expressing the pGL4-mut372 reporter system, in which the sequence pairing the miR-372 seed contains three mismatches and is thereby unable to sense miR-372. AGS-luc-PM372 cells express a low luciferase activity as compared to AGS-luc-Mut372 cells (5,611 ± 1,018 RLU/μg protein versus 1332,000 ± 178,000 RLU/μg protein), due to the miR-372-mediated post-transcriptional luc repression. AGS-luc-PM372 cells specifically respond to the TL372 at 2.5 μmol/l by a significant increase of the luciferase activity (Supplementary Figure S3a), whereas AGS-pGL4-Mut372 cells do not (Supplementary Figure S3b). These data suggest that 8-mer LNAs at micromolar range concentrations have entered freely into the cells to specifically inhibit the target miRNAs. Subsequently, an equimolar mix of TL372+TL373 induces a significant dose-dependent decrease in AGS growth rate (Supplementary Figure S3c) as compared to TLCo-treated cells, which grow at the same rate than untreated cells.
Seed-targeting 8-mer LNAs silence miR-372 and mir-373 in AGS cells grown in mice and inhibit tumor growth
We then determined whether the 8-mer LNAs could be used to target miR-372 and -373 in solid tumors in vivo and subsequently reduce their growth rate. Therefore, we subcutaneously engrafted the stable reporter cell lines AGS-luc-PM372 or AGS-luc-Mut372 into female NSG mice (n = 5, one tumor per mouse). The xenografts grew at a similar rate independently on the reporter they harbor (AGS-luc-PM372 and AGS-luc-mut372 tumor size: 119.6 mm3 ± 46.2 and 108.6 mm3 ± 61.2, respectively, after 2 weeks, P value = 0.22; 157.6 mm3 ± 42.3 and 150.5 ± 54.7 respectively after 4 weeks, P value = 0.43). Ten days after AGS cell grafting, increasing concentrations of an equimolar TL372+373 mix or TLCo were subcutaneously injected at the tumor periphery for 3 consecutive days. Both the size of the tumor and in vivo bioluminescence images were periodically recorded, starting at the day of the first injection (Figure 3). A noticeable delay was observed in the growth kinetics of the tumors treated with TL372+373 at 5 or 50 nmoles/mouse as compared to untreated or TLCo-treated ones, leading to a significant reduction of tumor size after 2 weeks following the first injection (Figure 3a). Bioluminescence images recorded at day 9 following the first injection show the luciferase activity appearing in the AGS-PM372 tumors upon treatment with TL372+373 but not with TLCo (Figure 3b and Figure 3c, E versus D), whereas in mock conditions luciferase activity of AGS-PM372 tumors remains undetectable as compared to that of AGS-mut372 tumors (Figure 3b and Figure 3c, C versus A). These data indicate that the injected TL372+373 may have reached the tumor cell cytoplasm to inhibit the function of the endogenous miR-372 and miR-373 levels. The TL372+373-mediated upregulation of the luciferase activity in the AGS-PM372 tumors likely results from the combined effects on de-repression of the luciferase reporter sensing miR-372, on the one side, and reduction of the tumor size, on the other side. These combined effects could explain why the luciferase activity recorded in TL372+373-treated mice bearing the AGS-Mut372 xenografts is lower than that of the untreated ones (Figure 3b and Figure 3c, B versus A).
Figure 3.
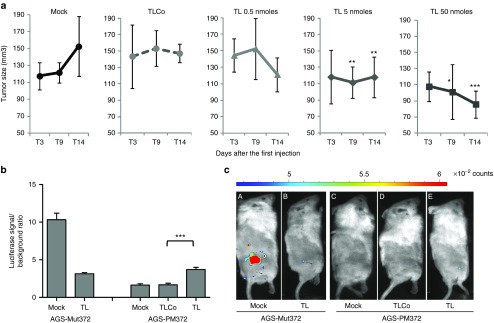
8-mer LNAs inhibit miR-372 in AGS cells grown in mice and delay tumor growth. (a) Growth kinetics of AGS tumors either untreated (mock) or treated with TLCo (5 nmol/mouse) or an equimolar TL372+TL373 mix (TL) at 0.5, 5, or 50 nmol/mouse. Tumors were measured the 3rd (T3), the 9th (T9), and the 14th (T14) day following the first injection and their size was expressed in mm3 (mean ± SD, n = 5). (b) Quantification of in vivo luciferase activity at T9 on AGS-PM372 or AGS-mut372 xenografts, either untreated (mock) or treated with TL or TLCo at 5 nanomoles/mouse: bars represent the mean ± SD (n = 5) of radiance (photons/s/cm2/steridian) of the region of interest relative to the background radiance (out of the region of interest). (c) Representative in vivo bioluminescence images of AGS-mut372 xenografts in mice either untreated (A) or TL-treated (B), and AGS-PM372 xenografts either untreated (C), TLCo- (D) or TL-treated (E) as in (B).
Two weeks post-treatment, the mice were euthanized. The residual tumors were removed and processed for further RNA analyses and histology. The northern blot analysis of miR-372 clearly shows a slower-migrating miR-372/TL372 heteroduplex in TL372+373-treated AGS xenografts, compared to TLCo or untreated tumors (Figure 4a, A). A similar band shift is observed on the northern blot analyzed for miR-373 expression in TL372+373-treated tumors and corresponds to the miR-373/TL373 heteroduplex (Supplementary Figure S4). The heteroduplex is formed in a TL372+373 dose-dependent manner at doses over 5 nanomoles/mouse (Figure 4a, B). On the contrary, the northern blot analysis of miR-21 does not show any band shift in the TL372+373-treated cells (Figure 4a, C and D), further stressing the specificity of miRNA targeting by 8-mer LNAs. Along with TL372+373-mediated miR-372 and miR-373 sequestrations and tumor growth inhibition, LATS2 is upregulated upon TL372+373 treatment, as compared to TLCo treatment, and accumulates in the carcinoma cell nucleus and cytoplasm (Figure 4b).
Figure 4.
8-mer LNAs stably sequester miR-372 in AGS xenografts and derepress LATS2. (a) Nondenaturing northern blot analysis of miR-372 (A,B) and miR-21 (C,D) in AGS-PM372 tumors treated with TLCo or an equimolar TL372+TL373 mix at 5 nanomoles/mouse (A,C) or with increasing doses (B,D). (b) Representative images of LATS2 immunostaining in AGS -PM372 tumors treated with either TLCo or TL372+373 at 5 nanomoles/mouse. Scale bars, 25 μm.
8-mer LNAs targeting miR-372 and mir-373 inhibit the growth of primary human gastric carcinoma xenografts
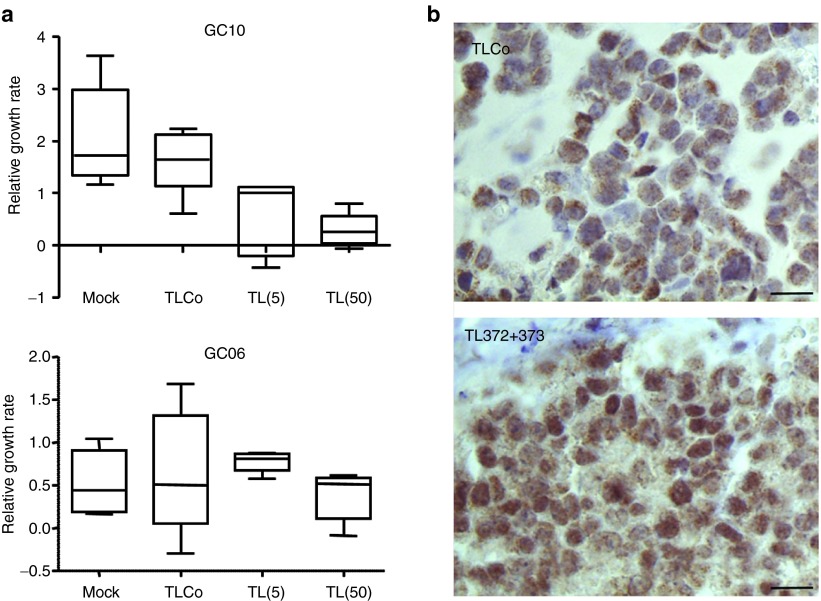
To assess whether the human gastric AGS cell line is relevant of other human gastric carcinoma, we selected two tumors in our collection of resected tumors from gastric cancer patients, to be xenografted in mice and treated with an equimolar TL372+373 mix. GC10 is a highly aggressive, fast growing gastric carcinoma of intestinal type; it expressed both miR-200b, which attests for its epithelial origin, and miR-372 in every epithelial cell, as shown by in situ hybridization (Supplementary Figure S5 left panels). GC06 is a gastric carcinoma of diffuse type that grows slower than GC10; it homogenously expressed miR-200b in every cell, but miR-372 only in some cell clusters (Supplementary Figure S5, right panels). Figure 5a shows that untreated GC10 tumors gain 200% volume in 2 weeks in average, whereas GC06 tumors gain only 60% volume during the same time, suggesting a causal relationship between the high miR-372 expression and rapid tumor growth. Indeed, TL372+373 treatment markedly inhibits GC10 growth at doses as low as 5 nanomoles per mouse. It is inefficient on GC06 (Figure 5a). TL372+373-treated GC10 xenografts exhibit an upregulated LATS2 expression as compared to TLCo-treated ones (Figure 5b).
Figure 5.
8-mer LNAs inhibit the growth of a miR-372-positive human gastric tumor. (a) TL372+373 (TL) dose-dependent effects on tumor growth: the horizontal bars inside the boxes indicate the median and the capped bars the minimal and the maximal data values (n = 5) of the relative tumor size increase during the 14 days following the first day of injection. (b) Representative images of LATS2 immunostaining in GC10 xenografts treated with either TLCo or an equimolar TL372+TL373 mix at 5 nanomoles/mouse. Scale bars, 25 μm.
Importantly, the subcutaneous injection of TL solution at the periphery of AGS, GC10 or GC6 tumors gave rise to no sign of inflammation/ulceration. The treated mice exhibited a good general state with no sign of distress, weight loss or reduced motility.
Discussion
MiRNA silencing by chemically modified oligonucleotides has become an important and widely used approach in studies aimed at unveiling miRNA functions, as well as in miRNA-based therapeutic strategies, when those miRNAs are aberrantly over-expressed in pathologies. Whereas satisfying oligonucleotide uptake can be accomplished in in vitro cultured cells using commercial transfection agents, efficient delivery of anti-miRNAs remains a critical factor for their successful use in vivo. We here applied an anti-miRNA strategy based on the use of seed-targeting 8-mer LNA oligonucleotides, originally developed by Obad et al.6, to antagonize the function of miR-372 and miR-373 in gastric carcinoma cells. Whereas these two miRNAs are specific of embryonic stem cells and not expressed in adult tissues, they may be expressed at high levels in some tumors of various tissue origins, in which they behave as oncomirs, silencing tumor suppressor genes such as the cell cycle regulator LATS2, and triggering cell growth.15,16,17,18,19,20,21,22 Using the AGS cell line as a model of gastric adenocarcinoma, in which miR-372 is the most expressed of its miRnome,11 we found that 8-mer LNAs complementary to the miR-372 and miR-373 seeds specifically antagonize the growth-promoting functions of these miRNAs and reduce the growth rate of both in vitro cultured cells and in vivo mouse xenografts. Moreover, they are also highly efficient on mouse xenografts of primary human gastric adenocarcinoma that express miR-372.
LNA oligonucleotides, in which the bicyclic furanose unit is locked in an RNA-mimicking sugar conformation, resemble natural nucleic acids with respect to Watson−Crick base-pairing and display outstanding hybridization affinity toward complementary single-stranded RNA or DNA.4 Accordingly, using TL372 targeting miR-372 as an example, we found that the melting temperature of the 8-mer LNA/miRNA duplex was raised by 5°C/base as compared to that of the 8-mer DNA/miRNA duplex. One could fear that the remarkable thermodynamic stability of the 8-mer LNAs for their sequence-specific targets may reach unspecific targets as well and affect their activity: this could explain why TLCo-transfected cells exhibit a slightly reduced growth rate, as observed in Figure 2c. However, the similar growth abilities of untreated cells and TLCo-treated cells without any transfection agent (Supplementary Figure S3c) exclude possible off-target effects of LNA oligonucleotides in these conditions, and the growth reduction in TLCo-transfected cells is likely due to the transfection procedure with lipofectamine rather than the LNA chemistry. The absence of off-target effects by LNA oligonucleotides has been previously reported and deeply analyzed for TLs in particular.4,6
The seed region comprises the nucleotides 2 to 8 of the miRNA, with nucleotides 2 to 7 being most critical for binding specificity.25 The efficiency of targeting the miRNA seed has been interpreted by the high accessibility of this particular region, exposed and preorganized in the Argonaute (Ago) complex to favor efficient pairing to the seed-match site in a target mRNA.26,27 Ago proteins form the functional core of the RNA-induced silencing complexes (RISC) that mediate RNA silencing in eukaryotes.4 8-mer LNAs have been shown to pair to mature miRNAs bound to Ago2.6 Thus TL372 and TL373 sequestering miR-372 and miR-373 within the Ago complex competes out the miR372 and -373 binding sites on the targeted mRNAs, among which the tumor suppressor LATS2, and alleviates their miRNA-dependent repression.
The length of 8-mer LNAs complementary to the miRNA nu 2–9 including the seed sequence has been found to be optimal to sequester the miRNA in competition with the targeted mRNA, 6-7-mer or 9-10-mer LNAs both being less or no efficient.6 Although miR-372 and miR-373 harbor the same nu 2–8 seed sequence and both target LATS2 through interactions at two sites in the LATS2 3′ UTR,11,15 their 5′ end differs at the nineth nucleotide, which pairs with the first nucleotide at the 5′ end of the tiny LNA. According to our analysis of the miRNA/8-mer LNA duplex formation, the 8-mer LNA targeting miR-372 is not able to sequester miR-373, and vice versa. In comparison, Obad et al.6 reported that a mismatch between the first nucleotide of TL21 and the 9th nu of miR-21 decreased by 60% its efficiency to derepress the miR-21 reporter. Thus, our data confirm that the optimal anti-miRNA activity of the 8-mer LNAs indeed requires the perfect pairing of all the eight nucleotides with the target miRNA. A 8-mer LNA targeting the seed of miR-17 would have been a more “universal” anti-miRNA sequestering also miR-372, -373, -93, and likely also miR-106b and miR-20a (Table 1), in addition to miR-17-5p. We choose to target miR-372 and -373 because, contrarily to miR-17-5p, they are not expressed in healthy adult tissues, and therefore their targeting should not lead to undesirable side effects on normal tissue, a potential risk targeting miR-17. Nevertheless, deleterious effects on healthy tissues of 8-mer LNAs targeting broadly expressed miRNA have not been found.6,10
An additional quality of 8-mer LNAs is the possibility to be delivered as simple unconjugated oligonuclotides without the use of any transfection agents. This property could be demonstrated in tissue culture, but requires micromolar ranges of TL concentrations instead of nanomolar concentrations in complex with a transfection agent such as lipofectamin. Nevertheless, micromolar concentrations are still acceptable and comparable to those used for pharmacological inhibitors of signaling pathways. The simple oligonucleotide delivery is highly valuable in vivo, and the antitumor properties of TL372+373 are emphasized in a human gastric carcinoma xenograft model in mice. They are associated with long-lasting tumor growth inhibition, likely related to LATS2 derepression for a couple of weeks after three daily topical injections. Although other miR-372/miR-373 targets may be similarly affected, LATS2 has been previously found to be necessary and sufficient to achieve the anti-miR-372/373-mediated growth inhibition of AGS cells.11 Of note, the efficient TL372+373 dose used in this study (namely 5 nanomoles per mouse (0.5 mg/kg)) is 50 times lower than used by Obad et al.6 in intravenous injections. Interestingly at a therapeutical point of view, the lack of general toxicity of TL372+373 is consistent with other data that have pointed out the safety of LNAs in mice as well as in nonhuman primates.7,8,9,10
Gastric cancers, which constitute the third cause of cancer-related mortality worldwide, are more often adenocarcinomas that derive from the malignant transformation of the gastric mucosa in the context of a chronic infection by Helicobacter pylori.28 Gastric adenocarcinomas exist in either diffuse or intestinal histological subtypes. Specific alterations in the miRNA expression pattern have been identified in human gastric adenocarcinoma since the earliest steps of tumorigenesis, and involve the oncomir miR-21 and miRNAs of the oncogenic miR-17~92 and miR-106~25 clusters.29,30 Despite the fact that, in our experience, the strong miR-372 or miR-373 expression is not a feature as common as that of those mentioned oncomirs (C. Staedel and C. Varon, unpublished results), it nevertheless confers a remarkably active growth potential to the tumor cells, as observed here for GC10 versus GC06 and by others in gastric cancer or in other localizations.15,16,17,18,19,20,21 As embryonic stem cell markers, miR-372 and miR-373 in tumors could be a signature of cancer stem cells (CSC), a tumor cell reservoir to be preferentially targeted in anticancer therapies, because they supply tumor growth and metastasis by their self-renewal capacities combined to their general resistance to chemo- and radiotherapy.12 Interestingly, CD44 is both a miR-373 target and a gastric CSC marker.22,31 LATS2 constitute the core of the Hippo pathway, which regulates organ size during development.32 The particular CD44/miR-372&373/LATS2 relationship would therefore deserve further exploration in the context of gastric adenocarcinoma.
In conclusion, our study support the importance of 8-mer seed-targeting LNAs as important tools to antagonize highly expressed oncomirs and derepress tumor suppressor genes to recover growth control. Targeting of miR-372 and -373 may be applicable to other cancer subsets depending on these oncomirs for their growth.
Materials and methods
Synthesis of LNA oligonucleotides. The tiny LNA were synthesized as fully phosphorothiolated oligonucleotides with eight consecutive LNAs on an automated Expedite 8909 DNA synthesizer (Dudelange, Luxembourg). For optimal synthesis yield, an additional 9th nucleobase at the 3′ end was a DNA instead a LNA (Supplementary Table S1). The LNA phosphoramidite solutions were prepared with commercially available LNA-amidites (ABz, GDMF, mCBz, T) from Exiqon (Vedbaek, Denmark) and diluted in anhydrous acetonitrile (Glen Research, Sterling, VA) and 25% of tetrahydrofuran (Sigma-Aldrich, Lyon, France) for LNA-mCBz. DNA SynBase CPG (1,000Å, loading 60–100 µmol/g, Link Technologies Bellshill, UK) was used as solid support. The phosphorothioate backbone of the oligonucleotides was synthesized using Sulfurizing Reagent II (Glen Research). After the synthesis, the oligonucleotides were cleaved from the supports and the nucleobases were deprotected with ammonium hydroxide at 55 °C for 4 hours. The oligonucleotides were then desalted and concentrated by 85% ethanol precipitation. Concentrations were determined by absorbance at 260 nm and the respective molar extinction coefficient. Their purity was assessed by high-pressure liquid chromatography (Supplementary Figure S6).
Determination of duplex melting temperature. The miR-372-3p strand (5′ AAAGUGCUGCGACAUUUGAGCGU3′) was synthesized in RNA/phosphodiester chemistry on the automated Expedite 8909 DNA synthesizer and purified on an 8% denaturing polyacrylamide gel using conventional procedures. MiR-372-3p, TL372, or TDNA372 in solution in RNAse-fre water were heated for 2 minutes at 90 °C and cooled for 5 minutes in ice, before being individually diluted in Tm-buffer (200 mmol/l NaCl, 0.2 mmol/l Ethylenediaminetetraacetic acid (EDTA), 20 mmol/l sodium cacodylate, pH 7.3) at a concentration of 4 μmol/l. The 4 μmol/l miR-372-3p solution was mixed with an equal volume of either 4 μmol/l TL372 or 4 μmol/l TDNA372 at room temperature. The duplex melting temperatures were determined using a Cary UV-Vis spectrophotometer equipped with a Peltier temperature controller (both from Agilent Technologies, Les Ulis, France). Measurements were performed in 0.2 ml quartz cuvettes containing 250 μl of the duplex solutions with a mineral oil overlay (200 μl). After an initial 15-minute equilibration, the temperature was ramped up from 20 to 90 °C and then down to 20 °C, at a rate of 0.3 °C/minute, recording absorption at 260 and 310 nm. Data were collected using the THERMAL software. The (A260 nm-A310 nm) curve of each duplex was corrected with that of the Tm-buffer alone, and the first derivative of the resulting curve was used to assess the Tm of the duplex.
Cell culture. All the tissue culture reagents were from Invitrogen (Illkirch, France). AGS gastric epithelial cell line (ATCC CRL 1739) was routinely grown in Dulbecco's modified Eagle medium/F-12 medium, supplemented with 10% heat-inactivated fetal bovine serum and 2 mmol/l L-glutamine at 37 °C in a humidified 5% CO2 atmosphere.
RNA extraction. Total RNA was extracted using Trizol reagent (Invitrogen, Illkirch, France), according to the manufacturer's protocol. Trizol was directly added onto the drained cell layers in cell culture wells, or onto the resected tumor pieces in a 2-ml Eppendorf tube containing a 5 mm steel bead. The tumor was completely disrupted after 90 seconds at 30 Hz in a Tissue-Lyser (Qiagen, Courtaboeuf, France). RNA concentrations were determined using a NanoDrop spectrophotometer (NanoDrop Technologies, Wilmington, DE). RNA quality was analyzed on a 2100 Bioanalyzer (Agilent Technologies, Les Ulis, France).
Quantitative RT-PCR. Total RNA (500 ng) was retro-transcribed in 10 μl using the miScipt Reverse Transcription kit (Qiagen), according to the manufacturer's instructions. Mature miRNA expressions were quantified on 1/200 RT reaction volume by real-time PCR using the 2× Quantitect SYBR Green PCR Master mix (Qiagen) with universal and miRNA-specific primers (both from Qiagen). Real-time PCRs were run on ROTORGENE (Qiagen). MiRNAs levels were normalized to the small noncoding RNA snor25 and U6, using the comparative Ct method.
Northern blots. Total RNA (10 µg) was resolved on a 20% nondenaturing polyacrylamide gel in 1× TBE and blotted onto nylon membranes (Hybond-N, GE Healthcare Little Chalfont, UK) according to standard procedures. Membranes were incubated with 5'-32P-radiolabeled LNA/DNA anti-miRNA probes (Supplementary Table S1) in 50% formamide, 5× SSPE (sodium chloride, sodium phosphate, EDTA), 5× Denhardt solution, 0.5% SDS (sodium dodecyl sulfate), 20 μg/ml salmon sperm DNA, at 42 °C overnight in a hybridization oven. After two washes for 5 minutes each in 2× SSC, 0.1% SDS at 42 °C (sodium chloride, sodium citrate), blots were analyzed by phospho-imaging (Molecular imager PharosFXplus, Biorad, Marnes-la-Coquette, France).
Luciferase reporter systems. The pGL3 and pRL-SV40 vectors harboring the firefly luciferase and the Renilla luciferase coding sequences, respectively, under the control of the SV40 promoter were from Promega. The pGL3-3'UTR-LATS2wt and pGL3-3'UTR-LATS2mut constructs were kindly provided by R. Agami (Amsterdam, The Nederlands). 15 The pGL4-PM372 and pGL4-Mut372 vectors were obtained upon the following cloning steps on the basis of the pGL4.15 vector (Promega, Charbonnieres, France) backbone harboring the firefly luciferase and hygromycin B phosphotransferase coding sequences: (i) insertion of the SV40 promoter retrieved by PCR from the pRL-SV40 vector between Bgl II and Hind III sites into the multi-cloning site upstream of the luciferase coding sequence; (ii) site-directed mutagenesis of the Xba I site at 3′ of the luciferase coding sequence into a Pca I site; (iii) insertion of a synthetic DNA fragment corresponding to Pac I site/single perfect-match target site for miR-372 (or single mutated target site)/SV40 late poly(A)region/BamH I site (synthesized by Millegen Biotechnologies, Labege, France) between the Pac I and BamH 1 sites downstream of the luciferase coding sequence. Molecular cloning was performed according to standard protocols and the final constructs were verified by sequencing.
Transient transfections. Cells were plated at 5 × 104 cells/well in a 24-well plate. Transfections were performed using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. Cells were cotransfected with 10 nmol/l of either 22-mer full-length anti-miRNA antisense (AS372, AS373), 22-mer scrambled oligonucleotides (sc372-373), 8-mer anti-miRNA LNA (TL372, TL373), or the 8-mer negative control (TLsc), mixed with 10 ng pRL-SV40 control vector (Promega) and 100 ng of either pGL3, pGL3-wt3'UTR-LATS2, pGL3-mut3'UTR-LATS2. Firefly and Renilla luciferase activites were measured 48 hours post-transfection using the Dual Luciferase Assay (Promega). Firefly luciferase activities were normalized for transfection efficiency by Renilla luciferase activity.
Western blot. Cells were washed twice with ice cold PBS and harvested in ice cold ProteoJET Mammalian Cell lysis reagent (Fermentas, Illkirch, France) supplemented with 5 mmol/l EDTA and 1× ProteoBlock Protease inhibitor cocktail (Fermentas). Proteins (40 μg) were separated by SDS-PAGE and western blot performed using Immobilon-P transfer membrane (Millipore, Molsheim, France), according to standard procedures. Anti-LATS2 (clone ST-3D10, Abnova, Fontenay-sous-Bois, France) and anti-α-tubulin (clone B-5-1-2, Sigma-Aldrich) antibodies were used at a 1:500 and 1:30,000 dilutions, respectively. Protein detection was carried out using HRP-conjugate anti-mouse IgG and enhanced chemoluminescence according to the manufacturer (Immobilon Western Chemiluminescent HRP substrate, Millipore). The luminescence on the blots was detected using a DS ImageQuant LAS (GE Healthcare) analyzer.
Cell growth. Cells were grown in a 96-well microplate, starting at 1,500 cells per well. Cell viability was assessed using the CellTiter96 AQueous One Solution Reagent (Promega), as recommended by the manufacturer. Absorbance at 492 nm was read on a Bio-Tek spectrophotometric microplate reader (Colmar, France).
Immunofluorescence. Cells were grown on glass coverslips, fixed with 3% buffered paraformaldehyde and permeabilized in Triton 0.2%. After blocking with 3% bovine serum albumin in phosphate-buffered saline (PBS) (PBS-BSA), they were stepwise incubated overnight at 8 °C with a rabbit anti-LATS2 antibody (Bethyl, Souffelweyersheim, France, 1/200 dilution), washed four times in PBS-BSA and then 30 min at room temperature with a secondary anti-rabbit IgG antibody coupled to Alexafluor488 (Invitrogen; 1/2,000 dilution). After extensive washes in PBS, the coverslips were mounted on glass slides using Slowfade reagent (Invitrogen) prior to imaging in epifluorescence on a Zeiss microscope (Marly le Roi, France).
Establishment of stable reporter AGS cell lines. Cells (105 per well) were plated in a 24-well plate and transfected the following day with 500 ng of either pGL4-PM372 or pGL4-Mut372 vectors using Fugene HP transfection reagent (3 μl/μg DNA; Promega) as recommended. Twenty-four hours post-transfection, the cells were detached and plated in a 10 cm diameter tissue culture plate in nonselective growth medium. After 24-hour growth, hygromycin B (Invitrogen) was added at a final concentration of 0.7 mg/ml to select the AGS cells having intregrated the luciferase reporter DNA. Stable pGL4-Mut372 reporter clones were chosen on the basis of the intensity of their firefly luciferase specific activity, and stable pGL4-PM372 reporter clones were chosen on the basis of the intensity of the luciferase activity upon specific derepression by anti-miR-372 antisense oligonucleotides.
In vivo experiments
Ethics statement. Approval was obtained from the French Committee of Genetic Engineering (approval number 4608) and the local Central Animal Facility Committee of the University of Bordeaux before initiation of the study. All animal experiments were performed in level 2 animal facilities of the University of Bordeaux, in accordance with national institutional guidelines and with the agreement of the local Ethic Committee on Animal Experiments CEEA50 of Bordeaux (agreement number 50120151-A).
Studies on fresh tumors from primary gastric adenocarcinoma cases were performed in agreement with the Direction for Clinical Research and the Tumor and Cell Bank of the Institut Bergonié of Bordeaux. Fresh tumors samples were collected from gastric surgical wastes from consenting patients (written consent), who underwent gastrectomy for gastric cancer. Activities of conservation and preparation of elements of human origin are declared at the French Ministry of Research (reference number DC-2008–412).
Mouse xenografts of AGS cells and in vivo imaging. Immunodeficient nonobese diabetous/shi-severe combined immunodeficiency/interleukin-2Rgammanull (NSG) mice under 2.5% isoflurane anesthesia (Belamont, Piramal Healthcare, Northumberland, UK) were injected subcutaneously into the right flank with 105 AGS reporter cells, GC10 or GC06 gastric tumor cells resuspended in 100 μl of 7 mg/ml Matrigel (BD Biosciences, Le Pont de Claix, France) in ice cold PBS. Tumor growth was periodically monitored by measuring the length (L) and width (W) of the tumor with a caliper, and tumor volume was calculated according to the formula ½(L × W2), as previously described.31 When the engrafted tumor was visible through the skin of the animal and reached ~100 mm3, 100 μl of 8-mer LNA solutions at the indicated concentrations were injected subcutaneously at the tumor periphery on 3 consecutive days. For AGS reporter cells, in vivo luciferase imaging was performed at the indicated periods of time upon LNA injection on isoflurane-anesthetized and shaved animals, previously injected intraperitoneally with 3 mg D-luciferin (Promega) per mouse in 100 μl PBS. In vivo luciferase activity was recorded within the region of interest in the flank of the animal, for 30 minutes following luciferin injection using a real time Photon IMAGER (Biospace, Nesles la Vallee, France). At the end of the experiments (4 weeks postengraftment), mice were sacrificed by cervical dislocation and tumors immediately harvested and processed for further analyses.
Primary human gastric adenocarcinoma-derived cells. GC10 and GC06 gastric adenocarcinoma xenografts were established in NSG mice from tumor samples collected from primary human gastric adenocarcinoma. Primary tumor samples were xenografted subcutaneously in isofluran-anesthesized NSG mice. Secondary tumors were amplified in mice by serial transplantation. Briefly, tumors were collected for cell isolation when reaching ~500 mm3: after mechanical mincing, samples were dissociated by collagenase IV and hyaluronidase (all from Sigma-Aldrich) as previously described.33 Viability was evaluated by trypan blue exclusion, before counting for analysis or resupension in matrigel for xenograft experiments. Histopathological characteristics were confirmed to be similar to those of the primary tumor after histological analysis of the tumor xenografts processed following standard procedures (hematoxylin and eosin, alcian blue and periodic acid Schiff stainings).34 Expression of human epithelial markers was verified by immunostaining of ESA (StemCell Technologies, Grenoble, France) and CD24 (BD Biosciences) and flow cytometry analysis as previously described.33 According to Lauren's classification, GC10 and GC06 are intestinal (moderately differentiated) and diffuse types, respectively, both infiltrating the sub-serosa.
Tumor immunohistochemistry (IHC) and in situ hybridization of miRNA. For histology processing, the tumor samples were fixed for 24 hours in 3.7% neutral-buffered formalin (Sigma), followed by standard histological processing and paraffin embedding. Sections of 3 µm thickness of paraffin-embedded tissues were used for standard IHC protocols with anti-LATS2 antibodies (Bethyl; 1/400 dilution) for 1 hour at room temperature, and then with labeled Polymer-Horse Radish Peroxidase anti-rabbit EnVision System (DAKO, les Ulis, France) for 30 minutes at room temperature. Immunolabeling was revealed by 10 minutes incubation in liquid substrate-diaminobenzidine-chromogen (DAKO) at room temperature. Slides were counterstained with hematoxylin, dehydrated, and mounted with Eukit-mounting medium (Labonord, Fontenay-sous-Bois, France).
For in situ hybridization of miR-372, deparaffinized and rehydrated tissue sections of 5 μm thickness were digested by Proteinase K (Invitrogen, 10 μg/ml in Tris 50 mmol/l pH 7.6) for 15 minutes at room temperature, and postfixed in 4% paraformaldehyde in PBS for 10 minutes. After three washes in PBS and one in SSC buffer 4×, the tissues were prehybridized for 2 hours at 52 °C in formamide 50%, SSC 4×, dextran sulfate 200 mg/ml, tRNA 500 μg/ml, and hybridized overnight at 52 °C in the same buffer containing 25 nmol/l anti-miR-372 DNA/LNA antisense (Supplementary Table S1) labeled at both 3′ and 5′ ends with digoxigenin. After two stringent washes in SSC 0.2× at 52 °C, the hybridized antisense was revealed with an anti-digoxigenin antibody coupled to alkaline phosphatase (Roche, Meylan, France, 1/400 dilution, overnight incubation at 8 °C), followed after four washes by a 2-hour incubation at room temperature in a 1-StepNBT/BCIP plus Suppressor solution (Pierce, Illkirch, France). Slides were dehydrated and mounted with Eukit-mounting medium.
Statistical analysis. Statistical significance (***P < 0.001, **P < 0.01, *P < 0.05) was calculated using the unpaired, one-tailed Student test.
SUPPLEMENTARY MATERIAL Table S1. Oligonucleotides used in this study. Figure S1. Determination of duplex melting temperature of the miR-372-3p in duplex with TL372 or TDNA372. Figure S2. Non denaturing northern blot analysis of miR-21 in AGS cells. Figure S3. Unassisted 8-mer LNAs inhibit miR-372 and miR-373 functions. Figure S4. Non denaturing northern blot analysis of miR-373 in AGS-PM372 tumors treated with TL372+373 or TLCo at 5 nanomoles/mouse. Figure S5. MiR-372 and miR-200b in situ hybridization. Figure S6. Chromatograms of TL372, TL373 and TLCo.
Acknowledgments
This work was supported by the SFR “Technology for Health” of Bordeaux, by the Conseil Regional d'Aquitaine (Grants # 20091301007 and 20091301027) and by the INSERM U869 (France). We thank all the authors' lab members, who contributed at developing the cell systems used in this study, as well as the technical team of the Animal Facilities of the University of Bordeaux. The authors declare no conflict of interest.
Supplementary Material
References
- Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39 Database issue:D152–D157. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntzinger E, Izaurralde E. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat Rev Genet. 2011;12:99–110. doi: 10.1038/nrg2936. [DOI] [PubMed] [Google Scholar]
- Di Leva G, Croce CM. miRNA profiling of cancer. Curr Opin Genet Dev. 2013;23:3–11. doi: 10.1016/j.gde.2013.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vester B, Wengel J. LNA (locked nucleic acid): high-affinity targeting of complementary RNA and DNA. Biochemistry. 2004;43:13233–13241. doi: 10.1021/bi0485732. [DOI] [PubMed] [Google Scholar]
- Thomas M, Lange-Grünweller K, Dayyoub E, Bakowsky U, Weirauch U, Aigner A, et al. PEI-complexed LNA antiseeds as miRNA inhibitors. RNA Biol. 2012;9:1088–1098. doi: 10.4161/rna.21165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obad S, dos Santos CO, Petri A, Heidenblad M, Broom O, Ruse C, et al. Silencing of microRNA families by seed-targeting tiny LNAs. Nat Genet. 2011;43:371–378. doi: 10.1038/ng.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garchow BG, Bartulos Encinas O, Leung YT, Tsao PY, Eisenberg RA, Caricchio R, et al. Silencing of microRNA-21 in vivo ameliorates autoimmune splenomegaly in lupus mice. EMBO Mol Med. 2011;3:605–615. doi: 10.1002/emmm.201100171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Roccaro AM, Rombaoa C, Flores L, Obad S, Fernandes SM, et al. LNA-mediated anti-miR-155 silencing in low-grade B-cell lymphomas. Blood. 2012;120:1678–1686. doi: 10.1182/blood-2012-02-410647. [DOI] [PubMed] [Google Scholar]
- Murphy BL, Obad S, Bihannic L, Ayrault O, Zindy F, Kauppinen S, et al. Silencing of the miR-17~92 cluster family inhibits medulloblastoma progression. Cancer Res. 2013;73:7068–7078. doi: 10.1158/0008-5472.CAN-13-0927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottiers V, Obad S, Petri A, McGarrah R, Lindholm MW, Black JC, et al. Pharmacological inhibition of a microRNA family in nonhuman primates by a seed-targeting 8-mer antimiR. Sci Transl Med. 2013;5:212ra162. doi: 10.1126/scitranslmed.3006840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belair C, Baud J, Chabas S, Sharma CM, Vogel J, Staedel C, et al. Helicobacter pylori interferes with an embryonic stem cell micro RNA cluster to block cell cycle progression. Silence. 2011;2:7. doi: 10.1186/1758-907X-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh MR, Lee Y, Kim JY, Kim SK, Moon SH, Lee JY, et al. Human embryonic stem cells express a unique set of microRNAs. Dev Biol. 2004;270:488–498. doi: 10.1016/j.ydbio.2004.02.019. [DOI] [PubMed] [Google Scholar]
- Judson RL, Babiarz JE, Venere M, Blelloch R. Embryonic stem cell-specific microRNAs promote induced pluripotency. Nat Biotechnol. 2009;27:459–461. doi: 10.1038/nbt.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Blelloch R. Cell cycle regulation by MicroRNAs in embryonic stem cells. Cancer Res. 2009;69:4093–4096. doi: 10.1158/0008-5472.CAN-09-0309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voorhoeve PM, le Sage C, Schrier M, Gillis AJ, Stoop H, Nagel R, et al. A genetic screen implicates miRNA-372 and miRNA-373 as oncogenes in testicular germ cell tumors. Cell. 2006;124:1169–1181. doi: 10.1016/j.cell.2006.02.037. [DOI] [PubMed] [Google Scholar]
- Palmer RD, Murray MJ, Saini HK, van Dongen S, Abreu-Goodger C, Muralidhar B, et al. Children's Cancer and Leukaemia Group Malignant germ cell tumors display common microRNA profiles resulting in global changes in expression of messenger RNA targets. Cancer Res. 2010;70:2911–2923. doi: 10.1158/0008-5472.CAN-09-3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KH, Goan YG, Hsiao M, Lee CH, Jian SH, Lin JT, et al. MicroRNA-373 (miR-373) post-transcriptionally regulates large tumor suppressor, homolog 2 (LATS2) and stimulates proliferation in human esophageal cancer. Exp Cell Res. 2009;315:2529–2538. doi: 10.1016/j.yexcr.2009.06.001. [DOI] [PubMed] [Google Scholar]
- Rippe V, Flor I, Debler JW, Drieschner N, Rommel B, Krause D, et al. Activation of the two microRNA clusters C19MC and miR-371-3 does not play prominent role in thyroid cancer. Mol Cytogenet. 2012;5:40. doi: 10.1186/1755-8166-5-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairo S, Wang Y, de Reyniès A, Duroure K, Dahan J, Redon MJ, et al. Stem cell-like micro-RNA signature driven by Myc in aggressive liver cancer. Proc Natl Acad Sci U S A. 2010;107:20471–20476. doi: 10.1073/pnas.1009009107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita S, Yamamoto H, Mimori K, Nishida N, Takahashi H, Haraguchi N, et al. MicroRNA-372 is associated with poor prognosis in colorectal cancer. Oncology. 2012;82:205–212. doi: 10.1159/000336809. [DOI] [PubMed] [Google Scholar]
- Zhang X, Li X, Tan Z, Liu X, Yang C, Ding X, et al. MicroRNA-373 is upregulated and targets TNFAIP1 in human gastric cancer, contributing to tumorigenesis. Oncol Lett. 2013;6:1427–1434. doi: 10.3892/ol.2013.1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Q, Gumireddy K, Schrier M, le Sage C, Nagel R, Nair S, et al. The microRNAs miR-373 and miR-520c promote tumour invasion and metastasis. Nat Cell Biol. 2008;10:202–210. doi: 10.1038/ncb1681. [DOI] [PubMed] [Google Scholar]
- Subramanyam D, Lamouille S, Judson RL, Liu JY, Bucay N, Derynck R, et al. Multiple targets of miR-302 and miR-372 promote reprogramming of human fibroblasts to induced pluripotent stem cells. Nat Biotechnol. 2011;29:443–448. doi: 10.1038/nbt.1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borgdorff V, Lleonart ME, Bishop CL, Fessart D, Bergin AH, Overhoff MG, et al. Multiple microRNAs rescue from Ras-induced senescence by inhibiting p21(Waf1/Cip1) Oncogene. 2010;29:2262–2271. doi: 10.1038/onc.2009.497. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Wang Y, Juranek S, Li H, Sheng G, Tuschl T, Patel DJ. Structure of an argonaute silencing complex with a seed-containing guide DNA and target RNA duplex. Nature. 2008;456:921–926. doi: 10.1038/nature07666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkayam E, Kuhn CD, Tocilj A, Haase AD, Greene EM, Hannon GJ, et al. The structure of human argonaute-2 in complex with miR-20a. Cell. 2012;150:100–110. doi: 10.1016/j.cell.2012.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cover TL, Blaser MJ. Helicobacter pylori in health and disease. Gastroenterology. 2009;136:1863–1873. doi: 10.1053/j.gastro.2009.01.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Li Z, Gao C, Chen P, Chen J, Liu W, et al. miR-21 plays a pivotal role in gastric cancer pathogenesis and progression. Lab Invest. 2008;88:1358–1366. doi: 10.1038/labinvest.2008.94. [DOI] [PubMed] [Google Scholar]
- Petrocca F, Visone R, Onelli MR, Shah MH, Nicoloso MS, de Martino I, et al. E2F1-regulated microRNAs impair TGFbeta-dependent cell-cycle arrest and apoptosis in gastric cancer. Cancer Cell. 2008;13:272–286. doi: 10.1016/j.ccr.2008.02.013. [DOI] [PubMed] [Google Scholar]
- Bessède E, Staedel C, Acuña Amador LA, Nguyen PH, Chambonnier L, Hatakeyama M, et al. Helicobacter pylori generates cells with cancer stem cell properties via epithelial-mesenchymal transition-like changes. Oncogene. 2014;33:4123–4131. doi: 10.1038/onc.2013.380. [DOI] [PubMed] [Google Scholar]
- Harvey KF, Zhang X, Thomas DM. The Hippo pathway and human cancer. Nat Rev Cancer. 2013;13:246–257. doi: 10.1038/nrc3458. [DOI] [PubMed] [Google Scholar]
- Ferrand J, Noël D, Lehours P, Prochazkova-Carlotti M, Chambonnier L, Ménard A, et al. Human bone marrow-derived stem cells acquire epithelial characteristics through fusion with gastrointestinal epithelial cells. PLoS One. 2011;6:e19569. doi: 10.1371/journal.pone.0019569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varon C, Dubus P, Mazurier F, Asencio C, Chambonnier L, Ferrand J, et al. Helicobacter pylori infection recruits bone marrow-derived cells that participate in gastric preneoplasia in mice. Gastroenterology. 2012;142:281–291. doi: 10.1053/j.gastro.2011.10.036. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.